Abstract
Sex steroid receptors have received much interest as potential mediators of human behaviors and mental disorders. Candidate gene association studies have identified about 50 genetic variants of androgen and estrogen receptors that correlate with human behavioral phenotypes. Because most of these polymorphisms lie outside coding regions, discerning their effect on receptor function is not straightforward. Thus, although discoveries of associations improve our ability to predict risk, they have not greatly advanced our understanding of underlying mechanisms. This article is intended to serve as a starting point for psychologists and other behavioral biologists to consider potential mechanisms. Here, I review associations between polymorphisms in sex steroid receptors and human behavioral phenotypes. I then consider ways in which genetic variation can affect processes such as mRNA transcription, splicing, and stability. Finally, I suggest ways that hypotheses about mechanism can be tested, for example using in vitro assays and/or animal models.
Keywords: androgen receptor, anxiety, association studies, depression, endophenotype, estrogen receptor, ESR1, ESR2, polymorphism, regulatory variation.
1. Introduction
The middleman between genotype and phenotype is often a hormone. Over the past two decades, more and more human behavioral phenotypes have been linked with hormone-related genes. Variation in sex steroid receptors in particular has been the subject of numerous studies; more than 50 polymorphisms in androgen or estrogen receptors have been associated with human behaviors or mental health outcomes. Most of these links have been revealed not by genome-wide association studies, but rather by targeted, hypothesis-driven approaches in which a single gene, or group of related genes, is considered. Genotype-phenotype associations are found by comparing allele frequencies between groups of individuals with vs. without the phenotype of interest or by correlating phenotypic expression with the number of copies of a certain allele. These types of studies tell us the extent to which a particular gene sequence predicts the expression of a phenotype, which can be a useful start to determining underlying mechanisms. When significant associations are found between a receptor polymorphism and a phenotype, authors typically conclude that the receptor is “involved” or “plays a role” in the phenotype. The trouble here is twofold: first, an association between genotype and phenotype could be mediated by any number of yet unknown factors, only some of which are biological, and does not necessarily indicate a direct relationship. Second, even if the effect is causal, the association itself tells us little about how a gene sequence contributes to behavior.
What do associations between polymorphisms and behavior actually tell us? Answering this question will require us to look under the hood, inside the black box. Efforts to move beyond associations will take advantage of tools that can reveal the impact of genetic variation at the molecular level—tools that are becoming more and more accessible to researchers in a variety of non-molecular fields. Functional studies, in which the effects of polymorphisms are tested experimentally, have been relatively slow to gain popularity in the area of sex steroid receptors and behavior. Below, I will review the known associations between sex steroid receptor polymorphisms and behavior in humans, discuss the mechanisms by which polymorphisms can, theoretically, affect gene function, and suggest ways that we might use sex steroid polymorphisms to understand how behavior is encoded in the genome.
Before proceeding, I should make clear that I have intentionally avoided the topic of effect sizes, or the extent to which a particular polymorphism predicts a behavioral phenotype. The fact that polymorphisms predict little variation has been covered extensively elsewhere (e.g., Cannon & Keller, 2006; Saltz, 2017). The predictive power of each polymorphism is not particularly relevant to my purposes here. Instead, I will focus on the following question: When an association (of any size) is found, to what extent can we infer that the receptor plays a role (of any size) in behavior? Jumping to such a conclusion glosses over perhaps too many steps in the path from genotype to phenotype. Given the state of current knowledge, the effect of a polymorphism on receptor functioning can be difficult to predict. The present review is a call for greater consideration of mechanisms, and for the type of research that will allow us to make stronger predictions and draw more rigorous conclusions.
2. Associations between polymorphisms and behavior in humans
2.1. What is a polymorphism?
A given location in the genome is polymorphic if at least two different sequences, or alleles, occur in the same population. For a site to be called polymorphic, the rarer allele must be found in at least 0.5–1% of the population; in other words, it must be frequent enough that it cannot be explained by a random mutation. Currently, the sex steroid receptor polymorphisms known to associate with behavior take one of two forms: single nucleotide polymorphism (SNP) or short tandem repeat (STR). In the case of SNPs, which are by far the most common type of polymorphism, variation is limited to a single base pair. For example, in a location where one allele may have an “A”, another may have a “C”. The overall length of the allele is not affected because one nucleotide is simply substituted for another. The human genome contains approximately 12 million SNPs (with frequencies above 0.5%), depending on the population (1000 Genomes Project Consortium, 2015).
A second major source of variation, STRs (also called microsatellites), are composed of short sequences, usually two or three nucleotides long, repeated over and over for a variable stretch. For example, the motif “CAG” might be repeated four times in some individuals (CAGCAGCAGCAG) whereas in others, this stretch could go on for 30–40 repetitions. Variation due to STRs is therefore represented in the number of repeats present in a particular allele. STRs, which are likely caused by errors during DNA replication, are common; they cover 1–3% of the human genome (Ellegren, 2004; Gymrek et al., 2016). STRs and SNPs are not the only types of polymorphisms, but they make up the majority of polymorphisms that have been linked to human behavioral phenotypes.
Polymorphisms can be a tool for linking genotype to phenotype. They represent genetic variation within a population that can be connected, via association studies, to phenotypic variation. Millions of polymorphisms in the human genome have been mapped and catalogued, and can be queried using online tools such as dbSNP (https://www.ncbi.nlm.nih.gov/projects/SNP). According to dbSNP, there are thousands of SNPs in the human genes for androgen and estrogen receptors (see also Gottlieb et al., 2012). Some polymorphisms have become more popular to study than others, however, and only a fraction have been investigated in the context of human behavior.
2.2. Androgen receptor
The gene for androgen receptor (AR), which in humans is located on the X chromosome, encodes a receptor with high affinity for the androgens testosterone and dihydrotestosterone. This receptor is likely to be the primary pathway by which testosterone affects behavior without being converted to estradiol. Its best-understood function is as a transcription factor that binds DNA and initiates the transcription of androgen-dependent genes. Nongenomic actions have also been described in a variety of tissues, including brain (reviewed by Foradori et al., 2008). The protein itself is large—more than 900 amino acids long—and like other steroid receptors, consists of three major functional domains (reviewed by Gao et al., 2005; see also Gottlieb et al., 2012). Exon 1 encodes a transactivation domain, which is necessary for the bound receptor to initiate transcription (Jenster et al., 1991). Exons two and three encode a DNA binding domain, which recognizes “response elements” in DNA –sequences that mark the genes regulated by AR and recruit the receptors. The remaining five exons encode the ligand-binding domain, which binds the hormone. Deletion of the ligand-binding domain can activate receptor activity as if hormone were bound, suggesting that when unbound, this domain may inhibit receptor function (Jenster et al., 1991). The ligand-binding domain also contains a conserved sequence necessary for association with the cell membrane during nongenomic actions (Pedram et al., 2007).
In humans, mutations in the AR gene can cause partial or complete androgen insensitivity syndrome (AIS), which results in an intersex or female phenotype in genetic males. Mutational analysis of AIS patients has shown that the genetic cause of the syndrome differs from family to family; in other words, it is not uniformly caused by a particular genetic event. Up to 40% of cases show no mutation in AR at all; of those that do, those mutations have occurred at hundreds of different locations spread throughout the gene (Gottlieb et al., 2012). Although it has been interesting and important to study these families and the effect each mutation has on phenotype, such mutations are not frequent enough in the population to qualify as polymorphisms and thus do not lend themselves well to case-control studies or other types of association studies of a single genetic site.
The overwhelming majority of studies associating true polymorphisms in AR with human behavior have focused on one or two STRs in exon 1. The first (rs193922933) consists of a CAG repeat in exon 1 (Fig. 1). Very long alleles (>38 CAG repeats) are linked to the neurological disease spinal bulbar muscular atrophy (La Spada et al., 1991); healthy individuals commonly have between 9 and 36 repeats. Because the relative length of the allele can be assessed simply by assessing the size of a PCR product on a gel, rather than expensive sequencing, genotyping is straightforward. As a result, this polymorphism has been associated with a wide variety of behavioral traits (see Table 1).
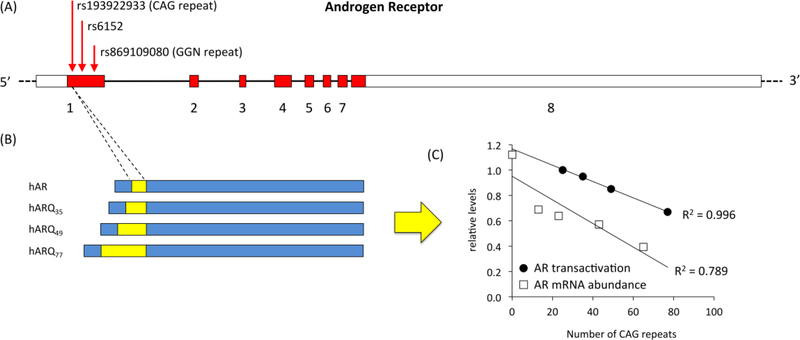
Fig. 1. Genetic polymorphisms in human AR.
(A) Polymorphisms that have been associated with behavioral phenotypes or outcomes are shown. Boxes indicate exons, and lines indicate introns (introns not drawn to scale). Exon numbering, coding regions (red) and UTRs (white) are based on the primary transcript (AR-001) as annotated in ENSEMBL. The positions of two tandem repeat polymorphisms and one single nucleotide polymorphism are indicated by arrows. Each polymorphism is labeled with the Reference SNP ID (rs) number assigned by dbSNP (NCBI). Locations of polymorphisms are approximate. (B) The greater the number of CAG repeats, the longer a stretch of glutamine residues in the translated protein (not to scale). This “poly-Q stretch” (yellow) contains 25 glutamines in the human wild-type AR (hAR). (C) Increased length of the poly-Q stretch inhibits the trans-regulatory activity of the AR protein, as evidenced by a decrease in the initiation of transcription in reporter assays in vitro (Chamberlain et al., 1994), as well as the abundance of AR mRNA (Choong et al.,1996).
Table 1.
Polymorphisms of AR that are associated with human behavioral phenotypes.
| Polymorphism | Class | Phenotype (trait or disorder) | Reference |
|---|---|---|---|
| rs193922933 | STR | Aggression | Butovskaya et al. (2012; 2015); Rajender et al. (2008); c.f. Jönsson et al. (2001) |
| Autism | Henningsson et al. (2009) | ||
| Cognitive impairment | Yaffe et al. (2003) | ||
| Depression |
Colangelo et al. (2007); Sankar et al. (2012); Schneider et al. (2011); Seidman et al. (2001); c.f. Schneider et al. (2013) |
||
| Extroversion | Westberg et al. (2009) | ||
| Hostility | Pivovarciova et al. (2016) | ||
| Hyperactivity | Butovskaya et al. (2012); Hurd et al. (2011); c.f., Jönsson et al. (2001) | ||
| Impulsivity | Aluja et al. (2011; 2015); Mettman et al, (2014) | ||
| Intellectual giftedness | Celec et al. (2013) | ||
| rs6152 | SNP | Autism | Henningsson et al. (2009) |
| rs869109080 | STR | Aggression | Comings et al. (2002) |
| Biological parents divorced | Comings et al. (2002) | ||
| Biological father absent | Comings et al. (2002) | ||
| Number of sex partners | Comings et al. (2002) | ||
| Sexual compulsivity | Comings et al. (2002) | ||
| Impulsivity | Aluja et al., (2012); Comings et al. (2002) |
SNP, single nucleotide polymorphism; STR, short tandem repeat
A similar but less well-studied polymorphism (rs869109080) consists of a series of repeats downstream in exon 1 (Fig. 1). The repeated motif is designated “GGN”, with the N standing for T or C (GGT and GGC code for the same amino acid). The repeat number tends to fall between 10 and 31; 86% of the European Caucasian population has either 23 or 24 repeats (Brockschmidt et al., 2007). Like the CAG repeat polymorphism, the GGN polymorphism predicts a number of traits such as aggression and impulsivity in men (Aluja et al., 2011; Comings et al., 2002). In some cases, the effect of this polymorphism is mediated by the effect of the other AR STR polymorphism, the CAG repeat (e.g. Aluja et al., 2011; Comings et al., 1999a), suggesting that the two interact to affect AR function.
Apart from the two STRs, few polymorphisms in AR have been associated with behavior. Because of the hypothesized link between androgens and autism, Henningsson et al. (2009) genotyped several hundred patients with autism, as well as controls, for both repeat polymorphisms and a SNP located between them (rs6152). In addition to associations with the repeat polymorphisms, they found a higher prevalence of the A (vs. G) allele of rs6152 in affected females. Overall, however, associations between AR polymorphisms and human behavioral phenotypes have been largely limited to the two repeat polymorphisms–likely both because the method of genotyping is well-worked out and widely available, and because the repeats occur in coding regions, meaning that their effects on receptor function are easier to conceptualize than for polymorphisms in non-coding regions (see section 4 below).
2.3. Estrogen receptors
Of the nuclear steroid receptors, estrogen receptors (ERs) are the most evolutionarily ancient (Thornton, 2001). Estrogens play key roles in reproductive behavior in all vertebrate taxa (reviewed by Maney, 2010), and in some taxa they drive sexual differentiation. Also relevant to ERs is testosterone, which can be aromatized to estradiol. Thus, ERs contribute to both androgen- and estrogen-dependent behaviors. ERs share many features with other steroid receptors; when bound to hormone, these receptors bind to DNA to initiate transcription of target genes. Their basic structure is similar to that of AR: a transactivation domain, a DNA binding domain, which in this case recognizes estrogen response elements in DNA, and a ligand-binding domain (Marino et al., 2006). ERs can also act via nongenomic mechanisms, for example through associations with membrane-bound receptors (Micevych & Mermelstein, 2008; Razandi et al., 1999).
The human genome contains two distinct genes for canonical estrogen receptors: ERα and ERß, located on chromosomes six and fourteen, respectively. Most research on these two receptors in animal models has been carried out on rodents, which, like all mammals and birds, express homologs of both. The gene for ERα (ESR1) was cloned and sequenced first (Greene et al., 1986), followed a decade later by ERß (ESR2; Mosselman et al., 1996). The two receptors have unique, albeit overlapping, distributions in the rodent brain and are thought to perform distinct functions (Ervin et al., 2015; Pfaff et al., 2011; Rissman, 2008). Early studies in mice lacking either ESR1 or ESR2 led to the preliminary conclusion that ERα was more important for reproduction, whereas ERß mediated non-reproductive functions (Ogawa et al., 1999; Rissman et al., 1999). It is currently understood that the two receptors can in fact modulate the same behaviors (reviewed by Tetel & Pfaff, 2010). In some cases, their effects are synergistic, in others clearly antagonistic (reviewed by Handa et al., 2011). Most comparisons of the two receptor subtypes have been carried out in rodent models, although there is a small literature in humans. The human hippocampus, for example, appears to express relatively high levels of ERß, which led to the hypothesis that this receptor may be important for memory and cognition (Osterlund et al., 2000). Overall, the complexity of the relationship between the two receptors precludes assigning distinct roles to each, especially with respect to behavioral phenotypes.
The literature on ESR1 polymorphisms is dominated by two SNPs: the PvuII polymorphism (rs2234693) and the XbaI polymorphism (rs9340799). These are known as “restriction fragment length polymorphisms” because they disrupt sites at which the restriction enzymes by the same names, PvuII or Xbal, cut DNA. Samples can be genotyped by amplifying the relevant portion of ESR1, digesting the product with the appropriate enzyme, and inspecting the resulting fragments on a gel. For example, a “C” substituted for the “T” in the sequence “CACCTG” renders the enzyme PvuII unable cut at that location, making the digestion products of one allele distinguishable from the products of the other allele. The C allele occurs with a frequency of 45% in the American Caucasian population (Database of Single Nucleotide Polymorphisms), so it is quite common. The XbaI polymorphism, for which the less prevalent allele is also common, similarly disrupts the restriction site for the enzyme XbaI.
The PvuII and XbaI polymorphisms were first associated with breast cancer and loss of bone density (e.g., Hill et al., 1989; Kobayashi et al., 1996). Associations with many behavioral phenotypes have now been discovered, for example with expression of anger (Vermeersch et al., 2013) and with episodic memory in women (Kravitz et al., 2006a; Sowers et al., 2006). These polymorphisms are also associated with clinical outcomes such as anxiety (Prichard et al., 2002), depression in women (e.g., Keyes et al., 2015), and obsessive compulsive disorder (Alonso et al., 2011). The PvuII and XbaI polymorphisms are located close together in an intron between exons 1 and 2 (Fig. 2), which complicates the assessment of their independent predictive power. When both are used in the same study, researchers often consider the haplotype, or the complement of both alleles, and investigate the interactions between the two (e.g., Prichard et al., 2002).
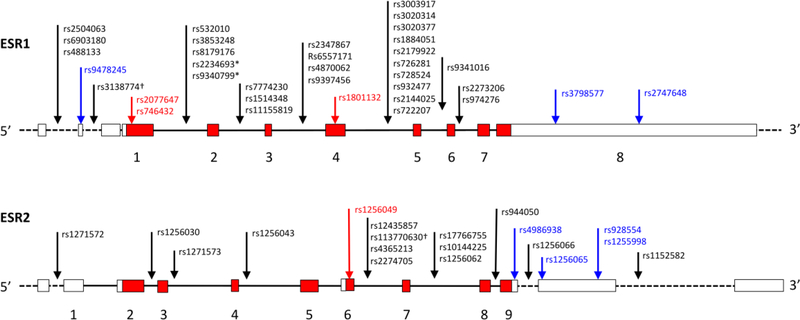
Fig. 2. Genetic polymorphisms in human ESR1 and ESR2.
Polymorphisms that have been associated with behavioral phenotypes or outcomes are shown. Boxes indicate exons, and lines indicate introns (introns not drawn to scale). Exon numbering, coding regions (red) and UTRs (white) are based on the primary transcripts (ESR1–001 and ESR2–001) as annotated in ENSEMBL. The dotted lines and unnumbered exons indicate introns and exons of alternative transcripts. The approximate positions of polymorphisms, most of which are single nucleotide polymorphisms, are indicated by arrows. Each polymorphism is labeled with the Reference SNP ID (rs) number assigned by dbSNP (NCBI). Black, blue, and red arrows indicate intronic, UTR, and synonymous (coding) variants, respectively. *PvuII and XbaI polymorphisms. †Repeat polymorphisms.
The third most popular ESR1 polymorphism for behavioral association studies consists of a dinucleotide (TA) repeat upstream of exon 1. The number of repeats ranges between nine and 27 and is bimodally distributed, with frequency peaks at 14 and 23 repeats (van Meurs et al., 2003). The number of repeats in this STR has been linked with behaviors such as aggression in men (Vaillancourt et al., 2012), harm avoidance (Gade-Andavolu et al., 2009), and non-conformism in women (Westberg et al., 2003) as well as to clinical outcomes such as psychoticism in women (Westberg et al., 2003), conduct disorder (Comings et al., 2000), and postpartum depression (Pinsonneault et al., 2013).
