Abstract
MicroRNAs (miRNAs) regulate many cellular events during brain development by interacting with hundreds of mRNA transcripts. However, miRNAs operate non-uniformly upon the transcriptional profile with an as yet unknown logic. Shortcomings in defining miRNA-mRNA networks are limited knowledge of in vivo miRNA targets, and their abundance in single cells. By combining multiple complementary approaches, AGO2-HITS-CLIP, single-cell profiling, and innovative computational analyses using bipartite and co-expression networks, we show that miRNA-mRNA interactions operate as functional modules that often correspond to cell-type identities and undergo dynamic transitions during brain development. These networks are highly dynamic during development and over the course of evolution. One such interaction is between radial glia-enriched ORC4 and miR-2115, a great ape specific miRNA, which appears to control radial glia proliferation rates during human brain development.
Recent studies utilizing single cell mRNA sequencing (scRNA-seq) to characterize cell-type diversity in tissues have highlighted the need for multi-modal analyses of cellular phenotypes by unbiased classification schemas, particularly in developing systems where complex gene regulatory networks control orthogonal sources of transcriptional variation, including morphology, physiology, maturation, differentiation, and spatial position1–4. While mRNA expression levels can be used directly to define putative cell types, unbiased clustering methods to infer cell identities and to determine the boundaries of these identities requires either prior knowledge or additional modalities. MicroRNAs (miRNAs) are an inherently complex network of interactions that can serve as an additional feature of cellular identity5 with important implications for protein expression. miRNAs have a role in fine-tuning signaling pathways related to corticogenesis and their altered expression has been associated with numerous neurological disorders (reviewed in 7). Changes in miRNA expression patterns, often of large magnitude, occur as defining decision nodes during cell differentiation6, suggesting that their cell type- specific abundance may represent an important parameter in cell type classification, and provide insights that extend beyond cell-type classification to the dynamic regulation of differentiation. The increase in miRNA numbers encoded in the genome as a function of organismal complexity implies that the emergence of novel cell types in the primate brain may be associated with increased numbers of cell type specific miRNAs in the brain. Previous studies ablating miRNA-processing enzyme Dicer1 emphasized the pleiotropic roles for this pathway related to tissue specificity, anatomical and cellular compartments, evolutionary relationships, developmental time points, and even specific cell types7–12, but the underlying framework for these differences is poorly understood. Profiling of miRNA abundance in developing human brain tissue samples suggested developmental regulation of miRNA expression13, but these studies could neither distinguish cell-type specific patterns of miRNA abundance, nor dynamic cell fate transitions during development at the single cell level. To characterize the in vivo miRNA-mRNA interactions during human brain development, and to contextualize these networks in the framework of developmental transitions and cell identity, we leveraged three complementary datasets: high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP)14 with an AGO2 antibody, simultaneous single cell profiling of mRNAs and miRNAs, and single-cell mRNA sequencing (scRNA-seq) data. Our study revealed a dynamic network involving cell-type specific enrichment of miRNA expression patterns across diverse cell types, and dynamic miRNA target acquisition and loss in which the population of targeted mRNAs keeps pace with the dynamics of tissue development, cell diversity, and lineage progression during human brain development.
Results
AGO2-HITS-CLIP identifies miRNA-mRNA interactions during prenatal human brain development
To identify the landscape of miRNA-mRNA interactions occurring in developing human brain in vivo, we performed high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) 14 with AGO2 (Fig.1). AGO2 bound profiles were generated for primary samples of the developing human brain from stages corresponding to peak neurogenesis (GW15 and 16.5 – early stage) and early gliogenesis (GW19–20.5 – late stage), nine samples in total were harvested from prefrontal cortex (PFC), motor cortex area one (M1), visual cortex area one (V1) and other regions (Supplementary Table 1). AGO2-bound miRNAs and mRNAs were identified after sequencing (Fig. 1a, Supplementary Table 1–2, see Methods for details). In total, 921 human miRNAs were detected and 10505 Ago2 binding sites were identified from both protein-coding genes and non-coding genes, including lncRNAs (Supplementary Table 2). Approximately 43% of sites were in three prime untranslated regions (3’UTR) and 27% of sites were in coding DNA sequence (CDS). For further analysis, we considered only sites identified in the CDS and the 3’UTR, reflecting canonical miRNA-mRNA interactions. We identified 3693 and 2705 genes at early and late stages of development, respectively, actively targeted by miRNAs through CDS or 3’UTR parts of the transcript (Supplementary Fig. 1). We validated a subset of the canonical AGO site interactions using luciferase reporter assays in human cells in vitro (Supplementary Figure 1, Supplementary Table 3). Among the detected interactions were previously validated ones, such as miR-9 with FOXG1 and HES1 and miR-210 with CDK7, thereby confirming the strength of the method.
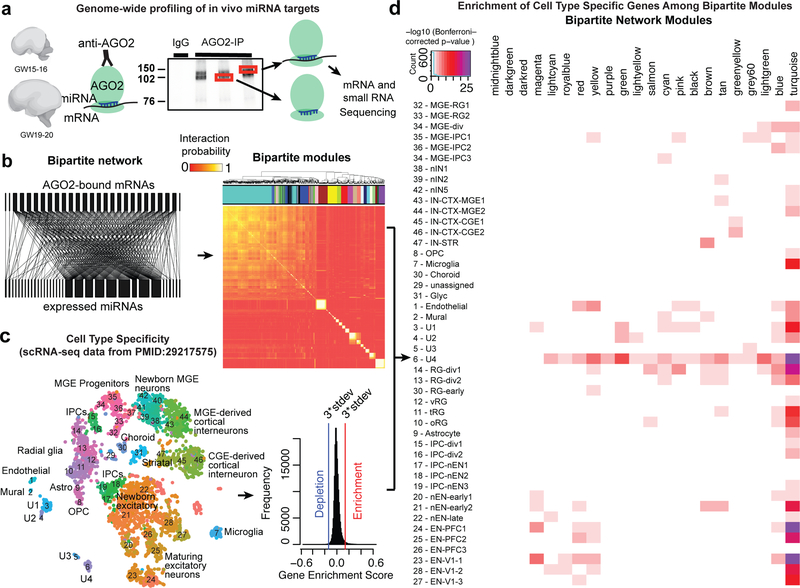
Fig. 1: High Throughput Profiling of miRNA-mRNA Interactions.
(a) Experimental design. Autoradiogram of 32P-labelled RNA tags crosslinked to AGO2 protein obtained from human prenatal brain homogenates. 110 kDa and 130 kDa bands are visible in samples with AGO2-immunoprecipitation as compared to IgG control. (b) The complete bipartite network analysis of miRNA-mRNA interactions shown as a correlation matrix, with bipartite network modules highlighted in colors above the heatmap, in the right panel and a segment of the bipartite network shown in the left panel that illustrates the inhomogeneity of the targeting miRNAs, the relative homogeneity of the targeted mRNAs and the modularity of the miRNA-mRNA network (c-d) Enrichment of bipartite modules according to cell-type identities. (c) Cellular specificity of genes expressed in the developing human brain according to published single-cell mRNA-sequencing dataset, with row names representing cell clusters described in the source study27, and also shown as a tSNE plot coloured by cluster identity. Pearson correlation above 3 times standard deviation (3*stdev) was considered as cutoff for defining enriched or depleted genes. (d) Enrichment of cell-type-specific genes among bipartite network modules. Heatmap demonstrates a significant association between the identified cell-types by scRNA-seq and the detected modules in the bipartite network. Enrichment scores represent Bonferroni-corrected –log10(p-value) calculated using one-sided Fisher’s exact test.
Unbiased enrichment analysis using the total expressed gene set in human developmental brain as the background, revealed that transcription factors, chromatin modifiers, and signaling pathway components were enriched among miRNA targets (Supplementary Table 4). Surprisingly, hundreds of in vivo miRNA targets we identified were well-established markers of distinct cell types15–17, regulators of neurogenesis, migration, axonogenesis, synaptogenesis, and neuronal subtype specification. Broadly, single miRNAs target many mRNAs and single mRNAs are targeted by far fewer miRNAs (Supplementary Fig. 2), representing a bipartite network of interactions between miRNAs and their direct target mRNAs (Fig. 1b). Using a bipartite community detection algorithm18, we revealed modules of miRNA-mRNA interactions (Fig. 1b, Supplementary Fig. 2–3, Supplementary Table 5, see Methods for details). Interestingly, the average abundance of a bound miRNA correlated negatively with the total number of different miRNAs bound in each module (Supplementary Fig. 4), suggesting that miRNA targeting utilizes two strategies: (a) target with one or a very few abundant miRNAs; (b) target with multiple low abundance miRNAs. To contextualize these interactions in the framework of cell diversity we projected bipartite graph modules onto cell-type specificity information, calculated from published scRNA-seq datasets (Fig. 1c-d, Supplementary Table 6). Our analysis revealed striking enrichment for cell-type specific transcripts among bipartite graph modules, suggesting that miRNAs acquire targets according to the cognate transcriptional landscape of individual cell-types.
