Abstract
Cell migration is a critical mechanism controlling tissue morphogenesis, epithelial wound healing and tumor metastasis. Migrating cells depend on orchestrated remodeling of the plasma membrane and the underlying actin cytoskeleton, which is regulated by the spectrin-adducin-based membrane skeleton. Expression of adducins is altered during tumorigenesis, however, their involvement in metastatic dissemination of tumor cells remains poorly characterized. This study investigated the roles of α-adducin (ADD1) and γ-adducin (ADD3) in regulating migration and invasion of non-small cell lung cancer (NSCLC) cells. ADD1 was mislocalized, whereas ADD3 was markedly downregulated in NSCLC cells with the invasive mesenchymal phenotype. CRISPR/Cas9-mediated knockout of ADD1 and ADD3 in epithelial-type NSCLC and normal bronchial epithelial cells promoted their Boyden chamber migration and Matrigel invasion. Furthermore, overexpression of ADD1, but not ADD3, in mesenchymal-type NSCLC cells decreased cell migration and invasion. ADD 1-overexpressing NSCLC cells demonstrated increased adhesion to the extracellular matrix (ECM), accompanied by enhanced assembly of focal adhesions and hyperphosphorylation of Src and paxillin. The increased adhesiveness and decreased motility of ADD 1-overexpressing cells were reversed by siRNA-mediated knockdown of Src. By contrast, the accelerated migration of ADD1 and ADD3-depleted NSCLC cells was ECM adhesion-independent and was driven by the upregulated expression of pro-motile cadherin-11. Overall, our findings reveal a novel function of adducins as negative regulators of NSCLC cell migration and invasion, which could be essential for limiting lung cancer progression and metastasis.
Keywords: membrane skeleton, invasion, focal adhesions, Src, actin cytoskeleton
1. INTRODUCTION
Cell migration is a critical mechanism that mediates plasticity of epithelial layers under normal conditions and in the disease state. For example, in the developing embryo and self-rejuvenating adult tissues, the coordinated migration of cells of different lineages plays key roles in the cell patterning, establishment of tissue boundaries and organ morphogenesis [1, 2]. Furthermore, cell migration is essential for repairing epithelial layers damaged by environmental and inflammatory factors [3, 4]. However, a dark side of epithelial cell motility is well recognized during tumorigenesis, where accelerated migration of cancer cells drives tumor dissemination and metastasis [5, 6].
Cell motility is accompanied by the dramatic remodeling of the cellular cortex, driven by the extension of plasma membrane protrusions at the migrating leading edge and membrane retraction at the trailing cell edge [7, 8]. The forces that enable such remodeling are generated by the cortical actomyosin cytoskeleton. Indeed, the formation of the membrane protrusions is known to be driven by the actin polymerization motor, whereas membrane retraction is mediated by the contractile activity of the non-muscle myosin II (NM II) motor [9, 10]. The efficient transduction of pushing and pulling forces from the actomyosin cytoskeleton to the plasma membrane requires physical interactions between the membrane and the underlying actin filaments [11, 12]. Therefore, the molecular complexes that mediate such interactions could serve as essential regulators of cell motility [13]. Although several mechanisms are known to connect plasma membrane to the actin cytoskeleton, the most common involves the spectrin-ankyrin-adducin membrane skeleton [14–16]. A core component of this membrane skeleton is a polymeric lattice composed of spectrin oligomers that lines the inner side of the plasma membrane. This spectrin lattice is linked to both transmembrane proteins and actin filaments, thereby servings as an essential interphase between the membrane and the actin cytoskeleton [14–16].
Adducins are important components of the spectrin-based membrane skeleton. They bind both spectrin oligomers and actin filaments, thereby promoting and stabilizing association of these cellular structures [17, 18], Furthermore, adducins could interact with the actin cytoskeleton in spectrin-independent fashion, by capping and bundling actin filaments [18–21], The adducin family is composed of three closely related members, α-adducin (ADD1), β-adducin (ADD2) and γ-adducin (ADD3) [22, 23], ADD1 and ADD3 are ubiquitously expressed in different mammalian tissues, whereas expression of ADD2 is limited to the brain and the hematopoietic system [24, 25], All three adducins have a similar molecular organization, which is characterized by a globular head domain at the N-terminus, a neck domain, and a tail domain at the C-terminus of the molecule [20, 24, 25], The neck and head domains possess oligomerization sites that facilitate formation of adducin heterodimers and heterotetramers [22, 23], The C-terminal tail mediates adducin interactions with main binding partners such as actin filaments, spectrins and transmembrane proteins [22, 23], It also contains a highly-conserved 22-residue MARCKS-related motif with phosphorylation sites for protein kinase A (PKA) and protein kinase C (PKC) [20, 22, 23], Phosphorylation of these MARCKS motifs was shown to disrupt interactions between adducins and both actin filaments and spectrin oligomers [20, 26–28].
Due to their involvement in the assembly of the cortical actin cytoskeleton and the spectrin-based membrane skeleton, adducins have multiple functional roles in different eukaryotic cells. For example, in epithelial cells, ADD1 and ADD3 control the formation of the lateral plasma membrane domain and regulate the dynamics of intercellular junctions [29–33], In neural tissue, adducins regulate synaptic plasticity and synaptic contacts [34, 35] and play a role in neuronal morphology [36].
Surprisingly, little is known about the roles and mechanisms of adducin-dependent regulation of cell motility. Several previous studies that focused on the roles of ADD1 in the migration of normal epithelial cells and cancer cells yielded controversial results with some studies describing ADD1 as a positive regulator [27, 37], while other suggesting it as a negative regulator of cell motility [38, 39]. The reason for this discrepancy is not clear. Importantly, the involvement of ADD3 in cell migration and invasion have not been previously investigated. Since expression and activity of adducins is altered in different tumors [40] it is essential to understand how these membrane skeleton proteins regulate migration and invasion of cancer cells and contribute to tumor metastasis.
The overall goal of this study was to understand the roles of adducins in the regulation of non-small cell lung cancer (NSCLC) cell motility in vitro. Clinical evidence suggests altered adducin expression and activity in lung cancer cells. A recent study demonstrated that oncogenic transcription factor ZNF322A upregulated ADD1 expression in a subset of NSCLC patients and connected this event to tumor growth and metastasis [37]. Another study documented hepatocyte growth factor-dependent phosphorylation of ADD1 and ADD3 in small cell lung cancer cells, which may promote lung cancer cell invasion [41]. Interestingly, accumulation of an alternative spliced, long isoform of ADD3 has been reported in NSCLC, although the functional significance of such tumor-related alternative splicing remains elusive [42]. Finally, loss of ADD1 was shown to impair the establishment of the basolateral plasma membrane in normal lung epithelial cells [29], which may affect cell surface expression of adhesion proteins and chemotactic receptors. In the present study, we found that adducins serve as negative regulators of NSCLC cell motility, acting via different mechanisms that involve modulation of cell-matrix adhesion and cellular level of cadherin-11.
2. MATERIALS AND METHODS
2.1. Human gene expression analysis
Gene expression profiles for human lung cancer samples were generated by The Cancer Genome Atlas (TCGA). We utilized the RSEM-quantified RNA-seq data for lung adenocarcinoma (LUAD) patients made available by the Broad GDAC Firehose repository (http://gdac.broadinstitute.org). This dataset includes 576 samples (515 primary solid tumors; 59 normal lung tissue). In order to determine whether ADD1 or ADD3 are differentially expressed between tumor and normal tissues we removed 58 cases with paired controls to create independent groups and performed a Wilcoxon rank sum test on the +1-shifted log2 values.
2.2. Antibodies and other reagents
The following monoclonal (mAb) and polyclonal (pAb) antibodies were used to detect cytoskeletal, focal adhesion and other proteins: anti-ADD1 pAb and ADD3 mAb (Santa Cruz Biotechnology, Dallas, TX); anti-FAK, paxillin, E-cadherin and vimentin mAbs (BD Biosciences, San Jose, CA); anti p-FAK, FLAG, p-paxillin, Src, p-Src, cadherin-11 and GAPDH pAbs (Cell Signaling, Beverly, MA); anti-N-cadherin pAb (Abcam, Cambridge, MA); anti-P-cadherin mAb (Millipore, Billerica, MA). Alexa Fluor-488-conjugated donkey anti-rabbit, Alexa Fluor-555-conjugated donkey anti-mouse secondary antibodies, and Alexa Fluor-488 or Alexa Fluor-555-labeled phalloidin were obtained from Thermo-Fisher Scientific (Waltham, MA). Horseradish peroxidase-conjugated goat anti-rabbit and anti-mouse secondary antibodies were obtained from Bio-Rad Laboratories. A functional inhibitory goat anti-human cadherin-11 antibody was obtained from R&D Systems (Minneapolis, MN) and control goat IgG was purchased from Jackson Immunoresearch Laboratories (West Grove, PA).
2.3. Cell Culture
Non-transformed and transformed HBEC3-KT and HBEC3-KTRL53Myc human bronchial epithelial cells were obtained from Dr. John D. Minna, The University of Texas Southwestern Medical Center. NCI-H1573, HCC4019, NCI-H1299, NCI-H2030, NCI-H1703 and NCI-H23 lung cancer cell lines were provided by Dr. Samir Hanash, The University of Texas MD Anderson Cancer Center, whereas 16HBE14o bronchial epithelial cells were provided by Dr. Dieter Gruenert, University of California San Francisco. NCI-H441 lung cancer cells were purchased from the American Type Culture Collection. 16HBE14o cells were cultured in MEM medium (Thermo-Fisher) supplemented with 10% fetal bovine serum (FBS), HEPES, penicillin and streptomycin. H441 cells were cultured in RPMI medium (Thermo-Fisher) supplemented with 5% FBS, dexamethasone (250 μg/ml, Sigma-Aldrich), insulin-transferrin-sodium selenite (Thermo-Fisher), penicillin and streptomycin. H1573, HCC4019, H1299, H2030, H1703 and H23 cells were cultured in RPMI medium supplemented with 10% FBS, HEPES, sodium pyruvate and antibiotics. HBEC3-KT and HBEC3-KTRL53Myc cells were cultured in Keratinocyte serum-free medium (Thermo-Fisher) without antibiotics. The cells were seeded on collagen-coated coverslips for immunolabeling experiments and on 6-well plastic plates for the functional and biochemical studies.
