Abstract
Phylogenies of mammals based on morphological data continue to show several major areas of conflict with the current consensus view of their relationships, which is based largely on molecular data. This raises doubts as to whether current morphological character sets are able to accurately resolve mammal relationships. We tested this under a hypothetical ‘best case scenario’ by using ancestral state reconstruction (under both maximum parsimony and maximum likelihood) to infer the morphologies of fossil ancestors for all clades present in a recent comprehensive DNA sequence-based phylogeny of mammals, and then seeing what effect the subsequent inclusion of these predicted ancestors had on unconstrained phylogenetic analyses of morphological data. We found that this resulted in topologies that are highly congruent with the current consensus phylogeny, at least when the predicted ancestors are assumed to be well preserved and densely sampled. Most strikingly, several analyses recovered the monophyly of clades that have never been found in previous morphology-only studies, such as Afrotheria and Laurasiatheria. Our results suggest that, at least in principle, improvements in the fossil record—specifically the discovery of fossil taxa that preserve the ancestral or near-ancestral morphologies of the nodes in the current consensus—may be sufficient to largely reconcile morphological and molecular estimates of mammal phylogeny, even using current morphological character sets.
Keywords: morphology, ancestors, fossil, phylogeny, mammals
1. Introduction
The evolutionary relationships of mammals have been a major focus of research within systematics for over a century [1–6]. In the last two decades, the increasing availability of molecular data has seen the emergence of a robust consensus phylogeny of extant mammals. This consensus indicates that several groupings of placental mammals proposed based on morphological data (such as ‘edentates’, ‘ungulates’ and ‘insectivorans’) are polyphyletic [4–6]. It also shows that living placentals are distributed among four major clades or ‘superorders’, probably reflecting their biogeographic history, namely Xenarthra, Afrotheria, Laurasiatheria and Euarchontoglires [4–6]. Arguably the most striking finding is that both Afrotheria and Laurasiatheria include ‘insectivoran-grade’ (afrotherian tenrecs and golden moles, laurasiatherian eulipotyphlans such as hedgehogs, shrews and moles), ‘ungulate-grade’ (afrotherian proboscideans, hyraxes and sea cows, laurasiatherian artiodactyls and perissodactyls) and myrmecophagous (afrotherian aardvarks, laurasiatherian pangolins) members. There is strong molecular evidence that Laurasiatheria and Euarchontoglires are sister taxa, forming Boreoeutheria [4–6], and it seems probable that Xenarthra and Afrotheria also form a clade, named Atlantogenata [4,7,8] (figure 1).
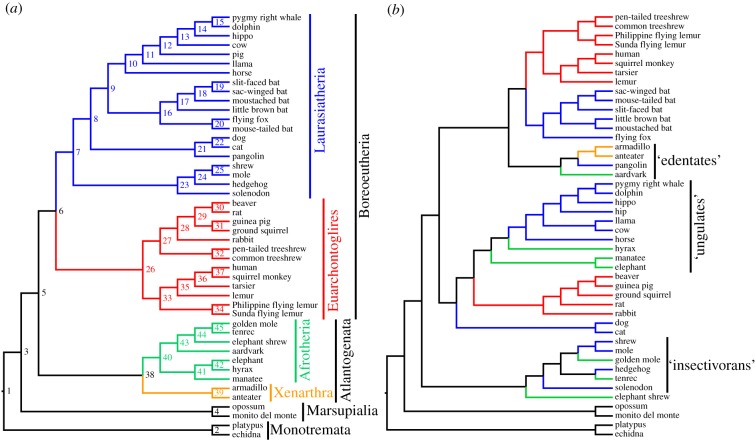
Figure 1.
Phylogenies of mammals based on (a) molecular and (b) morphological datasets. The topology shown in (a) is modified from fig. 1 of Meredith et al. [4] and is based on an 11 000 amino acid alignment from 26 gene fragments, whereas that in (b) is modified from electronic supplementary material, figure S2A of O'Leary et al. [3] and is based on 4541 morphological characters (fossil taxa have been deleted). Branches are colour coded according to their membership of the superorders Afrotheria, Xenarthra, Laurasiatheria and Euarchontoglires. Node numbers in (a) correspond to the predicted ancestors for those nodes in figure 2.
Recent morphology-only analyses of mammal relationships continue to show numerous areas of conflict with this consensus, and typically recover ‘insectivoran’, ‘ungulate’ and ‘edentate’ clades [3,9–11] (figure 1). ‘Total evidence’ analyses that combine morphological and molecular data are largely congruent with the consensus [3,9]. However, ‘artificial extinction’ (sensu [12,13]) or ‘pseudoextinction’ (sensu [14]) analyses of such total evidence datasets, in which selected extant taxa are treated as if they are fossils by deleting their molecular data, often fail to recover pseudoextinct placentals within their respective superorders [14] (but see [12,13]). This has led to suggestions that morphological data are ‘inadequate’ for accurately reconstructing the higher-level evolutionary relationships of mammals [10,14–16]. In particular, it raises questions as to whether genuine fossil taxa (for most of which molecular data are unlikely to ever become available) can be correctly placed within higher-level mammalian phylogeny, even when using a total evidence approach. This creates a dilemma, because the inclusion of fossil taxa may be critically important for phylogenetic comparative analyses [17–20], and yet such analyses assume that the fossil taxa are placed accurately in the phylogeny being used.
