Abstract
Structural variants can contribute to oncogenesis through a variety of mechanisms. Despite their importance, the identification of structural variants in cancer genomes remains challenging. Here, we present a framework that integrates optical mapping, high-throughput chromosome conformation capture (Hi-C), and whole genome sequencing to systematically detect SVs in a variety of normal or cancer samples and cell lines. We identify the unique strengths of each method and demonstrate that only integrative approaches can comprehensively identify structural variants in the genome. By combining Hi-C and optical mapping, we resolve complex SVs and phase multiple SV events to a single haplotype. Furthermore, we observe widespread structural variation events affecting the functions of non-coding sequences, including the deletion of distal regulatory sequences, alteration of DNA replication timing, and the creation of novel 3D chromatin structural domains. Our results indicate that non-coding structural variations may be underappreciated mutational drivers in cancer genomes.
Introduction
Structural variations (SVs), including inversions, deletions, duplications, and translocations, are a hallmark of most cancer genomes2. The discovery of recurrent SVs and their molecular consequences for gene organization and expression has greatly advanced our knowledge of oncogenesis. Numerous oncogenes have been identified as the products of recurrent translocations and have provided successful targets for drug therapies 1, 3-6, particularly for hematopoietic malignancies.
Despite their importance, identifying structural variations in cancer genomes remains challenging. G-band karyotyping has been the major method historically and is routinely performed in the clinic7. However, it is an inherently low resolution and low throughput method that cannot characterize extensively rearranged genomes. Microarrays are another commonly used method for detecting gains and losses of genetic material8, but they do not provide precise location of rearrangements and cannot detect balanced rearrangements. Targeted approaches such as fluorescence in situ hybridization (FISH) and PCR are also used extensively in the clinic. However, such methods require a priori knowledge of the rearrangements and hence are not suitable for de novo SV detection. Recently, high-throughput sequencing based methods, such as RNA-Seq and whole genome sequencing (WGS), have emerged as an attractive method for structural variant identification, and they can identify gene fusions and genomic rearrangements with high-resolution 9, 10,11, 12,13-16. Despite their success, these short reads-based approaches cannot effectively detect SVs in the repetitive regions in the genome and have limited power to resolve haplotype-resolved complex SVs.
Here we propose an integrative framework to comprehensively detect SVs by using a combination of technologies, including WGS, next-generation optical mapping (BioNano Irys), and high throughput chromosome conformation capture (Hi-C). In addition, we developed a novel algorithm that uses Hi-C data to detect SVs genome-wide. By integrating the results from different platforms, we compiled a list of high confidence SVs in eight human cancer cell lines (Table 1). We observed that optical mapping and Hi-C excelled at detecting large and complex structural alterations, whereas high coverage WGS was adept at identifying SVs with high resolution. We identified numerous instances of 3D genome organization alterations as a result of structural genome variation, such as the formation or dissolution of topologically associating domains (TADs), suggesting a critical role for structural variation in gene misregulation in oncogenesis.
Table 1.
Number of high-confidence large SVs in cancer and normal cells
| Confident calls of large intra-chr SVs ≥ 1Mb | inter-chr TLs | ||||
|---|---|---|---|---|---|
| Deletions | Duplications | Inversions | Unclassified SVs | ||
| NA12878 | 0 | 0 | 0 | 0 | 0 |
| T47D | 4 | 2 | 6 | 13 | 30 |
| Caki2 | 2 | 2 | 5 | 4 | 26 |
| K562 | 4 | 5 | 6 | 11 | 33 |
| A549 | 1 | 2 | 0 | 3 | 12 |
| NCI-H460 | 2 | 1 | 0 | 0 | 7 |
| SK-N-MC | 2 | 2 | 6 | 3 | 9 |
| PANC-1 | 3 | 0 | 0 | 4 | 14 |
| LNCaP | 3 | 0 | 0 | 4 | 9 |
Results
An integrated approach for structural variant detection
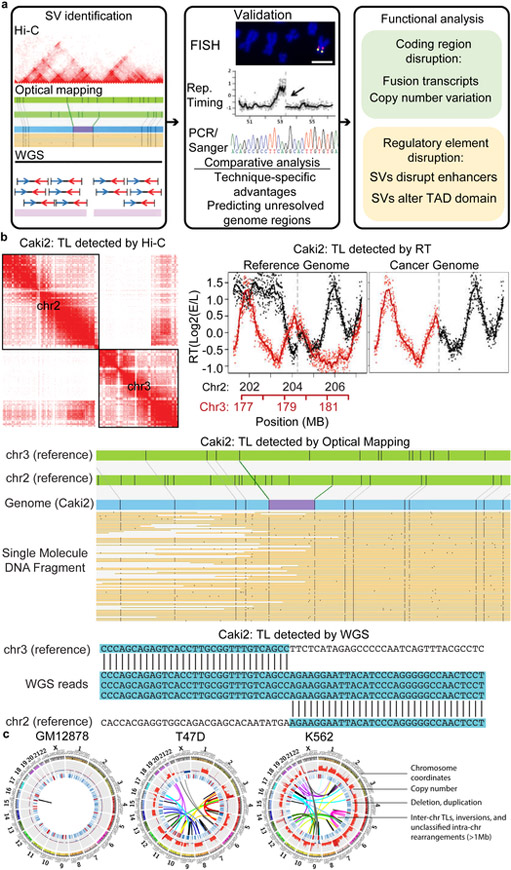
To evaluate the ability of different platforms in detecting structural variants, we compared whole-genome sequencing, optical mapping, and Hi-C data in eight cancer cell lines and one karyotypically normal control (GM12878) (Fig. 1a and Supplementary Table 1). We generated WGS data in seven cancer cell lines with an average coverage over 30X, and downloaded the data for LNCaP and GM12878 cells from a previous study17 and the Illumina Platinum Genome Dataset, respectively. We built an in-house pipeline that integrates the results from LUMPY, DELLY, and control-FREEC software18-20 for initial SV detection, and then performed extensive data filtering (Supplementary Fig. 1, Supplementary Table 2,3). Next, we performed optical mapping in the same nine cell lines with an average coverage of ~100X, the most extensive effort in cancer cells thus far. We used BioNano Refaligner 6119 and pipeline 6498 to conduct de novo assembly and SV detection, and we designed an in-house pipeline to perform further data filtering (Supplementary Fig. 2, Supplementary Table 4). Lastly, we performed Hi-C experiments in 14 cancer cell lines and analyzed an additional 21 previously published datasets 21-27. We developed a novel algorithm to use Hi-C data to identify re-arrangement events, including translocations, inversions, deletions, and tandem duplications (Supplementary Table 5, Supplementary Fig. 3 & 4). After comparing and merging the results from each platform, we identified thousands of insertions and deletions (>50bp), hundreds of tandem duplications and inter-chromosomal translocations, and tens of inversions (Supplementary Table 2). We compiled a list of high-confidence SVs that were predicted by at least two methods (Supplementary Table 6). An example is shown in Fig. 1b, where a translocation between chromosomes 2 and 3 in Caki2 cells was detected by all three methods. This translocation was also validated by observation of dramatic shifts in DNA replication timing profiles in the same region. Finally, we observed that the cancer genomes displayed many more rearrangement events compared with normal cells, as illustrated by circular genome structural profiles28 (Fig. 1c, Supplementary Fig. 5).
Figure 1 ∣. Overall strategy of SV detection in cancer genomes.
a. The pipeline of SV detection, validation, and functional analysis. b. An example of the same translocations detected by different technologies in Caki2 cells (hg38 coordinates: chr2:204,260,308 and chr3:179,694,900). c. WGS, Hi-C and optical mapping detect SVs at different scales. Hi-C can detect SVs genome-wide at a scale of up to chromosomal size, while optical mapping can detect SVs and build genome maps at ~10kb resolution. Combining Hi-C and optical mapping can resolve complex rearrangements and reconstruct local genome structure. WGS detects SVs at base pair resolution. d. Cancer genomes possess more CNVs and translocations in comparison with karyotypically normal GM12878 cells. Tracks from outer to inner circles are chromosome coordinates, copy number, duplications (red) and deletions (blue), and rearrangements including inversions, inter-chr translocations (TLs) and unclassified rearrangements. Outward red bars in CNV track indicate gain of copies (>2, 2-8 copies), and inward blue loss of copies (<2, 0–2 copies). CNVs are profiled by WGS with 50,000 bp bin size. Duplications, deletion, and TLs are detected by at least two methods from WGS, Irys, and Hi-C.
Detection of Large Scale Re-arrangements using Hi-C data
Several groups have reported unusual inter-chromosomal interactions in Hi-C data and suggested these signals are the results of SVs 21, 29-31, but to identify the breakpoints, they mainly relied on visual inspection 32, 33. Software tools have recently been developed to identify copy number alterations or inter-chromosomal translocations in Hi-C datasets32-35. However, to our knowledge, no algorithm has been developed that can use Hi-C for genome-wide detection of a full range of SVs, including deletions, inversions, tandem duplications, and inter-chromosomal translocations.