2.4. CRISPR-Cas9 mediated knockout of ADD1 and ADD3
A stable knockout of ADD1 or ADD3 in H1573 and 16HBE14o cells was carried out using a CRISPR-Cas9 technology. The guide oligonucleotide sequences used for knocking out ADD1 and ADD3 are shown in the Supplemental Table 1. The guide oligonucleotides were phosphorylated, annealed and cloned into the BsmBI site of a lentiCRISPR v2 vector (Addgene, 52961) according to a published protocol [43, 44], Obtained constructs were verified with sequencing. Transfer plasmids possessing annealed guide oligonucleotides were transformed into recombination-deficient Stbl3 bacteria and amplified plasmids were isolated from the bacteria using Qiagen midi prep plasmid isolation kit. Lentiviruses were produced by transfecting HEK-293T cells with the transfer lentiCRISPR v2 plasmids and packaging plasmids pLTR-G (Addgene, 17532) and pCD/NL-BH*DDD (Addgene, 17531). Viral supernatants were collected 48 and 72 h after transfection and used to infect H1573 and 16HBE14o cells. After 24 h of the infection, the lentivirus-containing medium were replaced with fresh cell culture medium containing puromycin (2 μg/ml for H1573 cells and 10 μg/ml forl6HBE14o cells) and puromycin-resistant cells were collected after 7 day selection.
2.5. Generation of adducin-overexpressing lung cancer cells
H1299 cell lines with stable overexpression of either ADD1, or ADD3 were generated using a lentiviral expression system. An expression pLKO. AS2.neomycin vector encoding FLAG-tagged wild-type ADD1 was obtained from Dr. Hong-Chen Chen, National Chung Hsing University, Taiwan [45], A cDNA encoding human ADD3 (clone BC062559 in the pOTB7 vector) was obtained from Transomic Technologies (Huntsville, AL) and the ADD3 insert was cloned into the pLKO.AS2.neo vector. All obtained constructs were verified by sequencing. H1299 cells grown till 70% confluency were infected with the lentiviral particle-containing supernatants. After 24 h infection, the medium was replaced with fresh cell culture medium containing 500 μ/ml neomycin and neomycin-resistant cells were selected for 7 days.
2.6. Cell fractionation and immunoblotting analysis
The nuclear and the cytoplasmic fractions of NSCLC cells were prepared using a NE-PER Nuclear Cytoplasmic Extraction Reagent kit (Thermo Fisher) according to manufacturer’s instructions. To prepare total cell lysates, cells were homogenized using a Dounce homogenizer in RIP A buffer (20 mM Tris, 50 mM NaCl, 2 mM EDTA, 2 mM EGTA, 1% sodium deoxycholate, 1% Triton X-100 (TX-100), and 0.1% SDS, pH 7.4) containing protease inhibitor cocktail and phosphatase inhibitor cocktails 2 and 3 (Sigma-Aldrich). The obtained total cell lysates were cleared by centrifugation (20 min at 14,000 × g), diluted with 2× SDS sample loading buffer and boiled. SDS-polyacrylamide gel electrophoresis was conducted using a standard protocol with equal amounts of total protein (10 or 20 μg) loaded per each lane. The separated proteins were transferred to nitrocellulose membranes, the membranes were blocked with either 5% non-fat milk for general protein detection, or with 3% bovine serum albumin (BSA) to detect phosphorylated proteins. The blocked membranes were incubated overnight with primary antibodies, exposed to HRP-conjugated secondary antibodies for 1 h, and the labeled proteins were visualized using a standard enhanced chemoluminescence solution and X-ray films.
2.7. Scratch wound assay
Confluent cell monolayers were mechanically wounded by making a thin scratch wound with a 200 μl pipette tip. The bottom of the well was marked to define the position of the wound and the monolayers were supplied with fresh cell culture medium. The images of a cell-free area at the marked region were acquired at the indicated times after wounding using an inverted bright field microscope equipped with a camera. The percentage of wound closure was calculated using an Image J software (NIH, Bethesda, MD).
2.8. Boyden chamber migration assay
Boyden chamber migration assay was performed using Transwell® 6. 5 mm membrane inserts with 8.0 μm pores (Coming Incorporated). The membrane inserts were coated with 15 μg/cm2of collagen I. Cells were detached from the plate using a TrypLE Express solution (Thermo-Fisher), resuspended in serum-free medium, and added to the Transwell upper chamber at the density of 6,000 cells per chamber. A complete cell culture medium containing 10% FBS as a chemoattractant was added to the lower chamber and cells were allowed to migrate for either 12 h, or 16 h at 37°C. Membrane inserts were fixed with methanol and non-migrated cells were removed from the top of the filter using a cotton swab. The cells remained at the bottom of the filter or within the membrane were labeled with DAPI nuclear stain, visualized by a fluorescence microscope and counted by using the Image J software.
2.9. Matrigel invasion assay
The Matrigel invasion assay was performed using commercially available BD Biocoat invasion chambers (BD Biosciences). Cells were detached, resuspended in serum-free medium, and added to the upper part of the invasion chamber at the density of 10,000 cells per chamber. A complete cell culture medium containing 10% FBS as a chemoattractant was added to the lower chamber and cells were allowed to invade Matrigel for 24 h at 37°C. The Matrigel plugs were washed with HBSS, fixed with methanol and non-migrated cells were removed from the top of the gel using cotton swabs. The invaded cells were stained with DAPI, visualized by a fluorescence microscope and counted by using the Image J program.
2.10. Immunofluorescence labeling and confocal microscopy
Cells cultured on collagen-coated coverslips were fixed with 4% PFA for 20 min at room temperature and permeabilized with 0.5% Triton X-100 for 5 min. Fixed and permeabilized cells were blocked with Hank’s Balanced Salt Saline (HBSS) containing 1% BSA for 1 h at room temperature, followed by sequential 1 h incubations with primary antibodies and Alexa Fluor-conjugated secondary antibodies diluted in the blocking solution. The immunolabeled coverslips were mounted using the ProLong® Gold Antifade reagent (Thermo-Fisher). Fluorescently labeled cell monolayers were examined using either a Zeiss LSM700 laser-scanning confocal microscope (Zeiss Microimaging, Thornwood, NY), or Leica SP8 confocal microscope (Wentzler, Germany). The images were processed using Zen Lite software (Carl Zeiss Microscopy LLC) and Adobe Photoshop.
2.11. Extracellular matrix adhesion assay
Cells were detached from the plate, counted with a hemocytometer, and resuspended in the complete medium. 10,000 cells were seeded to each well of a 24 well plate coated with either collagen I, or fibronectin and were allowed to adhere for 30 min at 37 °C. After incubation, unattached cells were aspirated and the wells were gently washed with HBSS buffer. The attached cells were fixed with methanol and stained using a DIFF stain kit (IMEB Inc., San Marcos, CA). Images of adherent cells were captured using a bright-field microscope and the number of adhered cells was determined using Image J software.
2.12. Cell proliferation assay
Cell proliferation was examined by the MTT assay involving the conversion of the water-soluble MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) into insoluble formazan. Cells were cultured on 98 well plates, and at different times post-plating, their medium was replaced with 100 μL of fresh culture medium containing 50 μg/ml of MTT. MTT added to 100 μL of medium alone was included as a negative control. Cells were incubated at 37°C for 4 h and the generated formazan was dissolved by adding 50 μL of DMSO with 10 min incubation at 37°C. The absorbance of formazan solution was measured using a standard plate reader at 540 nm.
2.13. RNA interference
siRNA-mediated knockdown of Src was performed by using a specific Dharmacon siRNA Smartpool (Thermo-Fisher) with non-targeting siRNA number 2 served as a control. Cadherin-11 was depleted by using individual siRNA duplexes and a negative siRNA control obtained from Qiagen. Cells were transfected using DharmaFECT 1 transfection reagent in Opti-MEM I medium (Thermo-Fisher) with final siRNA concentration for each target at 50 nM. Cells were used in experiments on days 3 and 4 posttransfection.
2.14. Statistical analysis:
All data are expressed as means ± standard error (se) from three biological replicates. Statistical analysis was performed by using a one-way ANOVA to compare the control and two experimental groups (knockout with two different adducin sgRNAs). A post-hoc t-test was used to compare controls with each adducin-depleted group. A two-tailed Student t-test was used to compare the results obtained with two experimental groups (control and adducin-overexpressing cells). We accounted for multiple comparisons by adjusting the significance level using a Bonferroni Correction. P values < 0.05 were considered statistically significant.
3. RESULTS
3.1. Adducins are either downregulated or mislocalized in mesenchymal-type lung cancer cells
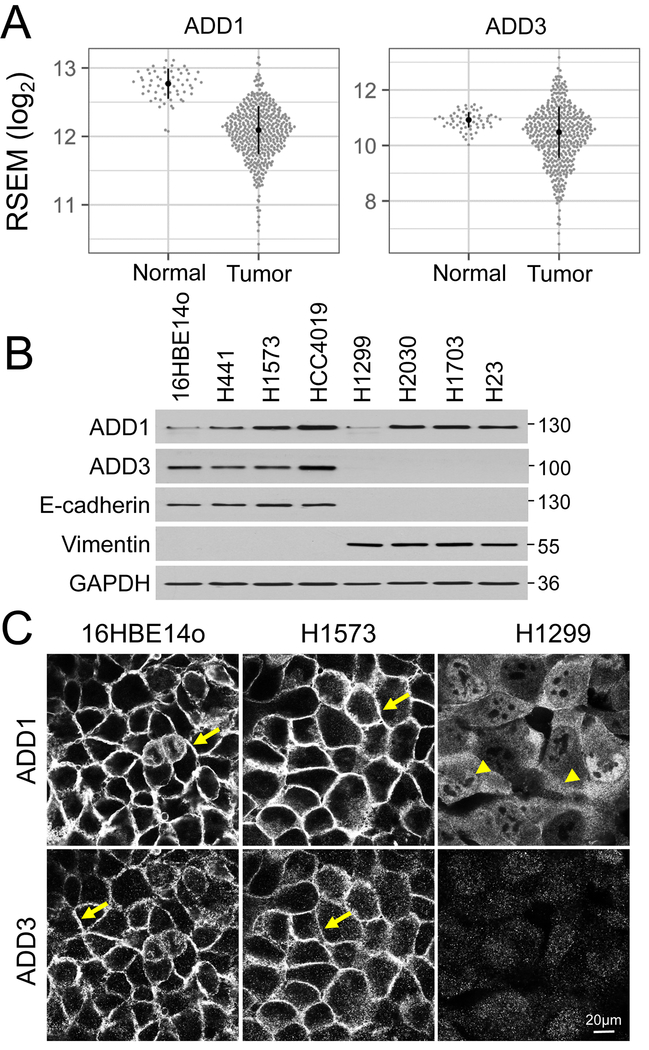
Despite early in vitro and animal model studies suggesting association of tumor progression with altered levels and localization of ADD1 and ADD3 [28, 46], little is known about expression of adducins in human cancers. Therefore, we used the TCGA database to examine expression of ADD1 and ADD3 in clinical samples of patients with NSCLC. Figure 1A demonstrates that mRNA levels were significantly lower in lung adenocarcinoma compared to normal lung tissue for both ADD1 (p < 10 −15) and ADD3 (p < 10 −5). Such expressional downregulation was especially vivid for ADD1. NSCLC is characterized by significant phenotypic heterogeneity, which can be grouped into three broad categories: epithelial, mesenchymal and epithelial-mesenchymal hybrid [47], The former category includes cells that preserve characteristics of well-differentiated normal pulmonary epithelium such as high E-cadherin expression, assembly of robust intercellular junctions and poor invasiveness. By contrast, mesenchymal-type NSCLC cells lost cell-cell contacts and acquired high invasiveness [47], Therefore, we next sought to compare expression and localization of adducins in a panel of NSCLC cells with either the epithelial (H441, H1573, HCC4019) or the mesenchymal (H1299, H2030, H1703, H23) phenotypes. The epithelial panel also included non-transformed 16HBE14o human bronchial epithelial cells [48], The epithelial and the mesenchymal phenotypes of these cells was confirmed by observing expression of either E-cadherin or vimentin, respectively (Fig. 1B). Immunoblotting analysis demonstrated that expression of ADD1 protein did not depend on cell phenotype, whereas the level of ADD3 protein was dramatically decreased in mesenchymal-type NSCLC cells (Fig. 1B). We also compared expression of adducins in the cell pair consisting of immortalized, non-transformed human bronchial epithelial cells, HBEC3-KT, and F1BEC3-KTRL53M cells fully transformed by p53 knockdown and overexpression of KRAS G12V and c-Myc [49], Transformation of HBEC cells caused a robust epithelial-to-mesenchymal transition that was accompanied by a selective downregulation of ADD3 expression (Suppl. Fig. 1 A).
