Significance
Atopic dermatitis (AD) is a common skin disease affecting children, but the mechanisms underlying the accompanying itch remain poorly understood. Indeed, treatments to alleviate itch in AD and the substantial morbidity it represents for patients are often unsuccessful. Here, we shed light on a form of AD associated with the loss of Tmem79 and provide insights into cellular and molecular players in this genetically driven form of the disease that has been described in mice and humans. We propose a link between this inherited form of AD and the recognized contribution of oxidative stress to this disease. Our analysis may suggest pharmacological strategies for treating patients who suffer from AD and bear mutations in Tmem79.
Keywords: itch, atopic dermatitis, oxidative stress, Tmem79, prostaglandin E2
Abstract
Atopic dermatitis (AD) is the most common skin disease in children. It is characterized by relapsing inflammation, skin-barrier defects, and intractable itch. However, the pathophysiology of itch in AD remains enigmatic. Here, we examine the contribution of Tmem79, an orphan transmembrane protein linked to AD in both mice and humans. We show that Tmem79 is expressed by both keratinocytes and sensory neurons, but that loss of keratinocytic Tmem79 is sufficient to elicit robust scratching. Tmem79−/− mice demonstrate an accumulation of dermal mast cells, which are diminished following chronic treatment with cyclooxygenase inhibitors and an EP3 receptor antagonist. In Tmem79−/− mice, mast cell degranulation produces histaminergic itch in a histamine receptor 1/histamine receptor 4 (H4R/H1R)-dependent manner that may involve activation of TRPV1− afferents. TMEM79 has limited sequence homology to a family of microsomal glutathione transferases and confers protection from cellular accumulation of damaging reactive species, and may thus play a role in regulating oxidative stress. In any case, mechanistic insights from this model suggest that therapeutics targeting PGE2 and/or H1R/H4R histaminergic signaling pathways may represent useful avenues to treat Tmem79-associated AD itch. Our findings suggest that individuals with mutations in Tmem79 develop AD due to the loss of protection from oxidative stress.
The development of atopic dermatitis (AD) is associated with a genetic predisposition toward immune dysregulation and skin-barrier defects as well as hypersensitivity to environmental factors and psychological triggers (1, 2). The canonical pathology of AD is often accompanied by an infiltrate of inflammatory cells (e.g., mast cells, eosinophils, and T lymphocytes) into the dermal layer of skin. In recent years, oxidative stress has also gained attention for its role in the pathogenesis of AD (3).
Several mouse models of AD have been characterized (4) including Tmem79−/− (or Matt−/−) mice. Mutations in Tmem79−/− mice underlie the matted hair and dermatitis phenotypes in flaky tail mice (Flgft) and lead to spontaneous AD characterized by skin-barrier defects, eczematous skin lesions, and severe scratching (5, 6). Importantly, a missense SNP in Tmem79 is associated with AD in human subjects (6). However, mechanisms contributing to itch in Tmem79-associated AD remain elusive. Furthermore, it is not known if Tmem79 plays a dominant role in keratinocytes, neurons, or other skin cells that results in the development of atopy. Here we leveraged Tmem79-null and -conditional knockout reporter mouse lines to explore the cellular and molecular mechanisms underlying itch in this model of AD.
Results and Discussion
Gross observation revealed that our de novo Tmem79−/− mouse line exhibits pelage (SI Appendix, Fig. S1A) consistent with a previously reported nonsense mutation (6). Additionally, we observed thickened dermis with immune cell infiltrate (SI Appendix, Fig. S1 B and C) and exaggerated scratching behavior (SI Appendix, Fig. S1 E–G), again consistent with previous reports (5, 6). Tmem79−/− mice are normal in weight (SI Appendix, Fig. S1D) and do not exhibit anxiety behavior as measured by the elevated plus maze (SI Appendix, Fig. S1H).
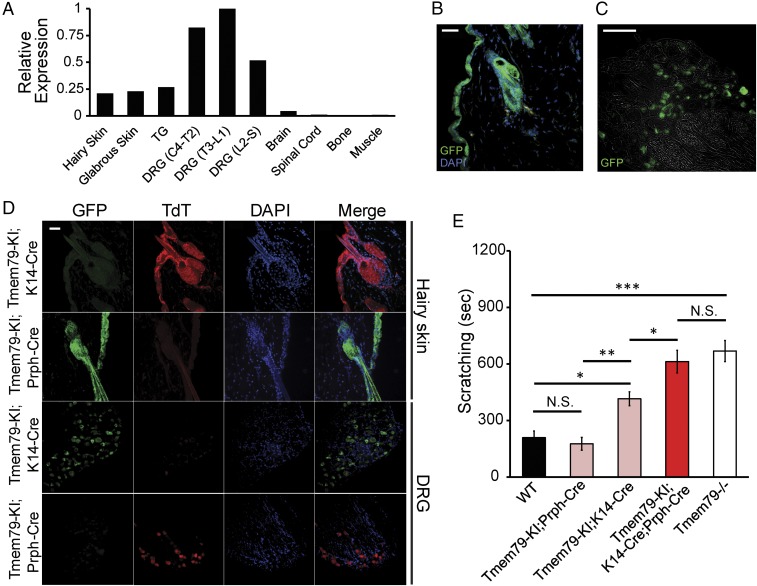
Scratching is typically preceded by pruritoception (i.e., activation of a subset of somatosensory neurons that trigger itch). We therefore asked if Tmem79 is expressed in neuronal tissues, in addition to skin as previously reported (5, 6). Indeed, Tmem79 initially came to our attention as a transcript that is preferentially expressed in somatosensory ganglia rather than in other neural tissues (SI Appendix, Fig. S2C). Consistent with this, we found that Tmem79 is transcribed in hairy and glabrous skin as well as trigeminal (TG) and dorsal root (DRG) sensory ganglia but not in brain or spinal cord (Fig. 1A). Expression by a subset of sensory neurons was confirmed by in situ hybridization histochemistry (SI Appendix, Fig. S2B) as well as by a reporter mouse line (Tmem79-KI) in which eGFP is produced from a Tmem79-2A-eGFP precursor protein (SI Appendix, Fig. S2A). In these mice, robust eGFP expression was observed in epidermal cells of hairy skin (Fig. 1B), cervix, prostate, and tongue but not in cells of the lung, adrenal medulla, brain, or spinal cord (SI Appendix, Fig. S2G). In hairy skin epithelium, eGFP was highly colocalized with keratin 14 (K14), a marker for keratinocytes (SI Appendix, Fig. S2H). Consistent with our transcriptomics, we found eGFP to be highly expressed in a subset of sensory neurons (Fig. 1C) of small-to-medium diameter (SI Appendix, Fig. S2F) and showed varying degrees of coexpression with the following markers: isolectin-B4 (IB4, 45%), NF200 (32%), peripherin (74%), TRPV1 (20%), calcitonin gene-related peptide (CGRP, 16%), substance P (SP, 32%), TRPA1 (24%), histamine receptor 1 (H1R, 61%), and histamine receptor 4 (H4R, 80%) (SI Appendix, Fig. S2 D and E). In conclusion, we find Tmem79 to be expressed in keratinocytes of the epidermis, epithelial tissues, and a subpopulation of somatosensory neurons.
Fig. 1.
Loss of Tmem79 in sensory neurons and keratinocytes produces itch. (A) Real-time qPCR analysis of Tmem79 expression relative to Rpl19 in tissues dissected from wild-type mice. The graph presents ΔΔCt values normalized to the tissue with greatest relative expression of Tmem79. ΔΔCt values were calculated from real-time PCR reactions run in triplicate from combined tissues from three wild-type mice. Tissues examined were hairy skin, glabrous (hairless) skin, TGs, DRG fourth cervical to second thoracic (DRG C4–T2), DRG third thoracic to first lumbar (DRG T3–L1), DRG second lumbar to sacral (DRG L2–S), brain, spinal cord, bone, and muscle. (B and C) Representative micrographs of tissue sections from Tmem79-KI reporter mice stained for immunoreactivity to GFP (green). (B) Hairy skin section (20 µm) with the superficial epidermis oriented to the left. Nuclei were counterstained using DAPI (blue). (Scale bar: 15 µm.) (C) DRG section (12 µm). The image is superimposed with brightfield micrograph. (Scale bar: 100 µm.) (D) Representative micrographs of hairy skin or DRG sections from Tmem79-KI;K14-Cre or Tmem79-KI;Prph-Cre mice stained for immunoreactivity to GFP (green) or TdT (red). Superficial epidermis is oriented to the left (1st Row, Tmem79-KI;K14-Cre) or the right (2nd Row, Tmem79-KI;Prph-Cre). Nuclei were counterstained with DAPI (blue). Merged images are shown in the far right column. (Scale bar: 15 µm.) (E) Quantified scratching behavior of adult wild-type, Tmem79-KI;Prph-Cre, Tmem79-KI;K14-Cre, Tmem79-KI;K14-Cre;Prph-Cre, and Tmem79−/− mice. Data shown are the mean number of seconds of scratching in multiple mice of each genotype ± SEM. n = 6 wild-type mice, 9 Tmem79-KI;Prph-Cre mice, 11 Tmem79-KI;K14-Cre mice, 5 Tmem79-KI;K14-Cre;Prph-Cre mice, and 12 Tmem79−/− mice; *P < 0.05, **P < 0.01, ***P < 0.001, one-way ANOVA with Holm–Sidak’s multiple comparisons correction; N.S., not significant.