A number of other SNPs in ESR1 have also been associated with behavioral phenotypes (Table 2). These SNPs are distributed throughout the gene, occurring in both coding and non-coding regions (Fig. 2). Even so, behavioral studies of the PvuII and XbaI polymorphisms outnumber the studies of other polymorphisms combined. The enthusiasm for these two SNPs has stemmed from the fact that the protocols for detecting them are well-established and that the AR literature has set a precedent for focusing on only one or two polymorphisms. The number of SNPs in ESR1 that have been investigated in the context of behavior, however, now greatly exceeds the number for AR.
Table 2.
Polymorphisms of ESR1 that are associated with human behavioral phenotypes.
| Polymorphism | Class | Phenotype (trait or disorder) | Reference |
|---|---|---|---|
| rs11155819 | SNP | Autistic-like traits | Chakrabarti et al. (2009) |
| rs1514348 | SNP | Alzheimer's disease | Ma et al. (2009); c.f.Goumidi et al. (2011) |
| rs1801132 | SNP | Abstractedness | Miller et al. (2010) |
| Alzheimer's disease | Ma et al. (2009) | ||
| Emotional stability | Miller et al. (2010) | ||
| Impression management | Miller et al. (2010) | ||
| rs1884051 | SNP | Abstractedness | Miller et al. (2010) |
| Harm avoidance | Miller et al. (2010) | ||
| Negative (harsh) parenting | Lahey et al. (2012) | ||
| Neuroticism | Miller et al. (2010) | ||
| Premenstrual dysphoric disorder | Huo et al. (2007) | ||
| rs2077647 | SNP | Alzheimer's disease | Ma et al. (2009; age of onset); Schupf et al. (2008); c.f. Goumidi et al. (2011) |
| rs1256062 | SNP | Childhood-onset mood disorder | Mill et al. (2008) |
| Postpartum depression | Pinsonneault et al. (2013) | ||
| rs2144025 | SNP | Various traits in bipolar disorder, ADHD, and schizophrenia | Pinsonneault et al. (2017) |
| rs2179922 | SNP | Episodic memory | Ma et al. (2014) |
| rs2234693 "PvuII" | SNP | Alzheimer's disease | Boada et al. (2012); Brandi et al. (1999); Corbo et al. (2006); Ji et al. (2000); Pan et al. (2014); Ryan et al. (2014); see also Xing et al. 2013 |
| Anger expression | Vermeersch et al. (2013) | ||
| Anxiety-related traits | Prichard et al. (2002) | ||
| Cognitive impairment | Yaffe et al. (2002) | ||
| Depression | Keyes et al. (2015); Kim et al. (2010); Ryan et al. (2011a); Tsai et al. (2003); Vermeersch et al. (2013); cf. Kravitz et al. (2006b) | ||
| Episodic memory | Kravitz et al. (2006a); Sowers et al. (2006) | ||
| Number of children | Corbo et al. (2007) | ||
| rs4986938 | SNP | Obsessive compulsive disorder | Alonso et al. (2011) |
| Phobia | Ryan et al. (2011b) | ||
| Schizophrenia | Weickert et al. (2008) | ||
| rs2273206 | SNP | Postpartum depression* | El-Ibiary et al. (2013) |
| rs2347867 | SNP | Alzheimer's disease | Ma et al. (2009) |
| Age at first birth | Barban et al. (2016) | ||
| rs2504063 | SNP | Voice recognition | Karlsson et al. (2016) |
| rs2747648 | SNP | Autistic-like traits | Zettergren et al. (2013); cf. Zettergren et al. (2016) |
| rs3003917 | SNP | Abstractedness | Miller et al. (2010) |
| Premenstrual dysphoric disorder | Huo et al. (2007) | ||
| rs3020314 | SNP | Abstractedness | Miller et al. (2010) |
| Harm avoidance | Miller et al. (2010) | ||
| Premenstrual dysphoric disorder | Huo et al. (2007) | ||
| rs3020377 | SNP | Abstractedness | Miller et al. (2010) |
| Harm avoidance | Miller et al. (2010) | ||
| Negative (harsh) parenting | Lahey et al. (2012) | ||
| Premenstrual dysphoric disorder | Huo et al. (2007) | ||
| rs3138774 "TA(n)" | STR | Aggression | Vaillancourt et al. (2012) |
| Anxiety-related traits | Comings et al (1999b); Prichard et al. (2002) | ||
| Conduct disorder | Comings et al. (2000) | ||
| Harm avoidance | Gade-Andavolu et al. (2009) | ||
| Non-conformism | Westberg et al. (2003) | ||
| Postpartum depression | Pinsonneault et al. (2013) | ||
| Psychoticism | Westberg et al. (2003) | ||
| rs3798577 | SNP | Anorexia nervosa | Versini et al. (2010); c.f. Slof-Op ‘t Landt et al., (2014) |
| rs3853248 | SNP | Alzheimer's disease (age of onset) | Ma et al. (2009) |
| rs4870062 | SNP | Abstractedness | Miller et al. (2010) |
| rs488133 | SNP | Obsessive compulsive disorder | Alonso et al. (2011) |
| rs532010 | SNP | Childhood-onset mood disorder | Mill et al. (2008) |
| rs6557171 | SNP | Alzheimer's disease | Ma et al. (2009) |
| rs6903180 | SNP | Obsessive compulsive disorder | Alonso et al. (2011) |
| rs722207 | SNP | Harm avoidance | Giegling et al. (2009) |
| rs726281 | SNP | Anorexia nervosa | Versini et al. (2010) |
| rs728524 | SNP | Cognitive impairment | Yaffe et al. (2009) |
| Perceptual speed | Kravitz (2006a) | ||
| rs746432 | SNP | Childhood-onset mood disorder | Mill et al. (2008) |
| rs7774230 | SNP | Autism-like traits | Chakrabarti et al. (2009) |
| rs8179176 | SNP | Cognitive impairment | Yaffe et al. (2009) |
| rs932477 | SNP | Episodic memory | Ma et al. (2014) |
| rs9340799 "Xbal" | SNP | Alzheimer's disease | Brandi et al. (1999); Corbo et al. (2006); Ji et al. (2000); Monastero et al. (2006); Pan et al. (2014) |
| Anger expression | Vermeersch et al. (2013) | ||
| Anxiety-related traits | Prichard et al. (2002) | ||
| Cognitive impairment | Olsen et al. (2006); Yaffe et al. (2002); Yaffe et al. (2009) | ||
| Depression | Keyes et al. (2015);Kim et al. (2010); Ryan et al. (2011a); Tsai et al. (2003) | ||
| Episodic memory | Kravitz et al. (2006a); Sowers et al. (2006) | ||
| Morphine consumption, postoperative | De Gregori et al. (2016) | ||
| Number of children | Corbo et al. (2007) | ||
| Obsessive compulsive disorder | Alonso et al. (2011) | ||
| Phobia | Ryan et al. (2011a) | ||
| rs9341016 | SNP | Episodic memory | Ma et al. (2014) |
| rs9397456 | SNP | Alzheimer's disease | Ma et al. (2009) |
| rs9478245 | SNP | Obsessive compulsive disorder | Alonso et al. (2011) |
| rs974276 | SNP | Harm avoidance | Giegling et al. (2008) |
SNP, single nucleotide polymorphism; STR, short tandem repeat
This association was marginal; it is included here because the polymorphism is mentioned in section 4.3 of the text
The second ER, encoded by the gene ESR2, is less well-studied thus far. ESR2 polymorphisms associated with behavior are concentrated on the 3’ end of the gene, downstream of exon 6 (Fig. 2). These polymorphisms have been associated with clinical outcomes such as anorexia nervosa (Scott-van Zeeland et al., 2014), autistic traits (Chakrabarti et al., 2009), cognitive impairment (Yaffe et al., 2009), and depression (Keyes et al., 2015; Ryan et al., 2011b). One of the most interesting polymorphisms, an STR with 19–35 repeats (rs113770630), has been associated with female-to-male transsexualism in two different studies (Fernández et al., 2014; Henningsson et al., 2005; c.f. Ujike et al., 2009); in neither of these studies was an association found with the CAG repeat in AR. Further downstream, in the 3’ untranslated region, the SNP rs928554 is associated with face recognition (Karlsson et al., 2016), intellectual giftedness (Celec et al., 2013) and sexual desire (Gunst et al., 2015).
2.4. Related genes
Because of the sheer number of studies linking them with behavioral phenotypes, this review focuses on polymorphisms in the canonical receptors for sex steroids. The sex steroid pathway includes other genes, and many association studies of sex steroid receptors have included other candidates. For example, SNPs in an evolutionary precursor to estrogen receptors, estrogen-related receptor gamma (Giguére, 2002) are associated with bipolar disorder (Jiang & Zhang, 2011) and substance abuse (Johnson et al., 2011; Kapoor et al., 2013). Although non-nuclear estrogen receptors such as GPER1 (formerly called GPR30) and Gq-mER would certainly be relevant to look at in the context of sex steroid receptors and behavior, they are understudied in that regard (Sundermann et al., 2010). More typically, when association studies of sex steroid receptors include other genes, they focus on steroid metabolic enzymes or factors known to interact with steroid receptors (e.g., Chakrabarti et al., 2009; Fernandez et al., 2014; Pinsonneault et al., 2013; Prichard et al., 2007; Yeung et al., 2016; Zettergren et al., 2013; Zhao et al., 2016). Associations between those genes and behavior are beyond the scope of this review; nonetheless, our interpretations of all candidate gene association studies and the conclusions we can draw from them are subject to the same caveats discussed below.
2.5. Interpreting association studies
Almost every paper cited in the tables contains a version of the line, “Our results show that receptor X may play a role in phenotype Y.” The interpretation of the study typically does not move beyond that, leaving open how a small change in gene sequence, which may or may not affect receptor function, could affect behavior. In order to gain the most information from association studies, more consideration should be given to mechanisms. How does a small change like a SNP ultimately influence receptor expression or function, and hence phenotype? Answering this question will ultimately require investigations at many levels, including gene transcription, transcript processing and stability, and ligand-receptor interactions. Only by looking carefully at how gene sequence affects receptor expression and function will we make meaningful progress toward understanding the role of polymorphisms in the evolution of behavior and the etiology of disease.
Before considering the impact of any polymorphism, it is important to acknowledge the correlational nature of association studies and the fact that the polymorphism itself may not drive associations. In other words, if a particular polymorphism is associated with a phenotype, it may simply be near another polymorphism that actually affects the phenotype. Because recombination rates can vary along any stretch of DNA, having a certain allele at one locus may significantly increase the odds of a certain allele at another, especially if the two genes are close together. This phenomenon, known as linkage disequilibrium, is an old problem in behavioral genetics because it limits the resolution with which one can map causal genes (see Wray, 2007). The human genome is structured into haplotype blocks, such that SNPs within the same block are highly likely to be inherited together (Gabriel et al., 2002). The ESR1 polymorphisms PvuII, XbaI, and the T(n) STR are close together and in linkage disequilibrium (Becherini et al., 2000; van Meurs et al., 2003). This issue is addressed, in some studies, by performing a linkage analysis to account for correlations among polymorphisms (e.g. Costas et al., 2009; Giegling et al., 2008; Goumidi et al., 2011; Huo et al., 2007; Karlsson et al., 2016; Ma et al., 2009; Pinsonneault et al., 2013; Versini et al., 2010; Weickert et al., 2008; Zhao et al., 2011). It is important to note that in many cases when polymorphisms are used to associate the “receptor” with a phenotype, researchers are not arguing that that polymorphism itself causes a change in expression or function. They are using it simply as a marker, which they assume is linked to something causal near that site. This approach represents a classic paradigm in quantitative genetics: mapping a polymorphism to a region of DNA, called a quantitative trait locus, without assuming it is causal for the trait. But as those associations are communicated by quantitative geneticists to behavioral endocrinologists, that detail can get lost in translation. Now that the complete sequence of these receptors is available from thousands of people and all common SNPs are presumably mapped, we can start to identify causal variants. To do so, we will need to test the impact of polymorphisms using experimental approaches.
3. Moving beyond association studies
3.1. The concept of endophenotypes
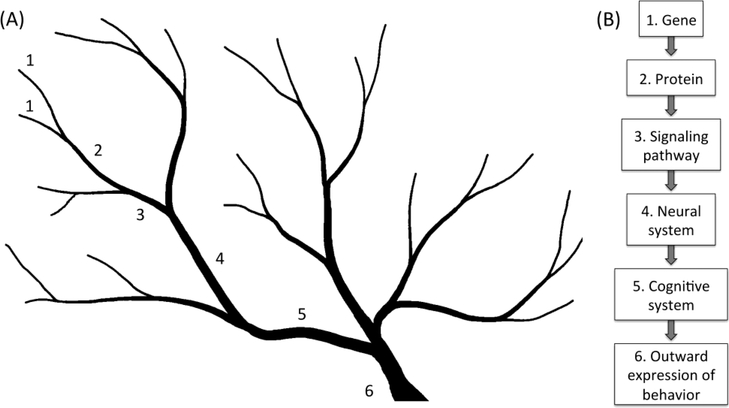
Genetic influences on behavior are complex. Phenotypic variation happens largely via the combined effects of numerous genetic polymorphisms, making it difficult to isolate and identify causal genetic variants. Early attempts to associate genetic variants with mental disorders, for example, famously failed to replicate (Sklar, 2002). To manage this complexity and design studies with greater power, Gottesman and Gould (2003) proposed breaking down the pathway from genotype to phenotype into intermediate levels, or “endophenotypes”, that are more feasible to study (see also Cannon & Keller, 2006). According to this framework, the pathway can be conceptualized as a watershed with small rivulets leading into larger streams, finally culminating in a major river (Fig. 3A). At each point where tributaries coalesce, noise is introduced into the pathway which becomes thunderous at the level of the river. Rather than quantifying the extent to which changes at the sources of the rivulets (the DNA sequences) predict the nature of the river (the phenotype), we should be asking how they predict the nature of the smaller streams into which they feed. In other words, the actual causal links between genes and behavior reside in relatively simpler intermediate levels that can be described quantitatively. To better understand an association between an ESR1 polymorphism and short-term memory loss, for example, we might ask whether the polymorphism predicts an endophenotype upstream, such as ERα expression in the hippocampus. If we start too far upstream, however, the endophenotype may be no better correlated with the polymorphism than with the phenotype itself (Flint & Munafò, 2007; Iacono et al., 2014). Such a result may tell us we are swimming up the wrong branch. It is important to ask how each endophenotype alters those immediately downstream, and so on, in order to finally understand what influences the river. When considering behavior, the endophenotypes might be conceptualized as gene, protein, signaling pathway, neural circuit, possibly cognition, and finally, behavior (Fig. 3B). This list is oversimplified and of course other levels would be appropriate to consider, depending on the context. But overall, this theoretical framework allows us to parse complexity and to ask questions about the effects of polymorphisms with greater power.
Fig. 3. The concept of endophenotypes.
(A) The watershed model of Cannon & Keller (2006) illustrates the pathway from genotype to phenotype. Specific variants (1) affect precisely defined endophenotypes such as the level of ESR1 expression in hippocampus (2) or downstream targets of estrogen receptor alpha (3). This endophenotype combines with others from different branches to influence broader endophenotypes such as the functioning of memory circuits (4) and subsequently, short-term memory (5). The endophenotype of short-term memory then in turn combines with still others to create behavioral phenotypes, such as dementia (6). (B) The pathway from gene to behavioral phenotype involves endophenotypes at many levels of biological organization.
The concept of endophenotypes was proposed in the 2000’s from the perspective of psychiatry and clinical psychology, and arose from the need to understand the genetic basis of mental disorders. The ideas were soon incorporated into a new NIMH-mandated framework for translational research on mental disorders, the Research Domain Criteria (RDoC; Insel et al., 2010), which encourages attention to underlying biology (Glatt & Lee, 2016). Around the same time, researchers in the field of molecular evolution were independently making a similar argument (Dean & Thornton, 2007; Dalziel et al., 2009; Wray, 2007). For example, Dean & Thornton (2007) hailed the arrival of a “functional synthesis” which would move the field of molecular evolution beyond gene associations to empirical studies of how genes affect function and fitness. They advocated an experimental approach to determine the effects of specific mutations on the function of encoded proteins, for example by synthesizing the mutated sequence and testing its effects on the properties of the resulting receptor. Along the same lines, Dalziel et al. (2009) argued that the impact of variation in candidate genes will be understood only through empirical, mechanistic studies at multiple levels of biological organization—for example genes, proteins, and biochemical networks. Only such an approach, which makes causal connections between each level and the level below, can explicitly connect genotype with phenotype.
Such an approach is best implemented by taking advantage of a priori knowledge about the function of proteins and their effects on phenotypic traits (Dalziel et al., 2009). Given the voluminous nature of our knowledge about the effects of sex steroids on behavior, what better molecules for this task than sex steroid receptors? The sequencing of these genes in thousands of human genomes, and the mapping of each human SNP, allows us to begin at gene sequence and work upward. In the sections below, I consider ways in which polymorphisms in the genes for sex steroid receptors might affect endophenotypes far below the level of behavior, for example receptor function and abundance. A truly integrated translational approach will ultimately involve consideration at additional levels downstream in the watershed, for example the effects of genetic changes on signaling pathways and neural circuits.
3.2. Variation in coding regions
Genes are composed of a number of elements, such as coding regions, untranslated regions, promoters, and introns. Because of this heterogeneous structure, the impact of a genetic polymorphism depends on its location within the gene. Polymorphisms within coding regions, in other words regions that directly encode the amino acid sequence of the protein, have the potential to cause changes in the protein’s structure and thus its function. Steroid receptors must interact not only with steroids, but also with a large number of other transcription factors and with DNA. Changes in the amino acid sequence can therefore affect function via many different mechanisms. Moreover, because steroid receptors are themselves transcription factors, functional changes could potentially affect the expression of all the genes targeted by the receptor elsewhere in the genome (see Carroll, 2005; Ketterson & Nolan, 1992).
The best-studied polymorphism in a sex steroid receptor, the CAG repeat in the AR gene, has a pronounced effect on the final AR protein. Each CAG codon, or triplet, encodes the amino acid glutamine, meaning that the series of repeats translates into a “polyglutamine stretch” (Fig. 1B). Many transcription factors contain such stretches, which typically exhibit high allelic variation (Gerber et al., 1994). They occur in the N-terminal domain, which activates transcription, and are widely thought to regulate the efficiency with which the protein is able to initiate the transcription of target genes. The effect of the AR polyglutamine stretch has been directly tested by performing experiments in cultured cells in vitro (e.g., Beilin et al., 2000; Chamberlain et al., 1994; Kazemi-Esfarjani et al., 1995; Tut et al., 1997). Chamberlain et al. (1994) constructed ARs that varied according to the position and the length of the stretch and tested their ability to activate transcription of a test gene, called a reporter gene, inserted downstream of androgen response elements. The number of glutamines in the stretch was negatively related, in a strikingly linear fashion, to transcription of the reporter gene (Fig. 1C). These results suggest that in vivo, individual variation in the length of the polyglutamine stretch could cause significant variation in the transcription of AR target genes. Although they cannot exactly mimic conditions in the brain, in vitro studies are an excellent first step toward making connections between receptor structure and the endophenotypes that influence behavior.
