Significance
We developed an approach for performing multicolor superresolution imaging by modulating excitation lasers at different frequencies, which we call frequency multiplexing (fm). We implemented this approach in two single-molecule localization microscopy modalities: fm-DNA-PAINT and fm-STORM. We show that fm-DNA-PAINT acquires multiple colors in the same time as single-color DNA-PAINT, dramatically improving image acquisition time without compromising the field of view or signal throughput. DNA-PAINT has several advantages over other single-molecule superresolution methods as it decouples fluorophore photophysics from photoswitching, is quantitative, and does not suffer from photobleaching. However, one major disadvantage of DNA-PAINT is that it is very time-consuming, making it difficult to multiplex. fm-DNA-PAINT overcomes this main limitation, making it amenable to multiplexed, high-throughput imaging.
Keywords: superresolution microscopy, DNA-PAINT, STORM, frequency multiplexing, multicolor imaging
Abstract
Recent advancements in single-molecule-based superresolution microscopy have made it possible to visualize biological structures with unprecedented spatial resolution. Determining the spatial coorganization of these structures within cells under physiological and pathological conditions is an important biological goal. This goal has been stymied by the current limitations of carrying out superresolution microscopy in multiple colors. Here, we develop an approach for simultaneous multicolor superresolution imaging which relies solely on fluorophore excitation, rather than fluorescence emission properties. By modulating the intensity of the excitation lasers at different frequencies, we show that the color channel can be determined based on the fluorophore’s response to the modulated excitation. We use this frequency multiplexing to reduce the image acquisition time of multicolor superresolution DNA-PAINT while maintaining all its advantages: minimal color cross-talk, minimal photobleaching, maximal signal throughput, ability to maintain the fluorophore density per imaged color, and ability to use the full camera field of view. We refer to this imaging modality as “frequency multiplexed DNA-PAINT,” or fm-DNA-PAINT for short. We also show that frequency multiplexing is fully compatible with STORM superresolution imaging, which we term fm-STORM. Unlike fm-DNA-PAINT, fm-STORM is prone to color cross-talk. To overcome this caveat, we further develop a machine-learning algorithm to correct for color cross-talk with more than 95% accuracy, without the need for prior information about the imaged structure.
Single-molecule-based superresolution microscopy is becoming a broadly used technique to investigate a multitude of biological processes with unprecedented spatial resolution. Extending its capabilities to multiple colors is important for determining the relationship among the spatial distribution and subcellular localization of different proteins. However, existing multicolor implementations for single-molecule superresolution microscopy have important caveats, including color cross-talk, unavailability of well-performing spectrally distinct photoswitchable fluorophores, and long acquisition times. One approach for multicolor superresolution microscopy uses fluorophore pairs in which the same reporter is coupled to different activators (1). In this case, the color is determined based on the wavelength of the activation laser. This approach is free from chromatic aberrations since the same reporter dye is always used, and hence it does not require image registration. In addition, the full camera field of view (FOV) is maintained over multiple colors. However, this is a sequential imaging approach, in which the image acquisition time scales with the number of colors needed. Moreover, it is prone to color cross-talk, as fluorophores can undergo spontaneous blinking or be activated by the “wrong” activation laser and thus localized during the wrong activation cycle (2). A second approach is sequential labeling and imaging, which reduces color cross-talk but at the expense of substantial time investment (3, 4). An alternative, third approach, which also reduces color cross-talk, is the use of spectrally distinct photoswitchable reporter dyes (2, 5–8). While the multiple colors can be imaged simultaneously to reduce time investment, it comes at the expense of a reduced FOV since it requires splitting the camera FOV into smaller subregions, one for each color to be detected, hence decreasing experimental throughput. In addition, there is a limited availability of spectrally distinct photoswitchable fluorophores that minimize color cross-talk while simultaneously maintaining favorable photoswitching properties in the same imaging buffer. As a result, sequential imaging approaches are often preferred at the expense of increased image acquisition time.
Fluorescent molecules can also be discriminated based on their emission spectra in a fourth class of multicolor methods that use spectral information (9, 10). In general, these approaches necessitate increasing molecular sparseness to avoid spatiospectral overlapping and in some cases splitting of the camera FOV into subregions, lengthening the acquisition time or reducing the available FOV. Importantly, spectral fluctuations inherent to single molecules (11) can limit the applicability of spectrally resolved superresolution approaches.
More recently, DNA point accumulation in nanoscale topography (PAINT) has emerged as a single-molecule localization microscopy method that uses oligo probes functionalized with spectrally distinct fluorophores. In this approach, freely diffusing labeled oligos (called the “imager” strand) transiently bind to the complementary (unlabeled) docking oligos functionalized to a secondary antibody, producing the required on/off switching for superresolution microscopy (12, 13). DNA-PAINT is quantitative (14) and does not suffer from photobleaching as there is continuous replenishment of imager strands from solution. Moreover, this approach is particularly amenable to multicolor superresolution imaging since the on/off “blinking” solely depends on the oligo binding properties such that conventional fluorophores can be used. However, these advantages come with the major shortcoming of requiring long image acquisition times (tens of minutes up to several hours per color). This drawback arises because the unbound diffusing imager strands contribute to background (15). The image acquisition time is related to the number of detected events per camera frame, which is proportional to the imager strand concentration (15). Since increasing the imager strand concentration leads to increased background, there is a practical limit to how fast images can be acquired with conventional DNA-PAINT (15). Typically, multicolor DNA-PAINT is performed sequentially by using imager strands labeled with the same fluorophore and with the target species labeled with orthogonal docking strands (12). Therefore, the time investment scales linearly with the number of colors in sequential multicolor DNA-PAINT.
