Significance
The tachykinin receptors are neuropeptide G-protein–coupled receptors (GPCRs) involved in pain and sensory transmission. Antagonists of the human NK1 tachykinin receptor are used to treat chemotherapy-induced nausea, and have also been intensively studied for other disorders including depression. We solved the crystal structure of the human NK1 receptor bound to a high-affinity antagonist. This structure of a tachykinin receptor reveals the molecular basis of antagonist interaction, and provides a framework for understanding ligand binding selectivity for this therapeutically important family of GPCRs.
Keywords: GPCR, tachykinin receptor, substance P, drug design, ligand recognition
Abstract
The NK1 tachykinin G-protein–coupled receptor (GPCR) binds substance P, the first neuropeptide to be discovered in mammals. Through activation of NK1R, substance P modulates a wide variety of physiological and disease processes including nociception, inflammation, and depression. Human NK1R (hNK1R) modulators have shown promise in clinical trials for migraine, depression, and emesis. However, the only currently approved drugs targeting hNK1R are inhibitors for chemotherapy-induced nausea and vomiting (CINV). To better understand the molecular basis of ligand recognition and selectivity, we solved the crystal structure of hNK1R bound to the inhibitor L760735, a close analog of the drug aprepitant. Our crystal structure reveals the basis for antagonist interaction in the deep and narrow orthosteric pocket of the receptor. We used our structure as a template for computational docking and molecular-dynamics simulations to dissect the energetic importance of binding pocket interactions and model the binding of aprepitant. The structure of hNK1R is a valuable tool in the further development of tachykinin receptor modulators for multiple clinical applications.
Tachykinin signaling is involved in a broad range of human physiology, and represents a therapeutic target for multiple pathologies ranging from inflammation and pain to cancer (1). Tachykinins occupy an important historical place in the study of gut–brain signaling, since substance P was first isolated as a peptide from horse brain that could induce vasodilation in rabbit intestine (2). The overall tachykinin system in mammals contains nine identified neuropeptides derived from three precursor genes that can interact with three receptor subtypes (1). Among the endogenous hormone agonists, substance P and the hemokinin/endokinin peptides bind preferentially to NK1R; neurokinin A (NKA) and elongated forms neuropeptide K and neuropeptide γ bind preferentially to NK2R; and neurokinin B (NKB) binds preferentially to NK3R. All of these validated tachykinin peptide agonists contain a conserved C-terminal motif F(F/V)GLM-NH2, which has also been characterized as the minimal active fragment of substance P (3).
The tachykinin receptors have a complex and overlapping pattern of expression throughout mammalian tissues. NK1R and NK2R are expressed in the central and peripheral nervous system, in the gastrointestinal system, and in immune cells (4, 5). An established function of NK1R signaling induced by substance P is to serve as a sensory mechanism for noxious stimuli. Thus, NK1R in the medulla controls emesis in response to ingestion of toxins, and substance P released from dorsal root ganglia neurons regulates pain transmission. NK1R and NK2R in peripheral tissues are involved in inflammatory processes, and expression of these G-protein–coupled receptors (GPCRs) is also up-regulated by inflammation (1). In contrast, expression of NK3R is largely restricted to the nervous system, and signaling induced by NKB regulates neurotransmitter systems (6) as well as secretion of reproductive hormones (GnRH and LH) (7).
Due to these diverse physiological roles, antagonists of the tachykinin receptors have been tested in clinical trials for neuropathic pain, major depressive disorder, chemotherapy-induced nausea and vomiting (CINV), irritable bowel syndrome (IBS), and schizophrenia (8). While the receptor subtypes must share structural features for ligand recognition based on the fact that they bind tachykinins containing a conserved C-terminal peptide motif, it has nonetheless been possible to develop subtype-selective antagonists that discriminate between them (4). Beyond emesis and pain, selective NK1R antagonists have been pursued in the clinic for IBS due to the proinflammatory role of substance P (1, 9). Selective NK2R antagonists have been pursued for depression and anxiety disorders, as well as IBS (10). Due to its interactions with dopaminergic neurons in the CNS, NK3R has been a clinical target for schizophrenia (11, 12). To date, the only tachykinin receptor ligands to be approved clinically are selective hNK1R antagonists for CINV, such as aprepitant (13). Despite good safety and tolerability, the preclinical promise of tachykinin receptor antagonists based on animal studies has not been matched by success in the clinic (8), especially for neuropsychiatric disorders (14). One contributing factor may be the complex overlap between expression patterns and functions of the receptors and neuropeptides. In CINV, where substance P plays a dominant role, selective NK1R blockade is sufficient. In other cases, such as IBS or depression, targeting multiple subtypes may be required for efficacy (8).
Further development of antagonists requires a deeper understanding of tachykinin signaling mechanisms underpinning disease, as well as elucidation of the molecular basis of ligand interaction and selectivity. As the primary endogenous target of substance P, hNK1R is the most extensively characterized tachykinin receptor. Using a protein engineering and lipidic cubic phase (LCP) crystallization strategy we previously developed for the orexin receptors (15), we solved the crystal structure of hNK1R in complex with the antagonist L760735 (16) at 3.4-Å resolution (Fig. 1 and SI Appendix, Table S1). With this structure, we modeled the binding mode of the clinically used drug aprepitant. Our crystal structure, modeling studies, and radioligand binding assays provide a foundation for understanding tachykinin receptor function at the atomic level.
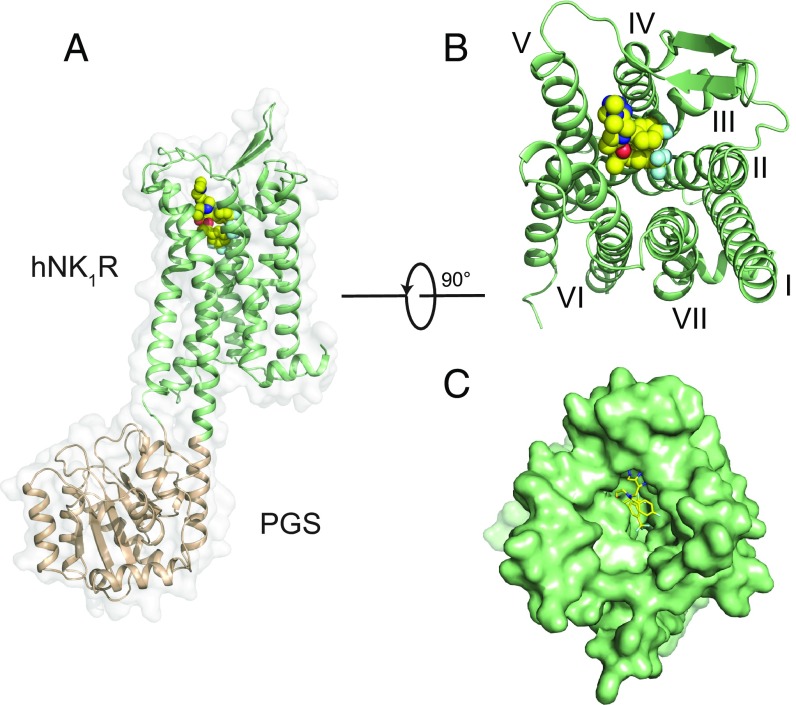
Fig. 1.
