Abstract
Background
Basal stem rot (BSR) caused by hemibiotroph Ganoderma boninense is a devastating disease resulting in a major loss to the oil palm industry. Since there is no physical symptom in oil palm at the early stage of G. boninense infection, characterisation of molecular defense responses in oil palm during early interaction with the fungus is of the utmost importance. Oil palm (Elaeis guineensis) seedlings were artificially infected with G. boninense inoculums and root samples were obtained following a time-course of 0, 3, 7, and 11 days-post-inoculation (d.p.i) for RNA sequencing (RNA-seq) and identification of differentially expressed genes (DEGs).
Results
The host counter-attack was evidenced based on fungal hyphae and Ganoderma DNA observed at 3 d.p.i which became significantly reduced at 7 and 11 d.p.i. DEGs revealed upregulation of multifaceted defense related genes such as PR-protein (EgPR-1), protease inhibitor (EgBGIA), PRR protein (EgLYK3) chitinase (EgCht) and expansin (EgEXPB18) at 3 d.p.i and 7 d.p.i which dropped at 11 d.p.i. Later stage involved highly expressed transcription factors EgERF113 and EgMYC2 as potential regulators of necrotrophic defense at 11 d.p.i. The reactive oxygen species (ROS) elicitor: peroxidase (EgPER) and NADPH oxidase (EgRBOH) were upregulated and maintained throughout the treatment period. Growth and nutrient distribution were probably compromised through suppression of auxin signalling and iron uptake genes.
Conclusions
Based on the analysis of oil palm gene expression, it was deduced that the biotrophic phase of Ganoderma had possibly occurred at the early phase (3 until 7 d.p.i) before being challenged by the fungus via switching its lifestyle into the necrotrophic phase at later stage (11 d.p.i) and finally succumbed the host. Together, the findings suggest the dynamic defense process in oil palm and potential candidates that can serve as phase-specific biomarkers at the early stages of oil palm-G. boninense interaction.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1594-9) contains supplementary material, which is available to authorized users.
Keywords: Early defense, Elaeis guineensis, Ganoderma boninense, Necrotrophic, Pathogenesis-related protein, Transcription factor
Background
Oil palm (Elaeis guineensis Jacq. Dura x Pisifera) is one of the main plantation crops in Malaysia and Indonesia and together these two countries contribute about 85 to 90% of global export [1, 2]. Palm oil which is recognised as one of the major sources of edible oil also serves as feedstock for oleochemicals and precursor for biodiesel fuel [3, 4]. The total export earnings from palm oil and palm oil products in Malaysia was reported at nearly USD18.5 billion [5]. Despite the huge export revenue from this commodity, oil palm plantation is facing major predicament due to basal stem rot (BSR) disease which hampers the oil palm production massively. BSR has been reported as a major threat in oil palm industry for over eight decades. It was estimated that in 2020, a total area of 443,430 ha or 65.6 millions of palm trees will be affected [6]. BSR is mainly caused by fungal infection on intact oil palm roots wherein the most prevalent species discovered was Ganoderma boninense [7–9]. BSR infects not only mature oil palm trees but also seedlings and younger plants where manifestation of the disease occurs earlier and more severe [8]. BSR is manifested by progressive decay of roots that disrupts water and nutrient transport to the upper part of mature oil palm trees which concomitantly will bring about frond wilting, yellowing of frond, un-opening of spear leaves and eventually resulting in stand collapse [10]. Regrettably, BSR-infected oil palms are symptomless during early stage of infection with the earliest symptom often observed on foliage when infection has progressed by 60–70% [11]. Once young oil palm plants show symptom of the disease they usually die within 1 or 2 years, while mature trees can survive for only another 3 or so years [12]. Thus, studies on early defense response are not just time and cost effective but provide insightful information on initiation of defense signaling networks upon recognition of pathogen.
Ganoderma spp. has been categorised as hemibiotrophs, with intermediate lifestyle of biotrophs and necrotrophs. Early stage of infection is the biotrophic phase whereby colonization of fungal on intact host plant cells takes place before initiating necrotrophic phase that involves extensive cell wall degradation [11]. Biotrophs survive by maintaining intact host cells for nutrient uptake, whilst necrotrophs involve killing of plant cells to infect and survive saprotrophically. Biotrophic infection is common during early interaction with pathogens whereby plant counteracts by enhancing production of reactive oxygen species (ROS) through an oxidative burst [13]. Consequently, plant executes programmed cell death (PCD) to restrict pathogen growth. This phenomenon is a form of hypersensitive response (HR) in which plant promotes cell death at and around the infection site [14]. Biotrophs utilize small amounts of cell wall degrading enzymes (CWDEs) to allow softening and loosening of cell wall without causing lethal effect to host cells [15]. Early defense response is also highly related to enhanced lignification of cell walls by plant to combat localized and controlled degradation of the cell wall by biotrophic fungi [16]. It is one of the strategies that plants employ to prevent penetration of pathogen’s toxins through cell wall degradation by CWDEs [17].
Regrettably, HR only induces transition of biotrophic to necrotrophic stage. Necrotrophs invade host tissues by extensively secreting CWDEs. The accessibility of CWDEs on cell wall is achieved by perception of necrotrophs to subvert host cell wall modification [18]. For instance, coactions of expansin and polygalacturonase which facilitate cell wall loosening were induced upon successful infection of necrotrophic pathogen Botrytis cinerea on Solanum lycopersicum [19]. Necrotroph also produces expansin-like protein to mediate penetration of hyphae [20]. Expansin-like protein provides protection for the hemibiotrophs, Fusarium graminearum from plant enzymatic degradation [21]. It has been postulated that biotrophic colonization is obligatory for hemibiotrophs to mediate successful infection while the time-period for switching from biotrophy to necrotrophy varies between pathogenic species [22]. Having intermediate lifestyle, hemibiotrophs may first overcome plant defense response during early colonization and subsequently deploy a more aggressive mode of attack for successful infection [23].
Studies on early interactions of plant-pathogens are crucial to allow screening for detection of potential threat of BSR especially on young palms. The present study attempts to investigate gene expression patterns in susceptible progeny of commercial oil palm (Dura x Pisifera) at early stages of G. boninense infection using high-throughput bioinformatics data (RNA-seq) via next generation sequencing (NGS) method. Despite the existence of a resistant variety (Zaire x Cameroon) [24], we decided to use the commercialized susceptible variety as it is vastly planted in oil palm plantations because this hybrid produces better yield performance compared to their parents [25, 26]. Previous study from our laboratory [27] using similar method and condition of treatment reported induced production of metabolites with anti-fungal properties in oil palm seedlings during early interaction (within a week of infection) with G. boninense suggesting activation of early defense responses in the host plant. Based on marker genes reported on biotrophs and necrotrophs, our study was able to differentiate the biotrophic stage before switching to necrotrophic phase which occurs later. The present work differs from previous reported studies which covered the later stages at three weeks post inoculation onwards [28, 29]. This work will enable a more complete understanding of oil palm defence response and is important for potentially early intervening strategies to protect the plant from severe infection.
Results
Preliminary screening of early defense response in G. boninense-infected oil palm roots
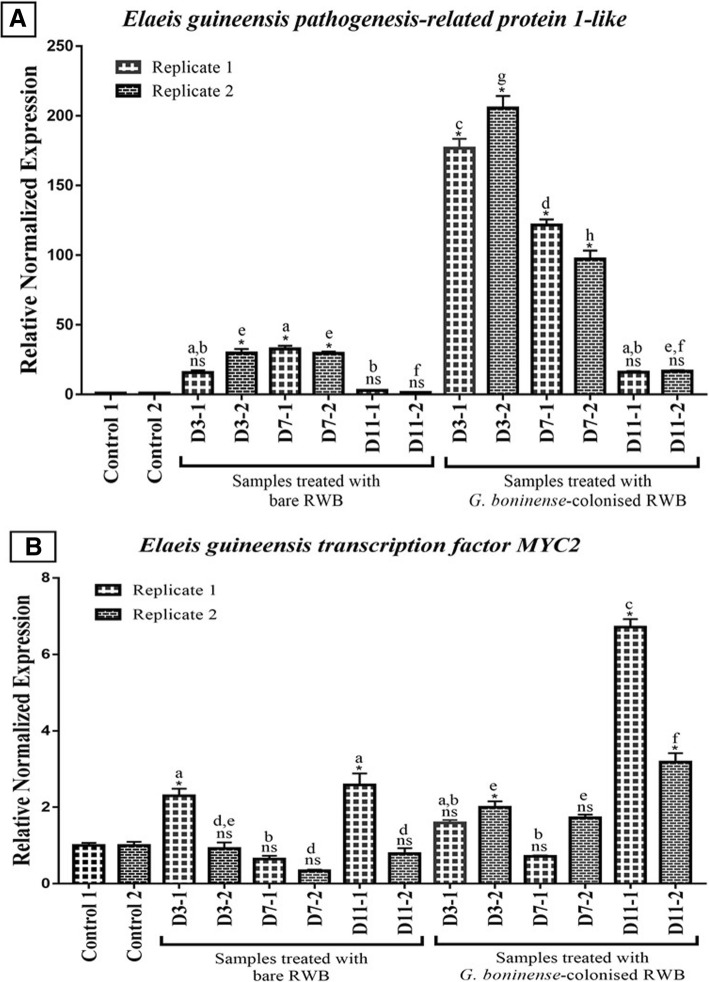
Eighty-four of 4-month-old oil palm seedlings were divided into two treatments which were inoculation with bare RWB (no fungal inoculum) as mock treatment (T0) and inoculation with RWB fully colonised with G. boninense (T1). Artificial infection of oil palm seedlings with G. boninense was performed via sitting-technique to mimic the mode of Ganoderma spp. infection through root contact with fungal mycelia [27]. T0 and T1 samples were harvested at 3, 7 and 11 d.p.i. while untreated seedlings were used as control. Our preliminary screening via real-time quantitative PCR (qPCR) of transcriptional regulation in oil palm-G. boninense interaction showed two distinct phases of fungal attack suggesting early (3 d.p.i) and later (11 d.p.i) defense mechanisms (Fig. 1). The expression of pathogenesis-related protein 1 (PR-1) and transcription factor MYC2 (MYC2) genes, which are common genetic biomarkers for biotic stress were analysed [30–34]. EgPR1 was highly expressed during the early phase of infection at 3 and 7 d.p.i before subsequently reduced at 11 d.p.i. Whereas EgMYC2 showed highest gene expression during later phase of infection at 11 d.p.i. Based on the preliminary screening, we suggested that there are two plausible phases of defense response primed by oil palm expressing at very early (3 d.p.i) and later (11 d.p.i) interaction with G. boninense. Hence, the same batch of root samples harvested at the time points (3, 7 and 11 d.p.i) were used for further transcriptomic analysis through high throughput NGS.
Fig. 1.
Preliminary screening of EgPR1 and EgMYC2 gene expression in G. boninense-infected oil palm roots. Histogram shows relative expressions of a pathogenesis-related protein 1 (EgPR1) and b transcription factor MYC2 (EgMYC2) genes at 3, 7, and 11 days-post-inoculation (d.p.i) compared to untreated control (c). The expressions of each gene were normalized by reference genes; GAPDH 2, NADH 5 and ß-actin expression levels. Data are expressed as the mean ± SEM of three individual technical replicates of each sample. Preliminary screening by qPCR was carried out on control and treated (T0 and T1) samples in two biological replicates (1 and 2). Each replicate consisted of pooled root from six plants. * P < 0.01 is significantly differed compared to corresponding control as assessed by one-way ANOVA analysis followed by Tukey’s test. ns is not significant. Different superscript letters between samples (within replicate) indicate significant different (P < 0.01) in mean values. RWB: Rubber wood block
Scanning electron microscopy and PCR using Ganoderma-specific primers performed on artificially infected oil palm roots
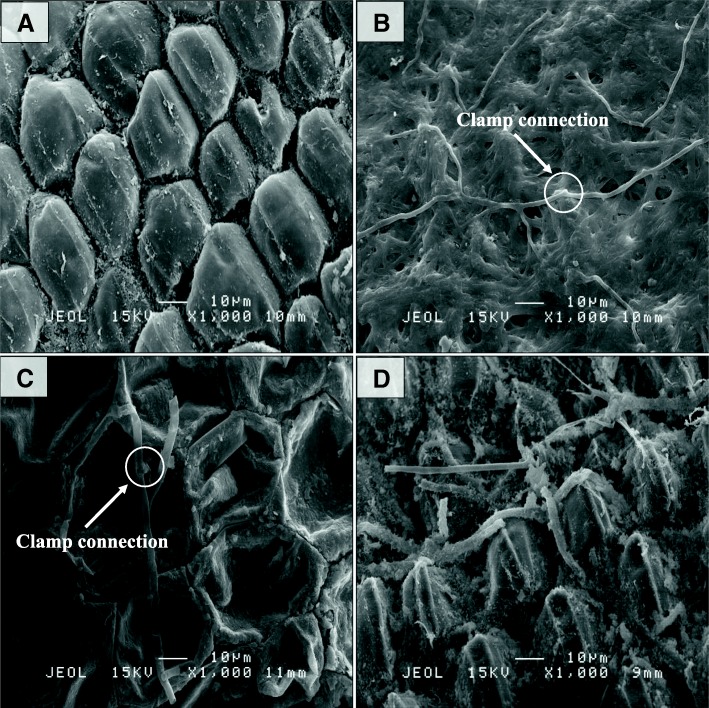
Scanning electron microscopy on the outer layer of oil palm roots revealed differences in morphology between control and T1 samples. The superficial layer of control samples was intact and healthy, whilst the cell walls of Ganoderma-treated (T1 samples) were observed to be shrunken with uneven shape that showed symptoms of necrosis or apoptosis as early as 3 d.p.i (Fig. 2). The fungal hyphae network was undetected on control samples while surprisingly thick multilayers of fungal hyphae was present on the surface of 3 d.p.i roots but significantly reduced on root samples harvested at 7 and 11 d.p.i.
Fig. 2.
Scanning electron microscopy (SEM) of control and T1 oil palm roots with Ganoderma boninense. Root of samples were imaged at different days of post inoculation (d.p.i): (a) 0 d.p.i, (b) 3 d.p.i, (c) 7 d.p.i and (d) 11 d.p.i
The presence of G. boninense DNA in oil palm roots at 3, 7 and 11 d.p.i of T1 samples was further validated by PCR using Ganoderma-specific PCR primers. Primers of G. boninense DNA were retrieved from Genebank (accession number KM015454.1) from sequence of G. boninense strain PER71 internal transcribed spacer 1 (ITS1), partial sequence; 5.8S ribosomal RNA gene, complete sequence; and internal transcribed spacer 2 (ITS2), partial sequence with the expected product length of 223 bp size. The gel electrophoresis image in Fig. 3 shows concentrated and clear band on lane T1D3 (normal PCR for 3 d.p.i root sample). Bands of amplicons were not detected on normal PCR for 7 and 11 d.p.i. root samples. Hence, nested PCR was performed using both 7 and 11 d.p.i samples to validate any trace amount of Ganoderma sp. DNA. Nested primers were generated from sequenced PCR product of 3 d.p.i sample and the resulting amplicon length was decreased to 102 bp. Nested PCR (Fig. 3) resulted in faint bands for both root samples of 7 d.p.i (lane T1D7n) and 11 d.p.i (T1D11n). No band was detected from untreated control (lane C) sample. The alignment of original G. boninense ITS1/2 sequence with sequenced amplicon of normal and nested PCR showed conserved sequence which confirmed that G. boninense fungal hyphae was present in all T1 samples (Additional file 1). Furthermore, homology searches of the sequenced amplicon with biological sequences in GenBank matched only with Ganoderma sp. sequences with ≥95% identity. Consistent with the microscopic data, it was confirmed that G. boninense hyphae at 3 d.p.i was abundant whilst much reduced in 7 and 11 d.p.i.
