Abstract
Muscle-enriched lamin-interacting protein (Mlip) is an alternatively spliced gene whose splicing specificity is dictated by tissue type. MLIP is most abundantly expressed in brain, cardiac, and skeletal muscle. In the present study, we systematically mapped the transcriptional start and stop sites of murine Mlip. Rapid amplification of cDNA ends (RACE) of Mlip transcripts from the brain, heart, and skeletal muscle revealed two transcriptional start sites (TSSs), exon 1a and exon 1b, and only one transcriptional termination site. RT-PCR analysis of the usage of the two identified TSSs revealed that the heart utilizes only exon 1a for MLIP expression, whereas the brain exclusively uses exon 1b TSS. Loss of Mlip exon 1a in mice resulted in a 7-fold increase in the prevalence of centralized nuclei in muscle fibers with the Mlip exon1a–deficient satellite cells on single fibers exhibiting a significant delay in commitment to a MYOD-positive phenotype. Furthermore, we demonstrate that the A-type lamin-binding domain in MLIP is encoded in exon 1a, indicating that MLIP isoforms generated with exon 1b TSS lack the A-type lamin-binding domain. Collectively these findings suggest that Mlip tissue-specific expression and alternative splicing play a critical role in determining MLIP's functions in mice.
Keywords: alternative splicing, brain, heart, gene expression, intracellular trafficking, protein-protein interaction, skeletal muscle, cellular localization, MLIP, transcriptional start site, muscle-enriched lamin-interacting protein, transcriptional regulation, splice variant, tissue-specific expression, protein isoform
Introduction
Muscle-enriched A-type lamin-interacting protein (Mlip)3 is encoded by a unique gene that is conserved among amniotes. Mlip was initially discovered through its interaction with A-type lamins (1) and with transcription factor ISL1 (2). The Mlip gene is most abundantly expressed in the heart and skeletal muscles with lower expression levels of Mlip also detected in other tissues (e.g. liver and brain) in both mouse and human (1, 3). Mlip is expressed in both the cytosol and nucleus of cells and colocalizes with A-type lamin and promyelocytic leukemia bodies in the nucleus (1). Despite recent studies that demonstrated that Mlip plays a crucial role in cardiac homeostasis and adaptation in response to stress (3–5), the exact molecular functions of MLIP remain unknown.
The investigation of MLIP's molecular function is complicated by the complex nature of the proteins: Mlip mRNA is alternatively spliced and gives rise to at least seven different mature mRNA and protein isoforms in the heart (1, 3). The splicing patterns and expression profile have not yet been characterized in the other tissues, but the pattern of MLIP isoform expression appears to be dependent on tissue type.
The aim of the present study was to investigate and define the expression of the Mlip gene and to characterize the different alternatively spliced forms in individual tissues. We report in this study the identification of a previously unknown second transcriptional start site for Mlip that is utilized primarily in the brain. Moreover, we demonstrate that cellular localization and functionality are dictated by alternative expression and splicing of MLIP isoforms.
Results
Tissue-specific alternative start site
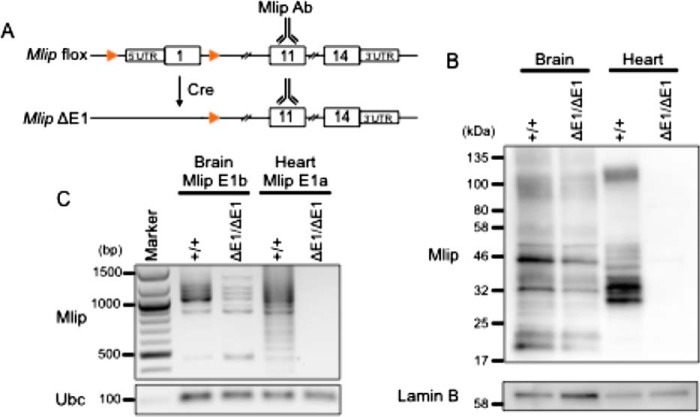
In a previous study, we described a Mlip knockout mouse model (3). This mouse model was genetically engineered using a recombinant strategy to delete the region encompassing the first exon (mCh9: 77,347,695–77,347,870) of the Mlip gene and its putative upstream promoter (Fig. 1A and Ref. 3). This deletion was sufficient to abolish the expression of MLIP in the heart. However, an MLIP protein, as detected by MLIP-specific antibodies raised against an epitope encoded within exon 11 (1), was still observed in the brain of these mice (referred to as ΔE1/ΔE1; Fig. 1B), suggesting the existence of at least one alternative transcription start site for the Mlip gene.
Figure 1.
MLIP expression in the heart and brain of Mlip+/+ and MlipΔE1/ΔE1 mice. A, generation of Mlip knockout mouse model (Mlip ΔE1) by deletion of exon 1 (E1) of the Mlip gene and its proximal promoter. Orange arrows indicate flox sites. Mlip Ab, polyclonal antibody targeting an epitope in exon 11 of the gene. B, Western blot with MLIP antibody of Mlip+/+ and MlipΔE1/ΔE1 brain and heart. The polyclonal MLIP antibodies are specific for an epitope encoded within exon 11. Lamin B was used as a loading control. C, PCR amplification of Mlip cDNA from brain and heart of Mlip+/+ and MlipΔE1/ΔE1 mice using forward primers specific for Mlip exon 1b or exon 1a, respectively, and exon 14 reverse primer. Ubc was used as a loading control.
Detailed analysis of the Mlip gene sequence and the interrogation of the Ensembl databases revealed the existence of another in-frame putative exon encompassing a start codon located in intron 1 of the Mlip gene. This putative alternative exon was referred to as exon 1b (mCh9: 77,251,747–77,251,841), exon 1a being the original first exon deleted in the MlipΔE1/ΔE1 mouse. This putative exon 1b had been computationally inferred and is supported at the exon level by Human and Vertebrate Analysis and Annotation (HAVANA) project data. Experimentally, however, neither mRNA nor EST sequencing has ever shown the actual usage of exon 1b in mRNA.
