ABSTRACT
Malaria remains the main cause of mortality and morbidity in the Central African Republic. However, the main malaria vectors remain poorly characterised, preventing the design of suitable control strategies. Here, we characterised the patterns and mechanisms of insecticide resistance in three important vectors from Bangui.
Mosquitoes were collected indoors, using electrical aspirators in July 2016 in two neighborhoods at Bangui. WHO bioassays performed, using F2An. gambiae sensu lato (s.l.), revealed a high level of resistance to type I (permethrin) and II (deltamethrin) pyrethroids and dichlorodiphenyltrichloroethane (< 3% mortality). Molecular analysis revealed the co-occurrence of Anopheles coluzzii (56.8 %) and An. gambiae s.s. (43.2%) within the An. gambiae complex. Anopheles funestus s.s. was the sole species belonging to An. funestus group. Both kdr-w (40% of homozygotes and 60% of heterozygotes/kdr-w/wild type) and kdr-e (37.5% of heterozygotes) mutations were found in An. gambiae. Contrariwise, only the kdr-w (9.5% homozygotes and 85.7% of heterozygotes) was detected in An. coluzzii. Quantitative RT-PCR showed that CYP6M2 and CYP6P3 are not upregulated in An. coluzzii from Bangui. Analysis of the sodium channel gene revealed a reduced diversity in An. coluzzii and An. gambiae s.s. In An. funestus s.s., the pyrethroid/DDT GSTe2 L119F resistance allele was detected at high frequency (54.7%) whereas a very low frequency for Rdl was observed. Polymorphism analysis of GSTe2 and GABA receptor gene in An. funestus revealed the presence of one resistant haplotype for each gene.
This study provides baseline information to help guide current and future malaria vector control interventions in CAR.
KEYWORDS: Malaria, vector, insecticide resistance mechanism, Central African Republic
1. Introduction
Despite the remarkable efforts in controlling malaria during the last 10 years, this disease continues to have a negative impact on people’s health and livelihoods in Africa. The World Health Organization (WHO) estimates that 216 million cases occurred globally in 2016, leading to 445,000 deaths, most of which were in children aged under 5 years in Africa [1]. In the Central African Republic (CAR), malaria is the most important disease responsible for 58% of all hospital consultation and a principal cause of death among children [2]. Malaria prevention mainly relies on insecticide-based interventions notably long lasting insecticidal nets (LLINs) and indoor residual sprays (IRS) [3]. However, the emergence of insecticide resistance in malaria vectors can compromise these control efforts. In Africa, several Anopheles species are implicated in malaria transmission. Among them, species such as An. gambiae sensu stricto (s.s.), An. arabiensis and An. coluzzii belonging to An. gambiae complex and An. funestus s.s. belonging to An. funestus group are implicated as major malaria vectors [4]. All these vectors have been found to be resistant to several insecticides belonging to four main classes (organophosphates, carbamates, organochlorines and pyrethroids) used in public health [5–7]. It has also been show on that several resistance mechanisms are involved in insecticide resistance to malaria vectors such as the target-site resistance and metabolic resistance. The most commonly reported target-site mutation described in An. gambiae s.s. and An. coluzzii, is the knockdown resistance (kdr) mutation which has two variants; L1014F (kdr-w) and L1014S (kdr-e) and confers resistance to pyrethroids and (dichlorodiphenyltrichloroethane) DDT [8,9]. Another mutation, the Ace1RG119S, confers resistance to carbamates and organophosphates [10]. In both An. gambiae s.s and An. coluzzii, metabolic resistance has been shown to be driven by multiple genes belonging to the cytochrome P450 monooxygenases (P450s) such as CYP6M2 and CYP6P3 [11–13] and glutathione S-transferases (GSTs) [14]. Contrary to An. gambiae s.s. or An. coluzzii, no kdr mutations have been reported in An. funestus s.s. despite the presence of some non-synonymous substitutions [7,15–17]. However, two target-site resistance markers have been described in this species, the N485I Ace-1 in southern African populations [18] and the Rdl A296S mutation mainly in West, Central and East Africa populations [19]. Metabolic resistance has been shown as the main mechanism driving pyrethroid and DDT resistance in this species [20–23]. The cytochrome P450s CYP6P9a, CYP6P9b and CYP6M7 and the glutathione S-transferases GSTe2 are the main resistance genes implicated in An. funestus s.s. [21,23,24]. It was notably demonstrated that the single amino acid change L119F in an up-regulated glutathione S-transferase gene, GSTe2, confers high levels of metabolic resistance to DDT and pyrethroids in the malaria vector An. funestus s.s. [23].
In the Central African Republic, the knowledge of malaria vectors remains limited. A recent study showing a multiple insecticide resistance in An. gambiae s.l. and An. funestus s.l. in Bangui [25,26]. To implement an optimal strategy to prevent malaria and reduce its burden in this country, up-to-date information is required about vector characterization as well as the molecular basis of insecticide resistance. Thus, we present here the characterisation of major malaria vectors in Bangui, the capital city of CAR including their resistance profiles to insecticides and the underlying resistance mechanisms.
2. Material and methods
2.1. Sampling sites and mosquito collection
Mosquitoes were sampled in July 2016 in Bangui (04°21ʹN, 18°33ʹE) in two neighbourhood, Gbanikola and Taoka St Paul. These locations are characterised by the presence of lakes, fishponds and polluted stagnant water. In each neighbourhood, mosquitoes were collected during six days in 30 houses per day from 7 am to 11 am. Houses were selected based on three main criteria: the proximity to the potential breeding sites, the type of building (houses without ceiling) and the physical state of bed-net or non-possession of mosquito nets. Blood-fed mosquitoes resting indoors were collected using electric aspirators and kept in cages. Mosquitoes were identified using morphological criteria described previously [27]. All female mosquitoes identified morphologically as belonging to the An. gambiae complex or to An. funestus group were kept in cardboard cups and fed with 10% sugar solution until they were fully gravid and ready to lay eggs. Females (F0) were allowed to oviposit individually using the forced-egg laying method as previously described [28]. After oviposition, the eggs (F1) were brought to the insectary at the Liverpool School of Tropical Medicine (LSTM) research Unit at OCEAC in Yaoundé, Cameroon where they were pooled together and reared to adults (F1). Larvae were fed with fish food (TetraMin Baby) and female adults were allowed to feed on blood from rabbits to induce eggs-laying. Due to the low number of adult (F1) obtained, mosquitoes were maintained in the insectary until F2 adult notably for An. gambiae s.l.. All the females (F0) that had laid eggs were put individually in the Eppendorf tubes containing desiccant (silica gel) and stored in −20 °C for molecular analysis.
