Abstract
Inteins are mobile genetic elements that are spliced out of proteins after translation. Some inteins contain a homing endonuclease (HEN) responsible for their propagation. Hedgehog/INTein (HINT) domains catalyzing protein splicing and their nested HEN domains are thought to be functionally independent because of the existence of functional mini-inteins without HEN domains. Despite the lack of obvious mutualism between HEN and HINT domains, HEN domains are persistently found at one specific site in inteins, indicating their potential functional role in protein splicing. Here we report crystal structures of inactive and active mini-inteins derived from inteins residing in the transcription factor IIB of Methanococcus jannaschii and Methanocaldococcus vulcanius, revealing a novel modified HINT fold that might provide new insights on the mutualism between the HEN and HINT domains. We propose an evolutionary model of inteins and a functional role of HEN domains in inteins.
Keywords: inteins, protein splicing, homing endonuclease, horizontal gene transfer
1. INTRODUCTION
RNA splicing and alternative splicing are highly regulated processes during gene expression in higher organisms, leading to diverse gene transcripts coding for multiple proteins from a single gene.1 Limited numbers of genes found in genomes of higher organisms could thus result in much larger proteomic diversity created by RNA alternative splicing. Indeed, a large fraction of the protein-coding genes of multicellular organisms is alternatively spliced.2 In addition to the molecular diversity created at the RNA level, alternative splicing at the protein level has been recently discovered, in which up to four different molecular species could be produced from intermolecular protein splicing between two precursor proteins (two coding genes).3 This alternative splicing at the protein level is mediated by another class of intervening sequences called inteins (internal protein) and is termed intein-mediated protein alternative splicing (iPAS).3 Inteins are parasitic genetic elements inserted into protein-coding genes without providing any benefits to host proteins, as well as to host organisms.4,5 Inteins catalyze self-removal from the intervening host proteins after protein translation, concomitantly producing the functional intein-less protein by introducing a peptide bond between the interrupted host protein fragments (Fig. 1).4,5 Until the discovery of iPAS6,7 protein splicing was thought to take place only as an intermolecular reaction or as a bi-molecular trans-reaction by split inteins. Inteins are particularly prevalent in archaea, present in half of their genomes, and have been found only in unicellular organisms.4,8 They have generally been considered to be pure parasitic proteins with no biological function, but several functional roles have been suggested for specific inteins, such as environmental sensors.9,10,11,12 The biological function of iPAS is not only unknown but also challenging to identify in nature because protein splicing does not leave any mark on the mature host proteins and is impossible to trace back to the originating genes from the alternatively spliced products.3
Figure 1:
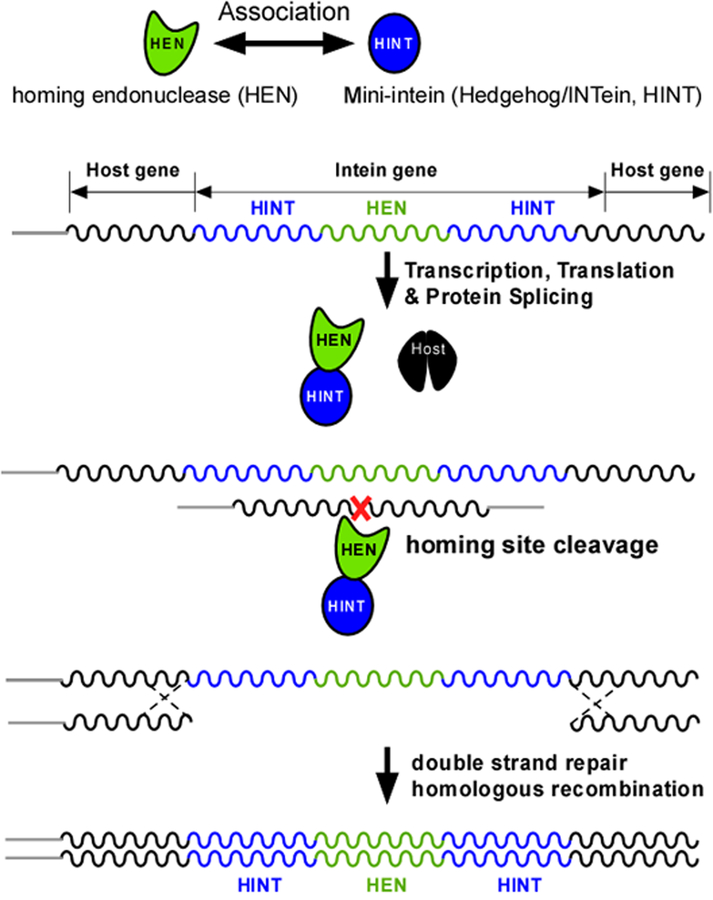
The homing mechanism of inteins. Host gene exons and products are in black. HEN stands for homing endonuclease (green). HINT stands for Hedgehog/INTein (HINT) domain (blue).
Protein splicing is catalyzed by inteins that share a common structural HINT (Hedgehog/INTein) fold.13 Many inteins are bi-functional, containing not only a HINT domain for protein splicing but also a HEN (homing endonuclease) domain, which is considered to be responsible for their propagation by horizontal gene transfer (HGT) (Fig. 1).5,14,15 The existence of natural mini-inteins without HEN domains and engineered functional mini-inteins without the nested HEN domains suggests that protein splicing and homing endonuclease domains are functionally independent.16,17,18,19 Inteins are found in conserved regions of their host proteins near the active sites. The insertion at the essential sites ensures the survival of inteins by making it difficult to remove them.5 Interestingly, HEN domains are found in only one specific site in many inteins, which also corresponds to the split site found in naturally split inteins. One of the remaining questions in the evolution of inteins is why HEN domains persist only at one specific insertion site when there is no mutualism between HEN and HINT domains (Fig. 1).20 Degenerated HEN domains without endonuclease activity persist against genetic drifts within inteins, suggesting that some parts of the HEN domain could be important for protein-splicing reaction by the intein, or contribute to its overall architecture.20,21
We previously found, that the Methanococcus jannaschii intein (MjaTFIIB) is very highly efficient in cis-splicing using an E. coli system.22 However, a MjaTFIIB mini-intein without the HEN domain turned out to be splicing-deficient, supporting the hypothesis that its HEN domain could play a critical role in the splicing process.20,21 Paradoxically, the inactive engineered MjaTFIIB mini-intein without the HEN domain could still induce iPAS in the presence of a split precursor protein containing the C-terminal 53-residue fragment of the MjaTFIIB intein.3 This observation contradicts any structural role of the HEN domain in MjaTFIIB intein because protein-splicing of the engineered MjaTFIIB mini-intein can be activated in the complete absence of the HEN domain by iPAS. Thus, the origin of the mutualism between HINT and HEN domains in MjaTFIIB intein, if any, is still enigmatic.
Here we report the structures of an inactive MjaTFIIB mini-intein and a partially active TFIIB mini-intein from Methanocaldococcus vulcanius M7 (Mvu), elucidated by X-ray crystallography. The structure of MjaTFIIB mini-intein revealed a novel HINT fold with an additional β-strand in the core of the HINT domain. Further, protein engineering of mini-inteins from MjaTFIIB and MvuTFIIB inteins indicated the importance of the length of the loop, which is not visible in the structure of the functional MvuTFIIB mini-intein. A structural comparison between the two mini-variants of the TFIIB inteins suggests the plasticity or flexibility in the HINT domains. Our results also indicate that there are dynamic features associated with the HINT domain of TFIIB inteins that significantly contribute to the protein-splicing activity. We discuss the functional and evolutionary roles of the HEN domains in inteins and propose a mutualism model between the HEN and HINT domains.