The other behaviorally relevant STR in AR, the GGN polymorphism, also occurs in the coding region of exon 1. This polymorphism may alter AR transactivation as well. Whereas the CAG repeats code for a stretch of glutamines, the GGN repeats encode a stretch of glycines. In vitro experiments have shown that transcription activity may depend on the length of the polyglycine stretch, but in a nonlinear manner (Ding et al., 2005; Gao et al., 1996; Lundin et al., 2007). Thus, the mechanism by which this STR affects receptor function may not be as straightforward.
As is the case for STRs, protein function can also be profoundly altered by SNPs. Nucleotide substitutions that alter a codon such that it codes for a different amino acid are called “non-synonymous”. Such alterations can change the physiochemical properties of the protein, impacting protein folding and the ability to bind to the ligand or DNA. Changes in protein sequence can also affect post-translational modifications such as phosphorylation and acetylation, which can alter receptor stability and activity (le Romancer et al., 2011), More often, however, SNPs in coding regions are “synonymous”, meaning that both alternative codons code for the same amino acid. Among the SNPs in the coding regions of sex steroid receptor genes, all that are known to be linked to behavioral phenotypes are synonymous (rs2077647, rs746432, and rs1801132 in ESR1; rs1256049 in ESR2, and rs6152 AR; see Fig. 2). The preponderance of synonymous changes, rather than non-synonymous, might arise because of strong selection pressure against alterations in the structure of highly pleiotropic genes. Synonymous changes are often said to be “silent” because they do not alter the sequence of the protein, at least not in the usual way (reviewed by Chamary et al., 2006). We will return in Section 4 to the issue of whether synonymous SNPs are actually silent.
Polymorphisms in coding regions can affect not only the sequence but also the abundance of mRNA or protein. For example, in an in vitro study of AR, Choong et al. (1996) showed a near-linear relationship between the number of CAG repeats and the levels of both the mRNA and protein (Fig. 1C). This finding emphasized the need to control for the amount of AR when assessing its activity in vitro, as well as the fact that even when polymorphisms alter protein sequence, they can also affect protein availability via lesser-known mechanisms. Some of these mechanisms are reviewed below.
3.3. Variation in non-coding regions
Most genetic variation associated with phenotypic traits or disorders, behavioral and otherwise, is not located in coding regions. Instead, it is more likely to be found in introns or between genes (Fraser, 2013; Hindorff et al., 2009). Genomic regions between coding regions have been called “junk DNA” (e.g., O’Brien, 1973). Even the term “non-coding” suggests that these sequences are less important. But the idea that variation in non-coding DNA can be important, particularly in the context of evolution, has been around for decades (Monod & Jacob, 1961; Britten & Davidson, 1971). In the 1970s, for example, King and Wilson (1975) argued that because protein sequences are nearly identical in humans and chimpanzees, phenotypic divergence between the two species must be attributable to differences in the regulation of gene expression. After this concept was proposed, other researchers reported evidence that differentiation of non-coding regions can, in fact, affect expression. Such variation has been called “regulatory variation” (Carroll, 2000; Stern, 2000), because, rather than causing variation in protein structure, it affects the regulation of protein abundance—i.e., the extent to which a particular gene is transcribed or translated, and under what circumstances.
The distinction between functionality and abundance is important in the context of sex steroid receptors because the receptors are themselves regulators – they bind to DNA sequences of other genes. It is therefore easy to conflate cis-regulatory variation, which occurs in non-coding regions of the receptor gene and affects expression of the receptor mRNA, with trans-regulatory effects, which alter the expression of steroid target genes elsewhere in the genome by affecting the abundance or function of the steroid receptor. It is not the case, for example, that transcriptional activation of the receptor protein is affected similarly by STRs in coding and non-coding regions alike, as has been argued by some authors. Cis-regulatory variation cannot alter the functionality of the final, translated receptor (but see section 4.3 below for an exception). Its power comes primarily from its ability to alter when, where, and how much of the receptor is produced, which of course can determine hormone sensitivity and, through feedback mechanisms, the level of the hormone itself. Indeed, some polymorphisms in sex steroid receptors are linked with plasma levels of sex steroid hormones (Westberg et al., 2001). Thus, cis-regulatory variation can have its own trans effects, adding further to the downstream effects of sex steroid receptor polymorphisms.
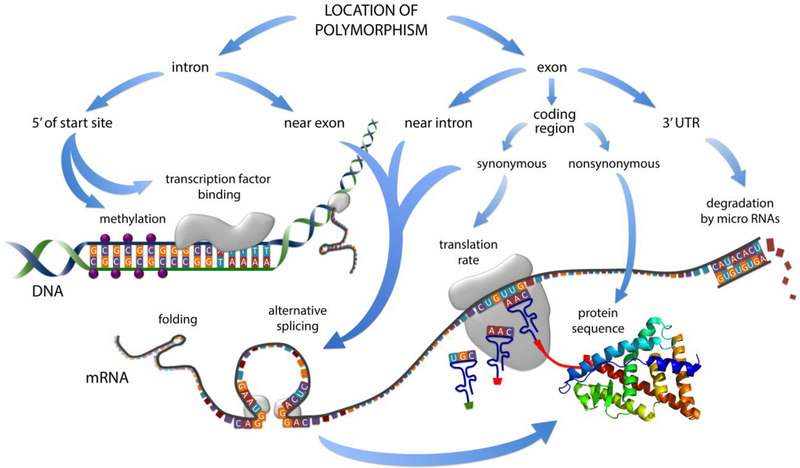
When polymorphisms in non-coding regions of sex steroid receptor genes are found to associate with behavioral phenotypes, authors are generally cautious about attributing behavioral effects to that polymorphism alone. Some authors consider the possibility of linkage with a causal genetic driver. Some authors cite molecular studies as evidence that a particular SNP affects gene expression (e.g., Alonso et al., 2011 cites Maruyama et al., 2000). Rarely, authors have performed their own functional studies to complement findings of associations (e.g., Maruyama et al., 2000; Weickert et al., 2008). More commonly, authors simply point out that more research is needed to understand the mechanisms that underlie gene-behavior associations. Performing such research can be a daunting undertaking because there are so many mechanisms by which variation in cis-regulatory sequences can lead to alterations in mRNA or protein expression. Sex steroid receptors in particular are extraordinarily complex, with multiple promoters, coding and noncoding exons, alternative splice variants, and isoforms (Pinsonneault et al., 2017). Below, I have reviewed the most obvious processes that can be affected by regulatory variation; those processes are also shown in Fig. 4. The next section is not meant to be an exhaustive review of these mechanisms, which would be impossible. Rather, it is intended as a jumping-off point as we begin to assess the meaning and impact of known variation in sex steroid receptor genes.
Fig. 4. The effect of a polymorphism depends on its location within a gene.
The above diagram can be used as a decision tree to identify some of the possible effects of a polymorphism with a known location. For example, a polymorphism in an intron 5’ of the start site could alter methylation of the promoter or the binding of transcription factors, thus affecting transcription rate. A nonsynonymous SNP in the coding region of an exon could affect protein sequence, thus affecting receptor function. A synonymous SNP in the coding region could affect translation rate and thus protein abundance, or splicing and thus protein sequence. This figure is meant as a starting point for considering mechanisms and does not depict all possible outcomes. For example, methylation and folding can be affected by variation at any location, and all of the processes depicted here may depend on the type of cell and/or region of the brain. Rendition of protein by Emw (2015).
4. Regulatory variation: Mechanisms and experimental approaches
4.1. Transcription factor binding sites
Transcription factors work by recruiting transcriptional activators or repressors to a gene or by altering the accessibility of that gene to those agents. Each transcription factor binds preferentially to a consensus sequence, or motif, usually located just upstream of the transcription start site—in other words, the promoter region. If a polymorphism disrupts that motif, the transcription factor may be less able to bind to the promoter and therefore less likely perform its regulatory function. Alternatively, a polymorphism may create a transcription factor binding site, thus inviting new regulatory control. Disruption or addition of transcription factor binding sites was one of the earliest-proposed models of how genetic variation leads to phenotypic variation (Monod & Jacob, 1961), and it remains one of the most popular. Some of the first work demonstrating this phenomenon was done in the context of hereditary diseases such as thalassemia, hemophilia, and some forms of cancer, which are associated with specific SNPs. Mutations in affected patients were found to alter the ability of transcription factors to bind to regulatory sequences in non-coding DNA, thereby either increasing or decreasing transcription of critical genes (Miller & Bieker, 1993; Reijnen et al., 1992; see also Deplancke et al., 2016; Funnell & Crossley, 2013). These findings laid the groundwork, both theoretically and technically, for the search for similar mechanisms underlying other kinds of traits.
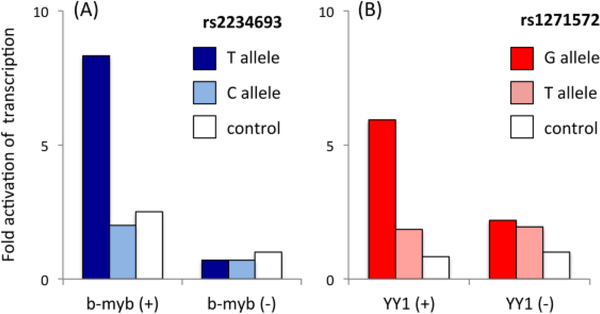
Several of the polymorphisms reviewed here (see Tables 2 and 3) disrupt transcription factor binding sites upstream of exon 1. For example, the ESR1 SNPs rs6903180, rs488133, and rs9478245, which are each associated with obsessive compulsive disorder (Alonso et al., 2011), occur within binding sites for the transcription factors myt1, NGF1C, and sry, respectively (Weickert et al., 2008). Whether a particular polymorphism is capable of affecting transcription can be tested using in vitro reporter assays. The variable sequence, presumably containing the promoter region, is inserted into a DNA construct upstream of a reporter gene such as luciferase or green fluorescent protein. When the construct is then introduced into cultured cells, the degree to which the promoter region drives transcription can be assessed by quantifying the resulting amount of reporter gene product. In this way, the effect of a polymorphism can be determined by comparing the level of transcription between alleles. For example, Herrington et al., (2002) compared the level of transcription activity driven by either allele of the ESR1 SNP rs2234693, the “PvuII” polymorphism; see Fig. 2, Table 2). Similarly, Chen et al. (2013) compared transcription rate between two alleles of rs1271572, a SNP in ESR2 (Fig. 2) associated with autistic traits (Chakrabarti et al., 2009), face recognition (Karlsson et al., 2016), and sexual desire (Gunst et al., 2015). In both cases, not only was one allele transcribed at significantly higher levels than the other, but that difference appeared to be attributable to differentiation of a transcription factor binding site. These two polymorphisms occur within binding motifs of the transcription factors myb-b and YY1, respectively. In the absence of the appropriate transcription factor, transcription of the two alleles was not significantly different in either case (Fig. 5). These studies illustrate the value of the in vitro approach, supplying convincing evidence that variation in sequence can have concrete, quantifiable effects on gene expression.
Table 3.
Polymorphisms of ESR2 that are associated with human behavioral phenotypes.
| Polymorphism | Class | Phenotype (trait or disorder) | Reference |
|---|---|---|---|
| rs10144225 | SNP | Semantic memory | Fehsel et al. (2016) |
| rs113770630 | STR | Alzheimer's disease | Forsell et al. (2001) |
| (D14S1026) | Depression | Geng at al. (2007); Takeo et al. (2005) | |
| Female-to-male transsexualism | Fernández et al. (2014); Henningsson et al. (2005); cf. Ujike (2009) | ||
| rs1152582 | SNP | Autistic-like traits | Chakrabarti et al. (2009) |
| rs12435857 | SNP | Alzheimer's disease | Zhao et al. (2011) |
| rs1255998 | SNP | Cognitive impairment | Yaffe et al. (2009) |
| rs1256030 | SNP | Cognitive impairment | Yaffe et al. (2009) |
| Face recognition | Karlsson et al. (2016) | ||
| rs1256043 | SNP | Alzheimer's disease | Pirskanen et al. (2005) |
| rs1256049 | SNP | Cognitive impairment | Ryan et al. (2013) |
| Depression | Ryan et al. (2011b) | ||
| Generalized anxiety disorder | Ryan et al. (2011a) | ||
| rs1256062 | SNP | Semantic memory, executive | Fehsel et al. (2016) |
| function | |||
| rs1256065 | SNP | Cognitive impairment | Yaffe et al. (2009) |
| rs1256066 | SNP | Anorexia nervosa | Scott-Van Zeeland et al. (2014) |
| rs1271572 | SNP | Autism-like traits | Chakrabarti et al. (2009) |
| Face recognition | Karlsson et al. (2016) | ||
| Sexual desire in women | Gunst et al. (2015) | ||
| rs1271573 | SNP | Alzheimer's disease | Pirskanen et al. (2005) |
| rs17766755 | SNP | Alzheimer's disease | Zhao et al. (2011); c.f. Goumidi et al. (2011) |
| rs2274705 | SNP | Semantic memory | Fehsel et al. (2016) |
| rs4365213 | SNP | Alzheimer's disease | Zhao et al. (2011) |
| rs4986938 | SNP | Alzheimer's disease | Zhao et al. (2012); c.f. Goumidi et al. (2011) |
| Cognitive impairment | Ryan et al. (2013) | ||
| Depression | Keyes et al. (2015); Ryan et al. (2011b) | ||
| Sexual desire | Gunst et al. (2015) | ||
| rs928554 | SNP | Face recognition | Karlsson et al. (2016) |
| Intellectual giftedness | Celec et al. (2013) | ||
| Sexual desire | Gunst et al. (2015) | ||
| rs944050 | SNP | Anorexia nervosa | Scott-Van Zeeland et al. (2014) |
SNP, single nucleotide polymorphism; STR, short tandem repeat
Fig. 5. Single nucleotide polymorphisms within transcription factor binding sites affect transcription rate in vitro.
rs2234693 (ESR1) and rs1271572 (ESR2) occur within binding motifs of transcription factors b-myb and YY1, respectively. Results from in vitro reporter assays show that the transcription rate of one allele is higher than the other, but only in the presence of the relevant transcription factor. (A) Herrington et al. (2002) demonstrated higher transcription rates for the T allele than the C allele of rs2234693 when the gene for b-myb was co-transfected into the cells. (B) Chen et al. (2013) showed higher levels of transcription of the G allele than the T allele of rs1271572, but when YY1 was knocked down, the difference was abolished. In both (A) and (B), the control condition is the level of transcription without either allele in the construct. All data are normalized to the control condition without transcription factor present.
Multiple tools (e.g. rSNP-MAPPER, TRANSFAC) are now available to help predict whether differentiation of DNA sequences affects transcription factor binding motifs, and it is becoming popular to include analysis of such in candidate gene association studies. The practice is controversial, however (Deplancke et al., 2016), because alteration of transcription factor binding sites does not automatically point to a causal mechanism. Binding of transcription factors is complex, and the effects of a polymorphism can be difficult to predict. A SNP within a particular binding motif may not affect local transcription at all; rather, it might affect expression of a relatively distant gene. In other words, associations between behavior and a SNP located within ESR1 may not actually be related to ERα at all. Second, the dynamics of transcription factor-DNA binding can be locally regulated and highly tissue-specific (reviewed by Deplancke et al., 2016); in other words, the degree to which a SNP affects transcription factor binding can depend on the cell type and even the brain region. Thus, even when the effect of a SNP is tested experimentally in vitro, studies of cell lines cannot tell us everything about what is going on locally in the brain. Finally, although more than a thousand transcription factors that bind DNA have been described in humans (Vaquerizas et al., 2009), there are undoubtedly others yet to be discovered. For all of these reasons, in vitro assays cannot show definitively that a particular polymorphism causes differential expression in vivo or that it affects behavior.
4.2. Epigenetic regulation
To be transcribed, DNA must be accessible to the proteins responsible for transcription, i.e., polymerases and other transcription machinery. The accessibility of DNA can be altered by epigenetic factors, such as acetylation of histones or methylation of the DNA itself. Methylation of the genes for sex steroid receptors mediates the influence of hormones and experience on the expression of those genes and is thought to contribute to sex differences in the brain and behavior (Champagne & Curley, 2009; Schwarz et al., 2010; Walker & Gore, 2017). Despite the name “epigenetic”, such mechanisms do depend, to a degree, on gene sequence (see Furey & Sethupathy, 2013). DNA methylation of promoter sequences, which can block transcription, occurs largely at cytosines followed by guanines, or “CpG” sites. Thus, the methylation status of a gene can theoretically be affected by genetic polymorphisms. STRs, particularly those such as the GGC repeat in AR, can dramatically change the methylation landscape because they can add or subtract many such sites. Methylation state can also be affected by SNPs; if a cytosine or guanine is substituted for a different nucleotide, a CpG site can be gained—or in the opposite scenario, lost—and a transcription factor binding site subsequently blocked or unblocked.
Of the more than 50 SNPs in sex steroid receptors reviewed here, (Tables 1, 2, and 3) 35% are associated with a loss, gain, or shift in CpG sites. The extent to which such changes actually affect expression is not well understood. A SNP in ESR1 (rs7766585) that causes a gain of a CpG site is highly associated with breast cancer (Harlid et al., 2011). It can be hard to detect associations between methylation and behavior, however, because the relevant methylation would presumably occur in brain tissue. The methylation state of any given locus can vary dramatically according to tissue type, meaning that a locus methylated in tissue easily obtainable from humans, such as blood or saliva, may not be methylated in brain (see Smith et al., 2015). Further, methylation can occur at locations other than CpG sites, making it difficult to predict which polymorphisms affect methylation state. The length of the CAG repeat in AR is correlated with the degree of nearby methylation, even though it does not itself contain CpG sites (Hickey et al., 2002; Vottero et al., 1999; cf. Calvo et al., 2000). Although genetic variation can help determine the likelihood that a particular site is methylated, it is perhaps not the most important driver of such. By definition, epigenetic modifications in the absence of underlying genetic variation can explain individual differences in phenotypic traits. They may therefore represent an important future avenue of research into phenotypic variation of sex steroid-dependent traits (see Champagne & Curley, 2009).
4.3. Alternative splicing
Almost all mRNA transcripts undergo splicing, a process that removes selected sequences such as introns. Because splice sites in mRNA are defined by the local sequence, a SNP could lead to large insertions or deletions in the final mRNA message. Introns could be left in the sequence, or exons skipped. In this way, regulatory variation can lead to alternative isoforms of the protein that vary in functionality. For example, in a schizophrenic patient with an aberrant isoform of ERα mRNA, part of intron 4 was not spliced out; the resulting sequence contained a premature stop codon which resulted in a truncated protein missing the estrogen binding domain (Weickert et al., 2008). More commonly, alternative splicing leads to missing exons; in some cases extra exons can result from the transcription of an intronic sequence. In either case, the functionality of the resulting translated protein can be difficult to predict. Not only can alternative isoforms lose function, but they can also inhibit functional isoforms by blocking DNA hormone response elements on DNA (Ohlsson et al., 1998). Alternative isoforms may even be able to initiate transcription without hormone or coactivators (Fuqua et al., 1991). Thus, alternative isoforms can have “dominant negative” or “dominant positive” effects in heterozygotes, and these effects may even go in opposite directions in different tissues of the same individual (Inoue et al., 2005). The functionality of alternative isoforms can be determined in vitro using reporter assays (see Section 3.2 above; Fig 1C), which test the ability of the receptor to initiate the transcription of target genes (e.g. Wieckert et al., 2008) or to interact with other proteins (e.g., Wong et al., 2011).
