Abstract
MicroRNAs, non-coding regulators of gene expression, are likely to function as important downstream effectors of many transcription factors including MYB. Optimal levels of MYB are required for transformation/maintenance of BCR-ABL-expressing cells. We investigated whether MYB silencing modulates microRNA expression in Philadelphia-positive (Ph+) leukemia cells and if MYB-regulated microRNAs are important for the “MYB addiction” of these cells. Thirty-five microRNAs were modulated by MYB silencing in lymphoid and erythromyeloid chronic myeloid leukemia-blast crisis BV173 and K562 cells; 15 of these were concordantly modulated in both lines. We focused on the miR-17-92 cluster because of its oncogenic role in tumors and found that: i) it is a direct MYB target; ii) it partially rescued the impaired proliferation and enhanced apoptosis of MYB-silenced BV173 cells. Moreover, we identified FRZB, a Wnt/β-catenin pathway inhibitor, as a novel target of the miR-17-92 cluster. High expression of MYB in blast cells from 2 Ph+leukemia patients correlated positively with the miR-17-92 cluster and inversely with FRZB. This expression pattern was also observed in a microarray dataset of 122 Ph+acute lymphoblastic leukemias. In vivo experiments in NOD scid gamma mice injected with BV173 cells confirmed that FRZB functions as a Wnt/β-catenin inhibitor even as they failed to demonstrate that this pathway is important for BV173-dependent leukemogenesis. These studies illustrate the global effects of MYB expression on the microRNAs profile of Ph+cells and supports the concept that the “MYB addiction” of these cells is, in part, caused by modulation of microRNA-regulated pathways affecting cell proliferation and survival.
Introduction
The Philadelphia chromosome (Ph) is the typical chromosomal abnormality of chronic myeloid leukemia (CML) patients.1 It is also detected in a subset of B-cell acute lymphoblastic leukemia (ALL), and less frequently in acute myeloid (AML) and mixed-phenotype acute (MPAL) leukemias.1 The hallmark of the Ph chromosome is the translocation of the proto-oncogene ABL1 from chromosome 9 to the breakpoint cluster region gene (BCR) on chromosome 22, generating the BCR-ABL1 fusion gene. Such a gene encodes the p190, p210 or the p230 BCR-ABL1 isoforms; these chimeric proteins have constitutively active tyrosine kinase activity and promote the aberrant activation of signaling pathways causing enhanced cell proliferation and resistance to cell death.2 We identified several transcription factors (TFs) whose expression/activity is regulated by BCR-ABL1 oncoproteins and is required for BCR-ABL1-dependent leukemogenesis.3–6 One such TF is MYB, the prototypical TF of the Myb family,7 essential for fetal and adult hematopoiesis8,9 and required for colony formation of myeloid leukemia blasts, a subset of T-cell leukemia, and BCR-ABL1-transformed myeloid and B cells.6,10–12 In vitro and in mice, BCR-ABL1-transformed cells are more dependent on MYB expression than their normal counterparts,6,12 supporting the concept that certain leukemic cells are “addicted” to MYB.10,11,13 This concept was validated in MLL-AF9-associated AML where partial and transient MYB suppression phenocopies MLL-AF9 withdrawal, eradicating aggressive AML in vivo without preventing normal myelopoiesis.14
MicroRNAs (miRNAs) are small molecules of approximately 22 nucleotides that reprogram gene expression, promoting mRNA degradation and blocking mRNA translation.15 MiRNAs may be especially important in regulating the expression of TFs such as MYB that has distinct biological effects in normal hematopoiesis and in leukemic cells based on its expression levels.15,16 Regulation of MYB expression through miRNAs has been reported previously.17–20 Levels of MYB expression may be differentially controlled by multiple miRNAs and, conversely, MYB could control the expression of different miRNAs9,17–21 to execute lineage-specific developmental choices at critical junctions during hematopoiesis. In particular, overexpression of miR-15 reduced MYB levels in vitro, suppressing erythroid and myeloid colony formation.17 MYB is a direct target of miR-150, playing a key role at different stages of B-cell development.18,20
To gain more information on the role of MYB-regulated miRNAs in leukemic cells, we investigated changes in miRNA levels induced by MYB silencing in Philadelphia-positive (Ph+) cells. We found that, upon MYB silencing, 15 miRNAs are modulated in K562 and in BV173 Ph+ cells. Among these, the miR-17-92 cluster was regulated transcriptionally by MYB through binding to its 5’ regulatory region. Restoring miR-17-92 expression in MYB-silenced BV173 cells partly rescued the reduced proliferation and enhanced apoptosis of these cells. The reduced expression of the miR-17-92 cluster in MYB-silenced Ph+ cells was associated with upregulation of FRZB, an inhibitor of the Wnt/β-catenin pathway, critical for the maintenance of BCR-ABL1-transformed stem cells.22
Methods
Cell lines
Philadelphia-positive BV173, SUP-B15 and K562 cells were used for the experiments performed in this study.
Culture condition, infection with viral vectors to obtain derivative cell lines, transfection, microarray and transcriptional profiling, cell proliferation, cell viability, cell cycle analysis, apoptosis assays, western blotting, RNA isolation and analysis by quantitative real-time PCR (qRT-PCR), chromatin immunoprecipitation (ChIP) assays and luciferase assay techniques are all described in the Online Supplementary Methods and Online Supplementary Table S1.
Details of statistical/bioinformatic analysis are also described in the Online Supplementary Appendix.
Patients
Bone marrow cells were obtained, after informed consent, from 2 Ph+ patients, one with CML-blast crisis with the p210 BCR-ABL isoform, and another with a de novo ALL with the p190 BCR-ABL isoform. In both cases, no additional chromosomal abnormalities were detected by cytogenetic analysis.
The study was approved by the Ethical Committee of the Regina Elena National Cancer Institute of Rome, in compliance with the Declaration of Helsinki.
In vivo studies assessing the effects of ectopic FRZB expression
Mice were injected in the tail vein with 2×106 BV173-ShMYB 7TFP pUltra-Empty Vector (EV) cells or BV173-ShMYB 7TFP pUltra-hot-FRZB cells (FRZB). Five weeks after the injection, the percentage of circulating leukemia cells was assessed by flow cytometry detection of peripheral blood GFP+mCherry+ cells using the LSR-Fortessa. Mice were sacrificed when moribund and the survival time recorded. For in vivo β-catenin activity analysis, 106 GFP+mCherry+ cells (estimated by flow cytometry) were purified from the bone marrow or the spleen of a mouse injected with EV-transduced or FRZB-expressing BV173 cells, lysed and analyzed for luciferase activity by using the Dual Luciferase Reporter Assay System (Cat. # E1910) and the signal was acquired using a Zylux Femtomaster FB 12 luminometer.
Details of the in vivo studies are available in the Online Supplementary Appendix.