Fig. 3.
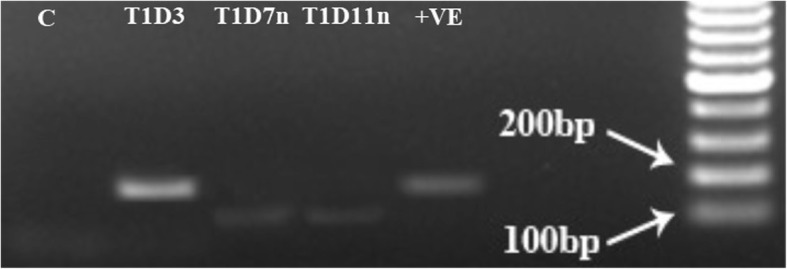
PCR amplification of Ganoderma species DNA using specific primer pairs of Ganoderma species. Lane C: uninoculated control; Lane T1D3: normal PCR for 3 d.p.i; Lane T1D7n: nested PCR for 7 d.p.i; lane T1D11n: nested PCR for 11 d.p.i; +VE: normal PCR for pure Ganoderma culture. The amplicon size for T1D3 and + VE are 223 bp whilst T1D7n and T1D11n amplicon size are 102 bp
Observation on extended period of infection resulted in wilted leaves and emergence of fruiting body (basidiomata) at the bole of the T1 samples at the 24th week post inoculation, indicating a well-established necrotrophic infection (Fig. 4). An excised bole of the plant showed a decayed region indicating symptoms of necrosis while uninfected plant was healthy without G. boninense mass or necrotic lesions. Based on the observation of fungal hyphae on the outer layer of infected oil palm roots, the thick hyphae multilayers on 3 d.p.i which significantly dropped at 7 and 11 d.p.i raised possibilities that the fungus had been weakened by the plant. However, emergence of basidiomata at chronic infection proved that the plant finally succumbed to the disease, thus it shows that the drop of fungal hyphae at early interaction (within the treatment period) is not an indicator that the plant had overcome the fungal threats. We hypothesize that the switching to a more aggressive mode of attack by the fungus plays a critical role. Hence, we profiled transcriptomes of oil palm roots during interactions with G. boninense at 3, 7, and 11 d.p.i. to test the hypothesis.
Fig. 4.
Signs and symptoms of Ganoderma boninense infection on oil palm seedlings. (a) healthy plant (uninfected) at 24 weeks after inoculation. (b) Appearance of G. boninense basidiomata on T1 oil palm stem base at 24 weeks after inoculation. (c) Stem base section of untreated control and (d) necrotic lesion (black arrows) in stem base of T1 oil palm at 24 weeks after inoculation
Transcriptomes of oil palm root during interactions with G. boninense at 3, 7, and 11 d.p.i
Biologically averaged samples are commonly practised in RNA-seq where pooled RNA samples, like in the present study from six oil palm seedlings were sequenced instead of individual samples. Biological averaging is not only cost efficient compared to mathematical averaging, but this method also could reduce the high biological variability which may be present among individual samples and raise the capability to detect differential gene expression between groups [35, 36].
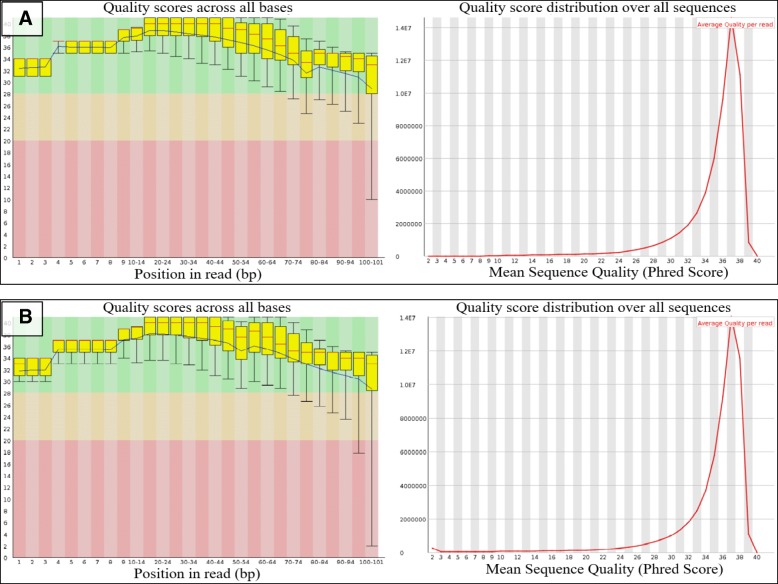
The RNA-seq generated 227,658,752 paired-end reads from the pooled two biological replicates of control, whilst the pooled biological replicates from 3, 7, and 11 d.p.i produced 227,400,216, 207,826,416 and 191,359,826 paired-end reads, respectively (Table 1) denoting that the pool of mRNAs in T1 samples decreased over time. Quality of the mRNA fragments from both biological replicates was measured using FastQC (Fig. 5a and b). An overview of quality values across all bases showed that the mean quality (blue line) for each base position lies in very good quality calls (green) region within the range of 28–38 quality scores. Furthermore, the quality score distribution graph showed that the highest number of sequences had mean sequence quality of 37. These paired-end reads were mapped to E. guineensis reference genome using Geneious for RNA-seq tool in the Geneious package. The percentages of mapped reads from samples were in the range of 54.14–60.21% with an average of 57.4%.
Table 1.
Summary of NGS data of T1 samples
| Replicate | Sample | Paired-end clean reads (fwd + rev) | Unmapped reads (%) | Mapped reads (%) | Contigs | GC (%) |
|---|---|---|---|---|---|---|
| 1 | Control | 115,806,338 | 47,978,266 (41.50%) | 67,828,072 (58.50%) | 68,042,571 | 50.4 |
| 3 d.p.i | 116,036,532 | 48,074,890 (43.17%) | 63,288,794 (56.83%) | 63,473,939 | 50.3 | |
| 7 d.p.i | 99,856,770 | 45,959,348 (46.03%) | 62,010,298 (53.97%) | 62,204,733 | 49.0 | |
| 11 d.p.i | 96,334,214 | 39,143,968 (40.63%) | 55,881,644 (59.37%) | 56,049,226 | 49.9 | |
| 2 | Control | 111,852,414 | 47,513,392 (42.48%) | 64,339,022 (57.52%) | 64,545,409 | 50.1 |
| 3 d.p.i | 111,363,684 | 50,771,616 (45.59%) | 65,264,916 (54.41%) | 65,460,036 | 49.6 | |
| 7 d.p.i | 107,969,646 | 42,963,067 (39.79%) | 56,893,703 (60.21%) | 57,072,615 | 48.8 | |
| 11 d.p.i | 95,025,612 | 39,734,017 (41.81%) | 56,600,197 (58.19%) | 56,777,437 | 49.3 |
Fig. 5.
Per base sequence quality of samples generated by FASTQC. Yellow boxes demonstrated base-calling quality scores across all (a) replicate 1; and (b) replicate 2 sequencing reads
Gene expression levels were calculated for each sample using transcript counts and presented in TPM unit [37]. Transcript counts is recommended in calculating the expression level of genes instead of reads or fragment counts since a single transcript can consists of multiple reads or fragments and yet they are not independent. Hence the accuracy of significance values is questionable. Normalization of transcript counts were performed using ‘Median of Gene Expression Ratios’ procedure which is identical to DESeq method [38].
Difference in transcriptomic expression of individual sequences between control and T1 samples at different time frames was searched via comparing their transcripts expression level to identify DEGs. A given gene is considered as DEGs if its expression difference complies to the cut-off values of log2 FC ≥ |1.0| and P-value < 0.01. All DEGs were clustered into upregulated and downregulated based on positive and negative values of log2 FC respectively. As depicted in Fig. 6, the number of unigenes of downregulated genes (4754 DEGs) was 1.25-fold higher than upregulated genes (3802 DEGs). Among the upregulated genes, the highest number of DEGs was from 3 d.p.i. whereas the highest number of DEGs in downregulated genes was from 7 d.p.i. Based on observations for genes in common (overlapped region), both groups showed that the overlapping region between 3 d.p.i and 7 d.p.i had the highest number of DEG unigenes, followed by the overlapping between 7 d.p.i and 11 d.p.i. The lowest number of genes found in common was between 11 d.p.i and 3 d.p.i.
Fig. 6.
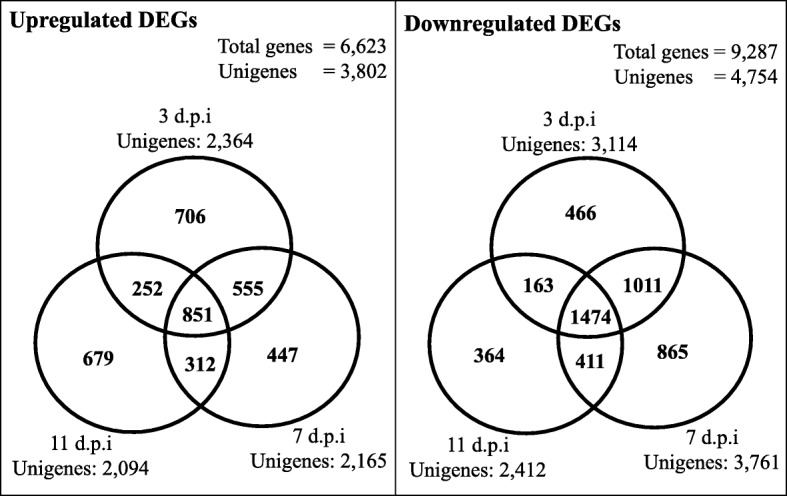
Venn Diagram of differentially-expressed genes in root of inoculated samples compared to uninoculated control samples. Genes were considered significantly upregulated or downregulated when their expression differences meet the cut-off values of fold change log2 ≥ |1.0| and P-value < 0.01
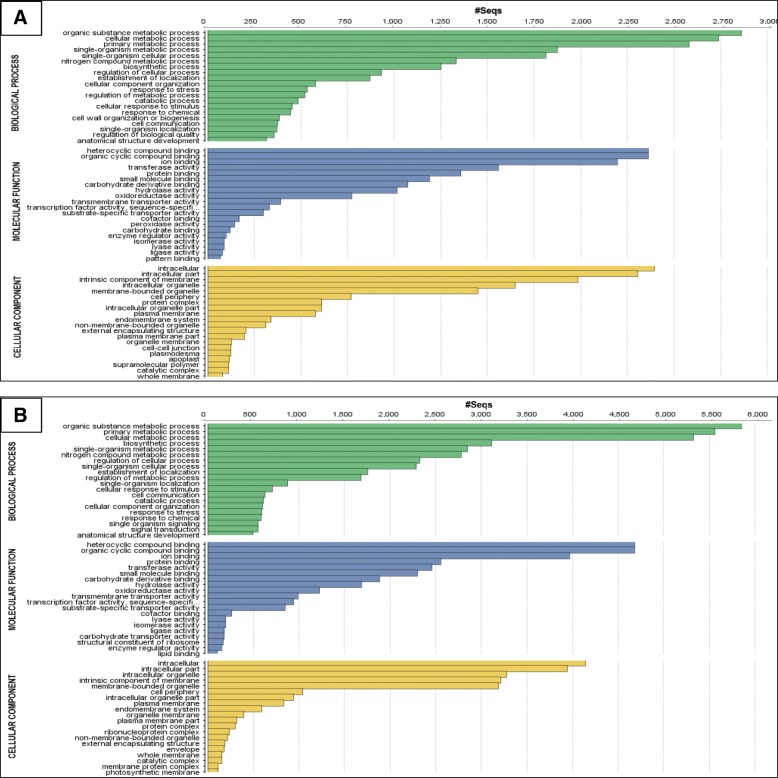
The upregulated and downregulated gene sequences were used to align with similar biological sequences using Basic Local Alignment Search Tool (BLAST) in database via the CloudBlast tools in Blast2GO. Top-hit species distribution revealed the best-aligned gene annotations of related plants with highest percentage of similarity and lowest e-value. With restriction to 20 blast hits and e-value cut-off of 1.0 × 10− 3, the most top-hit species was Elaeis guineensis with close to 9000 top-hits for upregulated genes and 15,000 top-hits for downregulated genes (Additional file 2). Phoenix dactylifera (date palm), and Elaeis oleifera (American oil palm) appeared as the second and third highest homolgy with maximum of ~ 150 top-hits. P. dactylifera is a close relative to E. guineensis, while E. oleifera is under the same genus Elaeis (tribe Cocoseae) in the family Arecaceae. The DEGs were annotated for Gene Ontology (GO) terms using Blast2Go Pro software. Fig. 7a and b show top 20 GO distribution (by level 3) by number of sequences of upregulated and downregulated DEGs respectively, which were categorized into biological process, molecular function, and cellular component. A single sequence could be present in more than one GO terms. Supplementary data showing statistics generated from blast and annotation method via Blast2Go Pro for quality control are available in Additional files 3 and 4 including annotation distribution, E-value distribution, sequence similarity distribution, and number of sequences with length.
Fig. 7.
Gene Ontology (GO) functional categorization of differentially-expressed genes (DEGs). The bar charts represent top 20 GO distribution by number of sequences of (a) upregulated and (b) downregulated DEGs in T1 samples of oil palm roots during early interaction (3, 7, and 11 d.p.i) with Ganoderma boninense compared to untreated control
The list of the top 20 GO of all categories was similar between upregulated and downregulated DEGs with exceptions for several terms. Furthermore, it was unsurprising to note that the number of downregulated DEGs were higher than the number of upregulated DEGs. However, ‘protein complex’, ‘non-membrane-bounded organelle’, and ‘external encapsulating structure’ from cellular component category have higher number of sequences in upregulated than downregulated DEGs. The majority of biological process GO terms either from upregulated or downregulated DEGs were related to various metabolic processes, while other possible defense-related GO terms include ‘response to stress’, ‘cell wall organization or biogenesis’, ‘biosynthetic process’, and ‘signal transduction’. Besides, molecular function GO terms exhibited multiple binding functions towards cyclic compounds, ions, proteins, enzymes, and metabolic substances. Different types of enzyme activities like transferase, hydrolase, oxidoreductase, lyase, isomerase, and ligase were also observed under this category. ‘Peroxidase activity’ from upregulated DEGs was not listed under downregulated DEGs for the top 20 GO terms, thus it showed that there was significant difference in the number of sequences from this enzyme function between upregulated and downregulated genes. The top 20 GO terms from cellular component category were related to intracellular parts, membranes and its components, spatially distinct organelles, ‘external encapsulating structure’, and ‘protein complex.
Enriched GO terms of upregulated and downregulated DEGs at 3, 7, and 11 d.p.i
Subsequently, Gene Set Enrichment Analysis (GSEA) was performed using Blast2GO Pro to discover enriched GO terms in biological systems of oil palm during G. boninense colonization represented by DEGs. The P-values of differential gene expression between T1 and control samples were adopted as numerical values for each functionally annotated DEGs to create a ranked list for enrichment analysis with cut-off value of 0.01. GSEA was performed at 3, 7, and 11 d.p.i in order to deduce oil palm defense management strategies at different time intervals during early interaction with the hemibiotroph. Regardless of GO category, analysis on upregulated and downregulated DEGs at 3, 7, and 11 d.p.i showed 23, 27, and 33 (upregulated) while 94, 96 and 77 (downregulated) enriched GO terms, respectively. Significant changes in gene expression in the host plant were observed at different time intervals of early interactions. Despite susceptible-type of oil palm seedlings were used, enormous genes involved in defense-related processes were upregulated and downregulated during interaction with G. boninense. The selected enriched GO terms of upregulated and downregulated genes were listed in Table 2 and Table 3 respectively. Seventy-two hours (3 d.p.i) of interactions between the host and hemibiotroph revealed enriched GO terms involved in response to stress, hormone-mediated signaling pathway, auxin-signaling and cation binding. Response to stress was also enriched in upregulated genes at 7 d.p.i, beside other terms such as mitotic cell cycle process, O-acyltransferase activity, kinesin complex and cytoplasmic vesicle. Whereas after 11 days of inoculation, significant upregulation of genes involved in oxidation-reduction process, acyl transferase activity, movement of cell or subcellular component was observed. Besides the upregulated genes, the oil palm orchestrated significant attenuation of gene expression in response to fungal threat. In downregulated genes, ion transport, autophagy, signalling receptor activity and regulation of localization GO terms were enriched throughout the time points. Transcription factors activity for sequence-specific DNA binding and response to chemical were significantly affected at 7 and 11 d.p.i. Carbohydrate transport, oxidoreductase activity, organelle fusion, monooxygenase activity and symporter activity were enriched at 3 d.p.i. Catabolic process, vesicle-mediated transport, integral component of plasma membrane were enriched at 7 d.p.i. Whereas, regulation of metabolic process, aromatic compound biosynthetic process and developmental process were enriched at 11 d.p.i.