More than 60 unique ERα mRNAs, missing one or more exons compared with the canonical receptor ESR1–001, have been isolated from human brain. Some authors have argued, in fact, that alternative transcripts far outnumber the canonical version in brain (Pinsonnault et al., 2017). The complement of distinct ERα mRNAs varies from person to person (Ishunina & Swaab, 2012; Perlman et al., 2005) and may affect risk for mental disorders. Schizophrenic patients were significantly less likely than healthy controls to express a full-length ERα (Weickert et al., 2008). On the other hand, the number of alternatively spliced variants was lower in Alzheimer’s patients than in a control population (Ishunina & Swaab, 2012).
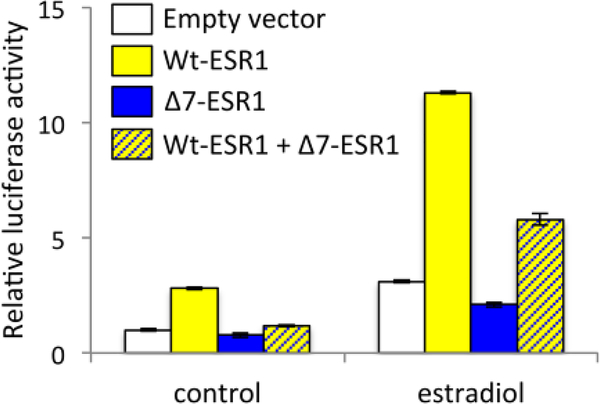
Weickert et al. (2008) performed an elegant analysis combining a candidate gene association study with experimental investigations of exon skipping. Using human brain tissue, they tested for SNPs that were predictive of alternative ERα transcripts. They identified a SNP (rs2773206) associated with an alternatively spliced transcript of ERα; people with the G allele of this SNP were more likely than those with the T allele to express an ERα variant missing exon 7 (Δ7-ESR1). Using an in vitro reporter assay, they then showed that the ERα isoform transcribed from this variant was unable to initiate transcription at estrogen response elements in DNA (Fig. 6). In fact, the isoform also inhibited the transcription activity of the full-length, wildtype ERα isoform. These results suggested that expression of the variant may impact ERα activity in vivo. When the researchers sequenced ERα variants in brain tissue from patients with mental disorders and from healthy controls, they found that Δ7-ESR1 was expressed in 80% of patients with major depression but only in 48% of controls. A few years later, El-Ibiary et al. (2013) discovered a nominal association between the predictive SNP and postpartum depression. Although the in vitro experiments of Weickert et al. do not tell us definitively that the SNP is causal for depression, they provide a much clearer picture about endophenotypes than is currently available for most sex steroid receptor polymorphisms associated with human behaviors.
Fig. 6. Deletion of exon 7 abolishes ERα transcription activity in vitro.
Cultured cells were transfected with plasmid vectors containing the genes for wild-type ESR1 (Wt-ESR1), ESR1 missing exon 7 (Δ7-ESR1), or both. Each plasmid also contained a luciferase reporter gene downstream of estrogen-response elements. Cells were then incubated for 24h in the presence or absence of 17ß-estradiol. Δ7-ESR1 was unable to initiate transcription of the luciferase reporter and attenuated the activity of Wt-ESR1. All values are normalized to the empty vector control. Redrawn from Weickert et al. (2008).
4.4. mRNA structure
To be translated into protein, mRNA must be relatively stable and accessible to translation machinery. Although most non-coding sequence is eliminated when introns are spliced out, the remaining sequence can nonetheless contain variation that affects mRNA stability and accessibility. A loss of stability can result in early degradation, which means less mRNA translated into protein. The effects of polymorphisms on thermodynamic stability can be modeled with software that predicts structure. Such analyses have shown, for example, that SNPs in ESR2 (rs2987983) and ESR1 (rs3798577) are likely and unlikely, respectively, to affect mRNA stability (Veronica et al., 2016; Haas et al., 2012). Sequence variation can cause changes in RNA folding and thus modify structural elements such as hairpins (Pinsonnault et al., 2017); variation can also affect RNA modifications such as methylation and RNA editing (the conversion of adenosine to inosine). These types of changes can alter the physical access of translation-related proteins and RNAs (Nainar et al., 2016), thus affecting translation rate.
4.5. microRNAs
One of the best-understood ways that polymorphisms can influence mRNA stability is by altering sites that interact with short, non-coding RNAs known as microRNAs (miRs). miRs bind to consensus sequences in the 3’ untranslated region (UTR) of mRNA to initiate its degradation. When an miR binding site is altered by a polymorphism, meaning that miRs are more or less likely to bind to that site, RNA abundance can be up- or down-regulated as a result (Kim & Bartel, 2009; Zhang et al., 2012). Several of the behaviorally-relevant polymorphisms in ESR1 and ESR2 are located in 3’ UTR, making them candidates for such regulation (Fig. 2). The ESR1 SNP rs2747648, which is associated with autism spectrum disorder (Zettegren et al., 2013) is predicted to affect the binding capacity of miR-453 (Tchatchou et al., 2009). Similarly, the ESR1 SNP rs3798577, which is associated with anorexia nervosa (Versini et al., 2010), occurs within a binding site for miR-122 (Haas et al., 2012). Whether these SNPs actually affect mRNA abundance is, however, not known.
The effect of a polymorphism on miR-associated regulation can be modeled in silico, then tested in vitro. For example, Adams et al. (2007) identified a C >T SNP in the 3’ UTR of ESR1 (rs9341070) that they predicted would affect the binding capacity of miR-206. When they tested constructs containing each variant in reporter assays, they found that activity from the T variant was lower and more sensitive to miR-206 regulation than the C allele. These results suggested that the T variant may reduce mRNA stability by introducing an miR-206 target sequence. Putnik et al. (2009) performed similar analyses on two behaviorally-relevant SNPs in the 3’UTR of ESR2 (rs4986938 and rs928554; see Table 3). They found that neither polymorphism significantly affected mRNA stability, and concluded that the association with phenotypes is more likely explained by linkage disequilibrium with causal SNPs.
Polymorphisms can occur not only in regions targeted by miRs, but also regions encoding the miRs themselves. Sequences encoding miRs are found in the introns and other regulatory sequences of many genes. Polymorphisms in these sequences could theoretically affect the stability of other mRNAs by altering the ability of the encoded miRs to interact with their targets. None of the polymorphisms in human sex steroid receptors, however, disrupt known miR coding sequences. According to miRBase.org , there are no known miR coding sequences in AR or ESR1. There is one near the 3’ end of ESR2, but this region does not contain known SNPs. Thus, at this time there is no evidence that polymorphisms in sex steroid receptor genes influence the expression of other genes by disrupting miR sequences. The study of miRs is relatively young, however, and new miRs are still being discovered at a rapid rate (e.g. Wake et al., 2016).
4.6. Codon optimality
Synonymous SNPs, in other words SNPs that occur within coding sequence but do not change the sequence of the resulting protein, represent the quintessential silent mutation. This sort of substitution does not alter the protein sequence because the new codon encodes the same amino acid as the old one; for example TTG and CTG both encode leucine. The T > C change in this case is not likely to be completely silent, however, because although it does not affect the protein sequence, it could affect the rate of translation and, ultimately, mRNA stability. Not all codons are translated at equal rates. There are multiple hypotheses about why (Plotkin & Kudla, 2011); translational efficiency may be an important cause. The rate of translation may be related to the availability of corresponding tRNA molecules, such that tRNAs are readily available for “optimal” codons but more rare for non-optimal ones (dos Reis et al., 2004; Pechmann & Frydman, 2013). According to this hypothesis, optimal codons are translated at a faster rate, which may also affect the overall stability of the mRNA (Presnyak et al., 2015). In the above example, the synonymous change from TTG to CTG, although “silent”, represents a change from a highly optimal, stable codon to a non-optimal, unstable one (Presnyak et al., 2015).
A single codon substitution is unlikely to meaningfully affect overall translation rate. It is interesting to note, however, that some of the SNPs in sex steroid receptors alter codons in the predicted direction, given the associations with behavioral phenotypes. The G > A change in the ESR2 polymorphism rs1256049 is in women associated with generalized anxiety disorder, depression, and mild cognitive decline (Ryan et al., 2011a; 2011b; 2013). The corresponding codon, GTA, is associated with lower mRNA stability than the original GTG (Presnyak et al., 2015), which is consistent with a protective role for estrogen in these conditions. The nonsynonymous SNP in AR, rs6152, swaps out the low-stability, non-optimal codon GAG for the more stable and optimal GAA, theoretically increasing the rate of synthesis for AR. This change is associated with increased risk of autism spectrum disorder in females, consistent with the widely hypothesized role for androgens in the etiology of this disorder (Henningsson et al., 2009). Two synonymous SNPs in ESR1, on the other hand, result in codons with about the same optimality and stability despite their associations with depression and Alzheimer’s disease. Isolated changes in codon usage are unlikely to account completely for associations between genotype and phenotype, but they may contribute to important variation by acting in concert with other mechanisms.
4.7. Region-specific expression
One of the most important factors to consider about regulatory variation is that its impact depends strongly on the local cellular environment. Because the local complement of transcription factors and miRs can vary dramatically according to cell phenotype (see Carroll, 2000; Deplancke et al., 2016; Hobert et al., 2004), the effects of a sequence polymorphism can also vary. In other words, the effects of a SNP on mRNA abundance in the amygdala may differ dramatically from those in cortex simply because the two brain regions express different complements of regulatory molecules, like transcription factors and miRs, that would interact with the affected sequence. Methylation and alternative splicing of sex steroid receptor genes and transcripts, respectively, can depend not only on brain region but also on age (Mott & Pak, 2013; Price et al., 2000; Schwarz et al., 2010). The main point here is that it can be difficult to predict the effect of a SNP on transcription in a particular type of tissue, because the same SNP could produce gain-of-function, loss-of-function, or be completely neutral depending on many tissue-specific factors. Those of us interested in behavior must analyze tissue collected from the relevant area of brain—a tall order when studying humans. To best understand how regulatory variation in sex steroid receptors contributes to behavioral phenotype, it will be necessary to make use of well-chosen animal models.
5. Animal models
5.1. Laboratory models
Animal models provide opportunities that human subjects cannot: a controlled environment, manipulation of the genes of interest, and extraction of any tissue to study endophenotypes. They have enabled the discovery of many causal links between expression of sex steroid receptors and behavior, which is a crucial step in the long and winding path from genotype to phenotype. Reviewing this literature is beyond our scope here, but it is worth briefly mentioning the animal models that have most informed our understanding of how variation in sex steroid receptors might contribute to phenotypic variation.
For studying the role of sex steroid receptors in behavior, the most commonly used models are testicular feminization mutants (Tfm) and knockout mice. The Tfm mutation was first described in rats (Bardin et al., 1970) and, shortly thereafter, in mice (Lyon & Hawkes, 1970). In both cases, animals carrying the Tfm allele were genetic males but exhibited a female-like external appearance, suggesting androgen insensitivity. Both mutations have been confirmed to reside within the gene for AR. In rats, a non-synonymous SNP in exon 5 causes an amino acid change in the ligand-binding domain, greatly reducing both androgen binding and receptor transactivation (Yarbrough et al., 1989). In mice, a deletion in exon 1 causes a shift in the translational reading frame, leading to a premature stop. The resulting truncated protein, which lacks both the DNA binding and the ligand binding domains (He et al., 1991), is non-functional.
For many years, Tfm rodents represented one of the only animal models in which the role of AR in a behavior could be tested; they were found to differ from wild-type animals with respect to spatial memory, anxiety-related behavior, aggression, social investigation and vocalization behavior (reviewed by Zuloaga et al., 2008). The models proved laborious to work with, however; because the mutations are located on the X chromosome, all male carriers are infertile and breeding programs are therefore difficult. This problem was solved with the advent of Cre-loxP technology, which allows editing of gene sequences such that females homozygous for a disrupted AR can be generated for breeding. Using this strategy, a number of AR knockout mice were developed to lack parts of exons 1, 2, or 3. With these newer models, the anatomical and behavioral phenotypes of Tfm mice were largely confirmed (reviewed by De Gendt & Verhoeven, 2012; Kerkhofs et al., 2009).
ERα knockout mice (ERαKO) were introduced by Lubahn et al. (1993). In these mice, ESR1 is disrupted by an insertion in the second exon. Although a truncated variant is expressed in brain, the full-length receptor is not (Shughrue et al., 2002). Early behavioral characterization of ERαKO mice revealed disruption of female sexual receptivity, masculine sexual behavior, male-male aggression, and maternal behavior (Ogawa et al., 1996; 1997; Rissman et al., 1997; Wersinger et al., 1997). Subsequent work suggested other phenotypic alterations, including effects on anxiety-like behavior, social discrimination, and cognitive function (Choleris et al., 2006; Fugger et al., 2000; Imwalle et al., 2002). A few years after the ERαKO mouse became available, Krege et al. (1998) introduced a line of mice lacking ESR2 expression (ERßKO). Unlike ERαKO mice, the ERßKOs engaged in nearly normal reproductive behaviors (Ogawa et al., 1999; c.f. Temple et al., 2003). The behavioral effects of ESR2 disruption in mice were more closely related to mood and cognition (reviewed by Bodo & Rissman, 2006; Rissman, 2008) and in females included alterations in social discrimination (Choleris et al., 2006), anxiety (Walf et al., 2009), and spatial cognition (Rissman et al., 2002). The AR and ER knockouts, which essentially changed the face of reproductive neuroendocrinology, quickly became so sophisticated that it is now possible to knock out a receptor selectively in neurons of a particular phenotype (De Gendt & Verhoeven, 2012; Mayer et al., 2010) or during a particular developmental period (e.g., Cheong et al., 2014). The utility and importance of these powerful models cannot be overestimated—transgenic mice will be major players in behavioral neuroendocrinology for the foreseeable future.
Studies on knockout mice are often cited in human association studies. For example, if a particular SNP in a sex steroid receptor is associated with memory, the report may review literature showing that knockout mice show similar deficits. Such comparisons are a bit tenuous, however, because in knockout mice the expression of the relevant gene is not simply altered—it is essentially abolished. Although insertions or nucleotide substitutions do sometimes abolish expression of functional sex steroid receptors in humans (Batch et al., 1992; Ahmed et al., 2000), none of the behaviorally relevant polymorphisms reviewed here (Figs. 1, 2; Tables 1, 2, 3) do so. Even unusually short or long polyglutamine stretches do not, alone, prevent receptor transactivation completely (McPhaul et al., 1991; Chamberlain et al., 1994). Knockout mice are thus not ideal for modeling the effects of functional receptor variants on behavioral phenotypes in humans. Other models will be necessary to recapitulate human biology.
It is technically possible to create “knockin” lines of animals that express human genetic variants. Nonetheless, few researchers have created such lines for genes relevant to behavioral disorders (see Chen et al., 2006; Mague et al., 2009). Using Cre-loxP technology, Robins and colleagues created a line of “humanized” mice that express exon 1 and surrounding intronic sequence of the human AR (Robins et al., 2008). The inserted sequences incorporate stretches of 12, 21, or 48 glutamines to mimic human variation. In males, the length of the polyglutamine stretch was inversely related to androgen-dependent measures such as body weight, seminal vesicle weight, prostate lobe weight, and plasma luteinizing hormone (Albertelli et al., 2006; Simanainen et al., 2011), suggesting that this STR polymorphism can alter the sensitivity of target organs to androgens. Surprisingly, however, the behavioral phenotype of these mice has yet to be characterized in detail. We do not know, for example, whether the polyglutamine stretch is associated with aggression, hyperactivity, or impulsivity, as it is in humans, or whether it affects performance on tests of depression-like behavior or cognitive impairment. Given the level of interest in this polymorphism in human behavioral phenotypes and disorders, more behavioral research with this model is warranted. Models of other polymorphisms can be developed as well. With the advent of CRISPR-Cas9 technology, which not only simplifies gene editing but extends the technique to a wider variety of model organisms (Hsu et al., 2014; Irion et al., 2014), we should expect to see increased availability of animals expressing human variants (see Zhu et al., 2016).
5.2. Animal models of naturally occurring genetic variation
Gene editing technology allows precise substitutions of human sequences into the genomes of non-human models, which presents tremendous opportunities for studies of mechanism. It also presents challenges because the edited sequences have been removed from their endogenous environment. It is important, therefore, to study variation in more natural contexts as well. Working with wild species, for example, allows us to take advantage of naturally occurring genetic diversity and individual differences in behavior. One such model is the white-throated sparrow (Maney, 2008; Maney & Goodson, 2011; Maney et al., 2015). In this songbird, a rearrangement of chromosome 2 (ZAL2m) has captured and subsequently driven the differentiation of genes inside it (Thomas et al., 2008). One of these genes is ESR1.
Differentiation of the genes within ZAL2m has led to differentiation of plumage and steroid-dependent behaviors. Birds of the white-striped (WS) morph (Fig. 7A), which are heterozygous for the rearrangement, exhibit higher levels of vocal aggression (Horton et al., 2014a). WS males engage in more risk-taking behavior, intruding into the territories of other males to attempt extra-pair copulations (Tuttle, 2003). Birds of the tan-striped (TS) morph, on the other hand, have two copies of the non-rearranged chromosome (ZAL2) and exhibit higher levels of parental provisioning (Horton et al., 2014a). TS males form stronger social attachments to their mates and offspring, performing more mate-guarding and parental provisioning than WS males (Horton et al., 2014b; Kopachena & Falls, 1993). Thus, the morphs differ with respect to a whole suite of steroid-dependent, correlated traits.
Fig. 7. The white-throated sparrow as a model of ERα polymorphism.
(A) white-throated sparrows occur in two plumage morphs known as white-striped (WS) and tan-striped (TS). (B) WS birds of both sexes engage in higher levels of singing in response to a simulated territorial intrusion. Data from Horton et al. (2014a). (C) WS birds are heterozygous for a series of nested inversions on chromosome 2, which have captured the gene ESR1. The rearranged chromosome is designated ZAL2m for metacentric (Thorneycroft, 1975; redrawn from Thomas et al., 2008; see also Horton et al., 2013). (D) Levels of ERa mRNA are much higher in nucleus taeniae of the amygdala (TnA) in WS than TS birds, and (E) that expression is correlated with songs given in response to simulated territorial intrusion (Horton et al., 2014b). (F) Regulatory variation in the promoter region of ESR1 causes the loss of two and the gain of six transcription factor binding sites on ZAL2m, relative to ZAL2. Open circles represent single nucleotide polymorphisms, close circles insertions, and gaps deletions. The ZAL2m sequence drives more expression of a luciferase reporter gene in vitro than the ZAL2 sequence. Redrawn from Horton et al. (2014b).
