Abstract
Solid-state NMR can elucidate membrane protein structures and dynamics in atomic detail to give mechanistic insights. By interrogating membrane proteins in phospholipid bilayers that closely resemble the biological membrane, solid-state NMR spectroscopists have revealed ion conduction mechanisms, substrate transport dynamics, oligomeric interfaces of seven-transmembrane-helix proteins, conformational plasticity that underlies virus-cell membrane fusions by complex protein machineries, and β-sheet folding and assembly by amyloidogenic proteins when bound to lipid membranes. These studies collectively show that membrane proteins exhibit extensive structural plasticity to carry out their functions. Solid-state NMR spectroscopy is ideally suited to elucidating this structural plasticity, local and global conformational dynamics, protein-lipid and protein-ligand interactions, and protonation states of polar residues. These mechanistically important structural studies are aided by new sensitivity enhancement techniques, resolution enhancement by ultrahigh magnetic fields, and the advent of 3D and 4D correlation techniques.
Membrane proteins carry out some of the most important biological functions such as transport of ions and metabolites and transduction of chemical signals and energy. Because of their lipid-associated nature, membrane proteins are difficult to crystallize and solubilize, thus posing significant challenges to structural biologists. So far most high-resolution membrane protein structures have come from X-ray crystallography, and cryo-electron microscopy has begun to contribute membrane protein structures in the 3–4 Å resolution range.
Compared to these techniques, solid-state NMR (SSNMR) spectroscopy provides unique, atomically detailed, dynamically sensitive, and biologically authentic information about membrane protein structure and dynamics. Two fundamental aspects of SSNMR spectroscopy make it especially well suited to membrane protein structural studies. First, the method probes chemical structure as well as angstrom-level three-dimensional structures by the local nature of nuclear spin interactions. The chemical shielding of nuclear spins by the surrounding electrons is sensitive to torsion angles, hydrogen bonding, and molecular packing. Thus, NMR chemical shifts are sensitive to even the most subtle structural changes in proteins. Dipolar couplings between nuclei such as 1H, 13C, 15N, 31P, 2H and 19F can be measured with ±0.1 Å precision for distances within 5 Å, and with ± 1 Å precision for distances between 5 and 15 Å. These nuclear spin interactions permit SSNMR studies of the active sites of membrane proteins in extraordinary detail. For example, protonation and deprotonation of titratable residues, motions of functionally important sidechains, and coordinated motions of entire domains for ligand transport, can be readily observed using a versatile array of SSNMR experiments. Second, SSNMR studies of membrane proteins is freed from the requirements of crystalline order and fast molecular tumbling, and can probe both rigid and partially dynamic proteins in lipid bilayers. Thus, membrane proteins can be studied in their native lipid environments. A variety of membranes can be used in SSNMR experiments: they can be lipid vesicles with compositions that mimic the biological membrane, whole cellular membranes, or bicelles with simple compositions. Since many membrane proteins exhibit structural plasticity depending on the lipid headgroup charge, cholesterol, membrane curvature, and other membrane properties, it is crucial to investigate membrane protein structures in native-like membranes by SSNMR.
SSNMR instrumentation and radiofrequency pulse sequences have recently seen dramatic advances in four areas: sensitivity enhancement by two orders of magnitude based on dynamic nuclear polarization from electrons (65); sensitivity enhancement by 1H-detected experiments under ultrafast (~100 kHz) magic-angle spinning (MAS) (4); resolution enhancement by ultrahigh magnetic fields (above 23.5 Tesla, or 1H Larmor frequency of 1.0 GHz); and resolution enhancement due to the advent of 3D and 4D correlation experiments (82; 85). These advances have allowed larger, multimeric, and more complex membrane proteins to be studied using SSNMR. Here we review SSNMR studies of several classes of membrane proteins that have yielded novel structural, dynamical and mechanistic information.
I. Ion Channels and Transporters
Ion channels conduct ions down their concentration gradients across the otherwise impermeable lipid membrane, using specific residues to control ion selectivity and channel opening and closing (90). Channels can be gated by a variety of stimuli such as membrane potential, pH, ligand, mechanical activity, and temperature (35). On the other hand, transporters move ligands against their electrochemical gradient by coupling conduction to an energetically favorable process, such as photon absorption, ATP hydrolysis, or movement of another ion down its concentration gradient (27). SSNMR has been used fruitfully to study pH-gated proton channels, pH and voltage-gated potassium channels, and multidrug-resistant transporters to reveal key mechanistic information about the ion conduction and substrate transport processes.
The Influenza M2 Proton Channel
The M2 protein of influenza A, B, and C viruses forms functionally homologous but sequence-distinct proton channels that are essential for the flu lifecycle. When a new virus is endocytosed into the host cell, the virus-envelope bound M2 activates in response to the low pH of the endosome and acidifies the virion, which in turn releases the viral ribonucleoprotein complex into the host cell. The wild-type M2 protein of influenza A virus, which is dominant in seasonal and pandemic flu outbreaks, is small (~100 residues) and modular, and is blocked by the amantadine class of antiviral drugs (6). The N-terminal ectodomain (50) facilitates protein incorporation into the virion (32; 68). The single-pass TM domain contains the proton-selective residue, a single histidine, and the gating residue, a tryptophan, in a conserved HxxxW motif (Fig. 1a). The AM2 TM domain shows similar drug-sensitive and pH-dependent proton conductance as the full-length protein (59), thus making the TM domain a relevant model system for full-length M2. The cytoplasmic domain contains an amphipathic helix (AH) that mediates membrane scission during virus budding (9; 76; 95), and a cytoplasmic tail that participates in matrix protein 1 (M1) recognition and virus assembly (62).
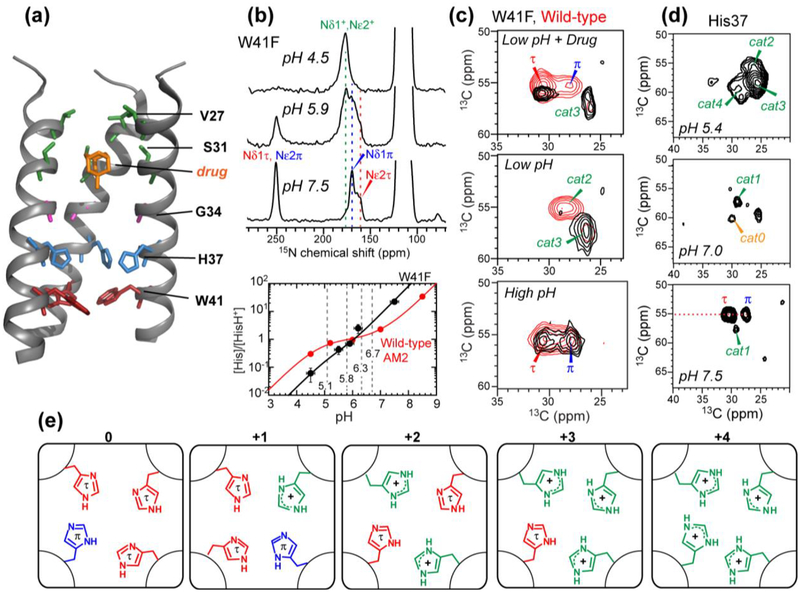
Figure 1.
Structural studies of the influenza M2 proton channel by SSNMR. (a) The TM domain structure with bound drug (PDB: 2KQT), solved in DMPC bilayers. Important functional residues are indicated. (b) Determination of His37 pKa’s from pH-dependent His37 sidechain 15N chemical shifts. The W41F mutant data is shown as an example. Titration curves yield the His37 pKa’s, which differ for the W41F mutant and wild-type channels. (c) Histidine Cα and Cβ chemical shifts from 2D 13C-13C correlation spectra. A W41F mutation (black spectra) changes the His37 chemical shifts compared to the wild-type (red spectra), indicating that His37 can be protonated from the C-terminus in the mutant. (d) 2D 13C-13C correlation spectra of cytoplasmic-containing M2 as a function of pH. Seven states of His37 are observed. (e) Proposed His37 tetrads with different charge states, explaining the resolved seven sets of histidine chemical shifts.
The TM helix conformation is sensitive to the membrane environment
Crystal structures, solid-state NMR, and solution NMR structures (69; 81) of M2 in various membrane-mimetic environments have been determined (36). Crystal structures of the TM domain have progressively improved in resolution from 3.5 Å in octylglucoside (OG) to 1.0 Å in lipid cubic phases (1; 84; 86). SSNMR studies of the TM domain in lipid bilayers have shown that the TM conformation and dynamics are sensitive to the membrane composition, pH, and drug binding. High membrane fluidity and negatively charged lipids increase helicity by promoting the cationic state of His37 (9; 39; 55), while thicker bilayers decrease the helix tilt angle (7; 17; 78). Adding cholesterol increases the helicity (54), immobilizes the four-helix bundle (57), and stabilizes the tetramer (14; 45). Drug binding causes a kink to the middle of the TM helix (8; 42). For a combined TM-AH construct, a solution NMR structure solved in DHPC micelles shows the AH to be separated from the TM helix by an unstructured loop (81), while a SSNMR orientational structure solved in DOPC/DOPE bilayers shows the AH to lie parallel to the membrane surface, connected to the TM helix by a tight turn (83). In comparison, a MAS SSNMR structure solved in DPhPC membranes shows a dimer of dimers structure with two distinct orientations and depths for the TM helices (3). This symmetry breaking, not seen in other structures, may be caused by the DPhPC lipid, whose methyl-rich acyl chains can cause significant negative curvature to the membrane. Taken together, these studies show that the TM and AH domains have significant conformational plasticity in response to the pH and membrane environment (110), thus structural comparisons of different M2 constructs should be made in the same lipid membranes and at the same pH.