Figure 1: Altered expression and localization of adducins in lung cancer cells.
(A) A comparative analysis of ADD1 and ADD3 mRNA expression levels in normal (n=59) and tumor (n=457) tissues from patients with lung adenocarcinoma. Error bars indicate the median absolute deviations. (B) Immunoblotting analysis of adducins expression in epithelial and mesenchymal-type non-small cell lung cancer cells. (C) Immunofluorescence labeling of ADD1 and ADD3 in non-tumorigenic bronchial epithelial cells (16HBE14o), epithelial-type (H1573) and mesenchymal-type (H1299) lung cancer cells. Arrows point on plasma membrane localization of ADD1 and ADD3 in epithelial-type cells. Arrowheads indicate cytoplasmic and nuclear localization of ADD1 in mesenchymal-type lung cancer cells. Scale bar, 20μm.
Next, we used immunofluorescence labeling and confocal microscopy to examine the localization of adducins in NSCLC cells. In agreement with previously published data [29, 31], both ADD1 and ADD3 predominantly localized to the lateral plasma membrane of confluent non-transformed lung epithelial cells and epithelial-type NSCLC cells (Fig. 1C & Suppl. Fig. 1B, arrows). By contrast, ADD1 was redistributed from the plasma membrane-associated pool into the cytoplasmic and nuclear pools in mesenchymal-type NSCLC cells and HBEC3-KTRL53M cells (Fig. 1C & Suppl. Fig. 1B, arrowheads). Consistent with our immunoblotting results, the intensity of ADD3 labeling was dramatically decreased in the mesenchymal-type NSCLC cells (Fig. 1C & Suppl. Fig. 1B). Together, these data indicate that epithelial-to-mesenchymal transition, which is characteristic of lung cancer progression, is accompanied by either down-regulation (ADD3) or mislocalization (ADD1) of adducins.
3.2. Downregulation of ADD1 and ADD3 enhances individual migration of NSCLC cells
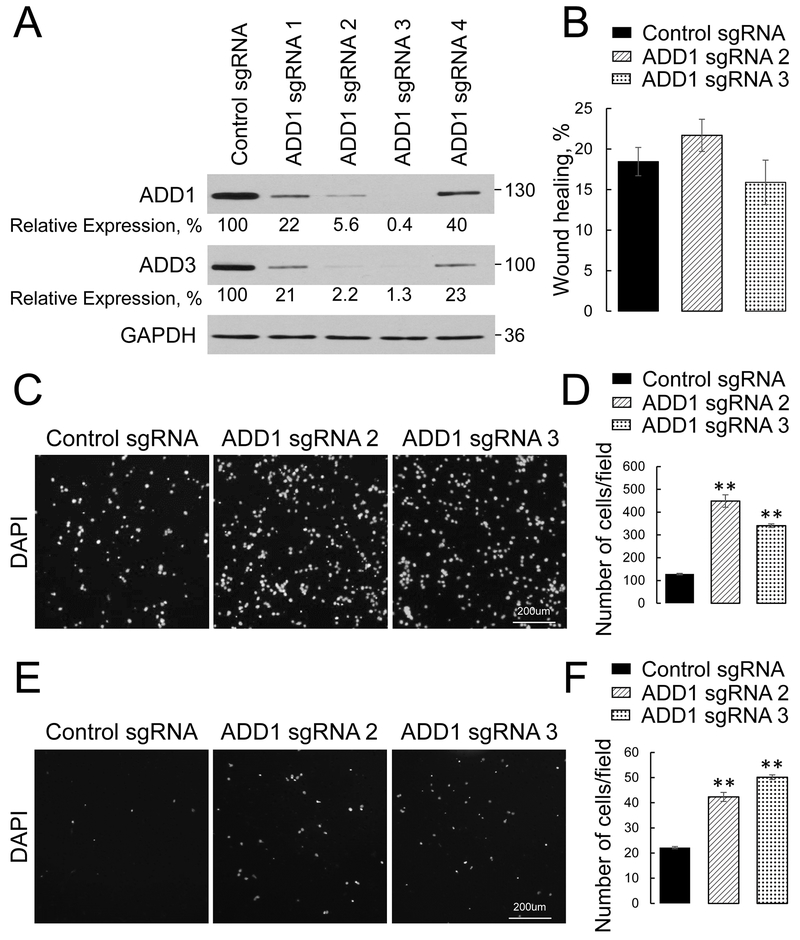
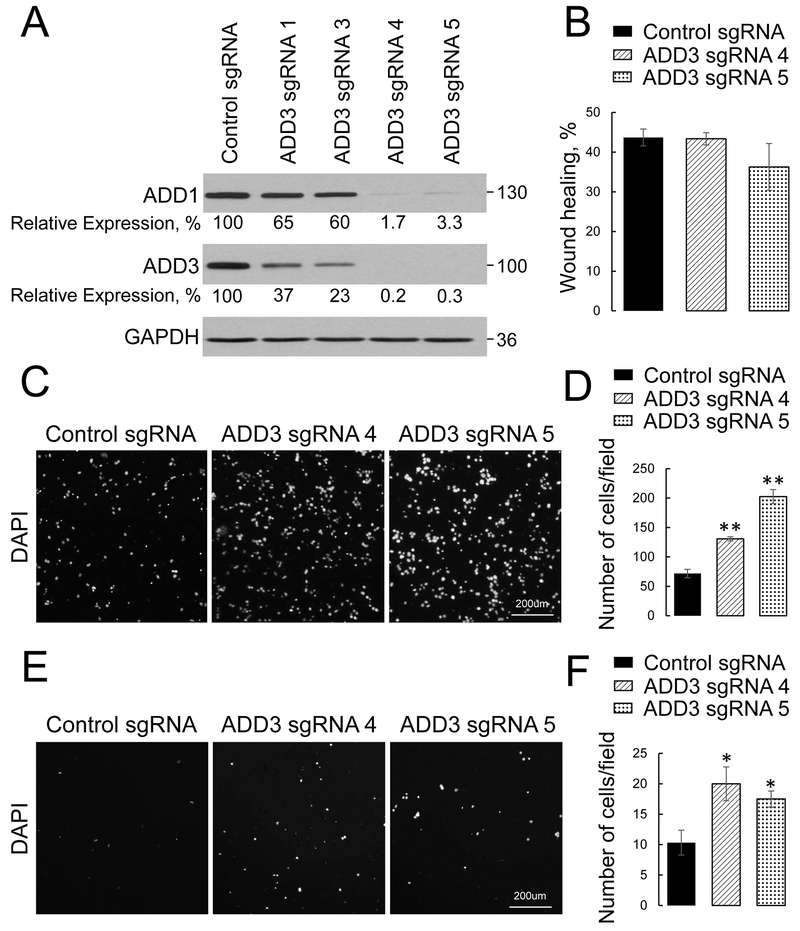
To investigate the roles of adducins in the regulation of lung cancer cell motility, we deleted either ADD1 or ADD3 in epithelial-type H1573 and 16HBE14o cells by using CRISPR/Cas9-mediated gene editing. Cells were transfected with either non-targeted guide RNA or four different small guide (sg) RNA oligonucleotides against each adducin isoform (ADD1 or ADD3), and stable knockout cell lines were obtained by puromycin treatment. This approach resulted in variable levels of adducin depletion (Fig. 2A, Fig. 3A, Suppl. Figs. 2 & 3). Two H1573 cell lines with the most efficient knockout of ADD1 (transfected with sgRNAs 2 and 3) and two stable cell lines with deepest ADD3 depletion (created by sgRNAs 4 and 5) were used for subsequent functional experiments. It is noteworthy that loss of ADD1 markedly decreased ADD3 protein level, and vice versa (Figs. 2 & 3; Suppl. Figs. 2 & 3). This phenomenon was previously described in adducin-depleted colonic epithelial cells [31] and tissues of either ADD1 or ADD3 null mice [50, 51] and could be due to stabilizing effects of the ADD1/ADD3 hetero-oligomerization, protecting these proteins from proteolytic degradation. Therefore, the outcome of either ADD 1 or ADD3 knockout in bronchial epithelial and lung cancer cells was a dual ADD1/ADD3 depletion.
Figure 2: Loss of ADD1 increases transfilter migration and Matrigel invasion of lung cancer cells.
ADD1 expression was down-regulated H1573 cells using CRISPR/Cas9-mediated gene editing. (A) Immunoblotting analysis shows co-depletion of ADD1 and ADD3 by 4 different ADD1 small guide (sg) RNAs. (B) Quantification of the planar migration of the control and ADDl-depleted H1573 cell monolayers at 24 h post-wounding. (C, D) Representative images and quantitative analysis of the DAPI-labeled control and ADD 1-deficient HI 573 cells after 16 h of transfilter migration in the Boyden chamber. (E, F) Representative images and quantitative analysis of the DAPI-labeled control and ADD 1-deficient H1573 cells after 24 h invasion into Matrigel. Data are presented as mean ± SE (n =3); **p < 0.005, as compared to the control sgRNA-transfected group.
Figure 3: Loss of ADD3 increases transfilter migration and Matrigel invasion of lung cancer cells.
ADD3 expression was down-regulated HI573 cells using CRISPR/Cas9-mediated gene editing. (A) Immunoblotting analysis shows co-depletion of ADD3 and ADD1 by 4 different ADD3 small guide (sg) RNAs. (B) Quantification of the planar migration of the control and ADD3-depleted HI573 cell monolayers at 24 h post-wounding. (C, D) Representative images and quantitative analysis of the DAPI-labeled control and ADD3-deficient HI 573 cells after 16 h of transfilter migration in the Boyden chamber. (E, F) Representative images and quantitative analysis of the DAPI-labeled control and ADD3-deficient HI573 cells after 24 h invasion into Matrigel. Data are presented as mean ± SE (n =3); *p < 0.05; **p < 0.005, as compared to the control sgRNA-transfected group.