All these caveats limit the practical application of multicolor superresolution imaging in biology. To overcome these limitations, we have developed an alternative multicolor superresolution imaging approach that exclusively relies on the excitation properties of fluorophores rather than their emission spectra. The method is based on frequency-encoded multiplexed excitation and color-blind detection and it was originally developed for high-throughput, low-cost DNA sequencing (16). Excitation modulation methods have been long used in conventional fluorescence microscopy and spectroscopy, typically implemented by modulating different excitations lasers and using spectral filter detection together with different detectors, one for each color to be detected (17–19). Recently, we demonstrated that such a modulated excitation scheme can be combined with color-blind detection in confocal mode to provide simultaneous multicolor imaging using a single detector (20). Here, we extended this approach to multicolor single-molecule superresolution imaging. We show that frequency multiplexed (fm) excitation combines the advantages of spectral detection with a simple optical configuration, without compromising the FOV and, importantly, decreasing the image acquisition time for multicolor DNA-PAINT superresolution microscopy.
Results
Optical Configuration and Analysis Concept.
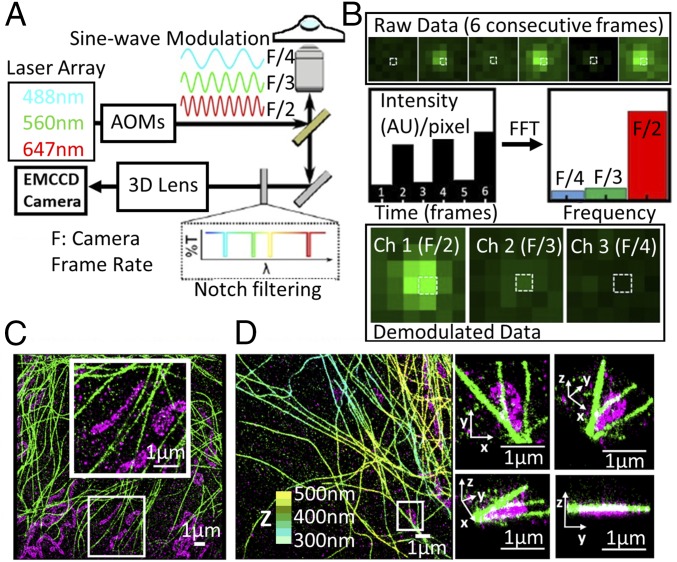
The method relies on the fact that individual fluorophores emit fluorescence directly proportional to their absorption cross-section at a given excitation wavelength. Thus, by modulating multiple excitation lasers at distinct frequencies, fluorophores with different excitation spectra will be excited to a varying degree according to their absorption cross-section at each given excitation wavelength. The total fluorescence emission from different fluorophores is collected on a single detector, in a color-blind fashion, and it is then demodulated using Fourier analysis in the frequency domain to retrieve the magnitude of the individual fluorophore signals for each color channel. For frequency multiplexing, the intensities of all of the excitation lasers were sinewave-modulated independently at their own unique frequencies using acousto-optic modulators (Fig. 1A). A frequency range of 50–10 Hz was used depending on the specific experimental configuration (SI Appendix, SI Methods). The different wavelengths were combined and coupled into the microscope objective through dichroic mirrors. The emitted fluorescence light was collected by the same objective and directed through a set of notch filters to solely reject the laser excitation wavelengths used in the experiment (Fig. 1A). This configuration yielded maximum signal throughput to the electron-multiplying CCD detector.
Fig. 1.
fm-DNA-PAINT concept. (A) Schematic of the microscope setup and imaging method. An example case of three illumination lasers are shown as sinewave-modulated at three different frequencies, F/2, F/3, and F/4, where F is the camera frame rate. (B) Representative example of data processing. (B, Top) SubROI of six consecutive frames with one fluorophore present. (B, Middle) Intensity evolution of the selected pixel (white box) in the time domain and amplitudes in the frequency domain after a FFT over the six frames. (B, Bottom) Resulting demodulated data split into the three different channels. (C) Two-Color, 2D fm-DNA-PAINT image of mitochondria (magenta) and microtubules (green). (D) Two-Color 3D fm-DNA-PAINT image of mitochondria and microtubules. Zooms on the right show 3D views of the white boxed region. Mitochondria are represented in magenta. For the microtubules, the color-coding indicates z-position (from 300 nm in light blue to 500 nm in yellow).
