Summary
Spore formation is essential for the bacterial pathogen and obligate anaerobe, Clostridioides (Clostridium) difficile, to transmit disease. Completion of this process depends on the mother cell engulfing the developing forespore, but little is known about how engulfment occurs in C. difficile. In Bacillus subtilis, engulfment is mediated by a peptidoglycan degradation complex consisting of SpoIID, SpoIIP, and SpoIIM, which are all individually required for spore formation. Using genetic analyses, we determined the functions of these engulfment-related proteins along with the putative endopeptidase, SpoIIQ, during C. difficile sporulation. While SpoIID, SpoIIP, and SpoIIQ were critical for engulfment, loss of SpoIIM minimally impacted C. difficile spore formation. Interestingly, a small percentage of ΔspoIID and ΔspoIIQ cells generated heat-resistant spores through the actions of SpoIIQ and SpoIID, respectively. Loss of SpoIID and SpoIIQ also led to unique morphological phenotypes: asymmetric engulfment and forespore distortions, respectively. Catalytic mutant complementation analyses revealed that these phenotypes depend on the enzymatic activities of SpoIIP and SpoIID, respectively. Lastly, engulfment mutants mislocalized polymerized coat even though the basement layer coat proteins, SpoIVA and SipL, remained associated with the forespore. Collectively, these findings advance our understanding of several stages during infectious C. difficile spore assembly.
Keywords: Clostridium difficile, engulfment, SpoIID, SpoIIP, SpoIIQ, coat localization
Introduction
The metabolically dormant spore form is the infectious particle of numerous Gram-positive bacterial pathogens such as Bacillus anthracis, Clostridium tetani, Clostridium perfringens, and Clostridioides (Clostridium) difficile (Swick et al., 2016). While different mechanisms control the induction of sporulation between these organisms (Al-Hinai et al., 2015), this developmental process appears to be ancient and highly conserved morphologically in monospore-forming species of the Bacilli and Clostridia families (Hutchison et al., 2014, Abecasis et al., 2013, Galperin et al., 2012). The first morphological event of endospore formation is asymmetric division, which creates a larger mother cell and smaller forespore (Tan & Ramamurthi, 2014). The mother cell encases the forespore using a phagocytic-like process known as engulfment, leaving the forespore suspended in the mother cell cytosol surrounded by two membranes. The outermost membrane comes from the mother cell, while the innermost membrane comes from the forespore. A thick layer of modified peptidoglycan known as the cortex is synthesized between the two membranes, while a series of proteinaceous shells known as the coat are deposited on the outermost forespore membrane (Driks & Eichenberger, 2016). The cortex maintains metabolic dormancy in spores (Setlow, 2014), while the coat protects against enzymatic and oxidative insults (Driks & Eichenberger, 2016).
Although the general morphological steps of sporulation appear to be conserved (Abecasis et al., 2013, Galperin et al., 2012), the biophysical mechanisms that control the assembly of spores remain unknown for most spore-forming organisms outside of the model organism Bacillus subtilis (Tan & Ramamurthi, 2014) where this process has been extensively studied. Notably, many of the proteins critical for sporulation in B. subtilis are conserved in the genomes of all spore-forming organisms (Abecasis et al., 2013, Galperin et al., 2012). However, recent studies in the healthcare-associated pathogen C. difficile indicate that gene conservation does not always predict functional conservation (Fimlaid et al., 2013, Fimlaid et al., 2015b, Pereira et al., 2013, Ribis et al., 2017, Serrano et al., 2016). Determining the mechanisms that control spore assembly in C. difficile could inform the development of therapies that can prevent infectious spore formation, which is essential for this obligate anaerobe to transmit disease (Deakin et al., 2012, Dembek et al., 2017) and cause high rates of recurrent infection (Lessa et al., 2015).
In this study, we assessed the requirement for proteins known to be essential for B. subtilis engulfment, namely SpoIID, SpoIIP, and SpoIIM (Lopez-Diaz et al., 1986, Smith et al., 1993, Frandsen & Stragier, 1995), during spore formation in C. difficile. These proteins are strictly conserved across spore-forming organisms (Abecasis et al., 2013, Galperin et al., 2012), suggesting that they play key roles during spore formation. However, the functions of these proteins, which we refer to as IID, IIP, and IIM, respectively, herein, have not been studied in spore-forming organisms beyond B. subtilis.
In B. subtilis, IIP is an autolysin (Aung et al., 2007, Chastanet & Losick, 2007) with both amidase and endopeptidase activity (Morlot et al., 2010), and IID is a lytic transglycosylase (Abanes-De Mello et al., 2002, Morlot et al., 2010) whose activity depends on IIP’s ability to remove stem peptides from peptidoglycan (Morlot et al., 2010) (Fig. 1A). Although B. subtilis IID activity requires IIP, B. subtilis IID also enhances IIP’s autolysin activity (Morlot et al., 2010) presumably through protein-protein interactions.
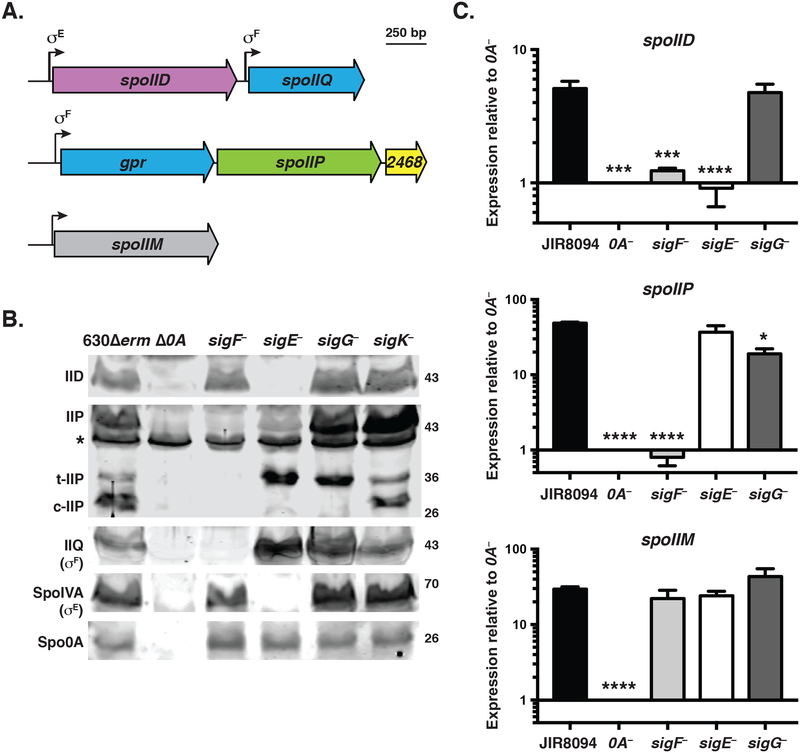
Fig. 1. Transcriptional regulation of spoIID, spoIIP, and spoIIM genes in C. difficile.
(A) Schematic of IID and IIP activities on peptidoglycan. Yellow hexagons (NAG) represent N-acetylglucosamine; blue hexagons (NAM) represent N-acetylmuramic acid, purple circles represent L-alanine, pink circles represent D-glutamic acid, grey circles represent m-2,6-diaminopimelic acid (DAP); green circles represent D-alanine; and the black line indicates cross-linking between DAP and D-ala residues. IIP has both endopeptidase and amidase activity (scissors), both of which are shown in the schematic. IID has lytic transglycoslase activity (pink triangles). (B) Schematic of spoIID (CD0126), spoIIP (CD2469), and spoIIM (CD1221) transcriptional units identified by RNA-Seq analyses (Fig. S1, (Fimlaid et al., 2013, Pishdadian et al., 2015)). Promoters are represented as bent arrows, and the sigma factor binding sites identified by Saujet et al. are shown above (Saujet et al., 2013). (C) qRT-PCR analyses of spoIID, spoIIP, and spoIIM transcription in JIR8094, spo0A::ermB (0A–), and sporulation sigma factor mutants. Transcript levels were normalized to the housekeeping gene, rpoB, using the standard curve method. The normalized transcript levels for each strain was compared to the normalized transcript level of spo0A–. Data represents the average of three independent biological replicates. Data were analyzed using a one-way ANOVA and Tukey’s test. (*, p < 0.05, ***, p < 0.005, ****, p < 0.0001). (D) Western blot analyses of IID and IIP in 630Δerm, Δ0A, and associated sporulation sigma factor mutants. While a clean deletion was made in spo0A, the sporulation sigma factor genes were disrupted using a Targetron insertion (Heap et al., 2007). The western blots shown are representative of the results of three independent biological replicates. Three separate isoforms were detected for IIP in wild type: full-length (IIP), truncated IIP (t-IIP), and cleaved IIP (c-IIP). The predicted MW of IIP is 38 kDa, and the MW of IIP lacking its signal peptide is 35 kDa. The asterisk denotes a non-specific protein recognized by the polyclonal anti-IIP antibodies. A faint cross-reacting band is also visible with the IID antibody. As controls for these analyses, western blotting was performed using anti-IIQ and anti-IVA antibodies, whose production depends on σF and σE, respectively. Spo0A (0A) was used as a proxy for measuring sporulation induction (Dembek et al., 2017, Putnam et al., 2013).
Similar to B. subtilis, C. difficile IID has lytic transglycosylase activity and preferentially degrades peptidoglycan strands whose peptide chains have been enzymatically removed in vitro (Nocadello et al., 2016). C. difficile IID has higher intrinsic activity on peptidoglycan whose peptide chains are still present in vitro than B. anthracis IID in vitro, suggesting that C. difficile IID may be less strict in its substrate requirement for denuded peptidoglycan strands relative to B. subtilis IID.
In B. subtilis, IID and IIP are transmembrane proteins (Frandsen & Stragier, 1995, Lopez-Diaz et al., 1986) that complex with the transmembrane scaffold protein IIM, which helps IID and IIP localize to the forespore membrane and mediate engulfment (Aung et al., 2007, Chastanet & Losick, 2007). This IID-IIM-IIP (DMP) complex is produced by the mother cell (Frandsen & Stragier, 1995, Rong et al., 1986, Smith & Youngman, 1993, Driks & Losick, 1991), and localize to the leading edge of the engulfing membrane along with the forespore peptidoglycan synthesis machinery; collectively these enzyme complexes generate the force necessary to drive engulfment to completion (Abanes-De Mello et al., 2002, Meyer et al., 2010, Ojkic et al., 2016). Loss of any one of the components of the DMP complex leads to severe engulfment defects (Abanes-De Mello et al., 2002, Frandsen & Stragier, 1995, Smith et al., 1993), including bulging of the forespore septum (Abanes-De Mello et al., 2002, Eichenberger et al., 2001, Frandsen & Stragier, 1995, Pogliano et al., 1999, Smith et al., 1993) where the peptidoglycan has been thinned (Rodrigues et al., 2013). Furthermore, loss of the entire DMP complex prevents engulfment from initiating such that a ΔIIDMP strain produces flat septa and disporic cells (Eichenberger et al., 2001).
B. subtilis engulfment is also facilitated by members of a conserved A-Q complex that connects the forespore and mother cell (Camp & Losick, 2009, Meisner et al., 2008, Morlot & Rodrigues, 2018) thought to form a channel also known as the “feeding tube” (Camp & Losick, 2009). The B. subtilis SpoIIQ (IIQ) and SpoIIIAH (IIIAH) constituents of this complex prevent back-sliding of the engulfing membrane and are sufficient to mediate engulfment using a ratchet mechanism if the peptidoglycan is enzymatically removed (Broder & Pogliano, 2006). In certain sporulation induction conditions, loss of IIQ reduces engulfment efficiency by ~10-fold (Londono-Vallejo et al., 1997, Sun et al., 2000), while in other conditions, it delays engulfment (Broder & Pogliano, 2006, Sun et al., 2000).
