Abstract
The mitochondrial genome (mtDNA) represents a tiny fraction of the whole genome, comprising just 16.6 kilobases encoding 37 genes involved in oxidative phosphorylation and the mitochondrial translation machinery. Despite its small size, much interest has developed in recent years regarding the role of mtDNA as a determinant of both aging and age-associated diseases. A number of studies have presented compelling evidence for key roles of mtDNA in age-related pathology, although many are correlative rather than demonstrating cause. In this review we will evaluate the evidence supporting and opposing a role for mtDNA in age-associated functional declines and diseases. We provide an overview of mtDNA biology, damage and repair as well as the influence of mitochondrial haplogroups, epigenetics and maternal inheritance in aging and longevity.
Keywords: Mitochondria, mtDNA, Lifespan, Aging
1. Introduction
Complex multicellular life owes its origin to the endosymbiosis of two primordial prokaryotes [1, 2]. The endosymbiont became the mitochondrion, providing the eukaryotic cell with abundant ATP through oxidative phosphorylation. While most mitochondrial proteins are now encoded in the nuclear genome, mitochondria have retained a portion of their own genome, the mitochondrial DNA (mtDNA). This is not only a legacy of their independent origin but seems to be essential for coupling oxidative phosphorylation to ATP synthesis [3]. In most metazoans, the mitochondrial genome has been compacted to a double-stranded circular molecule 11-28 kilobases with few intergenomic regions, no introns and overlap between some of the coding regions [4, 5]. Mammalian mitochondrial genomes are quite typical and consist of 16.6 kilobases which encode thirteen respiratory complex subunits as well as two ribosomal and twenty-two transfer RNA [6]. The thirteen protein-coding genes retained in the mtDNA encode subunits of complexes I, III, IV and V, which are the four complexes which pump protons and are partially imbedded in the inner mitochondrial membrane (Fig. 1). High GC content of the genes, hydrophobicity of the proteins and the ability to modulate mitochondria individually seem to be the driving forces behind retention of these genes in the mtDNA [7].
Figure 1.
The human mitochondrial genome consists of 16,569 bp encoding 13 respiratory complex subunits, 2 ribosomal RNAs and 22 transfer RNAs. The encoded proteins are all localized in the inner mitochondrial membrane and are some of the core subunits of respiratory complexes I, III, IV and V.
The past century has seen increasing understanding of how mitochondria not only enable our existence but also how disruption of their function may play an important role in age-related decline. Mitochondrial dysfunction has long been recognized as one of the hallmarks of aging [8] and theorized to play a causative role in aging pathology [9, 10]. It is well established that mitochondrial function deteriorates with age in different tissues such as heart [11], muscle [12] and liver [13]. However, the proximal cause of the decline in mitochondrial function remains elusive.
There are several roles which mtDNA may play in the pathology of aging as well as several age-related diseases which will be discussed in this review. These roles include changes in mtDNA copy number, epigenetic modifications and mtDNA mutations which have been linked with the age-associated functional decline of both the mitochondria and organism as a whole. In addition, alterations in the mtDNA genome have been linked with altered susceptibility to a wide range of age-associated diseases including Alzheimer’s disease, Parkinson’s disease, sarcopenia, heart failure and cancer. Variations in the mtDNA genome may also contribute to differences in lifespan between individuals as well as between different animal species. However, while there is a large body of correlative data linking mtDNA to aging and age-associated disease, demonstration of a direct role is lacking. This review will provide a breakdown of the relative strengths and weakness of the data supporting and opposing a role of mtDNA in the aging process.
2. Mitochondrial DNA structure and organization
The mammalian mitochondrial genome is a double-stranded supercoiled circular molecule of about 16.6 kb. It is composed of a heavy and light strand (based on the proportion of higher molecular weight nucleotides). The heavy strand contains the 2 rRNA sequences, 14 of the 22 mitochondrial tRNAs and all the protein-encoding genes with the exception of ND6 (Fig. 1). There is also a large non-coding region of about 1 kb that contains regulatory elements for the initiation and termination of transcription of both strands. The D-Loop (displacement loop) or control region is within this non-coding region and contains the OriH (heavy strand origin of replication), where replication is initiated. There is a second origin of replication in the light strand, the OriL [14] (Fig. 1).
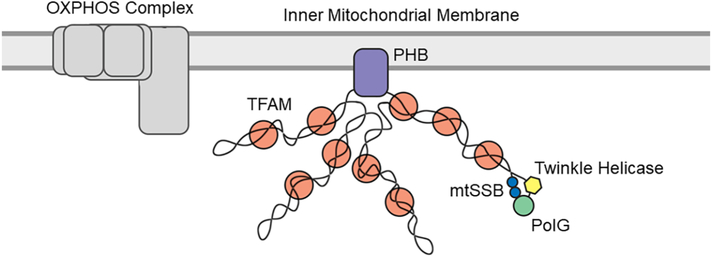
The mtDNA genome is packaged into nucleoids, which are dynamic structures that are normally formed by the mtDNA genome and the proteins involved in its replication and transcription. Although this structure provides a level of organization, it is not as tightly packed or organized as nuclear DNA (nDNA). The nucleoid proteins include polymerase-γ (PolG) (the mtDNA replicative polymerase), mtSSB (single stranded binding protein), twinkle (DNA helicase), Phb (prohibitin) and TFAM (transcription factor regulates mtDNA) [15]. The structure and organization of the mtDNA nucleoid is an emerging area of research with new proteins frequently being identified. The nucleoids are located on the matrix (interior) side of the inner mitochondria membrane in close proximity to the oxidative phosphorylation system (Fig. 2).
Figure 2.
The mitochondrial genome is located on the matrix side of the inner mitochondrial membrane and is packaged with numerous proteins which are still being discovered. These proteins are involved both in its replication, transcription and stability.
3. Mitochondrial DNA age-associated changes
Until recently, the study of mtDNA damage had been challenging due to the difficulty of detecting variations present in low abundance in any particular cell or tissue. In contrast to nuclear DNA, which is present in only two copies per cell, mtDNA can be present in hundreds to thousands of copies, and these do not always contain the same sequence. Mutated mtDNA can coexist with wild-type mtDNA in a condition called heteroplasmy. The development of next generation sequencing or ultra-deep sequencing has helped to characterize both mtDNA heteroplasmicity [16, 17] and the different types of damage and mutations that can be present in disease and aging. Some recent studies have suggested that heteroplasmy may serve a protective role since the ratio of mutant to wild-type DNA is key to the development of disease. This ratio is the determinant of the so called “Threshold Effect”; pathogenic mutations need to reach at least 70 to 90% of heteroplasmy to have a detrimental effect on mitochondrial function [18]. This threshold effect has been mimicked in a Drosophila model by manipulating a mitochondrial ribosomal gene, achieving different pathological phenotypic manifestations depending on the mutation dosage [19]. Therefore it is suggested that for mtDNA damage to produce detrimental effects, the mutation levels must reach high frequency within each cell, a process that would take many years to occur [20].
To examine this process in human aging, Christiansen and colleagues [21] along with Lee and colleagues [22] found declines in mtDNA copy number in whole blood to occur with age and lower mtDNA copy number to be associated with poorer health outcomes. This work was followed up Gu and colleagues who used available whole-genome sequencing data from peripheral blood mononuclear cells from more than 1500 women from two British cohorts. They found that aged individuals accumulated higher levels of mutant mtDNA than younger women and that these were associated with the appearance of age-related blood physiological markers [23]. A very recent study has found a positive correlation in elderly subjects between the abundance of the pathological A3243G mtDNA mutation and reduced strength, cognition, metabolism and cardiovascular fitness [24]. They also reported that the patients with the highest mutation burden presented a higher risk of dementia and stroke mortality [24].
The D-Loop is the region of mtDNA most prone to genome instability [25]. This is likely because it is the most exposed region as the primary replication initiation site. Studies have shown this region accrues high levels of DNA point mutations with age in a range of tissues including the brain, blood, skin, muscle and heart [26-29]. One of the most common mutations is a T414G transversion which has been found in up to 50% of mtDNA molecules of half of the individuals tested above 65 years of age [27]. A high frequency of mtDNA deletions in the D-Loop has also been observed in the central nervous system of 27-month-old rats and mice when compared to 7-month-old animals [30, 31].
Large mtDNA deletions have also been detected in human mtDNA during aging. A 5 kb deletion of mtDNA, which is usually associated with mitochondrial disease, has been reported to accumulate in the heart, brain and muscle of aging humans [32, 33]. There is evidence that mtDNA defects can clonally expand and create mosaic phenotypes in some tissues. Ragged red fibers (RRF) are muscles fibers with mitochondria deficient for cytochrome c oxidase (respiratory complex IV) activity, that can result from mutations or deletions in either mtDNA or nDNA. These fibers accumulate with age and are thought to be a contributing factor for sarcopenia (discussed in section 8.3) [33]. Further supporting the role of clonal expansion, Khrapko and colleagues found clonal mutant mtDNA that had expanded from an initial single mtDNA molecule in human buccal epithelium and cardiac muscle [29]. Similarly, Turnbull and colleagues reported a clonal expansion of mtDNA mutations in the colorectal epithelium with age, while the frequency of de novo mtDNA mutations did not increase, suggesting that clonal expansion is the driving force behind mitochondrial dysfunction with age [34]. Similar clonal expansion events have been observed in skin fibroblasts and blood of elderly subjects, producing an elevated load of heteroplasmic or even homoplasmic mutations [35]. In some cases, it is thought that the origin of clonal expansion could arise from stem-cell aging. It was shown within normal gastric epithelium that mtDNA mutations are present in its stem-cells which are then passed on to the differentiated progeny [36].
4. Mitochondria DNA damage and repair
The mechanism of mtDNA replication is controversial and new theories have emerged in recent years. The ‘Strand-Displacement model’ originally proposed in 1982 argues that replication displaces the lagging stand, leaving it unprotected. This could result in low stability and highly error prone replication that could increase the risk of single-strand breaks and misreads [37]. A newer theory proposes that mtDNA replication occurs through a bootlace model which involves the formation of RNA-DNA duplexes, with RNA hybridizing to the displaced lagging DNA strand and thereby preserving genome stability [38].
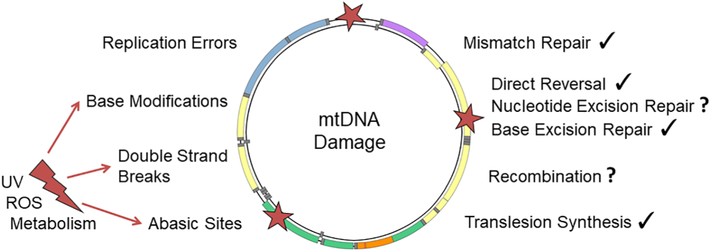
Mutations in mtDNA can originate from different causes; they can be maternally inherited or result from defects in the maintenance and repair systems. Mutations can also be generated by genotoxic agents such as ROS or UV irradiation. The mtDNA repair system plays an essential role in the repair of such mutations [39] (Fig. 3). More and more similarities are being found between the mtDNA and nDNA repair systems. Proofreading activity by the PolG 3’–5’ exonuclease is one of the most important systems in the maintenance of mtDNA integrity and correction of replication errors. Until recently, PolG was thought to be the sole DNA polymerase present in mitochondria, but the protein PrimPol has been identified as possessing roles in mtDNA translesion synthesis [40]. This enzyme is not only able to elongate DNA and to synthesize new DNA primers for PolG; it is also capable of skipping and copying ahead of DNA lesions [41]. PrimPol therefore provides a new translesion synthesis mechanism that was further confirmed by Pohjoismaki and colleagues [42]. While PrimPol-deficient mice showed a metabolic disease phenotype that appeared by middle-age and was associated with mitochondrial dysfunction (Chocron, Blanco, unpublished findings), the impact of this gene on aging has not yet been evaluated.
Figure 3.
Different types of damage-causing agents (ROS/UV/Metabolism) originating either from the external environment or from within the cell generate different types of damage that can each be repaired by a corresponding repair mechanism. Some of these mechanisms are unconfirmed in mitochondria and are thus depicted with a question mark (?).
Base excision repair (BER) is the best characterized DNA repair process in mitochondria and is the main process involved in the removal of oxidized bases such as 8-OHdG (8-hydroxydeoxyguanine). Very recently, Polymerase B (Pol Beta) has been found to be localized in mitochondria involved in BER mechanism [43, 44]. Mismatch repair, single strand breaks and double strand breaks repair have each been reported in mitochondria DNA [14] and Polymerase Q (Theta) has been suggested to have recently a role in these processes [45, 46]. Some studies have shown age-related declines in BER efficiency in mouse skeletal muscle but at the same time, increases in efficiency were seen in liver and heart [47]. In rat brains, BER activity was also reported to decrease with age with a concomitant decrease in expression of both PolG and 8-OHdG glycosylase, which is one of the BER enzymes [48]. However, it is unclear whether these changes contribute to age-related changes in functionality of these tissues. In addition, such observations have been limited to rodents and more work is needed to assess whether these changes are relevant to human aging.
In addition to proof-reading errors, deficits in mitophagy have been posited as a strong contributor to the accumulation of damaged mtDNA with age. Mitophagy is the process by which damaged mitochondria are engulfed by double-membrane vesicles called autophagosomes and destroyed [49]. In the best known mitophagic pathway, dysfunctional mitochondria cannot maintain membrane potential, resulting in the stabilization of PINK1 (PTEN-induced putative kinase 1), which then recruits Parkin to initiate mitophagy dependent on ATG (autophagy-related) proteins [49]. Mitophagy plays an important role in control and removal of mtDNA damage and in dealing with non-functional mitochondria which may contain excessive mtDNA damage [50]. It has been observed that mitophagy is disrupted in pathologies such as Alzheimer’s disease and other neurodegenerative disorders [51, 52] and is therefore being investigated as a potential therapeutic target [53]. In a fly Huntington disease model, PINK-1 overexpression has been reported to be neuroprotective [53]. Additionally, patients with early onset parkinsonism who carry a mutated PINK-1 protein harbor an increased level of mtDNA mutations [54]. It is worth noting that lifespan extending interventions such as caloric restriction and mTOR inhibition both lead to increased autophagy and mitophagy [55]. This supports the argument that the accumulation of mtDNA mutations with aging is due primarily to failure in the mitophagic processes to clear defective mtDNA rather than changes in rates of damage accrual [56].