Results
Modeling of MjaTFIIB mini-inteins
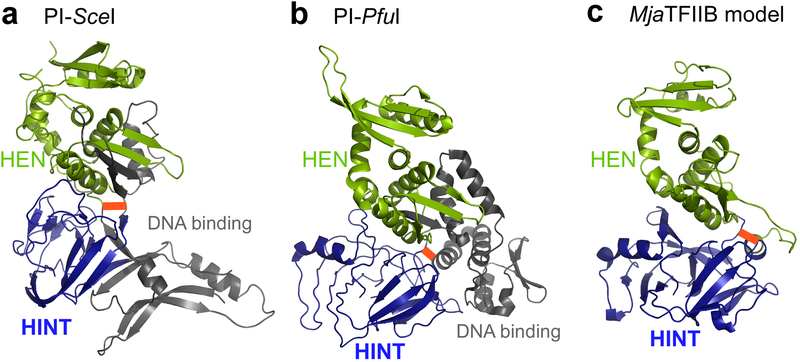
Small, highly efficient inteins without HEN domains are preferred as protein engineering tools, e.g., for protein ligation.18 TFIIB intein from Methanococcus jannaschii (MjaTFIIB intein) exhibits efficient cis-splicing activity in E. coli and is smaller than canonical inteins with a homing endonuclease (HEN) domain, such as SceVMA intein and PI-PfuI.22,23,24 Canonical inteins, exemplified by SceVMA intein, typically consist of about 450 residues because of the insertion of a LAGLIDADG-family homing endonuclease domain into the HINT (Hedgehog/INTein) fold.25 We were initially interested in rationally engineering robust MjaTFIIB mini-inteins with only the HINT domain, retaining the efficient splicing activity, based on the sequence homology. The homology search using the MjaTFIIB intein sequence identified a putative endonuclease domain within MjaTFIIB intein, but MjaTFIIB intein comprises only 335 residues, which is about 100 residues smaller than SceVMA intein. A BLAST search against the Protein Data Bank (PDB) identified PI-PkoII (PDB id: 2cw7) and PI-PfuI (PDB id: 1dq3) as the inteins with two highest homologies (Supplementary Fig. 1).24,26 We used PI-PfuI as the template to model a three-dimensional structure of MjaTFIIB intein for designing mini-inteins (Fig. 2). Mini-inteins containing only the HINT fold without deterioration of the protein-splicing activity have been successfully engineered by removing HEN domains, e.g., MtuRecA, SspDnaB, and NpuDnaB inteins, among others, suggesting that the HEN domains are not essential for protein splicing activity.16,18,19
Figure 2:
Design of MjaTFIIB mini-intein. Structures of PI-SceI (a), PI-PfuI (b), and the modeled full-length MjaTFIIB intein (c). HINT domains and HEN domains are colored in blue and green, respectively. The DNA binding domain of PI-SceI and the possible DNA-contacting domain of PI-PfuI are shown in gray. Red thick lines illustrate possible polypeptide linkers to detach HEN domains from HINT domains.
We first attempted to create a mini-intein from MjaTFIIB intein by retaining only the residues corresponding to the HINT fold, analyzing the homology model created from the alignment with PI-PfuI (Fig. 2; Supplementary Fig. 1). We expected that the inserted HEN domain (170 residues) might be safely removed from MjaTFIIB intein without disrupting the HINT fold, as it was successfully done with other inteins. However, the engineered MjaTFIIB mini-intein (MjaTFIIBΔ170 intein) turned out to be deficient in cis-splicing, although alternative protein splicing was observed with a C-terminal split fragment of MjaTFIIB intein by iPAS without the HEN domain (Fig. 3).3 This observation induced us to investigate the three-dimensional structure of MjaTFIIB intein further. This result might suggest that the three-dimensional structure could completely differ from other known inteins, from which HEN domains can be removed without affecting splicing activity. To understand the structural basis for the splicing deficiency of the engineered MjaTFIIB mini-intein (MjaTFIIBΔ170 intein), we attempted to determine the three-dimensional structures of various mini-inteins of MjaTFIIB intein and of its homolog.
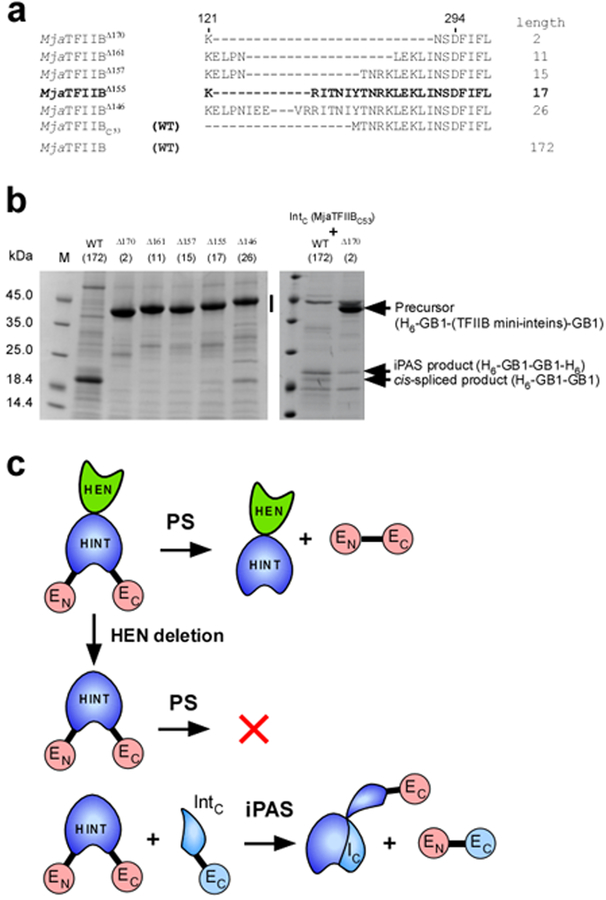
Figure 3:
SDS-PAGE analysis of cis-splicing and protein alternative splicing (iPAS) of the full-length MjaTFIIB intein and engineered MjaTFIIB mini-inteins. (a) The sequence alignment of the loop region among the engineered MjaTFIIB mini-inteins and the split intein. The numbers of amino acid residues deleted are indicated in superscript with the name of inteins. The lengths of the loop between residue 121 and 294 are indicated at the right side. WT stands for the wild-type sequence. C53 in subscript indicates the C-terminal 53 residue of the intein. (b) SDS-PAGE analysis of the elution fractions from IMAC purification using the N-terminal His-tag in the precursor protein for cis-splicing and alternative splicing. In the left panel, the vertical bar indicates the region where bands of unreacted cis-splicing precursor proteins are expected. The right panel shows SDS-PAGE analysis of the elution fractions from co-expression of the C-terminal split intein fragment (MjaTFIIBC53-GB1-H6) with the cis-splicing precursors indicated at the top. Arrows indicate, the position of a precursor protein, the band from iPAS product H6-GB1-GB1-H6, and the band corresponding to the cis-spliced product of H6-GB1-GB1. M stands for molecular weight markers. The numbers within brackets show the numbers of residues between residue 121 and 294. (c) A cartoon presentation of the effects of the HEN domain (in green) on cis-splicing by HINT domain (in blue) and co-expression of the C-terminal split intein fragment (IntC). HEN deletion results in a cis-splicing deficient intein, which can be partially activated by iPAS with an intein fragment (IC).
Deletion variants of MjaTFIIB inteins
We employed the strategy of engineering mini-inteins by eliminating their HEN domains based on sequence alignment that we used previously.18,27 However, the first engineered mini-intein derived from MjaTFIIB intein (MjaTFIIBΔ170) did not catalyze cis-splicing using a model system that utilized the B1 domain of IgG binding protein G (GB1) as flanking exteins (Fig. 3).3 We slightly modified the deletion in the loop where the presumed HEN domain located by extending the connecting loop length (Fig. 3). To our surprise, further elongation of the loop in the MjaTFIIB mini-inteins offered little improvement of the splicing activity, implying that the deficiency in protein splicing is not merely due to the insufficient loop length or constraints introduced by the deletion, but that other factors might play critical roles. This observation suggested that there could be some mutualism between the HEN and HINT domains (Fig. 3).
