 High affinity, cannabinoid type 2 receptor selective ligand.
High affinity, cannabinoid type 2 receptor selective ligand.
Abstract
Cannabinoid type 2 (CB2) receptor has been implicated in several diseases and conditions, however no CB2 receptor selective drugs have made it to market. The aim of this study was to develop fluorescent ligands as CB2 receptor tools, to enable an increased understanding of CB2 receptor expression and signalling and thereby accelerate drug discovery. Fluorescent ligands have been successfully developed for other receptors, however none with adequate subtype selectivity or imaging properties have been reported for CB2 receptor. A series of 1,8-naphthyridin-2-(1H)-one-3-carboxamides with linkers and fluorophores appended in the N1 and C3-positions were developed. Molecular modelling indicated the C3 cis-cyclohexanol-linked compounds directed the linker out of the CB2 receptor between transmembrane helices 1 and 7. Herein we report fluorescent ligand 32 (hCB2 pKi = 6.33 ± 0.02) as one of the highest affinity, selective CB2 receptor fluorescent ligands reported. Despite 32 displaying poor specific labelling of CB2 receptor, the naphthyridine scaffold with this linker remains highly promising for future development of CB2 receptor tools.
Introduction
Cannabinoid type 2 (CB2) receptor is a class A G protein-coupled receptor (GPCR) and is highly expressed in immune cells and lymphoid tissues,1 and in lower levels in the central nervous system.2 Along with cannabinoid type 1 (CB1) receptor, the endogenous agonists anandamide and 2-arachidonoylglycerol and various regulatory enzymes, CB2 receptor is part of the highly regulated endocannabinoid system.3 CB2 receptor modulates cytokine release and immune cell migration, thereby regulating immune responses and inflammatory pathways.4,5 CB2 receptor has been shown to play a role in neurodegenerative disorders,6 pain,7 atherosclerosis8 and cancer.9 As such, ligands of CB2 receptor hold promise as therapeutic interventions. However, to date no CB2 receptor selective drugs have made it to market, though a few have undergone clinical trials.10 Drug development efforts would be greatly aided by an increased understanding of CB2 receptor expression and signalling. The aim of this study was therefore to develop high affinity, selective CB2 receptor fluorescent ligands and evaluate these ligands as imaging tools. Fluorescent ligands have been successfully developed for other Class A GPCRs11 and used, for example, to visualise receptor,12 track receptor internalisation13,14 and to study single-cell ligand binding kinetics.15
In order to be useful imaging tools, fluorescent GPCR ligands require high affinity and selectivity, and should exhibit low levels of non-specific membrane interactions. A typical strategy for developing fluorescent ligands is to select a known high affinity and selective ligand as a scaffold to which a linker and fluorophore can be appended. It is crucial to identify a suitable position on the scaffold for linker attachment in order to retain affinity for the target receptor and minimise disruption of the binding orientation of the pharmacophore.11 It is particularly challenging to develop fluorescent ligands with minimal plasma membrane interactions for cannabinoid (CB) receptors due to the typically lipophilic nature of CB ligands.
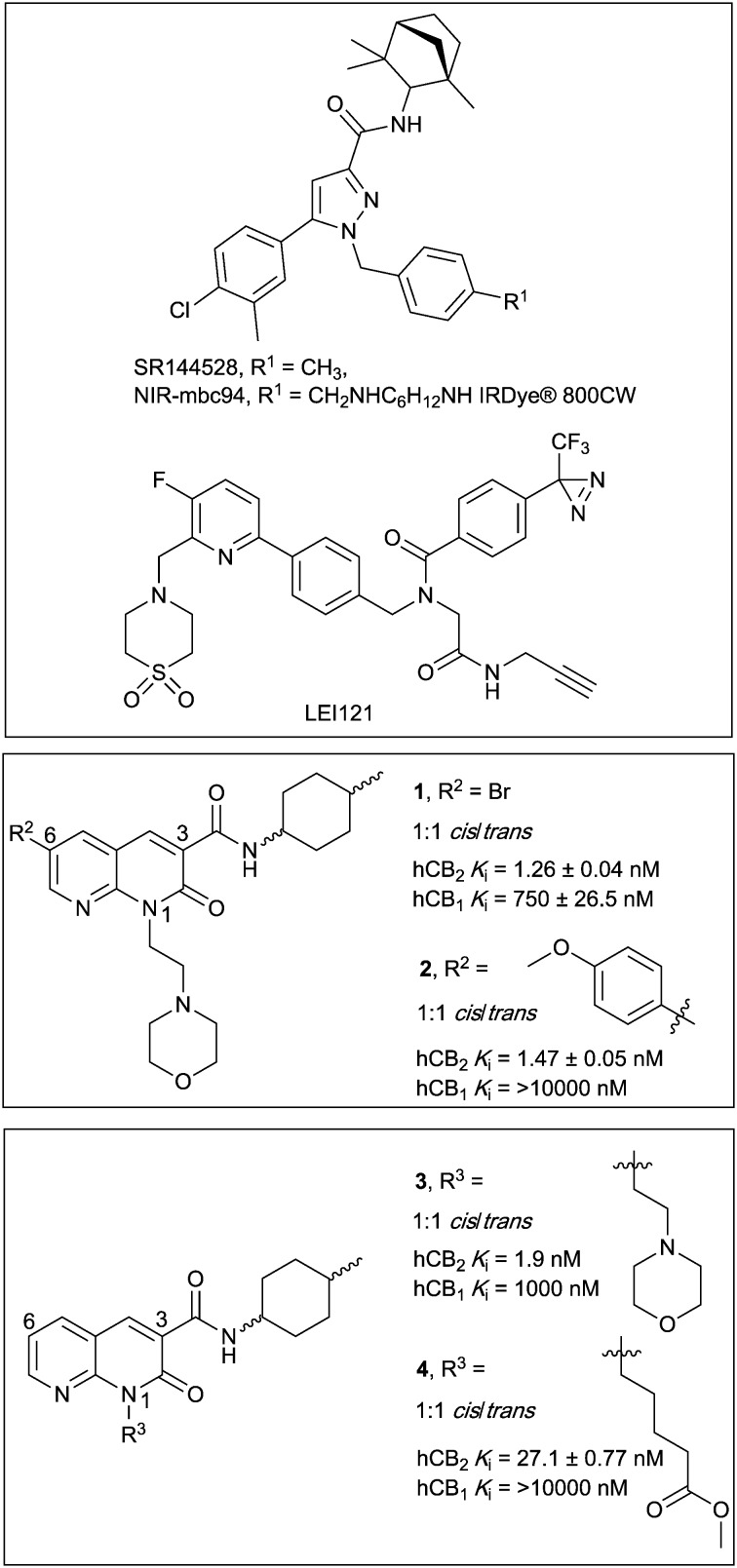
There have been several reports of fluorescent ligands for CB2 receptor, however, these lack either receptor subtype selectivity or display high levels of non-specific binding/interactions rendering these ligands unsuitable for use in techniques such as confocal microscopy.16–21 A scaffold based on SR144528 (Fig. 1) termed ‘mbc94’ has most commonly been utilised for development of CB2 receptor fluorescent ligands, for example NIR-mbc94 (Fig. 1, mCB2 receptor Ki = 260 nM).18 Fluorescent ligands based on an aminoalkylindole scaffold have also been attempted, however despite the ligand-linker conjugates retaining CB receptor affinity the fluorescent conjugates showed little CB receptor binding.22,23 The bifunctional, photoreactive scaffold LEI121 (Fig. 1) has recently been reported as an alternative strategy to a ‘pre-assembled’ CB2 receptor fluorescent ligand. Upon photoactivation, non-fluorescent LEI121 covalently bound to CB2 receptor, which was then labelled via reaction of an azide-fluorophore to the alkyne of LEI121.24 This is a promising strategy for interrogating CB2 receptor, however a ‘pre-assembled’ non-covalent fluorescent ligand is still very desirable for many competition-based and kinetic experiments.
Fig. 1. Diarylpyrazole SR144528, and 1,8-naphthyridin-2-(1H)-one-3-carboxamides 1–4, with previously reported binding affinities.25.
In this paper we report the development of CB2 receptor fluorescent ligands based on the 1,8-naphthyridin-2-(1H)-one-3-carboxamide scaffold. Many derivatives of this scaffold are reported to have very high affinity and subtype selectivity for CB2 receptor, (e.g.1 and 4,25Fig. 1) and there are structure–activity-relationships (SAR) reporting the effect of substitution at N1, C3 and C6 (ref. 26–29) (e.g.1–4,25Fig. 1). Additionally, the 1,8-naphthyridin-2-(1H)-one-3-carboxamide scaffold was selected because it is less lipophilic than many other cannabinoids (e.g. clog P of 2 is 2.93 compared to SR144528 clog P of 7.13), which is beneficial in terms of reducing the overall lipophilicity of a fluorescent ligand.
The N1, C3 and C6 naphthyridine positions were considered for linker and fluorophore attachment because there is SAR reported for these positions and all are synthetically accessible. Since SAR indicated groups of varying length and bulk were tolerated in the N1-position,25,29 including a ‘linker-like’ methyl valerate (4, Fig. 1), one series of conjugates were developed linked at this position. A range of factors were considered when analysing if the C3 cyclohexyl carboxamide position might be amenable to linker and fluorophore attachment. The stereochemistry of the 4-methylcyclohexyl moiety at the C3 carboxamide has been shown to be influential on CB2 receptor binding, with cis derivatives showing improved receptor affinity compared to trans derivatives.25,27 This sensitivity of the methylcyclohexyl group could translate to a position not tolerant of much chemical change/variation, however ligand docking of 1 into a CB2 receptor homology model (as is discussed for 28 in Modelling section) positioned the cyclohexyl group close to exiting CB2 receptor between transmembrane helix (TMH) 1 and TMH7. This therefore made the C3 cyclohexyl carboxamide an appealing linker attachment position, especially in light of previously reported molecular dynamics simulations indicating that cannabinoids may enter into CB receptors via the lipid membrane between TMH6 and TMH7 or between TMH1 and TMH7.30–32 Cyclohexanol and cyclohexylamine derivatives were designed to allow linker extension and both cis and trans isomers (of the cyclohexanol) were prepared since the previously established methylcyclohexyl cis/trans SAR could not be assumed to be the same.