In general, we should expect increased taxon sampling to improve phylogenetic accuracy [21–23]. In morphological and total evidence analyses, fossil taxa are likely to be particularly important: by exhibiting unique combinations of plesiomorphic and derived character states, they should help break up long morphological branches leading to extant taxa [22,24,25]. The most detailed morphological study of mammal phylogeny published to date is the character-rich ‘phenomic’ study of O'Leary et al. [3], but this is still highly incongruent with the consensus [10] (figure 1). However, O'Leary et al. [3] included only 40 fossil taxa, which is a tiny fraction of the number likely to have existed during the Mesozoic and Cenozoic [26]. A key question, then, is whether improving the taxon sampling of morphological analyses, particularly of fossil taxa, might be sufficient to resolve the conflict between morphological and molecular estimates of mammal phylogeny.
We investigate this by first reconstructing the expected morphological character states (based on the O'Leary et al. [3] matrix) of the fossil ancestors of all the clades present in a comprehensive, family-level phylogeny of extant mammals that is based on nuclear sequence data [4], and then testing what impact the inclusion of these predicted fossil ancestors has on the results of morphology-only analyses. In effect, our analyses represent a hypothetical ‘best case scenario’ for morphological studies of mammal phylogeny, in which we simulate the discovery of direct fossil ancestors.
If the inclusion of these predicted ancestors in morphology-only analyses is sufficient to result in phylogenies that are largely congruent with the consensus, then it suggests that the current conflict between morphological and molecular estimates of mammal phylogeny might be resolved by improvements in the fossil record and the addition of more fossil taxa to currently available morphological matrices. In turn, this would suggest that fossil taxa (including non-ancestral forms), for which molecular data are unavailable, could still be accurately placed within mammal phylogeny given sufficiently dense taxon sampling, and hence that phylogenies that include fossil and extant mammals may become sufficiently accurate for use in comparative analyses. Conversely, if morphology-only analyses continue to show major areas of conflict with the consensus, even under this hypothetical ‘best case scenario’, then it suggests that the conflict will not be resolved simply by improved taxon sampling, and that the higher-level relationships of fossil mammals inferred using current datasets and methods of analysis should not be viewed with confidence.
2. Material and methods
The morphological matrix used here is that of O'Leary et al. [3]; this ‘phenomic’ matrix is by far the most character-rich available for mammals, comprising 4541 characters scored for 46 extant and 40 fossil taxa. We modified the matrix by first merging the character scores for the fossil notoungulate Thomashuxleya externa with those from a more recent study [27], and then deleting 407 constant characters. This left a total of 4134 characters: 1170 cranial, 1311 dental, 890 postcranial and 763 soft tissue. For the ancestral state reconstructions (ASRs), all fossil taxa were deleted from the matrix, and we assumed the topology present in fig. 1 of Meredith et al. [4] (figure 1a). We used two different optimality criteria for inferring ASRs: maximum parsimony (MP) and maximum likelihood (ML). MP-ASRs for all nodes were calculated using the ‘trace all characters' command in Mesquite, whereas ML-ASRs were calculated in RAxML using the ‘-f A’ command (which calculates marginal ancestral states), assuming the MK + GAMMA model and applying the Lewis correction (absence of invariant sites) for ascertainment bias [28]. The collective MP-ASRs for each node were then added to the original matrix (i.e. with all extant and fossil taxa present), to act as predicted ancestors. The same was done for the ML-ASRs, resulting in two different matrices: one with predicted ancestors based on MP-ASRs, and one with predicted ancestors based on ML-ASRs.
Unless corrected for, ancestral state reconstruction does not take into account that certain characters should be scored as inapplicable given particular scores for other characters; for example, characters relating to specific dental features should be scored as inapplicable in a predicted ancestor if it is inferred as entirely lacking teeth. To correct for this, we downloaded the character ontology associated with the O'Leary et al. [3] matrix, which specifies which characters become inapplicable given other character scores, from the online Morphobank database (project 773). We then used a custom R script to apply this ontology to our MP-ASRs and ML-ASRs, ensuring the biological plausibility of our predicted ancestors.