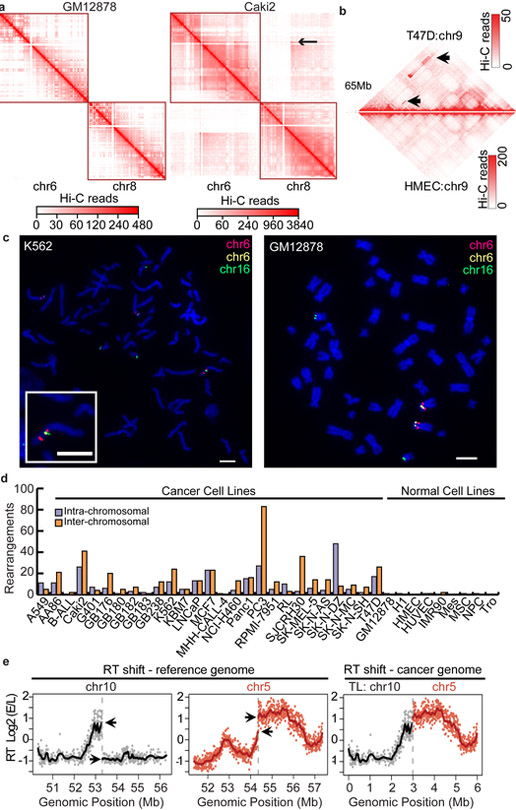
In a Hi-C experiment in karyotypically normal cells, inter-chromosomal interactions are rare (Left panel in Fig. 2a). However, this pattern does not hold in cancer cells. For example, in Caki2 cancer cells, we observed strong “inter-chromosomal” interactions (Right panel), which might be due to the fusion of chromosome 6 and chromosome 8. The challenge is to determine whether the increased signals are due to rearrangement or normal variation in 3D genome organization. We first developed probabilistic models for “normal” 3D genome organization features, including genomic distance between loci, TADs, A/B compartments, and the increased interactions between small chromosomes and between sub-telomeric regions (Supplementary Fig. 3, supplementary methods). In the event of a re-arrangement, the two re-arranged regions are genetically fused, altering the linear distance between loci. This leads to local clusters of deviations from the expected interaction frequencies, and such patterns can be used for SV detection (Fig. 2a,b). To systematically identify this signature, we developed an iterative approach to identify local clusters of interaction frequencies suggestive of the presence of rearrangements. The method progressively reduces the bin size to refine the resolution of breakpoints to as high as 1kb (Supplementary Fig. 6).
Figure 2 ∣. Detection of SVs using Hi-C in cancer genomes.
a,b. Inter-chromosomal (a) and intra-chromosomal rearrangements (b) detected by using Hi-C data (marked by arrow sign). In panel a, GM12878 heat maps are shown at 100kb resolution, and Caki2 are shown at 1Mb resolution c. A complex translocation (TL) (chr6-chr16-chr6) in K562 cells validated by fluorescence in situ hybridization (FISH). Similar results for FISH validation experiments were performed using 20 independent metaphase nuclei. Scale bars (white) represent 5μM. d. Number of inter-chromosomal and intra-chromosomal rearrangements detected by Hi-C in 29 cancer genomes and 9 normal genomes. e. An example of the impact of TLs on replication timing (RT). RT profiles of chr5 and chr10 of SK-N-MC, when plotted to the reference genome, show abrupt shifts at the TL breakpoints (←, left panels), and they are smoothly connected due to their juxtaposition in the cancer genome (right panel, normal chr10 is absent in SK-N-MC). Solid black (chr10) and red (chr5) lines indicate loess smoothened RT data. As RT experiments were designed for validation purposes, one replicate was performed for RT experiments.
We first evaluated our algorithm with a well characterized chronic myelogenous leukemia cell line (K562) and compared the results with the published karyotype. Of the 19 Hi-C predicted rearrangements, 11 can be confirmed and the remaining eight are novel 36. Since these eight events were found in both of the two replicate experiments that were performed in two independent laboratories, they are not likely a product of clonal evolution. Several of the events are complex re-arrangements: one event is between chromosome 16 and two different regions of chromosome 6 (Fig. 2c) and another is a re-arrangement involving chromosomes 1, 6, 18, and 20 (Supplementary Fig. 7). We performed FISH experiments to validate the novel predicted translocations, and 18 of the 19 predicted translocations using Hi-C data were validated by either FISH or previous karyotyping (Supplementary Table 7), suggesting that our algorithm can identify large-scale structural variation with high specificity.
To further evaluate the algorithm, we performed Hi-C in Tc1 cells (Supplementary Fig. 8a), which are a mouse ES cell line engineered to carry a copy of human chromosome 2137. In the process of establishing this cell line, human chromosome 21 was subject to gamma irradiation 37, leading to massive genomic re-arrangements, a subset of which have been previously identified using PCR and Sanger sequencing 38. We evaluated the sensitivity of our algorithm at various sequencing depths by sub-sampling, and found that the algorithm can achieve decent sensitivity with as few as 5-10 millions reads. The performance reaches a plateau at ~100 million sequencing read pairs with a sensitivity of 90% (Supplementary Fig. 8b). The predicted breakpoints are internally consistent when at least 50 million reads are available (Supplementary Fig. 8c,d). We noticed that sometimes Hi-C and WGS call breakpoints in the same regions but report different strandedness (Supplementary Fig. 8e-h). The discrepancies usually involve complex events, where Hi-C reports the larger scale SVs and WGS reports the smaller SVs for the sample complex event. To evaluate the effect of sample heterogeneity, we simulated mixed tumor/normal samples by combining Hi-C reads from K562 and GM12878 cells at different fractions while keeping the total sequencing depth at 100 million reads. We observe a limited loss of sensitivity even with tumor fractions as low as 30%, indicating that Hi-C based SV finding is robust to moderate sample heterogeneity (Supplementary Fig. 8i).
Finally, we expanded our Hi-C analysis to 27 cancer cell lines and 9 karyotypically normal lines (Fig. 2d). On average, we reported 25 re-arrangements in cancer cells and virtually no such events in normal cells, with an inter-chromosomal to intra-chromosomal rearrangements ratio of roughly 2:1 (424 vs 274 in all cell lines). Our algorithm appears to identify mostly large structural variants, with only 4.3% of intra-chromosomal SVs being less than 2Mb in size (Supplementary Fig. 8j). This is likely because it is challenging to distinguish the strong Hi-C signals as a result of structural variation from those strong local interaction signals within the same TAD.
Validation of Hi-C breakpoints by replication timing
Next, we compared our Hi-C defined breakpoints with altered DNA replication timing as an independent functional test. Eukaryotic genomes replicate via the synchronous firing of clusters of origins, which together produce multi-replicon domains, each of which complete replication in a short (45-60 min) burst during S-phase39, 40. Genome-wide profiling of replication timing reveals that these domains can be replicated at different times during S phase, with adjacent earlier and later replicating domains punctuated by regions of replication timing transition39, 40. Consequently, translocations that fuse domains of early and late replication can result in earlier replication of the late replicating domain and/or delayed replication of the early replicating domain41, 42. When mapped to the reference genome, these changes appear as abrupt shifts in replication timing profiles that have the potential to validate breakpoints (Fig. 2e, Supplementary Fig. 9a). Our Hi-C pipeline identified 249 translocations (at 10kb or 100kb resolution) in 10 cell lines with available replication timing. Among them, 75 translocations were associated with an abrupt shift in replication timing. Since an abrupt shift is only expected for translocations between domains that replicate at different times, we classified the genome into regions that are constitutively early replicating (CE), constitutively late replicating (CL) and regions that switch replication timing during development (S), using 48 replication timing profiles of non-cancerous cell lines and differentiation intermediates (Supplementary Methods, Supplementary Fig. 9b). Among the 249 translocations detected by Hi-C, 9 were CE to CL fusions and 32 were CE to CE or CL to CL fusions. As expected, an abrupt shift in timing was identified in CE to CL with a much higher frequency (~67%) than in CE to CE or CL to CL fusions (~13%) (Supplementary Fig. 9c). Translocations between CE were observed with a frequency three times higher than expected by chance (Supplementary Fig. 9c), which is consistent with previous reports linking chromosomal breakpoints to early replication and higher transcriptional activity45, 46. Overall, replication timing can provide functional validation of a specific class of translocation events that fuse regions that are replicated at different times in S phase.
Cross-platform comparison and integration of SV detection
To systematically evaluate the performance of different platforms, we compared the SVs predicted by Hi-C, optical mapping, WGS, fusion transcripts, karyotyping36, 47-54, and paired-end tag sequencing (PET-seq)55, 56 (Supplementary Fig. 10, Supplementary Table 8). We defined rearrangements detected by at least two different methods as high-confidence SVs. As a way to approximate sensitivity and specificity, we defined the contribution of a method as the fraction of high-confidence SVs that are detected by this method, and overlap rate refers to the proportion of SVs from one method that overlap with high-confidence SVs.
Overall, we observed that 20% of all inter-chromosomal translocations were identified by at least 2 platforms (Supplementary Fig. 11a-b). Compared with previously known karyotypes in each lineage, many of the observed translocations are novel. For example, 14 out of 26 translocations in T47D cells found in this study have not been reported before (Supplementary Fig. 11c). We selected eight of them for further validation, and all of them were confirmed by PCR (Supplementary Table 7). Hi-C is a method with significant contribution and high overlap rate (48% and 66%), and with better performance for inter-chromosomal translocations (53% and 66%) than intra-chromosomal SVs (43%, 71%) (Supplementary Table 9, Supplementary Fig. 12). Integration of Hi-C, optical mapping and WGS increases the overall contribution to 90% (their individual contributions are 48%, 40% and 64%, respectively). Karyotyping has a high overlap rate with the high confidence calls for all kinds of large SVs (88%) and relatively good contribution for inter-chr TLs (56%).
