CIPK25, a calcium-regulated protein kinase, plays an important role in the co-ordination of auxin and cytokinin signaling in root meristem development.
Keywords: Arabidopsis, auxin, CIPK25, cytokinin, meristem, PIN, root, SHY2
Abstract
Co-ordination of auxin and cytokinin activities determines root meristem size during post-embryonic development. Calcineurin B-like proteins (CBLs) and their interacting protein kinases (CIPKs) constitute signaling modules that relay calcium signals. Here we report that CIPK25 is involved in regulating the root meristem size. Arabidopsis plants lacking CIPK25 expression displayed a short root phenotype and a slower root growth rate with fewer meristem cells. This phenotype was rescued by restoration of CIPK25 expression. CIPK25 interacted with CBL4 and -5, and displayed strong gene expression in the flower and root, except in the cell proliferation domain in the root apical meristem. Its expression in the root was positively and negatively regulated by auxin and cytokinin, respectively. The cipk25 T-DNA insertion line was compromised in auxin transport and auxin-responsive promoter activity. The cipk25 mutant line showed altered expression of auxin efflux carriers (PIN1 and PIN2) and an Aux/IAA family gene SHY2. Decreased PIN1 and PIN2 expression in the cipk25 mutant line was completely restored when combined with a SHY2 loss-of-function mutation, resulting in recovery of root growth. SHY2 and PIN1 expression was partially regulated by cytokinin even in the absence of CIPK25, suggesting a CIPK25-independent cytokinin signaling pathway(s). Our results revealed that CIPK25 plays an important role in the co-ordination of auxin and cytokinin signaling in root meristem development.
Introduction
Roots exhibit indeterminate growth which is regulated by external factors (Schenk and Jackson, 2002; Chapman et al., 2012). Root cells are produced in the root apical meristem (RAM) region. The quiescent center (QC), a group of cells located at the rootward proximal border of the RAM, is considered as the organizing center. The cells that surround the QC constitute the stem cell niche (SCN) that gives rise to the different cell files of the root by cell proliferation. Each stem cell divides and the daughter cells undergo additional cycles of division and move longitudinally towards the shoot. Eventually, the proliferation rate of these cells decreases as they move upwards; at one point they show a very low probability of division and begin to elongate rapidly at the elongation zone (EZ). For practical convenience, the region between the QC and the first elongated cortex cell was considered as the RAM in several studies. A small domain of the RAM bordering the EZ shows a very low probability of cell division, but is yet to start cell elongation and is referred to as the transition domain (TD). The cells in this domain of the RAM grow at the same relative rate as the cells in the proliferative domain and pass to the EZ. As the rate of cell division is different for each cell type, the boundary of the TD towards the QC cannot be well demarcated (Verbelen et al., 2006; Dello Ioio et al., 2007, 2008; Perilli and Sabatini, 2010; Ivanov and Dubrovsky, 2013). Thus, a balance between cell division and differentiation is required for proper maintenance of meristem size.
Various phytohormones such as auxin, cytokinin, gibberellin, ethylene, and abscisic acid (ABA) play vital roles in maintenance of this balance (reviewed in Pacifici et al., 2015). Auxin promotes cell proliferation in the RAM in various ways including long- and short-distance signals mediated by the polar auxin transporters Pin-Formed (PIN), essential for creating auxin gradients (Tanaka et al., 2006). A high concentration of auxin promotes SCFTIR1-mediated degradation of the Aux/IAA family of transcriptional repressors that form heterodimeric complexes with auxin response factors (ARFs). Aux/IAA degradation releases ARFs from Aux/IAA-mediated repression, resulting in activation of auxin-responsive target genes (Kepinski and Leyser, 2005; Mockaitis and Estelle, 2008; Boer et al., 2014). On the other hand, cytokinin drastically reduces growth of the RAM primarily by promoting cell differentiation (Beemster and Baskin, 2000; Dello Ioio et al., 2008). Cytokinin signaling is mediated through a two-component system composed of membrane-localized Arabidopsis histidine kinases (AHKs) (Hwang and Sheen, 2001; Inoue et al., 2001; To and Kieber, 2008). The signal is transmitted through phosphorelay to activate Arabidopsis response regulators (ARRs) that promote expression of cytokinin target genes (To et al., 2004; To and Kieber, 2008). Transition from mitotic cells in the RAM to elongated cells in the EZ is accompanied by DNA polyploidization of cells (Dello Ioio et al., 2007; Ishida et al., 2010; Adachi et al., 2011). Cytokinin activates expression of CCS52A1, which promotes DNA polyploidization through ARR2, and thereby regulates meristem size (Takahashi et al., 2013). It is proposed that the position of the boundary between dividing and elongating zones is determined by the antagonistic interaction of auxin and cytokinin. Cytokinin-activated ARR1 directly activates expression of SHY2/IAA3, resulting in repression of ARF activity and down-regulation of auxin transporter PIN genes, causing suppression of cell division. On the other hand, a high concentration of auxin promotes degradation of SHY2 and releases ARFs from repression to promote PIN expression and cell division (Dello Ioio et al., 2008;Mockaitis and Estelle, 2008). It has recently been shown that in addition to cytokinin-mediated modulation of auxin distribution by regulating PIN expression, cytokinin-dependent auxin degradation also plays an important role in positioning the TD by creating a region of auxin minima (Di Mambro et al., 2017).
Calcium ions (Ca2+) plays a pivotal role in perception of external signals. An increase in free cytosolic Ca2+ ([Ca2+]cyt) is recognized by an array of Ca2+ sensors including calcineurin B-like proteins (CBLs). CBLs in concert with interacting kinases CIPKs (CBL-interacting protein kinases) relay the signal by phosphorylation of various substrates (Luan, 2009). Altered [Ca2+]cyt homeostasis inhibits root cell elongation (Bai et al., 2009; Wu et al., 2017). The role of Ca2+ and calcium-regulated kinases in auxin signaling has been described in several reports (Thimann and Schneider, 1939; Fuglsang et al., 2007; Duby et al., 2009; Monshausen et al., 2011; Takahashi et al., 2012; Li et al., 2016; Wang et al., 2016; Santin et al., 2017). In this study, we observed that Arabidopsis CIPK25 T-DNA insertion lines exhibited a short root phenotype resulting from reduced RAM size. Restoring expression of CIPK25 in one of these lines complemented the root phenotype, suggesting that this gene plays a role in root development. CIPK25 expression was responsive to external application of auxin and cytokinin. A mutant lacking CIPK25 expression displayed compromised auxin transport and low expression of PIN proteins. The short root phenotype of the cipk25 line was restored in the SHY2 loss-of-function background. Our data suggest that CIPK25 plays a role in balancing auxin and cytokinin signaling in root development.
Materials and methods
Plant materials, growth conditions, and treatments
Arabidopsis thaliana wild-type (ecotype Columbia; Col-0) and mutant lines (SALK_079011 and SALK_029271) with T-DNA insertions in the exon of AtCIPK25 (At5g25110) were procured from the Arabidopsis Biological Research Center. Homozygous lines with T-DNA insertions were screened using primers specific for T-DNA borders and the CIPK25 gene. Both SALK_079011 and SALK_029271 lines were used for the initial phenotyping study. The remaining experiments were performed with SALK_079011 which was designated as the cipk25 mutant. PPIN1::PIN1-GFP, DR5::GUS, and shy2-24 lines were kindly provided by Dr Jason W. Reed (University of North Carolina, USA). PPIN2::PIN2-GFP and arr1 arr12 lines were provided by Dr Ashverya Laxmi (National Institute of Plant Genome Research, India). The above-mentioned reporter lines were crossed with the cipk25 mutant. F3/F4 homozygous plants were selected by genotyping and used in the experiments. For stable line generation, 35S::CIPK25, PCIPK25::GUS, and PSHY2::GUS constructs were introduced into Col-0 plants by the Agrobacterium-mediated floral dip method as described before (Clough and Bent, 1998). The PCIPK25::CIPK25 construct was introduced into cipk25 mutant plants to generate the complementation line. T3 homozygous PSHY2::GUS-expressing plants were crossed with the cipk25 mutant for further studies. All seeds were surface-sterilized, stratified, and sown on half-strength Murashige and Skoog medium (1/2 MS) plates supplemented with 1% sucrose and 0.8% agar. Arabidopsis plants were grown on an agropeat:vermiculite (3:1) mixture in growth chambers with 120 µmol µm–2 s–1 light under a 16 h light/8 h dark cycle at 21 ± 1 °C. For root phenotyping, all plants were grown vertically on 1/2 MS medium plates as mentioned above. Root length was marked on plates from 4 days post-germination (4 dpg) onwards. Images of 10-day-old seedlings were captured for all the plates; root length and per day growth rate were measured for individual seedling using ImageJ software (Abramoff et al., 2004).
For analyzing fold expression of CIPK25 in response to hormones, Col-0 seedlings were transferred to liquid 1/2 MS medium, 12 h prior to the treatment for acclimatization. Auxin [indole acetic acid (IAA)] at 5 µM and t-zeatin at 5 µM final concentration were added to liquid medium. Root tissues were collected from treated seedlings at different time intervals. For CIPK25 expression studies at different developmental stages, root tissues were collected from seedlings grown on 1/2 MS plates. CIPK25 promoter activites were analyzed in PCIPK25::GUS lines in different tissues and at different developmental stages. Forty-five-day-old soil-grown plants were taken as matured plants. To determine promoter–reporter activity by staining, seedlings were grown on vertical 1/2 MS plates and treated with hormones for various periods before staining and microscopy.
Expression analysis by real-time PCR
Plant tissues were ground to a fine powder with liquid N2, and total RNA was isolated using the TRIzol Reagent (Invitrogen) according to the manufacturer’s protocol. DNA-free total RNA (1 µg) was converted into single-stranded cDNA using oligo(dT18) primers and a High Fidelity cDNA synthesis kit (Roche Diagnostics GmbH, Germany). Gene-specific primers were designed using the NCBI primer design tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast). For quantitative real-time PCR (qRT-PCR), primers were designed to produce 100–150 bp amplicons. qRT-PCR was done in optical 96-well plates on a Vii A 7 Real-Time PCR System (Applied Biosystems, CA, USA) using 2× Power SYBR Green PCR master mix (Applied Biosystems). A dissociation curve analysis was performed for all primer pairs, and all experimental samples yielded a single sharp peak at the amplicon’s melting temperature. ACTIN2 (At3g18780) was used as internal control. Fold induction values of target genes were calculated with the ΔΔCT equation (Rao et al., 2013) and are presented relative to the mRNA level of target genes in control conditions. Technical and biological replicates were used in all the assays. The primer pairs used are listed in Supplementary Table S1 at JXB online.
