Abstract
Recent evidence shows that cyclic GMP-AMP synthase (cGAS)/stimulator of interferon (IFN) genes (STING) signaling is essential for antitumor immunity by inducing the production of type I IFN and thus activating both innate and adaptive immunity based on gene knockout mouse models. However, the extensive detection of the expression of cGAS/STING signaling in human cancer and mining the roles of this signaling pathway in human cancer immunity have not been performed until now. In this study, we revealed that four key molecules (cGAS, STING, TANK binding kinase 1 [TBK1], and IFN regulatory factor 3 [IRF3]) in the cGAS/STING signaling are highly expressed in cancer tissues, and the expression levels of these genes are negatively correlated with their methylation levels in most of the detected cancer types. We also showed that highly upregulated cGAS/STING signaling is negatively correlated with the infiltration of immune cells in some tumor types, and consistent with these findings, we showed that a high level of cGAS/STING signaling predicts a poor prognosis in patients with certain cancers. This study suggests that it is necessary to deeply and fully evaluate the function of cGAS/STING signaling in cancer immunity and cancer progression before the application of the STING agonist-based anticancer immune therapy in the clinic.
Keywords: cGAS/STING, immune infiltration, prognosis, methylation, interferon
Graphical Abstract
Introduction
Although the crosstalk between the immune system and cancer has been documented for almost 60 years,1 the precise mechanism of how the immune system recognizes and kills cancer cells is unknown. Currently, it is well accepted that both innate and adaptive immunity play an important role in tumor immunosurveillance, and type 1 interferon (IFN) is critical for eliciting an effective antitumor immunity by bridging innate and adaptive immunity, because type 1 IFN, produced by antigen-presenting dendritic cells (DCs) in the tumor microenvironment, not only activates the innate immune response but also facilitates T cell cross-priming and infiltration.2 Numerous studies demonstrate that cytosolic double-strand DNA induces the expression of type 1 IFN in various cell types;3 however, the mechanism of type 1 IFN induction stimulated by cytosolic DNA was not truly revealed until 2008, when the stimulator of IFN genes (STING) was identified.4
Recent evidence demonstrates that cGAS (cyclic GMP-AMP synthase)/STING signaling is critical for the induction of type 1 IFN and plays an important role in the cancer immunity. Recent research shows that cytoplasmic dsDNA in cancer cells, caused by DNA virus infection, genomic DNA damage, or mitochondrial DNA leakage, binds to and then activates cGAS, an enzyme that catalyzes the production of cyclic GMP-AMP (cGAMP), which is a type of cyclic dinucleotide that binds to and activates STING.5 The activated STING changes its conformation to recruit TANK binding kinase 1 (TBK1) and then phosphorylates IFN regulatory factor 3 (IRF3). The phosphorylated IRF3 translocates into the nucleus and induces the production of type I IFN and other cytokines associated with immunity regulation.6
As a stimulator of type 1 IFN, the cGAS/STING pathway is reported to trigger a spontaneous antitumor T cell response in vivo,7 whereas the deficiency of STING or IRF3 shows an impaired spontaneous T cell response against tumors.8 Moreover, an intratumoral injection of a STING agonist promotes the infiltration of T cells into the tumor microenvironment in both murine melanoma and glioma models.9, 10 In most conditions, chemotherapy and radiotherapy destroy tumor cells by inducing DNA damage, and then they cause the release of the DNA fragments into the cytosol, thus activating the cGAS/STING pathway.8 There are some studies showing that a STING agonist also enhances the efficiency of immune-checkpoint blockade-based immunotherapy in some tumor types.11 However, STING-deficient hosts show a poorer treatment efficacy for immunotherapy and chemoradiation therapy.8 These studies imply that the cGAS/STING pathway plays an important role in antitumor immunity and cancer therapeutics.
Although it has been shown that the cGAS/STING pathway is essential for antitumor immunity in various mouse models, and there are limited studies suggesting that this signaling pathway is also an important regulator in certain human cancer types,12, 13 the extensive detection of the expression of cGAS/STING signaling in human cancer and mining the roles of this signaling pathway in human cancer immunity have not been performed until now. In this study, we detected the expression profiles of four key molecules (cGAS, STING, TBK1, and IRF3) in the cGAS/STING signaling pathway, explored their potential roles in the infiltration of immunity cells within human tumor tissues, and uncovered the value of predicting the prognosis of cancer patients by analyzing The Cancer Genome Atlas (TCGA) data in pan-cancer.