The ectodomain and cytoplasmic tail in M2 are dynamically disordered
2D and 3D correlation experiments on 13C, 15N-labeled full-length AM2 showed that the ectodomain and cytoplasmic tail are unstructured and highly dynamic. Resonances beyond the α-helical chemical shifts of the TM and AH residues show random coil 13C chemical shifts, narrow linewidths, and low order parameters, indicating that the two extramembrane domains of M2 are intrinsically disordered (54). Site-specifically labeled ectodomain peptides and cytoplasmic tail peptides were ligated to the central TM-AH domain and confirmed this dynamic disorder (50; 51). Interestingly, despite this dynamic disorder, the extra-membrane domains modulate the structure and function of the TM domain. Chemical shift measurements of His37 and other key TM residues indicate that the ectodomain shifts the TM equilibrium towards the drug-bound conformation even in the absence of drug, while the cytoplasmic tail shifts the protonation equilibria of His37 towards higher pKa’s (50). Thus, the ectodomain may preselect drug-binding competent conformation of the TM helix, while the cytoplasmic tail facilitates proton binding to His37, likely due to the acidic nature of the cytoplasmic tail.
Proton conduction mechanism and gating mechanism of M2
Solid-state NMR, MD simulations, and crystal structures that resolved water densities have provided rich information about how M2 is activated by low pH and how it conducts protons exclusively from the N-terminus to the C-terminus (72). 15N SSNMR spectra showed that the His37 imidazole ring shuttles protons into the virion (71) by microsecond-timescale protonation and deprotonation. This is manifested by pH-dependent histidine sidechain 15N and 13C chemical shifts and 15N chemical shift averaging at high temperature (23; 41; 103). The imidazole rings reorient at acidic pH to mediate proton transfer, as observed from motional averaging of the histidine sidechain dipolar couplings (40). Water-imidazole hydrogen bonding and proton exchange have been detected from water-imidazole 1H-15N correlation peaks (37; 63; 100).
The His37 tetrad protonates successively like a tetraprotic acid, with a maximum of four proton-dissociation constants (pKa’s). These pKa’s have been measured using 15N chemical shift spectra as a function of pH (Fig. 1b) under various conditions to investigate how proton shuttling depends on the protein structure and membrane environment. Comparison of wild type and mutant channels probed the effects of the gating tryptophan residue on proton shuttling (58; 61; 101; 104). Comparison of TM and longer constructs revealed the influence of extramembrane domains (55). Comparison of influenza A and B M2 probed the effects of the amino acid sequence on proton transfer (102), while comparison of TM peptides bound to different lipid membranes examined the influence of the lipid environment on proton relay (41; 43). These studies all show that the third protonation event, occurring between pH 5 and 6, causes the largest increase of proton conductance and is therefore responsible for channel activation, but the exact values of the pKa’s depend on the protein sequence and membrane composition. Cholesterol-containing membranes lower the pKa’s (41). Negatively charged lipids and the acidic cytoplasmic tail increase the pKa’s (11; 55), likely by better attracting protons to the membrane. BM2 shows lower pKa’s than AM2 (102), which may be due to faster proton relay to a second histidine that is C-terminal to the proton-selective histidine. When the gating Trp was mutated to Phe to allow bi-directional proton conductance, the His37 structural equilibrium was perturbed (Fig. 1c) (61). His37 shifted to the π tautomer and the neutral state at high pH, indicating faster deprotonation to the C-terminus, but became more cationic at low pH, indicating increased protonation from the C-terminus. Drug binding to this mutant channel left 50% of His37 cationic, in contrast to the wild-type, which became fully neutral upon drug binding. These results indicate that in wild-type M2, Trp41 blocks C-terminal acid activation of His37, causing asymmetric conductance of the channel (104).
The protonation equilibria of the proton-selective histidine can be described by a kinetic model that takes into account the asymmetric nature of proton shuttling (61):
| (1) |
Each equilibrium constant, measured from the 15N intensities of cationic and neutral histidines (Fig. 1b), is related to the rate constants as:
| (2) |
This kinetic model explains all SSNMR-detected chemical equilibria of His37 in M2 channels. Since four protonation equilibria lead to five charge states of the histidine tetrad, the structure of each histidine residue is expected to depend on the tetrad charge state. Indeed, seven sets of His37 13C and 15N chemical shifts have been resolved in cytoplasmic-tail-containing M2 (55) (Fig. 1d), and can be assigned to different charge states of the tetrad (Fig. 1e). Together, these SSNMR data indicate that the structural properties of M2 depend on the pH-sensitive His37 and the membrane composition.
Proton conduction requires histidine-water H-bonding, not imidazole-imidazolium H-bonding
Strong imidazole-imidazolium hydrogen bonds have been proposed for the histidine tetrad by Cross and coworkers to explain the conduction mechanism. The rationale for this model was to account for the +2 charge of the His37 tetrad near neutral pH (43), as the first histidine pKa measurement found high pKa’s (~8.2) for the first two protonation events. It was argued that a low-barrier hydrogen bond between a neutral imidazole and a cationic imidazolium can stabilize the +2 charge in the middle of the membrane. However, the energetic rationale for this model has since been removed by new experimental evidence. First, more accurate measurements of His37 pKa’s in cholesterol-containing membranes that mimic the virus envelope composition (57) found lower pKa’s (7.6 and 6.8) for the first two protonation events, indicating that the tetrad charge state is lower (about +1) at neutral pH (41; 55; 61; 102). Second, high-resolution crystal structures of the TM peptide showed a cluster of water molecules on both sides of the His37 tetrad, indicating proton delocalization among water molecules (1). So far, only a single 15N cross peak has been observed for the downfield 1H chemical shift in 2D 1H-15N correlation spectra, indicating that the hydrogen-bonded proton is not shared by two imidazole nitrogens, but by one nitrogen and one oxygen (63; 99). Moreover, at high temperature, the exchange-averaged 1H chemical shift that correlates with the imidazole 15N signal lies at the water chemical shift of ~ 5 ppm, indicating that the hydrogen-bonding partner of imidazole is water instead of another histidine (37). Finally, torsion angle measurements (40) and crystal structures (1) showed that the H37 (χ1, χ2) angles are about 180°, which prevent the imidazole N-H group from pointing to another imidazole in a geometry to form N-H…N hydrogen bonds.
Potassium Channels
K+ ion movement through potassium channels across lipid membrane underlies many biological processes such as electrical signaling in the nervous system and cardiac action potentials. While crystal structures of a number of potassium channels are available (16; 56; 111), SSNMR is well suited to characterizing the conformational dynamics and membrane-dependent structural variations of K+ channels. The prokaryotic K+ channel KcsA has been the main model system for SSNMR studies, because it is smaller than mammalian voltage-gated potassium (Kv) channels while retaining key ion selectivity and gating properties. A chimera between KcsA and Kv1.3, which grafts 11 residues in the turret region of Kv1.3 onto KcsA (Fig. 2a), has also served as a useful model for understanding toxin binding and lipid interaction of K+ channels (80).
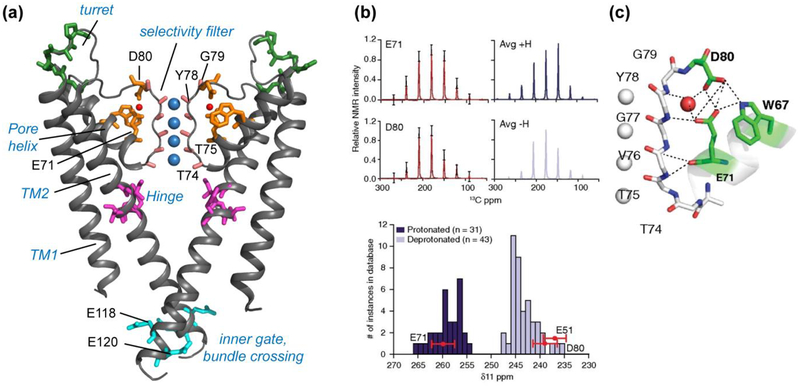
Figure 2.
KcsA structure investigated using SSNMR. (a) Key domains and functional residues in KcsA that have been studied (PDB: 1K4C). (b) Measured sidechain carboxyl chemical shift anisotropies (CSA) (black) of E71 and D80, with the simulated CSA patterns overlaid in red (5). The simulated spectra of average protonated (dark blue) and deprotonated (light blue) carboxyl CSA tensors are shown on the right, based on a database of known crystal structures. Compared to this database, the δ11 component of E71 matches with protonated (dark blue) carboxyls, while E51 and D80 δ11 components match with deprotonated (light blue) carboxyls, indicating an elevated pKa for E71. (c) The hydrogen-bonding network connecting the selectivity filter and the pore helix in the high K+ conductive state (PDB: 1K4C). Panels (b) and (c) reproduced with permission from Reference (5).
Potassium channels can be inactivated by two mechanisms: a fast process involving the intracellular part (N-type inactivation for Shaker channels), and a slower process involving the extracellular K+ ion selectivity filter (C-type inactivation) (38). KcsA is activated by a high concentration of intracellular protons, which causes protonation of E118 and E120 (87), but also undergoes C-type inactivation, which causes loss of ion binding sites at the extracellular selectivity filter. These two gates together can adopt four states (15), as defined by electrophysiological data, under different combinations of pH and K+ concentrations. The two longest-living states are the resting state, which is adopted when the intracellular gate is closed and the extracellular gate is open, and the inactivated state, adopted when the intracellular gate is open and the extracellular gate is closed. Only when both gates are open can K+ ions be conducted. The region near the extracellular inactivation gate has been extensively studied by SSNMR. From the N-terminus to the C-terminus, these segments consist of TM helix 1, which corresponds to the S5 helix of Kv channels, a water-exposed turret, a short pore helix, the selectivity filter, and TM helix 2, which corresponds to the S6 helix of Kv channels (Fig. 2a).
