Abstract
Interactions between biological membranes and disease-associated amyloids are well documented, and their prevalence suggests that an inherent affinity exists between these molecular assemblies. Our interest in the molecular origins of life have led us to investigate the nature of such interactions in the context of their molecular predecessors (i.e., vesicle-forming amphiphiles and small peptides). Under certain conditions, amyloidogenic peptides or fatty acids are each able to form ordered structures on their own; however, we report here on their cooperative assembly into novel, to our knowledge, highly ordered structures. We first examined an amyloidogenic eight-residue peptide, which forms amyloids at pH 11, yet because of its positive electrostatic character remains soluble at a neutral pH. In mixtures with simple fatty acids, this peptide is also able to form novel, to our knowledge, coaggregates at a neutral pH whose structures are sensitive to both the fatty acid concentration and identity. Below the critical vesicle concentration, the mixtures of fatty acid and peptide yield a flocculent precipitate with an underlying β-structure. Above the critical vesicle concentration, the mixtures yield a translucent precipitate that consists of tube-like structures. Small-angle x-ray scattering and fiber diffraction data were used to model their structures as hollow-core two-shell cylinders in which the inner shell is a bilayer of fatty acid and the outer shell alternates between amyloid and bilayers of fatty acid. The further analysis of decanoic acid with a panel of 13 other basic amyloidogenic peptides confirmed the general nature of the observed interactions. The cooperativity within this heterogeneous system is attributed to the structurally repetitive natures of the fatty acid bilayer and the cross-β-sheet motif, providing compatible scaffolds for attractive electrostatic interactions. We show these interactions to be mutually beneficial, expanding the phase space of both peptides and fatty acids while providing a simple yet robust physical connection between two distinct entities relevant for life.
Introduction
Historically, the “origin of life” field has been polarized by competing and seemingly exclusive hypotheses, such as replication first (1), metabolism first (2), and compartmentalization first (3) or RNA world (4), peptide world (5, 6), and amyloid world (7, 8, 9, 10, 11). It is evident that these hypotheses tend to oversimplify the problem and that at one point in the origin or evolution of early life, the primary molecules of biology, namely nucleic acids, amino acids, aliphatic compounds, and carbohydrates, became intimately intertwined. The emergence of these distinct molecular systems could have been sequential, separated in space, or all at once as it has been postulated recently (12, 13). Physical interactions between the distinct systems may have stabilized them, strengthened their replication potential, and/or enlarged their chemical and structural space in a way that would not be possible with a single type of biomolecule. Such mutually beneficial effects have been shown for DNA and peptide amyloids, for which DNA stabilizes the amyloid fibers, and the amyloid supports DNA hybridization (14). Furthermore, the encapsulation inside of vesicles by amphiphilic membrane-like structures may be beneficial to the encapsulated system by offering protection from hydrolysis and by preventing loss of the system through diffusion, as has been demonstrated for example by the chemical replication of RNA inside fatty acid vesicles (15) and by protein translation within vesicles that contain the 80 different macromolecular species required for translation (16).
It is well known that there is an inherent association of many disease-related amyloidogenic peptides with lipid bilayer membranes (17, 18, 19, 20). Such interactions may arise from the fact that both can form periodic structures at the subnanometer level, thereby enhancing the potential for cooperative interactions. Within the framework of our investigations on the origin of life, we hypothesized that fatty acids and amyloidogenic peptides, both arguably prebiotic entities, could have cooperative interactions that stabilize each other’s structures and thus enhance their potential to undergo Darwinian selection. The sequence space of amyloidogenic peptides and the structure space of their fibers as well as the structure and phase space of fatty acids could be expanded upon by cooperative interactions between peptides and fatty acids. Interactions of this sort in a simple system are demonstrated in the following.
Materials and Methods
Materials
Sodium decanoate (Tokyo Chemical Industry, Tokyo, Japan), sodium dodecanoate (Sigma-Aldrich, St. Louis, MO), nonanoic acid (NA) (Acros Organics, Morris Plains, NJ), decanol (DOH) (Sigma-Aldrich), sodium dodecyl sulfide (Sigma-Aldrich), Triton X-100 (Sigma-Aldrich), hydrochloric acid (Merck, Darmstadt, Germany), sodium hydroxide (VWR Chemicals, Leuven, Belgium), potassium phosphate monobasic (Acros Organics), sodium phosphate dibasic dodecahydrate (Acros Organics), sodium metaborate (Sigma-Aldrich), citric acid (Sigma-Aldrich), acetonitrile high-performance liquid chromatography (HPLC) gradient grade (Fisher Chemical, Loughborough, UK), trifluoroacetic acid (Fisher Chemical), proteinase K from Tritirachium album (Sigma-Aldrich), and phenylmethylsulfonyl fluoride (Biochemica, Darmstadt, Germany) were used without further purification.
The peptides (GL Biochem Ltd., Shanghai, China) were synthesized using standard fluorenylmethyloxycarbonyl chemistry on a rink amide resin. The crude peptides were purified to >98% by reverse-phase HPLC on a Kinetex C18 5-μm 100-Å 250 × 10 mm column (Phenomenex, Torrance, CA) in a gradient of acetonitrile in 0.1% trifluoroacetic acid (TFA). The purified peptides were quantitated by their calculated extinction coefficient at 214 nm (21). Pure peptides were stored in lyophilized aliquots and dissolved in water just before use.
Fatty acid solutions
Decanoate-decanoic acid (DA) and dodecanoate-dodecanoic acid (DDA) solutions were prepared by dissolving the corresponding sodium salts in phosphate buffer at room temperature or at 50°C for decanoate or dodecanoate, respectively, and adjusting the pH of each solution to the desired pH with 1 M HCl, using a SevenCompact Mettler Toledo (Columbus, OH) pH meter fitted with a Hamilton BioTrode (Reno, NV). After each addition of the pH titrant, the solutions were vortexed for 30 s to allow the solution to equilibrate before continuing the titration. After titration, the volume of the solution was adjusted to the desired concentration of fatty acid while keeping the concentration of phosphate constant at 50 mM. Nonanoate-NA solutions were prepared at room temperature by mixing NA in a molar ratio of 1:1 with sodium hydroxide and a volume of 0.5 M phosphate buffer equal to 0.1 of the final solution volume. Titration with HCl was carried out as for DA and DDA solutions. To prepare DA/DOH mixture, DOH was added to a sodium-decanoate solution and vortexed for 1 min. Subsequently, titration and volume adjustment were done as for the other solutions.
Amyloid formation
Peptides were dissolved in water and then diluted to 0.5 mg/mL in buffer to induce aggregation. The aggregation of VOVOVOVO-NH2 was induced in 50 mM borate buffer at pH 11, DFRFRF-NH2 was induced in 50 mM phosphate buffer at pH 7.8, and Ac-VDVDVDVDV-NH2 (V(DV)4) was induced in 50 mM sodium citrate buffer at pH 3.5. The samples were incubated at room temperature without agitation and analyzed after more than 24 h unless otherwise noted.
Formation of peptide-fatty acid mixtures
Directly before use, aliquots of lyophilized peptides were dissolved in water at 5 mg/mL and then added to a fatty acid solution in a 1:9 volume ratio to give a final peptide concentration of 0.5 mg/mL. After peptide addition, the solutions were vortexed for 30 s and then incubated overnight at room temperature for NA and DA solutions or at 50°C for DDA solutions.
CD
Peptide-fatty acid mixtures were analyzed by circular dichroism (CD) spectroscopy on a Jasco J-815 (Easton, MD) in a 0.1-mm quartz cell. The spectra were recorded from 260 to 190 nm at 50 nm/min, 2-nm band-pass, with a 2-s integration, and averaged over five repetitions. Measurements were done at room temperature for NA and DA solutions and at 50°C for DDA solutions.
CMCs and CVCs
We employed four different methods to estimate the fatty acid critical micelle concentration (CMC) and critical vesicle concentration (CVC) under the conditions used in this study. The results of these measurements are in the Supporting Materials and Methods and are summarized in Table 1.