Yeast two-hybrid assay
The CIPK25 full-length coding sequence (CDS) was fused to the GAL 4 DNA-binding domain in the pGBKT7-BD vector (Clontech Laboratories Inc.). The CDS of all CBLs (CBL1–CBL10) of Arabidopsis were cloned into the pGADT7-AD (Clontech Laboratories Inc.) to be expressed as a fusion with the GAL4 activation domain. The pGBKT7-BD and PGADT7-AD constructs were co-introduced into the Y2H Gold strain of yeast using the Matchmaker Gold yeast Two-Hybrid system kit (Clontech Laboratories Inc.). The yeast colonies harboring both the plasmids were selected on double drop-out (DDO) medium lacking leucine and tryptophan and were confirmed by colony PCR. Positive colonies selected on DDO plates were grown in liquid DDO media for 2 d, adjusted to an OD600 of 1, 0.1, and 0.01. A 10 µl aliquot of each dilution was spotted on quadruple drop-out medium (-Leu-Trp-Ade-His) with 2.5 mM 3-AT (3-amino-1,2,4-triazole). Plates were incubated at 30 °C for 5 d.
Transient expression of fusion proteins and microscopic visualization
The CIPK25 CDS was cloned between the NcoI and SpeI sites of the pCAMBIA1305 vector to generate overexpression lines. To generate the CIPK25 expression line under its native promoter, 2.6 kb of the promoter and coding region with the stop codon was amplified from genomic DNA and cloned in the SmaI site of the pBI101.2 promoter-less vector. For the promoter–β-glucuronidase (GUS) fusion construct, 2.6 kb fragments upstream of the translation start codon of the corresponding genes were amplified from genomic DNA and were cloned in the pBI101.2 vector at the BamHI site (for the CIPK25 promoter) and SalI/XbaI sites (SHY2 promoter). The 35S::CIPK25-YFP construct generated by Gateway Technology (Invitrogen) in the pEG101 binary vector was introduced into Agrobacterium tumefaciens (GV3101) and infiltrated in Nicotiana benthamiana leaves for transient expression. For agro infiltration, overnight-grown cell cultures were collected and resuspended in 1 ml of infiltration buffer (10 mM MES-KOH, pH 5.6, 10 mM MgCl2, and 150 µM acetosyringone) to an optical density at 600 nm (OD600) of 0.6. The working suspensions were prepared by mixing the Agrobacterium culture harboring the 35S::CIPK25-YFP construct with an organelle marker for the plasma membrane (PM-mCherry) (Nelson et al., 2007) in a 1:1 ratio, incubated at room temperature for 3 h, and infiltrated onto the abaxial surface of fully developed leaves. Plants were cultivated for 2–3 d under growth room conditions. Subcellular fractionation of CIPK25–yellow fluorescent protein (YFP)-expressing plants was done by using the Qproteome Cell Compartment kit (Qiagen GmbH, Germany) by following the manufacturer’s instructions. Cytosolic, membrane, and nuclear protein fractions were separated by SDS–PAGE followed by western blot, and probed with anti-green fluorescent protein (GFP) (Abcam, UK) antibody. The blotted membrane was probed with the antibodies against fraction-specific marker proteins, for example anti-histone H3 (nuclear fraction) (Sigma-Aldrich, Saint Louis, MO, USA), anti-plasma membrane H+ ATPase (membrane fraction), and anti-cytosolic fructose-1,6-bisphosphatase antibodies (cytosolic fraction) (Agrisera, Sweden).
For GUS reporter staining, Arabidopsis seedlings and different plant tissues were vacuum infiltrated with GUS staining solution [50 mM sodium phosphate buffer (pH 7.0), 2 mM EDTA, 0.12% Triton, 0.4 mM ferrocyanide, 0.4 mM ferricyanide, 1.0 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide cyclohexylammonium salt] for 5–15 min and incubated in the dark at 37 °C. The staining time varied between 2 h and 12 h depending on the tissue type. Tissues were cleared of chlorophyll by treatment with 70% ethanol at 65 °C for 1 h, and analyzed by light microscopy.
For bimolecular fluorescence complementation (BiFC) assay, the full-length cDNA fragments (without the stop codon) of CBL4, CBL5, and CBL9 were amplified using PCR primers having attB adaptors and cloned into the Gateway entry vector pDONR 207 by BP clonase reaction (Invitrogen). The CIPK25 CDS was cloned into pSITE-nEYFP-C1 (CD3-1648) (Martin et al., 2009) to generate the YFPN–CIPK25 construct. CBL4, CBL5, and CBL9 were cloned similarly into pSITE-cEYFP-N1 (CD3-1651) to generate CBL4–YFPC, CBL5–YFPC, and CBL9–YFPC constructs. Agrobacterium strain GV3101 cultures harboring BiFC constructs were infiltrated in N. benthamiana leaves as discussed above. For fluorescence quantification, 6 mm leaf discs were cut out of the infiltrated areas and put into a black 96-well plate. Fluorescence intensities of leaf discs were measured in a The FLUOstar® Omega plate reader (BMG LABTECH, Germany) using a 485 ± 12 nm excitation and a 520 nm emission filter. To examine root meristem cells, seedlings were incubated in 10 µg ml–1 propidium iodide for 5–10 min, mounted in water, and visualized by a Leica TCS SP5 (Leica Microsystems, Wetzlar, Germany) laser-scanning confocal microscope at 535 nm excitation and 617 nm emission for propidium iodide. GFP was visualized at 395 nm excitation and 509 nm emission in PPIN1::PIN1-GFP- and PPIN2::PIN2-GFP-expressing roots of Col-0, cipk25, and CIPK25OX seedlings. Agroinfiltrated N. benthamiana leaf cells for subcellular localization of CIPK25–YFP and plasma membrane marker fused with mCherry were analyzed at 514–527 nm and 587–610 nm for YFP and mCherry, respectively.
Site-directed mutagenesis, bacterial expression, and kinase assay
Site-directed mutagenesis was done by replacing threonine (T) with aspartic acid (D) at the 201th position in CIPK25 as described (Meena et al., 2015). The coding regions of CIPK25 and CIPK25T/D201 were cloned into the pGEX-4T2 vector (Amersham Pharmacia Biotech) at the SmaI site and transformed into Escherichia coli BL21 (DE3)-CodonPlus cells (Stratagene, La Jolla, CA, USA). Recombinant proteins were purified from the bacterial lysates by glutathione–Sepharose affinity chromatography as described before (Gong et al., 2002a, b). In vitro protein autophosphorylation assays were performed in a 40 µl reaction mixture comprising purified recombinant glutathione S-transferase (GST) proteins incubated in kinase buffer (10 µCi of [γ-32P]ATP, 10 µM ATP, 20 mM Tris–HCl pH 8.0, 5 mM MnCl2, 1 mM CaCl2, 0.1 mM EDTA, and 1 mM DTT) for 30 min at 30 °C. For substrate phosphorylation, MBP (myelin basic protein) was incubated with immunoprecipitated CIPK25 protein from Col-0 seedlings in kinase buffer. The reaction was stopped by addition of 4× SDS sample buffer. Reaction samples were fractionated by electrophoresis and analyzed by autoradiography. CIPK25 protein was detected by a specific antibody generated against the CIPK25 immunogenic peptide.
Auxin transport assay
Auxin transport was measured essentially as previously described (Shin et al., 2005). Five-day-old vertically grown seedlings were used for the experiment. Briefly, agar blocks of 1 mm in diameter containing 7.7 × 10−8 M [3H]IAA (Amersham) were applied at different positions. Agar blocks were placed at the root tip for basipetal (shootward) and the hypocotyl–root junction for acropetal (rootward) auxin transport. After incubation for 3 h, a 0.5 mm section of the root close to the agar block was removed under the microscope. Two consecutive 2 mm segments below the incision line were then collected separately from 15 roots and were pooled and placed into glass scintillation vials containing 5 ml of scintillation fluid. The radioactivities in these two pools of root segments were measured using a Perkin Elmer Tri-Carb 2800TR Scintillation counter (Waltham, MA, USA).
Statistical analysis
Statistical differences between different groups were detected by Student’s t-test and one-way ANOVA with post-hoc Student–Newman–Keuls (SNK) test in SigmaStat 2.03. Asterisks or different letters indicate significant differences between genotypes and treatments. Relative root growth rate in the presence of external IAA was given as the percentage growth rate (per day) of the same line in control conditions (without IAA).