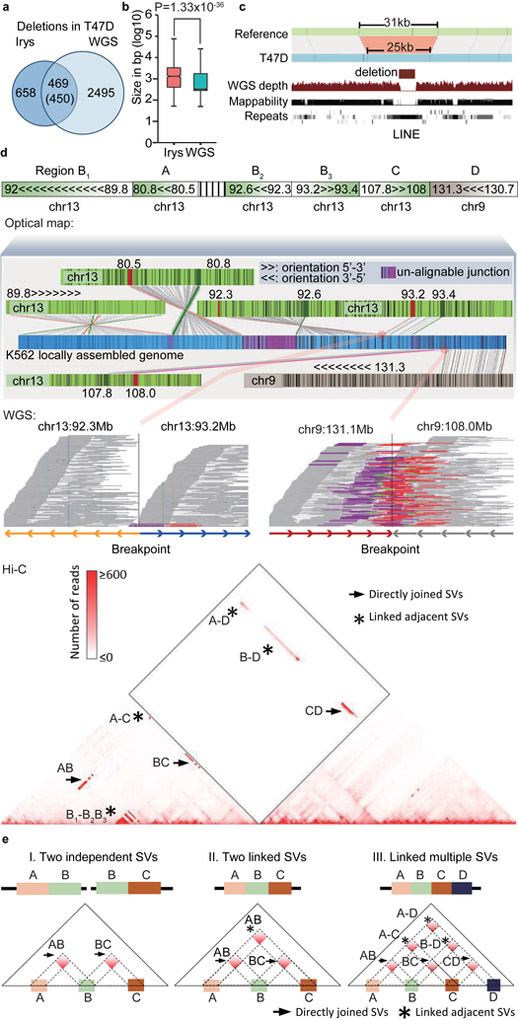
Next, we merged the results across different platforms in the same cell line into a final high-confidence SV list and refined the breakpoints using the highest resolution available (Supplementary Table 8, Supplementary Fig. 12c). More importantly, we resolved the SV type for a subset of unclassified large intra-chromosomal rearrangements detected by WGS and optical mapping. For example, Irys reported 24 unclassified intra-chr rearrangements (≥5Mb) in T47D cells. By comparing with Hi-C or WGS data, we were able to identify the SV types for 9 of them (37.5%).
We also identified thousands of gains or losses of genetic material by optical mapping and WGS in each cancer cell line. Optical mapping detects fewer but larger deletions than WGS (Supplementary Table 10). In T47D cells, WGS detected 2,943 deletions with a median size of 552 bp, while Irys reported 1,128 deletions with a median of 1,335 bp (Fig. 3a,b). 84% (2495/2943) of WGS-detected deletions are missed by Irys. Among them, 78% are smaller than 1Kb, which are likely to be missed by optical mapping, as its resolution is limited by the minimum distance between two nicking sites. 3% of the deletions predicted by Irys overlap with multiple smaller WGS deletions, and in those cases, the summed size of these WGS deletions are close to the Irys-detected deletion (Supplementary Fig. 13a-c, Supplementary Table 11). 58% of the Irys-detected deletions are not captured by WGS. We tested a subset of deletions detected by Irys, and 87.5% (14 of 16) were validated by PCR (Supplementary Table 7). Further, optical mapping can identify deletions within repetitive regions where WGS reads are not mapped (Fig. 3c) and in regions with lower mappability around the breakpoints (Supplementary Fig. 13d). We detected many megabase-scale deletions in the cancer cell lines. In contrast, the largest deletion we found in GM12878 cells is a 700kb event associated with potential V(D)J recombination (Supplementary Fig. 14). We found that WGS, Irys and Hi-C can detect different sets of inter-chromosomal and large-scale rearrangement (Supplementary Fig. 15). Besides mappability, we observed that both Hi-C and Irys are particularly powerful at detecting rearrangements involved with unalignable junctions (Supplementary Fig. 16a and b), which could come from a third chromosome that is too short to be recognized, the non-templated addition of bases to the genome, or exogenous DNA sequences such as that from viruses.
Figure 3 ∣. Comparison of SVs detected by different methods.
a. Overlap of deletions in T47D cells detected by optical mapping and WGS. b. Size distribution of deletions detected by optical mapping (n=1108) and WGS (n=2964, P = 1.33X10−36, two-sided Wilcoxon rank-sum test). For boxplots, the box represents the interquartile range (IQR), and the whiskers extend to 1.5 times the IQR or to the maximum/minimum if less than 1.5x IQR. c. Optical mapping detects a 6Kb deletion within chrX:96,041,289–96,072,340 that is missed by WGS. d. Reconstruction of the complex local structure of a derivative chromosome in K562 cells through integration of optical mapping, Hi-C and WGS. The rearranged allele consists of 5 regions: A (chr13:80.5–80.8Mb), B (chr13:89.7–93.3Mb), C (chr13:107.8–108Mb), D (chr9:130.7–131.3Mb), and an unalignable region. Further, segment B consists of three smaller regions (B1, B2, and B3 in the figure). We reconstructed a global view of the genome structures in this region by stitching several optical mapping contigs together (middle panel). Each junction of the optical mapping genome map can be validated by Hi-C data. WGS data can provide bp-resolution breakpoints for specific breakpoint junctions. Each line in the WGS panel represents a read pair. WGS reads that support the breakpoint site are marked as purple (forward strand) and red (reverse strand). e. Strategy of using Hi-C to reconstruct SVs. Hi-C shows increased interaction frequency if two translocated regions are directly joined (→) or if they are not immediately adjacent (*), but are linked to the same rearranged allele.
In summary, we found that an integrative approach combining complementary methods is essential to gain a more comprehensive understanding of structural variation in cancer genomes (Table 2). An example is in shown in Fig. 3d, where we use optical mapping to thread the putative local structure, WGS calls to pinpoint breakpoints, and the Hi-C data to validate the linkage of several adjacent rearrangements on the same allele (Fig. 3e, Supplementary Fig. 17).
Table 2.
Comparison of three methods
| WGS | Optical mapping (Irys) |
Hi-C | Optical mapping (Irys) +Hi-C |
Three methods |
||
|---|---|---|---|---|---|---|
| Resolution of breakpoint | 1bp resolution | √√ | √√ | |||
| 10kb resolution | √√ | √√ | √ | √√ | √√ | |
| Low-mappability region | Whole SV located inside a repeat | √ | √√ | √√ | √√ | |
| Breakpoint located inside a repeat | √ | √√ | √√ | √√ | √√ | |
| Estimate gap size | - | √√ | √√ | √√ | ||
| SV size | Global chromosomal alteration | √ | √√ | √√ | √√ | |
| Deletion | ≥1bp | ≥100bp | ≥1Mb | ≥100bp | ≥1bp | |
| Insertion | ≥1bp | ≥100bp | NA | ≥100bp | ≥1bp | |
| Inversion | ≥1bp | ≥70Kb | ≥1Mb | ≥10Kb | ≥1bp | |
| Inter-chr TL | ≥1bp | ≥100Kb | ≥10Kb | ≥10Kb | ≥1bp | |
| Complex SV | Overcome un-alignable junction | √ | √√ | √√ | √√ | |
| Link multiple SVs | √ | √√ | √√ | √√ | ||
| Reconstruct structure of complex SVs | √ | √ | √ | √√ |
Robust performance.
Potentially capable of detection depending on variables such as coverage, contig length & label density.
Better estimation of gaps in the human genome
We noticed that optical mapping can be used to better estimate the size of gap regions. We detected a number of deletions in multiple samples including GM12878 when we used the hg19 reference genome, but these deletions disappeared when we processed data with a more recent version of the reference genomes (GRCh38). Further investigation shows that many such “deletions” identified in the hg19 consist of gaps in the reference genome and that the size of these gaps have been corrected in the GRCh38 build. The corrected size in GRCh38 is very similar to our predictions (Supplementary Table 12). However, we noticed that there remain several such “deletions” over gap regions even in the GRCh38 build, indicating that either these gap sizes can be further refined or represent polymorphisms in the population. We compared our results with two recent studies that also re-estimated the genomic gaps in the GRCh38 reference57, 58. While our data show consistency to previous results overall (Supplementary Table 13), we do observe differences due to population polymorphisms, including a gap region where we reported a range of sizes between 889 bp to 1,535 bp across 9 different individual cell lines (the estimation is 1,299 bp by Pendleton et al. and 705 bp from Seo et al., respectively).
Functional consequences of structural variants in cancer genomes
To investigate the functional consequences of SVs, we first analyzed RNA-Seq data of 11 cancer cell lines to identify fused gene transcripts. We detected many RNA-Seq read pairs whose two ends are mapped to different chromosomes, crossing the translocation breakpoints identified in this study (Supplementary Table 14). We also discovered many novel fusion transcripts involving bona fide oncogenes, such as EVI1-CFAP70 in T47D cells. How these novel gene fusions events contribute to the oncogenic potential remains to be further investigated.
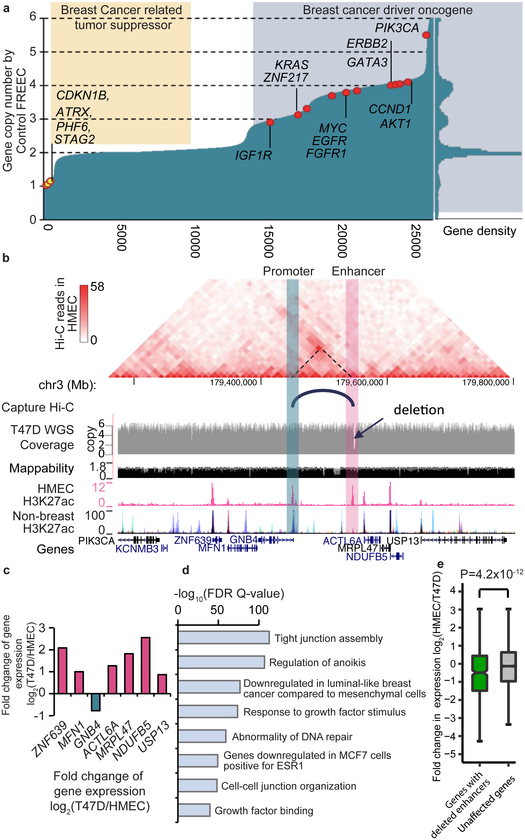
Copy number alterations (CNA) represent another class of genetic variation in cancer. We profiled the CNAs in the T47D breast cancer cell line and compared them with the WGS data of 560 breast cancer patients 16. Eight out of the top 10 frequently mutated oncogenes in patients were also amplified in T47D cancer cells, and tumor suppressor genes such as ATRX and CDKN1B displayed loss of copies (Fig. 4a), suggesting that T47D cells reflect the CNA landscape in breast cancer. We further compared the RNA-Seq data in T47D and HMEC (human mammary epithelial cells) and found that loss-of-heterozygosity (LOH) and homozygous deletions lead to significantly reduced gene expression, which was also observed in other cancer cell lines (Supplementary Fig. 18a-d). We found exon deletions in 25 COSMIC tumor-related genes, and the majority (76%) showed decreased transcription (Supplementary Fig. 18e). We noticed widespread amplification of known oncogenes (such as MYC) and loss of cell cycle checkpoint genes (such as CDKN2A/B, Supplementary Fig. 19). We found over 100 highly amplified (≥5 copies) or deleted genes in cancer cells that were not reported in COSMIC, suggesting their potential roles in cancer (Supplementary Fig. 20).