The C6 position was not selected for linker attachment based on SAR that showed that functional activity can be controlled by the C6 substituent. For example, compound 3 (Fig. 1) behaved as a CB2 receptor agonist in β-arrestin 2 and cAMP assays while 1 and 2 (Fig. 1) behaved as antagonists/inverse agonists in a β-arrestin 2 assay.25 It has been postulated with docking studies that this C6 substituent orientates deep into a receptor binding pocket and modulates a CWFP flexible hinge motif on TMH6.25 Linker substitution at C6 was therefore deemed most likely non-tolerable and likely would be detrimental to ligand affinity for CB2 receptor. This was also the reason that the two fluorescent ligand series (N1-linked and C3-linked) were therefore developed with a small C6 substituent present since the goal was to develop high affinity, CB2 receptor selective fluorescent ligands that do not activate CB2 receptor.
Results and discussion
Synthetic chemistry
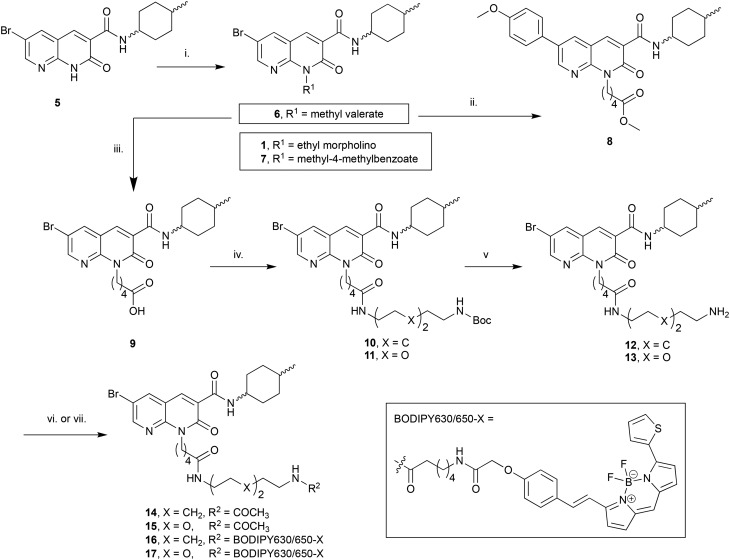
The N1-linked series was assembled as a 1 : 1 cis/trans mix at the C3 cyclohexyl carboxamide as the first goal was to determine if the N1-position was tolerant of linker and fluorophore attachment. Commercially available 2-amino-3-pyridinecarboxaldehyde was converted to 5 in 3 steps following previously reported syntheses.25 Alkylation of 5 with 4-(2-chloroethyl)morpholine hydrochloride afforded the previously reported 1 (ref. 25) (Scheme 1), which was used as a pharmacological control. Alkylation of 5 with methyl 5-bromovalerate or methyl 4-bromomethylbenzoate gave 6 or 7 in low yield, due to incomplete conversion of 5 and challenging separation of products from unreacted 5. Both 6 and 7 are amenable to linker extension following methyl ester deprotection, however it was decided to proceed with valerate linked-6 and only synthesise further benzoyl derivatives if 7 showed high affinity for CB2 receptor. Bromo-6 was subjected to Suzuki coupling with 4-methoxyphenylboronic acid to afford 8, to enable comparison of the C6 bromo to methoxyphenyl substituent.
Scheme 1. Reagents and conditions: (i) 4-(2-Chloroethyl)morpholine hydrochloride, methyl 5-bromovalerate, or methyl 4-bromomethylbenzoate, Cs2CO3, DMF, 50 °C, 12 h, 22–37%; (ii) 4-methoxyphenylboronic acid, Pd(OAc)2, Na2CO3, H2O : DMF 1 : 4 v:v, 110 °C, 3 h, 55%; (iii) 10% aq. NaOH, EtOH, 110 °C, 5 h, 43%; (iv) N-Boc-1,8-octanediamine or N-Boc-2,2′-(ethylenedioxy)diethylamine, DIPEA, HATU, DMF, rt, 14 h, 71–77%; (v) TFA, DCM, rt, 1 h, 86–91%; (vi) Ac2O, DIPEA, DCM, rt, 1 h, 95–96%; (vii) BODIPY 630/650-X-OSu, DIPEA, DMF, rt, 15 h, 88–92%. Compounds 1 : 1 cis/trans.
To further extend the distance of the naphthyridine core from the fluorophore the methyl ester of 6 underwent saponification to reveal carboxylic acid 9, which was coupled to either N-Boc-1,8-octanediamine or N-Boc-2,2′-(ethylenedioxy)diethylamine to yield 10 and 11 respectively. These alkyl and short PEG-like linkers of the same atom length were chosen as a way to compare different linker lipophilicity, since it was hypothesised that the lipophilic alkyl linker may be preferable for CB2 receptor affinity, whereas the PEG-like linker may impart more hydrophilicty (than the equivalent alkyl linker) to the overall fluorescent ligand and lead to better imaging properties (e.g. lower non-receptor specific membrane interactions). Boc deprotection of 10 and 11 yielded unprotected amines 12 and 13, which were then each acetylated using acetic anhydride to give 14 and 15. In a separate procedure these were reacted with 6-(((4,4-difluoro-5-(2-thienyl)-4-bora-3a,4a-diaza-sindacene-3-yl)styryloxy) acetyl) aminohexanoic acid, succinimidyl ester (BODIPY630/650-X-OSu) to yield fluorescent ligands 16 and 17. The BODIPY630/650-X fluorophore has been used successfully to develop fluorescent ligands for other Class A GPCRs,14,33,34 and has favourable properties (such as a good quantum yield, intense absorption and high chemical and photo stability) and as a red emitting fluorophore there is minimal detection interference from cellular autofluorescence.35 A red-shifted fluorescent ligand also allows co-localisation experiments with, for example, green-fluorescent-protein-tagged receptors or proteins, to be carried out.
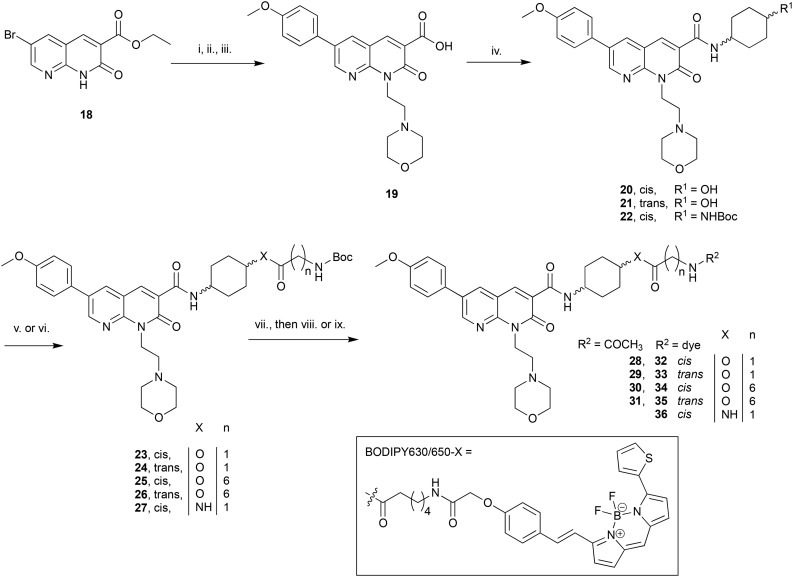
The C3-linked series was assembled in a different order to allow for more efficient variation of the C3 substituent. The previously reported 18 (ref. 25) was N1-alkylated, substituted at C6 with methoxyphenyl and the methyl ester saponified to afford 19 (Scheme 2). Carboxylic acid 19 was coupled in separate reactions to cis- or trans-4-aminocyclohexanol to give alcohols 20 and 21 respectively, or to 1-N-Boc-cis-1,4-cyclohexyldiamine to give cis-Boc-protected amine 22.
Scheme 2. Reagents and conditions: (i) 4-(2-Chloroethyl)morpholine hydrochloride, Cs2CO3, DMF, 50 °C, 12 h; (ii) 4-methoxyphenylboronic acid, Na2CO3, Pd(OAc)2, H2O, DMF, 110 °C, 3 h; (iii) 0.2 M aq. LiOH·H2O, THF, 0 °C, 1 h, 20% over three steps; (iv) cis-4-aminocyclohexanol or trans-4-aminocyclohexanol or 1-N-Boc-cis-1,4-cyclohexyldiamine, HATU, DIPEA, DMF, rt, 14 h, 79–82%; (v) N-Boc-glycine or N-Boc-7-aminoheptanoic acid, TFFH, Et3N, DMAP, DCM, rt, 14 h, 19–80%; (vi) 1.Trifluoroacetic acid (TFA), DCM, rt, 1 h, quantitative; 2. N-Boc-glycine, HATU, DIPEA, DMF, rt, 14 h, 31%; (vii) TFA, DCM, rt, 1 h, 45–57%; (viii) Ac2O, DIPEA, DCM, rt, 1 h, 96–98%; (ix) BODIPY 630/650-X-OSu, DIPEA, DMF, rt, 14 h, 72–97%.
The intention was to introduce an ether linkage via alkylation of alcohols 20 and 21. However, despite several attempts using alkyl bromides such as tert-butyl bromoacetate or 2-(Boc)-amino-ethylbromide with either NaH or CsCO3 as the base, varying equivalents of each base and addition order, and varying temperature the intended ether product was either detected only in trace amounts or not at all. An alternative synthetic strategy was envisaged, whereby the ether bond to the cyclohexyl could instead be preassembled and then coupled to 19. Attempts to alkylate carboxybenzyl and dibenzyl protected 4-aminocyclohexanol with various alkyl bromides using a range of bases (NaH, CsCO3, K2CO3, tBuOK and lithium bis(trimethylsilyl)amide), temperatures, solvents and reaction times were all unsuccessful. A Mitsunobu reaction to form the ether bond was not attempted due to the calculated pKa (∼15) of the cyclohexanol alcohol.