Our Tmem79-KI reporter mouse line also features an inverted floxed cassette (SI Appendix, Fig. S2A) that allows cell-specific targeting of Tmem79 deletion as indicated by the expression of TdTomato (TdT). As predicted, K14-TdT (Tmem79-KI:K14-Cre) and Prph-TdT (Tmem79-KI;Prph-Cre) mice showed robust TdT expression in the epidermis and sensory neurons, respectively, with no observable expression of eGFP (Fig. 1D). As further validation of faithful, cell-specific recombination, the population of sensory neurons expressing TdT in Prph-TdT mice matched the pattern of eGFP expression in Tmem79-KI animals (SI Appendix, Fig. S2F). We next leveraged these crosses to determine the contribution of Tmem79 in keratinocytes versus sensory neurons in regard to AD phenotypes. We found that while K14-TdT mice exhibit partially elevated scratching behavior and dermal thickening (SI Appendix, Fig. S5 F and G), only double-deletion, K14-TdT;Prph-TdT mice completely phenocopied Tmem79−/− scratching (Fig. 1E). These data suggest that while the loss of keratinocytic Tmem79 accounts for a substantial portion of scratching and AD in Tmem79-null mice, the loss of neuronal Tmem79 also contributes to the phenotype.
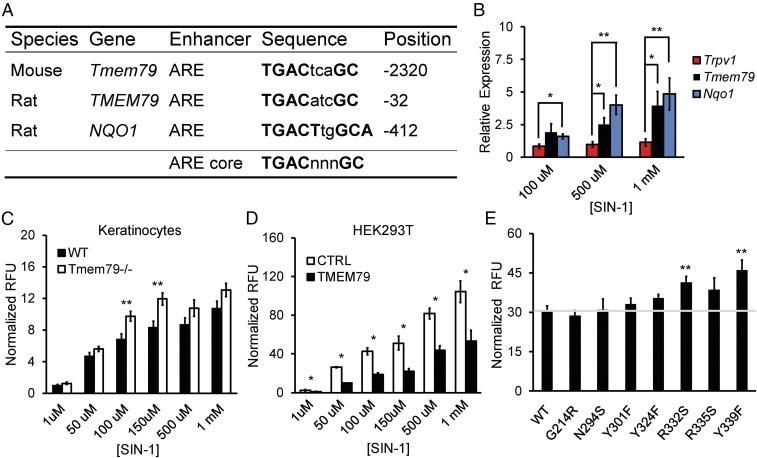
Previous work suggests that TMEM79 plays a role in secretory transport and that consequent defects in Tmem79 mutants produce AD due to deficiencies in skin-barrier function (5). However, TMEM79 shows limited, but intriguing, primary amino acid sequence similarity (∼30%) with membrane glutathione transferases (MAPEGs) such as MGST3 (7) or MGST1 (8–10) that detoxify reactive electrophiles (SI Appendix, Fig. S3 F–H) (6). In light of this, and our findings that Tmem79 is also expressed in sensory neurons, we asked if Tmem79 modulates oxidative stress in cellular assays. First, we found that acutely dissociated keratinocytes from Tmem79−/− mice accumulate higher levels of reactive species than do wild-type controls when challenged with moderate concentrations of the oxidant 3-Morpholinosydnonimine (SIN-1) (Fig. 2C). Similarly, heterologous expression of TMEM79 in HEK293T cells diminished the accumulation of reactive species in response to SIN-1 or sodium nitroprusside (but not tert-Butyl hydroperoxide or H2O2) (Fig. 2D and SI Appendix, Fig. S3 A–E). Furthermore, mutation of two putative active-site residues (R332S and Y339F) within a MAPEG-homologous region of TMEM79 eliminated the protective effect in this assay (Fig. 2E). Interestingly, the rat and mouse Tmem79 genes contain a canonical antioxidant response element sequence (Fig. 2A) (11–13). Consistent with this, we found that exposure of cultured rat sensory neurons to SIN-1 results in higher relative expression of Tmem79 mRNA than Trpv1 mRNA, which lacks the response element (Fig. 2B). Taken together, these data suggest that TMEM79 may function to neutralize reactive electrophiles, thus preventing cellular oxidative stress, a known contributor to the development of AD (3).
Fig. 2.
TMEM79 diminishes oxidative stress. (A) Table of antioxidant response elements (AREs) in rodent genes (Tmem79, TMEM79, and NQO1). Sequence identity and the position relative to the start codon for each gene are provided. The ARE core is shown for reference. (B) Real-time qPCR data for gene expression relative to Rpl19 in cultured P0 rat sensory neurons following 16-h incubation with concentrations of SIN-1. Data are shown as the mean relative expression ± SEM; n = 3 independent experiments with samples run in triplicate. **P < 0.01, *P < 0.05, two-tailed Holm t test. (C) Quantification of DCF fluorescence in acute cultures of keratinocytes from age-matched wild-type or Tmem79−/− mice following 30-min incubation with SIN-1 concentrations as noted. Data represent mean relative fluorescent units (RFU) normalized to baseline ± SEM; n = 7 mice with samples run in triplicate. **P < 0.01, two-tailed Student’s t test. (D) Quantification of DCF fluorescence in cultures of HEK293T cells expressing either control (CTRL) or mouse Tmem79 (TMEM79) vector following 30-min incubation with various SIN-1 concentrations. Data represent mean RFU normalized to baseline ± SEM; n = 3 or 4 experiments with samples run in triplicate. *P < 0.05, one-tailed Student’s t test. (E) Quantification of DCF fluorescence in cultures of HEK293T cells expressing vectors containing mouse Tmem79 (WT) or amino acid substitution mutants (G214R, N294S, Y301F, Y324F, R332S, R335S, and Y339F) following 30-min incubation in 150 µm of SIN-1. Data represent mean RFU normalized to baseline ± SEM. The horizontal gray line represents normalized RFU from mouse Tmem79 (WT) for reference. n = 5 experiments with samples run in triplicate. **P < 0.01, two-tailed Holm t test.
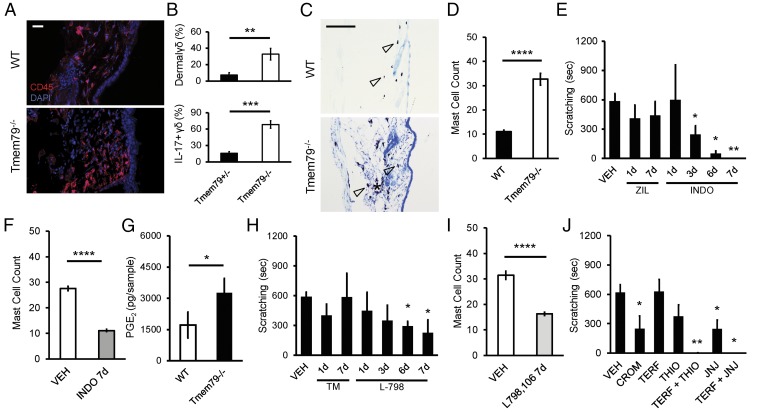
As noted, Tmem79−/− mice showed an increased presence of immune cells in the dermis as assessed by cytologic staining (SI Appendix, Fig. S1B) and CD45 immunoreactivity (Fig. 3A). Furthermore, flow cytometry (SI Appendix, Fig. S4A) revealed an expansion of IL-17–expressing γδ-T cells and mast cells (Fig. 3B and SI Appendix, Fig. S4 B–D) but no change in the number of neutrophils, eosinophils, or Ly6Chi or Ly6Clow monocytes in the skin of Tmem79−/− mice (SI Appendix, Fig. S4D). Histological inspection revealed an increase in mast cell count in Tmem79−/− dermis (Fig. 3 C and D). In some instances, degranulated mast cells were apparent at higher magnifications (SI Appendix, Fig. S5A), which may explain an underestimation of mast cell count by flow cytometry (SI Appendix, Fig. S4D). We similarly observed additional CD45+ immune cells (SI Appendix, Fig. S5C) and increased mast cell counts (SI Appendix, Fig. S5E) in the dermis of Tmem79-KI;K14-Cre mice. Of note, we failed to observe Tmem79 expression in the lung epithelium of Tmem79-KI reporter mice (SI Appendix, Fig. S2G) or any increased immune- or mast cell presence in sections of Tmem79−/− lung epithelium compared with wild-type mice (SI Appendix, Fig. S5B). Taken together, these observations suggest that the Tmem79−/− phenotype does not include airway inflammation characteristic of the “atopic march” seen in some cases of AD (14). These results suggest that TMEM79 dysregulation promotes the accumulation of IL-17–expressing γδ-T cells and mast cells which have been implicated in AD (15, 16) and pruritus (17–19).
Fig. 3.