The numbers of WS and TS birds are balanced in each population because of disassortative mating—nearly all mating pairs consist of one WS and one TS bird. Thus, this species essentially has four “sexes” in that individuals seek out partners of opposite sex and morph. This mating system confines ZAL2m to a near-constant state of heterozygosity reminiscent of the mammalian Y chromosome. This situation profoundly suppresses recombination (Thomas et al., 2008) and has led to the accumulation of SNPs. Two nonsynonymous mutations have occurred in the coding region of ESR1, but these changes are not expected to affect receptor function (Horton et al., 2014b). We have therefore turned our attention to the regulation of expression.
In a field study of breeding birds, we showed that expression of ESR1 differs between WS and TS birds in at least 10 brain regions, including nucleus taeniae of the amygdala (Horton et al., 2014a; Fig. 7D). Notably, levels of ESR1 mRNA in this region significantly predicted vocal aggression (Fig. 7E), suggesting a possible causal relationship between ERα and behavior. In contrast with the coding sequences, the promoter sequences upstream of the ERα start site contain substantial variation (Fig. 7F). This variation affects putative binding sites for transcription factors. The ZAL2m allele, for example, has gained a binding site for Pbx-1, a transcription factor that may regulate the expression of ESR1 in humans (Cheung et al., 2009). To test whether such variation drives variation in gene expression, we cloned the putative promoter regions and tested their ability to drive transcription of a luciferase reporter gene in cultured cells. We found that the ZAL2m and ZAL2 promoters do in fact drive transcription to different degrees (Horton et al., 2014b; Fig. 7F), which may explain differential ESR1 expression in the brain (Fig. 7D). Thus, we have shown that in this model, variation in the ESR1 sequence leads to variation in expression, and that variation in expression predicts variation in behavior. This approach illustrates an integrative strategy that can be used in other organisms to connect genotype to phenotype across multiple levels of biological organization.
One of the most interesting studies of AR polymorphism in non-humans was done by Konno et al. (2011) in Japanese Akito Inu dogs. Like humans, these dogs are polymorphic for an STR in exon 1. The number of CAG repeats, which was found to be 23, 24, or 26 in the three alleles, was inversely correlated with aggression as reported by the dogs’ owners. The AR polyglutamine polymorphism has been characterized in a number of other species, including non-human apes and equines. The length of the CAG stretch is significantly longer in bonobos, for example, than in their more aggressive congeners, chimpanzees (Garai et al., 2014). Similarly, the number of repeats in wild zebras was found to be lower than in domesticated horses, which are presumed to be less aggressive (Ito et al., 2015). Each of these results recapitulates those showing that in humans, the length of the repeat is inversely proportional to aggression (Butovskaya et al., 2012; 2015; Rajender et al., 2008; c.f. Jönsson et al., 2001). The number of repeats may have comparable effects on receptor function (Fig. 1C) in multiple species.
Because speciation alters the genome at many locations, interspecific comparisons, for example between chimpanzees and bonobos, make it somewhat difficult to relate behavior to a single polymorphism. It is perhaps more valuable to compare more recently diverged populations, for example geographically isolated populations of the same species—particularly species from which tissue can be collected to investigate endophenotypes. I’ll briefly discuss two good examples here: the dark-eyed junco and the prairie vole. Both species have been studied for decades in the context of behavioral neuroendocrinology. In both species, geographically separated subpopulations have recently diverged with respect to sex steroid-dependent behavioral phenotypes. In a population of juncos in South Dakota, for example, males have become larger, more ornamented, and more aggressive than males in an ecologically similar population in Virginia (Nolan et al., 2002; Bergeon Burns et al., 2013). The two populations did not differ with respect to plasma T (Bergeon Burns et al., 2013), suggesting differential sensitivity to androgens or estrogens at the level of receptors. In fact, the populations differed with respect to levels of AR mRNA in the ventromedial telencephalon, which includes nucleus taeniae of the amygdala (Bergeon Burns et al., 2013). Interestingly, the relationship between aggression and ERα mRNA in this brain region was positive for the Virginia population (see also Rosvall et al., 2012) but negative for the South Dakota one. Outside the brain, AR mRNA was higher in the pituitary in males of the Virginia population (Bergeon Burns et al., 2014); other differences in the steroidogenic pathway were recently described in testes (Rosvall et al., 2016). Overall, the work with juncos suggests that the mechanisms underlying steroid-dependent behaviors are plastic and subject to natural selection, and that divergence in behavioral phenotypes can and will be traced to specific, identifiable variation in steroid-related genes. Thus, this species is rapidly becoming an important model for understanding the impact of genetic diversity on behavioral diversity (see also McGlothlin & Ketterson, 2016).
The same comparative approach has been taken a bit further in a wild rodent, the prairie vole. Research on two study populations, one in Kansas and the other in Illinois, has revealed population-based differences in several behaviors in males. Although males in both populations are monogamous, those in Kansas are larger, wider-ranging, more aggressive, and less parental than their Illinois counterparts (reviewed by Cushing et al., 2001). These behavioral differences are correlated with expression of ERα in the medial amygdala; the number of cells immunopositive for ERα was higher in this region in the Kansas males than in the Illinois males (Cushing et al., 2004). This relationship appears to extend to another species of vole, the mandarin vole; ERα expression in the medial amygdala was higher in less parental, more aggressive males from a population in Xinzheng, China than in a more parental, less aggressive population in Chengcun (Wu et al., 2011). These correlations suggest, but do not definitively show, a causal relationship between ERα expression and behavior in voles. In a critical follow-up study, Cushing et al. (2008) used viral vectors to over-express ERα in the medial amygdala of male prairie voles from the more parental Illinois population. Artificial elevation of ERα expression inhibited parental behavior and stimulated interest in novel females; in other words, it induced behavior more typical of the Kansas males. This study, which remains among few of its kind in a vertebrate, showed strong evidence that phenotypic divergence between the populations is explained by divergence in ERα distribution. Levels of ERα in the medial amygdala depend, to a degree, on environmental factors (Perry et al., 2016) but may also be modulated by regulatory variation as described above in Section 4. To further connect gene sequence with behavior in voles, juncos, and other behaviorally diverging populations, regulatory variation in sex steroid receptors should be identified and its ability to alter gene expression tested in vitro.
Naturally-occurring variation, whether introduced by an inversion or by geographic separation, presents both opportunities and challenges. The rearrangement of chromosome 2 in white-throated sparrows contains about a thousand genes, all of which are inherited together as a supergene (Thomas et al., 2008; Thompson & Jiggins, 2014). This situation magnifies the problem of linkage; the effects of the genes located on that chromosomal branch of the watershed (Fig. 3A) cannot be separated, even with large sample sizes. Linkage is a problem also in the case of diverging populations because many genes are affected in addition to sex steroid receptors. Increasing sample size could theoretically diminish the problem of linkage in divergent populations of voles and juncos, but because animals must be observed and collected in the field, often at locations distant from the lab, such an approach is not very practical. The best approach will involve a combination of traditional and non-traditional models in both the lab and the field.
6. Conclusion
Understanding how behavior is encoded in the genome, and therefore how behavior evolves and how it is inherited, is one of the most interesting and important problems in psychology. Hormones and hormone pathways are uniquely positioned to shed a tremendous amount of light here. They are already established as powerful drivers of natural behavior; in other words, they mediate “some of the best phenotypic phenomena that can be analyzed” (Pfaff et al., 2011). From work with animal models, causal roles have been established for steroid hormone receptors in a variety of behaviors, both during development and in adulthood. In order for a SNP-behavior association to expand our knowledge into truly uncharted territory, we need to consider the factors that explain the association. Even if the goal is simply to use the SNP as a marker of risk, it is nonetheless beneficial to learn about the underlying mechanisms so that we can identify the most informative—ideally causal—markers.
Making causal connections between non-coding polymorphisms and behavior will require a multi-tier, experimental approach. Not all SNPs are in a position to affect gene expression. SNPs that actively regulate mRNA transcription and stability are likely to be located within stretches of ‘open’ chromatin, in other words within sequences that are accessible to transcription factors. Such areas can be identified using techniques such as DNase-seq, ATAC-seq, or FAIRE-seq (Tsompana & Buck, 2014). The variation must then be shown to affect mRNA abundance or protein function. This goal can be accomplished using in vitro approaches, such as the reporter assays described above (Figs. 5, 6, 7F). It is even possible to use high throughput reporter assays to identify new sequences regulating expression of genes of interest (Diao et al., 2016; Rajagopal et al., 2015). Reporter assays can establish that variation has the potential to regulate expression in brain tissue, but they come with the caveat that endogenous conditions are not easily simulated in a dish.
In vitro or in vivo, gene expression is far removed from behavioral phenotype. Expression must be connected with the behavior of interest. Proteins can be manipulated via knockdown or pharmacological interventions, and effects on phenotype shown (e.g. Cushing et al., 2008), but even that approach does not show direct evidence that a genetic change affects phenotype. Connecting specific genetic sequence with a behavioral phenotype requires a transgenic approach (Hoekstra & Coyne, 2007). The gold standard experiment is to replace the specific polymorphism in the genome, then test for phenotypic change (Wray, 2013). This goal can currently be accomplished in mice with Cre-loxP knockins (Glatt & Lee, 2016; Robins, 2008); the future of this field likely lies with the newer CRISPR-Cas9 technology (Hsu et al., 2014; Irion et al., 2014; Zhu et al., 2016) in a variety of non-traditional models.
Researchers working with humans have few options for performing functional studies. Nonetheless, if the hypothesis to be tested is whether the receptor plays a role in a human behavioral phenotype, steps can be taken to maximize the informativeness of associations and to draw stronger conclusions. First, since there are hundreds of polymorphisms in sex steroid receptors to choose from, we can focus first on those that have already been modeled in vitro or in vivo. Instead of looking to functional studies in which the gene has been entirely knocked out, for example, we should cite those in which the polymorphism itself is modeled—and encourage new studies on this topic. Second, we should directly address the fact that functional studies may not generalize across different types of tissue, for example from blood to brain. Studies are sorely needed in which the functional impact of polymorphisms, for example on transcription or methylation, is compared among tissue types in animal models. Such studies may lend credence to the typical conclusions drawn in the human literature, or at the very least they will show the extent to which those conclusions are warranted.
The path from SNP to behavior is long and circuitous. Small changes at the level of gene sequence have direct effects at the levels immediately above, e.g., transcription, but these effects can become difficult to detect at higher and higher levels (Fig. 3). At the level of behavioral phenotype, they often explain very little variation (Cannon & Keller, 2006; Saltz, 2017). Experimental manipulations of gene sequence in animal models have taught us that the effects of a polymorphism on lower-level endophenotypes can quickly lead to complex and even paradoxical effects at higher levels (Glatt & Lee, 2016). A polymorphism may produce both gain-of-function and loss-of-function in different tissues in the same individual, for example. For these reasons, it seems premature to use a polymorphism as a proxy for measuring the contribution of a receptor to a behavior. If we do make that leap, it is important to look down at what we are leaping over because that view might change everything.
Highlights.
More than 50 variants of sex steroid receptor genes are associated with human behavioral phenotypes.
Most of the polymorphisms do not occur in coding regions, making interpretation challenging.
Variation in non-coding regions can affect gene transcription via a variety of mechanisms.
Making use of in vitro assays and non-human models will advance understanding of these processes.
Acknowledgments
I am grateful to Chris Goode, Katie Grogan, Jenny Merritt, Rohan Palmer, Kim Wallen, and Soojin Yi for helpful discussions on this topic and for insightful comments on drafts. I also thank Michael Konomos for assisting with Fig. 4 and Jacques Balthazart for the invitation to write this article. The work on white-throated sparrows (Section 5 and Fig. 7) was funded by the National Institutes of Health (1R21MH082046; 1R01MH082833; 1R21NIMH102677) and the National Science Foundation (SMA-1306132 and IOS-0723805).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1000 Genomes Project Consortium. (2015). A global reference for human genetic variation. Nature, 526(7571), 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams BD, Furneaux H, & White BA (2007). The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-α (ERα) and represses ERα messenger RNA and protein expression in breast cancer cell lines. Molecular Endocrinology, 21(5), 1132–1147. [DOI] [PubMed] [Google Scholar]
- Ahmed SF, Cheng A, Dovey L, Hawkins JR, Martin H, Rowland J,… & Hughes IA (2000). Phenotypic features, androgen receptor binding, and mutational analysis in 278 clinical cases reported as androgen insensitivity syndrome. The Journal of Clinical Endocrinology & Metabolism, 85(2), 658–665. [DOI] [PubMed] [Google Scholar]
- Albertelli MA, Scheller A, Brogley M, & Robins DM (2006). Replacing the mouse androgen receptor with human alleles demonstrates glutamine tract length-dependent effects on physiology and tumorigenesis in mice. Molecular Endocrinology, 20(6), 1248–1260. [DOI] [PubMed] [Google Scholar]
- Alonso P, Gratacos M, Segalas C, Escaramis G, Real E, Bayes M,… & Menchon JM (2011). Variants in estrogen receptor alpha gene are associated with phenotypical expression of obsessive-compulsive disorder. Psychoneuroendocrinology, 36(4), 473–483. [DOI] [PubMed] [Google Scholar]
- Aluja A, García LF, Martí-Guiu M, Blanco E, García O, Fibla J, & Blanch À (2015). Interactions among impulsiveness, testosterone, sex hormone binding globulin and androgen receptor gene CAG repeat length. Physiology & Behavior, 147, 91–96. [DOI] [PubMed] [Google Scholar]
- Aluja A, García LF, Blanch A, & Fibla J (2011). Association of androgen receptor gene, CAG and GGN repeat length polymorphism and impulsive-disinhibited personality traits in inmates: the role of short–long haplotype. Psychiatric Genetics, 21(5), 229–239. [DOI] [PubMed] [Google Scholar]
- Barban N, Jansen R, de Vlaming R, Vaez A, Mandemakers JJ, Tropf FC,… & Tragante V (2016). Genome-wide analysis identifies 12 loci influencing human reproductive behavior. Nature Genetics, 48, 1462–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin CW, Bullock L, Schneider G, Allison JE, & Stanley AJ (1970). Pseudohermaphrodite rat: end organ insensitivity to testosterone. Science, 167(3921), 1136–1137. [DOI] [PubMed] [Google Scholar]
- Batch JA, Williams DM, Davies HR, Brown BD, Evans BAJ, Hughes IA, & Patterson MN (1992). Androgen receptor gene mutations identified by SSCP in fourteen subjects with androgen insensitivity syndrome. Human Molecular Genetics, 1(7), 497–503. [DOI] [PubMed] [Google Scholar]
- Becherini L, Gennari L, Masi L, Mansani R, Massart F, Morelli A,… & Brandi ML (2000). Evidence of a linkage disequilibrium between polymorphisms in the human estrogen receptor α gene and their relationship to bone mass variation in postmenopausal Italian women. Human Molecular Genetics, 9(13), 2043–2050. [DOI] [PubMed] [Google Scholar]
- Beilin J, Ball EM, Favaloro JM, & Zajac JD (2000). Effect of the androgen receptor CAG repeat polymorphism on transcriptional activity: specificity in prostate and non-prostate cell lines. Journal of Molecular Endocrinology, 25(1), 85–96. [DOI] [PubMed] [Google Scholar]
- Bergeon Burns CM, Rosvall KA, Hahn TP, Demas GE, Ketterson ED (2014). Examining sources of variation in HPG axis function among individuals and populations of the dark-eyed junco. Hormones & Behavior, 65, 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeon Burns CM, Rosvall KA, & Ketterson ED (2013). Neural steroid sensitivity and aggression: comparing individuals of two songbird subspecies. Journal of Evolutionary Biology, 26(4), 820–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boada M, Antunez C, López-Arrieta J, Caruz A, Moreno-Rey C, Ramírez-Lorca R,… & Martínez-Lage P (2012). Estrogen receptor alpha gene variants are associated with Alzheimer’s disease. Neurobiology of Aging, 33(1), 198.e15–198e.24. [DOI] [PubMed] [Google Scholar]
- Bodo C, & Rissman EF (2006). New roles for estrogen receptor β in behavior and neuroendocrinology. Frontiers in Neuroendocrinology, 27(2), 217–232. [DOI] [PubMed] [Google Scholar]
- Brandi ML, Becherini L, Gennari L, Racchi M, Bianchetti A, Nacmias B,… & Govoni S (1999). Association of the estrogen receptor α gene polymorphisms with sporadic Alzheimer’s disease. Biochemical and Biophysical Research Communications, 265(2), 335–338. [DOI] [PubMed] [Google Scholar]
- Britten RJ, & Davidson EH (1971). Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. The Quarterly Review of Biology, 46(2), 111–138. [DOI] [PubMed] [Google Scholar]
