Significance
The complexity of modern proteins makes the understanding of how proteins evolved from simple beginnings a daunting challenge. The Walker-A motif is a phosphate-binding loop (P-loop) found in possibly the most ancient and abundant protein class, so-called P-loop NTPases. By combining phylogenetic analysis and computational protein design, we have generated simple proteins, of only 55 residues, that contain the P-loop and thereby confer binding of a range of phosphate-containing ligands—and even more avidly, RNA and single-strand DNA. Our results show that biochemical function can be implemented in small and simple proteins; they intriguingly suggest that the P-loop emerged as a polynucleotide binder and catalysis of phosphoryl transfer evolved later upon acquisition of higher sequence and structural complexity.
Keywords: de novo protein design, protein evolution, Walker-A, RNA binding protein, conformational diversity
Abstract
Abundant and essential motifs, such as phosphate-binding loops (P-loops), are presumed to be the seeds of modern enzymes. The Walker-A P-loop is absolutely essential in modern NTPase enzymes, in mediating binding, and transfer of the terminal phosphate groups of NTPs. However, NTPase function depends on many additional active-site residues placed throughout the protein’s scaffold. Can motifs such as P-loops confer function in a simpler context? We applied a phylogenetic analysis that yielded a sequence logo of the putative ancestral Walker-A P-loop element: a β-strand connected to an α-helix via the P-loop. Computational design incorporated this element into de novo designed β-α repeat proteins with relatively few sequence modifications. We obtained soluble, stable proteins that unlike modern P-loop NTPases bound ATP in a magnesium-independent manner. Foremost, these simple P-loop proteins avidly bound polynucleotides, RNA, and single-strand DNA, and mutations in the P-loop’s key residues abolished binding. Binding appears to be facilitated by the structural plasticity of these proteins, including quaternary structure polymorphism that promotes a combined action of multiple P-loops. Accordingly, oligomerization enabled a 55-aa protein carrying a single P-loop to confer avid polynucleotide binding. Overall, our results show that the P-loop Walker-A motif can be implemented in small and simple β-α repeat proteins, primarily as a polynucleotide binding motif.
Although large and highly complex in structure and catalytic mechanism, modern proteins are thought to have evolved by duplication, fusion, and diversification of shorter polypeptides (1–4). The most conserved motifs in contemporary proteins are presumed to be relics of these simple, ancient beginnings. However, although the most archaic and functionally essential motifs may not have changed much, the structure and sequence context in which they currently reside fundamentally differs from the state in which they first emerged. Consequently, while in modern proteins these motifs are absolutely necessary, their function depends on a consortium of residues from the protein’s scaffold and its active-site pocket (5). How large the earliest proteins were, let alone what their composition, structure, or function was, are all unknown. Thus, reconstruction of historically relevant early protein forms is currently beyond reach. One can, however, attempt to obtain prototypes: proteins in which the presumed ancient motifs are implemented in a relatively rudimentary context, whereby biochemical function is mediated by these motifs on their own, in the absence of other functional motifs or an active-site pocket, and yet, the sequence, structure, and function of these prototypes relates to modern proteins (6–14). The ability to graft key functional motifs would also advance protein engineering. Protein scaffolds are routinely designed de novo, sometimes with no relation to existing structures. However, implementation of function, such as ligand binding, in a de novo-designed scaffold remains a challenge (15–19). To address these challenges, we have designed functional proteins harboring the P-loop Walker-A motif, arguably the most omnipresent and ancient function-mediating protein motif.
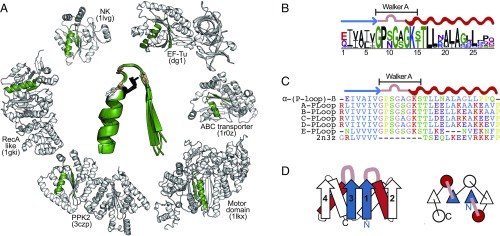
Systematic analyses of contemporary proteins have provided catalogs of ancient motifs, and the so-called Walker-A P-loop is consistently noted in these catalogs (20, 21), as are other widely present phosphate-binding loops, including the Rossmann fold’s P-loop (22, 23). P-loop–containing proteins were also unambiguously assigned to the last universal common ancestor (24–26). The Walker-A motif GxxGxGK[T/S] (27) typically binds the phosphate groups of phosphorylated ribonucleosides (NXPs) and catalyzes phosphoryl transfer. Beyond the Walker-A sequence, the P-loop motif also includes the flanking β-strand and α-helix (21, 22). This extended motif [hereinafter β-(P-loop)-α] is a key element of P-loop NTPases, the most abundant and diverse protein superfamily (28) constituting ≥10% of the predicted ORFs (29) (Fig. 1A). Structurally, the P-loop NTPases fold comprises a tandem repeat of βαβ elements arranged in a three-layered α/β/α sandwich architecture with the β-(P-loop)-α motif comprising the first β-α element. A key element of the P-loop is the backbone NH group, and in particular its second and third glycines, that forms a phosphate-binding nest, as demonstrated by the peptide SGAGKT weakly binding inorganic phosphate (30).
Fig. 1.
(A) Structural alignment of β-(P-loop)-α motifs of different P-loop NTPase (the canonical Walker-A residues: G1, G2, and G3 are in pink, K4 in black, and [T/S]5in white; G1-XX-G2-X-G3-K4-(S/T)5). (B) Sequence logo representing the inferred ancestral β-(P-loop)-α profile. (C) Sequence alignment of the most probable ancestral β-(P-loop)-α sequence and of the N-terminal segments of the PLoop designs. (D) Schematic representation of the secondary structure and topology of the PLoop designs. Helices are represented by rectangles or circles, strands by arrows or triangles, and the P-loops by pink segments. The flanking strand of the β-(P-loop)-α segment is in blue and the helix in red.
However, beyond the P-loop, additional functionally critical residues are located throughout the polypeptide chains of modern P-loop NTPases, including the Walker-B motif (27) and the residues that, together with the canonical T/S of the Walker-A motif, chelate the essential magnesium ion (31). An active-site pocket that excludes bulk water is also considered critical to function. Past studies tantalizingly indicated that ∼50-aa segments of P-loop NTPases exert ATP binding (6–8). However, an early attempt to graft the Walker-A P-loop onto a natural protein scaffold resembling the P-loop NTPase fold failed to yield NTP binding, let alone phosphoryl transfer (5). Here, by combining phylogenetic analysis and sequence-pattern recognition with computational protein design, we have generated de novo small and simple P-loop–containing β-α repeat proteins that confer binding of a range of phosphate-containing ligands, NTPs, as well as polynucleotides, in a context far simpler than contemporary P-loop NTPases.