Results
cGAS/STING Signaling Is Universally Elevated Based on the Expression of MB21D1, TMEM173, TBK1, and IRF3 in Pan-Cancer
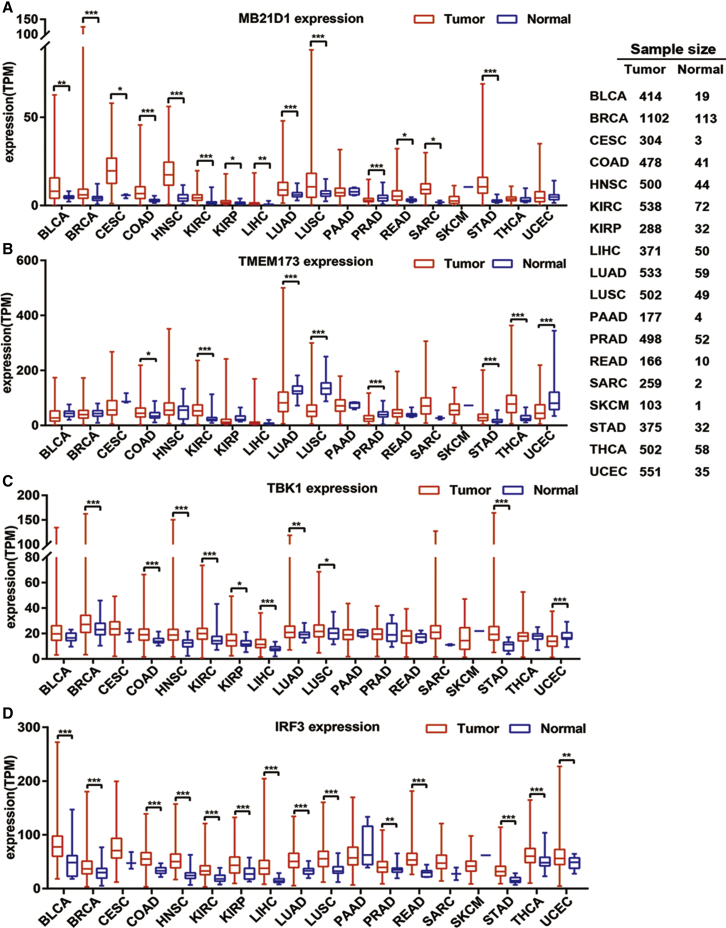
We first evaluated the expression change of four key molecules, MB21D1 encoding cGAS and TMEM173 encoding STING, TBK1, and IRF3, in the cGAS/STING signaling pathway by comparing their expression in both malignant tumor tissues and controlled normal tissues based on TCGA datasets containing 18 malignant tumor types. In the majority of the malignant tumor types, the expression of MB21D1 mRNA is significantly increased compared with that of the normal tissue (Figure 1A), with the exception that there is no significant difference for MB21D1 expression in pancreatic adenocarcinoma (PAAD), thyroid adenocarcinoma (THCA), skin cutaneous melanoma (SKCM), and uterine corpus endometrial carcinoma (UCEC), whereas it is downregulated only in prostate adenocarcinoma (PRAD) compared with normal tissues. The expression of TMEM173 in tumor tissues is upregulated significantly in colorectal carcinoma (COAD), kidney renal clear cell carcinoma (KIRC), stomach adenocarcinoma (STAD), and THCA, but is downregulated significantly in lung adenocarcinoma (LUAD), lung squamous carcinoma (LUSC), PRAD, and UCEC compared with normal control tissues (Figure 1B, p < 0.05). We revealed that the expression of TBK1 is also upregulated in more than half of the detected malignant tumor types (Figure 1C), including bladder carcinoma (BLCA), breast carcinoma (BRCA), COAD, head and neck squamous carcinoma (HNSC), KIRC, kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), LUAD, LUSC, and STAD, and it is downregulated only in UCEC compared with that of the normal control tissues. When investigating the expression of the IRF3 gene (Figure 1D), we found that it is upregulated in almost all of the detected cancer types, with the exception of PAAD and SKCM, compared with that of the normal control tissues. Considering the key roles of these molecules in cGAS/STING signaling, the elevated expression of these molecules in most of the detected cancer types implies that this signaling is universally elevated in pan-cancer.
Figure 1.
The Expression of the cGAS/STING Pathway in Different Types of Cancer
TCGA RNA-seq data were first TPM normalized, and the differential expression was assessed by an unpaired t test to generate a p value. (A) The expression of MB21D1 in pan-cancer. (B) The expression of TMEM173 in pan-cancer. (C) The expression of TBK1 in pan-cancer. (D) The expression of IRF3 in pan-cancer. *p < 0.05; **p < 0.01; ***p < 0.001.
The Promoter Methylation Levels of MB21D1, TMEM173, TBK1, and IRF3 Are Variant in Different Types of Malignant Tumors
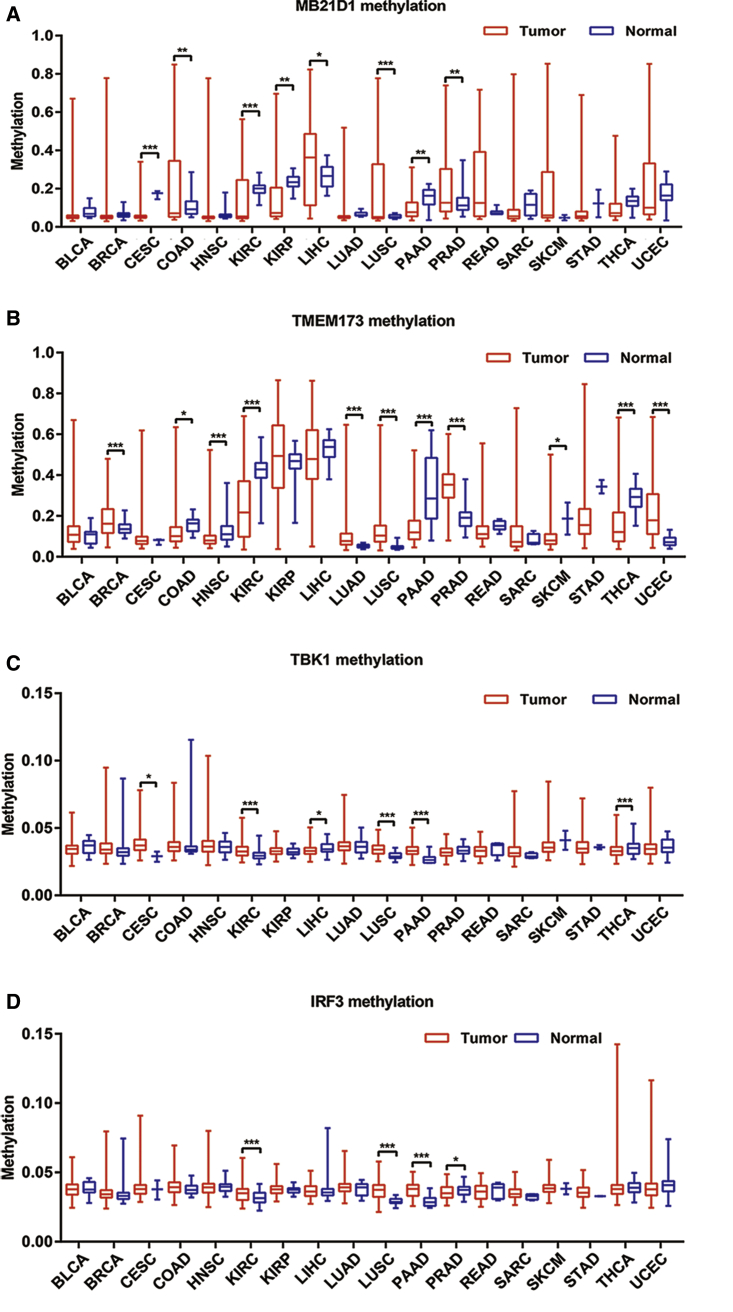
It has been reported that hypermethylation of the MB21D1 or TMEM173 gene promoter contributes to its downregulation in certain cancer types.14, 15 In this study, we revealed that cGAS/STING signaling is universally enhanced in pan-cancer due to the elevated expression of key molecules in this signaling pathway. To reveal the mechanism of the upregulation of these molecules, we investigated the methylation status of the promoters of the genes MB21D1, TMEM173, TBK1, and IRF3 by analyzing the MethHC database in pan-cancer. The results