The roles of adducins in regulating NSCLC cell motility were determined by examining three different modes of cell migration: collective cell migration during scratch wound closure, chemotactic transfilter migration of individual cells in the Boyden chamber, and cell invasion into Matrigel. Loss of ADD1 in H1573 cells had no effect on wound closure (Fig. 2B), but significantly accelerated transfilter migration and Matrigel invasion of these cells (Fig. 2C-F). Likewise, ADD3 knockout accelerated individual migration and invasion of NSCLC cells without affecting their wound closure (Fig. 3). Consistently, knockout of ADD1 and ADD3 accelerated transfilter migration, but not wound healing of non-tumorigenic 16HBE14o cells (Suppl. Figs. 2 & 3).
3.3. Overexpression of ADD1 inhibits migration and invasion of mesenchymal NSCLC cells
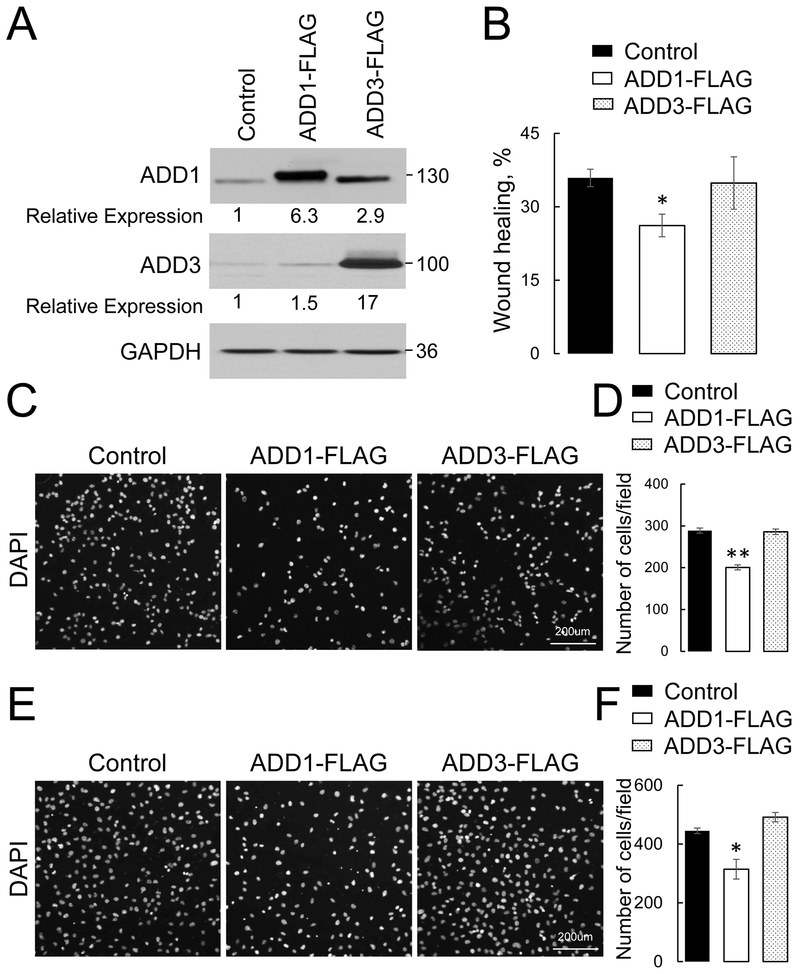
Next, a gain-of-function approach was used to collaborate the results obtained with adducins knockout in lung epithelial cells. H1299 cells, a known mesenchymal-type NSCLC cell line was chosen for stable overexpression of either FLAG-tagged ADD1, ADD3, or a control plasmid. Immunoblotting analysis demonstrated successful overexpression of either adducin isoform (Fig. 4A). Interestingly, while ADD3 overexpression also caused an approximately 3-fold increase in ADD1 level, the level of endogenous ADD3 was just marginally (~ 1.5 fold) increased in ADD1-overexpressing H1299 cells (Fig. 4A). This suggests that other mechanisms besides decreased protein stability could be responsible for low ADD3 expression in mesenchymal NSCLC cells. ADD1 overexpression significantly inhibited wound closure (Fig. 4B), transfilter migration (Fig. 4C, D) and Matrigel invasion (Fig. 4E, F) of H1299 cells. By contrast, ADD3 overexpression did not affect any of the examined modes of NSCLC cell migration (Fig. 4). Since both ADD1 and ADD3 knockouts decreased ADD1 expression and ADD1, but not ADD3 overexpression markedly elevated ADD1 level, results of our experiments suggest that ADD1 played a major role in suppressing migration and invasion of NSCLC cells.
Figure 4: Overexpression of ADD1 but not ADD3, inhibits lung cancer cell motility.
FLAG-tagged ADD1 or ADD3 were stably expressed in H1299 lung cancer cells using a lentiviral expression vector. (A) Immunoblotting analysis shows the levels of ADD1 and ADD3 proteins in the generated cell lines. (B) Quantification of the planar migration of the control, ADD1 or ADD3-overexpressing H1299 cell monolayers after 12 h of wound healing. (C, D) Representative images and quantitative analysis of the DAPI-labeled control, ADD1, or ADD3-overexpressing H1299 cells after 12 h transfilter migration in the Boyden chamber. (E, F) Representative images and quantitative analysis of the DAPI-labeled control, ADD1, or ADD3-overexpressing H1299 cells after 24 h invasion into Matrigel. Data are presented as mean ± SE (n =3); *p < 0.05; **p < 0.005, as compared to the control group.
3. 4. Overexpression of ADD1 increases ECM adhesion and stimulates focal adhesion assembly
The next series of experiments were designed to elucidate molecular mechanisms that underline the anti-migratory function of ADD1 in NSCLC cells. Since ADD1 was previously implicated in the control of mitosis [30, 45], we examined the possibility that adducins regulate cell migration indirectly, by altering cell proliferation. However, MTT assay of either ADD1- and ADD3-depleted, or ADD 1-overexpressing cells and their appropriate controls did not show significant effects of adducins on NSCLC cell proliferation (Suppl. Fig. 4). Adhesion to the ECM is known to be essential for individual and collective epithelial cell migration. Therefore, we asked if this process is affected by altered ADD1 expression. Overexpression of ADD1 caused an almost two-fold increase in H1299 cell adhesion to collagen I (Fig. 5A, B), and fibronectin (data not shown). By contrast, knockout of ADD1 and ADD3 did not change collagen or fibronectin adhesion of either H1573 or 16HBE14o cells (Suppl. Fig. 5 and data not shown). Interestingly, the increased ECM adhesion and decreased transfilter migration of ADD 1-overexpressed H1299 cells persisted even when ADD3 was also overexpressed (Suppl. Fig. 6), thereby arguing against a possibility that these are the neomorphic effects of the selective ADD1 overexpression.
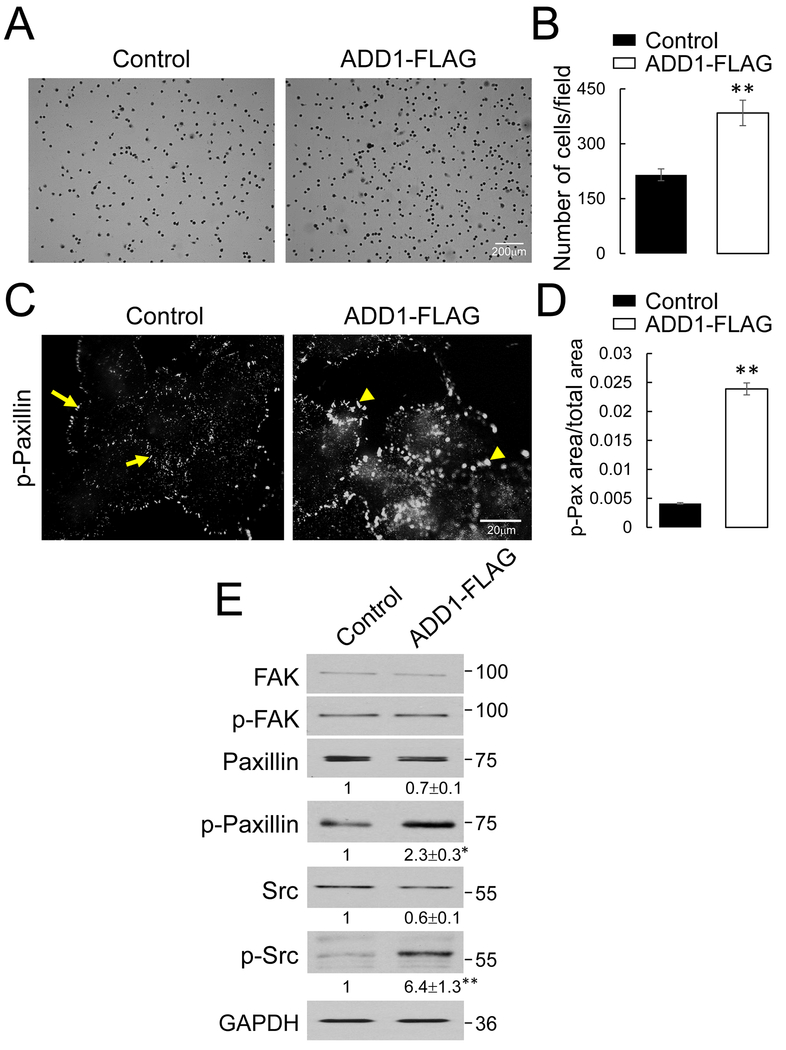
Figure 5: Overexpression of ADD1 increases extracellular matrix adhesion and promotes focal adhesion assembly in lung cancer cells.
(A, B) Representative images and quantification of 30 min adhesion to the collagen I matrix by the control and ADD 1-overexpressing H1299 cells. (C, D) Representative immunofluorescence images and quantitative analysis of phosphorylated (p) paxillin (Tyrll8) labeling in the control and ADD 1-overexpressing H1299 cells. Arrows indicate small peripherally located focal adhesions in the control cells. Arrowheads point at enlarged focal adhesions in ADD 1-overexpressing cells. (E) Immunoblotting analysis of expression and phosphorylation of major focal adhesion proteins in the control and ADD1-overexpressing H1299 cells. Data are presented as mean ± SE (n =3); *p < 0.05; **p < 0.005, as compared to the control group.