Since 20 and 21 were already prepared it was decided to proceed with formation of an ester bond to enable pharmacological evaluation of the C3-cyclohexyl-linker position, which could then be revisited for a more stable bond if the C3-position proved suitable. Activation of Boc-glycine or N-Boc-7-aminoheptanoic acid with tetramethylfluoroformamidinium hexafluorophosphate (TFFH) followed by addition of 20 or 21 yielded esters 23–26. TFFH was used rather than a carbodiimide reagent in an attempt to counteract the poor nucleophilicity of alcohols 20 and 21. Instead of using linkers analogous to the alkyl and PEG-like linkers of the N-substituted series, it was decided to use a commercially available single ‘glycine’ and a longer 7-aminoheptanoic acid linker to allow exploration of how varying linker length might effect receptor affinity. Boc-deprotection of 22 followed by HATU-mediated coupling of Boc-glycine afforded 27, the analogous compound to 23 but with an amide replacing the ester bond. Boc-deprotection of 23–27 followed by either acylation or reaction with BODIPY630/650-X-OSu gave 28–31 or 32–36 respectively.
Pharmacology
A radioligand competition binding screen was carried out to determine the ability of 10 μM of 1, 6–8, 14–17, 20, 21, 28–36 to displace [3H]CP-55 940 (2.5 nM or 1 nM) from CB2 or CB1 receptor (data not shown). Compounds that displaced [3H]CP55 940 by more than 50% were then evaluated at varying concentrations to determine pKi values (Tables 1 and 2). Compounds with pKi > 5 at CB2 receptor were also analysed for function at CB2 and CB1 receptors in a cAMP assay (Tables 1 and 2).
Table 1. Affinity and functional data of N1-linked naphthyridines.
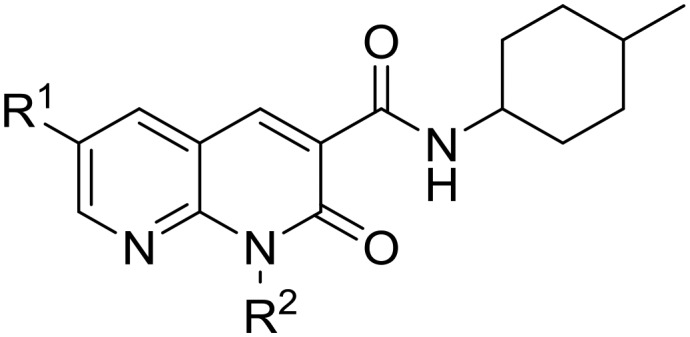
| |||||||||
| R 1 | R 2 | CB2 receptor |
CB1 receptor |
CB2 selectivity e | |||||
| pKi a (±SEM) | pIC50 b , c (±SEM) | E max b , d (% ± SEM) | pKi (±SEM) a | pIC50 (±SEM) b , c | E max b , d (% ± SEM) | ||||
| 1 | Br | Ethyl morpholino | 7.26 ± 0.04 | 7.41 ± 0.17 | 166 ± 5.6 | <5 | No response g | 89 ± 5.6 g | >182 |
| 6 | Br | (CH2)4CO2Me | 5.96 ± 0.04 | 6.24 ± 0.05 f | 161 ± 5.0 f | <5 | 5.73 ± 0.14 | 134 ± 4.4 | >9 |
| 7 | Br | Methyl-4-methylbenzoate | <5 | — | — | <5 | — | — | — |
| 8 | Me-OPh | (CH2)4CO2Me | 6.59 ± 0.05 | 7.19 ± 0.27 | 139 ± 4.2 | 5.06 ± 0.03 | 6.02 ± 0.25 | 126 ± 3.2 | 33 |
| 14 | Br | (CH2)4CO | <5 | — | — | <5 | — | — | — |
| NH(CH2)8NHAc | |||||||||
| 15 | Br | (CH2)4CO | <5 | — | — | <5 | — | — | — |
| NH(C2H4O)2NHAc | |||||||||
| 16 | Br | (CH2)4CO | <5 | — | — | <5 | — | — | — |
| NH(CH2)8NH-BODIPY-630/650-X | |||||||||
| 17 | Br | (CH2)4CO | <5 | — | — | <5 | — | — | — |
| NH(C2H4O)2NH-BODIPY-630/650-X | |||||||||
| SR144528 | — | 7.29 ± 0.03 | 6.90 ± 0.08 | 153 ± 3.8 | 5.40 ± 0.2 | No response g | 108 ± 3.6 g | 78 | |
aRadioligand binding performed with [3H]CP55 940 (2.5 nM) and HEK293-hCB2 or -hCB1 membranes. Data is the mean ± SEM of at least three individual experiments performed in triplicate. Compounds which at 10 μM significantly displaced [3H]CP55 940 but with <50% displacement are annotated as having pKi >5 M.
bcAMP levels measured in a BRET assay using a CAMYEL biosensor, in either HEK-293-hCB2 or hCB1 cells. Data is the mean ± SEM of at least three independent experiments conducted in duplicate.
cpIC50 calculated by concentration response.
d E max response (at 10 μM for compounds without pIC50 determined), normalised to basal (0%) and forskolin only (100%) levels of cAMP. Emax >100% is consistent with inverse agonism.
eCB2 receptor selectivity is calculated by: 10^(pKi CB2–pKi CB1) Naphthyridine compounds all 1 : 1 cis:trans mixture.
fExcept (6) which is two independent experiments performed in duplicate. ‘—’ Indicates that pIC50 or Emax was not determined.
gIndicates no significant difference from forskolin only (100%), indicating no measurable response.
Table 2. Affinity and functional data of cyclohexyl-linked naphthyridines.
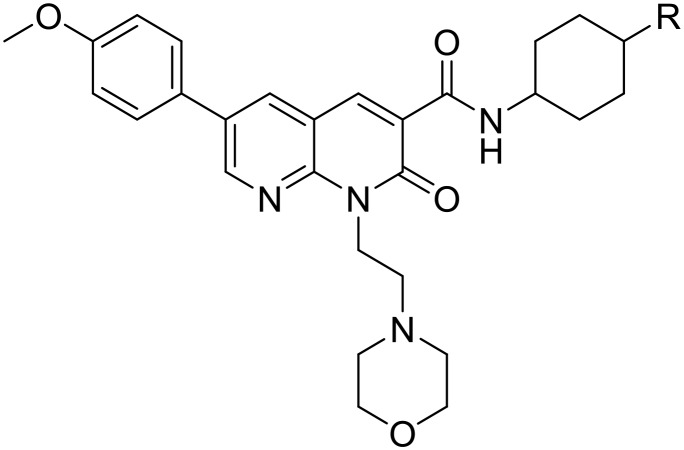
| |||||||||
| R | CB2 receptor |
CB1 receptor |
CB2 selectivity e | ||||||
| pKi a (±SEM) | pIC50 b , c (±SEM) | E max b , d (% ± SEM) | pKi a (± SEM) | pIC50 b , c (±SEM) | E max b , d (% ± SEM) | ||||
| 20 | cis | OH | 5.66 ± 0.07 | 6.74 ± 0.16 | 156 ± 5.5 | <5 | — | 117 ± 2.4 f | >4 |
| 21 | trans | OH | 5.47 ± 0.10 | — | — | No binding | — | — | — |
| 28 | cis | OC(O)CH2NHAc | 5.99 ± 0.03 | 6.99 ± 0.11 | 154 ± 5.4 | <5 | No response g | 115 ± 6.5 f , g | >9 |
| 29 | trans | OC(O)CH2NHAc | <5 | — | — | No binding | — | — | — |
| 30 | cis | OC(O)C6H12NHAc | 5.51 ± 0.02 | 5.81 ± 0.15 | 179 ± 12.1 | <5 | — | 130 ± 2.6 f | >3 |
| 31 | trans | OC(O)C6H12NHAc | 5.43 ± 0.06 | — | — | <5 | — | — | >2 |
| 32 | cis | OC(O)CH2NH-BODIPY630/650-X | 6.33 ± 0.02 | 6.72 ± 0.18 | 210 ± 15.7 | <5 | — | 117 ± 3.5 h | >21 |
| 33 | trans | OC(O)CH2NH-BODIPY630/650-X | 5.23 ± 0.06 | — | — | <5 | — | — g | >1.6 |
| 34 | cis | OC(O)C6H12NH-BODIPY 630/650-X | 5.11 ± 0.04 | — | — | <5 | — | — | >1.2 |
| 35 | trans | OC(O)C6H12NH-BODIPY 630/650-X | No binding | — | — | <5 | — | — | — |
| 36 | cis | NHC(O)CH2NH-BODIPY630/650-X | <5 | — | — | <5 | — | — | |
aRadioligand binding performed with [3H]CP55 940 (1 nM) and HEK293-hCB2 or -hCB1 membranes. Data is the mean ± SEM of at least three experiments performed in triplicate. Compounds which at 10 μM significantly displaced [3H]CP55 940 but with <50% displacement are annotated as having pKi >5 M.
bcAMP levels measured in a BRET assay using a CAMYEL biosensor, in either HEK-293-hCB2 or hCB1 cells. Data is the mean ± SEM of at least three independent experiments conducted in duplicate.
cpIC50 calculated by concentration response.
d E max response (at 10 μM for compounds without pIC50 determined.
eCB2 receptor selectivity is calculated by: 10^(pKi CB2–pKi CB1).
fExcept which are two independent experiments performed in duplicate. ‘—’ Indicates that pIC50 or Emax was not determined.
gIndicates no significant difference from forskolin only (100%), indicating no measurable response.
hExcept which is at 1 μM due to high non-specific effects at 10 μ, see ESI), normalised to basal (0%) and forskolin only (100%) levels of cAMP. Emax >100% is consistent with inverse agonism.
