Abstract
Cu/Zn Superoxide Dismutase (Sod1) is a highly conserved and abundant metalloenzyme that catalyzes the disproportionation of superoxide radicals into hydrogen peroxide and molecular oxygen. As a consequence, Sod1 serves dual roles in oxidative stress protection and redox signaling by both scavenging cytotoxic superoxide radicals and producing hydrogen peroxide that can be used to oxidize and regulate the activity of downstream targets. However, the relative contributions of Sod1 to protection against oxidative stress and redox signaling are poorly understood. Using the model unicellular eukaryote, Baker's yeast, we found that only a small fraction of the total Sod1 pool is required for protection against superoxide toxicity and that this pool is localized to the mitochondrial intermembrane space. On the contrary, we find that much larger amounts of extra-mitochondrial Sod1 are critical for peroxide-mediated redox signaling. Altogether, our results force the re-evaluation of the physiological role of bulk Sod1 in redox biology; namely, we propose that the vast majority of Sod1 in yeast is utilized for peroxide-mediated signaling rather than superoxide scavenging.
Keywords: Cu/Zn superoxide dismutase, Redox signaling, Superoxide, Hydrogen peroxide, Reactive oxygen species, Mitochondrial metabolism
1. Introduction
Superoxide (O2•−) and hydrogen peroxide (H2O2) are cytotoxic reactive oxygen species (ROS) that are also essential for the redox control of a multitude of physiological processes. O2•− toxicity is largely due to its ability to oxidize and inactivate [4Fe-4S] cluster-containing enzymes, which releases iron (Fe) in the process [1], [2], [3], [4]. The liberated Fe, upon complexation by appropriate ligands, promotes deleterious redox reactions, and in particular produces hydroxyl radicals (•OH) via the Fe-catalyzed reduction of H2O2 i.e. Haber-Weiss and Fenton reactions [2], [5]. Once formed, •OH indiscriminately oxidizes lipids, proteins, and nucleic acids, leading to membrane disruption, protein misfolding and aggregation, and DNA fragmentation, respectively. While O2•− itself is not likely to be a signaling molecule [6], it rapidly disproportionates into H2O2 (k ~ 105 M−1 s−1 at pH = 7.0), a well-established signaling molecule [6], [7], that can lead to the reversible oxidation of cysteine residues in a number of downstream targets [8], including phosphatases [9], [10], [11], kinases [12], [13], metabolic enzymes [14], and transcription factors [15], [16], to regulate protein activity.
The dual roles of O2•−/H2O2 in oxidative stress and redox signaling necessitates that the concentration and localization of these ROS are regulated in a manner that enables signaling but mitigates oxidative damage. In terms of localization, a number of metabolic sources of H2O2 and O2•− are present throughout the cell, including O2•−-generating NADPH oxidases (NOX) that have been found in the nucleus, endoplasmic reticulum (ER), cell membrane, and mitochondria [17], [18], mitochondrial respiratory Complexes I and III [19], [20], [21], and enzymes that release O2•− and/or H2O2, e.g. xanthine oxidase (cytosol) [22], [23], monoamine oxidase (mitochondria) [24], cytochrome P450's (ER) [25], and globins (cytosol) [26], [27]. In terms of concentration, detoxification systems have evolved to limit the levels of O2•−, e.g. superoxide dismutases (SODs), and H2O2, e.g. catalase (CAT), glutathione (GSH) peroxidases (GPx), and peroxiredoxins (Prx) [28]. Of these ROS scavenging systems, SODs, which catalyze the disproportionation of 2O2•− into H2O2 and O2, are unique in that they simultaneously affect both [O2•−] and [H2O2]. As a consequence, SODs play dual roles in both defending against O2•− toxicity and regulating H2O2-mediated redox signaling [4], [28].
Most eukaryotes express two intracellular SODs, a Mn-containing Sod2 that is exclusively localized to the mitochondrial matrix [29], and a highly abundant Cu/Zn Sod1 that is present virtually everywhere else [30], including the mitochondrial intermembrane space (IMS) [31], [32], nucleus [33], endoplasmic reticulum (ER) [34], and peroxisomes [35]. Sod1-deficient organisms, from yeast to mice, are oxidatively stressed and have reduced life spans. For example, SOD1-/- mice have a higher incidence of liver cancer, neuronal damage, and loss of muscle mass [36], [37], [38], [39], [40], [41]. Drosophila mutants of SOD1 are infertile and have dramatically reduced life spans [42]. In Saccharomyces cerevisiae (Baker's yeast), sod1∆ cells have defects in a number of metabolic pathways due to oxidative damage of critical [4Fe-4S] cluster containing enzymes [5], [43], [44], [45], as well as membrane and DNA fragmentation [33], [45], [46] due to increases in “free” or labile iron [47], [48], which promote hydroxyl radical formation [5]. In total, cell biological and biochemical studies across multiple organisms indicate Sod1 protects Fe-S cluster enzymes from O2•− damage and further oxidative stress due to Fe and •OH toxicity.
From the perspective of redox signaling, Sod1-derived H2O2 was found to regulate the oxidation of protein tyrosine phosphatases [11] and the tyrosine kinase growth factor receptor [49]. In addition, Sod1 was also found to provide a source of H2O2 that stabilizes a pair of plasma membrane casein kinases, Yck1 and Yck2, that control nutrient sensing and energy metabolism [50].
Sod1 is a highly abundant protein in various organisms [51], [52], and in yeast is present at concentrations of ~ 10–20 μM [53], [54], accounting for ~ 80–90% of total cellular Sod activity [55]. Given that Sod1 disproportionates O2•− at diffusion-limited rates (k ~ 109 M−1 s−1) [56], the rationale for producing such large quantities of Sod1 has been enigmatic. Moreover, the relative contributions of Sod1 towards protection against O2•− toxicity and H2O2-mediated redox signaling are not well understood [4]. Herein, using Baker's yeast as a eukaryotic model, we find that only a small fraction of total Sod1 is required for protection against O2•− toxicity and that this pool is localized to the mitochondrial intermembrane space (IMS). Instead, we find that much larger amounts of extra-mitochondrial Sod1 are critical for peroxide-mediated redox control of Yck1 signaling. Given that an exceedingly small fraction of Sod1 is required for protection against O2•− and much larger quantities are seemingly required for peroxide-mediated redox signaling, our results challenge us to re-evaluate the physiological role of bulk Sod1. We propose that yeast, and possibly other eukaryotic cells, express high levels of Sod1 to maintain appropriate peroxide fluxes to facilitate redox signaling, whereas superoxide detoxification can be handled by a relatively miniscule amount of Sod1.
