LUC7 proteins, which are subunits of the U1 snRNP, are important for plant development and stress resistance and control alternative splicing of hundreds of introns.
Abstract
Introns are removed by the spliceosome, a large macromolecular complex composed of five ribonucleoprotein subcomplexes (U snRNPs). The U1 snRNP, which binds to 5ʹ splice sites, plays an essential role in early steps of the splicing reaction. Here, we show that Arabidopsis thaliana LETHAL UNLESS CBC7 (LUC7) proteins, which are encoded by a three-member gene family in Arabidopsis, are important for plant development and stress resistance. We show that LUC7 is a U1 snRNP accessory protein by RNA immunoprecipitation experiments and LUC7 protein complex purifications. Transcriptome analyses revealed that LUC7 proteins are not only important for constitutive splicing, but also affect hundreds of alternative splicing events. Interestingly, LUC7 proteins specifically promote splicing of a subset of terminal introns. Splicing of LUC7-dependent introns is a prerequisite for nuclear export, and some splicing events are modulated by stress in a LUC7-dependent manner. Taken together, our results highlight the importance of the U1 snRNP component LUC7 in splicing regulation and suggest a previously unrecognized role of a U1 snRNP accessory factor in terminal intron splicing.
INTRODUCTION
Eukaryotic genes are often interrupted by noncoding sequences called introns that are removed from pre-mRNAs, while the remaining sequence, the exons, are joined together. This process, known as splicing, is an essential step before the translation of the mature mRNAs, and it offers a wide range of advantages for eukaryotic organisms. For instance, alternative splicing allows the production of more than one isoform from a single gene, expanding genome coding capacity (Kornblihtt et al., 2013; Reddy et al., 2013). Alternative splicing can also regulate gene expression by generating transcripts with premature termination codons and/or a long 3ʹ untranslated region, which may lead to RNA degradation via the nonsense-mediated decay (NMD) pathway (Kalyna et al., 2012; Drechsel et al., 2013; Shaul, 2015). Furthermore, splicing is usually coupled with other RNA processing events, such as 3ʹ end formation and RNA export to the cytosol (Kaida, 2016; Müller-McNicoll et al., 2016). In plants, alternative splicing contributes to essentially all aspects of development and stress responses (Carvalho et al., 2013; Staiger and Brown, 2013).
Intron removal is catalyzed by a large macromolecular complex, the spliceosome. The major U2 spliceosome is responsible for the removal of most introns, and a small fraction of introns is removed by the U12 minor spliceosome. The U2 spliceosome consists of five small ribonucleoprotein particles (U snRNPs): the U1, U2, U4, U5, and U6 snRNPs. Each U snRNP contains a heteroheptameric ring of Sm or Lsm proteins, snRNP-specific proteins, and a uridine-rich snRNA. Additional noncore spliceosomal proteins participate during the splicing reaction and may affect exon-intron recognition and splicing efficiency. The canonical splicing cycle starts with binding of the U1 snRNP to the 5ʹ splice site (5ʹss), followed by association of auxiliary proteins such as U2AF to the pre-mRNA, which facilitate the recognition of the 3ʹ splice site (3ʹss) and the branch point by other factors. The thereby formed complex E recruits the U2 snRNP to generate complex A. In the next step, a trimeric complex consisting of U4/U5/U6 snRNPs joins to form complex B. Several rearrangements and ejection of the U1 and U4 snRNP are necessary to generate a catalytically active splicing complex (Wahl et al., 2009; Will and Lührmann, 2011).
The fact that U1 snRNP is recruited to the 5ʹss in the initial step of splicing suggests that this complex is necessary for the correct 5ʹss selection. It has been shown that U1-deficient zebra fish mutants accumulate alternative spliced transcripts, indicating that the U1 snRNP indeed fulfills in some cases regulatory roles in splice site selection, though the U1 snRNP-mRNA interaction alone is not sufficient for productive splicing (Rösel et al., 2011). Although the spliceosome consists of stoichiometrically equal amounts of each subunit, the U1 snRNP is more abundant than all the other spliceosomal subcomplexes (Kaida et al., 2010; Kaida, 2016). One reason for this is that the U1 snRNP executes splicing-independent functions. The metazoan U1 snRNP, for instance, binds not only to the 5ʹss, but also throughout the nascent transcript, blocking a premature cleavage and polyadenylation (Kaida et al., 2010; Berg et al., 2012). Furthermore, the U1 snRNP is important in regulating promoter directionality and transcription in animals (Almada et al., 2013; Guiro and O’Reilly, 2015).
U1 snRNP complexes have been purified and characterized in yeast and human. The core U1 snRNP contains the U1 snRNA, Sm proteins, and three U1 core proteins (U1-70K, U1-A, and U1-C) (Gottschalk et al., 1998). In yeast, additional accessory proteins also belong to the U1 snRNP (LUC7p, Prp39p, Prp40p, Nam8p, Snu71p/RBM25, Prp42p, and Snu56p) (Neubauer et al., 1997; Gottschalk et al., 1998; Fortes et al., 1999b; Will and Lührmann, 2011). The majority of these accessory proteins have no counterpart in humans (Koncz et al., 2012; Kondo et al., 2015). In plants, most of the yeast U1 snRNP proteins are conserved, and some are encoded by small gene families, suggesting a similar or even more complex U1 snRNP composition than yeast (Wang and Brendel, 2004; Koncz et al., 2012; Reddy et al., 2013). In Arabidopsis thaliana, at least 10 accessory proteins are potentially part of the U1 snRNP (LUC7A, LUC7B, LUC7RL, PRP39A, PRP39B, PRP40A, PRP40B, PRP40C, and RBM25) (Wang and Brendel, 2004; Koncz et al., 2012; Reddy et al., 2013). Interaction studies also revealed that the U1 snRNP associates with serine-arginine (SR) proteins, indicating a complex mechanism for splice site selection that also involves non-snRNP proteins (Golovkin and Reddy, 1998; Cho et al., 2011).
The function of the plant U1 snRNP is not well characterized. This might be due to the fact that in Arabidopsis the core U1 snRNP components U1-70K and U1-C are single-copy genes and a complete knockout would likely cause severe mutant phenotypes or be lethal. Knockout of U1-A surprisingly produced viable mutants, suggesting that U1-A is not essential for Arabidopsis survival (Gu et al., 2018). PRP39, PRP40, and LETHAL UNLESS CBC7 (LUC7) are encoded by small gene families, which will require the generation of multiple mutants for functional studies. Some U1-specific Arabidopsis mutants have been characterized: Mutations in the accessory factor PRP39A cause delayed flowering, probably due to increased expression of the flowering time regulator FLOWERING LOCUS C, but the mutants do not exhibit severe development defects (Wang et al., 2007; Kanno et al., 2017). Nonetheless, prp39a mutants accumulate misspliced mRNAs containing mainly retained introns or skipped exons (Kanno et al., 2017). In a reverse-genetic approach, U1-70K expression was specifically reduced in flowers by an antisense RNA, and the resulting transgenic plants exhibit strong floral defects (Golovkin and Reddy, 2003). Moreover, a mutation in U1-A causes an altered salt stress response (Gu et al., 2018). Thus, despite evidence that U1 snRNP is essential for plant development and stress response, the functions of the U1 snRNP in regulating the transcriptome of plants are largely unknown. Other characterized factors in plants and metazoans, such as GEMIN2 or SRD2, are required for the functionality of all snRNPs and not specifically for U1 function (Ohtani and Sugiyama, 2005; Schlaen et al., 2015).
Here, we report on the functional characterization of Arabidopsis mutants impaired in U1 snRNP function. For this, we focused on the U1 snRNP component LUC7, which we show to be essential for normal plant development and stress resistance. Our whole-transcriptome analyses on the luc7 triple mutant show that impairments of LUC7 proteins affect constitutive and alternative splicing. Surprisingly, our results reveal the existence of transcripts in which terminal introns are preferentially retained in a LUC7-dependent manner. Unspliced LUC7-dependent introns cause a nuclear retention of the pre-mRNAs, and the splicing efficiency of LUC7-dependent introns can be modulated by stress. Our results suggest that the plant U1 snRNP component LUC7 regulates alternative splicing of pre-mRNAs and thereby has an influence on their nuclear export, which could be a mechanism to fine-tune gene expression under stress conditions.
RESULTS
LUC7 Proteins, a Family of Conserved Nuclear Zinc-Finger/Arginine-Serine Proteins, Redundantly Control Plant Development
LUC7 was identified in a screen for synthetic lethality in a yeast strain lacking the nuclear cap binding complex (CBC), which is involved in several RNA processing events (Fortes et al., 1999a; Rigaut et al., 1999; Gonatopoulos-Pournatzis and Cowling, 2014; Sullivan and Howard, 2016). LUC7 proteins carry a C3H- and a C2H2-type zinc-finger, which are located in the conserved LUC7 domain. In multicellular eukaryotes, LUC7 proteins usually also contain an additional C-terminal arginine/serine-rich (RS) domain, which is known to mediate protein-protein interactions (Puig et al., 2007; Webby et al., 2009; Heim et al., 2014). Arabidopsis contains three LUC7 genes (AthLUC7A, AthLUC7B, and AthLUC7RL), which are separated into two clades: LUC7A/B and LUC7RL (Figure 1A; Supplemental Figure 1). AthLUC7RL is more similar to its yeast homolog and lacks a conserved stretch of 80 amino acids of unknown function present in AthLUC7A and AthLUC7B (Supplemental Figure 1). A phylogenetic analysis revealed that algae contain a single LUC7 gene belonging to the LUC7RL clade, reinforcing the idea that LUC7RL proteins are closer to the ancestral LUC7 than LUC7A/B. In the moss Physcomitrella patens and in the fern Selaginella moellendorffii, one can find proteins belonging to both clades, suggesting that the split into LUC7RL and LUC7A/B occurred early during the evolution of land plants.
Figure 1.
Arabidopsis LUC7 Proteins Redundantly Control Plant Development.
(A) Phylogenetic analysis of LUC7 proteins in the plant kingdom using Saccharomyces cerevisiae as an external group. Sequences were aligned by Muscle and Maximum likelihood (PhYML) with 1000 bootstraps.
(B) Exon/intron structure of Arabidopsis LUC7A, LUC7B, and LUC7RL. Dotted lines indicate the positions of T-DNA insertions.
(C) Wild-type and luc7a-2 luc7b-1 luc7rl-1 (luc7a-2,b-1,rl-1) mutant plants growing for 21 d in long-day conditions (left) and for 45 d in standard greenhouse conditions (right).
(D) Length of the longest rosette leaf of 21-d-old wild-type, luc7 single, double, and triple mutant plants growing under long-day conditions. Leaves of 10 to 15 individual plants were measured. Dots indicate individual data points.
(E) Complementation of luc7a-2 luc7b-1 luc7rl-1 mutant by LUC7A, LUC7B, and LUC7RL genomic rescue constructs. Transformation of an “empty” binary vector served as a control. Two independent transgenic lines for each construct are shown.
To understand the function of the Arabidopsis U1 snRNP, we analyzed T-DNA insertion lines affecting LUC7 genes (Figure 1B). Single and double luc7 mutants were indistinguishable from wild-type plants (Supplemental Figure 2). However, the luc7 triple mutant exhibited a wide range of developmental defects, including dwarfism and reduced apical dominance (Figures 1C and 1D). To test whether the impairment of LUC7 functions was indeed responsible for the observed phenotypes, we reintroduced a wild-type copy of LUC7A, LUC7B, or LUC7RL in the luc7 triple mutant. Each of the LUC7 genes was sufficient to restore the growth phenotype of the luc7 triple mutant (Figure 1E), revealing that the phenotype observed in the luc7 triple mutant is due to loss of LUC7 function. These results further suggest that LUC7 genes act redundantly to control Arabidopsis growth and development.
