Abstract
The application of machine learning in medicine has been productive in multiple fields, but has not previously been applied to analyze the complexity of organ involvement by chronic graft-versus-host disease. Chronic graft-versus-host disease is classified by an overall composite score as mild, moderate or severe, which may overlook clinically relevant patterns in organ involvement. Here we applied a novel computational approach to chronic graft-versus-host disease with the goal of identifying phenotypic groups based on the subcomponents of the National Institutes of Health Consensus Criteria. Computational analysis revealed seven distinct groups of patients with contrasting clinical risks. The high-risk group had an inferior overall survival compared to the low-risk group (hazard ratio 2.24; 95% confidence interval: 1.36-3.68), an effect that was independent of graft-versus-host disease severity as measured by the National Institutes of Health criteria. To test clinical applicability, knowledge was translated into a simplified clinical prognostic decision tree. Groups identified by the decision tree also stratified outcomes and closely matched those from the original analysis. Patients in the high- and intermediate-risk decision-tree groups had significantly shorter overall survival than those in the low-risk group (hazard ratio 2.79; 95% confidence interval: 1.58-4.91 and hazard ratio 1.78; 95% confidence interval: 1.06-3.01, respectively). Machine learning and other computational analyses may better reveal biomarkers and stratify risk than the current approach based on cumulative severity. This approach could now be explored in other disease models with complex clinical phenotypes. External validation must be completed prior to clinical application. Ultimately, this approach has the potential to reveal distinct pathophysiological mechanisms that may underlie clusters. Clinicaltrials.gov identifier: NCT00637689.
Introduction
Stem cell transplantation is an important treatment for hematologic malignancies offering a potential cure and a treatment option for advanced disease. However, chronic graft-versus-host disease (GvHD) is a major cause of morbidity and mortality after a transplant.1 Chronic GvHD is a multisystem disease, however its current grading system categorizes disease compositely as mild, moderate or severe.2–4 The current grading system may overlook clinically relevant patterns of chronic GvHD organ scores. For example, a patient with severe skin sclerosis and a patient with highly elevated liver enzymes are both classified as having severe chronic GvHD, despite starkly different clinical manifestations of the disease.3
To date, it has not been straightforward to align the National Institutes of Health (NIH) overall severity classification system and biomarkers.5 There have been some associations between the severity of chronic GvHD, as determined by the NIH classification system (NIH-Severity) and biomarkers, but biomarkers have not been able to predict clinical outcomes as strongly in chronic GvHD as in acute GvHD.6–9 Previous analyses examined disease severity in individual organs and overall disease severity but have not combined organs for phenotypic clinical subgrouping.10 A phenotypic approach to classification has the potential to characterize the pathogenesis of chronic GvHD better. Furthermore, a computational workflow capable of analyzing patterns of chronic GvHD may also have the power to elucidate patterns in other diseases in oncology and throughout clinical medicine.
Machine learning and clustering techniques have successfully exposed patterns in medicine, including identifying breast cancer metastases and genetically targeted therapy for acute myeloid leukemia.11–15 Machine learning has the potential to find patterns in clinical data that may be missed by the human observer and traditional approaches alone.16 A potential advantage of machine learning approaches compared to traditional statistical approaches is that results can go beyond a preformed hypothesis allowing for discovery of novel associations and clusters.17 Additionally, with high-dimensional data, such as the types and grades of organ involvement in chronic GvHD, the multiple comparisons required in conventional statistics can lead to false-positives, whereas a machine learning-inspired approach allows for processing of multidimensional data.15,18,19 Furthermore, an algorithmic approach has outperformed traditional statistics in recent clinical studies.15,20
We used a computational approach to classify patients with chronic GvHD according to organ scores, identify phenotypic subgroups and stratify survival. We hypothesized that machine learning methods could identify distinct clusters of clinical phenotypes and survival patterns among patients with chronic GvHD.
Methods
Study population and chronic graft-versus-host disease assessment
Research was conducted with informed consent, Institutional Review Board approval and in accordance with the Declaration of Helsinki. The clinical data used were from 339 patients with incident chronic GvHD enrolled in the Chronic GvHD Consortium study, a pre-existing multicenter prospective observational clinical database.21 Incident disease was defined as new chronic GvHD within the 3 months preceding the first study visit and only adult patients (≥18 years of age) were included. The original cohort size was 341; three patients were excluded because of missing organ scores, leaving 339 patients in the final analysis.