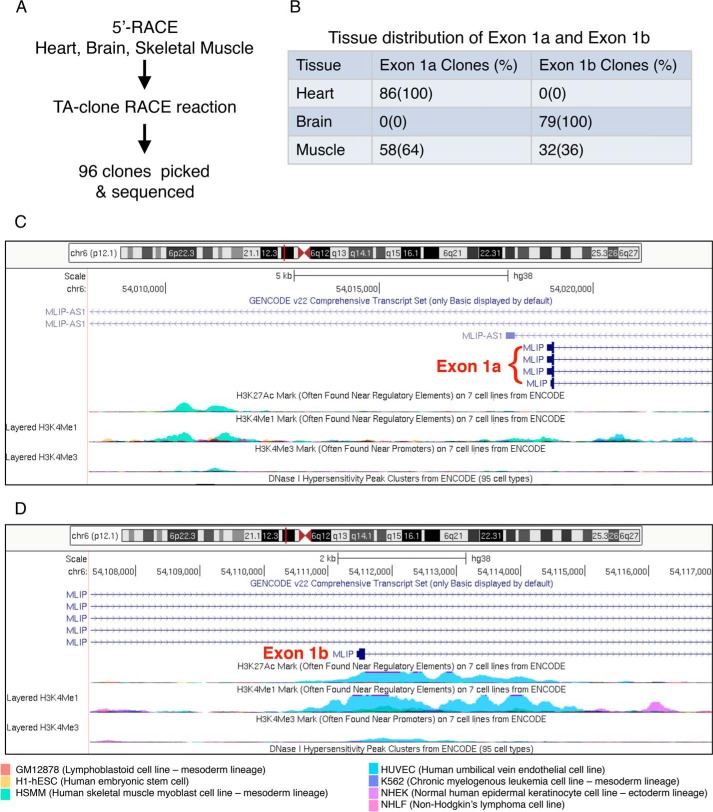
To confirm the existence of this alternative transcriptional start site in exon 1b, we performed 5′ rapid amplification of cDNA ends (RACE) on mRNA extracted from Mlip+/+ brain, skeletal muscle, and heart tissues (Fig. 2A) using primers anchoring in exon 2 or 3 of the Mlip gene. Sequencing of the clones generated revealed a 65-nucleotide sequence upstream of the start codon of exon 1b in the brain, corresponding to the 5′-untranslated region (UTR) of this Mlip mRNA (mCh9: 77,251,791–77,251,855). However, no positive clones were generated with mRNA extracted from Mlip+/+ heart tissue, suggesting that exon 1b was not expressed in the heart. Both exon 1a and exon 1b were observed in skeletal muscle (Fig. 2B). These findings were in accordance with the absence of MLIP expression in the MlipΔE1/ΔE1 heart and the detection of MLIP expression in the brain of MlipΔE1/ΔE1 mouse by Western blotting and PCR (Fig. 1, B and C).
Figure 2.
Identification of two Mlip alternative transcriptional start sites. A, 5′-RACE reactions targeting Mlip were performed on mRNA extracted from adult mouse heart, brain, and skeletal muscle and subsequently identified by DNA sequencing. B, tissue distribution of exon 1a and exon 1b identified from 96 clones from heart, brain, and skeletal muscle. C and D, display of UCSC Genome Browser (ENCODE project data) showing histone H3 epigenetic marks (H3K27Ac, H3K4Me1, and H3K4Me3; the height of the peak correlates with the strength of signal in a given cell line) centered on exon 1a (C) and exon 1b (D) of the Mlip gene in different cell lines.
The structure of the Mlip gene is conserved between mouse and human genomes in terms of number, size, and sequence of the 14 exons (Table 1). To further characterize exon 1b and its upstream 5′ region, we interrogated the Encyclopedia of DNA Elements (ENCODE) project, which provides informative data on epigenetic marks associated with DNA structure and accessibility of the human genome. These data revealed an enrichment of histone H3 epigenetic marks clustered between ∼1000 bp upstream and ∼3000 bp downstream of the start site in exon1b, especially acetylated lysine 27 (H3K27Ac) and monomethylated and trimethylated lysine 4 (H3K4Me1 and H3K4Me3, respectively). These marks were associated with clusters of DNase I hypersensitivity regions (Fig. 2C). Similar enrichment of histone H3 epigenetic marks was also found in the upstream region of exon 1a (Fig. 2D). Regulatory regions in general and promoters in particular have been shown to exhibit high sensitivity to DNA nucleases (6). Also, specific histone modifications such as H3K27Ac and H3K4Me1 are also associated with transcription factor–binding regions (i.e. enhancers and promoters). More importantly, H3K4Me3 is a predominant feature of active promoters (6). Therefore, in combination with the histone H3 modifications, the DNase I–sensitive pattern nearby the start site of exon 1b strongly suggests that this genomic region constitutes an alternative promoter or enhancer, driving the expression of the Mlip gene through the usage of exon 1b in a tissue-specific manner (brain and skeletal muscle).
Table 1.