- Brockschmidt FF, Nöthen MM, & Hillmer AM (2007). The two most common alleles of the coding GGN repeat in the androgen receptor gene cause differences in protein function. Journal of Molecular Endocrinology, 39(1), 1–8. [DOI] [PubMed] [Google Scholar]
- Butovskaya ML, Lazebny OE, Vasilyev VA, Dronova DA, Karelin DV, Mabulla AZ,… & Ryskov AP (2015). Androgen receptor gene polymorphism, aggression, and reproduction in Tanzanian foragers and pastoralists. PloS One, 10(8), e0136208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovskaya ML, Vasilyev VA, Lazebny OE, Burkova VN, Kulikov AM, Mabulla A,… & Ryskov AP (2012). Aggression, digit ratio, and variation in the androgen receptor, serotonin transporter, and dopamine D4 receptor genes in African foragers: the Hadza. Behavior Genetics, 42(4), 647–662. [DOI] [PubMed] [Google Scholar]
- Calvo RM, Asunción M, Sancho J, San Millán JL, & Escobar-Morreale HF (2000). The role of the CAG repeat polymorphism in the androgen receptor gene and of skewed X-chromosome inactivation, in the pathogenesis of hirsutism. The Journal of Clinical Endocrinology & Metabolism, 85(4), 1735–1740. [DOI] [PubMed] [Google Scholar]
- Cannon TD, & Keller MC (2006). Endophenotypes in the genetic analyses of mental disorders. Annual Review of Clinical Psychology, 2, 267–290. [DOI] [PubMed] [Google Scholar]
- Carroll SB (2000). Endless forms: the evolution of gene regulation and morphological diversity. Cell, 101(6), 577–580. [DOI] [PubMed] [Google Scholar]
- Carroll SB (2005). Evolution at two levels: on genes and form. PLoS Biology, 3(7), e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celec P, Tretinarova D, Minarik G, Ficek A, Szemes T, Lakatošová S,… & Ostatníková D (2013). Genetic polymorphisms related to testosterone metabolism in intellectually gifted boys. PloS One, 8(1), e54751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti B, Dudbridge F, Kent L, Wheelwright S, Hill‐Cawthorne G, Allison C,… & Baron‐Cohen S (2009). Genes related to sex steroids, neural growth, and social–emotional behavior are associated with autistic traits, empathy, and Asperger syndrome. Autism Research, 2(3), 157–177. [DOI] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, & Hurst LD (2006). Hearing silence: non-neutral evolution at synonymous sites in mammals. Nature Reviews Genetics, 7(2), 98–108. [DOI] [PubMed] [Google Scholar]
- Chamberlain NL, Driver ED, & Miesfeld RL (1994). The length and location of CAG trinucleotide repeats in the androgen receptor N-terminal domain affect transactivation function. Nucleic Acids Research, 22(15), 3181–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne FA, & Curley JP (2009). Epigenetic mechanisms mediating the long-term effects of maternal care on development. Neuroscience & Biobehavioral Reviews, 33(4), 593–600. [DOI] [PubMed] [Google Scholar]
- Chen L, Liang Y, Qiu J, Zhang L, Chen X, Luo X, & Jiang J (2013). Significance of rs1271572 in the estrogen receptor beta gene promoter and its correlation with breast cancer in a southwestern Chinese population. Journal of Biomedical Science, 20(1), 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Jing D, Bath KG, Ieraci A, Khan T, Siao CJ,… & Hempstead BL (2006). Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science, 314(5796), 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong RY, Porteous R, Chambon P, Ábrahám I, & Herbison AE (2014). Effects of neuron-specific estrogen receptor (ER) α and ERβ deletion on the acute estrogen negative feedback mechanism in adult female mice. Endocrinology, 155(4), 1418–1427. [DOI] [PubMed] [Google Scholar]
- Cheung CL, Chan BY, Chan V, Ikegawa S, Kou I, Ngai H,… & Sham PC (2009). Pre-B-cell leukemia homeobox 1 (PBX1) shows functional and possible genetic association with bone mineral density variation. Human Molecular Genetics, 18(4), 679–687. [DOI] [PubMed] [Google Scholar]
- Choleris E, Ogawa S, Kavaliers M, Gustafsson JÅ, Korach KS, Muglia LJ, & Pfaff DW (2006). Involvement of estrogen receptor α, β and oxytocin in social discrimination: a detailed behavioral analysis with knockout female mice. Genes, Brain and Behavior, 5(7), 528–539. [DOI] [PubMed] [Google Scholar]
- Choong CS, Kemppainen JA, Zhou ZX, & Wilson EM (1996). Reduced androgen receptor gene expression with first exon CAG repeat expansion. Molecular Endocrinology, 10(12), 1527–1535. [DOI] [PubMed] [Google Scholar]
- Colangelo LA, Sharp L, Kopp P, Scholtens D, Chiu BCH, Liu K, & Gapstur SM (2007). Total testosterone, androgen receptor polymorphism, and depressive symptoms in young black and white men: The CARDIA Male Hormone Study. Psychoneuroendocrinology, 32(8), 951–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comings DE, Gade‐Andavolu R, Gonzalez N, Wu S, Muhleman D, Blake H,… & Baker R (2000). Multivariate analysis of associations of 42 genes in ADHD, ODD and conduct disorder. Clinical Genetics, 58(1), 31–40. [DOI] [PubMed] [Google Scholar]
- Comings DE, Chen C, Wu S, & Muhleman D (1999a). Association of the androgen receptor gene (AR) with ADHD and conduct disorder. Neuroreport, 10, 1589–1592. [DOI] [PubMed] [Google Scholar]
- Comings DE, Muhleman D, Johnson P, & MacMurray JP (1999b). Potential role of the estrogen receptor gene (ESR1) in anxiety. Molecular Psychiatry, 4(4). [DOI] [PubMed] [Google Scholar]
- Corbo RM, Ulizzi L, Piombo L, Martinez-Labarga C, De Stefano GF, & Scacchi R (2007). Estrogen receptor alpha polymorphisms and fertility in populations with different reproductive patterns. Molecular Human Reproduction, 13(8), 537–540. [DOI] [PubMed] [Google Scholar]
- Corbo RM, Gambina G, Ruggeri M, & Scacchi R (2006). Association of estrogen receptor α (ESR1) PvuII and XbaI polymorphisms with sporadic Alzheimer’s disease and their effect on apolipoprotein E concentrations. Dementia and Geriatric Cognitive Disorders, 22(1), 67–72. [DOI] [PubMed] [Google Scholar]
- Costas J, Gratacòs M, Escaramís G, Martín-Santos R, de Diego Y, Baca-García E,… & Gutiérrez-Zotes A (2010). Association study of 44 candidate genes with depressive and anxiety symptoms in post-partum women. Journal of Psychiatric Research, 44(11), 717–724. [DOI] [PubMed] [Google Scholar]
- Cushing BS, Perry A, Musatov S, Ogawa S, & Papademetriou E (2008). Estrogen receptors in the medial amygdala inhibit the expression of male prosocial behavior. Journal of Neuroscience, 28(41), 10399–10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushing BS, Razzoli M, Murphy AZ, Epperson PM, Le WW, & Hoffman GE (2004). Intraspecific variation in estrogen receptor alpha and the expression of male sociosexual behavior in two populations of prairie voles. Brain Research, 1016(2), 247–254. [DOI] [PubMed] [Google Scholar]
- Cushing BS, Martin JO, Young LJ, & Carter CS (2001). The effects of peptides on partner preference formation are predicted by habitat in prairie voles. Hormones and Behavior, 39(1), 48–58. [DOI] [PubMed] [Google Scholar]
- Dalziel AC, Rogers SM, & Schulte PM (2009). Linking genotypes to phenotypes and fitness: how mechanistic biology can inform molecular ecology. Molecular Ecology, 18(24), 4997–5017. [DOI] [PubMed] [Google Scholar]
- Database of Single Nucleotide Polymorphisms (dbSNP). Bethesda (MD): National Center for Biotechnology Information, National Library of Medicine. (dbSNP Build ID: 149). Retrieved 1/2/2017 from: https://www.ncbi.nlm.nih.gov/projects/SNP/snp_summary.cgi. [Google Scholar]
- Davis JK, Mittel LB, Lowman JJ, Thomas PJ, Maney DL, Martin CL, NISC Comparative Sequencing Program, & Thomas JW (2011). Haplotype-based genomic sequencing of a chromosomal polymorphism in the white-throated sparrow (Zonotrichia albicollis). Journal of Heredity, 102, 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gendt K, & Verhoeven G (2012). Tissue-and cell-specific functions of the androgen receptor revealed through conditional knockout models in mice. Molecular and Cellular Endocrinology, 352(1), 13–25. [DOI] [PubMed] [Google Scholar]
- De Gregori M, Diatchenko L, Ingelmo PM, Napolioni V, Klepstad P, Belfer I,… & Normanno M (2016). Human genetic variability contributes to postoperative morphine consumption. The Journal of Pain, 17(5), 628–636. [DOI] [PubMed] [Google Scholar]
- Dean AM, & Thornton JW (2007). Mechanistic approaches to the study of evolution: the functional synthesis. Nature Reviews Genetics, 8(9), 675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Alpern D, & Gardeux V (2016). The genetics of transcription factor DNA binding variation. Cell, 166(3), 538–554. [DOI] [PubMed] [Google Scholar]
- Diao Y, Li B, Meng Z, Jung I, Lee AY, Dixon J,… & Ren B (2016). A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9-mediated genetic screening. Genome Research, 26(3), 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D, Xu L, Menon M, Reddy G, & Barrack ER (2005). Effect of GGC (glycine) repeat length polymorphism in the human androgen receptor on androgen action. The Prostate, 62(2), 133–139. [DOI] [PubMed] [Google Scholar]
- dos Reis M, Savva R, & Wernisch L (2004). Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Research, 32(17), 5036–5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Ibiary SY, Hamilton SP, Abel R, Erdman CA, Robertson PA, & Finley PR (2013). A pilot study evaluating genetic and environmental factors for postpartum depression. Innovations in Clinical Neuroscience, 10, 15–22. [PMC free article] [PubMed] [Google Scholar]
- Ellegren H (2004). Microsatellites: simple sequences with complex evolution. Nature Reviews Genetics, 5(6), 435–445. [DOI] [PubMed] [Google Scholar]
- Emw (2015). Own work, CC BY-SA 3.0, downloaded 23 May, 2017 from https://commons.wikimedia.org/w/index.php?curid=8767701.
- Ervin KS, Lymer JM, Matta R, Clipperton-Allen AE, Kavaliers M, & Choleris E (2015). Estrogen involvement in social behavior in rodents: rapid and long-term actions. Hormones and Behavior, 74, 53–76. [DOI] [PubMed] [Google Scholar]
- Fehsel K, Schikowski T, Jänner M, Hüls A, Voussoughi M, Schulte T,… & Krämer U (2016). Estrogen receptor beta polymorphisms and cognitive performance in women: associations and modifications by genetic and environmental influences. Journal of Neural Transmission, 123(12), 1369–1379. [DOI] [PubMed] [Google Scholar]
- Fernández R, Esteva I, Gómez‐Gil E, Rumbo T, Almaraz MC, Roda E,… & Pásaro E (2014). The (CA) n polymorphism of ERβ gene is associated with FtM transsexualism. The Journal of Sexual Medicine, 11(3), 720–728. [DOI] [PubMed] [Google Scholar]
- Flint J, & Munafò MR (2007). The endophenotype concept in psychiatric genetics. Psychological Medicine, 37(02), 163–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foradori CD, Weiser MJ, & Handa RJ (2008). Non-genomic actions of androgens. Frontiers in Neuroendocrinology, 29(2), 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsell C, Enmark E, Axelman K, Blomberg M, Wahlund LO, Gustafsson JÅ, & Lannfelt L (2001). Investigations of a CA repeat in the oestrogen receptor [beta] gene in patients with Alzheimer’s disease. European Journal of Human Genetics: EJHG, 9(10), 802. [DOI] [PubMed] [Google Scholar]
- Fraser HB (2013). Gene expression drives local adaptation in humans. Genome Research, 23(7), 1089–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugger HN, Foster TC, Gustafsson JÅ, & Rissman EF (2000). Novel effects of estradiol and estrogen receptor α and β on cognitive function. Brain Research, 883(2), 258–264. [DOI] [PubMed] [Google Scholar]
- Fuqua SA, Fitzgerald SD, Chamness GC, Tandon AK, McDonnell DP, Nawaz Z,… & McGuire WL (1991). Variant human breast tumor estrogen receptor with constitutive transcriptional activity. Cancer Research, 51(1), 105–109. [PubMed] [Google Scholar]
- Funnell AP, & Crossley M (2014). Hemophilia B Leyden and once mysterious cis-regulatory mutations. Trends in Genetics, 30(1), 18–23. [DOI] [PubMed] [Google Scholar]
- Furey TS, & Sethupathy P (2013). Genetics driving epigenetics. Science, 342(6159), 705–706. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B,… & Liu-Cordero SN (2002). The structure of haplotype blocks in the human genome. Science, 296(5576), 2225–2229. [DOI] [PubMed] [Google Scholar]
- Gade-Andavolu R, Macmurray J, Comings DE, Calati R, Chiesa A, & Serretti A (2009). Association between the estrogen receptor TA polymorphism and harm avoidance. Neuroscience Letters, 467(2), 155–158. [DOI] [PubMed] [Google Scholar]
- Gao T, Marcelli M, & McPhaul MJ (1996). Transcriptional activation and transient expression of the human androgen receptor. The Journal of Steroid Biochemistry and Molecular Biology, 59(1), 9–20. [DOI] [PubMed] [Google Scholar]
- Gao W, Bohl CE, & Dalton JT (2005). Chemistry and structural biology of androgen receptor. Chemical Reviews, 105(9), 3352–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garai C, Furuichi T, Kawamoto Y, Ryu H, & Inoue-Murayama M (2014). Androgen receptor and monoamine oxidase polymorphism in wild bonobos. Meta Gene, 2, 831–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng YG, Su QR, Su LY, Chen Q, Ren GY, Shen SQ,… & Xia GY (2007). Comparison of the polymorphisms of androgen receptor gene and estrogen α and β gene between adolescent females with first-onset major depressive disorder and controls. International Journal of Neuroscience, 117(4), 539–547. [DOI] [PubMed] [Google Scholar]
- Gerber HP, Seipel K, Georgiev O, Hofferer M, Hug M, Rusconi S, & Schaffner W (1994). Transcriptional activation modulation by homopolymeric glutamine and proline stretches. Science, 263(5148), 808–812. [DOI] [PubMed] [Google Scholar]
- Giegling I, Chiesa A, Calati R, Hartmann AM, Möller HJ, De Ronchi D,… & Serretti A (2009). Do the estrogen receptors 1 gene variants influence the temperament and character inventory scores in suicidal attempters and healthy subjects?. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 150(3), 434–438. [DOI] [PubMed] [Google Scholar]
- Giegling I, Rujescu D, Mandelli L, Schneider B, Hartmann AM, Schnabel A,… & Serretti A (2008). Estrogen receptor gene 1 variants are not associated with suicidal behavior. Psychiatry Research, 160(1), 1–7. [DOI] [PubMed] [Google Scholar]
- Giguére V (2002). To ERR in the estrogen pathway. TRENDS in Endocrinology & Metabolism, 13(5), 220–225. [DOI] [PubMed] [Google Scholar]
- Glatt CE, & Lee FS (2016). Common polymorphisms in the age of Research Domain Criteria (RDoC): Integration and translation. Biological Psychiatry, 79(1), 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II & Gould TD (2003). The endophenotype concept in psychiatry: etymology and strategic intentions. American Journal of Psychiatry, 160, 636–645. [DOI] [PubMed] [Google Scholar]
- Gottlieb B, Beitel LK, Nadarajah A, Paliouras M, & Trifiro M (2012). The androgen receptor gene mutations database: 2012 update. Human Mutation, 33(5), 887–894. [DOI] [PubMed] [Google Scholar]
- Goumidi L, Dahlman-Wright K, Tapia-Paez I, Matsson H, Pasquier F, Amouyel P,… & Meirhaeghe A (2011). Study of estrogen receptor-α and receptor-β gene polymorphisms on Alzheimer’s disease. Journal of Alzheimer’s Disease, 26(3), 431–439. [DOI] [PubMed] [Google Scholar]
- Greene GL, Gilna P, Waterfield M, Baker A, Hort Y, & Shine J (1986). Sequence and expression of human estrogen receptor complementary DNA. Science, 231(4742), 1150–1154. [DOI] [PubMed] [Google Scholar]
- Gunst A, Jern P, Westberg L, Johansson A, Salo B, Burri A,… & Santtila P (2015). A study of possible associations between single nucleotide polymorphisms in the estrogen receptor 2 gene and female sexual desire. The Journal of Sexual Medicine, 12(3), 676–684. [DOI] [PubMed] [Google Scholar]
- Gymrek M, Willems T, Guilmatre A, Zeng H, Markus B, Georgiev S,… & Erlich Y (2016). Abundant contribution of short tandem repeats to gene expression variation in humans. Nature Genetics, 48(1), 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas U, Sczakiel G, & Laufer S (2012). MicroRNA-mediated regulation of gene expression is affected by disease-associated SNPs within the 3′-UTR via altered RNA structure. RNA Biology, 9(6), 924–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa RJ, Ogawa S, Wang JM, & Herbison AE (2011). Roles for oestrogen receptor β in adult brain function. Journal of Neuroendocrinology, 24(1), 160–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlid S, Ivarsson MI, Butt S, Hussain S, Grzybowska E, Eyfjörd JE,… & Dillner J (2011). A candidate CpG SNP approach identifies a breast cancer associated ESR1‐SNP. International Journal of Cancer, 129(7), 1689–1698. [DOI] [PubMed] [Google Scholar]
- He WW, Kumar MV, & Tindall DJ (1991). A frame-shift mutation in the androgen receptor gene causes complete androgen insensitivity in the testicular feminized mouse. Nucleic Acids Research, 19(9), 2373–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrington DM, Howard TD, Brosnihan KB, McDonnell DP, Li X, Hawkins GA,… & Bleecker ER (2002). Common estrogen receptor polymorphism augments effects of hormone replacement therapy on E-selectin but not C-reactive protein. Circulation, 105(16), 1879–1882. [DOI] [PubMed] [Google Scholar]
- Henningsson S, Jonsson L, Ljunggren E, Westberg L, Gillberg C, Råstam M,… & Betancur C (2009). Possible association between the androgen receptor gene and autism spectrum disorder. Psychoneuroendocrinology, 34(5), 752–761. [DOI] [PubMed] [Google Scholar]
- Henningsson S, Westberg L, Nilsson S, Lundström B, Ekselius L, Bodlund O,… & Landén M (2005). Sex steroid-related genes and male-to-female transsexualism. Psychoneuroendocrinology, 30(7), 657–664. [DOI] [PubMed] [Google Scholar]
- Hickey T, Chandy A, & Norman RJ (2002). The androgen receptor CAG repeat polymorphism and X-chromosome inactivation in Australian Caucasian women with infertility related to polycystic ovary syndrome. The Journal of Clinical Endocrinology & Metabolism, 87(1), 161–165. [DOI] [PubMed] [Google Scholar]
- Hill SM, Fuqua SA, Chamness GC, Greene GL, & McGuire WL (1989). Estrogen receptor expression in human breast cancer associated with an estrogen receptor gene restriction fragment length polymorphism. Cancer Research, 49(1), 145–148. [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, & Manolio TA (2009). Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences, USA 106, 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O (2004). Common logic of transcription factor and microRNA action. Trends in Biochemical Sciences, 29(9), 462–468. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, & Coyne JA (2007). The locus of evolution: evo devo and the genetics of adaptation. Evolution, 61(5), 995–1016. [DOI] [PubMed] [Google Scholar]
- Horton BM, Moore IT, & Maney DL (2014a). New insights into the hormonal and behavioural correlates of polymorphism in white-throated sparrows. Animal Behaviour, 93, 207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton BM, Hudson WH, Ortlund EA, Shirk S, Thomas JW, Young ER, Zinzow-Kramer WM, & Maney DL (2014b). Estrogen receptor α polymorphism in a species with alternative behavioral phenotypes. Proceedings of the National Academy of Sciences, 111, 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton BM, Hu Y, Martin CL, Bunke BP, Matthews BS, Moore IT, Thomas JW, & Maney DL (2013). Behavioral characterization of a white-throated sparrow homozygous for the ZAL2m chromosomal arrangement. Behavior Genetics, 43, 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, & Zhang F (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell, 157(6), 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo L, Straub RE, Roca C, Schmidt PJ, Shi K, Vakkalanka R,… & Rubinow DR (2007). Risk for premenstrual dysphoric disorder is associated with genetic variation in ESR1, the estrogen receptor alpha gene. Biological Psychiatry, 62(8), 925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd PL, Vaillancourt KL, & Dinsdale NL (2011). Aggression, digit ratio and variation in androgen receptor and monoamine oxidase A genes in men. Behavior Genetics, 41(4), 543–556. [DOI] [PubMed] [Google Scholar]