Demographics and the patients’ characteristics were collected at enrollment and through abstraction from clinical charts (Online Supplementary Table S1). At enrollment, NIH 2005 consensus criteria scores from 0 (no involvement) to 3 (severely affected) were recorded for eye, liver, joint, mouth, gastrointestinal tract and lung. Symptom-based lung scores were used in the initial analysis. The percentage of the body surface area with erythema (% erythema) was measured. Skin sclerosis and fascia were assessed using Hopkins scores.22
Machine-learning workflow
Nine organ scores were analyzed via a computational workflow consisting of visualization of t-distributed stochastic neighbor embedding (viSNE) for dimensionality reduction,18,23 self-organizing maps (FlowSOM) for patient clustering24 and marker enrichment modeling (MEM) for feature enrichment scoring25,26 (Figure 1 and Online Supplementary Figure S1). viSNE is the visualization of an algorithm called t-distributed stochastic neighbor embedding (t-SNE). Therefore, on all viSNE maps the axes are called t-SNE1 and t-SNE2.23 The machine-learning algorithms are described in detail in the Online Supplementary Methods. NIH scores were squared prior to viSNE analysis and all scores were scaled from 0-1. FlowSOM clustering was done using t-SNE axes. Skin erythema and sclerosis were analyzed as separate skin features in order to capture type of skin involvement by chronic GvHD.
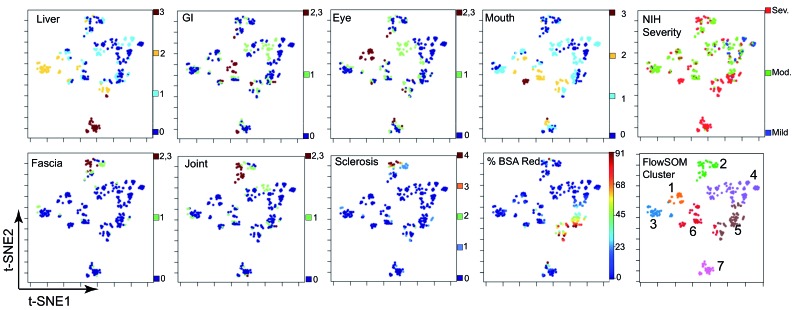
Figure 1.
A machine-learning workflow reveals clusters of patients with chronic graft-versus-host disease with shared organ involvement phenotypes. t-SNE/viSNE plots show organ scores (heat) for each patient (represented by a dot) on a scale where heat indicates organ involvement. Patients who are closer together are more similar while those who are farther apart are generally more different from each other. All organ domains shown were used to generate the viSNE plots, except National Institutes of Health-Severity which was not used as a parameter to generate the viSNE maps. FlowSOM clustering is shown (right) for the seven clusters of patients, with each cluster color overlaid as a dimension on the viSNE plot. For example, Cluster 7 is pink.
Lung scores did not contribute to patient clustering; lung was neither enriched nor negatively enriched in MEM analysis of organ scores (Online Supplementary Figure S2). Cluster stability analyses were used to determine optimal clustering parameters (Online Supplementary Methods). Analysis with lung excluded from the workflow increased cluster stability, so lung was dropped from the analysis and eight organ scores were used (Online Supplementary Figure S3). Cluster stability with six, seven and eight clusters was tested based on the appearance of seven clusters in viSNE plots (Online Supplementary Figure S4). FlowSOM was run to identify seven clusters, based on similar but increased stability with this parameter. MEM labels are reported as ▼or ▲ with OrganX where x represents a scale from −10 (most negatively enriched or ▼) to +10 (most enriched or ▲ ). Additional information on MEM and cluster stability validation is provided in the Online Supplementary Methods. De-identified data are available in FlowRepository (http://flowrepository.org/id/FR-FCM-ZYSU).
Risk analysis
Kaplan-Meier survival and Cox proportional hazards models were used to analyze overall survival as well as time from stem cell transplantation to development of chronic GvHD. The survival curve of each cluster was fitted using a Cox proportional hazards model and was compared to the survival curve of the whole cohort (Figure 2). The risk coefficient from the hazards model was used as a cluster risk score. Risk groups were stratified into low, intermediate and high based on a coefficient of risk of 0 representing the overall coefficient of risk for the whole cohort, with coefficients < −0.25 indicating low risk and coefficients >0.25 indicating high risk. Non-relapse mortality was analyzed in a competing-risk analysis with relapse as a competing risk. Additional information on the multivariate models is provided in the Online Supplementary Methods.
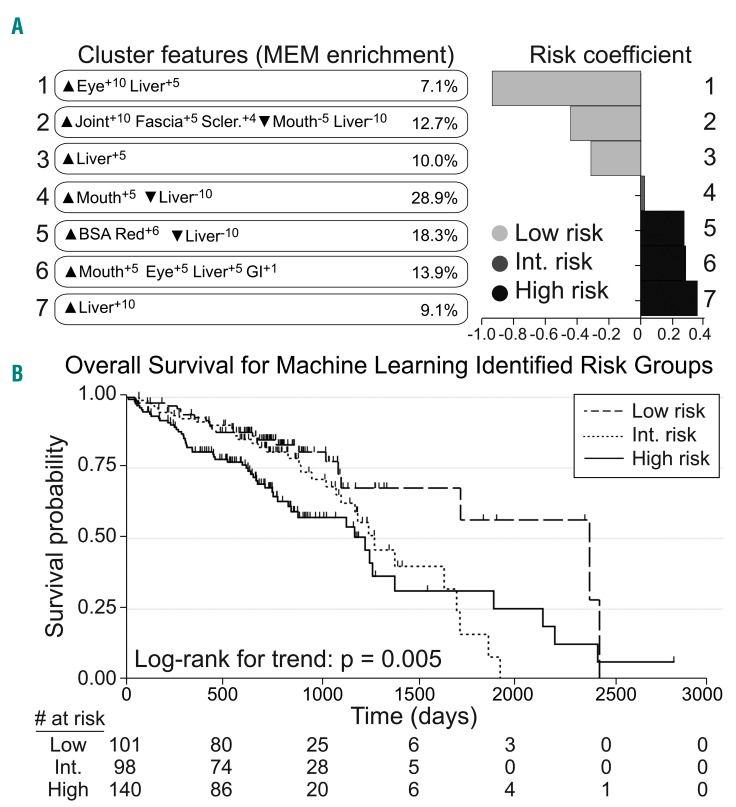
Figure 2.
