Abstract
Phosphoproteomics studies have reported phosphorylation at multiple sites within collagen, raising the possibility that these post-translational modifications regulate the physical or biological properties of collagen. In this study, molecular dynamics simulations and experimental studies were carried out on model peptides to establish foundational principles of phosphorylation of Ser residues in collagen. A (Gly-Xaa-Yaa)11 peptide was designed to include a Ser-containing sequence from type I collagen that was reported to be phosphorylated. The physiological kinase involved in collagen phosphorylation is not known. In vitro studies showed that a model kinase ERK1 (extracellular signal-regulated protein kinase 1) would phosphorylate Ser within the consensus sequence if the collagen-like peptide is in the denatured state but not in the triple-helical state. The peptide was not a substrate for FAM20C, a kinase present in the secretory pathway, which has been shown to phosphorylate many extracellular matrix proteins. The unfolded single chain (Gly-Xaa-Yaa)11 peptide containing phosphoSer was able to refold to form a stable triple helix but at a reduced folding rate and with a small decrease in thermal stability relative to the nonphosphorylated peptide at neutral pH. These biophysical studies on model peptides provide a basis for investigations into the physiological consequences of collagen phosphorylation and the application of phosphorylation to regulate the properties of collagen biomaterials.
Introduction
Collagen, an abundant protein in the extracellular matrix (ECM), is distinguished by the presence of a characteristic triple-helix structure, consisting of three tightly packed supercoiled polyproline-II-like chains. Type I is the most widespread type of collagen, forming fibrils in tendon, bone, skin, and many other tissues. The collagen triple-helix structure requires a high proportion of the imino acids Pro and hydroxyproline (abbreviated as Hyp or O) and the presence of Gly as every third residue, resulting in a (Gly-Xaa-Yaa)n repeating sequence (1, 2). While the collagen chain is unfolded in the endoplasmic reticulum (ER) during biosynthesis, the enzyme Pro 4-hydroxylase hydroxylates Pro residues in the Yaa position, and the resulting Hyp-4 residues stabilize the triple helix, provide intermolecular interactions within collagen fibrils, and are involved in collagen recognition (2, 3, 4, 5). Other post-translational modifications of unfolded collagen include the formation of hydroxylysine, glycosylation of hydroxylysine, and formation of Hyp-3 (6). After secretion, collagen molecules self-associate to form fibrils in the ECM and undergo covalent cross-linking mediated by the enzyme lysyl oxidase (7). Recently, another post-translational modification in collagen—the phosphorylation of Ser, Thr, and Tyr residues—has been firmly established by diverse phosphoproteomics studies (8). Phosphorylated Ser residues have been reported in type I collagen in tissues, cell cultures, and in a recombinant yeast expression system (8, 9, 10, 11, 12, 13, 14, 15). It has been suggested that phosphorylation/dephosphorylation may play a regulatory role in collagen processing, stability, assembly, degradation, or binding (8, 16). Further characterization of the structural basis of collagen phosphorylation and the effects of such phosphorylation on triple-helix properties will help clarify the biological role of this post-translational modification.
The kinases involved in collagen phosphorylation and the location of the phosphorylation process remain unknown. The unusual repeating (Gly-Xaa-Yaa)n amino acid sequence of collagen and its unique tightly packed triple-helical structure place structural constraints that could influence its ability to serve as a substrate for a kinase. Phosphorylation may be another post-translational modification that takes place in the ER while the collagen chain is still unfolded. Recently, a kinase in the secretory pathway, Fam20C, with a preference for S-X-E motifs, was found to be responsible for the phosphorylation of more than 100 secreted proteins (17). These results identify Fam20C as a major secretory pathway protein kinase, but thus far, collagen has not been reported as a substrate. It has also been suggested that collagen might be phosphorylated by kinases in the ECM after the triple-helical molecules have been secreted and formed fibrils because there are reports of kinases that are secreted in exosomes or present on the external surface of cell membranes (8).
Phosphorylated residues have been detected in collagen largely through phosphoproteomics studies (10, 11, 12, 13, 14, 15), and it is not clear if these residues are present in denatured and/or native triple-helical collagen. A recent study using host-guest collagen-like peptides showed that phosphoSer (abbreviated as pSer or J) and phosphoThr (pThr) in the Xaa position of the Gly-Xaa-Yaa triplet form a typical triple helix with an increased stability compared with nonphosphorylated control peptides (16). These studies confirm that phosphorylated residues do not interfere with the formation of a stable triple helix, and the authors proposed that phosphorylation may be a mechanism of controlling collagen triple-helix stability.
Here, we report studies on a peptide that models a Ser-containing sequence 544PGSP547 reported to be phosphorylated within type I collagen. The ability of ERK1 (extracellular signal-regulated protein kinase 1), a kinase with the appropriate consensus sequence (PGSP) to phosphorylate the Ser in this peptide, was investigated and indicated that the consensus sequence is a kinase substrate when the peptide is denatured, but not when it is in a triple-helical form. The effect of phosphorylation on triple-helix conformation, stability, and refolding were also examined for this peptide model: pSer resulted in a slight decrease in triple-helix stability and a somewhat slower rate of triple-helix folding at neutral pH. The experimental work was complemented by molecular dynamics (MD) simulation studies to clarify the molecular interactions involved.
Materials and Methods
General information
Phosphate-buffered saline (pH 7.4) was purchased from Thermo Fisher Scientific (Waltham, MA). ERK1 Kinase Enzyme System, ADP-Glo Kinase Assay, and the Kinase-Glo Max Luminescent Kinase Assay were purchased from Promega (Madison, WI). All other chemicals were purchased from Sigma-Aldrich (Natick, MA).
Peptides
All peptides were synthesized using FastMoc Chemistry by the Tufts University Core Facility (Medford, MA). pSer was incorporated during the synthesis process in peptide pSer546 in place of serine. Peptides were purified by HPLC, and identity was confirmed by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF).
Sample preparation
The synthetic peptides were dissolved in phosphate-buffered saline (pH 7.4) or 0.1 M acetic acid (pH 2.9). Peptide concentrations were determined by an ultraviolet-visible spectrophotometer (Aviv Biomedical, Lakewood, NJ) with an extinction coefficient of εTyrosine 280 = 1280 M−1cm−1. Samples were diluted to 97.0 μM for CD spectrometry and 600 μM for in vitro kinase reaction.
