Summary
When recombinant DNA technology was developed more than 40 years ago, no one could have imagined the impact it would have on both society and the scientific community. In the field of genetic engineering, the most important tool developed was the plasmid vector. This technology has been continuously expanding and undergoing adaptations. Here, we provide a detailed view following the evolution of vectors built throughout the years destined to study microorganisms and their peculiarities, including those whose genomes can only be revealed through metagenomics. We remark how synthetic biology became a turning point in designing these genetic tools to create meaningful innovations. We have placed special focus on the tools for engineering bacteria and fungi (both yeast and filamentous fungi) and those available to construct metagenomic libraries. Based on this overview, future goals would include the development of modular vectors bearing standardized parts and orthogonally designed circuits, a task not fully addressed thus far. Finally, we present some challenges that should be overcome to enable the next generation of vector design and ways to address it.
Introduction
In the past decades, plasmid vectors have become a pivotal tool in the field of molecular biology. They have allowed several major discoveries and have become essential tools in both basic and applied science by providing novel elements for accessing the molecular features of life. Plasmids have also been used worldwide for bolstering biotechnological advances, from the production of insulin by a recombinant strain of E. coli to treat diabetes (Johnson, 1982) to corn crops containing a Bacillus thuringiensis gene (Koziel et al., 1993). Joshua Lederberg coined the term ‘plasmid’ in his work on cytoplasmic heredity published in 1952 (Lederberg, 1952). This term has been widely accepted and used with the understanding that these genetic elements are not organelles, individual genes, parasites (viruses) or symbionts. The first time a plasmid was edited was in 1973 when researchers exchanged a gene for tetracycline resistance from pSC101 to a kanamycin one, becoming pSC102 (Cohen et al., 1973). Later, pBR322 was constructed and used as the base module for the engineering of a number of different genetic tools, many of them summarized in this review. Hence, plasmids became vectors, as means of transportation, for delivering and manipulating foreign DNA inside a host cell, starting the new era for molecular biology. The first edition of ‘Molecular cloning: the laboratory manual’, the reference book in most molecular biology laboratories, was published in 1982 and it marks the use of vectors as probably the most important tools for genetic manipulation. Therefore, scientists can now understand the behaviour, physiology, molecular mechanisms and gene expression patterns of cells and organisms, all due to increasing development of new molecular cloning strategies. A timeline for remarkable discoveries in vector design and technologies is presented in Fig. 1.
Figure 1.
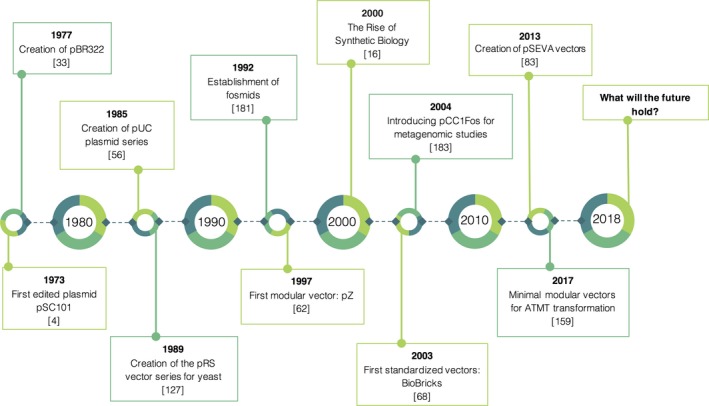
Timeline showing the most decisive breakthroughs regarding vector technology and design from 1970 until the present.
In general, vectors should have a set of characteristics that make them suitable for transformation and selection in the host organism. The first component is the origin of replication (ori) that will be recognized by the cellular replication machinery and will also define the number of copies of a given plasmid in the cell. Replication origins are usually recognized by their specific organism in what is called the narrow‐host‐range vectors, but there is also a category of broad‐host‐range vectors that contain origins capable of replicating in more than one species or genus, since they encode the protein that recognizes their own replication origin inside the plasmid (Durland et al., 1990). Furthermore, there is a series of vectors called ‘shuttle vectors’ that contain two different origins and two different selection markers so they can be transformed into two distinct organisms (Struhl et al., 1979). The differences between the ranges of hosts are assessed in Fig. 2A. It is important to notice that if the final plasmid has a proper origin of replication for the host, the genetic material inserted in the organisms is stable and can replicate autonomously. However, if no origin is available, the transformation requires the recombination of the vector into the host chromosome (e.g. in suicide plasmids), which necessarily leads to genome modification. Therefore, the existence of efficient origin of replications allows the decoupling between transformation and genome modification.
Figure 2.
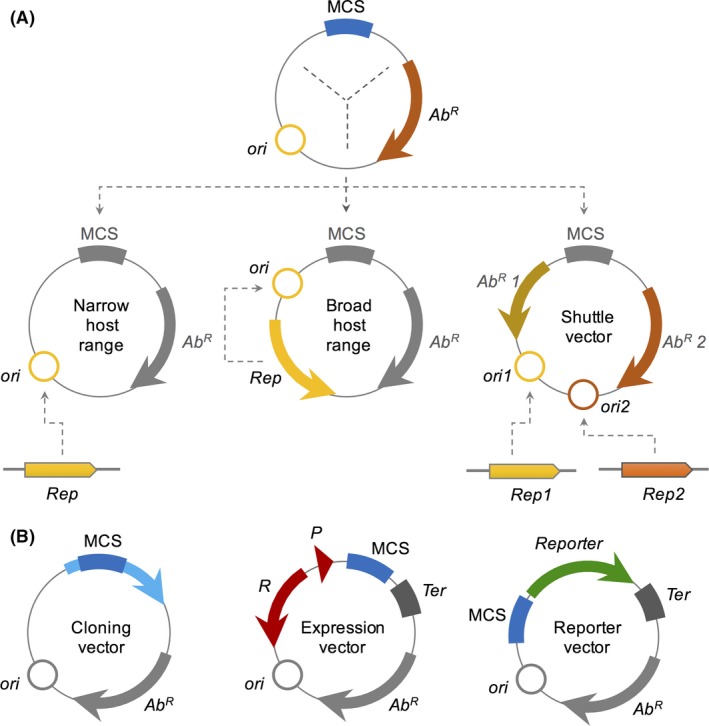
Most common bacterial plasmid architectures and categories.A. On the top, a minimal general architecture is formed by an origin of replication (ori), an antibiotic resistance marker (Ab R) and a multiple cloning site (MCS). From this general architecture, vectors can be categorized as narrow‐host‐range, broad‐host‐range and shuttle vectors. In the case of narrow‐host‐range, the ori relays on the replication machinery or a protein (Rep) provided by the host. In the case of broad‐host‐range vectors, the plasmid harbours its own Rep gene, which makes it mostly host‐independent. In the case of shuttle vectors, two ori regions (ori1 and ori2) are placed in the vector, each one being recognized by a specific Rep protein from different hosts (Rep1 and Rep2). In addition to that, two AbR (AbR 1 and AbR 2) are introduced to allow the selection in the appropriate host.B. Different functionalities of vectors. In cloning vectors, a MCS is usually located within a selection marker (such as the lacZα gene in pUC vectors) to allow the easy identification of plasmids with inserted fragments. In expression vectors, the MCS is preceded by an expression system (here shown as a regulated promoter P and its cognate regulator R) that allows the expression of the cloned fragment in response to a chemical inducer. Additionally, a strong terminator signal (Ter) is located after the MCS to ensure efficient transcriptional termination and increased plasmid stability. Finally, in a reporter vector, a reporter gene (such as a fluorescent protein, a luciferase coding gene or an enzyme‐coding gene such as lacZ) is flanked by an MCS and a strong terminator. In this system, the cloning of a promoter sequence in the MCS allows the investigation of promoter dynamics using the reporter gene of choice.
Another crucial component for a vector is the selection marker, which can be any gene allowing a selective advantage to the positive transformants, ranging from auxotrophy (corresponding to a metabolic enzyme missing in the host genome) to drug resistance (Gnügge and Rudolf, 2017). Also, a multiple cloning site (MCS) is usually added to facilitate cloning of the desired DNA, containing several sites recognized by different restriction enzymes. For simplicity, vectors can be divided into three typical classes most commonly used by researchers: cloning vectors, expression vectors and reporter vectors (Fig. 2B). Cloning vectors are the ones used to make numerous copies of a DNA of interest, keeping them stable inside a host organism (Rodriquez and Denhardt, 1988; Shizuya et al., 1992). Expression vectors are used to produce large amounts of a protein of interest; they usually contain a regulator and a target promoter that controls the expression of the gene encoding that protein (Stanley and Luzio, 1984; Terpe, 2006). Additionally, in reporter vectors, it is possible to place the promoter of the gene of interest to modulate a reporter protein, which can be fluorescent (Shaner et al., 2005), luminescent (Winson et al., 1998) or enzymatic, such as the ß‐galactosidase assay (Juers et al., 2012), among others. These reporter vectors allow in vivo analysis of gene expression kinetics throughout the organism's growth, allowing sophisticated studies even at the single‐cell level (Elowitz et al., 2002).
As new technologies and methodologies are surfacing, and researchers are now eager for fast, enhanced and easy‐to‐use molecular tools, mastering the principles and technologies of vector design has become a fundamental challenge. This is making room for the rise of an entirely novel discipline called synthetic biology (Rawis, 2000). This innovative field of study combines biological parts and modules to create more reliable and robust systems (Purnick and Weiss, 2009). The recent advances in DNA manipulation techniques such as automated DNA synthesis, sequencing and assembly have been combined with the synthetic biology framework, providing new perspectives on vector design and construction.
Not only did the majority of fundamental findings regarding molecular cloning arose from lessons given by microorganisms, but also there is an immense and much unexplored potential of those organisms in a wide range of applications such as biofuels and fine chemicals production (Sheldon, 2014; Jullesson et al., 2015; Kircher, 2015), biosensors (Courbet et al., 2015), bioremediation (Gavrilescu et al., 2015) and biomedical therapies (Din et al., 2016). Thus, efforts to understand and manipulate microorganisms are imperative, and as we acquire more knowledge, the classical genetic engineering approaches are no longer sufficient to answer all questions. Another challenge dwells in all the microorganisms that cannot be cultivated. Targeting that necessity, in 1998, Handelsman et al. launched a new approach called Metagenomics (Handelsman et al., 1998). The goal of metagenomics is to bypass the ‘pure culture paradigm’, which has blinded researchers from a genuine view of the microbial world for a long time. In fact, ‘meta’ is Greek for ‘transcendent’, meaning that pure genomics is not enough to understand all of Earth's diversity (Handelsman and Tiedje, 2007). To confront that task, several systems had to be adapted and created to clone DNA from environmental samples inside a cultivable host to search for genes of interest. This opens a new path towards the development of modern machinery for optimizing the prospection of novel biological parts through functional metagenomics (Guazzaroni et al., 2015; Alves et al., 2017a).
Despite the technological advances, the current vector architectures are still far from the conceptual framework. This is due to a number of caveats ranging from the intrinsic complexity of biological systems to a lack of solid standards for vector design. To address that, this review intends to condense most of the knowledge from the beginning of molecular cloning until the modern, advanced vectors used to transform microorganisms from bacteria to filamentous fungi, including their applications in metagenomics. In this sense, the present review aims to gather as much information regarding the available systems as possible, establishing a systematic overview of the uncontrolled expansion of genetic tools for microbiology accessible today, highlighting its current state along with the main challenges we still face. Thus, we provide an insight into how sophisticated current vectors are and glimpse into the new horizon of ever‐evolving tools we still need to generate.
Development of vectors to engineer bacteria
Extrachromosomal genetic elements, now widely known as plasmids, were first recognized in bacteria over 60 years ago (Cohen, 2013; Kado, 2014). The development of recombinant DNA techniques in the 1970s (Lobban and Kaiser, 1973; Novick et al., 1976; Bolivar et al., 1977; Sinsheimer, 1977; Cohen et al., 1992; Cohen, 2013; Kado, 2014) led to a multitude of possibilities for manipulating those natural plasmids, turning them into vector systems which could be useful in several applications. Early vector systems were based on natural ColE1 derivatives and were primarily restricted to E. coli owing to their replication machinery (Hershfield et al., 1974; Bolivar et al., 1977). The introduction of broad‐host‐range plasmids such as RK2 (from Pseudomonas aeruginosa; Stalker et al., 1981; Thomas, 1981) and RSF1010 (from Salmonella panama; de Graaff et al., 1978; Bagdasarian et al., 1981) made it possible to introduce recombinant DNA technologies into bacteria other than E. coli. In recent times, a number of vectors have been derived and gradually optimized (for properties such as size, stability and functionality) from those naturally occurring plasmids for a myriad of purposes.
One of the first and most significant artificial vectors developed was the pBR322 (Bolivar et al., 1977) which was derived from ColE1. This vector, still currently in use, was initially built for general cloning purposes and can be considered one of the most important bacterial vectors, since several other tools have been derived from it for a wide range of functions (Balbás et al., 1986; Rodriquez and Denhardt, 1988). Some structural and functional modifications include the addition of new restriction sites (Davison et al., 1984; Sambrook et al., 1989), differentiation or change of selection marker (Rao and Rogers, 1979; Herrin et al., 1982; Richardson et al., 1982), increased stability (Skogman et al., 1983; Summers and Sherratt, 1984; Zurita et al., 1984; Chiang and Bremer, 1988), change in the copy number (Twigg and Sherratt, 1980; Boros et al., 1984; Soberon et al., 1990), addition of a signal peptide to facilitate protein secretion (Villa‐Komaroff et al., 1978; Talmadge and Gilberg, 1980), and finally, change in the origin for one of a shuttle vector, allowing vector propagation in different hosts (Brückner, 1992). Nowadays, two important and widely used commercial cloning vectors are pJET1.2/blunt (Thermo Fisher Scientific) and pGEM‐T Easy (Promega Corporation), with features such as lethal phenotype and β‐galactosidase activity‐driven selection respectively. As these vectors can supply most requirements for simple cloning processes, few efforts have been made over the past years in the attempt to create novel tools for this task.
