Abstract
Natural brown–black eumelanin pigments confer structural coloration in animals and potently block ionizing radiation and antifungal drugs. These functions also make them attractive for bioinspired materials design, including coating materials for drug-delivery vehicles, strengthening agents for adhesive hydrogel materials, and free-radical scavengers for soil remediation. Nonetheless, the molecular determinants of the melanin “developmental road traveled” and the resulting architectural features have remained uncertain because of the insoluble, heterogeneous, and amorphous characteristics of these complex polymeric assemblies. Here, we used 2D solid-state NMR, EPR, and dynamic nuclear polarization spectroscopic techniques, assisted in some instances by the use of isotopically enriched precursors, to address several open questions regarding the molecular structures and associated functions of eumelanin. Our findings uncovered: 1) that the identity of the available catecholamine precursor alters the structure of melanin pigments produced either in Cryptococcus neoformans fungal cells or under cell-free conditions; 2) that the identity of the available precursor alters the scaffold organization and membrane lipid content of melanized fungal cells; 3) that the fungal cells are melanized preferentially by an l-DOPA precursor; and 4) that the macromolecular carbon- and nitrogen-based architecture of cell-free and fungal eumelanins includes indole, pyrrole, indolequinone, and open-chain building blocks that develop depending on reaction time. In conclusion, the availability of catecholamine precursors plays an important role in eumelanin development by affecting the efficacy of pigment formation, the melanin molecular structure, and its underlying scaffold in fungal systems.
Keywords: melanogenesis, fungi, biophysics, catecholamine, cell wall, nuclear magnetic resonance (NMR), polysaccharide, solid state NMR, electron paramagnetic resonance (EPR), melanin, melanization, pigment formation
Introduction
The diverse functional capabilities associated with natural brown–black eumelanin pigments range from structural coloration of bird feathers, to blockage of ionizing radiation and antifungal drugs, to energy transduction (1–3). These functions also make melanin-like polymer composites and structurally related polydopamines attractive platforms for bio-inspired materials design, where their applications include coatings for drug delivery vehicles and free radical scavengers for soil remediation (2, 4–7). Despite the evident versatility and practical importance of these materials, the molecular bases of melanin development and architecture have remained uncertain because of their insoluble, heterogeneous, and amorphous character.
Recent progress toward the understanding of eumelanin structure has exploited approaches that include magnetic resonance spectroscopy, stable isotope enrichment, computational modeling, and controlled chemical reactions to lay the groundwork for the current report: 1) eumelanin pigments synthesized within Cryptococcus neoformans fungal cells differ in molecular structure based on the availability of particular obligatory exogenous catecholamine precursors (8, 9); 2) the layered deposition of C. neoformans melanins relies upon a polysaccharide cell-wall scaffold (3, 10–12); 3) eumelanin pigments derived from cell-free or C. neoformans-based synthesis display a common indole-based aromatic core (13); 4) varying proportions of building block structures such as 5,6-dihydroxyindole (DHI),4 5,6-dihydroxyindole-2-carboxylic acid (DHICA), pyrrolecarboxylic acid, and uncyclized catecholamines can influence synthetic polydopamine and related supramolecular architectures along with their consequent optical and conductive properties (14–16).
C. neoformans melanizes in brain tissue during human infection, but the actual substrates for melanin synthesis are not known (17). Because brain tissues are rich in catecholamines (18), the presumption in the field is that they serve as the substrates for melanization (19). In fact, given that C. neoformans can use different substrates for melanization and that the brain contains different catecholamines, it is possible that multiple precursors are used in this process. Melanization in brain tissue has been proposed as a cause for reduced susceptibility to antifungal drugs, which could contribute to the difficulty in treating infection (20). Hence, it is important to study melanization with different precursors to gain insight into possible pathways to melanin synthesis in brain tissue.
Herein we use solid-state nuclear magnetic resonance (NMR), electron paramagnetic resonance (EPR), and dynamic nuclear polarization (DNP) methods to address the following open questions involving eumelanin molecular structure. 1) Does the catecholamine precursor structure also control melanin product outcomes during cell-free synthesis? 2) Does the availability of particular precursors impact the polysaccharide and lipid organization of the melanized fungal cell-wall scaffold? 3) Which catecholamines are the preferred biosynthetic precursors for melanization of C. neoformans cells? 4) Which melanin-building blocks and macromolecular organization can be identified as a function of reaction time in a cell-free system? With due apology to Robert Frost for adapting the title of his epic poem, the current work seeks to explore the “melanization road more traveled by” (21).
Results and discussion
Precursor identity alters pigment structure in cell-free as well as fungal melanization
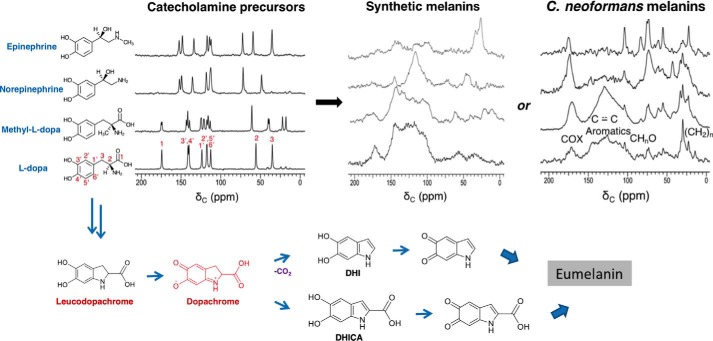
Given our prior observations of structurally diverse melanized cell-wall materials (“ghosts”) formed after provision of various obligatory exogenous catecholamine precursors to C. neoformans fungal cells (8, 9), we sought to distinguish several explanations for their differences in biosynthesis and macromolecular structure. These include the ability of the cell-wall scaffold to host the deposition of melanin pigments derived from each precursor, the intrinsically different polymerization pathways for the respective monomers, and some combination of these two rationales. Beginning with the l-DOPA, methyl-l-DOPA, (−)-norepinephrine (NorE), and (−)-epinephrine catecholamine precursors studied previously in this fungal system, the solid-state 13C NMR spectroscopic fingerprints of the starting materials and their respective insoluble solid products are compared in Fig. 1.
Figure 1.
From upper left to right are shown the 150 MHz solid-state CPMAS 13C NMR spectra of catecholamine precursors, the corresponding melanins obtained by cell-free autopolymerization, and the corresponding melanin ghosts obtained by fungal biosynthesis; below are the proposed structural intermediates in the conversion of l-DOPA to a melanin pigment. C. neoformans melanin spectra and the proposed synthetic scheme are adapted from Ref. 9. This research was originally published in Biochemistry. Chatterjee, S., Prados-Rosales, R., Frases, S., Itin, B., Casadevall, A., and Stark, R. E. Using solid-state NMR to monitor the molecular consequences of Cryptococcus neoformans melanization with different catecholamine precursors. Biochemistry. 2012; 51: 6080–6088. © American Chemical Society. Each chemical constituent contains the 13C isotope at natural abundance (1.1%).
As expected in conjunction with the formation of polymeric assemblies, the 13C cross-polarization-magic-angle spinning (CPMAS) spectra of the resulting (bio)synthetic pigments displayed altered resonance positions and line widths as compared with their respective small-molecule precursors. Although the structurally disordered amorphous melanin ghosts formed in both C. neoformans fungal cells and cell-free systems had typically broad resonances (9, 10, 22), we could nonetheless identify NMR signals attributable to carboxylate or amide structures (170–173 ppm), arenes and alkenes (110–160 ppm), alkoxy groups of the polysaccharide cell wall (55–105 ppm), and alkyl chains (20–40 ppm) (11).
Comparison of the products from cell-free and fungal melanin preparations revealed both commonalities and contrasts. Whereas the catecholamine precursors each had sharp 13C NMR spectral lines typical of crystalline solids, both the amorphous polymers resulting from autopolymerization and the C. neoformans melanin ghosts displayed broader features in the aromatic and aliphatic regions of their spectra. This common spectral appearance underscores the amorphous character of the solid aromatic melanin pigments and the melanized aliphatic cell walls that are formed by either synthetic method, regardless of whether enzymes or supporting scaffolds are present. We attribute the very broad aromatic spectral envelope of the pigment (110–160 ppm) to resonance overlap (chemical shift heterogeneity) rather than spin relaxation effects due to proximity to the free radicals present in these melanin samples (see EPR below), as many of the NMR resonances are better resolved in selectively 13C-labeled materials and with the benefit of two-dimensional NMR experiments (10, 11, 13, 22). Specifically, the broad appearance of aromatic resonances in the 13C spectra can reflect slightly dissimilar DHI, DHICA, and other possible structural building blocks; diverse inter-unit covalent connections; and/or heterogeneously aligned stacking of successive pigment layers (14).