Results
Differential expression of microRNAs in MYB-silenced Philadelphia-positive leukemic cells
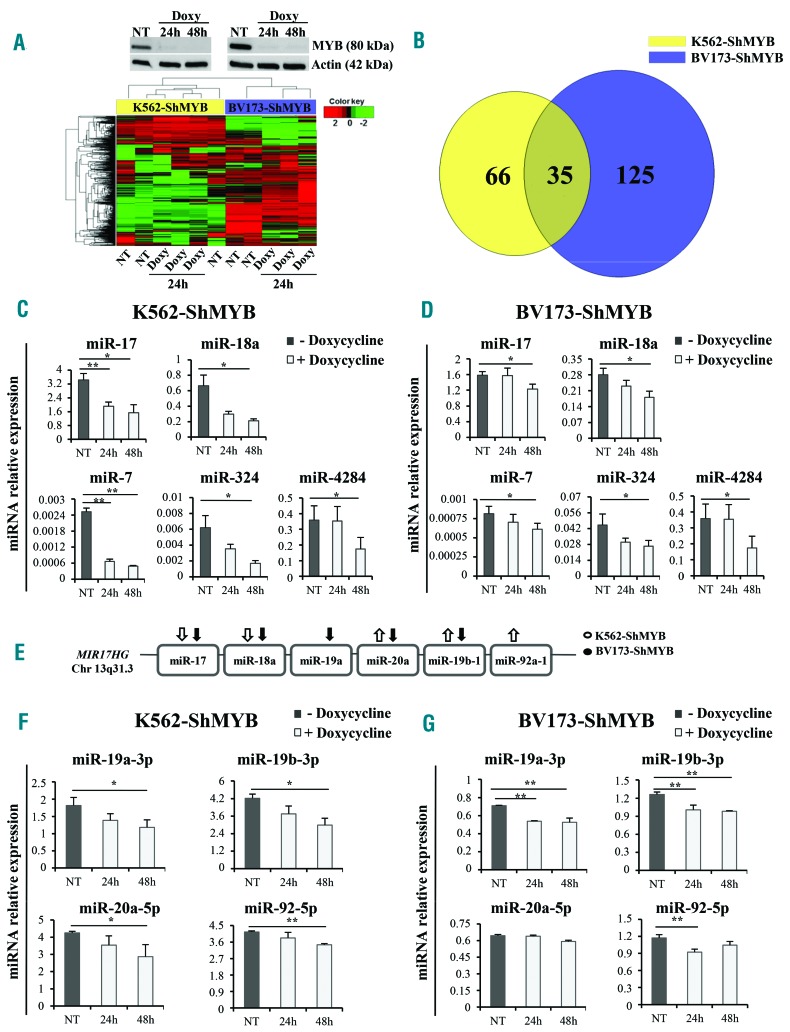
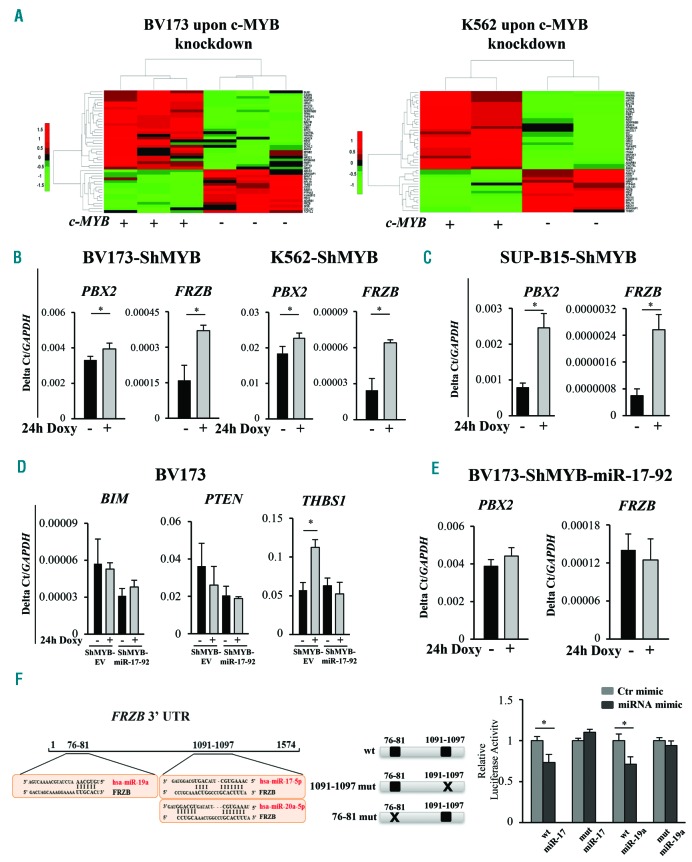
We showed previously that optimal levels of MYB expression are required for transformation and maintenance of BCR-ABL-expressing cells.6,12 Since miRNAs are exquisite regulators of gene expression, it is likely that MYB-regulated miRNAs are important for the “MYB addiction” of BCR-ABL-transformed cells. To this end, we performed microarray hybridization studies on RNA from the CML-lymphoid blast crisis BV173 and CML-erythromyeloid blast crisis K562 Ph+ cell lines transduced with the doxycycline (Doxy)-inducible lentiviral vector pLVTSH-MYB ShRNA (BV173-ShMYB and K562-ShMYB).23 Compared to untreated (not treated; NT) control cells, Doxy treatment essentially abolished MYB expression in BV173- and K562-ShMYB cells (Figure 1A, upper panel). Unsupervised hierarchical clustering analysis shows expression levels of 519 miRNAs in NT and Doxy-treated [24 hours (h)] BV173- and K562-ShMYB cells (Figure 1A, lower panel). Of these, 125 and 66 were differentially expressed (P≤0.05) in MYB-silenced BV173- and K562-ShMYB cells, respectively (Figure 1B). Of the 35 miRNAs whose expression was altered in both Ph+ cell lines, 15 were modulated concordantly (Online Supplementary Table S2) and 20 discordantly in the two lines (Online Supplementary Table S3).
