Abstract
Single-molecule fluorescence methods have illuminated the dynamics of the translational machinery. Structural and bulk biochemical experiments have provided detailed atomic and global mechanistic views of translation, respectively. Single-molecule studies of translation have bridged these views by temporally connecting the conformational and compositional states defined from structural data within the mechanistic framework of translation produced from biochemical studies. Here, we discuss the context for applying different single-molecule fluorescence experiments, and present recent applications to studying prokaryotic and eukaryotic translation. We underscore the power of observing single translating ribosomes to delineate and sort complex mechanistic pathways during initiation and elongation, and discuss future applications of current and improved technologies.
Proteins must be translated rapidly, with high fidelity and quality by the ribosome. Translation is thus a highly dynamic process with temporal changes in ligand stoichiometry on the ribosome, and conformational changes within ribosomes and ligands (Noller et al. 2017a,b; Rodnina et al. 2017). During initiation in all organisms (Merrick and Pavitt 2018; Rodnina 2018), ribosomal subunits must assemble at the correct start site on the messenger RNA (mRNA) defined by specific mRNA–ribosome interactions; this establishes the reading frame for translation of the correct protein sequence. On proper initiation, the assembled 70S or 80S ribosome with initiator transfer RNA (tRNA) base-paired with the start codon positioned in the peptidyl-tRNA site (P site) is competent for elongation of the polypeptide. During elongation (Dever et al. 2018; Rodnina 2018), the genetic code is read by cognate aminoacyl-tRNAs delivered to the aminoacyl-tRNA site (A site) as a complex with a GTPase elongation factor (EF-Tu in bacteria). This dynamic process of tRNA selection occurs with high fidelity (one wrong amino acid incorporated per 3000) (Loftfield 1963), aided by the free energy of GTP hydrolysis, ribosome and factor conformational changes, and induced fit with kinetic proofreading. Once a correct tRNA is accommodated into the peptidyl transferase center (PTC) of the ribosome, peptide bond formation occurs with subsequent conformational changes in the ribosome (intersubunit rotation) and tRNAs (hybrid state formation). The codon–anticodon pairs on the ribosome must be precisely and rapidly moved from the P and A sites to the exit (E site) and P site during the process of translocation, catalyzed by another GTPase elongation factor (EF-G in bacteria), which also resets the conformation of the ribosomal subunit and places the next codon in the A site. This elongation cycle repeats until a stop codon enters the A site, which signals release factors (RFs) to catalyze release of the polypeptide from the P-site tRNA during termination, and then prompts the ribosomal subunits to be split during recycling (Hellen 2018).
PROBING THE DYNAMICS OF TRANSLATION
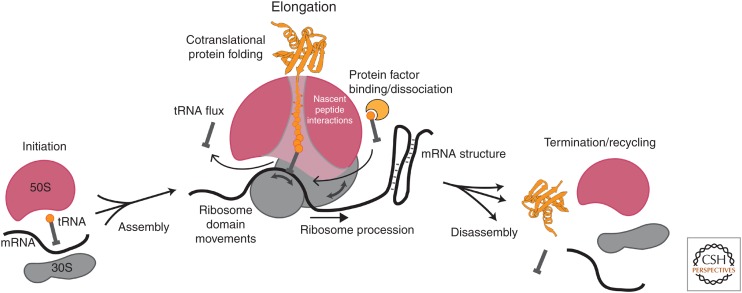
The basal process of translation is thus highly directional and dynamic. Even after the process of ribosome biogenesis, whereby ribosomal RNAs are transcribed, processed, and assembled with ribosomal proteins (Shajani et al. 2011), the ribosome undergoes numerous changes in conformation and composition during translation (Fig. 1). Ribosomal subunits assemble during initiation; tRNAs flux through the ribosome as it traverses a messenger RNA (mRNA) (Uemura et al. 2010), unfolding potential RNA structures (Wen et al. 2008), while spooling out a growing nascent polypeptide (which also may fold cotranslationally) during elongation (Thommen et al. 2017); and the disassembly of this translation complex demands coordinated binding and catalysis of many protein factors triggered by the stop codon signal (Petry et al. 2008; Sternberg et al. 2009; Dever and Green 2012; Koutmou et al. 2014; Peske et al. 2014; Prabhakar et al. 2017a). Underlying the process of translation are conformational dynamics of the ribosome and macromolecules that guide translation (Prabhakar et al. 2017b). Layered onto the basal process are regulatory events that modulate temporally the output of translation—the nature, rates, and efficiency of production of the protein product. An understanding of dynamics is thus required to delineate the mechanism of translation.
Figure 1.
Dynamic picture of translation. Translation is regulated through a complex network of interactions and movements of the ribosomal subunits, the translated messenger RNA (mRNA), the adaptor transfer RNAs (tRNAs), protein translation factors, and the growing nascent peptide chain. Key dynamic features of translation are highlighted.
Traditional study of dynamics involves measurements in bulk on large numbers of molecules in solution (Wintermeyer et al. 2004; Johansson et al. 2008). Chemical and biochemical kinetics methods have provided the lion's share of information on the time evolution of translation. Reactions are initiated by rapid mixing of the components, and a time-dependent signal is measured in a stopped-flow system either (1) in real time (e.g., fluorescence) to report compositional and conformational changes (Rodnina et al. 1994; Studer et al. 2003; Koutmou et al. 2014), or (2) using quenched-flow methods recording a chemical signal as a function of time to report chemical changes (peptide bond formation, GTP or ATP hydrolysis) (Bilgin et al. 1992; Pape et al. 1998). By using these approaches, the basic kinetic mechanisms of initiation (Antoun et al. 2003; Milon et al. 2012), elongation (Gromadski et al. 2002; Johansson et al. 2008), and termination/recycling (Karimi et al. 1999; Zavialov et al. 2001; Peske et al. 2005, 2014; Koutmou et al. 2014) in bacteria have been determined, and progress in understanding the special features of eukaryotic translation made (Shoemaker and Green 2011). More recently, ribosome profiling has revolutionized the study of translation in vivo, by quantifying the occupancy of ribosomes at specific positions on a given mRNA (Ingolia et al. 2009, 2018). These data, derived by deep sequencing of mRNA fragments protected by ribosomes from nuclease treatment, can be quantitatively treated to delineate translation rates and potential pausing sites during elongation (Brar and Weissman 2015).
From these data, the timescales of translation can be determined. Translation initiation occurs on a seconds-to-minutes timescale, whereas elongation rates are 1–20 amino acids per second (depending on in vivo or in vitro systems) (Young and Bremer 1976; Pedersen 1984; Pavlov and Ehrenberg 1996), and termination/recycling also takes seconds (Pavlov et al. 1997). This defines a key window of dynamic processes in the millisecond-to-second range to embrace translation and suggests approaches with corresponding time resolutions to investigate dynamics.
Structural biology has revolutionized the study of translation over the past two decades. Crystal structures of individual subunits and 70S bacterial ribosomes first revealed the architecture of the RNA–protein assemblies (Ban et al. 2000; Schluenzen et al. 2000; Wimberly et al. 2000). Subsequent crystallographic investigations revealed the basis for tRNA–mRNA decoding (Ogle and Ramakrishnan 2005), factor interaction sites (Gao et al. 2009), and GTPase activation of factors (Voorhees et al. 2010), and suggested mechanisms of initiation, translocation, and termination/recycling (Schmeing and Ramakrishnan 2009). In parallel, cryoelectron microscopy (cryoEM) has contributed enormously to our understanding of structure, in early years at low resolution, and more recently at atomic resolution (Jobe et al. 2018). CryoEM has allowed investigation of eukaryotic ribosomal complexes (yeast, human) involved in initiation (Passmore et al. 2007; Fernandez et al. 2013), elongation (Murray et al. 2016), termination (Taylor et al. 2012; Brown et al. 2015; Matheisl et al. 2015), and recycling (Becker et al. 2012). In addition, single-particle cryoEM methods (including parallel improvements in computation) have revealed conformational and compositional heterogeneity by sorting distinct classes of translational complexes (Llacer et al. 2015; Hussain et al. 2016). These structural studies highlight static conformational flexibility but lack a temporal dimension.