Unexpected modification of the HINT fold in the MjaTFIIBΔ155 mini-intein
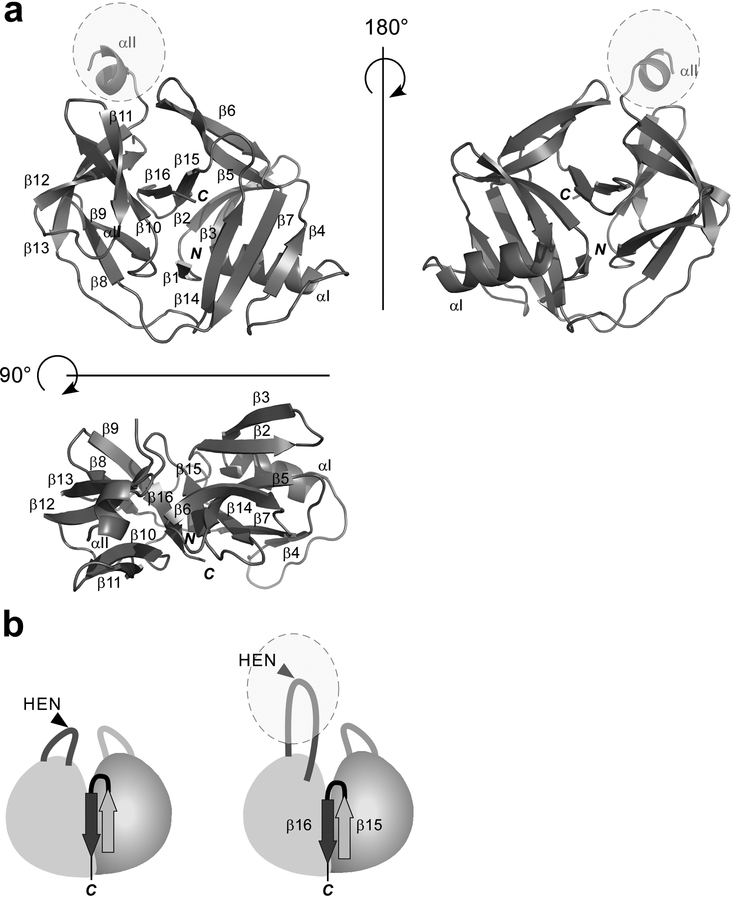
Despite many attempts to crystallize the full-length MjaTFIIB intein and various MjaTFIIB mini-inteins, we could obtain crystals for only one of them, namely MjaTFIIBΔ155 (Figs. 3 and 5). Even though MjaTFIIBΔ155 was deficient in cis-splicing in our model cis-splicing E. coli system, we succeeded in solving its three-dimensional structure at 2.0 Å resolution (Figs. 3 and 5). The structure of MjaTFIIBΔ155 that resulted from the final refinement contains two protein molecules, six dioxane molecules, one MES molecule, 14 glycerol molecules, ten ammonium ions, and 194 water molecules in the asymmetric unit. In chain A, 182 out of 185 amino acid residues have been modeled, but the first three N-terminal residues have not been included due to the complete lack of electron density. In chain B, 183 out of 185 amino acid residues have been modeled, with the first two N-terminal residues not visible. The structures of the two molecules in the asymmetric unit are very similar except for one loop region (residues 58–65), in which the difference is relatively large (3.8 Å for the Cα of Asn61). However, this loop region is involved in crystal contacts. The RMSD between these two monomers is 0.58 Å for the aligned backbone atoms.
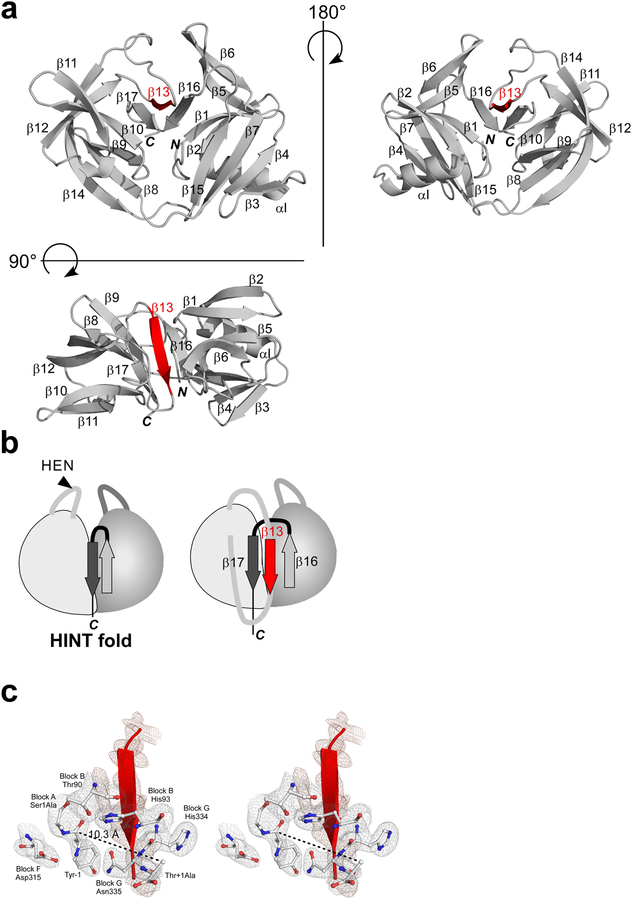
Figure 5:
The crystal structure of MjaTFIIB mini-intein (MjaTFIIBΔ155). (a) Schematic drawings of the crystal structure of MjaTFIIBΔ155 from three different orientations. The unusual β-strand (β13) insertion is colored in red. N and C stand for N- and C-termini, respectively. (b) Schematic illustrations of the canonical HINT fold and the novel HINT fold observed in MjaTFIIBΔ155 with the last two β strands (dark and light gray) and the unusual β-strand (β13) insertion (red). (c) A stereoview of the active site together with the inserted β−strand (β13) in red. The distance between the carbonyl carbon atom of Tyr-1 and Cβ atoms of Ala+1 is shown in dotted line. The final electron density map, contoured at 1.0 σ-level, is shown for the selected residues (in gray) and for the β13-strand (in dark pink).
Surprisingly, MjaTFIIBΔ155 revealed a novel, modified HINT fold, having an extra β-strand in the core near the splicing site (Figs. 4 and 5). The structure of MjaTFIIBΔ155 mini-intein can be superimposed well with the search model which was created based on PI-PfuII intein, confirming that it is similar to the HINT fold except for the inserted β-strand (β13) (Figs. 4 and 5).13,24 The RMSD between the search model and the monomer A of MjaTFIIBΔ155 is relatively large (2.9 Å for 628 pairs of the aligned backbone atoms) due to insertion of an additional β-strand. The loop in MjaTFIIBΔ155 mini-intein where a HEN domain typically locates included 17 residues between residues 121 and 294, which folded into a β-strand (β13) and formed an antiparallel β-sheet with β16 (Fig. 5a). The strand β13 is inserted into the core of the HINT fold between the last two β-strands, β16 and β17 (Fig. 5b). However, the distance between the carbonyl carbon atom of the N-terminal scissile bond and Cβ atom of the C-terminal nucleophilic residue of C-extein, which is mutated from Thr to Ala, is 10.3 Å. This distance is not much larger than the corresponding distances observed in a majority of the reported intein structures (8–9 Å), which typically have an open conformation (Fig. 5c).17,23,27 We initially assumed that this unusual β-strand insertion might inhibit the splicing activity by disrupting the active site coordination. However, deletion of this β-strand from MjaTFIIBΔ155 (e.g., MjaTFIIBΔ157) did not improve the activity of the MjaTFIIB mini-intein (Fig. 3). Therefore, we speculate that this unusual HINT fold of MjaTFIIBΔ155 is more likely to be accidentally produced as the lowest energy state due to the minimization engineering and does not represent the functionally relevant structure of MjaTFIIB intein. The interaction between the two pseudo-domains could be weak, thereby accommodating the insertion.
Figure 4:
The sequence comparison of the full-length MjaTFIIB intein and MjaTFIIBΔ155 mini-intein with the secondary structures. The secondary structures identified in the crystal structure of MjaTFIIB mini-intein (MjaTFIIBΔ155) are shown with arrows (β-sheets) and rectangles (helices). The region of a putative endonuclease domain of MjaTFIIB intein is in bold and underlined. The unique β-strand (β13) identified in the structure of MjaTFIIB mini-intein (MjaTFIIBΔ155) is indicated with arrow colored in red.
