Significance
It is thought that the distinct ether lipid membranes of archaea allow them to thrive in environmental extremes. However, it has been difficult to demonstrate this physiological role directly. Here, we identify a protein required for the biosynthesis of a unique archaeal lipid head group, calditol, whose production was considered to be restricted to a subset of archaeal thermoacidophiles. We show that deletion of this protein in Sulfolobus acidocaldarius prevents production of calditol-linked membrane lipids and, in turn, inhibits cell growth at extremely low pH. Our findings also suggest that archaea more broadly, like bacteria, employ radical S-adenosylmethionine proteins to modify membrane lipids in ways that confer protective effects when environmental conditions, such as pH, fluctuate significantly.
Keywords: calditol, glycerol dibiphytanyl glycerol tetraethers, GDGT, radical SAM, Sulfolobus
Abstract
Archaea have many unique physiological features of which the lipid composition of their cellular membranes is the most striking. Archaeal ether-linked isoprenoidal membranes can occur as bilayers or monolayers, possess diverse polar head groups, and a multiplicity of ring structures in the isoprenoidal cores. These lipid structures are proposed to provide protection from the extreme temperature, pH, salinity, and nutrient-starved conditions that many archaea inhabit. However, many questions remain regarding the synthesis and physiological role of some of the more complex archaeal lipids. In this study, we identify a radical S-adenosylmethionine (SAM) protein in Sulfolobus acidocaldarius required for the synthesis of a unique cyclopentyl head group, known as calditol. Calditol-linked glycerol dibiphytanyl glycerol tetraethers (GDGTs) are membrane spanning lipids in which calditol is ether bonded to the glycerol backbone and whose production is restricted to a subset of thermoacidophilic archaea of the Sulfolobales order within the Crenarchaeota phylum. Several studies have focused on the enzymatic mechanism for the synthesis of the calditol moiety, but to date no protein that catalyzes this reaction has been discovered. Phylogenetic analyses of this putative calditol synthase (Cds) reveal the genetic potential for calditol–GDGT synthesis in phyla other than the Crenarchaeota, including the Korarchaeota and Marsarchaeota. In addition, we identify Cds homologs in metagenomes predominantly from acidic ecosystems. Finally, we demonstrate that deletion of calditol synthesis renders S. acidocaldarius sensitive to extremely low pH, indicating that calditol plays a critical role in protecting archaeal cells from acidic stress.
One of the defining features that distinguishes archaea from both eukaryotes and bacteria is the structure of their membrane lipids (1, 2). Bacterial and eukaryotic membranes are composed of phospholipid bilayers made of fatty acid chains ester linked to glycerol-3-phosphate. Archaeal membranes have a similar overall amphiphilic structure but are composed of isoprenoidal chains ether bonded to glycerol-1-phosphate (3, 4). The majority of archaea can also generate membrane spanning monolayers known as glycerol dibiphytanyl glycerol tetraethers (GDGTs), which can contain cyclopentyl or cyclohexyl rings (SI Appendix, Fig. S1) (3, 4). In addition, archaea possess a diverse array of polar head groups, including phosphoesters of ethanolamine, serine, and myo-inositol, or glycosyl groups, such as glucose or galactose linked directly to the glycerol backbone (5, 6).
These unique archaeal membrane lipids are significant from several perspectives. From an evolutionary standpoint, the “lipid divide” between archaeal membranes and those of bacteria and eukaryotes raises interesting questions regarding the evolution of membrane lipid biosynthesis and what the membrane composition of the last universal common ancestor may have been (7–9). Geochemically, archaeal lipids detected in both modern environments and ancient sediments can function as taxonomic indicators and/or paleotemperature proxies (10–12). Archaeal biomarker studies have provided insight into a variety of ecological topics including the role of archaea in marine biogeochemistry, predictions of atmospheric and sea-surface temperatures in the Mesozoic and early Cenozoic, and the geologic history of archaea in ancient sediments (13, 14). However, deciphering the evolutionary implications of archaeal lipid biosynthesis, as well as being able to properly interpret archaeal lipid biomarkers, requires a full understanding of the biosynthetic pathways and the physiological roles of these various structures in extant archaea.
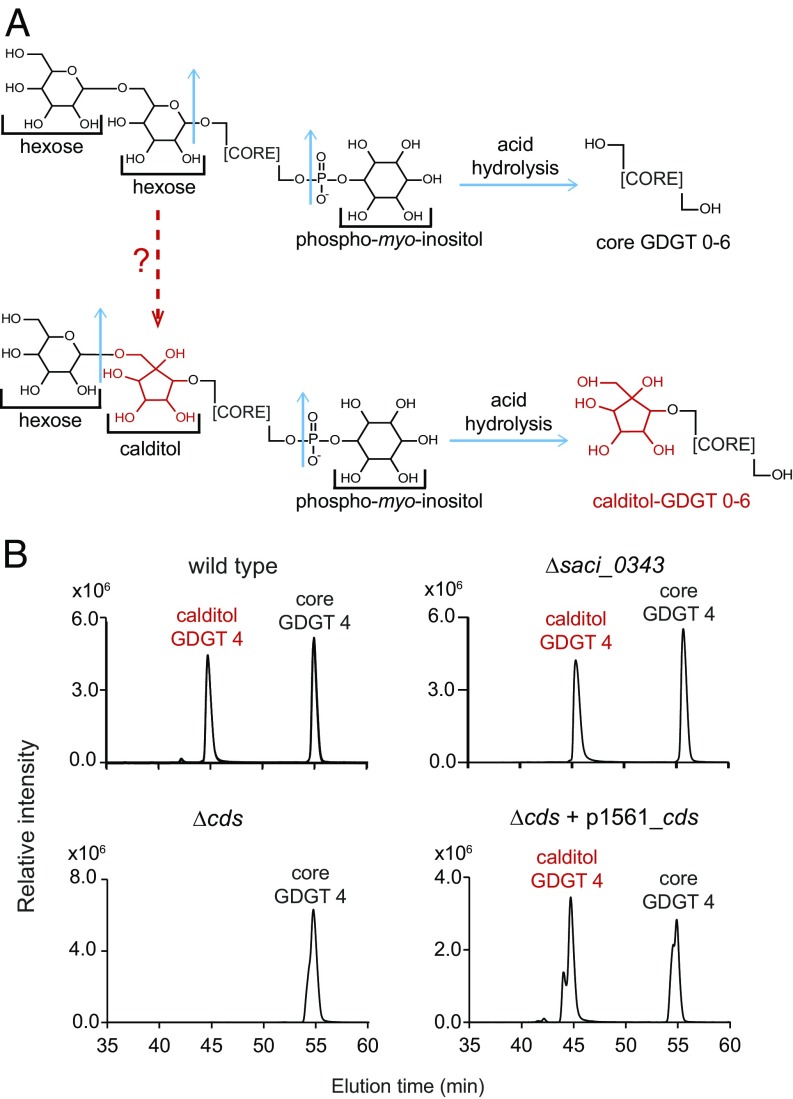
Studies of archaeal membrane lipid synthesis have revealed distinct proteins and biochemical reactions, but several open questions remain (4, 15). In particular, the production of a unique five-membered ring, calditol, that is ether-linked directly to the glycerol backbone of GDGTs is both an enzymatic and physiological mystery (Fig. 1) (16). Calditol-linked GDGTs were first discovered in Sulfolobus acidocaldarius, where they comprise up to 90% of the polar lipid fraction (17). Calditol–GDGTs have been detected subsequently in two other species of the Sulfolobales order within the Crenarchaeota phylum, Acidianus and Metallosphaera (18, 19). The enzyme(s) required for calditol synthesis has not been identified, although several structural and biosynthetic studies have validated the cyclic calditol structure (20–23) and demonstrated that calditol is derived from glucose (16, 24, 25).
