Abstract
The primary function of the genome is to store, propagate, and express the genetic information that gives rise to a cell’s architectural and functional machinery. However, the genome is also a major structural component of the cell. Besides its genetic roles, the genome affects cellular functions by nongenetic means through its physical and structural properties, particularly by exerting mechanical forces and by serving as a scaffold for binding of cellular components. Major cellular processes affected by nongenetic functions of the genome include establishment of nuclear structure, signal transduction, mechanoresponses, cell migration, and vision in nocturnal animals. We discuss the concept, mechanisms, and implications of nongenetic functions of the genome.
In eukaryotic cells, the genome is enclosed within the nucleus, where it is organized into chromatin fibers and separated from the cytoplasm by the nuclear membrane. The compaction of the chromatin fiber and its global organization within the confines of the nucleus play major roles in regulating the expression and propagation of the genetic information encoded in the genome (1, 2). The chromatin fiber shapes and controls the cell’s architectural and functional machinery by regulating gene expression. In addition to being the carrier of genetic information, the genome is also a major structural entity of the nucleus and, as such, affects cellular structure and function.
A diploid mammalian cell contains approximately 2 m of linear DNA, compressed in the form of chromatin into a nucleus of typically 10 mm in diameter. Chromatin is a linear polymer in which the DNA is wrapped around octameric histones to generate nucleosomes and form a fiber that resembles beads on a string. The chromatin fiber is further organized into several levels of higher-order structures by electrostatic, hydro-static, and elastic interactions along the fiber and between genome regions on distinct chromosomes (3) (Fig. 1). Short-range interactions among nucleosomes shape the linear chromatin fiber into higher-order domains, which are maintained by forces in the piconewton range (4). Longer-range interactions along the fiber, facilitated by chromatin-bound proteins, promote the formation of larger domains and ultimately give rise to whole chromosomes (5) (Fig. 1). The polymeric properties of the chromatin fiber and their role in generating and stabilizing higher-order chromatin organization are not fully understood, and multiple polymer folding and interaction models have been proposed to explain higher-order genome architecture (3, 6).
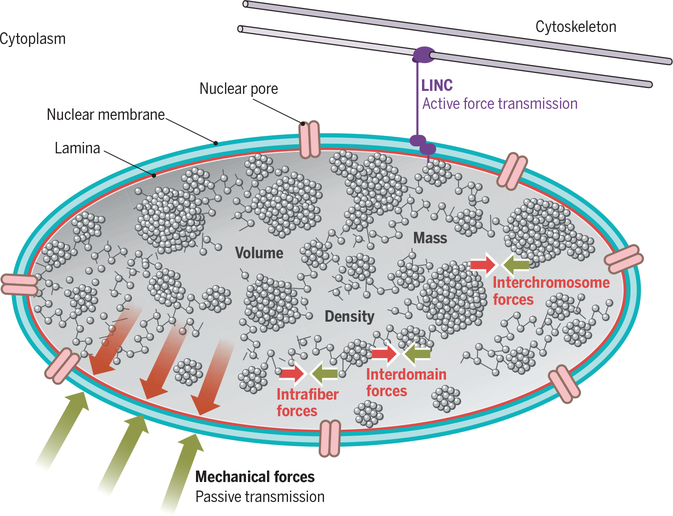
Fig. 1. The genome as a physical entity.
In eukaryotes, the genome is housed in the cell nucleus, which is separated from the cytoplasm by a double membrane (blue) supported by a network of intermediate filament proteins of the lamin family (red). The genome is a prominent physical entity with considerable mass, volume, and density. Transport between the nucleus and the cytoplasm occurs via nuclear pores (pink). DNA is folded into higher-order chromatin domains and ultimately chromosomes. The genome exerts, and is exposed to, mechanical forces transmitted into and out of the nucleus, either passively (red, green arrows) or actively via the membrane-spanning LINC protein complex (purple). The genome also exerts and is exposed to intranuclear forces (red, green arrows) via intrafiber, intrachromosomal, and interchromosomal interactions.
The physical features of the genome affect cellular structure and function. As a result of the mass of the genome, the inherent motion of the chromatin fiber (which undergoes fluctuations in the range of 1 to 2 μm) (6, 7), and biologically mediated chromatin reorganization events (for example, during remodeling of chromatin as part of activation of a gene), the genome exerts considerable mechanical forces, both within the nucleus and onto its cellular surroundings (Fig. 1). These mechanical forces affect cellular functions by nongenetic means. Dynamic interactions between neighboring chromatin domains, either on the same or on different chromosomes, generate intranuclear mechanical forces that propagate toward the cytoplasm, particularly at sites where chromatin contacts the nuclear envelope (Fig. 1). Conversely, the genome itself is exposed to mechanical forces emanating from the cytoplasm (8, 9), which may be transmitted either passively at contact points between the nuclear envelope and chromatin, or actively via the LINC (linker of nucleoskeleton and cytoskeleton) protein complex, which forms a bridge across the nuclear envelope that physically connects the genome with the cytoskeleton and transfers mechanical forces bidirectionally between the nucleus and the cytoplasm (10, 11) (Fig. 1).
An additional important nongenetic means by which the genome affects cellular processes is via its ability to serve as a binding platform for cellular components. Although this property is extensively used for genetic regulation by recruiting transcription factors to their regulatory sites (12), other proteins and large macromolecular complexes exploit binding to chromatin to perform their functions by nongenetic mechanisms. Prominent examples include the kinetochore, the nuclear pore complex, and the DNA repair machinery. The function of these cellular machineries is critically dependent on their binding to chromatin, but it is the physical properties of chromatin, rather than the genetic information encoded in the genome, that enables their function.
Whereas the genetic functions of the genome are well established, its nongenetic effects are poorly understood. The traditional genetic functions of the genome include storage, propagation, and transmission of the genetic material and rely on the use of the genetic information encoded in the DNA sequence. In contrast, the nongenetic functions of the genome do not use its genetic information and are mediated by the physical properties of genomes. Nongenetic functions of the genome are emerging in diverse biological processes, including nuclear assembly, response to mechanical forces, cell migration, cell cycle progression, cellular signaling, and even physiological functions such as vision in nocturnal animals. We discuss here the concept, mechanisms, and the biological implications of nongenetic functions of the genome.
