Abstract
The nucleic acids of Mycobacterium tuberculosis can be detected by intracellular DNA sensors, such as cyclic GMP-AMP synthase and absent in melanoma 2 (AIM2), which results in the release of type I IFN and the proinflammatory cytokine IL-1β. However, whether cross-talk occurs between AIM2–IL-1β and cyclic GMP-AMP synthase–type I IFN signaling upon M. tuberculosis infection in vivo is unclear. In this article, we demonstrate that mycobacterial infection of AIM2−/− mice reciprocally induces overreactive IFN-γ and depressive IFN-γ responses, leading to higher infection burdens and more severe pathology. We also describe the underlying mechanism whereby activated apoptosis-associated speck-like protein interacts with a key adaptor, known as stimulator of IFN genes (STING), and inhibits the interaction between STING and downstream TANK-binding kinase 1 in bone marrow–derived macrophages and bone marrow–derived dendritic cells, consequently reducing the induction of type I IFN. Of note, apoptosis-associated speck-like protein expression is inversely correlated with IFN-β levels in PBMCs from tuberculosis patients. These data demonstrate that the AIM2–IL-1β signaling pathway negatively regulates the STING–type I IFN signaling pathway by impeding the association between STING and TANK-binding kinase 1, which protects the host from M. tuberculosis infection. This finding has potential clinical significance. The Journal of Immunology, 2018, 200: 1016–1026.
Tuberculosis (TB) has existed for millennia and remains a major global health problem that now ranks above HIV/AIDS as a leading cause of death from infectious disease. One third of the world’s population has been infected with TB. Of these infected people, 1.4 million died in 2015 (1). The lack of effective vaccines and sensitive diagnostic methods has affected global control of the TB epidemic. The immune system plays critical roles in controlling TB infection (2). Therefore, the incompletely characterized immune responses of TB patients must be explored to enable the establishment of new vaccines, diagnostics, and treatments.
The innate immune system recognizes pathogen signatures, known as pathogen-associated molecular patterns (PAMPs), to sense pathogenic invasion by using germline-encoded pattern recognition receptors; the detection of foreign nucleic acids acting as classical PAMPs has evolved as a fundamental mechanism of host defense (3). Mycobacterium tuberculosis can activate the DNA-dependent cytosolic surveillance pathway (4). When the immune system of the host senses the invasion of M. tuberculosis, the DNA sensor cyclic GMP-AMP synthase (cGAS) binds to the genomic DNA (GD) of the pathogen and catalyzes the production of cyclic GMP-AMP (cGAMP). cGAMP then activates a downstream adaptor molecule known as stimulator of IFN genes (STING), which promotes the translocation of STING from the endoplasmic reticulum to the perinuclear microsomal compartments. After STING activation, downstream molecules, TANK-binding kinase 1 (TBK1) and IFN regulatory factor 3 (IRF3), are activated, which ultimately results in the release of type I IFN (5–7).
Although type I IFN plays an important role in the antiviral immune response, studies of patients and mice have highlighted the detrimental role of IFN-α/β during M. tuberculosis infection (8). Many reports have demonstrated that patients with active TB exhibit a prominent IFN-α/β–inducible transcription signature that correlates with the extent of radiographic disease and diminishes with successful treatment (9–11). Furthermore, disruption of the IFN-α/β receptor gene or Ab-mediated neutralization of IFN-α/β increases the survival rate of infected mice relative to wild-type (WT) control mice (12). Increased infection severity has also been demonstrated through direct instillation of IFN-α/β into the lungs and through administration of a TLR3 ligand that induces IFN-α/β (13, 14).
In addition to detection by the STING–type I IFN signaling pathway, GD of M. tuberculosis can be detected by absent in melanoma 2 (AIM2), another important DNA sensor. When AIM2 senses abnormal cytoplasmic DNA, it associates with adaptor molecules known as apoptosis-associated speck-like protein (ASC) and inactivated procaspase-1, forming a complex called the AIM2 inflammasome, which induces the production of proinflammatory cytokines, such as IL-1β and IL-18 (15–17). Unlike type I IFN, proinflammatory cytokines are conducive to the antibacterial immune response. For example, IL-1α/β–knockout mice develop significantly larger granulomatous lesions than WT mice postinfection with H37Rv (18). Furthermore, IL-1R−/− mice show increased mortality and enhanced mycobacterial outgrowth in their lungs and distant organs compared with normal mice (19). Additionally, the low production of IL-1β and IL-18 in AIM2−/− mice increases susceptibility to M. tuberculosis infection (20).
Because the balance between AIM2–IL-1β and cGAS–type I IFN signaling governs the outcome of M. tuberculosis infection, it is important to address whether there is a cross-talk between these two signaling pathways during M. tuberculosis infection in vivo. In this study, we demonstrated that the overreactive IFN-β and less protective IFN-γ responses were induced in AIM2−/− mice upon mycobacterial infection, leading to higher infection burdens and more severe pathology. Furthermore, our mechanistic study indicated that activated ASC interacted with STING and blocked the association between TBK1 and STING, thereby inhibiting the induction of type I IFN. Our study reveals a novel mechanism that is involved in the cross-talk between the AIM2–IL-1β and STING–type I IFN pathways during mycobacterial infection. This mechanism may have potential applications in the clinic.
Materials and Methods
Mice
AIM2−/− and Caspase-1−/− mice were obtained from The Jackson Laboratory. ASC−/− mice have been described previously (21). C57BL/6 WT mice were purchased from the Shanghai Laboratory Animal Center. All mice were housed in specific pathogen–free conditions.
Reagents
Polyinosinic-polycytidylic acid [poly(I:C)] and IFN stimulatory DNA (ISD) were purchased from Invitrogen. Poly(deoxyadenylic-thymidylic) acid [poly(dA:dT)], cGAMP, and LPS were purchased from Sigma. Genomic DNA from the spleens of C57BL/6 mice was obtained in-house using a DNeasy Blood & Tissue Kit (69504; QIAGEN). 150mer dsDNA was amplified by PCR from the β-actin gene and purified using a Plus DNA Clean/Extraction Kit (DP034P-300; GeneMark). The following Abs were used for the immunoblotting analysis or immunoprecipitation: anti-HA (CO-MMS-101R; Covance), anti-Flag (F3165; Sigma), anti–c-Myc (A02060; AmyJet Scientific), anti-STING (3337; Cell Signaling Technology), anti-TBK1 (3013S; Cell Signaling Technology), anti-phosphorylated TBK1 (5483S; Cell Signaling Technology), anti-IRF3 (sc-9082; Santa Cruz Biotechnology), and anti-phosphorylated IRF3 (4947s; Cell Signaling Technology). Anti-ASC Ab was produced by immunization of rabbits with ASC expressed by Escherichia coli. The following Abs were used for confocal microscopy: Alexa Fluor 488 goat anti-mouse IgG (A11034; Molecule Probes) and Cy3 goat anti-rabbit IgG (115–165-146; Jackson ImmunoResearch). Lipofectamine 2000 was obtained from Invitrogen. X-tremeGENE DNA transfection reagent was purchased from Roche. TurboFect Transfection Reagent was purchased from Thermo Fisher. A Dual-Luciferase Reporter Assay System was obtained from Promega. Protein A/G PLUS-Agarose Immunoprecipitation Reagent was purchased from Santa Cruz Biotechnology. Anti-HA and anti-FLAG beads were purchased from Sigma. Murine IFN-β (439408; BioLegend) and mouse IFN-r (P105796; R&D Systems) ELISA kits were used. All ASC truncations were generated by PCR and subcloned into a pcDNA3 vector. The other plasmids were generated or obtained as we described previously (22).