Figure 4 ∣. The impact of SVs on enhancers.
a. Copy number changes in T47D cells of Refseq genes, sorted by copy number. Genes that are frequently mutated in breast-cancer are labeled if they show amplification (red dots) or deletion (yellow dots). The right panel of this figure displays the density plot of gene copy numbers. b. A ~3.4kb deletion (chr3:179,546,826–179,550,207) in T47D overlaps an HMEC specific enhancer. Hi-C data from HMEC indicates that there is an interaction between the deleted enhancer and the promoter of gene GNB4. This enhancer-promoter linkage is also reported in GM12878 cells by the Capture Hi-C data. According to WGS data, the local region is amplified and has 6 copies in T47D cells, but the enhancer is deleted in 5 of the 6 copies. c. Compared with HMEC, all the genes in this region in T47D are up-regulated potentially due to the local amplification, except for GNB4, whose expression is reduced by ~50%. d. Functional pathway analysis of deleted enhancers (n=1859) by GREAT tool (P-value from two-sided Binomial test). e. Genes with deleted enhancers show reduced expression levels (two-sided Wilcoxon rank-sum test). Genes with exon deletions or copy number loss are excluded. 534 genes are linked by Capture Hi-C data to at least one deleted enhancer (green), and 10,677 genes are linked to enhancers that show no deletions (gray). For boxplots, the box represents the interquartile range (IQR), and the whiskers extend to 1.5 times the IQR or to the maximum/minimum if less than 1.5x IQR.
Deletions in cancer and normal cell lines differed in their likelihood of disrupting repetitive elements or functional elements. GM12878 cells are more enriched for deletions in repetitive elements when compared with cancer cell lines (70% vs 50%, expected value based on genomic background is 50%) (Supplementary Table 15). Interestingly, deletions of genes and enhancers are depleted in GM12878 cells relative to the genomic background (12 vs 60, empirical p value <0.001, Supplementary Table 16), while the cancer cell lines do not show such depletion of enhancer deletions (Supplementary Fig. 21a).
To identify deletions specific to cancer genomes, we compared the observed deletions with the Database of Genomic Variants (DGV), which compiles known polymorphic SVs identified by previous studies. 95% of the deletions identified in GM12878 cells have been previously reported, suggesting they are polymorphisms in the population. The fraction of polymorphic deletions in cancer cells is lower at 90% (Supplementary Fig. 18f, Supplementary Fig. 22a), likely due to the presence of somatic mutations. In total, cancer cells suffer a greater loss of genetic material compared with normal cells (Supplementary Fig. 22b). Further analysis shows that polymorphic deletions are enriched for repetitive elements (70% vs 50% genomic background) and depleted of exons (1.5% vs 4% genomic background) (Supplementary Fig. 22c-d). In the six cancer cell lines where we can find control cells with enhancer annotations, we found that the polymorphic deletions are resistant to enhancer loss (empirical P < 0.005 in all cell lines, Supplementary Fig. 21b). In contrast, the novel deletions are not enriched in repeats or depleted of enhancers or exons (Supplementary Fig. 21c and Supplementary Fig. 22). Instead, they are enriched in COSMIC tumor related genes (Supplementary Fig. 18f)59, suggesting that a subset of the deletions are potentially pathogenic. We confirmed that copy number changes detected by optical mapping and WGS are highly consistent (Supplementary Fig. 23).
Next, we investigated whether structural variants can influence the expression of cancer-related genes by disrupting distal regulatory elements. For this analysis, we focused on the comparison between T47D breast cancer cells and HMEC human mammary epithelial cells. We predicted enhancers in HMEC using H3K27ac data from the ENCODE Consortium and compared the enhancers with deleted regions in T47D to identify potential deleted enhancers in cancer cells (Supplementary Table 17). We show an example in Fig. 4b, where a 3.4kb deletion downstream of the gene GNB4 (G protein subunit Beta 4) overlaps with a breast-tissue specific enhancer. This region has six copies due to genomic amplification, five of which carry this deletion and only one copy of the enhancer remains undisrupted. Evidence by Hi-C in HMEC and capture Hi-C data60 suggests that GNB4 is potentially regulated by this enhancer. More importantly, it is the only gene in this region with decreased expression; the expression of the rest of genes in this region are highly upregulated possibly due to the increased copy number (Fig. 4c). Further, we found that globally, deleted enhancers are located near genes involved in breast cancer relevant pathways (Fig. 4d) and genes linked to these deleted enhancers show a reduced level of expression (Fig. 4e). Overall, these results suggest that deletions in cancer genomes may frequently affect enhancers and potentially contribute to oncogenesis.
The impact of structural variations on 3D genome organization
It has been shown that genetic mutations can disrupt TADs and create “neo-TADs” that lead to mis-regulated gene expression in developmental disorders61, 62. Several groups have also shown that alterations that affect TAD boundaries or CTCF binding sites at specific loci can create new chromatin structural domains leading to mis-regulation of nearby oncogenes through “enhancer hijacking”63-65. However, the extent to which SVs alter 3D genome structures genome-wide in cancer cells remains unclear.
Having identified structural variants in 20 cancer cell lines with Hi-C data, we systematically investigated the consequences of structural variation on TAD structure. We observed that neo-TADs are frequently formed as the result of large-scale genomic rearrangements in cancer cells. An example is shown in Fig. 5a, where the fusion between chromosome 9 and 18 forms a neo-TAD in PANC1 cells. Further, we found that many neo-TADs induced by SVs in cancer cells contain known cancer driver genes, such as MYC, TERT, ETV1, ETV4, and ERBB2 (Supplementary Fig. 24). To address whether neo-TAD formation is a general consequence of SV rearrangements in cancer genomes, we performed an aggregate analysis of all breakpoint-crossing Hi-C signals in each cell line. As shown in Fig. 5b, we observed that inter-chromosomal Hi-C signals form a sharp triangle shape (dashed line), suggesting the formation of a fusion-TAD as a result of the rearrangement (details in supplemental methods). This pattern was not observed when we performed the same analysis using shuffled TADs with randomized boundary positions (right panel in Fig.5b). These results indicate that structural variations in cancers can re-wire TAD structure and lead to TAD fusion and altered regulatory environments (Fig. 5c).
Figure 5 ∣. Rearrangements and TAD fusions.
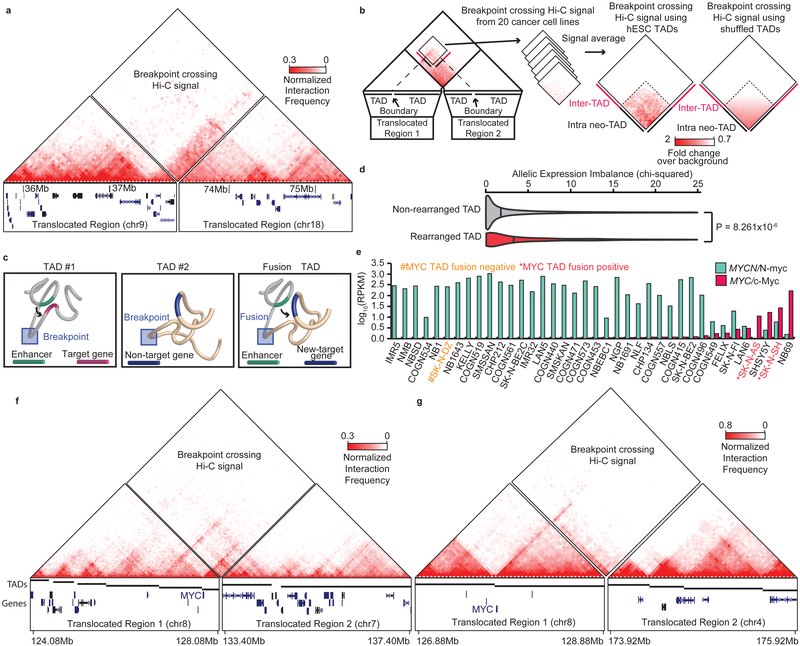
a. Fusion TAD formation as a result of a translocation in Panc-1 cells. The left box shows the rearranged region on chromosome 9, while the right box shows the rearranged region on chromosome 18. The breakpoint fusion lies in the middle. Triangle Hi-C heat maps show intra-chromosomal interactions. The diamond heat map shows the breakpoint crossing Hi-C signal, indicating the presence of a TAD fusion. b. Aggregate analysis of TAD fusions. Breakpoint crossing Hi-C signals were averaged and centered on bins between the nearest TAD boundaries (left) or shuffled TAD boundaries (right - randomization performed 1000 times). Dashed lines show expected neo-TAD borders based on the intersections of the nearest breakpoint proximal TAD boundaries. c. Model for neo-TAD formation. TADs are rearranged due to breaks and fusions, juxtaposing regulatory sequences with non-target genes. d. Violin plots showing the distribution of allelic expression bias for genes within rearranged (n=1004) or non-rearranged (n=74184) TADs. Vertical bars represent the median (p-value is from two-sided Wilcoxon rank-sum test). e. RNA-seq for MYCN/N-Myc (green) and MYC/c-Myc in neuroblastoma cell lines. Cell lines with TAD fusions at the MYC locus show high levels of MYC expression (marked in red), and the cell line that lacks a TAD fusion at the MYC locus lacks MYC expression (yellow). f. Hi-C data from SK-N-SH cells showing a TAD fusion at the MYC locus. g. Hi-C data in SK-N-AS cells showing a TAD fusion at the MYC locus.