The genome as a determinant of nuclear architecture
A major nongenetic function of the genome is its role in establishing and maintaining the overall structure of the eukaryotic cell nucleus (Fig. 2). This function is evident from in vivo and in vitro studies of the postmitotic assembly of the nuclear envelope, a key event in the cell cycle (13). During mitosis, the nuclear membrane disassembles and is absorbed by the endoplasmic reticulum (ER), and membrane fragments disperse in the dividing cell. As daughter cells are formed in telophase, the nucleus rapidly reassembles. During this stage, chromatin serves as a physical scaffold on which the nuclear membrane fragments are immobilized, facilitating their fusion into larger membrane sheets (13) (Fig. 2). Capture of nuclear membrane fragments, either directly by the genomic scaffold or via linker proteins, involves several inner nuclear membrane proteins including the lamin B receptor; the LEM (LAP2, emerin, MAN1) domain proteins LAP2b, emerin, and MAN1, which tether membrane fragments to chromatin via their interaction with heterochromatin protein HP1; and barrier-to-autointegration factor BAF (14–17). Direct experimental demonstration for a nongenetic scaffold function of the genome in establishing nuclear architecture comes from the finding that microinjection of bacteriophage λ DNA into Xenopus eggs leads to the formation of double-bilayer membranes around the injected genetic material (18). Similarly, nuclear membranes form in vitro around chromatin and even around purified DNA in mammalian or Xenopus whole-cell extracts (19, 20). Nuclear membranes assemble irrespective of DNA sequence or source of the genetic material used, demonstrating the nongenetic nature of this genome function.
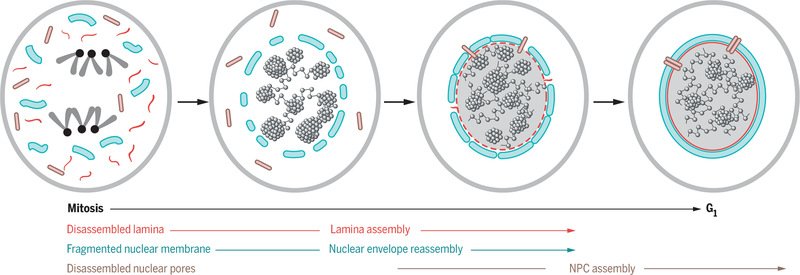
Fig. 2. The genome as a scaffold for nuclear assembly.
The cell nucleus disassembles during cell division. The genome serves as a critical scaffold to organize nuclear envelope fragments (blue) and nuclear pore components (pink) during formation of the nucleus in the two daughter cells.
The genome also plays a role in a later step of nuclear reassembly, during the formation of nuclear pore complexes (NPCs) (21, 22) (Fig. 2). NPCs are inserted into the reassembling nuclear membrane in telophase, as cells enter G1. In both mouse and Xenopus extracts, depletion of histone H3.3 or H4 prevents the formation of NPCs during postmitotic nuclear assembly, although nuclear membrane assembly occurs normally, directly demonstrating a scaffolding role for the genome in assembly of NPCs (21–23). The mechanism for this function of nucleosomes in NPC assembly is nongenetic and involves the nuclear pore protein ELYS, which initiates the assembly process by acting as a key adaptor protein between chromatin and the NPC and in this way initiates the assembly process (21–23). An even broader role of nucleosomes and chromatin in the formation and assembly of the nuclear periphery is suggested by the finding that in Xenopus the assembly of the nuclear lamina is incomplete in the absence of nucleosomes (21). Furthermore, the lysine demethylase LSD1, implicated in chromatin compaction, is required for the in vitro assembly of the nuclear envelope and the NPC in Xenopus extracts (24). These observations suggest that chromatin globally affects the assembly and structure of the nuclear envelope by nongenetic means.
A related function of the genome is its contribution to determining nuclear size. Although one mechanism for determining nuclear size appears to be the sensing of the nuclear/cytoplasmic ratio of soluble factors (25), several independent lines of observations suggest a critical role for global genome architecture, and for chromatin condensation in particular, in nuclear size control. In eukaryotic cells, chromatin condensation is mediated by the ubiquitous and abundant linker histone H1 family of chromatin binding proteins (26). A direct role for H1-mediated chromatin condensation in nuclear size determination is evident from observations in Tetrahymena, which contains a large macronucleus and a small micronucleus, each with a distinct H1 isoform. Depletion of either H1 isoform results in nuclear enlargement but affects only the nucleus in which the specific isoform is normally present (27). Similarly, RNA interference–mediated knockdown of condensins, which play key roles in preparing chromosomes for mitosis by facilitating their compaction (28), increases nuclear size in both mouse embryonic stem cells and human cells (29, 30). A physiological role for condensins in controlling nuclear size is also evident in mouse T cells, which fail to compact their nuclei and do not enter quiescence in the absence of condensin II (31). An additional architectural chromatin factor implicated in nuclear size control is the methyl-CpG binding protein MeCP2, whose loss in neurons, where it is expressed at high levels, leads to substantial nuclear shrinkage, possibly via its interplay with linker histone H1 (32). Taken together, these observations suggest that the global chromatin condensation status in a nucleus contributes to determining its size, evidently through nongenetic mechanisms.
The genome in the cellular response to mechanical forces
The accumulation of heterochromatin at the edge of the cell nucleus is an evolutionarily conserved feature of most eukaryotes (2). Targeting of compact heterochromatin to the nuclear periphery is generally thought to serve mainly as a means to facilitate gene silencing (33); however, several observations suggest that the peripheral sequestration of heterochromatin may also enhance the structural robustness of the nucleus and strengthen its ability to withstand physical challenges, such as mechanical forces exerted during cell migration or in mechanically active tissues (34, 35) (Fig. 3).
