Abstract
Targeted therapy with small molecules directed at essential survival pathways in leukemia represents a major advance, including the phosphatidylinositol-3′-kinase (PI3K) p110δ inhibitor idelalisib. Here, we found that genetic inactivation of p110δ (p110δD910A/D910A) in the Eμ-TCL1 murine chronic lymphocytic leukemia (CLL) model impaired B cell receptor signaling and B cell migration, and significantly delayed leukemia pathogenesis. Regardless of TCL1 expression, p110δ inactivation led to rectal prolapse in mice resembling autoimmune colitis in patients receiving idelalisib. Moreover, we showed that p110δ inactivation in the microenvironment protected against CLL and acute myeloid leukemia. After receiving higher numbers of TCL1 leukemia cells, half of p110δD910A/D910A mice spontaneously recovered from high disease burden and resisted leukemia rechallenge. Despite disease resistance, p110δD910A/D910A mice exhibited compromised CD4+ and CD8+ T cell response, and depletion of CD4+ or CD8+ T cells restored leukemia. Interestingly, p110δD910A/D910A mice showed significantly impaired Treg expansion that associated with disease clearance. Reconstitution of p110δD910A/D910A mice with p110δWT/WT Tregs reversed leukemia resistance. Our findings suggest that p110δ inhibitors may have direct antileukemic and indirect immune-activating effects, further supporting that p110δ blockade may have a broader immune-modulatory role in types of leukemia that are not sensitive to p110δ inhibition.
Keywords: Hematology, Immunology
Keywords: B cells, Leukemias, Protein kinases
Introduction
The development of chronic lymphocytic leukemia (CLL) is the consequence of malignant cell transformation, microenvironment-induced proliferation, and evasion of immune surveillance (1–3). Overwhelming evidence has highlighted the critical role that the leukemia microenvironment plays in the progression of CLL and other hematologic cancers. The leukemia microenvironment is composed of various cell types, including T cells, nurse-like cells, and stromal cells, the majority of which interact to collectively produce chemokines and cytokines to promote malignant cell tissue homing, retention, and proliferation (1). Another key aspect of the leukemia microenvironment is that CLL cells reshape the surrounding environment into a cancer-promoting and immunosuppressive milieu (1, 2). CLL cells can attract protective accessory cells by producing chemokines and cytokines (4–7). Furthermore, CLL cells evade immune destruction by expressing immunosuppressive molecules, secreting immunosuppressive cytokines and exosomes, increasing regulatory T cells (Tregs), and inducing myeloid-derived suppressor cells (MDSCs) (8–10).
The Bruton tyrosine kinase (BTK) inhibitor ibrutinib has been shown to have immunomodulatory functions through inhibiting direct tumor-mediated disease suppression, MDSC function, and regulatory B cell function (11–14). Through irreversibly inhibiting IL-2–inducible T cell kinase (ITK), ibrutinib can polarize CD4+ T cells to the antitumor Th1 lineage and expand effector T cells (15). Presumably through both its BTK-inhibiting and its ITK-inhibiting properties, ibrutinib enhances generation and function of chimeric antigen receptor (CAR) T cells and improves PD-1 blockade efficacy in several solid tumor models (13, 14). Acalabrutinib, despite lacking inhibitory activity against ITK, still can reverse CLL immune suppression by reducing CD200 and BTLA expression on CLL cells and PD-1 and CTLA4 expression on CD8+ T cells (13). The phosphatidylinositol-3′-kinase (PI3K) p110δ inhibitor idelalisib also appears to affect the immune compartment, as evidenced by the treatment-induced autoimmune side effects such as pneumonitis and colitis observed in patients receiving idelalisib therapy (16, 17). However, the precise mechanism of idelalisib-induced immune activation remains elusive.
PI3K p110δ, the catalytic isoform preferentially expressed in the hematopoietic system, contributes significantly to immune cell development and function (18–22). Activation of many membrane receptors of immune cells converges on the PI3K pathway (23, 24). In CLL cells, PI3K p110δ is abundantly expressed and constitutively activated (25, 26). Inhibitors of PI3K p110δ such as idelalisib or duvelisib have shown significant clinical activity in relapsed/refractory CLL (27, 28). While preclinical studies overwhelmingly focused on the effect of p110δ blockade in the leukemia cells, little is known about the effect of p110δ blockade in the neighboring cells of the microenvironment. p110δ kinase activity is important for the functions of T cells, natural killer cells, monocytes, macrophages, and dendritic cells in the CLL microenvironment (29). This raises the question of whether part of idelalisib’s therapeutic activity comes from inhibiting p110δ in these important surrounding accessory cells.
Emerging evidence has shown an immune-activating effect with pharmacologic p110δ inhibitors (16, 30–32). When given as initial therapy for CLL, idelalisib causes frequent hepatotoxicity associated with higher CD8+ T cell infiltration in the liver and lower numbers of Tregs in the blood (16). Murine and human CD8+ T cells treated with idelalisib in vitro exhibit an undifferentiated phenotype and persist longer after transfer into tumor-bearing mice, which leads to better antitumor immunity in both murine syngeneic and human xenograft tumor models (32). Coculture of CLL cells with T cells in the presence of idelalisib reduces CLL-induced T cell suppression (30). However, inhibitors have variable specificity, and the immune-activating effects of idelalisib could be confounded by the lowered disease burden. Therefore, the precise mechanism of p110δ blockade in the complexity of the leukemia microenvironment is best addressed using genetic animal models.
Mice with genetically inactivated p110δ (p110δD910A/D910A) have provided invaluable insight into the function of p110δ. p110δD910A/D910A mice have CD4+ T cells that are compromised in response to phytohemagglutinin and concanavalin A stimulation, and show dampened OT-II OVA-specific responses and impaired Th1/Th2 differentiation (33, 34). Despite the compromised T cell responses, p110δD910A/D910A mice are protected against a broad range of subcutaneously engrafted solid tumors (31). This is partly due to the impairment of Treg expansion and function that reverses antitumor immune tolerance and facilitates CD8+ T cell–mediated cytotoxicity (31). This antitumor immunity of p110δD910A/D910A mice can be recapitulated when solid tumor–implanted mice are treated with the p110δ inhibitor IC87114 (31). Tregs are found to be expanded in many types of leukemia, including CLL and acute myeloid leukemia (AML), and are associated with disease progression and poor prognosis (35–37). Therefore, it is reasonable to hypothesize that impaired Treg expansion could be beneficial in both CLL and AML.
However, owing to the immune-cell origin and the disseminated nature of hematologic malignancies, the mechanism of immune tolerance may be distinct from that in solid tumors. As a result of the poor vasculature of bulky tumors, immune cells have limited access to tumor cells. Solid tumors exhibit localized immune tolerance, and systemic immune dysfunction is only associated with advanced cancer. In contrast, the development of hematologic malignancies involves blood, bone marrow, and secondary lymphoid organs, where leukemia cells are in constant interaction with the normal immune cells. Moreover, many hematologic malignancies, including CLL, carry little neoantigen compared with many solid tumors, which further facilitates its immune tolerance (38). Systemic immune dysfunction is a hallmark of CLL, highlighted by the hypogammaglobulinemia and susceptibility to infections. CLL patients also exhibit profound T cell dysfunction, highlighted by increased T cell numbers yet severely compromised functions including proliferation and cytokine production (9, 39–42). In addition, accessory immune cells can interact with leukemic B cells as they would with normal B cells during normal immune responses. For example, binding between CD40 ligand (CD40L) on T cells and CD40 on B cells plays a critical role in generating humoral responses (43). However, this interaction can be exploited by CLL-B cells for leukemia survival and proliferation (44).
Notably, the Eμ-TCL1 (TCL1) mouse model, one of the most well-characterized CLL murine models (45–50), serves as an excellent tool for investigating immunomodulatory agents. Leukemia from Eμ-TCL1 mice represents unmutated IGHV CLL, the most aggressive form of CLL (45). Upon leukemia development, Eμ-TCL1 mice exhibit T cell defects similar to what is observed in CLL patients (46, 47, 49). These defects include aberrantly expressed genes for actin remodeling, impaired immunologic synapse formation, compromised T cell signaling, loss of T cell receptor diversity, clonal expansion of T cell populations, and an exhausted T cell phenotype (46, 47, 49). These T cell defects found in Eμ-TCL1 mice can be reversed by lenalidomide, an immunomodulatory agent used in CLL patients (42, 51).
PI3K p110δ inhibitors have shown broad activity in the treatment of hematologic malignancies. However, the role of p110δ inhibition in the leukemia microenvironment has not yet been addressed in a spontaneous, disseminated leukemia model. We sought to examine this using the Eμ-TCL1 transgenic mouse model of CLL and a mouse model of p110δ genetic inactivation (p110δD910A/D910A) to fully interrogate the effect of global and selective p110δ inhibition in the nonleukemic compartment in a complete immune microenvironment. These findings were confirmed in a second murine model of AML, providing evidence of the potential of p110δ inhibition in providing enhanced immune surveillance of leukemia.
Results
Global p110δ kinase inactivation significantly delays spontaneous leukemia development.
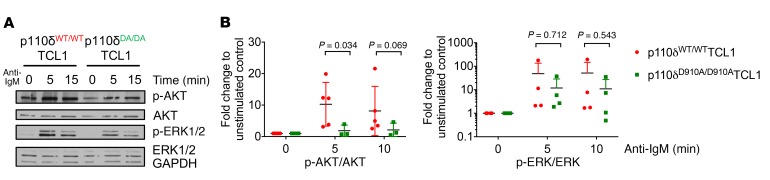
The Eμ-TCL1 transgenic mouse represents a model of IGHV unmutated CLL with epigenetic patterns, immune suppression characteristics, and response to pharmacologic agents similar to the human disease (45, 46, 49, 50, 52, 53). To assess the influence of p110δ loss of function on B cell receptor (BCR) signaling in this model, Eμ-TCL1 transgenic mice were crossed with p110δD910A/D910A (homozygous kinase-dead mutation) mice (18). TCL1 functions as an AKT kinase coactivator through binding and enhancing AKT kinase activity (52, 54). Since p110δD910A/D910A mice were reported to have compromised response to anti-IgM BCR cross-linking (18), we sought to examine whether the BCR signaling pathway of p110δD910A/D910A mice remains unresponsive upon TCL1 overexpression. B cells from spleen or bone marrow of 2- to 4-month-old p110δWT/WTTCL1 and p110δD910A/D910ATCL1 mice were subjected to varying durations of anti-IgM stimulation. Compared with p110δWT/WTTCL1, B cells from p110δD910A/D910ATCL1 mice showed significantly impaired AKT activation similar to what was previously reported in p110δD910A/D910A mice (Figure 1, A and B, and ref. 18). However, their changes in ERK1/2 and NF-κΒ signaling were not as profound (Figure 1, A and B, and Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI99386DS1). Similarly to p110δD910A/D910A, B cells from p110δD910A/D910ATCL1 mice also exhibited impaired migration toward CXCL13 but not toward CXCL12 (Supplemental Figure 1C and ref. 55).
Figure 1. Global p110δ kinase inactivation in Eμ-TCL1 mice partially impairs BCR signaling.
B cells were purified from spleens or bone marrow of 4-month-old p110δWT/WTTCL1 and p110δD910A/D910ATCL1 mice and stimulated with anti-IgM (10 μg/ml) for the indicated times. Cell lysates were immunoblotted for pATKS473 and total AKT. (A) The blots are representative of 4 independent experiments. (B) Densitometry analysis was conducted using ImageJ analysis software (NIH). All data were normalized to unstimulated control. Stimulated samples from 4 mice from each genotype were included for statistical analysis. ANOVA methods were used to compare condition means; data were log-transformed to stabilize variance. Bars represent mean ± SD.