Computational analysis of organ scores reveals phenotypic clusters of patients with chronic graft-versus-host disease who were stratified for overall survival. (A) Patients were grouped into seven clusters by the machine-learning workflow (Online Supplementary Figure S1) and described using marker enrichment modeling (MEM) labels (left), which captured features enriched (▲) or specifically lacking (▼) from each group relative to the others in the cohort. Risk coefficients (right) were then calculated for each group. Risk scores below −0.25 or above 0.25 were considered low and high risk, respectively, and 0 was the average risk for the cohort. Clusters 1-3 were lower risk, Cluster 4 was intermediate risk, and Clusters 5-7 were higher risk. (B) Overall survival probability was stratified for the patients with chronic graft-versus-host disease based on the low-, intermediate-, and high-risk clusters defined by the computational analysis.
Software
Analyses were conducted using Cytobank, R software version 3.4.2 for Mac, and STATA Version 14. A seed of 42 was used for the FlowSOM analyses.
Results
Patients’ organ scores
Three hundred and thirty-nine adult patients with chronic GvHD were analyzed, with predominantly intermediate (49.3%, n=167) and high (41.6%, n=141) overall NIH-Severity. Of these 339 patients, 338 had a malignancy as the indication for hematopoietic stem cell transplantation, with acute myeloid leukemia being the most common malignancy affecting 109 (32%) of the subjects. Additional characteristics are described in Online Supplementary Table S1. The organs involved by chronic GvHD at study entry by NIH criteria were the mouth (63%), gastrointestinal tract (37%), eye (43%), joint (24%), fascia (14%), skin by sclerosis (15%), skin by erythema (49%), and lung by symptom score (21%). Detailed organ scores are shown in Online Supplementary Table S2.
Unique chronic graft-versus-host disease phenotypes revealed by machine learning
Computational analysis of % erythema, eye, liver, gastrointestinal tract, fascia, joint, mouth, and sclerosis scores revealed seven groups of patients with different clinical phenotypes and risks (Online Supplementary Figure S1). viSNE analysis reduced the dimensionality of chronic GvHD organ scores, with patients who are more similar to each other shown closer together and patients who are more different from each other shown further apart on the scatterplot (Figure 1). For example, a group of patients emerged with involvement of fascia and joints as well as skin sclerosis. In FlowSOM clustering analysis, this group of patients was labeled as Cluster 2 (Figure 1).
FlowSOM clustering revealed a total of seven unique clusters of patients (Figures 1 and 2).
Cluster 1: ▲Eye+10 Liver+5 (7.1% of patients); unique in having predominantly ocular involvement, all with an NIH eye score of 3.
Cluster 2: ▲Joint+10, Fascia+5, Sclerosis+4, ▼Mouth−5, Liver−10 (12.7% of patients); a phenotype with enrichment for joint and fascia sclerosis, while specifically lacking mouth and liver GvHD.
Cluster 3: ▲Liver+5 (10.0% of patients); differentiated by moderate liver involvement, all patients with a NIH liver score of 2, while specifically lacking enrichment in other organ scores.
Cluster 4: ▲Mouth+5, ▼Liver−10 (28.9% of patients); enriched for mouth involvement, while lacking enrichment in other organ scores.
Cluster 5: ▲BSA Red+6, ▼Liver−10 (18.3% of patients); this cluster was differentiated by body surface area (BSA) involved by chronic GvHD.
Cluster 6: ▲Mouth+5, Eye+5, Liver+5, GI+1 (13.9% of patients); a phenotype enriched for mouth, eye, liver and gastrointestinal (GI) tract chronic GvHD.
Cluster 7: ▲Liver+10 (9.1% of patients); highly enriched for liver GvHD, all had NIH 3 liver scores while lacking specific involvement in other organ domains.
The meaning of positive liver enrichment differed between cluster groups. Cluster 7 differed from other clusters with liver enrichment by capturing patients with a liver score of 3 while Clusters 1, 3 and 6 had patients with liver scores of 1 and 2.