Table 2.
Enriched GO terms of upregulated DEG unigenes of T1 samples compared to untreated control
| 3 d.p.i | 7 d.p.i | 11 d.p.i | |||
|---|---|---|---|---|---|
| Biological Process | |||||
| GO:0006950 | response to stress | GO:0006950 | response to stress | GO:0055114 | oxidation-reduction process |
| GO:0009755 | hormone-mediated signaling pathway | GO:0030243 | cellulose metabolic process | GO:0009888 | tissue development |
| GO:0060918 | auxin transport | GO:1903047 | mitotic cell cycle process | GO:0006811 | ion transport |
| GO:0010817 | regulation of hormone levels | GO:0030154 | cell differentiation | GO:0051704 | multi-organism process |
| GO:0030001 | metal ion transport | GO:0051704 | multi-organism process | GO:0007010 | cytoskeleton organization |
| GO:0048646 | anatomical structure formation involved in morphogenesis | ||||
| Molecular Function | |||||
| GO:0016651 | Oxidoreductase activity, acting on NAD(P)H | GO:0046872 | metal ion binding | GO:0046872 | metal ion binding |
| GO:0010487 | thermospermine synthase activity | GO:0008374 | O-acyltransferase activity | GO:0043169 | cation binding |
| GO:0048037 | cofactor binding | GO:0010487 | thermospermine synthase activity | ||
| GO:0043169 | cation binding | GO:0050662 | coenzyme binding | ||
| GO:0016746 | transferase activity, transferring acyl groups | ||||
| GO:0006928 | movement of cell or subcellular component | ||||
| Cellular Component | |||||
| GO:0005829 | cytosol | GO:0005871 | kinesin complex | GO:0044446 | intracellular organelle part |
| GO:0031410 | cytoplasmic vesicle | ||||
Table 3.
Enriched GO terms of downregulated DEG unigenes of T1 samples compared to untreated control
| 3 d.p.i | 7 d.p.i | 11 d.p.i | |||
|---|---|---|---|---|---|
| Biological Process | |||||
| GO:0008643 | carbohydrate transport | GO:0006811 | ion transport | GO:0006811 | ion transport |
| GO:0016491 | oxidoreductase activity | GO:0005984 | disaccharide metabolic process | GO:0019222 | regulation of metabolic process |
| GO:0006914 | autophagy | GO:0006914 | autophagy | GO:0009059 | macromolecule biosynthetic process |
| GO:0048284 | organelle fusion | GO:0016192 | vesicle-mediated transport | GO:0006914 | autophagy |
| GO:0016071 | mRNA metabolic process | GO:0008380 | RNA splicing | GO:1902589 | single-organism organelle organization |
| GO:0032879 | regulation of localization | GO:0032879 | regulation of localization | GO:0019438 | aromatic compound biosynthetic process |
| GO:0006887 | Exocytosis | GO:1902456 | regulation of stomatal opening | GO:0032879 | regulation of localization |
| GO:0006811 | ion transport | GO:0042221 | response to chemical | GO:0032502 | developmental process |
| GO:0009056 | catabolic process | GO:0042221 | response to chemical | ||
| GO:0010817 | regulation of hormone levels | ||||
| Molecular Function | |||||
| GO:0022804 | active transmembrane transporter activity | GO:0038023 | signaling receptor activity | GO:0010857 | calcium-dependent protein kinase activity |
| GO:0004497 | monooxygenase activity | GO:0003700 | transcription factor activity, sequence-specific DNA binding | GO:0004872 | receptor activity |
| GO:0038023 | signaling receptor activity | GO:0005516 | calmodulin binding | GO:0003700 | transcription factor activity, sequence-specific DNA binding |
| GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | GO:0051119 | sugar transmembrane transporter activity | ||
| Cellular Component | |||||
| GO:0044459 | plasma membrane part | GO:0005887 | integral component of plasma membrane | ||
Significant changes of gene expressions involved in defense response, cell wall modification, growth, and metabolism in the host plant
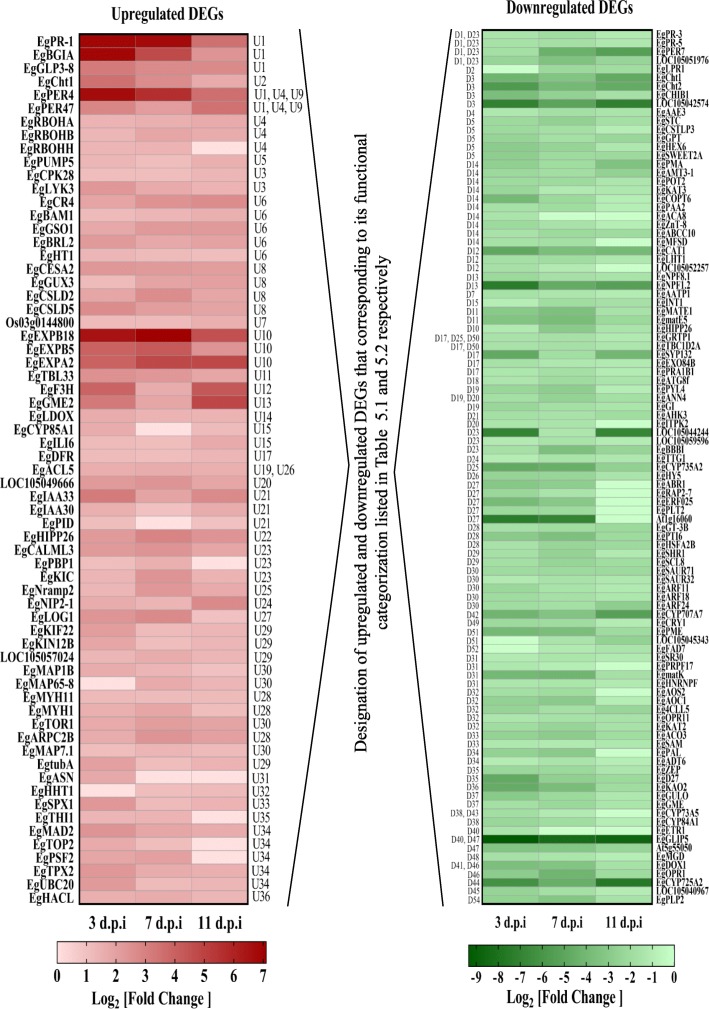
Analysis on differential expression of individual genes subset to the enriched GO terms has paved the way to observation of clusters of defense-related oil palm genes which either been activated or attenuated during interaction with G. boninense. Fold change of gene expression in T1 samples compared to control was applied to compute heatmap of selected significantly-expressed (P-value < 0.01) genes as shown in Fig. 8. Defense-related genes were the most affected by the host-fungus interaction whereby pathogenesis-related protein 1-like (EgPR-1), Glu S.griseus protease inhibitor-like (EgBGIA) and chitinases (EgCht) were significantly upregulated at 3 and 7 d.p.i and showed decreased in upregulation at 11 d.p.i compared to control. Other PR genes like germin-like proteins (EgGLP) and peroxidases (EgPER, an ROS scavenger) were both significantly upregulated and downregulated throughout the time points. Components of pattern-triggered immunity (PTI) signalling were shown to be significantly adjusted whereby lysM domain receptor-like kinase 3 (EgLYK3), pattern recognition receptor (PRR) protein which is involved in perception of fungal-derived chitin molecule also known as pathogen- or damage-associated molecular patterns (PAMP or DAMP), was found to be both upregulated and downregulated, but upregulated genes were higher in term of fold-change compared to downregulated genes. Other receptor-like kinases (RLKs) or receptor-like proteins were involved either in surveillance of bacterial PAMP or related to growth, reproduction, differentiation and homeostasis processes. Another member of PTI signalling, calcium-dependent protein kinase 28 (EgCPK28) was elevated at all time points.
Fig. 8.
Expression pattern of selected upregulated and downregulated differentially-expressed genes (DEGs) of enriched GO terms. The colour intensity of each gene is based on Log2 [fold change] values of DEGs in T1 samples of oil palm roots during early interaction (3, 7, and 11 d.p.i) with Ganoderma boninense compared to untreated control
As a mechanism to fortify the frontline barrier of defense in oil palm, genes associated with formation of primary and secondary cell wall and its modification were distinctively regulated in oil palm during interaction with G. boninense. The secondary cell wall biosynthetic genes, cellulose synthase A catalytic subunits (EgCESA) and cellulose synthase-like proteins (EgCSL) were only found in upregulated genes throughout all time points. Expansins are protein that regulate loosening and extension of cell wall. It was observed that both expansin A-like and expansin B-like were significantly upregulated in this study. Interestingly, expansin-B18-like (EgEXPB18) was outstandingly upregulated by 90- and 137-fold at 3 and 7 d.p.i respectively compared to the control. Expression of a gene involved in cutin, suberin, and wax biosynthesis, omega-hydroxypalmitate O-feruloyl transferase-like was elevated at later stage (7 and 11 d.p.i).
Several genes involved in lipid metabolism were significantly downregulated. A gene encoding a lipolytic enzyme, GDSL esterase/lipases 5 (EgGLIP5) showed the highest fold downregulation (745-fold compared to control) at 3 d.p.i and only slightly decreased from 7 to 11 d.p.i. Monogalactosyldiacylglycerol synthase 1 (EgMGD1) that catalyzes the synthesis of a galactolipid, monogalactosyldiacylglycerol (MGDG) was significantly downregulated by 3-fold. Auxin cellular level, signalling and movement are critical for root and shoot architecture, organ patterning and tissue differentiation. In our data, auxin-responsive proteins (EgIAA), repressor proteins in auxin signalling were upregulated whilst auxin response factor (EgARF), which mediates auxin-dependent transcriptional activation was downregulated at all time points. Furthermore, positive regulators of polar auxin efflux: putative auxin efflux carrier protein (EgPIN) and protein kinase PINOID (EgPID) were both upregulated thus may cause low cellular auxin level in the infected oil palm roots.
Genes involved in biosynthesis of secondary metabolites including flavonols, anthocyanidins, catechins and proanthocyanidins were upregulated but genes that confer biosynthesis of anthocyanins and fatty acid-derived secondary metabolites such as terpenes, terpenoids, and sphingolipids were downregulated. Furthermore, genes involved in biosynthesis of phytohormones like ethylene, jasmonate, L-ascorbate and gibberellin as well as their signalling pathway were downregulated at all time points. However, the downregulation was reduced at latter stage (11 d.p.i) for most of their biosynthetic genes such as jasmonate allene oxide cyclase 1, chloroplastic-like (EgAOC1) and 12-oxophytodienoate reductase 1-like (EgOPR1); ethylene 1-aminocyclopropane-1-carboxylate oxidase-like (EgACO); L-ascorbate L-gulonolactone oxidase-like (EgGULO); and ABA zeaxanthin epoxidase, chloroplastic-like (EgZEP).
Ion channels, multiple transport and carrier proteins for water, sugar, heavy-metal, drugs, ATP and ADP were downregulated throughout the experiments signifying transport of water and nutrients in oil palm was compromised during G. boninense attack. Nevertheless, a bHLH transcription factor FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (EgFIT) was significantly upregulated at 11 d.p.i rather than 3 and 7 d.p.i. It is an integral regulator in response to iron deficiency which upon activation will trigger downstream iron uptake genes, like ferric reduction oxidase 2 (FRO2) and ATPase AHA2 [39]. In our RNA-seq data, ferric reduction oxidase 2-like (EgFRO2) was not expressed in control samples but was induced in T1 samples. Genes related to vesicle trafficking, autophagy, and pre-mRNA splicing activity were downregulated.
The expression pattern of DEGs was validated through qPCR using the same samples that have been sequenced for RNA-seq data. Several genes mentioned earlier particularly that played crucial roles in defense response were analyzed and demonstrated consistent expression pattern with RNA-seq data (Table 4). The expression profiles (up- and down-regulation between time-points) were largely identical. Besides, 5 out of 8 samples tested showed that all T0 samples were not affected by the treatment and their expression levels were significantly different compared to the corresponding T1 samples. This is crucial for minimizing the abiotic stress effect on the T1 samples.
Table 4.