The fluorophores emitted light proportionally to their absorption cross-sections at each excitation wavelength. For demodulation, the intensity evolution of the fluorophore within a given frame window (m), ranging from four to six frames, was transformed to the frequency domain (SI Appendix, SI Methods). For a given frame window, m, the discrete Fourier transform generates m/2 frequency bins and therefore m/2 available color channels. Hence, a six-frame window enables three-color imaging (Fig. 1B). We implemented this fm excitation approach in two different modalities of superresolution microscopy: DNA-PAINT and STORM.
fm-DNA-PAINT.
For fm-DNA-PAINT, the intensity evolution of each pixel in the time domain, corresponding to six consecutive frames acquired at a camera exposure time t = 16 ms per frame (F = 60 Hz), was converted into amplitudes in the frequency domain by performing a fast Fourier transform (FFT). Those values were then assigned to the corresponding pixels on the demodulated images (one value per channel) (Fig. 1B). After demodulation, fluorophores were localized in their corresponding color channel, in which they were already separated spatially and spectrally. The demodulated image stack had an effective exposure time of m*t, with m = 6 and t = 16 ms, fulfilling the long exposure conditions of DNA-PAINT (i.e., ∼100 ms corresponding to an effective frame rate of 10 Hz). DNA-PAINT is particularly amenable to frequency multiplexing, since the fluorophore functionalized oligo stays bound to its complimentary oligo for a few hundred milliseconds. Hence, the bound fluorophore can be detected over multiple frames when imaged at the rate of 16 ms per frame, whereas the diffusing molecules are too dim after demodulation to be localized (SI Appendix, Fig. S1). As a result, the color assignment becomes unambiguous. Accordingly, two-color 2D and two-color 3D images of microtubules and mitochondria, using Cy5-equivalent and Cy3-equivalent as fluorophores, respectively, and imaged with fm-DNA-PAINT (Fig. 1 C and D) produced minimal color cross-talk (2.8% cross-talk from Cy5 into Cy3 channel and 0.8% cross-talk from Cy3 into Cy5 channel; SI Appendix, Fig. S2). If desired, the color cross-talk could be further reduced using a simple correction approach without compromising the overall detected number of localizations (SI Appendix, SI Methods and Fig. S2).
Localization precision is important for the final resolution in single-molecule-based localization microscopy. We thus compared the localization precision of fm-DNA-PAINT to conventional DNA-PAINT using two different methods (21–23) (SI Appendix, SI Methods and Fig. S3). Both methods showed that the localization precision of fm-DNA-PAINT was somewhat decreased (by a factor of ∼1.4–2) compared with conventional DNA-PAINT. This decrease is merely due to the fact that by modulating the excitation lasers, fluorophores are excited only half of the duration of a single frame, and thus emit roughly half of the photons compared with continuous excitation. The modest decrease in localization precision of fm-DNA-PAINT is largely compensated by its gain in image acquisition times (discussed below). Nonetheless, the fm-DNA-PAINT localization precisions were close to the reported values for other single-molecule localization microscopy methods (23–25). The localization precision can be improved by increasing the excitation laser powers so that the effective excitation is similar to that of conventional DNA-PAINT.
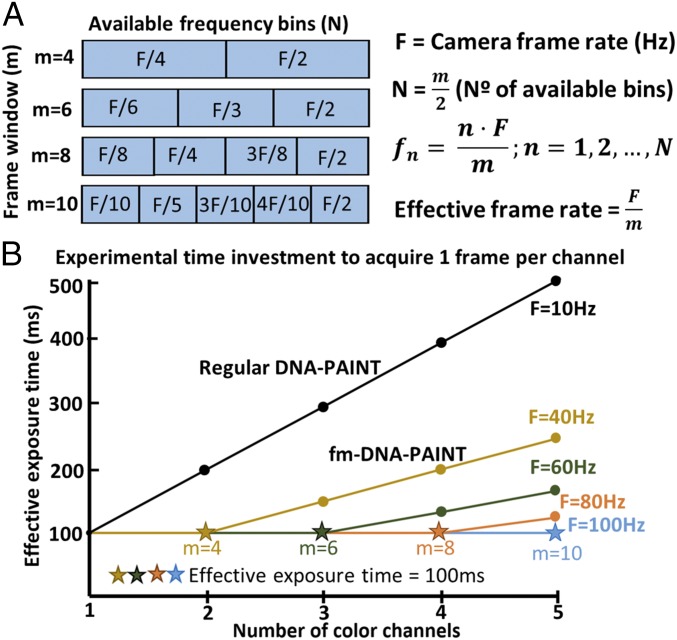
fm-DNA-PAINT is particularly powerful as it is much faster in terms of experimental time investment compared with conventional multicolor DNA-PAINT. Multicolor DNA-PAINT is typically performed sequentially and requires exchanging the imager oligo strands with orthogonal ones (12), which can be cumbersome, introduces drift, and adds additional time to the experiment. In fm-DNA-PAINT, multiple colors are obtained simultaneously and with an effective image acquisition time that is similar to that of one-color DNA-PAINT. There are some practical limits that determine the image acquisition time and the available number of color channels, such as the separation between the centers of the frequency bins used for modulation, the camera frame rate, the signal-to-noise ratio, and the overlap between the excitation spectra of fluorophores. The number of frequency bins fn (and hence the number of color channels) depends on the frame window size used for demodulation (m) (Fig. 2A). For example, a modest estimate of fn = 3 can be fit inside a demodulation frame window of m = six frames (Fig. 2A), which will maintain a good separation between the frequency bins. The effective frame rate depends on the camera frame rate F and the demodulation frame window m (Fig. 2A). For a frame rate of 60 Hz (used here), it is thus possible to image three colors with an effective frame rate of 10 Hz. This frame rate is equivalent to the one typically needed for acquiring a single-color conventional DNA-PAINT image, improving the imaging throughput by threefold (Fig. 2B). Since these acquisition settings do not reach the lower limit of photon collection for high localization precision (SI Appendix, Fig. S3), the number of channels can further be increased, while maintaining the same effective frame rate, by increasing F and m (Fig. 2B).