Inference of a β-(P-Loop)-α Sequence Prototype
In contemporary P-loop NTPases, the β-(P-loop)-α motif is found in extremely diverse protein families. Its sequence is highly variable, even in the canonical Walker-A positions. Nonetheless, a structural alignment of the β-(P-loop)-α motif identifies remarkable similarities (Fig. 1A). To derive a sequence profile that would represent a prototype of the last common ancestor of this motif, we extended several analyses that identified the β-(P-loop)-α as a primordial motif (20, 21, 32). Starting with five sequences originally identified by Walker et al. (27), we generated a sequence profile and systematically searched the National Center for Biotechnology Information Conserved Domain Database. Matching segments with known structure were used to identify the β-(P-loop)-α segment at a length of 27 residues. After filtering, an alignment of 3,775 segments was obtained (SI Appendix, Fig. S1A). A consensus prototype could be extracted from this alignment; however, sequence representation in databases is highly biased. We therefore applied ancestral inference, taking the phylogenetic relationship between protein families into consideration and minimizing biases. Although the aligned segment was short, the phylogenetic tree was largely monophyletic with respect to the known P-loop NTPase families (SI Appendix, Fig. S1B). The most probable ancestral amino acid was inferred in each position by maximum likelihood (33). To assess the robustness of inference, a sequence profile was built from multiple parallel inferences (SI Appendix, Fig. S1C).
The resulting profile logo is shown in Fig. 1B. The Walker-A sequence was unambiguously assigned, including in positions that are highly diverged in modern proteins (annotated as x, GxxGxGK[T/S]). The three residues following the Walker-A motif were also robustly assigned (positions 15–17 in Fig. 1B). In the remaining positions, several amino acids were predicted, yet mostly with a common physicochemical nature (e.g., at position 9 in Fig. 1B; N and S are both polar amino acids). Although not intended, the profile sequence is dominated by prebiotic amino acids [those obtained in spontaneous chemical reactions (34)], with the solely abiotic amino acid being the lysine of the Walker-A motif. The absence of aromatic amino acids, cysteines, and histidines is notable even in contemporary sequences (SI Appendix, Fig. S1D).
Engineering Simple Proteins Harboring the β-(P-Loop)-α Motif
Can the P-loop motif yield simple yet functional proteins? We first examined peptides whose sequences represented the most probable amino acids in the profile. These formed amyloid-like fibrils that changed in morphology upon ATP addition (SI Appendix, Fig. S2). However, we observed differences among preparations (35), and fibril formation is notoriously irreproducible. We further attempted to construct simple repeat proteins comprising two to four tandem repeats of the most probable β-(P-loop)-α ancestral sequence. However, these tandem repeat proteins were insoluble. We therefore turned to computational protein design that has also been applied for the reconstruction of ancient enzyme prototypes (11), including short, functional segments (12, 13). We used Rosetta folding simulations to integrate the sequences of the inferred β-(P-loop)-α segment into a suitable structural context provided by “ideal folds”: simple proteins that were de novo-designed based on set of rules relating secondary structure patterns to tertiary packing (15). These included two designs comprising four tandem β-α repeats with a three-layered α/β/α sandwich architecture: fold II, whose β-strands topology is symmetric (2-1-3-4; Flavodoxin/Rossmann-like fold; PDB ID code 2N3Z) and fold IV, with swapped β-strands topology (2-3-1-4; P-loop NTPases-like fold; PDB ID code 2LVB). These proteins were designed solely by packing criteria and, although they recapitulate architectures abundant in natural proteins, they show no detectable sequence homology to natural proteins (15).
The β-(P-loop)-α inferred sequence was readily incorporated into fold II by replacing the first and third β-α segments, and converged to stable structures with relatively few iterations and minimal sequence changes in the β-(P-loop)-α motif (Fig. 1 C and D). The remaining β-α repeats (second and fourth) were largely borrowed from the original de novo design fold. Overall, six predictions with the best Rosetta energy values were experimentally tested: five based on fold II (A-PLoop to E-PLoop) and only one based on fold IV (F-PLoop) (Table 1 and SI Appendix, Figs. S1 and S3 A–D). The simulations indicated that fold IV designs tended to switch topology toward fold II. The computation only optimized packing stability, whereas functional constrains, such as phosphate binding, were not modeled. Nonetheless, at least one characteristic of the P-loop was captured: the last two amino acids of the Walker-A motif, K[T/S], integrated into the flanking helix. In A–E designs, the P-loop’s backbone adopted a “double bent” configuration that is reminiscent of the natural loop conformation, while in design F the P-loop was modeled in a different configuration (SI Appendix, Fig. S3 A and B).
Table 1.
Summary of the properties of the designed PLoop proteins
| General properties | Structural properties | Binding properties | ||||||||
| PLoop | Ideal fold | MW (kDa) | Symmetry (%) | pI | Oligomerization | CD | Well-resolved NMR | ELISA | SPR | MST |
| A-PLoop | II | 11.12 | 50.9 | 9.5 | d/m | β/α | − | + | + | + |
| B-PLoop | II | 11.24 | 43.4 | 8.0 | d/m | β/α | − | + | + | + |
| C-PLoop | II | 11.13 | 38.2 | 9.7 | d/m | β/α | + | − | + | ND |
| D-PLoop | II | 11.22 | 42.9 | 9.7 | d/m | β/α | − | + | + | ND |
| E-PLoop | II | 10.82 | 50.0 | 4.4 | m | β/α | − | + | + | ND |
| F-PLoop | IV | 12.35 | 30.4 | 4.6 | m | β/α + random coil | − | − | ND | ND |
| 3N3Z (15) | II | 10.23 | 28.3 | 9.2 | m | β/α | + | − | − | − |
| 2LVB (15) | IV | 11.58 | 17.5 | 6.6 | m | β/α | + | − | ND | ND |
The second column shows the ideal fold used as a scaffold (15); the third, fourth, and fifth columns indicate the MW (excluding the His-tag), internal sequence symmetry, and the theoretical isoelectric point. The oligomerization state was determined by native MS and SDS/PAGE (d, dimer; m, monomer). Binding properties: ELISA detected binding of the PLoop designs to immobilized ssDNA via anti–His-tag antibodies; MST, microscale thermofluoresis with soluble fluorescently labeled ssDNA; ND, not determined; SPR, surface plasmon resonance detection of binding to immobilized ssDNA or RNA oligos.