Fig. 1.
The radical SAM protein encoded by cds is required for calditol synthesis. (A) Example of the polar head groups identified in archaeal membranes including the calditol moiety (shown in red). The top lipid is a dihexose phospho-myo-inositol GDGT, and the bottom lipid is monohexose calditol phospho-myo-inositol GDGT. The blue arrows represent where cleavage occurs when lipid extracts of S. acidocaldarius are acid-hydrolyzed, generating either a core GDGT or calditol–GDGT. (B) Merged extracted ion chromatograms (m/z 1456 and 1294) of acid hydrolyzed lipid fractions from S. acidocaldarius WT cells, Δsaci_0343 cells, Δcds (Δsaci_1489) cells, and Δcds cells complemented with the cds gene expressed on a self-replicating plasmid showing that the cds gene is required to produce calditol-linked GDGTs. For simplicity, only GDGT 4 core structures are shown. Total ion chromatograms showing all GDGT structures (GDGTs 0–6) can be found in SI Appendix, Fig. S2.
The physiological significance of calditol-linked membranes is also unknown. All calditol-producing archaea isolated thus far are thermoacidophiles with a pH range of 0.4–6 and a temperature range of 44 °C–85 °C (18, 19, 26). The ability of these calditol-producing archaea to grow in such environmental extremes has prompted the hypothesis that calditol-linked membranes contribute to their persistence under these conditions (25). Studies of the biophysical properties of various tetraether liposomes, as well as computer simulations of these membrane structures, have demonstrated their remarkable stability, low proton permeability, and tight membrane packing under various conditions (17). However, these studies utilized mixed GDGT structures and did not directly address how calditol head groups might contribute to these membrane properties.
Here, we utilized comparative genomics and gene deletion analyses to identify a gene locus encoding a radical S-adenosylmethionine (SAM) protein required for the formation of calditol in S. acidocaldarius. Bioinformatics analyses of this calditol synthase revealed the potential for calditol synthesis in uncultured archaeal phyla, as well as a predominant occurrence of this protein in thermoacidophilic archaea and in acidic ecosystems. Furthermore, we show that calditol is required for growth in S. acidocaldarius under extremely acidic conditions, directly demonstrating that calditol-linked lipids provide a protective role under environmentally relevant conditions.
Results
Identification of a Calditol Synthesis Protein in S. acidocaldarius.
Previous studies have proposed that a cyclase-like enzyme synthesized the formation of calditol through the C-4 oxidation of glucose in an “inositol-like” mechanism, followed by etherification to the core GDGT lipid (16, 24, 25). However, a second mechanism has been put forth that involves an unusual ring contraction of a glucose molecule linked to the GDGT core (27). This mechanism is similar to the proposed formation of the cyclitol ether side chain of bacteriohopanepolyols (BHPs). Synthesis of this BHP side chain requires a radical SAM protein encoded by the hpnJ gene (27, 28). Radical SAM enzymes catalyze a diverse set of difficult reactions through the generation of a 5-deoxyadenosyl radical (dAdo·) intermediate (29). Therefore, we hypothesized that a radical SAM protein may also be required to synthesize the calditol carbocycle observed in Sulfolobus membrane lipids.
We searched the S. acidocaldarius genome for all genes that encode proteins with a radical SAM motif [protein family (Pfam) identifier pfam04055] and identified 18 candidates (SI Appendix, Table S1). We used the Basic Local Alignment Search Tool (BLASTP) (30) to determine if any of these radical SAM proteins had homologs (e value < 1e−50; >30% identity) in other calditol-producing archaea (18, 19) but were absent from non–calditol-producing species (31–33). We utilized this stringent e value because radical SAM proteins are primarily identified by the short amino acid sequence motif CxxxCxxC. As a result, less-stringent BLAST analyses of these proteins may pull out other radical SAM proteins that are not involved in calditol synthesis. Using this cutoff, 3 of the 18 candidate proteins, Saci_0343, Saci_0344, and Saci_1489, were found only in the calditol-producing archaea, and we attempted to delete all three genes separately in S. acidocaldarius. We were able to construct markerless deletion mutants of saci_0343 and saci_1489 but not saci_0344.
The saci_0343 and saci_1489 mutants were each grown under standard conditions and their lipids were analyzed by liquid chromatography–mass spectrometry (LC-MS). To confirm the presence of calditol, cell pellets were acid-hydrolyzed, as calditol head groups are resistant to acid treatment, whereas phosphorylated and hexose head groups are not (Fig. 1) (20). Calditol-containing GDGTs and core GDGTs were detected in both the WT and Δsaci_0343 lipid extracts, indicating that the saci_0343 gene is not required for calditol synthesis (Fig. 1 and SI Appendix, Fig. S2). However, lipid extracts from Δsaci_1489 cells contained only core GDGTs. We were able to restore calditol production in the saci_1489 deletion mutant by introducing a copy of the saci_1489 gene on a self-replicating plasmid into this strain (Fig. 1). This process confirmed that the saci_1489 gene is required to synthesize calditol in S. acidocaldarius, and we renamed this locus cds for calditol synthase.