There is a nonlinear relationship between the strength of cell-ECM adhesion and the velocity of cell migration, where migration can be inhibited by both insufficient and very strong cell attachment to ECM [52, 53], Therefore, we rationalized that the observed hyperadhesiveness of an ADD 1-overexpressing H1299 cell is likely to contribute to their impaired motility. Cell adhesion to ECM depends on the assembly of specialized basal structures called ‘focal adhesions’ (FA) [54, 55]. Immunofluorescence labeling of a classical FA marker, phosphorylated (p) paxillin, revealed assembly of small FA localized to the edges of spreading control H1299 cells (Fig. 5C, arrows). ADD1 overexpression stimulated the formation of FA, which became enlarged and localized both at the periphery and the basal surface of spreading H1299 cells (Fig. 5C, arrowheads & 5D). Furthermore, immunoblotting analysis demonstrated that ADD1 overexpression significantly increased the levels of phosphorylated paxillin and phosphorylated (active) Src kinase (Fig. 5E), which is a key upstream regulator of FA assembly [56, 57].
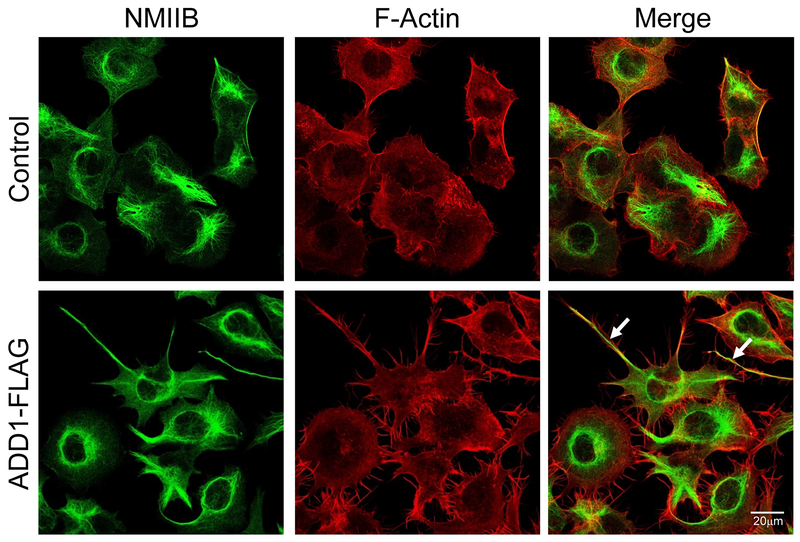
Since the formation of FA is known to be regulated by the actomyosin cytoskeleton, we next examined the cytoskeletal organization in control and ADD1-overexpressing H1299 cells by labeling filamentous (F) actin and an actin motor, non-muscle myosin IIB (NM IIB). Confocal microscopy revealed major morphological changes in ADD1-overexpressing cells, manifested by the formation of long peripheral protrusions enriched in F-actin and NM IIB (Fig. 6 arrows). To gain additional insights into the mechanisms that control formation of such protrusions, we performed live cell imaging of migrating cells. Control H1299 cells demonstrated a mesenchymal-type motility with the orchestrated formation of the migrating cellular front and detachment/forward translocation of the trailing cellular edge (Suppl. Movie 1). By contrast, ADD1 overexpressed cells failed to efficiently detach and retract the trailing edge resulting in the formation of long tail-like protrusions (Suppl. Movie 2). These results are consistent with the observed hyperadhesiveness of ADD1-overexpressed H1299 cells (Fig. 5).
Figure 6: ADD1 overexpression induces the formation of long actomyosin-rich protrusions.
A dual immunofluorescence analysis of F-actin (red) and non-muscle myosin IIB (NMIIB, green) in the control and ADD 1-overexpressing H1299 cells. Arrows show long protrusions enriched in F-actin and NMIIB induced by ADD1 expression.
Immunofluorescence labeling and confocal microscopy showed localization of exogenous ADD1 in the poorly-retractable F-actin rich protrusions (Suppl. Fig. 7A, arrows). However a significant fraction of the overexpressed ADD1 accumulated in the nuclei of H1299 cells (arrowheads), which was confirmed by the nuclear/cytoplasmic cell fractionation (Suppl. Fig. 7B). This prompted us to investigate whether the observed effects of ADD1 overexpression on H1299 cell adhesion and migration were mediated by its nuclear or non-nuclear fractions. Inhibition of the nuclear export by leptomycin B caused redistribution of peripheral ADD1 into the nucleus (Suppl. Fig. 8, arrows) and reversed the increased ECM adhesion of ADD 1-overexpressed H1299 cells (Suppl. Fig. B,C). Unfortunately, a long-time (~16 h) leptomycin treatment, which was required for examining cell migration, produced uninterpretable results by markedly inhibiting transfilter migration of control cells (data not shown). These results suggest that at least some effects of ADD 1-overexpression of the adhesion and migration of NSCLC cells could be mediated by the non-nuclear pool of ADD1.
3.5. Src inhibition reverses the increased adhesion and inhibited migration of ADD1-overexpressing lung cancer cells
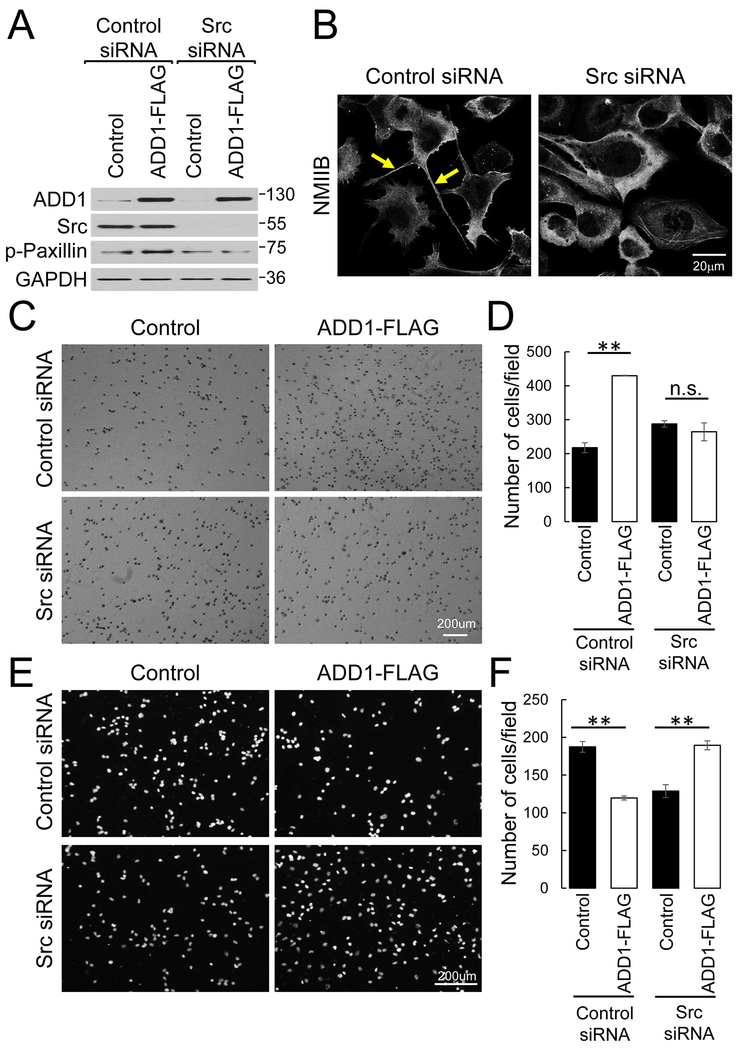
Next, we sought to elucidate if activation of Src is responsible for the increased ECM adhesion and attenuated motility of ADD 1-overexpressing NSCLC cells by using siRNA-mediated downregulation of Src expression (Fig. 7A). Depletion of Src decreased paxillin phosphorylation and inhibited the formation of non-retractable tails in ADD 1-overexpressing H1299 cells (Fig. 7A, B). Consistently, Src knockdown reversed the enhanced ECM adhesion of ADD 1-overexpressing cells without affecting adhesion of control cells (Fig. 7C, D). Interestingly, downregulation of Src had a complex effect on H1299 cell motility by decreasing transfilter migration of control cells, but increasing migration of ADD 1-overexpressing cells (Fig. 7E, F). This most likely reflects multiple roles of Src in the regulation of cell motility that include adhesion-dependent and independent mechanisms [56, 57], Together, our data demonstrate that ADD1 overexpression results in Src activation that increases ECM adhesion and impedes motility of NSCLC cells.
Figure 7: Downregulation of Src expression reverses the increased adhesion and attenuated motility of ADDl-overexpressing lung cancer cells.
Control and ADD 1-overexpressing H1299 cells were transfected with either Src or non-targeted siRNAs and were analyzed on day 3 posttransfection. (A) Immunoblotting analysis shows the expression of Src and p-paxillin in the control and ADDl-overexpressing cells transfected with either non-targeting or Src-specific siRNAs. (B) Immunolabeling analysis of NM IIB in ADDl-overexpressing lung cancer cells transfected with either control or Src-specific siRNAs. Arrows show long NM IIB-rich protrusions in ADD1-overexpressing cells that disappear after Src depletion. (C, D) Representative images and quantification of collagen I matrix adhesion of the control and ADD 1-overexpressing H1299 cells transfected with either control or Src-specific siRNAs. (E, F) Representative images and quantification analysis of transfilter migration of control and ADD 1-overexpressing H1299 cells transfected with either control or Src-specific siRNAs. Data are presented as mean ± SE (n = 3); **p < 0.005.
