Abstract
Gene expression is typically quantified as RNA abundance, which is influenced by both synthesis (transcription) and decay. Cytoplasmic decay typically initiates by deadenylation, after which decay can occur through any of three cytoplasmic decay pathways. Recent advances reveal several mechanisms by which RNA decay is regulated to control RNA abundance. mRNA can be post-transcriptionally modified, either indirectly through secondary structure or through direct modifications to the transcript itself, sometimes resulting in subsequent changes in mRNA decay rates. mRNA abundances can also be modified by tapping into pathways normally used for RNA quality control. Regulated mRNA decay can also come about through post-translational modification of decapping complex subunits. Likewise, mRNAs can undergo changes in subcellular localization (for example, the deposition of specific mRNAs into processing bodies, or P-bodies, where stabilization and destabilization occur in a transcript- and context-dependent manner). Additionally, specialized functions of mRNA decay pathways were implicated in a genome-wide mRNA decay analysis in Arabidopsis. Advances made using plants are emphasized in this review, but relevant studies from other model systems that highlight RNA decay mechanisms that may also be conserved in plants are discussed.
Keywords: mRNA decay, decapping, VCS, SOV, DIS3L2, P-bodies, gene expression
Introduction
This review examines cytoplasmic mRNA decay with a focus on how mRNA decay regulates transcript abundance. Typically, changes in RNA abundances are attributed to transcription; however, considerable evidence supports important contributions from mRNA decay and this review focuses on recent advances in this area. Failing to account for changes in mRNA abundance that arise from altered decay rates can compromise molecular strategies for improving agriculture and ignores interesting biological phenomena.
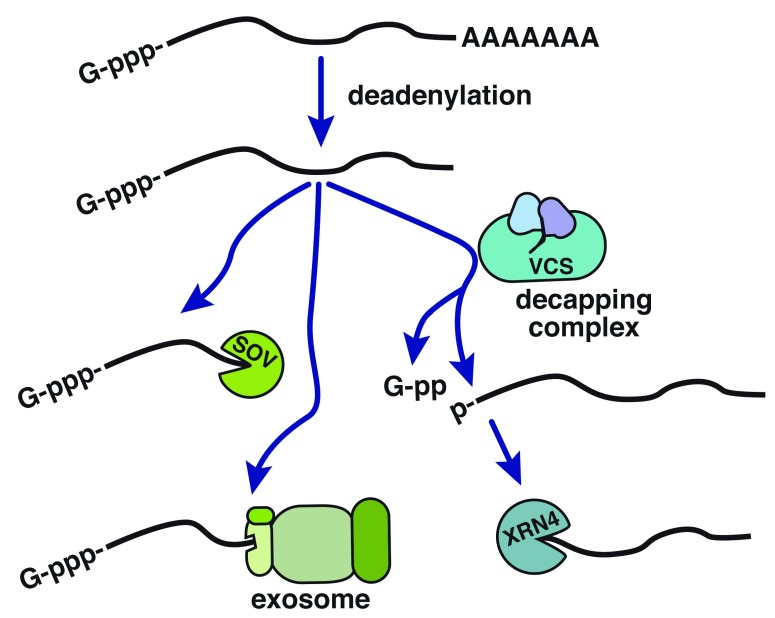
Two stability determinants protect mRNA from untimely degradation: (1) the 3′ polyadenosine (poly(A)) tail and (2) the 5′ 7-methylguanosine (m 7G) cap. mRNA decay is initiated by the removal of the 3′ poly(A) tail in a process called deadenylation 1 ( Figure 1). Further degradation can act at the newly deadenylated 3′ end through the activity of the RNA exosome, which has distinct nuclear and cytoplasmic RNA decay and processing functions 2. Alternatively, 3′→5′ decay can occur via SUPPRESSOR OF VARICOSE (SOV), which is also known as DIS3-like 3′-5′ exoribonuclease 2 (DIS3L2) in fungi and metazoans. To initiate 5′→3′ decay, the m 7G cap is removed by the decapping complex, resulting in a 5′ monophosphorylated mRNA that is vulnerable to digestion by the cytoplasmic eXoRiboNuclease 4 (XRN4; XRN1 in fungi and metazoans). All three of these RNA decay pathways are highly conserved in eukaryotic model systems, with the exception of Saccharomyces cerevisiae, which lacks homologs of the decapping complex scaffold, VARICOSE (VCS), and SOV/DIS3L2. Thus, advances in any model system are of potential importance to the field.
Figure 1. Schematic of the major cytoplasmic mRNA decay pathways.
Mature mRNAs are first deadenylated by removal of the poly(A) tail. Deadenylated transcripts can be degraded in either the 3′→5′ or 5′→3′ direction. 3′→5′ degradation can occur by activity of the RNA exosome or by SUPPRESSOR OF VARICOSE (SOV). For degradation to occur in the 5′→3′ direction, transcripts must first be stripped of their 5′ m 7G cap by the RNA decapping complex, and further decay occurs by XRN4. VCS, VARICOSE.
Deadenylation
Removal of the poly(A) tail of mRNA by deadenylases is the first and rate-limiting step in mRNA degradation 1. In plants, deadenylases include members of the CCR4-NOT complex, poly(A)-specific ribonuclease (PARN), and the poly(A) nuclease (PAN). CAF1 (of the CCr4-NOT complex) is a major deadenylase in plants, as loss-of-function mutants in this protein result in severely impaired mRNA decay 3. However, despite their importance for mRNA metabolism, the specificity of these three modes of deadenylation and their regulation are not well understood.
3′ to 5′ degradation
The RNA exosome is a large multi-subunit complex with both nuclear and cytoplasmic functions 2. Eukaryotic exosomes resemble the bacterial RNase PH and polynucleotide phosphorylase (PNPase) RNA decay complexes in that that they are large protein complexes with a barrel-like configuration 4. However, for bacterial PNPase, the barrel’s interior is the site of active RNA decay. This is in contrast to metazoan and fungal exosome core proteins, which lack catalytic activity, even though their sequences show some conservation 5, 6. In addition to the catalytically inactive central core (known as Exo9 for its nine subunits), the eukaryotic exosome contains peripheral subunits that carry out RNA processing and decay (for example, response regulator proteins 6 and 44 [Rrp6 and Rrp44]). A recent study showed that the Arabidopsis RRP41, an Exo9 subunit of the central barrel, retains catalytic activity 7, 8. This activity appears to extend through the entire plant lineage.
