Abstract
Different approaches have investigated the effects of different extracellular matrices (ECMs) and three-dimensional (3D) culture on islet function, showing encouraging results. Ideally, the proper scaffold should mimic the biochemical composition of the native tissue as it drives numerous signaling pathways involved in tissue homeostasis and functionality. Tissue-derived decellularized biomaterials can preserve the ECM composition of the native tissue making it an ideal scaffold for 3D tissue engineering applications. However, the decellularization process may affect the retention of specific components, and the choice of a proper detergent is fundamental in preserving the native ECM composition. In this study, we evaluated the effect of different decellularization protocols on the mechanical properties and biochemical composition of pancreatic ECM (pECM) hydrogels. Fresh porcine pancreas tissue was harvested, cut into small pieces, rinsed in water, and treated with two different detergents (sodium dodecyl sulfate [SDS] or Triton X-100) for 1 day followed by 3 days in water. Effective decellularization was confirmed by PicoGreen assay, Hoescht, and H&E staining, showing no differences among groups. Use of a protease inhibitor (PI) was also evaluated. Effective decellularization was confirmed by PicoGreen assay and hematoxylin and eosin (H&E) staining, showing no differences among groups. Triton-treated samples were able to form a firm hydrogel under appropriate conditions, while the use of SDS had detrimental effects on the gelation properties of the hydrogels. ECM biochemical composition was characterized both in the fresh porcine pancreas and all decellularized pECM hydrogels by quantitative mass spectrometry analysis. Fibrillar collagen was the major ECM component in all groups, with all generated hydrogels having a higher amount compared with fresh pancreas. This effect was more pronounced in the SDS-treated hydrogels when compared with the Triton groups, showing very little retention of other ECM molecules. Conversely, basement membrane and matricellular proteins were better retained when the tissue was pretreated with a PI and decellularized in Triton X-100, making the hydrogel more similar to the native tissue. In conclusion, we showed that all the protocols evaluated in the study showed effective tissue decellularization, but only when the tissue was pretreated with a PI and decellularized in Triton detergent, the biochemical composition of the hydrogel was closer to the native tissue ECM.
Impact Statement
The article compares different methodologies for the generation of a pancreas-derived hydrogel for tissue engineering applications. The biochemical characterization of the newly generated hydrogel shows that the material retains all the extracellular molecules of the native tissue and is capable of sustaining functionality of the encapsulated beta-cells.
Keywords: decellularization, hydrogel, diabetes, tissue engineering, extracellular matrix, cell encapsulation, pancreas
Introduction
Diabetes is a metabolic disorder that affects an estimated 30.3 million people of the United States (9.4%), making it the seventh leading cause of death (https://www.cdc.gov/diabetes/pdfs/data/statistics/national-diabetes-statistics-report.pdf). This disease ultimately results from insufficient function of pancreatic beta-cells, either due to autoimmune attack (in type 1 diabetes) or excessive insulin demand that exceeds compensatory insulin secretion (in type 2 diabetes).1 A major obstacle hampering progress in this field is the lack of understanding of the human beta-cell pathophysiology in an in vitro culture system, which can facilitate the development of alternative therapeutic products. Following isolation of human islets, it has been demonstrated that beta-cell death occurs when the islets are cultured in vitro.2 The main causes are loss of vasculature and subsequent hypoxia, loss of cell/cell interactions, as well as disruption of the peri-insular basal lamina and loss of cell/matrix interactions, leading to apoptosis.3–5
Recently, different approaches investigating the effects of the extracellular matrix (ECM) compositions and three-dimensional (3D) culture on islet function have shown encouraging results in terms of beta-cell survival and functionality when compared with standard culture conditions.6,7 In particular, basement membrane proteins have been shown to play an important role in maintaining beta-cell functionality, both in vivo8 and in vitro.9–12
Among the different scaffolds evaluated for 3D culture of islets or beta-cell lines, several decellularized tissues have recently been explored, including the pancreas,13–22 liver,23 lung,21 or small intestine submucosa.24,25 Tissue-derived biomaterials can closely mimic the microenvironment of the endogenous tissue, making them potentially more advantageous for tissue repair or in vitro models compared with other biologic or synthetic scaffolds.26–34 Sackett et al. recently developed a new protocol to successfully remove lipids from human pancreatic tissue and generate a pancreas-derived hydrogel. The authors showed that the hydrogel retained all the major ECM components of the native tissue and is capable of sustaining cell growth, both in vivo and in vitro, and could represent a valid platform for diabetes tissue engineering.22
In this regard, using human-derived decellularized hydrogel may have the advantage to better recapitulate the native ECM composition compared with nonhuman sources. However, the recruitment of pancreatic human tissue and the variability of human samples, such as seen with other decellularized tissues,35 could also be a disadvantage. The use of porcine tissue could represent a valid alternative due to the easier accessibility of this source and less animal to animal variability, in turn decreasing the batch-to-batch variability. Pancreas-derived scaffolds in particular have been generated using a combination of different detergents, such as Triton X-100,13–19 sodium dodecyl sulfate (SDS),13,17,18 and sodium deoxycholate,22 or enzymes, such as trypsin,19 as well as hyper/hypotonic solutions.20 However, to our knowledge, no studies have directly compared different detergents to generate an optimized protocol for the pancreas nor have they used reagents to prevent proteolytic degradation, or performed quantitative proteomic analysis to inform this process.
It has previously been shown with other tissues that the detergent and decellularization time can affect the retention and structural integrity of ECM proteins or the presence of residual detergent in the tissue.36–38 Therefore, to generate a tissue-derived scaffold that closely mimics the native microenvironment, retains the proper biochemical complexity and bioinductive properties of the native ECM, and sustains cell functionality, a proper evaluation of the ideal detergent, timing, tissue characteristics, and most importantly, an accurate characterization of ECM biochemical composition is fundamental. The pancreas also presents additional challenges compared with other tissues since it is rich in digestive enzymes, consisting mainly of different proteases,39 which could degrade the ECM during processing. The use of cold perfusion and protease inhibitors (PI) has been suggested as an advantageous approach to prevent damage to native ECM,40 and has been shown to better preserve the tissue ultrastructure.16 However, the effect of PI on ECM retention and composition has not been demonstrated.
The aim of this work was (1) to provide a new methodology to generate decellularized pancreas and further process it into a pancreatic ECM (pECM) hydrogel; (2) to quantitatively characterize the ECM biochemical composition of the decellularized pECM compared with native pancreas tissue and evaluate how different detergents and the use of a PI affect ECM retention; and (3) to evaluate the effects of the different processing methods on the gelation properties of generated hydrogels.
Materials and Methods
Generation of decellularized pECM hydrogel
Tissue collection and processing
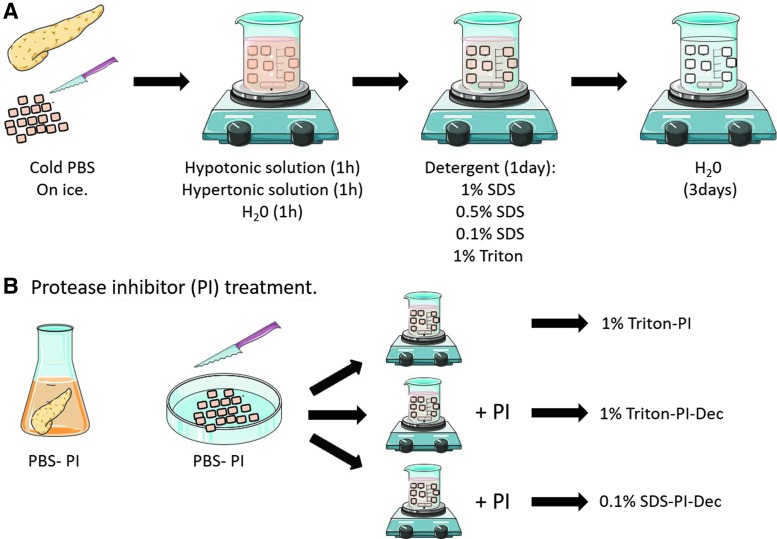
Pancreas tissue was obtained from Yorkshire farm pigs weighing ∼30–45 kg, immediately after euthanasia. The pancreas was briefly rinsed in water and stored in cold phosphate-buffered saline (PBS) on ice until tissue processing. Large vessels and connective tissue were removed, and the remaining tissue frozen at −20°C for at least 24 h. The frozen tissue is then thawed on ice and cut into small pieces, ∼2–3 mm in size (Figs. 1A and 2B). The tissue was weighed, and 25–30 g of tissue was placed into a 1 L sterile beaker for decellularization (Fig. 1A). Nondecellularized tissue was also collected and frozen in Tissue-Tek O.C.T. freezing medium (Sakura) for histological analysis as a “before decellularization” control sample.