LUC7 Is Important for ABA Signaling as Well as Cold and Salt Stress Responses
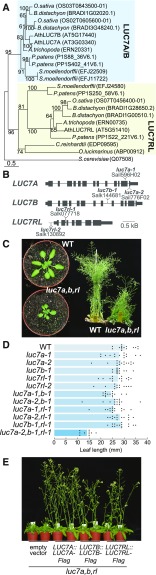
Splicing is essential for plant stress resistance, and mutants impaired in splicing are often hypersensitive to stress and to the stress hormone abscisic acid (ABA) (Filichkin et al., 2015; Zhan et al., 2015). In addition, global impairment of the splicing machinery elicits ABA signaling (AlShareef et al., 2017; Ling et al., 2017). To test whether LUC7 is important for plant stress resistance and ABA-mediated stress signaling, we analyzed growth parameters of the wild type, the luc7 triple mutant, and a luc7 rescue line in the presence of exogenous ABA or salt. A cotyledon greening assay showed that luc7 triple mutants exhibited hypersensitivity to exogenous ABA (Figures 2A and 2B), suggesting that LUC7 plays an important role in the ABA pathway. Furthermore, salt in the growth medium impaired root growth much more strongly in luc7 triple mutants than in the wild type or in a luc7 rescue line (Figures 2C and 2D). Similarly, cold temperatures strongly compromised the growth of luc7 triple mutant when compared with the wild type (Figure 2E). These results imply that functional LUC7 proteins are required for plant stress resistance and ABA responses.
Figure 2.
Arabidopsis LUC7 Is Involved in ABA Signaling and Abiotic Stress Responses.
(A) and (B) Wild type, luc7 triple mutant, and a luc7 complementation line (pLUC7A:LUC7A-YFP luc7a luc7b luc7rl) were grown on half-strength MS plates containing 1% sucrose and indicated amount of ABA. Seedling phenotypes (A) and quantification of seedlings with green cotyledons (B) are shown. Green cotyledons were scored 10 d after germination. One of two biological replicates is shown.
(C) and (D) Wild type, luc7 triple mutant, and a luc7 complementation line (pLUC7A:LUC7A-YFP luc7a luc7b luc7rl) germinating on half-strength MS vertical plates for 4 d were transferred on half-strength MS plates containing the indicated amount of NaCl. Phenotypes (C) and root growth quantification scored over 7 d (D) are shown. ANOVA was performed followed by Tukey post-hoc test for multiple comparison (Supplemental File 1). ****P < 0.0001; n.s., not significant.
(E) Gross phenotype of the wild type, luc7 triple mutant, and a luc7 complementation line grown at 21°C for 5 d and then, for cold stress, transferred to 8°C for 2 weeks.
LUC7 Is a U1 snRNP Component in Plants
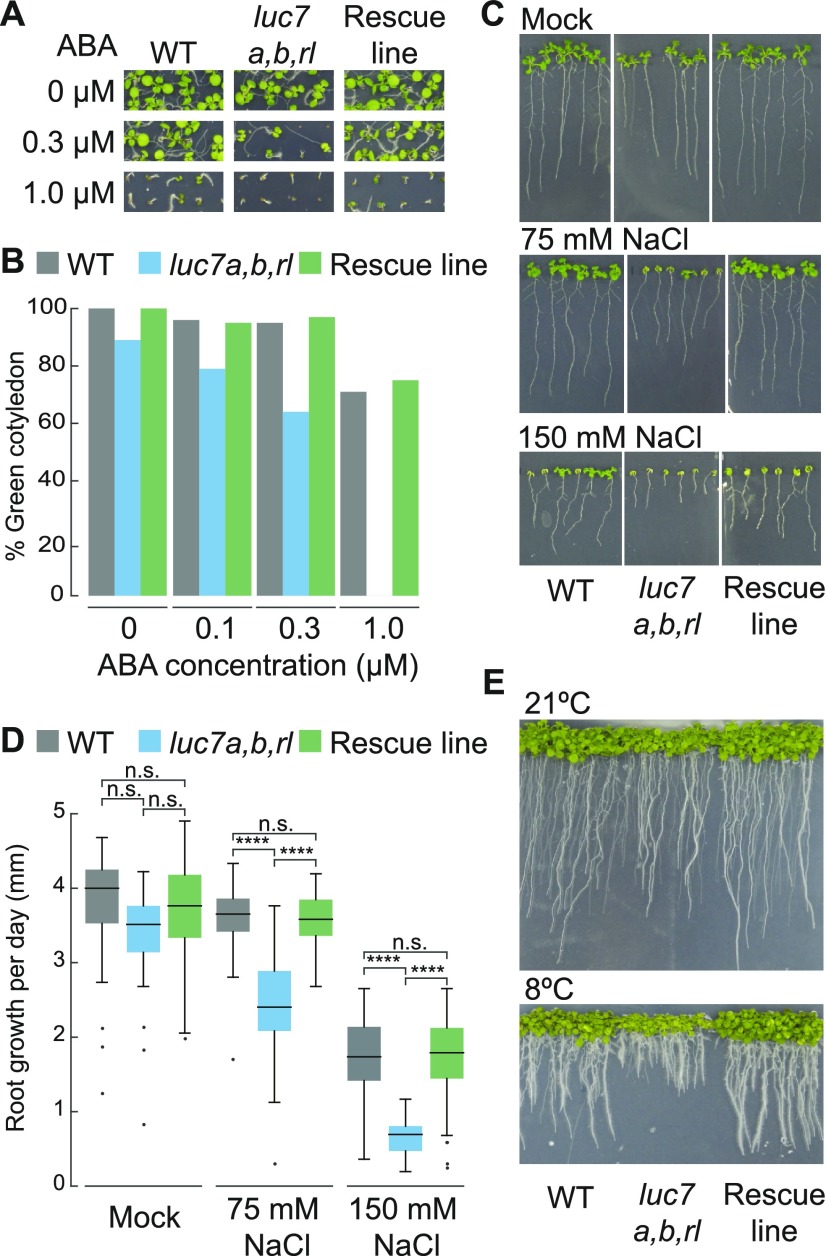
The composition of the U1 snRNP subcomplex is known in yeast and metazoans but not in plants (Will and Lührmann, 2011; Koncz et al., 2012). Therefore, we asked whether LUC7 is also a U1 component in Arabidopsis. Due to the fact that our genetic analyses of luc7 mutants suggested that LUC7 proteins act largely redundantly, we focused further analyses mainly on a single LUC7 protein, LUC7A. A protein that is part of the U1 complex is tightly associated with U1-specific components, such as the U1 snRNA. To test whether LUC7 is found in a complex with the U1 snRNA, we performed RNA immunoprecipitation experiments using a luc7 triple mutant carrying pLUC7A:LUC7A-YFP rescue construct (Supplemental Figure 3). Immunoprecipitation of LUC7A-YFP enriched the U1 snRNA more than 40-fold but did not enrich two unrelated, yet abundant RNAs, U3 snoRNA and ACTIN mRNA (Figure 3A). We also found small amounts of U2 snRNA to associate with LUC7A, which is in agreement with the fact that U1 and U2 snRNP directly interact to form spliceosomal complex A (Figure 3A). However, the amount of recovered U2 snRNA was more than 4-fold lower than that of the U1 snRNA (Figure 3A). These results strongly suggest that Arabidopsis LUC7 proteins are bona fide U1 snRNP components.
Figure 3.
Arabidopsis LUC7 Is an U1 snRNP Component.
(A) RNA immunoprecipitation using a pLUC7A:LUC7A-YFP luc7a luc7b luc7rl complementation line. Proteins were immunoprecipitated using GFP-affinity matrix and RNAs were extracted from the input and the immunoprecipitated fraction. U1, U2, U3 snRNAs, and ACTIN RNA were quantified using RT-qPCR. Enrichment of the respective RNA in pLUC7A:LUC7A-YFP luc7a luc7b luc7rl transgenic line was calculated compared with the wild type (negative control). Error bars denote the range of two biological replicates.
(B) Quantification of U1 and U2 snRNA levels in the wild type and luc7a luc7b luc7rl mutants. Total RNAs were isolated from 7-d-old seedlings, and RNA levels were analyzed by RT-qPCR using U1- or U2-specific oligonucleotides. Error bars denote the range of two biological replicates.
(C) Subcellular localization of LUC7A in pLUC7A:LUC7A-YFP luc7a luc7b luc7rl in Arabidopsis transgenic plants. Roots of 9-d-old seedlings were analyzed using confocal microscopy. Bar = 25 µm.
(D) U1-70K-mRFP and LUC7A-YFP or LUC7RL-YFP proteins were transiently expressed in N. benthamiana and their subcellular localization analyzed using confocal microscopy. Bars = 10 μm and 25 µm for upper and lower panels, respectively.
We then asked whether the integrity of the U1 snRNP is impaired in the luc7 triple mutant. Reduced U1 snRNP integrity would cause a change in the U1 snRNA levels since free U1 snRNA is unstable. We found that U1 snRNA levels did not change in the luc7 triple mutant (Figure 3B). This result suggests that LUC7 proteins are accessory U1 snRNP proteins that do not compromise the stability of the U1 snRNP core complex.
Next, we analyzed the subcellular localization of LUC7A and its colocalization with a core U1 snRNP subunit. LUC7A localized to the nucleus but not to the nucleolus in Arabidopsis plants containing the pLUC7A:LUC7A-YFP rescue construct (Figure 3C). In addition, LUC7A partially colocalized with U1-70K in the nucleoplasm when transiently expressed in Nicotiana benthamiana (Figure 3C). Similar results were obtained for LUC7RL, the Arabidopsis LUC7 most distant in sequence to LUC7A (Figure 3D). In plants, colocalization studies in protoplasts have shown that the core U1 components only partially colocalize as well (Lorković and Barta, 2008). These partial colocalizations suggest that plant U1 snRNP proteins may fulfill additional functions that could be similar to those observed in other eukaryotes (Workman et al., 2014).
To further test whether LUC7A associates in planta with known U1 snRNP components, we purified LUC7A-containing complexes. For this, we used pLUC7A:LUC7A-YFP-complemented lines and, as controls, wild-type plants and transgenic lines expressing free GFP (p35S:GFP). Immunoprecipitation followed by mass spectrometry analyses (IP-MS) were performed three to four times independently. We observed that the wild type often produced more background in MS analyses than the 35S:GFP line (Supplemental Data Set 1). Therefore, we used the wild type as a more stringent negative control (Table 1). Among all identified proteins, we considered LUC7 interactors those that were found in at least two independent experiments and that were at least three times more abundant in pLUC7A:LUC7A-YFP IPs than in wild type IPs. The IP-MS experiment does not provide information about whether these interactions are direct or bridged by other proteins, but it suggests that the proteins are found in common complexes.
