Abstract
Vaccines and antimicrobial drugs both impose strong selection for resistance. Yet only drug resistance is a major challenge for 21st century medicine. Why is drug resistance ubiquitous and not vaccine resistance? Part of the answer is that vaccine resistance is far less likely to evolve than drug resistance. But what happens when vaccine resistance does evolve? We review six putative cases. We find that in contrast to drug resistance, vaccine resistance is harder to detect and harder to confirm and that the mechanistic basis is less well understood. Nevertheless, in the cases we examined, the pronounced health benefits associated with vaccination have largely been sustained. Thus, we contend that vaccine resistance is less of a concern than drug resistance because it is less likely to evolve and when it does, it is less harmful to human and animal health and well-being. Studies of pathogen strains that evolve the capacity to replicate and transmit from vaccinated hosts will enhance our ability to develop next-generation vaccines that minimize the risk of harmful pathogen evolution.
Keywords: evolutionary rescue, pertussis, Streptococcus, HBV, Marek’s disease virus
Modern medicine and industrial animal farming greatly benefit from vaccines and antimicrobial drugs (1–3). But like all disease interventions targeted at pathogen fitness, drugs and vaccines may drive pathogen evolution that undermines their efficacy, threatening the sustainability of medical and agricultural gains. This is starkly obvious for antimicrobial drugs, with drug-resistant infections already responsible for well in excess of 100,000 deaths per year globally and a projected 10 million deaths per year by 2050 (4). In contrast, deaths due to vaccine-preventable diseases are almost entirely due to lack of access to vaccines, not vaccine resistance. Here we ask why drug resistance is one of the biggest global challenges of our era (4, 5) whereas vaccine resistance is not.
For the purposes of this paper, we use “resistance” to mean an evolutionarily acquired positive pathogen population growth in treated hosts. Defined this way, resistance is an increased ability of the pathogen to infect, replicate, and/or transmit from a treated host. Our definition of resistance therefore does not include cases of intrinsic resistance, sometimes referred to as insensitivity, where a pathogen was never susceptible to treatment. Insensitivity is no doubt important, but here we are interested in resistance that evolves in response to vaccine or drug pressure. Note that our definition of resistance is agnostic to the effects of resistance on the prevalence and severity of disease, an important distinction we return to below.
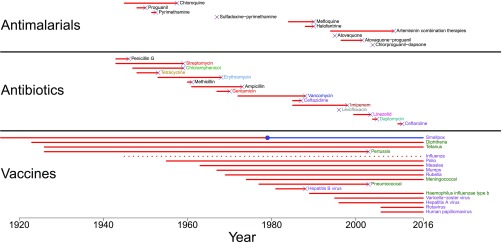
Vaccine resistance evolves less readily than drug resistance (Fig. 1). Elsewhere, we have argued that two key differences between drugs and vaccines explain why (6). The first is the timing of treatment, and the second is the multiplicity of target sites. Vaccines are used prophylactically, whereas drugs tend to be used therapeutically. This difference in timing means that, relative to drugs, vaccines tend to keep pathogens from ever achieving large population sizes within hosts. Resistance mutations are less likely to appear in small populations (7), and when such mutations appear and confer partial resistance within a host, they are unlikely to replicate to the large population sizes that are associated with onward transmission (6, 8). In addition, drugs tend to target pathogens in a single way (9) whereas vaccines tend to target pathogens in multiple ways by inducing host-specific antibody and/or T cell responses (10). This difference in the multiplicity of target sites means that relative to drugs, more mutations are likely needed to confer resistance to vaccines. Variability in immune responses between hosts (11) further implies that even if a pathogen variant were resistant to vaccine protection in one host, it may still be detected and killed in another vaccinated host.
Fig. 1.
Time between deployment of an intervention and the first documented failure in humans due to resistance (marked with “x”s). Different classes of antibiotic drugs are labeled in different colors. Viral vaccines are labeled in purple and bacterial vaccines are labeled in green. The circle for the smallpox vaccine denotes global eradication of the virus, which ended the opportunity for vaccine resistance to evolve. Influenza is shown as a dotted line to highlight that it is routinely changed in an attempt to match circulating virus strains. Note that serotype replacement is not shown. While many of the cases of antibiotic resistance and one case of vaccine resistance can be explained by horizontal gene transfer, horizontal gene transfer is not considered to be an important factor in the evolution of antimalarial, antitubercular, or antiviral drug resistance. Data for antimalarial drugs are from refs. 130–132. Data for antibiotic drugs are from ref. 132. Data for vaccines are from refs. 27, 48, and 133–136. Modified from ref. 6.
Together, these differences between drugs and vaccines mean that pathogen populations generate less variation for vaccine resistance than for drug resistance and that selection has fewer opportunities to act on any variation that is generated. These two factors synergistically slow the evolution of resistance to vaccines (6).
Thus, one reason why the evolution of drug resistance is more problematic than the evolution of vaccine resistance is that drug resistance is more likely to evolve. But what happens when vaccine resistance does evolve? Is it as harmful to human and animal well-being as drug resistance? To shed light on this, we briefly summarize the health impact of drug resistance and then contrast this with six cases in which the evolution of vaccine-resistant strains is strongly suspected.
Drug Resistance
The first naturally derived antibiotic, penicillin G, also known as benzylpenicillin, was serendipitously discovered in 1928 by Alexander Fleming (12), when he noted a halo around a fungal contaminant on his bacterial plates, indicating lysis. Efficacy in vivo was demonstrated a decade later, showing that injections of penicillin G could extend the life of mice challenged with lethal doses of bacteria (13). By 1946, penicillin G was widely available by prescription (14).
The first evidence that bacteria had potential to evolve resistance to penicillin G traces back to its original discovery by Fleming, when he noted differences in lysis between species of bacteria in vitro. Abraham and Chain (15) showed that at least some of those differences were due to the production of an enzyme now known to be beta-lactamase. Due to the common use of the drug, resistance to penicillin G quickly became widespread (14). For example, the majority of Staphylococcus aureus isolates in a British hospital were resistant by 1948 (16). Drug resistance is typically quantified by measuring minimum inhibitory concentrations (MIC) in vitro, but resistance can often be observed before such assays when patients either fail to respond to treatment or relapse during treatment (e.g., ref. 17).
The emergence of penicillin G–resistant infections prompted the development of penicillin derivatives resilient to beta-lactamase and with broader spectra of activity. It also prompted the discovery of alternative classes of antibiotics with fundamentally different mechanisms of action. Resistance, nevertheless, emerged against these derivatives and alternative antibiotic classes (18) (Fig. 1). Acquisition of antibiotic resistance has occurred through both horizontal gene transfer and de novo mutation (19, 20), and the mechanisms of resistance have been diverse, including the acquisition of enzymes that inactivate drugs, the modification of target site expression or drug-binding affinity, and the reduction of drug access to target sites (9).
Antibiotics are still highly effective in treating bacteria susceptible to particular drugs, but resistance is widespread, causing problems for public health. Currently, antibiotic resistance costs approximately $US 20 billion and involves 8 million extra hospital days per year in the United States alone (21). The rise of multidrug-resistant pathogens, including some resistant to all available antibiotics, is threatening to increase this burden. The consequences of such evolution can be severe. For example, multidrug-resistant Pseudomonas aeruginosa bloodstream infections result in 50% mortality compared with the 24% mortality of nonmultidrug-resistant infections (22). New measures to control antibiotic-resistant bacteria are therefore desperately needed. These include the development of new antibiotics, although new antibiotics are becoming increasingly difficult and expensive to discover and bring to market (23, 24). Moreover, it seems likely that resistance to next-generation antimicrobials will evolve just as readily as it has in the past.
One solution to the antibiotic-resistance crisis might be vaccination (23, 24). Vaccination can slow the spread of antibiotic resistance by reducing the need for the appropriate use of antibiotics (e.g., Haemophilus influenzae type b and Streptococcus pneumoniae), by reducing the inappropriate use of antibiotics for viral infections such as influenza virus, and by developing vaccine protection that targets antibiotic-resistant variants of a pathogen such as with S. pneumoniae vaccination (23, 24). But like drugs, vaccines also impose immense selection for resistance. We have already argued that vaccines are less likely to be undermined by resistance than are drugs, but what are the health consequences of vaccine resistance when it does evolve? Here we summarize six putative cases of vaccine resistance, three in human diseases and three in animal diseases. These cases are summarized in Table 1.
Table 1.