Different anatomical partitions differ in their preservation probability, at least in the form of associated remains: in general, soft tissue characters are the least likely to preserve, followed by postcranial characters (although certain postcranial elements, such as tarsals, may be relatively commonly preserved as isolated elements), then cranial, and lastly dental characters [29,30]. We further modified our matrices to take this into account: in the ‘all characters’ version of the matrix, we retained all character scores for the predicted ancestors; for ‘skeletal only’ we scored all soft tissue characters as unknown for the predicted ancestors; for ‘craniodental only’ we scored all soft tissue and postcranial characters as unknown for the predicted ancestors; for ‘dental only’, we only included dental character scores for the predicted ancestors. To further investigate the impact of fossil preservation, we used a custom R script to only retain character scores for the predicted ancestors that could be scored in (1) at least one of the 40 ‘real’ fossil taxa in the matrix, simulating a scenario in which the predicted ancestors are extremely well preserved (= ‘max preservation’), or (2) at least 50% (i.e. at least 20) of the ‘real’ fossil taxa, simulating a scenario in which the predicted ancestors show a ‘typical’ or ‘average’ degree of preservation (= ‘typical preservation’). Finally, we investigated the effect of incomplete sampling of predicted ancestors by creating versions of each matrix that include (1) predicted ancestors for all nodes, (2) predicted ancestors for nodes above the ordinal level only and (3) predicted ancestors for nodes representing the superorders and above (i.e. Xenarthra, Afrotheria, Laurasiatheria, Euarchontoglires, Atlantogeneta, Boreoeutheria, Placentalia, Marsupialia, Theria, Monotremata and Mammalia). All matrices are available in the electronic supplementary material, data file S1.
For the MP-ASRs, the original matrix and the different versions of the matrix with predicted ancestors included were analysed using MP, as implemented by TNT. Tree searches comprised new technology searches until the same minimum length was hit 100 times, followed by a traditional search with TBR branch-swapping among the trees already saved. All most parsimonious trees were summarized using strict consensus, and bootstrap support values were calculated as absolute frequencies using 500 replicates. For the ML-ASRs, the original matrix and the different versions of the matrix with predicted ancestors included were analysed using RAxML, again with the MK + GAMMA model and the Lewis correction for ascertainment bias. RAxML searches comprised 100 replicates of the default rapid hill-climbing algorithm. Non-parametric bootstrap support values were also calculated, with the number of replicates determined by the autoMRE criterion. Standard bootstrap values may be unduly conservative when one or more ‘rogue’ taxa are present, and so we also calculated support for all our MP and ML trees using the recently developed ‘transfer bootstrap expectation’ (TBE) method [31], using BOOSTER. All trees and associated support values are available in the electronic supplementary material (electronic supplementary material, data file S2).
After deleting all fossil taxa and predicted ancestors, the trees from all analyses were quantitatively compared with the same Meredith et al. [4] topology used to calculate the ASRs. We used the normalized Robinson–Foulds (= partition) metric (nRF), the SPR distance (SPRd) and the distortion coefficient (DC); the latter two metrics are less affected by shifts in the position of just one or a few taxa [32]. A greater degree of similarity between trees is indicated by values closer to 0 for nRF, but values closer to 1 for SPRd and DC. In the case of MP analyses that recovered more than one most parsimonious trees, we compared the individual most parsimonious trees, and also the strict consensus of these, to the Meredith et al. [4] topology.
3. Results and discussion
Strikingly, in several analyses, the inclusion of predicted ancestors was sufficient to result in phylogenies that are generally congruent with the consensus, particularly those that assumed well-preserved predicted ancestors (table 1 and figure 2). For example, for the MP analysis assuming ‘max preservation’ predicted ancestors, the strict consensus recovers the monophyly of Atlantogenata, Boreoeutheria and all four superorders (figure 2). Even when strict monophyly of the four superorders was not recovered, this was often due to only one or a few taxa being misplaced, as indicated by high SPRd and DC values (table 1). However, analyses that assume less well-preserved predicted ancestors (e.g. ‘dental only’, ‘typical preservation’) resulted in phylogenies that were much less congruent with the consensus (table 1), as did analyses that included only predicted ancestors for nodes above the ordinal level, or for superorders and above (electronic supplementary material, tables S3 and S4).
Table 1.
Summary of results of phylogenetic analyses with and without predicted ancestors (PAs) under maximum parsimony (MP) and maximum likelihood (ML). Fit relative to the molecular phylogeny of Meredith et al. was assessed using the normalized Robinson–Foulds metric (nRF), the SPR distance (SPRd) and the distortion coefficient (DC). nRF values closer to 0 and SPRd and DC values closer to 1 represent better fit. For the MP analyses that recovered more than one most parsimonious tree (MPT), the number of MPTs is indicated. Also indicated is whether the analyses recovered six superordinal clades found in the ‘molecular consensus’, with standard bootstrap (BS) and ‘transfer bootstrap expectation’ (TBE) values given in brackets.