Structural Characterization Reveals Folded and Stable yet Polymorphic Structures
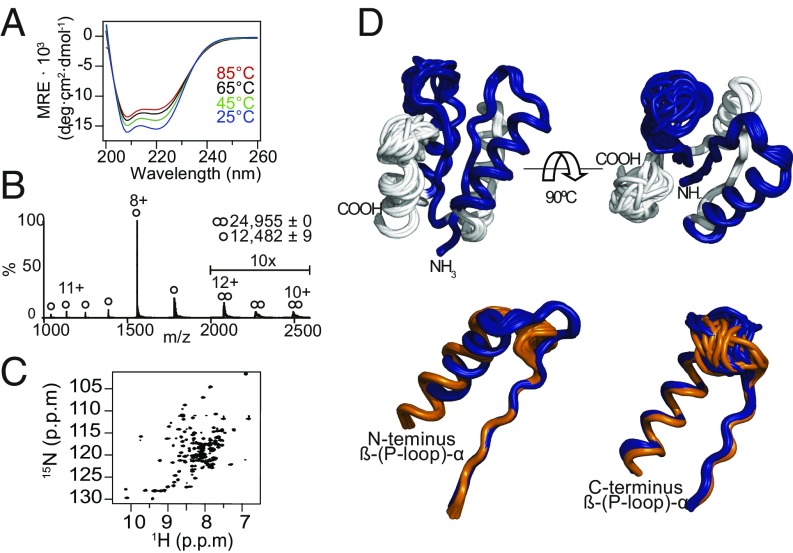
All six designs were expressed in soluble form and readily purified (SI Appendix, Material and Methods) but copurified with nucleic acids, which were removed by treatment with DNase or by additional chromatography steps. By circular dichroism (CD), A- to E-PLoops displayed characteristics of β-α proteins, as designed. However, the F-PLoop exhibited random coil features, in agreement with the difficulties to integrate two β-(P-loop)-α segments into fold IV (Fig. 2A and SI Appendix, Fig. S3E). Designs A–D exhibited no significant spectral changes at the highest temperature tested (85 °C), while design E exhibited partial yet reversible denaturation. Although designed as monomers, like the designed ideal folds (15), the A- to D-PLoop designs tended to oligomerize. Dimers were the dominating species in SDS/PAGE (SI Appendix, Fig. S3H); however, dimerization could be the outcome of the denaturing conditions. Native mass spectrometry (native MS) indicated monomer–dimer coexistence for designs A and B. Design C showed weaker dimerization propensity, and D–F were observed as monomers only (Fig. 2B, Table 1, and SI Appendix, Fig. S3F). The higher level of symmetry in the sequence of the PLoop designs compared with the original ideal folds (Table 1) likely promotes dimerization (36–38). Although not intended, dimer formation results in each molecule having four P-loops, enabling avidity to enhance potentially weak interactions (39). As shown below, these designs avidly bind polyvalent phosphate-containing ligands.
Fig. 2.
Structural characteristics of C-PLoop. (A) CD spectra demonstrate high thermostability. (B) Native MS analysis indicating monomers alongside a minor fraction of dimers. Noted are the measured MWs (the expected MW is 12,474 Da). (C) Two-dimensional 1H15N-HSQC spectra at 25 °C. (D) Structural models derived from NMR data. (Upper) The observed major C-PLoop conformation. (Lower) Alignment of the β-(P-loop)-α elements of the two identified conformations (major in blue, PDB ID code 6C2U, and minor in orange, PDB ID code 6C2V). RMSD values: 1.48 Å between the observed major conformation and the designed model, and 2.1 Å for the minor conformation.
All attempts to obtain diffracting crystals of these designed proteins failed. NMR (2D 1H-15N-heteronuclear single quantum coherence, HSQC) indicated structural polymorphism and partial order for all designs (SI Appendix, Fig. S3G), with the exception of the C-PLoop (Fig. 2C). It appears that although folded and stable, both the tertiary and quaternary structures of the PLoop designs are polymorphic. Native MS also indicated the presence of partially folded species (highly ionized species) compared with the better-packed C-PLoop (Fig. 2B and SI Appendix, Fig. S3F). The well-converged structures of C-PLoop, determined by NMR (Fig. 2D), indicated that C-PLoop’s secondary structure and α/β/α sandwich topology were largely as designed. However, two discrete coexisting conformations were identified in the NMR analysis in slow exchange with each other (PDB ID code of the major conformation, 6C2U, and of the minor, 6C2V), one of which significantly deviates from the design. In particular, the glycine-rich P-loops exhibited high flexibility, as anticipated by their solvent exposure and absence of interactions with scaffold. High flexibility was observed in other NTP-binding prototypes of ancient enzymes (7, 9, 40). The high conformational diversity was also reflected in the significantly higher backbone RMSD value of 1.61 Å among the populated conformations for the C-PLoop, compared with 0.53 Å for the ideal fold scaffold 3N3Z on which the C-PLoop was based.
The Designed PLoop Proteins Bind Phosphorylated Nucleoside Ligands
The functional diversity of P-loop NTPases is significant and includes kinases, chaperones, helicases, transporters, and other motor proteins. Nonetheless, conversion of NTP to NDP or NMP is common to all these enzymes (41, 42). Numerous Escherichia coli proteins can hydrolyze NTPs via phosphatase activity. We thus focused on the ligand-binding potential of the designed PLoop proteins, because binding is a stoichiometric event that can be unambiguously assigned to the designed protein without concerns for contaminating activities. Binding of phosphate and phosphate-containing ligands is a widespread feature of modern proteins (43–45) and presumably one of the elementary functions that linked RNA and ribunucleoside cofactors to the earliest proteins (10, 46–50). Furthermore, during purification, it became apparent that the PLoop designs bind nucleic acids and interact with triphosphate and hexametaphosphate (SI Appendix, Fig. S3I). We thus tested RNA, DNA, and ATP binding by applying different assays with immobilized and soluble ligands.
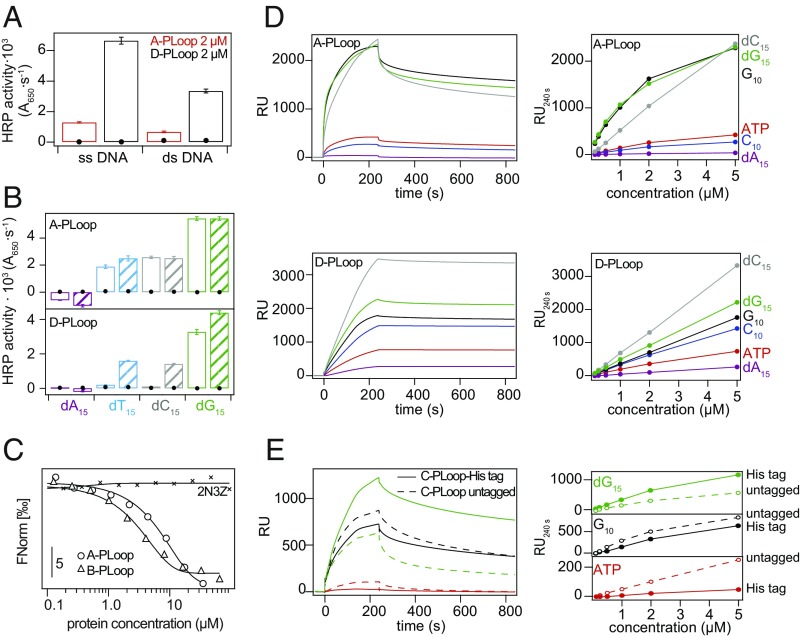
An ELISA was applied using immobilized single-strand DNA (ssDNA) and double-strand DNA (dsDNA) and detection via the designs’ His-tag. PLoops A, B, D, and E, exhibited binding to DNA at protein concentrations as low as 0.2 µM, while the ideal folds’ scaffolds showed no binding up to 5 µM protein. Binding to ssDNA was much stronger than to dsDNA (Fig. 3A and SI Appendix, Fig. S4A). Using ssDNA homo-oligomers, we observed the strongest binding to dG15, followed by dC15 and dT15, with no binding to dA15 (Fig. 3B and SI Appendix, Fig. S4B). Designs C and F failed to bind any of the tested ligands. In the case of the C-PLoop, the His-tag is likely sequestered (discussed below) while for the F-PLoop, the high degree of disorder is the likely reason.