showed that the promoter of the MB21D1 gene is significantly hypomethylated in cervical squamous carcinoma (CESC), KIRC, KIRP, and PAAD, but that it is significantly hypermethylated in COAD, LIHC, LUSC, and PRAD (Figure 2A). For the TMEM173 gene (Figure 2B), it is significantly hypomethylated in COAD, HNSC, KIRC, PAAD, SKCM, and THCA, but hypermethylated in BRCA, LUAD, LUSC, PRAD, and UCEC. Moreover, the methylation level of the TBK1 gene promoter is deficient in LIHC and THCA and is elevated in CESC, KIRC, LUSC, and PAAD (Figure 2C). Similarly, the methylation level of the IRF3 gene promoter is deficient only in PRAD but is elevated in KIRC, LUSC, and PAAD (Figure 2D).
Figure 2.
Promoter Methylation Level of the cGAS/STING Pathway in Different Types of Cancer
The p value was assessed by an unpaired t test. (A) Methylation level of MB21D1 gene promoter. (B) Methylation level of TMEM173 gene promoter. (C) Methylation level of TBK1 gene promoter. (D) Methylation level of IRF3 gene promoter. *p < 0.05; **p < 0.01; ***p < 0.001.
Next, we analyzed the correlation between the methylation and expression of these four genes in different tumor types. As expected, the methylation levels of the detected genes are generally negatively correlated with their expression levels. The correlation between the methylation and expression of the MB21D1 gene is significantly high in COAD and LUSC but was low in CESC, KIRC, KIRP, LIHC, PAAD, and PRAD (Table 1). For TMEM173, it is prominent in COAD, KIRC, LUSC, PAAD, PRAD, THCA, and UCEC and is low only in HNSC and SKCM (Table 1). The methylation and expression of TBK1 are weakly correlated in all of the methylation-differentiated tumors, and there is no correlation in KIRC (Table 1). Similarly, IRF3 shows a low correlation in all of the methylation-differentiated tumors (Table 1).
Table 1.
An Analysis of Correlation between the Expression and Methylation of Four Genes within the cGAS/STING Signaling Pathway
| Cancer | Genes |
|||
|---|---|---|---|---|
| MB21D1 | TMEM173 | TBK1 | IRF3 | |
| BLCA | −0.503*** | −0.568*** | −0.184*** | −0.268*** |
| BRCA | −0.303*** | −0.536*** | −0.220*** | −0.131*** |
| CESC | −0.259*** | −0.498*** | −0.337*** | −0.149** |
| CRC | −0.629*** | −0.669*** | −0.106* | −0.184*** |
| HNSC | −0.371*** | −0.413*** | −0.101* | −0.211*** |
| KIRC | −0.373*** | −0.663*** | −0.092 | −0.249*** |
| KIRP | −0.309*** | −0.767*** | −0.297*** | −0.209*** |
| LIHC | −0.429*** | −0.625*** | −0.123* | −0.333*** |
| LUAD | −0.567*** | −0.604*** | −0.244*** | −0.200*** |
| LUSC | −0.568*** | −0.519*** | −0.268*** | −0.199*** |
| PAAD | −0.408*** | −0.529*** | −0.261*** | −0.245*** |
| PRAD | −0.409*** | −0.762*** | −0.245*** | −0.205*** |
| SARC | −0.338*** | −0.619*** | −0.158* | −0.246*** |
| SKCM | −0.461*** | −0.360*** | −0.097* | −0.116* |
| STAD | −0.302*** | −0.415*** | −0.148** | −0.168** |
| THCA | −0.415*** | −0.791*** | −0.186*** | −0.145** |
| UCEC | −0.606*** | −0.574*** | −0.022 | −0.296*** |
The correlation between methylation and expression was analyzed by a Pearson correlation coefficient (|r| > 0.5 denoted a significant correlation, and |r| < 0.5 denoted a low correlation). The COAD and READ tumor samples were combined as colorectal cancer (CRC) samples. *p < 0.05), **p < 0.01), ***p < 0.001.
