Abstract
The RAS to extracellular signal–regulated kinase (ERK) signal transduction cascade is crucial to cell proliferation, differentiation, and survival. Although numerous growth factors activate the RAS-ERK pathway, they can have different effects on the amplitude and duration of the ERK signal and, therefore, on the biological consequences. For instance, nerve growth factor, which elicits a larger and more sustained increase in ERK phosphorylation in PC12 cells than does epidermal growth factor (EGF), stimulates PC12 cell differentiation, whereas EGF stimulates PC12 cell proliferation. Here, we show that protein arginine methylation limits the ERK1/2 signal elicited by particular growth factors in different cell types from various species. We found that this restriction in ERK1/2 phosphorylation depended on methylation of RAF proteins by protein arginine methyltransferase 5 (PRMT5). PRMT5-dependent methylation enhanced the degradation of activated CRAF and BRAF, thereby reducing their catalytic activity. Inhibition of PRMT5 activity or expression of RAF mutants that could not be methylated not only affected the amplitude and duration of ERK phosphorylation in response to growth factors but also redirected the response of PC12 cells to EGF from proliferation to differentiation. This additional level of regulation within the RAS pathway may lead to the identification of new targets for therapeutic intervention.
INTRODUCTION
A major challenge in cell signaling is to understand how different external cues and cell membrane receptors give rise to unique biological responses despite their promiscuous activation of shared pathways. For instance, although various growth factors initiate signaling through the same pathways (1), the biological consequences of the activation of a particular signaling pathway by different growth factors may differ. Many growth factors activate receptor tyrosine kinases (RTKs) to signal through the RAS (2) to RAF to mitogen-activated protein kinase (MAPK) signaling pathway. The extracellular signal–regulated protein kinase 1 and 2 (ERK1/2), MAPKs activated by phosphorylation and inactivated by dephosphorylation, play a prominent role in this pathway by phosphorylating transcription factors, cytoskeletal proteins, and enzymes (including other protein kinases) (3). Three different quantitative measures can be used to assess kinase signaling: signal amplitude (the peak response to a stimulus), duration (is the response transient or sustained?), and integral strength (integrated concentration of an active molecule, derived from the other two measures) (4, 5). From an oversimplified perspective, phosphorylation and dephosphorylation determine whether kinases are active or inactive; however, their subcellular distribution and, presumably, posttranslational modifications other than phosphorylation (6, 7) will influence the final biological outcomes. Signaling through the RAS-ERK1/2 pathway can be modulated at various levels; however, the activation of specific RAF isoforms, their homo- or heterodimerization with other isoforms, and their degradation are particularly relevant not only to the activation of ERK1/2 but also to determining the amplitude, duration, and integral strength of ERK1/2 phosphorylation (4, 5, 8–10).
Protein arginine methylation is increasingly being recognized for its role in regulating signal transduction, RNA processing, transcriptional activation, and DNA repair (11–13). The existence of a wide range of arginine-methylated substrates suggests that this eukaryotic modification may play a role as complex as that of phosphorylation and raises the possibility that these two regulatory mechanisms are somehow coordinated. Among the nine protein arginine (R) methyltransferases (PRMTs) in humans with a demonstrated physiological enzymatic activity (PRMT1 to 9) (11), PRMT5 was the first determined to catalyze the formation of symmetric dimethylarginines (sDMAs) on a Gly-Arg-Gly (GRG) motif (14). PRMT5 has been implicated in transcriptional regulation through histone methylation (15, 16) and methylation of the RNA polymerase II CTD phosphatase (FCP1) (17). It has also been implicated in promoting spliceosome assembly (18) and appears to be an HSP90 (heat shock protein 90 kD) client (19). Given these roles, it is unexpected that most PRMT5 is in the cytoplasm and not in the nucleus (20). However, PRMT5 was initially identified as a Janus kinase binding protein 1 (JBP1) (21), and it has also been found to interact with the death receptor for TRAIL (tumor necrosis factor–related apoptosis-inducing ligand) (22). Furthermore, PRMT5 is a component of the branch of the RAS signaling cascade implicated in regulating morphology in Schizosaccharomyces pombe and it positively modulates Shk1 [Ste20/p21-activated kinase (PAK) homolog] function (23), suggesting that PRMT5 may have unappreciated cytoplasmic functions.
Although the molecular machinery by which various growth factors control signal transduction has been extensively studied (1), the mechanism regulating signal amplitude in response to a given stimulus is largely unknown. Here, we show that arginine methylation of RAF proteins limits the ERK1/2 phosphorylation elicited by stimulation with certain growth factors and identify PRMT5 as the protein methyltransferase responsible for fine-tuning growth factor signals. PRMT5 forms a complex with RAF proteins and methylates them, decreasing their kinase activity and stability, thereby diminishing the amplitude of the ERK1/2 signal. Finally, we show that inhibiting methylation can alter growth factor–dependent biological responses, switching the response of PC12 cells to EGF from proliferation to differentiation by increasing the signal amplitude and prolonging its duration.
RESULTS
5′-Methylthioadenosine increases ERK1/2 signal amplitude in response to hepatocyte growth factor
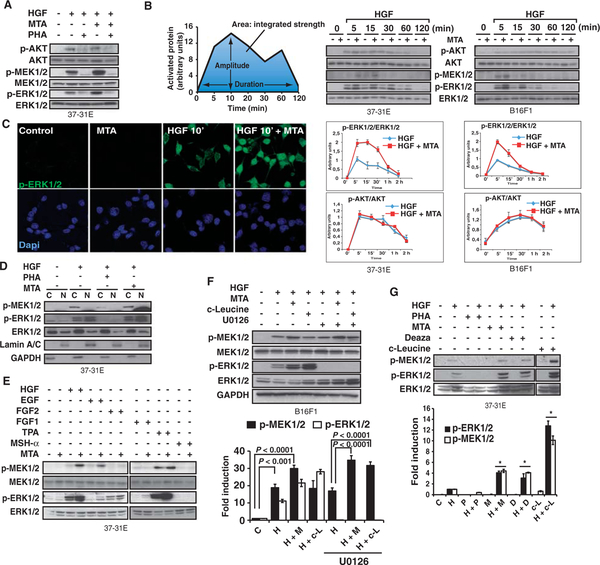
We observed that, in mouse melanoma cells, the methylation inhibitor 5′-methylthioadenosine (MTA) increased the degree of ERK1/2 phosphorylation in response to hepatocyte growth factor (HGF) treatment (Fig. 1A). Signaling through the HGF RTK c-Met activates both the RASERK1/2 and the phosphatidylinositol 3-kinase (PI3K)–AKT pathways (24); however, MTA did not appear to affect AKT phosphorylation (Fig. 1, A and B). We found that MTA specifically enhanced the amplitude of HGF-mediated ERK1/2 phosphorylation in various mouse and human cell lines (see below) and rat primary hepatocytes (fig. S1). MTA’s enhancement of ERK1/2 phosphorylation was observed 2 min after HGF stimulation (fig. S2A) and was still apparent 30 min after HGF stimulation (Fig. 1B). However, ERK1/2 phosphorylation began to decline 10 min after HGF treatment with or without MTA, indicating that phosphatase activity was unaltered (Fig. 1B). Indeed, activity of the nuclear phosphatase MKP1/2, which controls the amount of nuclear p-ERK1/2, increased with MTA treatment (fig. S2B). Consistent with this, we also observed that the translocation of phosphorylated ERK1/2 (p-ERK1/2) to the nucleus was intact in MTA-pretreated cells (Fig. 1, C and D).
Fig. 1.
MTA increases HGF-mediated ERK1/2 phosphorylation. (A) 37–31E melanoma cells were treated with PHA-665752 (PHA), MTA, or both under conditions of serum starvation, and then, after stimulation with HGF, total protein extracts were immunoblotted for p-ERK/2, ERK1/2, p-MEK1/2, MEK1/2, p-AKT, and total AKT. (B) MTA increases the HGF-dependent p-ERK1/2 signal amplitude. 37–31E and B16F1 melanoma cells were treated with MTA as in (A) and exposed to HGF for the indicated times. Graphs depict a schematic representation of the quantitative measures governing signal transduction and the quantification of the ERK1/2 and AKT activation profiles in response to HGF or HGF plus MTA from three independent experiments. (C) Immunofluorescence micrograph of p-ERK1/2 in 37–31E cells treated with HGF or MTA as indicated. (D) 37–31E cells were treated as in (A). Western blots show the cytoplasmic and nuclear distribution and phosphorylation of MEK1/2 and ERK1/2. Lamin A/C and GAPDH were used as markers of the fractions’ purity. (E) MTA-mediated effect on ERK1/2 and MEK1/2 phosphorylation is growth factor–specific. 37–31E cells were treated as in (A) with the indicated growth factors for 10 min. Total ERK1/2 is shown as a loading control. One representative experiment of three is shown. (F) B16F1 cells were serum-starved and treated for 14 hours with or without MTA or c-leucine and then treated for 5 min with HGF. U0126 was applied where indicated. p-MEK1/2, MEK1/2, p-ERK1/2, ERK1/2, and GAPDH were assessed by Western blot (n = 3 experiments). (G) Methylation inhibitors increase ERK1/2 signal amplitude in response to HGF. 37–31E melanoma cells were starved and treated for 14 hours with PHA, MTA, Deaza, or c-leucine. Then, cells were exposed to HGF for 10 min. p-MEK1/2, p-ERK1/2, and total ERK1/2 are shown in one representative experiment of three. Graph shows the quantification of p-MEK1/2 and p-ERK1/2. *P < 0.05 versus HGF-treated samples; n = 3 different experiments.