Each of the purified melanins also displayed notable aliphatic signals in the 20–40 and 55–75–ppm regions of their spectra. These features differ from spectra of the respective catecholamine precursors. Although the 30- and 72–ppm resonances of the fungal melanin preparations, for instance, could be derived from cell-membrane and cell-wall constituents in the melanin particles, this explanation cannot account for the same or similar features in spectra of the corresponding cell-free preparations. An alternative hypothesis that encompasses both preparation methods and has been proposed by our group for C. neoformans melanins (9, 22) in analogy with polydopamines (16) views the aliphatic structures as precursor moieties that remain uncyclized or partially polymerized, while nonetheless forming covalently bound polymers that can resist our exhaustive chemical and biochemical isolation protocol. This proposal of incompletely formed or “developing” aromatic melanin structures is revisited under “Molecular architectures of melanin biopolymers from contrasting synthetic and spectroscopic protocols include indole, pyrrole, open-chain, and newly proposed structures.”
Among the catecholamine precursors, l-DOPA and methyl-l-DOPA produced the most robust melanin pigment deposition, as evidenced by aromatic NMR signals arising from their indole-based structures, whereas epinephrine formed essentially no indole-like pigment in either cell-free or fungal reactions. These trends are in accord with prior C. neoformans preparations, which gave yields of 47, 61, 27, and 2% for melanin ghosts from l-DOPA, methyl-DOPA, norepinephrine, and epinephrine substrates, respectively (8). The results for norepinephrine were mixed: modest pigment signal intensity was evident in the fungal system and only a subset of aromatic features appeared in the 13C NMR spectrum of the corresponding cell-free melanin. As noted previously (9), even epinephrine-derived materials for which the pigment is not detected in the 13C spectrum can be assumed to possess cell walls that are partially melanized, because unmelanized cells cannot survive the harsh procedures used to isolate the ghosts.
Although only qualitative comparisons between cell-free and fungal preparations are possible from perusal of these standard one-dimensional natural-abundance spectroscopic fingerprints, the current findings align with and extend our recent demonstration of a common indole-based aromatic core in similar isotopically enriched l-tyrosine, l-DOPA, and dopamine-derived eumelanin systems using two-dimensional 13C–15N solid-state NMR correlations (13). The current finding of robust cell-free melanization with l-DOPA and methyl-l-DOPA bolsters our prior claim that formation of DHICA- and DHI-containing aromatic pigments requires stabilized cyclic dopachrome intermediate structures such as that illustrated in Fig. 1 (9). This hypothesis is also strengthened by the requirement of hydroxyl-substituted benzene or indole rings in fungal melanin precursor structures: C. neoformans can form aromatic pigments from serotonin and 4-hydroxyindole, to a lesser degree from 5-hydroxytryptophan and 5-hydroxyindole, but not from tryptophan itself (illustrated in Fig. S1).
The observation of similar chemical environments for the aromatic core in several pairs of cell-free versus C. neoformans melanins is surprising at first glance, given the reported involvement of the polysaccharide cell-wall scaffold in pigment deposition (24, 25). However, even in fungal cells the ∼200 Å thick melanin layers (26) should be comprised of numerous proximal aromatic pigment units, so that in both synthetic and biosynthetic systems any given unit is most likely to be surrounded by other pigment molecules. The predominance of pigment–pigment rather than pigment–cell-wall interactions had been validated in l-DOPA fungal melanin ghost preparations using 2D solid-state NMR experiments that identify pairs of 13C nuclei located within ∼6 Å of one another (11).
Precursor identity alters the structural order and lipid content of cellular scaffolds in fungal melanin ghosts
In addition to the role of precursor structure in determining the outcome of melanin (bio)synthesis, an understanding of the underlying melanized cell-wall architecture is critical for C. neoformans systems because pigment deposition is known to impact granule formation, cellular porosity, and drug resistance (26). A glucose-derived aliphatic scaffold is formed in advance of the melanin pigment in the C. neoformans fungal system (11). Moreover, the progressive development of closely packed granule layers within the melanized cells raises the question of whether these layers are simply cross-linked to one another or held together by a scaffold composed of pigments, proteins, polysaccharides, and/or lipids (26). Whatever the mechanism, it is important to note that a melanized cell wall must be sufficiently malleable to permit cell budding and morphological transitions such as hyphal formation (27). The importance of these considerations is underscored by reports of chitin synthase-deficient fungal cultures in which melanin pigments are produced but are poorly retained by the cell wall (24). Conversely, supplementation of C. neoformans cultures with the scaffold building block GlcNAc leads to diminished chitin-to-chitosan ratios and elevated pigment content in the resulting melanin ghosts (25).
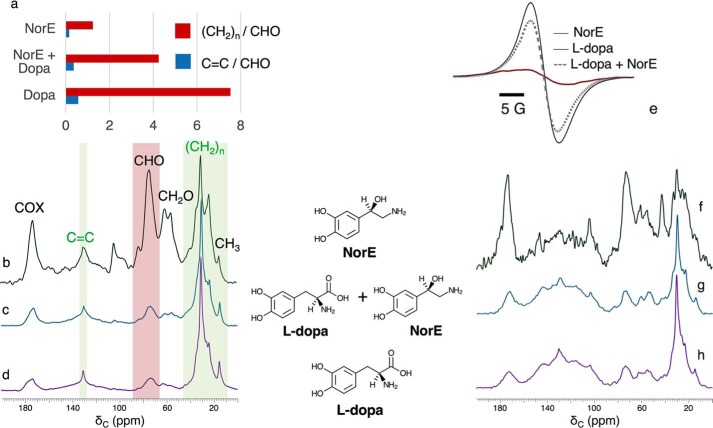
To test whether the hypothesized cell-wall scaffold and associated cellular constituents retained in the melanin ghosts differ in molecular composition, order, or organization according to the catecholamine from which the pigment is derived, we examined solid-state 13C NMR spectra of C. neoformans fungal cells grown with [U-13C6]glucose as demonstrated previously (10, 11). Notably, the constituents that resist our rigorous degradative treatments are precisely those cellular materials that are of interest because they are integrally associated with the melanin biopolymers. For pigments produced using l-DOPA or NorE (Fig. 2), each of the quantitatively reliable direct polarization (DPMAS) spectra shown in Fig. 2, b–d, displays 13C resonances attributable to cell-wall constituents (oxygen-linked carbons derived from β-glucans, chitin, chitosan, and mannans (28)) as well as membrane-derived glycerides (glycerol backbones, lipid head groups, and acyl chains) (10).
Figure 2.
150 MHz solid-state 13C magic-angle spinning (MAS NMR) spectra showing carbon-containing chemical moieties in C. neoformans melanized cell walls produced with different catecholamine precursors. Left, a–d: quantitatively reliable DPMAS NMR yields C = C/CHO and (CH2)n/CHO peak ratios for 13C-enriched membrane lipids and polysaccharides in melanin ghosts produced using d-[U-13C6]glucose and natural-abundance (1.1% 13C) catecholamine precursors. Peak assignments are based on published reports for fungal cell walls and mammalian phospholipids (10, 32), with C = C and (CH2)n groups that originate primarily from the membrane-derived functional groups marked in green. Right, e–h: comparative X-band EPR and cross-polarization (CPMAS) NMR spectra of melanins produced from natural-abundance glucose and catecholamine precursors. Each NMR spectrum is presented with the tallest peak set at full scale.
The NorE-derived fungal melanin stands out with respect to 13C spectral resolution, although the resonances are an order of magnitude broader than in native (unmelanized) Aspergillus fumigatus cells (29). The relatively sharp NorE melanin spectrum suggests fewer types of magnetically similar functional groups, a more ordered crystalline-like molecular architecture, and/or more motional averaging of the resonances. Whereas the modest EPR signal intensity displayed in Fig. 2e indicates fewer stable free radicals, diminished peak broadening would not be anticipated for the relatively distant aliphatic moieties. Motional averaging can also be viewed as unlikely, because 1 ms of cross-polarization suffices to build up nearly all of the signal intensity observed when [13]C DPMAS spectra are acquired with long recycle delays (data not shown).
Also of note in these melanin ghost 1D 13C NMR spectra are the relative integrated peak areas for primarily membrane lipid alkenes (C = C, 130 ppm) and polysaccharide oxymethines (CHO, 74 ppm). The C = C/CHO ratios are 0.57 and 0.15 for l-DOPA and NorE fungal melanins, respectively (Fig. 2a), suggesting that lipids are better retained in the ghosts by the well-melanized l-DOPA cell walls. An analogous trend involving (CH2)n/CHO ratios, 7.7 and 1.2, again supports a larger proportion of lipids retained by l-DOPA–melanized cell walls. Although such comparisons must be made with caution because few peaks represent a single aliphatic constituent, they support a model wherein the pigment traps a portion of the vesicle-derived lipids (30), rendering them less accessible to extraction and HCl boiling treatments.
