Abstract
Neuroinflammation and neurodegeneration are common during prion infection, but the mechanisms that underlie these pathological features are not well understood. Several components of innate immunity, such as Toll-like receptor (TLR) 4 and Complement C1q, have been shown to influence prion disease. To identify additional components of innate immunity that might impact prion disease within the central nervous system (CNS), we screened RNA from brains of pre-clinical and clinical 22L-infected mice for alterations in genes associated with innate immunity. Transcription of several genes encoding damage-associated molecular pattern (DAMP) proteins and receptors were increased in the brains of prion-infected mice. To investigate the role of some of these proteins in prion disease of the CNS, we infected mice deficient in DAMP receptor genes Tlr2, C3ar1, and C5ar1 with 22L scrapie. Elimination of TLR2 accelerated disease by a median of 10 days, while lack of C3aR1 or C5aR1 had no effect on disease tempo. Histopathologically, all knockout mouse strains tested were similar to infected control mice in gliosis, vacuolation, and PrPSc deposition. Analysis of proinflammatory markers in the brains of infected knockout mice indicated only a few alterations in gene expression suggesting that C5aR1 and TLR2 signaling did not act synergistically in the brains of prion-infected mice. These results indicate that signaling through TLR2 confers partial neuroprotection during prion infection.
Introduction
Prion diseases are fatal infections of the nervous system caused by the misfolding of the normal host protein (PrPc) into a self-propagating infectious conformer (PrPSc or PrPres) [1–3]. Prion disease results in gliosis, neuronal damage, and neuroinflammation [3–7], and proinflammatory cytokines and chemokines produced by microglia, astroglia, and other cell types in the brain increase as the disease progresses [6]. It remains unclear whether this neuroinflammation is beneficial or harmful to the prion-infected individual, but it has been proposed that microglia instigate the majority of the neuroinflammatory response [8].
Microglia serve many functions in the healthy CNS and are considered the first line of defense responding to infection and damaged tissue [9–13]. These glial cells constantly survey their microenvironment, using a cadre of receptors on their surface recognizing pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) to detect abnormalities such as infection or damage [13–16]. PAMPs include exogenous molecules derived from microorganisms (such as LPS, unmethylated CpG, or diacylated lipoproteins) that allow the host to distinguish “self” from “non-self”, while DAMPs include endogenous proteins, lipids, and nucleic acids released by damaged or dead cells [17–20]. These exogenous and endogenous signals, though of differing origins, often utilize overlapping recognition receptors to elicit an inflammatory response.
Recently, we identified microglia as critical in host defense during prion infection in the CNS [21]. Mice treated with the potent CSF-1R inhibitor PLX5622 to eliminate microglia are more susceptible to prion disease and succumb to disease 20 to 30 days faster than untreated mice. Moreover, PLX5622-treated mice accumulate PrPSc earlier and to a greater extent throughout the disease course. Thus, the neuroprotective effects of microglia might be due to microglial phagocytosis and clearance of PrPSc, resulting in extended survival. Signaling through DAMP receptors that are predominately expressed on the surface of microglia may contribute to this neuroprotective function.
There is ample evidence that the complement system is involved in extra-neural prion infection. Complement component C1q is essential for neuroinvasion when mice are inoculated intraperitoneally (ip) with prions, and prion interaction with C1q is required for follicular dendritic cell infection and transmission in the spleen [22]. Furthermore, elimination of component C3 or its receptor Cr2 delays neuroinvasion after ip inoculation [22–24]. In addition, mice deficient in complement receptors CD21/35 are partially resistant to terminal prion disease when infected ip [25,26]. Interestingly, a study using C5-deficient mice, which is a component of the complement membrane attack complex that disrupts targeted cell membranes, suggested that complement component C5 was not required for prion pathogenesis when mice were challenged either by intracerebral (ic) or ip routes [27].
Other experiments implicate Toll-like receptor (TLR) involvement in prion disease. Prion disease progression was delayed in mice injected with complete Freund’s adjuvant [28]. Conversely, preexposure of infected mice to LPS [29] or Poly[I:C] [30] worsened prion pathology. Immunization with unmethylated CpG DNA delayed prion disease [31], but these findings are controversial and possibly due to damage of peripheral lymphoid structures [32] and not a direct result of TLR9 signaling. Further studies in C3H/HeJ mice that have a constitutive defect in TLR4 signaling indicate TLR4 is neuroprotective, as C3H/HeJ mice reach clinical end-point faster than C3H/HeOuJ control mice when infected ic or ip with scrapie prions [33]. These results imply that selective TLR signaling may enhance a protective response in prion-infected mice and demonstrate that prion pathogenesis can be altered by host innate immune responses.
In the present paper, we screened for changes in expression of genes encoding DAMP-proteins and receptors in the brains of mice infected with the mouse-adapted scrapie strain 22L to identify potential innate immune pathways involved in prion disease. We identified increases in transcripts encoding DAMP receptors TLR2, C3aR1, and C5aR1 that are predominantly expressed by microglia [34]. Mice deficient in expression of these genes were then infected with prions and compared to infection in wild-type controls. Our results indicated that signaling through TLR2 prolonged survival, but deletion of C3aR1 and C5aR1 had no effect on prion infection.
Materials and methods
Ethics statement
All mice were housed at the Rocky Mountain Laboratories (RML) in an AAALAC-accredited facility in compliance with guidelines provided by the Guide for the Care and Use of Laboratory Animals (Institute for Laboratory Animal Research Council). Experimentation followed RML Animal Care and Use Committee approved protocol 2015-026E.
Mice and scrapie inoculations
Tlr2-/- (Jax# 004650), C3aR1-/- (Jax# 005712), C5aR1-/- (Jax# 006845) and BALB/cJ (Jax# 000651) mice were purchased from The Jackson Laboratory. C57BL/6 and C57BL/10 were obtained from colonies maintained at Rocky Mountain Laboratories. All mice were housed 3 to 4 animals per cage.
For scrapie inoculation, mice were anesthetized with isoflurane and inoculated ic. For the initial screening of changes in expression of genes encoding innate immune and DAMP proteins, C57BL/10 mice at 4–6 weeks of age were inoculated via the left and right hemispheres with 20 μl per hemisphere of a 1.0% w/v 22L scrapie brain homogenate stock (final of 8.0 x 105 LD50) or 1.0% w/v normal brain homogenate stock (mock control) in phosphate buffered balanced saline with 2% fetal bovine serum. Following infection, mice were euthanized at a pre-clinical time point (94 dpi) or monitored for onset of scrapie signs. Six to eight mice were euthanized at 94 (pre-clinical) and 131 (clinical) dpi and brains were collected.
For survival curve studies using C3aR1-/-, C5aR1-/-, and Balb/c mice, groups were inoculated ic in the left hemisphere at 4–6 weeks of age with our standard inoculum of 30 μl of a 1.0% w/v 22L scrapie brain homogenate stock, therefore each mouse received a dose of 6.0 x 105 LD50. In contrast, groups of 4–6 weeks old Tlr2-/- and C57BL/6 mice were inoculated ic in the left hemisphere with 30 μl of a 0.01% w/v 22L scrapie brain homogenate stock, thus each mouse in this experiment received a dose of 6.0 x 103 LD50. This lower LD50 was shown to be adequate in studies performed with C3H/HeJ mice to evaluate TLR4 in prion pathogenesis [33], and using this infectious dose allowed for better comparison between studies.
Mice were observed 2–3 times per week by observers blinded to the mouse genotype. Mice were euthanized when they reached an advanced stage of scrapie disease, characterized by an increased degree of somnolence, reluctance to move, muscle weakness (kyphosis, wasting), clear gait abnormalities, poor grooming habits, and an overall decrease in body condition. Brain tissue was dissected for future use in histology, western blot, or RNA gene expression assays. Survival curves were created using GraphPad Prism version 7, and statistical analysis was performed on the curves by Mantel-Cox log rank.
Preparation of brain homogenates for protein analysis from mice
Brains were homogenized (20% w/v) using a Mini Bead Beater (BioSpec Products) as previously described [6,35] in ice-cold Cell Lysis Buffer (Bio-Rad) supplemented with 2 X Complete EDTA-free protease inhibitor cocktail (Roche) and 1 X cell lysis factors 1 and 2 (Bio-Rad) consisting of sodium orthovanadate and sodium fluoride to prevent dephosphorylation of proteins. Homogenates were stored at -80°C until use.
