Abstract
Plasmacytoid dendritic cells (PDCs) are critical for defense against respiratory viruses because of their propensity to secrete high levels of type I interferons (IFN). The functions of PDCs in the lung can be influenced by airway epithelial cells. We examined the effect of human primary bronchial epithelial cells (PBECs) on PDC functions by performing RNA-sequencing of PDCs after co-culture with air liquid interface differentiated PBECs. Functional analysis revealed that PDCs co-cultured with PBECs displayed upregulation of type I IFN production and response genes. Upregulated transcripts included those encoding cytosolic sensors of DNA, ZBP-1,IRF-3, and NFkB as well as genes involved in amplification of the IFN response, such as IFNAR1, JAK/STAT, ISG15. In keeping with the RNA-seq data, we observe increased secretion of type I IFN and other cytokines in response to influenza in PDCs co-cultured with PBECs. The PDCs also primed Th1 responses in T cells. The enhanced response of PDCs co-cultured with PBECs was due to the action of growth factors, GMCSF, GCSF, and VEGF, which were secreted by PBECs on differentiation. These data highlight possible mechanisms to enhance the production of type-I IFN in the airways, which is critical for host defense against respiratory infections.
Introduction
Plasmacytoid dendritic cells (PDCs) are a subset of dendritic cells, which are found in circulation and other organs. They are distinguished by their capacity to produce copious amounts of type I interferons (IFN)1,2 via stimulation of Toll like receptor 7 and 9 with nucleic acids or viruses. Because of this property, they play a crucial role in fighting viral infections.1,2 However, the cytokine secretion by PDCs is not restricted to IFNs as they can also secrete other cytokines and chemokines including IL-6, IL-12, CXCL8, CXCL10, CCL3, and CCL4. Similar to conventional myeloid DCs, they also express MHC Class II and costimulatory molecules and migrate to prime T cell responses.3,4 Production of type I IFNs along with IL-12 by PDCs primes CD8+ T cells and IFN-γ secreting Th1 CD4+ T cells. PDCs can also induce T regulatory cells via induction expression of IDO.3,4 In addition, type I IFN production by PDCs enhances B cell activation, plasma cell generation and antibody secretion. TNF-related apoptosis inducing ligand (TRAIL) and granzyme B serve as immunoregulatory factors that endow PDCs with the capacity to kill tumor cells, induce apoptosis of infected CD4+ T cells and suppress T cell proliferation.3,4
PDCs also play an important role in pulmonary immune responses. PDCs in the lung have been reported to be immature, expressing low levels of MHC II and costimulatory molecules but high levels of PDL-1.5 These characteristics make them poor inducers of T cells and that is why they are thought to play an immune-regulatory role in the lung. Depletion of PDCs in a mouse model of asthma enhanced the inflammation against harmless antigens. Similarly, in acute lung injury (ALI) and acute respiratory distress syndrome (ARDS), activated PDCs are protective and have been shown to prevent recruitment of pro-inflammatory monocytes into the lung.6 In contrast, PDCs were reported to be key drivers of immune-inflammatory cascade during asthma exacerbations via boosting of Th2 responses.7 Thus, PDCs can be inflammatory or tolerogenic depending on the context. Activated PDCs are major drivers of protection against respiratory infections such as influenza and RSV by virtue of production of type I IFNs and other cytokines.4 The functions of lung DC are influenced by many soluble factors such as growth factors, chemokines, and cytokines that are released from different cells within the lung. These soluble factors regulate the intensity and duration of immune response by stimulating or inhibiting activation and function of DC. For example, previous studies from our group and others have demonstrated that PBECs enhance the inflammatory and immune-surveillance capacity of mDCs.8–10 However, their effect on PDCs is not elucidated. This knowledge is essential for manipulation of PDC response to respiratory pathogens. Here we examined the changes introduced in human PDCs by primary bronchial epithelial cells at gene and functional level.
Materials and methods
Blood donors
Peripheral blood samples were obtained from healthy volunteers (22–52 years), approved by the Institutional Review Board of the University of California (Irvine, CA, USA). Written, informed consent was obtained.
Primary bronchial epithelial cells (PBECs)
Primary bronchial epithelial cells (PBECs) [Air–liquid interface (ALI)-tested] from three normal, healthy individuals (27–45 years) were obtained from Lonza Inc. (Basel, Switzerland) and were differentiated at the ALI on transwell plates as described.8 Briefly, 5 × 104 PBECs per insert were seeded into 24-well transwell plate with apical chamber coated with the rat tail collagen (BD Biosciences, San Jose, CA, USA). Cells were seeded in 100 µl B-ALI™ growth medium onto the collagen coated trans well inserts; 500 µl of B- ALI™ growth medium was added to the basal chamber of wells containing the inserts. On day 3 after seeding, once the monolayer of PBEC was confluent, media were removed from the apical chamber and allowed for air–liquid differentiation. The media in the bottom chamber were changed every two days. Approximately 28 days post-differentiation, PBECs were tested for the presence of cilia by staining for beta-tubulin and Mucus secretion (data not shown).
Isolation and culture of human plasmacytoid (PDCs) with PBECs for RNA sequencing
PDCs were purified from the peripheral blood mononuclear cells (PBMCs) of young subjects by negative selection using a DC enrichment kit (Stemcell Sep, Vancouver). Enriched DC population was subsequently sorted using FACSARIA (BD Biosciences) to obtain a pure PDC population. Purity was above 92%. (CD123+/CD11c−) PDCs from different donors were added to the bottom chamber of the transwell with ALI-differentiated PBECs for 24 h. Subsequently, the PDCs were collected for RNA-Seq and other studies. PDCs cultured in PBEC medium without PBECs were used as controls.
RNA isolation from PDCs
Total RNA was isolated from cultured PDC cells from four individuals using FFPE RNA/DNA purification plus kit (Cat # 54300, Norgen Biotek Corp) with minor modifications. This kit is suitable for challenging samples such as Formalin Fixed Paraffin embedded tissue in, which the recovery of quality RNA is difficult. We applied this kit to isolate RNA from PDCs cells since the numbers of cells were small. The RNA quality and quantity was assessed using Agilent bioanalyzer and Qubit fluorimeter respectively. High yield and quality RNA was obtained.