A dream of structural biology is real-time observation of conformational states. Nuclear magnetic resonance (NMR) spectroscopy probes the dynamics of biological systems directly, either at fast (10−9 s), intermediate (10−6–10−3 s), or slow (seconds-to-minutes) timescales (Kovermann et al. 2016). Although NMR provides residue-level information on dynamic movements, correlated domain motion is more difficult to observe. Most critically, solution NMR is limited by size constraints to systems smaller than the ribosome (Huang and Kalodimos 2017). These size limitations can be overcome by application of challenging solid-state NMR measurements (Barbet-Massin et al. 2015; Kurauskas et al. 2016), but, unfortunately, NMR has not been a leading tool in delineating translational dynamics. Time-resolved crystallographic approaches are feasible, but require coordinated changes occurring within a crystalline lattice, often triggered by light (and limited to rather small conformational changes). Recent advances in rapid mixing have allowed a merging of cryoEM and rapid-mixing kinetics to observe the structural intermediates involved in ribosome recycling (Fu et al. 2016). Further development of time-resolved tools will remain a frontier of structural biology.
STUDYING TRANSLATION WITH SINGLE-MOLECULE FLUORESCENCE
Fluorescence spectroscopy provides a highly sensitive probe of biological dynamics. Using modern dyes, fluorescence from single dyes can be readily detected using standard optics and solid-state cameras (Betzig and Chichester 1993). Stable, bright fluorophores (Cy dyes, Alexa dyes) have been developed that are excited by standard lasers, with fluorescence emission at different visible wavelengths; these bright dyes emit millions of photons as they go through their photocycles before irreversible chemical reactions (photobleaching) that terminate fluorescence (Ha and Tinnefeld 2012). These properties have allowed application of single-molecule approaches over the past two decades (Michalet et al. 2003).
To use single-molecule fluorescence approaches, biological molecules must be fluorescently labeled with bright extrinsic fluorophores. These are usually the organic dyes discussed in the previous paragraph (Ha and Tinnefeld 2012), as fluorescent proteins tend to be dimmer with poorer photophysical properties for single-molecule dynamics studies. For nucleic acids, labeling can be achieved through chemical synthesis of oligonucleotides, hybridization of a labeled DNA or RNA to a target nucleic acid, or modification of thiols or primary amines placed within or at termini of an RNA or DNA (Solomatin and Herschlag 2009). For proteins, dyes can be attached to engineered cysteines using maleimide chemistry, often by mutagenesis to create a single-cysteine version (Nanda and Lorsch 2014). Alternatively, nonnatural amino acids can be incorporated, allowing use of orthogonal chemistries (e.g., CLICK). A number of additional tagging methods have been developed, which include a full protein or peptide domain that can be dye labeled (SNAP, HaloTag, ybbR) (Keppler et al. 2003; Yin et al. 2006; Los et al. 2008). For all these labeling approaches, the function of the modified protein must be tested, as modification of the protein (either the sequence, length, or dye labeling) may greatly perturb function. Such biochemical controls are absolutely essential for subsequent valid biophysical investigation.
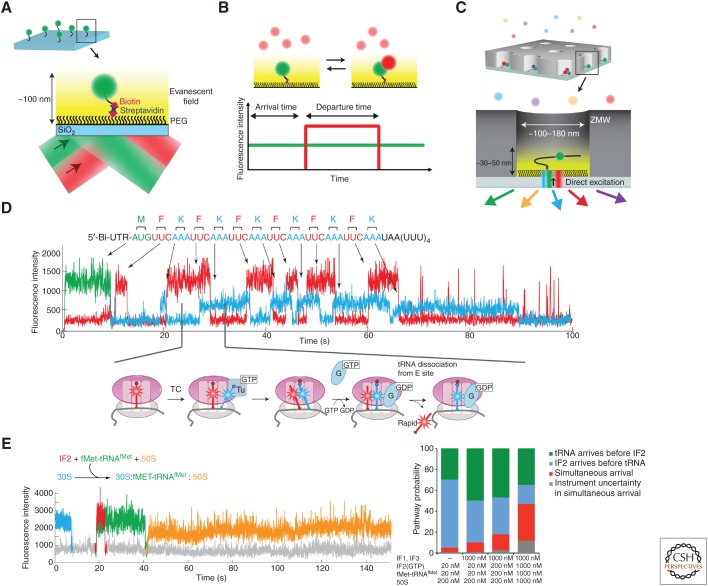
By observing the behavior of single molecules, complex dynamic behavior of biomolecular systems can be determined and sorted, free from ensemble averaging. Single-molecule fluorescence experiments are often performed with systems immobilized on surfaces, such that they can be observed for long time periods (seconds-to-hours) without diffusing away (Fig. 2A). These experiments harness surface chemistry to make the (usually) SiO2 surface inert to nonspecific binding of biomolecules, often using polyethylene glycol (PEG) derivatization of the surface, while using biotin–streptavidin interactions to provide specific, stable tethering of biomolecules to a surface (Fig 2A) (Ha 2001). The simplest single-molecule experiment monitors ligand binding and dissociation (Fig. 2B). A fluorescent ligand (labeled protein, DNA/RNA) is present in solution that can bind to a surface-immobilized biomolecule; fluorescence is normally excited using total internal reflectance (TIR), whereby a laser illuminates a quartz surface at a certain angle (dependent on the refractive index of the surface) to create an evanescent field that extends ∼100 nm above the surface, leading to wide spatial excitation. Binding of the fluorescent ligand to the surface-immobilized biomolecule leads to a burst in fluorescence, as the fluorophore is localized for a long period to emit photons. The free fluorescent molecule in solution is also excited but if the concentration is kept below ∼20–50 nm, the average occupancy of the fluorophore in this “excitation” volume caused by diffusion is less than one, leading to baseline fluorescence lower than the fluorescent burst. Using this experiment, the kinetics of ligand binding and dissociation can be measured at the single-molecule level by analyzing the statistical distribution of binding event lifetimes (dissociation rate), or times between events (association rate). Thus, simple compositional dynamics (ligand occupancy as a function of time) can be measured for biomolecular systems.
Figure 2.
Compositional studies of translation using single-molecule fluorescence. (A) Fluorescently labeled biomolecule (green) functionalized with a biotin is immobilized to the polyethylene glycol (PEG)-derivatized SiO2 surface through biotin–streptavidin interactions. Total internal reflectance (TIR) shown here produces an evanescent field, exciting only fluorophores within 100 nm from the surface. (B) Binding of a second labeled biomolecule (red) in solution to the immobilized biomolecule results in detection of red fluorescence burst, from which arrival and departure times can be measured. (C) Zero-mode waveguides (ZMWs) allow delivery of up to four different fluorescently labeled ligands at micromolar concentrations caused by its shorter excitation lengths with direct illumination by green (532 nm), red (642 nm), and cyan (488 nm) lasers. (D) 70S ribosomes with fMet-Cy3-tRNAfMet (green) were immobilized by the biotinylated mRNA and real-time transit of Phe-Cy5-tRNAPhe (red) and Lys-Cy2-tRNALys (cyan) ternary complexes (TCs) was monitored during translation (Uemura et al. 2010). (E) Tracking the binding events of Cy5-IF2 (red), fMet-Cy3-tRNAfMet (green), and Cy3.5-50S subunit (yellow) to the Alexa 488-30S-mRNA complex (cyan) (Tsai et al. 2012) revealed multiple pathways of translation initiation in bacteria (see text for details).
