Abstract
Poly(ADP-ribose) polymerase-1 (PARP-1) is a regulatory enzyme involved in many different processes of DNA and RNA metabolism, including DNA repair. Previously, PARP-1 was found capable of forming a covalent DNA-protein crosslink (DPC) at the apurinic/apyrimidinic (AP) site in double-stranded DNA. The C1´ atom of the AP site participates in Schiff base formation with a lysine side chain in PARP-1, and a covalent bond is formed upon reduction of the Schiff base. The PARP-1 DPC is formed in vivo where DPC formation correlates with AP site induction by a monofunctional alkylating agent. Here, we examined repair of PARP-1 DPCs in mouse fibroblasts and found that a proteasome inhibitor, MG-132, reduces repair resulting in accumulation of PARP-1 DPCs and increased alkylating agent cytotoxicity. Using a model DNA substrate mimicking the PARP-1 DPC after proteasomal degradation, we found that repair is completed by a sub-pathway of base excision repair (BER). Tyrosyl-DNA phosphodiesterase 1 was proficient in removing the ring-open AP site sugar at the phosphodiester linkage, leaving an intermediate for processing by other BER enzymes. The results reveal proteasomal degradation of the PARP-1 DPC is active in mouse fibroblasts and that a model repair intermediate is processed by the BER machinery.
Keywords: PARP-1 DPCs, pol β, TOP1, Tdp1, PNKP, APE1, base excision repair
1. Introduction
Cellular DNA is constantly damaged by endogenous DNA-damaging agents and environmental physical and chemical agents, including UV light and ionizing radiation [1–4]. These physical and chemical challenges can alter DNA structure due to base loss, chemical modifications, inter/intra-strand crosslinks, single- and double-strand breaks, among other lesions. The apurinic/apyrimidinic (AP) site is considered to be one of the most predominant lesions in the genome [5]. It is estimated to arise at a frequency of >10,000 per mammalian cell per day [6] and accumulates at a surprisingly high steady-state level in mammalian tissues [7, 8]. The AP site lesion is repaired mainly by base excision repair (BER). Unrepaired and persistent AP sites can have adverse consequences and may pose a formidable challenge to genome stability by interfering with biological processes [3, 6, 9, 10].
PARP-1 is considered to be a “first responder” and molecular sensor of DNA lesions, especially those containing AP sites and strand breaks that are common intermediates in the BER pathway [11, 12]. Upon binding to these lesions, PARP-1 becomes activated for synthesis of poly(ADP-ribose) (PAR), and PARP-1 can PARylate itself as well as other proteins involved in DNA metabolism [13]. During AP site repair, PARP-1 binds the lesion, conducts PARylation, and promotes recruitment of the BER scaffold protein X-ray cross-complementing protein 1 (XRCC1), as well as other BER enzymes [14, 15]. Thus, PARP-1 plays a critical role in protection of cells against adverse consequences of DNA lesions. Under conditions where AP sites persist in the DNA, due either to an overwhelming amount of DNA damage or inhibition of PARP-1 activity by an inhibitor (e.g., 4-AN, Olaparib, Veliparib, and Talazoparib [16, 17], PARP-1 may form a DNA-protein crosslink (DPC) at the AP site that is potentially cytotoxic to the cell if not repaired [18].
DPCs are formed in a multitude of ways, including by exposure to environmental genotoxicants, anticancer therapy, reactions of endogenous metabolites and abortive enzymatic activity [19–23]. In mammalian cells, there are two major categories of DPC formation, termed enzymatic and non-enzymatic covalent crosslinking. In the case of enzymatic DPC formation, enzymatic reactions that require a covalent transient intermediate between the DNA substrate and the enzyme can stall under certain conditions, and stable covalent crosslinking of the enzyme to DNA can occur. Examples of enzymes that become cross-linked to DNA in this way include DNA topoisomerases, AP lyases, DNA glycosylases, DNA endonucleases, DNA methyltransferases, and DNA polymerases, among others [18, 24–28]. A well-studied example of the enzymatic type of DPC formation is seen with DNA Topoisomerase 1 (TOP1). This enzyme nicks and religates one strand of DNA and, during strand cutting, forms an intermediate by covalently attaching to the 3´-end of DNA at the nick, while the DNA on the other side of the nick is now free to rotate relieving torsional stress. After DNA relaxation, the single-strand break is religated by TOP1. However, the ligation step is sensitive to inhibition if there is a structural distortion in the DNA. Such distortion may occur due to a nearby DNA lesion, including an AP site or bulky DNA adduct. Distortion may also occur with binding of a TOP1 inhibitor, such as those used in chemotherapy, where the inhibitor intercalates within the interface of the TOP1-DNA complex. Inhibition of the ligation step can result in persistent covalent crosslinking of TOP1 to its DNA substrate [24, 29, 30]. Another example of enzymatic DPC formation is seen with reactions requiring a transient Schiff base intermediate between the C1′ atom of deoxyribose in the AP site of DNA and a primary amine group of an enzyme; examples include AP lyases, DNA glycosylases and DNA polymerases [18, 31–33]. In these reactions, DPCs are formed when the transient Schiff base intermediate is reduced by a reducing agent or the sugar moiety in the AP site is damaged [18, 32]. We have shown that PARP-1 has weak AP site lyase activity and forms a Schiff base intermediate. PARP-1 also has intrinsic redox (oxidation-reduction) capacity that can reduce the Schiff base, leading to the PARP-1 DPC at AP sites in genomic DNA [18]. In a related form of DPC formation, the AP site is attacked by a lysine side chain in a physically adjacent protein molecule forming the Schiff base, as is the case with histone H4 in the nucleosome core particle [31]. However, in non-enzymatic DPCs, agents such as ionizing radiation, UV light, transition metals, formaldehyde as well as various other aldehydes, reactive oxygen and nitrogen species, and DNA helical alterations can induce crosslinking of proteins to DNA. It is noteworthy that DPCs are produced also endogenously by normal physiological processes [34]. For example, formaldehyde is a byproduct of histone demethylation, amino acid and one-carbon metabolism, AlkB-type DNA repair, and lipid peroxidation [34–38]. Formaldehyde is a potent crosslinking agent that is widely used in research to trap transient protein-DNA interactions in chromatin immunoprecipitation assays [39].
Thus, repair of all forms of DPCs is critical for maintaining genome integrity and it is known that DPCs can be resolved by BER, nucleotide excision repair, and by replication coupled proteolytic degradation [32, 40–42]. Recently, a specific type of protease-mediated DPC repair was reported in yeast and a homologue of this protease was found in higher eukaryotes [43–48]. However, there are no reports of PARP-1 DPC repair. Here, we provide evidence that the PARP-1 DPC is repaired by a proteasome-mediated BER sub-pathway in mammalian cells.