2.2. Insecticide susceptibility assay
Due to the low number of An. funestus s.l. collected, bioassays were carried out only with An. gambiae s.l. according to WHO procedures [29]. Impregnated papers with insecticides were provided by LSTM. F2 adults aged between two-five days with four replicates of 20 mosquitoes were exposed for one hour to insecticide impregnated papers. After exposure, mosquitoes were transferred into holding tubes (and allowed to feed on a 10% sugar solution) and mortality recorded 24 h later. Assays were performed in controlled conditions (at 28 ± 2 °C and a relative humidity of 80 ± 10%). Two insecticide classes used in public health were assessed: the pyrethroids (0.75% permethrin and 0.05% deltamethrin) and organochlorine (4% DDT). Each experiment included control mosquitoes exposed to untreated papers. Bioassay tests were concomitantly performed with the Ngousso susceptible reference strain An. coluzzii.
2.3. Molecular identification and Plasmodium infection rate
Genomic DNA was extracted in whole bodies of oviposited mosquitoes (F0) using the Livak protocol as previously described [30]. Different species within the An. gambiae complex and An. funestus group were determined as previously described [31,32]. DNA extracted in whole mosquitoes were used to screen the Plasmodium infection using the TaqMan assay as described by Bass and collaborators [33]. One µl of DNA sample was used as template in a 3-step PCR program with a denaturation at 95 °C for 10 min followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. The primer sequences used are: Falcip + _ TCTGAATACGAATGTC; OVM + _ CTGAATACAAATGCC; Plas-F_ GCTTAGTTACGATTAATAGGAGTAGCTTG and Plas R_ GAAAATCTAAGAATTTCACCTCTGACA. These primers were used together with two probes labelled with fluorophores: FAM to detect Plasmodium falciparum, and HEX to detect Plasmodium ovale, Plasmodium vivax and Plasmodium malariae. Plasmodium falciparum samples and a mix of P. ovale, P. vivax and P. malariae were used as positive controls.
2.4. Genotyping kdr and of Ace1RG119S mutations in An. gambiae s.s. and An. coluzzii
The presence of the target-site mutations Ace-1, L1014F (kdr-w) and L1014S (kdr-e) was determined in An. gambiae s.s. and An. coluzzii using TaqMan assay according to the protocols previously described [34,35]. Sequences of primers used are presented in Table S1. Ten μl volume containing 1 × Sensimix (Bioline), 80 × primer/probe mix and 1 μl template DNA were used. Probes were labelled with two specific fluorophores FAM and HEX, FAM to detect the resistant allele, HEX to detect the susceptible allele. The assay was performed on an Agilent MX3005 real-time PCR machine with cycling conditions of 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min.
2.5. Transcriptional profiling of two major resistance genes in An. coluzzii
Several P450 genes have been detected associated to insecticide resistance in Anopheles, but in this study we focused our analyses on CYP6M2 and CYP6P3 previously found associated to pyrethroid resistance in An. coluzzii and An. gambiae s.s [11,12]. including populations from Central Africa [14]. The expression profiles of these genes was assessed by quantitative real time (qRT)-polymerase chain reaction (PCR) as described previously [36]. Total RNA was extracted from three pools of ten F2 female mosquitoes for three conditions: unexposed to insecticides, permethrin-resistant, and full susceptible laboratory strain Ngousso (An. coluzzii). RNA extraction, cDNA synthesis and qRT-PCR reactions and analysis were performed as previously described [36] in three biological replicates. Primer sequences used are presented in Table S1. The relative expression and fold-change of the two target genes were calculated as previously. The relative expression and fold-change of the two target genes were calculated according to 2−ΔΔCT method, after normalization with reference genes Actin (AGAP000651_RA) and 40S ribosomal protein S7 (AGAP010592_RA) [14]. Differences in expression were statistically analysed using unpaired Student’s t-test.
2.6. Genotyping of resistance markers L119F-GSTe2 and A296S-RDL in An. funestus s.s
To assess the potential role of the L119F-GSTe2 mutation in conferring DDT resistance in An. funestus s.s. in CAR, an allelic specific (AS)-PCR assay (Tchouakui et al., unpublished) was used to genotype 51 F1 mosquitoes. The primers employed for the genotyping are presented in Table S1. Amplification was carried out using PCR parameters of 95 °C for 5 min; 30 cycles of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 45 s, final extension at 72 °C for 10 min.
The role of A296S-RDL mutation in dieldrin resistance was also assessed with a newly designed TaqMan assay and was used to genotype 51 F1 female mosquitoes as previously described [6]. Experiments were performed in 10 μl volume containing 1 × Sensimix (Bioline), 80 × primer/probe mix and 1 μl genomic DNA. The probes were labelled with two distinct fluorophores FAM and HEX, FAM to detect the resistant allele and HEX to detect the susceptible one. The assay was performed on an Agilent MX3005 real-time PCR machine with cycling conditions of 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min.
2.7. Sequencing of resistance markers
To detect potential signatures of selection acting on key resistance genes, the polymorphism of a fragment of voltage-gated sodium channel gene (VGSC), spanning intron 19 and exon 20 was assessed in ten F0An. gambiae s.s. and ten F0An. coluzzii as previously described [28]. The sequences of primers employed are presented in Table S1. PCR products were purified using the Exo-SAP protocol and directly sequenced. Similarly, the full-length of GSTe2 and a portion of the GABA receptor gene spanning the A296S RDL mutation were amplified in ten F0An. funestus s.s. as previously described [19,23] and directly sequenced after purification.
2.8. Sequence data analysis
Sequences were corrected manually using BioEdit software version 7.2.1 and aligned with ClustalW [37]. The number of haplotypes (h), the number of polymorphism sites (S), haplotype diversity (Hd), nucleotide diversity (π), synonymous mutations (syn) and nonsynonymous mutations (nonsyn) were computed with DnaSP 5.10 [38]. The statistical tests of Tajima [39], Fu and Li [40] were also estimated with DnaSP. Relationships between haplotypes were assessed by constructing a maximum likelihood phylogenetic tree using MEGA 7.0 [41]. Genealogical relationship between haplotype was assessed using TCS [42] and tcsBU [43] softwares. DNA sequences were submitted in GenBank database (accession numbers MG779881- MG779909).
3. Results
3.1. Anopheles composition and Plasmodium infection rate
In total 61 An. gambiae s.l. and 13 An. funestus s.l. were collected in Bangui after intense screening of the houses in the selected neighbourhoods in around 180 houses during 6 days. Polymerase chain reaction (PCR) for species identification of 37 An. gambiae s.l. females that laid eggs revealed the presence of An. coluzzii at 56.8 % (21/37) and An. gambiae s.s. at 43.2% (16/37) (Table 1). A Plasmodium infection rate of 33.3% (7/21) was observed in An. coluzzii and 25.0% (4/16) in An. gambiae s.s. all infected with P. falciparum (Table 1). All the specimens of An. funestus s.l. were detected as An. funestus s.s. with 38.4% (5/13) of P. falciparum infection rate.