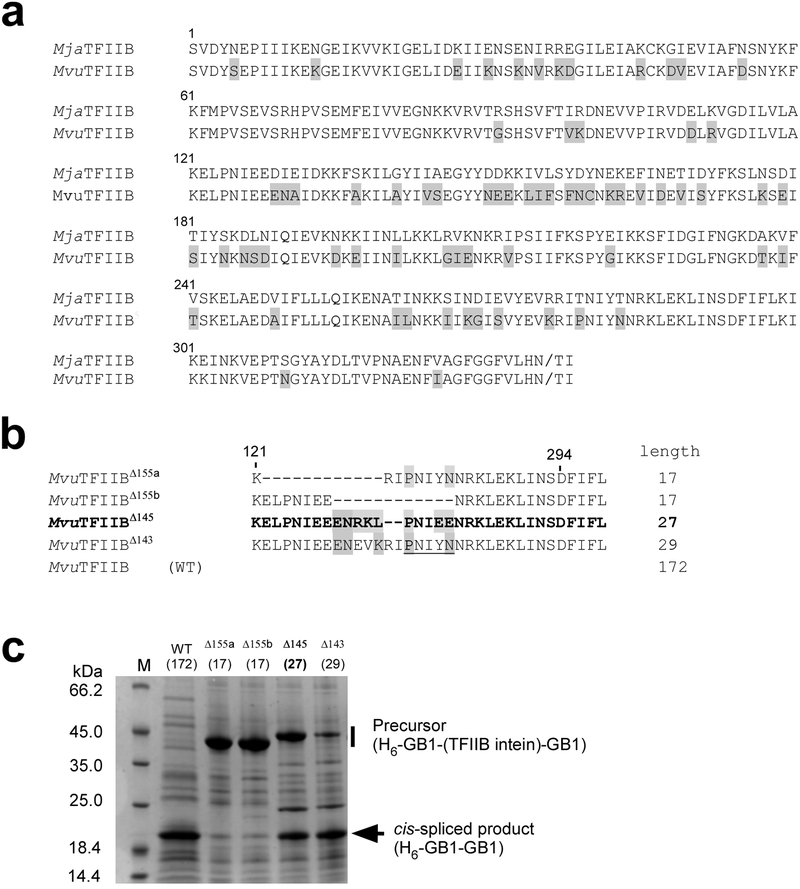
MvuTFIIB mini-intein (MvuTFIIBΔ145)
We still wanted to confirm whether the unexpected HINT fold of MjaTFIIBΔ155 was truly an accidentally trapped conformation resulting from the minimization of MjaTFIIB inteins, irrelevant for the splicing activity. To answer this question, we investigated another homologous TFIIB intein from Methanocaldococcus vulcanius M7 (MvuTFIIB intein), exhibiting sequence identity of 78.5% (Fig. 6a and Supplementary Fig. 1), with the hope of solving its crystal structure. We also found that MvuTFIIB mini-intein, e.g., MvuTFIIBΔ155 was inactive, like other MjaTFIIB mini-inteins with similar loop lengths, preserving the same intolerance of the HEN deletion observed for the MjaTFIIB intein (Fig. 6b and 6c). Unfortunately, we failed to obtain any diffracting crystals for the full-length MvuTFIIB intein and other MvuTFIIB mini-inteins except for MvuTFIIBΔ145. Importantly, this MvuTFIIBΔ145 mini-intein was at least notably active although the loop region, where the HEN domain was removed, contains an artificial sequence accidentally introduced during the cloning procedure (Fig. 6b). The HEN insertion loop contains 27 residues (between residues 121 and 294). The MvuTFIIB mini-intein with the 29-residue loop (143-residue deletion in the HEN region) in the same location (MvuTFIIBΔ143) was also partially active, suggesting that TFIIB mini-inteins need at least 27–29 residues in this region for cis-splicing activity. We determined the structure of MvuTFIIBΔ145 at 2.5 Å resolution (Fig. 7, Table 1). The overall structure of MvuTFIIBΔ145 reveals a canonical HINT fold of its two molecules in the asymmetric unit but does not share the unusual HINT fold of MjaTFIIBΔ155. The Ramachandran plot shows 93.5%, 4.1%, and 2.4% of all residues falling into the most favored, additionally allowed, and generously allowed regions, respectively (Table 1). Inferior statistics of this structure can be attributed to the poorly defined loop regions (Fig. 7a), presumably resulting in lower crystal quality that affected the resolution of diffraction data. We limited modeling of water molecules to only those that were located in very clear electron density. The longer loop at the deleted HEN region is mostly invisible and was thus not modeled, even though this longer loop was essential for the splicing activity. This observation suggests that this functionally required loop is flexible to the point of being disordered.
Figure 6:
MvuTFIIB intein and MvuTFIIB mini-inteins. (a) A comparison of the primary structures of MjaTFIIB and MvuTFIIB inteins. (b) The sequence alignment of the loop region of the engineered MvuTFIIB mini-inteins. The numbers of amino acid residues removed from the loop region are indicated in superscript together with the name of the intein. The lengths of the loop between residue 121 and 294 are indicated at the right side. (c) SDS-PAGE analysis of the elution fractions from IMAC using the N-terminal His-tag in the precursor protein. The vertical bar indicates the region where bands of unreacted precursor proteins are expected. An arrow indicates the band corresponding to the cis-spliced product of H6-GB1-GB1. M stands for molecular weight marker. Lengths of the loop in MvuTFIIB mini-inteins are shown at the top of each lane. Numbers within brackets indicate the numbers of the remaining residues between residues 121 and 294.
Figure 7:
The crystal structure of MvuTFIIB mini-intein (MvuTFIIBΔ145). (a) Schematic drawings of the crystal structure of MvuTFIIBΔ145 from three different orientations as Fig. 5. Shadowed dashed circles indicate the HEN-insertion region lacking the electron density. (b) Schematic illustrations of the canonical HINT fold and the HINT fold in MvuTFIIBΔ145 with the HEN insertion site indicated by arrowheads and the HEN insertion loop by a shadowed dashed circle.
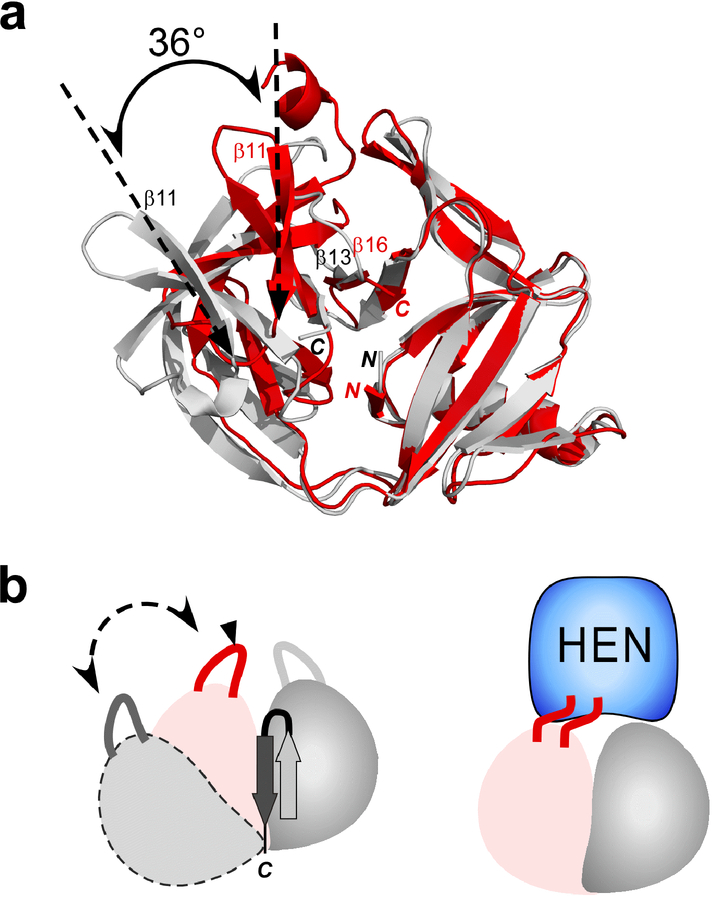
Comparison between MjaTFIIBΔ155 and MvuTFIIBΔ145
Importantly, the largest difference between the active MvuTFIIBΔ145 and inactive MjaTFIIBΔ155 is the unusual β-strand (β13) insertion found in the core of MjaTFIIBΔ155. The HINT fold can be divided into two pseudo-domains that presumably resulted from gene duplication during evolution.13 After superposition of the first pseudo domains (residues 1–75) of the two structures, the overlaid regions (residues 1–75) superimpose well, with RMSD of 0.51 Å for the backbone atoms. The last β-strand (β16) in MvuTFIIBΔ145 replaces the inserted β-strand (β13) between β16 and β17 in MjaTFIIBΔ155 (Figs. 7 and 8). Additionally, MvuTFIIBΔ145 assumes a more closed conformation with a rotation of 36° of the second domain, indicating plasticity between the two pseudo-domains (Fig. 8). However, the distance between the nitrogen atom of Cys1Ala and carbonyl carbon atom of the last residue of Asn is similar in the two structures (9.6 Å for MvuTFIIBΔ145, compared with 9.1 Å of MjaTFIIBΔ155). This result indicates the presence of an “open conformation” similar to many reported inteins structures, although MvuTFIIBΔ143 lacks the −1 and +1 residues.17,23,24,27 We believe that the structure of MvuTFIIBΔ145 represents better the functional state of the TFIIB inteins than that of MjaTFIIBΔ155.