In contrast with B. subtilis (Doan et al., 2009), C. difficile IIQ and IIIA complex components are required for engulfment completion (Fimlaid et al., 2015b, Serrano et al., 2016, Morlot & Rodrigues, 2018). Although B. subtilis and C. difficile IIQ both carry LytM (peptidase M23 family) zinc-binding cell wall endopeptidase domains, B. subtilis IIQ harbors a degenerate active site that cannot bind the zinc ion needed to form the catalytic core, whereas C. difficile IIQ has an intact site (Crawshaw et al., 2014) and binds Zn2+ (Serrano et al., 2016). These observations suggest that C. difficile IIQ could directly participate in peptidoglycan remodeling during engulfment.
Interestingly, the engulfment defects of C. difficile spoIIQ and spoIIIA engulfment mutants correlate with polymerized coat localization defects (Fimlaid et al., 2015b), prompting us to question whether similar coat abnormalities would be observed in all C. difficile engulfment mutants. We further wondered whether a C. difficile strain that fails to initiate engulfment would exacerbate the coat localization defects based on the observation that the B. subtilis coat morphogenetic protein, SpoVM, fails to localize specifically to the forespore in a ΔIIDPM mutant because it produces flat septa (Eichenberger et al., 2001) that lack positive curvature (Ramamurthi et al., 2009).
To address these questions, we used allelic exchange (Ng et al., 2013) to construct C. difficile mutants lacking the putative engulfment regulators, IID (CD0126), IIP (CD2469), and IIM (CD1221), and the known engulfment regulator and putative endopeptidase, IIQ (CD0125), singly and in combination. We then used these mutants to analyze the relationship between engulfment and coat protein localization during C. difficile spore formation.
Results
IID, IIP, and IIM are differentially regulated in C. difficile relative to B. subtilis
Before initiating these studies, we compared the regulation of C. difficile IID, IIP, and IIM genes relative to B. subtilis. In B. subtilis, IID, IIP, and IIM transcription is controlled by the mother cell-specific sporulation sigma factor E (σE, (Frandsen & Stragier, 1995, Rong et al., 1986, Smith & Youngman, 1993, Driks & Losick, 1991)). This regulation ensures that all three components of the peptidoglycan degradation machinery localize to the same mother cell-derived outer forespore membrane. B. subtilis IIP is also transcribed in the forespore due to read-through transcription from the upstream gpr gene, which is controlled by the forespore-specific sporulation sigma factor F (σF, (Dworkin & Losick, 2005, Frandsen & Stragier, 1995)). However, this read-through transcription is not necessary for IIP function in B. subtilis (Abanes-De Mello et al., 2002, Dworkin & Losick, 2005).
In C. difficile, spoIIP is also encoded downstream of σF-regulated gpr ((Fimlaid et al., 2013, Saujet et al., 2013), Fig. 1B). Notably, the intergenic distance between spoIIP and gpr in C. difficile and B. subtilis is 17 bp and 62 bp, respectively, suggesting that C. difficile spoIIP may be more dependent on σF for expression than in B. subtilis. To test this hypothesis, we analyzed previously published RNA-Seq data of sporulation sigma factor mutants during sporulation (Fig. S1A, (Fimlaid et al., 2013, Pishdadian et al., 2015)). These analyses confirmed that spoIIP expression requires σF, but not σE, in contrast with B. subtilis (Frandsen & Stragier, 1995) and are consistent with previously published C. difficile microarray data (Saujet et al., 2013). To confirm these observations, we performed qRT-PCR on a separate set of biological samples of JIR8094 sporulation sigma factor mutants. Consistent with the RNA-Seq data, spoIIP expression was largely undetectable in the sigF mutant (p < 0.0001, 66-fold difference), while the sigE mutant expressed spoIIP at levels comparable to wild type (Fig. 1C). spoIIP transcript levels were also reduced in the sigG mutant (p < 0.01, 3-fold difference), which may indicate that σG also activates spoIIP expression. Notably, the RNA-Seq analyses detected transcriptional reads spanning the intergenic region between spoIIP and gpr (Fig. S1A), suggesting that read-through transcription from the upstream gpr promoter can contribute to spoIIP expression as in B. subtilis (Abanes-De Mello et al., 2002, Dworkin & Losick, 2005).
We also analyzed SpoIIP levels in cell lysates prepared from sporulation sigma factor mutants induced to sporulate on 70:30 plates using Western blotting (Fig. 1D). However, for these analyses, we used sigma factor mutants constructed in the 630ΔermΔpyrE background, since this strain facilitates allele-coupled exchange and thus complementation from the chromosome (Ng et al., 2013). All strains from this point on derive from the 630ΔermΔpyrE parental strain. Consistent with the JIR8094 transcriptional data, IIP was detected in the 630Δerm sigE mutant but not the sigF mutant. Interestingly, three IIP isoforms were detected in both wildtype and sigK– cells: full-length (~43 kDa), truncated (t-IIP, ~36 kDa), and cleaved (c-IIP). Cleaved IIP was not observed in sigE– and sigG– cells, which may indicate that this isoform appears after engulfment completion, since sigE and sigG mutants have engulfment defects when grown on solid media (Fimlaid et al., 2013, Fimlaid et al., 2015b). While it was recently reported that a 630Δerm ΔsigG strain grown in broth culture completes engulfment based on FM4–64 staining (Dembek et al., 2017), these cells failed to exclude the Hoechst nucleoid stain, indicating that the engulfing membranes do not complete membrane fission (Doan et al., 2013, Fimlaid et al., 2013, Sharp & Pogliano, 1999, Sun et al., 2000). Regardless, since σF activity has been localized exclusively to the forespore in C. difficile (Pereira et al., 2013), our results strongly suggest that SpoIIP is primarily produced in the forespore rather than the mother cell.
In contrast with the regulation of C. difficile spoIIP, spoIID expression appeared to be controlled by σE based on the RNA-Seq (Fig. S1B, (Fimlaid et al., 2013, Pishdadian et al., 2015)) and microarray analyses (Saujet et al., 2013). Consistent with these findings a σE recognition sequence was detected in spoIID’s promoter region (Saujet et al., 2013), and qRT-PCR confirmed that spoIID expression depends on both σE (Fig 1C, p < 0.0005) and σF (p < 0.001). This regulation is consistent with σE activation being partially dependent on σF activation (Fimlaid et al., 2013, Saujet et al., 2013). Notably, even though spoIID transcript levels were reduced in the JIR8094 sigF mutant, IID protein was detectable in the sigF, but not the sigE, mutant in Western blot analyses in 630Δerm strains (Fig. 1D). Since σE activity has been localized exclusively to the mother cell (Pereira et al., 2013, Pishdadian et al., 2015), these results indicate that C. difficile IID is produced in the mother cell similar to B. subtilis (Driks & Losick, 1991, Rong et al., 1986).
The RNA-Seq analyses also revealed that spoIIM transcription required Spo0A (p < 0.0001), but not σF or σE, ((Fimlaid et al., 2013, Pishdadian et al., 2015), Fig. S1C) in contrast with its regulation by σE in B. subtilis (Smith & Youngman, 1993). Although differential expression of spoIIM was not detected in microarray expression data (Saujet et al., 2013), qRT-PCR analyses validated the regulation of spoIIM by Spo0A and not downstream sporulation sigma factors. Overall, these results suggest that IID, IIP, and IIM are produced in different compartments in C. difficile, with IIM being made in both the mother cell and forespore, IID in the mother cell, and IIP in the forespore. Since IID, IIP, and IIM form a complex in the mother cell in B. subtilis, our findings raise the possibility that IID, IIP, and/or IIM functions are not as tightly coordinated in C. difficile as they are in B. subtilis (Chastanet & Losick, 2007).
IID, IIP, IIQ, and IIM are differentially required for C. difficile spore formation
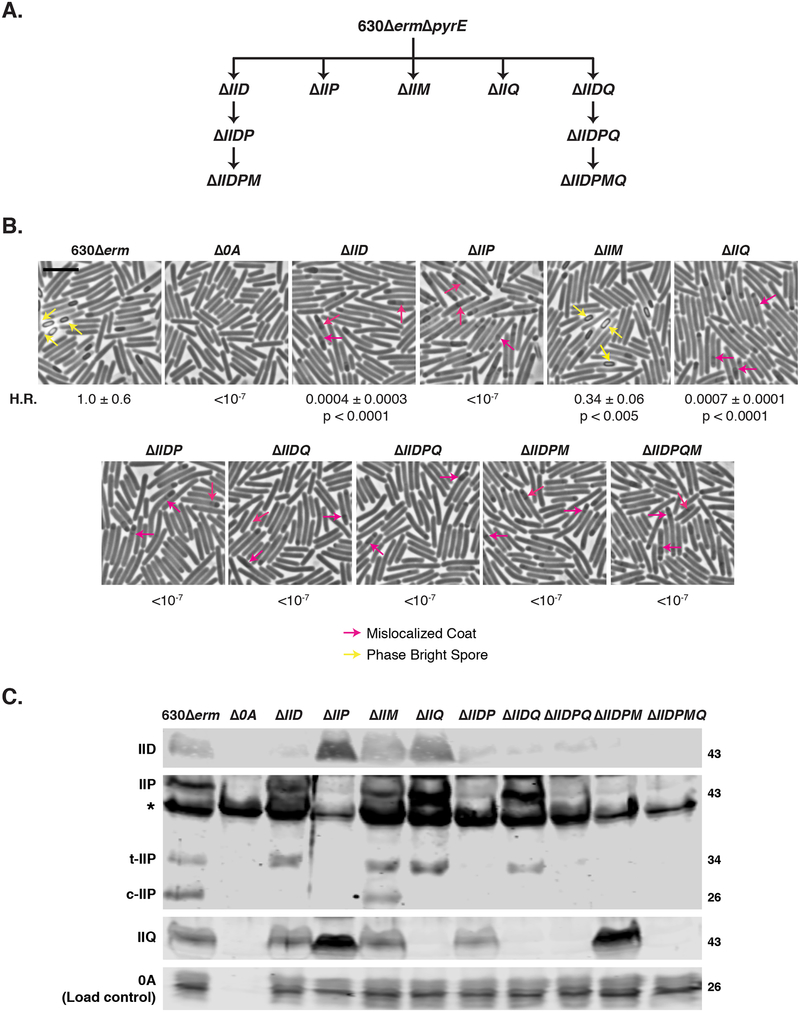
To gain insight into this question, we constructed clean deletions of spoIID, spoIIP, and spoIIM in 630ΔermΔpyrE (Fig. 2A) and restored the pyrE locus as previously described (Ng et al., 2013). We also constructed a clean deletion of spoIIQ to revisit IIQ’s role in regulating C. difficile engulfment, since we had previously constructed a spoIIQ Targetron mutant in the JIR8094 strain background (Fimlaid et al., 2015b). Gene deletions were confirmed by colony PCR using primers flanking the regions used for allelic exchange and an internal primer that binds within the region deleted (Fig. S2). We then assessed the functional requirement for IID, IIP, IIM, and IIQ during spore formation by measuring the heat resistance of sporulating cultures lacking these factors (Shen et al., 2016). Δspo0A was used as a negative control for these studies, since it cannot initiate sporulation (Deakin et al., 2012). ΔIIP failed to produce detectable heat-resistant spores, whereas ΔIID and ΔIIQ sporulating cells produced heat-resistant spores ~2000-fold less efficiently than wild type (Fig. 2B, p < 0.0001). Surprisingly, ΔIIM produced heat-resistant spores slightly less efficiently than wild type (~3-fold decrease, p < 0.005). Since IID, IIP, and IIM are each essential for B. subtilis spore formation (Smith et al., 1993, Londono-Vallejo et al., 1997, Lopez-Diaz et al., 1986), our results highlight differences in the requirement for IID, IIQ, and IIM in C. difficile relative to B. subtilis.
Fig. 2. Differential requirements for engulfment-related proteins during C. difficile spore formation.