Figure 1.
miRNA expression profile of MYB-silenced Philadelphia-positive (Ph+) leukemia cells and expression levels of miR-17-92 cluster members. (A) (Upper panels) Western blots of a representative experiment showing specific knockdown of MYB in Doxycycline (Doxy)-treated cells; (lower panel) heat map of differentially expressed miRNAs in Doxy-treated [24 hours (h)] K562-ShMYB and BV173-ShMYB cells. MiRNA expression levels are shown as color variations. Higher and lower values are represented by red and green points, respectively. Pairwise distances between rows and between columns were computed by Euclide distance metric. (B) Venn diagram of differentially expressed miRNAs: 35 miRNAs are commonly modulated in the indicated cell lines. (C and D) qRT-PCR of 5 selected miRNAs from the 15 miRNAs modulated in the same direction in untreated (Not Treated; NT) or Doxy-treated (24-48 h) K562- and BV173-ShMYB cells. Samples were normalized for RNU44 expression. Relative expression was calculated using the comparative Ct method. Data are the average of three independent experiments; error bars indicate SEM. P-values (*P≤0.05; **P≤0.01) were determined using Student t-test. (E) Schematic representation of members of miR-17-92 cluster included in the MIR17HG gene on Chr13q31.3. Arrows represent the direction of miRNA modulation based on the microarray experiment in K562-ShMYB (white) and BV173-ShMYB (black). (F and G) qRT-PCR of the indicated members of miR-17-92 cluster in NT or Doxy-treated (24-48 h) K562-ShMYB and BV173-ShMYB cells. Samples were normalized for RNU44 expression. QRT-PCR was performed in triplicate, including no-template controls. Relative expression was calculated using the comparative Ct method. Data are the average of three independent experiments; error bars indicate Standard Error of Mean. P-values were determined using Student t-test.*P≤0.05; **P≤0.01.
Real-time PCR analysis of differentially expressed miRNAs in doxycycline-treated BV173-, SUP-B15- and K562-ShMYB cells
To validate the results of the miRNA microarray analysis, expression levels of 5 miRNAs (miR-17, miR-18a, miR-7, miR-324 and miR-4284) down-regulated by MYB silencing in both cell lines were assessed by qRT-PCR. These miRNAs were selected based on the fold change of their expression in MYB-silenced cells and their role in tumors.24–28 In agreement with the microarray data, expression of all 5 miRNAs was significantly down-regulated after 48 h Doxy treatment (Figure 1C and D). Of note, levels of miR-17 and miR-7 were significantly down-regulated in K562-ShMYB cells after 24 h Doxy treatment (Figure 1C). Since miR-17 and miR-18a belong to the miR-17-92 cluster (Figure 1E) which is involved in BCR-ABL-dependent transformation,29 we also assessed levels of cluster members miR-19a-3p,-19b-3p,-20a-5p and miR-92-5p. These miRNAs were among those expressed in both cell lines in our microarray assay (Online Supplementary Tables S3 and S4). In contrast with the microarray data, qRT-PCR analysis showed that levels of all 4 miRNAs were down-regulated in Doxy-treated (48 h) K562-ShMYB cells, whereas only the expression of miR-19a-3p, miR-19b-3p and miR-92-5p (24 h) was decreased in Doxy-treated BV173-ShMYB cells (Figure 1F and G). These conflicting results may depend on the greater sensitivity of the qRT-PCR compared to the microarray assay. We also assessed the effects of MYB silencing on the expression of the miR-17-92 cluster in the Ph+ ALL cell line SUP-B15 which expresses the p190 BCR-ABL isoform. In this line, Doxy treatment (24 and 48 h) to silence MYB expression induced a statistically significant decrease of miR-17, miR-18a, miR-19a and miR-19b levels (Online Supplementary Figure S1). Specificity of the effects of MYB silencing on the expression of the miR-17-92 cluster were demonstrated by using a BV173 derivative line expressing a mutant MYB cDNA harboring synonymous point mutations in the sequence targeted by the MYB shRNA (shRNA-resi tant MYB BV173 cell line). Upon Doxy treatment to silence endogenous MYB expression, we found that, in contrast to the parental line (BV173-ShMYB), expression of members of miR-17-92 cluster was not modulated in the BV173 line expressing the MYB cDNA not targetable by the MYB ShRNA (Online Supplementary Figure S2). Thus, Doxy-induced changes in the expression of the miR-17-92 cluster are a specific consequence of MYB silencing.
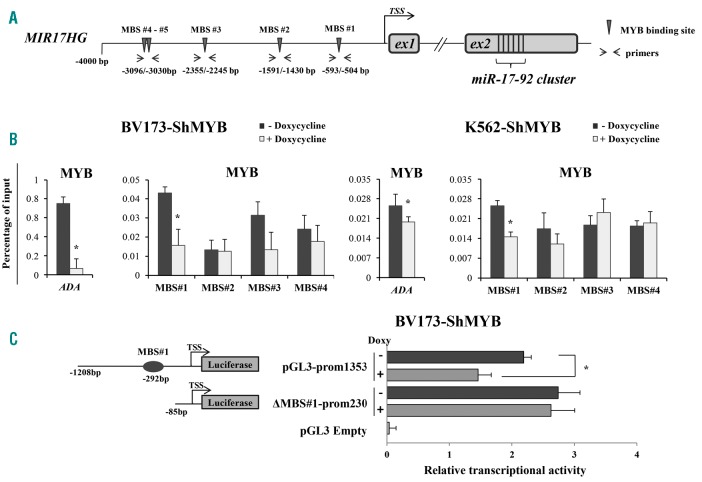
MYB binds the promoter of the miR-17-92 cluster
To investigate whether MYB could directly regulate transcription of the miR-17-92 cluster, we analyzed the MIR17HG promoter for the presence of putative MYB binding sites (MBS). Using MatInspector (www.genomatix.de/matinspector.html), we scanned 4000 bp upstream of the MIR17HG gene and identified several putative MBS (Figure 2A). We focused on 5 MBS with the highest matrix similarity score (Online Supplementary Table S5). Genomic positions of these MBSs relative to MIR17HG Transcriptional Start Site (TSS) are indicated in Figure 2A. To assess whether MYB binds these regions in vivo, ChIP assays were performed in NT and Doxy-treated BV173- and K562-ShMYB cells and de-cross-linked DNA amplified with primers flanking genomic regions that include putative MBS (Figure 2A). As a positive control, ChIP was performed using an MBS-containing segment of the adenosine deaminase gene (ADA), a known transcriptional target of c-MYB.30 MYB bound efficiently, in both untreated cell lines, to the promoter region that includes MBS#1, the site closest to the TSS of MIR17HG (Figure 2B); in contrast, reduced binding was detected at all other promoter segments (Figure 2B), especially in BV173 cells. Binding of MYB to the region of the miR-17-92 promoter that includes MBS#1 was markedly decreased upon Doxy treatment (72 h) of BV173- and K562-ShMYB cells (Figure 2B). As expected, MYB binding to the ADA promoter was also decreased (Figure 2B). To further investigate whether the miR-17-92 cluster is directly regulated by MYB we carried out luciferase assay using reporter plasmids with or without MBS#1 (PGL3-prom1353 and ΔMBS#1-prom230, respectively) (Figure 2C, left). We found that the luciferase activity of the ShMYB-BV173 cells transfected with the PGL3-prom1353 was decreased by approximately 33% after a 24 h Doxy treatment to silence MYB expression; in contrast, in cells transfected with the truncated ΔMBS#1-prom230 plasmid lacking MBS#1 there was only a 4% decrease of luciferase activity after Doxy treatment (Figure 2C, right). These data strongly suggest that MYB is important for the transcription of the MIR17HG locus.