A High Somatic Mutation Rate in the MB21D1, TMEM173, TBK1, and IRF3 Genes Is Present in Pan-Cancer
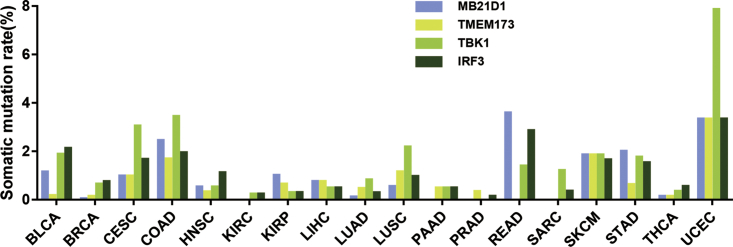
The impaired ability of STING to sense cGAMP caused by a SNP in the TMEM173 gene has been reported in a previous study,16 suggesting that gene mutations may also affect the activity of cGAS/STING signaling. Thus, we evaluated the mutation status of the MB21D1, TMEM173, TBK1, and IRF3 genes in pan-cancer. The result showed that various frequent mutations are present in the MB21D1, TMEM173, TBK1, and IRF3 genes in all the detected cancer types, and that the average mutation rate is comparable among the MB21D1, TMEM173, TBK1, and IRF3 genes. The top three highest mutation rates for the MB21D1 gene are present in COAD, READ, and UCEC, respectively (Figure 3), and the top three highest mutation rates for the TMEM173 gene are present in COAD, SKCM, and UCEC (Figure 3). Furthermore, the top three highest mutations rates for the TBK1 gene are present in CESC, COAD, and UCEC (Figure 3). For the IRF3 gene, the top three highest mutation rates are present in BLCA, READ, and UCEC (Figure 3). The highest mutation rates for the MB21D1, TMEM173, TBK1, and IRF3 genes are all present in UCEC, suggesting that cGAS/STING signaling in UCEC may be significantly impaired.
Figure 3.
Somatic Mutation Rate of the MB21D1, TMEM173, TBK1, and IRF3 Genes in Various Tumors
Although the mutation rates of these genes in the tumors are relatively high, because the mutated amino acids are not located in the functional regions of these genes, it appears that most of the mutations might not affect the ability of STING to bind cyclic diguanylate (CDN), the enzyme activity of cGAS, the kinase activity of TBK1, or the ability of IRF3 to act as a transcription factor.
Analyzing the Correlation between the Expression of the MB21D1, TMEM173, TBK1, or IRF3 Genes and Immune Infiltration in the Tumor Microenvironment in Pan-Cancer
Evidence from animal models has suggested that cGAS/STING signaling plays an important role in anticancer immunity. Thus, we evaluated the correlation between the expression of four key molecules in cGAS/STING signaling and immune cell infiltration within the tumor microenvironment, a key process of anticancer immunity in pan-cancer.
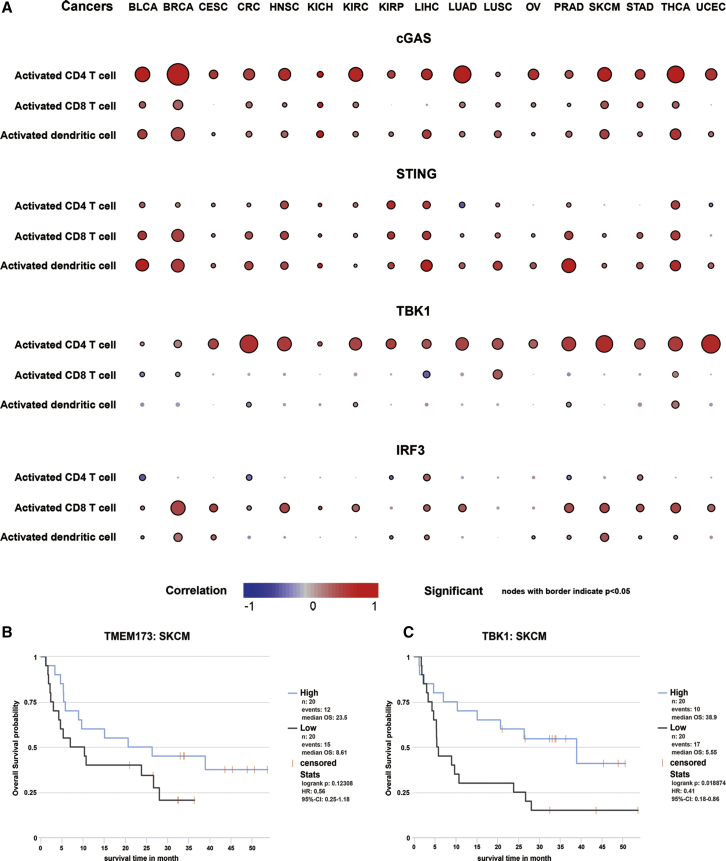
To investigate the impact of each gene on the infiltration of immune cells, we evaluated a total of 28 subpopulations of tumor-infiltrated leukocytes (TILs), including 16 adaptive immune cells and 12 innate immune cells. The correlation between the expression of MB21D1, TMEM173, TBK1, or IRF3 and these TILs is shown in Figures 4 and S1–S4, where the node size indicates the correlation significance and the color indicates the correlation degree. We found that the expression of MB21D1 mainly is associated with the infiltration of CD4 T cells, CD8 T cells, T helper cells, and natural killer T (NKT) cells in pan-cancer (Figure 4A; Figure S1), whereas TMEM173 is almost positively correlated with the infiltration of all immune cells in pan-cancer, especially in BLCA, BRCA, LIHC, PRAD, and THCA (Figure 4A; Figure S2). However, the effect of TBK1 and IRF3 on immune infiltration is mainly negative, and TBK1 expression is only positively correlated with the infiltration of CD4 T cells, including activated CD4 T cells, effector memory CD4 T cells, and type 2 T helper cells in pan-cancer (Figure 4A; Figure S3). By contrast, the expression of IRF3 is negatively correlated with the infiltration of effector memory CD4 T cells, memory B cell, type 2 T helper cell, and immature DC, but is positively correlated with the infiltration of activated CD8 T cells and CD56 dim natural killer cells in pan-cancer (Figure 4A; Figure S4). These results suggest that the role of cGAS/STING signaling in anticancer immunity is not consistent.