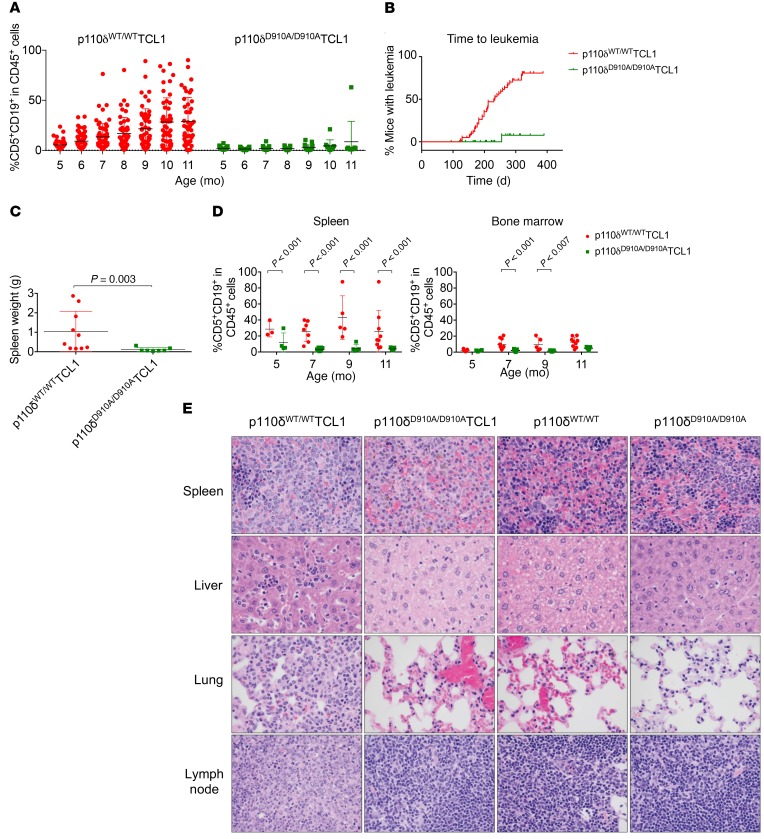
To understand the global effect of PI3K p110δ blockade in CLL pathogenesis in vivo, CLL disease development (presence of CD19/CD5-coexpressing cells in peripheral blood; ref. 45) was monitored in p110δWT/WTTCL1, p110δWT/D910ATCL1, and p110δD910A/D910ATCL1 mice by serial flow cytometry. The p110δWT/WTTCL1 mice demonstrated leukemia (≥10% CD19+CD5+ cells in the CD45+ population) in the blood at 5 months of age, with progressive expansion to over 80% of total lymphocytes by 7 months of age (Figure 2A). In contrast, leukemia cells in p110δD910A/D910ATCL1 mice were absent in the blood, except in 1 mouse that showed disease onset at the age of 9 months (out of 19 mice tested at that age) (Figure 2, A and B). p110δWT/D910ATCL1 mice did not appear to be different from p110δWT/WTTCL1 mice in terms of disease development, time to leukemia, or overall survival (Supplemental Figure 2, A–C), and therefore were not included in the subsequent investigations. Splenomegaly was observed in the p110δWT/WTTCL1 but not in the p110δD910A/D910ATCL1 mice (Figure 2C). The leukemia burden in the spleen and bone marrow was also examined at 7, 9, and 11 months in separate mice and demonstrated that the majority of p110δD910A/D910ATCL1 mice were nearly devoid of disease in spleen and bone marrow (Figure 2D). A detailed autopsy study at 9 months of age demonstrated that only 30% of p110δD910A/D910ATCL1 mice had disease, which was limited to spleen only (Figure 2E). In contrast, all p110δWT/WTTCL1 mice had histologic and/or gross evidence of lymphoid neoplasia in at least 1 of the organs, including spleen, lymph nodes, liver, kidney, and lung, and to a lesser extent in the heart and gastrointestinal tract (Figure 2E).
Figure 2. Global p110δ kinase inactivation delays spontaneous leukemia development.
(A) p110δ WT/WTTCL1 (n = 43–65) and p110δ D910A/D910ATCL1 (n = 9–39) mice were monitored monthly for leukemia development through immunophenotyping of CD5+CD19+ cells in the peripheral blood by flow cytometry. (B) Time to leukemia (leukemia is determined by having ≥10% CD5+CD19+ of total CD45+ cells in blood) of p110δWT/WTTCL1 (n = 61) and p110δD910A/D910ATCL1 mice (n = 44). (Log-rank test P < 0.001.) (C) Spleen weight from 9-month-old p110δWT/WTTCL1 and p110δD910A/D910ATCL1 mice. (Wilcoxon rank sum test P = 0.003.) (D) Lymphocytes from spleen and bone marrow were obtained from p110δWT/WTTCL1 (n = 3–10) and p110δD910A/D910ATCL1 mice (n = 4–11) 5, 7, 9, and 11 months of age. CD5+CD19+ cells were examined by flow cytometry. (Group comparisons from mixed-effects models using log-transformed data. P values within each organ were adjusted for multiple comparisons using Holm’s procedure.) Bars represent mean ± SD. (E) Histology of spleen, liver, and lung from p110δWT/WTTCL1 (n = 10) and p110δD910A/D910ATCL1 mice (n = 10) at 9 months of age. Original magnification, ×60.
p110δ kinase inactivation leads to severe autoimmune phenotype in Eμ-TCL1 mice.
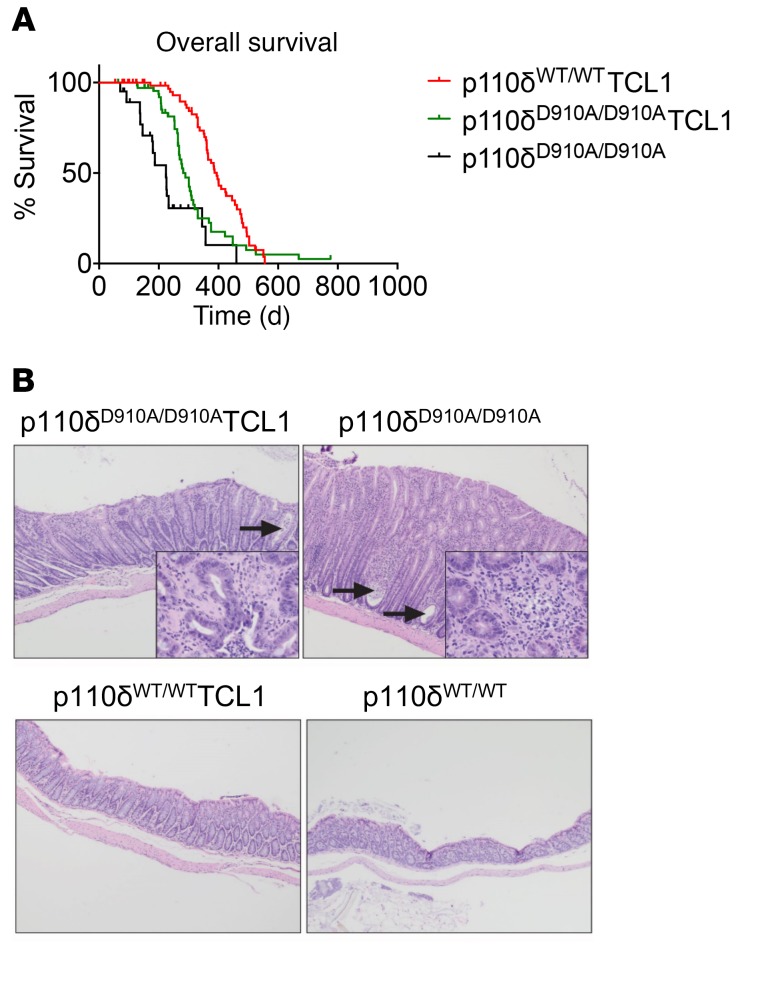
Although p110δD910A/D910ATCL1 mice had significantly less leukemia burden, their median survival was 289 days, significantly shorter than the 394 days for p110δWT/WTTCL1 mice (P < 0.001; Figure 3A). Similarly, p110δD910A/D910A mice exhibited shortened median survival of 224 days compared with reported 800-day median survival of normal C57BL/6J (B6) mice (56). p110δD910A/D910A mice have been previously described as developing spontaneous inflammatory bowel disease due to Treg impairment in maintaining peripheral tolerance and altered macrophage function, leading to a hyperactivated innate inflammatory response (21). Similarly, we found that p110δD910A/D910A and p110δD910A/D910ATCL1 mice showed evidence of neutrophilic and proliferative colitis occasionally with rectal prolapse and, to a lesser extent, enteritis and gastritis (Figure 3B).
Figure 3. p110δ kinase inactivation leads to severe autoimmune phenotype in Eμ-TCL1 mice.
(A) Overall survival of p110δWT/WTTCL1 (n = 88) and p110δD910A/D910ATCL1 mice (n = 79). (Log-rank test P < 0.001.) (B) Histology of colon in p110δD910A/D910ATCL1 (n = 10), p110δWT/WTTCL1 (n = 10), p110δD910A/D910A (n = 10), and p110δWT/WT (n = 10) mice at 9 months of age. Both p110δD910A/D910ATCL1 and p110δD910A/D910A demonstrated severe colitis, consisting of hyperplasia of the crypt epithelium, crypt abscesses (arrows), and numerous neutrophils within the lamina propria (inset). Colitis was not observed in p110δWT/WTTCL1 and p110δWT/WT mice. Original magnification, ×20; inset, ×40.
Microenvironment p110δ kinase inactivation protects against CLL.
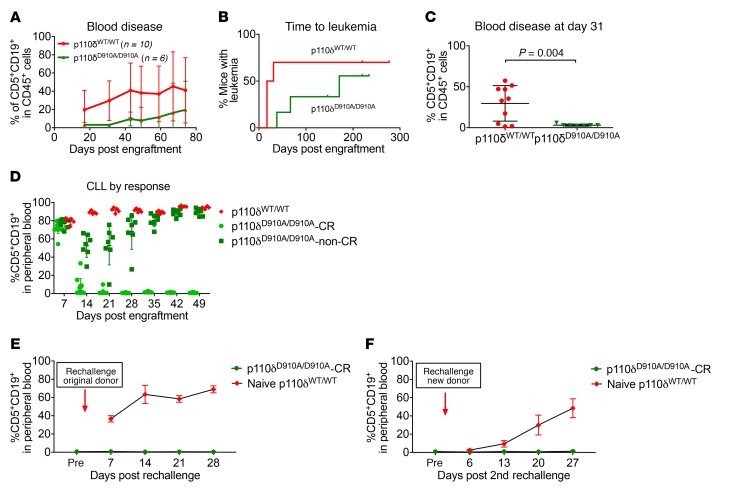
While an overwhelming number of in vitro studies have focused on p110δ blockade in CLL B cells, little is known about the effect of p110δ blockade on the microenvironment surrounding CLL or other disseminated leukemia models. Given that p110δD910A/D910A mice are resistant to subcutaneous engraftment of solid tumors (31), we sought to determine whether systemic tumor dissemination was impacted. To do this, we examined whether genetic p110δ inactivation in the nonleukemic compartment is protective against disseminated leukemia. Leukemia cells from Eμ-TCL1 mice with functional p110δ were adoptively transferred into recipient p110δWT/WT or p110δD910A/D910A mice as previously described (57). Compared with p110δWT/WT mice, p110δD910A/D910A mice showed more resistance to leukemia development. p110δD910A/D910A mice had 20% reduced disease incidence (3 of 6 mice in p110δD910A/D910A group vs. 7 of 10 mice in p110δWT/WT group) (Figure 4A), a trend toward longer time to leukemia (median: p110δD910A/D910A, 171 days, vs. p110δWT/WT, 24 days; P = 0.215) (Figure 4B), and lower peripheral disease burden (at day 31: p110δD910A/D910A, 3.1% [SD = 1.2%], vs. p110δWT/WT, 29.7% [SD = 21.7%]; Figure 4C). When this observation was repeated with a 4-fold greater number of tumor cells (2 × 107), p110δD910A/D910A mice developed a level of leukemia similar to that of the p110δWT/WT mice on day 7 (74.4% [SD = 6.5%] and 79.6% [SD = 3.6%], respectively; Figure 4D), but were found to spontaneously regress on day 14, with 12 of 19 mice completely clearing their leukemia by day 21 (Figure 4D). The peripheral blood disease in the completely recovered p110δD910A/D910A mice (p110δD910A/D910A-CR) remained low for the rest of the study, and spleen examination showed no signs of splenomegaly. On the contrary, the p110δWT/WT mice continued to progress until they reached a moribund state starting on day 49.
