Abstract
Metastasis virulence, a significant contributor to breast cancer prognosis, is influenced by environmental factors like diet. We previously demonstrated in an F2 mouse population generated from a cross between the M16i polygenic obese and MMTV-PyMT mammary cancer models that high fat diet (HFD) decreases mammary cancer latency and increases pulmonary metastases compared to a matched control diet (MCD). Genetic analysis detected eight modifier loci for pulmonary metastasis, and diet significantly interacted with all eight loci. Here, gene expression microarray analysis was performed on mam-mary cancers from these mice. Despite the substantial dietary impact on metastasis and its interaction with metastasis modifiers, HFD significantly altered the expression of only five genes in mammary tumors; four of which, including serum amyloid A (Saa), are downstream of the tumor suppressor PTEN. Conversely, HFD altered the expression of 211 hepatic genes in a set of tumor free F2 control mice. Independent of diet, pulmonary metastasis virulence correlates with mammary tumor expression of genes involved in endocrine cancers, inflammation, angiogenesis, and invasion. The most significant virulence-associated network harbored genes also found in human adipose or mammary tissue, and contained upregulated Vegfa as a central node. Additionally, expression of Btn1a1, a gene physically located near a putative cis-acting eQTL on chromosome 13 and one of the metastasis modifiers, correlates with metastasis virulence. These data support the existence of diet-dependent and independent cancer modifier networks underlying differential susceptibility to mammary cancer metastasis and suggest that diet influences cancer metastasis virulence through tumor autonomous and non-autonomous mechanisms.
Keywords: Btn1a1, eQTL, Mammary cancer, Nutrigenomics, Pulmonary metastasis, PyMT
Introduction
Breast cancer prognosis is largely determined by the level of metastasis and is influenced by non-genetic factors like diet, which is also a major contributor to obesity. A large prospective study of US women demonstrated that obese women in the highest quintile of body mass index (BMI) had twice the death rate from breast cancer as did women in the lowest BMI quintile [1], possibly due to a higher risk for metastasis [2]. The relationship between obesity and breast cancer risk has some genetic underpinnings; woman who have a family history of breast cancer are far more likely to develop breast cancer when obese rather than lean [3]. Obesity can be linked to virulent breast cancer through both proliferative and inflammatory mechanisms [4, 5]. For instance, adipocytes secrete the adipocytokine tumor necrosis factor (TNFα) and vascular endothelial growth factor (VEGF), which are both associated with breast cancer [4].
Much of the current research on breast cancer metastasis demonstrates an important role of angiogenesis and invasion. VEGF family members are associated with poor prognosis largely because of their angiogenic potency [6]. Further, studies of breast cancer metastatic invasion frequently include members of the matrix metallopeptidase (MMP) family, which remodel the primary tumor through intravasation and seed lung metastasis by mediating extravasation [7, 8]. Several obesity-modifying hormone pathways may interact in angiogenesis and invasion processes; for instance estradiol regulates MMP2 and tissue inhibitor of metalloproteinase 1 (TIMP1) [9]. While there have been efforts to characterize the mechanistic events driving metastasis and obesity-associated breast cancer, few have characterized the mechanistic relationships between obesity, breast cancer, and its metastasis.
In order to examine mechanisms underlying the interaction of obesity with breast cancer and its metastasis, we developed an obese mouse model of breast cancer metastasis by crossing M16i, a polygenic obesity line, with FVB/NJ-TgN(MMTV-PyMT)634Mul (PyMT) [10, 11]. PyMT develops aggressive mammary tumors with subsequent pulmonary metastasis and has primary tumor gene expression similar to the gene expression of luminal breast tumors in women [12, 13]. Mice from the resulting F2 mouse population co-segregating obesity quantitative trait loci (QTL) and the MMTV-PyMT transgene when fed a high fat diet (HFD) compared to those fed a matched control diet (MCD) had 8.6% increased body weight, 21.8% increased total body fat, decreased mammary cancer latency (3 days), 1.5 times larger mammary tumors, and 46–68% increased pulmonary metastases (dependent on method of measurement) [10, 11]. These substantial diet effects were seen despite that F2 mice on either diet were heavier than PyMT on these same diets [14].
Genome-wide single nucleotide polymorphism (SNP) analyses reveled a strong genetic role in modifying pulmonary metastases susceptibility of this model, and diet significantly interacted with novel pulmonary metastasis QTL at all eight modifier loci detected [10]. Since we previously showed that high fat diet is associated with increased mammary cancer metastasis and appears to modify genetic susceptibility to pulmonary metastasis, we investigated whether increased metastasis could be due to changes in the transcriptome of primary cancers. Proliferation, inflammation, angiogenesis, and invasion processes were all significantly evident. For instance, pulmonary metastasis migration-associated serum amyloid A (Saa2) was upregulated in mammary tumors by HFD. Additionally, the milk component gene butyrophilin (Btn1a1) was identified on the metastasis virulence bio-marker list, lies under a metastatic QTL that interacted with HFD, and mapped to an eQTL 7 cM away from its physical location on chromosome 13. These results suggest that Btn1a1 should be further examined as a biomarker of breast cancer metastasis risk among women consuming a high fat diet.
Materials and methods
Husbandry and specimen collection
An F2 population was developed by mating M16i and PyMT; full details of the generation and sampling of the F2 population are provided in Gordon et al. [11]. Briefly, F2 female mice hemizygous for PyMT received one of two synthetic purified diets, either HFD (n = 76 mice, D12451, Research Diets, New Brunswick, NJ) containing 45% of total calories from fat (36.3, 45.3, and 18.5% of which is saturated, monounsaturated, and polyunsaturated fats respectively), 20% from protein and 35% from carbohydrate, or MCD (n = 79 mice, D12450B, Research Diets) containing 10% of total calories from fat (25.1, 34.7, and40.2% of which is saturated, monounsaturated, and polyunsaturated fats respectively), 20% from protein and 70% from carbohydrates, at 4 weeks of age and thereafter ad libitum. Diet compositions maintained equivocal caloric content by increased lard (39% total calories) and maltodextrin 10 (10% total calories), and decreased sucrose (17% total calories) in the HFD compared to MCD (4, 3, and 34% respectively). Mice were palpated three times weekly beginning at 4 weeks of age. Pulmonary metastasis (MET) was evaluated on the whole lung superficially by counting the number of foci visible under a dissecting scope. Subsequently, three coronal nonadjacent sections of one lung lobe per animal were examined under 12 × magnification; the number of multicellular metastatic lesions observed per square micron of non-alveolar lung tissue was defined as the average pulmonary metastatic density (AMD). One axillary mammary tumor, which more closely resembles the anatomical position and underlying sub-structure of the human breast compared to the inguinal tumors, per mouse was flash frozen for microarray analyses (n = 64 and 67 mice fed HFD and MCD, respectively). Livers were collected from randomly selected PyMT negative, non-tumor bearing-F2 female sib-pairs fed opposing diets to validate that strong physiological effects of HFD typically seen in otherwise healthy mice were detectable by microarray (n = 12 mice fed HFD, n = 12 mice fed MCD).