Summary of vaccine-resistance case studies in main text
| Issues | Hepatitis B virus | S. pneumoniae | B. pertussis | Y. ruckeri | Avian metapneumovirus | Marek’s disease virus |
| Reason vaccine resistance was first suspected | Variation in key antigens | Variation in key antigens | Outbreaks in vaccinated populations | Outbreaks in vaccinated populations | Outbreaks in vaccinated populations | Outbreaks in vaccinated populations |
| Evidence of resistance | ||||||
| Direct experiments | Negative* | Not done | Positive* | Mixed | Positive | Positive |
| Molecular epidemiology | Positive | Positive | Positive | Not done | Not done | Not done |
| Mechanistic plausibility | Positive | Positive | Positive | Positive | Positive | Positive |
| Expert opinion | Positive | Positive | Positive | Positive | Positive | Positive |
| Putative route of resistance | Immune evasion | Immune evasion | Immune suppression and immune evasion | Loss of immune stimulation | Immune evasion | ?† |
| Impact of vaccine resistance on disease | ||||||
| Population level | Extremely small | Slight increase | ?‡ | ?§ | ?§ | Large increase¶ |
| Vaccinated host | Rare disease | Slight increase | ?‡ | Mixed evidence | Increase | Mild to severe |
| Unvaccinated host | ?# | Slight decrease | ?‖ | ? | No change | Very severe |
| Prompted development of vaccination strategies to combat resistance? | No | Yes | Unclear** | Yes | No | Yes |
Animals models, not natural hosts. For HBV, the putative vaccine-resistant strain was unable to infect vaccinated chimpanzees, even though it was able to replicate in unvaccinated animals. For B. pertussis, there was unambiguous evidence for prn−; ptxP3 evidence was mixed (main text).
Resistance correlates with virulence in unprotected hosts.
Debate over the degree to which resurgence in cases (Fig. 2) is attributable to vaccine resistance.
Despite lack of data, disease is presumably reduced given that vaccination is still widely used in the industry.
Vaccine-resistant isolates reduce survival and increase condemnation rates in flocks vaccinated with first-generation vaccines.
Vaccine-resistant isolates may have reduced pathogenicity due to fitness costs of resistance, but not obviously seen in the animal model (main text).
Some vaccine-resistant isolates may have increased virulence in unvaccinated hosts (main text).
New vaccines are in development but it is not clear that these are being developed to combat vaccine-resistant pathogen strains rather than, for example, waning immunity.
Vaccine Resistance
Hepatitis B Virus.
In 1967, Baruch Blumberg discovered hepatitis B virus (HBV), a causative agent of hepatitis, cirrhosis, and hepatocellular carcinoma that can be transmitted by infected blood and semen, including from mother to child during birth. A decade and a half later, a vaccine was licensed for use in humans composed of the hepatitis B surface antigen (HBsAg). Concerns that vaccine resistance might evolve arose from the discovery that a small fraction of unvaccinated patients with chronic liver disease had HBV DNA, but no detectable HBsAg (25, 26). These concerns were amplified by a large vaccine trial which detected virus in 44 vaccinated people (27). Viral typing from a subset of these individuals and their presumed infectious contacts showed that transmission to vaccinated hosts strongly correlated with a loss of monoclonal antibody binding to HBsAg (27).
Since then, numerous putative vaccine-resistant variants have been described (28, 29). Most are seen in newborns of HBV-infected mothers or in liver transplant patients, both situations where infection precedes the development of vaccine-induced immunity (30, 31). More rarely, people have become infected with HBV and developed acute illness after mounting a vaccine-induced immune response (e.g., ref. 32). Nevertheless, no putative alleles conferring vaccine resistance have yet become a public health concern (33). This could be because HBV has slow replacement dynamics (34), but available data suggest that, contrary to earlier reports (35, 36), putative vaccine-resistant variants are stable or even declining in vaccinated populations (33, 37–40). This might be because vaccine resistance is accompanied by large reductions in growth or transmission rates. It could also be that vaccine resistance is specific to the immune response of a particular host, and so a resistant pathogen might not be resistant in a new host. Consistent with that possibility, a putative vaccine-resistant strain was unable to infect vaccinated chimpanzees, despite being able to cause disease in seronegative chimpanzees (41, 42).
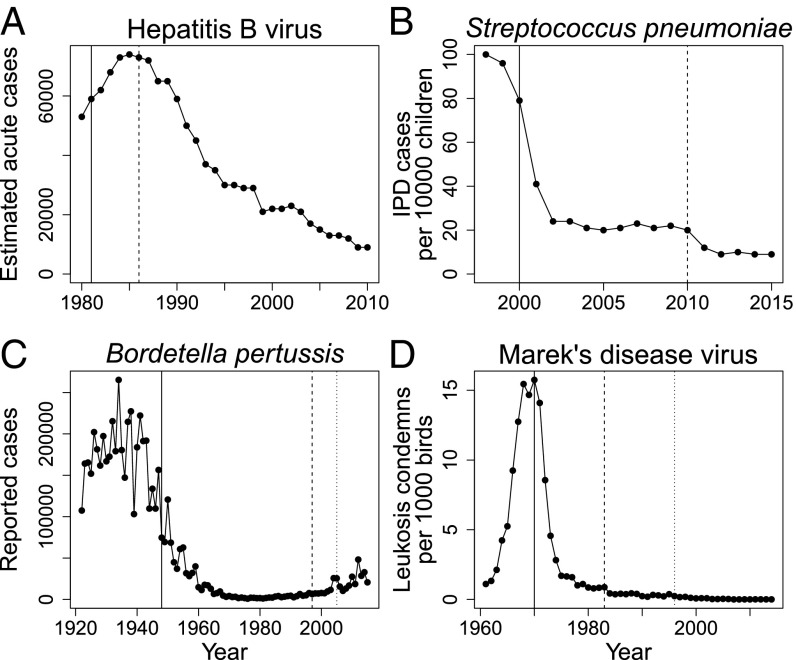
Rates of HBV infection and hepatocellular carcinoma are substantially down where the vaccine is in regular use (43) (Fig. 2). To the best of our knowledge, vaccine research in this system is focused on developing therapeutic vaccines rather than developing vaccines against vaccine-resistant isolates.
Fig. 2.
(A–D) Disease incidence before and after vaccine introductions for four of our case-study pathogens. Analogous data are unavailable for Y. ruckeri and avian metapneumovirus. All data reflect disease dynamics in the United States. Solid lines denote the approximate introduction of first-generation vaccines. Dashed and dotted lines denote the approximate introduction of second- and third-generation vaccines, respectively. Updates to the hepatitis B and pertussis vaccines were made in response to concerns of vaccine safety, not pathogen evolution. Disease dynamics are driven by many factors in addition to vaccines and vaccine resistance, and so not all declines in disease can be attributed to vaccination. We nevertheless note that in A–D, disease rates have never returned to prevaccination levels despite the emergence of vaccine resistance. Data in A–C are compiled from the Centers for Disease Control (137–139). Data in D are compiled from the USDA Poultry Slaughter Reports (107).
S. pneumoniae.
The bacterial pathogen S. pneumoniae is often found benignly colonizing the nasopharynx of healthy children, but it is also responsible for an array of illnesses in humans (44), a large set of which are referred to as invasive pneumococcal disease (IPD). Rates of IPD dropped substantially upon the introduction of pneumococcal conjugate vaccine 7 (PCV7) (45), a conjugate vaccine that induces immunity against the capsule polysaccharide of 7 of the >90 known serotypes of the bacteria. Even before the vaccine was first deployed, there was concern that the benefits of PCV7 might be quickly eroded by serotype replacement, in which nonvaccine serotypes increase in frequency following vaccination (46), especially since S. pneumoniae is naturally competent and serotype replacement was seen in clinical trials (47). Serotype surveillance therefore frequently followed vaccine rollout in a variety of locations.
Results varied slightly from study to study, but the balance of data showed strong impacts of serotype replacement on benign carriage rates and moderate but detectable impacts of serotype replacement on IPD rates (45). At least one serotype increase was facilitated by recombination between a vaccine-targeted and a nonvaccine-targeted serotype that resulted in capsule switching and thus serotype change (48). Presumably, this event and other serotype replacement events conferred resistance to vaccination (but see ref. 49).
After PCV7 vaccination was introduced, IPD rates continually declined without any indication that serotype replacement was causing year-over-year increases in total IPD rates (50) (Fig. 2). Nevertheless, an updated conjugate vaccine PCV13 was developed and introduced in 2010 to provide protection against the original seven serotypes and an additional six others. This update has prevented 30,000 extra IPD cases in its first 3 y of use (51).