Results
CIPK25 is involved in root meristem development
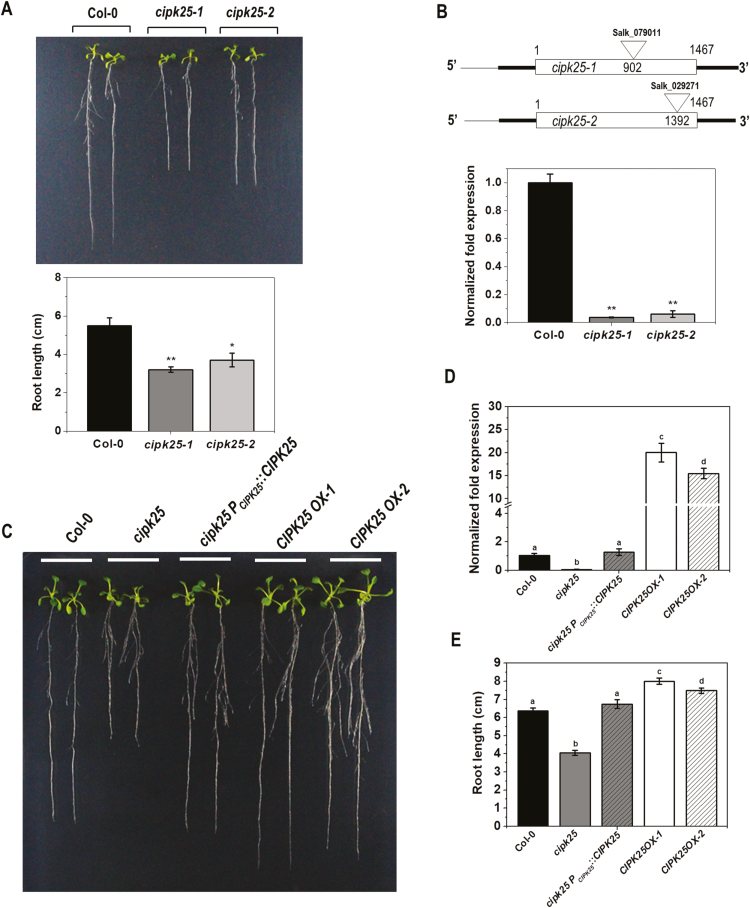
We previously reported that overexpression of the chickpea CIPK25 (CaCIPK25) in tobacco (Nicotiana tabacum) enhanced root growth (Meena et al., 2015). Arabidopsis CIPK25 (At5g25110.1) is an 1697 bp intron-less gene with a CDS of 1467 bp located between base pairs 8 657 626 and 8 659 322 on chromosome 5. Screening of T-DNA insertion lines (Alonso et al., 2003) to investigate the role of Arabidopsis CIPK25 identified two SALK lines, SALK_079011 (cipk25-1) and SALK_029271 (cipk25-2), with a T-DNA insertion in the CIPK25 coding region having a root length shorter by ~40% and ~32%, respectively, as compared with wild-type (Col-0) plants at the 8 dpg stage (Fig. 1A). Sequencing of PCR products with T-DNA- and gene-specific primers identified the T-DNA insertions at 902 bp and 1392 bp downstream of the translation initiation site in cipk25-1 and cipk25-2, respectively. Gene expression analyses by qRT-PCR showed that both mutant lines did not express a significant amount of CIPK25 mRNA as compared with that in Col-0 plants (Fig. 1B). The SALK_079011 line was used for further study and designated as cipk25. Expression of the CIPK25 full-length CDS under the 2.6 kb long native promoter in cipk25 plants (cipk25 PCIPK25::CIPK25) functionally complemented the short root phenotype as observed in plants at 10 dpg. CIPK25-overexpressing [under the Cauliflower mosaic virus (CaMV) 35S promoter] Col-0 lines (CIPK25OX, line 1 and 2) displayed ~20% longer roots (Fig. 1C–E), suggesting that CIPK25 is required for root development in Arabidopsis. CIPK25OX line 1 was used for further studies as it showed comparatively better expression and root growth than the other line.
Fig. 1.
The cipk25 mutant exhibits a shorter root length phenotype. (A) Primary root length phenotype of 8 dpg Col-0, cipk25-1 and cipk25-2 mutants grown vertically on 1/2 MS medium. The lower panel shows root length measurement of three replicate experiments with 15 seedlings for each replicate of each genotype. Error bars are ±SD. Statistically significant differences between root lengths of Col-0 and cipk25 mutants were analyzed by unpaired t-test: *P≤0.05, **P≤0.001. (B) Schematic representation of the CIPK25 gene structure and T-DNA insertion sites of cipk25-1 and cipk25-2 mutants. The lower panel shows the relative transcript level of CIPK25 in 8 dpg Col-0 and cipk25 mutants by qRT-PCR. ACTIN2 was used as the control. Error bars indicate ±SD of three replicates. Significant differences were calculated by unpaired t-test: **P≤0.001. (C) Root length phenotype of 10 dpg Col-0, cipk25, cipk25 PCIPK25::CIPK25, CIPK25OX-1, and CIPK25OX-2 lines. (D) CIPK25 relative transcript levels in 10 dpg Col-0, cipk25, cipk25 PCIPK25::CIPK25, CIPK25OX-1, and CIPK25OX-2 seedlings. ACTIN2 was used as the endogenous control. Different letters indicate significant differences (ANOVA; P≤0.05). (E) Root length comparison of 10 dpg Col-0, cipk25, cipk25 PCIPK25::CIPK25, CIPK25OX-1, and CIPK25OX-2 seedlings. Error bars are ±SD, n=30.
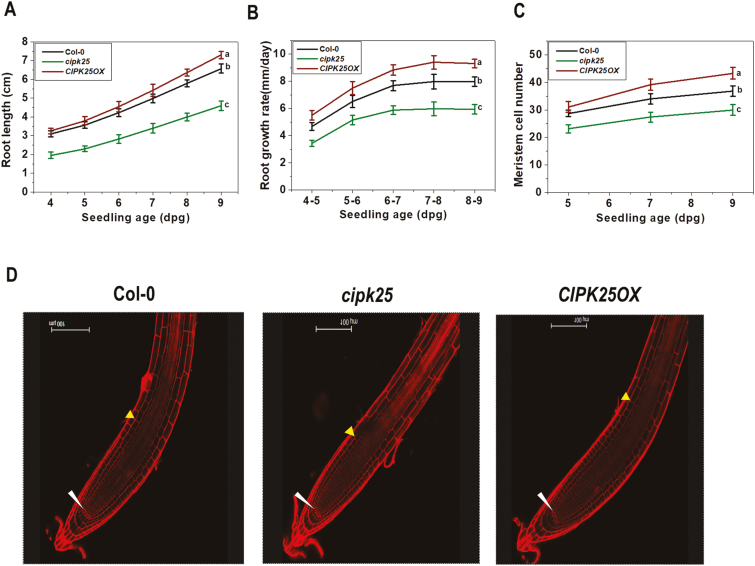
Root lengths and growth rates of the Col-0, cipk25, and CIPK25OX plants were compared over a period of 4–9 dpg. No apparent differences were observed in germination rates and germination periods. Measurement of five replicates of 15 seedlings each showed that the average root length of the mutant line was shorter by 30–36% as compared with the Col-0 plants, whereas that of the CIPK25OX line was >11% longer at 9 dpg (Fig. 2A). Root growth rate, as measured by the increase in the primary root length per day, of the mutant line was ~25% less than that of the Col-0 plants when measured between the fourth and fifth day (Fig. 2B). During the same period, the root growth rate of the CIPK25OX line was similar to that of the Col-0 plant. Root growth rates of cipk25 and CIPK25OX were ~26% less and ~17% more than that of the wild-type plants, respectively, when measured between the eighth and ninth day. To investigate the reason for the shorter root length in the mutant line, the number of root meristem cells was counted following the method of Perilli and Sabatini (2010). Propidium iodide-stained roots showed ~18–22% lower root meristem cell numbers in the cipk25 line as compared with the wild type at 5–9 dpg. During the same period, the root meristem cell numbers for the CIPK25OX line were higher by ~10–20%. However, organization of cells in the root meristematic zone was apparently unperturbed in the mutant line (Fig. 2C, D).
Fig. 2.
The cipk25 mutant exhibits a slower root growth rate and a lower number of root meristem cells. (A and B) Primary root length and growth rate (mm d–1) measurements of Col-0, cipk25, and CIPK25OX seedlings grown vertically on 1/2 MS medium. Values are ±SD of 75 samples for each genotype. Different letters indicate significant differences (ANOVA; P≤0.05) in seedlings at 9 and 8–9 dpg for root length and growth rate. (C) Meristem cell number count in Col-0, cipk25, and CIPK25OX in 5, 7, and 9 dpg seedlings. Error bars are ±SD of n=20. Different letters indicate statistically significant differences among 9 dpg seedlings (ANOVA; P≤0.05). (D) Propidium iodide-stained primary roots of 5 dpg Col-0, cipk25, and CIPK25OX seedlings. Lower and upper arrows indicate the quiescent center (QC) and the first elongated cortex cell, respectively.
CIPK25 encodes an active protein kinase and interacts with CBL4 and CBL5
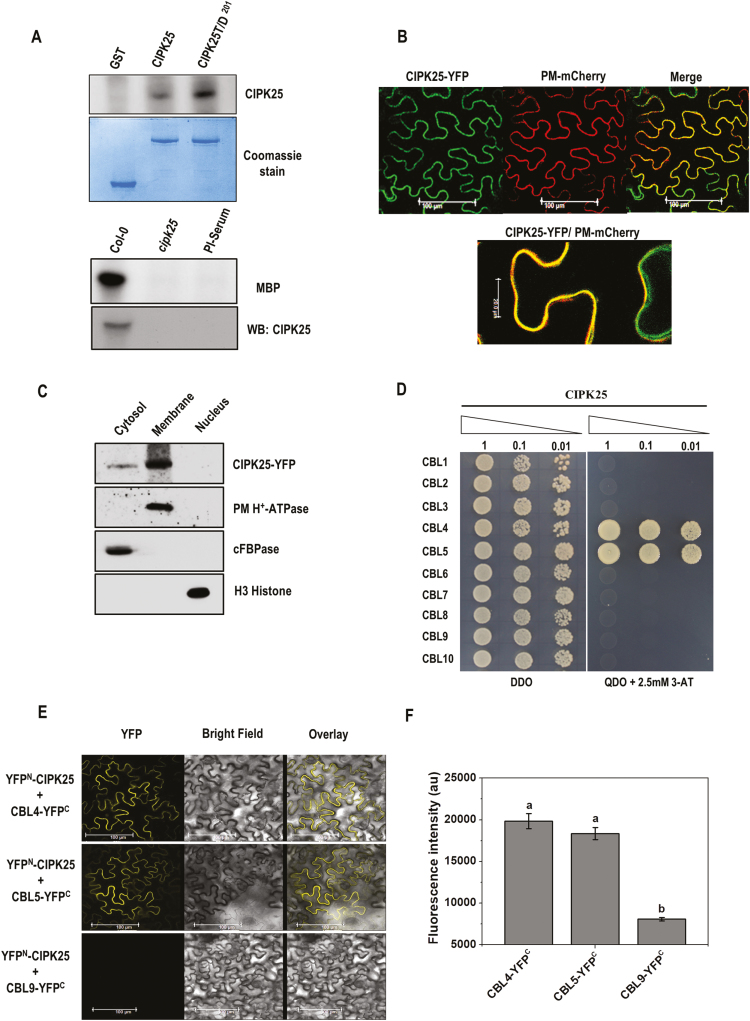
To investigate its biochemical character, CIPK25 protein fused with GST was expressed in bacteria. GST–CIPK25 protein showed autophosphorylation activity in an in vitro reaction. Replacement of a conserved threonine residue with aspartic acid in the activation loop has been shown to increase autophosphorylation activity for various CIPKs (Gong et al., 2002a, b). Similarly, substitution of the conserved Thr201 residue with aspartic acid resulted in an ~3-fold enhancement in the autophosphorylation activity of CIPK25. CIPK25 protein immunoprecipitated from Col-0 seedling protein extract showed in vitro kinase activity with MBP as substrate, implying that the plant CIPK25 protein is an active kinase (Fig. 3A). To explore the subcellular localization of CIPK25 protein, the CIPK25–YFP construct was agroinfiltrated in N. benthamiana leaves along with the plasma membrane marker PM-mCherry (Nelson et al., 2007). Fluorescence overlay showed that CIPK25 was localized in both the plasma membrane and the cytosol (Fig. 3B). To substantiate the localization of CIPK25, subcellular fractionation of transiently expressed CIPK25–YFP in N. benthamiana leaves was performed. The presence of CIPK25 was detected by western blot using anti-GFP antibody mostly in the membrane and much less in cytosolic fractions, but not in the nuclear fraction (Fig. 3C).