Multiple ligands can be tracked at higher free ligand concentration using different single-molecule configurations. We have applied nanophotonic devices called “zero-mode waveguides” (ZMWs) that reduce the excitation volume by orders of magnitude, allowing use of free ligand concentrations up to µm (Levene et al. 2003). This is critical in studying translation, in which many translational components are at the micromolar range to drive rapid association of ligands and efficient translation. The ZMWs are roughly cylindrical chambers 100–180 nm in diameter and ∼200 nm deep, with transparent quartz (with biotin-PEG) bottoms to immobilize and excite biomolecular and aluminum (or other conductor) side walls (Fig. 2C). Propagating laser excitation is quenched as it moves up the Z-axis of the ZMW, leading to high excitation only near the surface. We have applied a sophisticated optical system that allows detection of fluorescence with two laser excitation and four different emission channels (Chen et al. 2014). We can thus monitor, in real time, the ligand occupancy on a translation complex of up to four different ligands simultaneously. This allows determination of the relative order or coordination of ligand arrival and departures in translation, which we outline further in the next section.
EARLY COMPOSITIONAL STUDIES OF TRANSLATION USING OUR ZMW APPROACH
The power of this real-time compositional approach was first shown using labeled tRNAs during bacterial translation initiation and elongation. Using dye-labeled tRNAs (with dyes placed at the elbow position of the L-shaped tRNA, which does not interfere with function), we could track the arrival of tRNAs to an A-site codon, occupancy on the ribosome as the tRNA moves from A to P to E sites, and subsequent departure from the E site at each codon (Fig. 2D) (Uemura et al. 2010). The tRNA-binding events overlap, during two-tRNA occupancy, and from these data a codon-by-codon translation rate could be calculated. Importantly, these data showed a lack of coupling of A-site tRNA arrival with E-site tRNA departure.
We have applied similar approaches to determine the ligand-binding dynamics and pathways of translation initiation. We first investigated prokaryotic translation initiation, using labeled initiator tRNA, subunits, and initiation factor IF2 (Tsai et al. 2012). Using different color dyes on the different components, we could track the arrival of initiator tRNA, IF2, and 50S subunit simultaneously in real time (Fig. 2E). These experiments showed heterogeneous pathways for fMet-tRNAfMet and IF2 arrival, with the two components (which interact) arriving either sequentially or simultaneously on the 30S subunit. The simultaneous pathway occurs more frequently at higher concentrations of IF2 and tRNA. Both sequential and simultaneous pathways lead to efficient 50S subunit joining, with IF2 departing rapidly from the assembled 70S subunit. These experiments showed the power of tracking translational pathways with single-molecule approaches.
REAL-TIME TRACKING OF CONFORMATION DURING PROKARYOTIC TRANSLATION
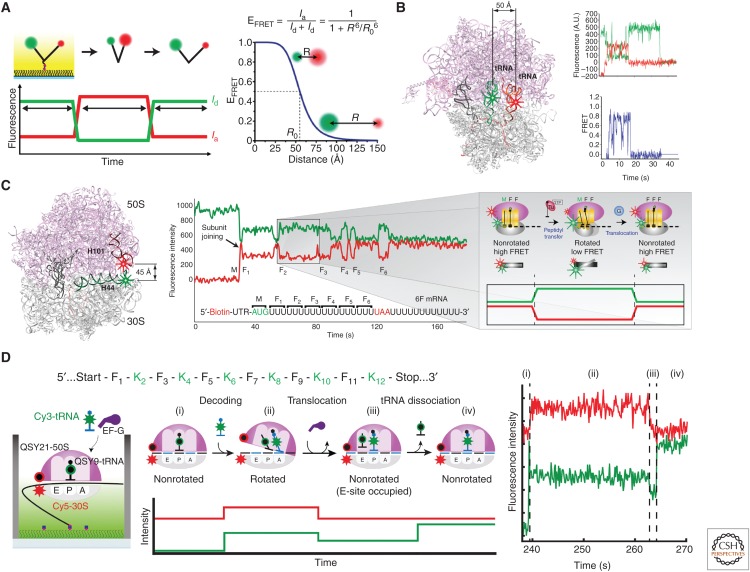
A powerful application of single-molecule fluorescence uses Förster resonance energy transfer (FRET) to probe conformational dynamics (Selvin 2000; Ha 2001). FRET relies on through-space, dipolar interactions between donor and acceptor dyes to allow energy transfer between the low-wavelength donor to higher-wavelength acceptor (Fig. 3A). The efficiency of the energy transfer depends on multiple factors (quantum yield of the donor, overlap of the emission spectrum of the donor with the absorbance spectrum of the acceptor, refractive index of the medium, dynamic orientations of the two dyes) but most critically the interdye distance, R, with 1/R6 dependence. For a given dye pair, this efficiency is given by the equation in Figure 3A, in which R0 is a characteristic of the dye pair (incorporating the parameters discussed above) and represents the distance separation for 50% energy transfer from donor to acceptor. With R0 values of typical FRET dye pairs (e.g., Cy3 and Cy5) at 40–50 Å, FRET efficiencies vary from 100% at interdye distances below 30 Å to zero beyond 80 Å; in between, FRET depends steeply on interdye distance, making FRET a powerful probe in this range. As such, these FRET measurements are best applied to larger systems with appropriate interdye distances. The ribosome, with its large geometry (250 Å sphere) is an outstanding system to apply FRET.
Figure 3.
Conformational studies of translation using single-molecule Förster resonance energy transfer (FRET). (A) With single-laser excitation of the donor dye, fluorescence intensities of donor (green, Id) and acceptor (red, Ia) dyes attached to a surface-immobilized biomolecular complex are measured to calculate the FRET efficiency (EFRET), which reports distance between the fluorophores (R) used to define conformational states of a biological process. (B) Cy3 and Cy5 labeling of P-site and A-site transfer RNAs (tRNAs), respectively, was used to follow the tRNA conformations during tRNA selection stage of elongation (Blanchard et al. 2004a). (C) (Left) Helix44 of 30S subunit was labeled with Cy3 and Helix101 of 50S subunit was labeled with Cy5, bringing the dyes within FRET distance to report intersubunit conformational changes (Marshall et al. 2008b). (Right) With immobilization of Cy3-30S (green) as a preinitiation complex and delivery of Cy5-50S (red) along with Phe-tRNAPhe and elongation factors, subunit joining is signaled by the simultaneous burst of Cy5 fluorescence and dip in Cy3 fluorescence at the beginning, and anticorrelated changes in the Cy3 and Cy5 fluorescence intensities correspond to transitions between the nonrotated and rotated conformational states evolved from cycles of peptidyl transfer and translocation during elongation. (D) Replacing the acceptor dyes in tRNA and ribosome labels in B and C with nonfluorescent quenchers (QSY9, QSY21, respectively) allowed simultaneous detection of ribosome intersubunit and tRNA conformations with Cy3 labeling on Lys-tRNALys and Cy5 labeling on the 30S subunit (Choi and Puglisi 2017).