5. The Mitochondrial Free Radical Theory of Aging (MFRTA)
Mitochondria are the major source of ROS production within the cell [57]. This led Denham Harman to propose the Mitochondrial Free Radical Theory of Aging (MFRTA) [9, 58], which posits that ROS produced by oxidative phosphorylation damages mitochondrial proteins, membranes and DNA [9]. This was proposed to cascade into a vicious cycle where mutations result in further mitochondrial dysfunction, that in turn lead to more ROS production, and eventually result in cell senescence or death [59].
Models of gain and lack of function of mitochondrial ROS scavenging enzymes have been generated in an attempt to test this theory. The results from these models have been contradictory, especially when evaluating their effects on lifespan (Table 1). Superoxide dismutases (SODs), catalase, thioredoxins (TRX), peroxiredoxins (PRX) and glutathione peroxidases (GPx) are the major antioxidant defense mechanisms within different cellular compartments. The effects of homozygous and heterozygous knockout or transgenic overexpression of various antioxidant enzymes in mice has been reviewed by Perez and colleagues [60]. Mitochondrial superoxide dismutase (SOD2), also known as manganese-dependent superoxide dismutase (MnSOD), reduces the highly reactive superoxide radical to hydrogen peroxide. SOD2 has been described to associate directly with respiratory complexes [61] to neutralize superoxide radicals where they are generated and is essential to the viability of many organisms, including yeast [62, 63], flies [64] and mice [65, 66]. In conflict with the critical role of SOD2 in removal of ROS from the mitochondria, many reports have shown limited effects of this protein on lifespan. In the nematode Caenorhabditis elegans deletion of any or even all of the SOD genes caused no reduction in lifespan under normal conditions (survival under oxidative stress was decreased) [67-69] and null mutations in SOD2 actually extended lifespan in these animals [68]. Mice heterozygous for a null SOD2 mutation did show increased levels of oxidative damage to both mtDNA and nDNA and had a much higher incidence of cancer. Significantly, however, both the mean and maximum lifespan of these mice was indistinguishable from wild-type and they showed no evidence of accelerated aging [70]. Conversely, overexpression of SOD2 either by itself or in combination with other antioxidant enzymes repeatedly showed no effect on lifespan in mice [60, 71]. Reports on the effect of SOD2 overexpression in Drosophila melanogaster have been conflicting [72-74]. Manipulations of other mitochondrial antioxidants have produced similar results and argue against a causative role of mitochondrial ROS in aging [60, 75, 76].
Table 1.
The effects on lifespan of knockdown (null), knockout (KO) and transgenic overexpression (OE) of mitochondrial oxidant scavenging and redox enzymes in C. elegans, D. melanogaster and M. musculus are summarized. In the two cases where a transgenic model was created but lifespan was not assayed, these studies are referenced.
| Effects of Mitochondrial Redox Enzyme Manipulation on Lifespan | |||
|---|---|---|---|
| C. elegans | D. melanogaster | M. musculus | |
| SOD2 null | Life extension [68] | Early lethality [64] | Lethal [65, 66] |
| SOD2 KD | No effect [210] | Life reduction [211] | No effect [70] |
| SOD2 OE | Life extension [212] | Life extension [72] | No effect [71] |
| GPx4 null | - | - | Lethal [213] |
| GPx4 KD | - | - | Life extension [76] |
| GPx4 OE | - | - | - |
| PRX3 null | - | - | Not reported [214] |
| PRX3 KD | - | - | - |
| PRX3 OE | - | - | - |
| TRX2 null | No effect [215] | - | Lethal [216] |
| TRX2 KD | - | - | Life reduction [60] |
| TRX2 OE | - | - | - |
| TXNRD2 null | No effect [215] | Lethal [217] | - |
| TXNRD2 KD | - | - | - |
| TXNRD2 OE | Not reported [215] | Life extension [84] | - |
An interesting exception is a study wherein catalase was ectopically expressed in the mouse mitochondria (catalase is normally found only in peroxisomes). These mice, known as mCAT mice, have been reported to live longer [77], maintain mitochondrial function into old age [78] and show a reduction in multiple age-related pathologies [79-81]. Recent studies have demonstrated that the benefits of mCAT overexpression in lifespan were related to both improved mitochondrial function and consequent attenuation of age-associated insulin resistance in muscle [78]. The ability of mitochondrial-targeted catalase to prevent age-related functional declines is curious, especially when manipulation of the mitochondria’s native H2O2 scavenger, GPx4, has had little to no effect on lifespan [60]. In contrast, mitochondrial-targeted catalase has shown no effect on lifespan in flies [73, 82]. The contribution of the mitochondrial thioredoxin system in this matter has also recently been evaluated. This system is involved in reducing the mitochondrial peroxirodoxin as well as both reducing oxidized proteins and signaling to apoptosis [83]. The rate limiting enzyme for this system is mitochondrial thioredoxin reductase (TXNRD2), which has been recently shown to be more abundant in longed-lived primate and rodent species [84]. While its role in mammalian aging is still under investigation, overexpression of TXNRD2 in flies increased median lifespan, suggesting a potential causative role in longevity [84].
While many studies focus on lifespan and pathology as endpoints, several have looked at the mtDNA for evidence as to whether or not ROS plays a major role in generating mutations. Oxidative damage can modify guanine nucleotides to 8-OHdG, which can then mis-pair with adenine and in one round of replication, lead to a TA transversion. Cytosine can also be oxidized and it is thought that this can lead to GC to AT transitions. Therefore GC to TA transversions and GC to AT transitions should be the most prevalent mutations resulting from oxidative damage [85]. Vermulst and colleagues studied the mutagenic contribution of ROS by comparing hearts derived from 26-month-old mice expressing ectopic mitochondria-targeted catalase (mCAT) and non-transgenic littermates. They found a 4-fold reduction in the mutation burden of mice expressing mCAT. In addition, they observed that GC to AT transitions were responsible for 81% of the mutation events in the aged control mice [86]. In contrast, oxidative stress is not thought to contribute greatly to age-related mtDNA mutations in Drosophila since only a small proportion of mutations were due to GC to TA transversions [87]. Other studies in aged human brains did not find significant changes on the mutations related to oxidative damage [88].
6. Mitochondrial DNA and progeria models
The majority of data linking mtDNA to aging is correlative. The closest evidence to a direct link between aging and mtDNA damage may be from progeria models. Here mutations which accelerate accumulation of mtDNA damage have been reported to produce aging-like phenotypes.
Goran-Larsson and colleagues designed the well-known mtDNA “mutator” mouse which contains a defect in the proof reading mtDNA Polymerase gamma exonuclease (PolGmut/mut) [89]. Prolla and colleagues also designed a mouse with a mutation in the same gene which produced deficits in proof reading capacity [90]. These mice possess a number of phenotypes which mimic aging, including rapid mortality; PolGmut/mut mice have a median lifespan of approximately 13 months. They also exhibit hair graying, hair loss, weight loss, reduced subcutaneous fat and kyphosis (curvature of the spine), all by approximately 9 months of age [89, 90]. However, other researchers have noted that PolGmut/mut mice harbor somatic point mutations an order of magnitude higher than what has been observed in aged human tissues [91]. In addition, the age-like phenotypes displayed by the PolGmut/mut mice include many disorders and traits which are not seen during ‘normal aging’ in mice, which calls into question whether the PolGmut/mut mouse does indeed represent a model of accelerated aging [92].
Other findings which argue against mtDNA damage causing accelerated aging include a study reporting that mice which are heterozygous for the PolG proofreading mutation continue to have over one order of magnitude higher levels of mitochondria DNA mutations than wild-type mice and yet exhibit a phenotype and lifespan little different than wild-type [86]. Moreover, aging wild-type mice also present a higher level of mutations than aged humans. This suggests that the multi-organ failure of the homozygous mutator mice at the early age of 5 months may be caused not by premature aging but by energetic deficiency. Evidence for energetic deficiency include heart hypertrophy and mosaic deficiencies for activity of complexes II and IV in skeletal muscle [89].
The PolG mutator mouse also presents evidence against the vicious cycle proposed by MFRTA. While they have an increased load of mtDNA mutations and deletions, they do not show higher levels of oxidative damage in proteins, lipids or DNA [93]. Heart-specific expression of the PolG mutation produced similar results that did not increase with age [94]. In addition, very recent data from the same group has reported that BER deficiency alone or combined with oxidative stress did not increase mtDNA mutation load in mice [95].
In contrast to the findings in mice there is limited human study evidence that mutation of mtDNA proofreading can accelerate onset of aging or age-related disease. There are several reported human progeroid mutations associated with nDNA proofreading and structural maintenance however there are no known human progeroid syndromes which impact mtDNA. Some studies have reported alterations in mitochondrial morphology and function under Hutchinson-Gilford progeria but no changes in mtDNA have been reported in these patients [96]. Mutation of PolG in humans is rare, there is one report of a dominant familial mutation in this gene amongst humans. This mutation produces mitochondrial dysfunction and progressive muscle weakness in adulthood but is not reported to produce progeria [97].
7. Mitochondrial DNA sequence and longevity: haplogroups and comparative biology
In all animals, the mitochondrial genome is significantly smaller than the nuclear genome and consequently has been sequenced in many more species. The human mitochondrial genome was sequenced [4] more than two decades before the nuclear genome and many studies have been done looking for associations between sequence polymorphisms and both disease and lifespan.
7.1. Mitochondrial haplogroups
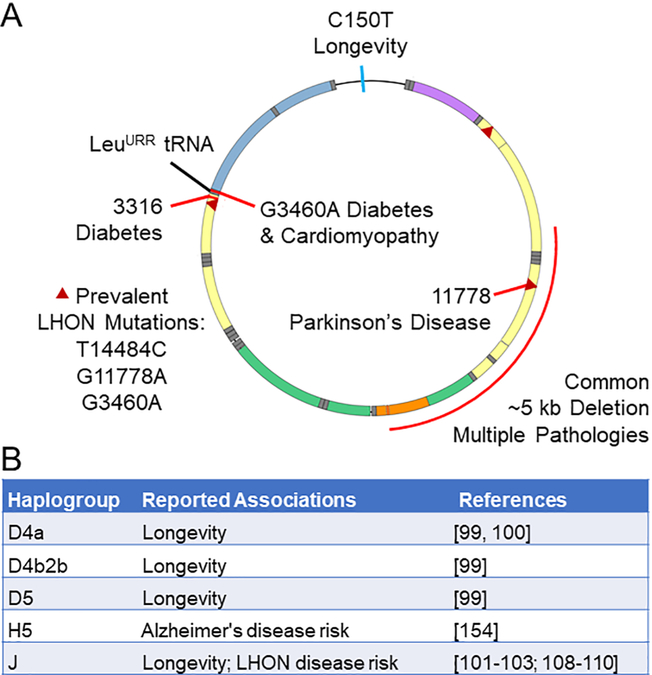
A haplotype is a group of alleles that are inherited from a single parent, such as the patrilineal inheritance of the Y-chromosome and the matrilineal inheritance of mtDNA. Neither chromosome is able to recombine and is thus passed along relatively unchanged for generations, with variations only introduced through mutations. Such mutations enable ancestry to be traced and haplogroups are defined as similar haplotypes which have diverged from a common ancestor. The identification of human mitochondrial haplogroups has revealed geographic associations and patterns of human migration and ancestry [98, 99]. Several mtDNA haplogroups have been reported to be associated with longevity in different populations (Fig. 4). The D4 and D5 haplogroups have been associated with longevity in the Japanese population [100] and the D4a sub-haplogroup in particular is enriched among Japanese aged over 105 years [101].
Figure 4.
Numerous associations have been found between specific mitochondrial mutations or haplogroups and longevity or disease. (A) In the control region, the C to T mutation at position 150 has been reported to correlate with longer lifespan. Mutations at positions 3243, 3316 and 11778 are associated with diabetes and cardiomyopathy, diabetes and Parkinson’s disease, respectively. Three common mutations that can cause Leber’s hereditary optic neuropathy are indicated (triangles). A common deletion of nearly 5 kb can contribute to multiple pathologies.(B) Several mitochondrial haplogroups have been found to correlate with either longer lifespan or disease, or in the case of haplogroup J, with both.
The J haplogroup is one of several present in populations throughout Europe and the Middle East, yet it associates with longevity only in specific sub-populations. It is over-represented among older northern Italian [102], Irish [103] and Finnish [104] cohorts but shows no correlation with longevity among Ashkenazi Jewish [105], Spanish [106] or southern Italian [107] cohorts. If the J haplogroup does contribute to longevity in a context-specific manner, it may be from allowing greater penetrance or manifestation of other mutations which themselves are beneficial in aging [108]. This theory arises from the converse observation: The J haplogroup has been reported to allow greater penetrance of pathological mutations and is associated with Leber’s hereditary optic neuropathy (LHON) [109-111] as well as optic neuritis in cases of multiple sclerosis [112]. Paradoxically, there is evidence to suggest that the same variations which allow for greater penetrance of LHON mutations may themselves be the variations which can lead to prolonged lifespan. Several of the variations associated with the J haplogroup would be expected to decrease functionality of complex I or III [113] and indeed, mitochondria with the J haplotype have significantly lower maximal oxygen consumption than those with the H haplotype [114].
Intriguingly, the H haplotype seems to be protective against LHON [111]. All this suggests that the J haplogroup produces a reduction in mitochondrial function that, by itself, promotes life extension. It is not a robust phenotype, however, and concurrent mutations affecting mitochondrial function lead to more severe pathology in this background. Another study supporting this hypothesis examined variations in human mitochondrial genes and found that mutations occurring in genes encoded complex I subunits can promote longevity, but not when they co-occur with impairment of complex III or V [115].