Bordetella pertussis.
Bordetella pertussis is the human bacterial pathogen responsible for whooping cough, a respiratory disease that is a substantial contributor to childhood mortality. Incidence of disease was greatly reduced in the mid- to late-20th century through mass vaccination, but when disease later resurged (52), vaccine resistance was posited as one of several explanations (53). Genetic differences between clones circulating before and after vaccine introduction (53), particularly at two key antigens, pertactin (prn) and pertussis toxin (54), heightened concerns about vaccine resistance. In vaccinated mice, this variation affected bacterial clearance rates (55), and in an epidemiological study, bacterial isolates with the same prn allele as the vaccine were underrepresented in vaccinated children relative to unvaccinated children (56).
Recently, isolates of B. pertussis entirely lacking prn expression (prn−) have been increasing in frequency (57–61) (but see ref. 62). In vaccinated mice, prn− strains persist longer than prn+ strains and outcompete prn+ strains during coinfection (63, 64), whereas in naive mice, prn+ strains outcompete prn− strains during coinfection (64). Similarly in humans, prn− strains are isolated more than twice as often from vaccinated than from unvaccinated patients (65).
A mutation that increases the production of pertussis toxin has also attracted considerable attention, the so-called ptxP3 allele (66). The ptxP3 allele has spread in vaccinated populations, and it is associated with increased hospitalizations, durations of stay, and deaths (66–70). These increases may be due to the immunosuppressive effects of the toxin (71–73). Investigations of the vaccine-break potential of ptxP3 in animal models have only just begun. However, experiments have shown that a ptxP3-bearing strain was a better colonizer of both vaccinated and unvaccinated mice than was a strain without the ptxP3 allele (74).
There is much debate about the role of bacterial evolution in the resurgence of whooping cough because the resurgence might be explained entirely by epidemiological or immunological factors (75, 76). If evolution is involved, the population-level benefits of vaccination are still far from completely eroded (Fig. 2). Moreover, vaccination substantially reduces the risk of severe disease, even when infected with prn−/ptxP3 strains (70). Current research into improving the efficacy of pertussis vaccines has been quite diverse, ranging from using live bacteria or inactivated cells to altering antigens, adjuvants, doses, number of boosters, and timing of boosters (77).
Yersinia ruckeri.
Yersinia ruckeri, the causative agent of enteric redmouth disease, is a bacterial pathogen of fish that can lead to substantial mortality in farmed salmonids. There are two biotypes, a motile type with a flagellum [biotype 1(Bt1)] and a nonmotile type that lacks a flagellum (Bt2) (78). A vaccine against enteric redmouth disease licensed in Europe in 1983 became the first commercially produced fish vaccine (79), and it was widely adopted by the aquaculture community. Outbreaks in vaccinated populations raised concerns about vaccine resistance, and because the outbreaks were often caused by Bt2 (80–82), concern was focused on the possibility that Bt2 was vaccine resistant. It is now known that vaccine protection is almost entirely due to IgM responses to the bacterial cell wall (83, 84) and that the cell wall does not necessarily differ between biotypes (85). Nevertheless, flagellin, a defining difference between Bt1 and Bt2, is an immune stimulator in fish (86), potentially providing a mechanistic explanation for differences in vaccine resistance between biotypes.
Experimental evidence that Bt2 is vaccine resistant is, however, mixed. Austin et al. (80) showed that commercial vaccines protected against disease less well for a European Bt2 strain than for a canonical Bt1 isolate, but this pattern reversed when using a noncommercial vaccine based on a European Bt1 strain. Tinsley et al. (87) similarly found no consistent pattern between vaccine efficacy and biotype. These experiments nevertheless show that bacterial isolates differ in their ability to cause disease in vaccinated hosts, and this difference must involve some trait not associated with the presence of a flagellum.
Given the lack of data on historical or current incidence of enteric redmouth disease and the lack of unbiased data on the relative prevalence of Bt1 and Bt2 isolates over time, we cannot assess how strongly vaccine resistance has impacted disease incidence or bacterial prevalence. In response to concerns of resistance, researchers have identified alternative routes of administration of existing vaccines that give better protective efficacy (88) and have developed new vaccines with improved efficacy against both Bt1 and Bt2 isolates (87).
Avian Metapneumovirus.
Avian metapneumovirus (AMPV) is a single-stranded negative-sense RNA virus that causes respiratory diseases in turkeys and other poultry (89). Various vaccines have been developed to control the four subtypes of this virus (90), but investigation into vaccine resistance began with the observation that even homologous vaccines gave imperfect protection in the field (91). To test whether this imperfect protection was a result of evolved vaccine resistance, Catelli et al. (92) tested whether the efficacy of vaccination differed among Italian isolates of AMPV in experimentally infected turkeys. Using one historical virus isolate and one isolate recently collected from a vaccinated flock with disease problems, Catelli et al. (92) indeed found evidence of vaccine resistance. In comparison with the historical isolate, the recent isolate caused more severe disease in vaccinated birds. Vaccinated birds also shed the recent isolate for longer than the historical isolate, but disease severity and shedding duration did not differ in unvaccinated birds (92). Further sequencing of virus isolates collected over two decades revealed substantial variation in the surface glycoprotein of the virus (93), a key antigen for vaccine-induced immune protection (94). Moreover, this sequence variation was located in putative T cell epitopes (93).
An isolate similar to the Italian vaccine-resistant isolate recently appeared in a vaccinated flock in Romania (95) and might be a harbinger of further spread. Despite these potential concerns, current vaccine research is largely geared toward reducing the rate of vaccine reversion to virulence and generating recombinant vaccines that protect against more than one disease (96, 97).
Marek’s Disease Virus.
Marek’s disease virus (MDV) is the causative agent of Marek’s disease (MD), a neoplastic disease of chickens with clinical signs that include immune suppression, paralysis, tumor formation, and death (98). Before the introduction of the first vaccines in 1970, outbreaks of MD would often wipe out entire flocks of chickens (99, 100). In addition, birds were frequently removed from the food chain (“condemned”) at the time of slaughter for traits closely associated with MD (101). The introduction of live-virus vaccines in 1970 substantially reduced both sources of loss, although the virus continued to circulate without causing clinical signs of disease (102). Fear of vaccine resistance began less than a decade later because of geographically clustered increases in MD-associated condemnation and because “vaccine breaks” occurred, where outbreaks of MD occurred in vaccinated flocks (103).
Laboratory infection experiments in chickens confirmed that vaccine resistance had indeed evolved. Initial studies showed that vaccine protection against disease was reduced for vaccine-break isolates relative to historical isolates and relative to isolates collected from farms without MD breaks (104, 105). A more recent study has shown that vaccine-break isolates have greater lifetime transmission potential than nonbreak isolates in vaccinated birds (106). The mechanistic basis of this resistance is still unknown, but vaccine-break isolates cause more severe disease in unvaccinated birds, suggesting that increases in traits associated with pathogenicity have facilitated vaccine resistance (98).
Despite the circulation of vaccine-break isolates, MD-associated condemnation rates have steadily declined at the national scale (107) (Fig. 2). Moreover, vaccinated birds infected with vaccine-break virus isolates fare better than unvaccinated birds infected with nonvaccine-break virus isolates (106), suggesting that the health benefits of vaccination were only partially eroded by evolution. Regardless, the scientific community responded to this evolution with the introduction of new generations of vaccines with improved efficacy (108, 109), and those are still in use today. Although MDV continues to circulate on commercial poultry farms (Fig. 3), MD is currently considered to be under control.
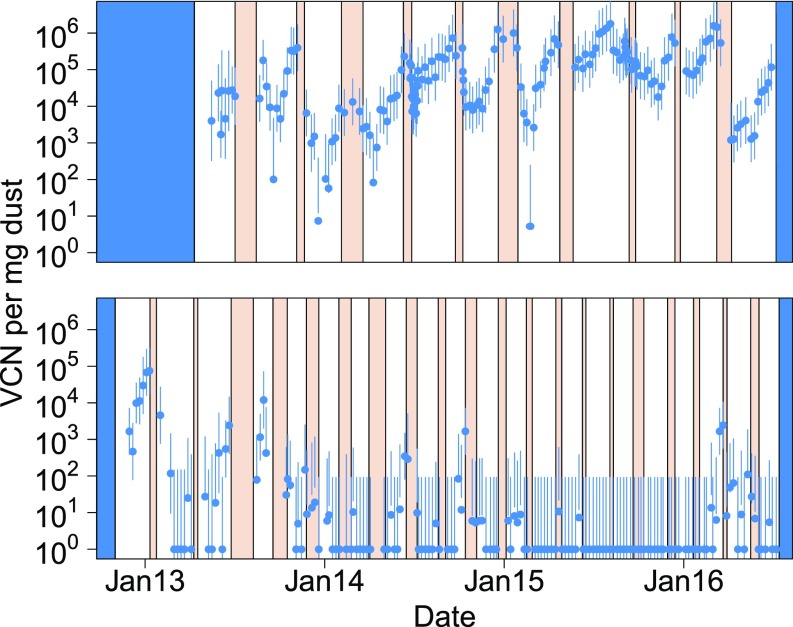
Fig. 3.