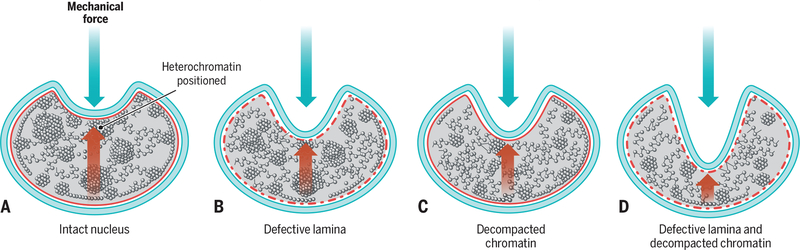
Fig. 3. Genome compactness enhances the sturdiness of the nucleus.
(A) Mechanical forces can deform the nucleus. (B to D) Nuclear deformations are larger in cells with defective lamina or decompacted chromatin and larger still in cells that have both defective lamina and decompacted chromatin. Size of red arrow indicates the relative intranuclear opposing force.
Initial evidence for a role of chromatin in conferring nuclear robustness comes from observations in the yeast Schizosaccharomyces pombe, where the condensed centromeric chromatin is clustered and positioned opposite the micro-tubule organizing center, the region of the nucleus that is subjected to the strongest mechanical forces generated in the cytoplasm (36–38). Mutations in the nuclear proteins HEH1, HEH2, and IMA1, which tether chromatin to the inner nuclear membrane, lower the ability of nuclei to withstand mechanical forces emanating from microtubules assembled in the cytoplasm, as demonstrated by decreased nuclear stiffness when challenged by optical tweezers (37, 38).
In vertebrate cells, the nucleoplasmic side of the nuclear membrane is lined by the nuclear lamina, a meshwork of intermediate filaments that supports and strengthens the nuclear membrane, thereby providing mechanical stability to the nucleus (Fig. 3). In mouse embryonic fibro-blasts, down-regulation of the Prdm3 and Prdm16 methyltransferases, which facilitate heterochromatic histone H3 Lys9 methylation and promote heterochromatinization, leads not only to decreased chromatin compaction but also to disruption of the nuclear lamina, invaginations of the nuclear envelope, and changes in nuclear shape (39). In addition, nuclear blebbing (deformation of the nucleus upon weakening of the nuclear lamina) and defects in the nuclear membrane have been observed in human laminopathies, diseases caused by mutations in the genes encoding the lamin proteins (11); similar defects are seen in tissue culture cells overexpressing HMGN5 (35), an architectural chromatin protein known to promote chromatin decompaction by reducing the ability of the linker histone H1 to bind to nucleosomes (40). In these cells, the levels of lamina components or histone modifications associated with condensed heterochromatin remain unaltered, which suggests that the defects in the nuclear membrane and lamina are directly due to changes in chromatin compaction.
An obvious interpretation of these findings is that chromatin decompaction likely alters the mechanical properties of the nucleus in a manner similar to that in cells with defective lamina components (41, 42). The notion that chromatin decondensation weakens the ability of the nucleus to withstand mechanical force is also supported by the finding that in Lmna−/− cells, which lack nuclear lamins A and C and already have a weakened lamina, chromatin decompaction synergistically reduces nuclear robustness (35) (Fig. 3). Although these observations point to an interplay between chromatin and the lamina in determining the mechanical properties of nuclei, the finding that nuclear stiffness changes upon alterations in the nuclear ionic environment without apparent changes to the lamina structure suggests that chromatin by itself contributes significantly, and directly, to determining the mechanical properties of nuclei (43).
Evidence for a physiological, nongenetic role for heterochromatin in supporting lamina integrity and nuclear robustness comes from analyses of transgenic mice. Global overexpression of HMGN5 leads to a hypertrophic heart harboring large cells with enlarged nuclei lacking visually detectable condensed heterochromatin, and to death within a few months of birth (35); this phenotype is similar to that of Lmna−/− mice (44). Transgenic mice that overexpress HMGN5 only in the heart are born with cardiomyocytes lacking heterochromatin, yet their heart and cardiomyocytes appear normal, their lamina is intact, and the transcription profile of their heart tissue is indistinguishable from that of wild-type mice. However, most animals die within 3 months of birth, showing marked cardiac hypertrophy with extremely large cells and nuclei in which the lamina is disrupted (35). These pathologies are likely caused by loss of heterochromatin, which gradually diminishes the ability of the nucleus to withstand the mechanical forces of the continuously beating heart (35).
The importance of genome condensation in the physiological response to mechanical stress is also evident in mouse melanoma cells, where forces generated by microtubule reassembly after chemically induced depolymerization lead to invagination of both the nuclear envelope and the underlying lamina opposite the microtubule organizing center (MTOC) (45). Imaging of live cells revealed a transient accumulation of heterochromatin at the membrane invagination sites located closest to the MTOC, which suggests that the deformations in the nuclear membrane trigger genome reorganizations, perhaps aimed at counteracting and minimizing deleterious effects of physical changes in the nuclear membrane (45).
Taken together, these observations suggest that targeting heterochromatin to the nuclear periphery not only serves a genetic function via silencing of subset of genes, but also strengthens the nuclear lamina and the nuclear membrane by nongenetic means, thereby supporting the structural integrity of the nucleus. This function of the genome may be especially crucial in cells exposed to mechanical stress, such as cardiomyocytes and migrating cells.
Genome organization in cell migration
Cell migration is involved in a wide range of biological processes including development, tumor progression, tissue renewal, and immune responses. The nucleus is the largest and stiffest cellular organelle, and its large size and rigidity limit the ability of cells to easily move through restricted spaces. Consequently, during cell migration, the nucleus is exposed to considerable mechanical stress as it undergoes changes in shape to facilitate passage through the narrow constrictions imposed by tissues (46) (Fig. 4). The integrity of the nuclear lamina and its components is known to play a major role in nuclear reshaping and its ability to withstand mechanical stress during migration. Changes in the nuclear lamina that increase nuclear stiffness lower the rate of cell migration, whereas decreased nuclear stiffness can reduce the viability of migrating cells (47, 48). Although the nuclear lamina plays a major role in affecting nuclear deformations during cell migration, additional factors, including the organization of the genome inside the nucleus, may affect this process (34, 49).