Fig. 3.
CIPK25, an active protein kinase, interacts with CBL4 and 5 in planta. (A) Autophosphorylation assay of bacterially expressed GST–CIPK25 and CIPK25T/D201 proteins. Below is the Coomassie blue-stained gel. The lower panel shows substrate phosphorylation of MBP by immunoprecipitated CIPK25 protein from Col-0 and cipk25 mutant seedlings. Western blot of CIPK25 is shown. (B) Visualization of CIPK25–YFP fusion protein in agroinfiltrated leaves of N. benthamiana co-infiltrated with the plasma membrane marker PM-mCherry. The magnified view of the merged image in the lower panel shows CIPK25 localization at the plasma membrane and cytoplasm. (C) Western blot showing subcellular fractions of CIPK25–YFP-expressing N. benthamiana leaves. The blot was probed with antibodies against GFP for CIPK25, plasma membrane (PM) H+-ATPase, cytosolic fructose-1,6-bisphosphatase (cFBPase), and H3 histone. (D) Yeast two-hybrid interaction study of CIPK25 and CBLs. Y2H gold yeast cells were co-transformed with pGBKT7-CIPK25 and pGADT7-CBL1-10, and auxotrophic selection was carried out to detect protein–protein interaction. (E) Interactions between CIPK25 and CBL4/CBL5 in planta were shown by BiFC assay. CIPK25 fused with the N-terminal domain of YFP (YFPN) and CBL4/CBL5/CBL9 fused with the C-terminal domain of YFP (YFPC) were transiently expressed in N. benthamiana by agroinfiltration. Images show YFP-mediated fluorescence derived from the protein–protein interaction. Scale bar=100 µm. Infiltration of CBL9–YFPC with YFPN–CIPK25 was used as the negative control. Expression of the proteins is shown in Supplementary Fig. S1. (F) Fluorescence intensity within leaf discs expressing YFPN–CIPK25 with CBL4–YFPC, CBL5–YFPC, or CBL9–YFPC. Different letters denote significant differences among samples based on Student–Newman–Keuls test (n=8, ANOVA; P≤0.05).
Multiple CBL proteins can interact with a specific CIPK depending on context, and this is thought to define distinct signaling pathways (Kolukisaoglu et al., 2004). A yeast two-hybrid assay was conducted to identify the interacting CBL partners of CIPK25. The full-length CIPK25 protein was found to interact only with CBL4 and CBL5 (Fig. 3D). In planta interaction of CIPK25 with CBL4 and CBL5 was investigated by BiFC assay in N. benthamiana leaves. Fluorescence due to reconstitution of YFP was observed when combinations of YFPN–CIPK25 with CBL4–YFPC or CBL5–YFPC were infiltrated in N. benthamiana leaves, whereas the combinations of YFPN–CIPK25 and CBL9–YFPC did not show any fluorescence (Fig. 3E). Expression of the fused protein in the infiltrated leaf was verified by western blot (see Supplementary Fig. S1). To assess the strength of BiFC signals from YFPN–CIPK25/CBL4–YFPC and YFPN–CIPK25/CBL5–YFPC pairs, the fluorescence signal intensity of the leaf discs infiltrated with the BiFC constructs were quantified using a spectrofluorometer. No significant differences were observed between the BiFC fluorescence signals of YFPN–CIPK25 with CBL4–YFPC and CBL5–YFPC, while the signals fromYFPN–CIPK25 and CBL9–YFPC were negligibly low (Fig. 3F).
Tissue-specific expression of CIPK25
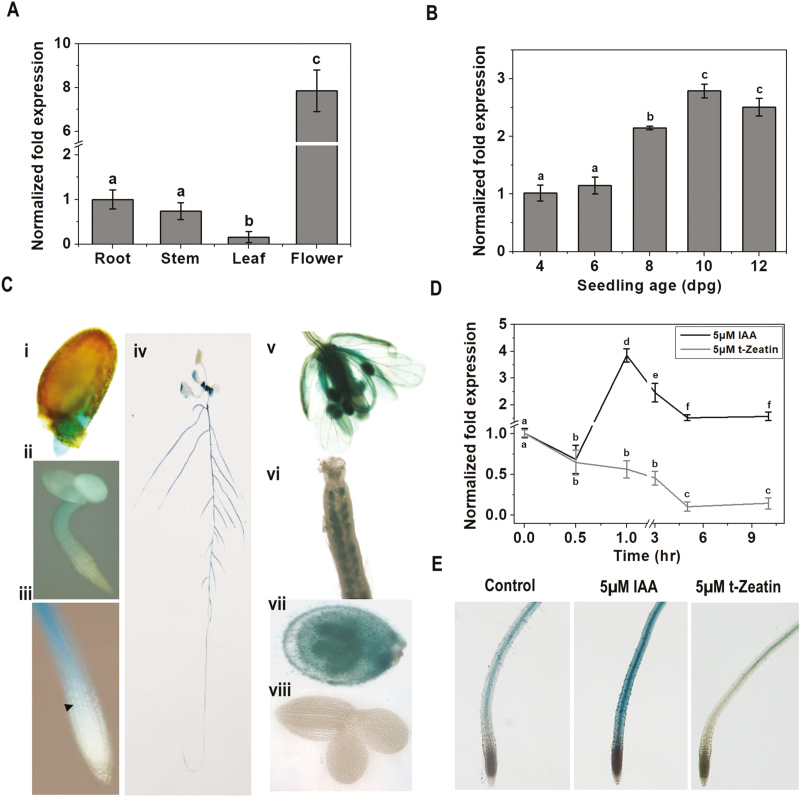
Expression of CIPK25 was investigated in different tissues and root growth stages, and in response to auxin and cytokinin treatments by qRT-PCR. Expression of CIPK25 was the lowest in leaves of the mature plant and highest in flowers, followed by the root and stem (Fig. 4A). Expression of CIPK25 in the root increased gradually from the fourth to the 10th day by ~3 fold, and decreased to ~2.5 fold on the 12th day (Fig. 4B). To investigate the in planta expression pattern, the GUS reporter gene was cloned under a 2.6 kb 5' upstream regulatory region (including the 5'-untranslated region) of CIPK25 (PCIPK25) and expressed in Arabidopsis. GUS activity could be detected throughout the radicles in the germinated seed. GUS activity was observed in cotyledons and hypocotyl, except in the proliferative region of the RAM in a 2-day-old seedling. In a 5-day-old seedling, GUS expression initiated at the TD and continued to the root–shoot junction. GUS activity in the aerial part of a 10-day-old seedling was not well pronounced where the blades of young leaves showed low levels of GUS expression. GUS activity progressively increased with age in the primary and lateral roots, except in the root tip. PCIPK25::GUS showed strong expression in the floral organs in adult plants. A high level of CIPK25 expression was detected in carpel, petals, and stamens. The ovule was dissected and a strong CIPK25 promoter activity was observed in ovule integuments, but not in the embryo (Fig. 4C). The plant growth hormone auxin (IAA) positively regulated CIPK25 expression in root. The transcript level was increased by ~4-fold within 1 h of treatment with 5 µM IAA and then slowly decreased to ~1.8-fold within 5 h and remained at a similar level till 10 h. In contrast, cytokinin (5 µM t-zeatin) treatment down-regulated CIPK25 gene expression by almost 10-fold within 5 h (Fig. 4D). Activation and repression of the CIPK25 promoter by auxin and cytokinin treatments was also validated by promoter–reporter assay (Fig. 4E).
Fig. 4.
CIPK25 gene expression profiles in different tissues, root growth stages, and in response to hormone treatments. CIPK25 gene expression in different tissues (A) and in different root growth stages (B) analyzed by qRT-PCR. ACTIN2 expression was used for normalization. Error bars show ±SD, n=3. Different letters indicate significant differences among samples (ANOVA; P≤0.05). (C) CIPK25 promoter activities were investigated with promoter::reporter gene (GUS) constructs expressed in transgenic plants. Histochemical GUS stainings is shown for (i) the radicle of a germinating seedling; (ii) a 2 dpg seedling; (iii) the primary root tip of a 5 dpg seedling (the arrow indicates the first elongated cortex cell); (iv) a 10 dpg seedling; (v) flower; (vi) gynoecium; (vii) immature seeds with embryo; (viii) embryo. (D) Fold change in CIPK25 expression as analyzed by qRT-PCR in roots of 10 dpg Col-0 seedlings treated with 5 µM IAA and 5 µM t-zeatin for different time intervals. ACTIN2 expression was used for normalization. Error bars show ±SD, n=3. Different letters indicate significant differences among samples (ANOVA; P≤0.05). (E) pCIPK25::GUS expression was visualized in primary roots of 10 dpg seedlings treated with 5 µM IAA for 1 h and with 5 µM t-zeatin for 5 h.