Validation of RNA-Seq data using
| Genes | Fold change of expression of | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RNA-seq data | qPCR | |||||||||||||||||
| in Ganoderma boninense-treated (T1 samples) compared to untreated control | in mock (T0 samples) compared to untreated control | |||||||||||||||||
| Replicate 1 | Replicate 2 | Replicate 1 | Replicate 2 | Replicate 1 | Replicate 2 | |||||||||||||
| 3 d.p.i | 7 d.p.i | 11 d.p.i | 3 d.p.i | 7 d.p.i | 11 d.p.i | 3 d.p.i | 7 d.p.i | 11 d.p.i | 3 d.p.i | 7 d.p.i | 11 d.p.i | 3 d.p.i | 7 d.p.i | 11 d.p.i | 3 d.p.i | 7 d.p.i | 11 d.p.i | |
| EgEXP18 | 157.59*a | 337.79*b | 25.99*c | 51.98*d | 55.72*d | 21.11*e | 112.05 ± 2.68*b | 105.28 ± 3.58*b | 62.21 ± 2.44*c | 119.44 ± 4.46*d,e | 120.39 ± 4.70*e | 10.36 ± 0.82d | 13.31 ± 1.42a | 11.08 ± 0.93a | 3.40 ± 0.53a | 14.97 ± 0.82d,e | 6.76 ± 0.65d | 2.88 ± 0.18d |
| EgPG | 9.19*a | 10.56*a | 3.03b | 6.50*c | 4.92*c | ND | 12.20 ± 1.22*a | 11.68 ± 1.24*a | 5.52 ± 0.79a,b | 21.51 ± 2.19*d | 18.76 ± 2.06*d | 8.76 ± 1.27c | 9.62 ± 1.53*a | 2.52 ± 0.33b | 2.31 ± 0.51b | 8.67 ± 1.40c | 2.43 ± 0.32c | 2.25 ± 0.48c |
| EgBGIA | 168.90*a | 29.86*b | 5.28c | 78.79*d | 24.25*e | ND | 140.66 ± 30.61*b | 29.46 ± 1.26a | 3.90 ± 1.17a | 147.81 ± 9.69*f | 16.34 ± 3.73d,e | 6.08 ± 0.66d | 23.81 ± 3.56a | 33.43 ± 2.21a | 0.30 ± 0.03a | 52.79 ± 5.05*c | 43.20 ± 4.04*c,e | 0.19 ± 0.05d |
| EgCht1 | 13.33*a | 10.40*a | 2.60*b | 10.70*c | 3.80*d | 3.90*d | 11.31 ± 0.44*e | 4.84 ± 0.48*a,b | 3.70 ± 0.08*b,c | 27.26 ± 1.80*j | 9.12 ± 0.31*g,h | 4.79 ± 0.11h,i | 5.62 ± 0.33*a | 2.13 ± 0.09c,d | 0.45 ± 0.02d | 45.12 ± 1.69*f | 11.00 ± 0.81*g | 2.11 ± 0.16i |
| EgERF113 | 2.05*a | 7.52a | 13.30*b | 3.65c | 13.64d | 45.30*e | 6.30 ± 0.20*b | 8.95 ± 0.50*c | 21.04 ± 0.58*d | 12.95 ± 0.85*f | 7.35 ± 0.27*f | 31.97 ± 2.26*g | 0.20 ± 0.01a | 0.06 ± 0.01a | 0.13 ± 0.01a | 1.37 ± 0.30e | 0.79 ± 0.07e | 0.70 ± 0.22e |
| EgFIT | 5.50a | 9.40*a | 22.20*b | 11.20*c | ND | 76.50*d | 8.40 ± 0.40*b | 2.90 ± 0.12a | 48.73 ± 2.12*c | 4.72 ± 0.54*e | 4.89 ± 0.34*e | 18.32 ± 0.45*f | 0.12 ± 0.02a | 0.55 ± 0.03a | 0.32 ± 0.05a | 1.28 ± 0.13d | 3.17 ± 0.13*d,e | 2.55 ± 0.47d |
| EgMTP10 | 2.60a | 2.50a | 8.30a | 5.60b | ND | 35.50*c | 8.18 ± 0.73a | 7.81 ± 0.96a | 172.68 ± 10.14*b | 3.31 ± 0.56c | 3.23 ± 2.71c | 25.03 ± 1.56*d | ND | 1.74 ± 0.10a | 0.45 ± 0.02a | 1.22 ± 0.08c | 2.47 ± 0.07c | 1.75 ± 0.22c |
| EgPIN8 | 2.30a | 2.20a | 3.00*a | 7.80*b | 5.40*b | 41.80*c | 75.66 ± 11.13a | 7.44 ± 2.00a | 1557.48 ± 33.63*b | 27.48 ± 3.21d,e | 57.97 ± 10.70*e | 165.38 ± 14.24*f | ND | ND | ND | 0.61 ± 0.05c | ND | ND |
EgEXP18: E. guineensis expansin-B18-like; EgPG: E. guineensis polygalacturonase-like; EgBGIA: E. guineensis glu S.griseus protease inhibitor-like; EgCht1: E. guineensis chitinase 1-like; EgERF113: E. guineensis ethylene-responsive transcription factor 113; EgMTP10: E. guineensis metal tolerance protein 10-like; EgPIN8: E. guineensis putative auxin efflux carrier component 8; EgFIT: E. guineensis FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR-like
qPCREach replicate consisted of pooled root from six plants. Pairwise comparison of RNA-seq data was evaluated according to cut-off values of log2 fold change (FC) ≥ |1.0| and P-value < 0.01. Data of qPCR are expressed as fold change mean ± SEM of three individual technical replicates of T0 and T1 samples compared to untreated control. The fold expressions of each gene were normalized by three reference genes; GAPDH 2, NADH 5 and ß-actin expression levels. Significant differences between qPCR groups were determined using one-way ANOVA analysis followed by Tukey’s test. * indicate significant different compared to corresponding control at: P < 0.01 and log2 FC ≥ |1.0| for RNA-seq; and P < 0.01 for qPCR. Different superscript letters (a-j) between samples (within replicate) indicate significant different at: P < 0.01 and log2 FC ≥ |1.0| for RNA-seq; and P < 0.01 in mean values for qPCR. ND: not detected
Discussion
Cell wall modification and production of antimicrobial compounds in plants are the non-specific preformed defense responses which act as the first barrier against pathogen [40]. Induced defense system for PTI in plant is initiated with the detection of DAMP or PAMP such as fungal chitin by PRR of the host [41, 42]. However, pathogens are able to suppress PTI when it successfully delivers the effectors leading to effector-triggered susceptibility. At a later phase of resistance, the pathogen effectors thereafter perceived by nucleotide-binding site leucine-rich repeat (NB-LRR) for a more specific response the so-called effector-triggered immunity (ETI). ETI is an exaggerated version of PTI which could bring response over the resistance threshold level that lead to HR. Through time-course transcriptome analysis of oil palm seedlings artificially inoculated with G. boninense, the occurrence of these responses was monitored based on transcriptome profiling at the early stages of interaction which was within 11 d.p.i.
PRR proteins are involved in surveillance of pathogens attack through recognition of their signature-pattern molecules known as PAMPs. Among the PRR proteins, the lysM domain receptor kinase has been recognized to play a role in the fungal perception [43]. In the present study, lysM domain receptor-like kinase 3 was significantly upregulated by 5-fold at 3 d.p.i with decrement at later time points. This will trigger PTI responses such as production of PR proteins, ROS and protease inhibitors during the biotrophic phase. In this study, the activation of PTI is further supported by upregulation of EgCDPK28 which serves as a mediator for PTI responses.
RLKs are widely known for their roles in development, cells differentiation and perception of stimuli [44]. In this report, we presented upregulation of CRINKLY-4 (EgCR-4) with ascending increment from 3 d.p.i until 11 d.p.i. CR-4 is one of the RLK mainly involved in roots stem cell differentiation and lateral roots formation [45]. CR-4 has been identified as one of several important extracellular domains of RLKs responsible in recognizing and perceiving diverse signals under both abiotic and biotic stresses [46]. It can be presumed that the stress signals are further transduced downstream to effector molecules via secondary signalling molecules, most commonly Ca2+ and ROS leading to orchestration of protein cascades to activate plant adaptation and/or defense responsive genes [47, 48].
PR-1 has been reported having prominent antifungal properties which combat fungal pathogens from further invading the host plant [49, 50]. Our discovery revealed significant upregulation of EgPR-1 gene at 3 and 7 d.p.i. while significantly reduced at 11 d.p.i. Several PR-1 family members are synthesized in response to pathogen attack specifically as components for the local HR and systemic acquired resistance (SAR). Recent discovery by Gamir et al. showed that PR-1 binds and sequesters sterols from the membranes of microbes [51]. Sterol-auxotroph oomycete, Phytophthora brassicae is sensitive to PR-1, whereas sterol-prototroph pathogens are sensitive to PR-1 only when the production of sterol is interrupted. High dose of PR-1, particularly from within vacuole has the capability to sequester more sterols compared to their biosynthetic capacity, thus achieving the antimicrobial effect in vivo. In this study, abundant G. boninense hyphae network was observed at 3 d.p.i. We are proposing that reduction of the hyphae network at the later stages (7 and 11 d.p.i) could be due to sequestration of their ergosterol by the highly expressed PR-1 which left the fungus sterol-deficient. Experimental evidences from Choon and colleagues proved that ergosterol is produced by G. boninense as their primary metabolite in primary cell wall development [52]. Several studies have shown that G. boninense colonization and its growth phase can be determined by measuring the concentration of ergosterol [53, 54].
ROS is a unique molecule that serves both physiological and stress-related functions by playing the role as signalling molecules for redox homeostasis and PTI responses [55, 56]. Peroxidases are another well-known PR-protein belonging to PR-9 family that are induced in plant host during pathogen infection [50]. Peroxidases are expressed in higher plants under colonization of fungi [57] and other microbes to limit pathogen spread by providing structural barrier and creating an extremely unpleasant environment via heavy production of ROS and reactive nitrogen species (RNS) [58] at the cell wall matrix level that promote HR and SAR. Intriguingly, the transcripts of the peroxidase family of genes such as EgPER3, EgPER4 and EgPER47 were being highly upregulated in oil palm host during Ganoderma attack from 3 d.p.i until 11 d.p.i.
Besides, the significantly upregulated NADPH oxidase: respiratory burst oxidase homolog proteins (EgRboHA and EgRboHB) observed in the present study, could have assisted in the establishment of HR via the synthesis of apoplastic ROS [59]. The finding was supported by our previous study which demonstrated effective hyphae penetration of Ganoderma spp. and plant’s cell wall degradation as early as 24 h-post-inoculation indicating involvement of ROS and strategized degradation of cell wall during biotrophic stage [27]. Nevertheless, the high dose of ROS which is toxic to plant cells promotes susceptibility to necrotrophs [60].
Development of secondary cell walls (SCWs) is essential for various physiological processes in plant including growth, seed dispersal, pollen release and fertilization as well as defense response against pathogens attack [61]. SCWs consist of cellulose, lignins, hemicelluloses and some proteins to structurally support plant as well as regulate water transport [61, 62]. In the present study, cellulose synthase complexes which comprised of different isoforms of cellulose synthases (EgCesAs) responsible in SCWs biosynthesis were upregulated as early as 3 d.p.i. We are also reporting for the first time on the involvement of Cobra-like 4 (EgCOBL-4) in defense response, a SCW biosynthetic gene which was upregulated at the later stage of infection against G. boninense (7 and 11 d.p.i). Arabidopsis COBL-4, ortholog of Brittle culm 1 has been reported to contribute in biogenesis of cellulose component as well as secondary cell wall thickening [63–65].
We also identified two out of four subfamilies of the transcripts for expansins (EgEXPA and EgEXPB) known to be responsible in cell wall expansion and loosening [66]. Expansins were mainly studied under abiotic stress due to water deficit [67–69]. Expansin has also been reported in cell wall alteration caused by flooding injury in soybean seedlings [70], thus it may be suggested that expansin was upregulated due to wounding by Ganoderma attack. A report has addressed regulation of expansin-like A2 against necrotrophic attack of B. cinerea on A. thaliana [71]. Our present data demonstrated 28-fold upregulation of expansin A2-like at 7 and 11 d.p.i. during G. boninense interaction. Two newly discovered expansins in the present study were expansin B18 (upregulated 90 and 137-fold) as well as expansin B5 (upregulated 15 and 19-fold) at 3 and 7 d.p.i. respectively. It was postulated that the ability of expansins to break noncovalent bonding of polysaccharides allow larger exposure of surface glucans of cellulose leading to cellulase enzymatic attack [72]. However, expansion and loosening of cell wall will increase susceptibility to necrotrophs infection.
During pathogenesis, pathogens secrete digestive proteases which facilitate degradation of plant proteins into smaller compounds beneficial as nutrient sources [73]. The proteolytic process is crucial for pathogen’s growth and cell proliferation within host cells. As pathogen proteases and their digested products are being administered, protease inhibitors (PIs) are released by host plant to inhibit the proteolytic enzyme as one of the resistance responses [74]. While PIs can be found naturally in plant to regulate many biological processes such as development [75] and abiotic stress induced-PCD [76], they are highly upregulated spatiotemporally during biotic stress [77–80]. PIs accumulate not only at the site of injury, but also at distal locations to prevent further protease digestive activities [74, 81]. Thus, protease inhibitors are recognized as one of the major inducible defenses to combat against phytopathogens [82, 83]. We are reporting for the first time a highly expressed EgBGIA from the less studied potato type 1 serine PIs family with 115-fold upregulation at 3 d.p.i. Interestingly, this PI was upregulated to the same level of PR-1 genes which is a prominent plant defense protein to combat fungal threats. Mostly produced by solanaceous plants, potato type 1 and II serine PIs have only been reported against herbivory attack [84], hence paving the way to further study on this gene, whether it has specific involvement in the responses against Ganoderma attack.
GDSL esterase/lipase is a lipolytic enzyme with conserved GDSL motif and wide substrate specificity. Arabidopsis GDSL lipase, AtGLIP1 was reported to have a positive effect in conferring resistance towards Alternaria brassicicola, while its homolog AtGLIP2 is involved in defense by inhibiting auxin response [85]. On the other hand, Gao and colleagues showed that rice infected with blast fungus Magnaporte oryzae treated with OsGLIP1/2-RNAi demonstrated reduced symptoms of disease, while OsGLIP1/2-overexpressed plant showed enhanced diseased symptoms. Thus, they proposed that OsGLIP1 and OsGLIP2 have negative regulatory role towards disease resistance in rice [86]. The dual positive and negative regulatory role indicates the diverse catalytic properties of GLIP1/2 in lipid metabolism. From our data, oil palm EgGLIP5 expression was significantly reduced during early interaction with G. boninense. It was interesting to report on this gene, but further experimental verification is needed to ensure if EgGLIP5 plays similar function as OsGLIP1/2 in plant immunity. Gao et al. also reported that high level of monogalactosyldiacylglycerol (MGDG) corresponds to overexpression of OsGLIP1 [86]. Although MGDG is abundant in leafy vegetables, varied total and relative contents are also observed in other plant parts [87]. We found that monogalactosyldiacylglycerol synthase 1 (EgMGD1) expression was also downregulated. Exogenous application of MGDG facilitated growth of pathogen signifying its negative role in rice immune response [86], whilst MGD1 is required as a positive regulator in Arabidopsis to induce SAR [88].
Due to its role as primary growth promoter, auxin or indole-acetic acid (IAA) has been shown to oppose the development of induced-resistance in plant against biotic and abiotic stresses while supporting disease manifestations in numerous plants. Auxin perception involves transport inhibitor response 1 (TIR1) and auxin signalling F-box protein 1, 2, and 3 (AFB1, AFB2, AFB3) as receptors which upon auxin signal will direct proteasomal degradation of Aux/IAA repressor proteins through ubiquitin ligase SKP-Cullin-F box, TRANSPORT INHIBITOR RESISTANT1/AUXIN SIGNALING F-BOX (SCFTIR1/AFB) complex and derepress ARF to regulate transcription of auxin-responsive proteins. Exogenous treatment of oligosaccharides on tobacco and Arabidopsis improved protection of these plants against B. cinerea, however early application with auxin restore their susceptibility [89, 90]. In our data, members of the Aux/IAA transcriptional repressors, auxin-responsive protein IAA33 and IAA30-like were upregulated at 3 d.p.i while the transcriptional activator EgARF4, EgARF11, EgARF18 and EgARF24 were downregulated at the same time point. This could suggest that auxin signalling pathway was inhibited throughout the treatment period which in turn compromised growth.
Apart from the inhibition of auxin signalling, our data also showed that polar auxin transport in root cells has been facilitated by the upregulation of putative auxin efflux carrier component 8 (EgPIN8) and protein kinase PINOID-like (EgPID). Based on accumulating research evidences, the plasma membrane-localized PIN are critical auxin efflux carrier component, while PID positively regulate polar trafficking of PIN [91]. Studies showed that over expression of PIN or PID strongly inhibits root hair growth, while exogenous auxin feed or application of PIN or PID inhibitors restores the growth [91]. Thus, exaggerated efflux of auxin from roots by the actions of EgPIN8 and EgPID may cause shortage of intracellular auxin which in turn suppressed auxin signalling and subsequently inhibited oil palm quaternary root growth during G. boninense interaction.
Gene expression of EgERF113 was highly elevated by approximately 29-fold suggesting recognition of necrotrophic attack at 11 d.p.i synergistic with large increment by 17-fold of EgPR-1. The results support recent studies claiming highly upregulation of transcription factor ERF113 in plant defense response against necrotrophs which subsequently promote PR-1 proteins [92, 93]. Overexpression of transcription factor ERF113 (RAP26.L) was reported to promote wound defense response triggered by jasmonate and ethylene [94]. The perception of JA-Ile induces interaction between its receptor CORONATINE INSENSITIVE 1 and JAZMONATE ZIM-DOMAIN proteins leads to the relieve of repression on MYC2 [95, 96], which explains upregulation of MYC2 at 11 d.p.i. MYC2 as well as MYC3 and MYC4 are essential in promoting accumulation of secondary metabolites during plant resistance against various pathogens [97]. MYCs are known as master regulator of JA expression under stress response but they differ in specificity depending on the spatiotemporal accumulation. Induced upregulation of EgMYC2 in our study supports the reports suggesting that MYC2 mediates JA-responsive genes against necrotrophic attack [30] predominantly in roots while MYC3 and MYC4 expressed mainly in aerial tissues [98–100]. We summarized the proposed functional categorization based on differentially expressed unigenes enriched in oil palm seedling roots during early interactions with G. boninense at 3, 7 and 11 d.p.i (Tables 5 and 6).
