Keywords: nerve regeneration, neural tube defects, spina bifida aperta, spinal cord, all-trans retinoic acid, O6-methylguanine DNA methyltransferase, gene expression, DNA methylation, promoter, bisulfite sequencing polymerase chain reaction, neural regeneration
Abstract
O6-methylguanine DNA methyltransferase (MGMT), a DNA repair enzyme, has been reported in some congenital malformations, but it is less frequently reported in neural tube defects. This study investigated MGMT mRNA expression and methylation levels in the early embryo and in different embryonic stages, as well as the relationship between MGMT and neural tube defects. Spina bifida aperta was induced in rats by a single intragastric administration of all-trans retinoic acid on embryonic day (E) 10, whereas normal control rats received the same amount of olive oil on the same embryonic day. DNA damage was assessed by detecting γ-H2A.X in spina bifida aperta rats. Real time-polymerase chain reaction was used to examine mRNA expression of MGMT in normal control and spina bifida aperta rats. In normal controls, the MGMT mRNA expression decreased with increasing embryonic days, and was remarkably reduced from E11 to E14, reaching a minimum at E18. In the spina bifida aperta model, γ-H2A.X protein expression was increased, and mRNA expression of MGMT was markedly decreased on E14, E16, and E18. Bisulfite sequencing polymerase chain reaction for MGMT promoter methylation demonstrated that almost all CpG sites in the MGMT promoter remained unmethylated in both spina bifida aperta rats and normal controls, and there was no significant difference in methylation level between the two groups on either E14 or E18. Our results show that DNA damage occurs in spina bifida aperta rats. The mRNA expression of MGMT is downregulated, and this downregulation is independent of promoter DNA methylation.
Chinese Library Classification No. R459.9; R394
Introduction
Neural tube defects (NTDs) are a series of severe congenital malformations in the central nervous system that result from defective neural tube closure during embryogenesis. NTDs are the second most common birth defect in the world, affecting 0.5–2 in every 1000 pregnancies (Greene et al., 2009). In China, the incidence of NTDs is high, particularly in Shanxi Province (Liu et al., 2016).
The aetiologies of NTDs are complicated, and both genetic and environmental factors have been implicated (Chen et al., 2010; Wilde et al., 2014). Over 100 candidate genes have been investigated for their association with NTDs (Boyles et al., 2005; Copp and Greene, 2010), but the mechanisms of neural tube defects are still unclear. Some studies have demonstrated that advanced glycation end products (Li et al., 2014a) or polycyclic aromatic hydrocarbon-DNA adducts (Yuan et al., 2013; Yi et al., 2015) mediate DNA damage that may be a significant contributor to neural tube defects. Furthermore, several DNA repair genes such as APEX1, XRCC1, XRCC3, XPD, hOGG1, hMLH1, and hMSH2 have been reported to be associated with NTDs (Olshan et al., 2005; Au et al., 2010; Liu et al., 2012). However, O6-methylguanine DNA methyltransferase (MGMT), a DNA repair enzyme, which includes the simplest of the human DNA repair responses, is less frequently reported in NTDs. In fact, only one study has reported this (Tran et al., 2012). MGMT has been reported in other congenital malformations; several SNPs within MGMT were examined for an association with increased risk of congenital heart disease (Chowdhury et al., 2012; Li et al., 2014b) and Beckwith-Wiedemann syndrome (Paganini et al., 2015). Prior studies have reported that lower MGMT expression levels are linked to both tumourigenesis and teratogenic drug susceptibility (Glassner et al., 1999; Kitajima et al., 2003; Bobola et al., 2007), and a loss of MGMT expression is almost entirely associated with the methylation of CpG islands in the promoter region of the MGMT gene (Minoo, 2013; Mokhtar et al., 2014; Toffolatti et al., 2014). Therefore, this study focused on alterations in MGMT expression or promoter methylation in an animal model of NTD.
Maternal all-trans retinoic acid (ATRA) administration has long been used to induce a rat model of foetal spina bifida aperta (SBA) for the study of NTDs (Diez-Pardo et al., 1995). Our previous studies used this model to explore the pathogenesis of SBA (Li et al., 2012; Wu et al., 2013), and we verified that this rat SBA model is similar to human NTDs (Cai et al., 2007). Because the role of MGMT in ATRA-induced SBA in rats has not been reported, the rat model of SBA was used in this study to investigate the expression patterns of MGMT in different embryonic stages.
Therefore, this study examined the potential influence of the expression pattern of MGMT in spinal tissues on neural tube closure in an ATRA-induced model of SBA in rats. We then investigated DNA methylation levels of the MGMT promoter to determine whether the expression of MGMT is controlled by methylation.
Materials and Methods
Animals and spinal cord preparation
Specific-pathogen-free female Wistar rats aged 10–12 weeks old and weighing 230–260 g were purchased from Liaoning Changsheng Biotechnology Co., Ltd., China [animal license number: SCXK (Liao) 2015-0001]. Forty-four pregnant rats were divided into two treatment groups. Rats in the SBA group (n = 23) received a single intragastric administration of ATRA (Sigma, St. Louis, MO, USA; 4% wt/vol in olive oil; 140 mg/kg body) via a single gavage feeding on embryonic day 10 (E10), as previously described (Danzer et al., 2005). Rats in the normal control group (n = 21) received the same amount of olive oil on the same embryonic day. All pregnant rats were euthanized with an overdose injection of 10% chloral hydrate on E11, E12, E14, E16, and E18; the foetuses were harvested immediately thereafter. Foetuses without defects in the ATRA treatment group were considered as the ATRA-treated control group for further analysis. For each analysis, spinal cords (from the inferior margin of the forelimb bud to the tail bud) were obtained from each group at each embryonic day from at least three dams. All experimental procedures were approved by the Animal Ethics Committee, Shengjing Hospital, China Medical University, China (approval No. 2015PS264K) on October 13, 2015.