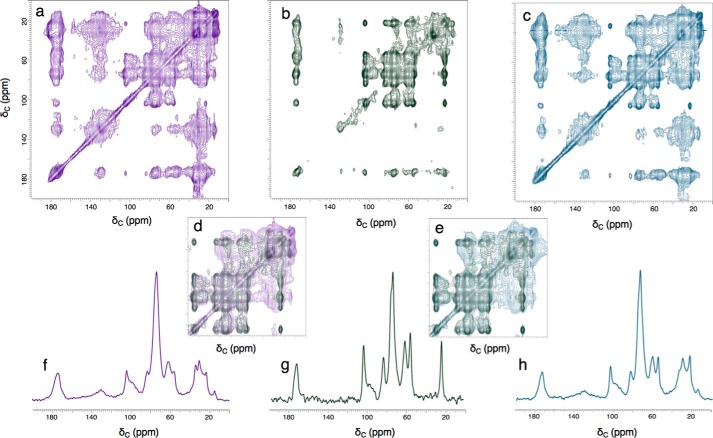
A more definitive picture of how the chemical moieties and associated cell-wall architectures depend on which melanin precursor was available can be seen by comparing the 2D 13C–13C dipolar correlation (DARR) spectra of C. neoformans ghost materials derived from [U-13C6]glucose and either unlabeled l-DOPA or NorE catecholamine precursors (Fig. 3). Many of the 13C chemical shift values and pairwise through-space carbon–carbon connectivities are maintained in each fungal melanin sample, a conclusion reinforced by the analogous overall spectroscopic similarities observed with 50-, 250-, and 500-ms DARR mixing times corresponding to carbon–carbon distances spanning ∼2–6 Å (Fig. S2). As shown previously using the l-DOPA precursor, both sets of 2D spectra identify proximal carbon pairs that are situated either within a given cell-wall sugar unit or on nearby polysaccharide and membranous constituents (11).
Figure 3.
Two-dimensional solid-state 13C–13C DARR NMR contour plots (56, 57) and 74-ppm ω1 cross-sections obtained from experiments conducted with 15 kHz MAS, a 1H frequency of 600 MHz, and a 500-ms mixing time for C. neoformans-melanized cell-wall ghosts produced with d- [U-13C6]glucose. Contours on the diagonal correspond to one-dimensional spectra such as those shown in Fig. 2; off-diagonal contours (cross-peaks or connectivities) correspond to pairwise through-space carbon–carbon proximities within ∼6 Å of one another. The indirect chemical shift dimension corresponding to the y axis of each plot is designated as ω1. Top row: a, ghosts produced with natural-abundance l-DOPA (adapted from Ref. 11) (This research was originally published in the Journal of Biological Chemistry. Chatterjee, S., Prados-Rosales, R., Itin, B., Casadevall, A., and Stark, R. E.. Solid-state NMR reveals the carbon-based molecular architecture of Cryptococcus neoformans fungal eumelanins in the cell wall. J. Biol. Chem. 2015; 290: 13779–13790. © the American Society for Biochemistry and Molecular Biology.); b, ghosts from natural-abundance NorE. c, ghosts produced with a 1:1 (mol/mol) mixture of l-DOPA and NorE. Middle row: d, upfield region of NorE overlaid with l-DOPA; e, upfield region of NorE overlaid with (l-DOPA + NorE). Bottom row, 74-ppm ω1 cross-sections: f, l-DOPA; g, NorE; h, (l-DOPA + NorE). Overall comparisons of the three 2D datasets are made by adjusting the vertical scale so that all significant cross-peaks are visible and the same numbers of contours are displayed for the major cell-wall resonance at 74 ppm. Comparisons of the three 1D cross-sections set the tallest 74-ppm peak at full scale.
Nonetheless, a close look at the 2D plots and selected 1D cross-sections in Fig. 3 reveals several key precursor-dependent contrasts among the [U-13C6]glucose-derived carbon spectra. First, the NorE melanin cross-peaks exhibit smaller line widths at half-height and stronger dipolar interactions with, e.g. the major polysaccharide carbons resonating at 74 ppm. Second, both the individual contour plots and the overlaid comparison of l-DOPA and NorE melanins show that the broad lipid cross-peaks to carbons resonating at ∼30, ∼130, and ∼173 ppm are greatly diminished if the precursor is NorE, thereby revealing prominent NMR signals at 21 and 172 pm that could arise from chitin polysaccharides (31, 32) present in fungal cell walls (33). Given the smaller C = C/CHO and (CH2)n/CHO ratios derived from quantitative DPMAS spectra (Fig. 2), we can attribute the dearth of through-space 13C–13C interactions to inefficient retention of membrane lipids. The modest amounts of both lipid and pigment constituents within the NorE-melanized cells can also explain the scaffold's near-crystalline–like molecular order. From a functional perspective, the enhanced molecular order, larger 13C–13C dipolar interactions, and absence of lipid cross-talk in the glucose-derived scaffold associated with the NorE precursor (Figs. 2b, and 3, b and g) correlate with less deposition of melanin (Fig. 1).
C. neoformans fungal cells are melanized preferentially by the l-DOPA precursor
Given that the identity of the catecholamine precursor exerts a significant impact on the product amount, the pigment molecular structure (Fig. 1), and the molecular order of the underlying aliphatic scaffold (Figs. 2 and 3), we supplied substrate pairs to the C. neoformans cells to investigate whether particular precursors were preferred or if pigments with unique macromolecular structures could be formed. Fig. 2, c, e, and g, illustrate the spectroscopic outcomes of a competition study conducted with l-DOPA and NorE, each of which individually deposits significant amounts of pigment and displays a broad aromatic resonance in the 13C NMR ghost spectrum (Fig. 1).
When a 1:1 (mol/mol) mixture of the l-DOPA and NorE catecholamines is used as the melanization substrate for C. neoformans cells, the resulting 13C NMR spectra (Fig. 2, c and g) bear a close resemblance to the l-DOPA fungal melanin itself and suggest that l-DOPA is the preferred substrate in C. neoformans cellular media. The DARR results of Fig. 3c confirm this similarity, revealing broad cross-peaks and diminished intensities relative to the diagonal peak when compared with NorE melanin; both spectral characteristics suggest an amorphous structure with sufficient lipid content or order to permit dipolar cross-talk. The hypothesis of preferential utilization of l-DOPA for melanin biosynthesis is also supported by the corresponding EPR spectra of Fig. 2e, which display an intrinsically weak signal for the NorE melanin but similar intensities and line widths at half-height for melanin ghosts from l-DOPA and the 1:1 precursor mixture. Taken together, these spectroscopic results support a mechanistic model in which norepinephrine does not compete with l-DOPA during polymerization to form an indole-based pigment, even though the C. neoformans fungus is capable of utilizing each substrate individually to achieve protective cell-wall melanization.
Although an analogous l-DOPA-epinephrine competition study yields pigment resonances with chemical shifts reminiscent of l-DOPA melanin, the d-[U-13C6]glucose-derived scaffold displays superior resolution in both 1D 13C and 2D DARR spectra as compared with the aliphatic constituents melanized using either catecholamine alone (Fig. S3). Thus the epinephrine alongside l-DOPA yields the hallmarks of pigment deposition in both the resulting 13C NMR and X-band EPR spectra, but they leave the aliphatic support scaffold surprisingly lipid deficient and well-ordered organizationally. These observations can be rationalized if association or covalent binding of the two precursors occur to expose hydrophilic -OH groups that facilitate pigment anchoring to the polysaccharide scaffold, whereas the vesicle-derived hydrophobic lipids are highly susceptible to extraction during the ghost isolation protocol.