Cell-type enrichment of miRNAs revealed by single-cell miRNA profiling
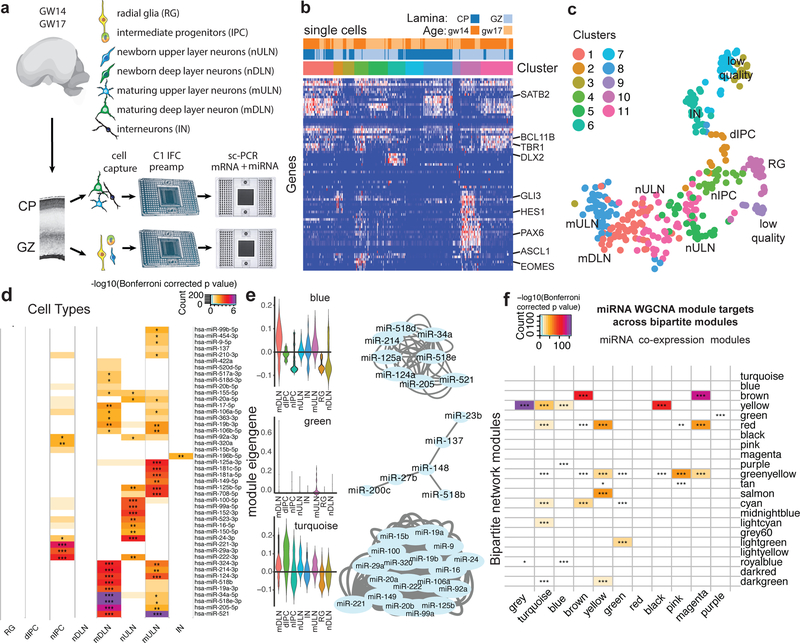
To further investigate how miRNA-mRNA interactions relate to the emerging diversity of cell types of the developing brain, we used an innovative protocol for combined detection miRNAs and mRNAs in the same single cells (Fig.2) using an automated microfluidic platform to perform automated cell capture, reverse transcription and targeted preamplification of mRNA and miRNA (Fig. 2a-c, Supplementary Table 7–8). In addition to long-established markers of distinct cell types in the developing cortex, we selected mRNA targets according to the specificity of their expression for distinct cell types (Fig. 2b) as determined in Pollen AA et al., 15. We profiled single cells isolated from human cortex samples at gestational week (GW) 14 (deep layer neurogenesis) and GW17 (upper layer neurogenesis). To enrich for progenitor cells and newborn neurons, we microdissected samples of the cortical germinal zone (GZ), and to capture maturing neuron populations and interneurons, we microdissected cortical plate regions (CP). In total, we retained data from 312 cells with more than 10 genes detected. Clustering analysis performed based on marker gene abundance revealed 11 clusters (Supplementary Table 7). We inferred the identities of individual cell clusters as radial glia, intermediate progenitors, upper and deep cortical layer neurons, and interneurons (Fig. 2b-c). Furthermore, spatial microdissections supported further refinement of our interpretations with respect to neuronal maturation state (newborn neurons captured from the GZ and maturing neurons captured from the CP)(Fig. 2c). Next, for every miRNA profiled, we quantified the abundance in every cell and calculated an expression enrichment score for every cell type (Fig. 2d). Surprisingly, the vast majority of miRNAs we profiled showed significant enrichment in at least one cell type, suggesting robust variation in miRNA abundance across closely related cells of the developing brain. For example, miR-221/222 and miR-92a were found enriched in cortical IPCs, in line with recent reports19 and consistent with their proposed roles in controlling proliferation20, while miR-124 was enriched in postmitotic neurons, consistent with its proneural role in development21. Furthermore, we grouped miRNAs according to the shared pattern of abundance across single cells using weighted gene co-expression network analysis (Supplementary Table 8). Some of these specific miRNA coexpression modules operate within specific cell types, whereas others are broadly distributed across multiple cell-types (Fig. 2e). Our analysis revealed dynamic changes in miRNA abundance in concordance with neuronal differentiation and maturation, a critical axis of transcriptional variation in the developing brain.
Fig. 2. Single-cell miRNA Expression Profiling Reveals Patterns of Cell-type Enrichment.
(a-c) sc-qPCR profiling of mRNA and miRNA abundance in the same cell. (a) Schematic outlining experimental approach and cell types expected to be enriched in microdissected brain regions. (b) Heatmap of mRNA target genes used to interpret cell identities. (c) tSNE plot of single-cell data generated using Seurat based on mRNA marker gene abundance. Colors represent unbiased clustering (see Methods for details). (d) Heatmap representing cell-type enriched miRNA expressions profiled (one-tailed U-test). Cell type enrichments were calculated for cells captured from 2 biologically independent specimens. (e) Weighted gene co-expression network analysis reveals modules of co-expressed miRNAs across single-cells profiled in a-b. Network plot shows miRNAs assigned to this network based on correlation of abundance across single cells. Module blue and turquoise are enriched in mDLN and IPC, respectively. As in (d), this calculation was based on values from cells captured from 2 biologically independent specimens. mDLN- maturing deep layer neurons, dIPC – dividing intermediate progenitor cells, nIPC – non-dividing intermediate progenitor cell, nULN- newborn upper layer neuron, IN – interneurons, mULN – maturing upper layer neurons, RG – radial glia, nDLN – newborn deep layer neurons. (f) Enrichment of targets of co-expressed miRNAs across bipartite network modules. Co-expression modules were calculated for two biologically independent specimens, while bipartite modules were calculated for n=9 biologically independent specimens. * p<0.05, ** p<0.01, *** p<0.001. Enrichment scores represent Bonferroni-corrected –log10(p-value) calculated using one-sided Fisher’s exact test.
Dual nature of miRNA-mRNA interactions
During mouse brain development, miRNAs are involved in regulating cell-type transitions8, but analogous regulatory mechanisms during human brain development have largely not been investigated. To address this limitation, we projected targets of miRNAs found to be co-expressed using sc-qPCR (Fig. 2e) onto bipartite co-regulatory modules inferred from HITS-CLIP (Fig. 1b). This analysis revealed a striking enrichment of targets of co-expressed miRNAs among bipartite network modules (Fig. 2f). Interestingly, we found examples of interactions where miRNAs were enriched in neurons (WGCNA module ‘blue’, Fig. 2e), and their targets fell into a regulatory module enriched for neuronal markers (bipartite module ‘yellow’, Fig. 1d, 2f). This interaction includes DNM1, a GTPase involved in synaptic vesicle recycling22, and NOVA1, a neuron specific RNA binding protein23. The presence of miRNAs and target mRNAs in the same cell type was also observed when we correlated the abundance of miRNAs and their targets across single cells (Supplementary Fig. 4b), and is consistent with the idea of cell type-specific interactions as has been reported. For example, enrichment of miR-92 and its direct target EOMES in the same cell type has been reported in the developing cerebral cortex 19,24.
In addition, we also found interactions, in which a miRNA co-expression module was enriched in neurons (WGCNA module ‘green’, Fig. 2e), but their targets show moderate enrichment for proliferating progenitors (bipartite module ‘lightgreen’, Fig. 1d, 2f). This interaction includes genes such as H2AFZ, involved in cell cycle regulation25, and PHGDH, a radial glia specific gene involved in L-serine biosynthesis26. Another example, miRNA co-expression module ‘turquoise’ is enriched in intermediate progenitor cells (Fig. 2e), whereas their targets are enriched in bipartite modules that include neuronal and radial glia genes, and genes expressed in both (Fig. 1d, 2f). For example, CC2D1B is expressed highly in radial glia and neurons27, and has been previously implicated in serotonergic signaling in neurons28, but also regulates EGFR expression and could regulate cell proliferation29.
