Abstract
A computer, trained to classify skin cancers using image analysis alone, can now identify certain cancers as successfully as can skin-cancer doctors. What are the implications for the future of medical diagnosis?
In the 1960s, the science-fiction television series Star Trek presented a vision of the future in which physician Dr Leonard McCoy used a portable diagnostic device, known as a tricorder, to assess the medical condition of Captain James Kirk and other Enterprise crew members. Although fanciful then, machines capable of the non-invasive diagnosis of human disease are becoming a reality, as are mobile devices that can image human skin to enable the identification of some cancers1,2. A study on page 115 by Esteva et al.3 has taken image-recognition technology to the next level by training a computer to classify digital images of skin lesions at least as accurately as can human skin-cancer specialists.
Skin-lesion diseases can be divided into three main groupings: non-proliferative lesions, for example inflammatory conditions such as acne; benign lesions, which are a type of cellular proliferation that does not pose a health threat; and malignant lesions of uncon-trolled proliferating cancer cells or metastatic cancer cells with the potential to migrate to other locations in the body, which require further medical attention. Although non-visual clues such as surface texture can aid the diagnosis of certain cancers, visual inspection is the primary means by which dermatologists categorize skin diseases. A previous study4 found moderately good to almost perfect agreement in the diagnosis of skin cancers, depending on the cancer type, whether der-matologists conducted a physical examination or studied a photographic image of the lesion. Therefore, image assessment can sometimes suffice for making an initial diagnosis or iden-tifying conditions that need further care. Such diagnoses can be confirmed in the clinic using direct assessment of lesions through biopsy, in which a tissue sample from the lesion is tested by a pathologist for cellular abnormalities by, for example, microscope-based observation.
Esteva et al. used an algorithmic technique known as deep learning to train a computer to develop artificial intelligence in pattern recognition, enabling the machine to analyse images and diagnose disease. There are probably many differences in the way a doctor and a computer would use visual analysis for diagnosis. For example, when diagnosing a malignant cancer called melanoma, a dermatologist often uses a set of criteria5 known as ABCDE (in which each letter stands for a characteristic to be assessed, such as A for asymmetrical lesion shape), and also relies on his or her previous experience to spot visual subtleties in the lesion. A trained computer does not necessarily mimic this decision-making approach. Instead, it identifies its own criteria for informative patterns associated with a disease and trains on a data set without using rules imposed by human methods of visualization. The computer can also assess image data that are imperceptible to the human eye.
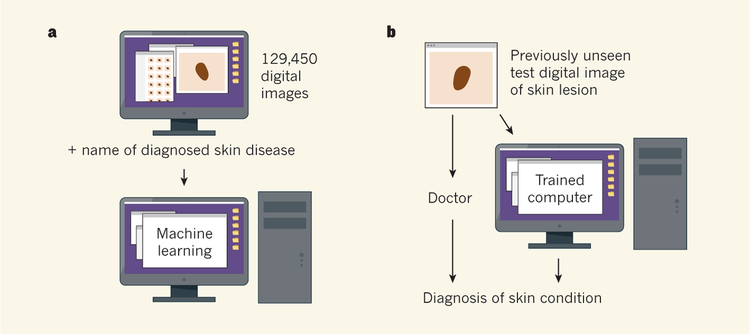
For computer training, the authors used a set of 129,450 images of skin lesions and the names of the conditions that each image represented, comprising 2,032 diagnosed skin diseases. The reference diagnoses were made by dermatologists, who classified lesions by non-invasive visual analysis or biopsy testing.
Esteva and colleagues then presented a set of previously unseen digital images of skin lesions to the trained computer and to 21 doctors, and queried whether the lesion in an image needed further medical attention. The diagnosis of these test images had been verified by biopsy testing. The computer diagnosed the images with a level of accuracy that was similar to or better than the dermatologists’ diagnoses (Fig. 1). (This contest between human and machine brings to mind the defeat of chess world champion Garry Kasparov by the Deep Blue computer in 1997.) Esteva et al. did not test whether the doctors’ diagnostic abilities varied depending on whether they assessed a lesion using a digital image or through a physical inspection.