| monophyletic? |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| analysis | nRF | SPRd | DC | Atlantogenata | Afrotheria | Xenarthra | Boreoeutheria | Euarchontoglires | Laurasiatheria |
| MP | |||||||||
| no PAs | 0.57 | 0.56 | 0.70 | no | no | yes (BS = 74; TBE = 87) | no | no | no |
| ‘all characters’ PAs MPTs (n = 24) strict consensus |
0.23 0.25 |
0.86 0.86 |
0.94 0.93 |
yes (BS < 50; TBE < 50) |
yes (BS < 50; TBE < 50) |
yes (BS = 62; TBE = 74) |
yes (BS < 50; TBE < 50) |
yes (BS = 84; TBE = 92) |
yes (BS < 50; TBE < 50) |
| ‘skeletal only’ PAs MPTs (n = 24) strict consensus |
0.23 0.25 |
0.86 0.86 |
0.94 0.93 |
yes (BS < 50; TBE < 50) |
yes (BS < 50; TBE < 50) |
yes (BS = 62; TBE = 74) |
yes (BS < 50; TBE < 50) |
yes (BS = 73; TBE = 86) |
yes (BS < 50; TBE < 50) |
| ‘craniodental only’ PAs MPTs (n = 12) strict consensus |
0.46 0.47 |
0.70 0.70 |
0.85 0.84 |
no |
no |
yes (BS < 50; TBE = 65) |
no |
yes (BS = 50; TBE = 74) |
no |
| ‘dental only’ PAs MPTs (n = 9) strict consensus |
0.57 0.56 |
0.60 0.63 |
0.73 0.73 |
no |
no |
yes (BS < 50; TBE = 58) |
no |
yes (BS < 50; TBE < 50) |
no |
| ‘max preservation’ PAs MPTs (n = 24) strict consensus |
0.23 0.25 |
0.86 0.86 |
0.94 0.93 |
yes (BS < 50; TBE < 50) |
yes (BS < 50; TBE < 50) |
yes (BS = 60; TBE = 73) |
yes (BS < 50; TBE < 50) |
yes (BS = 74; TBE = 86) |
yes (BS < 50; TBE < 50) |
| ‘typical preservation’ PAs MPTs (n = 2) strict consensus |
0.46 0.48 |
0.72 0.72 |
0.86 0.85 |
no |
no |
yes (BS < 50; TBE = 65) |
no |
yes (BS < 50; TBE = 59) |
no |
| ML | |||||||||
| no PAs | 0.59 | 0.56 | 0.70 | no | no | yes (BS = 96; TBE = 98) | no | no | no |
| ‘all characters’ PAs | 0.25 | 0.81 | 0.94 | yes (BS < 50; TBE = 94) | no | yes (BS = 89; TBE = 93) | no | no | yes (BS < 50; TBE = 90) |
| ‘skeletal only’ PAs | 0.27 | 0.81 | 0.93 | yes (BS < 50; TBE = 92) | no | yes (BS = 87; TBE = 92) | no | no | yes (BS < 50; TBE = 88) |
| ‘craniodental only’ PAs | 0.41 | 0.72 | 0.88 | no | yes (BS < 50; TBE = 65) | yes (BS = 87; TBE = 92) | no | no | no |
| ‘dental only’ PAs | 0.48 | 0.60 | 0.81 | no | no | yes (BS = 98; TBE = 99) | no | no | no |
| ‘max preservation’ PAs | 0.27 | 0.81 | 0.93 | yes (BS < 50; TBE = 92) | no | yes (BS = 89; TBE = 93) | no | no | yes (BS < 50; TBE = 88) |
| ‘typical preservation’ PAs | 0.43 | 0.74 | 0.87 | no | no | yes (BS = 75; TBE = 83) | no | no | no |
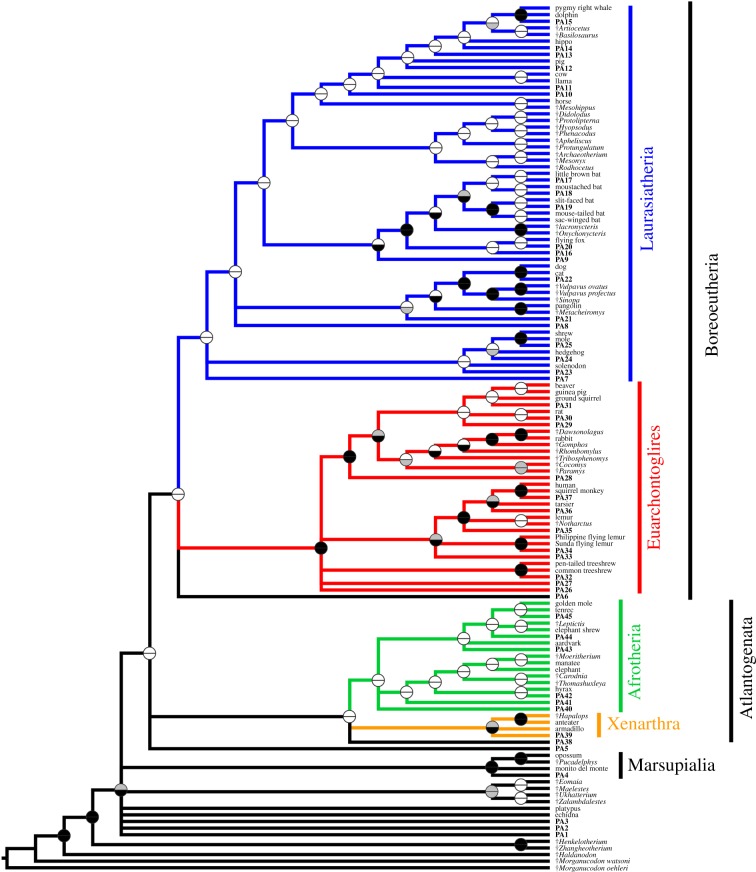
Figure 2.