Molecular architectures of melanin biopolymers from contrasting synthetic and spectroscopic protocols include indole, pyrrole, open-chain, and newly proposed structures
To augment our spectroscopic assessments of how catecholamine precursor availability impacts the carbon-based structures of both the resulting melanin pigment and the underlying cellular scaffold, we chose several readily available precursors enriched in NMR-active isotopes to probe the carbon- and nitrogen-containing heteronuclear molecular frameworks that are constructed during melanization. This set of studies builds on prior solid-state NMR findings: that l-DOPA or dopamine precursors yield progressively increasing amounts of pigment deposition in C. neoformans during cell culture lasting up to 14 days (11, 25); that synthetic polydopamines possess carbon and nitrogen moieties indicative of indole, pyrrole, and uncyclized catecholamine-building blocks (16); and that similar proximal carbon–carbon and carbon–nitrogen pairs of several structural types can be identified within fungal and synthetic melanins (11, 13). In the current investigation, two-dimensional double resonance solid-state NMR strategies were used to identify numerous 13C–15N pairs located 1–3 bonds from one another; the exquisite sensitivity of chemical shifts to molecular environment and the availability of statistically based spectral predictions derived from extensive NMR databases then allowed us to constrain the possible architectures of a range of eumelanin pigments: 1) l-[U-13C,15N]tyrosine-derived biopolymers at two developmental stages of their cell-free synthesis (developing and “mature” at 1.5 and 3.0 days, respectively); 2) mature products made with contrasting precursors and synthetic methods (cell-free l-[U-13C,15N]tyrosine and fungal [U-13C,15N]dopamine melanins). By adjusting the 13C–15N coherence transfer time, it was also possible to extend the reach of our spectroscopic experiments to distances corresponding to 2–3 chemical bond lengths. The goals were to evaluate possible structural hypotheses more rigorously and to gain new molecular insight into the developmental progression of melanization.
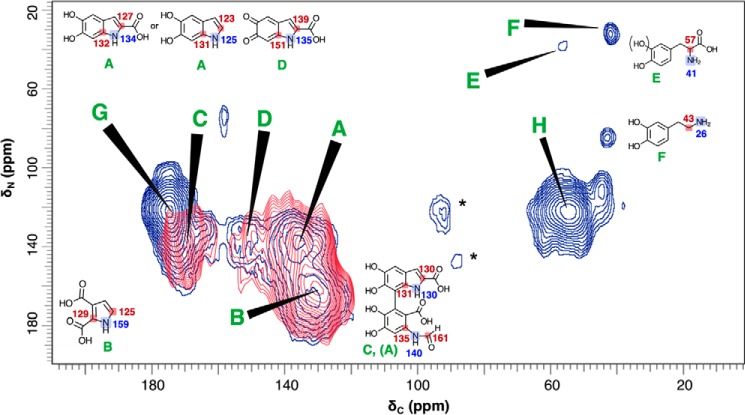
Fig. 4 illustrates 2D 13C–15N Z-filtered transferred echo double resonance (ZF-TEDOR) experiments (34, 35) that can correlate the NMR signals and reveal the respective chemical environments of proximal carbon–nitrogen pairs located within tailored distance ranges up to 5 Å, while avoiding possible dipolar dephasing effects from other than the isotopically enriched spins of interest. In the current study, we first compared the 2D spectra obtained with short (∼1.5 ms) TEDOR recoupling times for synthetic l-tyrosine melanins that are developing at 1.5 days versus mature for 3.0 days. When coupled with sensitivity-enhancing dynamic nuclear polarization (DNP) that was checked for uniform signal enhancements across the spectrum, this strategy enabled us to take a first atomic-scale look at the developmental course of solid-phase melanin polymer synthesis. The resulting through-space correlations identify 13C–15N pairs that are located within ∼1.5 Å of one another (35)5 and likely represent the directly bonded heteronuclei defining the polymeric melanin structure(s). (The pigmented layers evident in high-resolution TEM images of melanized fungal cells (36) are spaced at 3.7–4.0 Å (37), and thus not correlated in these experiments.)
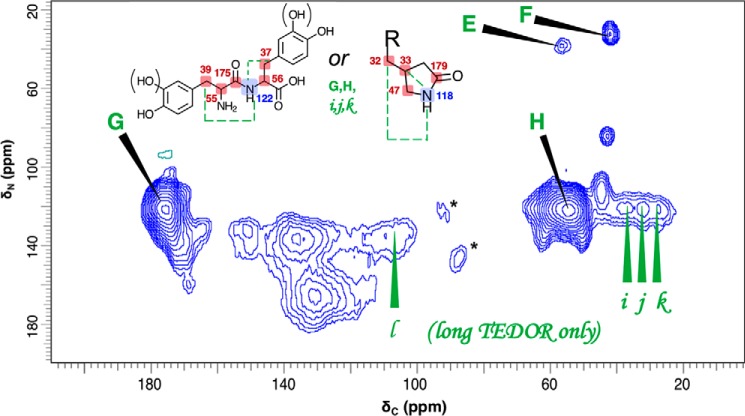
Figure 4.
Contour plots of 2D 13C–15N ZF-TEDOR short-recoupling results on samples from 1.5- versus 3.0-day cell-free melanization, respectively, with a l-[U-13C,15N]tyrosine precursor. The 1.5-day melanins were examined in a 10-h experiment (dark blue contours) run on a Bruker AV3 DNP spectrometer operating at a 1H frequency of 600 MHz, temperature of 100 K, magic-angle spinning frequency of 12.5000 ± 0.0005 kHz, and TEDOR evolution period of 1.6 ms corresponding to one-bond pairwise 13C–15N correlations. The 3.0-day melanins were examined with a 46-h experiment (red contours) run on a Varian (Agilent) DD2 spectrometer operating at a 1H frequency of 600 MHz, temperature of 234 K, magic-angle spinning frequency of 20.00 ± 0.02 kHz, and TEDOR evolution period of 1.4 ms. Spinning sidebands that are incidental to the structural analysis are denoted by asterisk symbols. The 13C–15N cross-peaks designated by uppercase letters are identified provisionally with directly bound C–N pairs within the illustrated chemical structures, drawing on the corresponding ACD-predicted chemical shifts and previously proposed melanin structural constituents (11, 13, 16, 41). The predicted chemical shift values, typically quoted with average error limits of 3–10 ppm, are shown for 13C (red) and 15N (blue).
The current findings indicate that closed-ring structures such as those illustrated in Fig. 4 are in place after 3.0 days of cell-free melanization (red contours). For instance, they support our prior heteronuclear double cross-polarization and proton-assisted recoupling (13) evidence for HN-C = moieties (peak A: 135 13C × 137 ppm 15N) of the DHICA and DHI building blocks typically invoked for eumelanin pigments (1, 38) and are consistent with the chemical shifts predicted semi-empirically with ACD software for a molecular architecture containing the elements displayed in Fig. 4. The TEDOR spectra also display a cross-peak (peak B: 130 × 165 ppm) previously proposed to be the HN-C = moiety of a pyrrole carboxylic acid (13, 16) that is thought to arise from the oxidative cleavage of DHI or DHICA units (16, 38). Additional degradative schemes have been proposed for synthetic polydopamine and a cell-free melanin derived from 5,6-dihydroxyindole (39, 40), in which the pyrrole moiety of an indole ring undergoes oxidative fission. These reports led us to propose the open-chain DHICA-derived compound shown in Fig. 4 to account for peak C (170 × 137 ppm) and a portion of the peak A intensity (135 × 137 ppm). Finally, peak D (150 × 131 ppm) can be assigned to an indolequinone (14, 38, 41). Correlations A, B, and C are also replicated for the polymer synthesized in C. neoformans fungal cells from a dopamine substrate (Fig. S4).
Even after just 1.5 days of reaction, the ZF-TEDOR spectrum of Fig. 4 displays peaks A (135 × 137 ppm) and B (130 × 165 ppm) assigned to the HN − C = proximities of the canonical closed-ring indole and pyrrole carboxylic acid melanin-building blocks (dark blue contours). Moreover, a striking set of additional carbon–nitrogen fingerprints is illustrated by peaks E and F, corresponding to two 13C–15N pairs with upfield shifts indicative of open-chain structures and attributable to incomplete indole cyclization of these developing pigments. As shown in Fig. 4, peak E (56 × 38 ppm) can be identified as the H2N − C-COOH of an uncyclized tyrosine or l-DOPA precursor, consistent with a semiempirical spectral prediction (57 × 41 ppm). Similar structures have been proposed as minor products from both 10-day C. neoformans melanin biosynthesis (9) and 3-day cell-free l-DOPA melanization (13). Peak F (42 × 33 ppm) exhibits chemical shifts in reasonable agreement with the predictions for a H2N − CH2- fragment in a dopamine-like structure (43 × 26 ppm) (42), although there is no straightforward rationale for its formation from l-DOPA in the absence of a decarboxylase enzyme. Novel structural proposals for G and H are described in conjunction with our multiple-bond TEDOR results below.