(A) Family tree of putative engulfment mutants. 630ΔermΔpyrE was the parental strain for making first-generation single gene deletions as well as the spoIID-spoIIQ double gene deletion mutant. Additional multiple gene deletions were constructed using either ΔIID and ΔIIDQ as the parental strains. (B) Phase-contrast microscopy analyses of wild type 630Δerm, Δspo0A (Δ0A), and mutants lacking putative engulfment regulators after 17 hrs of sporulation. Yellow arrows mark example phase-bright forespores, while pink arrows demarcate regions suspected to be mislocalized coat based on previous studies (Fimlaid et al., 2015b, Ribis et al., 2017). Heat resistance (H.R.) efficiencies were determined from 20–24 sporulating cultures and represent the mean and standard deviation for a given strain relative to wild type based on a minimum of three independent biological replicates. Statistical significance relative to wild type was determined using a one-way ANOVA and Tukey’s test. Scale bars represent 5 μm. The limit of detection of the assay is 10−6. (C) Western blot analyses of IID, IIP, and IIQ in mutants lacking putative engulfment regulators. Three separate isoforms were detected for IIP in wild type: full-length (IIP), truncated IIP (t-IIP), and cleaved IIP (c-IIP). The predicted MW of IIP is 38 kDa, and the MW of IIP lacking its signal peptide is 35 kDa. The asterisk denotes a non-specific protein recognized by the polyclonal anti-IIP antibodies. A faint cross-reacting band is also visible with the IID antibody. Spo0A was used as a proxy for measuring sporulation induction (Dembek et al., 2017, Putnam et al., 2013). The western blots shown are representative of the results of three independent biological replicates.
While our 630Δerm ΔspoIIQ strain generated heat-resistant spores less efficiently than wild type (~0.05%), loss of IIQ in this strain background has been previously reported to prevent heat-resistant spore formation (Dembek et al., 2015, Serrano et al., 2016). This discrepancy is likely due to differences in the sporulation method used (plate-based vs. liquid), since comparison of the heat resistance of sporulating ΔIIQ in liquid vs. plate culture revealed a 20-fold more severe defect during growth in broth with agitation relative to growth on plates (Fig. S3). However, we cannot rule out the contributions of slight differences in media, since Serrano et al. used SMC liquid media, and we use 70%SMC:30% BHIS plate media to induce sporulation. Regardless, analysis of the heritability of the ΔIIQ phenotype on 70:30 plates confirmed that this strain was not contaminated with wildtype spores (data not shown), and a spoIIQ targetron mutant in 630Δerm exhibited similar levels of heat resistance as the ΔIIQ strain (data not shown).
Loss of IID and IIQ, but not IID or IIQ alone, abrogates functional spore formation
Since the heat resistance phenotypes of C. difficile ΔIID and ΔIIQ strains were at least ~3-log less severe than C. difficile ΔIIP (Fig. 2B) and their B. subtilis counterparts (Abanes-De Mello et al., 2002, Smith et al., 1993), we wondered whether loss of both IID and IIQ exacerbate the heat resistance defects of IID and IIQ mutants. To test this possibility, we simultaneously deleted IID and IIQ, which are adjacent in the C. difficile genome (Fig. 2A). The resulting ΔIIDQ strain resembled ΔIIP in that it failed to produce detectable heat-resistant spores (Fig. 2B). This defect could be fully complemented by integrating IIDQ at the pyrE locus (Table 1). Importantly, complementation of ΔIIDQ with either IID or IIQ integrated in the pyrE locus restored heat resistance to levels close to or in excess of the individual ΔIIQ and ΔIID strains, respectively (Fig. 3). Taken together, these results indicate that, in a small subset of ΔIIQ cells (~0.05%), C. difficile IID facilitates sporulation completion; similarly, in a small subset of ΔIID cells (~0.05%), C. difficile IIQ can facilitate sporulation completion.
Table 1.
Sporulation efficiency of spoIIM and spoIIDQ complementation strains and spoIIP targetron mutant.
| Heat resistance efficiency | |
|---|---|
| 630Δerm | 1.0 ± 0.6 |
| Δspo0A | < 10−6 |
| ΔspoIIP | < 10−6 |
| spoIIP– | < 10−6 |
| ΔspoIIM | 0.5 ± 0.2 |
| ΔspoIIM/spoIIM | 0.7 ± 0.5 |
| ΔspoIIDQ | < 10−6 |
| ΔspoIIDQ/spoIIDQ | 0.7 ± 0.4 |
Heat resistance efficiencies represent the mean and standard deviation for a given strain relative to wild type based on a minimum of three biological replicates. Statistical significance relative to wild type was determined using a one-way ANOVA and Tukey’s test. Bold text indicates p < 0.0005.
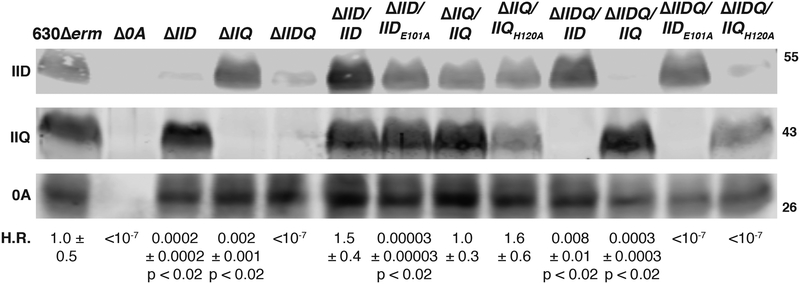
Fig. 3. IID can partially compensate for loss of IIQ and vice versa during C. difficile spore formation.
Heat resistance (H.R.) efficiencies shown are the mean and standard deviation for a given strain relative to wild type 630Δerm based on a minimum of three biological replicates. Statistical significance relative to wild type was determined using a one-way ANOVA and Tukey’s test. * p < 0.05, ** p < 0.01, **** p < 0.0001. The limit of detection of the assay is 10−6. Western blot analyses of ΔIID, ΔIIQ, and ΔIIDQ strains complemented with either IID or IIQ variants. Glu101 is the catalytic residue of IID (Nocadello et al., 2016), while His120 is necessary for Zn2+ binding by IIQ (Serrano et al., 2016) and its predicted endopeptidase activity (Crawshaw et al., 2014). The anti-IID polyclonal antibody detects a weakly cross-reactive band in the absence of IID. Spo0A was used as a proxy for measuring sporulation induction (Dembek et al., 2017, Putnam et al., 2013). The western blots shown are representative of the results from three independent biological replicates.
To test whether the enzymatic activities of C. difficile IID and IIQ are required for these compensatory phenotypes, we complemented ΔIIDQ with constructs encoding either IIDE101A or IIQH120A. Glutamate 101 is the catalytic residue of C. difficile IID and is required for its transglycosylase activity (Nocadello et al., 2016), and Histidine 120 is required for C. difficile IIQ to bind Zn2+ (Serrano et al., 2016) as well as for its predicted endopeptidase activity (Crawshaw et al., 2014). Complementation of ΔIIDQ with IIDE101A failed to restore even partial heat resistance to the parental ΔIIDQ strain, just as this construct failed to complement the heat resistance defect of ΔIID and may have even exacerbated its defect (Fig. 3). Importantly, IIDE101A was produced in the ΔIID and ΔIIDQ strain backgrounds at levels similar to wild type and the IID complementation strains (Fig. 3). Taken together, these results indicate that IID’s transglycosylase activity (Nocadello et al., 2016) is necessary for it to compensate for loss of IIQ.
Complementation of ΔIIDQ with IIQH120A also failed to restore heat-resistant spore formation to ΔIIDQ, even though this allele fully complemented the heat resistance defect of ΔIIQ (ΔIIQ/IIQH120A) consistent with previous analyses using multi-copy plasmid complementation (Fimlaid et al., 2015b). The inability of IIQH120A to increase heat-resistant spore formation in ΔIIDQ to the levels observed in the ΔIID single mutant could result from the reduced levels of IIQH120A observed in both ΔIIDQ/IIQH120A and ΔIIQ/IIQH120A relative to wild type and ΔIIQ/IIQ and ΔIIDQ/IIQ complementation strains (Fig. 3). Since analyses of a IIQH120S-SNAP fusion protein yielded similar decreases in IIQ levels and abrogated zinc binding in vitro (Serrano et al., 2016), zinc binding would appear to affect IIQ stability and/or production (Serrano et al., 2016).
Read-through transcription of IIP increases IIP levels in C. difficile
While we could readily complement the heat-resistant spore defects of ΔIID, ΔIIQ, ΔIIDQ strains expressing IID, IIQ, and IIDQ, respectively, from their native promoters in the pyrE locus (Table 1 and Fig. 3), we were unable to complement ΔIIP strains because we failed to clone IIP complementation constructs in E. coli despite numerous attempts. Since we could readily clone IIP complementation constructs harboring a predicted catalytic mutation, Glu309A (Chastanet & Losick, 2007, Rodrigues et al., 2013, Shida et al., 2001), in E. coli (Fig. S4), C. difficile IIP appears to be toxic in E. coli. Expression of the construct encoding IIPE309A from the pyrE locus of ΔIIP construct restored IIP production to wildtype levels when the complementation construct included the upstream gpr gene and its promoter but failed to restore heat-resistant spore formation to the ΔIIP mutant (Fig. S4). Since IIPE309A levels were considerably lower when the complementation construct included only the region 269 bp immediately upstream of IIPE309A, both the distal promoter (upstream of gpr) and a proximal promoter (immediately upstream of IIP) appear to contribute to IIP expression.
To independently test the requirement for C. difficile IIP during heat-resistant spore formation, we constructed a second IIP mutation using targetron mutagenesis (Heap et al., 2007). The resulting IIP::ermB mutant failed to produce detectable heat-resistant spores similar to ΔIIP (Table 1 and Fig. S4). Taken together, these results strongly suggest that IIP, and likely its catalytic activity (i.e. Glu309), are required for heat-resistant spore formation in C. difficile.
Loss of IID, IIP, and IIQ lead to severe defects in engulfment and coat localization
To test whether the severe heat resistance defects of IIP, IID, and IIQ mutants were due to defects in engulfment (Fig. 2B), we analyzed sporulating cultures of these mutants using phase-contrast microscopy. The ΔIID, ΔIIP, and ΔIIQ mutants exclusively produced cells with phase-dark forespore regions, whereas wildtype and ΔIIM strains produced phase-bright spores (Fig. 2B). These results indicate that IID, IIP, and IIQ mutants arrest at an early stage of sporulation. To test this possibility, we used the FM4–64 membrane and Hoechst nucleoid stains with fluorescence microscopy to better visualize sporulation progression in these mutant strains. These analyses revealed that none of the sporulating IID, IIP, and IIQ mutant cells (>200) quantified completed engulfment, whereas 43% of sporulating wildtype cells completed engulfment (Fig. S5). Despite this failure, engulfment appeared to qualitatively progress the furthest in the ΔIID mutant, whereas ΔIIQ and ΔIIP in particular exhibited little curvature in the forespore membrane. Unfortunately, quantifying engulfment progression in these strains proved difficult because of the apparent fragility of ΔIIP and ΔIIQ mutant cells (Fig. S5).
Consistent with our prior finding that a JIR8094 IIQ targetron mutant has coat mislocalization defects (Fimlaid et al., 2015b), the phase-contrast microscopy analyses suggested that 630Δerm ΔIID, ΔIIP, and ΔIIQ also have regions of mislocalized coat (Fig. 2B, pink arrows). To more confidently assess the engulfment and coat mislocalization defects revealed by our light microscopy analyses, we visualized sporulating clutures of 630Δerm ΔIID, ΔIIP, and ΔIIQ strains using transmission electron microscopy (TEM). Analysis of a minimum of 50 cells that had initiated sporulation by 23 hrs revealed that 100% of ΔIID and ΔIIP cells failed to complete engulfment (Fig. 4), while 98% of ΔIIQ cells failed to complete this process. Consistent with the heat resistance data (Fig. 2), loss of IIP led to the most severe engulfment defect, with the leading edge of ΔIIP cells advancing the least relative to ΔIID and ΔIIQ strains (Fig. 4). Interestingly, ΔIIM failed to complete engulfment in 14% of cells relative to 0% of wildtype cells, which may reflect ΔIIM’s 2–3-fold decrease in heat resistance relative to wild type (Fig. 2B and Table 1).