The other cytoplasmic 3′→5′ exonuclease, SOV/DIS3L2, is an exosome-independent enzyme that was first identified in Arabidopsis as an accession-specific suppressor of vcs mutants 9. It is a broadly conserved RNase II domain protein with highly conserved homologs in metazoans and fungi 10– 13. In these systems, SOV/DIS3L2 substrates include non-coding RNAs (ncRNAs), long non-coding RNAs (lncRNAs), microRNAs (miRNAs) and their precursors, and mRNAs 14– 17. Drosophila sov/dis3l2 knock-down lines show over-growth phenotypes 18, and mutations in the SOV/DIS3L2 gene in humans result in an embryonic-lethal cellular over-growth condition known as Perlman syndrome 19, 20. These severe phenotypes are in contrast to Arabidopsis, as several phenotypically normal accessions lack a functional SOV, including the Col-0 reference strain 10.
5′ to 3′ degradation
Removal of the 5′ m 7G cap of mRNAs is catalysed by DECAPPING2 (DCP2), which forms a heterodimer with its activator, DECAPPING1 (DCP1). In S. cerevisiae, dimerization of these two proteins is sufficient for decapping 21, but higher eukaryotes also require a scaffold protein, known as VCS (also referred to as human enhancer of decapping large subunit [HEDLS] or Ge-1 in other systems) 9, 22– 25. A notable feature of the mRNA decapping complex is that it can localize to cytoplasmic foci called processing bodies (P-bodies) 9, 26.
Decapped mRNAs are further degraded by XRN proteins, which also function in nuclear RNA metabolism (for example, RNA silencing, rRNA maturation, and transcription termination). XRN1 and XRN2 are the major 5′→3′ exonucleases in the fungal and metazoan cytoplasm and nucleus, respectively 27. Although plants do not possess an XRN1 ortholog, they do have an ortholog of XRN2, which is known as XRN4 and localizes to the cytoplasm and functions like XRN1 in other model systems 28.
The focus of this review is on regulation, a term we use in its strict sense: a tunable parameter that alters decay rates of specific mRNAs and results in changes in their abundance 29. Although endonucleolytic cleavage, which can be specified by small RNAs (including miRNAs), is an important regulatory process, space limitations prevent its inclusion here and so we refer readers to several excellent reviews 30– 33. We consider five emerging areas that reveal either regulation by decay or its potential: (1) RNA structure (covalent modifications and secondary structure), (2) RNA quality control (RQC), (3) regulation of mRNA decapping (including post-translational modifications of the decapping complex), (4) mRNA localization (to P-bodies, specifically), and (5) decay pathway interplay and the potential for an RNA to switch decay pathways.
1. RNA structure: covalent modifications and folding
Many RNA fates are determined by the activity of their binding proteins; different sets of proteins bind to RNAs as they progress from nascent transcripts through their eventual degradation. These proteins promote functions such as post-transcriptional processing, translation initiation, and targeting for decay. Recent progress in understanding the roles of covalent RNA modifications and their subsequent effects on the affinities of RNA-binding proteins and RNA secondary structure highlights their importance in regulating mRNA stability.
N6-methyladenosine modification of mRNA
N6-methyladenosine (m 6A) is the most prevalent reversible covalent mark on eukaryotic RNA and plays important roles in many steps of RNA metabolism, including mRNA decay 34. Dynamic m 6A modification has been demonstrated to be vital for development, most notably cell differentiation 35– 38. Enzymes that catalyse the addition and removal of this modification (known as writers and erasers, respectively) have been characterized. Regulatory outcomes arise through reader proteins, which consist of YT521-B homology (YTH) domain proteins that bind m 6A-modified RNAs.
m 6A modification of eukaryotic mRNA occurs adjacent to stop codons and transcription start sites and within 3′ untranslated regions (UTRs) 39. Transcriptome-wide studies of m 6A modifications in Arabidopsis suggest that plants additionally have m 6A sites adjacent to start codons 37, 40. Accordingly, there are plant-specific modification motifs (such as URUAY) that are methylated along with the general eukaryotic RRACH and RAC consensus sequences 41. In fungi and metazoans, the YTHDF (reader) proteins remove the bound m 6A-modified transcripts from the translational pool and initiate their decay by recruiting the CCR4-NOT deadenylase complex 42, 43. A similar modification—2′-O-dimethyladenosine (m 6Am)—at, or adjacent to, the 5′ cap also impacts decay rates by inhibiting the action of the decapping enzyme (DCP2), thereby slowing decay rates 44.
EVOLUTIONARILY CONSERVED C-TERMINAL REGION 2 and 3 (ECT2 and ECT3) are cytoplasmic m 6A readers in plants that share homology with human YTHDF proteins 45, 46. Roles for plant m 6A readers have been implicated by developmental defects in mutants: plants mutant for the m 6A readers ECT2 and ECT3 have defects in leaf and trichome development 41, 45, 46. Plants mutant for components of the m 6A methyltransferase (writer) complex have hypomethylated transcripts and display enlarged shoot apical meristems and organogenesis defects due to increased stability of their target transcripts 37, 38. Mutants with defects in the m 6A eraser ALKB HOMOLOG 10B ( ALKBH10B) have suppressed vegetative growth and a delayed transition to flowering because of global hypermethylation 47, which was associated with stabilization of transcripts encoding FLOWERING LOCUS T (FT) and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 and 9 (SPL3/9). Thus, contrary to animals, m 6A modification in plants appears to stabilize target transcripts. This is emphasized by the observation that faster RNA decay also occurred in ect2 reader mutants, indicating that the binding of ECT2 to m 6A-modified RNAs generally led to their stabilization 40, 41. Whether this pattern of stabilization by m 6A modification extends to all modified plant RNAs, and the mechanisms that bring about the differing responses in plants and other systems, will be an important topic of future exploration.