Table 1.
Summary of molecular characterization of An. coluzzii and An. gambiae s.s. from Bangui.
| Species | N | Pf positive | Kdr w-RR | Kdr w -RS | Kdr w -SS | Kdr e-RR | Kdr e-RS | kdre-SS | Ace 1-SS |
|---|---|---|---|---|---|---|---|---|---|
| An. coluzzii | 21 | 7 (33.3%) | 2 (9.5%) | 18 (85.7%) | 1 (4.8%) | 0 (0.0%) | 0 (0.0%) | 20 (100%) | 20 (100%) |
| An. gambiae | 16 | 4 (25.0%) | 6 (40.0%) | 9 (60.0%) | 0 (0.0%) | 0 (0.0%) | 6 (37.5%) | 10 (62.5%) | 10 (100%) |
N, number of specimens; Pf positive, Plasmodium falciparum infection rate; RS, heterozygote resistant; RR, homozygote resistant; SS, homozygote sensible.
3.2. Susceptibility to insecticides
A total of 241 adults F2An. gambiae s.l. mosquitoes were tested to assess susceptibility profile to permethrin, deltamethrin and DDT. Ngousso susceptible reference strain An. coluzzii was fully susceptible to all insecticides tested. The result revealed that mosquitoes collected in Bangui are fully resistant (0% of mortality) to both types of pyrethroids: deltamethrin and permethrin with no mortality recorded after 24h. This population was also highly resistant to DDT with less than 3% of mortality.
3.3. Genotyping of kdr and Ace-1 mutations in An. coluzzii and An. gambiae s.s
Genotyping of field collected mosquitoes revealed a high frequency of kdr-w mutation in An. coluzzii samples from Bangui with 52.3% for the 1014F resistant allele but with a predominance of heterozygote at 85.7% (18 of 21 mosquitoes) (1014F/L1014-RS), 9.5% (2 of 21 mosquitoes) homozygote resistant (1014F/F-RR) and 4.8% (1 of 21 mosquitoes) homozygote susceptible (1014/L-SS). Similarly, high frequency of kdr-w mutation was found in An. gambiae s.s. with 70% for the resistant 1014F allele obtained from 60.0% (9 of 15 mosquitoes) heterozygote RS and 40.0% (6/15) homozygote RR. In contrast, kdr-e mutation was reported only in An. gambiae s.s. always as heterozygote at 37.5% (6 of 16 mosquitoes) (Table 1). No specimen exhibited the Ace-1R allele mutation in both An. coluzzii and An. gambiae s.s. (Table 1).
3.4. Investigating the metabolic resistance in An. coluzzii
Transcriptional analysis of the candidate resistance genes CYP6M2 and CYP6P3 known to confer pyrethroid and/or DDT resistance in An. coluzzii using qRT-PCR showed that contrary to expectation both genes are not up-regulated in CAR with 0.92 and 0.60 fold change, respectively (Figure 1).
Figure 1.
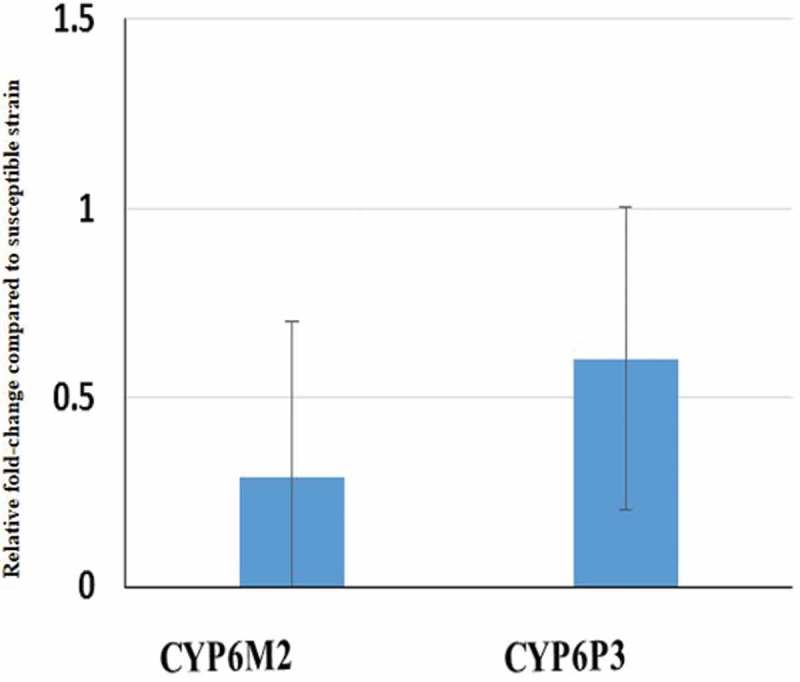
Differential expression of two resistance genes by qRT-PCR between permethrin-resistant An. coluzzii mosquitoes from Bangui and susceptible laboratory strain Ngousso.
Error bars represent standard deviation (n = 3).
3.5. Genetic variability of exon 20 in An. coluzzi and An. gambiae s.s
Sequencing analysis of a 512 bp fragment spanning intron 19 and exon 20 from 10 An. coluzzii revealed a low genetic diversity with a predominant haplotype (H1) containing the 1014F resistant allele at 65% (13/20). The genetic parameters are summarised in Table 2. No synonymous mutation was observed with a single nonsynonymous mutation corresponding to the 1014F. None of the specimen of An. coluzzii was detected carrying kdr-e mutation. Three haplotypes (H1, H4 and H5) were detected with 1014F mutation and two haplotypes (H2 and H3) carrying the wild susceptible allele (L1014) (Figure 2).
Table 2.
Summary statistics for polymorphism of some insecticide resistance markers in key malaria vectors from Bangui.
| Species | Gene | 2n | S | h | hd | Syn | Nonsyn | π(κ) | D | D* | F* |
|---|---|---|---|---|---|---|---|---|---|---|---|
| An. funestus s.s. | RDL | 18 | 6 | 7 | 0.771 | 0 | 1 | 0.002(1.294) | −0.849ns | −1.467 ns | −1.493 ns |
| GSTe2 | 18 | 14 | 11 | 0.882 | 4 | 7 | 0.007(3.366) | −0.865 ns | −0.125 ns | −0.389 ns | |
| An. gambiae s.s. | Exon 20 | 20 | 5 | 6 | 0.637 | 0 | 0 | 0.003(1.357) | −0.112 ns | 1.186 ns | 0.950 ns |
| An. coluzzii | Exon 20 | 20 | 5 | 5 | 0.568 | 0 | 1 | 0.002(1.231) | −0.385 ns | 0.387 ns | 0.197 ns |
N = number of sequences (2n); S, number of polymorphic sites; h, haplotype; Hd, haplotype diversity; Syn, Synonymous mutations; Nonsyn, Nonsynonymous mutations; π, nucleotide diversity (k = mean number of nucleotide differences); Tajima’s D and Fu and Li’s D* and F* statistics, ns, not significant.