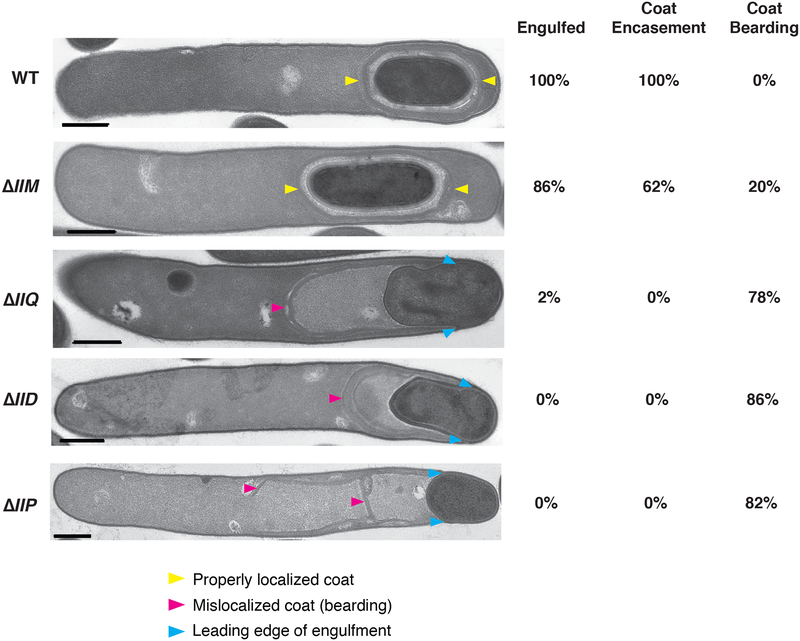
Fig. 4. Severe engulfment and coat localization defects in ΔIID, ΔIIQ, and ΔIIP mutants.
Transmission electron microscopy (TEM) analyses of wildtype 630Δerm, ΔIIM, ΔIIQ, ΔIID, and ΔIIP cultures after 23 hrs of sporulation induction. Strains are shown in decreasing order of engulfment completion. Scale bars represent 500 nm. Yellow arrows mark properly localized coat, and pink arrows mark mislocalized coat. Blue arrows mark the leading edge of the engulfing membranes in cells that fail to complete engulfment. The percentages shown are based on analyses of 50 cells for each strain with visible signs of sporulation from a single biological replicate. “Engulfed” indicates that the forespore was fully engulfed by the mother cell. Coat encasement refers to cells in which the coat surrounds the forespore, while coat bearding refers to cells where the polymerized coat sloughs off from the forespore.
In contrast with analyses of IID and IIP mutants in B. subtilis, no forespore bulging was observed in C. difficile ΔIID or ΔIIP mutants by TEM. Forespore bulging occurs when the forespore pushes into the mother cell through a small area of apparently weakened cell wall (Eichenberger et al., 2001, Frandsen & Stragier, 1995, Gutierrez et al., 2010) to balance the turgor pressure generated by chromosome packing into the small forespore (Lopez-Garrido et al., 2018). C. difficile ΔIIQ sporulating cells, nevertheless, produced forespore bulges in 8% of ΔIIQ sporulating cells analyzed, consistent with structured illumination microscopy (SIM) analyses of an independent ΔIIQ mutant in broth-grown cultures (Serrano et al., 2016).
A high frequency of coat mislocalization in engulfment-defective mutants (Fig. 4) was also observed, with polymerized coat appearing to slough off the forespore (previously termed “bearding”, (Ribis et al., 2017)) of ~80% ΔIID, ΔIIP, and ΔIIQ cells, and 16% of ΔIIM cells; coat “beards” were observed in all ΔIIM cells that failed to complete engulfment (14%, Figs. 4 and S6). The coat mislocalization phenotypes were the most severe in ΔIIP, with approximately one-third of cells generating “double” beards, in which the polymerized coat appeared to separate into two layers (Fig. S6). Approximately 10% of ΔIIQ cells produced “double” beards, while this phenotype was not observed in ΔIID cells..
We also assessed engulfment and coat localization phenotypes in double, triple, and quadruple mutants of IID, IIP, IIQ, and IIM (Fig. 2A). The ΔIIDQ mutant exhibited severe engulfment defects similar to the ΔIIP single mutant, with the polar septum exhibiting only a small amount of curvature (Fig. S6). Mutants missing IIP and additional engulfment-related factors resembled the single ΔIIP strain in terms of their limited septal curvature and high frequency of mislocalized polymerized coat (Fig. S6). Notably, deletion of all four genes (ΔIIDPMQ) did not generate a flat septum, in contrast with a B. subtilis ΔIIDMP strain (Eichenberger et al., 2001). While the reason for this difference is unclear, it remains possible that unidentified proteins may initiate engulfment in C. difficile ΔIIDPMQ.
IIP catalytic activity in the absence of IID can lead to asymmetric engulfment
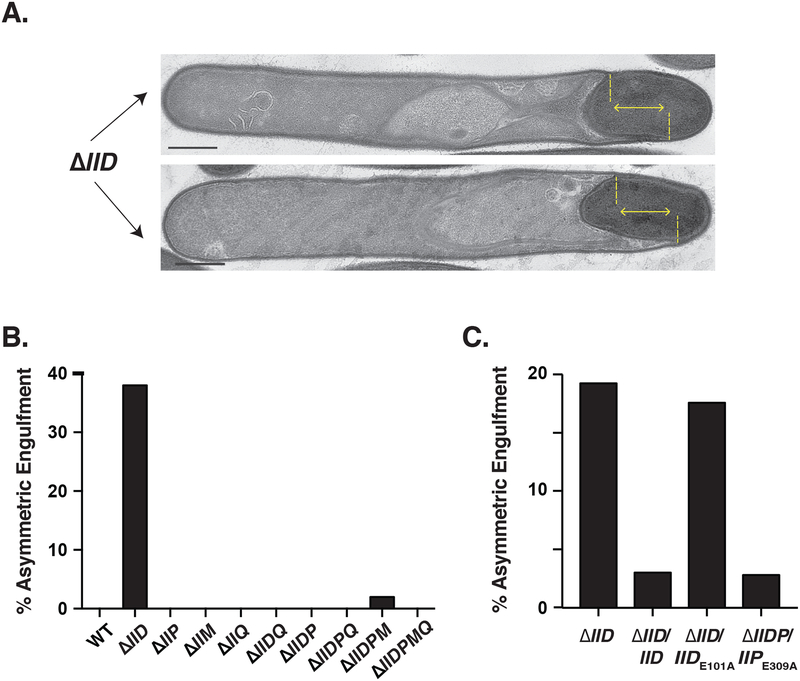
Although the curved septa of ΔIIDMPQ precluded determining the requirement for forespore curvature in localizing C. difficile coat basement layer proteins, we nevertheless identified phenotypes that were unique to single deletion strains relative to strains carrying multiple deletions. Engulfment proceeded unevenly in 36% of ΔIID cells, whereas this asymmetric engulfment phenotype was not observed in any other engulfment mutant strain aside from 2% of ΔIIDMP cells (Fig. 5). Asymmetric engulfment was also visible in fluorescence microscopy analyses of ΔIID using the FM4–64 membrane stain (Fig. S5). In B. subtilis, a IID mutant also exhibits asymmetric engulfment, with the leading edge that progresses furthest being enriched in IIP (Gutierrez et al., 2010). Since unequal IIP activity leads to asymmetric engulfment in B. subtilis (Abanes-De Mello et al., 2002), we tested whether a similar phenomenon could be occurring in C. difficile by comparing the engulfment frequency of ΔIIDP complemented with a IIPE309A catalytic mutant construct relative to ΔIID in an independent experiment. We were unable to complement the ΔIIDP mutant with the wildtype IIP allele because of the presumed toxicity issues of the C. difficile spoIIP gene in E. coli discussed earlier. We also tested whether the asymmetric engulfment phenotype of ΔIID cells was due to loss of IID transglycosylase activity by complementing ΔIID with wildtype IID or IIDE101A. Consistent with our prediction, complementation of the ΔIIDP double mutant with catalytically inactive IIP abrogated asymmetric engulfment, whereas complementation of ΔIID with catalytically inactive IID resulted in asymmetric engulfment at levels similar to the parental ΔIID (Figure 5). These results suggest that improper IIP activity leads to asymmetric engulfment when IID is absent or non-functional.
Fig. 5. Asymmetric engulfment in the absence of IID depends on IIP catalytic activity.
(A) Transmission electron microscopy (TEM) images of ΔIID cells undergoing asymmetric engulfment. The yellow lines highlight the uneven progression of the leading edge in the two cells shown. Scale bars represent 500 nm. (B) Percentage of cells that displayed asymmetric engulfment in the engulfment mutants constructed based on counts in a single biological replicate. (C) Percentage of cells showing asymmetric engulfment for IID and IIP catalytic mutant complementation strains using a single biological replicate. The TEM preparations counted in Fig, 5A are distinct from those in Fig. 5B. Percentages shown are based on analyses of 50 cells for each strain.
IID catalytic activity contributes to distortions in forespore morphology in ΔIIQ cells
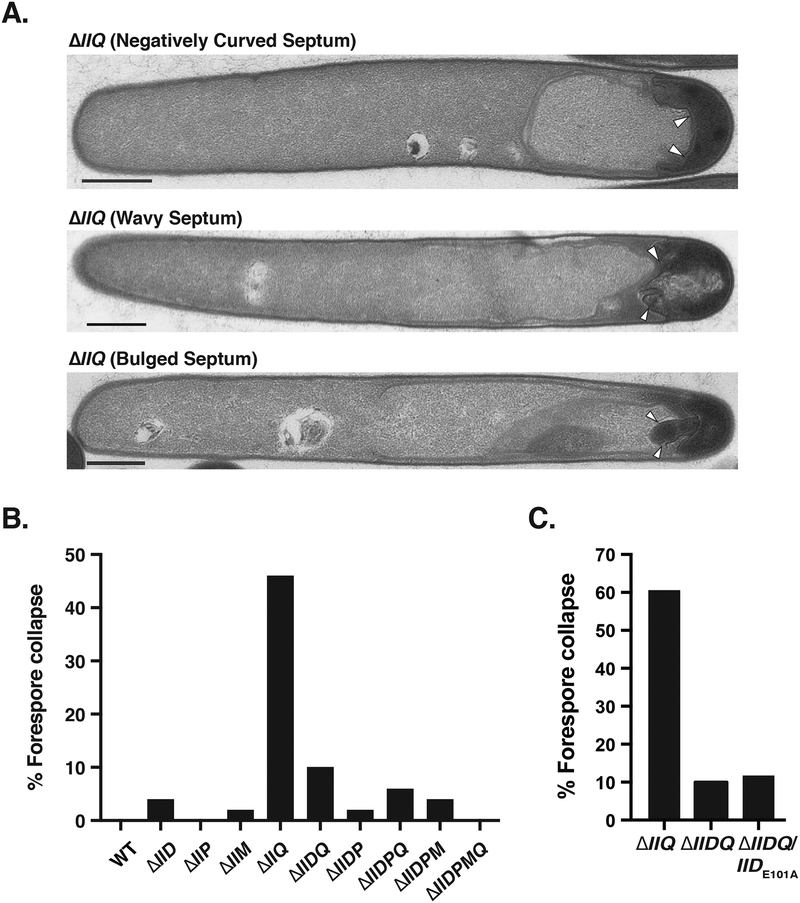
An even more striking phenotype was observed in the ΔIIQ single mutant relative to the other engulfment mutants: almost half of ΔIIQ cells exhibited forespore morphological defects, in which the forespore appeared wavy, invaginated upon itself, and/or bulged (Fig. 6). Interestingly, these distortions in forespore morphology were 5-fold less frequent in a C. difficile ΔIIDQ double mutant relative to the ΔIIQ single mutant (Fig. 6B). Based on this observation, we wondered whether IID activity could be contributing to these forespore morphological defects, so we complemented ΔIIDQ with IIDE101A. Loss of IID activity in the absence of ΔIIQ (ΔIIDQ/IIDE101A) reduced forespore distortions to levels identical to the parental ΔIIDQ strain (6-fold reduction, Fig. 6B), suggesting that thinning of the cell wall by IID when IIQ is absent can lead to forespore abnormalities in C. difficile.
