Abstract
Histology-directed imaging mass spectrometry (IMS) is a spatially-targeted IMS acquisition method informed by expert annotation that provides rapid molecular characterization of select tissue structures. The expert annotations are usually determined on digital whole slide images of histological stains where the staining preparation is incompatible with optimal IMS preparation, necessitating serial sections: one for annotation, one for IMS. Registration is then used to align staining annotations and IMS. Herein, we report a next-generation histology-directed platform implementing IMS-compatible autofluorescence (AF) microscopy taken prior to any staining or IMS. The platform enables two histology-directed workflows, one that improves the registration process between two separate tissue sections using automated, computational mono-modal AF-to-AF microscopy image registration, and a registration-free approach that utilizes AF directly to identify ROIs and acquire IMS on the same section. The registration approach is fully automated and delivers state of the art accuracy in histology-directed workflows for transfer of annotations (ca. 3-10 μm based on 4 organs from 2 species) while the direct AF approach is registration-free, allowing targeting of the finest structures visible by AF microscopy. We demonstrate the platform in biologically relevant case studies of liver stage malaria and human kidney disease with spatially targeted acquisition of sparsely distributed (composing less than one tenth of 1% of the tissue section area) malaria infected mouse hepatocytes, and glomeruli in the human kidney case study.
Introduction
Imaging mass spectrometry (IMS) is a technology used to collect mass spectrometry data of the distribution of analytes across a sample surface. Composing the MS signal into a map of ion intensity generates ion images.1 An important IMS application is the analysis of the thin tissue sections used in histopathology where IMS can augment the morphological information reported by microscopy with molecular information.2,3 Contrary to the cameras used in most forms of digital microscopy that record a complete viewport in one shot, most IMS platforms will probe a sample, using a desorption-ionization probe, in a point-by-point fashion or using sequential line-scans. This serial sampling approach results in long data acquisitions, which can be further compounded if high spatial resolution measurements (more IMS pixels) are required or high mass resolving power (more m/z bins) is needed (e.g., using FTICR or orbital trapping-based instrumentation). In contrast to acquiring full tissue section IMS images, histology-directed IMS offers a workflow where only specific regions of interest within the tissue sample are targeted for data acquisition. The supervised approach typically solicits expert annotation on digital whole slide microscopy images to decide which areas merit IMS thus avoiding acquisition across the entire tissue section.4 Altogether, histology-directed approaches leveraging domain-specific expertise can greatly reduce IMS data acquisition times while still providing molecular data relevant to the pathology being studied.
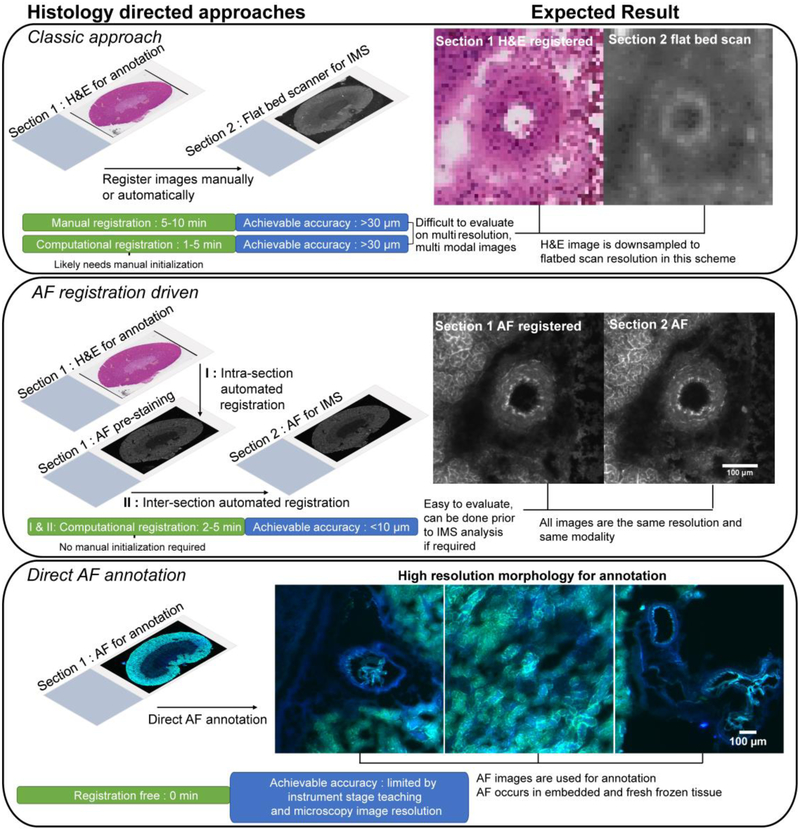
Histology-directed IMS has driven a number of studies with many focusing on diagnostic applications that take advantage of the rapid throughput.5-9 As domain-specific or medical expertise is often required to determine acquisition ROIs, remote annotation systems have been developed to facilitate the experiments.10 Despite powerful demonstrations of the technology11, the image registration process that maps expert annotations collected using stained serial images onto the target unstained IMS section remains an area where fundamental improvements can have a substantial impact. Impacts can be delivered in spatial accuracy, end user simplicity, and overall throughput of the workflow through automation. Image registration is the mathematical spatial transformation of the coordinate system of one image, or a set of images, to the coordinate space of a second image.12 The primary goal of image registration is to optimally align features from one image to those of another so that pixel-wise comparison of the content between the images becomes practical. In the context of histology-directed IMS, maintaining the spatial accuracy of the expert annotation after registration from an annotated tissue section to the IMS tissue section is critical and depends directly on the quality of the registration between the two tissue sections. Published histology-directed workflows (described in Figure 1, Classical Approach) suffer from uncertain spatial alignment due to the registration being performed, either automatically or manually, between a high-resolution microscopy image and a relatively coarse resolution image off a flatbed scanner used within instrument software to delineate the IMS section.4,10,11,13 If manual registration approaches are used with these same images (i.e., overlay of images in photo-editing software), there is significant labor on the part of the researcher and uncertainty remains.
Figure 1. Schematic of histology-driven workflows after tissue sectioning and prior to IMS sample preparation and data acquisition.
Section 1 and 2 refer to serial sections. The AF registration driven result images are after AF-to-AF registration. In the ‘Classic approach’ and ‘AF registration driven’ panels, images were taken from the same region and area for comparative purposes. Colors in the AF scan in ‘Direct AF annotation’ are from a two color channel image using DAPI and FITC filters.