Next, we investigated the impact of neo-TADs on gene expression. Across eight cancer cell lines, we observed that genes within TADs containing a re-arrangement show greater allelic bias than genes within non-rearranged TADs, suggesting that at least a subset of these events likely lead to altered gene expression in cis (Fig. 5d). We examined the Hi-C data in three neuroblastoma cell lines and compared the MYC gene expression. Among them, SK-N-DZ has high MYCN/N-myc expression, and the other two lines (SK-N-SH and SK-N-AS) have high MYC/c-Myc expression (Fig. 5e). Remarkably, in both of the two neuroblastoma cell lines that had high MYC expression (SK-N-AS and SK-N-SH), we identified the presence of translocations in the vicinity of the MYC gene. Copy number segmentation from the Cancer Cell Line Encyclopedia indicates that there is no MYC amplification in these two cell lines. Instead, we observed the formation of neo-TADs that encompass the MYC gene in both cases (Fig. 5f,g), suggesting that the formation of neo-TADs may be involved with the MYC activation. Determining whether any individual neo-TAD represents a recurrent alteration in a given cancer cell type, or how neo-TADs may ultimately contribute to oncogenesis, remains to be elucidated. However, our analysis suggests that creation of neo-TADs is a common consequence of re-arrangements in cancer genomes.
Discussion
Detecting structural variations in cancer genomes remains a challenge for geneticists and cancer biologists. Here we developed an algorithm that for the first time can use Hi-C data to identify a full range of SVs in cancer cells genome-wide. Our algorithm shows high accuracy for detecting inter-chromosomal translocations and large intra-chromosomal rearrangements, even with as little as ~1X genome coverage. Currently, our approach has limited power in detecting alterations less than 1Mb in size. On the other hand, we demonstrated that optical mapping excels at detecting complex SVs and resolving local genome structure but cannot detect small deletions and insertions (< 1kb). WGS has the highest resolution in detecting structural variation but is less successful in detecting SVs in poorly mappable regions of the genome or in resolving complex structural variants. Ultimately, only an integrative approach that employs complementary technologies can give the most comprehensive view of the cancer genome.
In examining regions affected by structural variants, we identified extensive deletions of distal enhancers, which are located in proximity to genes known to be mutated in cancer and important for pathways in cancer biology. To what extent such distal non-coding mutations are recurrent in cancer genomes remains unclear, but this represents an important, less explored aspect of cancer genomics. By analyzing the 3D genome structure surrounding the structural variants, we observed frequent creation of neo-TADs as a result of genomic rearrangements in cancer genomes. We have developed a web-based tool for users to visualize and examine such neo-TADs (available at the 3D Genome Browser). There has been ample evidence that the juxtaposition of active regulatory sequences to known oncogenes can contribute to tumorigenesis. Our results indicate that at least part of this effect may result from the creation of novel structural domains in cancer genomes. Whether all SVs generate fusion TADs, and the extent to which TAD fusion events are recurrent and act as driver mutations in cancer genomes will be an important question for future studies to address.
Online methods
Cell culture
K562 cells (ATCC CCL-243) were cultured in Iscove’s Modified Dulbecco’s Medium supplemented with 10% FBS and antibiotics. T47D cells (ATCC HTB-133), NCI-H460 cells (ATCC HTB-177), A549 cells (ATCC CCL-185), LNCaP (ATCC CRL-1740), and GM12878 cells (Coriell) were cultured in RPMI-1640 supplemented with 10% FBS and antibiotics, or 15% FBS and antibiotics (GM12878). Caki2 cells (ATCC HTB-47), G-401 cells (ATCC CRL-1441) were cultured in McCoy’s 5a Medium Modified supplemented with 10% FBS and antibiotics. PANC-1 cells (ATCC CRL-1469) were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% FBS and antibiotics. SK-N-MC (ATCC HTB-10), RPMI-7951 (ATCC HTB-66) cells were cultured in Eagle’s Minimum Essential Medium supplemented with 10% FBS and antibiotics. SK-N-AS cells (ATCC CRL-2137) were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% FBS, 0.1mM Non-Essential Amino Acids (Gibco) and antibiotics All cell lines cultured as part of ENCODE data generation (A549, Caki2, G401, LNCaP, NCI-H460, Panc1, RPMI-7951, SJCRH30, SK-MEL-5, SK-N-DZ, SK-N-MC,T47D) were cultured using standardized protocols, the details of which can be found through the ENCODE consortium website (https://www.encodeproject.org/).
Optical mapping experiments
10 million cells of T47D, Caki2, K562, SK-N-MC, A549, NCI-H460, PANC-1, and LNCaP were pelleted and then washed three times with PBS. Cells equivalent to 600ng of DNA were embedded in 2% Agarose (Bio-rad), solidified at 4°C for 45 minutes. Cells within plugs are lysed in 2ml cell lysis buffer (BioNano Genomics) containing 167ul proteinase K (Qiagen) for 48 hours, and washed twice with Tris-EDTA, pH 8 (TE) for 15 minutes per wash. DNA plugs were purified with 2ml 5% RNAase (Qiagen) for two hours, washed in TE for 15 minutes × 6 times, melted and equilibrated on 43°C for 45 minutes with 2ul of GELase (Epicentre). DNA was transferred onto a membrane floating in TE and concentrated by dialysis for 135 minutes. DNA was then equilibrated at room temperature overnight. 900ng DNA was digested by 30U nicking enzyme BspQ1 (New England Biolabs) in 1× buffer 3 (BioNano Genomic), 37 °C for 4 hours, and labeled with 1× labeling mix (BioNano Genomics) and 15U Taq polymerase (New England Biolabs) in 1× labeling buffer (BioNano Genomics) at 72°C for 60 minutes. Nick-labeled DNA was repaired in 1X repair mix (BioNano Genomics), 1× Thermo polymerase buffer (NEB), 50uM NAD+ (New England Biolabs), and 3ul 120U Taq DNA ligase (New England Biolabs) at 37°C for 30 minutes. DNA staining was finally performed with the final solution containing 1× flow buffer, 1× DTT (BioNano Genomics), and 3ul DNA stain (BioNano Genomics), in room temperature overnight. Optical mapping data collection: Each sample underwent in average 7 rounds of data collection on BioNano Irys platform to reach 100X reference coverage. For each round, 160ng prepared DNA was loaded to a BioNano Irys chip that contains two flow-cells, and each round contains 30 cycles of data collection.
Hi-C experiments and sequence read alignment
Hi-C in K562 and SK-N-AS cells was performed using the in situ Hi-C protocol 21 from 5 million cells using the MboI enzyme. Hi-C experiments in all ENCODE cells lines was performed using the original Hi-C protocol using the HindIII enzyme 66. Hi-C experiments were performed as biological replicates to ensure experimental reproducibility. Hi-C libraries were sequenced using Illumina HiSeq 2000 and HiSeq 2500 sequencing machines and processed to FASTQ files using standard processing pipelines. Read pairs were aligned independently using BWA-MEM to a custom GRCh38 genome assembly. The base for this assembly is available through the 1000 genomes consortium (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/reference/GRCh38_reference_genome) and contains “decoy” sequences representing common viral sequences and contigs assembled in certain individual genome assemblies but not found in the current reference. For our purposes, we also removed any alternate haplotype sequences from the reference. After initial alignment, individual reads were paired using a custom in house pipeline, and PCR duplicate reads were removed using Picard. The aligned sequences were then processed into raw Hi-C matrices using multiple bin sizes (1Mb, 100kb, 40kb, 10kb). For K562 and SK-N-AS libraries, these were also processed into 1kb matrices. This was not done for ENCODE cell line data, as HindIII is expected to cut every ~4kb in the genome, such that analyzing data with bin sizes below 4kb yields highly variable interaction matrices. We then computed the raw 1-dimensional coverage of bin and removed any bins in the bottom 2.5%. Hi-C matrices were then normalized using iterative correction 67, and we specifically retain the vector of intrinsic biases, q, for use in downstream breakpoint calling. Thus the normalized interaction frequency ni,j between any two bins i and j are given by the following equation:
where oi,j is the observed number of reads between bins i and j, and qi and qj are the biases of bins i and j, respectively.
For Hi-C data analyzed using 1Mb matrices, the A/B compartment patterns are a prominent feature of the data that complicate downstream breakpoint finding. To account for these features, we performed further processing of the 1Mb matrices. Specifically, we re-normalized the raw 1Mb matrices using iterative correction but with all intra-chromosomal interactions set to zero. We then performed principal component analysis on this balanced matrix. Specifically, using R, we computed the covariance matrix of our initial normalized matrix and then extracted the first eigen vector. We then computed a new matrix, D, representing the fraction of the original matrix derived from the first principal component by the following equation:
Where X is the original normalized matrix, v is the first eigenvector, and vT is the transposition of the first eigenvector. Each element bi,j of B represents the additive increase or decrease in interaction frequency due to A/B compartment patterns between bins i and j, and is used in modeling of interaction frequencies for breakpoint identification. For further details regarding the algorithms for SV detection using Hi-C data, please see the supplementary methods.