The success of the DNP-enhanced TEDOR experiments in suggesting possible noncanonical structures for 1.5-day developing synthetic melanins then encouraged us to augment our knowledge of the pigment's molecular architecture by identifying structural signatures that span distances corresponding to more than one chemical bond. Fig. 5 shows the 2D 13C–15N NMR through-space connectivities developed during 2.88 ms of coherence transfer for l-[U-13C,15N]tyrosine melanins after chemical reaction for 1.5 days, including spin pairs located within 2–3 bonds of one another; lowercase letters denote the subset of pairwise proximities observed exclusively with this longer coherence transfer period.
Figure 5.
Contour plot of 2D 13C–15N ZF-TEDOR long-recoupling results on a sample from 1.5-day cell-free melanization with a l-[U-13C,15N]tyrosine precursor. The experiment required 25 h on a Bruker AV3 DNP spectrometer operating at a 1H frequency of 600 MHz, temperature of 100 K, magic-angle spinning frequency of 12.5000 ± 0.0005 kHz, and TEDOR evolution period of 2.88 ms corresponding to 1- and 2–3 bond pairwise 13C–15N correlations. Spinning sidebands that are incidental to the structural analysis are denoted by asterisk symbols. The 13C–15N cross-peaks designated by uppercase letters are identified provisionally with directly bound C–N pairs within the illustrated chemical structures; pairwise proximities that were observed exclusively with the longer coherence transfer period are denoted with lowercase letters. These assignments draw on previously proposed melanin structural constituents (11, 13, 16, 41) and the corresponding ACD-predicted chemical shifts predicted with error limits of 3–10 ppm and shown for 13C (red) and 15N (blue).
Peaks G-H and i-k can be accommodated by at least two sets of chemical shift assignments summarized in Fig. 5, going beyond the commonly invoked melanin structural moieties but having precedents in related reports. Both structural hypotheses conform with the observation of a centrally located 15N resonance at ∼120 ppm. First, a dipeptide could possibly be formed between tyrosine and/or l-DOPA precursor units as shown and in analogy with the o-phenolic substrates implicated in enzyme-mediated insect sclerotization (43). This structural hypothesis and the supporting semiempirical predictions are consistent with five distinct 13C–15N proximities: directly bonded pairs (G observed at 176 × 122 ppm, predicted at 175 × 122 ppm; H observed at 55 × 122 ppm, predicted at 56 × 122 ppm) and remotely bonded pairs (i observed at 38 × 122 ppm, predicted at 55 × 122 ppm; j observed at 32 × 122 ppm, predicted at 39 × 122 ppm; k observed at 27 × 122 ppm, predicted at 37 × 122 ppm). Alternatively, a pyrrolidone could be formed by oxidative cleavage of an oxindole as proposed for polydopamine degradation products (16). This latter hypothesis accounts for two directly bonded pairs (G, H) and three remotely bonded pairs (i, j, k). Although neither oxindole intermediates nor pyrrolidone products have been reported previously in eumelanins, the ring cleavage required to produce such structures has been proposed in polydopamines (44). Finally, peak l is attributable to indole carbons 2–3 bonds from 15N in the DHI or DHICA units displayed in Fig. 4.
Taken together, the TEDOR studies provide an informative initial look at melanin pigment development, also providing a proof-of-principle for the exploitation of stable 13C and 15N isotopes in conjunction with multidimensional solid-state magnetic resonance to track the molecular architectures that characterize the maturation process of these intriguing pigments. These spectroscopic comparisons of proximal 13C–15N nuclear pairs reveal a diverse set of structures that can include open-chain (catecholamine), closed-ring (DHI, DHICA, and pyrrolidone), and possible peptide-linked catecholamines or oxindole-derived compounds at the 1.5-day stage of cell-free melanization but which “converge” to a simpler set of predominantly indole- and pyrrole-based polymers after 3.0 days of development. This temporal progression suggests that the open-chain entities should be attributed to incompletely reacted precursors that remain uncyclized rather than to indole breakdown products. Nonetheless, the diversity of possible cyclization and degradation steps that could occur between these developmental stages can be sorted out only upon systematic monitoring of the reaction times, oxidants, and isolation protocols that define the synthetic pathways. Such future studies also offer the promise of extending the methodology to the more complex fungal melanin systems described above.
Conclusions
The availability of particular catecholamine precursors was demonstrated to play an important role in eumelanin development in both C. neoformans and cell-free reactions, impacting the efficacy of pigment formation, the pigment molecular structure, and the underlying scaffold organization in fungal systems. Using solid-state NMR methodologies, we have been able to monitor the carbon–nitrogen architecture of this ubiquitous protective pigment at the atomic level and as a function of time during melanization.
Experimental procedures
Cell-free chemical synthesis of melanin pigments
Chemical reactions were carried out with l-DOPA (l-3,4-dihydroxyphenylalanine), methyl-l-DOPA, (−)-norepinephrine (NorE), and (−)-epinephrine precursors in separate experiments. These materials were purchased from Sigma. l- [U-13C,15N]Tyrosine was purchased from Cambridge Isotope Labs (Andover, MA); [U-13C,15N]dopamine was obtained from Medical Isotopes, Inc. (Pelham, NH).
Melanin pigments were produced under cell-free conditions by autopolymerization of the catecholamine precursors or tyrosinase-mediated polymerization of the tyrosine precursor as described previously (10, 11, 13, 45–47). Typically, 100 ml of a 5 mm precursor solution prepared in a pH 7.4 Tris-HCl buffer was stirred mechanically for 72 h in a beaker that was covered with punctured aluminum foil to allow aeration and kept at room temperature (∼295 K). The reaction setup was scaled up by a factor of two for the epinephrine precursor to obtain a sufficient yield of pigment for the subsequent physical studies. The reaction mixture was then brought to ∼pH 1 by addition of 6 m HCl and boiled for 20 min. After 15 min of centrifugation at 9000 rpm and 298 K, removal of the supernatant, and repeated washing with distilled water to reach a pH of ∼7, a solid precipitate was collected. After lyophilization, the sample container was sealed and stored at 277 K for future use.
C. neoformans melanin biosynthesis and isolation
Materials were purchased from Sigma unless otherwise noted. As described previously (8, 9, 13), fungal melanin biosynthesis was conducted using the serotype A H99 or serotype D 24067 strains of C. neoformans species complex (American Type Culture Collection 208821) with an obligatory precursor listed above, with dopamine, or with an indole compound such as 5-hydroxy-l-tryptophan, serotonin, 4-hydroxyindole, 5-hydroxyindole, or 5-hydroxyindole-3-acetic acid. A 1 mm solution of the precursor in defined minimal medium (29.4 mm KH2PO4, 15 mm d-glucose, 13 mm glycine, 10 mm MgSO4, and 3 μm thiamine at 303 K) was used to support 10 days of cellular growth in a rotatory shaker operating at 150 rpm. Either natural abundance or U-13C6-enriched glucose (Cambridge Isotope Labs) were used in the growth medium in separate experiments.
To isolate the melanized cell walls for biophysical study (8, 9, 13, 48, 49), a series of chemical treatments was used to remove the other cellular components. Materials were purchased from Sigma unless otherwise noted. First, cell pellets obtained by centrifugation at 2000 rpm were washed with ∼20 ml of phosphate-buffered saline (PBS) and suspended in 1.0 m sorbitol, 0.1 m sodium citrate, pH 5.5, solution, then incubated at 303 K for 24 h with 10 mg/ml of lysing enzymes from Trichoderma harzianum to remove the cell walls and generate melanized protoplasts. The pellet from centrifugation at 2000 rpm was washed repeatedly with PBS until the supernatant was nearly clear. The proteinaceous constituents were denatured by adding a 20-ml aliquot of 4.0 m guanidine thiocyanate to form a suspension that was incubated at room temperature for 12 h in a rocker (Shaker 35, Labnet, Woodbridge, NJ). The proteins were removed by collecting the resulting cell debris and washing 2–3 times with PBS, then incubating at 338 K for 4 h in 5 ml of buffer (10 mm Tris-HCl, pH 8.0, 5 mm CaCl2, 5% SDS) supplemented with 1 mg/ml of proteinase K (Roche Applied Science, Germany). The recovered cell debris was washed 2–3 times with PBS, then delipidated by three successive cycles of extraction using an 8:4:3 chloroform/methanol/saline mixture (50). Finally, the product was suspended in 20 ml of 6 m HCl and boiled for 1 h to hydrolyze any cellular contaminants associated with the melanin pigment. The suspension of black particles was dialyzed for 14 days against distilled water that was changed daily. The resulting melanized cell-wall particles (ghosts) were lyophilized and stored at room temperature for further spectroscopic examination.