Phylogeny of mammals based on maximum parsimony (MP) analysis of a modified version of the morphological dataset of O'Leary et al. [3] and with ‘max preservation’ predicted ancestors (PAs) added. Topology shown is a strict consensus of 64 most parsimonious trees. Ancestral states for the predicted ancestor were reconstructed using MP. Predicted ancestors are shown in bold, with numbers corresponding to the nodes for which they are ancestral, as shown in figure 1a. Fossil taxa are indicated with †. Circles at nodes indicate support, with the top half representing standard bootstrap values and the bottom half ‘transfer bootstrap expectation’ (TBE) values: black indicates greater than or equal to 70% support, grey 50–69% and white less than 50%.
Bootstrap values are low for most nodes in the MP analyses, even when using the TBE method (figure 2; electronic supplementary material, data file S2), but this might be expected given the inclusion of multiple fossil taxa (both ‘real’ fossil taxa and predicted ancestors) [25,33]. Support values were generally higher for the ML analyses, particularly when using the TBE method (figure 2; electronic supplementary material, data file S2), including for several of the placental superordinal clades, where recovered (table 1; electronic supplementary material, data file S2).
With all predicted ancestors included, the MP analyses recovered more of the placental superordinal clades than did the equivalent ML analyses (table 1). However, when comparing across all our MP and ML analyses (table 1; electronic supplementary material, tables S3 and S4), there was no significant difference in fit to the Meredith et al. topology between the two optimality criteria, except as measured by DC, where ML showed significantly better fit (electronic supplementary material, text file S5). Future studies could see whether data partitioning [34] improves the performance of ML. Another obvious area to explore would be the use of tip-dating approaches [35–37], with predicted ancestors assigned ages compatible with recent molecular clock analyses (e.g. [4]); recent work suggests that such approaches may be better able to identify cases of homoplastic resemblance than methods that do not incorporate temporal evidence [38].
We emphasize that our study represents a hypothetical ‘best case scenario’: we inferred the ancestral states of predicted ancestors using the same optimality criterion (either MP or ML using the MK + GAMMA model) that was subsequently used to analyse the matrix with the predicted ancestors added, and the predicted ancestors lack any apomorphies not present in their descendants (i.e. they represent ‘perfect’ ancestral morphologies). Both of these are unrealistic, or at least highly optimistic, assumptions (although direct ancestors may actually be relatively common in the fossil record [39,40]).
Nevertheless, we show that the inclusion of hypothetical ancestors predicted by the molecular consensus of placental phylogeny is sufficient to result in phylogenies that closely match this consensus, without the use of constraints or the addition of molecular data, at least when these predicted ancestors are assumed to be well preserved and densely sampled (figure 2 and table 1). Thus, there are at least hypothetical character combinations that can link morphologically disparate mammalian taxa, such as the ‘insectivoran-grade’, ‘ungulate-grade’ and myrmecophagous members of Afrotheria and Laurasiatheria. If genuine fossil taxa exhibit these character combinations, and are sufficiently well preserved, then their discovery and inclusion in phylogenetic analyses might be sufficient to largely resolve the current conflict between molecular and morphological analyses, even using morphological matrices that currently show extensive conflict with molecular data, such as that of O'Leary et al. [3]. Finding such well-preserved taxa may prove difficult, given that the mammal fossil record remains relatively poor [26], particularly for key regions such as Africa [41], and is likely to remain dominated by isolated dental remains [42,43]. However, recent discoveries show that progress is being made in this direction; for example, well-preserved remains of Ocepeia from the Palaeocene of Morocco reveal that it combines features of ‘insectivoran-grade’ and ‘ungulate-grade’ afrotherians [44], and other African fossils show that dental similarities between ‘ungulate-grade’ afrotherians and laurasiatherians are homoplastic [45].