Figure 4. Microenvironment p110δ kinase inactivation protects against CLL.
(A–C) p110δWT/WT (n = 10) and p110δD910A/D910A mice (n = 6) were engrafted with 5 × 106 Eμ-TCL1 leukemia cells through tail vein injection. Peripheral blood disease was monitored through immunophenotyping of CD5+CD19+ cells by flow cytometry. (A) Average weekly blood disease burden. Each data point represents mean ± SD. (B) Time to leukemia (leukemia is determined by having ≥10% CD5+CD19+ of total CD45+ cells in blood) of p110δWT/WT and p110δD910A/D910A mice. (Log-rank test P = 0.215.) (C) Individual blood disease on day 31. Bars represent mean ± SD. (P = 0.004 based on 2-sample t test with unequal variances.) (D–F) p110δWT/WT (n = 10) and p110δD910A/D910A mice (n = 7) were engrafted with 5 × 106 Eμ-TCL1 leukemia cells through tail vein injection. Peripheral blood disease was monitored through immunophenotyping of CD5+CD19+ cells by flow cytometry. (D) p110δWT/WT (n = 5) and p110δD910A/D910A mice (n = 19) underwent adoptive transfer of 2 × 107 Eμ-TCL1 leukemia cells. Blood disease was monitored weekly by flow cytometry. Bars represent mean ± SD. (E) Sixty-three days after the first transfer, p110δD910A/D910A mice that had regressed disease (n = 10) and naive p110δWT/WT (n = 5) were challenged with 2 × 107 leukemic cells of the same donor Eμ-TCL1 cells. Each data point represents mean ± SD. (F) Forty-two days after the second transfer, the remaining p110δD910A/D910A (n = 9) and new naive p110δWT/WT (n = 5) mice were challenged with 2 × 107 leukemic cells from a different Eμ-TCL1 donor. Each data point represents mean ± SD.
To further assess whether an antitumor immunologic response was present, on day 64 after the first transfer, p110δD910A/D910A-CR mice were rechallenged with the original Eμ-TCL1 donor to assess for an adaptive memory response. Wild-type naive mice were included as a positive control for successful adoptive transfer. For the first rechallenge, wild-type naive mice developed 63.4% (SD = 9.9%) leukemia in the blood by day 14, whereas p110δD910A/D910A-CR mice showed no sign of blood disease throughout the course of the experiment (Figure 4E). Day 50 after rechallenge, p110δD910A/D910A-CR mice were challenged again with a different Eμ-TCL1 leukemia donor. During the second rechallenge, wild-type naive mice developed 48.4% (SD = 10.4%) leukemia in the blood by day 27, while p110δD910A/D910A-CR mice again rejected the disease (Figure 4F).
p110δ kinase inactivation impedes leukemia via a T cell–dependent mechanism.
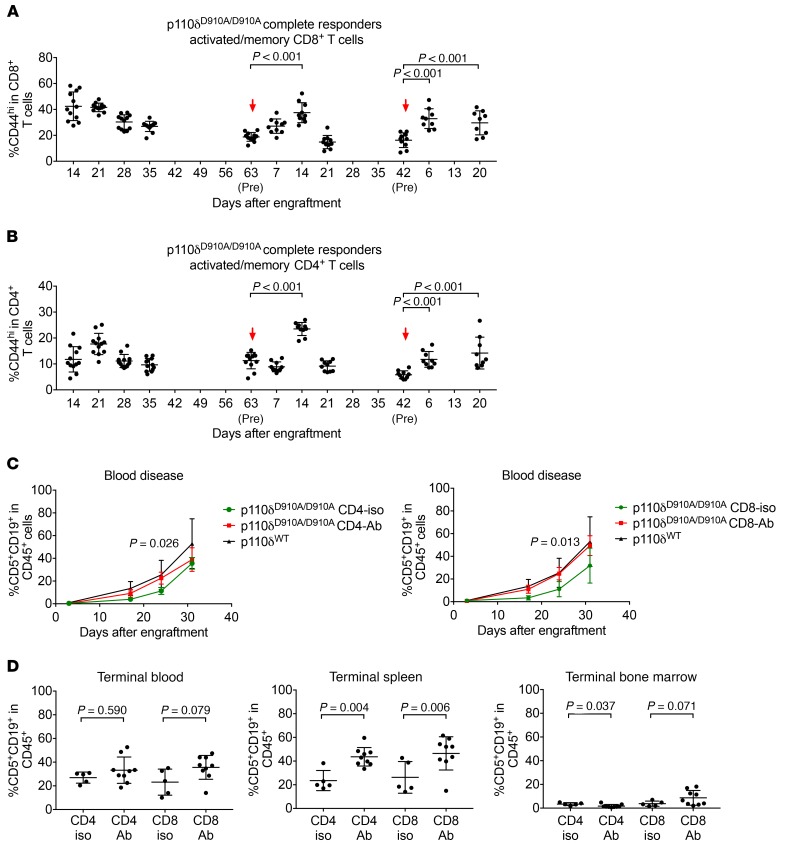
Interestingly, the p110δD910A/D910A mice that had leukemia regression and resisted rechallenge (Figure 4, E and F) showed expanded CD44+CD4+ and CD44+CD8+ T cells immediately after rechallenge (Figure 5, A and B). These data suggest that the leukemia resistance may be T cell mediated. Therefore, we sought to examine the dependence of leukemia resistance on CD4+ and CD8+ T cells in p110δD910A/D910A mice. p110δD910A/D910A mice were engrafted with leukemia and subjected to CD4+ or CD8+ T cell depletion. Confirmation of CD4+ or CD8+ depletion and blood disease burden were examined weekly (Supplemental Figure 3A). Day 17 after engraftment, p110δD910A/D910A mice that received corresponding isotype control developed leukemia significantly more slowly than p110δWT/WT mice as previously observed, whereas CD4+ or CD8+ depletion resulted in higher disease burden, to a level equivalent to that of p110δWT/WT mice (Figure 5C and Supplemental Figure 3, B and C). Upon euthanizing at day 37, higher splenic disease was found in both the CD4+ and the CD8+ depleted groups (Figure 5D). However, only CD8+ depletion resulted in higher disease in the bone marrow (Figure 5D).
Figure 5. p110δ kinase inactivation resists leukemia through a T cell–dependent mechanism.
(A and B) CD44+CD4+ (A) and CD44+CD8+ (B) T cells were examined in mice from Figure 3, D–F. Bars represent mean ± SD. Arrow denotes adoptive transfer of leukemia cells. A repeated-measures model was applied to the log-transformed data. For the first rechallenge, comparisons were made of day 7 and day 14 after rechallenge versus before rechallenge. For the second rechallenge, comparisons were made of day 6 and day 20 after rechallenge versus before rechallenge. (C) Eμ-TCL1 leukemia cells were adoptively transferred into p110δWT/WT (n = 5) and p110δD910A/D910A mice. p110δD910A/D910A mice were subjected to CD4-depleting (n = 9) or CD8-depleting (n = 9) antibody injections once every 3 days. Isotype controls (CD4 isotype n = 5, CD8 isotype n = 5) were included as controls. Group differences were estimated using mixed-effects models. Data represent mean ± SD. (D) Day 37 after the engraftment, p110δD910A/D910A mice in CD4 depletion, CD4 isotype, CD8 depletion, and CD8 isotype groups were euthanized. Spleen, bone marrow, and blood were analyzed for CD5+CD19+ leukemia cells. Bars represent mean ± SD. (Differences between antibody and corresponding isotype control groups were estimated using ANOVA based on the log-transformed data.)
To further investigate the different roles played by CD4+ and CD8+ T cells, recombination activating gene 1–knockout (RAG–/–) mice were reconstituted with CD3+, CD4+, and CD8+ T cells from p110δWT/WT and p110δD910A/D910A mice followed by Eμ-TCL1 leukemia engraftment. Eμ-TCL1 engraftment without T cell reconstitution was included as positive control. T cell population and leukemia burden were examined by flow cytometry (Supplemental Figure 4, A, E, and F). Day 21 after leukemia engraftment, both the p110δWT/WT-CD3+ group and the p110δD910A/D910A-CD3+ group showed less disease compared with the no–T cell group (Supplemental Figure 4A). For both p110δWT/WT and p110δD910A/D910A, the CD4+ T cell group had variable disease outcome, while the CD8+ T cell group had the least amount of disease compared with the T cell counterparts of the same genotype (Supplemental Figure 4A). When the differences between the 2 genotypes are compared, it appears that both p110δD910A/D910A CD3+ and CD8+ T cells led to less disease compared with their wild-type counterparts (Supplemental Figure 4, B–D).
p110δ kinase inactivation results in fewer antigen-experienced T cells.
To further elucidate the role of CD4+ and CD8+ T cells in leukemia resistance of p110δD910A/D910A mice, the effector and memory kinetics of these cells were examined in the context of leukemia regression. Disease regression in p110δD910A/D910A mice was recreated using secondary leukemia cells from p110δWT/WT mice previously engrafted as shown in Figure 4D. On day 14, among the 30 enrolled mice per group, 6 mice of the lowest and highest blood disease burden from each group were euthanized for further analysis. The mice with the highest and lowest disease burden from the p110δWT/WT group showed very similar levels of leukemia (80% [SD = 4.3%] vs. 85% [SD = 1.1%]) in the blood. In contrast, the 6 mice with lowest disease burden in the p110δD910A/D910A group resisted disease (40% [SD = 13.6]), and the 6 mice with highest disease burden did not (85% [SD = 2.5%]) (Figure 6A). Therefore, 12 p110δWT/WT mice were assigned into 1 group (p110δWT/WT), and p110δD910A/D910A were designated into responder (p110δD910A/D910A-R) and nonresponder (p110δD910A/D910A-NR) groups, respectively.