Together, these examples highlight dynamic rewiring of miRNA-mRNA interactions during neuronal differentiation and maturation in the developing human cerebral cortex. In particular, co-modularity miRNA-mRNA interactions as contextualized in the framework of gene and miRNA coexpression seems to follow at least two broad patterns: miRNAs are recruited in a cell type to repress genes not normally expressed in that cell type in some cases or miRNAs are expressed in a cell type to regulate the expression of genes expressed in that same cell type. Further refining these dual roles may emerge from higher resolution temporal data that synchronizes single cell developmental transitions with miRNA target degradation kinetics.
Dynamic changes in miRNA-mRNA network during development
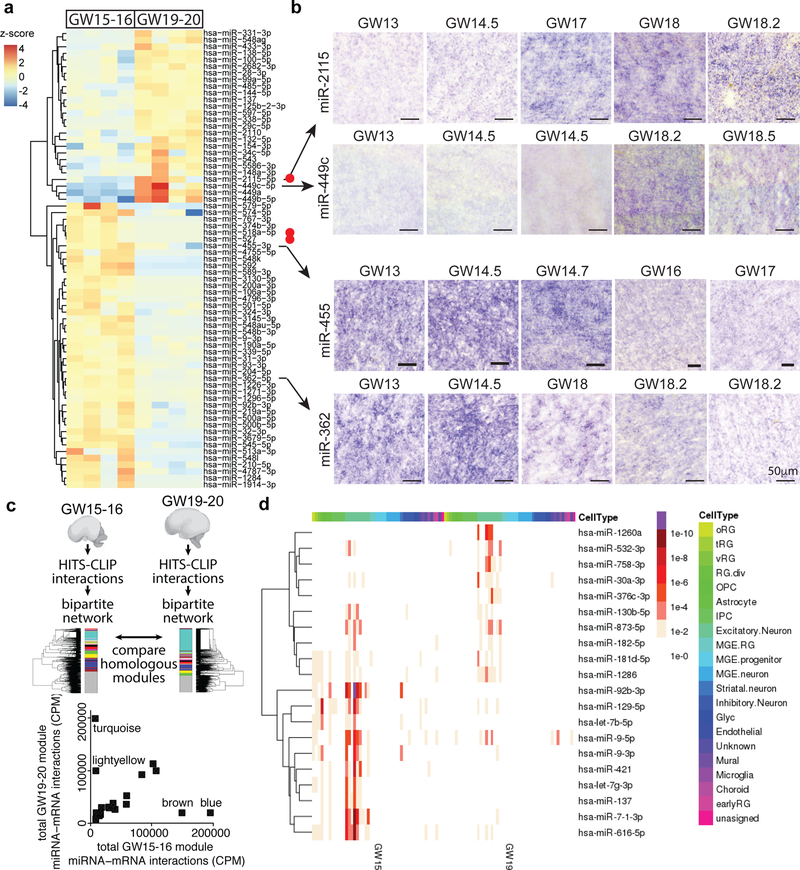
Next, we explored the temporal axis of miRNA-mRNA interactions. We compared the abundance of miRNAs at two stages of development - GW15–16.5 and GW19–20.5 (Fig.3) and found 69 differentially expressed miRNAs between these two stages including recently evolved miRNAs (Fig. 3a-b, Supplementary Fig. 5–11, and Supplementary Table 9). Two miRNAs, miR-449a and miR-449b-5p, which control mitotic spindle orientation during mammalian brain development 30,31 showed the highest overall fold change in expression level between GW15–16.5 and GW19–20.5. We confirmed the expression of miR-2115, miR-449c, miR-455 and miR-362 by in-situ hybridization (Fig. 3b, Supplementary Fig. 5–7). In addition, we also identified miRNAs, miR-1286, miR-142 and miR-548aa, as enriched in the occipital lobe compared to the frontal lobe (Supplementary Fig. 8–10, Supplementary Table 2) suggesting, in line with recent studies32, that miRNAs may regulate regionally divergent transcriptional states in the developing human cortex.
Fig. 3. Dynamic changes in miRNA regulatory networks during development.
(a) Differential expression analysis identifies miRNAs differentially expressed between GW15–16 and GW19–20 developing human cortex. Red dots indicate primate-specific miRNAs. Heatmap displays the expression z-score of each miRNA in that sample. Each column represents a specimen, and specimens are arranged according to age. (b) Validation of differentially expressed miRNAs by in-situ hybridization in the developing human neocortex sections. Images show staining in the outer sub-ventricular zone. For every specimen, the experiment was repeated three times in different sections with similar results. (c) Module preservation analysis demonstrates significant similarity between most modules obtained for networks generated across GW15–16 and across GW19–20 samples as well as a set of distinct interactions present predominantly at one stage (e.g. association of modules turquoise and lightyellow with GW19–20 and modules brown and blue with GW15–16). GW15–16 module names are used to compare GW15–16 modules with their homologues in GW19–20. (d) Stage-specific changes in miRNA targets according to their specificity to distinct cell types of the developing brain identified using single-cell RNA-seq27. Heatmap shows the enrichment p value of miRNAs in distinct cell-types in each stage of development.
By independently performing bipartite network analyses for samples at each of the two stages studied, we found a striking preservation of most co-regulatory modules, as well as a set of distinct interactions present predominantly at one stage (Fig. 3c, Supplementary Fig. 11a, Supplementary Table 10). Interestingly, many of these modules were also highly preserved when compared to adult human brain interactions previously surveyed using the same experimental strategy32,33 (Supplementary Fig. 11b). Together, our findings suggest that miRNA-mediated regulation forms a developmentally dynamic network of interactions related to cell type, developmental stage, and cortical area specificity.
Recent studies suggest that perturbations in miRNA expression may underlie human developmental neuropsychiatric disorders34,35, but the specific molecular consequences remain poorly understood. Interestingly, genes implicated in autism spectrum disorders (ASD), are enriched in the magenta module (Supplementary Fig. 12). In addition, we found that the expression of several miRNAs recently implicated in ASD34 was biased towards expression in excitatory neurons in the developing mid-gestational human samples (Supplementary Fig. 12), although their expression patterns may change over the course of brain development36. Genes targeted by miR-137 in developing brain differ greatly from targets identified in adult human brain tissue33, suggesting that in vivo target interactions of these miRNAs may also change substantially during development (Supplementary Table 11).
To explore this observation more broadly, we considered whether individual miRNAs dynamically change their target landscape during development. We intersected cell type specificity of miRNA targets at either stage of development and found miRNAs whose targets are enriched in one cell type during early development, and in a different cell type later in development (Fig. 3d). We found many miRNAs, like miR-181d-5p, miR-129–5p, miR-9 and miR-616–5p, whose targets were enriched in different cell types between early developmental stages and late developmental stages thereby indicating changing roles for these miRNAs during brain development. miR-1260a, miR-758–3p and miR-376c-3p targets were enriched in excitatory neurons only in late stages. miR-376c-3p is of interest because it can significantly enhance neural differentiation of in vitro pluripotent stem cell models37. Targets of miR-92b-3p, let-7–5p, miR-421, and miR-137 were enriched for radial glial markers or excitatory neurons only at early stages. Both miR-92b-3p and miR-130b-5p have been reported to be specifically associated with neural progenitors13. These examples further underscore the dynamic remodeling of miRNA interaction networks during development and suggest that further analysis of these interactions may reveal previously unappreciated cellular vulnerabilities of miRNA-mRNA interactions to disease mutations. Understanding cell-type-specific miRNA expression profiles, and their respective targets may highlight cellular patterns of selective vulnerability to disorders affecting miRNA expression by highlighting gene regulatory networks that might be perturbed in disease states.