RNA-Seq library constructions
The Illumina Truseq RNA Access library prep kit was used to study gene expression profiling in stimulated and unstimulated PDCs. The access method of Illumina is based on sequence specific capture of coding RNA. This method is suitable for wide range of RNA quality and low RNA input. Briefly, 50 ng of RNA was used for library preparation. The total RNA is fragmented into small pieces using divalent cation at 98o C. Then cDNA is synthesized using the cleaved RNA fragments using random priming during the first and the second synthesis. Sequencing adaptors are ligated to the dscDNA fragments. The quality of libraries was assessed after the first PCR amplification. The coding regions of the transcriptome are then captured from the library using sequence specific probes to create the final libraries (as described by the manufacturer). Samples were assayed after the second PCR amplification on the Agilent 2100 Bioanalyzer. Qualification assay (qPCR) was performed using KAPA SYBR Fast Library Quantification Kit (Illumina) Universal qPCR Mix ref # 0796014001. Samples were sequenced at 2 × 100 cycle at the UCI high throughput genomic core facility (https://biochemghtfuci.wordpress.com/). Sequencing reads were aligned to genome reference files from Ensembl using Strand NGS 3.1 (http://www.strand-ngs.com/) sequencing analysis package. Approximately 70–90% of mapped reads were aligned to protein coding regions as shown by the pie chart plot (Supplementary fig.1). Reads were filtered on duplicates to remove PCR artifacts. Reads with better mapping quality and base average were retained (we set the number of duplicated reads to be permitted at “0”) (Excel supplementary data). Deduplicated sequencing reads for each sample was quantified for gene expression, which is the measurement of the association of reads to genes and exons “raw reads”. To normalize the raw reads we used Reads Per Kilo Million (RPKM) method in Strand NGS program. We measured the variations in 5′ to 3′ end coverage along each transcript using gene body coverage analysis in Strand NGS. Uniform transcript coverage was observed among all samples. Pooled analysis was performed using the Audic Claverie Test (AC), which pools together raw counts across four PDCs and PDCs co-cultured with epithelial cells, separately, and tests the differences between them based on a Poisson assumption on distribution of counts. Multiple Testing Correction: Benjamini Hochberg FDR of 0.05 and the maximum p value cut off 0.05 was used (Excel supplementary data). The genes of AC test were ranked based on p value adjusted (FDR corrected) and cut off was set at p adj <0.001 to select for statistically significant differentially expressed genes between the PDCs co-cultured with epithelial cells and PDCs.
The methods used for RNA isolation and RNA seq library construction worked with samples with small number of cells.
Culture of PDCs with PBECs and Treg induction
For co-culture experiments purified PDCs were cultured without or with PBEC differentiated at ALI. After overnight co-culture, PDCs from both groups were collected and put together with purified CD4 T cells at a ratio of 1:10 (PDC:CD4 T) in serum free AIM V medium (Invitrogen) for 6 days. Subsequently, the cells were collected and stained with specific antibodies for CD4, CD25 and FoxP3 (RnD systems). Gated CD4 cells were analyzed for the expression of CD25 and FoxP3 using Flow jo. Supernatant collected was assayed for IFN-γ and IL-10 by ELISA.
Culture of PDCs with PBECs and stimulation with influenza
For co-culture experiments purified PDCs were cultured without or with PBEC differentiated at ALI. After overnight co-culture, PDCs from both groups were collected and stimulated with influenza (heat killed influenza A strain A/PR/8/34-(Flu, Charles river, North Franklin, CT)11 at 1 µg/ml) for 24 h.12 Culture supernatants were collected 24h post stimulation and run on Multiplex kit. The specific kit used was MILLIPLEX Human 30-Plex Cytokine Panel 1 (Cat# HCYTMAG-60K-PX30), which contains the following analytes: GM-CSF, G-CSF, IFNγ, IL-1α, IL-1rα, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12p70, IL-12p40, IL13, IL15, IL-17, MCP-1, TNFα, TNFβ, Eotaxin, type I IFN, IP-10, MIP-1α, MIP-1β, EGF, VEGF, and RANTES. The procedure followed was according to manufacturer’s protocol. Briefly, supernatant was mixed with premixed beads (30 cytokines) overnight and after incubation with detection Antibodies and Streptavidin-PE for 1 h each, the plate was run on Magpix to identify specific cytokines.
Culture of PDCs with PBECs/growth factors
Purified PDCs were cultured with and without GMCSF (10 ng/ml), G-CSF (10 ng/ml) or VEGF (10 ng/ml), IL-6(10 ng/ml), IL-8(10 ng/ml), MCP-1 (10 ng/ml) (RnD systems) or a cocktail of these in various combinations and stimulated with influenza for 24 h. Supernatant collected was assayed for inflammatory mediators with multiplex as described above.
Culture of PDCs with blocking antibodies
PDCs cultured without or with PBEC were stimulated with influenza in the presence or absence of specific blocking antibodies against GMCSF (Invitrogen), GCSF, VEGF (RnD systems) or a mixture of the three antibodies at 1 µg/ml. After 24 h supernatant was collected and assayed for type I IFN, TNF-α, IL-1β, IL-6 using multiplex. Percent inhibition in the antibody treated groups was calculated compared to control group without antibody treatment.
Assay of factors secreted by PBECs
PBECs from three different subjects were used for this experiment. PBECs were allowed to differentiate at ALI. The conditioned basal medium in the bottom chamber was collected once the AEC differentiation was complete. PBEC differentiation media alone was used as control. Multiplex detection of 30 factors (same as above) was performed to determine the factors secreted by PBECs using milliplex.
Statistical analyses
Statistical analysis for cell culture experiments was performed using GraphPad Prism (GraphPad Inc., San Diego, CA, USA). One way ANOVA followed by Tukeys multiple comparison test was used for the analysis. A p-value of <0.05 was considered statistically significant.