Figure 4.
The Correlation between the Expression of cGAS/STING Signaling and the Infiltration of Immune Cells
(A) The correlation between the expression of MB21D1, TMEM173, TBK1, IRF3, and the infiltration of activated CD4, CD8 T cell, and DC. Node color is determined by correlation, and node size indicates the significance of correlation. (B) The high expression of TMEM173 predicts a favorable prognosis of metastatic melanoma patients treated with monoclonal antibody against CTLA-4. (C) The high expression of TBK1 indicates a favorable prognosis of metastatic melanoma patients treated with monoclonal antibody against CTLA-4.
Because of that the T cell infiltration in the tumor microenvironment is closely associated with the efficiency of immune checkpoint blockade therapy; thus, we evaluated the impact of the expression of four key molecules in cGAS/STING signaling on the therapeutic effect of blocking immune checkpoint activity. By analyzing a transcriptome dataset from 40 patients with metastatic melanoma in a clinical trial to evaluate the benefit of CTLA-4 blockade, we revealed that patients with high expression of TMEM173 (Figure 4B) or TBK1 (Figure 4C) yield more considerable clinical benefit to anti-CTLA-4 therapy and had longer survival time than those patients with low expression of TMEM173 or TBK1, respectively, whereas the expression of MB21D1 or IRF3 has no impact on the clinical benefit for patients of CTLA-4 blockade (data not shown). This result is consistent with the observation that the expression of both TMEM173 and TBK1 is significantly correlated with the infiltration of CD4 T and CD8 T cells in melanoma (Figure 4A). Thus, we showed that the cGAS/STING signaling not only regulates immune cell infiltration, but also affects the benefit for cancer patients of immune checkpoint blockade and provides a theoretical basis for the combination of STING agonists and immunological checkpoint inhibitors.
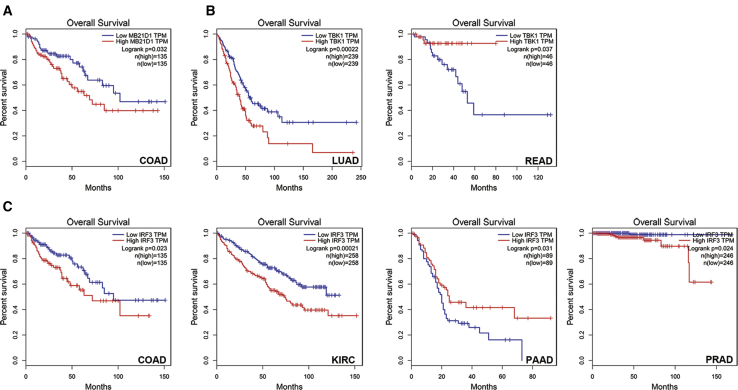
The Expression of the MB21D1, TMEM173, TBK1, and IRF3 Genes Predicts the Prognosis of Cancer Patients
By analyzing the correlation between key molecules of cGAS/STING signaling and the infiltration of individual types of immune cells, we showed that the expression of MB21D1 and TMEM173 is majorly correlated with the positive infiltration of adaptive and innate immune cells in pan-cancer, whereas the expression of TBK1 and IRF3 is negatively correlated with immune infiltration, which implies that MB21D1, TMEM173, TBK1, and IRF3 may play different roles in the prognosis of cancer patients. Thus, we evaluated the prediction value of the expression of the MB21D1, TMEM173, TBK1, and IRF3 genes in the prognosis of cancer patients. To our surprise, we found that, among the 18 cancer types, the expression levels of MB21D1 and TMEM173 fail to predict prognosis in any cancer type, with the exception that a high expression of MB21D1 predicts a poor prognosis in COAD patients (Figure 5A; Figures S5 and S6). A high expression of TBK1 in LUAD indicates a poor prognosis but predicts a good prognosis in READ patients (Figure 5B; Figure S7). By contrast, a high IRF3 expression predicts a poor prognosis in COAD, KIRC, and PRAD patients but indicates a good prognosis only in PAAD patients (Figure 5C; Figure S8).
Figure 5.
cGAS/STING Pathway Expression as a Prognostic Factor in Various Tumors
(A) MB21D1. (B) TBK1. (C) IRF3.
Discussion
Immunotherapy is an effective cancer therapy, and it was ranked number one among the 10 major scientific breakthroughs in 2013 by Science. Both adaptive immunity and innate immunity are targets of immunotherapy. The cGAS/STING signaling pathway has attracted massive interest in immunotherapy due to its positive effect on activating both the innate and the adaptive immune response. For instance, based on animal models, scientists suggest that the activation of this signaling pathway enhances the checkpoint blockade-based therapeutic effector and improves the effect of therapeutic cancer vaccination.17 However, current studies focusing on the role of this pathway in human malignant tumors are limited.