The cipk25 mutant showed compromised auxin transport and auxin-responsive promoter activity
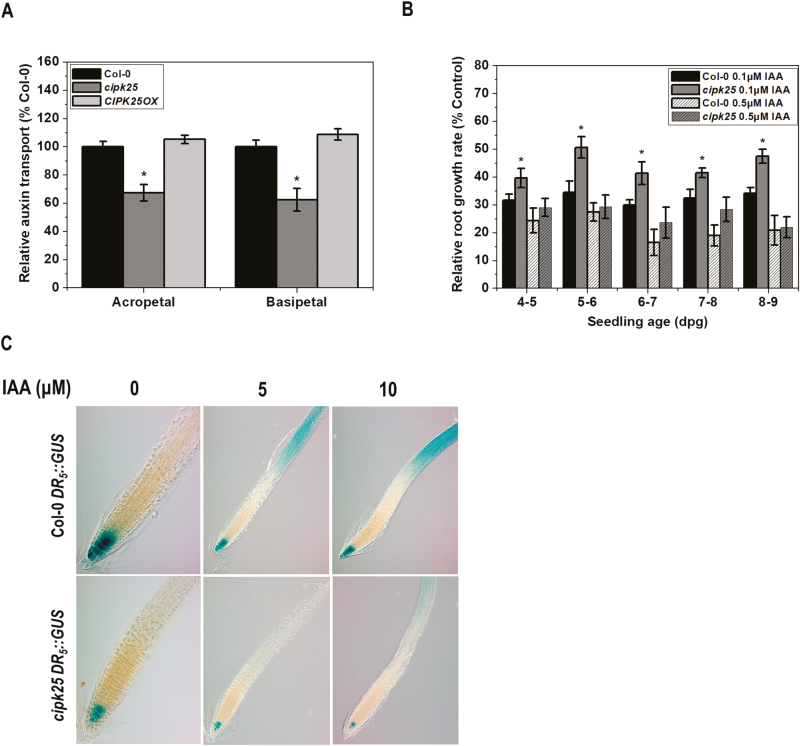
Cell division and patterning in the root are tightly controlled by the local concentration of auxin, which is regulated by a process commonly referred to as polar auxin transport. Acropetal (rootward) and basipetal (shootward) modes of auxin transport in root were assayed using radiolabeled IAA. The mutant line exhibited reduced acropetal and basipetal auxin transport by ~32% and ~38%, respectively, in comparison with Col-0 plants, while the CIPK25OX line displayed similar auxin transport to that in Col-0 by both modes (Fig. 5A).
Fig. 5.
Auxin transport and auxin-responsive promoter assays in cipk25. (A) Comparison of acropetal (rootward) and basipetal (shootward) root auxin transport in 5 dpg Col-0, cipk25, and CIPK25OX seedlings in a root segment 2 mm below (in acopetal) or above (in basipetal) the site of [3H]IAA application. Data are presented as the percentage of auxin transport relative to Col-0. Error bars are ±SD, t-test (*P≤0.05) of three measurements of 15 seedlings each. (B) Auxin-mediated root growth rate retardation of Col-0 and cipk25 in response to 0.1 µM and 0.5 µM auxin treatments. Col-0 and cipk25 mutant seedlings grown on 1/2 MS medium were transferred at 4 dpg to IAA-free or IAA-containing medium, and grown for an additional 5 d. Relative root growth rate in the presence of external IAA is presented as a percentage of the growth rate (per day) of the same line in control conditions (without IAA). The error bars represent ±SD, t-test (*P≤0.05), with three replicates of 10 seedlings each. (C) Auxin-responsive promoter activity using DR5::GUS in Col-0 and the cipk25 mutant. Seedlings at 5 dpg were treated with 5 µM and 10 µM IAA for 2 h and stained for GUS activity.
External treatment with auxin is known to retard root growth (Eliasson et al., 1989). To ascertain the effect of external auxin, 4-day-old seedlings of cipk25 and Col-0 were transferred to media containing 0.1 µM and 0.5 µM IAA, and incubated for 5 d. Exogenous application of 0.1 µM IAA caused relative root growth rate inhibition in Col-0 seedlings by 65–70%. In contrast, relative root growth rate inhibition of cipk25 seedlings was 48–60%, suggesting that the mutant was less responsive to auxin-mediated root growth retardation, and a higher concentration of auxin was required for a similar effect to that in Col-0. With higher concentration (0.5 µM) of IAA, the difference in relative root growth rate retardation between Col-0 and cipk25 was not significant (Fig. 5B).
Auxin-responsive gene expression in roots was assessed by the activity of the auxin-responsive promoter–reporter DR5::GUS. Col-0 plants with DR5::GUS were crossed with cipk25 to develop a homozygous mutant line expressing DR5::GUS (cipk25 DR5::GUS). GUS activity in the RAM of 5-day-old cipk25 seedlings was much lower than in Col-0, implying reduced auxin activity in the RAM of the mutant line. Further, external application of auxin (5 µM and 10 µM IAA for 2 h) caused weaker induction of the auxin-responsive promoter in the cipk25 root as compared with that in Col-0, suggesting lower auxin transport/signaling in the mutant line (Fig. 5C). A similar contrasting pattern of DR5::GUS activity was observed in the case of the agr1-5 pin2 mutant, which was impaired in basipetal auxin transport (Shin et al., 2005).
Reduced expression of auxin-responsive genes including auxin efflux carriers in the cipk25 root
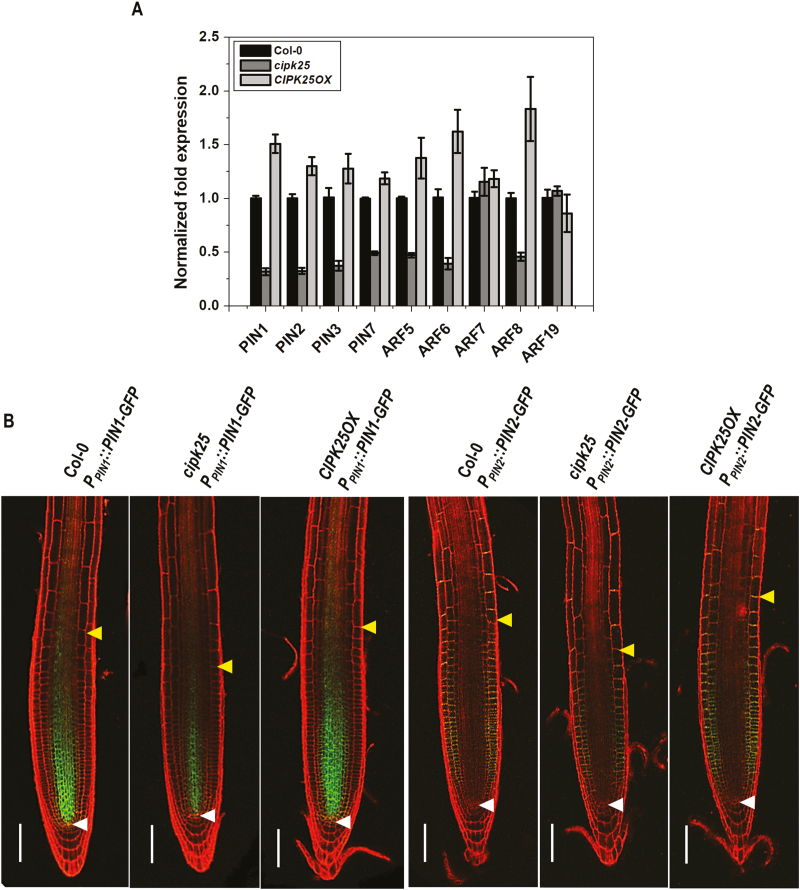
The PIN auxin efflux carriers control growth and patterning in Arabidopsis root (Blilou et al., 2005). ARFs, such as ARF6 and ARF8, were shown to be the positive regulators of adventitious root initiation (Gutierrez et al., 2012). MONOPTEROS/ARF5 is required for primary root formation and vascular development (Berleth and Jürgens, 1993; Przemeck et al., 1996). ARF7 and ARF19 were shown to regulate lateral root formation (Okushima et al., 2007). Expression levels of PINs and ARFs were analyzed by qRT-PCR in roots of Col-0, cipk25, and CIPK25OX plants (Fig. 6A). PIN1, PIN2, and PIN3 expression levels were down-regulated by ~3-fold in cipk25 seedlings, while an ~2-fold reduction was observed for PIN7. Out of five ARFs, expression levels of ARF5, ARF6, and ARF8 were down-regulated by >2-fold in cipk25 as compared with wild-type plants, whereas expression levels of ARF7 and ARF19, involved in lateral root formation, did not show a significant change in cipk25. ARF and PIN genes, which showed reduced expression in the cipk25 line, exhibited ~1.5- to 2-fold higher expression levels in the roots of the CIPK25OX line. Auxin is known to be transported through the central root tissue towards the tip and then redistributed to the cortical tissue and transported basipetally through the epidermis (Swarup and Bennett, 2003; Petrásek and Friml, 2009). The PIN protein auxin efflux carriers facilitate this polar auxin transport. PIN1 is thought to promote auxin movement towards the root tip (acropetal) (Blilou et al., 2005), whereas basipetal auxin transport is dependent on PIN2 (Müller et al., 1998; Abas et al., 2006). To examine the expression of PIN proteins in the absence of CIPK25 expression, cipk25 mutant lines were crossed with reporter lines expressing PIN1 and PIN2 proteins fused with GFP under their native promoters (PPIN1::PIN1-GFP and PPIN2::PIN2-GFP). Fluorescence imaging of roots of 5-day-old seedlings showed that expression of PIN1 protein was considerably reduced in the stele of the root in the cipk25 background. Similarly, the absence of CIPK25 caused a significant reduction in PIN2 protein expression in the epidermis. Expression of GFP-fused PIN1 and PIN2 was higher in CIPK25OX plants (Fig. 6B). All these results suggested that CIPK25 was required for optimal expression of some members of auxin efflux carriers and ARFs.
Fig. 6.
Expression of auxin response factors (ARFs) and efflux carriers (PINs) in cipk25. (A) Relative transcript levels of auxin response factors (ARF5, ARF6, ARF7, ARF8, and ARF19) and efflux carriers (PIN1, PIN2, PIN3, and PIN7) were analyzed by qRT-PCR in roots of 5 dpg Col-0, cipk25, and CIPK25OX seedlings. ACTIN2 expression was used for normalization. Error bars represent ±SD of three biological replicates with 15 seedlings for each genotype. (B) Visualization of expression of auxin efflux carriers PIN1 and PIN2. Fluorescence images from the GFP fusion proteins expressed through the native promoters of individual genes in the Col-0, cipk25, and CIPK25OX background. Seedlings at 5 dpg were stained with propidium iodide to explore meristem cell profiles and analyzed by confocal microscopy. Scale bars in confocal images=100 µm. Lower and upper arrows indicate the quiescent center (QC) and the first elongated cortex cell, respectively.