Fig. 2.
Multicolor fm-DNA-PAINT dramatically improves experimental time investment compared with sequential multicolor DNA-PAINT. (A) Scheme showing the available frequency bins fn (and thus the number of colors) for a given demodulation frame window size m. (B) Effective exposure time versus the number of color channels for different camera frame rates F. The black line shows conventional DNA-PAINT where multicolor imaging is performed sequentially, assuming an exposure time of 100 ms per color (i.e., F = 10 Hz).
The image acquisition time in DNA-PAINT depends on both the camera frame rate and the time needed to collect a sufficient number of localizations to satisfy the Nyquist criterion for high image resolution (26). DNA-PAINT does not suffer from photobleaching as there is continuous replenishment of imager strands from the solution. As a result, the number of localizations per frame is constant and hence the cumulative number of localizations increases linearly with the number of frames acquired. We experimentally verified that the cumulative number of localizations over time was similar for single-color fm-DNA-PAINT (16-ms exposure time, m = 6) and single-color conventional DNA-PAINT (100-ms exposure time) (SI Appendix, Fig. S4). Therefore, no additional time is needed in fm-DNA-PAINT to accumulate an equal number localizations compared with conventional DNA-PAINT.
To assess any potential limits in extending fm-DNA-PAINT to more colors, we first estimated the percentage of spatially overlapping point spread functions (PSFs) as the number of colors is increased. This control is needed since acquiring multiple colors simultaneously on a single detector can lead to crowding and spatial overlap between the fluorophores. Reducing the fluorophore density to minimize spatial overlap would jeopardize the advantages of the method, as it would result in longer image acquisition times to fully reconstruct a superresolution image. As expected, the probability of spatial overlap became significant for more than three colors (SI Appendix, Fig. S5A). To then assess the capability of our method to distinguish spectrally distinct fluorophores having full spatial overlap, we simulated three sinewave-modulated signals and combined them so that they spatially overlap on the same pixel and followed their time evolution (SI Appendix, Fig. S5B). In the frequency domain the amplitudes at the different modulation frequencies are fully distinguished and separated into each different component to retrieve their unique color (SI Appendix, Fig. S5B).
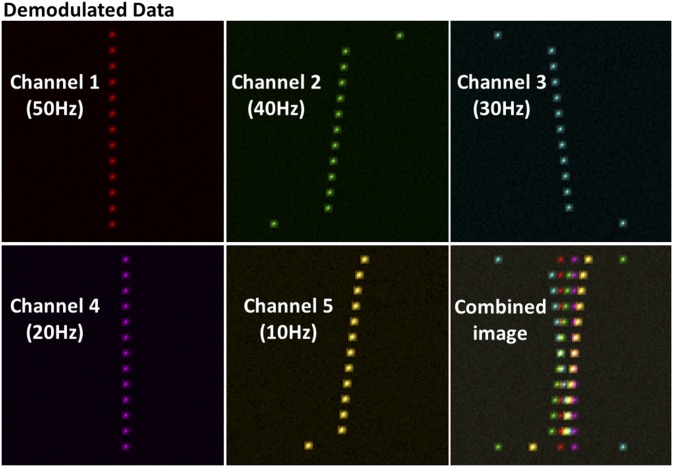
Since currently there are only two different types of commercially available DNA-PAINT antibodies, we generated synthetic data to further determine the ability of fm-DNA-PAINT to acquire more than two colors. The synthetic data were generated by taking as input a five-by-five-pixel subregion of interest (subROI) of a PSF from one frame of the single-color experimental data (SI Appendix, SI Methods). We created multiple PSFs over several consecutive frames to simulate the emission of a fluorophore under sinewave-modulated illumination. We first confirmed that the synthetic data generated for two different fluorophores faithfully represented our experimental data (SI Appendix, Fig. S5C). Having validated our approach, we next generated five-color synthetic data in which all five different fluorophores were spatially mixed together in a color-blind fashion and with spatial overlap (SI Appendix, SI Methods and Fig. S6). The synthetic data were then demodulated and separated into five different channels (Fig. 3). The fluorophores were correctly assigned to the corresponding channels, further demonstrating the capability of the method to separate spectrally distinct fluorophores in the presence of spatial overlap (Fig. 3). Importantly, these results also show that the increased fluorophore density resulting from multiple different fluorophores does not affect color discrimination of fm-DNA-PAINT, thus maintaining its advantages in terms of image acquisition speed.