Together, these data demonstrate that the methylation inhibitor MTA increases ERK1/2 signal amplitude in response to HGF. Furthermore, this seems to involve an increase in kinase activity upstream of ERK1/2, rather than a downstream decrease in phosphatase activity.
MTA-dependent augmentation of ERK1/2 phosphorylation is ligand-dependent and mediated by methylation inhibition
Next, we determined whether MTA enhanced the degree of ERK1/2 phosphorylation in response to other stimuli that activate the ERK1/2 signaling pathway. We found that MTA enhanced ERK1/2 phosphorylation mediated not only by treatment with HGF but also by exposure to the phorbol ester tetradecanoylphorbol-13-acetate (TPA), which activates the pathway in an RTK-independent manner, or epidermal growth factor (EGF) (showing limited amounts of p-ERK1/2 in response to EGF because of the low abundance of EGF receptors in the 37–31E melanoma cell line) (Fig. 1E). However, the ability of MTA to enhance ERK1/2 phosphorylation in response to either acidic or basic fibroblast growth factor (FGF1 and FGF2, respectively) depended on cell line (Fig. 1E and fig. S3A) and did not occur with melanocyte-stimulating hormone (MSH-α), which did not stimulate ERK1/2 phosphorylation (Fig. 1E). Similar results were obtained in human embryonic kidney (HEK) 293 cells and rat neuroblastoma (PC12) cells (fig. S3A).
This modulatory effect of MTA was also observed with MEK1/2 (mitogen-activated or extracellular signal–regulated protein kinase kinase 1 and 2), the kinases immediately upstream of ERK1/2 in the MAPK pathway (figs. S2A and S3D). Moreover, the increase in ERK1/2 phosphorylation by MTA upon growth factor stimulation was blocked by inhibition of MEK1/2 activity (Fig. 1F), suggesting that it did not involve crosstalk pathways that targeted ERK1/2 directly.
Next, we examined the effects of the protein methyltransferase inhibitors 3-deaza-adenosine (Deaza) and cycloleucine (c-leucine) (25) to confirm that the increase in signal amplitude in response to MTA indeed depended on inhibition of methylation (Fig. 1, F and G, and fig. S3, B to D). As expected, we did not observe any alteration in the translocation of p-ERK1/2 to the nucleus in either Deaza or c-leucine (fig. S3D). Together, these data indicate that protein methylation modulates signaling through the RAS pathway in response to specific growth factors upstream of MEK1/2.
Protein arginine methyltransferase PRMT5 limits ERK1/2 signal amplitude
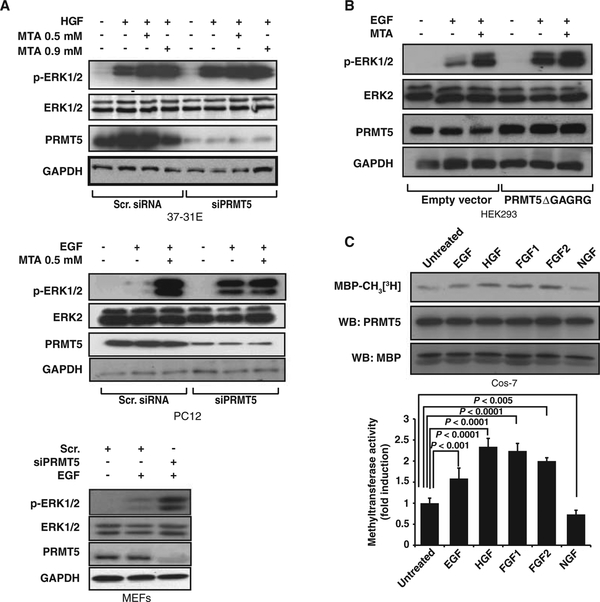
Our data suggested that the machinery responsible for this modulation of RAS pathway signaling was cytoplasmic. Whereas lysine methylation is involved in a wide range of nuclear processes (26–28), protein arginine methylation has been implicated in several cytoplasmic responses (13). Among the members of the PRMT family found in humans, only PRMT1, 3, 5, and 8 are found in the cytoplasm (29). We used a combination of genetic and functional analyses to eliminate Prmt1, Prmt3, and Prmt8 as possible candidates (fig. S4). Depletion of Prmt5 with small interfering RNA (siRNA) reproduced the MTA-dependent increase in growth factor–dependent ERK1/2 phosphorylation in both tumor and normal cells, and reduced or eliminated the effect of MTA, suggesting that MTA targeted PRMT5 (Fig. 2A). Moreover, overexpression of a catalytically inactive PRMT5 mutant (PRMT5DGAGRG) increased the degree of ERK1/2 phosphorylation in response to EGF (Fig. 2B).
Fig. 2.
PRMT5 depletion increases ERK1/2 phosphorylation in response to growth factors. (A) Knockdown of Prmt5 in 37–31E, PC12, or MEFs enhances the increase in ERK1/2 phosphorylation produced by growth factor treatment. Cells transfected with either scrambled siRNA (Scr.) or Prmt5 siRNA were serum-starved and exposed to MTA for 3 hours and then treated with HGF for 10 min. p-ERK1/2, ERK1/2, and PRMT5 abundance was assessed by Western blot. (B) Overexpression of the catalytically inactive PRMT5 mutant (PRMT5ΔGAGRG) in HEK293 cells reproduces the effects of MTA on ERK1/2 phosphorylation. Fortyeight hours after transfection with either empty vector or pEF2-PRMT5ΔGAGRG expression vector, cells were serumstarved and pretreated with MTA for 2 hours and then stimulated with HGF for 10 min. Cell lysates were assessed for p-ERK1/2, ERK1/2, PRMT5, and GAPDH by Western blotting. (C) Growth factors regulate PRMT5 methyltransferase activity. Cos-7 cells stably transfected with Flag-PRMT5 were serum-starved for 3 hours and then treated with the indicated growth factors for 10 min. After PRMT5 immunoprecipitation, its methyltransferase activity was measured in vitro. The3H radioactive signal incorporated in MBP was assessed autoradiographically after SDS-PAGE. Immunoprecipitated PRMT5 and MBP are shown as loading controls. Graph shows quantification of methyltransferase activity normalized by the amount of PRMT5 immunoprecipitated. P value was calculated with Student’s t test (n = 3 independent replicates).
Because PRMT5’s ability to affect ERK1/2 phosphorylation depended on the specific ligand used to activate MAPK signaling, we examined whether PRMT5 activity was itself regulated by growth factors. We found that PRMT5 activity was regulated by growth factors, increasing with EGF, HGF, FGF1, and FGF2 treatment and slightly decreasing with nerve growth factor (NGF) (Fig. 2C). Together, these data indicate that PRMT5 modulates ERK1/2 signal amplitude in response to specific growth factors.
PRMT5 colocalizes with RAF proteins and modulates their catalytic activity
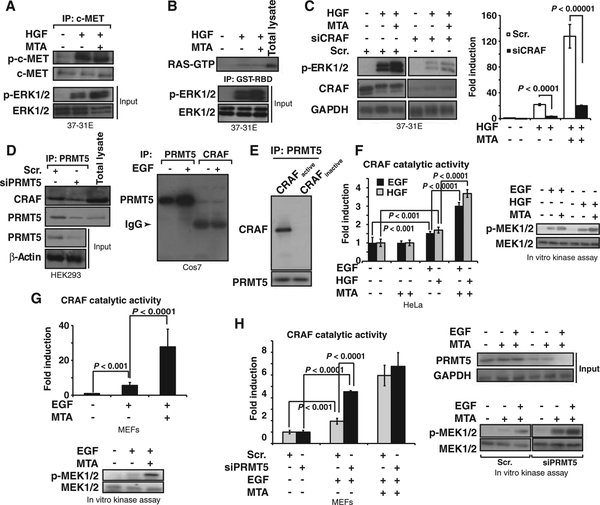
Our observations suggested that the putative PRMT5 target was within the RAS pathway and upstream of MEK1/2. MTA did not affect phosphorylation of the c-Met receptor (indicative of its activation) upon exposure to HGF (Fig. 3A), nor did it affect the amount of active RAS-GTP (guano-sine triphosphate) observed after HGF treatment (Fig. 3B). We also observed that MTA modulates the ERK1/2 signal amplitude in NRASQ61L mutant cells (which signal mainly through CRAF) (30), whereas this effect was not observed in BRAFV600E mutant cells, which signal independently of RAS (30) (fig. S5), supporting the RAS-dependent regulation of PRMT5. Moreover, HGF-dependent ERK1/2 phosphorylation was limited in cells in which CRaf was knocked down (fourfold less amount of p-ERK1/2), and the effect of MTA was nearly abolished (Fig. 3C), suggesting that CRAF participated in the methylation-mediated regulation of ERK1/2 activation. RAF proteins and PRMT5 are HSP90 clients (19, 31), indicating that they may be part of the same protein complexes. We found that PRMT5 was constitutively bound to CRAF and BRAF immunocomplexes (Fig. 3D and fig. S6A). The association of PRMT5 with CRAF was confirmed by immunolocalization experiments (fig. S6B). However, in vitro binding experiments using recombinant PRMT5 and recombinant full-length inactive glutathione S-transferase (GST)–CRAF or active GST-CRAF (N-terminal GST-tagged, residues 306 to end) showed that PRMT5 binds only to active CRAF (Fig. 3E).
Fig. 3.
