Abstract
Stem cell grafts from 10/10 HLA-matched unrelated donors are often mismatched for HLA-DP. In some patients, donor T-cell responses targeting the mismatched HLA-DP allele(s) have been found to induce a specific graft-versus-leukemia effect without coinciding graft-versus-host disease, whereas in other cases significant graft-versus-host disease occurred. Cell-lineage-specific recognition patterns within the allogeneic HLA-DP-specific donor T-cell repertoire could explain the differential clinical effects mediated by donor T cells after HLA-DP-mismatched allogeneic stem cell transplantation. To unravel the composition of the HLA-DP T-cell repertoire, donor T-cell responses were provoked by in vitro stimulation with allogeneic HLA-DP-mismatched monocyte-derived dendritic cells. A strategy including depletion of reactivity against autologous dendritic cells allowed efficient identification and enrichment of allo-reactive T cells upon stimulation with HLA-DP-mismatched dendritic cells. In this study we elucidated that the allogeneic HLA-DP-restricted T-cell repertoire contained T cells with differential cell-lineage-specific recognition profiles. As expected, some of the allogeneic HLA-DP-restricted T cells showed broad recognition of a variety of hematopoietic and non-hematopoietic cell types expressing the targeted mismatched HLA-DP allele. However, a significant proportion of the allogeneic HLA-DP-restricted T cells showed restricted recognition of hematopoietic cells, including primary malignant cells, or even restricted recognition of only myeloid cells, including dendritic cells and primary acute myeloid leukemia samples, but not of other hematopoietic and non-hematopoietic cell types. These data demonstrate that the allogeneic HLA-DP-specific T-cell repertoire contains T cells that show restricted recognition of hematopoietic cells, which may contribute to the specific graft-versus-leukemia effect without coinciding graft-versus-host disease.
Introduction
T-cell-depleted allogeneic stem-cell transplantation (alloSCT) can be applied in the treatment of patients suffering from hematologic malignancies.1–4 The aim of this approach is to replace the hematopoietic system of the patient (including the malignant cells) with a healthy hematopoietic system reconstituted from a stem cell graft of an HLA-matched donor.5–9 The depletion of T cells from the graft reduces the occurrence of acute graft-versus-host disease (GvHD) mediated by alloreactive donor T cells targeting normal, non-hematopoietic cells of the patient.3,10–12 However, donor T cells are essential in the beneficial graft-versus-leukemia (GvL) effect and T-cell immunity is also required for anti-viral protection.3,8,13 Therefore, scheduled donor lymphocyte infusions are often given to patients following T-cell-depleted alloSCT.4,14–17
The effect of the level of HLA matching on clinical outcome after alloSCT has been extensively studied and has led to a standardized donor-patient matching procedure in which HLA-DP is frequently not taken into account.18–20 The 10/10 HLA-matched stem cell grafts from unrelated donors are, therefore, often mismatched for one or two HLA-DP alleles. HLA-DP mismatches are in theory acceptable, as under non-inflammatory conditions expression of HLA-class II molecules is mainly restricted to hematopoietic cells. The balance between the induction of GvL and GvHD is one of the remaining challenges in treatment with (T-cell-depleted) alloSCT or donor lymphocyte infusion.14 Whether or not donor T cells targeting the mismatched HLA-DP allele(s) contribute to GvL and/or GvHD may depend on the magnitude, specificity and diversity of the allo-HLA-DP immune response as was illustrated for HLA-class I-restricted T-cell responses,21 and the tissue-specific pattern of HLA-DP expression.3,14,22–25 The magnitude of the T-cell response mounted against allo-HLA-DP molecules has been shown to be different depending on the specific HLA-DP allele(s) expressed in the donor and the patient.25,26 Differences at the amino acid sequence level have been found to influence the immunogenicity of specific HLA-DP molecules, probably caused by a different landscape of peptides (peptidome) presented in these HLA-DP molecules.26–28 Based on these insights, an algorithm has been proposed to predict the risk on an immune reaction in the host versus graft direction (rejection) and the graft versus host direction (GvL and/or GvHD), based on the immunogenicity of specific HLA-DP molecules and the differences between specific HLA-DP alleles.29 This has led to the distinction of two groups of HLA-DP mismatches, called the more tolerable, permissive HLA-DP mismatches that are predicted to induce T-cell responses with a lower amplitude, and the non-permissive mismatches that induce stronger T-cell responses.29–32 In addition to the specificity and magnitude of the allo-HLA-DP T-cell response, the pattern of expression of HLA-DP on patients’ tissues is decisive in the induction of GvL and/or GvHD. In some patients, profound CD4 T-cell responses targeting the mismatched allo-HLA-DP allele(s) have been found to be associated with the induction of different types of GvHD (e.g. skin GvHD, gut GvHD) mediated by recognition of inflamed HLA-class II-expressing non-hematopoietic tissues.23 In other patients specific GvL reactivity without coinciding GvHD mediated by allo-HLA-DP-reactive CD4 donor T cells was demonstrated. In these patients the allo-HLA-DP response appeared to be restricted to hematopoietic cells without cross-reactivity against non-hematopoietic tissues.22,24
To initiate the allo-HLA-DP-specific immune response in vivo, donor T cells most likely first have to encounter patient-derived HLA-DP-expressing antigen-presenting cells, such as dendritic cells (DC).33,34 Since DC are of hematopoietic origin, allo-HLA-DP T cells activated by DC will probably recognize antigens expressed by hematopoietic cells presented in the mismatched HLA-DP molecule.14 When the allo-HLA-DP-specific immune response is initiated by DC residing in inflamed HLA-DP-expressing non-hematopoietic tissue, the immune response is likely to be also directed against antigens expressed by non-hematopoietic cells presented in the mismatched HLA-DP molecule of the DC.35 The level of overlap between the antigens expressed by hematopoietic versus non-hematopoietic cells, will dictate the induction of a specific GvL response, a specific GvHD response, or a combination of both.3,14
In this study we analyzed the tissue/cell-lineage-specific recognition patterns within the allo-HLA-DP-specific T-cell repertoire provoked by stimulation with allogeneic HLA-DP-mismatched monocyte-derived DC. We observed that the allo-HLA-restricted T-cell repertoire contains T cells with a diverse spectrum of cell-lineage-specific recognition profiles, including T cells that show restricted recognition of hematopoietic cells, including primary malignant cells, or even T cells with myeloid-lineage-restricted recognition, including recognition of primary acute myeloid leukemia blasts.
Methods
Cell collection and preparation
The collection and preparation of cells is described in the Online Supplementary Appendix.