FIG. 1.
Overview of the experimental approaches used for the generation of a pancreas-derived hydrogel. (A) Overview of the decellularization steps; (B) overview of the protease inhibitor treatment. Color images available online at www.liebertpub.com/tec
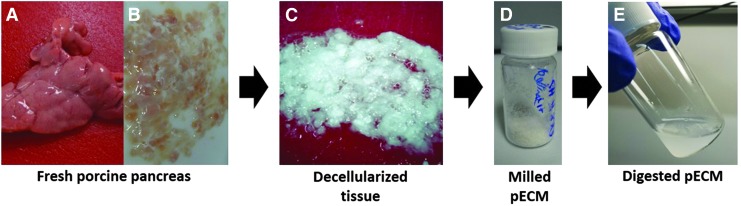
FIG. 2.
Representative images of pancreas decellularization. (A) Freshly harvested pancreas; (B) chopped tissue before decellularization; (C) decellularized pancreas; (D) lyophilized—milled pECM; (E) pepsin-digested pECM. pECM, pancreatic extracellular matrix. Color images available online at www.liebertpub.com/tec
Tissue decellularization
The tissue was stirred at 125 rpm in hypotonic solution (0.34 M NaCl; Fisher) followed by hypertonic solution (1.28 M NaCl) and Milli-Q water for 1 h each so that the final volume of tissue and solutions was 800 mL in each beaker (Fig. 1A). At the end of each washing step, the tissue was strained in an autoclaved fine mesh strainer, rinsed under water, and returned to the beaker for treatment. Decellularization was performed using four different conditions, and two different detergents were tested: 0.1%, 0.5%, and 1% wt./vol. SDS (Fisher) or 1% wt./vol. Triton X-100 (Sigma) in PBS (Fig. 1A). Penicillin and streptomycin (Invitrogen) were then added at a final concentration of 50 IU (international units)/mL. The tissue was decellularized for 24 h with a solution change after 12 ± 2 h. After the detergent treatment, the tissue was strained, rinsed thoroughly with water, placed into a new sterile beaker, and stirred for an additional 3 days in Milli-Q water (Fig. 1A). The solution was changed every 12 ± 2 h. As the pancreatic tissue clumped together during decellularization, it is important that the tissue is further cut into small pieces during the solution change to assure proper decellularization. After 3 days in water, the tissue was further rinsed in Milli-Q water and placed into an autoclaved 1 L bottle with water up to 800 mL total volume. The bottle was sealed, shaken vigorously for 30 s, and rinsed again in water. This process was repeated several times until no bubbles, indicative of residual detergent, were observed after shaking. The fully decellularized tissue (Fig. 2C) was then divided into 50 mL conical tubes and stored at −80°C. A few pieces of decellularized ECM were also frozen in Tissue-Tek O.C.T freezing medium for histological analysis. Frozen ECM was lyophilized and milled (No. 60 filter; Wiley Mini-Mill) to generate a fine powder for subsequent protease digestion (Fig. 2D). Decellularized tissues from four to six different pancreata were combined and milled together to reduce batch-to-batch variability and avoid tissue loss during milling. The milled lyophilized tissue was stored at −80°C. Four batches of pECM hydrogel were generated.
Tissue digestion
To generate a hydrogel form, the ECM was digested in pepsin (Sigma) for 48 h. Fresh pepsin was dissolved in 0.1 M HCl at 1 mg/mL final concentration through vortexing until no particles were visible and then sterile filtered using a 0.22 μm syringe filter (Millipore) (Fig. 2E). The ECM was aliquoted into 20 mL sterile scintillation vials (20–30 mg each vial), and the pepsin solution was added into each vial at a final concentration of 10 mg ECM/mL pepsin solution and stirred for 48 h. After digestion, the liquid ECM was neutralized to physiological pH using 1 M NaOH and 0.1 M HCl. Then, salt conditions were adjusted with 10 × PBS and the samples were diluted in water to a final concentration of 6 mg/mL and 1 × PBS.41 The digested, neutralized ECM was further aliquoted into 1.5 mL microcentrifuge tubes, frozen, lyophilized, and stored at −80°C for long-term storage. When ready to use, sterile water was added to the ECM at a desired concentration.
PI treatment
The effects of a PI on ECM retention were also evaluated. Freshly collected pancreas tissue was collected and rinsed briefly in water. The pancreas was then treated with 0.1 mM PI solution (Gabexate; MedChem Express) for 30 min and processed as previously described by chopping the tissue while still partly immersed in the PI buffer (Fig. 1B). The chopped tissue was then decellularized in 1% Triton (1% Triton-PI) as previously described. Alternatively, we also evaluated if using the PI for the entire decellularization process was more advantageous compared with the initial treatment only. In these cases, PI was added during the detergent and the 3-day water washing steps, and two different conditions were tested: 1% Triton (1% Triton-PI-Dec) and 0.1% SDS (0.1% SDS-PI-Dec). After decellularization, the ECM was then lyophilized, milled, digested, and stored at −80°C.
Decellularization evaluation
The effective decellularization and ECM characterization were evaluated through different assays as previously described.41
Hematoxylin and eosin staining
Fresh and decellularized pancreatic tissue was frozen in Tissue-Tek O.C.T. freezing medium and sectioned into 10-μm-thick slices. Hematoxylin and eosin (H&E) staining was performed to evaluate tissue morphology and the presence of hematoxylin-labeled nuclei for overall decellularization. The stained slides were imaged on a Leica Aperio ScanScope CS2 at 20 × magnification (Leica Biosystems).
Hoechst staining
Fresh and decellularized pancreatic tissue was also stained with Hoechst 33342 nuclear staining to further evaluate the presence of residual DNA. Tissue slices were hydrated 5 min in PBS before incubation with Hoechst (1 μg/mL) for 10 min. After incubation, the sections were washed once in PBS (5 min), rinsed in deionized water, and mounted with Fluoromount. Images were taken using a Zeiss fluorescence microscope (Carl Zeiss, Dublin, CA). Three images of each condition were taken and two pECM batches were tested.
Double-stranded DNA quantification
To further evaluate the presence of double-stranded DNA (dsDNA) in the decellularized ECM, DNA was extracted from 1 mg digested decellularized ECM using a standard DNA isolation kit (NucleoSpin; Macherey-Nagel, Bethlehem, PA). Briefly, the pECM aliquots were resuspended in proteinase K and lysis buffer T1 by pipetting the solubilized pECM several times. Effective material digestion was visually confirmed before extracting the dsDNA according to the manufacturer's datasheet. The amount of extracted DNA in a 100 μL sample was measured by PicoGreen fluorescent dye according to kit instructions. DNA was quantified for all batches (0.1% SDS-PI-Dec and 1% Triton-PI-Dec, n = 3; native pancreas, n = 2; all other groups, n = 4).