Quantitation of the anaphylatoxins C3a, C4a, and C5a
Enzyme-linked immunosorbent-assays (ELISA) for the quantitation of complement fragments C3a (catalog # 024347), C4a (catalog #024352), and C5a (catalog # 024362) were purchased from US Biologicals Life Sciences. All reagents were provided in the ELISA kits. Briefly, 100 μl of 10% brain homogenate (C4a and C5a) or 100 μl of 5.0% brain homogenate (C3a) was added to the well and incubated for 2 hours at 37°C. The liquid was removed and 100 μl of detection reagent A was added to the well and incubated for 1 hour at 37°C. Wells were aspirated and washed 3 times for 2 min each with 350 μl wash solution. The wash was removed and 100 μl of detection reagent B was added to the well and incubated for 30 minutes at 37°C. Wells were aspirated and washed 5 more times with wash solution. The wash was once again removed and 90 μl of 3,3′,5,5′-tetramethylbenzidine substrate was added to the well for 15 to 20 minutes. To stop the reaction, 50 μl of stop solution was added to the well. Plates were then read at 450 nm. Concentrations of bound C3a, C4a, or C5a were estimated by linear regression (GraphPad Prism version 7) using controls provided with the kit that were serially diluted 2-fold. Ranges of detection for C3a and C5a were 15.6–1000 pg/ml. Ranges for C4a were 31.2–2000 pg/ml. Determined concentrations in pg/ml of diluted brain homogenate were converted to pg/mg of brain tissue for comparison and presentation. Statistical analysis was performed by one-way analysis of variance (ANOVA) using GraphPad Prism version 7.
RNA isolation
Mouse brain halves were homogenized in 3.0 ml ZR RNA Buffer (Zymo Research) and stored up to 5 days at -80°C before processing. Total RNA was isolated using the Quick-RNA MidiPrep (Zymo Research) and treated with 4 Units of DNase I (Ambion) for 1 hour at 37°C. High-quality RNA was purified using the RNA Clean & Concentrator-100 (Zymo Research) per manufacturer’s instructions and eluted from the columns in 300–400 μl RNase-free water. For RNA isolation from the thalamus of 22L-infected or uninfected mice, thalamus tissue was excised and homogenized in 1.0 ml ZR RNA Buffer (Zymo Research) and stored up to 5 days at -80°C before processing. Total RNA was isolated using the Quick-RNAMiniPrep (Zymo Research), eluted with 75 ul nuclease-free water with 1 x RNase Inhibitor (SUPERase-In, Ambion). All RNA was stored at -80°C until use.
qRT-PCR analysis
For quantitative analysis of changes in transcription of genes encoding Complement and DAMP proteins and receptors, we compiled data using multiple qRT-PCR arrays available from Qiagen (Mouse G Protein Coupled Receptors, PAMM-3009ZE, Mouse Wound Healing PAMM-121ZE, Mouse Innate & Adaptive Immune Responses PAMM-052ZE, and Mouse Aging PAMM-178ZE). For quantitative analysis of changes in transcription of proinflammatory genes, we employed the Mouse Inflammatory Cytokine & Receptors PAMM-011ZE qRT-PCR pathway-focused array. For each array, 400 nanograms of high-quality RNA from each sample was reversed transcribed to synthesize cDNA using the RT2 First Stand Kit per manufacturer’s instructions (Qiagen). Each cDNA reaction was mixed with 2x RT2 SYBR Green Mastermix purchased from Qiagen with RNase-free water to a final volume of 1.3 ml. Ten microliters was then added to the appropriate well of a 384-well format plate. The analysis was carried out on an Applied Biosystems ViiA 7 Real-Time PCR System with a 384-well block using the following conditions: 1 cycle at 10 min, 95°C; 40 cycles at 15 s, 95°C then 1 min, 60°C with fluorescence data collection. Melting curves were generated at the end of the completed run to determine the quality of the reaction products. Raw threshold cycle (Ct) data was collected with a Ct of 35 as the cutoff. Ct data was analyzed using the web-based RT2 Profiler PCR Array Data Analysis (https://www.qiagen.com/us/shop/genes-and-pathways/data-analysis-center-overview-page/). All Ct values were normalized to the average of the Ct values for the housekeeping genes Actb, Gapdh, and Hsp90ab1. Changes in transcription were calculated by the software using the ΔΔCt based method. Statistical analysis was performed by unpaired Student’s t-test on the mean ΔCt values for each gene in the control and experimental groups, with a greater than 2-fold change and with p value of ≤ 0.05 considered significant. Each group consisted of a minimum of 3 or more independent RNA samples.
Immunoblotting for PrPres
For PrPres immunoblotting, tissue samples were analyzed as described previously [21,36,37]. Briefly, 0.18 or 0.36 mg of whole brain equivalents were treated with proteinase K, separated by SDS-PAGE, transferred to polyvinylidene difluoride, and probed with a 1:100 dilution of monoclonal human anti-PrP antibody D13 (kindly provided by R. Anthony Williamson). The secondary antibody was peroxidase-conjugated anti-human IgG used at 1:5,000 (Sigma), and immunoreactive bands were visualized using an enhanced chemiluminescence (ECL) detection system (GE Healthcare). Densitometry on unsaturated immunoreactive bands was performed on exposed film using the Bio-Rad ChemiDoc MP system. Adjusted volumes for immunoreactive bands were calculated by taking the total band volume and subtracting the global background using Image Lab software version 5.0 (Bio-Rad). Statistical analysis of densitometry by two-tailed Student t-test was performed using GraphPad Prism version 7.
Immunohistochemistry
Mice were euthanized, brains were removed, and half of the brain was placed in 3.7% phosphate buffered formalin for 3 to 5 days before dehydration and embedding in paraffin. Serial 5-μm sections were cut using a standard Leica microtome, placed on positively charged glass slides, and dried overnight at 56°C. Slides were stained with a standard protocol of hematoxylin and eosin (H&E) for observation of overall pathology. For the detection of microglia, sections were probed with antibodies against ionized calcium-binding adapter-1 (IBA-1). For astrocytes, sections were probed with antibodies to glial fibrillary acidic protein (GFAP). Slide processing was completed in a Discovery XT slide stainer (Ventana, Tucson, AZ) and read by microscopy as previously described [36,38].
Results
Initial screen for alterations in expression of DAMP/innate immune genes in 22L-infected C57BL/10 mice
In the present paper we used several PCR arrays with an expanded panel of genes to screen for changes in expression of DAMP/innate immune genes at pre-clinical (94 dpi) and clinical (131 dpi) times during 22L scrapie infection. Initially, we assayed RNA isolated from the thalamus at 60 dpi, since this region is an early site of pathology and agent replication. Several genes associated with the complement system (C4b, C1qa, C1qb, and C1qc) were increased in the thalamus at 60 dpi, and these genes were also increased in whole brain samples taken at later timepoints (Table 1). The complement-associated gene, C4b (C4 isoform B or C4 serum substance (C4Ss)), showed the highest fold-change in the thalamus and the brain. The increase in all the genes of the complement system reported herein have been detected during infection by other prion strains, including RML, 301V, and ME7 [39–42]. Thus, these changes are not specific to a single prion strain type.
Table 1. Screen for temporal changes in gene expression associated with the complement cascade and activation during infection with scrapie strain 22L.
| Gene | Thalamus 60 dpi | Brain 94 dpi | Brain 131 dpi | Complement component: | |||
|---|---|---|---|---|---|---|---|
| FC | P value | FC | P value | FC | P value | ||
| C4b (isoform B or C4Ss) | 2.9 | **** | 8.3 | *** | 16.4 | **** | C4B (serum substance) |
| C3 | -1.1 | Ns | 2.2 | ** | 13.6 | ** | C3 |
| C1qa | 2.6 | **** | 3.5 | *** | 11.5 | **** | C1, q subcomponent, alpha polypeptide |
| C1qb | 2.7 | **** | 3.6 | ** | 10.8 | **** | C1, q subcomponent, beta polypeptide |
| C1qc | 2.2 | **** | 2.9 | *** | 7.1 | **** | C1, q subcomponent, C chain |
| C1s1 | 1.2 | ns | 1.6 | ns | 2.7 | ns | C1, s subcomponent |
| C4a (isoform A or C4Slp) | 1.4 | ns | -1.2 | ns | -1.4 | ns | C4A (sex-limited protein) |
| Cfh | 1.7 | ns | 1.5 | ns | 1.9 | ns | factor h |
| Cfhr1 | 1.3 | ns | 1.6 | ns | -1.1 | ns | factor H-related 1 |
| Clu | 1.2 | ns | 1.2 | ns | 1.5 | ns | Clusterin (Apolipoprotein J) |
| Crp | ND | 1.1 | ns | -2.0 | ns | C-reactive protein, pentraxin-related | |
| Mbl2 | ND | 1.3 | ns | 2.0 | ns | Mannose-binding lectin (protein C) 2 | |
| Vtn | ND | 1.1 | ns | 1.1 | ns | Vitronectin | |
Fold Change (FC) in 22L C57BL/10 infected mice relative to uninfected C57BL/10 mice (n = 3 for all cohorts).