PGE2–EP3 interaction recruits dermal mast cells that drive histaminergic itch via H4R in Tmem79−/− mice. (A) Representative micrographs of stained hairy skin sections (20 µm) from age-matched wild-type (Upper) or Tmem79−/− (Lower) mice. Superficial epidermis is oriented to the right. Staining for immunoreactivity to CD45 (red) with nuclei counterstained using DAPI (blue). (Scale bar: 10 µm.) (B) Flow cytometry quantification of immune cells and cytokine expression in the hairy skin of littermate Tmem79+/− and Tmem79−/− mice. Data shown are the percentage of dermal immune cells that are γδ+ (Upper) and the percentage of dermal γδ+ cells that are also IL-17+ (Lower). Data shown are the mean percentages ± SEM from four Tmem79+/− and four Tmem79−/− mice. **P < 0.01, ***P < 0.001, one-tailed Student’s t test. (C) Representative micrographs of Toluidine blue O-stained hairy skin sections (20 µm) from age-matched wild-type (Upper) or Tmem79−/− (Lower) mice. Superficial epidermis is oriented to the right. Hollow arrowheads indicate examples of stained mast cells. (Scale bar: 100 µm.) (D) Quantification of mast cells from micrographs represented in C. Data shown are the mean mast cell count ± SEM from fields captured from wild-type or Tmem79−/− mice; n = 40 micrographs in wild-type mice and 47 micrographs in Tmem79−/− mice. ****P < 0.0001, two-tailed Student’s t test. (E) Quantified scratching behavior of adult Tmem79−/− mice following i.p. administration of vehicle (VEH), 20 mg/kg zileuton (ZIL), or 10 mg/kg indomethacin (INDO) for 1, 3, 6, or 7 consecutive days. Data shown are the mean number of seconds of scratching ± SEM from multiple mice (n = 4) in each treatment group. *P < 0.05, **P < 0.01, one-tailed Holm t test. (F) Quantification of mast cells from micrographs of stained hairy skin sections from mice receiving vehicle (VEH) or 10 mg/kg indomethacin (INDO) for seven consecutive days. Data shown are the mean mast cell count ± SEM; n = 43 micrographs from vehicle-treated mice and 85 micrographs from indomethacin-treated mice. ****P < 0.0001, two-tailed Student’s t test. (G) LC/MS/MS-based lipidomics was used to identify and quantify prostanoids, PGE2, and PGE2 metabolite content from lysed acutely dissociated keratinocytes from age-matched wild-type and Tmem79−/− mice. Deuterated internal standards were used to correct for extraction and processing recoveries. An AB Sciex QTRAP 4500 mass spectrometer was used for analyses; prostanoids were identified and quantitated using scheduled MRM with four to six specific transition ions for each analyte. Data represent the average ± SEM PGE2 content (quantified in picograms) from keratinocytes from weight-matched skin dissected from separate animals; n = 4 animals per group. *P < 0.05, two-tailed Student’s t test. (H) Quantified scratching behavior of adult Tmem79−/− mice following i.p. administration of vehicle (VEH), 5 mg/kg TM30089 (TM), or 100 mg/kg L-798,106 (L-798) for seven consecutive days. Data shown are the mean number of seconds of scratching ± SEM by multiple mice (n = 4) in each treatment group. *P < 0.05, one-tailed Holm t test. (I) Quantification of mast cells from micrographs of stained hairy skin sections from mice treated with vehicle (VEH) or 100 mg/kg L-796,106 (L-798) for seven consecutive days. Data shown are the mean mast cell count ± SEM. n = 54 micrographs from vehicle-treated mice and 62 micrographs from L-796,106–treated mice. ****P < 0.0001, two-tailed Student’s t test. (J) Quantified scratching behavior of adult Tmem79−/− mice following i.p. administration of vehicle (VEH), 20 mg/kg cromolyn sulfate (CROM), 20 mg/kg terfenidine (TERF), 20 mg/kg thioperamide (THIO), 20 mg/kg terfenidine + 20 mg/kg thioperamide (TERF + THIO), 20 mg/kg JNJ-7777120 (JNJ), or 20 mg/kg terfenidine + 20 mg/kg JNJ-7777120 (TERF + JNJ). Data shown are the mean number of seconds of scratching ± SEM from multiple mice (n = 4) in each treatment group. *P < 0.05, **P < 0.01, one-tailed Holm t test.
Eicosanoids (e.g., prostaglandins and leukotrienes) are produced by keratinocytes in response to cellular oxidative stress (20, 21) and could serve as mast cell chemoattractants (22). We therefore asked if these lipid mediators contribute to phenotypes associated with loss of Tmem79. Whereas daily administration of the 5-lipoxygenase inhibitor zileuton (20 mg/kg, i.p.) had no effect on scratching behavior, the cyclooxygenase inhibitor indomethacin (10 mg/kg, i.p.) abolished elevated scratching in Tmem79−/− after 7 d (Fig. 3E) while also reducing normal scratching in wild-type mice after 3 d (SI Appendix, Fig. S5J). Indomethacin also reduced dermal mast cell (Fig. 3F and SI Appendix, Fig. S5D) and CD45+ immune cell counts (SI Appendix, Fig. S5C), but not dermal thickness in Tmem79−/− mice (SI Appendix, Fig. S5 F and G), suggesting a prostanoid-independent mechanism. Taken together, these results show that scratching in Tmem79 mutants is dependent on the generation of an excess of endogenous prostanoids and that Tmem79-dependent itch is an exacerbation of normal baseline scratching behavior.
We next sought to determine which prostaglandin(s) may be mediating mast cell chemotaxis. Indomethacin prevents the formation of prostaglandin H2 (PGH2) and the downstream prostaglandins PGE2 and PGD2. Blockade of PGE2 signaling by the administration of an EP3 receptor antagonist (L-798,106, 100 mg/kg, i.p.) reduced scratching in Tmem79−/− mice (Fig. 3H). Furthermore, consistent with a known role for EP3R in PGE2-mediated chemoattraction (22), administration of this antagonist resulted in reduced dermal mast cell counts (Fig. 3I and SI Appendix, Fig. S5D) and in a slight reduction in CD45+ immune cells (SI Appendix, Fig. S5C), although dermal thickness was again unaffected (SI Appendix, Fig. S5 F and G). By contrast, a CRTH2/DP2 receptor antagonist (TM30089, 5 mg/kg, i.p.) had no measurable effect on these parameters, suggesting that PGD2 is not involved.
Do keratinocytes in the epidermis constitute a source of PGE2 in Tmem79−/− mice (20, 21, 23)? Indeed, we observed a specific increase in PGE2, but not in the prostanoid precursor PGH2 (SI Appendix, Fig. S5 H and I). LC-MS/MS–based lipidomic analyses confirmed a selective increase in PGE2 from Tmem79−/− keratinocytes with no change in PGE2 metabolites (13,14-dihydro-15-keto-PGE2, 13,14-dihydro-15 keto-PGA2, or bicyclo-PGE2) (Fig. 3G). Our analysis suggests that increased PGE2 levels are due to enhanced PGE2 synthesis rather than a general increase in COX enzymatic activity or decreased PGE2 turnover. These data are consistent with observations of increased PGE2 content in skin plaques from humans with AD (24) and suggest that EP3R antagonism may have a newfound application in treating AD-associated itch.
In light of these results, we asked if mast cell-mediated histamine release underlies scratching in Tmem79−/− mice. First, the administration of cromolyn sulfate, which prevents mast cell degranulation, diminished scratching in Tmem79−/− mice (Fig. 3J). Second, scratching in Tmem79−/− and Tmem79-KI;K14-Cre mice was diminished with the H4R antagonists thioperamide or JNJ-7777120, but not by the H1R antagonist terfenadine (Fig. 3J). Remarkably, dual administration of H1R and H4R antagonists had a synergistic effect in which scratching was completely abolished (Fig. 3J and SI Appendix, Fig. S5K). The s.c. administration of pruritogens (histamine or chloroquine) to Tmem79−/− mice did not further enhance scratching in these animals (SI Appendix, Fig. S8 A and B). Our data suggest that histamine from mast cells contributes to scratching in these mice primarily via activation of H4R, with some contribution from H1R. Because dual administration of histamine receptor antagonists completely abolished scratching, similar to COX inhibition in wild-type and Tmem79−/− mice, we suggest that Tmem79-dependent itch is an exacerbation of normal baseline scratching behavior. These findings provide further support for the use of a combination of H1R/H4R antagonists in treating dermatitis as gleaned from models of both atopic and allergic dermatitis (25–27).
H1R and H4R are expressed on somatosensory neurons (27) and mast cells (Fig. 4 A and B and SI Appendix, Fig. S6 A–C), raising the question of their location of action when producing AD phenotypes in Tmem79−/− mice. In light of our observations that (i) administration of H4R/H1R antagonists has no effect on mast cell degranulation (SI Appendix, Fig. S6D) and (ii) administration of H4R or H4R/H1R antagonists relieves scratching within 30 min, before H4R-mediated chemotaxis of mast cells would be blocked (28), we agree that the pruritic actions of histamine are likely to be working on histamine receptors located on nerve terminals (25, 27). Indeed, we find that a subset of DRG sensory neurons responds to 4-methyl histamine (4-MH), a selective H4R agonist (Fig. 4C and SI Appendix, Fig. S7A), and that these responses are blocked by JNJ-7777120 (SI Appendix, Fig. S7B). Taken together, these data suggest that H4R antagonists may reduce scratching in Tmem79−/− mice via their action on nerve terminals, as previously shown in models of induced pruritus (29).
Fig. 4.