Mycobacteria strain and culture conditions
Mycobacterium bovis bacillus Calmette-Guérin (BCG) Danish strain (ATCC 35733) was grown at 37°C in Middlebrook 7H9 broth or on Middlebrook 7H10 agar (both from Difco) supplemented with 10% oleic acid–albumin–dextrose–catalase–enriched Middlebrook (BD), 0.2% glycerol, and 0.05% Tween-80 for 3–4 wk. CFU of bacteria were counted.
Mouse infection
WT, AIM2−/−, and Caspase-1−/− mice were challenged by tail vein injection with 4 × 107 CFU attenuated M. bovis (BCG) in 200 μl of PBS per mouse (4–6 wk of age) at 0, 20, 30, and 40 d. Fifty days after the first injection, mice were sacrificed, and samples were harvested. The amounts of IFN-β and IFN-γ in the serum were detected as soon as possible. The lungs were homogenized in PBS and plated on 7H11 agar to determine the bacterial burden.
Histopathology analysis
The lungs of each mouse were excised and cut into two pieces: one was subjected to CFU counting, and one was fixed in a 4% neutral-buffered paraformaldehyde solution for 24 h. After paraffin embedding, a series of sections with a thickness of 4–7 μm were cut and stained with H&E using standard methods, and images were obtained using a microscope (BX51WI; Olympus).
Cell culture, transfection, and stimulation
Generation of bone marrow–derived macrophages (BMDMs) and bone marrow–derived dendritic cells (BMDCs) has been described previously (22). Briefly, bone marrow cells were flushed from the femurs and tibias of mice and subsequently depleted of RBCs with RBC lysing buffer. BMDCs were cultured at 1.5 × 106 cells per well in 24-well plates in RPMI 1640 supplemented with 10% (v/v) FBS, 20 ng/ml murine GM-CSF, and 2 mM L-glutamine. BMDMs were cultured at 3 × 106 cells per well in 12-well plates in DMEM supplemented with 10% (v/v) FBS and 20 ng/ml murine M-CSF. Fresh medium was added every 2 d. BMDCs and BMDMs were collected for further experiments on days 7 and 5, respectively. MEFs (American Type Culture Collection), RAW264.7 cells (Cell Bank of Chinese Academy of Sciences), and HEK293T cells (Cell Bank of Chinese Academy of Sciences) were cultured in humidified 5% CO2 at 37°C in DMEM supplemented with 10% (v/v) FBS, 0.5% penicillin (100 U/ml), and streptomycin (100 U/ml). X-tremeGENE was used for transient transfections of plasmid DNA into MEFs. Transfections of HEK293T cells were performed using TurboFect. Poly(dA:dT), ISD, cGAMP, GD, 150mer dsDNA, and poly(I:C) were transfected using Lipofectamine 2000, according to the manufacturer’s recommendations.
Generation of RAW-GFP and RAW-ASC cells
ASC was cloned into MSCV–IRES–GFP and transfected with pCMV/VSV-G and pKF3-RSV/Gag-Pol into HEK293T cells by calcium phosphate transfection. After 6 and 24 h, the medium was replaced with fresh DMEM containing 10% FBS. Forty-eight hours after the transfections, the retroviral supernatants were harvested, supplemented with 16 μg/ml Polybrene, and used to infect RAW264.7 cells. The cell culture plates were centrifuged at 1800 × g for 90 min at room temperature. After 12 h of incubation at 37°C, the retroviruses were removed, and the cells were cultured in fresh medium for another 12 h. Finally, the cells were collected to sort GFP+ cells using a FACSAria II (BD Biosciences) for culture and further experiments.
Real-time PCR
RNA that was extracted from the cells was reverse transcribed using a Prime Script RT Master Mix Kit (Takara), and quantitative PCR analysis was performed on an ABI Q6. All gene expression data are relative to HPRT. The following primers were used for PCR: IFN-β sp 5′-CCTGGAGCAGCTGAATGGAA-3′, as 5′-TTGAAGTCCGCCCTGTAGGT-3′ and HPRT sp 5′-TGCTCGAGATGTCATGAAGGAG-3′, as 5′-CAGAGGGCCACAATGTGATG-3′.
Immunoprecipitation and immunoblotting analysis
Immunoprecipitation and immunoblotting procedures have been described (22). Briefly, for exogenous coimmunoprecipitation experiments, HEK293T cells were transfected with various combinations of plasmids. Twenty-four hours after the transfections, cell lysates were prepared in lysis buffer and incubated with hemagglutinin (HA) beads overnight at 4°C. Complexes were washed three times with lysis buffer and analyzed by immunoblotting. For the endogenous coimmunoprecipitation experiments, BMDCs were stimulated with poly(dA:dT) for 6 h. Lysates were incubated with anti-ASC Ab or rabbit IgG Ab and analyzed by immunoblotting with anti-STING Ab.
Confocal microscopy
MEFs were transfected with the HA–ASC and Flag–STING plasmids. After 24 h, the cells were collected and seeded onto glass coverslips in 24-well plates overnight before stimulation with 1 μg/ml poly(dA:dT) or were left unstimulated for 4 h. After stimulation, cells were fixed with 4% paraformaldehyde in PBS, permeabilized with Triton X-100, and blocked with 1% BSA in PBS. Cells were stained with anti-HA mouse Ab and anti-FLAG rabbit Ab, followed by Cy3-conjugated anti-rabbit Ab and Alexa Fluor 488–conjugated anti-mouse IgG Ab, and counterstained for nucleic acids with DAPI. Finally, fluorescence images were captured with a Leica TCS SP5 MP laser confocal microscope.
Luciferase reporter gene assay
MEFs were cotransfected with an IFN-β, ISRE, or IFN-α4 luciferase reporter plasmid, a Renilla luciferase plasmid as an internal control, and the indicated expression plasmids. An empty control vector was added to equalize the amount of DNA added to each well of cells. Twenty-four hours after the transfection, cells were lysed, and the reporter activity was measured using a Dual-Luciferase Reporter Assay System (Promega).
Patients
PBMCs were isolated from EDTA-treated blood by Ficoll-Paque Plus density gradient centrifugation. TB patients were enrolled from the Shanghai Pulmonary Hospital (Shanghai, China). Age- and gender-matched healthy volunteers were recruited into this study who had no history or evidence of TB (negative T-SPOT for TB). None of the participants showed evidence of acute infection with hepatitis B virus, hepatitis C virus, HIV, or other infectious agent or of cancers. Informed consent was obtained from all participants.
Ethics statement
All experiments were carried out according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals with the approval of the Institutional Animal Care and Use Committee of the Institute Pasteur of Shanghai and the Institutional Animal Care and Use Committee of the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (approval number SIBCB-S303–1608-025). Protocols for the use of human blood samples for in vitro experimental procedures were evaluated and approved by the Institutional Review Board for Human Subjects Research and the Institutional Biosafety Committee at the Institute Pasteur of Shanghai, Shanghai Pulmonary Hospital. All studies were conducted in accordance with the amended Declaration of Helsinki (International Conference on Harmonisation-Good Clinical Practice) and consistent with the guidelines of the Office for Human Research Protections. All subjects were adults and signed written informed consent forms.
Statistical analysis
Statistical comparisons between the different treatments were performed using the unpaired Student t test. Correlations were performed using the Pearson test. Statistically significant p values are noted in the figures.
