Abstract
To assess the influence of microRNA-183 (miR-183) on the migration and invasion of A375 human melanoma cells, an A375 cell line with stable miR-183 overexpression or knockdown was constructed using lentiviral transfection. The change of miR-183 expression in these cells and in non-transfected controls was verified using reverse transcription-quantitative polymerase chain reaction. The impact of miR-183 on experimental A375 cell migration and invasion was assessed using a scratch and Transwell assay. The expression of Ezrin and matrix metalloprotease-9 (MMP-9), which are two mediator proteins that serve roles in tumor cell migration and invasion, were analyzed in each cell group via western blotting. The results of the present study indicated that miR-183 overexpression significantly inhibits A375 cell migration and invasion, which may be facilitated by miR-183 knockdown. Furthermore, Ezrin and MMP-9 protein levels were negatively associated with miR-183 expression, indicating that miR-183 may function as a tumor suppressor by inhibiting the expression of these two proteins. Additionally, miR-183 downregulation may be associated with the progression of melanoma.
Keywords: microRNA-183, Ezrin, matrix metalloproteinase-9, A375, melanoma, migration, invasion
Introduction
Melanoma is one of the fastest-growing malignancies worldwide (1,2), the incidence of which has continued to increase over the past several decades (2). Annually, this incidence increase varies among populations, ranging from 3 to 7%, which equates to a doubling of rates every 10 to 20 years (2). Although different treatment methods have been developed, metastasis remains a primary threat to patients with melanoma, resulting in high recurrence rates following therapy and a poor prognosis (3). Melanoma metastasis is a complex process, the mechanism of which is yet to be fully elucidated. However, several steps have been studied in the metastatic process of human melanoma, including dysregulated angiogenesis, angiotropism, cancer cell extravasation, immunoescape, epithelial-to-mesenchymal transition (EMT), capillary adhesion and colonization in specific organs (4,5). Cellular migration and invasion activity, which involve the dysregulation of multiple key adapters or signaling pathways, are considered vital determinants of melanoma metastasis and as such are attracting wide research interests (4,6).
In recent years, the dysregulation of Ezrin and matrix metalloprotease-9 (MMP-9) has been determined to be an important metastasis-associated mechanism in tumor cells of different tissue origins (7–12). Ezrin, a 69 kDa cytoplasmic peripheral membrane protein belonging to the Ezrin/radixin/moesin (ERM) protein family, is an essential factor for gynecologic cancer (13), osteosarcoma (14), rectal cancer (15,16) and melanoma (17) metastasis. Together with other ERM family proteins, Ezrin serves as an intermediate between cell membrane proteins and the actin cytoskeleton (18), regulating a series of cellular activities including cell survival, adhesion, migration and invasion (18,19). It has been confirmed in multiple different tumor models that Ezrin overexpression may contribute to tumor metastasis and invasion (20–24), potentially by inducing and promoting the EMT process (22,25–27). MMP-9 is a 92 kD matrixin (a type of enzyme), belonging to the zinc-metalloproteinase family, which functions to degrade the extracellular matrix (28). Mostly secreted as inactive pro-proteins and activated following cleavage by other extracellular proteinases, MMPs serve a role in extracellular matrix breakdown in certain physiological processes, including embryonic development, angiogenesis, wound healing and cell migration, as well as in some pathological progressions, including cancer cell invasion and metastasis (29). Emerging evidence has indicated that the overexpression or hyper-activation of MMP-9 contributes to tumor metastasis by facilitating tumor cell migration and invasion in different models (10,30,31).
Recently, abnormal changes in microRNA (miRNA or miR) levels have been proposed to be a primary mechanism for the dysregulation of effector proteins associated with tumor development (32–36). MiRNAs are small non-coding RNAs that serve a function in gene silencing via sequence-specific base pairing with the 3′ or 5′ untranslated regions of target mRNAs (37). This results in mRNA destabilization and degradation, thus serving as an important post-transcriptional regulatory mechanism (37). The dysregulation or loss of function of certain key regulatory miRNAs may therefore contribute to tumor development via the subsequential dysregulation of tumor suppressor proteins or oncoproteins (38). Several studies have indicated that a change in human miR-183 level is closely associated with tumor development (39–44). However, the influence of miR-183 expression change on melanoma is yet to be fully elucidated.
Previous studies have demonstrated that Ezrin and MMP-9 are direct targets of miR-183 regulation, such that a decrease of miR-183 causes the upregulation of Ezrin and MMP-9, which leads to the facilitated migration and invasion of tumor cells with different tissue origins (45–51). To assess the impact of miR-183 dysregulation in melanoma, the present study constructed an A375 human melanoma cell line with stable miR-183 overexpression or knockdown via lentiviral transfection with an miR-183 mimic or inhibitor, respectively. Changes in miR-183 expression in the miR-183 mimic, inhibitor and control group was verified using a reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay following total RNA isolation. Furthermore, the impact of miR-183 on the migration and invasion of A375 cells was evaluated using a scratch and Transwell assay, respectively. Ezrin and MMP-9 levels were analyzed using western blotting in each group. The results indicated that miR-183 functions as a tumor suppressor in melanoma by inhibiting tumor cell migration and invasion via the downregulation of Ezrin and MMP-9 protein and the deregulation of miR-183. Therefore, miR-183 may be an important factor for melanoma metastasis and development. These results may provide novel insights into the molecular mechanism of melanoma progression and may also elucidate a target for the development of novel gene therapy against melanoma and other metastatic tumors.