Roles of m 6A modification in many additional cellular responses, including viral responses in both plants and metazoans, have also been reported 48, 49. Teasing apart how cells integrate these reversible modifications and distinguish between selectively altered mRNA stability and other m 6A functions is likely to become a very interesting story.
Uridylation
RNAs are also modified on their 3′ ends, and poly(A) tail addition is the best-known example. Recent studies highlight the importance of another 3′ modification, uridylation, which is catalysed by TERMINAL URIDYLYLTRANSFERASES (TUTases). UTP:RNA URIDYLTRANSERASE (URT1) and HEN1 SUPPRESSOR 1 (HESO1) are the two major TUTases in Arabidopsis 50– 52. Both URT1 and HESO1 uridylate miRNAs. In metazoan and fungal systems, miRNA uridylation leads to destabilization via SOV/DIS3L2 11, 12, 53 but whether this is also the case in Arabidopsis is unknown.
mRNAs are also uridylated, which in fungal and metazoan systems is associated with destabilization. In Arabidopsis, URT1 is the major mRNA TUTase. It prevents trimming of the poly(A) tail and is also necessary to repair deadenylated RNAs 54. In other systems, mRNA uridylation has been linked to degradation by SOV/DIS3L2, including formation of SOV/DIS3L2-TUTase complexes 16, 55. However, whether mRNA uridylation in plants also leads to transcript destabilization is not known, perhaps because most Arabidopsis uridylation studies have used the Col-0 accession, which is an sov mutant 10. Finally, uridylation also tags the 5′ cleavage fragments of mRNAs that result from miRNA-induced cleavage. This feature promotes decay via RISC-INTERACTING CLEARING 3′-5′ EXORIBONUCLEASES1 and 2 (RICE1, 2) 56, which not only targets these fragments for decay but also appears to be important for allowing fast cycling of RISC complexes. In addition, non-stop decay (discussed in section 2, below) can eliminate 5′ cleavage products if miRNA or small interfering RNA (siRNA)-induced cleavage occurs in mRNA coding regions 57.
RNA secondary structure
The complex folded structures of RNAs can also have important implications for stability. Analyses of RNA secondary structure in Arabidopsis found patterns of structure distributions in mRNAs, including less structure in the UTRs than in coding regions, and the observation that more structure generally resulted in lower transcript stability 58– 60. The impact of RNA structure and protein binding on RNA decay was recently explored in the context of root epidermal development. RNA secondary structures that were specific for root-hair or non-root-hair fates were found, and proteins that bound to these cell type–specific folds were identified 61. Interestingly, one of these interactions was with SERRATE, a zinc finger domain protein with functions in miRNA biogenesis, splicing, and epigenetic silencing 62, and appears to contribute to root-hair fate selection by miRNA-independent stabilization of specific mRNAs. Folded structures can also be impacted by m 6A modification, which can act as a structural switch by disrupting local secondary structures to promote interaction with RNA-binding proteins. These m 6A switches are enriched in 3′ UTRs and near stop codons, and switches located in introns were shown to play a role in alternative splicing 39. The refinement of methods for determining in vivo RNA structure promises many more insights ahead 60, 63.
2. RNA quality control
mRNAs that potentially encode aberrant protein products, such as truncated proteins caused by premature termination codons (PTCs) or by physical impediments to translation, or that disrupt ribosome homeostasis (due to lack of a start codon) are potentially deleterious for normal cellular function. Cells avoid these problems by identifying and degrading the offending mRNAs in a series of reactions known generally as RQC. These pathways include nonsense-mediated decay (NMD), which degrades RNAs with an abnormally positioned PTC; non-stop decay, which degrades mRNAs that lack a stop codon; and no-go decay, which degrades mRNAs with stalled ribosomes.
These pathways, however, have the potential to go well beyond a protective function and can contribute to regulation of mRNA abundance 64. For example, alternative splicing can lead to mRNA isoforms, including ones with PTCs. In plants, environmental stresses lead to enhanced production of mRNA variants containing PTCs, and their subsequent degradation allows adaptive responses to the initial stress 65– 67. Similarly, NMD selectively regulates transcript abundance of isoforms arising from alternative transcription start sites 68 and can also function in transcript autoregulation 69.
Another mechanism of RQC is the production of siRNAs that direct ARGONAUT-induced cleavage and decay of corresponding mRNAs 70. The accumulation of siRNAs has been observed in Arabidopsis mRNA decapping mutants and has been shown to elicit the severe phenotypes of decapping mutants because mutations in RNA-DEPENDENT RNA POLYMERASE 6 ( rdr6), which is required for siRNA amplification, suppress this severe phenotype 71. This result highlights the importance of mRNA decapping in modulating RNA abundances. Similarly, siRNA accumulates in double mutants that lack both XRN4 and SKI2 (a component of the RNA exosome), and the resulting severe phenotype was similarly alleviated by loss of RDR6 function 72. Thus, in addition to potential specialized functions, the major cytoplasmic mRNA decay pathways prevent generation of siRNAs.
3. Regulation of mRNA decapping
Reversible phosphorylation
Each of the subunits of the mRNA decapping complex are subject to phosphorylation and these post-translational modification events have been implicated in regulating mRNA abundance 73, 74. DCP1 phosphorylation by MITOGEN-ACTIVATED PROTEIN KINASE 6 (MPK6) was shown to arise in response to dehydration. This modification is thought to slow mRNA decay, as transgenic lines with a non-phosphorylatable version of DCP1 showed slower decay of EXPL1 RNA. Furthermore, rapid phosphorylations of VCS and DCP2 were found in a phosphoproteomic analysis of rapid responses to osmotic stress 74. The major site of VCS phosphorylation is in its S-rich linker domain. Because DCP2 requires DCP1 for activation and VCS serves as their interaction scaffold, phosphorylation of the S-rich linker could alter DCP2 activation. Salt stress similarly leads to VCS phosphorylation but via the SNF1-RELATED KINASE, SnRK2G 75. VCS and SnSRKG2 show constitutive physical interaction, and salt stress led the SnSRK2G-VCS complex to relocate to P-bodies 75. VCS phosphorylation was also correlated with changes in RNA decay rates, as a set of VCS-dependent RNAs decayed faster in wild-type (WT) (Col-0) plants following exposure to salt stress. This correlation suggests that VCS phosphorylation led to faster decay of these RNAs. Important questions for the future include determining how phosphorylation of the mRNA decapping complex subunits causes changes in mRNA decay; for example, is substrate recruitment affected or are mRNA decapping kinetics altered?