Human and mouse MLIP gene structure
| Human position | Human | Mouse position | Mouse | |
|---|---|---|---|---|
| bp | bp | |||
| Exon 1a | 54,018,916–54,019,091 | 176 | 77,347,695–77,347,870 | 176 |
| Intron 1 | 92,357 | 96,125 | ||
| Exon 1b | 54,111,448–54,111,575 | 128 | 77,251,747–77,251,841 | 95 |
| Intron 2 | 9,871 | 8,019 | ||
| Exon 2 | 54,121,447–54,121,602 | 156 | 77,243,572–77,243,727 | 156 |
| Intron 3 | 2,870 | 3,787 | ||
| Exon 3 | 54,124,473–54,124,865 | 393 | 77,239,392–77,239,784 | 393 |
| Intron 4 | 11,849 | 22,343 | ||
| Exon 4 | 54,136,715–54,138,286 | 1572 | 77,231,029–77,229,485 | 1545 |
| Intron 5 | 10,769 | 16,510 | ||
| Exon 5 | 54,149,056–54,149,127 | 72 | 77,216,977–77,217,048 | 72 |
| Intron 6 | 11,239 | 26,239 | ||
| Exon 6 | 54,160,367–54,160,432 | 66 | 77,190,672–77,190,737 | 66 |
| Intron 7 | 83 | 81 | ||
| Exon 7 | 54,160,516–54,160,599 | 84 | 77,190,590–77,190,507 | 84 |
| Intron 8 | 140 | 123 | ||
| Exon 8 | 54,160,740–54,160,799 | 60 | 77,190,324–77,190,383 | 60 |
| Intron 9 | 8,728 | 8,927 | ||
| Exon 9 | 54,169,528–54,169,572 | 45 | 77,181,352–77,181,396 | 45 |
| Intron 10 | 20,297 | 7,348 | ||
| Exon 10 | 54,189,870–54,189,914 | 45 | 77,173,959–77,174,003 | 45 |
| Intron 11 | 12,190 | 9,065 | ||
| Exon 11 | 54,202,105–54,202,233 | 129 | 77,164,760–77,164,893 | 134 |
| Intron 12 | 28,480 | 26,192 | ||
| Exon 12 | 54,230,714–54,230,917 | 204 | 77,138,702–77,138,348 | 353 |
| Intron 13 | 26,390 | 25,298 | ||
| Exon13 | 54,257,308–54,257,361 | 54 | 77,113,048–77,112,974 | 75 |
| Intron 14 | 8,588 | 10,579 | ||
| Exon 14 | 54,265,950–54,266,043 | 94 | 77,102,393–77,102,081 | 313 |
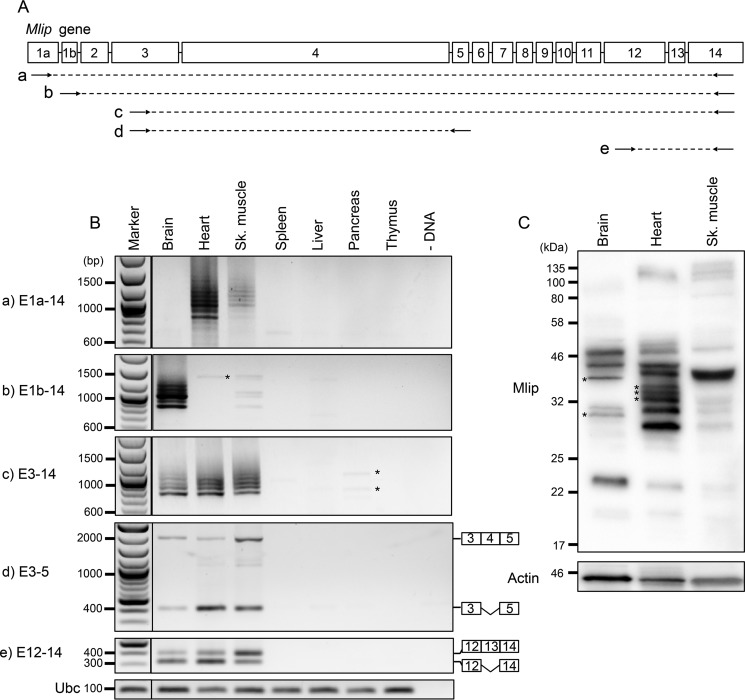
Based on these results, we hypothesized that the usage of these alternative first exons could be tissue-specific. To test this hypothesis, we performed PCR amplification of Mlip cDNA on different tissues of Mlip+/+ mice using forward primers specifically targeting these two exons (1a and 1b) and a reverse primer anchoring in exon 14 of the Mlip gene (Fig. 3A). All the PCR products were cloned and sequenced to verify the specificity of the products and determine the composition of the different splicing isoforms generated. The results indicated that the Mlip gene was only expressed in the brain, heart, and hind limb skeletal muscle tissues (Fig. 3B). The different combinations of primers revealed that the usage of exon 1a was restricted to the heart and skeletal muscle (Fig. 3B, panel a), whereas the expression of Mlip gene in the brain only occurred through the usage of exon 1b (Fig. 3B, panel b). Both exon 1a and exon 1b were detected in the skeletal muscle isolated from the gastrocnemius and soleus with exon 1a being preferentially used in these tissues. The sequencing results also confirmed that exons 1a and 1b were mutually exclusive (Fig. 2B), indicating that exon 1b was indeed an alternative start exon.
Figure 3.
MLIP tissue expression profile in Mlip+/+ mice. A and B, PCR amplification of Mlip cDNA on tissues of Mlip+/+ mice using different combinations of primers (arrows; A, a–e) targeting exons of the Mlip gene. *, nonspecific amplification. Ubc was used as a loading control. C, Western blot with MLIP antibody of MLIP+/+ brain, heart, and skeletal (Sk.) muscle. *, tissue-specific isoforms. Actin was used as a loading control.
The compilation of data retrieved from EST data sets suggested the existence of one Mlip isoform using a third alternative start site in exon 3, the 5′-UTR encompassing the sequence of exon 2, and a portion of intron 1 (mCh9: 77,340,416–77,340,627). To investigate whether this third start site drove Mlip expression in tissues in which exons 1a and 1b were not used (spleen, liver, pancreas, and thymus), we performed PCR amplification of Mlip cDNA on the different tissues of Mlip+/+ mice using primers anchoring in exon 3 and in exon 14 of the Mlip gene. The results of the PCR and sequencing of the PCR products confirmed that Mlip gene was not expressed in the spleen, liver, pancreas, and thymus. The faint bands visible on Fig. 3B were nonspecific artifacts of the PCR and did not match the Mlip gene sequence (Fig. 3B, panel c).
Tissue-specific splicing patterns
Beyond the identification of an alternative transcriptional start site (exon 1b), the systematic sequencing of all the Mlip PCR products revealed specific splicing patterns. The Ensembl database (data extracted from the UCSC Genome Browser, NCBI, and HAVANA project) described 15 different Mlip splice variants. Among them, 12 isoforms were predicted to be translated with 5′- and 3′-UTRs identified, and seven isoforms corresponded to complete mRNA sequences with both 5′-UTR and 3′-UTR. However, only three splice variants had a transcript (or full EST) supporting this observation, all of which included exon 1a. The other nine splice variants remained putative mRNA isoforms. The isoforms comprising exon 1b have been described in three putative splicing isoforms, two full sequences, and one partial sequence but were not supported by EST or mRNA data sets. The splicing pattern of the seven complete isoforms is predicted to be translated and to include exons 3, 9, 10, and 11. Our sequencing data also showed recurrent inclusion for these exons. The block of exons 9–11 was observed in more than 75% of all the sequences retrieved by PCR in the brain, heart, and skeletal muscle and was found in all the full-length isoforms in the heart and brain. Exon 3 represented the second most included exon after the exon 9–11 block and was found in the brain, heart, and skeletal muscle isoforms. Noteworthy, exon 3 was included in all the isoforms in which exon 2 was retained, suggesting that the inclusion of exon 2 conditioned the retention of exon 3.