Western blot assay
After samples were collected, spinal cord tissues were lysed with ice-cold radio-immunoprecipitation assay buffer (Solarbio, R0010, Beijing, China) supplemented with 1 mM of phenylmethanesulfonyl fluoride and centrifuged, and the supernatant was collected. Protein quantification was determined by the bicinchoninic acid assay. After separation on 13% sodium dodecyl sulphate-polyacrylamide gels, proteins were electrophoretically transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). The membrane was blocked with 5% bovine serum albumin in Tris-buffered saline containing 0.1% Tween-20 (TBST) at room temperature for 2 hours. Membranes were incubated with the primary antibody, mouse anti-γ-H2A.X (ab26350, Abcam Incorporation, Cambridge, MA, USA), at 1:1000 dilution in TBST + 1% bovine serum albumin overnight at 4°C, and then incubated with the secondary antibody, a goat anti-mouse horseradish peroxidase-conjugated antibody (ZDR-5307, ZSGB-bio, Beijing, China), at 1:2000 dilution at room temperature for 2 hours. Enhanced chemiluminescence (Millipore) was used to visualize the protein signals. The relative optical densities of the bands were measured using ImageJ software (NIH, Bethesda, MD, USA), with β-actin serving as the loading control.
Real time-polymerase chain reaction (RT-PCR) analysis
For mRNA expression analysis, total RNA was extracted from the spinal cords of five SBA foetuses and five control foetuses at each time point using TRIzol reagent (Applied Biosystems, Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocol. The RNA was quantified, and samples with an A260 nm/A280 nm ratio of less than 1.8 were discarded.
To perform the mRNA expression analysis, 1 μg of total RNA was transferred into cDNA using a Prime Script RT Reagent Kit with gDNA Eraser (TaKaRa Bio, Shiga, Japan). Reaction solutions (20 μL) were incubated at 37°C for 15 minutes, followed by 85°C for 5 seconds. Diluted cDNA (1:5) was subjected to RT-PCR using a Light Cycler 480 SYBR Green I Master (Roche Diagnostics GmbH, Mannheim, Germany). The 20 μL reaction solution contained 10 μL of 2 × SYBR Green Master Mix, 2 μL of the cDNA template, 1 μL of each primer (5 μM), 0.4 μL of ROXII, and 5.6 μL of RNase-free water. Primers for MGMT were designed using Primer Premier 5 software (Premier Company, Canada). Experiments were performed in triplicate on a PCR thermal cycler (Light Cycler 480 Real-time PCR System, Roche Diagnostics GmbH) as follows: pre-denaturation at 95°C for 30 seconds, 45 cycles at 95°C for 5 seconds and 60°C for 20 seconds. Expression values were normalized to the geometric means of the housekeeping genes β-actin and GAPDH. Relative mRNA expression levels were calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001). Detailed information for the primers used in this assay is included in Table 1.
Table 1.
Real time-polymerase chain reaction primers and products

Bisulfite treatment and DNA methylation analysis
After the mRNA expression analysis, genomic DNA was extracted from spinal cords using the DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany). Briefly, homogenized tissue was incubated in lysis buffer containing proteinase K and RNase at 56°C for 45 minutes. The DNA was eluted with TE buffer, then quantified using a NanoDrop ND-1000 (Thermo Scientific, Thermo Fisher Scientific, Waltham, MA, USA), and samples with an A260 nm/A280 nm ratio outside the range of 1.8–2.0 were discarded. The DNA was stored at −20°C for a subsequent experiment.
After genomic DNA extraction, 2 μg of genomic DNA from each sample was bisulfite-treated with the EpiTect Bisulfite Kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer’s instructions. Treated DNA was stored at −80°C until use.
Bisulfite sequencing PCR was used to investigate the methylation pattern of the MGMT promoter. The target region was a 283 bp section of the rat MGMT promoter CpG island containing 17 CpG sites. The bisulfite-treated DNA was then amplified by PCR. The primers were designed using Methprimer (Li and Dahiya, 2002). Bisulfite sequencing PCR primer sequences are shown in Table 2. The PCR cycles were as follows: pre-denaturation at 94°C for 5 minutes, denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 45 seconds. These steps were repeated for 34 cycles, followed by a final extension at 72°C for 7 minutes. The PCR products were run on 1.5% agarose gels with 10% ethidium bromide and visualized with ultraviolet light. The PCR products were purified using the QIAquick Gel Extraction Kit (Qiagen GmbH) and cloned into the pGEM-T vector (Promega, Madison, WI, USA). A minimum of five white colonies were selected for each allele using lacZ blue-white selection screening; automated sequencing was then performed using M13 primers. The methylation pattern of each allele was analyzed with the online software QUMA (http://quma.cdb.riken.jp/) (Kumaki et al., 2008).
Table 2.
BSP primers and products

Statistical analysis
All values are shown as the mean ± SEM. All statistical analyses were performed using GraphPad Prism version 5 software (GraphPad Software, Inc., La Jolla, CA, USA). Student’s t-test and one-way analysis of variance followed by Fisher’s least significant difference post hoc test were used for single and multiple comparisons, respectively. For all statistical analyses performed, P values less than 0.05 were considered significantly different.
Results
ATRA induces NTDs in rats
In the ATRA-treated group, 192 live embryos were harvested from 23 pregnant rats from E11, E12, E14, E16, and E18. There was approximately 11% incidence of dead or absorbed foetuses. SBA (61%; 118 of 192 live embryos) embryos were determined by gross morphologic examination under a stereomicroscope. Embryos with SBA were considered as the SBA group (n = 118). Embryos without defects were considered as the ATRA-treated control group (n = 62). The normal control group consisted of 167 embryos harvested from 21 olive oil-treated dams.
DNA damage analysis
To determine DNA damage, we assessed the γ-H2A.X (phosphorylated H2A.X on Ser139) level in foetuses with SBA. H2A.X is rapidly phosphorylated on serine 139 to form γ-H2A.X in response to DNA damage. Therefore, the level of γ-H2A.X is an indicator of DNA damage. Levels of γ-H2A.X were significantly increased in the spinal cords of foetuses with SBA induced by ATRA on E14 (P = 0.0174; Figure 1), indicating that DNA damage occurred in ATRA-induced SBA embryos. The expression levels of γ-H2A.X were not significantly different compared with the normal controls on E11, E16, and E18 (P = 0.9618, 0.6227, and 0.2740, respectively).