Decapping activators
The SM-LIKE (LSM) complex consists of seven RNA-binding subunits and produces distinct nuclear and cytoplasmic complexes 76– 78. The cytoplasmic complex binds to the 3′ termini of oligoadenylated and deadenylated mRNA and recruits the mRNA decapping complex. Genetic analysis of the LSM complex of Arabidopsis revealed that this complex functions in decay and is necessary for normal responses to abiotic stresses, including high salinity and cold temperatures, because it regulates the targeting of mRNAs to the decapping complex 79, 80. A functionally related protein, protein associated with topoisomerase II 1 (PAT1), also has diverse functions in post-transcriptional regulation of RNA, including decay 81. In Arabidopsis, PAT1 has been implicated in pathogen response–based changes in gene expression. It is phosphorylated by MPK4, which causes its localization to P-bodies in response to challenge by bacteria and where it activates decay of specific mRNAs 82.
4. P-body localization
P-bodies form through phase separation of intrinsically disordered regions of proteins, such as the decapping complex subunits, and RNA 83, 84. These concentrated collections of decay enzymes and mRNAs have been generally considered to expedite RNA decay. However, in yeast and drosophila, mRNA decapping does not require formation of these bodies 85, 86. Recent studies using Arabidopsis have advanced our understanding of how specific mRNAs come to be localized in P-bodies. One participating protein is SPIRRIG (SPI), a BEACH-domain protein that interacts with DCP1 87. SPI is required for localization of specific salt-response RNAs to P-bodies and for their stabilization. This compelling story is complicated by the multi-faceted functions of SPI, which also localizes to endosomes and is required for normal endosomal transport and vacuole morphology 88. The LSM proteins also have roles in localizing RNAs to P-bodies. These proteins interact to form heptameric rings, activate decapping, and associate with both mRNAs and the decapping complex in yeast 89. In Arabidopsis, the LSM proteins were shown to associate with stress-specific mRNAs and drive their localization into P-bodies, leading to faster transcript decay 80, 90. Although both SPI and LSM complexes move RNAs to P-bodies, their P-body localization has opposing effects on RNA stability. This raises many questions, including how P-bodies can be both stabilizing and destabilizing in an RNA-specific manner.
P-body studies using non-plant systems might offer some insight into these questions. A purification method was recently developed that allowed both proteomic and transcriptomic analyses of P-bodies isolated from human epithelial cells 86, 91, 92. This analysis confirmed P-body localization of mRNA decapping proteins, but there was no evidence that the localized mRNAs were undergoing decay. Instead, the results implicated P-bodies as an important site for translational arrest. However, whether P-bodies function similarly in all tissue types, and the extent to which human P-bodies can serve as a model for plants, needs to be determined. The stress-inducible P-bodies of yeast might be a more relevant model, even though S. cerevisiae lacks a homolog of the decapping complex scaffold, VCS. P-bodies of yeast have been shown to be sites of both decay and sequestration, and sequestered mRNAs can be restored to the translational pool 93. Understanding P-body functions and sorting out how P-body localization can lead to different RNA fates are important directions for future research.
5. Pathway interplay as a mechanism of selective regulation of mRNA decay
An under-explored aspect of mRNA decay is whether the three major cytoplasmic decay pathways ( Figure 1) have unique functional or regulatory significance. To identify their substrates, our lab carried out a genome-wide mRNA decay analysis using four Arabidopsis genotypes: a synthetic WT (Col-0 carrying a functional L. er SOV transgene), vcs and sov single mutants, and a vcs sov double mutant 94. Contributions of decapping (VCS) and SOV to the decay of mRNAs followed the assumption that mRNA substrates of decapping (VCS) and SOV would decay more slowly in vcs and sov mutants, respectively. We found that most RNAs decay by combined contributions of two or more pathways. While decapping (VCS) is required to sustain normal decay for 67% of the 17,293 analyzed RNAs, few were solely dependent on decapping for their decay. In addition, VCS-dependent RNAs tend to decay quickly, have abundances that are responsive to stress or developmental signals, and/or encode transcription factors 94. Decay of 22% of the analyzed transcripts was not attributable to either VCS (decapping) or SOV, suggesting a large role for the RNA exosome. In contrast to decapping (VCS)-dependent RNAs, putative exosome substrates were generally slow-decaying RNAs that encode proteins with housekeeping functions. Thus, both mRNA decapping and the RNA exosome are specialized in terms of mRNA substrate functions and decay rates.
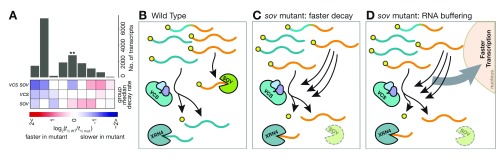
The search for mRNA substrates that decayed more slowly in sov mutants initially suggested that SOV/DIS3 contributes to decay of only about 9% of the analyzed transcripts 94. Curiously, 33% of these RNAs decay much faster in sov mutants than in WT. This faster decay comes from compensatory activity of the mRNA decapping complex, as indicated by slower decay of these same transcripts in vcs sov double mutants ( Figure 2A). Because many of the affected RNAs are not normally substrates of decapping, these findings suggest that transcripts that are normally substrates of SOV can become decapping substrates in its absence. Furthermore, the observation that mRNA decapping was associated with fast-decaying RNAs was supported because after these RNAs switched to the decapping pathway, their decay rates were much faster. We interpret this compensation by an alternate decay pathway as the activation of a feedback mechanism that compensates for the loss of SOV ( Figure 2B, C). Thus, triggering feedback results in a subset of the SOV substrates switching to decapping-mediated decay. This interpretation implicates that SOV actually contributes to decay of about 42% of the analyzed transcripts. Among the many questions raised by this analysis are whether the plasticity of mRNA decay pathways shown by SOV substrates extends more broadly across the transcriptome and whether pathway plasticity is used to regulate mRNA decay rates.