Solid-state NMR
NMR measurements were carried out on either of two spectrometers. At the City College of New York, a Varian (Agilent) VNMRS (DD1 or DD2 console) instrument operating at a 1H frequency of 600 MHz was equipped with a 1.6-mm HXY fastMAS probe containing 2–6 mg of each powdered sample and was typically spun at 15.00 ± 0.02 or 20.00 ± 0.02 kHz (Agilent Technologies, Santa Clara, CA). Experiments were run either at a nominal spectrometer set temperature of 298 K or regulated at 234 K using an FTS chiller (FTS Thermal Products, SP Scientific, Warminster, PA). Typical 90° pulse lengths for 1H, 13C, and 15N were 1.2, 1.3, and 2.65 μs, respectively. At the New York Structural Biology Center, a Bruker AV3 DNP spectrometer (Bruker BioSpin Corp., Billerica, MA), operating at a 1H NMR frequency of 600 MHz and a gyrotron frequency of 395 GHz, was equipped with a 3.2-mm E-free low-temperature MAS HCN probe. The customary sample preparation protocols for proteins were optimized for the very hydrophobic and amorphous melanin materials by decreasing the proportion of sample (∼3 mg in a 3.2-mm rotor) with respect to AmuPOL biradical solution (Cortecnet, Voisins-le-Bretonneux, France) and increasing the concentration of biradical (to 40 mm), to achieve better swelling and penetration of electrons within the sample of interest. The powdered sample was infused for 1 h with a 60:35:5 (v/v/v) mixture of 13C-depleted d6-glycerol/D2O/H2O containing the AmuPOL biradical. DNP experiments were run at a temperature of 100 K, making it possible to achieve enhancements in the signal-to-noise ratio (microwaves on/microwaves off) of ϵ = 120 for a [U-13C,15N]proline standard sample and 11 for l-[U-13C,15N]tyrosine synthetic melanin. The MAS rate was 12.500 ± 0.005 kHz; typical 90° pulse lengths were 2.5, 4.0, and 6.3 μs for 1H, 13C, and 15N, respectively.
For 1D CPMAS NMR using the 1.6-mm Varian probe, typical 1–2 ms cross-polarization times were used to transfer magnetization from 1H to 13C nuclear spin baths, including ∼20–50% linearly ramped radiofrequency (rf) field strengths (51) for 1H and a ∼50 kHz constant rf field for 13C. High-power heteronuclear proton decoupling (175–185 kHz) was achieved using the small phase incremental alternation (SPINAL) pulse sequence (52), and data were acquired with a 3-s recycle delay between successive acquisitions. 1D 13C-DPMAS spectra were acquired with spin echo experiments (53), 50-s recycle delays, and 106 kHz SPINAL proton decoupling. Spectral datasets were processed with 50–200 Hz of line broadening; chemical shifts were referenced externally to the methylene (-CH2-) group of adamantane (Sigma) at δC = 38.48 ppm (54) or calculated from 15N and 13C gyromagnetic ratios by IUPAC-specified procedures (55). Relative signal intensities for CHO, C = C, and (CH2)n chemical groups were determined using the GNU image manipulation program to compare pairs of the 13C NMR regions at 66–88, 127–134, and 10–46 ppm for each DPMAS spectrum. Semi-empirical 13C and 15N spectral simulations were conducted with ACD/NMR Processor Academic Edition (version 2017.1; Advanced Chemistry Development, Inc., Toronto, ON, Canada), which draws on a database of ∼250,000 chemical compounds.
Two-dimensional (2D) 13C–13C through-space correlation spectra were collected on 13C-enriched melanin samples using radiofrequency field-assisted diffusion mixing implemented in a DARR mixing experiment (56, 57) with a Varian HXY fastMAS probe as described previously (11). Briefly, separate experiments were conducted with mixing times of 50–500 ms, typical MAS rates of 15 kHz, 175–185 kHz 1H decoupling strengths during acquisition, and 64–256 scans. Proton irradiation of 15 kHz was applied during the DARR mixing period. 1H–13C cross-polarization used a carbon radiofrequency field corresponding to 50 kHz and a proton field that was ramped up to 90 kHz during mixing times between 1 and 3 ms in separate experiments. Typical spectral widths of 38–74 kHz in each 13C dimension, defined by 1024–2048 points (direct dimension), and 64 points (indirect dimension) were used. Phase-sensitive detection of the 2D spectra was accomplished with the time proportional phase incrementation method (23).
15N–13C spin correlations were obtained using ZF-TEDOR experiments (34, 35) conducted with typical 13C spectral widths between 75 and 100 kHz (500–667 ppm) and a 15N spectral width such that the dwell time corresponded to a multiple of the rotor period. For TEDOR experiments using the Varian DD2 system, the 15N pulse width was 2.8 μs (90°) and the 13C pulse width was 2.25 μs (90°). SPINAL decoupling (52) of 147 kHz was used during the 15N–13C mixing, evolution, and acquisition periods. An initial 1H–13C 1-ms 10% linearly ramped CP step with a 106 kHz midpoint was followed by 13C–15N dipolar recoupling using coherence transfer periods of 1.00 or 1.40 ms, corresponding to one-bond correlations between the nuclear pairs, respectively, estimated by measurements on a l-[U-13C5,15N2]glutamine standard sample. Spin coherences generated by 13C–13C J-evolution were removed by ∼147 kHz 1H rf fields during 50-μs Z-filter times. For TEDOR experiments conducted on the Bruker AV3 DNP system, we used a 15N pulse width of 6.5 (90°), 13C pulse width of 4.0 μs (90°), and SPINAL decoupling of 100 kHz during the 15N–13C mixing, evolution, and acquisition times. An initial 1H–13C 1.5-ms 20% tangentially ramped CP step with a 74 kHz midpoint was followed by 13C–15N dipolar recoupling using coherence transfer periods of 1.6 or 2.88 ms, corresponding to 1- and 2–3-bond correlations between the nuclear pairs. Spin coherences generated by 13C–13C J-evolution were removed by ∼12.5 kHz 1H rf fields during 80 μs Z-filter times.
For 2D ZF 13C–15N TEDOR experiments performed at 234 K on the 3-day cell-free melanized l-[U-13C,15N]tyrosine pigment with the Varian spectrometer, we typically used 42,000 scans (1,024 points in the direct dimension), 5 points spanning a 5-kHz sweep width (indirect dimension), and a 1-s recycle delay, thus requiring 46 h to acquire each dataset. For TEDOR of the 1.5-day cell-free melanized l-[U-13C,15N]tyrosine pigment carried out at 100 K on the Bruker spectrometer, we typically used 96–192 scans (direct dimension, 1572 points), 48 points (indirect dimension), and a 4-s recycle delay, thus requiring 5–10 h to acquire each dataset. For TEDOR of the 10-day fungal melanins derived from a [U-13C,15N]dopamine precursor, we used 512 scans (direct dimension) and 32 points (indirect dimension) on the Bruker spectrometer, with a total experiment time of 18 h. In each case, acquisition times in the indirect dimension were chosen to limit the total experiment duration; additional spectral resolution could not be obtained by acquiring more data points because the signal decay was already complete and the sweep width exceeded the relevant spectral window. An exponential apodization function with 100–400 Hz line broadening was used for both direct- and indirect-detected dimensions. The integrity of the 3-day cell-free melanized l-[U-13C,15N]tyrosine pigment sample was confirmed after a 4-day 2D NMR experiment by obtaining a 1D 13C spectrum.
EPR
Suspensions or solid samples of C. neoformans melanin ghosts (each 2 g/ml to permit quantitative comparisons) were placed in 4-mm precision-bore quartz EPR tubes and immediately frozen by immersion in liquid nitrogen. EPR spectra were recorded at 77 K using an X-band Bruker E500 ElexSys EPR spectrometer (Billerica, MA) equipped with an ER4122SHQE resonator cavity in which samples were held in a quartz immersion finger Dewar filled with liquid nitrogen. Data acquisition and manipulation were conducted using Bruker XeprView and WinEPR software. Typical experimental parameters were as follows: microwave frequency, 9.49 GHz; microwave power, 0.1 milliwatt; modulation amplitude, 1 G; modulation frequency, 100 kHz; scan rate, 1.2 G/s; conversion time, 163 ms; time constant, 1310 ms; number of averaged scans, 3.
Author contributions
S. C., A. C., and R. E. S. conceptualization; S. C., R. P.-R., S. T., V. C. P., C. C., B. I., and A. K. investigation; S. C., V. C. P., B. I., H. W., and R. E. S. methodology; S. C., R. P.-R., S. T., V. C. P., C. C., B. I., H. W., A. K., R. S. M., A. C., and R. E. S. writing-review and editing; C. C. and R. E. S. data curation; C. C. formal analysis; B. I., R. S. M., A. C., and R. E. S. resources; R. S. M., A. C., and R. E. S. supervision; A. C. and R. E. S. funding acquisition; A. C. and R. E. S. project administration; R. E. S. writing-original draft.