Figure 8:
Comparison of two structures of MjaTFIIBΔ155 and MvuTFIIBΔ145 mini-inteins. (a) A superposition of the two coordinates after fitting the backbone atoms of residues 1–75. The angle of β11 strand of both structures was derived from the superimposed structures and shown. Ribbon drawings of MvuTFIIBΔ145 and MjaTFIIBΔ155 are colored in red and gray, respectively. (b) Schematic illustrations of the two structures highlighting two pseudo-sub-domains and the movement observed for MjaTFIIBΔ155 (left) and a model with the HEN domain (right).
Discussion
The new crystal structures of the inactive MjaTFIIB mini-intein and partially active MvuTFIIB mini-intein (MjaTFIIBΔ155 and MvuTFIIBΔ145) shed new light on how HEN domains persist in inteins by providing a mutualism between HINT and HEN domains. Many canonical inteins contain a HEN domain that cleaves the DNA sequences near the intein insertion points. Such enzymatic activity has presumably played (or still plays) an important role in the propagation of intein genes by HGT (Fig. 1), similarly to other selfish gene elements such as intron-encoded homing endonucleases.28,29
Minimization engineering of TFIIB inteins by removing the HEN region resulted in unexpected splicing deficiency, unlike in other previously reported engineered mini-inteins.16,18,19 Nonetheless, the elucidated three-dimensional structures of the engineered TFIIB mini-inteins are in agreement with the structural requirement for active HINT domains (e.g., only 5.6 Å between the carbonyl carbon of residue 121 and nitrogen atom of residue 294 in the structure of MjaTFIIBΔ155 and 7.9 Å for MvuTFIIBΔ145). This agreement between the original homology model and the experimentally determined structure indicates that the distinct lowest energy status found in the crystal structure is not solely responsible for the splicing reaction. We speculate that the folding process and/or structural dynamics of the HEN domain in TFIIB inteins must play a critical role in protein splicing (Fig. 2). The engineered mini-inteins remain inactive despite further modifications of the connecting loop. A longer linker (at least 26 residues between residues 121 and 294) at the HEN insertion site was found to be required for restoring the partial activity of TFIIB inteins (Figs. 3 and 6). Despite the requirement of a longer linker for the function, these residues were invisible in the electron density of MvuTFIIBΔ145, suggesting that this region is disordered/flexible. This observation supports an interpretation that structural dynamics involved with the engineered longer linker and the original HEN domain might play an important role in protein splicing activity, rather than that some parts of the HEN domain contribute to the functional HINT domain architecture. The importance of structural dynamics rather than the structural integration of the HEN domain in the HINT domain could also explain the observed iPAS of MjaTFIIB mini-intein induced by the C-terminal 53-residue fragment of MjaTFIIB intein (MjaTFIIBC53) without any part of the HEN domain3 (Fig. 3). It is also in line with the engineered RecA mini-inteins, of which local dynamics could account for the difference in self-cleavage activity.33,34 A comparison between the inactive MjaTFIIBΔ155 and active MvuTFIIBΔ145 shows inter-domain flexibility between the two pseudo-domains of the HINT fold of TFIIB inteins (Fig. 8). The HEN domains of TFIIB inteins are likely to play a critical role in bringing the two pseudo-domains into an active conformation or/and controlling the concerted protein splicing reaction steps. Unlike other inteins, the HEN domain of TFIIB inteins might be essential for productive protein folding which is coupled with protein splicing reaction of the HINT domain or for structural dynamics necessary for protein splicing. In other words, the HEN domain of TFIIB inteins could be considered to have the maturase activity to assist proper folding of HINT domains, similar to HEN encoded RNA-maturase encoded in introns.35
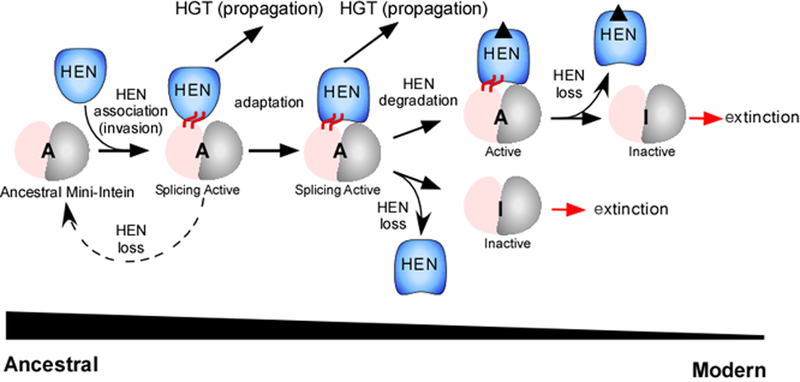
Our studies, as well as studies by others, postulate that HEN-containing inteins can be classified into at least two distinct classes. One of them is the group of inteins in which HEN and HINT domains are functionally independent and have developed little or no mutualism between them. In that case, the HEN domains can be easily removed without any loss of the protein splicing activity.36 One might consider that these inteins appeared by recent invasions of mini-inteins by a HEN domain (Fig. 9). Therefore, they are still mostly tolerant to the loss of HEN domains with no interference to protein splicing. The other class consists of inteins that have already developed some mutualism between the HINT and HEN domains, with their splicing activities becoming largely dependent on the existence of the inserted HEN domain. Adaptation of HEN domains to the invaded inteins could provide persistence or maintenance of HEN domains within inteins by mutualism. In the case of TFIIB intein, the function of HEN domain might be to assist folding of the HINT domain to a functional conformation of TFIIB inteins, thereby promoting protein splicing. This function is analogous to RNA-maturase as found in introns encoding HEN, which promotes intron splicing.35 For HINT domains, such mutualism could apparently ensure the propagation of intein genes by HGT.4,20 For HEN domains, mutualism could make it harder to eliminate them from intein genes, because a loss of the active or inactive HEN domain would lead to impaired splicing activity required for survival of host organisms, thereby ensuring the survival of the HEN domains in inteins. It might be possible to consider that these inteins have been invaded with a HEN domain much earlier and developed the mutualism by co-evolution (Fig. 9). In this scenario, naturally existing mini-inteins are possible survivors of ancestral mini-inteins that did not develop any mutualism with HEN domains during homing cycles and are still lacking a HEN domain (Fig. 9).
Figure 9:
An evolutionary model for the mutualism development between HEN and HINT domain during homing cycles. The nested HEN domains adapt with the inserted HINT domains so that HEN domains have become persistent by the developed mutualism.
To the best of our knowledge, all of the HEN-containing inteins share, without any exception, only one common insertion site of their HEN domains which also coincides with the naturally occurring split site (C35 site, according to the NpuDnaE-based numbering system that we previously proposed), even though HEN domains could, in principle, invade any sites of inteins during the homing cycle.39 It is still puzzling why there is only one HEN insertion site in inteins because there is no obvious selection mechanism that the HEN domain needs to locate at that particular place. Our experiments might suggest that the high conservation of the HEN insertion site among inteins is likely to due to the requirement for proper folding of HINT domains or for providing structural dynamics required for the protein-splicing reaction.