Results
RNA-seq data indicates PBECs induce tolerance in PDCs
The effect of PBECs on PDCs gene expression changes was investigated using the transwell co-culture system. To investigate the effect of AECs in the form of PBECs on PDCs, the PBECS were cultured at ALI till differentiation was achieved. PDCs purified from the blood were then added to the basal side of the monolayer to model the airways.13,14 An aliquot of the PDCs was also cultured without PBECs but with the media. Twenty-four later PDCs were collected and RNA was extracted. PDCs from four different individuals were used. The description is provided in supplementary Table 1. The viability was comparable between PDCs cultured with and without PBECs (data not shown). Because of the low quantity of RNA, we used the access method of library construction from Illumina, which is based on sequence specific capture of coding RNA. This method is suitable for wide range of RNA quality and low RNA input. Paired end RNA sequencing (RNA-Seq) was performed to determine the gene expression differences between PDC and PDCs co-cultured with epithelial cells. We observed a uniform reads coverage in the gene body for all RNA seq data plot. We did not observe very large increases or decreases in transcript abundance, which was to be expected because PDCs were exposed to PBECs and there was no stimulation with a pathogen.
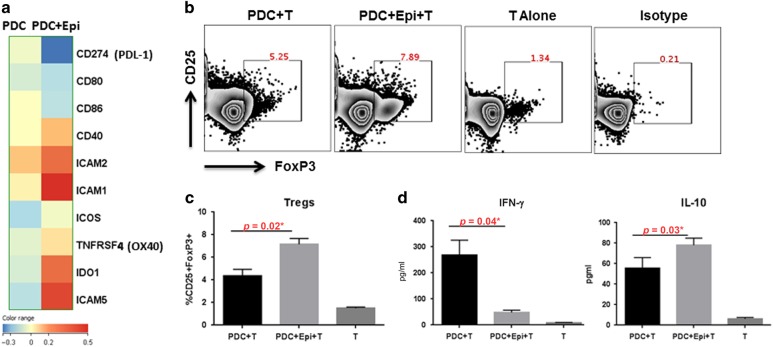
PDCs in the airways are reported to induce tolerance via the expression of IDO.15,16 We found increased expression of IDO-1 (Fig. 1a) in the PDCs cultured with PBECs compared to PDC alone. In addition, the expression of costimulatory molecules, CD80 and CD86 was decreased. However, expression of other costimulatory molecules including, CD40, OX40, ICOS, ICAM was upregulated. The expression of the inhibitory costimulatory molecule, PDL-1 was also decreased. The PDCs therefore displayed a mixed phenotype in terms of costimulatory molecules. We performed functional analysis to confirm whether the PDCs were tolerized by PBECs. PDCs exposed to PBECs induced significantly increased levels of CD4+CD25+FoxP3+ T regulatory cells to controls (p = 0.02), Fig. 1b, c). The secretion of IFN-γ was significantly decreased (p = 0.04) while IL-10 was increased (p = 0.03) in the PDCs exposed epithelial cell group compared to control. These data suggest that PBECs enhance the tolerogenic capacity of PDCs at homeostasis.
Fig. 1.
RNA-seq data indicates PBECs (Epi) induce tolerance in PDCs. RNA-seq of PDCs co-cultured with/without PBECs (Epi) was performed. a Heat map of differential expression of costimulatory and other molecules. b Zebra plots represent the induction of T regulatory cells by PDCs co-cultured with/without PBECs. c Bar graph depicts the average of similar experiments. d Bar graphs depict the level of IFN-γ and IL-10 in the supernatants. Data is mean +/− S.E. of six experiments of which three experiments were done where PDCs from three different subjects were cultured with one set of PBEC and the other three experiments were done, where PDCs from another three subjects were cultured with second set of PBEC
RNA-seq data indicates PBECs enhance the production and response of type I IFN in PDCs
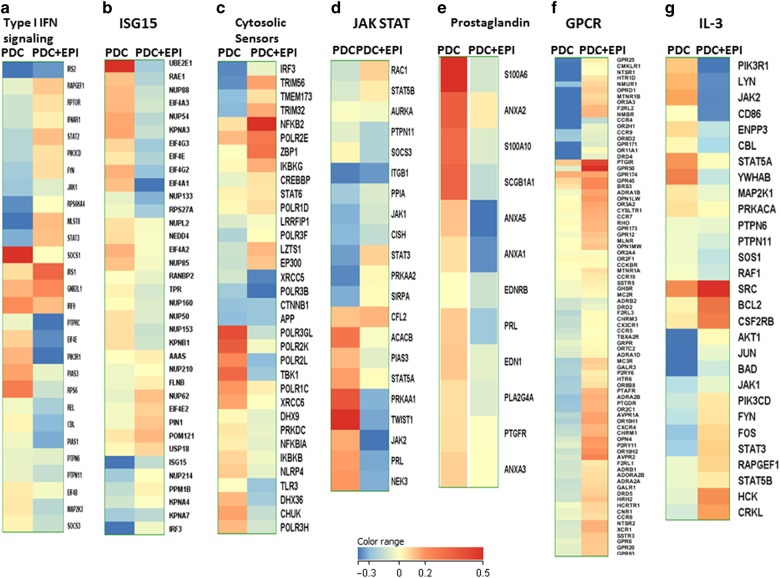
PDCs in the airways are also important for host defense against infections because of their capacity to secrete high levels of type I IFNs. Our results indicate that the major pathways altered in PDCs in response to PBECs are the type I IFN related pathways such as the type I IFN signaling, Interferon stimulated gene 15 (ISG15), cytosolic sensors of DNA as well as JAK-STAT (Fig. 2a–d). In the type I IFN pathway, the expression of IFNAR1 as well as JAK-STAT genes such as JAK1, STAT2, STAT3, which signify activation, are upregulated while SOCS1- and 3, which inhibit the pathway, are downregulated (Fig. 2a–d).17 Furthermore pathways involved in the interferon response such as ISG15 also displayed alterations after co-culture (Fig. 2b). Among the innate sensors, the expression of cytosolic DNA sensor, ZBP-118 was upregulated on PDCs after co-culture with APCs (Fig. 2c). The expression of IRF-3 and NFKB2, which are required for signaling via this receptor, was also upregulated. In contrast, the negative regulators of the pathway, NFKB1a, IKBKB are downregulated. Interestingly, the majority of the genes in the prostaglandin pathway which is a major negative regulator of IFN production were downregulated (Fig. 2e). In addition to IFN related pathways, G-protein coupled receptor (GPCR) signaling pathway also displayed major changes in PDCs co-cultured with PBECs compared to PDCs (Fig. 2f). Several of the chemokine receptor genes, such as CCR4, CCR5, CCR7, CCR10, CX3CR1, CXCR4 were upregulated indicating an activated state of PDCs. Genes in the IL-3 pathway also displayed changes (Fig. 2g). IL-3 is known to promote PDC activation and survival.19
Fig. 2.