Materials and methods
Cell culture
A375 cells (Chinese Academy of Sciences Cell Bank, Beijing, China) were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Biological Industries, Kibbutz Beit Haemek, Israel) and maintained at 37°C in a humidified incubator with 5% CO2, unless otherwise indicated.
MiR-183 overexpression and knockdown in A375 cells and the evaluation of miR-183 mRNA expression
The sequence of miR-183 is 5′-GUGAAUUACCGAAGGGCCAUAA-3′. MiR-183 overexpression and knockdown in A375 human melanoma cells was achieved using an miR-183 mimic, an miRNA mimic negative control (NC) [an insignificant short nucleotide sequence], an miR-183 inhibitor, an miRNA inhibitor NC and blank plasmids were all obtained from Guangzhou RiboBio Co., Ltd. (Guangzhou, China) following the manufacturer's protocol. Following transfection miR-183 (250 µg/ml) for 12 h by Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol, total RNA from each cell group was isolated via TRIzol-chloroform-isopropanol (TaKaRa, Bio, Inc., Otsu, Japan) extraction and miR-183 mRNA expression was assessed using RT-qPCR analysis with an All-in-One™ miRNA RT-qPCR detection kit (GeneCopoeia, Inc., Rockville, MD, USA) following the manufacturer's protocol. The sequences of miR-183 primers, forward: 5′-CGCGGTATGGCACTGGTAGA-3′, and reverse: 5′-AGTGCAGGGTCCGAGGTATTC-3′. MiR-183 expression levels were normalized to levels of U6 snRNA (5′-AAATTCGTGAAGCGTTCC-3′) using the 2−ΔΔCq method (52). Detection was achieved by SYBR green qPCR with the following conditions: 95°C for 10 min followed by 40 cycles of 95°C for 10 sec, 60°C for 20 sec and 72°C for 20 sec.
Scratch assay
A375 cells transfected with the miR-183 mimic, mimic NC, miR-183 inhibitor or inhibitor NC plasmid were cultured (37°C) overnight into confluent monolayers. Scratches were generated gently in each cell culture using a sterile 200 µl pipette tip. Cell migration towards the mid-line of the scratch was monitored at 0 and 24 h using a light microscope at ×100 magnification.
Transwell migration and invasion assays
Following pre-culture in serum-free RPMI-1640 medium for 24 h at 37°C, A375 cells with the aforementioned transfections were trypsinized, re-suspended in serum-free RPMI-1640 medium and diluted to a density of ~10,600 cells/ml. Cells (100 µl of each group) were then seeded in the inner chamber membrane of the Transwell apparatus. For the assessment of invasion activity, the inner chamber was coated with fibronectin under serum-free conditions and the uncoated inner chamber was used for the evaluation of migration activity. RPMI-1640 medium supplemented with 50% fetal bovine serum was added in the lower chamber of the Transwell apparatus in each group and cells were cultured for 24 h at 37°C in a humidified incubator with 5% CO2. A549 cells on the lower surface of the inner chamber were then gently rinsed with PBS and stained with crystal violet staining solution at room temperature for 10 min (Beyotime Institute of Biotechnology, Haimen, China) prior to being counted using light microscopy at ×200 magnification. Five randomly selected fields of view were selected.
Western blotting
A375 cells that underwent the aforementioned transfections were cultured in 6-well plates under normal culture conditions (37°C for 24 h). A375 cells were homogenized in lysis buffer [10 mM Tris base, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.5% NP-40, and a protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, Inc.)]. Protein concentrations were determined using a BCA assay kit (Thermo Fisher Scientific, Inc.) with bovine serum albumin (2 mg/ml) as a standard. The total protein of each cell group (20 µl) was separated using polyacrylamide gel (15%) electrophoresis under reducing conditions, followed by transferral onto nitrocellulose membranes. Following blocking at 4°C overnight with non-fat milk and preparation with PBS, membranes were probed with antibodies (incubated 16 h at 4°C) targeting human Ezrin (cat. no. ab40839; Abcam, Cambridge, UK; dilution of 1:2,000) and MMP-9 (cat. no. ab76003; Abcam; dilution of 1:1,000), with β-actin (cat. no. ab8227; Abcam; dilution of 1:2,000) as an internal reference. The membranes were then incubated with horseradish peroxidase conjugated secondary antibodies (dilution of 1:5,000, incubated 1 h at room temperature) (Goat Anti-Rabbit IgG H&L; cat. no. ab6702; Abcam) and colorized using an ECL substrate (Thermo Fisher Scientific, Inc.). The image of each sample was captured using chemo-fluorescence-sensitive film and the gray value of each band was analyzed. The quantity of target protein was evaluated by the ratio of its gray value to the internal reference using ImageJ software, version 1.48 (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
All experiments were independently repeated at least three times. Statistical analysis was performed using SPSS statistical software, version 17.0 (SPSS, Inc., Chicago, IL, USA). All statistical data were presented as the mean ± standard deviation. A Student's t-test was performed to assess significance and P<0.05 was considered to indicate a statistically significant difference.