Although not tested here, a similar principle may apply to other clades. If so, then we can be optimistic that we may be able to accurately infer a phylogeny for many clades using morphological data alone, given a sufficiently good fossil record. Improvements in phylogenetic methods will undoubtedly also play a role: these might include better models of morphological character evolution [46,47], clock models [35–37], methods that take into account character non-independence and saturation [48–50] or some combination of these. However, our results suggest that inclusion of fossil taxa may prove to be particularly important. In any case, arguments that morphological data are ‘inadequate’ for accurately inferring the phylogeny of mammals [14–16], or of the many other clades that currently show extensive morphological–molecular conflict (such as birds [51–54]), are at the very least premature.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Marcelo Sánchez-Villagra (University of Zürich) and Robert Asher (University of Cambridge) for discussion, and Maureen O'Leary (Stony Brook University) for assistance with Morphobank. Erin Saupe (University of Oxford), Robert Asher and an anonymous reviewer also gave helpful and constructive feedback that helped us to greatly improve the final paper.
Data accessibility
All data used in this study are available in the electronic supplementary material accompanying the article.
Authors' contributions
R.M.D.B. conceived of this study, developed methods, carried out analyses and drafted the manuscript. C.B. wrote R scripts, carried out analyses and helped draft the manuscript. Both authors gave final approval for publication.
Competing interests
We have no competing interests.
Funding
We received no funding for this study.
References
- 1.Gregory WK. 1910. The orders of mammals. Bull. Am. Mus. Nat. Hist. 27, 1–524. [Google Scholar]
- 2.Novacek MJ. 1992. Mammalian phylogeny: shaking the tree. Nature 356, 121–125. ( 10.1038/356121a0) [DOI] [PubMed] [Google Scholar]
- 3.O'Leary MA, et al. 2013. The placental mammal ancestor and the post-K-Pg radiation of placentals. Science 339, 662–667. ( 10.1126/science.1229237) [DOI] [PubMed] [Google Scholar]
- 4.Meredith RW, et al. 2011. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science 334, 521–524. ( 10.1126/science.1211028) [DOI] [PubMed] [Google Scholar]
- 5.Asher RJ, Bennett N, Lehmann T. 2009. The new framework for understanding placental mammal evolution. Bioessays 31, 853–864. ( 10.1002/bies.200900053) [DOI] [PubMed] [Google Scholar]
- 6.Springer MS, Stanhope MJ, Madsen O, de Jong WW.. 2004. Molecules consolidate the placental mammal tree. Trends Ecol. Evol. 19, 430–438. ( 10.1016/j.tree.2004.05.006) [DOI] [PubMed] [Google Scholar]
- 7.Liu L, et al. 2017. Genomic evidence reveals a radiation of placental mammals uninterrupted by the KPg boundary. Proc. Natl Acad. Sci. USA 114, E7282–E7290. ( 10.1073/pnas.1616744114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esselstyn JA, Oliveros CH, Swanson MT, Faircloth BC. 2017. Investigating difficult nodes in the placental mammal tree with expanded taxon sampling and thousands of ultraconserved elements. Genome Biol. Evol. 9, 2308–2321. ( 10.1093/gbe/evx168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asher RJ. 2007. A web-database of mammalian morphology and a reanalysis of placental phylogeny. BMC Evol. Biol. 7, 108 ( 10.1186/1471-2148-7-108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Springer MS, Meredith RW, Teeling EC, Murphy WJ. 2013. Technical comment on ‘The placental mammal ancestor and the post-K-Pg radiation of placentals'. Science 341, 613 ( 10.1126/science.1238025) [DOI] [PubMed] [Google Scholar]
- 11.Halliday TJ, Upchurch P, Goswami A. 2017. Resolving the relationships of Paleocene placental mammals. Biol. Rev. Camb. Philos. Soc. 92, 521–550. ( 10.1111/brv.12242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asher RJ, Hofreiter M. 2006. Tenrec phylogeny and the noninvasive extraction of nuclear DNA. Syst. Biol. 55, 181–194. ( 10.1080/10635150500433649) [DOI] [PubMed] [Google Scholar]
- 13.Pattinson DJ, Thompson RS, Piotrowski AK, Asher RJ. 2015. Phylogeny, paleontology, and primates: do incomplete fossils bias the tree of life? Syst. Biol. 64, 169–186. ( 10.1093/sysbio/syu077) [DOI] [PubMed] [Google Scholar]