Figure 2.
MYB binding to the promoter of the miR-17-92 cluster. (A) Schematic representation of 4000 bp regulatory regions upstream of the MIR17HG promoter. A transcription start site (TSS) is indicated. Arrows indicate the promoter region amplified by the specific primer pair used for qPCR amplification of immunoprecipitated chromatin. (B) ChIP analysis of the MIR17HG promoter using the indicated MYB antibody in untreated (Not Treated; NT) or Doxycycline (Doxy)-treated BV173-ShMYB and K562-ShMYB cells. Results of qPCR are analyzed with the comparative Ct method. Values of each immunoprecipitated sample are expressed as percentage relative to their respective input and by subtracting the values obtained in the negative controls (no antibody). Error bars represent Standard Error of Mean (SEM) (n=3); P-values (*P≤0.05) were determined using Student t-test. (C) (Left panel) Schematic representation of the reporter plasmids containing the MYB binding site (MBS) #1 (pGL3–prom1353) or its deletion mutant without the MBS#1 (ΔMBS#1-prom230). (Right panel) Dual luciferase assay performed in untreated or Doxy-treated BV173-ShMYB cells transfected with the pGL3–prom1353 or the ΔMBS#1-prom230 plasmid. Promoter activity of each plasmid was determined 48 hours (h) after transfection. Luciferase activity values were normalized for transfection efficiency according to the activity of a co-transfected Renilla luciferase plasmid. Data are the average of three independent experiments performed in duplicate; error bars indicate Standard Error of Mean (*P≤0.05).
Involvement of the miR-17-92 cluster in the “MYB addiction” of Ph+ leukemia cells
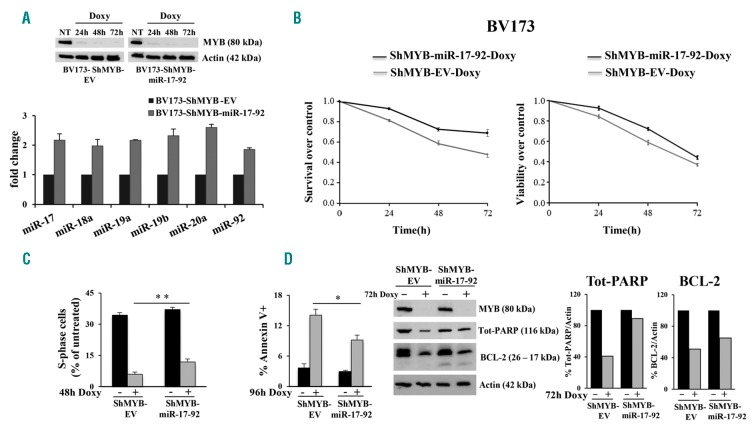
To investigate whether restoring expression of the miR-17-92 cluster affects the phenotype of MYB-silenced cells, we generated BV173-ShMYB cells over-expressing the miR-17-92 cluster and assessed proliferation and survival of these cells upon MYB silencing. These studies were not performed in K562 cells because the biological effects induced by MYB silencing in these cells were modest, compared to those in BV173 cells. Expression of MYB was suppressed in Doxy-treated BV173-ShMYB cells and in the miR-17-92 derivative line which exhibited increased expression of each member of the miR-17-92 cluster (Figure 3A). Compared to BV173-ShMYB-EV cells, the miR-17-92 over-expressing cell lines showed increased proliferation (P≤0.01) upon MYB silencing. This was evident after 24 h of Doxy treatment and persisted at 48 h and 72 h (Figure 3B, left). Likewise, viability of Doxy-treated BV173-ShMYB cells over-expressing the miR-17-92 cluster was significantly increased (P≤0.01) compared to that of Doxy-treated BV173-ShMYB-EV cells (Figure 3B, right). DNA content analysis revealed that Doxy-treated BV173-ShMYB cells over-expressing miR-17-92 have a greater proportion of S-phase cells than Doxy-treated BV173-ShMYB-EV cells (12% vs. 6% after 48 h Doxy treatment) (Figure 3C). In addition, cultures of Doxy-treated BV173-ShMYB over-expressing miR-17-92 cells had less apoptosis than Doxy-treated BV173-ShMYB-EV cells, as indicated by the lower frequency of Annexin V-positive cells (9% vs. 15%, after 96 h Doxy treatment) (Figure 3D, left) and the increased expression of uncleaved PARP and BCL-2 (48% and 14%, respectively) (Figure 3D, middle and right panels).
Figure 3.
Biological effects of over-expressed miR-17-92 cluster in MYB silenced Philadelphia-positive (Ph+) BV173 cells. (A) (Upper panel) Western blots of a representative experiment showing specific knockdown of MYB in Doxycycline (Doxy)-treated [24, 48 and 72 hours (h)] BV173-ShMYB cells; (lower panel) qRT-PCR of the indicated members of the miR-17-92 cluster in BV173-ShMYB-Empty Vector (EV) and the miR-17-92 over-expressing cells. Results are expressed as fold changes [mean±Standard Error of Mean (SEM) from three independent experiments] in miRNA expression in BV173-ShMYB-miR-17-92 cells as compared with values in BV173-ShMYB-EV cells. (B) MTT and ATPlite assays; data are the average of three independent experiments, and percentage of cell survival (left panel) and cell viability (right panel) were assessed at the indicated times of Doxy treatment. (C) Percentage of S-phase cells over control for untreated or Doxy-treated (48 h) BV173-ShMYB-EV and derivative miR-17-92 over-expressing lines (**P≤0.01). (D) (Left panel) Percentage of Annexin V for untreated or Doxy-treated (96 h) BV173-ShMYB-EV and derivative miR-17-92 over-expressing lines (*P≤0.05). (Middle panel) Western blot of a representative experiment of MYB, uncleaved PARP, BCL-2 and actin protein levels in BV173-ShMYB-EV and BV173-ShMYB-miR-17-92 over-expressing cells, 72 h after MYB silencing. (Right panel) Densitometric analysis by imageJ software. Actin was used as loading control within the same sample and expressed as fold changes compared to control.