ND = not determined
P values are calculated using the student t-test
* P value ≤ 0.05
** P value ≤ 0.01
*** P value ≤ 0.001
**** P value ≤ 0.0001, ns = not significant.
In contrast, during scrapie infection we saw no increase in expression of regulators of the complement cascade (Table 1, lower section). For example, genes encoding the complement regulators factor H (Cfh) and factor H-related protein 1 (Cfhr1), which influence the alternate pathway of complement activation by accelerating convertase decay [43], were unchanged. Furthermore, genes encoding the proteins Clusterin (Clu) and Vitronectin (Vtn) that prevent assembly of the complement membrane attack complex [43] were also unchanged. However, components of the complement cascade were increased early in the brain during 22L infection, thus complement activation in the brain may progress without a concomitant increase in regulatory mechanisms during prion disease.
During scrapie infection, we also identified increased expression of DAMP receptor genes in the brain (Table 2). Some of these alterations (i.e. Tlr2, Ddx58, and C3ar1) have been previously identified [39,42,44], but other changes reported here, such as Adora3, C5ar1, Tlr1 and Tlr8, have not been detected previously. Notably, Tlr1 and Tlr2 were significantly increased by 60 dpi in the thalamus, and Tlr2 had the highest fold change at clinical end-point (Table 2). Most of the increased DAMP-associated genes are likely to have been produced by microglia as they are predominantly expressed by microglia [34] in normal healthy brain (Table 2, underlined genes).
Table 2. Screen for temporal changes in gene expression of damage-associated molecular pattern (DAMP) recognition receptors and associated proteins during infection with scrapie strain 22L.
| Changed | ||||||||
| Gene | Thalamus 60 dpi | Brain 94 dpi | Brain 131 dpi | Description | Effect of loss on prion disease | |||
| FC | P value | FC | P value | FC | P value | |||
| Tlr2 | 2.5 | *** | 5.1 | **** | 20.6 | **** | Toll-like receptor 2 | Herein |
| Olr1 (Lox-1) | ND | - | 7.9 | **** | 18.9 | **** | Oxidized low density lipoprotein (lectin-like) receptor 1 | NA |
| Tlr1 | 3.1 | * | 2.1 | Ns | 7.5 | **** | Toll-like receptor 1 | NA |
| C3ar1 | 2.0 | *** | 3.1 | ** | 7.1 | *** | Complement component 3a receptor 1 | Herein |
| Ddx58 | 1.6 | Ns | 2.5 | *** | 6.3 | **** | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 (RIG-I) | NA |
| C5ar1 | 1.4 | Ns | 2.0 | ** | 4.5 | **** | Complement component 5a receptor 1 | Herein |
| Adora3 | ND | 2.0 | ** | 4.3 | ** | Adenosine A3 receptor | NA | |
| Cd14 | 2.4 | ** | 1.9 | ** | 4.3 | **** | CD14 Antigen | Delayed 8 days [82] |
| P2ry6 | ND | - | 2.1 | Ns | 4.1 | * | Pyrimidinergic receptor P2Y, G-protein coupled, 6 | NA |
| Tlr8 | 1.2 | Ns | 1.3 | Ns | 4.1 | * | Toll-like receptor 8 | NA |
| Tlr9 | 1.4 | Ns | 1.3 | * | 3.3 | **** | Toll-like receptor 9 | NA |
| C5ar2 | ND | - | 2.2 | * | 3.1 | ** | G protein-coupled receptor 77 | NA |
| Tlr4 | 1.6 | Ns | 2.1 | *** | 3.0 | *** | Toll-like receptor 4 | Accelerated 15–22 days [33] |
| Tlr7 | 2.0 | * | 1.5 | ns | 2.8 | * | Toll-like receptor 7 | NA |
| Nlrp3 | -1.3 | Ns | 1.2 | ns | 2.4 | *** | NLR family, pyrin domain containing 3 | No effect [83] |
| Tlr6 | 1.3 | Ns | 1.1 | ns | 2.4 | * | Toll-like receptor 6 | NA |
| MyD88 | 1.3 | Ns | 1.3 | ns | 2.3 | *** | Myeloid differentiation primary response gene 88 | No effect [51] |
| Tlr3 | 1.8 | ** | -1.1 | ns | 2.0 | * | Toll-like receptor 3 | NA |
| Unchanged | ||||||||
| Gene | Thalamus 60 dpi | Brain 94 dpi | Brain 131 dpi | Description | Effect of loss on prion disease | |||
| FC | P value | FC | P value | FC | P value | |||
| Adora1 | ND | - | 1.1 | ns | -1.1 | ns | Adenosine A1 receptor | NA |
| Adora2a | ND | - | -1.1 | ns | -1.2 | ns | Adenosine A2a receptor | NA |
| Adora2b | ND | - | 1.1 | ns | -1.1 | ns | Adenosine A2b receptor | NA |
| Nod1 | -1.5 | ns | 1.2 | ns | 1.6 | ns | Nucleotide-binding oligomerization domain containing 1 | NA |
| Nod2 | -1.6 | ns | 1.6 | ns | 1.7 | ns | Nucleotide-binding oligomerization domain containing 2 | NA |
| Tlr5 | -1.3 | ns | 1.6 | ns | 1.1 | ns | Toll-like receptor 5 | NA |
| P2ry2 | ND | - | 1.6 | ns | 1.2 | ns | Purinergic receptor P2Y, G-protein coupled 2 | NA |
| P2ry4 | ND | - | -1.0 | ns | 2.0 | ns | Pyrimidinergic receptor P2Y, G-protein coupled, 4 | NA |
| P2ry10 | ND | - | 1.3 | ns | 2.2 | ns | Purinergic receptor P2Y, G-protein coupled 10 | NA |
| P2ry12 | ND | - | 1.0 | ns | 1.4 | ns | Purinergic receptor P2Y, G-protein coupled 12 | NA |
| P2ry13 | ND | - | 1.0 | ns | 1.9 | ns | Purinergic receptor P2Y, G-protein coupled 13 | NA |
| P2ry14 | ND | - | 1.1 | ns | 1.5 | ns | Purinergic receptor P2Y, G-protein coupled, 14 | NA |
Fold Change (FC) in 22L C57BL/10 infected mice relative to uninfected C57BL/10 mice (n = 4 for all cohorts).
P values are calculated using the student t-test
* P value ≤ 0.05
** P value ≤ 0.01
*** P value ≤ 0.001
**** P value ≤ 0.0001, ns = not significant.
ND, not determined. NA, not yet assessed during prion disease.
Underlined genes are predominantly expressed by microglia in the normal adult mouse brain [34].
Assessment of potential ligands in the brain for targeted DAMP receptors
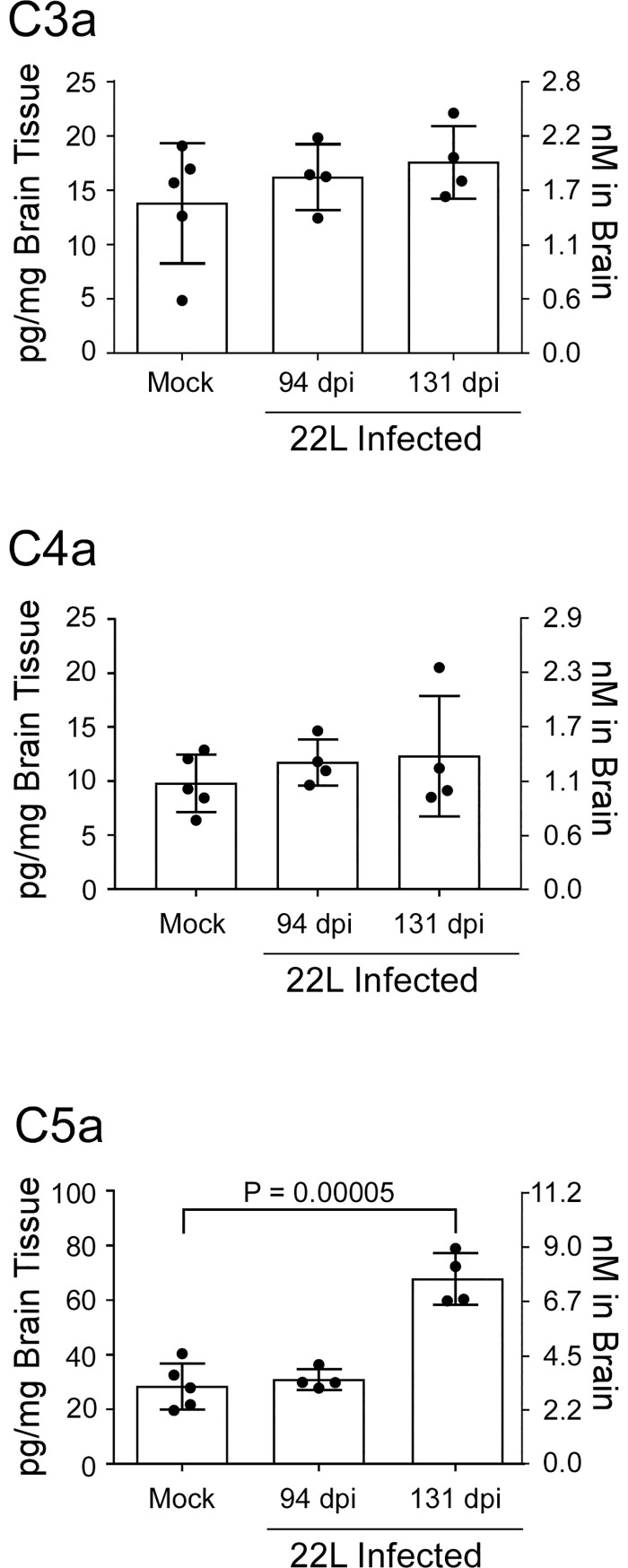
During complement activation components C3, C4, and C5 are cleaved. The large fragments are functional proteins that are either incorporated into convertase enzymes or make up a component of the membrane attack complex. The smaller fragments C3a, C4a, and C5a are collectively referred to as anaphylatoxins, and in the case of C3a and C5a these cleavage fragments act as ligands for the innate immune receptors C3aR1 and C5aR1, respectively [43,45,46]. We assessed changes in the concentration of complement fragments C3a, C4a, and C5a in mouse brain homogenates at pre-clinical and clinical times during 22L infection (Fig 1).