Machine-learning clusters were stable
In a cluster stability analysis involving four additional runs of viSNE and FlowSOM using the same organ features, five of the seven clusters were highly stable (Online Supplementary Figure S5). Stability was defined as having a median f-measure ≥0.85. Stable clusters had phenotypically similar MEM labels between replications of analysis as well. Clusters 2-5 and 7 were highly stable. Clusters 1 and 6 were unstable with low reproducibility between replications of analysis.
Clusters of patients identified by machine learning had different overall survival
Overall survival probability was stratified for chronic GvHD patients identified in low-risk (Clusters 1-3), intermediate-risk (Cluster 4), and high-risk groups (Cluster 5-7) defined by computational analysis (Figure 2). Time from the development of chronic GvHD to death differed between the high-risk group and the low-risk group [hazard ratio (HR)=2.24; 95% confidence interval (95% CI: 1.36-3.68); P=0.002) and between the intermediate-risk group and the low-risk group (HR=1.70; 95% CI: 0.99- 2.94; P=0.055).
Survival differences were not explained by NIH-Severity alone. When NIH-Severity was viewed on the viSNE scatter plot, clusters varied in NIH-Severity. For example, Cluster 2 patients had a combination of moderate and severe chronic GvHD (Figure 1). Additionally, when overall survival of all patients was stratified by NIH-Severity in a Kaplan-Meier analysis, NIH-Severity did not significantly stratify overall survival (log-rank for trend: P=0.08) (Online Supplementary Figure S6).
A physician-driven decision tree recapitulates machine-learning clusters
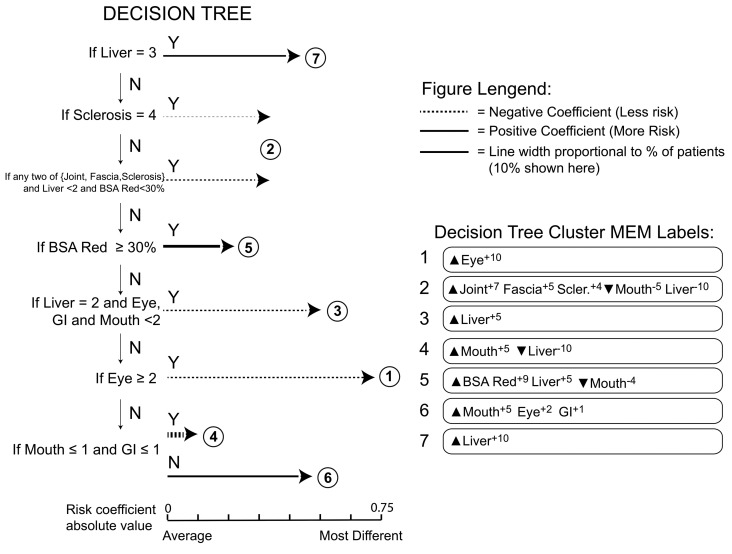
To test clinical applicability, a decision tree was developed to classify patients into the seven clusters (Figure 3). The decision tree was based on expert physicians’ interpretation of the organs that were found together in the machine-learning workflow. The decision tree was constructed through observation of viSNE scatter plots and MEM labels from the clusters of patients identified by the machine learning (Figures 1 and 2A). Patients’ outcomes were not considered in developing the decision tree. This decision tree asks a series of seven questions and can phenotype patients in as few as one question for patients in Cluster 7.
Figure 3.
A simple, physician-driven decision tree defines chronic graft-versus-host disease phenotypes. A decision tree designed to separate patients into groups with similar phenotypes and clinical risks as those revealed by the machine-learning approach in Figure 1 is shown. The decision tree is read from the top down and sequentially identifies and segregates patients in the most phenotypically distinct clusters (Y=Yes, N=No). Patients meeting the criteria at the decision point are assigned to that cluster and patients who do not meet the criteria are further advanced in the tree logic. Each circled number represents a cluster of patients. For cluster 2, two decision points were used to identify patients (arrows above and below the encircled 2). The length of the horizontal arrow is proportional to the risk coefficient and the width of the arrow is proportional to the percentage of patients in this cohort who were assigned to the cluster.
The decision tree successfully identified the seven clusters of patients, with highly similar phenotypes to those of the original analysis (Figure 3). Specifically, Clusters 3, 4 and 7 had identical phenotypes by MEM labels when compared with the original machine-learning analysis (Figure 2). The remaining clusters had similar MEM labels to those of the original machine-learning analysis.