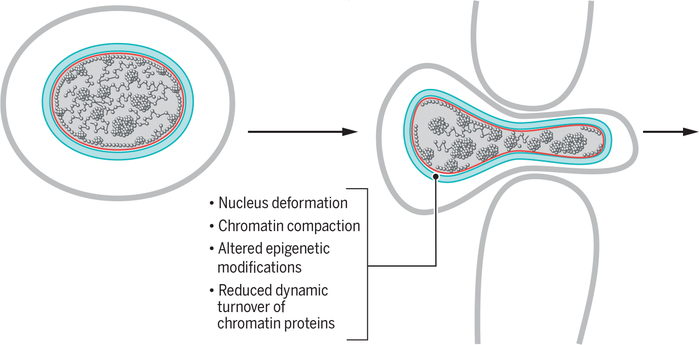
Fig. 4. Nuclear rearrangements during cellular migration.
Cellular migration through narrow spaces leads to deformation of the nucleus, structural changes in the nuclear envelope (blue) and lamina (red), and genome reorganization.
A major factor affecting genome organization is the dynamic binding of linker H1 variants to chromatin (50). H1 variant molecules continually cycle between chromatin-bound and unbound states, and the length of their chromatin residence time is related to the degree of chromatin compaction, with shorter residence times in de-compacted chromatin (50). In mouse melanoma cells, migration cues result in an increase in the time that H1 variants remain bound to chromatin, leading to an increase in chromatin compaction. Migrating melanoma cells also exhibit increased levels of histone H3 Lys9 trimethylation, histone H3 Lys27 trimethylation, and histone H4 Lys20 methylation—histone modifications that mark transcriptionally silent or condensed heterochromatin (51)—as well as a decreased rate of nuclease digestion, an assay for chromatin compaction (52). It thus appears that cell migration is associated with chromatin compaction. Conversely, chromatin decompaction reduces the rate of cell migration. These effects are seen in the presence of transcriptional inhibitors, which suggests that the lower rate of migration is mainly due to alteration in chromatin organization rather than to transcriptional responses (53), strongly pointing to nongenetic mechanisms. A direct causal link between chromatin condensation and enhanced cell migration is also seen during migration of immune T cells and their extravasation into inflamed tissues (54). T cell migration is facilitated by integrin-induced recruitment of the histone methyltransferase G9a to the nuclear periphery, where it methylates specific residues in histone H3, thereby promoting chromatin condensation and alterations in the physical properties of the nucleus that facilitate T cell migration (54).
Changes in chromatin condensation and cell migration have also been seen in studies aimed at understanding the biological consequences of alteration in histone modifications during tumor invasion (55) and during development (56). In these processes, the effects of several histone modifications on cell migration were attributed to a genetic mechanism via their role in control of gene expression programs. However, it is also possible that some of the observed migration defects were due to epigenetically induced changes in global chromatin organization rather than to altered gene expression patterns. The decrease in nuclear size and the increase in the stability of the nuclear lamina resulting from chromatin condensation likely minimize potential damage during nuclear reshaping, thereby enhancing the migration ability of cells (34).
Chromatin-mediated signaling
The complex higher-order topology of the genome serves as an efficient binding platform for a wide range of cellular components. Although binding of chromatin remodelers and transcription factors to the genome leads to changes in gene expression patterns, other binding events occur in the genome that do not result in gene regulation but elicit downstream effects by non-genetic means. In particular, chromatin binding has been implicated in activation of cellular signaling pathways (Fig. 5).
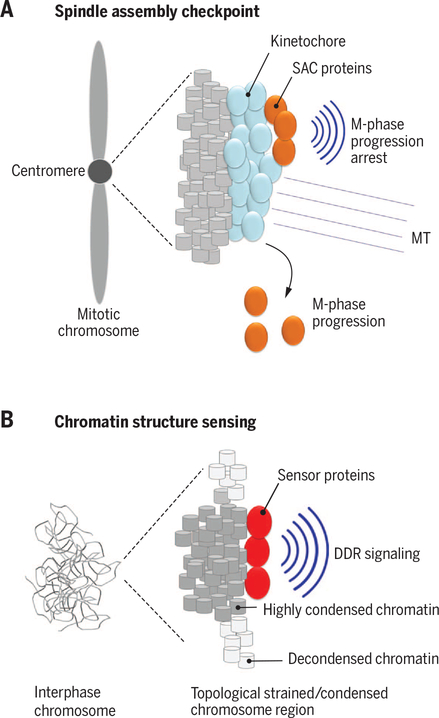
Fig. 5. The genome as a signaling platform.
(A) Proteins of the spindle assembly checkpoint (SAC; orange) bind via the kinetochore (blue) to mitotic chromosomes at centromeres. Upon binding of microtubules (MT) to kinetochores, SAC proteins are released and promote M-phase progression. (B) Topologically strained or highly condensed chromosome regions can be recognized by DNA damage sensors (red) and activate DNA damage response (DDR) pathways.
A prominent example of chromatin-mediated signaling is the spindle checkpoint response (57). Segregation of mitotic chromosomes is mediated by the spindle microtubules, which attach to the chromosomes to align them at the metaphase plate. The attachment of microtubules occurs at the kinetochore, a complex proteinaceous structure that forms at the centromere, a specialized region of chromosomes, which is characterized by the presence of specific core histone proteins, CENP-A in mammals and Cse4 in Saccharomyces cerevisiae (57). The spindle checkpoint is a signaling cascade that emanates from the kinetochore and involves the recruitment of a complex set of kinetochore-associated proteins, such as BUB1–3 and MAD1–3, to chromatin (Fig. 5A). Association of these proteins with chromatin inhibits the activity of the anaphase-promoting complex (APC), which controls the destruction of mitotic cyclins and thus controls exit from mitosis. As long as the checkpoint proteins are bound to the kinetochore, APC is inhibited and cells do not proceed through mitosis. Proper attachment of microtubules to all kinetochores, most likely via sensing of microtubule tension, generates a signal to activate APC by releasing bound proteins, thus triggering progression into anaphase (Fig. 5A). In parallel, the protein phosphatase PP1-Sds22 binds to kinetochores when they reach the cortex in late anaphase, and appears to trigger subsequent cytokinesis (58). Both of these signaling pathways rely on, and are triggered by, the regulated binding of signaling components to chromatin in specialized regions of the chromosome (57, 58). Indeed, recent studies demonstrate that loss of methylation of H3 Lys9 at the centromere leads to defects in both sister chromatid cohesion and kinetochore attachment, ultimately causing chromosomal instability, a major driver of tumor progression (59).