To achieve high accuracy in registration and simultaneously improve the automation and in turn the speed of histology-directed workflows, we have developed next-generation histology-directed methods guided by autofluorescence (AF) as well as an open source software to perform all necessary computational tasks. Fundamentally, AF microscopy is wide-field microscopy of endogenous fluorophores that provides preparation-less microscopy-grade imaging of tissue sections.14 Most importantly for histology directed workflows, AF microscopy can be acquired prior to any other sample preparation such as IMS or histological staining. In this study, two AF guided histology-directed IMS workflows are demonstrated. The workflows were designed specifically to improve the spatial accuracy of the workflow and overall efficiency of the user experience (Figure 1 – Registration-driven and Direct AF annotation).
Incorporating AF images into the registration process avoids the difficulty of mapping a flat-bed scan image of the IMS destined tissue section (often coarse resolution) directly to the stained microscopy used for annotations (Figure 1 Classical Approach). Inter-section registration accuracy is shown to be substantially augmented when registration is between AF microscopy images (AF-to-AF) of the same resolution and mono-modal intensity patterns. The improved accuracy for spatially targeted IMS data acquisition that this workflow provides is demonstrated in a challenging case study of individual analysis of liver stage malaria mouse hepatocytes. Malaria is caused by the Plasmodium species that enters the human host when an infected mosquito takes a blood meal. Plasmodium sporozoites enter the blood stream and travel to the liver where they invade a host hepatocyte and undergo a program of rapid division before entering the bloodstream and becoming symptomatic.15 Studying the malaria parasite at this stage is analytically challenging as infection rates are low and consequently infected hepatocytes are sparsely distributed in the liver (less than 1% in our example).16 Thus, traditional bulk measurements of the liver provide limited information regarding biological changes that occur within the infected hepatocytes. This work represents the first report of IMS in Plasmodium-infected liver and thus paves the way for critical biochemical insights of the alterations that occur during the liver stage of malaria infection.
While the registration-driven workflow advances the traditional setting for histology-directed IMS acquisition, the direct AF annotation workflow delivers a new histology-driven setup where no external stain or registration is required (Figure 1 Direct AF annotation). Instead, the morphological information reported by AF microscopy is used to recognize and annotate tissue features. This novel approach addresses critical challenges occasionally encountered in serial section workflows where some tissue types exhibit morphological change on the order of a single histological section (5-20 μm), effectively resulting in the loss of targeted tissue structures and no inability to target tissue structures below finest registration performance. As an example of how AF can help avoid such challenges, we demonstrate the annotation and IMS of glomeruli in human kidney tissue directly from AF images with the aim of driving insight into chronic kidney disease (CKD). CKD is characterized by a loss in renal function from progressive scarring that affects all structures (tubules, glomeruli, etc.) of the kidney.17 Molecular characterization of CKD’s progression has high potential to elucidate underlying pathological processes and in biomarker discovery. However, efforts to molecularly image kidney structures like glomeruli by IMS in a great enough number to statistically capture the intra- and inter-patient variation in human samples is hindered by long data acquisition times. The demonstrated direct AF annotation workflow effectively circumvents prohibitively long acquisition times and provided 10 μm IMS resolution images of more than 100 glomeruli from a single tissue section in under 40 minutes on a high speed MALDI-TOF instrument.
Finally, although this work focuses on MALDI IMS, the described workflows are equally applicable to other IMS ionization modalities or any workflow that requires spatial sampling such as laser capture micro-dissection, genomics, or liquid surface sampling.
Materials & Methods
Chemicals and reagents
1,5-diaminonapthalene (DAN) MALDI matrix was acquired from Sigma Aldrich (St. Louis, MO).
Mosquito rearing and P. yoelii sporozoite production for malarial model
Female 6-8 week-old Swiss Webster mice (Harlan, Indianapolis, IN, USA) were injected with blood stage P. yoelii (17XNL) parasites to begin the growth cycle at the Center for Infectious Disease Research (Seattle, Washington). Animal handling was conducted according to the Institutional Animal Care and Use Committee-approved protocols. Infected mice were fed to female Anopheles stephensi mosquitoes after gametocyte exflagellation was observed. Sporozoites were isolated from salivary glands of the mosquitoes at 14 days post-blood meal by manual dissection. All work in animals was performed according to protocols and SOPs approved by the Center for Infectious Disease Research IACUC.
Animal & mammalian tissue samples
Rat brain, rat kidney, mouse brain, and mouse spleen tissues were purchased from BioreclamationIVT (Hicksville, NY). Fresh frozen human kidney samples were provided by the cooperative human tissue network (CHTN) - Western Division at Vanderbilt University Medical Center (Nashville, TN).
To obtain P. yoelii-infected livers, female BALB/cAnNHsd (Harlan) mice were retro-orbitally injected with 106 sporozoites. At 44 hours post-injection, mice were euthanized by CO2 asphyxiation, livers were extracted and snap frozen in liquid nitrogen, and stored at −80°C until shipment. Livers were sent to the Vanderbilt University Mass Spectrometry Research Center on dry ice. All work in animals was performed according to protocols and SOPs approved by the Center for Infectious Disease Research IACUC.
Brightfield and Autofluorescence microscopy
All AF images were captured on a Nikon Eclipse 90i (Nikon Instruments Inc., Melville, NY) microscope at 10x with a pixel resolutions of 0.92 μm / pixel. DAPI (ex. = 340-380, em. = 435-485), FITC (ex. = 465-495, em. = 515-555), and TRITC (ex. = 528-553, em. = 590-650) (all wavelengths in nm) specific epifluorescence filters were used to cover a range of emission and excitation bands for AF. AF exposure times varied in most instances around 150 ms exposure time, with 8.0x analog gain used. The supplemental methods in the Supporting Information has more details on the AF microscopy considerations for registration and annotation.
Brightfield RGB images of unstained and stained tissues were acquired using a Leica SCN-400 (Leica Biosystems, Buffalo Grove, IL) slide scanner at 10x (1 μm/pixel).
Image preprocessing and registration
FIJI ImageJ18,19 was used for visualization and annotation of microscopy images.