FRET energy transfer rates are rapid (nanosecond timescale), so FRET provides a powerful probe of conformational dynamics in translation. Changes in interdye distances from a conformational change in a two-state process appear as abrupt changes in single-molecule FRET trajectories that are readily analyzed. We initially characterized tRNA dynamics on the bacterial ribosome using FRET between A- and P-site tRNAs (Fig. 3B) (Blanchard et al. 2004b), as tRNAs were thought to change configurations on the ribosome during translation. Using Cy3 donor dye on the P-site tRNA and Cy5 acceptor dye on the A-site tRNA, FRET monitored the arrival of Cy5-(acceptor) labeled tRNA as a ternary complex with EF-Tu in the A site, and subsequent steps of GTP hydrolysis by EF-Tu, accommodation and conformational changes post-peptide bond formation. Using these experiments, we observed the dynamic pathway of tRNA selection, how near cognate tRNAs are rejected, and how drugs perturb the conformational dynamics of tRNA selection (Blanchard et al. 2004a). Cognate aminoacyl-tRNAs rapidly proceed through an initial low FRET through GTPase activation (medium FRET) to accommodated states (high FRET), whereas near-cognate tRNAs are rejected at earlier steps and rarely proceed to accommodation. Drugs, such as tetracycline, disrupt these processes; with tetracycline blocking the final position of the codon–anticodon on the small subunit, only brief, low FRET-binding events of cognate ternary complexes were observed. Small molecule drugs are powerful disrupters of ribosome dynamics.
FRET has been used to observe many features of ribosome function. With ribosome structures as blueprints, FRET probes can be placed at appropriate points in the ribosome. Ribosomal proteins can be labeled using single-cysteine mutants or more sophisticated tagging methods (peptide tags, nonnatural amino acid incorporation), and incorporated into ribosomes during biogenesis. The labeled ribosomal protein can be incorporated into ribosomal particles that have single protein deletions in certain cases (Ermolenko et al. 2007) or by in vitro reconstitution (Hickerson et al. 2005). Ribosomal RNA has been labeled using mutations of ribosomal RNA elements to allow hybridization of labeled oligonucleotides (Dorywalska et al. 2005). In all cases, the function of the labeled ribosomes must be tested.
Using labeled bacterial ribosomes, we and others have investigated intersubunit conformation (Ermolenko et al. 2007; Marshall et al. 2008b), head movement of the ribosome (Hickerson et al. 2005), and dynamics of the L1 stalk region (Fei et al. 2008; Cornish et al. 2009). By using our labeling scheme (helix44 on the small subunit, helix101 on the large), the intersubunit rotational state of the ribosome as it passes from initiation, to elongation, to termination, and then to recycling, has been tracked (Marshall et al. 2008b, 2009; Prabhakar et al. 2017a). The two most populated intersubunit states involve the nonrotated and rotated conformations distinguished by rotation of 3°–10° (Valle et al. 2003). The two states interconvert on peptide bond formation or translocation driven by EF-G (or the action of IF2-GTP or RF3-GTP); the nature of the barrier height of this conformational change remains to be determined. However, this conformational signal allowed tracking of translation. Initiation is signaled by the arrival of FRET (subunits join) and elongation by cycles of high to lower to high FRET, representing one round of tRNA selection, peptide bond formation, and translocation (Fig. 3C). We have used these signals to watch translation in real time (Aitken and Puglisi 2010).
These FRET measurements provide time-resolved conformational information on the ribosome during translation. However, all FRET measurements suffer from the sensitivity of FRET to parameters beyond interdye distance (dye mobility, photophysics of the dyes, the interdye medium). Thus, conformational information should be obtained from multiple perspectives using multiple labeling schemes, with each FRET measurement at best representing a single distance within the complexity of the translational apparatus. In addition, conformational changes can be correlated to ligand arrival and departure, but this is challenging with normal FRET as a result of cross talk of the individual dyes required to measure FRET and ligand arrival. By swapping the acceptor dye with a nonfluorescent acceptor that can absorb the energy transfer via FRET but release it as heat and not light, the normal acceptor dye (e.g., Cy5) can be used to label the ligand. We first showed this quencher approach in tracking the ribosome intersubunit conformations during translation (Chen et al. 2012). Using this “one-color” FRET, correlated conformational changes with ligand arrival and departure have been measured, and reviewed extensively elsewhere (See Fig. 4) (Chen et al. 2013; Prabhakar et al. 2017b).
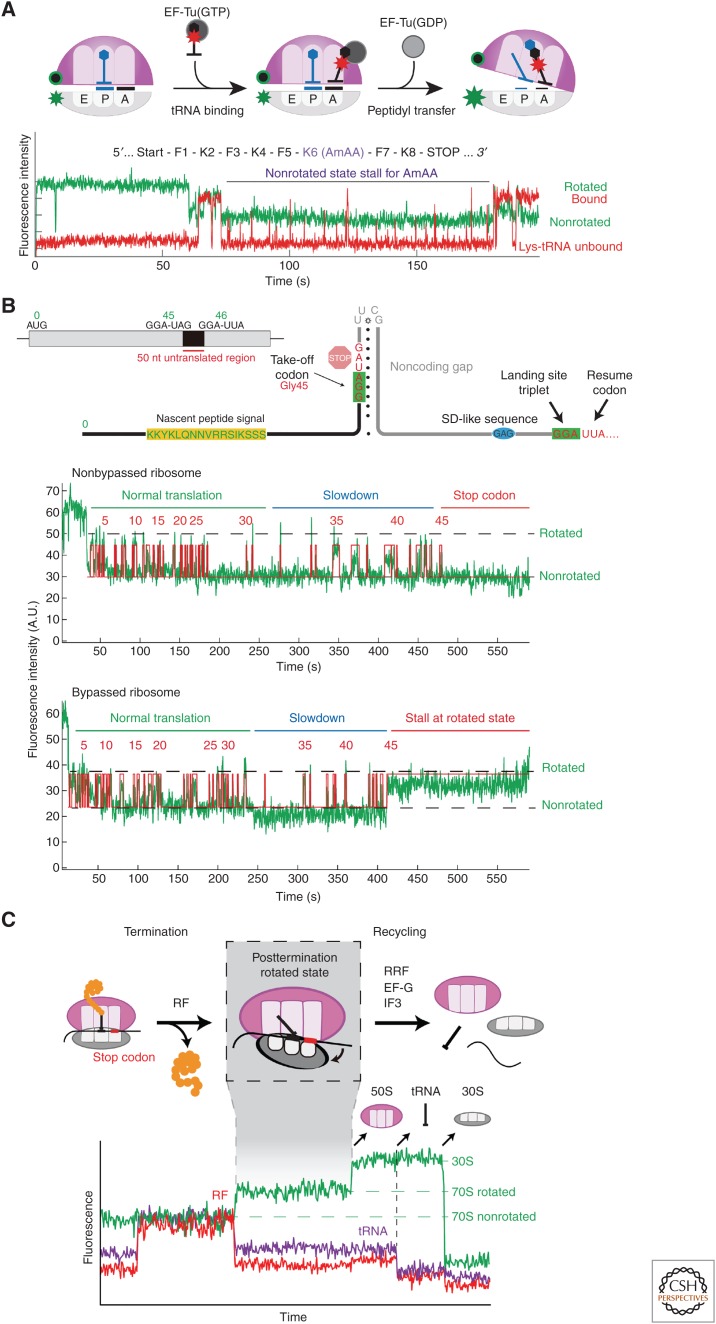
Figure 4.