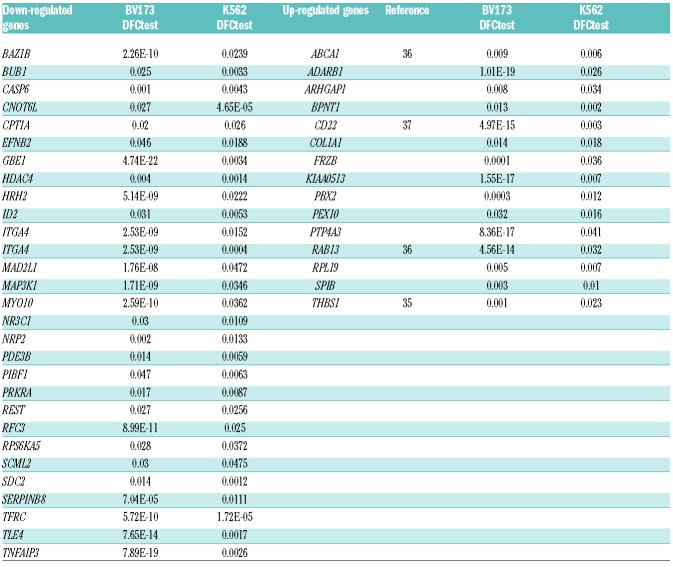
Integrative analysis of gene expression profiles of MYB-silenced cells and predicted miRNA-regulated genes identifies novel putative miR-17-92 targets
We used gene expression profiling of MYB-silenced cells to identify MYB target genes potentially regulated by the miR-17-92 cluster. The miRWalk 2.0 database was used to investigate potential interactions of the miR-17-92 cluster with genes regulated by MYB silencing in BV173 and K562 cells. From this analysis, we found that 44 genes modulated by MYB silencing (15 up-regulated and 29 down-regulated) are predicted targets of the miR-17-92 cluster (Table 1 and Figure 4A). We focused on the up-regulated genes since the decreased expression of the miR- 17-92 cluster in MYB-silenced cells should increase the levels of its putative targets. Thus, we performed qRT-PCR to assess the expression of two candidate targets, PBX2 and FRZB, involved in the regulation of proliferation and apoptosis.31,32 Such analysis revealed a statistically significant (P≤0.05) increase of PBX2 and FRZB expression in MYB-silenced Ph+ ALL BV173 and SUP-B15 or K562 cells (Figure 4B and C).
Table 1.
Predicted down-and up-regulated target genes in BV173-ShMYB and K562-ShMYB cells after gene expression and miR-17-92 cluster analyses.
Figure 4.
Transcriptional analysis and evaluation of mRNA expression levels of miR-17-92 cluster target genes. (A) Unsupervised hierarchical clustering of common deregulated genes from gene expression analysis of parental and MYB-silenced BV173 and K562 cells. (B and C) qRT-PCR of PBX2 and FRZB expression levels upon MYB knockdown [24 hours (h)] of the indicated Philadelphia-positive (Ph+) ShMYB cell lines. Results are mean of three experiments. Error bars indicate Standard Error of Mean (SEM). (D) Analysis of mRNA expression levels, using SYBR Green-based qRT-PCR, of BIM, PTEN and THBS1 in untreated and Doxy-treated BV173-ShMYB-Empty Vector (EV) and ShMYB-miR-17-92 cells. Results are mean of three experiments. Error bars indicate SEM. (E) Quantification by SYBR Green-based qRT-PCR of PBX2 and FRZB mRNA in untreated and Doxy-treated BV173-ShMYB-miR-17-92 cells. Values are reported as 2-ΔCt. GAPDH gene expression was used as endogenous control. Error bars indicate SEM (n=3). (F) (Left panel) Schematic representation of 3’UTRs of FRZB gene with putative binding sites for miR-17-92 cluster. (Right panel) Schematic representation of reporter plasmids containing the wild-type (wt) or mutant (76-81 mut, 1091-1097 mut of miR-17-92-binding sequences) FRZB 3’UTR. Dual Luciferase assay in recipient cells co-transfected with luciferase reporter vectors containing the wt-3’UTR FRZB or the indicated FRZB mutant and either the hsa-miR-17, the hsa-miR-19a mimics or a control (Ctr)-mimic RNA. Firefly luciferase activity of each sample was normalized by Renilla luciferase activity. Results are expressed as fold activation relative to the basal activity of the control mimic (ctr-mimic). (*P≤0.05). The normalized luciferase activity, set as mean of at least three independent experiments performed in duplicate, is shown. Error bars represent the mean±SEM (n=3).
FRZB is a potential effector of the miR-17-92 cluster in the “MYB addiction” of Ph+ leukemia cells
The oncogenic effect of the miR-17-92 cluster is caused by the co-operation of its members in targeting tumor-suppressive pathways.28,33 Several studies have shown that the miR-17-92 cluster directly targets “pro-apoptotic” genes such as Phosphatase and tensin homolog (PTEN), the apoptosis facilitator BCL2L11 (BIM) and the anti-angiogenic factor thrombospondin-1 (THBS1) in normal lymphopoiesis,34–37 in MYC-driven lymphomas38 and in immunodeficiency or lymphoproliferative states.39 To assess whether the expression of validated miR-17-92 targets is modulated by miR-17-92 overexpression, qRT-PCR experiments were performed in Doxy-treated BV173-ShMYB-EV cells and in the miR-17-92 over-expressing line. After 24 h Doxy treatment, levels of BIM and PTEN mRNA were essentially identical in both BV173-ShMYB- EV and BV173-ShMYB-miR-17-92 over-expressing cells compared to those in NT cells (Figure 4D). In contrast, THBS1 mRNA levels showed an increase (P≤0.05) in Doxy-treated BV173-ShMYB-EV cells compared to untreated cells, and such an increase was blocked by over- expression of the miR-17-92 cluster (Figure 4D). Expression levels of BIM, PTEN and THBS1 mRNA, after 24 h Doxy treatment, were assessed also in the SUP-B15 ShMYB cells analysis. This revealed a statistically significant (P≤0.05) increase of BIM and THBS1 in MYB-silenced SUP-B15 cells compared to untreated cells (Online Supplementary Figure S3).
The expression of p21 and E2F1 genes, two other experimentally validated miR-17-92 targets,40 was also assessed in MYB-silenced BV173 cells. MYB silencing induced an increase in the expression of p21 but this increase was not blocked by overexpression of the miR-17-92 cluster. In contrast, expression of E2F1 was down-modulated after MYB silencing and was not affected by overexpression of the miR-17-92 cluster (Online Supplementary Figure S4). These results suggest that MYB silencing modulates p21 and E2F1 expression independently of its effect on the miR-17-92 cluster expression.