Structural features of hNK1R; (A) Global structure of the hNK1R-PGS fusion protein from side view. The hNK1R is represented as a green cartoon, with the PGS domain (wheat) fused between TMs 5 and 6. L760735 is shown as spheres with yellow carbons. (B) Extracellular view of hNK1R, with PGS domain removed. (C) Surface representation as in B.
Results
The overall architecture of hNK1R is shown in Fig. 1. Compared with OX2R (15) in the same β-branch of the rhodopsin GPCR family, the root-mean-squared deviation (RMSD) over 258 Cα atoms is 1.7 Å. For superposition with the inactive state of the β2-adrenergic receptor (β2AR) (17), the rmsd is 2.1 Å over 237 Cα atoms, with a similar conformation at the intracellular surface (SI Appendix, Fig. S3). The conformation of hNK1R in this structure is therefore consistent with the activity of bound L760735 as an hNK1R antagonist (16). Residues 170–181 in the second extracellular loop (ECL2) form a β-hairpin structure (Fig. 1 A and B), a common feature of peptide-activated GPCRs (15). This β-hairpin element forms a conserved disulfide bond with TM3 that is required for substance P binding (18), and ECL2 residues Glu172, Met174, Met181, and Ile182 have been shown to participate in recognition of the neuropeptide (19–21).
Bound Conformation of the Antagonist and Conformational Bias.
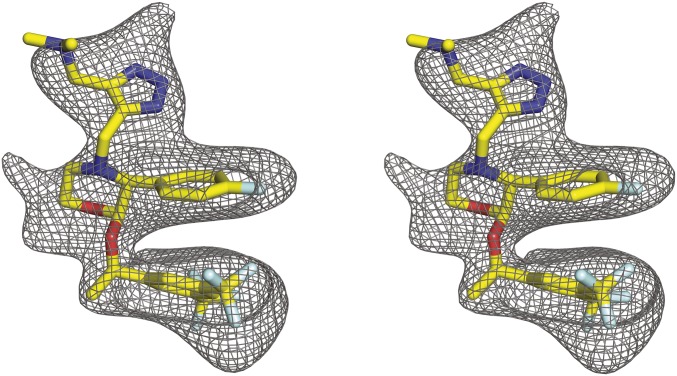
Our structure was refined using a resolution cutoff of 3.4 Å, due to challenges with signal-to-noise heterogeneity in our multicrystal dataset (SI Appendix, Table S1). Nonetheless, the position and conformation of L760735 is well defined by the ligand OMIT and 2Fo-Fc electron density maps (Fig. 2 and SI Appendix, Fig. S4A). The antagonist binds in a narrow pocket (Fig. 1C), spanning 14 Å perpendicular to the membrane and adopting a three-layered architecture with amine-substituted triazole most extracellular, morpholine and fluorophenyl moieties in the middle, and 3,5-trifluroromethyl-benzylether at the base (Fig. 3 A and B). The fluorophenyl and 3,5-trifluroromethyl-benzylether moieties form an intramolecular π-π stacking interaction, which is also plausible for other active analogs profiled in structure-activity relationship studies (22, 23). L760735 was initially developed to improve the aqueous solubility of aprepitant (16). The chemical structures of these two compounds are very similar, with the only difference being that aprepitant has a 3-oxo-substituted triazole instead of the amine-substituted triazole in L760735 (Fig. 3C). The 3D structure of aprepitant determined by small-molecule crystallography (24) is similar to the conformation of L760735 in the receptor–ligand complex, implying that these ligands can adopt a low-energy bound-like conformation on their own before engaging with the receptor. A similar phenomenon was seen in our structural characterization of the orexin receptors bound to the insomnia drug suvorexant (15), where the entropy cost of binding is reduced by restriction of the conformational space of the free ligand.
Fig. 2.
Stereoview of polder OMIT map (46) for the ligand L760735 (yellow sticks). The contour level is 3.1σ.
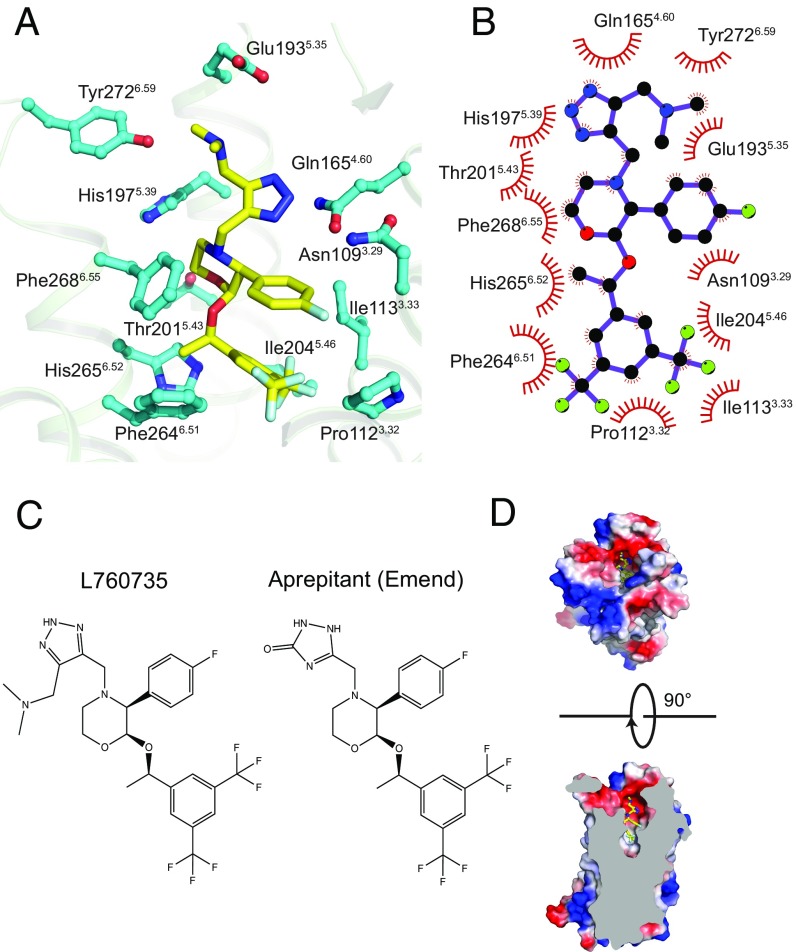
Fig. 3.