On the other hand, gradual development and optimization of expression vectors have potentially allowed the biosynthesis of any type of heterologous proteins in vivo. In this context, the pUC‐plasmid series for expression (Norrander et al., 1983; Hanna et al., 1984; Yanisch‐Perron et al., 1985), one of the pBR322 derivatives, marked the timeline of expression vectors. The pUC‐series vectors are mainly composed of a lac promoter–operator and require compatible hosts for α‐complementation (blue/white screening system that allows recovering of functional β‐galactosidase LacZ), providing a positive selection for recombinants. In the same way cloning vectors were successively derived from each other, many expression vectors were derived from pUC‐series as the lacUV5 mutant (Rodriquez and Denhardt, 1988), which contains just two base pair mutations in the −10 hexamer of the classical lac promoter and the tac hybrid promoter (Rodriquez and Denhardt, 1988). Many promoter/operator modifications were made during this time for addressing specific needs in protein expression systems. In addition to pUC‐based vectors, a number of other expression vectors were constructed from the trp‐promoter, carrying different segments of the trp operon (Enger‐Valk et al., 1980; Hallewell and Emtage, 1980). Nowadays, besides pUC18 and pUC19, the pET‐series (Novagen, Madison, WI, USA) vectors, which were also derived from pBR322 (Ramos et al., 2004) and pGEX (GE Healthcare Life Sciences), are widely used because they are high copy number expression vectors that contain protein tags that facilitate the subsequent purification of the desired protein. These two vectors are best suited for quick and easy heterologous protein expression.
Although the diversity of bacterial vectors has enormously increased during the decades following the discovery of the recombinant DNA technology, the design and architecture of those tools occurred rather unsystematically. Vectors with similar functions were generated with different standards between laboratories, hindering data comparison and requiring fastidious efforts for re‐cloning sequences into available vectors. In addition, although a few plasmids were developed for different hosts, most of them were focused on the organism model E. coli, which is not ideal for many biotechnological applications (e.g. fine chemical production, biodegradation and environmental release). Furthermore, the lack of standards for minimal genetic units such as promoters, terminators and replication origins (usually based on the amplification of naturally occurring sequences) generated vectors with long and unnecessary sequences, sometimes bearing undesired features such as common restriction sites and cryptic functions, which could interfere with the plasmid integrity/stability (Summers and Sherratt, 1984). Thus, despite the massive amount of information accumulated regarding bacterial vectors, the field has drifted towards structural diversification rather than to a more unified standard.
The era of modular vectors in bacteria
In the recent years, further advances in molecular cloning process, computational methods and DNA sequencing automation have gradually allowed the scientific community to overcome the challenges of vector design. A cornerstone in generating a standardized system was the creation of pZ vectors in 1997 for the study of transcriptional regulatory elements in bacteria (Lutz and Bujard, 1997). In this work, Lutz and Bujard (1997) introduced the term ‘modularity’ in vector design. Modularity has been widely used in studies of technological and organizational systems. Product systems are deemed ‘modular’, for example, when they can be decomposed into a number of components that may be mixed and matched in a variety of configurations (Baldwin and Clark, 2000). In this sense, the pZ vectors were organized into three main functional modules, physically separated by specific restriction sites: (i) antibiotic resistance module, (ii) origin of replication module and (iii) expression/transcriptional regulated module. Each part of the system could be easily shifted by a functional variant and combined to the next one, allowing the generation of a wide range of testable vectors. The result was a library of unique vectors composed of different components, yet bearing the same architecture. The central point of this system was to evaluate the efficiency of previously described transcriptional elements (Knaus and Bujard, 1990; Skerra, 1994; Guzman et al., 1995) in regard to plasmid features such as copy number and antibiotic resistance. The use of the promoters controlled by elements of the lac, ara or tet operon (Tn10) and therefore induced by IPTG, arabinose and tetracycline, respectively, generated strongly repressible promoters that could regulate expression levels up to 5000‐fold. In addition, using different origins of replication was shown to shift expression tightness (Lutz and Bujard, 1997). Therefore, those findings represented a major advance not just in the study of gene regulation, but also in the study of how the components or functional modules of a vector system affect each other (Fig. 3).
Figure 3.
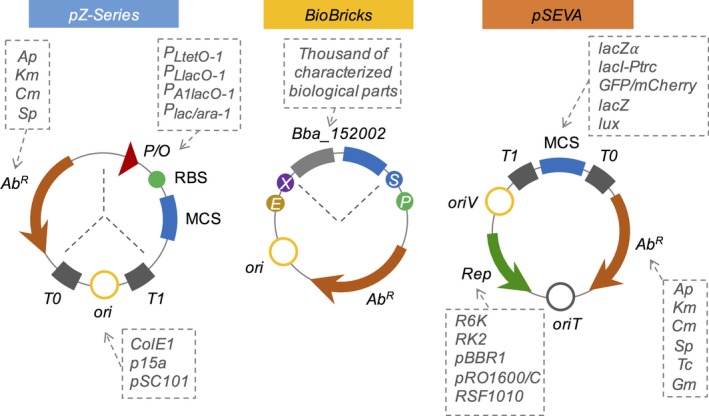
New generation of modular vectors. The pZ‐series represented remarkable progress in the new generation of vectors. In this set‐up, three modules are combined to generate a collection of vectors. In the original design, three narrow‐host‐range origins were combined with four antibiotic resistance markers and a few expression systems and reporters (Lutz and Bujard, 1997). In the BioBrick platform, a very interactive set‐up is used to assemble complex circuits by the joint of prefix and suffix fragments using only four restriction enzymes (Eco RI, XbaI, SpeI and PstI). This platform is merged with a collection of thousands of well‐characterized biological parts and is widely used by the iGEM community (Knight, 2003; Shetty et al., 2008). In the SEVA platform, broad‐host‐range origins of replication (represented by the oriV and Rep elements) are selected, allowing the replication of the plasmids in multiple Gram‐negative bacteria. This is combined with a number of antibiotic resistance markers and many functional systems at the MCS regions (some examples are shown in the figure). Another exclusive feature of this platform is the existence of a transference origin (oriT) to allow the mobilization of the plasmids to the target bacteria using conjugation (Silva‐Rocha et al., 2013).
A few years after Lutz and Bujard's work, despite the rather tumultuous generation of cloning vectors, a new conceptual framework was conceived, combining the technological advances in molecular biology and the hierarchical abstraction and rational design concepts from Engineering Sciences. It was called synthetic biology (Andrianantoandro et al., 2006) and its main rationale was to shift molecular biology from traditional ‘copy and paste’ methods to a more high‐throughput, concise and design‐oriented background for generating novel biological functions. As highlighted by Lutz and Bujard's studies (Lutz and Bujard, 1997) and by many others afterwards, each part of the vector system matters, and in order to standardize biological parts, the generation of vectors with easily interchangeable modules would be essential. Thus, it did not take long for the first library of standardized vectors and biological parts to be established. In 2003, the BioBrick standard (BBF RFC 10) was proposed by Knight (2003), employing standard suffix and prefix sequences that flanked every designed biological part (see Fig. 3). This method was further optimized (Canton et al., 2008; Shetty et al., 2008) and has allowed an easy process for joining parts by standard restriction‐ligation methods and hierarchical assembly (Shetty et al., 2011). Further efforts driven by the dissemination of the community‐based iGEM competition have allowed the development of a repository of standard biological parts and novel standards for vector assembly (Endy, 2005; Knight, 2007; Müller et al., 2009; Røkke et al., 2014).
Although the BioBricks standard was essential for establishing functional units in synthetic biology such as promoters, terminators and genes, it did not provide many flexible modules (e.g. no easily and independent interchangeable modules for origins of replication and antibiotic resistance genes). Furthermore, the traditional restriction enzyme‐based method required the previous removal of standard recognition sites from the biological part and generated an 8‐bp scar during the assembly process, which could destabilize the plasmidial system (Anderson et al., 2010; Ellis et al., 2011; Yao et al., 2013). In order to overcome these issues, a few other systems with the same basic architecture were designed, such as the Bgl Bricks (Anderson et al., 2010; Lee et al., 2011), the iBricks (Liu et al., 2014) and the epathBricks (Xu and Koffas, 2013), the latter being the most modular system among them. The pBAM1 vectors (Martínez‐García et al., 2011), a modular remake of the original mini‐Tn5 transposon vector concept, and its more versatile and successful successor, the Standard European Vector Architecture (SEVA; Silva‐Rocha et al., 2013), also use a set of restriction sites to standardize DNA assembly (see Fig. 3). The SEVA collection differs from the majority of assembly methods in that it is more correctly described as a modular standard. This describes a set of criteria for the physical assembly of plasmids according to a three‐component architecture: an origin of replication segment, a selection marker segment and a cargo segment (Silva‐Rocha et al., 2013). These segments are flanked by insulator sequences and are assembled together with a set of rare restriction sites (Fig. 3). Although some optimizations were made over the years, such as the SEVA linkers (Kim et al., 2016), the SEVAs still suffer from a few issues such as rare restriction enzyme sites, non‐optimized synthetic parts and a lack of standardization for some of its functional parts.
While the rationales behind traditional restriction site‐based assembly methods support modularity, their limitations have led several research groups in the synthetic biology community to ‘trade‐in’ standardization and modularity for ‘bespoke’ assembly methods that enable one‐pot assembly of multiple DNA parts. Those methods for à la carte vector assembly have become increasingly popular along the last decade due to their versatility, low cost and high speed for simultaneous assemblies. Although very diversified, the most common methods are Gateway (Invitrogen, 2011), Golden Gate (Engler et al., 2009), MoClo (Weber et al., 2011), Gibson (Gibson et al., 2009), Slic (Li and Elledge, 2007), Cpec (Quan and Tian, 2009), Slice (Zhang et al., 2012) and Paperclip (Trubitsyna et al., 2014). For a more detailed review, see (Kelwick et al., 2014; Casini et al., 2015). However, it is important to highlight that although vector modularity has gradually shifted towards novel high‐throughput assembly methods, well‐defined modular vectors are still essential tools for many applications. Here, we can emphasize the use of modular vectors for the exploration of genetic functional space in bacteria (Westmann et al., 2018), the functional re‐wiring of genetic features and the generation of novel biological functions. The concept of modular vectors has also inspired the development of plasmid collections for specific bacterial hosts such as Pseudomonas putida, Clostridium spp., Cyanobacteria spp., Bacillus spp. and Geobacillus spp. (Heap et al., 2009; Radeck et al., 2013, 2017; Silva‐Rocha et al., 2013; Taton et al., 2014; Wright et al., 2015; Reeve et al., 2016; Popp et al., 2017), which become essential for establishing novel chassis in synthetic biology.
Thus, we have reached a state in which modular vector or high‐throughput assembled constructs, from vectors to genomes, can be easily designed and assembled through computational and experimental tools (Casini et al., 2015; Woodruff et al., 2017). However, it is clear that this powerful design and build process was not followed by the standardization of biological parts that ultimately compose those systems (Decoene et al., 2017). The lack of biological information regarding the behaviour of simple genetic features such as promoters, terminators and origins of replication hinders the potential of engineering living organisms. Furthermore, we are currently experiencing the age of emergent phenomena in synthetic biology (Pósfai et al., 2006; Kwok, 2010; Chen et al., 2015; Monteiro et al., 2017), an ubiquitous property of life, highlighted decades ago by systems biology (Bhalla and Iyengar, 1999; Kitano, 2002). In this context, the most well‐characterized biological part might exhibit unpredictable behaviour when combined with other parts or when exposed to different compositional contexts such as the constraints imposed by vector biophysical structure/architecture or the combination of functional regulatory cis‐elements (Cardinale and Arkin, 2012; Goñi‐Moreno et al., 2017; Maria Oliveira Monteiro et al., 2017; Yeung et al., 2017). Therefore, the new frontier in vector design is to understand the constraints imposed by its minimal parts on a systemic context, allowing the fine‐tuning of its functionalities.
Moreover, an important step is to ensure that the massive amounts of data generated on biological parts and devices do not end up disconnected, and for that, standardization in data sharing is needed. To this end, various data registries and repositories of parts and devices have already been established and are curated regularly. Noteworthy examples are the Virtual Parts Repository (URL: http://sbol.ncl.ac.uk:8081/; Cooling et al., 2010), the Registry of Standard Biological Parts (URL: http://parts.igem.org/; Endy, 2005), the Joint BioEnergy Institute's Inventory of Composable Elements (JBEI‐ICE; URL: https://acs-registry.jbei.org; Huynh and Tagkopoulos, 2016), the Standard European Vector Architecture 2.0 database (SEVA‐DB 2. 0, URL: http://seva.cnb.csic.es/; Martínez‐Garćía et al., 2015), and newly the Plant Associated and Environmental Microbes Database (PAMDB; URL: http://genome.ppws.vt.edu/cgi-bin/MLST/home.pl). Some repositories, such as the Registry of Standard Biological Parts, have also implemented quality control checks.
The crude, yet essential, generation of bacterial vectors back in the early days has been continuously refined over the years, following the development of novel technologies and molecular techniques. The subsequent advent of synthetic biology and novel assembly methods has driven the field of bacterial vector design to a whole new level. The previously proposed concept of modularity was finally incorporated into biological engineering, allowing a number of standard collections to be derived from it. Currently, vector modularity, although essential, is at stake with the novel high‐throughput technologies for à la carte DNA assembly (see Fig. 3). Still, directed approaches such as the rigorous standardization of biological parts, the comprehension of emergent properties on genetic circuits and the expansion of biological functions to a wide range of chassis will constitute the next frontier for advancing bacterial vector design.
Evolution of vector engineering for fungi
Tools for yeast transformation
Circular DNA was already known to be present in prokaryotes, as already mentioned, over 60 years ago. In eukaryotes, more precisely in Saccharomyces cerevisiae, circular DNA was first discovered in mitochondria, and it took several years for researchers to realize this was too prokaryotic DNA (Margulis and Chapman, 1998). A non‐mitochondrial natural plasmid DNA was first described in 1971 (Guerineau et al., 1971). Three years later, researchers found an antibiotic resistance gene present in those plasmids and realized they could behave just like the bacterial ones allowing DNA cloning (Guerineau et al., 1974). Soon after, yeast transformation was described as well as integration of the genes in the yeast genome (Hinnen et al., 1978; Cameron et al., 1983). Guerineau's group noticed that yeast plasmids had a length of about 2 μm, consequently, they were named 2μ. In the years that followed, many researchers began constructing vectors for yeast manipulation, many of which used the 2μ origin (Struhl et al., 1979; Ferguson et al., 1981).
Plasmids used to transform Saccharomyces can be divided into three groups: yeast centromeric plasmids (YCps), Yeast Episomal plasmids (YEps) and Yeast Integrative plasmids (Yips). The YCps need autonomously replicating sequences (ARS) and centromeric sequences (CEN) where kinetochore complexes attach, thus behaving like a microchromosome (Clarke and Carbon, 1980; Westermann et al., 2007). The YEps are based on the endogenous 2μ plasmid mentioned above with addition of a bacterial origin of replication and selection marker, yeast selection marker and the expression cassette. Still, the YIps need to have homology sequences so they can integrate in the yeast genome via homologous recombination (Gnügge and Rudolf, 2017). For all of them, the selection marker is usually auxotrophic, which somewhat restricts their use because there is usually only URA3 (encoding orotidine‐5′‐phosphate decarboxylase), LEU2 (encoding 3‐isopropylmalate dehydrogenase), HIS3 (encoding imidazoleglycerol‐phosphate dehydratase) and TRP1 (encoding phosphoribosylanthranilate isomerase) options, besides the need for a strain with the original gene deleted. Generally, all series of vectors present the three types; in a matter of deciding which methodology to use, the biological question being examined should be considered (Fig. 4).