Figure 1.
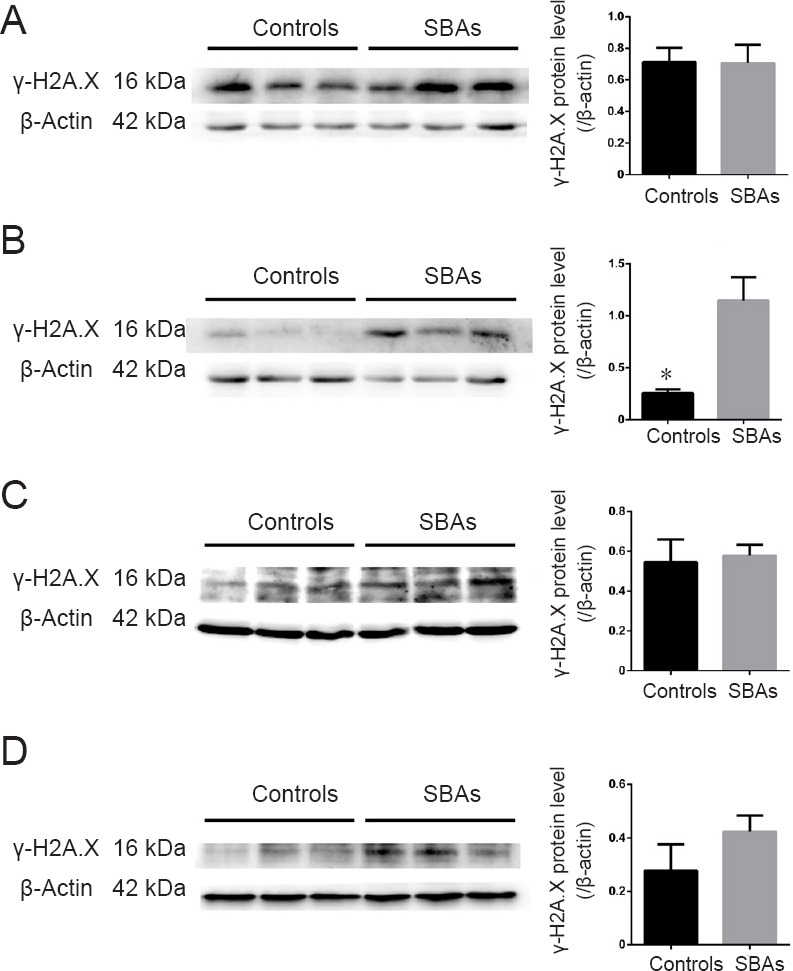
Western blot assay of γ-H2A.X protein expression in SBA embryos and controls.
(A–D) Western blot results of γ-H2A.X protein expression in individual embryos on E11, E14, E16, and E18. There was a significant difference in the relative protein expression of γ-H2A.X in SBAs and controls on E14. *P < 0.05, vs. control. Data are represented as the mean ± SEM (n = 3 per group at each time point; Student’s t-test). MGMT: O6-methylguanine DNA methyltransferase; SBA: spina bifida aperta; E: embryonic day.
mRNA expression of MGMT in SBA and control embryos
To investigate whether the expression of MGMT is disrupted in embryos with ATRA-induced SBA, MGMT mRNA expression in the spinal cords of E11, E12, E14, E16, and E18 embryos was quantified via RT-PCR. In the normal control group, there was a trend of decreased MGMT mRNA expression with increased embryonic days. MGMT mRNA expression was dramatically reduced from E11 to E14 (0.29-fold; P = 0.0483), remained essentially stable from E14 to E18, and reached a minimum on E18 (vs. E11, 0.46-fold; P = 0.0031). Spinal cords from SBA embryos had a statistically significant reduction in MGMT mRNA expression compared with normal controls on E14, E16, and E18.
MGMT mRNA expression was reduced 0.33-fold (P = 0.0029) on E14, 0.31-fold (P = 0.0425) on E16, and 0.30-fold (P = 0.0386) on E18 (Figure 2A).
Figure 2.
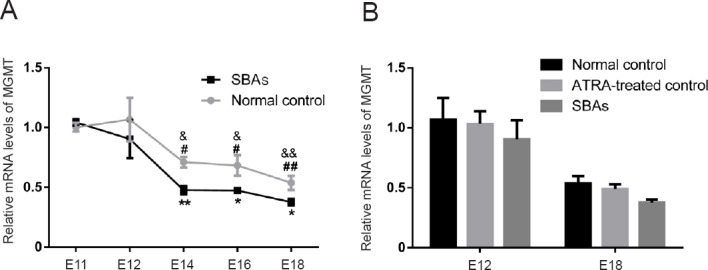
RT-PCR analysis of the DNA repair gene MGMT.
(A) Expression profiles of MGMT in normal control (grey, n = 5 at each time point) and SBA (black, n = 5 at each time point) spinal cords, as detected by RT-PCR. #P < 0.05, ##P < 0.01, vs. the E11 control group; &P < 0.05, &&P < 0.01, vs. the E12 control group (Student’s t-test); *P < 0.05, **P < 0.01, vs. normal control group; Data are shown as the mean ± SEM (n = 5 at each time point; one-way analysis of variance followed by Fisher’s least significant difference post hoc test). (B) RT-PCR analysis of MGMT in normal control, ATRA-treated control and SBAs group on E12 and E18. Data are shown as the mean ± SEM (n = 5 at each time point, Student’s t-test). MGMT: O6-methylguanine DNA methyltransferase; SBA: spina bifida aperta; RT-PCR: real time-polymerase chain reaction; E: embryonic day.