L760735 interaction with hNK1R. (A) All residues within 4 Å of L760735 are depicted with cyan carbons. (B) Two-dimensional schematic of contacts between L760735 and hNK1R, produced in Ligplot (65); (C) Chemical structures of aprepitant and L760735. (D, Top) Extracellular view, receptor surface is colored according to electrostatic potential. (D, Bottom) Side view, cut through the receptor.
Overlap Between Substance P and Antagonist Binding Sites.
Previous studies identified multiple residues involved in the binding of substance P (25–27) and synthetic small-molecule antagonists (27–31). Mutagenesis and photo-cross-linking experiments showed that substance P interacts with the extracellular surface including the N terminus and ECL2, as well as residues His1083.28, Phe2686.55, and Tyr2877.35 in the transmembrane (TM) region (19) (Ballesteros–Weinstein numbering). Of these residues, His1083.28 and Phe2686.55 are within 4 Å of L760735 in the crystal structure (Fig. 3 A and B). Our radioligand binding experiments (below) show that mutation of additional residues at the antagonist binding site also diminishes substance P binding (Table 1). The overlap between the observed binding contacts for L760735 and previously characterized mutations altering substance P binding implies that the complexed neuropeptide extends into the same deep cavity within the TM region. Many of the 12 residues lining the binding pocket (within 4 Å of the ligand) were also identified as important for interaction with other nonpeptide antagonists, implying that chemically diverse hNK1R inhibitors bind to the same site. For example, CP96345 and CP99994 were found to interact with Gln1654.60, His1975.39, and His2656.52 (27, 31), residues that also contact L760735 in the crystal structure.
Table 1.
Ligand binding properties of wild-type (WT) hNK1R and binding pocket point mutants, using [3H]-substance-P as a radioligand
| Construct | Kd, nM | Ratio of Kd | pKi (L760735) | Ratio of Ki |
| WT | 2.15 0.22 | 1.0 | 8.08 0.06 | 1.0 |
| E193A | 3.64 0.37 | 1.7* | 7.66 0.05 | 2.6 |
| Y196A | >10,000 | ND | ND | ND |
| H197A | 2.94 0.38 | 1.4* | 7.76 0.05 | 2.1 |
| H265A | 5.35 0.47 | 2.5 | 6.59 0.05 | 31 |
| Y272A | 4.51 0.55 | 2.1 | 7.60 0.08 | 3.1 |
ND, not determined. Ratios of Kd and Ki are for mutants over WT values
These differences did not rise to statistical significance judged by one-way ANOVA with Dunnett’s posttest (compared with WT, P < 0.05).
Mutagenesis of Binding Site Residues.
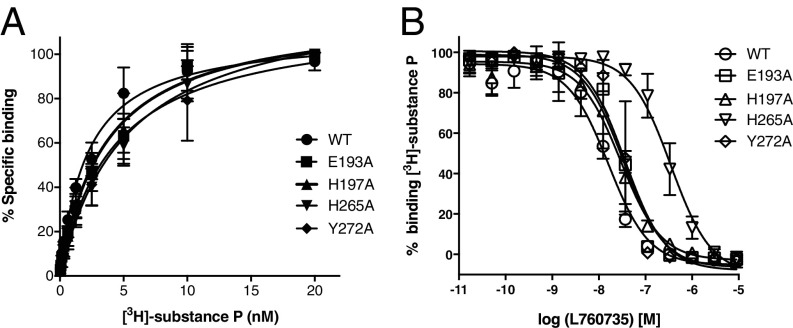
Hydrophobic contacts predominate between hNK1R and L760735 in the buried deeper part of the interface, while polar residues contact the ligand at the extracellular surface. We carried out mutagenesis experiments to validate the importance of several of these interactions. His1975.39 packs against the morpholine ring of the antagonist (Fig. 3 A and B), and mutation of this residue to alanine reduced the affinity for L760735 twofold (Fig. 4 and Table 1). For the antagonists CP96345 and CP99994, the H1975.39A mutation had a larger effect, decreasing affinity 70-fold and 10-fold, respectively (31). His2656.52 forms an edge-to-face interaction with the 3,5-trifluoromethyl-benzylether group, and the mutation H2656.52A leads to a 31-fold reduction in L760735 affinity and a smaller 2.5-fold reduction for substance P (Fig. 4 and Table 1). The Y1965.38A mutation leads to a total loss of detectable [3H]-substance P binding, indicating that while this residue contacts the antagonist, it is also important for the structural integrity of the orthosteric site. As seen by electrostatic surface calculation (Fig. 3D), hNK1R has an electronegative groove at the extracellular end of the pocket formed by Glu183ECLII, Tyr1925.34, Glu1935.35, and Tyr2726.59. The mutations E1935.35A and Y2726.59A decrease the L760735 binding affinity 2.6-fold and threefold, respectively (Fig. 4 and Table 1), indicating that interactions with the antagonist’s dimethylamino group contribute modestly to binding. These same mutations were shown to decrease the binding affinity of other hNK1R antagonists such as CP96345 and CP99994 (31).
Fig. 4.
Binding of substance P and L760735 to wild-type hNK1R and point mutants. (A) Saturation binding isotherms for [3H]-substance P. (B) Competition binding assay for L760735, using [3H]-substance P as a probe. Error bars denote SE (n = 3 separate assays, each in duplicate).
Docking of Antagonists.