Results
Analysis of miR-183 expression following transfection
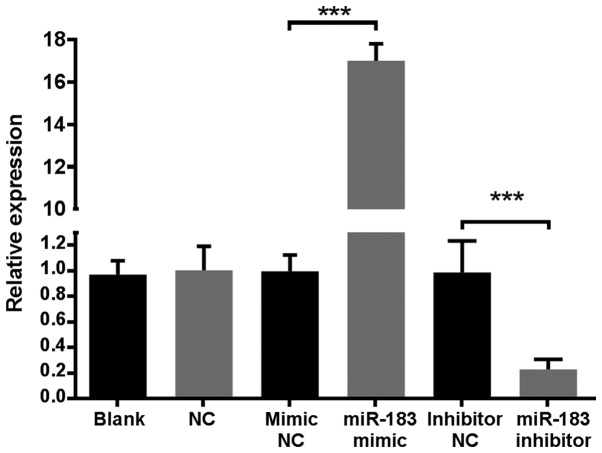
To determine the influence of increased and decreased miR-183 expression in human melanoma, A375 cells were transfected with miR-183 mimic and miR-183 inhibitor sequences, which served as the experimental groups. MiRNA mimic and inhibitor NC sequences were utilized as internal reference groups, using a lentiviral plasmid transfection system. A375 cells that were non-transfected and transfected with blank plasmids were used as external references. MiR-183 expression in each group 12 h following transfection was analyzed using RT-qPCR following total RNA isolation. The results demonstrated that the expression of miR-183 in the miR-183 mimic group was >16-fold higher compared with all control groups, while the miR-183 inhibitor group was 5-fold lower compared with control groups (Fig. 1). These results indicated that the established cell line was stable. MiR-183 levels in the mimic and inhibitor NC groups were similar to those of the blank and NC groups, indicating that transfection alone did not affect miR-183 expression. Additionally, no notable morphological changes were observed among A375 cells in the blank, NC, mimic NC and inhibitor NC groups (data not shown). Therefore, A375 cells in mimic NC and inhibitor NC groups alone were utilized as NCs in future experiments.
Figure 1.
Comparison of miR-183 expression in A375 human melanoma cells with different transfections. The blank group consisted of non-transfected A375 cells. A375 cells transfected with blank plasmids were utilized as the NC. Data are presented as fold change relative to the mean expression of the NC group. ***P<0.001. miR, microRNA; NC, negative controls.
MiR-183 inhibits the migration of A375 cells in vitro
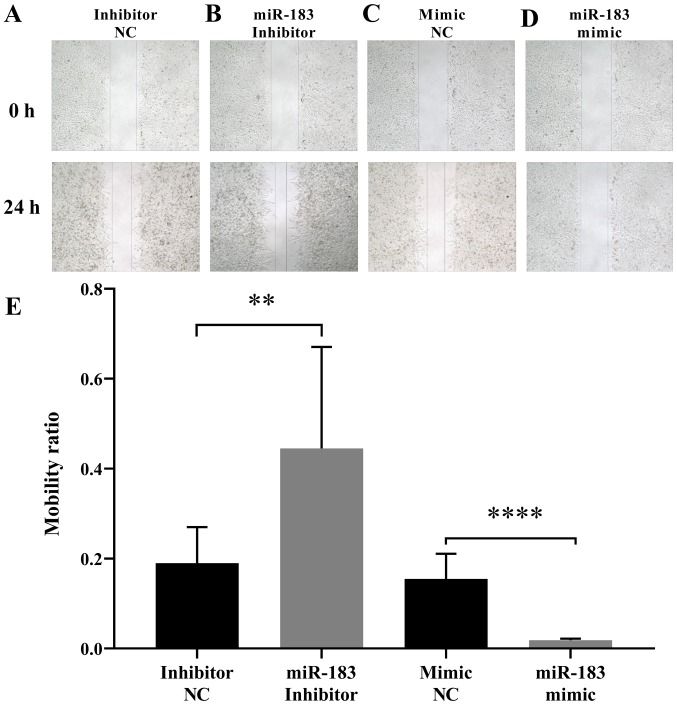
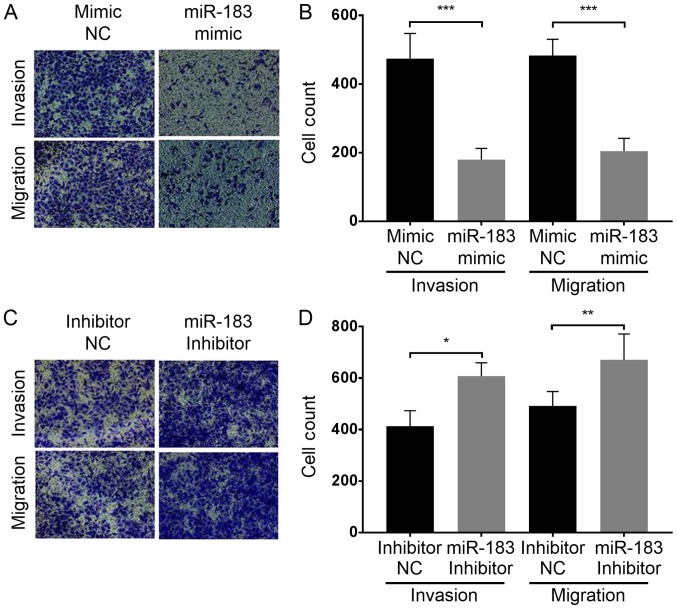
To assess the influence of miR-183 expression changes on A375 cell migration, a scratch assay was performed as aforementioned. The results demonstrated that the artificial upregulation of miR-183 expression significantly suppressed A375 cell migration, whereas miR-183 downregulation promoted A375 cell migration when compared with their respective controls (Fig. 2). This indicated that miR-183 inhibits the migration of A375 cells in vitro. To further evaluate the influence of miR-183 expression change on A375 cell migration and invasion, a Transwell assay was performed with each group of cells as aforementioned. The results demonstrated that miR-183 overexpression in the miR-183 mimic group significantly inhibited the migration and invasion of A375 cells in vitro compared with the mimic NC group (Fig. 3A and B). Furthermore, miR-183 knockdown in the inhibitor group promoted A375 cell migration and invasion compared with the inhibitor NC group (Fig. 3C and D). Taken together, this indicates that miR-183 suppresses A375 cell migration and invasion in vitro, and migration and invasion are increased by the downregulation of miR-183.