Results
Mycobacterial infection of AIM2−/− mice reciprocally induces overreactive IFN-β and depressive IFN-γ responses, leading to higher infection burdens and more severe pathology
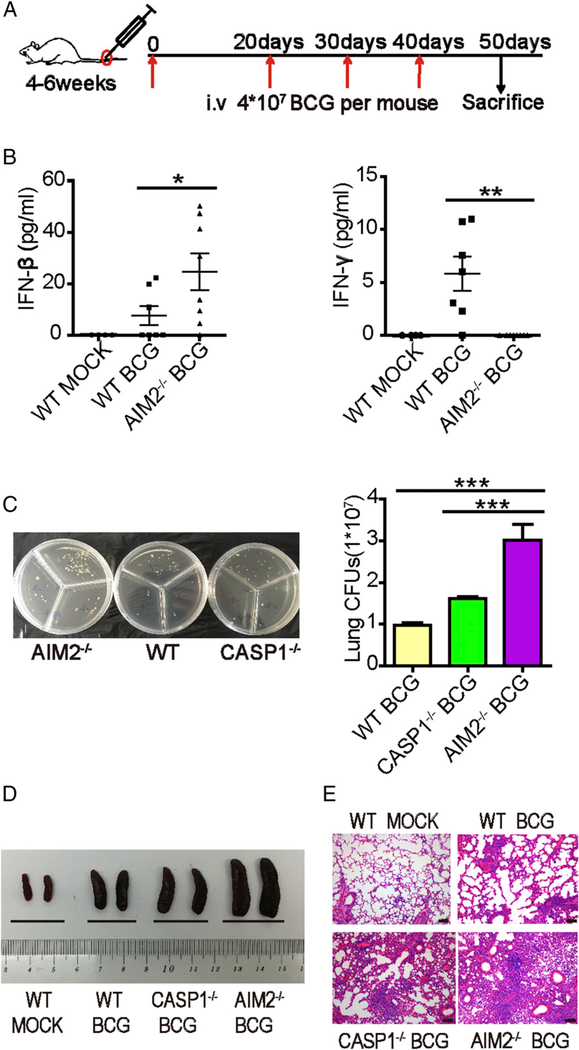
To explore whether the AIM2–IL-1β signaling pathway affected the STING–type I IFN signaling pathway during mycobacterial infection in vivo, AIM2−/− (Supplemental Fig. 1A, 1B) and WT mice were infected with attenuated M. bovis (BCG) (Fig. 1A), and their levels of protective IFN-γ, bacilli-enhancing IFN-β, and BCG infection burdens, as well as the severity of tissue pathology, were assessed. Caspase-1−/− mice (Supplemental Fig. 1D) were used as the control for AIM2-deficient mice in the context of BCG infection based on the involvement of the AIM2/ASC/caspase-1 inflammasome complex that is capable of regulating TB infection (20). We used BCG infection for two reasons: 1) BCG infection and vaccination reportedly induce a strong IFN-γ response and immunity (23), and potential overproduction of IFN-β in infected AIM2−/− mice may antagonize IFN-γ, leading to a loss of immunity or a different outcome and 2) high levels of mycobacterial DNA can be generated by repeated BCG infections, making it possible to optimally uncover the DNA sensor–driven IFN-β function and mechanisms in vivo during repeated BCG infections. Moreover, BCG could readily allow us to conduct proof-of-concept and mechanistic studies at Animal Biosafety Level 2, without the need for expensive or highly regulated Animal Biosafety Level 3 facilities. Interestingly, serum IFN-β levels were significantly higher in AIM2−/− mice than in WT control mice (Fig. 1B), suggesting that AIM2 deficiency might enhance the induction of type I IFN during mycobacterial infection in vivo. Surprisingly, BCG infection of AIM2−/− mice with overreactive IFN-β almost shut down the protective IFN-γ response, whereas WT mice exhibited robust IFN-γ production in the serum (Fig. 1B). The strong inverse correlation between IFN-β and IFN-γ in WT and AIM2−/− mice suggested that IFN-β overproduction might help to antagonize the protective IFN-γ response in mycobacterial infection (24). In contrast to AIM2−/− mice, Caspase-1−/− controls did not exhibit significantly higher serum IFN-β levels than WT mice after BCG infection, whereas AIM2−/− and Caspase-1−/− mice produced less IL-1β than WT animals (Supplemental Fig. 2). Notably, BCG-infected AIM2−/− mice showed significantly higher levels of mycobacterial burdens in their lung tissues compared with WT and Caspase-1−/− controls (Fig. 1C). Moreover, BCG infection of AIM2−/− mice caused a more striking splenomegaly compared with that of WT and Caspase-1−/− mice (Fig. 1D). Consistently, BCG-infected AIM2−/− mice showed more severe lung pathology, which was characterized by apparent infiltration of inflammatory exudates/cells in the lung tissues, compared with WT and Caspase-1−/− controls (Fig. 1E). Taken together, these results demonstrated that BCG infection of AIM2−/− mice reciprocally induced overreactive IFN-β and depressive IFN-γ responses, leading to higher infection burdens and more severe pathology compared with WT and Caspase-1−/− controls.
FIGURE 1.
AIM2-mediated inhibition of IFN-β production is necessary for controlling M. bovis (BCG) infection in vivo. WT, AIM2−/−, and Caspase-1−/− mice were infected with 4×107 CFU BCG four times. (A) Schematic diagram of the infection with BCG. (B) Levels of IFN-β and IFN-γ in the sera of WT and AIM2−/− mice, with or without BCG infection. (C) Bacterial CFU counts in the homogenates of lungs from BCG-infected WT, AIM2−/−, and Caspase-1−/− mice in a representative of two independent experiments. Five to six mice per group were used in the experiments, and two AIM2−/− mice died with extremely high CFU counts (outlier not included). Similar significance trends for comparisons between AIM2−/− and control groups were seen in the other experiment. (D) Gross pathology features of the spleens of two representative mice from each group infected with BCG. (E) Histopathology of the lungs from mice infected with BCG. Sections were stained with H&E and assessed. Scale bars, 100 μm. Data are representative of at least two independent experiments. The results are shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, Student t test.