Marek’s disease virus concentration in dust over time on two different broiler chicken farms where all birds were vaccinated against Marek’s disease (140). Infection is transmitted through inhalation of virus-contaminated dust, and so these plots show a proxy for force of infection over time. Birds were reared in discrete nonoverlapping cohorts within each farm. White intervals denote periods when birds were present, red intervals denote periods between chicken flocks when birds were absent, and blue intervals denote periods of no surveillance. Error bars show 95% confidence intervals. Vaccine protection is imperfect such that vaccinated birds may still become infected and transmit, although at reduced rates (141, 142). Even in the presence of vaccination (bivalent vaccination on these farms), the virus is able to amplify within some chicken flocks. Nevertheless, vaccination was effective in the sense that no mortality or production losses due to Marek’s disease were documented on either farm during the surveillance period. Reprinted from ref.143. VCN, virus genome copy number.
Discussion
Drug resistance almost always evolves in response to the widespread use of drugs, but vaccine resistance does not (Fig. 1). Vaccine resistance is less likely to emerge than drug resistance because unlike drugs, vaccines are used prophylactically and they target pathogens in multiple ways simultaneously, which together drastically reduce the chance that resistance will emerge (6). There are a handful of documented cases where vaccines have lacked the benefits of these features and where putative vaccine-resistant strains have been subject to empirical analysis. We summarized six, three from farm animals and three from humans.
Several patterns emerged (Table 1). First, in four of the six, vaccine resistance was first suspected because of disease outbreaks in vaccinated populations. This contrasts with penicillin G where the first evidence of resistance came from studies performed in vitro before the drug was even approved for use (15). In two cases, S. pneumoniae and HBV, vaccine resistance was discovered by targeted surveillance before there were outbreaks of disease. This early detection was no doubt possible because these two vaccines seemed likely to prompt resistance evolution, each inducing immunity against a single antigenic target. A similar argument might be made for the detection of prn− variants in B. pertussis. We thus conclude that a narrow target makes a vaccine prone to drive the evolution of resistance as expected (6), but it also makes resistance easy to detect, meaning that resistance can be monitored and responses can potentially be implemented before disease resurgence (e.g., PCV13 for S. pneumoniae).
Second, the emergence of vaccine resistance is confirmed in various ways. In contrast, suspected cases of antimicrobial resistance are usually easily confirmed using in vitro assays. In Table 1, we have separated the evidence of vaccine resistance into direct experiments (measures of the performance of putative vaccine-resistant strains in vaccinated hosts), molecular epidemiology (surveillance of putative vaccine-resistant strains in vaccinated and unvaccinated hosts or in pre- and postvaccine eras), mechanistic plausibility, and expert opinion. This order is intentional to depict the rough hierarchy of evidential quality. For example, a single experiment performed under field-like conditions can demonstrate vaccine resistance (e.g., refs. 92 and 106), whereas a plausible mechanism for resistance may be better thought of as hypothesis generating (e.g., Bt2 of Y. ruckeri). We note that for obvious reasons, the confirmation of resistance to human vaccines frequently relies on epidemiological data and lacks experimental data, whereas for animal vaccines the reverse applies.
Third, vaccine resistance can be achieved by either active immunosuppression (turning down host defenses) or immune evasion (avoiding detection). Pathogens have evolved a vast array of countermeasures against natural host immunity (110, 111) and it seems likely that many of these could be deployed against vaccine-induced immunity. It will be especially interesting to see the countermeasures pathogens evolve in response to next-generation vaccines. Many vaccines of the future will likely induce responses not naturally experienced by target pathogens. Presumably those pathogens may evolve escape or evasion responses they do not currently possess. Many routes to resist vaccine-induced immunity are possible, not least mechanisms analogous to those that commonly confer drug resistance: the production of enzymes that degrade effector molecules, altered target sites, altered expression of target sites, and reduced access to target sites (9).
Fourth, our case studies show reduction of disease even in the presence of vaccine resistance (Fig. 2). This is an interesting contrast with drugs, where the therapeutic gains in a patient can be completely nullified by resistance. We can imagine several potential explanations for this observation. None are mutually exclusive. First, resistant pathogen strains might still be evolving or spreading, thus requiring more time to fully erode the benefits of vaccination. Second, vaccine-resistant strains might never fully replace vaccine-sensitive pathogen strains, for example, due to very high fitness costs, competitive interactions, or heterogeneity in selection among hosts or geographic locations. Third, vaccine resistance might be incomplete, for example, if some degree of protection persists against vaccine-resistant strains. Finally, the mechanisms by which vaccines protect against disease (pathology) often differ from those that protect against infection and transmission (112). If so, vaccines could continue to deliver antidisease protection even when hosts become infected with vaccine-resistant strains (see Fig. 3, for an example). Perhaps vaccine-induced immunity nudges host responses away from trajectories associated with immune pathology (113).
Finally, vaccine resistance has prompted research and development into new vaccination strategies for only three of the case studies (Table 1). Vaccine research is ongoing for all six, but vaccine resistance seems not to be the major motivator for HBV, B. pertussis, or AMPV. This contrasts strongly with drugs, where development pipelines exist primarily to combat the evolution of resistance (114).
In the interest of brevity, we focused on six of the best-documented cases of vaccine resistance. Other putative cases include avian influenza (115), avian reovirus (116), Corynebacterium diphtheria (117), feline calicivirus (118), H. influenzae (119), infectious bursal disease virus (120), Neisseria meningitidis (121), Newcastle disease virus (122), and porcine circovirus type 2 (123). Still more cases may have gone undocumented, particularly in agriculture where autogenous vaccines are frequently developed and deployed when commercial vaccines are unavailable or give inadequate protection against disease (124). Like the cases we did discuss, all of these deserve further work. We would be particularly interested in seeing whether epidemiological factors, such as host population density or host species, correlate with the emergence of vaccine resistance. We note that there may also be as yet unnoticed cases of vaccine resistance; vaccine protection against infection and transmission is difficult to measure (125) and usually not the primary trait of interest with regard to human or animal health.
Conclusions
We have argued that vaccine resistance is substantially less problematic for human and animal health than is drug resistance because vaccine resistance is far less likely to evolve, and when it has evolved, the pronounced health benefits associated with vaccination have largely been retained. This provides yet another reason why vaccines are an important part of the solution to drug resistance (23, 24). However, we do not want to give the impression that the possibility of vaccine resistance can be safely ignored. First, it is unclear to what extent past successes are a guide to future performance. If next-generation vaccines target single antigens and/or fail to generate sterilizing immunity and so allow onward transmission, vaccine resistance could become more common (6). Second, vaccine resistance is in some ways harder to respond to than drug resistance. Population-level protection against a vaccine-resistant pathogen may require large-scale catch-up campaigns, whereas drug resistance can be handled by swapping drugs at the time of treatment. Third, it is clear from the literature that vaccine resistance is not just about immune evasion; it can involve other phenotypes like immune suppression and faster replication to outrun vaccine-induced immunity (106, 126, 127). Some of these phenotypes could cause more severe disease in unvaccinated individuals (128, 129). While this is a minor inconvenience in agriculture when every animal can be vaccinated, the evolution of that sort of vaccine resistance would be a substantial concern in human populations where universal vaccination is not achievable.