Fig 1. Determination of anaphylatoxin concentration in brain homogenates from 22L-infected mice.
The concentration of C3a, C4a, and C5a were determined by ELISA in the brains of mice at preclinical (94 dpi) and at clinical (131 dpi) time points after scrapie infection. Mock infected brain homogenate served as a control. Scrapie-infected cohorts were compared to mock-infected controls using a one-way ANOVA.
The concentration of C3a or C4a did not significantly change in the brains of infected mice relative to mock. In contrast, the concentration of C5a in brain more than doubled during the course of prion disease. Although C5 is not required for prion disease [27], the production of this cleavage product might influence innate immune signaling during the disease process.
Mice deficient in Tlr2, but not C3ar1 or C5ar1, are more susceptible to prion disease
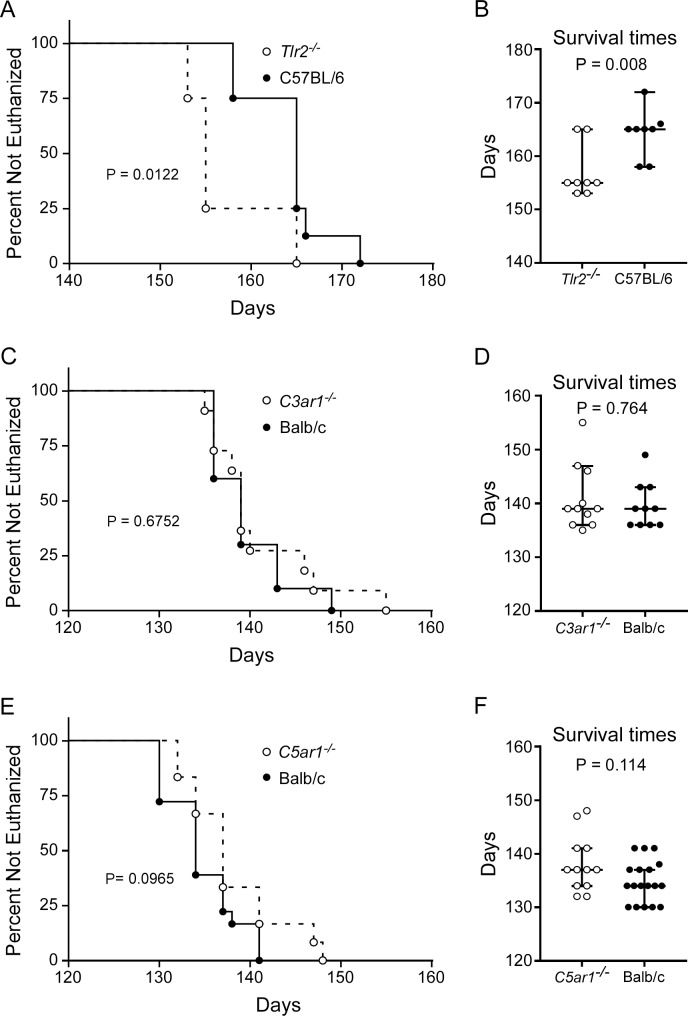
Though transcription of cell surface receptors such as TLR2, C3aR1, and C5aR1 are increased in brain prior to clinical onset (Table 2), their influence on prion pathogenesis has not been investigated. Furthermore, there is mounting evidence that the TLRs and complement receptors like C5aR1 and C3aR1 act synergistically to promote a heightened inflammatory response [47–49]. Though complement components C3 and C5 are not required for prion pathogenesis, the G protein-coupled receptors C3aR1 and C5aR1 might influence prion disease by interacting with other unidentified ligands to elicit a response. To assess the influence of these individual innate immune receptors on CNS prion pathogenesis, we used ic inoculation of 22L scrapie in mice deficient in TLR2 (Tlr2-/-), C3aR1 (C3ar1-/-), or C5aR1 (C5ar1-/-).
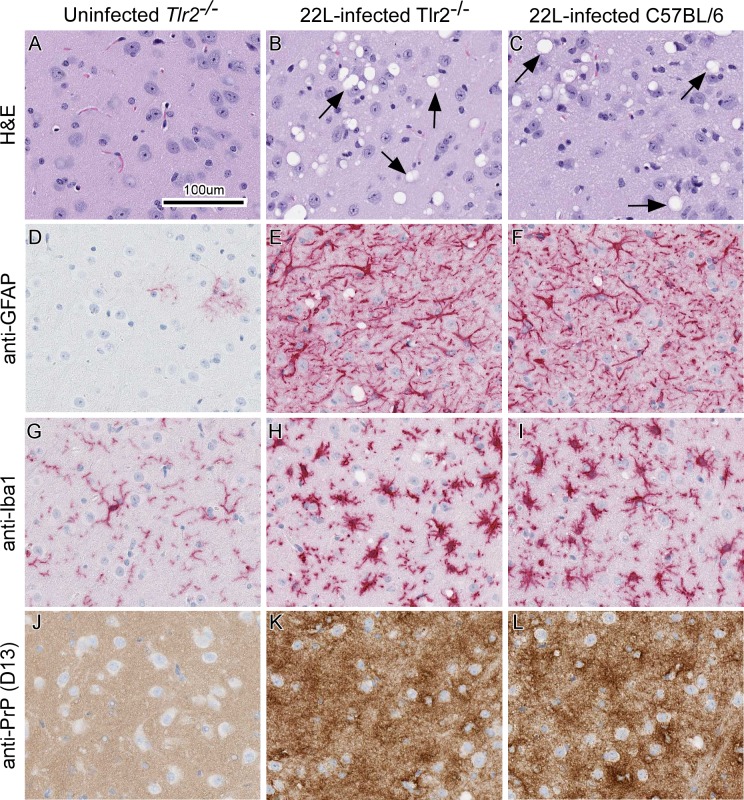
Absence of Tlr2 expression resulted in a slight (median = 10 days), but statistically significant acceleration in time to euthanasia due to advanced clinical signs compared to similarly infected C57BL/6 controls (Fig 2A and 2B). This suggested that TLR2 signaling was partially protective during prion infection. Histopathological comparison of scrapie-infected Tlr2-/- mice to infected control mice indicated that there was no difference in astrogliosis, microgliosis, vacuolation, or PrPSc deposition in the brains of mice that were clinical (Fig 3). Our overall results with Tlr2-/- mice were analogous to previous studies using C3H/HeJ mice with impaired TLR4 signaling [33]. Together these results indicated that signaling through DAMP receptors TLR2 and TLR4, which are predominantly expressed by microglia [34], contribute to neuroprotection during scrapie infection.
Fig 2. Survival of Tlr2-/-, C3ar1-/-, and C5ar1-/- mice relative to control mice infected with scrapie strain 22L.