2. Results
2.1. The vast majority of Sod1 is dispensable for protection against superoxide toxicity
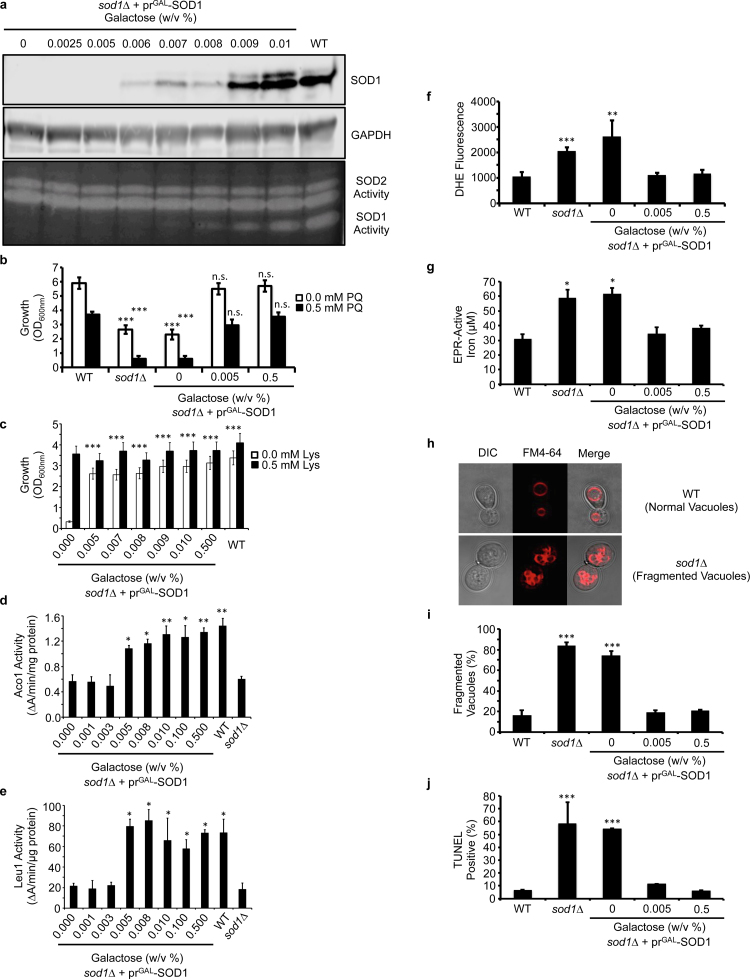
In atmospheric oxygen (21% O2), sod1∆ cells exhibit a number of markers of O2•− toxicity. This includes elevated [O2•−] [57], O2•−-mediated inactivation of a number of [4Fe-4S] enzymes [5], [45], [58], including aconitase (Aco1), isopropylmalate isomerase (Leu1), and homoaconitase (Lys4), increased labile Fe due to its release from oxidized Fe-S clusters [45], [47], [48], and vacuolar [46] and DNA [33] fragmentation due to deleterious Fe-mediated redox reactions. Collectively, these defects lead to reduced aerobic growth [55], decreased lifespan [59], and a number of metabolic defects, including perturbations to redox homeostasis [44], energy metabolism [50], [60] and a number of amino acid auxotrophies [5], [43], [44], [45], e.g. defects in the biosynthesis of leucine (due to inhibition of Leu1), lysine (due to inhibition of Lys4), and methionine (due to reduced pentose phosphate pathway activity and NADPH). Using the galactose-inducible GAL1 promoter to drive SOD1 expression in the background of sod1∆ cells, we sought to determine the amount of Sod1 required to rescue various cell-wide markers of oxidative stress. As shown in Fig. 1a, titration of galactose (GAL) resulted in undetectable (0.000–0.005% w/v GAL), intermediate (0.006% –0.008% w/v GAL), and high (> .01% w/v GAL) levels of Sod1 expression and activity. High concentrations of GAL (> 0.01%) consistently resulted in near WT-levels of Sod1 expression and activity. Most interestingly, only 0.005% GAL, a concentration that results in the induction of an undetectable amount of Sod1 activity and polypeptide (Fig. 1a), rescues major hallmarks of O2•− toxicity, including sensitivity to paraquat, a O2•−-generating agent (Fig. 1b), lysine auxotrophy (Fig. 1c), the activity of mitochondrial and cytosolic [4Fe-4S] cluster containing enzymes, Aco1 (Fig. 1d) and Leu1 (Fig. 1e), respectively, cellular [O2•−] as measured by dihydroethidium (DHE) fluorescence (Fig. 1f), electron paramagnetic resonance (EPR)-detectable labile Fe pools (Fig. 1g), vacuolar fragmentation as imaged by FM4–64 (Fig. 1h and i), and DNA damage as assessed by terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick end labeling (TUNEL) (Fig. 1j). Parenthetically, it is important to note that at the excitation and emission wavelengths chosen to measure DHE fluorescence, there are contributions from both superoxide specific, e.g. 2-hydroxyethidium, and non-specific, e.g. ethidium or ethidium/ethidine dimers, DHE oxidation products [61]. Thus, while our DHE fluorescence measurements are not specific for superoxide per se, the differences in DHE fluorescence we observe reflect Sod1-dependent DHE oxidation products. Altogether, these results indicate that the vast majority of Sod1 is dispensable for protection against superoxide toxicity.
Fig. 1.
The vast majority of Sod1 is dispensable for protection against superoxide toxicity. (a) Titration of galactose (GAL) into cultures of sod1::LEU2 (sod1∆) cells expressing the GAL1 driven Sod1 expression vector (prGAL-SOD1; pAR1026) results in the expression of low (≤ .005% GAL), intermediate (0.006–0.008% GAL), and high (≥ 0.009% GAL) expression and activity of Sod1. The immunoblots and activity gels depicted are representative of multiple trials across different batches of media. (b) Paraquat (PQ) sensitivity of WT and sod1∆ cells compared to sod1∆ + prGAL-SOD1 cells expressing none (0% GAL), low (0.005% GAL) or high (0.5% GAL) levels of Sod1 as measured by solution turbidity. (c) Lysine (Lys) auxotrophy, (d) aconitase (Aco1) activity, and (e) isopropylmalate isomerase (Leu1) activity of sod1∆ + prGAL-SOD1 cells is measured as a function of Sod1 expression and compared to WT and/or sod1∆ cells. (f) DHE detectable superoxide is monitored in WT, sod1∆, and sod1∆ + prGAL-SOD1 cells expressing none (0% GAL), low (0.005% GAL), or high (0.5% GAL) Sod1. (g) EPR detectable labile Fe, (h and i) FM4–64 visualized vacuolar fragmentation, and (j) DNA damage using the TUNEL assay was monitored in WT, sod1∆, or sod1∆ + prGAL-SOD1 cells expressing none (0% GAL), low (0.005% GAL), or high (0.5% GAL) Sod1. In panels i and j, approximately ~100 cells were counted from each culture condition in triplicate and scored for (i) having single or multiple fragmented vacuoles, as depicted in panel h, or (j) being non-fluorescent or fluorescent in the FITC channel. Error bars indicate the average ± s.d. of triplicate (b, c, f, i, j) or duplicate (d, e, g) independent cultures. The statistical significance relative to WT (b, f, g, i, j) or sod1∆ + prGAL-SOD1 cells cultured with 0% GAL (c, d, e) is indicated by asterisks using an ordinary one-way ANOVA with Dunnett's post-hoc test. * P < 0.05, ** P < 0.01, *** P < 0.0001, n.s. = not significant.
2.2. IMS-targeted Sod1 is sufficient to protect against cell-wide markers of O2− toxicity
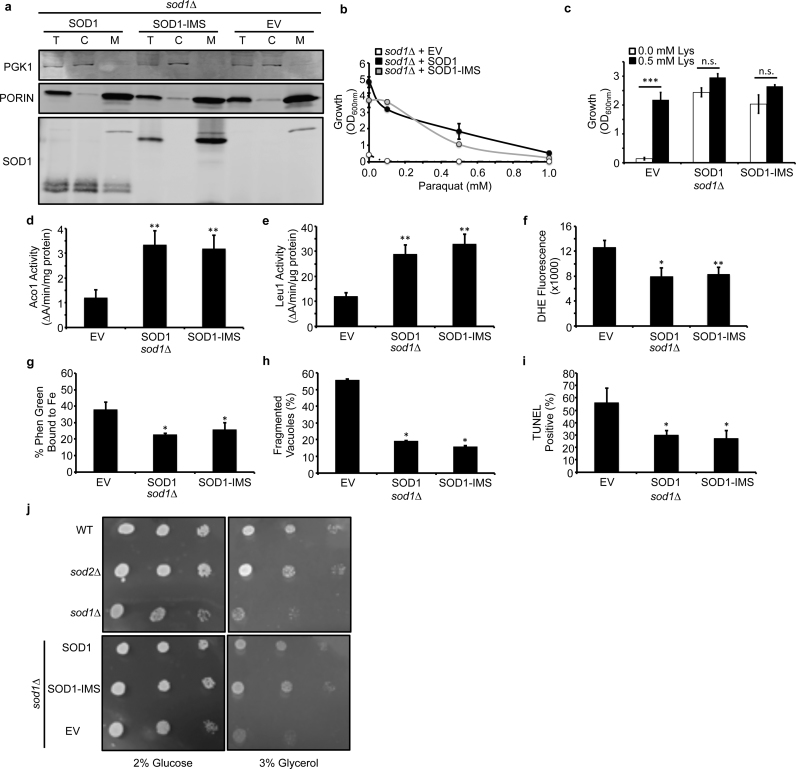
We next sought to determine if the localization of Sod1 is important for protection against cell-wide markers of superoxide toxicity. Mitochondria are a major source of ROS and O2− due to electron leakage during cellular respiration, and in particular from Complex III, which can release O2•− into the mitochondrial matrix and IMS [19]. Deletion of Sod1, which is in-part localized to the mitochondrial IMS [31], [32], but not Sod2, which exclusively resides in the mitochondrial matrix, results in lysine auxotrophy due to the O2•−-dependent inhibition of matrix-localized homoaconitase (Lys4). This suggests that O2•− leakage into the IMS occurs to a greater extent than into the matrix and the ultimate source of matrix O2•− is from the IMS. In order to determine the extent to which IMS-localized Sod1 protects against oxidative stress, including in the mitochondrial matrix, an allele of Sod1 that is exclusively targeted to the IMS, SOD1-IMS, due to fusion of the Sco2 IMS localization sequence [50], [62] was expressed in sod1∆ cells (Fig. 2a). Interestingly, we found that SOD1-IMS rescues cell-wide markers of O2•−-toxicity, including paraquat sensitivity (Fig. 2b), lysine auxotrophy (Fig. 2c), the activity of mitochondrial and cytosolic [4Fe-4S] cluster containing enzymes, Aco1 (Fig. 2d) and Leu1 (Fig. 2e), respectively, cellular [O2•−] as measured by dihydroethidium (DHE) fluorescence (Fig. 2f), Phen Green-detectable labile Fe (Fig. 2g), vacuolar fragmentation as imaged by FM4–64 (Fig. 2h), and DNA damage as assessed by terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick end labeling (TUNEL) (Fig. 2i). The effects of SOD1-IMS on defending against O2•− toxicity are distinct from mitochondrial matrix localized Sod2. Unlike sod1∆ cells, sod2∆ mutants do not exhibit lysine, leucine, or methionine auxotrophies, stunted aerobic growth, or growth defects on respiratory carbon sources, e.g. 3% glycerol (Fig. 2j). Notably, SOD1-IMS expression is sufficient to rescue growth of sod1∆ cells on glycerol (Fig. 2j). Altogether, these results indicate that IMS-localized Sod1 alone can protect against cell-wide superoxide toxicity and that the source of matrix and extra-mitochondrial O2•− is from the IMS.