N1-linked naphthyridines
The N1-linked naphthyridine with the highest affinity for CB2 receptor (Table 1) was the previously reported N-ethyl morpholino 1. In our hands, 1 had a pKi of 7.26 ± 0.04, which was equivalent to that measured for SR144528 (pKi 7.29 ± 0.03). This was a slightly different result to that previously reported wherein 1 had a ∼0.5 log unit lower affinity than SR144528 in their study (Fig. 1 and ref. 25). Exchange of the ethyl morpholino group (1) with a methyl benzoate (7) greatly reduced CB2 receptor binding, which was somewhat surprising since there are literature reports of similar naphthyridine scaffolds with N1-aromatic groups such as benzyl and p-fluorobenzyl with nanomolar CB2 receptor affinity.25,27 The affinities of N1-valerate linked 6 and 8 for CB2 receptor were both poorer than the previously reported N1-valerate 4 (Fig. 1),25 although 4 is unsubstituted at the C6 position. Unfortunately, extension of the N1-linker (14, 15) and attachment of the BODIPY 630/650 fluorophore to this extended linker resulted in little affinity for CB2 receptor. N1-linked 1, 6–8 and 14–17 were also tested for CB1 receptor affinity and, in agreement with previously reported naphthyridine compounds, none showed appreciable binding.
The function of N1-linked naphthyridines 1, 6 and 8 was analysed using a bioluminescence resonance energy transfer (BRET) biosensor to measure modulation of forskolin-stimulated cyclic adenosine monophosphate (cAMP) at CB2 and CB1 receptors (Table 1). All three (1, 6, 8) were found to increase the level of cAMP, consistent with inverse agonism for CB2 receptor, in agreement with the literature data for 1 and the reported trend that substitution at the C6 position results in inverse agonism.25 The most potent for CB2 receptor was 1, followed by 8, while 6 was the least potent. A much lower potency (6 and 8) or no response (1) was measured for CB1 receptor. It was concluded from these results (Table 1) that linker and fluorophore substitution at the naphthyridine N1-position is unlikely to lead to fluorescent ligands with useful affinity at CB2 receptor.
Cyclohexyl-linked naphthyridines
The cis isomer showed higher affinity for CB2 receptor than the analogous trans isomer across all cyclohexyl derivatives tested (Table 2), sometimes as much as 1 log unit (28vs.29 and 32vs.33). This trend is in agreement with cis-4-methylcyclohexyl derivatives previously reported to have higher affinity for CB2 receptor than trans analogues.25,27 There was a dramatic loss in CB2 receptor affinity for cyclohexanol derivatives 20 and 21 compared to previously reported 1 : 1 cis:trans methylcyclohexyl 2 (ref. 25) (Table 1) despite the relatively small chemical difference of an alcohol or methyl moiety. The acetylated aminoheptanoate-linked 30 and 31 also showed comparable poor affinity for CB2 receptor. Cis-glycine linked-28 showed a 0.5 log unit improvement in CB2 receptor affinity over the analogous cis-aminoheptanoate 30. All cyclohexyl-linked compounds showed little or no affinity for CB1 receptor.
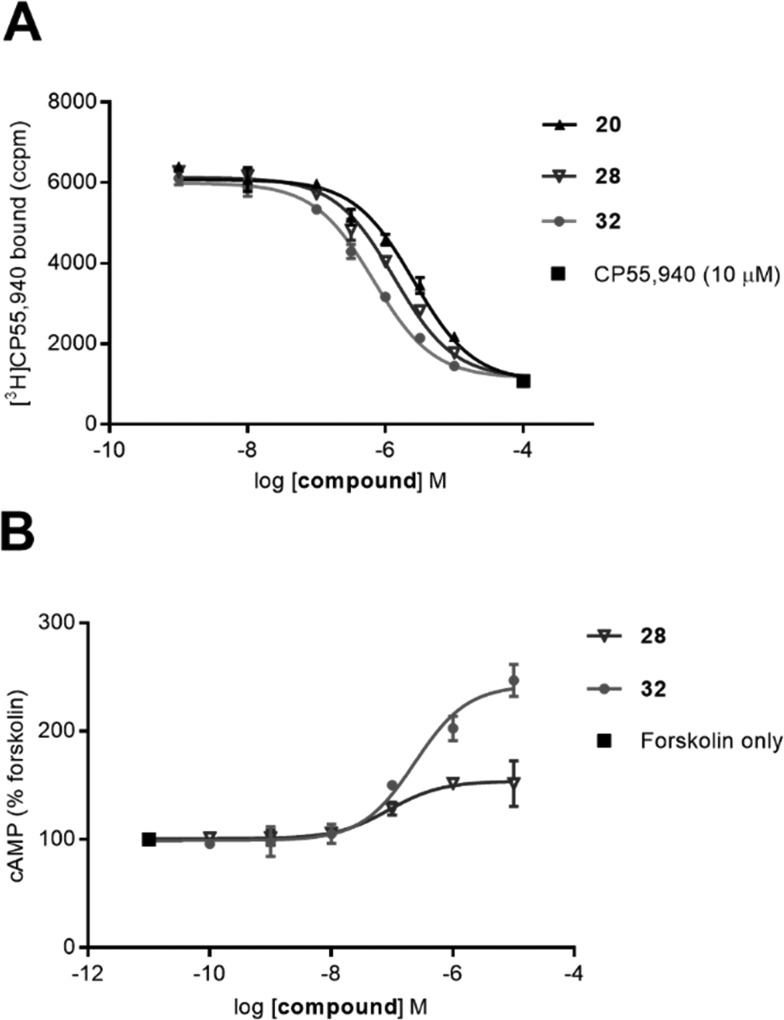
Amongst all the cyclohexyl-linked naphthyridines, the highest CB2 receptor affinity (and the only fluorescent ligand with any appreciable affinity) was measured for fluorescent ligand 32 (pKi = 6.33 ± 0.02 at hCB2 receptor and >21-fold selectivity over CB1 receptor). It is interesting that the larger, BODIPY 630/650-contianing 32 showed approximately 0.3 log unit better affinity for CB2 receptor than the corresponding truncated 28 without the fluorophore (Fig. 2A), implying the BODIPY 630/650 moiety contributed favourably to binding. Similar observations that Class A GPCR fluorescent ligands have higher affinity for the receptor than just the core ligand and/or ligand-linker have also been reported in the literature.33,34 Despite fluorescent ligand 36 differing from 32 only by replacement of the ester in 32 with an amide, 36 showed minimal binding (pKi < 5). This could be due to the lack of flexibility of the amide in comparison to the ester, restricting the movement of 36 and preventing favourable positioning of the linker and fluorophore relative to the naphthyridine core ligand. However, evaluation of shorter precursors to 36 would be required to draw conclusions.
Fig. 2. (A) Competition radioligand binding assay of 20, 28 and 32 at hCB2 receptor, using [3H]CP55 940. Data shown is a single experiment conducted in triplicate with error bars showing ±SD; representative of 3 independent experiments. (B) Concentration response curve of 28 and 32 measuring forskolin (5 μM) stimulated cAMP. Area under the curve values from kinetic data were normalised so that forskolin-only response = 100% and basal (vehicle) response = 0%. Representative data from a single experiment conducted in duplicate, with error bars showing ± SD.
The ability of cyclohexyl-linked naphthyridines with micromolar or nanomolar affinity for CB2 receptor to modulate cAMP was determined in a BRET cAMP assay (Table 2). All tested compounds reduced basal signalling (i.e. increased cAMP levels) consistent with inverse agonism at CB2 receptor. The inverse agonist function observed for these cyclohexyl-linked compounds, all which contain a C6-p-methoxy benzyl substituent, aligns with literature reports of inverse agonism/antagonism for C6-substituted 1,8-naphthyridin-2(1H)-one-3-carboxamides (as also described for compounds in Table 1). The potency of 20, 28, 30 and 32 (Fig. 2B shows 28, 32) was determined using concentration response assays at CB2 receptor (and at CB1 receptor for 32 only). It is interesting that despite 20, 28 and 32 all having between a 1–1.5 log unit weaker affinity for CB2 receptor compared to SR144528 (Table 1), 20, 28, 32 and SR144528 all showed similar pIC50 values at CB2 receptor. Fluorescent ligand 32 appeared to display a greater Emax at CB2 receptor (hCB2 pIC50 = 6.72 ± 0.18, Emax = 210 ± 15.7) compared to SR144528 (hCB2 pIC50 = 6.90 ± 0.08, Emax = 153 ± 3.8) and 32 was much less potent at CB1 receptor (hCB1 pIC50 = 5.26 ± 0.15, Emax = 157 ± 2.1). Compounds analysed for potency in the cAMP BRET assay (Tables 1 and 2) were also screened at 10 μM and 1 μM in the parental HEK cells that lack CB receptors (refer to ESI†). A small effect was observed in the parental HEK cells using fluorescent ligand 32 at 10 μM but not at 1 μM (Table S1†). However, due to the low potency of 32 the 10 μM data point of the concentration response assay at CB2 receptor could not be excluded and therefore the calculated potency of 32 reported at CB2 receptor is only an estimate. In addition, the higher Emax of fluorescent ligand 32 at CB2 receptor compared to SR144528 could be influenced the small non-receptor mediated effect at 10 μM, thus illustrating the importance of wild type controls to verify receptor-mediated responses.