The decision tree stratifies patients’ outcomes independently of NIH-Severity
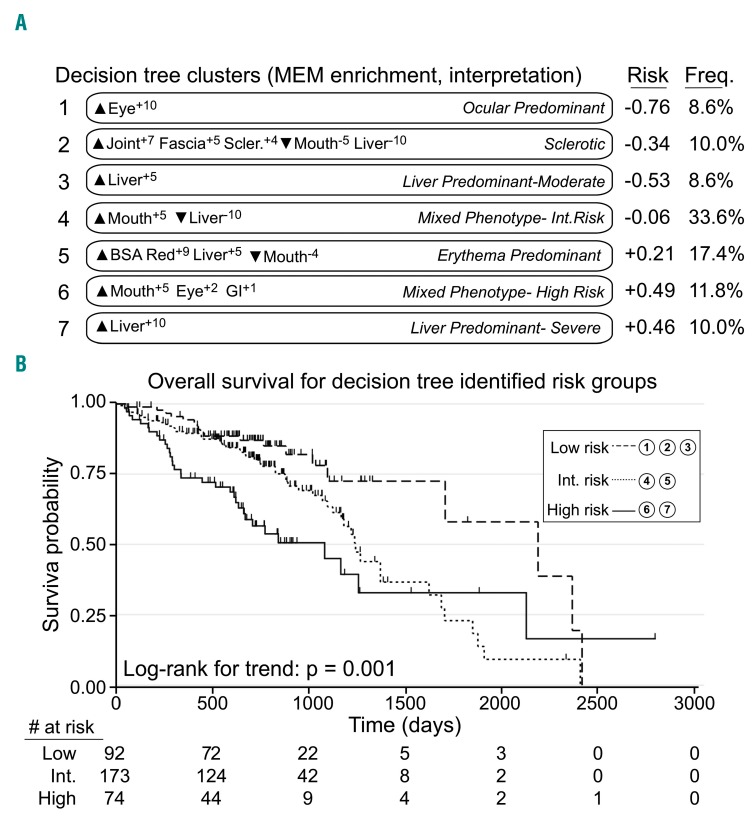
Decision-tree-determined risk groups stratified survival. Patients in decision-tree-derived Clusters 1 (ocular predominant phenotype), 2 (sclerotic phenotype) and 3 (liver predominant-moderate phenotype) were classified as low risk based on Cox proportional hazards risk coefficients (Figure 4). Patients in decision-tree-derived Clusters 4 (mixed-phenotype intermediate risk) and 5 (erythema predominant phenotype) were classified as intermediate risk, while patients in Clusters 6 (mixed phenotype-high risk phenotype) and 7 (liver predominant-severe phenotype) were classified as high risk. Patients in the high- and intermediate-risk groups had significantly shorter overall survival than those in the low-risk group (HR=2.79; 95% CI: 1.58-4.91; P<0.001 and HR=1.78; 95% CI: 1.06-3.01; P=0.03, respectively (Figure 4). Decision-tree-determined cluster risk groups were also significantly associated with non-relapse mortality (P=0.03).
Figure 4.
A simple, physician-driven decision tree created groups of patients with chronic graft-versus-host disease that were similar to computational patient clusters and stratified for overall survival. (A) Cluster numbers, newly calculated marker enrichment modeling (MEM) labels, phenotype interpretations (italics), risk coefficients, and group frequencies (n=339) are shown for the new groups of patients defined using the decision tree in Figure 3. MEM labels and risk were calculated as before (Figure 1 and Methods). Phenotype interpretations were assigned by expert physicians based on analysis of MEM labels and risk. Decision tree groups 1-3 were lower risk, groups 4-5 were intermediate risk, and groups 6-7 were higher risk. (B) Overall survival probability was stratified for patients with chronic graft-versus-host disease identified in the low-, intermediate-, and high-risk groups defined by the physician-driven decision tree.
In a multivariate Cox proportional hazards model for overall survival, decision-tree-identified risk groups and platelet counts from 0-590 days were associated with survival (intermediate-risk: HR=1.83; 95% CI; P=0.03, high-risk: HR=2.65; 95% CI: 1.42-4.94; P=0.002; platelet count: HR=3.10; 95% CI: 1.77-5.42; P<0.0001). NIH-Severity was not predictive of survival (moderate: HR=1.49; 95% CI: 0.66-3.38; P=0.34; severe: HR=1.71; 95% CI: 0.75-3.90; P=0.20). A model of decision-tree risk group and NIH-Severity alone showed no statistically significant interaction between these variables. The association between platelet counts and machine-learning-defined clusters is illustrated in Online Supplementary Figure S7.
Individual decision-tree clusters had differential disease trajectories
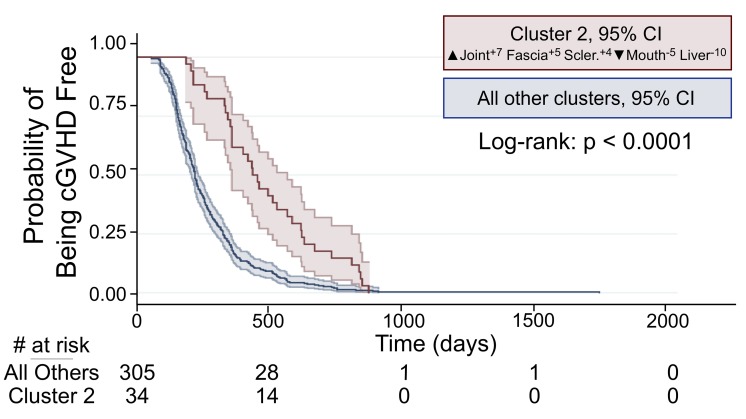
Outcomes and clinical trajectories in the decision-tree-identified clusters were compared. Patients in Cluster 2, a sclerotic phenotype with ▲Joint+7, Fascia+5, Sclerosis+4, ▼ Mouth−5, Liver−10, accounting for 10% of patients, had a significantly longer time from stem cell transplantation to chronic GvHD onset (log-rank: P<0.0001) (Figure 5).