7.2. Single nucleotide polymorphisms in the mitochondrial genome
A mutation from C to T at position 150 of the mtDNA control region (C150T) (Fig. 4) has been found to associate with longer lifespan across multiple populations [103, 116-118] although not without exception [119]. Analysis of haplogroup-matched fibroblasts with and without the C150T transition revealed no association with respiratory capacity nor mtDNA copy number but did correlate with reduced ROS generation as measured by CM-H2DCFDA (which fluoresces upon reaction with thiol groups within the cell) [120]. Considering all the evidence against a causative role for ROS in aging (discussed previously), this result is surprising. However, as the authors of the study point out, there were various background mutations in the cell lines which may have contributed to the phenotypes observed and until methodology exists to make targeted point mutations in mtDNA, establishing the effects of such mutations may remain elusive [120]. It is also possible that the C150T mutation might lead to different phenotypes in other tissues under different metabolic requirements.
7.3. Insights from comparative mitochondrial biology
Several studies have gone beyond comparing mtDNA sequences among the human population to compare across species. This does add to the difficulty of finding differences that actually influence lifespan and are not merely an unrelated feature of a particular clade. With this caveat, however, comparative biology allows one to compare across a much broader range of lifespans and potentially discover novel adaptations that we have not evolved.
In the proteins encoded in mammalian mtDNA, methionine abundance was found to negatively correlates with lifespan [121]. This correlation extended to nuclear-encoded respiratory subunits residing in the inner mitochondrial membrane as well. Interestingly, encoding for methionine seems to have been enriched in the genes of shorter-lived mammals (which tend to produce more ROS [122]), rather than selectively reduced in longer-lived [121]. This is likely a strategy to combat the excess ROS produced in the mitochondria as methionine residues react readily and then can be repaired by methionine sulfoxide reductases [123-125]. As the authors point out, this argues against a causative role for ROS in aging but rather that species which produce high levels have adapted to deal with it [121].
A study of the mtDNA molecule itself revealed that, among mammals, the relative abundance of cytosine on the L-strand correlates with longer lifespan. Conversely, thymine evinced a negative correlation. The authors speculated that this could be due to a decreased incidence of TT dimers which can be modified into cyclobutene pyrimidine dimers, which the mitochondria may not be able to repair [126].
More broadly, among metazoans, mitochondrially encoded cysteine has been found to negatively correlate with species lifespan [126, 127], although this association was substantially diminished under correction for phylogenetic contrast [121]. This depletion is specific to components of respiratory complex I [128] and supports other studies that demonstrate a key role for complex I in determining lifespan [129]. Separate from lifespan determination, cysteine is also specifically depleted in complex IV of aerobic organisms relative to anaerobic [128].
Birds are of particular interest in the comparative biology study of aging because, relative to mammals, many of them live much longer than one would predict based on their size and metabolic rate [130, 131]. Additionally, fibroblasts from multiple bird species have demonstrated longer survival under conditions of oxidative stress [132, 133]. A feature of the mitochondrial genome of many bird species is a duplicated or bi-directional control region [134]. This duplication has apparently evolved independently several times in the relatively long-lived Psittaciformes (parrots and cuckatoos) [135] and Passeriformes (songbirds) [136] and has been associated with lifespan across several avian families [137]. It has been reported that the number and length of direct repeats in mtDNA negatively correlate with lifespan in mammals [138]. Initiating replication in multiple directions or from multiple locations along the mitochondrial genome may reduce the incidence of deletions that are theorized to result from these repeats [139]. In a study where human, mouse, quail and pigeon cells were all driven to replicative senescence, the avian cells not only underwent more doublings but, in contrast to the mammalian cells, did not show an accumulation of mutations in their mtDNA [140]. Further experiments pointed to a key role for autophagy in promoting replicative lifespan [140]. Whether the absence of mtDNA mutations resulted from characteristics of the genome itself or through elimination of mutated copies through mitophagy or some other mechanism requires further study, as indeed, does whether improved maintenance of mtDNA contributes to the lifespan of cells or birds.
8. The role of the mitochondrial genome in age-associated diseases
A number of studies have linked accumulation of mtDNA mutations and deletions with progression of a wide assortment of age-associated diseases (Table 2). In this section, the relative contribution of mtDNA to several of these diseases will be evaluated.
Table 2.
Changes in mtDNA and mitochondrial function are summarized in the age-related diseases discussed in section 8.
| mtDNA in Age-Related Diseases | ||
|---|---|---|
| Disease | Rodent / Cell Culture | Humans |
| Parkinson’s disease | • Complex I inhibitors induce parkinsonian symptoms in rodents [142, 143] | • Reduced complex I activity [144] • mtDNA deletions in the striatum [145] and substantia niagra [146] significantly higher in Parkinson’s disease than aging alone. • mtDNA polymerase PolG mutations [148] • G11778A mtDNA mutation, which impairs complex I, has been reported to cause Parkinson’s disease [147]. |
| Alzheimer’s disease | • Mitochondrial dysfunction in APP mice [151] • Mutation of mtDNA proof reading accelerates amyloid pathology [218]. • mtDNA deletions produce mitochondrial dysfunction but do not alter Aβ plaque formation [219]. |
• Complex IV dysfunction [149, 150] • Increased mtDNA mutations [152] • Increased mtDNA in cytoplasm [153] • Increased mtDNA oxidative damage [154, 220] • No difference in mtDNA damage between healthy elderly patients and elderly patients with AD [155] • H5 haplogroup enriched in AD patients [157] |
| Sarcopenia | • Mitochondrial abnormalities and mtDNA deletions correlate with age-related muscle loss [158]. • Clonal expansion of mtDNA deletions occur in muscles with age [158]. |
• Fibers deficient in complex IV activity increase with age and possess mtDNA deletions [159]. • mtDNA deletions accumulate in muscle with age, eventually accounting for >90% of total mtDNA [161]. |
| Heart failure | • Cardiac specific deletion of DNase II causes mortality, myocarditis and cardiomyopathy under heart pressure overload [168]. • Depletion of mtDNA nucleoid proteins causes mtDNA escape to the cytosol, triggering proinflammatory response [169]. |
• The mtDNA A3243G mutation leads to cardiomyopathy [163]. • OPA1, involved in mitochondrial fusion dynamics, is diminished in heart disease patients and OPA1 mutations are linked with a decline in mtDNA and age-related cardiac dysfunction [164]. • Increased circulating extracellular mtDNA in patients with coronary heart disease [165, 166]. |
| Diabetes | • High glucose increases mtDNA damage [171]. • Transfer of mtDNA from a mouse tumor into mouse embryonic cells predisposed mice to diabetes, lymphoma and metastasis [178]. |
• Elevated mtDNA damage [170] • Lower mtDNA copy number [177] • Increased circulating extracellular mtDNA in patients with diabetes [167] • Mutation at mtDNA position 3243 is linked with diabetes and deafness [172, 173]. • Mutation at mtDNA position 3316 increases diabetes risk [174]. • Large mtDNA deletions associated with familial diabetes [175, 176]. |
| Cancer | • mtDNA mutations reported in esophageal, endometrial [181, 182], colorectal [183], prostate [221], breast [222], ovarian [223], gastric [224], hepatocellular [225], pancreatic [226] and lung cancers [227] as well as several others (reviewed in [228]) | |
8.1. Parkinson’s disease
Mitochondria dysfunction is strongly linked with Parkinson’s disease. This connection was originally found upon observations that MPP+ (an inhibitor of mitochondrial complex I) produced parkinsonian symptoms in drug users [141] and was replicated in rodent models [142, 143]. Lower complex I activity was found in the substantia nigra of patients who had Parkinson’s disease than controls [144].
Levels of mtDNA deletions have been reported in the striatum of patients with Parkinson’s disease to be significantly higher than that seen under normal aging [145]. Similar increases in mtDNA deletions were reported in the substantia niagra under both aging and Parkinson’s disease with greater prevalence under the latter. Many of the mutations appear linked with respiratory chain deficiencies [146]. The G11778A mtDNA mutation, which impairs a subunit of mitochondrial complex I, has been reported to produce parkinsonism although it also produces a range of other disorders as well [147] (Fig. 4). Mutations in the mtDNA polymerase PolG have also been reported to co-segregate with Parkinson’s disease in some familial studies [148].
8.2. Alzheimer’s disease
A broad range of studies have reported mitochondrial dysfunction to occur in Alzheimer’s disease, including reduced activity of cytochrome c oxidase (respiratory complex IV) [149, 150]. In addition, mitochondrial dysfunction has been widely reported both in human and animal studies as an early event in Alzheimer’s progression [151]. However, whether mitochondrial dysfunction is a key driver of Alzheimer’s pathology or just a symptom of the disease remains unclear, as does the relative role of mtDNA damage. While some studies have shown that mtDNA damage increases with age and to a greater extent in Alzheimer’s disease [152-154], others have not shown an increase beyond that seen with aging [155]. A study from Khan and colleagues has reported a causative relationship between mtDNA damage in Alzheimer’s disease and mitochondrial function. They created cybrids cells using the platelet mitochondria of 5 sporadic Alzheimer’s disease subjects and 5 age-matched neurologically normal subjects. They reported the cells created using mitochondria derived from Alzheimer’s disease patients had mitochondria dysfunction along with increased production of β-amyloid [156]. Independent of the role of mtDNA damage in Alzheimer’s disease, it has been reported that the mtDNA haplogroup H5 is represented at a threefold higher frequency in patients with Alzheimer’s disease [157] (Fig. 4) and may therefore be a risk factor for development of the disease.
8.3. Sarcopenia
Sarcopenia is the progressive decline in muscle mass and function over the course of aging. There is a reasonable body of evidence implicating mitochondria dysfunction as a causative factor of this muscle loss. Wanagat and colleagues reported a 33% decline in muscle mass with age in rats with an associated 30% decline in muscle fiber number. The group reported that this occurred in parallel with a significant elevation in mitochondrial abnormalities and an increase in mtDNA deletions. Significantly, the group reported that muscle fibers harboring mtDNA deletions displayed a higher frequency of muscle atrophy, fiber splitting and oxidative damage than fibers without mtDNA deletions [158]. Though far from conclusive, these findings support a causative link between mtDNA deletions and sarcopenia. Further investigation into mitochondrial dysfunction in skeletal muscle aging led to observations of an age-related increase in ragged red fibers (RRF), which stain red under incubation with Gomori Trichrome and represent fibers deficient in the cytochrome c oxidase activity. Such fibers appear at around age 40 and become more prevalent with age. At later ages researchers report that almost all RRF fibers are possess a large mitochondrial DNA deletion and this deletion is correlated with deficiency in cytochrome c oxidase activity. This suggests occurrence of mtDNA deletions are an important factor in sarcopenia through are likely driven by other changes to muscle fiber [159]. Other researchers likewise noted an enrichment of mtDNA changes in RRF but noted that the pattern of mtDNA changes was distinct in most individuals [160]. Studies of these changes in rats support an argument that the accumulation of mtDNA deletions represent a clonal expansion of mitochondria carrying large sequence deletions through the mechanism behind this expansion remains unclear [158]. It has been reported that mtDNA deletions accumulate in muscle with age to highly detrimental levels, eventually accounting for >90% of total mtDNA [161].
8.4. Heart failure
In contrast to sarcopenia, there has been very little data characterizing whether prevalence of mtDNA damage or mutations in cardiac tissue increases with age and plays a role in age-related increases in heart disease prevalence. Conversely, there is a reasonable level of evidence supporting an important role for mtDNA in heart function as many mtDNA mutations result in changes to cardiac function (reviewed in [162]). As an example, the mtDNA A3243G mutation in the LeuURR tRNA gene (Fig. 4) impairs translation of respiratory complex subunits, resulting in disrupted glucose metabolism and ultimately cardiomyopathy [163]. The nuclear-encoded gene optic atrophy 1 (OPA1), involved in mitochondrial fusion dynamics, has been found to be diminished in heart disease patients and mutation of this gene has been reported to cause both a decline in mtDNA and age-related cardiac dysfunction although it is unclear whether these are causally linked [164]. While it is unclear if mtDNA damage or mutation has a role in driving age-related cardiac dysfunction, there is a reasonable body of evidence showing that alterations in mtDNA levels are an important part of cardiac disease progression. Epidemiological studies have reported a significant decline in mtDNA levels in leukocytes and peripheral blood cells with coronary heart disease and have argued that there is a dose response relationship for risk of coronary heart disease; they also report mtDNA content to be impacted by smoking [165, 166]. It has been reported that accumulation of circulating extracellular mtDNA, likely caused as a product of cell death and necrosis, is elevated in patients with diabetes and is further elevated in diabetic patients with coronary heart disease. Levels of free mtDNA are argued by this group to be a predictor for developing coronary heart failure in patients with diabetes [167]. Oka and colleagues argue that the elevation of circulating free mtDNA is not simply a predictor of coronary heart disease but a contributing factor. The group argued that circulating mtDNA which escapes degradation of by the authophagial system would trigger an inflammatory response in heart tissue producing myocarditis and dilated cardiomyopathy. In support of this they showed that cardiac specific deletion of DNase II (a critical enzyme in removal of mtDNA in apoptotic cells) caused mortality, myocarditis and cardiomyopathy under heart pressure overload but no changes in cardiac function under normal conditions. They went on to show that these effects were ablated by inhibition of the TLR9 inflammatory pathway, suggesting that the toxic effect of free mtDNA was driven by recruitment of pro-inflammatory pathways [168]. The increase in free mtDNA may be partly driven by changes in the mtDNA nucleoid; there is a study reporting that depletion of certain mtDNA nucleoid proteins such as TFAM causes mtDNA escape to the cytosol triggering pro-inflammatory response through type I interferon responses [169]. This suggests that in situations of acute mitochondria damage, mtDNA may serve as a signaling molecule to potentiate immune defense mechanisms.