We next aimed to determine if the cds mutant was capable of producing any hexose GDGT lipids or if the loss of calditol synthesis prevented GDGT glycosylation. To do so, both cds mutant and WT lipids were extracted from freeze-dried cells without prior acid hydrolysis to prevent the loss of the hexose head groups. These analyses revealed that the cds mutant still produced glycosylated GDGTs (SI Appendix, Figs. S3 and S4). In addition, derivatization of these nonhydrolyzed lipid extracts confirmed that the sugar moieties present in the membranes of the cds mutant were composed of only hexose sugars and not calditol. For example, the dihexose GDGT 4 in the cds mutant extracts and the monohexose calditol–GDGT 4 in the WT extracts both eluted at 38.5 min and both had a molecular mass ion of 1636 Da in the nonderivatized forms. However, acetylation of the lipid extracts demonstrated that the dihexose GDGT 4 from the cds mutant had a molecular ion of 1,971 Da, whereas the monohexose calditol–GDGT 4 from the WT extract had a molecular ion of 2,013 Da (SI Appendix, Figs. S3 and S4). This increase of 42 Da is due to the extra acetylated hydroxyl group on the calditol moiety that is absent in the hexose sugar. These data confirm that although the cds mutant does not synthesize calditol–GDGTs, it does still produce glycosylated GDGTs and raises the possibility that a hexose-linked lipid may be the precursor substrate for the calditol synthase.
Distribution of Calditol Synthase Genes in Genomes and Metagenomes.
Calditol production has been reported in only three species of the Sulfolobales order (18, 19, 26). To determine if any other archaea were capable of producing calditol-linked GDGTs, we searched for Cds homologs in the Joint Genome Institute Integrated Microbial Genomes & Microbiomes (JGI IMG/M) (34) database and in the National Center for Biotechnology Information (NCBI) nonredundant protein sequences collection. A BLASTP search returned 513 radical SAM protein homologs with an e value of 1e−50 or lower and with at least 30% amino acid identity. All 513 sequences were aligned and, after reducing protein redundancy, the final alignment had a total of 59 sequences with 16 sequences from genomes of cultured archaea, 34 sequences from environmental metagenomes, and 9 sequences from metagenome acquired genomes (MAGs) or single-cell acquired genomes (SAGs) (SI Appendix, Table S2).
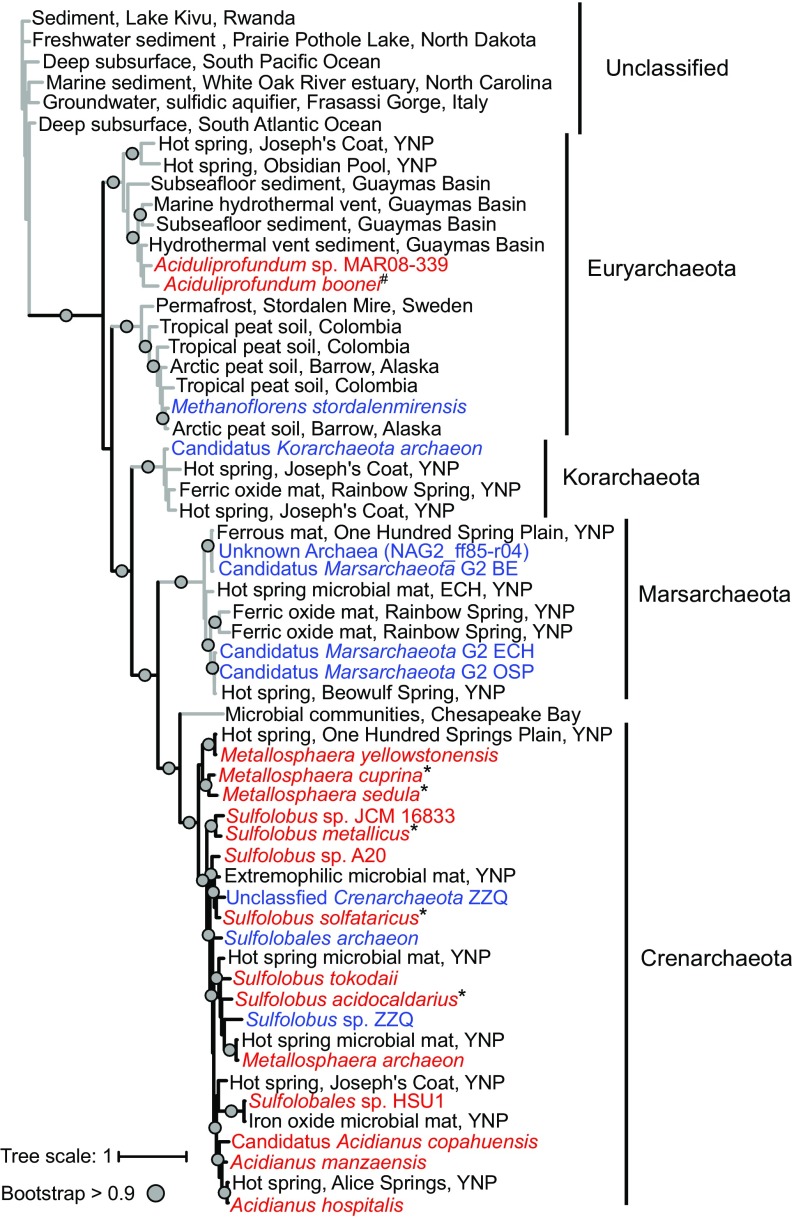
An unrooted phylogenetic tree of this alignment revealed five distinct clades representing four phyla: Crenarchaeota, Marsarchaeota, Korarchaeota, and Euryarchaeota (Fig. 2). A sixth clade was composed of environmental sequences only and did not cluster with any known archaeal species. The majority of Cds homologs from cultured organism genomes were restricted to the three known Sulfolobales species with all of the Sulfolobus (82 total), Acidianus (4 total), and Metallosphaera (12 total) genomes in the IMG/M database harboring a copy. Cds homologs were not present in any other order of the Crenarchaeota phylum. The non-Sulfolobales Cds homologs were primarily from MAGs and SAGs and not from cultured organisms (35–37). Lipid analyses have not been carried out directly from these uncultured organisms, and so it not yet possible to correlate the occurrence of Cds with calditol production in these phyla. However, one Euryarchaeota species in our tree, Aciduliprofundum boonei, has been cultured from deep-sea sediments and shown to not produce any calditol-linked GDGTs (38, 39). Cds homologs were also identified in a variety of environmental metagenomes, including terrestrial hot springs, hydrothermal vents, permafrost, and peat soil (SI Appendix, Table S3). The majority of the environmental metagenomes with a Cds homolog are from acidic ecosystems in which the reported pH ranges from 1 to 4, although some sites had a more neutral upper pH limit of 7 (35, 37, 40, 41).
Fig. 2.