To probe the molecular mechanisms of antagonist binding to hNK1R, we performed Glide docking (32) for L760735 and aprepitant in both neutral and positively charged states (Fig. 5). We calculated pKa values of 7.4 for L760735 and 5.2 for aprepitant (SI Appendix, Fig. S5 and Methods), predicting that ∼70% of free L760735 is protonated at pH 7.0 while free aprepitant is largely neutral. For L760735 in both neutral and charged forms (Fig. 5 A and B), the docking scores and rmsds between the top poses and the crystal structure were very similar: −9.50 vs. −9.37 kcal/mol (docking scores) and 1.83 vs. 1.73 Å (rmsds). The major differences in these models relative to the crystal structure are the hydrogen bonds between ligand and receptor (to Glu1935.35 and Tyr272 for neutral L760735 and to Glu1935.35 for charged L760735). Such interactions are likely to be weak in such a solvent-exposed environment, which may account for the lack of hydrogen bonds evident in the refined crystal structure, as well as the modest effect of mutating these residues (Table 1). For aprepitant (Fig. 5 C and D), the top pose for the neutral form has a docking score of −9.12 kcal/mol and rmsd 1.27 Å; however, for the charged form the second-best pose is closer to the crystal structure, with a docking score of −8.91 kcal/mL and rmsd 1.97 Å (compared with 3.67 Å for the top pose). These flexible docking calculations validate the crystallographic model and predict that aprepitant binds similarly compared with L760735; however, they do not fully address whether the ligands prefer to bind in their positively charged or neutral forms.
Fig. 5.
Top flexible docking poses for L760735 and aprepitant in both neutral and protonated states. The L760735 ligand in the crystal structure is shown as orange sticks, while the docked ligands are shown as yellow sticks. Residues that form hydrogen bonds (dashed lines) with L760735 and aprepitant are shown as cyan sticks. (A) L760735 (neutral, ΔGglide = −9.50, rmsd = 1.83 Å, best pose); (B) L760735 (charged, ΔGglide = −9.37, rmsd = 1.73 Å, best pose); (C) aprepitant (neutral, ΔGglide = −9.12, rmsd = 1.27 Å, best pose); (D) aprepitant (charged, ΔGglide = −8.91, rmsd = 1.97 Å, second best pose). Glide docking scores are in kcal/mol.
Molecular-Dynamics Simulations and Binding Energy Prediction.
To further dissect contributions to receptor-ligand binding, we performed molecular-dynamics (MD) simulations (SI Appendix, Fig. S6) for both L760735 and aprepitant complexes in explicit lipid bilayers (Methods). The starting conformations of hNK1R/L760735 in both the charged and neutral forms came from the crystal structure, while the starting conformations of hNK1R/aprepitant came from superimposing the ligand nonhydrogen atoms with those of L760735. For both ligands, the neutral form adopts a more stable conformation similar to the crystal structure along a 125-ns MD trajectory. This is particularly true with aprepitant, where the protonated ligand No-Fit rmsds (calculated directly for the heavy atoms after hNK1R structures are aligned) are much larger than the corresponding LS-Fit rmsds (simple least-squares fitting for the heavy atoms). This difference indicates that the protonated aprepitant undergoes larger translational/rotational movements in the binding pocket during the simulation.
We used data from these MD simulations to predict the binding free energy of ligands for hNK1R. MM-PBSA (molecular mechanics–Poisson-Boltzmann surface area) binding free-energy calculations and MM-GBSA (mechanics-generalized Born surface area) free-energy decompositions were performed for the collected snapshots after the MD trajectories were stabilized (Methods). Neutral forms of L760735 and aprepitant are predicted to bind with significantly higher affinity to hNK1R than their corresponding charged forms. The MM-PBSA binding free energies are −4.15 ± 0.06 and −6.07 ± 0.55 for neutral L760735 and aprepitant, respectively. In contrast, the MM-PBSA binding free energies for the charged forms of L760735 and aprepitant are 0.20 ± 0.37 and −2.5 ± 0.05 kcal/mol, respectively. This calculation predicts that while the receptor has an electronegative surface at the extracellular apex of the binding site, the ligands prefer to bind in a neutral deprotonated form due to solvent exposure of the ionizable sidechains and ligand functional groups. Our MM-PBSA calculations also predict that aprepitant is more potent than L760735, consistent with experiments (33). Hotspots for L760735 and aprepitant identified through MM/GBSA free-energy decomposition (SI Appendix, Fig. S7) show that neutral L760735 (SI Appendix, Fig. S5A) and aprepitant (SI Appendix, Fig. S5C) have very similar profiles, with 8 out of 10 top contributing residues in common.
Discussion
Our human NK1R crystal structure represents an atomic model for the tachykinin receptor family, and provides a foundation for understanding receptor–ligand interaction and selectivity. In line with previous biochemical data, our structure and mutagenesis data show that antagonists and substance P compete for an overlapping surface on the receptor. The L760735 antagonist is oriented roughly perpendicular to the membrane, with its three-layered architecture making strong interactions with residues on TMs 3, 5, and 6 (Figs. 1B and 3A) analogous to ligands for the biogenic amine receptors (17). Many antagonists of hNK1R have a basic amine substituent thought to form electrostatic interactions with the receptor; however, our docking and MD simulations indicate that L760735 and aprepitant prefer to bind in their neutral forms, which helps to explain the possibility of developing antagonists lacking such an ionizable basic group (34). At the bottom of the antagonist binding pocket lies W2616.48, a conserved residue whose movement is involved in activation for multiple class A GPCRs (35). The indole sidechain of W2616.48 is 4.1 Å away from the 3,5-trifluroromethyl-benzyl group of the antagonist, and our MD simulations indicate that this interaction contributes to the stability of the ligand-bound complex (SI Appendix, Fig. S7). Due to the ligand orientation and steric occlusion by receptor sidechains, the antagonist does not make strong contacts with TMs 2 and 7. This arrangement allows the antagonist to bind without orthosteric pocket contraction, a common step in GPCR activation (36). These features of the orthosteric region of hNK1R may underlie the dearth of small-molecule agonists for this receptor (37), a persistent challenge for many neuropeptide GPCRs.