Fig. 6. IID catalytic activity in the absence of IIQ can induce distortions in forespore morphology.
(A) Transmission electron microscopy (TEM) images of ΔIIQ cells exhibiting different types of forespore distortions, in which the polar septum either became invaginated, wavy, or the forespore cytosol appeared to leak into the mother cell cytosol (bulged septum). The latter phenotype was observed in 8% of ΔIIQ cells analyzed. (B) Percentage of cells with forespore morphological abnormalities in the engulfment mutants constructed. (C) and (D) Percentage of cells showing forespore morphological abnormalities in ΔIIQ, ΔIIDQ, ΔIIDP, and double mutants complemented with catalytic mutant complementation constructs. The samples analyzed in Fig. 6B, 6C, and 6D derive from independent cultures and TEM preparations processed on different days. Percentages shown are based on analyses of a minimum of 50 cells.
To test whether IIP activity contributed to the forespore distortions in the ΔIIQ mutant, we deleted spoIIP from the ΔIIQ strain and complemented this mutant with a construct encoding IIPE309A. Loss of either IIP or IIP catalytic activity in the ΔIIQ mutant resulted in a ~2-fold decrease in forespore morphological defects relative to the parental ΔIIQ strain (Fig. 6D), suggesting that both IID and IIP activity contributes to the forespore distortions of ΔIIQ cells.
Basement layer coat proteins adhere to the forespore in engulfment mutants
While the multiple gene deletion strains yielded insight into the individual functions of IID, IIP, and IIQ, the original goal of constructing these mutants was to test whether C. difficile coat proteins can localize to the forespore if the septum is flat. Although this was not possible because the septa of C. difficile ΔIIDPM and ΔIIDPMQ stays curved (Fig. S6) unlike B. subtilis ΔIIDMP (Chastanet & Losick, 2007, Ramamurthi et al., 2009), we could still ask whether C. difficile coat proteins exhibit differential localization patterns within engulfment mutants. For example, some coat proteins might stay associated with the forespore rather than localize to the polymerized coat visible by phase-contrast microscopy and TEM (Figs. 2 and 4). We hypothesized that C. difficile coat proteins in the innermost basement layer (McKenney et al., 2012) would associate with the forespore. This hypothesis was based on the observation that the B. subtilis basement layer protein SpoVM preferentially localizes to the forespore by recognizing its positive membrane curvature (Ramamurthi et al., 2009), while its binding partner, SpoIVA, localizes to the forespore as a single focus in the absence of SpoVM (Ramamurthi et al., 2006).
To test this hypothesis, we assessed the localization patterns of mCherry fusions to the basement layer proteins, SpoIVA (IVA) and SipL, and the outer coat protein, CotE, in wildtype, ΔIIM, and ΔIIP strains. We chose to analyze SpoIVA and SipL because they are required for polymerized coat to localize around the forespore in C. difficile (Putnam et al., 2013), while CotE is a surface-accessible coat protein (Hong et al., 2017, Permpoonpattana et al., 2013) that we have shown mislocalizes to coat “beards” in a IIQ mutant (Fimlaid et al., 2015b). The ΔIIM and ΔIIP strain backgrounds were chosen because they exhibit varying degrees of engulfment defects: ΔIIM has subtle defects in engulfment and coat localization in TEM analyses, while ΔIIP exhibits the most severe engulfment and coat localization defects (Figs. 4 and S6). We used mCherry fusions instead of SNAP fusions because mCherry-IVA and SipL-mCherry fusions complemented ΔIVA (Ribis et al., 2017) and ΔsipL strains (Fig. S7), respectively, more efficiently than SNAP-tag fusions (data not shown).
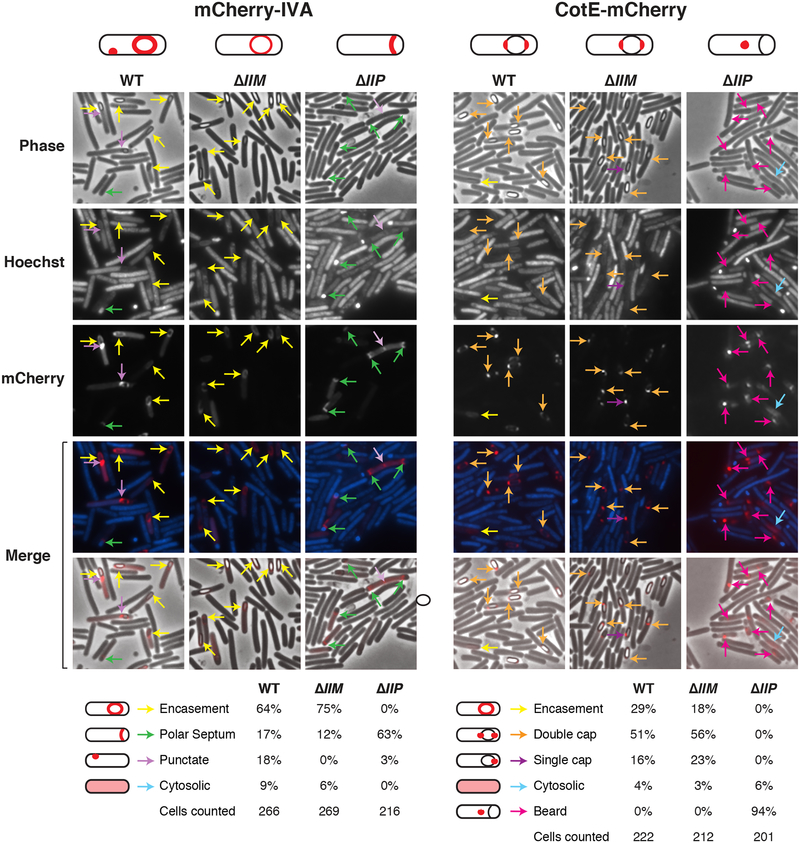
Sporulating cultures were stained with Hoechst to visualize the nucleoid region (Fimlaid et al., 2013, Ribis et al., 2017) and analyzed by phase-contrast and fluorescence microscopy. If engulfment fails to complete, the forespore stains intensely with nucleoid dyes, since the dyes are excluded from the forespore following membrane fission, and the forespore is a double membrane-bound protoplast in the mother cell (Doan et al., 2013, Gutierrez et al., 2010, Pogliano et al., 1999). Consistent with previously published analyses, mCherry-IVA concentrated around the forespore of wildtype cells ((Ribis et al., 2017), Fig. 7, yellow arrows). A similar localization pattern was observed in ΔIIM cells, although the fluorescent signal was muted relative to wild type even though it produced wildtype levels of mCherry-IVA (Fig. S8). In engulfment-defective ΔIIP, the mCherry-IVA signal concentrated along the presumed polar septum in 61% of cells based on Hoecsht labeling of the forespore nucleoid (Fig. 7, green arrows), implying that this basement layer protein can adhere to the forespore rather than to the mislocalized coat. Occasionally, mCherry-IVA formed punctate foci on the forespore or on the mother cell membrane in wildtype cells either (purple arrows, 15%) and ΔIIP cells (5%) but not in ΔIIM (Fig. 7). Cytosolic mCherry-IVA signal was observed in all three strains, since mCherry is released during proteolysis of the fusion protein (Fig. S8) as previously reported (Ribis et al., 2017).
Fig. 7. Localization of mCherry fusions to the basement layer protein, IVA, and outer coat layer protein, CotE.
Fluorescence microscopy analyses of WT, ΔIIM, and ΔIIP cells producing mCherry fusions to (A) IVA and (B) CotE at 23 hrs post sporulation induction. Phase-contrast (phase) microscopy was used to visualize sporulating cells. The Hoechst-stained nucleoid is shown in blue, and mCherry fluorescence is shown in red. Note that engulfment completion excludes Hoechst from staining the forespore (Pogliano et al., 1999). The merge of Hoechst and mCherry (top) and phase-contrast and mCherry (bottom) is shown. Yellow arrows denote encasement of the forespore; green arrows highlight staining along the presumed polar septum (based on Hoechst labeling); light purple arrows highlight punctate foci in the mother cell; orange arrows indicate staining at both forespore poles (double cap); blue arrows point to CotE-mCherry localization to single foci on the forespore; and pink arrows indicate localization to phase-dark “beards.” mCherry-IVA localization analyses were performed in the presence of a wildtype copy of IVA, since the fusion protein does not efficiently encase the forespore unless a wildtype copy of IVA is present (Ribis et al., 2017). Schematics depicting the primary localization pattern of mCherry-IVA and CotE-mCherry in each strain background are shown above, and the percent for the given phenotypes is shown below; the total number of cells from which these percentages were derived is also listed.
To localize SipL-mCherry in the ΔIIM and ΔIIP backgrounds, we constructed ΔsipLΔIIM and ΔsipLΔIIP double mutants so that the SipL-mCherry fusion protein would be the only version of SipL present in these cells. This was necessary because producing SipL-mCherry in the presence of wildtype SipL increases the cytosolic fluorescent signal (M. Touchette, unpublished data), presumably due to displacement of SipL-mCherry from the forespore by wildtype SipL. SipL-mCherry localized almost exclusively around the forespore of ΔsipL/sipL-mCherry (90%) and slightly less frequently in ΔsipLΔIIM/sipL-mCherry (74%, Fig. S9), consistent with the minor engulfment defects reported for the ΔIIM strain (Fig. 3). In contrast, SipL-mCherry was distributed along the presumed polar septum of ΔsipLΔIIP/sipL-mCherry in 45% of cells, and punctate foci were observed more frequently in the ΔIIP (~10%) relative to the wildtype or ΔIIM strain backgrounds (Fig. S9). ΔsipLΔIIP/sipL-mCherry cells also exhibited higher cytosolic fluorescence relative to ΔsipL/sipL-mCherry and ΔsipLΔIIM/sipL-mCherry. This cytosolic signal likely results from higher levels of unincorporated SipL-mCherry being present in ΔIIP mutant cells due to its engulfment defect.
Polymerized coat “beards” of engulfment mutants contain the outer coat protein, CotE
While the mCherry-IVA and SipL-mCherry fusion proteins appeared to concentrate along the forespore-mother cell interface of most ΔIIP cells (Figs. 7 and S9), the CotE-mCherry fusion protein localized mainly as punctate foci in ΔIIP cells at sites distal from the presumed polar septum (95%, Fig. 7). These foci overlapped with regions with coat “bearding” visible by phase-contrast microscopy; in contrast, in wildtype and ΔIIM cells, CotE-mCherry localized primarily to either the polar caps of the forespore or a single focus at one of the poles (70–80%, Fig. 7). The capped localization pattern is consistent with studies of CotE-SNAP in JIR8094 cells grown on solid media (Fimlaid et al., 2015b), although CotE-SNAP grown in broth culture appears to encase the forespore more uniformly (Pereira et al., 2013, Serrano et al., 2016).
Taken together, the fluorescent protein localization studies indicate that basement layer coat proteins can associate with the presumed polar septum of ΔIIP engulfment mutants, whereas outer coat proteins localize to regions of displaced, polymerized coat (known as bearding). They further suggest that the basement layer proteins, IVA and SipL, are not incorporated in the mislocalized polymerized coat of engulfment mutants, implying that IVA and SipL have mechanisms for staying adhered to the available forespore surface.
Discussion
Collectively, our analyses have identified both conserved and differential functions for the strictly conserved sporulation proteins IID, IIP, IIM, and IIQ during C. difficile spore formation relative to B. subtilis. While IIP is essential for engulfment in both organisms (Figs. 2–4 and S4), IIQ is more important for C. difficile engulfment (Figs. 2 and 4) than B. subtilis engulfment, as observed previously (Fimlaid et al., 2015b, Londono-Vallejo et al., 1997, Serrano et al., 2016, Sun et al., 2000). In contrast, the spore formation defect of a B. subtilis IID mutant is more severe than a C. difficile IID mutant by ~100-fold (Figs. 2 and 4, (Lopez-Diaz et al., 1986)). The biggest difference in the requirement between these two organisms was observed for IIM, which is essential in B. subtilis (Smith et al., 1993) but largely dispensable in C. difficile (Figs. 2, 4 and S6). IIM is essential for engulfment in B. subtilis because it binds the IIP peptidoglycan hydrolase, recruits IIP to the polar septum (Aung et al., 2007, Chastanet & Losick, 2007) and then forms a mother cell-specific peptidoglycan degradation complex with IID (Aung et al., 2007, Chastanet & Losick, 2007, Morlot et al., 2010, Rodrigues et al., 2013, Sun et al., 2000) that synergistically degrades the cell wall and drives engulfment to completion (Abanes-De Mello et al., 2002, Morlot et al., 2010, Ojkic et al., 2016).