Image registration was performed with the Insight Segmentation and Registration Toolkit (ITK)20 and elastix21 libraries wrapped to Python3 using a suite of in-house scripts for SimpleITK22 and simpleElastix23 packages. Image transformations were developed using linear transformations (rigid & affine), and non-linear (b-spline) transformation models using a multi-resolution scheme (10 progressive resolutions) optimized by stochastic gradient descent with mutual information as the metric. After transformation, images were interpolated to a grid using nearest neighbor interpolation. The supplemental methods has a deeper look into the optimized parameters. Scripts and software used throughout this work are available at http://github.com/NHPatterson/regtoolboxmsrc. Documents on the use of the software for these workflows will be also be made available on the GitHub.
MALDI imaging mass spectrometry
Mass spectrometry
MALDI time-of-flight (TOF) IMS was performed using a Bruker rapifleX TissueTyper equipped with a Gaussian 355 nm Nd:YAG laser operating in reflectron mode with a detection mass range between 600-1700 Da. The laser was operating in the ‘Single’ setting with rapifleX beam scanning on and equal to IMS raster pitch for each experiment, defining an approximately square pixel at each position. Five hundred laser shots were fired per position. MALDI TOF delayed extraction time, extraction voltages, and laser power settings (35% of global attenuater) were optimized for sensitivity. Internal calibration was performed using common, high intensity negative mode lipids.
Sample preparation and experiments
Mouse liver and human kidney were sectioned at 8 and 12 μm, respectively, using a Leica CM3050 S Cryostat (Leica Biosystems, Buffalo Grove, IL). In both IMS experiments, DAN was used as the MALDI matrix. DAN matrix sublimation was done according to the protocol from Thomas et. al.24 To avoid degradation of lipids in the tissue samples, all samples were analyzed the day of cutting and microscopy.25 AF microscopy was performed immediately after sectioning and 15-30 minutes in a vacuum desiccator. AF was not found to rapidly decay when tissues were stored out of light. Lab observation shows AF signal is robust over months with and without freezing (data not shown). However, IMS signal is known to rapidly decay without proper storage, and was the limiting sample handling factor driving rapid data acquisition.
Data analysis
IMS data was exported to the imzML format26 and loaded into R via the Cardinal package.27 Data was TIC normalized and Cardinal’s ‘simple’ peak picking was used to select for signal to noise greater than 3 and a minimum frequency of 1%. Custom scripts were used to coerce the position data of each non-contiguous IMS ROI into a grid for rapid, convenient visualization of all ROIs. These scripts will eventually be made available at the previously mentioned GitHub. Figures and overlays were prepared in ImageJ through export of ion images from R. Frequency histograms in the kidney data analysis were plotted using the mean of the each glomerulus to show global glomerulus change for a given signal.
Results and Discussion
AF registration-driven histology-directed experiments
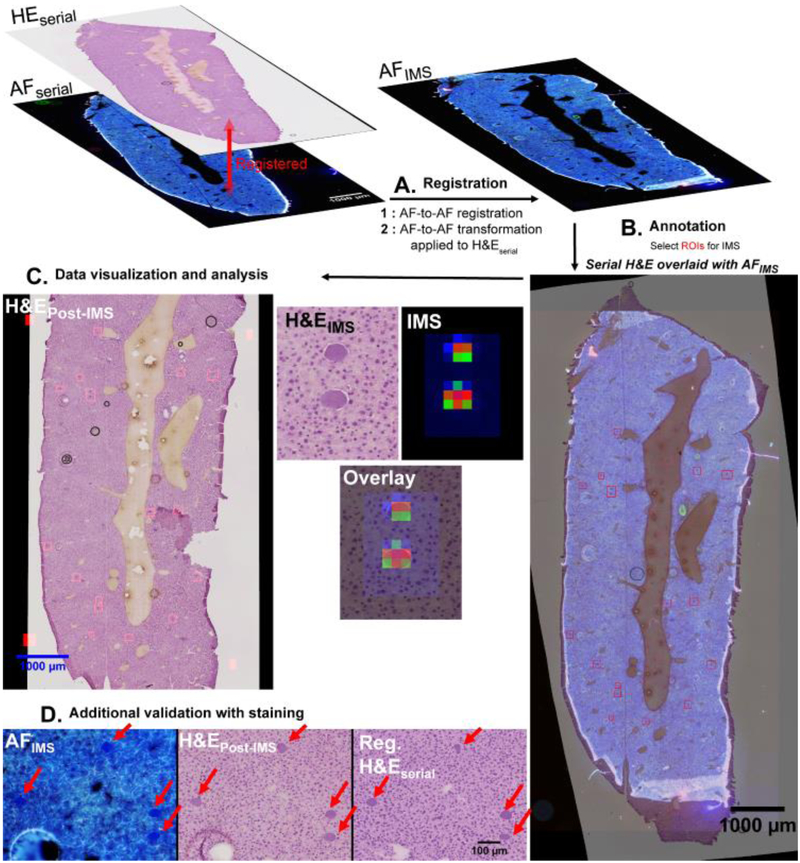
AF intermediated registration is demonstrated to address accuracy and speed issues (fully automated) of histology-directed workflows, specifically as an IMS-compatible microscopy modality for mono-modal high-resolution image registration of serial sections (Figure S1-4). This work was partly inspired by the use of AF and Raman microscopy for cellular segmentation and cancer detection28, however, in this work AF microscopy is not used in the data analysis, but as a means to achieve non-destructive, high resolution, IMS compatible imaging for histology-directed IMS. Figure 2 shows an outline of the registration-driven workflow using a malarial mouse liver example. In this process, multiple registrations are concatenated to project a staining image onto the coordinate space of the target tissue section AF image without sacrificing microscopy grade spatial resolution. Comparative registration-driven histology-directed IMS approaches use a combination of high and low resolution images, and less robust and less evaluable multi-modal registrations.11,29 In approaches with mixed image resolutions, the inability to resolve tissue structures in the low resolution images used for IMS acquisition means that these areas cannot be successfully registered and targeted for IMS. While the AF microscopy does add time, with length dependent on the microscope employed, only a small amount of user time is needed to set up the scans and registrations. A deeper discussion of this and a step-by-step time analysis for this workflow is available in the Supporting Information around Figure S6.
Figure 2. Full workflow for registration-driven histology-directed experiments and data analysis strategies.
A. First, intra-section registration is performed between H&Eserial and AFserial. After intra-section registration, inter-section registration is performed between AFserial & AFIMS. The developed inter-section transformation is then used to transform H&Eserial, thus projecting H&Eserial onto AFIMS. B. With H&Eserial projected onto the section destined for IMS (AFIMS), annotations can be made to the H&E for IMS acquisition. Additionally, the AF provides morphological information that may sometimes be used to verify annotations by overlay of AFIMS and the registered H&Eserial. C. Data is acquired from the section only in the annotated areas. D. After IMS acquisition, the AFIMS section can be stained to compare inter-section H&Es directly. H&Eserial = H&E staining image from the annotation serial section. AFserial = AF image from the annotation serial section. AFIMS = AF image from section destined for IMS. H&EPost-IMS = H&E staining image from the section where IMS was performed, stained after IMS data acquisition.