Fig. 3.
Simulations demonstrate the extendibility of multicolor fm-DNA-PAINT to five-color channels. Demodulated frames for each channel from a five-color synthetic image generated with camera frame rate F = 100 Hz and fi of 50, 40, 30, 20, and 10 Hz, assuming that the fluorophores have minimal spectral overlap (similar to the two-color experimental data with Cy5 and Cy3). The overlapped (combined) image is shown in the lower right.
Despite correct color discrimination, the spatial overlap between fluorophores could still distort the reconstructed PSF on the demodulated data if there is partial spectral overlap between the fluorophores. For instance, in our experiments, Cy5 absorbs ∼10% of the 561-nm laser. This additional amplitude in the “wrong” modulation frequency will perturb the reconstructed PSF, affecting the accuracy of the corresponding localization. To determine the magnitude of this effect, we generated semisynthetic stacks of images taking as input the experimental PSFs obtained from the two-color fluorophores (SI Appendix, SI Methods). In these image stacks, we kept the spatial positions of the PSFs belonging to one fluorophore constant and shifted the PSFs of the second fluorophore (Dshift), allowing for spatial overlap (SI Appendix, Figs. S7 and S8A). The semisynthetic stacks were demodulated and the centers of the PSFs of the shifted fluorophores were localized. We then computed the distances from the localized x and y positions to their actual simulated positions (Drelative) (SI Appendix, Fig. S8A). Drelative was only slightly affected (SI Appendix, Fig. S8 B and C, Insets) with an overall effect for Cy5 of 10.6 nm and 5.9 nm for spatial overlapping and nonoverlapping fluorophores, respectively, and 6.8 nm and 2.1 nm for Cy3. Thus, overall the distortions due to spatial overlap led to a localization error of ∼5 nm for both channels, which was smaller than the average localization precision (SI Appendix, Fig. S3). We also determined if changes in the relative brightness of one fluorophore with respect to the other fluorophore has an impact on the localization error in the presence of spatial overlap (SI Appendix, Fig. S9 A and B). Again, the error in localization was minor compared with spatially nonoverlapping fluorophores for all relative intensity values.
fm-STORM.
STORM is another commonly used single-molecule localization microscopy method, which relies on the use of buffers containing reducing agents and oxygen scavengers to induce photoswitching in organic fluorophores (21). We next set out to implement fm in STORM (fm-STORM) using a four-frame window for the demodulation. In this case, unlike fm-DNA-PAINT, the demodulation was performed after the fluorophores were localized in each frame. We determined the localization precision of fm-STORM from the standard deviation (SD) of the localized positions of the same fluorophore over consecutive frames. The average localization precision was around 18 nm for both fluorophores, close to typical values reported for conventional STORM (24) (SI Appendix, Fig. S10).
The stochastic nature of photoswitching in organic fluorophores led to more ambiguity in assigning a unique color to each fluorophore in fm-STORM compared with fm-DNA-PAINT, giving rise to higher cross-talk between color channels. This ambiguity is due to the broad on/off blinking dynamics of the fluorophores, which is difficult to control and leads to fluorophores’ often being detected in fewer than four consecutive frames. Indeed, color cross-talk quantification showed that around 12% of A647 localizations and 10% of Cy3B localizations were misassigned to the wrong color channel (SI Appendix, Fig. S11).
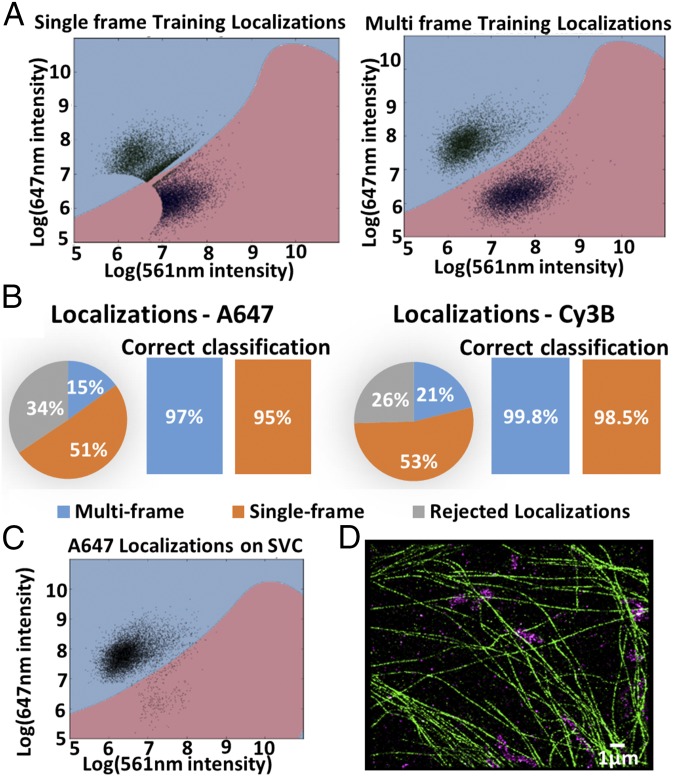
To correct for the fm-STORM–associated cross-talk between color channels, we developed a machine-learning algorithm. The algorithm relies on the use of training data consisting of biological samples labeled with a single fluorophore and imaged with fm-STORM (SI Appendix, Fig. S12). The x–y positions of fluorophores in the training data were first determined and localizations were classified as single or multiframe localizations depending on whether they appeared only in one frame or in multiple subsequent frames (SI Appendix, Fig. S12). Around 50% of localizations were initially characterized as single-frame localizations. The localized x–y position of each fluorophore was then used to define a subROI of four by four pixels and the FFT was performed for this subROI to determine the amplitudes belonging to each different frequency bin. These data were used to train the support vector classifier (SVC) to build decision boundaries separating the multiple color channels (Fig. 4A and SI Appendix, SI Methods and Fig. S12). In practice, a percentage of single-frame localizations (30–40%) was in fact multiframe localizations that failed to be localized in one or more frames by the localization algorithm. Hence, these localizations could correctly be classified after the FFT as they appeared with higher intensity in one frequency bin (Fig. 4A). The true single-frame localizations (∼30% of all localizations) were rejected, as they could not be accurately classified (SI Appendix, SI Methods). Once the decision boundaries were built we used them to classify the fluorophores in a multicolor experiment.