H4R receptor-mediated calcium influx in DRG sensory neurons and Tmem79−/− histaminergic itch is TRPV1 independent. (A) Representative micrographs of fluorescent colabeling of 12-µm DRG sections from adult wild-type (Left and Middle) or Tmem79-KI (Right) mice. Sections were stained for immunoreactivity to H4R or GFP (green) and binding to IB4 or for immunoreactivity to NF200, TRPV1, SP, or H4R as indicated (red). (Scale bar: 50 µm.) (B) Quantification of colabeling from micrographs depicted in A. Colabeling is shown as a proportion of total H4R+ and marker+ neurons. Proportions were calculated from counts taken from at least eight micrographs. (C–E) Normalized F340/F380 calcium imaging traces from adult DRG neurons. (C) Averaged responses to the application of 500 µm 4-MH or 1 µm capsaicin (CAP). Neurons were identified at the conclusion of each experiment by a positive response to elevated extracellular potassium (150 mM KCl, K+). Each trace represents a distinct subpopulation of responders: 4-MH (purple), 4-MH + CAP (magenta), CAP (red), or K+ only (gray). (Inset) The pie chart shows the percentage of each subpopulation from seven independent experiments (4-MH, 77/483, 15.9%; 4-MH+CAP, 45/483, 9.3%; CAP, 103/483, 21.3%; K+, 258/483, 53.4%). (Scale bar: 60 s.) (D and E) Responses to the application of 500 µm 4-MH and 10 µm AMG (D) or 1 mM A967079 (E). Neurons were identified at the conclusion of each experiment by a positive response to elevated extracellular potassium (150 mM KCl, K+). Each trace represents a distinct responder from a single experiment. (Scale bar: 60 s.) (F) Quantified scratching behavior of adult Tmem79−/− mice following i.p. administration of vehicle (VEH), 30 mg/kg AMG9810, or 50 mg/kg A967079. Data shown are the mean number of seconds ± SEM of scratching from multiple mice in each treatment group (n = 9 vehicle-treated mice, 4 AMG9810-treated mice, 21 A967079-treated mice). *P < 0.05, one-tailed Holm t test. (G) Quantified scratching behavior of adult Tmem79−/− mice 1–14 d after intrathecal administration of vehicle (I.T. VEH) or 10 µg capsaicin (I.T. CAP) and i.p. administration of 50 mg/kg A967079 (I.T. CAP + A967079). Data shown are the mean seconds of scratching ± SEM from multiple mice in each treatment group (n = 7 intrathecal vehicle-treated mice, 7 intrathecal capsaicin-treated mice, 4 intrathecal capsaicin + A967079-treated mice). **P < 0.01, one-tailed Holm t test. (H) Proposed model: Loss of keratinocytic Tmem79 results in the accumulation of reactive species (RS) stimulating the production of PGE2. PGE2 from keratinocytes serves to attract mast cells to the dermis of Tmem79−/− mice through activation of EP3. Mast cells in the dermis degranulate, releasing histamine and activating H4R on nonpeptidergic C-fibers that are TRPV1− and presumably have lost Tmem79.
Last, we sought to determine the major sensory neural pathway that transduces these histaminergic signals. Since TRPV1 (25, 27, 30–32) and TRPA1 (33) have been implicated in the transmission of itch signals, we asked if either of these channels is required in this model of AD. To this end, we ablated central projections of TRPV1+ neurons [which encompasses most TRPA1+ fibers (34–36)] using intrathecal administration of capsaicin (I.T. CAP). Although it severely attenuated TRPV1- and TRPA1-mediated nocifensive behaviors (SI Appendix, Fig. S8 F–H), I.T. CAP administration had no effect on Tmem79−/− scratching (Fig. 4G). Consistent with a TRPV1-independent mechanism, we found that H4R+ neurons are mostly nonpeptidergic with only partial (<40%) coexpression of TRPV1 and that a majority (>85%) were also positive for Tmem79 (Fig. 4 A and B). Furthermore, we found that sensory neurons that respond to 4-MH are capsaicin insensitive (Fig. 4C). Additionally, responses to 4-MH persist in the presence of the selective TRPV1 or TRPA1 antagonists AMG9810 (AMG) or A967079, respectively (Fig. 4 D and E and SI Appendix, Fig. S7 H–J), despite their dependence on extracellular calcium (SI Appendix, Fig. S7 C–G and J).
Consistent with these observations, we found that administration of a TRPV1 antagonist (AMG9810, 30 mg/kg, i.p.) to Tmem79−/− mice did not diminish scratching (Fig. 4F) despite blocking the nocifensive behaviors elicited by intraplantar injection of capsaicin or elevated surface temperatures (SI Appendix, Fig. S8 C and D). Administration of a TRPA1 antagonist (A967079, 50 mg/kg, i.p.) produced a partial reduction in scratching (Fig. 4F), which also persisted following I.T. CAP treatment (Fig. 4G). This reduction following A967079 administration was not observed in Tmem79−/−;Trpa1−/− double-knockout animals, which were indistinguishable from Tmem79−/− mice (SI Appendix, Fig. S8E), ruling out an off-target effect. These results suggest that TRPA1 contributes in some manner to the itch phenotype, which is revealed by acute, on-target drug action, but is masked by compensation in the unconditional gene knockout. Future experiments will clarify if TRPA1 ontribution arises from peripheral terminals of TRPV1+/TRPA1+ neurons, TRPV1−/TRPA1+ neurons, or nonneuronal TRPA1.
These data support a model of itch transmission in Tmem79−/− mice that does not depend on canonical TRPV1+/TRPA1+ central projections (30), but instead involves nonpeptidergic, TRPV1−/TRPA1−, H4R+ C fibers. The high coexpression of Tmem79 on H4R+ neurons may help explain the contribution of neuronal Tmem79 to scratching behavior (Fig. 1E), although the precise mechanism remains unclear. The observed partial contribution of TRPA1 could derive from its activation by reactive electrophiles on peripheral nociceptor terminals with consequent localized release of proinflammatory mediators and vasoactive substances (37). Expression and activation of TRPA1 on keratinocytes has been suggested to mediate the release of interleukins, mast cell degranulation, and the development of allergic dermatitis (38), representing another potential mechanism whereby this channel contributes to Tmem79−/−-associated itch.
Tmem79-deficient mice provide a robust and compelling model of AD. Our results demonstrate that the main somatosensory component of AD, namely itch, is mediated through a keratinocyte-, PGE2/EP3-, mast cell-, and histamine-dependent signaling pathway (Fig. 4H). While the biochemical function of TMEM79 remains uncertain, our findings lead us to propose that it plays a role in scavenging reactive electrophiles or other deleterious products of oxidative stress that, when left unchecked, foster a proinflammatory state resulting in the observed phenotypes. Our genetic models also suggest that Tmem79 expressed in keratinocytes affords the bulk of this proposed protective function, although expression in sensory nerve fibers contributes as well. Indeed, this is consistent with a model in which TMEM79 neutralizes freely diffusible endogenous irritants in a paracrine manner, irrespective of their cellular origin. In any case, Tmem79−/− mice represent a tractable and robust model for studying mechanisms of AD and itch and for evaluating possible treatments. In this regard, we show that interfering with PGE2 synthesis/signaling or histaminergic signaling through H4 receptors, in addition to TRPA1 antagonism, are efficacious in treating the debilitating pruritic component of AD. More broadly, our work suggests that enhancing the activity of glutathione transferases or other mechanisms to reduce oxidative stress may be useful strategies for protecting against the development of AD and other inflammatory pain signals.
Materials and Methods
Animal Use, Husbandry, and Genotyping.
Mice were bred and housed in accordance with University of California, San Francisco (UCSF) Institutional Animal Care guidelines. Two to five mice were housed together on a 12-h light/dark schedule with constant access to food and water. Tmem79−/− mice were generated by ES electroporation and homologous recombination of a pBS targeting vector containing the Tmem79 gene lacking exon 2 with the start codon. The loss of exon 2 in Tmem79−/− mice was confirmed using the following primers: Tmem79−/− forward: 5′-AGC CTC CCA TTC CAA AGC-3′ and reverse: 5′-AGT CGT GCT GCT TCA TGT G-3′. Tmem79-KI mice were generated by Ingenious Targeting Laboratory using ES electroporation and homologous recombination of a targeting vector containing the Tmem79 coding sequence fused to eGFP via a 2A peptide. The vector also features and an inverted floxed (Lox71 and Lox66) tandem dimer tomato (TdT) that is knocked in to replace the Tmem79 coding sequence at the start codon following Cre recombination. Mismatched lox sequences prohibit further recombination of the locus. The genotype of Tmem79-KI mice was confirmed using the following primers: Tmem79-KI forward: 5′-CTG ATA TAC TGG TTG ACC TTT GCT-3′ and reverse: 5′-CCA GGC CTA CAA CTG TTC CA-3′. Tmem79-KI mice were bred to Tg(KRT14-cre)1Amc/J (K14-Cre; strain 004782; Jackson Laboratory) or Peripherin–Cre (Prph–Cre) mice (39) to produce Tmem79-KI;K14-Cre, Tmem79-KI;Prph-Cre, or Tmem79-KI;K14-Cre;Prph-Cre conditional-knockin mice. Cre alleles were detected and differentiated using the following primers: K14-Cre forward: 5′-TTC CTC AGG AGT GTC TTC GC-3′ and reverse: 5′-GTC CAT GTC CTT CCT GAA GC-3′; Prph-Cre forward: 5′-TGA AGT CAG GCC AGT GGG AG-3′ and reverse: 5′-CCT GAT CCT GGC AAT TTC GGC T-3′.
RT-PCR from Whole Tissues.