SHY2 loss of function rescued the short root phenotype of cipk25
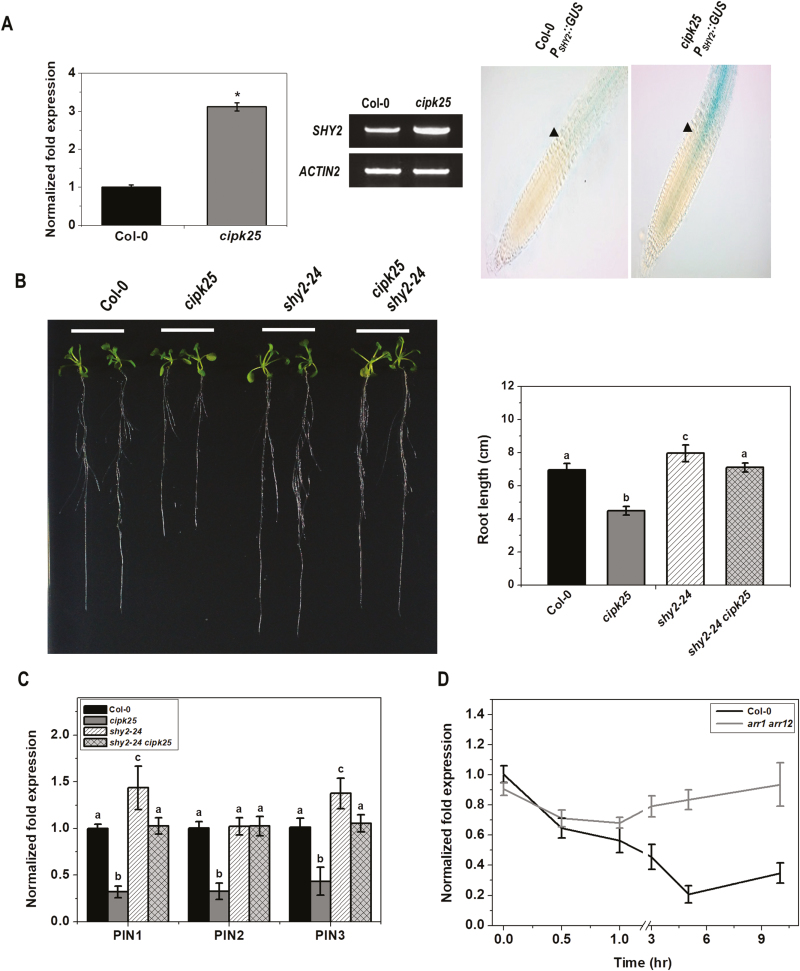
The number of cells in the root meristem is regulated by auxin and cytokinin, which promote cell division and cell elongation, respectively. SHY2/IAA3 was proposed to act as a point of intersection of auxin- and cytokinin-mediated signaling to regulate root meristem size. Cytokinin represses PIN1, PIN3, and PIN7 expression to influence root meristem maintenance in a SHY2-dependent manner (Tian et al., 2002; Blilou et al., 2005; Vieten et al., 2005; Dello Ioio et al., 2008). As CIPK25 was found to influence root meristem size and PIN expression, the SHY2 expression level was investigated in cipk25 and was found to be ~3-fold higher than in the roots of wild-type seedlings. This observation was substantiated by in planta expression of the SHY2 promoter–GUS fusion construct (PSHY2::GUS). The GUS reporter gene was cloned under a 2.6 kb long SHY2 promoter and was expressed in wild-type Arabidopsis. It was crossed with the cipk25 line to generate the cipk25 PSHY2::GUS line. The SHY2 promoter showed a very weak activity in the TD and upwards in the wild-type background, whereas a high SHY2 promoter activity was observed in the cipk25 background (Fig. 7A).
Fig. 7.
Genetic interaction of CIPK25 and SHY2. (A) Relative transcript abundance of SHY2 in 5 dpg Col-0 and cipk25 mutant roots was analyzd by qRT-PCR and semi-quantitative RT-PCR. ACTIN2 was used as the internal control. The right-hand panel shows histochemical GUS staining of primary roots of 5 dpg seedlings expressing the GUS reporter under the SHY2 promoter. The arrow shows the first elongated cortex cell. (B) Primary root length phenotype of 10 dpg Col-0, cipk25, shy2-24, and cipk25 shy2-24 mutants grown vertically on 1/2 MS medium. The right-hand panel shows root length measurement. Error bars represent ±SD, n=24. (C) Relative transcript levels of auxin efflux carriers were analyzed by qRT-PCR in roots of 5 dpg Col-0, cipk25, shy2-24, and cipk25 shy2-24 seedlings. Different letters indicate statistically significant differences by ANOVA, P≤0.05. Error bars are ±SD, n=3. (D) qRT-PCR analysis of the expression levels of CIPK25 in roots of 10 dpg Col-0 and arr1 arr12 seedlings treated with 5 µM t-zeatin for different time intervals. ACTIN2 expression was used as the internal control. Error bars are ±SD, n=3.
We explored the influence of SHY2 on CIPK25 expression by assessing it in the SHY2 loss-of-function mutant shy2-24. In shy2-24, a stop codon was introduced at amino acid position 61, which caused truncation of the protein, and the mutant was considered as a null mutant for SHY2 (Tian and Reed, 1999). CIPK25 expression in shy2-24 was equivalent to that in Col-0 (Supplementary Fig. S2). To investigate the genetic interaction between CIPK25 and SHY2, a cipk25 shy2-24 double mutant was generated. The short root phenotype of the cipk25 plants was completely recovered in the cipk25 shy2-24 mutant, resulting in a similar root length to that of the wild-type plants (Fig. 7B). PIN1, PIN2, and PIN3 expression levels in cipk25 shy2-24 were found to be equivalent to those of the wild-type plants (Fig. 7C), indicating that CIPK25 is genetically epistatic to SHY2. Cytokinin treatment down-regulated expression of CIPK25 (Fig. 4D). Cytokinin control of root meristem size is mediated through ARR1 and ARR12 expression in the TD (Dello Ioio et al., 2007, 2008). Absence or overexpression of CIPK25 did not cause appreciable changes in ARR1 and ARR12 expression (Supplementary Fig. S3). Moreover, cytokinin treatment did not change CIPK25 expression in the arr1 arr12 background, suggesting that cytokinin-mediated down-regulation of CIPK25 expression was dependent on ARR1/ARR12 (Fig. 7D). An analysis of the CIPK25 promoter using PlantPan2.0 (Chow et al., 2015) showed a number of potential ARR1- and ARR12-binding sites (Supplementary Table S2). All these observations suggested an involvement of CIPK25 in cytokinin signaling mediated through ARR1/ARR12.
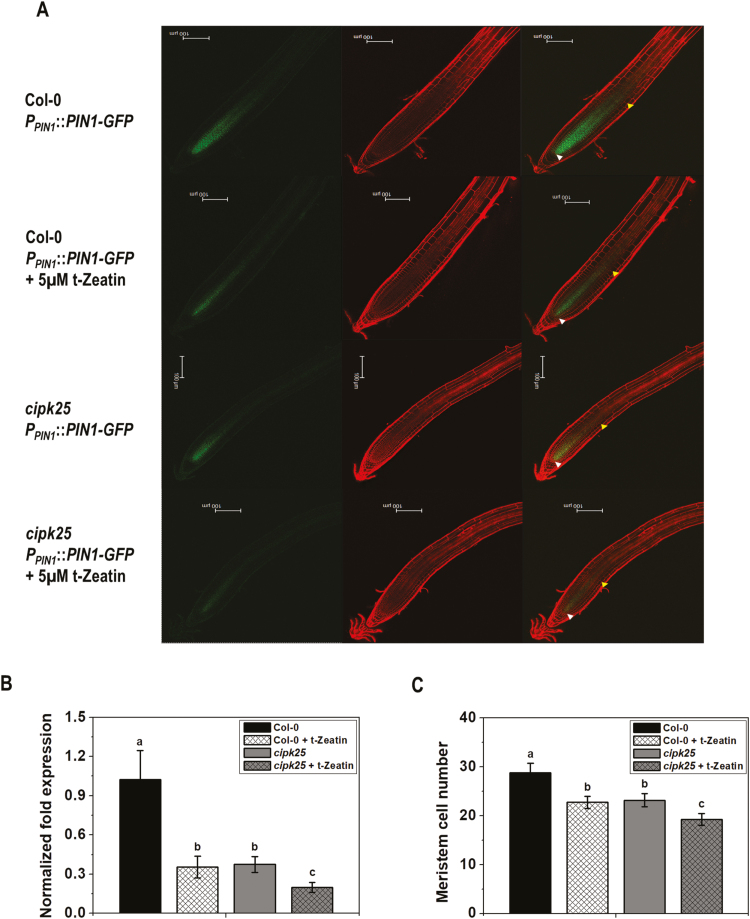
Cytokinin-mediated down-regulation of PIN1 expression in roots was shown to be dependent on SHY2 (Dello Ioio et al., 2008). To determine whether down-regulation of PIN1 expression by cytokinin is completely dependent on CIPK25, wild-type (Col-0) and cipk25 plants expressing PPIN1::PIN1-GFP were treated with 5 μM t-zeatin. Cytokinin treatment resulted in marked down-regulation of PIN1 protein expression in the wild-type background. A reduction in PIN1 expression in cipk25 roots in control conditions was already shown earlier. Treatment with cytokinin further reduced PIN1 expression in the cipk25 root (Fig. 8A). This observation was substantiated by PIN1 gene expression in the cipk25 root. Cytokinin treatment resulted in an ~3-fold reduction in PIN1 expression in the wild-type root. A similar (~3-fold lower) PIN1 expression level was observed in control cipk25. Cytokinin treatment caused further reduction in the PIN1 expression level in cipk25 by 1.8-fold (Fig. 8B). Additional reduction in meristematic cell number in cipk25 after cytokinin treatment corroborated reduced PIN1 expression (Fig. 8C). This result suggested that cytokinin regulates root meristem size by both CIPK25-dependent and -independent pathways.
Fig. 8.