A related function of chromatin-bound proteins is in defining the spatial orientation of the mitotic spindle and the cleavage furrow. Mitotic chromosomes generate from their surface a gradient of the guanosine triphosphate–bound form of Ran (Ras-related nuclear protein), which is implicated in nuclear transport during inter-phase and spindle assembly in mitosis (60). The gradient is generated by retention of the Ran exchange factor RCC1 on the mitotic chromosomes via binding to chromatin (61). The Ran gradient promotes spindle assembly and also influences spindle positioning via displacement of the LGN protein, which is involved in centering the spindle and maintaining its localization (62). Similarly, chromatin serves as a regulatory binding platform for a group of proteins referred to as chromosome passenger complex (CPC) proteins. These proteins are essential for accurate progression of cells through mitosis and associate with condensed chromosomes during early stages of mitosis (63). Their controlled release from chromatin at late stages of mitosis is a key regulator step in cell division, as it allows the CPC proteins to associate with the central spindle and the mid-body where they play critical roles in formation of the cleavage furrow and abscission (63).
A nongenetic signaling activity of chromatin is also evident from several findings indicating that the condensation status of chromatin can be sensed by components of the DNA damage response (DDR) machinery (64, 65) (Fig. 5B). The essential cell cycle checkpoint kinase ATR is thought to promote the detachment of chromatin from the nuclear envelope during replication and to ease the topological strain on the chromatin fiber (64, 66). Recent observations suggest that ATR is activated by changes in chromatin structure induced by mechanical and topological strain, such as unwinding during replication (64). In normally growing cells, ATR localizes in part to the nuclear envelope, but it also accumulates at the nuclear periphery in response to various stressors including osmotic stress and mechanical force (64). ATR activation during mechanical strain is independent of DNA damage, which suggests that the kinase does not sense DNA damage but detects topological changes in chromatin or the nuclear membrane. Sensing of the chromatin condensation status may be of physiological relevance when chromosomes begin to compact during mitotic prophase, because inhibition of ATR in early mitosis delays full chromatin condensation, leading to delayed nuclear envelope breakdown and progression through mitosis (64). Independent evidence that changes in chromatin structure can trigger DDR signaling cascades comes from experiments in which highly compacted chromatin domains were generated by stably tethering heterochromatin binding factors such as heterochromatin protein HP1 to chromatin (65), which led to ATM- and ATR-dependent activation of canonical DNA damage responses, even in the absence of detectable DNA damage (65). Similarly, DDR activation without DNA damage also occurs upon global decondensation of chromatin (67).
These observations of ATR signaling and chromatin-mediated DDR induction in response to structural changes of chromatin suggest that the DDR machinery not only recognizes DNA breaks but may also be able to detect changes in chromatin structure. Sensing of chromatin structure by the DDR machinery may be an integral function of the regular DDR cascade, because recent analysis by live-cell imaging demonstrates that chromatin domains flanking the site of DNA damage undergo sequential decondensation and recondensation (68, 69). It is thus tempting to speculate that chromatin structure serves as a signal to alert the DDR machinery to damage occurring in the DNA sequence, and that the DDR machinery not only repairs damaged genome regions but also continuously surveys the physical organization of the genome.
Chromatin organization as a facilitator of nocturnal vision
A striking example of a nongenetic role of the genome that affects the biological function of a tissue rather than that of single cells is seen in the physiology of vision in nocturnal animals. Extensive comparative analysis of the global organization of the genome in rod photoreceptor cells from evolutionarily distant species revealed a remarkable correlation between heterochromatin organization and the capacity of animals for night vision (70). Whereas in most cell types and species the highly condensed heterochromatin regions of the genome are found at the nuclear periphery, in rod cells of nocturnal animals the orientation is inverted, with most heterochromatin congregated in the nuclear interior (70). The mechanism for this orientation involves the lamin A/C proteins and the inner nuclear membrane protein LBR (lamin B receptor), which appear to tether heterochromatin to the periphery (71). The likely evolutionary basis driving the inversion of heterochromatin location in nocturnal animals are the resulting beneficial changes of the optical properties of the rod cells, because the inverted pattern of heterochromatin location leads to a considerably higher refractive index at the center of the nucleus, thereby reducing light scattering and enhancing the focusing of light onto the photoreceptor plane (70). It thus appears that in the case of mammalian night vision, evolutionary pressure has selected for a physical, nongenetic, rather than a genetic, property of the genome.
Outlook
The genome has traditionally been thought to exert its function exclusively by genetic means. However, it is increasingly apparent that genomes also affect and regulate a large array of vital cellular and physiological processes through their physical properties via nongenetic means. Given that only a relatively small fraction of the genome is transcribed into protein-coding or noncoding RNAs, it is intriguing to entertain the possibility that the presence of large non-transcribed regions is not only a remnant of evolutionary genetic reshuffling events but may also serve to enhance its nongenetic functions by increasing the physical mass of the genetic material. It is notable that several of the cellular processes affected by nongenetic functions are intimately linked to mitosis, including postmitotic assembly of the nuclear membrane, chromatin-initiated spindle checkpoint signaling, and chromatin condensation, allowing for the possibility that nongenetic genome functions are particularly relevant for ensuring proper cell cycle progression. It is highly likely that additional unanticipated nongenetic functions of the genome will be discovered. For example, chromatin and chromatin binding proteins are already known to signal not only within the confines of the cell but also extra-cellularly, where they can function as immune triggers and are involved in the etiology of lupus erythematosus (72), enhance the sensing of cytoplasmic DNA in innate immune recognition (73, 74), and can act as cytokines in apoptosis, inflammation, and infection (75–78).