Recently evolved miR-2115 regulates cell cycle in human radial glia
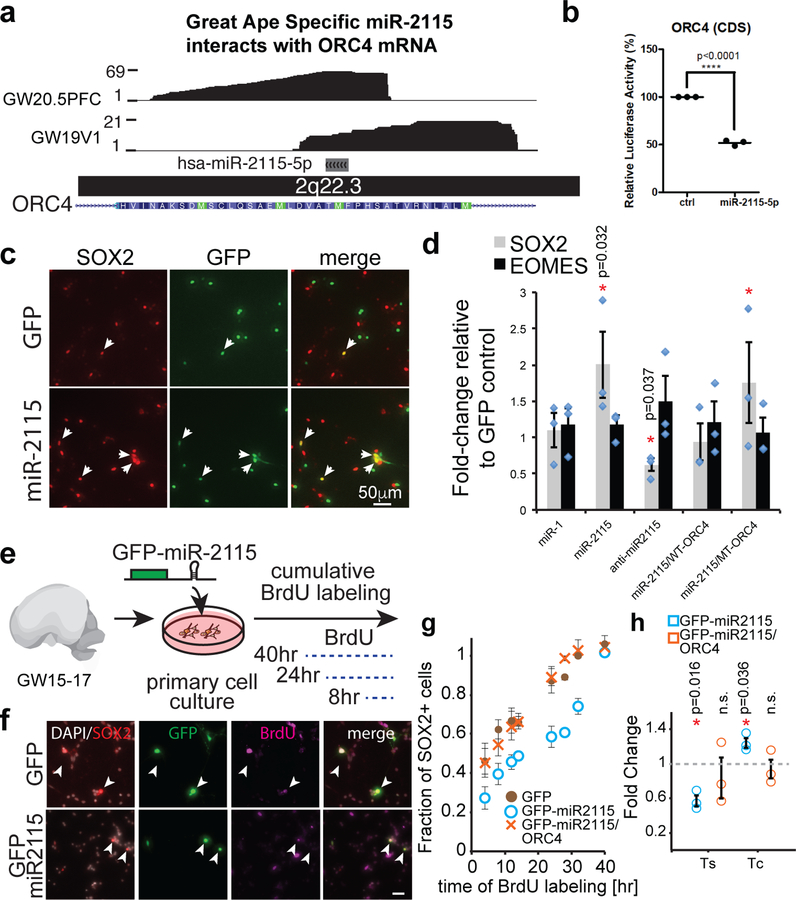
The developmental transition between GW15–16.5 and GW19–20.5 coincides with changes in proliferation rates of radial glia, and depletion of proliferative capacity in the human ventricular zone38. Among the top five miRNAs differentially expressed between these stages, a great ape specific miRNA, miR-2115, was prominently upregulated at GW19–20 in the germinal zones (Fig. 3a-b, Supplementary Fig. 5, 7). Among miR-2115 targets, ORC4, a known regulator of DNA replication39, was enriched in radial glia at early stages of development 15,17, and is a member of the turquoise module which is enriched for GW19–20.5 HITS-CLIP interactions within a segment corresponding to a putative miR-2115 response element (Fig. 3c, Supplementary Table 11). Mutations in ORC4 are linked to Meier-Gorlin syndrome, which is frequently associated with microcephaly, suggesting that this gene may play an important role in normal brain development40. We hypothesized that miR-2115 acts through a radial glia enriched gene regulatory network involving ORC4 to regulate cell cycle dynamics and thereby influences cortical progenitor cell function. To test this hypothesis, we first confirmed the binding of the ORC4 miRNA response element and miR-2115 using a reporter assay and inhibitor studies (Fig. 4a-d, Supplementary Fig. 1) as well as a nearly zero p value using the highly stringent target prediction algorithm PACCMIT-CDS41. Next, we overexpressed a synthetic mmu-miR-2115 (see Methods) in developing mouse cortex and found an increased proportion of radial glia, but fewer radial glia in mitosis, among the electroporated cells (Supplementary Fig. 13). Similarly, manipulation of miR-2115 expression influenced the development of human primary radial glia cells in vitro. Both overexpression and inhibition of miR-2115 changed the proportion of cells expressing SOX2, indicating a possible role for this miRNA in proliferation or differentiation (Fig. 4c-d, Supplementary Figure 13b). The phenotype was rescued by the addition of WT-ORC4 reporter construct expressed together with miR-2115 (Fig. 4d).
Fig. 4. miRNAs contribute to cell-type-specific function.
(a) HITS-CLIP data tracks showing reads spanning a predicted miR-2115 response element in the CDS of ORC4 mRNA. (b) Luciferase reporter assay demonstrating functionality of miR-2115 interaction through the CDS site identified in a) (**** p<0.0001, unpaired two-sided t-test), n=3 biologically independent experiments. Center value represents mean and error bars represent standard error of the mean. (c-d) miRNA-2115 influences radial glia development. (c) Primary radial glia were transfected with miR-2115 expression plasmid and the cultures were immunostained for markers of radial glia (SOX2) and intermediate progenitors (EOMES). (d) Quantification of immunopositive cells (N = 3 biological replicates). “miR-1” – GFP-miR1 small RNA overexpression control construct, “miR-2115” – GFP-miR2115 overexpression construct, “WT-ORC4” – reporter construct with wild type miR-2115 response element containing sequence, “MT-ORC4” – reporter construct lacking miR-2115 response element, “anti-miR-2115” – miR-2115 inhibitor co-transfected with GFP expression construct. All constructs and reagents are described in the Methods. *-p<0.05, two-sided student’s t-test. Center values represent mean and error bars represent standard error of the mean. (e) Experimental design of cumulative BrdU labeling in human cells in vitro performed to assess the impact of miR-2115 on progenitor proliferation. (f) Immunostaining of human cultured cells. Arrowheads indicate GFP and SOX2 double positive cells. Scale bar 25μm (g-h) Quantification of BrdU labeling of SOX2 positive cells (g) and estimates of S-phase length (Ts) and cell cycle length (Tc). Center values represent mean and error bars represent standard error of the mean. (h) Relative to control conditions (N = 3 specimens). *-p<0.05, two-sided student’s t-test. Center values represent mean and error bars represent standard error of the mean.
To more specifically test for a possible cell cycle phenotype, we performed a cumulative BrdU incorporation assay (Fig. 4e-g) in human radial glia using GFP-miR2115 construct. The change in the cell cycle rate caused by miR-2115 overexpression was rescued by the addition of ORC4 protein expression construct (full-length) (Fig. 4h). This showed that miR-2115 expression regulates normal cell cycle duration in human radial glia by controlling ORC4 protein levels. Together, these findings suggest that miR-2115 emerged recently in evolution and integrated into post-transcriptional regulatory networks controlling cell cycle dynamics during human cortical development.
Discussion
Our study revealed several distinct mechanisms by which miRNA regulatory pathways contribute to human brain development. We developed a new single cell profiling approach for combined mRNA and miRNA profiling in the same cell. Using this approach, we examined cell type-specific patterns of miRNA abundance, which revealed highly dynamic changes in miRNA expression, even among the closely related cells of the developing brain. Our findings support the emerging view that many miRNAs are expressed in cell type specific patterns42,43. To gain insight into miRNA-mRNA interactome remodeling during cell type transitions, we intersected high throughput profiling of miRNA-mRNA interactions with cell type specific gene expression profiles. Many miRNAs, including those expressed in multiple cell types, regulate the expression of cell type specific genes. Dynamic changes in cellular transcriptomes occurring during developmental lineage progression are likely controlled through a variety of regulatory networks involving transcription factors, signaling pathways, as well as post-transcriptional, epigenetic, and epitranscriptomic mechanisms. Our analysis further emphasizes the contribution of miRNAs to vast majority of such networks, including genes regulating excitatory neuron laminar and projection fates, such as regulators of callosal projection neurons (BHLHE22, SATB244), corticothalamic neurons (TBR145), and interneurons (LHX6, DLX546). Dissecting the implications of these interactions for neuronal subtype specification will require highly multiplexed approaches for functional validation.
Furthermore, by projecting cell-type-specific miRNA and mRNA expression patterns against the modular framework of the bipartite network of miRNA-mRNA interactions, our study revealed dynamic developmental remodeling of miRNA-mRNA interaction networks, involving conserved and recently evolved miRNAs, as well as cell-type-specific miRNA regulatory networks operating in the developing human brain (Supplementary Table 12). Comprehensive understanding of cell-type-specific miRNA-mRNA interactions may reveal previously unappreciated patterns of selective vulnerability of cell-types in neurodevelopmental disorders, including Autism Spectrum Disorders.
This multi-modal approach to cell-type identity revealed an additional regulatory element introduced into radial glial cells. The expression of a great-ape specific miRNA (miR-2115) targets the more ancient gene (ORC4). By locating the target site in the conserved coding sequence (CDS) without the same necessity for co-evolution of the target site as the presumably less constrained 3’ UTR, greater evolvability of this regulatory interaction is possible. Functional sites have been extensively reported in the CDS47–50 and PAR-CLIP experiments in the human demonstrate that the numbers of CDS and 3′ UTR are similar48, a finding supported by predictive algorithms49. The functional data here on the control of the cell cycle duration in radial glia is consistent with the microcephalic primordial dwarfism phenotype of Meier-Gorlin syndrome (MGORS2) due to mutations in ORC4. These individuals have lissencephaly and hypoplastic frontal lobes among other structural abnormalities.