2. Materials and methods
2.1. Cell culture and cytotoxicity measurements
PARP-1+/+ and PARP-1−/− spontaneously immortalized mouse embryonic fibroblasts (MEFs) were obtained from Dr. Josianne Ménissier-de Murcia (CNRS, Illikirch-Graffenstaden, France). These cells were cultured at 37 °C in a 10% CO2 incubator in Dulbecco’s modified Eagle’s medium containing L-glutamine and 10% FBS. Cells were routinely tested and found to be free of mycoplasma contamination. Cytotoxicity of methyl methanesulfonate (MMS) and the effect of MG-132 was determined by growth inhibition assays as described previously [49]. PARP-1+/+ and PARP-1−/− cells were seeded in 6-well dishes at a density of 4 × 104 cells/well. The following day they were treated for one hour with a range of concentrations of MMS in the absence or presence of 300 nM MG-132. After washing, cells were further incubated as appropriate with MG-132 for a further 23 h. After washing again, cells were incubated until 80% confluence of untreated wells, then nuclei were harvested (triplicate wells for each drug concentration) following cell lysis using hypotonic solution and detergent [50]. Results were expressed as percentage of control cells without any treatment.
2.2. Isolation and quantification of PARP-1 DPCs by RADAR (rapid approach to DNA adduct recovery) assay
To study PARP-1 DPCs, total genomic DNA was isolated using a modification of the RADAR assay protocol, as previously described [25]. Briefly, PARP-1+/+ or PARP-1−/− MEFs (1 × 106 per well) were treated with MMS (3 mM for 1 h) in the presence or absence of 4-AN or MG-132 (10 μM for a total of 2 h). Cells were washed and lysed with 1 ml of a mixture of DNAzol (Invitrogen) plus 1% Sarkosyl (Sigma-Aldrich). For each type of cell treatment, genomic DNA was isolated from 3 wells. The lysate (1 ml) from one well was transferred to an Eppendorf tube, and the genomic DNA was precipitated by addition of 0.5 ml 100% ethanol, followed by 30 min incubation at −30 °C and centrifugation at 12,000 rpm for 15 min at 4 °C. The DNA pellets were washed three times with ice-cold 75 % ethanol and collected by centrifugation. The DNA pellet was immediately resuspended in 150 μl of freshly prepared 8 mM NaOH and the liquid from three wells was combined for each sample. All DNA samples were gently rocked overnight at 4 °C for complete solubilization. The next day, DNA samples were treated with RNase A (10 μg/ml) for 30 min at 37 °C followed by sonication (20 s at 7 Watts) (Fisher Scientific Series 60 Sonic Dismembrator Model F60). Samples then were centrifuged for 15 min at 12,000 rpm and the supernatant was removed. The DNA concentration of each sample was quantified by measuring florescence of DNA-bound PicoGreen dye (Life Technology), as recommended by the manufacturer’s instructions.
2.3. Analysis of PARP-1 DPC by slot blot
For immunoblotting, samples of genomic DNA were diluted in 200 μl 1X TBE and applied to a nitrocellulose membrane (Bio-Rad) using a vacuum slot-blot manifold (Bio-Rad). To reduce sample viscosity and to avoid membrane clogging, aliquots of genomic DNA were treated with 20 units of benzonase nuclease (Sigma-Aldrich) for 30 min at 37 °C prior to loading onto nitrocellulose membrane. The membrane was then blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.1% (v/v) Tween 20 (TBS-T) and then probed either with anti-PARP-1 (BD Pharmingen), anti-HA antibody (Cell Signaling Technology) or with anti-ubiquitin (Abcam) antibody, as indicated in the figure legends. Goat anti-mouse or goat anti-rabbit IgG conjugated to horseradish peroxidase (Bio-Rad) was used as secondary antibody, and immobilized horseradish peroxidase activity was detected by enhanced chemiluminescence (Thermo Fisher Scientific).
2.4. Repair of DPCs in vitro using a model DNA substrate and purified BER enzymes
A 15-mer DNA oligonucleotide containing 5´-FAM and 3´-C7-NH2 was annealed with a 5´-phosphorylated 18-mer down-stream oligonucleotide and a 34-mer complementary template strand. Thus, the model substrate mimics DNA with the 3´-deoxyribose moiety linked to the remnant short peptide after proteosome activity on the PARP-1 DPC. The reaction mixture (10 μl) containing 50 mM Tris-HCl, pH 7.5, 20 mM KCl, 0.5 mM EDTA, 2 mM DTT, and 250 nM duplex DNA was assembled on ice, as indicated in the figure legends. As specified, reactions were initiated by the addition of enzymes: tyrosyl-DNA phosphodiesterase 1 (Tdp1) Polynucleotide kinase 3´-phosphatase (PNKP) (100 nM), AP endonuclease 1 (APE1) (500 nM), or DNA polymerase β (pol β) (25 nM). In the Tdp1 case as specified in the legends, the reaction mixture contained 5.5 mM EDTA; in the cases of PNKP and APE1, the reaction mixture was supplemented with 5 mM MgCl2; and in the case of pol β, the reaction mixture also contained 5 mM MgCl2 and 25 μM dCTP. Incubation was for 20 min at 37 °C.
In separate experiments, removal of the 3´-C7-NH2 group was examined. The reaction conditions were as described above. In this case, the reaction mixture was incubated with increasing concentrations of Tdp1 (25, 50, 100, 200 and 500 nM) or APE1 (500 nM) for 20 min at 37 °C, as indicated in the figure legend. All protein dilutions were made in a dilution buffer containing 50 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 2 mM DTT, 150 mM NaCl, 0.1 mg/bovine serum albumin and 20% glycerol. The reaction was terminated by addition of an equal volume of DNA gel loading buffer (95% formamide, 20 mM EDTA, 0.02% bromophenol blue, and 0.02% xylene cyanol). After incubation at 75 °C for 2 min, the reaction products were separated by electrophoresis in a 16% polyacrylamide gel containing 8 M urea in 89 mM Tris-HCl, pH 8.8, 89 mM boric acid, and 2 mM EDTA. A Typhoon PhosphorImager was used for gel scanning and imaging, and the data were analyzed by ImageQuant software.