-
1)
The fluorescence intensity of hydrophobic dyes as well as their fluorescence polarization can be used to measure the phase transitions of amphiphiles (22, 23). We chose 1,6-diphenyl-1,3,5-hexatriene (DPH) as a reporter molecule because both its fluorescence intensity and polarization are sensitive to amphiphile aggregation (24). DPH is minimally soluble in water and even at 5 μM remains mostly in aggregates that have low fluorescence intensity but a relatively high fluorescence anisotropy. Above the CMC, micelles of the amphiphiles can solubilize some of the DPH leading to a moderate increase of intensity with a concomitant decrease in the fluorescence anisotropy. However, we found that DPH is significantly more soluble in vesicles than in micelles, such that above the CVC, the fluorescence increases much more quickly with fatty acid concentration, and there is another concomitant decrease in anisotropy. Thus, the CMC is taken as the concentration at which the DPH fluorescence anisotropy begins to decrease, and the CVC is taken as the concentration at which the anisotropy falls to its lowest value. This fluorescence anisotropy CMC determination method was also tested with the non-vesicle-forming amphiphiles sodium dodecyl sulfate (SDS) and Triton X-100, and the results (Fig. S1) are in good agreement with the literature values (25).
-
2)
Light scattering has also been shown to correlate with amphiphile aggregation (26). Below the CMC, the scattered light intensity is stable; however, when the concentration of fatty acids reached the CMC, instead of a stable increase in scattered light, we observed large fluctuations in the light intensity. This would indicate that small numbers of larger aggregates are also transiently forming at this concentration. Therefore, we interpret the onset of the intensity fluctuations as an upper limit for the fatty acid CMC. To corroborate this data, we measured the CMC for SDS by the same method, which yielded results that are close to the published values (25) (Fig. S1 B). The CVC of the fatty acid cannot be measured by this method because of the instability of the light intensity above CMC. Measurements were performed on a Wyatt DynaPro dynamic light-scattering (DLS) instrument (Santa Barbara, California), and each sample was filtered with 0.02-μm Anotop 10-membrane filter (Whatman, Maidstone, UK). Only the scattered intensity and not the correlation function was used in the analysis.
-
3)
The onset of turbidity, measured as absorbance at 400 nm, was used to evaluate the CVC of the fatty acid solutions (27). The measurements were performed on a Jasco V-650 with a 1-cm pathlength at room temperature for NA and DA and at 50°C for DDA solutions.
-
4)
The change in the 1H-NMR signal intensity of the methylene groups of fatty acids (NA: δH = 1.16 ppm; DA: δH = 1.20 ppm; DDA: δH = 1.40 ppm) was employed to find the transition between the micellar and vesicular states of amphiphiles. Below the CVC, the 1H signals are directly proportional to the fatty acid concentration. Above the CVC, the 1H signal intensity decreases with increasing concentration because of the large correlation time of the very large vesicles yielding fast spin relaxation. The CMC of fatty acids as well as SDS (Fig. S1) is not detected via this signal intensity method most likely because a fast exchange between monomers and micelles and a rather short correlation time of the fatty acids in the micelles and thus little change to their spin relaxation rate compared to monomers. Spectra were measured with Bruker Avance III 700-MHz and Bruker Avance III HD 600-MHz spectrometers (Fällanden, Switzerland), both equipped with triple resonance TCI CryoProbes (Bruker). Samples were in 10% (v/v) D2O at 24.85°C for NA and DA and at 50°C for DDA. Data were processed and analyzed by TopSpin 3.5 (Bruker).
Table 1.
Measured CMC and CVC Ranges for NA, DA, and DDA Solutions at pH 7.8
| Fatty Acid | CMC |
CVC |
Literature |
||||
|---|---|---|---|---|---|---|---|
| Fluorescence (mM) | DLSa | ||||||
| NMR | Turbidity | Fluorescence | CMCb (mM) | CVCb (mM) | |||
| NA | 50–70 | 50–70 mM | NDc | NDc | 150–200 mM | 110 | 85 |
| DA | 20–22 | 22–30 mM | 49–52 mM | 55–60 mM | 55–60 mM | 28.8 | 43 |
| DDA | 3.6–4.5 | NMd | 4.7–5.6 mM | 5 mM | 5–6 mM | 1.58 | 22.5 |
| SDS | 7–8 | 8–8.5 mM | no transition to vesicles | 8.2 | – | ||
| Triton-X | 0.25–0.30 | – | no transition to vesicles | 0.25 | – | ||
CMC was determined by the concentration at which signal began to fluctuate widely.
Published CMC and CVC values were determined under different conditions and at different pH (29, 42) and are tabulated to emphasize the importance of doing the measurements under the conditions used in this study.
No transition was detected with this technique.
DDA solutions were not measured by DLS due to issues with solidification during samples preparation.
All signal versus concentration data were analyzed using R software. Critical concentrations were determined from the NMR intensity data by fitting the linear portion of the trends and using the break from linearity as the CVC. For the fluorescence, light scattering, and turbidity data, the CMC and CVC are reported as a range between the data points at which the trend in the data changed.
Fluorescence anisotropy
The fluorescence anisotropy of 5 μM DPH in various fatty acid solutions was measured on an ISS K2 spectrofluorometer (ISS, Champaign, IL) with 1-cm pathlength and excitation/emission wavelengths of 330/480 nm, respectively. The samples were prepared by diluting 5 μL of a 1 mM DPH solution in acetonitrile into 1 mL of the sample to be analyzed and by vortexing for 15 min. NA and DA samples were prepared and measured at room temperature, whereas DDA samples were kept at 50°C. The G-factor was measured independently for each sample, and the recorded anisotropy values are the average of 10 measurements.
Cryo-EM and negative stain transmission electron microscopy
For cryo-electron microscopy (cryo-EM), 3 μL of respective solutions were applied to a Quantifoil R2/2 Cu 300 grid and bloated for 10 s with bloat force 20 and flash frozen in liquid ethane using an FEI Vitrobot (Quantifoil Micro Tools GmbH, Großlöbichau, Germany). The images were recorded with an FC20 cryo-FEI Tecnai electron microscope (FEI, Hillsboro, Oregon). The widths of the striations were determined using ImageJ software based on the fast Fourier transform of the regions containing the striations. For each fatty acid sample, multiple regions of three independent images were analyzed. The values reported are the mean and SD of 5, 8, and 11 measurements for NA, DA, and DDA, respectively. Samples for negative stain transmission electron microscopy (TEM) were prepared by applying the solutions to a CF400-Cu grids and staining with phosphotungstic acid (2% solution at pH 7.2). Images were recorded with a Morgagni 268 FEI electron microscope (FEI).
ATR-FTIR
Samples were centrifuged at 25 kg for 5 min, and pellets were resuspended in 4 μL of supernatant and placed on the diamond attenuated total reflectance (ATR) cell of Bruker Alpha-P ATR-FTIR (Fourier transform infrared) spectrometer. Before the measurements, samples were air dried at room temperature. Fatty acid solutions without peptide were also measured as a control. An empty cell (air) was used as a blank spectrum.
X-ray diffraction
Samples were centrifuged at 130 kg with a Beckman Coulter Airfuge Ultracentrifuge (Brea, CA), and the pellets were transferred into 0.3-mm glass capillaries, which were then flame-sealed to prevent drying. Alternatively, fibers in the samples were aligned by pipetting 4 μL of the mixture in the gap formed by two glass rods spaced ∼2–3 mm apart and dried overnight in a parafilm-sealed petri dish. Scattering data were recorded with a Rigaku MicroMax-007HF (Tokyo, Japan) source on a Mar345 image plate detector (MarXperts, Norderstedt, Germany) at room temperature with a 1-h exposure. The scattering from an empty capillary and air were also measured and used for background subtraction. The diffraction patterns of the samples in capillaries were radially integrated with FIT2D software (28) to yield a plot of the scattered intensity as a function of d-spacing. The images of the diffraction patterns of the aligned samples are displayed with Adxv software.
SAXS
Small-angle x-ray scattering (SAXS) experiments were performed using a Rigaku MicroMax-002+ equipped with a microfocused beam (40 W; 45 kV; 0.88 mA) with the λCuKα = 0.15418-nm radiation collimated by three pinhole collimators (0.4, 0.3, and 0.8 mm) to obtain direct information on the scattering patterns. The SAXS intensity was collected by a two-dimensional Triton-200 gas-filled x-ray detector (20 cm diameter; 200-μm resolution). An effective scattering vector range of 0.05 nm−1 < q < 2 nm−1 was obtained, where q is the scattering wave vector defined as q = 4π sin θ/λCuKα with a scattering angle of 2θ.
