This review provides an overview of the influence of the gut microbiome on host health with a focus on immunomodulation and discusses strategies for manipulating the gut microbiome for the management or prevention of chronic inflammatory conditions.
Abstract
Over the past decade, our view of human-associated microbes has expanded beyond that of a few species toward an appreciation of the diverse and niche-specialized microbial communities that develop in the human host with chronological age. The largest reservoir of microbes exists in the distal gastrointestinal tract, both in the lumen, where microbes facilitate primary and secondary metabolism, and on mucosal surfaces, where they interact with host immune cell populations. While local microbial-driven immunomodulation in the gut is well described, more recent studies have demonstrated a role for the gut microbiome in influencing remote organs and mucosal and hematopoietic immune function. Unsurprisingly, therefore, perturbation to the composition and function of the gut microbiota has been associated with chronic diseases ranging from gastrointestinal inflammatory and metabolic conditions to neurological, cardiovascular, and respiratory illnesses. Considerable effort is currently focused on understanding the natural history of microbiome development in humans in the context of health outcomes, in parallel with improving our knowledge of microbiome–host molecular interactions. These efforts ultimately aim to develop effective approaches to rehabilitate perturbed human microbial ecosystems as a means to restore health or prevent disease. This review details the role of the gut microbiome in modulating host health with a focus on immunomodulation and discusses strategies for manipulating the gut microbiome for the management or prevention of chronic inflammatory conditions.
Introduction to the field of microbiome research
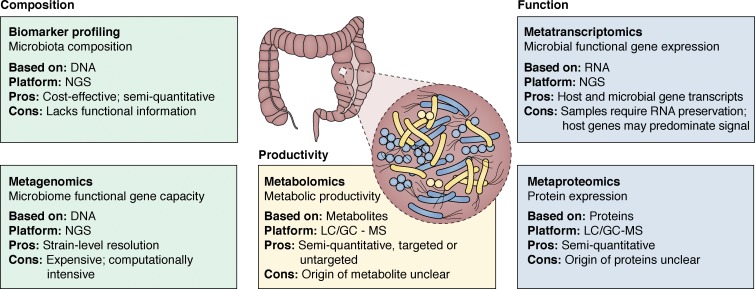
Over evolutionary time, humans have developed symbiotically with a diversity of environmental microbial species (Sender et al., 2016). Appreciation of the diversity of human-associated microbes, their functional gene capacity, and the breadth of biochemicals they generate in and on the human body has only relatively recently been appreciated, in large part due to advances in next-generation, high-throughput sequencing and mass-spectrometry platforms (Fig. 1). Sequencing platforms permit assessment of microbiota composition via biomarker gene sequencing, for example, sequencing and classification of 16S ribosomal RNA or the interspacer region amplicon pools to assess the identity and distribution of bacteria or fungi, respectively. These communities may then be interrogated to identify signature microbiota features associated with indications of interest. As the field moves toward a functional understanding of the human microbiome, efforts to investigate the functional gene capacity and transcriptional activity of microbiomes have advanced via shotgun sequencing of extracted DNA (metagenomics) or RNA (metatranscriptomics/RNA Seq), respectively. In parallel, detection and identification of microbiome-associated products, including metabolites and proteins, have been facilitated by targeted and untargeted mass spectrometry. These advances in high-throughput technologies have not only facilitated analyses of the composition and function of human-associated microbiomes, but they have also begun to reveal a diverse array of microbial-derived products that facilitate interspecies interactions in the human host (Rooks and Garrett, 2016; Cohen et al., 2017).
Figure 1.
Tools for analyses of the human gut microbiome. Microbiome studies are facilitated by next-generation sequencing (NGS) and liquid/gas chromatography (LC/GC) mass spectrometry (MS) platforms that permit analysis of composition, function, and productivity of the microbiome. Ideally, these approaches are applied in parallelto provide the most comprehensive view of host microbiomes.
Niche specificity of the microbiome
Through the application of high-throughput assays, it has become apparent that the healthy human superorganism comprises a series of niche-specific microbiomes across distinct body habitats (Costello et al., 2009; Caporaso et al., 2011). On the skin, for example, factors such as substrate and water availability as well as pH strongly correlate with the presence of habitat-specific microbiome compositions that are relatively consistent across healthy individuals (Oh et al., 2014). This is also true of the gastrointestinal (GI) tract, where distinct microbiome assemblies exist in various intestinal compartments (Donaldson et al., 2016). According to a recent study of infant microbiome development at multiple body sites, microbial niche specificity is evident as early as 6 wk of age (Chu et al., 2017), implicating early life factors in shaping niche-specific microbial assembly in infancy. Among the factors influencing early-life microbiome assembly is pioneer microbial colonization, which likely influences niche-specific microbiome development via local nutrient utilization and/or production of molecules that dictate interspecies compatibility and competitive colonization (Livingston et al., 2012; Verster et al., 2017; Ferretti et al., 2018).
Microbes are masters at sensing and responding to their environment; this is achieved via production of a range of biochemicals, from small diffusible quorum-sensing molecules that facilitate microbe–microbe–host communications (Schaefer et al., 2008; Papenfort and Bassler, 2016) to complex natural products such as macrolides and polyketides, many of which have potent antimicrobial and immunomodulatory activities (Donia and Fischbach, 2015). This biochemical arsenal facilitates microbial competitive colonization, selective inclusion or exclusion, and modification of the activities of other species in the local environment (Fan et al., 2015; Rangan et al., 2016; Thiemann et al., 2017). This concept was elegantly demonstrated in a recent study in which oral supplementation of antimicrobial-treated mice with a consortium of four bacterial species (Bacteroides sartorii, Parabacteroides distasonis, and Clostridium cluster XIVa members Clostridium bolteae and Blautia producta) prevented colonization by vancomycin-resistant Enterococcus faecium (VRE; Caballero et al., 2017). Critically, multilevel cooperation between bacterial species within this consortium was necessary to support colonization of the murine intestine with B. producta, the species that directly inhibited growth of pathogenic VRE. Such instances of microbial competitive colonization may be enhanced by microbial manipulation of host immunity, which acts as a reinforcing selective pressure on microbes in a given niche. Indeed, a recent study of the skin microbiota supports this concept; early life introduction of Staphylococcus epidermidis to the developing hair follicles of mice induced a wave of regulatory T (T reg) cells (Scharschmidt et al., 2015, 2017) and facilitated competitive colonization by S. epidermidis to the exclusion of other bacterial species (Naik et al., 2015; Nakatsuji et al., 2017). In healthy humans, relatively stable, niche-specific microbiomes have been described (Oh et al., 2014; Donaldson et al., 2016; Proctor and Relman, 2017), reinforcing the notion that local microenvironmental conditions, and perhaps a small number of keystone organisms that induce specific nutritional and/or immunological conditions at distinct body sites, promote niche-specialized microbiomes.
Variability of the gut microbiome
Although at higher levels of classification, distributions of organisms at specific body sites are highly conserved, there remains a high degree of interindividual variability at the species and strain level—an important feature rarely captured in biomarker-based microbiota profiling studies. Several studies have reported associations between host genetics and the microbiome (Blekhman et al., 2015; Bonder et al., 2016; Turpin et al., 2016). In mice, the relationships between host genotype and the gut microbiota have been demonstrated in genetically distinct mouse strains, in which independent quantitative trait loci analysis revealed 169 joint quantitative trait loci intervals that were significantly associated with the abundance of specific microbial taxa in the gut (Snijders et al., 2016). However, in humans, the relationship between host genotype and the microbiota appears less strong. While there is relatively greater similarity in microbial community membership within the intestinal microbiome among family members (Schloss et al., 2014; Korpela et al., 2018) and in monozygotic, compared with dizygotic, twins (Goodrich et al., 2014), this relative similarity may be confounded by common environmental exposures. Indeed, divergence of microbial functional modules in the gut microbiota (Xie et al., 2016), in parallel with changes in immune status (Brodin et al., 2015), have been observed when monozygotic twins reside in distinct environments. A more recent study of >1,000 subjects corroborated this observation by demonstrating that environmental exposures, including diet, dominate genetic factors in shaping gut microbiota (Rothschild et al., 2018). Moreover, inclusion of microbial factors improved prediction accuracy for many host traits, including glucose and obesity measures, compared with models that exclusively used host genetics and environmental data (Rothschild et al., 2018). Thus, combinatorial algorithms that consider microbial and host genetics in addition to environmental exposures may offer the most prediction accuracy for a range of host traits.