RNA isolation and microarray analyses
RNA from both axillary mammary tumor and liver of individual mice was isolated by TRIzol reagent (Invitrogen, Carlsbad, CA), and amplified using the Illumina® TotalPrep RNA Amplification kit, both according to manufacturer’s instructions (Ambion, Austin, TX). A solution containing 1.5 μg of highly purified biotinylated cRNA was applied to the Illumina Mouse 6 Sentrix array (version 1, Illumina, San Diego, CA) surface and hybridized at 55°C for 17.5 h. Following the hybridization period arrays were placed in High Temperature Wash Buffer (Illumina) for 10 min, E1BC Buffer (Illumina) for 5 min, 100% ethanol for 10 min, E1BC Buffer (Illumina) for 2 min, Block E1 Buffer for 10 min, and rocked with 2 ml of streptavidin-Cy3 (1 mg/ml in Block E1 Buffer, Illumina) for 10 min. Arrays were then washed in E1BC Buffer, dried, and evaluated on an Illumina Bead Scanner.
Microarray data processing
Raw data containing ~46,000 probe sets were log transformed and then normalized using a combination of the Loess and Quantile methods available in the R-based Lumi evaluation program for Illumina expression data [15]. Loess followed by Quantile normalization of identical pooled mammary tumor RNA ran across 11 chips produced a R2 value of 0.95. In order to eliminate transcripts that were not significantly expressed above the background signal, data were filtered at an Illumina detection score of0.95 and above.
Statistical analyses
Log transformed normalized data for all samples were run with 1,000 permutations at a false discovery rate q-value <0.05 to correct for multiple comparisons in SAM software using two class-unpaired analyses [16, 17]. We examined the effect of diet in tumors and livers by looking for significantly differential gene expression between HFD and MCD. To examine the relationship between metastasis virulence and gene expression in primary cancers, we used a multi-tiered approach independent of diet. Samples were ordered such that 25% (n = 33) of the total samples had no surface- or section-metastatic lesions and the longest tumor onset. These 33 samples were paired with 33 samples that had the most surface metastatic lesions detected (MET66), or with 33 samples that had the most sectional metastatic lesions detected (AMD66). Further analysis examined only those genes common to both MET66 and AMD66 with also known expression in human-mammary and/or adipose-tissues. We previously found there was not a strong relationship between tumor size and metastatic burden therefore metastasis data has not been normalized by tumor burden [18].
Functional analyses
Significant genes and their fold change values that were generated in SAM were imported into Ingenuity Pathways Analysis (IPA) 6.5–1602 (Redwood City, CA). RefSeq identifiers and their corresponding fold changes were mapped to corresponding gene objects in the IPA Knowledge Base (IPAKB). The curated IPAKB was used to generate functional analyses of significantly differential gene expression. IPAKB can also identify common pharmacological interactions through curation of Food & Drug Association data on approved pharmaceuticals, and of the National Institute of Health service ClinicalTrials.gov. The significance of functions and diseases to the gene set was determined by Fisher’s exact test to calculate the probability (P-value, or P) that each biological function and/or disease assigned to the gene set was due to chance alone.
Candidate gene evaluation
Normalized expression profiles were analyzed with the F2 inbred/Co-dominant Marker Analysis option of the web-based program QTL Express [19], fitting one expression QTL (eQTL) per chromosome. The genetic model included the additive plus dominance effects and fitted replicate and diet as fixed effects. The resulting expression loci were classified in one of two categories, “cis” acting if they mapped within 10 cM of the physical location of the actual gene they represent; otherwise they were classified as “trans” acting. A genome-wide significance threshold for eQTL effects (LOD = 3.5) was estimated using permutation testing with 1,000 iterations [20].
Results
HFD alters expression of a limited repertoire of genes in mammary cancers
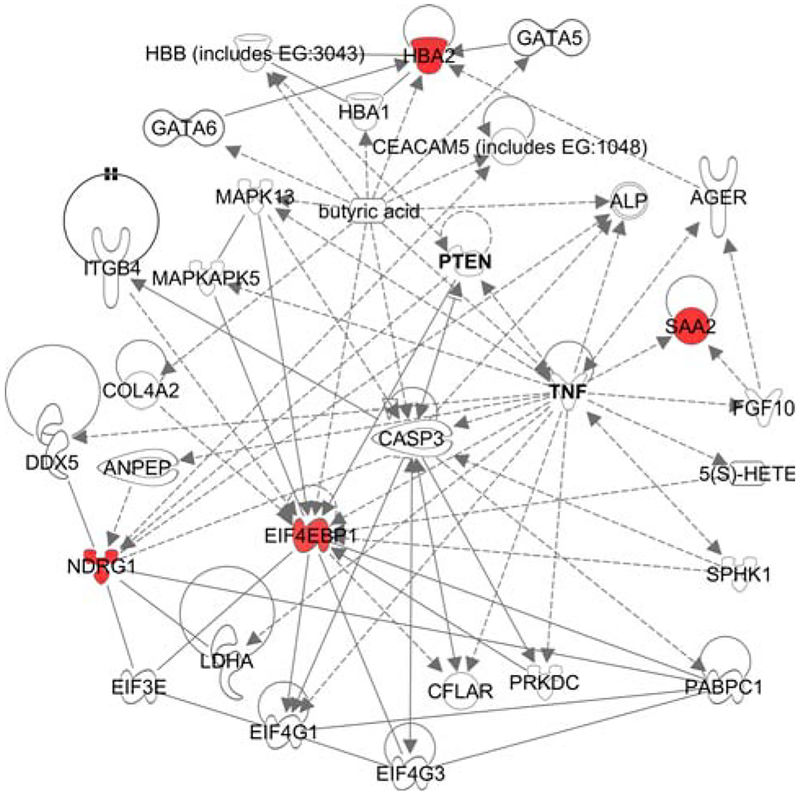
Genes differentially expressed between mammary primary cancers from mice fed HFD versus those from mice fed MCD were identified through comparison of microarray gene expression profiles. We previously demonstrated the influence of HFD on tumor-latency, -size, and -metastasis in this F2 population [10]. Despite the profound HFD effect, only five genes were significantly upregulated in mammary cancers of mice fed HFD compared to those fed MCD, four of which were connected within a common network (Fig. 1, Q = 0, Mon1a not shown). Reflective of their significant association with cancer (P < 0.001), the four genes of this network are downstream PTEN and/or TNF (Fig. 1).