- 14.Springer MS, Burk-Herrick A, Meredith R, Eizirik E, Teeling E, O'Brien SJ, Murphy WJ. 2007. The adequacy of morphology for reconstructing the early history of placental mammals. Syst. Biol. 56, 673–684. ( 10.1080/10635150701491149) [DOI] [PubMed] [Google Scholar]
- 15.Springer MS, Meredith RW, Eizirik E, Teeling E, Murphy WJ. 2008. Morphology and placental mammal phylogeny. Syst. Biol. 57, 499–503. ( 10.1080/10635150802164504) [DOI] [PubMed] [Google Scholar]
- 16.Springer MS, Emerling CA, Meredith RW, Janecka JE, Eizirik E, Murphy WJ. 2017. Waking the undead: implications of a soft explosive model for the timing of placental mammal diversification. Mol. Phylogenet. Evol. 106, 86–102. ( 10.1016/j.ympev.2016.09.017) [DOI] [PubMed] [Google Scholar]
- 17.Slater GJ, Harmon LJ, Alfaro ME. 2012. Integrating fossils with molecular phylogenies improves inference of trait evolution. Evolution 66, 3931–3944. ( 10.1111/j.1558-5646.2012.01723.x) [DOI] [PubMed] [Google Scholar]
- 18.Rabosky DL. 2010. Extinction rates should not be estimated from molecular phylogenies. Evolution 64, 1816–1824. ( 10.1111/j.1558-5646.2009.00926.x) [DOI] [PubMed] [Google Scholar]
- 19.Rabosky DL. 2016. Challenges in the estimation of extinction from molecular phylogenies: a response to Beaulieu and O'Meara. Evolution 70, 218–228. ( 10.1111/evo.12820) [DOI] [PubMed] [Google Scholar]
- 20.Crisp MD, Trewick SA, Cook LG. 2011. Hypothesis testing in biogeography. Trends Ecol. Evol. 26, 66–72. ( 10.1016/j.tree.2010.11.005) [DOI] [PubMed] [Google Scholar]
- 21.Heath TA, Zwickl DJ, Kim J, Hillis DM. 2008. Taxon sampling affects inferences of macroevolutionary processes from phylogenetic trees. Syst. Biol. 57, 160–166. ( 10.1080/10635150701884640) [DOI] [PubMed] [Google Scholar]
- 22.Wiens JJ. 2005. Can incomplete taxa rescue phylogenetic analyses from long-branch attraction? Syst. Biol. 54, 731–742. ( 10.1080/10635150500234583) [DOI] [PubMed] [Google Scholar]
- 23.Zwickl DJ, Hillis DM. 2002. Increased taxon sampling greatly reduces phylogenetic error. Syst. Biol. 51, 588–598. ( 10.1080/10635150290102339) [DOI] [PubMed] [Google Scholar]
- 24.Gauthier J, Kluge AG, Rowe T. 1988. Amniote phylogeny and the importance of fossils. Cladistics 4, 105–209. ( 10.1111/j.1096-0031.1988.tb00514.x) [DOI] [PubMed] [Google Scholar]
- 25.Horovitz I. 1999. A phylogenetic study of living and fossil platyrrhines. Am. Mus. Novit. 3269, 1–40. [Google Scholar]
- 26.Marshall CR. 2017. Five palaeobiological laws needed to understand the evolution of the living biota. Nat. Ecol. Evol. 1, 165 ( 10.1038/s41559-017-0165) [DOI] [PubMed] [Google Scholar]
- 27.Carrillo JD, Asher RJ.. 2017. An exceptionally well-preserved skeleton of Thomashuxleya externa (Mammalia, Notoungulata), from the Eocene of Patagonia, Argentina. Palaeontol. Electron. 20.2.34A, 1–33. [Google Scholar]
- 28.Lewis PO. 2001. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 50, 913–925. ( 10.1080/106351501753462876) [DOI] [PubMed] [Google Scholar]
- 29.Sansom RS, Wills MA, Williams T. 2017. Dental data perform relatively poorly in reconstructing mammal phylogenies: morphological partitions evaluated with molecular benchmarks. Syst. Biol. 66, 813–822. ( 10.1093/sysbio/syw116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sansom RS, Wills MA. 2013. Fossilization causes organisms to appear erroneously primitive by distorting evolutionary trees. Sci. Rep. 3, 2545 ( 10.1038/srep02545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemoine F, Domelevo Entfellner JB, Wilkinson E, Correia D, Davila Felipe M, De Oliveira T, Gascuel O.. 2018. Renewing Felsenstein's phylogenetic bootstrap in the era of big data. Nature 556, 452–456. ( 10.1038/s41586-018-0043-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goloboff PA, Torres A, Arias JS. 2018. Weighted parsimony outperforms other methods of phylogenetic inference under models appropriate for morphology. Cladistics 34, 407–437. ( 10.1111/cla.12205) [DOI] [PubMed] [Google Scholar]
- 33.Cobbett A, Wilkinson M, Wills MA. 2007. Fossils impact as hard as living taxa in parsimony analyses of morphology. Syst. Biol. 56, 753–766. ( 10.1080/10635150701627296) [DOI] [PubMed] [Google Scholar]
- 34.Clarke JA, Middleton KM. 2008. Mosaicism, modules, and the evolution of birds: results from a Bayesian approach to the study of morphological evolution using discrete character data. Syst. Biol. 57, 185–201. ( 10.1080/10635150802022231) [DOI] [PubMed] [Google Scholar]