Since our goal was to investigate novel miR-17-92 targets, potentially involved in the “MYB addiction” of Ph+ leukemia cells, we focused on FRZB because ectopic expression of the miR-17-92 cluster blocked the increased expression of FRZB mRNA but not of PBX2 mRNA (Figure 4E) induced by MYB silencing in BV173-ShMYB cells (Figure 4B). FRZB is the founding member of the secreted Frizzled-related protein (SFRP) family of Wnt inhibitors32,41 and suppresses Wnt signaling thus preventing the accumulation of β-catenin into the nucleus.42 Then, we used miRwalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mir-walk2/), TargetScan 5.2 (http://www.targetscan.org), and miRanda (http://www.microrna.org/microrna) algorithms to assess the presence of putative miR-17-92-binding sites within the 3’untranslated region (3’UTR) of FRZB-mRNA. This analysis identified one putative miR-17-92 binding site for miR-19a (seed sequences: 76-81 bp) and one for miR-17 and -20a (seed sequences:1091-1097 bp) (Figure 4F, left panel). To assess whether FRZB is a direct target of miR-17-92, a human FRZB 3’UTR fragment containing wild-type or mutated miR-17 or miR-19a seed sequences (Figure 4F, middle panel) was cloned downstream of the firefly luciferase reporter gene and co-transfected with miR-17 or miR-19a mimics in 293T cells. The relative luciferase activity of the reporter with wild-type 3’UTR was decreased by 27% upon expression of the miR-17 mimic and by 29% upon expression of the miR-19a mimic; in contrast, there was no decrease in luciferase activity of the mutant reporter (Figure 4F, right panel), suggesting that FRZB is a direct target of miR-17 as well as of miR-19a.
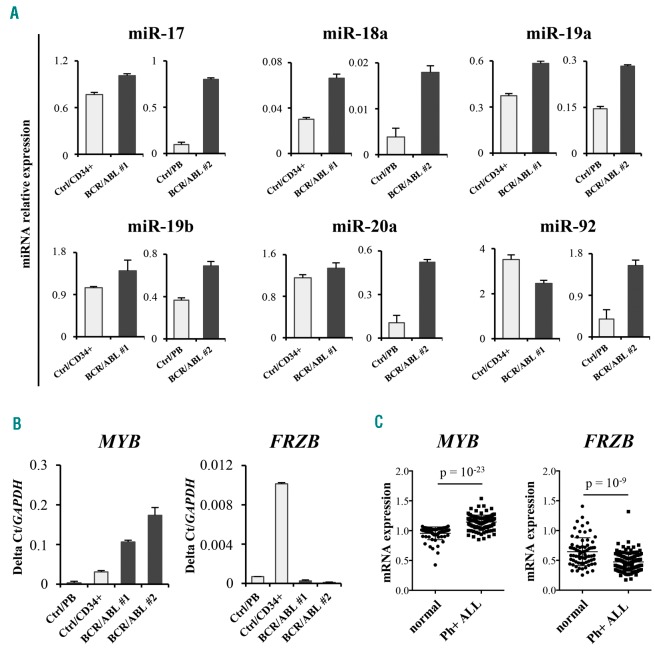
To investigate whether FRZB has a role as a miR-17-92 target gene in the “MYB addiction” of BCR-ABL-transformed cells, we assessed the relative expression of FRZB, the miR-17-92 cluster and MYB in blast cells from 2 Ph+ leukemia patients (n=1: p210BCR/ABL CML-myeloid blast crisis; n=1: p190BCR/ABL ALL). High expression of the miR-17-92 cluster correlated with that of MYB and was more abundant than in CD34+ or peripheral blood mononuclear cells from healthy donors (Figure 5A and B, left panel). In contrast, levels of FRZB were much higher in cells from healthy donors than in blast cells from the Ph+ leukemia patients (Figure 5B, right panel). In agreement with these findings, we found that, in a microarray dataset of 122 Ph+ ALL samples, MYB mRNA levels were more abundant in Ph+ ALL cells compared to normal B cells, while the opposite was found for FRZB expression (Figure 5C).
Figure 5.
Expression of the miR-17-92 cluster and its target FRZB correlates with MYB levels in Philadelphia-positive (Ph+) acute lymphoblastic leukemia (ALL) cells. (A) MiR-17-92 expression levels evaluated by stem-loop qRT-PCR in primary leukemia cells (Patient 1: p210BCR/ABL chronic myeloid leukemia (CML)-myeloid blast crisis) compared to normal CD34+ cells from a healthy subject [Control (Ctrl) CD34+] and Patient 2 (p190BCR/ABL ALL) compared to normal peripheral blood mononuclear (PBMC) cells (Ctrl/PB). Samples were normalized for RNU44 small-nucleolar RNA expression using the comparative Ct method. Data are the average of three experiments; error bars indicate Standard Deviation (SD). (B) mRNA quantification of MYB and FRZB, by SYBR Green-based qRT-PCR, in Patient 1 (p210BCR/ABL CML-myeloid blast crisis) and Patient 2 (p190BCR-ABL ALL) compared to normal CD34+ cells and PBMC cells from healthy donors (Ctrl/CD34+ and Ctrl/PB), respectively. Values are reported as 2-ΔCt normalizing to GAPDH gene expression. (C) mRNA expression by microarray of MYB or FRZB in normal B cells or Ph+ ALL cells. (Values represent the sum of all probes signals for each gene and are derived from dataset GSE13159).
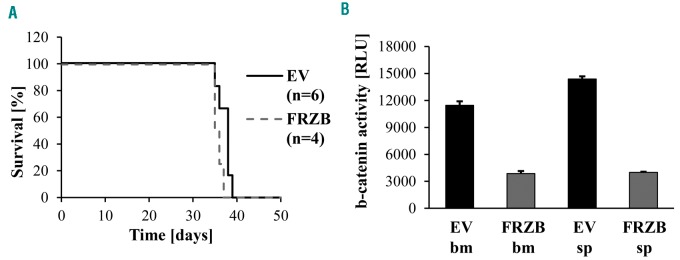
To investigate directly whether expression of FRZB has a negative effect for leukemia development, NOD scid gamma (NSG) mice were injected with EV-transduced or FRZB-expressing BV173 cells carrying the β-catenin-Luc reporter plasmid and assessed for overall survival. Survival of the two groups was identical (Figure 6A); however, β-catenin activity was markedly reduced in BV173 cells isolated from the bone marrow or spleen of a mouse injected with FRZB-expressing compared to EV-trans-duced cells (Figure 6B). These data suggest that leukemia induced by Ph+ BV173 cells is β-catenin-independent but do not exclude the possibility that FRZB-dependent regulation of β-catenin activity is important for leukemia induced by primary Ph+ ALL cells.
Figure 6.
Effect of FRZB expression on leukemogenesis and β-catenin activity of Philadelphia-positive (Ph+) BV173 cells. (A) Survival of mice injected with 2×106 BV173-ShMYB 7TFP pUltra-Empty Vector (EV) or BV173-ShMYB 7TFP pUltra-hot-FRZB cells (FRZB). (B) Luciferase reporter assay for β-catenin activity in GFP+ cells isolated from the bone marrow (bm) or spleen (sp) of a NOD scid gamma (NSG) mouse injected with (EV)- or FRZB-BV173 cells and sacrificed when terminally ill.