Induction of allo-HLA-DP-specific immune responses
CD14-depleted donor peripheral blood mononuclear cells were stimulated with irradiated (25 Gy) HLA-DP mismatched allogeneic DC (alloDC) in a 10:1, responder T-cell to stimulator-cell ratio. The cells were cultured in Iscove modified Dulbecco medium (IMDM) containing 10% heat-inactivated ABOS supplemented with interleukin-7 (10 ng/mL, Miltenyi Biotec), interleukin-15 (0.1 ng/mL, Miltenyi Biotec) and interleukin-2 (50 IU/mL, Novartis Sandoz Pharmaceuticals). To activate and remove auto-reactive T cells, cell cultures were stimulated after 14 days with autologous DC (autoDC), followed by depletion of these reactive T cells around 36 h later based on activation marker (CD137) expression using CD137-allophycocyanin (BD Pharmingen, San Diego, CA, USA), APC-beads (Miltenyi Biotec) and MACS LD columns (Miltenyi Biotec) according to the manufacturer’s instructions. To activate allo-HLA-DP-reactive T cells, the negative fraction was subsequently specifically restimulated with the allogeneic HLA-DP-mismatched DC. After approximately 36 h, HLA-DP-mismatched-DC-reactive CD4 T cells were quantified and clonally isolated by single cell flow cytometric cell sorting based on CD137 expression using a FACSAria (BD Biosciences, San Jose, CA, USA). The time-point for CD137 isolation was chosen based on our previous work, including an unpublished clinical trial [administration of leukemia-reactive donor T cells after alloSCT or donor lymphocyte infusion to patients with persistent or relapsed mature B-cell neoplasms with blood and/or bone marrow involvement (EudraCT number 2012-003691-40)] and our experience with the isolation of virus-specific CD4 and CD8 T cells.36,37 Unstimulated peripheral blood mononuclear cells and autoDC-stimulated peripheral blood mononuclear cells from the same responder cells were used as controls to determine the gating strategy. The T-cell clones were expanded using allogeneic feeder mixture consisting of IMDM containing 5% heat-inactivated ABOS and 5% heat-inactivated fetal calf serum supplemented with 5× irradiated (35 Gy) allogeneic feeder cells, 0.5× irradiated (60 Gy) allogeneic Epstein-Barr virus-transformed lymphoblastoid cell line (EBV-LCL), 100 IU/mL interleukin-2, and 800 ng/mL phytohemagglutinin (PHA-HA16, Oxoid, Altrincham, UK).
Generation and culture of stimulator cells for functional analyses
The method of generating and culturing stimulator cells is described in the Online Supplementary Appendix.
Recognition assay
The procedure for testing the recognition profiles of expanded T-cell clones is described in the Online Supplementary Appendix.
Flow cytometric analyses
Information on the flow cytometric analyses performed is provided in the Online Supplementary Appendix.
Approval by ethics committee
Donors and patients had given written informed consent to the storage of biomaterials in the LUMC Biobank and the use of these materials was approved by the institutional medical ethical committee (protocol number B 16.039).
Results
Depletion of reactivity against autologous dendritic cells allows efficient identification and enrichment of allo-reactive T cells upon stimulation with HLA-DP-mismatched dendritic cells
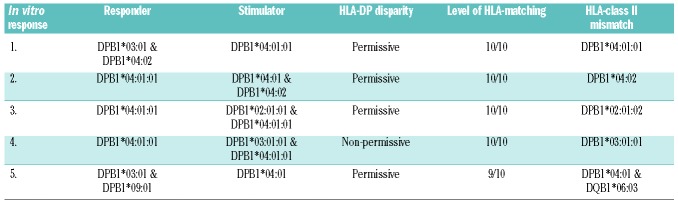
To study the composition of the allo-HLA-DP-specific T-cell repertoire, HLA-DP-mismatched alloDC were used to provoke allo-HLA-DP-reactive T-cell responses from healthy donor peripheral blood mononuclear cells. Responder/stimulator pairs with various HLA-DP mismatches were selected (Table 1): two pairs with a targeted HLA-DP*04:01 or HLA-DP*04:02 mismatch using HLA-DP*04:02- or HLA-DP*04:01-expressing responder cells, respectively, classified as minimal, permissive mismatches31 (pairs 1 and 2), one pair with a targeted HLA-DP mismatch classified as permissive (pair 3), one pair with a targeted HLA-DP mismatch classified as non-permissive (pair 4), and one 9/10 HLA-matched pair with an HLA-DP mismatch classified as permissive and an additional HLA-DQB1 mismatch (pair 5).
Table 1.
HLA-DP typing of the responder/stimulator pairs.
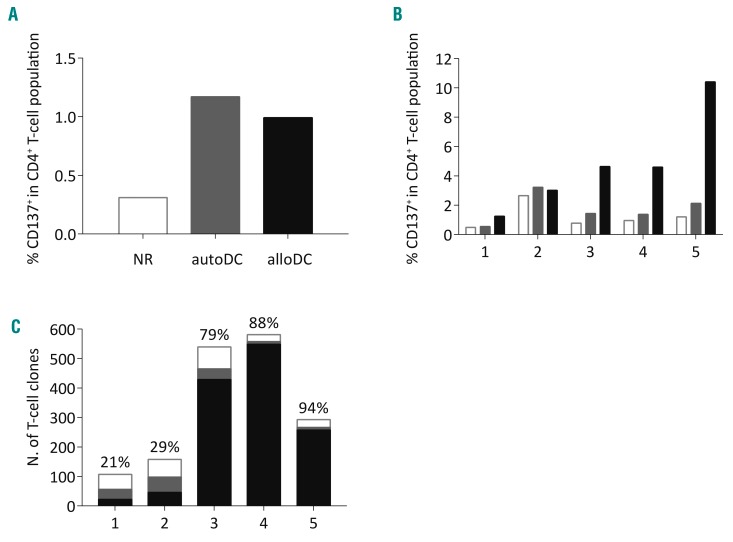
Since we previously illustrated that stimulation with alloDC results in coinciding stimulation of reactivity against autoDC,38 we first investigated whether this would hamper the isolation of allo-HLA-DP reactive T cells. Donor peripheral blood mononuclear cells were stimulated with HLA-DP-mismatched alloDC, cultured for 2 weeks, and then restimulated with either autoDC or alloDC. Flow cytometric analysis of the expression of the activation marker CD137 on CD4 T cells showed that upon (re)stimulation with autoDC and alloDC similar frequencies of CD4 T cells were activated (Figure 1A), indicating that a depletion step for the autoDC reactivity could allow more efficient identification and enrichment of allo-HLA-DP reactive T cells.
Figure 1.