RNA-seq data indicates PBECs (Epi) enhance the production and responses of type I IFN in PDCs. Heat maps depict all the genes in various pathways displaying significant changes in PDCs after co-culture with PBECs. a Type I IFN signaling; b ISG15; c Cytosolic sensors; d JAK/STAT; e Prostaglandin. f GPCR; g IL-3. Color range depicts log 2 fold changes in expression between PDC and PDC + epi. The values are based on AC test results with a FDR of p < 0.001
In summary, RNA seq data suggests that the capacity of PDCs to produce and amplify type I IFN production is upregulated in PDCs after co-culture with PBECs.
PBECs enhance the cytokine and chemokine response of PDCs to influenza
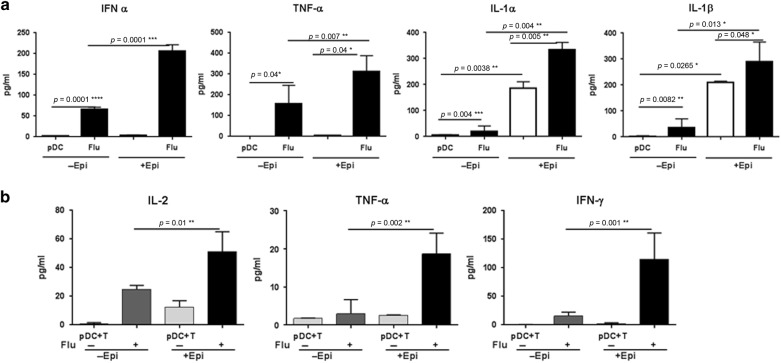
To confirm the RNA-seq data we performed functional assays to determine the alterations in the capacity of PDCs to produce type I IFN after exposure to PBECs. Briefly, PDCs co-cultured with/without PBECs differentiated at ALI were stimulated with influenza and cytokines and chemokines were assayed in the supernatants. Conditioned PBEC medium was used as control. PDCs exposed to PBECs secreted significantly higher levels of type I IFNs (p = 0.0001) on stimulation with influenza as compared to PDCs cultured alone (Fig. 3a), which is in keeping with the RNA-seq data. TNF-α secretion was also significantly increased (p = 0.007) in influenza stimulated AEC-exposed PDCs as compared to unexposed PDC (Fig. 3a). Secretion of IL-1α (p = 0.004) and IL-1β (p = 0.01) was also increased significantly in PDCs co-cultured with PBECs with and without stimulation with influenza (Fig. 3a). No type I IFN, TNF-α, IL-1α, and IL-1β was detected in the conditioned medium. Secretion of other mediators including anti-inflammatory cytokine, IL-10 was below the limit of detection (data not shown). These data indicate that PBECs enhance the inflammatory response of PDCs to pathogens as suggested by the RNA-seq data.
Fig. 3.
PBECs (Epi) enhance the cytokine and chemokine response of PDCs to influenza. PDCs co-cultured with/without PBECs (Epi) were stimulated with influenza virus and production of cytokines and chemokines was assayed by magpix. a Cytokines increased in PDCs coc-cultured with PBECs after stimulation with influenza; b Cytokines secreted by CD4 T cells primed with PDCs co-cultured with/without PBECs. Data is mean +/− S.E. of six experiments of which three experiments were done where PDCs from three different subjects were cultured with one set of PBEC and the other three experiments were done where PDCs from another three subjects were cultured with second set of PBEC
To confirm that the AEC-exposed PDCs were indeed primed to induce enhanced responses to influenza, the PDCs co-cultured with/without PBECs and influenza were cultured with purified T cells for five days and secretion of T cell specific cytokines was determined by multiplex. Culture of T cells with PDC exposed to PBECs and stimulated with influenza induced significantly higher level of IL-2 (p = 0.01) as compared to PDCs stimulated with influenza without AEC exposure (Fig. 3b). Enhanced IL-2 secretion indicated increased proliferation. In keeping with the increased proliferation, the levels of TNF-α (p = 0.002) and IFN-γ (p = 0.001) secretion by these T cells was also significantly enhanced (Fig. 3b). Other cytokines such as IL-10, IL-17, IL-4, IL-5 were not induced by influenza primed PDCs and were comparable between the T cells cultured with PDC with and without PBECs (data not shown). These results suggest that PBECs enhance the inflammatory response of PDCs to influenza, which leads to increased induction of Th1 responses.