Figure 1 |. Accurate cancer diagnosis by computer.
a, Esteva et al.3 trained a computer to recognize skin diseases using a deep-learning approach in which the computer was presented with 129,450 digital images of medically diagnosed samples. b, To test the diagnostic accuracy level of the trained machine, the computer and 21 skin specialists were presented with previously unseen images representing verified examples of benign or malignant skin disease arising from two cell types. The machine was as successful as or better than the doctors at diagnosing whether or not the images represented conditions that might need further medical attention.
The test images used were examples of two categories of benign and malignant lesions. One category was melanocytic lesions (derived from pigmented skin cells known as melanocytes), including moles and melanoma. The other category was predominantly keratinocytic lesions (derived from skin cells called keratinocytes), such as benign seborrhoeic keratosis and non-melanocytic carcinomas. However, some thorny issues that can plague dermatologists remain to be addressed when assessing the computer’s diagnostic abilities. For example, the authors did not report investigating whether the computer could distinguish between similar-looking diseases such as melanoma and benign seborrhoeic keratosis. And how accurately might the computer perform in distinguishing between an amelanocytic (non-pigmented) melanoma and a malignant carcinoma?
The training set of images used by Esteva et al. was about 100 times larger than any reported previously6 for such approaches, and this might explain the machine’s success. There could be room for improvement. As more data are added to such a system, the machine learns as its mistakes are corrected and its performance subsequently improves. Esteva and colleagues’ work represents a first point along a line of improvement, not a peak. The authors used a model algorithm called Inception v3, and new programs and algorithms are now available that offer improved training time and accuracy.
However, an algorithm is only as accurate as its reference information. If the machine diagnoses a lesion as malignant but the biopsy-confirmed classification of the lesion by a pathologist was non-malignant, this would be an ‘incorrect’ machine diagnosis. But what about cases in which the machine rather than the pathologist is correct? The relative diagnostic accuracies of the machine and the human could be tested by tracking how diagnosed lesions progressed over time.
An obvious potential societal benefit of artificial intelligence in diagnostic technology would be improved access to high-quality health care. A smartphone app involving this technology might enable effective, easy and low-cost medical assessments of more individuals than is possible with existing medical-care systems. Using skin-cancer detection as a proof-of-principle, other medical fields that rely on doctors for image-based cancer diagnoses, such as radiology, could also be transformed.
However, diagnostics driven by artificial intelligence might have unintended adverse consequences. Would medical staff become mere technicians responding to a machine’s diagnostic decisions, perhaps with the power occasionally to override the computer? And if medical examinations begin to rely on patient self-identification of suspicious lesions, would individuals at high-risk of skin cancer be more likely to opt out of regular full skin screening in a doctor’s surgery that could save their lives?
Accurately and effortlessly diagnosing cancer during the early disease stages, when the chances of a cure are optimal, has long seemed a possibility closer to the world of science fiction than to reality. Yet perhaps it won’t be too long before there is a real smartphone equivalent of the Star Trek tricorder. We should be prepared, and perhaps steel ourselves, to boldly take this technology to a place where no technology has gone before.
Contributor Information
SANCY A. LEACHMAN, Department of Dermatology, Melanoma Research Program, Knight Cancer Institute, Oregon Health and Science University, Portland, Oregon 97239, USA.
GLENN MERLINO, Laboratory of Cancer Biology and Genetics, Center for Cancer Research, National Cancer Institute, Bethesda, Maryland 20892, USA..
References
- 1.Maier T et al. J. Eur. Acad. Dermatol. Venereol 29, 663–667 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Gareau DS et al. Exp. Dermatol 10.1111/exd.13250 (2016). [DOI]
- 3.Esteva A et al. Nature 542, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warshaw EM, Gravely AA & Nelson DB J. Am. Acad. Dermatol 72, 426–435 (2015). [DOI] [PubMed] [Google Scholar]
- 5.American Academy of Dermatology Ad Hoc Task Force for the ABCDEs of Melanoma. J. Am. Acad. Dermatol 72, 717–723 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Carrera C et al. JAMA Dermatol 152, 798–806 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]