The dispensability of IIM during C. difficile engulfment was surprising, given the strict conservation of IIM in spore-forming organisms (Abecasis et al., 2013, Galperin et al., 2012). However, our results suggest a possible mechanism for why the membrane scaffolding function identified in B. subtilis would appear to be dispensable in C. difficile. Whereas B. subtilis IID, IIP, and IIM form a complex in the mother cell membrane, C. difficile IID and IIP would appear to be inserted into opposing membranes based on transcriptional and Western blot analyses (Fig. 1 and 8). Despite this difference in topology, C. difficile may nevertheless coordinate the enzymatic activities of C. difficile IID and IIP because C. difficile IIP is processed (Figs. 2 and S4), which we speculate liberates IIP’s enzymatic domain into the intermembrane space where it can degrade peptidoglycan in concert with IID in the outer forespore membrane (Fig. 8). Consistent with this model, Dembek et al. observed that the extracellular domains of C. difficile IID and IIP physically interact (P. Salgado, accompanying manuscript) similar to B. subtilis IID and IIP (Aung et al., 2007, Chastanet & Losick, 2007). However, unlike C. difficile IIP, B. subtilis IIP does not undergo processing (Aung et al., 2007, Chastanet & Losick, 2007, Rodrigues et al., 2013). Validating the interaction between C. difficile IID and IIP in sporulating cells, along with identifying the protease(s) that mediate C. difficile IIP processing, would allow this model to be directly tested.
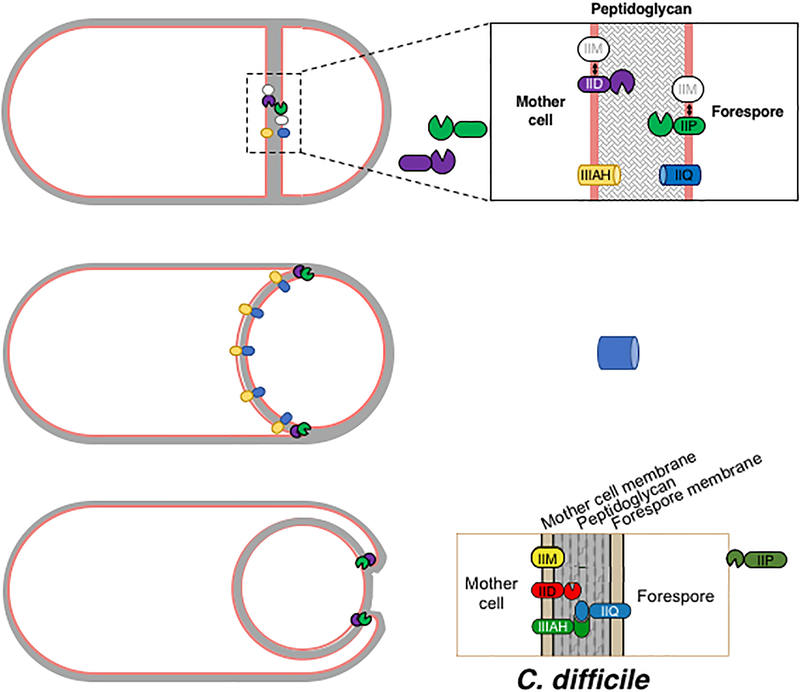
Fig. 8. Model for C. difficile engulfment.
Following asymmetric engulfment and activation of σF and σE, IIM is inserted into both the mother cell and forespore membranes, since it is produced upon Spo0A activation (Fig. 1); IIP and IIQ are inserted into the forespore membrane; and IID and IIIAH are inserted into the mother cell membrane. IID and IIP may associate with IIM, but this interaction is not critical for their function (Fig. 2). IIP appears to undergo site-specific processing (Figs. 1 and 2), which could liberate its hydrolase domain into the inter-membrane space where it can bind to the IID transglycosylase domain. Based on studies in B. subtilis, the IID-IIP degradation machine localizes to the leading edge, thins the peptidoglycan, and likely promotes binding between IIQ and IIIAH (Abanes-De Mello et al., 2002, Blaylock et al., 2004). The interaction between these two proteins is predicted to function like a Brownian ratchet in C. difficile to facilitate engulfment completion analogous to B. subtilis (Broder & Pogliano, 2006).
Another possible reason for why C. difficile IIM is not essential for engulfment is that an as-yet-unidentified factor functionally substitutes for loss of IIM. B. subtilis employs multiple mechanisms to ensure the proper localization and function of both the DMP engulfment complex (Aung et al., 2007, Chastanet & Losick, 2007) and A-Q complex (Fredlund et al., 2013, Rodrigues et al., 2016, Rodrigues et al., 2013). Indeed, the latter complex enhance the robustness of the engulfment process in B. subtilis (Broder & Pogliano, 2006, Sun et al., 2000).
Notably, our deletion analyses revealed that C. difficile IID and IIQ, respectively, could partially compensate for loss of IIQ and IID, respectively, in a small fraction of cells (Figs. 2 and 3). While the mechanism(s) underlying this partial compensation are unknown, one possibility is that C. difficile IIQ and IID independently stimulate (directly or indirectly) IIP’s peptidoglycan hydrolase activities so that IIP can drive engulfment completion in a small fraction of ΔIID and ΔIIQ sporulating cells, respectively. Consistent with this hypothesis, loss of C. difficile IIP completely abrogates engulfment, whereas loss of either IID or IIQ, but not both, permits spore formation in ~0.05% of sporulating cells (Figs. 2 and S6). Furthermore, C. difficile IIP activity in a ΔIID single mutant, but not a ΔIIDQ double mutant, can drive engulfment forward, albeit unevenly, since we observed asymmetric engulfment in ΔIID but not ΔIIDQ cells (Fig. 5). In addition, B. subtilis IID stimulates IIP activity in vitro (Morlot et al., 2010), so a similar scenario could occur in C. difficile. Lastly, a IIQH120A mutation, which prevents C. difficile IIQ from binding to Zn2+ (Serrano et al., 2016), blocks engulfment completion in ΔIID cells (Fig. 3, ΔIIDQ/IIQH120A). IIQH120A may be insufficient to drive engulfment to completion in some ΔIID cells because IIQH120A is present at lower levels (Fig. 3) and/or because it fails to bind Zn2+. Interestingly, in B. subtilis the degenerate LytM domain of IIQ, which cannot bind Zn2+, recruits additional proteins (Rodrigues et al., 2016, Rodrigues et al., 2013). Biochemical analyses of IIP activity in the presence of IID or IIQ would allow this model to be directly tested.
Another possible explanation for how IIQ in particular can partially substitute for loss of IID in C. difficile is that the IIQ-IIIAH complex (Fimlaid et al., 2015b, Serrano et al., 2016) functions as a ratchet similar to B. subtilis (Broder & Pogliano, 2006) to prevent back-sliding of the engulfing membrane (Broder & Pogliano, 2006, Ojkic et al., 2014), thus bypassing the need for IID in some cells. Regardless of whether this model holds true, it is still unclear why IIQ is more essential for C. difficile engulfment than B. subtilis. One possibility for why C. difficile IIQ is defective in engulfment is that ΔIIQ cells have mis-regulated IID (and to a lesser extent IIP) activities that contribute to the forespore distortions (ΔIIDQ/IIDE101A and ΔIIPQ/IIPE309A, Fig. 6). Further analyses are needed to identify the mechanisms underlying the forespore distortions in ΔIIQ cells and whether they are specific to loss of A-Q complex components or to IIQ alone. Indeed, recent studies in B. subtilis suggest that turgor pressure exerted by the translocated chromosome (or lack there-of) could contribute to the forespore distortions (Lopez-Garrido et al., 2018).
Another major difference between C. difficile and B. subtilis engulfment was our finding that C. difficile ΔIIDPM and ΔIIDPMQ sporulating cells produce curved polar septa (Fig. S6), in contrast with the flat septa of B. subtilis ΔIIDPM cells (Eichenberger et al., 2001, Ramamurthi et al., 2009). Again, it is unclear why these differences are observed, but an additional peptidoglycan remodeling system initiating engulfment in C. difficile might be at play or outward pressure from the forespore nucleoid (Lopez-Garrido et al., 2018) could cause bending of the C. difficile septum as discussed above.
While the curved septa of the C. difficile ΔIIDPM mutants did not allow us to test whether positive curvature is necessary for coat morphogenetic protein localization, the ΔIIP mutant nevertheless helped establish that polymerized coat, including the outer coat protein CotE, adheres poorly to the forespore membrane in the absence of engulfment (Figs. 4 and 7). In contrast, the basement layer coat morphogenetic proteins, IVA and SipL, remain associated with the forespore region of most ΔIIP sporulating cells (Figs. 7 and S9). Thus, mechanisms appear to exist for localizing and retaining these basement layer proteins to the forespore surface even in the absence of engulfment. Whatever mechanism is at work, it likely does not depend on the coat morphogenetic protein, SpoVM, which retains SpoIVA on the forespore in B. subtilis (Ramamurthi et al., 2006, Wang et al., 2009). SpoVM is dispensable for this process in C. difficile (Ribis et al., 2017), so the mechanism by which SpoIVA and SipL remain adhered to the forespore of engulfment mutants is unclear. Possible mechanisms include that C. difficile IVA and possibly SipL intrinsically recognize the forespore or are recruited and retained by as-yet-identified factors.
Given that the outer coat protein CotE is displaced from the forespore in engulfment mutants (Fig. 7), whereas the basement layer proteins, SpoIVA and SipL, are retained, interactions between SpoIVA and/or SipL and outer coat proteins would appear to be weak. Indeed, polymerized coat can be loosely associated with the forespore even in wildtype C. difficile, where separation of the coat and cortex region is visible in some wildtype cells ((Ribis et al., 2017), Figs. 4 and S6) and purified spores (Fimlaid et al., 2015a). Furthermore, the finding that outer coat proteins readily detach from partially engulfed forespores suggests that coat proteins need to fully encase the spore and covalently link the coat polymer ends together to remain associated with the forespore.
Collectively, our results advance our understanding of how C. difficile sporulating cells complete engulfment, reveal differential phenotypes for mutants lacking individual engulfment factors (e.g. asymmetric phenotype and distortions in forespore morphologies), and establish differential requirements for coat protein localization during spore formation in C. difficile. They also raise questions as to how IID, IIP, and IIQ coordinate engulfment in C. difficile. Future localization and interaction studies will provide much needed insight into this question. Studies of engulfment in other spore-forming organisms will also reveal the extent to which diverse mechanisms have evolved to mediate this important morphological event.
Experimental Procedures
Bacterial strains and growth conditions.
630ΔermΔpyrE (Ng et al., 2013) was used as the parental strain for Targetron-based gene disruption (Heap et al., 2007) and pyrE-based allele-coupled exchange (ACE, (Ng et al., 2013)). C. difficile strains are listed in Table S1 and were grown on BHIS agar (Sorg & Dineen, 2009) supplemented with taurocholate (TA, 0.1% w/v; 1.9 mM), kanamycin (50 μg/mL), cefoxitin (8 μg/mL), FeSO4 (50 μM), and/or erythromycin (10 μg/mL) as needed. C. difficile defined media (CDDM, (Cartman & Minton, 2010)) was supplemented with 5-fluoroorotic acid (5-FOA) at 2 mg/mL and uracil at 5 μg/mL as needed for ACE. Cultures were grown under anaerobic conditions using a gas mixture containing 85% N2, 5% CO2, and 10% H2.