Fig. 2.
IMS-targeted Sod1 is sufficient to protect against cell-wide markers of superoxide toxicity. (a) Immunoblots of whole cell extracts (T), cytosolic (C), or mitochondrial (M) fractions from sod1::kanMX4 (sod1∆) cells expressing empty vector (EV; pRS415), WT SOD1 (SOD1; pRS415-SOD1), or mitochondrial IMS targeted SCO2-SOD1 (SOD1-IMS; pRS415-SCO2-SOD1). The purity of subcellular fractions was assessed by probing for cytosolic or mitochondrial marker proteins, Pgk1 and Porin, respectively. (b) Paraquat (PQ) sensitivity, (c) lysine (Lys) auxotrophy, (d) aconitase (Aco1) activity, (e) isopropylmalate isomerase (Leu1) activity, (f) DHE-detectable superoxide, (g) Phen-Green detectable labile Fe, (h) vacuolar fragmentation, and (i) TUNEL-detected DNA fragmentation in sod1∆ cells expressing EV, SOD1, or SOD1-IMS. (j) Respiratory (3% glycerol) versus fermentative (2% glucose) growth of WT, sod1::kanMX4, sod2::kanMX4, sod1::kanMX4 + pRS415, sod1::kanMX4 + pRS415-SOD1, sod1::kanMX4 + pRS415-SCO2-SOD1 was tested by spotting 104, 103, and 102 cells on YP media plates containing 2% glucose or 3% glycerol for 3 days. In panels h and i, approximately ~100 cells were counted from each culture condition in triplicate and scored for (h) having single or multiple fragmented vacuoles, as depicted in Fig. 1h, or (i) being non-fluorescent or fluorescent in the FITC channel. Error bars indicate the average ± s.d. of triplicate independent cultures. The statistical significance relative to EV cells is indicated by asterisks using an ordinary one-way ANOVA with Dunnett's post-hoc test or a Bonferroni test for the indicated pairwise comparisons in panel c. * P < 0.05, ** P < 0.01, *** P < 0.0001, n.s. = not significant.
2.3. High concentrations of Sod1 are required for Yck1 signaling
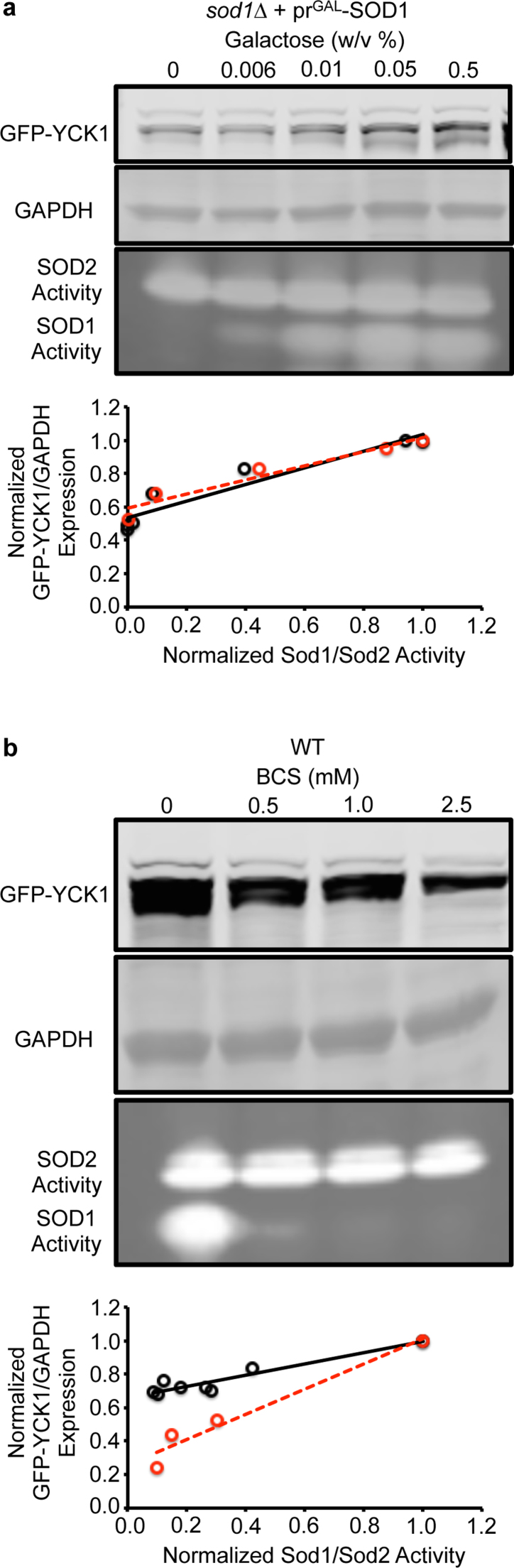
Given that the vast majority of extra-mitochondrial Sod1 in yeast is apparently dispensable for protection against superoxide toxicity, we next sought to determine the relative contribution of Sod1 towards H2O2-mediated redox signaling. In Saccharomyces cerevisiae, the only known case of Sod1-mediated redox signaling to date involves a pathway in which Sod1 derived H2O2 regulates the stability of a pair of plasma membrane tethered casein kinases, Yck1 and Yck2, that integrate nutrient sensing with energy metabolism [50]. Sod1, which physically associates with the C-terminus of Yck1, produces a local flux of H2O2 that prevents the degradation of Yck1. In the absence of Sod1, its substrate, O2•−, or a O2•−-generating NADPH oxidase, Yno1 [63], Yck1 is degraded through a mechanism that is currently unknown [50]. The loss of Yck1 shifts energy metabolism from fermentation to respiration. Notably, IMS-localized Sod1 does not contribute towards the regulation of Yck1 stability [50]. Most interestingly, unlike numerous hallmarks of superoxide-toxicity, Yck1 stability is very sensitive to Sod1 expression. Titration of Sod1 using the GAL-inducible SOD1 expression system results in a positive correlation between Sod1 activity and Yck1 expression at Sod1 levels that exceed the minimal threshold required to protect against superoxide toxicity (Fig. 3a). Similarly, inhibiting Sod1 activity using the copper chelator BCS results in a dose-dependent decrease in Yck1 expression (Fig. 3b). Altogether, we find that redox signaling via the Sod1/H2O2/Yck1 signaling axis is far more sensitive to fluctuations in bulk Sod1 activity than protection against cell-wide markers of superoxide toxicity, which only requires a vanishingly low amount of Sod1 activity.
Fig. 3.
Yck1 expression is more sensitive to fluctuations in Sod1 expression and activity than various markers of superoxide toxicity. (a) Titration of galactose (GAL) into cultures of sod1::LEU2 (sod1∆) cells co-expressing the GAL1 driven Sod1 expression vector (prGAL-SOD1; pAR1026) and the TEF1 driven GFP-Yck1 expression vector (pAR113) results in a positive correlation between Sod1 activity and Yck1 expression. (b) Titration of the copper chelator, BCS, into cultures of sod1::LEU2 cells expressing the TEF1 driven GFP-Yck1 expression vector (pAR113) results in a positive correlation between Sod1 activity and Yck1 expression. In the correlation plots, relative GFP-Yck1 expression is measured as the ratio of GFP-Yck1 to GAPDH signal intensities as determined from densitometry of the immunoblots. The ratios are normalized to the maximal ratio observed with 0.5% GAL. Sod1 activity is expressed as the ratio of BCS sensitive Sod1 activity to BCS-insensitive Sod2 activity as determined from densitometry of the activity gels. The ratios are normalized to the maximal ratio observed in each experimental set, i.e. 0.5% GAL in GAL titrations or 0 mM BCS in BCS titrations. In panel a, the Pearson's correlation coefficients (r) for Trial 1 (red circles) and Trial 2 (black circles) are r = 0.935 (p < .01) and r = 0.906 (p < .001), respectively. In panel b, the Pearson's correlation coefficients (r) for Trial 1 (red circles) and Trial 2 (black circles) are r = 0.955 (p < .05) and r = 0.888 (p < .001), respectively. Immunoblots and Sod activity gels are shown for Trial 1, which are representative of all trials.
3. Discussion
Since the seminal discovery of Sod1 in 1969 [30], there was great controversy surrounding its proposed physiological function in O2•− scavenging due to the low reactivity of O2•− with various biomolecules, e.g. nucleic acids, proteins, and lipids, and the short lifetime of O2•− due to its rapid un-catalyzed disproportionation, k ~ 105 M−1 s−1 at pH 7.0 [64], [65]. In fact, it was proposed that Sod1 had other unknown functions in biology and that it coincidentally catalyzed O2•− disproportionation [65]. This controversy was largely put to rest with the realization that [4Fe-4S] cluster containing enzymes are primary targets of O2•−, which can oxidize and destroy Fe-S clusters with rate constants up to 107 M−1 s−1 [1], [2], [3], [4]. In fact, most of the pathological hallmarks of Sod1 deletion and O2•− toxicity are due to the diminished activity of certain Fe-S cluster enzymes and inhibition of the corresponding metabolic pathways they operate in, increased labile Fe due to the destruction of Fe-S clusters, and Fe-mediated oxidative stress that results in the damage of lipids, proteins, and nucleic acids. However, given that Sod1 is amongst the most abundant proteins, constituting as much as 0.5% of total yeast protein [66], and disproportionates O2•− at diffusion-limited rates (k ~ 109 M−1 s−1), the rationale for producing large quantities of Sod1 has been a mystery. Indeed, herein, using the model unicellular eukaryote, Saccharomyces cerevisiae (Baker's yeast), we find that the vast majority of Sod1 is dispensable for protection against numerous cell-wide markers of O2•− toxicity, including cellular [O2•−], loss of Fe-S cluster enzyme activity, increased labile Fe, and vacuolar and DNA damage (Fig. 1).