PBECs secrete various growth factors, cytokines, and chemokines on differentiation at ALI
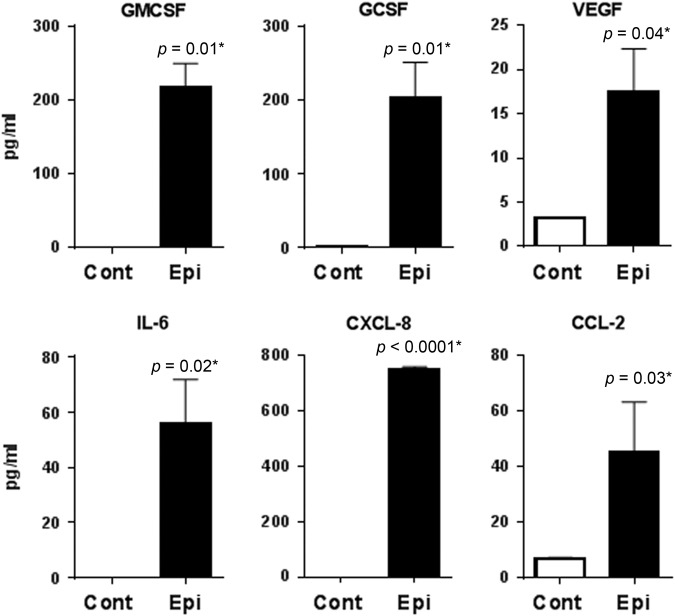
Next, we examined the mechanisms by which PBECs crosstalk with PDCs. The PDCs are cultured in the bottom chamber of the transwell and are therefore not in direct contact with PBECs suggesting that the observed enhanced response of PDCs/PBECs co-culture might be a consequence of some soluble factors secreted by PBECs. Therefore, in order to investigate the mechanisms used by PBECs to prime PDCs, a multiplex analysis was performed to determine the soluble factors in the conditioned medium from PBECs differentiated at the ALI. The medium for differentiation was used as control. Remarkably, out of 30 mediators tested (see methods) we observed only six factors, which were significantly upregulated in the medium of ALI differentiated PBECs (Fig. 4). Differentiated PBECs secreted significant levels of growth factors G-CSF (p = 0.01), GMCSF (p = 0.01) and VEGF (p = 0.04). Secretion of cytokines, IL-6 (p = 0.02) as well as chemokines, CXCL-8 (IL-8, p < 0.0001), CCL-2 (p = 0.03) was also upregulated. ALI differentiated PBECs might be priming the PDCs via secretion of the growth factors and the cytokines, chemokines.
Fig. 4.
PBECs (Epi) secrete various growth factors on differentiation at ALI. Primary bronchial epithelial cells (PBECs, Epi) were differentiated at air liquid interface (ALI). Supernatant collected after differentiation was assayed for various proteins using magpix. Histograms depict the growth and other factors, which displayed significant increase over media alone. Data is mean +/− S.E. of four experiments. PBECs from three different subjects were used for this assay
Growth factors secreted by PBECs prime the PDC responses to influenza
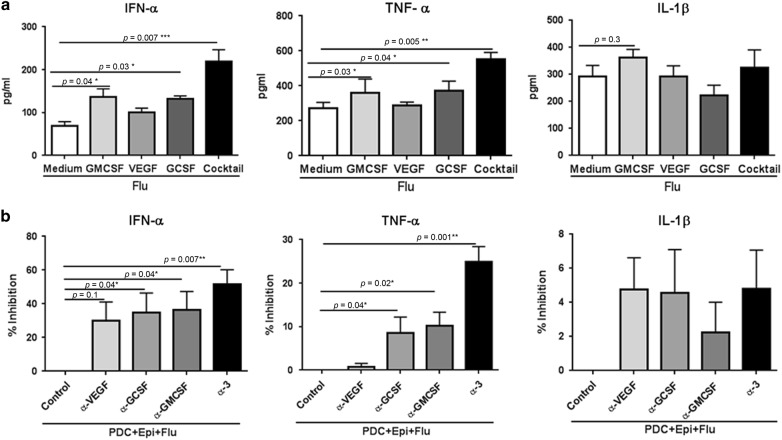
To identify the specific mechanism that leads to PDC priming in presence of PBECs, the effect of the factors found to be significantly enhanced in differentiated PBECs were explored. Initial experiments were performed with three cocktails, growth factor and cytokine, chemokine cocktail (GMCSF, VEGF, GCSF, IL-6, IL-8, CCL-2), growth factor cocktail (GMCSF, VEGF and GCSF) and cytokine, chemokine cocktail (IL-6, IL-8, CCL-2). Purified PDCs were cultured in the presence or absence of these cocktails and activated with influenza. Culture of PDCs with combination cocktail as well as growth factor cocktail enhanced the secretion of type I IFN while the cytokine, chemokine cocktail had no significant effect (supplementary fig. 2). Further experiments were then done with growth factors. We tested the individual effects of the growth factors to determine whether this was due to a single factor or a synergistic effect of all 3 factors. Both GCSF (p = 0.03), GMCSF (p = 0.04) led to increase in type I IFN secretion by PDCs in the response to influenza (Fig. 5a). The increase by VEGF was not significant (p = 0.1). The secretion of type I IFN was maximal when a cocktail of all three growth factors was used indicating a synergistic effect. Both GMCSF and GCSF were also able to induce significantly increased (p = 0.03-GMCSF; p = 0.04-GCSF; Fig. 5a) levels of TNF-α. TNF-α was also significantly increased in the cocktail (p = 0.005). IL-1β displayed no significant increase with the growth factors as well as the cocktail suggesting that the growth factors do not induce IL-1β in PDCs.
Fig. 5.