Extensive chemical shift mapping of KcsA using 2D, 3D and 4D correlation SSNMR gave a wealth of information about the conformational equilibria among the four states and the structural coupling between the two gates. Bhate et al compared the acid-induced inactivated state of the gate with low [K+]-induced collapsed state of the protein (5) using an E71A mutant, which does not undergo C-type inactivation. Chemical shift data indicate that the mutant cannot access the collapsed state even when [K+] is low, suggesting that the acid-induced C-type inactivated state is structurally similar to the low [K+]-induced collapsed state. Between low and high K+ concentrations, the wild-type protein exhibits chemical shift changes of the E71 sidechain carboxyl, which is involved in a hydrogen-bonding network with water, D80 and W67. The sidechain 13CO chemical shift tensors of E71 and D80 (Fig. 2b) were measured from sideband intensities and showed that the E71 carboxyl is protonated even at pH 7.5, indicating an unusually high pKa, while the D80 sidechain is deprotonated. Therefore, the protonation state of E71 is unchanged between high and low [K+], but the hydrogen-bonding network between the pore-helix and selectivity filter is altered, likely because of differences in K+ ion binding at the selectivity filter.
Chemical shift comparisons between the high [K+]/neutral pH state, the low [K+]/neutral pH state, and the high [K+]/low pH state gave information about structural similarities and differences among these states (105). Extensive chemical shift changes were observed in the hinge, pH sensor and selectivity filter regions at low [K+]/neutral pH, when the channel occupies the collapsed state, compared to the high [K+]/neutral pH state. These structural changes were mirrored at high [K+]/low pH, indicating that there is allosteric coupling between the two modes of inactivation: the pH-dependent protonation change at E118 and E120 and the [K+]-dependent C-type inactivation.
In addition to pH and K+ concentration, the lipid composition of the membrane affects the KcsA conformational equilibria. Varying the lipid headgroup charge allowed all four gating states to be accessed (92). Cardiolipin and phosphatidylphosphate (PA) were found to increase the probability of the open-conductive state, while neutral lipids and high pH shifted the equilibrium towards one of the three non-conducting conformations. Residues that are diagnostic of these conformational states are found in the turret (e.g. Ala50) and the pore helix (e.g. Glu71). The lipid sensitivity of the protein structure was also examined by comparing the structures of KcsA and the KcsA-Kv1.3 chimera. The chimera contains a charge mutation in the pore helix, where a cationic R64 is changed to an anionic D64. The chimera shows cross peaks between D64 and R89, indicating a salt bridge, but no cross peak with lipids, while KcsA shows Ser/Thr cross peaks with asolectin lipids, indicating lipid binding (97). MD simulations suggest direct protein-lipid interactions that are affected by the electrostatic properties of the turret residues (91). The turret also shows significant structural plasticity. At high [K+]/low pH, long-range cross peaks such as D53-T85 are lost compared to the high [K+]/high pH state, indicating that the turret and the neighboring TM1 and TM2 helices undergo conformational changes to cause C-type inactivation.
Transporters
Transporters are chiefly responsible for multidrug resistance of cells, because small, hydrophobic, and positively charged drug molecules can be actively exported out of cells by these proteins in a non-specific manner (34). The guiding transporter hypothesis is that these proteins undergo conformational changes between at least two states, facing inward and outward, to move substrate molecules. EmrE is a small multidrug resistance transporter, and couples the import of two protons with the export of one substrate molecule from the cytoplasm to the periplasm (Fig. 3a). The protein contains four TM helices and forms an antiparallel, asymmetric dimer. TM helices 1–3 in each subunit are responsible for drug and proton antiport, while the fourth TM helix serves as the dimerization unit (Fig. 3b).
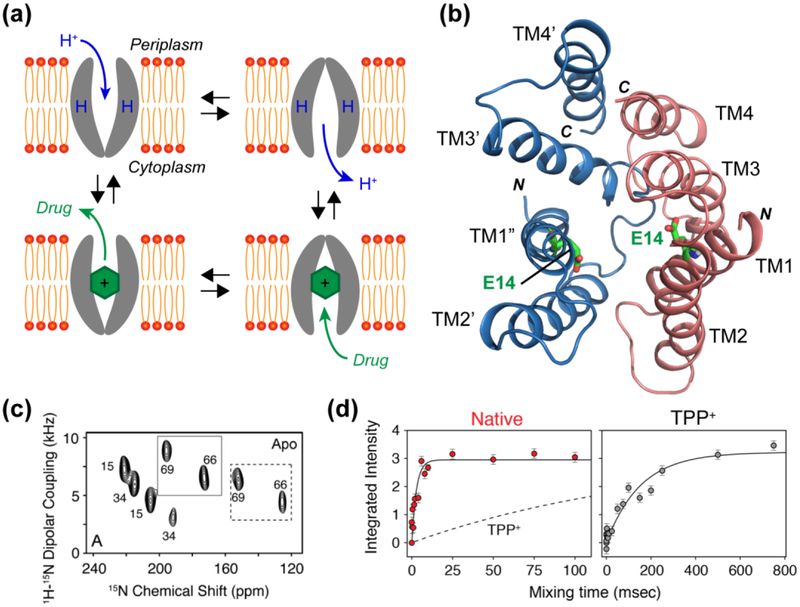
Figure 3.
(a) Single-site alternating-access model for substrate transport by EmrE. The transporter is only accessible to one side of the membrane at a time, and a conformational change is needed to switch between the two sides. (b) Model of the asymmetric antiparallel dimer structure of EmrE (PDB: 3B5D), in which E14 affects the conformational exchange rate. (c) Representative 2D PISEMA spectra of 15N-Val labeled EmrE in oriented bicelles. Most residues show peak doubling, indicating two distinct orientations relative to the bilayer normal (28). (d) Millisecond-timescale conformational change measured by static 15N exchange NMR on substrate-free (red) and substrate (TPP+)-bound EmrE (grey) (10). The exchange presumably reflects inter-conversion between the inward-facing and outward-facing states, and is ~50 times faster for the ligand-free protein than the TPP+-bound protein. Panel (c) reproduced with permission from Reference (28). Panel (d) reproduced with permission from Reference (10).
Solid-state and solution NMR experiments have described the topological structure, conformational dynamics, and mechanism of action of EmrE. Using oriented-membrane SSNMR experiments, Gayen et al showed that the four TM helices in the two subunits have different orientations relative to the bilayer normal. In magnetically oriented bicelles, each residue exhibits two sets of signals in 2D polarization inversion spin exchange at the magic angle (PISEMA) spectra, which correlate the 15N-1H dipolar coupling with 15N chemical shifts (28) (Fig. 3c). This peak doubling was found for both ligand-free and ligand-bound proteins, indicating that the two subunits are asymmetric. Chemical cross-linking experiments showed that the two subunits are antiparallel. Binding of a substrate, tetraphenylphosphonium (TPP+), at a saturating molar ratio of one ligand per dimer affected the orientation of TM3 in both subunits, indicating that ligand binding changes the protein conformation. It is noteworthy that oriented-membrane SSNMR is especially suited for investigating such asymmetric membrane proteins, since the structural asymmetry is directly encoded in the orientation-dependent NMR frequencies, but is not inherently detectable in isotropic chemical shift spectra that are measured under MAS.
After establishing the asymmetric and antiparallel topology of the EmrE dimer, it is important to characterize the protein conformational motion that underlies substrate transport. Here, solution NMR (64), oriented-membrane SSNMR, and MAS SSNMR (10) have all been used to probe EmrE motion. The chemical shifts and linewidths of EmrE bound to isotropic and oriented bicelles both showed that the apo protein undergoes conformational exchange at a rate of ~350 s−1 at 37°C, while the TPP+-bound protein has 50-fold slower motion (10) (Fig. 3d). Fitting the linewidths as a function of temperature gave an activation energy of 28±5 kcal/mol, which is consistent with the expected free energy change when the hydrophobic area between the two subunits changes during the inward-outward conformational conversion. The activation energy suggests that three out of four TM helices are involved in the conformational exchange, while TM4 is unchanged. The motional rates measured in isotropic bicelles by solution NMR agree well with rates measured from bilayer samples using 15N exchange NMR (10), indicating that EmrE motion is preserved between the two environments. The NMR-detected slow rate of 3.2–4.9 s−1 (37–45°C) is overall consistent with the turnover rate of < 1 s−1 for TPP+.
EmrE operates in a single-site alternating-access fashion, where a critical E14 mediates both proton import and substrate efflux (Fig. 3a). The ligand interaction and protonation behavior of E14 have been studied using SSNMR and mutagenesis. Two sets of E14 chemical shifts were observed that differ between the apo and ligand-bound states, indicating that this residue has two conformations in the dimer, and substrate binding creates additional states that are in slow exchange (52). The close proximity of E14 to the substrate TPP+ was demonstrated using dynamic nuclear polarization (DNP) experiments that enhance the 13C sensitivity (67). Cross peaks between 13C-labeled TPP+ aromatic carbons and the sidechain carboxyl of deprotonated E14 were observed in 2D 13C-13C correlation spectra. Recent studies also showed that the EmrE conformational motion is pH-dependent. The 15N NMR-detected exchange rate is faster (~220 s−1) at low pH when E14 is protonated than at high pH (~40 s−1) where E14 is unprotonated (29). Thus, the E14 protonation state affects the inward-outward conversion rate. These data also indicate that E14 has an elevated pKa of 7.0 compared to the bulk pKa, reminiscent of the pKa increase of E71 in KcsA.