Structural variant detection and filtration from whole genome sequencing
SV detection
Structural variants were detected by three independent pipelines. In the first pipeline, paired-end sequencing reads were first aligned by BWA-MEM (v0.7.15-r1140) to a GRCh38 human reference genome (version GCA000001405.015) with alternate haplotypes removed. Duplicate reads were removed by Picard. Reads with a mapping quality of at least 20 were retained for SV detection. SV calls were generated from this mapped data using Delly (v0.7.7) with default parameters (-q 20). Delly detects deletions, inversions, tandem duplications, insertions, and inter-chromosomal translocations.
In the second pipeline, paired-end reads were processed by the Speedseq framework. Paired-end reads were aligned to the GRCh38 reference genome using BWA-MEM in the same manner as the first pipeline. Duplicated reads are removed by SAMBLASTER (v0.1.24). Discordant and split reads were extracted by SAMBLASTER for SV detection. SV calls were generated using Lumpy (v0.2.13) with default parameters (speedseq sv -g -t 64 -x). Lumpy reports SVs as deletions, inversions, duplications, inter-chromosomal translocations, and unresolved break ends. In both pipelines, telomeric, centromeric, and 12 heterochromatic regions are masked for SV detection using blacklisted regions provided by the Delly software.
Copy number profiles were generated using Control-FREEC [61](v11.0). For all cell lines, we used a set of common parameters (ploidy = 2 for normal cells NA12878, pseudodiploid cells SK-N-MC, and hypotriploid cells T47D, A549, LNCaP, NCI-H460 and Caki2; ploidy = 3 for triploid cells K562 and hypertriploid cells PANC-1), breakPointThreshold = 0.8, coefficientOfVariation=0.062, mateOrientation = FR). For A549, Caki2, LNCAP, NCI-H460, and PANC-1, sex was set to “XY”; for K562, NA12878, SK-N-MC, and T47D, sex was set to “XX”. Predicted copy number for each 50,000bp bin was used for making Circos plots. Regions with copy loss (copy number equal to 0 or 1) that are not captured by SV detection using Delly or Lumpy (by exclusion of those reciprocally overlap by at least 50% with deletions called by Delly and Lumpy) were included in the set of detected deletions.
SV filtration
To reduce false positive calls, the following filtration steps were applied for Delly and Lumpy SV calls. First, we require all SV calls to be supported by at least three split reads (SR) or three spanning paired-end reads (PE). Insertions or deletions less than 50 bp are removed, as are SV that map to chromosome Y or to the mitochondrial genome. SV calls from Delly and Lumpy are then merged, and only SVs that are identified by both methods are retained. We used separate criteria to call SVs overlapping between the two methods depending on the type of SV. For deletions, calls were merged between the two pipelines if they had an reciprocal overlap (RO) ≥ 50%. We used the coordinates provided by Lumpy for this merged deletion set. For inversions, calls identified by both Lumpy and Delly were merged if they had an RO ≥ 0.9. The final merged coordinates were based on the coordinates from the Lumpy calls. Translocations were merged between the two pipelines if the paired break ends mapped within +/− 50 bp of each other and if the strand of the break ends matched. The final coordinates were based on the calls from Lumpy. Regions annotated as insertions were identified by Delly alone, since Lumpy does does not annotate SVs as insertions. No specific filtration for insertion was applied.
Additional filtration was applied to specific types of SVs. For deletions, we removed deletions that have at least 50% reciprocal overlap (RO ≥ 50%) with known gap regions (+-50bp), or at least 1bp overlap with centromere regions (+/− 1kb). Recurrent deletions that are larger than 1Mb and present in more than one cell line with an RO ≥ 99.9% are removed. Large deletions (≥100kb) that do not show consistent decrease of read depth compared with adjacent regions are also removed (less than one difference of read depth between deletions and flanking 10Kb regions). For inversions, recurrent inversions that are longer than 100Kb and are present in more than one cell line (defined by RO ≥99.9%) are removed. For translocations, recurrent translocations that are present in more than one cell line (defined by both break ends being within +/− 50pb) are filtered out.
We required a minimal number of supporting reads (SR+PE) for translocation calls that we varied according to the sequencing depth and the ploidy of the WGS sample. (Cells with polyploidy can harbor an SV in only one copy of the DNA so that the SV is only present in a small fraction WGS reads.) Due to high sequencing coverage (~80X) in LNCAP sample, we only keep translocations with at least 15 supporting reads (PE+SR). For GM12878 cells (coverage of 50X), since they are diploid, we use a more stringent filter of 20 supporting reads, with at least two being split reads. For all other cell lines, which have similar read depth and ploidy, we require at least five supporting reads to call a translocation. We further compiled a list of high-coverage regions (coverage > 500X) in NA12878 which are largely characterized by repetitive genomic elements. In our initial analysis, we observed that such regions have high rates of translocation calls. However, given their extreme outlier coverage and association with repetitive elements, these are most likely simply anomalous alignments. We filtered out translocations whose breakpoint ends are located in those regions. In addition, for unclassified intra-chromosomal rearrangements called by Lumpy, we removed calls with a quality score less than 100. Finally, for tandem duplications, we require 10 supporting reads for LNCAP and 5 for GM12878 and 3 supporting reads for all other samples.
Detection of structural variants based on optical mapping
Cell line or sample-specific genomic maps are generated through de novo assembly of DNA optical reads using BioNano Refaligner 6119 and pipeline 6498. We require that DNA reads be no shorter than 150Kb with at least 9 labels per molecule, and the signal to noise ratio no less than 2.75, while the maximum backbone intensity is 0.6. The assembly pipeline was applied with the following parameters: iterations: 5; initial assembly P value threshold: 1e-11; extension and refinement P value threshold: 1e-11. De novo assembly noise are specifically: False positive density/100Kb:1.0; False negative rate:0.1; SiteSD:0.15; ScalingSD:0; RelativeSD: 0.03; ResolutionSD: 0.25.
SV detection is performed after the completion of de novo assembly by comparing assembled contigs to the GRCh38 reference genome GRCh38 using the built-in module runSV. All centromere regions are skipped during SV identification. Deletions, insertions and inversions are detected with the default settings using a p-value threshold of 1e-12. In the default output, any intra-chromosomal SVs larger than 5Mb are defined as “unclassified” intra-chromosomal rearrangements. Unclassified intra-chromosomal rearrangements and inter-chromosomal translocations are detected using a less stringent P-value threshold of 1e-8.
For further details on filtration and classification of SVs detected by optical mapping, please see the supplementary information file.
Profiling of gene copies using optical mapping
In order to identify genes that had undergone copy number alterations, we compared copy number profiles from optical mapping in the 4 primary normal tissues and 8 cancer cell lines with gene lists from RefSeq Gene annotation. The longest isoform was used for characterization of copy number changes. For each gene, the average copy number profiles of each 50kb bin spanned by the gene was considered as the copy number of that gene. The CNV of genes were also profiled by WGS normalized coverage (Control-FREEC) in T47D and Caki2 for differential gene expression analysis.
Re-prediction of gap sizes
To gain a list of candidate unresolved gap regions, recurrent deletions detected by optical mapping at least twice in cancer cells lines and at least once in normal cells were collected from 12 samples, including 8 cancer cell lines (T47D, Caki2, K562, A549, NCI-H460, PANC-1, LNCaP, SK-N-MC) and 4 normal cells (GM12878, 3078entB, 3045entB, and 3391entB). Recurrent deletions were then intersected with hg19 gaps using bedtools. Only gaps where at least 80% of the gap overlap with a deletion and the gap accounts for at least 30% of the deletion are retained for gap size re-estimation. When using hg19 as the reference genome, the gap size was predicted by subtracting the deletion size from gap size in hg19. To evaluate the predictions, the gap regions were lifted over to GRCh38, and the sizes of the same regions in GRCh38 were compared with our prediction and the size in hg19. Some gaps will ultimately have a negative value, meaning that the size of the deletion is shorter than annotated gap in the reference genome, potentially due to the variation across populations.
To predict the size of unresolved gaps in GRCh38, we repeated our analysis of deletions overlapping gap regions using GRCh38 as the reference genome as described above. In some cases, the re-estimated size of the same gap could vary among different cell lines, and the degree of variation is relatively small with respect to the overall change of perceived scale of gap size. Therefore, we report the median, the maximum, and minimal gap size of each gap from our estimation, as this variation can represent polymorphisms of gap sizes in the population. We then annotate what genes are spanned by those adjusted gaps and could be affected by intersecting re-estimated gaps with gene list in GRCh38. We further compare our gap size predictions in GRCh38 with results from previous publications 57, 58.
Genome-wide DNA replication timing
Genome-wide replication timing was measured in A549, Caki2, G401, NCI-H460, SK-N-MC, T47D and LNCaP using the Repli-seq method68. Briefly, asynchronously cycling cells were pulse labeled with the nucleotide analog 5-bromo-2-deoxyuridine (BrdU). The cells were then sorted into early and late S-phase fractions on the basis of DNA content using flow cytometry. BrdU-labeled DNA from each fraction was immunoprecipitated (BrdU IP), amplified and sequenced using Illumina HiSeq 2500. Replication timing was then measured as log2 ratio of early over late reads in 5kb bins. For K562, MCF7 and SK-N-SH cell lines, raw data for 6-fraction Repli-seq was downloaded from the ENCODE portal. The data was transformed to match the early/late repli-seq by combining G1, S1 and S2 fractions to represent early S phase and S3, S4 and G2 fractions to represent the late S phase. Smoothed replication timing profiles around the breakpoints were produced by loess smoothing replication timing data separately for the upstream and the downstream segments from the breakpoints predicted by Hi-C (Fig. 1b, Fig. 2e).