Discussion
The expression of MYB is critical for the proliferation and survival of many leukemic cells, including BCR-ABL1-transformed myeloid and lymphoid cells;6,12 however, the mechanisms responsible for the “MYB addiction” of these cells are only partially understood.
In this study, we assessed the miRNA expression profile of MYB-silenced BV173 and K562 CML-blast crisis cells with the goal of identifying miRNAs whose modulation might explain the impaired proliferation and survival associated with MYB knockdown in BCR-ABL1-transformed lymphoid or myeloid precursors.6,12 Interestingly, MYB appears to have broad effects, directly or indirectly, on the levels of miRNAs since approximately 24% and 13% of those expressed in BV173 and K562 cells, respectively, were modulated by MYB silencing. Although many miRNAs regulated by MYB exhibited changes in both cell lines, a high number of the modulated miRNAs exhibited cell-type specificity.
We speculated that those modulated by MYB in a cell-type specific manner may regulate pathways required for more specialized cell functions, while those regulated in both cell lines may be involved in more general biological processes, such as cell proliferation and survival. Within the miRNAs regulated by MYB in both cell lines, we focused on the miR-17-92 cluster because of its oncogenic role in many tumors,28,43,44 its involvement in BCR-ABL1-transformed cells,45 and its regulation by MYC,44 a known MYB target.46 We found that MYB bound directly to the miR-17-92 promoter, suggesting that its effects on the expression of several members of the miR-17-92 cluster are direct, although an indirect effect through other transcription factors (eg. c-Myc) and/or co-activators cannot be excluded.47 On the other hand, silencing MYB alone does not abolish expression of the miR-17-92 cluster, suggesting that other transcription factors also regulate the expression of the miR-17-92 cluster in BCR-ABL1-transformed cells.16 Compared to control cells, MYB-silenced BV173 cells exhibit a marked inhibition of cell growth which is due to cell-cycle arrest and induction of apoptosis.48 Thus, we asked whether restoring expression of the miR-17-92 cluster would rescue the impaired growth of MYB-silenced BV173 cells. Ectopic expression of the miR-17-92 cluster caused an increase in the S phase fraction and a decrease in the apoptosis of MYB-silenced BV173 cells, but the effect was modest. This is not surprising, since silencing MYB expression induces global changes in miRNA and mRNA levels causing an impaired proliferation and survival that cannot be rescued by expression of the miR-17-92 cluster alone. The expression of some established targets of the miR-17-92 cluster (e.g. p21 and E2F1) was also markedly modulated by MYB silencing; however, restoring the targets of the miR-17-92 cluster did not change the effects on such expression induced by MYB silencing, strongly suggesting that the predominant mechanism of MYB regulation of these two genes is miR-17-92-independent. In contrast, ectopic expression of miR-17-92 completely blocked the upregulation of THBS1, a known miR-17-92 target,37 and of FRZB, a novel candidate for miR-17-92 inhibition, which is induced by MYB silencing. FRZB functions as an inhibitor of the Wnt/β-catenin signaling pathway which is activated in CML stem cells/early progenitors and is important for their proliferation and survival.22,42 However, ectopic expression of FRZB in BV173 cells, when injected in NSG mice, had no effect on their survival, in spite of a marked inhibition of β-catenin activity.
These data suggest that BV173 cells induce leukemia in mice through β-catenin-independent mechanisms but do not exclude the possibility that FRZB-dependent regulation of β-catenin activity is important for leukemia induced by primary Ph+ ALL cells.
In summary, this study illustrates the global effects of MYB expression on the miRNA profile of Ph+ leukemic cells and supports the concept that the “MYB addiction” of Ph+ BV173 cells is, in part, caused by modulation of miRNA-regulated pathways affecting cell proliferation and survival.
Supplementary Material
Acknowledgments
This article is dedicated to the memory of Professor Franco Mandelli for his life-long dedication to research on hematological malignancies and patients’ care. “The results achieved are not a treasure to be defended but a wealth to be transmitted”. The authors would like to thank Dr Scott M. Hammond from University of North Carolina for a kind gift of the MIR17HG promoter plasmids and Dr. Thomas Gonda for the pLVTSH ShMYB lentivirus.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/1/82
Funding
This work was supported, in part, by NCI grant CA167169 to BC, Italian Association for Cancer Research (AIRC) grant to GB and by Funds Celgene protocol (08/CE/R/15) to FP. GR is a PhD student at University of Rome “Sapienza” and an Intramural funds (Hematologic Tumors) fellow. MS is a recipient of an Intramural fellowship.
References
- 1.Kang ZJ, Liu YF, Xu LZ, et al. The Philadelphia chromosome in leukemogenesis. Chin J Cancer. 2016;35:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li S, Ilaria RL, Jr, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999; 189(9):1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perrotti D, Cesi V, Trotta R, et al. BCR-ABL suppresses C/EBPalpha expression through inhibitory action of hnRNP E2. Nat Genet. 2002;30(1):48–58. [DOI] [PubMed] [Google Scholar]
- 4.Soliera AR, Mariani SA, Audia A, et al. Gfi-1 inhibits proliferation and colony formation of p210BCR/ABL-expressing cells via transcriptional repression of STAT 5 and Mcl-1. Leukemia. 2012;26(7):1555–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soliera AR, Lidonnici MR, Ferrari-Amorotti G, et al. Transcriptional repression of c-Myb and GATA-2 is involved in the biologic effects of C/EBPalpha in p210BCR/ABL-expressing cells. Blood. 2008;112(5):1942–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lidonnici MR, Corradini F, Waldron T, Bender TP, Calabretta B. Requirement of c-Myb for p210 (BCR/ABL)-dependent transformation of hematopoietic progenitors and leukemogenesis. Blood. 2008; 111(9):4771–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008; 8(7):523–534. [DOI] [PubMed] [Google Scholar]
- 8.Lieu YK, Reddy EP. Conditional c-myb knockout in adult hematopoietic stem cells leads to loss of self-renewal due to impaired proliferation and accelerated differentiation. Proc Natl Acad Sci USA. 2009; 106(51): 21689–21694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianchi E, Bulgarelli J, Ruberti S, et al. MYB controls erythroid versus megakaryocyte lineage fate decision through the miR-486-3p-mediated downregulation of MAF. Cell Death Differ. 2015;22(12):1906–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calabretta B, Sims RB, Valtieri M, et al. Normal and leukemic hematopoietic cells manifest differential sensitivity to inhibitory effects of c-myb antisense oligodeoxynu-cleotides: an in vitro study relevant to bone marrow purging. Proc Natl Acad Sci USA. 1991;88(6):2351–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lahortiga I, De Keersmaecker K, Van Vlierberghe P, et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat Genet. 2007;39(5):593–595. [DOI] [PubMed] [Google Scholar]