Structural characterization
Circular dichroism (CD) spectra of peptides were carried out on an Aviv Model 430 CD spectrometer (Aviv Biomedical). Wavelength scans were collected from 260 to 190 nm at different temperatures (0, 30, 34, 37, and 70°C). Melting curves were obtained by measuring the CD signal at 225 nm from 0 to 70°C with an average heating rate of 0.1°C min–1 (3). Kinetic analysis of collagen refolding was performed by denaturing the peptides at 70°C for 20 min, followed by injection of the peptides into a cuvette equilibrated at the folding temperature (0, 30, 34, and 37°C). The signal at 225 nm was monitored with a time constant of 2 s and time interval of 10 s. The halftime of refolding t1/2 was defined as the time at which the fraction folded reached 0.5 (18).
In vitro phosphorylation assay
In vitro phosphorylation of peptide Ser546 using ERK1 kinase was performed based on the ERK1 Kinase Assay Protocol (Promega) with the following modifications. All ERK1 kinase assays were carried out in reaction solution containing 40 mM Tris (pH 7.5), 20 mM MgCl2, 0.1 mg mL–1 bovine serum albumin, 50 μM dithiothreitol, and 1% dimethyl sulfoxide. Various levels of substrates (0–600 μM), ATP concentrations (100–200 μM), ERK1 kinase concentrations (0–550 ng), incubation time (20 min to overnight), and reaction temperatures (30–37°C) were tested for the optimal reaction conditions. Both the native Ser546 peptide taken directly from 4°C storage and the denatured Ser546 peptide after heating to 70°C for 20 min were used as substrates. The reaction was initiated by the addition of an appropriate amount of ERK1 and terminated by the addition of EDTA to 40 mM.
Substrate kinetics was determined according to the ADP-Glo Kinase Assay Protocol (Promega), using twofold serial dilutions of substrates, fixed amounts of ATP (100 μM), and ERK1 kinase (20 ng). Similar experimental conditions were used for titration of ERK1 kinase, in the presence of increasing concentration of ERK1 at fixed concentrations of ATP (100 μM) and Ser546 peptide (40 μM). Phosphorylation efficiency was assessed by determination of the amount of ADP generated in the reaction relative to the total amount of ATP. Km and EC50 were calculated by using Prism software.
Fam20C kinase activity assay was carried out using the Kinase-Glo Max Luminescent Kinase Assay Protocol, using 100 μM concentration of peptide substrates and fixed amounts of ATP (1 μM). Fam20C kinase was purified as reported previously, and a total of 100 ng of kinase was used per kinase assay reaction, and KKIEKFQSEEQQQ (β28–40) peptide substrate was used as a positive control (19). Kinase reaction was incubated at 30°C for 30–60 min, and luminescence counting was carried out in a Tecan M200 plate reader (Männedorf, Switzerland). Data were reported as the inverse of the signal relative to control.
Mass spectrometry
The synthetic peptides and the phosphorylation products were analyzed with a MALDI-TOF Microflex LT instrument (Bruker, Billerica, MA) in positive ion mode. Samples were mixed with MALDI matrix (a saturated sinapinic acid solution in 50% (v/v) acetonitrile and 0.3% (v/v) trifluoroacetic acid) in a 1:4 ratio, and 1 μL of the peptide-containing matrix was pipetted onto a MALDI target plate and allowed to dry, as previously described (20). It was noted that each peptide produced multiple peaks, one at the expected mass of the peptide as well as a second peak at a higher molecular weight that corresponded to the mass of the peptide plus an associated cation (either sodium or potassium).
MD simulations of collagen-like triple helices
To investigate the effects of serine phosphorylation on the triple-helical structure of type I collagen, MD simulations were performed for the model α1(I)3 homotrimer collagen peptide and its phosphorylated versions. The sequences of three triplets (residues 542–550, underlined below) were retrieved from the UniProt database from the human α1(I) chain (P02452) (21) and flanked by GPO stabilizing triplets at both ends. The serine at position 543 was changed to alanine. The wild-type (WT) sequence used in the simulations was POGPOGPOGPOGAPGSPGPDGPOGPOGPOGPOG.
The initial triple-helical structures for the WT collagen peptides were built using the triple-helical collagen building script (22). The α1(I) chains were capped with an acetyl group and NH2 group in the N- and C-termini, respectively. In addition to the WT system, phosphorylated collagen model peptides were also simulated by using the UCSF Chimera package (23) to make atom additions. The serine at position 546 (bolded above) was phosphorylated, and both acidic and neutral conditions were simulated.
All simulations were performed with GROMACS 4.6.7 (24) using the AMBER99SB force field (25, 26) and TIP3P water model (27). For each collagen triple helix, the starting structure was energy minimized in vacuum using the steepest descent algorithm for a maximum of 2000 steps. All heavy atoms of the triple helix were position restrained with a force constant of 1000 kJ/mol/nm2. The vacuum-minimized structure was then solvated in a rectangular water box such that no collagen atom was closer than 1.5 nm to the edges of the box; the long axis of the collagen triple helix was parallel to the z axis of the box. Na+ ions were added to neutralize the system. The system was then further energy minimized using the steepest descent algorithm for a maximum of 5000 steps. In this step, the periodic boundary condition was applied, and a cutoff value of 0.8 nm was used for both the short-range Coulombic and the van der Waals interactions. The long-range Coulombic interaction was treated with the particle mesh Ewald algorithm (28). The Fourier spacing for particle mesh Ewald was set to 0.12 nm, and the cubic interpolation was also implemented. Long-range dispersion correction for energy was applied to account for the truncation of the van der Waals interactions. During the minimization, the heavy atoms of the triple helix remained restrained.
The well-minimized system was then subjected to a two-stage equilibration process. In the first stage of equilibration, the system was annealed from 5 to 300 K over 20 ps and was then equilibrated at 300 K for 30 ps in an NVT (isochoric-isothermal) ensemble. The temperature of the system was maintained using the v-rescale thermostat with a time constant of 0.1 ps. The system then underwent the second stage of equilibration for 500 ps in an NPT (isobaric-isothermal) ensemble. The pressure of the system was maintained at 1.0 bar using the isotropic Berendsen barostat with a time constant of 2.0 ps and an isothermal compressibility of 4.5 × 10−5 bar−1. The temperature was maintained at 300 K using the Nosé-Hoover thermostat with a time constant of 1.0 ps. To alleviate the “hot solvent/code solute” problem (29, 30, 31), separate thermostats were applied to the collagen and the solvent individually. During this two-stage equilibration, all the heavy atoms of the peptide remained restrained with a force constant of 1000 kJ/mol/nm2. The bond lengths for the peptide were constrained using the LINCS algorithm (32). The geometry of the water was constrained using the SETTLE algorithm. All the equilibrations were performed with a 2-fs time step using the leapfrog algorithm. After equilibration, the 100-ns production run at 1.0 bar and 300 K was performed. During the production, only the first and last Cα atoms on each chain of the triple helix (six atoms in total) were restrained with a force constant of 10 kJ/mol/nm2.