In this study, we investigated the expression of four key molecules in cGAS/STING signaling in pan-cancer. We found that the expression of the MB21D1, TMEM173, TBK1, and IRF3 genes is significantly upregulated in almost all of the detected cancer types, which implies that cGAS/STING signaling may be activated in pan-cancer. Specifically, the expression levels of these four key molecules are all upregulated in COAD, KIRC, and STAD, whereas the level at least one of these four molecules is increased in BLCA, BRCA, CESC, HNSC, KIPR, LIHC, READ, THCA, SKCM, and SARC. Notably, the expression levels of the MB21D1, TMEM173, and TBK1 genes are not altered, but the expression of IRF3 is downregulated in PAAD, suggesting that the immunity status in PAAD may be suppressed; this finding deserves further experiments for validation. For LUAD and LUSC, although most of these molecules are upregulated, the expression of TMEM173 is decreased, suggesting a different immune status for these cancer types.
Furthermore, we found that hypomethylation is more frequently observed in the GC island within these genes’ promoters. This observation is consistent with the result that most of the four key molecules are upregulated in pan-cancer. However, inconsistencies between the expression and the promoter methylation status of the genes are also found. For example, the expression of TBK1 and IRF3 is significantly upregulated, whereas the promoters of these genes are also hypermethylated in KIRC and LUSC tissues compared with that in normal tissues. In another case, the promoter of MB21D1 is hypermethylated, but it is highly expressed in LIHC and LUSC tissues compared with that in normal tissues. The mRNA level of the genes may also be regulated by other factors, such as microRNAs (miRNAs) and RNA binding proteins;18, 19 thus, the posttranscriptional regulation may contribute to revealing the aberrant expression of these genes in certain cancer tissues. In addition to the expression level, we also detected the mutation status of these four molecules in cGAS/STING signaling. Although we find that cGAS-STING signaling presents a higher mutation rate in a variety of tumors, most of these mutations are not in the functional region, suggesting that these mutations may not affect their functions, and thus mutations may not be the main reason for the aberrant regulation of cGAS/STING activity in pan-cancer.
According to our study, the cGAS/STING signaling pathway might be activated in most cancer types, which is unexpected because, on the one hand, it is widely accepted that this signaling is a tumor suppressor,20 and on the other hand, several previous studies reveal that this signaling pathway is suppressed in some tumor tissues.15 However, a very recent study reports that STING signaling may mediate the metastasis of cancer cells. Bakhoum et al.21 found that the genomic instability of the tumor cells results in the activation of STING signaling in response to cytosolic DNA, and the activation of STING signaling activates downstream noncanonical nuclear factor κB (NF-κB) signaling, which then promotes the metastasis of cancer cells. A very recent study reported that cGAS overexpression enhances the double-strand DNA damage and results in genome instability in a lung cancer model, and induces malignant transformation, stimulates the proliferation in vitro, and accelerates tumor growth in vivo of lung cancer cells.22 As a hallmark of cancer, genomic instability drives tumor evolution by activating STING signaling and promoting cancer progression, which may explain why this signaling is universally elevated in pan-cancer. Additionally, it has been reported that the activation of cGAS/STING signaling stimulates the expression of PD-L1 in cancer cells, which mediates the immune evasion of cancer cells.23 These studies suggest that cGAS/STING signaling activation can promote cancer progression.
By analyzing the relationship between the expression of key molecules in cGAS/STING signaling and the signature of different immune cells, we revealed that only the expression level of TMEM173 is positively correlated with the infiltration of most immune cells, whereas the expression levels of MB21D1, TBK1, or IRF3 are only positively correlated with the infiltration of certain immune cell types but were negatively correlated with the infiltration of other immune cell types in pan-cancer. In the past few years, various STING agonists have been developed to improve anticancer immunity. For example, directly injecting synthetic CDN into mice tumors activates STING and the innate immune system, which then triggers a series of cascade reactions and activates T cells against tumors.24 However, a recent study found that STING agonists induce cell death in T cells by activating cell stress.25 Similarly, in another study, the proliferation of T lymphocytes is impaired upon constitutive STING activation; this process is dependent on NF-κB and results from STING relocalization to the Golgi apparatus after activation.26 These new findings suggest that cGAS/STING signaling may impair the adaptive immune system, and our results also show that a high expression of cGAS/STING signaling components is negatively correlated with the infiltration of certain immune cells. Thus, the relationship between the activity of cGAS/STING signaling and the immune infiltration is more complicated than what we currently know.
Conclusions
Collectively, we have for the first time revealed that cGAS/STING signaling is highly expressed in pan-cancer tissues. We also show that highly upregulated cGAS/STING signaling is negatively correlated with the infiltration of immune cells in some tumor types, and consistent with these findings, we showed that a high level of cGAS/STING signaling predicts a poor prognosis in patients with some cancers. This study suggests that it is necessary to deeply and fully evaluate the functions of cGAS/STING signaling in cancer immunity and cancer progression before the application of STING agonist-based anticancer immune therapy in the clinic.