Fig. 4.
Machine-learning algorithm effectively corrects for color cross-talk in fm-STORM. (A) Training dataset plots with decision boundaries, localized in one frame (Left) and in multiple frames (Right). (B) Pie chart shows percentage of single, multiframe, and rejected localizations for Alexa Fluor 647 and Cy3B. Bar plots show percentage of correctly assigned single and multiframe localizations. (C) Alexa Fluor 647 localizations on SVC decision boundary plot. (D) Two-color image of microtubules (green) and mitochondria (magenta) labeled with Cy3B and Alexa Fluor 647, respectively.
To validate the algorithm for two-color fm-STORM, we labeled individual structures (mitochondria or microtubules) in single color with the photoswitchable fluorophores Cy3B and/or A647 and imaged them with fm-STORM with the two laser intensities modulated at different frequencies (647 nm at 45 Hz and 561 nm at 22.5 Hz). After classification using the decision boundary, 97% of localizations from multiframes were correctly classified for the A647 channel, and this number increased up to nearly 100% in the case of the Cy3B channel (Fig. 4 B–D). The decision boundaries could also be generated using training data from three different fluorophores (ATTO488, Cy3B, and A647), demonstrating that this approach can be readily extended to more than two colors (SI Appendix, Fig. S13). Overall, the image acquisition times of fm-STORM are comparable to or even faster than sequential multicolor imaging with activator–reporter pairs (Discussion), while the cross-talk correction can be performed with better accuracy using the machine-learning algorithm. Typically, 10–30% of cross-talk is present in the standard activator–reporter approaches, depending on the fluorophores used and the labeling density (1, 3) (SI Appendix, Fig. S14). To correct for this cross-talk, statistical methods have been developed (27), but their performance often depends on the amount of cross-talk and the spatial separation between the different structures in the image. For the structures imaged here, standard cross-talk correction could correctly classify up to only 88% of localizations (SI Appendix, Fig. S14).
Discussion
We have developed a multicolor superresolution method that relies entirely on the excitation rather than emission properties of fluorophores and implemented it in two modalities of single-molecule-based superresolution microscopy: fm-DNA-PAINT and fm-STORM. DNA-PAINT offers multiple advantages in terms of increased localization precision, minimal photobleaching, image quantification, and multicolor capabilities compared with other single-molecule superresolution approaches (12, 14). However, these advantages come at the expense of image acquisition time, making conventional DNA-PAINT extremely slow for multicolor superresolution or for multiplexed, high-throughput applications (28). The temporal bottleneck arises because of the need of detecting true binding events in a background of diffusing molecules. Faster camera speeds or increased concentration of imager strands in the imaging buffer will lead to an effective increase in background and/or false localizations (15). Recently, the background problem was addressed by combining FRET with DNA-PAINT (15). This approach significantly increases the image acquisition speed, but it also makes DNA-PAINT prone to photobleaching and dependent on the photophysics of donor–acceptor dyes. Our approach of fm-DNA-PAINT preserves all of the advantages of conventional DNA-PAINT, and, importantly, it can acquire multiple colors in the same amount of time needed to acquire a single-color conventional DNA-PAINT image.
Importantly, fm-DNA-PAINT does not require color cross-talk correction or the rejection of localizations due to color misassignment since the long binding times (hundreds of milliseconds) of the imager oligos make color discrimination much less ambiguous. In our current implementation, the number of colors was only limited by the commercially available oligo-coupled antibodies, and as such there is no fundamental limit to extend this approach to more colors with the only requirement that the fluorophores be preferentially excited at one unique excitation wavelength. Indeed, our simulations show that even in the case of substantial spatial overlap among fluorophores for a five-color acquisition, the demodulation step properly assigns each fluorophore to the correct color channel, with minimal distortion to the PSF from the spatial overlap and minimal error in localization.