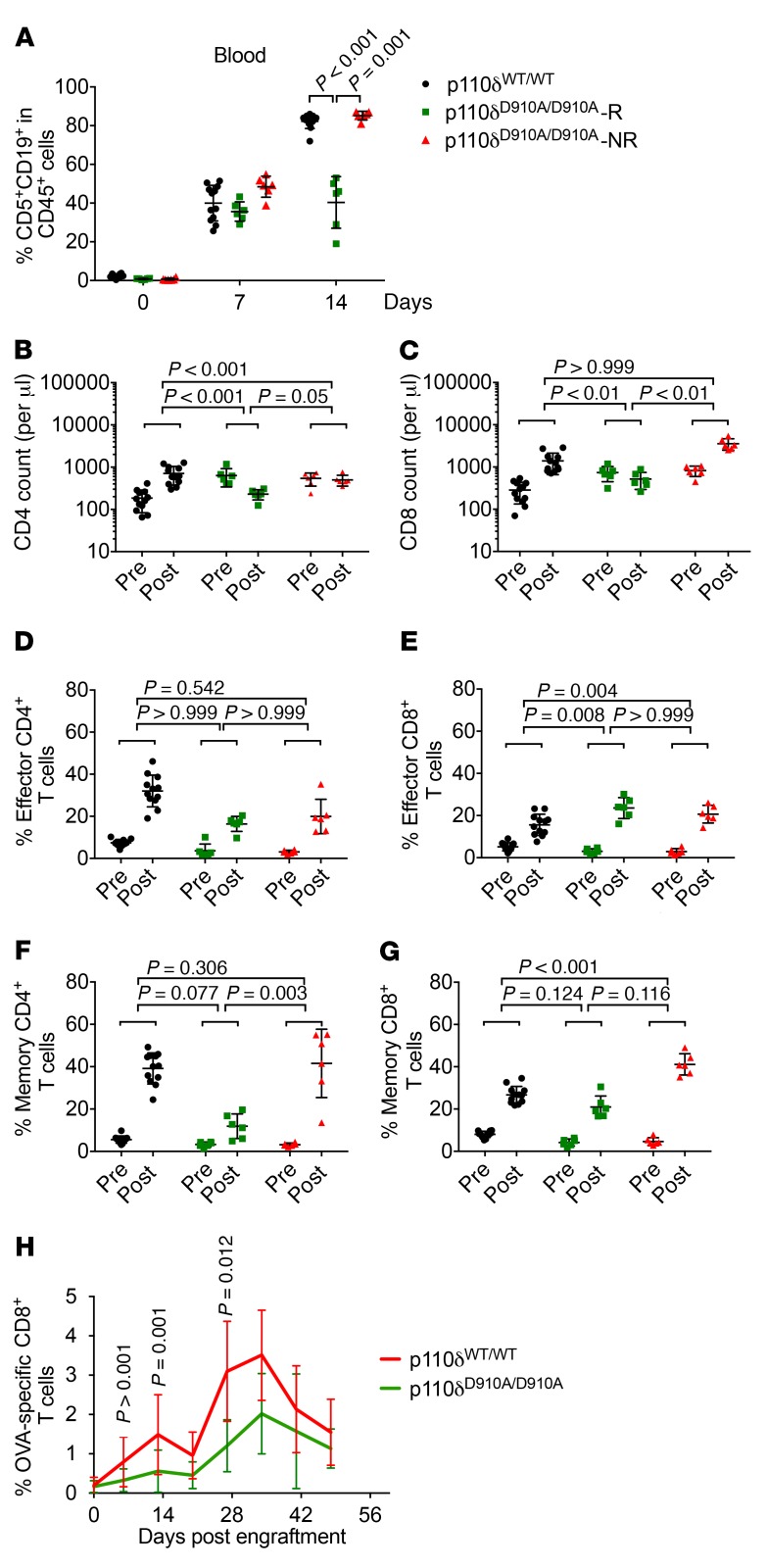
Figure 6. Impact of p110δ inactivation on T cell antileukemia response.
p110δWT/WT and p110δD910A/D910A mice after adoptive transfer with 2 × 107 secondary Eμ-TCL1 leukemia cells. (A) Percentage CD5+CD19+ of CD45+ cells was examined by flow cytometry on days 0, 7, and 14. (B–G) Absolute number of CD4+ T cells (B), absolute number of CD8+ T cells (C), effector CD4+ T cells (percentage CD44loCD62LhiCD4+ of CD4+ cells) (D), effector CD8+ T cells (percentage CD44loCD62LhiCD8+ of CD8+ cells) (E), memory CD4+ T cells (percentage CD44hiCD62LhiCD4+ of CD4+ cells) (F), and memory CD8+ T cells (percentage CD44hiCD62LhiCD8+ of CD8+ cells) (G) were examined before the engraftment (Pre) and day 14 after the engraftment (Post). Black circles, p110δWT/WT; green squares, p110δD910A/D910A-R; red triangles, p110δD910A/D910A-NR. Comparisons were based on separate mixed-effects models (log-transformed data); P values were adjusted using Holm’s procedure. (H) OVA-TCL1 cells were adoptively transferred into p110δWT/WT and p110δD910A/D910A mice; OVA-specific CD8+ T cells were examined weekly using fluorescent tagged OVA tetramer by flow cytometry. Bars represent mean ± SD. (Group comparisons are based on mixed-effects models on log-transformed data; P values were adjusted for multiple comparisons using Holm’s procedure; 9 total time points examined.)
In agreement with a previous report (58), both CD4+ and CD8+ T cells expanded in p110δWT/WT mice, whereas they either remained the same or showed a slight decrease in the p110δD910A/D910A-R mice (Figure 6, B and C). The frequency of effector (CD44hiCD62Llo) and memory (CD44hiCD62Lhi) CD4+ and CD8+ T cells expanded in all groups at varying degrees (Figure 6, D–G). Gating strategy is in included in Supplemental Figure 5. The degree of expansion in effector (CD44hiCD62Llo) CD4+ T cells did not differ among the groups (Figure 6D), whereas effector (CD44hiCD62Llo) CD8+ T cells expanded to a higher degree in p110δD910A/D910A-R mice, suggesting the potential role of effector CD8+ T cells in disease clearance (Figure 6E). On the other hand, memory (CD44hiCD62Lhi) CD4+ T cells expanded to a higher degree in p110δWT/WT and p110δD910A/D910A-NR diseased mice (Figure 6F), which likely associates with disease burden. In contrast, the expansion of memory (CD44hiCD62Lhi) CD8+ T cells does not seem to correlate with genotype or disease burden (Figure 6G).
p110δ kinase inactivation results in fewer antigen-specific CD8+ T cells.
To further examine the effect of p110δ inhibition on the development or recruitment of antigen-specific antitumor CD8+ T cells, MHC I ovalbumin-expressing Eμ-TCL1 (OVA-TCL1) leukemia cells were engrafted into p110δWT/WT and p110δD910A/D910A mice. Tumor-infiltrating OVA-specific CD8+ T cells in the blood were examined weekly using a fluorescent tetramer recognizing MHC I specific for OVA antigen. Paradoxically, p110δD910A/D910A mice had diminished OVA+CD8+ T cells as compared with p110δWT/WT mice (Figure 6H and Supplemental Figure 6).
p110δ kinase inactivation reverses immune suppression in leukemia mice.
Effective antitumor immunity is often suppressed by immune checkpoint molecules and/or Tregs (59). p110δD910A/D910A mice were reported to have impaired Treg suppression that unleashes CD8+ cytotoxic T cells in solid tumor–bearing mice (31). Given that p110δD910A/D910A mice resist leukemia despite compromised antigen-specific CD8+ response, we sought to examine the immune-suppressive profile of p110δD910A/D910A mice in the context of leukemia.
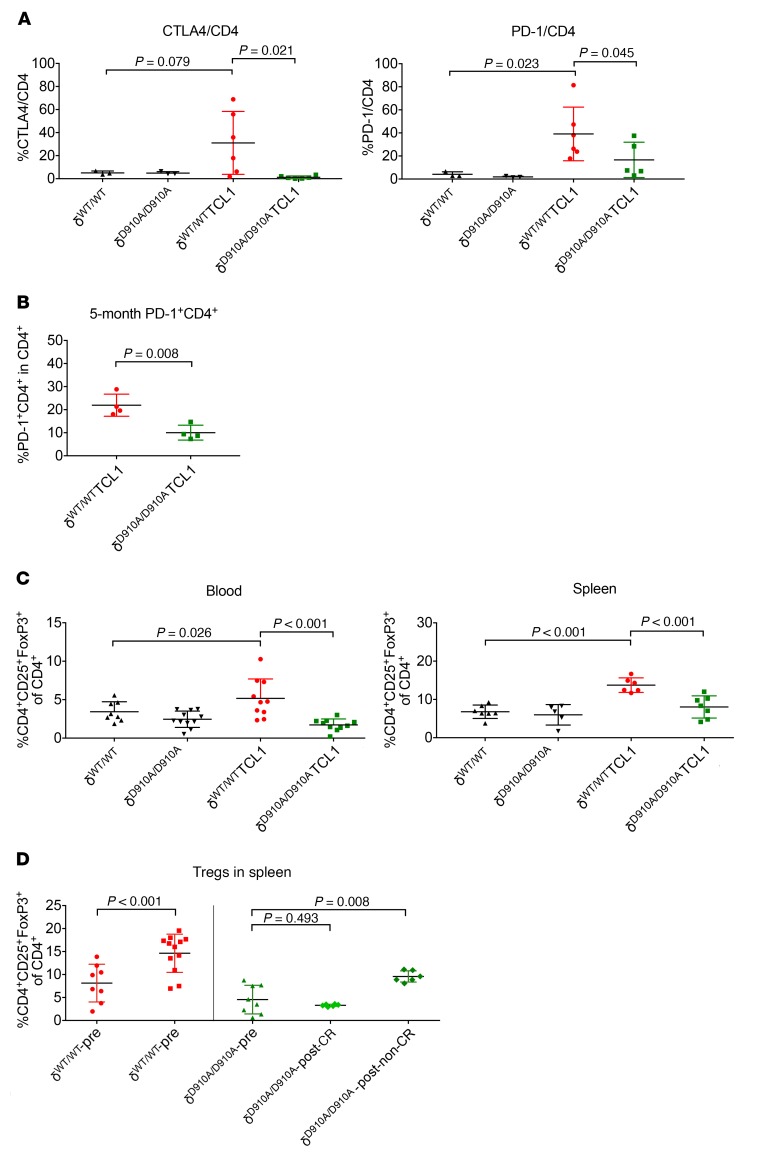
T cell defects in CLL are characterized by the exhausted phenotype, overexpression of checkpoint molecules, and decreased effector function (9, 41, 42). At 11 months of age, p110δD910A/D910ATCL1 mice showed significantly fewer CTLA4+CD4+ and PD-1+CD4+ T cells compared with p110δWT/WTTCL1 mice (Figure 7A). However, the lack of immune checkpoint molecule expression could be due to the lower disease burden. We therefore examined blood and spleen samples from preleukemic 5-month-old mice. p110δWT/WTTCL1 mice still expressed a higher frequency of PD-1+CD4+ T cells in the spleen, despite similar leukemia burden (Figure 7B and Supplemental Figure 7A). To better isolate the T cell effect, these markers were examined in the adoptive transfer study shown in Figure 4D. Immediately after engraftment on day 7, p110δD910A/D910A mice showed a higher frequency of CTLA4+CD4+ and PD-1+CD4+ T cells (Supplemental Figure 7B). However, from day 14 and throughout the disease course, the frequency of CTLA4+CD4+ and PD-1+CD4+ T cells correlated with disease burden in both p110δWT/WT and p110δD910A/D910A mice, as seen in Figure 4D. A similar correlation was also found in the adoptive transfer study shown in Figure 6.
Figure 7. p110δ kinase inactivation reverses immune suppression in leukemia mice.
(A) Blood from 11-month-old p110δWT/WT, p110δD910A/D910A, p110δWT/WTTCL1, and p110δD910A/D910ATCL1 mice was examined for immune checkpoint CTLA4 and PD-1 expression on CD4+ cells by flow cytometry. (Separate ANOVA models were applied to each outcome. P values within each outcome were adjusted for multiple comparisons using Holm’s procedure.) (B) Blood from 5-month-old p110δWT/WTTCL1 and p110δD910A/D910ATCL1 mice was examined for immune checkpoint PD-1 expression on CD4+ cells and leukemia burden by flow cytometry. (Group means were compared using 2-sample t tests with unequal variances.) (C) Relative Tregs (CD4+CD25+FoxP3+ of CD4+ cells) in blood and spleen of 9-month-old p110δWT/WT, p110δD910A/D910A, p110δWT/WTTCL1, and p110δD910A/D910ATCL1 mice were examined by flow cytometry. (Separate ANOVA models were applied to each outcome. P values within each outcome were adjusted for multiple comparisons using Holm’s procedure.) (D) Frequency of Tregs (CD4+CD25+FoxP3+ of CD4+ cells) in spleen was examined in p110δWT/WT and p110δD910A/D910A mice on day 14 after adoptive transfer with 2 × 107 Eμ-TCL1 secondary leukemia cells. (Differences were estimated using ANOVA.) Bars represent mean ± SD.