A much larger class of transporters, ATP-binding cassette (ABC) transporters, has also been studied by SSNMR. ABC transporters utilize the energy from ATP hydrolysis to transport substrate molecules. The transporter associated with antigen processing (TAP) is a 150 kDa heterodimeric protein that binds viral peptide antigens of >8 amino acids that are processed by the proteasome. TAP delivers the antigenic peptides to the endoplasmic reticulum as part of the adaptive immune response. Binding of an antigenic peptide to human TAP was studied using 13C, 15N-labeled peptide and unlabeled TAP (53). The 13C chemical shifts of the labeled peptide were detected using double-quantum (DQ) experiments, which suppressed the natural abundance TAP signals. The bound peptide was found to be extended, spanning a length of 2.5 nm. Docking of this structure onto TAP suggested the binding site to be a negatively charged pocket at the dimer interface. These DQ 13C experiments required DNP to achieve high sensitivity, since the large molecular weight of the TAP limited the mass of the labeled antigenic peptide that can be used in the study.
II. Seven-Transmembrane-Helix (7TM) Proteins
While the small ion channels and transporters that have been characterized by SSNMR so far are marked by significant water-exposed protein surfaces and extensive structural plasticity, 7TM helix proteins are nearly diametrically opposite in both respects. Their water-exposed surface area is a small fraction of the whole protein and is mostly restricted to short loops between TM helices, and their structures are relatively insensitive to the membrane composition. So far, the 7TM proteins that have been studied by SSNMR fall into two classes: microbial rhodopsins and eukaryotic G-protein coupled receptors (GPCRs). Microbial rhodopsins serve as light-driven proton pumps, with bacteriorhodopsin (bR) and proteorhodopsin (PR) as prime examples, photosensors such as sensory rhodopsin, and light-gated non-specific cation channels such as channelrhodopsins (ChR) (22). These microbial rhodopsins all contain a retinal chromophore, whose photocycle is intimately related to the protein function. In comparison, eukaryotic GPCRs use ligand binding from outside the cell to activate an array of cellular responses (70), and are the target of ~40% of pharmaceutical drugs.
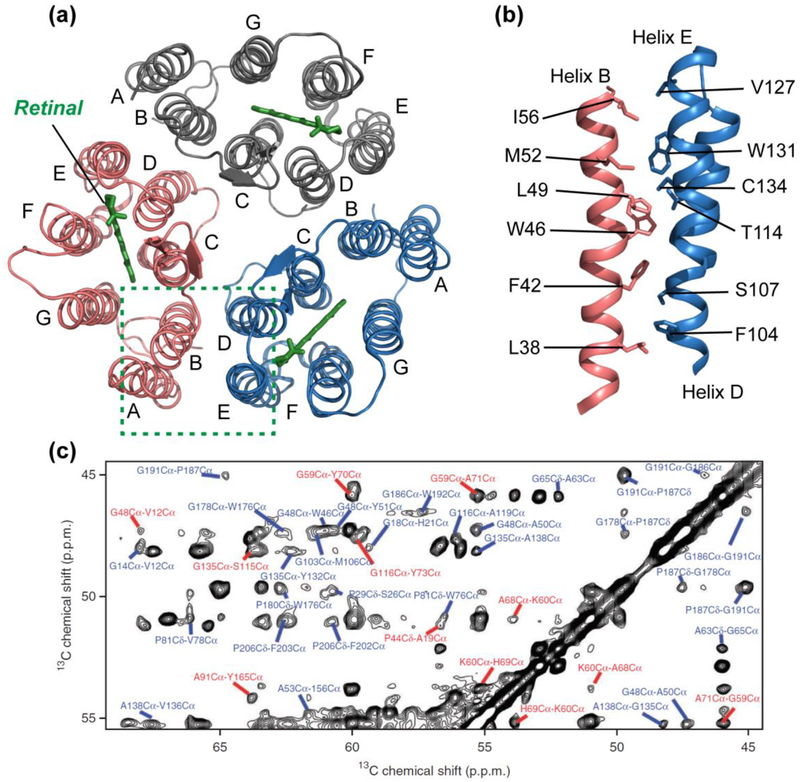
Because of the dominance of protein-protein interaction over protein-lipid and protein-water interactions, 7TM proteins typically give much narrower SSNMR spectral linewidths than small ion channels and transporters. This structural property, together with sparse isotopic labeling, enabled the determination of the atomic-resolution structure of Anabaena sensory rhodopsin (ASR), a cyanobacterial photoreceptor (94). Chemical-shift based torsion angle restraints and inter-residue cross-peaks as distance constraints were measured to determine the structure of each protein monomer (Fig. 4a). Since ASR forms trimers in lipid membranes, the intermolecular interface was determined by covalent attachment of a nitroxide spin label to S26C in helix A and measurement of paramagnetic relaxation enhancement (PRE) of neighboring spins. The PRE data showed that helices A and B of one subunit are packed against helices D and E of the adjacent subunit (93) (Fig. 4b). This intermolecular helix packing is the same as that of bR, but the trimer interface is rich with aromatic residues in ASR, in contrast to the methyl-rich interface in bR. The higher aromatic content of the ASR trimer interface was proposed to explain the higher stability of ASR in detergents. The chemical shifts of ASR bound to DMPC/DMPA liposomes were found to be very similar to the chemical shifts of protein bound to E. coli lipids (96), indicating that the ASR structure is mostly independent of the membrane composition, in contrast to the conformational plasticity of single-span TM proteins such as influenza M2.
Figure 4.
Structure of microbial rhodopsin ASR determined by SSNMR. (a) Structure of the trimeric retinal-bound protein in DMPC/DMPA liposomes, with the retinal cofactors shown in green (PDB: 2M3G). (b) Side view of residues that line the trimer interface between helix B of one monomer and helices D and E of the adjacent monomer. The high aromatic content at the interface contributes to the high stability of the ASR trimer compared with the prototypical microbial rhodopsin, bR (94). (c) Representative 2D 13C-13C proton-driven spin-diffusion (PDSD) correlation spectrum of membrane-bound ASR with a mixing time of 500 ms, showing interhelical (red) and intrahelical (blue) cross peaks as structural restraints. Panel (c) reproduced with permission from Reference (94).
The importance of understanding the oligomeric structure of 7TM proteins is further exemplified by the fact that microbial rhodopsins can adopt multiple oligomeric states. A case in point is green PR, which can form pentamers or hexamers depending on the protein to lipid ratio and membrane composition. Using DNP SSNMR and mixtures of 13C and 15N-labeled proteins, Glaubitz and coworkers measured intermolecular contacts (60). 2D 15N-13C transferred-echo double-resonance (TEDOR) spectra of pentameric PR showed cross peaks between Arg/Lys and Asp/Glu, indicating two salt bridges at the pentameric interface. Point mutations of these charged residues disrupted the pentameric assembly to favor monomeric or hexameric PR, confirming the importance of these salt bridges for the oligomeric assembly.
III. β-barrel Outer Membrane Proteins
While α-helical TM proteins are found in cytoplasmic membranes, β-barrel membrane proteins are found in the outer membranes of Gram-negative bacteria and in eukaryotic mitochondria membranes. These outer membrane proteins (OMPs) carry out many functions such as ion and metabolite transport, recognition of eukaryotic cells during infection, outer membrane biogenesis, and structural roles (46).
The voltage-dependent anion channel (VDAC) is a mitochondrial β-barrel protein that regulates metabolite exchange, calcium homeostasis, and mitochondria-mediated apoptosis. SSNMR has been used to investigate VDAC isoform 1 (VDAC1) in lipid bilayers to understand protein-lipid interactions and motions of specific protein domains. Static 2H spectra of d54-DMPC indicate that VDAC1 caused an 8 °C increase and broadening of the lipid phase transition temperature (19). 2D 13C-13C correlation spectra of 13C, 15N-labeled VDAC1 in 2D crystals of DMPC showed little changes in cross-peak positions or intensities between 0 °C and 30 °C, indicating that the protein conformation and dynamics are unaffected by the membrane phase transition. In comparison, 2D crystals of DPhPC lipids affected some of the cross-peak positions and enhanced the spectral resolution, indicating that this lipid affects the protein structure, reminiscent of the large effect of DPhPC on the influenza M2 structure (3). Other experiments investigated the N-terminal structure of VDAC1, which was proposed to be important for channel gating. 2D 13C-13C proton assisted recoupling (PAR) spectra detected cross peaks between S193 and the N-terminal residue A14, supporting the position of the N-terminus inside the β-barrel (18). Phosphorylation or mutation of S193 to Glu has been proposed to close the channel, possibly due to structural changes in the N-terminus. However, mutation of S193 to Glu or Ala showed only minor chemical shift perturbations for the N-terminal residues, and functional assays indicate that the mutant has similar conductance as the wild-type channel, thus refuting this model. Extensive backbone and sidechain resonance assignments for VDAC1 in 2D crystals of DMPC bilayers (20) found that the β-strand residues in the protein show very similar structure to LDAO-micelle bound VDAC1, while loops and solvent-exposed residues exhibit larger chemical shift differences.
IV. Viral Fusion Proteins
Enveloped viruses enter cells by fusing the virus envelope with the target cell membrane using viral fusion proteins. These proteins respond to environmental cues such as low pH and receptor binding to undergo a cascade of conformational changes to pull the two membranes into close proximity, cause membrane curvature and dehydration, and eventually merge the two membranes (98). The influenza virus hemagglutinin (HA), human immunodeficiency virus (HIV) gp41, and paramyxovirus (PIV) F, all share the common features of a predominantly α-helical water-soluble ectodomains and trimeric organization of the pre-fusion and post-fusion states. They also contain two important membrane-interacting domains, an N-terminal fusion peptide (FP) that inserts into the target membrane during early stages of fusion, and a C-terminal transmembrane domain (TMD) that remains anchored in the virus envelope. In the post-fusion state, the ectodomain of the protein forms a trimer of hairpins, or six-helix bundle (6HB), which presumably brings the hydrophobic FP and TMD together in the same membrane.