Figure 2.
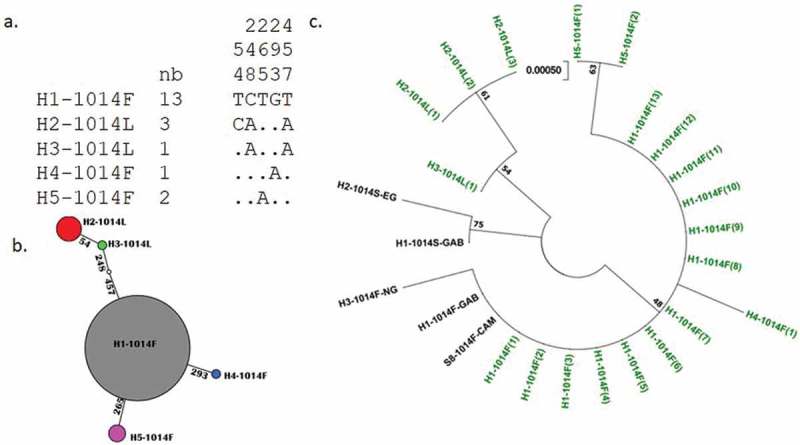
Genetic diversity pattern of fragment of VGSC spanning exon 20 in An. coluzzii from Bangui. a) Haplotype diversity patterns of the 512 bp fragment in Bangui. b) TCS and tcsBU haplotype network showing a low polymorphism of the exon 20 fragment with low number of mutational steps between haplotypes. c) Molecular phylogenetic analysis by maximum likelihood method based on the Tamura 3-parameter model. In green represent the haplotype detected in this study.
Similar analysis from ten An. gambiae s.s., evidenced also a low genetic diversity with a major haplotype (H1) carrying the 1014F resistant allele at 60% (12/20) (Figure 3). The genetic parameters are summarised in Table 2. In six haplotypes detected in An. gambiae s.s., kdr-w mutation (1014F) was detected in two haplotypes (H1and H5) and kdr-e mutation (1014S) in four haplotypes (H2, H3, H4 and H6) (Figure 3, Table 2). Neutrality test of Tajima’s D had a negative value in both species but it was not significant in either species.
Figure 3.
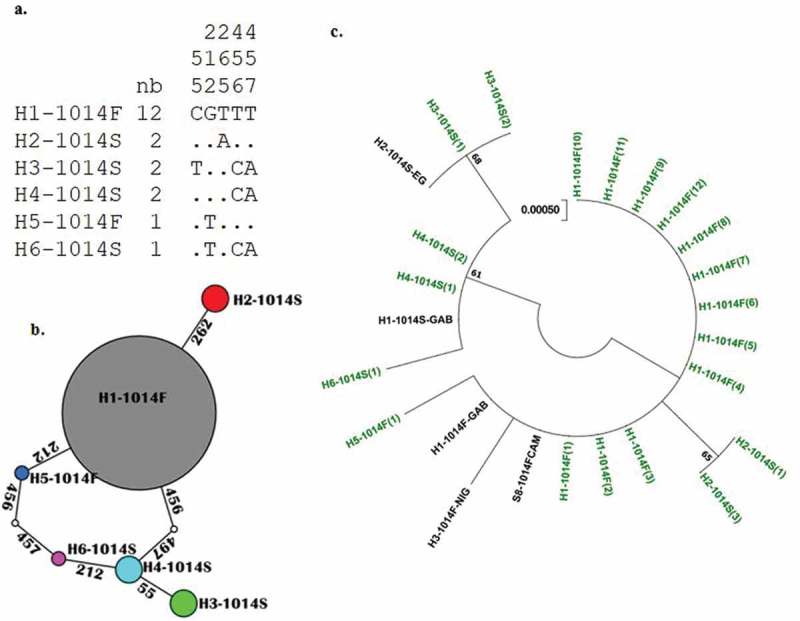
Genetic diversity pattern of fragment of VGSC spanning exon 20 in An. gambiae s.s. from Bangui. a) Haplotype diversity patterns of the 512 bp fragment in Bangui. b) TCS and tcsBU haplotype network showing a high polymorphism of the exon 20 fragment with high number of mutational steps between haplotypes. c) Molecular Phylogenetic analysis by maximum likelihood method based on the Tamura 3-parameter model. In green represent the haplotype detected in this study.
Phylogenetic analysis of the six haplotypes found in An. gambiae s.s. was performed using previous sequences published in GenBank originating from Cameroon, Nigeria, Equatorial Guinea and Gabon (Table S2) using the Maximum Likelihood method (Figure 3(c)). The results indicated that the dominant haplotype (H1) matched with that found across Africa suggesting extensive gene flow in An. gambiae s.s. populations across the continent. Overall, this analysis detected two distinct haplotype groups: the haplotypes carrying 1014F resistance mutation and the haplotypes carrying 1014S mutation.
Similar phylogenetic analysis in An. coluzzii also revealed the existence of two distinct groups: one including all the haplotypes carrying the resistant mutation 1014F and the other made up of haplotypes of the susceptible allele 1014L.
Genealogical relationships between haplotypes are presented in Figures 2 and 3. In An. gambiae s.s., haplotype H2-1014S and H5-1014F derived from single mutational steps from the dominant haplotype H1-1014F. H6-1014S and H3-1014S resulted from three mutational steps and H5-1014S from two mutational steps (Figure 3(b)). In An. coluzzii, three haplotypes H1, H4 and H5 carrying the 1014F mutation were separated by one mutational step. Haplotype H2-1014L is the most isolated to the dominant haplotype H1-1014F with three mutational steps (Figure 2(b)).
3.6. Genotyping of GSTe2 and Rdl in An. funestus s.s
The genotyping of L119F-GSTe2 and A296S-Rdl with TaqMan in 51 non-exposed F1 revealed a contrasting result (Figure 3) with a higher frequency of the 119F resistance allele for GSTe2 but very low frequency for the resistant 296S allele for the Rdl mutation. The distribution of the genotypes for the L119F-GSTe2 marker was 30.23% homozygotes (13 of 43 mosquitoes) and 48.84% heterozygotes (21 of 43 mosquitoes) of (L119F). In contrast, the analysis of frequency of A296S-Rdl mutation revealed no homozygote resistant allele and only one heterozygote allele (1 of 51 mosquitoes; 1.90%) was detected (Figure 4).