Reactivities within allo-reactive T-cell responses provoked by stimulation with HLA-DP-mismatched dendritic cells. (A) Frequencies (%) of CD137+ cells in the CD4+ T-cell population at 36 h after restimulation [white bars = responder cells not restimulated (NR), gray bars = responder cells stimulated with autologous CD14-derived dendritic cells (autoDC), black bars = responder cells restimulated with allogeneic CD14-derived dendritic cells (alloDC)]; representative example. (B) Frequencies (%) of CD137+ T cells in the CD4+ T-cell populations of responses 1-5 at 36 h after restimulation of the responder cells that underwent a previous depletion step of autoDC reactive T cells (white bars = NR responder cells, gray bars = autoDC-stimulated responder cells, black bars = alloDC-restimulated responder cells). (C) Reactivity of the expanded T-cell clones from the allo-reactive T-cell responses 1-5 (n=107, 158, 540, 581, and 293, respectively) (white bars = non-reactive T-cell clones, gray bars = autoDC reactive T-cell clones, black bars = alloDC reactive T-cell clones).
Therefore, a strategy was developed using initial stimulation with HLA-DP-mismatched alloDC and subsequent culture for 2 weeks, followed by a stimulation step with autoDC and a depletion step using magnetic bead separation (MACS) based on the expression of CD137 on autoDC reactive T cells. The depleted fraction (CD137−) was subsequently restimulated with alloDC to specifically activate allo-reactive T cells (Online Supplementary Figure S1). For four of the five pairs the frequencies of CD137-expressing CD4 T cells were consistently higher upon specific restimulation with the HLA-DP-mismatched alloDC compared to the non-restimulated and autoDC-stimulated controls, although there was biological variability in the magnitude of the immune responses (Figure 1B). These data indicate that this strategy using a depletion step for autoDC reactivity allows more specific identification of allo-reactive T cells.
Next, CD4 T cells expressing CD137 upon restimulation with HLA-DP-mismatched alloDC were sorted as single cells per well, expanded and screened (n=1679) for allo-reactivity by interferon-γ enzyme-linked immunosorbent assay upon stimulation with alloDC, using autoDC as controls (response 4, as a representative example, is shown in Online Supplementary Figure S2). The clonality of expanded T-cell clones was confirmed by T-cell receptor Vβ repertoire analysis (data not shown). For responses 3-5 the majority of expanded T-cell clones were found to be allo-reactive (79-94%, n=1235). For responses 1 and 2, in which minimal, permissive HLA-DP mismatches were targeted resulting in less profound immune responses, only a limited proportion of the expanded clones were found to be allo-reactive (21-29%, n=68). A small proportion of the clones showed no reactivity or showed coinciding reactivity against the autoDC (Figure 1C).
The allo-HLA-DP-specific T-cell repertoire provoked by in vitro stimulation with HLA-DP-mismatched dendritic cells contains T cells that selectively recognize dendritic cells, but not Epstein-Barr-transformed lymphoblastoid cell lines
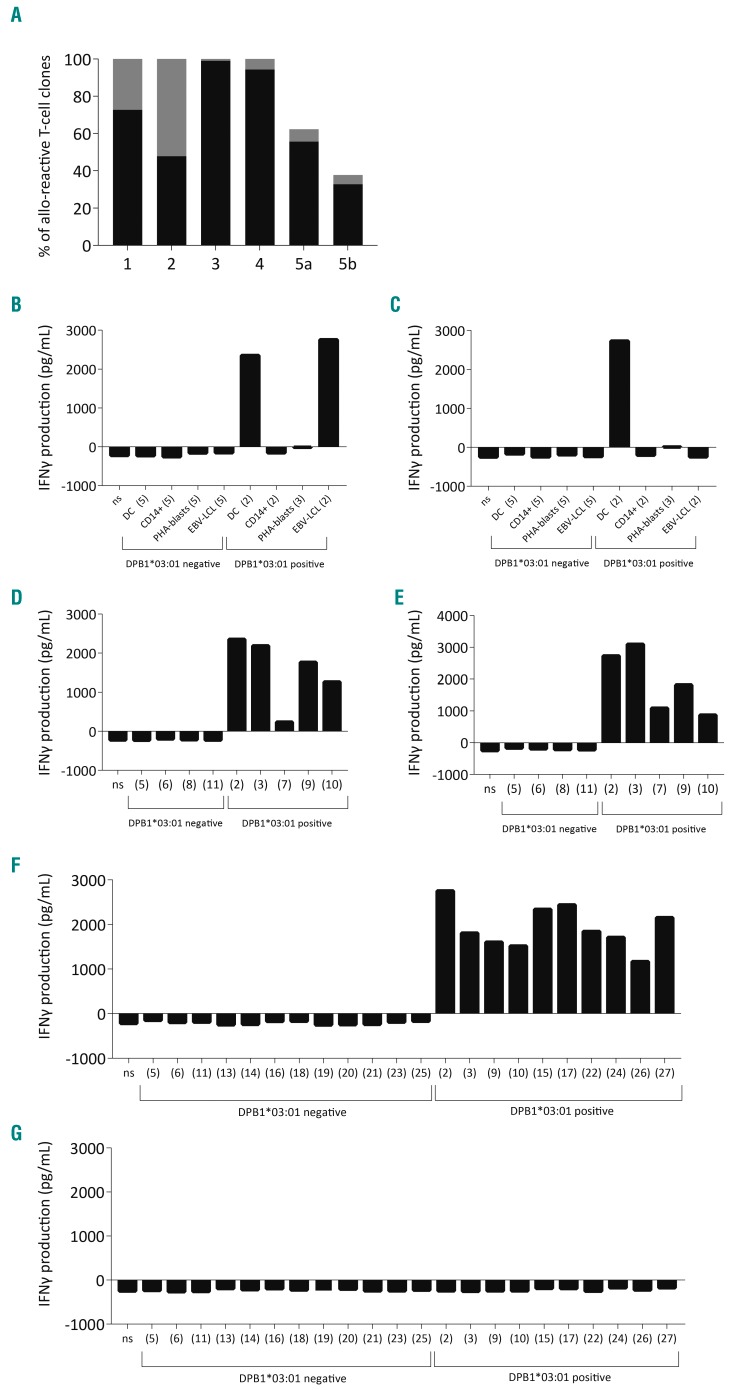
To investigate the HLA-DP restriction of the allo-reactive CD4 T-cell clones, clones (n=1303) were tested in a stimulation assay against third-party DC and EBV-LCL expressing the mismatched HLA-DP alleles (Online Supplementary Figure S3A) that were targeted in the immune responses, using autoEBV-LCL, autoDC and alloDC as controls. The majority (48-99%, median 73%, black bars in Figure 2A) of the allo-reactive T-cell clones showed HLA-DP-restricted reactivity demonstrated by recognition of alloDC and third-party EBV-LCL expressing the targeted mismatched HLA-DP allele (response 4, as a representative example, is shown in Online Supplementary Figure S3B). A representative HLA-DPB1*03:01-restricted T-cell clone is shown that only recognizes DC and EBV-LCL, but no other hematopoietic cells (CD14+ and PHA-blasts) (Figure 2B). However, a significant proportion (1-52%, median 7%, gray bars in Figure 2A) of the allo-reactive T-cell clones recognized alloDC, but showed no reactivity against the third-party EBV-LCL expressing the targeted mismatched HLA-DP allele (response 4, as a representative example, is shown in Online Supplementary Figure S3C). Selected T-cell clones from this group were tested against third-party DC and showed recognition of these stimulator cells by production of interferon-γ (response 4, as a representative example, is shown in Online Supplementary Figure S3D). A representative HLA-DPB1*03:01-restricted T-cell clone that recognizes only DC and no other hematopoietic cells is shown (Figure 2C). To confirm the specific HLA-DP restriction, T-cell clones were tested against K562 cell lines transduced with specific HLA-DP alleles or with empty vector (mock) (Online Supplementary Figure S4A). The T-cell clones only produced interferon-γ against the K562 transduced with the target HLA-DP allele (HLA-DPB1*03:01; representative T-cell clones, response 4 in Online Supplementary Figure S4B).