MTA and PRMT5 depletion increases CRAF’s catalytic activity. (A) 37–31E cells were serum-starved, pretreated with MTA for 3 hours, and then treated with HGF for 5 min. c-Met was immunoprecipitated from total lysates. Western blot shows the amount of p-c-Met in the immunocomplexes. p-ERK1/2 and ERK1/2 levels in the samples used in the immunoprecipitation are shown (Input). (B) RAS activation is not affected by MTA. 37–31E cells were treated as in (A). RAS-GTP was isolated by affinity chromatography with GST-RBD (GST–RAS binding domain of CRAF). Complexes were separated by SDS-PAGE. Western blot shows the amount of RAS-GTP. p-ERK1/2 and ERK1/2 demonstrate the activation of the pathway in the samples used. (C) 37–31E cells transfected with CRaf siRNA were serumstarved and pretreated with MTA for 2 hours and then triggered with HGF for 10 min. Western blot shows CRAF and p-ERK1/2. P value was calculated with Student’s t test (n = 3 different experiments). (D) PRMT5 was immunoprecipitated from total lysates of HEK293 cells transfected with either scrambled (Scr.) or PRMT5 siRNA. Western blots show PRMT5 and CRAF in the immunocomplexes and PRMT5 in the initial total lysates (left panel). Endogenous PRMT5 and CRAF were immunoprecipitated from lysates of Cos-7 cells untreated or treated with EGF. Western blots assess the presence of PRMT5 in the immunocomplexes. (E) PRMT5 binds to active CRAF. Recombinant PRMT5 was incubated with either full-length inactive GST-CRAF or active GST-RAF (N-terminal GST-tagged, residues 306 to end). Western blot shows CRAF after PRMT5 immunoprecipitation. Immunoprecipitated PRMT5 is shown as loading control. (F) MTA increases growth factor–induced CRAF catalytic activity. HeLa cells transiently transfected with Flag-CRAFWT were serum-starved, treated with MTA for 3 hours, and then treated with either EGF or HGF for 10 min. Immunoprecipitated Flag-CRAF was used to perform an in vitro kinase assay. Graph shows CRAF catalytic activity (fold induction over untreated cells) in a coupled RAF-MEK-ERK-MBP radioactive assay under the different conditions (left). Western blots were performed against p-MEK under the same conditions (right). P value was calculated with Student’s t test (n = 3 replicates). (G) MTA increases the endogenous CRAF catalytic activity in response to growth factors. MEFs were starved and treated with MTA for 3 hours, and cells were exposed to EGF for 10 min. Immunoprecipitated endogenous CRAF was used to perform an in vitro kinase assay as in (F). Graphs show the CRAF catalytic activity fold induction over untreated cells. A radioactive assay and Western blots show the p-MEK levels induced by immunoprecipitated CRAF. Bars indicate the SD. P value was calculated with Student’s t test (n = 3 replicates). (H) 37–31E cells transfected with either scrambled (Scr.) or Prmt5 siRNA were treated as in (F). Graph shows the kinase activity of endogenous CRAF. Western blots show the p-MEK levels induced by immunoprecipitated CRAF. PRMT5 protein levels in total lysates used in the immunoprecipitation are shown. Bars indicate the SD. P value was calculated with Student’s t test (n = 3 replicates).
To determine whether the catalytic activity of RAF family proteins was modulated by posttranslational methylation, we measured their in vitro kinase activity after immunoprecipitation from cells treated with growth factors, MTA, or both. The in vitro kinase activity of CRAF immunoprecipitated from HeLa cells transfected with Flag-CRAFWT and treated with HGF or EGF was significantly increased by MTA (120%; P < 0.0001), whereas MTA had no effect on CRAF activity in cells that had not been treated with growth factor (Fig. 3F). MTA had the same effect on endogenous CRAF immunoprecipitated from mouse embryo fibroblasts (MEFs) treated with EGF (Fig. 3G; or HeLa and HEK293 cells treated with EGF or HGF, fig. S8C). The catalytic activity of CRAF immunoprecipitated from cells treated with EGF was also increased significantly by Prmt5 knockdown with siRNA (P < 0.001) (Fig. 3H). Posttranslational methylation also enhanced the in vitro catalytic activity of BRAF (figs. S7 and S8B) and ARAF (fig. S8A) immunoprecipitated from HeLa and HEK293 cells treated with growth factors. Thus, these data indicate that MTA and PRMT5 modulate the kinase activity of RAF proteins induced by growth factors.
CRAFR563K and BRAFR671K mutants are more stable than the corresponding wild-type proteins and show increased kinase activity in response to growth factors
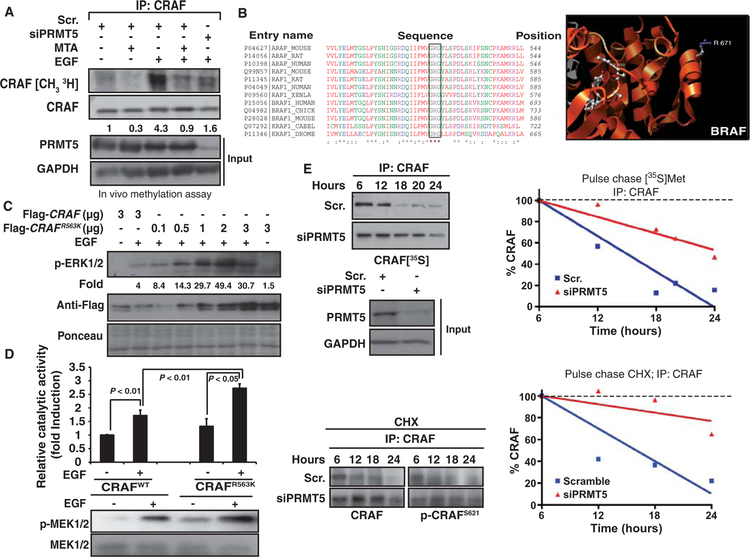
In vivo metabolic labeling of cells showed that growth factor treatment stimulated the methylation of CRAF and BRAF and that this was prevented by MTA treatment or Prmt5 knockdown (Fig. 4A and fig. S9A). PRMT5 methylates arginine residues within GRG motifs (11), and, of all components of the RAS pathway, only the RAF proteins contained a GRG motif. The GRG motif was present in all RAF family members analyzed and was conserved from Caenorhabditis elegans to humans (Fig. 4B). Moreover, according to the BRAF three-dimensional (3D) structure, BRAFR671 should be available for modification by methylation (Fig. 4B).
Fig. 4.
Arginine-to-lysine mutation in the GRG motif of RAF proteins promotes their stability and amplifies their increase in kinase activity in response to growth factors. (A) In vivo methylation assay in PC12 cells. Cells transiently transfected with either scrambled or Prmt5 siRNA and metabolically labeled ([3H]methionine) were serum-starved and pretreated with MTA, and then cells were exposed to EGF for 10 min. After immunoprecipitation of endogenous CRAF, the3H radioactive signal incorporated in CRAF was assessed autoradiographically after SDS-PAGE. The same membranes were assessed for CRAF (loading control). PRMT5 in total lysates is shown in the lower panel. (B) Sequence alignment of RAF isoforms from different species showing the conserved GRG motif (left). A 3D image made with UCSF Chimera (http://www.cgl.ucsf.edu/chimera) and the 1UWH file from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (http://www.pdb.org) show the location of the Arg671 residue in the BRAF 3D structure (right). (C) HEK293 cells were transiently transfected with Flag-CRAFWT or Flag-CRAFR563K as indicated. Western blot shows p-ERK1/2 and abundance of transfected proteins. Quantification of p-ERK1/2 normalized by Ponceau S staining is shown (fold induction with respect to Flag-CRAFWT–transfected untreated cells). (D) Catalytic activity of CRAFR563K mutant compared to that of the wild-type isoform. HeLa cells were transfected with Flag-CRAFWT or one-sixth of the amount of the Flag-CRAFR563K mutant. CRAF catalytic activity was assessed in immunoprecipitates. Graph shows the CRAF catalytic activity (fold induction over untreated cells) in a coupled RAF-MEK-ERK-MBP radioactive assay under the different conditions (top). P value was calculated with Student’s t test (n = 3 replicates). Western blots show p-MEK under the same conditions (lower panel). (E) SKMel147 cells were transiently transfected with scrambled siRNA (Scr.) or PRMT5 siRNA (siPRMT5) for 24 hours. Cells were pulse-labeled with [35S]Met and re-collected over a 24-hour time course (upper left panels). Cell lysates were harvested and immunoprecipitated with antibody directed against CRAF; after SDS-PAGE separation and capture of the35S signal, CRAF was quantitated by autoradiography. Left, data presented as autoradiogram; right, data presented graphically. Cells were treated with CHX (5 μg/ml) and collected at the indicated time points (lower panel). Immunoprecipitation of CRAF was performed from 500 μg of total protein. Western blots show CRAF and p-CRAFS621. Graph shows CRAF quantifications. Dashed line represents 100%. Amounts of PRMT5 are shown in the input samples.