Decellularized tissue characterization
Gelation assay
Each batch of ECM was evaluated for its ability to form a gel under appropriate conditions. The digested lyophilized pECM aliquots were resuspended in sterile Milli-Q water at a final concentration of 6 mg/mL and final volume of 400 μL in a 1.5 mL microcentrifuge tube. pECM rehydration in water was performed on ice and the solution was pipetted up and down every 5 min until no ECM particles were visible (30–45 min in total). The solubilized pECM was then incubated for 1 h or overnight (16 ± 2 h) at 37°C. Gelation was evaluated by flipping the tube upside down and observing motion of a gel (with minimal displacement) versus a liquid solution (with fluid motion).
Turbidimetric assay
Turbidimetric gelation kinetics was performed as previously described.42 pECM hydrogels were resuspended in water at 6 mg/mL as previously described and 100 μL was pipetted into each well of a 96-well plate. The plate was then placed into a preheated (37°C) spectrophotometer (Tecan Infinite M200 Pro), absorbance was measured at 405 nm every minute for 90 min, and data were collected with the i-control 2.0 software. Data were normalized using Equation (1), where A is the absorbance at a specific time point, A0 is the minimum absorbance, and Amax is the maximum absorbance.
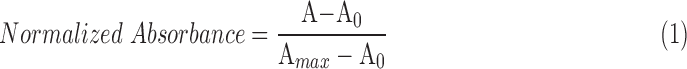 |
Normalized data were used to calculate the parameters of gelation speed (S), t1/2, and tlag. S was generated by identifying the growth region of the gelation profile and determining the slope of a linear approximation. t1/2 was defined as the time at which the absorbance value is 50% of the maximum absorbance. tlag was defined as the time required for gelation to begin and was determined by calculating the x-intercept of the linear region of the curve. Samples from each of the protocol conditions were tested in triplicate and averaged. Two different pECM batches were tested.
Sulfated glycosaminoglycan quantification
The retention of sulfated glycosaminoglycans (sGAGs) was evaluated by 1,9-dimethylmethylene blue dye (DMMB) as previously described.41 pECM samples were resuspended at 6 mg/mL and run in triplicate samples of 100 μL per microcentrifuge tube. A standard curve was generated using chondroitin sulfate dilutions from 0 to 50 μg and used to determine sGAG concentration in the samples. Absorbance was read at 656 nm using a Synergy 4 Multi-Mode Microplate Reader (BioTek). sGAG content was quantified for each batch (n = 3 0.1% SDS-PI-Dec and 1% Triton-PI-Dec, n = 2 native pancreas; n = 4 all other groups).
Quantitative mass spectrometry analysis
The biochemical composition of the decellularized pECM was evaluated by liquid chromatography/tandem mass spectrometry (LC-MS/MS) analysis as previously described.35 Briefly, three batches of pECM were pooled together and chemically digested in 100 mM cyanogen bromide (CNBr) in 86% trifluoroacetic acid (TFA). After neutralization in 1 M Tris-HCl pH 8.0 and protein quantification, 50 μg of protein was digested according to the FASP protocol using a 10 kDa molecular weight cutoff filter and supplemented with 13C6-labeled QconCAT peptides as previously described.43 High pH reversed-phase chromatography was performed on a Gemini C18, the collected fractions were pooled into 6 fractions, and Global LC-MS/MS was performed using the LQT Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). Selected reaction monitoring (SRM) was performed on the QTRAP® 5500 triple quadrupole mass spectrometer (AB Sciex) coupled with an Eksigent nanoLC-2D and Agilent 1200 LC system. Experimental condition and data analysis were performed as previously described.43 The generated data were searched against the UniProtKB/Swiss-Prot Sus scrofa protein sequence database, and peptide identifications were accepted at >99.0% confidence interval or >95.0% with at least two peptides per protein resulting in a false discovery rate (FDR) of 0.1%. Skyline v2.2 software was used for data processing,44 and quantification was based on the ratio of the 12C peptide to the corresponding 13C peptide from QconCATs.43
Mechanical properties
The gel stiffness was measured on a parallel plate rheometer (ARG2 Rheometer; TA Instruments). Five hundred microliters of resuspended digested ECM sample (6 mg/mL) was added to a 4 mL scintillation vial (Thermo Scientific), incubated overnight at 37°C, and used for mechanical testing. The parallel plate rheometer was set at 37°C and a gap height of 1200 μm. The storage and loss moduli were measured by an oscillatory frequency sweep from 0.25 to 100 rad/s.
Cell viability assay
Hydrogel biocompatibility was tested using the INS1 rat beta-cell line. The cells were stained with calcein-AM cell viability dye (25 nm; eBioscience) for 30 min, resuspended in 6 mg/mL pECM hydrogel, incubated for 10 min on ice, and four images were taken per condition using a fluorescence microscope (Carl Zeiss). Cells resuspended in media only were used as a control. The percentage of viable cells was determined by counting the stained cells over the total number of cells. Biocompatibility assays were performed for every batch of decellularized pECM (n = 4).
3D in vitro assay
Based on the gelation assay and mass spectrometry analysis, only the pECM generated from pancreata treated with PI after tissue harvesting and processing only, and decellularized in 1% Triton (1%-Triton-PI), was evaluated for 3D encapsulation experiments.
Cell culture
The rat insulinoma cell line INS1832/13 was used to evaluate whether the generated pECM hydrogel supported 3D cell encapsulation and functionality in vitro. The cells were cultured in RPMI 1640 with 11.1 mM d-glucose and 2 mM l-glutamine (Gibco, Life Technologies), supplemented with 10% fetal bovine serum (Gibco), 100 U/mL penicillin/streptomycin (Gibco, Life Technologies), 10 mM HEPES (Gibco), 1 mM sodium pyruvate (Corning Cellgro), and 50 μM β-mercaptoethanol (Gibco). Cells were cultured in 100 mm Petri dishes and incubated at 37°C in a humidified air atmosphere containing 5% CO2.
Cell encapsulation
For the 3D culture encapsulation, pECM was resuspended in sterile water at a final concentration of 6 mg/mL as previously described. INS1 cells, 2.5 × 105, were resuspended in 25 μL pECM and incubated at 37°C for 45 min to allow gelation of cell-pECM mixture before adding the cell culture media. Cells at passages 15–20 were used for all experiments.
Cell viability assay
To evaluate cell viability and morphological changes over time, INS1-encapsulated cells were stained with calcein-AM cell viability dye (25 nm; eBioscience) and Hoechst 33342 nuclear staining (1 μg/mL) for 30 min at 37°C. After incubation, the gels were washed with PBS and imaged immediately under a Leica confocal microscope after 1, 4, 7, and 10 days in culture. Twenty-five z-stack images were taken for each gel. Cellular density in each image was calculated using ImageJ software analysis and averaged to determine cellular density within the hydrogel. To evaluate any differences in cellular density within each hydrogel, five z-stack images from the top, cross-section, or bottom view were used. Each experiment was performed in triplicate and two different batches were tested.