- Iacono WG, Vaidyanathan U, Vrieze SI, & Malone SM (2014). Knowns and unknowns for psychophysiological endophenotypes: integration and response to commentaries. Psychophysiology, 51(12), 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwalle DB, Scordalakes EM, & Rissman EF (2002). Estrogen receptor α influences socially motivated behaviors. Hormones and Behavior, 42(4), 484–491. [DOI] [PubMed] [Google Scholar]
- Inoue S, Ogawa S, Horie K, Hoshino S, Goto W, Hosoi T,… & Ouchi Y (2000). An estrogen receptor β isoform that lacks exon 5 has dominant negative activity on both ERα and ERβ. Biochemical and Biophysical Research Communications, 279(3), 814–819. [DOI] [PubMed] [Google Scholar]
- Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K,… & Wang P (2010). Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. American Journal of Psychiatry, 167(7), 748–751. [DOI] [PubMed] [Google Scholar]
- Insel TR, & Fernald RD (2004). How the brain processes social information: searching for the social brain. Annual Review of Neuroscience, 27, 697–722. [DOI] [PubMed] [Google Scholar]
- Irion U, Krauss J, & Nüsslein-Volhard C (2014). Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development, 141(24), 4827–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishunina TA, & Swaab DF (2012). Decreased alternative splicing of estrogen receptor-α mRNA in the Alzheimer’s disease brain. Neurobiology of Aging, 33(2), 286–296. [DOI] [PubMed] [Google Scholar]
- Ito H, Langenhorst T, Ogden R, & Inoue-Murayama M (2015). Androgen receptor gene polymorphism in zebra species. Meta Gene, 5, 120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenster G, van der Korput HA, van Vroonhoven C, van der Kwast TH, Trapman J, & Brinkmann AO (1991). Domains of the human androgen receptor involved in steroid binding, transcriptional activation, and subcellular localization. Molecular Endocrinology, 5(10), 1396–1404. [DOI] [PubMed] [Google Scholar]
- Ji Y, Urakami K, Wada-Isoe K, Adachi Y, & Nakashima K (2000). Estrogen receptor gene polymorphisms in patients with Alzheimer’s disease, vascular dementia and alcohol-associated dementia. Dementia and Geriatric Cognitive Disorders, 11(3), 119–122. [DOI] [PubMed] [Google Scholar]
- Jiang Y, & Zhang H (2011). Propensity score based nonparametric test revealing genetic variants underlying bipolar disorder. Genetic Epidemiology, 35(2), 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Drgon T, Walther D, & Uhl GR (2011). Genomic regions identified by overlapping clusters of nominally-positive SNPs from genome-wide studies of alcohol and illegal substance dependence. PloS One, 6(7), e19210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson EG, von Gertten C, Gustavsson JP, Yuan QP, Lindblad-Toh K, Forslund K,… & Schalling M (2001). Androgen receptor trinucleotide repeat polymorphism and personality traits. Psychiatric Genetics, 11(1), 19–23. [DOI] [PubMed] [Google Scholar]
- Kapoor M, Wang JC, Wetherill L, Le N, Bertelsen S, Hinrichs AL,… & Harari O (2013). A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. Human Genetics, 132(10), 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson S, Henningsson S, Hovey D, Zettergren A, Jonsson L, Cortes DS,… & Westberg L (2016). Social memory associated with estrogen receptor polymorphisms in women. Social Cognitive and Affective Neuroscience, 2016, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemi-Esfarjani P, Trifiro MA, & Pinsky L (1995). Evidence for a repressive function of the long polyglutamine tract in the human androgen receptor: possible pathogenetic relevance for the (CAG) n-expanded neuropathies. Human Molecular Genetics, 4(4), 523–527. [DOI] [PubMed] [Google Scholar]
- Kerkhofs S, Denayer S, Haelens A, & Claessens F (2009). Androgen receptor knockout and knock-in mouse models. Journal of Molecular Endocrinology, 42(1), 11–17. [DOI] [PubMed] [Google Scholar]
- Ketterson ED, & Nolan V Jr. (1992). Hormones and life histories: an integrative approach. The American Naturalist, 140, S33–S62. [DOI] [PubMed] [Google Scholar]
- Keyes K, Agnew-Blais J, Roberts AL, Hamilton A, De Vivo I, Ranu H, & Koenen K (2015). The role of allelic variation in estrogen receptor genes and major depression in the Nurses Health Study. Social Psychiatry and Psychiatric epidemiology, 50(12), 1893–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, & Bartel DP (2009). Allelic imbalance sequencing reveals that single-nucleotide polymorphisms frequently alter microRNA-directed repression. Nature Biotechnology, 27(5), 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Pae CU, Kim MR, Min JA, Kim KH, Lee CU,… & Paik IH (2010). Association between estrogen receptor gene polymorphisms and depression in postmenopausal women: a preliminary study. Psychiatry Investigation, 7(3), 224–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, & Wilson AC (1975). Evolution at two levels in humans and chimpanzees. Science, 188(4184), 107–116. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Inoue S, Hosoi T, Ouchi Y, Shiraki M, & Orimo H (1996). Association of bone mineral density with polymorphism of the estrogen receptor gene. Journal of Bone and Mineral Research, 11(3), 306–311. [DOI] [PubMed] [Google Scholar]
- Konno A, Inoue-Murayama M, & Hasegawa T (2011). Androgen receptor gene polymorphisms are associated with aggression in Japanese Akita Inu. Biology Letters, 7(5), 658–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopachena JG, & Falls JB (1993). Aggressive performance as a behavioral correlate of plumage polymorphism in the white-throated sparrow (Zonotrichia albicollis). Behaviour, 124(3), 249–266. [Google Scholar]
- Kravitz HM, Meyer PM, Seeman TE, Greendale GA, & Sowers MR (2006a). Cognitive functioning and sex steroid hormone gene polymorphisms in women at midlife. The American Journal of Medicine, 119(9), S94–S102. [DOI] [PubMed] [Google Scholar]
- Kravitz HM, Janssen I, Lotrich FE, Kado DM, & Bromberger JT (2006b). Sex steroid hormone gene polymorphisms and depressive symptoms in women at midlife. The American Journal of Medicine, 119(9), S87–S93. [DOI] [PubMed] [Google Scholar]
- Krege JH, Hodgin JB, Couse JF, Enmark E, Warner M, Mahler JF,… & Smithies O (1998). Generation and reproductive phenotypes of mice lacking estrogen receptor β. Proceedings of the National Academy of Sciences, 95(26), 15677–15682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahey BB, Michalska KJ, Liu C, Chen Q, Hipwell AE, Chronis-Tuscano A,… & Decety J (2012). Preliminary genetic imaging study of the association between estrogen receptor-α gene polymorphisms and harsh human maternal parenting. Neuroscience Letters, 525(1), 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH 1991. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature, 352:77–79. [DOI] [PubMed] [Google Scholar]
- Le Romancer M, Poulard C, Cohen P, Sentis S, Renoir JM, & Corbo L (2011). Cracking the estrogen receptor’s posttranslational code in breast tumors. Endocrine Reviews, 32(5), 597–622. [DOI] [PubMed] [Google Scholar]
- Lubahn DB, Moyer JS, Golding TS, Couse JF, Korach KS, & Smithies O (1993). Alteration of reproductive function but not prenatal sexual development after insertional disruption of the mouse estrogen receptor gene. Proceedings of the National Academy of Sciences, 90(23), 11162–11166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin KB, Giwercman A, Dizeyi N, & Giwercman YL (2007). Functional in vitro characterisation of the androgen receptor GGN polymorphism. Molecular and Cellular Endocrinology, 264(1), 184–187. [DOI] [PubMed] [Google Scholar]
- Lyon MF, & Hawkes SG (1970). X-linked gene for testicular feminization in the mouse. Nature, 227, 1217–1219. [DOI] [PubMed] [Google Scholar]
- Ma SL, Tang NLS, Leung GTY, Fung AWT, & Lam LCW (2014). Estrogen receptor α polymorphisms and the risk of cognitive decline: A 2-Year follow-up study. The American Journal of Geriatric Psychiatry, 22(5), 489–498. [DOI] [PubMed] [Google Scholar]
- Ma SL, Tang NL, Tam CW, Lui VW, Lau ES, Zhang YP,… & Lam, L. C. (2009). Polymorphisms of the estrogen receptor α (ESR1) gene and the risk of Alzheimer’s disease in a southern Chinese community. International Psychogeriatrics, 21(5), 977–986. [DOI] [PubMed] [Google Scholar]
- Mague SD, Isiegas C, Huang P, Liu-Chen LY, Lerman C, & Blendy JA (2009). Mouse model of OPRM1 (A118G) polymorphism has sex-specific effects on drug-mediated behavior. Proceedings of the National Academy of Sciences, 106(26), 10847–10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney DL (2010) Female sexual behavior: Hormonal basis in non-mammalian vertebrates In: Breed MD and Moore J, (eds.) Encyclopedia of Animal Behavior, volume 1, pp. 697–703 Oxford: Academic Press. [Google Scholar]
- Maney DL (2008). Endocrine and genomic architecture of life history trade-offs in an avian model of social behavior. General and Comparative Endocrinology, 157, 275–282. [DOI] [PubMed] [Google Scholar]
- Maney DL & Goodson JL (2011). Neurogenomic mechanisms of aggression in songbirds. Advances in Genetics, 75, 83–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney DL, Horton BM, & Zinzow-Kramer WM (2015). Estrogen receptor alpha as a mediator of life-history trade-offs. Integrative and Comparative Biology, 55, 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney DL, Lange HS, Raees MQ, Reid AE, & Sanford SE (2009). Behavioral phenotypes persist after gonadal steroid manipulation in white-throated sparrows. Hormones & Behavior, 55, 113–120. [DOI] [PubMed] [Google Scholar]
- Marino M, Galluzzo P, & Ascenzi P (2006). Estrogen signaling multiple pathways to impact gene transcription. Current Genomics, 7(8), 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama H, Toji H, Harrington CR, Sasaki K, Izumi Y, Ohnuma T, Arai H, Yasuda M, Tanaka C, Emson PC, Nakamura S, & Kawakami H (2000). Lack of an association of estrogen receptor alpha gene polymorphisms and transcriptional activity with Alzheimer disease. Archives of Neurology, 57, 236–240. [DOI] [PubMed] [Google Scholar]
- Mayer C, Acosta-Martinez M, Dubois SL, Wolfe A, Radovick S, Boehm U, & Levine JE (2010). Timing and completion of puberty in female mice depend on estrogen receptor α-signaling in kisspeptin neurons. Proceedings of the National Academy of Sciences, 107(52), 22693–22698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MM (2011). A lumpers versus splitters approach to sexual differentiation of the brain. Frontiers in Neuroendocrinology, 32(2), 114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlothlin JW & Ketterson ED (2016). Hormonal pleiotropy and the evolution of correlated traits In Ketterson ED, Atwell J (Eds.), Snowbird: Integrative biology and evolutionary diversity in the junco. University of Chicago Press, Chicago, pp. 100–119. [Google Scholar]
- McPhaul MJ, Marcelli M, Tilley WD, Griffin JE, Isidro-Gutierrez RF, & Wilson JD (1991). Molecular basis of androgen resistance in a family with a qualitative abnormality of the androgen receptor and responsive to high-dose androgen therapy. Journal of Clinical Investigation, 87(4), 1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettman DJ, Butler MG, Poje AB, Penick EC, & Manzardo AM (2014). A preliminary case study of androgen receptor gene polymorphism association with impulsivity in women with alcoholism. Advances in Genomics and Genetics, 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micevych PE, Mermelstein PG (2008). Membrane estrogen receptors acting through metabotropic glutamate receptors: an emerging mechanism of estrogen action in brain. Molecular Neurobiology, 38, 66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill J, Kiss E, Baji I, Kapornai K, Daróczy G, Vetró Á,… & Barr C (2008). Association study of the estrogen receptor alpha gene (ESR1) and childhood‐onset mood disorders. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 147(7), 1323–1326. [DOI] [PubMed] [Google Scholar]
- Miller IJ, & Bieker JJ (1993). A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Molecular and Cellular Biology, 13(5), 2776–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A, Vo H, Huo L, Roca C, Schmidt PJ, & Rubinow DR (2010). Estrogen receptor alpha (ESR-1) associations with psychological traits in women with PMDD and controls. Journal of psychiatric research, 44(12), 788–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J, & Jacob F (1961). General conclusions: teleonomic mechanisms in cellular metabolism, growth, and differentiation In Cold Spring Harbor Symposia on Quantitative Biology (Vol. 26, pp. 389–401), Cold Spring Harbor Laboratory Press. [DOI] [PubMed] [Google Scholar]
- Monastero R, Cefalu AB, Camarda C, Noto D, Camarda LK, Caldarella R,… & Camarda R (2006). Association of estrogen receptor α gene with Alzheimer’s disease: A case-control study. Journal of Alzheimer’s Disease, 9(3), 273–278. [DOI] [PubMed] [Google Scholar]
- Mosselman S, Polman J, & Dijkema R (1996). ERβ: identification and characterization of a novel human estrogen receptor. FEBS letters, 392(1), 49–53. [DOI] [PubMed] [Google Scholar]
- Mott NN, & Pak TR (2013). Estrogen signaling and the aging brain: context-dependent considerations for postmenopausal hormone therapy. ISRN Endocrinology, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nainar S, Marshall PR, Tyler CR, Spitale RC, & Bredy TW (2016). Evolving insights into RNA modifications and their functional diversity in the brain. Nature Neuroscience, 19(10), 1292–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan V, Ketterson ED, Cristol DA, Rogers CM, Clotfelter ED, Titus RC, Schoech SJ, Snajdr E, 2002. Dark-eyed junco (Junco hyemalis) In: Poole A (Ed.), The Birds of North America Online. Cornell Lab of Ornithology, Ithaca, NY. USA. [Google Scholar]
- O’Brien SJ (1973). On estimating functional gene number in eukaryotes. Nature, 242(115), 52–54. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Chan J, Chester AE, Gustafsson JÅ, Korach KS, & Pfaff DW (1999). Survival of reproductive behaviors in estrogen receptor β gene-deficient (βERKO) male and female mice. Proceedings of the National Academy of Sciences, 96(22), 12887–12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Lubahn DB, Korach KS, & Pfaff DW (1997). Behavioral effects of estrogen receptor gene disruption in male mice. Proceedings of the National Academy of Sciences, 94(4), 1476–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Taylor JA, Lubahn DB, Korach KS, & Pfaff DW (1996). Reversal of sex roles in genetic female mice by disruption of estrogen receptor gene. Neuroendocrinology, 64(6), 467–470. [DOI] [PubMed] [Google Scholar]
- Ohlsson H, Lykkesfeldt AE, Madsen MW, & Briand P (1998). The estrogen receptor variant lacking exon 5 has dominant negative activity in the human breast epithelial cell line HMT-3522S1. Cancer Research, 58(19), 4264–4268. [PubMed] [Google Scholar]
- Olsen L, Rasmussen HB, Hansen T, Bagger YZ, Tankó LB, Qin G,… & Werge T (2006). Estrogen receptor alpha and risk for cognitive impairment in postmenopausal women. Psychiatric Genetics, 16(2), 85–88. [DOI] [PubMed] [Google Scholar]
- Osterlund MK, Gustafsson JA, Keller E, & Hurd YL (2000). Estrogen receptor beta (ERbeta) messenger ribonucleic acid (mRNA) expression within the human forebrain: distinct distribution pattern to ERalpha mRNA. Journal of Clinical Endocrinology and Metabolism, 85, 3840–3846. [DOI] [PubMed] [Google Scholar]
- Pan Y, Li Y, & Shen H (2014). Meta-analysis of the association between polymorphisms of estrogen receptor α genes rs9340799 and rs2234693 and Alzheimer’s Disease: Evidence from 23 articles. American Journal of Alzheimer’s Disease and Other Dementias, 29, 704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, & Frydman J (2013). Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nature Structural & Molecular Biology, 20(2), 237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Sainson RC, Kim JK, Hughes CC, & Levin ER (2007). A conserved mechanism for steroid receptor translocation to the plasma membrane. Journal of Biological Chemistry, 282(31), 22278–22288. [DOI] [PubMed] [Google Scholar]
- Perlman WR, Matsumoto M, Beltaifa S, Hyde TM, Saunders RC, Webster MJ,… & Weickert CS (2005). Expression of estrogen receptor alpha exon-deleted mRNA variants in the human and non-human primate frontal cortex. Neuroscience, 134(1), 81–95. [DOI] [PubMed] [Google Scholar]
- Perry AN, Carter CS, & Cushing BS (2016). Chronic social isolation enhances reproduction in the monogamous prairie vole (Microtus ochrogaster). Psychoneuroendocrinology, 68, 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff D, Waters E, Khan Q, Zhang X, & Numan M (2011). Minireview: estrogen receptor-initiated mechanisms causal to mammalian reproductive behaviors. Endocrinology, 152(4), 1209–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirskanen M, Hiltunen M, Mannermaa A, Helisalmi S, Lehtovirta M, Hänninen T, & Soininen H (2005). Estrogen receptor beta gene variants are associated with increased risk of Alzheimer’s disease in women. European Journal of Human Genetics, 13(9), 1000–1006. [DOI] [PubMed] [Google Scholar]
- Pinsonneault JK, Frater JT, Kompa B, Mascarenhas R, Wang D, & Sadee W (2017). Intronic SNP in ESR1 encoding human estrogen receptor alpha is associated with brain ESR1 mRNA isoform expression and behavioral traits. PloS One, 12(6), e0179020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsonneault JK, Sullivan D, Sadee W, Soares CN, Hampson E, & Steiner M (2013). Association study of the estrogen receptor gene ESR1 with postpartum depression—a pilot study. Archives of Women’s Mental Health, 16(6), 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pivovarciova A, Durdiakova J, Babinska K, Kubranska A, Vokalova L, Minarik G,… & Ostatnikova D (2016). Testosterone and Androgen Receptor Sensitivity in Relation to Hyperactivity Symptoms in Boys with Autism Spectrum Disorders. PloS One, 11(2), e0149657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin JB, & Kudla G (2011). Synonymous but not the same: the causes and consequences of codon bias. Nature Reviews Genetics, 12(1), 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N,… & Coller J (2015). Codon optimality is a major determinant of mRNA stability. Cell, 160(6), 1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RH, Lorenzon N, & Handa RJ (2000). Differential expression of estrogen receptor beta splice variants in rat brain: identification and characterization of a novel variant missing exon 4. Molecular Brain Research, 80(2), 260–268. [DOI] [PubMed] [Google Scholar]
- Prichard ZM, Jorm AF, Mackinnon A, & Easteal S (2007). Association analysis of 15 polymorphisms within 10 candidate genes for antisocial behavioural traits. Psychiatric Genetics, 17(5), 299–303. [DOI] [PubMed] [Google Scholar]