AIM2 deficiency enhances STING-driven type I IFN signaling upon BCG infection or stimulation with various DNA forms in BMDCs and BMDMs
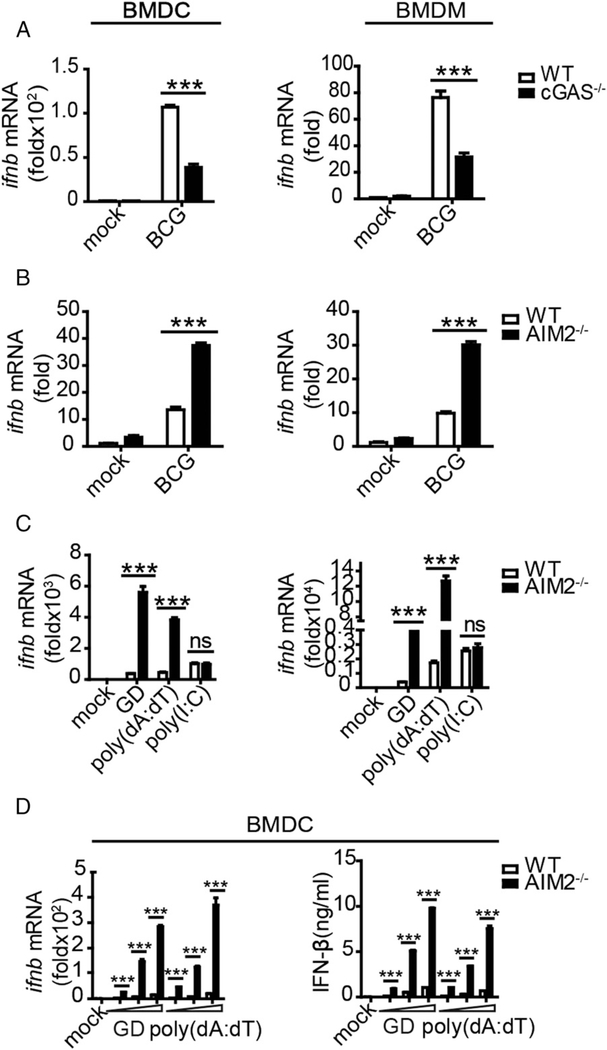
We aimed to dissect the signaling cascade whereby AIM2 deficiency during BCG infection led to an overreactive IFN-β response and exacerbated pathology. Because macrophages and dendritic cells in the lungs are major target cells for M. tuberculosis and BCG during the early stage of infection (2), we used BMDMs and BMDCs for the mycobacterial DNA–mediated signaling studies. We first examined whether BCG, similar to M. tuberculosis bacilli, could induce IFN-β production through activation of the cGAS–STING–TBK1 signaling pathway. Indeed, BCG behaved like M. tuberculosis in terms of cGAS-driven production of IFN-β, which was reflected by a significant reduction in BCG-stimulated production of IFN-β after cGAS was knocked down in BMDCs or BMDMs (Fig. 2A). The in vivo data led us to speculate that the AIM2 inflammasome signaling pathway might inhibit the production of IFN-β upon BCG infection. To test this speculation, BMDCs and BMDMs from WT and AIM2−/− mice were stimulated with BCG in culture and assessed for an IFN-β response. We found that BCG infection of AIM2-deficient BMDCs and BMDMs also upregulated IFN-β (Fig. 2B), thus recapitulating the in vivo finding in BCG-infected AIM2−/− mice (Fig. 1B). To examine this further, we determined whether other common forms of DNA, such as poly(dA:dT) and murine-derived GD, promoted IFN-β production based on a similar effect that was previously reported with tumor-derived DNA (25). We observed that stimulation of AIM2-deficient BMDCs and BMDMs with GD and poly(dA:dT), but not with the RNA sensor ligand poly(I:C), similarly enhanced the production of IFN-β and related cytokines (i.e., IFN-α1 and IP-10) (Fig. 2C, Supplemental Fig. 3A, 3B). Moreover, the enhancing effect in the absence of AIM2 was dependent on the DNA concentration at the IFN-β mRNA and protein levels (Fig. 2D). These data suggested that the AIM2 inflammasome signaling pathway negatively regulated STING-driven production of type I IFN upon stimulation with BCG infection or various DNA forms.
FIGURE 2.
AIM2 deficiency enhances the induction of IFN-β upon BCG infection or stimulation with various DNA forms in BMDCs and BMDMs. BMDCs and BMDMs from WT, cGAS−/− (A), and AIM2−/− mice (B) were infected with BCG (multiplicity of infection = 10) for 6 h, and IFN-β transcripts were measured by quantitative real-time PCR (qRT-PCR). (C) BMDCs and BMDMs from WT and AIM2−/− mice were stimulated with GD (1.5 μg/ml), poly(dA:dT) (1.5 μg/ml), and poly(I:C) (2 μg/ml), and IFN-β was measured by qRT-PCR 8 h after transfections. (D) BMDCs from WTand AIM2−/− mice were stimulated with 0.5, 1, or 2 μg/ml DNA for 10 h, and IFN-β was measured by qRT-PCR and ELISA. Data are representative of at least three independent experiments. The results are shown as mean ± SEM. ***p < 0.001, Student t test. ns, not significant.
Activated ASC negatively regulates the production of IFN-β
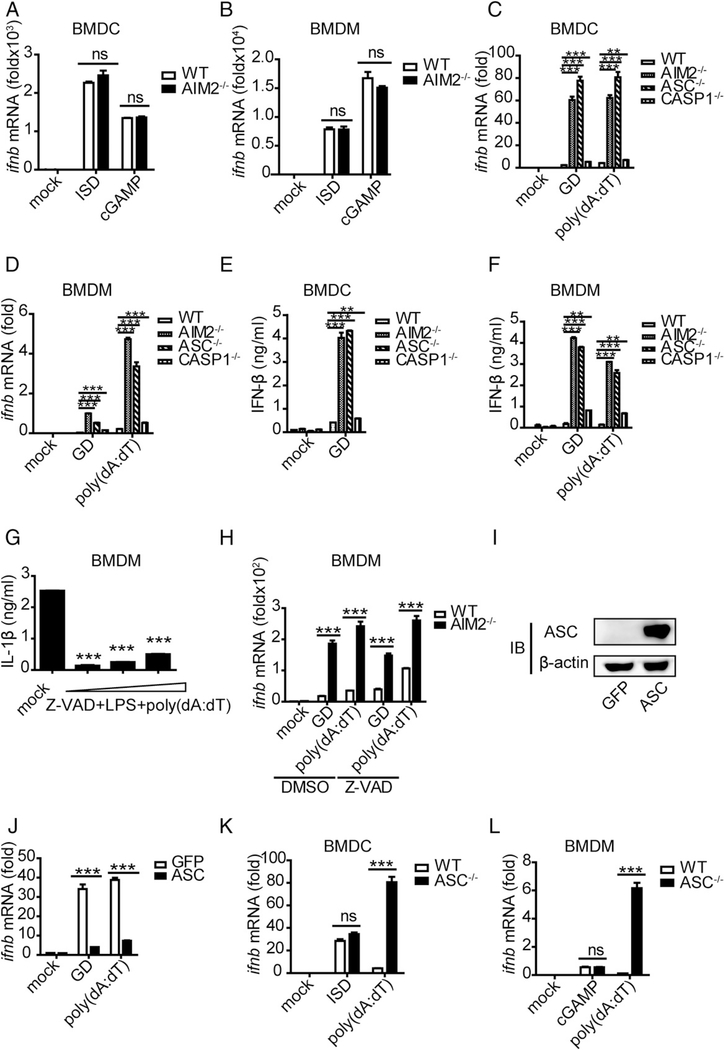
We examined whether AIM2-related inhibition of IFN-β induction in response to stimulation with BCG and various DNA forms was dependent on the AIM2 sensor itself or on the downstream inflammasome signaling of AIM2 in the context of the STING pathway. Interestingly, when STING-activating ISD (a 45-bp DNA) and cGAMP (the catalytic product of cGAS) were used to stimulate WT and AIM2−/− BMDCs and BMDMs, no difference in the induction of IFN-β was observed (Fig. 3A, 3B). Given that ISD and cGAMP activate only STING signaling and that poly(dA:dT) stimulates STING signaling and AIM2 inflammasome signaling (Supplemental Fig. 4A) (26), we speculated that the AIM2 inflammasome signaling pathway might be involved in the negative regulation of IFN-β. To identify potential regulatory molecules in the AIM2 inflammasome signaling pathway, we used B-form DNA, poly(dA:dT), or GD to activate downstream molecules in BMDCs and BMDMs from WT, AIM2−/−, ASC−/− (Supplemental Fig. 1C), and Caspase-1−/− mice. We found that IFN-β was increased only 2-fold in caspase-1–deficient cells compared with WT cells. In contrast, >10-fold increases in IFN-β were similarly observed in AIM2-deficient and ASC-deficient cells at the mRNA and protein levels (Fig. 3C-F). Notably, these results were consistent with previous reports (25). To confirm the role of caspase-1 in IFN-β production, we incubated WT and AIM2−/− BMDCs with Z-VAD-FAM, a caspase inhibitor, and then stimulated them with DNA. A concentration of 50 μM was chosen as the final working concentration based on its inhibitory efficiency of IL-1β (Fig. 3G). Although Z-VAD-FAM–mediated inhibition in WT BMDMs moderately upregulated IFN-β, it did not have a significant impact on the remarkably high levels of IFN-β in AIM2−/− BMDMs (Fig. 3H), suggesting that caspase-1/IL-1β or IL-18 might not be the major negative-regulatory molecules of IFN-β. This prompted the hypothesis that ASC, the upstream molecule, might be responsible for the negative regulation. To test this hypothesis, we overexpressed ASC in RAW264.7 cells and stimulated them with B-form DNA (Fig. 3I, Supplemental Fig. 3C). We found that overexpression of ASC led to marked decreases in IFN-β and a related cytokine, IP-10, compared with RAW264.7 control cells without ASC overexpression (Fig. 3J, Supplemental Fig. 3D). Similar to the setting of AIM2 deficiency, stimulation of the cGAS–STING pathway with ISD or cGAMP in ASC−/− BMDCs did not alter IFN-β production, whereas poly(dA:dT) stimulation in the absence of ASC led to remarkable increases in IFN-β (Fig. 3K, 3L). Taken together, these results suggest that the activated form of ASC may inhibit the production of IFN-β upon DNA stimulation.