Survival curves comparing 22L-infected control mice (filled circles) to mice deficient in Tlr2 (A and B, open circles), C3ar1 (C and D, open circles), or C5ar1 (E and F, open circles). Infected mice were monitored by observers blinded to the genotype for clinical signs of scrapie requiring euthanasia. Data in panels A, C, and E are the percentage of animals not euthanized versus days post-infection (Days). The day each individual mouse was euthanized, the median day of euthanasia for each mouse cohort (line), and the 95% confidence interval (bars) are presented in panels B, D, and F. Mice in panels A and B were inoculated with 100-fold less 22L than C3ar1-/- or C5ar1-/- mice; therefore, the longer incubation periods seen with these mice were not unexpected. Statistical analysis in panels A, C, and E was performed using Mantel–Cox log-rank analysis comparing infected knockout mice to infected control mice. Statistical analysis in panels B, D, and F was performed using Mann-Whitney test comparing infected knockout mice to infected control mice.
Fig 3. Representative neuropathology and immunohistochemical assessment of gliosis and PrP deposition in thalamic brain sections from uninfected and 22L-infected Tlr2-/- mice compared to 22L-infected C57BL/6 controls.
Tlr2-/- mice were uninfected (panels A, D, G, and J) or infected with scrapie strain 22L (panels B, E, H, and K). For comparison C57BL/6 mice were also infected with strain 22L (panels C, F, I, and L). Sections of thalamus were stained by H&E (A-C) or probed with antibodies that recognize GFAP (D—F), Iba1 (G—I), and PrP (J—L). Representative images of the thalamus are shown for all at the same scale as indicated in panel A. Black arrows in panels B and C indicate vacuoles present in the neuropil of 22L-infected mice. Similar histopathological results were seen when 22L-infected C3ar1-/- and C5ar1-/- mice were compared to infected Balb/c control mice.
Unlike Tlr2-/- mice, deletion of C3ar1 or C5ar1 had no detectable effect on survival in 22L-infected mice relative to infected Balb/c mice used as controls (Fig 2C–2F). Infected C3ar1-/-, C5ar1-/-, and Balb/c mice were histologically indistinguishable in astrogliosis, microgliosis, vacuolation, and PrPSc deposition in the brain. Moreover, these histological observations were also indistinguishable from those of infected Tlr2-/- and C57BL/6 mice in shown Fig 3. These results suggest that the loss of C3aR1 or C5aR1 signaling does not greatly influence scrapie pathogenesis.
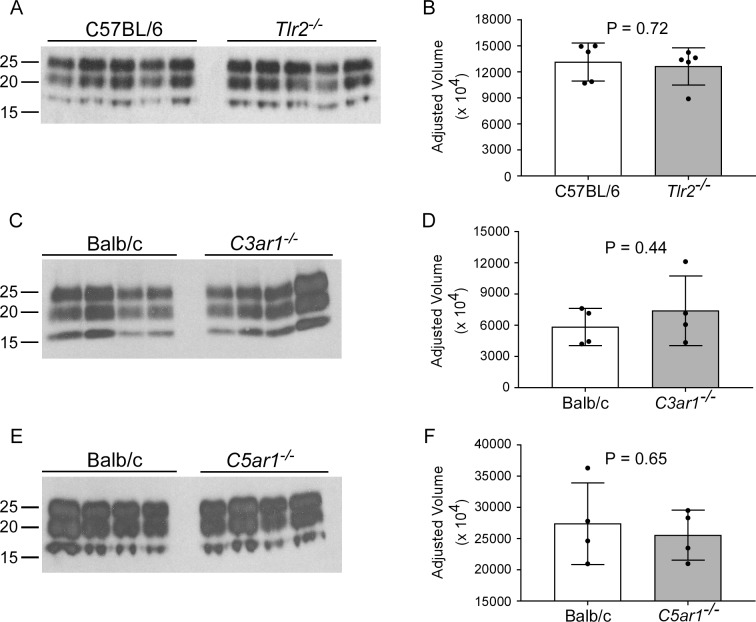
Additional assessment of disease-associated protease resistant PrP (PrPres) accumulation was performed by immunoblot. Densitometry of immunoblots of 22L-infected mouse brain homogenates indicated PrPres accumulated to similar levels in all knockout mice relative to control mice (Fig 4). These results agreed with our histological examination of brain sections stained for PrP (Fig 3J, 3K, and 3L) and suggested that deletion of Tlr2, C3ar1, or C5ar1 did not affect PrP deposition.
Fig 4. Western blot and densitometry of PrPres in the brains of knockout mice and control mice infected with 22L scrapie.
Tlr2-/- (A and B), C3ar1-/- (C and D), and C5ar1-/- (E and F) mice with appropriate control mice were infected with scrapie strain 22L and euthanized at clinical end-point. Brain homogenates were prepared and digested with proteinase K, proteins separated by SDS-PAGE, and transferred to PVDF membranes. Blocked membranes were probed with anti-PrP (D13) and bands were visualized by chemiluminescence. Panels A, C, and E were 2-minute exposures to film using ECL Western Blotting substrate. Densitometry on each lane was performed to assess differences between the groups, and plots of the adjusted volumes (see Methods) are shown to the right of the corresponding immunoblots (panels B, D, and F). Statistical analysis was performed using a two-tailed t-test comparing knockout mice to control mice. P values are as indicated.
Transcription of proinflammatory genes is largely unaffected by loss of Tlr2 or C5aR1 expression during prion infection
TLR2 and C5aR1 are involved with inflammatory signaling cascades that can act independently or synergistically [47–49]. Therefore, using the Mouse Inflammatory Cytokine & Receptors array (Qiagen) we assessed 84 genes associated with inflammation by qRT-PCR to quantify potential changes in proinflammatory expression during prion infection with the deletion of Tlr2 or C5ar1. Only three genes were significantly altered, albeit slightly, in the brains of infected Tlr2-/- mice relative to infected control mice (Table 3). Comparing 22L-infected C5ar1-/- to infected control mice, the expression of only five genes was slightly increased in mutant mice (Table 4). Of note, the genes that were altered in C5aR1-/- mice differed from those in the Tlr2-/- mice (compare Tables 3 and 4). Furthermore, most of the pro-inflammatory genes found on this array, which we have previously identified to be increased with 22L infection (such as Cxcl10, Ccl8, Tnf, IL1a, and IL1b) [6,21,38,50], were similarly increased in the absence of TLR2 or C5aR1 signaling during scrapie infection. These data suggest that the majority of the ongoing inflammatory response in the CNS during prion disease is not reliant solely on signaling through DAMP receptors TLR2 or C5aR1.
Table 3. Proinflammatory genes altered in brains of 22L-infected Tlr2-/- mice relative to 22L-infected C57BL/6 mice at the terminal timepoint.
| Gene | FC | P value | Description |
|---|---|---|---|
| Ccl2 | 2.0 | ** | Chemokine (C-C motif) ligand 2 |
| Cxcl11 | -2.1 | * | Chemokine (C-X-C motif) ligand 11 |
| IL4 | -3.2 | * | Interleukin 4 |
Fold Change (FC) in infected Tlr2-/- mice (n = 4) relative to infected C57BL/10 mice (n = 4).
P values are calculated using the Student t-test
* P value ≤ 0.05
** P value ≤ 0.01.
See Supplemental data for full analysis.
Table 4. Proinflammatory genes altered in brains of 22L-infected C5ar1-/- mice relative to 22L-infected Balb/c mice at the terminal timepoint.
| Gene | FC | P value | Description |
|---|---|---|---|
| Ccl9 | 2.2 | ** | Chemokine (C-C motif) ligand 9 |
| Ccr2 | 2.3 | * | Chemokine (C-C motif) receptor 2 |
| Csf2 | 3.7 | * | Colony stimulating factor 2 (granulocyte-macrophage) |
| Cxcl13 | 3.5 | ** | Chemokine (C-X-C motif) ligand 13 |
| Tnfsf11 | 3.6 | * | Tumor necrosis factor (ligand) superfamily, member 11 |
Fold Change (FC) in infected C5ar1-/- mice (n = 4) relative to infected Balb/c mice (n = 4).
P values are calculated using the Student t-test
* P value ≤ 0.05
** P value ≤ 0.01.
See Supplemental data for full analysis.
Discussion
We have identified numerous innate immune receptors that are increased early in the thalamus and in the brain prior to clinical signs of prion disease (Table 2). Only a few of these innate immune receptors have been studied in the context of prion infection. Thus, our aim was to assess the influence of TLR2 and the anaphylatoxin receptors, C3aR1 and C5aR1, during infection with scrapie strain 22L. Our results with Tlr2-/- mice were similar to those of Spinner et al. using prion-infected C3H/HeJ mice that lack TLR4 signaling [33]. Loss of either TLR2 or TLR4 accelerates the disease. The influence of TLR2 and TLR4 in prion infection appears to be MyD88-independent because deletion of MyD88, an adaptor molecule often shared between TLRs, does not alter the course of prion pathogenesis [51]. Thus, the protective aspects of TLR2 and TLR4 signaling might be coordinated through a different adaptor molecule like the TIR domain-containing adaptor protein inducing IFN-β (TRIF). This is supported by experiments using mice with deletion of interferon regulatory factor 3 (Irf3) [52], a key transcription factor of the TRIF-dependent pathway. Prion infection of Irf3-/- mice results in acceleration of disease onset [52], which is similar to our results with Tlr2-deficient mice and to studies performed in C3H/HeJ mice [33].