Escherichia coli strains for HB101/pRK24-based conjugations and BL21(DE3)-based protein production are listed in Table S1. E. coli strains were grown at 37˚C, shaking at 225 rpm in Luria-Bertani broth (LB). The media was supplemented with chloramphenicol (20 μg/mL), ampicillin (50 μg/mL), or kanamycin (30 μg/mL) as indicated.
E. coli strain construction.
All primers are listed in Table S2. Details of E. coli strain construction are provided in the Supplementary Text S1. All plasmid constructs were cloned into DH5α and sequenced confirmed using Genewiz. Plasmids to be conjugated into C. difficile were transformed into HB101/pRK24, while plasmids used for antibody production were transformed into BL21(DE3).
C. difficile strain mutant construction.
Targetron-based gene disruption using pJS107-spoIIP was performed as previously described (Fimlaid et al., 2013, Heap et al., 2007). Primer pair #2297 and 2316 was used to screen isolated erythromycin-resistant colonies for Targetron (2 kB) insertions in spoIIP. Allele-coupled exchange (ACE, (Ng et al., 2013)) was used as previously described (Donnelly et al., 2017) to construct the clean deletions of spoIID, spoIIP, spoIIM, spoIIQ, and spoIIDQ in 630ΔermΔpyrE. The primer pairs used to screen isolated FOA-resistant, uracil auxotroph colonies for each deletion are shown in Fig. S2 as are the internal primers used to validate the deletion strains. To construct the strains carrying multiple gene deletions, the ΔIIDΔpyrE and ΔIIDQΔpyrE strains were used as the parental mating strains as shown in Fig. 2. At least two clones of each deletion strain were phenotypically characterized prior to restoring the pyrE locus using pMTL-YN1C.
The pyrE locus was restored using pMTL-YN1C and pMTL-YN1C-based complementation constructs as previously described (Donnelly et al., 2017). Two independent clones of each complementation strain were phenotypically characterized.
Plate-based Sporulation.
C. difficile strains were grown from glycerol stocks overnight on BHIS plates containing TA (0.1% w/v). These cultures were used to inoculate liquid BHIS cultures, which were grown to stationary phase and back-diluted 1:50 into BHIS. The cultures were grown until they reached an OD600 between 0.35 and 0.7 after which 120 μL was used to inoculate 70:30 agar plates (40 mL, (Putnam et al., 2013)). Sporulation was induced on this media for 17–24 hrs. The 17 hr timepoint was used to analyze cultures by phase-contrast microscopy and harvest samples for Western blot analyses, since the levels of engulfment-related proteins were maximally detected at this timepoint.
Liquid-culture sporulation.
C. difficile strains were grown from glycerol stocks overnight on BHIS plates containing TA (0.1% w/v). These cultures were used to inoculate liquid BHIS cultures, which were grown to stationary phase and back-diluted 1:200 into SMC broth ((Permpoonpattana et al., 2011), 90 g Bacto-peptone, 5 g Protease peptone, 0.5 g (NH4)2SO4, 1.5 g Tris) and grown for 24h. Cells were imaged by phase-contrast microscopy at the 23h time point.
Heat resistance assay on sporulating cells.
Heat-resistant spore formation was measured in sporulating C. difficile cultures after 20–24 hrs as previously described (Shen et al., 2016). The heat resistance efficiency represents the average ratio of heat-resistant cells for a given strain relative to the average ratio determined for wild type based on a minimum of three biological replicates. The ratios obtained for each strain and replicate are shown in Table S3. Statistical significance was determined using a one-way ANOVA and Tukey’s test.
Quantitative RT-PCR (qRT-PCR).
Transcript levels for spoIID, spoIIP, spoIIM, and rpoB were determined using cDNA templates prepared from three independent biological replicates of wildtype JIR8094, spo0A–, sigF–, sigE–, and sigG– strains as previously described (Fimlaid et al., 2015b). Briefly, the indicated strains were induced to sporulate for 17 hrs on 70:30 media and then RNA was harvested from these cultures. The RNA samples used for qRT-PCR samples were distinct from those used in the RNA-Seq analyses (Pishdadian et al., 2015) shown in Fig. S1. Primer pairs #1558 and 1658, #1560 and 1561, and #1563 and 1565 were used to amplify spoIID, spoIIP, and spoIIM, respectively, while the housekeeping gene rpoB-specific primers have been previously described (Pishdadian et al., 2015). qRT-PCR analyses were performed as previously described (Pishdadian et al., 2015) using SYBR Green to quantify transcript levels for spoIID, spoIIP, and spoIIM, and rpoB. Transcript levels were normalized to rpoB using the standard curve method. The ratio of normalized transcript levels for a given strain relative to the normalized transcript levels in the spo0A mutant was then determined (Fig. 1C).
Antibody production.
The anti-IID and anti-IIP antibodies used in this study were raised against His6-IIDΔ27aa and ΔIIPΔ27aa-His6, respectively, in rabbits by Cocalico Biologicals (Reamstown, PA). These proteins lack N-terminal transmembrane sequences and were purified from E. coli strains #1366 and 1845 using Ni2+-affinity resin as previously described (Adams et al., 2013).
FM4–64 fluorescence microscopy.
Live cell fluorescence microscopy was performed using Hoechst 33342 (Molecular Probes, 15 μg/mL) and 1 μg/mL FM4–64 membrane stain (Molecular Probes) as previously described (Fimlaid et al., 2013, Fimlaid et al., 2015b). Briefly, 23 hr sporulating cultures of C. difficile strains were harvested into 1 mL PBS, pelleted, and resuspended into 100 μL PBS containing the dyes listed above. Live bacterial suspensions (4 μL) were added to a freshly prepared 1% agarose pad.
DIC and fluorescence microscopy was performed using a Nikon PlanApo Vc 100x oil immersion objective (1.4 NA) on a Nikon Eclipse Ti2000 epifluorescence microscope. Multiple fields for each sample were acquired with an EXi Blue Mono camera (QImaging) with a hardware gain setting of 1.0 using the NIS-Elements software (Nikon). Images were subsequently imported into Adobe Photoshop CC 2015 for minimal adjustments in brightness/contrast levels and pseudocoloring. The percentage of sporulating cells of wildtype, ΔIID, ΔIIP, and ΔIIQ that had completed engulfment was determined from 6–10 fields from one to two biological replicates. A minimum of 200 cells were counted per strain.
TEM analyses.
Sporulating cultures (23 hrs) were fixed and processed for electron microscopy by the University of Vermont Microscopy Center as previously described (Putnam et al., 2013). A minimum of 50 full-length sporulating cells were used for phenotype counting. While the phenotype counting for many of the strains shown are based on analyses of a single biological replicate, several of the strains were analyzed two or three times, namely ΔIIQ, ΔIID, and ΔIIDQ. The reproducibility of these replicates combined with the wildtype and catalytic complementation analyses gave greater confidence in the results obtained for a single biological replicate. Nevertheless, we cannot rule out the possibility that analyzing additional biological replicates could yield more variable results.
mCherry fluorescence microscopy.
Live cell fluorescence microscopy was performed using Hoechst 33342 (Molecular Probes, 15 μg/mL) and mCherry protein fusions. Samples were prepared on agarose pads as previously described (Fimlaid et al., 2015b), and samples were imaged 30 min after harvesting to allow for mCherry fluorescence signal reconstitution in the anaerobically grown bacteria. Sporulating cells were exposed to ambient oxygen for a maximum of 90 min to minimize DNA fragmentation; no cell lysis was observed under these conditions. DIC and fluorescence microscopy were performed using a Nikon 60x oil immersion objective (1.4 NA) on a Nikon 90i epifluorescence microscope. A CoolSnap HQ camera (Photometrics) was used to acquire multiple fields for each sample in 12 bit format using NIS-Elements software (Nikon). The Texas Red channel was used to acquire images after a 100–400 ms exposure (100 ms for SipL-mCherry; ~200 ms for mCherry-IVA; 400 ms for CotE-mCherry); 100 ms exposures were used to visualize the Hoechst stain; and ~10–20 ms exposures were used for phase-contrast microscopy. Ten Mhz images were subsequently imported into Adobe Photoshop CC 2015 for minimal adjustments in brightness/contrast levels and pseudocoloring. Counting of fluorescence localization phenotypes was done on a minimum of 200 sporulating cells from two images from independent replicates of each strain.
Western blot analyses.
Samples for immunoblotting were prepared as previously described (Putnam et al., 2013). Briefly, sporulating cell pellets were resuspended in 100 μL of PBS, and 50 μL samples were freeze-thawed for three cycles and then resuspended in 100 μL EBB buffer (8 M urea, 2 M thiourea, 4% (w/v) SDS, 2% (v/v) β-mercaptoethanol). The samples were boiled for 20 min, pelleted, re-suspended in the same buffer to maximize protein solubilization, boiled for another 5 min and then pelleted. Samples were resolved on 12% SDS-PAGE gels, transferred to Immobilon-FL PVDF membrane, blocked in Odyssey® Blocking Buffer with 0.1% (v/v) Tween 20. Rabbit anti-SpoIID, rabbit anti-SpoIIP, rabbit anti-SpoIIQ (Fimlaid et al., 2015b), and mouse anti-Spo0A (Fimlaid et al., 2013) antibodies were used at 1:1000 dilutions; mouse anti-SpoIVA (Kevorkian et al., 2015), rabbit anti-SipL (Putnam et al., 2013), and rabbit anti-CotE (Fimlaid et al., 2013) antibodies were used at 1:2500 dilutions; and rabbit anti-mCherry (Abcam) was used at a 1:2000 dilution. IRDye 680CW and 800CW infrared dye-conjugated secondary antibodies were used at a 1:25,000 dilution, and blots were imaged on an Odyssey LiCor CLx. Western blots were performed on sporulating samples derived from three independent biological replicates.
Supplementary Material
Acknowledgments
We would like to thank N. Bishop and J. Schwarz for excellent assistance in preparing samples for transmission electron microscopy throughout this study; K. Schutz for excellent technical assistance in conducting the qRT-PCR analyses; D. Weiss and C. Ellermeier for providing the codon-optimized mCherry construct; A. Camilli for access to the Nikon microscope; N. Minton (U. Nottingham) for generously providing us with access to the 630ΔermΔpyrE strain and pMTL-YN1C and pMTL-YN3 plasmids for allele-coupled exchange (ACE); M. Dembek for directly providing these materials to us and sharing his specific protocols on ACE; and P. Salgado for sharing unpublished data and helpful discussions.
Research in this manuscript was funded by Award Number R01AI22232 from the National Institutes of Allergy and Infectious Disease (NIAID) to A.S. A.S. is a Pew Scholar in the Biomedical Sciences supported by The Pew Charitable Trusts and a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease supported by the Burroughs Wellcome Fund. The content is solely the responsibility of the author(s) and does not necessarily reflect the views of the Pew Charitable Trusts, Burroughs Wellcome, NIAID, or the National Institutes of Health. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
A.S. has a paid consultancy for BioVector, Inc., a diagnostic start-up.