Results
Peptide design
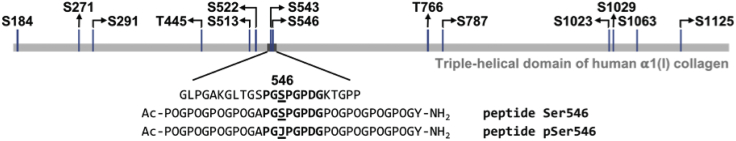
Our goal was to design a peptide that contained a well-documented phosphorylation site in the Gly-Xaa-Yaa region of the α1(I) chain of type I collagen. Reported phosphorylated locations in the α1(I) chain of type I collagen include 12 Ser and 2 Thr sites within the Gly-Xaa-Yaa domain (Fig. 1), but only a subset of these phosphorylation sites was reported in any given publication. Phosphorylation was observed during expression of a relatively short human type I collagen fragment (α1(I) residues 531–631) in a yeast expression system (9). In the purified recombinant fragment, which was not triple helical, Olsen et al. (9) observed phosphorylation at Ser546 (∼50% of expressed protein), together with a smaller amount of protein (∼10%), with phosphorylation at both Ser546 and the preceding Ser543. Residue Ser546 was also reported to be phosphorylated in a HeLa cell culture system (14) and human lung cancer tumor tissues (12). Because of these multiple independent reports, this Ser546 residue was selected as a model phosphorylation site within type I collagen.
Figure 1.
Reported phosphorylation sites in the triple-helical domain (residues 179–1192) of α1 chain of human type I collagen. Phosphorylation of S543 and S546 was found in the recombinant human gelatin expressed by yeast (9); pSer at 184, 291, and 787 sites was reported in rat organs and tissues (11); phosphorylation of S271, S291, T445, S543, S546, S1063, and S1125 was observed in human tumor tissues (12, 13, 15); and phosphorylation of S513, S522, S543, S546, T766, S1023, S1029, and S1125 was reported in HeLa cell lines (10, 14). The sequence around residue Ser546 is shown, together with the design of peptides to model the Ser546 site. To see this figure in color, go online.
A collagen-like peptide with Gly as every third residue and a high imino acid content was synthesized with two design principles: 1) to include the immediate sequence surrounding residue Ser546 in the human α1(I) collagen chain and 2) to form a triple helix with a thermal stability near 30–40°C that would allow for assessing phosphorylation in both monomer and triple-helical states using an in vitro kinase reaction. The original 542GSPGSPGPD550 sequence was modified from GSPGSPGPD by GAPGSPGPD to eliminate any possibility of secondary phosphorylation at Ser543 (9) (Fig. 1). Within a collagen molecule, the Pro residues in the Yaa positions would be largely hydroxylated to Hyp in the ER before triple-helix formation (6), but Pro residues PGSP were incorporated in the model peptide because of the well-defined PxSP consensus sequence for kinases (33). To encourage triple-helix formation, the sequence GAPGSPGPD was flanked by stabilizing (POG)4 triplets to produce peptide Ser546, Ac-(POG)4APGSPGPDG(POG)4Y-NH2 (Fig. 1). Peptide Ser546 is predicted to have a thermal stability of 35.9°C based on the collagen stability calculator (34). A homologous peptide was also synthesized, which contains pSer at residue 546, and this is designated as peptide pSer546 (Fig. 1).
MD simulations of the collagen triple helix with pSer
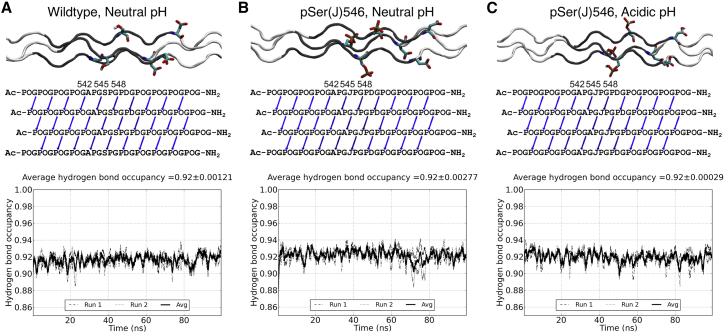
MD simulations were carried out to investigate whether the bulky and charged pSer within the Gly-Xaa-Yaa repeating sequence of peptide pSer546 could be incorporated into a standard triple helix. The sequence of the phosphorylated peptide was constructed into a triple helix (20). MD simulations were carried out for 100 ns at 300 K using the Amber99SB force field and TIP3P water model, following a previously described simulation protocol (3, 20). At neutral pH, the phosphate group of pSer is fully deprotonated, with a −2e charge, and the Asp side chain in the triplet following the GSP has a −1e charge, raising the possibility of repulsive electrostatic interactions between them. The phosphorylated peptide was also simulated under acidic conditions in which each protonated phosphate group of pSer had a reduced net charge of −1e, and each protonated side chain of Asp had no net charge.
The structures reached at the end of the MD simulations indicated that pSer within a Gly-pSer-Pro sequence can be incorporated into a typical triple helix, preserving standard interchain Gly NH···O=C (Xaa) hydrogen bond formation under both physiological (charge −2e) and acidic (charge −1e) conditions. The significant solvent exposure of side chains of the Xaa residues in the triple helix (35) and the positioning of the three phosphate groups ∼108° apart on the surface of the triple-helix cylinder appear to minimize negative steric and charge interactions (Fig. 2, top). For all three systems, the characteristic interchain hydrogen bonding pattern of the triple helix was maintained in the simulations (Fig. 2, middle). The average hydrogen bond occupancies were all at or above 0.90 in the human region, indicative of stable interchain hydrogen bonding, with slight differences in occupancy from bond to bond. The hydrogen bond occupancies of the three systems as a function of time were found to be stable throughout the simulation time (Fig. 2, bottom), indicating that the hydrogen bond patterns present in the triple-helical peptides reached a steady state and experienced minimal fluctuations during the simulation. The distances between the Ser and the Asp side chains during the simulations were also examined. No significant change in the average distance was observed when the Ser was phosphorylated, but there were small changes in the distribution of distances.