Fig. 1.
Top network depicting the influence of high fat diet on mammary tumor gene expression (Ingenuity Pathway Analysis Knowledge Base). One axillary mammary tumor per mouse was used for microarray analyses (n = 64 and 67 microarrays of mammary tumor RNA from mice fed HFD and MCD, respectively). Gene networks depict the direct physical (solid) or indirect (dashed) interactions of molecules and genes’ products, including those genes not identified as significant on the microarrays, thus networks are ranked such that the highest ranked network contains the highest number of significantly expressed genes. Red nodes denote significant upregulation comparing high fat diet-relative to matched control diet-fed mice (Q = 0)
To determine if the increased gene expression associated with HFD might interact with existing cancer therapies, we used the IPAKB to identify several pharmaceuticals that target products of HFD-induced genes. Hyperphosphorylation of eukaryotic translation initiation factor 4E binding protein 1 (EIF4EBP1) is induced by the breast cancer therapeutic paclitaxel [21], and SCIO-469 blocks synthesis of TNFA, VEGF, and IL1B by inhibiting a MAPK14 complex that binds and phosphorylates EIF4EBP1 (clinicaltrials.gov ID NCT00744432). Given Eif4ebp1 was upregulated by HFD in mammary cancers here, the efficacy of paclitaxel and SCIO-469 may be enhanced by greater abundance of their substrate in persons eating a HFD.
HFD causes extensive changes in liver gene expression
To demonstrate that the modest HFD effect on mammary cancer gene expression was not due to microarray technical issues, we examined global gene expression of livers from wild-type F2 littermates of the MMTV-PyMT transgenic mice, because obesity resulting from HFD is closely associated with substantial liver transcriptional changes [22, 23]. Livers from mice fed HFD were compared to livers from mice fed MCD and significant gene expression differences were identified (Q < 0.05). A total of 211 genes met this criterion, of which 116 were significantly downregulated and 95 were upregulated (Supplemental Table 1). As expected, HFD deregulated numerous biological and toxicological functions in liver including carbohydrate-, nucleic acid-, and lipid-metabolism (Supplemental Table 2, P < 0.05). Despite the non-transgenic status of these mice, diet was associated with hepatic expression changes of 36 genes that are significantly associated with cancer phenotypes (P < 0.01).
Diet-independent transcriptional changes in primary cancers associated with metastasis
To investigate the relationship between gene expression in primary mammary cancers and metastasis, we identified genes with a Q < 0.05 by comparing global gene expression of primary cancers from mice with aggressive metastasis (MET66 and AMD66) to those from mice with no metastasis. A total of 478 genes met this criterion in MET66, of which 271 were significantly downregulated and 207 were upregulated (Supplemental Table 3). A total of 212 genes met this criterion in AMD66, of which 140 were significantly downregulated and 72 were upregulated (Supplemental Table 4). Together, all significant genes found in each MET66 and AMD66 (n = 690 genes) were examined in IPA for functional analyses.
The top three most significant function and disease classes of metastasis-associated genes were endocrine system disorders, metabolic disease and cancer (P < 0.05). Tumorigenesis (101 genes), neoplasia (98 genes), and cell death (94 genes) were the processes most populated by genes differentially regulated distinguished by metastasis (Supplemental Table 5, P < 0.05). Many tissue remodeling activities, including that of the hematological system and connective tissue, were also significantly altered based upon metastasis (P < 0.05). However, only inflammation was evident among significant biological functions, diseases, and canonical pathways (e.g. glucocorticoid-, inter-feron-, and platelet derived growth factor-signaling canonical pathways, P < 0.05).
Development of candidate transcriptional biomarkers of metastasis
Given the large number of genes involved in various processes of metastasis, we restricted our analysis to transcripts that might later serve as biomarkers of human breast cancer metastasis risk. First, a list of candidate metastasis virulence biomarkers was developed from the significant gene lists generated by AMD66 (n = 147 biomarker filter eligible genes) and MET66 (n = 420 biomarker filter eligible genes) groups. The list of candidate biomarkers was developed by including only those genes whose expression occurs in mammary or adipose tissues of humans. Because AMD and MET are measurements of a similar phenotype, we further reduced the candidate biomarker list to those genes common to both gene sets (n = 128 genes). This list of candidate biomarkers was then subjected to IPA Core Analysis using the IPAKB as the Reference Set to assess significance of functions, pathways, and toxicological analyses.
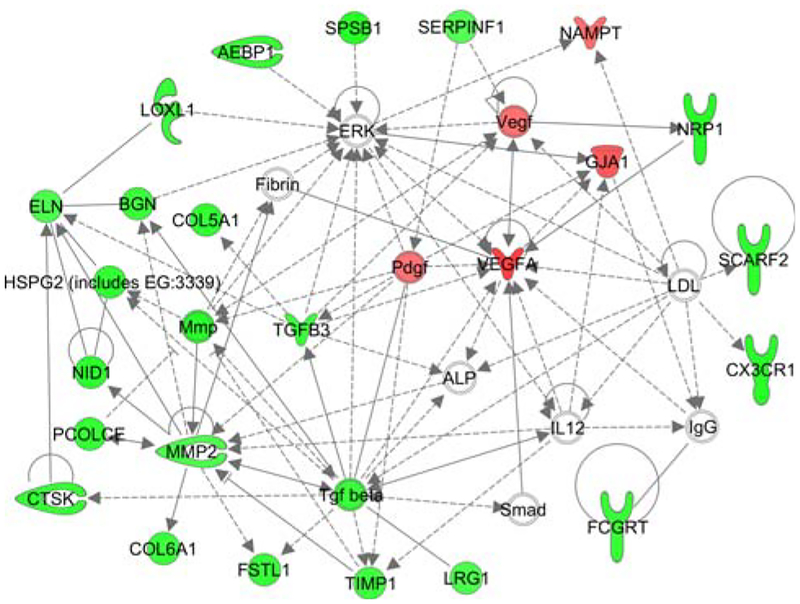
Tumorigenesis (36 genes), cell death (36 genes), neoplasia (35 genes), genetic disorder (34 genes), and proliferation (32 genes) were the function- and disease-categories most populated in the candidate biomarker list (Supplemental Table 6). The top network predicted by IPAKB is enriched in candidate biomarker genes involved in cardiovascular disease, organismal injury and abnormalities, as well as cardiovascular development and function (P < 0.001, Fig. 2). A focal gene of the top biomarker network is upregulated Vegfa, whose product is implicated in many metastasis processes, including angiogenesis, and invasion (Fig. 2, Supplemental Table 6) [24]. Other candidate metastasis virulence biomarkers are involved in angiogenesis of blood vessels (nine genes, P < 10−4) and angiogenesis-related processes including neovascularization (six genes, P < 10−4), as well as endothelial cell-migration (nine genes, P < 10−4) and proliferation (six genes, P \ 0.01, Supplemental Table 6). Further, molecules of the biomarker list were significantly associated with invasion (12 molecules, P < 0.001) and its processes, such as chemotaxis (10 molecules, P < 0.01), and breast cancer cell migration (six genes, P < 10−4, Supplemental Table 6). The biomarker genes also associated with other diseases with an etiological basis in obesity, e.g. diabetes (15 genes, P < 10−6), hypertension (six genes, P < 0.01) and atherosclerosis (ten genes, P < 10−6, Supplemental Table 6).