Acknowledgments
We thank A. Graham and R. Regoes for comments on earlier drafts of this paper. This work was funded by the Institute of General Medical Sciences (R01GM105244) and the joint NSF–NIH–US Department of Agriculture Ecology and Evolution of Infectious Diseases program. This paper was written while A.F.R. was supported on sabbatical by the ETH Zurich. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Arias E, Heron M, Xu J. Division of Vital Statistics . United States Life Tables, 2012. Vol 65. US Department of Health and Human Services, Centers for Disease Control and Prevention; and Prevention; Hyattsville, MD: 2016. pp. 46–47. [PubMed] [Google Scholar]
- 2.Cromwell GL. Why and how antibiotics are used in swine production. Anim Biotechnol. 2002;13:7–27. doi: 10.1081/ABIO-120005767. [DOI] [PubMed] [Google Scholar]
- 3.Morrow C, Fehler F. Marek’s disease: A worldwide problem. In: Davison F, Nair V, editors. Marek’s Disease: An Evolving Problem. Elsevier; San Diego: 2004. pp. 49–61. [Google Scholar]
- 4.O’Neill J. 2015 Tackling a global health crisis: Initial steps. The Review on Antimicrobial Resistance Chaired by Jim O’Neill. Available at https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=1&ved=0ahUKEwj41c7f4YXbAhUEheAKHYHlD8wQFggpMAA&url=https%3A%2F%2Famr-review.org%2Fsites%2Fdefault%2Ffiles%2FReport-52.15.pdf&usg=AOvVaw2qPCJBHD2IuAa5Xju5N31i. Accessed May 16, 2018.
- 5.Davies SC. Annual Report of the Chief Medical Officer. Vol 2 Department of Health; London: 2013. [Google Scholar]
- 6.Kennedy DA, Read AF. Why does drug resistance readily evolve but vaccine resistance does not? Proc R Soc Lond B Biol Sci. 2017;284:20162562. doi: 10.1098/rspb.2016.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orr HA, Unckless RL. The population genetics of evolutionary rescue. PLoS Genet. 2014;10:e1004551. doi: 10.1371/journal.pgen.1004551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Préziosi MP, Halloran ME. Effects of pertussis vaccination on transmission: Vaccine efficacy for infectiousness. Vaccine. 2003;21:1853–1861. doi: 10.1016/s0264-410x(03)00007-0. [DOI] [PubMed] [Google Scholar]
- 9.Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 10.Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poulsen TR, Meijer PJ, Jensen A, Nielsen LS, Andersen PS. Kinetic, affinity, and diversity limits of human polyclonal antibody responses against tetanus toxoid. J Immunol. 2007;179:3841–3850. doi: 10.4049/jimmunol.179.6.3841. [DOI] [PubMed] [Google Scholar]
- 12.Fleming A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Bull World Health Organ. 1929;79:780–790. [PMC free article] [PubMed] [Google Scholar]
- 13.Chain E, et al. Penicillin as a chemotherapeutic agent. Lancet. 1940;236:226–228. [Google Scholar]
- 14.Lobanovska M, Pilla G. Penicillin’s discovery and antibiotic resistance: Lessons for the future? Yale J Biol Med. 2017;90:135–145. [PMC free article] [PubMed] [Google Scholar]
- 15.Abraham EP, Chain E. An enzyme from bacteria able to destroy penicillin. Nature. 1940;146:837. [PubMed] [Google Scholar]
- 16.Barber M, Whitehead JEM. Bacteriophage types in penicillin-resistant staphylococcal infection. Br Med J. 1949;2:565–569. doi: 10.1136/bmj.2.4627.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woods RJ, Read AF. Clinical management of resistance evolution in a bacterial infection: A case study. Evol Med Public Health. 2015;2015:281–288. doi: 10.1093/emph/eov025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ventola CL. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm Ther. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 19.Abel zur Wiesch P, Kouyos R, Engelstädter J, Regoes RR, Bonhoeffer S. Population biological principles of drug-resistance evolution in infectious diseases. Lancet Infect Dis. 2011;11:236–247. doi: 10.1016/S1473-3099(10)70264-4. [DOI] [PubMed] [Google Scholar]
- 20.Read AF, Woods RJ. Antibiotic resistance management. Evol Med Public Health. 2014;2014:147. doi: 10.1093/emph/eou024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fair RJ, Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect Medicin Chem. 2014;6:PMC.S14459. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tumbarello M, et al. Multidrug-resistant Pseudomonas aeruginosa bloodstream infections: Risk factors and mortality. Epidemiol Infect. 2011;139:1740–1749. doi: 10.1017/S0950268810003055. [DOI] [PubMed] [Google Scholar]
- 23.Lipsitch M, Siber GR. How can vaccines contribute to solving the antimicrobial resistance problem? MBio. 2016;7:e00428-16. doi: 10.1128/mBio.00428-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jansen KU, Knirsch C, Anderson AS. The role of vaccines in preventing bacterial antimicrobial resistance. Nat Med. 2018;24:10–19. doi: 10.1038/nm.4465. [DOI] [PubMed] [Google Scholar]
- 25.Bréchot C, et al. Hepatitis B virus DNA in patients with chronic liver disease and negative tests for hepatitis B surface antigen. New Engl J Med. 1985;312:270–276. doi: 10.1056/NEJM198501313120503. [DOI] [PubMed] [Google Scholar]
- 26.Wands JR, et al. Identification and transmission of hepatitis B virus-related variants. Proc Natl Acad Sci USA. 1986;83:6608–6612. doi: 10.1073/pnas.83.17.6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carman WF, et al. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336:325–329. doi: 10.1016/0140-6736(90)91874-a. [DOI] [PubMed] [Google Scholar]
- 28.François G, Kew M, Van Damme P, Mphahlele MJ, Meheus A. Mutant hepatitis B viruses: A matter of academic interest only or a problem with far-reaching implications? Vaccine. 2001;19:3799–3815. doi: 10.1016/s0264-410x(01)00108-6. [DOI] [PubMed] [Google Scholar]
- 29.Coppola N, et al. Clinical significance of hepatitis B surface antigen mutants. World J Hepatol. 2015;7:2729–2739. doi: 10.4254/wjh.v7.i27.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pawlotsky JM. The concept of hepatitis B virus mutant escape. J Clin Virol. 2005;34:S125–S129. doi: 10.1016/s1386-6532(05)80021-6. [DOI] [PubMed] [Google Scholar]
- 31.Gerlich WH. Prophylactic vaccination against hepatitis B: Achievements, challenges and perspectives. Med Microbiol Immun. 2015;204:39–55. doi: 10.1007/s00430-014-0373-y. [DOI] [PubMed] [Google Scholar]
- 32.Luongo M, et al. Acute hepatitis B caused by a vaccine-escape HBV strain in vaccinated subject: Sequence analysis and therapeutic strategy. J Clin Virol. 2015;62:89–91. doi: 10.1016/j.jcv.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 33.Romanò L, et al. Hepatitis B vaccination: Are escape mutant viruses a matter of concern? Hum Vacc Immunother. 2015;11:53–57. doi: 10.4161/hv.34306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson JN, Nokes DJ, Carman WF. The predicted pattern of emergence of vaccine-resistant hepatitis B: A cause for concern? Vaccine. 1999;17:973–978. doi: 10.1016/s0264-410x(98)00313-2. [DOI] [PubMed] [Google Scholar]
- 35.Hsu HY, Chang MH, Liaw SH, Ni YH, Chen HL. Changes of hepatitis B surface antigen variants in carrier children before and after universal vaccination in Taiwan. Hepatology. 1999;30:1312–1317. doi: 10.1002/hep.510300511. [DOI] [PubMed] [Google Scholar]
- 36.Hsu HY, Chang M, Ni Y, Chen HL. Survey of hepatitis B surface variant infection in children 15 years after a nationwide vaccination programme in Taiwan. Gut. 2004;53:1499–1503. doi: 10.1136/gut.2003.034223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu HY, et al. No increase in prevalence of hepatitis B surface antigen mutant in a population of children and adolescents who were fully covered by universal infant immunization. J Infect Dis. 2010;201:1192–1200. doi: 10.1086/651378. [DOI] [PubMed] [Google Scholar]
- 38.Yimnoi P, et al. A molecular epidemiological study of the hepatitis B virus in Thailand after 22 years of universal immunization. J Med Virol. 2016;88:664–673. doi: 10.1002/jmv.24368. [DOI] [PubMed] [Google Scholar]