Figure 2. Loss of SUPPRESSOR OF VARICOSE (SOV) induces RNA decay feedback in Arabidopsis.
( A) Heat map depicts RNA decay rates, relative to the wild type, and histogram indicates the degree to which each pattern was represented. Bar with two asterisks indicates RNAs with VARICOSE (VCS)-dependent faster decay rates in sov mutants. ( B) Diagram of VCS and SOV decay in wild type. Yellow circles represent the 5′ m 7G cap, blue RNAs decay by mRNA decapping, orange RNAs by SOV, and blue-orange gradient colored RNAs are substrates of both pathways. ( C) In sov mutants, some RNAs that are normally substrates of SOV instead decay by mRNA decapping, and they decay faster. ( D) In sov mutants, faster-decay RNAs maintain a normal abundance, indicating transcriptional feedback, which is also called RNA buffering.
The sov-triggered feedback also appears to result in mRNA buffering, as indicated by near WT abundances despite much faster decay 94. This requires a commensurate increase in transcription and thus communication from cytoplasmic decay to the transcriptional machinery ( Figure 2D). A similar feedback pathway that coordinates transcription and decay has been described in yeast 95, 96. This RNA buffering system appears to explain why some Arabidopsis accessions tolerate mutations in SOV/DIS3L2.
Outlook
RNA decay pathways are highly conserved across eukaryotes, and research using Arabidopsis continues to contribute strongly to this field, as the genetic resources for studying mRNA decay in Arabidopsis make it an outstanding choice. However, many mRNA decay studies appear to be technically and computationally challenged. Selection of time points can have enormous implications on outcomes and accordingly should be selected on the basis of mRNA half-life range. Similarly, quantitative approaches to data analysis can compromise data outcomes. Pools of RNA change over time and can lead to the false impression that the abundance of stable RNAs increase. Furthermore, slight differences in measured decay rates can be difficult to assess. We have generated an R package in Bioconductor (RNAdecay) that, among other things, assists with normalization and allows statistical comparisons between treatments. This resource is freely available through Bioconductor ( https://bioconductor.org/packages/release/bioc/html/RNAdecay.html).
Recent discoveries have led to the identification of novel regulatory mechanisms for mRNA decay, including uridylation, methylation, and the potential for mRNAs to switch between decay pathways. Similarly, recent discoveries have led to the reconsideration of some past concepts, including P-bodies and the functional consequences of localized mRNAs. However, most studies address the behavior of only a few mRNAs, cell types, or a single condition, limiting the generality of outcomes. As costs for genome-wide approaches continue to decline, we can look forward to a clearer picture of the flexibility of mRNA stability, mechanisms of stability control, and positioning decay in the overall control of mRNA abundance.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Dominique Gagliardi, Institut de Biologie Moleculaire des Plantes (IBMP), CNRS, University of Strasbourg, Strasbourg, France
Daniel Silhavy, Department of Genetics, Agricultural Biotechnology Institute, Gödöllő, Hungary
Brian D Gregory, Department of Biology, University of Pennsylvania School of Arts and Sciences, Philadelphia, USA
Funding Statement
This work was supported by National Science Foundation grant MCB-1616779 to LES.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 3 approved]
References
- 1. Decker CJ, Parker R: A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7(8):1632–43. 10.1101/gad.7.8.1632 [DOI] [PubMed] [Google Scholar]
- 2. Januszyk K, Lima CD: The eukaryotic RNA exosome. Curr Opin Struct Biol. 2014;24:132–40. 10.1016/j.sbi.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chou WL, Huang LF, Fang JC, et al. : Divergence of the expression and subcellular localization of CCR4-associated factor 1 (CAF1) deadenylase proteins in Oryza sativa. Plant Mol Biol. 2014;85(4–5):443–58. 10.1007/s11103-014-0196-7 [DOI] [PubMed] [Google Scholar]
- 4. Lykke-Andersen S, Brodersen DE, Jensen TH: Origins and activities of the eukaryotic exosome. J Cell Sci. 2009;122(Pt 10):1487–94. 10.1242/jcs.047399 [DOI] [PubMed] [Google Scholar]
- 5. Liu Q, Greimann JC, Lima CD: Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127(6):1223–37. 10.1016/j.cell.2006.10.037 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 6. Dziembowski A, Lorentzen E, Conti E, et al. : A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14(1):15–22. 10.1038/nsmb1184 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 7. Sikorska N, Zuber H, Gobert A, et al. : RNA degradation by the plant RNA exosome involves both phosphorolytic and hydrolytic activities. Nat Commun. 2017;8(1):2162. 10.1038/s41467-017-02066-2 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 8. Chekanova JA, Shaw RJ, Wills MA, et al. : Poly(A) tail-dependent exonuclease AtRrp41p from Arabidopsis thaliana rescues 5.8 S rRNA processing and mRNA decay defects of the yeast ski6 mutant and is found in an exosome-sized complex in plant and yeast cells. J Biol Chem. 2000;275(42):33158–66. 10.1074/jbc.M005493200 [DOI] [PubMed] [Google Scholar]
- 9. Goeres DC, Van Norman JM, Zhang W, et al. : Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell. 2007;19(5):1549–64. 10.1105/tpc.106.047621 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 10. Zhang W, Murphy C, Sieburth LE: Conserved RNaseII domain protein functions in cytoplasmic mRNA decay and suppresses Arabidopsis decapping mutant phenotypes. Proc Natl Acad Sci U S A. 2010;107(36):15981–5. 10.1073/pnas.1007060107 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 11. Lubas M, Damgaard CK, Tomecki R, et al. : Exonuclease hDIS3L2 specifies an exosome-independent 3′-5′ degradation pathway of human cytoplasmic mRNA. EMBO J. 2013;32(13):1855–68. 10.1038/emboj.2013.135 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 12. Malecki M, Viegas SC, Carneiro T, et al. : The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 2013;32(13):1842–54. 10.1038/emboj.2013.63 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 13. Lv H, Zhu Y, Qiu Y, et al. : Structural analysis of Dis3l2, an exosome-independent exonuclease from Schizosaccharomyces pombe. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 6):1284–94. 10.1107/S1399004715005805 [DOI] [PubMed] [Google Scholar]