Figure 4.
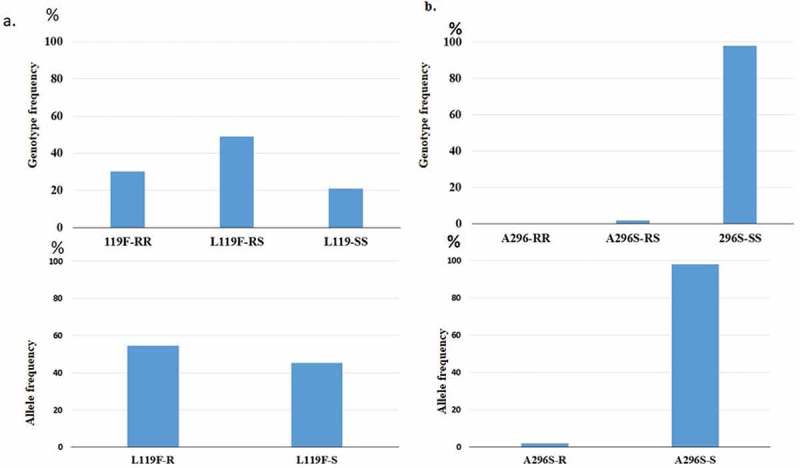
Distribution of resistance markers in An. funestus s.s. from Bangui. a) Frequency of the three genotypes of the L119F-GSTe2 conferring DDT resistance; b) is for A296S-rdl mutation conferring dieldrin resistance.
3.7. Genetic variability of GSTe2 and Rdl in An. funestus s.s
Polymorphism analysis of 444 bp of GSTe2 and 574 bp of GABA receptor gene was performed for nine specimens of F0An. funestus s.s. successfully amplified. The GABA receptor gene presented six substitution sites, seven haplotypes, no synonymous mutations and only one nonsynonymous mutation (A296S) resulting in high haplotype diversity (hd = 0.771) (Table 2). The haplotype H7 was the only resistant haplotype (1 of 18) which is in accord with genotyping assay.
Similar analysis with the GSTe2 gene indicated a high level of polymorphism compared to RDL gene with 14 polymorphic sites, 11 haplotypes, and four synonymous and seven nonsynonymous sites. As with RDL gene, one haplotype (H10) was found resistant but with the difference that the resistant haplotype is the dominant (6/18) (Figure 5).
Figure 5.
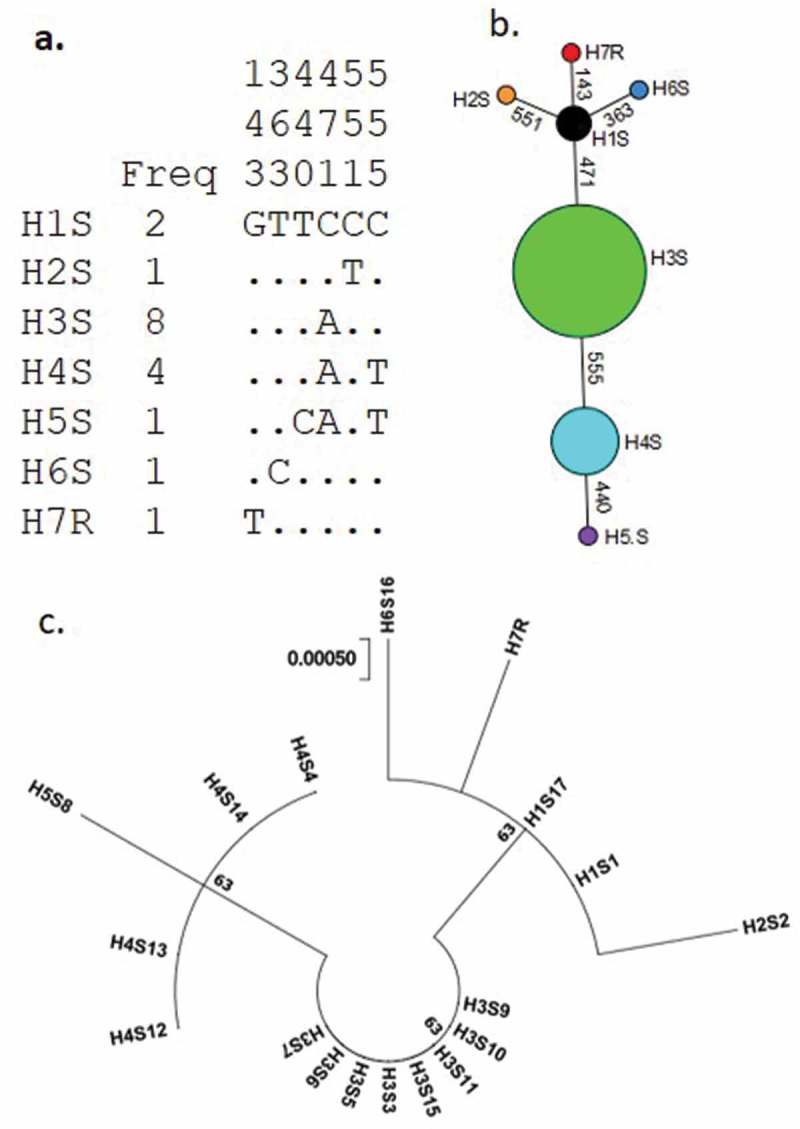
Genetic diversity pattern of rdl gene in An. funestus from Bangui. a) Haplotype diversity patterns of the 574 bp fragment in Bangui. b) TCS and tcsBU haplotype network showing a low polymorphism of the rdl gene fragment with low number of mutational steps between haplotypes. c) Molecular phylogenetic analysis by maximum likelihood method based on the Hasegawa-Kishino-Yano model.
Neutrality tests revealed negative values of Tajima’s D and Fu and Li’s D* and F* statistics for both genes but none of these values was statistically significant (Table 2).
Phylogenetic analysis of 18 DNA sequences from Rdl gene in An. funestus s.s. collected in Bangui detected three distinct groups (Figure 5(c)). Genealogical relationships between haplotypes using TCS software revealed that haplotypes H2, H6, H7 and H3 are separated from a common ancestor, haplotype H1, by one mutational step. Haplotype H5 is the most isolated to ancestor haplotype with three mutational steps (Figure 5(b)).
A similar analyses was performed with GSTe2 sequences including previously detected haplotypes across Africa [23]. The dominant haplotype (H10R) carrying 119F mutation matched with the resistant haplotype found in Benin, Ghana, and Cameroon (Figure 6(c)). TCS network showed that haplotypes H10 and H11 are mostly related to theancestor haplotype and are separated by two mutational steps. Haplotype H2 is the most isolated from the core group and separated by seven mutational steps (Figure 6(b)).