FIGURE 3.
The activated form of ASC performs an inhibitory role on IFN-β upon DNA ligand stimulation. (A and B) BMDCs and BMDMs isolated from WT and AIM2−/− mice were stimulated with ISD (1.5 μg/ml) and cGAMP (4 μg/ml), and IFN-β was measured by quantitative real-time PCR (qRT-PCR) 8 h after transfection. (C and D) BMDCs and BMDMs isolated from WT, AIM2−/−, ASC−/−, and Caspase-1−/− mice were stimulated with GD (1.5 μg/ml) and poly(dA:dT) (1.5 μg/ml), and IFN-β was measured by qRT-PCR 10 h after transfection. (E and F) BMDCs and BMDMs isolated from WT, AIM2−/−, ASC−/−, and Caspase-1−/− mice were stimulated with GD (1.5 μg/ml) and poly(dA:dT) (1.5 μg/ml), and IFN-β was measured by ELISA 10 h after transfection. (G) BMDMs isolated from WT mice were stimulated with different concentrations of a caspase inhibitor (Z-VAD-FAM) for 1 h and then stimulated with LPS for 2 h before being stimulated with poly(dA:dT) (1.5 μg/ml); IL-1β was measured by ELISA 6 h after transfection. (H) BMDMs isolated from WT and AIM2−/− mice were stimulated with DMSO or a caspase inhibitor (Z-VAD-FAM) for 1 h and then stimulated with GD (1.5 μg/ml) and poly(dA:dT) (1.5 μg/ml). IFN-β was measured by qRT-PCR 8 h after transfection. (I) Immunoblot of ASC and β-actin in RAW-GFP and RAW-ASC cell lysates. (J) RAW-GFP and RAW-ASC cells were stimulated with GD (1.5 μg/ml) and poly(dA:dT) (1.5 μg/ml), and IFN-β was measured by qRT-PCR 8 h after transfection. (K and L) BMDCs and BMDMs isolated from WT and ASC−/− mice were stimulated with ISD (1.5 μg/ml), cGAMP (4 μg/ml), and poly(dA:dT) (1.5 μg/ml), and IFN-β was measured by qRT-PCR 8 h after transfection. Data are representative of at least two independent experiments. The results are shown as mean ± SEM. **p < 0.01, ***p < 0.001, Student t test. ns, not significant.
Activated ASC targets STING to inhibit the induction of IFN-β
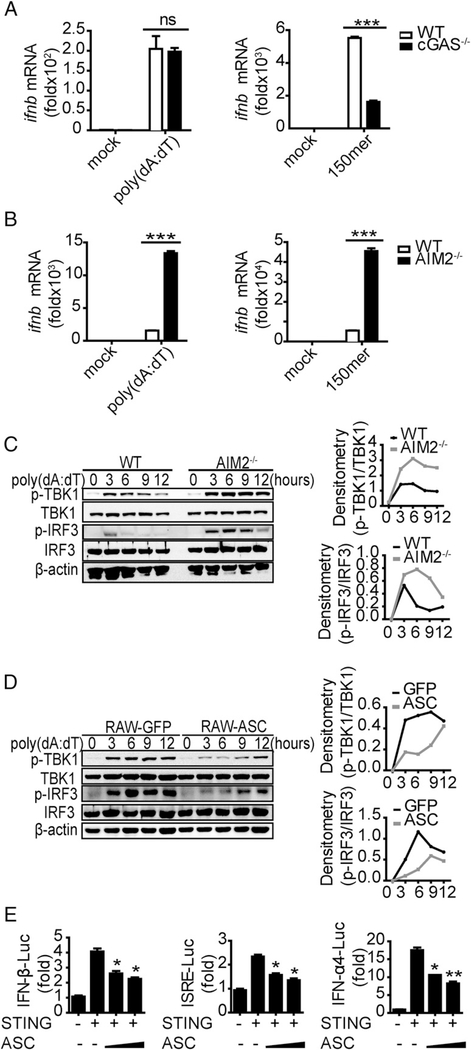
Several DNA sensors, including cGAS, DDX41, and IFI16 (27–29), have been discovered. To examine whether the DNA sensors were the target molecules that were negatively regulated, we stimulated WTand AIM2−/− BMDCs with poly(dA:dT) and synthetic 150mer dsDNA, with recognition of the former being independent of cGAS but recognition of the latter being dependent on cGAS in BMDCs (Fig. 4A, 4B). Induction of IFN-β was strongly increased in AIM2-deficient cells (Fig. 4A, 4B), suggesting that the target was not at the DNA sensor level but was likely downstream of the DNA sensors. To identify the potential target, we performed Western blots to assess activation of two downstream molecules: TBK1 and IRF3. Higher levels of phosphorylated TBK1 and IRF3, which are the activated forms of both molecules, were promoted by AIM2 deficiency relative to control (Fig. 4C). Consistent with the results from BMDCs, overexpression of ASC in RAW264.7 cells inhibited phosphorylation of TBK1 and IRF3 after stimulation with B-form DNA (Fig. 4D), indicating that the target might be upstream of TBK1 and IRF3. Thus, we hypothesized that ASC might target STING to inhibit the production of IFN-β upon DNA stimulation. As shown in Fig. 4E, exogenous expression of ASC indeed inhibited STING-induced IFN-β, ISRE, and IFN-α4 activation in a dose-dependent manner. Taken together, these data showed that activated ASC targeted STING to inhibit DNA-triggered IFN-β induction.
FIGURE 4.
ASC negatively regulates IFN-β production by affecting STING function. (A and B) BMDCs isolated from WT, cGAS−/−, and AIM2−/− mice were stimulated with poly(dA:dT) (1.5 μg/ml) and 150mer dsDNA (1.5 μg/ml), and IFN-β was measured by quantitative real-time PCR (qRT-PCR) 8 h after transfection. Immunoblot of p-TBK1, total TBK1, p-IRF3, total IRF3, and b-actin in WT and AIM2−/− BMDC lysates (C) or RAW-GFP and RAW-ASC cell lysates (D) that were stimulated with poly(dA:dT) (1.5 μg/ml) for the indicated times. (E) Luciferase activity of MEFs that were cotransfected with the IFN-β luciferase reporter (0.4 μg/ml), ISRE luciferase reporter (0.4 μg/ml), or IFN-α4 luciferase reporter (0.4 μg/ml) and Flag-STING (0.4 μg/ml) and the empty vector or increasing amounts of HA-ASC. Data are representative of at least two independent experiments. The results are shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, Student t test. ns, not significant.