TLR2 can form a homodimer or a heterodimer with TLR1 or TLR6, increasing its repertoire of interactive ligands [53–56]. The involvement of microglial TLR2-ligand interaction in neurodegenerative diseases has become increasingly apparent. For example, TLR2 has been implicated as a primary receptor for amyloid β peptide [57], and this interaction is thought to facilitate an increase in neuroinflammation during Alzheimer’s disease. Furthermore, α-synuclein released from neurons can stimulate proinflammatory gene expression by microglia specifically through the interaction of α-synuclein and TLR2 [58,59], suggesting that TLR2 signaling may be involved in many neurodegenerative diseases including Parkinson’s disease and dementia with Lewy bodies.
To date, the ability of TLR2 to interact with PrPSc has not been addressed, but it is interesting that Tlr2 and Tlr1 expression was increased early in the thalamus in our study. This might suggest the formation of a TLR1/TLR2 heterodimer by microglia in response to prion infection, although we have no direct evidence for heterodimer formation at this time. It is noteworthy that mixed glia cultures upregulate Tlr1 and Tlr2 when exposed to recombinant prion fibrils, and preexposure of mixed neuronal and glia cultures to recombinant prion fibrils reduces prion replication in these cultures [60]. It is possible that a similar mechanism is involved during prion infection in vivo, where signaling through the TLR1/TLR2 heterodimer might reduce prion replication enough to extend survival in infected mice.
C5a is considered an extremely potent inflammatory effector that influences many cell types by increasing chemotaxis [61], augmenting vasodilation [62,63], and enhancing chemokine release [64–67]. Signaling through C5aR1 via the ligand C5a is known to contribute to neuroinflammation and pathology in neurogenerative diseases [68–71], and high concentrations of C5a can compromise the innate immune response [72–74]. The average concentration of C5a in the brain during prion infection reaches 7.6 nM by our estimate (Fig 1), and one might hypothesize that at this level C5a could contribute to the dysfunction seen in microglial phagocytosis during prion disease [75]. Surprisingly, deletion of C5, the protein that is cleaved to generate C5a, does not affect scrapie survival or vacuolation [27], and our results indicate deletion of the C5a receptor C5ar1 also has no measurable effect on prion disease. One explanation is that elevated C5a in the brain may only play a minor role in prion pathogenesis, which is not detectable in our experiments. Alternatively, many of the other innate immune receptor-ligand systems that are likely induced during prion infection might compensate for C5aR1 deficiency.
TLR2 and C5aR1 have been shown to act synergistically, resulting in crosstalk between the cellular pathways leading to enhanced inflammation and pathology [47–49]. For example, exposure of cultured human monocytes to LPS and C5a greatly enhances IL-6 and TNF production relative to LPS alone [76]. Considering this crosstalk and that Tlr2, C5ar1, and C5a were increased in the brain, we hypothesized that loss of either of the receptors would influence gene expression of an overlapping subset of proinflammatory markers during prion disease. Interestingly, this was not the case since the list of modestly altered proinflammatory genes from the 22L-infected Tlr2-/- and similarly infected C5ar1-/- mice were mutually exclusive (compare Tables 3 and 4). Furthermore, proinflammatory genes like IL1a and Tnf that are increased during prion infection [6,36,38] were similarly increased in 22L-infected Tlr2-/- or C5ar1-/- mice relative to infected control mice. From these experiments, there are two possible conclusions: 1) one pathway may compensate for the loss of the other, or 2) crosstalk between these pathways is unlikely in the brain during prion disease.
Various branches of the innate immune response are activated in neurodegenerative conditions including multiple sclerosis, Alzheimer’s disease, and traumatic brain injury [15,77–80]. Similarly, genes associated with complement, TLRs, and scavenging receptors are increased early and remain elevated during prion infection [4,81]. Early pathological changes in prion disease are likely due, in part, to an innate immune response to multiple DAMPs induced by damaged cells. Whether this damage is caused by direct interaction with PrPSc or by some other means is unclear. What is certain is that PrPSc associates with both neurons and astrocytes [38], and that DAMPs are potentially released by these infected or damaged cells. Activation of many of the DAMP receptors leads to a similar outcome, the translocation of NF-kB into the nucleus. Therefore, loss of any one receptor may be compensated by another intact system. Thus, it is not surprising that using any single innate immune receptor deletion mutant might yield, at best, only partial protection from prion infection. Alternative approaches such as network analysis to identify and alter “signaling bottlenecks” may be necessary to fully understand the involvement of DAMPs and the innate immune system in the brain during prion pathogenesis.
Supporting information
(XLSX)
(XLSX)
Acknowledgments
We thank Jeffrey Severson for assistance with animal husbandry; Nancy Kurtz and Lori Lubke for assistance with immunohistochemical procedures; and Drs. Karin Peterson, Sonja Best, and Suzette Priola for critical reading of the manuscript. This research was supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases.
Data Availability
All relevant data are in the paper and its Supporting Information files.
Funding Statement
JAC, BR, KW, and BC were supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Colby DW, Prusiner SB. Prions. Cold Spring Harb Perspect Biol. 2011; 3(1):a006833 10.1101/cshperspect.a006833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguzzi A, Falsig J. Prion propagation, toxicity and degradation. Nat Neurosci. 2012; 15(7):936–939. 10.1038/nn.3120 [DOI] [PubMed] [Google Scholar]
- 3.Caughey B, Baron GS, Chesebro B, Jeffrey M. Getting a grip on prions: oligomers, amyloids, and pathological membrane interactions. Annu Rev Biochem. 2009; 78:177–204. 10.1146/annurev.biochem.78.082907.145410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aguzzi A, Zhu C. Microglia in prion diseases. J Clin Invest. 2017; 127(9):3230–3239. 10.1172/JCI90605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booth S, Bowman C, Baumgartner R, Sorensen G, Robertson C, et al. Identification of central nervous system genes involved in the host response to the scrapie agent during preclinical and clinical infection. J Gen Virol. 2004; 85:3459–3471. 10.1099/vir.0.80110-0 [DOI] [PubMed] [Google Scholar]
- 6.Carroll JA, Striebel JF, Race B, Phillips K, Chesebro B. Prion infection of mouse brain reveals multiple new upregulated genes involved in neuroinflammation or signal transduction. J Virol. 2015; 89(4):2388–2404. 10.1128/JVI.02952-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams A, Lucassen PJ, Ritchie D, Bruce M. PrP deposition, microglial activation, and neuronal apoptosis in murine scrapie. Exp Neurol. 1997; 144(2):433–438. 10.1006/exnr.1997.6424 [DOI] [PubMed] [Google Scholar]
- 8.Vincenti JE, Murphy L, Grabert K, McColl BW, Cancellotti E, et al. Defining the Microglia Response during the Time Course of Chronic Neurodegeneration. J Virol. 2015; 90(6):3003–3017. 10.1128/JVI.02613-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong S, Stevens B. Microglia: Phagocytosing to Clear, Sculpt, and Eliminate. Dev Cell. 2016; 38(2):126–128. 10.1016/j.devcel.2016.07.006 [DOI] [PubMed] [Google Scholar]
- 10.Harry GJ. Microglia during development and aging. Pharmacol Ther. 2013; 139(3):313–326. 10.1016/j.pharmthera.2013.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nau R, Ribes S, Djukic M, Eiffert H. Strategies to increase the activity of microglia as efficient protectors of the brain against infections. Front Cell Neurosci. 2014; 8:138 10.3389/fncel.2014.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kofler J, Wiley CA. Microglia: key innate immune cells of the brain. Toxicol Pathol. 2011; 39(1):103–114. 10.1177/0192623310387619 [DOI] [PubMed] [Google Scholar]