To investigate whether the reduction of MGMT gene expression is ATRA-dependent, MGMT mRNA expression was analyzed in embryos harvested from ATRA-treated dams that had no abnormal spinal cord phenotype (the ATRA-treated control group). The gene expression of MGMT was compared between the normal control and ATRA-treated control groups, and between ATRA-treated control and SBA groups on E12 and E18. No significant differences in MGMT levels were detected between normal controls and ATRA-treated controls (E12, P = 0.8738; E18, P = 0.5218), and there was no difference between the ATRA-treated controls and the SBA group (E12, P = 0.3736; E18, P = 0.4236) (Figure 2B). These quantitative RT-PCR results demonstrate that the reduction in MGMT gene expression was not ATRA-dependent, and that downregulation of MGMT was indeed associated with ATRA-induced SBA.
DNA methylation analysis of the MGMT promoter in SBA cases
We next tested whether DNA methylation affected MGMT gene expression. The promoter CpG island region of the MGMT gene contains 17 CpG sites, as shown in Figure 3. To determine the MGMT promoter methylation status, a bisulfite sequencing PCR analysis was performed on a subset of five SBA cases and five controls on E14 and E18. Figures 4 and 5 show the results of the promoter methylation analysis in these two groups on E14 and E18, respectively. We could only detect the methylation levels of 16 CpG sites, because the tenth CpG site was a SNP (rs107272582 C/G) that contained a C to G transversion. MGMT promoter methylation levels in the control and SBA groups were 0.0 ± 0.0% and 0.2638 ± 0.1619%, respectively, on E14 (Figure 6A), and 1.000 ± 0.4898% and 1.812 ± 1.009%, respectively, on E18 (Figure 6B). On both E14 and E18, MGMT promoter methylation appeared to increase in the SBA group compared with controls; however, no significant difference was found between SBA embryos and controls on E14 or E18 (P = 0.18 and P = 0.49, respectively).
Figure 3.

Map of the MGMT promoter region.
This is a sketch map of the CpG site in the promoter region of the MGMT gene. The horizontal line represents the MGMT promoter region, from ch1: 216,161,172- ch1: 216,161,454. Vertical lines represent the CpG sites. Star: SNPs at CpG locations (rs107272582 C/G and rs104902905 C/T, successively). MGMT: O6-methylguanine DNA methyltransferase; SNPs: single nucleotide polymorphisms.
Figure 4.

Methylation status analysis of the MGMT promoter in SBA and control animals on E14.
Methylation analysis of the MGMT promoter in E14 spinal cords (n = 5) by BSP in normal control and SBA animals. Each of lines represents a single clone. ●: Methylated CpGs; ○: unmethylated CpGs; N: control; S: SBA. MGMT: O6-methylguanine DNA methyltransferase; SBA: spina bifida aperta; BSP: bisulfite sequencing PCR; E: embryonic day.
Figure 5.

Methylation status analysis of the MGMT promoter in SBA and control animals on E18.
Methylation analysis of the MGMT promoter in E18 spinal cords (n = 5) by BSP in normal control and SBA animals. Each of lines represents a single clone. ●: Methylated CpGs; ○: unmethylated CpGs; N: control; S: SBA. MGMT: O6-methylguanine DNA methyltransferase; SBA: spina bifida aperta; BSP: bisulfite sequencing PCR; E: embryonic day.
Figure 6.
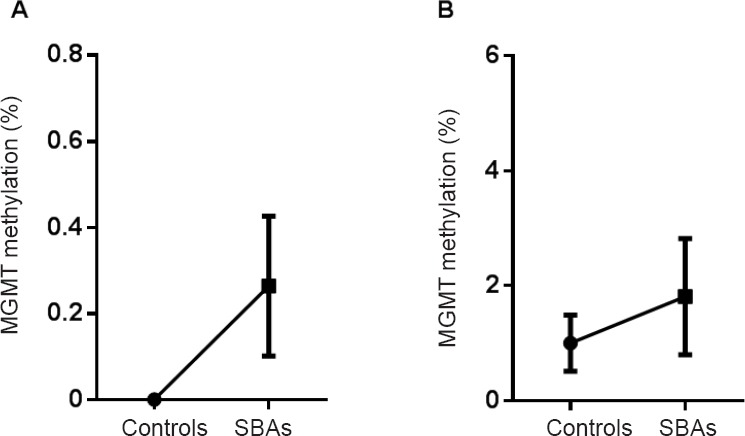
Methylation levels of the MGMT promoter in the SBA and normal control groups on E14 and E18.
(A, B) Error bar plot showing mean methylation levels at E14 (A) and E18 (B) in spinal cords (n = 5 per group at each time point) between SBA and control groups for MGMT methylation analysis by BSP. The bottom and top of the bar represent the 25th and 75th percentiles (the lower and upper quartiles, respectively), and the band near the middle of the bar represents the 50th percentile (the median). MGMT: O6-methylguanine DNA methyltransferase; SBA: spina bifida aperta; BSP: bisulfite sequencing PCR; E: embryonic day.
Discussion
The aetiologies of human NTDs are complicated and varied (Li et al., 2018). DNA damage is a risk factor for NTDs. In the present study, the expression of γ-H2A.X was increased on E14 in ATRA-treated SBA embryos compared with normal controls, confirming that DNA damage occurred in ATRA-induced SBA embryos.