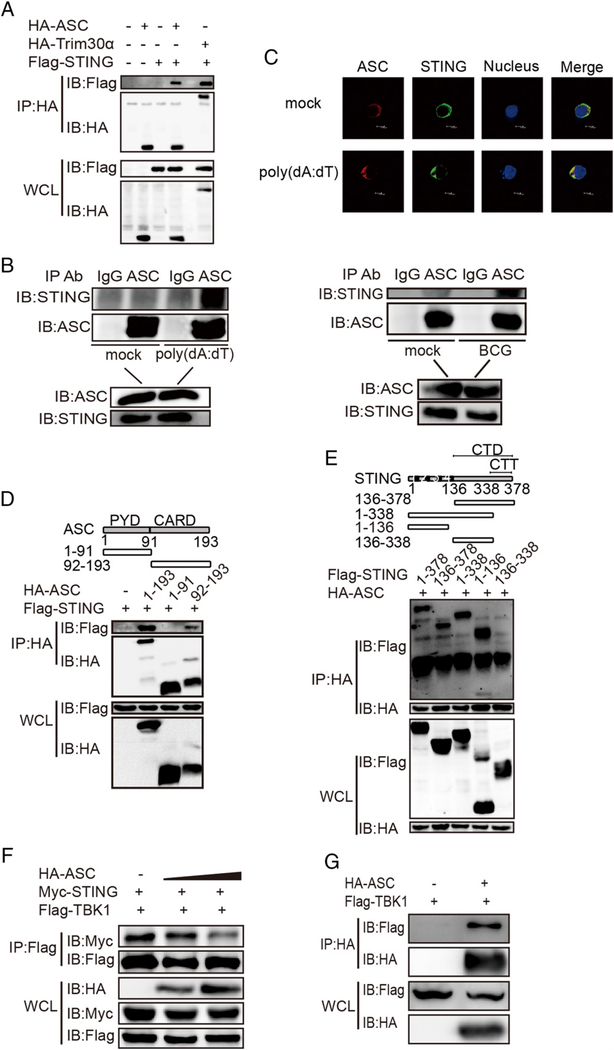
ASC interacts with STING
To investigate how ASC targeting of STING resulted in the DNA-mediated inhibition of IFN-β, coimmunoprecipitation assays were performed. The results showed that ASC interacted with STING when cotransfected into HEK293T cells (Fig. 5A). Trim30α interaction with STING, which we reported previously (22), was used as a positive control. We then verified that endogenous ASC could indeed interact with STING in BMDCs upon poly(dA:dT) stimulation or BCG infection (Fig. 5B). After 3′HA–ASC and 5′Flag–STING were expressed in HEK293T cells, confocal microscopy revealed that the two proteins colocalized without stimulation and that the colocalization tended to accumulate after poly(dA:dT) stimulation (Fig. 5C). To explore the domains that governed the association between ASC and STING, Flag-tagged full-length STING and two ASC truncations were expressed in HEK293T cells. Coimmunoprecipitation assays showed that only the ASC–CARD domain, which might not affect the activation of ASC, bound to STING (Fig. 5D). This is consistent with findings that activated ASC played the negative-regulatory role. Unlike ASC, STING is a membrane protein that contains four transmembrane domains. After expressing an HA-tagged full-length ASC protein and various STING truncations in HEK293T cells, we found that the N-terminal transmembrane domains (1–136) or the CTT domain (338–378) of STING were required for ASC–STING interaction (Fig. 5E). Because the C terminus (341–379) of STING binds to and activates TBK1 to phosphorylate IRF3 upon DNA ligand stimulation (30), we asked whether the interaction between ASC and STING impeded the interaction between STING and TBK1. To test this supposition, we co-overexpressed STING and TBK1 in HEK293T cells without ASC expression or with gradient ASC expression. The results showed that addition of ASC decreased the association between STING and TBK1 and that the inhibitory effect of ASC on the STING–TBK1 association was gradient dependent (Fig. 5F). Further evidence that ASC could also interact with TBK1 was found for this phenomenon (Fig. 5G). These results indicated that ASC might interfere with the STING–TBK1 association, thus inhibiting the production of IFN-β.
FIGURE 5.
ASC interacts with STING. (A) Immunoblot analysis of lysates from HEK293T cells that were cotransfected with the Flag vector or the Flag-STING vector and the HA-tagged vector, HA-ASC vector, or HA-Trim30a vector, followed by immunoprecipitation (IP) with anti-HA Ab and immunoblotting with anti-Flag Ab. (B) Immunoblot analysis of lysates of BMDCs that were mock treated or stimulated with poly(dA:dT) (1 μg/ml) or infected with BCG (multiplicity of infection = 10) for 6 h, followed by immunoprecipitation with anti-IgG Ab or anti-ASC Ab and immunoblotting with anti-STING Ab. (C) Confocal microscopy of MEFs that were transfected for 4 h with Flag-tagged STING and HA-tagged ASC and then mock treated or stimulated for 4 h with poly(dA:dT) (2 μg/ml). Immunofluorescence was performed using anti-HA Ab (red), anti-Flag Ab (green), and DAPI. (D) Immunoblot analysis of lysates from HEK293T cells that were cotransfected with Flag-STING and HA-tagged ASC or HA-tagged ASC truncations, followed by immunoprecipitation (IP) with anti-HA Ab and immunoblot analysis with anti-Flag Ab. (E) Immunoblot analysis of lysates from HEK293T cells cotransfected with HA-tagged ASC and Flag-STING or Flag-STING truncations, followed by immunoprecipitation (IP) with anti-HA Ab and immunoblot analysis with anti-Flag Ab. (F) Immunoblot analysis of lysates from HEK293T cells cotransfected with Myc-STING and Flag-TBK1 plus the HA vector or HA-ASC in gradient concentrations, followed by immunoprecipitation (IP) with anti-Flag Ab and immunoblot (IB) analysis with anti-Myc Ab. (G) Immunoblot analysis of lysates from HEK293T cells cotransfected with the Flag vector or Flag-TBK1 and HA-ASC, followed by immunoprecipitation (IP) with anti-HA Ab and immunoblot (IB) analysis with anti-Flag Ab. Data are representative of at least two independent experiments.
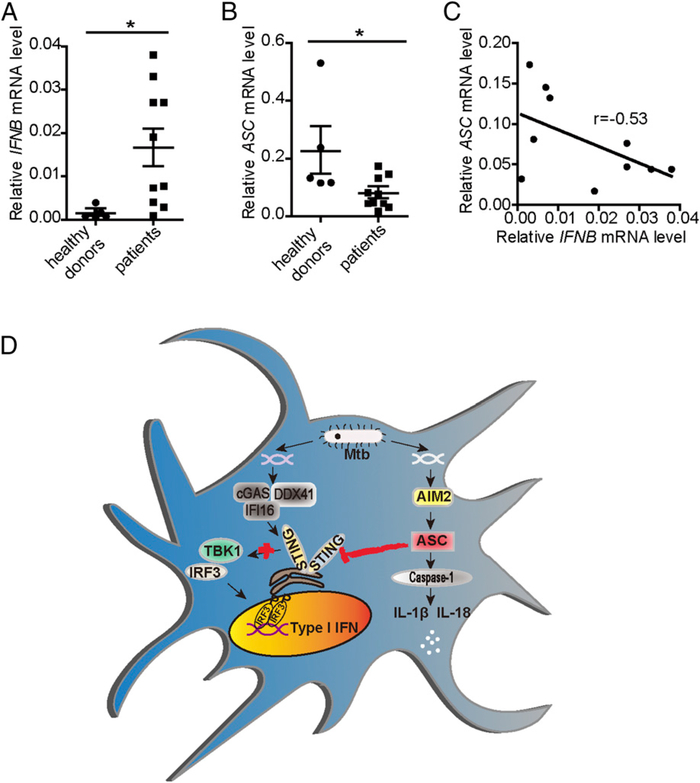
ASC expression is inversely correlated with IFN-β levels in PBMCs of TB
Previous studies have shown that active TB patients exhibit a prominent IFN-α/β-inducible transcription signature. Consistent with these findings, we observed higher IFN-β expression in PBMCs of TB patients compared with healthy donors (Fig. 6A). To identify whether the inhibitory role of ASC on the induction of IFN-β was involved in this phenomenon, we simultaneously measured the expression of ASC. Interestingly, lower expression of ASC was detected in TB patients than in healthy donors (Fig. 6B). Additionally, an inverse correlation was detected between IFN-β levels and ASC levels in TB patients (Fig. 6C), indicating that downregulated expression of ASC might contribute, in part, to the detrimental IFN-α/β–inducible transcriptional signature of TB patients.