Cds homologs are found primarily in acidophilic archaea and acidic environments. Unrooted maximum-likelihood phylogenetic tree of Cds homologs (e value < 1e−50; protein identity, >30%) identified in cultured archaeal genomes (red), SAGs or MAGs (blue), or metagenomes (black). A total of 513 homologs were identified, and after decreasing sequence redundancy in the alignment, 59 Cds homologs were used to generate the tree. Black lines indicate clades for which organisms have been tested and shown to produce calditol–GDGTs (*), and gray lines indicate clades for which lipid analyses have not been undertaken or lipid analyses did not show any calditol–GDGT production (#). Gray circles indicate branches that have bootstrap values greater than 0.9, and the scale bar represents one change per nucleotide site.
Calditol Containing Lipids Provide a Protective Effect Under Acidic Stress.
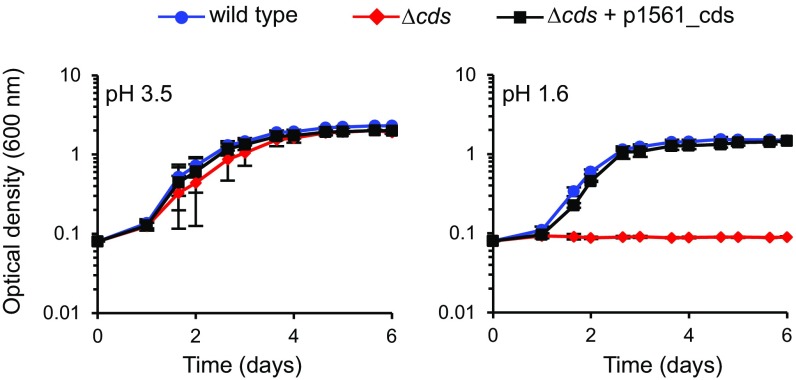
The distribution of environmental Cds homologs in primarily acidic environments (Fig. 2), as well as the inability to cleave calditol from the core GDGT structure through acid hydrolysis (Fig. 1), prompted us to hypothesize that calditol may provide a protective effect during growth at low pH. To test this hypothesis, we monitored the growth of the S. acidocaldarius cds mutant and the WT strain by optical density under varying pH conditions. The growth of the Δcds mutant was similar to the WT strain when grown under the optimal pH of 3.5 (Fig. 3). However, when the cells were grown at pH 1.6, the WT exhibited normal growth, whereas the cds deletion mutant did not grow at all. Growth at pH 1.6 was restored in the Δcds complemented mutant strain, indicating that the calditol-linked membrane lipids were essential under these conditions.
Fig. 3.
Calditol is required for growth at low pH in S. acidocaldarius. Growth of S. acidocaldarius wild type (blue circles), Δcds (red diamonds), and complemented Δcds (black squares) over 6 d at 75 °C at pH 3.5 (Left) or pH 1.6 (Right) showing that the calditol deletion mutant is unable to grow at pH 1.6. Growth is restored at pH 1.6 in Δcds when a copy of the cds gene is provided in trans. One representative growth curve done in triplicate is shown, and error bars indicate SDs.
Discussion
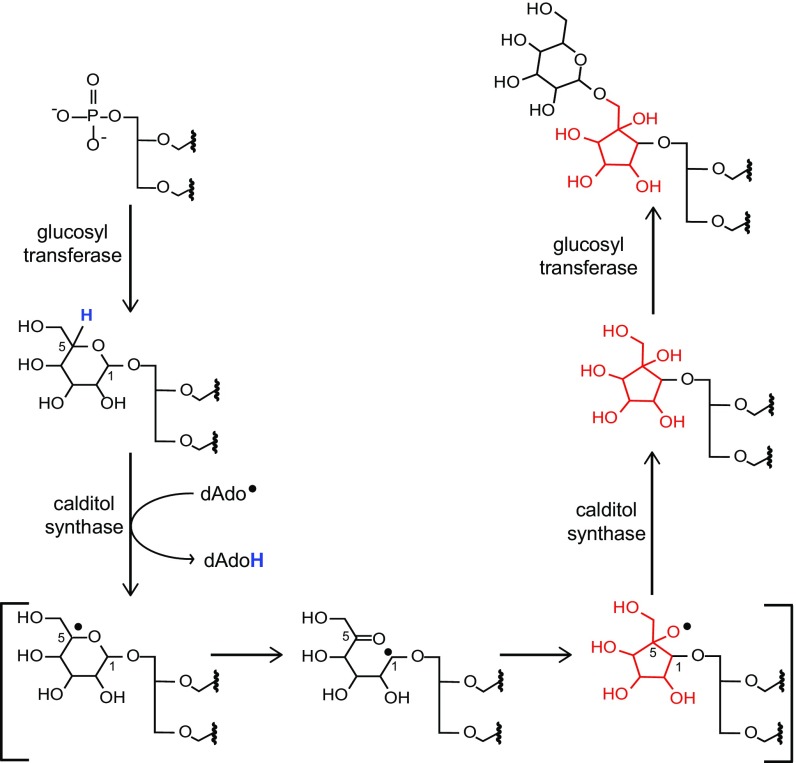
Our discovery of a calditol synthase and subsequent characterization of a calditol synthase mutant provides insight into an unusual membrane modification that was discovered in archaea over 40 years ago (21). The biosynthesis of calditol has been the focus of numerous studies, but the exact biochemical mechanism involved in generating this structure has remained elusive. Previous experiments with 14C- and deuterium-labeled glucose and galactose in Sulfolobus species demonstrated that the production of the calditol carbocycle occurs through a new C–C bond formation between C-1 and C-5 of glucose (16, 24, 25). This process is accompanied by an inversion of the stereochemistry of the C-4 hydroxyl and no hydrogen loss at C-2, C-3, C-4, or C-6 (16, 24, 25). These findings led to the proposal that calditol was most likely formed through a catalytic oxidoreduction at C-4 and that the formation of calditol occurs before etherification to the core GDGT (16, 25). However, the requirement of a radical SAM protein for calditol synthesis indicates that a radical mechanism could be involved instead. Specifically, we hypothesize that the calditol synthase generates a deoxyadenosyl radical that extracts a hydrogen from the C-5 position of a monohexose substrate (Fig. 4). Subsequent migration of this radical results in the conversion of the hexose ring into the five-membered ring structure of calditol. This type of ring contraction is unusual, but, in two instances, radical SAM chemistry has been implicated in this type of reaction: the biosynthesis of the cyclitol ether side chain of BHPs in Burkholderia cenocepacia and in the biosynthesis of 7-deazapurines in Bacillus subtilis (28, 42). In addition, we demonstrate that the cds mutant still produces glycosylated GDGTs, suggesting that a hexose bound glycerol ether lipid could be the substrate for the Cds protein. It should be noted that our genetic analyses do not rule out the possibility that calditol is synthesized before etherification and that a separate enzyme is required to transfer calditol to a diether or tetraether core lipid. Subsequent in vitro studies are needed to demonstrate this proposed radical mechanism directly, and our discovery of the putative enzyme that carries out this reaction makes these studies finally possible.
Fig. 4.
Proposed radical mechanism for calditol synthesis. A monohexose lipid is first synthesized by a glucosyltransferase that would be the substrate for the calditol synthase. Cds subsequently generates a dAdo• radical that extracts a hydrogen from the C-5 of the monohexose, producing a radical at this position. Migration of this radical would result in ring breakage at C-1, subsequent C–C bond formation between C-1 and C-5, and quenching of the final radical on the oxygen molecule. A second glucosyltransferase would subsequently add a hexose group to calditol to generate a monohexose calditol head group.
Our bioinformatics analyses of Cds also allows us to further explore the taxonomic distribution and phylogenetics of calditol synthesis. Previous lipid analyses have restricted calditol synthesis to the Sulfolobales order of the Crenarchaeota, and we observe that same distribution of the calditol synthase in genomes of cultured organisms. In addition, the phylogenetic grouping of Sulfolobales Cds homologs is congruent with the 16S phylogeny of Sulfolobales species (35), suggesting that calditol synthesis was acquired via vertical transfer in the Sulfolobales order. However, our metagenomic analyses identified homologs in other archaeal phyla, such as the Marsarchaeota and Korarchaeota, suggesting that calditol synthesis may have a more complex evolutionary history. The Marsarchaeota are a newly discovered archaeal phylum identified through single-cell genome sequencing of enrichments from acidic, high-temperature iron oxide microbial mats found in Yellowstone National Park (YNP) (35). Korarchaeota have also been detected through 16S rDNA analyses in a variety of terrestrial hot springs, including several from YNP, although these organisms typically compose a minor component of the microbial community (36). The Marsarchaeota and the Korarchaeota have been shown to co-occur in these environments with the Sulfolobales Crenarchaeota (Metallosphaera, Sulfolobus, and Acidianus) (43). The spatial proximity of these archaeal species may have potentially fostered the horizontal transfer of calditol-synthesis genes across these phyla. However, our assessment of whether the Marsarchaeota and Korarchaeota Cds homologs are true calditol synthases is constrained by the fact that species from these phyla have not been cultured or had their lipids analyzed. As more archaeal cultures and genomes become available, however, we will be able to better decipher the distribution of calditol synthesis and the evolutionary history of this pathway.
A subset of the Cds homologs identified in our search clustered with Euryarchaeota species, a finding that was surprising, as calditol production has not been observed in this phylum. One clade was restricted to acidic soil samples from arctic and tropical environments and included a homolog identified in the MAG of the permafrost methanogen Methanoflorens storedalenmirensis (37). To date, no cultured methanogens have been shown to produce calditol, and M. storedalenmirensis has not been isolated in culture to undertake lipid analyses. The second clade of putative Euryarchaeota Cds homologs includes metagenomes sequences from subsurface sediments from hydrothermal vents and from two cultured hydrothermal vent species of Aciduliprofundum. As mentioned before, lipid analyses of A. boonei did not reveal any calditol-containing ether lipids (39), suggesting that radical SAMs in this particular clade may be distinct from Cds and are perhaps carrying out a different function in these organisms.
In terms of the physiological role of calditol-linked tetraethers, it has been suggested that calditol provides a protective effect under extreme environmental conditions (17). However, there has been no in vivo experimental evidence to demonstrate this proposal directly. Here, we show that inhibition of calditol synthesis hinders the viability of S. acidocaldarius under extremely low pH conditions. Interestingly, deletion of calditol does not affect cell growth at the organisms’ optimal pH of 3.5 but rather only when the cells are exposed to a significant drop in pH from 3.5 to 1.6. We presume that the glycosylation of the core tetraethers observed in the cds mutant provides sufficient protection at pH 3.5 (44). However, at pH 1.6, the glyosidic bonds are less stable than the ether bond of calditol (25, 45). Exposure to this low pH could result in a breakdown of the hexose head groups in the cds mutant, and this reduction in glycosylation most likely leads to increased membrane permeability. In addition, our identification of putative Cds homologs in a variety of metagenomes from acidic ecosystems suggests that the protective effect observed in our physiological experiments with S. acidocaldarius may be relevant to other species. The ability to synthesize calditol could has evolved as a response to dramatic shifts in pH: many of the geothermal pools and sediments from which calditol-producing archaea have been discovered range dramatically in their pH values (46). Introducing calditol into their membranes may lead to a competitive advantage as calditol-producing cells would be poised to withstand a dramatic drop in pH in their surroundings. It would be interesting to test this hypothesis further either by developing methods to delete or prevent calditol synthesis in other calditol-producing archaea or by expressing cds in non–calditol-producing archaea, and determining if the induction of calditol synthesis in these organisms results in survival at lower pH.
Taken together, our results highlight the effectiveness of combining a bioinformatics approach with gene deletion and lipid analyses to address pertinent outstanding questions regarding archaeal membrane biosynthesis, function, and evolution. More broadly, we demonstrate the significant role that radical-mediated reactions, particularly those involving radical SAM proteins, may have in the end stages of lipid biosynthesis that could enhance the physiological flexibility of microbes. Previously, we have shown that structural modifications of bacterial hopanoids by radical SAM enzymes are important in withstanding various environmental stressors, including extreme changes in pH (28, 47, 48). The radical SAM-dependent formation of calditol in archaea demonstrates that these radical-mediated reactions might, in fact, be quite prevalent as a means to fine-tune lipid structures to enhance a microbe’s environmental flexibility. Given that radical mechanisms have been proposed as one potential option for other membrane modifications in archaea, namely the formation of GDGTs with cyclized biphytanyl moieties (49, 50), the possibility exists that further evidence will emerge demonstrating that membrane modifications by SAM-dependent enzymes is a common adaptation strategy in a broader swath of archaea.
Methods
Microbial Strains, Media, and Growth Conditions.
Strains used in this study are listed in SI Appendix, Table S4. Escherichia coli strains were cultured in lysogeny broth (LB) at 37 °C with shaking at 225 rpm. S. acidocaldarius strains were cultured at 75 °C in Brock medium supplemented with 0.1% N-Z-Amine and 0.2% sucrose, and buffered with 5 mM sodium citrate (26, 51). The pH was adjusted with sulfuric acid to pH 3.5 or 1.6, and cultures were aerated on a magnetic stir plate with stirring at 200 rpm. The S. acidocaldarius MW001 parent and mutant strains are uracil auxotrophs and were supplemented with 10 μg/mL uracil. Complemented strains (containing pSVA1561 plasmids) were supplemented with 0.4% maltose to induce protein expression. LB was supplemented, if necessary, with 100 μg/mL ampicillin or 50 μg/mL kanamycin for E. coli. For growth on solid medium, LB was solidified with 1.5% agar and Brock medium with 0.6% Gelrite.