HPLC analysis
To quantify the amount of peptide and fatty acid in the distinct aggregates, 100-μL samples containing aggregates (either flocculent or translucent) of OVOVOVOV-NH2 (subsequently referred to as (OV)4, in which O stands for ornithine) and fatty acids were centrifuged in a tared tube. The masses of the pellets were determined and then dissolved in 500 μL of 8 M guanidine solution. Supernatants were analyzed without dilutions for NA and DA, and the DDA supernatant was diluted twofold with 5% acetonitrile in H2O. Reversed-phase analyses were performed on a Kinetex C8 2.6-μm 50 × 4.6 mm column connected to an Agilent 1200 HPLC (Santa Clara, CA) system with an autosampler and diode array detector. The (OV)4 peptide and fatty acid were resolved by a linear gradient of acetonitrile with 0.1% TFA (5–98%) at 3 mL/min. The injected amount of (OV)4 was quantified based on its peak area and calculated extinction coefficient at 214 nm (19) of 6797 M−1cm−1, and fatty acids were quantified based on a calibration with known quantities of material. The pellet mass was mostly due to water (dried pellets had negligible mass), so the volume of the pellet was calculated from the mass of the wet pellet. This volume was then used to correct for the amount of material in the pellet fraction that is due to the soluble material trapped in the pellet.
Protease sensitivity assay
The stability of (OV)4 in phosphate buffer and in the translucent peptide/DA mixture at pH 7.8 was probed by its resistance to digestion by the nonspecific peptidase, proteinase K. Standard solutions of (OV)4 alone and (OV)4 with 72 mM DA were incubated for 4 h at room temperature with 50 ng/mL proteinase K, at which time the protease was deactivated with the addition of 0.1 mM phenylmethylsulfonyl fluoride. For HPLC analyses, the DA/(OV)4 sample was diluted 1:4 (v/v) with 8 M guanidine, and the (OV)4 alone sample was diluted 1:4 (v/v) with H2O/acetonitrile/TFA (98:2:0.1 v/v). Solutions were vortexed for 30 min and analyzed by HPLC.
Results and Discussion
Amyloidogenic peptide and fatty acid characterizations
To investigate the interactions between peptides and fatty acids with a focus on a cooperative amyloid fibril and membrane bilayer formation, we designed the amyloidogenic peptide (OV)4. It was predicted that the positive charge of (OV)4 at a neutral pH would prevent self-aggregation while allowing for favorable electrostatic interactions with the carboxylate headgroup of fatty acids. Indeed, at pH 7.8, the peptide is soluble up to 5 mg/mL, and at 2 mg/mL, it adopts a random coil-like conformation, whereas at pH 11, above the pKa of ornithine, (OV)4 readily forms a β-structured aggregate (Fig. S2). We characterized the interaction of (OV)4 with three simple vesicle-forming fatty acids: NA, DA, and DDA. These fatty acids form vesicles in water at a pH near their pKa (29) but remain monomeric and micellar at a pH much above their pKa. Because it was expected that cooperativity based on the electrostatic interactions between the peptide and fatty acid would be more pronounced at a pH above the fatty acid pKa, we chose to study the interactions at pH 7.8 (pKa of NA, DA, and DDA is 7.0, 7.3, and 7.5, respectively). Because both their CMC as well as the CVC are sensitive to pH, buffer strength, and solution composition, we carefully measured these critical concentrations for each fatty acid in phosphate buffer at the working pH of 7.8 (Figs. S3–S6; Table 1). It is evident from the values in Table 1 that under the conditions used here (phosphate buffer, pH > pKa), the CMC and CVC values deviate from reported values measured at their respective pKas. This deviation is consistent with the known pH dependence of the fatty acid CVC and was confirmed by our measurement of the CVC of DA at two different pH values (7.4 and 7.8; Fig. S6, B and C). Additionally, at a pH significantly above their pKa, fatty acids do not form vesicular assemblies as determined for DA at pH 8.2 (Fig. S5 D) or for NA at our standard condition of pH 7.8 (Figs. S5 A and S6 A).
Cooperative interactions between (OV)4 and fatty acids
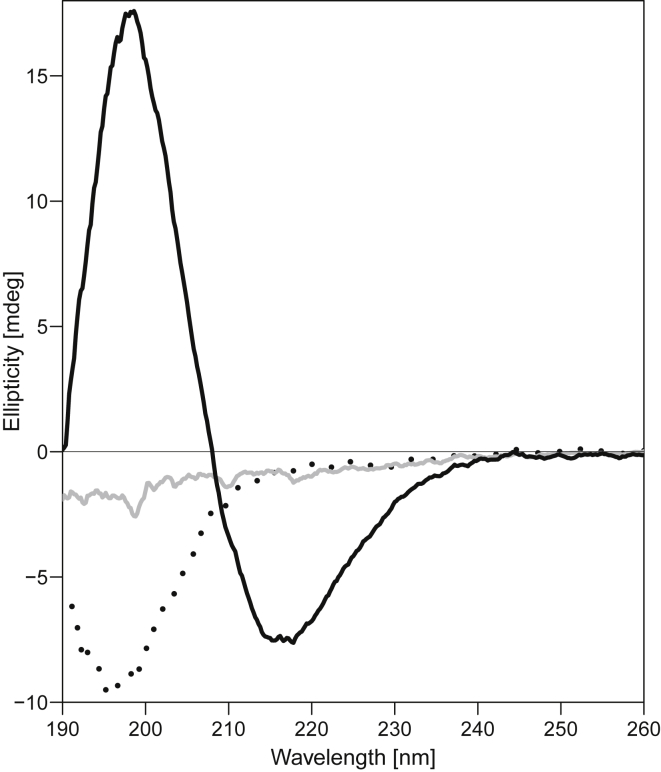
To search for cooperative interactions, we analyzed a concentration series of the fatty acids mixed with 575 μM (0.5 mg/mL) (OV)4 at pH 7.8. Upon mixing of the peptide and fatty acid, many samples form an immediate precipitate. At the higher fatty acid concentrations, this initial precipitate appears to go back into solution over a period of minutes to hours. This phenomenon was observed for all three fatty acids, and the CD spectra of these mixtures reveal two fatty acid concentration-dependent transitions separating three distinct states of the peptide (Figs. 1 and S7). These data point to a correlation between the transition concentrations observed in the CD spectra and the CMC and CVC values for the fatty acids (illustrated in Fig. 2). Below the CMC, the peptide-fatty acid mixture is transparent, and the CD spectra are indicative of a random coil-like conformation as it is in the absence of fatty acids. Near their CMC, fatty acids mixed with (OV)4 produce a flocculent precipitate, and the CD spectra of the mixtures are featureless, indicating that the peptide is contained in this flocculent precipitate. At any concentration above the CVC, the mixtures become translucent after an initial formation of precipitate, and the CD spectra indicate that the peptide adopts a β-strand conformation. That this trend is dependent on the fatty acid concentration and not the peptide concentration is supported by the observation of β-structure at a range of peptide concentrations (Fig. S8). To further characterize the mixtures, we centrifuged the translucent β-structured DA sample at 130 kg and found that the CD signal was absent in the soluble fraction. Thus, the β-structured material in the translucent samples above the fatty acid CVC is also an aggregate but one that is small enough to absorb the ultraviolet light. The loss of the CD signal in the concentration range between the CMC and CVC is likely due to the large size of the flocculent aggregates that can no longer absorb light. Therefore, the flocculent precipitate was further characterized by ATR-FTIR. The FTIR spectra had absorbance bands near 1625 cm−1, indicating that the peptide in the flocculent precipitate was also in a β-strand conformation (Fig. S9).
Figure 1.
The concentration dependence of the CD spectra of DA solutions with (OV)4. The CD spectra of three solutions of 575 μM (OV)4 with either 1.8 mM DA (dotted line), 18 mM DA (solid gray line), or 72 mM DA (solid black line) are shown. At 1.8 mM DA, the CD spectrum is random coil like; at 18 mM DA, it is nearly featureless; and at 72 mM, it is β-sheet like.
Figure 2.