In addition to specific miRNA coexpression modules associated with specific cell types (Fig 2e), many miRNAs are expressed across a variety of cell types and in so doing, operate within diverse trans- environments of cell type specific transcriptomes. In these latter cases, some miRNA modules can be broadly distributed across multiple cell-types (Fig. 2e). These latter modules may represent shared functionalities across different cell types or poorly resolved intermediate states. Setting boundaries for cell clusters is a statistical matter, which can continue to accrue additional support with more miRNA expression data. miRNA abundance is not easily measured with commonly used single cell RNA sequencing platforms. With more highly resolved single cell quantification novel cell identities may become apparent or cell type boundaries may emerge as less bistable and rather depend upon an analog model.
The developing primate brain forms a tiered structure through and within which precursor cells migrate and acquire a series of identities ultimately leading to the diverse terminal identities of their progeny. Mitosis is a pivotal event in which the progeny reproduce the parental identity, acquire new identities or produce divergent identities in the two daughter cells. Mitosis wipes the slate of the parental profile and novel cell identities can arise in the daughter cells through transcription factor expression. miRNAs as less potent drivers of cell identity than transcription factors, but as critical tuners of precise and robust identities assume an expression profile tightly linked to cell identity. When serial identities emerge as cells migrate to their architectonic destinations in the absence of mitosis, environmental influences and intrinsic mechanisms drive neurons to their terminal identities. Interestingly, precursors that differentiate in the absence of mitosis must utilize miRNAs differently than in stably differentiated cells in which their prominent role is homeostatic to buffer change. This strategic difference in the role of miRNAs—to foster change rather than stability—suggests further realms of cellular systems control over cell identity.
Methods
Dissociation cell culture
Cortical cells were dissociated using Papain (Worthington labs) and cultured on matrigel (BD Biosciences) coated tissue culture treated plates. Cells were plated at approximately 100,000–200,000 cells per well of a 12 well plate. Culture media used in this experiment consisted of DMEM (Invitrogen, 11965) supplemented with N2 (Invitrogen, 12587–010) and B27 (17502–048), as well as Penicillin and Streptomycin, but no serum. At the time of plating, culture media was spiked with recombinant human FGF-basic (10ng/ml, Peprotech, AF-100–18B). Approximately 24–48 hours after plating, cells were transfected with plasmids using Lipofectamine 2000 (Life Technologies) following manufacturer protocol. BrdU (Sigma) was diluted in the culture media for dissociated cells (DMEM, supplemented with B27 and N2, with Penicillin-Streptomycin) at 50 ?g/ml.
Sample Collection
All mice in this study were obtained from Simonsen Laboratories and maintained according to protocols approved by the Institutional Animal Care and Use Committee at UCSF. De-identified human tissue samples were collected with patient consent in strict observance of the legal and institutional ethical regulations from elective pregnancy termination. Protocols were approved by the Human Gamete, Embryo, and Stem Cell Research Committee (UCSF institutional review board) at the University of California, San Francisco. Sample processing for dissociation as well as fixation, cryosectioning, and long-term storage was performed as described before 15.
In-utero electroporation
Survival in utero surgery was performed in strict observance of protocols and recommendations approved by the Institutional Animal Care and Use Committee at UCSF. Plasmids were injected at approximately 1.5 ?g/?l as described before 24,47. Although the ORC4 protein sequence is highly conserved, the miR-2115–5p MRE sequence is not fully conserved in mouse. We generated a mutant miR-2115–5p hairpin sequence (mmu-miR-2115) whose seed would be complementary to the mouse ORC4 mRNA coding sequence at the site homologous to the human miR-2115–5p MRE.
Plasmid Constructs
gBlocks gene fragments for the respective target sites or miRNAs (table S3) with the restriction site(s) at their ends were purchased from IDT. Restriction site cloning was performed using the standard method. Full length ORC4 expression plasmid was generated using gene synthesis using a full length human ORC4 and cloned into CAG-IRES-GFP vector using the GeneArt service (Thermo Fisher).
Luciferase Activity Assay
Luciferase activity assay was performed as described earlier 48 in HEK293 cells. The target sites were cloned in psiCHECK2 (Promega) plasmid (table S3) and miR-2115 and miR-9 were cloned in pCAG-GFP (Addgene 11150) plasmid (Table S2). miRNA mimics (Life Technologies) were used for other miRNA assays. For ORC4, wild type MRE reporter was named “WT-ORC4”, and a reporter lacking the miR2115 MRE was called “MT-ORC4”.
Fluorescence Reporter Expression Measurement
Fluorescence reporters were generated by cloning the respective target site, as indicated above, into the NotI site of the GFP expression vector (Addgene plasmid 11153), and validated by Sanger sequencing. For each culture condition, 100ng of plasmid was transfected into primary dissociated human cells in culture, alone or together with LNA anti-miRNA inhibitor (Qiagen) (50pmol). Mutant reporters were generated as gBlocks from IDT lacking the miRNA response element being tested. After 72 hours in culture, cells were lightly fixed in 4% PFA, followed by imaging of fluorescence using SP5 confocal microscope at constant laser power and detector conditions. Approximately 10 fields of view were considered at random, and all cells in every field of view were used to quantify the average intensity of fluorescence signal (per pixel) across four conditions for every miRNA:mRNA interaction examined: control reporter with wild type MRE, control reporter with wild type MRE together with anti-miRNA inhibitor, reporter lacking an MRE, and a reporter lacking an MRE together with anti-miRNA inhibitor. For wild type and mutant reporter, we calculated the ratio of average fluorescence per pixel with and without anti-miR inhibitor. The experiment was repeated for cells derived from three independent biological specimens (N=3, at GW14, GW18, and GW20), except for miR-2115-ORC4 CDS interactions which was tested in additional three biological specimens (GW15, 17, 19). For experiments presented in Figure S1g, loading control CAG-dsRed expression plasmid (100ng) was co-transfected with the reporter, to increase the accuracy of our quantification efforts. dsRed immunofluorescence was to normalize the GFP fluorescence signal in every cell analyzed.
In-situ hybridization
In situ hybridization in primary tissue sections was performed as described before 49, with the exception that we did not perform a probe linearization step. DIG-conjugated probes for miRNA detection were purchased from Exiqon or Qiagen.
Immunofluorescence
Thin 20 μm cryosections were collected on superfrost slides (VWR) using Leica CM3050S cryostat. Immunohistochemistry based detection of specific antigens was performed according to standard protocols. In short, heat-mediated antigen retrieval was performed in 10mM sodium citrate for 15 min. Cells were permeabilized in phosphate buffered saline (pH = 7.4, PBS) supplemented with 2% Triton X-100. Blocking buffer consisted of PBS supplemented with 10% donkey serum, 0.2% gelatin and 2% Triton X-100. The antibodies used in this study included chicken anti-GFP (1:1000, Aves Labs GFP-1020), rabbit anti-PAX6 (1:300, Covance prb-278p), and mouse anti-pHH3 (1:100, Abcam ab1791). These antibodies were used previously in human tissue50. Secondary antibodies used were from Life Technologies. Nuclei were counterstained with DAPI (Sigma).
After cell fixation, BrdU epitope was unmasked using 2N hydrochloric acid, neutralized using 0.1M boric acid, and stained using a rat anti-BrdU [BU1/75 (ICR1)] antibody (1:50, Abcam ab6326). This antibody was used previously in human tissue15. Coverslips were mounted with Aqua-mount (Lerner Laboratories)
Images were collected with a Leica DMI 4000B microscope using a Leica DFC295 camera Leica TCS SP5 X Confocal microscope. Quantification of immune-positive cells was performed in Adobe Photoshop. Mouse embryonic electroporation cortical staining was quantified as described before 24. Quantification results for every biological replicate (embryo) represent an average of quantification across three non-adjacent sections. Replicates were drawn from at least two independent litters. Quantification of BrdU incorporation into SOX2 positive primary cells in culture was performed by imaging randomly selected fields in the well using tile-scanning feature. GFP positive cells were first evaluated for expression of SOX2, and only after that, BrdU immunoreactivity was assessed. Between 100 and 200 SOX2/GFP double-positive cells were evaluated per well. Quantification of cell cycle phenotypes was performed by fitting linear regression, and cell cycle parameters were calculated as described before 51.