Cytokinin-responsive PIN1 expression in cipk25. (A) Fluorescence images of auxin efflux carrier PIN1–GFP fusion protein driven by its native promoter in Col-0 and cipk25 seedlings in control conditions and in response to cytokinin treatment (5 μM t-zeatin, 6 h). Roots of 5 dpg seedlings were stained with propidium iodide (PI) for visualizing root cells. Lower and upper arrows indicate the quiescent center (QC) and the first elongated cortex cell, respectively. Scale bar=100 µm. (B) Relative transcript levels of PIN1 were analyzed by qRT-PCR in roots of 5 dpg Col-0 and cipk25 seedlings treated or not with cytokinin (5 μM t-zeatin, 6 h). Error bars are ±SD of three biological replicates, ANOVA, P≤0.05. (C) Meristem cell number count in 5 dpg Col-0 and cipk25 seedlings in control conditions and in response to cytokinin treatment (5 μM t-zeatin, 12 h). Values are the mean ±SD of 20 samples each. Different letters indicate significant differences among samples (ANOVA; P≤0.05).
Discussion
Post-embryonic root growth is maintained by the RAM. Highly proliferative cells of the RAM move longitudinally towards the shoot and eventually differentiate. Hence, cell division and differentiation rates must be equal in order to maintain root meristem size and ultimately overall root development. It has been proposed that a balance of auxin and cytokinin signaling regulates the transition of dividing cells to differentiated cells by activating expression of SHY2 at the TD, which subsequently decreases expression of PIN genes (Dello Ioio et al., 2008) by suppressing auxin signaling and defines the position of the TD by reducing auxin transport and, additionally, by auxin degradation (Di Mambro et al., 2017). It was reported that SHY2 expression in the root was minimal at 3 dpg in an actively growing meristem and reached its highest level at 5 dpg, when the meristem attained its final size (Moubayidin et al., 2010). By the same analogy, SHY2 expression must also be restricted to maintain an equal rate of cell division and rate of transition to elongation. Our results suggest that CIPK25 negatively regulates SHY2 expression to maintain root meristem size. CIPK25 expression initiated at the TD and was intense in the differentiated zone, and its expression level was highest when SHY2 attained its highest expression level after the establishment of the meristem. SHY2 expression is induced by cytokinin treatment in an ARR1-dependent manner (Moubayidin et al., 2010), while cytokinin treatment decreased CIPK25 expression in an ARR1/ARR12-dependent manner. A high SHY2 expression was observed in the absence of CIPK25. All of these observations suggested an antagonistic relationship between SHY2 and CIPK25. However, application of cytokinin caused a further reduction in PIN1 expression and meristem size in the cipk25 mutant, suggesting that cytokinin also functions in CIPK25-independent pathways. The CIPK25-dependent pathway seems to be totally dependent on SHY2, as a SHY2 loss-of-function mutant complemented the short root phenotype of cipk25 and restored expression of PIN1, PIN2, and PIN3. Therefore, CIPK25 may be considered as one of the candidate molecules that balance activities of auxin and cytokinin during root development.
As reported before (Schmid et al., 2005), CIPK25 displayed the highest expression in flowers (Fig. 4). Interestingly, like ARF6 and ARF8, CIPK25 was also expressed in the stamen and gynoecium, and its expression was greatly reduced in arf6 arf8 flowers (Reeves et al., 2012). ARF6 and ARF8 showed reduced expression in cipk25 roots. ARF6 and ARF8 are required for flower maturity and growth of the ovule integument (Nagpal et al., 2005; Wu et al., 2006). CIPK25 also displayed a high expression in ovule integuments. We did not observe any abnormal phenotype in the flowers of cipk25 plants. This might be due to the fact that expression of ARF6 and ARF8 was only reduced, but not abolished, in cipk25.
CBL4 and CBL5 interacted with CIPK25 in yeast two-hybrid and in planta BiFC assays. It is known from several previous reports that an individual CBL protein can interact with multiple CIPKs and vice versa, depending on context (Luan, 2009). It is believed that this flexibility of interaction and their subcellular localizations are crucial in sensing and responding to specific signals. CBL4 was previously shown to interact with CIPK24 and CIPK6 in the context of salinity stress to modulate the activity of the Na+/H+ antiporter (Qiu et al., 2002) and plasma membrane targeting of the potassium channel AKT2 (Held et al., 2011). Both CBL4 and CBL5 possess conserved domains for myristoylation and acylation to localize in cell membranes (Batistič et al., 2010; Saito et al., 2018). Arabidopsis CIPK25 protein, although it possesses an MG-motif, does not undergo N-myristoylation (Saito et al., 2018). It is possible that a major pool of the expressed Arabidopsis CIPK25 protein interacted with the corresponding CBL(s) of N. benthamiana and was targeted to the plasma membrane. CBL sequences possess evolutionarily conserved motifs (Mohanta et al., 2015). Cross-species interaction of CIPKs and CBLs was reported before (Tripathi et al., 2009; Wang et al., 2012). Plasma membrane localization might be essential for CIPK25 function. CIPK11 interacts with CBL5, and myristoylation and acylation of CBL5 were shown to be required for the CBL5–CIPK11 complex to phosphorylate the guard cell anion channel SLAC1 (Saito et al., 2018).
Auxin and cytokinin together regulate various aspects of a plant’s growth and development. The balance between their interactions is maintained by negative regulation of one signaling pathway by the other. On the other hand, they regulate each other’s biosynthesis (Nordström et al., 2004; Jones et al., 2010). The cytokinin biosynthetic gene ATP/ADP isopentyl transferase 5 (IPT5) is rapidly induced by auxin and further induced in the SHY2 loss-of-function background, whereas auxin-dependent induction of PIPT5::GUS was totally abolished in the SHY2 gain-of-function background (Dello Ioio et al., 2008). Relative enhancement of cytokinin activity at the root transition domain might reduce CIPK25 expression, leading to increased SHY2 expression resulting in reduced auxin signaling. We propose CIPK25 functions genetically above SHY2. Cytokinin-mediated suppression of CIPK25 expression was dependent on ARR1/ARR12. ARR1 is able to regulate SHY2 expression directly by binding to and activating the SHY2 promoter (Dello Ioio et al., 2008). It is noteworthy that cytokinin was able to suppress PIN1 expression even in the absence of CIPK25, suggesting a partial role for CIPK25 in balancing auxin–cytokinin signaling in root meristem development. Presently, it is not clear how CIPK25 functions in auxin/cytokinin signaling. Auxin and cytokinin are involved in many interconnected developmental processes through various cross-connected signaling pathways. Therefore, a role for CIPK25 in other pathways related to root development cannot be ruled out.
It is known that extracellular application of calcium inhibits cell growth in the TD (Ishikawa and Evans, 1992; Legué et al., 1997; Fasano et al., 2002). Ca2+ plays a vital role in polarity and rate of auxin transport through PIN proteins by modulating endo- and exocytosis and the direction of PIN localization in the plasma membrane (reviewed in Vanneste and Friml, 2013). In this study, we proposed that a Ca2+-regulated protein kinase, CIPK25, is involved in regulation of root meristem size by regulating auxin transport through modulating PIN protein expression.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primers used in this study.
Table S2. A list of auxin- and cytokinin-responsive elements in the CIPK25 promoter.
Fig. S1. Detection of YFPN–CIPK25, CBL4–YFPC, and CBL9–YFPC fusion proteins in BiFC assay.
Fig. S2. qRT-PCR analysis of the expression levels of CIPK25 in the shy2-24 line.
Fig. S3. Relative transcript abundance of ARR1 and ARR12 in cipk25 and CIPK25OX seedlings.
Acknowledgements
This project was funded by National Institute of Plant Genome Research (NIPGR), an autonomous institute funded by the Department of Biotechnology (DBT), Government of India, and by the DBT under the Next Generation Challenge Programme on Chickpea Genomics (grant no. BT/PR12919/AGR/02/676/2009 from 2009–14). The authors acknowledge Dr J.W. Reed and Dr Punita Nagpal of the Department of Biology, University of North Carolina, Chapel Hill, USA (UNC) for their scientific and technical suggestions and the plant lines, and Dr J. Vadassery, NIPGR, for critical reading of the manuscript. MKM, NKV, and VT acknowledge the Council for Scientific and Industrial Research (CSIR), India and DBT, respectively, for fellowships. The authors are grateful to the DBT-eLibrary Consortium (DeLCON) for providing access to e-Resources. They also acknowledge the central instrument facility at NIPGR for instrumentation support.