DNA repair is a direct response to DNA damage. DNA repair genes are known play a role in human NTDs (Olshan et al., 2005; Liu et al., 2012), but little is known about whether DNA repair genes are involved in ATRA-induced rat models of NTDs. DNA repair genes are crucial for proper embryogenesis and central nervous system development. DNA repair pathways in mammalian cells include damage reversal by MGMT, mismatch repair, nucleotide excision repair, base nucleotide repair, and DNA double-strand break repair (Vlachostergios et al., 2009). The proper functioning of DNA repair mechanisms plays a major role in normal cellular activities and embryogenesis by protecting the genome from endogenous metabolites or exogenous agents that can cause DNA damage. In mice in which a DNA repair gene (such as Rad17 or MGMT) has been knocked out, embryonic lethality occurs, demonstrating that DNA repair enzymes are essential for mammalian development (Glassner et al., 1999; Budzowska et al., 2004). In addition, the elevated expression of DNA repair genes in enriched mouse neural crest cells during early embryogenesis may contribute to maintaining DNA integrity (Albino et al., 2011). We established the expression pattern of MGMT in the neural tube development of normal rat embryos. Our results demonstrated that there was a trend towards decreased MGMT expression with increasing embryonic days in the normal control group. MGMT expression was dramatically reduced from E11 to E14 and remained essentially stable from E14 to E18, suggesting that MGMT is overexpressed during the early stages of embryogenesis to reduce any potential genotoxic damage. MGMT is a DNA repair gene that is known to protect DNA from alkylating agents. The methylating agents methylazoxymethanol and nitrogen mustard can seriously interfere with brain neuron development and motor function in Mgmt–/– mice (Kisby et al., 2009). Some studies have found that epigenetic modification can lead to the loss of MGMT expression in a number of human cancers (Lind et al., 2004; Bugni et al., 2009; Su et al., 2012). In agreement with these previous studies, we found that MGMT mRNA expression was reduced in the SBA group compared with normal controls; moreover, there was a statistically significant difference on E14–E18. Furthermore, no significant differences were detected in MGMT mRNA levels between normal controls and ATRA-treated controls on E12 and E18. These data suggest that the reduction in MGMT mRNA expression is not ATRA-dependent, and downregulation of MGMT is indeed associated with NTDs in the ATRA-induced SBA rat model. Because neural tube closure begins and ends quite early in development, at approximately E9–E12, a perturbation in neural tube or neural crest cells can be considered relevant only if it can be shown to occur at the time of neural tube closure. However, there was no difference between the groups in MGMT expression at E11 and E12. Differences between the groups were discovered at E14–E18, and the findings may thus be associated with NTD progression rather than initiation. If the downregulation of MGMT gene expression is associated with NTD progression, these results may provide new insights for the prognosis and treatment of NTDs. In contrast to the significant decrease in MGMT expression on E14, the level of γ-H2A.X was dramatically increased, indicating that DNA damage occurred on E14 without the repair effects of MGMT. Because MGMT inhibition can induce cell apoptosis (Konduri et al., 2009; Song et al., 2015), and increased cell apoptosis has been found in the neuroepithelial cells of the lumbosacral neural tube of ATRA-induced SBA rats (Wei et al., 2012), we conclude that the downregulation of MGMT may lead to further apoptosis in lesioned areas in SBA rats.
Because both environmental and genetic factors are thought to be involved in the development of SBA, the observed change in MGMT expression might be due to a number of causes, including genetic mutation, promoter hypermethylation, suppressed transcription factors, altered histone methylation and acetylation, or gene silencing via miRNA/siRNA. DNA methylation, which can be viewed as a link between environmental factors and the control of gene expression, is a heritable and reversible epigenetic process that has been linked to NTDs. A previous study has shown that inactivation of the methyltransferase Dnmt3b disturbs genome-wide de novo methylation and causes a variety of developmental defects in mice, including NTDs (Dean et al., 2005). An in vitro analysis of cultured chicken embryos demonstrated that disrupted DNA methylation might prevent the proper closure of the neural tube (van der et al., 2008). More recent studies have demonstrated that global DNA hypomethylation (Chen et al., 2010), and hypomethylation of LINE-1 (Wang et al., 2010, 2015), are each related to an increased risk of NTDs. We investigated the methylation status of all CG sites in the MGMT promoter CpG island and examined whether the downregulation of MGMT is determined by promoter DNA methylation. However, there were no significant differences in MGMT promoter methylation between SBA and control embryos. The decreased MGMT observed in ATRA-induced SBA in rats thus cannot be explained by DNA methylation in the promoter region. Some scholars have reported that the overexpression of wild-type P53 in vivo can inhibit the transcriptional activity of MGMT (Srivenugopal et al., 2001; Grombacher et al., 1998) without affecting promoter methylation, and ATRA is known to be able to upregulate p53 expression (Lu et al., 2013). These previous findings may explain our results, at least in part. However, the interaction between MGMT and p53 in ATRA-induced SBA in rat embryos requires further exploration. Our results may also be due to the small sample size, which needs to be verified in the future with a larger sample size.
Tran et al. (2012) found that high MGMT expression is associated with NTDs in humans, particularly in female embryos. However, our study showed that MGMT expression is reduced in ATRA-induced SBA in rats. The divergent results may be due to the fact that we are using a non-human animal model of NTD. Further studies are necessary to understand the divergent results of these two studies. Tran et al. (2012) also found that hypomethylation was related to an increased risk for NTDs – more precisely, for cephalic malformations – but that in spina bifida cases, the average MGMT methylation level was not significantly different than that in the controls. This finding is consistent with our results.
One limitation in the current study is that we focused on DNA methylation in the MGMT promoter region only. DNA methylation beyond the CpG island could also play a role in gene expression (Edgar et al., 2014), and we may have overlooked CpG sites that fall outside the CpG island. Additionally, recent findings revealed that non-CpG methylation, of CHH and CHG (where H is A, T, or C), could also influence the gene expression of MGMT (Saikia et al., 2017). Apart from changes in DNA methylation, there are many mechanisms for decreased gene expression, including miRNA/siRNA expression and histone modifications. The miR221/222 can act on MGMT, and the downregulation of MGMT is associated with elevated expression of miR221/222 (Quintavalle et al., 2013). The mechanism of reduced MGMT mRNA expression in ATRA-induced SBA requires further exploration. Another limitation of our study is that protein expression and activity of MGMT in ATRA-induced SBAs were not detected. The protein expression and activity of MGMT, and post-translational modifications that may affect MGMT expression in the developing embryo, will be explored in our future research.