Our calculations for spatial overlap errors were based on experimental data of Cy3- and Cy5-equivalent fluorophores, which have some spectral overlap (SI Appendix, Fig. S15). Fluorophores with greater overlap in their excitation spectra can be used in fm-DNA-PAINT, but as the spectral overlap increases their discrimination in the frequency domain becomes more challenging, especially when spatial overlap is also present, in which case PSF distortions may become significant. Spectral overlap will ultimately lead to an increase in color cross-talk, necessitating correction algorithms like the machine-learning algorithm developed here or advanced unmixing algorithms (20). The need to correct for color cross-talk also has an impact on the total imaging time, since cross-talk algorithms typically discard a portion of localizations and more localizations must be accumulated to get to the same final Nyquist resolution. That being said, since DNA-PAINT uses conventional fluorophores, it is possible to choose up to five different fluorophores with spectral separations comparable to those between Cy3 and Cy5 (SI Appendix, Fig. S15). As a result, we expect that fm-DNA-PAINT can easily multiplex five different colors in one shot, dramatically enhancing the throughput of this method. This enhanced throughput should not only improve multicolor superresolution imaging but also other single-molecule-based techniques that depend on multiplexing, including bar-coding approaches such as MERFISH (29) or Oligo-PAINT (30).
We also showed that our frequency multiplexing scheme can be implemented for multicolor STORM. fm-STORM was prone to color cross-talk due to the stochasticity of fluorophore photoswitching. The machine-learning algorithm we developed could correct for color cross-talk with high accuracy (>95% correctly classified fluorophores). This algorithm should be applicable to correct for color cross-talk that arises in emission-based superresolution microscopy when fluorophores with overlapping emission spectra are used (31, 32), as long as the training dataset is generated using the specific experimental configuration, making it a versatile approach for correcting color cross-talk in superresolution imaging.
The speed of fm-STORM is an improvement over the sequential, activator–reporter scheme of multicolor STORM imaging (1, 2). The latter also uses a frame window of four to six frames to ensure that fluorophores belonging to one activator–reporter pair fully switch off before the fluorophores belonging to the next activator–reporter pair are switched on. This approach is needed to reduce color cross-talk. However, in the activator–reporter multicolor STORM case the colors are acquired sequentially (four- to six-frame window per color channel), whereas in fm-STORM multiple colors can be acquired simultaneously within the same frame window. Both methods lead to similar levels of color cross-talk and require correction algorithms that discard a similar percentage of localizations. Although from the technical point of view fm can be readily implemented in both DNA-PAINT and STORM, controlling the photophysical properties of multiple fluorophores under a single buffer solution remains a challenge. The working principle of DNA-PAINT makes this technique much more amenable to fm and thus we expect fm-DNA-PAINT to become highly useful to the community.
Methods
Description of the sample preparation and imaging conditions for fm-DNA-PAINT and fm-STORM can be found in SI Appendix, SI Methods. The fm approach was implemented around a custom-built inverted Nikon Eclipse Ti microscope and described in SI Appendix, SI Methods. Procedures on data analysis for fm-DNA-PAINT, estimation of localization precision, and generation of synthetic and semisynthetic data are extensively described in SI Appendix, SI Methods. Implementation of the machine-learning algorithm for cross-talk correction in fm-STORM was performed in Python and is described in SI Appendix, SI Methods and the software is deposited on GitHub (https://github.com/PabloAu/Excitation-multiplexed-multicolor-super-resolution-imaging-with-fm-DNA-PAINT-and-fm-STORM).
Supplementary Material
Acknowledgments
We thank Dr. Joseph S. Borbely (Measurement Standards Laboratory of New Zealand) for all the technical support he provided throughout this project. This work was supported by the Fundació Privada Cellex, Generalitat de Catalunya through the CERCA program, Spanish Ministry of Economy and Competitiveness (“Severo Ochoa” Programme for Centres of Excellence in R&D Grants SEV-2015-0522, FIS2015-63550-R, and FIS2017-89560-R), the European Union Seventh Framework Programme under European Research Council Grants 337191- MOTORS and European Union’s Horizon 2020 Research and Innovation Programme (CellViewer 686637) (M.L.), the EC-Marie Sklodowska-Curie COFUND action ICFONest+ GA 609416 (to E.T.G. and J.J.O.), and EC-Marie Sklodowska-Curie Individual Fellowship VCSD G.A. 656873 (to J.J.O.).
Footnotes
Conflict of interest statement: A patent application is under preparation and will be filed.
This article is a PNAS Direct Submission.
Data deposition: The software for the study has been deposited on GitHub and is available at https://github.com/PabloAu/Excitation-multiplexed-multicolor-super-resolution-imaging-with-fm-DNA-PAINT-and-fm-STORM.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804725115/-/DCSupplemental.