2.5. Establishment of a stable cell line overexpressing HA-tagged ubiquitin
The HA-tag containing a full-length cDNA of human ubiquitin was amplified from vector “HA-ubiquitin” (Addgene plasmid ID: 18712) using primer pairs UBB-F (5´-AAAATCTAGAGCCACCATGTACCCATACGATG-3´, XbaI site is indicated in italics) and UBB-R (5´-CCGTGAATTCCTAATAACCACCTCTCAGACG-3´, EcoRI site is indicated in italics). Then, the product was gel-purified and ligated into the XbaI/EcoRI sites of vector pCDH-CMV-MCS-EF1-puro (System Biosciences, Mountain View, CA) to obtain the lentivirus plasmid pCDH-HA-ubiquitin. The lentivirus particles were prepared and packaged as described previously [51]. Briefly, 293T cells were co-transfected with pCDH-HA-ubiquitin, pMD2.G (Addgene plasmid ID: 12259) and psPAX2 (Addgene plasmid ID: 12260) using lipofectamine 2000. Two days later, the lentivirus supernatant was collected and used for the infection of PARP-1+/+ cells at MOI=1. Twenty-four hours after infection, cells were grown in selection medium containing 2 μg/ml puromycin for 7 days to eliminate untransformed cells. The puromycin resistant cells were then diluted and separated into a 96-well plate with only one cell in each well. After selection for an additional 4 weeks, western blotting was used for detection of the ubiquitin expression level in the surviving clones, as follows: 25 μg extract protein was loaded and separated by a 15% Criterion Tris-HCl Gel (Bio-Rad), and the samples were then transferred onto a nitrocellulose membrane (Life Technologies). The membrane was immunoblotted with mouse monoclonal anti-HA antibody (Cell Signaling Technology) at a dilution of 1:1000, and the immunoblot was processed as above.
3. RESULTS
3.1. PARP-1 DPC formation in mouse fibroblast genomic DNA
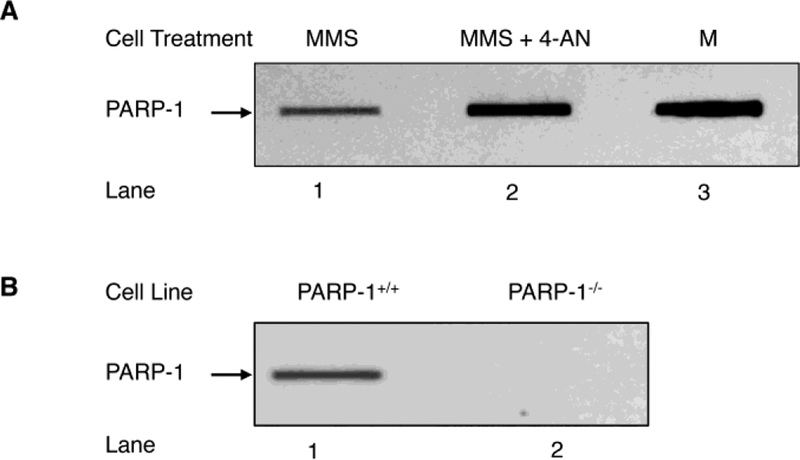
It is known that MEFs exhibit strong cytotoxicity when treated with a monofunctional DNA methylating agent in combination with a PARP inhibitor [52–54]. The DNA lesions produced by these methylating agents are removed by DNA glycosylases leaving AP sites in double stranded DNA that are repaired by BER. The AP site containing repair intermediate is recognized by PARP-1 [55], and the PARylation required for efficient BER is induced by APE1 strand incision. However, in the presence of a PARP inhibitor, DNA lesion repair is blocked and higher levels of PARP-1 DPCs are found in genomic DNA [18, 56]. To further examine PARP-1 DPCs in the current work, we adopted an alternate quantification procedure termed “rapid approach to DNA adduct recovery” (RADAR) [25] and applied it to evaluate PARP-1 DPCs in wild-type and PARP-1 null MEF cells (Fig. 1). Cells were treated with MMS or MMS plus the PARP-1 inhibitor 4-AN, and DNA samples containing PARP-1 DPCs were isolated; the genomic DNA concentration was assessed as described (Fig. S1). Quantification of these PARP-1 DPCs revealed they were more abundant in cells treated with the combination of MMS plus 4-AN (Fig. 1A) than with MMS alone. PARP-1 inhibitor alone modestly increased PARP-1 DPCs in wild-type MEF cells that were “untreated cells,” but the effect was slightly more visible in cells with a BER gene deficiency (i.e., in the presence of a PARP-1 inhibitor, Fig. S2). Finally, in control experiments, PARP-1 DPCs were not found in PARP-1 null cells, as expected (Fig. 1B).
Fig. 1.
Detection of PARP-1 DPCs in genomic DNA by the RADAR method. (A) PARP-1 DPCs are formed as a function of AP sites in the genomic DNA. PARP-1+/+ MEFs were treated with MMS (lane 1) or MMS plus 4-AN (lane 2), as described under “Materials and Methods”. An equal amount of the genomic DNA (5 μg) isolated from PARP-1+/+ cells treated with MMS or with MMS plus 4-AN was applied into slot blot wells, as indicated. The blot was probed with anti-PARP-1 antibody. Lane 3, represents a sample of purified PARP-1 used as a positive control (M). (B) Requirement of PARP-1 expression for detection of PARP-1 DPC formation in cells. Genomic DNA (5 μg) from MMS treated PARP-1+/+ (lane 1) or PARP-1−/− (lane 2) MEFs was used for slot blot analysis as in (A). PARP-1−/− cells failed to show PARP-1 DPCs (lane 2), indicating PARP-1 DPC is specific to expression of PARP-1. The results shown are representative of at least three independent experiments.
3.2. Stabilization of PARP-1 DPCs by the proteasome inhibitor MG-132 is associated with increased MMS sensitivity in MEF cells
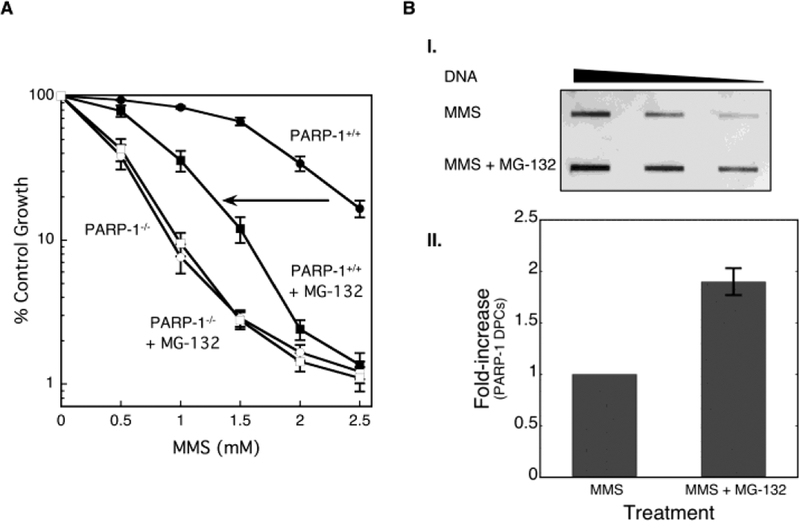
As illustrated in studies of TOP1 DPCs, repair occurs by a proteasome-dependent pathway in which the covalently crosslinked enzyme is ubiquitinated and then degraded by the proteasome [40, 57, 58]. To address whether proteasome-dependent degradation of PARP-1 DPCs is involved in their repair and also in MMS-induced cytotoxicity, the sensitivity to MMS was examined in combination with a proteasome inhibitor (MG-132) in PARP-1+/+ and PARP-1−/− MEF cells. The results shown in Figure 2A reveal that PARP-1+/+ cells treated with MMS in combination with MG-132 are more sensitive than cells treated with MMS alone (Fig. 2A). In contrast, MG-132 treatment failed to exert an effect on MMS sensitivity in PARP-1−/− cells. These results were consistent with the hypothesis that PARP-1 DPCs induce cytotoxicity. Thus, an increased cell-killing effect is observed in PARP-1+/+ cells when the proteasome-mediated DPC repair pathway is inhibited by MG-132; while a similar effect of MG-132 is not observed in the absence of PARP-1 expression. Next, we confirmed the presence of a higher level of PARP-1 DPCs in cells treated with MMS plus MG-132 (Figs. 2B and S3), but not in cells treated with MMS alone. Repair of PARP-1 DPCs in MMS treated MEF cells was evident, whereas the DPCs remained stable in the presence of the proteasome inhibitor (Fig. S3).