Figure 2.
Scratch assay to compare human melanoma A375 cell migration with different transfections. Representative results of A375 cells transfected with the (A) miRNA inhibitor NC (Inhibitor NC) and the (B) miR-183 inhibitor. Representative results of A375 cells transfected with the (C) miRNA mimic NC and the (D) miR-183 mimic. All images were obtained using a light microscope at 0 or 24 h following the scratch assay. The magnification of the images was ×100. (E) Statistical analysis of the scratch assay results at 24 h. The mobility ratio of each experiment was measured by dividing the scratch width at 24 h by that at 0 h. **P<0.01; ****P<0.0001. miRNA and miR, microRNA; NC, negative controls.
Figure 3.
Transwell assay to compare human melanoma A375 cell migration and invasion with different transfections. (A and C) Representative results of the Transwell assay. Cells on the lower surface of the inner chamber membrane were stained with crystal violet prior to gentle rinsing with PBS. (B and D) Statistical analysis of experimental group cell counts. A total of five microscopic fields of view (at ×200 magnification) were randomly selected from each inner membrane and cells were counted. *P<0.05; **P<0.01; ***P<0.001. NC, negative control; miR, microRNA.
Western blot analysis of Ezrin and MMP-9 in miR-183 overexpression or knockdown A375 cells
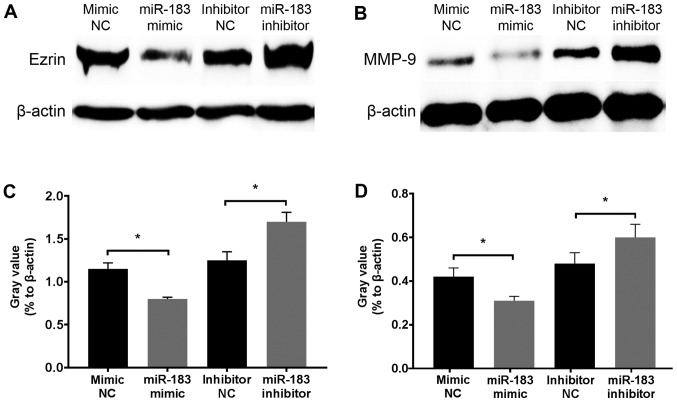
To determine the inhibitory mechanism of miR-183, changes in Ezrin and MMP-9 protein expression in A375 cells were assessed at 12 h following transfection with an miR-183 mimic or inhibitor. Western blotting revealed that Ezrin and MMP-9 were downregulated and upregulated following miR-183 overexpression and knockdown in vitro, respectively (Fig. 4). Considering the well-established association of Ezrin and MMP-9 with the migratory and invasive activity of melanoma, and that these proteins act as direct targets of miR-183 (45,46,49,53), the results demonstrated that miR-183 inhibits A375 cell migration and invasion potentially via the downregulation of Ezrin and MMP-9 expression.
Figure 4.
Comparison of Ezrin and MMP-9 protein expression in A375 human melanoma cells with different transfections. Representative western blotting results of (A) Ezrin and (B) MMP-9 protein expression 12 h following transfection in each experimental group. β-actin was utilized as an internal reference. Statistical analysis of (C) Ezrin and (D) MMP-9 protein expression in each experimental group. Data were acquired by measuring the gray value of each bar from the results of western blotting and were presented as the fold change relative to that of β-actin. *P<0.05. MMP-9, matrix metalloprotease-9; NC, negative controls; miR, microRNA.
Discussion
The results of the present study support the theory that miR-183 may serve as a tumor suppressor in melanoma. The migratory activity and invasiveness of A375 human melanoma cells was demonstrated to be negatively associated with miR-183 expression. Furthermore, Ezrin and MMP-9, which have been previously regarded as important promoters of melanoma metastasis (54–59), were revealed to be negatively regulated by miR-183, indicating that miR-183 inhibits A375 human melanoma cell migration and invasion, possibly through the downregulation of Ezrin and MMP-9 expression. Thus, the abnormal decrease of miR-183 in melanoma may cause Ezrin and MMP-9 overexpression, resulting in an increased metastatic activity. Several studies have revealed that miR-183 may exhibit either pro- or anti-metastatic functions in different tumor cells (60–65). More specifically, miR-183 has been considered to promote metastasis in most tumor models, including breast cancer (66), medulloblastoma (67), hepatocellular carcinoma (68), follicular thyroid carcinoma (43), esophageal cancer (69), gastric cancer (70), pancreatic cancer (71) and synovial sarcoma (72), while inhibiting metastasis in lung (73), colon (74) and ovarian cancer (75). This is possibly due to the heterogeneity of cancer and the differences in ontogenesis of different types of cancer.