Table 5.
Proposed functional categorization of upregulated DEG unigenes at different time points compared to untreated control
| 3 d.p.i | 7 d.p.i | 11 d.p.i |
|---|---|---|
| [U1] Pathogenesis-related protein activity, [U2] defense against chitin-containing fungal pathogens | [U1] Pathogenesis-related protein activity, [U2] defense against chitin-containing fungal pathogens | [U1] Pathogenesis-related protein activity, [U2] defense against chitin-containing fungal pathogens |
| U3] Pattern recognition receptor activity and PAMP-triggered immunity (PTI) signalling | [U3] Pattern recognition receptor activity and PAMP-triggered immunity (PTI) signalling | [U3] Pattern recognition receptor activity and PAMP-triggered immunity (PTI) signalling |
| [U4] ROS production, [U5] scavenging activity | [U4] ROS production, [U5] scavenging activity | [U4] ROS production, [U5] scavenging activity |
| [U6] Signal transduction involve in growth, development, reproduction, and differentiation | [U6] Signal transduction involve in growth, development, reproduction, and differentiation | [U6] Signal transduction involve in growth, development, reproduction, and differentiation |
| Cell wall formation: [U7] Primary, [U8] Secondary | Cell wall formation: [U7] Primary, [U8] Secondary | Cell wall formation: [U7] Primary, [U8] Secondary |
| Cell wall modification: [U9] lignin degradation, [U10] loosening and extension, [U11] O-acetylation of cell wall polymers | Cell wall modification: [U9] lignin degradation, [U10] loosening and extension, [U11] O-acetylation of cell wall polymers | Cell wall modification: [U9] lignin degradation, [U10] loosening and extension, [U11] O-acetylation of cell wall polymers |
| Biosynthesis of secondary metabolites: [U12] flavonols, anthocyanidins, catechins and proanthocyanidins, [U13] ascorbate, [U14] anthocyanidins, [U15] brassinosteroid biosynthesis and signalling, [U17] flavonoid metabolism | Biosynthesis of secondary metabolites: [U12] flavonols, anthocyanidins, catechins and proanthocyanidins, [U13] ascorbate, [U14] anthocyanidins, [U16] brassinosteroid signalling, [U17] flavonoid metabolism | Biosynthesis of secondary metabolites: [U12] flavonols, anthocyanidins, catechins and proanthocyanidins, [U13] ascorbate, [U18] ubiquinone, other terpenoid-quinone, phenylpropanoids, [U15] brassinosteroid biosynthesis and signalling, [U17] flavonoid metabolism |
| [U19] Repression of early auxin response genes, [U20] auxin transport, [U21] regulation of auxin signalling | [U19] Repression of early auxin response genes, [U20] auxin transport, [U21] regulation of auxin signalling | [U19] Repression of early auxin response genes, [U20] auxin transport, [U21] regulation of auxin signalling |
| Binding protein and transport: [U22] heavy-metal, [U23] calcium, [U24] water, [U25] iron | Binding protein and transport: [U22] heavy-metal, [U23] calcium, [U24] water, [U25] iron | Binding protein and transport: [U22] heavy-metal, [U23] calcium, [U24] water, [U25] iron |
| [U26] Negative regulation in the proliferation of xylem vessels | [U26] Negative regulation in the proliferation of xylem vessels | [U26] Negative regulation in the proliferation of xylem vessels |
| [U27] Conversion of gibberellin and cytokinin from inactive form into bioactive form | [U27] Conversion of gibberellin and cytokinin from inactive form into bioactive form | |
| [U28] Cytoskeleton organization: [U29] Kinesin, [U30] microtubule | [U28] Cytoskeleton organization: [U29] Kinesin, [U30] microtubule | [U28] Cytoskeleton organization: [U29] Kinesin, [U30] microtubule |
| [U31] Nitrogen assimilation, distribution and remobilization within the plant | U33] Adaptation to phosphate starvation | [U32] Biosynthesis of cuticular wax and suberin |
| [U33] Adaptation to phosphate starvation | U34] Cell cycle process | [U33] Adaptation to phosphate starvation |
| U34] Cell cycle process | [U35] Protection from oxidative damage | U34] Cell cycle process |
| [U35] Protection from oxidative damage | ||
| [U36] Fatty acid oxidation | [U36] Fatty acid oxidation | [U36] Fatty acid oxidation |
Table 6.
Proposed functional categorization of downregulated DEG unigenes at different time points compared to untreated control
| 3 d.p.i | 7 d.p.i | 11 d.p.i |
|---|---|---|
| [D1] ROS scavenging activity | [D1] ROS scavenging activity, [D2] oxidative stress response | |
| [D3] Defense against chitin- and glucan-containing and [D4] oxalate-producing fungal pathogens | [D3] Defense against chitin- and glucan-containing and [D4] oxalate-producing fungal pathogens | [D3] Defense against chitin- and glucan-containing and [D4] oxalate-producing fungal pathogens |
| Transport: [D5] sugar, [D6] water, [D7] ATP, [D9] protein, [D10] heavy-metal, [D11] drug, [D12] amino acid, [D13] peptide, [D14] ion | Transport: [D12] amino acid, [D13] peptide, [D6] water, [D14] ion, [D5] sugar, [D7] ATP, [D11] drug, [D15] inositol, [D16] various | Transport: [D5] sugar, [D14] ion, [D10] heavy-metal, [D15] inositol, [D12] amino acid, [D7] ATP, [D11] drugs, [D6] water, [D13] peptide |
| [D17] Vesicle trafficking | [D17] Vesicle trafficking | [D17] Vesicle trafficking |
| [D18] Autophagy | [D18] Autophagy | [D18] Autophagy |
| [D19] Signal transduction: [D20] osmotic response, [D21] histidine kinase, [D22] phosphatidylinositol signalling | [D19] Signal transduction: [D21] histidine kinase, [D20] osmotic response, [D22] phosphatidylinositol signaling | [D19] Signal transduction: [D20] osmotic response, [D21] histidine kinase |
| [D23] Pathogenesis-related protein activity | [D23] Pathogenesis-related protein activity | [D23] Pathogenesis-related protein activity |
| Growth: [D24] trichome and root hair development, [D25] homeostasis | Growth: [D24] trichome and root hair development, [D25] homeostasis | Growth: [D24] trichome and root hair development, [D25] homeostasis |
| Transcription factor activity in regulating [D26] photomorphogenesis, [D27] ethylene-responsive genes, [D28] defense response, [D28a] biotic and abiotic stress response, [D29] growth and development | Transcription factor activity in regulating [D26] photomorphogenesis, [D27] ethylene-responsive genes, [D28] defense response, [D28a] biotic and abiotic stress response, [D29] growth and development | Transcription factor activity in regulating [D26] photomorphogenesis, [D28] defense response, [D28a] biotic and abiotic stress response, [D29] growth and development |
| [D30] Auxin responsive genes | [D30] Auxin responsive genes | [D30] Auxin responsive genes |
| [D31] Pre-mRNA splicing activity | [D31] Pre-mRNA splicing activity | [D31] Pre-mRNA splicing activity |
| Biosynthesis of [D32] jasmonate, [D33] ethylene, [D34] salicylate, [D35] ABA, [D36] gibberellin, [D37] L-ascorbate and [D38] phenylpropanoids. Phytohormones signalling pathway: [D39] jasmonate, [D40] ethylene, [D41] ABA | Biosynthesis of [D32] jasmonate, [D33] ethylene, [D34] salicylate, [D35] ABA, [D36] gibberellin, [D37] L-ascorbate and [D38] phenylpropanoids. Phytohormones signalling pathway: [D39] jasmonate, [D40] ethylene, [D41] ABA | Biosynthesis of [D32] jasmonate, [D33] ethylene, [D35] ABA, [D36] gibberellin and [D37] L-ascorbate. Phytohormones signalling pathway: [D40] ethylene, [D41] ABA |
| [D42] Oxidative degradation of abscisic acid | [D42] Oxidative degradation of abscisic acid | [D42] Oxidative degradation of abscisic acid |
| Biosynthesis of secondary metabolites: [D43] anthocyanins, tocopherols, terpenes, terpenoids, oxylipins and sphingolipids, [D44] taxols, [D45] pterostilbene | Biosynthesis of secondary metabolites: [D44] taxols, [D45] pterostilbene, [D46] oxylipins | Biosynthesis of secondary metabolites: [D44] taxols, [D46] oxylipins, [D45] pterostilbene |
| [D47] Lipid metabolism activity that confer negative regulation in resistance towards fungal pathogen | [D47] Lipid metabolism activity that confer negative regulation in resistance towards fungal pathogen | [D47] Lipid metabolism activity that confer negative regulation in resistance towards fungal pathogen |
| [D48] Biosynthesis of structural component of photosynthetic membrane | [D48] Biosynthesis of structural component of photosynthetic membrane | [D50] GTPase-activating protein for Rab family protein |
| [D49] Photoreceptor activity | [D49] Photoreceptor activity | |
| [D50] GTPase-activating protein for Rab family | [D50] GTPase-activating protein for Rab family | [D50] GTPase-activating protein for Rab family |
| [D51] Cell wall modification | [D51] Cell wall modification | [D51] Cell wall modification |
| [D52] Biosynthesis of 16:3 and 18:3 fatty acids | [D52] Biosynthesis of 16:3 and 18:3 fatty acids | |
| [D54] Non-specific lipolytic acyl hydrolase activity | [D54] Non-specific lipolytic acyl hydrolase activity | [D54] Non-specific lipolytic acyl hydrolase activity |
The molecular and physiological evidences regarding transition from biotrophy to necrotrophy are still awaiting elucidation. This brings up the question on how long should the biotrophic phase be before the transition? Apparently, the biotrophic phase needs to keep progressing until the host defense is overwhelmed. In M. oryzae-rice and C. graminicola-maize pathosystems, the establishment of disease is favoured even though the fungi were not able to dampen the magnitude of defense at the early stages of interaction [23]. Hence, C. graminicola and M. oryzae presumably could endure the elevated host defense until the point when they change to the necrotrophic mode. Based on our observation in oil palm during Ganoderma attack, upregulation of important genes involved in defense responses such as PR-proteins (EgPR-1), protease inhibitor (EgBGIA), PRR proteins (EgLYK3) and chitinases (EgCht) was observed at 3 and 7 d.p.i before dropping to insignificant level at 11 d.p.i, suggesting the occurrence of the biotrophic phase whereby multifaceted plant defense responses were deployed to counteract the G. boninense attack. The subsequent reduction in the defense response suggests switching to necrotrophic phase by the fungus which was essential for successful infection. The result agrees with report suggesting suppression of pathogen-responsive genes by transcription factor MYC2 during necrotrophic attack [30]. Furthermore, significant upregulation of the EgFIT at later phase (11 d.p.i) and minor induction of EgFRO2 could be another clue for necrotrophic phase that caused disturbance in iron uptake. EgFIT is a central transcription factor required in upregulation of iron deficiency responses in root of Arabidopsis [101] hence suggested iron deprivation at later phase of the infection in the oil palm root. Despite other pathosystems showing distinct transition period (i.e. C. graminicola only took 72 h post infection to begin necrotrophy on maize), development and spread of the fungi in the host plant may vary across different species and rely on the infection conditions.
Vargas et al. hypothesized that increasing pressure by plant defense responses during biotrophy has augmented pathogen to shift into necrotrophy [23]. Genes involved in ROS production: EgPER and EgRBOH were upregulated and maintained throughout the treatment period in the present study may cause overwhelming ROS accumulation thus underwent self-propagation causing cell damage which promotes necrotrophic infection denoting transition from biotrophy [102]. Besides, the loosening and expansion of oil palm cell wall by expansin may contribute to increase susceptibility to necrotrophs.
At the necrotrophic phase, we found out that CR-4 playing an important role in pathogen perception in oil pam. The oil palm then deploys another set of defense response against the necrotrophic attack that includes fortification of cell wall as well as rapid and significant upregulation of transcription factors. Transcription factor EgMYC2 is known to regulate defense response against necrotroph [30]. EgERF113 is proposed based on the present study as a novel transcription factor involved in biotic stress responses. The most commonly reported ERF transcription factors associated with biotic stress however, are ERF1 and ERF2 which are activated through ethylene and jasmonate signalling pathways [103–105]. It is evident that the oil palm finally succumbed to chronic infection. Schematic diagram on the proposed defense mechanism in oil palm during transition from the biotrophic to the necrotrophic phase is depicted in Fig. 9.
Fig. 9.
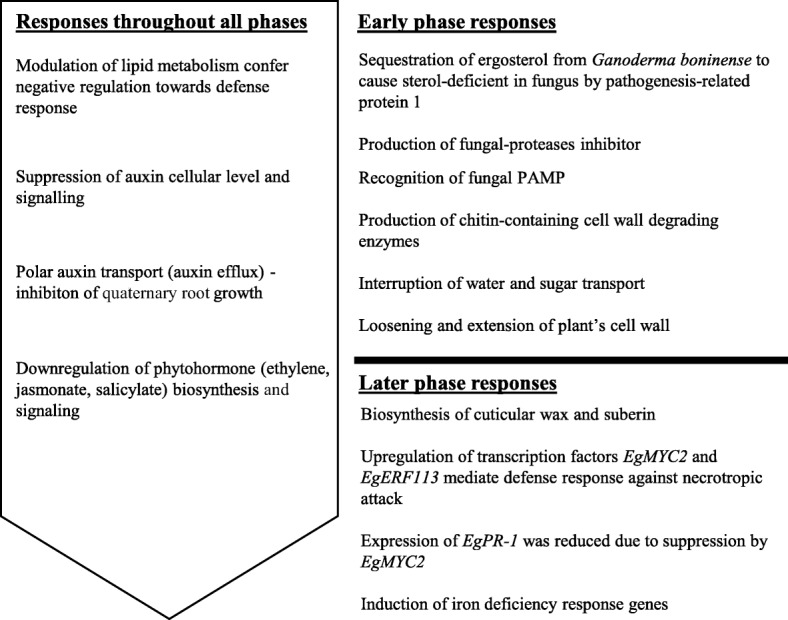
Proposed summary of defense-related events in oil palm roots during early interaction with Ganoderma boninense. Early phase responses (biotrophic phase) are the summary of events which occurred at 3 and 7 d.p.i while later stage responses (necrotrophic phase) are events occurring at 11 d.p.i based on analysis of DEGs
Conclusions
Observations on early interaction between oil palm and G. boninense at different time points provided insights on early defense mechanism in oil palm to overcome the fungal threats even though the plant finally succumbed to BSR. Based on the evidences of the current study, several suggestions can be made: (1) the thick multilayer of Ganoderma hyphae observed at 3 d.p.i on oil palm root surface was significantly reduced at 7 and 11 d.p.i, indicating that the plant was likely to overcome the infection, however emergence of chronic-stage fruiting bodies indicated that the plant defense was overwhelmed by the fungus; (2) the fungus had possibly established a biotrophic relationship with the oil palm at early phase (3 and 7 d.p.i) of interaction as evidenced by significant upregulation of defense-related genes; and (3) the fungus may have switched its lifestyle to necrotrophy at later phase (11 d.p.i) of colonization whereby the elevated expression of the same defense-related genes was significantly reduced. The increasing pressure by plant defense responses during biotrophy could have triggered the transition as well as the overproduction of ROS which caused cellular damage and subsequent promotion of necrotrophic lifestyle to the fungus. The data provides evidence supporting the hemibiotrophic nature of this pathogen and it showed that practising hemibiotrophic routine is always an advantage for phytopathogen over the host. Analysis on DEGs revealed potential candidate genes to be further elucidated which can serve as phase-specific biomarkers at the early stages of oil palm-Ganoderma interaction.