Ligand 32 is one of the highest affinity CB2 receptor selective fluorescent ligands reported in the literature to date. It is not possible to meaningfully compare the affinity of 32 to other fluorescent ligands reported in the literature due to different experimental conditions used to measure binding. For example, the affinity of fluorescent ligand ‘NIR760-Q’ (Kd = 75.5 ± 28.0 nM) was determined in Jurkat cells using a fluorescence saturation binding assay.20 Fluorescent ligand ‘NMP6’ had a reported affinity for hCB2 receptor of Ki = 387 nM using CHO-K1 cells, but with no SEM or Kd value provided for the competing radioligand utilised ([3H]CP55 940).16
Molecular modelling
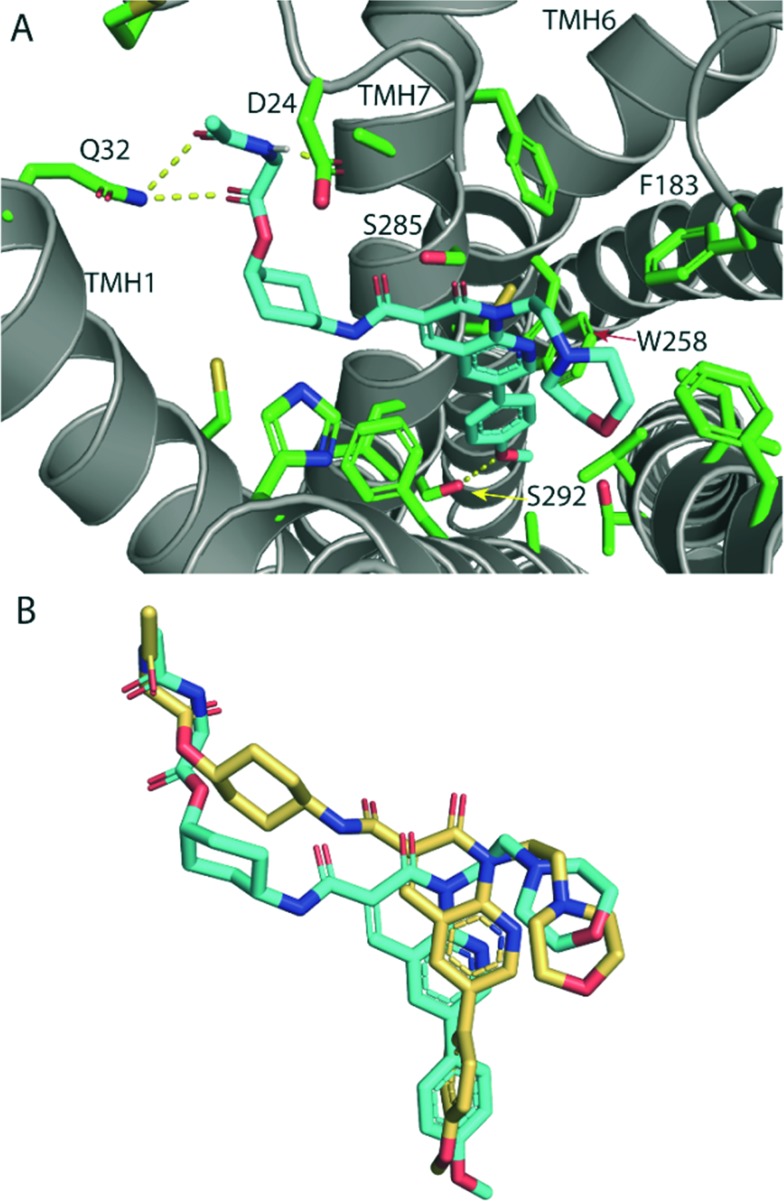
The high CB2 receptor affinity of fluorescent ligand 32 validates the cyclohexyl position as suitable for linker and fluorophore extension. To explore how the ligand-linker of the cyclohexyl-substituted naphthyridines may interact with the receptor, ligand docking studies were performed using a homology model of CB2 receptor, that was generated using the crystal structure of antagonist bound CB1 receptor (PDB:; 5TGZ).36 To improve docking accuracy, the linker conjugate 28 rather than fluorescent ligand 32 was used. The lowest energy and most consistent docking pose showed the glycine linker of 28 exiting the receptor between TMH1 and TMH7 (Fig. 3A), thus potentially illustrating why the cyclohexyl group is a favourable linker position – allowing an exit point for the linker and fluorophore whilst allowing the core ligand to bind deep within the orthosteric pocket. This is especially interesting in light of previously reported molecular dynamics simulations where a potential ligand entry pathway between TMH1 and TMH7 was identified for anandamide and tetrahydrocannabinol binding to CB1 receptor,32 as well as with other fluorescent ligand docking studies at CB2 receptor.17,23 The phenyl group of 28 was positioned deep in the hydrophobic region of the binding pocket, in a similar pose to other previously reported docking studies with this naphthyridine scaffold.25 The methoxy group showed a hydrogen bond with S292, whilst the glycine linker hydrogen bonded with D24 of the amino-terminus and Q32 of TMH1. Favourable placement of hydrogen bond donors and acceptors in the ligand's linker that allow interactions with the polar residues located around the proposed ligand binding pocket entry/exit between TMH1 and TMH7 may be significant for receptor affinity.
Fig. 3. (A) Lowest energy docking pose of glycine-linked cis-28 (cyan carbon atoms) in hCB2 receptor homology model (grey ribbons). Hydrogen bonding (yellow dotted line) is shown between the methoxy phenyl oxygen and S292, the ester carbonyl and the glycine amide with Q32 and between the glycine amide and D24. The glycine linker is directed out of the receptor binding pocket between TMH1 and TMH7. Side chains of selected residues within 4 Å of 28 are shown as sticks (green). (B) Overlay of the lowest energy docking poses of cis-28 (cyan) and the analogous trans-29 (gold) docked into hCB2 receptor homology model (receptor is hidden for clarity), showing that trans-29 is shifted upwards in the binding pocket, which resulted in loss of hydrogen bonding with receptor residues.
The cis isomers of the cyclohexyl-linked series consistently showed higher affinities for CB2 receptor than the trans isomers therefore this was explored further through ligand docking studies. In contrast to cis-28, trans-29 showed an altered pose, in which the whole ligand was shifted upwards in the binding pocket (Fig. 3B). Cyclohexyl-linked trans-29 did not show any hydrogen bonds between the methoxy and S292 or the glycine linker and the residues around the binding pocket exit of TMH1 and TMH7, which may explain the reduced affinity of the trans derivatives.
Cellular imaging
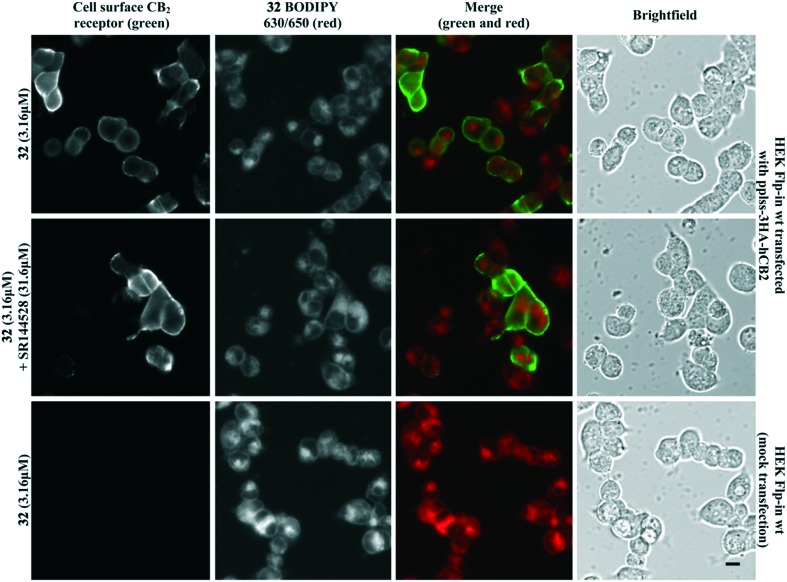
Fluorescent ligand 32 showed promise as an imaging tool due to high affinity for CB2 receptor, selectivity over CB1 receptor, and a similar pIC50 to SR144528. Unfortunately, 32 did not show ideal properties in imaging experiments (Fig. 4). There was little co-localisation of CB2 receptor (as detected by antibody labelling) with 32, pre-incubation of cells with a high concentration of non-fluorescent high affinity ligand SR144528 did not appear to prevent or reduce 32 labelling, and application of 32 to cells not transfected with CB2 receptor still showed a high level of fluorescence. Cell-associated fluorescence arising from 32 was readily detectable within two minutes of incubation, and although fluorescence intensity increased with longer incubation (tested up to 30 minutes) the pattern of staining was equivalent (data not shown). Similarly, incubation with a greater (10 μM) or lesser (1 μM) concentration of 32 influenced the overall intensity of fluorescence but not the subcellular distribution of fluorescence (data not shown). It was concluded that the ester bond of 32 was stable for the duration of the imaging experiments, based on the SAR from 32 and truncated analogues (Table 2, in particular 20 compared to 32) and from reverse phase high performance liquid chromatography (RP-HPLC) experiments that indicated 32 remained stable with no ester bond cleavage (data not shown). Therefore, reasons for the poor CB2 receptor imaging properties of 32 are likely due to high levels of membrane association and non-CB2 receptor associated intracellular accumulation.
Fig. 4. HEK Flp-in wt cells transiently transfected with pplss-3HA-hCB2 or mock transfection, preincubated with SR144528 or vehicle for 30 min, then treated with 32 + SR144528 or vehicle (2 min followed by 3 washes). Cell surface CB2 receptor visualised using mouse anti-HA and Alexa 488-conjugated goat anti-mouse. Scale bar 10 μM. Images representative of n = 3 experiments.
Conclusions
A library of 1,8-naphthyridin-2(1H)-one-3-carboxamides with linker and fluorophore substitution at the N1 and C3-carboxamide cyclohexyl position were designed, synthesised and pharmacologically evaluated. The N1 position was not tolerant of linker and fluorophore attachment, however a high affinity, selective CB2 receptor fluorescent inverse agonist (32) was developed via a C3-carboxamide-cyclohexanol linkage. Despite imaging studies that showed high levels of non-CB2 receptor-specific fluorescence, 32 remains a promising lead for future fluorescent ligand development because of the affinity and selectivity of this large ligand. Molecular modelling and SAR showed the C3-carboxamide-cyclohexyl position is an excellent position for linker and fluorophore attachment.