Figure 6.
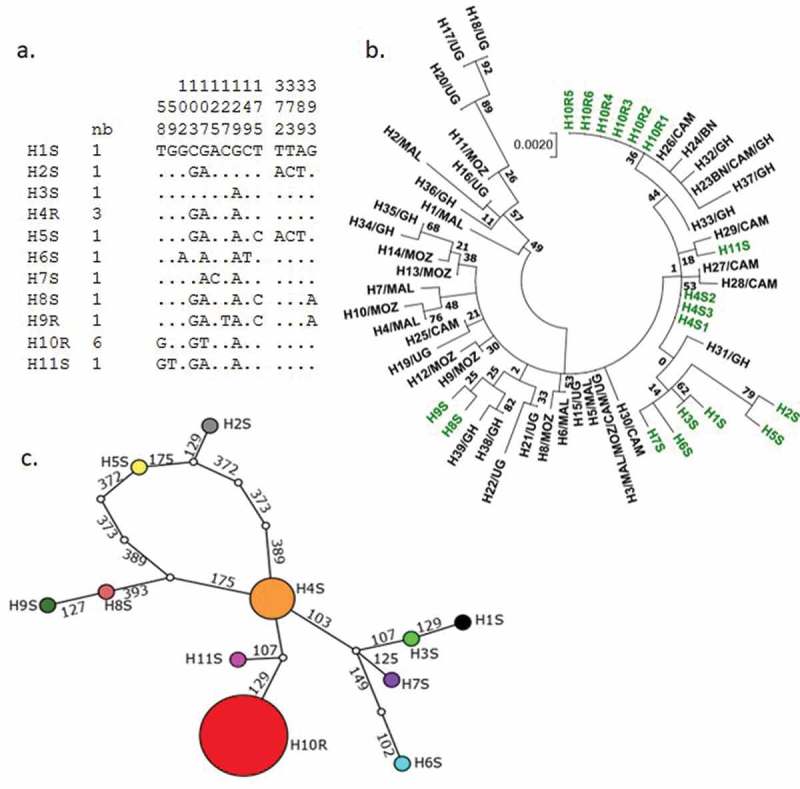
Genetic diversity pattern of GSTe2 gene in An. funestus from Bangui. a) Haplotype diversity patterns of the 444 bp fragment in Bangui. b) TCS and tcsBU haplotype network showing a low polymorphism of the GSTe2 gene fragment with low number of mutational steps between haplotypes. c) Molecular phylogenetic analysis by maximum likelihood method. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. In green represent the haplotype detected in this study. BN, Benin; CAM, Cameroon; GH, Ghana; MAL, Malawi; MOZ, Mozambique; UG, Uganda.
4. Discussion
4.1. Malaria vector composition and their contribution in Plasmodium transmission
This study has revealed the presence of three malaria vectors, An. gambiae s.s., An. coluzzii and An. funestus s.s., in two neighbourhoods of Bangui in July 2016 with An. coluzzii being the most abundant species. This is in contrast with the previous work done at Bangui where An. coluzzii was not collected from seven districts in Bangui [25]. However, our findings are consistent with previous results in Central Africa notably in Cameroon showing that An. coluzzii is the most abundant Anopheles species in the urban area due to the greater tolerance of this species to pollution [44–46].
High P. falciparum infection rate was observed in all these vectors examined. This indicates the high burden of malaria at Bangui. Meanwhile, it is important to highlight that high infection rate found in this study may be due to the fact that the parasite DNA was extracted from whole mosquitoes and not just from head and thorax. Nevertheless, other studies performed with whole mosquitoes have detected significantly lower infection rate such as in Malawi in southern Africa where Riveron et al. [47] only found an infection rate of 4.8% or across Uganda and Kenya where the rate varied from 4.2 to 10.4% [48]. Therefore, it is likely that the high infection rates in the three species reflect a high level of malaria transmission in the area although further studies across the city with larger samples will help to confirm it.
4.2. Anopheles gambiae s.s. and An. culuzzii are highly resistant to pyrethroids and DDT
Both species were fully resistant to both types of pyrethroid: deltamethrin (type II) and permethrin (type I), and highly resistant to DDT. These findings are in contrast with the results of previous study performed at Bangui in 2014 indicating the moderate level of resistance to deltamethrin with mortality rate varying from 68.1% to 74.6% [25]. The high pyrethroid resistance reported in this study in Bangui An. gambiae s.l. populations could be of great concern for malaria prevention since this insecticide class is the only one used for bed net impregnation. A high level of resistance to DDT has been repeatedly reported in Central Africa [7,14] even though this insecticide is no longer used for malaria control in the region. It is possible that this higher level of resistance is linked to over-expression of other detoxification genes including other cytochrome P450 genes not tested here or even o gluthione-S transferases such as GSTe2 as seen in An. funestus. Furthermore, it also possible that presence of additional kdr mutations such as the N1575Y could be enhancing the resistance phenotype as this mutation has been shown to provide an additive resistance in combination with the L1014F mutation [49] as also recently shown in An. gambiae from Mali [50]. More surveys with greater sample sizes are needed to establish the extent and distribution of this resistance in Bangui and across CAR.
4.3. Molecular mechanism involved to insecticide resistance
The full resistance to deltamethrin and permethrin and the high resistance to DDT in An. gambiae s.l. correlated with high frequency of the L1014F kdr mutation. Indeed, both An. coluzzii and An. gambiae s.s. present a high frequency of the L1014F kdr mutation whereas, the L1014S kdr mutation was detected only in An. gambiae s.s. The presence of both type of kdr mutations have been previously reported in An. coluzzii and An. gambiae s.s. in Central Africa [51,52]. The higher frequency of heterozygotes is contrary to the previous results from Bangui indicating rather higher frequency of resistant homozygous allele of kdr-w, in Gbanikolla (the same location we sampled) with 100% of homozygotes resistant [25]. None of the specimens tested had the Ace-1 mutation. This is in accordance with previous results obtained in Central Africa notably in CAR [25].
The exploration of the metabolic resistance mechanisms performed through the transcription profiling of the common major resistance genes CYP6M2 and CYP6P3 surprisingly revealed that both genes are not up regulated in An. coluzzii suggesting that they play no role in the observed resistance in Bangui. This result contrasts with previous reports in Central Africa notably in Cameroon showing that these genes are the main drivers of metabolic resistance to pyrethroid and DDT in An. gambiae s.s. and An. coluzzii [14,52]. However, other P450 genes like CYP9K1, CYP6Z1 and CYP325A [53,54] have been also detected associated with insecticide resistance in Anopheles but these genes were not tested in this study. Further studies are needed to elucidate the molecular basis of pyrethroid/DDT resistance in CAR.