Fig. 3.
Polynucleotide and ATP binding properties of A- to D-PLoops. (A) Binding to 287-nt ssDNA and the corresponding dsDNA, determined by ELISA. Black circles mark the 2N3Z control that showed no binding. (B) ELISA at 0.2 μM (plain bars) and 1 μM (striated bars) protein concentrations with immobilized homomeric ssDNA (black circles mark the 2N3Z control). (C) MST profiles with fluorescently labeled dC15. The lines designate a fit to a sigmoidal binding curve. (D) SPR sensograms at 5-μM protein concentration on ssDNA, RNA, and ATP. (Right) Maximal RU values at different protein concentrations. (E) Effect of the His-tag on C-PLoop’s binding properties. The His-tag may trigger formation of high oligomeric forms upon binding causing a slow and variable association phase (SI Appendix, Fig. S6). SPR sensograms at 5-μM protein concentration over several immobilized biotinylated ligands. (Right) Maximal RU values at different protein concentration.
Similar binding patterns were observed by surface plasmon resonance (SPR), thus eliminating the need for antibody binding to the His-tag epitopes and also demonstrating binding to RNA (Fig. 3D and SI Appendix, Fig. S4C). The C-PLoop exhibited binding by SPR (Fig. 3D and SI Appendix, Fig. S4C), suggesting that inaccessibility of the His-tag hindered its ELISA signal. Binding to ATP was also detected by SPR using a biotinylated analog (Fig. 3D and SI Appendix, Fig. S4C). The SPR binding kinetics were highly complex with multiple association phases and partial dissociation within the experimental time-scale, suggestive of multiple conformations with different affinities, and structural rearrangements induced upon binding (supported by fitting of individual phases) (SI Appendix, Fig. S4 D–N and Tables S2–S5). In most likelihood, initial fast binding of monomeric and dimeric forms is followed by conformational rearrangements and oligomerization, resulting in very slow dissociation.
Immobilization of ligands, as applied for ELISA and SPR, probably increases affinity due to polyvalency, especially given the designs’ oligomerization tendency. Binding to soluble ATP by the C-PLoop was therefore tested by 1H-NMR titrations (SI Appendix, Fig. S5). Additionally, binding of the A- and B-PLoop was established with soluble fluorescently labeled dC15 oligonucleotide using microscale thermofluoresis (MST), indicating binding of the PLoop designs with micromolar affinity and no detectable binding by the ideal fold itself (PBD ID code 2N3Z) (Fig. 3C). We also tested a tag-free version of the C-PLoop. This construct exhibited binding and, as indicated by SPR, with distinctly faster dissociation rates (Fig. 3E), suggesting that the His-tag promotes higher oligomeric forms. A lower occurrence of dimers was also observed by native MS; nonetheless, when mixing the tagged and untagged variants at a 1:1 ratio, mixed dimers were observed implying similar structures for both constructs (SI Appendix, Fig. S6). Taking these data together, we find that while the His-tag might promote higher-order quaternary structures, phosponucleoside binding occurs independently of its presence.
Binding Is Magnesium-Ion Independent and Does Not Require Overall Positive Charge
In contemporary NTPases, the P-loop binds the β- and γ-phosphate of the bound NTP (27, 42). However, the phosphate group is also coordinated to a divalent cation, typically magnesium, that is essential for enzymatic function (exceptions are known; e.g., ref. 51). The magnesium ion is coordinated by the hydroxyl of Ser/Thr of the Walker-A motif, and by one or more residues from other parts of the protein (31). None of these additional auxiliary residues are present in our PLoop proteins. Accordingly, EDTA was routinely used in all SPR binding experiments, including when ATP binding was tested, and neither magnesium ions nor EDTA affected the ELISA signal (SI Appendix, Fig. S7A). Magnesium-independent binding of ATP was observed in segments taken from extant P-loop NTPases (6, 7) and in other NTP-binding prototypes (9); however, binding occurred at pH 4, where the phosphate’s negative charge is reduced by protonation. In contrast, we observed ATP binding at pH 7.4.
Overall, the data from ELISA, SPR, and MST indicate Kd values for phosponucleoside ligands in the low micromolar range, while no binding was observed with 2N3Z, the designed ideal scaffold that does not contain the P-loop motif. Notably, binding of the PLoop designs occurred despite negative surface charge. Designs A-, C-, and D-PLoop had a high positive pI (9.5–9.7). However, design B was closer to neutrality (pI = 8) and design E-PLoop was acidic (pI = 4.4). Binding of the latter two was distinctly weaker in ELISA tests; however, the SPR signals were well above background (no protein or 2N3Z) and only few-fold weaker than for A- and D-PLoops (Table 1 and SI Appendix, Fig. S4). This suggests that binding was not primarily driven by nonspecific electrostatic interactions.