To investigate the role of this residue in the activation of ERK1/2 upon growth factor triggering, we generated the equivalent CRAFR563K mutant. Transient transfection of the same DNA quantity of Flag-CRAFR563K or Flag-CRAFwt in HEK293 cells resulted in higher amounts of Flag-CRAFR563K protein, indicating that the CRAFR563K mutant was more stable than its wild-type counterpart (Fig. 4C). Cells transfected with one-sixth of the amount of Flag-CRAFR563K compared with Flag-CRAFwt to compensate for protein expression abundance showed more p-ERK1/2 than did Flag-CRAFwt–transfected cells in response to EGF (14.3-fold versus 4-fold compared to Flag-CRAFwt–transfected and untreated cells) (Fig. 4C). In agreement with this, the CRAFR563K mutant showed significantly increased catalytic activity in response to the growth factor compared with that of the wild-type variant (62.9 ± 1.2% more activity; P < 0.01) (Fig. 4D). These results suggest that methylation of CRAFR563 could decrease the stability of active CRAF molecules and consequently decrease their signal output. To confirm that methylation reactions contributed to CRAF degradation upon RAS pathway activation, we measured CRAF stability in NRASQ61L mutant SKMel147 melanoma cells with or without PRMT5 depletion or in the presence or absence of MTA (Fig. 4E and fig. S9B). Inhibition of protein methylation or PRMT5 knockdown stabilized CRAF [~50% more stable with a half-life (t1/2) of ~12 to 15 hours in PRMT5 siRNA–transfected cells than in cells transfected with scrambled siRNA] (Fig. 4E). These data correlated with the phosphorylation state of CRAF Ser621, a residue whose phosphorylation prevents proteasomal degradation of CRAF (32) (Fig. 4E and fig. S9B). The notion that a subset of RAF molecules from the small fraction of the total pool mobilized to signal are subjected to this regulation was indirectly supported by the phosphorylation code of RAF proteins 10 min after growth factor triggering. MTA-pretreated cells compared with cells treated only with growth factor showed increased amounts of p-CRAFS338, a residue related to its active conformation; decreased amounts of p-CRAFS289,296,301, residues related to the inactivation of CRAF; and increased amounts of p-CRAFS621 (fig. S9C). Similarly, the BRAFR671K mutant had increased kinase activity and stability compared to wild-type BRAF, and depletion of PRMT5 increased the stability of the wild-type protein (fig. S10, A to C). PRMT5 knockdown did not affect ARAF stability in SKMel147 cells (fig. S10D), although ARAF’s catalytic activity in response to EGF and HGF was increased by MTA in HEK293 and HeLa cells (fig. S8A).
Together, these results indicate that the arginine residue within the GRG motif regulates the kinase activity and stability of activated BRAF and CRAF and consequently modulates ERK1/2 signal amplitude.
PRMT5 methylates CRAFR563 after growth factor treatment
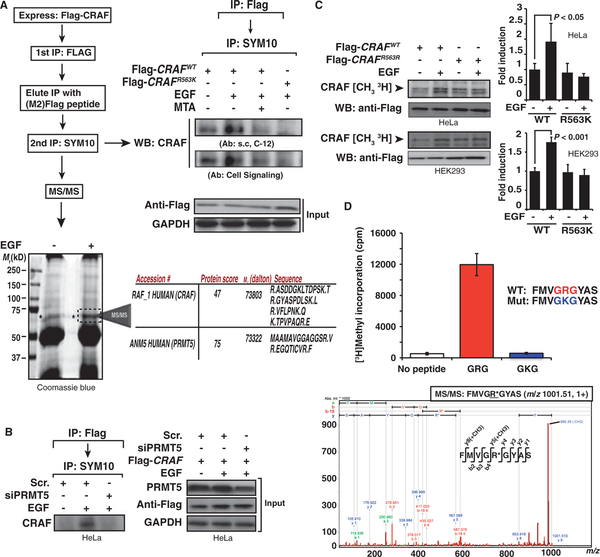
Our data suggested that CRAF was methylated on the arginine within the GRG motif. Only a small fraction of the total RAF proteins in a cell participates in signaling response to a given stimulus (fig. S11A) (33). To identify methylated molecules and determine whether CRAF was methylated on CRAFR563K, we enriched the population of activated CRAF proteins by performing two consecutive immunoprecipitations. We transfected HeLa cells with either Flag-CRAFWT or Flag-CRAFR563K and, after treating them with EGF in the presence or absence of MTA, immunoprecipitated CRAF with an anti-Flag antibody. The resulting immunocomplexes were eluted with a Flag peptide, and a second immunoprecipitation was performed with an antibody directed against anti–symmetric methylarginines (SYM10). Then, Western blot and mass spectrometry analyses were used to detect CRAF in the anti-methylarginine immunocomplexes.
EGF treatment increased the amount of CRAF detected by Western blot analysis in the anti-methylarginine immunocomplexes, whereas treatment with the combination of EGF and MTA decreased it (Fig. 5A). We could not detect CRAF in anti-methylarginine immunocomplexes from cells transfected with Flag-CRAFR563K. We scaled up the experiment to identify CRAF by mass spectrometry. Analysis of the immunoprecipitated proteins within the band showing an appropriate molecular size confirmed the presence of CRAF and also identified PRMT5 (~70 kD) (Fig. 5A). PRMT5 knockdown abolished the EGF-mediated increase in CRAF abundance in the immunocomplexes (Fig. 5B). The identity of the methylated residue was confirmed by in vivo methylation experiments performed on HeLa and HEK293 cells and showed that the Flag-CRAFR563K mutant was not methylated in response to growth factor treatment (Fig. 5C). Furthermore, in vitro methylation assays showed that PRMT5 methylated Arg563 of a CRAF-derived peptide containing the GRG (Fig. 5D), which differs from the comparable regions of ARAF and BRAF by a single amino acid (fig. S11B). Together, these data indicate that growth factors stimulate methylation of CRAFR563 by PRMT5.
Fig. 5.
PRMT5 methylates CRAF on Arg563. (A) HeLa cells were transfected with either Flag-CRAFWT or Flag-CRAFR563K for 48 hours. Cells were starved and treated with MTA for 2 hours and then treated with EGF for 10 min. Subsequently, Flag-tagged CRAF proteins were immunoprecipitated and the complexes were eluted with M2 peptide. A second immunoprecipitation was performed on the eluates with the anti-sDMA antibody SYM10. The immunocomplexes were analyzed for the presence of CRAF by Western blot and mass spectrometry (MS). Two different antibodies against CRAF were used. Coomassie blue staining of the gel showing the band corresponding to the size of CRAF (*) analyzed by liquid chromatography–mass spectrometry is shown. Table shows the sequences of the identified peptides corresponding to the indicated proteins (lower panel). (B) PRMT5 depletion inhibits CRAF methylation. HeLa cells transfected with either scrambled or PRMT5 siRNA for 48 hours and Flag-CRAFWT for an extra 24 hours were starved for 2 hours and treated with EGF. Lysates were processed according to the protocol described in (A). Western blot shows the abundance of CRAF. Right panel shows PRMT5 in the initial total lysates. (C) CRAFR563K is not methylated after treatment with growth factors. HeLa or HEK293 cells transfected with either Flag-CRAFWT or Flag-CRAFR563K were metabolically labeled with [3H]methionine and treated with EGF for 10 min. After immunoprecipitation with the α-Flag antibody, the3H radioactive signal incorporated in CRAF was assessed autoradiographically after SDS-PAGE. Immunoprecipitated CRAF is shown as loading control. Graph shows quantification of CRAF isoform methylation relative to untreated cells. P value was calculated with Student’s t test (n = 3 replicates). (D) CRAF peptide containing GRG motif is methylated in vitro. In vitro methylation assay with immunoprecipitated PRMT5 from EGF-treated Cos-7 cells stably transfected with PRMT5. Assay was performed adding either no peptide, FMVGRGYAS peptide, or FMVGKGYAS mutant peptide as a control for specificity of arginine methylation. The fragmentation spectrum of the methylated peptide identified by MALDI-TOF/TOF mass spectrometry is shown in the bottom panel. m/z, mass/charge ratio.
Inhibition of methylation switches the response to growth factors from proliferation to differentiation
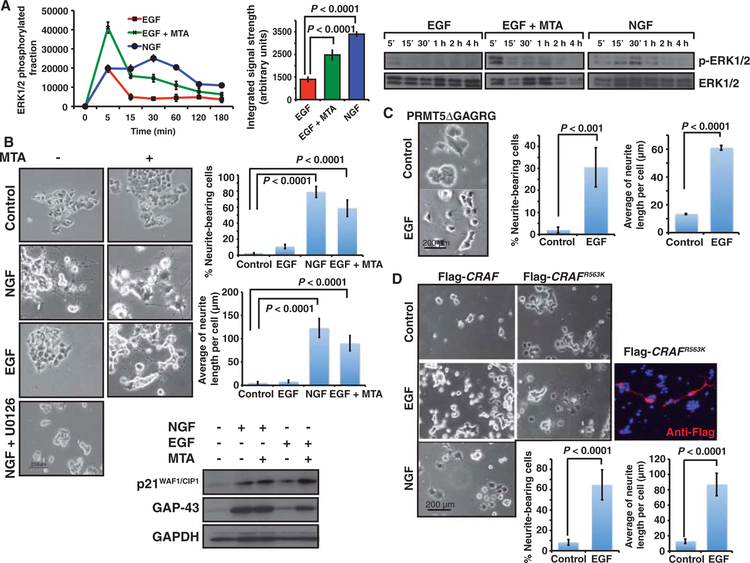
Both NGF and EGF stimulate the RAS-MAPK pathway in PC12 cells, but they have different effects on ERK1/2 activation: EGF elicits a transient phosphorylation of ERK1/2, whereas NGF elicits a sustained activation of ERK1/2 (34). This differential activation of ERK1/2 by NGF or EGF translates into distinct biological responses: NGF promotes PC12 cell differentiation, whereas EGF promotes their proliferation (35). We wondered whether the restriction in ERK1/2 signal amplitude and integral strength after PRMT-mediated methylation could affect cellular behavior in response to a particular ligand. To test this, we attempted to mimic the differentiating effect of NGF on PC12 cells with EGF in combination with a methyltransferase inhibitor. As previously noted, both MTA and Prmt5 deletion enhanced EGF-mediated ERK1/2 phosphorylation in PC12 cells (Fig. 2A and fig. S3A). When we compared the effects of EGF, NGF, and EGF plus MTA on ERK1/2 phosphorylation in PC12 cells over a 3-hour time course, we observed that EGF induced a transient phosphorylation of ERK1/2, whereas NGF elicited a sustained signal with a total signal strength 3.2 ± 0.1–fold higher than that induced by EGF (P < 0.0001). MTA pre-treatment of EGF-treated cells induced an increase in ERK1/2 signal amplitude (2.1 ± 0.9–fold versus EGF) with an integrated signal strength 2.21 ± 0.95–fold higher than that induced by EGF alone (P < 0.0001) (Fig. 6A). Furthermore, PC12 cells treated with EGF plus MTA differentiated in a manner almost indistinguishable from that of cells treated with NGF [58.6 ± 10.3% versus 78.9 ± 6.9% neurite-bearing cells (P < 0.0001), with an average neurite length of 90.3 ± 16.4 mm versus 123.2 ± 20.2 mm (P < 0.0001), respectively], whereas MTA alone or EGF treatment had almost no effect on neurite outgrowth [2.3 ± 0.7 and 9.1 ± 0.1% of neurite-bearing cells (P < 0.0001), with an average neurite length of 5.5 ± 2.3 and 8.4 ± 2.9 μm, respectively] (Fig. 6C and fig. S13). These morphological changes correlated with the abundance of the differentiation markers p21WAF1/CIP1 and GAP-43 (Fig. 6C). PC12 cells transfected with the catalytically inactive PRMT5 mutant of (PRMT5ΔGAGRG) also differentiated in response to EGF [30.5 ± 8.9% of neurite-bearing cells in PRMT5 mutant–transfected cells versus 2 ± 1.3% in nontransfected cells (P < 0.001), with an average length of 60.9 ± 1.7 μm] (Fig. 6D).