Protein engineering of mini-inteins from HEN-containing inteins as demonstrated in this article could reveal the evolutionary history of individual inteins and might be able to provide some hints for the primeval functions of ancestral inteins, the emergence of protein-splicing phenomenon, and naturally occurring iPAS phenomena, if any. A better understanding of the evolutionary aspects of individual inteins might assist efficient usage of protein splicing and protein alternative splicing as protein-engineering tools for controlling protein functions, targeting inteins as drug targets, and creating molecular diversities on the protein level.40
Methods
Construction of vectors of MjaTFIIB mini-inteins for cis-splicing tests
The plasmid (pSKDuet20) for the full-length MjaTFIIB intein was previously reported and used as a template for MjaTFIIB mini-inteins.22 MjaTFIIBΔ170 (pSADuet760) was constructed by inverse PCR with the following oligonucleotides: I292: 5’-TTTAAGAATATGAAATCAGAATTCTTTGCTAAAAC and I291: 5’-GATATATTAGTTTTAGCAAAGAATTCTGATTTCAT.
Similarly, MjaTFIIBΔ161 (pBHDuet45), MjaTFIIBΔ157 (pSADuet777), MjaTFIIBΔ155 (pSADuet779), and MjaTFIIBΔ146 (pBHDuet61) mini-inteins were constructed by inverse PCR using pSKDuet20 as a template and pairs of the following oligonucleotides: I532: 5’-AAAAGAATTGCCGAATACCAATAGAAAACTCGAAAA and I533: 5’-CGAGTTTTCTATTGGTATTCGGCAATTCTTTTGC, I319: 5’- CAAAAGAATTGCCGAATCTCGAAAAACTTATAAAC and I320: 5’- TTTATAAGTTTTTCGAGATTCGGCAATTCTTTTGC, I338: 5’- ATTAGTTTTAGCAAAAAGAATAACAAATATATATACC and I339: 5’- TATTTGTTATTCTTTTTGCTAAAACTAATATATCTC, and I581: 5’- GAATATTGAAGAAGAGAATGAAGTAAAGAGAATACCC and I582: 5’- GGGTATTCTCTTTACTTCATTCTCTTCTTCAATATTC, respectively.
Construction of vectors of MvuTFIIB mini-inteins for cis-splicing tests
The gene of the full-length MvuTFIIB intein (pBHDuet33) was amplified from the genomic DNA of Methanocaldococcus vulcanius M7 (DSM-12094) using the following two oligonucleotides of I510: 5’- ACGGATCCTACAGTGTTGATTATAGCGAACC and I511: 5’- TCTGGTACCGTGGATGGTGTTGTGTAAAAC and cloned between BamHI and KpnI sites of pSKDuet16. MvuTFIIBΔ155a (pBHDuet173) was constructed from pBHDuet33 using the two oligonucleotides of I877: 5’- TATACTGGTTTTAGCAAAACGAATACCCAATATATATAAC and I878: 5’- GTTATATATATTGGGTATTCGTTTTGCTAAAACCAGTATA. Plasmids for MvuTFIIBΔ155b (pBHDuet50K) and MvuTFIIBΔ145 (pBHDuet50F) were constructed using the two oligonucleotides of I543: 5’- GCCGAATATTGAAGAAAATAGAAAACTCGAAAAAC and I544: 5’- TCGAGTTTTCTATTTTCTTCAATATTCGGCAATTC by inverse PCR amplification of pBHDuet33 as the template. pBHDuet50K was accidentally created by incorporation of the oligonucleotides twice. MvuTFIIBΔ143 (pBHDuet64) was constructed from pBHDuet33 using the two oligonucleotides of I581: 5’- GAATATTGAAGAAGAGAATGAAGTAAAGAGAATACCC and I582: 5’- GGGTATTCTCTTTACTTCATTCTCTTCTTCAATATTC.
Analysis of cis-splicing by mini-inteins
Cis-splicing by MjaTFIIB and MvuTFIIB mini-inteins was analyzed by expressing the constructs described above. E. coli strain ER2566 (New England Biolabs) was transformed with each plasmid carrying a mini-intein and plated on LB-agar plates supplemented with 25 μg/ml kanamycin at 37 °C. 5 ml of LB-medium supplemented with a final concentration of 25 μg/ml kanamycin was inoculated with a single colony and incubated with vigorous shaking at 250 rpm overnight at 37 °C. 5 ml of the overnight culture was diluted into 45 ml of fresh LB-medium supplemented with a final concentration of 25 μg/ml kanamycin and incubated at 37 °C with shaking at 250 rpm. When OD600 reached 0.6, the mini-intein was induced with a final concentration of 1 mM isopropyl-β-D-thiogalactoside (IPTG) for 3 hours at 37 °C. The induced cells were harvested by a 10-minutes centrifugation at 4000 rpm, 4 °C and re-suspended in 4 ml of 50 mM sodium phosphate buffer (pH 8.0) and 300 mM NaCl. The half of the re-suspended cells was lysed by sonication. The His-tagged protein was purified using a Ni-NTA spin column according to the manufacturer’s protocol (Qiagen). The elution from the spin-column was diluted with two-fold SDS loading buffer containing 1 mM dithiothreitol (DTT) and analyzed on 18% SDS polyacrylamide gels after Coomassie Blue R (GE Healthcare Life Sciences) staining.
Cloning, expression, and purification of MjaTFIIBΔ155 mini-intein
The gene of MjaTFIIBΔ155 mini-intein with C1A mutation for structure determination was amplified from pSADuet779 as the template using the two oligonucleotides of HK803: 5’- ATGGATCCGGTGGATATGCTGTTGATTACAACGAAC and HK804: 5’- TCGGTACCTTAGGCGTTGTGTAATACAAATCCTC, and cloned between BamHI and KpnI site of pHYRSF53, resulting in plasmid pSCFRSF131 bearing MjaTFIIBΔ155 as His-tagged SUMO fusion protein.42
E. coli strain ER2566 (New England Biolabs) was transformed with the plasmid pSCFRSF131 carrying H6-SUMO-MjaTFIIBΔ155 (C1A). 50 ml of LB-medium supplemented with a final concentration of 25 μg/ml kanamycin was inoculated with a single colony and incubated with vigorous shaking at 250 rpm overnight, at 30 °C. The overnight culture was diluted into 2 liters of fresh LB-medium supplemented with a final concentration of 25 μg/ml kanamycin and incubated at 37 °C with shaking at 250 rpm. When OD600 reached 0.6, MjaTFIIBΔ155 was induced with a final concentration of 1 mM IPTG for 3 hours at 37 °C. The induced cells were harvested by a 10-minutes centrifugation at 1500 rpm, 4 °C and re-suspended in 15 ml with lysis buffer (50 mM sodium phosphate buffer (pH 8.0) and 300 mM NaCl). The cells were flash-frozen in liquid nitrogen, and stored at −74 °C. The SUMO-fusion was purified by immobilized metal ion affinity chromatography (IMAC) using a 5 ml HisTrap FF column (GE Healthcare Life Sciences) following the previously published protocol for purification of the SUMO-fusion proteins.43 MjaTFIIBΔ155 mini-intein with C1A mutation was collected from flow-through fractions from the second IMAC after Ulp1 protease digestion and dialyzed against 2 liters of MilliQ water overnight at 4 °C. The protein was concentrated to 447 μM using an ultracentrifugation device, and flash-frozen in liquid nitrogen for storage at −74 °C.
Cloning, expression, and purification of MvuTFIIBΔ145 mini-intein for structure determination
The gene of MvuTFIIBΔ145 mini-intein with C1A mutation was amplified by PCR from the pBHDuet50F plasmid using the two oligonucleotides of I583: 5’- ATGGATCCGGTGGTTACGCTGTTGATTATAGCGAACC and HK804: 5’- TCGGTACCTTAGGCGTTGTGTAATACAAATCCTC. The amplified gene was inserted into pHYRSF53 using BamHI and KpnI sites to make the SUMO-fusion protein, resulting in pBHRSF63.42
The SUMO-fusion bearing MvuTFIIBΔ145 mini-intein with C1A mutation was expressed and purified following the protocol above.43 The protein was further purified by gel filtration chromatography. The protein solution was concentrated using an ultracentrifugation device to a volume of 2 ml and loaded onto Superdex75 size exclusion chromatography column (GE Healthcare Life Sciences) with Tris-buffered saline (TBS) buffer (pH 7.4). The mono-disperse peak fractions containing MvuTFIIBΔ155b were dialyzed against two liters of MilliQ water overnight at 4 °C. The protein was concentrated to 802 μM using an ultracentrifugation device, and flash-frozen in liquid nitrogen for storage at −74 °C.