Table 1. Selected Potential LUC7A Interacting Proteins Identified in Immunoprecipitation Experiments Followed by MS Analysis.
| Identified Protein | Aver. No. of Peptides IP LUC7 | Aver. No. of Peptides IP Wild Type | Seq. Coverage (%) IP LUC7 | Seq. Coverage (%) IP Wild Type | MW (kD) |
|---|---|---|---|---|---|
| U1 snRNP components | |||||
| LUC7A | 45.8 | 0 | 71.9 | 0 | 47.4 |
| U1-70K | 5.0 | 0 | 13.3 | 0 | 50.4 |
| SmB-a; SmB-b | 2.5 | 0 | 9.8 | 0 | 27.0 |
| U1A | 2.0 | 0 | 7.7 | 0 | 28.1 |
| Splicing-related | |||||
| U2AF35A; U2AF35B | 3.5 | 1.0 | 11.9 | 4.3 | 34.6 |
| SCL30A; SCL33 | 2.8 | 0 | 12.3 | 0 | 20.2 |
| SR45 | 1.8 | 0 | 5.2 | 0 | 44.6 |
| RSZ21; RSZ22; RSZ22A | 1.8 | 0 | 8.0 | 0 | 22.5 |
| SR30 | 1.0 | 0 | 3.9 | 0 | 29.1 |
| RS2Z33 | 1.0 | 0 | 2.9 | 0 | 32.9 |
| SCL30 | 0.8 | 0 | 2.8 | 0 | 29.6 |
| U2AF65A | 0.8 | 0 | 1.7 | 0 | 60.7 |
| Kinases | |||||
| SRPK4 | 8.5 | 0 | 16.2 | 0 | 59.4 |
| SRPK3 | 4.3 | 0 | 8.9 | 0 | 61.2 |
| Proteins linked to 3′ end processing/poly(A) binding | |||||
| SPT6L (AT1g63210, AT1g65440) | 8.5 | 1.0 | 4.8 | 0.9 | 185.0 |
| Poly(A)-binding protein RBP47C | 1.5 | 0.3 | 4.0 | 0.7 | 48.6 |
| La-related protein 6B (LARP6) | 2.3 | 0.7 | 5.5 | 1.9 | 60.6 |
The MS analysis revealed that LUC7 is indeed found in a complex with core U1 snRNP proteins U1-A and U1-70K (Table 1; Supplemental Data Set 1). Moreover, we detected peptides corresponding to the spliceosomal complex E components U2AF35 and U2AF65, further suggesting that LUC7 proteins are involved in very early steps of the splicing cycle (Table 1; Supplemental Data Set 1). Additional proteins known to be involved in splicing and general RNA metabolism, including several SR proteins (SR30, SCL30A, and SCL33), SR45, SERRATE (SE), and the CBC component ABH1/CBP80, were found in LUC7A complexes (Table 1; Supplemental Data Set 1). To validate the LUC7 complex purification experiment, we confirmed by in planta coimmunoprecipitation the interactions of LUC7 with SE and ABH1/CBP80, two interactors containing, respectively, high and low number of peptides in the MS analyses (Supplemental Figure 4). Interestingly, we also identified regulatory proteins in LUC7A-containing complexes, among them several kinases, and proteins involved in 3ʹ end processing (Table 1; Supplemental Data Set 1). One of the potential interactors, SPT6L, has been shown in independent experiments to associate with LUC7 proteins and other U1-specific proteins in an SPT4 complex (Antosz et al., 2017).
LUC7 Effects on the Arabidopsis Coding and Noncoding Transcriptome
To identify misregulated genes and misspliced transcripts in luc7 mutants, we performed an RNA-sequencing (RNA-seq) analysis with three biological replicates. We decided to use 7-d-old wild-type and luc7 triple mutant seedlings because at this age they are morphologically similar; therefore, changes in transcript levels and splicing patterns most likely reflect changes caused by LUC7 impairments and not altered development and/or different morphology (Supplemental Figure 5). We sequenced between 22.1 and 27.6 million reads per library.
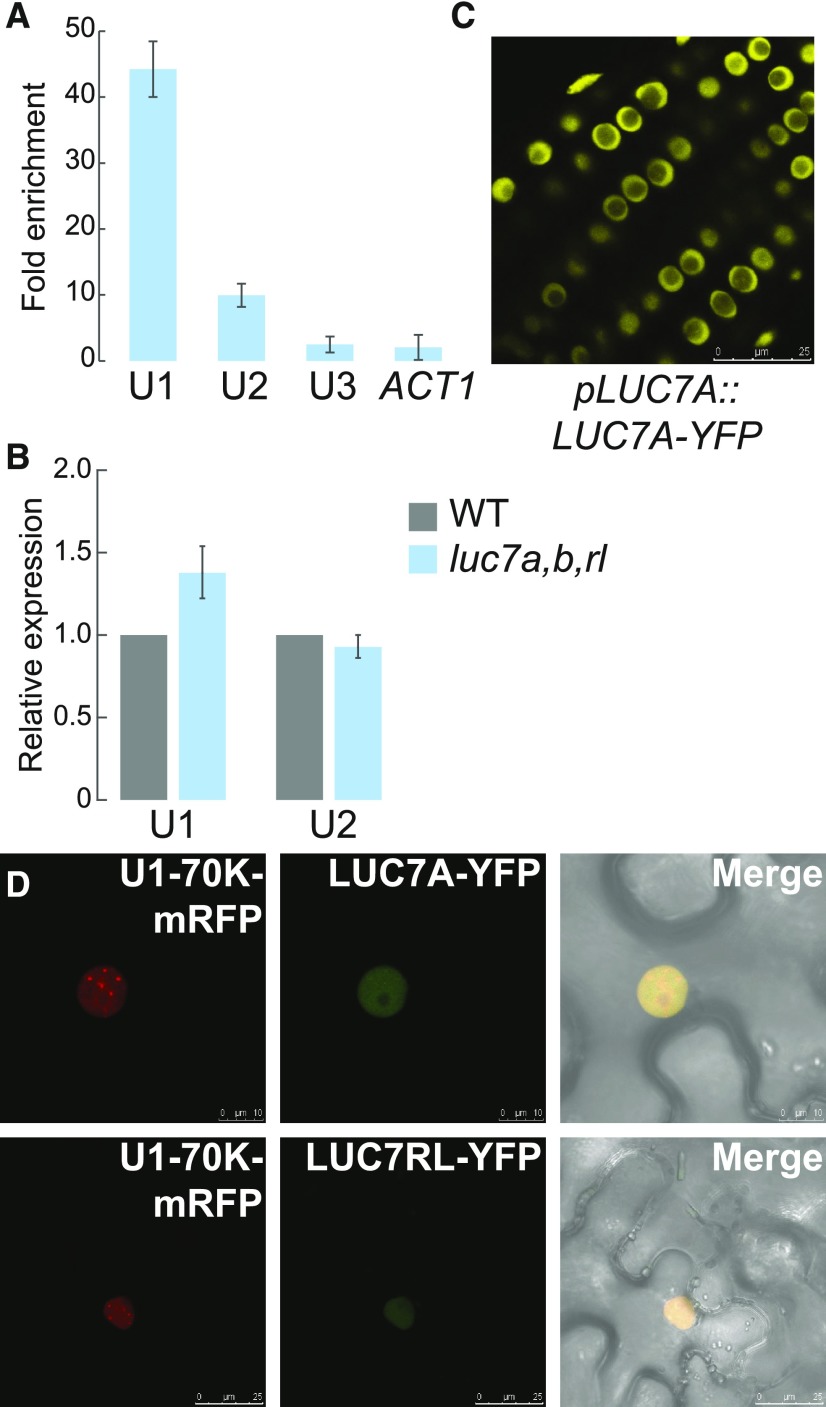
Analysis of differentially expressed genes revealed that 840 genes are upregulated and 703 are downregulated in luc7 triple mutant when compared with the wild type (Supplemental Data Sets 2 and 3). The majority of genes that change expression were protein-coding genes (Figure 4A). Nevertheless, noncoding RNAs (ncRNAs) were significantly enriched among the LUC7 affected genes (P < 0.05, hypergeometric test), although the overall number of ncRNAs affected in luc7 triple mutant was relatively small (Figures 4A and 4B; Supplemental Data Set 4). Previous studies implied that the U1 snRNP regulates microRNA (miRNA) biogenesis (Bielewicz et al., 2013; Schwab et al., 2013; Knop et al., 2016; Stepien et al., 2017). However, the expression of MIRNA genes was not affected in luc7 triple mutant (Figure 4A; Supplemental Data Set 4). In addition, quantification of mature miRNA levels revealed that all four tested miRNAs did not change abundance in luc7 triple mutants (Figure 4C). These results show that LUC7 proteins affect the expression of protein-coding genes and a subset of ncRNAs, but do not support a role for LUC7 in the processing of the analyzed MIRNAs.
Figure 4.
Mutations in LUC7 Result in Misexpression of Protein-Coding and Noncoding Genes, but Not in MIRNA Genes.
(A) Differentially expressed genes from RNA-seq experiments with the wild type and luc7a luc7b luc7rl mutants were classified in protein coding genes, ncRNAs, miRNAs, and transposons (Supplemental File 1).
(B) RT-qPCR analysis of selected ncRNA in the wild type and luc7a luc7b luc7rl. Differentially expressed ncRNAs identified by RNA-seq were analyzed by RT-qPCR. Total RNAs were isolated from 7-d-old seedlings. Error bars denote se (n = 3).
(C) RT-qPCR analysis of four selected miRNAs in the wild type and luc7a luc7b luc7rl. Total RNAs were isolated from 7-d-old seedlings. Error bars denote se (n = 3).
Arabidopsis LUC7 Functions Are Important for Constitutive and Alternative Splicing
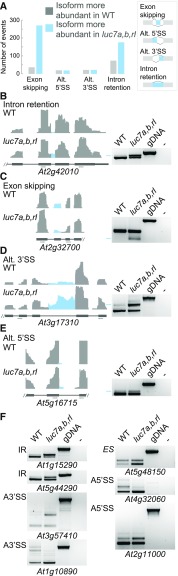
Because LUC7 proteins are U1 snRNP components, we asked whether misspliced transcripts accumulate in the luc7 triple mutant. In total, we identified 640 differential splicing events in luc7 triple mutant compared with the wild type (Supplemental Data Set 5). Only 17 of these alternative splicing events occurred in mRNAs whose expression also differed between the luc7 triple mutant and the wild type (Supplemental Data Sets 6 and 7). Hence, the splicing differences found were mainly not due to changes in transcript abundance. Among the differential splicing events, we detected a large number of intron retention events in the luc7 triple mutant (Figure 5A). RT-PCR experiments with oligonucleotides flanking selected intron retention events confirmed the RNA-seq data (Figure 5B). These results suggest that lack and/or impairment of the U1 snRNP components LUC7 disturb intron recognition and, thus, splicing. Additionally, we identified a large number of exon skipping events in the luc7 triple mutant. Exon skipping is also a major outcome of impairing U1 snRNP function (Rösel et al., 2011; Antosz et al., 2017). These defects are most likely caused by changes in the interaction network among the U2 snRNP and U1 snRNP components, due to absence/impairment of LUC7 (Morcos, 2007). Furthermore, we detected several cases of alternative 5ʹss and 3ʹss selection in luc7 triple mutant (Figures 5A to 5F).
Figure 5.
Global Analysis of Splicing Defects in luc7 Triple Mutant.
(A) Classification of splicing events changes in luc7 triple mutant compared with the wild type.
(B) to (F) Coverage plots and RT-PCR validation experiments for selected splicing events in the wild type and luc7 triple mutant. Genomic DNA (gDNA) or water (−) served as a control. Primer positions are indicated with gray arrows. IR, intron retention; ES, exon skipping; Alt.3ʹSS, alternative 3ʹ splicing site; Alt.5ʹSS, alternative 5ʹ splicing site.
Some splicing defects observed in luc7 triple mutant generated transcript variants that did not exist in the wild type (e.g., At2g32700; Figure 5C). In these cases, LUC7 proteins affect the splicing of an intron that is most likely constitutively removed in wild-type plants. In other instances, when compared with the wild type, the luc7 triple mutant lacked specific mRNA isoforms (e.g., At1g10980 and At4g32060) or the ratio of different mRNA isoforms was altered (e.g., At3g17310, At5g16715, At5g48150, and At2g11000) (Figures 5D to 5F). In such cases, LUC7 proteins affect splicing events that are subjected to alternative splicing in wild-type plants. These results show that LUC7 proteins are involved in both constitutive and alternative splicing in Arabidopsis.