8.5. Diabetes
Variations in mtDNA are reported in patients with diabetes, including mutations, deletions and changes in mtDNA abundance, all argued to be driven by oxidative damage [170]. This is supported by studies showing that exposure to high glucose results in an accumulation of mtDNA damage which is blocked by overexpression of the antioxidant scavenger MnSOD [171]. However, it is unclear from such studies whether elevated mtDNA damage is just a symptom of the disease or has pathological consequences. It is plausible that mutations in mtDNA-encoded genes would produce further dysfunction in glucose metabolism and exacerbate diabetes progression, but this is difficult to directly test.
In support of a causative role for mtDNA in diabetes, there are a number of familial studies which show mtDNA mutations to predispose individuals to development of the disease. The most well described of these is a substitution at mtDNA position 3243, in the gene for LeuURR tRNA (Fig. 4). This mutation results in impaired respiratory complex translation, diabetes and deafness [172, 173]. Similar deficits have also been noted with mutation of position 3316, in the gene encoded the complex I subunit ND1 [174] (Fig. 4). Increased diabetes risk has also been shown in a number of mtDNA deletion mutations, including a familial form of diabetes which cosegregates with a 10.4 kb mtDNA deletion [175] and a 7.6 kb mtDNA deletion producing Wolfram syndrome with accompanying respiratory enzyme dysfunction and diabetes [176]. In support of a causative role for mtDNA changes in diabetes, a longitudinal study reported that pre-diabetic individuals who went on to develop diabetes had lower mtDNA quantities than controls who did not, suggesting that a decline in mtDNA quantity may contribute to diabetes development [177].
A study by Hashizume and colleagues is one of the few demonstrations of a direct link between mtDNA damage and diabetes: The group transferred mtDNA from a mouse tumor cell with a mutation thought to induce metastasis through excessive ROS production. This mtDNA was introduced into mouse embryonic cells, creating a strain of animals predisposed to diabetes as well as lymphoma and metastasis [178]. More research is needed to determine whether mitochondrial deficits consequent of mtDNA make a significant contribution to primarily age-associated diabetes development.
8.6. Cancer
Otto Warburg was the first to propose that cancer cells make a metabolic switch driven in part by mitochondrial dysfunction [179]. This is thought to enable cancer cells to undergo rapid growth [180]. As a consequence of this, many studies have searched for evidence of mitochondrial genome instability in cancer. Esophageal and endometrial carcinomas have indeed been associated with the appearance of mtDNA point mutations, especially in the D-Loop region [181, 182]. Strikingly, in a study of human colorectal cancer, it was reported that tumor tissue had a lower random mutation frequency than non-neoplasic tissue [183]. The authors argued that mtDNA genome stability in such tumors may have risen due to lower ROS production and therefore less oxidative damage. However, more studies are needed to address whether this represents an oddity of colorectal cancer or a feature of multiple malignant tumor types.
9. Mitochondrial DNA epigenetics
The role of epigenetics in mitochondrial DNA and aging is a controversial topic. Several studies have identified DNA methyltransferases which possess mitochondrial localization tags and have been found in mitochondria, most notably Dnmt1 and Dnmt3a [184-186]. Some studies have reported alteration in expression of these enzymes over the course of age. A study looking at 4- and 24-month-old rats showed region specific changes in expression of these enzymes in different regions of the rat brain [187]. In humans, hypermethylation of the D-loop and ND6 have been suggested to contribute to pathology in diabetes [188, 189] and non-alcoholic fatty liver disease [190], respectively. Using retinal endothelial cells, one group found that high glucose treatment increased Dnmt1 mitochondrial localization and binding to the D-Loop, leading to cytosine methylation and eventual apoptosis. Significantly, both cytosine methylation and apoptosis were attenuated by Dnmt1 inhibition [189]. In humans, mtDNA methylation has been found to vary by tissue [191] while any changes with aging appear to be highly variable [192]. However, there is controversy regarding what role DNA methyltransferases play in mitochondria in vivo [193, 194] and whether measures of mtDNA methylation are accurate or primarily experimental artifacts [195, 196].
10. Mitochondrial DNA maternal inheritance: aged mothers
In mammals, mtDNA inheritance is solely maternal [197]. It was previously thought to be a simple consequence of the oocyte providing the cytoplasm to the newly forming embryo but is increasingly evident to be the result of a selective process that eliminates paternal mitochondria. Endonuclease G was recently described in the breakdown of paternal mitochondria during fertilization in C. elegans [198]. It has long been known that fertility decreases with age, and therefore awareness of maternal age and its influence on the quality of the mtDNA being inherited has risen. Inheritance of mutated mtDNA can also lead to the so-called mitochondrial diseases.
In 2000, Brenner and colleagues found that a higher percentage of single human oocytes aged mothers (36 to 42 years old) contained the mtDNA T414G transversion point mutation present in the control region (D-loop) as compared to younger mothers (26 to 35) using regular Sanger sequencing [199]. Thanks to ultra-deep sequencing, Makova and colleagues were able to conduct a population study in mother-child pairs and scored different mtDNA copy variants (heteroplasmicity) with an allele frequency higher than 1%. They found a positive association between the amount of heteroplasmicity in a child and maternal age at fertilization. This is normally due to the appearance of point mutations, suggesting that older mothers accumulate more mutations in their germ-line, likely attributable to oocyte aging [200]. They additionally found that the older mothers accumulated more mutations in their somatic tissues relative to their children than the younger mothers. In addition, they found the number of point heteroplasmicities to triple over 30 years of life.
Goran Larson and colleagues used mice heterozygous for the mtDNA mutator allele (Polgmut/+) to maternally transmit mtDNA mutations [201]. Through doing this, they observed several mild aging phenotypes even though they had a wild-type nDNA. They also observed reduced fertility at earlier ages.
The above findings suggest a double impact of maternal age effects: A decline in fertility with age and the transfer of higher levels of mtDNA mutations. Fertility and embryo quality have also been studied in the context of assisted reproductive methods. The cells that surround oocytes at ovulation, human cumulus granulosa cells (CGC) were studied for their influence in oocyte quality. mtDNA copy number on these cells was indeed found to be positively correlated to oocyte quality [202]. However, it has also been reported that the quantity of CGC mtDNA is negatively influenced by BMI and smoking, potentially suggesting that the individual’s health is important to maintain healthier mitochondria. Moreover, a new bovine model to study the effect of both age and ovarian stimulation found that ovarian aging, but not ovarian hormonal stimulation, increased the number of oocytes with mtDNA deletions [203].
11. Conclusions
Changes in mtDNA over the course of aging are well supported in the literature. Such changes include an accumulation of oxidized base pairs, base mismatches, strand breaks and deletions. These changes have been linked with mitochondrial dysfunction. However, most studies linking mtDNA damage to aging are correlative and those studies which have attempted to manipulate accumulation of mtDNA damage have a number of confounding issues which make interpretation difficult. For this reason, the role of mtDNA damage as a driving force for age-related functional decline remains controversial.
There is also a reasonable body of evidence for specific mutations and mtDNA haplogroups as predictors of both lifespan and risk of various age-associated disease including cancer, diabetes, heart failure, sarcopenia and Parkinson’s disease. However, for many of these diseases, most reports on a role for mtDNA haplogroups and other sequence variations in aging and lifespan determination are again correlative.
The ability to make targeted sequence alterations in mtDNA – as is routinely done in nDNA with techniques such as CRISPR-Cas9 [204, 205] – would be a significant advancement in the field and allow direct testing for a causative role of mtDNA variations and mutations in determining lifespan. Several studies have reported success with various techniques in eliminating specific mtDNA from heteroplasmic cells [206-208], However, difficulty in targeting RNA to mitochondria and uncertainty whether mtDNA can undergo homologous recombination in many species present significant obstacles to making specific point mutations in the mitochondrial genome [209].
In summation, the role of mtDNA in aging and age-associated disease remains a fascinating topic with a large body of correlative data suggesting a key role. However, the lack of methods and techniques to directly manipulate the role of mtDNA in this area continues to present a substantial challenge.
Supplementary Material
Highlights.
mtDNA can be present in hundreds to thousands of copies where mutated mtDNA can coexist with wild-type mtDNA in a condition called heteroplasmy. Many of these heteroplasmic mutations have been correlated with and aging and age-associated diseases.
mtDNA has it’s own set of repair mechanisms. Though some evidence suggests these may play a role in aging and age-associated diseases the more research is needed.
Although ROS is a driver of damage to mtDNA a mounting body of evidence against and a contradictory body of evidence in support have made the Mitochondrial Free Radical Theory of aging an increasingly controversial concept.
A number of mtDNA haplogroups and SNPs have been found within humans which can either slow aging or trigger age-related diseases.
mtDNA mutations and deletions have been identified as drivers of a range of Parkinson’s disease Alzheimer’s disease, cancer, sarcopenia, heart disease and diabetes.
Maternal age can affect transmission of mtDNA heteroplasmicities
Funding
This research was supported by a Voelcker Foundation Young Investigator Award, the Glenn Foundation for Medical Research and AFAR Grant for Junior Faculty program, and the UTHSCSA Nathan Shock Pilots grants program to AMP. Additional support was provided through a National Institute of Health (NIH) T32 postdoctoral training grant [AG021890-14] to EM.
Abbreviations
- 8-OHdG
- ATP
- BER
- DNA
- GPx
- kb
- LHON
- mCAT
- MnSOD
- MRFTA
- mtDNA
- mTOR
- nDNA
- OXPHOS
- PINK-1
- PolG
- PRX
- RNA
- ROS
- RRF
- rRNA
- SOD
- tRNA
- TRX
- TXNRD
- UV
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Sagan L, On the origin of mitosing cells, J Theor Biol, 14 (1967) 255–274. [DOI] [PubMed] [Google Scholar]
- [2].Lane N, Martin W, The energetics of genome complexity, Nature, 467 (2010) 929–934. [DOI] [PubMed] [Google Scholar]
- [3].Allen JF, Why chloroplasts and mitochondria contain genomes, Comp Funct Genomics, 4 (2003) 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG, Sequence and organization of the human mitochondrial genome, Nature, 290 (1981) 457–465. [DOI] [PubMed] [Google Scholar]
- [5].Gissi C, Iannelli F, Pesole G, Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species, Heredity, 101 (2008) 301–320. [DOI] [PubMed] [Google Scholar]
- [6].Kolesnikov AA, Gerasimov ES, Diversity of mitochondrial genome organization, Biochemistry. Biokhimiia, 77 (2012) 1424–1435. [DOI] [PubMed] [Google Scholar]
- [7].Johnston IG, Williams BP, Evolutionary Inference across Eukaryotes Identifies Specific Pressures Favoring Mitochondrial Gene Retention, Cell Syst, 2 (2016) 101–111. [DOI] [PubMed] [Google Scholar]
- [8].Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G, The hallmarks of aging, Cell, 153 (2013) 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Harman D, The biologic clock: the mitochondria?, Journal of the American Geriatrics Society, 20 (1972) 145–147. [DOI] [PubMed] [Google Scholar]
- [10].Bratic A, Larsson NG, The role of mitochondria in aging, The Journal of clinical investigation, 123 (2013) 951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hoppel CL, Lesnefsky EJ, Chen Q, Tandler B, Mitochondrial Dysfunction in Cardiovascular Aging, Adv Exp Med Biol, 982 (2017) 451–464. [DOI] [PubMed] [Google Scholar]
- [12].Waltz TB, Fivenson EM, Morevati M, Li C, Becker KG, Bohr VA, Fang EF, Sarcopenia, aging and prospective interventional strategies, Curr Med Chem, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ogrodnik M, Miwa S, Tchkonia T, Tiniakos D, Wilson CL, Lahat A, Day CP, Burt A, Palmer A, Anstee QM, Grellscheid SN, Hoeijmakers JHJ, Barnhoorn S, Mann DA, Bird TG, Vermeij WP, Kirkland JL, Passos JF, von Zglinicki T, Jurk D, Cellular senescence drives age-dependent hepatic steatosis, Nature communications, 8 (2017) 15691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kazak L, Reyes A, Holt IJ, Minimizing the damage: repair pathways keep mitochondrial DNA intact, Nat Rev Mol Cell Biol, 13 (2012) 659–671. [DOI] [PubMed] [Google Scholar]