Materials and Methods
Data Obtaining from TCGA, MethHC, and cBioPortal Databases
We obtained 18 kinds of tumor mRNA RNA-Seq-HTSeq-fragments per kilobase of exon model per million mapped reads (FPKM) data from TCGA database27 to further analyze the expression of cGAS/STING signaling; then we used trans per million (TPM) to standardize these data for a better comparison. An unpaired t test was applied to determine the difference between the gene expression in the tumor and normal tissue. The methylation levels were obtained from MethHC (http://methhc.mbc.nctu.edu.tw/php/index.php),28 an online database of DNA methylation and gene expression in human cancer, which comprises 6,548 DNA methylation data generated by the Illumina Human Methylation 450K BeadChip and 12,567 mRNA and miRNA expression data generated by RNA-sequencing (RNA-seq)/miRNA-seq in 18 human cancers. We downloaded the promoter methylation data of MB21D1, TMEM173, TBK1, and IRF3 in 18 kinds of tumors. In addition, the p value was calculated by an unpaired t test. To determine the correlation between methylation and expression, we screened tumors with statistically significant different methylation levels and analyzed the correlation between methylation and the expression of target genes in different cancer types using an online database cBioPortal. The cBioPortal (http://www.cbioportal.org) for Cancer Genomics provides visualization, analysis, and downloading of large-scale cancer genomics datasets.29, 30 We used the plot function of this website, choosing the profile mRNA expression (RNA Seq V2 RSEM) and Illumina human methylation 450 (HM450) to calculate the Pearson coefficient to achieve a correlation analysis. In addition, |r| > 0.5 was a significant correlation, and |r| < 0.5 was a low correlation. Additionally, the somatic mutation data from various tumors were also downloaded from TCGA. The present study was approved by the Ethics Committee of Harbin Medical University (Heilongjiang, China).
GEPIA Survival Analysis
The prognostic value of the expression of MB21D1, TMEM173, TBK1, and IRF3 mRNA was evaluated using the online database GEPIA. GEPIA (Gene Expression Profiling Interactive Analysis)31 is a web-based tool that delivers fast and customizable functionalities based on TCGA and genotype-tissue expression (GTEx) data, which contains the gene expression data and survival information of a number of cancer types. To analyze the overall survival (OS) of the patients, we set the median expression as the expression threshold to split the patient samples into high-expression and low-expression groups, and used a Kaplan-Meier survival plot with the hazard ratio (HR), a 95% confidence interval (CI), and a log rank test p value.
Analyzing the Correlation between the Infiltration of Immune Cells and the Expression of Key Molecules in cGAS/STING Signaling
The RNA-seq data of TCGA tumors across 24 cancer types were downloaded from the GEO database (GEO: GSE62944). All of the raw data were reprocessed using the R package “Rsubread” by aligning the fastq files downloaded from the Cancer Genomics Hub32 so that the expression value of the genes could be compared between the different samples. A list of 782 immune marker genes, corresponding to 28 tumor-infiltrating immune subpopulations, was from a previous study.33 The infiltration level of each immune subpopulation was estimated by the single sample gene set enrichment analysis (ssGSEA) method using the R package “GSVA,” as previously described.33 The COAD and READ tumor samples were combined as colorectal cancer (CRC) samples. To determine the association between gene expression and immune cells, the Pearson correlation coefficient was computed between the gene expression and the enrichment score of each immune subpopulation. The prognostic value of the genes was evaluated using a Cox proportional hazards regression in each cancer type, and the overall HR was estimated by a fixed-effects meta-analysis as a pan-cancer prognostic value.
Analyzing the Correlation between the cGAS/STING Pathway and the Clinical Outcome of CTLA-4 Blockade
In order to obtain the relationship between the cGAS/STING pathway and the clinical response of immune checkpoint blockade therapy, we analyzed the correlation between the expression of four key molecules in cGAS/STING signaling and the clinical benefit for patients with metastatic melanoma to ipilimumab, a monoclinic antibody against CTLA-4.34 We obtained 40 tumor samples with pretreatment transcriptome data and then divided these patients into high-expression and low-expression groups according to the median expression of these genes, respectively, and assessed the OS of patients by using a Kaplan-Meier survival plot with the HR, 95% CI, and log rank test p value.
Author Contributions
X.L. and D.H. conceived and designed the experiments. X.A., Y.Z., T.Z., G.W., M.Z., J.L., H.J., S.L., S.Y., D.X., Z.L., T.W., Y.H., L.Z., W.Y., and R.Z. performed the experiments and analyzed the data. X.A. and X.L. wrote the paper. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant 31701153). The data used and analyzed during the current study are available from the corresponding author on reasonable request.
Footnotes
Supplemental Information includes eight figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.11.003.
Contributor Information
Dapeng Hao, Email: dapeng.hao@gmail.com.
Xiaobo Li, Email: lixiaobo@ems.hrbmu.edu.cn.