An important goal in GPCR drug development is to understand the molecular basis of ligand selectivity, which is highly relevant to the tachykinin receptor family (8). SI Appendix, Fig. S8 shows alignment of the three human receptor subtypes, and SI Appendix, Fig. S9 illustrates the conservation of binding residues mapped onto the hNK1R orthosteric site. As in most GPCRs, the sequence identity between subtypes decreases when moving from the membrane-embedded region to the ECL region. Among the interacting residues with L760735 (Fig. 3 A and B), the only diverging positions across all subtypes are 5.35 and 6.59, with hNK2R and hNK3R having a less electronegative surface at the extracellular end of the pocket. In hNK2R, Glu1935.35 and Tyr2726.59 are leucine and serine, respectively. In hNK3R, these residues are histidine and alanine (SI Appendix, Figs. S8 and S9). Although L760935 and aprepitant likely bind in their neutral forms, our binding data still show that Glu1935.35 and Tyr2726.59 contribute to L760935 affinity (Fig. 4 and Table 1). These contributions may involve hydrogen bonding (Fig. 5 and SI Appendix, Fig. S7), although the lack of such interactions in the crystal structure points to van der Waals and solvent-mediated effects. The divergence in electrostatic surface properties in this region between the three tachykinin receptors presents opportunities for optimizing selectivity and rationally building subtype-selective ligands.
There are more than 2,200 reported ligands targeting the tachykinin receptors (38). The cocrystallized L760735 antagonist and aprepitant share a highly similar chemical structure and represent a small subset of ligands targeting these GPCRs. Further understanding the diversity of ligand recognition and subtype selectivity in this family would require structures of each tachykinin receptor subtype bound to multiple representative chemotypes. To understand hNK1R activation, we need structures of the receptor occupied by substance P in an active conformation (e.g., with G protein bound). These efforts will build on the inactive state structure reported here.
Methods
Cloning, Expression, and Purification.
The hNK1R cDNA was subcloned into a modified pFastBac 1 vector (Invitrogen) with the hemagglutinin signal sequence followed by an FLAG tag (DYKDDDDA) at the N terminus. A deca-histidine tag was added at the C terminus for purification. For successful crystallization, residues 228–237 in the intracellular loop 3 were replaced (15) by the Pyrococcus abysii glycogen synthase domain, and residues 347–407 at the C terminus were removed. Finally, residues 1–21 at the N terminus were omitted during optimization of crystal quality for diffraction. The resulting construct was transfected into Sf9 cells to produce recombinant baculovirus using the Bac-to-Bac system (Invitrogen). For large-scale purification, cultures at a density of 3 × 106 cells per milliliter were infected with 1 μM L760735 (Tocris) added to the media. After 48 h, cells were harvested and stored at −80 °C for future use.
For purification, Sf9 cells were lysed in a hypotonic buffer containing 10 mM Tris⋅HCl pH 7.4, 1 mM EDTA, 160 μg/mL benzamidine, 100 μg/mL leupeptin, 2 mg/mL iodoacetimide, and 1 μM L760735 for 30 min at 4 °C. Lysed membrane was resuspended and dounce homogenized in a buffer containing 50 mM Tris⋅HCl pH 7.4, 500 mM NaCl, 1% (wt/vol) n-dodecyl-β-d-maltopyranoside (DDM, Anatrace), 0.2% Na Cholate, 0.2% cholesteryl hemisuccinate (CHS), 10% glycerol, 2 mg/mL iodoacetamide, and 5 μM L760735. Solubilization proceeded for 1 h at 4 °C, followed by ultracentrifugation at 100,000 × g for 30 min at 4 °C. After supplementing with 20 mM imidazole, the supernatant was incubated with Ni-NTA agarose beads (GE Healthcare) in batch-binding mode for 4 h at 4 °C. After binding, the Ni-NTA beads were transferred into a glass column. Then beads were washed with 10 volumes of wash buffer: 50 mM Tris⋅HCl pH 7.4, 500 mM NaCl, 0.05% DDM, 0.01% Na Cholate, 0.01% CHS, 5% glycerol, 50 mM imidazole, 5 μM ligand (L760735). Receptor was eluted with 5 volumes of elution buffer: 50 mM Tris⋅HCl pH 7.4, 500 mM NaCl, 0.05% DDM, 0.01% Na Cholate, 0.01% CHS, 5% glycerol, 200 mM imidazole, 5 μM L760735. Then, 2 mM calcium was added to the Ni eluate, which was applied to a 3-mL column of M1 anti-FLAG affinity beads (Sigma). Detergent was exchanged on the M1 beads from DDM to 0.05% lauryl maltose neopentyl glycol (Anatrace). Receptor was eluted from M1 beads with buffer containing 200 μg/mL FLAG peptide plus 5 mM EDTA. Finally, the receptor was concentrated and purified over a Superdex 200 size exclusion column (GE Healthcare). The single peak corresponding to monomeric receptor was collected and the purity was evaluated by SDS/PAGE (SI Appendix, Fig. S1).
Crystallization and Diffraction Data Collection.
Purified protein was concentrated to 30–50 mg/mL using a Vivaspin concentrator with a 100-kDa molecular weight cutoff (Sartorius), and crystallized by LCP. Concentrated receptor was reconstituted into a lipid mixture containing 90% (wt/wt) monoolein and 10% (wt/wt) cholesterol (Sigma) at mass ratio 2:3. Crystallization experiments were carried out in 96-well glass sandwich plates (Molecular Dimensions) using a Gryphon LCP crystallization robot (Art Robbins Instruments), comprising 40 nl LCP overlaid with 800 nl precipitant solution. Plates were incubated at 20 °C. Crystals were typically observed in 1 d, and reached their full size in 1 wk. Improved crystals grew in conditions consisting of 100 mM sodium citrate pH 5.4, 30% PEG300, 200 mM potassium nitrate, 2% 2,5-Hexanediol. (SI Appendix, Fig. S2). Crystals were cryoprotected in crystallization mother liquid, harvested with 100-μm MiTeGen loops and immediately flash frozen in liquid nitrogen.
Diffraction data were collected at the 23ID-D beamline at the Advanced Photon Source, Argonne National Laboratory. All data were acquired using a 20-μm collimated minibeam with X-ray energy 12 keV, and captured on a Pilatus3 6M detector (Dectris). For each crystal, 25 images were collected with 0.4° oscillation and 1-s exposure, without attenuation of the beam. A full dataset was attained by merging datasets from 36 crystals. HKL3000 (39) was employed to index, integrate, and scale data and perform anisotropy analysis and truncation. The resolution limit was set at 3.4 Å according to the CC1/2 method. The resolution limits along the a*, b*, and c* directions after anisotropy truncation during scaling with HKL3000 were 3.9 Å, 3.7 Å and 3.4 Å, respectively.
Structure Determination and Refinement.