Fig. 6.
Modulation of signal amplitude affects the biological response to growth factors. PC12 cells were used as a model for differentiation or proliferation in response to NGF or EGF, respectively. (A) Time course of ERK1/2 phosphorylation in response to EGF, EGF + MTA, or NGF. Graph represents the averaged activation profiles of three replicates. (B) Treatment of PC12 cells with the combination of EGF and MTA promotes neuronal differentiation. PC12 cells were treated with either MTA, EGF, MTA + EGF, NGF, or NGF + U0126. Representative images of three different experiments after 96 hours of treatment are shown. Neurite outgrowth and length were scored and plotted after 7 days. Data are means ± SD from n = 3 experiments. (C) Overexpression of catalytically inactive PRMT5ΔGAGRG promotes neuron-like differentiation of PC12 cells in response to EGF. Neurite outgrowth and length were scored and plotted after 4 days. Data are means ± SD from n = 3 experiments. (D) PC12 cells transiently transfected with Flag-CRAFR563K mutant differentiate in response to EGF. Cells transfected with either Flag-CRAFWT (1.2 μg) or Flag-CRAFR563K (0.2 μg) were treated with EGF or NGF for 96 hours. Representative pictures are shown. Transfected cells were identified with an anti-Flag antibody to detect Flag-CRAFR563K (red staining). DNA appears stained in blue (DAPI). Neurite outgrowth and length in transfected cells were scored and plotted after 4 days. Data are means ± SD from at least 100 transfected cells.
As noted, CRAFR563K increased ERK1/2 phosphorylation in HEK293 cells, thereby mimicking the response to growth factors in PRMT5-depleted cells (Fig. 4C). We found that PC12 cells expressing the CRAFR563K mutant underwent differentiation when treated with EGF [64.8 ± 14.7% versus 8.2 ± 2.8% neurite-bearing cells (P < 0.0001), with an average neurite length of 87 ± 3 μm in EGF-treated CRAFR563K-expressing cells (P < 0,0001)] (Fig. 6E). We also observed this effect in PC12 cells transfected with the BRAFR671K mutant, although this mutant, which has a higher basal kinase activity, increased the number of neurite-bearing cells even in the absence of EGF [87.3 ± 13.4% EGF-treated BRAFR671K-expressing cells versus 30.2 ± 9.2% EGF-treated BRAFWT-transfected cells (P < 0.0001), with an average neurite length of 90.5 ± 21.7 μm and 51.1 ± 9.4 μm, respectively] (fig. S12). Moreover, the degree of BRAF methylation induced by NGF versus EGF (fig. S9A) correlated with the activation of PRMT5 activity in response to growth factors (Fig. 2C) and the sustained p-ERK1/2 signal promoted by NGF (Fig. 6A). Together, our data indicate that restriction of ERK1/2 signal amplitude through the methylation of RAF proteins represents a central mechanism in determining the response to growth factors.
DISCUSSION
Activation of the MAPKs ERK1/2 through the RAS signaling pathway plays a crucial role in cell proliferation, differentiation, survival, and oncogenic transformation. Various growth factors that activate this pathway elicit distinct biological responses, with the temporal pattern of ERK1/2 phosphorylation determining which response occurs (4, 5, 36–38). Here, we identify methylation of RAF proteins as a mechanism for modulating RAS-ERK1/2 pathway signaling in response to growth factors. We identify PRMT5 as the arginine methyltransferase involved in limiting ERK1/2 phosphorylation and identify a specific PRMT5 methylation motif (GRG) that is conserved in RAF proteins during evolution. The arginine residue in this motif is involved in the limitation of the kinase activity and the stability of CRAF, thereby contributing to ERK1/2 signal output. We found that inhibiting methylation could redirect growth factor–elicited responses from proliferation to differentiation, indicating that methylation plays an important role in the response to growth factors. The deregulation of this mechanism might be particularly relevant in a number of solid tumors and hematologic malignancies deficient in the methylthioadenosine phosphorylase gene (MTAP) (39, 40). MTAP regulates the intracellular amounts of naturally occurring MTA, which accumulates and is secreted from MTAP-deficient cells (41, 42).
The ability of methylation to modify the response to growth factors was RAS-ERK1/2 pathway–specific, because PI3K-AKT signaling was not affected. Activation of PI3K by HGF occurs in a RAS-independent manner (24), supporting the specificity of the pathway modulation. It is well accepted that, in general, kinases control the amplitudes more than the duration of signals, whereas phosphatases tend to control both (4, 5). Three metrics describe the transient ERK1/2 phosphorylation profile: am plitude, duration, and integrated output (4, 5). Most reactions in the pathway do not affect any of these. RAF proteins are involved in the molecular processes that were identified as important to defining the final amplitude, duration, and integrated response of the signaling output (4, 5). Our data indicate that RAF methylation appears to be a critical posttranslational modification controlling downstream activity, supporting the role of RAF proteins in regulating ERK1/2 phosphorylation profiles. In some cases, the ERK1/2 activation profile also might be determined by the preferential use of a RAF isoform (that is, the preferential signaling of FGF through ARAF) (fig. S8) or by the specific signaling protein complexes used by a particular receptor in a particular cell type.
Whereas lysine methylation is mainly confined to the nucleus, methylation of arginine residues appears to occur throughout the cell (12, 13). Although PRMT1 is responsible for 85% of methylation reactions (43), our results identify PRMT5 as the enzyme involved in restricting ERK1/2 phosphorylation. Thus, PRMT5 limits ERK1/2 signal amplitude in response to growth factors, suggesting that methylation could facilitate the appropriate cellular response to a specific stimulus.
The Shk1 protein kinase, a homolog of Saccharomyces cerevisiae Ste20 and mammalian PAKs, is an essential component of a Ras- and Cdc42-dependent signaling cascade required for cell viability, normal morphology, and MAPK-mediated sexual responses in the fission yeast (23). Ternary complexes between Skb1 (PRMT5 yeast homolog), Shk1 (PAK yeast homolog), and Cdc42 have been described (23), supporting the possible involvement of PRMT5 in the RAS signaling pathway. Indeed, we found that the activity of PRMT5 was regulated by growth factors. RAF activation by RAS is a highly complex process (10, 44). We found that either inhibition of protein methylation or depletion of PRMT5 enhanced the induction of CRAF’s catalytic activity by specific growth factors, identifying a previously unknown level of regulation for RAF proteins. The basal activity of CRAF proteins is not affected by either MTA or PRMT5 depletion, indicating that modulation of CRAF kinase activity by PRMT5 depends on RAS activation. Thus, we observed that MTA modulates the ERK1/2 signal amplitude in NRASQ61L mutant cells and not in BRAFV600E mutant cells, which signal independently of RAS (30) (fig. S5). In vitro experiments demonstrate that PRMT5 binds to active CRAF and not inactive CRAF. These data confirm the requirement for RAS (a ligand-dependent process) to engage and probably activate PRMT5 in the downstream signaling machinery. The role of protein arginine methylation as posttranslational modification controlling RAS ERK1/2 signaling pathway is supported by the detection of PRMT5 and CRAF in the same complexes. Indeed, PRMT5 has been described as an HSP90 client, found in association with CRAF or BRAF (19), fostering the notion that these molecules can exist within the same protein complex.
PRMT5, the most highly conserved methyltransferase, methylates arginines symmetrically within GRG motifs (11). Our results indicate that PRMT5 methylates CRAF at Arg563. In agreement with a role for PRMT5 in limiting ERK1/2 output in RAS pathway signaling, CRAF methylation is increased by exposure to growth factors, and depletion of PRMT5 or mutation of CRAFR563 abolishes this effect. Conservative substitution of this arginine with a lysine increased the stability of CRAF and BRAF, suggesting that methylation of this arginine decreases the stability of active RAF molecules. Consistent with this mechanism, the basal activity of CRAFR563K mutant does not differ from that of wild-type CRAF; however, ERK1/2 phosphorylation in response to a growth factor is exaggerated. These data, and similar results obtained with BRAF, suggest that methylation targets active RAF molecules for degradation and that lack of methylation increases the stability of the RAF active molecules, thereby increasing the RAF signal. Consistent with this, we found that CRAF protein was stabilized in the presence of MTA and was highly phosphorylated at Ser621, a modification that has been related to CRAF stabilization (32).