Figure 2.
(A) Molecular dynamics (MD) simulation results of the α1(I)3 wild-type (WT) Ser546 peptide at neutral pH, (B) α1(I)3 pSer546 at neutral pH, and (C) α1(I)3 pSer546 at acidic pH. Top: A side view of the triple helices in the end of the simulations is shown. The GPO triplets are colored white, and human sequence triplets are colored gray; Ser546/pSer546 and Asp are drawn in sticks. Middle: A diagram of the average occupancy of interchain NH···CO hydrogen bonds within the peptide is shown. In the sequences, phosphorylated serines are designated as J. Arrows are used to indicate hydrogen bonds, defined as a donor-acceptor distance <3.5 Å and a hydrogen-donor-acceptor angle <30° and based on the average of two 100 ns MD runs. Bottom: Occupancy of the interchain NH···CO hydrogen bonds in the human sequence region as a function of simulation time is shown. Hydrogen bond occupancies as a function of time during each of the two MD runs are depicted as dashed and dotted, respectively; the average of the two runs is shown in a solid line. A running average with a sliding window of 1 ns is used. To see this figure in color, go online.
Structural consequences of pSer on collagen triple-helix conformation, stability, and folding
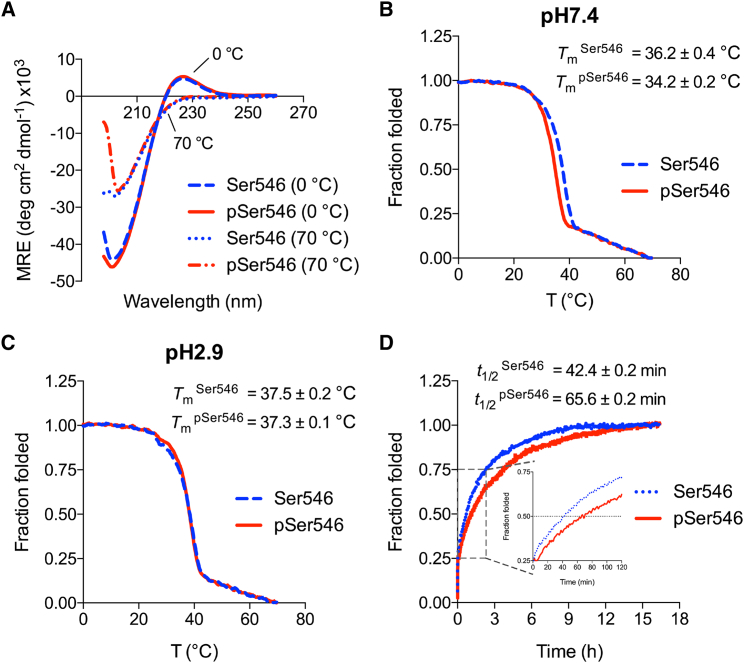
At both neutral and acidic pH values, CD spectroscopy of peptide Ser546 showed the characteristic collagen maximum at 225 nm and minimum near 198 nm at low temperatures (Fig. 3 A), indicative of the triple-helix conformation. The 225-nm maximum was not present at high temperatures when the triple helix was dissociated into monomeric unfolded chains. At pH 7.4, a sharp thermal transition was observed for peptide Ser546 at an apparent Tm = 36.2 ± 0.4°C (Fig. 3 B), in close agreement with the Tm = 35.9°C value predicted from the collagen stability calculator. At pH 2.9, the apparent thermal stability of Ser546 peptide was slightly increased to 37.5 ± 0.2°C (Fig. 3 C), consistent with the previously reported effect of a loss of charge on the Asp side chain (36).
Figure 3.
(A) Circular dichroism (CD) spectra of Ser546 (dashed blue line) and pSer546 (solid red line) peptides in neutral pH phosphate buffer at 0°C, indicating their triple-helical conformation, and at 70°C (Ser546, dotted blue line; pSer546, dashed red line), showing their unfolded state. CD temperature scans of Ser546 (dashed blue line) and pSer546 (solid red line) at (B) neutral pH and (C) pH 2.9 are shown. (D) Refolding profiles of Ser546 (dotted blue line) and pSer546 (solid red line) at 0°C (pH 7.4). Data are representative of three independent experiments and depict mean ± SEM. To see this figure in color, go online.
The phosphorylated peptide pSer546 also showed a typical triple-helix CD spectrum at both neutral and acidic pH values. At pH 7.4, the apparent Tm value of peptide pSer546 was found to be 34.2 ± 0.2°C, showing that the presence of pSer led to a small degree of destabilization compared with peptide Ser546 (Fig. 3 B). The apparent thermal stability of pSer546 peptide showed an increase to 37.3 ± 0.1°C at pH 2.9 (Fig. 3 C), so there was no significant difference in the thermal stability of Ser546 and pSer546 under acidic conditions. Factors that could affect the stability at acidic pH include the reduction of charge on the phosphate group from −2e to −1e and/or the neutralization of the Asp side chain.
To investigate whether the phosphorylation of monomer Gly-Xaa-Yaa chains affects the kinetics of triple-helix folding, the refolding of peptide Ser546 and peptide pSer546 was compared. The peptides were heated to 70°C for 20 min to denature the triple helices into unfolded single chains, followed by refolding studies at 0°C in which efficient refolding was found to occur. The unfolded peptide containing pSer is capable of refolding to form a triple helix, but the refolding rate is slower than seen for the unphosphorylated peptide (t1/2 = 42.4 ± 0.2 min for the Ser546 peptide, and t1/2 = 65.6 ± 0.2 min for the pSer546 peptide at c = 96.9 μM (pH 7.4); Fig. 3 D).
Enzymatic phosphorylation of collagen peptides
The physiological kinase responsible for collagen phosphorylation is not known, but in vitro experiments were carried out to determine whether well-defined kinases will recognize a Ser-containing consensus sequence within the collagen Gly-Xaa-Yaa repeating sequence in the native state and/or in the denatured state. Examination of the Human Protein Reference Database (37) and Phosphorylation Site Database indicated that the collagen sequence surrounding Ser546 meets the consensus criteria for four kinases: ERK1, ERK2, GSK3, and CDK5. Substrates containing PxS/TP (33), a sequence present in peptide Ser546 (PGSP), are preferentially phosphorylated by ERK1 (38), and this kinase was selected as a model kinase predicted to phosphorylate in vitro the Ser546 site, which has been shown to be phosphorylated in vivo.