- 14. Ustianenko D, Hrossova D, Potesil D, et al. : Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA. 2013;19(12):1632–8. 10.1261/rna.040055.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Łabno A, Warkocki Z, Kuliński T, et al. : Perlman syndrome nuclease DIS3L2 controls cytoplasmic non-coding RNAs and provides surveillance pathway for maturing snRNAs. Nucleic Acids Res. 2016;44(21):10437–53. 10.1093/nar/gkw649 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 16. Reimão-Pinto MM, Manzenreither RA, Burkard TR, et al. : Molecular basis for cytoplasmic RNA surveillance by uridylation-triggered decay in Drosophila. EMBO J. 2016;35(22):2417–34. 10.15252/embj.201695164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pirouz M, Du P, Munafò M, et al. : Dis3l2-Mediated Decay Is a Quality Control Pathway for Noncoding RNAs. Cell Rep. 2016;16(7):1861–73. 10.1016/j.celrep.2016.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 18. Towler BP, Jones CI, Harper KL, et al. : A novel role for the 3′-5′ exoribonuclease Dis3L2 in controlling cell proliferation and tissue growth. RNA Biol. 2016;13(12):1286–99. 10.1080/15476286.2016.1232238 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 19. Chang HM, Triboulet R, Thornton JE, et al. : A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497(7448):244–8. 10.1038/nature12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Astuti D, Morris MR, Cooper WN, et al. : Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet. 2012;44(3):277–84. 10.1038/ng.1071 [DOI] [PubMed] [Google Scholar]
- 21. LaGrandeur TE, Parker R: Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 1998;17(5):1487–96. 10.1093/emboj/17.5.1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deyholos MK, Cavaness GF, Hall B, et al. : VARICOSE, a WD-domain protein, is required for leaf blade development. Development. 2003;130(26):6577–88. 10.1242/dev.00909 [DOI] [PubMed] [Google Scholar]
- 23. Xu J, Yang JY, Niu QW, et al. : Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell. 2006;18(12):3386–98. 10.1105/tpc.106.047605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fenger-Grøn M, Fillman C, Norrild B, et al. : Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20(6):905–15. 10.1016/j.molcel.2005.10.031 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 25. Yu JH, Yang WH, Gulick T, et al. : Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA. 2005;11(12):1795–802. 10.1261/rna.2142405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sheth U, Parker R: Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300(5620):805–8. 10.1126/science.1082320 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 27. Chang JH, Xiang S, Tong L: 5′-3′ Exoribonucleases.In Ribonucleases.(Springer, Berlin, Heidelberg).2011;167–192. 10.1007/978-3-642-21078-5_7 [DOI] [Google Scholar]
- 28. Kastenmayer JP, Green PJ: Novel features of the XRN-family in Arabidopsis: evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc Natl Acad Sci U S A. 2000;97(25):13985–90. 10.1073/pnas.97.25.13985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pichersky E: Is the concept of regulation overused in molecular and cellular biology? Plant Cell. 2005;17(12):3217–8. 10.1105/tpc.105.038968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fouracre JP, Poethig RS: The role of small RNAs in vegetative shoot development. Curr Opin Plant Biol. 2016;29:64–72. 10.1016/j.pbi.2015.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 31. Rogers K, Chen X: Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013;25(7):2383–99. 10.1105/tpc.113.113159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Borges F, Martienssen RA: The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol. 2015;16(12):727–41. 10.1038/nrm4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rodriguez RE, Schommer C, Palatnik JF: Control of cell proliferation by microRNAs in plants. Curr Opin Plant Biol. 2016;34:68–76. 10.1016/j.pbi.2016.10.003 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 34. Roignant JY, Soller M: m 6A in mRNA: An Ancient Mechanism for Fine-Tuning Gene Expression. Trends Genet. 2017;33(6):380–90. 10.1016/j.tig.2017.04.003 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 35. Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. : Stem cells. m 6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347(6225):1002–6. 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- 36. Batista PJ, Molinie B, Wang J, et al. : m 6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15(6):707–19. 10.1016/j.stem.2014.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bodi Z, Zhong S, Mehra S, et al. : Adenosine Methylation in Arabidopsis mRNA is Associated with the 3' End and Reduced Levels Cause Developmental Defects. Front Plant Sci. 2012;3:48. 10.3389/fpls.2012.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shen L, Liang Z, Gu X, et al. : N 6-Methyladenosine RNA Modification Regulates Shoot Stem Cell Fate in Arabidopsis. Dev Cell. 2016;38(2):186–200. 10.1016/j.devcel.2016.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 39. Liu N, Dai Q, Zheng G, et al. : N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518(7540):560–4. 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 40. Luo GZ, MacQueen A, Zheng G, et al. : Unique features of the m 6A methylome in Arabidopsis thaliana. Nat Commun. 2014;5:5630. 10.1038/ncomms6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wei LH, Song P, Wang Y, et al. : The m 6A Reader ECT2 Controls Trichome Morphology by Affecting mRNA Stability in Arabidopsis. Plant Cell. 2018;30(5):968–85. 10.1105/tpc.17.00934 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 42. Du H, Zhao Y, He J, et al. : YTHDF2 destabilizes m 6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. 