Molecular Cloning.
All plasmids and oligonucleotides used in this study are described in SI Appendix, Tables S5 and S6. Details of cloning and gene deletion procedures are described in SI Appendix, SI Methods.
Construction of S. acidocaldarius Mutants and Complementation.
S. acidocaldarius markerless gene deletions were constructed through homologous recombination-mediated allelic replacement as previously described by Wagner et al. (51) with minor modifications outlined in SI Appendix. For complementation, the S. acidocaldarius MW001 parent strain was transformed with the empty vector pSVA1561, and Δsaci_1489 mutant was transformed with either the empty vector or the complementation plasmid pSVA1561-saci_1489. All strains were cultured in 100 mL of Brock medium supplemented with 0.1% N-Z-Amine, 0.2% sucrose, and 0.4% maltose at 75 °C. Cells were harvested for lipid extraction at stationary phase.
Growth Curves.
Fifty-milliliter starter cultures of S. acidocaldarius Δsaci_1489 strains harboring pSVA1561 or pSVA1561-saci_1489 and the MW001 parent strain harboring pSVA1561 were grown to midexponential phase at 75 °C. The starter cultures were used to inoculate 100 mL of Brock medium supplemented with 0.1% N-Z-Amine, 0.2% sucrose, 0.4% maltose, and buffered with 5 mM sodium citrate in a 250-mL flask (1% inoculum; in triplicate). The pH of the growth medium was adjusted with sulfuric acid to pH 3.5 or pH 1.6 and monitored throughout growth to ensure no significant change in pH occurred. Cultures were set on magnetic stir plate with stirring 200 rpm within an oven at 75 °C. Growth for 6 d was monitored by measuring the absorbance at 600 nm of a 200-μL aliquot in a Synergy 2 Microplate Reader (BioTek).
Lipid Extractions and Analyses.
S. acidocaldarius cultures (45 mL) were harvested for lipid analysis by centrifugation at 11,000 × g for 10 min, and pellets were stored at −20 °C before extraction. A modified ultrasonic extraction method (52) was used to extract both intact polar and core GDGTs as described in SI Appendix, SI Methods. Lipid analysis by LC-MS was performed with an Agilent 1290 series ultraperformance liquid chromatography system coupled to an Agilent 6530 quadrupole time-of-flight (qTOF) mass spectrometer through an Agilent jet stream dual electrospray ionization (AJS-ESI) interface. The ESI drying gas (N2) temperature was set at 300 °C, the N2 flow rate was 8 L min−1, and the nebulizer gas (N2) pressure was 35 psi. The qTOF parameters were set to the following in auto MS/MS scanning mode with a first stage of MS (MS1) mass range of m/z 100–3,000 and a second stage of MS (MS2) mass range of m/z 50–3,000: capillary voltage, 3.5 kV; fragmentor voltage, 175 V; skimmer voltage, 65 V; and octopole voltage, 750 V.
LC methods applied in this work were modified from Zhu et al. (53). To separate calditol–GDGTs and core GDGTs, samples were dissolved in methanol and injected with a volume of 10 μL onto an Agilent Poroshell 120 EC-C18 column (1.8 µm; 2.1 × 150 mm) maintained at 35 °C. Mobile phase was at a flow rate of 0.2 mL min−1, first isocratically with 100% A for 5 min, followed by a gradient to 40% B at 20 min, and to 70% B at 60 min, and finally reequilibrated with 100% A for 10 min, where the eluent A was 100:0.04:0.10 of methanol/formic acid/14.8 M NH3(aq.) and B was 100:0.04:0.10 of 2-propanol/formic acid/14.8 M NH3(aq.). In later experiments, the separation of acetylated compounds was achieved with a shorter column (1.8 µm; 4.6 × 50 mm; Agilent Zorbax Eclipse XDB-C18) maintained at 35 °C. The same eluent A and B were applied but with a different gradient at a flow rate of 0.4 mL min−1, first isocratically with 100% A for 3 min, followed by a gradient to 40% B at 20 min, and to 90% B at 45 min, and then to 100% B at 46 min and hold for 9 min, and finally reequilibrated with 100% A for 10 min.
Bioinformatics Analyses.
S. acidocaldarius radical SAM proteins were identified by searching this genome for the radical SAM protein family identifier pfam04055 through the JGI IMG/M portal (https://img.jgi.doe.gov) (34). BLASTP searches (e value < 1e−50; 30% identity) of each of these 18 radical SAM proteins were done to identify homologs present in other calditol-producing archaea (Acidianus hospitalis and Metallosphaera sedula) that were absent from non–calditol-producing species (Thermoproteus tenax, Haloferax volcanii, and Methanosarcina barkeri). Homologs of Cds were identified through BLASTP searches (e value < 1e−50; 30% identity) of the JGI genomic, MAG, and SAG databases (76,964 genomes) and the NCBI nonredundant protein sequences database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Metagenomic homologs of Cds were also identified through BLASTP searches (e value < 1e−50; 20% identity; >300 amino acids) of the JGI aquatic and terrestrial metagenomic databases (10,096 metagenomes). Protein sequences were aligned via Multiple Sequence Comparison by Log-Expectation (54) using Geneious (Biomatters Limited). Redundant sequences were removed from alignments using the ExPASy decrease redundancy web tool (https://web.expasy.org/decrease_redundancy) (55). Maximum-likelihood trees were constructed using the LG model, 4 gamma rate categories, 10 random starting trees, nearest-neighbor-interchange branch swapping, and substitution parameters estimated from the data (56). All tree files were imported and edited in the Interactive Tree of Life online tool (https://itol.embl.de) (57).
Supplementary Material
Acknowledgments
We thank Dr. Sonja-Verena Albers and Alexander Wagner for the gift of Sulfolobus acidocaldarius MW001 and plasmids to genetically manipulate this organism. We also thank Dr. Emily Matys for technical assistance, members of P.V.W.’s group for helpful discussions, and two anonymous reviewers for their constructive comments. This study was funded by grants from the Simons Foundation Collaboration on the Origins of Life (to P.V.W. and R.E.S.). X.-L.L. was supported by startup funding from the Department of Geology and Geophysics, University of Oklahoma.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814048115/-/DCSupplemental.