As SODs are the only enzymes that simultaneously affect both [O2•−] and [H2O2], they play dual roles in defending against O2•− toxicity and regulating H2O2-mediated redox signaling. While the bulk of Sod1 is dispensable for protection against O2•− toxicity in yeast, we find that Sod1-mediated peroxide regulation of Yck1 and Yck2 is far more sensitive to fluctuations in Sod1 expression and activity (Fig. 3). However, if anything, logic dictates that Sod1 would have more influence on superoxide scavenging than peroxide-mediated signaling given that it facilitates the production of 1 H2O2 molecule per 2 O2•− molecules. This paradox can be resolved by considering that the biological targets of O2•− toxicity are limited in scope, primarily Fe-S proteins, necessitating that very little Sod1 is required to protect against O2•− damage. In contrast, peroxide-mediated signaling may require large amounts of Sod1 in order to ensure that a sufficient concentration is present in locations proximal to sites of O2•− generation, e.g. NADPH oxidases, so as to provide an adequate flux of H2O2 for the redox control of downstream targets. Directly testing this hypothesis is not trivial due to the technical challenges associated with measuring localized pools of H2O2. Moreover, given that sod1∆ cells exhibit profound metabolic changes, including increased rates of respiration, it is difficult to parse apart contributions arising from the production of H2O2 directly from Sod1 versus various metabolic sources that are affected by Sod1 expression, e.g. electron transport chain.
In light of the fact that vanishingly little Sod1 is required for defense against O2•− toxicity, the primary physiological role of Sod1 as a O2•− scavenger may need to be re-considered in yeast, and potentially other cell types and organisms; its role in redox signaling [11], [50], or non-redox related functions, e.g. Cu buffering [66], [67], [68] or as a transcription factor [33], may account for the function of most Sod1 in cells. An alternative rationale that may account for cells maintaining a high level of Sod1 is that it is required to protect against pathological conditions that transiently increase O2•− burdens. Indeed, Sod1 over-expression can protect against the oxidative stress associated with post-ischemic injury in mouse models [69]. In this context, our results from yeast suggest that in the absence of redox stress and O2•− toxicity, the majority of Sod1 is more vital for functions unrelated to its role in superoxide scavenging. However, when cells are oxidatively stressed, larger amounts of Sod1 that are otherwise utilized for redox signaling or non-redox functions can “moonlight” as a O2•− scavenger.
Another interesting outcome of our study is that Sod1 localized to the IMS is sufficient to protect against cell-wide markers of O2•− toxicity in yeast. This result suggests that different pools of Sod1 may have very different physiological functions. For instance, IMS-localized Sod1 may be critical for protection against O2•− toxicity whereas extra-mitochondrial Sod1 may be more important for non-O2•− scavenging related functions, such as mediating H2O2-based redox signaling or acting as a transcription factor. The dual roles of Sod1 in redox signaling and protecting against O2•− toxicity in different locales may necessitate the existence of mechanisms to dynamically regulate the localization and/or activity/function of Sod1. Indeed, in human cell lines, it was recently found that acetylation of Sod1 at K122, which is in-part regulated by SIRT5, regulates the partitioning of Sod1 between the cytosol and mitochondrial IMS, which in-turn affects respiratory vs. fermentative energy metabolism [70]. In yeast and human cell lines, it was found that in response to H2O2, the cell cycle checkpoint regulating Mec1/ATM effector Dun1/Cds1 kinase phosphorylates Sod1 at S60 and S99 to trigger its nuclear import to regulate gene transcription [33]. In addition, it was recently demonstrated that Sod1 is reversibly phosphorylated at S39 in yeast or T40 in human cell lines by the nutrient sensing mTORC1 to regulate redox homeostasis and adaptation to changes in nutrient availability [71].
Why can IMS-localized Sod1 protect against cell-wide markers of O2•−-toxicity? First, our data suggests that the source of matrix and extra-mitochondrial O2•− originates in the IMS. How then can O2•− in the IMS damage biomolecules in other compartments, including the mitochondrial matrix cytosol, vacuole, and nucleus? One possibility is that O2•− or HO2• in the IMS diffuses into the matrix and cytosol, possibly through membrane channels [72], damaging Fe-S enzymes that reside in these locations [58]. The liberated labile Fe can then be bound and trafficked in a manner that enables it to catalyze deleterious redox reactions throughout the cell. Our results support prior observations that ROS derived from Complex III exerts its influence on extra-mitochondrial targets [73], [74].
Sod1 catalyzed O2•− disproportionation is paramount to its biological function. However, herein we propose that the majority of Sod1 in yeast, and possibly other cell types, is more vital as a source of H2O2 for redox signaling, and potentially other non-redox functions, than for scavenging O2•−. The concentration and spatio-temporal distribution of O2•− encountered in various cells and organisms in vivo are still not well understood and will further define the relative roles of Sod1 in redox signaling and oxidative stress protection. Moreover, the identification of proteome-wide redox-targets of Sod1-derived H2O2 will be a new frontier in the cell biology of Sod1. Our results will have implications for understanding the basic redox biology of Sod1 and better inform the treatment of diseases in which Sod1 or redox homeostasis can be targeted, including certain cancers [11], [75], neurodegenerative disorders, e.g. amyotrophic lateral sclerosis (ALS) [76], [77], and aging [78], [79].
4. Methods
4.1. Chemicals, media components, and immunological reagents
Dihydroethidium (Cat. # 50-850-563), FM-4-64 (Cat # T-3166), and Phen Green SK diacetate (Cat. # P-14313) was purchased from Thermo Fisher Scientific. Paraquat dichloride (Cat # 856177-1G) was purchased from Sigma-Aldrich. Yeast nitrogen base and SC dropout mixtures were purchased from Sunrise Science Products. The Yeast Mitochondria Isolation kit was purchased from Bio Vision (Cat. # K259-50). Rabbit polyclonal antibodies against GAPDH (Cat. # 89348-232) and GFP (Cat. # 89362-978) were purchased from VWR. Rabbit polyclonal antibody against PGK1 (Cat. #PA528612) and mouse monoclonal antibody against Porin (Cat. #459500) were purchased from Thermo Fisher and Invitrogen, respectively. A previously described rabbit polyclonal antibody against Sod1 was obtained from the laboratory of Valeria Culotta (Johns Hopkins University) [50].
4.2. Yeast strains, plasmids, and growth
S. cerevisiae strains used in this study were derived from BY4741 (MATa, his3∆1, leu2∆0, met15∆0, ura3∆0). sod1::LEU2 and sod1::kanMX4 strains were generated by knocking out SOD1 using the previously described deletion plasmids, pKS1 [67] and pJAB002 [80].
The GAL1 driven SOD1 expression plasmid (pAR1026) was constructed by PCR amplification of the SOD1 open reading frame from BY4741 genomic DNA with primers that introduced flanking 5′ and 3′ SpeI and BamHI sites, respectively. The SOD1 amplicon was sub-cloned into the SpeI and BamHI sites of pRS316-GAL1 [81] to generate pAR1026. The TEF1-driven GFP-Yck1 expression construct (pAR113) was previously reported [50]. Expression constructs for wild type SOD1 (pRS415-SOD1) and IMS localized SCO2-SOD1 (pRS415-SCO2-SOD1), which are both driven by the native SOD1 promoter, were previously described and were provided by the laboratory of Professor Dennis Thiele (Duke University) [62].
Yeast transformations were performed by the lithium acetate procedure [82]. Strains were maintained at 30 °C on either enriched yeast extract-peptone based medium supplemented with 2% glucose (YPD), or synthetic complete medium (SC) supplemented with 2% glucose and the appropriate drop-out mixture to maintain selection. For all experiments, cells were streaked from − 80 °C glycerol stocks onto solid agar media plates and pre-cultured in an anaerobic chamber (Coy laboratories) maintained with an atmosphere of 95% N2 and 5% H2. Anaerobically grown cells required supplementing YPD or SC media with 15 mg/L of ergosterol and 0.5% Tween-80 (YPDE or SCE, respectively) [83].