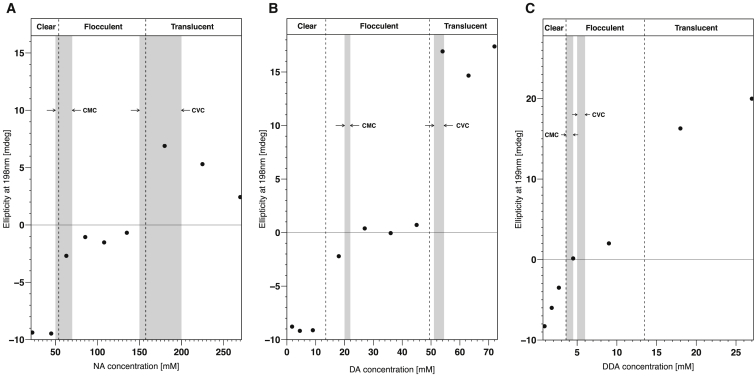
Correlation of (OV)4 secondary structure with fatty acid CMC and CVC. The β-strand secondary structure of the peptide in the fatty acid mixtures as monitored by its CD signal at 198 nm is plotted against the fatty acid concentration in solutions of NA (A), DA (B), and DDA (C). Negative values are associated with a random coil-like structure of the peptide and positive values with a β-sheet secondary structure. The dashed lines roughly demarcate the boundaries between the visible changes in the mixtures from clear solution to flocculent precipitate to translucent solution with increasing concentration of fatty acid. The gray bands represent the ranges of CMC and CVC values for the fatty acids as measured independently in the conditions used in this study (see Figs. S3–S6) and indicate that the transitions of peptide/fatty acids mixtures occur at very similar concentration ranges as do the phase transitions of the pure fatty acids themselves. The deviation of the DDA mixtures from this trend can be explained by the fact that its concentration is low enough that the binding of DDA to the 575 μM (OV)4 consumes a significant amount of the fatty acid, effectively shifting the flocculent-translucent boundary.
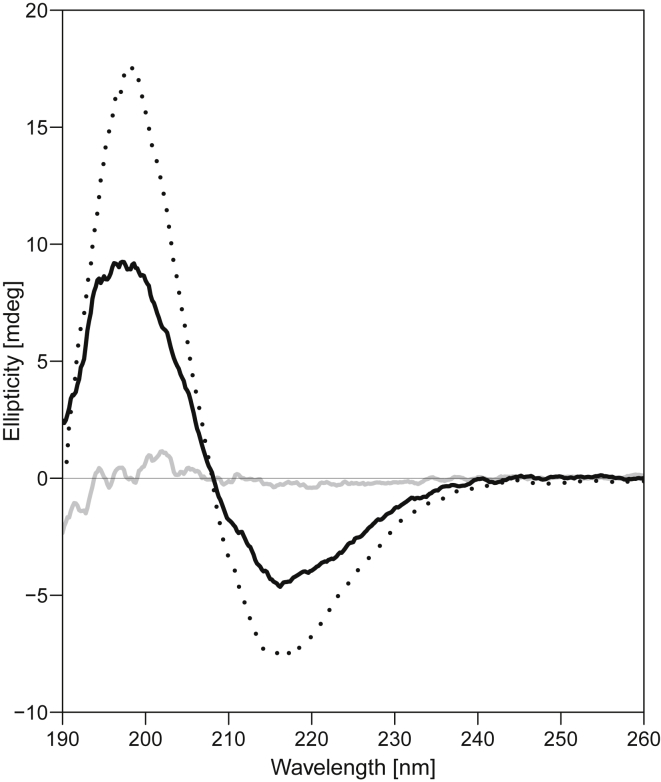
The data illustrated in Fig. 2 suggest that the interaction between fatty acid micelles with (OV)4 leads to the flocculent precipitate, whereas fatty acid vesicles are required to form the translucent precipitate. This correlation was further probed by modulating the CVC of DA through the addition of DOH. It is known that the CVC of fatty acid solutions are lowered in mixtures that contain small amounts of the corresponding fatty alcohol (29), and we found that a 10:1 DA/DOH mixture at pH 7.8 has a CVC between 20 and 22 mM (Fig. S10), close to the reported value of ∼20 mM (29) and in contrast to the CVC of 50 mM in absence of fatty alcohol (Table 1). In the DA/DOH mixtures, the transition from flocculent to translucent precipitate and the corresponding changes in the CD spectra were both shifted to lower fatty acid concentration (Figs. 3 and S11), further supporting the correlation between the CVC and the formation of the translucent precipitate. The appearance of the flocculent precipitate above the measured CVC for DDA (Fig. 2 C) could be because the CMC and CVC of DDA are not far apart and so low that the binding of DDA to the 575 μM (OV)4 consumes a significant amount of the fatty acid, thereby effectively shifting the flocculent-translucent boundary by a few mM above the CVC. Such a shift was also detected for DA/DOH mixture; however, it would be less noticeable for NA and DA mixtures because of their relatively large CVC values.
Figure 3.
Fatty alcohol lowers both CVC and the concentration at which translucent precipitate is formed. Depicted are three CD spectra for 575 μM solutions of (OV)4 with either 72 mM DA (dotted line), 36 mM DA (solid gray line), or a 10:1 DA/DOH mixture with 34.65 mM total amphiphiles (solid black line). The β-sheet-like spectrum absent in the 36 mM DA solution due to the formation of the flocculent precipitate is present in the lower concentration DA solution with a 1:10 molar ratio of fatty alcohol. This amount of DOH decreases the CVC of the system from ∼55 to ∼20 mM (Fig. S10).
To test the hypothesis that electrostatic interactions are a major driving force in the observed interactions, we prepared similar mixtures of the acidic amyloidogenic peptide V(DV)4 with DA. In contrast to (OV)4, V(DV)4 forms amyloids at pH 3.5, whereas at pH 7.8, it remains soluble and adopts a random coil-like conformation (Fig. S12). The mixtures of V(DV)4 with DA solutions spanning both its CMC and CVC displayed no change in peptide conformation (Fig. S12) and no visible changes in the solutions. The lack of interactions between V(DV)4 and DA indicates that assemblies of fatty acids with (OV)4 are likely driven by ionic interactions.
Structural characterization of peptide-fatty acid coaggregates
While analyzing the different peptide-fatty acid aggregation states, we found that their formation is quickly reversible. Upon dilution of the mixtures with buffer, the precipitates dissolve, and the diluted samples have random coil-like CD spectra. This complicated our measurement of the fatty acid and peptide content in the aggregates because we could not simply rinse the aggregates but rather had to subtract the amount of soluble peptide and fatty acid remaining in the wet pellet (see Materials and Methods). This analysis showed that the flocculent and translucent aggregates do in fact contain fatty acids in addition to the peptide. For NA and DA, the molar ratio of fatty acid/peptide in the flocculent precipitates is ∼3, whereas in the translucent precipitate, the ratio is ∼7 (Fig. S13). DDA precipitates could not be reproducibly assayed because of the high melting temperature of DDA. The doubling of the fatty acid content in the translucent precipitate compared to the flocculent precipitate would be consistent with a conversion from a monolayer to a bilayer structure. This is particularly noteworthy in light of the facts that a micelle-to-vesicle conversion is also a change from a monolayer to bilayer and that the formation of the two types of precipitates is correlated with the fatty acid CMC and CVC. We also measured the fluorescence anisotropy of DPH in the translucent precipitate and in the pure DA vesicles. The fluorescence anisotropy of DPH in the translucent precipitate (0.055) is higher than the respective value measured for DPH in the DA vesicles alone (0.034) suggesting that the fatty acid in the translucent precipitate is more rigid than the vesicles. Furthermore, the fact that the fluorescence intensity of DPH in DA vesicles and in the DA-peptide translucent aggregates were comparable suggests that there is in fact a fatty acid bilayer in the translucent precipitate.