3.6. Upregulation of cadherin-11 mediates the increased motility of adducin-depleted lung cancer cells
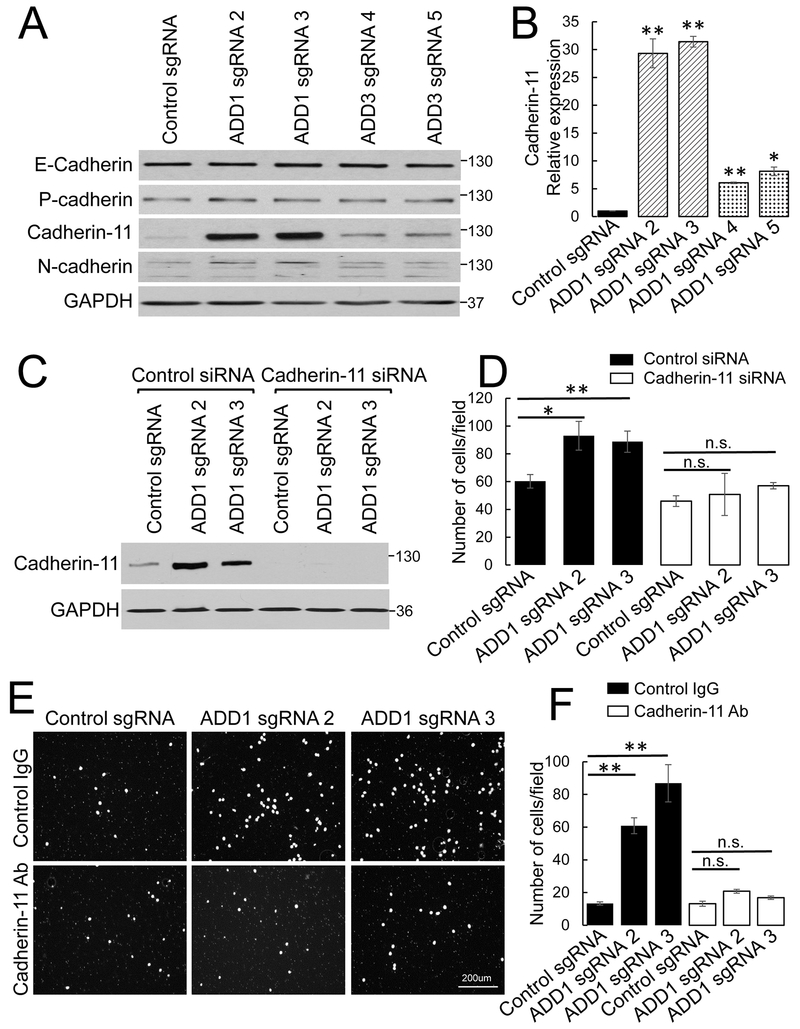
In the final series of experiments, we investigated the mechanisms that mediate ECM adhesion-independent acceleration of motility of adducin-depleted NSCLC cells. Since adducins were previously implicated in the regulation of cadherin-based intercellular junctions and epithelial differentiation [31, 33, 58, 59] we asked if the loss of adducins could trigger the phenotypic alterations of NSCLC cells. Immunoblotting analysis was used to examine the expression of major epithelial and mesenchymal cadherins in control and adducin-depleted H1573 cells. While the levels of epithelial-type E- and P-cadherins were not affected by ADD1 and ADD3 knockout, expression of mesenchymal-type cadherin-11 was dramatically (up to 31-fold) upregulated in ADD1-deficient and in less extent (6–8 fold) in ADD3 knockout cells (Fig. 8 A,B). Consistently, cadherin-11 protein expression was downregulated in ADD1-overexpressing H1299 cells (data not shown). To examine the functional roles of cadherin-11 overexpression in accelerated motility of adducin-depleted H1573 cells, cadherin-11 functions were inhibited by two different approaches. One approach involved siRNA-mediated knockdown of cadherin-11 (Fig. 8C, D), while the other approach used an anti-cadherin-11 inhibitory antibody (Fig. 8E, F). Both approaches produced consistent results by selectively attenuating transfilter migration of ADD1-deficient H1573 cells, while having little effects on the motility of control cells (Fig. 8C-F). Similar results were obtained when cadherin-11 was inhibited in ADD3-deficient H1573 cells (Suppl. Fig. 9). Together, our results demonstrate that upregulation of cadherin-11 expression plays a major role in the accelerated motility of adducin-deficient NSCLC cells.
Figure 8: Inhibition of cadherin-11 reverses the increased motility of ADD 1-deficient lung cancer cells.
(A, B) Representative immunoblots showing expression of different cadherins and densitometric quantification of the cadherin-11 protein level in the control, ADD1, or ADD3-depleted H1573 cells. (C, D) Cadherin-11 was transiently depleted by siRNA in control and ADD 1-deficient H1573 cells. (C) Immunoblotting analysis shows the efficiency of cadherin-11 depletion. (D) Results of the transfilter migration assay of the control and ADD 1-deficient H1573 cells with and without cadherin-11 depletion. (E, F) Transfilter migration data for the control and ADD 1-deficient H1573 cells treated with either anti-cadherin-11 antibody or control IgG. Data are presented as mean ± SE (n =3); *p < 0.05; **p < 0.005.
4. DISCUSSION
4.1. Altered expression and localization of adducins in NSCLC and other cancers
Adducins are scaffolding proteins, best known for their roles in regulating the organization and dynamics of the plasma membrane-associated cytoskeleton [22, 23]. They are dysregulated in different types of solid tumors, although functional roles of adducins in tumorigenesis remain poorly characterized [40]. The present study identifies adducins as potent inhibitors of NSCLC cell motility and reveals non-canonical mechanisms of their actions involving the regulation of ECM adhesion signaling and cell-cell adhesion proteins. Importantly, our data suggest that functional activities of adducins are impaired in NSCLC cells. First, mRNA expression of ADD1 and ADD3 was significantly decreased in lung adenocarcinoma patient samples as compared to normal human lung tissue (Fig. 1A). Second, the level of ADD3 protein was dramatically decreased in mesenchymal NSCLC cells, the most aggressive and metastatic type of lung cancer (Fig. 1B, C; Suppl. Fig. 1). Finally, ADD1 was translocated from the cell cortex into cytoplasmic and nuclear compartments of mesenchymal-type NSCLC cells (Fig. 1B, C; Suppl. Fig. 1). Such mislocalization of ADD1 could be due to phosphorylation by oncogenic kinases that weaken its interactions with cortical actin filaments and spectrin oligomers, although this suggestion has not been proved experimentally. Since deletion of ADD1 and ADD3 potently promoted migration and invasion of NSCLC and non-transformed lung epithelial cells (Figs. 2 & 3; Suppl. Figs. 2 & 3), it is reasonable to suggest that adducin dysfunctions due to either expressional downregulation or mislocalization contribute to the increased motility of mesenchymal NSCLC cells. Importantly, previous studies demonstrated that loss of adducins blocked the formation of the lateral plasma membrane domain and impaired assembly of intercellular junctions in normal epithelial and cancer cells [29–31, 33]. These findings highlight another possible consequence of adducin dysfunctions in lung cancer, which is destabilization of intercellular contacts. Collectively, our present and published data suggest that the net effect of the impaired functional activity of adducins would be increased metastatic dissemination of NSCLC cells. This suggestion could be extended to other types of cancers characterized by the diminished expression and altered localization of adducins. Indeed, loss of ADD3 was reported in human glioma patients [60, 61], whereas ADD1 was downregulated in ovarian cancer cell lines as compared to normal human ovarian surface epithelium [39]. Additional evidence were provided by animal cancer model studies where decreased expression and mislocalization of ADD1 and ADD3 were found in rat renal carcinoma [28, 46] and alternative splicing of ADD3 was associated with highly metastatic murine breast tumor [62]. While these clinical and experimental evidence suggest that adducins could act as a general suppressor of tumor metastasis, our study provides the first mechanistic insights that may explain the antimetastatic activity of these proteins.
4.2. ADD1 is a potent suppressor of lung cancer cell motility
Our findings primarily implicate ADD1 in the regulation of lung cancer cell motility, while providing suggestive evidence about anti-migratory activity of ADD3. Indeed, knockout of either ADD1 or ADD3 that increased migration and invasion of normal lung epithelial and NSCLC cell, resulted in simultaneous down-regulation of both adducin isoforms (Figs. 2 & 3; Suppl. Figs. 2 & 3). On the other hand, ADD1 but not ADD3 overexpression inhibited the motility of mesenchymal NSCLC cells (Fig. 4). It is generally believed that ADD1 and ADD3 stabilize each other by forming heterodimers or heterotetramers [22, 23] and our present and previous results with genetic depletion of different adducins support this postulate [31]. However, under certain conditions, ADD1 appears to be stable in the absence of other adducin isoforms. For example, mesenchymal NSCLC cells demonstrated high expression of ADD1 along with the very low level of ADD3 and undetectable ADD2 protein (Fig. 1; Suppl. Fig. 1 and data not shown). Overexpression of exogenous ADD1 in these cells just marginally increased ADD3 level (Fig. 4A). Furthermore, expression of the ADD1 protein was minimally altered in the kidneys of mice with either individual ADD3 knockout or a dual loss of ADD2 and ADD3 [51]. Hence, it is reasonable to suggest that ADD1 in addition to canonical heterooligomerization with ADD3 or ADD2, is capable of forming homooligomers and functioning independently from other adducin isoforms. Further studies are needed to compare the functional activities of homooligomers and heterooligomers of adducins.
4.3. High expression of ADD1 suppresses lung cancer cell motility by strengthening ECM adhesion
We found that ADD1 suppressed lung epithelial cell and NSCLC cell motility via two different mechanisms, depending on the cellular level of this protein. High ADD1 expression inhibited cell migration by increasing the avidity of ECM adhesion, while moderate ADD1 expression attenuated cell migration by downregulating expression of the promigratory cadherin-11 (Figs. 5 & 8). Both mechanisms are novel and have not been previously associated with functional activities of adducins. The observed different modes of ADD1 actions are not surprising, given its known roles as the membrane skeleton component and the actin-binding protein involving in multiple interactions [22, 23]. The outcomes of such interactions are likely to be an assembly of different cytoskeletal complexes, which abundancy and functions depend on the cellular level of ADD1.
Our data suggest that high ADD1 expression creates a strong ECM adhesion associated with the increased FA signaling events, such as Src activation (Figs. 5 & 7). Such enhanced FA are likely to be resistant to disassembly, thereby impeding detachment of the trailing cellular edge and attenuating cell motility (Fig. 6; Suppl. Movies 1 & 2). Similar formation of large integrin-based FA was previously reported in rat renal epithelial cells after expression of an ADD1 point mutant associated with human hypertension [63]. While we did not investigate the precise mechanisms that underline the increased Src activation and the enhanced FA assembly in ADD1-overexpressing cells, these events are likely to be related to the adducin-dependent reorganization of the actomyosin cytoskeleton. Indeed, ADD1-overexpressing NSCLC cells were characterized by the peripheral protrusions that contained prominent actomyosin bundles (Fig. 6). These findings are in agreement to previous studies showing that adducins promoted actin filament assembly in intestinal and renal epithelial cells [31, 63] and that loss of ADD1 increased F-actin dynamics in the neuronal growth cone [36]. Furthermore, the described effects of ADD1 on the actin cytoskeleton in live cells are consistent with the known ability of adducins to bundle and cap actin filaments in cell-free systems [18–21]. Therefore, a functional outcome of the ADD1 overexpression could be creation of stable F-actin bundles. On the basal side of the cells, these F-actin bundles promote FA assembly and increase cell ECM attachment [54, 55, 64]. It is unclear why the formation of peripheral F-actin bundles requires the high level of ADD1 expression. One possibility is that some optimal actin-ADD1 stoichiometry should be achieved, while another possibility is that ADD1 competes with other highly-expressed actin-bundling and capping proteins for binding to actin filaments.