Methods
Host plant and fungal inoculums preparation
Ganoderma boninense strain PER 71, an aggressive fungal pathogen causing BSR was obtained from Ganoderma and Diseases Research for Oil Palm (GanoDROP) Unit, Biology Division, Malaysian Palm Oil Board (MPOB) [106]. The fungus was isolated and purified from an infected oil palm in United Plantation Teluk Intan, Perak, Malaysia as described in Sundram et al. [107]. Four-month-old seedlings of susceptible oil palm (Elaeis guineensis Jacq. Dura x Pisifera), purchased from Sime Darby Plantation, Banting, Malaysia, were used as host plants. The seedlings were planted in Cobalt-60 (60Co) gamma radiation-sterilized (40 kGy) soil mix consisting of topsoil, peat and sand (3:2:1) placed in inert clay pot/vase and irrigated twice a day. Freshly prepared malt extract agar (Merck) was added onto sterile rubber wood block (RWB, 6 cm × 6 cm × 6 cm in dimension) and autoclaved at 121 °C for 30 mins, before inoculation with one-week-old G. boninense PER 71 cultured in potato dextrose agar (Difco). Inoculated RWBs were incubated at room temperature in the dark for 4 weeks to be fully colonised by the Ganoderma inoculum.
Inoculation of G. boninense on oil palm seedlings (artificial infection)
A total of 84 of four-month-old oil palm seedlings (including control) were tested with two different treatments: inoculation with bare RWB (without fungal inoculum) as mock treatment (hereafter referred to as T0) and inoculation with RWB fully colonised with G. boninense (hereafter referred to as T1). Throughout the treatment, all seedlings were arranged in a complete randomised design under conditionally-controlled plant house and watered twice daily using distilled water. Destructive sampling consisting of two biological replicates was performed at 3, 7 and 11 d.p.i (according to preliminary screening). Untreated seedlings were used as control. Each replicate consisted of pooled root samples from six randomly picked oil palm seedlings. The root samples were flash-frozen in liquid nitrogen and kept in − 80 °C until further use. Several untreated and T1 seedlings were kept for extended period for observation of chronic infection.
Artificial infection of G. boninense on oil palm seedlings were carried out using the method described by Idris et al. [106]. For inoculated samples, the colonised RWBs were placed in direct contact with the entire roots of the plant seedlings and were placed in clay pots which had been quarter-filled with the soil mixture. Soil was added until bole of the seedlings were fully covered.
For NGS, control and T1 samples were analysed for DEGs between different time points. DEGs of interest from the NGS data were validated using qPCR which also include the T0 samples for validation of G. boninense effect on oil palm gene expression.
RNA extraction
Total RNA of all samples were extracted using the method described by [108] with minor modifications. Two grams of harvested root samples were ground to fine powder with mortar and pestle in liquid nitrogen. Six millilitre of RNA extraction buffer (50 mM Tris-HCl pH 9.0, 150 mM lithium chloride, 5 mM ethylenediaminetetraacetic acid pH 8.0, 5% (w/v) sodium dodecyl sulfate, 2 mM aurintricarboxylic acid) was freshly prepared and 0.4% of 2-mercaptoethanol was added into the buffer prior to use. Equal volume of phenol/chloroform (1:1) was added, and the mixture was centrifuged at 18,514 g for 30 mins at 25 °C. The aqueous phase was collected and transferred into a new tube. The addition of phenol/chloroform was repeated. Then, 6 mL of chloroform:isoamyl alcohol (24:1) was added and the tube was centrifuged at 18,514 g for 30 mins at 25 °C. The aqueous phase was collected and transferred into a new tube. Lithium chloride (8 M) was added to obtain a final concentration of 2 M and the mixture was kept overnight in 4 °C. After overnight incubation, the mixture was centrifuged at 12,857 g for 30 mins at 4 °C. The pellet was washed twice with molecular grade 90% ethanol and centrifuged at 12,857 g for 10 mins at 4 °C. The pellet was dried using Concentrator 5301 (Eppendorf, Germany). RNA pellet was dissolved in ultrapure nuclease-free water and kept in − 80 °C until further use. All centrifugations were performed using Centrifuge 5810R (Eppendorf, Germany).
mRNA library construction and sequencing
Prior to mRNA library preparation, the RNA Integrity Number (RIN) of each sample was measured by Agilent 2100 Bioanalyzer (Agilent, USA) wherein only samples with RIN value of > 7.5 were accepted for sequencing. The mRNA library was constructed using the Illumina TruSeq RNA Library Prep Kit (Illumina, USA). Reads with an average length of 101 bp was used for sequencing on an Illumina HiSeq 2000 (Illumina, USA). Paired-end (2 × 100 bp) reads with an average length of 101 bp were sequenced by Illumina HiSeq 2000 system (Illumina, USA) at Macrogen, Korea. As pre-processing steps, the sequenced reads were saved in FASTQ format to determine the quality scores across all bases of short 101 bp paired-end reads using FastQC software.
Genome assembly and identification of differentially expressed genes
The mRNA fragments were then mapped to Elaeis guineensis coding sequences as reference genome (retrieved from www.ebi.ac.uk/genomes) through Geneious software version 9.1.5 (Biomatters Ltd.). From align/assemble tools, Geneious for RNA-Seq was used as mapper with medium-low sensitivity using clean reads before mapping. Upon completion of the mapping step, the transcript abundance of each sample was calculated as transcript per kilobase million (TPM). To analyse the alteration in gene expression of infected oil palm compared to uninfected control, genes expressed from G. boninense-infected samples at each time point were compared to the genes expressed in absolute control sample. DEGs were evaluated according to stringent cut-off values of log2 fold change (FC) ≥ |1.0| and P-value < 0.01. DEGs that met the stringent cut off-values from comparative analysis between control and T1 samples of all time points were clustered according to upregulated or downregulated genes. The data of the sequenced mRNA have been deposited at European Nucleotide Archive under the accession number PRJEB27915.
Scanning electron microscopy
Scanning electron microscopy was performed according to in-house method of Microscopy Unit, Institute of Bioscience, Universiti Putra Malaysia. Uninoculated and G. boninense-inoculated oil palm root seedlings were sliced into 1 cm3 using clean blades. Samples were fixed in 4% glutaraldehyde for 2 days at 4 °C and washed with 0.1 M sodium cacodylate buffer thrice for 30 mins each. Post-fixation was carried out in osmium tetroxide for 2 h at 4 °C, followed by dehydration through a graded acetone series (35, 50, 75, and 95% for 30 mins and 100% for 1 h, with three changes of acetone). The samples were then transferred into specimen vials and placed in critical dryer (LEICA EM CPD030) for about 30 mins. Samples were mounted onto stub and sputtered with colloidal silver and gold (BALTEC SC030) prior to viewing under a scanning electron microscopy (XL30 ESEM, Philips).
GO and gene set enrichment analysis of DEGs
The DEGs were adopted for sequence homology searches (NCBI blast+) with biological sequences in CloudBlast database of Blast2GO with subset to Viridiplantae (taxa: 33090) [109]. Blastx program were executed in protein database, with limit to 20 blast hit results and restricted to maximum E-value of 0.001. Functional annotation and gene ontology of the DEGs were retrieved using default parameters in Blast2GO package and classified into biological process, molecular function and cellular component. Enrichment analysis of upregulated and downregulated GO terms was carried out via Gene Set Enrichment Analysis (GSEA) by applying the P-value of differential expression analysis as reference.
DNA extraction from infected root tissue and pure culture of G. boninense
Mycelia of G. boninense PER 71 were streaked and inoculated into 150 mL freshly made potato dextrose broth (Difco) in 250 mL conical flasks. The cultures were incubated for a week using benchtop incubator shaker SI-600 (Lab Companion, Korea) at 37 °C with agitation at 150 rpm. The grown mycelia were rinsed using distilled water, filtered using filter papers and promptly grounded using mortar and pestle in N2 suspension. Powdered form mycelia were stored in − 80 °C and ready to be used for DNA extraction. DNA of G. boninense from 200 mg of mycelia and infected root tissues were extracted using Prescott and Martin, (1987) method [108] with minor modifications. DNA pellet of both G. boninense and all infected samples were dissolved in ultrapure nuclease-free water and stored in − 20 °C until further use.
Validation of G. boninense presence within T1 oil palm seedlings via PCR and nested PCR
The DNA obtained from G. boninense PER 71 was used as control to validate the presence of infection within the roots of T1 samples. DNA of mycelia was amplified using primers of G. boninense strain PER71 internal transcribed spacer 1 (ITS1), partial sequence; 5.8S ribosomal RNA gene, complete sequence; and internal transcribed spacer 2 (ITS2), partial sequence with genebank accession number KM015454.1. Product length was expected to be 223 bp. Sequence of primer is listed in Table 7. Tubes containing reaction mixtures of DNA (5 ng), primers (5 ng each) and 2X KAPA Hifi HotStart Readymix (12.5 μL) were inserted in thermocycler (MyCycler™ Thermal Cycler System with Gradient Option, Bio-Rad, USA) with cycling parameters of 95 °C for 5 mins; 40 cycles of 94 °C for 35 s, 35 s at 63 °C, 40 s at 72 °C; and 72 °C for 10 mins. The amplicon of the PCR was sent for sequencing (Apical Scientific, Malaysia). Nested PCR was performed for 7 and 11 d.p.i root samples by using PCR product of 3 d.p.i root sample as template. Nested primer pair of the sequenced product was designed using Primer3 (v.0.4.0) based on certain criteria such as annealing temperature of primer pairs support separation of both PCRs based on the given parameter and high sensitivity of primers for improved detection threshold of nested PCR. Sequences of nested primers are listed in Table 7. The cycling parameters were similar to previous PCR with the exception of annealing temperature of 62 °C. Amplified products were dyed with 1.5 μL bromophenol blue dye and electrophoresed in 1% agarose gel. The gel was stained initially using FloroSafe DNA Stain and visualized using Gel Doc™ XR+ Imager (Bio-Rad, USA).
Table 7.
Primers used for validation of Ganoderma boninense DNA of oil palm roots
| Primer ID | Accession No. | Sense sequence (5′-3′) | Antisense sequence (5′-3′) |
|---|---|---|---|
| C (control) | KM015454.1 | CAACGGATCTCTTGGCTCTC | GCCGATCAATAAAAGACCGA |
| T1D3 (3 d.p.i) | KM015454.1 | CAACGGATCTCTTGGCTCTC | GCCGATCAATAAAAGACCGA |
| T1D7n (7 d.p.i) | – | GATCGGCTCCTCTCAAATGC | CGGTTAGAAGCTCGCCAAAC |
| T1D11n (11 d.p.i) | – | GATCGGCTCCTCTCAAATGC | CGGTTAGAAGCTCGCCAAAC |
| +VE | – | GATCGGCTCCTCTCAAATGC | CGGTTAGAAGCTCGCCAAAC |
Note: C - primer for nested PCR from untreated control sample
T1D3 - primer for normal PCR from 3 day-post-inoculation sample
T1D7n* - primer for nested PCR from 7 day-post-inoculation sample
T1D11n* - primer for nested PCR from 11 day-post-inoculation sample
+ve -primer for normal PCR from Ganoderma boninense PER71 pure culture
* retrieved from sequenced PCR amplicon of T1D3 sample
Experimental validation with qPCR
All primers encoding EgPR-1, EgEXP18, EgPG, EgMYC2, EgBGIA, EgMTP10, EgCht1, EgERF113, EgPIN8 and EgFIT (Table 8) were designed using Primer3 (v.0.4.0) for qPCR. Preliminary screening of defense-related genes and validation of genes expression were performed using qPCR Green Master Mix LRox (2x) according to manufacturer’s protocol (Biotechrabbit GmbH, Germany). The qPCR cycling parameters were set as follow; 1 cycle of 95 °C for 3 mins, 40 cycles of 95 °C for 15 s and 62 °C for 30 s, followed by melt curve at 65 °C to 95 °C (5 s for every increment of 0.5 °C). Analysis of the qPCR were performed using Bio-Rad CFX Manager (Bio-Rad, USA). The specificity of each primer pair was verified by melt curve analysis. Stability of five endogenous controls (Ubiquitin, Manganese Superoxide dismutase (MSD), GAPDH 2, β-actin and NADH 5) was tested over all samples (control, mock and treated). Expression levels of all analyzed genes were normalized against the expression level of three most stable reference genes which were GAPDH 2, β-actin and NADH 5. Using more than one reference genes as normalization factor for qPCR data is needed to avoid the drawbacks of single gene normalization error [110]. Primers for NADH5 and β-actin were designed based on Kwan et al. [111]. PCR efficiency, R2 value and slope value for the three reference genes which fall within acceptable range [112] were tabulated in Additional file 5. All assays were performed in three individual technical replicates of samples and non-template control was included.
Table 8.
Primers used for quantitative real-time PCR analysis
| Primers name | Sense sequence (5′-3′) | Antisense sequence (5′-3′) |
|---|---|---|
| EgGAPDH 2 | GAAGGTCATCATATCTGCTCCC | CATCAACAGTCTTCTGAGTGGC |
| EgNADH 5 | GCTCCCCTTTATTTGAATACCC | AATAGTTAGAGATGCCGCAAGC |
| Egβ-Actin | GAGAGAGCGTGCTACTCATCTT | CGGAAGTGCTTCTGAGATCC |
| EgPR-1 | GTCAGGCAGCTCAACTTCAC | TCGAACTTGAACTGGGTCGA |
| EgPG | CTGGAGTGAAGATTAGTCAGGTG | ACAGAACTAGAGGCAGTAACATG |
| EgEXP18 | ATGGCTACTTCTCTCCTGGC | CTTGATCCACAGCATTGCGA |
| EgMYC2 | CTCAATCAGAGATTCTACGCCC | CCTTGAGGGTATCAACTTGGC |
| EgBGIA | ATGCACTGGGAAGAGCTCAT | GATGCCATCTTTGTCCACCC |
| EgCht1 | AGCTCATCACTGTTCGACCA | CAAGAAAGCAGCGATCTCCC |
| EgFIT | GTGAAGTTGGAGTGCAGCAA | TCGCTGTCATCTCGAACTCA |
| EgMTP10 | TTGGCAGTTATCGCTTCCAC | TGCAGACCAAGTGTAGCCAT |
| EgPIN8 | GGTGGTGCTCGTATTGTGTC | CGAACCCTCCATGATGCTTG |
| EgERF113 | AGCAGCACTAAAGTTCAAAGGC | GAATAAGGTCTGGGTAGGAGGG |
Statistical analysis
According to RNA-seq data, DEGs were determined following cut off-values of log2 FC ≥ |1.0| and P-value < 0.01. The expression of each genes from qPCR analysis was normalized by three reference genes; GAPDH 2, NADH 5 and ß-actin expression levels. Expression levels were expressed as the mean ± SEM of three individual technical replicates of each sample. P < 0.01 denoted significant different between groups as assessed by one-way ANOVA analysis followed by Tukey’s test.
Additional files
Alignment of Ganoderma boninense PER71 ITS1/2 sequence with sequenced amplicon of normal and nested PCR. Nested primers were generated from sequenced PCR product of 3 days post infected oil palm root sample. Result showed conserved sequence which confirmed that G. boninense fungal hyphae were present in all T1 samples. (JPG 235 kb)
Top-hit species distribution of best-aligned gene annotations with highest percentage of similarity and lowest e-value. With restriction to 20 blast hits and e-value cut-off of 0.001, Elaeis guineensis was the most top-hit species with close to 9000 top-hits for upregulated genes and 15,000 top-hits for downregulated genes. (JPG 173 kb)
Statistics for blast and annotation procedures generated by Blast2Go Pro package from upregulated genes. (A) Annotation distribution; (B) E-value distribution; (C) Sequence similarity distribution; (D) Number of sequence with length. (JPG 118 kb)
Statistics for blast and annotation procedures generated by Blast2Go Pro package from downregulated genes. (E) Annotation distribution; (F) E-value distribution; (G) Sequence similarity distribution; (H) Number of sequence with length. (JPG 125 kb)
PCR efficiency, R2 value and slope value for the three reference genes. (PPTX 38 kb)
Acknowledgements
Not applicable.