The influence of diet on microbiota composition and function is well established (David et al., 2014; Desai et al., 2016; Sonnenburg et al., 2016; Wu et al., 2016; Kamiya et al., 2018). Beyond diet, a number of other factors have been associated with variability in microbiota composition or function, including sex hormones (Fransen et al., 2017), treatment with antibacterial or antifungal agents (Reijnders et al., 2016; Wheeler et al., 2016), pharmaceuticals such as proton pump inhibitors (Imhann et al., 2016), xenobiotics (Maurice et al., 2013), environmental toxicants (Lu et al., 2014; Allais et al., 2016), and the number of prescription drugs consumed (Ticinesi et al., 2017). Emerging evidence suggests that these factors collectively shape the gut microbiome throughout an individual’s lifespan, resulting in a unique and personalized microbial fingerprint (Franzosa et al., 2015).
Development of the human gut microbiome
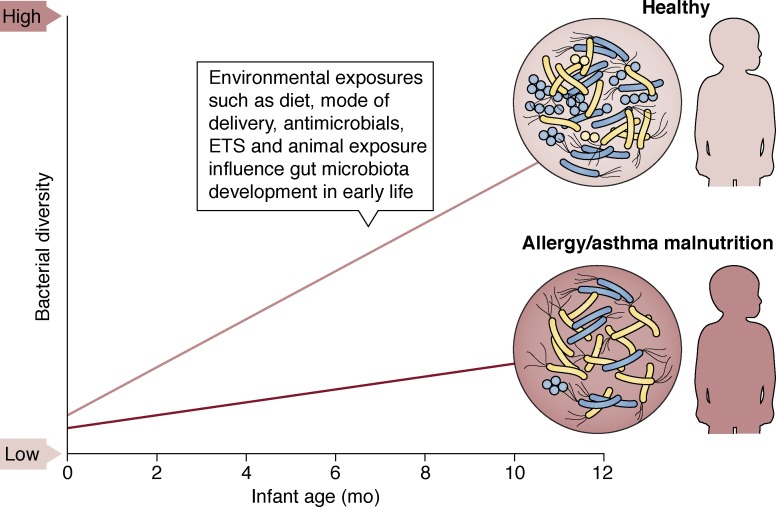
There is growing evidence that exposure to microbes begins in utero, based on studies using DNA-based assays that detected bacteria in placental material (Aagaard et al., 2014), amniotic fluid (Collado et al., 2016), and meconium, which forms in utero (Gosalbes et al., 2013; Nagpal et al., 2016; Durack et al., 2018; Ferretti et al., 2018). Although these studies are limited by the fact that DNA-based assays do not provide evidence of viable microbial cells, they offer initial indications that human–microbial encounters commence during gestation. Following delivery, the repertoire of bacteria (Yatsunenko et al., 2012; Durack et al., 2018; Stokholm et al., 2018) and both prokaryotic and eukaryotic viruses (Lim et al., 2015) expands, increasing the functional gene capacity of the gut microbiome. In parallel, fungal diversity contracts, likely owing to unfavorable selective pressures in the developing gut (Fujimura et al., 2016). A number of factors, including the mode of delivery (Bäckhed et al., 2015; Levin et al., 2016; Korpela et al., 2018), early-life diet (Bäckhed et al., 2015; Bokulich et al., 2016; Stokholm et al., 2016), antibiotic use (Bokulich et al., 2016), pet exposure (Fujimura et al., 2016; Levin et al., 2016; Durack et al., 2018), sex (Fujimura et al., 2016), and maternal health (Chu et al., 2017; Durack et al., 2018; Stokholm et al., 2018) have been linked to distinct gut microbiome compositions at discrete time points and to variation in microbial successional trajectories during this critical window of development (Fig. 2). Evidence that these early-life microbial colonization patterns and successional trajectories are relevant for subsequent health outcomes is growing. For example, 1-mo-old infants with altered microbiota composition and metabolic function were found to be at significantly higher risk of developing atopy (allergic sensitization) and asthma in childhood (Fujimura et al., 2016). In addition, delayed gut microbiome diversification over the first year of life has been linked to allergy, asthma (Durack et al., 2018; Stokholm et al., 2018), and malnutrition (Subramanian et al., 2014).
Figure 2.
The infant gut bacterial microbiome rapidly diversifies over the first year of life in healthy infants but is delayed in those who develop allergy or asthma or who are malnourished. A number of pre-, peri-, and postnatal environmental exposures are known to modulate risk for childhood disease, e.g., formula feeding, antimicrobial use, and exposure to environmental tobacco smoke (ETS) or animals. These same exposures also relate to gut microbiome composition at discrete developmental time points and to successional trajectories in early life.
The rapid expansion of bacterial diversity observed in the first year of life slows considerably by 3 yr of age (Yatsunenko et al., 2012; Vatanen et al., 2016), and by 5 yr of age the composition of the gut microbiome becomes more stable, attributable, at least in part, to a large proportion of stably maintained Bacteroides. Nonetheless, the gut microbiome in children is less diverse and functionally distinct from that of healthy adults (Cheng et al., 2016). Gut microbiota complexity (number of taxa and functional genes) typically reaches adult levels by preadolescence (7–12 yr of age), but microbial communities at this age remain taxonomically and functionally distinct from those of adults, with relatively lower levels of Bacteroides and higher levels of Bifidobacterium (Hollister et al., 2015).
In adulthood, despite the taxonomic uniqueness and personal nature of the human gut microbiome (Faith et al., 2013), its functional attributes are relatively consistent across healthy adult populations (Turnbaugh et al., 2009). Some of the core functional pathways in the adult gut microbiome include those involved in carbohydrate and amino acid metabolism, fermentation, and oxidative phosphorylation (Turnbaugh et al., 2009; Qin et al., 2010; Yatsunenko et al., 2012). In the elderly, gut microbiomes become compositionally unstable and less diverse (Odamaki et al., 2016), a phenomenon that has been linked with increasing frailty (Jackson et al., 2016) and declining immune function (Claesson et al., 2012). In older populations, low gut microbiota richness, a proxy for loss of microbial species and their repertoire of functional genes, appears to be a predictor of mortality (Ticinesi et al., 2017), whereas enrichment of certain bacteria, including Akkermansia and Bifidobacterium, is associated with longevity (Biagi et al., 2016). A direct link between age-related gut microbiota changes and age-associated systemic inflammation has been established in a recent study, in which cohousing germ-free (GF) mice with old, but not young, microbiome-replete mice increased intestinal permeability and age-related inflammation (Thevaranjan et al., 2017). Thus, the evidence to date indicates that age-associated changes in the gut microbiome parallel fluctuations in immune status (Simon et al., 2015), and this represents an important consideration in studies evaluating human health or when considering microbiota manipulation for prevention or treatment of specific illnesses.