- [15].Gilkerson R, Bravo L, Garcia I, Gaytan N, Herrera A, Maldonado A, Quintanilla B, The mitochondrial nucleoid: integrating mitochondrial DNA into cellular homeostasis, Cold Spring Harb Perspect Biol, 5 (2013) a011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Just RS, Irwin JA, Parson W, Mitochondrial DNA heteroplasmy in the emerging field of massively parallel sequencing, Forensic Sci Int Genet, 18 (2015) 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Huang T, Next generation sequencing to characterize mitochondrial genomic DNA heteroplasmy, Curr Protoc Hum Genet, Chapter 19 (2011) Unit19 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rossignol R, Faustin B, Rocher C, Malgat M, Mazat JP, Letellier T, Mitochondrial threshold effects, Biochem J, 370 (2003) 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Toivonen JM, Manjiry S, Touraille S, Alziari S, O'Dell KM, Jacobs HT, Gene dosage and selective expression modify phenotype in a Drosophila model of human mitochondrial disease, Mitochondrion, 3 (2003)83–96. [DOI] [PubMed] [Google Scholar]
- [20].Pinto M, Moraes CT, Mechanisms linking mtDNA damage and aging, Free Radic Biol Med, 85 (2015) 250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mengel-From J, Thinggaard M, Dalgard C, Kyvik KO, Christensen K, Christiansen L, Mitochondrial DNA copy number in peripheral blood cells declines with age and is associated with general health among elderly, Hum Genet, 133 (2014) 1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lee JW, Park KD, Im JA, Kim MY, Lee DC, Mitochondrial DNA copy number in peripheral blood is associated with cognitive function in apparently healthy elderly women, Clin Chim Acta, 411 (2010) 592–596. [DOI] [PubMed] [Google Scholar]
- [23].Zhang R, Wang Y, Ye K, Picard M, Gu Z, Independent impacts of aging on mitochondrial DNA quantity and quality in humans, BMC Genomics, 18 (2017) 890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tranah GJ, Katzman SM, Lauterjung K, Yaffe K, Manini TM, Kritchevsky S, Newman AB, Harris TB, Cummings SR, Mitochondrial DNA m.3243A > G heteroplasmy affects multiple aging phenotypes and risk of mortality, Sci Rep, 8 (2018) 11887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rothfuss O, Gasser T, Patenge N, Analysis of differential DNA damage in the mitochondrial genome employing a semi-long run real-time PCR approach, Nucleic acids research, 38 (2010) e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jazin EE, Cavelier L, Eriksson I, Oreland L, Gyllensten U, Human brain contains high levels of heteroplasmy in the noncoding regions of mitochondrial DNA, Proc Natl Acad Sci U S A, 93 (1996) 12382–12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G, Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication, Science, 286 (1999) 774–779. [DOI] [PubMed] [Google Scholar]
- [28].Wang Y, Michikawa Y, Mallidis C, Bai Y, Woodhouse L, Yarasheski KE, Miller CA, Askanas V, Engel WK, Bhasin S, Attardi G, Muscle-specific mutations accumulate with aging in critical human mtDNA control sites for replication, Proc Natl Acad Sci U S A, 98 (2001) 4022–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nekhaeva E, Bodyak ND, Kraytsberg Y, McGrath SB, Van Orsouw NJ, Pluzhnikov A, Wei JY, Vijg J, Khrapko K, Clonally expanded mtDNA point mutations are abundant in individual cells of human tissues, Proc Natl Acad Sci U S A, 99 (2002) 5521–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].McInerny SC, Brown AL, Smith DW, Region-specific changes in mitochondrial D-loop in aged rat CNS, Mech Ageing Dev, 130 (2009) 343–349. [DOI] [PubMed] [Google Scholar]
- [31].Piko L, Hougham AJ, Bulpitt KJ, Studies of sequence heterogeneity of mitochondrial DNA from rat and mouse tissues: evidence for an increased frequency of deletions/additions with aging, Mech Ageing Dev, 43 (1988) 279–293. [DOI] [PubMed] [Google Scholar]
- [32].Cortopassi GA, Arnheim N, Detection of a specific mitochondrial DNA deletion in tissues of older humans, Nucleic Acids Res, 18 (1990) 6927–6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fayet G, Jansson M, Sternberg D, Moslemi AR, Blondy P, Lombes A, Fardeau M, Oldfors A, Ageing muscle: clonal expansions of mitochondrial DNA point mutations and deletions cause focal impairment of mitochondrial function, Neuromuscular disorders : NMD, 12 (2002) 484–493. [DOI] [PubMed] [Google Scholar]
- [34].Greaves LC, Nooteboom M, Elson JL, Tuppen HA, Taylor GA, Commane DM, Arasaradnam RP, Khrapko K, Taylor RW, Kirkwood TB, Mathers JC, Turnbull DM, Clonal expansion of early to mid-life mitochondrial DNA point mutations drives mitochondrial dysfunction during human ageing, PLoS Genet, 10 (2014) e1004620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kang E, Wang X, Tippner-Hedges R, Ma H, Folmes CD, Gutierrez NM, Lee Y, Van Dyken C, Ahmed R, Li Y, Koski A, Hayama T, Luo S, Harding CO, Amato P, Jensen J, Battaglia D, Lee D, Wu D, Terzic A, Wolf DP, Huang T, Mitalipov S, Age-Related Accumulation of Somatic Mitochondrial DNA Mutations in Adult-Derived Human iPSCs, Cell Stem Cell, 18 (2016) 625–636. [DOI] [PubMed] [Google Scholar]
- [36].McDonald SA, Greaves LC, Gutierrez-Gonzalez L, Rodriguez-Justo M, Deheragoda M, Leedham SJ, Taylor RW, Lee CY, Preston SL, Lovell M, Hunt T, Elia G, Oukrif D, Harrison R, Novelli MR, Mitchell I, Stoker DL, Turnbull DM, Jankowski JA, Wright NA, Mechanisms of field cancerization in the human stomach: the expansion and spread of mutated gastric stem cells, Gastroenterology, 134 (2008) 500–510. [DOI] [PubMed] [Google Scholar]
- [37].Clayton DA, Replication of animal mitochondrial DNA, Cell, 28 (1982) 693–705. [DOI] [PubMed] [Google Scholar]
- [38].Holt IJ, Jacobs HT, Unique features of DNA replication in mitochondria: a functional and evolutionary perspective, Bioessays, 36 (2014) 1024–1031. [DOI] [PubMed] [Google Scholar]
- [39].Gredilla R, Bohr VA, Stevnsner T, Mitochondrial DNA repair and association with aging--an update, Exp Gerontol, 45 (2010) 478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Garcia-Gomez S, Reyes A, Martinez-Jimenez MI, Chocron ES, Mouron S, Terrados G, Powell C, Salido E, Mendez J, Holt IJ, Blanco L, PrimPol, an archaic primase/polymerase operating in human cells, Mol Cell, 52 (2013) 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Martinez-Jimenez MI, Garcia-Gomez S, Bebenek K, Sastre-Moreno G, Calvo PA, Diaz-Talavera A, Kunkel TA, Blanco L, Alternative solutions and new scenarios for translesion DNA synthesis by human PrimPol, DNA Repair (Amst), 29 (2015) 127–138. [DOI] [PubMed] [Google Scholar]
- [42].Torregrosa-Munumer R, Forslund JME, Goffart S, Pfeiffer A, Stojkovic G, Carvalho G, Al-Furoukh N, Blanco L, Wanrooij S, Pohjoismaki JLO, PrimPol is required for replication reinitiation after mtDNA damage, Proc Natl Acad Sci U S A, 114 (2017) 11398–11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sykora P, Kanno S, Akbari M, Kulikowicz T, Baptiste BA, Leandro GS, Lu H, Tian J, May A, Becker KA, Croteau DL, Wilson DM 3rd, Sobol RW, Yasui A, Bohr VA, DNA polymerase beta participates in mitochondrial DNA repair, Mol Cell Biol, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kaufman BA, Van Houten B, POLB: A new role of DNA polymerase beta in mitochondrial base excision repair, DNA Repair (Amst), 60 (2017) A1–A5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wisnovsky S, Sack T, Pagliarini DJ, Laposa RR, Kelley SO, DNA Polymerase theta Increases Mutational Rates in Mitochondrial DNA, ACS Chem Biol, 13 (2018) 900–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wisnovsky S, Jean SR, Kelley SO, Mitochondrial DNA repair and replication proteins revealed by targeted chemical probes, Nat Chem Biol, 12 (2016) 567–573. [DOI] [PubMed] [Google Scholar]
- [47].Szczesny B, Tann AW, Mitra S, Age- and tissue-specific changes in mitochondrial and nuclear DNA base excision repair activity in mice: Susceptibility of skeletal muscles to oxidative injury, Mech Ageing Dev, 131 (2010) 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chen D, Cao G, Hastings T, Feng Y, Pei W, O'Horo C, Chen J, Age-dependent decline of DNA repair activity for oxidative lesions in rat brain mitochondria, J Neurochem, 81 (2002) 1273–1284. [DOI] [PubMed] [Google Scholar]
- [49].Ding WX, Yin XM, Mitophagy: mechanisms, pathophysiological roles, and analysis, Biol Chem, 393 (2012) 547–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Shokolenko IN, Wilson GL, Alexeyev MF, Persistent damage induces mitochondrial DNA degradation, DNA Repair (Amst), 12 (2013) 488–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Onyango IG, Dennis J, Khan SM, Mitochondrial Dysfunction in Alzheimer's Disease and the Rationale for Bioenergetics Based Therapies, Aging Dis, 7 (2016) 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Nissanka N, Moraes CT, Mitochondrial DNA damage and reactive oxygen species in neurodegenerative disease, FEBS Lett, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Khalil B, El Fissi N, Aouane A, Cabirol-Pol MJ, Rival T, Lievens JC, PINK1-induced mitophagy promotes neuroprotection in Huntington's disease, Cell Death Dis, 6 (2015) e1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Piccoli C, Ripoli M, Quarato G, Scrima R, D'Aprile A, Boffoli D, Margaglione M, Criscuolo C, De Michele G, Sardanelli A, Papa S, Capitanio N, Coexistence of mutations in PINK1 and mitochondrial DNA in early onset parkinsonism, J Med Genet, 45 (2008) 596–602. [DOI] [PubMed] [Google Scholar]
- [55].Johnson SC, Rabinovitch PS, Kaeberlein M, mTOR is a key modulator of ageing and age-related disease, Nature, 493 (2013) 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gaziev AI, Abdullaev S, Podlutsky A, Mitochondrial function and mitochondrial DNA maintenance with advancing age, Biogerontology, 15 (2014) 417–438. [DOI] [PubMed] [Google Scholar]
- [57].Balaban RS, Nemoto S, Finkel T, Mitochondria, oxidants, and aging, Cell, 120 (2005) 483–495. [DOI] [PubMed] [Google Scholar]
- [58].Harman D, Aging: a theory based on free radical and radiation chemistry, Journal of gerontology, 11 (1956) 298–300. [DOI] [PubMed] [Google Scholar]
- [59].Beckman KB, Ames BN, The free radical theory of aging matures, Physiological reviews, 78 (1998) 547–581. [DOI] [PubMed] [Google Scholar]
- [60].Perez VI, Bokov A, Van Remmen H, Mele J, Ran Q, Ikeno Y, Richardson A, Is the oxidative stress theory of aging dead?, Biochimica et biophysica acta, 1790 (2009) 1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Suthammarak W, Somerlot BH, Opheim E, Sedensky M, Morgan PG, Novel Interactions between Mitochondrial Superoxide Dismutases and the Electron Transport Chain, Aging cell, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Longo VD, Gralla EB, Valentine JS, Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo, The Journal of biological chemistry, 271 (1996) 12275–12280. [DOI] [PubMed] [Google Scholar]
- [63].Wawryn J, Krzepilko A, Myszka A, Bilinski T, Deficiency in superoxide dismutases shortens life span of yeast cells, Acta Biochim Pol, 46 (1999) 249–253. [PubMed] [Google Scholar]
- [64].Duttaroy A, Paul A, Kundu M, Belton A, A Sod2 null mutation confers severely reduced adult life span in Drosophila, Genetics, 165 (2003) 2295–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Lebovitz RM, Zhang H, Vogel H, Cartwright J Jr., Dionne L, Lu N, Huang S, Matzuk MM, Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice, Proceedings of the National Academy of Sciences of the United States of America, 93 (1996) 9782–9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Huang TT, Carlson EJ, Kozy HM, Mantha S, Goodman SI, Ursell PC, Epstein CJ, Genetic modification of prenatal lethality and dilated cardiomyopathy in Mn superoxide dismutase mutant mice, Free radical biology & medicine, 31 (2001) 1101–1110. [DOI] [PubMed] [Google Scholar]
- [67].Doonan R, McElwee JJ, Matthijssens F, Walker GA, Houthoofd K, Back P, Matscheski A, Vanfleteren JR, Gems D, Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans, Genes & development, 22 (2008) 3236–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Van Raamsdonk JM, Hekimi S, Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans, PLoS genetics, 5 (2009) e1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Van Raamsdonk JM, Hekimi S, Superoxide dismutase is dispensable for normal animal lifespan, Proceedings of the National Academy of Sciences of the United States of America, 109 (2012) 5785–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Van Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, Thorpe SR, Alderson NL, Baynes JW, Epstein CJ, Huang TT, Nelson J, Strong R, Richardson A, Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging, Physiol Genomics, 16 (2003) 29–37. [DOI] [PubMed] [Google Scholar]