Details of the registration-driven workflow
The computational workflow consists of several steps: (1) Linear multi-modal registration of the annotated staining image to its underlying AF image (Intra-section registration) (Fig 2. A-(1), left). (2) Linear followed by linear then non-linear registration of mono-modal AF images from the serial sections (AF-to-AF registration, inter-section) (Fig 2. A-(2), right, Video S1). Performing this registration AF-to-AF allows mono-modal registration for the more difficult inter-section registration task. (3) The staining layer registered described in 1) is projected onto the serial section on which IMS will be performed using the transformation obtained by AF-to-AF image registration in (2) (Fig. 2-B). Annotations can be made before registration and passed along the registration path of 1) and 2) (made possible by all registrations being preserved as mathematical models) or made with the staining projected onto the AF image. The latter approach allows for direct comparison of the stain and underlying AF, where AF may give additional visual cues about the presence of targeted histological structures (Fig 2 B & C). In this example an H&E image is used for annotation, however it is worth noting that AF measurements are non-destructive and are compatible with any histological or immuno-histological stain desired for annotation.
Quantitative evaluation of global AF-to-AF registration accuracy on test tissues
The quantitative performance of the AF-to-AF registration workflow is evaluated on sets of images from 4 organs from mouse and rat by calculating the Euclidean distance between ~30 corresponding points placed by 3 independent observers. This metric was used rather than the algorithm optimization metric for the reasons outlined by Rohlfing.30 Table 1 lists statistics for the distances after registration. Overall, an almost four-fold improvement in mean distance (40 to 10 μm) was found when using a linear + nonlinear registration scheme versus linear alone when considering all 5 test cases. However, there are points that do not register well in every example set, as reflected in the median and range values of Table 1 where median values < 10 μm for every example dataset, but the range maximum distance value in the range remains high. These are a combination of less performant registration, annotation error, and sample preparation artefacts.
Table 1. Median, range, mean, and standard deviation of landmark distances for various tissues after linear and linear + non-linear registration.
For each organ and species combination, two serial sections were prepared. Mu = mouse.
| Sample | ||||||
|---|---|---|---|---|---|---|
| Mu Brain | Mu Liver | Mu Spleen | Rat Brain | Rat Kidney | Mean | |
| No. of Point Pairs | 25 | 18 | 30 | 30 | 30 | 26.6 |
| Median Dist. Linear (μm) | 37.1 | 24.23 | 73.44 | 33.99 | 13.69 | 36.49 |
| Median Dist. Linear + Non-linear (μm) | 8.14 | 6.44 | 9.35 | 9.51 | 6.52 | 7.99 |
| Range Dist. Linear (μm) | (7.16, 84.03) | (6.02, 54.24) | (18.81, 164.08) | (3.58, 178.78) | (3.53, 79.53) | |
| Range Dist. Linear + Non-linear (μm) | (1.69, 58.12) | (1.5, 21.21) | (2.54, 41.14) | (2.54, 40.6) | (1.58, 28.5) | |
| Mean Dist. Linear (μm) | 37.88 ± 19.93 | 27.78 ± 16.09 | 73.7 ± 32.22 | 46.73 ± 40.33 | 17.39 ± 14.82 | 40.69 ± 19.2 |
| Mean Dist. Linear + Non-linear (μm) | 12.91 ± 12.36 | 7.99 ± 4.01 | 12.07 ± 9.97 | 11.56 ± 9.1 | 8.99 ± 6.69 | 10.7 ± 1.88 |
| Mean Dist. Fold change | 2.93 ± 1.61 | 3.47 ± 4.01 | 6.11 ± 3.23 | 4.04 ± 4.43 | 1.93 ± 2.22 | 3.7 ± 3.1 |
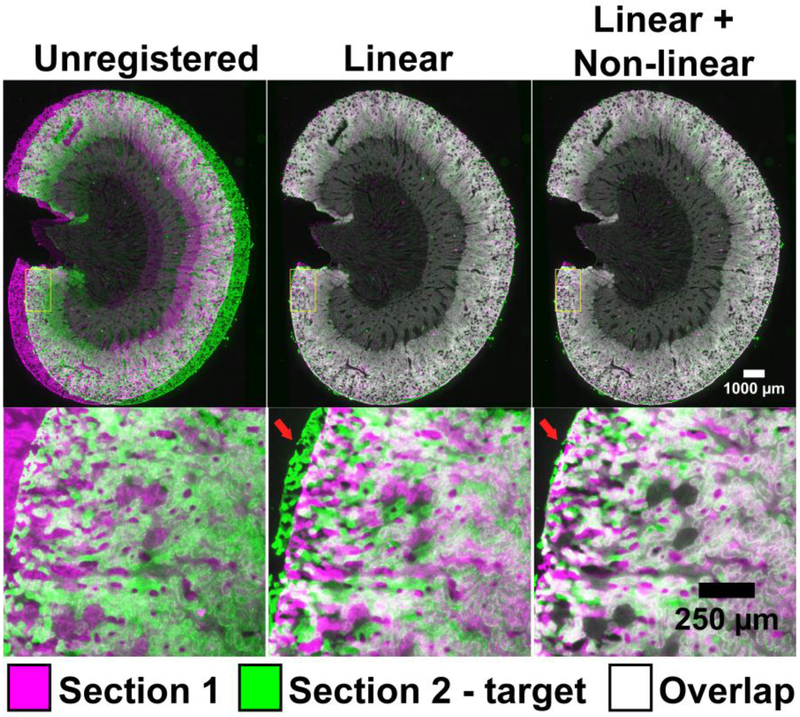
Figure S7-9 shows landmark selection and registration accuracy across three of the five test cases as well as visualizes the achieved point distances after registration. Figure 3 shows whole and high-magnification images of rat kidney before and after registration where the target image is in green, the source (warped) image is magenta, and overlap appears as white. In practical terms, the linear transformation grossly aligns the morphology while the non-linear registration allows for localized image warping, which is often necessary to account for cutting artefacts and complex tissue heterogeneity. Despite achieving gross alignment using only a linear transformation, a major misalignment is visible near one of the edges at higher magnification (Fig. 3 – Linear), however, this is visually well corrected after non-linear registration as shown in Fig. 3 - Nonlinear. Importantly, comparison with previous methods with mixed resolution images is difficult. Figure S10 shows a rat kidney H&E image scanned with a microscope slide scanner (annotation image) and the same section scanned with a flat-bed scanner. Cellular features are entirely lost at the maximum scan resolution of the flat-bed scanner, thus annotation for evaluation is not possible. Indeed, the true resolving power on a flat-bed scan is estimated to be 25-35 μm, already above the accuracy of the AF microscopy registration. The registration would also be between an H&E microscopy scan and a flat-bed scan of an unstained tissue section, furthering the difference.