The realization that the physical properties of genomes have a substantial impact on a diverse set of cellular functions calls for several avenues of further investigation. For one, it will be critical to determine the biophysical properties of the genome in intact cells. A key step in characterizing nongenetic functions of the genome will be the development of live-cell and intravital approaches to measure the type and magnitude of forces the genome exerts on its surroundings, as well as the forces to which the genome is exposed, in intact cells under various physiological and environmental conditions. Likewise, novel experimental and computational methods need to be developed to probe, simulate, and quantify the physical properties of the nuclear environment and to determine how forces are transmitted from and to the genome. Because many of the nongenetic functions of the genome involve interaction with structural elements of the nucleus, such as the lamina or the nuclear pore complex, further investigation of these interplays should be useful in characterizing new genome functions. A particularly intriguing aspect of nongenetic genome function is its role in signaling events where the genome seems to serve as an assembly platform for signaling complexes. Characterization of chromatin-associated signaling complexes and of the physiological consequences of their interactions with the genome will be of considerable interest.
In addition to measuring the physical properties of the genome, it will be equally, if not more, important to develop methods to manipulate nongenetic genome features in intact cells to test their functional consequences, akin to the use of overexpression and gene inhibition to probe genetic functions of the genome. Nanomanipulation devices and light-induced optogenetic methods may be promising approaches to alter the physical properties of the genome in single cells. Ideally, these methods will be applicable to whole-animal studies so as to allow testing of nongenetic genome functions in physiological settings.
The observations discussed here highlight the emerging notion that the functions of the genome are substantially more diverse than merely the regulation of gene expression, and that the genome’s physical nongenetic properties play a central role in cellular function. Although decades of studies on the regulation of gene expression have led to some of the most revolutionary advances in biology, our understanding the genome will be incomplete without elucidation of these nongenetic genome functions.
ACKNOWLEDGMENTS
We thank members of the Misteli lab and P. Oberdoerffer for comments on the manuscript; G. Gerlitz (Ariel University, Israel) and members of the Bustin laboratory for informative discussions on various topics of this review; and T. Ried and R. Lefcoe for illustrations. Work in the authors’ laboratories is supported by the NIH Intramural Research Program, National Cancer Institute, Center for Cancer Research.
REFERENCES AND NOTES
- 1.Cavalli G, Misteli T, Functional implications of genome topology. Nat. Struct. Mol. Biol 20, 290–299 (2013). doi: 10.1038/nsmb.2474; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bickmore WA, The spatial organization of the human genome. Annu. Rev. Genomics Hum. Genet 14, 67–84 (2013). doi: 10.1146/annurev-genom-091212-153515; [DOI] [PubMed] [Google Scholar]
- 3.Ozer G, Luque A, Schlick T, The chromatin fiber: Multiscale problems and approaches. Curr. Opin. Struct. Biol 31, 124–139 (2015). doi: 10.1016/j.sbi.2015.04.002; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui Y, Bustamante C, Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc. Natl. Acad. Sci. U.S.A 97, 127–132 (2000). doi: 10.1073/pnas.97.1.127; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosa A, Topological constraints and chromosome organization in eukaryotes: A physical point of view. Biochem. Soc. Trans 41, 612–615 (2013). doi: 10.1042/BST20120330; [DOI] [PubMed] [Google Scholar]
- 6.Pederson T, King MC, Marko JF, Forces, fluctuations, and self-organization in the nucleus. Mol. Biol. Cell 26, 3915–3919 (2015). doi: 10.1091/mbc.E15-06-0357; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorski SA, Dundr M, Misteli T, The road much traveled: Trafficking in the cell nucleus. Curr. Opin. Cell Biol 18, 284–290 (2006). doi: 10.1016/j.ceb.2006.03.002; [DOI] [PubMed] [Google Scholar]
- 8.Swift J, Discher DE, The nuclear lamina is mechano-responsive to ECM elasticity in mature tissue. J. Cell Sci 127, 3005–3015 (2014). doi: 10.1242/jcs.149203; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maizels Y, Gerlitz G, Shaping of interphase chromosomesby the microtubule network. FEBS J 282, 3500–3524 (2015). doi: 10.1111/febs.13334; [DOI] [PubMed] [Google Scholar]
- 10.Tapley EC, Starr DA, Connecting the nucleus to the cytoskeleton by SUN-KASH bridges across the nuclear envelope. Curr. Opin. Cell Biol 25, 57–62 (2013). doi: 10.1016/j.ceb.2012.10.014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gruenbaum Y, Foisner R, Lamins: Nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu. Rev. Biochem 84, 131–164 (2015). doi: 10.1146/annurev-biochem-060614-034115; [DOI] [PubMed] [Google Scholar]
- 12.Voss TC, Hager GL, Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet 15, 69–81 (2014). doi: 10.1038/nrg3623; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wandke C, Kutay U, Enclosing chromatin: Reassemblyof the nucleus after open mitosis. Cell 152, 1222–1225 (2013). doi: 10.1016/j.cell.2013.02.046; [DOI] [PubMed] [Google Scholar]
- 14.Gorjánácz M et al. , Caenorhabditis elegans BAF-1 andits kinase VRK-1 participate directly in post-mitotic nuclear envelope assembly. EMBO J 26, 132–143 (2007). doi: 10.1038/sj.emboj.7601470; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segura-Totten M, Kowalski AK, Craigie R, Wilson KL, Barrier-to-autointegration factor: Major roles in chromatin decondensation and nuclear assembly. J. Cell Biol 158, 475–485 (2002). doi: 10.1083/jcb.200202019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vlcek S, Korbei B, Foisner R, Distinct functions of the unique C terminus of LAP2alpha in cell proliferation and nuclear assembly. J. Biol. Chem 277, 18898–18907 (2002). doi: 10.1074/jbc.M200048200; [DOI] [PubMed] [Google Scholar]