Fig. 2.
Top network of metastasis virulence biomarkers (Ingenuity Pathway Analysis Knowledge Base). The gene expression data from individual mammary tumor samples were ordered such that 25% (33 samples) of the total samples came from mice with no surface- or section-metastatic lesions and the longest tumor onset. Gene expression data from these 33 samples served as the reference group when paired with the gene expression data of 33 mammary tumor samples from mice that had the most sectional metastatic lesions detected (AMD66), and when paired with the gene expression data of 33 mammary tumor samples from mice that had the most superficial metastatic lesions detected (MET66). The differential expression of genes common to both the AMD66 and MET66 analyses was further restricted to genes expressed in human adipose or breast to define the set of metastasis virulence biomarkers. Gene networks depict the direct physical (solid) or indirect (dashed) interactions of molecules and genes’ products, including those genes not identified as significant on the microarrays, thus networks are ranked such that the highest ranked network contains the highest number of significantly expressed genes. Red nodes denote significant upregulation in high virulent mammary tumors relative to non-virulent mammary tumors (Q < 0.05). Green nodes denote significant downregulation in high virulent mammary tumors relative to non-virulent mammary tumors (Q < 0.05)
To determine if products of the candidate transcriptional biomarkers correlate with existing therapies, we used the IPAKB to identify those biomarkers whose expression changed in direction as predicted by their clinical target. Vegf and Vegfa are upregulated in the biomarker list, and the latter is also a drug target for endocrine-, epithelial-, and meta-static-cancers, including breast cancer. Another upregulated gene in the biomarker list, endothelin receptor type B (Ednrb), is inhibited by atrasentan. Atrasentan is in phase II and III trials to treat various cancers as well as endothelial dys-function (clinicaltrials.gov ID NCT00046943).
BTN1A1 as a candidate therapeutic target for metastasis
Given the non-invasive benefit of nipple aspirate as a potential biomarker of breast cancer metastasis risk, we further investigated the candidate biomarker list for milk fat components. Btn1a1, a major protein associated with milk fat, expression was significantly lower in primary cancers associated with metastasis compared to those without metastasis (P < 0.01). Similarly, a node just upstream of two genes upregulated by HFD in mammary tumors is butyric acid (Fig. 1), another bioactive component of milk fat. Further, both Btn1a1 and butyric acid are implicated in breast cancer [25, 26].
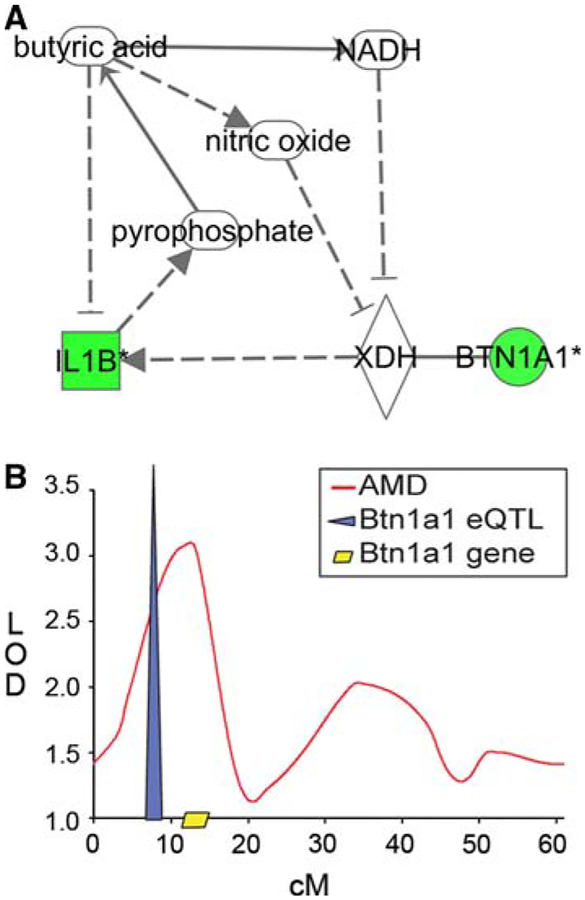
To evaluate the plausibility of these correlations, the IPAKB path explorer tool was used to determine whether butyric acid and BTN1A1 are functionally related. Through a number of pathways, butyric acid and BTN1A1 appear to be involved in a negative feedback loop (Fig. 3a). Butyric acid is one molecule upstream of BTN1A1 binding partner, xanthine dehydrogenase (XDH, Fig. 3a) [27]. The BTN1A1-XDH complex may be inhibited by butyric acid through the production of angiogenic-NADH and nitric oxide [28–31]. Butyric acid decreases interleukin 1 beta (IL1b) expression [32], which would also serve as negative feedback to the BTN1A1-XDH complex, normally stimulated by IL1b [33]. Because IL1b decreases inorganic pyrophosphate production, which synthesizes butyric acid, BTN1A1-XDH stimulus decreases butyric acid.
Fig. 3.