References
- 1.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woese CR, Magrum LJ, Fox GE. Archaebacteria. J Mol Evol. 1978;11:245–251. doi: 10.1007/BF01734485. [DOI] [PubMed] [Google Scholar]
- 3.Kates M. The biochemistry of archaea (Archaeabacteria) In: Kushner DJ, Matheson AT, editors. Membrane Lipids of Archaea. Elsevier Science; Amsterdam: 1993. pp. 261–295. [Google Scholar]
- 4.Koga Y, Morii H. Biosynthesis of ether-type polar lipids in archaea and evolutionary considerations. Microbiol Mol Biol Rev. 2007;71:97–120. doi: 10.1128/MMBR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kates M. Diether and tetraether phospholipids and glycolipids as molecular markers for archaebacteria (archaea) In: Eganhouse RP, editor. Molecular Markers in Environmental Geochemistry. Vol 671. American Chemical Society; Washington, DC: 1997. pp. 35–48. [Google Scholar]
- 6.Morii H, Eguchi T, Koga Y. In vitro biosynthesis of ether-type glycolipids in the methanoarchaeon Methanothermobacter thermautotrophicus. J Bacteriol. 2007;189:4053–4061. doi: 10.1128/JB.01875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eme L, Spang A, Lombard J, Stairs CW, Ettema TJG. Archaea and the origin of eukaryotes. Nat Rev Microbiol. 2017;15:711–723. doi: 10.1038/nrmicro.2017.133. [DOI] [PubMed] [Google Scholar]
- 8.Koga Y. From promiscuity to the lipid divide: On the evolution of distinct membranes in archaea and bacteria. J Mol Evol. 2014;78:234–242. doi: 10.1007/s00239-014-9613-4. [DOI] [PubMed] [Google Scholar]
- 9.Lombard J, López-García P, Moreira D. The early evolution of lipid membranes and the three domains of life. Nat Rev Microbiol. 2012;10:507–515. doi: 10.1038/nrmicro2815. [DOI] [PubMed] [Google Scholar]
- 10.Hinrichs KU, Hayes JM, Sylva SP, Brewer PG, DeLong EF. Methane-consuming archaebacteria in marine sediments. Nature. 1999;398:802–805. doi: 10.1038/19751. [DOI] [PubMed] [Google Scholar]
- 11.Schouten S, Hopmans EC, Schefuss E, Damste JSS. Distributional variations in marine crenarchaeotal membrane lipids: A new tool for reconstructing ancient sea water temperatures? Earth Planet Sci Lett. 2002;204:265–274. [Google Scholar]
- 12.Pearson A. 2014. Lipidomics for geochemistry. Treatise on Geochemistry, Organic Geochemistry, eds Holland HD, Turekian KK (Elsevier Science, Oxford), 2nd Ed, Vol 12, pp 291–336.
- 13.Summons RE, Lincoln SA. Biomarkers: Informative molecules for studies in geobiology. In: Knoll AH, Canfield DE, Konhauser KO, editors. Fundamentals of Geobiology. 1st Ed. Blackwell Publishing; West Sussex, UK: 2012. pp. 269–296. [Google Scholar]
- 14.Tierney JE. GDGT thermometry: Lipid tools for reconstruicting paleotemperatures. In: Ivany LC, Huber BT, editors. Reconstructing Earth’s Deep-Time Climate–The State of the Art in 2012. Vol 18. Paleontological Society; Ithaca, NY: 2012. pp. 115–131. [Google Scholar]
- 15.Caforio A, Driessen AJM. Archaeal phospholipids: Structural properties and biosynthesis. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:1325–1339. doi: 10.1016/j.bbalip.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Yamauchi N, Kamada N, Ueoka H. The possibility of involvement of “cyclase” enzyme of the calditol carbocycle with broad substrate specificity in Sulfolobus acidcaldarius, a typical thermophilic archaea. Chem Lett. 2006;35:1230–1231. [Google Scholar]
- 17.Chong PL, Ayesa U, Daswani VP, Hur EC. On physical properties of tetraether lipid membranes: Effects of cyclopentane rings. Archaea. 2012;2012:138439. doi: 10.1155/2012/138439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber G, Spinnler C, Gambacorta A, Stetter KO. Metallosphaera sedula gen, and sp. nov. Represents a new genus of aerobic, metal-mobilizing, thermoacidophilic archaebacteria. Syst Appl Microbiol. 1989;12:38–47. [Google Scholar]
- 19.Plumb JJ, Haddad CM, Gibson JA, Franzmann PD. Acidianus sulfidivorans sp. nov., an extremely acidophilic, thermophilic archaeon isolated from a solfatara on Lihir Island, Papua New Guinea, and emendation of the genus description. Int J Syst Evol Microbiol. 2007;57:1418–1423. doi: 10.1099/ijs.0.64846-0. [DOI] [PubMed] [Google Scholar]
- 20.Blériot Y, Untersteller E, Fritz B, Sinaÿ P. Total synthesis of calditol: Structural clarification of this typical component of archaea order Sulfolobales. Chemistry. 2002;8:240–246. doi: 10.1002/1521-3765(20020104)8:1<240::aid-chem240>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 21.Langworthy TA, Smith ME, Mayberry WR. Long-chain glycerol diether and polyol dialkyl glycerol triether lipids of Sulfolobus acidocaldarius. J Bacteriol. 1972;112:1193–1200. doi: 10.1128/jb.119.1.106-116.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicolaus B, et al. Calditol tetraether lipids of the archaebacterium Sulfolobus solfataricus. Biosynthetic studies. Biochem J. 1990;266:785–791. [PMC free article] [PubMed] [Google Scholar]
- 23.Sugai A, et al. The structure of the core polyol of the ether lipids from Sulfolobus acidocaldarius. Lipids. 1995;30:339–344. doi: 10.1007/BF02536042. [DOI] [PubMed] [Google Scholar]
- 24.Gambacorta A, et al. Biosynthesis of calditol, the cyclopentanoid containing moiety of the membrane lipids of the archaeon Sulfolobus solfataricus. Tetrahedron Lett. 2002;43:451–453. [Google Scholar]
- 25.Yamauchi N, Ueoka H, Kamada N, Murae T. Resemblance of carbocycle formation from carbohydrates between archaea and Eucarya/Eubacteria. Biosynthesis of calditol, the characteristic lipid-content molecule in Sulfolobus acidocaldarius. Bull Chem Soc Jpn. 2004;77:774–778. [Google Scholar]
- 26.Brock TD, Brock KM, Belly RT, Weiss RL. Sulfolobus: A new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Mikrobiol. 1972;84:54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- 27.Vincent SP, Sinaÿ P, Rohmer M. Composite hopanoid biosynthesis in Zymomonas mobilis: N-acetyl-D-glucosamine as precursor for the cyclopentane ring linked to bacteriohopanetetrol. Chem Commun (Camb) 2003:782–783. doi: 10.1039/b212685k. [DOI] [PubMed] [Google Scholar]