- 12.Waldron T, De Dominici M, Soliera AR, et al. c-Myb and its target Bmi1 are required for p190BCR/ABL leukemogenesis in mouse and human cells. Leukemia. 2012;26(4):644–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ratajczak MZ, Hijiya N, Catani L, et al. Acute- and chronic-phase chronic myelogenous leukemia colony-forming units are highly sensitive to the growth inhibitory effects of c-myb antisense oligodeoxynucleotides. Blood. 1992;79(8):1956–1961. [PubMed] [Google Scholar]
- 14.Zuber J, Rappaport AR, Luo W, et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011;25(15):1628–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El Gazzar M, McCall CE. MicroRNAs regulatory networks in myeloid lineage development and differentiation: regulators of the regulators. Immunol Cell Biol. 2012;90(6): 587–593. [DOI] [PubMed] [Google Scholar]
- 16.Organista-Nava J, Gomez-Gomez Y, Illades- Aguiar B, Leyva-Vazquez MA. Regulation of the miRNA expression by TEL/AML1, BCR/ABL, MLL/AF4 and TCF3/PBX1 onco-proteins in acute lymphoblastic leukemia (Review). Oncol Rep. 2016;36(3):1226–1232. [DOI] [PubMed] [Google Scholar]
- 17.Zhao H, Kalota A, Jin S, Gewirtz AM. The c-myb proto-oncogene and microRNA-15a comprise an active autoregulatory feedback loop in human hematopoietic cells. Blood. 2009;113(3):505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao C, Calado DP, Galler G, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007; 131(1):146–159. [DOI] [PubMed] [Google Scholar]
- 19.Fahl SP, Crittenden RB, Allman D, Bender TP. c-Myb is required for pro-B cell differentiation. J Immunol. 2009;183(9):5582–5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia P, Frampton J. Hematopoietic lineage commitment: miRNAs add specificity to a widely expressed transcription factor. Dev Cell. 2008;14(6):815–816. [DOI] [PubMed] [Google Scholar]
- 21.Vargova K, Curik N, Burda P, et al. MYB transcriptionally regulates the miR-155 host gene in chronic lymphocytic leukemia. Blood. 2011;117(14):3816–3825. [DOI] [PubMed] [Google Scholar]
- 22.Hu Y, Chen Y, Douglas L, Li S. Beta-Catenin is essential for survival of leukemic stem cells insensitive to kinase inhibition in mice with BCR-ABL-induced chronic myeloid leukemia. Leukemia. 2009; 23(1):109–116. [DOI] [PubMed] [Google Scholar]
- 23.Drabsch Y, Hugo H, Zhang R, et al. Mechanism of and requirement for estrogen-regulated MYB expression in estrogen-receptor-positive breast cancer cells. Proc Natl Acad Sci USA. 2007;104(34):13762–13767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horsham JL, Kalinowski FC, Epis MR, Ganda C, Brown RA, Leedman PJ. Clinical Potential of microRNA-7 in Cancer. J Clin Med. 2015;4(9):1668–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blume CJ, Hotz-Wagenblatt A, Hüllein J, et al. p53-dependent non-coding RNA networks in chronic lymphocytic leukemia. Leukemia. 2015;29(10):2015–23. [DOI] [PubMed] [Google Scholar]
- 26.Kuo WT, Yu SY, Li SC, et al. MicroRNA-324 in Human Cancer: miR-324-5p and miR-324-3p Have Distinct Biological Functions in Human Cancer. Anticancer Res. 2016;36(10):5189–5196. [DOI] [PubMed] [Google Scholar]
- 27.Tamaddon G, Geramizadeh B, Karimi MH, Mowla SJ, Abroun S. miR-4284 and miR-4484 as Putative Biomarkers for Diffuse Large B-Cell Lymphoma. Iran J Med Sci. 2016;41(4):334–339. [PMC free article] [PubMed] [Google Scholar]
- 28.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20(12):1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scherr M, Elder A, Battmer K, et al. Differential expression of miR-17~92 identifies BCL2 as a therapeutic target in BCR-ABL-positive B-lineage acute lymphoblastic leukemia. Leukemia. 2014;28(3):554–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ess KC, Whitaker TL, Cost GJ, Witte DP, Hutton JJ, Aronow BJ. A central role for a single c-Myb binding site in a thymic locus control region. Mol Cell Biol. 1995; 15(10):5707–5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelosi A, Careccia S, Lulli V, et al. miRNA let-7c promotes granulocytic differentiation in acute myeloid leukemia. Oncogene. 2013;32(31):3648–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Surana R, Sikka S, Cai W, et al. Secreted frizzled related proteins: Implications in cancers. Biochim Biophys Acta. 2014; 1845(1):53–65. [DOI] [PubMed] [Google Scholar]
- 33.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mavrakis KJ, Wolfe AL, Oricchio E, et al. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol. 2010;12(4):372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dews M, Homayouni A, Yu D, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38(9):1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mo W, Zhang J, Li X, et al. Identification of novel AR-targeted microRNAs mediating androgen signalling through critical pathways to regulate cell viability in prostate cancer. PLoS One. 2013;8(2):e56592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Psathas JN, Doonan PJ, Raman P, Freedman BD, Minn AJ, Thomas-Tikhonenko A. The Myc-miR-17-92 axis amplifies B-cell receptor signaling via inhibition of ITIM proteins: a novel lymphomagenic feed-forward loop. Blood. 2013;122(26):4220–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9(4):405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inomata M, Tagawa H, Guo YM, Kameoka Y, Takahashi N, Sawada K. MicroRNA-17-92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood. 2009;113(2):396–402. [DOI] [PubMed] [Google Scholar]
- 41.Leyns L, Bouwmeester T, Kim SH, Piccolo S, De Robertis EM. Frzb-1 is a secreted antagonist of Wnt signaling expressed in the Spemann organizer. Cell. 1997; 88(6):747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005; 434(7035): 843–850. [DOI] [PubMed] [Google Scholar]
- 43.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005; 435(7043):828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010; 42(8):1348–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venturini L, Battmer K, Castoldi M, et al. Expression of the miR-17-92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood. 2007;109(10):4399–4405. [DOI] [PubMed] [Google Scholar]
- 46.Cogswell JP, Cogswell PC, Kuehl WM, et al. Mechanism of c-myc regulation by c-Myb in different cell lineages. Mol Cell Biol. 1993;13(5):2858–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ji M, Rao E, Ramachandrareddy H, et al. The miR-17-92 microRNA cluster is regulated by multiple mechanisms in B-cell malignancies. Am J Pathol. 2011; 179(4):1645–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Dominici M, Porazzi P, Soliera AR, et al. Targeting CDK6 and BCL2 exploits the “MYB addiction” of Ph+ acute lymphoblastic leukemia. Cancer Res. 2018;78(4):1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.