Figure 2.
Frequencies of HLA-class II-restricted CD4 T-cell clones. (A) Frequencies (%) of T-cell clones showing reactivity against allogeneic CD14-derived dendritic cells (alloDC) and third-party Epstein-Barr virus-transformed lymphoblastoid cell lines (EBV-LCL) expressing the targeted mismatched HLA-DP allele (black bars) or against alloDC but not against third-party EBV-LCL expressing the targeted mismatched HLA-DP allele (gray bars) in responses 1-5 (n=22, 46, 429, 549, 157, respectively). Pair 5 was a 9/10 HLA match containing an additional HLA-DQ mismatch; the HLA-DP-restricted T-cell clones are shown in bar 5a and the HLA-DQ-restricted T-cell clones in bar 5b. (B) Interferon-gamma (IFNγ) production by a representative HLA-DPB1*03:01-restricted T-cell clone (from response 4) after overnight incubation with hematopoietic stimulator cells (DC, CD14+ cells, PHA-blasts and EBV-LCL). The clone only shows reactivity against alloDC and alloEBV-LCL. (C) Reactivity of a representative HLA-DPB1*03:01-restricted T-cell clone (from response 4) against only alloDC. (D) The representative HLA-DPB1*03:01-restricted T-cell clone from Figure 2B tested against a panel of third-party DC. (E) The representative HLA-DPB1*03:01-restricted T-cell clone from Figure 2C was also tested against third-party DC. (F) The representative clone of Figure 2B shows reactivity against a panel of third-party EBV-LCL, but (G) the clone of Figure 2C does not show reactivity against third-party EBV-LCL. The number between brackets indicates the individual from whom the stimulator cells were derived.
To analyze whether the differential cell-recognition patterns of the allo-HLA-DP reactive T-cell clones is caused by recognition of a polymorphic antigen (minor histocompatibility antigen), a larger panel of DC and EBV-LCL was used as stimulator cells. The allo-reactive T-cell clones that showed HLA-DP-restricted reactivity against both DC and EBV-LCL manifested HLA-DP-restricted recognition of all DC (Figure 2D; representative HLA-DPB1*03:01-restricted clone) and EBV-LCL (Figure 2F; representative HLA-DPB1*03:01-restricted clone) expressing the target HLA-DP allele, indicating that most likely a monomorphic peptide is recognized. The allo-reactive T-cell clones that showed HLA-DP-restricted reactivity against DC but not against EBV-LCL showed HLA-DP-restricted recognition of the extended panel of DC (Figure 2E; representative HLA-DPB1*03:01-restricted clone) but not of the extended panel of EBV-LCL (Figure 2G; representative HLA-DPB1*03:01-restricted clone). Furthermore, the restricted recognition of DC but not of EBV-LCL from the same individual (e.g. DC and EBV-LCL from individuals 2, 3, 9 and 10 in Figure 2E,G) indicates that this restricted recognition profile is most likely caused by recognition of a cell-lineage-specific monomorphic peptide in the context of the allo-HLA-DPB1*03:01 molecule.
In response 5, with an additional HLA-DQB1 mismatch, 38% of the allo-reactive clones did not show HLA-DP-restricted reactivity against EBV-LCL expressing the mismatched HLA-DP molecules. Testing of these clones against a panel of DC and EBV-LCL expressing the mismatched HLA-DQB1*06:03 allele revealed that 33% of the allo-reactive clones exerted HLA-DQB1-restricted reactivity against DC and EBV-LCL, and 5% against DC, but not against EBV-LCL (bar 5b in Figure 2A; reactivity of repre sentative HLA-DQB1*06:03-restricted T-cell clones shown in Online Supplementary Figure S5). These results illustrate that the allo-HLA-DP and allo-HLA-DQ T-cell repertoire provoked by stimulation of donor T cells with HLA-class II-mismatched DC contains T-cell clones that show HLA-DP- or HLA-DQ-restricted recognition of DC, but not of EBV-LCL expressing the targeted mismatched HLA-DP or HLA-DQ allele.