Fig. 2.
Stabilization of PARP-1 DPCs by the proteasome inhibitor MG-132. (A) Effect of the proteasome inhibitor MG-132 on cellular sensitivity to MMS. PARP-1+/+ and PARP-1−/− MEFs were treated with MMS for 1 h in the presence or absence of MG-132 for 24 h. Cytotoxicity was determined by growth inhibition assays as outlined under “Materials and Methods”. The MG-132-mediated sensitization in PARP-1+/+ MEFs is indicated by the arrow. (B, I.) Stabilization of PARP-1-DPCs by the proteasome inhibitor MG-132. PARP-1+/+ MEFs were treated with MMS or MMS plus MG-132, as described under “Materials and Methods”. Genomic DNA was prepared and analyzed, as described. Two-fold dilutions (high to low) of genomic DNA were applied into slot blot wells and probed with anti-PARP-1 antibody. (B, II.) Quantification of PARP-1 DPC formation as a function of the proteasome inhibitor MG-132. Fold-increase in PARP-1 DPCs in the presence of MG-132 is plotted; PARP-1 DPCs in MMS treated cells were set to a value of 1.0. The data are the average from four independent experiments, the error bar represents mean ± SD.
3.3. The PARP-1 DPC is ubiquitinated
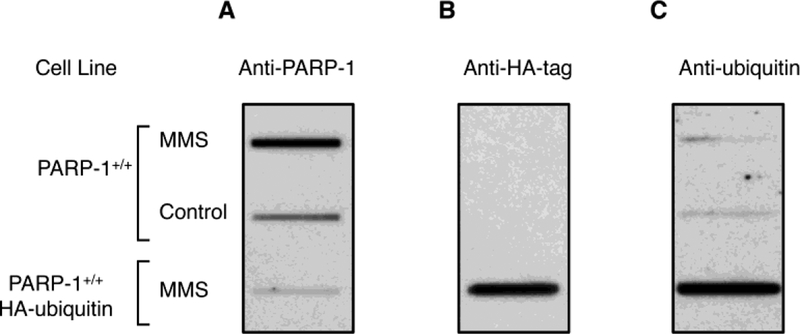
Proteasome-mediated repair requires that DPCs become ubiquitinated prior to initiation of proteasome proteolysis [40, 57]. To address the question of whether or not PARP-1 DPCs are subject to ubiquitination, stable cell lines were established with HA-ubiquitin expression in the wild-type (PARP-1+/+) MEF cell background. Expression of PARP-1 was lower in these cells than in control cells (Fig. S4A). The same immunoblot was stripped and then probed with anti-ubiquitin antibody. The antibody detected monoubiquitinated PARP-1 and higher molecular mass proteins representing polyubiquitinated PARP-1 in the extract from cells expressing HA-ubiquitin, but not in the extract from control cells (Fig. S4B). Therefore, MEF cells expressing HA-ubiquitin were employed to enhance the sensitivity of measurement of ubiquitinated PARP-1 DPCs. Cells with or without HA-ubiquitin were treated with MMS and followed by genomic DNA isolation. PARP-1 DPCs were detected by immunoblotting with anti-PARP-1, anti-HA or anti-ubiquitin antibody. As expected, a higher level of PARP-1 DPCs was observed in the MMS treated wild-type (PARP-1+/+) cells than in the untreated wild-type cells, and a relatively lower level of ubiquitinated DPCs was observed in both MMS treated and untreated cells. Interestingly, a much lower level of PARP-1 DPCs was observed in the HA-ubiquitin expressing cells (Fig. 3A), suggesting that expression of HA-ubiquitin promoted repair of the PARP-1 DPCs. Ubiquitination of the residual PARP-1 DPCs was confirmed by immunoblotting the same membrane with anti-HA antibody (Fig. 3B), and reactivity of the anti-HA antibody was not observed in the control cells, i.e., PARP-1+/+ cells (Fig. 3B). Ubiquitination of the residual DPCs also was confirmed by probing with anti-ubiquitin antibody (Fig. 3C). These results provide evidence for PARP-1 DPC ubiquitination in vivo. Repair of PARP-1 DPCs was more evident in cells expressing HA-ubiquitin than in MMS treated cells alone (Fig. S5).
Fig. 3.
Evidence of ubiquitination of PARP-1 DPCs. Genomic DNA was isolated from PARP-1+/+ cells or a stable PARP-1+/+ cell line expressing HA-ubiquitin. Both cell lines were treated with MMS, as described in “Materials and Methods,” and genomic DNA was isolated after 2 h. Representative results from two independent experiments are shown. For each sample, 5 μg genomic DNA was applied into slot blot wells. Control represents cells without MMS. The immunoblot was probed either with anti-PARP-1 (A), anti-HA (B), or anti-ubiquitin (C) antibody. Strong reactivity to HA-tag (B) or ubiquitin (C) was observed in the cells expressing HA-ubiquitin, whereas no reactivity to the anti-HA antibody was observed in cells that were not transfected with HA-ubiquitin. A relatively low level of PARP-1 DPCs was found in the cell line expressing HA-ubiquitin (A). In panel C, a relatively low level of ubiquitination of the PARP-1 DPCs was found in the PARP-1+/+ cell line.