To our knowledge, this is the first study to verify that DNA damage occurs in ATRA-induced SBA embryos, indicating that DNA damage and DNA repair responses are involved in SBA in rats. Moreover, this study explored the expression pattern of the DNA repair gene MGMT in neural tube development in normal rat foetuses and established a relationship between MGMT mRNA downregulation and the risk of NTD in ATRA-induced SBA in rat foetuses. The downregulated MGMT might be involved in NTD progression. It is important to determine the effect of MGMT on NTD and to investigate the mechanism of its downregulation for the treatment and prognosis of NTD. However, this downregulation of MGMT is not caused by a change in the methylation level of its promoter. Further studies will continue to investigate the mechanism of MGMT mRNA downregulation in ATRA-induced SBA in rat foetuses.
Additional file: Open peer review reports 1 (108.7KB, pdf) and 2 (108.7KB, pdf) .
Acknowledgments:
We are very grateful to Hui Gu, Xiao-Wei Wei, Dan Liu, Lian-Shuai Zhao, Chao-Nan Zhang, Li-Zhu Chen, Song-Ying Cao, and Tian-Chu Huang in the Key Laboratory for Congenital Malformation of Health Ministry, Shengjing Hospital, China Medical University, for their experimental technique guidance.
Footnotes
Conflicts of interest: The authors declare that there is no duality of interest associated with this manuscript.
Financial support: This study was supported by the National Natural Science Foundation of China, No. 81671469, 81171072 (to ZWY); the National Basic Research Program of China (973 Program), No. 2013CB945402 (to ZWY); the Program for Liaoning Innovative Research Team in University of China, No. LT2013016 (to ZWY). The funding sources had no role in study conception and design, data analysis or interpretation, paper writing or deciding to submit this paper for publication.
Institutional review board statement: All experimental procedures and protocols were approved by the Animal Ethics Committee, Shengjing Hospital, China Medical University (approval number: 2015PS264K) on October 13, 2015. All experimental procedures described here were in accordance with the National Institutes of Health (NIH) guidelines for the Care and Use of Laboratory Animals.
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open peer reviewers: Shen Lin, Temple University Health System, USA; Chun Liu, Michigan State University, USA.
P-Reviewer: Lin S; C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Gardner B, Stow A, Qiu Y, Song LP; T-Editor: Liu XL
Funding: This study was supported by the National Natural Science Foundation of China, No. 81671469, 81171072 (to ZWY); the National Basic Research Program of China (973 Program), No. 2013CB945402 (to ZWY); the Program for Liaoning Innovative Research Team in University of China, No. LT2013016 (to ZWY).
References
- 1.Albino D, Brizzolara A, Moretti S, Falugi C, Mirisola V, Scaruffi P, Di Candia M, Truini M, Coco S, Bonassi S, Tonini GP. Gene expression profiling identifies eleven DNA repair genes down-regulated during mouse neural crest cell migration. Int J Dev Biol. 2011;55:65–72. doi: 10.1387/ijdb.092970da. [DOI] [PubMed] [Google Scholar]
- 2.Au KS, Ashley-Koch A, Northrup H. Epidemiologic and genetic aspects of spina bifida and other neural tube defects. Dev Disabil Res Rev. 2010;16:6–15. doi: 10.1002/ddrr.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bobola MS, Blank A, Berger MS, Silber JR. O6-methylguanine-DNA methyltransferase deficiency in developing brain: implications for brain tumorigenesis. DNA Repair. 2007;6:1127–1133. doi: 10.1016/j.dnarep.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyles AL, Hammock P, Speer MC. Candidate gene analysis in human neural tube defects. Am J Med Genet C Semin Med Genet. 2005;135C:9–23. doi: 10.1002/ajmg.c.30048. [DOI] [PubMed] [Google Scholar]
- 5.Budzowska M, Jaspers I, Essers J, de Waard H, van Drunen E, Hanada K, Beverloo B, Hendriks RW, de Klein A, Kanaar R, Hoeijmakers JH, Maas A. Mutation of the mouse Rad17 gene leads to embryonic lethality and reveals a role in DNA damage-dependent recombination. EMBO J. 2004;23:3548–3558. doi: 10.1038/sj.emboj.7600353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bugni JM, Meira LB, Samson LD. Alkylation-induced colon tumorigenesis in mice deficient in the Mgmt and Msh6 proteins. Oncogene. 2009;28:734–741. doi: 10.1038/onc.2008.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai W, Zhao H, Guo J, Li Y, Yuan Z, Wang W. Retinoic acid-induced lumbosacral neural tube defects: myeloschisis and hamartoma. Childs Nerv Syst. 2007;23:549–554. doi: 10.1007/s00381-006-0289-y. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Guo J, Lei Y, Zou J, Lu X, Bao Y, Wu L, Wu J, Zheng X, Shen Y, Wu BL, Zhang T. Global DNA hypomethylation is associated with NTD-affected pregnancy: a case-control study. Birth Defects Res A. 2010;88:575–581. doi: 10.1002/bdra.20670. [DOI] [PubMed] [Google Scholar]
- 9.Chowdhury S, Hobbs CA, MacLeod SL, Cleves MA, Melnyk S, James SJ, Hu P, Erickson SW. Associations between maternal genotypes and metabolites implicated in congenital heart defects. Mol Genet Metab. 2012;107:596–604. doi: 10.1016/j.ymgme.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Copp AJ, Greene ND. Genetics and development of neural tube defects. J Pathol. 2010;220:217–230. doi: 10.1002/path.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danzer E, Schwarz U, Wehrli S, Radu A, Adzick NS, Flake AW. Retinoic acid induced myelomeningocele in fetal rats: characterization by histopathological analysis and magnetic resonance imaging. Exp Neurol. 2005;194:467–475. doi: 10.1016/j.expneurol.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 12.Dean W, Lucifero D, Santos F. DNA methylation in mammalian development and disease. Birth Defects Res C. 2005;75:98–111. doi: 10.1002/bdrc.20037. [DOI] [PubMed] [Google Scholar]