Growth factors secreted by PBECs prime the PDC responses to influenza. Purified PDCs were stimulated with influenza in the presence of growth factors, GMCSF, GCSF, VEGF or a cocktail of three. a Bar graphs depict the cytokines secreted by PDCs as assayed by magpix assay. Data is mean +/− S.E. of five experiments with PDCs from different subjects. b PDCs were cultured with PBEC conditioned medium and stimulated with influenza (Flu) in the presence of antibodies to GMCSF, GCSF, VEGF (α-GMCSF, α-GCSF, α-VEGF) or a combination of all three antibodies (α−3). Bar graphs depict percent inhibition of the cytokines secreted by PDCs. Data is mean +/− S.E. of five experiments of which three experiments were done where PDCs from three different subjects were cultured with PBEC conditioned media from one set of PBEC and the other two experiments were done where PDCs from another two subjects were cultured with conditioned media from second set of PBEC
To further confirm the action of the growth factors, experiments using blocking antibodies against GMCSF, VEGF and GCSF as well as a combination of all three antibodies were performed. PDCs cultured in the presence of PBEC conditioned medium were stimulated with influenza in the presence of the antibodies and percent inhibition of type I IFN, TNF-α and IL-1β secretion by PDCs was calculated. As is evident from Fig. 5b, antibodies against GMCSF (Mean +/− S.E. = 152+/−28.3 pg/ml) and GCSF (Mean +/− S.E. = 161+/−29.6 pg/ml) were able to significantly block (approximately 33%; p = 0.04 for both α-GMCSF and α-GCSF) type I IFN secretion as compared to influenza stimulated pDC (Mean+/−S.E. = 232+/−53.9 pg/ml) cultured in PBEC conditioned medium without antibodies. Inhibition of TNF-α by these antibodies was more modest (around 10%) but both GMCSF (Mean +/− S.E. = 277+/−17.8 pg/ml; p = 0.02) and GCSF (Mean +/-S.E. = 269+/−20.2 pg/ml; p = 0.04) displayed significant blockade (control mean +/-S.E. = 340+/−14.2 pg/ml). VEGF antibodies were unable to inhibit type I IFN (Mean+/-S.E. = 234+/−52.8 pg/ml) or TNF-α (Mean +/−S.E. = 353+/−20.1 pg/ml) (Fig. 5b). A combination of all three antibodies displayed maximal inhibition for type I IFN (Mean +/−S.E. = 87.07 +/−20.2 pg/ml; p = 0.007; approximately 55–60%) and TNF-α (Mean +/-S.E. = 155+/−33.3 pg/ml; p = 0.005; approximately 25–30%). None of the antibodies including the combination were able to inhibit IL-1β secretion. No inhibition was observed by the antibodies when PDCs were cultured without the conditioned media and stimulated with influenza (data not shown).
These data suggest that though GMCSF and GCSF can enhance the induction of type I IFN and TNF-α in PDCs, the synergistic action of all three factors is really what is responsible for the increase of these cytokines observed in PDC cocultured with PBECs.
Discussion
Type I IFN secretion by PDCs plays a critical role in fighting viral infection at the respiratory mucosa. Here we demonstrate for the first time that the capacity of PDCs to secrete type I IFN is enhanced by growth factors secreted by PBECs.
Previous studies suggest that PDCs may play a tolerogenic role in the respiratory mucosa via induction of T regulatory cells.5,20 A very recent study in mice by Lynch et al.21 has demonstrated that depletion of PDCs early in life enhanced features of asthma such as airway hyper-responsiveness, allergic inflammation and airway remodeling due to an impaired ability to expand Tregs and maintain tolerance in the airways. Our data indicates that exposure to PBECs induces tolerance in PDCs at homeostasis but concomitantly also primes PDCs to enhance their response to infections (Figs. 1, 2). The induction of Tregs by PDCs is enhanced which could be due to the increased expression of IDO. The factors which lead to the increase in tolerogenic capacity of PDCs remain to be explored.
In keeping with the activation genes in RNA-seq, we also observed increased secretion of type I IFN from influenza stimulated PDCs co-cultured with PBECs. The expression of TLR9 and ZBP1 was upregulated. ZBP1 has recently been shown to be the innate sensor of influenza virus.22 It signals via IRF3 and NFkB both of, which were upregulated in PDCs exposed to PBECs. The RNA-seq data did not display significant changes in pathways known to induce IFN secretion in PDCs such as the IRF7.23 However, increased NFkB related genes may play a role in TLR-dependent IFN production.24,25 Interestingly, PGE2 has been reported to be a negative regulator of type I IFN secretion by human DCs upon stimulation with TLRs26 and prostaglandin pathway was downregulated in PDCs exposed to PBECs (Fig. 2e). The expression of another gene, FcɛRIγ/FCE1A is significantly downregulated in PDCs co-cultured with PBECs and it has been demonstrated to inhibit TLR mediated type I IFN production in human PDCs via ITAM-mediated signals.27 Similarly SOCS1, a key negative regulator of MyD88-dependent type I IFN signaling28,29 is also downregulated in PBECs exposed PDCs. In addition to negative regulators, PDCs co-cultured with also PBECs displayed increased expression of genes involved in type-I-IFN signaling. Type I IFN can act both in an autocrine and a paracrine manner.30 It can amplify its own secretion via autocrine actions. Increased expression of IFNAR1 would enhance the autocrine action of IFNs. Therefore, PBECs seem to prime PDCs to enhance type I IFN secretion. This is supported by the increase in the genes of the ISG15 pathway. ISG15 is not only an interferon response pathway31 but is also reported to bind to IRF-3 and enhance IFN responses.32,33 Furthermore, ISG15 pathway can also affect important signaling pathways such as IFN, NFκB, and JNK pathways, all of, which are upregulated in PDCs after exposure to PBECs.34 Together the gene expression changes suggest that PBECs enhance the capacity of PDCs to produce type I IFN via inhibiting the negative regulators as well as by enhancing the expression of molecules, which can amplify the production of type I IFNs. We had observed similar priming of mDCs by PBECs earlier.8,35 Even though RNA-seq data suggests that PDCs co-cultured with PBECs express type I IFN at homeostasis, we were unable to detect type I IFN in our supernatants without stimulation with influenza. This could be due to lack of sensitivity of the ELISA which may not be able to detect the various type I IFN subtypes. It is also possible that the type I IFN is bound to the receptor and therefore not detectable. High level of type I IFN secretion in the absence of infection is not desirable since its upregulation under such conditions is usually associated with autoimmunity. The tolerogenic nature of PDCs at homeostasis may therefore prevent secretion of the type I IFN to prevent inflammation. It is also possible that the changes visible at gene level are not transcribed to proteins as it is well established that only about 40% of genes are transcribed to proteins. This is especially true for cytokines.