- [71].Jang YC, Perez VI, Song W, Lustgarten MS, Salmon AB, Mele J, Qi W, Liu Y, Liang H, Chaudhuri A, Ikeno Y, Epstein CJ, Van Remmen H, Richardson A, Overexpression of Mn superoxide dismutase does not increase life span in mice, The journals of gerontology. Series A, Biological sciences and medical sciences, 64 (2009) 1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Sun J, Folk D, Bradley TJ, Tower J, Induced overexpression of mitochondrial Mn-superoxide dismutase extends the life span of adult Drosophila melanogaster, Genetics, 161 (2002) 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Mockett RJ, Sohal BH, Sohal RS, Expression of multiple copies of mitochondrially targeted catalase or genomic Mn superoxide dismutase transgenes does not extend the life span of Drosophila melanogaster, Free radical biology & medicine, 49 (2010) 2028–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Curtis C, Landis GN, Folk D, Wehr NB, Hoe N, Waskar M, Abdueva D, Skvortsov D, Ford D, Luu A, Badrinath A, Levine RL, Bradley TJ, Tavare S, Tower J, Transcriptional profiling of MnSOD-mediated lifespan extension in Drosophila reveals a species-general network of aging and metabolic genes, Genome biology, 8 (2007) R262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ranjan M, Gruber J, Ng LF, Halliwell B, Repression of the mitochondrial peroxiredoxin antioxidant system does not shorten life span but causes reduced fitness in Caenorhabditis elegans, Free radical biology & medicine, 63 (2013) 381–389. [DOI] [PubMed] [Google Scholar]
- [76].Ran Q, Liang H, Ikeno Y, Qi W, Prolla TA, Roberts LJ 2nd, Wolf N, Van Remmen H, Richardson A, Reduction in glutathione peroxidase 4 increases life span through increased sensitivity to apoptosis, The journals of gerontology. Series A, Biological sciences and medical sciences, 62 (2007) 932–942. [DOI] [PubMed] [Google Scholar]
- [77].Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, Van Remmen H, Wallace DC, Rabinovitch PS, Extension of murine life span by overexpression of catalase targeted to mitochondria, Science, 308 (2005) 1909–1911. [DOI] [PubMed] [Google Scholar]
- [78].Lee HY, Choi CS, Birkenfeld AL, Alves TC, Jornayvaz FR, Jurczak MJ, Zhang D, Woo DK, Shadel GS, Ladiges W, Rabinovitch PS, Santos JH, Petersen KF, Samuel VT, Shulman GI, Targeted expression of catalase to mitochondria prevents age-associated reductions in mitochondrial function and insulin resistance, Cell metabolism, 12 (2010) 668–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Ge X, Pettan-Brewer C, Morton J, Carter K, Fatemi S, Rabinovitch P, Ladiges WC, Mitochondrial catalase suppresses naturally occurring lung cancer in old mice, Pathobiology of aging & age related diseases, 5 (2015) 28776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Dai DF, Santana LF, Vermulst M, Tomazela DM, Emond MJ, MacCoss MJ, Gollahon K, Martin GM, Loeb LA, Ladiges WC, Rabinovitch PS, Overexpression of catalase targeted to mitochondria attenuates murine cardiac aging, Circulation, 119 (2009) 2789–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Treuting PM, Linford NJ, Knoblaugh SE, Emond MJ, Morton JF, Martin GM, Rabinovitch PS, Ladiges WC, Reduction of age-associated pathology in old mice by overexpression of catalase in mitochondria, The journals of gerontology. Series A, Biological sciences and medical sciences, 63 (2008) 813–822. [DOI] [PubMed] [Google Scholar]
- [82].Mockett RJ, Bayne AC, Kwong LK, Orr WC, Sohal RS, Ectopic expression of catalase in Drosophila mitochondria increases stress resistance but not longevity, Free radical biology & medicine, 34 (2003)207–217. [DOI] [PubMed] [Google Scholar]
- [83].Huang Q, Zhou HJ, Zhang H, Huang Y, Hinojosa-Kirschenbaum F, Fan P, Yao L, Belardinelli L, Tellides G, Giordano FJ, Budas GR, Min W, Thioredoxin-2 inhibits mitochondrial reactive oxygen species generation and apoptosis stress kinase-1 activity to maintain cardiac function, Circulation, 131 (2015) 1082–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Pickering AM, Lehr M, Gendron CM, Pletcher SD, Miller RA, Mitochondrial thioredoxin reductase 2 is elevated in long-lived primate as well as rodent species and extends fly mean lifespan, Aging cell, 16 (2017) 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Wang D, Kreutzer DA, Essigmann JM, Mutagenicity and repair of oxidative DNA damage: insights from studies using defined lesions, Mutat Res, 400 (1998) 99–115. [DOI] [PubMed] [Google Scholar]
- [86].Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA, Mitochondrial point mutations do not limit the natural lifespan of mice, Nat Genet, 39 (2007) 540–543. [DOI] [PubMed] [Google Scholar]
- [87].Itsara LS, Kennedy SR, Fox EJ, Yu S, Hewitt JJ, Sanchez-Contreras M, Cardozo-Pelaez F, Pallanck LJ, Oxidative stress is not a major contributor to somatic mitochondrial DNA mutations, PLoS genetics, 10 (2014) e1003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Kennedy SR, Salk JJ, Schmitt MW, Loeb LA, Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage, PLoS Genet, 9 (2013) e1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG, Premature ageing in mice expressing defective mitochondrial DNA polymerase, Nature, 429 (2004) 417–423. [DOI] [PubMed] [Google Scholar]
- [90].Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA, Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging, Science, 309 (2005) 481–484. [DOI] [PubMed] [Google Scholar]
- [91].Khrapko K, Kraytsberg Y, de Grey AD, Vijg J, Schon EA, Does premature aging of the mtDNA mutator mouse prove that mtDNA mutations are involved in natural aging?, Aging Cell, 5 (2006) 279–282. [DOI] [PubMed] [Google Scholar]
- [92].Miller RA, Evaluating evidence for aging, Science, 310 (2005) 441–443; author reply 441–443. [DOI] [PubMed] [Google Scholar]
- [93].Thompson LV, Oxidative stress, mitochondria and mtDNA-mutator mice, Exp Gerontol, 41 (2006) 1220–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Mott JL, Zhang D, Stevens M, Chang S, Denniger G, Zassenhaus HP, Oxidative stress is not an obligate mediator of disease provoked by mitochondrial DNA mutations, Mutat Res, 474 (2001) 35–45. [DOI] [PubMed] [Google Scholar]
- [95].Kauppila JHK, Bonekamp NA, Mourier A, Isokallio MA, Just A, Kauppila TES, Stewart JB, Larsson NG, Base-excision repair deficiency alone or combined with increased oxidative stress does not increase mtDNA point mutations in mice, Nucleic Acids Res, 46 (2018) 6642–6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Xiong ZM, Choi JY, Wang K, Zhang H, Tariq Z, Wu D, Ko E, LaDana C, Sesaki H, Cao K, Methylene blue alleviates nuclear and mitochondrial abnormalities in progeria, Aging Cell, 15 (2016) 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Van Goethem G, Dermaut B, Lofgren A, Martin JJ, Van Broeckhoven C, Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions, Nat Genet, 28 (2001) 211–212. [DOI] [PubMed] [Google Scholar]
- [98].Merriwether DA, Clark AG, Ballinger SW, Schurr TG, Soodyall H, Jenkins T, Sherry ST, Wallace DC, The structure of human mitochondrial DNA variation, J Mol Evol, 33 (1991) 543–555. [DOI] [PubMed] [Google Scholar]
- [99].Torroni A, Wallace DC, Mitochondrial DNA variation in human populations and implications for detection of mitochondrial DNA mutations of pathological significance, Journal of bioenergetics and biomembranes, 26 (1994) 261–271. [DOI] [PubMed] [Google Scholar]
- [100].Alexe G, Fuku N, Bilal E, Ueno H, Nishigaki Y, Fujita Y, Ito M, Arai Y, Hirose N, Bhanot G, Tanaka M, Enrichment of longevity phenotype in mtDNA haplogroups D4b2b, D4a, and D5 in the Japanese population, Hum Genet, 121 (2007) 347–356. [DOI] [PubMed] [Google Scholar]
- [101].Bilal E, Rabadan R, Alexe G, Fuku N, Ueno H, Nishigaki Y, Fujita Y, Ito M, Arai Y, Hirose N, Ruckenstein A, Bhanot G, Tanaka M, Mitochondrial DNA haplogroup D4a is a marker for extreme longevity in Japan, PloS one, 3 (2008) e2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].De Benedictis G, Rose G, Carrieri G, De Luca M, Falcone E, Passarino G, Bonafe M, Monti D, Baggio G, Bertolini S, Mari D, Mattace R, Franceschi C, Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans, FASEB journal : official publication of the Federation of American Societies for Experimental Biology, 13 (1999) 1532–1536. [DOI] [PubMed] [Google Scholar]
- [103].Ross OA, McCormack R, Curran MD, Duguid RA, Barnett YA, Rea IM, Middleton D, Mitochondrial DNA polymorphism: its role in longevity of the Irish population, Experimental gerontology, 36 (2001) 1161–1178. [DOI] [PubMed] [Google Scholar]
- [104].Niemi AK, Hervonen A, Hurme M, Karhunen PJ, Jylha M, Majamaa K, Mitochondrial DNA polymorphisms associated with longevity in a Finnish population, Hum Genet, 112 (2003) 29–33. [DOI] [PubMed] [Google Scholar]
- [105].Shlush LI, Atzmon G, Weisshof R, Behar D, Yudkovsky G, Barzilai N, Skorecki K, Ashkenazi Jewish centenarians do not demonstrate enrichment in mitochondrial haplogroup J, PloS one, 3 (2008) e3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Pinos T, Nogales-Gadea G, Ruiz JR, Rodriguez-Romo G, Santiago-Dorrego C, Fiuza-Luces C, Gomez-Gallego F, Cano-Nieto A, Garatachea N, Moran M, Angel Martin M, Arenas J, Andreu AL, Lucia A, Are mitochondrial haplogroups associated with extreme longevity? A study on a Spanish cohort, Age, 34 (2012) 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Dato S, Passarino G, Rose G, Altomare K, Bellizzi D, Mari V, Feraco E, Franceschi C, De Benedictis G, Association of the mitochondrial DNA haplogroup J with longevity is population specific, Eur J Hum Genet, 12 (2004) 1080–1082. [DOI] [PubMed] [Google Scholar]
- [108].de Benedictis G, Carrieri G, Varcasia O, Bonafe M, Franceschi C, Inherited variability of the mitochondrial genome and successful aging in humans, Annals of the New York Academy of Sciences, 908 (2000) 208–218. [DOI] [PubMed] [Google Scholar]
- [109].Hudson G, Carelli V, Spruijt L, Gerards M, Mowbray C, Achilli A, Pyle A, Elson J, Howell N, La Morgia C, Valentino ML, Huoponen K, Savontaus ML, Nikoskelainen E, Sadun AA, Salomao SR, Belfort R Jr., Griffiths P, Man PY, de Coo RF, Horvath R, Zeviani M, Smeets HJ, Torroni A, Chinnery PF, Clinical expression of Leber hereditary optic neuropathy is affected by the mitochondrial DNA-haplogroup background, American journal of human genetics, 81 (2007) 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Torroni A, Petrozzi M, D'Urbano L, Sellitto D, Zeviani M, Carrara F, Carducci C, Leuzzi V, Carelli V, Barboni P, De Negri A, Scozzari R, Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484, American journal of human genetics, 60 (1997) 1107–1121. [PMC free article] [PubMed] [Google Scholar]
- [111].Howell N, Herrnstadt C, Shults C, Mackey DA, Low penetrance of the 14484 LHON mutation when it arises in a non-haplogroup J mtDNA background, American journal of medical genetics. Part A, 119A (2003) 147–151. [DOI] [PubMed] [Google Scholar]
- [112].Reynier P, Penisson-Besnier I, Moreau C, Savagner F, Vielle B, Emile J, Dubas F, Malthiery Y, mtDNA haplogroup J: a contributing factor of optic neuritis, Eur J Hum Genet, 7 (1999) 404–406. [DOI] [PubMed] [Google Scholar]
- [113].Rose G, Passarino G, Carrieri G, Altomare K, Greco V, Bertolini S, Bonafe M, Franceschi C, De Benedictis G, Paradoxes in longevity: sequence analysis of mtDNA haplogroup J in centenarians, Eur J Hum Genet, 9 (2001) 701–707. [DOI] [PubMed] [Google Scholar]
- [114].Martinez-Redondo D, Marcuello A, Casajus JA, Ara I, Dahmani Y, Montoya J, Ruiz-Pesini E, Lopez-Perez MJ, Diez-Sanchez C, Human mitochondrial haplogroup H: the highest VO2max consumer--is it a paradox?, Mitochondrion, 10 (2010) 102–107. [DOI] [PubMed] [Google Scholar]
- [115].Raule N, Sevini F, Li S, Barbieri A, Tallaro F, Lomartire L, Vianello D, Montesanto A, Moilanen JS, Bezrukov V, Blanche H, Hervonen A, Christensen K, Deiana L, Gonos ES, Kirkwood TB, Kristensen P, Leon A, Pelicci PG, Poulain M, Rea IM, Remacle J, Robine JM, Schreiber S, Sikora E, Eline Slagboom P, Spazzafumo L, Antonietta Stazi M, Toussaint O, Vaupel JW, Rose G, Majamaa K, Perola M, Johnson TE, Bolund L, Yang H, Passarino G, Franceschi C, The co-occurrence of mtDNA mutations on different oxidative phosphorylation subunits, not detected by haplogroup analysis, affects human longevity and is population specific, Aging cell, 13 (2014) 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Guney O, Ak H, Atay S, Ozkaya AB, Aydin HH, Mitochondrial DNA polymorphisms associated with longevity in the Turkish population, Mitochondrion, 17 (2014) 7–13. [DOI] [PubMed] [Google Scholar]
- [117].Niemi AK, Moilanen JS, Tanaka M, Hervonen A, Hurme M, Lehtimaki T, Arai Y, Hirose N, Majamaa K, A combination of three common inherited mitochondrial DNA polymorphisms promotes longevity in Finnish and Japanese subjects, Eur J Hum Genet, 13 (2005) 166–170. [DOI] [PubMed] [Google Scholar]
- [118].Zhang J, Asin-Cayuela J, Fish J, Michikawa Y, Bonafe M, Olivieri F, Passarino G, De Benedictis G, Franceschi C, Attardi G, Strikingly higher frequency in centenarians and twins of mtDNA mutation causing remodeling of replication origin in leukocytes, Proceedings of the National Academy of Sciences of the United States of America, 100 (2003) 1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Pan H, Kong QP, Cheng YT, Lian SG, Yang J, Gao SJ, Xu LY, Zhang YP, Absence of association between mitochondrial DNA C150T polymorphism and longevity in a Han Chinese population, Experimental gerontology, 46 (2011) 511–515. [DOI] [PubMed] [Google Scholar]