Genotyping of L119F-GSTe2 and A296S-Rdl mutations revealed a contrasting result with a high frequency of the resistant allele of GSTe2 and only one heterozygote allele of Rdl. This finding matches with sequencing data which detected only one resistant haplotype (1/18) for Rdl whereas the predominant GSTe2 haplotype was found in resistant mosquitoes. This observation suggests higher resistant of An. funestus s.s. from Bangui to DDT and possibly to pyrethroids. Indeed, up-regulation for GSTe2 gene combined with the presence of the L119F-GSTe2 mutation have been strongly associated with DDT resistance but also to pyrethroid resistance [23]. The haplotype H10 carrying 119F-GSTe2 mutation, which has been shown to play a significant role in DDT resistance in West-Central Africa [7,23], was found in high frequency. The 119F resistant allele at Bangui was probably selected during the intense DDT spraying between 1950s-1960s as part of the malaria elimination campaign as suggested previously [55]. Another possibility is that the 119F allele could also have been further selected by pyrethroid-based interventions as it was demonstrated that GSTe2 can metabolize permethrin as previously suggested [23].
5. Conclusion
This study shows that three Anopheles species: An. gambiae s.s., An. coluzzii and An. funestus s.s. are present in Bangui and can contribute to malaria transmission in this city. Full resistance to deltamethrin and permethrin, which are used for impregnating bed net, could pose a serious threat for malaria control in Bangui. Further study is required to assess the vector composition and their distribution in entire CAR as well as the resistance status and the resistance mechanisms involved. This will be needed to better manage the insecticide resistance and develop alternative malaria control strategies in Central Africa.
Funding Statement
This work was supported by the Wellcome Trust under Grant 083515/Z/07/Z to CSW and 204862/Z/16/Z to BK.
Acknowledgments
We thank the population of Bangui for their collaboration during filed investigation.
Disclosure statement
No potential conflict of interest was reported by the authors.
Ethics approval
Verbal consents were obtained from the house owners for the mosquito collection.
Supplementary material
Supplementary data for this article can be accessed here.
References
- [1].WHO World malaria report 2017. Geneva: World Health Organization; 2017. [Google Scholar]
- [2].MSP Rapport ministère de la santé de la République Centrafricaine 2015. Plan de transition du secteur santé 2015–2016; 2015[cited 2017 February08] p. 81 Available from:http://www.nationalplanningcycles.org/sites/default/files/planning_cycle_repository/central_african_republic/rca_-ptss_v_definitive_1.pdf
- [3].WHO World malaria report 2015. Geneva: World Health Organization; 2015. [Google Scholar]
- [4].Sinka ME, Bangs MJ, Manguin S, et al The dominant Anopheles vectors of human malaria in Africa, Europe and the Middle East: occurrence data, distribution maps and bionomic precis. Parasit Vectors. 2010;3:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Edi CV, Koudou BG, Jones CM, et al. Multiple-insecticide resistance in Anopheles gambiae mosquitoes, Southern Cote d’Ivoire. Emerg Infect Dis. 2012;18(9):1508–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Djouaka R, Riveron JM, Yessoufou A, et al Multiple insecticide resistance in an infected population of the malaria vector Anopheles funestus in Benin. Parasit Vectors. 2016;9(1):453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Menze BD, Riveron JM, Ibrahim SS, et al. Multiple insecticide resistance in the malaria vector Anopheles funestus from Northern Cameroon is mediated by metabolic resistance alongside potential target site insensitivity mutations. PLoS One. 2016;11(10):e0163261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ranson H, Jensen B, Vulule JM, et al. Identification of a point mutation in the voltage-gated sodium channel gene of Kenyan Anopheles gambiae associated with resistance to DDT and pyrethroids. Insect Mol Biol. 2000;9(5):491–497. [DOI] [PubMed] [Google Scholar]
- [9].Martinez-Torres D, Chandre F, Williamson MS, et al. Molecular characterization of pyrethroid knockdown resistance (kdr) in the major malaria vector Anopheles gambiae s.s. Insect Mol Biol. 1998;7(2):179–184. [DOI] [PubMed] [Google Scholar]
- [10].Weill M, Malcolm C, Chandre F, et al. The unique mutation in ace-1 giving high insecticide resistance is easily detectable in mosquito vectors. Insect Mol Biol. 2004;13(1):1–7. [DOI] [PubMed] [Google Scholar]
- [11].Djouaka RF, Bakare AA, Coulibaly ON, et al. Expression of the cytochrome P450s, CYP6P3 and CYP6M2 are significantly elevated in multiple pyrethroid resistant populations of Anopheles gambiae s.s. from Southern Benin and Nigeria. BMC Genomics. 2008;9:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Edi CV, Djogbenou L, Jenkins AM, et al CYP6 P450 enzymes and ACE-1 duplication produce extreme and multiple insecticide resistance in the malaria mosquito Anopheles gambiae. PLoS Genet. 2014;10(3):e1004236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Muller P, Warr E, Stevenson BJ, et al Field-caught permethrin-resistant Anopheles gambiae overexpress CYP6P3, a P450 that metabolises pyrethroids. PLoS Genet. 2008;4(11):e1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fossog Tene B, Poupardin R, Costantini C, et al. Resistance to DDT in an urban setting: common mechanisms implicated in both M and S forms of Anopheles gambiae in the city of Yaounde Cameroon. PLoS One. 2013;8(4):e61408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Okoye PN, Brooke BD, Koekemoer LL, et al. Characterisation of DDT, pyrethroid and carbamate resistance in Anopheles funestus from Obuasi, Ghana. Trans R Soc Trop Med Hyg. 2008;102(6):591–598. [DOI] [PubMed] [Google Scholar]
- [16].Mulamba C, Riveron JM, Ibrahim SS, et al. Widespread pyrethroid and DDT resistance in the major malaria vector Anopheles funestus in East Africa is driven by metabolic resistance mechanisms. PLoS One. 2014;9(10):e110058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Irving H, Wondji CS.. Investigating knockdown resistance (kdr) mechanism against pyrethroids/DDT in the malaria vector Anopheles funestus across Africa. BMC Genet. 2017;18(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ibrahim SS, Ndula M, Riveron JM, et al. The P450 CYP6Z1 confers carbamate/pyrethroid cross-resistance in a major African malaria vector beside a novel carbamate-insensitive N485I acetylcholinesterase-1 mutation. Mol Ecol. 2016;25(14):3436–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wondji CS, Dabire RK, Tukur Z, et al. Identification and distribution of a GABA receptor mutation conferring dieldrin resistance in the malaria vector Anopheles funestus in Africa. Insect Biochem Mol Biol. 2011;41(7):484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wondji CS, Coleman M, Kleinschmidt I, et al. Impact of pyrethroid resistance on operational malaria control in Malawi. Proc Natl Acad Sci USA. 2012;109(47):19063–19070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Riveron JM, Irving H, Ndula M, et al. Directionally selected cytochrome P450 alleles are driving the spread of pyrethroid resistance in the major malaria vector Anopheles funestus. Proc Natl Acad Sci USA. 2013;110(1):252–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Christian RN, Matambo TS, Spillings BL, et al. Age-related pyrethroid resistance is not a function of P450 gene expression in the major African malaria vector, Anopheles funestus (Diptera: culicidae). Genet Mol Res. 2011;10(4):3220–3229. [DOI] [PubMed] [Google Scholar]