Supplementary Material
Acknowledgments
The 600 MHz NMR facilities used in this work are operated by The City College and the CUNY Institute for Macromolecular Assemblies, with additional infrastructural support provided by National Institutes of Health Grant 3G12MD007603-30S2 from the National Institute on Minority Health and Health Disparities of the National Institutes of Health. R. E. S. is a member of the New York Structural Biology Center (NYSBC); data collection at that facility was made possible by a grant from the New York State Office of Science, Technology and Academic Research, an National Institutes of Health Office of Research Infrastructure Program Facility Improvement Grant CO6RR015495, and an National Institutes of Health Equipment Grant S10RR029249.
This work was supported by National Institutes of Health Grant R01-AI052733. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4.
V. C. Phan, personal communication.
- DHI
- 5,6-dihydroxyindole
- DHICA
- 5,6-dihydroxyindole-2-carboxylic acid
- DNP
- dynamic nuclear polarization
- CPMAS
- 13C cross-polarization-magic-angle spinning
- DPMAS
- direct polarization-magic-angle spinning
- ZF-TEDOR
- Z-filtered transferred echo double resonance
- DNP
- dynamic nuclear polarization
- NorE
- (−)-norepinephrine
- SPINAL
- small phase incremental alternation
- DARR
- dipolar-assisted rotational resonance
- DOPA
- dihydroxyphenylalanine.
References
- 1. d'Ischia M., Napolitano A., Pezzella A., Meredith P., and Sarna T. (2009) Chemical and structural diversity in eumelanins: unexplored bio-optoelectronic materials. Angew. Chem. Int. Ed. 48, 3914–3921 10.1002/anie.200803786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Simon J. D., and Peles D. N. (2010) The red and the black. Accts. Chem. Res. 43, 1452–1460 10.1021/ar100079y [DOI] [PubMed] [Google Scholar]
- 3. Eisenman H. C., and Casadevall A. (2012) Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 93, 931–940 10.1007/s00253-011-3777-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meredith P., Bettinger C. J., Irimia-Vladu M., Mostert A. B., and Schwenn P. E. (2013) Electronic and optoelectronic materials and devices inspired by nature. Rep. Prog. Phys. 76, 034501 10.1088/0034-4885/76/3/034501 [DOI] [PubMed] [Google Scholar]
- 5. Panzella L., Gentile G., D'Errico G., Della Vecchia N. F., Errico M. E., Napolitano A., Carfagna C., and D'Ischia M. (2013) Atypical structural and π-electron features of a melanin polymer that lead to superior free-radical-scavenging properties. Angew. Chem. Int. Ed. 52, 12684–12687 10.1002/anie.201305747 [DOI] [PubMed] [Google Scholar]
- 6. Ju K.-Y., Lee Y., Lee S., Park S. B., and Lee J.-K. (2011) Bioinspired polymerization of dopamine to generate melanin-like nanoparticles having an excellent free-radical-scavenging property. Biomacromolecules 12, 625–632 10.1021/bm101281b [DOI] [PubMed] [Google Scholar]
- 7. Buszman E., Pilawa B., and Witoszy T. (2003) Effect of Zn2+ and Cu2+ on free radical properties of melanin from Cladosporium cladosporioides. Appl. Magn. Reson. 24, 401–407 10.1007/BF03166942 [DOI] [Google Scholar]
- 8. Garcia-Rivera J., Eisenman H. C., Nosanchuk J. D., Aisen P., Zaragoza O., Moadel T., Dadachova E., and Casadevall A. (2005) Comparative analysis of Cryptococcus neoformans acid-resistant particles generated from pigmented cells grown in different laccase substrates. Fung. Gen. Biol. 42, 989–998 10.1016/j.fgb.2005.09.003 [DOI] [PubMed] [Google Scholar]
- 9. Chatterjee S., Prados-Rosales R., Frases S., Itin B., Casadevall A., and Stark R. E. (2012) Using solid-state NMR to monitor the molecular consequences of Cryptococcus neoformans melanization with different catecholamine precursors. Biochemistry 51, 6080–6088 10.1021/bi300325m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhong J., Frases S., Wang H., Casadevall A., and Stark R. E. (2008) Following fungal melanin biosynthesis with solid-state NMR: biopolymer molecular structures and possible connections to cell-wall polysaccharides. Biochemistry 47, 4701–4710 10.1021/bi702093r [DOI] [PubMed] [Google Scholar]
- 11. Chatterjee S., Prados-Rosales R., Itin B., Casadevall A., and Stark R. E. (2015) Solid-state NMR reveals the carbon-based molecular architecture of Cryptococcus neoformans fungal eumelanins in the cell wall. J. Biol. Chem. 290, 13779–13790 10.1074/jbc.M114.618389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nosanchuk J. D., Stark R. E., and Casadevall A. (2015) Fungal melanin: what do we know about structure? Front. Microbiol. 6, 1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chatterjee S., Prados-Rosales R., Tan S., Itin B., Casadevall A., and Stark R. E. (2014) Demonstration of a common indole-based aromatic core in natural and synthetic eumelanins by solid-state NMR. Org. Biomol. Chem. 12, 6730–6736 10.1039/C4OB01066C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Adhyaru B. B., Akhmedov N. G., Katritzky A. R., and Bowers C. R. (2003) Solid-state cross-polarization magic angle spinning 13C and 15N NMR characterization of Sepia melanin, Sepia melanin free acid and Human hair melanin in comparison with several model compounds. Magn. Res. Chem. 41, 466–474 10.1002/mrc.1193 [DOI] [Google Scholar]
- 15. Meng S., and Kaxiras E. (2008) Theoretical models of eumelanin protomolecules and their optical properties. Biophys. J. 94, 2095–2105 10.1529/biophysj.107.121087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Della Vecchia N. F., Avolio R., Alfè M., Errico M. E., Napolitano A., and d'Ischia M. (2013) Building-block diversity in polydopamine underpins a multifunctional eumelanin-type platform tunable through a quinone control point. Adv. Funct. Materials 23, 1331–1340 10.1002/adfm.201202127 [DOI] [Google Scholar]
- 17. Nosanchuk J. D., Rosas A. L., Lee S. C., and Casadevall A. (2000) Melanisation of Cryptococcus neoformans in human brain tissue. Lancet 355, 2049–2050 10.1016/S0140-6736(00)02356-4 [DOI] [PubMed] [Google Scholar]
- 18. Crawford T. B., and Yates C. M. (1970) A method for the estimation of the catecholamines and their metabolites in brain tissue. J. Phamacol. 38, 56–71 10.1111/j.1476-5381.1970.tb10336.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Polacheck I., Platt Y., and Aronovitch J. (1990) Catecholamines and virulence of Cryptococcus neoformans. Infect. Immun. 58, 2919–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grossman N. C., and Casadevall A. (2017) Physiological differences in Cryptococcus neoformans strains in vitro versus in vivo and their effects on antifungal susceptibility. Antimicrob. Agents Chemother. 61, e02108–02116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frost R. (1920) The road not taken. in Mountain Interval, Henry Holt and Company, New York [Google Scholar]
- 22. Tian S., Garcia-Rivera J., Yan B., Casadevall A., and Stark R. E. (2003) Unlocking the molecular structure of fungal melanin using 13C biosynthetic labeling and solid-state NMR. Biochemistry 42, 8105–8109 10.1021/bi0341859 [DOI] [PubMed] [Google Scholar]
- 23. States D. J., Haberkorn R. A., and Ruben D. J. (1982) A two-dimensional nuclear Overhauser experiment with pure absorption phase in four quadrants. J. Magn.Reson. 48, 286–292 10.1016/0022-2364(82)90279-7 [DOI] [Google Scholar]