4.4. Adducins are novel regulators of cadherin-11 expression in lung cancer cells
One of the most surprising findings of this study is that adducins modulate NSCLC cell motility by regulating expression of cadherin-11. Indeed, loss of ADD1 and ADD3 resulted in marked increase of cadherin-11 expression in epithelial-type H1573 cells, which played a causal role in the accelerated motility of adducin-depleted cells (Fig. 8, Suppl. Fig. 9). Furthermore, ADD1 overexpression downregulated cadherin-11 expression in mesenchymal H1299 cells (data not shown), which could synergize with the increased ECM adhesion in attenuating motility of these cells. Cadherin-11 is a member of the cadherin family of adhesion proteins and is known to be essential for embryonic development and tumorigenesis [65], Cadherin-11 is highly expressed in mesenchymal cells and is upregulated in different solid tumors [66–68], Importantly, several studies demonstrated that cadherin-11 drives cancer cell motility [66–68], which is consistent with the cadherin-11 activity observed in adducin-depleted NSCLC cells (Fig. 8). How does cadherin-11 accelerate motility of cancer cells remain poorly understood, although overexpression of this protein in mesenchymal stem cells and fibroblasts was shown to dramatically affect the contractile properties of the cells and production of ECM proteins [69, 70].
The observed regulation of cadherin-11 expression in NSCLC cells represents an unusual activity of adducins. Only one previous study documented decreased expression of adhesion-related proteins, synaptopodin, and alpha-actinin, in podocytes of ADD2-null mice [71], The mechanisms of adducin-dependent regulation of protein expression await further investigations, however, at least two possibilities could be envisioned. One possibility is that adducins affect cadherin-11 expression indirectly, by modulating cytoskeleton-dependent activity of serum response factor (SRF). An alternative possibility would be a direct effect of ADD1 on cadherin-11 transcription in the nucleus. The former mechanism is based on a recent report that cadherin-11 expression is controlled by SRF during mesenchymal stem cell differentiation [69], On the other hand, SRF is a transcriptional factor, which activity is regulated by the actin cytoskeleton and specifically by the availability of monomeric actin [72, 73], Since altered adducin expression could shift the ratio of monomeric-to-polymeric actin in epithelial cells [31], this may result in altered SRF activity and SRF-dependent expression of cadherin-11. The possibility of direct adducin-dependent regulation of cadherin-11 expression is based on the fact that adducins possess nuclear localization signals and could translocate into the nucleus (Fig. 1; Suppl. Fig. 7) [30, 74], Interestingly, nuclear retention of ADD1 was shown to be significantly higher as compared to the ADD3 isoform [74], which is consistent with its superior ability to repress cadherin-11 expression (Fig. 8). In the nucleus, ADD1 was shown to interact with RNA polymerase and a transcription factor, ZNF331, however, the functional consequences of such interactions have not been explored [74].
4.5. Conclusion
In conclusion, the results of our study demonstrate that adducins, especially ADD1, act as potent inhibitors of migration and invasion of NSCLC cells and non-transformed bronchial epithelial cells. Furthermore, we identified two distinct mechanisms of adducin-dependent regulation of cell motility depending on the level of ADD1 expression and involving modulation of ECM adhesion and regulation of cadherin-11expression. Since adducins are either downregulated or functionally inhibited by pro-oncogenic signaling pathways, dysfunction of these membrane skeleton proteins is likely to contribute to the metastatic dissemination of lung cancer cells. Additional studies are warranted to dissect possible roles and mechanisms of adducins in tumor progression and metastasis.
Supplementary Material
Highlights.
Development of non-small cell lung cancer is accompanied by the decreased expression and mislocalization of adducins
Depletion of ADD1 and ADD3 accelerates migration and invasion of NSCLC cells, whereas ADD1 overexpression inhibits cell motility
ADD1 overexpression attenuates lung cancer cell migration by activating Src and enhancing cell-matrix adhesion
Loss of ADD1 and ADD3 stimulates lung cancer cell motility in adhesion-independent fashion, by upregulating cadherin-11 expression
Acknowledgments
We thank Drs. Samir M. Hanash, John Minna and Hong-Chen Chen for providing valuable reagents for this study. Microscopy was performed at the VCU Department of Anatomy and Neurobiology Microscopy Facility, supported, in part, with funding from the National Institute of Health (NIH)-NINDS Center core grant 5P30NS047463 and NIH-NCI Cancer Center Support Grant P30CA016059. Some imaging experiments were performed using a Leica SP8 confocal microscope that was purchased with funding from the National Institutes of Health SIG grant S10-OD019972. This work was supported by the NIH-NTDDK grant RO1 DK108278 to A.I.I.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Navis A, Nelson CM, Pulling together: Tissue-generated forces that drive lumen morphogenesis, Semin Cell Dev Biol, 55 (2016) 139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wang S, Sekiguchi R, Daley WP, Yamada KM, Patterned cell and matrix dynamics in branching morphogenesis, J Cell Biol, 216 (2017) 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Abreu-Blanco MT, Watts JJ, Verboon JM, Parkhurst SM, Cytoskeleton responses in wound repair, Cell Mol Life Sci, 69 (2012) 2469–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Krafts KP, Tissue repair: The hidden drama, Organogenesis, 6 (2010) 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lambert AW, Pattabiraman DR, Weinberg RA, Emerging Biological Principles of Metastasis, Cell, 168 (2017) 670–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Paul CD, Mistriotis P, Konstantopoulos K, Cancer cell motility: lessons from migration in confined spaces, Nat Rev Cancer, 17 (2017) 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Carlier MF, Pernier J, Montaville P, Shekhar S, Kuhn S, D. Control of polarized assembly of actin filaments in cell motility, Cell Mol Life Sci, 72 (2015) 3051–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM, Mechanical integration of actin and adhesion dynamics in cell migration, Annu Rev Cell Dev Biol, 26 (2010) 315–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Murrell M, Oakes PW, Lenz M, Gardel ML, Forcing cells into shape: the mechanics of actomyosin contractility, Nat Rev Mol Cell Biol, 16 (2015) 486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pollard TD, Borisy GG, Cellular motility driven by assembly and disassembly of actin filaments, Cell, 112 (2003) 453–465. [DOI] [PubMed] [Google Scholar]
- [11].Mattila PK, Batista FD, Treanor B, Dynamics of the actin cytoskeleton mediates receptor cross talk: An emerging concept in tuning receptor signaling, J Cell Biol, 212 (2016) 267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Salbreux G, Charras G, Paluch E, Actin cortex mechanics and cellular morphogenesis, Trends Cell Biol, 22 (2012) 536–545. [DOI] [PubMed] [Google Scholar]
- [13].Kapus A, Janmey P, Plasma membrane—cortical cytoskeleton interactions: a cell biology approach with biophysical considerations, Compr Physiol, 3 (2013) 1231–1281. [DOI] [PubMed] [Google Scholar]
- [14].Baines AJ, The spectrin-ankyrin-4.1-adducin membrane skeleton: adapting eukaryotic cells to the demands of animal life, Protoplasma, 244 (2010) 99–131. [DOI] [PubMed] [Google Scholar]
- [15].Bennett V, Healy J, Membrane domains based on ankyrin and spectrin associated with cell-cell interactions, Cold Spring Harb Perspect Biol, 1 (2009) a003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Machnicka B, Grochowalska R, Boguslawska DM, Sikorski AF, Lecomte MC, Spectrin-based skeleton as an actor in cell signaling, Cell Mol Life Sci, 69 (2012) 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li X, Matsuoka Y, Bennett V, Adducin preferentially recruits spectrin to the fast growing ends of actin filaments in a complex requiring the MARCKS-related domain and a newly defined oligomerization domain, J Biol Chem, 273 (1998) 19329–19338. [DOI] [PubMed] [Google Scholar]
- [18].Mische SM, Mooseker MS, Morrow JS, Erythrocyte adducin: a calmodulin-regulated actin-bundling protein that stimulates spectrin-actin binding, J Cell Biol, 105 (1987) 2837–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kuhlman PA, Hughes CA, Bennett V, Fowler VM, A new function for adducin. Calcium/calmodulin-regulated capping of the barbed ends of actin filaments, J Biol Chem, 271 (1996) 7986–7991. [DOI] [PubMed] [Google Scholar]
- [20].Matsuoka Y, Li X, Bennett V, Adducin is an in vivo substrate for protein kinase C: phosphorylation in the MARCKS-related domain inhibits activity in promoting spectrin-actin complexes and occurs in many cells, including dendritic spines of neurons, J Cell Biol, 142 (1998) 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Taylor KA, Taylor DW, Formation of two-dimensional complexes of F-actin and crosslinking proteins on lipid monolayers: demonstration of unipolar α-actinin-F-actin crosslinking, Biophys J, 67 (1994) 1976–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Franco T, Low PS, Erythrocyte adducin: a structural regulator of the red blood cell membrane, Transfus Clin Biol, 17 (2010) 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Matsuoka Y, Li X, Bennett V, Adducin: structure, function and regulation, Cell Mol Life Sci, 57 (2000) 884–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dong L, Chapline C, Mousseau B, Fowler L, Ramsay K, Stevens JL, Jaken S, 35H, a sequence isolated as a protein kinase C binding protein, is a novel member of the adducin family, J Biol Chem, 270 (1995) 25534–25540. [DOI] [PubMed] [Google Scholar]
- [25].Joshi R, Gilligan DM, Otto E, McLaughlin T, Bennett V, Primary structure and domain organization of human α and β adducin, J Cell Biol, 115 (1991) 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Barkalow KL, Italiano JE Jr., Chou DE, Matsuoka Y, Bennett V, Hartwig JH, α-adducin dissociates from F-actin and spectrin during platelet activation, J Cell Biol, 161 (2003) 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen CL, Hsieh YT, Chen HC, Phosphorylation of adducin by protein kinase Cdelta promotes cell motility, J Cell Sci, 120 (2007) 1157–1167. [DOI] [PubMed] [Google Scholar]
- [28].Fowler L, Dong L, Bowes RC 3rd, van de Water B, Stevens JL, Jaken S, Transformation-sensitive changes in expression, localization, and phosphorylation of adducins in renal proximal tubule epithelial cells, Cell Growth Differ, 9 (1998) 177–184. [PubMed] [Google Scholar]
- [29].Abdi KM, Bennett V, Adducin promotes micrometer-scale organization of P2-spectrin in lateral membranes of bronchial epithelial cells, Mol Biol Cell, 19 (2008) 536–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chen CL, Lin YP, Lai YC, Chen HC, α-Adducin translocates to the nucleus upon loss of cell-cell adhesions, Traffic, 12 (2011) 1327–1340. [DOI] [PubMed] [Google Scholar]