Exons 4, 12, and 13 described in the Ensembl database were not found in our sequencing data and could be due to lower abundance of these Mlip isoforms in which these exons were retained. To confirm the existence of these exons, we performed PCR amplification of Mlip cDNA using different combinations of primers (Fig. 3A, panels d and e). The results indicated that exons 4, 12, and 13 were transcribed in the brain, heart, and skeletal muscle (Fig. 3B, panels d and e) and could be either retained or spliced out. The retention of exon 4 appeared to be higher in the brain and skeletal muscle compared with the heart (ratio of included versus excluded: 1.1, 0.1, and 0.9 in the brain, heart, and skeletal muscle, respectively). The inclusion of exon 13 was also predominant in the skeletal muscle when compared with the other two tissues (ratio of included versus excluded: 0.4, 0.5, and 1.8 in the brain, heart, and skeletal muscle, respectively).
Despite a lower expression level of Mlip in skeletal muscle, a similar splicing pattern was observed when exon 1a was used to drive the expression of Mlip (Fig. 3B, panel a). Similarly, the splicing profile was identical in the brain, heart, and skeletal muscle for the PCR products obtained using primers sitting in exons 3 and 14 (Fig. 3B, panel c), suggesting that the splicing of the distal portion of Mlip (downstream of exon 3) was the same in these three tissues. The difference in the size of the bands (∼50–100 bp) suggested that one or two short exons were spliced out in the different isoforms (exons 5–8 and exon 13).
In accordance with the observed mRNA splicing pattern, the Mlip protein isoforms detected by Western blotting were comparable in the heart and skeletal muscle (Fig. 3C). However, we identified four isoforms that were specifically expressed in the brain or the heart: two isoforms (∼38.6 and ∼30.4 kDa) were restricted to the brain, and two distinct isoforms (∼35.5 and ∼33.9 kDa) were confined to the heart (Fig. 3C, *), suggesting tissue-specific functions associated with these isoforms.
Using the protein structure prediction algorithm RaptorX, we investigated whether the splicing of Mlip exons would affect the three-dimension structure of the MLIP protein. The overall structure of the MLIP amino acid sequence of all the different isoforms (deletion of one exon at a time) was predicted to be disordered (78–87% of the residues depending on the isoforms) as reported previously (1). Approximately 97–99% of the amino acid chain is predicted to form loops without specific secondary structure. Only a portion of the amino acid sequence encoded by exon 4 of the Mlip gene was predicted to fold in an organized structure (β-sheets). This structured domain was modeled on the crystal structure of the secreted proline-rich antigen MTC28 from Mycobacterium tuberculosis (p value, 5.92E−22) based on sequence similarity.
The in silico modeling of the different Mlip spliced forms did not provide critical information regarding Mlip structure and function; therefore, we next sought to elucidate the impact of the alternative splicing upon Mlip isoform localization. Due to the absence of isoform-specific antibodies, we chose to address this question through the expression of synthetic Mlip isoforms in vitro and cellular subfractionation of heart samples.
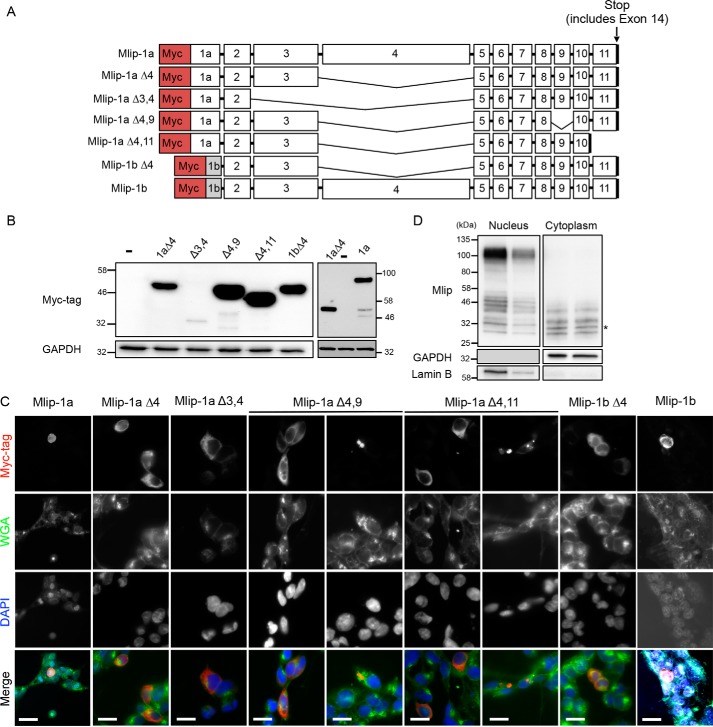
Because of the high frequency of retention of exon 3 and the exon 9–11 block in the different Mlip splice variants, we hypothesized that these exons could impact MLIP's function, stability, and localization in cells. To test this hypothesis, we expressed synthetic human Myc-tagged MLIP “full length” including exon 1a (MLIP-1a) or exon 1b (MLIP-1b), MLIP including exon 1b (MLIP-1bΔ4), and different MLIP splice variants deleted of exon 3, exon 9, or exon 11 (Fig. 4A) into 293 cells. Exon 14 contains a stop codon and 3′-UTR, and therefore the stop codon of the synthetic MLIP cDNAs represents exon 14 without the 3′-UTR. The distal exons of the MLIP gene (exons 12 and 13) were not included in these synthetic isoforms as they were not observed in 3′-RACE–derived clones from heart, brain, or skeletal muscle. Exons 12 and 13 were only detected by RT-PCR when one of the DNA primers targets exon 12 (Fig. 3B, panel e). Despite a lower expression level of MLIP-Δ3,4 (Fig. 4B), the localization of the different synthetic MLIP isoforms was similar for the different isoforms and mainly cytosolic in 293 cells (Fig. 4C). The inclusion of exon 4, an exon that harbors a nuclear localization sequence (NLS) (1), resulted in nuclear localization of MLIP (Fig. 4C). In cells transfected with MLIP-Δ4,9 and MLIP-Δ4,11, the recombinant protein was found in cytosolic aggregate-like structures, evocative of lysosomes and suggesting that both exons 9 and 11 could be essential for proper folding of MLIP proteins.
Figure 4.
Cellular localization of MLIP isoforms. A, schematic representation of the different synthetic human Myc-tagged Mlip isoforms used to transfect 293 cells. B, Western blot with Myc tag antibody. GAPDH was used as a loading control. C, immunostaining of 293 cells with Myc tag antibody (red), wheat germ agglutinin (WGA; green), and 4′,6-diamidino-2-phenylindole (DAPI; blue). Scale bars, 20 μm. D, Western blot with MLIP antibody of nuclear and cytoplasmic fractions isolated from MLIP+/+ heart. GAPDH and lamin B were used as cellular fractionation controls. *, cytoplasmic-specific isoform.