The structure of hNK1R-PGS was solved by molecular replacement with Phaser (40) in Phenix (41). PGS [Protein Data Bank (PDB) ID code 2BFW] and the hOX1R 7TM domain (PDB ID code 4ZJ8) were used as independent search models. The resulting solution contained one receptor and fusion protein per asymmetric unit. The model was built by autobuilding with Buccaneer (42) and iterative rebuilding in Coot (43). Initial refinement was performed with Phenix. Translation-libration-screw (TLS) refinement was employed to model atomic displacement factors with TLS groups generated by Phenix (41). The final refinement was carried out with Refmac (44) with Babinet scaling and B-factor values restrained with a weight of 0.1. Initial coordinates and refinement parameters for the ligand were prepared with the Grade web server (www.globalphasing.com). MolProbity (45) was used to evaluate the final structure. The MolProbity score was 1.74, placing the structure in the 100th percentile in the range of 3.15–3.65 Å. The statistics for data collection and refinement are included in SI Appendix, Table S1. Figures were prepared with Pymol (https://www.pymol.org/2/). Representative views of the 2Fo − Fc electron density map are shown in SI Appendix, Fig. S4, and the polder OMIT map (46) calculated for the ligand in Phenix (41) is shown in Fig. 2. Atomic coordinates and structure factors have been deposited in the PDB under accession code 6E59 (47).
Saturation and Competition Binding.
Membranes of wild-type hNK1R and point mutants were prepared from Sf9 cells, and aliquots in assay buffer (20 mM Hepes pH 8.0, HBSS) were stored at −80 °C. Radioligand binding assays were performed on a vacuum manifold. Saturation binding assay was carried out by incubating membranes (typically 2–15 μg) with different concentrations of [3H]-substance P in a final volume of 250 μL HBSS buffer with 20 mM Hepes pH 8.0 and 0.5% BSA. The full name of the radioligand is “substance-P (9-Sar, 11-Met(O2)), [2-Prolyl-3,4-3H]” (44.4 Ci/mmol; Perkin-Elmer). Nonspecific binding was determined with reactions containing 1 μM CP99994 (Tocris). Binding proceeded for 1 h at 4 °C. Reactions were passed through Whatman GF/C filters presoaked in assay buffer plus 0.5% polyethyleninine (Sigma P-3143) to retain membranes and remove unbound ligand. After washing with cold HBSS buffer three times, bound radioactivity was quantified with a scintillation counter. For competition binding experiments, membranes (typically 2–15 μg) were incubated with 2 nM [3H]-substance P and serially diluted competitor ligand (L760735). The reaction protocol and workup were the same as for the saturation binding assay. All binding studies were carried out as three independent experiments, each performed in duplicate. Data analysis and fitting was performed with Prism (GraphPad Software).
Molecular Docking and MD Simulations.
The pKa calculations were performed using Epik (Schrodinger) (48). Molecular docking for the two antagonists was performed using Glide (Schrodinger) following standard protocols (32). For each MD simulation, CHARMM-GUI (www.charmm-gui.org) was used to add a POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) lipid bilayer, counterions, and water molecules. All MD systems have one copy of hNK1R, one copy of the ligand, 240 POPC, 51 K+, 59 Cl−, and 17,862 TIP3P (49) water molecules. For the force-field parameters, the partial atomic charges of two ligands were derived by using the RESP (50) program to fit the HF/6–31G* electrostatic potentials generated with the GAUSSIAN 16 software package. The other force-field parameters came from GAFF (51) in AMBER 14 (52). The residue topologies for ligands were prepared using the Antechamber module (53) in AMBER 14. The AMBER FF14SB (54) and LIPID14 (55) force fields were used to model proteins and lipids, respectively. MD simulations were performed to produce isothermal–isobaric ensembles using the modified pmemd.cuda program in AMBER 14. The particle mesh Ewald method (56) was used to calculate the full electrostatic energy of a unit cell in a macroscopic lattice of repeating images. All bonds were constrained using the SHAKE algorithm (57) in both the minimization and MD simulation stages. For each MD simulation, the system was first relaxed to remove any possible steric clashes by a set of 10,000-step minimizations with the main chain atoms restrained using the harmonic restraint force constants decreased from 20 to 10, 5, and 1 kcal/mol/Å2, progressively. After that, the system was further relaxed by a 10,000-step minimization without any constraints or restraints. There were three phases for the subsequent MD simulations: the relaxation phase, the equilibrium phase, and the sampling phase. In the relaxation phase, the simulation system was heated up progressively from 50 to 250 K at steps of 50 K. At each temperature, a 1-ns MD simulation was run without any restraints or constraints. In the following equilibrium phase, the system was equilibrated at 298 K, 1 bar for 20 ns. Finally, a 100-ns MD simulation was performed at 298 K, 1 bar to produce constant temperature and pressure ensembles. In total, 1,000 snapshots were recorded from the last simulation. Two hundred snapshots were evenly selected from the sampling phase for the MM-PBSA binding free-energy calculation. For the crystal structure of hNK1R/L76035, 3,000 snapshots were collected and 600 were evenly selected for the MM-PBSA binding free-energy calculation. Temperature was regulated using Langevin dynamics (58) with a collision frequency of 5 ps−1; pressure was regulated using the isotropic position scaling algorithm with the pressure relaxation time set to 1.0 ps; integration of the equations of motion was conducted at a time step of 1 fs for the relaxation phase and 2 fs for the equilibrium and sampling phases.
MM-PBSA and MM-GBSA.
For each MD snapshot, the molecular mechanical (MM) energy (EMM) and the MM-PBSA or MM-GBSA solvation free energy were calculated (59–61) with the following key parameters: external dielectric constant, ∼80; internal dielectric constant, ∼1; and the surface tension for estimating the nonpolar solvation energy, ∼0.054. The Parse radii (62) were used in the MM-PB/SA solvation calculation using the Delphi package (compbio.clemson.edu/delphi). The entropic term was estimated using a method described previously (63). Free-energy decompositions were performed for all of the snapshots collected in the sampling phases of the MD simulations. The interaction energies between each residue and the ligand were calculated using an MM-GBSA solvation model developed previously (64).
Supplementary Material
Acknowledgments
We thank the staff of the GM/CA-CAT beamline 23ID at the Advanced Photon Source (APS) for support during data collection. This project was supported by an Edward Mallinckrodt, Jr. Foundation Scholar Award (to D.M.R.), the Welch Foundation [Grant I-1770 (to D.M.R.)], and the National Institutes of Health [R01-GM117080 (to D.B.), R01-GM079383 and R21-GM097617 (to J.W.), and R01-NS103939 (to D.M.R.)]. APS is a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory (DE-AC02-06CH11357).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6E59).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812717115/-/DCSupplemental.