Figure 3. Visual evaluation of registration accuracy of AF rat kidney images.
Visualization of the registration results at low (top panels) and high magnification (bottom panels). The greyscale AF images are color-coded as magenta and green for display. Linear here refers to an affine registration, non-linear refers to a non-linear b-spline registration following a linear affine registration.
Quantitative accuracy evaluation for targeting individual infected hepatocytes in liver-stage malarial model
With the registration accuracy offered by AF as an intermediate modality, it becomes possible to perform histology-driven IMS in advanced scenarios and at tissue structure scales that hereto would not have been feasible or practical. The novel approach is demonstrated using a malarial model driven by Plasmodium yoelii-infected mouse hepatocytes, the main parenchymal cell of the liver. In malaria, the liver is the first site of invasion in the mammalian host, with Plasmodium dividing up to 100,000 times prior to escaping the hepatocyte into the bloodstream where the disease becomes symptomatic.15 Infected hepatocytes range in 10-50 μm in size with a round or oval shape and have a strong hematoxylin staining in H&E (see panels in Figure 2). Video S1 shows the iterative registration progress for the AF-to-AF image registration in this case study where the image colored in green is the target AF image and the magenta image that is warped is the source AF image. As registration progresses, one sees a linear alignment of gross structure then a non-linear alignment of fine structure which is especially visible near the edges of the section in the last 5 seconds of the video. We further supplemental our landmark distance evaluation with a validation of the AF-to-AF registration performed after data acquisition using binary masks of the infected hepatocyte ROIs selected on the H&E stainings, shown in Video S2, Figure S11 & 12, further discussed in detail in the supporting information. In this video and Figure S11, the top window is an overlay of high magnification micrographs of the registration process applied H&E images, but note that this is registration between AF-to-AF images that is applied to the H&E images post-hoc and not registration based on the H&E images themselves. The bottom frame of Video S2 shows binary masks of the infected hepatocyte ROIs color coded by target image (green) and source image (magenta) and overlap shown in white subjected to the same registration. It is clear that as the registration progresses, there is increased overlap of the annotated ROIs. Importantly, > 50% overlap is achieved for every infected hepatocyte present across both tissue sections. Perfect overlap is not expected as the data is from serial sections. In this instance, the target ROIs composes 0.05% of the tissue section’s area yet structure-to-structure overlap is achieved for each ROI with AF-to-AF registration. When expanding this quantitative analysis to 3 biological and 3 technical replicates, a DICE coefficient of 0.53 is obtained for 188 infected hepatocyte ROIs spread across the 9 serial tissue section pairs (Table S1). A deeper discussion of this method and results as well as a competitive test of AF-to-AF registration against non-destructive unstained brightfield and H&E is available in Figures S13-14 and Tables S1-2 in the Supporting Information section Grading accuracy of the annotation overlap of the infected mouse hepatocytes.
Registration-driven histology-directed IMS of individual infected hepatocytes in liver-stage malarial model
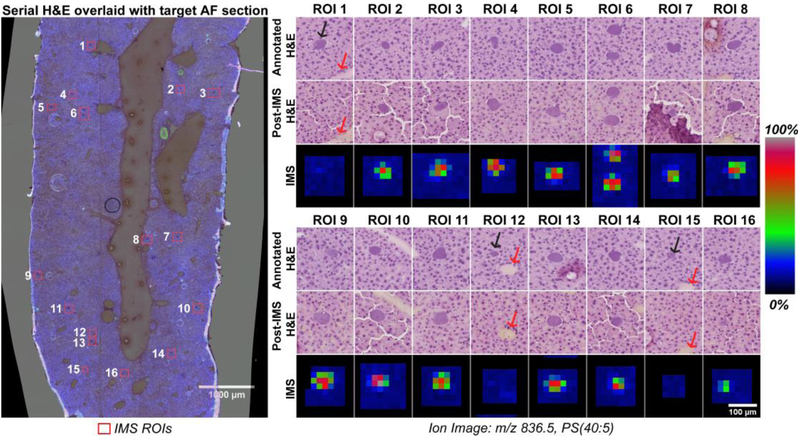
Fourteen individual plasmodia were imaged with relatively high spatial resolution IMS (20 μm). The data resulted in 812 IMS pixels and a run time of less than 2 minutes. For comparison, acquiring the entire tissue section at 20 μm would require ~82,000 pixels and an hour on the same instrument, and considerably longer on a slower MALDI instrument. Quantitatively, histology-directed IMS enabled an 82-fold reduction in acquired pixels versus non-targeted IMS for same information about these specific ROIs. Figure 4 presents all 16 IMS ROIs in the studied tissue section. The annotated serial section H&E is shown paneled with the post-acquisition H&E alongside the ion image for m/z 836.5, putatively identified through mass search of the LIPID MAPS database as a Phosphoserine phospholipid with a fatty acid tails totaling 40 carbons with 5 unsaturations (PS(40:5)). Figure 4 shows a false positive annotation in ROIs 1, 12 and 15 where a malarial plasmodium was not present as revealed by both IMS signal and the post-acquisition H&E staining. The black arrows on Figure 4 indicate the infected hepatocyte that was annotated but absent in the target section. The red arrows point to other morphology that is persistent across the serial sections and demonstrates conclusively that the false positive is not due to poor registration. These false positives highlight an unavoidable problem in the registration approach that is the loss of a given substructure from one serial section to the next. However, even in this case where infected mouse hepatocytes are generally only 1.5 to 4 times larger than the inter-section distance, only 3 such misses were counted among the annotations.
Figure 4. IMS data at 20 μm of P. yoelii infected hepatocytes targeted using serial section AF registration.