Although crystal structures of the water-soluble ectodomain of a number of viral fusion proteins have been determined (33), the structures of the membrane-interacting FP and TMD have been much more elusive. SSNMR and solution NMR studies have been conducted extensively to characterize the structures and lipid interactions of these hydrophobic fusion protein domains. A common observation that has emerged from these studies is that the FPs and TMDs adopt different conformations depending on the membrane conditions and pH. Chemical shift measurements indicate that the HIV and influenza FPs are predominantly α-helical in negatively charged lipids and cholesterol-free membranes, but convert to the β-sheet conformation in cholesterol-containing membranes (26; 30; 31; 79). The parainfluenza virus 5 (PIV5) FP exhibits α-helical conformation in negatively charged lipids with and without cholesterol, but shifts to the β-sheet structure in negative-curvature lipids such as DOPC and DOPE (106; 107). This membrane-curvature induced conformational change was also observed for the PIV5 TMD, which adopts a sheet-helix-sheet structure in the most negative-curvature membrane (DOPE) but a predominantly helical structure in lamellar membranes, regardless of the membrane surface charge and cholesterol content (109).
Lipid mixing assays, mutagenesis, and measurement of membrane curvature and dehydration, shed light on the relevance of the α-helical and β-sheet conformations for membrane fusion. For the PIV5 FP and TMD, 31P NMR and small-angle X-ray scattering data show that the β-sheet conformation induces negative Gaussian curvature to phosphatidylethanolamine (PE) lipids (106; 109), which is important in hemifusion intermediates. 2D 31P-1H correlation spectra correlating the lipid headgroup with water 1H chemical shifts further indicate that the β-sheet rich conformation causes membrane dehydration compared to the helix-containing membrane. These data suggest that the β-sheet conformation is more important for fusion. For the HIV and influenza FP, both helical and sheet conformations were found to be fusogenic from lipid mixing assays, but fusion activity correlates with other structural features. 13C-15N and 13C-31P rotational-echo double-resonance (REDOR) data showed that β-strand FP of HIV gp41 associates in an antiparallel fashion (79) and is inserted into intermediate depths of the lipid membrane (26), with the deeper insertion correlating with stronger fusogenicity.
The influenza FP forms a helix-turn-helix motif in cholesterol-free membranes, with the angle between the two helices sensitive to pH. 13C-15N REDOR distance measurements between G16 and F9 and between A5 and M17 found that the peptide backbone adopts a mixture of closed and semi-closed states (31). Greater fusogenicity was observed at low pH where the semi-closed population is higher, suggesting that the larger ratio of hydrophobic to hydrophilic surface areas promotes fusion, possibly by stabilizing the membrane curvature needed for fusion (30).
Recent SSNMR experiments have increasingly focused on larger constructs of viral fusion proteins to address the question of whether the FP and TMD structural information obtained from peptide studies is preserved in the intact protein, and how this structure fits into the context of coordinated conformational changes of these multi-domain proteins. HIV gp41 has a relatively small ectodomain, and the FP has been successfully ligated to the two coiled coil segments (NHR and CHR) to probe the post-fusion state, while ligation of FP with NHR alone serves as a model of the prehairpin structure (75; 77). When the FP is tethered to both NHR and CHR, the resulting FP-hairpin was found to cause rapid fusion of anionic vesicles at low pH but not at high pH, nor to neutral vesicles, indicating that electrostatic interactions between the lipids and the ectodomain have significant effects on fusion. Chemical shift measurements indicate that the hairpin increased the helical propensity of the FP compared to the separate FP peptide, but the FP β-sheet remains antiparallel in the long construct, as shown by intermolecular 13CO-13CO dipolar coupling measurements (77). Thus, the ectodomain does not change the oligomeric assembly of the FP. Since the full-length protein forms parallel coiled coils for the ectodomain, the antiparallel FP structure found from the SSNMR data was explained as interdigitation of at least two trimers in the membrane.
Compared to HIV, the PIV5 fusion protein has a much larger ectodomain, thus the post-fusion state was investigated using a chimera that links the hydrophobic FP and TMD through a short and flexible linker (108). Interestingly, chemical shifts from 2D correlation spectra indicate that both domains exhibit stable α-helical conformations in all membranes examined, without a shift to the β-sheet conformation in negative-curvature lipids. This implies that each hydrophobic domain exerts an influence on the conformation of the other domain. However, no inter-domain cross peaks were observed in 2D correlation spectra up to a mixing time of 1.0 s, indicating that these FP and TMD helices are not tightly packed as the ectodomain 6HB. The chimera is well inserted into the membrane without perturbing the lamellar structure, which also differs from the property of the separate peptides. These results illustrate the dramatic difference in the structural properties and lipid interactions between the spatially separate peptides and the combined protein, indicating that the early fusion states of the protein have very distinct structures from the post-fusion structure. Future studies will be important for delineating the atomic details and membrane interactions of the prefusion and postfusion states of these fusion machineries.
V. Membrane-interacting amyloid-forming proteins: Aβ and α-synuclein
Although atomic-resolution structures have been determined for a number of amyloid fibrils using SSNMR (13; 89), recent research increasingly suggests that the neurotoxic agents in some of the most common neurodegenerative diseases may be prefibrillar and oligomeric states of these proteins rather than mature fibrils. For the Alzheimer’s disease (AD) Aβ peptide, one hypothesis of the mechanism of toxicity is Aβ interaction with lipid membranes (49): the membrane surface was proposed to speed up fibril formation compared to aqueous solution, and this faster aggregation may in turn disrupt the membrane integrity. An atomic-resolution structure of Aβ40 bound to DMPC/DMPG membranes was recently reported (66). The lipid-bound Aβ fibrils exhibit the same general β-loop-β architecture as the solution fibrils, but the C-terminal strand has a significant kink, which is absent in the solution fibril. How this β-loop-β binds to and self-assembles in the lipid membrane is of significant interest. Binding and fibril formation depend on the peptide-lipid mixing protocol. Direct titration of monomeric Aβ to lipid vesicles gives slower fibrilization (days) and less membrane disruption, while peptide that is mixed with lipids in organic solvents first before switching to aqueous solution caused β-sheet formation within hours (73; 74). These differences in fibril-forming kinetics were manifested in thioflavin-T fluorescence, circular dichroism, 31P NMR spectra, and 2D 13C-13C correlation spectra. The lipid composition affects Aβ binding to the membrane and folding: negatively charged lipids such as POPG and thin bilayers such as DLPC facilitate fibril formation (48). The slower folding of titrated Aβ allowed the comparison of the folding rates of different domains of Aβ: A30 and L34, which reside in the C-terminal hydrophobic core, showed no chemical shift changes after 2 days, while D23, S26 in the loop region showed chemical shift changes over the entire incubation period (73). The depth of insertion and membrane topology of the Aβ peptide were measured using 13C-31P REDOR experiments, which showed that the L17-L34 segment has close contact with lipid headgroups (2; 73). Complementary to the SSNMR data of membrane-bound Aβ fibrils, solution NMR studies of Aβ in earlier stages of binding to zwitterionic lipids showed that the region between K16 and E22 has a conserved α-helical structure while the rest of the peptide is disordered (47). Many questions remain open about the precise structure of lipid-bound Aβ fibrils, the mechanism of fibril formation in the membrane, and whether membrane pores are induced by these fibrils.
The 140-residue protein α-synuclein (αS) is intrinsically disordered in solution and can misfold into parallel in-register β-sheets, which are the major constituent of Lewy bodies in Parkinson’s disease (PD) (88). αS can also bind membranes, and this binding is implicated in the physiological function of αS in regulating synaptic vesicle clustering, recycling and neurotransmitter release (25). SSNMR and solution NMR have been used to characterize the propensities of different regions of αS to bind lipid membranes and adopt defined secondary structures. The protein consists of a long N-terminal domain, a central non-amyloid-β component (NAC) region, which is the core of the β-sheet fibril in the aggregated state (88), and a disordered C-terminus. 2D 13C-13C correlation spectra of POPC/POPA-bound αS followed the structural evolution of αS in the course of several days (12). αS exhibits α-helical chemical shifts for N-terminal and NAC-enriched Ala, Thr, and Val residues up to a day, after which the chemical shifts progressively change to β-sheet values and become stable after ~11 days (12). In contrast, fibrilization from solution did not show the α-helical intermediate. The depth of insertion of αS into the POPC/POPA bilayers was probed using 1H-13C 2D correlation experiments with 1H spin diffusion (44). Comparison of water-protein and lipid-protein 1H-13C cross peaks indicates that αS is well exposed to water but far from the lipid acyl chains at short incubation times, but the lipid-protein cross peaks become equal to water-protein cross peaks after 3 days, concomitant with the dominance of the β-sheet conformation. These results indicate that membrane-bound αS evolves from a surface-bound α-helical structure for the N-terminal and NAC domains to an inserted β-sheet structure, with the end point of the structure similar to fibrils formed in solution.