- 39.Klushkina VV, et al. Impact of universal hepatitis B vaccination on prevalence, infection-associated morbidity and mortality, and circulation of immune escape variants in Russia. PLoS One. 2016;11:e0157161. doi: 10.1371/journal.pone.0157161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aghasadeghi MR, et al. Low prevalence of hepatitis B vaccine escape mutants among individuals born after the initiation of a nationwide vaccination program in Iran. Arch Virol. 2016;161:3405–3411. doi: 10.1007/s00705-016-3050-1. [DOI] [PubMed] [Google Scholar]
- 41.Ogata N, Zanetti AR, Yu M, Miller RH, Purcell RH. Infectivity and pathogenicity in chimpanzees of a surface gene mutant of hepatitis B virus that emerged in a vaccinated infant. J Infect Dis. 1997;175:511–523. doi: 10.1093/infdis/175.3.511. [DOI] [PubMed] [Google Scholar]
- 42.Ogata N, et al. Licensed recombinant hepatitis B vaccines protect chimpanzees against infection with the prototype surface gene mutant of hepatitis B virus. Hepatology. 1999;30:779–786. doi: 10.1002/hep.510300309. [DOI] [PubMed] [Google Scholar]
- 43.Trépo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet. 2014;384:2053–2063. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- 44.Ghaffar F, Friedland IR, McCracken GH., Dynamics of nasopharyngeal colonization by Streptococcus pneumoniae. Pediatr Infect Dis J. 1999;18:638–646. doi: 10.1097/00006454-199907000-00016. [DOI] [PubMed] [Google Scholar]
- 45.Weinberger DM, Malley R, Lipsitch M. Serotype replacement in disease after pneumococcal vaccination. Lancet. 2011;378:1962–1973. doi: 10.1016/S0140-6736(10)62225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lipsitch M. Bacterial vaccines and serotype replacement: Lessons from Haemophilus influenzae and prospects for Streptococcus pneumoniae. Emerg Infect Dis. 1999;5:336–345. doi: 10.3201/eid0503.990304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Obaro SK, Adegbola RA, Banya WAS, Greenwood BM. Carriage of pneumococci after pneumococcal vaccination. Lancet. 1996;348:271–272. doi: 10.1016/s0140-6736(05)65585-7. [DOI] [PubMed] [Google Scholar]
- 48.Brueggemann AB, Pai R, Crook DW, Beall B. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 2007;3:e168. doi: 10.1371/journal.ppat.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hausdorff WP, Hoet B, Schuerman L. Do pneumococcal conjugate vaccines provide any cross-protection against serotype 19A? BMC Pediatr. 2010;10:4. doi: 10.1186/1471-2431-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pilishvili T, et al. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J Infect Dis. 2010;201:32–41. doi: 10.1086/648593. [DOI] [PubMed] [Google Scholar]
- 51.Moore MR, et al. Effect of use of 13-valent pneumococcal conjugate vaccine in children on invasive pneumococcal disease in children and adults in the USA: Analysis of multisite, population-based surveillance. Lancet Infect Dis. 2015;15:301–309. doi: 10.1016/S1473-3099(14)71081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cherry JD. Epidemic pertussis in 2012–The resurgence of a vaccine-preventable disease. New Engl J Med. 2012;367:785–787. doi: 10.1056/NEJMp1209051. [DOI] [PubMed] [Google Scholar]
- 53.van der Zee A, Vernooij S, Peeters M, van Embden J, Mooi FR. Dynamics of the population structure of Bordetella pertussis as measured by IS 1002-associated RFLP: Comparison of pre-and post-vaccination strains and global distribution. Microbiology. 1996;142:3479–3485. doi: 10.1099/13500872-142-12-3479. [DOI] [PubMed] [Google Scholar]
- 54.Mooi FR, et al. Polymorphism in the Bordetella pertussis virulence factors p. 69/pertactin and pertussis toxin in The Netherlands: Temporal trends and evidence for vaccine-driven evolution. Infect Immun. 1998;66:670–675. doi: 10.1128/iai.66.2.670-675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gzyl A, et al. Sequence variation in pertussis S1 subunit toxin and pertussis genes in Bordetella pertussis strains used for the whole-cell pertussis vaccine produced in Poland since 1960: Efficiency of the DTwP vaccine-induced immunity against currently circulating B. pertussis isolates. Vaccine. 2004;22:2122–2128. doi: 10.1016/j.vaccine.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 56.Mastrantonio P, et al. Antigenic variants in Bordetella pertussis strains isolated from vaccinated and unvaccinated children. Microbiology. 1999;145:2069–2075. doi: 10.1099/13500872-145-8-2069. [DOI] [PubMed] [Google Scholar]
- 57.Bouchez V, et al. First report and detailed characterization of B. pertussis isolates not expressing pertussis toxin or pertactin. Vaccine. 2009;27:6034–6041. doi: 10.1016/j.vaccine.2009.07.074. [DOI] [PubMed] [Google Scholar]
- 58.Barkoff AM, et al. Appearance of Bordetella pertussis strains not expressing the vaccine antigen pertactin in Finland. Clin Vaccine Immunol. 2012;19:1703–1704. doi: 10.1128/CVI.00367-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Otsuka N, et al. Prevalence and genetic characterization of pertactin-deficient Bordetella pertussis in Japan. PLoS One. 2012;7:e31985. doi: 10.1371/journal.pone.0031985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lam C, et al. Rapid increase in pertactin-deficient Bordetella pertussis isolates, Australia. Emerg Infect Dis. 2014;20:626–633. doi: 10.3201/eid2004.131478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pawloski LC, et al. Prevalence and molecular characterization of pertactin-deficient Bordetella pertussis in the United States. Clin Vaccine Immunol. 2014;21:119–125. doi: 10.1128/CVI.00717-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hiramatsu Y, et al. Significant decrease in pertactin-deficient Bordetella pertussis isolates, Japan. Emerg Infect Dis. 2017;23:699–701. doi: 10.3201/eid2304.161575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hegerle N, Dore G, Guiso N. Pertactin deficient Bordetella pertussis present a better fitness in mice immunized with an acellular pertussis vaccine. Vaccine. 2014;32:6597–6600. doi: 10.1016/j.vaccine.2014.09.068. [DOI] [PubMed] [Google Scholar]
- 64.Safarchi A, et al. Pertactin negative Bordetella pertussis demonstrates higher fitness under vaccine selection pressure in a mixed infection model. Vaccine. 2015;33:6277–6281. doi: 10.1016/j.vaccine.2015.09.064. [DOI] [PubMed] [Google Scholar]
- 65.Martin SW, et al. Pertactin-negative Bordetella pertussis strains: Evidence for a possible selective advantage. Clin Infect Dis. 2015;60:223–227. doi: 10.1093/cid/ciu788. [DOI] [PubMed] [Google Scholar]
- 66.Mooi FR, et al. Bordetella pertussis strains with increased toxin production associated with pertussis resurgence. Emerg Infect Dis. 2009;15:1206–1213. doi: 10.3201/eid1508.081511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Advani A, Gustafsson L, Carlsson R, Donnelly D, Hallander HO. Clinical outcome of pertussis in Sweden: Association with pulsed-field gel electrophoresis profiles and serotype. APMIS. 2007;115:736–742. doi: 10.1111/j.1600-0463.2007.apm_628.x. [DOI] [PubMed] [Google Scholar]
- 68.Bart MJ, et al. Global population structure and evolution of Bordetella pertussis and their relationship with vaccination. MBio. 2014;5:e01074. doi: 10.1128/mBio.01074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Octavia S, et al. Newly emerging clones of Bordetella pertussis carrying prn2 and ptxP3 alleles implicated in Australian pertussis epidemic in 2008–2010. J Infect Dis. 2012;205:1220–1224. doi: 10.1093/infdis/jis178. [DOI] [PubMed] [Google Scholar]
- 70.Clarke M, et al. The relationship between Bordetella pertussis genotype and clinical severity in Australian children with pertussis. J Infect. 2016;72:171–178. doi: 10.1016/j.jinf.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 71.Carbonetti NH, Artamonova GV, Van Rooijen N, Ayala VI. Pertussis toxin targets airway macrophages to promote Bordetella pertussis infection of the respiratory tract. Infect Immun. 2007;75:1713–1720. doi: 10.1128/IAI.01578-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kirimanjeswara GS, Agosto LM, Kennett MJ, Bjornstad ON, Harvill ET. Pertussis toxin inhibits neutrophil recruitment to delay antibody-mediated clearance of Bordetella pertussis. J Clin Invest. 2005;115:3594–3601. doi: 10.1172/JCI24609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mielcarek N, et al. Homologous and heterologous protection after single intranasal administration of live attenuated recombinant Bordetella pertussis. Nat Biotechnol. 1998;16:454–457. doi: 10.1038/nbt0598-454. [DOI] [PubMed] [Google Scholar]