Funding
This work was supported by Ministry of Education, Malaysia under NanoMalaysia Institute for Innovative Technology (NanoMITe) Consortium projects 2015–2020 (5526302). NS was supported by MyPhD scholarship from Ministry of Higher Education, Malaysia. MB was supported by Graduate Research Fellowship from Universiti Putra Malaysia. G. boninense PER 71 culture was provided by Dr. Idris Abu Seman from Malaysian Palm Oil Board (MPOB). Malaysian Nuclear Agency has provided Cobalt-60 gamma radiation service for the research purposes.
Availability of data and materials
The data of sequenced mRNA are available in the European Nucleotide Archive (ENA) under the accession number PRJEB27915.
Abbreviations
- AFB
Auxin signalling F-box protein
- BSR
Basal stem rot
- COBL-4
Cobra-like 4
- CR-4
CRINKLY-4
- CWDEs
Cell wall degrading enzymes
- d.p.i
Days-post-inoculation
- DAMP
Damage-associated molecular pattern
- DEG
Differentially expressed genes
- ERF
Ethylene-responsive transcription factor
- ETI
Effector-triggered immunity
- EXP
Expansin
- FC
Fold change
- FIT
Fer-like iron deficiency-induced transcription factor
- FRO2
Ferric reduction oxidase 2
- GLIP
GDSL esterase/ lipase
- GO
Gene Ontology
- GSEA
Gene set enrichment analysis
- HR
Hypersensitive response
- IAA
Indole-acetic acid
- Ile
Isoleucine
- ITS
Internal transcribed spacer
- JA
Jasmonate
- MGDG
Monogalactosyldiacylglycerol
- MYC
Transcription factor MYC
- NGS
Next-generation sequencing
- PAMP
Pathogen-associated molecular pattern
- PCD
Programmed cell death
- PCR
Polymerase chain reaction
- PER
Peroxidase
- PI
Protease inhibitor
- PID
Protein kinase PINOID
- PIN
Auxin efflux carrier
- PR
Pathogenesis-related protein
- PRR
Pattern recognition receptor
- PTI
Pattern-triggered immunity
- RBOH
Respiratory burst oxidase homolog
- RIN
RNA Integrity Number
- RLKs
Receptor-like kinases
- RNA-seq
RNA sequencing
- RNS
Reactive nitrogen species
- ROS
Reactive oxygen species
- RWB
Rubber wood block
- SAR
Systemic acquired resistance
- SCFTIR1/AFB
SKP-Cullin-F box, transport inhibitor resistant1/ auxin signaling f-box
- SCWs
Secondary cell walls
- TIR
Transport inhibitor response
- TPM
Transcript per kilobase million
Author’s contributions
SA designed and supervised the studies carried out by MB, NS and RR. MB and NS contributed equally in this study. MB performed the bioinformatics analysis of transcriptome data. MB, NS and RR contributed in DEGs analysis and validation of RNA-seq using qPCR. MB and NS prepared while SA reviewed and edited the manuscript. All authors read and approved the final version of the submitted manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohammad Nazri Abdul Bahari, Email: nazrischolar@gmail.com.
Nurshafika Mohd Sakeh, Email: shafikams@gmail.com.
Siti Nor Akmar Abdullah, Phone: +603-89464260, Email: snaa@upm.edu.my.
Redzyque Ramza Ramli, Email: ramzaeight@gmail.com.
Saied Kadkhodaei, Email: s_kadkhodaei@yahoo.com.
References
- 1.Kadarusman YB, Herabadi AG. Improving sustainable development within Indonesian palm oil: the importance of the reward system. Sust Dev. 2018. 10.1002/sd.1715.
- 2.Varkkey H, Tyson A, Choiruzzad SAB. Palm oil intensification and expansion in Indonesia and Malaysia: environmental and socio-political factors influencing policy. Forest Policy Econ. 2018;92:148–159. doi: 10.1016/j.forpol.2018.05.002. [DOI] [Google Scholar]
- 3.Stichnothe H, Schuchardt F, Rahutomo S. European renewable energy directive: critical analysis of important default values and methods for calculating greenhouse gas (GHG) emissions of palm oil biodiesel. Int J LCA. 2014;19(6):1294–1304. doi: 10.1007/s11367-014-0738-x. [DOI] [Google Scholar]
- 4.Kurnia J, Jangam S, Akhtar S. Advances in biofuel production from oil palm and palm oil processing wastes: a review. Biofuel Res J. 2016;3(1):332–346. doi: 10.18331/BRJ2016.3.1.3. [DOI] [Google Scholar]
- 5.Malaysian Palm Oil Board (MPOB) monthly export of oil palm products – 2017. http://bepi.mpob.gov.my/index.php/en/statistics/export/180-export-2017/811-monthly-export-of-oil-palm-products-2017.html. Accessed 10 April 2018.
- 6.Roslan A, Idris A. Economic impact of Ganoderma incidence on Malaysian oil palm plantation–a case study in Johor. Oil Palm Industry Econ J. 2012;12(1):24–30. [Google Scholar]
- 7.Moncalvo JM. Systematics of Ganoderma. In: Flood J, Bridge PD, Holderness M, editors. Ganoderma diseases of perennial crops. Wallingford: CAB International; 2000. pp. 23–45. [Google Scholar]
- 8.Susanto A, Sudharto P, Purba R. Enhancing biological control of basal stem rot disease (Ganoderma boninense) in oil palm plantations. Mycopathologia. 2005;159(1):153–157. doi: 10.1007/s11046-004-4438-0. [DOI] [PubMed] [Google Scholar]
- 9.Rees RW, Flood J, Hasan Y, Cooper RM. Effects of inoculum potential , shading and soil temperature on root infection of oil palm seedlings by the basal stem rot pathogen Ganoderma boninense. Plant Pathol. 2007;56(5):862–870. doi: 10.1111/j.1365-3059.2007.01621.x. [DOI] [Google Scholar]
- 10.GaitFee C. Management of Ganoderma diseases in oil palm plantations. Planter. 2011;87(1022):325–339. [Google Scholar]
- 11.Chong KP, Dayou J, Alexander A. Pathogenic nature of Ganoderma boninense and basal stem rot disease. In: Chong KP, Dayou J, Alexander A, editors. Detection and control of Ganoderma boninense in oil palm crop. Cham: Springer; 2017. p. 5–12.
- 12.Corley RHV, Tinker PB. Vegetative propagation and biotechnology. In: Corley RHV, Tinker PB, editors. The oil palm: Wiley; 2003. p. 201–15.
- 13.Morkunas I, Ratajczak L. The role of sugar signaling in plant defense responses against fungal pathogens. Acta Physiol Plant. 2014;36(7):1607–1619. doi: 10.1007/s11738-014-1559-z. [DOI] [Google Scholar]
- 14.Lam E, Kato N, Lawton M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature. 2001;411(6839):848. doi: 10.1038/35081184. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Z, Liu H, Wang C, Xu JR. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics. 2013;14(1):274. doi: 10.1186/1471-2164-14-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellincampi D, Cervone F, Lionetti V. Plant cell wall dynamics and wall-related susceptibility in plant–pathogen interactions. Front Plant Sci. 2014;5:228. doi: 10.3389/fpls.2014.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sattler S, Funnell-Harris D. Modifying lignin to improve bioenergy feedstocks: strengthening the barrier against pathogens? Front Plant Sci. 2013;4:70. doi: 10.3389/fpls.2013.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hok S, Attard A, Keller H. Getting the most from the host: how pathogens force plants to cooperate in disease. Mol Plant-Microbe Interact. 2010;23(10):1253–1259. doi: 10.1094/MPMI-04-10-0103. [DOI] [PubMed] [Google Scholar]
- 19.Cantu D, Blanco-Ulate B, Yang L, Labavitch JM, Bennett AB, Powell AL. Ripening-regulated susceptibility of tomato fruit to Botrytis cinerea requires NOR but not RIN or ethylene. Plant Physiol. 2009;150(3):1434–1449. doi: 10.1104/pp.109.138701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pazzagli L, Seidl-Seiboth V, Barsottini M, Vargas WA, Scala A, Mukherjee PK. Cerato-platanins: elicitors and effectors. Plant Sci. 2014;228:79–87. doi: 10.1016/j.plantsci.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 21.Quarantin A, Glasenapp A, Schäfer W, Favaron F, Sella L. Involvement of the Fusarium graminearum cerato-platanin proteins in fungal growth and plant infection. Plant Physiol Biochem. 2016;109:220–229. doi: 10.1016/j.plaphy.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Kabbage M, Yarden O, Dickman MB. Pathogenic attributes of Sclerotinia sclerotiorum: switching from a biotrophic to necrotrophic lifestyle. Plant Sci. 2015;233:53–60. doi: 10.1016/j.plantsci.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 23.Vargas WA, Martín JMS, Rech GE, Rivera LP, Benito EP, Díaz-Mínguez JM, Thon MR, Sukno SA. Plant defense mechanisms are activated during biotrophic and necrotrophic development of Colletotricum graminicola in maize. Plant Physiol. 2012;158(3):1342–1358. doi: 10.1104/pp.111.190397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Idris AS, Kushairi A, Ismail S, Ariffin D. Selection for partial resistance in oil palm progenies to Ganoderma basal stem rot. J Oil Palm Res. 2004;16(2):12–18. [Google Scholar]
- 25.Sustainable Oil Palm Farming / Tenera, Dura, and Pisifera. http://akvopedia.org/wiki/Sustainable_Oil_Palm_Farming_/_Tenera,_Dura,_and_Pisifera. Accessed 10 April 2018.
- 26.Kushairi A, Jalani BS, Rajanaidu N, Zakri AH. Variation in Dura × Pisifera planting materials, 1: bunch yield. Elaeis. 1994;6:14–24. [Google Scholar]
- 27.Nusaibah SA, Akmar ASN, Idris AS, Sariah M, Pauzi ZM. Involvement of metabolites in early defense mechanism of oil palm (Elaeis guineensis Jacq.) against Ganoderma disease. Plant Physiol Biochem. 2016;109:156–165. doi: 10.1016/j.plaphy.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 28.Ho CL, Tan YC, Yeoh KA, Ghazali AK, Yee WY, Hoh CC. De novo transcriptome analyses of host-fungal interactions in oil palm (Elaeis guineensis Jacq.) BMC Genomics. 2016;17(1):66. doi: 10.1186/s12864-016-2368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho CL, Tan YC, Yeoh KA, Lee WK, Ghazali AK, Yee WY, Hoh CC. Transcriptional response of oil palm (Elaeis guineensis Jacq.) inoculated simultaneously with both Ganoderma boninense and Trichoderma harzianum. Plant Gene. 2018;13:56–63. doi: 10.1016/j.plgene.2018.01.003. [DOI] [Google Scholar]
- 30.Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pozo MJ, Van Der Ent S, Van Loon LC, Pieterse CM. Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol. 2008;180(2):511–523. doi: 10.1111/j.1469-8137.2008.02578.x. [DOI] [PubMed] [Google Scholar]
- 32.Hiruma K, Nishiuchi T, Kato T, Bednarek P, Okuno T, Schulze-Lefert P, Takano Y. Arabidopsis ENHANCED DISEASE RESISTANCE 1 is required for pathogen-induced expression of plant defensins in nonhost resistance, and acts through interference of MYC2-mediated repressor function. Plant J. 2011;67(6):980–992. doi: 10.1111/j.1365-313X.2011.04651.x. [DOI] [PubMed] [Google Scholar]
- 33.Kloppholz S, Kuhn H, Requena N. A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol. 2011;21(14):1204–1209. doi: 10.1016/j.cub.2011.06.044. [DOI] [PubMed] [Google Scholar]
- 34.Liu G, Kennedy R, Greenshields DL, Peng G, Forseille L, Selvaraj G, Wei Y. Detached and attached Arabidopsis leaf assays reveal distinctive defense responses against hemibiotrophic Colletotrichum spp. Mol Plant-Microbe Interact. 2007;20(10):1308–1319. doi: 10.1094/MPMI-20-10-1308. [DOI] [PubMed] [Google Scholar]
- 35.Kendziorski C, Irizarry RA, Chen KS, Haag JD, Gould MN. On the utility of pooling biological samples in microarray experiments. Proc Natl Acad Sci U S A. 2005;102(12):4252–4257. doi: 10.1073/pnas.0500607102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biswas S, Agrawal YN, Mucyn TS, Dangl JL, Jones CD. Biological averaging in RNA-seq. Quant. Methods arXiv 2013;1309.0670 [q-bio.QM].
- 37.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theor Biosci. 2012;131(4):281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 38.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colangelo EP, Guerinot ML. The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell. 2004;16(12):3400–12. doi: 10.1105/tpc.104.024315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Senthil-Kumar M, Mysore KS. Nonhost resistance against bacterial pathogens: retrospectives and prospects. Annu Rev Phytopathol. 2013;51:407–427. doi: 10.1146/annurev-phyto-082712-102319. [DOI] [PubMed] [Google Scholar]
- 41.Nürnberger T, Brunner F, Kemmerling B, Piater L. Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev. 2004;198(1):249–266. doi: 10.1111/j.0105-2896.2004.0119.x. [DOI] [PubMed] [Google Scholar]
- 42.Zipfel C, Felix G. Plants and animals: a different taste for microbes? Curr Opin Plant Biol. 2005;8(4):353–360. doi: 10.1016/j.pbi.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Zipfel C. Plant pattern-recognition receptors. Trends Immunol. 2014;35(7):345–351. doi: 10.1016/j.it.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Nikonorova N, Vu LD, Czyzewicz N, Gevaert K, De Smet I. A phylogenetic approach to study the origin and evolution of the CRINKLY4 family. Front Plant Sci. 2015;6:880. doi: 10.3389/fpls.2015.00880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stahl Y, Wink R, Ingram G, Simon R. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol. 2009;19(11):909–914. doi: 10.1016/j.cub.2009.03.060. [DOI] [PubMed] [Google Scholar]
- 46.Shiu S, Bleecker A. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132(2):530–543. doi: 10.1104/pp.103.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaid N, Pandey PK, Tuteja N. Lectin receptor-like kinases and their emerging role in abiotic stress tolerance. In: Tuteja N, Gill SS, editors. Abiotic stress response in plants. Weinheim: Wileh-VCH. 2016. pp. 203–216. [Google Scholar]
- 48.Choi WG, Miller G, Wallace I, Harper J, Mittler R, Gilroy S. Orchestrating rapid long-distance signaling in plants with Ca2+, ROS and electrical signals. Plant J. 2017;90(4):698–707. doi: 10.1111/tpj.13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niderman T, Genetet I, Bruyere T, Gees R, Stintzi A, Legrand M, Fritig B, Mosinger E. Pathogenesis-related PR-1 proteins are antifungal. Isolation and characterization of three 14-kilodalton proteins of tomato and of a basic PR-1 of tobacco with inhibitory activity against Phytophthora infestans. Plant Physiol. 1995;108(1):17–27. doi: 10.1104/pp.108.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Loon L, Rep M, Pieterse C, Pieterse C. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- 51.Gamir J, Darwiche R, Hof P, Choudhary V, Stumpe M, Schneiter R, Mauch F. The sterol-binding activity of PATHOGENESIS-RELATED PROTEIN 1 reveals the mode of action of an antimicrobial protein. Plant J. 2017;89(3):502–509. doi: 10.1111/tpj.13398. [DOI] [PubMed] [Google Scholar]
- 52.Choon RLT, Sariah M, Mariam S. Ergosterol from the soilborne fungus Ganoderma boninense. J Basic Microbiol. 2012;52(5):608–612. doi: 10.1002/jobm.201100308. [DOI] [PubMed] [Google Scholar]
- 53.Gao Y, Chen T, Breuil C. Ergosterol—a measure of fungal growth in wood for staining and pitch control fungi. Biotechnol Tech. 1993;7(9):621–626. doi: 10.1007/BF00151858. [DOI] [Google Scholar]
- 54.Muniroh MS, Sariah M, Abidin MZ, Lima N, Paterson RRM. Rapid detection of Ganoderma-infected oil palms by microwave ergosterol extraction with HPLC and TLC. J Microbiol Methods. 2014;100:143–147. doi: 10.1016/j.mimet.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 55.Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9(10):490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 56.Trchounian A, Petrosyan M, Sahakyan N. Plant cell redox homeostasis and reactive oxygen species. In: Gupta DK, Palma JM, Corpas FJ, editors. Redox state as a central regulator of plant-cell stress responses. Cham: Springer; 2016. p. 25–50.