3.4. Repair of the PARP-1 DPC in vitro
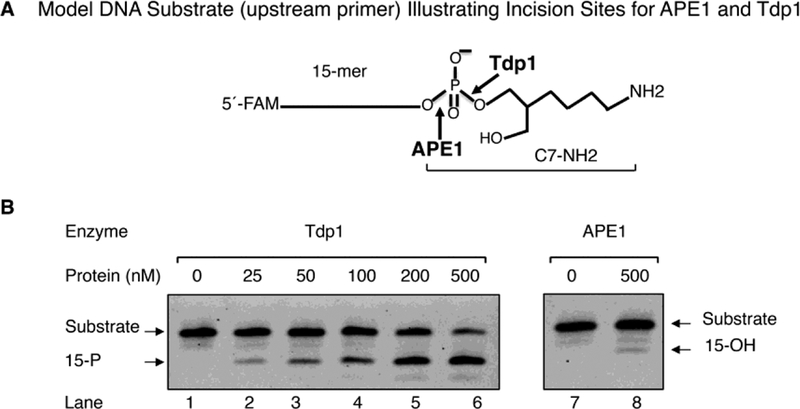
To examine repair of PARP-1 DPCs in vitro, we used a model DNA substrate that has features similar to the proteasomal proteolytic product of the PARP-1 DPC (Figs. 4, S7 and S8A). The double-stranded DNA model substrate had a one nucleotide gap; the O3´ atom in the gap contained a phosphate group linked to ring-opened deoxyribose and a short carbon chain attached to the reduced Schiff base NH2 group (i.e., termed C7-NH2). Potential incision sites for APE1 and Tdp1 in the model substrate are illustrated (Figs. 4A, and S8C). This model substrate was first used in a comparative assessment of purified Tdp1 and APE1 (Fig. 4). The reaction mixtures contained either increasing concentrations of Tdp1 or a large excess of APE1 (500 nM). The results revealed that Tdp1 cleaved the substrate in an enzyme concentration-dependent fashion (Fig. 4B, lanes 2 to 6); in contrast, APE1 showed negligible activity (Fig. 4B, lanes 7–8), in spite of the presence of a potential APE1 incision site.
Fig. 4.
Comparison of Tdp1 and APE1 activities on the model DNA substrate. (A) Schematic of the model DNA substrate. This substrate mimics post-proteasomal treatment of a PARP-1 DPC. The 15-mer strand DNA has FAM at the 5´-end. (B) Representative phosphorimage of incision activities of Tdp1 (lanes, 1–6) and APE1 (lanes, 7–8). The incision reactions were performed as described under “Materials and Methods.” Reactions were initiated by the addition of different concentrations of Tdp1 (25 to 500 nM, lanes 2–6) or APE1 (500 nM, lane 8). Incubation was 20 min at 37 °C. Representative results from three independent experiments are shown. The migration positions of the substrate and the products with 3´-PO4 (15-P) and 3´-OH (15-OH), respectively, are indicated.
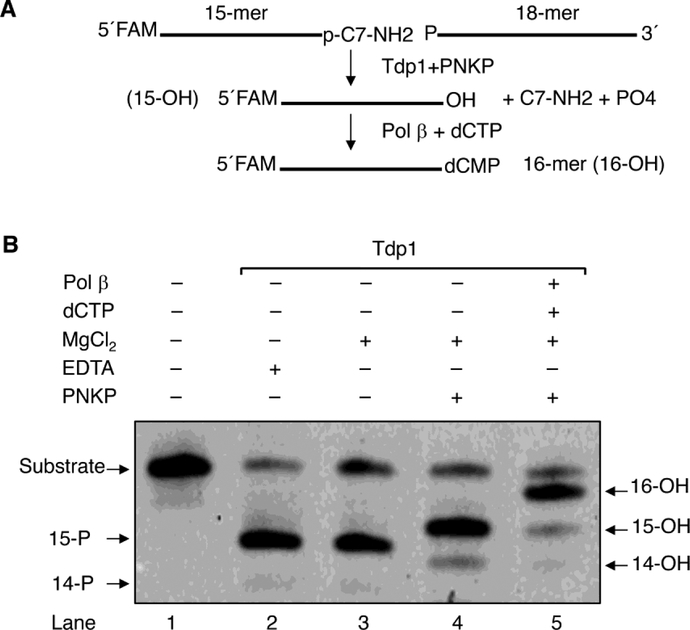
Next, we examined activities on the model substrate with two additional purified BER enzymes, PNKP and pol β (Fig. 5). Tdp1, as expected, did not require Mg+2 for activity and produced the O3´-PO4-containing 15-mer DNA product upon incision releasing C7-NH2 (Fig. 5, lanes 2 and 3). Addition of PNKP to the reaction mixture, along with MgCl2, resulted in removal of the O3´-PO4 group, generating the 15-mer DNA with O3´, the substrate for gap filling DNA synthesis (Fig. 5, lane 4). Finally, with addition of pol β and dCTP, incorporation of dCMP occurred producing the 16-mer product (Fig. 5, lane 5).
Fig. 5.
Repair of PARP-1 DPC by purified BER enzymes. (A) The repair scheme of a model DNA substrate that mimics the remnant of post-proteasomal degradation of the PARP-1 DPC at the 3´-end in a one-nucleotide gap opposite template G. For clarity, only the top strand of the DNA is shown. The O3´-end trimming and gap filling reactions are illustrated. (B) Representative phosphorimage of repair of the model substrate by BER enzymes. The repair reactions were performed as described under “Materials and Methods.” Incubation was 20 min at 37 °C. The migration positions of the substrate and the products with 3´-PO4 (14-P), 3´-PO4 (15-P), or 3´-OH (14, 15, and 16-OH) are indicated. The reaction products as illustrated are shown in lanes 1–5. Typical results from three independent experiments are shown.
In light of the results described in Figures 4 and 5, we examined whether the PARP-1 DPC could be repaired by Tdp1 or APE1 without prior protease action and whether the DPC after partial digestion with trypsin could be repaired by Tdp1 or APE1. To this end, we isolated PARP-1 DPCs formed in vitro using 5´−32P-labeled 34 bp DNA containing an AP site along with purified PARP-1 (Fig. S6A); formation of the PARP-1 DPC and its expected sensitivity to proteinase K digestion were confirmed by SDS-PAGE analysis (Fig. S6B, lanes 1 and 2). The PARP-1 DPC was subjected to incubation with Tdp1 or APE1, but both enzymes failed to act on the intact PARP-1 DPC (not shown). Next, we digested the PARP-1 DPC with trypsin and then added Tdp1 or APE1 to the reaction mixture before extending the incubation. Under these conditions, the PARP-1 DPC was partially digested by trypsin (Fig S6B, lanes 3–5) generating mainly a 27 kDa XL-tryptic peptide. However, the incubation with both Tdp1 and APE1 failed to produce any change in the gel profile (Fig. S6B, lanes 4–5). From these results, we concluded that the tryptic peptide attached to the DNA was large enough to cause hinderance for Tdp1 or APE1 incision of the phosphodiester bond at the 3´-end of DNA (Figs. 4A, and S8). Therefore, we chose a model DNA substrate to investigate repair of DPCs, as described in Figures 4 and 5.