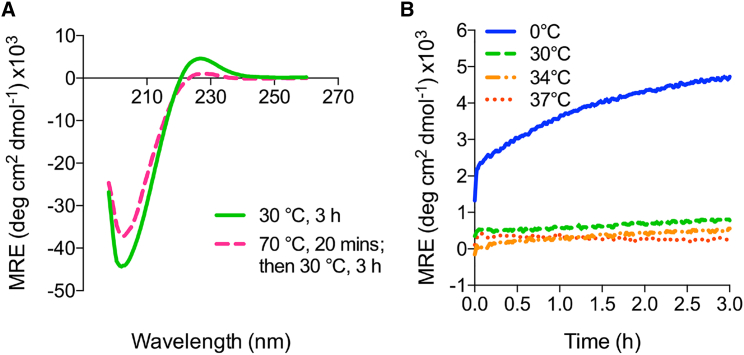
The recommended temperature for ERK1 is 30°C (enzyme activity assay protocol; Promega). After a 3-h incubation at 30°C, the CD spectrum of peptide Ser546 (Fig. 4 A) was indistinguishable from its CD spectrum at 0°C (Fig. 3 A), showing that a fully triple-helical state is maintained. Thus, kinase assays could be carried out at 30°C for peptide Ser546 in the well-defined native state. To define conditions such that the peptide would remain in the unfolded state during the reaction, the Ser546 peptide was denatured by heating to 70°C for 20 min, and the ellipticity at 225 nm was monitored after the sample was cooled to different temperatures (Fig. 4 B). At 0°C, the Ser546 peptide quickly began to reform a triple helix, whereas very little refolding was observed at 30, 34, or 37°C over a 3-h period. Therefore, heating to 70°C and then cooling to 30°C were conditions that would keep the peptide in its unfolded form for the kinase assay.
Figure 4.
Establishment of conditions for the native and denatured states of peptide Ser546 for phosphorylation assays. (A) CD spectra of peptide Ser546 after 3 h of incubation at 30°C (green solid line), showing it is indistinguishable from the spectrum at 0°C (Fig. 3A) and maintains the native state; CD spectrum of peptide Ser546 after heating to 70°C for 20 min followed by 3 h of incubation at 30°C (pink dashed line), showing the peptide remains in the denatured state. (B) The refolding profiles of peptide Ser546 heated to 70°C for 20 min, followed by refolding at 0°C (blue solid line), 30°C (green dashed line), at 34°C (orange dash-dotted line), and 37°C (dark orange dotted line); the ellipticity at 225 nm was monitored with time to follow folding. To see this figure in color, go online.
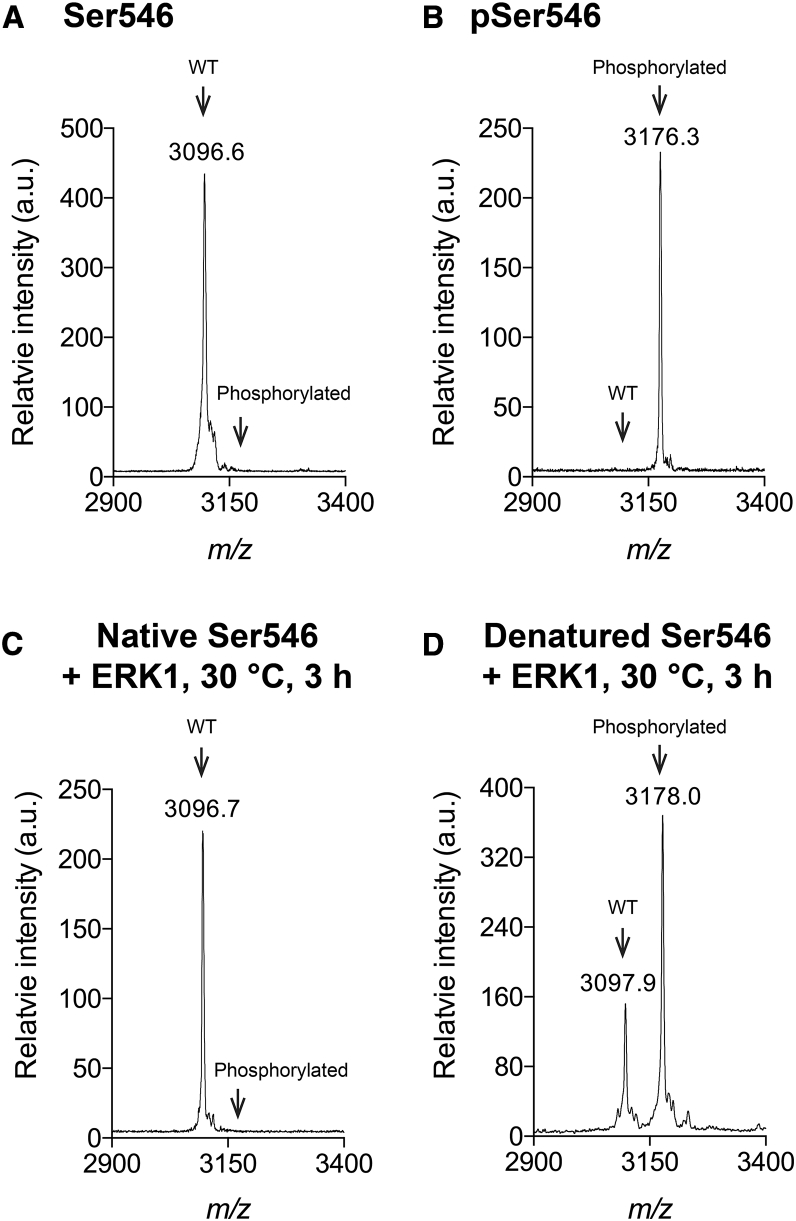
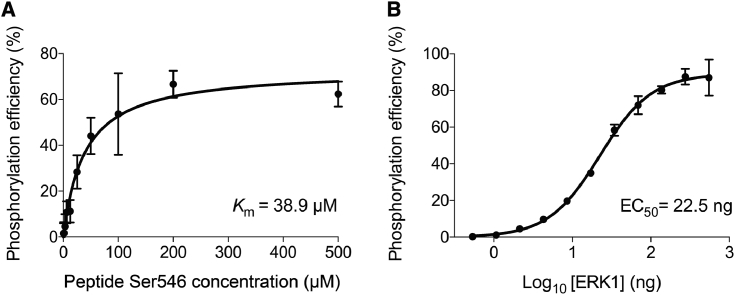
Mass spectroscopy (MALDI-TOF) was used initially to qualitatively examine whether the Ser546 peptide was a substrate for ERK1 because the nonphosphorylated peptide can be clearly distinguished from the phosphorylated one (Fig. 5, A and B). When the native triple-helical peptide Ser546 was incubated with ERK1 at 30°C for 3 h, no phosphorylation was observed regardless of the length of reaction time (up to 14 h), substrate concentration, ATP concentration, or kinase concentration (Fig. 5 C). To determine if the denatured peptide is a substrate, the Ser546 peptide was first heated to 70°C for 20 min to dissociate the triple helix, and then it was incubated with ERK1 for 3 h at 30°C in the unfolded state. In this case, phosphorylation of Ser was observed (Fig. 5 D). The amount of phosphorylation depended upon the incubation time (maximum at 3 h), ATP concentration, and peptide concentration. Although mass spectroscopy is not quantitative, it is estimated that ∼60% of peptide Ser546 is phosphorylated when 100 μM peptide was incubated with 100 μM ATP and 20 ng ERK1 (Fig. 5 D). Experiments carried out at 34 and 37°C were all consistent with significant phosphorylation when the Ser546 peptide was in the unfolded state (data not shown). Thus, the consensus sequence PGSP was phosphorylated by ERK1 in the denatured but not in the native triple-helical state. Luminescence was applied to the ERK1-denatured Ser546 peptide system to obtain more quantitative data on the phosphorylation reaction. As shown in Fig. 6 A, the Km value was determined to be 38.9 μM. With increasing kinase concentration, the phosphorylation efficiency reached 90% (Fig. 6 B).