Experimental
Chemistry
Chemicals were purchased from Sigma Aldrich, Merck or AK Scientific and BODIPY 630/650-X-OSu from Life Technologies. Anhydrous grade solvents were used when a dry atmosphere was required. Unless stated, all reactions were carried out at room temperature (rt) under atmospheric pressure. Thin layer chromatography was carried out on 0.2 mm aluminium-backed silica gel plates 60 F254 and visualised using UV light (λ = 254 nM, 365 nM) and potassium permanganate. Flash column chromatography used 40–63 μm silica. RP-HPLC was carried out on an Agilent 1260 Infinity system with a YMC C8 5 μm (150 × 4.6 mm) or YMC C8 5 μm (150 × 10 mm) column, and mobile phases A: H2O (0.05% TFA) and B: 9 : 1 acetonitrile (ACN) : H2O (0.05% TFA). Analytical RP-HPLC retention times were determined using the method –5% B/A 0–1 min, linear gradient of 5–95% B 1–27 min, 95% B 27–28 min, linear gradient of 95–5% B 28–30 min, 5% B/A 30–34 min. All compounds analysed for biological activity were >95% purity by analytical RP-HPLC UV detection at 254, 380 and 550 nm. All compounds HPLC purified as the TFA salt were neutralised using an Amberlyst A21 ion exchange resin before biological testing. High resolution electrospray ionisation mass spectra (HRMS-ESI) were obtained on a microTOFQ mass spectrometer. Proton and carbon nuclear magnetic resonance (NMR) spectra were obtained on a 400 MHz or a 500 MHz Varian MR spectrometer. Chemical shifts are listed on the δ scale in ppm, spectra are referenced to CDCl3, MeOD-d4 or DMSO-d6 residual solvent. Coupling constants (J) are recorded in Hertz (Hz) with signals assigned as: s, singlet; d, doublet; t, triplet; q, quartet; br, broad; or m, multiplet.
Synthesis of fluorescent ligand 32 is detailed below. Synthesis and characterisation of all other compounds is detailed in the ESI.†
6-(4-Methoxyphenyl)-1-[2-(morpholin-4-yl)ethyl]-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxylic (19)
To a stirred solution of 18 (ref. 25) (6.08 g, 20.5 mmol) in anhydrous DMF (150 mL) was added caesium carbonate (18.67 g, 57.3 mmol). The reaction mixture was stirred for 1 h then 4-(2-chloroethyl)morpholine hydrochloride (7.62 g, 40.1 mmol) was added and the mixture was heated to 50 °C and stirred for 12 h. The DMF was evaporated under reduced pressure and then saturated aq. NaHCO3 (80 mL) was added to the residue, which was then extracted with DCM (3 × 80 mL). The combined organics were washed with H2O (2 × 80 mL), dried over MgSO4, filtered and evaporated under reduced pressure. Precipitation in ACN yielded a 1 : 3 mixture (5.17 g) of the ethyl ester (ethyl 6-bromo-1-[2-(morpholin-4-yl)ethyl]-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxylate) and the carboxylic acid (6-bromo-1-[2-(morpholin-4-yl)ethyl]-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxylic acid) as a yellow solid. This 1 : 3 mixture (4.07 g), 4-methoxyphenylboronic acid (2.03 g, 13.3 mmol), Na2CO3 (2.83 g, 26.7 mmol) were dissolved in H2O (25 mL) and DMF (100 mL). Pd(OAc)2 (23 mg, 0.10 mmol) was added and the reaction heated to 110 °C and stirred for 3 h. After cooling to rt, aq. HCl was added until pH 1–2, H2O (100 mL) added, and extracted with DCM (3 × 100 mL). The combined organics were washed with H2O (2 × 150 mL) and sat. aq. NaCl (150 mL), dried over MgSO4, filtered and evaporated under reduced pressure. The residue was washed with EtOH, filtered and the solid dried under reduced pressure yielding a 1 : 15 mixture (1.99 g) of ethyl 6-(4-methoxyphenyl)-1-[2-(morpholin-4-yl)ethyl]-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxylate and 6-(4-methoxyphenyl)-1-[2-(morpholin-4-yl)ethyl]-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxylic acid, as a yellow solid. A stirred solution of the 1 : 15 mixture (1.9 g) in THF (30 mL) was cooled to 0 °C and 0.2 M LiOH·H2O (49 mL) was added dropwise. The reaction was stirred at 0 °C for 1 h and then quenched with a biphase of 0.2 M aq. HCl/EA (1 : 1 v:v, 200 mL). The aqueous layer was extracted with DCM (10 × 100 mL), dried over MgSO4, filtered and evaporated under reduced pressure to yield 19 (1.66 g, 4.1 mmol, 20% over three steps) as a yellow solid. 1H NMR (400 MHz, CDCl3) δ 8.97 (d, J = 2.4 Hz, 1H, ArH), 8.93 (s, 1H, ArH), 8.22 (d, J = 2.4 Hz, 1H, ArH), 7.58–7.51 (m, 2H, ArH MeOP[combining low line]h[combining low line]), 7.08–7.01 (m, 2H, ArH MeOP[combining low line]h[combining low line]), 4.86 (t, J = 6.8 Hz, 2H, N1–CH2), 3.87 (s, 3H, O–CH3), 3.77–3.64 (m, 4H, O–CH2 morpholino), 2.93–2.83 (m, 2H, N1–CH2CH[combining low line]2[combining low line]), 2.82–2.56 (m, 4H, N–CH2 morpholino). 13C NMR (101 MHz, CDCl3) δ 164.63, 164.56, 160.41, 152.08, 148.49, 144.86, 135.94, 133.49, 128.33, 128.14, 119.28, 115.08, 115.07, 66.93, 56.00, 55.59, 53.96, 39.37. HRMS-ESI calculated for C22H24N3O5 [M + H]+ 410.1710, found m/z 410.1701.
cis-N-(4-Hydroxycyclohexyl)-6-(4-methoxyphenyl)-1-[2-(morpholin-4-yl)ethyl]-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxamide (20)
To a stirred solution of 19 (800 mg, 2.0 mmol) in anhydrous DMF (40 mL) was added DIPEA (1 mL, 5.9 mmol) and HATU (743 mg, 2.0 mmol). The mixture was stirred for 5 min, then cis-4-aminocyclohexanol (889 mg, 5.9 mmol) and DIPEA (1 mL, 5.9 mmol) were added and the mixture stirred for 42 h. The reaction mixture was diluted with NaHCO3 (50 mL) and the aqueous phase extracted with DCM (4 × 50 mL). The combined organics were washed with H2O (2 × 60 mL), dried over MgSO4, filtered and evaporated under reduced pressure. Precipitation in ACN yielded 20 (784 mg, 1.5 mmol, 79%) as a yellow solid. 1H NMR (400 MHz, CDCl3) δ 9.95 (d, J = 7.8 Hz, 1H, NH), 8.91 (s, 1H, ArH), 8.88 (d, J = 2.4 Hz, 1H, ArH), 8.16 (d, J = 2.4 Hz, 1H, ArH), 7.55 (d, J = 8.5 Hz, 2H, ArH MeOPh), 7.04 (d, J = 8.5 Hz, 2H, ArH MeOPh), 4.78 (t, J = 7.1 Hz, 2H, N–CH2), 4.18–4.08 (m, 1H, CH), 3.96–3.88 (m, 1H, CH), 3.87 (s, 3H, O–CH3), 3.69 (t, J = 4.6 Hz, 4H, O–CH2 morpholino), 2.74 (t, J = 7.1 Hz, 2H, N–CH2), 2.63 (t, J = 4.5 Hz, 4H, N–CH2 morpholino), 1.94–1.68 (m, 8H, CH2). 13C NMR (101 MHz, CDCl3) δ 162.56, 162.15, 160.08, 150.57, 148.68, 142.09, 135.40, 132.27, 128.89, 128.24, 123.35, 114.93, 114.77, 67.23, 67.16, 56.04, 55.57, 54.04, 46.58, 39.09, 31.50, 27.64. HRMS-ESI calculated for C28H35N4O5 [M + H]+ 507.2602, found m/z 507.2552. Analytical RP-HPLC Rt = 14.13 min.
cis-4-[6-(4-Methoxyphenyl)-1-[2-(morpholin-4-yl)ethyl]-2-oxo-1,2-dihydro-1,8-naphthyridine-3-amido]cyclohexyl 2-{[(tert-butoxy)carbonyl]amino}acetate (23)
A stirred solution of Boc-glycine (17 mg, 99 μmol) and TFFH (26 mg, 99 μmol) in anhydrous DCM (3 mL) was cooled to 0 °C and Et3N (69 μL, 0.49 mmol) was added. The mixture was warmed to rt and stirred for 30 min and then 20 (50 mg, 99 μmol) and DMAP (1.2 mg, 9.9 μmol) were added. The mixture was stirred for 14 h and then evaporated under reduced pressure. The residue was taken up in EA (3 mL) and washed with H2O (3 × 3 mL) and sat. aq. NaCl (3 mL), dried over MgSO4, filtered and evaporated under reduced pressure. The residue was purified by flash silica column chromatography (100% EA) to yield 23 (7.4 mg, 11.1 μmol, 19%) as a yellow solid. 1H NMR (400 MHz, CDCl3) δ 9.89 (d, J = 7.7 Hz, 1H, NH), 8.95–8.86 (m, 2H, ArH), 8.17 (d, J = 2.4 Hz, 1H, ArH), 7.62–7.47 (m, 2H, ArH MeOP[combining low line]h[combining low line]), 7.10–6.99 (m, 2H, ArH MeOP[combining low line]h[combining low line]), 5.07–4.99 (m, 2H, NH, CH), 4.80 (t, J = 7.1 Hz, 2H, N1–CH[combining low line]2[combining low line]), 4.20–4.07 (m, 1H, CH), 3.93 (d, J = 5.7 Hz, 2H, CH2), 3.88 (s, 3H, O–CH3), 3.76–3.63 (m, 4H, N–CH2 morpholino), 2.76 (t, J = 7.1 Hz, 2H, N1–CH2CH[combining low line]2[combining low line]), 2.70–2.58 (m, 4H, O–CH2 morpholino), 1.98–1.71 (m, 8H, CH2), 1.47 (s, 9H, tBu CH3). 13C NMR (101 MHz, CDCl3) δ 169.94, 162.61, 162.24, 160.13, 150.68, 148.69, 142.25, 135.45, 132.38, 128.87, 128.27, 123.24, 114.97, 114.79, 80.11, 70.85, 67.18, 56.06, 55.59, 54.07, 46.82, 42.82, 39.17, 28.53, 28.49, 27.74 (one quaternary carbon not observed). HRMS-ESI calculated for C35H46N5O8 [M + H]+ 664.3341, found 664.3343.