- Prichard Z, Jorm AF, Prior M, Sanson A, Smart D, Zhang Y,… & Easteal S (2002). Association of polymorphisms of the estrogen receptor gene with anxiety‐related traits in children and adolescents: A longitudinal study. American Journal of Medical Genetics, 114(2), 169–176. [DOI] [PubMed] [Google Scholar]
- Putnik M, Zhao C, Gustafsson JÅ, & Dahlman-Wright K (2009). Effects of two common polymorphisms in the 3’untranslated regions of estrogen receptor β on mRNA stability and translatability. BMC Genetics, 10(1), 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal N, Srinivasan S, Kooshesh K, Guo Y, Edwards MD, Banerjee B,… & Sherwood RI (2016). High-throughput mapping of regulatory DNA. Nature Biotechnology, 34(2), 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajender S, Pandu G, Sharma JD, Gandhi KPC, Singh L, & Thangaraj K (2008). Reduced CAG repeats length in androgen receptor gene is associated with violent criminal behavior. International Journal of Legal Medicine, 122(5), 367–372. [DOI] [PubMed] [Google Scholar]
- Razandi M, Pedram A, Greene GL, & Levin ER (1999). Cell membrane and nuclear estrogen receptors (ERs) originate from a single transcript: studies of ERα and ERβ expressed in Chinese hamster ovary cells. Molecular Endocrinology, 13(2), 307–319. [DOI] [PubMed] [Google Scholar]
- Reijnen MJ, Sladek FM, Bertina RM, & Reitsma PH (1992). Disruption of a binding site for hepatocyte nuclear factor 4 results in hemophilia B Leyden. Proceedings of the National Academy of Sciences, 89(14), 6300–6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissman EF (2008). Roles of oestrogen receptors α and β in behavioural neuroendocrinology: beyond Yin/Yang. Journal of Neuroendocrinology, 20(6), 873–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissman EF, Heck AL, Leonard JE, Shupnik MA, & Gustafsson JÅ (2002). Disruption of estrogen receptor β gene impairs spatial learning in female mice. Proceedings of the National Academy of Sciences, 99(6), 3996–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissman EF, Wersinger SR, Fugger HN, & Foster TC (1999). Sex with knockout models: behavioral studies of estrogen receptor α. Brain Research, 835(1), 80–90. [DOI] [PubMed] [Google Scholar]
- Rissman EF, Early AH, Taylor JA, Korach KS, & Lubahn DB (1997). Estrogen receptors are essential for female sexual receptivity. Endocrinology, 138(1), 507–510. [DOI] [PubMed] [Google Scholar]
- Robins DM, Albertelli MA, & O’Mahony OA (2008). Androgen receptor variants and prostate cancer in humanized AR mice. The Journal of Steroid Biochemistry and Molecular Biology, 108(3), 230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosvall KA, Burns CMB, Jayaratna SP, & Ketterson ED (2016). Divergence along the gonadal steroidogenic pathway: Implications for hormone-mediated phenotypic evolution. Hormones and Behavior, 84, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosvall KA, Burns CB, Barske J, Goodson JL, Schlinger BA, Sengelaub DR, & Ketterson ED (2012). Neural sensitivity to sex steroids predicts individual differences in aggression: implications for behavioural evolution. Proceedings of the Royal Society of London B: Biological Sciences, 279(>1742), 3547–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Różycka A, Słopień R, Słopień A, Dorszewska J, Seremak-Mrozikiewicz A, Lianeri M,… & Drews K (2016). The MAOA, COMT, MTHFR and ESR1 gene polymorphisms are associated with the risk of depression in menopausal women. Maturitas, 84, 42–54. [DOI] [PubMed] [Google Scholar]
- Ryan J, Carriere I, Carcaillon L, Dartigues JF, Auriacombe S, Rouaud O,… & Ancelin ML (2014). Estrogen receptor polymorphisms and incident dementia: the prospective 3C study. Alzheimer’s & Dementia, 10(1), 27–35. [DOI] [PubMed] [Google Scholar]
- Ryan J, Carrière I, Amieva H, Rouaud O, Berr C, Ritchie K,… & Ancelin ML (2013). Prospective analysis of the association between estrogen receptor gene variants and the risk of cognitive decline in elderly women. European Neuropsychopharmacology, 23(12), 1763–1768. [DOI] [PubMed] [Google Scholar]
- Ryan J, Scali J, Carrière I, Peres K, Rouaud O, Scarabin PY,… & Ancelin ML (2011a). Oestrogen receptor polymorphisms and late-life depression. The British Journal of Psychiatry, 199(2), 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan J, Scali J, Carrière I, Scarabin PY, Ritchie K, & Ancelin ML (2011b). Estrogen receptor gene variants are associated with anxiety disorders in older women. Psychoneuroendocrinology, 36(10), 1582–1586. [DOI] [PubMed] [Google Scholar]
- Saltz JB, Hessel FC, & Kelly MW (2017). Trait correlations in the genomics era. Trends in Ecology & Evolution, in press. [DOI] [PubMed] [Google Scholar]
- Sankar JS, & Hampson E (2012). Testosterone levels and androgen receptor gene polymorphism predict specific symptoms of depression in young men. Gender Medicine, 9(4), 232–243. [DOI] [PubMed] [Google Scholar]
- Schneider G, Zitzmann M, Gromoll J, Ladwig KH, & Berger K (2013). The relation between sex hormone levels, the androgen receptor CAGn-polymorphism and depression and mortality in older men in a community study. Psychoneuroendocrinology, 38(10), 2083–2090. [DOI] [PubMed] [Google Scholar]
- Schneider G, Nienhaus K, Gromoll J, Heuft G, Nieschlag E, & Zitzmann M (2011). Depressive symptoms in men aged 50 years and older and their relationship to genetic androgen receptor polymorphism and sex hormone levels in three different samples. The American Journal of Geriatric Psychiatry, 19(3), 274–283. [DOI] [PubMed] [Google Scholar]
- Schupf N, Lee JH, Wei M, Pang D, Chace C, Cheng R,… & Silverman W. (2008). Estrogen receptor-α variants increase risk of Alzheimer’s disease in women with Down syndrome. Dementia and Geriatric Cognitive Disorders, 25(5), 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Nugent BM, & McCarthy MM (2010). Developmental and hormoneinduced epigenetic changes to estrogen and progesterone receptor genes in brain are dynamic across the life span. Endocrinology, 151(10), 4871–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Van Zeeland AA, Bloss CS, Tewhey R, Bansal V, Torkamani A, Libiger O,… & Smith EN (2014). Evidence for the role of EPHX2 gene variants in anorexia nervosa. Molecular Psychiatry, 19(6), 724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman SN, Araujo AB, Roose SP, & McKinlay JB (2001). Testosterone level, androgen receptor polymorphism, and depressive symptoms in middle-aged men. Biological Psychiatry, 50(5), 371–376. [DOI] [PubMed] [Google Scholar]
- Shughrue PJ, Askew GR, Dellovade TL, & Merchenthaler I (2002). Estrogen-binding sites and their functional capacity in estrogen receptor double knockout mouse brain. Endocrinology, 143(5), 1643–1650. [DOI] [PubMed] [Google Scholar]
- Simanainen U, Brogley M, Gao YR, Jimenez M, Harwood DT, Handelsman DJ, & Robins DM (2011). Length of the human androgen receptor glutamine tract determines androgen sensitivity in vivo. Molecular and Cellular Endocrinology, 342(1), 81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P (2002). Linkage analysis in psychiatric disorders: the emerging picture. Annual Review of Genomics and Human Genetics, 3(1), 371–413. [DOI] [PubMed] [Google Scholar]
- Slof‐Op’t Landt MC, Furth EF, Meulenbelt I, Bartels M, Hottenga JJ, Slagboom PE, & Boomsma DI (2014). Association study of the estrogen receptor I gene (ESR1) in anorexia nervosa and eating disorders: no replication found. International Journal of Eating Disorders, 47(2), 211–214. [DOI] [PubMed] [Google Scholar]
- Smith AK, Kilaru V, Klengel T, Mercer KB, Bradley B, Conneely KN,… & Binder EB (2015). DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 168(1), 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers MR, Wilson AL, Karvonen-Gutierrez CA, & Kardia SR (2006). Sex steroid hormone pathway genes and health-related measures in women of 4 races/ethnicities: the Study of Women’s Health Across the Nation (SWAN). The American Journal of Medicine, 119(9), S103–S110. [DOI] [PubMed] [Google Scholar]
- Stern DL (2000). Perspective: evolutionary developmental biology and the problem of variation. Evolution, 54(4), 1079–1091. [DOI] [PubMed] [Google Scholar]
- Sundermann EE, Maki PM, & Bishop JR (2010). A review of estrogen receptor α gene (ESR1) polymorphisms, mood, and cognition. Menopause (New York, NY), 17(4), 874–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchatchou S, Jung A, Hemminki K, Sutter C, Wappenschmidt B, Bugert P,… & Ditsch N (2009). A variant affecting a putative miRNA target site in estrogen receptor (ESR) 1 is associated with breast cancer risk in premenopausal women. Carcinogenesis, 30(1), 59–64. [DOI] [PubMed] [Google Scholar]
- Temple JL, Scordalakes EM, Bodo C, Gustafsson JÅ, & Rissman EF (2003). Lack of functional estrogen receptor β gene disrupts pubertal male sexual behavior. Hormones and Behavior, 44(5), 427–434. [DOI] [PubMed] [Google Scholar]
- Tetel MJ, & Pfaff DW (2010). Contributions of estrogen receptor-α and estrogen receptor-β to the regulation of behavior. Biochimica et Biophysica Acta (BBA)-General Subjects, 1800(10), 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JW, Cáceres M, Lowman JJ, Morehouse CB, Short ME, Baldwin EL, Maney DL, & Martin CL (2008). The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotrichia albicollis) is a complex rearrangement and suppressor of recombination. Genetics, 179, 1455–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MJ, & Jiggins CD (2014). Supergenes and their role in evolution. Heredity, 113(1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JW (2001). Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proceedings of the National Academy of Sciences, 98(10), 5671–5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorneycroft HB (1975). A cytogenetic study of the white-throated sparrow, Zonotrichia albicollis (Gmelin). Evolution, 611–621. [DOI] [PubMed] [Google Scholar]
- Tsai SJ, Wang YC, Hong CJ, & Chiu HJ (2003). Association study of oestrogen receptor α gene polymorphism and suicidal behaviours in major depressive disorder. Psychiatric Genetics, 13(1), 19–22. [DOI] [PubMed] [Google Scholar]
- Tsompana M, & Buck MJ (2014). Chromatin accessibility: a window into the genome. Epigenetics & Chromatin, 7(1), 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tut TG, Ghadessy FJ, Trifiro MA, Pinsky L, & Yong EL (1997). Long polyglutamine tracts in the androgen receptor are associated with reduced trans-activation, impaired sperm production, and male infertility. Journal of Clinical Endocrinology & Metabolism, 82(11), 3777–3782. [DOI] [PubMed] [Google Scholar]
- Tuttle EM (2003). Alternative reproductive strategies in the white-throated sparrow: behavioral and genetic evidence. Behavioral Ecology, 14(3), 425–432. [Google Scholar]
- Ujike H, Otani K, Nakatsuka M, Ishii K, Sasaki A, Oishi T,… & Kimata Y (2009). Association study of gender identity disorder and sex hormone-related genes. Progress in Neuro-Psychopharmacology and Biological Psychiatry, 33(7), 1241–1244. [DOI] [PubMed] [Google Scholar]
- Vaillancourt KL, Dinsdale NL, & Hurd PL (2012). Estrogen receptor 1 promoter polymorphism and digit ratio in men. American Journal of Human Biology, 24(5), 682–689. [DOI] [PubMed] [Google Scholar]
- van Meurs JB, Schuit SC, Weel AE, van der Klift M, Bergink AP, Arp PP,… & van Leeuwen JP (2003). Association of 5′ estrogen receptor alpha gene polymorphisms with bone mineral density, vertebral bone area and fracture risk. Human Molecular Genetics, 12(14), 1745–1754. [DOI] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, & Luscombe NM (2009). A census of human transcription factors: function, expression and evolution. Nature Reviews Genetics, 10(4), 252–263. [DOI] [PubMed] [Google Scholar]
- Vermeersch H, T’Sjoen G, Kaufman JM, & Van Houtte M (2013). ESR1 polymorphisms, daily hassles, anger expression, and depressive symptoms in adolescent boys and girls. Hormones and Behavior, 63(3), 447–453. [DOI] [PubMed] [Google Scholar]
- Veronica M, Ali A, Venkateshwari A, Mamata D, & Nallari P (2016). Association of estrogen and progesterone receptor gene polymorphisms and their respective hormones in uterine leiomyomas. Tumor Biology, 37(6), 8067–8074. [DOI] [PubMed] [Google Scholar]
- Versini A, Ramoz N, Le Strat Y, Scherag S, Ehrlich S, Boni C,… & Gorwood P (2010). Estrogen receptr 1 gene (ESR1) is associated with restrictive anorexia nervosa. Neuropsychopharmacology, 35(8), 1818–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vottero A, Stratakis CA, Ghizzoni L, Longui CA, Karl M, & Chrousos GP (1999). Androgen receptor-mediated hypersensitivity to androgens in women with nonhyperandrogenic hirsutism: Skewing of X-chromosome inactivation. The Journal of Clinical Endocrinology & Metabolism, 84(3), 1091–1095. [DOI] [PubMed] [Google Scholar]
- Wake C, Labadorf A, Dumitriu A, Hoss AG, Bregu J, Albrecht KH,… & Myers RH (2016). Novel microRNA discovery using small RNA sequencing in post-mortem human brain. BMC Genomics, 17(1), 776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walf AA, Koonce C, Manley K, & Frye CA (2009). Proestrous compared to diestrous wildtype, but not estrogen receptor beta knockout, mice have better performance in the spontaneous alternation and object recognition tasks and reduced anxiety-like behavior in the elevated plus and mirror maze. Behavioural Brain Research, 196(2), 254–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DM, & Gore AC (2017). Epigenetic impacts of endocrine disruptors in the brain. Frontiers in Neuroendocrinology, 44, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert CS, Miranda-Angulo AL, Wong J, Perlman WR, Ward SE, Radhakrishna V,… & Kleinman JE (2008). Variants in the estrogen receptor alpha gene and its mRNA contribute to risk for schizophrenia. Human Molecular Genetics, 17(15), 2293–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wersinger SR, Sannen K, Villalba C, Lubahn DB, Rissman EF, & De Vries GJ (1997). Masculine sexual behavior is disrupted in male and female mice lacking a functional estrogen receptor α gene. Hormones and Behavior, 32(3), 176–183. [DOI] [PubMed] [Google Scholar]
- Westberg L, Henningsson S, Landén M, Annerbrink K, Melke J, Nilsson S,… & Eriksson E (2009). Influence of androgen receptor repeat polymorphisms on personality traits in men. Journal of Psychiatry & Neuroscience, 34(3), 205. [PMC free article] [PubMed] [Google Scholar]
- Westberg L, Melke J, Landén M, Nilsson S, Baghaei F, Rosmond R,… & Eriksson E (2003). Association between a dinucleotide repeat polymorphism of the estrogen receptor alpha gene and personality traits in women. Molecular Psychiatry, 8(1), 118–122. [DOI] [PubMed] [Google Scholar]
- Westberg L, Baghaei F, Rosmond R, Hellstrand M, Landén M, Jansson M,… & Eriksson E (2001). Polymorphisms of the androgen receptor gene and the estrogen receptor β gene are associated with androgen levels in women. The Journal of Clinical Endocrinology & Metabolism, 86(6), 2562–2568. [DOI] [PubMed] [Google Scholar]
- Wong J, Woon HG, & Weickert CS (2011). Full length TrkB potentiates estrogen receptor alpha mediated transcription suggesting convergence of susceptibility pathways in schizophrenia. Molecular and Cellular Neuroscience, 46(1), 67–78. [DOI] [PubMed] [Google Scholar]
- Wray GA (2013). Genomics and the evolution of phenotypic traits. Annual Review of Ecology, Evolution, and Systematics, 44, 51–72. [Google Scholar]
- Wray GA (2007). The evolutionary significance of cis-regulatory mutations. Nature Reviews Genetics, 8(3), 206–216. [DOI] [PubMed] [Google Scholar]
- Wu R, Yuan A, Yuan Q, Guo R, Tai F, Song Z, & Yu C (2011). Comparison of sociability, parental care and central estrogen receptor alpha expression between two populations of mandarin voles (Microtus mandarinus). Journal of Comparative Physiology A, 197(3), 267–277. [DOI] [PubMed] [Google Scholar]
- Xing Y, Jia JP, Ji XJ, & Tian T (2013). Estrogen associated gene polymorphisms and their interactions in the progress of Alzheimer’s disease. Progress in Neurobiology, 111, 53–74. [DOI] [PubMed] [Google Scholar]
- Yaffe K, Lindquist K, Sen S, Cauley J, Ferrell R, Penninx B,… & Cummings SR (2009). Estrogen receptor genotype and risk of cognitive impairment in elders: findings from the Health ABC study. Neurobiology of Aging, 30(4), 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe K, Edwards ER, Lui LY, Zmuda JM, Ferrell RE, & Cauley JA (2003). Androgen receptor CAG repeat polymorphism is associated with cognitive function in older men. Biological Psychiatry, 54(9), 943–946. [DOI] [PubMed] [Google Scholar]
- Yaffe K, Lui LY, Grady D, Stone K, & Morin P (2002). Estrogen receptor 1 polymorphisms and risk of cognitive impairment in older women. Biological Psychiatry, 51(8), 677–682. [DOI] [PubMed] [Google Scholar]
- Yarbrough WG, Quarmby VE, Simental JA, Joseph DR, Sar M, Lubahn DB,… & Wilson EM (1990). A single base mutation in the androgen receptor gene causes androgen insensitivity in the testicular feminized rat. Journal of Biological Chemistry, 265(15), 8893–8900. [PubMed] [Google Scholar]
- Yeung SLA, Jiang C, Cheng KK, Zhang W, Lam TH, Leung GM, & Schooling CM (2016). Genetically predicted 17beta-estradiol, cognitive function and depressive symptoms in women: A Mendelian randomization in the Guangzhou Biobank Cohort Study. Preventive Medicine, 88, 80–85. [DOI] [PubMed] [Google Scholar]
- Zettergren A, Karlsson S, Hovey D, Jonsson L, Melke J, Anckarsäter H,… & Westberg L (2016). Further investigations of the relation between polymorphisms in sex steroid related genes and autistic-like traits. Psychoneuroendocrinology, 68, 1–5. [DOI] [PubMed] [Google Scholar]
- Zettergren A, Jonsson L, Johansson D, Melke J, Lundström S, Anckarsäter H,… & Westberg L (2013). Associations between polymorphisms in sex steroid related genes and autistic-like traits. Psychoneuroendocrinology, 38(11), 2575–2584. [DOI] [PubMed] [Google Scholar]
- Zhang W, Edwards A, Zhu D, Flemington EK, Deininger P, & Zhang K (2012). miRNA-mediated relationships between Cis-SNP genotypes and transcript intensities in lymphocyte cell lines. PLoS One, 7(2), e31429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JV, Lam TH, Jiang C, Cherny SS, Liu B, Cheng KK,… & Schooling CM (2016). A Mendelian randomization study of testosterone and cognition in men. Scientific Reports, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Lee JH, Pang D, Temkin A, Park N, Janicki SC,… & Schupf N (2011). Estrogen receptor-Beta variants are associated with increased risk of Alzheimer’s disease in women with down syndrome. Dementia and Geriatric Cognitive Disorders, 32(4), 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QM, Ko KA, Ture S, Mastrangelo MA, Chen MH, Johnson AD,… & Lowenstein CJ (2016). Novel thrombotic function of a human SNP in STXBP5 revealed by CRISPR/Cas9 gene editing in mice. Arteriosclerosis, Thrombosis, and Vascular Biology, ATVBAHA-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuloaga DG, Puts DA, Jordan CL, & Breedlove SM (2008). The role of androgen receptors in the masculinization of brain and behavior: What we’ve learned from the testicular feminization mutation. Hormones and Behavior, 53(5), 613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]