- 13.Diez-Pardo JA, Marino JM, Baoquan Q, Delgado-Baeza E, Fernaneez A, Morales MC, Tovar JA. Neural tube defects: an experimental model in the foetal rat. Eur J Pediatr Surg. 1995;5:198–202. doi: 10.1055/s-2008-1066204. [DOI] [PubMed] [Google Scholar]
- 14.Edgar R, Tan PP, Portales-Casamar E, Pavlidis P. Meta-analysis of human methylomes reveals stably methylated sequences surrounding CpG islands associated with high gene expression. Epigenet Chromatin. 2014;7:28. doi: 10.1186/1756-8935-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glassner BJ, Weeda G, Allan JM, Broekhof JL, Carls NH, Donker I, Engelward BP, Hampson RJ, Hersmus R, Hickman MJ, Roth RB, Warren HB, Wu MM, Hoeijmakers JH, Samson LD. DNA repair methyltransferase (Mgmt) knockout mice are sensitive to the lethal effects of chemotherapeutic alkylating agents. Mutagenesis. 1999;14:339–347. doi: 10.1093/mutage/14.3.339. [DOI] [PubMed] [Google Scholar]
- 16.Greene ND, Stanier P, Copp AJ. Genetics of human neural tube defects. Hum Mol Genet. 2009;18:R113–R129. doi: 10.1093/hmg/ddp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grombacher T, Eichhorn U, Kaina B. p53 is involved in regulation of the DNA repair gene O6-methylguanine-DNA methyltransferase (MGMT) by DNA damaging agents. Oncogene. 1998;17:845–851. doi: 10.1038/sj.onc.1202000. [DOI] [PubMed] [Google Scholar]
- 18.Kisby GE, Olivas A, Park T, Churchwell M, Doerge D, Samson LD, Gerson SL, Turker MS. DNA repair modulates the vulnerability of the developing brain to alkylating agents. DNA Repair. 2009;8:400–412. doi: 10.1016/j.dnarep.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitajima Y, Miyazaki K, Matsukura S, Tanaka M, Sekiguchi M. Loss of expression of DNA repair enzymes MGMT, hMLH1, and hMSH2 during tumor progression in gastric cancer. Gastric Cancer. 2003;6:86–95. doi: 10.1007/s10120-003-0213-z. [DOI] [PubMed] [Google Scholar]
- 20.Konduri SD, Ticku J, Bobustuc GC, Sutphin RM, Colon J, Isley B, Bhakat KK, Srivenugopal KS, Baker CH. Blockade of MGMT expression by O6 benzyl guanine leads to inhibition of pancreatic cancer growth and induction of apoptosis. Clin Cancer Res. 2009;15:6087–6095. doi: 10.1158/1078-0432.CCR-09-0887. [DOI] [PubMed] [Google Scholar]
- 21.Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008;36:W170–W175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Gao F, Ma L, Jiang J, Miao J, Jiang M, Fan Y, Wang L, Wu D, Liu B, Wang W, Lui VC, Yuan Z. Therapeutic potential of in utero mesenchymal stem cell (MSCs) transplantation in rat foetuses with spina bifida aperta. J Cell Mol Med. 2012;16:1606–1617. doi: 10.1111/j.1582-4934.2011.01470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li LC, Dahiya R. Meth Primer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 24.Li M, Cleves MA, Mallick H, Erickson SW, Tang X, Nick TG, Macleod SL, Hobbs CA National Birth Defect Prevention Study. A genetic association study detects haplotypes associated with obstructive heart defects. Hum Genet. 2014b;133:1127–1138. doi: 10.1007/s00439-014-1453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li R, Yang P, Chen X, Wang L. Maternal serum AGEs levels in pregnancies associated with neural tube defects. Int J Dev Neurosci. 2014a;33:57–61. doi: 10.1016/j.ijdevneu.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Li RL, Zhao WW, Gao BY. Advanced glycation end products induce neural tube defects through elevating oxidative stress in mice. Neural Regen Res. 2018;13:1368–1374. doi: 10.4103/1673-5374.235249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lind GE, Thorstensen L, Lovig T, Meling GI, Hamelin R, Rognum TO, Esteller M, Lothe RA. A CpG island hypermethylation profile of primary colorectal carcinomas and colon cancer cell lines. Mol cancer. 2004;3:28. doi: 10.1186/1476-4598-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Zhang L, Li Z, Jin L, Zhang Y, Ye R, Liu J, Ren A. Prevalence and trend of neural tube defects in five counties in Shanxi province of Northern China 2000 to 2014. Birth Defects Res A. 2016;106:267–274. doi: 10.1002/bdra.23486. [DOI] [PubMed] [Google Scholar]
- 29.Liu Z, Wang Z, Li Y, Ouyang S, Chang H, Zhang T, Zheng X, Wu J. Association of genomic instability, and the methylation status of imprinted genes and mismatch-repair-genes, with-neural tube defects. Eur J Hum Genet. 2012;20:516–520. doi: 10.1038/ejhg.2011.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Lu J, Zhang F, Yuan Y, Ding C, Zhang L, Li Q. All-trans retinoic acid upregulates the expression of p53 via Axin and inhibits the proliferation of glioma cells. Oncol Rep. 2013;29:2269–2274. doi: 10.3892/or.2013.2391. [DOI] [PubMed] [Google Scholar]
- 32.Minoo P. Toward a molecular classification of colorectal cancer: the role of MGMT. Front Oncol. 2013;3:266. doi: 10.3389/fonc.2013.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mokhtar M, Kondo K, Namura T, Ali AH, Fujita Y, Takai C, Takizawa H, Nakagawa Y, Toba H, Kajiura K, Yoshida M, Kawakami G, Sakiyama S, Tangoku A. Methylation and expression profiles of MGMT gene in thymic epithelial tumors. Lung Cancer. 2014;83:279–287. doi: 10.1016/j.lungcan.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 34.Olshan AF, Shaw GM, Millikan RC, Laurent C, Finnell RH. Polymorphisms in DNA repair genes as risk factors for spina bifida and orofacial clefts. Am J Med Genet A. 2005;135:268–273. doi: 10.1002/ajmg.a.30713. [DOI] [PubMed] [Google Scholar]