4. Discussion
In this study, we found that the bulky PARP-1 DNA-protein crosslink, arising by reduction of the Schiff base lyase reaction intermediate, can be repaired by proteasomal degradation coupled with Tdp1-mediated BER. This repair pathway appears similar to that established for the TOP1 DNA-protein crosslink, except for the important difference that the DNA 3´-end linkage is to the AP site sugar phosphate and lysine primary amine in the case of PARP-1, instead of a tyrosine hydroxyl in the case of TOP1. Interestingly, inhibition of proteasomal degradation and accumulation of the PARP-1 DPC correlates with increased cell death after MMS treatment. DNA-protein crosslinks are bulky lesions that occur naturally and upon exposure to various stressors including environmental toxicants, endogenous metabolites such as formaldehyde, and chemotherapeutic agents such as camptothecin. DPCs can impose steric hindrance to DNA transactions such as replication, transcription, and repair, and DPC removal is important in preserving genome stability. Production and repair of DPCs may be robust events in cells, and work is underway to appreciate their biological significance. PARP-1 is considered to be a “first responder” for several types of DNA lesions, especially the single strand break and AP site [12, 14, 15, 55, 59, 60]. Endogenously formed AP sites are common in mammalian cells. Therefore, it is not surprising that AP sites are a source of a fraction of the DPCs in the cell, and this includes histone H4 DPCs formed in nucleosomes [61]. Upon binding to the AP site, PARP-1 becomes activated for PAR synthesis after APE1 strand incision [55]. During this process, PARP-1 can become covalently adducted to the AP site forming a DPC in vitro as well as in vivo [18]. The level of PARP-1 DPCs increases when the BER process is interrupted, either due to a deficiency in a repair gene product or when cells are challenged with an alkylating agent, either alone or in combination with a PARP-1 inhibitor [18, 56].
The aim of the present study was to investigate how PARP-1 DPCs are repaired. First, we confirmed PARP-1 DPC formation in vivo using an enhanced method for their isolation termed RADAR. The results revealed PARP-1 DPCs are generated as a function of BER intermediate accumulation in genomic DNA, and in control experiments this was found to be dependent on PARP-1 expression, as expected (Fig. 1). In order to identify a repair pathway for PARP-1 DPC removal, we assessed involvement of proteasome-mediated degradation of DPCs, since this pathway had been implicated in TOP1 DPC repair and is well known for repair of other types of DPCs [57, 58, 62–64]. After proteasome-mediated degradation of a DPC, a small peptide may remain covalently adducted to DNA. For repair of the TOP1 DPC, this intermediate is processed by Tdp1 that hydrolyzes the phosphodiester bond between the remnant TOP1 peptide and DNA [65]. Our results strongly imply that a proteasome-mediated degradation pathway is involved in PARP-1 DCP removal, since cells treated with the proteasome inhibitor MG-132 accumulated more DPCs, compared with untreated cells (Fig. 2).
The proteasome inhibitor MG-132 has been widely used to reveal a proteasome-mediated repair pathway for the DPC lesion [57, 58]. In fact, the results with proteasome inhibitor MG-132 are in line with our experimental observations. Involvement of a proteasome-mediated pathway for PARP-1 DPC repair was further corroborated in experiments where HA-tagged-ubiquitin was expressed in PARP-1+/+ cells; ubiquitination is required when DPCs are processed by the proteasome-mediated degradation pathway. In HA-tagged-ubiquitin expressing cells, the level of PARP-1 DPCs was diminished compared with that in control cells (Fig. 3). This suggested that highly ubiquitinated PARP-1 DPCs are repaired at a faster rate than in control cells. In addition, wild-type MEFs were more sensitive to MMS in the presence of MG-132, whereas cells lacking PARP-1 showed no effect of the proteasome inhibitor. A likely trigger for this MG-132 mediated potentiation of cytotoxicity appears to be the inhibition of PARP-1 DPC repair, and this interpretation is consistent the higher level of DPCs detection in cells incubated with MG-132 [62].
In assessing PARP-1 DPC repair in vitro, we were unable to isolate sufficient PARP-1 DPCs from MEFs to test the hypothesis that DPCs are repaired by BER, in partnership with proteasomal degradation. To overcome this limitation, we chose to use a model DNA substrate that has features similar to the proteasomal proteolytic product of the PARP-1 DPC (Fig. S7). The model oligonucleotide substrate has potential incision sites for APE1 and Tdp1; the substrate contains a 3´-end ring-opened deoxyribose and three extra carbon groups linked to an NH2 group at the end (Fig. S8). APE1 failed to incise this substrate, whereas Tdp1 was active on the substrate leaving a phosphate group at the O3´ atom. The phosphate group was then removed by PNKP (Fig. 5). Based on these results, the active site of Tdp1 can accommodate a ring-opened sugar phosphate group, and this was not surprising since Tdp1 is known to handle a broad range of DNA substrates [66]. Taken together, our results shed new light on MG-132 proteasome inhibition and stabilization of the PARP-1 DPC in cells, showing that a proteasome-mediated BER pathway operates to eliminate PARP-1 DPCs.
Supplementary Material
Highlights.
PARP-1 DPCs are formed as a results of AP sites accumulation in genomic DNA.
Inhibition of proteasome-mediated DPC repair pathway by MG-132 results in cell cytotoxicity.
PARP-1 DPCs are repaired primarily by APE1-independent BER pathway.
Acknowledgements
We thank Yesenia Rodriguez and Cristina Nadalutti for critical reading of the manuscript.
Funding
This work was supported in part by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences, Project Numbers Z01ES050158 and Z01ES050159.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest:
The authors declare that there are no conflicts of interest.