Figure 4.
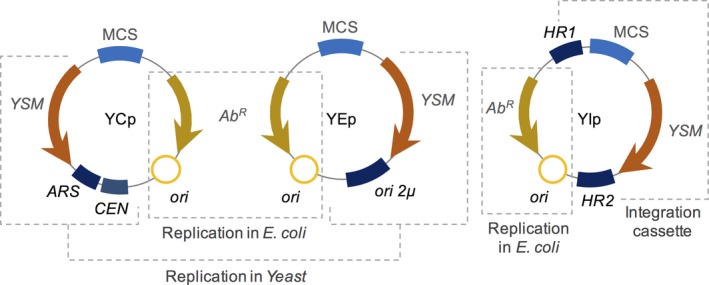
Modular vectors designed for yeast. The yeast centromeric plasmids (YCps) harbour the autonomously replicating sequences (ARS) and centromeric sequences (CEN), which allows the vectors to behave as mini‐chromosomes. The Yeast Episomal plasmids (YEps) are endowed with the 2μ origin of replication and are similar to plasmids in bacteria. In the case of Yeast Integrative plasmids (Yips), homologous regions (labelled as HR1 and HR2) to the host chromosome allow the integration of the target region through homologous recombination events. In all vectors, the yeast selection marker (YSM) represents a gene that allows the selection of transformants harbouring the vectors. In all cases, specific regions for replication of the bacterial host (usually E. coli) and the region required for replication or integration in yeast are highlighted.
Despite the fact that it was Hinnen and colleagues that developed a transformation method for yeast using bacterial vectors in 1978, the vectors containing the ability to replicate in both E. coli and S. cerevisiae were developed a year later by Struhl et al. (1979). Shuttle vectors, as they are called now, are still the preferred method for yeast transformation, and they are constantly evolving. Ferguson et al. (1981) developed the series of pRC1, pRC2 and pRC3 derived from pKC7 (which was originally derived from pBR322). Gietz and Sugino (1988) built the plasmids YCplac, YEplac and YIplac based on pUC19 in 1988. Around the same time, Ma et al. (1987) developed a method of easy recombination of plasmid parts and established the series of shuttle vectors YCp400 and YEp400. All of these vectors were considerably large (some more than 10 kb) and had no more than 10 unique restriction sites for cloning. However, in 1989, there was the breakthrough for yeast scientists: the pRS series (Sikorski and Hieter, 1989). These vectors were made small (around 5 kb), with several restriction sites (around 13 unique sites each), four different selection markers to choose from, and since they were based on pBLUESCRIPT, positive transformants could be selected by colour in E. coli. pRS vectors were such a turning point in this field that new designs for yeast tools were very limited for almost twenty years afterwards, with scientists only adding some adaptations such as new resistance markers (Chee and Haase, 2012), and these tools are still frequently used and adapted (Avalos et al., 2013).
Nevertheless, in the last 10 years, huge improvements have been made in yeast molecular tools. The plasmids are now smaller and have the feature of recycling selection markers to overcome the lack of variability of those. In 2007, the pAG series was created, still based on the pRS series, containing more than 200 options of YCps, YEps and Yips vectors for cloning, expression and also presenting reporter genes such as GFP and dsRed (Alberti et al., 2007). In 2011, Fang et al. (2011) developed a collection called pXP based on pUC18, with the advantage of a selection marker flanked by loxP sites, meaning it can be recycled using loxP/Cre technology (Güldener et al., 1996). Three years later, in 2014, Jensen and his group designed the EasyClone: a set of vectors that can integrate three cassettes at a time carrying up to two genes each and showed that this is more homogeneous and stable than expressing more than one gene in episomal vectors (Jensen et al., 2014). A variation of EasyClone called EasyClone2.0, published in 2015, has the very unique feature of using dominant markers for selection instead of auxotrophy, so it can be applied to prototrophic strains (Stovicek et al., 2015). A year later, the EasyCloneMulti collection was created, complementing the latter two. EasyCloneMulti integrates into long terminal repeats (LTR) of Ty retrotransposon sequences, enabling multiple integrations throughout the yeast genome (Maury et al., 2016). All of these three series of EasyClone vectors contain USER cloning sites (Bitinaite et al., 2007) as a facilitator strategy and present the benefit of loxP/Cre recycling systems as well.
At last, the pRG series was generated very recently by Gnügge et al. (2016). This collection comprises vectors of all types – YEps, YCps and Yips – exhibiting the unlimited benefit of modularity, which means that all parts of the vectors are flanked by restriction sites and can be interchanged or substituted, as mentioned earlier. The systems also allow multiple integrations and their resistance markers are auxotrophic. The integration vectors of both EasyClone and pRG series use a double cross‐over mechanism to integrate into the genome, thus determining the stability of the insert inside the chromosome. Table 1 summarizes the most important tools for yeast manipulation from 2007 to 2017. Likewise, Gnügge and Rudolf (2017) also described, in an extended review, most of the shuttle vectors available for yeast.
Table 1.
Plasmid vectors used for fungal transformation
| Plasmid series name | Fungal selection marker | Approximate average vector size | Features | Fungi type | References |
|---|---|---|---|---|---|
| pAG | HIS3, LEU2, TRP1, URA3 | 7 kb | More than 200 options; contains fluorescence reporters | Yeast | Alberti et al. (2007) |
| pXP | HIS3, LEU2, MET15, TRP1, URA3 | 5 kb | Recycling of selection markers by loxP/Cre technology | Yeast | Fang et al. (2011) |
| EasyClone | HIS3, LEU2, LYS5, URA3 | 6 kb | Multiple integrations; recycling of markers. | Yeast | Jensen et al. (2014) |
| EasyClone2.0 | amds, ble, dsd, hph, kan, nat | 6 kb | Compatible with prototrophic strains; recycling of markers | Yeast | Stovicek et al. (2015) |
| EasyCloneMulti | Kl.URA3‐degradation signal | 6 kb | Integrates into Ty sequences; recycling of markers. | Yeast | Maury et al. (2016) |
| pRG | HIS3, LEU2, LYS2, MET15, URA3 | 6 kb | Modular design; multiple integrations; recycling of markers | Yeast | Gnügge et al. (2016) |
| pWEF | hph | 12 kb | Binary vector | Filamentous | Lv et al. (2012) |
| pDESTR | hph | 5 kb | Gene targeting and disruption | Filamentous | Abe et al. (2006) |
| pCBGW‐GFP | hph | 8 kb | Expression vector | Filamentous | Zhu et al. (2009) |
| pGWB2‐GFP | hph | Not shown | Binary vector | Filamentous | Zhu et al. (2009) |
| pEX1 and pEX2 | pyrG | 10 kb | Binary vector | Filamentous | Nguyen et al. (2016) |
| pBI‐hph | hph | 15 kb | Binary vector | Filamentous | Zhong et al. (2007) |
| pALS‐1 | qa‐2+ | 13 kb | Tested in N. crassa. | Filamentous | Sthol and Lambowitz (1983) |
In contrast with the classical vectors and taking advantage of the CRISPR/Cas feature of not needing a selection marker, Mans et al. (2015), Generoso et al. (2016), Shi et al. (2016) and Apel et al. (2017) developed tools for efficiently editing the S. cerevisiae genome. Similarly, the EasyClone series already has a new version of CRISPR/Cas vectors named EasyClone‐MarkerFree (Jessop‐Fabre et al., 2016). These are whole new strategies that can also be availed by researchers to study the characteristics of yeast cells.
Saccharomyces cerevisiae is a model organism with a very well‐annotated genome, thus it has been essential for the evolution of the ever‐growing field of genetic engineering (Nielsen et al., 2013). However, genetic tools such as vectors and integrating cassettes have been developed for non‐conventional yeast as well, considering their rising importance in biotechnology, such as Pichia pastoris (Cereghino and Cregg, 2000), Kluveromyces lactis (Van Ooyen et al., 2006) and Yarrowia lipolytica (Bredeweg et al., 2017). For an expanded review of non‐conventional yeasts, tools refer to Wagner and Alper (2016). Undoubtedly, efforts to construct yeast tools have largely focused on Saccharomyces for decades. However, the need for alternative and more adapted species of yeasts, for example Kluyveromyces marxianus that is thermo‐tolerant and can be used in bioreactors (Nambu‐Nishida et al., 2017), is accelerating the search for adaptable genetic systems. Considering this, we are probably only a few years away from achieving a robust yeast vector. These vectors are becoming smaller, easy to manipulate, modular and capable of multiple integrations. Additionally, they have options for recycling selection markers and most of them currently contain several restriction sites as cloning options. Still, the ideal framework would be a standard vector that could work in several (if not all) yeast species.
Tools for filamentous fungi
Since genetically manipulating filamentous fungi is considerably more complex than manipulating yeast, much discussion has been raised to improve the molecular tools for genetic transformation of filamentous species. In this sense, some efforts have been made in this field to unravel the genetic mechanisms for successful cell transformation using fungal plasmids. A remarkable study in Neurospora crassa, a model fungus for genetic research, has helped determine how plasmids could be extended to successful fungal manipulation. The shuttle vector pALS‐1, originally described by Sthol and Lambowitz (1983), is a 13.1 kb recombinant plasmid that replicates in both N. crassa and E. coli; it is based on the backbone of the mitochondrial plasmid P405‐Labelle and on the E. coli plasmid pBR325, and also contains the Neurospora qa‐2+ selection gene (Sthol and Lambowitz, 1983). This vector was one of the landmark genetic tools for fungi manipulation because it was described as one of the first vectors shown to replicate autonomously in the nucleus or in the cytosol of a filamentous fungus cell, which highlighted the field of fungal genetics and spread opportunities for creating versatile tools for this purpose (Sthol and Lambowitz, 1983).
Years later, in Trichoderma reesei, the most utilized fungus for cellulase production, Steiger et al. (2011) obtained a successful genetic transformation system that favoured homologous recombination using a loxP/Cre system for creating gene deletion with pMS plasmids vectors (Steiger et al., 2011). In this same fungus, Lv et al. (2012) reported the construction of two expression vectors, pWEF31 and pWEF32, with the cellobiohydrolase gene I (cbhI) promoter regulating the expression of a reporter fluorescent red protein (Lv et al., 2012) and demonstrating an effective Agrobacterium tumefacien‐mediated transformation (ATMT) process (Fig. 5). Despite the promising outcomes observed in T. reesei, further studies are required to more deeply elucidate its genetic mechanisms for transformation effectiveness, showing that creating new plasmid vectors is pivotal for functional genomic studies in this very relevant species.
Figure 5.
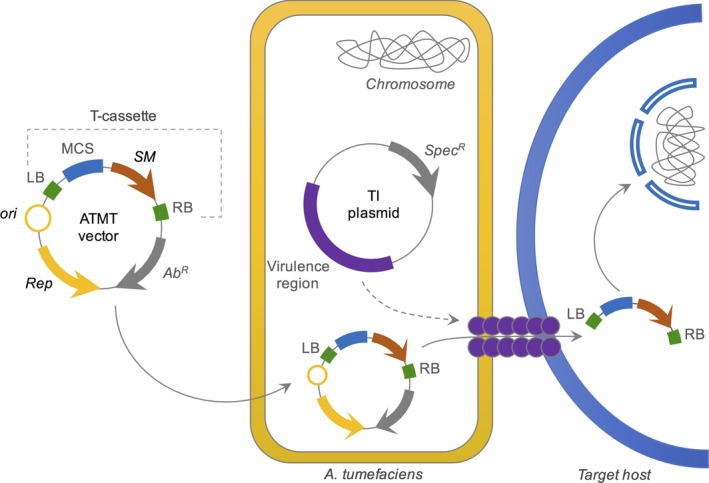
Minimal genetic tools based on Agrobacterium tumefacien‐mediated transformation (ATMT). ATMT vectors are based on broad‐host‐range plasmids and harbour a T‐cassette, which is composed of a MCS and a selection marker (SM) flanked by the left and right borders (LB and RB) required for the recognition of the A. tumefaciens machinery. Once inserted in the proper A. tumefaciens strain harbouring the TI plasmid (which expresses the components for T‐cassette mobilization), this vector can be used to introduce the T‐cassette into hosts such as fungi and plants.
Some fungal plasmids have been created towards multifunctional roles in fungal transformation, from increasing of gene copy number to integrative recombination. In this sense, a series of plasmids have been constructed based on pDONR vectors (Gateway) through Gateway cloning technology using λ phage integrase proteins and attachment regions for recombination (Schorbele et al., 2013). These vectors were designed to attend different conditions and applications in fungal species, and for that reason, it allows selection by nutritional and drug resistance markers, showing to be a promising and a versatile genetic tool for genetically manipulating distinct fungi (Schorbele et al., 2013).
Gateway technology has been also utilized to create several other vectors. For example, the vector pTROYA allowed the utilization of a fungal vector for RNA interference approaches to construct the PAC1 mutant strain in Colletotrichum gloeosporioides and it was a potential tool for screens in non‐sequenced organisms (Shafran et al., 2008). Additionally, Gateway approaches were also used for developing the pDESTR vector, a plasmid based on the backbone of pGEM‐T easy (Promega) without its MCS region plus a Gateway cassette and a hygromycin resistance gene sequence, created to accomplish gene targeting and disruption in filamentous species (Abe et al., 2006). Zhu et al. (2009) used Gateway for the construction of expression vectors for ATMT (Zhu et al., 2009). Studies like this one that perform fungal cell transformation through ATMT are getting more visibility in the scientific community due to the higher transformant frequency when compared to usual methods like protoplast fusion. Other binary plasmids have been reported for ATMT processes, such as the vectors pEX1 and pEX2, with auxotrophic selection for pyrG from Aspergillus oryzae (Nguyen et al., 2016), and pBI‐hph, with selection for hygromycin (Zhong et al., 2007).
New advances have continuously been reported concerning vectors used for Agrobacterium transformation. Recently, the creation of a series of vectors was described for fungal transformation through ATMT based on the plant binary vector pCAMBIA2200, named by Nishikawa et al. (2016) as pFungiway. These vectors were created for two distinct purposes: to guarantee the expression of a gene under the regulation of a constitutive promoter and to promote the negative regulation of target gene expression through RNAi (Nishikawa et al., 2016). More recently, a set of synthetic, minimal and modular binary vectors for multiple transfer of T‐DNA was published in 2017 (Pasin et al., 2017). Despite being applied in plants, this is a frontier technology that can be expanded and adapted for fungal transformation in the future. Regardless, Table 1 summarizes some of the most important vectors built for fungal transformation.