- 17.Güttinger S, Laurell E, Kutay U, Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol 10, 178–191 (2009). doi: 10.1038/nrm2641; [DOI] [PubMed] [Google Scholar]
- 18.Forbes DJ, Kirschner MW, Newport JW, Spontaneous formation of nucleus-like structures around bacteriophage DNA microinjected into Xenopus eggs. Cell 34, 13–23 (1983). doi: 10.1016/0092-8674(83)90132-0; [DOI] [PubMed] [Google Scholar]
- 19.Burke B, Gerace L, A cell free system to study reassembly of the nuclear envelope at the end of mitosis. Cell 44, 639–652 (1986). doi: 10.1016/0092-8674(86)90273-4; [DOI] [PubMed] [Google Scholar]
- 20.Hartl P, Olson E, Dang T, Forbes DJ, Nuclear assembly with lambda DNA in fractionated Xenopus egg extracts: An unexpected role for glycogen in formation of a higher order chromatin intermediate. J. Cell Biol 124, 235–248 (1994). doi: 10.1083/jcb.124.3.235; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zierhut C, Jenness C, Kimura H, Funabiki H, Nucleosomal regulation of chromatin composition and nuclear assembly revealed by histone depletion. Nat. Struct. Mol. Biol 21, 617–625 (2014). doi: 10.1038/nsmb.2845; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoue A, Zhang Y, Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat. Struct. Mol. Biol 21, 609–616 (2014). doi: 10.1038/nsmb.2839; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doucet CM, Talamas JA, Hetzer MW, Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell 141, 1030–1041 (2010). doi: 10.1016/j.cell.2010.04.036; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schooley A, Moreno-Andrés D, De Magistris P, Vollmer B,Antonin W, The lysine demethylase LSD1 is required for nuclear envelope formation at the end of mitosis.J. Cell Sci 128, 3466–3477 (2015). doi: 10.1242/jcs.173013; [DOI] [PubMed] [Google Scholar]
- 25.Jevtić P, Edens LJ, Vuković LD, Levy DL, Sizing and shaping the nucleus: Mechanisms and significance. Curr. Opin. Cell Biol 28, 16–27 (2014). doi: 10.1016/j.ceb.2014.01.003; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hergeth SP, Schneider R, The H1 linker histones: Multifunctional proteins beyond the nucleosomal core particle. EMBO Rep 16, 1439–1453 (2015). doi: 10.15252/embr.201540749; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen X, Yu L, Weir JW, Gorovsky MA, Linker histones are not essential and affect chromatin condensation in vivo. Cell 82, 47–56 (1995). doi: 10.1016/0092-8674(95)90051-9; [DOI] [PubMed] [Google Scholar]
- 28.Bell JC, Straight AF, Condensing chromosome condensation. Nat. Cell Biol 17, 964–965 (2015). doi: 10.1038/ncb3212; [DOI] [PubMed] [Google Scholar]
- 29.Fazzio TG, Panning B, Condensin complexes regulate mitotic progression and interphase chromatin structure in embryonic stem cells. J. Cell Biol 188, 491–503 (2010). doi: 10.1083/jcb.200908026; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.George CM, Bozler J, Nguyen HQ, Bosco G, Condensins are required for maintenance of nuclear architecture. Cells 3, 865–882 (2014). doi: 10.3390/cells3030865; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rawlings JS, Gatzka M, Thomas PG, Ihle JN, Chromatin condensation via the condensin II complex is required for peripheral T-cell quiescence. EMBO J 30, 263–276 (2011). doi: 10.1038/emboj.2010.314; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yazdani M et al. , Disease modeling using embryonic stem cells: MeCP2 regulates nuclear size and RNA synthesis in neurons. Stem Cells 30, 2128–2139 (2012). doi: 10.1002/stem.1180; [DOI] [PubMed] [Google Scholar]
- 33.Bickmore WA, van Steensel B, Genome architecture: Domain organization of interphase chromosomes. Cell 152, 1270–1284 (2013). doi: 10.1016/j.cell.2013.02.001; [DOI] [PubMed] [Google Scholar]
- 34.Gerlitz G, Bustin M, The role of chromatin structure in cell migration. Trends Cell Biol 21, 6–11 (2011). doi: 10.1016/j.tcb.2010.09.002; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furusawa T et al. , Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat. Commun 6, 6138 (2015). doi: 10.1038/ncomms7138; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Funabiki H, Hagan I, Uzawa S, Yanagida M, Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol 121, 961–976 (1993). doi: 10.1083/jcb.121.5.961; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King MC, Drivas TG, Blobel G, A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell 134, 427–438 (2008). doi: 10.1016/j.cell.2008.06.022; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schreiner SM, Koo PK, Zhao Y, Mochrie SG, King MC, The tethering of chromatin to the nuclear envelopesupports nuclear mechanics. Nat. Commun 6, 7159 (2015). doi: 10.1038/ncomms8159; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinheiro I et al. , Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 150, 948–960 (2012). doi: 10.1016/j.cell.2012.06.048; [DOI] [PubMed] [Google Scholar]
- 40.Rochman M et al. , The interaction of NSBP1/HMGN5with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol. Cell 35, 642–656 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK,Wilson KL, The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol 6, 21–31 (2005). doi: 10.1038/nrm1550; [DOI] [PubMed] [Google Scholar]
- 42.Lammerding J et al. , Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest 113, 370–378 (2004). doi: 10.1172/JCI200419670; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ,Discher DE, Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. U.S.A 104, 15619–15624 (2007). doi: 10.1073/pnas.0702576104; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nikolova V et al. , Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice.J. Clin. Invest 113, 357–369 (2004). doi: 10.1172/JCI200419448; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerlitz G, Reiner O, Bustin M, Microtubule dynamics alter the interphase nucleus. Cell. Mol. Life Sci 70, 1255–1268 (2013). doi: 10.1007/s00018-012-1200-5; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedl P, Wolf K, Lammerding J, Nuclear mechanicsduring cell migration. Curr. Opin. Cell Biol 23, 55–64 (2011). doi: 10.1016/j.ceb.2010.10.015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harada T et al. , Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J. Cell Biol 204, 669–682 (2014). doi: 10.1083/jcb.201308029; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swift J et al. , Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 341, 1240104 (2013). doi: 10.1126/science.1240104; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolf K et al. , Physical limits of cell migration: Control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol 201, 1069–1084 (2013).doi: 10.1083/jcb.201210152; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Catez F, Ueda T, Bustin M, Determinants of histone H1 mobility and chromatin binding in living cells. Nat. Struct. Mol. Biol 13, 305–310 (2006). doi: 10.1038/nsmb1077; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strahl BD, Allis CD, The language of covalent histone modifications. Nature 403, 41–45 (2000). doi: 10.1038/47412; [DOI] [PubMed] [Google Scholar]