10.1038/ncomms12626 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 43. Wang X, Lu Z, Gomez A, et al. : N 6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–20. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 44. Mauer J, Luo X, Blanjoie A, et al. : Reversible methylation of m 6A m in the 5' cap controls mRNA stability. Nature. 2017;541(7637):371–5. 10.1038/nature21022 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 45. Arribas-Hernández L, Bressendorff S, Hansen MH, et al. : An m 6A-YTH Module Controls Developmental Timing and Morphogenesis in Arabidopsis. Plant Cell. 2018;30(5):952–67. 10.1105/tpc.17.00833 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 46. Scutenaire J, Deragon JM, Jean V, et al. : The YTH Domain Protein ECT2 Is an m 6A Reader Required for Normal Trichome Branching in Arabidopsis. Plant Cell. 2018;30(5):986–1005. 10.1105/tpc.17.00854 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 47. Duan HC, Wei LH, Zhang C, et al. : ALKBH10B Is an RNA N 6-Methyladenosine Demethylase Affecting Arabidopsis Floral Transition. Plant Cell. 2017;29(12):2995–3011. 10.1105/tpc.16.00912 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 48. Martínez-Pérez M, Aparicio F, López-Gresa MP, et al. : Arabidopsis m 6A demethylase activity modulates viral infection of a plant virus and the m 6A abundance in its genomic RNAs. Proc Natl Acad Sci U S A. 2017;114(40):10755–60. 10.1073/pnas.1703139114 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 49. Tan B, Gao SJ: RNA epitranscriptomics: Regulation of infection of RNA and DNA viruses by N 6 -methyladenosine (m 6 A). Rev Med Virol. 2018;28(4):e1983. 10.1002/rmv.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 50. Sement FM, Ferrier E, Zuber H, et al. : Uridylation prevents 3' trimming of oligoadenylated mRNAs. Nucleic Acids Res. 2013;41(14):7115–27. 10.1093/nar/gkt465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ren G, Chen X, Yu B: Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr Biol. 2012;22(8):695–700. 10.1016/j.cub.2012.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao Y, Yu Y, Zhai J, et al. : The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol. 2012;22(8):689–94. 10.1016/j.cub.2012.02.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tu B, Liu L, Xu C, et al. : Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis. PLoS Genet. 2015;11(4):e1005119. 10.1371/journal.pgen.1005119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zuber H, Scheer H, Ferrier E, et al. : Uridylation and PABP Cooperate to Repair mRNA Deadenylated Ends in Arabidopsis. Cell Rep. 2016;14(11):2707–17. 10.1016/j.celrep.2016.02.060 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 55. Lin CJ, Wen J, Bejarano F, et al. : Characterization of a TUTase/RNase complex required for Drosophila gametogenesis. RNA. 2017;23(3):284–96. 10.1261/rna.059527.116 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 56. Zhang Z, Hu F, Sung MW, et al. : RISC-interacting clearing 3'- 5' exoribonucleases (RICEs) degrade uridylated cleavage fragments to maintain functional RISC in Arabidopsis thaliana. eLife. 2017;6: pii: e24466. 10.7554/eLife.24466 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 57. Szádeczky-Kardoss I, Csorba T, Auber A, et al. : The nonstop decay and the RNA silencing systems operate cooperatively in plants. Nucleic Acids Res. 2018;46(9):4632–48. 10.1093/nar/gky279 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 58. Li F, Zheng Q, Vandivier LE, et al. : Regulatory impact of RNA secondary structure across the Arabidopsis transcriptome. Plant Cell. 2012;24(11):4346–59. 10.1105/tpc.112.104232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ding Y, Tang Y, Kwok CK, et al. : In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505(7485):696–700. 10.1038/nature12756 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 60. Bevilacqua PC, Ritchey LE, Su Z, et al. : Genome-Wide Analysis of RNA Secondary Structure. Annu Rev Genet. 2016;50:235–66. 10.1146/annurev-genet-120215-035034 [DOI] [PubMed] [Google Scholar]
- 61. Foley SW, Gosai SJ, Wang D, et al. : A Global View of RNA-Protein Interactions Identifies Post-transcriptional Regulators of Root Hair Cell Fate. Dev Cell. 2017;41(2):204–220.e5. 10.1016/j.devcel.2017.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 62. Ma Z, Castillo-González C, Wang Z, et al. : Arabidopsis Serrate Coordinates Histone Methyltransferases ATXR5/6 and RNA Processing Factor RDR6 to Regulate Transposon Expression. Dev Cell. 2018;45(6):769–784.e6. 10.1016/j.devcel.2018.05.023 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 63. Bevilacqua PC, Assmann SM: RNA structure: A LASER-focused view into cells. Nat Chem Biol. 2018;14(3):200–1. 10.1038/nchembio.2570 [DOI] [PubMed] [Google Scholar]
- 64. Peccarelli M, Kebaara BW: Regulation of natural mRNAs by the nonsense-mediated mRNA decay pathway. Eukaryotic Cell. 2014;13(9):1126–35. 10.1128/EC.00090-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sureshkumar S, Dent C, Seleznev A, et al. : Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nat Plants. 2016;2(5):16055. 10.1038/nplants.2016.55 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 66. Filichkin SA, Cumbie JS, Dharmawardhana P, et al. : Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Mol Plant. 2015;8(2):207–27. 10.1016/j.molp.2014.10.011 [DOI] [PubMed] [Google Scholar]
- 67. Kwon YJ, Park MJ, Kim SG, et al. : Alternative splicing and nonsense-mediated decay of circadian clock genes under environmental stress conditions in Arabidopsis. BMC Plant Biol. 2014;14:136. 10.1186/1471-2229-14-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kurihara Y, Makita Y, Kawashima M, et al. : Transcripts from downstream alternative transcription start sites evade uORF-mediated inhibition of gene expression in Arabidopsis. Proc Natl Acad Sci U S A. 2018;115(30):7831–6. 10.1073/pnas.1804971115 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 69. Nyikó T, Auber A, Szabadkai L, et al. : Expression of the eRF1 translation termination factor is controlled by an autoregulatory circuit involving readthrough and nonsense-mediated decay in plants. Nucleic Acids Res. 2017;45(7):4174–88. 10.1093/nar/gkw1303 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 70. Liu L, Chen X: RNA Quality Control as a Key to Suppressing RNA Silencing of Endogenous Genes in Plants. Mol Plant. 2016;9(6):826–36. 10.1016/j.molp.2016.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Martínez de Alba AE, Moreno AB, Gabriel M, et al. : In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res. 2015;43(5):2902–13. 10.1093/nar/gkv119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang X, Zhu Y, Liu X, et al. : Plant biology. Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science. 2015;348(6230):120–3. 10.1126/science.aaa2618 [DOI] [PubMed] [Google Scholar]