To further investigate the cellular distribution of the different endogenous isoforms of MLIP, we performed subcellular fractionation of mouse heart samples followed by Western blot analysis. The results presented in Fig. 4D show subtle differences in localization of the different isoforms between nucleus and cytoplasm. Two of the four isoforms detected in the cytoplasmic fraction were not found in the nuclear fraction (∼30 and ∼33 kDa, respectively). The calculated size indicated an ∼2-kDa difference between these cytoplasmic isoforms and the closest nuclear isoforms (i.e. ∼Δ60 bp, the size of the smallest exons of the Mlip gene), suggesting that the splicing could influence the subcellular localization of MLIP. Although we cannot exclude that these differences could be due to post-translational modifications of MLIP, the MLIP isoforms detected by Western blotting match the PCR products obtained previously (Fig. 3B). The highest molecular mass isoform (∼100 kDa) correspond to MLIP isoforms in which exon 4 was retained.
Increased prevalence of centralized nuclei in skeletal muscle of mice lacking MLIP exon 1a
Previous studies reported that the loss of MLIP from mouse hearts results in a baseline hyperactivation of the Akt and mTOR pathways. These mice are susceptible to insult, such as in a pressure overload model, in which the MLIP-deficient hearts have a reduced capacity for beneficial remodeling, resulting in the early onset of heart failure(3). To unravel the potential biological role of MLIP in skeletal muscle, we have begun to characterize the skeletal muscle phenotype of MLIP exon 1a–deleted mice. A reduction, but not a complete loss, in MLIP protein expression was observed in the skeletal muscle of these MLIP-deficient mice (Fig. S1). These hypomorphs appear to be normal. However, a striking feature in the skeletal muscle of these MLIP exon 1a–null mice was a large number (23.2 versus 3.3% in control muscle) of fibers that have centralized nuclei in the hind leg muscle indicative of ongoing muscle repair that is a classic signature in the mdx mouse model (Fig. S1B). Furthermore, we observed a significant delay in satellite cell commitment to MYOD-positive cells at 72 h in the fibers isolated from MLIP exon 1a–null mice (Fig. S1C and Table S1).
Interaction with lamin A/C is exon 1a–specific
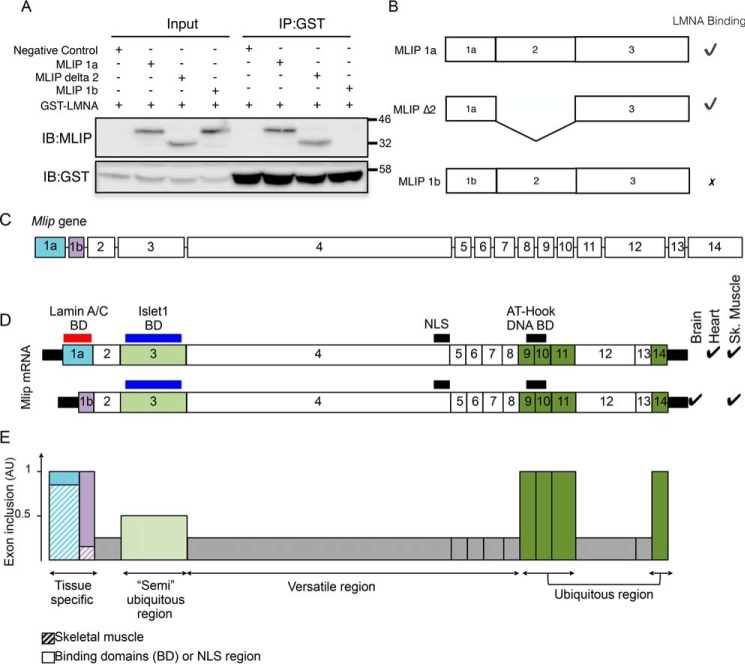
Using a yeast two-hybrid interaction screen, MLIP was initially discovered through its interaction with A-type lamin with six independent MLIP clones identified, all containing exon 1a and exon 2 (1). To determine whether MLIP's interaction with A-type lamin depends on exon 1a of MLIP, we generated and purified recombinant MLIP proteins fused to hexahistidine from bacteria with either exon 1a or 1b present (Fig. 5A). These proteins were incubated with purified GST-LMNA in various combinations (Fig. 5A). Complexes were precipitated with GST-Sepharose beads and resolved by SDS-PAGE. Western blot analysis with MLIP or GST antibodies revealed that A-type lamin–binding domain resides within exon 1a of MLIP and that MLIP 1b isoform does not interact with A-type lamin (Fig. 5, A and B).
Figure 5.
Structure, expression and splicing of the Mlip gene. A, Mlip exon 1a encodes an A-type lamin interaction domain. Purified recombinant MLIP isoforms and lamin A fusion proteins were incubated together. Complexes were immunoprecipitated and resolved by SDS-PAGE. B, schematic summary of MLIP–LMNA interaction assay. C, structure of the Mlip gene. The size of the boxes, corresponding to the exons, is proportional to the exon's size in bp. The lines (representing the introns) are not representative of the actual size of the introns in the genome. D, using alternative start sites located in exon 1a or exon1b, the gene gives rise to different mRNA expressed in a tissue-specific manner. ✓, tissue in which the given mRNA is expressed; BD, known binding domain. E, inclusion of the exons in the mRNA based on sequencing data. The gray boxes represent the exons largely spliced out (versatile regions), exon 3 (light green box) is included in about 50% of all the mRNA (“semi”-ubiquitous region), and exons 9–11 and exon 14 (dark green) constitute a ubiquitous region of the gene.
Discussion
In the present report we show that (i) the Mlip gene is exclusively expressed in the brain, heart, and skeletal muscle; (ii) its expression in these tissues is regulated by a tissue-specific transcriptional start site with exon 1a restricted to muscle (heart and skeletal muscle) and exon 1b used in the brain and skeletal muscle; (iii) the genomic DNA sequences upstream of both exons 1a and 1b are decorated with epigenetic marks suggesting active promoter regions; and (iv) the alternative splicing patterns of Mlip mRNA define versatile and stable regions of the protein likely to modulate MLIP's interactions and functions in the different tissues. (v) Loss of Mlip exon 1a in mice results in a 7-fold increase in the prevalence of centralized nuclei in muscle fibers with the MLIP exon1a–deficient satellite cells presenting with a significant delay in commitment to a MYOD-positive phenotype. Finally, (vi) MLIP 1a–containing isoforms interact with A-type lamin, whereas the brain-specific MLIP 1b isoforms do not interact with A-type lamin.