AGO2-HITS-CLIP
We have chosen AGO2 as a target for HITS-CLIP experiment as most other studies did for miRNA target identification because among the Argonaute family of proteins, AGO2 has slicer activity, which is not the case for AGO1, AGO3 or AGO4 in mammals52,53. This mechanism of mRNA destruction by miRNA binding is both prevalent and implements a more rapid and more readily detectable effect on downstream pathways. Moreover, we chose AGO2 because the data would be suitable for comparison with the previously generated HITS-CLIP data in the adult brain33, which we analyzed to gain insights into the preservation of the miRNA - mRNA interaction modules in Supplementary Figure S11b. The experiments were performed as described earlier14 except for a few modifications. Monoclonal anti-Ago2 antibody (Sigma, 11A9 clone, SAB4200085) was used to perform immunoprecipitation of protein on protein G dynabeads (Invitrogen, 100–03D). For the negative control, goat anti-rat IgG antibody (Sigma, A9037) was used. RNase dilution of 1:50,000 was used after optimization. Primary tissue samples at stages corresponding to peak neurogenesis (gestational weeks GW 15–16.5), and at stages corresponding to upper layer neurogenesis and early gliogenesis were analyzed. Sample quality and data processing metrics were comparable to previously published results for adult human brain HITS-CLIP study 33. 3258 genes were actively targeted by miRNAs through protein coding (CDS) or 3’ untranslated regions (3’UTR) of the transcript. Strikingly, close to 80% of gene targets were detected in at least two samples, suggesting that we recovered the majority of in vivo mRNA targets.
Library Preparation for Sequencing RNA tags on Ion-Torrent
For library preparation, all steps were performed as described before 14, except that the primers with adapter sequences were modified according to the Ion Torrent sequencing platform.
Primers with Adapter sequences -
DSFP5 – 5’- CCATCTCATCCCTGCGTGTCTCCGACTCAGAGGGAGGACGATGCGG −3’
DSFP3 – 5’- CCTCTCTATGGGCAGTCGGTGATCCGCTGGAAGTGACTGACAC −3’
The bands corresponding to AGO2:miRNA:target complexes (130 kDa) were cut. miRNA libraries and target mRNA libraries were made separately.
Single-cell qPCR Analysis
The capture of single-cells was done using the C1 Single-Cell Auto Prep Integrated Fluidic Circuit (IFC), which uses a microfluidic chip to capture the cells, perform lysis, reverse transcription and cDNA amplification in nano-liter reaction volumes of miRNA and mRNA species at the same time. The details of the cell capture protocol used are described in protocol 100–6667 at http://www.fluidigm.com/. During the reverse transcription step, miRNAs are reverse-transcribed to cDNA using stem loop RT primers from the Megaplex RT primer pool (Life Technologies) that are specific for mature miRNA species and reagents from the Single-cell-to-CT kit (Life Technologies). mRNA species are reverse-transcribed at the same time during this process using mRNA primers which are present in the Single-Cell VILO RT. Megaplex primers and mRNA primers are added at the recommended concentrations. The choice of the sc-PCR microRNA targets was constrained by the pool of miRNA pre-enrichment primers from Life Technologies, and by the cost of Taqman primers. Therefore, the decision to include a primer for particular miRNA was driven by a combination of expression analysis, literature search. In particular, we leveraged published expression profiling data from the developing human or non-human primate brain tissue33,54–57.
During the PCR step, products are uniformly amplified from cDNA templates using the Megaplex PreAmp Primers (Life Technologies), a pool of DELTAgene primers and Single-Cell Preamp mix from the Ambion Single-Cell-to-CT kit (Life Technologies).
The cycling conditions were as followed: for reverse transcription, primers were annealed at 16C for 2min, followed by extension at 42C for 1min and extension at 50C for 1min. This cycle was repeated 40 times, followed by a hold step at 85C for 5min. For preamplification, polymerase was activated at 95C for 10min, followed by incubation at 55C for 2min and 72C for 2min. Preamplification cycling steps were performed by denaturing step at 95C for 15min followed by extension at 60C for 4min. At the end, the reaction was incubated at 99.9C for 10min, and the reaction was cooled to 4C.
After pre-amplification PCR, the amplicons were diluted 1:4 with C1 DNA Dilution Reagent (Fluidigm 100–5317) and stored in −20C until needed. qPCR was carried out using the 96.96 dynamic array (Fluidigm Corporation) following the manufacturer’s protocol (100–3909 and 100–9792). Gene expression analysis was done using the Fluidigm Real-Time PCR Analysis Software (v.3.0.2). Ct values were obtained and then square root normalized to stabilize the variance.
Clustering of single-cells was performed using a recently developed method combining Louvain clustering of single-cell sample coordinates with Jaccard distance metric 58. PCA dimensionality reduction and clustering was performed in the space of the following genes meaningful for cell type classification:
| CORO1C | FAM107A | GLI3 | CLU | CENPF | PROM1 | NES | C3 |
| SATB2 | ALDOA | AUTS2 | EOMES | TBR1 | SOW10 | MOXD1 | CSF3R |
| TAGLN3 | SEMA3C | ELAVL2 | GRIA2 | CLDN11 | TJP1 | PDGFD | PPP1R17 |
| DDAH1 | TGFBR1 | NOVA1 | BCL11B | GAD1 | OLIG1 | NEUROG2 | NEUROD6 |
| ETV5 | DLX2 | RBFOX1 | CNTNAP2 | PALLD | MKI67 | PAX6 | GFAP |
| ADRA2A | FBXO32 | GPX3 | KIF26B | SLC1A3 | PARD6A | VIM | PARD3 |
| CBX1 | DLL1 | CNN3 | NR4A2 | TFAP2C | SOX2 | PDGFRA | PTPRZ1 |
| MEF2C | DLX5 | FGF12 | ERBB4 | ASPM | OLIG2 | BTG2 | TNC |
| TUBB3 | ASCL1 | ROBO2 | HES1 | HES5 | SLA | SOX5 | DLX6-AS1 |
T-stochastic neighbor embedding (tSNE) was used to visualize cells in two dimensions.
Computational Analyses
Preprocessing and Mapping of AGO2-HITS-CLIP Tags
Barcodes were identified and reads were separated into each sample. Adapter sequences at both ends of reads were removed using Cutadapt 59. Trimmed reads were mapped to the human genome (hg19) with novoalign (http://www.novocraft.com/). Identical alignments were collapsed in each sample to remove PCR replicates. Strand specific read coverage was then calculated using the alignments from each sample.
miRNA profiling and Differential Expression Analysis
Adapter-trimmed AGO2 reads from miRNA libraries were mapped to human miRNA precursors from miRBase version 21 by using miRdeep2 60. DESeq2 was used to identify differentially expressed miRNAs between 2 developmental stages 61.
Peak Calling and Identification of Clusters/AGO Footprint
Piranha and zero-truncated negative binomial model (ZTNB) were used to calculate the significance of read coverage at each mapped genomic position in each samples 33,62. In summary, the read heights for each mapped genomic position were assumed to be sampled from an underlying ZTNB distribution, and parameters for ZTNB probability density functions were estimated using read height measured at all positions of the genome. P-values were calculated as the probability of observing a read height as large as the height in question and assigned to each position. We used Fisher’s method to calculate the joined p-values at each genomic position across all samples. Positions with a joined FDR <5% were deemed significant. Significant positions within 50 nts of one another were merged into a single contiguous interval as single AGO binding site, and resulting regions of less than 50 nts were symmetrically extended to 50 nts as accounting for the AGO binding footprint 63. AGO binding sites were then annotated according to their overlapping gene structures from GENCODE annotation Version 19.
Identification of miRNAs for each AGO binding sites
In order to identify miRNAs that bind each AGO site on mRNAs and lncRNAs, all types of canonical binding sites including 7mer-1A,7mer-m8 and 8mer 64 for all prenatal brain expressed miRNAs were searched within the full length AGO binding sites defined as above. miRNAs with miRNA recognition elements (MREs) within each AGO binding site were counted.
WGCNA Analysis
To detect groups of co-expressed miRNAs, we used the WGCNA R package 65. Cells analyzed using sc-qPCR were included in this analysis. To intersect miRNA co-expression module targets with bipartite network annotations (Fig. 2f), for every miRNA co-expression module we took the union of HITS-CLIP predicted targets of the miRNAs in that module.
Cell-type-specificity and Enrichment Analysis for miRNAs in single-cell data
We performed one-tailed Wilcox/Mann-Whitney-U Test to detect differential enrichment of each miRNA in each cell-type against the other cell-types. A miRNA is defined to be enriched in a cell-type, if it is expressed significantly higher in that cell-type compared to the other cells in that population (adjusted p<0.05).