MiR-183 is located and transcribed in cluster with miR-96 and miR-182, which exerts various functions (40). It regulates multiple mRNA targets; however, not all targets have been fully elucidated (38). Further studies are thus required to assess the transcriptional regulation of the miR-183-96-182 cluster under tumor pathological conditions. Considering the complexity of cytokinetic regulation, proteomic methods that assess the function and regulatory network of miR-183 target proteins, and particularly their interaction with cytoskeleton regulators and integrins, which interact with the extracellular matrix, are being employed (42). However, further studies are required to obtain a better understanding of the regulatory mechanism of miR-183. This may provide novel insights into the development of anti-metastasis gene therapy.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YSZ and GQW completed all the experiments and the statistical analysis. YSZ wrote and revised the manuscript. All authors read and approved the final the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Ward EM, Johnson CJ, Cronin KA, Ma J, Ryerson B, Mariotto A, Lake AJ, Wilson R, Sherman RL, et al. Annual report to the nation on the status of cancer, 1975-2014, featuring survival. J Natl Cancer Inst. 2017;109 doi: 10.1093/jnci/djx030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nikolaou V, Stratigos AJ. Emerging trends in the epidemiology of melanoma. Br J Dermatol. 2014;170:11–19. doi: 10.1111/bjd.12492. [DOI] [PubMed] [Google Scholar]
- 3.Damsky WE, Theodosakis N, Bosenberg M. Melanoma metastasis: New concepts and evolving paradigms. Oncogene. 2014;33:2413–2422. doi: 10.1038/onc.2013.194. [DOI] [PubMed] [Google Scholar]
- 4.Zbytek B, Carlson JA, Granese J, Ross J, Mihm MC, Jr, Slominski A. Current concepts of metastasis in melanoma. Expert Rev Dermatol. 2008;3:569–585. doi: 10.1586/17469872.3.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen DX, Bos PD, Massagué J. Metastasis: From dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 6.Bravo-Cordero JJ, Hodgson L, Condeelis J. Directed cell invasion and migration during metastasis. Curr Opin Cell Biol. 2012;24:277–283. doi: 10.1016/j.ceb.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long ZY, Wang TH. Advances of the role of Ezrin in migration and invasion of breast cancer cells. Sheng Li Ke Xue Jin Zhan. 2016;47:21–26. (In Chinese) [PubMed] [Google Scholar]
- 8.Liu HY, Gu WJ, Wang CZ, Ji XJ, Mu YM. Matrix metalloproteinase-9 and −2 and tissue inhibitor of matrix metalloproteinase-2 in invasive pituitary adenomas: A systematic review and meta-analysis of case-control trials. Medicine. 2016;95:e3904. doi: 10.1097/MD.0000000000003904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Candido S, Abrams SL, Steelman LS, Lertpiriyapong K, Fitzgerald TL, Martelli AM, Cocco L, Montalto G, Cervello M, Polesel J, et al. Roles of NGAL and MMP-9 in the tumor microenvironment and sensitivity to targeted therapy. Biochim Biophys Acta. 2016;1863:438–448. doi: 10.1016/j.bbamcr.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Pârvănescu V, Georgescu M, Georgescu I, Șurlin V, Pătraşcu Ș, Picleanu AM, Georgescu E. The role of matrix metalloproteinase-9 (MMP-9) as a prognostic factor in epithelial and lymphatic neoplasia. Chirurgia. 2015;110:506–510. [PubMed] [Google Scholar]
- 11.Li J, Wei K, Yu H, Jin D, Wang G, Yu B. Prognostic value of Ezrin in various cancers: A systematic review and updated meta-analysis. Sci Rep. 2015;5:17903. doi: 10.1038/srep17903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao DH, Zhu J, Wang WB, Dong F, Zhang Q, Fan HW, Zhang JZ, Wang YM. Correlations of ezrin expression with pathological characteristics and prognosis of osteosarcoma: A meta-analysis. ScientificWorldJournal. 2014;2014:837543. doi: 10.1155/2014/837543. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Choi SD, Fadiel A, Naftolin F. Erratum to: Ezrin is an essential marker for metastasis of gynecologic cancer. J Menopausal Med. 2016;22:188. doi: 10.6118/jmm.2016.22.3.188. [DOI] [Google Scholar]
- 14.Khanna C, Wan X, Bose S, Cassaday R, Olomu O, Mendoza A, Yeung C, Gorlick R, Hewitt SM, Helman LJ. The membrane-cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nat Med. 2004;10:182–186. doi: 10.1038/nm982. [DOI] [PubMed] [Google Scholar]
- 15.Korkeila EA, Syrjänen K, Bendardaf R, Laulajainen M, Carpén O, Pyrhönen S, Sundström J. Preoperative radiotherapy modulates ezrin expression and its value as a predictive marker in patients with rectal cancer. Hum Pathol. 2011;42:384–392. doi: 10.1016/j.humpath.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Patara M, Santos EM, de Almeida Coudry R, Soares FA, Ferreira FO, Rossi BM. Ezrin expression as a prognostic marker in colorectal adenocarcinoma. Pathol Oncol Res. 2011;17:827–833. doi: 10.1007/s12253-011-9389-4. [DOI] [PubMed] [Google Scholar]