- [23].Riveron JM, Yunta C, Ibrahim SS, et al A single mutation in the GSTe2 gene allows tracking of metabolically based insecticide resistance in a major malaria vector. Genome Biol. 2014;15(2):R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Riveron JM, Ibrahim SS, Chanda E, et al. The highly polymorphic CYP6M7 cytochrome P450 gene partners with the directionally selected CYP6P9a and CYP6P9b genes to expand the pyrethroid resistance front in the malaria vector Anopheles funestus in Africa. BMC Genomics. 2014;15:817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ole Sangba ML, Sidick A, Govoetchan R, et al. Evidence of multiple insecticide resistance mechanisms in Anopheles gambiae populations in Bangui, Central African Republic. Parasit Vectors. 2017;10(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sangba ML, Deketramete T, Wango SP, et al. Insecticide resistance status of the Anopheles funestus population in Central African Republic: a challenge in the war. Parasit Vectors. 2016;9:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gillies MT, De Meillon B. The anophelinae of Africa south of the Sahara (Ethiopian Zoogeographical Region). Johannesbourg: The South African Institute for Medical Research; 1968;54. [Google Scholar]
- [28].Morgan JC, Irving H, Okedi LM, et al. Pyrethroid resistance in an Anopheles funestus population from Uganda. PLoS One. 2010;5(7):e11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].WHO Test procedures for insecticide resistance monitoring in malaria vector, bio-efficacy and persistence of insecticides on treated surfaces. Geneva: WHO/CDS/CPC/MAL/;2013 [Google Scholar]
- [30].Livak KJ. Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics. 1984;107(4):611–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Santolamazza F, Mancini E, Simard F, et al. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar J. 2008;7:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Koekemoer LL, Kamau L, Hunt RH, et al. A cocktail polymerase chain reaction assay to identify members of the Anopheles funestus (Diptera: culicidae) group. Am J Trop Med Hyg. 2002;66(6):804–811. [DOI] [PubMed] [Google Scholar]
- [33].Bass C, Nikou D, Blagborough AM, et al. PCR-based detection of Plasmodium in Anopheles mosquitoes: a comparison of a new high-throughput assay with existing methods. Malar J. 2008;7:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bass C, Williamson MS, Wilding CS, et al. Identification of the main malaria vectors in the Anopheles gambiae species complex using a TaqMan real-time PCR assay. Malar J. 2007;6:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bass C, Nikou D, Vontas J, et al. Development of high-throughput real-time PCR assays for the identification of insensitive acetylcholinesterase (ace-1R) in Anopheles gambiae. Pestic Biochem Physiol. 2010;96(2):80–85. [Google Scholar]
- [36].Kwiatkowska RM, Platt N, Poupardin R, et al. Dissecting the mechanisms responsible for the multiple insecticide resistance phenotype in Anopheles gambiae s.s., M form, from Vallee du Kou, Burkina Faso. Gene. 2013;519(1):98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. [DOI] [PubMed] [Google Scholar]
- [39].Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3):585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133(3):693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9(10):1657–1659. [DOI] [PubMed] [Google Scholar]
- [43].Murias Dos Santos A, Cabezas MP, Tavares AI, et al. tcsBU: a tool to extend TCS network layout and visualization. Bioinformatics. 2016;32(4):627–628. [DOI] [PubMed] [Google Scholar]
- [44].Kamdem C, Fouet C, Gamez S, et al. Pollutants and insecticides drive local adaptation in African malaria mosquitoes. Mol Biol Evol. 2017;34:1261–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kamdem C, Tene Fossog B, Simard F, et al Anthropogenic habitat disturbance and ecological divergence between incipient species of the malaria mosquito Anopheles gambiae. PLoS One. 2012;7(6):e39453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tene Fossog B, Antonio-Nkondjio C, Kengne P, et al. Physiological correlates of ecological divergence along an urbanization gradient: differential tolerance to ammonia among molecular forms of the malaria mosquito Anopheles gambiae. BMC Ecol. 2013;13:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Riveron JM, Chiumia M, Menze BD, et al. Rise of multiple insecticide resistance in Anopheles funestus in Malawi: a major concern for malaria vector control. Malar J. 2015;14:344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mulamba C, Irving H, Riveron JM, et al. Contrasting Plasmodium infection rates and insecticide susceptibility profiles between the sympatric sibling species Anopheles parensis and Anopheles funestus s.s: a potential challenge for malaria vector control in Uganda. Parasit Vectors. 2014;7:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jones CM, Liyanapathirana M, Agossa FR, et al. Footprints of positive selection associated with a mutation (N1575Y) in the voltage-gated sodium channel of Anopheles gambiae. Proc Natl Acad Sci USA. 2012;109(17):6614–6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Mavridis K, Wipf N, Muller P, et al. Detection and monitoring of insecticide resistance mutations in Anopheles gambiae: individual vs pooled specimens. Genes. 2018;9(10): 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Nwane P, Etang J, Chouasmall yi UM, etal. kdr-based insecticide resistance in Anopheles gambiae s.s populations in. BMC Res Notes. 2011;4:463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Antonio-Nkondjio C, Tene Fossog B, Kopya E, et al. Rapid evolution of pyrethroid resistance prevalence in Anopheles gambiae populations from the cities of Douala and Yaounde (Cameroon). Malar J. 2015;14:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Main BJ, Everitt A, Cornel AJ, et al. Genetic variation associated with increased insecticide resistance in the malaria mosquito, Anopheles coluzzii. Parasit Vectors. 2018;11(1):225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Nikou D, Ranson H, Hemingway J. An adult-specific CYP6 P450 gene is overexpressed in a pyrethroid-resistant strain of the malaria vector, Anopheles gambiae. Gene. 2003;318:91–102. [DOI] [PubMed] [Google Scholar]
- [55].Kamgang B, Yougang AP, Tchoupo M, et al. Temporal distribution and insecticide resistance profile of two major arbovirus vectors Aedes aegypti and Aedes albopictus in Yaounde, the capital city of Cameroon. Parasit Vectors. 2017;10(1):469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