Interesting structural differences were found when αS is bound to small unilamellar vesicles (SUV’s) that mimic the composition (DOPE, DOPC, and DOPS) and curvature of synaptic vesicles. Chemical shifts measured from 2D 13C-13C and 15N-13C correlation spectra indicate that the SUV-bound αS has a continuous and rigid α-helical structure for residues 6–25 (24; 25), and this domain is strongly associated with the membrane. The N-terminal α-helix is relatively independent of the lipid composition (21), which is consistent with the POPC/POPA study using large unilamellar vesicles. In comparison, residues 26–97 exhibit intermediate dynamics and cannot be detected in cross polarization (CP) and insensitive nuclei enhanced by polarization transfer (INEPT) spectra, in contrast to the POPC/POPA results. These intermediate dynamics are proposed to be relevant for αS’s function in anchoring two synaptic vesicles. The depth of insertion of αS in SUV’s was probed using PRE experiments (24). Gadolinium salts of PE-DTPA lipids caused stronger PRE broadening of hydrophilic residues than hydrophobic residues of the N-terminal α-helix, indicating a membrane surface location, while 16-doxyl-PC with a paramagnetic doxyl in the lipid tails did not cause line broadening of protein residues, indicating that αS does not insert deeply into these curved SUVs. This result again differs from the topology of αS in large membrane vesicles. Thus, both the conformation and membrane topology of αS are sensitive to the membrane composition and curvature.
Conclusion
Two common themes about membrane protein structures have emerged from SSNMR studies of membrane proteins in the last 10 years. First, the protonation state of polar residues is critical for the structure and function of many membrane proteins, as exemplified by the proton-selective histidine in the influenza M2 protein, glutamic acid residues in the pH sensor of KcsA, and the substrate-binding glutamate in EmrE. SSNMR is ideal for elucidating the protonation state and hydrogen bonding of these residues and will continue to make unique and insightful discoveries in this area. Second, structural plasticity is extensive for membrane proteins whose function requires substrate movement or membrane remodeling. α-helical ion channels and transporters exhibit both localized motion of functional residues and coordinated motions of protein backbones. The former is exemplified by the proton-selective histidine and gating tryptophan of influenza M2, while the latter is showcased by the EmrE transporter. This plasticity is manifested especially strikingly in viral fusion proteins, whose two hydrophobic domains can undergo dramatic helix-sheet transitions due to changes of the membrane lipid composition. Even for larger 7TM helix proteins, the oligomeric structure can vary, as seen in the case of proteorhodopsin. The factor that affects membrane protein structure most significantly is the lipid composition of the membrane. Thus, choosing the lipid composition that best mimics the natural membrane in which a protein carries out its function is crucial for obtaining biologically relevant structural information. As SSNMR techniques increasingly mature and become well adopted to determine high-resolution structures of membrane proteins, elucidating protein conformational dynamics and conformational plasticity, unraveling protein-lipid interactions, and illuminating how membrane proteins interact with ions, drug molecules, ligands, and other proteins, will become ever more important areas of discovery. For revealing the complex structural and dynamical basis of membrane protein function, SSNMR remains an unrivaled technique.
Abbreviations:
- DHPC
1,2-dihexanoyl-sn-glycero-3-phosphocholine
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- DOPE
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- DPhPC
1,2-diphytanoyl-sn-glycero-3-phosphocholine
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DMPA
1,2-dimyristoyl-sn-glycero-3-phosphate
- DMPG
1,2-dimyristoyl-sn-glycero-3-phospho-(1’-rac-glycerol)
- DLPC
1,2-dilinoleoyl-sn-glycero-3-phosphocholine
- DOPS
1,2-dioleoyl-sn-glycero-3-phosphocholine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac-glycerol)
- POPA
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate
- PE-DTPA
1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-diethylenetriaminepentaacetic acid
- 16-doxyl-PC
1-palmitoyl-2-stearoyl-[16-doxyl]-sn-glycero-3-phosphocholine
- PDSD
proton-driven spin-diffusion
- PISEMA
polarization inversion spin exchange at the magic angle
- TEDOR
transferred-echo double-resonance
- PAR
proton assisted recoupling
- REDOR
rotational-echo double-resonance
- INEPT
insensitive nuclei enhanced by polarization transfer
References
- 1.Acharya A, Carnevale V, Fiorin G, Levine BG, Polishchuk A, et al. 2010. Structural mechanism of proton transport through the influenza A M2 protein, Proc. Natl. Acad. Sci. U.S.A 107:15075–15080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akinlolu R, Nam M, Qiang W. 2015. Competition between Fibrillation and Induction of Vesicle Fusion for the Membrane-Associated 40-Residue b-Amyloid Peptides, Biochemistry 54:3416–3419 [DOI] [PubMed] [Google Scholar]
- 3.Andreas LB, Reese M, Eddy MT, Gelev V, Ni QZ, et al. 2015. Structure and Mechanism of the Influenza A M218–60 Dimer of Dimers, J. Am. Chem. Soc 137:14877–14886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbet-Massin E, Pell AJ, Retel JS, Andreas LB, Jaudzems K, et al. 2014. Rapid proton-detected NMR assignment for proteins with fast magic angle spinning, J. Am. Chem. Soc 136:12489–12497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhate MP, McDermott AE. 2012. Protonation state of E71 in KcsA and its role for channel collapse and inactivation, Proc. Natl. Acad. Sci. U.S.A 109:15265–15270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bright RA, Medina MJ, Xu X, Perez-Oronoz G, Wallis TR, et al. 2005. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern, Lancet 366:1175–1181 [DOI] [PubMed] [Google Scholar]
- 7.Cady SD, Goodman C, Tatko C, DeGrado WF, Hong M. 2007. Determining the orientation of uniaxially rotating membrane proteins using unoriented samples: a 2H, 13C, and 15N solid-state NMR investigation of the dynamics and orientation of a transmembrane helical bundle, J. Am. Chem. Soc 129:5719–5729 [DOI] [PubMed] [Google Scholar]
- 8.Cady SD, Schmidt-Rohr K, Wang J, Soto CS, DeGrado WF, Hong M. 2010. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers, Nature 463:689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cady SD, Wang T, Hong M. 2011. Membrane-Dependent Effects of a Cytoplasmic Helix on the Structure and Drug Binding of the Influenza Virus M2 Protein, J. Am. Chem. Soc:in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho MK, Gayen A, Banigan JR, Leninger M, Traaseth NJ. 2014. Intrinsic Conformational Plasticity of Native EmrE Provides a Pathway for Multidrug Resistance, J. Am. Chem. Soc 136:8072–8080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colvin MT, Andreas LB, Chou JJ, Griffin RG. 2014. Proton association constants of His 37 in the Influenza-A M218–60 dimer-of-dimers, Biochemistry 53:5987–5994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comellas G, Lemkau L, Zhou D, George J, Rienstra C. 2012. Stuctural Intermediates During a-Synuclein Fibrillogenesis on Phospholipid Vesicles, J. Am. Chem. Soc 134:5090–5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comellas G, Rienstra CM. 2013. Protein structure determination by magic-angle spinning solid-state NMR, and insights into the formation, structure, and stability of amyloid fibrils, Annu. Rev. Biophys 42:515–536 [DOI] [PubMed] [Google Scholar]
- 14.Cristian L, Lear JD, DeGrado WF. 2003. Use of thiol-disulfide equilibria to measure the energetics of assembly of transmembrane helices in phospholipid bilayers., Proc. Natl. Acad. Sci. U.S.A 100:14772–14777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuello LG, Jogini V, Cortes DM, Perozo E. 2010. Structural mechanism of C-type inactivation in K(+) channels, Nature 466:203–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doyle DA, Cabral JM, Pfuetzner RA, Kuo A, Gulbis JM, et al. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity, Science 280:69–77 [DOI] [PubMed] [Google Scholar]
- 17.Duong-Ly KC, Nanda V, DeGrado WF, Howard KP. 2005. The conformation of the pore region of the M2 proton channel depends on lipid bilayer environment, Protein Sci 14:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eddy MT, Andreas L, Teijido O, Su Y, Clark L, et al. 2015. Magic angle spinning nuclear magnetic resonance characterization of voltage-dependent anion channel gating in two-dimensional lipid crystalline bilayers, Biochemistry 54:994–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eddy MT, Ong TC, Clark L, Teijido O, van der Wel PC, et al. 2012. Lipid dynamics and protein-lipid interactions in 2D crystals formed with the beta-barrel integral membrane protein VDAC1, J. Am. Chem. Soc 134:6375–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eddy MT, Su Y, Silvers R, Andreas L, Clark L, et al. 2015. Lipid bilayer-bound conformation of an integral membrane beta barrel protein by multidimensional MAS NMR, J. Biomol. NMR 61:299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eichmann C, Campioni S, Kowal J, Maslennikov I, Gerez J, et al. 2016. Preparation and Characterization of Stable a-Synuclein Lipoprotein Particles, J. Biol. Chem 291:8516–8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ernst OP, Lodowski DT, Elstner M, Hegemann P, Brown LS, Kandori H. 2014. Microbial and Animal Rhodopsins: Structures, Functions, and Molecular Mechanisms, Chem. Rev 114:126–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu R, Miao Y, Qin H, Cross TA. 2016. Probing Hydronium Ion Histidine NH Exchange Rate Constants in the M2 Channel via Indirect Observation of Dipolar-Dephased 15N Signals in Magic-Angle-Spinning