Figure 5.
Time from stem cell transplantation to chronic graft-versus-host disease in decision tree Cluster 2 versus other clusters. Patients in decision-tree-identified Cluster 2-sclerotic phenotype had a significantly longer time from stem cell transplantation to chronic graft-versus-host disease (cGvHD) when compared to patients in all other clusters.
Worse overall survival was observed for patients in the decision-tree-derived Cluster 7, a liver predominant-severe phenotype, ▲Liver+10 (HR=1.72; 95% CI: 1.01-2.93; P=0.04) compared with patients in other clusters. Cluster 6, a mixed phenotype, ▲Mouth+5 Eye+2 GI+1, was a novel group with worse overall survival, found after ruling out the other phenotypes in the decision tree (HR=1.75; 95% CI: 1.02-2.98; P=0.04).
Decision-tree reliability and cluster-risk stability
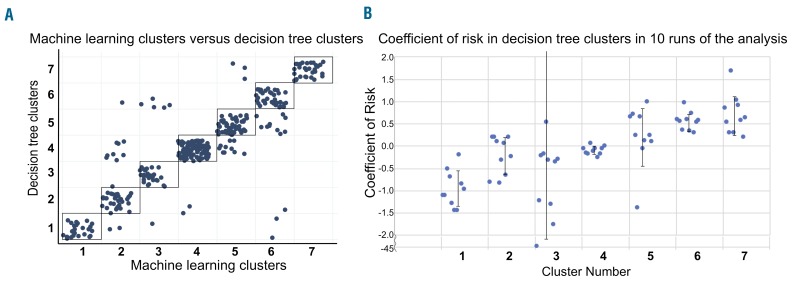
There was 86.1% concordance between clusters identified through machine learning and those identified through the decision tree (Figure 6). Bootstrapping indicated stability of risk coefficients in all but one cluster, with all clusters, except Cluster 3, having a standard deviation of risk coefficients <0.7 on ten runs of analysis (Figure 6).
Figure 6.
The physician-driven decision tree recapitulates the machine-learning workflow and finds clusters with stable risk. (A) A scatter plot shows the same patients in groups resulting from the decision tree (y-axis) or computational analysis (x-axis). Patients within or touching the black boxes were those with the same group classification in both workflows (86% of patients, n=339). (B) Bootstrapping analysis revealed stability of cluster risk across ten decision-tree analysis runs using 130 of 339 randomly sampled patients. The coefficient of risk was calculated for each run of the analysis for each cluster. The standard deviation of the ten coefficients of risks was calculated and was <0.7 for all clusters, except Cluster 3.
Discussion
Seven unique chronic GvHD patients’ phenotypes were revealed through a machine-learning workflow and successfully recapitulated with a clinically applicable decision-tree tool. The revealed groups of patients were stratified for overall survival and a unique sclerotic phenotype with different time from stem cell transplantation to development of chronic GvHD was found. The clusters of patients we describe may overcome the limitations of the current NIH classification system of disease severity which does not account for combinations of organ involvement and did not stratify survival in this cohort.
The process of applying this computational workflow to chronic GvHD patients yielded clinically applicable insights. Training analyses revealed that symptom-based lung score did not contribute to clustering and that cluster stability was improved without the lung score (Online Supplementary Figures S2 and S3). In the NIH symptom-based lung score, a score from 0-3 is assigned based on the degree of activity needed to cause dyspnea with a requirement for oxygen being scored 3.3 The fact that this symptom-based lung score did not contribute to patient clustering may be due to the subjective nature of the score and suggests that it reflects overall well-being rather than organ-specific involvement. However, it is important to note that the NIH symptom-based lung score has been associated with patients’ outcomes, including non-relapse mortality and overall survival, in an analysis that also included chronic GvHD Consortium patients.27
Clusters of patients identified by the computational workflow were associated with different clinical risk, demonstrated by differences in overall survival. Clusters of patients in the high-risk group were enriched for skin and liver involvement. A skin score of 3 and liver score of 3 have previously been shown to be associated with non-relapse mortality in an analysis that included patients in this cohort.10
Groups identified by the decision tree continued to stratify survival, with patients in the intermediate-risk group having a 1.8-fold higher risk of mortality compared to those in the low-risk group and patients in the high-risk group having a 2.8-fold higher risk of mortality. Individual high-risk clusters, i.e., Clusters 6 and 7, also independently stratified overall survival when identified by the decision tree. Importantly, the decision tree stratified risk of mortality independently of previously defined risk factors for chronic GvHD, including NIH-Severity. Notably, platelet count was a risk factor that continued to stratify risk significantly. Overall, the decision tree has the potential to be applied in the clinical setting to assess patients’ phenotypes, once further validation in prospective, independent cohorts has been completed. Additionally, this decision tree can be applied in the research setting to large cohorts of patients.