Glucose-stimulated insulin secretion assay
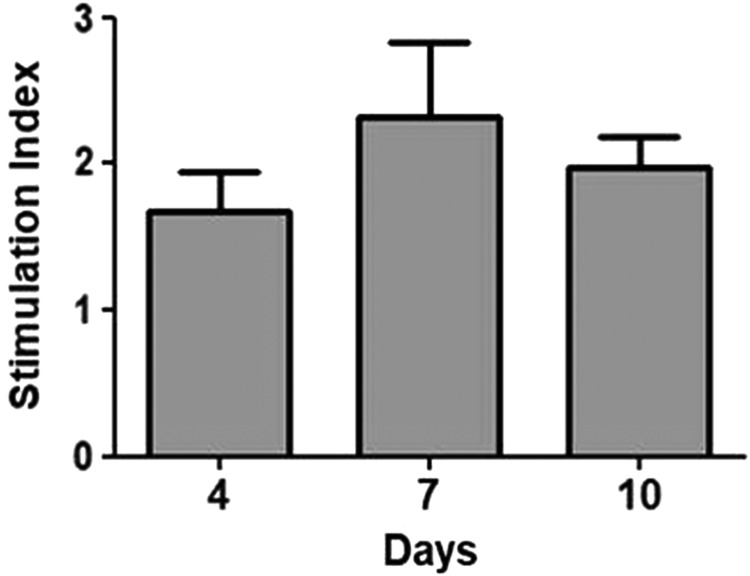
To evaluate if the hydrogel would sustain functionality of encapsulated INS1 cells, a glucose-stimulated insulin secretion (GSIS) assay was performed on days 4, 7, and 10 after encapsulation. Cells were washed briefly with PBS and starved for 1 h in low-glucose Krebs/Ringer Buffer (KRB) (130 mM NaCl, 5 mM KCl, 1.2 mM CaCl2, 1.2 mM MgCl2, 1.2 mM KH2PO4, 20 mM HEPES pH 7.4, 25 mM NaHCO3, 0.1% bovine serum albumin, and 2.8 mM glucose in H2O) at 37°C, followed by a 1-h stimulation in low-glucose KRB (2.8 mM glucose) and then a 1-h stimulation in high-glucose KRB (16.8 mM glucose). Both low- and high-glucose KRB buffers were collected after stimulation and stored at −20°C. New culture media were added to encapsulated cells. Insulin concentration was assessed using the mouse ultrasensitive enzyme-linked immunosorbent assay (Alpco) according to the manufacturer's protocol. Cell functionality was expressed as stimulation index (SI) and calculated by dividing the insulin concentration obtained after high-glucose stimulation over the insulin concentration of the low-glucose stimulation. Four gels were tested for each experiment, and 4 different experiments were performed.
Data analysis
All data are expressed as mean ± standard deviation. An unpaired Student's t-test was performed to compare two groups. One-way analysis of variance and Bonferroni post hoc test were used for multiple group comparisons. A p-value of <0.05 was considered significant.
Results
Pancreas decellularization
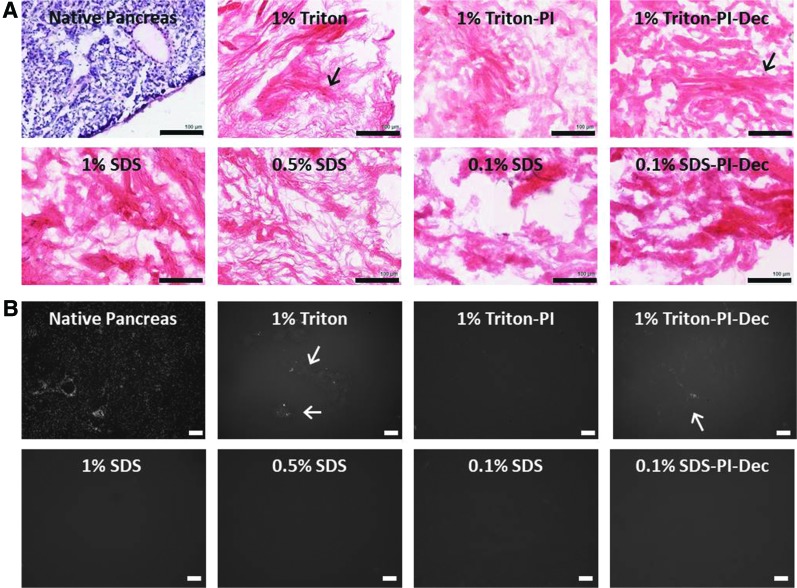
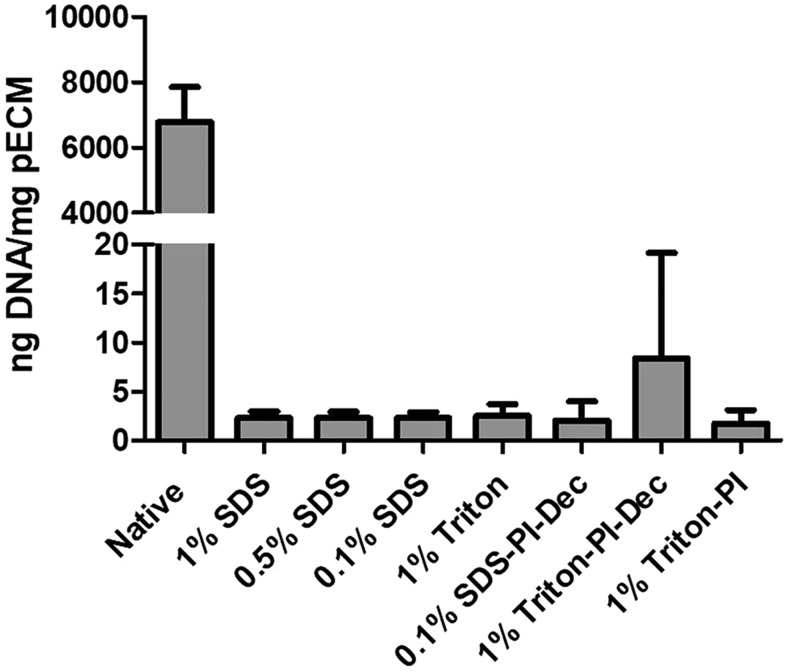
The effective decellularization of the pancreas tissue was first evaluated visually and then accessed via H&E staining and a PicoGreen assay to determine residual DNA content. Following decellularization, all the experimental groups analyzed lost the characteristic red/pink color of the fresh pancreas tissue, and the remaining ECM appeared white, indicative of a successful decellularization (Fig. 2A–C). Macroscopic observations were then further confirmed by the H&E and Hoechst nuclear staining analysis. No major histochemical differences were observed among the different groups (Fig. 3A, B). However, occasionally, the pancreas tissue treated with 1% Triton showed some hematoxylin- or Hoechst-stained areas that may be indicative of some residual DNA (Fig. 3A, B). The residual DNA content was further quantified by PicoGreen assay, both on predigested and postdigested pECM. No significant differences in DNA content were observed among groups in the predigested pECM. However, before the pepsin digestion step, the pancreas treated with 1% SDS showed a lower DNA content and less batch-to-batch variability when compared with other treatments (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/tec). After pepsin digestion, DNA content was further reduced and was similar among all the groups (1% Triton: 2.51 ± 1.18; 1% SDS: 2.03: ± 0.30; 0.5% SDS: 2.28 ± 0.66; 0.1% SDS 2.31 ± 0.60.18 ng DNA/mg pECM) (Fig. 4). Similar results were obtained when the pancreas was processed in the presence of PI, either when it was used during tissue processing and harvesting (1% Triton-PI: 1.71 ± 1.34 ng DNA/mg pECM) or for the entire length of the decellularization process (1% Triton-PI-Dec: 8.38 ± 10.77; 0.1% SDS-PI-Dec: 2.04 ± 1.97 ng DNA/mg pECM) (Fig. 4).
FIG. 3.
Histological characterization of fresh and decellularized pancreas. (A) Representative images of H&E staining of fresh and decellularized pancreas. (B) Representative images of Hoechst staining of fresh and decellularized pancreas. Arrow indicates potential residual DNA occasionally observed in Triton-treated samples. Scale bar = 100 μm. H&E, hematoxylin and eosin. Color images available online at www.liebertpub.com/tec
FIG. 4.
PicoGreen assay of pECM after pepsin digestion. PicoGreen assay of pECM after pepsin digestion. SDS-PI-Dec 0.1% and 1% Triton-PI-Dec, n = 3; native pancreas, n = 2; all other groups, n = 4. PI, protease inhibitor; SDS, sodium dodecyl sulfate.