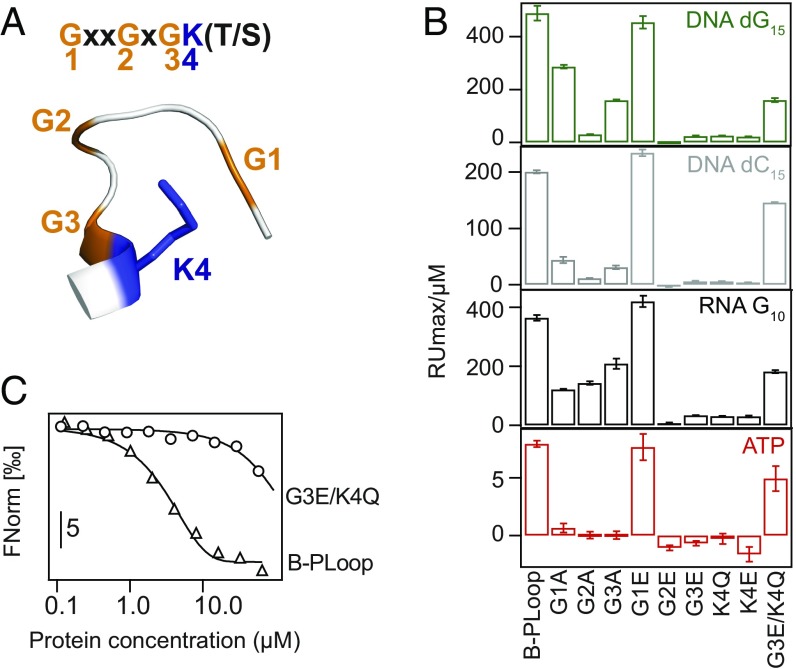
Binding Involves the Key P-Loop Residues
Next we sought to confirm that the P-loop is the key mediator of binding. A set of mutants of the P-loop’s most conserved residues was generated. Mutations of the P-loop’s glycines to alanine were examined (mutated residues numbered as G1xxG2xG3K4[S/T]) (Fig. 4A). However, alanine mutants can retain function in P-loop NTPases (52), so the potentially more perturbing mutations to glutamic acid were also examined. The lysine was mutated to both glutamate and glutamine, as the latter was also reported to diminish ATPase activity (53). Finally, a double mutant of the third glycine and the lysine (G3E/K4Q) was tested. All these mutants expressed well and their CD spectra suggested unperturbed secondary structure (SI Appendix, Fig. S7B). However, nearly all mutations significantly reduced binding, both in SPR and ELISA; as expected, mutations to Glu had a generally larger impact compared with Ala mutations (Fig. 4B and SI Appendix, Fig. S7C). Notably, binding did not decrease when the first glycine was mutated, not even to glutamic acid. Indeed, the second and third glycines are considered the primary requisite for the P-loop’s phosphate nest binding mode (30), and in modern P-loop NTPases, the first glycine rarely plays a direct role in phosphate binding (SI Appendix, Fig. S1D). A marked decrease in binding of the double G3E/K4Q mutant to soluble ssDNA was also observed (Fig. 4C). However, some P-loop mutants retained considerable poly-dG and poly-G binding (Fig. 4B). This suggests that in addition to the phosphate groups, the bases, especially guanine, may contribute to binding. However, what makes poly-G a preferred ligand remains unclear at this point; guanine is the most hydrophilic base, and the stacking potential of adenine (weakest binding) is higher (54).
Fig. 4.
P-loop residues mediate ligand binding. (A) Schematic representation of the Walker-A motif. (B) Binding of B-PLoop and its mutants to ssDNA, RNA, and ATP, assayed by SPR. Shown are the initial slopes derived from plots of maximal RU versus protein concentration (as in Fig. 3D). (C) MST profiles of the titration of dC15 with B-PLoop and its double mutant. The lines represent a fit to sigmoidal binding curves.
The C-PLoop Design Appears to Promote ATP Hydrolysis
We observed binding to a variety of phosphate-containing ligands, not only ATP but also RNA and ssDNA, and sought to verify that the phosphate group of these ligands is directly involved in binding. To this end, we monitored changes in the 31P-NMR spectrum of ATP upon addition of the C-PLoop (for 1H-NMR titrations, see SI Appendix, Fig. S10). Upon addition of the C-PLoop, ATP’s γ- and β-phosphates exhibited only minor shifts. However, over time a peak corresponding to free phosphate and two peaks corresponding to ADP appeared in the spectra. Furthermore, when ATP was incubated with the C-PLoop–G3E/K4Q mutant, ATP remained stable (Fig. 5A). The C-PLoop’s detected activity, although faster than the spontaneous ATP hydrolysis (55), was extremely slow—approximately one ATP hydrolyzed per protein molecule per 30 min—and even a minuscule contamination of an E. coli enzyme could account for such low activity. However, the same level of ATP hydrolysis was retained upon two further purification steps, while the C-PLoop–G3E/K4Q expressed and purified using the very same protocol still showed no hydrolysis (Fig. 5A).
Fig. 5.
C-PLoop mediates ATP hydrolysis. (A) 31P-NMR of 0.5 mM ATP in 20 mM Mes, 50 mM NaCl, pH 6 at room temperature (Top); the same solution with 1 mM C-PLoop (Middle) or C-PLoop-G3E/K4Q (Bottom). The C-PLoop constructs were purified by His-tag chromatography, anion exchange, and gel filtration. (B) The rates of ATP, ADP, and AMP hydrolysis upon incubation with 62.5 μM C-PLoop or C-PLoop–G3E/K4Q, or with no protein. Rates were measured at at 45 °C, pH 6.0; the lines represent the fit to a single exponential and the apparent rate constants are annotated. (C) Addition of either 1 mM MgCl2 or of 1 mM EDTA did not affect ATP hydrolysis by C-PLoop (rates measured at 45 °C, pH 6.0, after 240 min, with 80 μM protein). (D) SPR sensograms of two C-PLoop preparations (with immobilized dG15, as in Fig. 3D). Shown in green are injections of 0.125, 0.25, 0.5, 1, 2, and 5 μM protein. The inactive C-PLoop–G3E/K4Q mutant at 5 μM is shown in red (the close to baseline traces indicate no binding).
ATP hydrolysis in the presence of the C-PLoop showed distinct characteristics. The nucleoside had no significant effect, as different NTPs and dNTPs were hydrolyzed at very similar rates (SI Appendix, Fig. S8A). Furthermore, the C-PLoop also hydrolyzed ADP and AMP, yet at increasingly slower rates (Fig. 5B). The most distinctive characteristic was the lack of dependence on magnesium or other divalent ions. Addition of either Mg2+ or EDTA had no effect on ATP hydrolysis by the C-PLoop (Fig. 5C). This result matches the observed Mg2+ independency of binding of ATP and other phosphate ligands (see above).
Upon multiple repeated attempts to reproduce the ATP hydrolysis activity, we noticed that, in general, the C-PLoop preparations at the University of Washington consistently showed ATP hydrolysis at the same magnitude (including when produced there by M.L.R.R.) while at the Weizmann Institute, preparations exhibited low or no activity. We suspected that this variability could relate to structural polymorphism, including the variability in oligomerization states. Indeed, the ATP hydrolyzing C-PLoop samples were predominantly monomeric, as indicated by 2D 1H15N-HSQC spectra (Fig. 2), and showed a profoundly different ssDNA binding profile (very fast dissociation with overall weak binding). In contrast, samples that exhibited weak or no ATP hydrolysis exhibited very slow dissociation and high-affinity ssDNA binding (SI Appendix, Fig. S9 A and B). Furthermore, mutating the key P-loop residues resulted in a parallel loss of ssDNA binding (Fig. 5D) and of the ATPase activity (SI Appendix, Fig. S9A). By native MS, alongside monomers, dimers and a subpopulation of partially unfolded states (as can be seen by the broad distribution of charge states at the lower m/z) could be observed in the C-PLoop preparation that failed to hydrolyze ATP, while in the active sample the C-PLoop sample appeared solely as monomers (SI Appendix, Fig. S9C). Overall, the C-PLoop can be trapped in different structural forms that exhibit distinctly different binding and ATP hydrolysis patterns. However, their structure’s characteristic and what triggers one from the other remains unknown to us. Similarly, the presence of a contaminating enzyme, although unlikely, cannot be completely ruled out at this stage (we ordered, for example, a synthetic protein but obtained a heterogeneous sample from which we could not purify a peptide corresponding to the C-PLoop). Nonetheless, taken together, our results unambiguously indicate magnesium-independent NTP binding, and also suggest that the P-loop Walker-A motif grafted in a simple context may also promote NTP hydrolysis.