The allo-HLA-DP-specific T-cell repertoire comprises T cells with different cell-lineage-restricted recognition patterns
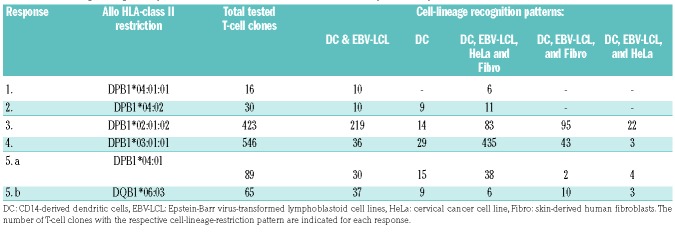
To further investigate the presence of T cells with a cell-lineage-specific recognition pattern within the allo-HLA-DP- and allo-HLA-DQ-specific T-cell repertoires, the reactivity pattern of the majority of allo-HLA-DP-and allo-HLA-DQ-restricted T-cell clones (n=1104, some T-cell clones were discarded because of contamination) recognizing alloDC, with or without recognition of third-party EBV-LCL, was analyzed using a panel of skin-derived fibroblasts and HeLa cells expressing the targeted mismatched HLA-DP alleles (Online Supplementary Figure S6A). DC and EBV-LCL were used as controls. A significant proportion (12-63%, median 53%) of the allo-HLA-DP-restricted T-cell clones showed restricted reactivity against DC, with (first recognition profile column in Table 2) or without (second recognition profile column in Table 2) EBV-LCL recognition, but not against fibroblasts or HeLa cells expressing the targeted mismatched HLA-DP allele, whereas the other HLA-DP-restricted clones showed a broad recognition pattern against all stimulator cells expressing the mismatched HLA-DP allele (20-80%, median 38%, third recognition profile column in Table 2), or against a selection of the tested stimulator cells (0-28%, median 7%, fourth and fifth recognition profile columns in Table 2) (the reactivity of representative clones of response 4 is shown in Online Supplementary Figure S6B-F). A selection of allo-HLA-DP-restricted T-cell clones was tested for their cytotoxic capacity and was found to be cytotoxic with variable efficiency (data not shown). Similar reactivity patterns were observed for the HLA-DQB1-restricted clones derived from the immune response mounted in response 5 (Table 2). These data illustrate that the allo-HLA-DP-and allo-HLA-DQ-specific T-cell repertoire contains T cells with various cell-lineage-specific recognition pat terns, as well as T cells that recognize all cells expressing the targeted mismatched HLA-DP or HLA-DQ allele, irrespectively of their cell lineage.
Table 2.
Cell-lineage recognition patterns of HLA-class II-restricted T-cell clones provoked by HLA-class II mismatched dendritic cells.
The allo-HLA-DP specific T-cell repertoire contains T cells that are able to recognize primary hematopoietic malignant cells, without coinciding reactivity against non-hematopoietic cells
To investigate whether the allo-HLA-DP-specific T-cell repertoire contains T cells that harbor the potential to recognize primary malignant cells without recognition of various non-hematopoietic cells, a selected number of T-cell clones (response 2, n=9; response 3, n=6; response 4, n=10; and response 5a, n=10) were tested against a panel of third-party primary malignant hematopoietic cell samples (acute myeloid leukemia, 13 samples; B-cell acute lymphoblastic leukemia, 9 samples; and chronic lymphocytic leukemia, 6 samples) expressing the targeted mismatched allele (HLA-DP expression shown in Online Supplementary Figure S7A,B), and against a panel of non-hematopoietic cell lines derived from different GvHD target organs (skin-derived fibroblasts, HeLa cells, colon carcinoma, and biliary epithelial cell lines) expressing the targeted mismatched HLA-DP allele. The HLA-DP-restricted T-cell clones that previously showed a broad recognition pattern recognized all target-HLA-DP-expressing malignant and non-hematopoietic cell types (clone response 4, as a representative example, is shown in Online Supplementary Figure S7C-I).
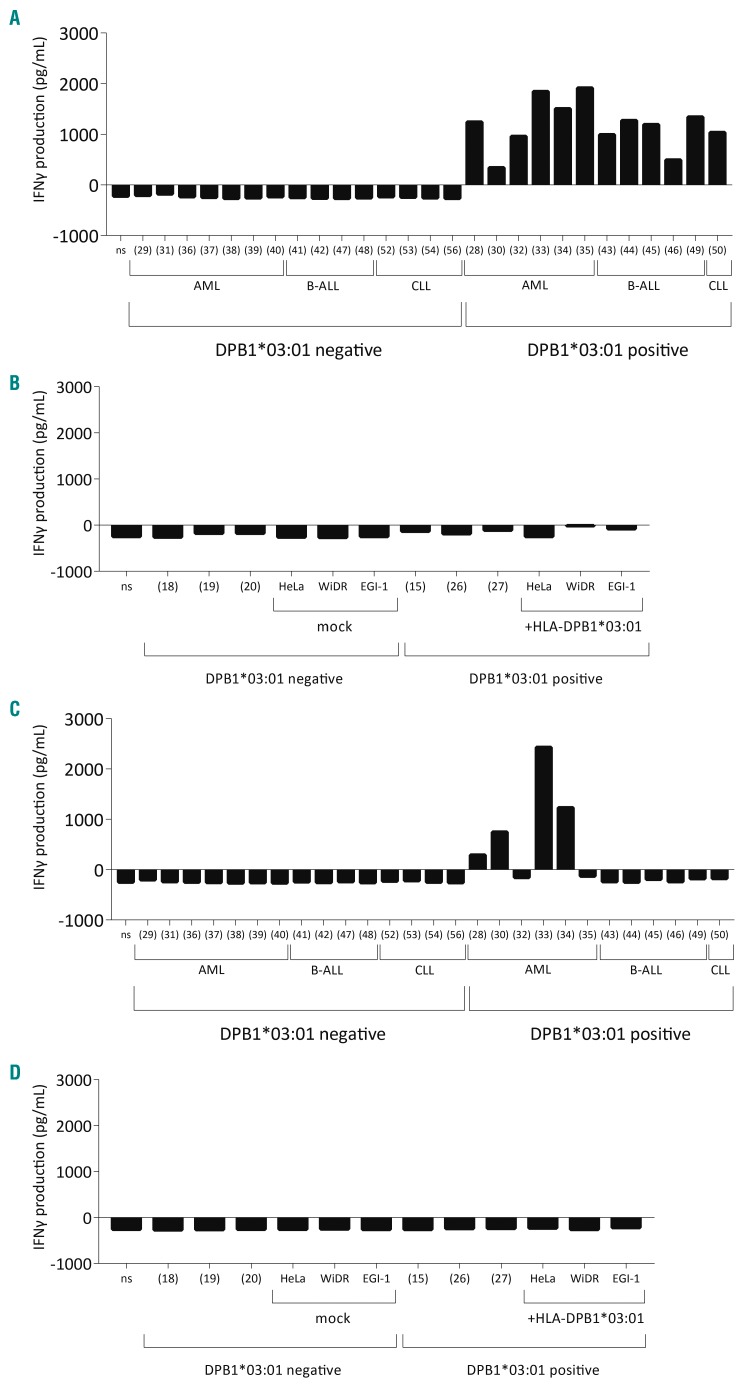
The HLA-DP-restricted T-cell clones that showed recognition of DC and EBV-LCL recognized malignant hematopoietic cell subsets of different lineages (representative HLA-DPB1*03:01-restricted clone in Figure 3A), but showed no reactivity against skin-derived fibroblasts and different non-hematopoietic cells expressing the target HLA-DP molecule (representative HLA-DPB1*03:01-restricted clone in Figure 3B). The HLA-DP-restricted T-cell clones that previously showed restricted recognition of DC, but not of EBV-LCL, showed only recognition of acute myeloid leukemia, but not of malignant B cells (e.g. chronic lymphocytic leukemia or B-cell acute lymphoblastic leukemia) (a representative clone from response 4 is shown in Figure 3C and from responses 2 and 5 in Online Supplementary Figure S8) and non-hematopoietic cells (e.g. fibroblasts and cell lines) (a representative clone from response 4 is shown in Figure 3D).