Recent work on bacterial translation involved use of intersubunit dye–quencher labeling (Cy3B-30S and BHQ-2-50S; see Fig. 3C for labeling positions) to track ribosome conformation (green signal). (A) Monitoring Lys-Cy5-tRNALys-binding dynamics (red) on ribosomes with Nm-modified mRNAs (Choi et al. 2018) shows multiple short Cy5 pulses representing tRNA rejection events, explaining the long ribosome stall in nonrotated state. (B) Tracking translation of gene 60 mRNA revealed that the subset of ribosomes that bypassed stalled at the take-off Gly-45 codon in the rotated state. (C) Simultaneously tracking the ribosome intersubunit conformation (green), Cy5-RF binding (red), and P-site tRNA occupancy (Phe-Cy5.5-tRNAPhe, violet) during translation termination and recycling (Prabhakar et al. 2017a) helped resolve the posttermination rotated state, a key intermediate preceding subunit splitting step (catalyzed by RRF and EF-G) and subsequent 30S complex disassembly (performed by IF3).
Recently, we expanded this quencher approach to monitor two conformational changes simultaneously (Choi and Puglisi 2017). We used two donor–quencher dye pairs, and designed the separation of the pairs to be greater than 100 Å to avoid cross talk. As such, we could monitor intersubunit conformation directly correlated to tRNA–tRNA conformation during translation. Our results allowed us to show that, as expected from structural data, these conformational fluctuations were correlated. Using this approach, we identified a novel intermediate intersubunit rotational state when three tRNAs reside in the A, P, and E sites. This three-tRNA state slows elongation until E-site tRNA is spontaneously released, allowing EF-G to drive translocation. Further structural characterization is needed to confirm this state, but these results highlight the future interplay of structural and single-molecule approaches.
Biological machines generate force during their function. Using optical trapping methods, the forces generated by these machines can be measured or perturbed in real time, providing a direct view of their mechanochemical cycle. In an optical trapping experiment, biological systems are tethered to large beads, which can be manipulated and trapped by a strong laser beam. As forces are generated, the bead is pulled out of the beam, allowing measurement of the force generated by, or application of force upon, the system. Traditionally, optically trapping has been applied to protein motors (myosins, kinesins, gyrases) and polymerases, because the rigidity of nucleic acids allows more facile trapping and monitoring of forces. It is far more challenging to apply these approaches to translation, which occurs on a flexible single-stranded RNA. By circumventing these difficulties through clever experimental and instrument designs, Bustamante, Tinoco, and colleagues have elegantly explored the mechanisms of ribosome movement during translocation (Wen et al. 2008) and protein folding (Kaiser et al. 2011) on the ribosome. These experiments showed how ribosomes efficiently unfold mRNA structures, and revealed the forces generated by both the ribosome and protein folding within the ribosomal peptide exit tunnel. These force measurements can be combined with fluorescence and/or FRET to provide combined force and conformation/composition information. Such experiments will be powerful probes of translational processes.
RECENT WORK ON BACTERIAL TRANSLATION
To understand the applications of single-molecule fluorescence methods to translation, we highlight below some recent examples from our own work; this, of course, is completely arbitrary, and ignores the outstanding work performed by many other groups through the years (Gonzalez, Blanchard, Cooperman, Goldman, and others). Our goal is not to be comprehensive—many recent reviews have covered the field extensively—but to give examples of how these methods can provide insights into aspects of translation (Marshall et al. 2008a; Blanchard 2009; Perez and Gonzalez 2011; Petrov et al. 2012).
Effect of mRNA Chemical Modifications on Elongation Dynamics
Recent transcriptome-wide mapping revealed the prevalence of different RNA chemical modifications within the coding sequences of cellular mRNAs (Roundtree et al. 2017). These findings suggested that these modifications may modulate the decoding steps of elongation. We investigated this phenomenon by testing the effects of two frequently occurring modifications, N6-methyladenosine (m6A) and 2′-O-methylation (Nm), on the dynamics of elongation. By simultaneously tracking binding of labeled tRNA (Cy5) and ribosome conformation (Cy3B-quencher) in our single-molecule translation system (Fig. 4A), combined with bulk biochemical and structural studies, we discovered that the two mRNA modifications perturb different steps of the tRNA-decoding mechanism of elongation. The methylated adenosine of the m6A-containing codon destabilizes the codon–anticodon interactions critical in the initial tRNA selection step, thus increasing the probability of tRNA rejection in the early stage of decoding (Choi et al. 2016). Methylation of the 2′OH of the ribose sugar in Nm, however, does not disrupt codon–anticodon base-pairing; instead, it sterically blocks interactions that allow formation of the correct geometry of ribosomal monitoring bases required to activate the GTPase of EF-Tu, a requisite for successful tRNA accommodation (Choi et al. 2018). In addition to mechanistic differences, Nm results in an enormous slowdown in translation of the codon (1500-fold) in contrast to the modest effects of m6A (18-fold). This disparity may signify diversity in regulatory elements to tune the local translation elongation rates, which could in turn modulate coupled cotranslational processes like protein folding (Kim et al. 2015).
Ribosome and mRNA Structural Rearrangements during Bypassing
Programmed translational bypassing, in which a ribosome hops over a long stretch of mRNA nucleotides during elongation, remains one of the most unusual and understudied translational phenomena. The best-documented example of this occurs in bacteria when the Escherichia coli ribosome translates and bypasses a 50-nt stretch of the T4 bacteriophage gene 60 mRNA from the Gly-45 (take-off) codon to another Gly (landing) codon. Two key stimulatory elements that initiate this process are an upstream nascent peptide signal and an mRNA hairpin structure at the take-off site with a UUCG tetraloop (Fig. 4B). The mechanism of stimulation by these elements as well as correct ribosome landing remained unclear. Interested in how these elements changed the translation elongation dynamics, we tracked gene 60 translation using our single-molecule intersubunit dye-quencher system (Chen et al. 2015). Measuring local elongation rates, we observed a gradual slowdown in elongation as the ribosome approached the bypass site. After incorporation of the Gly-tRNAGly at the take-off codon, 35% of the ribosomes experienced a 10- to 20-fold longer pause in the rotated state, while the rest translocated to the adjacent stop-46 codon and stopped translating. For the 35% that paused at codon Gly-45, elongation continued afterward, representing the subset of ribosomes that successfully bypassed. This long, noncanonical rotated-state pause became the centerpiece of an updated mechanistic model. The model describes the initial slowdown at codons 40–45 to be caused by the peptide interacting with and/or folding within the ribosome peptide exit tunnel. This results in buildup of tension until the rotated state pause at Gly-45 occurs, producing weakened ribosome–tRNA–mRNA interactions key to promoting bypassing. At this point, the takeoff hairpin is partially melted within the mRNA channel, so EF-G-catalyzed translocation then leads to refolding of the UUCG tetraloop that then threads mRNA in the 5′ direction, initiating bypassing. Tracking the ribosome position relative to the landing site with ribosome–mRNA FRET showed that the ribosome reaches near the landing site rapidly but spends most of the long rotated-state pause trekking a short distance to find the appropriate landing site. Through this work, we laid a mechanistic framework that places the newly discovered stalled translational state as the focal point and highlights the important coupling of ribosome and mRNA structural rearrangements to successful bypassing.
Dynamic Coupling of Prokaryotic Translation Termination and Ribosome Recycling
The ribosome undergoes multiple cycles of elongation as it decodes every codon with the exception of the three stop codons (UAG, UGA, UAA), which signal the closing events of nascent peptide release (termination) and ribosome disassembly (recycling) (see Rodnina 2018). Termination begins with the recruitment of two classes of RFs. The class I RF (RF1 and RF2 in bacteria) recognizes the stop codon and catalyzes the release of the peptide. Bacterial class II RF, or RF3, is a nonessential (Grentzmann et al. 1994; O'Connor 2015) GTPase factor that accelerates the dissociation of class I RF from the ribosome (Freistroffer et al. 1997), but its general role in translation is still uncertain. Thus, the molecular events that occur after peptide release are still unclear. Eventually, the posttermination ribosome complex is disassembled through a recycling mechanism involving ribosome recycling factor (RRF), elongation factor EF-G, and initiation factor IF3. This mechanism was still unclear owing to the difficulty in resolving transient intermediates of the process.