NMR, J. Am. Chem. Soc 138:15801–15804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fusco G, De Simone A, Gopinath T, Vostrikov V, Vendruscolo M, et al. 2014. Direct Observation of the Three Regions in a-Synuclein that Determine its Membrane-Bound Behaviour, Nat. Comm 5:3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fusco G, Pape T, Stephens A, Mahou P, Costa A, et al. 2016. Structural Basis of Synaptic Vesicle Assembly Promoted by a-Synuclein, Nat. Comm 7:12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gabrys C, Qiang W, Sun Y, Xie L, Schmick S, Weliky D. 2013. Solid-State Nuclear Magnetic Resonance Measurements of HIV Fusion Peptides 13CO to Lipid 31P Proximities Support Similar Partially Inserted Membrane Locations of the a Helical and b Sheet Peptide Structures, J. Phys. Chem. A 117:9848–9859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gadsby DC. 2009. Ion channels versus ion pumps: the principal difference, in principle, Nat. Rev. Mol. Cell. Biol 10:344–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gayen A, Banigan JR, Traaseth NJ. 2013. Ligand-Induced Conformational Changes of the Multidrug Resistance Transporter EmrE Probed by Oriented Solid-State NMR Spectroscopy, Angew. Chem. Int. Ed 52:10321–10324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gayen A, Leninger M, Traaseth NJ. 2016. Protonation of a glutamate residue modulates the dynamics of the drug transporter EmrE, Nat. Chem. Biol 12:141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghosh U, Xie L, Jia L, Liang S, Weliky D. 2015. Closed and Semiclosed Interhelical Structures in Membrane vs Closed and Open Structures in Detergent for the Influenza Virus Hemagglutinin Fusion Peptide and Correlation of Hydrophobic Surface Area with Fusion Catalysis, J. Am. Chem. Soc 137:7548–7551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh U, Xie L, Weliky D. 2013. Detection of Closed Influenza Virus Hemagglutinin Fusion Peptide Structures in Membranes by Backbone 13CO-14N Rotational-Echo Double-Resonance Solid-State NMR, J. Biomol. NMR 55:139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grandea AG, Olsen OA, Cox TC, Renshaw M, Hammond PW, et al. 2010. Human antibodies reveal a protective epitope that is highly conserved among human and nonhuman influenza A viruses, Proc. Natl. Acad. Sci. U.S.A 107:12658–12663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrison SC. 2008. Viral membrane fusion, Nat. Struct. Mol. Biol 15:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higgins CF. 2007. Multiple molecular mechanisms for multidrug resistance transporters, Nature 446:749–757 [DOI] [PubMed] [Google Scholar]
- 35.Hille B 1992. Ionic channels of excitable membranes Sundeland: Sinauer Associates Inc. [Google Scholar]
- 36.Hong M, DeGrado WF. 2012. Structural basis for proton conduction and inhibition by the influenza M2 protein, Protein Sci 21:1620–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong M, Fritzsching KJ, Williams JK. 2012. Hydrogen-bonding partner of the proton-conducting histidine in the influenza M2 proton channel revealed from 1H chemical shifts, J. Am. Chem. Soc 134:14753–14755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoshi T, Zagotta WN, Aldrich RW. 1991. 2 Types of Inactivation in Shaker K+ Channels - Effects of Alterations in the Carboxy-Terminal Region, Neuron 7:547–556 [DOI] [PubMed] [Google Scholar]
- 39.Hu F, Luo W, Cady SD, Hong M. 2011. Conformational plasticity of the influenza A M2 transmembrane peptide in lipid bilayers under varying pH, drug binding and membrane thickness, Biochim. Biophys. Acta 1808:415–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu F, Luo W, Hong M. 2010. Mechanisms of proton conduction and gating by influenza M2 proton channels from solid-state NMR, Science 330:505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu F, Schmidt-Rohr K, Hong M. 2012. NMR Detection of pH-dependent histidine-water proton exchange reveals the conduction mechanism of a transmembrane proton channel, J. Am. Chem. Soc 134:3703–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu J, Asbury T, Achuthan S, Li C, Bertram R, et al. 2007. Backbone structure of the amantadine-blocked trans-membrane domain M2 proton channel from Influenza A virus, Biophys. J 92:4335–4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu J, Fu R, Nishimura K, Zhang L, Zhou HX, et al. 2006. Histidines, heart of the hydrogen ion channel from influenza A virus: Toward an understanding of conductance and proton selectivity, Proc. Natl. Acad. Sci. U.S.A 103:6865–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huster D, Yao XL, Hong M. 2002. Membrane Protein Topology Probed by 1H Spin Diffusion from Lipids Using Solid-State NMR Spectroscopy, J. Am. Chem. Soc 124:874–883 [DOI] [PubMed] [Google Scholar]
- 45.Kim SS, Upshur MA, Saotome K, Sahu ID, McCarrick RM, et al. 2015. Cholesterol-Dependent Conformational Exchange of the C-Terminal Domain of the Influenza A M2 Protein, Biochemistry 54:7157–7167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koebnik R, Locher KP, Van Gelder P. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell, Mol. Microbiol 37:239–253 [DOI] [PubMed] [Google Scholar]
- 47.Korshavn K, Bhunia A, Lim M, Ramamoorthy A. 2016. Amyloid-b Adopts a Conserved, Partially Folded Structure Upon Binding to Zwitterionic Lipid Bilayers Prior to Amyloid Formation, Chem. Commun 52:882–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korshavn K, Satriano C, Lin Y, Zhang R, Dulchavsky M, et al. 2017. Reduced Lipid Bilayer Thickness Regulates the Aggregation and Cytotoxicity of Amyloid-b, J. Biol. Chem 292:4638–4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kotler S, Walsh P, Brender J, Ramamoorthy A. 2014. Differences between Amyloid-b Aggregation in Solution and on the Membrane: Insights inot Elucidation of the Mechanistic Details of Alzheimer’s Disease, Chem. Soc. Rev 43:6692–6700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwon B, Hong M. 2016. The Influenza M2 Ectodomain Regulates the Conformational Equilibria of the Transmembrane Proton Channel: Insights from Solid-State Nuclear Magnetic Resonance, Biochemistry 55:5387–5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwon B, Tietze D, White PB, Liao SY, Hong M. 2015. Chemical Ligation of the Influenza M2 Protein for Solid-State NMR Characterization of the Cytoplasmic Domain Structure, Protein Sci 24:1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lehner I, Basting D, Meyer B, Haase W, Manolikas T, et al. 2008. The key residue for substrate transport (Glu14) in the EmrE dimer is asymmetric, J. Biol. Chem 283:3281–3288 [DOI] [PubMed] [Google Scholar]
- 53.Lehnert E, Mao JF, Mehdipour AR, Hummers G, Abele R, et al. 2016. Antigenic Peptide Recognition on the Human ABC Transporter TAP Resolved by DNP-Enhanced Solid-State NMR Spectroscopy, J. Am. Chem. Soc 138:13967–13974 [DOI] [PubMed] [Google Scholar]
- 54.Liao SY, Fritzsching KJ, Hong M. 2013. Conformational analysis of the full-length M2 protein of the influenza A virus using solid-state NMR, Protein Sci 22:1623–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao SY, Yang Y, Tietze D, Hong M. 2015. The influenza M2 cytoplasmic tail changes the proton-exchange equilibria and the backbone conformation of the transmembrane histidine residue to facilitate proton conduction, J. Am. Chem. Soc 137:6067–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Long SB, Tao X, Campbell EB, MacKinnon R. 2007. Atomic Structure of a Voltage-Dependent K+ Channel in a Lipid Membrane-Like Environment, Nature 450:376–383 [DOI] [PubMed] [Google Scholar]
- 57.Luo W, Cady SD, Hong M. 2009. Immobilization of the Influenza A M2 Transmembrane Peptide in Virus-Envelope Mimetic Lipid Membranes: A Solid-State NMR Investigation, Biochemistry 48:6361–6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo W, Mani R, Hong M. 2007. Sidechain conformation and gating of the M2 transmembrane peptide proton channel of influenza A virus from solid-state NMR, J. Phys. Chem 111:10825–10832 [DOI] [PubMed] [Google Scholar]
- 59.Ma CL, Polishchuk AL, Ohigashi Y, Stouffer AL, Schon A, et al. 2009. Identification of the functional core of the influenza A virus A/M2 proton-selective ion channel, Proc. Natl. Acad. Sci. U.S.A 106:12283–12288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maciejko J, Mehler M, Kaur J, Lieblein T, Morgner N, et al. 2015. Visualizing Specific Cross-Protomer Interactions in the Homo-Oligomeric Membrane Protein Proteorhodopsin by Dynamic-Nuclear-Polarization-Enhanced Solid-State NMR, J. Am. Chem. Soc 137:9032–9043 [DOI] [PubMed] [Google Scholar]
- 61.Mandala VS, Liao SY, Kwon B, Hong M. 2017. Structural Basis for Asymmetric Conductance of the Influenza M2 Proton Channel Investigated by Solid-State NMR Spectroscopy, J. Mol. Biol 429:2192–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCown MF, Pekosz A. 2006. Distinct domains of the influenza a virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production, J. Virol 80:8178–8189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miao Y, Fu R, Zhou HX, Cross TA 2015. Dynamic Short Hydrogen Bonds in Histidine Tetrad of Full-Length M2 Proton Channel Reveal Tetrameric Structural Heterogeneity and Functional Mechanism, Structure 23:2300–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morrison EA, DeKoster GT, Dutta S, Vafabakhsh R, Clarkson MW, et al. 2011. Antiparallel EmrE exports drugs by exchanging between asymmetric structures, Nature 481:45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ni QZ, Daviso E, Can TV, Markhasin E, Jawla SK, et al. 2013. High Frequency Dynamic Nuclear Polarization, Acc. Chem. Res 46:1933–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niu Z, Zhao W, Zhang Z, Xiao F, Tang X, Yang J. 2014. The molecular structure of Alzheimer β-amyloid fibrils formed in the presence of phospholipid vesicles, Angew. Chem. Int. Ed. Engl 53:9294–9297 [DOI] [PubMed] [Google Scholar]