The use of differential transcription start sites (TSS) and alternative splicing is a strategy commonly used in higher eukaryotes to increase protein complexity without expanding the genome's size (for a review, see Ref. 7). It has been estimated that there are, on average, four TSSs per gene in the human genome (8, 9). A number of transcriptome-wide studies have also shown that the use of alternative TSS is highly tissue-specific (8, 10, 11).
Within the Mlip gene, we demonstrated the existence of two TSSs located in distinct, mutually exclusive exons. The usage of these TSSs proved to be tissue-specific (i.e. a TSS in exon 1a in the heart and a TSS in exon 1b in the brain). In skeletal muscle, the expression of Mlip is driven by both TSSs (1a and 1b), and whether the expression of the brain-specific exon 1b occurs primarily in neuromuscular plates (innervation) remains to be determined. There is no sequence homology between exons 1a and 1b or with another region of the genome (in both human and mouse genomes), which suggests that exons 1a and 1b did not originate from a duplication of one another or other specific sequences in the genome (transposition). The usage of exon 1a in the heart and exon 1b in the brain strongly suggests specific function(s) provided to the MLIP isoforms by exon 1a or 1b in these two tissues.
It has been estimated that about 95% of mammalian genes use alternative splicing as a regulatory mechanism (12). In humans, the overall ratio of transcripts over protein-coding genes revealed an average of four transcripts per gene with up to 25 different transcripts arising from a single gene through alternative splicing (13). Alternative splicing is involved in the regulation of a myriad of cellular and tissue processes (cell proliferation, differentiation, apoptosis, etc.) by allowing changes in cellular localization, modulation of functional or regulatory domains modifying protein–protein binding, and enzymatic properties among others (for a review, see Ref. 14).
MLIP's protein structure is predicted to be disordered, but the complex arrangement of the Mlip exons through alternative splicing is likely to generate different binding domains and functions for the various MLIP isoforms (Fig. 5, C–E). For instance, exon 3, which encompasses the Islet1-binding domain, is spliced out in some isoforms, suggesting a way to modulate MLIP's interaction with Islet1 and possibly Islet1's function through MLIP's splicing. Likewise, A-type lamin–binding domain of MLIP resides within exon 1a, indicating that MLIP interacts with A-type lamin solely in the heart and skeletal muscle but not in the brain (which expresses MLIP 1b isoform only) (Fig. 5, D and E).
Also, the high retention of particular exons suggests that these exons or regions encode amino acid sequences necessary for MLIP's structure and function (Fig. 5E). In particular, the systematic inclusion of the exons that constitute the distal portion of the gene (exons 9–11) together with the ability to form aggregate-like structures when exons 9 and 11 are deleted from MLIP isoforms suggests that this region might be essential for MLIP's proper folding through inhibition of aggregation of otherwise disordered protein structure. This region also encodes the putative AT-hook DNA-binding domain of MLIP (2), which is found in all MLIP isoforms (Fig. 5E). However, the putative NLS sequence, encoded by the largest exon (exon 4), is spliced out in many isoforms of MLIP. Therefore, even if the putative AT-hook DNA-binding domain of MLIP is harbored by all MLIP isoforms (exons 9–11), the absence of NLS due to splicing of exon 4 would, in theory, prevent or limit MLIP localization in the nucleus (as seen in Fig. 4, C and D), thereby abolishing its potential interaction with DNA. Noteworthy, however, a large number of small MLIP isoforms were also detected in the nuclei-enriched fraction, which suggests that MLIP could be imported into the nucleus independently of the NLS sequence. Whether this process involves the formation of heterodimers between different MLIP isoforms or other mechanisms remain to be elucidated.
The similar isoform patterns found in the heart and skeletal muscle suggest that MLIP plays comparable functions in these two types of muscles. Importantly, however, a number of MLIP isoforms were specifically detected in the heart (Fig. 3C), suggesting that some MLIP isoforms might also have tissue-specific functions. We recently reported (3) that MLIP was involved in cardiac adaptation to stress via proper integration of Akt/mTOR signaling pathways. MLIP's regulation of Akt/mTOR activity is cardiac-specific and does not occur in skeletal muscle. The exact nature of MLIP's interaction (physical and functional) with the Akt/mTOR pathways in the heart remains unknown, but one may hypothesize that it is carried out by these cardiac-specific isoforms of MLIP. Conversely, the two isoforms explicitly expressed in the brain (Fig. 2C) as well as the exclusive use of exon 1b evokes an original role of MLIP in this tissue.
We made several confounding observations during our extensive phenotyping of the Mlip gene–targeted model that may originate in part from the alternative promotors associated with exon 1a and exon 1b. Furthermore, we have demonstrated that the usage of these two start sites for Mlip is tissue-specific in that the heart exclusively uses the 1a start site, whereas the brain only uses the 1b start site. However, in skeletal muscle, both start sites (1a and 1b) were found to be in use.
In conclusion, MLIP's molecular function remains to be elucidated. However, our results show a highly complex expression of the Mlip gene that leads to up to 10 different protein isoforms and suggests tissue-specific roles.
Materials and methods
Animals
All the mice were studied according to protocols approved by the Canadian Council on Animal Care's Guide to the Care and Use of Experimental Animals and the Animals for Research Act.
MlipEΔ1/EΔ1 knockout mice were generated and described previously (3). Briefly, Mlip exon 1 and putative proximal promoter were flanked by loxP sequences. Mice bearing this mutant Mlip allele (designated Mlipfl/+) were crossed with transgenic C57BL/6J CMV-Cre mice to generate MlipEΔ1/EΔ1 knockout mice (Fig. 1).
mRNA analysis and cloning of PCR products
Brain, heart, skeletal muscle (gastrocnemius), spleen, liver, pancreas, and thymus were dissected and snap frozen in liquid nitrogen. Total RNA extraction was performed with TRIzol as described previously (1). Total RNA was reverse transcribed using Transcriptor reverse transcriptase (Roche Diagnostic) and oligo(dT)15 as recommended by the manufacturer. PCRs were carried out with standard Taq DNA polymerase (New England Biolabs) following the manufacturer's instructions. The sequences of oligonucleotides used are listed in Table S1.