Using single-molecule real-time tracking of all four stages of translation, we resolved a key intermediate at the transition from termination to recycling and delineated the timings of events during recycling (Prabhakar et al. 2017a). As our intersubunit dye–quencher system monitored initiation and elongation, we tracked binding of Cy5-labeled class I RF during termination (Fig. 4C). We discovered that after successful peptide release, class I RF dissociation triggers an intersubunit rotation from the nonrotated to the rotated state. This posttermination rotated state represents an unstable intermediate that either undergoes uncatalyzed ribosomal subunit splitting or RF rebinding. We found that RRF binds to this intermediate to block RF from rebinding and forms the ribosomal substrate for EF-G, to catalyze subunit disassembly through GTP hydrolysis. IF3 binds to the remaining 30S•tRNA•mRNA complex to induce dissociation of the deacylated tRNA, followed by rapid dissociation of the 30S subunit from the mRNA. By simultaneously tracking ribosome conformation and occupancy of tRNA and protein factors, most of the events of termination and recycling were temporally resolved to bring light to the dynamic coupling between these processes. This method will be powerful in answering further questions surrounding termination and recycling, such as the role of RF3 in this mechanism.
RECENT WORK ON EUKARYOTIC TRANSLATION
The mechanistic pathways by which eukaryotic translation initiation occurs remain unclear and are an outstanding target for single-molecule analyses. However, single-molecule experiments on eukaryotic translation are challenged by the complexity of the process, and the practicality of labeling factors, ligands, and ribosomes for tracking. We have applied a variety of approaches to obtain labeled versions of initiation factors and ribosomes in yeast and human translation systems. These include yeast genetic approaches (Petrov and Puglisi 2010) or CRISPR knockouts (Fuchs et al. 2015) to obtain pure populations of mutant ribosomes, which can be labeled using nucleic acids or labeled proteins added back to the mutants. We have also developed genetic systems for labeling critical initiation factors (such as eIF3) in humans and have shown the power of using these labeled reagents to track translation initiation processes directly (Johnson et al. 2017).
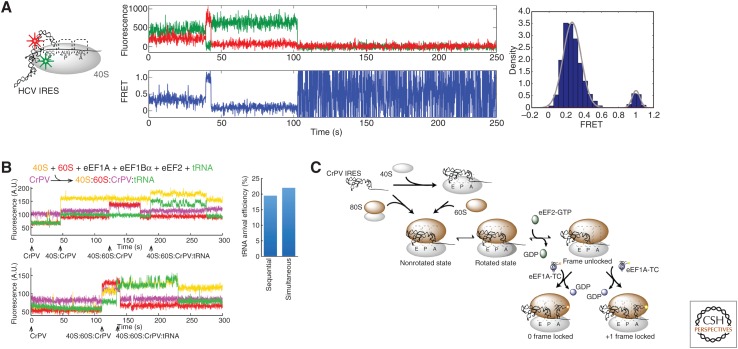
We first applied single-molecule approaches to the much simpler internal ribosome entry site (IRES)-mediated mechanisms, which require a small subset of factors. We showed that human 40S subunits can form dynamic complexes with the hepatitis C virus (HCV) IRES by using FRET between the IRES RNA and labeled ribosomal protein eS25 on the 40S subunit (Fig. 5A) (Fuchs et al. 2015). We determined the pathways by which the cricket paralysis virus (CrPV) IRES initiates translation directly without initiation factors or initiator tRNA (Petrov et al. 2016). Using direct tracking of 40S and 60S subunits and tRNAs, we could determine the distinct pathways by which the IRES initiates (Fig. 5B). In addition to an expected sequential pathway in which 40S subunits bind to the IRES RNA, followed by 60S subunit binding, we observed a significant population of direct 80S ribosome arrival (simultaneous 40S and 60S binding) to an IRES. Both pathways were equally functional in subsequent elongation, which required the continued presence of eEF2•GTP to allow the first elongator tRNA to bind. We also uncovered heterogeneity in reading frame establishment, with ribosomes initiating on the IRES in both the 0 and +1 frame. Even for a simple system like the CrPV IRES, single-molecule approaches revealed the complex pathways by which initiation occurs (Fig. 5C).
Figure 5.
Recent work on eukaryotic translation. (A) Conformational dynamics of hepatitis C virus (HCV) internal ribosome entry site (IRES) bound to human 40S subunit was monitored using Cy3-Cy5 Förster resonance energy transfer (FRET) (Fuchs et al. 2015), detecting two different conformational states important in the pathway of HCV IRES initiation. (B) Tracking real-time assembly of yeast Cy3.5-40S (yellow), yeast Cy5-60S (red), and Phe-Cy3-tRNAPhe (green) on immobilized Cy5.5-CrPV IRES (violet) (Petrov et al. 2016) led to discovery of parallel pathways of CrPV IRES initiation. (C) Petrov et al. (2016) also found that ribosomes initiate on the CrPV IRES in both 0 and +1 reading frames, another pathway branchpoint in the mechanism of CrPV IRES initiation.
CONCLUDING REMARKS
Single-molecule approaches have provided a structurally dynamic view of translation, complementing biochemical and structural experiments. Single-molecule measurements have connected structural states, determined by X-ray crystallography and cryoEM, into a more coherent pathway of states during normal initiation, elongation, and termination. In short, these methods have animated the static pictures from structural biology and identified transient states that are difficult to capture by these methods. By tracking single ribosomes, the simple, linear view of translation has been adjusted. These methods have parsed distinct, parallel pathways in noncanonical translation during initiation, frameshifting, and bypassing, which directly determine the timing and outcome of protein synthesis. Despite these advances, many open questions remain. In bacterial translation, the interplay of the nascent protein chain and its folding with translational dynamics is a fascinating frontier of investigation. The dynamics of multiple ribosomes (polysomes) on an mRNA, and whether there is communication between ribosomes, should be probed. Eukaryotic translation and its regulation remain a challenging and central topic that has only recently been explored. New methods that access faster timescale dynamics and embrace greater complexity will be needed to elucidate the dynamic role of translation in human health and disease.
ACKNOWLEDGMENTS
Single-molecule research in the Puglisi group is funded by National Institutes of Health (NIH) Grants GM51266, GM113078, AI047365, AI099506, and GM082545. A.P. is funded by NIH Molecular Biophysics Training Grant (T32-GM008294) and a Stanford Interdisciplinary Graduate Fellowship.