CD4+CD25+FoxP3+ Tregs from blood and spleen of p110δWT/WTTCL1 and p110δD910A/D910ATCL1 mice were examined at 9 months of age. p110δD910A/D910ATCL1 exhibited significantly fewer Tregs compared with p110δWT/WTTCL1 mice (Figure 7C). Splenic Tregs were also examined in naive and postengraftment p110δWT/WT and p110δD910A/D910A mice, as shown in Figure 4D. Tregs were found to be higher in p110δWT/WT mice before engraftment and significantly expanded after engraftment (Figure 7D). However, they were lower in p110δD910A/D910A mice before engraftment and either remained the same in mice that regressed or slightly expanded in mice that progressed on disease (Figure 7D). To examine whether the Tregs were functionally impaired in p110δD910A/D910A mice, splenic CD4+CD25+ T cells were purified from p110δWT/WT and p110δD910A/D910A mice at baseline and cocultured with splenic p110δWT/WT CD4+CD25– T cells in the presence of anti-CD3/anti-CD28 Dynabeads. The purities of CD4+CD25– and CD4+CD25+ T cells were confirmed with intranuclear FoxP3 staining (Supplemental Figure 8A). In accordance with a previous report, p110δWT/WT CD4+CD25+ T cells suppressed CD4+CD25– T cell proliferation at a higher ratio, whereas p110δD910A/D910A CD4+CD25+ T cells failed to do so (Supplemental Figure 8B and ref. 60). To further investigate whether lack of Treg suppression and expansion are responsible for leukemia resistance, p110δD910A/D910A mice were reconstituted with p110δWT/WT Tregs followed by leukemia transfer. Day 27 after leukemia engraftment, p110δD910A/D910A mice receiving p110δWT/WT Tregs showed an abundant level of leukemia, whereas p110δD910A/D910A mice left unengrafted or engrafted with p110δD910A/D910A Tregs had no evidence of disease (Supplemental Figure 8C).
p110δ kinase inactivation in the microenvironment is also effective against myeloid leukemia.
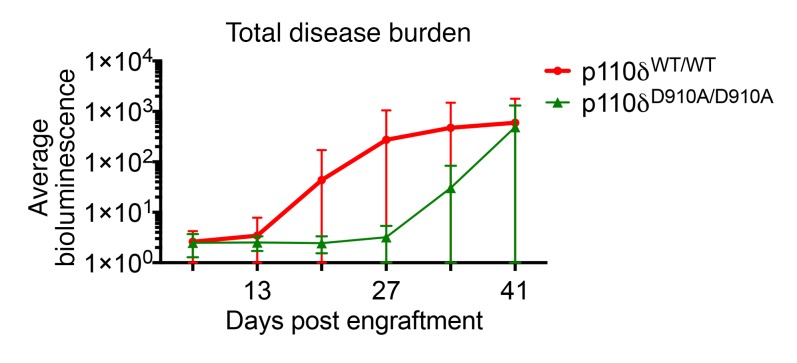
Acute myeloid leukemia (AML) is the most common adult leukemia of nonlymphocyte lineage and has a poor outcome with standard therapies (61). To examine whether the protective effect of p110δ inactivation is observed across lineages to myeloid leukemia, an aggressive murine AML cell line of C57BL/6 background (Supplemental Figure 10), C1498-luciferase, was engrafted into p110δWT/WT and p110δD910A/D910A recipient mice. Disease burden was measured by whole-body luciferase activity using an In Vivo Imaging System (IVIS) (Supplemental Figure 9) and quantified for average fluorescence intensity. p110δD910A/D910A mice showed less leukemia incidence (p110δWT/WT, 7 of 10, vs. p110δD910A/D910A, 3 of 10) and reduced overall disease burden compared with the wild-type controls (Figure 8).
Figure 8. p110δ kinase inactivation in the microenvironment is also effective against myeloid leukemia.
p110δWT/WT (n = 10) and p110δD910A/D910A (n = 10) mice were engrafted with C1498-luciferse cells through tail vein injection; the leukemia burden was examined by the amount of bioluminescence with an IVIS imager. Overall tumor kinetics is quantified by average bioluminescence of every mouse in each group. Data represent mean ± SD.
Discussion
In this study, we identified, using genetic animal models, that the function of p110δ, both in leukemia B cells and in the nonleukemic microenvironment, is critical for CLL pathogenesis. Global inactivation of p110δ in the Eμ-TCL1 murine CLL model blocks leukemia progression in blood and major lymphoid organs. B cells from p110δD910A/D910ATCL1 mice still exhibit partially impaired BCR signaling and migratory function. Inactivation of p110δ in the nonleukemic microenvironment also protects against leukemia through a T cell–dependent mechanism. Despite dampened antigen-specific CD8+ T cell responses, p110δ-inactivated T cells remain cytotoxic to leukemia cells. It is possible that p110δ inactivation impairs Treg expansion followed by enhanced host antitumor immunity. Reconstituted p110δD910A/D910A mice with p110δWT/WT Tregs reversed such disease resistance. Nonetheless, p110δ inactivation introduces side effects such as colitis, suggesting autoimmunity. These murine studies further confirmed that the colitis is an on-target side effect of idelalisib treatment in patients with CLL and non-Hodgkin’s lymphoma (NHL). Finally, we demonstrate that the antitumor benefits of p110δ inactivation are not lineage specific, as it also impacted engraftment and dissemination of murine AML. Taken together, our study supports that p110δ inhibition presents an attractive therapeutic option in multiple cancer types, and that p110δ inhibitors may be used for immunomodulation in disseminated models of cancer.
The Eμ-TCL1 murine CLL model is by far the most validated and characterized spontaneous in vivo model of human CLL (46, 49, 50, 52, 53). Extensive studies on BCR signaling and microenvironment regulation have been performed using this model. Since the development of inhibitors of kinases associated with the BCR signalosome, many genetic models emerged to investigate the mechanism of these agents (57, 62–65). Interestingly, multiple kinases belonging to the BCR signalosome have dual functions in both the leukemic and the nonleukemic cell compartments. For example, PKCβ is required by both CLL cells and bone marrow cells to maintain leukemogenesis (62, 63). Additionally, LYN–/–/TCL1 mice have delayed leukemia onset, and LYN–/– mice are resistant to leukemia engraftment (64). Despite the FDA approval of idelalisib in relapsed and refractory CLL, the exact mechanism of p110δ remains elusive. Many in vitro characterizations highlight the importance of p110δ in BCR signaling (66–69). However, the role of p110δ outside of leukemic cells remains unclear. Here, we provide what is to our knowledge the first direct evidence that p110δ activity is critical for both the leukemic and nonleukemic compartments. Compared with XID/TCL1 or LYN–/–/TCL1 (57, 64), p110δD910A/D910ATCL1 resulted in much more dramatic disease suppression (Figure 2A). In addition, we show that p110δ is required for T cell immune tolerance. p110δ inactivation suppresses Treg expansion (Figure 7, C and D), which correlates with disease progression in this model. Our data validate that p110δ blockade in CLL is efficacious and provide evidence that p110δ blockade can be immunomodulatory.
p110δD910A/D910A mice have been reported to have compromised BCR signaling (18), and our data show that despite the TCL1 transgene, B cells from p110δD910A/D910ATCL1 mice still exhibit partially compromised BCR signaling and impaired migration toward CXCL13 (Figure 1 and Supplemental Figure 1). However, we cannot exclude the effect of p110δ blockade in the microenvironment in the global inactivation model, since p110δD910A/D910A mice resist leukemia engraftment (Figure 4). To further confirm the precise role of p110δ in the B cell compartment, it would be of interest to generate B cell–specific p110δ inactivation. In addition, given that p110δD910A/D910ATCL1 mice have sustained p110δ inhibition prior to disease development and TCL1 acts in the exact pathway downstream of PI3K signaling, it would be of interest to create an inducible model where p110δ kinase inactivation could be turned on upon disease development. Additional CLL murine models should also be tested to validate these findings.
The Eμ-TCL1 transgenic mouse is also valuable for investigating CLL immune dysfunction. Upon disease development, Eμ-TCL1 mice or syngeneic mice with adoptively transferred leukemia cells develop T cell defects and profound immunodeficiency closely resembling what is observed in CLL patients (47, 49, 70). However, few therapeutic agents have been developed that target the leukemia microenvironment. Emerging studies have revealed the critical roles T cells play in the pathogenesis of CLL. For example, CLL cells are known to induce the generation and/or accumulation of memory CD4+ T cells. Despite being an important component of the immune system, T cells can be exploited by CLL cells for their own survival. Owing to the weak antigen stimulation, CLL can induce naive T cells to become central memory T cells. These central memory T cells can then migrate toward the lymphoid niches because of their high CXCR5 expression and provide costimulatory signaling to CLL cells through CD40L and cytokine production (71). Effector memory T cells can also contribute to CLL progression through IL-4 and IFN-γ production. Given that p110δ is abundantly expressed in T lymphocytes and plays critical roles in T cell biology, we suspect that inactivation of p110δ in T cells would compromise their ability to support CLL. In accordance with our hypothesis, we found that p110δD910A/D910A mice resisted leukemia development (Figure 4) and generated significantly fewer memory CD4+ T cells (Figure 6F).
From our observation, it first seemed that both CD4+ T cells and CD8+ T cells are critical for the leukemia resistance in p110δD910A/D910A mice (Figure 5D). However, depleting CD4+ T cells in Eμ-TCL1 mice also leads to higher leukemia burden (72). All of this is contradictory to what was observed in mice bearing solid tumors. Depleting CD4+ T cells in 4T1 tumor–bearing p110δD910A/D910A mice did not accelerate disease (31). Depletion of CD4+ T cells in wild-type C3B6F1 mice inoculated with Ag104Ld tumor resulted in complete tumor rejection (73). We hypothesize that CD4+ T cells are critical as T helper cells at the initial antigen recognition/presentation stage. In the case of solid tumor, CD4+ Tregs accumulate in a confined location as disease progresses, effectively suppressing antitumor CD8+ T cells. Depleting CD4+ T cells under this circumstance removes Tregs from the suppression. In 4T1 tumor–bearing p110δD910A/D910A mice, in which Treg expansion was already impaired (31), CD4+ T cell depletion would not affect disease course. In disseminated leukemia, it is possible that CD8+ T cells rely on CD4+ T cells’ help. Therefore, leukemia-bearing mice depleted of CD4+ T cells would suffer from the loss of T helper cells rather than benefit from the removal of Tregs. This hypothesis is also supported by our observation that transferring either p110δWT/WT or p110δD910A/D910A CD3+ or CD4+ T cells with Eμ-TCL1 leukemia into RAG–/– mice slows down disease compared with transferring leukemia alone (Supplemental Figure 4A).
Owing to the direct antitumor cytotoxicity of CD8+ T cells, it is not surprising to see that depleting CD8+ T cells in p110δD910A/D910A mice bearing either Eμ-TCL1 leukemia or 4T1 solid tumor led to higher disease burden (Figure 5, C and D, and ref. 31). Moreover, depletion of CD8+ T cells rather than CD4+ T cells in p110δD910A/D910A mice reversed leukemia resistance in the bone marrow compartment (Figure 5D). In contrast, replenishing RAG–/– mice with either p110δWT/WT or p110δD910A/D910A CD8+ T cells leads to the least amount of disease in comparison with their same-genotype counterparts (Supplemental Figure 4, A and C). All of this further highlights the importance of CD8+ T cells in the antitumor immunity of p110δD910A/D910A mice.