- 74.Safarchi A, et al. Better colonisation of newly emerged Bordetella pertussis in the co-infection mouse model study. Vaccine. 2016;34:3967–3971. doi: 10.1016/j.vaccine.2016.06.052. [DOI] [PubMed] [Google Scholar]
- 75.Burns DL, Meade BD, Messionnier NE. Pertussis resurgence: Perspectives from the working group meeting on pertussis on the causes, possible paths forward, and gaps in our knowledge. J Infect Dis. 2014;209:S32–S35. doi: 10.1093/infdis/jit491. [DOI] [PubMed] [Google Scholar]
- 76.Bolotin S, Harvill ET, Crowcroft NS. What to do about pertussis vaccines? Linking what we know about pertussis vaccine effectiveness, immunology and disease transmission to create a better vaccine. Pathog Dis. 2015;73:ftv057. doi: 10.1093/femspd/ftv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Plotkin SA. The pertussis problem. Clin Infect Dis. 2014;58:830–833. doi: 10.1093/cid/cit934. [DOI] [PubMed] [Google Scholar]
- 78.Davies RL, Frerichs GN. Morphological and biochemical differences among isolates of Yersinia ruckeri obtained from wide geographical areas. J Fish Dis. 1989;12:357–365. [Google Scholar]
- 79.Stevenson RM. Immunization with bacterial antigens: Yersiniosis. Dev Biol Stand. 1997;90:117–124. [PubMed] [Google Scholar]
- 80.Austin DA, Robertson PAW, Austin B. Recovery of a new biogroup of Yersinia ruckeri from diseased rainbow trout (Oncorhynchus mykiss, Walbaum) Syst Appl Microbiol. 2003;26:127–131. doi: 10.1078/072320203322337416. [DOI] [PubMed] [Google Scholar]
- 81.Fouz B, Zarza C, Amaro C. First description of non-motile Yersinia ruckeri serovar I strains causing disease in rainbow trout, Oncorhynchus mykiss (Walbaum), cultured in Spain. J Fish Dis. 2006;29:339–346. doi: 10.1111/j.1365-2761.2006.00723.x. [DOI] [PubMed] [Google Scholar]
- 82.Arias CR, et al. First report of Yersinia ruckeri biotype 2 in the USA. J Aquat Anim Health. 2007;19:35–40. doi: 10.1577/H06-011.1. [DOI] [PubMed] [Google Scholar]
- 83.Evenhuis JP, et al. Transfer of serum and cells from Yersinia ruckeri vaccinated doubled-haploid hot creek rainbow trout into outcross F1 progeny elucidates mechanisms of vaccine-induced protection. Dev Comp Immunol. 2014;44:145–151. doi: 10.1016/j.dci.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 84.Welch TJ, LaPatra S. Yersinia ruckeri lipopolysaccharide is necessary and sufficient for eliciting a protective immune response in rainbow trout (Oncorhynchus mykiss, Walbaum) Fish Shellfish Immunol. 2016;49:420–426. doi: 10.1016/j.fsi.2015.12.037. [DOI] [PubMed] [Google Scholar]
- 85.Welch TJ, et al. Independent emergence of Yersinia ruckeri biotype 2 in the United States and Europe. Appl Environ Microbiol. 2011;77:3493–3499. doi: 10.1128/AEM.02997-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wangkahart E, Scott C, Secombes CJ, Wang T. Re-examination of the rainbow trout (Oncorhynchus mykiss) immune response to flagellin: Yersinia ruckeri flagellin is a potent activator of acute phase proteins, anti-microbial peptides and pro-inflammatory cytokines in vitro. Dev Comp Immunol. 2016;57:75–87. doi: 10.1016/j.dci.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 87.Tinsley JW, Lyndon AR, Austin B. Antigenic and cross-protection studies of biotype 1 and biotype 2 isolates of Yersinia ruckeri in rainbow trout, Oncorhynchus mykiss (Walbaum) J Appl Microbiol. 2011;111:8–16. doi: 10.1111/j.1365-2672.2011.05020.x. [DOI] [PubMed] [Google Scholar]
- 88.Chettri JK, et al. Comparative evaluation of administration methods for a vaccine protecting rainbow trout against Yersinia ruckeri O1 biotype 2 infections. Vet Immunol Immunopathol. 2013;154:42–47. doi: 10.1016/j.vetimm.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 89.Easton AJ, Domachowske JB, Rosenberg HF. Animal pneumoviruses: Molecular genetics and pathogenesis. Clin Microbiol Rev. 2004;17:390–412. doi: 10.1128/CMR.17.2.390-412.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cook J. Avian pneumovirus infections of turkeys and chickens. Vet J. 2000;160:118–125. doi: 10.1053/tvjl.2000.0486. [DOI] [PubMed] [Google Scholar]
- 91.Banet-Noach C, Simanov L, Laham-Karam N, Perk S, Bacharach E. Longitudinal survey of avian metapneumoviruses in poultry in Israel: Infiltration of field strains into vaccinated flocks. Avian Dis. 2009;53:184–189. doi: 10.1637/8466-090408-Reg.1. [DOI] [PubMed] [Google Scholar]
- 92.Catelli E, et al. Field avian metapneumovirus evolution avoiding vaccine induced immunity. Vaccine. 2010;28:916–921. doi: 10.1016/j.vaccine.2009.10.149. [DOI] [PubMed] [Google Scholar]
- 93.Cecchinato M, et al. Avian metapneumovirus (AMPV) attachment protein involvement in probable virus evolution concurrent with mass live vaccine introduction. Vet Microbiol. 2010;146:24–34. doi: 10.1016/j.vetmic.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 94.Naylor CJ, Ling R, Edworthy N, Savage CE, Easton AJ. Avian metapneumovirus SH gene end and G protein mutations influence the level of protection of live-vaccine candidates. J Gen Virol. 2007;88:1767–1775. doi: 10.1099/vir.0.82755-0. [DOI] [PubMed] [Google Scholar]
- 95.Franzo G, et al. First report of avian metapneumovirus subtype B field strain in a Romanian broiler flock during an outbreak of respiratory disease. Avian Dis. 2017;61:250–254. doi: 10.1637/11557-121216-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 96.Sun J, et al. Methyltransferase-defective avian metapneumovirus vaccines provide complete protection against challenge with the homologous Colorado strain and the heterologous Minnesota strain. J Virol. 2014;88:12348–12363. doi: 10.1128/JVI.01095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hu H, Roth JP, Zsak L, Yu Q. Engineered Newcastle disease virus expressing the F and G proteins of AMPV-C confers protection against challenges in turkeys. Sci Rep. 2017;7:4025. doi: 10.1038/s41598-017-04267-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Osterrieder N, Kamil JP, Schumacher D, Tischer BK, Trapp S. Marek’s disease virus: From miasma to model. Nat Rev Microbiol. 2006;4:283–294. doi: 10.1038/nrmicro1382. [DOI] [PubMed] [Google Scholar]
- 99.Davison F, Nair V. Use of Marek’s disease vaccines: Could they be driving the virus to increasing virulence? Expert Rev Vaccines. 2005;4:77–88. doi: 10.1586/14760584.4.1.77. [DOI] [PubMed] [Google Scholar]
- 100.Biggs PM, Nair V. The long view: 40 years of Marek’s disease research and avian pathology. Avian Pathol. 2012;41:3–9. doi: 10.1080/03079457.2011.646238. [DOI] [PubMed] [Google Scholar]
- 101.Sharma JM. Laboratory diagnosis. In: Payne LN, editor. Marek’s Disease: Scientific Basis and Methods of Control. Martinus Nijhoff; Boston: 1985. pp. 151–175. [Google Scholar]
- 102.Purchase HG, Okazaki W, Burmester BR. Long-term field trials with the herpesvirus of turkeys vaccine against Marek’s disease. Avian Dis. 1972;16:57–71. [PubMed] [Google Scholar]
- 103.Witter RL. Historic incidence of Marek’s disease as revealed by condemnation statistics. In: Silva RF, Cheng HH, Coussens PM, Lee LF, Velicer LF, editors. Current Research on Marek’s Disease: Proceedings of the 5th International Symposium on Marek’s Disease. American Association of Avian Pathologists; Kennett Square, PA: 1996. pp. 501–508. [Google Scholar]
- 104.Witter RL. Characteristics of Marek’s disease viruses isolated from vaccinated commercial chicken flocks: Association of viral pathotype with lymphoma frequency. Avian Dis. 1983;27:113–132. [PubMed] [Google Scholar]
- 105.Witter RL. Increased virulence of Marek’s disease virus field isolates. Avian Dis. 1997;41:149–163. [PubMed] [Google Scholar]