Adult mice were euthanized by CO2 asphyxiation, and tissues were dissected into TRIzol reagent (Invitrogen) and were kept on ice until processed. Tissues were homogenized using a T25 Ultra-Turrax homogenizer (IKA), and RNA was extracted and purified using TRIzol and chloroform extraction according to the recommendations of the TRIzol reagent manufacturer. Following extraction of RNA, the aqueous phase was cleaned using RNA Clean and Concentrator (Zymo), and RNA was eluted into RNase-free water. The quality and concentration of RNA was determined using the Agilent 2100 Bioanalyzer. Samples were used for downstream applications only if the RNA integrity number was greater than 7. Isolated RNA was stored at −80 °C until used for RT-PCR. For each tissue, equivalent RNA content was used to carry out first-strand cDNA synthesis using SuperScript III (Thermo Fisher) according to the manufacturer’s recommendations. First-strand cDNA synthesis reactions for each tissue were carried out in triplicate and were combined. Following cDNA synthesis, RT-PCR reactions were run in triplicate using SYBR Green dye (Applied Biosciences) and Stratagene Mx3005P. Average cycle threshold (Ct) values were calculated, and relative expression of Tmem79 was determined using Rpl19 as a control. Primers were determined to have similar efficiencies across a series of seven twofold dilutions. Melt curves were assessed to ensure amplification of single product. Primers used were Tmem79 forward: 5′-CTT GGT TTA CAC CCT TCG CT-3′ and reverse 5′-GTA TAT CAG CCG AGA TAT GGC A-3′; Rpl19 forward: 5′-GAT TGA CCG CCA TAT GTA TCA CAG-3′ and reverse: 5′-ACA GAG TCT TGA TGA TCT CTT CCT-3′.
RT-PCR from Cultured Rat Sensory Neurons.
P0 rats were euthanized by cooling and decapitation. TG were dissected from rats and incubated in 1 mg/mL collagenase (Worthington Biochemical) and then in 0.05% trypsin-EDTA with phenol red (25300054; Thermo Fisher) for 10 min each. TG were transferred to complete DMEM [DMEM (Thermo Fisher) with 10% FBS and 1× penicillin/streptomycin antibiotic] and then were carefully passed through the tips of P1000 and P200 Pipetman pipettes (Gilman) to mechanically triturate and dissociate the ganglia. Dissociated neurons were plated on coverslips overnight (O/N) at 37 °C and 5% CO2. After O/N incubation, neurons were incubated in SIN-1(BML-CN245-0020; Enzo Life Science) in complete DMEM for 16 h. Following incubation, neurons were collected in an excess of TRIzol, and RT-PCR was carried out as described above. Primers used were Tmem79 forward: 5′-CTT GGT TTA CAC CCT TCG CT-3′ and reverse: 5′-GTA TAT CAG CCG AGA TAT GGC A-3′; Trpv1 forward: 5′-GGA TGA GGT GAA CTG GACTAC-3′ and reverse: 5′-GTG CTA TGC CTA TCT CGA GTG-3′; Nqo1 forward: 5′-GAG CCC GGA TAT TGT AGC TG-3′ and reverse: 5′-CTT CTT ATT CTG GAA AGG ACC C-3′; Rpl19 forward: 5′-AGC CTG TGA CTG TCC ATT CC-3′ and reverse: 5′-GGC AGT ACC CTT CCT CTT CC-3′.
RNA Sequencing.
Sequencing libraries were prepared as described (40). Briefly, cDNA libraries were prepared from poly(A)+ RNA using the Illumina mRNA-Seq Sample Prep Kit (RS-100-0801) according to the manufacturer’s instructions. Libraries were then sequenced on the Illumina Genome Analyzer II using two 36-cycle sequencing kits (FC-104-3002) to read 80 nt of sequence from a single end of each insert, by standard protocols. Between 2.4 million and 12.5 million inserts were sequenced for each sample. Sequences were aligned to the mouse reference genome mm9 using TopHat v 07.0 (Trapnell Laboratory, Center for Bioinformatics and Computational Biology, University of Maryland) and Bowtie v0.9.7 (Trapnell Laboratory, Center for Bioinformatics and Computational Biology, University of Maryland), and the number of sequences aligned to Tmem79 was determined for each neuronal tissue type using Cufflink (Trapnell Laboratory, Center for Bioinformatics and Computational Biology, University of Maryland).
Tissue Dissection and Cryosectioning.
Adult mice were deeply anesthetized with pentobarbital and then were perfused transcardially with 10 mL of PBS followed by 10 mL of 10% neutral buffered formalin (NBF). Tissues were dissected, postfixed in 10% NBF at 4 °C O/N, and then cryoprotected in PBS with 30% (wt/vol) sucrose O/N at 4 °C. Just before sectioning, tissue samples were embedded in OCT compound (VWR) at −20 °C. DRGs and all other tissues were sectioned with a Leica CM3050 S cryostat at 12 µm and 20 µm, respectively, and then were thaw-captured on Diamond White Glass slides (Globe Scientific). Slides were stored at −20 °C until use. Slides were used within 2 wk of processing to produce optimal signals.
In Situ Hybridization of Tissue Sections.
Tissue sections were processed using the ViewRNA ISH Tissue 1-plex or 2-plex Assay Kits (Affymetrix) following the manufacturer’s recommendations including frozen tissue modifications. Affymetrix was commissioned to design a type 1 probe set to mouse Tmem79 (NM_024246.5) and a type 6 probe set to mouse Trpa1 (NM_177781.4). For two-plex processing, endogenous alkaline phosphatase was inactivated by incubation in H2O with 0.1 M HCl and 300 mM NaCl. For one- and two-plex protocols, the H&E counterstaining procedure was omitted. Images were acquired with a Leica DMRB microscope and a DFC500 digital camera using Leica Application Suite v3.5.0 and then were further analyzed using ImageJ software (NIH).
Immunohistochemical Staining of Tissue Sections.
Slides containing sections of DRG or dorsal skin were blocked for 1 h at RT in PBS with 0.1% (vol/vol) Triton X-100 (Sigma) and 10% normal goat serum (NGS). Slides were briefly rinsed once with PBS with 0.1% Triton X-100 and 2.5% NGS and were incubated O/N with primary antibody in PBS with 0.1% Triton X-100 and 2.5% NGS at 4 °C. Following O/N incubation, slides were washed three times with vigorous agitation (2 min each washing) in fresh PBS, rinsed once with PBS with 0.1% Triton X-100, and were incubated in secondary antibody for 2 h in PBS with 0.1% Triton X-100 at RT in the dark. Following incubation, slides were washed three times with vigorous agitation (2 min each washing) in fresh PBS and were mounted in ProLong Gold antifade reagent with DAPI (Life Technologies) under a coverslip. Slides were stored covered at RT until used for imaging. Epifluorescent images were acquired with a Leica DMRB microscope mounted with a DFC500 digital camera using Leica Application Suite v3.5.0 or with an Infinity 3-3UR Monochrome CCD digital camera using Infinity Analyze software. Confocal images were acquired with a Nikon Ti inverted microscope equipped with an Andor Borealis CSU-W1 spinning-disk confocal system, an Andor four-line laser launch (100 mW at 405, 561, and 640 nm; 150 mW at 488 nm), a Lumencor Spectra X six-channel LED illuminator, a Sutter excitation and emission filter wheel, and an Andor DU-888 EMCCD camera using Micro-Manager software (Open Imaging). Following capture, all images were further analyzed using ImageJ software. The following primary antibodies were used: chicken anti-GFP (1:500; A10262; Thermo Fisher), rabbit anti-RFP (1:500; 600-401-379 which recognizes tdT; Rockland), rat anti-CD45 (1:500; 103101; BioLegend), rabbit anti-H1R (1:100; AHR-001; Alomone Labs), rabbit anti-H4R (1:1,000; Biorbyt), guinea pig anti-TRPV1 (1:1,000; D.J. laboratory, UCSF), guinea pig anti-substance P (1:5,000; gift of John Maggio, University of Cincinnati, Cincinnati), mouse anti-NF200 (1:10,000; N5389; Sigma), rabbit anti-CGRP (1:10,000; T-4031.400; Peninsula Labs), rabbit anti-peripherin (1:1,000; SAB4502419; Sigma), and rabbit anti-TH (1:5,000; EP1532Y; Abcam). Fluorophore-conjugated secondary antibodies raised in goat against mouse, rat, rabbit, chicken, or guinea pig were used, as appropriate (1:1,000; Alexa-Fluor 488, 568, or 596; Life Technologies). To identify IB4-binding cells, biotinylated IB4 (1:1,000; B-1205; Vector Labs) and fluorophore-conjugated streptavidin (1:1,000; Alexa-Fluor 488; Life Technologies) were used in place of primary and secondary antibodies. Mouse anti-K14 primary antibody (ab7800; Abcam) was preconjugated to Alexa-Fluor 594 using the APEX Alexa-Fluor 594 Antibody Labeling Kit according to the manufacturer’s recommendations and was used at 1:100.
Immunohistochemical Staining of Transiently Transfected HEK293T Cells.