Btn1a1-gene expression network and quantitative trait locus in metastatic mammary cancer. a Btn1a1, significantly downregulated by metastasis, and binding partner Xdh are regulated in a feedback loop with butyric acid, implicated in high fat diet effects on mammary tumors (Fig. 1) through IL1B, also downregulated by metastasis (P < 0.05). Gene networks depict the direct physical (solid) or indirect (dashed) interactions of molecules and genes’ products, including those genes not identified as significant on the microarrays, thus networks are ranked such that the highest ranked network contains the highest number of significantly expressed genes. Green nodes denote significant downregulation in high virulent mammary tumors relative to non-virulent mammary tumors (Q < 0.05). b Btn1a1 lies within the 95% confidence interval of a QTL (LOD = 3.09) associated with the trait average pulmonary metastatic density only among mice fed high fat diet (11). Variation in mammary tumor Btn1a1 expression (n = 131 mice) was partially explained by a significant expression quantitative trait locus (n = 148 mice) also within the 95% confidence interval of the quantitative trait locus that describes the average pulmonary metastatic density × high fat diet interaction and near the physical location of the Btn1a1 gene
We extended our evaluation of BTN1A1 using eQTL analysis. We examined SNP markers across chromosome 13, and identified an eQTL on chromosome 13 near the Btn1a1 locus that regulates a significant amount of variation in Btn1a1 mRNA abundance, suggesting the presence of a cis-acting eQTL (Fig. 3b, LOD = 7.39). Further, Btn1a1 colocalizes with a previously detected modifier for AMD (LOD = 3.09) that was only significant in mice fed HFD (Fig. 3b) [10].
Discussion
We previously demonstrated that mice fed HFD had substantially early tumor onset, increased mammary tumor weight, and increased metastatic virulence in an obese mouse model of breast cancer metastasis based on the MMTV-PyMT transgene [10]. Yet the modest number of genes for which HFD significantly altered expression in primary tumors was unexpected given the strong HFD × QTL effects seen on mammary cancer metastasis phenotypes in this F2 mouse population [10]. This result may be explained by: (1) the few but highly significant transcript changes due to diet may have been sufficient to drive metastasis (Fig. 1); (2) non-autonomous systemic changes in extra-mammary sites (Supplemental Table 2); and/or (3) interaction between transcripts involved in the diet network (Fig. 1) and transcripts involved in the metastasis networks (Supplemental Tables 3, 4, Fig. 3). Although HFD may have changed the expression of few genes because of experimental design limitations, the effect of HFD on the liver transcriptome strongly indicates that the experimental design was adequate to detect diet effects.
The expression of few genes in primary tumors change in response to diet
The upregulated diet-associated genes have an association with cancer (Fig. 1, P < 0.001), perhaps because they are downstream from PTEN and/or TNF, both of which are implicated in advanced epithelial cancers and insulin resistance [34, 35]. Further, upregulated Eif4ebp1 is joined with PTEN in the phosphatidylinositol 3-kinase (PI3K)/thymoma viral proto-oncogene (AKT) canonical pathway, a pathway that promotes both obesity and cancer metastasis through the regulation of glucose uptake and cellular proliferation [36]. Among genes for which HFD significantly altered their expression, only Mon1a does not have an existing link with cancer. Although MON1A is poorly characterized, it is involved in macrophage iron loading [37], and thus may interact with the hemoglobin complex, which was also upregulated by HFD. Notwithstanding these associations with cancer processes, the modest number of HFD-induced changes in mammary cancer gene expression was surprising given the robust effects of HFD on tumor-latency, -size, -metastasis, and -modifier interaction in this F2 population [10, 11].
Non-tumor-autonomous actions of diet may influences metastasis
Diet may have exerted its effects on metastasis through systemic changes in extra-mammary sites, such as the hepatic induction of the PI3K/AKT pathway genes associated with metastatic potential in the liver (P < 0.01, Supplemental Tables 1, 2). In both hepatic and mammary tumor tissues, the PI3K/AKT pathway was significantly altered by HFD (P < 0.05). A candidate PI3K signaling molecule driving such cancer non-autonomous action may be catenin beta 1 (Ctnnb1). Ctnnb1 was upregulated in livers from mice fed HFD (P \ 0.01), and is implicated in breast cancer and its invasion [38, 39]. Further, CTNNB1 binds N-myc downstream regulated gene 1 (NDRG1) [40], which was upregulated by HFD in mammary tumors (Supplemental Table 2, Fig. 1). Cancer non-autonomous mechanisms may extend beyond the PI3K/AKT pathway. In liver from the non-transgenic F2 mice and elsewhere [41], HFD significantly upregulated hepatic ectonucleotide pyrophosphatase/phosphodiesterase 2 (Enpp2, P < 0.05), a cell membrane enzyme associated with invasion and metastasis (Supplemental Tables 1, 2) [42]. These observations suggest that part of the effect of HFD on cancer metastasis may be through cancer non-autonomous mechanisms.
Potential interactions between diet- and metastasis-networks
Saa2 and Saa3 were significantly downregulated in our previous functional genomic analysis of the PyMT model on FVB/J background compared to other strains of the PyMT model [43]. Yet here Saa2 was upregulated by HFD, suggesting HFD and/or polygenic obesity can overturn the negative regulation of SAA family expression associated with this genetic cancer model. Indeed, SAA2 varies according to the metastatic potential of mouse models of breast cancer and is part of a gene expression signature that distinguishes breast cancer patient outcomes across independent breast cancer datasets [44].
While the mechanism of SAA2 on metastasis has not yet been determined, the involvement of SAA2 in NFKB signaling in mammary epithelium has recently been demonstrated [45]. Here, the second most significant network of the candidate biomarkers of metastasis has several up-regulated molecules in direct interaction with NFKB. Given the mechanism of SAA3 seeding metastasis in the pre-metastatic lung is attributed to regulation of chemoattractant secretion and resulting NFKB-mediated cell migration [46], diet-induced SAA2 may interact with the metastasis-induced NFKB pathway to increase meta-static virulence.
Milk fat components are another compelling link between the influences of diet and metastasis on mammary tumor gene expression. We found significantly lower expression of Btn1a1, a major component of milk fat droplets, in our candidate biomarkers of metastasis (P < 0.01, Supplemental Tables 3, 4). Further, our QTL analyses suggest that Btn1a1 may be one of potentially multiple modifiers of the AMD locus. Consistent with this finding, decreased Btn1a1 expression was identified as part of the high virulent signature that previously characterized the effects of the MMTV-PyMT mammary cancer model [43], and has also been associated with metastatic breast cancer in humans [26].