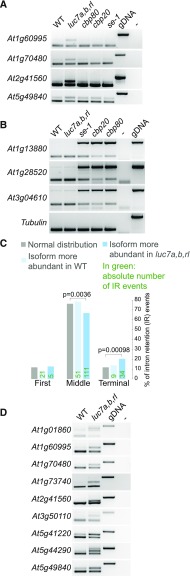
Next, we checked whether splicing changes observed in the luc7 triple mutant are due to the loss of only a specific LUC7 gene or whether LUC7 genes act redundantly. To test this, we analyzed the splicing pattern of selected mRNAs in luc7 single, double, and triple mutants. Some splicing defects were detectable even in luc7 single mutants (Supplemental Figure 6), but the degree of missplicing increased in luc7 double and triple mutants, suggesting that LUC7 proteins act additively at these introns (e.g., At5g16715). Some splicing defects occurred only in the luc7 triple mutant, implying that LUC7 proteins act redundantly to ensure splicing of these introns (e.g., At1g60995). Other splicing defects could more likely be due to the lack of LUC7A/B or LUC7RL. For instance, intron removal of At2g42010 relied more strongly on LUC7RL, while removal of an intron in At5g41220 preferentially depends on LUC7A/LUC7B (Supplemental Figure 6). These findings suggest that Arabidopsis LUC7 genes function redundantly, additively, or specifically to ensure proper splicing.
LUC7 Proteins Are Preferentially Involved in the Removal of Terminal Introns
In yeast, LUC7 connects the CBC with the U1 snRNP, and this interaction is important for the correct 5ʹss selection (Fortes et al., 1999b). In plants, the CBC associates with SE to ensure splicing of cap-proximal first introns (Laubinger et al., 2008; Raczynska et al., 2010, 2014). In addition, we show here that LUC7A form complexes with SE and ABH1/CBP80, one of the CBC components (Supplemental Data Set 1 and Supplemental Figure 4). To investigate the relationship between LUC7 and the CBC/SE in plants, we analyzed the splicing patterns of LUC7 dependent introns in cbc mutants (cbp20 and cbp80) and se-1 by RT-PCR. All tested introns retained in luc7 triple mutant were correctly spliced in cbc and se mutants (Figure 6A). Conversely, first introns that were partially retained in cbp20, cbp80, and se-1 mutants were completely removed in the luc7 triple mutant (Figure 6B). These observations suggest that the functions of LUC7 and CBC/SE in splicing of the selected introns do not overlap.
Figure 6.
LUC7 Proteins Have a Pronounced Effect on Terminal Intron Splicing.
(A) RT-PCR analysis of LUC7-dependent introns in the wild type, luc7 triple mutant, and cbp80, cbp20, and se-1 mutants. RT-PCR analyses were performed using oligonucleotides flanking the LUC7-dependent splicing event.
(B) RT-PCR analysis of CBC/SE-dependent introns in the wild type, luc7 triple mutant, and cbp80, cbp20, and se-1 mutants. RT-PCR analyses were performed using oligonucleotides flanking the CBC/SE-dependent splicing event.
(C) Classification of intron retention according to the intron position (first, middle, or terminal). Only genes with three or more introns were considered for this analysis. A Fisher’s exact test was performed for the statistical analysis (Supplemental File 1).
(D) RT-PCR analysis of genes carrying retained terminal introns in the wild type and luc7 triple mutants. RT-PCR analyses were performed using oligonucleotides flanking the LUC7-dependent splicing event.
Next, we asked whether LUC7 has a preference for promoting splicing of cap-proximal first introns as it has the CBC/SE complex. We classified retained introns in the luc7 triple mutant according to their position within the gene (first, middle, or last introns). Only genes with at least three introns were considered for this analysis. We found a significant increase in retained terminal introns, but not first introns, in luc7 triple mutant (Figure 6C). Although the total number of retained introns was higher among middle introns, the relative amount of retained middle introns in luc7 triple mutant was significantly reduced (Figure 6C). Retention of terminal introns in the luc7 triple mutant was confirmed by RT-PCR analysis (Figure 6D). In summary, our data revealed that (1) CBC/SE acts independently of LUC7 in splicing of cap-proximal introns and that (2) LUC7 proteins play an important role in the removal of certain terminal introns.
mRNAs Harboring Unspliced LUC7-Dependent Introns Remain in the Nucleus and Escape NMD
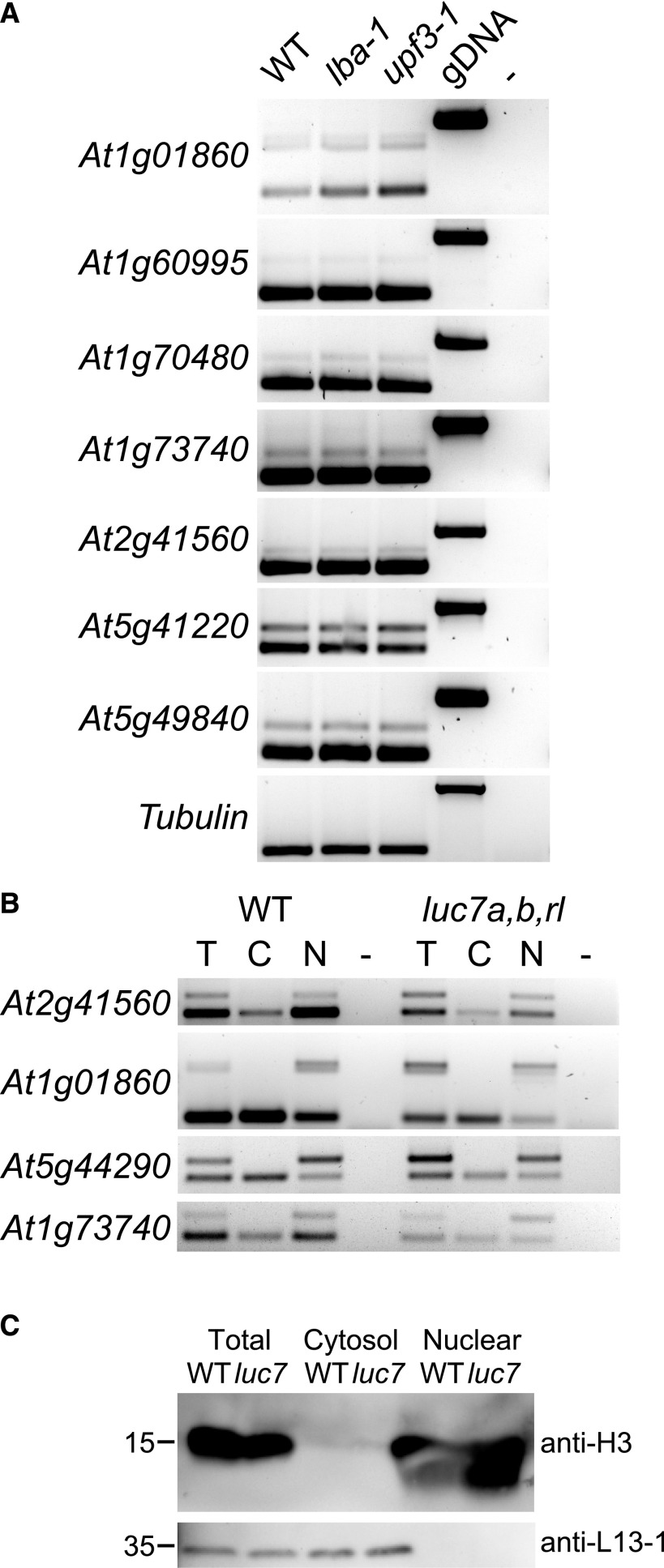
We were further interested in determining the characteristics and possible functions of LUC7-dependent introns. When introns are retained, the resulting mRNA can contain a premature stop codon and a long 3ʹ untranslated region, which are hallmarks of NMD targets (Kalyna et al., 2012; Drechsel et al., 2013; Shaul, 2015). To check whether mRNAs containing a retained LUC7-dependent intron are NMD substrates, we analyzed their splicing patterns in two mutants impaired in NMD, lba-1 and upf3-1. If unspliced isoforms were indeed NMD targets, we would expect their abundance to be increased in NMD mutants. Interestingly, we did not observe any change between the wild type and upf mutants (Figure 7A). Thus, we conclude that the tested LUC7-dependent introns do not trigger degradation via the NMD pathway.
Figure 7.
mRNAs Containing Retained LUC7-Dependent Introns Are NMD-Insensitive and Remain Nuclear.
(A) RT-PCR analysis of LUC7-dependent introns in the wild type and NMD mutants (lba1 and upf3-1).
(B) Splicing patterns of mRNAs isolated from total (T), cytosolic (C), and nuclear (N) fractions. RT-PCR analyses were performed using oligonucleotides flanking the LUC7-dependent splicing event.
(C) Immunoblot analysis of proteins isolated from total, cytosolic, and nuclear fractions. Blots were probed with antibodies against histone H3 and a ribosomal protein, L13-1.
NMD occurs in the cytoplasm, and RNAs can escape NMD by not being exported from the nucleus to the cytosol (Göhring et al., 2014). We therefore checked in which cellular compartment mRNAs with spliced and unspliced LUC7-dependent introns accumulate. To do this, we isolated total, nuclear, and cytosolic fractions from wild-type and luc7 triple mutant plants and performed RT-PCR analyses (Figure 7B). The purity of cytosolic and nuclear fractions was monitored by immunoblot analysis using antibodies against histone H3 (specific for nuclear fractions) and a 60S ribosomal protein (L13-1, specific for cytosolic fractions) (Figure 7C). Spliced mRNA isoforms accumulated in the cytosol, whereas mRNAs containing the unspliced LUC7-dependent introns were found in nuclear fractions (Figure 7B). These results suggest that splicing of the analyzed LUC7-dependent introns is essential for mRNA export to the cytosol.
Splicing of LUC7-Dependent Introns Can Be Modulated by Stress
Our results revealed that a subset of alternatively spliced introns requires LUC7 proteins for efficient splicing and that splicing of these introns is a prerequisite for nuclear export. This mechanism could serve as a nuclear quality control step to prevent the premature export of unspliced mRNAs. Interestingly, a Gene Ontology (GO) analysis of genes containing LUC7-dependent introns indicated an enrichment for stress-related genes (Supplemental Figure 7). This prompted us to speculate that nuclear retention of mRNAs could be exploited as a regulatory mechanism to fine-tune gene expression under stress conditions.
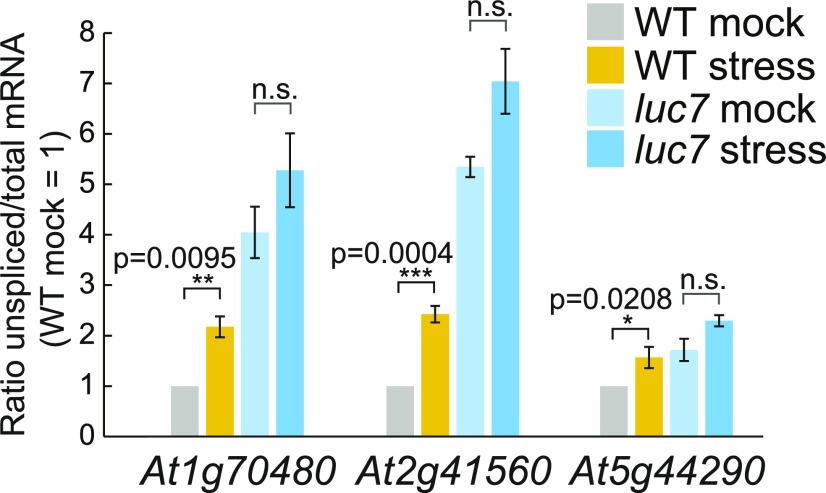
To test this hypothesis, we investigated the splicing of some LUC7-dependent terminal introns in the wild type under stress condition. We chose cold stress because luc7 triple mutants are cold-sensitive (Figure 2), and, in addition, it was suggested that U1 snRNP functionality is impaired under cold conditions (Schlaen et al., 2015). To quantify the amount of unspliced isoforms in cold conditions, we designed qPCR primers specific to unspliced isoforms and the total mRNA pool, as well as to calculate the relative amount of mRNA carrying unspliced LUC7-dependent introns compared with the total mRNA pool. mRNAs of At1g70480, At2g41560, and At5g44290 significantly accumulated unspliced isoforms in response to cold treatment in wild-type plants, demonstrating that cold stress modulates the splicing efficiency of these LUC7-dependent introns (Figure 8). Furthermore, the amount of unspliced mRNA in luc7 triple mutants did not differ significantly between mock and stress conditions (Figure 8). This observation suggests that LUC7 is directly involved in the regulation of intron splicing under stress conditions and that LUC7 might be a target for stress response pathways.