- 106.Read AF, et al. Imperfect vaccination can enhance the transmission of highly virulent pathogens. PLoS Biol. 2015;13:e1002198. doi: 10.1371/journal.pbio.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kennedy DA, Dunn JR, Dunn PA, Read AF. An observational study of the temporal and spatial patterns of Marek’s-disease-associated leukosis condemnation of young chickens in the United States of America. Prev Vet Med. 2015;120:328–335. doi: 10.1016/j.prevetmed.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Calnek BW, Schat KA, Peckham MC, Fabricant J. Field trials with a bivalent vaccine (HVT and SB-1) against Marek’s disease. Avian Dis. 1983;27:844–849. [PubMed] [Google Scholar]
- 109.Witter RL. 4th International Symposium on Marek’s Disease, 19th World’s Poultry Congress. Vol 1. World’s Poultry Science Association; Amsterdam: 1992. Safety and comparative efficacy of the CVI988/Rispens vaccine strain; pp. 315–319. [Google Scholar]
- 110.Finlay BB, McFadden G. Anti-immunology: Evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 111.Schmid-Hempel P. Parasite immune evasion: A momentous molecular war. Trends Ecol Evol. 2008;23:318–326. doi: 10.1016/j.tree.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 112.Plotkin SA. Correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47:401–409. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 113.Graham AL, Allen JE, Read AF. Evolutionary causes and consequences of immunopathology. Annu Rev Ecol Evol Syst. 2005;36:373–397. [Google Scholar]
- 114.Luepke KH, et al. Past, present, and future of antibacterial economics: Increasing bacterial resistance, limited antibiotic pipeline, and societal implications. Pharmacotherapy. 2017;37:71–84. doi: 10.1002/phar.1868. [DOI] [PubMed] [Google Scholar]
- 115.Lee CW, Senne DA, Suarez DL. Effect of vaccine use in the evolution of Mexican lineage H5N2 avian influenza virus. J Virol. 2004;78:8372–8381. doi: 10.1128/JVI.78.15.8372-8381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lu H, et al. Isolation and molecular characterization of newly emerging avian reovirus variants and novel strains in Pennsylvania, USA, 2011–2014. Sci Rep. 2015;5:14727. doi: 10.1038/srep14727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Soubeyrand B, Plotkin SA. Microbial evolution (communication arising): Antitoxin vaccines and pathogen virulence. Nature. 2002;417:609–610. doi: 10.1038/417609b. [DOI] [PubMed] [Google Scholar]
- 118.Radford AD, Dawson S, Coyne KP, Porter CJ, Gaskell RM. The challenge for the next generation of feline calicivirus vaccines. Vet Microbiol. 2006;117:14–18. doi: 10.1016/j.vetmic.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 119.Ribeiro GS, et al. Prevention of Haemophilus influenzae type b (Hib) meningitis and emergence of serotype replacement with type a strains after introduction of Hib immunization in Brazil. J Infect Dis. 2003;187:109–116. doi: 10.1086/345863. [DOI] [PubMed] [Google Scholar]
- 120.Van Den Berg TP. Acute infectious bursal disease in poultry: A review. Avian Pathol. 2000;29:175–194. doi: 10.1080/03079450050045431. [DOI] [PubMed] [Google Scholar]
- 121.Kertesz DA, Coulthart MB, Ryan JA, Johnson WM, Ashton FE. Serogroup B, electrophoretic type 15 Neisseria meningitidis in Canada. J Infect Dis. 1998;177:1754–1757. doi: 10.1086/517439. [DOI] [PubMed] [Google Scholar]
- 122.van Boven M, et al. Herd immunity to Newcastle disease virus in poultry by vaccination. Avian Pathol. 2008;37:1–5. doi: 10.1080/03079450701772391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Franzo G, Tucciarone CM, Cecchinato M, Drigo M. Porcine circovirus type 2 (PCV2) evolution before and after the vaccination introduction: A large scale epidemiological study. Sci Rep. 2016;6:39458. doi: 10.1038/srep39458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chambers MA, Graham SP, La Ragione RM. 2016. Challenges in veterinary vaccine development and immunization. Vaccines for Veterinary Diseases, Vaccine Design: Methods and Protocols, ed Thomas S (Humana Press, New York. ), Vol 2, pp 3–35. [DOI] [PMC free article] [PubMed]
- 125.Halloran ME, Haber M, Longini IM., , Struchiner CJ. Direct and indirect effects in vaccine efficacy and effectiveness. Am J Epidemiol. 1991;133:323–331. doi: 10.1093/oxfordjournals.aje.a115884. [DOI] [PubMed] [Google Scholar]
- 126.Mackinnon MJ, Read AF. Immunity promotes virulence evolution in a malaria model. PLoS Biol. 2004;2:E230. doi: 10.1371/journal.pbio.0020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Barclay VC, et al. The evolutionary consequences of blood-stage vaccination on the rodent malaria Plasmodium chabaudi. PLoS Biol. 2012;10:e1001368. doi: 10.1371/journal.pbio.1001368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gandon S, Mackinnon MJ, Nee S, Read AF. Imperfect vaccines and the evolution of pathogen virulence. Nature. 2001;414:751–756. doi: 10.1038/414751a. [DOI] [PubMed] [Google Scholar]
- 129.Boots M. The need for evolutionarily rational disease interventions: Vaccination can select for higher virulence. PLoS Biol. 2015;13:e1002236. doi: 10.1371/journal.pbio.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hyde JE. Drug-resistant malaria. Trends Parasitol. 2005;21:494–498. doi: 10.1016/j.pt.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Read AF, Huijben S. Evolutionary biology and the avoidance of antimicrobial resistance. Evol Appl. 2009;2:40–51. doi: 10.1111/j.1752-4571.2008.00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McClure NS, Day T. A theoretical examination of the relative importance of evolution management and drug development for managing resistance. Proc R Soc Lond B Biol Sci. 2014;281:20141861. doi: 10.1098/rspb.2014.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hilleman MR. Six decades of vaccine development – A personal history. Nat Med. 1998;4:507–514. doi: 10.1038/nm0598supp-507. [DOI] [PubMed] [Google Scholar]
- 134.Plotkin SL, Plotkin SA. A short history of vaccination. In: Plotkin SA, Orenstein WA, Offit PA, editors. Vaccines. 5th Ed. Elsevier; Philadelphia: 2008. pp. 1–16. [Google Scholar]
- 135.Hegerle N, et al. Evolution of French Bordetella pertussis and Bordetella parapertussis isolates: Increase of Bordetellae not expressing pertactin. Clin Microbiol Infect. 2012;18:E340–E346. doi: 10.1111/j.1469-0691.2012.03925.x. [DOI] [PubMed] [Google Scholar]
- 136.Hambrosky J, Kroger A, Wolfe C, editors. Centers for Disease Control and Prevention. Epidemiology and Prevention of Vaccine-Preventable Diseases. 13th Ed Public Health Foundation; Washington DC: 2015. [Google Scholar]
- 137.Centers for Disease Control and Prevention 2017 Pertussis cases by year (1922–2015). Available at https://www.cdc.gov/pertussis/surv-reporting/cases-by-year.html. Accessed November 28, 2017.
- 138.Centers for Disease Control and Prevention 2017 Surveillance and reporting. Available at https://www.cdc.gov/pneumococcal/surveillance.html. Accessed November 28, 2017.
- 139.Centers for Disease Control and Prevention 2017 Historical reported cases and estimates of viral hepatitis. Available at https://www.cdc.gov/hepatitis/statistics/incidencearchive.htm. Accessed November 28, 2017.
- 140.Kennedy DA, et al. Industry-wide surveillance of Marek’s disease virus on commercial poultry farms. Avian Dis. 2017;61:153–164. doi: 10.1637/11525-110216-Reg.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Witter RL, Solomon JJ, Champion LR, Nazerian K. Long-term studies of Marek’s disease infection in individual chickens. Avian Dis. 1971;15:346–365. [PubMed] [Google Scholar]
- 142.Islam AFMF, Walkden-Brown SW, Groves PJ, Underwood GJ. Kinetics of Marek’s disease virus (MDV) infection in broiler chickens 1: Effect of varying vaccination to challenge interval on vaccinal protection and load of MDV and herpesvirus of Turkey in the spleen and feather dander over time. Avian Pathol. 2008;37:225–235. doi: 10.1080/03079450701802230. [DOI] [PubMed] [Google Scholar]
- 143.Kennedy DA, Dunn PA, Read AF. Modeling Marek’s disease virus transmission: A framework for evaluating the impact of farming practices and evolution. Epidemics. 2018;23:85–95. doi: 10.1016/j.epidem.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]