- [31].Naydenov NG, Ivanov AI, Adducins regulate remodeling of apical junctions in human epithelial cells, Mol Biol Cell, 21 (2010) 3506–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Naydenov NG, Ivanov AI, Spectrin-adducin membrane skeleton: A missing link between epithelial junctions and the actin cytoskeletion?, Bioarchitecture, 1 (2011) 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rotzer V, Breit A, Waschke J, Spindler V, Adducin is required for desmosomal cohesion in keratinocytes, J Biol Chem, 289 (2014) 14925–14940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bednarek E, Caroni P, β-Adducin is required for stable assembly of new synapses and improved memory upon environmental enrichment, Neuron, 69 (2011) 1132–1146. [DOI] [PubMed] [Google Scholar]
- [35].Fu C, Xu J, Li RJ, Crawford JA, Khan AB, Ma TM, Cha JY, Snowman AM, Pletnikov MV, Snyder SH, Inositol Hexakisphosphate Kinase-3 Regulates the Morphology and Synapse Formation of Cerebellar Purkinje Cells via Spectrin/Adducin, J Neurosci, 35 (2015) 11056–11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Leite SC, Sampaio P, Sousa VF, Nogueira-Rodrigues J, Pinto-Costa R, Peters LL, Brites P, Sousa MM, The Actin-Binding Protein α-Adducin Is Required for Maintaining Axon Diameter, Cell Rep, 15 (2016) 490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jen J, Lin LL, Chen HT, Liao SY, Lo FY, Tang YA, Su WC, Salgia R, Hsu CL, Huang HC, Juan HF, Wang YC, Oncoprotein ZNF322A transcriptionally deregulates α-adducin, cyclin D1 and p53 to promote tumor growth and metastasis in lung cancer, Oncogene, 35 (2016) 2357–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fukata Y, Oshiro N, Kinoshita N, Kawano Y, Matsuoka Y, Bennett V, Matsuura Y, Kaibuchi K, Phosphorylation of adducin by Rho-kinase plays a crucial role in cell motility, J Cell Biol, 145 (1999)347–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Syed V, Zhang X, Lau KM, Cheng R, Mukheijee K, Ho SM, Profiling estrogen-regulated gene expression changes in normal and malignant human ovarian surface epithelial cells, Oncogene, 24 (2005) 8128–8143. [DOI] [PubMed] [Google Scholar]
- [40].Luo C, Shen J, Adducin in tumorigenesis and metastasis, Oncotarget, 8 (2017) 48453–48459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ma PC, Tretiakova MS, Nallasura V, Jagadeeswaran R, Husain AN, Salgia R, Downstream signalling and specific inhibition of c-MET/HGF pathway in small cell lung cancer: implications for tumour invasion, Br J Cancer, 97 (2007) 368–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Langer W, Sohler F, Leder G, Beckmann G, Seidel H, Grone J, Hummel M, Sommer A, Exon array analysis using re-defined probe sets results in reliable identification of alternatively spliced genes in non-small cell lung cancer, BMC Genomics, 11 (2010) 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sanjana NE, Shalem O, Zhang F, Improved vectors and genome-wide libraries for CRISPR screening, Nat Methods, 11 (2014) 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F, Genome-scale CRISPR-Cas9 knockout screening in human cells, Science, 343 (2014) 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chan PC, Hsu RY, Liu CW, Lai CC, Chen HC, Adducin-1 is essential for mitotic spindle assembly through its interaction with myosin-X, J Cell Biol, 204 (2014) 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Fowler L, Everitt J, Stevens JL, Jaken S, Redistribution and enhanced protein kinase C-mediated phosphorylation of α- and γ-adducin during renal tumor progression, Cell Growth Differ, 9(1998)405–413. [PubMed] [Google Scholar]
- [47].Schliekelman MJ, Taguchi A, Zhu J, Dai X, Rodriguez J, Celiktas M, Zhang Q, Chin A, Wong CH, Wang H, McFerrin L, Selamat SA, Yang C, Kroh EM, Garg KS, Behrens C, Gazdar AF, Laird-Offringa IA, Tewari M, Wistuba II, Thiery JP, Hanash SM, Molecular portraits of epithelial, mesenchymal, and hybrid states in lung adenocarcinoma and their relevance to survival, Cancer Res, 75 (2015) 1789–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cozens AL, Yezzi MJ, Kunzelmann K, Ohrui T, Chin L, Eng K, Finkbeiner WE, Widdicombe JH, Gruenert DC, CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells, Am J Respir Cell Mol Biol, 10 (1994) 38–47. [DOI] [PubMed] [Google Scholar]
- [49].Sato M, Larsen JE, Lee W, Sun H, Shames DS, Dalvi MP, Ramirez RD, Tang H, DiMaio JM, Gao B, Xie Y, Wistuba II, Gazdar AF, Shay JW, Minna JD, Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations, Mol Cancer Res, 11 (2013)638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Robledo RF, Ciciotte SL, Gwynn B, Sahr KE, Gilligan DM, Mohandas N, Peters LL, Targeted deletion of α-adducin results in absent β- and γ-adducin, compensated hemolytic anemia, and lethal hydrocephalus in mice, Blood, 112 (2008) 4298–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sahr KE, Lambert AJ, Ciciotte SL, Mohandas N, Peters LL, Targeted deletion of the γ-adducin gene (Add3) in mice reveals differences in α-adducin interactions in erythroid and nonerythroid cells, Am J Hematol, 84 (2009) 354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gupton SL, Waterman-Storer CM, Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration, Cell, 125 (2006) 1361–1374. [DOI] [PubMed] [Google Scholar]
- [53].Naydenov NG, Feygin A, Wang L, Ivanov AI, N-ethylmaleimide-sensitive factor attachment protein a (aSNAP) regulates matrix adhesion and integrin processing in human epithelial cells, J Biol Chem, 289 (2014) 2424–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Burridge K, Guilluy C, Focal adhesions, stress fibers and mechanical tension, Exp Cell Res, 343 (2016) 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ciobanasu C, Faivre B, Le Clainche C, Integrating actin dynamics, mechanotransduction and integrin activation: the multiple functions of actin binding proteins in focal adhesions, Eur J Cell Biol, 92 (2013) 339–348. [DOI] [PubMed] [Google Scholar]
- [56].Boateng LR, Huttenlocher A, Spatiotemporal regulation of Src and its substrates at invadosomes, Eur J Cell Biol, 91 (2012) 878–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sirvent A, Urbach S, Roche S, Contribution of phosphoproteomics in understanding SRC signaling in normal and tumor cells, Proteomics, 15 (2015) 232–244. [DOI] [PubMed] [Google Scholar]
- [58].Tanaka S, Sumioka T, Fujita N, Kitano A, Okada Y, Yamanaka O, Flanders KC, Miyajima M, Saika S, Suppression of injury-induced epithelial-mesenchymal transition in a mouse lens epithelium lacking tenascin-C, Mol Vis, 16 (2010) 1194–1205. [PMC free article] [PubMed] [Google Scholar]
- [59].Wu J, Masci PP, Chen C, Chen J, Lavin MF, Zhao KN, β-Adducin siRNA disruption of the spectrin-based cytoskeleton in differentiating keratinocytes prevented by calcium acting through calmodulin/epidermal growth factor receptor/cadherin pathway, Cell Signal, 27 (2015) 15–25. [DOI] [PubMed] [Google Scholar]
- [60].Huang H, Colella S, Kurrer M, Yonekawa Y, Kleihues P, Ohgaki H, Gene expression profiling of low-grade diffuse astrocytomas by cDNA arrays, Cancer Res, 60 (2000) 6868–6874. [PubMed] [Google Scholar]
- [61].van den Boom J, Wolter M, Kuick R, Misek DE, Youkilis AS, Wechsler DS, Sommer C, Reifenberger G, Hanash SM, Characterization of gene expression profiles associated with glioma progression using oligonucleotide-based microarray analysis and real-time reverse transcription-polymerase chain reaction, Am J Pathol, 163 (2003) 1033–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Dutertre M, Lacroix-Triki M, Driouch K, de la Grange P, Gratadou L, Beck S, Millevoi S, Tazi J, Lidereau R, Vagner S, Auboeuf D, Exon-based clustering of murine breast tumor transcriptomes reveals alternative exons whose expression is associated with metastasis, Cancer Res, 70 (2010) 896–905. [DOI] [PubMed] [Google Scholar]
- [63].Tripodi G, Valtorta F, Torielli L, Chieregatti E, Salardi S, Trusolino L, Menegon A, Ferrari P, Marchisio PC, Bianchi G, Hypertension-associated point mutations in the adducin a and β subunits affect actin cytoskeleton and ion transport, J Clin Invest, 97 (1996) 2815–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Babbin BA, Koch S, Bachar M, Conti MA, Parkos CA, Adelstein RS, Nusrat A, AT. Ivanov, Non-muscle myosin IIA differentially regulates intestinal epithelial cell restitution and matrix invasion, Am J Pathol, 174 (2009) 436–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Alimperti S, Andreadis ST, CDH2 and CDH11 act as regulators of stem cell fate decisions, Stem Cell Res, 14 (2015) 270–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Birtolo C, Pham H, Morvaridi S, Chheda C, Go VL, Ptasznik A, Edderkaoui M, Weisman MH, Noss E, Brenner MB, Larson B, Guindi M, Wang Q, Pandol SJ, Cadherin-11 Is a Cell Surface Marker Up-Regulated in Activated Pancreatic Stellate Cells and Is Involved in Pancreatic Cancer Cell Migration, Am J Pathol, 187 (2017) 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Schulte JD, Srikanth M, Das S, Zhang J, Lathia JD, Yin L, Rich JN, Olson EC, Kessler JA, Chenn A, Cadherin-11 regulates motility in normal cortical neural precursors and glioblastoma, PLoS One, 8 (2013) e70962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Yao J, Deng B, Zheng L, Dou L, Guo Y, Guo K, miR-27b is upregulated in cervical carcinogenesis and promotes cell growth and invasion by regulating CDH11 and epithelial-mesenchymal transition, Oncol Rep, 35 (2016) 1645–1651. [DOI] [PubMed] [Google Scholar]
- [69].Alimperti S, You H, George T, Agarwal SK, Andreadis ST, Cadherin-11 regulates both mesenchymal stem cell differentiation into smooth muscle cells and the development of contractile function in vivo, J Cell Sci, 127 (2014) 2627–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Row S, Liu Y, Alimperti S, Agarwal SK, Andreadis ST, Cadherin-11 is a novel regulator of extracellular matrix synthesis and tissue mechanics, J Cell Sci, 129 (2016) 2950–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ferrandi M, Cusi D, Molinari I, Del Vecchio L, Barlassina C, Rastaldi MP, Schena FP, Macciardi F, Marcantoni C, Roccatello D, Peters LL, Armelloni S, Min L, Giardino L, Mattinzoli D, Camisasca C, Palazzo F, Manunta P, Ferrari P, Bianchi G, α- and β-Adducin polymorphisms affect podocyte proteins and proteinuria in rodents and decline of renal function in human IgA nephropathy, J Mol Med, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lechuga S, Baranwal S, Li C, Naydenov NG, Kuemmerle JF, Dugina V, Chaponnier C, Ivanov AI, Loss of γ-cytoplasmic actin triggers myofibroblast transition of human epithelial cells, Mol Biol Cell, 25 (2014) 3133–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Posern G, Treisman R, Actin’ together: serum response factor, its cofactors and the link to signal transduction, Trends Cell Biol, 16 (2006) 588–596. [DOI] [PubMed] [Google Scholar]
- [74].Liu CM, Hsu WH, Lin WY, Chen HC, Adducin family proteins possess different nuclear export potentials, J Biomed Sci, 24 (2017) 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.