Crystallization of MjaTFIIBΔ155 and MvuTFIIBΔ145 mini-inteins
447 μM solution of MjaTFIIBΔ155 and 802 μM solution of MvuTFIIBΔ145 were used for crystallization trials. Drops of 200 nl (100 nl of protein solution and 100 nl of screen solution) were set up in 96-well MRC (Molecular Dimensions) crystallization plates using a Mosquito LCP® (TTP Labtech, UK). Helsinki Random I and II (HRI and HRII) screens (http://www.biocenter.helsinki.fi/bi/xray/automation/services.html), which are the local modifications of the classic sparse matrix screens yielded initial hits.44 Optimization grid screens were designed based on the initial hits and crystal growth was improved. The final growth conditions for the diffracting crystals were 0.1 M MES buffer (pH 6.5), 10% dioxane, and 1.6 M ammonium sulfate for MjaTFIIBΔ155, and 0.2 M calcium chloride and 20 % PEG 3350 for MvuTFIIBΔ145. 25% glycerol was added for MjaTFIIBΔ155 on top of the drop, which served as a cryoprotectant when flash-freezing crystals in liquid nitrogen. For MvuTFIIBΔ145 sufficient cryoprotection was obtained with 20% PEG 3350 present in crystallization drop.
Diffraction data collection and processing
Diffraction data for the crystal of MjaTFIIBΔ155 mini-intein were collected in a single pass on beamline I04 at the Diamond Light Source, Oxfordshire, and were subsequently indexed, integrated, and scaled to 2.0 Å resolution using the program XDS.45, 46 Crystal parameters and data processing statistics are listed in Table 1. Diffraction data for the crystal of MvuTFIIBΔ145 mini-intein were collected in a single pass on beamline ID30A-3 at the European Synchrotron Research Facility (ESRF), Grenoble and were subsequently indexed, integrated, and scaled to 2.5 Å resolution.47
Structure determination and refinement
The structures of MjaTFIIBΔ155 and MvuTFIIBΔ145 were solved by molecular replacement. The search model used for MjaTFIIBΔ155 was based on the coordinates the intein part of the homing endonuclease II (PDB ID: 2cw8). Since the intein is present in this structure as two separate segments joined by the extein, a model of the single-chain target protein was constructed with the program Sculptor.48 The sequence of this model was mutated to that of MjaTFIIBΔ155, and the resulting coordinates were subjected to restrained molecular dynamics with Rosetta. Since the sequence identity between MjaTFIIB and the homing endonuclease II is only 31%, molecular replacement runs that used either this starting model, or unmodified and modified structures of several inteins, were initially unsuccessful. A correct solution was only obtained with the help of the program MR_Rosetta coupled to the Phenix package.49,50 The model was adjusted with Coot followed by rounds of refinement with Phenix.50,51 The quality of the final structure was validated by the MolProbity webserver (Table 1).52
The structure of molecule A of MjaTFIIB intein, with the sequence adjusted with Sculptor to that of MvuTFIIB, was used as a starting model for molecular replacement. The structure was solved with Phenix and improved with MR_Rosetta, yielding a solution consisting of two molecules in the asymmetric unit, with the R of 0.286 and Rfree of 0.377, with several loops still missing.49,50 Further refinement was performed with Refmac5 from CCP4 package, and the model was rebuilt with Coot and validated with MolProbity (Table 1).51,52,53,54
Homology modeling of the full-length MjaTFIIB intein
The three-dimensional model of the full-length MjaTFIIB intein was built by SwissModel online server (https://swissmodel.expasy.org/) using PI-PfuI (PDB ID: 1dq3) as a template model.55
Accession numbers
Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 5o9j for the MjaTFIIB intein, 5o9i for the MvuTFIIB intein.
Supplementary Material
Acknowledgements
We thank B. Haas and S. Jääskeläinen for technical help in preparation of proteins and plasmids. We thank S. Mäki and Dr. K. Kogan for technical help at the crystallization facility. This work was supported in part by the Academy of Finland (137995, 1277335) and Biocenter Finland for the crystallization and NMR facilities at the Institute of Biotechnology, by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and with Federal funds from the National Cancer Institute, NIH, under Contract No. HHSN261200800001E (to M.L.). The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U. S. Government.
Abbreviations:
- IPTG
isopropyl-β-D-1-thiogalactopyranoside
- Mja
Methanococcus jannaschii
- Mvu
Methanocaldococcus vulcanius M7
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- RMSD
root mean square deviation
- PTS
protein trans-splicing
- Ulp1
Ubiquitin-like-specific protease 1
- DTT
dithiothreitol
- TBS
Tris-buffered saline
- HEN
homing endonuclease
- HINT
Hedgehog/INTein
- HGT
horizontal gene transfer
- DOD
dodecapeptide
- PDB
Protein Data Bank
- GB1
B1 domain of IgG binding protein G
- IMAC
immobilized metal ion affinity chromatography
- iPAS
intein-mediated protein alternative splicing
- ESRF
European Synchrotron Radiation Facility
References
- 1.Keren H, Lev-Maor G & Ast G (2010). Alternative splicing and evolution: diversification, exon definition and function. Nat. Rev. Genet 11, 345–355. [DOI] [PubMed] [Google Scholar]
- 2.Black DL (2000). Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 103, 367–370. [DOI] [PubMed] [Google Scholar]
- 3.Aranko AS, Oeemig JS, Kajander T, & Iwaï H (2013). Intermolecular domain swapping induces intein-mediated protein alternative splicing. Nat. Chem. Biol 9, 616–622. [DOI] [PubMed] [Google Scholar]
- 4.Gogarten JP, Senejani AG, Zhaxybayeva O, Olendzenski L & Hilario E (2002). Inteins: Structure, function, and evolution. Annual Review of Microbiology. 56, 263–287. [DOI] [PubMed] [Google Scholar]
- 5.Paulus H (2000). Protein splicing and related forms of protein autoprocessing. Annu. Rev. Biochem 69, 447–496. [DOI] [PubMed] [Google Scholar]
- 6.Kawasaki M, Satow Y, Ohya Y & Anraku Y (1997). Protein splicing in the yeast Vma1 protozyme: evidence for an intramolecular reaction. FEBS Lett. 412, 518–520. [DOI] [PubMed] [Google Scholar]
- 7.Wu H, Hu Z & Liu XQ (1998). Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. U. S. A 95, 9226–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Novikova O, Jayachandran P, Kelley DS, Morton Z, Merwin S, Topilina NI et al. (2016). Intein clustering suggests functional importance in different domains of life. Mol. Biol. Evol 33, 783–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swithers KS, Soucy SM & Gogarten JP (2012). The role of reticulate evolution in creating innovation and complexity. Int. J. Evol. Biol 2012, ID 418964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callahan BP, Topilina NI, Stanger MJ, Van Roey P & Belfort M (2011). Structure of catalytically competent intein caught in a redox trap with functional and evolutionary implications. Nat. Struct. Mol. Biol 18, 630–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Topilina NI, Novikova O, Stanger M, Banavali NK & Belfort M (2015). Post-translational environmental switch of RadA activity by extein-intein interactions in protein splicing. Nucleic Acids Res. 43, 6631–6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Topilina NI, Green GM, Jayachandran P, Kelley DS, Stanger M, Piazza CL, Nayak S & Belfort M (2015). SufB intein of Mycobacterium tuberculosis as a sensor for oxidative and nitrosative stresses. Proc. Natl. Acad. Sci. U. S. A 112, 10348–10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall TM, Porter JA, Young KE, Koonin EV, Beachy PA & Leahy DJ (1997). Crystal structure of a Hedgehog autoprocessing domain: homology between Hedgehog and self-splicing proteins. Cell. 91, 85–97. [DOI] [PubMed] [Google Scholar]