Figure 8.
Splicing of LUC7-Dependent Introns Can Be Modulated by Stress.
Seven-day-old wild-type and luc7 triple mutant seedlings were exposed to cold for 60 min. Splicing ratios (unspliced/total RNA) of three genes featuring a LUC7-dependent intron was analyzed by RT-qPCR. Three biological replicates were performed and t tests were employed for statistical analyses (Supplemental File 1). Error bars denote se of the mean.
Stress-Regulated Retention of Terminal Introns Is Widespread under Cold and Salt Stress
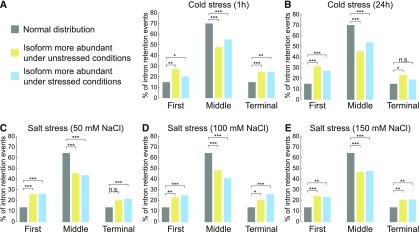
To investigate whether terminal intron retention is a general feature of plant stress transcriptomes, we performed a global analysis of intron retention under stress conditions. We made use of publicly available RNA-seq data sets from Arabidopsis plants treated with cold and salt stress (Ding et al., 2014; Schlaen et al., 2015). We identified stress-regulated intron retention events, filtered for genes containing three or more introns, and assigned retained introns based on their position within the transcript (first, middle, or terminal intron). The relative amounts of first, middle, and terminal introns among retained introns under stress conditions were compared with the relative amounts of introns among all expressed genes. Intron retention under cold and salt stress occurred significantly more often in terminal introns than one would expect by chance (Figures 9A to 9E). Also, first introns were more prone for splicing regulation under cold and salt stress (Figures 9A to 9E). These results show that first and terminal introns are preferentially subjected to alternative splicing regulation under stress conditions. Thus, our data strongly suggest that regulated splicing of first and terminal introns widely contributes to shaping plant transcriptomes under stress conditions.
Figure 9.
Salt and Cold Stress Significantly Affect Retention of First and Terminal Introns.
Classification of intron retention events in cold- (A) and salt-treated (C) plants. RNA-seq experiments from Arabidopsis plants treated with cold for 1 h (A) and 24 h (B), or 50 mM (C), 100 mM (D), or 150 mM (E) NaCl were analyzed for intron retention events. The relative amount of retained/removed first, middle, and terminal intron in stress samples was compared with the relative amount of first, middle, and last intron among all expressed genes (normal distribution). Only genes with three or more introns were considered for this analysis. A Fisher’s exact test was performed for the statistical analysis (Supplemental File 1). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; n.s., not significant.
DISCUSSION
Functions of the Arabidopsis U1 snRNP Component LUC7
For this study, we generated an Arabidopsis triple mutant deficient in the U1 snRNP components LUC7 and dissected the genome-wide effects of LUC7 impairments on the Arabidopsis transcriptome. Our results show that LUC7 proteins are bona fide U1 components that act redundantly for the most part. The reduction of U1 function in the luc7 triple mutant affects constitutive splicing. A large number of introns are retained in the luc7 triple mutant, suggesting that without proper recognition of the 5ʹss, splicing of the affected introns is impaired.
Our results also show that exon skipping occurs more often in the luc7 triple mutant, revealing that a functional plant U1 snRNP containing LUC7 is essential for exon definition. In addition, we show that LUC7 proteins affect alternative splicing by influencing events of alternative 5ʹ and 3ʹ splice sites. This implies that the U1 snRNP not only affects 5ʹss usage but might also indirectly regulate usage of 3ʹss via its interaction with U2AFs and the U2 snRNP (Hoffman and Grabowski, 1992; Shao et al., 2012).
The effects of LUC7 proteins on regulating the Arabidopsis transcriptome are likely to be underestimated because misspliced mRNAs in luc7 mutants may contain hallmarks of NMD and therefore be rapidly turned over and escape detection. Analysis of luc7 mutants combined with mutations in NMD factors would help to uncover the full set of splicing events affected by LUC7. Furthermore, our RNA-seq experiments revealed that, while the chosen luc7rl allele is a RNA-null allele, the luc7a and luc7b alleles still produced mRNAs that might be translated into truncated proteins (Supplemental Figure 8). Hence, we cannot exclude that a true luc7 null mutant might exhibit even more severe mutant phenotypes and splicing defects. One also has to consider that U1 snRNP-independent splicing has been described in animals, indicating that not all introns require the U1 complex for efficient intron removal (Fukumura et al., 2009). The degree of U1-independent splicing in plants remains to be elucidated.
Duplications among genes encoding for U1 snRNP proteins, such as the LUC7 genes, open up possibilities for sub- and neofunctionalization of U1 accessory proteins. Furthermore, the Arabidopsis genome encodes 14 potential U1 snRNAs, which differ slightly in sequence (Wang and Brendel, 2004). Therefore, the plant U1 snRNP presumably does not exist as a single complex, but rather as different subcomplexes exhibiting distinct specificities and functions. In metazoans, the existence of at least four different U1 snRNP subcomplexes has been suggested (Hernández et al., 2009; Guiro and O’Reilly, 2015). Specific combinations of plant U1 protein family members and U1 snRNAs could generate an even higher number of such U1 subcomplexes, which could be responsible for specific splicing events. Our results show that LUC7 proteins can act redundantly but can also fulfill specific functions, suggesting that LUC7 complexes act specifically on certain pre-mRNAs. In this regard, it is important to note that an additional short stretch of amino acids separates the two zinc-finger domains in LUCA and LUC7B (Supplemental Figure 1). Changing the space between RNA binding domains affects substrate specificities and could explain different specificities among LUC7 proteins (Chen and Varani, 2013).
LUC7 Function in Terminal Intron Splicing
The luc7 triple mutant showed a significantly higher retention rate for terminal introns compared with first or middle introns. This was surprising, as LUC7 was initially found to act in concert with the CBC, a complex involved in the removal of cap-proximal introns (Lewis et al., 1996; Laubinger et al., 2008). We found LUC7 in a complex with the CBC and the CBC-associated protein SE in Arabidopsis. However, functional significance of LUC7-CBC/SE interaction remains to be established.
Often, the removal of terminal introns is coupled to polyadenylation (Cooke et al., 1999; Cooke and Alwine, 2002; Rigo and Martinson, 2008). Interestingly, we detected components involved in RNA 3ʹ end processing or poly(A) binding as part of LUC7A complexes, suggesting that such interactions may contribute to the specific functions of LUC7 in terminal intron splicing. We found LUC7 in complexes with RBP47C, a poly(A) binding protein of unknown function (Rigaut et al., 1999), and LARP6, which is targeted to 3ʹ ends of mRNA through interaction with POLYA BINDING PROTEIN2 (Morcos, 2007). Currently, the function of these LUC7 complex components in plants is not known, but future studies will help to determine the role of these proteins in terminal intron splicing. It is noteworthy that we also found an SPT6-like transcription factor associated with LUC7 complexes. SPT6/SPT6L and LUC7 proteins copurify in a SPT4 containing-complex, which is involved in transcriptional elongation (Antosz et al., 2017). SPT6 binds the pol II C-terminal domain phosphorylated at serine 2 (Ser2P), which accumulates at 3ʹ ends of genes (Kaplan et al., 2000; Sun et al., 2010). This raises the possibility that LUC7 proteins are guided to 3ʹ ends of genes through interactions with SPT6 and transcription elongation complexes.
In general, our analysis suggests that first and terminal introns are hotspots for regulated splicing under stress conditions. First and last introns are close to the 5ʹ cap and the poly(A) tail, respectively. These positions might offer more possibilities for splicing regulation via crosstalk between the splicing machinery and factors involved in capping and cleavage/polyadenylation. Future identification of cis- and trans-regulatory factors involved in the regulation of terminal intron splicing will shed additional light on this layer of gene expression.
Possible Functions of Regulated Intron Retention for Plant Stress Responses
We found that splicing of LUC7-dependent introns is required for the export of mRNAs from the nucleus to the cytosol. The fact that we cannot detect unspliced transcript in the cytosol suggests a nuclear retention mechanism for such mRNAs. One possibility is that LUC7-dependent introns might contain binding sites for specific trans-regulatory factors that, upon binding, inhibit export. POLYPYRIMIDINE TRACT BINDING PROTEIN1 (PTB1) is a candidate for such a trans-regulatory protein because binding of PTB1 to introns represses nuclear export of certain RNAs (Yap et al., 2012; Roy et al., 2013). In animals, for instance, non-neuronal cells are known to inhibit the expression of neuronal proteins by repressing their transcripts splicing of 3ʹ terminal introns through Ptbp1 binding (Yap et al., 2012).
Nuclear retention of unspliced mRNAs might be a much more general mechanism to escape NMD and to regulate gene expression (Marquez et al., 2015; Wong et al., 2016). In plants, some specific transcript isoforms have been detected only in the nucleus and not in the cytosol (Göhring et al., 2014; Hartmann et al., 2018). In addition, in metazoans, intron retention might have a more general role in regulating gene expression (Yap et al., 2012; Braunschweig et al., 2014; Pimentel et al., 2016; Naro et al., 2017). The so-called detained introns, which are evolutionary conserved, are NMD insensitive, and retained in the nucleus (Boutz et al., 2015). The functional importance of intron retention was also suggested in the fern Marsilea vestita, in which many mRNAs contain introns that are only spliced shortly before gametophyte development (Boothby et al., 2013).
We found that splicing of LUC7 dependent introns can be modulated by cold stress. Because retention of these introns causes nuclear trapping, it is tempting to speculate that environmental cues affect splicing and nuclear retention of mRNAs. Such a mechanism would regulate the amount of translatable mRNAs in the cytosol in a cost-efficient and rapid manner. Since the stress-dependent regulation of splicing of LUC7-dependent introns is lost in luc7 triple mutants, one can expect that LUC7 function might be regulated under stress conditions. Interestingly, the RS domains of LUC7 proteins are phosphorylated, and we identified three kinases as potential LUC7A interactors (Heazlewood et al., 2008; Durek et al., 2010). In addition, stress signaling triggered by the phytohormone ABA causes differential phosphorylation of several splicing factors (Umezawa et al., 2013; Wang et al., 2013). Whether stress-induced changes in phosphorylation play a role in regulating LUC7 proteins and whether the LUC7-interacting kinases here identified are involved in this process remain to be elucidated.
METHODS
Plant Material and Growth Conditions
All mutants were in the Columbia-0 (Col-0) background. luc7a-1 (SAIL_596_H02), luc7a-2 (SAIL_776_F02), luc7b-1 (SALK_144681), luc7rl-1 (SALK_077718), and luc7rl-2 (SALK_130892C) were isolated by PCR-based genotyping (Supplemental Data Set 8). luc7 double and triple mutants were generated by crossing individual mutants. All other mutants used in this study (abh1-285, cbp20-1, se-1, lba-1, and upf3-1) were described elsewhere (Prigge and Wagner, 2001; Papp et al., 2004; Hori and Watanabe, 2005; Yoine et al., 2006; Laubinger et al., 2008). The line expressing GFP was generated using the vector pBinarGFP and was kindly provided by Andreas Wachter (Wachter et al., 2007). For complementation analyses, pLUC7A:LUC7A-FLAG, pLUC7B:LUC7B-FLAG, pLUC7RL:LUC7RL-FLAG, and pLUC7A:LUC7A-YFP constructs were introduced in luc7 triple mutant by Agrobacterium tumefaciens-mediated transformation (Clough and Bent, 1998).