Figure 3.
Differential recognition of primary malignant and non-hematopoietic cells by allo-HLA-DP-specific T cells. (A) Interferon-gamma (IFNγ) production by a representative HLA-DPB1*03:01-restricted T-cell clone (derived from response 4) when stimulated with a variety of primary malignant cells and (B) not when stimulated with non-hematopoietic tissue cell lines. This recognition pattern was seen for the tested T-cell clones of response 2 (n=3), response 3 (n=3), response 4 (n=1) and response 5a (n=5). (C) IFNγ production by a representative HLA-DPB1*03:01-restricted T-cell clone (derived from response 4) when stimulated with primary acute myeloid leukemic blasts but not when stimulated with other primary malignant cells or (D) other non-hematopoietic cells. This recognition pattern was seen for the tested T-cell clones of response 2 (n=3), response 4 (n=6) and response 5a (n=3). The number between brackets indicates the individual from whom the stimulator cells were derived. AML: acute myeloid leukemia; B-ALL: B-cell acute lymphoblastic leukemia; CLL: chronic lymphocytic leukemia.
Since the level of recognition of the different primary acute myeloid leukemia samples by these clones was rather heterogeneous, flow cytometric analysis was performed to measure the surface expression of HLA-DP and adhesion molecule CD54 on the leukemic blasts (Online Supplementary Figures S7 and S9). For some samples (e.g. samples 37 and 40, Online Supplementary Figure S8D,F) the absence of recognition was correlated with the lack of proper HLA-DP surface expression. However, sample 35 (Online Supplementary Figure S8B) was not recognized despite high surface HLA-DP expression. Moreover, the maturation state (e.g. co-expression of maturation markers) of this specific acute myeloid leukemia sample was not found to be different from that of other samples that were properly recognized (data not shown). These data indicate that the level of recognition is likely to be determined by a combination of factors, including the level of HLA-DP expression, the expression of adhesion molecules (e.g. CD54), and the expression of the respective antigen.
These data illustrate that the allo-HLA-DP-specific T-cell repertoire comprises T cells with the capacity to recognize primary malignant cells, without recognition of non-hematopoietic tissue cells.
Discussion
In this study we demonstrate that the allo-HLA-DP-specific donor T-cell repertoire provoked by stimulation with HLA-DP-mismatched DC contains a variety of cell-lineage specificities. We found T cells that recognize all cells expressing the targeted mismatched HLA-DP allele, irrespective of the cell lineage, T cells with the capacity to recognize hematopoietic cells, including primary malignant cells without recognition of non-hematopoietic tissue cell lines, and also T cells that show restricted recognition of myeloid cells, including DC and primary acute myeloid leukemia samples, but not of cells of other hematopoietic and non-hematopoietic cell lineages. Similar cell-lineage-specific recognition patterns were found for allo-HLA-DQ-restricted T-cell clones isolated from a 9/10 HLA-matched pair, containing an additional HLA-DQ mismatch. Although the amplitude of the T-cell response is lower in the permissive HLA-DP responses compared to the non-permissive HLA-DP responses, a variety of cell-lineage-specific reactivity patterns was also found in the allo-HLA-DP-specific T-cell repertoire for the permissive HLA-DP responses.
It has been shown in patients receiving alloSCT and/or donor lymphocyte infusion from HLA-DP-mismatched donors, that the mismatched allo-HLA-DP alleles were targeted by CD4 T cells, resulting in the induction of GvHD in different organs (e.g. skin GvHD, and gut GvHD) indicating an upregulation of HLA-class II on inflamed non-hematopoietic tissues.23 However, in other patients allo-HLA-DP-reactive CD4 donor T cells induced specific GvL reactivity without coinciding GvHD, suggesting a hematopoietic-restricted HLA-DP response without cross-reactivity against non-hematopoietic tissues and/or absence of expression of HLA-class II on non-hematopoietic tissues in these cases.22,24 These clinical observations can be explained by different factors, the first being that the tissue restricted reactivity is dictated by the specificities present in the allo-HLA-DP T-cell repertoire.
The allo-HLA-DP-specific response is expected to be ini tiated in secondary lymphoid organs, most likely upon stimulation with patient-derived HLA-DP-expressing antigen-presenting cells of hematopoietic origin (DC).33,34 The allo-HLA-DP-restricted T cells activated by DC probably recognize antigens expressed by hematopoietic cells and presented in the mismatched HLA-DP molecule.14 This process could lead to an immune response skewed towards restricted recognition of hematopoietic cells, explaining the clinical observation in some patients of GvL reactivity without coinciding GvHD mediated by allo-HLA-DP-reactive CD4 donor T cells.22,24 In this study we showed that the allo-HLA-DP-restricted T-cell repertoire provoked by in vitro stimulation of donor T cells with HLA-DP-mismatched DC contains a broad spectrum of T-cell specificities. The restricted recognition of hematopoietic cells (e.g. DC and EBV-LCL) could indicate that in vivo T cells with comparable recognition profiles could contribute to a GvL effect in patients with HLA-DP-expressing myeloid or B-cell malignancies.24,39 On the other hand, the allo-HLA-DP-specific immune response can also be initiated by DC residing in inflamed HLA-DP-expressing non-hematopoietic tissues. If the DC in inflamed tissues are cross-presenting antigens from the damaged surrounding environment, allo-HLA-DP-restricted T cells provoked by these DC are more likely to be directed against antigens also expressed by non-hematopoietic cells and presented in the mismatched HLA-DP molecule.35 Most likely, the magnitude of the allo-HLA-DP response and, thereby, the absolute number of allo-reactive T cells as well as the recognition profile of the induced T cells will determine the balance between GvL and GvHD induction.