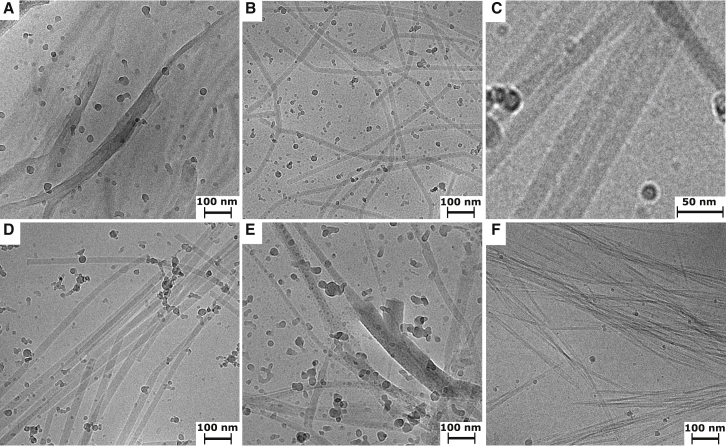
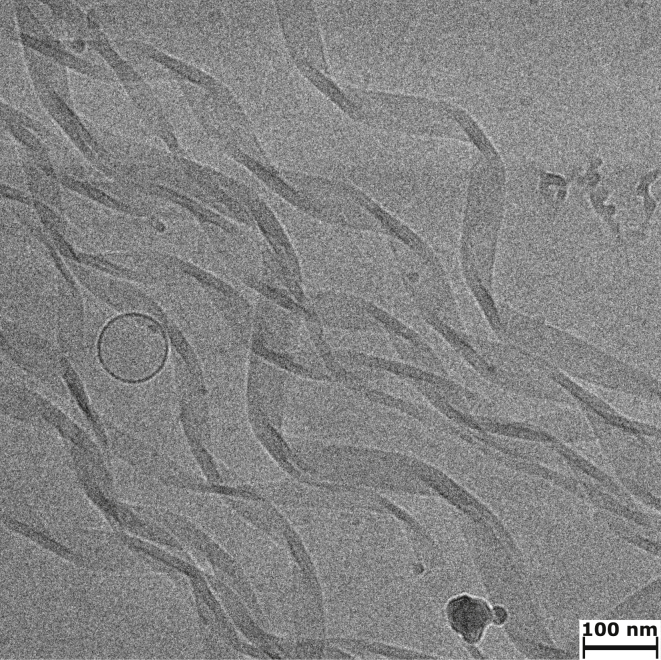
To characterize the mesoscopic structure of peptide-fatty acid mixtures, we analyzed the flocculent and translucent precipitates by cryo-EM. The DA-(OV)4 flocculent precipitate was generally too dense to get much information from the EM images; however, in a few instances, we could detect fibril-like structures, suggesting that the β-structure detected by FTIR is in fact an amyloid (Fig. 4 A). The translucent precipitates of all the mixtures consisted primarily of long, straight structures that are suggestive of ribbons, rods, or tubes. In the case of the DA-(OV)4 mixture, these straight structures have a uniform width of ∼25 nm and lengths of up to several microns (Fig. 4 B). Further cryo-TEM analysis with a tilted sample indicates that the structures in the translucent DA-(OV)4 precipitate are actually not flat ribbons but rather rod or tube like (Fig. S14). The translucent NA-(OV)4 precipitate has similar structures, in line with its similar fatty acid content; however, instead of a uniform width, there is a narrow distribution ranging from 20 to 30 nm. In contrast, the DDA-(OV)4 mixtures gave structures with a much larger variation in width from 15 to 80 nm (Fig. 4, D and E). The rod- or tube-like structures in the translucent precipitates are unlike typical amyloid fibrils, which are often found to be composed of bundles of protofilaments that can be either twisted, straight, or displaying some other combination of quaternary structures (30). In fact, such filament bundling is detected in the pure (OV)4 fibrils formed at pH 11 without fatty acid (Fig. 4 F). Interestingly, the tube-like structures in the translucent precipitates often have a detectable striation that indicates that they may be composed of many smaller fibrils that have assembled into a larger ordered structure (Fig. 4 C) such as a nanotube, as has been observed for other amyloids (31, 32, 33). The widths of the striations are correlated with the fatty acid chain length, and, as estimated by Fourier transform of the images, are 2.40 ± 0.11, 2.84 ± 0.11, and 3.46 ± 0.06 nm for structures assembled with NA, DA, and DDA, respectively (see Materials and Methods).
Figure 4.
Morphology of fatty-acid-(OV)4 precipitates compared to pure (OV)4 amyloids. Cryo-EM micrographs of the flocculent precipitate (A) and translucent precipitate (B) formed in mixtures of DA and (OV)4. The flocculent precipitate is usually too thick for observing detailed morphological features, but in a few instances like the one depicted in (A), fibrillar structures are visible. The translucent precipitate is characterized by straight fiber-like structures that have a uniform width of ∼25 nm and lengths of up to several microns. In many of the tube-like structures, there is a visible striation, indicating that they may be composed of an alignment of smaller fibrils (C). The translucent precipitates of (OV)4 mixtures with NA (D) and DDA (E) also reveal fiber-like structures; however, they present a distribution of widths. The NA samples have a narrow distribution, whereas the DDA/(OV)4 mixtures have structures with a much larger variation in width. The amyloid fibrils of (OV)4 that form at pH 11 (F) are characterized by many straight fibers that form bundles of varying numbers of protofilaments.
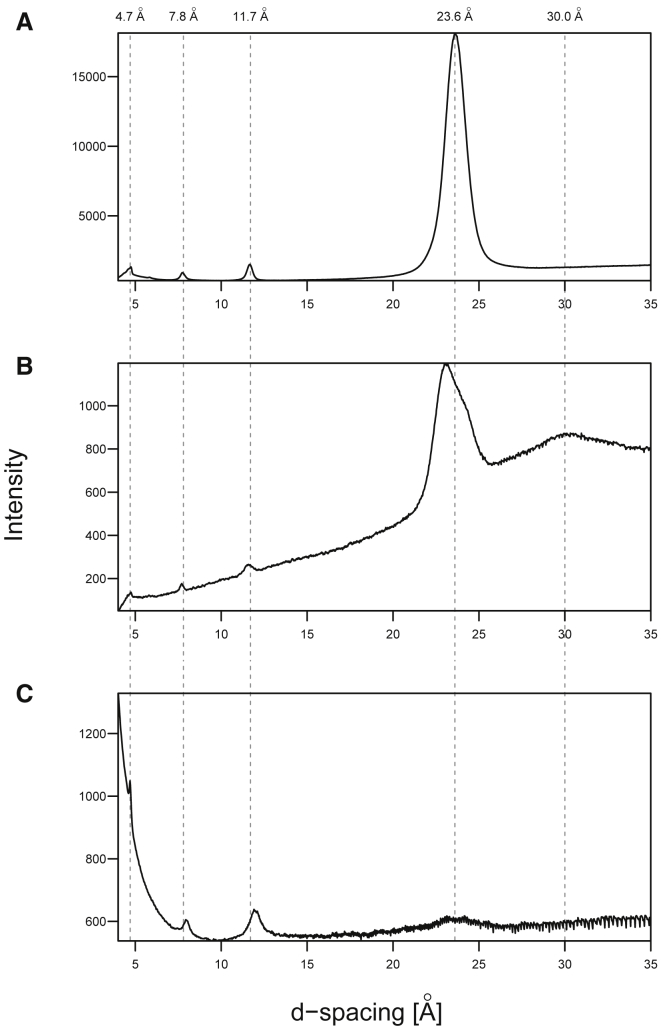
The peptide-fatty acid precipitates were further characterized by x-ray diffraction, which revealed some similarities between the flocculent and translucent precipitates that are also shared with the (OV)4 pH 11 fibrils. These include a reflection at 4.7 Å, which corresponds to the spacing between strands in a β-sheet. The other similarities include reflections near but not all identically at 7.8 and 11.7 Å (Fig. 5). The DA-(OV)4 precipitates have as well an intense reflection near 24 Å that is mostly absent in the pH 11 fibrils of the peptide without DA. Unique to the fatty acid translucent precipitate is a broad reflection at 30 Å, which may be indicative of a fatty acid bilayer. The samples could be aligned upon drying, which allowed us to confirm the underlying cross-β structure. The x-ray diffraction of aligned samples of translucent aggregates and the pH 11 fibrils both had the signature cross-β pattern (Fig. S15). The diffraction of dried and aligned DA revealed that the strongest reflections from the aligned translucent sample arise from the excess DA that dried along with the precipitate. Still, we could detect weaker reflections (i.e., 11.4, 7.7 Å, and meridional reflection at 4.7 Å) uniquely in the aligned translucent precipitate, which are indicative of the presence of cross-β structure. The relatively low intensity of the peptide-derived reflection is not surprising considering that the translucent aggregate samples have a 150-fold molar excess of DA compared to peptide.
Figure 5.