Together, the feedback loop of Btn1a1 with butyric acid and the chromosomal associations of Btn1a1 are suggestive of a mechanistic relationship between HFD-induced obesity and mammary metastasis virulence. Both BTN1A1 and butyric acid reside on the cell surface of mammary alveolar epithelial cells and are regulated by angiogenic Vegf (Fig. 2) [27, 47]. Butyric acid is currently thought to inhibit breast cancer through histone deacetylase (HDAC) inhibition [25]. There is little known about the biological activity of BTN1A1, however its binding partner XDH increases secretion of MMP2 (Fig. 2), which has been shown to increase metastasis through the degradation of extracellular matrix [48]. Elsewhere XDH activation of NFKB mediates angiogenesis [49]. Thus the Btn1a1-butyric acid feedback loop may be influencing metastasis through epigenetic-, motility- and angiogenic-activity.
Given the active chemotherapeutic research of butyric acid as a HDAC inhibitor, and the downregulation of Btn1a1 reported here, our findings indicate that high levels of Btn1a1 expression may protect rather than promote of breast cancer. BTN1A1 protein was successfully measured in human nipple aspirate fluid, but was not deemed a suitable biomarker of cancer risk only because no studies demonstrated a correlation between BTN1A1 and cancer at that time [50]. Consequently, Btn1a1 levels in cancer patients treated by HDAC inhibitors may merit monitoring. Investigation of the biological function of differential Btn1a1 expression in mouse models of mammary tumor metastasis is underway.
Study limitations
A role of decreased carbohydrates cannot be eliminated as an influencing factor of gene expression- and metastatic-effects of HFD seen here because HFD was formulated to have the same caloric density as MCD through a relative decrease in carbohydrates. However, diets matched for every nutrient besides fat would require differences in caloric density. Additionally, total caloric intake and total fat are highly correlated in freely eating mammals [51], thus even if caloric intake was evaluated here, it would be difficult to separate the effect of fat from the effect of total calories. Further, while diets high in calories, fat, and carbohydrates are associated with obesity, it is obesity and weight gain, and not any dietary factor, that consistently correlates with breast cancer risk [52].
Similarly, HFD may have increased mammary tumor gene expression at a different time or through subtle, undetectable changes in gene expression in molecules upstream of those altered by HFD. TNFα, PTEN, and butyric acid are examples of molecules directly upstream of at least two genes that HFD upregulated, and are also associated with breast cancer. We also cannot exclude the possibility that the strong influence of HFD on metastasis phenotypes seen in this mouse population was mediated through gene expression changes in pulmonary metastasis. While the relative contribution of adipocyte- and vascular cell-expression is intrinsic to the total heterogeneous mammary tumor expression profiles presented here, without microdissection, the contribution of adipocytes and vascular cells to changes in gene expression cannot be specifically evaluated. However, the high R2 value of identical, pooled, and normalized mammary tumor RNA ran across 11 microarray chips suggests minimal technical error. The strong influence of metastatic virulence on differential mammary tumor gene expression further indicates minimal technical error. As expected, these data indicate that HFD substantially altered liver gene transcription, and further suggest that cancer non-autonomous changes may contribute to the effects of HFD on mammary cancer in MMTV-PyMT transgenic mice.
Conclusion
A diet high in fat is becoming a more prevalent occurrence [53]. The resulting elevation in obesity prevalence is a substantial public health concern in part because of its association with breast cancer morbidity and mortality. Our data suggest that diet may increase breast cancer metastatic virulence through multifactoral cancer-autonomous and non-autonomous effects on cell migration, angiogenesis, and extracellular matrix breakdown. Transcript changes due to HFD, to metastasis, and to their interaction illuminate the influence of the primary mam-mary tumor on pulmonary metastatic virulence. Yet transcript changes in the liver indicate that HFD influenced metastasis through systemic, non-autonomous mechanisms as well. Our data suggest that the complex relationships between diet, mammary carcinogenesis, and its metastasis will become clearer if greater focus is placed on understanding the role of extra-mammary sites in carcinogenesis and metastasis.
Supplementary Material
Acknowledgments
This work was partially funded by a grant from the NCI-MMHCC (U01CA105417) to D.P. and D.W.T.; Department of Defense Fellowship (BC050873) to M.L.; National Institute of Environmental Health Sciences Training Grant (T32ES007126); NICHD Training Grant (5T32 HD049311); and by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The authors acknowledge support from the Animal Metabolism and Phenotyping Core Facility within UNC’s Clinical Nutrition Research Unit funded by NIH-NIDDK (DK056350). The authors are grateful to Anita Ferrell, Jackie Potts, and Barry Simpson for assistance with mouse dissections, and to Peter Sørensen for assistance with LUMI.
Abbreviations
- AMD
Average pulmonary metastatic density
- AKT
Thymoma viral proto-oncogene
- BMI
Body mass index
- BTN1A1
Butyrophilin, subfamily 1, member A1
- CTNNB1
Catenin (cadherin associated protein) beta 1
- EDNRB
Endothelin receptor type
- BEIF4EBP1
Eukaryotic translation initiation factor 4E binding protein 1
- ENPP2
Ectonucleotide pyrophosphatase/phosphodiesterase 2
- eQTL
Expression quantitative trait locus
- F2
Filial 2
- HDAC
Histone deacetylase
- HFD
High fat diet
- IL1b
Interleukin 1β
- IPA
Ingenuity pathways analysis
- IPAKB
Ingenuity pathways analysis knowledge base
- MCD
Matched control diet
- MET
Superficial pulmonary metastasis
- MMP
Matrix metallopeptidase
- MMTV-PyMT
FVB/N-Tg(MMTV-PyVT)634Mul/J
- MON1A
MON1 homolog A
- NADH
Nicotinamide adenine dinucleotide
- NDRG1
N-myc downstream regulated gene 1
- NFKB
Nuclear factor of kappa light polypeptide gene enhancer in B-cells
- P
P-value
- PI3K
Phosphatidylinositol 3-kinase
- PTEN
Phosphatase and tensin homolog
- QTL
Quantitative trait loci
- RNA
Ribonucleic acid
- SAA
Serum amyloid A
- SNP
Single nucleotide polymorphism
- TIMP1
Tissue inhibitor of metalloproteinase 1
- TNFα
Tumor necrosis factor alpha
- VEGF
Vascular endothelial grow factor
- XDH
Xanthine dehydrogenase
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10585-009-9302-7) contains supplementary material, which is available to authorized users.