Classification of human genome into constitutive/switching regions
48 human replication timing datasets (ENCODE, www.replicationdomain.com) were used for the annotation of the human genome into constitutive/switching regions. The datasets were windowed into 50 Kb bins. Then the following criteria were used for the annotation. A threshold of above 0.15 was used to identify early replicating bins and below −0.15 was used to identify a late replicating bin for each dataset. If a bin was early in 2 or more cell types and late in 2 or more cell types, those bins were classified as “Switching” (S). The remaining bins were then evaluated as being either “Constitutive Early” (CE), “Constitutive Late” (CL) or left un-classified (N/A). If a bin was early in at least 46 out of 48 cell types, it was classified as CE. If a bin was late in at least 46 out of 48 cell types, it was classified as CL.
Quantifying abrupt shifts in RT
Genome-wide replication timing profiles in cancer genomes show several abrupt shifts in replication timing associated with translocations. We sought to quantify the frequency of these abrupt shifts. To this end we made a pipeline to detect abrupt shifts next to translocations identified by Hi-C. For each predicted translocation, un-smoothed RT data in 5kb bins from +/− 200kb of the breakpoint was used to scan for abrupt shifts. A span of +/− 200kb was chosen because the resolution of Hi-C translocation calls started at 100kb. Then for every 5kb bin, the difference between the median of the preceding 20 bins and succeeding 20 bins were calculated. Outliers were removed from this metric by a median filter (span=5). Then a threshold of 0.6 was used to determine the presence/absence of an abrupt shift. While the threshold was chosen empirically, the results showed the same trend across a wide range of thresholds.
Fusion transcripts
We downloaded paired-end RNA-seq data for 14 cell lines from the ENCODE project, European Nucleotide Archive (ENA), or Sequence Read Archive (SRA) databases (Supplementary Table 1). We used three different pipelines (Tophat-Fusion [v2.1.0]69, Star-Fusion [v1.1.0]70, and EricScript [v0.5.5]71 to identify fusion transcripts. For Tophat-Fusion, paired-end reads were aligned to a GRCh38 reference genome (version GCA000001405.015) to identify fusion events. Tophat-Fusion was run on the following parameters: “--no-coverage-search -r 50 --mate-std-dev 80 --max-intron-length 100000 --fusion-min-dist 1000 --fusion-anchor-length 13”. Tophat-Fusion outputs a list of potential fusion events, which were then processed by Tophat-fusion-post to filter out false positives by aligning sequences flanking fusion junctions against BLAST databases. Fusion events were further filtered requiring at least three split reads or three spanning read pairs. In Star-Fusion, a built-in GRCh38 reference genome with Gencode v26 annotation was used. Fusion transcripts were detected by Star-Fusion with default parameters. To reduce false positives, fusion events with a Fusion Fragment Per Million total reads (FFPM) less than 0.1 were removed. EricScript detects fusion transcripts by aligning the reads to a pre-built reference transcriptome (Ensembl Version 84) provided by the authors. Candidate fusions are further required to be supported by at least three spanning read pairs and three split-reads. We also included a fourth set of fusion transcripts from Kljin et al72. The final set of fusion transcripts was obtained by considering the union of fusion calls from the three pipelines and the fourth set of fusion events identified by Kljin et al.
Identification of allelic imbalance in expression.
To evaluate the effects of TAD fusion events on altered gene expression in cis, we tested whether TADs containing rearrangements showed different patterns of allele specific gene expression compared to TADs that lack rearrangements. For each cell line where we had WGS (T47D, Caki2, K562, A549, NCI-H460, PANC-1, LNCaP, SK-N-MC), we aligned RNA-seq data to the genome using STAR. We then implemented the WASP pipeline (PMID: 26366987) for filtering and re-aligning reads to identify reads that show inherent allelic mapping biases. We then computed the number of reads that aligned to each allele at each single nucleotide variant within an exon of any GENCODE gene using samtools mpileup. The number of reads aligning to each allele was normalized by the total number of reads (RPM), to account for sequencing depth differences between cell lines. To compute the degree of bias in expression between alleles, we used a simple chi-squared statistic. To account for potential differences in copy number between alleles, the expected value of the chi-squared statistic for each SNV was derived from the observed ratio of coverage between alleles from WGS. Specifically, the expected value for each allele was calculated as the fraction of reads from WGS aligning to that allele multiplied by the sum of the RNA-seq RPM values across both alleles.
Gene ontology analysis of deleted enhancers
To perform ontology analysis of enhancer deletions, the locations of high confidence deletions in T47D cells was intersected with H3K27ac defined enhancers in HMEC cells. After removal of duplicates, the loci of deleted enhancer were lifted-over from hg38 to hg19 and gene ontology analysis was performed by GREAT using the hg19 reference as background [76] (GREAT requires the use of the hg19 reference). Association rule was set as “Basal plus extension”, with “proximal 5.0kb upstream”, “1.0 kb downstream”, and “plus Distal: up to 1000.0kb”.
Differential gene expression from gene dosage or enhancer deletion
To evaluate the effects of gene dosage and enhancer deletions on gene expression, we evaluated the expression of genes in T47D or Caki2 cell lines where we detected copy number alterations of the gene itself or of linked enhancers. For T47D, we used RNA-seq data from HMEC cells as a normal control, and for Caki2, we used RNA-seq data from primary kidney tissue as a normal control. We downloaded FASTQ files of paired-end RNA-seq data from T47D, HMEC, Caki2, and primary kidney from the SRA database or ENCODE. Each sample contained two replicates. The raw reads were aligned, and differential expression analysis was performed using Tophat and cufflinks . For analyzing the impact of gene dosage on expression, we grouped genes into 4 classes: homozygous deletions (0 copy), genes with LOH (1 copy), normal genes (2 copies), and amplified genes (≥3 copies) according to CNV profiles from WGS. We calculated the expression (FPKM) fold change of all genes in each category relative to the control sample.
For analyzing the impact of enhancer deletion of gene expression, we first filtered genes and removed those with deletions of exons or entire genes to control for deletions that the impact of gene dosage on expression. We further filter genes and focus only on the 9672 genes with evidence of expression in HMEC cells (FPKM ≥ 1). Enhancers were annotated enhancers as homozygous deletion or LOH based on WGS coverage, and were examined for linkage to filtered genes from significant interactions identified by capture Hi-C in GM12878 cells. The expression fold change in expression between T47D and HMEC cells was then computed for the 530 genes with a copy number loss of linked enhancers was compared with 9142 unaffected genes using the Wilcoxon test.
TAD fusions
To evaluate the effects of SVs on TAD structure, we analyzed breakpoint crossing Hi-C signal. Our initial observations identified cases where the nearest TAD boundaries to the breakpoint were being “fused” together to create a new TAD. To evaluate whether such TAD fusion events were generally the case, we analyzed whether the breakpoint crossing Hi-C signal between the nearest TAD boundaries showed a local enrichment, which is characteristic of “normal” TADs.
We begin this analysis with a list of breakpoints within each cell type. For each breakpoint, we identified the nearest breakpoint proximal TAD boundary based on TAD calls from H1 hESCs. We chose TAD calls from H1 hESCs as we wanted to use TAD calls from an independent, non-rearranged cell type, in case the rearrangement was altering TAD calls within the rearranged cell line. We should note that TAD calls are highly stable between cell types, such that these results are similar regardless of the source of the TAD calls. We then identified the predicted “peak” of the TAD “triangle” by identifying the bin representing the interaction between each of the nearest breakpoint proximal TAD boundaries. The bin representing the interaction between each of the breakpoint proximal TAD boundaries was then considered as the center of a sub-matrix. We calculated the average interaction frequency of all bins within the 41×41 bin sub-matrix centered on the TAD boundary interacting bin. Each bin was then normalized to this average interaction frequency, such that the new sub-matrix would represent a fold change above the average value in the sub-matrix. This was then log-transformed (with a pseudocount of 1 added to avoid taking the log of zero and to minimize the effects of noisy low frequency interactions). The reason for normalizing to mean of the submatrix is to account for the differences in interaction frequencies that would be expected due to genomic distance alone. In other words, without normalizing to the central bin, the aggregated Hi-C data would be dominated by short distance interactions. The log-fold change sub-matrix was then averaged for all breakpoints in all cell types, yielding a single aggregate log fold-change sub-matrix. For display purposes, this was then exponentiated to represent these values again as a fold change. This process was also applied to a random set of TAD boundaries. Random TAD boundaries analysis was performed by first randomly permuting the TAD boundaries from H1 hESCs, using the following the following approach: for TADs on chromosomes affected by SVs, we generate a random number between 1 and the size of the chromosome where it is located. This number is then added to the start and end coordinates of every TAD on the chromosome. If the randomly generated TAD is larger than the size of the chromosome, the size of the chromosome in base-pairs is then subtracted. This is done to preserve to observed size and spacing of TADs in the random dataset to limit any artifacts or bias of randomization. This set of permuted TADs was then used for the input into the same process as described to evaluate the chromatin interactions across the breakpoints. We want to point out that the only data that is being randomized are the positions of TADs, and the SVs and chromatin interaction maps used for the plot are both from the true cancer cell lines in this study. This randomization was repeated 1,000 times.
Statistics
We use the Wilcoxon rank-sum test for comparison of distributions between two groups, as this is a non-parametric distribution that does not make underlying assumptions of normality. We also use permutation to calculate empirical P-values, which does not make any assumptions on the underlying distribution of the data.
Data Availability:
Hi-C and replication timing data generated in this study have been deposited to ENCODE portal and can be accessed without restrictions (http://encodeproject.org/). Details of specific accession numbers for each dataset can be found in the supplementary method section.