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Aitken CE, Puglisi JD. 2010. Following the intersubunit conformation of the ribosome during translation in real time. Nat Struct Mol Biol 17: 793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoun A, Pavlov MY, Andersson K, Tenson T, Ehrenberg M. 2003. The roles of initiation factor 2 and guanosine triphosphate in initiation of protein synthesis. EMBO J 22: 5593–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Barbet-Massin E, Huang CT, Daebel V, Hsu ST, Reif B. 2015. Site-specific solid-state NMR studies of “trigger factor” in complex with the large ribosomal subunit 50S. Angew Chem Int Ed Engl 54: 4367–4369. [DOI] [PubMed] [Google Scholar]
- Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, et al. 2012. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482: 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Chichester RJ. 1993. Single molecules observed by near-field scanning optical microscopy. Science 262: 1422–1425. [DOI] [PubMed] [Google Scholar]
- Bilgin N, Claesens F, Pahverk H, Ehrenberg M. 1992. Kinetic properties of Escherichia coli ribosomes with altered forms of S12. J Mol Biol 224: 1011–1027. [DOI] [PubMed] [Google Scholar]
- Blanchard SC. 2009. Single-molecule observations of ribosome function. Curr Opin Struct Biol 19: 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. 2004a. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol 11: 1008–1014. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Kim HD, Gonzalez RL Jr, Puglisi JD, Chu S. 2004b. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci 101: 12893–12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Weissman JS. 2015. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol 16: 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Shao S, Murray J, Hegde RS, Ramakrishnan V. 2015. Structural basis for stop codon recognition in eukaryotes. Nature 524: 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Tsai A, Petrov A, Puglisi JD. 2012. Nonfluorescent quenchers to correlate single-molecule conformational and compositional dynamics. J Am Chem Soc 134: 5734–5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Petrov A, Tsai A, O'Leary SE, Puglisi JD. 2013. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat Struct Mol Biol 20: 718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Dalal RV, Petrov AN, Tsai A, O'Leary SE, Chapin K, Cheng J, Ewan M, Hsiung PL, Lundquist P, et al. 2014. High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Proc Natl Acad Sci 111: 664–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Coakley A, O'Connor M, Petrov A, O'Leary SE, Atkins JF, Puglisi JD. 2015. Coupling of mRNA structure rearrangement to ribosome movement during bypassing of non-coding regions. Cell 163: 1267–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Puglisi JD. 2017. Three tRNAs on the ribosome slow translation elongation. Proc Natl Acad Sci 114: 13691–13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Ieong KW, Demirci H, Chen J, Petrov A, Prabhakar A, O'Leary SE, Dominissini D, Rechavi G, Soltis SM, et al. 2016. N6-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat Struct Mol Biol 23: 110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Indrisiunaite G, DeMirci H, Ieong KW, Wang J, Petrov A, Prabhakar A, Rechavi G, Dominissini D, He C, et al. 2018. 2′-O-methylation in mRNA disrupts tRNA decoding during translation elongation. Nat Struct Mol Biol 25: 208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T. 2009. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci 106: 2571–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Green R. 2012. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol 4: a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dever TE, Dinman JD, Green R. 2018. Translation elongation and recoding in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorywalska M, Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. 2005. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res 33: 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. 2007. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol 370: 530–540. [DOI] [PubMed] [Google Scholar]
- Fei J, Kosuri P, MacDougall DD, Gonzalez RL Jr. 2008. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell 30: 348–359. [DOI] [PubMed] [Google Scholar]
- Fernandez IS, Bai XC, Hussain T, Kelley AC, Lorsch JR, Ramakrishnan V, Scheres SHW. 2013. Molecular architecture of a eukaryotic translational initiation complex. Science 342: 1240585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistroffer DV, Pavlov MY, MacDougall J, Buckingham RH, Ehrenberg M. 1997. Release factor RF3 in E. coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J 16: 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Kaledhonkar S, Borg A, Sun M, Chen B, Grassucci RA, Ehrenberg M, Frank J. 2016. Key intermediates in ribosome recycling visualized by time-resolved cryoelectron microscopy. Structure 24: 2092–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Petrov AN, Marceau CD, Popov LM, Chen J, O'Leary SE, Wang R, Carette JE, Sarnow P, Puglisi JD. 2015. Kinetic pathway of 40S ribosomal subunit recruitment to hepatitis C virus internal ribosome entry site. Proc Natl Acad Sci 112: 319–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. 2009. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326: 694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G, Brechemier-Baey D, Heurgue V, Mora L, Buckingham RH. 1994. Localization and characterization of the gene encoding release factor RF3 in Escherichia coli. Proc Natl Acad Sci 91: 5848–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski KB, Wieden HJ, Rodnina MV. 2002. Kinetic mechanism of elongation factor Ts-catalyzed nucleotide exchange in elongation factor Tu. Biochemistry 41: 162–169. [DOI] [PubMed] [Google Scholar]
- Ha T. 2001. Single-molecule fluorescence resonance energy transfer. Methods 25: 78–86. [DOI] [PubMed] [Google Scholar]
- Ha T, Tinnefeld P. 2012. Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu Rev Phys Chem 63: 595–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hellen CUT. 2018. Translation termination and ribosome recycling in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickerson R, Majumdar ZK, Baucom A, Clegg RM, Noller HF. 2005. Measurement of internal movements within the 30S ribosomal subunit using Forster resonance energy transfer. J Mol Biol 354: 459–472. [DOI] [PubMed] [Google Scholar]
- Huang C, Kalodimos CG. 2017. Structures of large protein complexes determined by nuclear magnetic resonance spectroscopy. Annu Rev Biophys 46: 317–336. [DOI] [PubMed] [Google Scholar]
- Hussain T, Llacer JL, Wimberly BT, Kieft JS, Ramakrishnan V. 2016. Large-scale movements of IF3 and tRNA during bacterial translation initiation. Cell 167: 133–144.e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Ingolia NT, Hussmann JA, Weissman JS. 2018. Ribosome profiling: Global views of translation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Jobe A, Liu Z, Gutierrez-Vargas C, Frank J. 2018. New insights into ribosome structure and function. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Lovmar M, Ehrenberg M. 2008. Rate and accuracy of bacterial protein synthesis revisited. Curr Opin Microbiol 11: 141–147. [DOI] [PubMed] [Google Scholar]
- Johnson AG, Petrov AN, Fuchs G, Majzoub K, Grosely R, Choi J, Puglisi JD. 2017. Fluorescently-tagged human eIF3 for single-molecule spectroscopy. Nucleic Acids Res 46: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser CM, Goldman DH, Chodera JD, Tinoco I Jr, Bustamante C. 2011. The ribosome modulates nascent protein folding. Science 334: 1723–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi R, Pavlov MY, Buckingham RH, Ehrenberg M. 1999. Novel roles for classical factors at the interface between translation termination and initiation. Mol Cell 3: 601–609. [DOI] [PubMed] [Google Scholar]
- Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. 2003. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol 21: 86–89. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Yoon JS, Shishido H, Yang Z, Rooney LA, Barral JM, Skach WR. 2015. Protein folding. Translational tuning optimizes nascent protein folding in cells. Science 348: 444–448. [DOI] [PubMed] [Google Scholar]
- Koutmou KS, McDonald ME, Brunelle JL, Green R. 2014. RF3:GTP promotes rapid dissociation of the class 1 termination factor. RNA 20: 609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovermann M, Rogne P, Wolf-Watz M. 2016. Protein dynamics and function from solution state NMR spectroscopy. Q Rev Biophys 49: e6. [DOI] [PubMed] [Google Scholar]
- Kurauskas V, Crublet E, Macek P, Kerfah R, Gauto DF, Boisbouvier J, Schanda P. 2016. Sensitive proton-detected solid-state NMR spectroscopy of large proteins with selective CH3 labelling: Application to the 50S ribosome subunit. Chem Commun (Camb) 52: 9558–9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW. 2003. Zero-mode waveguides for single-molecule analysis at high concentrations. Science 299: 682–686. [DOI] [PubMed] [Google Scholar]