Disease trajectory differed in the decision-tree-identified clusters, most notably for Clusters 2, 6 and 7. The time from stem cell transplantation to development of chronic GvHD was different in Cluster 2, a sclerotic phenotype. This is a clinically relevant and potentially biologically distinct cluster of patients. Longer time to chronic GvHD development is a known clinical finding in patients with sclerotic chronic GvHD.5,28 Previous work defined patients with sclerotic chronic GvHD as having at least one of the following: sclerosis, fascia or joint involvement.29,30 This literature did not comment on the sclerotic phenotype as one with “de-enrichment” of liver and mouth involvement or take into account the combination of multiple sclerotic features.29,30 The combination of enriched and de-enriched features we describe may enable better association with biomarkers and treatment response.
Cluster 6, a mixed phenotype, high-risk cluster, was a novel high-risk cluster revealed by the decision tree. This cluster was defined by enrichment for mouth, eye, and gastrointestinal tract involvement. Notably, this cluster required the highest number of questions on the decision tree to reach, indicating that it was poorly defined and required that other clusters were ruled out to find patients in this phenotypic group. Patients in this cluster had significantly worse overall survival when compared to all those in all other clusters combined. A caveat is that, in stability analysis of the machine-learning workflow, Cluster 6 was not highly stable, but it did recur through all repetitions of analysis (Online Supplementary Figure S5). The combination of these areas of organ involvement has not been previously cited as a risk factor for adverse outcomes in chronic GvHD and should be further explored through cellular analyses for biomarkers and evaluated in continued validation cohorts.
Patients in Cluster 7 derived from the decision tree, a liver predominant-severe phenotype, also had a different disease trajectory when compared to patients in other clusters in that they had a significantly worse overall survival than patients in all other clusters combined. This decision-tree-derived cluster is supported by previous research showing that severe elevation of liver enzymes is a known risk factor for adverse outcomes in chronic GvHD.10
Prognostication by clustering is distinct from prognostication by individual organ scores alone. For example, in the machine-learning analysis, Cluster 5 lacked liver involvement and was a high-risk cluster, while high-risk Cluster 6 and Cluster 7 were specifically enriched for liver involvement. This supports the concept that this single organ score does not confer unidirectional low or high risk within the clusters. Furthermore, Liver+5 enrichment was seen in multiple low-risk clusters and one high-risk cluster. Clustering is unique in that it is not an individual organ score or characteristic but rather combinations of organ involvement and the specific absence of organ involvement that drive cluster formation and likely prognosis. Another example of this is that mouth enrichment was seen in both an intermediate-risk cluster (Cluster 4) and high-risk cluster (Cluster 6). Cluster 6, a high-risk cluster, comprises mouth, eye and liver enrichment; these individual enrichment types appear in low-risk clusters but it is perhaps the combination that makes this a high-risk cluster. However, we cannot rule out that gastrointestinal tract enrichment, uniquely present in Cluster 6, is not the driving force of adverse outcomes.
A limitation of the machine-learning approach is that it is not possible to add new patients to this analysis without shifting the current clusters. This was overcome by the decision-tree approach. Validation with an external cohort as well as comparison with other risk stratification tools for chronic GvHD31 should further strengthen the findings of the computational and decision-tree analyses. We were unable to analyze whether clusters predicted response to therapy, as this was an observational cohort in which patients were on any systemic therapy at study entry. Thus, treatment response is an outcome of interest in assessing the utility of machine learning for chronic GvHD outcome stratification. An external validation cohort is pending for this analysis. External validation of machine-learning approaches is the gold standard, and external validation is necessary prior to clinical application of the findings.
These results have the potential to be applied to stratify risk in the clinical setting, enhance the current chronic GvHD classification system, refine inclusion criteria for phase 2 trials, and guide biomarker discovery for more specific therapeutic targets. The distillation of machine-learning knowledge into a decision tree increases the feasibility of clinical application of the clusters. However, the clusters have not been externally validated, and this step should be explored before clinical application.
Lastly, this a flexible machine learning-inspired work- flow with numerous potential applications. The stability of the clusters suggests that this approach will be highly useful in revealing groups not only for this disease but for others that have complex phenotypes. Although the endpoint for this analysis was overall survival, this workflow could be applied to explore whether clusters of patients differ in treatment response or composite chronic GvHD endpoints, such as failure-free survival. Additionally, this workflow has the potential to be applied to other human diseases with complex classification systems such as myelodysplastic syndrome and brain tumors. This approach may change the classification of human disease by revealing otherwise unapparent, clinically relevant patterns.