References
- Abas L, Benjamins R, Malenica N, Paciorek T, Wiśniewska J, Wirniewska J, Moulinier-Anzola JC, Sieberer T, Friml J, Luschnig C. 2006. Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nature Cell Biology 8, 249–256. [DOI] [PubMed] [Google Scholar]
- Abramoff MD, Magalhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics International 11, 36–42. [Google Scholar]
- Adachi S, Minamisawa K, Okushima Y, et al. 2011. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proceedings of the National Academy of Sciences, USA 108, 10004–10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Bai L, Zhang G, Zhou Y, Zhang Z, Wang W, Du Y, Wu Z, Song CP. 2009. Plasma membrane-associated proline-rich extensin-like receptor kinase 4 a novel regulator of Ca signalling is required for abscisic acid responses in Arabidopsis thaliana. The Plant Journal 60, 314–327. [DOI] [PubMed] [Google Scholar]
- Batistic O, Waadt R, Steinhorst L, Held K, Kudla J. 2010. CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. The Plant Journal 61, 211–222. [DOI] [PubMed] [Google Scholar]
- Beemster GT, Baskin TI. 2000. Stunted plant 1 mediates effects of cytokinin, but not of auxin, on cell division and expansion in the root of Arabidopsis. Plant Physiology 124, 1718–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleth T, Jürgens G. 1993. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development 118, 575–587. [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44. [DOI] [PubMed] [Google Scholar]
- Boer DR, Freire-Rios A, van den Berg WA, et al. 2014. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156, 577–589. [DOI] [PubMed] [Google Scholar]
- Chapman N, Miller AJ, Lindsey K, Whalley WR. 2012. Roots, water, and nutrient acquisition: let’s get physical. Trends in Plant Science 17, 701–710. [DOI] [PubMed] [Google Scholar]
- Chow CN, Zheng HQ, Wu NY, Chien CH, Huang HD, Lee TY, Chiang-Hsieh YF, Hou PF, Yang TY, Chang WC. 2015. PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Research 44, D1154–D1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. 2007. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Current Biology 17, 678–682. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322, 1380–1384. [DOI] [PubMed] [Google Scholar]
- Di Mambro R, De Ruvo M, Pacifici E, et al. 2017. Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proceedings of the National Academy of Sciences, USA 114, 7641–7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duby G, Poreba W, Piotrowiak D, Bobik K, Derua R, Waelkens E, Boutry M. 2009. Activation of plant plasma membrane H+-ATPase by 14-3-3 proteins is negatively controlled by two phosphorylation sites within the H+-ATPase C-terminal region. Journal of Biological Chemistry 284, 4213–4221. [DOI] [PubMed] [Google Scholar]
- Eliasson L, Bertell G, Bolander E. 1989. Inhibitory action of auxin on root elongation not mediated by ethylene. Plant Physiology 91, 310–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano JM, Massa GD, Gilroy S. 2002. Ionic signaling in plant responses to gravity and touch. Journal of Plant Growth Regulation 21, 71–88. [DOI] [PubMed] [Google Scholar]
- Fuglsang AT, Guo Y, Cuin TA, et al. 2007. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. The Plant Cell 19, 1617–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D, Gong Z, Guo Y, Chen X, Zhu JK. 2002a Biochemical and functional characterization of PKS11, a novel Arabidopsis protein kinase. Journal of Biological Chemistry 277, 28340–28350. [DOI] [PubMed] [Google Scholar]
- Gong D, Zhang C, Chen X, Gong Z, Zhu JK. 2002b Constitutive activation and transgenic evaluation of the function of an arabidopsis PKS protein kinase. Journal of Biological Chemistry 277, 42088–42096. [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Mongelard G, Floková K, et al. 2012. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. The Plant Cell 24, 2515–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held K, Pascaud F, Eckert C, et al. 2011. Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Research 21, 1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J. 2001. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389. [DOI] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. 2001. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409, 1060–1063. [DOI] [PubMed] [Google Scholar]
- Ishida T, Adachi S, Yoshimura M, Shimizu K, Umeda M, Sugimoto K. 2010. Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development 137, 63–71. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Evans ML. 1992. Induction of curvature in maize roots by calcium or by thigmostimulation: role of the postmitotic isodiametric growth zone. Plant Physiology 100, 762–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov VB, Dubrovsky JG. 2013. Longitudinal zonation pattern in plant roots: conflicts and solutions. Trends in Plant Science 18, 237–243. [DOI] [PubMed] [Google Scholar]
- Jones B, Gunnerås SA, Petersson SV, Tarkowski P, Graham N, May S, Dolezal K, Sandberg G, Ljung K. 2010. Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. The Plant Cell 22, 2956–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451. [DOI] [PubMed] [Google Scholar]
- Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J. 2004. Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL–CIPK signaling networks. Plant Physiology 134, 43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legué V, Blancaflor E, Wymer C, Perbal G, Fantin D, Gilroy S. 1997. Cytoplasmic free Ca2+ in Arabidopsis roots changes in response to touch but not gravity. Plant Physiology 114, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Zhao C, Zhang Y, Wang X, Wang X, Wang J, Wang F, Bi Y. 2016. Calcium alleviates cadmium-induced inhibition on root growth by maintaining auxin homeostasis in Arabidopsis seedlings. Protoplasma 253, 185–200. [DOI] [PubMed] [Google Scholar]
- Luan S. 2009. The CBL–CIPK network in plant calcium signaling. Trends in Plant Science 14, 37–42. [DOI] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM. 2009. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. The Plant Journal 59, 150–162. [DOI] [PubMed] [Google Scholar]
- Meena MK, Ghawana S, Dwivedi V, Roy A, Chattopadhyay D. 2015. Expression of chickpea CIPK25 enhances root growth and tolerance to dehydration and salt stress in transgenic tobacco. Frontiers in Plant Science 6, 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M. 2008. Auxin receptors and plant development: a new signaling paradigm. Annual Review of Cell and Developmental Biology 24, 55–80. [DOI] [PubMed] [Google Scholar]
- Mohanta TK, Mohanta N, Mohanta YK, Parida P, Bae H. 2015. Genome-wide identification of Calcineurin B-Like (CBL) gene family of plants reveals novel conserved motifs and evolutionary aspects in calcium signaling events. BMC Plant Biology 15, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen GB, Miller ND, Murphy AS, Gilroy S. 2011. Dynamics of auxin-dependent Ca2+ and pH signaling in root growth revealed by integrating high-resolution imaging with automated computer vision-based analysis. The Plant Journal 65, 309–318. [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S. 2010. The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Current Biology 20, 1138–1143. [DOI] [PubMed] [Google Scholar]
- Müller A, Guan C, Gälweiler L, Tänzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K. 1998. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO Journal 17, 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, et al. 2005. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132, 4107–4118. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nordström A, Tarkowski P, Tarkowska D, Norbaek R, Astot C, Dolezal K, Sandberg G. 2004. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin–cytokinin-regulated development. Proceedings of the National Academy of Sciences, USA 101, 8039–8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. 2007. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. The Plant Cell 19, 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacifici E, Polverari L, Sabatini S. 2015. Plant hormone cross-talk: the pivot of root growth. Journal of Experimental Botany 66, 1113–1121. [DOI] [PubMed] [Google Scholar]
- Perilli S, Sabatini S. 2010. Analysis of root meristem size development. Methods in Molecular Biology 655, 177–187. [DOI] [PubMed] [Google Scholar]
- Petrásek J, Friml J. 2009. Auxin transport routes in plant development. Development 136, 2675–2688. [DOI] [PubMed] [Google Scholar]
- Przemeck GK, Mattsson J, Hardtke CS, Sung ZR, Berleth T. 1996. Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200, 229–237. [DOI] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK. 2002. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proceedings of the National Academy of Sciences, USA 99, 8436–8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao X, Huang X, Zhou Z, Lin X. 2013. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostatistics, Bioinformatics and Biomathematics 3, 71–85. [PMC free article] [PubMed] [Google Scholar]
- Reeves PH, Ellis CM, Ploense SE, et al. 2012. A regulatory network for coordinated flower maturation. PLoS Genetics 8, e1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Hamamoto S, Moriya K, et al. 2018. N-myristoylation and S-acylation are common modifications of Ca2+-regulated Arabidopsis kinases and are required for activation of the SLAC1 anion channel. New Phytologist 218, 1504–1521. [DOI] [PubMed] [Google Scholar]
- Santin F, Bhogale S, Fantino E, Grandellis C, Banerjee AK, Ulloa RM. 2017. Solanum tuberosum StCDPK1 is regulated by miR390 at the posttranscriptional level and phosphorylates the auxin efflux carrier StPIN4 in vitro, a potential downstream target in potato development. Physiologia Plantarum 159, 244–261. [DOI] [PubMed] [Google Scholar]
- Schenk HJ, Jackson RB. 2002. Rooting depths lateral root spreads and below-ground/above-ground allometries of plants in water-limited ecosystems. Journal of Ecology 90, 480–494. [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. 2005. A gene expression map of Arabidopsis thaliana development. Nature Genetics 37, 501–506. [DOI] [PubMed] [Google Scholar]
- Shin H, Shin HS, Guo Z, Blancaflor EB, Masson PH, Chen R. 2005. Complex regulation of Arabidopsis AGR1/PIN2-mediated root gravitropic response and basipetal auxin transport by cantharidin-sensitive protein phosphatases. The Plant Journal 42, 188–200. [DOI] [PubMed] [Google Scholar]
- Swarup R, Bennett M. 2003. Auxin transport: the fountain of life in plants?Developmental Cell 5, 824–826. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Hayashi K, Kinoshita T. 2012. Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiology 159, 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Kajihara T, Okamura C, Kim Y, Katagiri Y, Okushima Y, Matsunaga S, Hwang I, Umeda M. 2013. Cytokinins control endocycle onset by promoting the expression of an APC/C activator in Arabidopsis roots. Current Biology 23, 1812–1817. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Dhonukshe P, Brewer PB, Friml J. 2006. Spatiotemporal asymmetric auxin distribution: a means to coordinate plant development. Cellular and Molecular Life Sciences 63, 2738–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimann KV, Schneider CA. 1939. The relative activities of different auxins. American Journal of Botany 26, 328–333. [Google Scholar]
- Tian Q, Reed JW. 1999. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126, 711–721. [DOI] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW. 2002. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. The Plant Cell 14, 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ. 2004. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. The Plant Cell 16, 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JP, Kieber JJ. 2008. Cytokinin signaling: two-components and more. Trends in Plant Science 13, 85–92. [DOI] [PubMed] [Google Scholar]
- Tripathi V, Parasuraman B, Laxmi A, Chattopadhyay D. 2009. CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. The Plant Journal 58, 778–790. [DOI] [PubMed] [Google Scholar]
- Vanneste S, Friml J. 2013. Calcium: the missing link in auxin action. Plants (Basel, Switzerland) 2, 650–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbelen JP, De Cnodder T, Le J, Vissenberg K, Baluska F. 2006. The root apex of Arabidopsis thaliana consists of four distinct zones of growth activities: meristematic zone, transition zone, fast elongation zone and growth terminating zone. Plant Signaling & Behavior 1, 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wisniewska J, Benková E, Benjamins R, Beeckman T, Luschnig C, Friml J. 2005. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132, 4521–4531. [DOI] [PubMed] [Google Scholar]
- Wang F, Chen ZH, Liu X, Colmer TD, Zhou M, Shabala S. 2016. Tissue-specific root ion profiling reveals essential roles of the CAX and ACA calcium transport systems in response to hypoxia in Arabidopsis. Journal of Experimental Botany 67, 3747–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RK, Li LL, Cao ZH, Zhao Q, Li M, Zhang LY, Hao YJ. 2012. Molecular cloning and functional characterization of a novel apple MdCIPK6L gene reveals its involvement in multiple abiotic stress tolerance in transgenic plants. Plant Molecular Biology 79, 123–135. [DOI] [PubMed] [Google Scholar]
- Wu J, Wang Y, Kim SG, Jung KH, Gupta R, Kim J, Park Y, Kang KY, Kim ST. 2017. A secreted chitinase-like protein (OsCLP) supports root growth through calcium signaling in Oryza sativa. Physiologia Plantarum 161, 273–284. [DOI] [PubMed] [Google Scholar]
- Wu MF, Tian Q, Reed JW. 2006. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133, 4211–4218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.