CRISPR‐Cas9‐based vectors are also feasible in fungal genetic manipulation. In this concern, Nødvig et al. (2015) created four new vectors derived from the pFC330 vector with distinct fungal selectable markers (pyrG, arbB, bleR or hygR; Nødvig et al., 2015). At this point, authors successfully demonstrated the genetic transformation and genome engineering of Aspergillus species, suggesting that CRISPR‐Cas9 tools are efficient but still could be enhanced.
Filamentous fungi are by far the toughest organisms to transform and manipulate among all microbes (Ruiz‐Díez, 2002). Even with the advances towards the state‐of‐the‐art vector creation, fungal plasmids used for genetic engineering are still poorly understood, unreliable and inefficient, especially when compared to bacterial ones. Still, the increasing development of synthetic biology studies focused on fungi has strongly contributed to the rising and constant necessity of creating new tools for fungal applications in several areas of molecular biology (Amores et al., 2016). New forms of vector design, plus characterization and standardization of their parts, are crucially needed to promote a better understanding of fungal molecular mechanisms.
Engineering new vector for Metagenomics
The term ‘metagenome’ was introduced in 1998 by Handelsman et al. (1998), and since its first use, this methodology became a powerful tool for analysing composite genomes of microbial communities and their potential products for novel biotechnological and pharmaceutical applications. In addition, the use of functional metagenomics has been shown to be effective for identifying new enzymes, antibiotics and other molecules derived from a variety of environments without the need for isolating and cultivating microorganisms in the laboratory (Courtois et al., 2003; Ferrer et al., 2012; Pozo et al., 2012; Thompson et al., 2013; Yang et al., 2016).
Despite the great potential of metagenomic approaches, some barriers have limited the discovery of new genes. The probability of identifying a particular gene depends on multiple factors that are intrinsically linked: the host‐vector system, the size of the gene, the recovered metagenomic DNA abundance, the screening method and the efficiency of heterologous expression of the gene in a substitute host (the most frequently used is E. coli; Gabor et al., 2004; Villegas and Kropinski, 2008; Uchiyama and Miyazaki, 2009). According to Gabor et al. (2004), only 40% of enzymatic activities can be recovered from random cloning in E. coli. This might be due to a number of factors that exist in heterologous genes that differ from those used by E. coli, preventing the host cell expression machinery from recognizing these signals, such as differences in translation initiation codons; in E. coli, the preferred translation initiation codon is AUG, whereas in some organisms, GUG and UUG are preferred (Villegas and Kropinski, 2008). In addition, differences in codon usage, promoters for transcription and ribosomal binding sites may lead to no detectable expression of the target genes. Furthermore, incorrect protein folding, toxicity of the gene product and an inability to secrete the gene product may be obstacles to identifying new genes in functional screenings (Ekkers et al., 2012). Choosing an appropriate vector for constructing a metagenomic library depends on various factors, such as the DNA size of the expected target compound of interest, whether a small‐ or large‐insert library is considered, the hosts that will be used in the screenings, as well as the screening tests (Ekkers et al., 2012).
Plasmid vectors are usually chosen to generate small‐insert metagenomic libraries (< 10 kb average insert size), becoming an appropriate tool to identify single gene products encoded by small DNA sequences, such as enzymes and antibiotic resistance genes (Guazzaroni et al., 2010). Regulating the expression level of cloned genes is possible by using inducible promoters upstream of the DNA insertion site or by choosing a suitable plasmid copy number, avoiding high expression rates of toxic genes or inclusion body formation of target proteins, and low expression rates that could prevent detection in functional screenings (Hudson, 2001). Large‐insert metagenomic libraries are usually generated using cosmid or fosmid vectors and provide a more efficient method to identify complete operons and biosynthetic clusters of genes. Cosmids are vectors capable of carrying DNA inserts of about 30–40 kb in size that contain the cos site of λ phage for packing inserted metagenomic DNA in λ phages (Hohn and Collins, 1980). This type of vector can replicate in suitable hosts since it carries a proper origin of replication, but the lack of a copy number control mechanism usually decreases cosmid stability (Haley, 1988; Cheng et al., 2017). Fosmid vectors are similar to cosmids, but they use an E. coli F‐factor origin of replication, making the fosmid copy number highly regulated to 1 or 2 copies in the cell and replication is restricted to E. coli (Kim et al., 1992; Fig. 6). This fact could avoid gene toxicity interference in metagenomic tests; also, fosmids are capable of carrying large‐insert DNA sizes of about 40–50 kb (Santana‐pereira and Liles, 2017). One example of a regularly used fosmid is the pCC1FOS (Epicentre, Madison, Wisconsin), a commercial cloning vector that allows copy number to be controlled through the CopyControl Cloning System. The pCC1FOS fosmid was first introduced in metagenomic studies in 2004 in a proof‐of‐concept report using a (meta)genomic library from Collimonas fungivorans (Leveau et al., 2004). Yet, for even larger fragments, bacterial artificial chromosomes (BACs) are used. These vectors are modified plasmids that contain an origin of replication derived from the E. coli F‐factor and can stably maintain and replicate inserts ranging from 100 kb to 220 kb, as well as inserts of more than 300 kb, and are usually used in E. coli (Shizuya et al., 1992; Beja et al., 2000).
Figure 6.
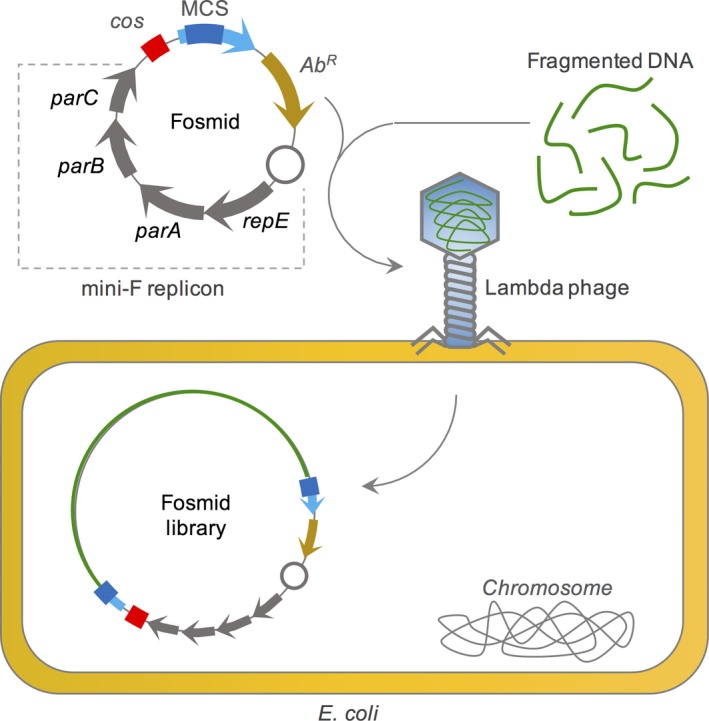
Tools based on minimal fosmids for cloning large DNA fragments. Fosmids are plasmid vectors based on the mini‐F origin of replication that harbour, in addition to MCS and AbR, a cos site for DNA packing in lambda phages. This allows the cloning of very long DNA fragments (up to 50 kb) by ligation and packing into empty lambda phages. The packed DNA is then used to infect E. coli host strains, resulting in the construction of genomic or metagenomic DNA libraries.
Functional metagenomics: using broad‐host‐range vectors to enhance the probability of identifying target genes
An alternative to overcoming the limitations of metagenomic approaches and to enhance the discovery of new biocatalysts and molecules of interest is the use of alternative host organisms (besides E. coli) to perform the functional screening tests. For this, shuttle vectors and broad‐host‐range vectors have been mostly used. This type of vector (see Fig. 2) has been largely used in functional metagenomics, increasing the efficacy of identifying target activities (Table 2). Vectors used in metagenomic screening in alternative hosts contain a single broad‐host origin oriV or a multiple oriV, which permit its replication in E. coli and other hosts, or integrative‐based systems that allow integration of the metagenomic DNA into the chromosome of the screening host (Cheng et al., 2017).
Table 2.
Multiple host‐vector systems for metagenomic DNA cloning and functional screening
| Library vector | Origin type/Vector type | Vector size | Relevant characteristics | Average insert size | Environment | Screening hosts | Library size (Mb or Gb)/Number of clones | Gene/function target | References |
|---|---|---|---|---|---|---|---|---|---|
| pLF61 | Shuttle/plasmid | 5.4 kb | PGK promoter; URA3 gene; origins from yeast 2 μm plasmid E. coli vector pUC19 | 1–3 kb | Soil eukaryote organisms (metatranscriptome) | S. cerevisiae W303 | 1.75 × 106 | Oligopeptide transporters | Damon et al. (2011) |
| pMM436 | Shuttle/cosmid | Not shown | ColE1 origin for E. coli and integrative attB for S. lividans; PacI restriction site flanking attB integration site for recovering metagenomic DNA | 30–40 kb | Libraries I and II different soil samples |
S. lividans
∆act ∆red |
I) 367.5 Mb/10 500 II) 2485 Mb/71 000 |
I) Haemolytic activity II) Pigmentation producing |
McMahon et al. (2012) |
| pCT3FK | Shuttle/fosmid | 11.6 kb | RK2 origin, pyr (chromosomal surroundings of the pyr locus allow homologous recombination in T. thermophilus), KmR | 35–40 kb | Water, sediment, biofilm from hot springs | T. thermophilus BL03, E. coli EPI300 | ~8000 | Esterase‐active enzymes | Leis et al. (2015) |
| pCT3FK | Shuttle/fosmid | 11.6 kb | RK2 origin, pyr (chromosomal surroundings of the pyr locus were inserted in the vector to allow homologous recombination in T. thermophilus), KmR | 50 kb | S. thermophila (genomic library) | E. coli K12 and T. thermophilus HB27 | 2.9 Mb/192 | Xylanase activity | Angelov et al. (2009) |
| pLAFR3 | Broad‐host‐range/cosmid | 22 kb | pLAFR1 with HaeII fragment (MCS and a‐complementation for ß‐galactosidase activity), TcR, cos site, RK2 origin | 25 kb | Wastewater treatment plant anaerobic sludge digestor | E. coli and R. leguminosarum bv. Viciae | 2750 Mb/110 000 | Dehydrogenase genes | Wexler et al. (2005) |
| pGNS‐BAC‐1 | Shuttle/BAC | 11.9 kb | Two origins of replication (i.e. F and RK2), oriV origin, arabinose‐inducible plasmid copy number, CmR. | 80 kb | Forest soil | Clones from E. coli were electroporated into S. marcescens, V. cholerae, E. nimipressuralis | Not shown | Not shown | Kakirde et al. (2011) |
| pOS700I | Shuttle/cosmid | Not shown | AmpR, cos sequence, pOS700I integrative in S. lividans via attB site and int gene from the Streptomyces integrative element pSAM2, allowing site‐specific integration of pOS700I in many Streptomyces species | 50 kb | Soil from an arable field | E. coli/S. lividans (only to check phenotypes found in E. coli) | 250 Mb/5000 | Polyketide synthase genes | Courtois et al. (2003) |
| pWEB436 | Shuttle/cosmid | Not shown | ColE1 origin, cos site, AmpR, ΦC31 integration system for integration in Streptomyces, ApraR, oriT from the lncP plasmid | 40–50 kb | Texas desert soil | Streptomyces albus | 1.5 million | Polyketide synthase genes | Iqbal et al. (2016) |
| pSrpsL14 | Shuttle/BAC | Not shown | ColE1 origin, pSG5 origin of Streptomyces; AmpR (E. coli); ThioR (Streptomyces); GmR (E. coli and Streptomyces) | 50–100 kb | Soil | S. lividans ∆act ∆red, P. putida MBD1 | Not shown | Antibacterial and antifungal activities | Martinez et al. (2004) |
| pKS13S | Broad‐host‐range/cosmid | 21.7 kb | RK2, TcR, cos site, RK2 oriT. | 25 kb | Oil‐contaminated soil | P. putida G7K2 and KTSK2 | 294 Mb/24 000 | Naphthalene‐catabolic genes | Ono et al. (2007) |
| pJWC1 | Broad‐host‐range/cosmid | 14 kb | trfA, RK2 oriT, RK2, TcR, ApR, RK2 stability locus (0.8 kb). | High‐molecular weight | Deciduous forest topsoil, creek bed mud/sediment and sand/clay‐covered cold desert soil | Agrobacterium tumefaciens LBA4404, Burkholderia graminis C4D1M, Caulobacter vibrioides CB15, Escherichia coli EC100, Pseudomonas putida KT2440, Ralstonia metallidurans CH34 | 750 000 | Pigmentation producing and antibacterial activity | Craig et al. (2010) |
| pJWC1 | Broad‐host‐range/cosmid | 14 kb | trfA, oriT (RK2 origin of transfer), RK2, TcR, ApR, RK2 stability locus (0.8 kb) | High‐molecular weight | Soil | Ralstonia metallidurans CH34 | 575 000 | Metabolites from pigmented compounds and antibacterial | Craig et al. (2010) |
| pKS13S | Broad‐host‐range/cosmid | 21.7 kb | RK2 origin, TcR, cos site, RK2 oriT | 25 kb | Artificially polluted soil (with biphenyl, phenanthrene, carbazole and 3‐chlorobenzoate) | P. putida KTSK2 and P. putida G7NAD2 | 5.2 Gb/208 000 | Oxygenase genes | Nagayama et al. (2015) |
| pJC8 | Broad‐host‐range/cosmid | 13 kb | TcR, GmR, Rk2, oriT, Gateway attL sites used for recombination transfer of cosmid inserts into other destination vectors | 33 kb | Soil from the wheat field | P. putida PpUW2 (PHA‐) | 362 Gb/9 × 106 | Polyhydroxy‐alkanoate synthases genes | Cheng and Charles (2016) |
| pRS44 | Broad‐host‐range/fosmid and BAC | 10.3 kb | CmR, KmR, stabilization element parDE (from RK2), oriT, Lac system for blue/white screening, ori2 origin (from the F plasmid) oriV from RK2 | 35 kb | Marine sediment | Pseudomonas fluorescens and Xanthomonas campestris | 20 000 | Not shown | Aakvik et al. (2009) |
A variety of studies have found that using diverse hosts when screening metagenomics libraries can increase the discovery rate of active clones (Table 2). Metagenomic libraries are often directly constructed in E. coli due to the higher number of transformants obtained when compared with the low rate of transformants achieved in other alternative hosts. Thus, to perform screenings in alternative hosts, a common method is to use shuttle or broad‐host‐range vectors for library construction in E. coli and then to transfer and screening these libraries in other host organisms. For instance, Damon et al. (2011) used the pFL61 yeast‐E. coli plasmid shuttle vector to construct a small‐insert library from soil eukaryote DNA using a metatranscriptomic approach. Performing functional complementation of a yeast mutant defective in a di/tripeptide, they identified a novel family of oligopeptide transporters expressed by fungi. Also, the pMycoFos fosmid shuttle vector has been used to allow transfer of a butane‐oxidizing strain Nocardioides CF8 DNA from E. coli EPI300 to Mycobacterium spp (Ly et al., 2011). In addition, McMahon et al. (2012) developed a shuttle cosmid vector for E. coli and Streptomyces lividans and used an optimized S. lividans strain for screening. In another study, Martinez et al. (2004) used a pSrps14 shuttle BAC vector and showed different functions among E. coli, P. putida and S. lividans expressing heterologous metagenomic genes.