The gut microbiome in immunity and homeostasis
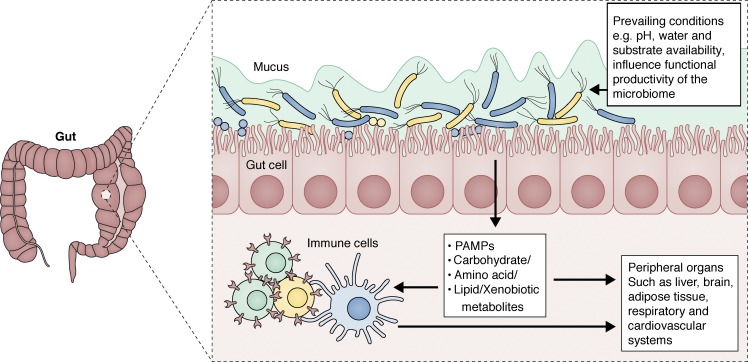
In healthy humans, the gut microbiome has coevolved to exist in a state of mutually beneficial symbiosis with its host (Fig. 3) and encodes a breadth of functional genes that dwarfs that of the human genome. A study cataloging functional genes in the human gut microbiome identified as many as 9.9 million unique microbial genes across 1,200 healthy subjects on three different continents (Li et al., 2014). Among the established microbial functions known to be expressed by human gut microbiomes are catalytic pathways for the metabolism of complex carbohydrates that produce short chain fatty acids (SCFAs; Table 1), anti-inflammatory, anti-proliferative lipids that represent an essential energy source for GI epithelial cells (Kelly et al., 2015). Other microbial-derived bioactive metabolites include essential vitamins, such as K (Karl et al., 2017) and B (Magnúsdóttir et al., 2015); hormones (Yatsunenko et al., 2012; Yan et al., 2016); neurological signaling molecules, such as serotonin (Yano et al., 2015; Romano et al., 2017), which may be derived via microbial metabolism of tryptophan (Table 1); and a large suite of other natural products, many of which have as-yet-undefined functions (Donia and Fischbach, 2015; Guo et al., 2017). Given the increasing appreciation for the diversity of functional genes encoded and bioactive biochemicals produced by the mammalian coevolved gut microbiome, it is unsurprising that critical aspects of physiological development have been linked to the microbiome. Indeed, juvenile GF mice commonly exhibit stunted growth and reduced somatotropic insulin-like growth factor 1 (IGF1) activity, which is restored by GI colonization by commensal bacteria (Schwarzer et al., 2016).
Figure 3.
In healthy adults, the gut microbiome exists in a state of mutual symbiosis with its host. The environment of the gut dictates both the composition and functional productivity of the adult gut microbiota, which may interact with the host through presentation of various ligands such as pathogen-associated molecular patterns (PAMPs) and production of metabolites, e.g., SCFAs. These molecules modulate immune homeostasis in the GI tract and at remote mucosal surfaces and organs via their entry into the circulation.
Table 1. Examples of gut microbiota–derived metabolites and their beneficial effects on human health.
| Metabolite/pathway | Microbial agent | Health benefits |
|---|---|---|
| Butyrate (carbohydrate metabolism) | Clostridia (clusters IV and XIVa) | Increased intestinal barrier function (Kelly et al., 2015; Zheng et al., 2017) |
| F. prausnitzii | Modulate intestinal macrophage function (Chang et al., 2014) | |
| Eubacterium spp. | Regulation of colonic T reg cell homeostasis (Furusawa et al., 2013; Smith et al., 2013) | |
| Roseburia spp. | Induction of tolerogenic DCs that polarize naive CD4+ T cells toward IL-10–producing T reg cells (Kaisar et al., 2017) | |
| Coprococcus catus | Suppression of colonic inflammation (Singh et al., 2014; Simeoli et al., 2017) | |
| Anaerostipes hadrus | Improvements in insulin sensitivity (Khan and Jena, 2016) | |
| Propionate (carbohydrate metabolism) | Bacteroides spp. | Regulation of colonic T reg cell homeostasis (Furusawa et al., 2013; Smith et al., 2013) |
| Blautia obeum | Suppression of colonic inflammation (Tong et al., 2016) | |
| C. catus | Decreased innate immune responses to microbial stimulation (Ciarlo et al., 2016) | |
| Roseburia inulinivorans | Protection from allergic airway inflammation (Trompette et al., 2014) | |
| P. copri | Improvements in insulin sensitivity and weight control in obese mice (den Besten et al., 2015) | |
| Alistipes putredinis | ||
| Dialister invisus | ||
| A. muciniphila | ||
| Eubacterium hallii | ||
| Indole (tryptophan metabolism) | A variety of bacteria possessing tryptophanase, including: | |
| Lactobacillus spp. | Maintenance of host–microbe homeostasis at mucosal surfaces via IL-22 (Zelante et al., 2013) | |
| B. longum | Increased barrier function (Bansal et al., 2010) | |
| B. fragilis | Modulation of host metabolism (Chimerel et al., 2014) | |
| P. distasonis | ||
| Clostridium bartlettii | ||
| E. hallii | ||
| E. coli | ||
| I3A (tryptophan metabolism) | Lactobacillus spp. | Maintenance of mucosal homeostasis and intestinal barrier function via increased IL-22 production (Zelante et al., 2013) |
| Protection against intestinal inflammation in mouse models of colitis (Lamas et al., 2016) | ||
| IPA (tryptophan metabolism) | Clostridium sporogenes | Maintenance of intestinal barrier function and mucosal homeostasis (Venkatesh et al., 2014) |
| Increased production of antioxidant and neuroprotectant products (Hwang et al., 2009) | ||
| HYA (lipid metabolism) | Lactobacillus spp. | Maintenance of intestinal barrier function (Miyamoto et al., 2015) |
| Decreased inflammation (Kaikiri et al., 2017) | ||
| Increased intestinal IgA production (Kaikiri et al., 2017) | ||
| CLA (lipid metabolism) | Lactobacillus spp. | Decreased inflammation (Viladomiu et al., 2016) |
| Bifidobacterium spp. | Reduced adiposity (Segovia et al., 2017) | |
| F. prausnitzii | Improved insulin sensitivity (Garibay-Nieto et al., 2017) |
CLA, conjugated linoleic acid; HYA, 10-hydroxy-cis-12-octadecoate (linoleic acid derivative); I3A, indole-3-aldehyde; IPA, indole-3-propionate.
Of equal importance to the microbiome’s effects on host physiology is its role in the development and maintenance of local (Hooper et al., 2001; Levy et al., 2015) and systemic innate and adaptive immune function (Schirmer et al., 2016). Immune dysfunction is increasingly identified as a component of a range of chronic diseases, including obesity (Winer et al., 2016), metabolic syndrome (Richard et al., 2017), multiple sclerosis (MS; Kallaur et al., 2017), and autism spectrum disorders (ASD; Careaga et al., 2017), among others. Notably, all of these disorders have also been shown to associate with perturbations of the gut microbiome, generally associated with loss of microbial diversity and, particularly, with depletion of specific bacteria, including Akkermansia and Faecalibacterium, that are thought to promote immune tolerance (Sokol et al., 2008; Schneeberger et al., 2015; Rossi et al., 2016; Ottman et al., 2017). Evidence for the key role of microbes in shaping immune function is illustrated in studies of GF mice, which have underdeveloped innate and adaptive immune systems (Khosravi et al., 2014) and reduced expression of antimicrobial peptides (Natividad et al., 2013) and IgA (Hapfelmeier et al., 2010) necessary for clearance of pathogenic microbes from the gut lumen (Moor et al., 2017). Additionally, GF animals possess fewer T reg cells (Atarashi et al., 2011; Ohnmacht et al., 2015), lack regulatory gut CD4+CD8αα+ double-positive intra-epithelial lymphocytes (Cervantes-Barragan et al., 2017), have compromised innate lymphoid cell function (Hepworth et al., 2013), and are more susceptible to microbial infections. Thus, the confluence of data from human and murine studies indicates that the gut microbiome plays a key role in promoting appropriate physiological and immune development.
Both the diversity and redundancy in immune system activation by human gut microbial symbionts is well accepted (Geva-Zatorsky et al., 2017). A variety of gut bacteria, including Bifidobacterium adolescentis and segmented filamentous bacteria, have been identified as key modulators of GI T helper 17 (Th17) cells (Ivanov et al., 2009; Tan et al., 2016; Geva-Zatorsky et al., 2017), instrumental in the maintenance of barrier function and protection against pathogenic microbes (Pandiyan et al., 2011; Wang et al., 2014b). Clostridium species, including those belonging to clusters IV, XIVa, and XVIII, induce CD4+Foxp3+ T reg cells, via production of SCFAs (Atarashi et al., 2013; Sefik et al., 2015; Geva-Zatorsky et al., 2017). Surface polysaccharide A of Bacteroides fragilis has been shown to bind to Toll-like receptor 2 on dendritic cells (DCs), which then induce the production of the anti-inflammatory cytokine IL-10 by T reg cells (Dasgupta et al., 2014) and promote immune tolerance (An et al., 2014). Although the field is still nascent, it is already clear that a broad range of microbial species and their associated products modulate discrete features of adaptive and innate immunity, which offers a novel opportunity for microbial-based approaches for prevention or treatment of inflammatory conditions.