Figure 5.
Mass spectroscopy of (A) Ser546 control peptide; (B) pSer546 control peptide; (C) incubation of native triple-helical Ser546 peptide with ERK1 kinase for 3 h at 30°C; and (D) incubation of denatured peptide Ser546 with ERK1 for 3 h at 30°C.
Figure 6.
Phosphorylation efficiency of Ser546 peptide using ERK1 kinase was assessed by determination of the amount of ADP generated in the reaction with the ADP-Glo assay method. (A) Titration of Ser546 peptide contained 20 ng of ERK1 kinase, 100 μM ATP, and a serial dilution of Ser546 substrate. (B) The optimal ERK1 amount for phosphorylation of Ser546 (100 μM ATP, 40 μM Ser546 substrate). Data are representative of three independent experiments and presented as mean ± SD of six replicates.
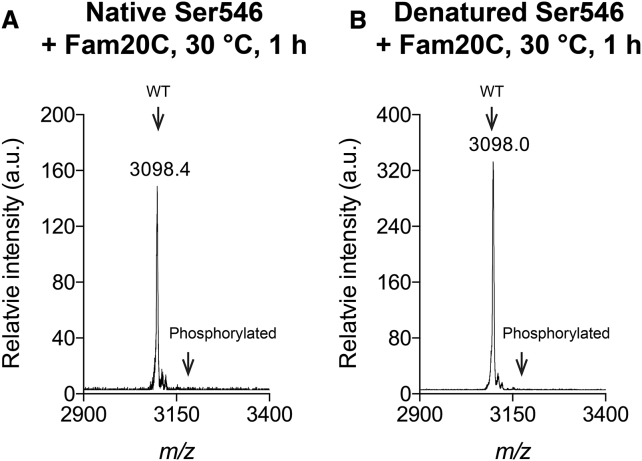
The search for a physiological kinase that phosphorylates collagen is ongoing, and one promising candidate is the ER/Golgi kinase Fam20C, which has been shown to phosphorylate more than 100 ECM secreted proteins (17). Fam20C was tested here for its ability to phosphorylate the collagen peptide Ser546. Fam20C was incubated with the Ser546 peptide in either its native or denatured state, but no phosphorylation was observed under conditions in which a positive control peptide KKIEKFQSEEQQQ (β28–40) was phosphorylated (data not shown). Both MALDI-TOF (Fig. 7) and luminescent assays (data not shown) gave negative results, with no phosphorylation seen.
Figure 7.
Mass spectroscopy of phosphorylation products of Ser546 peptides by Fam20C. (A) The incubation of native triple-helical Ser546 peptide with Fam20C kinase for 1 h at 30°C; (B) the incubation of denatured peptide Ser546 with Fam20C for 1 h at 30°C.
Discussion
Although phosphoproteomics studies have indicated the presence of phosphorylated Ser and Thr residues in the collagen Gly-Xaa-Yaa sequence, little is known about their physiological formation and function. The residue focused on here, Ser546, is the only type I collagen site whose phosphorylation has been characterized on a purified protein level in a denatured recombinant fragment expressed in a yeast system (9) as well as being reported by phosphoproteomics studies of a HeLa cell culture (14) and human lung tumors (12).
MD simulation and experimental studies on a peptide containing immediate surrounding residues from the α1 chain of human type I collagen indicate that the presence of pSer at the reported phosphorylation site 546 allows the formation of a stable triple helix that maintains all standard interchain hydrogen bonds. CD studies indicated the peptide with pSer had a Tm value slightly lower than the unphosphorylated peptide at neutral pH. The small amount of destabilization could reflect repulsion between the phosphate group and the negatively charged Asp residue in the adjacent triplet. The increased distances expected for such repulsion were not clearly observed in MD simulations. However, phosphorylation of Ser residues led to a change of the distribution of its χ1 conformers, which affected and complicated the interpretation of the pSer-Asp side-chain distances. It is also possible that the force filed parameters may not be accurate enough to predict the small differences in stability seen by CD. In contrast to the small amount of destabilization due to pSer in our peptide, a recent article showed that phosphorylation of Ser or Thr in the host-guest peptide system, (Pro-Hyp-Gly)4-pSer-Hyp-Gly-(Pro-Hyp-Gly)3 and (Pro-Hyp-Gly)4-pThr-Hyp-Gly-(Pro-Hyp-Gly)3, increased the stability of the triple helix (16). In addition, there was evidence of electrostatic stabilization when a Lys was introduced two residues away from the pSer (POG)3PKGJOG(POG)3, consistent with previously studied charged-pair interactions (39). NMR studies on these host-guest collagen peptides with pSer or pThr led to constrained models, indicating electrostatic interactions form the molecular basis for the stability (16). The stabilizing effect of the pSer in a simple host-guest sequence compared with the destabilizing effect seen when there are natural type I neighboring triplets indicates that the specific sequence environment influences how phosphorylation impacts the triple helix. One limitation of our study is the homotrimeric nature of the pSer peptide, with a pSer residue in each of the three chains. Type I collagen is a heterotrimer, with two α1 chains and one α2 chain, and the development of new approaches may allow the synthesis of heterotrimers with one or two chains with pSer in a triple helix as better models (40). In addition, there is no information on the degree of phosphorylation of any residue in collagen, so it is likely that sites may only be partially phosphorylated.