All plants were grown on soil in long-day conditions (16 h light/8 h dark) under fluorescent tubes (120–160 µmol/m2/s) at 20°C/18°C day/night. The size of luc7 mutants was assessed by measuring the longest rosette leaf after 21 d. For all molecular studies, seeds were surface-sterilized, plated on 0.5× MS medium with 0.8% phytoagar, and grown for 7 d in continuous light at 22°C. For the cold treatment, plates with Arabidopsis seedlings were transferred to ice water for 60 min. For the root growth assay, 4-d-old seedlings growing on vertical plates were transferred to mock plates or plates containing indicated amount of NaCl and grown for more 11 d vertically. Root growth rate per day was assessed by measuring in ImageJ the root length in the days 2 and 9 after transfer. For ABA sensitivity assays, seedlings were grown for 10 d on 0.5× MS plates supplemented with 0.8% phytoagar, 1% sucrose, and indicated amounts of ABA (+) (Sigma–Aldrich; A4906). For cold stress experiments, seeds were grown at 20°C for 5 d and then transferred to 8°C for 2 weeks. We analyzed at least 10 individual plants or for all experiments or pooled 10 individual plants for one biological replicate. Two or three biological replicates were analyzed for all experiments. We consider a biological replicate a measurement of completely independent pools of plants grown side-by-side, or pools of plants not grown side-by-side, but under the exact same environmental conditions.
Plasmid Construction and Transient Expression Analyses
For the expression of C-terminal FLAG- and YFP-tagged LUC7 proteins expressed from their endogenous regulatory elements, 2100, 4120, and 2106 bp upstream of the ATG start codon of LUC7A, LUC7B, and LUC7RL, respectively, to the last coding nucleotide were PCR-amplified and subcloned in pCR8/GW/TOPO (Invitrogen). Oligonucleotides are listed in Supplemental Data Set 8. Entry clones were recombined with pGWB10 and pGWB540 using Gateway LR clonase II (Invitrogen) to generate binary plasmids containing pLUC7A:LUC7A-FLAG, pLUC7B:LUC7B-FLAG, pLUC7RL:LUC7RL-FLAG, and pLUC7A:LUC7A-YFP. For the colocalization studies, entry vector containing the coding sequence of U1-70k was recombined with pGWB654 for the expression of p35S:U1-70k-mRFP (Nakagawa et al., 2007). Agrobacterium-mediated transient transformation of Nicotiana benthamiana plants was conducted as following. Overnight Agrobacterium culture were diluted in 1:6 and grown for 4 h at 28°C. After centrifugation, pellets were resuspended in infiltration medium (10 mM MgCl2, 10 mM MES-KOH, pH 5.6, and 100 µM acetosyringone). The OD600 was adjusted to 0.6 to 0.8, and samples were mixed when required. N. benthamiana were infiltrated and subcellular localization was checked after 3 d. Subcellular localization of fluorescent proteins was analyzed by confocal microscopy using a Leica TCS SP8.
Phylogenetic Analysis
AthLUC7A (AT3G03340) protein sequence was analyzed in Interpro (https://www.ebi.ac.uk/interpro/) to retrieve the Interpro ID for the conserved Luc7-related domain (IPR004882). The sequence for Saccharomyces cerevisiae (strain ATCC 204508_S288c) was obtained in Interpro. Plants sequences were extracted using BioMart selecting for the protein domain IPR004882 on Ensembl Plants (http://plants.ensembl.org/). The following genomes were included in our analyses: Amborella trichopoda (AMTR1.0 [2014-01-AGD]), Arabidopsis thaliana (TAIR10 [2010-09-TAIR10]), Brachypodium distachyon (v1.0), Chlamydomonas reinhardtii (v3.1 [2007-11-ENA]), Physcomitrella patens (ASM242v1 [2011-03-Phypa1.6]), Selaginella moellendorffii (v1.0 [2011-05-ENA]), Oryza sativa Japonica (IRGSP-1.0), and Ostreococcus lucimarinus genes (ASM9206v1). The phylogenetic analysis was performed in Seaview (version 4.6.1) using Muscle for sequence alignment (Supplemental Data Set 9). Maximum likelihood (PhYML) was employed with 1000 bootstraps (Gouy et al., 2010).
RNA Extractions, RT-PCR, and RT-qPCR
RNA extractions were performed with Direct-zol RNA MiniPrep Kit (Zymo Research). Total RNAs were treated with DNase I and cDNA synthesis performed with RevertAid First Strand cDNA synthesis kit (Thermo Scientific) using oligo(dT) primers or a mixture of hexamer and miRNA-specific stem-loop primers (Supplemental Data Set 8). Standard PCRs for the splicing analysis were performed with DreamTaq DNA Polymerase (Thermo Scientific). RT-qPCR was performed using the Maxima SYBR Green (Thermo Scientific) in a Bio-Rad CFX 384. For all qPCR-primers, primer efficiencies were determined by a serial dilution of cDNA template. The relative expressions were calculated using the 2(−ΔΔCT) method with PP2A or ACTIN as control. For the RT-qPCR to detect splicing ratio changes under cold condition, the ratio 2ˆ(−ΔCTunspliced)/2ˆ(−ΔCTtotal RNA) was calculated separately for each replicate and t test was performed before calculating the relative to wild-type mock. Oligonucleotides are listed in Supplemental Data Set 8. For RNA-seq analysis, poly(A) RNAs were enriched from 4 µg of total RNAs using NEBNext Oligo d(T)25 Magnetic Beads (New England Biolabs). The libraries were prepared using ScriptSeq Plant Leaf kit (Epicentre) following the manufacturer’s instructions. Single-end sequencing was performed on an Illumina HiSeq 2000. Sequencing data were deposited at Gene Expression Omnibus under accession number GSE98779.
RNA-Seq Libraries: Mapping, Differential Expression Analysis, and Splicing Analysis
RNA-seq reads for each replicate were aligned against the Arabidopsis reference sequence (TAIR10) using TopHat (v2.0.10, -p2, -a 10, -g 10, -N 10,–read-edit-dist 10,–library-type fr-secondstrand,–segment-length 31, -G TAIR10.gff). Next, cufflinks (version 2.2.1) was used to extract FPKM counts for each expressed transcript generating a new annotation file (transcripts.gtf), where the coordinates of each expressed transcript can be found. Cuffcompare (version 2.2.1) was then used to generate a non-redundant annotation file containing all reference transcripts in addition to new transcripts expressed in at least one of the nine samples (cuffcmp.combined.gtf). The differential expression analysis was performed with cuffdiff (version 2.2.1) between wild type/luc7 triple using the annotation file generated by cuffcompare (false discovery rate [FDR] < 0.05 and fold change > 2). For the splicing analysis, the same alignment files generated by TopHat, and annotation files generated by cuffcompare (cuffcmp.combined.gtf) were used as input for MATS (version 3.0.8) in order to test for differentially spliced transcripts (Shen et al., 2014).
Global Analysis of Intron Regulation under Stress Conditions
For the analyses of intron retention under stress conditions, published data sets were analyzed (accession numbers SRP035234 and SRP049993) (Ding et al., 2014; Schlaen et al., 2015). Reads were aligned to the Arabidopsis Ensembl3 33 genome and to the annotation GTF file (ftp://ftp.ensemblgenomes.org/pub/release33/plants/fasta/arabidopsis_thaliana/dna/Arabidopsis_thaliana.TAIR10.dna.toplevel.fa.gz, ftp://ftp.ensemblgenomes.org/pub/release-33/plants/gtf/arabidopsis_thaliana/Arabidopsis_thaliana.TAIR10.33.gtf.gz) using TopHat2 applying following parameters: tophat2 -p 10 -i 10 -I 1000 -G Arabidopsis_thaliana.TAIR10.33.gtf Arabidopsis_thaliana.TAIR10.dna.toplevel.fa. After alignment, mock-treated samples were used to generate an expressed background for the respective data set using featureCounts from the Rsubread package (Kim et al., 2013; Liao et al., 2013; Kersey et al., 2016). Read numbers and gene lengths per gene (featureCounts -T 6 -R -p -F GTF -J -G Arabidopsis_thaliana.TAIR10.dna.toplevel.fa -a Arabidopsis_thaliana.TAIR10.33.gtf) were collected and TPM values were calculated using an in-house script. Log2 transformed values of expressed genes were visualized with ggplot2 version 2.1.0, and based on the density plot, threshold of expressed genes was defined as TPMexpressed > 0.6. Intron retention events were identified using rMATS with the following parameters: python RNASeq-MATS.py -b1 untreated.bam -b2 treated.bam -gtf Arabidopsis_thaliana.TAIR10.33.gtf -o output_dir -t paired -len 101 (Shen et al., 2014). After filtering the outputs (P value < 0.05, FDR < 0.05), we categorized introns based on their position and annotation. In cases of a few ambiguous hits, they were manually recategorized. For the categorization of introns in first, middle, and terminal introns, we used the GTF annotation file (Ensembl 33) and selected genes with three or more introns and TPM > 0.6. The intron distribution of all expressed genes served as a background reference, to which the distribution of retained introns under stress conditions was compared. To test for significance of changes in intron distribution, Fisher’s exact test was employed.
Subcellular Fractionation
Two grams of seedlings were ground in N2 liquid and resuspended in 4 mL of Honda buffer (0.44 M sucrose, 1.25% Ficoll 400, 2.5% Dextran T40, 20 mM HEPES KOH, pH 7.4, 10 mM MgCl2, 0.5% Triton X-100, 5 mM DTT, 1 mM PMSF, and protease inhibitor cocktail [Roche] supplemented with 40 units/mL of Ribolock). The homogenate was filtered through two layers of Miracloth, which was washed with 1 mL of Honda buffer. From the filtrate, 300 μL was removed as “total” fraction and kept on ice. Filtrates were centrifuged at 1500g for 10 min, 4°C for pelleting nuclei, and supernatants were transferred to a new tube. Supernatants were centrifuged at 13,000g, 4°C, 15 min, and 300 μL was kept on ice as cytoplasmic fraction. Nuclei pellets were washed five times in 1 mL of Honda buffer (supplemented with 8 units/mL of Ribolock, with centrifugation at 1800g for 5 min. The final pellet was resuspended in 300 μL of Honda buffer. To all the fractions (total, cytoplasmic, and nuclei), 900 μL of TRI Reagent (Sigma-Aldrich) was added. After homogenization, 180 μL of chloroform was added and samples were incubated at room temperature for 10 min. After centrifugation at 17,000g for 20 min at 4°C, the aqueous phase was transferred to a new tube and RNA extracted with Direct-zol RNA MiniPrep Kit (Zymo Research). The organic phase was collected and proteins were isolated according to manufacturer’s instructions. cDNA synthesis with random primers was performed as above. Proteins extracted were analyzed by standard immunoblot techniques using the following antibodies: H3 (∼17 kD; catalog no. ab1791; Abcam) and 60S ribosomal (∼23,7–29 kD; L13; catalog no. AS132650; Agrisera).