Code availability
Code for Hi-C based structural variant identification can be accessed through Github (https://github.com/dixonlab/hic_breakfinder). We used publically available software for whole genome sequencing structural variant detection (LUMPY, DELLY, control-FREEC). We used BioNano Refaligner 6119 and pipeline 6498 for structural variant detection from optical mapping experiments. Custom data processing scripts can be made available upon request.
Supplementary Material
Acknowledgements:
This work was supported by NIH grants R35GM124820, R01HG009906, and U01CA200060 (F.Y.), R24DK106766 (R.C.H. and F.Y.), GM083337 (D.M.G.), GM085354 (D.M.G.), DK107965 (D.M.G.), U54HG004592 (J.D. and J.A.S.), HG003143 and DK107980 (J. D.), U41HG007000 (W.S.N.), and DP5OD023071 (J. Dixon). This work was also supported by European Research Council (D.T.O., C.E. #615584), Cancer Research UK (D.T.O., C.E. #20412 & 22398), Wellcome Trust (#84459 to D.T.O. and C.E.), Wellcome Trust (#106985/Z/15/Z to SH). J.D. is an investigator of the Howard Hughes Medical Institute. J.R.D. is also supported by the Leona M. and Harry B. Helmsley Charitable Trust grant #2017-PG-MED001. F.A. was supported by Institute Leadership Funds from La Jolla Institute for Allergy and Immunology. F.Y. is also supported by Leukemia Research Foundation and Penn State Clinical and Translational Science Institute. We thank the ENCODE Data Coordination Center for helping with Hi-C and replication time data deposition. We would also like to thank Jan Karlseder and Nausica Arnault for help with FISH experiments.
Footnotes
URLs
ENCODE: http://encodeproject.org/; Replication timing data: http://replicationdomain.com/; 3D genome browser: http://3dgenome.org/;
Competing interests
The authors declare no competing interests.
Reference:
- 1.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Futreal PA et al. A census of human cancer genes. Nat Rev Cancer 4, 177–183 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soda M et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Kwak EL et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363, 1693–1703 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rowley JD Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 243, 290–293 (1973). [DOI] [PubMed] [Google Scholar]
- 6.Kantarjian H et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med 346, 645–652 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Wan TS Cancer cytogenetics: methodology revisited. Ann Lab Med 34, 413–425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zack TI et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet 45, 1134–1140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mardis ER & Wilson RK Cancer genome sequencing: a review. Hum Mol Genet 18, R163–168 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inaki K et al. Transcriptional consequences of genomic structural aberrations in breast cancer. Genome Res 21, 676–687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maher CA et al. Transcriptome sequencing to detect gene fusions in cancer. Nature 458, 97–101 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J et al. INTEGRATE: gene fusion discovery using whole genome and transcriptome data. Genome Res 26, 108–118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell PJ et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet 40, 722–729 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alkan C, Coe BP & Eichler EE Genome structural variation discovery and genotyping. Nat Rev Genet 12, 363–376 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peifer M et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature 526, 700–704 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nik-Zainal S et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu H et al. Integrative Analysis Reveals the Transcriptional Collaboration between EZH2 and E2F1 in the Regulation of Cancer-Related Gene Expression. Mol Cancer Res 14, 163–172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Layer RM, Chiang C, Quinlan AR & Hall IM LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 15, R84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rausch T et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, i333–i339 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boeva V et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28, 423–425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao SS et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon JR et al. Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z et al. The properties of genome conformation and spatial gene interaction and regulation networks of normal and malignant human cell types. PLoS One 8, e58793 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barutcu AR et al. Chromatin interaction analysis reveals changes in small chromosome and telomere clustering between epithelial and breast cancer cells. Genome Biol 16, 214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barutcu AR et al. RUNX1 contributes to higher-order chromatin organization and gene regulation in breast cancer cells. Biochim Biophys Acta 1859, 1389–1397 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taberlay PC et al. Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations. Genome Res 26, 719–731 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Y et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell 162, 900–910 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krzywinski M et al. Circos: an information aesthetic for comparative genomics. Genome Res 19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burton JN et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol 31, 1119–1125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engreitz JM, Agarwala V & Mirny LA Three-dimensional genome architecture influences partner selection for chromosomal translocations in human disease. PLoS One 7, e44196 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naumova N et al. Organization of the mitotic chromosome. Science 342, 948–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seaman L et al. Nucleome Analysis Reveals Structure-Function Relationships for Colon Cancer. Mol Cancer Res 15, 821–830 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harewood L et al. Hi-C as a tool for precise detection and characterisation of chromosomal rearrangements and copy number variation in human tumours. Genome Biol 18, 125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu HJ & Michor F A computational strategy to adjust for copy number in tumor Hi-C data. Bioinformatics 32, 3695–3701 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakraborty A & Ay F Identification of copy number variations and translocations in cancer cells from Hi-C data. Bioinformatics (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naumann S, Reutzel D, Speicher M & Decker HJ Complete karyotype characterization of the K562 cell line by combined application of G-banding, multiplex-fluorescence in situ hybridization, fluorescence in situ hybridization, and comparative genomic hybridization. Leuk Res 25, 313–322 (2001). [DOI] [PubMed] [Google Scholar]
- 37.O’Doherty A et al. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science 309, 2033–2037 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gribble SM et al. Massively parallel sequencing reveals the complex structure of an irradiated human chromosome on a mouse background in the Tc1 model of Down syndrome. PLoS One 8, e60482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhind N & Gilbert DM DNA replication timing. Cold Spring Harb Perspect Biol 5, a010132 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dileep V, Rivera-Mulia JC, Sima J & Gilbert DM Large-Scale Chromatin Structure-Function Relationships during the Cell Cycle and Development: Insights from Replication Timing. Cold Spring Harb Symp Quant Biol 80, 53–63 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Pope BD et al. Replication-timing boundaries facilitate cell-type and species-specific regulation of a rearranged human chromosome in mouse. Hum Mol Genet 21, 4162–4170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryba T et al. Abnormal developmental control of replication-timing domains in pediatric acute lymphoblastic leukemia. Genome Res 22, 1833–1844 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dileep V et al. Topologically associating domains and their long-range contacts are established during early G1 coincident with the establishment of the replication-timing program. Genome Res 25, 1104–1113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivera-Mulia JC et al. Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome Res 25, 1091–1103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sima J & Gilbert DM Complex correlations: replication timing and mutational landscapes during cancer and genome evolution. Curr Opin Genet Dev 25, 93–100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiarle R et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 147, 107–119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Struski S et al. Identification of chromosomal loci associated with non-P-glycoprotein-mediated multidrug resistance to topoisomerase II inhibitor in lung adenocarcinoma cell line by comparative genomic hybridization. Genes Chromosomes Cancer 30, 136–142 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Strefford JC et al. A combination of molecular cytogenetic analyses reveals complex genetic alterations in conventional renal cell carcinoma. Cancer Genet Cytogenet 159, 1–9 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Peng KJ et al. Characterization of two human lung adenocarcinoma cell lines by reciprocal chromosome painting. Dongwuxue Yanjiu 31, 113–121 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Beheshti B, Karaskova J, Park PC, Squire JA & Beatty BG Identification of a high frequency of chromosomal rearrangements in the centromeric regions of prostate cancer cell lines by sequential giemsa banding and spectral karyotyping. Mol Diagn 5, 23–32 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Liu J et al. Modeling of lung cancer by an orthotopically growing H460SM variant cell line reveals novel candidate genes for systemic metastasis. Oncogene 23, 6316–6324 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Espino PS, Pritchard S, Heng HH & Davie JR Genomic instability and histone H3 phosphorylation induction by the Ras-mitogen activated protein kinase pathway in pancreatic cancer cells. Int J Cancer 124, 562–567 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Sirivatanauksorn V et al. Non-random chromosomal rearrangements in pancreatic cancer cell lines identified by spectral karyotyping. Int J Cancer 91, 350–358 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Rondon-Lagos M et al. Differences and homologies of chromosomal alterations within and between breast cancer cell lines: a clustering analysis. Mol Cytogenet 7, 8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hillmer AM et al. Comprehensive long-span paired-end-tag mapping reveals characteristic patterns of structural variations in epithelial cancer genomes. Genome Res 21, 665–675 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hampton OA et al. Long-range massively parallel mate pair sequencing detects distinct mutations and similar patterns of structural mutability in two breast cancer cell lines. Cancer Genet 204, 447–457 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pendleton M et al. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat Methods 12, 780–786 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seo JS et al. De novo assembly and phasing of a Korean human genome. Nature 538, 243–247 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Forbes SA et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 43, D805–811 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mifsud B et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nature genetics 47, 598–606 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Franke M et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538, 265–269 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Lupianez DG et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hnisz D et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Northcott PA et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511, 428–434 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weischenfeldt J et al. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat Genet 49, 65–74 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lieberman-Aiden E et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Imakaev M et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods 9, 999–1003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marchal C et al. Genome-wide analysis of replication timing by next-generation sequencing with E/L Repli-seq. Nat Protoc 13, 819–839 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim D & Salzberg SL TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 12, R72 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haas B et al. STAR-Fusion: Fast and Accurate Fusion Transcript Detection from RNA-Seq. bioRxiv (2017). [Google Scholar]
- 71.Benelli M et al. Discovering chimeric transcripts in paired-end RNA-seq data by using EricScript. Bioinformatics 28, 3232–3239 (2012). [DOI] [PubMed] [Google Scholar]
- 72.Klijn C et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol 33, 306–312 (2015). [DOI] [PubMed] [Google Scholar]
- 73.Trapnell C et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Hi-C and replication timing data generated in this study have been deposited to ENCODE portal and can be accessed without restrictions (http://encodeproject.org/). Details of specific accession numbers for each dataset can be found in the supplementary method section.