On the other hand, several studies using broad‐host‐range vectors also have been reported in metagenomics screenings. For instance, Craig et al. (2010) described the construction of a cosmid library from soil samples in a broad‐host‐range vector (pJWC1), which was screened for antibacterial activity, altered pigmentation and altered colony morphology in six different Proteobacteria: A. tumefaciens, Burkholderia graminis, Caulobacter vibrioides, E. coli, P. putida and Ralstonia metallidurans. The screenings in E. coli identified two clones displaying antibiosis activity, but no clones displayed either pigmentation or morphology alterations, and the screenings in other hosts identifying a number of other target characteristics. This indicates that the same metagenomic library can yield different results depending on the expression host used (Craig et al., 2010).
Moreover, Nagayama et al. (2015) constructed a metagenomic library from artificially polluted soil samples using a broad‐host‐range vector (pKS13S) based on the RK2 origin of replication, showing different success rate when analysed the library in P. putida, E. coli and B. multivorans. Leis et al. (2015) used a T. thermophilus/E. coli shuttle fosmid vector (pCT3FK; Angelov et al., 2009) to generate a large‐insert metagenomic library and performed screenings for lipolytic activities in E. coli and T. thermophilus HB2 (using a multiple clean deletion mutant T. thermophilus) which lacks several characterized extracellular and putative esterase‐encoding genes. They found two thermostable a/b‐fold hydrolase enzymes with high amino acid sequence similarity to already characterized enzymes in E. coli screening. In contrast, they found six fosmids that conferred lipolytic activities to T. thermophilus.
Additionally, in the last years, diverse efforts have been made to produce new vectors displaying relevant characteristics, and synthetic biology has become a powerful tool for this (Guazzaroni et al., 2015; Alves et al., 2017b). For instance, Bryksin and Matsumura (2010) described an engineered broad‐host‐range origin of replication (pWV01 RCR) used to create the high copy number vector pBAV1K‐T5, which can replicate in different Gram‐negative and Gram‐positive bacterial species. In another study, Terrón‐González et al. (2013) developed vectors and specialized E. coli strains as improved metagenomic DNA heterologous expression systems, which was based on the T7 RNA‐polymerase and the lambda phage transcription anti‐termination protein N (Terrón‐González et al., 2013). However, the approaches developed are limited to E. coli as a host. Another alternative to overcome the low expression of heterologous genes in functional metagenomics was proposed by Gaida et al. (2015). Authors created E. coli strains expressing heterologous sigma factors that are able to recognize heterologous promoters from metagenomic and genomic DNA libraries. The study showed that RpoD from Lactobacillus plantarum can initiate transcription from all sources of tested DNA. Moreover, the use of modular vectors, such as the pSEVA vectors listed above (Silva‐Rocha et al., 2013), confers diverse benefits for constructing metagenomic libraries. Yet, besides all the advantageous characteristics already mentioned, these vectors only allow the cloning of small metagenomic DNA fragments (up to 10 kb) and are not suitable for constructing large‐insert DNA libraries, which are essential for functional recovery of complete biosynthetic pathways involved in producing bioactive compounds (as in the case of non‐ribosomal peptide synthetases, polyketide synthases and terpene synthases genes, among others). Therefore, since metagenomics is a rising field, innovations in tools to facilitate manipulation and promote the discovery of functional genes in environmental samples are still needed.
Challenges and perspectives in vector design
When considering the tremendous advances in vector engineering, it becomes evident that even the sophisticated set of tools constructed so far cannot solve all the bottlenecks existing in the field. In this section, we present some of the challenges that need to be taken into account when designing the next generation of modular genetic tools (Fig. 7).
Figure 7.
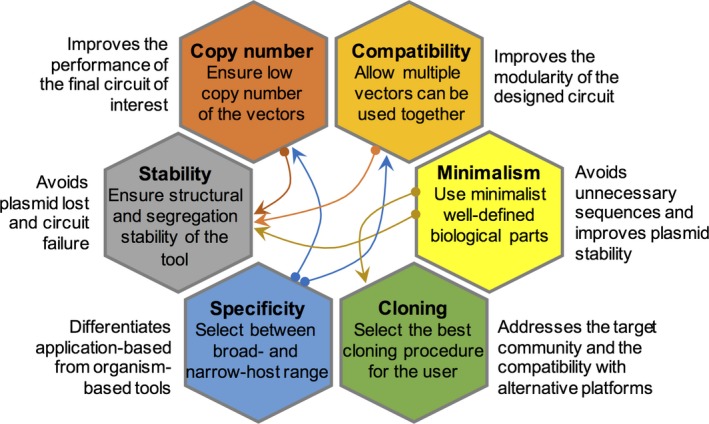
Critical features to consider for efficient vector engineering. In this schematic illustration, outside hexagons represent the main impact of each step on the effectiveness of the tool. Arrows connecting hexagons indicate which features significantly impact each other.
Copy number control
While plasmid vectors represent an easy way to insert and test synthetic circuits in microorganisms, they usually replicate, generating more copies per cell and more chromosomes. Indeed, it has been increasingly recognized that circuits implemented in low copy exhibit enhanced performance compared to those placed in multicopy (Lee et al., 2016). Since vector engineering usually requires the modification of the ori of replication or of its surrounding area, special care must be taken to determine if those changes modify the copy number of the final vector. In this context, some very elegant approaches using fluorescent activated cell sorting (FACS) and digital PCR have been established to assay variations and plasmid copy number (Jahn et al., 2016), and variations of these methods are highly recommended in new vector design projects.
Plasmid incompatibility
The use of multiple plasmids to implement complex synthetic circuits is a very attractive approach since it allows the optimization of the whole system in a modular way. However, while bacterial plasmids use diverse mechanisms for autonomous DNA replication, some of these require the same host machinery. As a result, many origins of replications belong to the same incompatibility group, which means that they cannot be stably maintained simultaneously in the same host (Novick, 1987). Therefore, it is imperative to consider plasmid incompatibility groups when designing novel genetic tools. Alternatively, it would be possible to create orthogonal origins of replication through engineering Rep proteins, but this type of approach has not been reported yet.
Plasmid structural and segregation stability
Two fundamental features of plasmid biology that are virtually neglected in recent engineering approaches are the structural and segregation stability of the vectors. These features were intensively investigated in the 1980s, as researchers reported that many natural plasmids presented spontaneous loss during cell division (segregation instability) or displayed profound rearrangements in their structures and loss of DNA segments (structural stability; Maschke et al., 1992). The former process would occur due to defects during replication of the plasmid DNA and to the absence of the specific segment of the plasmid responsible for efficient partition of vectors to the daughter cells (Nordström and Austin, 1989). The latter, though, could be due to many reasons, from an excess of homologous sequences in the plasmid backbone to the high‐level expression of toxic genes in the plasmid, which may result in a fitness advantage for those cells that harbour mutant versions of the systems with the deleted gene (Ehrlich et al., 1991). When designing new synthetic vectors, it is important to ensure that the final tool is stable enough to guarantee the performance of the final circuit of interest.
Use of minimalist, fully characterized parts
The use of minimalist DNA fragments is also a good practice in vector design to allow the final tool to be as minimal as possible. This is manageable for bacterial plasmids but is not trivial for vectors designed for yeast and filamentous fungi, for example, where there is a lack of consistent information regarding minimal regulatory elements. For those cases, the characterization of the individual biological parts is crucial for the use of the appropriate fragments and to ensure the reliability of the final tool.
Universal versus case‐specific platforms
The dualism between universality vs. specificity is best represented by broad‐ and narrow‐host‐range vectors. This is particularly important as the field of synthetic biology moves into real applications where non‐model organisms may be required (Bassalo et al., 2016). While designing narrow‐host‐range platforms allows the construction of tools that meet the specific requirements of the hosts, broad‐host‐range vectors allow the user to easily switch the host without requiring the reconstruction of the circuit of interest. Yet, the way each host interacts with the genetic elements of the tools can vary drastically, which could impair the functioning of, for example, some regulatory elements of the vector. In that sense, the new generation of universal tools should consider the use of orthogonal elements to ensure the efficient recognition of such regulatory elements in the targeted hosts. Examples of this have been the recent engineering of synthetic promoters efficiently recognized by Gram‐negative and Gram‐positive bacteria, as well as by yeast (Yang et al., 2018).
Selection of appropriate circuit‐cloning methods
When designing novel genetic tools, it is imperative to consider the final target community and their preferences. While the initial progress in plasmid engineering was built upon the use of restriction enzymes and DNA ligase, restriction‐free methods are becoming more and more popular in the synthetic biology community (Ellis et al., 2011). Moreover, the continuous decline of DNA synthesis costs will certainly lead to a future where synthetic biology projects will rely on complex circuits encoded in several kb of DNA constructed by de novo synthesis. Looking further forward, an immediate need for a valuable genetic tool is to reach virtually any user and be based in the most straightforward cloning procedures.
Concluding remarks
Bacteria and fungi are multifaceted organisms containing several layers of complexity. Here, we have briefly reviewed the major events that have shaped the field of vector design for all those microorganisms over the past decades. In summary, the two major requirements for exceptional vectors are versatility and modularity. Undoubtedly, most tools were built based on the model organisms from each Kingdom: E. coli and S. cerevisiae. However, we realize now that those technologies are on an inevitable path to expand to other microorganisms due to their enormous importance in health and industry. Broad‐host‐range vectors and shuttle vectors are part of that solution. Ideally, the same tool should work for both the model and the other target organism. Versatility would benefit not only fundamental science but would also help the search for new metagenomic products. Yet, modularity, as stated several times throughout the review, is the ultimate stage we must reach to enter the new era of synthetic biology‐based vectors. To achieve that, we need a complete and extensive characterization and standardization of all biological parts, either inside or outside biological systems. Perhaps we will not find an ‘one vector for them all’ solution, where a single platform can be used for any organism of interest. Therefore, we anticipate a situation where basic design rules are generated in as many model organisms as possible and then are applied to new organisms using DNA synthesis technologies that are continuously decreasing in cost.
Acknowledgements
The authors are thankful to laboratory members and to the anonymous reviewers for insightful comments on this manuscript.
Microbial Biotechnology (2019) 12(1), 125–147
Funding information
This work was supported by the São Paulo Research Foundation – FAPESP [Young Research Award 2012/22921‐8 to RS‐R; 2015/04309‐1 to M‐EG; Master Fellowships 2016/03763‐3 to LCN; 2016/05472‐6 to CAW; PhD Fellowships 2017/17924‐1 to LMS; 2016/06323‐4 to LFA; 2016/19179‐9 to LMOM].