FIGURE 6.
ASC expression is inversely correlated with IFN-β levels in PBMCs from TB patients. IFN-β and ASC expression in PBMCs from healthy donors (n = 5) and TB patients (n = 10) was examined by quantitative real-time PCR (qRT-PCR) (A and B), and their correlation was assessed with the Pearson test (C). (D) GD of M. tuberculosis can be recognized by the AIM2–IL-1β signaling pathway and the STING–type I IFN signaling pathway. Activated ASC interacts with STING and blocks the activation of downstream molecules TBK1 and IRF3, thus inhibiting the induction of IFN-β. The results are shown as mean ± SEM. *p < 0.05, Student t test.
Discussion
Although the AIM2–IL-1β and STING–type I IFN signaling pathways are activated by the GD of M. tuberculosis, the roles of these pathways are protective (AIM2–IL-1β) and detrimental (STING–type I IFN) to the host against infection. We found that AIM2 deficiency reciprocally induced overreactive IFN-β and depressive IFN-γ responses in vivo, leading to higher infection burdens and more severe pathology. While exploring the potential mechanism behind this phenomenon, we discovered that the interaction between activated ASC and STING impeded the association of STING with its downstream molecule, TBK1, which ultimately inhibited the induction of type I IFN. IL-1β and type I IFN represent two major counterregulatory classes of inflammatory cytokines that control the outcome of M. tuberculosis infection (31). Therefore, it is physiologically significant that the IL-1β signaling pathway negatively regulates the type I IFN signaling pathway during M. tuberculosis infection. Based on our data, we believe that activation of the AIM2 inflammasome protects the host from M. tuberculosis infection on one hand due to the induction of IL-1β and on the other hand due to downregulation of the type I IFN induction via cross-talk through ASC and STING interaction. Importantly, ASC expression is inversely correlated with the levels of IFN-β in PBMCs from TB patients, which emphasizes the clinical significance of the inhibitory role of ASC on IFN-β production.
Our results suggest that overproduction of type I IFNs leads to a reduced IFN-γ response during M. tuberculosis infection. The reciprocal IFN-β overproduction and IFN-γ depression during mycobacterial infection of AIM2−/− mice are consistent with the recent observation that IFN-β counterregulates the IFN-γ response upon infection with various M. tuberculosis strains (32). Our data also support recent reports implicating that type I IFNs impair the Th1 response and inhibit IFN-γ–induced antimicrobial responses in macrophages/monocytes in virulent M. tuberculosis infection (13, 33).
In addition to the involvement of the AIM2 inflammasome and type I IFN signaling pathways in M. tuberculosis detection, another pathway is relevant: autophagy. Although one report suggested that AIM2-inhibited autophagy increases M. bovis survival in vitro (34), it is conceivable that activated ASC-mediated inhibition of IFN-β promotes resistance to M. tuberculosis in vivo. Therefore, additional studies are needed to elucidate the relationship among these three pathways. It will also be interesting to determine whether single nucleotide polymorphisms of ASC in TB patients correlate with changes in type I IFN. In addition to M. tuberculosis, another intracellular bacterium, Listeria monocytogenes, can induce the production of type I IFN and IL-1β in a DNA-dependent manner. Type I IFN is detrimental, and IL-1β is beneficial (35–37). Therefore, it would be helpful to determine whether the mechanism in our study applies to L. monocytogenes infection in vivo.
Intracellular DNAs that behave like classical PAMPs are involved in various innate immune response to viruses and intracellular bacteria and function during conditions of sterile inflammation, autoimmunity, and cancers (38). Because immune sensing of DNA plays critical roles in the early resistance to infections and subsequent bridge toward activation of adaptive immune responses, searching for DNA sensors that are involved in this process is especially important. Multiple DNA sensors, including DAI, AIM2, RNA PolIII, LRRFIP1, DHX9/DHX36, IFI16, Ku70, DDX41, DNA-PK, MRE11, cGAS, and Rad50, have been identified, and their functions have been studied (39). However, how these DNA sensors act and influence each other remain unclear. The relationship that exists between the STING–type I IFN and AIM2 inflammasome signaling pathways, which are the two major pathways that are activated by DNA ligands, might be the most important to elucidate. A recently published article reported that inflammasome activation regulates type I IFN responses to DNA virus infection, which is presumably due to caspase-1–mediated cleavage of cGAS (40). Although our data showed that the production of IFN-β increased in Caspase-1−/− mice compared with WT mice, the extent of the increase was far below that of AIM2−/− and ASC−/− mice, which was consistent with previous studies (25). Therefore, it is likely that more than one mechanism is involved in the cross-talk between these two pathways. Given the different stimuli, cell types, and animal models that we used, it is possible that the dominant mechanisms underlying the regulatory effect of the AIM2 inflammasome on the type I IFN response differed based on the specific conditions used. And further studies are needed. Additionally, the blocked induction of downstream IFN-β by ASC interaction with STING prompts us to ask whether the STING pathway affects ASC-associated pathways, such as the AIM2 inflammasome and NLRP3 inflammasome signaling pathways. If so, exploring whether the effect is negative or positive and the pathological conditions in which this mechanism functions is meaningful.
Supplementary Material
Acknowledgments
We thank Shanghai Pulmonary Hospital for samples from TB patients.
This work was supported by grants from the Ministry of Science and Technology of China (2016YFA0502202/2016YFA0502204), the National Natural Science Foundation of China (31230024), and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000).
Abbreviations
- AIM2
absent in melanoma 2
- ASC
apoptosis-associated speck-like protein
- BCG
bacillus Calmette–Guérin
- BMDC
bone marrow–derived dendritic cell
- BMDM
bone marrow–derived macrophage
- cGAMP
cyclic GMP-AMP
- cGAS
cyclic GMP-AMP synthase
- GD
genomic DNA
- HA
hemagglutinin
- IRF3
IFN regulatory factor 3
- ISD
IFN stimulatory DNA
- PAMP
pathogen-associated molecular pattern
- poly(dA:dT)
poly(deoxyadenylic-thymidylic) acid
- poly(I:C)
polyinosinic-polycytidylic acid
- STING
stimulator of IFN genes
- TB
tuberculosis
- TBK1
TANK-binding kinase 1
- WT
wild-type
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.World Health Organization. Global tuberculosis report 2015. Available at: http://www.tbonline.info/posts/2016/10/16/global-tb-report-2016/.