Early-life microbiome and immune maturation
The evidence that microbes are important symbiotic modulators of physiological, metabolic, and immunological function in the mammalian host has expanded over the past decade (Lloyd-Price et al., 2016). Thus, the early-life gut microbiome has formed the focus of intense research (Chung et al., 2012; Schnupf et al., 2015; Gensollen et al., 2016; Ferretti et al., 2018; Pan et al., 2018). Recent data indicate that the developing gut microbiota of human infants affects the progression of intestinal mucosal IgA responses (Planer et al., 2016), and perturbations to these nascent microbial communities cause long-lasting metabolic and immunological dysregulation (Ruiz et al., 2017; Lynn et al., 2018). For instance, macrolide treatment of conventional, neonatal mice perturbs their gut microbiota, with depletion of Bifidobacterium and segmented filamentous bacteria, resulting in decreased numbers of intestinal CD4+IL-17A+ lymphocytes and fecal IgA concentrations, persistent features not observed in neonatal GF or conventional adult mice (Ruiz et al., 2017).
Development of the gut microbiome in early life appears to adhere to ecological principles of primary succession, in which the initial (pioneer) organisms colonize a previously pristine habitat, influence ecosystem conditions, and dictate subsequent accumulation of species in the niche via competitive colonization. Initial colonization by distinct microbial species in the nascent human gut may thereby induce discrete immune and physiological conditions (Fig. 2) that affect health outcomes later in life. Support for this concept comes from recent studies showing, for example, that risk of childhood atopy and asthma was linked to the composition and metabolic activity of the gut microbiome at 1 mo of age (Fujimura et al., 2016). Longitudinal birth cohort studies, such as a large study of Swedish mother–infant dyads, have also indicated that infants delivered by cesarean section and formula-fed in early life exhibit distinct neonatal gut microbiome features that persist throughout the first year of life (Bäckhed et al., 2015) and significantly reduce vertical transmission (Korpela et al., 2018) compared with infants born vaginally. The observation that cesarean section and formula-fed babies develop along a distinct microbial successional trajectory suggests that founder microbial populations that develop in utero could be usurped by more competitive microbial strains encountered in early postnatal life, whose colonization is reinforced by the selective effects of diet. Likewise, babies at high risk for asthma development exhibit distinct bacterial microbiota in their meconium and delayed diversification of their gut microbiota over the first year of life (Durack et al., 2018). Encouragingly, this delayed diversification was partially remedied with daily, postpartum Lactobacillus rhamnosus supplementation, which was associated with a restructured gut microbiota composition and metabolic output more consistent with that of low-risk infants (Durack et al., 2018), providing evidence that gut microbiota development and metabolism may be reengineered via microbial intervention. However, this commercially available probiotic does not engraft in the human gut microbiome, and the beneficial effects of supplementation were lost upon cessation of probiotic administration. Although a relatively new concept in terms of applicability to the human microbiome, recent data in the field suggest that ecological principles may offer a useful theoretical framework to understand human microbiome development in early life. Improved understanding of microbial competitive colonization, engraftment, and succession during specific stages of human development may inform rational design of microbiome-targeted therapeutics aimed at reengineering microbial community composition and metabolic output, and ultimately at preventing chronic disease in later life.
Microbiome perturbations, immune dysfunction, and chronic disease
Inflammatory bowel disease (IBD)
Perturbations to the composition and function of bacterial and fungal gut microbiota have been implicated in various forms of IBD, including Crohn’s disease (CD) and ulcerative colitis (UC; Sokol et al., 2008, 2017; Kudelka et al., 2016; Lamas et al., 2016; Mar et al., 2016; Schirmer et al., 2018). Both conditions exhibit loss of enteric bacterial diversity (Gevers et al., 2014; Mar et al., 2016) and expansion of specific bacterial clades such as Enterobacteriaceae (Gevers et al., 2014). Expansion of Enterobacteriaceae has been associated with new-onset CD, and targeted depletion of this bacterial family ameliorates intestinal inflammation in mice (Zhu et al., 2018). Additionally, loss of certain symbiotic taxa such as Faecalibacterium prausnitzii has been related to recurrence of CD, and supplementation of mice with this organism reduced inflammation in a model of chemically induced colitis (Sokol et al., 2008), suggesting an anti-inflammatory role for this mucosal-associated enteric symbiont. More recently, a bioactive microbial anti-inflammatory molecule was identified in supernatant of cultured F. prausnitzii (Quévrain et al., 2016) and is thought to be responsible, at least partly, for reducing intestinal inflammation. Indeed, supplementing mice with Lactococcus lactis expressing the F. prausnitzii microbial anti-inflammatory molecule alleviated intestinal inflammation (Quévrain et al., 2016; Breyner et al., 2017). A number of studies have indicated that helminth infection abrogates IBD (Broadhurst et al., 2010), including one study showing that helminth infection promotes the diversification of gut microbiota and enrichment of anti-inflammatory Clostridiales in mice genetically predisposed to disease (Ramanan et al., 2016). Although there is limited evidence that helminth introduction ameliorates IBD in humans, these observations support the concept that enteric bacterial microbiota manipulation holds potential for the management or prevention of IBD.
Microbial-derived products found in stool of patients with UC have been shown to promote inflammation. Human DCs stimulated with fecal water from the stool of patients with UC increased the ratio of Th2 to Th1 cells upon coculture with autologous naive T cells (Mar et al., 2016). The degree of Th2 induction was related to the specific fecal microbiota composition and to the degree of disease severity (Mar et al., 2016). Specifically, microbiota associated with the most severely ill patients exhibited expansions of Bacteroides and Candida (Mar et al., 2016), which were associated with a distinct profile of microbial ligands and metabolic products in the fecal microbiome of these patients. Additionally, independent studies have indicated that both the gut microbiota of patients with IBD and that of colitis-susceptible Card9−/− mice exhibit reductions in the concentration of tryptophan-derived indole derivatives that act as aryl hydrocarbon receptor ligands and promote IL-22 production (Lee et al., 2011; Lamas et al., 2016). Transfer of feces from IL-22−/− (Zenewicz et al., 2013) or Card9−/− mice to wild-type GF recipients increased their susceptibility to colitis (Lamas et al., 2016), and supplementation with three Lactobacillus strains capable of metabolizing tryptophan, or treatment with an aryl hydrocarbon receptor agonist, restored intestinal IL-22 production and attenuated intestinal inflammation (Lamas et al., 2016). The protective effect of Lactobacillus was also observed in another experimental model of colitis, in which Lactobacillus murinus enrichment was associated with expansion of T reg cells and protection (Tang et al., 2015). Hence dysbiosis associated with loss of intestinal bacteria capable of modulating intestinal inflammation further suggests that microbiome manipulation, particularly supplementation with strains that restore depleted microbial functions in diseased patients, represents a plausible avenue of therapeutic intervention.
Persistent antibiotic-induced colitis
Antibiotic-induced alterations in the gut microbiome and its associated metabolome reduce colonization resistance against the spore-forming, toxin-producing pathogen Clostridium difficile (Theriot et al., 2014), a causative agent of antibiotic-associated colitis. Germination and overgrowth of this pathogen are thought to be facilitated by a reduction of microbiota-derived secondary bile acid metabolites that inhibit spore germination (Buffie et al., 2015). Using mathematical modeling guided by analysis of the microbiota of hospitalized patients, a recent study identified Clostridium scindens as an intestinal bacterial species that dehydroxylates bile acid 7α, thereby mediating colonization resistance against C. difficile (Buffie et al., 2015). These results are encouraging first steps for exploring novel preventive strategies, such as prophylactic microbial-based supplements, for populations at risk of developing antibiotic-associated colitis.
Atopic asthma
The rapid increase in asthma prevalence in industrialized nations over the past several decades, particularly among pediatric populations, cannot be explained by genetic risk factors alone and is thought to be related to altered environmental exposures associated with Western lifestyles. A number of validated epidemiological observations have implicated early life environmental exposures in increased risk for childhood asthma. Many of these exposures are known to shape the nascent gut microbiome, including cesarean birth (Dominguez-Bello et al., 2010; Jakobsson et al., 2014; Bäckhed et al., 2015; Stokholm et al., 2016; Chu et al., 2017), antimicrobial administration (Johnson et al., 2005; Korpela et al., 2016; Ahmadizar et al., 2017; Yamamoto-Hanada et al., 2017), formula feeding, exposure to furred pets, and environmental toxins (Levin et al., 2016; O’Connor et al., 2018).
Further evidence for an intricate relationship between environmental exposure, the gut microbiome, and allergic airway disease comes from an expanding body of work (Fujimura and Lynch, 2015; Durack et al., 2016), particularly those utilizing experimental animal models. For example, treating neonatal mice with vancomycin was shown to diminish gut microbial diversity, alter metabolite profiles, exacerbate Th2 responses, and increase susceptibility to allergic lung inflammation (Russell et al., 2012; Cait et al., 2018). Supplementation with SCFAs ameliorated airway inflammation in these mice, with the mechanism attributed to decreased activity of T cells and DCs, decreased numbers of IL-4–producing CD4+ T cells, and reduced levels of circulating IgE (Cait et al., 2018). Oral supplementation of mice with house dust from homes with dogs also reduced allergic airway sensitization to cockroach antigen (Fujimura et al., 2014), consistent with the observation that children who grow up with dogs are less likely to develop atopy (Ownby et al., 2002). The gut microbiota in mice treated with dog-associated house dust was found to be enriched with Lactobacillus johnsonii, and oral supplementation with this species protected mice against airway allergen challenge and respiratory viral infection, with reduced airway concentrations of IL-4, IL-5, IL-13, and IL-17 and increased numbers of T reg cells (Fujimura et al., 2014). More recently, protection mediated by oral L. johnsonii supplementation was linked to alterations in circulating immunomodulatory metabolites, including anti-inflammatory ω-3 fatty acids, such as docosahexanoic acid (Fonseca et al., 2017). That study attributed protection in part to reduced activation of DCs, as DCs exposed to serum from Lactobacillus-supplemented, respiratory syncytial virus–infected mice (or to docosahexanoic acid) dampened cytokine production by the DCs upon viral infection (Fonseca et al., 2017). Reduced allergic airway inflammation is also seen in mice fed a high-fiber diet, which is exclusively metabolized by microbes to produce SCFAs, with evidence for functional reprogramming of hematopoietic cells (Trompette et al., 2014). Collectively, these observations indicate that the gut microbiome is plastic and that its related metabolic products have the capacity to suppress inflammation at distal mucosal sites, such as the lungs, and to reprogram bone marrow–derived immune cells.
Early-life microbiota dysbiosis has also been observed in infants at risk for asthma development later in life. Bacterial depletion and fungal expansion were observed in the gut microbiome of at-risk infants at 1 mo of age, with a fecal metabolic profile characterized by depletion of ω-3 fatty acids and prostaglandin precursors (Fujimura et al., 2016). The soluble products of the gut microbiota from these high-risk infants induced Th2 cell expansion, increased IL-4 expression, and decreased T reg cell populations ex vivo, the latter attributed in part to enrichment of the oxylipin, 12,-13-DiHOME (Fujimura et al., 2016). The concept that microbial products associated with a disease-susceptible microbiome drive heritable immune dysfunction was supported by a study showing that fecal microbiota transfer from infants at high risk for asthma into GF mice resulted in exacerbated allergic lung inflammation in their adult offspring (Arrieta et al., 2015). Lung inflammation was alleviated when the offspring mice were supplemented with bacterial taxa depleted in the microbiota of high-risk infants, suggesting a causal role of these bacteria in averting allergic inflammation (Arrieta et al., 2015).
Observations from independent, geographically distinct birth cohorts encompassing various socioeconomic and racial groups consistently report depletion of symbiotic bacterial taxa, such as Faecalibacterium, Akkermansia, and Lachnospira, in infants at risk of developing atopy or asthma (Arrieta et al., 2015; Fujimura et al., 2016; Durack et al., 2018; Stokholm et al., 2018). These studies add to the mounting evidence that the early-life gut microbiome contributes to risk of childhood asthma development, highlighting the neonatal period as an opportune time for preventive microbial interventions in high-risk infants.
Behavioral disorders
The gut microbiota has recently emerged as an important mediator of biochemical signaling between the GI tract and the central nervous system, more commonly referred to as the gut–brain axis (Fung et al., 2017). As such, a number of behavioral disorders have been associated with perturbed enteric microbiota, including ASD in children (Li et al., 2017; Strati et al., 2017). ASD-associated gut microbiota exhibit increased bacterial diversity (Finegold et al., 2010) and enrichment of Collinsella, Corynebacterium, and Lactobacillus and the fungal pathobiont Candida relative to unaffected siblings (Strati et al., 2017). These microbial perturbations are associated with an altered fermentative profile, with significantly higher concentrations of SCFAs and ammonia (Wang et al., 2012), metabolites that at high concentrations are considered neurotoxic and may promote the adverse neurological effects associated with ASD (Morland et al., 2018).
Given that gut microbiota development in infancy has been linked to maternal health (Chu et al., 2017; Durack et al., 2018; Stokholm et al., 2018), it is perhaps unsurprising that maternal immune activation (Hsiao et al., 2013; Brown et al., 2014; Lee et al., 2015; Kim et al., 2017) and obesity (Buffington et al., 2016) during pregnancy are associated with features of ASD in offspring, with murine studies suggesting that these processes are governed by the maternal intestinal microbiota (Kim et al., 2017). Feeding pregnant mice a high-fat diet (HFD) induces behavioral alterations in their offspring, in which a number of bacterial taxa including Lactobacillus reuteri are depleted from the gut microbiota, and levels of oxytocin in the hypothalamus are decreased (Buffington et al., 2016). These behavioral changes could be reversed by cohousing the affected mice with those fed a regular diet or by supplementing them with L. reuteri (Buffington et al., 2016), a species that promotes oxytocin levels (Poutahidis et al., 2013).
Chronic stress, which is associated with despair behavior in mice, also alters gut microbiota composition. Chronically stressed mice show reduced fecal peroxide levels, L. reuteri depletion in their microbiota, and profound changes in serum metabolites, particularly those involved in the tryptophan–kynurenine metabolism pathway (Marin et al., 2017). These mice also have increased intestinal expression of IDO1, the main enzyme responsible for metabolizing tryptophan to kynurenine and thus preventing its conversion to serotonin. Supplementation with L. reuteri relieved signs of depression and restored fecal peroxide levels in stressed animals (Marin et al., 2017). An exploratory study in which fecal transfer was used to restore the perturbed gut microbiota in children with ASD showed promising improvements in GI and behavioral symptoms (Kang et al., 2017), although these effects need to be validated in larger trials. Collectively, these findings point to an important role for the gut microbiota in neuro-behavioral modulation and hint at the potential efficacy of microbial restoration as a means of treating or preventing behavioral disorders. However, much more work is needed to elucidate the underlying mechanisms before manipulation of the gut microbiota can be considered for prevention or management of these disorders.
Obesity and type 2 diabetes mellitus (T2DM)
Obesity and T2DM are interrelated and associated with dysbiotic gut microbiota (Qin et al., 2012; Karlsson et al., 2013; Le Chatelier et al., 2013; Forslund et al., 2015) and with significantly reduced bacterial complexity (Turnbaugh et al., 2009; Karlsson et al., 2013; Le Chatelier et al., 2013). A decrease in the relative abundance of Bacteroides spp. is particularly striking in obesity (Ley et al., 2005; Andoh et al., 2016). A causal role for the gut microbiome in obesity is supported by studies showing that GF mice reconstituted with feces from monozygotic twins discordant for obesity recapitulates the metabolic phenotype (including obesity status) of the donor (Ridaura et al., 2013). Bacterial species (specifically Bacteroides spp.) associated with the lean phenotype seemed to dominate over those associated with obesity, as cohousing mice harboring “lean” microbiota with those harboring “obese” microbiota prevented weight gain and obesity-associated metabolic phenotypes in the latter. Similarly, transplantation of fecal microbiota from healthy lean donors to obese patients resulted in improved insulin sensitivity (Vrieze et al., 2012). Insulin resistance has been linked to differences in the gut microbiome and microbe-associated increases in serum concentrations of branched-chain amino acids (Pedersen et al., 2016).
Conversely, an abundance of a mucin-degrading gut bacteria, Akkermansia muciniphila, is associated with increased metabolic health in obese patients (Dao et al., 2016) and in mice fed a HFD (Schneeberger et al., 2015). Supplementation with A. muciniphila of HFD-induced obese mice reduced circulating LPS levels, enhanced lipid oxidation (Everard et al., 2013), improved glucose tolerance, and reduced systemic inflammation (Shin et al., 2014). Prebiotic plant polyphenols that enrich for A. muciniphila showed similar beneficial effects (Anhê et al., 2015). The benefits of A. muciniphila supplementation have been attributed in part to a bacterial outer membrane protein that interacts with Toll-like receptor 2 and restores gut barrier function (Plovier et al., 2017). However, such activity is very likely strain-specific and dependent on the local environment and coassociated microbial peers, since enrichment of A. muciniphila has also been observed in diabetic Chinese patients compared with healthy controls (Qin et al., 2012).
Lastly, prebiotic fermentable dietary fiber has been shown to protect mice against HFD-induced metabolic syndrome in a microbiota-dependent manner (Schroeder et al., 2018; Zou et al., 2018) that was not dependent on SCFAs (Zou et al., 2018) but rather on microbiota-mediated restoration of IL-22 production and enterocyte function (Zou et al., 2018), leading to reduced intestinal permeability (Schroeder et al., 2018). Interestingly, supplementation of HFD-fed mice with Bifidobacterium longum also reversed mucus abnormalities by stimulation of mucin secretion (Schroeder et al., 2018), suggesting that probiotic and prebiotic treatments may prevent distinct HFD-induced defects in the intestinal mucus layer. Nondigestible fiber has been shown to induce clinically relevant metabolic improvements, coupled with expansion of SCFA-producing enteric microbial species, in Asian patients with T2DM (Zhao et al., 2018).
Cardiovascular disease
Microbial metabolism of dietary choline and carnitine, which comprise a large component of a Western diet, has been shown to increase risk of cardiovascular disease (Iqbal et al., 2008; Estruch et al., 2018). Metabolism of these compounds produces trimethylamine (TMA), which is oxidized in the liver to trimethylamine-N-oxide, an amine oxide associated with development of atherosclerosis (Wang et al., 2011, 2015; Koeth et al., 2013; Zhu et al., 2016). The production of TMA is catalyzed by microbial TMA lyase (Koeth et al., 2013), and inhibition of this enzyme reduced atherosclerotic plaque development in susceptible Apoe−/− mice (Wang et al., 2015). Additionally, supplementation of atherosclerosis-prone mice with A. muciniphila protected against atherosclerosis development induced by feeding a Western diet (Li et al., 2016a). Indeed, mice harboring high levels of choline-metabolizing bacteria are more susceptible to diet-induced metabolic disease (Romano et al., 2017). Modulation of microbial metabolism through dietary intervention or direct supplementation might therefore provide an effective strategy, either alone or in combination with established therapies, for prevention of cardiovascular diseases.
Autoimmunity
The frequency of autoimmune conditions has increased over the past several decades (Patterson et al., 2009; Hunter et al., 2017; Magyari and Koch-Henriksen, 2017). Molecular mimicry of human antigens by the microbiota is one potential mechanism by which the immune system is reprogrammed, resulting in autoimmune tissue damage (Cusick et al., 2012). Variation in microbial LPS immunogenicity has been proposed as one alternative facet that contributes to type 1 diabetes mellitus (T1DM) pathogenesis. In a study of Northern European infants, enrichment of Bacteroides spp., particularly Bacteroides dorei, was observed in the first three years of life in Finnish and Estonian infants, who exhibit a high prevalence of T1DM compared with Russian infants (Vatanen et al., 2016). Within this cohort, a positive correlation between the abundance of these bacteria at the phylum level and serum insulin autoantibody levels was observed. This association was attributable to the unique structure of LPS from genus Bacteroides that inhibited immune stimulation induced by Escherichia coli LPS, which the authors suggest might interfere with normal immune education and increase susceptibility to autoimmunity (Vatanen et al., 2016). In an independent human study, intestinal microbiota alterations, including loss of bacterial diversity, preceded the onset of metabolic symptoms associated with T1DM (Kostic et al., 2015). Additionally, antibiotic-induced dysbiosis altered microbial lipid metabolism and suppressed enteric Th17 and T reg cell populations, leading to increased incidence of T1DM-like disease in nonobese diabetic mice (Livanos et al., 2016).
Rheumatoid arthritis (RA) is a systemic autoimmune inflammatory condition that manifests in joint damage. The observation that GF mice are protected from development of experimental arthritis (Wu et al., 2010) suggests a possible role for the microbiome in the pathogenesis of this disease. Patients with RA show expansion of enteric Prevotella copri (Scher et al., 2013), and autoantigens in these patients exhibit high homology to Prevotella-associated peptides (Pianta et al., 2017). Molecular mimicry of RA-associated antigens by the gut microbiota has also been reported in a metagenomic study of RA patients (Zhang et al., 2015). However, whether precision editing of the microbiota could alleviate clinical symptoms in patients with RA remains to be elucidated.
Gut microbiota perturbation has also been described in adult (Cekanaviciute et al., 2017) and pediatric (Tremlett et al., 2016b) patients with MS. In pediatric MS patients, bacterial microbiota richness was positively associated with circulating pro-inflammatory Th17 cells, whereas the relative abundance of SCFA-producing Bacteroidetes inversely correlated with systemic inflammatory markers (Tremlett et al., 2016a). GF mice humanized with fecal microbiota from MS patients developed more severe experimental autoimmune encephalomyelitis associated with lower frequencies of T reg cells (Cekanaviciute et al., 2017). These initial observations implicate the gut microbiome in disease pathogenesis, but additional studies are necessary to delineate the specific microbial strains and products that confer these phenotypes.
Gut microbiota in cancer immunotherapy
Recent evidence has emphasized the importance of enteric microbiota in modulating response to various forms of cancer immunotherapy (Roy and Trinchieri, 2017; Zitvogel et al., 2018). Cancer patients who responded to anti–programmed cell death protein 1 (PD1) therapy have been shown to harbor more diverse gut microbiota compared with nonresponders (Gopalakrishnan et al., 2018; Routy et al., 2018), who were consistently depleted for bacterial taxa generally associated with health, including Akkermansia, Faecalibacterium, and Bifidobacterium (Gopalakrishnan et al., 2018; Matson et al., 2018; Routy et al., 2018). Improved efficacy of anti-PD1 treatment, with increased antitumor T cell responses, was seen in mice transplanted with fecal microbiota from responding patients (Matson et al., 2018) and in mice supplemented with bacterial taxa depleted in nonresponders (Routy et al., 2018). Although preliminary, these observations suggest that the gut microbiota might offer the opportunity to identify patients who are more likely to respond to treatment, to enhance existing therapeutics, and to develop novel therapeutic strategies.
The microbiome as therapeutic target
Given the tight interplay between enteric microbial symbionts and host immunity, efforts have focused on implementing various strategies targeting the gut microbiota to manage or prevent chronic inflammatory disease. Clinical approaches to modify gut microbiota generally focus on depleting overabundant members or overall microbial load using antibiotics or antifungal agents, modulation through diet manipulation, or supplementation with live microbes (single or mixed species). More recently, fecal microbial transplantation (FMT) has been used in a range of infectious, neurological, and GI conditions, with promising outcomes. Although antimicrobial drugs are not generally considered appropriate for long-term management of chronic conditions, given the need for repeat dosing and concerns about emergence of antimicrobial resistance, early data indicate that administration of antimicrobial drugs, as a means to perturb pathogenic microbiomes before FMT or microbial supplementation, enhances engraftment of beneficial species and improves treatment efficacy (Keshteli et al., 2017).
Dietary interventions
Although diet strongly shapes the composition and function of the gut microbiota, only a small number of controlled clinical dietary intervention studies targeting the human gut microbiota have been reported (Cotillard et al., 2013; Brahe et al., 2015; Thompson et al., 2017). Recently, a diet rich in fiber was shown to significantly improve glucose control and promote a healthier metabolic profile in T2DM patients (Zhao et al., 2018). Other studies showed marked interindividual variation in postprandial glycemic responses after consumption of identical meals (Zeevi et al., 2015). Consideration of microbial composition alongside known T2DM risk factors (e.g., body mass index, fasting glucose levels) enabled accurate glucose response prediction, thus allowing design of more effective, personalized diets for improved glycemic control (Zeevi et al., 2015).
Multispecies microbial supplements
Despite encouraging evidence from animal models in which inflammatory conditions were successfully treated via gut microbiota manipulation (Table 2), data from human trials is less conclusive. Findings from recent work showed that the ability of an introduced microbe to successfully colonize the gut depends on the structure of the resident gut microbiota (Zhang et al., 2016; Zmora et al., 2018), a factor that has so far been overlooked in microbial intervention–based clinical trials and that might explain discrepancies in animal models and human trials.
Table 2. Evidence for gut microbiota modulation strategies in preventing chronic inflammatory conditions derived from animal models.
| Bacterial strain | Chronic inflammatory condition |
|---|---|
| A. muciniphila | Ameliorates HFD-induced obesity and insulin resistance (Everard et al., 2013; Plovier et al., 2017) |
| Protects against atherosclerosis by decreasing gut permeability and preventing endotoxemia-induced inflammation (Li et al., 2016a) | |
| Improves efficacy of anti-cancer immunotherapy (Routy et al., 2018) | |
| B. fragilis | Restores the integrity of the intestinal barrier and ameliorates autistic-like behavior (Hsiao et al., 2013) |
| Suppresses neuro-inflammation in experimental model of MS (Wang et al., 2014a) | |
| B. adolescentis | Ameliorates HFD-induced colitis (Lim and Kim, 2017) |
| Bifidobacterium animalis spp. lactis | Restores the integrity of the intestinal barrier, thus reducing bacterial translocation and insulin sensitivity (Amar et al., 2011) |
| B. animalis spp. lactis and LGG | Improves HFD-induced obesity and insulin resistance (Alard et al., 2016) |
| B. longum | Reduces anxiety-like behavior in a mouse model of chemically induced colitis (Bercik et al., 2011) |
| Restores the integrity of colonic mucus layer impaired by HFD (Schroeder et al., 2018) | |
| Bifidobacterium pseudocatenulatum | Reduces weight gain, body fat, fasting glucose, and insulin resistance in mice with HFD-induced obesity (Zhao et al., 2018) |
| Christensenella minuta | Reduces weight gain in mice inoculated with obese human fecal samples (Goodrich et al., 2014) |
| C. scindens | Protects against antibiotic-induced C. difficile colitis (Buffie et al., 2015) |
| F. prausnitzii | Alleviates inflammation in chemically induced models of colitis (Sokol et al., 2008; Quévrain et al., 2016; Breyner et al., 2017) |
| L. johnsonii | Provides airway protection against allergen challenge and respiratory virus infection (Fujimura et al., 2014; Fonseca et al., 2017) |
| L. murinus | Protects from dextran sodium sulfate–induced colitis by promoting increase in T reg cells in the colon (Tang et al., 2015) |
| L. plantarum | Recovers growth stunting in juvenile GF mice associated with somatotropic axis signaling (Schwarzer et al., 2016) |
| Lowers fasting blood glucose and triglyceride levels (Toshimitsu et al., 2017) | |
| L. reuteri | Restores social behavior in offspring of dams fed HFD (Buffington et al., 2016) |
| Restores despair behavior induced by exposure to chronic stress via suppression of tryptophan/kynurenine metabolism (Marin et al., 2017) | |
| L. reuteri, L. murinus, and Lactobacillus taiwanensis | Attenuates intestinal inflammation in experimental model of colitis (Lamas et al., 2016) |
| LGG | Ameliorates ASD-like behavior by modulating the expression of GABA receptors in the brain (Bravo et al., 2011) |
| Lactobacillus sakei | Improves HFD-induced obesity and hyperglycemia by reducing inflammation and increasing the expression of colon tight junction proteins (Lim et al., 2016) |
Nonetheless, intervention with a multispecies consortium of bacteria was shown to be effective in the induction and maintenance of remission in patients with UC (Mimura et al., 2004; Sood et al., 2009), but not in those with CD (Fedorak et al., 2015). Four months of daily supplementation with the same consortium led to significant improvements in fatty liver disease severity, reductions in body mass index, and increases in glucagon-like peptide levels in children with obesity (Alisi et al., 2014). Meta-analysis of intervention trials in which various combinations of bacterial strains were administered to adults with T2DM showed moderate improvements in hyperglycemia (Samah et al., 2016). Moreover, probiotic supplementation during the first 27 d of life reduced risk of islet autoimmunity in a large multicenter prospective cohort study of children at high genetic risk for T1DM, compared with no supplementation or supplementation later in infancy (Uusitalo et al., 2016).
Reduced allergic sensitization was also demonstrated in a meta-analysis of randomized trials of early-life oral microbial interventions (Elazab et al., 2013). Additionally, in a recent follow up of a trial in which children with peanut allergies were randomized to receive either placebo or oral peanut immunotherapy combined with daily L. rhamnosus GG (LGG) for 18 mo, 67% of children in the immunotherapy/probiotic group remained desensitized to peanuts 4 yr after the intervention compared with 4% in the placebo group (Hsiao et al., 2017). This observation provides promising results for the long-term efficacy of a combined intervention approach in preventing food allergies; however, as the trial lacked control groups that received either oral immunotherapy or microbial supplementation alone, it remains to be determined whether the observed beneficial effect is attributable to the immunotherapy, the microbial supplementation, or both. Nevertheless, these observations are encouraging and warrant further controlled trials.
Despite promising outcomes of early-life oral microbial supplementation on reduction of atopic sensitization in children, trials have not shown benefits on development of atopic asthma or wheeze (Elazab et al., 2013). A recent study examining the effect of early-life (birth to 6 mo of age) LGG supplementation in infants at high risk for asthma found no significant effect of LGG on the development of asthma at 3 yr of age (Cabana et al., 2017). However, a subsequent analysis showed that LGG supplementation partially abrogated delayed gut microbiota bacterial diversification and resolved metabolic dysfunction observed in placebo-supplemented high-risk infants (Durack et al., 2018). Furthermore, the associated products of LGG-supplemented fecal microbiomes induced significantly more T reg cells ex vivo. However, LGG colonization and associated benefits of metabolic reprogramming were not sustained (Durack et al., 2018), perhaps suggesting the need for longer-term or earlier (e.g., during pregnancy) intervention or use of a multispecies microbial cocktail with enhanced capacity for competitive colonization of the neonatal gut to achieve long-term efficacy. Despite the negative outcome of this trial, it verifies the plastic nature of the early-life gut microbiome, demonstrates that microbial supplementation is safe and well tolerated in infancy, and provides encouraging data suggesting that gut microbiome–mediated metabolism may be reengineered to promote induction of immune tolerance.
Intervention studies involving live microbial supplementation have shown encouraging results, although more attention to microbial strain selection based on functional attributes, defined timing or duration of supplementation, and/or tailoring of the supplemented organisms to the endogenous gut microbiome of the recipient may significantly improve efficacy in future studies. Ongoing studies are focused on understanding the basis of microbe–microbe interactions in order to identify discrete gut microbiomes that more readily respond to specific microbial interventions.