References
- Aakvik, T. , Degnes, K. F. , Dahlsrud, R. , Schmidt, F. , Dam, R. , Yu, L. , et al (2009) A plasmid RK2‐based broad‐host‐range cloning vector useful for transfer of metagenomic libraries to a variety of bacterial species. FEMS Microbiol Lett 296: 149–158. [DOI] [PubMed] [Google Scholar]
- Abe, A. , Elegado, E.B. , and Sone, T. (2006) Construction of pDESTR, a Gateway vector for gene disruption in filamentous fungi. Curr Microbiol 52: 210–215. [DOI] [PubMed] [Google Scholar]
- Alberti, S. , Gitler, A.D. , and Lindquist, S. (2007) A suite of Gateway cloning vectors for high‐throughput genetic analysis in Saccharomyces cerevisiae . Yeast 24: 913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves, L.F. , Silva‐Rocha, R. , and Guazzaroni, M.‐E. (2017a) Enhancing metagenomic approaches through synthetic biology In Functional Metagenomics: Tools and Applications. Trevor C. Charles, Mark R. Liles. and Angela Sessitsch. (ed). Cham, Switzerland: Springer International Publishing, pp. 75–94. [Google Scholar]
- Alves, L.D.F. , Silva‐Rocha, R. , and Guazzaroni, M.‐E. (2017b) Enhancing metagenomic approaches through synthetic biology In Functional Metagenomics: Tools and Applications María‐Eugenia Guazzaroni. Charles T.C., Liles M.R., and Sessitsch A. (eds). Berlin, Germany: Springer International Publishing, pp. 1–14. [Google Scholar]
- Amores, G. , Guazzaroni, M.‐E. , Arruda, L. , and Silva‐ Rocha, R. (2016) Recent progress on systems and synthetic biology approaches to engineer fungi as microbial cell factories. Curr Genomics 17: 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.C. , Dueber, J.E. , Leguia, M. , Wu, G.C. , Goler, J.A. , Arkin, A.P. , and Keasling, J.D. (2010) BglBricks: a flexible standard for biological part assembly. J Biol Eng 4: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianantoandro, E. , Basu, S. , Karig, D.K. , and Weiss, R. (2006) Synthetic biology: new engineering rules for an emerging discipline. Mol Syst Biol 2: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelov, A. , Mientus, M. , Liebl, S. , and Liebl, W. (2009) A two‐host fosmid system for functional screening of (meta)genomic libraries from extreme thermophiles. Syst Appl Microbiol 32: 177–185. [DOI] [PubMed] [Google Scholar]
- Apel, A.R. , D'Espaux, L. , Wehrs, M. , Sachs, D. , Li, R.A. , Tong, G.J. , et al (2017) A Cas9‐based toolkit to program gene expression in Saccharomyces cerevisiae . Nucleic Acids Res 45: 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avalos, J.L. , Fink, G.R. , and Stephanopoulos, G. (2013) Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched‐chain alcohols. Nat Biotechnol 31: 335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdasarian, M. , Lurz, R. , Rückert, B. , Franklin, F.C. , Bagdasarian, M.M. , Frey, J. , and Timmis, K.N. (1981) Specific‐purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010‐derived vectors, and a host‐vector system for gene cloning in Pseudomonas. Gene 16: 237–247. [DOI] [PubMed] [Google Scholar]
- Balbás, P. , Soberón, X. , Merino, E. , Zurita, M. , Lomeli, H. , Valle, F. , et al (1986) Plasmid vector pBR322 and its special‐purpose derivatives ‐ a review. Gene 50: 3–40. [DOI] [PubMed] [Google Scholar]
- Baldwin, C.Y. , and Clark, K.B. (2000) Design Rules: The Power of Modularity (vol 1). Cambridge, MA, USA: MIT Press. [Google Scholar]
- Bassalo, M.C. , Liu, R. , and Gill, R.T. (2016) Directed evolution and synthetic biology applications to microbial systems. Curr Opin Biotechnol 39: 126–133. [DOI] [PubMed] [Google Scholar]
- Beja, O. , Suzuki, M.T. , Koonin, E.V. , Aravind, L. , Hadd, A. , Nguyen, L.P. , et al (2000) Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ Microbiol 2: 516–529. [DOI] [PubMed] [Google Scholar]
- Bhalla, U.S. , and Iyengar, R. (1999) Emergent properties of networks of biological signaling pathways. Science 283: 381–387. [DOI] [PubMed] [Google Scholar]
- Bitinaite, J. , Rubino, M. , Varma, K.H. , Schildkraut, I. , Vaisvila, R. , and Vaiskunaite, R. (2007) USER friendly DNA engineering and cloning method by uracil excision. Nucleic Acids Res 35: 1992–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar, F. , Rodriguez, R.L. , Greene, P.J. , Betlach, M.C. , Heyneker, H.L. , Boyer, H.W. , et al (1977) Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2: 95–113. [PubMed] [Google Scholar]
- Boros, I. , Pósfai, G. , and Venetianer, P. (1984) High‐copy‐number derivatives of the plasmid cloning vector pBR322. Gene 30: 257–260. [DOI] [PubMed] [Google Scholar]
- Bredeweg, E.L. , Pomraning, K.R. , Dai, Z. , Nielsen, J. , Kerkhoven, E.J. , and Baker, S.E. (2017) A molecular genetic toolbox for Yarrowia lipolytica . Biotechnol Biofuels 10: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner, R. (1992) A series of shuttle vectors for Bacillus subtilis and Escherichia coli . Gene 122: 187–192. [DOI] [PubMed] [Google Scholar]
- Bryksin, A.V. , and Matsumura, I. (2010) Rational design of a plasmid origin that replicates efficiently in both gram‐positive and gram‐negative bacteria. PLoS ONE 5: 463–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, J.R. , Philippsen, P. , and Davis, R.W. (1983) Analysis of chromosomal integration and deletions of yeast plasmids. Nucleic Acids Res 1: 1429–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton, B. , Labno, A. , and Endy, D. (2008) Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol 26: 787–793. [DOI] [PubMed] [Google Scholar]
- Cardinale, S. , and Arkin, A.P. (2012) Contextualizing context for synthetic biology ‐ identifying causes of failure of synthetic biological systems. Biotechnol J 7: 856–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casini, A. , Storch, M. , Baldwin, G.S. , and Ellis, T. (2015) Bricks and blueprints: methods and standards for DNA assembly. Nat Rev Mol Cell Biol 16: 568–576. [DOI] [PubMed] [Google Scholar]
- Cereghino, J.L. , and Cregg, J.M. (2000) Heterologous protein expression in the methylotrophic yeast Pichia pastoris . FEMS Microbiol Rev 24: 45–66. [DOI] [PubMed] [Google Scholar]
- Chee, M.K. and Haase, S.B. (2012) New and redesigned pRS plasmid shuttle vectors for genetic manipulation of Saccharomyces cerevisiae . G3: Genes ‐ Genomes ‐ Genetics 2, 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Kim, J.K. , Hirning, A.J. , Josi, K. , and Bennett, M.R. (2015) Emergent genetic oscillations in a synthetic microbial consortium. Science 349: 986–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J. , and Charles, T. C. (2016) Novel polyhydroxyalkanoate copolymers produced in Pseudomonas putida by metagenomic polyhydroxyalkanoate synthases. Appl Microbiol Biotechnol 100: 7611–7627. [DOI] [PubMed] [Google Scholar]
- Cheng, J. , Lam, K.N. , Engel, K. , Hall, M. , Neufeld, J.D. , and Charles, T.C. (2017) Metagenomic cosmid libraries suitable for functional screening in proteobacteria In Functional Metagenomics: Tools and Applications. Charles T.C., Liles M.R., and Sessitsch A. (eds). Cham, Switzerland: Springer, pp. 1–11. [Google Scholar]
- Chiang, C.S. , and Bremer, H. (1988) Stability of pBR322‐derived plasmids. Plasmid 20: 207–220. [DOI] [PubMed] [Google Scholar]
- Clarke, L. , and Carbon, J. (1980) Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 287: 504–509. [DOI] [PubMed] [Google Scholar]
- Cohen, S.N. (2013) DNA cloning: a personal view after 40 years. Proc Natl Acad Sci USA 110: 15521–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, S.N. , Chang, A.C.Y. , Boyer, H.W. , and Helling, R.B. (1973) Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci USA 70: 3240–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, S.N. , Chang, A.C. , Boyer, H.W. , and Helling, R.B. (1992) Construction of biologically functional bacterial plasmids in vitro. 1973. Biotechnology 24: 188–192. [PubMed] [Google Scholar]
- Cooling, M.T. , Rouilly, V. , Misirli, G. , Lawson, J. , Yu, T. , Hallinan, J. , and Wipat, A. (2010) Standard virtual biological parts: a repository of modular modeling components for synthetic biology. Bioinformatics 26: 925–931. [DOI] [PubMed] [Google Scholar]
- Courbet, A. , Endy, D. , Renard, E. , Molina, F. and Bonnet, J. (2015) Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates. Sci Transl Med 7, 289ra83. [DOI] [PubMed] [Google Scholar]
- Courtois, S. , Cappellano, C.M. , Ball, M. , Francou, F. , Normand, P. , Martinez, A. , et al (2003) Recombinant environmental libraries provide access to microbial diversity for drug discovery from natural products recombinant environmental libraries provide access to microbial diversity for drug discovery from natural products. Appl Environ Microbiol 69: 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, J.W. , Chang, F.‐Y. , Kim, J.H. , Obiajulu, S.C. , and Brady, S.F. (2010) Expanding small‐molecule functional metagenomics through parallel screening of broad‐host‐range cosmid environmental DNA libraries in diverse proteobacteria. Appl Environ Microbiol 76: 1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damon, C. , Vallon, L. , Zimmermann, S. , Haider, M.Z. , Galeote, V. , Dequin, S. , et al (2011) A novel fungal family of oligopeptide transporters identified by functional metatranscriptomics of soil eukaryotes. ISME J 5: 1871–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison, J. , Heusterspreute, M. , Merchez, M. , and Brunel, F. (1984) Vectors with restriction‐site banks I. pJRD158, a 3903‐bp plasmid containing 28 unique cloning sites. Gene 28: 311–318. [DOI] [PubMed] [Google Scholar]
- Decoene, T. , De Paepe, B. , Maertens, J. , Coussement, P. , Peters, G. , De Maeseneire, S.L. , and De Mey, M. (2017) Standardization in synthetic biology: an engineering discipline coming of age. Crit Rev Biotechnol 38: 647–656. [DOI] [PubMed] [Google Scholar]
- Din, M.O. , Danino, T. , Prindle, A. , Skalak, M. , Selimkhanov, J. , Allen, K. , et al (2016) Synchronized cycles of bacterial lysis for in vivo delivery. Nature 536: 81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durland, R.H. , Toukdarian, A. , Fang, F. , and Helinski, D.R. (1990) Mutations in the trfA replication gene of the broad‐host‐range plasmid RK2 result in elevated plasmid copy numbers. J Bacteriol 172: 3859–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich, S.D. , Bruand, C. , Sozhamannan, S. , Dabert, P. , Gros, M.F. , Jannière, L. , and Gruss, A. (1991) Plasmid replication and structural stability in Bacillus subtilis. Res Microbiol 142: 869–873. [DOI] [PubMed] [Google Scholar]
- Ekkers, D.M. , Cretoiu, M.S. , Kielak, A.M. , and Van Elsas, J.D. (2012) The great screen anomaly‐a new frontier in product discovery through functional metagenomics. Appl Microbiol Biotechnol 93: 1005–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, T. , Adie, T. , and Baldwin, G.S. (2011) DNA assembly for synthetic biology: from parts to pathways and beyond. Integr Biol 3: 109. [DOI] [PubMed] [Google Scholar]
- Elowitz, M.B. , Levine, A.J. , Siggia, E.D. , and Batzer, A.G. (2002) Stochastic gene expression in a single cell. Science 297: 1183–1187. [DOI] [PubMed] [Google Scholar]
- Endy, D. (2005) Foundations for engineering biology. Nature 438: 449–453. [DOI] [PubMed] [Google Scholar]
- Enger‐Valk, B.E. , Heyneker, H.L. , Oosterbaan, R.A. , and Pouwels, P.H. (1980) Construction of new cloning vehicles with genes of the tryptophan operon of Escherichia coli as genetic markers. Gene 9: 69–85. [DOI] [PubMed] [Google Scholar]
- Engler, C. , Gruetzner, R. , Kandzia, R. , and Marillonnet, S. (2009) Golden gate shuffling: a one‐pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE 4: e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, F. , Salmon, K. , Shen, M.W.Y. , Aeling, A.K. , Ito, E. , Irwin, B. , et al (2011) A vector set for systematic metabolic engineering in Saccharomyces cerevisae. Yeast 28: 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, J. , Groppe, J.C. , and Reed, S.I. (1981) Construction and characterization of three yeast‐Escherichia coli shuttle vectors designed for rapid subcloning of yeast genes on small DNA fragments. Gene 16: 191–197. [DOI] [PubMed] [Google Scholar]
- Ferrer, M. , Ghazi, A. , Beloqui, A. , Vieites, J.M. , López‐Cortés, N. , Marín‐Navarro, J. , et al (2012) Functional metagenomics unveils a multifunctional glycosyl hydrolase from the family 43 catalysing the breakdown of plant polymers in the calf rumen. PLoS ONE 7: e38134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabor, E.M. , Alkema, W.B.L. , and Janssen, D.B. (2004) Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ Microbiol 6: 879–886. [DOI] [PubMed] [Google Scholar]
- Gaida, S.M. , Sandoval, N.R. , Nicolaou, S.A. , Chen, Y. , Venkataramanan, K.P. , and Papoutsakis, E.T. (2015) Expression of heterologous sigma factors enables functional screening of metagenomic and heterologous genomic libraries. Nat Commun 6: 7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilescu, M. , Demnerová, K. , Aamand, J. , Agathos, S. , and Fava, F. (2015) Emerging pollutants in the environment: present and future challenges in biomonitoring, ecological risks and bioremediation. New Biotechnol 32: 147–156. [DOI] [PubMed] [Google Scholar]
- Generoso, W.C. , Gottardi, M. , Oreb, M. , and Boles, E. (2016) Simplified CRISPR‐Cas genome editing for Saccharomyces cerevisiae . J Microbiol Methods 127: 203–205. [DOI] [PubMed] [Google Scholar]