One goal of this study was to compare in vitro phosphorylation of a collagen peptide in a well-defined triple-helical state versus a well-defined denatured state, and the design of peptide Ser546, with a Tm ∼36°C, allowed such studies to be carried out. Our results showed that ERK1 did not phosphorylate the Ser546 collagen peptide in the triple-helical state, indicating that the consensus sequence is not accessible to the kinase in the rigid triple-helical molecule. In contrast, the kinase ERK1 recognized and phosphorylated the PGSP consensus sequence in the Ser546 collagen peptide when the substrate was in a single-chain denatured state. Similar results, with the consensus sequence accessible to ERK1 in the unfolded but not the triple-helical state, were found for a recombinant collagen containing four triplets around Ser546. An early in vitro study in 1984 by Glass and May (41) showed that a different kinase, protein kinase A (cyclic AMP-dependent protein kinase), efficiently phosphorylated a single-chain (denatured) peptide modeling residues 98–110 of the chick α1(I) chain, whereas protein kinase A did not phosphorylate native type I collagen (42). It remains to be seen if this selective phosphorylation of denatured collagen is a general feature of all kinases that can recognize consensus sequences in collagen and, more importantly, in the yet-to-be-identified physiological kinase.
Attempts were made to move from the model ERK1 kinase to a more biologically relevant kinase. Data from cell cultures for which the ATP concentration outside the cells would be very low suggest that collagen is phosphorylated as it transits the secretory pathway in these cases (10, 14), and a likely scenario would involve 1 of the 13 identified secretory pathway kinases being the physiologically relevant kinase. The most well characterized of these kinases is Fam20C (17), but, in our experiments, this kinase did not phosphorylate peptide Ser 546 in either the denatured or triple-helical form. An important caveat to these experiments is that previous studies have shown that Fam20C efficiently and almost exclusively phosphorylates S-X-E sites within peptides in vitro; phosphorylation of nonconsensus sequences, such as those found in peptide Ser546, has only been observed within the context of a larger protein, preferably within the lumen of the secretory pathway of a cell. There are three SGE sites within the collagen α1(I) chain, but these sites have not yet been reported to be phosphorylated. An interesting kinase candidate, if not Fam20C, could possibly be Fjx1. Fjx1 was described as a Ser kinase that could phosphorylate cadherin domains in flies (17); however, there have not been any targets identified for the mammalian ortholog Fjx1.
Although the kinase for collagen phosphorylation is not known, the recent phosphoproteomic reports of phosphorylated collagen in tissues and in cell cultures suggest that phosphorylated collagen is present in the ECM and may occur in the secretory pathway (8, 11, 13, 14). If phosphorylation occurs on denatured or partially unfolded collagen chains, our studies suggest that phosphorylation of a Ser will delay but not prevent the individual Gly-Xaa-Yaa chains folding to form a stable triple helix. Thus, it is possible that phosphorylated collagen chains form phosphorylated triple helices that are secreted and perhaps even incorporated into fibrils. Alternatively, there has been an increased understanding of the importance of collagen degradation during ECM remodeling and tissue damage related to cancer, myocardial infarction, and fibrosis (43). Phosphorylation could take place on such unfolded collagen substrates in the ECM, perhaps catalyzed by kinases that are secreted in exosomes. However, such a mechanism would be dependent on achieving a sufficiently high ATP concentration in the ECM.
This study establishes some of the basic principles of collagen phosphorylation, indicating that a well-defined kinase can recognize consensus sequences in unfolded collagen chains in vitro and that phosphorylated collagen peptide chains are capable of folding to form stable triple-helical molecules. These results will serve as a basis for interpreting physiological studies and for using phosphorylation/dephosphorylation as a regulatory tool for collagen and gelatin-based biomaterials.
Author Contributions
Y.Q., D.L.K., Y.-S.L., E.P., and B.B. designed the study and wrote the article. Y.Q., H.Y., A.M., E.P., Y.-S.L., and B.B. analyzed and critically evaluated the results. Y.Q. and E.P. performed experiments with in vitro ERK1 kinase phosphorylation and characterization of peptides. H.Y. and A.M. carried out MD simulations. Y.Q., S.B., S.E.W. and J.E.D. performed the Fam20C kinase activity assays. All authors read and approved the final version of the manuscript.
Acknowledgments
We thank Dr. David Wilbur from Tufts University Department of Chemistry for the generous use of the MALDI-TOF mass spectrometer.
This work was supported by National Institutes of Health grants GM 60048 (B.B.), DK 018849-41 (J.E.D.), and DK 018024-43 (J.E.D.), the Tufts Start-Up Fund (Y.-S.L.), and the Knez Family Faculty Investment Fund (Y.-S.L).
Editor: Nancy Forde.
Contributor Information
David L. Kaplan, Email: david.kaplan@tufts.edu.
Barbara Brodsky, Email: barbara.brodsky@tufts.edu.