References
- 1.Steinhoff MS, von Mentzer B, Geppetti P, Pothoulakis C, Bunnett NW. Tachykinins and their receptors: Contributions to physiological control and the mechanisms of disease. Physiol Rev. 2014;94:265–301. doi: 10.1152/physrev.00031.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.V Euler US, Gaddum JH. An unidentified depressor substance in certain tissue extracts. J Physiol. 1931;72:74–87. doi: 10.1113/jphysiol.1931.sp002763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michelot R, Mayer M, Magneney S, Thierry J, Potier P. Substance P and Neurokinins. Springer; New York: 1987. Activity of the C-terminal part in tachykinins on guinea pig ileum and trachea preparations; pp. 158–160. [Google Scholar]
- 4.Almeida TA, et al. Tachykinins and tachykinin receptors: Structure and activity relationships. Curr Med Chem. 2004;11:2045–2081. doi: 10.2174/0929867043364748. [DOI] [PubMed] [Google Scholar]
- 5.Douglas SD, Leeman SE. Neurokinin-1 receptor: Functional significance in the immune system in reference to selected infections and inflammation. Ann N Y Acad Sci. 2011;1217:83–95. doi: 10.1111/j.1749-6632.2010.05826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langlois X, Wintmolders C, te Riele P, Leysen JE, Jurzak M. Detailed distribution of neurokinin 3 receptors in the rat, guinea pig and gerbil brain: A comparative autoradiographic study. Neuropharmacology. 2001;40:242–253. doi: 10.1016/s0028-3908(00)00149-0. [DOI] [PubMed] [Google Scholar]
- 7.Lehman MN, Coolen LM, Goodman RL. Minireview: Kisspeptin/neurokinin B/dynorphin (KNDy) cells of the arcuate nucleus: A central node in the control of gonadotropin-releasing hormone secretion. Endocrinology. 2010;151:3479–3489. doi: 10.1210/en.2010-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quartara L, Altamura M, Evangelista S, Maggi CA. Tachykinin receptor antagonists in clinical trials. Expert Opin Investig Drugs. 2009;18:1843–1864. doi: 10.1517/13543780903379530. [DOI] [PubMed] [Google Scholar]
- 9.O’Connor TM, et al. The role of substance P in inflammatory disease. J Cell Physiol. 2004;201:167–180. doi: 10.1002/jcp.20061. [DOI] [PubMed] [Google Scholar]
- 10.Lecci A, Capriati A, Maggi CA. Tachykinin NK2 receptor antagonists for the treatment of irritable bowel syndrome. Br J Pharmacol. 2004;141:1249–1263. doi: 10.1038/sj.bjp.0705751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meltzer HY, Arvanitis L, Bauer D, Rein W. Meta-Trial Study Group Placebo-controlled evaluation of four novel compounds for the treatment of schizophrenia and schizoaffective disorder. Am J Psychiatry. 2004;161:975–984. doi: 10.1176/appi.ajp.161.6.975. [DOI] [PubMed] [Google Scholar]
- 12.Dawson LA, et al. In vitro and in vivo characterization of the non-peptide NK3 receptor antagonist SB-223412 (talnetant): Potential therapeutic utility in the treatment of schizophrenia. Neuropsychopharmacology. 2008;33:1642–1652. doi: 10.1038/sj.npp.1301549. [DOI] [PubMed] [Google Scholar]
- 13.Tattersall FD, et al. The novel NK1 receptor antagonist MK-0869 (L-754,030) and its water soluble phosphoryl prodrug, L-758,298, inhibit acute and delayed cisplatin-induced emesis in ferrets. Neuropharmacology. 2000;39:652–663. doi: 10.1016/s0028-3908(99)00172-0. [DOI] [PubMed] [Google Scholar]
- 14.Griebel G, Holsboer F. Neuropeptide receptor ligands as drugs for psychiatric diseases: The end of the beginning? Nat Rev Drug Discov. 2012;11:462–478. doi: 10.1038/nrd3702. [DOI] [PubMed] [Google Scholar]
- 15.Yin J, Mobarec JC, Kolb P, Rosenbaum DM. Crystal structure of the human OX2 orexin receptor bound to the insomnia drug suvorexant. Nature. 2015;519:247–250. doi: 10.1038/nature14035. [DOI] [PubMed] [Google Scholar]
- 16.Harrison T, et al. An orally active, water-soluble neurokinin-1 receptor antagonist suitable for both intravenous and oral clinical administration. J Med Chem. 2001;44:4296–4299. doi: 10.1021/jm0109558. [DOI] [PubMed] [Google Scholar]
- 17.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 18.Elling CE, Raffetseder U, Nielsen SM, Schwartz TW. Disulfide bridge engineering in the tachykinin NK1 receptor. Biochemistry. 2000;39:667–675. doi: 10.1021/bi991777b. [DOI] [PubMed] [Google Scholar]
- 19.Valentin-Hansen L, et al. Mapping substance P binding sites on the neurokinin-1 receptor using genetic incorporation of a photoreactive amino acid. J Biol Chem. 2014;289:18045–18054. doi: 10.1074/jbc.M113.527085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lequin O, et al. Involvement of the second extracellular loop (E2) of the neurokinin-1 receptor in the binding of substance P. Photoaffinity labeling and modeling studies. J Biol Chem. 2002;277:22386–22394. doi: 10.1074/jbc.M110614200. [DOI] [PubMed] [Google Scholar]
- 21.Macdonald D, et al. Photoaffinity labeling of mutant neurokinin-1 receptors reveals additional structural features of the substance P/NK-1 receptor complex. Biochemistry. 2001;40:2530–2539. doi: 10.1021/bi001880x. [DOI] [PubMed] [Google Scholar]
- 22.Lowe JA, III, et al. N-alkyl quinuclidinium substance P antagonists. Bioorg Med Chem Lett. 1994;4:1153–1156. [Google Scholar]
- 23.Swain CJ, et al. Identification of a series of 3-(benzyloxy)-1-azabicyclo[2.2.2]octane human NK1 antagonists. J Med Chem. 1995;38:4793–4805. doi: 10.1021/jm00024a007. [DOI] [PubMed] [Google Scholar]
- 24.Hale JJ, et al. Structural optimization affording 2-(R)-(1-(R)-3, 5-bis(trifluoromethyl)phenylethoxy)-3-(S)-(4-fluoro)phenyl-4- (3-oxo-1,2,4-triazol-5-yl)methylmorpholine, a potent, orally active, long-acting morpholine acetal human NK-1 receptor antagonist. J Med Chem. 1998;41:4607–4614. doi: 10.1021/jm980299k. [DOI] [PubMed] [Google Scholar]
- 25.Fong TM, Huang RR, Strader CD. Localization of agonist and antagonist binding domains of the human neurokinin-1 receptor. J Biol Chem. 1992;267:25664–25667. [PubMed] [Google Scholar]