- 28.Schmerk CL, et al. Elucidation of the Burkholderia cenocepacia hopanoid biosynthesis pathway uncovers functions for conserved proteins in hopanoid-producing bacteria. Environ Microbiol. 2015;17:735–750. doi: 10.1111/1462-2920.12509. [DOI] [PubMed] [Google Scholar]
- 29.Broderick JB, Duffus BR, Duschene KS, Shepard EM. Radical S-adenosylmethionine enzymes. Chem Rev. 2014;114:4229–4317. doi: 10.1021/cr4004709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Rosa M, et al. A range of ether core lipids from the methanogenic archaebacterium Methanosarcina barkeri. Biochim Biophys Acta. 1986;875:487–492. [Google Scholar]
- 32.Sprott GD, et al. Novel polar lipids of halophilic eubacterium Planococcus H8 and archaeon Haloferax volcanii. Biochim Biophys Acta. 2003;1633:179–188. doi: 10.1016/j.bbalip.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Thurl S, Schafer W. Lipids from the sulfur-dependent archaebacterium Thermoproteus tenax. Biochim Biophys Acta. 1988;961:253–261. [Google Scholar]
- 34.Markowitz VM, et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014;42:D560–D567. doi: 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jay ZJ, et al. Marsarchaeota are an aerobic archaeal lineage abundant in geothermal iron oxide microbial mats. Nat Microbiol. 2018;3:732–740. doi: 10.1038/s41564-018-0163-1. [DOI] [PubMed] [Google Scholar]
- 36.Miller-Coleman RL, et al. Korarchaeota diversity, biogeography, and abundance in Yellowstone and Great Basin hot springs and ecological niche modeling based on machine learning. PLoS One. 2012;7:e35964. doi: 10.1371/journal.pone.0035964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mondav R, et al. Discovery of a novel methanogen prevalent in thawing permafrost. Nat Commun. 2014;5:3212. doi: 10.1038/ncomms4212. [DOI] [PubMed] [Google Scholar]
- 38.Reysenbach AL, et al. A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nature. 2006;442:444–447. doi: 10.1038/nature04921. [DOI] [PubMed] [Google Scholar]
- 39.Schouten S, Baas M, Hopmans EC, Reysenbach AL, Damsté JS. Tetraether membrane lipids of Candidatus “Aciduliprofundum boonei”, a cultivated obligate thermoacidophilic euryarchaeote from deep-sea hydrothermal vents. Extremophiles. 2008;12:119–124. doi: 10.1007/s00792-007-0111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burgess EA, Unrine JM, Mills GL, Romanek CS, Wiegel J. Comparative geochemical and microbiological characterization of two thermal pools in the Uzon Caldera, Kamchatka, Russia. Microb Ecol. 2012;63:471–489. doi: 10.1007/s00248-011-9979-4. [DOI] [PubMed] [Google Scholar]
- 41.Teske A, et al. The Guaymas Basin hiking guide to hydrothermal mounds, chimneys, and microbial mats: Complex seafloor expressions of subsurface hydrothermal circulation. Front Microbiol. 2016;7:75. doi: 10.3389/fmicb.2016.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCarty RM, Krebs C, Bandarian V. Spectroscopic, steady-state kinetic, and mechanistic characterization of the radical SAM enzyme QueE, which catalyzes a complex cyclization reaction in the biosynthesis of 7-deazapurines. Biochemistry. 2013;52:188–198. doi: 10.1021/bi301156w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inskeep WP, et al. Phylogenetic and functional analysis of metagenome sequence from high-temperature archaeal habitats demonstrate linkages between metabolic potential and geochemistry. Front Microbiol. 2013;4:95. doi: 10.3389/fmicb.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimada H, Nemoto N, Shida Y, Oshima T, Yamagishi A. Effects of pH and temperature on the composition of polar lipids in Thermoplasma acidophilum HO-62. J Bacteriol. 2008;190:5404–5411. doi: 10.1128/JB.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiang Q, Lee YY, Torget RW. Kinetics of glucose decomposition during dilute-acid hydrolysis of lignocellulosic biomass. Appl Biochem Biotechnol. 2004;113–116:1127–1138. doi: 10.1385/abab:115:1-3:1127. [DOI] [PubMed] [Google Scholar]
- 46.McCarthy S, et al. Expanding the limits of thermoacidophily in the archaeon Sulfolobus solfataricus by adaptive evolution. Appl Environ Microbiol. 2015;82:857–867. doi: 10.1128/AEM.03225-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Welander PV, et al. Identification and characterization of Rhodopseudomonas palustris TIE-1 hopanoid biosynthesis mutants. Geobiology. 2012;10:163–177. doi: 10.1111/j.1472-4669.2011.00314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welander PV, Summons RE. Discovery, taxonomic distribution, and phenotypic characterization of a gene required for 3-methylhopanoid production. Proc Natl Acad Sci USA. 2012;109:12905–12910. doi: 10.1073/pnas.1208255109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eguchi T, Nishimura Y, Kakinuma K. Importance of the isopropylidene terminal of geranylgeranyl group for the formation of tetraether lipid in methanogenic archaea. Tetrahedron Lett. 2003;44:3275–3279. [Google Scholar]
- 50.Galliker P, Gräther O, Rümmler M, Fitz W, Arigoni D. New structural and biosynthetic aspects of the unusual core lipids from archaebacteria. In: Kräutler B, Arigoni D, Golding BT, editors. Vitamin B12 and B12-Proteins. Wiley-VCH; Weinheim, Germany: 1998. pp. 447–458. [Google Scholar]
- 51.Wagner M, et al. Versatile genetic tool box for the Crenarchaeote Sulfolobus acidocaldarius. Front Microbiol. 2012;3:214. doi: 10.3389/fmicb.2012.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huguet C, Martens-Habbena W, Urakawa H, Stahl DA, Ingalls AE. Comparison of extraction methods for quantitative analysis of core and intact polar glycerol dialkyl glycerol tetraethers (GDGTs) in environmental samples. Limnol Oceanogr Methods. 2010;8:127–145. [Google Scholar]
- 53.Zhu C, et al. Comprehensive glycerol ether lipid fingerprints through a novel reversed phase liquid chromatography-mass spectrometry protocol. Org Geochem. 2013;65:53–62. [Google Scholar]
- 54.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Artimo P, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 57.Letunic I, Bork P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.