- 52.Weintraub H, Groudine M, Chromosomal subunits in active genes have an altered conformation. Science 193, 848–856 (1976). doi: 10.1126/science.948749; [DOI] [PubMed] [Google Scholar]
- 53.Gerlitz G, Bustin M, Efficient cell migration requires global chromatin condensation. J. Cell Sci 123, 2207–2217 (2010). doi: 10.1242/jcs.058271; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X et al. , Integrin a4b1 controls G9a activity that regulates epigenetic changes and nuclear properties required for lymphocyte migration. Nucleic Acids Res (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hieda M, Matsuura N, Kimura H, Histone modifications associated with cancer cell migration and invasion. Methods Mol. Biol 1238, 301–317 (2015). doi: 10.1007/978-1-4939-1804-1_16; [DOI] [PubMed] [Google Scholar]
- 56.Hu N, Strobl-Mazzulla PH, Bronner ME, Epigenetic regulation in neural crest development. Dev. Biol 396, 159–168 (2014). doi: 10.1016/j.ydbio.2014.09.034; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.London N, Biggins S, Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol 15, 736–747 (2014). doi: 10.1038/nrm3888; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodrigues NT et al. , Kinetochore-localized PP1-Sds22 couples chromosome segregation to polar relaxation. Nature 524, 489–492 (2015). doi: 10.1038/nature14496; [DOI] [PubMed] [Google Scholar]
- 59.Tanno Y et al. , The inner centromere-shugoshin network prevents chromosomal instability. Science 349, 1237–1240 (2015). doi: 10.1126/science.aaa2655; [DOI] [PubMed] [Google Scholar]
- 60.Kalab P, Weis K, Heald R, Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science 295, 2452–2456 (2002). doi: 10.1126/science.1068798; [DOI] [PubMed] [Google Scholar]
- 61.Li HY, Wirtz D, Zheng Y, A mechanism of coupling RCC1 mobility to RanGTP production on the chromatin in vivo.J. Cell Biol 160, 635–644 (2003). doi: 10.1083/jcb.200211004; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kiyomitsu T, Cheeseman IM, Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol 14, 311–317 (2012). doi: 10.1038/ncb2440; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carmena M, Wheelock M, Funabiki H, Earnshaw WC,The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol 13, 789–803 (2012). doi: 10.1038/nrm3474; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar A et al. , ATR mediates a checkpoint at the nuclear envelope in response to mechanical stress. Cell 158, 633–646 (2014). doi: 10.1016/j.cell.2014.05.046; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burgess RC, Burman B, Kruhlak MJ, Misteli T, Activation of DNA damage response signaling by condensed chromatin. Cell Rep 9, 1703–1717 (2014). doi: 10.1016/j.celrep.2014.10.060; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bermejo R et al. , The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 146, 233–246 (2011). doi: 10.1016/j.cell.2011.06.033; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bakkenist CJ, Kastan MB, DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421, 499–506 (2003). doi: 10.1038/nature01368; [DOI] [PubMed] [Google Scholar]
- 68.Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD, DNA double-strand breaks promote methylation of histoneH3 on lysine 9 and transient formation of repressive chromatin. Proc. Natl. Acad. Sci. U.S.A 111, 9169–9174 (2014). doi: 10.1073/pnas.1403565111; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khurana S et al. , A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell Rep 8, 1049–1062 (2014). doi: 10.1016/j.celrep.2014.07.024; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Solovei I et al. , Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137, 356–368 (2009). doi: 10.1016/j.cell.2009.01.052; [DOI] [PubMed] [Google Scholar]
- 71.Solovei I et al. , LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152, 584–598 (2013). doi: 10.1016/j.cell.2013.01.009; [DOI] [PubMed] [Google Scholar]
- 72.Podolska MJ, Biermann MH, Maueröder C, Hahn J,Herrmann M, Inflammatory etiopathogenesis of systemic lupus erythematosus: An update. J. Inflamm. Res 8, 161–171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deng L et al. , STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity 41, 843–852 (2014).doi: 10.1016/j.immuni.2014.10.019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.S. R. Woo, L. Corrales, T. F. Gajewski, The STING pathway and the T cell-inflamed tumor microenvironment. Trends Immunol 36, 250–256 (2015). doi: 10.1016/j.it.2015.02.003; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu Z, Huang Y, Mao P, Zhang J, Li Y, Sepsis and ARDS: The dark side of histones. Mediators Inflamm 2015, 205054 (2015). doi: 10.1155/2015/205054; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bianchi ME, Manfredi A, Chromatin and cell death. Biochim. Biophys. Acta 1677, 181–186 (2004). doi: 10.1016/j.bbaexp.2003.10.017; [DOI] [PubMed] [Google Scholar]
- 77.Yang D et al. , High-mobility group nucleosome-binding protein 1 acts as an alarmin and is critical for lipopolysaccharide-induced immune responses. J. Exp. Med 209, 157–171 (2012). doi: 10.1084/jem.20101354; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Andersson U, Tracey KJ, HMGB1 is a therapeutic target for sterile inflammation and infection. Annu. Rev. Immunol 29, 139–162 (2011). doi: 10.1146/annurev-immunol-030409-101323; [DOI] [PMC free article] [PubMed] [Google Scholar]