We also find that growth factors, GMCSF, GCSF, and VEGF secreted by PBECs help prime PDC responses to influenza (Fig. 5). Interestingly, these factors signal via the JAK-STAT pathway, which displays significant upregulation in the RNA-seq data (Fig. 2d). Some of the growth factors have already been reported to modulate DC function. For example, knockdown of GCSF receptor in DCs has been reported to reduce the expression of at least pro-inflammatory cytokines and antigen presenting molecules, while the expression of PDL1 and PDL2 was increased.36 GMCSF is also considered as a DC activation agent37 and has been demonstrated to enhance the IFN-α production from PDCs in lupus patients.38 Moreover, GMCSF secretion by kidney epithelial cells has also been reported to enhance maturation and type I IFN production in PDCs.39 Additionally, GMCSF has been shown to synergize with TLR-dependent microbial stimuli to upregulate of pro-inflammatory cytokines.40 Interestingly, BDCA-4, which functions as a marker for PDCs is a receptor on endothelial and tumor cells for vascular endothelial growth factor (VEGF-A).41 Studies examining the effect of VEGF on lung DC subtypes in VEGF-transgenic mice have reported an increase in lung mDC and PDC populations.42 An increase in inflammatory responses of mDC was also reported in the same study. The effect of VEGF on PDC responses has not been examined. Our results indicate a possible synergistic effect of VEGF with other growth factors on type I IFN production by PDCs.
The present study only examines effect of soluble factors from PBECs on PDC functions. Information regarding cell to cell contact between PBECs and PDC is missing which can also play an important role in modulation the functions of PDCs. This is because existing technology only allows the differentiation of PBECs at ALI on a transwell insert which prevents contact with PDCs. Development of systems which can allow PBEC differentiation on surfaces which are capable of providing cell to cell contact would be very valuable in studying such interactions.
In summary, RNA- seq data indicates that exposure of PDCs to PBECs both tolerizes and activates PDCs. PBECs exposed PDCs induce increased levels of Tregs at homeostasis. However, the PDCs are also primed by PBECs to respond to infections. In keeping with the RNA-seq data, we observe increased secretion of type I IFN and other cytokines in response to influenza in PDCs co-cultured with PBECs. The PDCs also primed Th1 responses in T cells. The enhanced response of PDCs co-cultured with PBECs was due to the action of growth factors, GMCSF, GCSF and VEGF, which were secreted by PBECs on differentiation. To our knowledge this is the first report, which investigates the crosstalk between human PDCs and PBECs. These data highlight possible mechanisms to enhance the production of type-I IFN in the airways, which is critical for host defense against respiratory infections.
Electronic supplementary material
Acknowledgements
This study was supported by grant from the NIH AG045216 (to AA), and from the National Center for Research Resources and the National Center for Advancing Translational Sciences # UL1 TR000153. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We are grateful to ICTS UC Irvine for providing the blood samples.
Author contributions
F.R. performed sequencing analysis, N.J. performed the functional studies, S.A. sorted pDCs for sequencing and helped perform functional studies, H.N. helped in cell purification, M.M. helped in sequencing analysis, A.A. wrote the manuscript and supervised the experiments. All authors helped in the discussing and editing of manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Farah Rahmatpanah, Sudhanshu Agrawal, Natasha Jaiswal
Change history
1/31/2019
The original version of this Article contained an error in the spelling of the author Hannah Nguyen, which was incorrectly given as Hannah Ngyuen. This has now been corrected in both the PDF and HTML versions of the Article.
Electronic supplementary material
The online version of this article (10.1038/s41385-018-0097-1) contains supplementary material, which is available to authorized users.
References
- 1.Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 2008;8:594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- 2.Swiecki M, Colonna M. Unraveling the functions of plasmacytoid dendritic cells during viral infections, autoimmunity, and tolerance. Immunol. Rev. 2010;234:142–162. doi: 10.1111/j.0105-2896.2009.00881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swiecki M, Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat. Rev. Immunol. 2015;15:471–485. doi: 10.1038/nri3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Heer HJ, et al. Essential role of lung plasmacytoid dendritic cells in preventing asthmatic reactions to harmless inhaled antigen. J. Exp. Med. 2004;200:89–98. doi: 10.1084/jem.20040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venet F, Huang X, Chung CS, Chen Y, Ayala A. Plasmacytoid dendritic cells control lung inflammation and monocyte recruitment in indirect acute lung injury in mice. Am. J. Pathol. 2010;176:764–773. doi: 10.2353/ajpath.2010.090765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chairakaki AD, et al. Plasmacytoid dendritic cells drive acute asthma exacerbations. J. Allergy Clin. Immunol. 2017;S0091-6749:31586–5. doi: 10.1016/j.jaci.2017.08.032. [DOI] [PubMed] [Google Scholar]
- 8.Agrawal S, et al. Airway epithelial cells enhance the immunogenicity of human myeloid dendritic cells under steady state. Clin. Exp. Immunol. 2017;189:279–289. doi: 10.1111/cei.12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rate A, Bosco A, McKenna KL, Holt PG, Upham JW. Airway epithelial cells condition dendritic cells to express multiple immune surveillance genes. PLoS One. 2012;7:e44941. doi: 10.1371/journal.pone.0044941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rate A, Upham JW, Bosco A, McKenna KL, Holt PG. Airway epithelial cells regulate the functional phenotype of locally differentiating dendritic cells: implications for the pathogenesis of infectious and allergic airway disease. J. Immunol. 2009;182:72–83. doi: 10.4049/jimmunol.182.1.72. [DOI] [PubMed] [Google Scholar]