Supplemental Information
References
- 1.Prehn R.T. Tumor specific immunity to nonviral tumors. Proc. Can. Cancer Conf. 1963;5:387–395. [PubMed] [Google Scholar]
- 2.Bose D. cGAS/STING pathway in cancer: Jekyll and Hyde story of cancer immune response. Int. J. Mol. Sci. 2017;18:E2456. doi: 10.3390/ijms18112456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rivera Vargas T., Benoit-Lizon I., Apetoh L. Rationale for stimulator of interferon genes-targeted cancer immunotherapy. Eur. J. Cancer. 2017;75:86–97. doi: 10.1016/j.ejca.2016.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ran Y., Shu H.B., Wang Y.Y. MITA/STING: a central and multifaceted mediator in innate immune response. Cytokine Growth Factor Rev. 2014;25:631–639. doi: 10.1016/j.cytogfr.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo S.R., Corrales L., Gajewski T.F. The STING pathway and the T cell-inflamed tumor microenvironment. Trends Immunol. 2015;36:250–256. doi: 10.1016/j.it.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woo S.R., Fuertes M.B., Corrales L., Spranger S., Furdyna M.J., Leung M.Y., Duggan R., Wang Y., Barber G.N., Fitzgerald K.A. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demaria O., De Gassart A., Coso S., Gestermann N., Di Domizio J., Flatz L., Gaide O., Michielin O., Hwu P., Petrova T.V. STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc. Natl. Acad. Sci. USA. 2015;112:15408–15413. doi: 10.1073/pnas.1512832112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohkuri T., Ghosh A., Kosaka A., Zhu J., Ikeura M., David M., Watkins S.C., Sarkar S.N., Okada H. STING contributes to antiglioma immunity via triggering type I IFN signals in the tumor microenvironment. Cancer Immunol. Res. 2014;2:1199–1208. doi: 10.1158/2326-6066.CIR-14-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu J., Kanne D.B., Leong M., Glickman L.H., McWhirter S.M., Lemmens E., Mechette K., Leong J.J., Lauer P., Liu W. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci. Transl. Med. 2015;7:283ra52. doi: 10.1126/scitranslmed.aaa4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song S., Peng P., Tang Z., Zhao J., Wu W., Li H., Shao M., Li L., Yang C., Duan F. Decreased expression of STING predicts poor prognosis in patients with gastric cancer. Sci. Rep. 2017;7:39858. doi: 10.1038/srep39858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu S., Concha-Benavente F., Shayan G., Srivastava R.M., Gibson S.P., Wang L., Gooding W.E., Ferris R.L. STING activation enhances cetuximab-mediated NK cell activation and DC maturation and correlates with HPV+ status in head and neck cancer. Oral Oncol. 2018;78:186–193. doi: 10.1016/j.oraloncology.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia T., Konno H., Barber G.N. Recurrent loss of STING signaling in melanoma correlates with susceptibility to viral oncolysis. Cancer Res. 2016;76:6747–6759. doi: 10.1158/0008-5472.CAN-16-1404. [DOI] [PubMed] [Google Scholar]
- 15.Xia T., Konno H., Ahn J., Barber G.N. Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Rep. 2016;14:282–297. doi: 10.1016/j.celrep.2015.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shu C., Yi G., Watts T., Kao C.C., Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat. Struct. Mol. Biol. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y.J. STINGing the Tumor’s immune evasion mechanism. OncoImmunology. 2018;7:e1083673. doi: 10.1080/2162402X.2015.1083673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang T., Wang G., Hao D., Liu X., Wang D., Ning N., Li X. Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer. Mol. Cancer. 2015;14:125. doi: 10.1186/s12943-015-0402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsuchiya Y., Nakajima M., Takagi S., Taniya T., Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66:9090–9098. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- 20.Bhatelia K., Singh A., Tomar D., Singh K., Sripada L., Chagtoo M., Prajapati P., Singh R., Godbole M.M., Singh R. Antiviral signaling protein MITA acts as a tumor suppressor in breast cancer by regulating NF-κB induced cell death. Biochim. Biophys. Acta. 2014;1842:144–153. doi: 10.1016/j.bbadis.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Bakhoum S.F., Ngo B., Laughney A.M., Cavallo J.A., Murphy C.J., Ly P., Shah P., Sriram R.K., Watkins T.B.K., Taunk N.K. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018;553:467–472. doi: 10.1038/nature25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H., Zhang H., Wu X., Ma D., Wu J., Wang L., Jiang Y., Fei Y., Zhu C., Tan R. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature. 2018;563:131–136. doi: 10.1038/s41586-018-0629-6. [DOI] [PubMed] [Google Scholar]
- 23.He L., Xiao X., Yang X., Zhang Z., Wu L., Liu Z. STING signaling in tumorigenesis and cancer therapy: A friend or foe? Cancer Lett. 2017;402:203–212. doi: 10.1016/j.canlet.2017.05.026. [DOI] [PubMed] [Google Scholar]
- 24.Corrales L., Glickman L.H., McWhirter S.M., Kanne D.B., Sivick K.E., Katibah G.E., Woo S.R., Lemmens E., Banda T., Leong J.J. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 2015;11:1018–1030. doi: 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larkin B., Ilyukha V., Sorokin M., Buzdin A., Vannier E., Poltorak A. Cutting edge: activation of STING in T cells induces type I IFN responses and cell death. J. Immunol. 2017;199:397–402. doi: 10.4049/jimmunol.1601999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerboni S., Jeremiah N., Gentili M., Gehrmann U., Conrad C., Stolzenberg M.C., Picard C., Neven B., Fischer A., Amigorena S. Intrinsic antiproliferative activity of the innate sensor STING in T lymphocytes. J. Exp. Med. 2017;214:1769–1785. doi: 10.1084/jem.20161674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M., Cancer Genome Atlas Research Network The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang W.Y., Hsu S.D., Huang H.Y., Sun Y.M., Chou C.H., Weng S.L., Huang H.D. MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 2015;43:D856–D861. doi: 10.1093/nar/gku1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahman M., Jackson L.K., Johnson W.E., Li D.Y., Bild A.H., Piccolo S.R. Alternative preprocessing of RNA-sequencing data in The Cancer Genome Atlas leads to improved analysis results. Bioinformatics. 2015;31:3666–3672. doi: 10.1093/bioinformatics/btv377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., Hackl H., Trajanoski Z. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 34.Van Allen E.M., Miao D., Schilling B., Shukla S.A., Blank C., Zimmer L., Sucker A., Hillen U., Foppen M.H.G., Goldinger S.M. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.