- [120].Chen A, Raule N, Chomyn A, Attardi G, Decreased reactive oxygen species production in cells with mitochondrial haplogroups associated with longevity, PloS one, 7 (2012) e46473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Aledo JC, Li Y, de Magalhaes JP, Ruiz-Camacho M, Perez-Claros JA, Mitochondrially encoded methionine is inversely related to longevity in mammals, Aging cell, 10 (2011) 198–207. [DOI] [PubMed] [Google Scholar]
- [122].Barja G, Rate of generation of oxidative stress-related damage and animal longevity, Free radical biology & medicine, 33 (2002) 1167–1172. [DOI] [PubMed] [Google Scholar]
- [123].Bender A, Hajieva P, Moosmann B, Adaptive antioxidant methionine accumulation in respiratory chain complexes explains the use of a deviant genetic code in mitochondria, Proceedings of the National Academy of Sciences of the United States of America, 105 (2008) 16496–16501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Schindeldecker M, Moosmann B, Protein-borne methionine residues as structural antioxidants in mitochondria, Amino Acids, 47 (2015) 1421–1432. [DOI] [PubMed] [Google Scholar]
- [125].Levine RL, Mosoni L, Berlett BS, Stadtman ER, Methionine residues as endogenous antioxidants in proteins, Proceedings of the National Academy of Sciences of the United States of America, 93 (1996) 15036–15040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Lehmann G, Budovsky A, Muradian KK, Fraifeld VE, Mitochondrial genome anatomy and species-specific lifespan, Rejuvenation research, 9 (2006) 223–226. [DOI] [PubMed] [Google Scholar]
- [127].Moosmann B, Behl C, Mitochondrially encoded cysteine predicts animal lifespan, Aging cell, 7 (2008)32–46. [DOI] [PubMed] [Google Scholar]
- [128].Schindeldecker M, Stark M, Behl C, Moosmann B, Differential cysteine depletion in respiratory chain complexes enables the distinction of longevity from aerobicity, Mechanisms of ageing and development, 132 (2011) 171–179. [DOI] [PubMed] [Google Scholar]
- [129].Miwa S, Jow H, Baty K, Johnson A, Czapiewski R, Saretzki G, Treumann A, von Zglinicki T, Low abundance of the matrix arm of complex I in mitochondria predicts longevity in mice, Nature communications, 5 (2014) 3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Speakman JR, Body size, energy metabolism and lifespan, The Journal of experimental biology, 208 (2005) 1717–1730. [DOI] [PubMed] [Google Scholar]
- [131].Tacutu R, Craig T, Budovsky A, Wuttke D, Lehmann G, Taranukha D, Costa J, Fraifeld VE, de Magalhaes JP, Human Ageing Genomic Resources: integrated databases and tools for the biology and genetics of ageing, Nucleic acids research, 41 (2013) D1027–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Harper JM, Wang M, Galecki AT, Ro J, Williams JB, Miller RA, Fibroblasts from long-lived bird species are resistant to multiple forms of stress, The Journal of experimental biology, 214 (2011) 1902–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Pickering AM, Lehr M, Kohler WJ, Han ML, Miller RA, Fibroblasts From Longer-Lived Species of Primates, Rodents, Bats, Carnivores, and Birds Resist Protein Damage, The journals of gerontology. Series A, Biological sciences and medical sciences, 70 (2015) 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Reyes A, Yang MY, Bowmaker M, Holt IJ, Bidirectional replication initiates at sites throughout the mitochondrial genome of birds, The Journal of biological chemistry, 280 (2005) 3242–3250. [DOI] [PubMed] [Google Scholar]
- [135].Schirtzinger EE, Tavares ES, Gonzales LA, Eberhard JR, Miyaki CY, Sanchez JJ, Hernandez A, Mueller H, Graves GR, Fleischer RC, Wright TF, Multiple independent origins of mitochondrial control region duplications in the order Psittaciformes, Mol Phylogenet Evol, 64 (2012) 342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Caparroz R, Rocha AV, Cabanne GS, Tubaro P, Aleixo A, Lemmon EM, Lemmon AR, Mitogenomes of two neotropical bird species and the multiple independent origin of mitochondrial gene orders in Passeriformes, Mol Biol Rep, (2018). [DOI] [PubMed] [Google Scholar]
- [137].Skujina I, McMahon R, Lenis VP, Gkoutos GV, Hegarty M, Duplication of the mitochondrial control region is associated with increased longevity in birds, Aging, 8 (2016) 1781–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Samuels DC, Mitochondrial DNA repeats constrain the life span of mammals, Trends Genet, 20 (2004) 226–229. [DOI] [PubMed] [Google Scholar]
- [139].Khaidakov M, Species-specific lifespans: Can it be a lottery based on the mode of mitochondrial DNA replication?, Mechanisms of ageing and development, 155 (2016) 1–6. [DOI] [PubMed] [Google Scholar]
- [140].Strecker V, Mai S, Muster B, Beneke S, Burkle A, Bereiter-Hahn J, Jendrach M, Aging of different avian cultured cells: lack of ROS-induced damage and quality control mechanisms, Mechanisms of ageing and development, 131 (2010) 48–59. [DOI] [PubMed] [Google Scholar]
- [141].Langston JW, Ballard P, Tetrud JW, Irwin I, Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis, Science, 219 (1983) 979–980. [DOI] [PubMed] [Google Scholar]
- [142].Heikkila RE, Nicklas WJ, Vyas I, Duvoisin RC, Dopaminergic toxicity of rotenone and the 1-methyl-4-phenylpyridinium ion after their stereotaxic administration to rats: implication for the mechanism of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine toxicity, Neuroscience letters, 62 (1985) 389–394. [DOI] [PubMed] [Google Scholar]
- [143].Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT, Chronic systemic pesticide exposure reproduces features of Parkinson's disease, Nat Neurosci, 3 (2000) 1301–1306. [DOI] [PubMed] [Google Scholar]
- [144].Schapira AH, Cooper JM, Dexter D, Jenner P, Clark JB, Marsden CD, Mitochondrial complex I deficiency in Parkinson's disease, Lancet, 1 (1989) 1269. [DOI] [PubMed] [Google Scholar]
- [145].Ikebe S, Tanaka M, Ohno K, Sato W, Hattori K, Kondo T, Mizuno Y, Ozawa T, Increase of deleted mitochondrial DNA in the striatum in Parkinson's disease and senescence, Biochem Biophys Res Commun, 170 (1990) 1044–1048. [DOI] [PubMed] [Google Scholar]
- [146].Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, Taylor RW, Turnbull DM, High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease, Nat Genet, 38 (2006) 515–517. [DOI] [PubMed] [Google Scholar]
- [147].Simon DK, Pulst SM, Sutton JP, Browne SE, Beal MF, Johns DR, Familial multisystem degeneration with parkinsonism associated with the 11778 mitochondrial DNA mutation, Neurology, 53 (1999) 1787–1793. [DOI] [PubMed] [Google Scholar]
- [148].Luoma P, Melberg A, Rinne JO, Kaukonen JA, Nupponen NN, Chalmers RM, Oldfors A, Rautakorpi I, Peltonen L, Majamaa K, Somer H, Suomalainen A, Parkinsonism, premature menopause, and mitochondrial DNA polymerase gamma mutations: clinical and molecular genetic study, Lancet, 364 (2004) 875–882. [DOI] [PubMed] [Google Scholar]
- [149].Parker WD Jr., Filley CM, Parks JK, Cytochrome oxidase deficiency in Alzheimer's disease, Neurology, 40 (1990) 1302–1303. [DOI] [PubMed] [Google Scholar]
- [150].Kish SJ, Bergeron C, Rajput A, Dozic S, Mastrogiacomo F, Chang LJ, Wilson JM, DiStefano LM, Nobrega JN, Brain cytochrome oxidase in Alzheimer's disease, J Neurochem, 59 (1992) 776–779. [DOI] [PubMed] [Google Scholar]
- [151].Hauptmann S, Scherping I, Drose S, Brandt U, Schulz KL, Jendrach M, Leuner K, Eckert A, Muller WE, Mitochondrial dysfunction: an early event in Alzheimer pathology accumulates with age in AD transgenic mice, Neurobiol Aging, 30 (2009) 1574–1586. [DOI] [PubMed] [Google Scholar]
- [152].Casoli T, Di Stefano G, Spazzafumo L, Balietti M, Giorgetti B, Giuli C, Postacchini D, Fattoretti P, Conti F, Contribution of non-reference alleles in mtDNA of Alzheimer's disease patients, Ann Clin Transl Neurol, 1 (2014) 284–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Hirai K, Aliev G, Nunomura A, Fujioka H, Russell RL, Atwood CS, Johnson AB, Kress Y, Vinters HV, Tabaton M, Shimohama S, Cash AD, Siedlak SL, Harris PL, Jones PK, Petersen RB, Perry G, Smith MA, Mitochondrial abnormalities in Alzheimer's disease, J Neurosci, 21 (2001) 3017–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Mecocci P, MacGarvey U, Beal MF, Oxidative damage to mitochondrial DNA is increased in Alzheimer's disease, Ann Neurol, 36 (1994) 747–751. [DOI] [PubMed] [Google Scholar]
- [155].Lin MT, Simon DK, Ahn CH, Kim LM, Beal MF, High aggregate burden of somatic mtDNA point mutations in aging and Alzheimer's disease brain, Hum Mol Genet, 11 (2002) 133–145. [DOI] [PubMed] [Google Scholar]
- [156].Khan SM, Cassarino DS, Abramova NN, Keeney PM, Borland MK, Trimmer PA, Krebs CT, Bennett JC, Parks JK, Swerdlow RH, Parker WD Jr., Bennett JP Jr., Alzheimer's disease cybrids replicate beta-amyloid abnormalities through cell death pathways, Ann Neurol, 48 (2000) 148–155. [PubMed] [Google Scholar]
- [157].Santoro A, Balbi V, Balducci E, Pirazzini C, Rosini F, Tavano F, Achilli A, Siviero P, Minicuci N, Bellavista E, Mishto M, Salvioli S, Marchegiani F, Cardelli M, Olivieri F, Nacmias B, Chiamenti AM, Benussi L, Ghidoni R, Rose G, Gabelli C, Binetti G, Sorbi S, Crepaldi G, Passarino G, Torroni A, Franceschi C, Evidence for sub-haplogroup h5 of mitochondrial DNA as a risk factor for late onset Alzheimer's disease, PLoS One, 5 (2010) e12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Wanagat J, Cao Z, Pathare P, Aiken JM, Mitochondrial DNA deletion mutations colocalize with segmental electron transport system abnormalities, muscle fiber atrophy, fiber splitting, and oxidative damage in sarcopenia, FASEB J, 15 (2001) 322–332. [DOI] [PubMed] [Google Scholar]
- [159].Pesce V, Cormio A, Fracasso F, Vecchiet J, Felzani G, Lezza AM, Cantatore P, Gadaleta MN, Age-related mitochondrial genotypic and phenotypic alterations in human skeletal muscle, Free Radic Biol Med, 30 (2001) 1223–1233. [DOI] [PubMed] [Google Scholar]
- [160].Cormio A, Milella F, Vecchiet J, Felzani G, Gadaleta MN, Cantatore P, Mitochondrial DNA mutations in RRF of healthy subjects of different age, Neurobiol Aging, 26 (2005) 655–664. [DOI] [PubMed] [Google Scholar]
- [161].Bua E, Johnson J, Herbst A, Delong B, McKenzie D, Salamat S, Aiken JM, Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers, American journal of human genetics, 79 (2006) 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Bates MG, Bourke JP, Giordano C, d'Amati G, Turnbull DM, Taylor RW, Cardiac involvement in mitochondrial DNA disease: clinical spectrum, diagnosis, and management, Eur Heart J, 33 (2012) 3023–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Lindroos MM, Parkka JP, Taittonen MT, lozzo P, Karppa M, Hassinen IE, Knuuti J, Nuutila P, Majamaa K, Myocardial glucose uptake in patients with the m.3243A > G mutation in mitochondrial DNA, J Inherit Metab Dis, 39 (2016) 67–74. [DOI] [PubMed] [Google Scholar]
- [164].Chen L, Liu T, Tran A, Lu X, Tomilov AA, Davies V, Cortopassi G, Chiamvimonvat N, Bers DM, Votruba M, Knowlton AA, OPA1 mutation and late-onset cardiomyopathy: mitochondrial dysfunction and mtDNA instability, J Am Heart Assoc, 1 (2012) e003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Chen S, Xie X, Wang Y, Gao Y, Xie X, Yang J, Ye J, Association between leukocyte mitochondrial DNA content and risk of coronary heart disease: a case-control study, Atherosclerosis, 237 (2014) 220–226. [DOI] [PubMed] [Google Scholar]
- [166].Liu LP, Cheng K, Ning MA, Li HH, Wang HC, Li F, Chen SY, Qu FL, Guo WY, Association between peripheral blood cells mitochondrial DNA content and severity of coronary heart disease, Atherosclerosis, 261 (2017) 105–110. [DOI] [PubMed] [Google Scholar]
- [167].Liu J, Cai X, Xie L, Tang Y, Cheng J, Wang J, Wang L, Gong J, Circulating Cell Free Mitochondrial DNA is a Biomarker in the Development of Coronary Heart Disease in the Patients with Type 2 Diabetes, Clin Lab, 61 (2015) 661–667. [DOI] [PubMed] [Google Scholar]
- [168].Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, Oyabu J, Murakawa T, Nakayama H, Nishida K, Akira S, Yamamoto A, Komuro I, Otsu K, Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure, Nature, 485 (2012) 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, Kaech SM, Smiley JR, Means RE, Iwasaki A, Shadel GS, Mitochondrial DNA stress primes the antiviral innate immune response, Nature, 520 (2015) 553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Fetterman JL, Holbrook M, Westbrook DG, Brown JA, Feeley KP, Breton-Romero R, Linder EA, Berk BD, Weisbrod RM, Widlansky ME, Gokce N, Ballinger SW, Hamburg NM, Mitochondrial DNA damage and vascular function in patients with diabetes mellitus and atherosclerotic cardiovascular disease, Cardiovasc Diabetol, 15 (2016) 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Madsen-Bouterse SA, Zhong Q, Mohammad G, Ho YS, Kowluru RA, Oxidative damage of mitochondrial DNA in diabetes and its protection by manganese superoxide dismutase, Free Radic Res, 44 (2010) 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Kadowaki T, Kadowaki H, Mori Y, Tobe K, Sakuta R, Suzuki Y, Tanabe Y, Sakura H, Awata T, Goto Y, et al. , A subtype of diabetes mellitus associated with a mutation of mitochondrial DNA, N Engl J Med, 330 (1994) 962–968. [DOI] [PubMed] [Google Scholar]
- [173].Reardon W, Ross RJ, Sweeney MG, Luxon LM, Pembrey ME, Harding AE, Trembath RC, Diabetes mellitus associated with a pathogenic point mutation in mitochondrial DNA, Lancet, 340 (1992) 1376–1379. [DOI] [PubMed] [Google Scholar]