We tested the hypothesis that protein methylation is a critical posttranslational modification in growth factor–elicited biological responses with the PC12 model of cellular differentiation (45). A particular growth factor will not only initiate the signals that lead to activation of a kinase (thereby affecting signal amplitude) but also activate the mechanisms that deactivate that signal (thereby affecting signal duration). When PRMT5 is inhibited in PC12 cells, the increase in ERK1/2 phosphorylation in response to EGF (signal amplitude) overrides the phosphatase activity triggered by EGF, resulting in a long-lasting signal that reaches the threshold required to elicit neuronal differentiation. Consistent with the proposed mechanism, NGF induced less methylation of BRAF [the major RAF isoform participating in the PC12 differentiation process (34)] than did EGF in wild-type cells, but similar to that induced by EGF in Prmt5-depleted cells (fig. S9A). CRAFR563K and BRAFR671K mutants also promoted PC12 cell differentiation in response to EGF, supporting the critical role of RAF proteins in controlling the ERK1/2 phosphorylation profile and, consequently, the biological outcome.
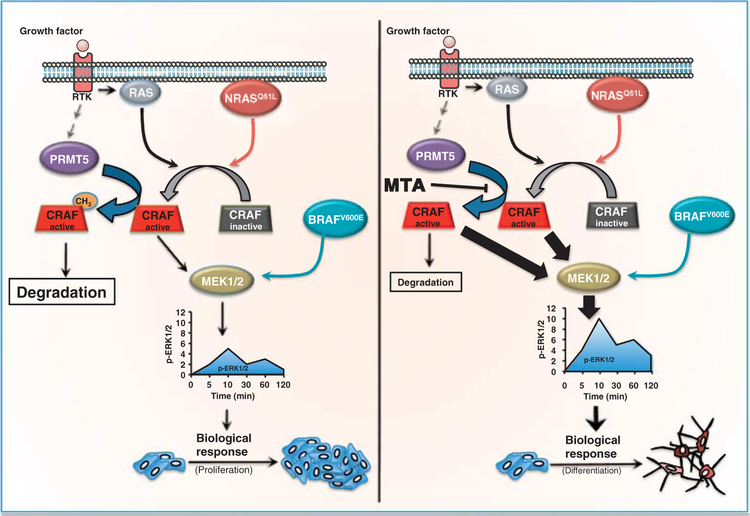
In summary, we show that protein arginine methylation limits signal transduction in response to growth factors. We demonstrate that PRMT5 mediates the methylation of RAF, thereby limiting downstream activity in the RAS-MAPK pathway (Fig. 7). This provides a mechanism whereby cells can modulate the temporal profile of ERK1/2 phosphorylation. Because cancer cells rely heavily on oncogenic signaling through the RASERK1/2 pathway (46), this additional level of regulation may provide additional targets for therapeutic intervention.
Fig. 7.
Model for the limitation of the ERK1/2 signal by PRMT5. Growth factors or mutated RAS activate CRAF and thereby the MAPK signaling pathway. PRMT5 promotes CRAF degradation and limits its catalytic activity, reducing the activation of downstream kinases, such as MEK1/2 and ERK1/2. In contrast, PRMT5 does not affect downstream signals in cells with BRAFV600E, which are independent of RAS. Decreasing PRMT5 activity (pharmacologically or by its knockdown) increases the stability of activated CRAF, increasing the amplitude of the ERK1/2 signal and the integrated signal strength and consequently affecting the evoked biological response.
MATERIALS AND METHODS
Reagents
c-Met inhibitor PHA-665752 (200 nM) was from Pfizer. MTA, Deaza, c-leucine, cycloheximide (CHX), chloramphenicol, anti-Flag (M2), and anti-Flag (M2) resin were purchased from Sigma-Aldrich. Anti-MKP1 (v-15), anti–lamin A/C (N-18), anti-PRMT5 (C-20), anti-PRMT1 (B-2), c-Met, anti-CRAF (C-12), anti–GAP-43 (B-5), and anti-ERK2 (C-14) were purchased from Santa Cruz Biotechnology. Anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase), used to demonstrate equal loading in Western blots, was from Trevigen. Anti–MAPK p-ERK1/2, anti–p-MEK1/2, anti–p-AKT, anti-ERK1/2, anti-MEK1/2, anti-AKT, anti–p-c-Met (Tyr1234/1235), and U0126 were from Cell Signaling. Anti-RAS anti-p21WAF1/CIP1 and anti-Sam68 were from BD Biosciences, Pharmingen. L-[methyl-3 H]Methionine (70 to 85 Ci/mmol), S-adenosyl- L-[methyl-3H]methionine, and [35 S]methionine were obtained from GE Healthcare.
Constructs
pcDNA3.1-Flag-CRAF was provided by A. Shaw (Washington University, St. Louis, Missouri). pEF2-Flag-PRMT5 and pEF2-PRMT5ΔGAGRG mutant expression vectors were a gift from S. Petska (R. W. Johnson Medical School, New Brunswick, New Jersey). pcDNA3.1-Flag-CRAFR563K and pLNCX2-myc-BRAFR671K mutants were generated with a QuikChange Site-Directed Mutagenesis kit from Stratagene.
Cell culture, transfection, and growth factor stimulation
The 37–31E and 37–31T mouse melanoma cell lines have been previously described (47). B16F1, HEK293, PC12, U87, A375, and Cos-7 cells were obtained from the American Type Culture Collection. B16F1, HEK293, A375, and U87 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) containing 10% fetal bovine serum (FBS) (Invitrogen) and penicillin/streptomycin (P/S) [P (100 U/ml)/S (100 μg/ml)] (Invitrogen). Cos-7 cells were grown in RPMI 1640 (Invitrogen) plus 10% FBS and P/S. PC12 cells were maintained in F12K medium (Invitrogen) containing 10% horse serum (Invitrogen), 5% FBS, and P/S. SKMel147 human melanoma cells were a gift from M. S. Soengas (Centro Nacional de Investigaciones Oncológicas, Madrid, Spain). UACC903 human melanoma cells were a gift from P. Pollock (TGen). Wild-type MEFs were provided by M. Baccarini (Max F. Perutz Laboratories, Austria). For growth factor treatments, cells were starved for 3 hours or overnight depending on cell requirements. MTA (0.4 mM) and Deaza (20 mM) were added 3 hours before addition of growth factors. c-Leucine (20 mM) was added overnight before growth factor treatment. Cells were stimulated with HGF (40 ng/ml), FGF1 (40 ng/ml), and FGF2 (40 ng/ml) (R&D Systems), EGF (50 ng/ml) (Millipore), insulin (100 nM), MSH-α (100 nM), NGF, or TPA (200 nM) (Sigma-Aldrich) and harvested at the times indicated. Constructs were transfected into Cos-7, PC12, B16F1, or HEK293 cells with Lipofectamine (Invitrogen), following the manufacturer’s recommended protocol.
Isolation of primary rat hepatocytes
Primary rat hepatocytes were isolated as previously described (48). Cells were plated onto collagen-coated six-well dishes (type I collagen from rat tail; Collaborative Biomedical) at 106 cells per dish. Cultures were maintained in minimal essential medium supplemented with 5% FBS, non- essential amino acids, 2 mM glutamine, and antibiotics (Invitrogen). After a 6-hour incubation, the culture medium was removed and replaced with the same medium without FBS. Where indicated, cells were pre-treated with MTA for 3 hours and then exposed to HGF for various periods of time.
Western blot and immunoprecipitation
Cells were lysed with M-PER (Pierce) in the presence of protease inhibitor cocktail (Roche). Fifty micrograms of protein was separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) using 4 to 20% or 12% gels (Invitrogen). Proteins were transferred to polyvinylidene difluoride membranes and probed with primary antibodies. Proteins were visualized with secondary antibodies coupled to horseradish peroxidase enhanced chemiluminescence (GE Healthcare) or SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Immunoprecipitations were performed in M-PER lysis buffer (Pierce) after the addition of Protein A/G Agarose PLUS (Santa Cruz Biotechnology) or anti–Flag resin (Sigma-Aldrich). Samples were fractionated and visualized as described above. After stripping with 5 mM tris-HCl (pH 2), membranes were blocked and incubated with other antibodies as indicated.
Quantification of p-ERK1/2 (fold induction with respect to the untreated cells) in Fig. 4C and fig. S10A was performed by dividing the densitometric value obtained for p-ERK1/2 by the densitometric value obtained from all the proteins (Ponceau S staining) from the same lane. Then, the resulting number was divided by the number obtained in the untreated cells transfected with the wild-type isoform.
Immunofluorescence
Cells were grown on glass coverslips. Briefly, after treatment, cells were washed in cold phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde in PBS for 10 min, and permeabilized with 0.1% Triton X-100 in PBS for 5 min. Cells were blocked with PBS containing 3% bovine serum albumin (BSA) and then incubated with the primary antibody overnight at 4°C. Proteins were visualized by confocal microscopy (Espectral FV1000, Olympus) with fluorescein isothiocyanate–or phycoerythrinconjugated secondary antibodies. DAPI (4′,6-diamidino-2-phenylindole) was used for nuclear staining.