References
- [1].Lindahl T, DNA repair enzymes, Annu. Rev. Biochem, 51 (1982) 61–87. [DOI] [PubMed] [Google Scholar]
- [2].Lindahl T, Wood RD, Quality control by DNA repair, Science, 286 (1999) 1897–1905. [DOI] [PubMed] [Google Scholar]
- [3].Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE, Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage, Proc. Natl. Acad. Sci, 96 (1999) 13300–13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Slupphaug G, Kavli B, Krokan HE, The interacting pathways for prevention and repair of oxidative DNA damage, Mutation research, 531 (2003) 231–251. [DOI] [PubMed] [Google Scholar]
- [5].Nakamura J, Mutlu E, Sharma V, Collins L, Bodnar W, Yu R, Lai Y, Moeller B, Lu K, Swenberg J, The endogenous exposome, DNA Repair (Amst), 19 (2014) 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lindahl T, Instability and decay of the primary structure of DNA, Nature, 362 (1993) 709–715. [DOI] [PubMed] [Google Scholar]
- [7].Chastain PD 2nd, Nakamura J, Rao S, Chu H, Ibrahim JG, Swenberg JA, Kaufman DG, Abasic sites preferentially form at regions undergoing DNA replication, FASEB J, 24 (2010) 3674–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nakamura J, Swenberg JA, Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues, Cancer Res, 59 (1999) 2522–2526. [PubMed] [Google Scholar]
- [9].Loeb LA, Apurinic sites as mutagenic intermediates, Cell, 40 (1985) 483–484. [DOI] [PubMed] [Google Scholar]
- [10].Barnes DE, Lindahl T, Sedgwick B, DNA repair, Curr. Opin. Cell Biol, 5 (1993) 424–433. [DOI] [PubMed] [Google Scholar]
- [11].Hassa PO, Hottiger MO, The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases, Front. Biosci, 13 (2008) 3046–3082. [DOI] [PubMed] [Google Scholar]
- [12].Schreiber V, Dantzer F, Ame JC, de Murcia G, Poly(ADP-ribose): novel functions for an old molecule, Nat. Rev. Mol. Cell Biol, 7 (2006) 517–528. [DOI] [PubMed] [Google Scholar]
- [13].D’Amours D, Desnoyers S, D’Silva I, Poirier GG, Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions, Biochem. J, 342 (Pt 2) (1999) 249–268. [PMC free article] [PubMed] [Google Scholar]
- [14].Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A, In situ analysis of repair processes for oxidative DNA damage in mammalian cells, Proc. Natl. Acad. Sci. U. S. A, 101 (2004) 13738–13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gassman NR, Wilson SH, Micro-irradiation tools to visualize base excision repair and single-strand break repair, DNA Repair (Amst), 31 (2015) 52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ganguly B, Dolfi SC, Rodriguez-Rodriguez L, Ganesan S, Hirshfield KM, Role of Biomarkers in the Development of PARP Inhibitors, Biomark Cancer, 8 (2016) 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG, PARP inhibition: PARP1 and beyond, Nat. Rev. Cancer, 10 (2010) 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Prasad R, Horton JK, Chastain PD 2nd, Gassman NR, Freudenthal BD, Hou EW, Wilson SH, Suicidal cross-linking of PARP-1 to AP site intermediates in cells undergoing base excision repair, Nucleic Acids Res, 42 (2014) 6337–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Henner WD, Grunberg SM, Haseltine WA, Enzyme action at 3’ termini of ionizing radiation-induced DNA strand breaks, J. Biol. Chem, 258 (1983) 15198–15205. [PubMed] [Google Scholar]
- [20].Henner WD, Rodriguez LO, Hecht SM, Haseltine WA, gamma Ray induced deoxyribonucleic acid strand breaks. 3’ Glycolate termini, J. Biol. Chem, 258 (1983) 711–713. [PubMed] [Google Scholar]
- [21].Datta K, Purkayastha S, Neumann RD, Pastwa E, Winters TA, Base damage immediately upstream from double-strand break ends is a more severe impediment to nonhomologous end joining than blocked 3’-termini, Radiat. Res, 175 (2011) 97–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Speit G, Schutz P, Merk O, Induction and repair of formaldehyde-induced DNA-protein crosslinks in repair-deficient human cell lines, Mutagenesis, 15 (2000) 85–90. [DOI] [PubMed] [Google Scholar]
- [23].Lai Y, Yu R, Hartwell HJ, Moeller BC, Bodnar WM, Swenberg JA, Measurement of Endogenous versus Exogenous Formaldehyde-Induced DNA-Protein Crosslinks in Animal Tissues by Stable Isotope Labeling and Ultrasensitive Mass Spectrometry, Cancer Res, 76 (2016) 2652–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hsiang YH, Hertzberg R, Hecht S, Liu LF, Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I, J. Biol. Chem, 260 (1985) 14873–14878. [PubMed] [Google Scholar]
- [25].Kiianitsa K, Maizels N, A rapid and sensitive assay for DNA-protein covalent complexes in living cells, Nucleic Acids Res, 41 (2013) e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].McCullough AK, Dodson ML, Lloyd RS, Initiation of base excision repair: glycosylase mechanisms and structures, Annu. Rev. Biochem, 68 (1999) 255–285. [DOI] [PubMed] [Google Scholar]
- [27].Piersen CE, Prasad R, Wilson SH, Lloyd RS, Evidence for an imino intermediate in the DNA polymerase beta deoxyribose phosphate excision reaction, J. Biol. Chem, 271 (1996) 17811–17815. [DOI] [PubMed] [Google Scholar]
- [28].Prasad R, Liu Y, Deterding LJ, Poltoratsky VP, Kedar PS, Horton JK, Kanno S, Asagoshi K, Hou EW, Khodyreva SN, Lavrik OI, Tomer KB, Yasui A, Wilson SH, HMGB1 is a cofactor in mammalian base excision repair, Mol. Cell, 27 (2007) 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hsiang YH, Liu LF, Identification of mammalian DNA topoisomerase I as an intracellular target of the anticancer drug camptothecin, Cancer Res, 48 (1988) 1722–1726. [PubMed] [Google Scholar]
- [30].Pourquier P, Pommier Y, Topoisomerase I-mediated DNA damage, Adv. Cancer Res, 80 (2001) 189–216. [DOI] [PubMed] [Google Scholar]
- [31].Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM, Rapid DNA-protein cross-linking and strand scission by an abasic site in a nucleosome core particle, Proc. Natl. Acad. Sci, 107 (2010) 22475–22480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Quinones JL, Demple B, When DNA repair goes wrong: BER-generated DNA-protein crosslinks to oxidative lesions, DNA Repair (Amst), 44 (2016) 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Quinones JL, Thapar U, Yu K, Fang Q, Sobol RW, Demple B, Enzyme mechanism-based, oxidative DNA-protein cross-links formed with DNA polymerase beta in vivo, Proc. Natl. Acad. Sci, 112 (2015) 8602–8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Swenberg JA, Lu K, Moeller BC, Gao L, Upton PB, Nakamura J, Starr TB, Endogenous versus exogenous DNA adducts: their role in carcinogenesis, epidemiology, and risk assessment, Toxicol. Sci, 120 Suppl 1 (2011) S130–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kooistra SM, Helin K, Molecular mechanisms and potential functions of histone demethylases, Nat. Rev. Mol. Cell Biol, 13 (2012) 297–311. [DOI] [PubMed] [Google Scholar]