- 17.Ilmonen S, Vaheri A, Asko-Seljavaara S, Carpen O. Ezrin in primary cutaneous melanoma. Mod Pathol. 2005;18:503–510. doi: 10.1038/modpathol.3800429. [DOI] [PubMed] [Google Scholar]
- 18.Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: Integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 19.McClatchey AI. Merlin and ERM proteins: Unappreciated roles in cancer development? Nat Rev Cancer. 2003;3:877–883. doi: 10.1038/nrc1213. [DOI] [PubMed] [Google Scholar]
- 20.Li M, Feng YM, Fang SQ. Overexpression of ezrin and galectin-3 as predictors of poor prognosis of cervical cancer. Braz J Med Biol Res. 2017;50:e5356. doi: 10.1590/1414-431x20165356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horwitz V, Davidson B, Stern D, Trope CG, Tavor Re'em T, Reich R. Ezrin is associated with disease progression in ovarian carcinoma. PLoS One. 2016;11:e0162502. doi: 10.1371/journal.pone.0162502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong J, Di C, Piao J, Sun J, Han L, Chen L, Yan G, Lin Z. Ezrin contributes to cervical cancer progression through induction of epithelial-mesenchymal transition. Oncotarget. 2016;7:19631–19642. doi: 10.18632/oncotarget.7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McRobert EA, Bach LA. Ezrin contributes to impaired podocyte migration and adhesion caused by advanced glycation end products. Nephrology. 2016;21:13–20. doi: 10.1111/nep.12526. [DOI] [PubMed] [Google Scholar]
- 24.Piao J, Liu S, Xu Y, Wang C, Lin Z, Qin Y, Liu S. Ezrin protein overexpression predicts the poor prognosis of pancreatic ductal adenocarcinomas. Exp Mol Pathol. 2015;98:1–6. doi: 10.1016/j.yexmp.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Lin Z, Chen B, Chen S, Jiang Z, Zhou T, Hou Z, Wang Y. Ezrin/NF-κB activation regulates epithelial-mesenchymal transition induced by EGF and promotes metastasis of colorectal cancer. Biomed Pharmacother. 2017;92:140–148. doi: 10.1016/j.biopha.2017.05.058. [DOI] [PubMed] [Google Scholar]
- 26.He J, Ma G, Qian J, Zhu Y, Liang M, Yao N, Ding Q, Chen L, Liu X, Xia T, et al. Interaction between ezrin and cortactin in promoting epithelial to mesenchymal transition in breast cancer cells. Med Sci Monit. 2017;23:1583–1596. doi: 10.12659/MSM.904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lan M, Kojima T, Murata M, Osanai M, Takano K, Chiba H, Sawada N. Phosphorylation of ezrin enhances microvillus length via a p38 MAP-kinase pathway in an immortalized mouse hepatic cell line. Exp Cell Res. 2006;312:111–120. doi: 10.1016/j.yexcr.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 28.Dang B, Duan X, Wang Z, He W, Chen G. A therapeutic target of cerebral hemorrhagic stroke: Matrix metalloproteinase-9. Curr Drug Targets. 2017;18:1358–1366. doi: 10.2174/1389450118666170427151657. [DOI] [PubMed] [Google Scholar]
- 29.Boziki M, Grigoriadis N. An update on the role of matrix metalloproteinases in the pathogenesis of multiple sclerosis. Med Chem. 2018;14:155–169. doi: 10.2174/1573406413666170906122803. [DOI] [PubMed] [Google Scholar]
- 30.Banday MZ, Sameer AS, Mir AH, Mokhdomi TA, Chowdri NA, Haq E. Matrix metalloproteinase (MMP) −2, −7 and −9 promoter polymorphisms in colorectal cancer in ethnic Kashmiri population - A case-control study and a mini review. Gene. 2016;589:81–89. doi: 10.1016/j.gene.2016.05.028. [DOI] [PubMed] [Google Scholar]
- 31.Gong L, Wu D, Zou J, Chen J, Chen L, Chen Y, Ni C, Yuan H. Prognostic impact of serum and tissue MMP-9 in non-small cell lung cancer: A systematic review and meta-analysis. Oncotarget. 2016;7:18458–18468. doi: 10.18632/oncotarget.7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yonemori K, Kurahara H, Maemura K, Natsugoe S. MicroRNA in pancreatic cancer. J Hum Genet. 2017;62:33–40. doi: 10.1038/jhg.2016.59. [DOI] [PubMed] [Google Scholar]
- 33.Xue J, Yang J, Luo M, Cho WC, Liu X. MicroRNA-targeted therapeutics for lung cancer treatment. Expert Opin Drug Discov. 2017;12:141–157. doi: 10.1080/17460441.2017.1263298. [DOI] [PubMed] [Google Scholar]
- 34.Manasa VG, Kannan S. Impact of microRNA dynamics on cancer hallmarks: An oral cancer scenario. Tumour Biol. 2017;39:1010428317695920. doi: 10.1177/1010428317695920. doi: 10.1177/1010428317695920. [DOI] [PubMed] [Google Scholar]
- 35.Latchana N, Ganju A, Howard JH, Carson WE., III MicroRNA dysregulation in melanoma. Surg Oncol. 2016;25:184–189. doi: 10.1016/j.suronc.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 36.Kanekura K, Nishi H, Isaka K, Kuroda M. MicroRNA and gynecologic cancers. J Obstet Gynaecol Res. 2016;42:612–617. doi: 10.1111/jog.12995. [DOI] [PubMed] [Google Scholar]