Hydrogel characterization
Gelation assay and mechanical properties
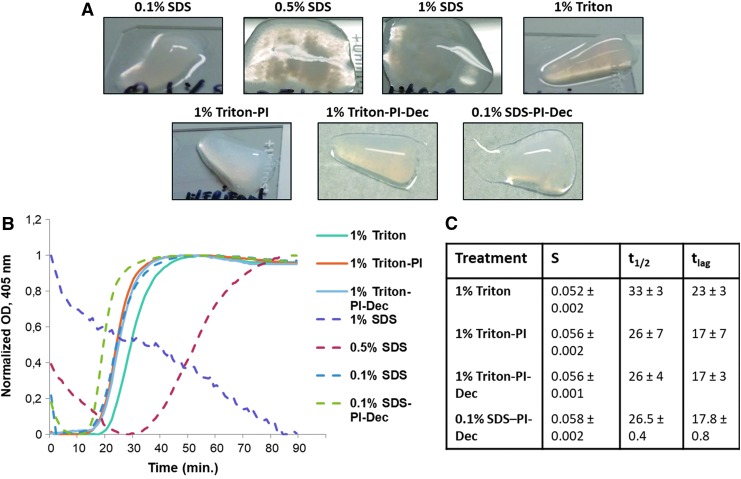
The ability of the different pECM hydrogels to gel under appropriate conditions was also evaluated. pECM was resuspended in water at 6 mg/mL and incubated at 37°C for 1 h. Only the pECM decellularized with the 1% Triton and 0.1% SDS group was capable of self-assembling into a firm gel with the 0.1% SDS groups showing also a small liquid fraction (Fig. 5A). Conversely, the pECM decellularized using 0.5% SDS showed only partial gelation with the majority of the material staying in a liquid form, and the 1% SDS group did not self-assemble (Fig. 5A). Similar results were observed when PI was used, with both Triton groups capable of forming a firm gel (Fig. 5A), and the SDS group showing only partial gelation (Fig. 5A). Increasing matrix concentration up to 10 mg/mL or incubation time up to 16 h did not result in any noticeable differences compared with 6 mg/mL pECM (data not shown). We further evaluated gelation kinetics of the different groups. Our data showed that all Triton and 0.1% SDS groups exhibited a sigmoidal curve in the gelation assay, similar to the curve usually observed with collagen, and with minor differences among groups. In particular, both Triton and PI samples showed a comparable gelation speed and lag time (time to upslope), while the 1% Triton group showed a trend of a slightly longer lag time, but with similar gelation speed (Fig. 5B, C). All SDS-treated groups showed a reduction in normalized absorbance at the beginning of the assay. The reduction was marginal in both 0.1% SDS groups and was followed by a sigmoidal curve with similar lag time compared with 1% Triton-PI-treated groups and a slightly higher gelation speed in the 0.1% SDS-PI group. Conversely, the normalized absorbance decreased up to 30 min from the beginning of the assay in the 0.5% SDS group, followed by a sigmoidal curve with much lower gelation speed compared with the Triton and 0.1% SDS groups (Fig. 5B), probably indicative of the partial gelation that we observed in this experimental group. Gelation kinetics of the 1% SDS group confirmed lack of gelation with the normalized absorbance values that decreased throughout the experiment (Fig. 5B). Based on these results, and the biochemical characterization of different hydrogels, only the rheological properties of the pECM-decellularized hydrogel using 1% Triton and PI after harvesting and processing (1% Triton-PI), used in the 3D in vitro encapsulation, were further analyzed, resulting in a storage modulus of 21.44 ± 0.77 Pa.
FIG. 5.
(A) Gelation assay of 6 mg/mL pECM hydrogels incubated at 37°C for 1 h. (B) Representative turbidimetric gelation kinetics of pECM hydrogels and (C) the calculated gelation speed (S), time to reach 50% of the maximum adsorbance (t1/2), and time to upslope (tlag) of two different batches of decellularized pancreatic hydrogels. Color images available online at www.liebertpub.com/tec
pECM biochemical characterization
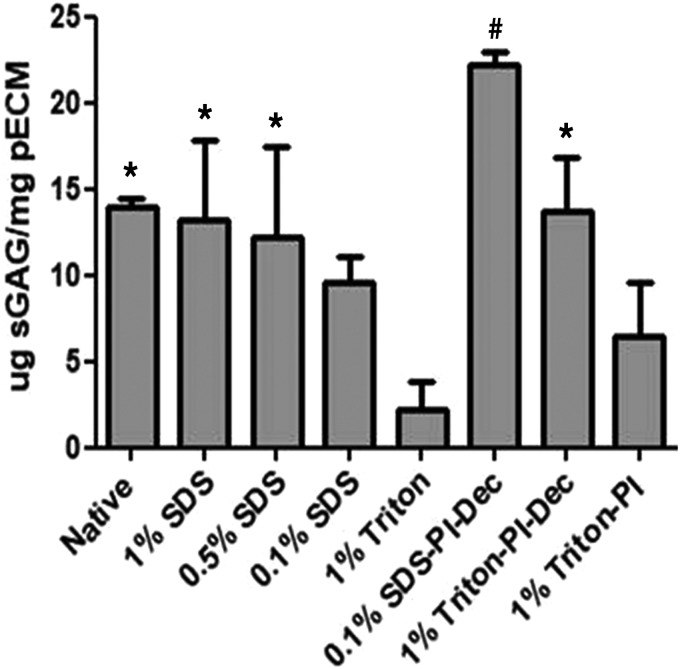
sGAG retention was evaluated with a DMMB assay. All groups analyzed retained sGAGs, with the 1% and 0.5% SDS-treated groups showing a significantly higher amount of the polysaccharides compared with the 1% Triton group (2.18 ± 1.68 μg sGAG/mg pECM). Interestingly, 0.5% SDS (12.23 ± 5.18 μg sGAG/mg pECM) and 1% SDS (13.26 ± 4.52 μg sGAG/mg pECM) groups showed a higher retention compared with the 0.1% SDS group (9.58 ± 1.48 μg sGAG/mg pECM), although not significant (Fig. 6). A similar trend was also observed when the tissue was harvested and treated with PI. The treatment resulted in an overall increase of the sGAG retention when compared with tissue harvested and processed without PI. The SDS-PI-Dec group also showed a significant increase compared with 1% Triton-PI (22.24 ± 0.66 vs. 6.44 ± 3.11 μg sGAG/mg pECM, respectively) and 1% Triton, while a nonsignificant increase compared with 1% Triton-PI-Dec (13.76 ± 3.18 μg sGAG/mg pECM) was also observed.
FIG. 6.
Sulfated glycosaminoglycan retention in the decellularized pECM hydrogels. One percent, 0.5%, and 0.1% SDS, and 1% Triton, n = 4; 1% Triton-PI, 0.1% SDS-PI-Dec, and 0.1% Triton-PI-Dec, n = 3; native pancreas, n = 2; *p ≤ 0.05 versus 1% Triton; #p ≤ 0.05 versus 1% Triton, 0.1% SDS, and 1% Triton-PI.