HEK293T cells were grown to confluence in a 10-cm plate and were split 1:15 into six wells of a six-well plate containing poly-l-lysine (Sigma)–coated 8-mm round coverslips. Cells were allowed to adhere for 2 h at 37 °C and 5% CO2 and then were transfected as directed with Lipofectamine 3000 (Thermo Fisher) and pcDNA3.1+ vectors containing the coding sequence for Tmem79 linked to HA via Gly-Ala on either the N or C terminus (HA coding sequence: 5′-TACCCATACGATGTTCCAGATTACGCT-3′). Following transfection, cells were allowed to incubate O/N at 37 °C and 5% CO2; then coverslips were carefully transferred to slides. To prepare the FIX− condition, slides were washed once with PBS (note: all washes were conducted using gentle pipetting) and then were blocked with PBS for 1 h with 0.1% Triton-X 100 and 10% NGS at RT. Following blocking, slides were rinsed once with PBS, incubated in primary antibody (rabbit anti-HA; 1:500; H6908; Sigma) for 1 h in PBS with 0.1% Triton-X 100 and 2.5% NGS at RT and then were rinsed three times with PBS. After rinsing, slides were incubated with a secondary antibody (1:1,000; Alexa-Fluor 594; Life Technologies) for 30 min in PBS with 0.1% Triton-X 100 at RT, were rinsed three times with PBS, and were mounted in ProLong Gold antifade reagent with DAPI (Life Technologies) under a larger coverslip. To prepare the FIX+ condition, slides were fixed in PBS with 10% NBF for 10 min and were rinsed twice with PBS before undergoing the FIX− steps outlined above, concluding with the placement of a larger coverslip. Slides were stored in the dark at RT until used for imaging. Images were acquired using a Nikon Ti inverted microscope equipped with an Andor Borealis CSU-W1 spinning-disk confocal system, an Andor four-line laser launch (100 mW at 405, 561, and 640 nm; 150 mW at 488 nm), a Lumencor Spectra-X six-channel LED illuminator, a Sutter excitation and emission filter wheel, and an Andor DU-888 EM CCD camera using Micromanager software (Open Imaging) and then were further processed using ImageJ software.
Histological and Cytological Staining.
Dorsal hairy skin or lung tissues were dissected and prepared as noted above and then were stained with Toluidine blue O or H&E. For Toluidine blue O staining, a stock solution was prepared by dissolving 1 g of Toluidine blue O (Sigma) into 70% ethanol. Fresh Toluidine blue working solution was prepared by diluting the stock solution 1:10 into 1% sodium chloride in distilled water. The working solution was adjusted to pH 2.4. Slides were rehydrated in tap water (40 dips) and then were stained for 2 min in working solution. Slides were rinsed in tap water (30 dips) and were cleared in 100% ethanol (30 dips) followed by xylene (6 min). For histology, slides were mounted in DPX Mountant (Sigma) under a coverslip and were allowed to set O/N at RT in the dark until used for imaging. For H&E staining, slides were rehydrated in tap water (40 dips) and were incubated twice (2 min each incubation) in hematoxylin solution (Gill no. 2; Sigma). Slides were blued in tap water (40 dips), dipped twice in eosin (Eosin Y Solution, alcoholic, with phyloxine; Sigma). Following eosin staining, slides were rinsed in tap water (30 dips) and were cleared in 100% ethanol (30 dips) followed by xylene (30 dips). For histology, slides were mounted in DPX Mountant (Sigma) under a coverslip and were allowed to set O/N at RT in the dark. Slides were stored covered at RT until used for imaging. Images were acquired with a Nikon Eclipse E600 and an Infinity 3-6URC Color CCD digital camera using Infinity Analyze software. Mast cells in the dermis were counted in each unique field captured from sections of dorsal hairy skin. Degranulated mast cells were counted as a percentage of total (degranulated + nondegranulated) mast cells in each unique field captured from sections of dorsal hairy skin following treatment with vehicle or drug. Dermal thickness was calculated in each unique field as the average of 15 measurements made from deepest portion of the stratum basale to the most superficial portion of the hypodermis as identified by adipocytes.
Cell Preparation from Tissues and Stimulation for Intracellular Cytokine Staining.
Single-cell suspensions of skin draining lymph nodes were mechanically dissociated through a 100-µm filter, and dorsal skin (2.5 cm2) was processed as previously described (41). Single cells were washed in tissue-culture medium and filtered. Cells were counted using an automated cell counter (NucleoCounter NC-200; ChemoMetec) to determine the absolute number of specific cell populations per unit area. Single cells (2–3 × 106) were stained for flow cytometry or cultured for intracellular cytokine staining using a phorbol12-myristate13-acetate and ionomycin cell-stimulation mixture (Tonbo Biosciences).
Flow Cytometry.
Single-cell suspensions were pelleted, and cells were washed and stained with Ghost Viability dye (Tonbo Biosciences) in PBS. Following a wash in PBS, cells were stained for surface markers in PBS containing 2% FCS. For intracellular staining, cells were fixed and permeabilized with a FoxP3 buffer set (eBioscience). Samples were run on a Fortessa analyzer (BD Biosciences) in the UCSF Flow Cytometry Core and were collected using FACS Diva software (BD Biosciences). Flow cytometry data were analyzed using FlowJo software (TreeStar). Fluorophore-conjugated antibodies specific for mouse surface and intracellular antigens were purchased from eBioscience, BD Biosciences, and BioLegend. The following antibodies and clones were used: anti-Ly6G (1A8), anti-CD11b (M1/70), anti-CD11c (HL3), anti-MHC class II (M5/114.15.2), anti-Ly6C (HK1.4), anti–IL-17 (SHLR17), anti–IFN-γXMG1.2), anti-CD3 (145-2C11), anti-CD4 (RM4-5), anti-CD8 (SK1), anti-TCRγδ), anti-CD45 (30-F11), Siglec-F (E50-2440), anti-CD117 (2B8), anti-FcεR1 (MAR-1), and anti-FoxP3 (FJK-16s).
Protein Sequence Alignment and Topology Prediction.
The TMEM79 sequence was obtained from the National Center for Biotechnology Information. Blastp was used to query the TMEM79 sequence against reference proteins in the Mus musculus protein database. Blastp query of TMEM79 revealed a conserved domain found in Membrane-Associated Proteins in Eicosanoid and Glutathione metabolism (MAPEG, pfam01124). Protein sequences encoded by mouse MAPEG family members (Mgst1, Pges1, Mgst3, Flap, Mgst2, and Ltc4s) were aligned to TMEM79 using ClustalW2 multiple sequence-alignment software. TMEM79 protein topology was predicted using TMpred software.
Heterologous Expression of Tmem79 and Mutant Generation.
For reactive species assays, Tmem79 was transiently expressed in HEK293T cells using Lipofectamine 3000 (Thermo Fisher). A QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent) was used to generate amino acid substitutions in the coding sequence of Tmem79 in pcDNA3.1+.
DCF Assay in HEK293T Cells.
HEK293T cells were grown to confluence in a 10-cm plate and then were split 1:15 into six wells of a six-well plate. Cells were allowed to adhere for 2 h and then were transfected with 5 μg of plasmid (2.5 μg of mCherry and 2.5 μg of either empty pcDNA3.1+ or pcDNA3.1+ containing the coding sequence for Tmem79 or Tmem79 amino acid substitution mutants). Cells were incubated for >24 h in transfection medium at 37 °C and 5% CO2. Following incubation, cells were detached using 0.05% trypsin-EDTA with phenol red and gentle pipetting, resuspended in complete DMEM, and plated in a poly-l-lysine–coated 96-well plate (354640; Corning). Cells were allowed to adhere O/N at 37 °C and 5% CO2 and then were washed twice in Ringer’s solution (280 mM NaCl, 10 mM KCl, 4 mM CaCl2⋅0.2H2O, 4 mM MgCl2⋅0.6H2O, 20 mM d-glucose, 20 mM Hepes, pH 7.4) and incubated in 5 μM DCF (cell-permeant 2,7-dichlorodihydrofluorescein diacetate or H2DCFDA; D399; Thermo Fisher) for 10 min at 37 °C and 5% CO2. Cells were washed twice in Ringer’s solution and then were introduced to oxidant in Ringer’s solution. Immediately following incubation of cells in oxidant, plates were read (excitation at 495 nm/emission at 529 nm) at 37 °C using a BioTek H4 plate reader (Thermo Fisher). Fluorescence readings were collected from wells starting at time 0 and continuing every 10 min for 1 h. Oxidants were hydrogen peroxide (516813; Sigma), tert-butyl hydroperoxide (416665; Sigma), sodium nitroprusside (71780; Sigma), and SIN-1 (BML-CN245-0020; Enzo Life Science). N-acetyl cysteine (A7250; Sigma) was added during 10 min DCF loading.
Acute Keratinocyte Dissociation and ELISA for Prostaglandins.
Adult mice were euthanized by CO2 asphyxiation and decapitation. Hair was cut from mice dorsal and ventral surfaces using hair trimmers (Oster). A 3 × 2 cm area of trimmed skin was dissected, and submucosa was removed using blunt dissection. Skins were incubated floating on the surface of 0.25% trypsin-EDTA with phenol red (Thermo Fisher) for 1 h at 37 °C and 5% CO2. Following incubation, skins were transferred to complete DMEM, and keratinocytes were dissociated into solution by scraping the surface of the segment with the blunted edge of a pair of forceps. Dermis from each skin was discarded, and the remaining solution and particulates were collected. The suspension was shaken vigorously by hand for 30 s to homogenize and dissociate keratinocytes and then was filtered using a 100-μm cell strainer (BD). For prostaglandin quantification using ELISA, keratinocytes dissociated from age- and sex-matched wild-type or Tmem79−/− mice were incubated for an additional 30 min at 37 °C in complete DMEM, sonicated on ice for 30 s (10 s on/10 s off), and spun at 8,000 × g at 4 °C to pellet debris. Supernatant was collected and kept on ice until processing using ELISA kits for quantification of PGE2 (514010; Cayman Chemical) or PGH2 (CEA612Ge; Cloud Clone) according to the manufacturer’s guidelines.
Lipidomics.