For experiments involving the titration of SOD1 using the GAL1 driven SOD1 expression plasmid, pAR1026, cells were cultured aerobically in SC-URA, with 2% raffinose and the indicated galactose concentrations. For typical experiments involving the IMS-targeted SCO2-SOD1 expression plasmid, cells were cultured aerobically in SC-LEU, with 2% raffinose. In all cases, cells were seeded at an OD600 nm ~ .01 and cultured for 14–17 h to a density of OD600nm ~ 1.0 at 30 °C in a shaking incubator (220 RPM). Following growth, cells were processed as described below for immunoblotting, enzyme assays, EPR spectroscopy, or measurements of labile Fe, superoxide, DNA damage, or vacuolar fragmentation. For all experiments, Sod1 activity and/or expression was assessed as described below. All experiments were conducted using biological replicates arising from duplicate or triplicate independent cultures of multiple clones. While the data reported in the figures reflect biological replicates from single experimental trials, all of the data has been re-produced on multiple occasions in independent experimental trials.
4.3. Cell fractionation
Mitochondria were isolated using the Yeast Mitochondria Isolation Kit (Bio Vision) according to the manufacturer's specifications. For this purpose, sod1∆ cells expressing an empty vector (pRS415), SOD1 (pRS415-SOD1) or IMS localized SCO2-SOD1 (pRS415-SCO2-SOD1) were grown in 20 mL, SC-2% glucose cultures to a density of OD600 nm = 1.0. 4 × 108 cells of each strain were harvested and washed in ice-cold ultra pure water prior to fractionation. After fractionation, volumes corresponding to 2.5% of the whole cell extract and cytosolic fractions, and 10% of the mitochondrial fraction were assessed for Pgk1, Sod1, and Porin expression by SDS-PAGE and immunoblotting. Pgk1 and Sod1 were probed with anti-PGK1 (1:1000) and anti-SOD1 (1:5000) polyclonal antibodies and detected using a goat anti-rabbit secondary antibody conjugated to a 680 nm emitting fluorophore (Biotium). Porin was probed with an anti-Porin (1:5000) monoclonal antibody and detected using a goat anti-mouse secondary antibody conjugated to a 797 nm emitting fluorophore (Thermo Fisher).
4.4. Immunoblotting
~ 2 × 108 cells were harvested, washed in ice-cold Milli-Q water, and lysed in two pellet volumes of lysis buffer (10 mM sodium phosphate, 50 mM sodium chloride, 5 mM EDTA, 1.0% Triton X-100, 1 mM PMSF and a protease inhibitor cocktail (GBiosciences) as described previously [84]. Lysis was achieved at 4 °C using one pellet volume of zirconium oxide beads and a bead beater (Bullet Blender, Next Advance) on a setting of 8 for 3 min [84]. Lysate protein concentrations were determined by the Bradford method (Bio-rad) and 14% tris-glycine gels (Invitrogen) were employed for SDS-PAGE [84]. Anti-GFP (1:4000), anti-GAPDH (1:4000), or anti-Sod1 (1:5000) polyclonal antibodies and a goat anti-rabbit secondary antibody conjugated to a 680 nm emitting fluorophore (Biotium) were used to probe for GFP-Yck1, GAPDH, or Sod1, respectively. All gels were imaged on a LiCOR Odyssey Infrared imager [50], [55], [84].
4.5. Enzyme assays
SOD activity analysis was carried out by native PAGE and nitroblue tetrazolium staining as described previously [50], [85], [86] on exponential phase cultures grown to a final OD600 nm = 1.0 in SC, 2% raffinose media containing the indicated concentration of galactose. Yeast cells were washed with ultra pure H2O, resuspended in lysis buffer and lysed as described in the section on immunblotting. Protein samples (~ 10–30 μg) were separated in 14% native PAGE gels. Sod1 activity was visualized by staining gels with 2.43 mM nitro blue tetrazolium chloride (Sigma), 0.14 M riboflavin-50-phosphate (Sigma) and 28 mM TEMED (Bio-Rad) for 60 min at room temperature in darkness. To visualize Sod1 activity, gels were rinsed with water twice and exposed to light.
For aconitase (Aco1p) and isopropylmalate isomerase (Leu1p) activity assays, cells were subjected to ZrO bead lysis in 50 mM MES, 100 mM KCl, 0.1% Triton X-100, pH 7.0 under a nitrogen atmosphere in a COY chamber. Aco1p and Leu1p activity was determined spectrophotometrically as described previously [55], [58] using a Biotek Synergy Mx multi-modal plate. The assay mixture contained 50–300 μg of lysate protein in 200 μL of a buffer containing 50 mM tris(hydroxymethyl) aminomethane (Tris)-HCl, pH 7.4, and 100 mM NaCl and supplemented with either 0.5 mM cis-aconitate (Aco1 activity) or 0.5 mM citraconitate (Leu1 activity). Activities were determined by monitoring the disappearance of cis-aconitate (Aco1p) or citraconitate (Leu1p) at 240 or 235 nm, respectively, over the course of 5 min.
4.6. Superoxide measurements
Superoxide levels were measured by monitoring the fluorescence of DHE stained cells (λex = 485 nm, λem = 620 nm) similarly to what was described previously [50], [57]. Briefly, 1 × 107 cells were harvested from duplicate or triplicate cultures, resuspended and incubated in 500 μL of fresh media containing 50 μM DHE for 20 min in the dark, washed twice with PBS solution, and fluorescence recorded in a Biotek Synergy Mx multi-modal plate reader.
4.7. Detection of labile Fe using EPR spectroscopy
EPR detection of labile Fe in yeast was accomplished as described previously [45], [48], [87], but with the following modifications. 50 mL cultures of sod1∆ cells expressing prGAL-SOD1 seeded at a density of OD600 nm = .01 were grown in 250 mL Erlenmeyer flasks containing SC, 2% raffinose media with the indicated galactose concentration. Cultures were grown for 16 h to an OD600 nm ~ 1. Cells were washed 2x with 10 mL of cold ultrapure H2O and 1x with 10 mL cold 20 mM Tris-Cl, pH 7.4 on ice. Finally, the cells were resuspended in 500 μL of cold 20 mM Tris-Cl, pH 7.4, containing 10% glycerol and transferred into an EPR tube. The sample was flash frozen in liquid N2 and stored at − 80 °C until EPR measurements were performed. Spectra were recorded with a Bruker EMX X-band spectrometer equipped with an ESR900 continuous flow cryostat (Oxford Instruments, Concord, MA) at 70 K. The parameters for EPR were as follows: center field, 1560G; sweep width, 500 G; frequency 9.45 GHz; microwave power, 31 mW; attenuation, 10 dB; modulation amplitude, 20 G; modulation frequency, 100 kHz; receiver gain, 2·105; sweep time, 20.97 s; time constant, 81.92 ms; resolution, 2048 points; number of scans, 16. Fe(III) desferrioxamine (DFO) standards were prepared over a range of concentrations in 20 mM Tris HCl, 1 mM DFO, 10% glycerol, pH 7.4. The Fe(III) signal at g = 4.3 was analyzed with the Xenon software (Bruker) and used for quantitation of EPR-detectable iron levels. Calculation of cellular EPR-detectable Fe(III) was performed as described previously [87].
4.8. Detection of labile Fe with Phen Green SK
Labile Fe was detected as described previously using Phen Green SK (λex = 488 nm, λem = 530 nm), a fluorescent probe for divalent metals that is quenched upon Fe2+ binding [88], [89], [90]. 5 × 107 cells were resuspended in 300 μL of phosphate buffered saline (PBS). The cell suspension was incubated with 3 μL of a 2 mM Phen Green SK DMSO stock solution at 30 °C for 15 min in the dark. Cells were then washed with PBS, and split into 3 × 100 μL aliquots, and were treated with 1 μL of H2O, 1 μL of a 200 mM aqueous stock solution of 1,10-phenantroline, a ferrous iron chelator, or treated with 1 μL of a 10 mM aqueous stock solution of ferrous ammonium sulfate, and incubated in the dark for 10 min, prior to recording fluorescence. Phen Green fluorescence was recorded in a Biotek Synergy Mx multi-modal plate reader. After subtracting the background fluorescence of unlabeled cells, the percentage of Phen Green bound to Fe2+ (% Bound) was calculated using the following formula:
where, F is Phen Green fluorescence intensity in the test sample, Fmin is the Phen Green fluorescence intensity when it is not bound to Fe, and Fmax is the Phen Green fluorescence intensity when it is saturated with Fe. Fmin is determined by recording Phen Green fluorescence in cells incubated with the iron chelator, 1,10-phenantroline. Fmax is determined by recording Phen Green fluorescence in cells incubated with ferrous ammonium sulfate.
4.9. Vacuolar fragmentation
Vacuolar fragmentation was assessed as previously described [45]. Briefly, 2 × 107 cells were resuspended in 50 μL of fresh growth media, typically SC media with 2% raffinose and appropriate galactose concentration. The cell suspension was incubated with 1 μL of a 2 mM DMSO stock solution of FM4–64 at 30 °C for 20 min in the dark. The cells were then pelleted, washed with fresh media, and resuspended in 5 mL of fresh SC media with 2% raffinose and appropriate galactose concentration. The cells were cultured for an additional 1.5 h at 30 °C, shaking at 220 RPM in the dark. The cells were then washed with PBS and resuspended in PBS to a density of 2 × 108 cells mL−1. 3 μL of the cell suspension were placed on a glass slide and imaged using a Zeiss LSM 700 microscope equipped with a 63×, 1.4 numerical aperture oil objective, using the using the Texas Red channel. Approximately ~ 100 cells were counted from each culture condition in triplicate and scored for having either single vacuoles or multiple fragmented vacuoles as indicated in Fig. 1j.