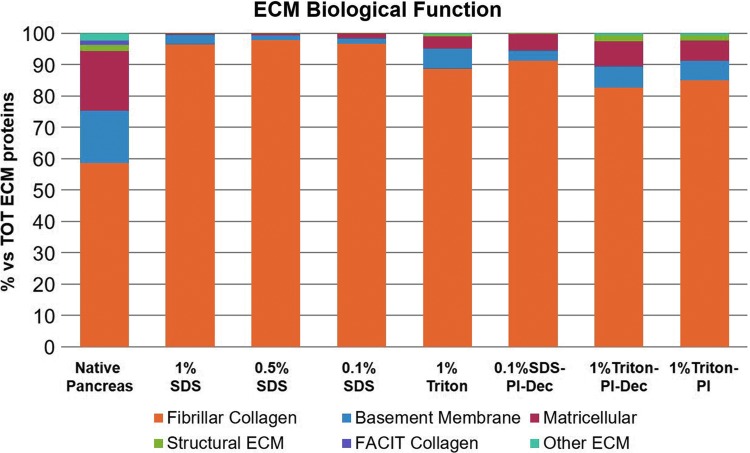
A quantitative evaluation of the biochemical composition of the different hydrogels was performed using quantitative mass spectrometry analysis and compared with fresh pancreas. The ECM proteins detected were further classified into six major categories based on their ECM biological function (Fig. 7, Supplementary Tables S1 and S2). Fibrillar collagens, mainly collagen I and V, were the most abundant ECM proteins in all groups, accounting for ∼90% or more of all ECM proteins (Fig. 7 and Supplementary Fig. S2). When compared with fresh tissue, the amount of fibrillar collagen was significantly higher in the decellularized hydrogels, indicating that the decellularization process significantly reduced other ECM protein classes, particularly basement membrane and matricellular proteins (Fig. 7). Among the basement membrane proteins, laminins were the most affected by decellularization, while the amount of collagen IV was comparable with the native tissue (Supplementary Fig. S3). The other major class of ECM proteins identified both in the native and decellularized pancreas consisted of matricellular proteins, particularly collagen VI (Fig. 7 and Supplementary Fig. S4), also reduced after decellularization. When comparing the different detergents used, all SDS-treated groups showed a severe reduction of both basement membrane and matricellular proteins. In particular, laminins were the most affected and were not detected in any SDS group analyzed. Interestingly, the use of PI had a beneficial effect on ECM retention, both on basement membrane and matricellular proteins (Fig. 7 and Supplementary Figs. S3 and S4). Laminin concentrations in the PI-treated Triton samples doubled compared with 1% Triton only (Supplementary Fig. S3), and a similar trend was also observed with collagen VI (Supplementary Fig. S4), although still reduced after decellularization compared with the native tissue. No major differences were observed when the PI was used only during harvesting and processing of the tissue (1% Triton-PI) or for the entire decellularization process (1% Triton-PI-Dec), suggesting that using PI during all of the decellularization procedure did not have any advantage compared with initial PI treatment only.
FIG. 7.
Quantitative proteomic analysis of the decellularized pECM hydrogels and native pancreas. ECM proteins were classified into six categories based on their biological function. Three pECM batches of each condition were pooled together for the proteomic analysis. Color images available online at www.liebertpub.com/tec
Viability assay
Biomaterial cytotoxicity was evaluated by incubation of pECM hydrogels with calcein-AM-labeled INS1 cells. All analyzed groups showed a good biocompatibility with cell viability ranging from ∼74% in the 0.5% and 1% SDS groups to ∼98% in the PBS and 1% Triton groups (Supplementary Fig. S5). However, when compared among different treatments, cellular viability in both 0.5% and 1% SDS groups was significantly lower compared with all Triton-treated groups and a nonsignificant decrease compared with 0.1% SDS (Supplementary Fig. S5).
3D in vitro assay
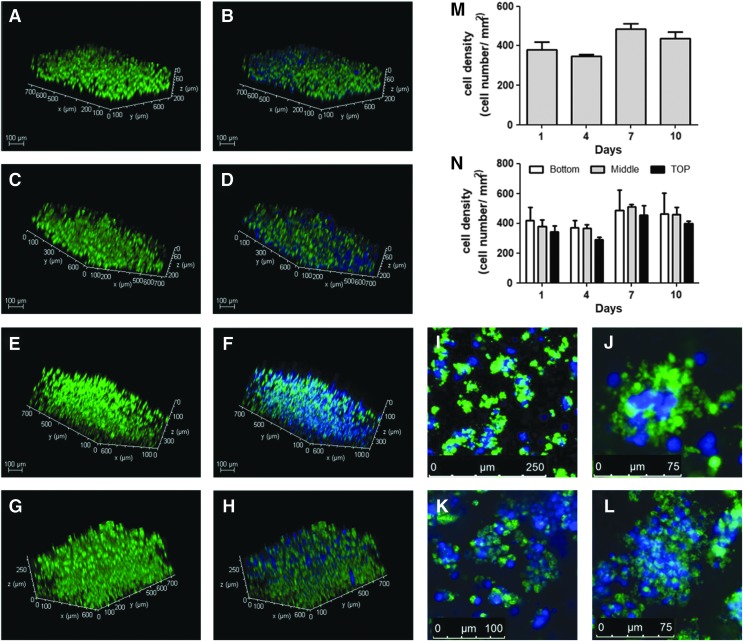
Among all the tested experimental conditions, only Triton-treated samples were capable of self-assembling into a firm gel, and the use of PI was required to preserve the ECM biochemical composition. Since no major differences were observed if the PI was used only at the initial stage or during the entire decellularization process, only the samples treated with PI after harvesting the tissue and during tissue processing (1% Triton-PI group) were used for 3D encapsulation studies to evaluate whether the hydrogel could be used for long-term 3D in vitro studies and to assess if the material was capable of sustaining functionality of the cultured cells. Cell morphology and density were evaluated by labeling INS1-encapsulated cells at 1, 4, 7, and 10 days in culture. After 1 day, a good distribution of the cells throughout the scaffold was observed, although cellular density on the top of the gel was slightly lower compared with the bottom (Fig. 8A–H, P). Over time, a nonsignificant increase in cell density was also seen, most noticeably after 7 and 10 days (Fig. 8E–H, O), with the INS1 cells appearing to organize in spherical clusters (Fig. 8I, L, M, N). We also evaluated whether the biomaterial was capable of sustaining functionality of the encapsulated cells. A GSIS assay was performed after 4, 7, and 10 days and showed an SI around 2 (Fig. 9), which indicated that the 3D encapsulation and culture preserved cell functionality. Most importantly, a slight increase, although nonsignificant, in SI was also observed after 7 and 10 days in culture, which showed that the hydrogel was capable of sustaining cell functionality of the encapsulated cells for an extended culture time.
FIG. 8.
Representative fluorescent images of calcein-AM-labeled INS1 cells encapsulated in 1% Triton-PI pECM hydrogel. Viability assay showed a homogeneous cell distribution throughout the scaffold after 1 (A, B), 4 (C, D), 7 (E, F), and 10 (G, H) days in culture (N). Cell density slightly increased after 7 days (E–H, M), indicative of cell proliferation, and clusters of encapsulated cells were present after 7 (I, J) and 10 (K, L) days. Color images available online at www.liebertpub.com/tec
FIG. 9.
Glucose-stimulated insulin secretion assay of INS1 cells encapsulated in 6 mg/mL 1% Triton-PI pECM; n = 4.
Discussion
For a successful tissue engineering application, the choice of the proper material is crucial since the microenvironment is an important determinant of cellular responses including proliferation, differentiation, and functionality. In this regard, tissue-derived biomaterials, composed of an array of different ECM proteins, have the advantage of recapitulating the biochemical signals and bioactive stimuli of the native tissue ECM.45 However, to preserve these cues, each decellularization protocol should be tailored to the specific characteristics of the tissue of interest, and the effects of different detergents on ECM protein retention should be evaluated. A good balance between an effective decellularization, which is critical to reduce risk of an adverse cellular response, and retention of the biochemical composition and production costs should always be taken into account when generating ECM-derived scaffolds.