- [174].Nakagawa Y, Ikegami H, Yamato E, Takekawa K, Fujisawa T, Hamada Y, Ueda H, Uchigata Y, Miki T, Kumahara Y, et al. , A new mitochondrial DNA mutation associated with non-insulin-dependent diabetes mellitus, Biochem Biophys Res Commun, 209 (1995) 664–668. [DOI] [PubMed] [Google Scholar]
- [175].Ballinger SW, Shoffner JM, Hedaya EV, Trounce I, Polak MA, Koontz DA, Wallace DC, Maternally transmitted diabetes and deafness associated with a 10.4 kb mitochondrial DNA deletion, Nat Genet, 1 (1992) 11–15. [DOI] [PubMed] [Google Scholar]
- [176].Rotig A, Cormier V, Chatelain P, Francois R, Saudubray JM, Rustin P, Munnich A, Deletion of mitochondrial DNA in a case of early-onset diabetes mellitus, optic atrophy, and deafness (Wolfram syndrome, MIM 222300), J Clin Invest, 91 (1993) 1095–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Lee HK, Song JH, Shin CS, Park DJ, Park KS, Lee KU, Koh CS, Decreased mitochondrial DNA content in peripheral blood precedes the development of non-insulin-dependent diabetes mellitus, Diabetes Res Clin Pract, 42 (1998) 161–167. [DOI] [PubMed] [Google Scholar]
- [178].Hashizume O, Shimizu A, Yokota M, Sugiyama A, Nakada K, Miyoshi H, Itami M, Ohira M, Nagase H, Takenaga K, Hayashi J, Specific mitochondrial DNA mutation in mice regulates diabetes and lymphoma development, Proc Natl Acad Sci U S A, 109 (2012) 10528–10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Warburg O, On respiratory impairment in cancer cells, Science, 124 (1956) 269–270. [PubMed] [Google Scholar]
- [180].Liberti MV, Locasale JW, The Warburg Effect: How Does it Benefit Cancer Cells?, Trends Biochem Sci, 41 (2016) 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Kumimoto H, Yamane Y, Nishimoto Y, Fukami H, Shinoda M, Hatooka S, Ishizaki K, Frequent somatic mutations of mitochondrial DNA in esophageal squamous cell carcinoma, Int J Cancer, 108 (2004) 228–231. [DOI] [PubMed] [Google Scholar]
- [182].Liu VW, Yang HJ, Wang Y, Tsang PC, Cheung AN, Chiu PM, Ng TY, Wong LC, Nagley P, Ngan HY, High frequency of mitochondrial genome instability in human endometrial carcinomas, Br J Cancer, 89 (2003) 697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Ericson NG, Kulawiec M, Vermulst M, Sheahan K, O'Sullivan J, Salk JJ, Bielas JH, Decreased mitochondrial DNA mutagenesis in human colorectal cancer, PLoS Genet, 8 (2012) e1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM, DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria, Proc Natl Acad Sci U S A, 108 (2011) 3630–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Wong M, Gertz B, Chestnut BA, Martin LJ, Mitochondrial DNMT3A and DNA methylation in skeletal muscle and CNS of transgenic mouse models of ALS, Front Cell Neurosci, 7 (2013) 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Saini SK, Mangalhara KC, Prakasam G, Bamezai RNK, DNA Methyltransferase1 (DNMT1) Isoform3 methylates mitochondrial genome and modulates its biology, Scientific reports, 7 (2017) 1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Dzitoyeva S, Chen H, Manev H, Effect of aging on 5-hydroxymethylcytosine in brain mitochondria, Neurobiol Aging, 33 (2012) 2881–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Zheng LD, Linarelli LE, Liu L, Wall SS, Greenawald MH, Seidel RW, Estabrooks PA, Almeida FA, Cheng Z, Insulin resistance is associated with epigenetic and genetic regulation of mitochondrial DNA in obese humans, Clin Epigenetics, 7 (2015) 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Mishra M, Kowluru RA, Epigenetic Modification of Mitochondrial DNA in the Development of Diabetic Retinopathy, Invest Ophthalmol Vis Sci, 56 (2015) 5133–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Pirola CJ, Gianotti TF, Burgueno AL, Rey-Funes M, Loidl CF, Mallardi P, Martino JS, Castano GO, Sookoian S, Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease, Gut, 62 (2013) 1356–1363. [DOI] [PubMed] [Google Scholar]
- [191].Devall M, Smith RG, Jeffries A, Hannon E, Davies MN, Schalkwyk L, Mill J, Weedon M, Lunnon K, Regional differences in mitochondrial DNA methylation in human post-mortem brain tissue, Clin Epigenetics, 9 (2017) 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Mawlood SK, Dennany L, Watson N, Dempster J, Pickard BS, Quantification of global mitochondrial DNA methylation levels and inverse correlation with age at two CpG sites, Aging, 8 (2016) 636–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].van der Wijst MG, van Tilburg AY, Ruiters MH, Rots MG, Experimental mitochondria-targeted DNA methylation identifies GpC methylation, not CpG methylation, as potential regulator of mitochondrial gene expression, Scientific reports, 7 (2017) 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Bellizzi D, D'Aquila P, Scafone T, Giordano M, Riso V, Riccio A, Passarino G, The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern, DNA Res, 20 (2013) 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Mechta M, Ingerslev LR, Fabre O, Picard M, Barres R, Evidence Suggesting Absence of Mitochondrial DNA Methylation, Front Genet, 8 (2017) 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Liu B, Du Q, Chen L, Fu G, Li S, Fu L, Zhang X, Ma C, Bin C, CpG methylation patterns of human mitochondrial DNA, Scientific reports, 6 (2016) 23421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Hecht NB, Liem H, Kleene KC, Distel RJ, Ho SM, Maternal inheritance of the mouse mitochondrial genome is not mediated by a loss or gross alteration of the paternal mitochondrial DNA or by methylation of the oocyte mitochondrial DNA, Dev Biol, 102 (1984) 452–461. [DOI] [PubMed] [Google Scholar]
- [198].Zhou Q, Li H, Li H, Nakagawa A, Lin JL, Lee ES, Harry BL, Skeen-Gaar RR, Suehiro Y, William D, Mitani S, Yuan HS, Kang BH, Xue D, Mitochondrial endonuclease G mediates breakdown of paternal mitochondria upon fertilization, Science, 353 (2016) 394–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Barritt JA, Cohen J, Brenner CA, Mitochondrial DNA point mutation in human oocytes is associated with maternal age, Reprod Biomed Online, 1 (2000) 96–100. [DOI] [PubMed] [Google Scholar]
- [200].Rebolledo-Jaramillo B, Su MS, Stoler N, McElhoe JA, Dickins B, Blankenberg D, Korneliussen TS, Chiaromonte F, Nielsen R, Holland MM, Paul IM, Nekrutenko A, Makova KD, Maternal age effect and severe germ-line bottleneck in the inheritance of human mitochondrial DNA, Proc Natl Acad Sci U S A, 111 (2014) 15474–15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Ross JM, Stewart JB, Hagstrom E, Brene S, Mourier A, Coppotelli G, Freyer C, Lagouge M, Hoffer BJ, Olson L, Larsson NG, Germline mitochondrial DNA mutations aggravate ageing and can impair brain development, Nature, 501 (2013) 412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Desquiret-Dumas V, Clement A, Seegers V, Boucret L, Ferre-L'Hotellier V, Bouet PE, Descamps P, Procaccio V, Reynier P, May-Panloup P, The mitochondrial DNA content of cumulus granulosa cells is linked to embryo quality, Hum Reprod, 32 (2017) 607–614. [DOI] [PubMed] [Google Scholar]
- [203].Hammond ER, Green MP, Shelling AN, Berg MC, Peek JC, Cree LM, Oocyte mitochondrial deletions and heteroplasmy in a bovine model of ageing and ovarian stimulation, Mol Hum Reprod, 22 (2016) 261–271. [DOI] [PubMed] [Google Scholar]
- [204].Ford K, McDonald D, Mali P, Functional Genomics via CRISPR-Cas, Journal of molecular biology, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Perez Rojo F, Nyman RKM, Johnson AAT, Navarro MP, Ryan MH, Erskine W, Kaur P, CRISPR-Cas systems: ushering in the new genome editing era, Bioengineered, 9 (2018) 214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Bacman SR, Williams SL, Pinto M, Peralta S, Moraes CT, Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs, Nature medicine, 19 (2013) 1111–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Yang Y, Wu H, Kang X, Liang Y, Lan T, Li T, Tan T, Peng J, Zhang Q, An G, Liu Y, Yu Q, Ma Z, Lian Y, Soh BS, Chen Q, Liu P, Chen Y, Sun X, Li R, Zhen X, Liu P, Yu Y, Li X, Fan Y, Targeted elimination of mutant mitochondrial DNA in MELAS-iPSCs by mitoTALENs, Protein & cell, 9 (2018) 283–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Gammage PA, Gaude E, Van Haute L, Rebelo-Guiomar P, Jackson CB, Rorbach J, Pekalski ML, Robinson AJ, Charpentier M, Concordet JP, Frezza C, Minczuk M, Near-complete elimination of mutant mtDNA by iterative or dynamic dose-controlled treatment with mtZFNs, Nucleic acids research, 44 (2016) 7804–7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Gammage PA, Moraes CT, Minczuk M, Mitochondrial Genome Engineering: The Revolution May Not Be CRISPR-Ized, Trends Genet, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Yang W, Li J, Hekimi S, A Measurable increase in oxidative damage due to reduction in superoxide detoxification fails to shorten the life span of long-lived mitochondrial mutants of Caenorhabditis elegans, Genetics, 177 (2007) 2063–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Paul A, Belton A, Nag S, Martin I, Grotewiel MS, Duttaroy A, Reduced mitochondrial SOD displays mortality characteristics reminiscent of natural aging, Mech Ageing Dev, 128 (2007) 706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Cabreiro F, Ackerman D, Doonan R, Araiz C, Back P, Papp D, Braeckman BP, Gems D, Increased life span from overexpression of superoxide dismutase in Caenorhabditis elegans is not caused by decreased oxidative damage, Free radical biology & medicine, 51 (2011) 1575–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Yant LJ, Ran Q, Rao L, Van Remmen H, Shibatani T, Belter JG, Motta L, Richardson A, Prolla TA, The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults, Free radical biology & medicine, 34 (2003) 496–502. [DOI] [PubMed] [Google Scholar]
- [214].Li L, Shoji W, Takano H, Nishimura N, Aoki Y, Takahashi R, Goto S, Kaifu T, Takai T, Obinata M, Increased susceptibility of MER5 (peroxiredoxin III) knockout mice to LPS-induced oxidative stress, Biochem Biophys Res Commun, 355 (2007) 715–721. [DOI] [PubMed] [Google Scholar]
- [215].Cacho-Valadez B, Munoz-Lobato F, Pedrajas JR, Cabello J, Fierro-Gonzalez JC, Navas P, Swoboda P, Link CD, Miranda-Vizuete A, The characterization of the Caenorhabditis elegans mitochondrial thioredoxin system uncovers an unexpected protective role of thioredoxin reductase 2 in beta-amyloid peptide toxicity, Antioxid Redox Signal, 16 (2012) 1384–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Nonn L, Williams RR, Erickson RP, Powis G, The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice, Molecular and cellular biology, 23 (2003) 916–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Missirlis F, Ulschmid JK, Hirosawa-Takamori M, Gronke S, Schafer U, Becker K, Phillips JP, Jackle H, Mitochondrial and cytoplasmic thioredoxin reductase variants encoded by a single Drosophila gene are both essential for viability, J Biol Chem, 277 (2002) 11521–11526. [DOI] [PubMed] [Google Scholar]
- [218].Kukreja L, Kujoth GC, Prolla TA, Van Leuven F, Vassar R, Increased mtDNA mutations with aging promotes amyloid accumulation and brain atrophy in the APP/Ld transgenic mouse model of Alzheimer's disease, Mol Neurodegener, 9 (2014) 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Pinto M, Pickrell AM, Fukui H, Moraes CT, Mitochondrial DNA damage in a mouse model of Alzheimer's disease decreases amyloid beta plaque formation, Neurobiol Aging, 34 (2013) 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Wang J, Xiong S, Xie C, Markesbery WR, Lovell MA, Increased oxidative damage in nuclear and mitochondrial DNA in Alzheimer's disease, J Neurochem, 93 (2005) 953–962. [DOI] [PubMed] [Google Scholar]
- [221].Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC, mtDNA mutations increase tumorigenicity in prostate cancer, Proc Natl Acad Sci U S A, 102 (2005) 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Tan DJ, Bai RK, Wong LJ, Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer, Cancer Res, 62 (2002) 972–976. [PubMed] [Google Scholar]
- [223].Liu VW, Shi HH, Cheung AN, Chiu PM, Leung TW, Nagley P, Wong LC, Ngan HY, High incidence of somatic mitochondrial DNA mutations in human ovarian carcinomas, Cancer Res, 61 (2001) 5998–6001. [PubMed] [Google Scholar]
- [224].Zhao YB, Yang HY, Zhang XW, Chen GY, Mutation in D-loop region of mitochondrial DNA in gastric cancer and its significance, World J Gastroenterol, 11 (2005) 3304–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Nomoto S, Sanchez-Cespedes M, Sidransky D, Identification of mtDNA mutations in human cancer, Methods Mol Biol, 197 (2002) 107–117. [DOI] [PubMed] [Google Scholar]
- [226].Jones JB, Song JJ, Hempen PM, Parmigiani G, Hruban RH, Kern SE, Detection of mitochondrial DNA mutations in pancreatic cancer offers a "mass"-ive advantage over detection of nuclear DNA mutations, Cancer Res, 61 (2001) 1299–1304. [PubMed] [Google Scholar]
- [227].Sanchez-Cespedes M, Parrella P, Nomoto S, Cohen D, Xiao Y, Esteller M, Jeronimo C, Jordan RC, Nicol T, Koch WM, Schoenberg M, Mazzarelli P, Fazio VM, Sidransky D, Identification of a mononucleotide repeat as a major target for mitochondrial DNA alterations in human tumors, Cancer Res, 61 (2001) 7015–7019. [PubMed] [Google Scholar]
- [228].Chatterjee A, Mambo E, Sidransky D, Mitochondrial DNA mutations in human cancer, Oncogene, 25 (2006) 4663–4674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.