4.10. TUNEL assays
TUNEL assays were conducted as previously described [33]. Yeast cells were fixed in 4% p-formaldehyde at room temperature for 30 min. The cells were then washed three times with PBS. The cell pellet was then re-suspended in PBS and digested with 300 μg/mL of Zymolyase 100 T at 37 °C for 60 min. After 60 min, 10 μL of the cell suspension was applied to a clean glass slide and dried at 37 °C for 30 min. The slides were rinsed with PBS and incubated in a permeabilization solution (0.1% Triton X-100% and 0.1% sodium citrate) on ice for 2 min. The slides were then rinsed twice with PBS. The TUNEL reaction mixture (50 μL of enzyme solution and 450 μL of Label solution; In Situ Cell Death Detection Kit, Roche Diagnostics) was applied to the slides and incubated in the dark for 60 min. The cells labeled with fluorescein-dUTP were imaged using a Zeiss LSM 700 microscope equipped with a 63×, 1.4 numerical aperture oil objective. Approximately ~ 100 cells were counted from each culture condition in triplicate and scored for being either non-fluorescent or fluorescent using the FITC channel.
Acknowledgements
We thank Professor Valeria Culotta (Johns Hopkins University) for the Sod1 antibody and Professor Dennis Thiele (Duke University) for the Sco2-Sod1 expression constructs. We acknowledge support from the U.S. National Institutes of Health (GM118744 to A.R.R. and GM112919 to F.W.O.), the Blanchard Professorship (to A.R.R.) and start-up funding from the Georgia Institute of Technology (to A.R.R.).
References
- 1.Flint D.H., Tuminello j.F., Emptage M.H. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J. Biol. Chem. 1993;268:22369–22376. [PubMed] [Google Scholar]
- 2.Liochev S.I., Fridovich I. Superoxide and iron: partners in crime. IUBMB Life. 1999;48:157–161. doi: 10.1080/713803492. [DOI] [PubMed] [Google Scholar]
- 3.Imlay J.A. Pathways of oxidative damage. Annu. Rev. Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y., Branicky R., Noe A., Hekimi S. Superoxide dismutases: dual roles in controlling ros damage and regulating ros signaling. J. Cell Biol. 2018;217:1915–1928. doi: 10.1083/jcb.201708007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilinski T., Krawiec Z., Liczmanski L., Litwinska J. Is hydroxyl radical generated by the fenton reaction in vivo? Biochem. Biophys. Res. Commun. 1985;130:533–539. doi: 10.1016/0006-291x(85)90449-8. [DOI] [PubMed] [Google Scholar]
- 6.Forman H.J., Maiorino M., Ursini F. Signaling functions of reactive oxygen species. Biochemistry. 2010;49:835–842. doi: 10.1021/bi9020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veal E.A., Day A.M., Morgan B.A. Hydrogen peroxide sensing and signaling. Mol. Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 8.Veal E., Day A. Hydrogen peroxide as a signaling molecule. Antioxid. Redox Signal. 2011;15:147–151. doi: 10.1089/ars.2011.3968. [DOI] [PubMed] [Google Scholar]
- 9.Meng T.C., Fukada T., Tonks N.K. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol. Cell. 2002;9:387–399. doi: 10.1016/s1097-2765(02)00445-8. [DOI] [PubMed] [Google Scholar]
- 10.Denu J.M., Tanner K.G. Specific and reversible inactivation of protein tyrosine phosphatases by hydrogen peroxide: evidence for a sulfenic acid intermediate and implications for redox regulation. Biochemistry. 1998;37:5633–5642. doi: 10.1021/bi973035t. [DOI] [PubMed] [Google Scholar]
- 11.Juarez J.C., Manuia M., Burnett M.E., Betancourt O., Boivin B., Shaw D.E., Tonks N.K., Mazar A.P., Donate F. Superoxide dismutase 1 (Sod1) is essential for H2O2-mediated oxidation and inactivation of phosphatases in growth factor signaling. Proc. Natl. Acad. Sci. USA. 2008;105:7147–7152. doi: 10.1073/pnas.0709451105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paulsen C.E., Truong T.H., Garcia F.J., Homann A., Gupta V., Leonard S.E., Carroll K.S. Peroxide-dependent sulfenylation of the Egfr catalytic site enhances kinase activity. Nat. Chem. Biol. 2011;8:57–64. doi: 10.1038/nchembio.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keyes J.D., Parsonage D., Yammani R.D., Rogers L.C., Kesty C., Furdui C.M., Nelson K.J., Poole L.B. Endogenous, regulatory cysteine sulfenylation of Erk kinases in response to proliferative signals. Free Radic. Biol. Med. 2017;112:534–543. doi: 10.1016/j.freeradbiomed.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peralta D., Bronowska A.K., Morgan B., Doka E., Van Laer K., Nagy P., Grater F., Dick T.P. A proton relay enhances H2O2 sensitivity of gapdh to facilitate metabolic adaptation. Nat. Chem. Biol. 2015;11:156–163. doi: 10.1038/nchembio.1720. [DOI] [PubMed] [Google Scholar]
- 15.Dinkova-Kostova A.T., Holtzclaw W.D., Cole R.N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., Talalay P. Direct evidence that sulfhydryl groups of keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang D.D., Lo S.C., Cross J.V., Templeton D.J., Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Block K., Gorin Y. Aiding and abetting roles of nox oxidases in cellular transformation. Nat. Rev. Cancer. 2012;12:627–637. doi: 10.1038/nrc3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shanmugasundaram K., Nayak B.K., Friedrichs W.E., Kaushik D., Rodriguez R., Block K. Nox4 functions as a mitochondrial energetic sensor coupling cancer metabolic reprogramming to drug resistance. Nat. Commun. 2017;8:997. doi: 10.1038/s41467-017-01106-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller F.L., Liu Y., Van Remmen H. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J. Biol. Chem. 2004;279:49064–49073. doi: 10.1074/jbc.M407715200. [DOI] [PubMed] [Google Scholar]
- 20.Muller F.L., Liu Y., Abdul-Ghani M.A., Lustgarten M.S., Bhattacharya A., Jang Y.C., Van Remmen H. High rates of superoxide production in skeletal-muscle mitochondria respiring on both complex I- and complex II-linked substrates. Biochem. J. 2008;409:491–499. doi: 10.1042/BJ20071162. [DOI] [PubMed] [Google Scholar]
- 21.Lustgarten M.S., Bhattacharya A., Muller F.L., Jang Y.C., Shimizu T., Shirasawa T., Richardson A., Van Remmen H. Complex I generated, mitochondrial matrix-directed superoxide is released from the mitochondria through voltage dependent anion channels. Biochem.Biophys. Res Commun. 2012;422:515–521. doi: 10.1016/j.bbrc.2012.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ichikawa M., Nishino T., Nishino T., Ichikawa A. Subcellular localization of xanthine oxidase in rat hepatocytes: high-resolution immunoelectron microscopic study combined with biochemical analysis. J. Histochem. Cytochem. 1992;40:1097–1103. doi: 10.1177/40.8.1619276. [DOI] [PubMed] [Google Scholar]
- 23.Jarasch E.D., Grund C., Bruder G., Heid H.W., Keenan T.W., Franke W.W. Localization of xanthine oxidase in mammary-gland epithelium and capillary endothelium. Cell. 1981;25:67–82. doi: 10.1016/0092-8674(81)90232-4. [DOI] [PubMed] [Google Scholar]
- 24.Pizzinat N., Copin N., Vindis C., Parini A., Cambon C. Reactive oxygen species production by monoamine oxidases in intact cells. Naunyn Schmiede. Arch. Pharmacol. 1999;359:428–431. doi: 10.1007/pl00005371. [DOI] [PubMed] [Google Scholar]
- 25.Hrycay E.G., Bandiera S.M. Involvement of cytochrome P450 in reactive oxygen species formation and cancer. Adv. Pharmacol. 2015;74:35–84. doi: 10.1016/bs.apha.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Wallace W.J., Houtchens R.A., Maxwell J.C., Caughey W.S. Mechanism of autooxidation for hemoglobins and myoglobins. promotion of superoxide production by protons and anions. J. Biol. Chem. 1982;257:4966–4977. [PubMed] [Google Scholar]
- 27.De Henau S., Tilleman L., Vangheel M., Luyckx E., Trashin S., Pauwels M., Germani F., Vlaeminck C., Vanfleteren J.R., Bert W., Pesce A., Nardini M., Bolognesi M., De Wael K., Moens L., Dewilde S., Braeckman B.P. A redox signalling globin is essential for reproduction in Caenorhabditis elegans. Nat. Commun. 2015;6:8782. doi: 10.1038/ncomms9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lei X.G., Zhu J.H., Cheng W.H., Bao Y., Ho Y.S., Reddi A.R., Holmgren A., Arner E.S. Paradoxical roles of antioxidant enzymes: basic mechanisms and health implications. Physiol. Rev. 2016;96:307–364. doi: 10.1152/physrev.00010.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisiger R.A., Fridovich I. Mitochondrial superoxide dismutase. J. Biol. Chem. 1973;248:4793–4796. [PubMed] [Google Scholar]