In our work, we investigated the effects of different detergents and the use of a proteinase inhibitor on the mechanical properties and biochemical composition of a pECM hydrogel, and quantitatively evaluated how different decellularization treatments affect these parameters. We investigated two different detergents, Triton X-100 and SDS, at different concentrations (0.1%, 0.5%, and 1% wt./vol.). All the tested conditions were effective in decellularizing pancreatic tissue as shown by the H&E and Hoechst staining (Fig. 3) and residual DNA quantification (Fig. 4). Higher concentrations of SDS demonstrated greater efficiency in cellular removal compared with Triton X-100 groups, as demonstrated by the lower amount of residual DNA and less batch-to-batch variability, although not significant compared with other groups (Supplementary Fig. S1). However, all groups showed very low residual DNA content, which is further reduced after pepsin digestion (Fig. 4). Residual DNA has been observed in many laboratory and commercially available decellularized products,46 indicating that the presence of very small traces of DNA does not impact the potential benefits of these products both for research and therapeutic uses. All SDS treatments had a detrimental influence on the retention of different ECM proteins and the ability of hydrogels to form a gel under appropriate conditions. Only hydrogels generated using 0.1% SDS showed good gelation with only a minimal amount of liquid fraction (Fig. 5), while 0.5% SDS only showed a partial gelation. Conversely, all Triton- treated groups were able to form a firm gel. When looking into ECM protein retention, all analyzed groups showed that the decellularization process negatively affected the retention of basement membrane and matricellular proteins. Only in the recently published study by Sackett et al. were the basement membrane proteins retained when the pancreatic tissue was decellularized and processed in a hydrogel form.22 In other previously published studies,17,21 a limited retention of collagen IV was reported, while laminin proteins were either not detected or investigated. In our study, only hydrogels treated with 1% Triton showed retention of laminin proteins, despite being lower when compared with native tissue (Supplementary Fig. S3). A similar trend was also observed with matricellular proteins and specifically with collagen VI (Supplementary Fig. S4). On the contrary, all SDS groups showed higher retention of sGAGs compared with the Triton-treated samples, with a higher concentration of sGAGs observed in the hydrogels treated with 0.5% and 1% SDS. However, this may be also due to some residual SDS in these groups rather than a real increase in sGAG retention since the DMMB assay could potentially detect the sulfated groups present in the SDS. This discrepancy could also be further supported by the significantly lower cell survival in the 0.5% and 1% SDS groups compared with Triton-treated hydrogels and by the comparable, if not higher, amount of sGAG in the SDS groups to native tissue.
Besides the choice of detergent and decellularization length, it is also fundamental to take into account the specific characteristics of the tissue of interest when generating a tissue-derived hydrogel. The pancreas is an organ rich in proteases that start self-digesting the tissue as soon as the tissue is harvested.39 We therefore also investigated if the use of gabexate, a serine PI used therapeutically in the treatment of pancreatitis, could have an effect on the retention of the ECM proteins. The use of PI did not have any influence on the decellularization of the pancreatic tissue, but it had a positive influence on the retention of sGAGs (Fig. 6), and basement membrane and matricellular proteins (Fig. 7), indicating that preventing tissue self-digestion immediately postharvesting has a positive effect on ECM protein retention. Interestingly, we did not observe any major differences when PI was used for the entire length of the decellularization process or just during tissue collection and processing, indicating that the inhibition of the pancreatic proteases soon after tissue harvesting may be sufficient to partly preserve the retention of these proteins during decellularization. However, we cannot exclude that the detergents could inhibit the activity of the inhibitor during decellularization and thereby lower the PI efficacy in preventing ECM digestion by the proteases. Similarly, it may also be possible that the endogenous proteases could be inactivated (or washed out of the tissue after cell permeabilization) during the initial washing steps and detergent treatments, making the use of the PI during decellularization unnecessary.
Finally, we also showed that the generated hydrogel could be used for 3D encapsulation of INS1 cells over an extended period of time. Cellular density and distribution were homogeneous in the scaffold throughout the culture period, with a nonsignificant increase in cellular density observed after 7 and 10 days in culture. Moreover, we also showed that the hydrogel supported the functionality of the encapsulated cells up to 10 days in culture, as demonstrated by the GSIS assay (Fig. 9), although further studies are needed to evaluate if the generated pECM hydrogel is advantageous compared with other biomaterials.
In conclusion, we showed a new approach to generate a pECM hydrogel that more closely mimics the native pECM, as demonstrated by the retention of all ECM protein classes present in the native tissue. We also showed that the choice of the detergent is fundamental in preserving the biochemical composition of the native tissue and the capability to form a hydrogel under appropriate conditions. Therefore, it is imperative that an effective decellularization, despite being the most important parameter, should also be accompanied with a proper ECM biochemical composition evaluation to create a tissue-derived scaffold that recapitulates the native tissue. Moreover, we also showed that tissue-specific characteristics, such as the presence of proteases in the pancreas, should also be taken into account in the development of an effective decellularization protocol.
Supplementary Material
Acknowledgments
This research was performed using resources and/or funding provided by the NIDDK-supported Human Islet Research Network (HIRN, RRID:SCR_014393; 1UC4DK104202 to K.L.C.), Juvenile Diabetes Research Foundation (JDRF) postdoctoral fellowships 3-PDF-2014-193-A-N (M.W.) and 3-PDF-2017-386-A-N (K.V.N.N.), and the John G. Davies Endowed Fellowship in Pancreatic Research (M.W.). Dr. Hughes receives support from the Chao Family Comprehensive Cancer Center (CFCCC) through an NCI Center Grant, P30A062203, and technology development award R33CA183685.
Disclosure Statement
No competing financial interests exist.
References
- 1. Cnop M., Welsh N., Jonas J.C., Jörns A., Lenzen S., and Eizirik D.L. Mechanisms of pancreatic beta-cell death in type 1 and type 2 diabetes: many differences, few similarities. Diabetes 54(Suppl. 2), S97, 2005 [DOI] [PubMed] [Google Scholar]
- 2. Paraskevas S., Maysinger D., Wang R., Duguid T.P., and Rosenberg L. Cell loss in isolated human islets occurs by apoptosis. Pancreas 20, 270, 2000 [DOI] [PubMed] [Google Scholar]
- 3. Ilieva A., Yuan S., Wang R.N., Agapitos D., Hill D.J., and Rosenberg L. Pancreatic islet cell survival following islet isolation: the role of cellular interactions in the pancreas. J Endocrinol 161, 357, 1999 [DOI] [PubMed] [Google Scholar]
- 4. Rosenberg L., Wang R., Paraskevas S., and Maysinger D. Structural and functional changes resulting from islet isolation lead to islet cell death. Surgery 126, 393, 1999 [PubMed] [Google Scholar]
- 5. Wang R.N., and Rosenberg L. Maintenance of beta-cell function and survival following islet isolation requires re-establishment of the islet-matrix relationship. J Endocrinol 163, 181, 1999 [DOI] [PubMed] [Google Scholar]
- 6. Stendahl J.C., Kaufman D.B., and Stupp S.I. Extracellular matrix in pancreatic islets: relevance to scaffold design and transplantation. Cell Transplant 18, 1, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng J.Y., Raghunath M., Whitelock J., and Poole-Warren L. Matrix components and scaffolds for sustained islet function. Tissue Eng Part B Rev 17, 235, 2011 [DOI] [PubMed] [Google Scholar]
- 8. Nikolova G., Jabs N., Konstantinova I., et al. The vascular basement membrane: a niche for insulin gene expression and Beta cell proliferation. Dev Cell 10, 397, 2006 [DOI] [PubMed] [Google Scholar]