- 11.Prakash S, Agrawal S, Cao JN, Gupta S, Agrawal A. Impaired secretion of interferons by dendritic cells from aged subjects to influenza: role of histone modifications. Age. 2013;35:1785–1797. doi: 10.1007/s11357-012-9477-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sridharan A, et al. Age-associated impaired plasmacytoid dendritic cell functions lead to decreased CD4 and CD8 T cell immunity. Age. 2011;33:363–376. doi: 10.1007/s11357-010-9191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blank F, et al. Macrophages and dendritic cells express tight junction proteins and exchange particles in an in vitro model of the human airway wall. Immunobiology. 2011;216:86–95. doi: 10.1016/j.imbio.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Rothen-Rutishauser BM, Kiama SG, Gehr P. A three-dimensional cellular model of the human respiratory tract to study the interaction with particles. Am. J. Respir. Cell Mol. Biol. 2005;32:281–289. doi: 10.1165/rcmb.2004-0187OC. [DOI] [PubMed] [Google Scholar]
- 15.Araujo EF, et al. Tolerogenic plasmacytoid dendritic cells control paracoccidioides brasiliensis infection by inducting regulatory T cells in an IDO-dependent manner. PLoS Pathog. 2016;12:e1006115. doi: 10.1371/journal.ppat.1006115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lippens C, et al. IDO-orchestrated crosstalk between pDCs and Tregs inhibits autoimmunity. J. Autoimmun. 2016;75:39–49. doi: 10.1016/j.jaut.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kesavardhana S, et al. ZBP1/DAI ubiquitination and sensing of influenza vRNPs activate programmed cell death. J. Exp. Med. 2017;214:2217–2229. doi: 10.1084/jem.20170550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grouard G, et al. The enigmatic plasmacytoid T cells develop into dendritic cells with interleukin (IL)-3 and CD40-ligand. J. Exp. Med. 1997;185:1101–1111. doi: 10.1084/jem.185.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matta BM, Castellaneta A, Thomson AW. Tolerogenic plasmacytoid DC. Eur. J. Immunol. 2010;40:2667–2676. doi: 10.1002/eji.201040839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch JP, et al. Plasmacytoid dendritic cells protect from viral bronchiolitis and asthma through semaphorin 4a-mediated T reg expansion. J. Exp. Med. 2018;215:537–557. doi: 10.1084/jem.20170298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuriakose T, et al. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 2016;1:aag2045. doi: 10.1126/sciimmunol.aag2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 24.Engel A, Barton GM. Compartment-specific control of signaling from a DNA-sensing immune receptor. Sci. Signal. 2010;3:pe45. doi: 10.1126/scisignal.3150pe45. [DOI] [PubMed] [Google Scholar]
- 25.Birmachu W, et al. Transcriptional networks in plasmacytoid dendritic cells stimulated with synthetic TLR 7 agonists. BMC Immunol. 2007;8:26. doi: 10.1186/1471-2172-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabricius D, et al. Prostaglandin E2 inhibits IFN-alpha secretion and Th1 costimulation by human plasmacytoid dendritic cells via E-prostanoid 2 and E-prostanoid 4 receptor engagement. J. Immunol. 2010;184:677–684. doi: 10.4049/jimmunol.0902028. [DOI] [PubMed] [Google Scholar]
- 27.Cao W, et al. Plasmacytoid dendritic cell-specific receptor ILT7-Fc epsilonRI gamma inhibits Toll-like receptor-induced interferon production. J. Exp. Med. 2006;203:1399–1405. doi: 10.1084/jem.20052454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prele CM, et al. SOCS1 regulates the IFN but not NFkappaB pathway in TLR-stimulated human monocytes and macrophages. J. Immunol. 2008;181:8018–8026. doi: 10.4049/jimmunol.181.11.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu X, et al. Cross-regulation of two type I interferon signaling pathways in plasmacytoid dendritic cells controls anti-malaria immunity and host mortality. Immunity. 2016;45:1093–1107. doi: 10.1016/j.immuni.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S, et al. Self-priming determines high type I IFN production by plasmacytoid dendritic cells. Eur. J. Immunol. 2014;44:807–818. doi: 10.1002/eji.201343806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giannakopoulos NV, et al. Proteomic identification of proteins conjugated to ISG15 in mouse and human cells. Biochem. Biophys. Res. Commun. 2005;336:496–506. doi: 10.1016/j.bbrc.2005.08.132. [DOI] [PubMed] [Google Scholar]
- 32.Shi HX, et al. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol. Cell. Biol. 2010;30:2424–2436. doi: 10.1128/MCB.01466-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dos Santos PF, Mansur DS. Beyond ISGlylation: functions of free intracellular and extracellular ISG15. J. Interferon Cytokine Res. 2017;37:246–253. doi: 10.1089/jir.2016.0103. [DOI] [PubMed] [Google Scholar]
- 34.Lu G, et al. ISG15 enhances the innate antiviral response by inhibition of IRF-3 degradation. Cell Mol. Biol. 2006;52:29–41. [PubMed] [Google Scholar]
- 35.Agrawal Anshu. Dendritic Cell-Airway Epithelial Cell Cross-Talk Changes with Age and Contributes to Chronic Lung Inflammatory Diseases in the Elderly. International Journal of Molecular Sciences. 2017;18(6):1206. doi: 10.3390/ijms18061206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geng D, Joshi SK, Podolsky R, She JX. GCSF receptor regulates antigen uptake and expression of cytokines and costimulatory molecules in dendritic cells. Mol. Immunol. 2007;44:521–529. doi: 10.1016/j.molimm.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 37.Ghirelli C, Zollinger R, Soumelis V. Systematic cytokine receptor profiling reveals GM-CSF as a novel TLR-independent activator of human plasmacytoid predendritic cells. Blood. 2010;115:5037–5040. doi: 10.1182/blood-2010-01-266932. [DOI] [PubMed] [Google Scholar]
- 38.Eloranta ML, et al. Regulation of the interferon-alpha production induced by RNA-containing immune complexes in plasmacytoid dendritic cells. Arthritis Rheum. 2009;60:2418–2427. doi: 10.1002/art.24686. [DOI] [PubMed] [Google Scholar]
- 39.Ruben JM, et al. Human plasmacytoid dendritic cells acquire phagocytic capacity by TLR9 ligation in the presence of soluble factors produced by renal epithelial cells. Kidney Int. 2018;93:355–364. doi: 10.1016/j.kint.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 40.Min L, et al. Cutting edge: granulocyte-macrophage colony-stimulating factor is the major CD8+T cell-derived licensing factor for dendritic cell activation. J. Immunol. 2010;184:4625–4629. doi: 10.4049/jimmunol.0903873. [DOI] [PubMed] [Google Scholar]
- 41.Dzionek A, et al. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J. Immunol. 2000;165:6037–6046. doi: 10.4049/jimmunol.165.11.6037. [DOI] [PubMed] [Google Scholar]
- 42.Chapoval SP, et al. Lung vascular endothelial growth factor expression induces local myeloid dendritic cell activation. Clin. Immunol. 2009;132:371–384. doi: 10.1016/j.clim.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.