- 37.D'Angelo B, Benedetti E, Cimini A, Giordano A. MicroRNAs: A puzzling tool in cancer diagnostics and therapy. Anticancer Res. 2016;36:5571–5575. doi: 10.21873/anticanres.11142. [DOI] [PubMed] [Google Scholar]
- 38.Lima CR, Gomes CC, Santos MF. Role of microRNAs in endocrine cancer metastasis. Mol Cell Endocrinol. 2017;456:62–75. doi: 10.1016/j.mce.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 39.Moridikia A, Mirzaei H, Sahebkar A, Salimian J. MicroRNAs: Potential candidates for diagnosis and treatment of colorectal cancer. J Cell Physiol. 2018;233:901–913. doi: 10.1002/jcp.25801. [DOI] [PubMed] [Google Scholar]
- 40.Ma Y, Liang AJ, Fan YP, Huang YR, Zhao XM, Sun Y, Chen XF. Dysregulation and functional roles of miR-183-96-182 cluster in cancer cell proliferation, invasion and metastasis. Oncotarget. 2016;7:42805–42825. doi: 10.18632/oncotarget.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimono Y, Mukohyama J, Nakamura S, Minami H. MicroRNA regulation of human breast cancer stem cells. J Clin Med. 2015;5(pii):E2. doi: 10.3390/jcm5010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dambal S, Shah M, Mihelich B, Nonn L. The microRNA-183 cluster: The family that plays together stays together. Nucleic Acids Res. 2015;43:7173–7188. doi: 10.1093/nar/gkv703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wojtas B, Ferraz C, Stokowy T, Hauptmann S, Lange D, Dralle H, Musholt T, Jarzab B, Paschke R, Eszlinger M. Differential miRNA expression defines migration and reduced apoptosis in follicular thyroid carcinomas. Mol Cell Endocrinol. 2014;388:1–9. doi: 10.1016/j.mce.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 44.Zhang QH, Sun HM, Zheng RZ, Li YC, Zhang Q, Cheng P, Tang ZH, Huang F. Meta-analysis of microRNA-183 family expression in human cancer studies comparing cancer tissues with noncancerous tissues. Gene. 2013;527:26–32. doi: 10.1016/j.gene.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Ruan H, Liang X, Zhao W, Ma L, Zhao Y. The effects of microRNA-183 promots cell proliferation and invasion by targeting MMP-9 in endometrial cancer. Biomed Pharmacother. 2017;89:812–818. doi: 10.1016/j.biopha.2017.02.091. [DOI] [PubMed] [Google Scholar]
- 46.Fan D, Wang Y, Qi P, Chen Y, Xu P, Yang X, Jin X, Tian X. MicroRNA-183 functions as the tumor suppressor via inhibiting cellular invasion and metastasis by targeting MMP-9 in cervical cancer. Gynecol Oncol. 2016;141:166–174. doi: 10.1016/j.ygyno.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J, Zuo J, Lei M, Wu S, Zang X, Zhang C. Ezrin promotes invasion and migration of the MG63 osteosarcoma cell. Chin Med J. 2014;127:1954–1959. [PubMed] [Google Scholar]
- 48.Mu Y, Zhang H, Che L, Li K. Clinical significance of microRNA-183/Ezrin axis in judging the prognosis of patients with osteosarcoma. Med Oncol. 2014;31:821. doi: 10.1007/s12032-013-0821-3. [DOI] [PubMed] [Google Scholar]
- 49.Cao LL, Xie JW, Lin Y, Zheng CH, Li P, Wang JB, Lin JX, Lu J, Chen QY, Huang CM. miR-183 inhibits invasion of gastric cancer by targeting Ezrin. Int J Clin Exp Pathol. 2014;7:5582–5594. [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu J, Feng Y, Ke Z, Yang Z, Zhou J, Huang X, Wang L. Down-regulation of miR-183 promotes migration and invasion of osteosarcoma by targeting Ezrin. Am J Pathol. 2012;180:2440–2451. doi: 10.1016/j.ajpath.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 51.Zhao H, Guo M, Zhao G, Ma Q, Ma B, Qiu X, Fan Q. miR-183 inhibits the metastasis of osteosarcoma via downregulation of the expression of Ezrin in F5M2 cells. Int J Mol Med. 2012;30:1013–1020. doi: 10.3892/ijmm.2012.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Wang G, Mao W, Zheng S. MicroRNA-183 regulates Ezrin expression in lung cancer cells. FEBS Lett. 2008;582:3663–3668. doi: 10.1016/j.febslet.2008.09.051. [DOI] [PubMed] [Google Scholar]
- 54.Hsu YY, Shi GY, Kuo CH, Liu SL, Wu CM, Ma CY, Lin FY, Yang HY, Wu HL. Thrombomodulin is an ezrin-interacting protein that controls epithelial morphology and promotes collective cell migration. FASEB J. 2012;26:3440–3452. doi: 10.1096/fj.12-204917. [DOI] [PubMed] [Google Scholar]
- 55.Brambilla D, Fais S. The Janus-faced role of ezrin in ‘linking’ cells to either normal or metastatic phenotype. Int J Cancer. 2009;125:2239–2245. doi: 10.1002/ijc.24734. [DOI] [PubMed] [Google Scholar]
- 56.Federici C, Brambilla D, Lozupone F, Matarrese P, de Milito A, Lugini L, Iessi E, Cecchetti S, Marino M, Perdicchio M, et al. Pleiotropic function of ezrin in human metastatic melanomas. Int J Cancer. 2009;124:2804–2812. doi: 10.1002/ijc.24255. [DOI] [PubMed] [Google Scholar]