Keratinocyte lysates were collected as described above; then cold methanol was added immediately to stabilize lipid mediators. Samples were stored at −80 °C until processing. Lipid mediators and metabolites were extracted through C-18 solid-phase columns and quantified via LC-MS/MS as described previously (42–44). Deuterated internal standards (PGE2-d4, LTB4-d4, 15-HETE-d8, LXA4-d5, DHA-d5, and AA-d8) were added to samples to calculate extraction recovery. The LC-MS/MS system consisted of an Agilent 1200 Series HPLC, Luna C18 column (Phenomenex), and AB Sciex QTRAP 4500 mass spectrometer. Analysis was carried out in negative ion mode, and lipid mediators were quantitated using the scheduled multiple reaction monitoring (MRM) mode using multiple specific transition ions.
Dichlorofluorescein Assay in Dissociated Keratinocytes.
Following dissociation and filtration using a cell strainer as described above, keratinocytes were pelleted at 425 × g and then were washed once in complete DMEM and once in Ringer’s solution. Following the washes, keratinocytes were resuspended in Ringer’s solution and were incubated in 5 μM dichlorofluorescein (DCF) for 10 min at 37 °C and 5% CO2. Keratinocytes were pelleted at 425 × g, resuspended in 1.5 mL Ringer’s solution, and incubated in SIN-1. Immediately following the incubation of cells in SIN-1, plates were read (excitation at 495 nm/emission at 529 nm) at 37 °C using a BioTek H4 plate reader (Thermo Fisher). Fluorescence readings were collected from wells starting at time 0 and continuing every 1 min for 1 h.
Primary Cultures of Adult DRG Neurons and Calcium Imaging.
Glass coverslips (8 mm; Electron Microscopy Sciences) were washed briefly with 70% ethanol, dried with a Kimwipe (Kimberly-Clark), and were coated with 40 μL of 10 μg/mL laminin (23017-015; Thermo Fisher) in 0.01% poly-L-lysine (P4707; Sigma) and incubated for >1 h at 37 °C and 5% CO2. Coverslips were then rinsed once with complete DMEM. Adult mice were euthanized by CO2 asphyxiation and decapitation, and vertebral columns were dissected. DRGs from a single mouse were dissected and combined in 1 mL of collagenase I (1 mg/mL in Ringer’s solution) with DNase I (150 K/mL; DN25; Sigma) on ice. DRGs in 1 mL collagenase were incubated for 20 min in a 37 °C water bath; then supernatant was removed without disturbing the loose DRG pellet. DRGs were resuspended without trituration in 1 mL of 0.25% trypsin-EDTA with phenol red (Thermo Fisher) with DNase I (150 K/mL) and were incubated for 20 min in a 37 °C water bath. Supernatant was removed without disturbing the loose DRG pellet. DRGs were resuspended in 1 mL of complete DMEM (preheated to 37 °C) with DNase I (150 K/mL) and were triturated 30× each with a P1000 and P200 Pipetman to dissociate neurons into the solution. The homogenate was gently loaded onto the surface of 3 mL of 15% Percoll (vol/vol) Ringer’s solution in a 15-mL Falcon tube (Corning) and was spun at 1,300 × g at 4 °C for 10 min. The supernatant was removed without disturbing the pellet. The pellet was completely resuspended in 300 μL complete DMEM, and 25 μL of suspended neurons were plated onto prepared glass coverslips (one mouse produced 12 coverslips) and were allowed to plate O/N at 37 °C and 5% CO2. Following O/N plating, Fura-2 AM working solution [10 μL of 1 mg/mL Fura-2 AM dye (F1201; Life Technologies) and 1 μL of 20% pluronic acid in 1 mL Ringer’s solution] was prepared. One to two hours before imaging, 25 μL of Fura-2 AM working solution was added to coverslips; then neurons were allowed to load in the dark at RT. Coverslips containing Fura-2 AM–loaded neurons were placed in a laminar flow system with bath solutions controlled by a ValveBank8 Perfusion System (Automate Scientific) and driven by two synchronized Dynamax peristaltic pumps (Rainin). Sequential excitation of Fura at 340 and 380 nm in neurons was conducted using a Lambda LS Illuminator with a 10-2 Controller (Sutter). Emission signals were captured using an ORCA-ER camera and controller (Hamamatsu) mounted on a Nikon Eclipse TE200 microscope. Data were digitized and analyzed using MetaFluor software (Molecular Devices).
Statistics and Experimental Design.
Sample sizes for experiments were chosen as the minimum number of independent observations required for statistically significant results, based on previous experience with these assays. No statistical methods were used to predetermine sample size. Numbers of micrographs, replicates, or animals used for each experiment are reported in the figure legends. For behavioral experiments, animals were randomly chosen for experimental cohorts, conditions were compared within the same experimental time course, and experimenters were double-blinded to experimental conditions. Animal genotype was tracked by ear tags, and genotype unblinding occurred after analysis was complete.
Data were analyzed using Prism 6 (GraphPad) or Microsoft Excel software and significance testing with a Student’s t test, Holm t test, or one-way ANOVA with Holm–Sidak’s multiple comparisons correction, as noted in the figure legends. Tests were either one- or two-tailed as indicated in the figure legends. The number of experiments and significance are reported in the figure legends. All significance tests were justified as appropriate given the experimental design and the nature of the comparisons. With our statistical tests, we assume equal variance and normal distributions of data, which are common assumptions made for significance testing within these experimental paradigms as previously published by our group and others.
Animal Behavior and Pharmacology.
Behavioral experiments were approved by the UCSF Institutional Animal Care and Use Committee and were in accordance with the NIH Guide for the Care and Use of Laboratory Animals (45) and the recommendations of the International Association for the Study of Pain. For scratching assays, mice were allowed to acclimate in clear, ventilated cylindrical chambers for 30 min. Chambers were covered by a clear lid, and opaque barriers were placed between chambers to prohibit any visual interference among mice. Following acclimation, behavior was captured for 30 min using a HDR-CX160 High-Definition Handycam Camcorder (Sony); then scratching behavior was quantified from recordings. Scratching behavior was defined as oral contact, including licking or biting, with the dorsal or ventral hairy skin including upper portions of limbs. To assess the effects of pharmacologic compounds on scratching behavior, mice were i.p. administered drug or vehicle [10% DMSO (vol/vol) and 10% Tween 20 (vol/vol) in sterile normal saline] (doses are indicated in figure legends) by a blinded operator following the 30-min acclimation period. Behavior was captured for 30–60 min following drug or vehicle administration, and scratching behavior was quantified by a blinded observer from recordings. For experiments involving s.c. injection of histamine or chloroquine, mice were administered drug or vehicle by a blinded operator following the 30-min acclimation period, and behavior was captured immediately for 20 min. Scratching behavior in these two assays was defined as forepaw or hindpaw contact at or near the s.c. injection site and was quantified by a blinded observer from recordings. For experiments involving intraplantar injection of capsaicin or mustard oil, mice were administered drug by an operator following the 30-min acclimation period, and behavior was captured immediately for 10 min (capsaicin) or 20 min (mustard oil). Paw licking was quantified by a blinded observer from recordings. For studies of intrathecal capsaicin administration, mice (20–40 g) were anesthetized with 1.5% isoflurane (vol/vol) and injected intrathecally with 5 μL capsaicin (10 μg) or vehicle [10% ethanol (vol/vol) and 10% Tween 80 (vol/vol) in sterile normal saline] using a Hamilton syringe at the level of the pelvic girdle (46). Behavioral assays were performed 1–14 d after capsaicin injection. Anxiety-related behavior was assessed using the elevated plus maze as previously described (47). For the hot plate assay, latency to nocifensive responses (flinching, jumping, or licking) for each mouse was calculated as an average of three trials (60-s maximum and at 10-min intervals) in response to indicated surface temperatures (ThermoBrite; Abbott Molecular).
Supplementary Material
Acknowledgments
We thank Diana Bautista, Allan Basbaum, Ralph Marcucio, and members of the D.J. laboratory for their insights, thoughtful discussion, and assistance surrounding this work. We also thank J. Poblete, J. Bráz, B. Zirak, S. Chen, X. Wang, C. Solorzano, and F. Cevikbas for technical advice and assistance. This work was supported by Ruth Kirschstein NIH Predoctoral Fellowship DE023476 (to J.J.E.), Career Development Award AR070910 (to A.M.), and NIH Grants NS081115 and NS105038 (to D.J.) and EY026082 (to K.G.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 12851.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814132115/-/DCSupplemental.