Correlation with target mRNA levels
To correlate the relative abundance of co-expressed miRNAs and their targets across the major cell-types of the developing brain, we calculated the average module eigen-gene for modules detected in fig. S2 across radial glia, intermediate progenitors, neurons and interneurons. In parallel, we used published scRNA-seq data 38 and the cell-type assignments therein for radial glia, interneurons, intermediate progenitors, and excitatory cortical neurons, to calculate cell-type-wise average expression level of the genes identified as AGO2 bound miRNA targets as well as non-targets. We then correlated the average module eigen-gene with all of the gene targets predicted by HITS-CLIP and non-targets, and we calculated the average correlated for each in Fig. 2f. To compare the average correlations, we first converted correlation to z-scores using Fisher transformation. The differences between average z-scores (for targets and non-targets) were divided by the joint standard errors and significance was calculated based on normal distribution.
Gene Enrichment Analysis
Transcriptome data from similar prenatal brain tissues 66 was downloaded and the expressed genes were used as a better background set for gene enrichment analysis. The p-value was calculated with hypergeometric test, and adjusted using the Benjamini-Hochberg (BH) method (unless otherwise mentioned). To formally demonstrate that cell-type-specific genes are regulated by miRNAs, we used published scRNA-seq datasets to calculate cell-type-specificity scores using ideal vector correlation for every miRNA target identified by HITS-CLIP (Supplementary Table 7). We defined a gene to be a signature of a cell-type if its Pearson correlation is 3*stdev higher than the mean Pearson correlation score of all genes in all cell types (histogram in Fig. 1C) 15,27.
To calculate the overlap with Autism Spectrum Disorder (ASD) genes, we used genes associated with ASD annotated by SFARI Gene67. A module is defined to be enriched in ASD if the FDR corrected p value of hypergeometric test is less than 0.05 (the total number of expressed genes in prenatal brain tissues is used as the background as well).
Bipartite Community Detection Analysis
An unweighted (binary) bipartite network was constructed such that there exists an edge between each miRNA-mRNA pair if and only if such interaction is detected by AGO2-HITS-CLIP. First, this network was shown to be scale-free (P < 0.001), and then by generating random networks while constraining the number of edges and nodes to the original miRNA-mRNA network and calculating Barber`s modularity score 68 (Null distribution), the miRNA-mRNA network shows to be significantly modular (P for permutation test < 2e-16). Label Propagation followed by the Bipartite Recursively Induced Modularity (LP-BRIM) algorithm 18 was used for community detection in this bipartite network. Due to stochasticity of this method, we obtained robust communities by repeating LP-BRIM 2500 times and determining the overlap among all the iterations.
Bipartite network construction
The bipartite networks were generated using all detected target genes (3463 nodes in mode I) and all miRNA (514 nodes in mode II). An edge exists between only one node in mode I and one node in mode II if such interaction is present in HITS-CLIP data set (Table S2) (no edge is allowed between nodes in mode I (or mode II)). Hence in this study, three distinct networks were built using GW15–16, GW19–20 and the combined GW15–16/19–20 HITS-CLIP data set with 31859, 20734, and 36176 total edges, respectively.
Bipartite network modularity statistics
First, the R “bipartite” package 69 was used to show that the constructed network is scale-free. For consistency with the literature definition of scale-free networks, we showed that this network significantly obeys power law, truncated power law, and exponential distribution (fig. S11 A and B, P < 0.001). Next to demonstrate that these networks are significantly modular, we first generated a null distribution by randomly shuffling the edges between the nodes and calculating Barber`s modularity score. Finally, we showed that the network is significantly modular using a permutation test (P < 2e-16).
Bipartite community detection
We used R “lpbrim” package 70 to detect communities in the described networks. Due to stochasticity of the method, we ran the community detection algorithm 25 times and obtained the best solution among all 25 runs (with maximum Barber`s modularity score). We repeated this procedure 100 times, and found the consensus clustering as follows:
We first defined the Overlap Rate Matrix (ORM) as a symmetric N by N matrix where N is the total number of miRNA and target genes combined. Each entry ORMij shows the probability (or rate) of which target gene (or miRNA) i lies within the same cluster as target gene (or miRNA) j upon 100 runs.
The obtained ORM was further clustered using hierarchical clustering (fig. S11 C and D). Each obtained diagonal block after hierarchical clustering represents one community (contains both target genes and miRNAs that have significant intra-modular interactions compared to their inter-modular interactions with target genes (or miRNAs) in other communities.).
Finally, dynamic branch cutting implemented in “dynamicTreeCut” R package was used for tree cutting and assigning cluster ids to each node (fig. S11 E and F).
We repeated this procedure to show that the obtained clusters are robust and reproducible (fig. S11 C and D).
Bipartite module preservation analysis
In order to determine whether each identified module in GW15–16 network is statistically preserved in GW19–20, we first find the closest module in GW19–20 to GW15–16 in terms of the number of shared nodes in that module. Then, using the hypergeometric test, module preservation statistics were obtained and corrected using the BH method for multiple comparison. As shown in Fig. 3F, two homolog modules in GW15–16 and GW19–20 have similar interaction levels suggesting preserved topology of modules as well. Note that names of GW15–16 module colors are used to label Fig. 3F.
Inference of evolutionary history of miR-2115
To infer the evolutionary history of miR2115, we used genome sequences obtained from the UCSC Genome Browser (the most recent genome assemblies used). Alignments were done using the Geneious bioinformatics platform (version 9.1.8). Human SPINK8 gene sequence is annotated with introns, exons, CDS and UTR regions and miR-2115 location according to “NCBI RefSeq” track on the UCSC Genome Browser. SPINK8 gene orthologs were located in chimpanzee, gorilla, orangutan, and gibbon genomes using “Other RefSeq” track on UCSC Genome Browser. We excluded Bonobo because of the poor quality of genome assembly at the area of interest. Primate SPINK8 sequences were aligned with human SPINK8 sequences individually using the MUSCLE Alignment algorithm. Primate SPINK8 sequences were annotated with introns, exons, CDS and UTR regions and miR-2115 location according to alignment with the annotated human SPINK8 sequence. Originally primate SPINK8 genes were annotated according to the “Other RefSeq” track on UCSC Genome Browser but this yielded varied and unreliable results. Presence or absence of miR-2115 was determined based on alignment of human miR-2115 to the orthologous primate sequence (with mature transcript and seed region taken into consideration). Annotated human, chimpanzee, gorilla, orangutan, and gibbon SPINK8 intron 3/4 (intron miR-2115 is located in) sequences were aligned together to visualize changes in intron sequences between species. Boundaries of insertions and deletions in SPINK8 intron 3/4 occurring between species defined based off evolutionarily chronological alignments of SPINK8 intron 3/4 sequences (e.g. gibbon and orangutan alignment, orangutan and gorilla alignment, etc.). Evolution of the SPINK8 intron 3/4 was predicted using the fewest number of mutations that would give rise to observed insertions and deletions.
Statistics
Statistical tests, sample sizes and assumptions are indicated in each corresponding figure legend, except for bipartite network analysis, which is described above. Data distribution was assumed to be normal but this was not formally tested.
Across all experiments and quantification, each tissue specimen was considered a biological replicate. Although we did not use formal methods of randomization, quantification of immunostaining was always performed on ‘blind’, such that identity of sample and condition was not known to the person performing the quantification.
Across all experiments, specimens with low viability of cells after plating, and specimens that have been contaminated with bacteria, or specimes for which we could not perform the entire experiment were not included in the analysis and not reported. For mouse embryos, we selected embryos with highest quality of electroporation based on visual assessment under a stereotaxic fluorescence microscope. For animal and human embryonic tissue, we did not examine the embryos for sex in these experiments.
No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications33. Across all experiments, data distribution was assumed to be normal but this was not formally tested.
Supplementary Material
Acknowledgement
The authors thank Shaohui Wang, Carmen Sandoval-Espinosa, Elmer Guzman, Aparna Bhaduri, Nianzhen Li for providing research resources, technical help, and helpful comments during manuscript preparation. N.R. acknowledges the support (Ramanujan Fellowship SB/S2/RJN-030/2017) from Science and Engineering Research Board, Department of Science & Technology, India. This research was supported by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (KSK), NIH awards U54NS100717 (KSK), MH105989 (ARK) and R01NS075998 (A.R.K). This work was supported by a grant from the Simons Foundation/SFARI 491371 (TJN).
Footnotes
The authors declare no competing interests.
Accession code
The data used in this study are available as part of the publicly available Gene Expression Omnibus database under the accession number GSE107468.
Code Availability
Scripts used in data analysis for this manuscript can be found at: GitHub: https://github.com/mgolkaram/Nature-Neuroscience-2018-miRNA-mRNA-paper.
Other details
For other details regarding study design, please refer to the Life Sciences Reporting Summary associated with this manuscript.