RAF kinase assays
Coupled RAF kinase assays were performed with immunoprecipitated ARAF, BRAF, or CRAF as previously described (49). Briefly, immunoprecipitated CRAF was incubated with 20 μl of MKK buffer [30 mM tris (pH 7.5), 0.1 mM EDTA, 10 mM MgCl2, 0.1% (v/v) Triton X-100, 5 mM NaF, 0.2 mM Na3VO4, 0.3% (v/v) β-mercaptoethanol, GST-MKK (6.5 μg/ml), GST-ERK (100 μg/ml), and 0.8 mM adenosine 5′-triphosphate (ATP)] and incubated at 30°C for 10 to 30 min. Reactions were terminated by the addition of 20 μl of KILL buffer [30 mM tris (pH 7.5), 6 mM EDTA, 0.1% (v/v) Triton X-100, 5 mM NaF, 0.2 mM Na3VO4, and 0.3% (v/v) β-mercaptoethanol]. Five microliters of reaction supernatant was incubated in triplicate with 25 μl of myelin basic protein (MBP) buffer [50 mM tris (pH 7.5), 0.1 mM EDTA, 10 mM MgCl2, 0.1% (v/v) Triton X-100, 5 mM NaF, 0.2 mM Na3VO4, 0.1 mM ATP, 0.3% β-mercaptoethanol, BSA (200 μg/ml), MBP (1 mg/ml), 50 μCi [γ−32P]ATP (5000 Ci/mmol)] for 10 min before blotting onto P81 paper and placing in a 25 mM orthophosphate solution. P81 paper was washed extensively and the amount of [γ−32P]ATP incorporated into MBP was measured by Cherkenov counting. All experiments were performed in triplicate. ERK kinase activity was measured with MBP as a substrate. When MEK was used as the unique CRAF substrate, the kinase assays were resolved by SDS-PAGE and Western blots were performed against p-MEK1/2 and MEK1/2. All experiments were performed at least in triplicate.
Nuclear extracts
Nuclear extracts were obtained with an N-PER kit (Pierce) following the manufacturer’s instructions.
Pulse chase
Nontransfected and PRMT5 siRNA–transfected HeLa cells were incubated for 30 min in a CO2 incubator in 5 ml of DMEM without Met and Cys. Then, 2 ml of fresh DMEM without Met and Cys containing 10% dialyzed FBS and [35S]methionine (250 μCi/ml) were added to each dish and incubated for 30 min. Cells were washed once with warm PBS, incubated with complete DMEM, and then chased for 24 hours. In experiments performed with CHX, cells were treated with CHX (5 μg/ml) and chased at the times indicated. Immunoprecipitation of the indicated proteins was performed with 500 μg of total protein.
In vivo methylation assays
In vivo methylation assays were performed as described (50, 51). Briefly, cells were incubated with CHX (100 μg/ml) and chloramphenicol (40 g/ml) in complete medium for 30 min. The medium was then replaced with DMEM without methionine, supplemented with penicillin, streptomycin, and 10% fetal calf serum containing L-[methyl-3 H]methionine (10 μCi/ml), and the methyl donor and cells were incubated for an additional 3 hours in the presence of the same protein synthesis inhibitors. The cells were lysed in 10 mM tris-HCl–100 mM NaCl–2.5 mM MgCl2 (pH 7.4) containing 0.5% Triton X-100, 1% aprotinin, leupeptin (2 μg/ml), pepstatin A (2 μg/ml), and 0.1 mM dithiothreitol (DTT), and proteins of interest were immunoprecipitated with the appropriate antibodies. For experiments with PC12 cells, siRNA transfection was performed 50 to 60 hours before the experiment. For experiments with HeLa or HEK293 cells, complementary DNA (cDNA) constructs and siRNAs were transfected 24 hours before metabolic labeling.
In vitro methylation assays
Flag-PRMT5 was expressed in HeLa cells and immunopurified with Flag resin (Sigma-Aldrich). Enzymes immobilized in the resin were then incubated with either 4 μg of MBP (Sigma-Aldrich), 100 μg of unmodified CRAF peptide (FMVGRGYAS), or 100 μg of modified CRAF peptide (FMVGKGYAS) (GeneScript Inc.) in the presence of 10 μCi of S-adenosyl-L-[methyl-3 H]methionine for 1 hour at 30°C in a final volume of 50 μl of methylation buffer [100 mM tris-HCl (pH 8.0), 1 mM EDTA, and 1 mM DTT]. Peptides were purified with ZipTip C18 Pipette Tips (Millipore). Five micrograms of peptide was loaded onto P81 papers, washed, and counted by liquid scintillation. Ten micrograms of peptide was analyzed by matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF)/TOF mass spectrometry on an Autoflex instrument (Bruker). When MBP was used as substrate, the whole reaction was resolved by SDS-PAGE. Then, the3H signal was captured by exposing the gels to a film.
In vitro binding assays
Recombinant PRMT5 (2 μg; Novus Biologicals) and recombinant full-length inactive GST-CRAF (2 μg; Sigma-Aldrich) or active GST-CRAF (N-terminal GST-tagged, residues 306 to end) (2 μg; GeneScript Inc.) were incubated for 1 hour at 30°C in a final volume of 50 μl of methylation buffer [100 mM tris-HCl (pH8.0), 1 mM EDTA, and 1 mMDTT]. Then, PRMT5 was immunoprecipitated and CRAF was detected by immunoblotting.
Phosphatase activity assays
The MAPK phosphatases 1 and 2 (MPK1/2) were immunoprecipitated from nuclear extracts, and phosphatase activity was measured with Ser/Thr Phosphatase Assay Kit 1 (K-R-pT-I-R-R) (Millipore) according to the manufacturer’s protocol.
siRNA transfection
PRMT1 (52), PRMT5 (53), CRAF (30), and scrambled siRNA were purchased from Dharmacon. Cells were transfected with siRNAs (100 nM final concentration) using Lipofectamine 2000. The assays were performed 48 to 72 hours after transfection.
PC12 cell differentiation assay
The assay was performed as previously described (54). Briefly, PC12 cells were treated with EGF, NGF, EGF plus MTA, or NGF plus U0126 for 7 days. Cells with neurites >20 μm in length were considered neurite-bearing. At least 30 fields measured at 20× magnification were counted per condition. Average neurite length was calculated with ImageJ64 software where at least 100 cells from each experimental condition were scored from randomly chosen fields.
Liquid chromatography–mass spectrometry analysis
Samples of interest were separated on a 10% SDS-PAGE gel, and the gel was stained with colloidal Coomassie blue. Protein bands of interest were cut from the gel and digested with modified porcine trypsin (Trypsin Gold, Promega). Digests were analyzed on an Esquire HCT IT mass spectrometer (Bruker) coupled to a nano-HPLC (high-performance liquid chromatography) system (Ultimate, LC Packings) as described (55).
Statistics
All quantitative results are presented as the means ± SD of independent experiments. Statistical differences between two groups of data were analyzed by Student’s t test. After testing the nonparametric distribution of PC12 neurite-outgrowth quantification data, we performed a Mann-Whitney test to analyze the statistical differences among the different groups.
Supplementary Material
Acknowledgments:
We thank J. Seoane, J. C. Rodriguez-Manzanque, and C. Jhappan for useful discussions and reading the manuscript.
Funding: This work was supported in part by Instituto de Salud Carlos III grants PI-050227 and PI-080653 (J.A.R.) and ISCIII-RTICC RD06/00200061 PI070392 and PI10/00038 (M.A.A.), Marie Curie Reintegration grant MIRG-CT-2005–029135 (J.A.R.), Fundación Mutua Madrileña and Fundación Segunda Opinión en Oncología (FUSEON) (J.A.R.), Ministerio de Educación y Ciencia SAF 2004–03538 (M.A.A.), UTE project CIMA (M.A.A.), and the Intramural Research Program of the NIH, National Cancer Institute, and Center for Cancer Research (G.M.).
Footnotes
Competing interests: The authors declare that they have no competing interests.
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/4/190/ra58/DC1
Fig. S1. MTAincreasesERK1/2 phosphorylation in response toHGFin primary rat hepatocytes.
Fig. S2. Modulation of HGF induced MEK1/2-ERK1/2 signaling by MTA in melanoma cells.
Fig. S3. MTA modulates ERK1/2 phosphorylation upon specific growth factor treatment in human cells, and protein methylation inhibition modulates ERK1/2 signal amplitude in response to HGF.
Fig. S4. PRMT1, PRMT3, and PRMT8 are not responsible for the MTA-mediated effect.
Fig. S5. MTA signal amplitude modulation of ERK1/2 is RAS activation–dependent.
Fig. S6. CRAF and BRAF immunoprecipitate and colocalize with PRMT5 in the cytoplasm.
Fig. S7. MTA regulates growth factor–induced BRAF kinase activity.
Fig. S8. Differential activation of RAF isoforms by growth factors.
Fig. S9 MTA treatment inhibits CRAF degradation in SKMel147 cells.
Fig. S10. BRAFR671K mutant shows the same response as CRAFR563K. ARAF stability is not regulated by MTA in SKMel147 cells.
Fig. S11. A small fraction of total CRAF is recruited to the membrane in response to growth factor signaling. Detection by MALDI of both the unmethylated and the methylated CRAF peptides.
Fig. S12. BRAF is in vivo methylated. BRAFR671K mutant mimics the MTA-induced effects on PC12 cells.
Fig. S13. PC12 differentiation assay.
Methods
References
REFERENCES AND NOTES
- 1.Lewis TS, Shapiro PS, Ahn NG, Signal transduction through MAP kinase cascades. Adv. Cancer Res. 74, 49–139 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Human gene symbols are italicized, with all letters in uppercase. Protein designations are the same as the gene symbol, but are not italicized; all letters are in uppercase. mRNAs and cDNAs use the same formatting conventions as the gene symbol. Mouse and rat gene symbols are italicized, with only the first letter in uppercase and the remaining letters in lowercase. Protein designations are the same as the gene symbol, but are not italicized, all uppercase letters.