- 57.Sasaki K, Iwai T, Hiraga S, Kuroda K, Seo S, Mitsuhara I, Miyasaka A, Iwano M, Ito H, Matsui H, Ohashi Y. Ten rice peroxidases redundantly respond to multiple stresses including infection with rice blast fungus. Plant Cell Physiol. 2004;5(10):1442–1452. doi: 10.1093/pcp/pch165. [DOI] [PubMed] [Google Scholar]
- 58.Passardi F, Cosio C, Penel C, Dunand C. Peroxidases have more functions than a Swiss army knife. Plant Cell Rep. 2005;24(5):255–265. doi: 10.1007/s00299-005-0972-6. [DOI] [PubMed] [Google Scholar]
- 59.Yoshioka H, Numata N, Nakajima K, Katou S, Kawakita K, Rowland O, Jones JD, Doke N. Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell. 2003;15:706–718. doi: 10.1105/tpc.008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abdullah SNA, Akhtar MS. Plant and necrotrophic fungal pathogen interaction: mechanism and mode of action. In: Hakeem KR, Akhtar MS, Abdullah SNA, editors. Plant, soil and microbes. Cham: Springer; 2016. pp. 29–53. [Google Scholar]
- 61.MacMillan CP, Birke H, Chuah A, Brill E, Tsuji Y, Ralph J, Dennis E, Llewellyn D, Pettolino FA. Tissue and cell-specific transcriptomes in cotton reveal the subtleties of gene regulation underlying the diversity of plant secondary cell walls. BMC Genomics. 2017;18(1):539. doi: 10.1186/s12864-017-3902-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cosgrove DJ, Jarvis MC. Comparative structure and biomechanics of plant primary and secondary cell walls. Front Plant Sci. 2012;3:204. doi: 10.3389/fpls.2012.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown D, Zeef L, Ellis J, Goodacre R, Turner SR. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell. 2005;17(8):2281–2295. doi: 10.1105/tpc.105.031542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sato K, Suzuki R, Nishikubo N, Takenouchi S, Ito S, Nakano Y, Nakaba S, Sano Y, Funada R, Kajita S, Kitano H. Isolation of a novel cell wall architecture mutant of rice with defective Arabidopsis COBL4 ortholog BC1 required for regulated deposition of secondary cell wall components. Planta. 2010;232(1):257–270. doi: 10.1007/s00425-010-1171-4. [DOI] [PubMed] [Google Scholar]
- 65.Liu L, Shang-Guan K, Zhang B, Liu X, Yan M, Zhang L, Shi Y, Zhang M, Qian Q, Li J, Zhou Y. Brittle Culm1, a COBRA-like protein, functions in cellulose assembly through binding cellulose microfibrils. PLoS Genet. 2013;9(8):e1003704. doi: 10.1371/journal.pgen.1003704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marowa P, Ding A, Kong Y. Expansins: roles in plant growth and potential applications in crop improvement. Plant Cell Rep. 2016;35(5):949–965. doi: 10.1007/s00299-016-1948-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jones L, McQueen-Mason S. A role for expansins in dehydration and rehydration of the resurrection plant Craterostigma Plantagineum. FEBS Lett. 2004;559(1–3):61–65. doi: 10.1016/S0014-5793(04)00023-7. [DOI] [PubMed] [Google Scholar]
- 68.Guo W, Zhao J, Li X, Qin L, Yan X, Liao H. A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J. 2011;66(3):541–552. doi: 10.1111/j.1365-313X.2011.04511.x. [DOI] [PubMed] [Google Scholar]
- 69.Li F, Han Y, Feng Y, Xing S, Zhao M, Chen Y, Wang W. Expression of wheat expansin driven by the RD29 promoter in tobacco confers water-stress tolerance without impacting growth and development. J Biotechnol. 2013;163(3):281–291. doi: 10.1016/j.jbiotec.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 70.Nanjo Y, Nakamura T, Komatsu S. Identification of indicator proteins associated with flooding injury in soybean seedlings using label-free quantitative proteomics. J Proteome Res. 2013;12(11):4785–4798. doi: 10.1021/pr4002349. [DOI] [PubMed] [Google Scholar]
- 71.Abuqamar S, Ajeb S, Sham A, Enan MR, Iratni R. A mutation in the expansin-like A2 gene enhances resistance to necrotrophic fungi and hypersensitivity to abiotic stress in Arabidopsis thaliana. Mol Plant Pathol. 2013;14(8):813–827. doi: 10.1111/mpp.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cosgrove DJ. Plant expansins: diversity and interactions with plant cell walls. Curr Opin Plant Biol. 2015;25:162–172. doi: 10.1016/j.pbi.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H, Child M, Bogyo M. Proteases as regulators of pathogenesis: examples from the Apicomplexa. Biochim Biophys Acta. 2012;1824(1):177–185. doi: 10.1016/j.bbapap.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seow-Neng C, Bakar NA, Mahmood M, Chai-Ling H, Shaharuddin NA. Alternative strategy in crop protection: protease inhibitors from turmeric. In: Abdullah SNA, Ho CL, Wagstaff C. crop improvement: Cham: Springer International Publishing; 2017. p. 253–70.
- 75.Rustgi S, Boex-Fontvieille E, Reinbothe C, von Wettstein D, Reinbothe S. Serpin1 and WSCP differentially regulate the activity of the cysteine protease RD21 during plant development in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2017;201621496. [DOI] [PMC free article] [PubMed]
- 76.Petrov V, Hille J, Mueller-Roeber B, Gechev T. ROS-mediated abiotic stress-induced programmed cell death in plants. Front Plant Sci. 2015;6:69. doi: 10.3389/fpls.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dutt S, Gaur V, Taj G, Kumar A. Differential induction of two different cystatin genes during pathogenesis of Karnal bunt (Tilletia indica) in wheat under the influence of jasmonic acid. Gene. 2012;506(1):253–260. doi: 10.1016/j.gene.2012.06.028. [DOI] [PubMed] [Google Scholar]
- 78.Hartl M, Giri AP, Kaur H, Baldwin IT. The multiple functions of plant serine protease inhibitors: defense against herbivores and beyond. Plant Signal Behav. 2011;6(7):1009–1011. doi: 10.4161/psb.6.7.15504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bode RF, Halitschke R, Kessler A. Herbivore damage-induced production and specific anti-digestive function of serine and cysteine protease inhibitors in tall goldenrod, Solidago altissima L. (Asteraceae) Planta. 2013;237(5):1287–1296. doi: 10.1007/s00425-013-1845-9. [DOI] [PubMed] [Google Scholar]
- 80.Müller V, Bonacci G, Batthyany C, Amé MV, Carrari F, Gieco J, Asis R. Peanut seed cultivars with contrasting resistance to Aspergillus parasiticus colonization display differential temporal response of protease inhibitors. Phytopathology. 2017;107(4):474–482. doi: 10.1094/PHYTO-09-16-0346-R. [DOI] [PubMed] [Google Scholar]
- 81.Bangrak P, Chotigeat W. Molecular cloning and biochemical characterization of a novel cystatin from Hevea rubber latex. Plant Physiol Biochem. 2011;49(3):244–250. doi: 10.1016/j.plaphy.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Y, Lubberstedt T, Xu M. The genetic and molecular basis of plant resistance to pathogens. J Genet Genomics. 2013;40(1):23–35. doi: 10.1016/j.jgg.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 83.Habib H, Fazili K. Plant protease inhibitors: a defense strategy in plants. Biotechnol Mol Biol Rev. 2007;2(3):68–85. [Google Scholar]
- 84.Dunse KM, Stevens JA, Lay FT, Gaspar YM, Heath RL, Anderson MA. Coexpression of potato type I and II proteinase inhibitors gives cotton plants protection against insect damage in the field. Proc Natl Acad Sci. 2010;107(34):15011–15015. doi: 10.1073/pnas.1009241107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee DS, Kim BK, Kwon SJ, Jin HC, Park OK. Arabidopsis GDSL lipase 2 plays a role in pathogen defense via negative regulation of auxin signaling. Biochem Biophys Res Commun. 2009;379(4):1038–1042. doi: 10.1016/j.bbrc.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 86.Gao M, Yin X, Yang W, Lam SM, Tong X, Liu J, Wang X, Li Q, Shui G, He Z. GDSL lipases modulate immunity through lipid homeostasis in rice. PLoS Pathog. 2017;13(11):e1006724. doi: 10.1371/journal.ppat.1006724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Christensen LP. Galactolipids as potential health promoting compounds in vegetable foods. Recent Pat Food Nutr Agric. 2009;1(1):50–58. doi: 10.2174/2212798410901010050. [DOI] [PubMed] [Google Scholar]
- 88.Gao QM, Yu K, Xia Y, Shine MB, Wang C, Navarre D, Kachroo A, Kachroo P. Mono-and digalactosyldiacylglycerol lipids function nonredundantly to regulate systemic acquired resistance in plants. Cell Rep. 2014;9(5):1681–1691. doi: 10.1016/j.celrep.2014.10.069. [DOI] [PubMed] [Google Scholar]
- 89.Ferrari S, Galletti R, Pontiggia D, Manfredini C, Lionetti V, Bellincampi D, Cervone F, DeLorenzo G. Transgenic expression of a fungal endo-polygalacturonase increases plant resistance to pathogens and reduces auxin sensitivity. Plant Physiol. 2008;146:669–681. doi: 10.1104/pp.107.109686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Savatin DV, Ferrari S, Sicilia F, De Lorenzo G. Oligogalacturonide-auxin antagonism does not require posttranscriptional gene silencing or stabilization of auxin response repressors in Arabidopsis. Plant Physiol. 2011;157(3):1163–1174. doi: 10.1104/pp.111.184663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee SH. Cho HT. PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells. Plant Cell. 2006;18(7):1604–1616. doi: 10.1105/tpc.105.035972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang H, Lin J, Chang Y, Jiang C. Comparative transcriptomic analysis reveals that ethylene/H2O2-mediated hypersensitive response and programmed cell death determine the compatible interaction of sand pear and Alternaria alternata. Front Plant Sci. 2017;8:195. doi: 10.3389/fpls.2017.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao Y, Chang X, Qi D, Dong L, Wang G, Fan S, Jiang L, Cheng Q, Chen X, Han D, Xu P. A novel soybean ERF transcription factor, GmERF113, increases resistance to Phytophthora sojae infection in soybean. Front Plant Sci. 2017;8:229. doi: 10.3389/fpls.2017.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Asahina M, Azuma K, Pitaksaringkarn W, Yamazaki T, Mitsuda N, Ohme-Takagi M, Yamaguchi S, Kamiya Y, Okada K, Nishimura T, Koshiba T. Spatially selective hormonal control of RAP2. 6L and ANAC071 transcription factors involved in tissue Reunion in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108(38):16128–16132. doi: 10.1073/pnas.1110443108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell. 2009;21(8):2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhu Z, An F, Feng Y, Li P, Xue L, Mu A. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108(30):12539–12544. doi: 10.1073/pnas.1103959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schweizer F, Fernández-Calvo P, Zander M, Diez-Diaz M, Fonseca S, Glauser G, Lewsey MG, Ecker JR, Solano R, Reymond P. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell. 2013;25(8):3117–3132. doi: 10.1105/tpc.113.115139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico C, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, Pauwels L. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23(2):701–715. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kazan K, Manners JM. MYC2: the master in action. Mol Plant. 2013;6(3):686–703. doi: 10.1093/mp/sss128. [DOI] [PubMed] [Google Scholar]
- 100.Gasperini D, Chételat A, Acosta I, Goossens J, Pauwels L, Goossens A, Dreos R, Alfonso E, Farmer EE. Multilayered organization of jasmonate signalling in the regulation of root growth. PLoS Genet. 2015;11(6):e1005300. doi: 10.1371/journal.pgen.1005300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P. FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett. 2004;577(3):528–534. doi: 10.1016/j.febslet.2004.10.062. [DOI] [PubMed] [Google Scholar]
- 102.Rossi FR, Krapp AR, Bisaro F, Maiale SJ, Pieckenstain FL, Carrillo N. Reactive oxygen species generated in chloroplasts contribute to tobacco leaf infection by the necrotrophic fungus Botrytis cinerea. Plant J. 2017;92(5):761–773. doi: 10.1111/tpj.13718. [DOI] [PubMed] [Google Scholar]
- 103.Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 2003;15(1):165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K. Repressor-and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 2005;139(2):949–959. doi: 10.1104/pp.105.068544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Khan NA, Khan MIR, Ferrante A, Poor P. Ethylene: a key regulatory molecule in plants. Front Plant Sci. 2017;8:1782. doi: 10.3389/fpls.2017.01782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Idris AS, Kushairi D, Ariffin D, Basri MW. Technique for inoculation of oil palm germinated seeds with Ganoderma. MPOB Inf Ser. 2006;314:1–4. [Google Scholar]
- 107.Sundram S, Meon S, Seman IA, Othman R. Symbiotic interaction of endophytic bacteria with arbuscular mycorrhizal fungi and its antagonistic effect on Ganoderma boninense. J Microbiol. 2011;49(4):551. doi: 10.1007/s12275-011-0489-3. [DOI] [PubMed] [Google Scholar]
- 108.Prescott A, Martin C. A rapid method for the quantitative assessment of levels of specific mRNAs in plants. Plant Mol Biol Report. 1986;4(4):219–224. doi: 10.1007/BF02675414. [DOI] [Google Scholar]
- 109.Conesa A, Götz S, García-Gómez J, Terol J. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 110.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):research0034–research0031. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kwan YM, Meon S, Ho CL, Wong MY. Selection of reference genes for quantitative real-time PCR normalization in Ganoderma-infected oil palm (Elaeis guineensis) seedlings. Australasian Plant Path. 2016;45(3):261–268. doi: 10.1007/s13313-016-0417-4. [DOI] [Google Scholar]
- 112.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of Ganoderma boninense PER71 ITS1/2 sequence with sequenced amplicon of normal and nested PCR. Nested primers were generated from sequenced PCR product of 3 days post infected oil palm root sample. Result showed conserved sequence which confirmed that G. boninense fungal hyphae were present in all T1 samples. (JPG 235 kb)
Top-hit species distribution of best-aligned gene annotations with highest percentage of similarity and lowest e-value. With restriction to 20 blast hits and e-value cut-off of 0.001, Elaeis guineensis was the most top-hit species with close to 9000 top-hits for upregulated genes and 15,000 top-hits for downregulated genes. (JPG 173 kb)
Statistics for blast and annotation procedures generated by Blast2Go Pro package from upregulated genes. (A) Annotation distribution; (B) E-value distribution; (C) Sequence similarity distribution; (D) Number of sequence with length. (JPG 118 kb)
Statistics for blast and annotation procedures generated by Blast2Go Pro package from downregulated genes. (E) Annotation distribution; (F) E-value distribution; (G) Sequence similarity distribution; (H) Number of sequence with length. (JPG 125 kb)
PCR efficiency, R2 value and slope value for the three reference genes. (PPTX 38 kb)
Data Availability Statement
The data of sequenced mRNA are available in the European Nucleotide Archive (ENA) under the accession number PRJEB27915.