REFERENCES
- 1.Tasic B Single cell transcriptomics in neuroscience: cell classification and beyond. Current opinion in neurobiology 50, 242–249, doi: 10.1016/j.conb.2018.04.021 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Griffiths JA, Scialdone A & Marioni JC Using single-cell genomics to understand developmental processes and cell fate decisions. Molecular systems biology 14, e8046, doi: 10.15252/msb.20178046 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanay A & Regev A Scaling single-cell genomics from phenomenology to mechanism. Nature 541, 331–338, doi: 10.1038/nature21350 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng H & Sanes JR Neuronal cell-type classification: challenges, opportunities and the path forward. Nature reviews. Neuroscience 18, 530–546, doi: 10.1038/nrn.2017.85 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Kosik KS MicroRNAs and cellular phenotypy. Cell 143, 21–26, doi: 10.1016/j.cell.2010.09.008 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Monticelli S et al. MicroRNA profiling of the murine hematopoietic system. Genome biology 6, R71, doi: 10.1186/gb-2005-6-8-r71 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fineberg SK, Kosik KS & Davidson BL MicroRNAs potentiate neural development. Neuron 64, 303–309, doi: 10.1016/j.neuron.2009.10.020 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Volvert ML, Rogister F, Moonen G, Malgrange B & Nguyen L MicroRNAs tune cerebral cortical neurogenesis. Cell death and differentiation 19, 1573–1581, doi: 10.1038/cdd.2012.96 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berezikov E et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120, 21–24, doi: 10.1016/j.cell.2004.12.031 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Kapsimali M et al. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome biology 8, R173, doi: 10.1186/gb-2007-8-8-r173 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baudet ML et al. miR-124 acts through CoREST to control onset of Sema3A sensitivity in navigating retinal growth cones. Nature neuroscience 15, 29–38, doi: 10.1038/nn.2979 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein E et al. Dicer is essential for mouse development. Nature genetics 35, 215–217, doi: 10.1038/ng1253 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Jonsson ME et al. Comprehensive analysis of microRNA expression in regionalized human neural progenitor cells reveals microRNA-10 as a caudalizing factor. Development 142, 3166–3177, doi: 10.1242/dev.122747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore MJ et al. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nature protocols 9, 263–293, doi: 10.1038/nprot.2014.012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollen AA et al. Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67, doi: 10.1016/j.cell.2015.09.004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camp JG et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proceedings of the National Academy of Sciences of the United States of America 112, 15672–15677, doi: 10.1073/pnas.1520760112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller JA et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206, doi: 10.1038/nature13185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X & Murata T Community detection in large-scale bipartite networks. Information and Media Technologies 5, 184–192 (2010). [Google Scholar]
- 19.Florio M et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347, 1465–1470, doi: 10.1126/science.aaa1975 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Yu B et al. miR-221 and miR-222 promote Schwann cell proliferation and migration by targeting LASS2 after sciatic nerve injury. Journal of cell science 125, 2675–2683, doi: 10.1242/jcs.098996 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Maiorano NA & Mallamaci A Promotion of embryonic cortico-cerebral neuronogenesis by miR-124. Neural development 4, 40, doi: 10.1186/1749-8104-4-40 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boumil RM et al. A missense mutation in a highly conserved alternate exon of dynamin-1 causes epilepsy in fitful mice. PLoS genetics 6, doi: 10.1371/journal.pgen.1001046 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buckanovich RJ, Yang YY & Darnell RB The onconeural antigen Nova-1 is a neuron-specific RNA-binding protein, the activity of which is inhibited by paraneoplastic antibodies. The Journal of neuroscience : the official journal of the Society for Neuroscience 16, 1114–1122 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nowakowski TJ et al. MicroRNA-92b regulates the development of intermediate cortical progenitors in embryonic mouse brain. Proceedings of the National Academy of Sciences of the United States of America 110, 7056–7061, doi: 10.1073/pnas.1219385110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magri L et al. c-Myc-dependent transcriptional regulation of cell cycle and nucleosomal histones during oligodendrocyte differentiation. Neuroscience 276, 72–86, doi: 10.1016/j.neuroscience.2014.01.051 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawakami Y et al. Impaired neurogenesis in embryonic spinal cord of Phgdh knockout mice, a serine deficiency disorder model. Neuroscience research 63, 184–193, doi: 10.1016/j.neures.2008.12.002 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Nowakowski TJ et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323, doi: 10.1126/science.aap8809 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadjighassem MR et al. Human Freud-2/CC2D1B: a novel repressor of postsynaptic serotonin-1A receptor expression. Biological psychiatry 66, 214–222, doi: 10.1016/j.biopsych.2009.02.033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deshar R, Cho EB, Yoon SK & Yoon JB CC2D1A and CC2D1B regulate degradation and signaling of EGFR and TLR4. Biochemical and biophysical research communications 480, 280–287, doi: 10.1016/j.bbrc.2016.10.053 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Fededa JP et al. MicroRNA-34/449 controls mitotic spindle orientation during mammalian cortex development. The EMBO journal 35, 2386–2398, doi: 10.15252/embj.201694056 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J et al. Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proceedings of the National Academy of Sciences of the United States of America 111, E2851–2857, doi: 10.1073/pnas.1407777111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sousa AMM et al. Molecular and cellular reorganization of neural circuits in the human lineage. Science 358, 1027–1032, doi: 10.1126/science.aan3456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boudreau RL et al. Transcriptome-wide discovery of microRNA binding sites in human brain. Neuron 81, 294–305, doi: 10.1016/j.neuron.2013.10.062 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu YE, Parikshak NN, Belgard TG & Geschwind DH Genome-wide, integrative analysis implicates microRNA dysregulation in autism spectrum disorder. Nature neuroscience 19, 1463–1476, doi: 10.1038/nn.4373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abu-Elneel K et al. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics 9, 153–161, doi: 10.1007/s10048-008-0133-5 (2008). [DOI] [PubMed] [Google Scholar]
- 36.He M et al. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron 73, 35–48, doi: 10.1016/j.neuron.2011.11.010 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J et al. A reciprocal antagonism between miR-376c and TGF-beta signaling regulates neural differentiation of human pluripotent stem cells. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 28, 4642–4656, doi: 10.1096/fj.13-249342 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Nowakowski TJ, Pollen AA, Sandoval-Espinosa C & Kriegstein AR Transformation of the Radial Glia Scaffold Demarcates Two Stages of Human Cerebral Cortex Development. Neuron 91, 1219–1227, doi: 10.1016/j.neuron.2016.09.005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guernsey DL et al. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nature genetics 43, 360–364, doi: 10.1038/ng.777 (2011). [DOI] [PubMed] [Google Scholar]
- 40.de Munnik SA et al. Meier-Gorlin syndrome: growth and secondary sexual development of a microcephalic primordial dwarfism disorder. American journal of medical genetics. Part A 158A, 2733–2742, doi: 10.1002/ajmg.a.35681 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Marin RM, Sulc M & Vanicek J Searching the coding region for microRNA targets. Rna 19, 467–474, doi: 10.1261/rna.035634.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramachandran Iyer EP et al. Barcoded oligonucleotides ligated on RNA amplified for multiplex and parallel in-situ analyses. bioRxiv, doi: 10.1101/281121 (2018). [DOI] [PMC free article] [PubMed]
- 43.Faridani OR et al. Single-cell sequencing of the small-RNA transcriptome. Nat Biotechnol 34, 1264–1266, doi: 10.1038/nbt.3701 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Srinivasan K et al. A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proceedings of the National Academy of Sciences of the United States of America 109, 19071–19078, doi: 10.1073/pnas.1216793109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hevner RF et al. Tbr1 regulates differentiation of the preplate and layer 6. Neuron 29, 353–366 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Marin O, Anderson SA & Rubenstein JL Origin and molecular specification of striatal interneurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 20, 6063–6076 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forman JJ, Legesse-Miller A & Coller HA A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proceedings of the National Academy of Sciences of the United States of America 105, 14879–14884, doi: 10.1073/pnas.0803230105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hafner M et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141, doi: 10.1016/j.cell.2010.03.009 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schnall-Levin M, Zhao Y, Perrimon N & Berger B Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3’UTRs. Proceedings of the National Academy of Sciences of the United States of America 107, 15751–15756, doi: 10.1073/pnas.1006172107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schnall-Levin M et al. Unusually effective microRNA targeting within repeat-rich coding regions of mammalian mRNAs. Genome research 21, 1395–1403, doi: 10.1101/gr.121210.111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.