The β-(P-Loop)-α Segment: A Simpler Precursor
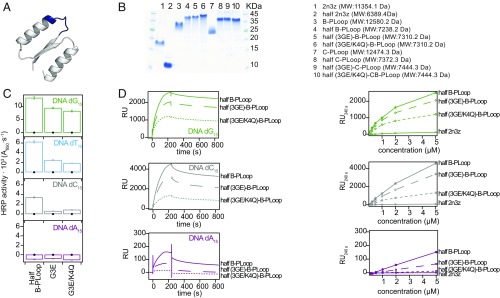
The PLoop prototypes described herein exhibit rudimentary features, foremost simplicity of sequence and structure, and high internal symmetry. The latter suggests the emergence from a shorter peptide via duplication and fusion (1, 37, 56). We thus sought to identify fragments of the PLoop proteins that might be folded and functional via self-assembly. To this end, we examined the N-terminal halves of the B- and C-PLoop: the ancestral β-(P-loop)-α segment followed by just one structural β-α segment borrowed from the designed ideal fold (55 aa in total) (Fig. 6A). The N-terminal half of the initial scaffold, 2N3Z, was constructed as control.
Fig. 6.
Half–B-PLoop and its mutants. (A) Structural cartoon of the 55-aa half–B-PLoop (grafted from the Rosetta design model of the intact protein). The P-loop Walker-A motif is in blue. (B) SDS/PAGE of intact 2N3Z, B- and C-PLoop, and of their corresponding halves. (C) ELISA at 0.1-μM protein concentration with ssDNA (black circles mark the half-2N3Z control). (D) SPR sensograms of half–B-PLoop and it mutants at 5-μM protein concentration on ssDNA. (Right) Maximal RU values at different protein concentrations, including of the control half-2N3Z.
The half–B- and half–C-PLoop were likely isolated as tetramers as judged by SDS/PAGE, while half-2N3Z remained a monomer (Fig. 6B) [molecular weights (MWs) were confirmed by MALDI-TOFF mass spectrometry] (SI Appendix, Fig. S10 A–G). Although the half–B- and half–C-PLoop expressed as soluble proteins, they precipitated following purification and had to be stored in 1 M arginine. Upon dilution from these storage solutions, the half–B-PLoop showed the same binding pattern as the intact B-PLoop: stronger binding to dG15, followed by dC15 and dT15, and no binding to dA15 (Figs. 5D and 6C). Furthermore, as observed in the intact B-PLoop (Fig. 4), binding was significantly reduced in the single (G3E) and double P-loop mutant (G3E/K4Q) (Fig. 6 C and D) and no binding was detected with the control half-2N3Z. However, the half–C-PLoop exhibited weak, nonspecific binding ssDNA (SI Appendix, Fig. S10H).
Concluding Remarks
Our results indicate that the P-loop Walker-A motif can exhibit distinct and potentially beneficial biochemical function on its own with no auxiliary residues and in a structural context far simpler than today’s P-loop NTPases. Whether these P-loop prototypes bear resemblance to the historical prelast universal common ancestor P-loop NTPase ancestors is a scientifically irrelevant question because there is currently no way of addressing it. However, our results confirm that the P-loop Walker-A motif can confer relevant biochemical functions in a context much simpler than todays’ proteins: in proteins comprising 55 residues and composed almost exclusively of abiotic amino acids, and in the absence of other functional motifs and an active-site pocket.
Our work follows previous descriptions of relatively short segments that recapitulate functional elements of modern proteins (6–12). We observed two notable differences between our P-loop prototype proteins and modern P-loop NTPases. First, as observed with other prototypes (6, 7, 9), while in modern NTPases magnesium ions are essential, our PLoop prototypes avidly bind phosphate-containing ligands, including ATP, without magnesium. Second, while binding and hydrolysis of phosphorylated nucleosides (NTPs) is the hallmark of modern P-loop NTPases, we have uniquely observed that the P-loop prototypes not only interact with NTPs, but also and foremost, avidly bind RNA and ssDNA. This raises the tantalizing possibility that early P-loop proteins emerged in a context of polynucleotide binding, and RNA in particular (57). Although some potential to hydrolyze NTP might be attributed to our P-loop prototypes, the far more efficient enzymatic NTPase functions we see today were likely acquired at a later stage when higher sequence and structural complexity evolved, including the acquisition of a magnesium-binding site and an active-site cavity. In accordance with the functional differences and lack of magnesium coordination, the C-PLoop’s NMR structure indicates a conformation that differs from the P-loop of today’s NTPases. However, ligand binding is likely inducing structural rearrangements of the P-loops themselves as well as of the scaffold (58). The backbone differences between the two coexisting C-PLoop conformations and the observation of dimers in native MS both indicate structural polymorphism. The complex binding kinetics with polyvalent ligands also suggest conformational changes, including oligomerization, upon binding. Finally, we observed two C-PLoop forms that, although soluble, do not readily interchange and exhibit distinctly different ssDNA binding and ATP hydrolysis.
Structural polymorphism and self-assembly may enable the P-loop prototypes to exert avid phosphor ligands binding despite their simplicity, in the absence of magnesium, and despite a configuration that differs from contemporary enzymes. Emergence of large, complex enzymes from a simple beginning is also supported by the observation of a 55-aa fragment that comprises the β-(P-loop)-α segment followed by just one additional β-α segment. Self-assembly, possibly also via hetero-oligomerization (4), and the resulting avidity because of multiple P-loops, could enable polyphosphate-ligand binding in this rudimentary context. The bases seem to provide additional weak interactions that jointly result in avid binding to polynucleotides. Further research may reveal how these simple P-loop prototypes exert function, and whether more complex forms with higher binding affinity and specificity, and higher ATPase activity, could be constructed.
Materials and Methods
The β-(P-loop)-α inferred sequence was incorporated into ideal folds II or IV, by replacing the original first and third β-α segments, and the sequence of the resulting chimeric proteins was optimized with RosettaRemodel. The designed proteins were expressed in E. coli, purified via a C-terminal 6His-tag, followed by ion-exchange chromatography (exceptions are specified) and structurally characterized by NMR, CD, and native MS. Their functionality was examined with a range of binding assays, including ELISA and SPR (using biotinylated ligands such as ssDNA, RNA, or ATP, immobilized to streptavidin-coated surfaces), MST (with fluorescently labeled ssDNA), and proton NMR (with intact ATP), and enzymatic assays using thin-layer chromatography (detection by UV absorbance of NTPs and their hydrolysis products) and P31 NMR. The N-terminal halves of the B- and C-PLoop were expressed and purified as above, and characterized by MALDI-TOFF MS, ELISA, and SPR. Further details are provided in SI Appendix, Material and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Irina Shin, Dr. Aharon Rabinkov, and Dr. Yael Fridmann Sirkis for assistance with SPR and MST instrumentation; and Dr. Mark Karpasas for assistance with MALDI-TOFF MS experiments. Funding by the Israel Science Foundation (Grant 980/14) and the Sasson & Marjorie Peress Philanthropic Fund are gratefully acknowledged. D.S.T. is the Nella and Leo Benoziyo Professor of Biochemistry. M.L.R.R. received support from the Koshland Foundation, a McDonald-Leapman grant, and the Ramon Areces Foundation. Work in the G.V. group was supported by the NSF and the NIH (Grants R35 GM126942 and RO1 GM103834).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6C2U and 6C2V).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812400115/-/DCSupplemental.