X-ray diffraction of fatty acid-(OV)4 precipitates compared to pure (OV)4 amyloids. The x-ray diffraction images of flocculent (A) and translucent (B) precipitates from DA-(OV)4 mixtures at 18 mM DA and 63 mM DA, respectively, are shown. (C) The diffraction of (OV)4 amyloids formed at pH 11 in the absence of fatty acids is shown. All of the various aggregates have a reflection at 4.7 Å corresponding to the spacing between strands in a β-sheet. The other similarities include reflections near 7.8 and 11.7 Å. The DA/(OV)4 precipitates also have an intense reflection around 24 Å that is mostly absent in the pH 11 amyloid fibrils that lack DA. Unique to the translucent precipitate is a broad low-resolution reflection at 30 Å. The diffraction patterns were radially integrated with the FIT2D software tο yield a plot of the scattered intensity as a function of d-spacing.
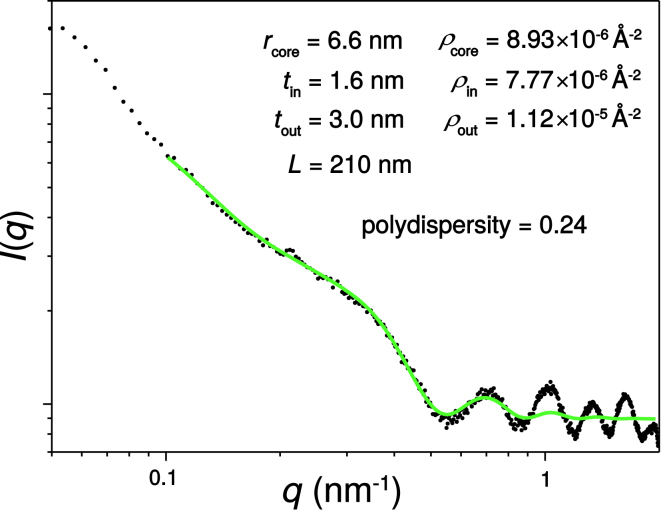
To better characterize the shape of the tube-like structures, SAXS experiments were conducted on a DA-(OV)4 precipitate formed from 1 mM (OV)4 and 63 mM DA at pH 7.8. The dispersion clearly showed a characteristic intensity profile as indicated in Fig. 6, which was fitted following the hollow-core two-shell cylinder form factor P(q) model (see Supporting Materials and Methods) to extract the core radius, its polydispersity and both the inner and outer shell thicknesses (32). Also fitted were the scattering length densities, ρ, of the core and inner and outer shells. The fitted scattering data resulted in a main core radius value of rcore = 6.57 nm (polydispersity = 0.24) and thicknesses of tin = 1.62 nm and tout = 2.98 nm. These values match well the dimensions obtained from molecular simulations for the thickness of an interdigitated DA bilayer (1.62 nm) and the length of a fully extended (OV)4 peptide (2.95 nm) (Fig. S16 A). Thus, the diameter of the core two-shell cylindrical objects calculated from the fitted parameters is D = 2(rcore + tin + tout) = 22.3 ± 1.6 nm and is in good agreement with the cryo-EM images for which a diameter of 25 nm was measured (Fig. 4, B and C). The fitted value of ρcore (8.93 × 10−6 Å−2) is close to that of water (9.44 × 10−6 Å−2), whereas ρin (7.77 × 10−6 Å−2) is less than water and close to that of DA (8.47 × 10−6 Å−2), and ρout (1.12 × 10−5 Å−2) is higher than water and corresponds to a density of 1.21 g/mL for a molecule with a molecular formula similar to (OV)4 or its 1:1 mixture with DA (Fig. 6). The fitted parameters request the presence of two domains, which are depicted in Fig. S16 and are modeled as follows: 1) the inner shell consisting of a series of interdigitated and deprotonated DA molecules (orange rectangular region) and 2) the outer shell comprising the (OV)4 peptide in a cross-β arrangement, exposing the amide-terminus to the outside of the tube and connected by ionic interactions between the ornithine and the deprotonated interdigitated DA molecules to the next radially distributed β-sheet (depicted in Fig. S16 B). The inner and outer shells are connected via ionic interactions between the deprotonated DA molecules and the protonated N-terminus of the (OV)4 peptide (Fig. S16 A).
Figure 6.
A one-dimensional SAXS intensity profile for the DA-(OV)4 translucent precipitate. The intensity of the scattering (I) as a function of the scattering wave vector (q) is plotted with the fit to the hollow-core two-shell cylinder form factor P(q) (see Supporting Materials and Methods) in green. The fitted parameters for this form factor are listed in the figure. To see this figure in color, go online.
In more detail, the model of these supramolecular colloidal aggregates is based on the self-assembly of the (OV)4 peptide as it would be expected to occur in amyloid fibrils, by means of hydrogen bonding between the amide moieties from parallel peptide backbones and a hydrophobic face rich in valine residues that is buried between two opposing sheets. This amyloid core has a hydrophilic, positively charged face on which all ornithine residues can ionically interact with the free carboxylates from DA molecules at pH 7.8. The DA in turn forms an interdigitated bilayer, creating the next interface for the interaction with another amyloid core (Fig. S16 B). Another interdigitated DA bilayer also interacts with the protonated N-termini of the (OV)4 peptides on the β-sheet edge constituting the inner shell (orange rectangle in Fig. S16 A). Thus, the outer-shell building unit consists of a cross-β entity composed of two β-sheets stabilized by van der Waals interactions between the valine residues with an approximated interstrand distance of 3.9 Å (blue rectangular region with two green lines indicating the β-sheet front edge) and the hydrogen bonding between strands with a 4.7-Å distance between strands. This cross-β entity is connected to another cross-β entity via the protonated ornithine residues ionically interacting with the previously described deprotonated and interdigitated DA bilayer (yellow rectangular region). Thus, the radially repeating unit results in an average distance of 2.89 nm (Fig. S16, C and D), which matches well the 2.85-nm distance between striations detected on the surface of the objects visualized in cryo-EM images (Fig. 4 C) and could be the source of the broad 30-Å reflection in the unaligned sample (Fig. 5 B). An overview of the core two-shell model is depicted in Fig. S16 in which the cross-β entities are radially distributed as “paddle wheels” with the β-strands pointing out from the center and the valine and ornithine residues projecting out of the planes into different domains (amyloid core or fatty acid bilayer) due to the alternating amino acid sequence and the specific geometry of the parallel β-sheet secondary structure (34). This model predicts a molar ratio of peptide/DA of 1:6, which is close to the HPLC estimated value of 1:7.
General nature of cooperative interactions between amyloidogenic peptides and fatty acids
Having observed some variation in aggregate structure with fatty acid chain length, we also wanted to explore the importance of the peptide sequence. Therefore, we characterized the interactions of 13 additional amyloidogenic peptides with DA. These peptides were chosen to have an alternating hydrophobic/hydrophilic sequence with primarily basic hydrophilic residues to encourage the formation of an amphipathic β-strand and ionic interactions with the fatty acid. Mixtures of the 13 peptides at pH 7.8 were analyzed at three different concentrations of DA (4.5, 18, and 72 mM), spanning the monomeric, micellar, and vesicular states of DA. The structural changes of peptides were monitored by CD and ATR-FTIR spectroscopy. The results of the measurements and the information about appearance of the mixtures are presented in Table 2 (and Table S1 with links to individual spectra). It is clear that the structures and appearances of many peptides are altered in the presence of DA. Interestingly, at 18 mM DA (below its CVC), most of the peptides form a flocculent precipitate, and the CD signal is lost. Still, there were usually clear changes in the CD spectra as a function of DA concentration. The secondary structure of peptides with multiple phenylalanine residues could not be assigned by their CD spectra alone because the contribution from the phenylalanine side chain overwhelms the backbone spectrum. However, ATR-FTIR spectra measured for the mixtures with 18 and 72 mM DA revealed that most but not all of the peptides in the precipitates with DA have β-strand-like secondary structure. We further characterized a pair of peptides of identical composition but slightly different sequence (i.e., DFRFRF-NH2 and RFRFDF-NH2). The peptide with an N-terminal aspartate has a significant change in the CD spectra in the presence of 72 mM DA, whereas the second peptide does not show such a structural change (Tables 2 and S1). For a deeper insight into this difference, their mesoscopic structures were characterized by both cryo- and negative-stain TEM. The mixture of 72 mM DA with DFRFRF-NH2 contains helical ribbon structures (Fig. 7), whereas in the absence of DA, the peptide forms the bundles of twisted protofilaments that are characteristic of amyloids fibrils (Fig. S17). The sequence isomer RFRFDF-NH2 on the other hand does not form any regular structures with or without DA that could be characterized by EM (Fig. S17). Thus, the extent of both assembly into ordered aggregates and the structure of the resulting aggregates are a function of not only the fatty acid identity and concentration but also of the peptide sequence.