- 73. Xu J, Chua NH: Dehydration stress activates Arabidopsis MPK6 to signal DCP1 phosphorylation. EMBO J. 2012;31(8):1975–84. 10.1038/emboj.2012.56 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 74. E Stecker K, Minkoff BB, Sussman MR: Phosphoproteomic Analyses Reveal Early Signaling Events in the Osmotic Stress Response. Plant Physiol. 2014;165(3):1171–87. 10.1104/pp.114.238816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Soma F, Mogami J, Yoshida T, et al. : ABA-unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat Plants. 2017;3:16204. 10.1038/nplants.2016.204 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 76. Bouveret E, Rigaut G, Shevchenko A, et al. : A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19(1):1661–71. 10.1093/emboj/19.7.1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chowdhury A, Tharun S: Activation of decapping involves binding of the mRNA and facilitation of the post-binding steps by the Lsm1-7-Pat1 complex. RNA. 2009;15(10):1837–48. 10.1261/rna.1650109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chowdhury A, Mukhopadhyay J, Tharun S: The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13(7):998–1016. 10.1261/rna.502507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Golisz A, Sikorski PJ, Kruszka K, et al. : Arabidopsis thaliana LSM proteins function in mRNA splicing and degradation. Nucleic Acids Res. 2013;41(12):6232–49. 10.1093/nar/gkt296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Perea-Resa C, Carrasco-López C, Catalá R, et al. : The LSM1-7 Complex Differentially Regulates Arabidopsis Tolerance to Abiotic Stress Conditions by Promoting Selective mRNA Decapping. Plant Cell. 2016;28(2):505–20. 10.1105/tpc.15.00867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Marnef A, Standart N: Pat1 proteins: a life in translation, translation repression and mRNA decay. Biochem Soc Trans. 2010;38(6):1602–7. 10.1042/BST0381602 [DOI] [PubMed] [Google Scholar]
- 82. Roux ME, Rasmussen MW, Palma K, et al. : The mRNA decay factor PAT1 functions in a pathway including MAP kinase 4 and immune receptor SUMM2. EMBO J. 2015;34(5):593–608. 10.15252/embj.201488645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Protter DSW, Rao BS, van Treeck B, et al. : Intrinsically Disordered Regions Can Contribute Promiscuous Interactions to RNP Granule Assembly. Cell Rep. 2018;22(6):1401–12. 10.1016/j.celrep.2018.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 84. Banani SF, Rice AM, Peeples WB, et al. : Compositional Control of Phase-Separated Cellular Bodies. Cell. 2016;166(3):651–63. 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 85. Franks TM, Lykke-Andersen J: The control of mRNA decapping and P-body formation. Mol Cell. 2008;32(5):605–15. 10.1016/j.molcel.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tutucci E, Vera M, Biswas J, et al. : An improved MS2 system for accurate reporting of the mRNA life cycle. Nat Methods. 2018;15(1):81–9. 10.1038/nmeth.4502 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 87. Steffens A, Bräutigam A, Jakoby M, et al. : The BEACH Domain Protein SPIRRIG Is Essential for Arabidopsis Salt Stress Tolerance and Functions as a Regulator of Transcript Stabilization and Localization. PLoS Biol. 2015;13(7):e1002188. 10.1371/journal.pbio.1002188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Steffens A, Jakoby M, Hülskamp M: Physical, Functional and Genetic Interactions between the BEACH Domain Protein SPIRRIG and LIP5 and SKD1 and Its Role in Endosomal Trafficking to the Vacuole in Arabidopsis Front Plant Sci. 2017;8:1969. 10.3389/fpls.2017.01969 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 89. He W, Parker R: Functions of Lsm proteins in mRNA degradation and splicing. Curr Opin Cell Biol. 2000;12(3):346–50. 10.1016/S0955-0674(00)00098-3 [DOI] [PubMed] [Google Scholar]
- 90. Perea-Resa C, Hernández-Verdeja T, López-Cobollo R, et al. : LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. Plant Cell. 2012;24(12):4930–47. 10.1105/tpc.112.103697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hubstenberger A, Courel M, Bénard M, et al. : P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol Cell. 2017;68(1):144–157.e5. 10.1016/j.molcel.2017.09.003 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 92. Horvathova I, Voigt F, Kotrys AV, et al. : The Dynamics of mRNA Turnover Revealed by Single-Molecule Imaging in Single Cells. Mol Cell. 2017;68(3):615–625.e9. 10.1016/j.molcel.2017.09.030 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 93. Brengues M, Teixeira D, Parker R: Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310(5747):486–9. 10.1126/science.1115791 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 94. Sorenson RS, Deshotel MJ, Johnson K, et al. : Arabidopsis mRNA decay landscape arises from specialized RNA decay substrates, decapping-mediated feedback, and redundancy. Proc Natl Acad Sci U S A. 2018;115(7):E1485–E1494. 10.1073/pnas.1712312115 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 95. Sun M, Schwalb B, Pirkl N, et al. : Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol Cell. 2013;52(1):52–62. 10.1016/j.molcel.2013.09.010 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 96. Haimovich G, Medina DA, Causse SZ, et al. : Gene expression is circular: Factors for mRNA degradation also foster mRNA synthesis. Cell. 2013;153(5):1000–11. 10.1016/j.cell.2013.05.012 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation