Abstract
In vivo analyses of the occurrence, subcellular localization, and dynamics of protein–protein interactions (PPIs) are important issues in functional proteomic studies. The bimolecular fluorescence complementation (BiFC) assay has many advantages in that it provides a reliable way to detect PPIs in living cells with minimal perturbation of the structure and function of the target proteins. Previously, to facilitate the application of the BiFC assay to genome-wide analysis of PPIs, we generated a collection of yeast strains expressing full-length proteins tagged with the N-terminal fragment of Venus (VN), a yellow fluorescent protein variant, from their own native promoters. In the present study, we constructed a VC (the C-terminal fragment of Venus) fusion library consisting of 5671 MATα strains expressing C-terminally VC-tagged proteins (representing ∼91% of the yeast proteome). For genome-wide analysis of protein homomer formation, we mated each strain in the VC fusion library with its cognate strain in the VN fusion library and performed the BiFC assay. From this analysis, we identified 186 homomer candidates. We further investigated the functional relevance of the homomerization of Pln1, a yeast perilipin. Our data set provides a useful resource for understanding the physiological roles of protein homomerization. Furthermore, the VC fusion library together with the VN fusion library will provide a valuable platform to systematically analyze PPIs in the natural cellular context.
Most proteins work with other proteins in cells, and thus, finding the interaction partners of a target protein is a good way to understand the function of the target protein. For this reason, researchers developed various methods to detect protein–protein interactions (PPIs). In recent decades, several large-scale analyses of PPIs have been performed in the eukaryotic model organism Saccharomyces cerevisiae using the yeast two-hybrid method (Uetz et al. 2000), tandem affinity purification–mass spectrometry (TAP-MS) analysis (Ho et al. 2002; Krogan et al. 2006), and the protein fragment complementation assay (Tarassov et al. 2008).
Among the several kinds of PPIs, protein homomerization holds a special place. A protein homomer is a complex formed by two or more identical subunits. Homomerization is one of the simplest and most efficient ways for cells to regulate the function, concentration, and structure of proteins. It can help to minimize the genome size while providing the advantages of modular complex formation, and it has been reported that more than one-third of human enzymes function as homomers (Marianayagam et al. 2004). Protein homomerization offers several advantages. First, proteins obtain new binding sites with increased specificity in homomeric states. Many DNA-binding proteins in S. cerevisiae are known to bind DNA through sites expanded by their homomerization (Ellenberger et al. 1992; Hong et al. 2008). Second, cells can build up large protein complexes by homomerization with a relatively small number of genes (Goodsell and Olson 2000). Third, proteins can go through functional transitions depending on their homomeric states. For example, Escherichia coli UvrD acts as either a helicase or a translocase depending on whether they are homomers or monomers, respectively (Niedziela-Majka et al. 2007). Homomerization can also influence protein modifications. S. cerevisiae Hxk2 exists in either monomeric or dimeric form, but only monomeric Hxk2 is phosphorylated in response to a carbon source (Randez-Gil et al. 1998). Despite the profound physiological importance of protein homomerization, there has been no attempt to systematically identify protein homomers at a genome-wide level to date.
The bimolecular fluorescence complementation (BiFC) assay is a method for investigating PPIs by using fluorescent proteins (Kerppola 2008). In the BiFC assay, a fluorescent protein is split into two fragments that cannot fluoresce themselves, and each fragment is tagged to the target protein. If two target proteins come into close proximity by physical interaction, then the two tagged fluorescent fragments also become close to each other, and the interaction of the target proteins can be detected by the recovered fluorescence. The BiFC assay is a good way to detect PPIs in the cellular context with minimal disturbance. Furthermore, not only the occurrence but also the subcellular localization of PPIs can be examined by the BiFC assay. Because of these advantages, the BiFC assay has been used for large-scale PPI studies in various species (Klopffleisch et al. 2011; Snider et al. 2013). To facilitate genome-wide analysis of PPIs using the BiFC technique, our laboratory constructed the VN fusion library in a previous study (Sung et al. 2013). The VN fusion library is a collection of yeast strains in which VN, the N-terminal fragment of the Venus protein (Nagai et al. 2002), is tagged at the C termini of all yeast open reading frames (ORFs). The VN fusion library consists of 5911 VN-tagged MATa strains with a coverage of 95% of all ORFs in S. cerevisiae. We demonstrated the utility of the VN fusion library by systematically analyzing the interactome of the small ubiquitin-related modifier. In the present study, we generated a collection of yeast MATα strains expressing full-length proteins tagged with VC, the C-terminal fragment of Venus, and screened protein homomer candidates at a genome-wide scale using the VC and VN fusion libraries. As far as we know, this is the first report that describes a genome-wide screen for protein homomers.
Results
Construction of the VC fusion library
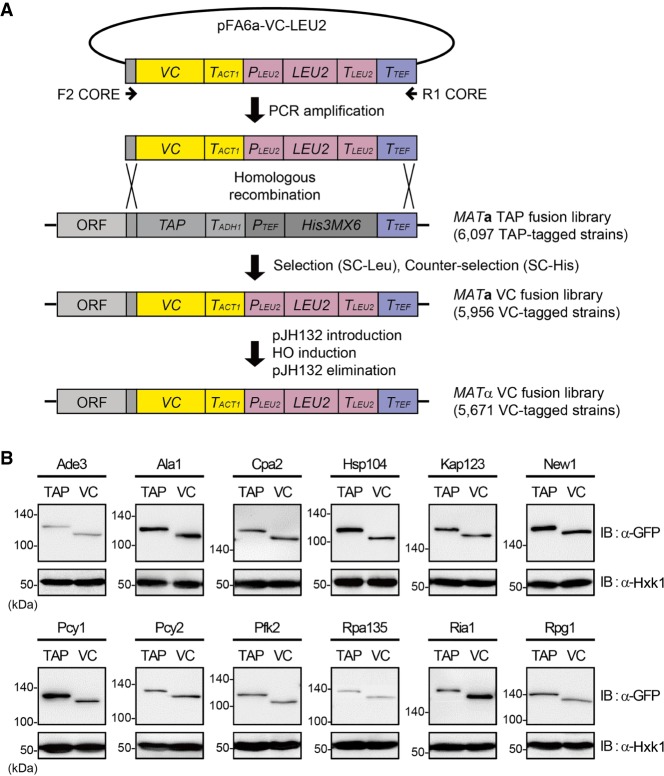
To construct the VC fusion library, we employed the same method used in the previous study (Sung et al. 2013) with some modifications. First, we constructed the plasmid pFA6a-VC-LEU2 that can be used for switching C-terminally tagged epitopes to the VC tag (Fig. 1A), based on an epitope-switching strategy (Sung et al. 2008). A DNA fragment containing the VC tag and LEU2 marker sequences was amplified by PCR using pFA6a-VC-LEU2 as a template and a set of the universal primers F2CORE and R1CORE (Fig. 1A; Supplemental Table S1). The resulting PCR products were transformed into each strain of the TAP fusion library (Ghaemmaghami et al. 2003), which consists of 6097 MATa strains with chromosomal C-terminally TAP-tagged ORFs that encompass 98% of all ORFs. Transformants were subjected to medium selection (SC-Leu) and counter-selection (SC-His) to verify successful tag switching from TAP to VC at the corresponding locus. Following this procedure, we obtained 5956 VC-tagged MATa strains with a coverage of 95% of all ORFs (Supplemental Table S2).
Figure 1.
Construction of the MATα VC fusion library and confirmation of proper tagging. (A) Scheme of the construction of the MATα VC fusion library. A DNA fragment containing the VC tag and LEU2 marker sequences was amplified by PCR using the pFA6a-VC-LEU2 vector as the template and the F2CORE and R1CORE primers, and substituted for the C-terminal TAP tag sequence in the chromosome of each strain in the TAP fusion library by homologous recombination. A chromosomally VC-tagged MATα strain collection was obtained by using pJH132, as described in Methods. LEU2, PLEU2, TLEU2, TACT1, TADH1, and TTEF represent Candida glabrata LEU2, C. glabrata LEU2 promoter, C. glabrata LEU2 terminator, S. cerevisiae ACT1 terminator, S. cerevisiae ADH1 terminator, and Ashbya gossypii TEF terminator, respectively. (B) Confirmation of proper epitope switching by western blot analysis. Selected strains from the TAP fusion library and the VC fusion library were lysed and immunoblotted using an anti-GFP antibody. Hexokinase was used as a loading control. The names of proteins and the tagged epitopes are indicated at the top of each panel. The positions of molecular-weight markers are indicated on the left of each blot.
Because the VN fusion library consists of MATa strains (Sung et al. 2013), the above-constructed VC-tagged MATa strains cannot be mated with the VN fusion library strains. To switch the mating type of VC-tagged strains from MATa to MATα, we introduced the plasmid pJH132 carrying a galactose-inducible homothallic switching endonuclease (HO) gene (Herskowitz and Jensen 1991) into each VC-tagged strain. After mating-type switching, pJH132 was eliminated from cells to prevent further unintended mating-type switching. Through these steps, we finally obtained the VC fusion library consisting of 5671 VC-tagged MATα strains that cover 91% of all ORFs (Supplemental Table S3). Western blot analysis of some strains confirmed successful tag switching from TAP to VC (Fig. 1B).
Genome-wide screen for protein homomers using the VC and VN fusion libraries
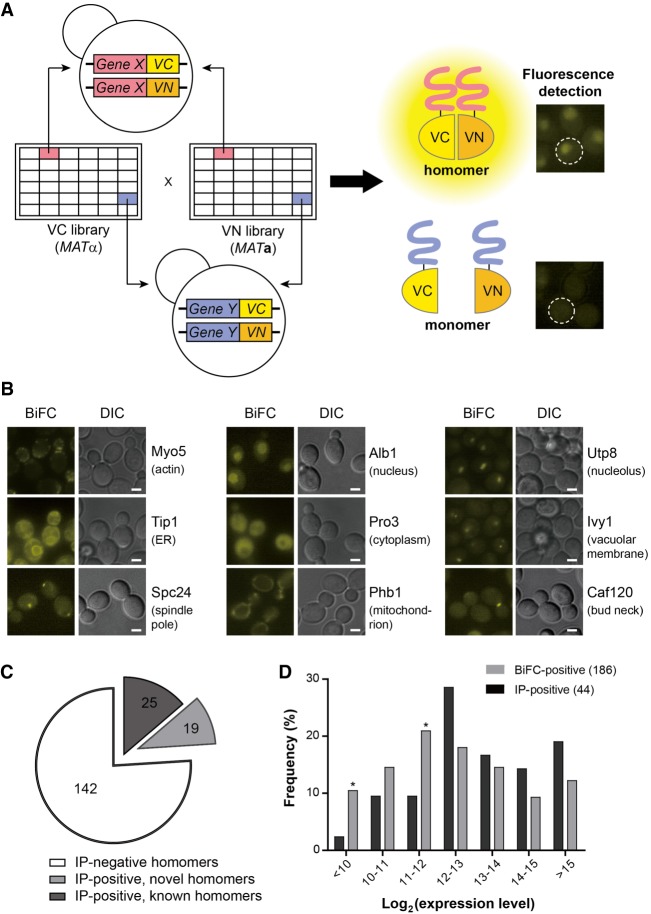
Next, we set out to identify protein homomers by a genome-wide BiFC assay. We mated each strain in the MATα VC fusion library with its cognate strain in the MATa VN fusion library and analyzed the resulting diploid strains by fluorescence microscopy (Fig. 2A). Of 5597 strains examined, 630 showed BiFC signals above the background level. To identify false-positives arising from the self-assembly of VN and VC, we mated a MATa strain expressing VN alone, without any fusion partner, to the 630 VC-tagged strains identified above and performed the BiFC assay. Of the 630 strains, 444 showed fluorescence above the background level and were regarded as false-positives (Supplemental Fig. S1; Supplemental Table S4). Most of the 444 strains exhibited fluorescence signals of the same subcellular location in both screens (Supplemental Fig. S1A), while 16 showed signals localized to different cellular sites (Supplemental Fig. S1B). Consequently, we finally obtained 186 protein homomer candidates (Fig. 2B; Supplemental Table S5).
Figure 2.
Genome-wide screen for protein homomers using the MATα VC fusion library and the MATa VN fusion library. (A) Scheme of the genome-wide BiFC analysis of protein homomers. Each strain in the MATα VC fusion library was mated with its cognate strain in the MATa VN fusion library. The resulting diploid cells expressing both the VC and VN fusion proteins were analyzed by fluorescence microscopy. (B) Representative BiFC images of homomer candidates with diverse subcellular localizations (Huh et al. 2003). The names of proteins and their localizations are designated on the right of each image. Scale bars, 2 µm. (C) Venn diagram depicting the numbers of homomer candidates detected by Co-IP and the BiFC assay. For detailed data from the Co-IP assay, see Supplemental Figure S2. (D) Bar graph depicting the comparison of the expression levels of homomer candidates detected by Co-IP (black bars) and the BiFC assay (gray bars). The expression level of each protein was obtained from the Yeast GFP Fusion Localization Database (https://yeastgfp.yeastgenome.org). Asterisks indicate significant enrichment in BiFC compared with Co-IP (P < 0.05; hypergeometric test).
To validate the homomer candidates identified above, we performed a coimmunoprecipitation (Co-IP) assay using HA- and GFP-tagged strains. Of the 186 homomer candidates, we could detect Co-IP bands with 44 proteins, of which 19 were novel homomer candidates (Fig. 2C; Supplemental Fig. S2). We failed to detect Co-IP bands for 142 proteins, presumably because the homomeric forms of these proteins are present at levels too low to be detected by western blotting or are severely lost during the immunoprecipitation procedure due to weak and transient interactions. Proteins with positive Co-IP results tended to have a high expression level, while the BiFC-positive homomer candidates exhibited a more evenly distributed expression level (Fig. 2D), suggesting that our approach is reliable for identifying protein homomers regardless of their expression levels or interaction strengths. We also examined the possibility that stable BiFC complex formation might cause lengthened duration of protein homomerization and lead to phenotypic change. However, we could detect little, if any, phenotypic change in cells expressing BiFC complexes (Supplemental Fig. S3), suggesting that the effect of BiFC complex formation on protein function is negligible for most proteins.
We noticed that some known homomers were not identified in our screen. There are several possible explanations. First, because we regarded any VC fusion proteins showing fluorescence with free VN as false-positives during the self-assembly analysis, true homomers may have been eliminated at this step. Indeed, among the 444 homomer candidates designated false-positives, 171 are known homomers reported in previous studies (Supplemental Table S4). Second, because it is estimated that BiFC can occur when VN and VC are fused to positions that are separated by a distance no greater than ∼10 nm (Hu et al. 2002), some homomers may be subject to topological constraints during the formation of BiFC complexes. We chose Rad52 to check this possibility. Rad52, a recombinase involved in the repair of DNA double-strand breaks, has been reported to form a homomeric ring (Shinohara et al. 1998), but a BiFC signal for Rad52 was not detected in our screen. We tagged VN and VC at the N terminus of Rad52 and performed the BiFC assay using four combinations. As shown in Supplemental Figure S4, only N-terminally VN- and VC-tagged Rad52 clearly exhibited a nuclear BiFC signal, suggesting that C-terminally tagged Rad52 has topological constraints preventing the BiFC complex formation. Given that we used C-terminally VN- or VC-tagged strains in our screen, it is reasonable to assume that several true homomers did not show positive BiFC signals due to topological constraints. Third, there is a possibility that C-terminal tagging of some proteins with VN or VC may disturb proper expression or folding of the fusion proteins and consequently lead to failure to make BiFC signals, as discussed later.
Characterization of protein homomers
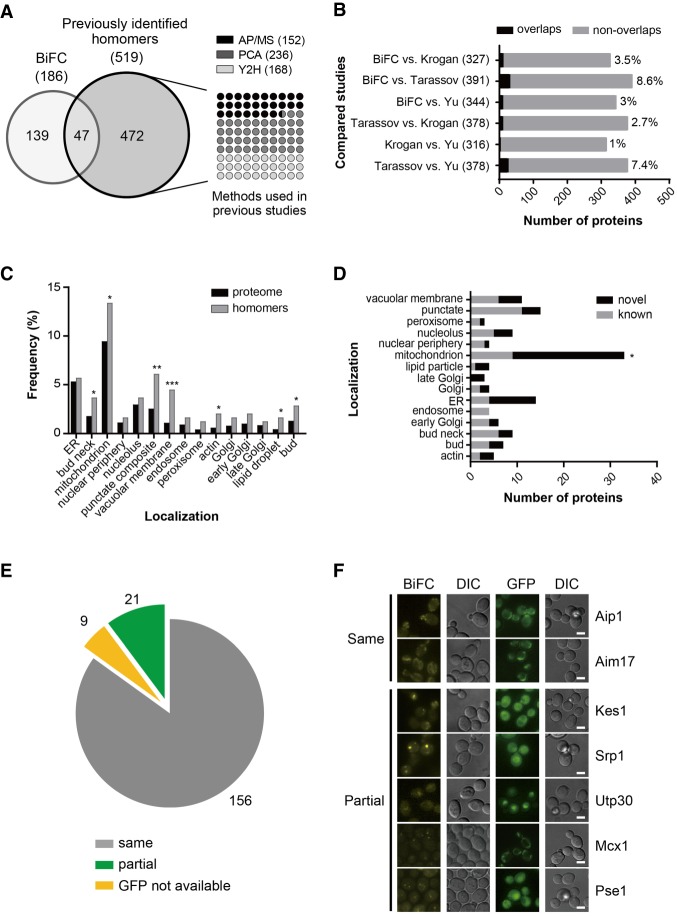
In this study, we identified 186 protein homomers, with 144 (77%) soluble proteins and 42 (23%) membrane proteins. Among the 186 protein homomers, 82 are known to form homomers according to the Saccharomyces Genome Database (https://www.yeastgenome.org, as of June 14th, 2017), and 104 are novel homomers (Supplemental Table S5). Although there has never been an attempt to systematically screen protein homomers at a genome-wide scale, 519 homomer candidates have been reported in previous studies to define the protein interactome in S. cerevisiae (Krogan et al. 2006; Tarassov et al. 2008; Yu et al. 2008). Among the 186 protein homomers identified in this study, 47 overlapped with 519 homomer candidates in previous data sets, but the remaining 139 were defined only in this study (Fig. 3A). Our data set showed a modest concordance with previous reports from Krogan et al. (2006) (4%), Tarassov et al. (2008) (9%), and Yu et al. (2008) (3%) (Fig. 3B). This small percentage of overlap was similarly observed in comparisons between other studies. The identification of 104 new protein homomers in this study reflects the difference in the employed experimental methods between this study and previous studies, and thus, our data set appears to be highly complementary to the existing data sets.
Figure 3.
Characterization of protein homomer candidates. (A, left) Venn diagram depicting the comparison of homomer candidates identified in this study and those identified in three previous analyses. (Right) Analysis methods used in previous studies and the number of homomer candidates detected by each method. AP/MS, PCA, and Y2H represent affinity purification–mass spectrometry (Krogan et al. 2006), the protein complementation assay (Tarassov et al. 2008), and the yeast two-hybrid assay (Yu et al. 2008), respectively. (B) Bar graph depicting the comparison between the data set in this study and each of the three previous data sets. Black and gray bars indicate the number of homomer candidates that overlap and do not overlap, respectively. The percentage of concordance between data sets is indicated on the right of the bars. Numbers in parentheses indicate the total number of homomer candidates identified in two studies. (C) Subcellular localization of homomer candidates identified in this study. Black bars indicate whole proteins for which subcellular localization has been annotated in the Yeast GFP Fusion Localization Database (https://yeastgfp.yeastgenome.org), and gray bars represent homomer candidates identified in this study. Asterisks indicate significant differences compared with the proteome (Fisher's exact test): (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (D) Localization distribution of known and novel homomers. Gray and black bars indicate the number of known and novel homomers, respectively. Asterisks indicate significant differences compared with known homomers (Fisher's exact test): (*) P < 0.05. (E) Venn diagram depicting the comparison between the BiFC signal and the GFP signal of each homomer candidate. Proteins with identical BiFC and GFP signal patterns are shown as a gray pie slice. Proteins showing the BiFC signal as foci or partially localized within the GFP signal are indicated as a green pie slice. Proteins for which subcellular localization has not been annotated in the Yeast GFP Fusion Localization Database are shown as a yellow pie slice. (F) Representative fluorescence images of proteins showing the same or partial localization between the BiFC and GFP signals. Scale bars, 2 µm.
The subcellular region in which the BiFC signal is detected represents the localization of the corresponding protein homomer. According to the Yeast GFP Fusion Localization Database (Huh et al. 2003; https://yeastgfp.yeastgenome.org), the protein homomers identified in this study were present at various subcellular regions (Fig. 3C). Protein homomers were significantly enriched in the bud neck (P = 0.0399), mitochondrion (P = 0.0444), vacuolar membrane (P = 0.0001), actin (P = 0.0122), lipid droplet (P = 0.017), and bud (P = 0.0363). Known and novel homomers were distributed evenly across various organelles except the mitochondrion, where novel homomers were highly enriched (P = 0.0138) (Fig. 3D). Comparison of protein homomers identified in this study with positive/negative reference sets (Supplemental Table S6, S7), adaptins (Supplemental Table S8), and p24 family proteins (Supplemental Table S9) revealed the suitability of BiFC assay to detect protein homomerization especially in membrane-bound organelles or vesicles. Among the six adaptins (Apl1-6) acting in vesicle transport between organelles, three (Apl3, Apl4, and Apl6) were previously reported to be homomers (Babu et al. 2012; Schlecht et al. 2012), and four (Apl2, Apl4, Apl5, and Apl6) were detected in our BiFC screen, including two new homomer candidates (Supplemental Table S8). Also, among the eight p24 family proteins acting in the vesicle formation (Pastor-Cantizano et al. 2016), five (Emp24, Erp1, Erp3, Erp5, and Erv25) were previously reported to be homomers (Miller et al. 2005; Tarassov et al. 2008; Babu et al. 2012), and three of them (Emp24, Erp1, and Erv25) were detected in our BiFC screen (Supplemental Table S9).
Gene Ontology (GO) analysis revealed that the protein homomers identified in this study can be categorized into a variety of biological processes (Supplemental Table S10) and are significantly enriched in the biological processes of transport and cellular component organization. This finding is reasonable given that many transport-related proteins, such as channel proteins or vesicle proteins, form homomers to provide space and specificity for their cargo molecules (Agre and Kozono 2003; MacKinnon 2003).
Next, we compared the subcellular localization of the BiFC and GFP signals of protein homomers. Among 186 protein homomers, 156 showed identical localization patterns between the BiFC and GFP signals (Fig. 3E,F; Supplemental Table S5). For 21 proteins, the BiFC signal for homomerization appeared as foci or was partially localized within a region where the corresponding GFP fusion protein was detected (Supplemental Fig. S5). This observation suggests that only a small fraction of these proteins exist as homomers, while the rest stay in monomeric forms. It is also likely that the subcellular localization of some protein homomers is regulated differently from that of the monomeric forms.
Changes in protein homomerization upon nitrogen starvation
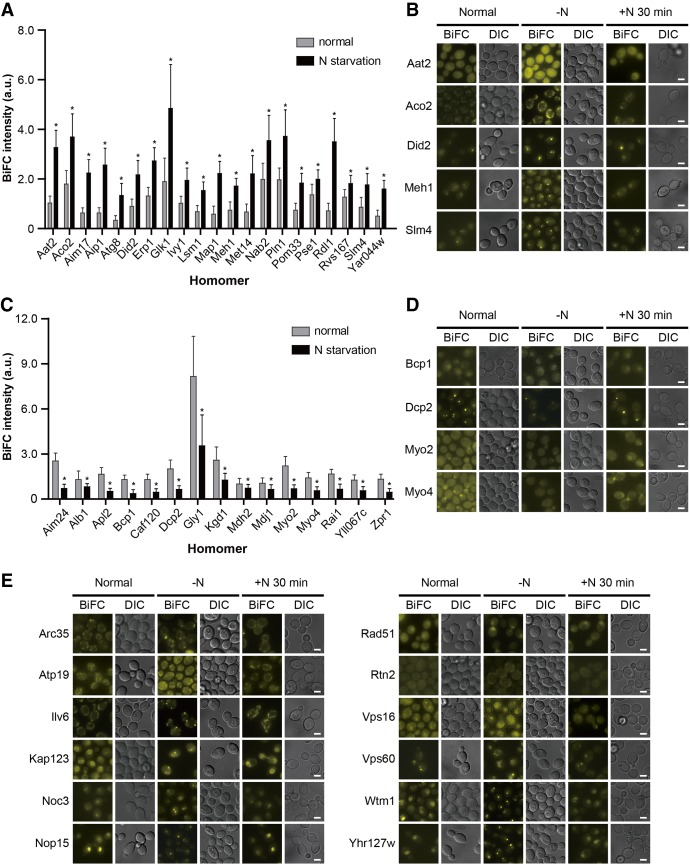
Nitrogen is an essential element in every living organism, and its deprivation induces an overall rearrangement of gene expression and signaling pathways in yeast (Gasch et al. 2000; Kamada et al. 2004; Conway et al. 2012), resulting in various phenomena, such as autophagy, filamentous growth, and the accumulation of specific macromolecules (Gimeno et al. 1992; Schulze et al. 1996; Parrou et al. 1999; Onodera and Ohsumi 2005). To investigate whether protein homomerization is influenced by nitrogen starvation, we incubated yeast cells expressing VN/VC-tagged protein homomers in nitrogen-deprived medium and examined changes in the BiFC signal pattern. We found that the BiFC signals of 21 proteins were significantly increased under nitrogen starvation, while those of 15 proteins were decreased (Fig. 4A–D; Supplemental Fig. S6; Supplemental Table S11). The GFP signal also increased in four (Aim17, Met14, Rdl1, and Rvs167) of 21 proteins showing an increased BiFC signal under nitrogen starvation, and seven (Apl2, Bcp1, Caf120, Gly1, Mdh2, Myo2, and Myo4) of 15 proteins with a decreased BiFC signal under nitrogen starvation also showed a decrease in the GFP signal. This observation suggests that changes in the BiFC signal intensity of the above 11 proteins under nitrogen starvation are due to their altered expression levels. It is likely that the other 25 proteins, which showed changes in the BiFC signal intensity but not in the GFP signal intensity under nitrogen starvation, experience alterations in their homomeric states by nitrogen deprivation. We also performed a flow cytometric analysis for 16 proteins with increased or decreased BiFC signal intensity under nitrogen starvation. The BiFC signal intensity of all tested 16 proteins showed a consistent pattern between flow cytometry and fluorescence microscopy (Supplemental Fig. S7), regardless of their subcellular localization, supporting the reliability of the BiFC assay in monitoring changes in the homomeric states of proteins in nitrogen-deprived conditions.
Figure 4.
Changes in protein homomerization upon nitrogen starvation. (A,C) Protein homomers showing increased (A) or decreased (C) BiFC signals upon nitrogen starvation. Gray and black bars indicate the mean BiFC signal intensity of protein homomers under the normal conditions and nitrogen starvation, respectively. Error bars, SD. Asterisks indicate significant differences compared with the normal conditions (Student's t-test): (*) P < 0.05. For detailed information, see also Supplemental Table S11. (B,D,E) Representative BiFC images of protein homomers showing increased (B), decreased (D), or localization-changed (E) BiFC signals upon nitrogen starvation and restored signals after transfer to nitrogen-containing medium. Scale bars, 2 µm.
Among the 186 protein homomers, 12 exhibited changes in the BiFC signal localization under nitrogen starvation (Fig. 4E; Supplemental Table S12). Nine (Arc35, Atp19, Ilv6, Kap123, Noc3, Nop15, Rad51, Wtm1, and Yhr127w) of 12 proteins localized to punctate foci under nitrogen starvation. The GFP signal localization of 12 proteins was similarly changed by nitrogen deprivation (Supplemental Fig. S8), suggesting that changes in the BiFC signal localization of 12 proteins under nitrogen starvation are caused by translocation of the proteins and are not related to alterations in their homomeric states. Next, we examined whether the BiFC signals of protein homomers showing changes in the BiFC signal intensity (36 proteins) or localization (12 proteins) under nitrogen starvation are restored after reincubation in nitrogen-containing medium. We observed that all 48 proteins restored their signal intensity or localization (Fig. 4B,D,E; Supplemental Fig. S6), suggesting that the BiFC complex formation has little effect on the protein dynamics that occur in response to environmental changes.
Enhanced homomerization of Pln1 under nitrogen starvation
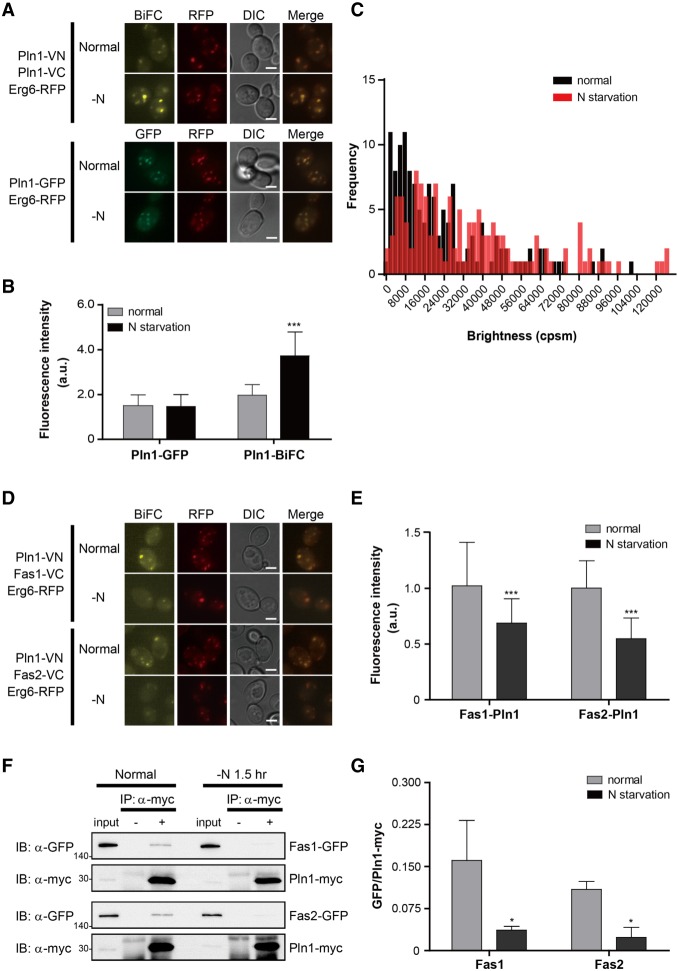
Pln1 (Ykr046c) is a lipid droplet protein (Athenstaedt et al. 1999), and its homomerization has not been reported before. C-terminally VN- and VC-tagged Pln1 showed a clear BiFC signal in our screen, and both the BiFC and GFP signals of Pln1 overlapped with that of RFP-tagged Erg6, a lipid droplet marker protein (Fig. 5A). The BiFC signal of Pln1 is not likely to be caused by nonspecific interaction between Pln1 monomers clustered in close proximity because VN-tagged Scs3, Sei1, and Yft2, which act with Pln1 in lipid droplet biogenesis (Gao et al. 2017), could not produce BiFC signals with VC-tagged Pln1 (Supplemental Fig. S9). The BiFC signal of Pln1 significantly increased under nitrogen starvation (Figs. 4A, 5A,B; Supplemental Table S11). This increase in the BiFC signal seems to be due to increased homomerization of Pln1, rather than increased expression of Pln1, because the signal intensity of Pln1-GFP did not increase under the same condition (Fig. 5A,B).
Figure 5.
Homomerization of Pln1 and its interactors. (A) Representative BiFC and GFP images of Pln1 under the normal conditions and nitrogen starvation. Erg6-RFP was used as a lipid droplet marker. Scale bars, 2 μm. (B) Quantification of the fluorescence intensity of cells in A. Gray and black bars indicate the fluorescence intensity of cells under the normal conditions and nitrogen starvation, respectively. Error bars, SD. Asterisks indicate significant differences compared with the normal conditions (Student's t-test): (***) P < 0.001. (C) PCH analysis of Pln1-GFP. The brightness (counts per second per molecule [cpsm]) of Pln1-GFP under the normal conditions and nitrogen starvation was counted and shown as a histogram. Black and red bars indicate the frequency of Pln1-GFP fluorescence under the normal conditions and nitrogen starvation, respectively. One hundred forty cells were counted under each condition. Statistical significance was determined by Student's t-test (P = 0.0097). (D) The BiFC assay for the interaction of Pln1 with Fas1 and Fas2. Erg6-RFP was used as a lipid droplet marker. Scale bars, 2 μm. (E) Quantification of the fluorescence intensity of cells in D. Gray and black bars indicate the fluorescence intensity of cells under the normal conditions and nitrogen starvation, respectively. Error bars, SD. Asterisks indicate significant differences compared with the normal conditions (Student's t-test): (***) P < 0. 001. (F) Co-IP assay to validate the interaction of Pln1 with Fas1 and Fas2. Co-IP assay was performed using the Myc-tagged Pln1 expressed under the ADH1 promoter and GFP-tagged Fas1 and Fas2. The positions of molecular-weight markers (in kDa) are indicated on the left of each blot. (G) Quantification of Co-IP in F. The ratios of GFP-tagged proteins co-precipitated with Pln1-Myc were calculated by using ImageJ (Schneider et al. 2012). Values represent the average of three independent experiments; error bars, SD. Asterisks indicate significant differences compared with the normal conditions (Student's t-test): (*) P < 0.05.
We performed biochemical assays such as cross-linking and Co-IP to confirm the homomerization of Pln1 but failed to see any positive results. We inferred that Pln1 homomers are rather weakly or transiently formed, preventing their detection by conventional biochemical methods. To verify the homomerization of Pln1 and its increase under nitrogen starvation, we performed a fluorescence correlation spectroscopy (FCS) and photon counting histogram (PCH) analysis with C-terminally GFP-tagged Pln1. FCS and PCH analysis are well suited for examining the biophysical properties of protein complexes in living cells and have been successfully used to determine the state of protein homomerization (Rigler et al. 1993; Chen et al. 2002; Saffarian et al. 2007; Malengo et al. 2008). While free GFP exhibited a clear, predominant single peak in PCH analysis, Pln1-GFP showed more than one peak (Supplemental Fig. S10), implying that Pln1 can exist as both monomers and homomers. Under nitrogen starvation, the PCH of Pln1-GFP slightly moved to the right (Fig. 5C). This observation is consistent with the increase in the BiFC signal of Pln1 under the same conditions and suggests that the population of Pln1 homomers is increased by nitrogen deprivation.
Involvement of Pln1 in lipid metabolism
In an attempt to define the function of Pln1, we performed a BiFC screen to identify its interactors. Based on previous studies, we suspected its role in respiration (Brown et al. 2000) or lipid metabolic processes (Athenstaedt et al. 1999; Currie et al. 2014), and we selected 44 proteins that are key components of the electron transfer chain, mitochondrial dynamics, or phospholipid/sterol synthesis (Supplemental Table S13). Forty-four VC-tagged strains from the VC fusion library were mated with a MATa strain expressing C-terminally VN-tagged Pln1, and the resulting diploids were analyzed by fluorescence microscopy. Among the 44 candidates, three (Fas1, Fas2, and Slc1) showed positive BiFC signals with Pln1-VN (Fig. 5D; Supplemental Fig. S11). Fas1 and Fas2 are the β and α subunits of the fatty acid synthase (FAS) complex (Schweizer et al. 1986; Mohamed et al. 1988), and Slc1 is an acyltransferase acting in phosphatidic acid synthesis (Nagiec et al. 1993; Athenstaedt and Daum 1999). Although Fas1 and Fas2 are known to be present in the cytoplasm (Huh et al. 2003), interaction between Pln1 and Fas1 or Fas2 occurred on lipid droplets (Fig. 5D,E). We performed a Co-IP assay using GFP-tagged Fas1, Fas2, and Slc1 and Myc-tagged Pln1 to verify their interactions. As shown in Figure 5F and Supplemental Figure S11, all three proteins were efficiently coprecipitated with Pln1. Furthermore, the proportion of Pln1 physically interacting with Fas1 and Fas2 significantly decreased under nitrogen starvation, as demonstrated by both the BiFC assay (Fig. 5D,E; Supplemental Fig. S12) and Co-IP assay (Fig. 5F,G), suggesting that Pln1 can form a complex with FAS components, and the formation of this complex depends on nutrient availability.
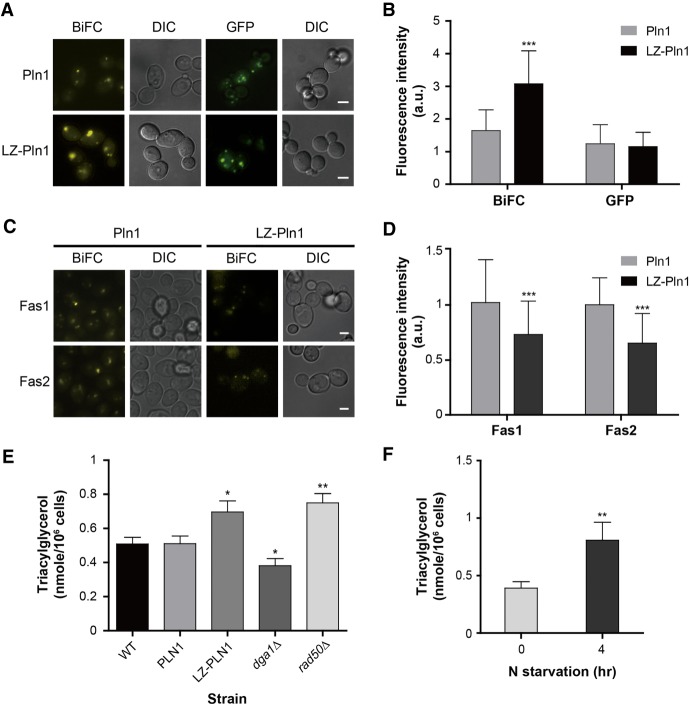
Given that Fas1, Fas2, and Slc1 are crucial effectors in lipid metabolism (Raychaudhuri et al. 2012), we hypothesized that Pln1 may have a function in the same pathway. A recent study also proposed that Pln1 is a yeast perilipin functioning in neutral lipid homeostasis (Gao et al. 2017). To evaluate the physiological effect of Pln1 homomerization, we constructed yeast strains expressing Pln1 tagged with the leucine zipper (LZ) motif at its N terminus and with VC, VN, or GFP at its C terminus. LZ-Pln1 showed a significantly increased BiFC signal compared with that of Pln1 (Fig. 6A,B), indicating that LZ-Pln1 has a higher tendency to dimerize. The GFP signal intensity of LZ-Pln1 was similar to that of Pln1, indicating that LZ tagging does not disturb the expression or stability of Pln1. The BiFC signal intensity between LZ-Pln1 and Fas1 or Fas2 was lower than that between Pln1 and the FAS components (Fig. 6C,D), suggesting that LZ-Pln1 forms a weak interaction with Fas1 and Fas2, as Pln1 does under nitrogen starvation. In addition, cells expressing LZ-Pln1 showed a significantly increased triacylglycerol (TAG) level compared with that of cells expressing Pln1 (Fig. 6E). Recently, it was reported that TAG accumulates upon nitrogen starvation in various species (Breuer et al. 2012; Lopez Garcia de Lomana et al. 2015). Consistent with these reports, we also observed a significantly increased TAG level under nitrogen starvation in S. cerevisiae (Fig. 6F). Taken together, our results suggest that the state of Pln1 homomerization is related to the regulation of intracellular TAG level in response to nutrient availability.
Figure 6.
Involvement of Pln1 in lipid metabolism. (A) Representative BiFC and GFP images of cells expressing Pln1 and LZ-Pln1. Scale bars, 2 μm. LZ-Pln1 represents Pln1 tagged with the leucine zipper (LZ) motif. (B) Quantification of the fluorescence intensity of cells in A. Gray and black bars indicate the fluorescence intensity of cells expressing Pln1 and LZ-Pln1, respectively. Error bars, SD. Asterisks indicate significant differences compared with Pln1 (Student's t-test): (***) P < 0.001. (C) Representative BiFC image of cells expressing Pln1- or LZ-Pln1-VN and Fas1- or Fas2-VC. Scale bars, 2 μm. (D) Quantification of the fluorescence intensity of cells in C. Gray and black bars indicate the fluorescence intensity of cells expressing Pln1 and LZ-Pln1, respectively. Quantitative and statistical analysis was performed as described in B. (E) Measurement of TAG contents. TAG was measured as described in the Supplemental Material. Values represent the average of eight independent experiments; error bars, SD. dga1Δ and rad50Δ cells were used as a negative and a positive control, respectively (Oelkers et al. 2002; Kanagavijayan et al. 2016). Asterisks indicate significant differences compared with wild-type cells (Student's t-test): (*) P < 0.05, (**) P < 0.01. (F) TAG contents of cells under nitrogen starvation. The TAG levels in wild-type cells under the normal conditions and nitrogen starvation were measured. Values represent the average of three independent experiments; error bars, SD. Asterisks indicate significant differences compared with the normal conditions (Student's t-test): (**) P < 0.01.
Discussion
In this study, we performed a genome-wide screen for protein homomers using the VC and VN fusion libraries. There are some caveats for using the VC and VN fusion libraries. Although the VC and VN fusion libraries cover >90% of all ORFs in S. cerevisiae, it does not reflect the actual number of proteins that can be successfully analyzed for their interactors. C-terminal tagging can perturb proper folding and function of some proteins. Additionally, some proteins are expressed in precursor forms and need to be cleaved for complete maturation, and C-terminally tagged VC or VN can be eliminated in this process. These proteins cannot generate BiFC signals even though they exist in sufficient amounts and form actual interactions. It is known that many proteins localized to the cell wall (van Berkel et al. 1994), peroxisome (Gould et al. 1990), and endoplasmic reticulum (Pelham et al. 1988) contain C-terminal targeting signals. The C-terminal fusion of a tag can cause mislocalization of these proteins through steric hindrance or interruption of critical C-terminal localization/retention sequences. Huh et al. (2003) identified 37 cell wall proteins, nine HDEL proteins, eight PTS1-containing proteins, and 11 fatty-acylated proteins that are mislocalized by the C-terminal fusion of GFP. Glycosylphosphatidylinositol (GPI)-anchored proteins have a conserved GPI anchoring signal at the C-terminal region, which is recognized and removed by GPI transamidase before attachment to GPI anchors (Pittet and Conzelmann 2007). Because tagging at the C termini of GPI-anchored proteins usually causes their mislocalization, internal tags have been used for studying these proteins (De Groot et al. 2003; Du et al. 2012; Zordan et al. 2015). For at least the above proteins, our strategy using C-terminal VC and VN fusions is not likely to provide valid data in PPI analysis. Indeed, of the 22 putative GPI-anchored proteins (De Groot et al. 2003), 21 did not show any BiFC signals in our screen, except Ddr2.
Furthermore, given that we used C-terminally VN- or VC-tagged strains in our screen, it is reasonable to assume that several true homomers, e.g., Rad52 and Cet1 (Sung and Huh 2007), did not show positive BiFC signals due to the topological constraints. It is estimated that BiFC can occur when the fluorescent protein fragments are fused to positions that are separated by up to a distance of ∼10 nm, provided that there is sufficient flexibility to allow association of the fragments (Hu et al. 2002). Based on this estimation, it is likely that we obtained negative BiFC signals for protein homomers of which the C-terminal ends stay away (>10 nm) from each other. The future construction and utilization of N-terminally VN- and VC-tagged strain collections will alleviate topological problems in genome-wide BiFC screens and greatly reduce false-negative results.
In this study, we adopted the self-assembly screen strategy because it is practically impossible to construct a specific negative control mutant defective in homotypic interaction for each of a large number of homomer candidates. In the self-assembly analysis, 16 homomer candidates showed a different localization pattern between the BiFC signal of homomer and that of self-assembly (Supplemental Fig. S1B; Supplemental Table S4). Among them, eight (Agp1, Atp14, Mup1, Pst2, Sec63, Sec7, Sna3, and Vps4) are known homomers, suggesting high enrichment of true homomers in this population. Given this, it is highly probable that the rest of them may also form homomers. Even with the stringent self-assembly screen, our data set is highly complementary to the existing data sets, and thus, combining the BiFC assay with other methods, such as Co-IP, TAP/MS, and the yeast two-hybrid assays, would help reduce the number of false-positives. For example, we anticipate that screening 444 false-positives by Co-IP assay would recover many true homomers.
It is noteworthy that two components of the EGO complex, Meh1 and Slm4 (Binda et al. 2009), showed increased BiFC signals upon nitrogen starvation (Fig. 4A,B). Given that the homomeric state of Slm4 has been reported to be important for the regulation of TORC1 signaling (Zhang et al. 2012), it will be interesting to examine whether Meh1 homomerization also influences TORC1 signaling and how the increase in homomerization of Meh1 and Slm4 is related to the regulation of TORC1 signaling. Several proteins involved in the multivesicular body (MVB) pathway, such as Did2 and Vps60 (Rue et al. 2008), are also among the candidates showing BiFC signal changes. The MVB pathway is required for starvation-induced degradation of plasma membrane proteins (Jones et al. 2012), and its proper activation is related to autophagy (Lee et al. 2007). Homomerization of these proteins may be an indication of the MVB pathway activation. As such, analysis of the dynamics of protein homomerization under stress conditions raises many research issues and challenges, which need to be addressed.
Our observations of the increase in Pln1 homomerization and the decrease in the interaction of Pln1 with FAS components under nitrogen starvation give new insights into the possible role of Pln1, especially about its relation to TAG accumulation under nitrogen starvation, which was also observed in many species (Breuer et al. 2012; Lopez Garcia de Lomana et al. 2015). Although we detected homomerization of Pln1 in living cells by the BiFC and PCH assays, we failed to confirm it by biochemical assays using cell-free extracts. This failure could be due to the dimensionality effects related to Pln1 localization. Localization of Pln1 on the lipid droplet surface may enhance its homomeric interaction by restricting its search space, as described in a recent study (Yogurtcu and Johnson 2018), and may help visualize its homomerization. Indeed, in an attempt to find a crucial domain for Pln1 homomerization, we observed that the N-terminal truncated Pln1 was abnormally localized to the cytoplasm and did not show a BiFC-positive homomeric signal (Supplemental Fig. S13). The physiological effect and its mechanism of perturbed Pln1 homomerization remain unclear and require further study. Aside from this, as far as we know, Pln1 is the only lipid droplet protein that physically interacts with Fas1 and Fas2. Further investigation into how and why Pln1 interacts with FAS components and Slc1 will help increase our knowledge about the regulation of lipid metabolism. Understanding the functional role of Pln1 in lipid metabolism will also be useful in bioengineering applications, given that increasing lipid production in microorganisms has long been a challenge (Brennan and Owende 2010; Zeng et al. 2011).
Methods
Yeast strains and culture conditions
S. cerevisiae strains used in this study are listed in Supplemental Table S14. Yeast cells were grown at 30°C in YPD (1% yeast extract, 2% peptone, and 2% glucose) or synthetic complete (SC) medium lacking appropriate amino acids for selection (Sherman 2002). For solid media, 2% agar was added. See Supplemental Methods for more details.
Construction of plasmids
The oligonucleotide primers used in this study are listed in Supplemental Table S1. For the construction of pFA6a-VC-LEU2 vector, the ∼1900-bp PCR product of LEU2 obtained using pCgLEU2 as a template and the ∼250-bp PCR product of Ashbya gossypii TEF terminator obtained using pFA6a-GFP-His3MX6 (Longtine et al. 1998) as a template, and with the two PCR products as templates, the ∼2150-bp PCR product was obtained as previously described (Horton 1997). Next, the obtained ∼2150-bp PCR product was digested with BamHI and PmeI and ligated into the BglII-PmeI–digested pFA6a-VC-KlURA3 (Sung et al. 2008), generating pFA6a-VC-LEU2. See Supplemental Methods for more details.
Construction of the VC-tagged strain collection
A chromosomally VC-tagged MATa strain collection was generated from the TAP-tagged collection (Ghaemmaghami et al. 2003) using the epitope switching strategy (Sung et al. 2008). An epitope switching module containing the VC tag and LEU2 marker sequences was amplified by PCR using pFA6a-VC-LEU2 as a template and a universal primer set F2CORE and R1CORE. The resulting module was transformed into each strain of the TAP-tagged strain collection, which consists of 6097 MATa strains with chromosomal C-terminally TAP-tagged ORFs. Transformed cells were spread on SC-Leu plates and incubated for 3 d at 30°C. To confirm correct switching from the TAP tag to the VC tag, colonies selected on SC-Leu medium were counter-selected on SC-His medium. For a chromosomally VC-tagged MATα strain collection, pJH132 vector (Herskowitz and Jensen 1991) was transformed into each strain of the VC-tagged MATa strain collection. Transformed cells were spread on SC-Ura plates and incubated for 3 d at 30°C. Transformants selected on SC-Ura plates were grown in SC medium with raffinose overnight and then transferred to SC medium with galactose, followed by incubation for 4 h. An aliquot of each strain was spread on SC-Ura plates, and six colonies per strain were scored after 3 d. MATα cells were selected by the capability of mating with the reference mating type tester strain, and then pJH132 vector was removed by 5-FOA treatment (Boeke et al. 1987).
Microscopic analysis and fluorescence quantification
For a genome-wide screen of protein homomers, each strain of the VC-tagged MATα strain collection was grown in YPD medium and mated with its cognate strain of the VN-tagged MATa strain collection. Diploid cells were then microscopically analyzed in 96-well glass-bottomed microplates (MGP096, Matrical Bioscience). Microscopy was performed on a Nikon eclipse E1 microscope with a Plan Fluor 100×/1.30 NA oil immersion objective. Quantification of fluorescent images was performed using the NIS-Elements imaging software (Nikon). See Supplemental Methods for more details.
Co-IP assay
Diploid cells expressing GFP- and HA- or Myc-tagged target proteins were used for Co-IP assay. Cell extracts were prepared as previously described (Sung et al. 2008). Proteins were loaded on SDS-PAGE gels and detected with a HRP-conjugated anti-GFP antibody (SC-9996 HRP, Santa Cruz Biotechnology), a HRP-conjugated anti-Myc antibody (SC-40 HRP, Santa Cruz Biotechnology), or a HRP-conjugated anti-HA antibody (SC-7392 HRP, Santa Cruz Biotechnology). See Supplemental Methods for more details.
Western blot analysis
Cell extracts were prepared as previously described (Sung et al. 2008). SDS-PAGE and western blot analysis were performed using standard methods with a HRP-conjugated anti-mouse IgG antibody (A9044, Sigma-Aldrich), a HRP-conjugated anti-rabbit IgG antibody (A6154, Sigma-Aldrich), a HRP-conjugated anti-GFP antibody (SC-9996 HRP, Santa Cruz Biotechnology), a HRP-conjugated anti-Myc antibody (SC-40 HRP, Santa Cruz Biotechnology), a HRP-conjugated anti-HA antibody (SC-7392 HRP, Santa Cruz Biotechnology), a HRP-conjugated anti-GFP antibody (600-103-215, Rockland), and a rabbit anti-hexokinase antibody (H2035-03, United States Biological).
Flow cytometry
Flow cytometry of cells was performed based on the description in the previous study (Murozuka et al. 2013) using BD FACSCanto II (BD Biosciences) equipped with BD FACSDiva 8.0.1 software. Samples were prepared as described in the Supplemental Material. One hundred thousand cells were analyzed at each condition; intensities of fluorescence of cells were plotted on a histogram using FITC (525 nm); and the geometric mean of fluorescence intensity was obtained using BD FACSDiva software. See Supplemental Methods for more details.
FCS and PCH analyses
FCS and PCH measurements were all performed at 25°C with a LSM 780 confocal microscope (Carl Zeiss) equipped with GaAsP multichannel spectral detectors as previously described (Herrick-Davis et al. 2013; Kim et al. 2017). Cells grown to OD600 = 1.0 at 30°C in YPD medium were diluted to OD600 = 0.1 in SC medium. After 4 h of incubation, FCS and PCH measurements on yeast cells were recorded sequentially 10 times with a duration of 5 sec. One hundred forty cells were measured for each experiment. See Supplemental Methods for more details.
TAG quantification assay
Intracellular TAG was quantified by using the Triglyceride Assay Kit - Quantification (ab65336, Abcam) according to the manufacturer's instructions. Detection was performed using the FlexStation 3 microplate reader (Molecular Devices), and the absorbance at 570 nm was measured. The net absorbance was obtained by subtracting the absorbance of the blank and then normalized by the number of cells. See Supplemental Methods for more details.
Data access
The protein interactions from this study have been submitted to the IMEx Consortium (https://www.imexconsortium.org/) through DIP (https://dip.doe-mbi.ucla.edu) under the record no. IM-26661.
Supplementary Material
Acknowledgments
We thank members of the Huh laboratory for helpful discussions. We also thank the Confocal Microscope core facility at the Convergence Medicine Research Center (CREDIT), Asan Institute for Life Sciences, for technical support and instrumentation. This work was supported by the National Research Foundation of Korea grants (2012R1A2A2A01047175, 2015R1A2A1A01007871, 2018R1A2B2009169) funded by the Ministry of Education, Science and Technology, Republic of Korea.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.231860.117.
References
- Agre P, Kozono D. 2003. Aquaporin water channels: molecular mechanisms for human diseases. FEBS Lett 555: 72–78. 10.1016/S0014-5793(03)01083-4 [DOI] [PubMed] [Google Scholar]
- Athenstaedt K, Daum G. 1999. Phosphatidic acid, a key intermediate in lipid metabolism. Eur J Biochem 266: 1–16. 10.1046/j.1432-1327.1999.00822.x [DOI] [PubMed] [Google Scholar]
- Athenstaedt K, Zweytick D, Jandrositz A, Kohlwein SD, Daum G. 1999. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J Bacteriol 181: 6441–6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu M, Vlasblom J, Pu S, Guo X, Graham C, Bean BD, Burston HE, Vizeacoumar FJ, Snider J, Phanse S, et al. 2012. Interaction landscape of membrane–protein complexes in Saccharomyces cerevisiae. Nature 489: 585–589. 10.1038/nature11354 [DOI] [PubMed] [Google Scholar]
- Binda M, Péli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C. 2009. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell 35: 563–573. 10.1016/j.molcel.2009.06.033 [DOI] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol 154: 164–175. 10.1016/0076-6879(87)54076-9 [DOI] [PubMed] [Google Scholar]
- Brennan L, Owende P. 2010. Biofuels from microalgae: a review of technologies for production, processing, and extractions of biofuels and co-products. Renew Sustain Energy Rev 14: 557–577. 10.1016/j.rser.2009.10.009 [DOI] [Google Scholar]
- Breuer G, Lamers PP, Martens DE, Draaisma RB, Wijffels RH. 2012. The impact of nitrogen starvation on the dynamics of triacylglycerol accumulation in nine microalgae strains. Bioresour Technol 124: 217–226. 10.1016/j.biortech.2012.08.003 [DOI] [PubMed] [Google Scholar]
- Brown MP, Grundy WN, Lin D, Cristianini N, Sugnet CW, Furey TS, Ares M Jr, Haussler D. 2000. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc Natl Acad Sci 97: 262–267. 10.1073/pnas.97.1.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Müller JD, Ruan Q, Gratton E. 2002. Molecular brightness characterization of EGFP in vivo by fluorescence fluctuation spectroscopy. Biophys J 82: 133–144. 10.1016/S0006-3495(02)75380-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway MK, Grunwald D, Heideman W. 2012. Glucose, nitrogen, and phosphate repletion in Saccharomyces cerevisiae: common transcriptional responses to different nutrient signals. G3 (Bethesda) 2: 1003–1017. 10.1534/g3.112.002808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie E, Guo X, Christiano R, Chitraju C, Kory N, Harrison K, Haas J, Walther TC, Farese RV Jr. 2014. High confidence proteomic analysis of yeast LDs identifies additional droplet proteins and reveals connections to dolichol synthesis and sterol acetylation. J Lipid Res 55: 1465–1477. 10.1194/jlr.M050229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot PW, Hellingwerf KJ, Klis FM. 2003. Genome-wide identification of fungal GPI proteins. Yeast 20: 781–796. 10.1002/yea.1007 [DOI] [PubMed] [Google Scholar]
- Du Y, Pattnaik AK, Song C, Yoo D, Li G. 2012. Glycosyl-phosphatidylinositol (GPI)-anchored membrane association of the porcine reproductive and respiratory syndrome virus GP4 glycoprotein and its co-localization with CD163 in lipid rafts. Virology 424: 18–32. 10.1016/j.virol.2011.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger TE, Brandl CJ, Struhl K, Harrison SC. 1992. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: crystal structure of the protein-DNA complex. Cell 71: 1223–1237. 10.1016/S0092-8674(05)80070-4 [DOI] [PubMed] [Google Scholar]
- Gao Q, Binns DD, Kinch LN, Grishin NV, Ortiz N, Chen X, Goodman JM. 2017. Pet10p is a yeast perilipin that stabilizes lipid droplets and promotes their assembly. J Cell Biol 216: 3199–3217. 10.1083/jcb.201610013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257. 10.1091/mbc.11.12.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. 2003. Global analysis of protein expression in yeast. Nature 425: 737–741. 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68: 1077–1090. 10.1016/0092-8674(92)90079-R [DOI] [PubMed] [Google Scholar]
- Goodsell DS, Olson AJ. 2000. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct 29: 105–153. 10.1146/annurev.biophys.29.1.105 [DOI] [PubMed] [Google Scholar]
- Gould SJ, Keller GA, Schneider M, Howell SH, Garrard LJ, Goodman JM, Distel B, Tabak H, Subramani S. 1990. Peroxisomal protein import is conserved between yeast, plants, insects and mammals. EMBO J 9: 85–90. 10.1002/j.1460-2075.1990.tb08083.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick-Davis K, Grinde E, Cowan A, Mazurkiewicz JE. 2013. Fluorescence correlation spectroscopy analysis of serotonin, adrenergic, muscarinic, and dopamine receptor dimerization: the oligomer number puzzle. Mol Pharmacol 84: 630–642. 10.1124/mol.113.087072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I, Jensen RE. 1991. Putting the HO gene to work: practical uses for mating-type switching. Methods Enzymol 194: 132–146. 10.1016/0076-6879(91)94011-Z [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183. 10.1038/415180a [DOI] [PubMed] [Google Scholar]
- Hong M, Fitzgerald MX, Harper S, Luo C, Speicher DW, Marmorstein R. 2008. Structural basis for dimerization in DNA recognition by Gal4. Structure 16: 1019–1026. 10.1016/j.str.2008.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton RM. 1997. In vitro recombination and mutagenesis of DNA. SOEing together tailor-made genes. Methods Mol Biol 67: 141–149. 10.1385/0-89603-483-6:141 [DOI] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798. 10.1016/S1097-2765(02)00496-3 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Jones CB, Ott EM, Keener JM, Curtiss M, Sandrin V, Babst M. 2012. Regulation of membrane protein degradation by starvation-response pathways. Traffic 13: 468–482. 10.1111/j.1600-0854.2011.01314.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Sekito T, Ohsumi Y. 2004. Autophagy in yeast: a TOR-mediated response to nutrient starvation. Curr Top Microbiol Immunol 279: 73–84. [DOI] [PubMed] [Google Scholar]
- Kanagavijayan D, Rajasekharan R, Srinivasan M. 2016. Yeast MRX deletions have short chronological life span and more triacylglycerols. FEMS Yeast Res 16: fov109 10.1093/femsyr/fov109 [DOI] [PubMed] [Google Scholar]
- Kerppola TK. 2008. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys 37: 465–487. 10.1146/annurev.biophys.37.032807.125842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kwon S, Nam SH, Jung JW, Kang M, Ryu J, Kim JE, Cheong JG, Cho CY, Kim S, et al. 2017. Dynamic and coordinated single-molecular interactions at TM4SF5-enriched microdomains guide invasive behaviors in 2- and 3-dimensional environments. FASEB J 31: 1461–1481. 10.1096/fj.201600944RR [DOI] [PubMed] [Google Scholar]
- Klopffleisch K, Phan N, Augustin K, Bayne RS, Booker KS, Botella JR, Carpita NC, Carr T, Chen JG, Cooke TR, et al. 2011. Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol Syst Biol 7: 532 10.1038/msb.2011.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643. 10.1038/nature04670 [DOI] [PubMed] [Google Scholar]
- Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. 2007. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol 17: 1561–1567. 10.1016/j.cub.2007.07.029 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lopez Garcia de Lomana A, Schauble S, Valenzuela J, Imam S, Carter W, Bilgin DD, Yohn CB, Turkarslan S, Reiss DJ, Orellana MV, et al. 2015. Transcriptional program for nitrogen starvation-induced lipid accumulation in Chlamydomonas reinhardtii. Biotechnol Biofuels 8: 207 10.1186/s13068-015-0391-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon R. 2003. Potassium channels. FEBS Lett 555: 62–65. 10.1016/S0014-5793(03)01104-9 [DOI] [PubMed] [Google Scholar]
- Malengo G, Andolfo A, Sidenius N, Gratton E, Zamai M, Caiolfa VR. 2008. Fluorescence correlation spectroscopy and photon counting histogram on membrane proteins: functional dynamics of the glycosylphosphatidylinositol-anchored urokinase plasminogen activator receptor. J Biomed Opt 13: 031215 10.1117/1.2940570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marianayagam NJ, Sunde M, Matthews JM. 2004. The power of two: protein dimerization in biology. Trends Biochem Sci 29: 618–625. 10.1016/j.tibs.2004.09.006 [DOI] [PubMed] [Google Scholar]
- Miller JP, Lo RS, Ben-Hur A, Desmarais C, Stagljar I, Noble WS, Fields S. 2005. Large-scale identification of yeast integral membrane protein interactions. Proc Natl Acad Sci 102: 12123–12128. 10.1073/pnas.0505482102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed AH, Chirala SS, Mody NH, Huang WY, Wakil SJ. 1988. Primary structure of the multifunctional α subunit protein of yeast fatty acid synthase derived from FAS2 gene sequence. J Biol Chem 263: 12315–12325. [PubMed] [Google Scholar]
- Murozuka E, Hanisch S, Pomorski TG, Jahn TP, Schjoerring JK. 2013. Bimolecular fluorescence complementation and interaction of various Arabidopsis major intrinsic proteins expressed in yeast. Physiol Plant 148: 422–431. 10.1111/j.1399-3054.2012.12000.x [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20: 87–90. 10.1038/nbt0102-87 [DOI] [PubMed] [Google Scholar]
- Nagiec MM, Wells GB, Lester RL, Dickson RC. 1993. A suppressor gene that enables Saccharomyces cerevisiae to grow without making sphingolipids encodes a protein that resembles an Escherichia coli fatty acyltransferase. J Biol Chem 268: 22156–22163. [PubMed] [Google Scholar]
- Niedziela-Majka A, Chesnik MA, Tomko EJ, Lohman TM. 2007. Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro. J Biol Chem 282: 27076–27085. 10.1074/jbc.M704399200 [DOI] [PubMed] [Google Scholar]
- Oelkers P, Cromley D, Padamsee M, Billheimer JT, Sturley SL. 2002. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J Biol Chem 277: 8877–8881. 10.1074/jbc.M111646200 [DOI] [PubMed] [Google Scholar]
- Onodera J, Ohsumi Y. 2005. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem 280: 31582–31586. 10.1074/jbc.M506736200 [DOI] [PubMed] [Google Scholar]
- Parrou JL, Enjalbert B, Plourde L, Bauche A, Gonzalez B, Francois J. 1999. Dynamic responses of reserve carbohydrate metabolism under carbon and nitrogen limitations in Saccharomyces cerevisiae. Yeast 15: 191–203. [DOI] [PubMed] [Google Scholar]
- Pastor-Cantizano N, Montesinos JC, Bernat-Silvestre C, Marcote MJ, Aniento F. 2016. p24 family proteins: key players in the regulation of trafficking along the secretory pathway. Protoplasma 253: 967–985. 10.1007/s00709-015-0858-6 [DOI] [PubMed] [Google Scholar]
- Pelham HR, Hardwick KG, Lewis MJ. 1988. Sorting of soluble ER proteins in yeast. EMBO J 7: 1757–1762. 10.1002/j.1460-2075.1988.tb03005.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittet M, Conzelmann A. 2007. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1771: 405–420. 10.1016/j.bbalip.2006.05.015 [DOI] [PubMed] [Google Scholar]
- Randez-Gil F, Sanz P, Entian KD, Prieto JA. 1998. Carbon source-dependent phosphorylation of hexokinase PII and its role in the glucose-signaling response in yeast. Mol Cell Biol 18: 2940–2948. 10.1128/MCB.18.5.2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Young BP, Espenshade PJ, Loewen C Jr. 2012. Regulation of lipid metabolism: a tale of two yeasts. Curr Opin Cell Biol 24: 502–508. 10.1016/j.ceb.2012.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigler R, Mets Ü, Widengren J, Kask P. 1993. Fluorescence correlation spectroscopy with high count rate and low background: analysis of translational diffusion. Eur Biophys J 22: 169–175. 10.1007/BF00185777 [DOI] [Google Scholar]
- Rue SM, Mattei S, Saksena S, Emr SD. 2008. Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol Biol Cell 19: 475–484. 10.1091/mbc.e07-07-0694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffarian S, Li Y, Elson EL, Pike LJ. 2007. Oligomerization of the EGF receptor investigated by live cell fluorescence intensity distribution analysis. Biophys J 93: 1021–1031. 10.1529/biophysj.107.105494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlecht U, Miranda M, Suresh S, Davis RW, St Onge RP. 2012. Multiplex assay for condition-dependent changes in protein–protein interactions. Proc Natl Acad Sci 109: 9213–9218. 10.1073/pnas.1204952109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze U, Liden G, Nielsen J, Villadsen J. 1996. Physiological effects of nitrogen starvation in an anaerobic batch culture of Saccharomyces cerevisiae. Microbiology 142: 2299–2310. 10.1099/13500872-142-8-2299 [DOI] [PubMed] [Google Scholar]
- Schweizer M, Roberts LM, Holtke HJ, Takabayashi K, Hollerer E, Hoffmann B, Muller G, Kottig H, Schweizer E. 1986. The pentafunctional FAS1 gene of yeast: its nucleotide sequence and order of the catalytic domains. Mol Gen Genet 203: 479–486. 10.1007/BF00422073 [DOI] [PubMed] [Google Scholar]
- Sherman F. 2002. Getting started with yeast. Methods Enzymol 350: 3–41. 10.1016/S0076-6879(02)50954-X [DOI] [PubMed] [Google Scholar]
- Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. 1998. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells 3: 145–156. 10.1046/j.1365-2443.1998.00176.x [DOI] [PubMed] [Google Scholar]
- Snider J, Hanif A, Lee ME, Jin K, Yu AR, Graham C, Chuk M, Damjanovic D, Wierzbicka M, Tang P, et al. 2013. Mapping the functional yeast ABC transporter interactome. Nat Chem Biol 9: 565–572. 10.1038/nchembio.1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung MK, Huh WK. 2007. Bimolecular fluorescence complementation analysis system for in vivo detection of protein–protein interaction in Saccharomyces cerevisiae. Yeast 24: 767–775. 10.1002/yea.1504 [DOI] [PubMed] [Google Scholar]
- Sung MK, Ha CW, Huh WK. 2008. A vector system for efficient and economical switching of C-terminal epitope tags in Saccharomyces cerevisiae. Yeast 25: 301–311. 10.1002/yea.1588 [DOI] [PubMed] [Google Scholar]
- Sung MK, Lim G, Yi DG, Chang YJ, Yang EB, Lee K, Huh WK. 2013. Genome-wide bimolecular fluorescence complementation analysis of SUMO interactome in yeast. Genome Res 23: 736–746. 10.1101/gr.148346.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. 2008. An in vivo map of the yeast protein interactome. Science 320: 1465–1470. 10.1126/science.1153878 [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. 2000. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627. 10.1038/35001009 [DOI] [PubMed] [Google Scholar]
- van Berkel MA, Caro LH, Montijn RC, Klis FM. 1994. Glucosylation of chimeric proteins in the cell wall of Saccharomyces cerevisiae. FEBS Lett 349: 135–138. 10.1016/0014-5793(94)00631-8 [DOI] [PubMed] [Google Scholar]
- Yogurtcu ON, Johnson ME. 2018. Cytosolic proteins can exploit membrane localization to trigger functional assembly. PLoS Comput Biol 14: e1006031 10.1371/journal.pcbi.1006031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. 2008. High-quality binary protein interaction map of the yeast interactome network. Science 322: 104–110. 10.1126/science.1158684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Danquah MK, Chen XD, Lu Y. 2011. Microalgae bioengineering: from CO2 fixation to biofuel production. Renew Sustain Energy Rev 15: 3252–3260. 10.1016/j.rser.2011.04.014 [DOI] [Google Scholar]
- Zhang T, Peli-Gulli MP, Yang H, De Virgilio C, Ding J. 2012. Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the EGO complex to activate TORC1. Structure 20: 2151–2160. 10.1016/j.str.2012.09.019 [DOI] [PubMed] [Google Scholar]
- Zordan RE, Beliveau BJ, Trow JA, Craig NL, Cormack BP. 2015. Avoiding the ends: internal epitope tagging of proteins using transposon Tn7. Genetics 200: 47–58. 10.1534/genetics.114.169482 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.