- 57.Kim A, Im M, Yim NH, Ma JY. Reduction of metastatic and angiogenic potency of malignant cancer by Eupatorium fortunei via suppression of MMP-9 activity and VEGF production. Sci Rep. 2014;4:6994. doi: 10.1038/srep06994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee KR, Lee JS, Kim YR, Song IG, Hong EK. Polysaccharide from Inonotus obliquus inhibits migration and invasion in B16-F10 cells by suppressing MMP-2 and MMP-9 via downregulation of NF-κB signaling pathway. Oncol Rep. 2014;31:2447–2453. doi: 10.3892/or.2014.3103. [DOI] [PubMed] [Google Scholar]
- 59.Tang ZY, Liu Y, Liu LX, Ding XY, Zhang H, Fang LQ. RNAi-mediated MMP-9 silencing inhibits mouse melanoma cell invasion and migration in vitro and in vivo. Cell Biol Int. 2013;37:849–854. doi: 10.1002/cbin.10107. [DOI] [PubMed] [Google Scholar]
- 60.Cheung CC, Lun SW, Chung GT, Chow C, Lo C, Choy KW, Lo KW. MicroRNA-183 suppresses cancer stem-like cell properties in EBV-associated nasopharyngeal carcinoma. BMC Cancer. 2016;16:495. doi: 10.1186/s12885-016-2525-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song C, Zhang L, Wang J, Huang Z, Li X, Wu M, Li S, Tang H, Xie X. High expression of microRNA-183/182/96 cluster as a prognostic biomarker for breast cancer. Sci Rep. 2016;6:24502. doi: 10.1038/srep24502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu C, Deng X, Wu J, Zhang J, Yang H, Fu S, Zhang Y, Han Y, Zou Y, Chen Z, et al. MicroRNA-183 promotes migration and invasion of CD133+/CD326+ lung adenocarcinoma initiating cells via PTPN4 inhibition. Tumour Biol. 2016;37:11289–11297. doi: 10.1007/s13277-016-4955-8. [DOI] [PubMed] [Google Scholar]
- 63.Miao F, Zhu J, Chen Y, Tang N, Wang X, Li X. MicroRNA-183-5p promotes the proliferation, invasion and metastasis of human pancreatic adenocarcinoma cells. Oncol Lett. 2016;11:134–140. doi: 10.3892/ol.2015.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu F, Zhang H, Su Y, Kong J, Yu H, Qian B. Up-regulation of microRNA-183-3p is a potent prognostic marker for lung adenocarcinoma of female non-smokers. Clin Transl Oncol. 2014;16:980–985. doi: 10.1007/s12094-014-1183-9. [DOI] [PubMed] [Google Scholar]
- 65.Zhou T, Zhang GJ, Zhou H, Xiao HX, Li Y. Overexpression of microRNA-183 in human colorectal cancer and its clinical significance. Eur J Gastroenterol Hepatol. 2014;26:229–233. doi: 10.1097/MEG.0000000000000002. [DOI] [PubMed] [Google Scholar]
- 66.Li P, Sheng C, Huang L, Zhang H, Huang L, Cheng Z, Zhu Q. MiR-183/-96/-182 cluster is up-regulated in most breast cancers and increases cell proliferation and migration. Breast Cancer Res. 2014;16:473. doi: 10.1186/s13058-014-0473-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weeraratne SD, Amani V, Teider N, Pierre-Francois J, Winter D, Kye MJ, Sengupta S, Archer T, Remke M, Bai AH, et al. Pleiotropic effects of miR-183~96~182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol. 2012;123:539–552. doi: 10.1007/s00401-012-0969-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li ZB, Li ZZ, Li L, Chu HT, Jia M. MiR-21 and miR-183 can simultaneously target SOCS6 and modulate growth and invasion of hepatocellular carcinoma (HCC) cells. Eur Rev Med Pharmacol Sci. 2015;19:3208–3217. [PubMed] [Google Scholar]
- 69.Yang M, Liu R, Li X, Liao J, Pu Y, Pan E, Yin L, Wang Y. miRNA-183 suppresses apoptosis and promotes proliferation in esophageal cancer by targeting PDCD4. Mol Cells. 2014;37:873–880. doi: 10.14348/molcells.2014.0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li C, Deng L, Zhi Q, Meng Q, Qian A, Sang H, Li X, Xia J. MicroRNA-183 functions as an oncogene by regulating PDCD4 in gastric cancer. Anticancer Agents Med Chem. 2016;16:447–455. doi: 10.2174/1871520615666150914114237. [DOI] [PubMed] [Google Scholar]
- 71.Zhang YY, Feng HM. MEG3 suppresses human pancreatic neuroendocrine tumor cells growth and metastasis by down-regulation of Mir-183. Cell Physiol Biochem. 2017;44:345–356. doi: 10.1159/000484906. [DOI] [PubMed] [Google Scholar]
- 72.Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010;70:9570–9580. doi: 10.1158/0008-5472.CAN-10-2074. [DOI] [PubMed] [Google Scholar]
- 73.Yang CL, Zheng XL, Ye K, Ge H, Sun YN, Lu YF, Fan QX. MicroRNA-183 acts as a tumor suppressor in human non-small cell lung cancer by down-regulating MTA1. Cell Physiol Biochem. 2018;46:93–106. doi: 10.1159/000488412. [DOI] [PubMed] [Google Scholar]
- 74.Gao P, He M, Zhang C, Geng C. Integrated analysis of gene expression signatures associated with colon cancer from three datasets. Gene. 2018:654–695. doi: 10.1016/j.gene.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 75.Li J, Liang S, Jin H, Xu C, Ma D, Lu X. Tiam1, negatively regulated by miR-22, miR-183 and miR-31, is involved in migration, invasion and viability of ovarian cancer cells. Oncol Rep. 2012;27:1835. doi: 10.3892/or.2012.1744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.