Table 2.
Characterization of Structural Changes for 14 Peptides in the Presence of DA
| Sequence | 0 mM DA |
4.5 mM DA |
18 mM DA |
72 mM DA |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Appearance | CD | Appearance | CD | Appearance | CD | ATR-FTIR | Appearance | CD | ATR-FTIR | |
| RFRFRFR-NH2 | C | max@ 220 nm | F | max@ 220 nm | F | - | – | F | - | β+ |
| FRFRFR-NH2 | C | max@ 220 nm | C | max@ 220 nm | F | - | – | F | max@ 220 nm | – |
| DFRFRF-NH2 | F | max@ 220 nm | F | - | F | - | β+ | F | min@ 215 nm | β2+ |
| RFRFDF-NH2 | C | max@ 220 nm | C | max@ 220 nm | F | - | β+ | T | max@ 220 nm | – |
| RFRFR-NH2 | C | max@ 220 nm | C | max@ 220 nm | T | - | – | F | - | – |
| RFRFD-NH2 | C | max@ 220 nm | C | max@ 220 nm | C | max@ 220 nm | – | NC | max@ 220 nm | – |
| Ac-VOVAVOVAV-NH2 | F | min@ 220 nm | F | min@ 220 nm | F | min@ 220 nm | β2+ | F | β | β2+ |
| Ac-V-{Dab}-VAV-{Dab}-VAV-NH2 | F | min@ 190–220 nm | F | min@ 197 and 220 nm | F | min@ 197 and 220 nm | β2+ | F | β | β2+ |
| VOVOVOVOV-NH2 | C | RC | F | RC | F | - | β2+ | F | β | β2+ |
| V-{Dab}-V-{Dab}-V-{Dab}-V-{Dab}-V-NH2 | F | min@ 220 nm | F | min@ 220 nm | F | - | β2+ | F | min@ 220 nm | β2+ |
| V-{Dab}-V-{Dab}-V-{Dab}-V-NH2 | C | RC | F | RC | F | - | β+ | F | min@ 205 nm | β2+ |
| {Dab}-V-{Dab}-V-{Dab}-V-{Dab}-V-NH2 | F | min@ 225 nm | C | min@ 225 nm | F | - | β2+ | T | min@ 220 nm | β+ |
| {Dab}-V-{Dab}-V-{Dab}-V-NH2 | C | RC | C | RC | F | - | – | T | β | – |
| (OV)4 | C | RC | C | RC | F | - | β2+ | T | β | β+ |
This table summarizes the data that are displayed in Table S1 with its associated spectra. β, β-strand structure of peptide; β+, weak signal; β2+, strong signal near 1625 cm−1; C, clear solution; {Dab}, diaminobutyric acid; F, flocculent precipitate; max@, wavelength of maximal ellipticity in the spectra that does not match any typical secondary structure; min@, wavelength of minimal ellipticity in the spectra that does not match any typical secondary structure; NC, no visible change in appearance; O, ornithine; RC, random coil-like structure; T, translucent precipitate; -, signal too weak to interpret; –, no signal.
Figure 7.
A cryo-EM image of DFRFRF-NH2 peptide mixture with 72 mM DA at pH 7.8. The twisted ribbons resulting in the shape of helices are formed in these conditions. The widths of the ribbons as well as the tightness of helices varies between each assembly. A vesicle of decanoic acid (DA) is also visible in the image.
Mutualism in peptide-fatty acid interactions
Aside from the presented expansion of the phase and structure space in mixtures of peptide and fatty acid, we tested whether the interactions between peptide and fatty acid provided some other potential benefit to the molecules. In particular, we measured the stability of the peptide against hydrolysis by the nonspecific protease proteinase K as well as the thermal stability of the fatty acid bilayer. Proteinase K is commonly used to probe the domain structure and solvent accessibility of proteins. Treatment of the (OV)4 fatty acid translucent precipitate with 50 ng/mL proteinase K for 4 h did not produce any detectable degradation of the peptide. However, under the same conditions, the soluble (OV)4 without DA was 50% digested (Fig. S18). Additionally, the thermal stability of the DA bilayer was increased in the presence of (OV)4. To demonstrate this, we monitored the fluorescence of DPH as a function of temperature and found that in the translucent precipitate its fluorescence intensity decreases by 25.5% from 25 to 60°C, whereas in DA vesicles under the same conditions, it decreases by 42% (Table S2). This difference is consistent with a greater thermal stability of the intercalated DA bilayer in the coaggregate and may be explained by the greater rigidity of this bilayer monitored by DPH fluorescence anisotropy. Thus, the fatty acid not only increases the pH range at which (OV)4 is able to aggregate, but just as importantly, it produces a novel structure, to our knowledge, that protects the peptide from hydrolysis. Conversely, the peptide changes the structure of the fatty acid aggregates, increases the pH range (at least for NA) at which aggregation occurs, and enhances the stability of the bilayer.
Conclusions
Viewed from a prebiotic perspective, the increase of not only the structural space of these molecules but also their stability and the area of the environmental conditions under which they are capable of forming repetitive structures can be regarded as selective advantages. That these cooperative interactions are not simply a unique feature of the (OV)4 sequence is confirmed by the characterization of 13 additional peptides. Despite the variety of sequences we tested, the phase of the fatty acid remains correlated with structural changes for many of the peptides. We observed thereby a variety of aggregate structures, including uniform- and non-uniform-width tubes, helical ribbons, and straight and twisted fibers, whose formation are sensitive to the peptide sequence as well as the type and concentration of fatty acid. In most of the cases, the formation of coaggregate or the change in peptide structure is correlated to the critical aggregation concentrations (CMC and CVC) of the fatty acid. Although the fatty acids used in this study have CVCs that are higher than their expected concentration in a prebiotic milieu, there are many reasons to expect that they preceded biological lipids (35, 36). Therefore, at some point in time, either in the origin or early evolution of life, the interactions that we describe here could have been relevant.
Considering the replicative properties of both amyloids and fatty acids (37, 38), cooperative interactions of the kind we report here may have led to new functions, some of which could be beneficial to the propagation of either one or both of these molecules. These functions may for example include novelcatalytic activities, as has been reported for numerous amyloidogenic peptides (39). In addition, the rapid and reversible soluble-aggregate nature of the peptide-fatty acid interactions may be a functionally relevant property of the coaggregates because amyloids are usually rather stable (8), and reversible aggregation is more often associated with biologically functional amyloids (40, 41). The cooperative interaction that we describe here would be therefore one way to explain the original connection between peptides and fatty acids that eventually led to cellular life.
Author Contributions
All authors contributed to the writing of the manuscript. R.B., W.K., R.R., and J.G. conceived and designed the experiments. R.B., W.K., and A.S.-F. carried out the experiments.
Acknowledgments
We thank the ETH Scientific Center for Optical and Electron Microscopy (ScopeM) for their assistance as well as Dr. Marc Leibundgut of ETH Zurich for providing us access to the IMBB x-ray facility.
Editor: James Shorter.
Footnotes
Supporting Materials and Methods, eighteen figures, and two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)31223-2.
Contributor Information
Roland Riek, Email: roland.riek@phys.chem.ethz.ch.
Jason Greenwald, Email: jason.greenwald@phys.chem.ethz.ch.