- 30.McCord J.M., Fridovich I. Superoxide dismutase: an enzymic function for Erythrocuprein (Hemocuprein) J. Biol. Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 31.Okado-Matsumoto A., Fridovich I. Subcellular distribution of superoxide dismutase (Sod) in rat liver. Cu/Zn sod in mitochondria. J. Biol. Chem. 2001;276:38388–38393. doi: 10.1074/jbc.M105395200. [DOI] [PubMed] [Google Scholar]
- 32.Sturtz L.A., Diekert K., Jensen L.T., Lill R., Culotta V.C. A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, Ccs, localize to the ntermembrane space of mitochondria. J. Biol. Chem. 2001;276:38084–38089. doi: 10.1074/jbc.M105296200. [DOI] [PubMed] [Google Scholar]
- 33.Tsang C.K., L., Y, Thomas J., Zhang Y., Zheng X.F. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat. Commun. 2014;5:3446. doi: 10.1038/ncomms4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeuchi H., Kobayashi Y., Ishigaki S., Doyu M., Sobue G. Mitochondrial localization of mutant superoxide dismutase 1 triggers caspase-dependent cell death in a cellular model of familial amyotrophic lateral sclerosis. J. Biol. Chem. 2002;277:50966–50972. doi: 10.1074/jbc.M209356200. [DOI] [PubMed] [Google Scholar]
- 35.Islinger M., Li K.W., Seitz J., Volkl A., Luers G.H. Hitchhiking of Cu/Zn superoxide dismutase to peroxisomes–evidence for a natural piggyback import mechanism in mammals. Traffic. 2009;10:1711–1721. doi: 10.1111/j.1600-0854.2009.00966.x. [DOI] [PubMed] [Google Scholar]
- 36.Elchuri S., Oberley T.D., Qi W., Eisenstein R.S., Jackson Roberts L., Van Remmen H., Epstein C.J., Huang T.T. Cuznsod deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene. 2005;24:367–380. doi: 10.1038/sj.onc.1208207. [DOI] [PubMed] [Google Scholar]
- 37.Jackson M.J. Lack of cuznsod activity: a pointer to the mechanisms underlying age-related loss of muscle function, a commentary on "absence of cuzn superoxide dismutase leads to elevated oxidative stress and acceleration of age-dependent skeletal muscle atrophy". Free. Radic. Biol. Med. 2006;40:1900–1902. doi: 10.1016/j.freeradbiomed.2006.02.022. [DOI] [PubMed] [Google Scholar]
- 38.Muller F.L., Song W., Liu Y., Chaudhuri A., Pieke-Dahl S., Strong R., Huang T.T., Epstein C.J., Roberts L.J., 2nd, Csete M., Faulkner J.A., Van Remmen H. Absence of cuzn superoxide dismutase leads to elevated oxidative stress and acceleration of age-dependent skeletal muscle atrophy. Free Radic. Biol. Med. 2006;40:1993–2004. doi: 10.1016/j.freeradbiomed.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 39.Jang Y.C., Lustgarten M.S., Liu Y., Muller F.L., Bhattacharya A., Liang H., Salmon A.B., Brooks S.V., Larkin L., Hayworth C.R., Richardson A., Van Remmen H. Increased superoxide in vivo accelerates age-associated muscle atrophy through mitochondrial dysfunction and neuromuscular junction degeneration. FASEB J. 2010;24:1376–1390. doi: 10.1096/fj.09-146308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sentman M.L., Granstrom M., Jakobson H., Reaume A., Basu S., Marklund S.L. Phenotypes of mice lacking extracellular superoxide dismutase and copper- and zinc-containing superoxide dismutase. J. Biol. Chem. 2006;281:6904–6909. doi: 10.1074/jbc.M510764200. [DOI] [PubMed] [Google Scholar]
- 41.Reaume A.G., Elliott J.L., Hoffman E.K., Kowall N.W., Ferrante R.J., Siwek D.F., Wilcox H.M., Flood D.G., Beal M.F., Jr., R.H.B, Scott R.W., Snider W.D. Motor neurons in Cu/Zn-sod superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat. Genet. 1996;13:43–47. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 42.Phillips J., Campbell S., Michard D., Charbonneau M., Hilliker A. Null mutations of copper/zinc superoxide in drosophila confer hypersensitivity to paraquat and reduced longevity. Proc. Natl. Acad. Sci. USA. 1989;83:3820–3824. doi: 10.1073/pnas.86.8.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang E., Kosman D. O2-dependent methionine auxotrophy in Cu,Zn superoxide dismutase deficient mutants of Saccharomyces cerevisiae. J. Bacteriol. 1990;172:1840–1845. doi: 10.1128/jb.172.4.1840-1845.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slekar K.H., Kosman D., Culotta V.C. The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J. Biol. Chem. 1996;271:28831–28836. doi: 10.1074/jbc.271.46.28831. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez R.J., Srinivasan C., Munroe W.H., Wallace M.A., Martins J., Kao T.Y., Le K., Gralla E.B., Valentine J.S. Exogenous manganous ion at millimolar levels rescues all known dioxygen-sensitive phenotypes of yeast lacking cuznsod. J. Biol. Inorg. Chem. 2005;10:913–923. doi: 10.1007/s00775-005-0044-y. [DOI] [PubMed] [Google Scholar]
- 46.Corson L.B., Folmer J., Strain J.S., Culotta V.C., Cleveland D.W. Oxidative stress and iron are implicated in fragmenting vacuoles of Saccharomyces cerevisiae lacking Cu,Zn superoxide dismtuase. J. Biol. Chem. 1999;274:27590–27596. doi: 10.1074/jbc.274.39.27590. [DOI] [PubMed] [Google Scholar]
- 47.Freitas J.M.D., Liba A., Meneghini R., Valentine J.S., Gralla E.B. Yeast lacking Cu-Zn superoxide dismutase show altered iron homeostasis. role of oxidative stress in iron metabolism. J. Biol. Chem. 2000;275:11645–11649. doi: 10.1074/jbc.275.16.11645. [DOI] [PubMed] [Google Scholar]
- 48.Srinivasan S.C., Liba A., Imlay J.A., Valentine J.S., Gralla E.B. Yeast lacking superoxide dismutase(s) show elevated levels of "free iron" as measured by whole cell electron paramagnetic resonance. J. Biol. Chem. 2000;275:29187–29192. doi: 10.1074/jbc.M004239200. [DOI] [PubMed] [Google Scholar]
- 49.Sakanyan V., Hulin P., Alves de Sousa R., Silva V.A., Hambardzumyan A., Nedellec S., Tomasoni C., Loge C., Pineau C., Roussakis C., Fleury F., Artaud I. Activation of EGFR by small compounds through coupling the generation of hydrogen peroxide to stable dimerization of Cu/Zn Sod1. Sci. Rep. 2016;6:21088. doi: 10.1038/srep21088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reddi A.R., Culotta V.C. Sod1 integrates signals from oxygen and glucose to repress respiration. Cell. 2013;152:224–235. doi: 10.1016/j.cell.2012.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Halliwell B., Gutteridge J.M.C. Free Radicals in Biology and Medicine. 4 ed. Oxford Biosciences; 2007. [Google Scholar]
- 52.Pardo C.A., Xu Z., Borchelt D.R., Price D.L., Sisodia S.S. Superoxide dismutase is an abundant component in cell bodies, dendrites, and axons of motor neurons and in a subset of other neurons. Proc. Natl. Acad. Sci. USA. 1995;92:954–958. doi: 10.1073/pnas.92.4.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O'Shea E.K., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 54.Kulak N.A., Pichler G., Paron I., Nagaraj N., Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 55.Reddi A.R., Culotta V.C. Regulation of manganese antioxidants by nutrient sensing pathways in Saccharomyces cerevisiae. Genetics. 2011;189:1261–1270. doi: 10.1534/genetics.111.134007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klug D., Rabani J., Fridovich I. A drect demonstration of the catalytic action of superoxide dismutase through the use of pulse radiolysis. J. Biol. Chem. 1972;247:4839–4842. [PubMed] [Google Scholar]
- 57.Neklesa T.K., Davis R.W. Superoxide anions regulate Torc1 and its ability to bind Fpr1: rapamycin complex. Proc. Natl. Acad. Sci. USA. 2008;105:15166–15171. doi: 10.1073/pnas.0807712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wallace M.A., Liou L.L., Martins J., Clement M.H., Bailey S., Longo V.D., Valentine J.S., Gralla E.B. Superoxide inhibits 4fe-4s cluster enzymes involved in amino acid biosynthesis. Cross-compartment protection by cuzn-superoxide dismutase. J. Biol. Chem. 2004;279:32055–32062. doi: 10.1074/jbc.M403590200. [DOI] [PubMed] [Google Scholar]
- 59.Longo V.D., Gralla E.B., Valentine J.S. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae: mitochondrial production of toxic oxygen species in vivo. J. Biol. Chem. 1996;271:12275–12280. doi: 10.1074/jbc.271.21.12275. [DOI] [PubMed] [Google Scholar]