- 2.O’Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, and Berry MP 2013. The immune response in tuberculosis. Annu. Rev. Immunol 31: 475–527. [DOI] [PubMed] [Google Scholar]
- 3.Cai X, Chiu YH, and Chen ZJ 2014. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol. Cell 54: 289–296. [DOI] [PubMed] [Google Scholar]
- 4.Manzanillo PS, Shiloh MU, Portnoy DA, and Cox JS 2012. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe 11: 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins AC, Cai H, Li T, Franco LH, Li XD, Nair VR, Scharn CR,Stamm CE, Levine B, Chen ZJ, and Shiloh MU 2015. Cyclic GMP-AMP synthase is an innate immune DNA sensor for Mycobacterium tuberculosis. Cell Host Microbe 17: 820–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wassermann R, Gulen MF, Sala C, Perin SG, Lou Y, Rybniker J,Schmid-Burgk JL, Schmidt T, Hornung V, Cole ST, and Ablasser A 2015. Mycobacterium tuberculosis differentially activates cGAS- and inflammasome-dependent intracellular immune responses through ESX-1. Cell Host Microbe 17: 799–810. [DOI] [PubMed] [Google Scholar]
- 7.Watson RO, Bell SL, MacDuff DA, Kimmey JM, Diner EJ, Olivas J,Vance RE, Stallings CL, Virgin HW, and Cox JS 2015. The cytosolic sensor cGAS detects Mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host Microbe 17: 811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNab F, Mayer-Barber K, Sher A, Wack A, and O’Garra A 2015. Type Iinterferons in infectious disease. Nat. Rev. Immunol 15: 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T,Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, et al. 2010. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466: 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cliff JM, Lee JS, Constantinou N, Cho JE, Clark TG, Ronacher K,King EC, Lukey PT, Duncan K, Van Helden PD, et al. 2013. Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. J. Infect. Dis 207: 18–29. [DOI] [PubMed] [Google Scholar]
- 11.Maertzdorf J, Repsilber D, Parida SK, Stanley K, Roberts T, Black G,Walzl G, and Kaufmann SH 2011. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun. 12: 15–22. [DOI] [PubMed] [Google Scholar]
- 12.Manca C, Tsenova L, Freeman S, Barczak AK, Tovey M, Murray PJ,Barry C, and Kaplan G 2005. Hypervirulent M. tuberculosis W/Beijing strains upregulate type I IFNs and increase expression of negative regulators of the Jak-Stat pathway. J. Interferon Cytokine Res 25: 694–701. [DOI] [PubMed] [Google Scholar]
- 13.Manca C, Tsenova L, Bergtold A, Freeman S, Tovey M, Musser JM,Barry CE III, Freedman VH, and Kaplan G 2001. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-α/β. Proc. Natl. Acad. Sci. USA 98: 5752–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonelli LR, Gigliotti Rothfuchs A, Gonçalves R, Roffê E, Cheever AW, Bafica A, Salazar AM, Feng CG, and Sher A 2010. Intranasal Poly-IC treatment exacerbates tuberculosis in mice through the pulmonary recruitment of a pathogenpermissive monocyte/macrophage population. J. Clin. Invest 120: 1674–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bürckstümmer T, Baumann C, Blüml S, Dixit E, Dürnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, and Superti-Furga G 2009. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol 10: 266–272. [DOI] [PubMed] [Google Scholar]
- 16.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, and Alnemri ES 2009. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458: 509–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G,Caffrey DR, Latz E, and Fitzgerald KA 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458: 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamada H, Mizumo S, Horai R, Iwakura Y, and Sugawara I 2000. Protectiverole of interleukin-1 in mycobacterial infection in IL-1 α/β double-knockout mice. Lab. Invest 80: 759–767. [DOI] [PubMed] [Google Scholar]
- 19.Juffermans NP, Florquin S, Camoglio L, Verbon A, Kolk AH, Speelman P,van Deventer SJH, and van der Poll T 2000. Interleukin-1 signaling is essential for host defense during murine pulmonary tuberculosis. J. Infect. Dis 182: 902–908. [DOI] [PubMed] [Google Scholar]
- 20.Saiga H, Kitada S, Shimada Y, Kamiyama N, Okuyama M, Makino M,Yamamoto M, and Takeda K 2012. Critical role of AIM2 in Mycobacterium tuberculosis infection. Int. Immunol 24: 637–644. [DOI] [PubMed] [Google Scholar]
- 21.Mariathasan S, Newton K, Monack DM, Vucic D, French DM,Lee WP, Roose-Girma M, Erickson S, and Dixit VM 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430: 213–218. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Lian Q, Yang B, Yan S, Zhou H, He L, Lin G, Lian Z, Jiang Z,and Sun B 2015. TRIM30a is a negative-feedback regulator of the intracellular DNA and DNA virus-triggered response by targeting STING. PLoS Pathog. 11: e1005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waeckerle-Men Y, Bruffaerts N, Liang Y, Jurion F, Sander P, Kündig TM, Huygen K, and Johansen P 2013. Lymph node targeting of BCG vaccines amplifies CD4 and CD8 T-cell responses and protection against Mycobacterium tuberculosis. Vaccine 31: 1057–1064. [DOI] [PubMed] [Google Scholar]
- 24.Platanias LC 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol 5: 375–386. [DOI] [PubMed] [Google Scholar]
- 25.Corrales L, Woo SR, Williams JB, McWhirter SM, Dubensky TW Jr., and Gajewski TF 2016. Antagonism of the STING pathway via activation of the AIM2 inflammasome by intracellular DNA. J. Immunol 196: 3191–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z,Horvath G, Rathinam VA, Johnstone RW, et al. 2012. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 36: 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun L, Wu J, Du F, Chen X, and Chen ZJ 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339: 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S,Sirois CM, Jin T, Latz E, Xiao TS, et al. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol 11: 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Yuan B, Bao M, Lu N, Kim T, and Liu YJ 2011. The helicaseDDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol 12: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka Y, and Chen ZJ 2012. STING specifies IRF3 phosphorylation byTBK1 in the cytosolic DNA signaling pathway. Sci. Signal 5: ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayer-Barber KD, Andrade BB, Oland SD, Amaral EP, Barber DL,Gonzales J, Derrick SC, Shi R, Kumar NP, Wei W, et al. 2014. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature 511: 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dey B, Dey RJ, Cheung LS, Pokkali S, Guo H, Lee J-H, and Bishai WR 2015. A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat. Med 21: 401–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ordway D, Henao-Tamayo M, Harton M, Palanisamy G, Troudt J, Shanley C, Basaraba RJ, and Orme IM 2007. The hypervirulent Mycobacterium tuberculosis strain HN878 induces a potent TH1 response followed by rapid down-regulation. J. Immunol 179: 522–531. [DOI] [PubMed] [Google Scholar]
- 34.Liu C, Yue R, Yang Y, Cui Y, Yang L, Zhao D, and Zhou X 2016. AIM2 inhibits autophagy and IFN-β production during M. bovis infection. Oncotarget 7: 46972–46987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Auerbuch V, Brockstedt DG, Meyer-Morse N, O’Riordan M, and Portnoy DA 2004. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J. Exp. Med 200: 527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carrero JA, Calderon B, and Unanue ER 2004. Type I interferonsensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J. Exp. Med 200: 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA,Zarnegar B, Perry AK, Nguyen BO, Lane TF, Taniguchi T, et al. 2004. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J. Exp. Med 200: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M,Hoshino K, Wagner H, Takeda K, and Akira S 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408: 740–745. [DOI] [PubMed] [Google Scholar]
- 39.Unterholzner L 2013. The interferon response to intracellular DNA: why somany receptors? Immunobiology 218: 1312–1321. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Ning X, Gao P, Wu S, Sha M, Lv M, Zhou X, Gao J, Fang R,Meng G, et al. 2017. Inflammasome activation triggers caspase-1-mediated cleavage of cGAS to regulate responses to DNA virus infection. Immunity 46: 393–404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.