- 35.Paganini L, Carlessi N, Fontana L, Silipigni R, Motta S, Fiori S, Guerneri S, Lalatta F, Cereda A, Sirchia S, Miozzo M, Tabano S. Beckwith-Wiedemann syndrome prenatal diagnosis by methylation analysis in chorionic villi. Epigenetics. 2015;10:643–649. doi: 10.1080/15592294.2015.1057383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quintavalle C, Mangani D, Roscigno G, Romano G, Diaz-Lagares A, Iaboni M, Donnarumma E, Fiore D, De Marinis P, Soini Y, Esteller M, Condorelli G. MiR-221/222 target the DNA methyltransferase MGMT in glioma cells. PLoS One. 2013;8:e74466. doi: 10.1371/journal.pone.0074466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saikia S, Rehman AU, Barooah P, Sarmah P, Bhattacharyya M, Deka M, Deka M, Goswami B, Husain SA, Medhi S. Alteration in the expression of MGMT and RUNX3 due to non-CpG promoter methylation and their correlation with different risk factors in esophageal cancer patients. Tumor Biol. 2017;39:1010428317701630. doi: 10.1177/1010428317701630. [DOI] [PubMed] [Google Scholar]
- 38.Song T, Li H, Tian Z, Xu C, Liu J, Guo Y. Disruption of NF-kappaB signaling by fluoxetine attenuates MGMT expression in glioma cells. Oncotargets Ther. 2015;8:2199–2208. doi: 10.2147/OTT.S85948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srivenugopal KS, Shou J, Mullapudi SR, Lang FF, Jr, Rao JS, Ali-Osman F. Enforced expression of wild-type p53 curtails the transcription of the O(6)-methylguanine-DNA methyltransferase gene in human tumor cells and enhances their sensitivity to alkylating agents. Clin Cancer Res. 2001;7:1398–1409. [PubMed] [Google Scholar]
- 40.Su Y, Xu H, Xu Y, Yu J, Xian Y, Luo Q. Azacytidine inhibits the proliferation of human promyelocytic leukemia cells (HL60) by demethylation of MGMT, DAPK and p16 genes. Hematology. 2012;17:41–46. doi: 10.1179/102453312X13221316477624. [DOI] [PubMed] [Google Scholar]
- 41.Toffolatti L, Scquizzato E, Cavallin S, Canal F, Scarpa M, Stefani PM, Gherlinzoni F, Dei Tos AP. MGMT promoter methylation and correlation with protein expression in primary central nervous system lymphoma. Virchows Arch. 2014;465:579–586. doi: 10.1007/s00428-014-1622-6. [DOI] [PubMed] [Google Scholar]
- 42.Tran S, Wang L, Le J, Guan J, Wu L, Zou J, Wang Z, Wang J, Wang F, Chen X, Cai L, Lu X, Zhao H, Guo J, Bao Y, Zheng X, Zhang T. Altered methylation of the DNA repair gene MGMT is associated with neural tube defects. J Mol Neurosci. 2012;47:42–51. doi: 10.1007/s12031-011-9676-2. [DOI] [PubMed] [Google Scholar]
- 43.van der Linden IJ, Heil SG, van Egmont Petersen M, van Straaten HW, den Heijer M, Blom HJ. Inhibition of methylation and changes in gene expression in relation to neural tube defects. Birth Defects Res A. 2008;82:676–683. doi: 10.1002/bdra.20509. [DOI] [PubMed] [Google Scholar]
- 44.Vlachostergios PJ, Patrikidou A, Daliani DD, Papandreou CN. The ubiquitin-proteasome system in cancer, a major player in DNA repair. Part 2: transcriptional regulation. J Cell Mol Med. 2009;13:3019–3031. doi: 10.1111/j.1582-4934.2009.00825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Chang S, Guan J, Shangguan S, Lu X, Wang Z, Wu L, Zou J, Zhao H, Bao Y, Qiu Z, Niu B, Zhang T. Tissue-specific methylation of long interspersed nucleotide element-1 of homo sapiens (L1Hs) during human embryogenesis and roles in neural tube defects. Curr Mol Med. 2015;15:497–507. doi: 10.2174/1566524015666150630130229. [DOI] [PubMed] [Google Scholar]
- 46.Wang L, Wang F, Guan J, Le J, Wu L, Zou J, Zhao H, Pei L, Zheng X, Zhang T. Relation between hypomethylation of long interspersed nucleotide elements and risk of neural tube defects. Am J Clin Nutr. 2010;91:1359–1367. doi: 10.3945/ajcn.2009.28858. [DOI] [PubMed] [Google Scholar]
- 47.Wei X, Li H, Miao J, Zhou F, Liu B, Wu D, Li S, Wang L, Fan Y, Wang W, Yuan Z. Disturbed apoptosis and cell proliferation in developing neuroepithelium of lumbo-sacral neural tubes in retinoic acid-induced spina bifida aperta in rat. Int J Dev Neurosci. 2012;30:375–381. doi: 10.1016/j.ijdevneu.2012.03.340. [DOI] [PubMed] [Google Scholar]
- 48.Wilde JJ, Petersen JR, Niswander L. Genetic, epigenetic, and environmental contributions to neural tube closure. Annu Rev Genet. 2014;48:583–611. doi: 10.1146/annurev-genet-120213-092208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu LN, Wei XW, Fan Y, Miao JN, Wang LL, Zhang Y, Wu D, Yuan ZW. Altered expression of 14-3-3zeta protein in spinal cords of rat fetuses with spina bifida aperta. PLoS One. 2013;8:e70457. doi: 10.1371/journal.pone.0070457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yi D, Yuan Y, Jin L, Zhou G, Zhu H, Finnell RH, Ren A. Levels of PAH-DNA adducts in cord blood and cord tissue and the risk of fetal neural tube defectsin a Chinese population. Neurotoxicology. 2015;46:73–78. doi: 10.1016/j.neuro.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan Y, Jin L, Wang L, Li Z, Zhang L, Zhu H, Finnell RH, Zhou G, Ren A. Levels of PAH-DNA adducts in placental tissue and the risk of fetal neural tube defects in a Chinese population. Reprod Toxicol. 2013;37:70–75. doi: 10.1016/j.reprotox.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