PCR products were cloned into pGEM®-T vector (Promega) and transformed into NEB 10-β competent Escherichia coli as directed by the manufacturers. Positive clones were sequenced at the Centre for Applied Genomics, SickKids Hospital, Toronto, Ontario, Canada, and sequences were analyzed with UCSC Genome Browser.
5′ rapid amplification of cDNA ends
5′-RACE was performed on cDNA isolated from mouse brain, heart, skeletal muscle (gastrocnemius), liver, and pancreas as described in the 5′-RACE kit, second generation (Roche Applied Sciences), using primers specific to Mlip exons 1 and 3.
Protein analysis
For Western blot analyses, proteins were extracted from frozen total brain, heart, and skeletal muscle (gastrocnemius) as described previously (1). Proteins were resolved by SDS-PAGE. Membranes were hybridized with lamin B (Santa Cruz Biotechnology, sc-6216; 1:1000), actin, and MLIP-C antibodies (custom-made; 1:5000). Signals were developed using enhanced chemiluminescence reagent (SuperSignal West Femto, Thermo Scientific).
Cell culture
The mouse HL1 cell line was cultured at 37 °C in an atmosphere of 5% CO2 in Claycomb medium (Sigma) supplemented with 10% fetal bovine serum, 1% (v/v) penicillin-streptomycin, and 2 mm l-glutamine. The HEK293 cell line was cultured at 37 °C in an atmosphere of 5% CO2 in Dulbecco's modified Eagle's medium (Thermo Fisher) supplemented with 10% fetal bovine serum, 1% (v/v) penicillin-streptomycin, and 2 mm l-glutamine.
Whole-cell extract preparation for immunoprecipitation and Western blotting
Proteins were extracted using lysis buffer containing 50 mm Tris-HCl (pH 8.0), 200 mm NaCl, 20 mm NaF, 20 mm β-glycerol phosphate, 0.5% Nonidet P-40, 0.1 mm Na3Vo4, 1 mm DTT, 1× protease inhibitor mixture (Roche Applied Science; one tablet/7.0 ml), and phosphatase inhibitor mixtures (Sigma; 0.1 ml/7.0 ml). Cells were scraped from dishes, and lysates were pipetted into 1.5-ml centrifuge tubes. Lysates were incubated on ice for 20 min and cleared by centrifugation at 10,000 × g for 10 min at 4 °C. Supernatants were collected, and protein concentrations were determined using the Bradford protein assay (Bio-Rad). Proteins were resolved on a 15% 0.75-mm-thick SDS-Tris-glycine polyacrylamide gel (200 V) and transferred to polyvinylidene difluoride membranes (Millipore) for 1 h at 100 V at 4 °C. Membranes were blocked in 5% nonfat milk dissolved in Tris-buffered saline + 0.05% Tween 20. MLIP-mediated immunoprecipitations were performed as described previously (1).
For Western blot analysis, MLIP antibodies were raised in rabbits against a synthetic peptide, N-LRKDEEVYEPNPFSKYL-C (21st Century Biochemicals) (1). The following primary antibody concentrations were used: MLIP rabbit antibody at 1:50,000, myogenin mouse monoclonal IgG at 1:1,000 (Clone F5D, sc12732, Santa Cruz Biotechnology), anti-α-tubulin mouse monoclonal at 1:5,000 (T9026, Sigma, and MYOD mouse monoclonal IgG at 1:500 (Clone 5.8A, sc32758, Santa Cruz Biotechnology). Membranes were then incubated with horseradish peroxidase–conjugated anti-rabbit or anti-mouse IgG secondary antibody (Santa Cruz Biotechnology). Immunoblot signals were detected with a SuperSignal West Pico chemiluminescence kit (Thermo Scientific) and visualized on X-ray film (Sigma).
Single fiber isolation and immunocytochemistry
As previously described (15), single muscle fibers were isolated from the extensor digitorum longus muscle of 6–8-week-old control or MLIP exon 1a–null mice and cultured in floating conditions in fiber media (Dulbecco's modified Eagle's media, 15% (v/v) fetal bovine serum, and 2% (v/v) chick embryo extract).
Recombinant proteins and pulldown
Construction of the bacterial expression plasmids for MLIP proteins and lamin A and purification of the recombinant protein were carried out as described previously (1). Briefly, the different Mlip cDNAs were subcloned into the His tag fusion vector pET100D (Life Technologies), and lamin A (amino acids 1–230) was subcloned into the GST fusion vector pGEX-2T (GE Healthcare). Plasmids were transformed independently into the bacterial strain BL21 DE3 (pLysS) and induced with 250 μm isopropyl 1-thio-d-galactopyranoside for 2 h during exponential growth phase. Cells were lysed in 50 mm Tris-HCl (pH 7.5), 500 mm NaCl, and 0.1% Triton X-100, and the lysates were centrifuged at 100,000 × g for 45 min. The His6-MLIP supernatants were loaded onto a Ni2+-nitrilotriacetic acid column (GE Healthcare); washed with 50 mm Tris-HCl (pH 7.5), 500 mm NaCl, and 25 mm imidazole buffer; and eluted with a linear gradient from 25 to 250 mm imidazole in the presence of 50 mm Tris-HCl (pH 7.5) and 500 mm NaCl. The GST-lamin bacterial supernatants were run across a GSTrap 4B column (GE Healthcare) and washed, and the GST-lamin was eluted with 50 mm Tris-HCl and 20 mm reduced GSH, pH 8.0. The fractions containing the His6-tagged MLIP proteins or GST-lamin were then dialyzed against 10 mm phosphate buffer (pH 7.4), 50 mm NaCl, and 0.05% Triton X-100. The purity of each fusion protein was 95% as determined by Coomassie Blue–stained protein gels. Various combinations of His6-MLIP and GST-lamin recombinant proteins were mixed together and incubated at room temperature for 30 min in 10 mm phosphate buffer (pH 7.4), 50 mm NaCl, and 0.05% Triton X-100. Ni2+-nitrilotriacetic acid-Sepharose beads were added to each reaction. Complexes were washed in phosphate buffer (pH 7.4), 50 mm NaCl, and 0.05% Triton X-100; eluted by the addition of SDS-PAGE loading buffer; and resolved by SDS-PAGE. Western blot analysis was performed using anti-GST (Cell Signaling Technology), anti-Myc tag (Cell Signaling Technology), and anti-MLIP polyclonal antibodies. A 1:10 dilution of the total starting material was run on the same gel. The assay was repeated two additional times with similar results.