References
- 1.Eck RV, Dayhoff MO. Evolution of the structure of ferredoxin based on living relics of primitive amino acid sequences. Science. 1966;152:363–366. doi: 10.1126/science.152.3720.363. [DOI] [PubMed] [Google Scholar]
- 2.Söding J, Lupas AN. More than the sum of their parts: On the evolution of proteins from peptides. BioEssays. 2003;25:837–846. doi: 10.1002/bies.10321. [DOI] [PubMed] [Google Scholar]
- 3.Romero Romero ML, Rabin A, Tawfik DS. Functional proteins from short peptides: Dayhoff’s hypothesis turns 50. Angew Chem Int Ed Engl. 2016;55:15966–15971. doi: 10.1002/anie.201609977. [DOI] [PubMed] [Google Scholar]
- 4.Setiyaputra S, Mackay JP, Patrick WM. The structure of a truncated phosphoribosylanthranilate isomerase suggests a unified model for evolution of the (βα)8 barrel fold. J Mol Biol. 2011;408:291–303. doi: 10.1016/j.jmb.2011.02.048. [DOI] [PubMed] [Google Scholar]
- 5.Cronet P, Bellsolell L, Sander C, Coll M, Serrano L. Investigating the structural determinants of the p21-like triphosphate and Mg2+ binding site. J Mol Biol. 1995;249:654–664. doi: 10.1006/jmbi.1995.0326. [DOI] [PubMed] [Google Scholar]
- 6.Chuang WJ, Abeygunawardana C, Gittis AG, Pedersen PL, Mildvan AS. Solution structure and function in trifluoroethanol of PP-50, an ATP-binding peptide from F1ATPase. Arch Biochem Biophys. 1995;319:110–122. doi: 10.1006/abbi.1995.1272. [DOI] [PubMed] [Google Scholar]
- 7.Chuang WJ, Abeygunawardana C, Pedersen PL, Mildvan AS. Two-dimensional NMR, circular dichroism, and fluorescence studies of PP-50, a synthetic ATP-binding peptide from the beta-subunit of mitochondrial ATP synthase. Biochemistry. 1992;31:7915–7921. doi: 10.1021/bi00149a024. [DOI] [PubMed] [Google Scholar]
- 8.Fry DC, Kuby SA, Mildvan AS. NMR studies of the MgATP binding site of adenylate kinase and of a 45-residue peptide fragment of the enzyme. Biochemistry. 1985;24:4680–4694. doi: 10.1021/bi00338a030. [DOI] [PubMed] [Google Scholar]
- 9.Mullen GP, Vaughn JB, Jr, Mildvan AS. Sequential proton NMR resonance assignments, circular dichroism, and structural properties of a 50-residue substrate-binding peptide from DNA polymerase I. Arch Biochem Biophys. 1993;301:174–183. doi: 10.1006/abbi.1993.1130. [DOI] [PubMed] [Google Scholar]
- 10.Carter CW., Jr Urzymology: Experimental access to a key transition in the appearance of enzymes. J Biol Chem. 2014;289:30213–30220. doi: 10.1074/jbc.R114.567495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Rodriguez L, et al. Functional class I and II amino acid-activating enzymes can be coded by opposite strands of the same gene. J Biol Chem. 2015;290:19710–19725. doi: 10.1074/jbc.M115.642876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pham Y, et al. A minimal TrpRS catalytic domain supports sense/antisense ancestry of class I and II aminoacyl-tRNA synthetases. Mol Cell. 2007;25:851–862. doi: 10.1016/j.molcel.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 13.Li L, Weinreb V, Francklyn C, Carter CW., Jr Histidyl-tRNA synthetase urzymes: Class I and II aminoacyl tRNA synthetase urzymes have comparable catalytic activities for cognate amino acid activation. J Biol Chem. 2011;286:10387–10395. doi: 10.1074/jbc.M110.198929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Francklyn C, Carter CW., Jr Aminoacylating urzymes challenge the RNA world hypothesis. J Biol Chem. 2013;288:26856–26863. doi: 10.1074/jbc.M113.496125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koga N, et al. Principles for designing ideal protein structures. Nature. 2012;491:222–227. doi: 10.1038/nature11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhlman B, et al. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 17.Walsh ST, Cheng H, Bryson JW, Roder H, DeGrado WF. Solution structure and dynamics of a de novo designed three-helix bundle protein. Proc Natl Acad Sci USA. 1999;96:5486–5491. doi: 10.1073/pnas.96.10.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dahiyat BI, Mayo SL. De novo protein design: Fully automated sequence selection. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 19.Huang PS, Boyken SE, Baker D. The coming of age of de novo protein design. Nature. 2016;537:320–327. doi: 10.1038/nature19946. [DOI] [PubMed] [Google Scholar]
- 20.Zheng Z, Goncearenco A, Berezovsky IN. Nucleotide binding database NBDB—A collection of sequence motifs with specific protein-ligand interactions. Nucleic Acids Res. 2016;44:D301–D307. doi: 10.1093/nar/gkv1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alva V, Söding J, Lupas AN. A vocabulary of ancient peptides at the origin of folded proteins. eLife. 2015;4:e09410. doi: 10.7554/eLife.09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laurino P, et al. An ancient fingerprint indicates the common ancestry of Rossmann-fold enzymes utilizing different ribose-based cofactors. PLoS Biol. 2016;14:e1002396. doi: 10.1371/journal.pbio.1002396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bork P, Koonin EV. A P-loop-like motif in a widespread ATP pyrophosphatase domain: Implications for the evolution of sequence motifs and enzyme activity. Proteins. 1994;20:347–355. doi: 10.1002/prot.340200407. [DOI] [PubMed] [Google Scholar]
- 24.Harris JK, Kelley ST, Spiegelman GB, Pace NR. The genetic core of the universal ancestor. Genome Res. 2003;13:407–412. doi: 10.1101/gr.652803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koonin EV. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol. 2003;1:127–136. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- 26.Aravind L, Anantharaman V, Koonin EV. Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: Implications for protein evolution in the RNA. Proteins. 2002;48:1–14. doi: 10.1002/prot.10064. [DOI] [PubMed] [Google Scholar]
- 27.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma BG, et al. Characters of very ancient proteins. Biochem Biophys Res Commun. 2008;366:607–611. doi: 10.1016/j.bbrc.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 29.Koonin EV, Wolf YI, Aravind L. Protein fold recognition using sequence profiles and its application in structural genomics. Adv Protein Chem. 2000;54:245–275. doi: 10.1016/s0065-3233(00)54008-x. [DOI] [PubMed] [Google Scholar]
- 30.Bianchi A, Giorgi C, Ruzza P, Toniolo C, Milner-White EJ. A synthetic hexapeptide designed to resemble a proteinaceous P-loop nest is shown to bind inorganic phosphate. Proteins. 2012;80:1418–1424. doi: 10.1002/prot.24038. [DOI] [PubMed] [Google Scholar]