- Llacer JL, Hussain T, Marler L, Aitken CE, Thakur A, Lorsch JR, Hinnebusch AG, Ramakrishnan V. 2015. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol Cell 59: 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftfield RB. 1963. The frequency of errors in protein biosynthesis. Biochem J 89: 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, et al. 2008. HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol 3: 373–382. [DOI] [PubMed] [Google Scholar]
- Marshall RA, Aitken CE, Dorywalska M, Puglisi JD. 2008a. Translation at the single-molecule level. Annu Rev Biochem 77: 177–203. [DOI] [PubMed] [Google Scholar]
- Marshall RA, Dorywalska M, Puglisi JD. 2008b. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci 105: 15364–15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RA, Aitken CE, Puglisi JD. 2009. GTP hydrolysis by IF2 guides progression of the ribosome into elongation. Mol Cell 35: 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheisl S, Berninghausen O, Becker T, Beckmann R. 2015. Structure of a human translation termination complex. Nucleic Acids Res 43: 8615–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Merrick WC, Pavitt GD. 2018. Protein synthesis in eukaryotic cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a32714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X, Kapanidis AN, Laurence T, Pinaud F, Doose S, Pflughoefft M, Weiss S. 2003. The power and prospects of fluorescence microscopies and spectroscopies. Annu Rev Biophys Biomol Struct 32: 161–182. [DOI] [PubMed] [Google Scholar]
- Milon P, Maracci C, Filonava L, Gualerzi CO, Rodnina MV. 2012. Real-time assembly landscape of bacterial 30S translation initiation complex. Nat Struct Mol Biol 19: 609–615. [DOI] [PubMed] [Google Scholar]
- Murray J, Savva CG, Shin BS, Dever TE, Ramakrishnan V, Fernandez IS. 2016. Structural characterization of ribosome recruitment and translocation by type IV IRES. eLife 5: e13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda JS, Lorsch JR. 2014. Labeling of a protein with fluorophores using maleimide derivitization. Methods Enzymol 536: 79–86. [DOI] [PubMed] [Google Scholar]
- Noller HF, Lancaster L, Mohan S, Zhou J. 2017a. Ribosome structural dynamics in translocation: Yet another functional role for ribosomal RNA. Q Rev Biophys 50: e12. [DOI] [PubMed] [Google Scholar]
- Noller HF, Lancaster L, Zhou J, Mohan S. 2017b. The ribosome moves: RNA mechanics and translocation. Nat Struct Mol Biol 24: 1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M. 2015. Interactions of release factor RF3 with the translation machinery. Mol Genet Genomics 290: 1335–1344. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V. 2005. Structural insights into translational fidelity. Annu Rev Biochem 74: 129–177. [DOI] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV. 1998. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J 17: 7490–7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. 2007. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26: 41–50. [DOI] [PubMed] [Google Scholar]
- Pavlov MY, Ehrenberg M. 1996. Rate of translation of natural mRNAs in an optimized in vitro system. Arch Biochem Biophys 328: 9–16. [DOI] [PubMed] [Google Scholar]
- Pavlov MY, Freistroffer DV, MacDougall J, Buckingham RH, Ehrenberg M. 1997. Fast recycling of Escherichia coli ribosomes requires both ribosome recycling factor (RRF) and release factor RF3. EMBO J 16: 4134–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S. 1984. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J 3: 2895–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez CE, Gonzalez RL Jr. 2011. In vitro and in vivo single-molecule fluorescence imaging of ribosome-catalyzed protein synthesis. Curr Opin Chem Biol 15: 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F, Rodnina MV, Wintermeyer W. 2005. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol Cell 18: 403–412. [DOI] [PubMed] [Google Scholar]
- Peske F, Kuhlenkoetter S, Rodnina MV, Wintermeyer W. 2014. Timing of GTP binding and hydrolysis by translation termination factor RF3. Nucleic Acids Res 42: 1812–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov A, Puglisi JD. 2010. Site-specific labeling of Saccharomyces cerevisiae ribosomes for single-molecule manipulations. Nucleic Acids Res 38: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov A, Chen J, O'Leary S, Tsai A, Puglisi JD. 2012. Single-molecule analysis of translational dynamics. Cold Spring Harb Perspect Biol 4: a011551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov A, Grosely R, Chen J, O'Leary SE, Puglisi JD. 2016. Multiple parallel pathways of translation initiation on the CrPV IRES. Mol Cell 62: 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S, Weixlbaumer A, Ramakrishnan V. 2008. The termination of translation. Curr Opin Struct Biol 18: 70–77. [DOI] [PubMed] [Google Scholar]
- Prabhakar A, Capece MC, Petrov A, Choi J, Puglisi JD. 2017a. Post-termination ribosome intermediate acts as the gateway to ribosome recycling. Cell Rep 20: 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar A, Choi J, Wang J, Petrov A, Puglisi JD. 2017b. Dynamic basis of fidelity and speed in translation: Coordinated multistep mechanisms of elongation and termination. Protein Sci 26: 1352–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Rodnina MV. 2018. Translation in prokaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Fricke R, Wintermeyer W. 1994. Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry 33: 12267–12275. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Fischer N, Maracci C, Stark H. 2017. Ribosome dynamics during decoding. Philos Trans R Soc Lond B Biol Sci 372: 20160182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, He C. 2017. Dynamic RNA modifications in gene expression regulation. Cell 169: 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, et al. 2000. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell 102: 615–623. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Ramakrishnan V. 2009. What recent ribosome structures have revealed about the mechanism of translation. Nature 461: 1234–1242. [DOI] [PubMed] [Google Scholar]
- Selvin PR. 2000. The renaissance of fluorescence resonance energy transfer. Nat Struct Biol 7: 730–734. [DOI] [PubMed] [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR. 2011. Assembly of bacterial ribosomes. Annu Rev Biochem 80: 501–526. [DOI] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. 2011. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci 108: E1392–E1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomatin S, Herschlag D. 2009. Methods of site-specific: Labeling of RNA with fluorescent dyes. Methods Enzymol 469: 47–68. [DOI] [PubMed] [Google Scholar]
- Sternberg SH, Fei J, Prywes N, McGrath KA, Gonzalez RL Jr. 2009. Translation factors direct intrinsic ribosome dynamics during translation termination and ribosome recycling. Nat Struct Mol Biol 16: 861–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer SM, Feinberg JS, Joseph S. 2003. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol 327: 369–381. [DOI] [PubMed] [Google Scholar]
- Taylor D, Unbehaun A, Li W, Das S, Lei J, Liao HY, Grassucci RA, Pestova TV, Frank J. 2012. Cryo-EM structure of the mammalian eukaryotic release factor eRF1-eRF3-associated termination complex. Proc Natl Acad Sci 109: 18413–18418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thommen M, Holtkamp W, Rodnina MV. 2017. Co-translational protein folding: Progress and methods. Curr Opin Struct Biol 42: 83–89. [DOI] [PubMed] [Google Scholar]
- Tsai A, Petrov A, Marshall RA, Korlach J, Uemura S, Puglisi JD. 2012. Heterogeneous pathways and timing of factor departure during translation initiation. Nature 487: 390–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD. 2010. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature 464: 1012–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. 2003. Locking and unlocking of ribosomal motions. Cell 114: 123–134. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. 2010. The mechanism for activation of GTP hydrolysis on the ribosome. Science 330: 835–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, Bustamante C, Tinoco I. 2008. Following translation by single ribosomes one codon at a time. Nature 452: 598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339. [DOI] [PubMed] [Google Scholar]
- Wintermeyer W, Peske F, Beringer M, Gromadski KB, Savelsbergh A, Rodnina MV. 2004. Mechanisms of elongation on the ribosome: Dynamics of a macromolecular machine. Biochem Soc Trans 32: 733–737. [DOI] [PubMed] [Google Scholar]
- Yin J, Lin AJ, Golan DE, Walsh CT. 2006. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat Protoc 1: 280–285. [DOI] [PubMed] [Google Scholar]
- Young R, Bremer H. 1976. Polypeptide-chain-elongation rate in Escherichia coli B/r as a function of growth rate. Biochem J 160: 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavialov AV, Buckingham RH, Ehrenberg M. 2001. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell 107: 115–124. [DOI] [PubMed] [Google Scholar]