- 35.Pyron RA. 2011. Divergence time estimation using fossils as terminal taxa and the origins of Lissamphibia. Syst. Biol. 60, 466–481. ( 10.1093/sysbio/syr047) [DOI] [PubMed] [Google Scholar]
- 36.Ronquist F, Klopfstein S, Vilhelmsen L, Schulmeister S, Murray DL, Rasnitsyn AP. 2012. A total-evidence approach to dating with fossils, applied to the early radiation of the Hymenoptera. Syst. Biol. 61, 973–999. ( 10.1093/sysbio/sys058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C, Stadler T, Klopfstein S, Heath TA, Ronquist F. 2016. Total-evidence dating under the fossilized birth-death process. Syst. Biol. 65, 228–249. ( 10.1093/sysbio/syv080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee M.SY, Yates AM. 2018. Tip-dating and homoplasy: reconciling the shallow molecular divergences of modern gharials with their long fossil record. Proc. R. Soc. B 285, 20181071 ( 10.1098/rspb.2018.1071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foote M. 1996. On the probability of ancestors in the fossil record. Paleobiology 22, 141–151. ( 10.1017/S0094837300016146) [DOI] [Google Scholar]
- 40.Gavryushkina A, Heath TA, Ksepka DT, Stadler T, Welch D, Drummond AJ. 2017. Bayesian total-evidence dating reveals the recent crown radiation of penguins. Syst. Biol. 66, 57–73. ( 10.1093/sysbio/syw060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seiffert ER. 2010. Chronology of Paleogene mammal localities. In Cenozoic mammals of Africa (eds Sanders WJ, Werdelin L), pp. 19–26. Berkeley, CA: University of California Press. [Google Scholar]
- 42.Kielan-Jaworowska Z, Cifelli RL, Luo Z.-X. 2004. Mammals from the age of dinosaurs: origins, evolution, and structure. New York, NY: Columbia University Press. [Google Scholar]
- 43.Rose KD. 2006. The beginning of the age of mammals. Baltimore, MD: Johns Hopkins University Press. [Google Scholar]
- 44.Gheerbrant E, Amaghzaz M, Bouya B, Goussard F, Letenneur C. 2014. Ocepeia (Middle Paleocene of Morocco): the oldest skull of an afrotherian mammal. PLoS ONE 9, e89739 ( 10.1371/journal.pone.0089739) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gheerbrant E, Filippo A, Schmitt A. 2016. Convergence of Afrotherian and Laurasiatherian ungulate-like mammals: first morphological evidence from the Paleocene of Morocco. PLoS ONE 11, e0157556 ( 10.1371/journal.pone.0157556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pyron RA. 2017. Novel approaches for phylogenetic inference from morphological data and total-evidence dating in squamate reptiles (lizards, snakes, and amphisbaenians). Syst. Biol. 66, 38–56. ( 10.1093/sysbio/syw068) [DOI] [PubMed] [Google Scholar]
- 47.Wright AM, Lloyd GT, Hillis DM. 2016. Modeling character change heterogeneity in phylogenetic analyses of morphology through the use of priors. Syst. Biol. 65, 602–611. ( 10.1093/sysbio/syv122) [DOI] [PubMed] [Google Scholar]
- 48.Davalos LM, Velazco PM, Warsi OM, Smits PD, Simmons NB. 2014. Integrating incomplete fossils by isolating conflicting signal in saturated and non-independent morphological characters. Syst. Biol. 63, 582–600. ( 10.1093/sysbio/syu022) [DOI] [PubMed] [Google Scholar]
- 49.Herrera JP, Davalos LM. 2016. Phylogeny and divergence times of lemurs inferred with recent and ancient fossils in the tree. Syst. Biol. 65, 772–791. ( 10.1093/sysbio/syw035) [DOI] [PubMed] [Google Scholar]
- 50.Billet G, Bardin J. In press. Serial homology and correlated characters in morphological phylogenetics: modeling the evolution of dental crests in placentals. Syst. Biol. ( 10.1093/sysbio/syy071) [DOI] [PubMed] [Google Scholar]
- 51.Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM, Lemmon AR. 2015. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526, 569–573. ( 10.1038/nature15697) [DOI] [PubMed] [Google Scholar]
- 52.Jarvis ED, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346, 1320–1331. ( 10.1126/science.1253451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livezey BC, Zusi RL. 2007. Higher-order phylogeny of modern birds (Theropoda, Aves: Neornithes) based on comparative anatomy. II. Analysis and discussion. Zool. J. Linn. Soc. 149, 1–95. ( 10.1111/j.1096-3642.2006.00293.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayr G. 2008. Avian higher-level phylogeny: well-supported clades and what we can learn from a phylogenetic analysis of 2954 morphological characters. J. Zool. Syst. Evol. Res. 46, 63–72. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study are available in the electronic supplementary material accompanying the article.