- Gibson, D.G. , Young, L. , Chuang, R.‐Y. , Venter, J.C. , Hutchison, C.A. , and Smith, H.O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D. , and Sugino, A. (1988) New yeast‐Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six‐base pair restriction sites. Gene 74: 527–534. [DOI] [PubMed] [Google Scholar]
- Gnügge, R. , and Rudolf, F. (2017) Saccharomyces cerevisiae Shuttle vectors. Yeast 26: 545–551. [DOI] [PubMed] [Google Scholar]
- Gnügge, R. , Liphardt, T. , and Rudolf, F. (2016) A shuttle vector series for precise genetic engineering of Saccharomyces cerevisae. Yeast 33: 83–98. [DOI] [PubMed] [Google Scholar]
- Goñi‐Moreno, Á. , Benedetti, I. , Kim, J. , and De Lorenzo, V. (2017) Deconvolution of gene expression noise into spatial dynamics of transcription factor‐promoter interplay. ACS Synth Biol 6: 1359–1369. [DOI] [PubMed] [Google Scholar]
- de Graaff, J. , Crosa, J.H. , Heffron, F. , and Falkow, S. (1978) Replication of the nonconjugative plasmid RSF1010 in Escherichia coli K‐12. J Bacteriol 134: 1117–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guazzaroni, M.‐E. , Golyshin, P.N. , and Ferrer, M. (2010) Analysis of complex microbial communities through metagenomic survey In Metagenomic: Theory, Methods and Applications. Marco D. (ed). Norfolf, UK: Claster Academic Press, pp. 55–78. [Google Scholar]
- Guazzaroni, M.‐E. , Silva‐Rocha, R. , and Ward, R.J. (2015) Synthetic biology approaches to improve biocatalyst identification in metagenomic library screening. Microb Biotechnol 8: 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerineau, M. , Grandchamp, C. , Paoletti, C. , and Slonimski, P. (1971) Characterization of a new class of circular DNA molecules in yeast. Biochem Biophys Res Commun 42: 550–557. [DOI] [PubMed] [Google Scholar]
- Guerineau, M. , Slonimski, P.P. , and Avner, P.R. (1974) Yeast Episome: oligomycin resistance associated with a small covalently closed non‐mitochondrial circular DNA. Biochem Biophys Res Commun 61: 462–469. [DOI] [PubMed] [Google Scholar]
- Güldener, U. , Heck, S. , Fiedler, T. , Beinhauer, J. , and Hegemann, J.H. (1996) A new efficient gene disruption cassette for repeateduse in budding yeast. Nucleic Acids Res 24: 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman, L.M. , Belin, D. , Carson, M.J. , and Beckwith, J. (1995) Tight regulation, modulation, and high‐level expression by vectors containing the arabinose P(BAD) promoter. J Bacteriol 177: 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley, J.D. (1988) Cosmid library construction. Methods Mol Biol 4: 257–283. [DOI] [PubMed] [Google Scholar]
- Hallewell, R.A. , and Emtage, S. (1980) Plasmid vectors containing the tryptophan operon promoter suitable for efficient regulated expression of foreign genes. Gene 9: 27–47. [DOI] [PubMed] [Google Scholar]
- Handelsman, J. , Tiedje, J. , and National Research Council (US) Committee on Metagenomics: Challenges and Functional, and Applications (2007) The New Science of Metagenomics: Revealing the Secrets of our Microbial Planet. Washington, DC, USA: National Academies Press. [Google Scholar]
- Handelsman, J. , Rondon, M.R. , Brady, S.F. , Clardy, J. , and Goodman, R.M. (1998) Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol 5: R245–R249. [DOI] [PubMed] [Google Scholar]
- Hanna, Z. , Fregeau, C. , Préfontaine, G. , and Brousseau, R. (1984) Construction of a family of universal expression plasmid vectors. Gene 30: 247–250. [DOI] [PubMed] [Google Scholar]
- Heap, J.T. , Pennington, O.J. , Cartman, S.T. , and Minton, N.P. (2009) A modular system for Clostridium shuttle plasmids. J Microbiol Methods 78: 79–85. [DOI] [PubMed] [Google Scholar]
- Herrin, G.L. , Russell, D.R. , and Bennett, G.N. (1982) A stable derivative of pBR322 conferring increased tetracycline resistance and increased sensitivity to fusaric acid. Plasmid 7: 290–293. [DOI] [PubMed] [Google Scholar]
- Hershfield, V. , Boyer, H.W. , Yanofsky, C. , Lovett, M.A. , and Helinski, D.R. (1974) Plasmid ColEl as a molecular vehicle for cloning and amplification of DNA. Proc Natl Acad Sci USA 71: 3455–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen, A. , Hicks, J.B. , and Fink, G.R. (1978) Transformation of yeast. Proc Natl Acad Sci USA 75: 1929–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn, B. , and Collins, J. (1980) A small cosmid for efficient cloning of large DNA fragments. Gene 11: 291–298. [DOI] [PubMed] [Google Scholar]
- Hudson, T.J. (2001) Construction of small‐insert libraries from genomic DNA. Curr Protoc Hum Genet 1: 2.1.1–2.1.6. [DOI] [PubMed] [Google Scholar]
- Huynh, L. , and Tagkopoulos, I. (2016) A parts database with consensus parameter estimation for synthetic circuit design. ACS Synth Biol 5: 1412–1420. [DOI] [PubMed] [Google Scholar]
- Iqbal, H. A. , Low‐Beinart, L. , Obiajulu, J. U. , and Brady, S. F. (2016) Natural product discovery through improved functional metagenomics in Streptomyces. J Am Chem Soc 138: 9341–9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Invitrogen (2011) Gateway recombination cloning technology. Integr VLSI J 1: 1–15. [Google Scholar]
- Jahn, M. , Vorpahl, C. , Hübschmann, T. , Harms, H. , and Müller, S. (2016) Copy number variability of expression plasmids determined by cell sorting and droplet digital PCR. Microb Cell Fact 15: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, N.B. , Strucko, T. , Kildegaard, K.R. , David, F. , Maury, J. , Mortensen, U.H. , et al (2014) EasyClone: method for iterative chromosomal integration of multiple genes in Saccharomyces cerevisiae . FEMS Yeast Res 14: 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop‐Fabre, M.M. , Jakočiūnas, T. , Stovicek, V. , Dai, Z. , Jensen, M.K. , Keasling, J.D. , and Borodina, I. (2016) EasyClone‐MarkerFree: a vector toolkit for marker‐less integration of genes into Saccharomyces cerevisiae via CRISPR‐Cas9. Biotechnol J 11: 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, I.S. (1982) Human insulin from recombinant DNA technology. Science 219: 632–637. [DOI] [PubMed] [Google Scholar]
- Juers, D.H. , Matthews, B.W. , and Huber, R.E. (2012) LacZ β‐galactosidase: structure and function of an enzyme of historical and molecular biological importance. Protein Sci 21: 1792–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullesson, D. , David, F. , Pfleger, B. , and Nielsen, J. (2015) Impact of synthetic biology and metabolic engineering on industrial production of fine chemicals. Biotechnol Adv 33: 1395–1402. [DOI] [PubMed] [Google Scholar]
- Kado, C.I. (2014) Historical events that spawned the field of plasmid biology. Microbiol Spectr 2: 3–11. [DOI] [PubMed] [Google Scholar]
- Kakirde, K. S. , Wild, J. , Godiska, R. , Mead, D. A. , Wiggins, A. G. , Goodman, R. M. , et al (2011) Gram negative shuttle BAC vector for heterologous expression of metagenomic libraries. Gene 475: 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelwick, R. , MacDonald, J.T. , Webb, A.J. , and Freemont, P. (2014) Developments in the tools and methodologies of synthetic biology. Front Bioeng Biotechnol 2: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, U. , Shizuya, H. , De Jong, P.J. , Birren, B. , and Simon, M.I. (1992) Stable propagation of cosmid sized human DNA inserts in an F factor based vector. Nucleic Acids Res 20: 1083–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H. , Cavaleiro, A.M. , Rennig, M. , and Nørholm, M.H.H. (2016) SEVA Linkers: a versatile and automatable DNA backbone exchange standard for synthetic biology. ACS Synth Biol 5: 1177–1181. [DOI] [PubMed] [Google Scholar]
- Kircher, M. (2015) Sustainability of biofuels and renewable chemicals production from biomass. Curr Opin Chem Biol 29: 26–31. [DOI] [PubMed] [Google Scholar]
- Kitano, H. (2002) Systems biology: a brief overview. Science 295: 1662–1664. [DOI] [PubMed] [Google Scholar]
- Knaus, R. , and Bujard, H. (1990) Principles governing the activity of E. coli promoters In Nucleic Acids and Molecular Biology. Eckstein F., and Lilley D.M.J. (eds). Heidelberg, Germany: Springer, Berlin, pp. 110–122. [Google Scholar]
- Knight, T. (2003) Idempotent vector design for standard assembly of biobricks.
- Knight, T. (2007) Draft standard for biobrick biological parts. BioBricks Found. Req. Comments.
- Koziel, M.G. , Beland, G.L. , Bowman, C. , Carozzi, N.B. , Crenshaw, R. , Crossland, L. , et al (1993) Field performance of elite transgenic maize plants expressing an insecticidal protein derived from Bacillus thuringiensis . Biotechnology 11: 194–200. [Google Scholar]
- Kwok, R. (2010) Five hard truths for synthetic biology. Nature 463: 288–290. [DOI] [PubMed] [Google Scholar]
- Lederberg, J. (1952) Cell genetics and hereditary symbiosis. Physiol Rev 32: 403–430. [DOI] [PubMed] [Google Scholar]
- Lee, T. , Krupa, R.A. , Zhang, F. , Hajimorad, M. , Holtz, W.J. , Prasad, N. , et al (2011) BglBrick vectors and datasheets: a synthetic biology platform for gene expression. J Biol Eng 5: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.W. , Gyorgy, A. , Cameron, D.E. , Pyenson, N. , Choi, K.R. , Way, J.C. , et al (2016) Creating single‐copy genetic circuits. Mol Cell 63: 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leis, B. , Angelov, A. , Mientus, M. , Li, H. , Pham, V.T.T. , Lauinger, B. , et al (2015) Identification of novel esterase‐active enzymes from hot environments by use of the host bacterium Thermus thermophilus . Front Microbiol 6: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveau, J.H.J. , Gerards, S. , De Boer, W. , and Van Veen, J.A. (2004) Phylogeny‐function analysis of (meta)genomic libraries: screening for expression of ribosomal RNA genes by large‐insert library fluorescent in situ hybridization (LIL‐FISH). Environ Microbiol 6: 990–998. [DOI] [PubMed] [Google Scholar]
- Li, M.Z. , and Elledge, S.J. (2007) Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods 4: 251–256. [DOI] [PubMed] [Google Scholar]
- Liu, J.K. , Chen, W.H. , Ren, S.X. , Zhao, G.P. , and Wang, J. (2014) IBrick: a new standard for iterative assembly of biological parts with homing endonucleases. PLoS ONE 9: e110852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobban, P.E. , and Kaiser, A.D. (1973) Enzymatic end‐to end joining of DNA molecules. J Mol Biol 78: 453–471. [DOI] [PubMed] [Google Scholar]
- Lutz, R. , and Bujard, H. (1997) Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1‐I2 regulatory elements. Nucleic Acids Res 25: 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, D. , Wang, W. , and Wei, D. (2012) Construction of two vectors for gene expression in Trichoderma reesei . Plasmid 67: 67–71. [DOI] [PubMed] [Google Scholar]
- Ly, M.A. , Liew, E.F. , Le, N.B. , and Coleman, N.V. (2011) Construction and evaluation of pMycoFos, a fosmid shuttle vector for Mycobacterium spp. with inducible gene expression and copy number control. J Microbiol Methods 86: 320–326. [DOI] [PubMed] [Google Scholar]
- Ma, H. , Kunes, S. , Schatz, P.J. , and Botstein, D. (1987) Plasmid construction by linker‐assisted homologous recombination in yeast. Gene 58: 201–216. [DOI] [PubMed] [Google Scholar]
- Mans, R. , van Rossum, H.M. , Wijsman, M. , Backx, A. , Kuijpers, N.G.A. , van den Broek, M. , et al (2015) CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae . FEMS Yeast Res 15: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis, L. , and Chapman, M.J. (1998) Endosymbioses: cyclical and permanent in evolution. Trends Microbiol 6: 342–345. [DOI] [PubMed] [Google Scholar]
- Maria Oliveira Monteiro, L. , Magalhães Arruda, L. and Silva‐Rocha, R. (2017) Emergent properties in complex synthetic bacterial promoters. [DOI] [PubMed]
- Martinez, A. , Kolvek, S.J. , Yip, C.L.T. , Hopke, J. , Brown, K.A. , MacNeil, I.A. , and Osburne, M.S. (2004) Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple expression hosts. Appl Environ Microbiol 70: 2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐García, E. , Calles, B. , Arévalo‐Rodríguez, M. , and de Lorenzo, V. (2011) pBAM1: an all‐synthetic genetic tool for analysis and construction of complex bacterial phenotypes. BMC Microbiol 11: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐Garćía, E. , Aparicio, T. , Goñi‐Moreno, A. , Fraile, S. , and De Lorenzo, V. (2015) SEVA 2.0: an update of the Standard European Vector Architecture for de‐/re‐construction of bacterial functionalities. Nucleic Acids Res 43: D1183–D1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maschke, H.E. , Kumar, P.K.R. , Geiger, R. , and Schügerl, K. (1992) Plasmid instabilities of single and three‐plasmid systems in Escherichia coli during continuous cultivation. J Biotechnol 24: 235–251. [DOI] [PubMed] [Google Scholar]
- Maury, J. , Germann, S.M. , Baallal Jacobsen, S.A. , Jensen, N.B. , Kildegaard, K.R. , Herrgàrd, M.J. , et al (2016) EasyCloneMulti: a set of vectors for simultaneous and multiple genomic integrations in saccharomyces cerevisiae. PLoS ONE 11: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon, M.D. , Guan, C. , Handelsman, J. , and Thomas, M.G. (2012) Metagenomic analysis of Streptomyces lividans reveals host‐dependent functional expression. Appl Environ Microbiol 78: 3622–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro, L.M.O. , Arruda, L.M. , and Silva‐Rocha, R. (2017) Emergent properties in complex synthetic bacterial promoters. ACS Synth. Biol 7: 602–612. [DOI] [PubMed] [Google Scholar]
- Müller, K. , Arndt, K. , iGEM 2007 Team Freiburg , and Grünberg, R. (2009) BBF RFC 25: Fusion Protein (Freiburg) Biobrick assembly standard. BioBricks Found. Req. Comments.