References
- 1.Bella J. Collagen structure: new tricks from a very old dog. Biochem. J. 2016;473:1001–1025. doi: 10.1042/BJ20151169. [DOI] [PubMed] [Google Scholar]
- 2.Brodsky B., Persikov A.V. Molecular structure of the collagen triple helix. Adv. Protein Chem. 2005;70:301–339. doi: 10.1016/S0065-3233(05)70009-7. [DOI] [PubMed] [Google Scholar]
- 3.Yigit S., Yu H., Brodsky B. Mapping the effect of Gly mutations in collagen on α2β1 integrin binding. J. Biol. Chem. 2016;291:19196–19207. doi: 10.1074/jbc.M116.726182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kramer R.Z., Venugopal M.G., Berman H.M. Staggered molecular packing in crystals of a collagen-like peptide with a single charged pair. J. Mol. Biol. 2000;301:1191–1205. doi: 10.1006/jmbi.2000.4017. [DOI] [PubMed] [Google Scholar]
- 5.Burjanadze T.V. Hydroxyproline content and location in relation to collagen thermal stability. Biopolymers. 1979;18:931–938. doi: 10.1002/bip.1979.360180413. [DOI] [PubMed] [Google Scholar]
- 6.Gjaltema R.A., Bank R.A. Molecular insights into prolyl and lysyl hydroxylation of fibrillar collagens in health and disease. Crit. Rev. Biochem. Mol. Biol. 2017;52:74–95. doi: 10.1080/10409238.2016.1269716. [DOI] [PubMed] [Google Scholar]
- 7.Yamauchi M., Taga Y., Terajima M. Analysis of collagen and elastin cross-links. Methods Cell Biol. 2018;143:115–132. doi: 10.1016/bs.mcb.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Yalak G., Olsen B.R. Proteomic database mining opens up avenues utilizing extracellular protein phosphorylation for novel therapeutic applications. J. Transl. Med. 2015;13:125. doi: 10.1186/s12967-015-0482-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olsen D., Jiang J., Polarek J.W. Expression and characterization of a low molecular weight recombinant human gelatin: development of a substitute for animal-derived gelatin with superior features. Protein Expr. Purif. 2005;40:346–357. doi: 10.1016/j.pep.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 10.Olsen J.V., Vermeulen M., Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 11.Lundby A., Secher A., Olsen J.V. Quantitative maps of protein phosphorylation sites across 14 different rat organs and tissues. Nat. Commun. 2012;3:876. doi: 10.1038/ncomms1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schweppe D.K., Rigas J.R., Gerber S.A. Quantitative phosphoproteomic profiling of human non-small cell lung cancer tumors. J. Proteomics. 2013;91:286–296. doi: 10.1016/j.jprot.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mertins P., Yang F., Carr S.A. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol. Cell. Proteomics. 2014;13:1690–1704. doi: 10.1074/mcp.M113.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma K., D’Souza R.C., Mann M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014;8:1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 15.Mertins P., Mani D.R., Carr S.A., NCI CPTAC Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Acevedo-Jake A.M., Ngo D.H., Hartgerink J.D. Control of collagen triple helix stability by phosphorylation. Biomacromolecules. 2017;18:1157–1161. doi: 10.1021/acs.biomac.6b01814. [DOI] [PubMed] [Google Scholar]
- 17.Tagliabracci V.S., Wiley S.E., Dixon J.E. A single kinase generates the majority of the secreted phosphoproteome. Cell. 2015;161:1619–1632. doi: 10.1016/j.cell.2015.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshizumi A., Fletcher J.M., Brodsky B. Designed coiled coils promote folding of a recombinant bacterial collagen. J. Biol. Chem. 2011;286:17512–17520. doi: 10.1074/jbc.M110.217364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao J., Tagliabracci V.S., Dixon J.E. Crystal structure of the golgi casein kinase. Proc. Natl. Acad. Sci. USA. 2013;110:10574–10579. doi: 10.1073/pnas.1309211110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chhum P., Yu H., Brodsky B. Consequences of glycine mutations in the fibronectin-binding sequence of collagen. J. Biol. Chem. 2016;291:27073–27086. doi: 10.1074/jbc.M116.753566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apweiler R., Bairoch A., Yeh L.S. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rainey J.K., Goh M.C. An interactive triple-helical collagen builder. Bioinformatics. 2004;20:2458–2459. doi: 10.1093/bioinformatics/bth247. [DOI] [PubMed] [Google Scholar]
- 23.Pettersen E.F., Goddard T.D., Ferrin T.E. UCSF chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 24.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 25.Wang J.M., Cieplak P., Kollman P.A. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 26.Hornak V., Abel R., Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Gunsteren W.F., Berendsen H.J., Postma J.P. Computer simulation of the dynamics of hydrated protein crystals and its comparison with x-ray data. Proc. Natl. Acad. Sci. USA. 1983;80:4315–4319. doi: 10.1073/pnas.80.14.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 29.Cheng A.L., Merz K.M. Application of the Nose-Hoover chain algorithm to the study of protein dynamics. J. Phys. Chem. 1996;100:1927–1937. [Google Scholar]
- 30.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 31.Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984;52:255–268. [Google Scholar]
- 32.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 33.Songyang Z., Lu K.P., Cantley L.C. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Persikov A.V., Ramshaw J.A., Brodsky B. Prediction of collagen stability from amino acid sequence. J. Biol. Chem. 2005;280:19343–19349. doi: 10.1074/jbc.M501657200. [DOI] [PubMed] [Google Scholar]
- 35.Jones E.Y., Miller A. Analysis of structural design features in collagen. J. Mol. Biol. 1991;218:209–219. doi: 10.1016/0022-2836(91)90885-a. [DOI] [PubMed] [Google Scholar]
- 36.Chan V.C., Ramshaw J.A., Brodsky B. Positional preferences of ionizable residues in Gly-X-Y triplets of the collagen triple-helix. J. Biol. Chem. 1997;272:31441–31446. doi: 10.1074/jbc.272.50.31441. [DOI] [PubMed] [Google Scholar]
- 37.Amanchy R., Periaswamy B., Pandey A. A curated compendium of phosphorylation motifs. Nat. Biotechnol. 2007;25:285–286. doi: 10.1038/nbt0307-285. [DOI] [PubMed] [Google Scholar]
- 38.Roskoski R., Jr. ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol. Res. 2012;66:105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 39.Fallas J.A., Dong J., Hartgerink J.D. Structural insights into charge pair interactions in triple helical collagen-like proteins. J. Biol. Chem. 2012;287:8039–8047. doi: 10.1074/jbc.M111.296574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jalan A.A., Hartgerink J.D. Simultaneous control of composition and register of an AAB-type collagen heterotrimer. Biomacromolecules. 2013;14:179–185. doi: 10.1021/bm3015818. [DOI] [PubMed] [Google Scholar]
- 41.Glass D.B., May J.M. In vitro phosphorylation of a synthetic collagen peptide by cyclic AMP-dependent protein kinase. Coll. Relat. Res. 1984;4:63–74. doi: 10.1016/s0174-173x(84)80029-1. [DOI] [PubMed] [Google Scholar]
- 42.Glass D.B., McPherson J.M. In vitro phosphorylation of type I collagen by cyclic AMP-dependent protein kinase. J. Biol. Chem. 1986;261:5674–5679. [PubMed] [Google Scholar]
- 43.Hwang J., Huang Y., Li Y. In situ imaging of tissue remodeling with collagen hybridizing peptides. ACS Nano. 2017;11:9825–9835. doi: 10.1021/acsnano.7b03150. [DOI] [PMC free article] [PubMed] [Google Scholar]