- 26.Lundstrom K, et al. Effect of single point mutations of the human tachykinin NK1 receptor on antagonist affinity. Eur J Pharmacol. 1997;337:73–81. doi: 10.1016/s0014-2999(97)01226-0. [DOI] [PubMed] [Google Scholar]
- 27.Turcatti G, et al. Characterization of non-peptide antagonist and peptide agonist binding sites of the NK1 receptor with fluorescent ligands. J Biol Chem. 1997;272:21167–21175. doi: 10.1074/jbc.272.34.21167. [DOI] [PubMed] [Google Scholar]
- 28.Fong TM, et al. Amino-aromatic interaction between histidine 197 of the neurokinin-1 receptor and CP 96345. Nature. 1993;362:350–353. doi: 10.1038/362350a0. [DOI] [PubMed] [Google Scholar]
- 29.Gether U, Nilsson L, Lowe JA, 3rd, Schwartz TW. Specific residues at the top of transmembrane segment V and VI of the neurokinin-1 receptor involved in binding of the nonpeptide antagonist CP 96,345 [corrected] J Biol Chem. 1994;269:23959–23964, and erratum (1994) 269:32708. [PubMed] [Google Scholar]
- 30.Cascieri MA, et al. Characterization of the interaction of diacylpiperazine antagonists with the human neurokinin-1 receptor: Identification of a common binding site for structurally dissimilar antagonists. Mol Pharmacol. 1995;47:660–665. [PubMed] [Google Scholar]
- 31.Greenfeder S, et al. Two related neurokinin-1 receptor antagonists have overlapping but different binding sites. Bioorg Med Chem. 1998;6:189–194. doi: 10.1016/s0968-0896(97)10019-0. [DOI] [PubMed] [Google Scholar]
- 32.Friesner RA, et al. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 33.Hanessian S, et al. Design and synthesis of potential dual NK(1)/NK(3) receptor antagonists. Bioorg Med Chem Lett. 2014;24:510–514. doi: 10.1016/j.bmcl.2013.12.033. [DOI] [PubMed] [Google Scholar]
- 34.Hoffmann T, et al. Design and synthesis of a novel, achiral class of highly potent and selective, orally active neurokinin-1 receptor antagonists. Bioorg Med Chem Lett. 2006;16:1362–1365. doi: 10.1016/j.bmcl.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 35.Holst B, et al. A conserved aromatic lock for the tryptophan rotameric switch in TM-VI of seven-transmembrane receptors. J Biol Chem. 2010;285:3973–3985. doi: 10.1074/jbc.M109.064725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rasmussen SGF, et al. Structure of a nanobody-stabilized active state of the β(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cappelli A, et al. A non-peptide NK1 receptor agonist showing subpicomolar affinity. J Med Chem. 2004;47:1315–1318. doi: 10.1021/jm034219a. [DOI] [PubMed] [Google Scholar]
- 38.Vass M, et al. Chemical diversity in the G protein-coupled receptor superfamily. Trends Pharmacol Sci. 2018;39:494–512. doi: 10.1016/j.tips.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cowtan K. Fitting molecular fragments into electron density. Acta Crystallogr D Biol Crystallogr. 2008;64:83–89. doi: 10.1107/S0907444907033938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liebschner D, et al. Polder maps: Improving OMIT maps by excluding bulk solvent. Acta Crystallogr D Struct Biol. 2017;73:148–157. doi: 10.1107/S2059798316018210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin J, et al. 2018 Crystal structure of the human NK1 tachykinin receptor. Protein Data Bank. Available at https://www.rcsb.org/structure/6E59. Deposited July 20, 2018.
- 48.Shelley JC, et al. Epik: A software program for pK(a) prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des. 2007;21:681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- 49.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 50.Bayly CI, Cieplak P, Cornell W, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J Phys Chem. 1993;97:10269–10280. [Google Scholar]
- 51.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 52.Case DA, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 54.Maier JA, et al. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dickson CJ, et al. Lipid14: The amber lipid force field. J Chem Theory Comput. 2014;10:865–879. doi: 10.1021/ct4010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Darden T, Perera L, Li L, Pedersen L. New tricks for modelers from the crystallography toolkit: The particle mesh Ewald algorithm and its use in nucleic acid simulations. Structure. 1999;7:R55–R60. doi: 10.1016/s0969-2126(99)80033-1. [DOI] [PubMed] [Google Scholar]
- 57.Miyamoto S, Kollman PA. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem. 1992;13:952–962. [Google Scholar]
- 58.Larini L, Mannella R, Leporini D. Langevin stabilization of molecular-dynamics simulations of polymers by means of quasisymplectic algorithms. J Chem Phys. 2007;126:104101. doi: 10.1063/1.2464095. [DOI] [PubMed] [Google Scholar]
- 59.Hou T, Wang J, Li Y, Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model. 2011;51:69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hou T, Wang J, Li Y, Wang W. Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized Born surface area methods. II. The accuracy of ranking poses generated from docking. J Comput Chem. 2011;32:866–877. doi: 10.1002/jcc.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu L, Sun H, Li Y, Wang J, Hou T. Assessing the performance of MM/PBSA and MM/GBSA methods. 3. The impact of force fields and ligand charge models. J Phys Chem B. 2013;117:8408–8421. doi: 10.1021/jp404160y. [DOI] [PubMed] [Google Scholar]
- 62.Sitkoff D, Sharp KA, Honig B. Accurate calculation of hydration free energies using macroscopic solvent models. J Phys Chem. 1994;98:1978–1988. [Google Scholar]
- 63.Wang J, Hou T. Develop and test a solvent accessible surface area-based model in conformational entropy calculations. J Chem Inf Model. 2012;52:1199–1212. doi: 10.1021/ci300064d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hawkins GD, Cramer CJ, Truhlar DG. Parametrized models of aqueous free energies of solvation based on pairwise descreening of solute atomic charges from a dielectric medium. J Phys Chem. 1996;100:19824–19839. [Google Scholar]
- 65.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.