cis-4-[6-(4-Methoxyphenyl)-1-[2-(morpholin-4-yl)ethyl]-2-oxo-1,2-dihydro-1,8-naphthyridine-3-amido]cyclohexyl 2-[6-(2-{4-[(E)-2-[2,2-difluoro-4-(thiophen-2-yl)-1λ4,3-diaza-2λ4-boratricyclo[7.3.0.03,7]dodeca-1(12),4,6,8,10-pentaen-12-yl]ethenyl]phenoxy}acetamido)hexanamido]acetate (32)
A solution of 23 (7.0 mg, 10.4 μmol) was dissolved in DCM (0.8 mL) and TFA (0.2 mL) was added. After 1 h stirring, the reaction mixture was evaporated under N2 stream, followed by reduced pressure. The crude was purified by semi-preparative RP-HPLC to yield the TFA salt cis-4-[6-(4-methoxyphenyl)-1-[2-(morpholin-4-yl)ethyl]-2-oxo-1,2-dihydro-1,8-naphthyridine-3-amido]cyclohexyl 2-aminoacetate (4.3 mg, 4.75 μmol, 45%) as a yellow solid. Analytical RP-HPLC Rt = 12.83 min. To a solution this TFA salt (2.8 mg, 3.1 μmol) in anhydrous DMF (500 μL), was added a solution of DIPEA (1.7 μL, 10 μmol) in DMF (128 μL), then BODIPY 630/650-X-OSu (1.25 mg, 1.89 μmol) in DMF (100 μL). The mixture was swirled, left for 12 h, then evaporated under reduced pressure. The crude product was purified by semi-preparative RP-HPLC and passed through an Amberlyst A21 ion exchange resin to yield 32 (2.03 mg, 1.83 μmol, 97%) as a bright blue solid. HRMS-ESI calculated for C59H64BF2N8O9S [M + H]+ 1109.4582, found m/z 1109.4556. Analytical RP-HPLC Rt = 20.47 min.
Pharmacology
Radioligand binding assays
Cell membranes were isolated from HEK (human embryonic kidney) 293 cells (ATCC, Manassas, VA, USA; #CRL-1573) stably transfected with either hCB1 or hCB2 receptors37,38 and binding assays carried out as previously described with minor modifications.39 Briefly, membranes were resuspended in binding buffer (50 mM HEPES, 1 mM MgCl2, 1 mM CaCl2, 0.2% [w/v] fatty acid free bovine serum albumin [FAF BSA; MP Biomedicals, Auckland, New Zealand], pH 7.4) to give a final assay concentration of 10 μg per well (Table 1) or 7.5 μg per well (Table 2) for hCB2 or 7.5 μg per well (Table 1) or 5 μg per well (Table 2) for hCB1. Compounds (stocks at 10 mM in DMSO, except CP55 940 (Cayman Chemical, Michigan, USA) which was 10 mM in EtOH) were serially diluted using binding buffer containing the requisite amount of EtOH and DMSO to maintain equivalent vehicle levels throughout the dilution series and between all compounds. For vehicle control points, binding buffer containing matched concentrations of EtOH and DMSO was used in place of test ligands. [3H]CP55 940 (PerkinElmer, Waltham, MA, USA) was used at a final concentration of 2.5 nM (Table 1) or 1 nM (Table 2). V-Bottom plates containing hCB2 or hCB1 membranes, [3H]CP55 940 and ligand (or CP55 940 or vehicle) were incubated at 30 °C for 1 h prior to harvesting and washing on filter plates (treated with PEI to minimise non-specific binding of the ligand), drying, incubation with scintillation fluid and detection. Binding experiments were performed a minimum of three independent times in technical triplicate. Data was analysed with GraphPad Prism 7 (GraphPad Software, Inc., San Diego, CA, USA) and competition binding curves fit by nonlinear regression using one site competition binding. Dissociation constants (pKi) of compounds were determined using [3H]CP55 940 Kd = 2 nM (hCB1) or 3 nM (hCB2), and are expressed as mean ± standard error of the mean (SEM). In cases where less than 50% displacement of [3H]CP55 940 was observed with 10 μM compound, affinity of the compound was deemed too low to be able to generate an accurate competition binding curve. Therefore, a one sample t-test (P < 0.05) was used to determine if there was significant difference between displacement in the absence (vehicle normalised to 0%) and presence of compound (with CP55 940 normalised to 100%); if so, the ligand was determined to have a pKi <5, otherwise it was determined to show no significant binding.
cAMP assays
A BRET assay was carried out to measure forskolin-stimulated cellular cAMP in the appropriate HEK 293 cells transfected with a plasmid that encodes for the cAMP biosensor YFP-Epac-RLuc (CAMYEL) as previously described.38 Briefly, one or two days prior to transfection, HEK 3HA-hCB1 pEF4A (same as binding), HEK-Flp HA-hCB2 pcDNA5/FRT (generated as previously described40), HEK wild type (WT) (ATCC, #CRL-1573) or HEK-Flp WT (Invitrogen, Carlsbad, CA, USA) cells, were seeded in 10 cm dishes.
Cells were transfected with 5 μg of pcDNA3L-His-CAMYEL plasmid (ATCC) using 30 μg of linear PEI (molecular weight 25 kDa; Polysciences, Warrington, PA, USA) in 150 mM NaCl. After 24 h, transfected cells were plated in poly-d-lysine (PDL) (0.05 mg mL–1 in PBS; Sigma-Aldrich, St Louis, MO, USA) treated 96-well solid white flat bottom polystyrene TC-treated microplates (Corning) at a density of 60–80 000 cells per well in Dulbecco's Modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS; New Zealand-origin, Moregate Biotech, Brisbane, Australia). After 16 h, cells were serum-starved in Hank's balanced salt solution (HBSS, Thermo Fisher Scientific, Waltam, MA, USA) containing 1 mg mL–1 FAF BSA, pH 7.4 for 30 min. Cells were then treated with 7.5 μM coelenterazine-h (Nanolight Technology, Pinetop, AZ, USA) for 5 min, followed by addition of ligand or matched vehicle in HBSS plus 1 mg mL–1 FAF BSA and 5 μM forskolin (Cayman Chemical, Michigan, USA). A LUMIstar plate reader (BMG Labtech, Ortenberg, German) was used to immediately measure emission signals at 37 °C following ligand addition, which were simultaneously detected at 460/25 nM (Renilla luciferase, RLuc) and 535/25 nM (yellow fluorescent protein, YFP). Assays were carried out a minimum of three times (except where stated) in duplicate. Data analysis was performed using GraphPad Prism, with sigmoidal concentration response curves fit by nonlinear regression using values normalised to the vehicle (0%) or forskolin (100%) values for individual experiments. A t-test (P < 0.05) was used to determine if there was a significant difference in response for compounds at 10 μM in WT HEK cells to determine receptor mediated signalling.
Molecular modelling
The CB2 receptor homology model was generated using MODELLER 9.15 (ref. 41) using the structure of the antagonist-bound CB1 receptor (PDB ID: ; 5TGZ) as a template, based on a modified sequence alignment between hCB1 and hCB2 receptors from the T-Coffee server.42 Three dimensional models of ligands were generated using Avogadro 1.2 (ref. 43) and minimised using the universal force field. Ligand docking was performed using GOLD v5.5 (CCDC Software)44 centred on Ser285 extending for a distance of up to 15 Å and visualised in PyMOL (The PyMOL Molecular Graphics System, Version 1.8.6.0 Schrödinger, LLC.).
Cellular imaging
HEK Flp-in wt cells were seeded at a density of 30 000 cells per well in PDL treated Nunc™ 96-well black optical-bottom plates (Thermo Fisher Scientific). Approximately 24 h after seeding, cells were transfected with 125 ng per well of pplss-3HA-hCB2 or empty pcDNA 3.1 (for mock transfected cells) using Lipofectamine® 2000 Transfection Reagent (0.5 μL per well). All drugs and reagents for imaging assays were prepared in HBSS supplemented with 1 mg mL–1 BSA. After expressing for 18–24 h, medium was aspirated and cells incubated with mouse monoclonal anti-HA.11 (Clone 16B12, BioLegend, San Diego, CA, USA) diluted 1 : 500, for 30 min at room temperature. Cells were then briefly washed and co-incubated with Alexa Fluor® 488-conjugated goat anti-mouse secondary antibody (Thermo Fisher Scientific) diluted 1 : 300, and 31.6 μM SR144528 (kindly gifted by Roche; Basel, Switzerland) or Vehicle for 30 min at room temperature. Following a brief wash, cells were then treated with 3.16 μM 32 and SR144528 or Vehicle for 2 min, followed by 3 washes. Cells were then imaged using an ImageXpress® Micro XLS Widefield Microscope (Molecular Devices, Sunnyvale, CA, USA) (20× objective).