Contributor Information
Michele La Merrill, Department of Preventive Medicine, Mount Sinai School of Medicine, Box 1057, New York, NY 10029, USA; Curriculum in Toxicology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA; Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Ryan R. Gordon, Department of Nutrition, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA
Kent W. Hunter, Laboratory of Cancer Biology and Genetics, NIH/NCI, Bethesda, MD 20892, USA
David W. Threadgill, Curriculum in Toxicology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA; Center for Environmental Health and Susceptibility, Lineberger Cancer Center and Carolina Center for Genome Sciences, University of North Carolina at Chapel Hill, Chapel Hill,NC 27599, USA.
Daniel Pomp, Curriculum in Toxicology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA; Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA; Department of Nutrition, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA; Center for Environmental Health and Susceptibility, Lineberger Cancer Center and Carolina Center for Genome Sciences, University of North Carolina at Chapel Hill, Chapel Hill,NC 27599, USA.
References
- 1.Calle EE, Rodriguez C, Walker-Thurmond K et al. (2003) Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med 348(17):1625–1638 [DOI] [PubMed] [Google Scholar]
- 2.Berclaz G, Li S, Price KN et al. (2004) Body mass index as a prognostic feature in operable breast cancer: the International Breast Cancer Study Group experience. Ann Oncol 15(6):875–884 [DOI] [PubMed] [Google Scholar]
- 3.Carpenter CL, Ross RK, Paganini-Hill A et al. (2003) Effect of family history, obesity and exercise on breast cancer risk among postmenopausal women. Int J Cancer 106(1):96–102 [DOI] [PubMed] [Google Scholar]
- 4.Rose DP, Komninou D, Stephenson GD (2004) Obesity, adipocytokines, and insulin resistance in breast cancer. Obes Rev 5(3):153–165 [DOI] [PubMed] [Google Scholar]
- 5.Lorincz AM, Sukumar S (2006) Molecular links between obesity and breast cancer. Endocr Relat Cancer 13(2):279–292 [DOI] [PubMed] [Google Scholar]
- 6.Mohammed RA, Green A, El-Shikh S et al. (2007) Prognostic significance of vascular endothelial cell growth factors -A, -C and -D in breast cancer and their relationship with angio- and lymphangiogenesis. Br J Cancer 96(7):1092–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minn AJ, Gupta GP, Padua D et al. (2007) Lung metastasis genes couple breast tumor size and metastatic spread. Proc Natl Acad Sci USA 104(16):6740–6745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta GP, Nguyen DX, Chiang AC et al. (2007) Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature 446(7137):765–770 [DOI] [PubMed] [Google Scholar]
- 9.Nilsson UW, Garvin S, Dabrosin C (2007) MMP-2 and MMP-9 activity is regulated by estradiol and tamoxifen in cultured human breast cancer cells. Breast Cancer Res Treat 102(3):253–261 [DOI] [PubMed] [Google Scholar]
- 10.Gordon RR, Hunter KW, La Merrill M et al. (2008) Genotype X diet interactions in mice predisposed to mammary cancer: II. Tumors and metastasis. Mamm Genome 19(3):179–189 [DOI] [PubMed] [Google Scholar]
- 11.Gordon RR, Hunter KW, Sorensen P et al. (2008) Genotype X diet interactions in mice predisposed to mammary cancer. I. Body weight and fat. Mamm Genome 19(3):163–178 [DOI] [PubMed] [Google Scholar]
- 12.Guy CT, Cardiff RD, Muller WJ (1992) Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol 12(3):954–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herschkowitz JI, Simin K, Weigman VJ et al. (2007) Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 8(5):R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.La Merrill M, Baston DS, Denison MS et al. (2009) Mouse breast cancer model-dependent changes in metabolic syndrome-associated phenotypes caused by maternal dioxin exposure and dietary fat. Am J Physiol Endocrinol Metab 296(1):E203–E210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du P, Kibbe WA, Lin SM (2008) Lumi: a pipeline for processing Illumina microarray. Bioinformatics 24(13):1547–1548 [DOI] [PubMed] [Google Scholar]
- 16.Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98(9):5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Storey JD (2002) A direct approach to false discovery rates. J Roy Stat Soc Ser B 64:479–498 [Google Scholar]
- 18.Lifsted T, Le Voyer T, Williams M et al. (1998) Identification of inbred mouse strains harboring genetic modifiers of mammary tumor age of onset and metastatic progression. Int J Cancer 77(4):640–644 [DOI] [PubMed] [Google Scholar]
- 19.Seaton G, Haley CS, Knott SA et al. (2002) QTL express: mapping quantitative trait loci in simple and complex pedigrees. Bioinformatics 18(2):339–340 [DOI] [PubMed] [Google Scholar]
- 20.Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138(3):963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenberg VL, Zimmer SG (2005) Paclitaxel induces the phosphorylation of the eukaryotic translation initiation factor 4E-binding protein 1 through a Cdk1-dependent mechanism. Oncogene 24(30):4851–4860 [DOI] [PubMed] [Google Scholar]
- 22.Li H, Xie Z, Lin J et al. (2008) Transcriptomic and metabonomic profiling of obesity-prone and obesity-resistant rats under high fat diet. J Proteome Res 7(11):4775–4783 [DOI] [PubMed] [Google Scholar]
- 23.Morgan K, Uyuni A, Nandgiri G et al. (2008) Altered expression of transcription factors and genes regulating lipogenesis in liver and adipose tissue of mice with high fat diet-induced obesity and nonalcoholic fatty liver disease. Eur J Gastroenterol Hepatol 20(9):843–854 [DOI] [PubMed] [Google Scholar]
- 24.Oshima RG, Lesperance J, Munoz V et al. (2004) Angiogenic acceleration of Neu induced mammary tumor progression and metastasis. Cancer Res 64(1):169–179 [DOI] [PubMed] [Google Scholar]
- 25.De los Santos M, Martinez-Iglesias O, Aranda A (2007) Anti-estrogenic actions of histone deacetylase inhibitors in MCF-7 breast cancer cells. Endocr Relat Cancer 14(4):1021–1028 [DOI] [PubMed] [Google Scholar]