- 13.Marin-Teva JL, Cuadros MA, Martin-Oliva D, Navascues J. Microglia and neuronal cell death. Neuron Glia Biol. 2011; 7(1):25–40. 10.1017/S1740925X12000014 [DOI] [PubMed] [Google Scholar]
- 14.Falsig J, van Beek J, Hermann C, Leist M. Molecular basis for detection of invading pathogens in the brain. J Neurosci Res. 2008; 86(7):1434–1447. 10.1002/jnr.21590 [DOI] [PubMed] [Google Scholar]
- 15.Shastri A, Bonifati DM, Kishore U. Innate immunity and neuroinflammation. Mediators Inflamm. 2013; 2013:342931 10.1155/2013/342931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kettenmann H, Hanisch UK, Noda M, Verkhratsky A. Physiology of microglia. Physiol Rev. 2011; 91(2):461–553. 10.1152/physrev.00011.2010 [DOI] [PubMed] [Google Scholar]
- 17.Piccinini AM, Midwood KS. DAMPening inflammation by modulating TLR signalling. Mediators Inflamm. 2010; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang D, Kang R, Coyne CB, Zeh HJ, Lotze MT. PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunol Rev. 2012; 249(1):158–175. 10.1111/j.1600-065X.2012.01146.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohl J. The role of complement in danger sensing and transmission. Immunol Res. 2006; 34(2):157–176. 10.1385/IR:34:2:157 [DOI] [PubMed] [Google Scholar]
- 20.Tsan MF, Gao B. Endogenous ligands of Toll-like receptors. J Leukoc Biol. 2004; 76(3):514–519. 10.1189/jlb.0304127 [DOI] [PubMed] [Google Scholar]
- 21.Carroll JA, Race B, Williams K, Striebel J, Chesebro B. Microglia Are Critical in Host Defense Against Prion Disease. J Virol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein MA, Kaeser PS, Schwarz P, Weyd H, Xenarios I, et al. Complement facilitates early prion pathogenesis. Nat Med. 2001; 7(4):488–492. 10.1038/86567 [DOI] [PubMed] [Google Scholar]
- 23.Mabbott NA, Bruce ME, Botto M, Walport MJ, Pepys MB. Temporary depletion of complement component C3 or genetic deficiency of C1q significantly delays onset of scrapie. Nat Med. 2001; 7(4):485–487. 10.1038/86562 [DOI] [PubMed] [Google Scholar]
- 24.Michel B, Ferguson A, Johnson T, Bender H, Meyerett-Reid C, et al. Complement protein C3 exacerbates prion disease in a mouse model of chronic wasting disease. Int Immunol. 2013; 25(12):697–702. 10.1093/intimm/dxt034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michel B, Ferguson A, Johnson T, Bender H, Meyerett-Reid C, et al. Genetic depletion of complement receptors CD21/35 prevents terminal prion disease in a mouse model of chronic wasting disease. J Immunol. 2012; 189(9):4520–4527. 10.4049/jimmunol.1201579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kane SJ, Swanson E, Gordon EO, Rocha S, Bender HR, et al. Relative Impact of Complement Receptors CD21/35 (Cr2/1) on Scrapie Pathogenesis in Mice. mSphere. 2017; 2(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mabbott NA, Bruce ME. Complement component C5 is not involved in scrapie pathogenesis. Immunobiology. 2004; 209(7):545–549. 10.1016/j.imbio.2004.06.003 [DOI] [PubMed] [Google Scholar]
- 28.Tal Y, Souan L, Cohen IR, Meiner Z, Taraboulos A, et al. Complete Freund's adjuvant immunization prolongs survival in experimental prion disease in mice. J Neurosci Res. 2003; 71(2):286–290. 10.1002/jnr.10474 [DOI] [PubMed] [Google Scholar]
- 29.Cunningham C, Wilcockson DC, Campion S, Lunnon K, Perry VH. Central and systemic endotoxin challenges exacerbate the local inflammatory response and increase neuronal death during chronic neurodegeneration. J Neurosci. 2005; 25(40):9275–9284. 10.1523/JNEUROSCI.2614-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Field R, Campion S, Warren C, Murray C, Cunningham C. Systemic challenge with the TLR3 agonist poly I:C induces amplified IFNalpha/beta and IL-1beta responses in the diseased brain and exacerbates chronic neurodegeneration. Brain Behav Immun. 2010; 24(6):996–1007. 10.1016/j.bbi.2010.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sethi S, Lipford G, Wagner H, Kretzschmar H. Postexposure prophylaxis against prion disease with a stimulator of innate immunity. Lancet. 2002; 360(9328):229–230. 10.1016/S0140-6736(02)09513-2 [DOI] [PubMed] [Google Scholar]
- 32.Heikenwalder M, Polymenidou M, Junt T, Sigurdson C, Wagner H, et al. Lymphoid follicle destruction and immunosuppression after repeated CpG oligodeoxynucleotide administration. Nat Med. 2004; 10(2):187–192. 10.1038/nm987 [DOI] [PubMed] [Google Scholar]
- 33.Spinner DS, Cho IS, Park SY, Kim JI, Meeker HC, et al. Accelerated prion disease pathogenesis in Toll-like receptor 4 signaling-mutant mice. J Virol. 2008; 82(21):10701–10708. 10.1128/JVI.00522-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014; 34(36):11929–11947. 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tribouillard-Tanvier D, Striebel JF, Peterson KE, Chesebro B. Analysis of protein levels of 24 cytokines in scrapie agent-infected brain and glial cell cultures from mice differing in prion protein expression levels. J Virol. 2009; 83(21):11244–11253. 10.1128/JVI.01413-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tribouillard-Tanvier D, Race B, Striebel JF, Carroll JA, Phillips K, et al. Early cytokine elevation, PrPres deposition, and gliosis in mouse scrapie: no effect on disease by deletion of cytokine genes IL-12p40 and IL-12p35. J Virol. 2012; 86(19):10377–10383. 10.1128/JVI.01340-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meade-White K, Race B, Trifilo M, Bossers A, Favara C, et al. Resistance to chronic wasting disease in transgenic mice expressing a naturally occurring allelic variant of deer prion protein. J Virol. 2007; 81(9):4533–4539. 10.1128/JVI.02762-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carroll JA, Striebel JF, Rangel A, Woods T, Phillips K, et al. Prion Strain Differences in Accumulation of PrPSc on Neurons and Glia Are Associated with Similar Expression Profiles of Neuroinflammatory Genes: Comparison of Three Prion Strains. PLoS Pathog. 2016; 12(4):e1005551 10.1371/journal.ppat.1005551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang D, Lee IY, Yoo H, Gehlenborg N, Cho JH, et al. A systems approach to prion disease. Mol Syst Biol. 2009; 5:252 10.1038/msb.2009.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skinner PJ, Abbassi H, Chesebro B, Race RE, Reilly C, et al. Gene expression alterations in brains of mice infected with three strains of scrapie. BMC Genomics. 2006; 7:114 10.1186/1471-2164-7-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riemer C, Neidhold S, Burwinkel M, Schwarz A, Schultz J, et al. Gene expression profiling of scrapie-infected brain tissue. Biochem Biophys Res Commun. 2004; 323(2):556–564. 10.1016/j.bbrc.2004.08.124 [DOI] [PubMed] [Google Scholar]
- 42.Xiang W, Windl O, Wunsch G, Dugas M, Kohlmann A, et al. Identification of differentially expressed genes in scrapie-infected mouse brains by using global gene expression technology. J Virol. 2004; 78(20):11051–11060. 10.1128/JVI.78.20.11051-11060.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010; 11(9):785–797. 10.1038/ni.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moody LR, Herbst AJ, Yoo HS, Vanderloo JP, Aiken JM. Comparative prion disease gene expression profiling using the prion disease mimetic, cuprizone. Prion. 2009; 3(2):99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee H, Whitfeld PL, Mackay CR. Receptors for complement C5a. The importance of C5aR and the enigmatic role of C5L2. Immunol Cell Biol. 2008; 86(2):153–160. 10.1038/sj.icb.7100166 [DOI] [PubMed] [Google Scholar]
- 46.Klos A, Tenner AJ, Johswich KO, Ager RR, Reis ES, et al. The role of the anaphylatoxins in health and disease. Mol Immunol. 2009; 46(14):2753–2766. 10.1016/j.molimm.2009.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hajishengallis G, Lambris JD. More than complementing Tolls: complement-Toll-like receptor synergy and crosstalk in innate immunity and inflammation. Immunol Rev. 2016; 274(1):233–244. 10.1111/imr.12467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hajishengallis G, Lambris JD. Crosstalk pathways between Toll-like receptors and the complement system. Trends Immunol. 2010; 31(4):154–163. 10.1016/j.it.2010.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hovland A, Jonasson L, Garred P, Yndestad A, Aukrust P, et al. The complement system and toll-like receptors as integrated players in the pathophysiology of atherosclerosis. Atherosclerosis. 2015; 241(2):480–494. 10.1016/j.atherosclerosis.2015.05.038 [DOI] [PubMed] [Google Scholar]