- 14.Nogami S, Satow Y, Ohya Y & Anraku Y (1997). Probing novel elements for protein splicing in the yeast Vma1 protozyme: a study of replacement mutagenesis and intragenic suppression. Genetics. 147, 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gimble FS & Thorner J (1992). Homing of a DNA endonuclease gene by meiotic gene conversion in Saccharomyces cerevisiae. Nature. 357, 301–306. [DOI] [PubMed] [Google Scholar]
- 16.Derbyshire V, Wood DW, Wu W, Dansereau JT, Dalgaard JZ & Belfort M (1997). Genetic definition of a protein-splicing domain: functional mini-inteins support structure predictions and a model for intein evolution. Proc. Natl. Acad. Sci. U. S. A 94, 11466–11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klabunde T, Sharma S, Telenti A, Jacobs WR & Sacchettini JC (1998). Crystal structure of GyrA intein from Mycobacterium xenopi reveals structural basis of protein splicing. Nat. Struct. Biol 5, 31–36. [DOI] [PubMed] [Google Scholar]
- 18.Aranko AS, Oeemig JS, Zhou D, Kajander T, Wlodawer A & Iwaï H (2014). Structure-based engineering and comparison of novel split inteins for protein ligation. Mol. Biosyst 10, 1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu H, Xu MQ & Liu XQ (1998). Protein trans-splicing and functional mini-inteins of a cyanobacterial dnaB intein. Biochim. Biophys. Acta 1387, 422–432. [DOI] [PubMed] [Google Scholar]
- 20.Barzel A, Naor A, Privman E, Kupiec M & Gophna U (2011). Homing endonucleases residing within inteins: evolutionary puzzles awaiting genetic solutions. Biochem. Soc. Trans 39, 169–173. [DOI] [PubMed] [Google Scholar]
- 21.Koufopanou V & Burt A (2005). Degeneration and domestication of a selfish gene in yeast: molecular evolution versus site-directed mutagenesis. Mol. Biol. Evol 22, 1535–1538. [DOI] [PubMed] [Google Scholar]
- 22.Ellilä S, Jurvansuu JM & Iwaï H (2011). Evaluation and comparison of protein splicing by exogenous inteins with foreign exteins in Escherichia coli. FEBS lett. 585, 3471–3477. [DOI] [PubMed] [Google Scholar]
- 23.Mizutani R, Nogami S, Kawasaki M, Ohya Y, Anraku Y & Satow Y (2002). Protein-splicing reaction via a thiazolidine intermediate: crystal structure of the VMA1-derived endonuclease bearing the N and C-terminal propeptides. J. Mol. Biol 316, 919–929. [DOI] [PubMed] [Google Scholar]
- 24.Ichiyanagi K, Ishino Y, Ariyoshi M, Komori K & Morikawa K (2000). Crystal structure of an archaeal intein-encoded homing endonuclease PI-PfuI. J. Mol. Biol 300, 889–901. [DOI] [PubMed] [Google Scholar]
- 25.Moure CM, Gimble FS & Quiocho FA (2002). Crystal structure of the intein homing endonuclease PI-SceI bound to its recognition sequence. Nat. Struct. Biol 9, 764–770. [DOI] [PubMed] [Google Scholar]
- 26.Matsumura H, Takahashi H, Inoue T, Yamamoto T, Hashimoto H, Nishioka M et al. (2006). Crystal structure of intein homing endonuclease II encoded in DNA polymerase gene from hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1. Proteins. 63, 711–715. [DOI] [PubMed] [Google Scholar]
- 27.Oeemig JS, Zhou D, Kajander T, Wlodawer A & Iwaï H (2012). NMR and crystal structures of the Pyrococcus horikoshii RadA intein guide a strategy for engineering a highly efficient and promiscuous intein. J Mol Biol. 421, 85–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okuda Y, Sasaki D, Nogami S, Kaneko Y, Ohya Y & Anraku Y (2003). Occurrence, horizontal transfer and degeneration of VDE intein family in Saccharomycete yeasts. Yeast. 20, 563–573. [DOI] [PubMed] [Google Scholar]
- 29.Swithers KS, Senejani AG, Fournier GP & Gogarten JP (2009). Conservation of intron and intein insertion sites: implications for life histories of parasitic genetic elements. BMC Evol. Biol 9, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naor A, Altman-Price N, Soucy SM, Green AG, Mitiagin Y, Turgeman-Grott I et al. , (2016). Impact of a homing intein on recombination frequency and organismal fitness. Proc. Natl. Acad. Sci. U. S. A 113, E4654–4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saves I, Morlot C, Thion L, Rolland JL, Dietrich J & Masson JM (2002). Investigating the endonuclease activity of four Pyrococcus abyssi inteins. Nucleic Acids Res. 30, 4158–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Posey KL, Koufopanou V, Burt A & Gimble FS (2004). Evolution of divergent DNA recognition specificities in VDE homing endonucleases from two yeast species. Nucleic Acids Res. 32, 3947–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cronin M, Coolbaugh MJ, Nellis D, Zhu J, Wood DW, Nussinov R, et al. (2015). Dynamics differentiate between active and inactive inteins. Eur. J. Med. Chem 91, 51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du Z, Liu Y, Ban D, Lopez MM, Belfort M & Wang C (2010). Backbone dynamics and global effects of an activating mutation in minimized Mtu RecA inteins. J. Mol. Biol 400, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wenzlau JM, Saldanha RJ, Butow RA & Perlman PS (1989). A latent intron-encoded maturase is also an endonuclease needed for intron mobility. Cell 56, 421–430. [DOI] [PubMed] [Google Scholar]
- 36.Gogarten JP & Hilario E (2006). Inteins, introns, and homing endonucleases: recent revelations about the life cycle of parasitic genetic elements. BMC Evol. Biol 6, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perler FB (1999). A natural example of protein trans-splicing. Trends Biochem. Sci 24, 209–211. [DOI] [PubMed] [Google Scholar]
- 38.Pietrokovski S (2001). Intein spread and extinction in evolution. Trends Genet. 17, 465–472. [DOI] [PubMed] [Google Scholar]
- 39.Aranko AS, Wlodawer A & Iwaï H (2014). Nature’s recipe for splitting inteins. Protein Eng. Des. Sel 27, 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lennon CW & Belfort M (2017). Inteins. Curr. Biol 27, R204–R206. [DOI] [PubMed] [Google Scholar]
- 41.Brünger AT (1992). The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 355, 472–475. [DOI] [PubMed] [Google Scholar]
- 42.Muona M, Aranko AS & Iwaï H (2008). Segmental isotopic labelling of a multidomain protein by protein ligation by protein trans-splicing. Chembiochem. 9, 2958–2961. [DOI] [PubMed] [Google Scholar]
- 43.Guerrero F, Ciragan A & Iwaï H (2015). Tandem SUMO fusion vectors for improving soluble protein expression and purification. Protein Expr. Purif 116, 42–49. [DOI] [PubMed] [Google Scholar]
- 44.Cudney R, Patel S, Weisgraber K, Newhouse Y, & McPherson A (1994). Screening and optimization strategies for macromolecular crystal growth. Acta Crystallogr. D Biol. Crystallogr 50, 414–423. [DOI] [PubMed] [Google Scholar]
- 45.Theveneau P, Baker R, Barrett R, Beteva A, Bowler MW, Carpentier P, et al. (2013). The upgrade programme for the structural biology beamlines at the european synchrotron radiation facility – high throughput sample evaluation and automation. J. Phys. Conf. Ser 425, 012001 doi: 10.1088/1742-6596/425/1/012001. [DOI] [Google Scholar]
- 46.Kabsch W (2010). XDS. Acta Crystallogr. D 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allan DR, Collins SP, Evans G, Hall D, McAuley K, Owen RL, et al. (2015). Status of the crystallography beamlines at Diamond Light Source. Eur. Phys. J. Plus 130, 56. [Google Scholar]
- 48.Bunkóczi G & Read RJ (2011). Improvement of molecular-replacement models with Sculptor. Acta Crystallogr. D Biol. Crystallogr 67, 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DiMaio F, Terwilliger TC, Read RJ, Wlodawer A, Oberdorfer G, Wagner U, et al. (2011). Increasing the radius of convergence of molecular replacement by density and energy guided protein structure optimization. Nature. 473, 540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, et al. (2002). PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954. [DOI] [PubMed] [Google Scholar]
- 51.Emsley P, Lohkamp B, Scott WG & Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, et al. (2011). REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arnold K, Bordoli L, Kopp J & Schwede T (2006). The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 22, 195–201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.