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by a University of Otago Research Grant, the School of Pharmacy, University of Otago and the Maurice Wilkins Centre for Molecular Biodiscovery. A. C. was supported by a University of Otago Doctoral Scholarship. We thank Christa Macdonald for her technical assistance with pharmacological assay preparations.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c8md00448j
References
- Galiegue S., Mary S., Marchand J., Dussossoy D., Carriere D., Carayon P., Bouaboula M., Shire D., Fur G. L., Casellas P. Eur. J. Biochem. 1995;232:54–61. doi: 10.1111/j.1432-1033.1995.tb20780.x. [DOI] [PubMed] [Google Scholar]
- Chen D.-J., Gao M., Gao F.-F., Su Q.-X., Wu J. Nature. 2017;38:312–316. doi: 10.1038/aps.2016.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katona I., Freund T. F. Annu. Rev. Neurosci. 2012;35:529–558. doi: 10.1146/annurev-neuro-062111-150420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrini M. E., Hong S., Stierle A., Stierle D., Stella N., Roberts K., Jaffar Z. Allergy. 2017;72:937–947. doi: 10.1111/all.13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte C., Blanchet M.-R., Laviolette M., Flamand N. Cell. Mol. Life Sci. 2016;73:4449–4470. doi: 10.1007/s00018-016-2300-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassano T., Calcagnini S., Pace L., De Marco F., Romano A., Gaetani S. Front. Neurosci. 2017;11:30. doi: 10.3389/fnins.2017.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Thoresen L., Jung J.-K., Zhu X., Thatte J., Solomon M., Gaidarov I., Unett D. J., Yoon W. H., Barden J., Sadeque A., Usmani A., Chen C., Semple G., Grottick A. J., Al-Shamma H., Christopher R., Jones R. M. ACS Med. Chem. Lett. 2017;8:1309–1313. doi: 10.1021/acsmedchemlett.7b00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone F., Mach F., Vuilleumier N., Montecucco F. Curr. Med. Chem. 2014;21:4046–4058. doi: 10.2174/0929867321666140915141332. [DOI] [PubMed] [Google Scholar]
- Fraguas-Sánchez A. I., Martín-Sabroso C., Torres-Suárez A. I. Br. J. Pharmacol. 2018;175:2566–2580. doi: 10.1111/bph.14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghazadeh Tabrizi M., Baraldi P. G., Borea P. A., Varani K. Chem. Rev. 2016;116:519–560. doi: 10.1021/acs.chemrev.5b00411. [DOI] [PubMed] [Google Scholar]
- Vernall A. J., Hill S. J., Kellam B. Br. J. Pharmacol. 2014;171:1073–1084. doi: 10.1111/bph.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam R., Gondin A. B., Canals M., Kellam B., Briddon S. J., Graham B., Scammells P. J. J. Med. Chem. 2018;61:1316–1329. doi: 10.1021/acs.jmedchem.7b01811. [DOI] [PubMed] [Google Scholar]
- Tabor A., Möller D., Hübner H., Kornhuber J., Gmeiner P. Sci. Rep. 2017:1–11. doi: 10.1038/s41598-017-11436-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddart L. A., Vernall A. J., Briddon S. J., Kellam B., Hill S. J. Neuropharmacology. 2015;98:68–77. doi: 10.1016/j.neuropharm.2015.04.013. [DOI] [PubMed] [Google Scholar]
- Stoddart L. A., Kilpatrick L. E., Hill S. J. Trends Pharmacol. Sci. 2017:1–12. doi: 10.1016/j.tips.2017.10.006. [DOI] [PubMed] [Google Scholar]
- Petrov R. R., Ferrini M. E., Jaffar Z., Thompson C.C. M.M., Roberts K., Diaz P. Bioorg. Med. Chem. Lett. 2011;21:5859–5862. doi: 10.1016/j.bmcl.2011.07.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Couce L., Martín-Fontecha M., Palomares Ó., Mestre L., Cordomí A., Hernangomez M., Palma S., Pardo L., Guaza C., López-Rodríguez M. L., Ortega-Gutiérrez S. Angew. Chem., Int. Ed. 2012;51:6896–6899. doi: 10.1002/anie.201200467. [DOI] [PubMed] [Google Scholar]
- Sexton M., Woodruff G., Horne E. A., Lin Y. H., Muccioli G. G., Bai M., Stern E., Bornhop D. J., Stella N. Chem. Biol. 2011;18:563–568. doi: 10.1016/j.chembiol.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Shao P., Bai M. Bioconjugate Chem. 2013;24:1907–1916. doi: 10.1021/bc400328m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Shao P., Zhang S., Ling X., Bai M. J. Biomed. Opt. 2014;19:076016–076018. doi: 10.1117/1.JBO.19.7.076016. [DOI] [PubMed] [Google Scholar]
- Ling X., Zhang S., Shao P., Li W., Yang L., Ding Y., Xu C. Biomaterials. 2015;57:169–178. doi: 10.1016/j.biomaterials.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A. S., Doughty S. W., Kendall D. A., Kellam B. Bioorg. Med. Chem. Lett. 2005;15:3758–3762. doi: 10.1016/j.bmcl.2005.05.049. [DOI] [PubMed] [Google Scholar]
- Cooper A. G., MacDonald C., Glass M., Hook S., Tyndall J. D. A., Vernall A. J. Eur. J. Med. Chem. 2018;145:770–789. doi: 10.1016/j.ejmech.2017.11.076. [DOI] [PubMed] [Google Scholar]
- Soethoudt M., Stolze S. C., Westphal M. V., van Stralen L., Martella A., van Rooden E. J., Guba W., Varga Z. V., Deng H., van Kasteren S. I., Grether U., Ijzerman A. P., Pacher P., Carreira E. M., Overkleeft H. S., Ioan-Facsinay A., Heitman L. H., Van der Stelt M. J. Am. Chem. Soc. 2018;140:6067–6075. doi: 10.1021/jacs.7b11281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi V., Hurst D. P., Shore D. M., Bertini S., Ehrmann B. M., Allarà M., Lawrence L., Ligresti A., Minutolo F., Saccomanni G., Sharir H., Macchia M., Di Marzo V., Abood M. E., Reggio P. H., Manera C. J. Med. Chem. 2014;57:8777–8791. doi: 10.1021/jm500807e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manera C., Benetti V., Castelli M. P., Cavallini T., Lazzarotti S., Pibiri F., Saccomanni G., Tuccinardi T., Vannacci A., Martinelli A., Ferrarini P. L. J. Med. Chem. 2006;49:5947–5957. doi: 10.1021/jm0603466. [DOI] [PubMed] [Google Scholar]
- Manera C., Saccomanni G., Adinolfi B., Benetti V., Ligresti A., Cascio M. G., Tuccinardi T., Lucchesi V., Martinelli A., Nieri P., Masini E., Di Marzo V., Ferrarini P. L. J. Med. Chem. 2009;52:3644–3651. doi: 10.1021/jm801563d. [DOI] [PubMed] [Google Scholar]
- Manera C., Saccomanni G., Malfitano A. M., Bertini S., Castelli F., Laezza C., Ligresti A., Lucchesi V., Tuccinardi T., Rizzolio F., Bifulco M., Di Marzo V., Giordano A., Macchia M., Martinelli A. Eur. J. Med. Chem. 2012;52:284–294. doi: 10.1016/j.ejmech.2012.03.031. [DOI] [PubMed] [Google Scholar]
- Manera C., Malfitano A. M., Parkkari T., Lucchesi V., Carpi S., Fogli S., Bertini S., Laezza C., Ligresti A., Saccomanni G., Savinainen J. R., Ciaglia E., Pisanti S., Gazzerro P., Di Marzo V., Nieri P., Macchia M., Bifulco M. Eur. J. Med. Chem. 2015;97:10–18. doi: 10.1016/j.ejmech.2015.04.034. [DOI] [PubMed] [Google Scholar]
- Kimura T., Cheng K., Rice K. C., Gawrisch K. Biophys. J. 2009;96:4916–4924. doi: 10.1016/j.bpj.2009.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst D. P., Grossfield A., Lynch D. L., Feller S., Romo T. D., Gawrisch K., Pitman M. C., Reggio P. H. J. Biol. Chem. 2010;285:17954–17964. doi: 10.1074/jbc.M109.041590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakowiecki J., Filipek S. J. Chem. Inf. Model. 2016;56:2457–2466. doi: 10.1021/acs.jcim.6b00499. [DOI] [PubMed] [Google Scholar]
- Vernall A. J., Stoddart L. A., Briddon S. J., Ng H. W., Laughton C. A., Doughty S. W., Hill S. J., Kellam B. Org. Biomol. Chem. 2013;11:5673–5682. doi: 10.1039/c3ob41221k. [DOI] [PubMed] [Google Scholar]
- Vernall A. J., Stoddart L. A., Briddon S. J., Hill S. J., Kellam B. J. Med. Chem. 2012;55:1771–1782. doi: 10.1021/jm201722y. [DOI] [PubMed] [Google Scholar]
- Ni Y., Wu J. Org. Biomol. Chem. 2014;12:3774–3791. doi: 10.1039/c3ob42554a. [DOI] [PubMed] [Google Scholar]
- Hua T., Vemuri K., Pu M., Qu L., Han G. W., Wu Y., Zhao S., Shui W., Li S., Korde A., Laprairie R. B., Stahl E. L., Ho J.-H., Zvonok N., Zhou H., Kufareva I., Wu B., Zhao Q., Hanson M. A., Bohn L. M., Makriyannis A., Stevens R. C., Liu Z.-J. Cell. 2016;167:750–755.e14. doi: 10.1016/j.cell.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsey N. L., Goodfellow C. E., Dragunow M., Glass M. Biochim. Biophys. Acta, Mol. Cell Res. 2011;1813:1554–1560. doi: 10.1016/j.bbamcr.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Cawston E. E., Redmond W. J., Breen C. M., Grimsey N. L., Connor M., Glass M. Br. J. Pharmacol. 2013;170:893–907. doi: 10.1111/bph.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPartland J. M., MacDonald C., Young M., Grant P. S., Furkert D. P., Glass M. Cannabis Cannabinoid Res. 2017;2:87–95. doi: 10.1089/can.2016.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soethoudt M., Grether U., Fingerle J. U. R., Grim T. W., Fezza F., de Petrocellis L., Ullmer C., Usler B. R. A., Perret C., van Gils N., Finlay D., MacDonald C., Chicca A., Gens M. D., Stuart J., de Vries H., Mastrangelo N., Xia L., Alachouzos G., Baggelaar M. P., Martella A., Mock E. D., Deng H., Heitman L. H., Connor M., Di Marzo V., Gertsch J. U. R., Lichtman A. H., Maccarrone M., Pacher P., Glass M., Van der Stelt M. Nat. Commun. 2016;8:1–14. doi: 10.1038/ncomms13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A., Blundell T. L. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Notredame C., Higgins D. G., Heringa J. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Hanwell M. D., Curtis D. E., Lonie D. C., Vandermeersch T., Zurek E., Hutchison G. R. J. Cheminf. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G., Willett P., Glen R. C., Leach A. R., Taylor R. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.