References
- Abanes-De Mello A, Sun YL, Aung S & Pogliano K, (2002) A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev 16: 3253–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis AB, Serrano M, Alves R, Quintais L, Pereira-Leal JB & Henriques AO, (2013) A genomic signature and the identification of new sporulation genes. J Bacteriol 195: 2101–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams CM, Eckenroth BE, Putnam EE, Doublie S & Shen A, (2013) Structural and functional analysis of the CspB protease required for Clostridium spore germination. PLoS Pathog 9: e1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hinai MA, Jones SW & Papoutsakis ET, (2015) The Clostridium sporulation programs: diversity and preservation of endospore differentiation. Microbiol Mol Biol Rev 79: 19–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung S, Shum J, Abanes-De Mello A, Broder DH, Fredlund-Gutierrez J, Chiba S & Pogliano K, (2007) Dual localization pathways for the engulfment proteins during Bacillus subtilis sporulation. Mol Microbiol 65: 1534–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaylock B, Jiang X, Rubio A, Moran CP Jr. & Pogliano K, (2004) Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev 18: 2916–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broder DH & Pogliano K, (2006) Forespore engulfment mediated by a ratchet-like mechanism. Cell 126: 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp AH & Losick R, (2009) A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev 23: 1014–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartman S & Minton N, (2010) A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile. Appl Environ Microbiol 76: 1103–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastanet A & Losick R, (2007) Engulfment during sporulation in Bacillus subtilis is governed by a multi-protein complex containing tandemly acting autolysins. Mol Microbiol 64: 139–152. [DOI] [PubMed] [Google Scholar]
- Crawshaw AD, Serrano M, Stanley WA, Henriques AO & Salgado PS, (2014) A mother cell-to-forespore channel: current understanding and future challenges. FEMS Microbiol Lett 358: 129–136. [DOI] [PubMed] [Google Scholar]
- Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, Wren BW, Fairweather NF, Dougan G & Lawley TD, (2012) The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun 80: 2704–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembek M, Barquist L, Boinett CJ, Cain AK, Mayho M, Lawley TD, Fairweather NF & Fagan RP, (2015) High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio 6: e02383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembek M, Willing SE, Hong HA, Hosseini S, Salgado PS & Cutting SM, (2017) Inducible Expression of spo0A as a Universal Tool for Studying Sporulation in Clostridium difficile. Front Microbiol 8: 1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T, Coleman J, Marquis KA, Meeske AJ, Burton BM, Karatekin E & Rudner DZ, (2013) FisB mediates membrane fission during sporulation in Bacillus subtilis. Genes Dev 27: 322–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T, Morlot C, Meisner J, Serrano M, Henriques A, Moran C & Rudner D, (2009) Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly ML, Li W, Li YQ, Hinkel L, Setlow P & Shen A, (2017) A Clostridium difficile-Specific, Gel-Forming Protein Required for Optimal Spore Germination. mBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driks A & Eichenberger P, (2016) The Spore Coat. Microbiol Spectr 4. [DOI] [PubMed] [Google Scholar]
- Driks A & Losick R, (1991) Compartmentalized expression of a gene under the control of sporulation transcription factor sigma E in Bacillus subtilis. Proc Natl Acad Sci U S A 88: 9934–9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin J & Losick R, (2005) Developmental commitment in a bacterium. Cell 121: 401–409. [DOI] [PubMed] [Google Scholar]
- Eichenberger P, Fawcett P & Losick R, (2001) A three-protein inhibitor of polar septation during sporulation in Bacillus subtilis. Mol Microbiol 42: 1147–1162. [DOI] [PubMed] [Google Scholar]
- Fimlaid KA, Bond JP, Schutz KC, Putnam EE, Leung JM, Lawley TD & Shen A, (2013) Global Analysis of the Sporulation Pathway of Clostridium difficile. PLoS Genet 9: e1003660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimlaid KA, Jensen O, Donnelly ML, Francis MB, Sorg JA & Shen A, (2015a) Identification of a Novel Lipoprotein Regulator of Clostridium difficile Spore Germination. PLoS Pathog. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimlaid KA, Jensen O, Donnelly ML, Siegrist MS & Shen A, (2015b) Regulation of Clostridium difficile Spore Formation by the SpoIIQ and SpoIIIA Proteins. PLoS Genet 11: e1005562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen N & Stragier P, (1995) Identification and characterization of the Bacillus subtilis spoIIP locus. J Bacteriol 177: 716–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredlund J, Broder D, Fleming T, Claussin C & Pogliano K, (2013) The SpoIIQ landmark protein has different requirements for septal localization and immobilization. Mol Microbiol 89: 1053–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Mekhedov SL, Puigbo P, Smirnov S, Wolf YI & Rigden DJ, (2012) Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation-specific genes. Environ Microbiol 14: 2870–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez J, Smith R & Pogliano K, (2010) SpoIID-mediated peptidoglycan degradation is required throughout engulfment during Bacillus subtilis sporulation. J Bacteriol 192: 3174–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap JT, Pennington OJ, Cartman ST, Carter GP & Minton NP, (2007) The ClosTron: a universal gene knock-out system for the genus Clostridium. J Microbiol Methods 70: 452–464. [DOI] [PubMed] [Google Scholar]
- Hong HA, Ferreira WT, Hosseini S, Anwar S, Hitri K, Wilkinson AJ, Vahjen W, Zentek J, Soloviev M & Cutting SM, (2017) The Spore Coat Protein CotE Facilitates Host Colonisation by Clostridium difficile. J Infect Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison EA, Miller DA & Angert ER, (2014) Sporulation in Bacteria: Beyond the Standard Model. Microbiol Spectr 2. [DOI] [PubMed] [Google Scholar]
- Kevorkian Y, Shirley DJ & Shen A, (2015) Regulation of Clostridium difficile spore germination by the CspA pseudoprotease domain. Biochimie. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN & McDonald LC, (2015) Burden of Clostridium difficile infection in the United States. N Engl J Med 372: 825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londono-Vallejo JA, Frehel C & Stragier P, (1997) SpoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol Microbiol 24: 29–39. [DOI] [PubMed] [Google Scholar]
- Lopez-Diaz I, Clarke S & Mandelstam J, (1986) spoIID operon of Bacillus subtilis: cloning and sequence. J Gen Microbiol 132: 341–354. [DOI] [PubMed] [Google Scholar]
- Lopez-Garrido J, Ojkic N, Khanna K, Wagner FR, Villa E, Endres RG & Pogliano K, (2018) Chromosome Translocation Inflates Bacillus Forespores and Impacts Cellular Morphology. Cell 172: 758–770 e714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney PT, Driks A & Eichenberger P, (2012) The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J, Wang X, Serrano M, Henriques A & Moran C, (2008) A channel connecting the mother cell and forespore during bacterial endospore formation. Proc Natl Acad Sci 105: 15100–15105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Gutierrez J, Pogliano K & Dworkin J, (2010) Cell wall synthesis is necessary for membrane dynamics during sporulation of Bacillus subtilis. Mol Microbiol 76: 956–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlot C & Rodrigues CD, (2018) The New Kid on the Block: A Specialized Secretion System during Bacterial Sporulation. Trends Microbiol 26: 663–676. [DOI] [PubMed] [Google Scholar]
- Morlot C, Uehara T, Marquis KA, Bernhardt TG & Rudner DZ, (2010) A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis. Genes Dev 24: 411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng YK, Ehsaan M, Philip S, Collery MM, Janoir C, Collignon A, Cartman ST & Minton NP, (2013) Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles. PLoS One 8: e56051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocadello S, Minasov G, Shuvalova LS, Dubrovska I, Sabini E & Anderson WF, (2016) Crystal Structures of the SpoIID Lytic Transglycosylases Essential for Bacterial Sporulation. J Biol Chem 291: 14915–14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojkic N, Lopez-Garrido J, Pogliano K & Endres RG, (2014) Bistable forespore engulfment in Bacillus subtilis by a zipper mechanism in absence of the cell wall. PLoS Comput Biol 10: e1003912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojkic N, Lopez-Garrido J, Pogliano K & Endres RG, (2016) Cell-wall remodeling drives engulfment during Bacillus subtilis sporulation. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira FC, Saujet L, Tome AR, Serrano M, Monot M, Couture-Tosi E, Martin-Verstraete I, Dupuy B & Henriques AO, (2013) The Spore Differentiation Pathway in the Enteric Pathogen Clostridium difficile. PLoS Genet 9: e1003782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permpoonpattana P, Phetcharaburanin J, Mikelsone A, Dembek M, Tan S, Brisson M-C, La Ragione R, Brisson A, Fairweather N, Hong H & Cutting S, (2013) Functional characterization of Clostridium difficile spore coat proteins. J Bacteriol 195: 1492–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permpoonpattana P, Tolls E, Nadem R, Tan S, Brisson A & Cutting S, (2011) Surface layers of Clostridium difficile endospores. J Bacteriol 193: 6461–6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pishdadian K, Fimlaid KA & Shen A, (2015) SpoIIID-mediated regulation of sigma(K) function during Clostridium difficile sporulation. Mol Microbiol 95: 189–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J, Osborne N, Sharp MD, Abanes-De Mello A, Perez A, Sun YL & Pogliano K, (1999) A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol Microbiol 31: 1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam EE, Nock AM, Lawley TD & Shen A, (2013) SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins. J Bacteriol 195: 1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Clapham KR & Losick R, (2006) Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol Microbiol 62: 1547–1557. [DOI] [PubMed] [Google Scholar]
- Ramamurthi KS, Lecuyer S, Stone HA & Losick R, (2009) Geometric cue for protein localization in a bacterium. Science 323: 1354–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribis JW, Ravichandran P, Putnam EE, Pishdadian K & Shen A, (2017) The Conserved Spore Coat Protein SpoVM Is Largely Dispensable in Clostridium difficile Spore Formation. mSphere 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues CD, Henry X, Neumann E, Kurauskas V, Bellard L, Fichou Y, Schanda P, Schoehn G, Rudner DZ & Morlot C, (2016) A ring-shaped conduit connects the mother cell and forespore during sporulation in Bacillus subtilis. Proc Natl Acad Sci U S A 113: 11585–11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues CD, Marquis KA, Meisner J & Rudner DZ, (2013) Peptidoglycan hydrolysis is required for assembly and activity of the transenvelope secretion complex during sporulation in Bacillus subtilis. Mol Microbiol 89: 1039–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong S, Rosenkrantz MS & Sonenshein AL, (1986) Transcriptional control of the Bacillus subtilis spoIID gene. J Bacteriol 165: 771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saujet L, Pereira FC, Serrano M, Soutourina O, Monot M, Shelyakin PV, Gelfand MS, Dupuy B, Henriques AO & Martin-Verstraete I, (2013) Genome-wide analysis of cell type-specific gene transcription during spore formation in Clostridium difficile. PLoS Genet 9: e1003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Crawshaw AD, Dembek M, Monteiro JM, Pereira FC, Pinho MG, Fairweather NF, Salgado PS & Henriques AO, (2016) The SpoIIQ-SpoIIIAH complex of Clostridium difficile controls forespore engulfment and late stages of gene expression and spore morphogenesis. Mol Microbiol 100: 204–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P, (2014) Spore Resistance Properties. Microbiol Spectr 2. [DOI] [PubMed] [Google Scholar]
- Sharp MD & Pogliano K, (1999) An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc Natl Acad Sci U S A 96: 14553–14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A, Fimlaid KA & Pishdadian K, (2016) Inducing and Quantifying Clostridium difficile Spore Formation. Methods Mol Biol 1476: 129–142. [DOI] [PubMed] [Google Scholar]
- Shida T, Hattori H, Ise F & Sekiguchi J, (2001) Mutational analysis of catalytic sites of the cell wall lytic N-acetylmuramoyl-L-alanine amidases CwlC and CwlV. J Biol Chem 276: 28140–28146. [DOI] [PubMed] [Google Scholar]
- Smith K, Bayer ME & Youngman P, (1993) Physical and functional characterization of the Bacillus subtilis spoIIM gene. J Bacteriol 175: 3607–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K & Youngman P, (1993) Evidence that the spoIIM gene of Bacillus subtilis is transcribed by RNA polymerase associated with sigma E. J Bacteriol 175: 3618–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg JA & Dineen SS, (2009) Laboratory maintenance of Clostridium difficile. Curr Protoc Microbiol Chapter 9: Unit 9A 1. [DOI] [PubMed] [Google Scholar]
- Sun YL, Sharp MD & Pogliano K, (2000) A dispensable role for forespore-specific gene expression in engulfment of the forespore during sporulation of Bacillus subtilis. J Bacteriol 182: 2919–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swick MC, Koehler TM & Driks A, (2016) Surviving Between Hosts: Sporulation and Transmission. Microbiol Spectr 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan IS & Ramamurthi KS, (2014) Spore formation in Bacillus subtilis. Env Microbiol 6: 212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Isidro AL, Domingues L, Eskandarian HA, McKenney PT, Drew K, Grabowski P, Chua MH, Barry SN, Guan M, Bonneau R, Henriques AO & Eichenberger P, (2009) The coat morphogenetic protein SpoVID is necessary for spore encasement in Bacillus subtilis. Mol Microbiol 74: 634–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.