RNA Immunoprecipitation
RNA immunoprecipitation using the wild type and a pLUC7A:LUC7A-eYFP rescue line was performed as described elsewhere with minor modifications (Rowley et al., 2013; Xing et al., 2015). Isolated nuclei were sonicated in nuclear lysis buffer in a Covaris E220 (duty cycle, 20%; peak intensity, 140; cycles per burst, 200; cycle time, 3ʹ). RNAs were extracted using RNeasy Plant Mini Kit (Qiagen) following the manufacturer’s instructions. The RNA were treated with DNaseI (Thermo Scientific) and samples were split in half for the (-)RT reaction. cDNA syntheses were performed with SuperScript III reverse transcriptase (Invitrogen). RT-qPCRs were performed with QuantiNova SYBRR Green PCR (Qiagen).
GO Analysis
GO analysis was performed in Bar Utoronto (http://bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer.cgi).
Protein Complex Purification and MS Analysis
LUC7A immunoprecipitation was performed using a complemented line pLUC7A:LUC7A-eYFP (line 20.3.1) and a transgenic p35S:GFP and wild type as controls. Four independent biological replicates were performed. Seedlings (4 g) were ground in N2 liquid and resuspended in 1 volume of extraction buffer (50 mM Tris-Cl, pH 7.5, 100 mM NaCl, 0.5% Triton X-100, 5% glycerol, 1 mM PMSF, 100 µM MG132, Complete Protease Inhibitor Cocktail EDTA-free [Roche], and plant-specific protease Inhibitor [Sigma-Aldrich P9599]). After thawing, samples were incubated on ice for 30 min, centrifuged at 3220 rcf for 30 min at 4°C, and filtrated with two layers of Miracloth. For each immunoprecipitation, 20 μL of GFP-trap (Chromotek) was washed twice with 1 mL of washing buffer (50 mM Tris-Cl, pH 7.5, 100 mM NaCl, and 0.2% Triton X-100) and once with 0.5 mL of IP buffer. For each replicate, the same amount of plant extracts (∼5 mL) was incubated with GFP-trap on a rotating wheel at 4°C for 3 h. Samples were centrifuged at 800 to 2000 rcf for 1 to 2 min and the supernatant discarded. GFP beads were resuspended in 1 mL of washing buffer, transferred into a new tube, and washed four to five times. Then, beads were resuspended in ∼40 μL of 2× Laemmli buffer and incubated at 80°C for 10 min. Short gel purifications (SDS-PAGE) were performed and gels slices were digested with trypsin. LC-MS/MS analyses were performed in two mass spectrometers. Samples from R10 to R14 were analyzed on a Proxeon Easy-nLC coupled to Orbitrap Elite (method: 90 min, Top10, HCD). Samples from R15 to R17 were analyzed on a Proxeon Easy-nLC coupled to OrbitrapXL (method: 90 min, Top10, CID). Samples from R18 to R20 analysis on a Proxeon Easy-nLC coupled to OrbitrapXL (method: 130 min, Top10, CID). All the replicates were processed together on MaxQuant software (version 1.5.2.8. with integrated Andromeda Peptide search engine) with a setting of 1% FDR, and the spectra were searched against an Arabidopsis Uniprot database (UP000006548_3702_complete_20151023.fasta). All peptides identified are listed in Supplemental Data Set 1, and raw data were deposited publically (accession PXD006127). For coimmunoprecipitation experiments shown in Supplemental Figure 4, experiments were conducted as described above and immunoprecipitated protein fractions were analyzed using SDS-PAGE followed by detection with GFP-specific (catalog no. ab290; Abcam), SE-specific (catalog no. AS09532A; Agrisera), and CBP80-specific (catalog no. AS09531, Agrisera) antibodies. An HRP-conjugated antibody (catalog no. AS163152; Agrisera) and the ECL Prime Western Blotting System (catalog no. RPN2232; Merck) were used for visualization according to the manufacturer’s instructions.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: LUC7A, AT3G03440; LUC7B, AT5G17440; LUC7RL, AT5G51410; GSE98779; PXD006127, SRP035234; and SRP049993.
Supplemental Data
Supplemental Figure 1. Alignment of Arabidopsis and S. cerevisiae LUC7 proteins.
Supplemental Figure 2. Growth phenotypes of wild-type, luc7 single, double, and triple mutants.
Supplemental Figure 3. Growth phenotypes of wild-type, luc7 triple mutants, and luc7 triple mutants containing a pLUC7A:LUC7A-YFP rescue construct.
Supplemental Figure 4. Interaction test of LUC7A with SE and CBP80.
Supplemental Figure 5. Growth phenotypes of wild-type and luc7 triple mutant seedlings.
Supplemental Figure 6. RT-PCR splicing analysis in wild-type, luc7 single, double, and triple mutants.
Supplemental Figure 7. GO analysis of genes with mRNAs that retained introns in luc7 triple mutants.
Supplemental Figure 8. Analysis of LUC7 mRNA accumulation in the wild type and luc7a luc7b luc7rl mutants.
Supplemental Data Set 1. List of proteins identified in LUC7A IP-MS experiments.
Supplemental Data Set 2. Genes upregulated in luc7 triple mutants.
Supplemental Data Set 3. Genes downregulated in luc7 triple mutants.
Supplemental Data Set 4. Characterization of genes differentially expressed in luc7 triple mutants.
Supplemental Data Set 5. All significant splicing changes detected in luc7 triple mutants.
Supplemental Data Set 6. Genes alternatively spliced and upregulated in luc7 mutants.
Supplemental Data Set 7. Genes alternatively spliced and downregulated in luc7 mutants.
Supplemental Data Set 8. List of all oligonucleotides.
Supplemental Data Set 9. Text file of the alignment used for the phylogenetic analysis shown in Figure 1A.
Supplemental File 1. Statistical analyses.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (LA2633-4/1 to S.L.), by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Brazil) for doctoral fellowship (to M.d.F.A.), by the Max Planck Society (to K.S.), and by the Max Planck Society Chemical Genomics Centre through its supporting companies AstraZeneca, Bayer CropScience, Bayer Healthcare, Boehringer-Ingelheim, and Merck (to S.L.). We thank Andreas Wachter (ZMBP, University of Tuebingen, Germany), Amanda Kaye-Riepel, and members of the lab for critical reading of the manuscript, Marjori Matzke for discussions on the preprint version of this article, Christa Lanz for her invaluable help with Illumina sequencing, Johanna Schröter and her team for excellent care of our plants, Anja Hoffmann for excellent technical assistance, and Andreas Wachter (ZMBP, University of Tuebingen, Germany), Tsuyoshi Nakagawa (Department of Molecular and Functional Genomics, Center for Integrated Research in Science, Shimane University, Matsue, Japan), and the Nottingham Arabidopsis Stock Centre for providing seeds and DNA constructs.
AUTHOR CONTRIBUTIONS
M.d.F.A. and S.L. designed research. M.d.F.A., A.G.F.-M., I.D.-B., and S.L. performed experiments. E.-M.W., E.X.S., M.d.F.A., A.G.F.-M., I.D.-B., B.M., K.S., and S.L. analyzed the data. M.d.F.A. and S.L. wrote the manuscript with contributions from all authors.
Footnotes
Articles can be viewed without a subscription.
References
- Almada A.E., Wu X., Kriz A.J., Burge C.B., Sharp P.A. (2013). Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499: 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AlShareef S., Ling Y., Butt H., Mariappan K.G., Benhamed M., Mahfouz M.M. (2017). Herboxidiene triggers splicing repression and abiotic stress responses in plants. BMC Genomics 18: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antosz W., et al. (2017). The composition of the Arabidopsis RNA polymerase II transcript elongation complex reveals the interplay between elongation and mRNA processing factors. Plant Cell 29: 854–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg M.G., Singh L.N., Younis I., Liu Q., Pinto A.M., Kaida D., Zhang Z., Cho S., Sherrill-Mix S., Wan L., Dreyfuss G. (2012). U1 snRNP determines mRNA length and regulates isoform expression. Cell 150: 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielewicz D., Kalak M., Kalyna M., Windels D., Barta A., Vazquez F., Szweykowska-Kulinska Z., Jarmolowski A. (2013). Introns of plant pri-miRNAs enhance miRNA biogenesis. EMBO Rep. 14: 622–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothby T.C., Zipper R.S., van der Weele C.M., Wolniak S.M. (2013). Removal of retained introns regulates translation in the rapidly developing gametophyte of Marsilea vestita. Dev. Cell 24: 517–529. [DOI] [PubMed] [Google Scholar]
- Boutz P.L., Bhutkar A., Sharp P.A. (2015). Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 29: 63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U., Barbosa-Morais N.L., Pan Q., Nachman E.N., Alipanahi B., Gonatopoulos-Pournatzis T., Frey B., Irimia M., Blencowe B.J. (2014). Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 24: 1774–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho R.F., Feijão C.V., Duque P. (2013). On the physiological significance of alternative splicing events in higher plants. Protoplasma 250: 639–650. [DOI] [PubMed] [Google Scholar]
- Chen Y., Varani G. (2013). Engineering RNA-binding proteins for biology. FEBS J. 280: 3734–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S., Hoang A., Sinha R., Zhong X.Y., Fu X.D., Krainer A.R., Ghosh G. (2011). Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. USA 108: 8233–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cooke C., Alwine J.C. (2002). Characterization of specific protein-RNA complexes associated with the coupling of polyadenylation and last-intron removal. Mol. Cell. Biol. 22: 4579–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke C., Hans H., Alwine J.C. (1999). Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol. Cell. Biol. 19: 4971–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F., Cui P., Wang Z., Zhang S., Ali S., Xiong L. (2014). Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genomics 15: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel G., Kahles A., Kesarwani A.K., Stauffer E., Behr J., Drewe P., Rätsch G., Wachter A. (2013). Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 25: 3726–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durek P., Schmidt R., Heazlewood J.L., Jones A., MacLean D., Nagel A., Kersten B., Schulze W.X. (2010). PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 38: D828–D834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin S., Priest H.D., Megraw M., Mockler T.C. (2015). Alternative splicing in plants: directing traffic at the crossroads of adaptation and environmental stress. Curr. Opin. Plant Biol. 24: 125–135. [DOI] [PubMed] [Google Scholar]
- Fortes P., Bilbao-Cortés D., Fornerod M., Rigaut G., Raymond W., Séraphin B., Mattaj I.W. (1999b). Luc7p, a novel yeast U1 snRNP protein with a role in 5′ splice site recognition. Genes Dev. 13: 2425–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P., Kufel J., Fornerod M., Polycarpou-Schwarz M., Lafontaine D., Tollervey D., Mattaj I.W. (1999a). Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol. Cell. Biol. 19: 6543–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumura K., Taniguchi I., Sakamoto H., Ohno M., Inoue K. (2009). U1-independent pre-mRNA splicing contributes to the regulation of alternative splicing. Nucleic Acids Res. 37: 1907–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhring J., Jacak J., Barta A. (2014). Imaging of endogenous messenger RNA splice variants in living cells reveals nuclear retention of transcripts inaccessible to nonsense-mediated decay in Arabidopsis. Plant Cell 26: 754–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin M., Reddy A.S. (1998). The plant U1 small nuclear ribonucleoprotein particle 70K protein interacts with two novel serine/arginine-rich proteins. Plant Cell 10: 1637–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin M., Reddy A.S. (2003). Expression of U1 small nuclear ribonucleoprotein 70K antisense transcript using APETALA3 promoter suppresses the development of sepals and petals. Plant Physiol. 132: 1884–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonatopoulos-Pournatzis T., Cowling V.H. (2014). Cap-binding complex (CBC). Biochem. J. 457: 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A., Tang J., Puig O., Salgado J., Neubauer G., Colot H.V., Mann M., Séraphin B., Rosbash M., Lührmann R., Fabrizio P. (1998). A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA 4: 374–393. [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. (2010). SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Gu J., Xia Z., Luo Y., Jiang X., Qian B., Xie H., Zhu J.K., Xiong L., Zhu J., Wang Z.Y. (2018). Spliceosomal protein U1A is involved in alternative splicing and salt stress tolerance in Arabidopsis thaliana. Nucleic Acids Res. 46: 1777–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiro J., O’Reilly D. (2015). Insights into the U1 small nuclear ribonucleoprotein complex superfamily. Wiley Interdiscip. Rev. RNA 6: 79–92. [DOI] [PubMed] [Google Scholar]