- Nagayama, H. , Sugawara, T. , Endo, R. , Ono, A. , Kato, H. , Ohtsubo, Y. , et al (2015) Isolation of oxygenase genes for indigo‐forming activity from an artificially polluted soil metagenome by functional screening using Pseudomonas putida strains as hosts. Appl Microbiol Biotechnol 99: 4453–4470. [DOI] [PubMed] [Google Scholar]
- Nambu‐Nishida, Y. , Nishida, K. , Hasunuma, T. , and Kondo, A. (2017) Development of a comprehensive set of tools for genome engineering in a cold‐ A nd thermo‐tolerant Kluyveromyces marxianus yeast strain. Sci Rep 7: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, K.T. , Ho, Q.N. , Pham, T.H. , Phan, T.‐N. , and Tran, V.‐T. (2016) The construction and use of versatile binary vectors carrying pyrG auxotrophic marker and fluorescent reporter genes for Agrobacterium‐mediated transformation of Aspergillus oryzae. World J Microbiol Biotechnol 32: 204. [DOI] [PubMed] [Google Scholar]
- Nielsen, J. , Larsson, C. , van Maris, A. , and Pronk, J. (2013) Metabolic engineering of yeast for production of fuels and chemicals. Curr Opin Biotechnol 24: 398–404. [DOI] [PubMed] [Google Scholar]
- Nishikawa, R. , Yoshida, M. , Noda, T. , Okuhara, T. , Taguchi, G. , Inatomi, S. , and Shimosaka, M. (2016) pFungiway: a series of plasmid vectors used for gene manipulation in fungi. Ann Microbiol 66: 825–832. [Google Scholar]
- Nødvig, C.S. , Nielsen, J.B. , Kogle, M.E. , and Mortensen, U.H. (2015) A CRISPR‐Cas9 system for genetic engineering of filamentous fungi. PLoS ONE 10: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström, K. , and Austin, S.J. (1989) Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet 23: 37–69. [DOI] [PubMed] [Google Scholar]
- Norrander, J. , Kempe, T. , and Messing, J. (1983) Construction of improved M13 vectors using oligodeoxynucleotide‐directed mutagenesis. Gene 26: 101–106. [DOI] [PubMed] [Google Scholar]
- Novick, R.P. (1987) Plasmid incompatibility. Microbiol Rev 51: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick, R.P. , Clowes, R.C. , Cohen, S.N. , Curtiss, R. , Datta, N. , and Falkow, S. (1976) Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol Rev 40: 168–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, A. , Miyazaki, R. , Sota, M. , Ohtsubo, Y. , Nagata, Y. , and Tsuda, M. (2007) Isolation and characterization of naphthalene‐catabolic genes and plasmids from oil‐contaminated soil by using two cultivation‐independent approaches. Appl Microbiol Biotechnol 74: 501–510. [DOI] [PubMed] [Google Scholar]
- Pasin, F. , Bedoya, L.C. , Bernabé‐Orts, J.M. , Gallo, A. , Simón‐Mateo, C. , Orzaez, D. , and García, J.A. (2017) Multiple T‐DNA delivery to plants using novel mini binary vectors with compatible replication origins. ACS Synth Biol 6: 1962–1968. [DOI] [PubMed] [Google Scholar]
- Popp, P.F. , Dotzler, M. , Radeck, J. , Bartels, J. and Mascher, T. (2017) The Bacillus BioBrick Box 2.0: expanding the genetic toolbox for the standardized work with Bacillus subtilis . Sci Rep 7, 15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai, G. , Plunkett, G. , Fehér, T. , Frisch, D. , Keil, G.M. , Umenhoffer, K. , et al (2006) Emergent properties of reduced‐genome Escherichia coli . Science 312: 1044–1046. [DOI] [PubMed] [Google Scholar]
- Pozo, M.V.Del. , Fernández‐arrojo, L. , Gil‐martínez, J. , Montesinos, A. , Chernikova, T.N. , Nechitaylo, T.Y. , et al (2012) Microbial β ‐glucosidases from cow rumen metagenome enhance the saccharification of lignocellulose in combination with commercial cellulase cocktail. Biotechnol Biofuels 5: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnick, P.E.M. , and Weiss, R. (2009) The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol 10: 410–422. [DOI] [PubMed] [Google Scholar]
- Quan, J. , and Tian, J. (2009) Circular polymerase extension cloning of complex gene libraries and pathways. PLoS ONE 4: e6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radeck, J. , Kraft, K. , Bartels, J. , Cikovic, T. , Dürr, F. , Emenegger, J. , et al (2013) The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis . J Biol Eng 7: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radeck, J. , Meyer, D. , Lautenschläger, N. , and Mascher, T. (2017) Bacillus SEVA siblings: a Golden Gate‐based toolbox to create personalized integrative vectors for Bacillus subtilis . Sci Rep 7: 14134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, C.R.R. , Abreu, P.A.E. , Nascimento, A.L.T.O. , and Ho, P.L. (2004) A high‐copy T7 Escherichia coli expression vector for the production of recombinant proteins with a minimal N‐terminal his‐tagged fusion peptide. Braz J Med Biol Res 37: 1103–1109. [DOI] [PubMed] [Google Scholar]
- Rao, R.N. , and Rogers, S.G. (1979) Plasmid pKC7: a vector containing ten restriction endonuclease sites suitable for cloning DNA segments. Gene 7: 79–82. [DOI] [PubMed] [Google Scholar]
- Rawis, R.L. (2000) “Synthetic biology” makes its debut. Chem Eng News 78: 49–53. [Google Scholar]
- Reeve, B. , Martinez‐Klimova, E. , de Jonghe, J. , Leak, D.J. , and Ellis, T. (2016) The geobacillus plasmid set: a modular toolkit for thermophile engineering. ACS Synth Biol 5: 1342–1347. [DOI] [PubMed] [Google Scholar]
- Richardson, M.A. , Mabe, J.A. , Beerman, N.E. , Nakatsukasa, W.M. , and Fayerman, J.T. (1982) Development of cloning vehicles from the Streptomyces plasmid pFJ103. Gene 20: 451–457. [DOI] [PubMed] [Google Scholar]
- Rodriquez, R.L. , and Denhardt, D.T. (1988) Vectors: A Survey of Molecular Cloning Vectors and Their Uses. London, UK: Butterworth‐Heinemann. [Google Scholar]
- Røkke, G. , Korvald, E. , Pahr, J. , Øyås, O. , and Lale, R. (2014) BioBrick assembly standards and techniques and associated software tools. Methods Mol Biol 1116: 1–24. [DOI] [PubMed] [Google Scholar]
- Ruiz‐Díez, B. (2002) A Review: strategies for the transformation of filamentous fungi. J Appl Microbiol 92: 189–195. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. , and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Santana‐pereira, A.L.R. , and Liles, M.R. (2017) Challenges and opportunities in discovery of secondary metabolites using a functional metagenomic approach In Functional Metagenomics: Tools and Applications. Charles T., Liles M., and Sessitsch A. (eds). Cham, Switzerland: Springer, pp. 119–138. [Google Scholar]
- Schorbele, T.J. , Nguyen‐Coleman, C.K. , and May, G.S. (2013) Genetic engineering in filamentous fungi. Fungal Genet Biol 00: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafran, H. , Miyara, I. , Eshed, R. , Prusky, D. , and Sherman, A. (2008) Development of new tools for studying gene function in fungi based on the Gateway system. Fungal Genet Biol 45: 1147–1154. [DOI] [PubMed] [Google Scholar]
- Shaner, N.C. , Steinbach, P.A. , and Tsien, R.Y. (2005) A guide to choosing fluorescent proteins. Nat Methods 2: 905–909. [DOI] [PubMed] [Google Scholar]
- Sheldon, R.A. (2014) Green and sustainable manufacture of chemicals from biomass: state of the art. Green Chem 16: 950–963. [Google Scholar]
- Shetty, R.P. , Endy, D. , and Knight, T.F. (2008) Engineering BioBrick vectors from BioBrick parts. J Biol Eng 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty, R. , Lizarazo, M. , Rettberg, R. , and Knight, T.F. (2011) Assembly of BioBrick standard biological parts using three antibiotic assembly. Methods Enzymol 498: 311–326. [DOI] [PubMed] [Google Scholar]
- Shi, S. , Liang, Y. , Zhang, M.M. , Ang, E.L. , and Zhao, H. (2016) A highly efficient single‐step, markerless strategy for multi‐copy chromosomal integration of large biochemical pathways in Saccharomyces cerevisiae . Metab Eng 33: 19–27. [DOI] [PubMed] [Google Scholar]
- Shizuya, H. , Birren, B. , Kim, U. , Mancino, V. , Slepak, T. , Tachiiri, Y. , and Simon, M. (1992) Cloning and stable maintenance of 300‐kilobase‐pair fragments of human DNA in Escherichia coli using an F‐factor‐based vector. Proc Natl Acad Sci USA 89: 8794–8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S. , and Hieter, P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva‐Rocha, R. , Martínez‐García, E. , Calles, B. , Chavarría, M. , Arce‐Rodríguez, A. , De Las Heras, A. , et al (2013) The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res 41: 666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinsheimer, R.L. (1977) Recombinant DNA. Annu Rev Biochem 46: 415–438. [DOI] [PubMed] [Google Scholar]
- Skerra, A. (1994) Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli . Gene 151: 131–135. [DOI] [PubMed] [Google Scholar]
- Skogman, G. , Nilsson, J. , and Gustafsson, P. (1983) The use of a partition locus to increase stability of tryptophan‐operon‐bearing plasmids in Escherichia coli . Gene 23: 105–115. [DOI] [PubMed] [Google Scholar]
- Soberon, X. , Covarrubias, L. , and Bolivar, F. (1990) Construction and characterization of new cloning vehicles IV. Deletion derivatives of pBR322 and pBR325. Gene 9: 287–305. [DOI] [PubMed] [Google Scholar]
- Stalker, D.M. , Thomas, C.M. , and Helinski, D.R. (1981) Nucleotide sequence of the region of the origin of replication of the broad host range plasmid RK2. Mol Gen Genet 181: 8–12. [DOI] [PubMed] [Google Scholar]
- Stanley, K.K. , and Luzio, J.P. (1984) Construction of a new family of high efficiency bacterial expression vectors: identification of cDNA clones coding for human liver proteins. EMBO J 3: 1429–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger, M.G. , Vitikainen, M. , Uskonen, P. , Brunner, K. , Adam, G. , Pakula, T. , et al (2011) Transformation system for hypocrea jecorina (Trichoderma reesei) that favors homologous integration and employs reusable bidirectionally selectable markers. Appl Environ Microbiol 77: 114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sthol, L.L. , and Lambowitz, A.M. (1983) Construction of a shuttle vector for the filamentous fungus Neurospora crassa. Proc Natl Acad Sci USA 80: 1058–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stovicek, V. , Borja, G.M. , Forster, J. , and Borodina, I. (2015) EasyClone 2.0: expanded toolkit of integrative vectors for stable gene expression in industrial Saccharomyces cerevisiae strains. J Ind Microbiol Biotechnol 42: 1519–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl, K. , Stinchcomb, D.T. , Scherer, S. , and Davis, R.W. (1979) High‐frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci USA 76: 1035–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers, D.K. , and Sherratt, D.J. (1984) Multimerization of high copy number plasmids causes instability: cole 1 encodes a determinant essential for plasmid monomerization and stability. Cell 36: 1097–1103. [DOI] [PubMed] [Google Scholar]
- Talmadge, K. , and Gilberg, W. (1980) Construction of plasmid vectors with unique PstI cloning sites in a signal sequence coding region. Gene 12: 235–241. [DOI] [PubMed] [Google Scholar]
- Taton, A. , Unglaub, F. , Wright, N.E. , Zeng, W.Y. , Paz‐Yepes, J. , Brahamsha, B. , et al (2014) Broad‐host‐range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Res 42: e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpe, K. (2006) Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol 72: 211–222. [DOI] [PubMed] [Google Scholar]
- Terrón‐González, L. , Medina, C. , Limón‐Mortés, M.C. , and Santero, E. (2013) Heterologous viral expression systems in fosmid vectors increase the functional analysis potential of metagenomic libraries. Sci Rep 3: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C.M. (1981) Complementation analysis of replication and maintenance functions of broad host range plasmids RK2 and RP1. Plasmid 5: 277–291. [DOI] [PubMed] [Google Scholar]
- Thompson, C.E. , Beys‐da‐Silva, W.O. , Santi, L. , Berger, M. , Vainstein, M.H. , Guima Rães, J.A. , and Vasconcelos, A.T.R. (2013) A potential source for cellulolytic enzyme discovery and environmental aspects revealed through metagenomics of Brazilian mangroves. AMB Express 3: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trubitsyna, M. , Michlewski, G. , Cai, Y. , Elfick, A. , and French, C.E. (2014) PaperClip: rapid multi‐part DNA assembly from existing libraries. Nucleic Acids Res 42: e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg, A.J. , and Sherratt, D. (1980) Trans‐complementable copy‐number mutants of plasmid ColE1 [24]. Nature 283: 216–218. [DOI] [PubMed] [Google Scholar]
- Uchiyama, T. , and Miyazaki, K. (2009) Functional metagenomics for enzyme discovery: challenges to efficient screening. Curr Opin Biotechnol 20: 616–622. [DOI] [PubMed] [Google Scholar]
- Van Ooyen, A.J.J. , Dekker, P. , Huang, M. , Olsthoorn, M.M.A. , Jacobs, D.I. , Colussi, P.A. , and Taron, C.H. (2006) Heterologous protein production in the yeast Kluyveromyces lactis . FEMS Yeast Res 6: 381–392. [DOI] [PubMed] [Google Scholar]
- Villa‐Komaroff, L. , Efstratiadis, A. , Broome, S. , Lomedico, P. , Tizard, R. , Naber, S.P. , et al (1978) A bacterial clone synthesizing proinsulin. Proc Natl Acad Sci USA 75: 3727–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas, A. , and Kropinski, A.M. (2008) An analysis of initiation codon utilization in the Domain Bacteria ‐ concerns about the quality of bacterial genome annotation. Microbiology 154: 2559–2661. [DOI] [PubMed] [Google Scholar]
- Wagner, J.M. , and Alper, H.S. (2016) Synthetic biology and molecular genetics in non‐conventional yeasts: current tools and future advances. Fungal Genet Biol 89: 126–136. [DOI] [PubMed] [Google Scholar]
- Weber, E. , Engler, C. , Gruetzner, R. , Werner, S. , and Marillonnet, S. (2011) A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 6: e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann, S. , Drubin, D.G. , and Barnes, G. (2007) Structures and functions of yeast kinetochore complexes. Annu Rev Biochem 76: 563–591. [DOI] [PubMed] [Google Scholar]
- Westmann, C.A. , Alves, L.F. , Silva‐Rocha, R. , and Guazzaroni, M.‐E. (2018) Mining novel constitutive promoter elements in soil metagenomic libraries in Escherichia coli . Front Microbiol 9: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler, M. , Bond, P. L. , Richardson, D. J. , and Johnston, A. W. (2005) A wide host‐range metagenomic library from a waste water treatment plant yields a novel alcohol/aldehyde dehydrogenase. Environ Microbiol 7: 1917–1926. [DOI] [PubMed] [Google Scholar]
- Winson, M.K. , Swift, S. , Hill, P.J. , Sims, C.M. , Griesmayr, G. , Bycroft, B.W. , et al (1998) Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini‐Tn 5 constructs. FEMS Microbiol Lett 163: 193–202. [DOI] [PubMed] [Google Scholar]
- Woodruff, L.B.A. , Gorochowski, T.E. , Roehner, N. , Mikkelsen, T.S. , Densmore, D. , Gordon, D.B. , et al (2017) Registry in a tube: multiplexed pools of retrievable parts for genetic design space exploration. Nucleic Acids Res 45: 1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, O. , Delmans, M. , Stan, G.‐B. , and Ellis, T. (2015) GeneGuard: a modular plasmid system designed for biosafety. ACS Synth Biol 4: 307–316. [DOI] [PubMed] [Google Scholar]
- Xu, P. , and Koffas, M.A.G. (2013) Assembly of multi‐gene pathways and combinatorial pathway libraries through ePathBrick vectors. Methods Mol Biol 1073: 107–129. [DOI] [PubMed] [Google Scholar]
- Yang, C. , Xia, Y. , Qu, H. , Li, A.‐D. , Liu, R. , Wang, Y. , and Zhang, T. (2016) Discovery of new cellulases from the metagenome by a metagenomics‐guided strategy. Biotechnol Biofuels 9: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. , Liu, Q. , Zhang, Y. , Du, G. , Chen, J. , and Kang, Z. (2018) Construction and characterization of broad‐spectrum promoters for synthetic biology. ACS Synth Biol 7: 287–291. [DOI] [PubMed] [Google Scholar]
- Yanisch‐Perron, C. , Vieira, J. , and Messing, J. (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33: 103–119. [DOI] [PubMed] [Google Scholar]
- Yao, A.I. , Fenton, T.A. , Owsley, K. , Seitzer, P. , Larsen, D.J. , Sit, H. , et al (2013) Promoter element arising from the fusion of standard BioBrick parts. ACS Synth Biol 2: 111–120. [DOI] [PubMed] [Google Scholar]
- Yeung, E. , Dy, A.J. , Martin, K.B. , Ng, A.H. , Del Vecchio, D. , Beck, J.L. , et al (2017) Biophysical constraints arising from compositional context in synthetic gene networks. Cell Syst 5: 11–24. e12. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Werling, U. , and Edelmann, W. (2012) SLiCE: a novel bacterial cell extract‐based DNA cloning method. Nucleic Acids Res 40: e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Y.H. , Wang, X.L. , Wang, T.H. , and Jiang, Q. (2007) Agrobacterium‐mediated transformation (AMT) of Trichoderma reesei as an efficient tool for random insertional mutagenesis. Appl Microbiol Biotechnol 73: 1348–1354. [DOI] [PubMed] [Google Scholar]
- Zhu, T. , Wang, W. , Yang, X. , Wang, K. , and Cui, Z. (2009) Construction of two Gateway vectors for gene expression in fungi. Plasmid 62: 128–133. [DOI] [PubMed] [Google Scholar]
- Zurita, M. , Bolivar, F. , and Soberón, X. (1984) Construction and characterization of new cloning vehicles VII. Construction of plasmid pBR327par, a completely sequenced, stable derivative of pBR327 containing the par locus of pSC101. Gene 28: 119–122. [DOI] [PubMed] [Google Scholar]