Left: Overlay of H&E from serial section and the AF of the IMS section. Numbers correspond to the panels on the right. ROIs are sorted by y-coordinate. Right: Ion images of m/z 836.5, PS(40:5), from individual acquisition ROIs displayed with the corresponding H&Es, the Annotated H&E is the image used for annotation blind to the target section histology while the Post-IMS H&E serves as a validation after IMS. ROIs 1, 12 & 15 contained an infected hepatocyte on the annotation serial section that was not present in serial for IMS. The black arrows highlight the annotated infected hepatocyte while the red arrows highlight surrounding morphology that is visible in the registered H&E images to indicate the loss of the hepatocyte is not registration error.
In comparison, a more traditional IMS approach to somewhat rapidly acquire a larger portion of the infected areas would be to image large subsections of the tissue. However, the number of infected sites imaged will be limited due to the spatial sparsity inherent to the histological structure of interest. In the liver-stage malarial Plasmodia case, the targeted ROIs are sparsely distributed (they compose 0.05% of the total area of the tissue section), suggesting that even a larger but not whole section traditional imaging ROI will only capture a limited number at the cost of substantial instrument time at the higher resolutions needed to spatially resolve the infected hepatocyte.
Direct autofluorescence annotation histology-directed experiments
Motivation of the direct annotation workflow
The second approach to histology-directed experiments using AF is to directly use the rich morphological information provided by AF microscopy images to identify ROIs. Direct annotation avoids complex registration and issues inherent to serial sections altogether, allowing targeting of any structure visible by AF microscopy. Thus in this approach, the limit on targeting is limited to the coordination between MS instrument stage and the annotation image.
Direct AF annotation workflow applied to the analysis of human kidney glomeruli
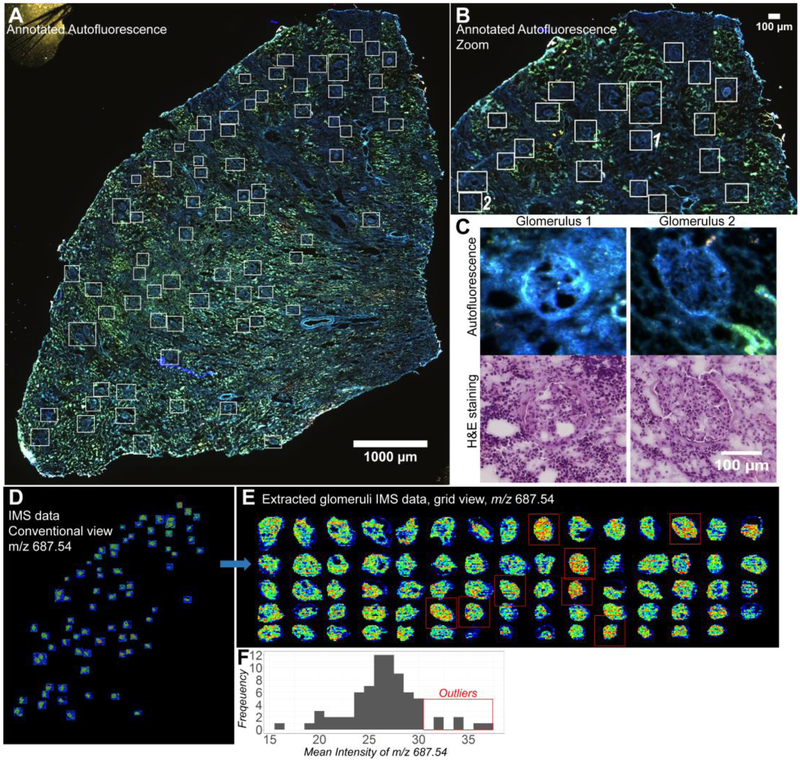
Glomeruli are the filtration units of the kidney and have important implications in kidney disease.31 For instance, chronic disruption of their function can lead to end stage renal disease.32 In Figure 5 A, the result of our direct AF annotation workflow applied to human kidney glomeruli is shown. Glomeruli have a distinct AF profile in both health and disease. In normal kidney, glomeruli display minimal AF signal and are visible as a ‘negative’ in the AF image. However, in kidney disease where the glomeruli begin to become sclerotic, collagen is accumulated, and strong AF from near-UV excitation is observed as shown in the sample in Figure 5. Given glomeruli’s AF signature in health and disease, registration to a separate annotation section is not necessary. Instead, the AF image can be annotated directly for histology-specific IMS acquisition. Figure 5A shows an AF image with annotated glomeruli in white boxes. In this case, IMS was performed at 10 μm spatial resolution using the Bruker RapiFlex TissueTyper on the glomeruli ROIs, generating 105,442 IMS pixels in total. Imaging the entire tissue section with the same instrumental settings and at the same spatial resolution would require ~1,000,000 IMS pixels or 10-fold more and would engender extremely long processing times and a large data footprint. Figure 5B shows a zoomed-in view of the same kidney section, showing the AF-described glomeruli in more detail. A zoom-in of two individual glomeruli is presented in Figure 5C, demonstrating the AF intensity and shape features that make glomerulus annotation facile. Below the AF in Fig 5C is a registered H&E stain of the same section (acquired post-IMS) and the same glomeruli, to allow comparison between the AF morphology and a conventional H&E staining of a glomerulus as a post-hoc validation.
Figure 5. Direct histology-directed IMS on human kidney glomeruli guided by AF microscopy.
A: Microscopy image of the whole tissue section with white boxes indicating glomeruli annotations. B: Zoom of the top portion of the tissue showing the distinct AF profile of the glomeruli. C: High magnification of two glomeruli and their registered H&E image taken after IMS acquisition. D: Conventional view of the IMS data as it would appear directly from the instrument of the ion image at m/z 687.54. E: IMS data of individual glomeruli size sorted and placed on a grid for rapid and convenient visualization. Extractions were made following the high accuracy IMS-microscopy registration workflow and markup of the staining in conjunction with the AF data. F: Frequency histogram of the glomeruli level mean intensity for m/z 687.54. Red boxes in E indicate high intensity outliers.