- 24. Banks I. R., Specht C. A., Donlin M. J., Gerik K. J., Levitz S. M., and Lodge J. K. (2005) A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryotic Cell 4, 1902–1912 10.1128/EC.4.11.1902-1912.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Camacho E. C., Chrissian C., Cordero R. J. B., Liporagi-Lopes L., Stark R. E., and Casadevall A. (2017) N-Acetylglucosamine supplementation affects Cryptococcus neoformans cell wall composition and melanin architecture. Microbiology 163, 1540–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eisenman H. C., Nosanchuk J. D., Webber J. B., Emerson R. J., Camesano T. A., and Casadevall A. (2005) Microstructure of cell wall-associated melanin in the human pathogenic fungus. Biochemistry 44, 3683–3693 10.1021/bi047731m [DOI] [PubMed] [Google Scholar]
- 27. Henson J. M., Butler M. J., and Day A. W. (1999) The dark side of the mycelum: melanins of phytopathogenic fungi. Annu. Rev. Phytopathol. 37, 447–471 10.1146/annurev.phyto.37.1.447 [DOI] [PubMed] [Google Scholar]
- 28. Free S. J. (2013) Fungal cell wall organization and biosynthesis. Adv. Genet. 81, 33–82 [DOI] [PubMed] [Google Scholar]
- 29. Kang X., Kirui A., Musznski A., Widanage M. C. D., Chen A., Azadi P., Wang P., Mentink-Vigier F., and Wang T. (2018) Molecular architecture of fungal cell walls revealed by solid-state NMR. Nat. Commun. 9, 2747–2766 10.1038/s41467-018-05199-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eisenman H. C., Frases S., Nicola A. M., Rodrigues M. L., and Casadevall A. (2009) Vesicle-associated melanization in Cryptococcus neoformans. Microbiology 155, 3860–3867 10.1099/mic.0.032854-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fukamizo T., Kramer K. J., Mueller D. D., Schaefer J., Garbow J., and Jacob G. S. (1986) Analysis of chitin structure by nuclear magnetic resonance spectroscopy and chitinolytic enzyme digestion. Arch. Biochem. Biophys. 249, 15–26 10.1016/0003-9861(86)90555-2 [DOI] [PubMed] [Google Scholar]
- 32. Li K. L., Tihal C. A., Guo M., and Stark R. E. (1993) Multinuclear and magic-angle spinning NMR investigations of molecular organization in phospholipid-triglyceride aqueous dispersions. Biochemistry 32, 9926–9935 10.1021/bi00089a008 [DOI] [PubMed] [Google Scholar]
- 33. Gow N. A. R, Latge J.-P., and Munro C. A. (2017) The fungal cell wall: structure, biosynthesis, and function. Microbiol Spectr. 5, 10.1128/microbiolspec.FUNK-0035-2016 10.1128/microbiolspec.FUNK-0035-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hing A. W., Vega S., and Schaefer J. (1992) Transferred-echo double-resonance NMR. J. Magn. Reson. 96, 205–209 10.1016/0022-2364(92)90305-Q [DOI] [Google Scholar]
- 35. Jaroniec C. P., Filip C., and Griffin R. G. (2002) 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon-nitrogen distances in uniformily 13C,15N-labeled solids. J. Am. Chem. Soc. 124, 10728–10742 [DOI] [PubMed] [Google Scholar]
- 36. Eisenman H. C., Mues M., Weber S. E., Frases S., Chaskes S., Gerfen G., and Casadevall A. (2007) Cryptococcus neoformans laccase catalyses melanin synthesis from both d- and l-DOPA. Microbiology 153, 3954–3962 10.1099/mic.0.2007/011049-0 [DOI] [PubMed] [Google Scholar]
- 37. Watt A. A. R., Bothma J. P., and Meredith P. (2009) The supramolecular structure of melanin. Soft Matter 5, 3754–3760 10.1039/b902507c [DOI] [Google Scholar]
- 38. Ito S. (2003) A chemist's view of melanogenesis. Pigm. Cell Res. 16, 230–236 10.1034/j.1600-0749.2003.00037.x [DOI] [PubMed] [Google Scholar]
- 39. Crescenzi O., Kroesche C., Hoffbauer W., Jansen M., Napolitano A., Prota G., and Peter M. G. (1994) Synthesis of dopamines labelled with 13C in the α or β-side chain position and their application to structural studies on melanins by solid-state NMR spectroscopy. Liebigs Ann. Chem. 6, 563–567 10.1002/jlac.199419940606 [DOI] [Google Scholar]
- 40. Napolitano A. P., A; Prota G., Seraglia R., and Traldi P. (1996) Structural analysis of synthetic melanins from 5,6-dihydroxyindole by matrix-assisted laser desorption/ionization mass spectrometry. Rapid. Commun. Mass Spectrom. 10, 468–472 10.1002/(SICI)1097-0231(19960315)10:4%3C468::AID-RCM506%3E3.0.CO%3B2-6 [DOI] [Google Scholar]
- 41. Prota G. (1992) Melanins and melanogenesis, pp. 88–118, Academic Press, San Diego, CA [Google Scholar]
- 42. Proks V., Brus J., Pop-Georgievski O., Večerníková E., Wisniewski W., Kotek J., Urbanová M., and Rypáček F. (2013) Thermal-induced transformation of polydopamine structures: an efficient route for the stabilization of the polydopamine surfaces. Macromol. Chem. Phys. 214, 499–507 10.1002/macp.201200505 [DOI] [Google Scholar]
- 43. Kramer K. J., Hopkins T. L., and Schaefer J. (1995) Mini-review: applications of solids NMR to the analysis of insect sclerotized structures. Insect Biochem. Mol. Biol. 25, 1067–1080 10.1016/0965-1748(95)00053-4 [DOI] [Google Scholar]
- 44. Alfieri M. L., Micillo R., Panzella L., Crescenzi O., Oscurato S. L., Maddalena P., Napolitano A., Ball V., and d'Ischia M. (2018) Structural basis of polydopamine film formation: probing 5,6-dihydroxyindole-based eumelanin type units and the porphyrin issue. ACS Appl. Mater. Interfaces 10, 7670–7680 10.1021/acsami.7b09662 [DOI] [PubMed] [Google Scholar]
- 45. Aime S., Fasano M., Bergamasco B., Lopiano L., and Quattrocolo G. (1996) Nuclear magnetic resonance spectroscopy characterization and iron content determination of human mesencephalic neuromelanin. Adv. Neurol. 69, 263–270 [PubMed] [Google Scholar]
- 46. Jastrzebska M., Kocot A., and Tajber L. (2002) Photoconductivity of synthetic dopa-melanin polymer. J. Photochem. Photobiol. B 66, 201–206 10.1016/S1011-1344(02)00268-3 [DOI] [PubMed] [Google Scholar]
- 47. Ghiani S., Baroni S., Burgio D., Digilio G., Fukuhara M., Martino P., Monda K., Nervi C., Kiyomine A., and Aime S. (2008) Characterization of human hair melanin and its degradation products by means of magnetic resonance techniques. Magn. Reson. Chem. 46, 471–479 10.1002/mrc.2202 [DOI] [PubMed] [Google Scholar]
- 48. Rosas A. L., Nosanchuk J. D., Feldmesser M., Cox G. M., McDade H. C., and Casadevall A. (2000) Synthesis of polymerized melanin by Cryptococcus neoformans in infected rodents. Infect. Immun. 68, 2845–2853 10.1128/IAI.68.5.2845-2853.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Y., Aisen P., and Casadevall A. (1996) Melanin, melanin “ghosts,” and melanin composition in Cryptococcus neoformans. Infect. Immun. 64, 2420–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Folch J., Lees M., and Sloan Stanley G. H. (1957) A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 51. Metz G., Wu X., and Smith S. O. (1994) Ramped-amplitude cross polarization in magic-angle-Spinning NMR. J. Magn. Reson. A 110, 219–227 10.1006/jmra.1994.1208 [DOI] [Google Scholar]
- 52. Fung B. M., Khitrin A. K., and Ermolaev K. (2000) An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Res. 142, 97–101 10.1006/jmre.1999.1896 [DOI] [PubMed] [Google Scholar]
- 53. Hahn E. (1950) Spin Echoes. Phys. Rev. 80, 58–59 10.1103/PhysRev.80.58 [DOI] [Google Scholar]
- 54. Morcombe C. R., and Zilm K. W. (2003) Chemical shift referencing in MAS solid state NMR. J. Magn. Res. 162, 479–486 10.1016/S1090-7807(03)00082-X [DOI] [PubMed] [Google Scholar]
- 55. Harris R. K., Becker E. D., Cabral de Menezes S. M., Goodfellow R., and Granger P. (2001) NMR nomenclature: nuclear spin properties and conventions for chemical shifts. Pure Appl. Chem. 73, 1795–1818 10.1051/pac200173111795 [DOI] [PubMed] [Google Scholar]
- 56. Takegoshi K., Nakamura S., and Terao T. (2001) 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 344, 631–637 10.1016/S0009-2614(01)00791-6 [DOI] [Google Scholar]
- 57. Morcombe C. R., Gaponenko V., Byrd R. A., and Zilm K. W. (2004) Diluting abundant spins by isotope edited radiofrequency field assisted diffusion. J. Am. Chem. Soc. 126, 7196–7197 10.1021/ja047919t [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.