- 67.Ong YS, Lakatos A, Becker-Baldus J, Pos KM, Glaubitz C. 2013. Detecting substrates bound to the secondary multidrug efflux pump EmrE by DNP-enhanced solid-state NMR, J. Am. Chem. Soc 135:15754–15762 [DOI] [PubMed] [Google Scholar]
- 68.Park EK, Castrucci MR, Portner A, Kawaoka Y. 1998. The M2 ectodomain is important for its incorporation into influenza A virions, J. Virol 72:2449–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pielak RM, Chou JJ. 2011. Influenza M2 proton channels, Biochim. Biophys. Acta Biomem 1808:522–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pierce KL, Premont RT, Lefkowitz RJ. 2002. Seven-transmembrane receptors, Nat. Rev. Mol. Cell. Biol 3:639–650 [DOI] [PubMed] [Google Scholar]
- 71.Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, et al. 1997. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity, Proc. Natl. Acad. Sci. U.S.A 94:11301–11306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pinto LH, Lamb RA. 2006. The M2 Proton Channels of Influenza A and B Viruses, J. Biol. Chem 281:8997–9000 [DOI] [PubMed] [Google Scholar]
- 73.Qiang W, Akinlolu R, Nam M, Shu N. 2014. Structural Evolution and Membrane Interaction of the 40-Residue b Amyloid Peptides: Differences in the Initial Proximity between Peptides and the Membrane Bilayer Studied by Solid-State Nuclear Magnetic Resonance Spectroscopy, Biochemistry 53:7503–7514 [DOI] [PubMed] [Google Scholar]
- 74.Qiang W, Yau W-M, Schulte J. 2015. Fibrillation of b Amyloid Peptides in the Presence of Phospholipid Bilayers and the Consequent Membrane Disruption, Biochim. Biophys. Acta Biomem 1848:266–276 [DOI] [PubMed] [Google Scholar]
- 75.Ratnayake P, Sackett K, Nethercott M, Weliky D. 2015. pH-Dependent Vesicle Fusion Induced by the Ectodomain of the Human Immunodeficiency Virus Membrane Fusion Protein gp41: Two Kinetically Distinct Processes and Fully-Membrane-Associated gp41 with Predominant b Sheet Fusion Peptide Conformation, Biochim. Biophys. Acta Biomem 1848:289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rossman JS, Jing X, Leser GP, Lamb RA. 2010. Influenza virus M2 protein mediates ESCRT-independent membrane scission, Cell 142:902–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sackett K, Nethercott M, Zheng Z, Weliky D. 2014. Solid-State NMR Spectroscopy of the HIV gp41 Membrane Fusion Protein Supports Intermolecular Antiparallel b Sheet Fusion Peptide Structure in the Final Six-Helix Bundle State, J. Mol. Biol 426:1077–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saotome K, Duong-Ly KC, Howard KP. 2015. Influenza A M2 protein conformation depends on choice of model membrane, Biopolymers 104:405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schmick S, Weliky D. 2010. Major Antiparallel and Minor Parallel b Sheet Populations Detected in the Membrane-Associated Human Immunodeficiency Virus Fusion Peptide, Biochemistry 49:10623–10635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schneider R, Ader C, Lange A, Giller K, Hornig S, et al. 2008. Solid-state NMR spectroscopy applied to a chimeric potassium channel in lipid bilayers, J. Am. Chem. Soc 130:7427–7435 [DOI] [PubMed] [Google Scholar]
- 81.Schnell JR, Chou JJ. 2008. Structure and mechanism of the M2 proton channel of influenza A virus, Nature 451:591–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sergeyev IV, Itin B, Rogawski R, Day LA, McDermott AE. 2017. Efficient assignment and NMR analysis of an intact virus using sequential side-chain correlations and DNP sensitization, Proc. Natl. Acad. Sci. U.S.A 114:5171–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sharma M, Yi M, Dong H, Qin H, Peterson E, et al. 2010. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer, Science 330:509–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, et al. 2008. Structural basis for the function and inhibition of an influenza virus proton channel, Nature 451:596–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tang M, Comellas G, Rienstra CM. 2013. Advanced solid-state NMR approaches for structure determination of membrane proteins and amyloid fibrils, Acc. Chem. Res 46:2080–2088 [DOI] [PubMed] [Google Scholar]
- 86.Thomaston JL, Alfonso-Prieto M, Woldeyes RA, Fraser JS, Klein ML, et al. 2015. High-resolution structures of the M2 channel from influenza A virus reveal dynamic pathways for proton stabilization and transduction, Proc. Natl. Acad. Sci. U.S.A 112:14260–14265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thompson AN, Posson DJ, Parsa PV, Nimigean CM. 2008. Molecular mechanism of pH sensing in KcsA potassium channels, Proc. Natl. Acad. Sci. U.S.A 105:6900–6905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tuttle M, Comellas G, Nieuwkoop A, Covell D, Berthold D, et al. 2016. Solid-State NMR Structure of a Pathogenic Fibril of Full-Length Human a-Synuclein, Nat. Struct. Mol. Biol 23:409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tycko R, Wickner RB. 2013. Molecular structures of amyloid and prion fibrils: consensus versus controversy, Acc. Chem. Res 46:1487–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Unwin N 1989. The Structure of Ion Channels in Membranes of Excitable Cells, Neuron 3:665–676 [DOI] [PubMed] [Google Scholar]
- 91.van der Cruijsen EA, Nand D, Weingarth M, Prokofyev A, Hornig S, et al. 2013. Importance of lipid-pore loop interface for potassium channel structure and function, Proc. Natl. Acad. Sci. U.S.A 110:13008–13013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van der Cruijsen EAW, Prokofyev AV, Pongs O, Baldus M. 2017. Probing Conformational Changes during the Gating Cycle of a Potassium Channel in Lipid Bilayers, Biophys. J 112:99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang S, Munro RA, Kim SY, Jung KH, Brown LS, Ladizhansky V. 2012. Paramagnetic relaxation enhancement reveals oligomerization interface of a membrane protein, J. Am. Chem. Soc 134:16995–16998 [DOI] [PubMed] [Google Scholar]
- 94.Wang SL, Munro RA, Shi LC, Kawamura I, Okitsu T, et al. 2013. Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein, Nature Methods 10:1007-+ [DOI] [PubMed] [Google Scholar]
- 95.Wang T, Cady SD, Hong M. 2012. NMR Determination of Protein Partitioning into Membrane Domains with Different Curvatures and Application to the Influenza M2 Peptide, Biophys. J 102:787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ward ME, Wang SL, Munro R, Ritz E, Hung I, et al. 2015. In Situ Structural Studies of Anabaena Sensory Rhodopsin in the E. coli Membrane, Biophys. J 108:1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Weingarth M, Prokofyev A, van der Cruijsen EA, Nand D, Bonvin AM, et al. 2013. Structural determinants of specific lipid binding to potassium channels, J. Am. Chem. Soc 135:3983–3988 [DOI] [PubMed] [Google Scholar]
- 98.White JM, Delos SE, Brecher M, Schornberg K. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme, Crit. Rev. Biochem. Mol. Biol 43:189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.White PB, Hong M. 2015. (15)N and (1)H Solid-State NMR Investigation of a Canonical Low-Barrier Hydrogen-Bond Compound: 1,8-Bis(dimethylamino)naphthalene, J. Phys. Chem. B 119:11581–11589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Williams JK, Hong M. 2014. Probing membrane protein structure using water polarization transfer solid-state NMR, J. Magn. Reson 247:118–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Williams JK, Schmidt-Rohr K, Hong M. 2015. Aromatic spectral editing techniques for magic-angle-spinning solid-state NMR spectroscopy of uniformly (13)C-labeled proteins, Solid State Nucl. Magn. Reson 72:118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Williams JK, Tietze D, Lee M, Wang J, Hong M. 2016. Solid-State NMR Investigation of the Conformation, Proton Conduction, and Hydration of the Influenza B Virus M2 Transmembrane Proton Channel, J. Am. Chem. Soc 138:8143–8155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Williams JK, Tietze D, Wang J, Wu Y, DeGrado WF, Hong M. 2013. Drug-Induced Conformational and Dynamical Changes of the S31N Mutant of the Influenza M2 Proton Channel Investigated by Solid-State NMR, J. Am. Chem. Soc 135:9885–9897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Williams JK, Zhang Y, Schmidt-Rohr K, Hong M. 2013. pH-Dependent Conformation, Dynamics, and Aromatic Interaction of the Gating Tryptophan Residue of the Influenza M2 Proton Channel from Solid-State NMR, Biophys. J 104:1698–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wylie BJ, Bhate MP, McDermott AE. 2014. Transmembrane allosteric coupling of the gates in a potassium channel, Proc. Natl. Acad. Sci. U.S.A 111:185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yao H, Hong M. 2014. Conformation and Lipid Interaction of the Fusion Peptide of the Paramyxovirus PIV5 in Anionic and Negative-Curvature Membranes from Solid-State NMR, J. Am. Chem. Soc 136:2611–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yao H, Hong M. 2013. Membrane-Dependent Conformation, Dynamics, and Lipid Interactins of the Fusion Peptide of the Paramyxovirus PIV5 from Solid-State NMR, J. Mol. Biol 425:563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yao H, Lee M, Liao S, Hong M. 2016. Solid-State Nuclear Magnetic Resonance Investigation of the Structural Topology and Lipid Interactions of a Viral Fusion Protein Chimera Containing the Fusion Peptide and Transmembrane Domain, Biochemistry 55:6787–6800 [DOI] [PubMed] [Google Scholar]
- 109.Yao H, Lee M, Waring A, Wong G, Hong M. 2015. Viral Fusion Protein Transmembrane Domain Adopts b-Strand Structure to Facilitate Membrane Topological Changes for Virus-Cell Fusion, Proc. Natl. Acad. Sci. U.S.A 112:10926–10931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhou HX, Cross TA. 2013. Influences of membrane mimetic environments on membrane protein structures, Annu. Rev. Biophys 42:361–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution, Nature 414:43–48 [DOI] [PubMed] [Google Scholar]