- 3.Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ, Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell 90, 859–869 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Hornberg JJ, Binder B, Bruggeman FJ, Schoeberl B, Heinrich R, Westerhoff HV, Control of MAPK signalling: From complexity to what really matters. Oncogene 24, 5533–5542 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Hornberg JJ, Bruggeman FJ, Binder B, Geest CR, de Vaate AJ, Lankelma J, Heinrich R, Westerhoff HV, Principles behind the multifarious control of signal transduction. ERK phosphorylation and kinase/phosphatase control. FEBS J. 272, 244–258 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Perkins ND, Post-translational modifications regulating the activity and function of the nuclear factor kB pathway. Oncogene 25, 6717–6730 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Okada T, Masuda T, Shinkai M, Kariya K, Kataoka T, Post-translational modification of H-Ras is required for activation of, but not for association with, B-Raf. J. Biol. Chem 271, 4671–4678 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, Morales T, Aliagas I, Liu B, Sideris S, Hoeflich KP, Jaiswal BS, Seshagiri S, Koeppen H, Belvin M, Friedman LS, Malek S, RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 464, 431–435 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N, RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464, 427–430 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, Marais R, Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 140, 209–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McBride AE, Silver PA, State of the Arg: Protein methylation at arginine comes of age. Cell 106, 5–8 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Bedford MT, Richard S, Arginine methylation an emerging regulator of protein function. Mol. Cell 18, 263–272 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Bedford MT, Clarke SG, Protein arginine methylation in mammals: Who, what, and why. Mol. Cell 33, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, Clarke S, PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem 276, 32971–32976 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Lee YH, Stallcup MR, Minireview: Protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol. Endocrinol 23, 425–433 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richard S, Morel M, Cleroux P, Arginine methylation regulates IL-2 gene expression: A role for protein arginine methyltransferase 5 (PRMT5). Biochem. J 388, 379–386 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amente S, Napolitano G, Licciardo P, Monti M, Pucci P, Lania L, Majello B, Identification of proteins interacting with the RNAPII FCP1 phosphatase: FCP1 forms a complex with arginine methyltransferase PRMT5 and it is a substrate for PRMT5-mediated methylation. FEBS Lett. 579, 683–689 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Friesen WJ, Wyce A, Paushkin S, Abel L, Rappsilber J, Mann M, Dreyfuss G, A novel WD repeat protein component of the methylosome binds Sm proteins. J. Biol. Chem 277, 8243–8247 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Maloney A, Clarke PA, Naaby-Hansen S, Stein R, Koopman JO, Akpan A, Yang A, Zvelebil M, Cramer R, Stimson L, Aherne W, Banerji U, Judson I, Sharp S, Powers M, deBilly E, Salmons J, Walton M, Burlingame A, Waterfield M, Workman P, Gene and protein expression profiling of human ovarian cancer cells treated with the heat shock protein 90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 67, 3239–3253 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Rho J, Choi S, Seong YR, Cho WK, Kim SH, Im DS, PRMT5, which forms distinct homo-oligomers, is a member of the protein-arginine methyltransferase family. J. Biol. Chem 276, 11393–11401 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Pollack BP, Kotenko SV, He W, Izotova LS, Barnoski BL, Pestka S, The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J. Biol. Chem 274, 31531–31542 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Tanaka H, Hoshikawa Y, Oh-hara T, Koike S, Naito M, Noda T, Arai H, Tsuruo T, Fujita N, PRMT5, a novel TRAIL receptor-binding protein, inhibits TRAIL-induced apoptosis via nuclear factor-κB activation. Mol. Cancer Res. 7, 557–569 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Gilbreth M, Yang P, Wang D, Frost J, Polverino A, Cobb MH, Marcus S, The highly conserved skb1 gene encodes a protein that interacts with Shk1, a fission yeast Ste20/PAK homolog. Proc. Natl. Acad. Sci. U.S.A 93, 13802–13807 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF, Met, metastasis, motility and more. Nat. Rev. Mol. Cell Biol. 4, 915–925 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Martínez-Chantar ML, Latasa MU, Varela-Rey M, Lu SC, García-Trevijano ER, Mato JM, Avila MA, L-methionine availability regulates expression of the methionine adenosyltransferase 2A gene in human hepatocarcinoma cells: Role of S-adenosylmethionine. J. Biol. Chem 278, 19885–19890 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D, Regulation of p53 activity through lysine methylation. Nature 432, 353–360 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Martin C, Zhang Y, The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6, 838–849 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, Viale A, Reinberg D, Wülfing C, Tarakhovsky A, Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell 121, 425–436 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Krause CD, Yang ZH, Kim YS, Lee JH, Cook JR, Pestka S, Protein arginine methyltransferases: Evolution and assessment of their pharmacological and therapeutic potential. Pharmacol. Ther 113, 50–87 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Dumaz N, Hayward R, Martin J, Ogilvie L, Hedley D, Curtin JA, Bastian BC, Springer C, Marais R, In melanoma RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res. 66, 9483–9491 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Sharp SY, Prodromou C, Boxall K, Powers MV, Holmes JL, Box G, Matthews TP, Cheung KM, Kalusa A, James K, Hayes A, Hardcastle A, Dymock B, Brough PA, Barril X, Cansfield JE, Wright L, Surgenor A, Foloppe N, Hubbard RE, Aherne W, Pearl L, Jones K, McDonald E, Raynaud F, Eccles S, Drysdale M, Workman P, Inhibition of the heat shock protein 90 molecular chaperone in vitro and in vivo by novel, synthetic, potent resorcinylic pyrazole/isoxazole amide analogues. Mol. Cancer Ther. 6, 1198–1211 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Noble C, Mercer K, Hussain J, Carragher L, Giblett S, Hayward R, Patterson C, Marais R, Pritchard CA, CRAF autophosphorylation of serine 621 is required to prevent its proteasome-mediated degradation. Mol. Cell 31, 862–872 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hallberg B, Rayter SI, Downward J, Interaction of Ras and Raf in intact mammalian cells upon extracellular stimulation. J. Biol. Chem 269, 3913–3916 (1994). [PubMed] [Google Scholar]
- 34.Kao S, Jaiswal RK, Kolch W, Landreth GE, Identification of the mechanisms regulating the differential activation of the MAPK cascade by epidermal growth factor and nerve growth factor in PC12 cells. J. Biol. Chem 276, 18169–18177 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Traverse S, Gomez N, Paterson H, Marshall C, Cohen P, Sustained activation of the mitogen-activated protein (MAP) kinase cascade may be required for differentiation of PC12 cells. Comparison of the effects of nerve growth factor and epidermal growth factor. Biochem. J 288, 351–355 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall CJ, Specificity of receptor tyrosine kinase signaling: Transient versus sustained extracellular signal–regulated kinase activation. Cell 80, 179–185 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Tombes RM, Auer KL, Mikkelsen R, Valerie K, Wymann MP, Marshall CJ, McMahon M, Dent P, The mitogen-activated protein (MAP) kinase cascade can either stimulate or inhibit DNA synthesis in primary cultures of rat hepatocytes depending upon whether its activation is acute/phasic or chronic. Biochem. J 330, 1451–1460 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH, Mitogen-activated protein (MAP) kinase pathways: Regulation and physiological functions. Endocr. Rev 22, 153–183 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, Chen ZH, Savarese TM, Codeletion of the genes for p16INK4, methylthioadenosine phosphorylase, interferon-α1, interferon-β1, and other 9p21 markers in human malignant cell lines. Cancer Genet. Cytogenet 86, 22–28 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Behrmann I, Wallner S, Komyod W, Heinrich PC, Schuierer M, Buettner R, Bosserhoff AK, Characterization of methylthioadenosin phosphorylase (MTAP) expression in malignant melanoma. Am. J. Pathol 163, 683–690 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Avila MA, Garcia-Trevijano ER, Lu SC, Corrales FJ, Mato JM, Methylthioadenosine. Int. J. Biochem. Cell Biol. 36, 2125–2130 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Stevens AP, Dettmer K, Wallner S, Bosserhoff AK, Oefner PJ, Quantitative analysis of 5′-deoxy-5′-methylthioadenosine in melanoma cells by liquid chromatography-stable isotope ratio tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 876, 123–128 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, Clarke S, Herschman HR, PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem 275, 7723–7730 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Wellbrock C, Karasarides M, Marais R, The RAF proteins take centre stage. Nat. Rev. Mol. Cell Biol. 5, 875–885 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Santos SD, Verveer PJ, Bastiaens PI, Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat. Cell Biol. 9, 324–330 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Roberts PJ, Der CJ, Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 26, 3291–3310 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Esteve-Puig R, Canals F, Colomé N, Merlino G, Recio JA, Uncoupling of the LKB1-AMPKa energy sensor pathway by growth factors and oncogenic BRAFV600E. PLoS One 4, e4771 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.García-Trevijano ER, Martínez-Chantar ML, Latasa MU, Mato JM, Avila MA, NO sensitizes rat hepatocytes to proliferation by modifying S-adenosylmethionine levels. Gastroenterology 122, 1355–1363 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, Marais R; Cancer Genome Project, Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116, 855–867 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Liu Q, Dreyfuss G, In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol 15, 2800–2808 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abramovich C, Yakobson B, Chebath J, Revel M, A protein-arginine methyltransferase binds to the intracytoplasmic domain of the IFNAR1 chain in the type I interferon receptor. EMBO J. 16, 260–266 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, David M, Arginine methylation of STAT1 modulates IFNa/b-induced transcription. Cell 104, 731–741 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, La Thangue NB, Arginine methylation regulates the p53 response. Nat. Cell Biol. 10, 1431–1439 (2008). [DOI] [PubMed] [Google Scholar]
- 54.New L, Li Y, Ge B, Zhong H, Mansbridge J, Liu K, Han J, SB203580 promotes EGF-stimulated early morphological differentiation in PC12 cell through activating ERK pathway. J. Cell. Biochem 83, 585–596 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Colomé N, Collado J, Bech-Serra JJ, Liiv I, Anton LC, Peterson P, Canals F, Jaraquemada D, Alvarez I, Increased apoptosis after autoimmune regulator expression in epithelial cells revealed by a combined quantitative proteomics approach. J. Proteome Res. 9, 2600–2609 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.