Author contributions
M.-E. C. and P. G. B. conceptualization; M.-E. C., S. A. Deeke, S. A. Dick, and P. G. B. data curation; M.-E. C., S. A. Deeke, S. A. Dick, L. A. M., and P. G. B. formal analysis; M.-E. C., E. M., C. L. R., J. J. W., L. A. M., and P. G. B. supervision; M.-E. C., S. A. Deeke, S. A. Dick, Z. J. A. V.-B., G. T., R. G., E. M., C. L. R., J. J. W., and P. G. B. investigation; M.-E. C. and P. G. B. methodology; M.-E. C. and P. G. B. writing-original draft; M.-E. C. and P. G. B. project administration; M.-E. C., Z. J. A. V.-B., G. T., R. G., E. M., J. J. W., L. A. M., and P. G. B. writing-review and editing; L. A. M. and P. G. B. funding acquisition.
Supplementary Material
This work was supported by the Canadian Institutes of Health Research (to P. G. B. & L. A. M.) and an Ontario Graduate Scholarship in Science and Technology fellowship (to J. J. W.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Fig. S1 and Table S1.
- MLIP
- muscle-enriched A-type lamin-interacting protein
- RACE
- rapid amplification of cDNA ends
- TSS
- transcriptional start site
- EST
- expressed sequence tag
- mCh9
- mouse chromosome 9
- H3K27Ac
- histone H3 with acetylated lysine 2
- H3K4Me1
- histone H3 with monomethylated lysine 4
- H3K4Me3
- histone H3 with trimethylated lysine 4
- NLS
- nuclear localization signal
- mTOR
- mechanistic target of rapamycin
- LMNA
- lamin A
- GST
- glutathione S-transferase
- E1
- exon 1.
References
- 1. Ahmady E., Deeke S. A., Rabaa S., Kouri L., Kenney L., Stewart A. F., and Burgon P. G. (2011) Identification of a novel muscle A-type lamin-interacting protein (MLIP). J. Biol. Chem. 286, 19702–19713 10.1074/jbc.M110.165548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang Z.-P., Young Seok H., Zhou B., Chen J., Chen J.-F., Tao Y., Pu W. T., and Wang D.-Z. (2012) CIP, a cardiac Isl1-interacting protein, represses cardiomyocyte hypertrophy. Circ. Res. 110, 818–830 10.1161/CIRCRESAHA.111.259663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cattin M.-E., Wang J., Weldrick J. J., Roeske C. L., Mak E., Thorn S. L., DaSilva J. N., Wang Y., Lusis A. J., and Burgon P. G. (2015) Deletion of MLIP (muscle-enriched A-type lamin-interacting protein) leads to cardiac hyperactivation of Akt/mammalian target of rapamycin (mTOR) and impaired cardiac adaptation. J. Biol. Chem. 290, 26699–26714 10.1074/jbc.M115.678433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang Z.-P., Kataoka M., Chen J., Wu G., Ding J., Nie M., Lin Z., Liu J., Hu X., Ma L., Zhou B., Wakimoto H., Zeng C., Kyselovic J., Deng Z.-L., et al. (2015) Cardiomyocyte-enriched protein CIP protects against pathophysiological stresses and regulates cardiac homeostasis. J. Clin. Investig. 125, 4122–4134 10.1172/JCI82423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Esslinger U., Garnier S., Korniat A., Proust C., Kararigas G., Müller-Nurasyid M., Empana J.-P., Morley M. P., Perret C., Stark K., Bick A. G., Prasad S. K., Kriebel J., Li J., Tiret L., et al. (2017) Exome-wide association study reveals novel susceptibility genes to sporadic dilated cardiomyopathy. PLoS One 12, e0172995 10.1371/journal.pone.0172995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Klerk E., and 't Hoen P. A. (2015) Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet. 31, 128–139 10.1016/j.tig.2015.01.001 [DOI] [PubMed] [Google Scholar]
- 8. FANTOM Consortium and the RIKEN PMI and CLST (DGT), Forrest A. R., Kawaji H., Rehli M., Baillie J. K., de Hoon M. J., Haberle V., Lassmann T., Kulakovskiy I. V., Lizio M., Itoh M., Andersson R., Mungall C. J., Meehan T. F., and Schmeier S. (2014) A promoter-level mammalian expression atlas. Nature 507, 462–470 10.1038/nature13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wakamatsu A., Kimura K., Yamamoto J., Nishikawa T., Nomura N., Sugano S., and Isogai T. (2009) Identification and functional analyses of 11,769 full-length human cDNAs focused on alternative splicing. DNA Res. 16, 371–383 10.1093/dnares/dsp022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Hoon M., and Hayashizaki Y. (2008) Deep cap analysis gene expression (CAGE): genome-wide identification of promoters, quantification of their expression, and network inference. Biotechniques 44, 627–632 10.2144/000112802 [DOI] [PubMed] [Google Scholar]
- 11. Hestand M. S., Klingenhoff A., Scherf M., Ariyurek Y., Ramos Y., van Workum W., Suzuki M., Werner T., van Ommen G.-J., den Dunnen J. T., Harbers M., and 't Hoen P. A. (2010) Tissue-specific transcript annotation and expression profiling with complementary next-generation sequencing technologies. Nucleic Acids Res. 38, e165 10.1093/nar/gkq602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pan Q., Shai O., Lee L. J., Frey B. J., and Blencowe B. J. (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- 13. Djebali S., Davis C. A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., Xue C., Marinov G. K., Khatun J., Williams B. A., Zaleski C., et al. (2012) Landscape of transcription in human cells. Nature. 489, 101–108 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kelemen O., Convertini P., Zhang Z., Wen Y., Shen M., Falaleeva M., and Stamm S. (2013) Function of alternative splicing. Gene 514, 1–30 10.1016/j.gene.2012.07.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dick S. A., Chang N. C., Dumont N. A., Bell R. A., Putinski C., Kawabe Y., Litchfield D. W., Rudnicki M. A., and Megeney L. A. (2015) Caspase 3 cleavage of Pax7 inhibits self-renewal of satellite cells. Proc. Natl. Acad. Sci. U.S.A. 112, E5246–E5252 10.1073/pnas.1512869112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.