After AF-based annotation, IMS measurements are acquired in the designated ROIs. These resulting IMS data for ion m/z 687.54, putatively identified as a phosphoethanolamine (PE) with 36 carbons and 1 unsaturation on the fatty acid chains (PE-Cer(36:1)) is shown in Fig. 5D (in an untransformed coordinate space). Ceramide dysregulation has been implicated in kidney disease.33 Since the ROIs are scattered throughout the tissue section, presenting an easily interpretable visualization of ion distribution can be difficult. This is addressed in Figure 5E by placing the ROIs into a closely spaced grid for clearer visualization. Extraction of solely the glomeruli data highlights some limitations to this approach as it is currently implemented. First, rectangular bounding boxes were used for ROI selection because they are quicker to draw than polygons. Second, the bounding boxes were extended beyond the glomeruli to give a margin of error to the instrument spatial teaching accuracy (the accuracy with which the instrument stage can pinpoint the annotated areas or likewise, the accuracy with which one can register stage coordinates to image coordinates), an aspect that is difficult to quantitatively assess prior to measurement. Unknown teaching accuracy is circumvented by expanding the acquisition ROI rectangles in all directions and acquiring more data than would be truly necessary to only capture the glomeruli. Quantitatively, this approach results in 32,163 pixels of the 105,442 pixels (~30%) actually overlapping with glomeruli. Avoiding the work-around for this lingering systemic challenge could again reduce acquisition from 10-fold to 30-fold.
As each glomerulus is indexed by sample origin and glomeruli number, intensity patterns can be searched visually or with statistical approaches. The latter is demonstrated in Figure 5F, where an ion intensity histogram for ion m/z 687.54 (PE-Cer(36:1)) is shown. Exploring the frequency distribution of the mean intensities recorded for m/z 687.54 across the different glomeruli highlights some that are of high intensity (noted with a red box in Figure 5E). Multimodal approaches like this leverage the advantages of IMS in semi-quantitative chemical measurement together with qualitative information gained from high spatial resolution microscopy, and hint at substantial potential for more elaborate and deeper histological and pathological analyses in the future.
Conclusion
Within this work we demonstrate two histology-directed workflows utilizing AF microscopy. The first workflow uses mono-modal AF-to-AF registration to project annotations onto an untouched target section destined for IMS. The second workflow forgoes complex registration and directly annotates images based on morphology-rich AF images. The increased number of individual substructures that can be accurately targeted and measured through these approaches can aid in performing statistical characterization of tissue structures while concurrently realizing exceptionally high throughput.
Registration of AF with a classical staining of the same section allows us to establish annotation through traditional histological approaches. Improving the accuracy of histology-directed registration through AF allows us to probe even very small structures that are sparsely distributed. In the liver stage malaria case study, the targeted ROIs are between 10-40 μm in diameter, and it was essential to have high accuracy registration. Our accuracy across the mouse liver, was 8 μm on average with many hepatocyte landmarks registered within 5 μm of the ground truth. As further validation, we found that every annotated infected hepatocyte ROI had an overlap greater than 50% after registration. In total through this approach, we targeted individual cells that compose less than 1% of the total tissue area without a single miss that was not due to loss of the targeted structure across serial sections.
A critical juncture may be reached in histology-directed IMS approaches when sub-micron spatial resolution IMS becomes routine and individual cells of interest are probed for subcellular molecular content by IMS. At these resolutions having a microscopy grade view of tissue structure will be an important aid in targeting. We demonstrate the potential of direct AF annotation on larger structures (glomeruli) and note that AF microscopy is microscopy resolution and should be able to meet the demands of single cell or subcellular targeting in tissue. Using a direct AF annotation approach of human kidney glomeruli, we achieved a 10-fold time reduction to produce a full profile of almost all glomeruli within the target tissue section at high IMS spatial resolution (10 μm) and used the resultant data to identify outliers based on a glomerulus associated signal. Such rapid profiling has implications in both large tissue studies and for clinical adaptation.
Going forward with this in mind, it may be important for AF microscopy build a morphological base of knowledge to relate to traditional and routine histological analyses. Registration of the AF to such sources will be integral to transfer of domain-specific histological knowledge to AF microscopy. Given the wealth of knowledge available on stained tissue section histology, connecting AF to this resource to glean knowledge is fortunately feasible and could have a major impact in histology-directed IMS.
Supplementary Material
References
- (1).Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS,Caprioli RM; Farmer TB; Gile J Anal. Chem 1997, 69, 4751–4760. [DOI] [PubMed] [Google Scholar]
- (2).Integrating histology and imaging mass spectrometry,Chaurand P; Schwartz SA; Billheimer D; Xu BJ; Crecelius A; Caprioli RM Anal. Chem 2004, 76, 1145–1155. [DOI] [PubMed] [Google Scholar]
- (3).Image fusion of mass spectrometry and microscopy: a multimodality paradigm for molecular tissue mapping,Van de Plas R; Yang JH; Spraggins J; Caprioli RM Nat. Methods 2015, 12, 366–U138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).A novel histology-directed strategy for MALDI-MS tissue profiling that improves throughput and cellular specificity in human breast cancer,Cornett DS; Mobley JA; Dias EC; Andersson M; Arteaga CL; Sanders ME; Caprioli RM Mol. Cell. Proteomics 2006, 5, 1975–1983. [DOI] [PubMed] [Google Scholar]