References
- 1.Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates M, Dempsey GT, Chen KH, Zhuang X. Multicolor super-resolution fluorescence imaging via multi-parameter fluorophore detection. ChemPhysChem. 2012;13:99–107. doi: 10.1002/cphc.201100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tam J, Cordier GA, Borbely JS, Sandoval Álvarez A, Lakadamyali M. Cross-talk-free multi-color STORM imaging using a single fluorophore. PLoS One. 2014;9:e101772. doi: 10.1371/journal.pone.0101772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valley CC, Liu S, Lidke DS, Lidke KA. Sequential superresolution imaging of multiple targets using a single fluorophore. PLoS One. 2015;10:e0123941. doi: 10.1371/journal.pone.0123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endesfelder U, et al. Chemically induced photoswitching of fluorescent probes–A general concept for super-resolution microscopy. Molecules. 2011;16:3106–3118. doi: 10.3390/molecules16043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lampe A, Haucke V, Sigrist SJ, Heilemann M, Schmoranzer J. Multi-colour direct STORM with red emitting carbocyanines. Biol Cell. 2012;104:229–237. doi: 10.1111/boc.201100011. [DOI] [PubMed] [Google Scholar]
- 8.van de Linde S, et al. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat Protoc. 2011;6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- 9.Shechtman Y, Weiss LE, Backer AS, Lee MY, Moerner WE. Multicolour localization microscopy by point-spread-function engineering. Nat Photonics. 2016;10:590–594. doi: 10.1038/nphoton.2016.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Kenny SJ, Hauser M, Li W, Xu K. Ultrahigh-throughput single-molecule spectroscopy and spectrally resolved super-resolution microscopy. Nat Methods. 2015;12:935–938. doi: 10.1038/nmeth.3528. [DOI] [PubMed] [Google Scholar]
- 11.Piatkowski L, Gellings E, van Hulst NF. Broadband single-molecule excitation spectroscopy. Nat Commun. 2016;7:10411. doi: 10.1038/ncomms10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jungmann R, et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat Methods. 2014;11:313–318. doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schnitzbauer J, Strauss MT, Schlichthaerle T, Schueder F, Jungmann R. Super-resolution microscopy with DNA-PAINT. Nat Protoc. 2017;12:1198–1228. doi: 10.1038/nprot.2017.024. [DOI] [PubMed] [Google Scholar]
- 14.Jungmann R, et al. Quantitative super-resolution imaging with qPAINT. Nat Methods. 2016;13:439–442. doi: 10.1038/nmeth.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auer A, Strauss MT, Schlichthaerle T, Jungmann R. Fast, background-free DNA-PAINT imaging using FRET-based probes. Nano Lett. 2017;17:6428–6434. doi: 10.1021/acs.nanolett.7b03425. [DOI] [PubMed] [Google Scholar]
- 16.Lewis EK, et al. Color-blind fluorescence detection for four-color DNA sequencing. Proc Natl Acad Sci USA. 2005;102:5346–5351. doi: 10.1073/pnas.0501606102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Åslund N, Carlsson K. Confocal scanning microfluorometry of dual-labelled specimens using two excitation wavelengths and lock-in detection technique. Micron. 1993;24:603–609. [Google Scholar]
- 18.Hwang J, et al. Frequency- and spectrally-encoded confocal microscopy. Opt Express. 2015;23:5809–5821. doi: 10.1364/OE.23.005809. [DOI] [PubMed] [Google Scholar]
- 19.Zhao M, Li Y, Peng L. Parallel excitation-emission multiplexed fluorescence lifetime confocal microscopy for live cell imaging. Opt Express. 2014;22:10221–10232. doi: 10.1364/OE.22.010221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garbacik ET, Sanz-Paz M, Borgman KJE, Campelo F, Garcia-Parajo MF. Frequency-encoded multicolor fluorescence imaging with single-photon-counting color-blind detection. Biophys J. 2018;115:725–736. doi: 10.1016/j.bpj.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith CS, Joseph N, Rieger B, Lidke KA. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat Methods. 2010;7:373–375. doi: 10.1038/nmeth.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Y, et al. Quantifying and optimizing single-molecule switching nanoscopy at high speeds. PLoS One. 2015;10:e0128135. doi: 10.1371/journal.pone.0128135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang B, Bates M, Zhuang X. Super-resolution fluorescence microscopy. Annu Rev Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dani A, Huang B, Bergan J, Dulac C, Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beghin A, et al. Localization-based super-resolution imaging meets high-content screening. Nat Methods. 2017;14:1184–1190. doi: 10.1038/nmeth.4486. [DOI] [PubMed] [Google Scholar]
- 29.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beliveau BJ, et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc Natl Acad Sci USA. 2012;109:21301–21306. doi: 10.1073/pnas.1213818110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gunewardene MS, et al. Superresolution imaging of multiple fluorescent proteins with highly overlapping emission spectra in living cells. Biophys J. 2011;101:1522–1528. doi: 10.1016/j.bpj.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winterflood CM, Platonova E, Albrecht D, Ewers H. Dual-color 3D superresolution microscopy by combined spectral-demixing and biplane imaging. Biophys J. 2015;109:3–6. doi: 10.1016/j.bpj.2015.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.