- 26.Woelfle U, Cloos J, Sauter G et al. (2003) Molecular signature associated with bone marrow micrometastasis in human breast cancer. Cancer Res 63(18):5679–5684 [PubMed] [Google Scholar]
- 27.McManaman JL, Palmer CA, Wright RM et al. (2002) Functional regulation of xanthine oxidoreductase expression and localization in the mouse mammary gland: evidence of a role in lipid secretion. J Physiol 545(Pt 2):567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rinaldo JE, Clark M, Parinello J et al. (1994) Nitric oxide inactivates xanthine dehydrogenase and xanthine oxidase in inter-feron-gamma-stimulated macrophages. Am J Respir Cell Mol Biol 11(5):625–630 [DOI] [PubMed] [Google Scholar]
- 29.Park JW, Chun YS, Kim MS et al. (1998) Metabolic modulation of cellular redox potential can improve cardiac recovery from ischemia-reperfusion injury. Int J Cardiol 65(2):139–147 [DOI] [PubMed] [Google Scholar]
- 30.Hewett P, Popplewell A, Finney H et al. (1999) Changes in microvessel endothelial cell gene expression in an in vitro human breast tumour endothelial cell model. Angiogenesis 3(3):221–229 [DOI] [PubMed] [Google Scholar]
- 31.Thomsen LL, Miles DW (1998) Role of nitric oxide in tumour progression: lessons from human tumours. Cancer Metastasis Rev 17(1):107–118 [DOI] [PubMed] [Google Scholar]
- 32.Joseph J, Mudduluru G, Antony S et al. (2004) Expression profiling of sodium butyrate (NaB)-treated cells: identification of regulation of genes related to cytokine signaling and cancer metastasis by NaB. Oncogene 23(37):6304–6315 [DOI] [PubMed] [Google Scholar]
- 33.Kocic G, Vlahovic P, Dordevic V et al. (1995) Effects of growth factors on the enzymes of purine metabolism in culture of regenerating rat liver cells. Arch Physiol Biochem 103(6): 715–719 [DOI] [PubMed] [Google Scholar]
- 34.Rosner M, Hanneder M, Siegel N et al. (2008) The mTOR pathway and its role in human genetic diseases. Mutat Res 659(3):284–292 [DOI] [PubMed] [Google Scholar]
- 35.Ikubo M, Wada T, Fukui K et al. (2008) Impact of lipid phosphateases SHIP2 and PTEN on the time- and Akt isoform-specific amelioration of TNFalpha-induced insulin resistance in 3T3-L1 adipocytes. Am J Physiol Endocrinol Metab (in press) [DOI] [PubMed] [Google Scholar]
- 36.Gingras AC, Kennedy SG, O’Leary MA et al. (1998) 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev 12(4):502–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang F, Paradkar PN, Custodio AO et al. (2007) Genetic variation in Mon1a affects protein trafficking and modifies macrophage iron loading in mice. Nat Genet 39(8):1025–1032 [DOI] [PubMed] [Google Scholar]
- 38.Adam L, Vadlamudi RK, McCrea P et al. (2001) Tiam1 overexpression potentiates heregulin-induced lymphoid enhancer factor-1/beta-catenin nuclear signaling in breast cancer cells by modulating the intercellular stability. J Biol Chem 276(30):28443–28450 [DOI] [PubMed] [Google Scholar]
- 39.Michaelson JS, Leder P (2001) Beta-catenin is a downstream effector of Wnt-mediated tumorigenesis in the mammary gland. Oncogene 20(37):5093–5099 [DOI] [PubMed] [Google Scholar]
- 40.Tu LC, Yan X, Hood L et al. (2007) Proteomics analysis of the interactome of N-myc downstream regulated gene 1 and its interactions with the androgen response program in prostate cancer cells. Mol Cell Proteomics 6(4):575–588 [DOI] [PubMed] [Google Scholar]
- 41.Maxwell KN, Soccio RE, Duncan EM et al. (2003) Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. J Lipid Res 44(11):2109–2119 [DOI] [PubMed] [Google Scholar]
- 42.Nam SW, Clair T, Campo CK et al. (2000) Autotaxin (ATX), a potent tumor motogen, augments invasive and metastatic potential of ras-transformed cells. Oncogene 19(2):241–247 [DOI] [PubMed] [Google Scholar]
- 43.Qiu TH, Chandramouli GV, Hunter KW et al. (2004) Global expression profiling identifies signatures of tumor virulence in MMTV-PyMT-transgenic mice: correlation to human disease. Cancer Res 64(17):5973–5981 [DOI] [PubMed] [Google Scholar]
- 44.Lukes L, Crawford NP, Walker R et al. (2009) The origins of breast cancer prognostic gene expression profiles. Cancer Res 69(1):310–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kho Y, Kim S, Yoon BS et al. (2008) Induction of serum amyloid A genes is associated with growth and apoptosis of HC11 mam-mary epithelial cells. Biosci Biotechnol Biochem 72(1):70–81 [DOI] [PubMed] [Google Scholar]
- 46.Hiratsuka S, Watanabe A, Sakurai Y et al. (2008) The S100A8-serum amyloid A3-TLR4 paracrine cascade establishes a premetastatic phase. Nat Cell Biol 10(11):1349–1355 [DOI] [PubMed] [Google Scholar]
- 47.Rossiter H, Barresi C, Ghannadan M et al. (2007) Inactivation of VEGF in mammary gland epithelium severely compromises mammary gland development and function. FASEB J 21(14): 3994–4004 [DOI] [PubMed] [Google Scholar]
- 48.Galli A, Svegliati-Baroni G, Ceni E et al. (2005) Oxidative stress stimulates proliferation and invasiveness of hepatic stellate cells via a MMP2-mediated mechanism. Hepatology 41(5):1074–1084 [DOI] [PubMed] [Google Scholar]
- 49.Shenkar R, Schwartz MD, Terada LS et al. (1996) Hemorrhage activates NF-kappa B in murine lung mononuclear cells in vivo. Am J Physiol 270(5 Pt 1):L729–L735 [DOI] [PubMed] [Google Scholar]
- 50.Varnum SM, Covington CC, Woodbury RL et al. (2003) Proteomic characterization of nipple aspirate fluid: identification of potential biomarkers of breast cancer. Breast Cancer Res Treat 80(1):87–97 [DOI] [PubMed] [Google Scholar]
- 51.Willett W, Stampfer MJ (1986) Total energy intake: implications for epidemiologic analyses. Am J Epidemiol 124(1):17–27 [DOI] [PubMed] [Google Scholar]
- 52.Michels KB, Mohllajee AP, Roset-Bahmanyar E et al. (2007) Diet and breast cancer: a review of the prospective observational studies. Cancer 109(12 Suppl):2712–2749 [DOI] [PubMed] [Google Scholar]
- 53.Schrauwen P, Westerterp KR (2000) The role of high-fat diets and physical activity in the regulation of body weight. Br J Nutr 84(4):417–427 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.