References
- 1.Sidbury R, et al. American Academy of Dermatology Guidelines of care for the management of atopic dermatitis: Section 3. Management and treatment with phototherapy and systemic agents. J Am Acad Dermatol. 2014;71:327–349. doi: 10.1016/j.jaad.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weidinger S, Novak N. Atopic dermatitis. Lancet. 2016;387:1109–1122. doi: 10.1016/S0140-6736(15)00149-X. [DOI] [PubMed] [Google Scholar]
- 3.Ji H, Li XK. Oxidative stress in atopic dermatitis. Oxid Med Cell Longev. 2016;2016:2721469. doi: 10.1155/2016/2721469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin H, He R, Oyoshi M, Geha RS. Animal models of atopic dermatitis. J Invest Dermatol. 2009;129:31–40. doi: 10.1038/jid.2008.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sasaki T, et al. A homozygous nonsense mutation in the gene for Tmem79, a component for the lamellar granule secretory system, produces spontaneous eczema in an experimental model of atopic dermatitis. J Allergy Clin Immuno. 2013;132:1111–1120.e4. doi: 10.1016/j.jaci.2013.08.027. [DOI] [PubMed] [Google Scholar]
- 6.Saunders SP, et al. Tmem79/Matt is the matted mouse gene, is a predisposing gene for atopic dermatitis in human subjects. J Allergy Clin Immunol. 2013;132:1121–1129. doi: 10.1016/j.jaci.2013.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jakobsson PJ, Mancini JA, Riendeau D, Ford-Hutchinson AW. Identification and characterization of a novel microsomal enzyme with glutathione-dependent transferase and peroxidase activities. J Biol Chem. 1997;272:22934–22939. doi: 10.1074/jbc.272.36.22934. [DOI] [PubMed] [Google Scholar]
- 8.Morgenstern R, Meijer J, Depierre JW, Ernster L. Characterization of rat-liver microsomal glutathione S-transferase activity. Eur J Biochem. 1980;104:167–174. doi: 10.1111/j.1432-1033.1980.tb04412.x. [DOI] [PubMed] [Google Scholar]
- 9.Morgenstern R, DePierre JW, Ernster L. Activation of microsomal glutathione S-transferase activity by sulfhydryl reagents. Biochem Biophys Res Commun. 1979;87:657–663. doi: 10.1016/0006-291x(79)92009-6. [DOI] [PubMed] [Google Scholar]
- 10.Letelier ME, et al. Comparative effects of superoxide anion and hydrogen peroxide on microsomal and cytosolic glutathione S-transferase activities of rat liver. Biol Trace Elem Res. 2010;134:203–211. doi: 10.1007/s12011-009-8461-3. [DOI] [PubMed] [Google Scholar]
- 11.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 12.Jowsey IR, Jiang Q, Itoh K, Yamamoto M, Hayes JD. Expression of the aflatoxin B1-8,9-epoxide-metabolizing murine glutathione S-transferase A3 subunit is regulated by the Nrf2 transcription factor through an antioxidant response element. Mol Pharmacol. 2003;64:1018–1028. doi: 10.1124/mol.64.5.1018. [DOI] [PubMed] [Google Scholar]
- 13.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: Reassessment of the ARE consensus sequence. Biochem J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bantz SK, Zhu Z, Zheng T. The atopic march: Progression from atopic dermatitis to allergic rhinitis and asthma. J Clin Cell Immunol. 2014;5:202. doi: 10.4172/2155-9899.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu FT, Goodarzi H, Chen HY. IgE, mast cells, and eosinophils in atopic dermatitis. Clin Rev Allergy Immunol. 2011;41:298–310. doi: 10.1007/s12016-011-8252-4. [DOI] [PubMed] [Google Scholar]
- 16.Kawakami T, Ando T, Kimura M, Wilson BS, Kawakami Y. Mast cells in atopic dermatitis. Curr Opin Immunol. 2009;21:666–678. doi: 10.1016/j.coi.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin X, et al. Elevated levels of mast cells are involved in pruritus associated with polycythemia vera in JAK2V617F transgenic mice. J Immunol. 2014;193:477–484. doi: 10.4049/jimmunol.1301946. [DOI] [PubMed] [Google Scholar]
- 18.Yamaura K, Doi R, Suwa E, Ueno K. Repeated application of glucocorticoids exacerbate pruritus via inhibition of prostaglandin D2 production of mast cells in a murine model of allergic contact dermatitis. J Toxicol Sci. 2012;37:1127–1134. doi: 10.2131/jts.37.1127. [DOI] [PubMed] [Google Scholar]
- 19.Gerber PA, et al. Preliminary evidence for a role of mast cells in epidermal growth factor receptor inhibitor-induced pruritus. J Am Acad Dermatol. 2010;63:163–165. doi: 10.1016/j.jaad.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 20.Hu YP, et al. Reactive oxygen species mediated prostaglandin E2 contributes to acute response of epithelial injury. Oxid Med Cell Longev. 2017;2017:4123854. doi: 10.1155/2017/4123854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahn SM, et al. Fructose-1,6-diphosphate attenuates prostaglandin E2 production and cyclo-oxygenase-2 expression in UVB-irradiated HaCaT keratinocytes. Br J Pharmacol. 2002;137:497–503. doi: 10.1038/sj.bjp.0704896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halova I, Draberova L, Draber P. Mast cell chemotaxis–Chemoattractants and signaling pathways. Front Immunol. 2012;3:119. doi: 10.3389/fimmu.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravid A, Shenker O, Buchner-Maman E, Rotem C, Koren R. Vitamin D induces cyclooxygenase 2 dependent prostaglandin E2 synthesis in HaCaT keratinocytes. J Cell Physiol. 2016;231:837–843. doi: 10.1002/jcp.25132. [DOI] [PubMed] [Google Scholar]
- 24.Fogh K, Herlin T, Kragballe K. Eicosanoids in skin of patients with atopic dermatitis: Prostaglandin E2 and leukotriene B4 are present in biologically active concentrations. J Allergy Clin Immunol. 1989;83:450–455. doi: 10.1016/0091-6749(89)90132-2. [DOI] [PubMed] [Google Scholar]
- 25.Thurmond RL, Gelfand EW, Dunford PJ. The role of histamine H1 and H4 receptors in allergic inflammation: The search for new antihistamines. Nat Rev Drug Discov. 2008;7:41–53. doi: 10.1038/nrd2465. [DOI] [PubMed] [Google Scholar]
- 26.Ohsawa Y, Hirasawa N. The role of histamine H1 and H4 receptors in atopic dermatitis: From basic research to clinical study. Allergol Int. 2014;63:533–542. doi: 10.2332/allergolint.13-RA-0675. [DOI] [PubMed] [Google Scholar]
- 27.Rossbach K, et al. Histamine H1, H3 and H4 receptors are involved in pruritus. Neuroscience. 2011;190:89–102. doi: 10.1016/j.neuroscience.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Hofstra CL, Desai PJ, Thurmond RL, Fung-Leung WP. Histamine H4 receptor mediates chemotaxis and calcium mobilization of mast cells. J Pharmacol Exp Ther. 2003;305:1212–1221. doi: 10.1124/jpet.102.046581. [DOI] [PubMed] [Google Scholar]
- 29.Dunford PJ, et al. Histamine H4 receptor antagonists are superior to traditional antihistamines in the attenuation of experimental pruritus. J Allergy Clin Immunol. 2007;119:176–183. doi: 10.1016/j.jaci.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 30.Shim WS, et al. TRPV1 mediates histamine-induced itching via the activation of phospholipase A2 and 12-lipoxygenase. J Neurosci. 2007;27:2331–2337. doi: 10.1523/JNEUROSCI.4643-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jian T, et al. TRPV1 and PLC participate in histamine H4 receptor-induced itch. Neural Plast. 2016;2016:1682972. doi: 10.1155/2016/1682972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wouters MM, et al. Histamine receptor H1-mediated sensitization of TRPV1 mediates visceral hypersensitivity and symptoms in patients with irritable Bowel syndrome. Gastroenterology. 2016;150:875–887.e9. doi: 10.1053/j.gastro.2015.12.034. [DOI] [PubMed] [Google Scholar]
- 33.Wilson SR, et al. The ion channel TRPA1 is required for chronic itch. J Neurosci. 2013;33:9283–9294. [Google Scholar]
- 34.Shields SD, Cavanaugh DJ, Lee H, Anderson DJ, Basbaum AI. Pain behavior in the formalin test persists after ablation of the great majority of C-fiber nociceptors. Pain. 2010;151:422–429. doi: 10.1016/j.pain.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Usoskin D, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 36.Li CL, et al. Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res. 2016;26:83–102. doi: 10.1038/cr.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bautista DM, Pellegrino M, Tsunozaki M. TRPA1: A gatekeeper for inflammation. Annu Rev Physiol. 2013;75:181–200. doi: 10.1146/annurev-physiol-030212-183811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang J, et al. TRPA1 mediated aggravation of allergic contact dermatitis induced by DINP and regulated by NF-κB activation. Sci Rep. 2017;7:43586. doi: 10.1038/srep43586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scherrer G, et al. VGLUT2 expression in primary afferent neurons is essential for normal acute pain and injury-induced heat hypersensitivity. Proc Natl Acad Sci USA. 2010;107:22296–22301. doi: 10.1073/pnas.1013413108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gracheva EO, et al. Molecular basis of infrared detection by snakes. Nature. 2010;464:1006–1011. doi: 10.1038/nature08943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gratz IK, et al. Cutting edge: Self-antigen controls the balance between effector and regulatory T cells in peripheral tissues. J Immunol. 2014;192:1351–1355. doi: 10.4049/jimmunol.1301777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leedom AJ, Sullivan AB, Dong B, Lau D, Gronert K. Endogenous LXA4 circuits are determinants of pathological angiogenesis in response to chronic injury. Am J Pathol. 2010;176:74–84. doi: 10.2353/ajpath.2010.090678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sapieha P, et al. 5-Lipoxygenase metabolite 4-HDHA is a mediator of the antiangiogenic effect of ω-3 polyunsaturated fatty acids. Sci Transl Med. 2011;3:69ra12. doi: 10.1126/scitranslmed.3001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Moltke J, et al. Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature. 2012;490:107–111. doi: 10.1038/nature11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed The National Academies; Washington, DC: 2011. [Google Scholar]
- 46.Solorzano C, et al. Primary afferent and spinal cord expression of gastrin-releasing peptide: Message, protein, and antibody concerns. J Neurosci. 2015;35:648–657. doi: 10.1523/JNEUROSCI.2955-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walf AA, Frye CA. The use of the elevated plus maze as an assay of anxiety-related behavior in rodents. Nat Protoc. 2007;2:322–328. doi: 10.1038/nprot.2007.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.