Moreover, it was also reported that p110δD910A/D910A mice generated a comparable number of OVA-specific CD8+ T cells to that seen in p110δWT/WT mice upon subcutaneous inoculation of OVA-expressing Lewis lung carcinoma cells (LLC-OVA) (31). However, we found that upon adoptive transfer of OVA-TCL1 cells, p110δD910A/D910A mice developed approximately half the number of OVA-specific CD8+ T cells compared with p110δWT/WT mice (Figure 6H and Supplemental Figure 6). This may contradict our finding that CD8+ T cell–replenished RAG–/– mice had the least amount of disease. The discrepancy of these data may be due to the disseminated nature of leukemia inducing different host antitumor immunity (74) and tolerance. Localized tumors are susceptible to the immune surveillance enforced by tumor-infiltrating lymphocytes; however, the immunosuppressive microenvironment may prevent local activation or effector function. In contrast, disseminated tumors like leukemia are more vulnerable to immune surveillance once in the lymphatic/blood circulation, which may place less pressure for immune activation.
Although inactivation of p110δ in mice partially dampens CD4+ and CD8+ T cell function, these mice remain resistant to Leishmania major infection and a broad range of solid tumor engraftments (31, 75). This is due to severely suppressed Treg expansion and defects in suppressive activity. In agreement with previous reports, we also found that Treg expansion was impaired in p110δD910A/D910ATCL1 mice and p110δD910A/D910A mice into which Eμ-TCL1 leukemia cells were adoptively transferred (Figure 7D). Replenishing p110δD910A/D910A mice with p110δWT/WT Tregs reversed the leukemia resistance (Supplemental Figure 8C), providing direct evidence that the lack of Treg expansion protects p110δD910A/D910A mice from developing leukemia. Treg expansion is found in CLL and AML patients and associated with progressive disease. Our finding suggests that p110δ blockade could be used as an immune-modulatory agent for these diseases.
One major consequence of p110δ inhibition seen in the clinic is autoimmunity (16). Despite low leukemic incidence, p110δD910A/D910ATCL1 mice had shortened overall survival due to autoimmune colitis (Figure 3). It has been previously described that p110δD910A/D910A mice spontaneously develop colitis due to an exaggerated mucosal and systemic Th1/Th17 cytokine profile and altered macrophage function (60). We observed neutrophilic colitis accompanied by rectal prolapse in both p110δD910A/D910A and p110δD910A/D910ATCL1 mice. Both the Treg suppression and colitis symptoms are observed in CLL patients with idelalisib treatment and associated with liver toxicity (16). Our genetic model, therefore, provides strong and precise evidence that the gastrointestinal consequence of idelalisib is an on-target side effect of p110δ blockade.
This autoimmune phenotype in the mouse model, however, presents a significant confounding factor in the disease tracking of p110δD910A/D910ATCL1. It remains unknown how the disease might develop in p110δD910A/D910ATCL1 mice if they are free of autoimmune complication. Compelling evidence suggests that the microbiota helps to shape the immune system as a whole (76). Germ-free (GF) mice that lack intestinal microbiota are reported to have defective immune response (77, 78). Dysbiosis, which is characterized by a less diverse and less stable microbiota, can lead to systemic activation of Th17 cells and effector T cells (79, 80). The colitis observed in p110δD910A/D910A mice is believed to be caused by the presence of enteric microbiota dysbiosis (81). p110δD910A/D910A mice that were housed GF did not develop histologic colitis up to 30 weeks of age. However, 7 days upon transfer to traditional housing, p110δD910A/D910A mice showed significant colonic inflammation and an increased colitis score (81). Given the increasing evidence suggesting the association between microbiota and antitumor immunity (80), it is reasonable to hypothesize that the enhanced antitumor immunity in p110δD910A/D910A mice may be associated with colitis symptoms. It is fascinating for us to observe that compared with p110δWT/WT or p110δD910A/D910A-non-CR mice, p110δD910A/D910A-CR mice have a higher plasma IL-17 level and a lower IL-10 level (data not shown). They are also the only group that had detectable plasma IL-12p70 (data not shown). These cytokine profiles are consistent with what was observed in p110δD910A/D910A mice that developed microbiota-dependent colitis due to the defects in macrophages (81). Because the experiment was short-term, we did not observe any apparent colitis. However, it would be of interest to examine the antitumor immunity of p110δD910A/D910A mice in GF condition to test whether the leukemia resistance associates with microbiota-dependent colitis. On a separate note, it seems interesting that p110δD910A/D910ATCL1 mice gain survival advantages over p110δD910A/D910A mice. While p110δD910A/D910ATCL1 mice appear to have a similar level of colitis to p110δD910A/D910A mice at the time of assessment, it remains to be explored whether the presence of TCL1 transgene delays the onset of colitis symptoms.
Given that p110δ is also abundantly expressed in myeloid cells (82, 83), it is possible that inactivation of p110δ affected their crosstalk with leukemia. It has been shown that myeloid-derived suppressor cells (MDSCs) from solid tumor–bearing p110δD910A/D910A mice are less suppressive than their wild-type counterparts (31). However, it was not clear whether it was an intrinsic difference in these myeloid cells or an indirect result of less disease burden. The same study also reported that RAG–/– p110δD910A/D910A mice bearing 4T1 solid tumor exhibited similar primary tumor burden but fewer metastatic lesions, suggesting that inactivation of p110δ in non–B/T cell lineage is not sufficient to delay tumor progression (31). It would be interesting for future studies to determine whether inactivation of p110δ in the myeloid compartment contributes to leukemia resistance.
In summary, we believe that our study provides the first genetic evidence confirming the functional role of p110δ in CLL initiation and progression and T cell tumor immune tolerance. Additionally, this finding is reproducible in another distinct disseminated model of AML. These findings improve our understanding of the mechanism of action of p110δ small-molecule inhibitors. Furthermore, these findings suggest the immunoregulatory potential of p110δ inhibitors and highlight the importance of prophylactic care for on-target side effects associated with p110δ blockade.
Methods
Supplemental Methods are available online with this article; https://doi.org/10.1172/JCI99386DS1
Cells and reagents.
The C1498-luciferase cell line was a gift from Bruce R. Blazar (University of Minnesota, Minneapolis, Minnesota, USA). Antibodies against cell surface markers or intracellular proteins were from: BD Biosciences: CD45 (30-F11), CD19 (1D3), CD5 (53-7.3), CD3 (134-2C11), CD4 (RM4-5), CD8a (53-6.7), CD25 (PC61), CTLA4 (UC10-4B9), PD-1 (J43), PD-L1 (MIH5), CD44 (IM7), CD62L (MEL-14); eBioscience: FoxP3 (FJK-16s). Antibodies for depletion of CD4 (GK1.5) or CD8 (53-6.72) cells were from Bio X Cell. Isotype control antibodies were LTF-2 and 2A3 from Bio X Cell. CountBright beads were obtained from Invitrogen and used per the manufacturer’s instructions.
Mice.
All animal experiments were performed under a protocol approved by the Ohio State University Institutional Laboratory Animal Care and Use Committee. Eμ-TCL1 transgenic mice and PI3K p110δ mice were provided by Carlo M. Croce (The Ohio State University, Columbus, Ohio, USA) and Bart Vanhaesebroeck (University College London, London, UK), respectively. Eμ-TCL1–transgenic mice on a C57BL/6J (B6) background have been previously described (45). This mouse strain has been extensively backcrossed with the B6 mouse to obtain a Eμ-TCL1–expressing mouse on a pure B6 background. PI3K p110δD910A mice have been previously described (18) and were backcrossed on C57BL/6J for more than 12 generations. To generate p110δD910A/D910ATCL1 mice, homozygous B6/Eμ-TCL1 mice were crossed with homozygous p110δD910A mice to generate p110δWT/D910ATCL1 mice. p110δWT/D910ATCL1 mice were further crossed with each other to generate p110δWT/WTTCL1 mice and p110δD910A/D910ATCL1 mice. For survival studies, mice were euthanized when they reached early-removal criteria by animal protocol, including lethargy, difficulty walking due to spleen size, or loss of 20% body weight. Animals were evaluated daily, with formal measurements of weight weekly during engraftment studies and biweekly or monthly for long-term studies. For experiments characterizing disease compartments, mice were euthanized at stated ages, and blood from heart puncture, bone marrow, and spleen samples were collected for further analysis.
Adoptive transfer studies.
For Eμ-TCL1 engraftment studies, leukemic spleen lymphocytes were obtained from B6/Eμ-TCL1 mice with high white blood cell counts and splenomegaly. Spleens were manually disrupted, and spleen lymphocyte single cells were obtained by homogenization of the tissue and filtering of cells through a 100-μm nylon cell strainer (BD Falcon, Heidelberg, Germany). Red cells were lysed with RBC lysis buffer (eBioscience). Flow cytometry was used to confirm that lymphocytes were leukemia, and then spleen lymphocytes were resuspended in sterile PBS and injected through a lateral tail vein. For p110δD910A/D910A and B6 engraftments, 5 × 106 to 2 × 107 cells were injected into each mouse, with equal amounts used for each individual experiment. Submandibular bleeding was used to obtain blood for smears and flow cytometry to evaluate disease progression. For C1498-luciferase cell adoptive transfer, leukemia burden was determined via luciferase activity measured by an In Vivo Imaging System (IVIS) imager (PerkinElmer).
In vivo CD4+ and CD8+ T cell depletion study.
In vivo antibody depletion studies were initiated 10 days after leukemia adoptive transfer; antibodies were administered through i.p. injection of 0.5 mg of antibodies to CD4 or CD8 (or antibody isotype controls) every 3 days, for the duration of the study.
Flow cytometry.
Peripheral blood leukemia involvement was performed by staining of fresh whole blood with anti–mouse CD45–APC, anti–mouse CD5–FITC, and anti–mouse CD19–PE (all from BD Biosciences). Absolute counts of CLL cells were determined through gating on live CD45+CD5+CD19+ cells and counting beads (CountBright absolute counting beads; Invitrogen) following the manufacturer’s recommendations. Flow cytometry was performed on a Beckman Coulter FC-500 flow cytometer or a BD LSRFortessa, and Kaluza software was used for data analysis.
Isolation of murine cells.
Purification of murine B cells was performed by negative selection using an EasySep Mouse B Cell Isolation Kit according to the manufacturer’s instruction.
BCR cross-linking experiments and immunoblot analysis.
Murine spleen white blood cells were either left unstimulated or stimulated with F(ab′)2 goat anti-mouse IgM (Jackson ImmunoResearch Laboratories; Dianova). Whole cell lysates were prepared as previously described by our group (84). Equivalent amounts of protein were loaded onto polyacrylamide gels and transferred onto nitrocellulose membranes. Following antibody incubations, proteins were detected with chemiluminescent substrate (SuperSignal; Pierce). Phospho-AKT (Ser473) (D9W9U; 12694), total AKT (polyclonal; 9272), phospho–p44/42 MAPK (ERK1/2) (Thr202/Tyr204) (polyclonal; 9101), total ERK1/2 (polyclonal; 9102), phospho-IκBα (Ser32/36) (5A5; 9246), and total IκBα (112B2; 9247) antibodies were obtained from Cell Signaling Technology.
Histopathology.
Spleen, liver, heart, lungs, kidneys, gastrointestinal tract, sternal bone marrow, thymus, and mesenteric lymph nodes were collected following natural death or euthanasia due to fulfillment of early-removal criteria. Tissues were examined grossly and microscopically. Tissues were fixed in 10% neutral-buffered formalin, paraffin-embedded, and sectioned at 3 μm onto glass slides with H&E staining by routine methodology. Sternums (for bone marrow) were decalcified in 10% formic acid solution for 24 hours immediately prior to paraffin embedding.
Statistics.