- Hartmann L., Wießner T., Wachter A. (2018). Subcellular compartmentation of alternatively spliced transcripts defines SERINE/ARGININE-RICH PROTEIN30 expression. Plant Physiol. 176: 2886–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood J.L., Durek P., Hummel J., Selbig J., Weckwerth W., Walther D., Schulze W.X. (2008). PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res. 36: D1015–D1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim A., Grimm C., Müller U., Häußler S., Mackeen M.M., Merl J., Hauck S.M., Kessler B.M., Schofield C.J., Wolf A., Böttger A. (2014). Jumonji domain containing protein 6 (Jmjd6) modulates splicing and specifically interacts with arginine-serine-rich (RS) domains of SR- and SR-like proteins. Nucleic Acids Res. 42: 7833–7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández H., Makarova O.V., Makarov E.M., Morgner N., Muto Y., Krummel D.P., Robinson C.V. (2009). Isoforms of U1-70k control subunit dynamics in the human spliceosomal U1 snRNP. PLoS One 4: e7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B.E., Grabowski P.J. (1992). U1 snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev. 6: 2554–2568. [DOI] [PubMed] [Google Scholar]
- Hori K., Watanabe Y. (2005). UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 43: 530–540. [DOI] [PubMed] [Google Scholar]
- Kaida D. (2016). The reciprocal regulation between splicing and 3′-end processing. Wiley Interdiscip. Rev. RNA 7: 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D., Berg M.G., Younis I., Kasim M., Singh L.N., Wan L., Dreyfuss G. (2010). U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M., et al. (2012). Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40: 2454–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T., Lin W.D., Fu J.L., Chang C.L., Matzke A.J.M., Matzke M. (2017). A genetic screen for pre-mRNA splicing mutants of Arabidopsis thaliana identifies putative U1 snRNP components RBM25 and PRP39a. Genetics 207: 1347–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan C.D., Morris J.R., Wu C., Winston F. (2000). Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 14: 2623–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P.J., et al. (2016). Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res. 44: D574–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop K., Stepien A., Barciszewska-Pacak M., Taube M., Bielewicz D., Michalak M., Borst J.W., Jarmolowski A., Szweykowska-Kulinska Z. (2016). Active 5′ splice sites regulate the biogenesis efficiency of Arabidopsis microRNAs derived from intron-containing genes. Nucleic Acids Res. pii: gkw895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Dejong F., Villacorta N., Szakonyi D., Koncz Z. (2012). The spliceosome-activating complex: molecular mechanisms underlying the function of a pleiotropic regulator. Front. Plant Sci. 3: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y., Oubridge C., van Roon A.M., Nagai K. (2015). Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. eLife 4: 4:e04986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt A.R., Schor I.E., Alló M., Dujardin G., Petrillo E., Muñoz M.J. (2013). Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 14: 153–165. [DOI] [PubMed] [Google Scholar]
- Laubinger S., Sachsenberg T., Zeller G., Busch W., Lohmann J.U., Rätsch G., Weigel D. (2008). Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 8795–8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.D., Izaurralde E., Jarmolowski A., McGuigan C., Mattaj I.W. (1996). A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 10: 1683–1698. [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. (2013). The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y., et al. (2017). Pre-mRNA splicing repression triggers abiotic stress signaling in plants. Plant J. 89: 291–309. [DOI] [PubMed] [Google Scholar]
- Lorković Z.J., Barta A. (2008). Role of Cajal bodies and nucleolus in the maturation of the U1 snRNP in Arabidopsis. PLoS One 3: e3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez Y., Höpfler M., Ayatollahi Z., Barta A., Kalyna M. (2015). Unmasking alternative splicing inside protein-coding exons defines exitrons and their role in proteome plasticity. Genome Res. 25: 995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcos P.A. (2007). Achieving targeted and quantifiable alteration of mRNA splicing with Morpholino oligos. Biochem. Biophys. Res. Commun. 358: 521–527. [DOI] [PubMed] [Google Scholar]
- Müller-McNicoll M., Botti V., de Jesus Domingues A.M., Brandl H., Schwich O.D., Steiner M.C., Curk T., Poser I., Zarnack K., Neugebauer K.M. (2016). SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 30: 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., et al. (2007). Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100. [DOI] [PubMed] [Google Scholar]
- Naro C., Jolly A., Di Persio S., Bielli P., Setterblad N., Alberdi A.J., Vicini E., Geremia R., De la Grange P., Sette C. (2017). An orchestrated intron retention program in meiosis controls timely usage of transcripts during germ cell differentiation. Dev. Cell 41: 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer G., Gottschalk A., Fabrizio P., Séraphin B., Lührmann R., Mann M. (1997). Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc. Natl. Acad. Sci. USA 94: 385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani M., Sugiyama M. (2005). Involvement of SRD2-mediated activation of snRNA transcription in the control of cell proliferation competence in Arabidopsis. Plant J. 43: 479–490. [DOI] [PubMed] [Google Scholar]
- Papp I., Mur L.A., Dalmadi A., Dulai S., Koncz C. (2004). A mutation in the Cap Binding Protein 20 gene confers drought tolerance to Arabidopsis. Plant Mol. Biol. 55: 679–686. [DOI] [PubMed] [Google Scholar]
- Pimentel H., Parra M., Gee S.L., Mohandas N., Pachter L., Conboy J.G. (2016). A dynamic intron retention program enriched in RNA processing genes regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 44: 838–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge M.J., Wagner D.R. (2001). The Arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13: 1263–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Bragado-Nilsson E., Koski T., Séraphin B. (2007). The U1 snRNP-associated factor Luc7p affects 5′ splice site selection in yeast and human. Nucleic Acids Res. 35: 5874–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczynska K.D., Simpson C.G., Ciesiolka A., Szewc L., Lewandowska D., McNicol J., Szweykowska-Kulinska Z., Brown J.W., Jarmolowski A. (2010). Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 38: 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczynska K.D., Stepien A., Kierzkowski D., Kalak M., Bajczyk M., McNicol J., Simpson C.G., Szweykowska-Kulinska Z., Brown J.W., Jarmolowski A. (2014). The SERRATE protein is involved in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 42: 1224–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A.S., Marquez Y., Kalyna M., Barta A. (2013). Complexity of the alternative splicing landscape in plants. Plant Cell 25: 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17: 1030–1032. [DOI] [PubMed] [Google Scholar]
- Rigo F., Martinson H.G. (2008). Functional coupling of last-intron splicing and 3′-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage. Mol. Cell. Biol. 28: 849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösel T.D., Hung L.H., Medenbach J., Donde K., Starke S., Benes V., Rätsch G., Bindereif A. (2011). RNA-seq analysis in mutant zebrafish reveals role of U1C protein in alternative splicing regulation. EMBO J. 30: 1965–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley M.J., Böhmdorfer G., Wierzbicki A.T. (2013). Analysis of long non-coding RNAs produced by a specialized RNA polymerase in Arabidopsis thaliana. Methods 63: 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D., Bhanja Chowdhury J., Ghosh S. (2013). Polypyrimidine tract binding protein (PTB) associates with intronic and exonic domains to squelch nuclear export of unspliced RNA. FEBS Lett. 587: 3802–3807. [DOI] [PubMed] [Google Scholar]
- Schlaen R.G., Mancini E., Sanchez S.E., Perez-Santángelo S., Rugnone M.L., Simpson C.G., Brown J.W., Zhang X., Chernomoretz A., Yanovsky M.J. (2015). The spliceosome assembly factor GEMIN2 attenuates the effects of temperature on alternative splicing and circadian rhythms. Proc. Natl. Acad. Sci. USA 112: 9382–9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Speth C., Laubinger S., Voinnet O. (2013). Enhanced microRNA accumulation through stemloop-adjacent introns. EMBO Rep. 14: 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W., Kim H.S., Cao Y., Xu Y.Z., Query C.C. (2012). A U1-U2 snRNP interaction network during intron definition. Mol. Cell. Biol. 32: 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul O. (2015). Unique aspects of plant nonsense-mediated mRNA decay. Trends Plant Sci. 20: 767–779. [DOI] [PubMed] [Google Scholar]
- Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y. (2014). rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 111: E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D., Brown J.W. (2013). Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25: 3640–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien A., Knop K., Dolata J., Taube M., Bajczyk M., Barciszewska-Pacak M., Pacak A., Jarmolowski A., Szweykowska-Kulinska Z. (2017). Posttranscriptional coordination of splicing and miRNA biogenesis in plants. Wiley Interdiscip. Rev. RNA 8: 10.1002/wrna.1403. [DOI] [PubMed] [Google Scholar]
- Sullivan C., Howard P.L. (2016). The diverse requirements of ARS2 in nuclear cap-binding complex-dependent RNA processing. RNA Dis. 3: e1376. [Google Scholar]
- Sun M., Larivière L., Dengl S., Mayer A., Cramer P. (2010). A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II C-terminal repeat domain (CTD). J. Biol. Chem. 285: 41597–41603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Takahashi F., Anderson J.C., Ishihama Y., Peck S.C., Shinozaki K. (2013). Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6: rs8. [DOI] [PubMed] [Google Scholar]
- Wachter A., Tunc-Ozdemir M., Grove B.C., Green P.J., Shintani D.K., Breaker R.R. (2007). Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell 19: 3437–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M.C., Will C.L., Lührmann R. (2009). The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Wang B.B., Brendel V. (2004). The ASRG database: identification and survey of Arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol. 5: R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Tian Q., Hou Z., Mucha M., Aukerman M., Olsen O.A. (2007). The Arabidopsis thaliana AT PRP39-1 gene, encoding a tetratricopeptide repeat protein with similarity to the yeast pre-mRNA processing protein PRP39, affects flowering time. Plant Cell Rep. 26: 1357–1366. [DOI] [PubMed] [Google Scholar]
- Wang P., Xue L., Batelli G., Lee S., Hou Y.J., Van Oosten M.J., Zhang H., Tao W.A., Zhu J.K. (2013). Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 110: 11205–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webby C.J., et al. (2009). Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325: 90–93. [DOI] [PubMed] [Google Scholar]
- Will C.L., Lührmann R. (2011). Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. pii: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J.J., Au A.Y., Ritchie W., Rasko J.E. (2016). Intron retention in mRNA: No longer nonsense: Known and putative roles of intron retention in normal and disease biology. BioEssays 38: 41–49. [DOI] [PubMed] [Google Scholar]
- Workman E., Veith A., Battle D.J. (2014). U1A regulates 3′ processing of the survival motor neuron mRNA. J. Biol. Chem. 289: 3703–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D., Wang Y., Hamilton M., Ben-Hur A., Reddy A.S. (2015). Transcriptome-wide identification of RNA targets of Arabidopsis SERINE/ARGININE-RICH45 uncovers the unexpected roles of this RNA binding protein in RNA processing. Plant Cell 27: 3294–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K., Lim Z.Q., Khandelia P., Friedman B., Makeyev E.V. (2012). Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 26: 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoine M., Ohto M.A., Onai K., Mita S., Nakamura K. (2006). The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 47: 49–62. [DOI] [PubMed] [Google Scholar]
- Zhan X., Qian B., Cao F., Wu W., Yang L., Guan Q., Gu X., Wang P., Okusolubo T.A., Dunn S.L., Zhu J.K., Zhu J. (2015). An Arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and ABA responses. Nat. Commun. 6: 8139. [DOI] [PMC free article] [PubMed] [Google Scholar]