- 60.Sehati S., Clement M.H., Martins J., Xu L., Longo V.D., Valentine J.S., Gralla E.B. Metabolic alterations in yeast lacking copper-zinc superoxide dismutase. Free Radic. Biol. Med. 2011;50:1591–1598. doi: 10.1016/j.freeradbiomed.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zielonka J., Vasquez-Vivar J., Kalyanaraman B. Detection of 2-hydroxyethidium in cellular systems: a unique marker product of superoxide and hydroethidine. Nat. Protoc. 2008;3:8–21. doi: 10.1038/nprot.2007.473. [DOI] [PubMed] [Google Scholar]
- 62.Wood L.K., Thiele D.J. Transcriptional activation in yeast in response to copper deficiency involves copper-zinc superoxide dismutase. J. Biol. Chem. 2009;284:404–413. doi: 10.1074/jbc.M807027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rinnerthaler M., Buttner S., Laun P., Heeren G., Felder T.K., Klinger H., Weinberger M., Stolze K., Grousl T., Hasek J., Benada O., Frydlova I., Klocker A., Simon-Nobbe B., Jansko B., Breitenbach-Koller H., Eisenberg T., Gourlay C.W., Madeo F., Burhans W.C., Breitenbach M. Yno1p/Aim14p, a nadph-oxidase ortholog, controls extramitochondrial reactive oxygen species generation, apoptosis, and actin cable formation in yeast. Proc. Natl. Acad. Sci. USA. 2012;109:8658–8663. doi: 10.1073/pnas.1201629109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sawyer D.T., Valentine J.S. How super is superoxide? Acc. Chem. Res. 1981;14:393–400. [Google Scholar]
- 65.Fee J.A. Is superoxide important in oxygen poisoning? Trends Biochem. Sci. 1982;3:84–86. [Google Scholar]
- 66.Corson L.B., Strain J., Culotta V.C., Cleveland D.W. Chaperone-facilitated copper binding Is a property common to several classes of familial amyotrophic lateral sclerosis-linked superoxide dismutase mutants. Proc. Natl. Acad. Sci. USA. 1998;95:6361–6366. doi: 10.1073/pnas.95.11.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Culotta V.C., Joh H.D., Lin S.J., Slekar K.H., Strain J. A physiological role for Saccharomyces cerevisiae copper/zinc superoxide dismutase in copper buffering. J. Biol. Chem. 1995;270:29991–29997. doi: 10.1074/jbc.270.50.29991. [DOI] [PubMed] [Google Scholar]
- 68.Petrovic N., Comi A., Ettinger M.J. Copper incorporation into superoxide dismutase in Menkes lymphoblasts. J. Biol. Chem. 1996;271:28335–28340. doi: 10.1074/jbc.271.45.28335. [DOI] [PubMed] [Google Scholar]
- 69.Wang P., Chen H., Qin H., Sankarapandi S., Becher M.W., Wong P.C., Zweier J.L. Overexpression of human copper, zinc-superoxide dismutase (Sod1) prevents postischemic injury. Proc. Natl. Acad. Sci. USA. 1998;95:4556–4560. doi: 10.1073/pnas.95.8.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Banks C.J., Rodriguez N.W., Gashler K.R., Pandya R.R., Mortenson J.B., Whited M.D., Soderblom E.J., Thompson J.W., Moseley M.A., Reddi A.R., Tessem J.S., Torres M.P., Bikman B.T., Andersen J.L. Acylation of superoxide dismutase 1 (Sod1) at K122 governs sod1-mediated inhibition of mitochondrial respiration. Mol. Cell Biol. 2017;37 doi: 10.1128/MCB.00354-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsang C.K., Chen M., Cheng X., Qi Y., Chen Y., Das I., Li X., Vallat B., Fu L.W., Qian C.N., Wang H.Y., White E., Burley S.K., Zheng X.F.S. Sod1 phosphorylation by Mtorc1 couples nutrient sensing and redox regulation. Mol. Cell. 2018;70:502–515. doi: 10.1016/j.molcel.2018.03.029. (e508) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Han D., Antunes F., Canali R., Rettori D., Cadenas E. Voltage-dependent anion channels control the release of the superoxide anion from mitochondria to cytosol. J. Biol. Chem. 2003;278:5557–5563. doi: 10.1074/jbc.M210269200. [DOI] [PubMed] [Google Scholar]
- 73.Tormos K.V., Anso E., Hamanaka R.B., Eisenbart J., Joseph J., Kalyanaraman B., Chandel N.S. Mitochondrial complex iii ros regulate adipocyte differentiation. Cell Metab. 2011;14:537–544. doi: 10.1016/j.cmet.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hamanaka R.B., Chandel N.S. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem. Sci. 2010;35:505–513. doi: 10.1016/j.tibs.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Glasauer A., Sena L.A., Diebold L.P., Mazar A.P., Chandel N.S. Targeting Sod1 reduces experimental non-small-cell lung cancer. J. Clin. Investig. 2014;124:117–128. doi: 10.1172/JCI71714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rowan A. Sod1 targeting in Als. Nat. Rev. Neurosci. 2004;5:676. [Google Scholar]
- 77.Tokuda E., Furukawa Y. Copper homeostasis as a therapeutic target in amyotrophic lateral sclerosis with Sod1 mutations. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17050636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y., Unnikrishnan A., Deepa S.S., Liu Y., Li Y., Ikeno Y., Sosnowska D., Van Remmen H., Richardson A. A new role for oxidative stress in aging: the accelerated aging phenotype in sod1(-/)(-) mice is correlated to increased cellular senescence. Redox Biol. 2017;11:30–37. doi: 10.1016/j.redox.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Blander G., de Oliveira R.M., Conboy C.M., Haigis M., Guarente L. Superoxide dismutase 1 knock-down induces senescence in human fibroblasts. J. Biol. Chem. 2003;278:38966–38969. doi: 10.1074/jbc.M307146200. [DOI] [PubMed] [Google Scholar]
- 80.Leitch J.M., Li C.X., Baron J.A., Matthews L.M., Cao X., Hart P.J., Culotta V.C. Post-translational modification of Cu/Zn superoxide dismutase under anaerobic conditions. Biochemistry. 2012;51:677–685. doi: 10.1021/bi201353y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chernoff Y.O., Newnam G.P., Kumar J., Allen K., Zink A.D. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone Ssb in formation, stability, and toxicity of the [Psi] prion. Mol. Cell Biol. 1999;19:8103–8112. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gietz R.D., Schiestl R.H. Applications of high efficiency lithium acetate trasformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast. 1991;7:253–263. doi: 10.1002/yea.320070307. [DOI] [PubMed] [Google Scholar]
- 83.Ness F., Achstetter T., Duport C., Karst F., Spagnoli R., Degryse E. Sterol uptake in Saccharomyces cerevisiae heme auxotrophic mutants is affected by ergosterol and oleate but not by almitoleate or by sterol esterification. J. Bacteriol. 1998;180:1913–1919. doi: 10.1128/jb.180.7.1913-1919.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hanna D.A., Harvey R.M., Martinez-Guzman O., Yuan X., Chandrasekharan B., Raju G., Outten F.W., Hamza I., Reddi A.R. Heme dynamics and trafficking factors revealed by genetically encoded fluorescent heme sensors. Proc. Natl. Acad. Sci. USA. 2016;113:7539–7544. doi: 10.1073/pnas.1523802113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Flohe L., Otting F. Superoxide dismutase assays. In: Packer L., editor. Methods in Enzymology: Oxygen Radicals in Biological Systems. Academic press; New York: 1984. pp. 93–104. [DOI] [PubMed] [Google Scholar]
- 86.Luk E., Yang M., Jensen L.T., Bourbonnais Y., Culotta V.C. Manganese activation of superoxide dismutase 2 in the mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:22715–22720. doi: 10.1074/jbc.M504257200. [DOI] [PubMed] [Google Scholar]
- 87.Srinivasan C., Gralla E.B. Measurement of "free" or electron paramagnetic resonance-detectable iron in whole yeast cells as indicator of superoxide stress. Methods Enzymol. 2002;349:173–180. doi: 10.1016/s0076-6879(02)49333-0. [DOI] [PubMed] [Google Scholar]
- 88.Petrat F., Rauen U., de Groot H. Determination of the chelatable iron pool of isolated rat hepatocytes by digital fluorescence microscopy using the fluorescent probe, phen green Sk. Hepatology. 1999;29:1171–1179. doi: 10.1002/hep.510290435. [DOI] [PubMed] [Google Scholar]
- 89.Irazusta V., Moreno-Cermeno A., Cabiscol E., Ros J., Tamarit J. Major targets of iron-induced protein oxidative damage in frataxin-deficient yeasts are magnesium-binding proteins. Free Radic. Biol. Med. 2008;44:1712–1723. doi: 10.1016/j.freeradbiomed.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 90.Fernandez-Castane A., Li H., Thomas O.R.T., Overton T.W. Flow cytometry as a rapid analytical tool to determine physiological responses to changing O2 and iron concentration by magnetospirillum gryphiswaldense strain Msr-1. Sci. Rep. 2017;7:13118. doi: 10.1038/s41598-017-13414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]