It has been shown in vivo that the magnitude of the allo-HLA-DP response is affected by the specific HLA-DP allele(s) expressed in the donor and patient.27,28 In the case of permissive HLA-DP mismatches it has been demonstrated in vivo29–32 and in vitro,40 in line with the results of our current study, that the HLA-DP response is of a lower amplitude and therefore could result in a more tolerable response than in the non-permissive HLA-DP-mismatched combinations. However, in vitro HLA-DP-specific T-cell responses showed immunogenicity of HLA-DP alleles in both permissive and non-permissive mismatched pairs.39,41 If the HLA-DP alleles and the peptidomes presented in the HLA-DP alleles are similar between donor and patient, a large proportion of the allo-HLA-DP-specific T cells is likely to be deleted during negative selection of self-reactive T cells in the thymus of the donor.42 This may explain the lower magnitude of the allo-HLA-DP-specific immune responses in permissive HLA-DP-mismatched donor/patient pairs.
Donor allo-HLA-DP-restricted CD4 T cells that target peptides expressed in non-self HLA-DP molecules in hematopoietic malignancies may specifically contribute to the GvL response after alloSCT. However, the occurrence of GvHD after HLA-DP-mismatched alloSCT and donor lymphocyte infusion remains a challenge, also in permissive HLA-DP-mismatched donor/patient pairs. The aim of our study was to elucidate the HLA-DP response in the donor T-cell repertoire when exposed to HLA-DP-mismatched antigen-presenting cells, similar to the clinical setting in HLA-DP-mismatched alloSCT. New strategies resulting in the enrichment of hematopoiesis-restricted allo-HLA-DP-specific T cells from the donor repertoire for adoptive T-cell therapy may increase the likelihood of inducing a GvL effect without provoking severe GvHD. For example, the current procedure could be optimized by including a depletion step to remove not only auto-reactive T cells but also T cells reactive against non-hematopoietic tissue. Although the use of HLA-DP-mismatched DC from patients will give a range of antigen specificities, including specificities against polymorphic antigens, the dependency on sufficient material from patients to generate CD14-derived DC restricts applicability. Therefore, the sophisticated approach of Herr and colleagues, who used autoDC transfected with allo-HLA-DP-encoding RNA as stimulator cells, shows an alternative way of provoking allo-HLA-DP-restricted donor T cells with different tissue specificities.43 Another strategy to obtain a hematopoiesis-specific T-cell product is to isolate HLA-DP-restricted T-cell receptors from our T-cell clones that showed a more hematopoiesis-specific or myeloid-lineage-specific recognition profile and perform T-cell receptor gene transfer.
Supplementary Material
Acknowledgments
The research in this manuscript was financially supported by the Dutch Cancer Society (project UL 2013-5989).
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/1/197
References
- 1.Miller JS, Warren EH, van den Brink MR, et al. NCI First International Workshop on The Biology, Prevention, and Treatment of Relapse After Allogeneic Hematopoietic Stem Cell Transplantation: report from the Committee on the Biology Underlying Recurrence of Malignant Disease following Allogeneic HSCT: graft-versus-tumor/leukemia reaction. Biol Blood Marrow Transplant. 2010;16(5):565–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von dem Borne PA, Starrenburg CW, Halkes SJ, et al. Reduced-intensity conditioning allogeneic stem cell transplantation with donor T-cell depletion using alemtuzumab added to the graft (’Campath in the bag’). Curr Opin Oncol. 2009;21(Suppl 1): S27–29. [DOI] [PubMed] [Google Scholar]
- 3.Falkenburg JH, Jedema I. Allo-reactive T cells for the treatment of hematological malignancies. Mol Oncol. 2015;9(10):1894–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barge RM, Osanto S, Marijt WA, et al. Minimal GVHD following in-vitro T cell-depleted allogeneic stem cell transplantation with reduced-intensity conditioning allowing subsequent infusions of donor lymphocytes in patients with hematological malignancies and solid tumors. Exp Hematol. 2003;31(10):865–872. [DOI] [PubMed] [Google Scholar]
- 5.Barrett AJ. Mechanisms of the graft-versus-leukemia reaction. Stem Cells. 1997;15(4): 248–258. [DOI] [PubMed] [Google Scholar]
- 6.Falkenburg JH, Warren EH. Graft versus leukemia reactivity after allogeneic stem cell transplantation. Biol Blood Marrow Transplant. 2011;17(1 Suppl):S33–38. [DOI] [PubMed] [Google Scholar]
- 7.Horowitz MM, Gale RP, Sondel PM, et al. Graft-versus-leukemia reactions after bone marrow transplantation. Blood. 1990;75(3): 555–562. [PubMed] [Google Scholar]
- 8.Kolb HJ. Graft-versus-leukemia effects of transplantation and donor lymphocytes. Blood. 2008;112(12):4371–4383. [DOI] [PubMed] [Google Scholar]
- 9.Riddell SR, Berger C, Murata M, Randolph S, Warren EH. The graft versus leukemia response after allogeneic hematopoietic stem cell transplantation. Blood Rev. 2003;17(3):153–162. [DOI] [PubMed] [Google Scholar]
- 10.Ferrara JL, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet. 2009;373(9674):1550–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Markey KA, MacDonald KP, Hill GR. The biology of graft-versus-host disease: experimental systems instructing clinical practice. Blood. 2014;124(3):354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nash RA, Storb R. Graft-versus-host effect after allogeneic hematopoietic stem cell transplantation: GVHD and GVL. Curr Opin Immunol. 1996;8(5):674–680. [DOI] [PubMed] [Google Scholar]
- 13.Barge RM, Starrenburg CW, Falkenburg JH, Fibbe WE, Marijt EW, Willemze R. Long-term follow-up of myeloablative allogeneic stem cell transplantation using Campath “in the bag” as T-cell depletion: the Leiden experience. Bone Marrow Transplant. 2006;37(12):1129–1134. [DOI] [PubMed] [Google Scholar]
- 14.Falkenburg JHF, Jedema I. Graft versus tumor effects and why people relapse. Hematology Am Soc Hematol Educ Program. 2017;2017(1):693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eefting M, de Wreede LC, Halkes CJM, et al. Multi-state analysis illustrates treatment success after stem cell transplantation for acute myeloid leukemia followed by donor lymphocyte infusion. Haematologica. 2016;101(4):506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eefting M, Halkes CJ, de Wreede LC, et al. Myeloablative T cell-depleted alloSCT with early sequential prophylactic donor lymphocyte infusion is an efficient and safe post-remission treatment for adult ALL. Bone Marrow Transplant. 2014;49(2):287–291. [DOI] [PubMed] [Google Scholar]