- (5).Imaging mass spectrometry assists in the classification of diagnostically challenging atypical Spitzoid neoplasms,Lazova R; Seeley EH; Kutzner H; Scolyer RA; Scott G; Cerroni L; Fried I; Kozovska ME; Rosenberg AS; Prieto VG; Shehata BM; Durham MM; Henry G; Rodriguez-Peralto JL; Riveiro-Falkenbach E; Schaefer JT; Danialan R; Fraitag S; Vollenweider-Roten S; Sepehr A; Sangueza M; Hijazi N; Corredoira Y; Kowal R; Harris OM; Bravo F; Boyd AS; Gueorguieva R; Caprioli RM J. Am. Acad. Dematol 2016, 75, 1176–1186 e1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Histology-directed and imaging mass spectrometry: An emerging technology in ectopic calcification,Taverna D; Boraldi F; De Santis G; Caprioli RM; Quaglino D Bone 2015, 74, 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Invasive mucinous cystic neoplasms of the pancreas,Baker ML; Seeley ES; Pai R; Suriawinata AA; Mino-Kenudson M; Zamboni G; Kloppel G; Longnecker DS Exp. Mol. Pathol 2012, 93, 345–349. [DOI] [PubMed] [Google Scholar]
- (8).Lung cancer diagnosis from proteomic analysis of preinvasive lesions,Rahman SM; Gonzalez AL; Li M; Seeley EH; Zimmerman LJ; Zhang XJ; Manier ML; Olson SJ; Shah RN; Miller AN; Putnam JB; Miller YE; Franklin WA; Blot WJ; Carbone DP; Shyr Y; Caprioli RM; Massion PP Cancer Res. 2011, 71, 3009–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Mass Spectrometry Imaging Can Distinguish on a Proteomic Level Between Proliferative Nodules Within a Benign Congenital Nevus and Malignant Melanoma,Lazova R; Yang Z; El Habr C; Lim Y; Choate KA; Seeley EH; Caprioli RM; Li YQ Am. J. Dermatopathol 2017, 39, 689–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Pathology interface for the molecular analysis of tissue by mass spectrometry,Norris JL; Tsui T; Gutierrez DB; Caprioli RM J. Pathol. Inform 2016, 7, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Histology-Guided High-Resolution Matrix-Assisted Laser Desorption Ionization Mass Spectrometry Imaging,Heijs B; Abdelmoula WM; Lou S; Briaire-de Bruijn IH; Dijkstra J; Bovee JV; McDonnell LA Anal. Chem 2015, 87, 11978–11983. [DOI] [PubMed] [Google Scholar]
- (12).Image registration,Fitzpatrick JM; Hill DL; Maurer CR Jr Handbook of medical imaging 2000, 2, 447–513. [Google Scholar]
- (13).MALDI Imaging Mass Spectrometry Profiling of N-Glycans in Formalin-Fixed Paraffin Embedded Clinical Tissue Blocks and Tissue Microarrays,Powers TW; Neely BA; Shao Y; Tang HY; Troyer DA; Mehta AS; Haab BB; Drake RR Plos One 2014, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Autofluorescence spectroscopy and imaging: a tool for biomedical research and diagnosis,Croce AC; Bottiroli G Eur. J. Histochem, 2014, 58, 2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Malaria: Biology and Disease,Cowman AF; Healer J; Marapana D; Marsh K Cell 2016, 167, 610–624. [DOI] [PubMed] [Google Scholar]
- (16).Selection and refinement: the malaria parasite's infection and exploitation of host hepatocytes,Kaushansky A; Kappe SH Curr. Opin. Microbiol 2015, 26, 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Mechanisms of progression of chronic kidney disease,Fogo AB Pediatr. Nephrol 2007, 22, 2011–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Fiji: an open-source platform for biological-image analysis,Schindelin J; Arganda-Carreras I; Frise E; Kaynig V; Longair M; Pietzsch T; Preibisch S; Rueden C; Saalfeld S; Schmid B; Tinevez JY; White DJ; Hartenstein V; Eliceiri K; Tomancak P; Cardona A Nat. Methods 2012, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).The ImageJ ecosystem: An open platform for biomedical image analysis,Schindelin J; Rueden CT; Hiner MC; Eliceiri KW Mol. Reprod. Dev 2015, 82, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Engineering and algorithm design for an image processing Api: a technical report on ITK--the Insight Toolkit,Yoo TS; Ackerman MJ; Lorensen WE; Schroeder W; Chalana V; Aylward S; Metaxas D; Whitaker R Stud. Health Technol. Inform 2002, 85, 586–592. [PubMed] [Google Scholar]
- (21).elastix: a toolbox for intensity-based medical image registration,Klein S; Staring M; Murphy K; Viergever MA; Pluim JP IEEE Trans. Med. Imaging 2010, 29, 196–205. [DOI] [PubMed] [Google Scholar]
- (22).The Design of SimpleITK,Lowekamp BC; Chen DT; Ibanez L; Blezek D Front. Neuroinform 2013, 7, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).SimpleElastix: A user-friendly, multi-lingual library for medical image registration, Marstal K; Berendsen F; Staring M; Klein S In WBIR Conference: Las Vegas, Nevada, USA, 2016. [Google Scholar]
- (24).Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: enhanced information in both positive and negative polarities after 1,5-diaminonapthalene deposition,Thomas A; Charbonneau JL; Fournaise E; Chaurand P Anal. Chem 2012, 84, 2048–2054. [DOI] [PubMed] [Google Scholar]
- (25).Monitoring time-dependent degradation of phospholipids in sectioned tissues by MALDI imaging mass spectrometry,Patterson NH; Thomas A; Chaurand P J. Mass Spectrom 2014, 49, 622–627. [DOI] [PubMed] [Google Scholar]
- (26).imzML--a common data format for the flexible exchange and processing of mass spectrometry imaging data,Schramm T; Hester A; Klinkert I; Both JP; Heeren RM; Brunelle A; Laprevote O; Desbenoit N; Robbe MF; Stoeckli M; Spengler B; Rompp A J. Proteomics 2012, 75, 5106–5110. [DOI] [PubMed] [Google Scholar]
- (27).Cardinal: an R package for statistical analysis of mass spectrometry-based imaging experiments,Bemis KD; Harry A; Eberlin LS; Ferreira C; van de Ven SM; Mallick P; Stolowitz M; Vitek O Bioinformatics 2015, 31, 2418–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Diagnosis of tumors during tissue-conserving surgery with integrated autofluorescence and Raman scattering microscopy,Kong K; Rowlands CJ; Varma S; Perkins W; Leach IH; Koloydenko AA; Williams HC; Notingher I Proc. Natl. Acad. Sci. U. S. A 2013, 110, 15189–15194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Fourier Transform Infrared Microscopy Enables Guidance of Automated Mass Spectrometry Imaging to Predefined Tissue Morphologies,Rabe JH; D, A. S.; Schulz S; Munteanu B; Ott M; Ochs K; Hohenberger P; Marx A; Platten M; Opitz CA; Ory DS; Hopf C Sci. Rep 2018, 8, 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Image Similarity and Tissue Overlaps as Surrogates for Image Registration Accuracy: Widely Used but Unreliable,Rohlfing T IEEE Trans. Med. Imaging 2012, 31, 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Endothelin and endothelin antagonists in chronic kidney disease,Kohan DE; Barton M Kidney Int. 2014, 86, 896–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Endothelin antagonism for patients with chronic kidney disease: still a hope for the future,Schneider MP; Mann JF Nephrol. Dial. Transpl 2014, 29, I69–I73. [DOI] [PubMed] [Google Scholar]
- (33).Targeted Lipidomic and Transcriptomic Analysis Identifies Dysregulated Renal Ceramide Metabolism in a Mouse Model of Diabetic Kidney Disease,Sas KM; Nair V; Byun J; Kayampilly P; Zhang H; Saha J; Brosius FC 3rd; Kretzler M; Pennathur S J. Proteomics Bioinf 2015, Suppl 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.