- [36].Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B, Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage, Nature, 419 (2002) 174–178. [DOI] [PubMed] [Google Scholar]
- [37].Lu K, Gul H, Upton PB, Moeller BC, Swenberg JA, Formation of hydroxymethyl DNA adducts in rats orally exposed to stable isotope labeled methanol, Toxicol. Sci, 126 (2012) 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].O’Brien PJ, Siraki AG, Shangari N, Aldehyde sources, metabolism, molecular toxicity mechanisms, and possible effects on human health, Crit. Rev. Toxicol, 35 (2005) 609–662. [DOI] [PubMed] [Google Scholar]
- [39].Jackson V, Studies on histone organization in the nucleosome using formaldehyde as a reversible cross-linking agent, Cell, 15 (1978) 945–954. [DOI] [PubMed] [Google Scholar]
- [40].Duxin JP, Dewar JM, Yardimci H, Walter JC, Repair of a DNA-protein crosslink by replication-coupled proteolysis, Cell, 159 (2014) 346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Nakano T, Morishita S, Katafuchi A, Matsubara M, Horikawa Y, Terato H, Salem AM, Izumi S, Pack SP, Makino K, Ide H, Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein crosslinks, Mol. Cell, 28 (2007) 147–158. [DOI] [PubMed] [Google Scholar]
- [42].Reardon JT, Cheng Y, Sancar A, Repair of DNA-protein cross-links in mammalian cells, Cell Cycle, 5 (2006) 1366–1370. [DOI] [PubMed] [Google Scholar]
- [43].Stingele J, Bellelli R, Alte F, Hewitt G, Sarek G, Maslen SL, Tsutakawa SE, Borg A, Kjaer S, Tainer JA, Skehel JM, Groll M, Boulton SJ, Mechanism and Regulation of DNA-Protein Crosslink Repair by the DNA-Dependent Metalloprotease SPRTN, Mol. Cell, 64 (2016) 688–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Stingele J, Bellelli R, Boulton SJ, Mechanisms of DNA-protein crosslink repair, Nat. Rev. Mol. Cell Biol, 18 (2017) 563–573. [DOI] [PubMed] [Google Scholar]
- [45].Stingele J, Habermann B, Jentsch S, DNA-protein crosslink repair: proteases as DNA repair enzymes, Trends Biochem. Sci, 40 (2015) 67–71. [DOI] [PubMed] [Google Scholar]
- [46].Stingele J, Schwarz MS, Bloemeke N, Wolf PG, Jentsch S, A DNA-dependent protease involved in DNA-protein crosslink repair, Cell, 158 (2014) 327–338. [DOI] [PubMed] [Google Scholar]
- [47].Vaz B, Popovic M, Ramadan K, DNA-Protein Crosslink Proteolysis Repair, Trends Biochem. Sci, 42 (2017) 483–495. [DOI] [PubMed] [Google Scholar]
- [48].Lopez-Mosqueda J, Maddi K, Prgomet S, Kalayil S, Marinovic-Terzic I, Terzic J, Dikic I, SPRTN is a mammalian DNA-binding metalloprotease that resolves DNA-protein crosslinks, Elife, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Horton JK, Joyce-Gray DF, Pachkowski BF, Swenberg JA, Wilson SH, Hypersensitivity of DNA polymerase beta null mouse fibroblasts reflects accumulation of cytotoxic repair intermediates from site-specific alkyl DNA lesions, DNA Repair (Amst), 2 (2003) 27–48. [DOI] [PubMed] [Google Scholar]
- [50].Butler WB, Preparing nuclei from cells in monolayer cultures suitable for counting and for following synchronized cells through the cell cycle, Anal. Biochem, 141 (1984) 70–73. [DOI] [PubMed] [Google Scholar]
- [51].Caglayan M, Horton JK, Dai DP, Stefanick DF, Wilson SH, Oxidized nucleotide insertion by pol beta confounds ligation during base excision repair, Nat Commun, 8 (2017) 14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kedar PS, Stefanick DF, Horton JK, Wilson SH, Increased PARP-1 association with DNA in alkylation damaged, PARP-inhibited mouse fibroblasts, Mol. Cancer Res, 10 (2012) 360–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Horton JK, Wilson SH, Predicting enhanced cell killing through PARP inhibition, Mol. Cancer Res, 11 (2013) 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, Pommier Y, Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors, Cancer Res, 72 (2012) 5588–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Khodyreva SN, Prasad R, Ilina ES, Sukhanova MV, Kutuzov MM, Liu Y, Hou EW, Wilson SH, Lavrik OI, Apurinic/apyrimidinic (AP) site recognition by the 5’-dRP/AP lyase in poly(ADP-ribose) polymerase-1 (PARP-1), Proc. Natl. Acad. Sci, 107 (2010) 22090–22095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Horton JK, Stefanick DF, Prasad R, Gassman NR, Kedar PS, Wilson SH, Base excision repair defects invoke hypersensitivity to PARP inhibition, Mol. Cancer Res, 12 (2014) 1128–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Desai SD, Li TK, Rodriguez-Bauman A, Rubin EH, Liu LF, Ubiquitin/26S proteasome-mediated degradation of topoisomerase I as a resistance mechanism to camptothecin in tumor cells, Cancer Res, 61 (2001) 5926–5932. [PubMed] [Google Scholar]
- [58].Desai SD, Liu LF, Vazquez-Abad D, D’Arpa P, Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin, J. Biol. Chem, 272 (1997) 24159–24164. [DOI] [PubMed] [Google Scholar]
- [59].Dantzer F, de La Rubia G, Menissier-De Murcia J, Hostomsky Z, de Murcia G, Schreiber V, Base excision repair is impaired in mammalian cells lacking Poly(ADP-ribose) polymerase-1, Biochemistry, 39 (2000) 7559–7569. [DOI] [PubMed] [Google Scholar]
- [60].Liu L, Kong M, Gassman NR, Freudenthal BD, Prasad R, Zhen S, Watkins SC, Wilson SH, Van Houten B, PARP1 changes from three-dimensional DNA damage searching to one-dimensional diffusion after auto-PARylation or in the presence of APE1, Nucleic Acids Res, 45 (2017) 12834–12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Oleinick NL, Chiu SM, Ramakrishnan N, Xue LY, The formation, identification, and significance of DNA-protein cross-links in mammalian cells, Br J Cancer Suppl, 8 (1987) 135–140. [PMC free article] [PubMed] [Google Scholar]
- [62].Zhang HF, Tomida A, Koshimizu R, Ogiso Y, Lei S, Tsuruo T, Cullin 3 promotes proteasomal degradation of the topoisomerase I-DNA covalent complex, Cancer Res, 64 (2004) 1114–1121. [DOI] [PubMed] [Google Scholar]
- [63].Mao Y, Desai SD, Ting CY, Hwang J, Liu LF, 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes, J. Biol. Chem, 276 (2001) 40652–40658. [DOI] [PubMed] [Google Scholar]
- [64].Lin CP, Ban Y, Lyu YL, Desai SD, Liu LF, A ubiquitin-proteasome pathway for the repair of topoisomerase I-DNA covalent complexes, J. Biol. Chem, 283 (2008) 21074–21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Pommier Y, Barcelo JM, Rao VA, Sordet O, Jobson AG, Thibaut L, Miao ZH, Seiler JA, Zhang H, Marchand C, Agama K, Nitiss JL, Redon C, Repair of topoisomerase I-mediated DNA damage, Prog. Nucleic Acid Res. Mol. Biol, 81 (2006) 179–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Interthal H, Chen HJ, Champoux JJ, Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages, J. Biol. Chem, 280 (2005) 36518–36528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.