Supplementary Material
Acknowledgments
This work was supported by funding from The Vanderbilt Medical Scholars Program and Vanderbilt Ingram Cancer Center. We thank Dr. Yan Guo for helpful discussions, Allison Greenplate and Benjamin Reisman for helpful insight on figure design, and Dr. Paul Martin for his critical review of the manuscript.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/1/189
References
- 1.Arora M, Cutler CS, Jagasia MH, et al. Late Acute and chronic graft-versus-host disease after allogeneic hematopoietic Cell Transplantation. Biol Blood Marrow Transplant. 2016;22(3):449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Socié G, Ritz J. Current issues in chronic graft-versus-host disease. Blood. 2014;124(3):374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filipovich AH, Weisdorf D, Pavletic S, et al. National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease: I. Diagnosis and Staging Working Group report. Biol Blood Marrow Transplant. 2005;11(12):945–956. [DOI] [PubMed] [Google Scholar]
- 4.Jagasia MH, Greinix HT, Arora M, et al. National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease: I. The 2014 Diagnosis and Staging Working Group report. Biol Blood Marrow Transplant. 2015;21(3):389–401.e381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooke KR, Luznik L, Sarantopoulos S, et al. The biology of chronic graft-versus-host disease: a task force report from the National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease. Biol Blood Marrow Transplant. 2017;23(2): 211–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu J, Storer BE, Kushekhar K, et al. Biomarker panel for chronic graft-versus-host disease. J Clin Oncol. 2016;34(22):2583–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartwell MJ, Ozbek U, Holler E, et al. An early-biomarker algorithm predicts lethal graft-versus-host disease and survival. JCI Insight. 2017;2(3):e89798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Major-Monfried H, Renteria AS, Pawarode A, et al. MAGIC biomarkers predict long-term outcomes for steroid-resistant acute GVHD. Blood. 2018;131(25):2846–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paczesny S, Krijanovski OI, Braun TM, et al. A biomarker panel for acute graft-versus-host disease. Blood. 2009;113(2):273–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inamoto Y, Martin PJ, Storer BE, et al. Association of severity of organ involvement with mortality and recurrent malignancy in patients with chronic graft-versus-host disease. Haematologica. 2014;99(10): 1618–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ting DSW, Cheung CY, Lim G, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318(22):2211–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Liang R, Zhou T, et al. Identification and validation of asthma phenotypes in Chinese population using cluster analysis. Ann Allergy Asthma Immunol. 2017;119(4):324–332. [DOI] [PubMed] [Google Scholar]
- 14.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318(22):2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SI, Celik S, Logsdon BA, et al. A machine learning approach to integrate big data for precision medicine in acute myeloid leukemia. Nat Commun. 2018;9(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deo RC. Machine learning in medicine. Circulation. 2015;132(20):1920–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diggins KE, Ferrell PB, Jr, Irish JM. Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods. 2015;82:55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Maaten LHG. Visualizing high-dimensional data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 19.Saeys Y, Van Gassen S, Lambrecht BN. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat Rev Immunol. 2016;16(7):449–462. [DOI] [PubMed] [Google Scholar]
- 20.Singal AG, Mukherjee A, Elmunzer BJ, et al. Machine learning algorithms outperform conventional regression models in predicting development of hepatocellular carcinoma. Am J Gastroenterol. 2013;108(11):1723–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chronic GVHD Consortium. Rationale and design of the chronic GVHD cohort study: improving outcomes assessment in chronic GVHD. Biol Blood Marrow Transplant. 2011;17(8):1114–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inamoto Y, Pidala J, Chai X, et al. Joint and fascia manifestations in chronic graft-versus-host disease and their assessment. Arthritis Rheumatol. 2014;66(4):1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amir E-aD, Davis KL, Tadmor MD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nature Biotechnology. 2013;31(6):545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Gassen S, Callebaut B, Van Helden MJ, et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015;87(7):636–645. [DOI] [PubMed] [Google Scholar]
- 25.Diggins KE, Greenplate AR, Leelatian N, Wogsland CE, Irish JM. Characterizing cell subsets using marker enrichment modeling. Nat Methods. 2017;14(3):275–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diggins KE, Gandelman JS, Roe CE, Irish JM. Generating quantitative cell identity labels with marker enrichment modeling (MEM). Curr Protoc Cytom. 2018;83: 10.21.11–10.21.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer J, Williams K, Inamoto Y, et al. Pulmonary symptoms measured by the national institutes of health lung score predict overall survival, nonrelapse mortality, and patient-reported outcomes in chronic graft-versus-host disease. Biol Blood Marrow Transplant. 2014;20(3):337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitko CL, White ES, Baird K. Fibrotic and sclerotic manifestations of chronic graft versus host disease. Biol Blood Marrow Transplant. 2012;18(1 Suppl):S46–S52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inamoto Y, Storer BE, Petersdorf EW, et al. Incidence, risk factors, and outcomes of sclerosis in patients with chronic graft-versus-host disease. Blood. 2013;121(25):5098–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inamoto Y, Martin PJ, Flowers MED, et al. Genetic risk factors for sclerotic graft-versus-host disease. Blood. 2016;128(11):1516–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arora M, Klein JP, Weisdorf DJ, et al. Chronic GVHD risk score: a Center for International Blood and Marrow Transplant Research analysis. Blood. 2011;117(24): 6714–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.