- 31.Frasch WD. The participation of metals in the mechanism of the F(1)-ATPase. Biochim Biophys Acta. 2000;1458:310–325. doi: 10.1016/s0005-2728(00)00083-9. [DOI] [PubMed] [Google Scholar]
- 32.Goncearenco A, Berezovsky IN. Prototypes of elementary functional loops unravel evolutionary connections between protein functions. Bioinformatics. 2010;26:i497–i503. doi: 10.1093/bioinformatics/btq374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Z, Kumar S, Nei M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics. 1995;141:1641–1650. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Longo LM, Blaber M. Protein design at the interface of the pre-biotic and biotic worlds. Arch Biochem Biophys. 2012;526:16–21. doi: 10.1016/j.abb.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 35.Wellner A. 2013. Mechanisms of protein sequence divergence and incompatibility. PhD dissertation (Weizmann Institute of Science, Rehovot, Israel)
- 36.Levy ED, Teichmann S. Structural, evolutionary, and assembly principles of protein oligomerization. Prog Mol Biol Transl Sci. 2013;117:25–51. doi: 10.1016/B978-0-12-386931-9.00002-7. [DOI] [PubMed] [Google Scholar]
- 37.Smock RG, Yadid I, Dym O, Clarke J, Tawfik DS. De novo evolutionary emergence of a symmetrical protein is shaped by folding constraints. Cell. 2016;164:476–486. doi: 10.1016/j.cell.2015.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia-Seisdedos H, Empereur-Mot C, Elad N, Levy ED. Proteins evolve on the edge of supramolecular self-assembly. Nature. 2017;548:244–247. doi: 10.1038/nature23320. [DOI] [PubMed] [Google Scholar]
- 39.Rufo CM, et al. Short peptides self-assemble to produce catalytic amyloids. Nat Chem. 2014;6:303–309. doi: 10.1038/nchem.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sapienza PJ, Li L, Williams T, Lee AL, Carter CW., Jr An ancestral tryptophanyl-tRNA synthetase precursor achieves high catalytic rate enhancement without ordered ground-state tertiary structures. ACS Chem Biol. 2016;11:1661–1668. doi: 10.1021/acschembio.5b01011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramakrishnan C, Dani VS, Ramasarma T. A conformational analysis of Walker motif A [GXXXXGKT (S)] in nucleotide-binding and other proteins. Protein Eng. 2002;15:783–798. doi: 10.1093/protein/15.10.783. [DOI] [PubMed] [Google Scholar]
- 42.Saraste M, Sibbald PR, Wittinghofer A. The P-loop—A common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 43.Hirsch AK, Fischer FR, Diederich F. Phosphate recognition in structural biology. Angew Chem Int Ed Engl. 2007;46:338–352. doi: 10.1002/anie.200603420. [DOI] [PubMed] [Google Scholar]
- 44.Parca L, Mangone I, Gherardini PF, Ausiello G, Helmer-Citterich M. Phosfinder: A web server for the identification of phosphate-binding sites on protein structures. Nucleic Acids Res. 2011;39:W278–W282. doi: 10.1093/nar/gkr389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parca L, Gherardini PF, Helmer-Citterich M, Ausiello G. Phosphate binding sites identification in protein structures. Nucleic Acids Res. 2011;39:1231–1242. doi: 10.1093/nar/gkq987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goncearenco A, Berezovsky IN. Protein function from its emergence to diversity in contemporary proteins. Phys Biol. 2015;12:045002. doi: 10.1088/1478-3975/12/4/045002. [DOI] [PubMed] [Google Scholar]
- 47.Gray MJ, et al. Polyphosphate is a primordial chaperone. Mol Cell. 2014;53:689–699. doi: 10.1016/j.molcel.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carter CW, Jr, et al. The Rodin-Ohno hypothesis that two enzyme superfamilies descended from one ancestral gene: An unlikely scenario for the origins of translation that will not be dismissed. Biol Direct. 2014;9:11. doi: 10.1186/1745-6150-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cammer S, Carter CW., Jr Six Rossmannoid folds, including the class I aminoacyl-tRNA synthetases, share a partial core with the anti-codon-binding domain of a class II aminoacyl-tRNA synthetase. Bioinformatics. 2010;26:709–714. doi: 10.1093/bioinformatics/btq039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carter CW. What RNA world? Why a peptide/RNA partnership merits renewed experimental attention. Life (Basel) 2015;5:294–320. doi: 10.3390/life5010294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinreb V, Carter CW., Jr Mg2+-free Bacillus stearothermophilus tryptophanyl-tRNA synthetase retains a major fraction of the overall rate enhancement for tryptophan activation. J Am Chem Soc. 2008;130:1488–1494. doi: 10.1021/ja076557x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deyrup AT, Krishnan S, Cockburn BN, Schwartz NB. Deletion and site-directed mutagenesis of the ATP-binding motif (P-loop) in the bifunctional murine ATP-sulfurylase/adenosine 5′-phosphosulfate kinase enzyme. J Biol Chem. 1998;273:9450–9456. doi: 10.1074/jbc.273.16.9450. [DOI] [PubMed] [Google Scholar]
- 53.Korangy F, Julin DA. Enzymatic effects of a lysine-to-glutamine mutation in the ATP-binding consensus sequence in the RecD subunit of the RecBCD enzyme from Escherichia coli. J Biol Chem. 1992;267:1733–1740. [PubMed] [Google Scholar]
- 54.Guckian KM, et al. Factors contributing to aromatic stacking in water: Evaluation in the context of DNA. J Am Chem Soc. 2000;122:2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stockbridge RB, Wolfenden R. The intrinsic reactivity of ATP and the catalytic proficiencies of kinases acting on glucose, N-acetylgalactosamine, and homoserine: A thermodynamic analysis. J Biol Chem. 2009;284:22747–22757. doi: 10.1074/jbc.M109.017806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Höcker B. Design of proteins from smaller fragments-learning from evolution. Curr Opin Struct Biol. 2014;27:56–62. doi: 10.1016/j.sbi.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 57.Kovacs NA, Petrov AS, Lanier KA, Williams LD. Frozen in time: The history of proteins. Mol Biol Evol. 2017;34:1252–1260. doi: 10.1093/molbev/msx086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clarke J, Pappu RV. Editorial overview: Protein folding and binding, complexity comes of age. Curr Opin Struct Biol. 2017;42:v–vii. doi: 10.1016/j.sbi.2017.03.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.