- 17.Yun HD, Waller EK. Finding the sweet spot for donor lymphocyte infusions. Biol Blood Marrow Transplant. 2013;19(4):507–508. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Vina MA, Klein JP, Haagenson M, et al. Multiple mismatches at the low expression HLA loci DP, DQ, and DRB3/4/5 associate with adverse outcomes in hematopoietic stem cell transplantation. Blood. 2013;121(22):4603–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreau P, Cesbron A. HLA-DP and allogeneic bone marrow transplantation. Bone Marrow Transplant. 1994;13(6):675–681. [PubMed] [Google Scholar]
- 20.Petersdorf EW, Smith AG, Mickelson EM, et al. The role of HLA-DPB1 disparity in the development of acute graft-versus-host disease following unrelated donor marrow transplantation. Blood. 1993;81(7):1923–1932. [PubMed] [Google Scholar]
- 21.van Bergen CA, van Luxemburg-Heijs SA, de Wreede LC, et al. Selective graft-versus-leukemia depends on magnitude and diversity of the alloreactive T cell response. J Clin Invest. 2017;127(2):517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rutten CE, van Luxemburg-Heijs SA, Halkes CJ, et al. Patient HLA-DP-specific CD4+ T cells from HLA-DPB1-mismatched donor lymphocyte infusion can induce graft-versus-leukemia reactivity in the presence or absence of graft-versus-host disease. Biol Blood Marrow Transplant. 2013;19(1):40–48. [DOI] [PubMed] [Google Scholar]
- 23.Stevanovic S, van Bergen CA, van Luxemburg-Heijs SA, et al. HLA class II upregulation during viral infection leads to HLA-DP-directed graft-versus-host disease after CD4+ donor lymphocyte infusion. Blood. 2013;122(11):1963–1973. [DOI] [PubMed] [Google Scholar]
- 24.Rutten CE, van Luxemburg-Heijs SA, Griffioen M, et al. HLA-DP as specific target for cellular immunotherapy in HLA class II-expressing B-cell leukemia. Leukemia. 2008;22(7):1387–1394. [DOI] [PubMed] [Google Scholar]
- 25.Petersdorf EW, Malkki M, O’HUigin C, et al. High HLA-DP expression and graft-versus-host disease. N Engl J Med. 2015;373(7):599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fleischhauer K, Beelen DW. HLA mismatching as a strategy to reduce relapse after alternative donor transplantation. Semin Hematol. 2016;53(2):57–64. [DOI] [PubMed] [Google Scholar]
- 27.Cesbron A, Moreau P, Cheneau ML, et al. Crucial role of the third and fourth hyper-variable regions of HLA-DPB1 allelic sequences in primary mixed-lymphocyte reaction: application in allogeneic bone marrow transplantation. Transplant Proc. 1993;25(1 Pt 2):1232–1233. [PubMed] [Google Scholar]
- 28.Nicholson I, Varney M, Kanaan C, et al. Alloresponses to HLA-DP detected in the primary MLR: correlation with a single amino acid difference. Hum Immunol. 1997;55(2):163–169. [DOI] [PubMed] [Google Scholar]
- 29.Zino E, Frumento G, Marktel S, et al. A T-cell epitope encoded by a subset of HLA-DPB1 alleles determines nonpermissive mismatches for hematologic stem cell transplantation. Blood. 2004;103(4):1417–1424. [DOI] [PubMed] [Google Scholar]
- 30.Fleischhauer K, Locatelli F, Zecca M, et al. Graft rejection after unrelated donor hematopoietic stem cell transplantation for thalassemia is associated with nonpermissive HLA-DPB1 disparity in host-versus-graft direction. Blood. 2006;107(7):2984–2992. [DOI] [PubMed] [Google Scholar]
- 31.Fleischhauer K, Shaw BE, Gooley T, et al. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol. 2012;13(4): 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleischhauer K, Shaw BE. HLA-DP in unrelated hematopoietic cell transplantation revisited: challenges and opportunities. Blood. 2017;130(9):1089–1096. [DOI] [PubMed] [Google Scholar]
- 33.Ni K, O’Neill HC. The role of dendritic cells in T cell activation. Immunol Cell Biol. 1997;75(3):223–230. [DOI] [PubMed] [Google Scholar]
- 34.Reddy P, Maeda Y, Liu C, Krijanovski OI, Korngold R, Ferrara JL. A crucial role for antigen-presenting cells and alloantigen expression in graft-versus-leukemia responses. Nat Med. 2005;11(11):1244–1249. [DOI] [PubMed] [Google Scholar]
- 35.Shlomchik WD. Graft-versus-host disease. Nat Rev Immunol. 2007;7(5):340–352. [DOI] [PubMed] [Google Scholar]
- 36.Jedema I, van de Meent M, Pots J, Kester MG, van der Beek MT, Falkenburg JH. Successful generation of primary virus-specific and anti-tumor T-cell responses from the naive donor T-cell repertoire is determined by the balance between antigen-spe cific precursor T cells and regulatory T cells. Haematologica. 2011;96(8):1204–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wehler TC, Nonn M, Brandt B, et al. Targeting the activation-induced antigen CD137 can selectively deplete alloreactive T cells from antileukemic and antitumor donor T-cell lines. Blood. 2007;109(1):365–373. [DOI] [PubMed] [Google Scholar]
- 38.Lam TS, van de Meent M, Falkenburg JF, Jedema I. Monocyte-derived dendritic cells can induce autoreactive CD4(+) T cells showing myeloid lineage directed reactivity in healthy individuals. Eur J Immunol. 2015;45(4):1030–1042. [DOI] [PubMed] [Google Scholar]
- 39.Rutten CE, van Luxemburg-Heijs SA, van der Meijden ED, et al. HLA-DPB1 mismatching results in the generation of a full repertoire of HLA-DPB1-specific CD4+ T cell responses showing immunogenicity of all HLA-DPB1 alleles. Biol Blood Marrow Transplant. 2010;16(9):1282–1292. [DOI] [PubMed] [Google Scholar]
- 40.Sizzano F, Zito L, Crivello P, et al. Significantly higher frequencies of alloreactive CD4+ T cells responding to nonpermissive than to permissive HLA-DPB1 T-cell epitope disparities. Blood. 2010;116(11): 1991–1992. [DOI] [PubMed] [Google Scholar]
- 41.Rutten CE, van Luxemburg-Heijs SA, van der Meijden ED, et al. Both permissive and nonpermissive HLA-DPB1 mismatches can induce polyclonal HLA-DPB1 specific immune responses in vivo and in vitro. Blood. 2010;115(1):151–153. [DOI] [PubMed] [Google Scholar]
- 42.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. [DOI] [PubMed] [Google Scholar]
- 43.Herr W, Eichinger Y, Beshay J, et al. HLA-DPB1 mismatch alleles represent powerful leukemia rejection antigens in CD4 T-cell immunotherapy after allogeneic stem-cell transplantation. Leukemia. 2017;31(2):434–445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.