Supporting Material
References
- 1.Orgel L.E. Evolution of the genetic apparatus. J. Mol. Biol. 1968;38:381–393. doi: 10.1016/0022-2836(68)90393-8. [DOI] [PubMed] [Google Scholar]
- 2.Wächtershäuser G. Before enzymes and templates: theory of surface metabolism. Microbiol. Rev. 1988;52:452–484. doi: 10.1128/mr.52.4.452-484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segré D., Ben-Eli D., Lancet D. The lipid world. Orig. Life Evol. Biosph. 2001;31:119–145. doi: 10.1023/a:1006746807104. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert W. Origin of life: the RNA world. Nature. 1986;319:618. [Google Scholar]
- 5.Oba T., Fukushima J., Ikehara K. Catalytic activities of [GADV]-peptides. Formation and establishment of [GADV]-protein world for the emergence of life. Orig. Life Evol. Biosph. 2005;35:447–460. doi: 10.1007/s11084-005-3519-5. [DOI] [PubMed] [Google Scholar]
- 6.Rode B.M. Peptides and the origin of life. Peptides. 1999;20:773–786. doi: 10.1016/s0196-9781(99)00062-5. [DOI] [PubMed] [Google Scholar]
- 7.Maury C.P.J. Self-replicating protein conformations and information transfer: the adaptive β-sheet model. Biosci. Hypotheses. 2008;1:82–89. [Google Scholar]
- 8.Greenwald J., Riek R. Biology of amyloid: structure, function, and regulation. Structure. 2010;18:1244–1260. doi: 10.1016/j.str.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Greenwald J., Riek R. On the possible amyloid origin of protein folds. J. Mol. Biol. 2012;421:417–426. doi: 10.1016/j.jmb.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 10.Carny O., Gazit E. A model for the role of short self-assembled peptides in the very early stages of the origin of life. FASEB J. 2005;19:1051–1055. doi: 10.1096/fj.04-3256hyp. [DOI] [PubMed] [Google Scholar]
- 11.Maury C.P. Self-propagating β-sheet polypeptide structures as prebiotic informational molecular entities: the amyloid world. Orig. Life Evol. Biosph. 2009;39:141–150. doi: 10.1007/s11084-009-9165-6. [DOI] [PubMed] [Google Scholar]
- 12.Patel B.H., Percivalle C., Sutherland J.D. Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nat. Chem. 2015;7:301–307. doi: 10.1038/nchem.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szostak J.W. An optimal degree of physical and chemical heterogeneity for the origin of life? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011;366:2894–2901. doi: 10.1098/rstb.2011.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun S., Humphreys C., Dale T.C. Amyloid-associated nucleic acid hybridisation. PLoS One. 2011;6:e19125. doi: 10.1371/journal.pone.0019125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adamala K., Szostak J.W. Nonenzymatic template-directed RNA synthesis inside model protocells. Science. 2013;342:1098–1100. doi: 10.1126/science.1241888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira de Souza T., Stano P., Luisi P.L. The minimal size of liposome-based model cells brings about a remarkably enhanced entrapment and protein synthesis. Chembiochem. 2009;10:1056–1063. doi: 10.1002/cbic.200800810. [DOI] [PubMed] [Google Scholar]
- 17.Kayed R., Sokolov Y., Glabe C.G. Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J. Biol. Chem. 2004;279:46363–46366. doi: 10.1074/jbc.C400260200. [DOI] [PubMed] [Google Scholar]
- 18.Butterfield S.M., Lashuel H.A. Amyloidogenic protein-membrane interactions: mechanistic insight from model systems. Angew. Chem. Int. Ed. Engl. 2010;49:5628–5654. doi: 10.1002/anie.200906670. [DOI] [PubMed] [Google Scholar]
- 19.Quist A., Doudevski I., Lal R. Amyloid ion channels: a common structural link for protein-misfolding disease. Proc. Natl. Acad. Sci. USA. 2005;102:10427–10432. doi: 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynolds N.P., Soragni A., Seeger S. Mechanism of membrane interaction and disruption by α-synuclein. J. Am. Chem. Soc. 2011;133:19366–19375. doi: 10.1021/ja2029848. [DOI] [PubMed] [Google Scholar]
- 21.Kuipers B.J., Gruppen H. Prediction of molar extinction coefficients of proteins and peptides using UV absorption of the constituent amino acids at 214 nm to enable quantitative reverse phase high-performance liquid chromatography-mass spectrometry analysis. J. Agric. Food Chem. 2007;55:5445–5451. doi: 10.1021/jf070337l. [DOI] [PubMed] [Google Scholar]
- 22.Chaudhuri R., Guharay J., Sengupta P.K. Fluorescence polarization anisotropy as a novel tool for the determination of critical micellar concentrations. J. Photochem. Photobiol. Chem. 1996;101:241–244. [Google Scholar]
- 23.Thorsteinsson M.V., Richter J., DePhillips P. 5-Dodecanoylaminofluorescein as a probe for the determination of critical micelle concentration of detergents using fluorescence anisotropy. Anal. Biochem. 2005;340:220–225. doi: 10.1016/j.ab.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X., Jackson J.K., Burt H.M. Determination of surfactant critical micelle concentration by a novel fluorescence depolarization technique. J. Biochem. Biophys. Methods. 1996;31:145–150. doi: 10.1016/0165-022x(95)00032-m. [DOI] [PubMed] [Google Scholar]
- 25.Chattopadhyay A., London E. Fluorimetric determination of critical micelle concentration avoiding interference from detergent charge. Anal. Biochem. 1984;139:408–412. doi: 10.1016/0003-2697(84)90026-5. [DOI] [PubMed] [Google Scholar]
- 26.Khougaz K., Gao Z., Eisenberg A. Determination of the critical micelle concentration of block copolymer micelles by static light scattering. Macromolecules. 1994;27:6341–6346. [Google Scholar]
- 27.Apel C.L., Deamer D.W., Mautner M.N. Self-assembled vesicles of monocarboxylic acids and alcohols: conditions for stability and for the encapsulation of biopolymers. Biochim. Biophys. Acta. 2002;1559:1–9. doi: 10.1016/s0005-2736(01)00400-x. [DOI] [PubMed] [Google Scholar]
- 28.Hammersley A.P., Svensson S.O., Hausermann D. Two-dimensional detector software: from real detector to idealised image or two-theta scan. High Press. Res. 1996;14:235–248. [Google Scholar]
- 29.Monnard P.A., Deamer D.W. Preparation of vesicles from nonphospholipid amphiphiles. Methods Enzymol. 2003;372:133–151. doi: 10.1016/S0076-6879(03)72008-4. [DOI] [PubMed] [Google Scholar]
- 30.Usov I., Mezzenga R. Correlation between nanomechanics and polymorphic conformations in amyloid fibrils. ACS Nano. 2014;8:11035–11041. doi: 10.1021/nn503530a. [DOI] [PubMed] [Google Scholar]
- 31.Lara C., Handschin S., Mezzenga R. Towards lysozyme nanotube and 3D hybrid self-assembly. Nanoscale. 2013;5:7197–7201. doi: 10.1039/c3nr02194g. [DOI] [PubMed] [Google Scholar]
- 32.Mehta A.K., Lu K., Lynn D.G. Facial symmetry in protein self-assembly. J. Am. Chem. Soc. 2008;130:9829–9835. doi: 10.1021/ja801511n. [DOI] [PubMed] [Google Scholar]
- 33.Reches M., Gazit E. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science. 2003;300:625–627. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- 34.Sánchez-Ferrer A., Adamcik J., Mezzenga R. Controlling supramolecular chiral nanostructures by self-assembly of a biomimetic β-sheet-rich amyloidogenic peptide. ACS Nano. 2018;12:9152–9161. doi: 10.1021/acsnano.8b03582. [DOI] [PubMed] [Google Scholar]
- 35.Oro J. Chemical synthesis of lipids and the origin of life. J. Biol. Phys. 1995;20:135–147. [Google Scholar]
- 36.Schrum J.P., Zhu T.F., Szostak J.W. The origins of cellular life. Cold Spring Harb. Perspect. Biol. 2010;2:a002212. doi: 10.1101/cshperspect.a002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rout S.K., Friedmann M.P., Greenwald J. A prebiotic template-directed peptide synthesis based on amyloids. Nat. Commun. 2018;9:234. doi: 10.1038/s41467-017-02742-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu T.F., Szostak J.W. Coupled growth and division of model protocell membranes. J. Am. Chem. Soc. 2009;131:5705–5713. doi: 10.1021/ja900919c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zozulia O., Dolan M.A., Korendovych I.V. Catalytic peptide assemblies. Chem. Soc. Rev. 2018;47:3621–3639. doi: 10.1039/c8cs00080h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bergman P., Roan N.R., Münch J. Amyloid formation: functional friend or fearful foe? J. Intern. Med. 2016;280:139–152. doi: 10.1111/joim.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maji S.K., Perrin M.H., Riek R. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kimura Y., Hosoda Y., Matsuno R. Physico-chemical properties of fatty acids for assessing the threshold concentration to enhance the absorption of a hydrophilic substance. Biosci. Biotechnol. Biochem. 1998;62:443–447. doi: 10.1271/bbb.62.443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.