- 9. Weber L.M., Hayda K.N., and Anseth K.S. Cell-matrix interactions improve beta-cell survival and insulin secretion in three-dimensional culture. Tissue Eng Part A 14, 1959, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Delalat B., Rojas-Canales D.M., Rasi Ghaemi S., et al. A combinatorial protein microarray for probing materials interaction with pancreatic islet cell populations. Microarrays (Basel) 5, 21, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davis N.E., Beenken-Rothkopf L.N., Mirsoian A., et al. Enhanced function of pancreatic islets co-encapsulated with ECM proteins and mesenchymal stromal cells in a silk hydrogel. Biomaterials 33, 6691, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yap W.T., Salvay D.M., Silliman M.A., et al. Collagen IV-modified scaffolds improve islet survival and function and reduce time to euglycemia. Tissue Eng Part A 19, 2361, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goh S.K., Bertera S., Olsen P., et al. Perfusion-decellularized pancreas as a natural 3D scaffold for pancreatic tissue and whole organ engineering. Biomaterials 34, 6760, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mirmalek-Sani S.H., Orlando G., McQuilling J.P., et al. Porcine pancreas extracellular matrix as a platform for endocrine pancreas bioengineering. Biomaterials 34, 5488, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peloso A., Urbani L., Cravedi P., et al. The human pancreas as a source of protolerogenic extracellular matrix scaffold for a new-generation bioartificial endocrine pancreas. Ann Surg 264, 169, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elebring E., Kuna V.K., Kvarnström N., and Sumitran-Holgersson S. Cold-perfusion decellularization of whole-organ porcine pancreas supports human fetal pancreatic cell attachment and expression of endocrine and exocrine markers. J Tissue Eng 8, 2041731417738145, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chaimov D., Baruch L., Krishtul S., Meivar-Levy I., Ferber S., and Machluf M. Innovative encapsulation platform based on pancreatic extracellular matrix achieve substantial insulin delivery. J Control Release 257, 91, 2017 [DOI] [PubMed] [Google Scholar]
- 18. Napierala H., Hillebrandt K.H., Haep N., et al. Engineering an endocrine Neo-Pancreas by repopulation of a decellularized rat pancreas with islets of Langerhans. Sci Rep 7, 41777, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katsuki Y., Yagi H., Okitsu T., et al. Endocrine pancreas engineered using porcine islets and partial pancreatic scaffolds. Pancreatology 16, 922, 2016 [DOI] [PubMed] [Google Scholar]
- 20. De Carlo E., Baiguera S., Conconi M.T., et al. Pancreatic acellular matrix supports islet survival and function in a synthetic tubular device: in vitro and in vivo studies. Int J Mol Med 25, 195, 2010 [PubMed] [Google Scholar]
- 21. Jiang K., Chaimov D., Patel S.N., et al. 3-D physiomimetic extracellular matrix hydrogels provide a supportive microenvironment for rodent and human islet culture. Biomaterials 2018. Epub ahead of print; [DOI: 10.1016/j.biomaterials.2018.08.057] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sackett S.D., Tremmel D.M., Ma F., et al. Extracellular matrix scaffold and hydrogel derived from decellularized and delipidized human pancreas. Sci Rep 8, 10452, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu T., Zhu M., Guo Y., et al. Three-dimensional culture of mouse pancreatic islet on a liver-derived perfusion-decellularized bioscaffold for potential clinical application. J Biomater Appl 30, 379, 2015 [DOI] [PubMed] [Google Scholar]
- 24. Xiaohui T., Wujun X., Xiaoming D., et al. Small intestinal submucosa improves islet survival and function in vitro culture. Transplant Proc 38, 1552, 2006 [DOI] [PubMed] [Google Scholar]
- 25. Lakey J.R., Woods E.J., Zieger M.A., et al. Improved islet survival and in vitro function using solubilized small intestinal submucosa. Cell Tissue Bank 2, 217, 2001 [DOI] [PubMed] [Google Scholar]
- 26. Uygun B.E., Soto-Gutierrez A., Yagi H., et al. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat Med 16, 814, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Petersen T.H., Calle E.A., Zhao L., et al. Tissue-engineered lungs for in vivo implantation. Science 329, 538, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sellaro T.L., Ranade A., Faulk D.M., et al. Maintenance of human hepatocyte function in vitro by liver-derived extracellular matrix gels. Tissue Eng Part A 16, 1075, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karabekmez F.E., Duymaz A., and Moran S.L. Early clinical outcomes with the use of decellularized nerve allograft for repair of sensory defects within the hand. Hand (N Y) 4, 245, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wicha M.S., Lowrie G., Kohn E., Bagavandoss P., and Mahn T. Extracellular matrix promotes mammary epithelial growth and differentiation in vitro. Proc Natl Acad Sci U S A 79, 3213, 1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Flynn L.E. The use of decellularized adipose tissue to provide an inductive microenvironment for the adipogenic differentiation of human adipose-derived stem cells. Biomaterials 31, 4715, 2010 [DOI] [PubMed] [Google Scholar]
- 32. Gaetani R., Yin C., Srikumar N., et al. Cardiac-derived extracellular matrix enhances cardiogenic properties of human cardiac progenitor cells. Cell Transplant 25, 1653, 2016 [DOI] [PubMed] [Google Scholar]
- 33. Seif-Naraghi S.B., Singelyn J.M., Salvatore M.A., et al. Safety and efficacy of an injectable extracellular matrix hydrogel for treating myocardial infarction. Sci Transl Med 5, 173ra25, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ungerleider J.L., Johnson T.D., Hernandez M.J., et al. Extracellular matrix hydrogel promotes tissue remodeling, arteriogenesis, and perfusion in a rat hindlimb ischemia model. JACC Basic Transl Sci 1, 32, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Johnson T.D., Hill R.C., Dzieciatkowska M., et al. Quantification of decellularized human myocardial matrix: a comparison of six patients. Proteom Clin Appl 10, 75, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. White L.J., Taylor A.J., Faulk D.M., et al. The impact of detergents on the tissue decellularization process: a ToF-SIMS study. Acta Biomater 50, 207, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Syed O., Walters N.J., Day R.M., Kim H.-W., and Knowles J.C. Evaluation of decellularization protocols for production of tubular small intestine submucosa scaffolds for use in oesophageal tissue engineering. Acta Biomater 10, 5043, 2014 [DOI] [PubMed] [Google Scholar]
- 38. González-Andrades M., Carriel V., Rivera-Izquierdo M., et al. Effects of detergent-based protocols on decellularization of corneas with sclerocorneal limbus. Evaluation of regional differences. Transl Vis Sci Technol 4, 13, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hegyi P., and Petersen O.H. The exocrine pancreas: the acinar-ductal tango in physiology and pathophysiology. Rev Physiol Biochem Pharmacol 165, 1, 2013 [DOI] [PubMed] [Google Scholar]
- 40. Gilbert T.W., Sellaro T.L., and Badylak S.F. Decellularization of tissues and organs. Biomaterials 27, 3675, 2006 [DOI] [PubMed] [Google Scholar]
- 41. Ungerleider J.L., Johnson T.D., Rao N., and Christman K.L. Fabrication and characterization of injectable hydrogels derived from decellularized skeletal and cardiac muscle. Methods 84, 53, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Freytes D.O., Martin J., Velankar S.S., Lee A.S., and Badylak S.F. Preparation and rheological characterization of a gel form of the porcine urinary bladder matrix. Biomaterials 29, 1630, 2008 [DOI] [PubMed] [Google Scholar]
- 43. Hill R.C., Calle E.A., Dzieciatkowska M., Niklason L.E., and Hansen K.C. Quantification of extracellular matrix proteins from a rat lung scaffold to provide a molecular readout for tissue engineering. Mol Cell Proteomics 14, 961, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. MacLean B., Tomazela D.M., Shulman N., et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Agmon G., and Christman K.L. Controlling stem cell behavior with decellularized extracellular matrix scaffolds. Curr Opin Solid State Mater Sci 20, 193, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gilbert T.W., Freund J.M., and Badylak S.F. Quantification of DNA in biologic scaffold materials. J Surg Res 152, 135, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.