- 50.Carroll JA, Race B, Phillips K, Striebel JF, Chesebro B. Statins are ineffective at reducing neuroinflammation or prolonging survival in scrapie-infected mice. J Gen Virol. 2017; 98(8):2190–2199. 10.1099/jgv.0.000876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prinz M, Heikenwalder M, Schwarz P, Takeda K, Akira S, et al. Prion pathogenesis in the absence of Toll-like receptor signalling. EMBO Rep. 2003; 4(2):195–199. 10.1038/sj.embor.embor731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishibashi D, Atarashi R, Fuse T, Nakagaki T, Yamaguchi N, et al. Protective role of interferon regulatory factor 3-mediated signaling against prion infection. J Virol. 2012; 86(9):4947–4955. 10.1128/JVI.06326-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, et al. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci U S A. 2000; 97(25):13766–13771. 10.1073/pnas.250476497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takeuchi O, Kawai T, Muhlradt PF, Morr M, Radolf JD, et al. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int Immunol. 2001; 13(7):933–940. [DOI] [PubMed] [Google Scholar]
- 55.Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, et al. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol. 2002; 169(1):10–14. [DOI] [PubMed] [Google Scholar]
- 56.Morr M, Takeuchi O, Akira S, Simon MM, Muhlradt PF. Differential recognition of structural details of bacterial lipopeptides by toll-like receptors. Eur J Immunol. 2002; 32(12):3337–3347. [DOI] [PubMed] [Google Scholar]
- 57.Liu S, Liu Y, Hao W, Wolf L, Kiliaan AJ, et al. TLR2 is a primary receptor for Alzheimer's amyloid beta peptide to trigger neuroinflammatory activation. J Immunol. 2012; 188(3):1098–1107. 10.4049/jimmunol.1101121 [DOI] [PubMed] [Google Scholar]
- 58.Kim C, Ho DH, Suk JE, You S, Michael S, et al. Neuron-released oligomeric alpha-synuclein is an endogenous agonist of TLR2 for paracrine activation of microglia. Nat Commun. 2013; 4:1562 10.1038/ncomms2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dzamko N, Gysbers A, Perera G, Bahar A, Shankar A, et al. Toll-like receptor 2 is increased in neurons in Parkinson's disease brain and may contribute to alpha-synuclein pathology. Acta Neuropathol. 2017; 133(2):303–319. 10.1007/s00401-016-1648-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kang SG, Kim C, Cortez LM, Carmen Garza M, Yang J, et al. Toll-like receptor-mediated immune response inhibits prion propagation. Glia. 2016; 64(6):937–951. 10.1002/glia.22973 [DOI] [PubMed] [Google Scholar]
- 61.Shin HS, Snyderman R, Friedman E, Mellors A, Mayer MM. Chemotactic and anaphylatoxic fragment cleaved from the fifth component of guinea pig complement. Science. 1968; 162(3851):361–363. [DOI] [PubMed] [Google Scholar]
- 62.Schumacher WA, Fantone JC, Kunkel SE, Webb RC, Lucchesi BR. The anaphylatoxins C3a and C5a are vasodilators in the canine coronary vasculature in vitro and in vivo. Agents Actions. 1991; 34(3–4):345–349. [DOI] [PubMed] [Google Scholar]
- 63.Marceau F, Hugli TE. Effect of C3a and C5a anaphylatoxins on guinea-pig isolated blood vessels. J Pharmacol Exp Ther. 1984; 230(3):749–754. [PubMed] [Google Scholar]
- 64.Riedemann NC, Guo RF, Sarma VJ, Laudes IJ, Huber-Lang M, et al. Expression and function of the C5a receptor in rat alveolar epithelial cells. J Immunol. 2002; 168(4):1919–1925. [DOI] [PubMed] [Google Scholar]
- 65.Cavaillon JM, Fitting C, Haeffner-Cavaillon N. Recombinant C5a enhances interleukin 1 and tumor necrosis factor release by lipopolysaccharide-stimulated monocytes and macrophages. Eur J Immunol. 1990; 20(2):253–257. 10.1002/eji.1830200204 [DOI] [PubMed] [Google Scholar]
- 66.Jauneau AC, Ischenko A, Chan P, Fontaine M. Complement component anaphylatoxins upregulate chemokine expression by human astrocytes. FEBS Lett. 2003; 537(1–3):17–22. [DOI] [PubMed] [Google Scholar]
- 67.Monsinjon T, Gasque P, Chan P, Ischenko A, Brady JJ, et al. Regulation by complement C3a and C5a anaphylatoxins of cytokine production in human umbilical vein endothelial cells. FASEB J. 2003; 17(9):1003–1014. 10.1096/fj.02-0737com [DOI] [PubMed] [Google Scholar]
- 68.Woodruff TM, Ager RR, Tenner AJ, Noakes PG, Taylor SM. The role of the complement system and the activation fragment C5a in the central nervous system. Neuromolecular Med. 2010; 12(2):179–192. 10.1007/s12017-009-8085-y [DOI] [PubMed] [Google Scholar]
- 69.Fonseca MI, Ager RR, Chu SH, Yazan O, Sanderson SD, et al. Treatment with a C5aR antagonist decreases pathology and enhances behavioral performance in murine models of Alzheimer's disease. J Immunol. 2009; 183(2):1375–1383. 10.4049/jimmunol.0901005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ager RR, Fonseca MI, Chu SH, Sanderson SD, Taylor SM, et al. Microglial C5aR (CD88) expression correlates with amyloid-beta deposition in murine models of Alzheimer's disease. J Neurochem. 2010; 113(2):389–401. 10.1111/j.1471-4159.2010.06595.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hernandez MX, Jiang S, Cole TA, Chu SH, Fonseca MI, et al. Prevention of C5aR1 signaling delays microglial inflammatory polarization, favors clearance pathways and suppresses cognitive loss. Mol Neurodegener. 2017; 12(1):66 10.1186/s13024-017-0210-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huber-Lang MS, Younkin EM, Sarma JV, McGuire SR, Lu KT, et al. Complement-induced impairment of innate immunity during sepsis. J Immunol. 2002; 169(6):3223–3231. [DOI] [PubMed] [Google Scholar]
- 73.Guo RF, Riedemann NC, Bernacki KD, Sarma VJ, Laudes IJ, et al. Neutrophil C5a receptor and the outcome in a rat model of sepsis. FASEB J. 2003; 17(13):1889–1891. 10.1096/fj.03-0009fje [DOI] [PubMed] [Google Scholar]
- 74.Flierl MA, Schreiber H, Huber-Lang MS. The role of complement, C5a and its receptors in sepsis and multiorgan dysfunction syndrome. J Invest Surg. 2006; 19(4):255–265. 10.1080/08941930600778263 [DOI] [PubMed] [Google Scholar]
- 75.Hughes MM, Field RH, Perry VH, Murray CL, Cunningham C. Microglia in the degenerating brain are capable of phagocytosis of beads and of apoptotic cells, but do not efficiently remove PrPSc, even upon LPS stimulation. Glia. 2010; 58(16):2017–2030. 10.1002/glia.21070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seow V, Lim J, Iyer A, Suen JY, Ariffin JK, et al. Inflammatory responses induced by lipopolysaccharide are amplified in primary human monocytes but suppressed in macrophages by complement protein C5a. J Immunol. 2013; 191(8):4308–4316. 10.4049/jimmunol.1301355 [DOI] [PubMed] [Google Scholar]
- 77.Lehnardt S. Innate immunity and neuroinflammation in the CNS: the role of microglia in Toll-like receptor-mediated neuronal injury. Glia. 2010; 58(3):253–263. 10.1002/glia.20928 [DOI] [PubMed] [Google Scholar]
- 78.Amor S, Woodroofe MN. Innate and adaptive immune responses in neurodegeneration and repair. Immunology. 2014; 141(3):287–291. 10.1111/imm.12134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Amor S, Peferoen LA, Vogel DY, Breur M, van der Valk P, et al. Inflammation in neurodegenerative diseases—an update. Immunology. 2014; 142(2):151–166. 10.1111/imm.12233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Amor S, Puentes F, Baker D, van der Valk P. Inflammation in neurodegenerative diseases. Immunology. 2010; 129(2):154–169. 10.1111/j.1365-2567.2009.03225.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bradford BM, Mabbott NA. Prion disease and the innate immune system. Viruses. 2012; 4(12):3389–3419. 10.3390/v4123389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sakai K, Hasebe R, Takahashi Y, Song CH, Suzuki A, et al. Absence of CD14 delays progression of prion diseases accompanied by increased microglial activation. J Virol. 2013; 87(24):13433–13445. 10.1128/JVI.02072-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nuvolone M, Sorce S, Schwarz P, Aguzzi A. Prion pathogenesis in the absence of NLRP3/ASC inflammasomes. PLoS One. 2015; 10(2):e0117208 10.1371/journal.pone.0117208 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are in the paper and its Supporting Information files.