For Western blot data (Figure 1B, Supplemental Figure 1B, and Supplemental Figure 8C), ANOVA methods were used to compare group means; outcome data were log-transformed to stabilize variances. Mixed-effects models were used for Figure 2C, Figure 4A, Figure 5, A–C, Figure 6, A–G, and Supplemental Figure 1C, to account for correlations among observations from the same subject; as with the Western blot data, outcomes were first log-transformed. Estimated differences with 95% confidence intervals were obtained from the models, and P values were adjusted for multiple comparisons using Holm’s procedure. For experiments described in Figure 4C, Figure 5D, Figure 7, A–D, and Supplemental Figure 8B, 2-sided 2-sample t tests using unequal variances or ANOVA methods were used to compare group means. The nonparametric Wilcoxon rank sum test was used to compare groups for Supplemental Figure 4, A and B. P values were adjusted for multiple comparisons using Holm’s procedure.
For the mouse survival models described in Figure 2B, Figure 3A, Figure 4B, and Supplemental Figure 2, B and C, estimated times to leukemia development or overall survival (OS) were obtained using the Kaplan-Meier method, and the log-rank test evaluated differences between curves. Time to leukemia was defined as the date of engraftment to the date of flow cytometry showing CD5/CD19 in more than 10% of lymphocytes. OS was defined as the time from either birth or development of leukemia (noted in each individual experiment) until death. Spleen weight (Figure 2C) was compared between groups using the Wilcoxon rank sum test. All analyses were performed using SAS/STAT software (SAS version 9.4 for Windows, SAS Institute Inc.). P values less than 0.05 were considered significant.
Study approval.
All animal studies were approved by the Institutional Animal Care and Use Committees at The Ohio State University (Columbus, Ohio, USA).
Author contributions
SD contributed to the design and performance of the experiments, data analysis, and writing of the manuscript. BKH processed pathology samples and acquired and analyzed pathology data. EYH performed the flow sort and acquired and analyzed data. JTG performed the tritiated thymidine incorporation assay and acquired and analyzed data. AML performed the statistical analysis and interpreted data. MT and RLW contributed to mouse colony maintenance and blood and tissue harvesting. ML, NM, JRB critically reviewed the manuscript and helped with data interpretation. AJJ and JCB designed the study, secured funding, directed the research, analyzed data, and provided overall guidance of the project and manuscript.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Carlo Croce for providing the Eμ-TCL1 mice, Bart Vanhaesebroeck for providing the PI3K p110δD910A/D910A mice, and Bruce R. Blazar for providing the C1498-luciferase cell line. This work was supported by grants from the NIH (R01CA159296, R01CA213442, R35CA197734, and F30CA225070-01A1), the Leukemia & Lymphoma Society, the American Society of Hematology, and the Pelotonia Fellowship Program, as well as Michael and Judy Thomas, the D. Warren Brown Foundation, and the Four Winds Foundation.
Version 1. 11/19/2018
Electronic publication
Version 2. 01/02/2019
Print issue publication
Funding Statement
R01CA159296, R01CA213442, and R35 CA197734
Footnotes
Conflict of interest: JRB receives research funding and consulting fees from Gilead Sciences and Sun Pharmaceutical, and consulting fees from Janssen and AbbVie. AJJ is an employee of Janssen Research and Development.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(1):122–136.https://doi.org/10.1172/JCI99386.
Contributor Information
Shuai Dong, Email: dong.209@osu.edu.
Bonnie K. Harrington, Email: harrington.120@osu.edu.
Eileen Y. Hu, Email: eileen.hu@osumc.edu.
Joseph T. Greene, Email: joseph.greene@osumc.edu.
Amy M. Lehman, Email: amy.lehman@osumc.edu.
Minh Tran, Email: minh.tran@osumc.edu.
Meixiao Long, Email: meixiao.long@osumc.edu.
Natarajan Muthusamy, Email: raj.muthusamy@osumc.edu.
Jennifer R. Brown, Email: jennifer_brown@dfci.harvard.edu.
Amy J. Johnson, Email: ajohn118@its.jnj.com.
John C. Byrd, Email: john.byrd@osumc.edu.
References
- 1.Burger JA. Nurture versus nature: the microenvironment in chronic lymphocytic leukemia. Hematology Am Soc Hematol Educ Program. 2011;2011:96–103. doi: 10.1182/asheducation-2011.1.96. [DOI] [PubMed] [Google Scholar]
- 2.Purroy N, Wu CJ. Coevolution of leukemia and host immune cells in chronic lymphocytic leukemia. Cold Spring Harb Perspect Med. 2017;7(4):a026740. doi: 10.1101/cshperspect.a026740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kipps TJ, et al. Chronic lymphocytic leukaemia. Nat Rev Dis Primers. 2017;3:16096. doi: 10.1038/nrdp.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burger JA, et al. High-level expression of the T-cell chemokines CCL3 and CCL4 by chronic lymphocytic leukemia B cells in nurselike cell cocultures and after BCR stimulation. Blood. 2009;113(13):3050–3058. doi: 10.1182/blood-2008-07-170415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghia P, et al. Chronic lymphocytic leukemia B cells are endowed with the capacity to attract CD4+, CD40L+ T cells by producing CCL22. Eur J Immunol. 2002;32(5):1403–1413. doi: 10.1002/1521-4141(200205)32:5<1403::AID-IMMU1403>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 6.Zucchetto A, et al. Monocytes/macrophages but not T lymphocytes are the major targets of the CCL3/CCL4 chemokines produced by CD38(+)CD49d(+) chronic lymphocytic leukaemia cells. Br J Haematol. 2010;150(1):111–113. doi: 10.1111/j.1365-2141.2010.08152.x. [DOI] [PubMed] [Google Scholar]
- 7.Maffei R, et al. The monocytic population in chronic lymphocytic leukemia shows altered composition and deregulation of genes involved in phagocytosis and inflammation. Haematologica. 2013;98(7):1115–1123. doi: 10.3324/haematol.2012.073080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giannopoulos K, et al. Characterization of regulatory T cells in patients with B-cell chronic lymphocytic leukemia. Oncol Rep. 2008;20(3):677–682. [PubMed] [Google Scholar]
- 9.Riches JC, et al. T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production. Blood. 2013;121(9):1612–1621. doi: 10.1182/blood-2012-09-457531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jitschin R, et al. CLL-cells induce IDOhi CD14+HLA-DRlo myeloid-derived suppressor cells that inhibit T-cell responses and promote TRegs. Blood. 2014;124(5):750–760. doi: 10.1182/blood-2013-12-546416. [DOI] [PubMed] [Google Scholar]
- 11.Stiff A, et al. Myeloid-derived suppressor cells express bruton’s tyrosine kinase and can be depleted in tumor-bearing hosts by ibrutinib treatment. Cancer Res. 2016;76(8):2125–2136. doi: 10.1158/0008-5472.CAN-15-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondo K, et al. Ibrutinib modulates the immunosuppressive CLL microenvironment through STAT3-mediated suppression of regulatory B-cell function and inhibition of the PD-1/PD-L1 pathway. Leukemia. 2018;32(4):960–970. doi: 10.1038/leu.2017.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long M, et al. Ibrutinib treatment improves T cell number and function in CLL patients. J Clin Invest. 2017;127(8):3052–3064. doi: 10.1172/JCI89756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruella M, et al. The addition of the BTK inhibitor ibrutinib to anti-CD19 chimeric antigen receptor T cells (CART19) improves responses against mantle cell lymphoma. Clin Cancer Res. 2016;22(11):2684–2696. doi: 10.1158/1078-0432.CCR-15-1527. [DOI] [PubMed] [Google Scholar]
- 15.Dubovsky JA, et al. Ibrutinib is an irreversible molecular inhibitor of ITK driving a Th1-selective pressure in T lymphocytes. Blood. 2013;122(15):2539–2549. doi: 10.1182/blood-2013-06-507947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lampson BL, et al. Idelalisib given front-line for treatment of chronic lymphocytic leukemia causes frequent immune-mediated hepatotoxicity. Blood. 2016;128(2):195–203. doi: 10.1182/blood-2016-03-707133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coutre S, et al. Safety of idelalisib in B-cell malignancies: integrated analysis of eight clinical trials. J Clin Oncol. 2015;33(15_suppl):e18030 [Google Scholar]
- 18.Okkenhaug K, et al. Impaired B and T cell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science. 2002;297(5583):1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- 19.Guo H, Samarakoon A, Vanhaesebroeck B, Malarkannan S. The p110 delta of PI3K plays a critical role in NK cell terminal maturation and cytokine/chemokine generation. J Exp Med. 2008;205(10):2419–2435. doi: 10.1084/jem.20072327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim N, et al. The p110delta catalytic isoform of PI3K is a key player in NK-cell development and cytokine secretion. Blood. 2007;110(9):3202–3208. doi: 10.1182/blood-2007-02-075366. [DOI] [PubMed] [Google Scholar]
- 21.Uno JK, et al. Altered macrophage function contributes to colitis in mice defective in the phosphoinositide-3 kinase subunit p110δ. Gastroenterology. 2010;139(5):1642–1653.e1. doi: 10.1053/j.gastro.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji H, et al. Inactivation of PI3Kgamma and PI3Kdelta distorts T-cell development and causes multiple organ inflammation. Blood. 2007;110(8):2940–2947. doi: 10.1182/blood-2007-04-086751. [DOI] [PubMed] [Google Scholar]
- 23.Herman SE, Johnson AJ. Molecular pathways: targeting phosphoinositide 3-kinase p110-delta in chronic lymphocytic leukemia. Clin Cancer Res. 2012;18(15):4013–4018. doi: 10.1158/1078-0432.CCR-11-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okkenhaug K, Burger JA. PI3K signaling in normal B cells and chronic lymphocytic leukemia (CLL) Curr Top Microbiol Immunol. 2016;393:123–142. doi: 10.1007/82_2015_484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ringshausen I, et al. Constitutively activated phosphatidylinositol-3 kinase (PI-3K) is involved in the defect of apoptosis in B-CLL: association with protein kinase Cdelta. Blood. 2002;100(10):3741–3748. doi: 10.1182/blood-2002-02-0539. [DOI] [PubMed] [Google Scholar]
- 26.Clayton E, et al. A crucial role for the p110delta subunit of phosphatidylinositol 3-kinase in B cell development and activation. J Exp Med. 2002;196(6):753–763. doi: 10.1084/jem.20020805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furman RR, et al. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med. 2014;370(11):997–1007. doi: 10.1056/NEJMoa1315226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flinn IW, et al. Duvelisib, a novel oral dual inhibitor of PI3K-δ,γ, is clinically active in advanced hematologic malignancies. Blood. 2018;131(8):877–887. doi: 10.1182/blood-2017-05-786566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chantry D, et al. p110δ, a novel phosphatidylinositol 3-kinase catalytic subunit that associates with p85 and is expressed predominantly in leukocytes. J Biol Chem. 1997;272(31):19236–19241. doi: 10.1074/jbc.272.31.19236. [DOI] [PubMed] [Google Scholar]
- 30.Hock BD, MacPherson SA, McKenzie JL. Idelalisib and caffeine reduce suppression of T cell responses mediated by activated chronic lymphocytic leukemia cells. PLoS One. 2017;12(3):e0172858. doi: 10.1371/journal.pone.0172858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali K, et al. Inactivation of PI(3)K p110δ breaks regulatory T-cell-mediated immune tolerance to cancer. Nature. 2014;510(7505):407–411. doi: 10.1038/nature13444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowers JS, et al. PI3Kδ inhibition enhances the antitumor fitness of adoptively transferred CD8+ T cells. Front Immunol. 2017;8:1221. doi: 10.3389/fimmu.2017.01221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jou ST, et al. Essential, nonredundant role for the phosphoinositide 3-kinase p110delta in signaling by the B-cell receptor complex. Mol Cell Biol. 2002;22(24):8580–8591. doi: 10.1128/MCB.22.24.8580-8591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okkenhaug K, Patton DT, Bilancio A, Garçon F, Rowan WC, Vanhaesebroeck B. The p110delta isoform of phosphoinositide 3-kinase controls clonal expansion and differentiation of Th cells. J Immunol. 2006;177(8):5122–5128. doi: 10.4049/jimmunol.177.8.5122. [DOI] [PubMed] [Google Scholar]
- 35.Szczepanski MJ, et al. Increased frequency and suppression by regulatory T cells in patients with acute myelogenous leukemia. Clin Cancer Res. 2009;15(10):3325–3332. doi: 10.1158/1078-0432.CCR-08-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Q, et al. Program death-1 signaling and regulatory T cells collaborate to resist the function of adoptively transferred cytotoxic T lymphocytes in advanced acute myeloid leukemia. Blood. 2010;116(14):2484–2493. doi: 10.1182/blood-2010-03-275446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beyer M, et al. Reduced frequencies and suppressive function of CD4+CD25hi regulatory T cells in patients with chronic lymphocytic leukemia after therapy with fludarabine. Blood. 2005;106(6):2018–2025. doi: 10.1182/blood-2005-02-0642. [DOI] [PubMed] [Google Scholar]
- 38.Fabbri G, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med. 2011;208(7):1389–1401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goolsby CL, Kuchnio M, Finn WG, Peterson L. Expansions of clonal and oligoclonal T cells in B-cell chronic lymphocytic leukemia are primarily restricted to the CD3(+)CD8(+) T-cell population. Cytometry. 2000;42(3):188–195. doi: 10.1002/1097-0320(20000615)42:3<188::AID-CYTO5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 40.Van den Hove LE, et al. Peripheral blood lymphocyte subset shifts in patients with untreated hematological tumors: evidence for systemic activation of the T cell compartment. Leuk Res. 1998;22(2):175–184. doi: 10.1016/S0145-2126(97)00152-5. [DOI] [PubMed] [Google Scholar]
- 41.Görgün G, Holderried TA, Zahrieh D, Neuberg D, Gribben JG. Chronic lymphocytic leukemia cells induce changes in gene expression of CD4 and CD8 T cells. J Clin Invest. 2005;115(7):1797–1805. doi: 10.1172/JCI24176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramsay AG, et al. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. J Clin Invest. 2008;118(7):2427–2437. doi: 10.1172/JCI35017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casamayor-Palleja M, Khan M, MacLennan IC. A subset of CD4+ memory T cells contains preformed CD40 ligand that is rapidly but transiently expressed on their surface after activation through the T cell receptor complex. J Exp Med. 1995;181(4):1293–1301. doi: 10.1084/jem.181.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pascutti MF, et al. IL-21 and CD40L signals from autologous T cells can induce antigen-independent proliferation of CLL cells. Blood. 2013;122(17):3010–3019. doi: 10.1182/blood-2012-11-467670. [DOI] [PubMed] [Google Scholar]
- 45.Bichi R, et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci U S A. 2002;99(10):6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gorgun G, et al. E(mu)-TCL1 mice represent a model for immunotherapeutic reversal of chronic lymphocytic leukemia-induced T-cell dysfunction. Proc Natl Acad Sci U S A. 2009;106(15):6250–6255. doi: 10.1073/pnas.0901166106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hofbauer JP, et al. Development of CLL in the TCL1 transgenic mouse model is associated with severe skewing of the T-cell compartment homologous to human CLL. Leukemia. 2011;25(9):1452–1458. doi: 10.1038/leu.2011.111. [DOI] [PubMed] [Google Scholar]
- 48.Widhopf GF, et al. ROR1 can interact with TCL1 and enhance leukemogenesis in Eμ-TCL1 transgenic mice. Proc Natl Acad Sci U S A. 2014;111(2):793–798. doi: 10.1073/pnas.1308374111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gassner FJ, et al. Chronic lymphocytic leukaemia induces an exhausted T cell phenotype in the TCL1 transgenic mouse model. Br J Haematol. 2015;170(4):515–522. doi: 10.1111/bjh.13467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson AJ, et al. Characterization of the TCL-1 transgenic mouse as a preclinical drug development tool for human chronic lymphocytic leukemia. Blood. 2006;108(4):1334–1338. doi: 10.1182/blood-2005-12-011213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramsay AG, Evans R, Kiaii S, Svensson L, Hogg N, Gribben JG. Chronic lymphocytic leukemia cells induce defective LFA-1-directed T-cell motility by altering Rho GTPase signaling that is reversible with lenalidomide. Blood. 2013;121(14):2704–2714. doi: 10.1182/blood-2012-08-448332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pekarsky Y, et al. Tcl1 enhances Akt kinase activity and mediates its nuclear translocation. Proc Natl Acad Sci U S A. 2000;97(7):3028–3033. doi: 10.1073/pnas.97.7.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen SS, et al. Epigenetic alterations in a murine model for chronic lymphocytic leukemia. Cell Cycle. 2009;8(22):3663–3667. doi: 10.4161/cc.8.22.9957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaudio E, et al. Tcl1 interacts with Atm and enhances NF-κB activation in hematologic malignancies. Blood. 2012;119(1):180–187. doi: 10.1182/blood-2011-08-374561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reif K, Okkenhaug K, Sasaki T, Penninger JM, Vanhaesebroeck B, Cyster JG. Cutting edge: differential roles for phosphoinositide 3-kinases, p110γ and p110δ, in lymphocyte chemotaxis and homing. J Immunol. 2004;173(4):2236–2240. doi: 10.4049/jimmunol.173.4.2236. [DOI] [PubMed] [Google Scholar]
- 56.Yuan R, et al. Aging in inbred strains of mice: study design and interim report on median lifespans and circulating IGF1 levels. Aging Cell. 2009;8(3):277–287. doi: 10.1111/j.1474-9726.2009.00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woyach JA, et al. Bruton’s tyrosine kinase (BTK) function is important to the development and expansion of chronic lymphocytic leukemia (CLL) Blood. 2014;123(8):1207–1213. doi: 10.1182/blood-2013-07-515361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Göthert JR, et al. Expanded CD8+ T cells of murine and human CLL are driven into a senescent KLRG1+ effector memory phenotype. Cancer Immunol Immunother. 2013;62(11):1697–1709. doi: 10.1007/s00262-013-1473-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Curiel TJ. Regulatory T cells and treatment of cancer. Curr Opin Immunol. 2008;20(2):241–246. doi: 10.1016/j.coi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patton DT, et al. Cutting edge: the phosphoinositide 3-kinase p110 delta is critical for the function of CD4+CD25+Foxp3+ regulatory T cells. J Immunol. 2006;177(10):6598–6602. doi: 10.4049/jimmunol.177.10.6598. [DOI] [PubMed] [Google Scholar]
- 61.De Kouchkovsky I, Abdul-Hay M. ‘Acute myeloid leukemia: a comprehensive review and 2016 update’. Blood Cancer J. 2016;6(7):e441. doi: 10.1038/bcj.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holler C, et al. PKCbeta is essential for the development of chronic lymphocytic leukemia in the TCL1 transgenic mouse model: validation of PKCbeta as a therapeutic target in chronic lymphocytic leukemia. Blood. 2009;113(12):2791–2794. doi: 10.1182/blood-2008-06-160713. [DOI] [PubMed] [Google Scholar]
- 63.Lutzny G, et al. Protein kinase c-β-dependent activation of NF-κB in stromal cells is indispensable for the survival of chronic lymphocytic leukemia B cells in vivo. Cancer Cell. 2013;23(1):77–92. doi: 10.1016/j.ccr.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nguyen PH, et al. LYN kinase in the tumor microenvironment is essential for the progression of chronic lymphocytic leukemia. Cancer Cell. 2016;30(4):610–622. doi: 10.1016/j.ccell.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 65.Nganga VK, et al. Accelerated progression of chronic lymphocytic leukemia in Eμ-TCL1 mice expressing catalytically inactive RAG1. Blood. 2013;121(19):3855–3366. doi: 10.1182/blood-2012-08-446732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Herman SE, et al. Phosphatidylinositol 3-kinase-δ inhibitor CAL-101 shows promising preclinical activity in chronic lymphocytic leukemia by antagonizing intrinsic and extrinsic cellular survival signals. Blood. 2010;116(12):2078–2088. doi: 10.1182/blood-2010-02-271171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoellenriegel J, et al. The phosphoinositide 3′-kinase delta inhibitor, CAL-101, inhibits B-cell receptor signaling and chemokine networks in chronic lymphocytic leukemia. Blood. 2011;118(13):3603–3612. doi: 10.1182/blood-2011-05-352492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balakrishnan K, et al. The phosphoinositide-3-kinase (PI3K)-delta and gamma inhibitor, IPI-145 (Duvelisib), overcomes signals from the PI3K/AKT/S6 pathway and promotes apoptosis in CLL. Leukemia. 2015;29(9):1811–1822. doi: 10.1038/leu.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fiorcari S, et al. The PI3-kinase delta inhibitor idelalisib (GS-1101) targets integrin-mediated adhesion of chronic lymphocytic leukemia (CLL) cell to endothelial and marrow stromal cells. PLoS One. 2013;8(12):e83830. doi: 10.1371/journal.pone.0083830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McClanahan F, et al. Mechanisms of PD-L1/PD-1-mediated CD8 T-cell dysfunction in the context of aging-related immune defects in the Eμ-TCL1 CLL mouse model. Blood. 2015;126(2):212–221. doi: 10.1182/blood-2015-02-626754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 72.Kocher T, et al. CD4+ T cells, but not non-classical monocytes, are dispensable for the development of chronic lymphocytic leukemia in the TCL1-tg murine model. Leukemia. 2016;30(6):1409–1413. doi: 10.1038/leu.2015.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu P, et al. Intratumor depletion of CD4+ cells unmasks tumor immunogenicity leading to the rejection of late-stage tumors. J Exp Med. 2005;201(5):779–791. doi: 10.1084/jem.20041684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mohme M, Riethdorf S, Pantel K. Circulating and disseminated tumour cells — mechanisms of immune surveillance and escape. Nat Rev Clin Oncol. 2017;14(3):155–167. doi: 10.1038/nrclinonc.2016.144. [DOI] [PubMed] [Google Scholar]
- 75.Liu D, Uzonna JE. The p110 delta isoform of phosphatidylinositol 3-kinase controls the quality of secondary anti-Leishmania immunity by regulating expansion and effector function of memory T cell subsets. J Immunol. 2010;184(6):3098–3105. doi: 10.4049/jimmunol.0903177. [DOI] [PubMed] [Google Scholar]
- 76.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535(7610):75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 77.Johansson ME, et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe. 2015;18(5):582–592. doi: 10.1016/j.chom.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Spiljar M, Merkler D, Trajkovski M. The immune system bridges the gut microbiota with systemic energy homeostasis: focus on TLRs, mucosal barrier, and SCFAs. Front Immunol. 2017;8:1353. doi: 10.3389/fimmu.2017.01353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17(4):219–232. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 80.Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 2018;33(4):570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Steinbach EC, et al. Innate PI3K p110δ regulates Th1/Th17 development and microbiota-dependent colitis. J Immunol. 2014;192(8):3958–3968. doi: 10.4049/jimmunol.1301533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vanhaesebroeck B, et al. P110δ, a novel phosphoinositide 3-kinase in leukocytes. Proc Natl Acad Sci U S A. 1997;94(9):4330–4335. doi: 10.1073/pnas.94.9.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sujobert P, et al. Essential role for the p110delta isoform in phosphoinositide 3-kinase activation and cell proliferation in acute myeloid leukemia. Blood. 2005;106(3):1063–1066. doi: 10.1182/blood-2004-08-3225. [DOI] [PubMed] [Google Scholar]
- 84.Lapalombella R, et al. Tetraspanin CD37 directly mediates transduction of survival and apoptotic signals. Cancer Cell. 2012;21(5):694–708. doi: 10.1016/j.ccr.2012.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.