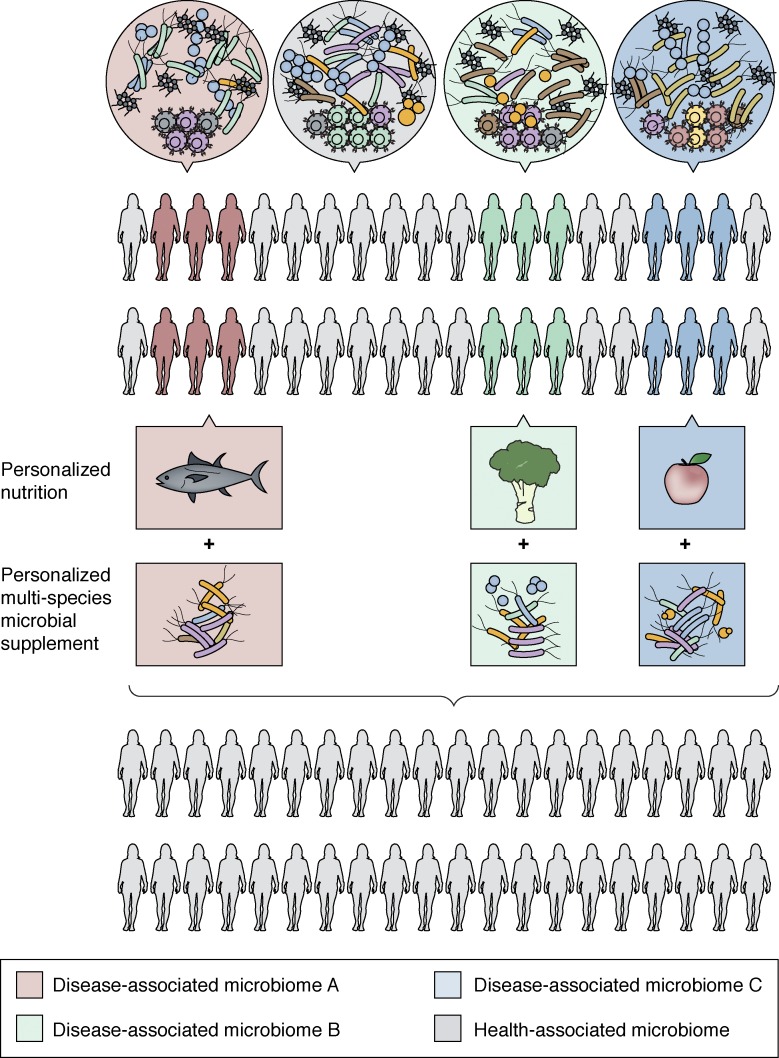
There is also a great need for tailored interventions that consider the microbiological individuality of the recipient. Such approaches are likely to result in a transition away from historically used probiotic strains, which are frequently poorly adapted to the enteric microenvironment, toward personalized multispecies microbial consortia sourced from healthy human enteric ecosystems. To further enhance efficacy of microbial supplementation, nutritional support in the form of targeted dietary modifications, tailored to the specific substrate requirements of the supplemented microbes, should also be considered. Such an integrated approach to microbiome-based therapeutics (Fig. 4), built on independent observations in the field of gut microbiome research, may offer more effective, predictable, and sustainable microbial restitution in cases of chronic disease in which microbiome perturbation and functional gene loss are prominent features.
Figure 4.
A strategic framework for a personalized integrated approach to microbiome manipulation. Due to microbial heterogeneity across populations, personalized nutrition in combination with the administration of live, functionally defined microbial strains to reengineer microbiome composition, functional gene capacity, and metabolic output may prove most effective in rehabilitating perturbed gut microbiomes for effective disease prevention or management.
FMT
FMT has proven effective in >90% of patients with recurrent antibiotic-resistant C. difficile infection (van Nood et al., 2013). The success rate of FMT administration for other chronic intestinal inflammatory conditions such as IBD, however, is more modest, with clinical remission being less predictable (Colman and Rubin, 2014). More recently, FMT has been applied to chronic inflammatory conditions affecting organs distal to the GI tract. A small trial of children aged 7–16 yr diagnosed with ASD showed that FMT improved both GI and ASD behavioral symptoms, in concert with changes to their gut microbiota composition, which was sustained 2 mo after intervention (Kang et al., 2017). In a separate study, FMT from lean donors improved insulin sensitivity in obese men with metabolic syndrome, and this improvement was linked to changes in plasma metabolites, in particular increased gamma-aminobutyric acid (GABA), tryptophan, and phenylalanine (Kootte et al., 2017). GABA has been associated with regulation of metabolism in mouse models (Meng et al., 2016) and with successful (albeit short-lived) engraftment of lean-donor FMT and improved health in men with metabolic syndrome (Kootte et al., 2017), suggesting a role for GABA in glucose metabolism in humans. Studies focused on understanding the dynamics of donor microbiota colonization following single-donor FMT have demonstrated varying degrees of microbial transfer in different recipients (Li et al., 2016b), which was later shown to depend on the abundance and composition of bacteria in both the donor and pre-FMT recipient (Smillie et al., 2018). These results suggest that, as for other organ transplants, gut microbiome compatibility is an important, but currently overlooked, factor that needs to be considered in future trials.
The risks associated with FMT are not completely defined. Donor material is prescreened for known infectious agents, but there remains a risk of transferring to the recipient phenotypes that are clinically silent in the donor. For this reason alone, the FMT approach may not be feasible for infants at high risk for developing chronic conditions, and alternative interventions with quality-controlled microbes from human sources may prove more appropriate for such vulnerable populations. In a recent proof-of-principle study, the gut microbiota of cesarean section–delivered infants was manipulated to more closely resemble that of vaginally delivered newborns by swabbing the neonate with gauze inoculated with the mother’s vaginal microbiota (Dominguez-Bello et al., 2016). These results demonstrate that vaginal microbes typically underrepresented in babies delivered by cesarean section can be partially and safely restored at birth, although the long-term health consequences of these interventions remain to be determined.
Microbial transplantation is proving useful as a means of manipulating the gut microbiome for management or prevention of certain illnesses or disorders, although many more well-designed studies are needed. More importantly, early studies have provided insights into the critical organisms and biomolecules that mediate efficacy and those that impede it. As the field progresses, it is likely that this approach will be surpassed by more precise and integrated multispecies microbial/diet-based interventions tailored to the individual and their specific microbial perturbation (Fig. 4).
Summary and perspectives
Sequencing, mass spectrometry, and a variety of in vitro and in vivo model approaches have led to the observation that gut microbiome perturbation is evident in a variety of human diseases that manifest either locally in the gut or remotely at discrete organ and mucosal sites. In specific cases, gut microbiome perturbations are evident years in advance of disease development, offering biomarkers for early detection of disease risk and opportunities for preventive interventions. Paramount to all of these observations is the increasing evidence for microbial causality in disease development and identification of specific mechanisms of pathogenesis, the latter identifying novel microbial targets for therapeutic development. Positive results have been observed for some disease indications following efforts to rehabilitate perturbed gut microbiota via dietary intervention, microbial supplementation, or FMT. These efforts provide proof of principle that the gut microbiome represents a viable therapeutic target, with the opportunity to develop more refined and integrated approaches for disease management, prevention, or cure. Despite the promise, the gut microbiome field has much to do, including large and longitudinal integrative studies of humans to include all (bacterial, fungal, and viral) members of the gut microbiome and their products in the context of objective immune and physiological measurements and clinical outcomes. Complementing such investigations with mechanistic studies is critical to move the field beyond description and toward a molecular understanding of how microbe–microbe–host interactions influence human health over the course of life and is key to realizing the full potential of the field.
Acknowledgments
This work was supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases (grant AI113916 to S.V. Lynch).
S.V. Lynch is a founder, board member, and consultant for Siolta Therapeutics and is on the Scientific Advisory Board of Bloom Science. J. Durack declares no competing financial interests.
Author contributions: J. Durack and S.V. Lynch conceived and wrote this review article.
References
- Aagaard K., Ma J., Antony K.M., Ganu R., Petrosino J., and Versalovic J.. 2014. The placenta harbors a unique microbiome. Sci. Transl. Med. 6:237ra65 10.1126/scitranslmed.3008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadizar F., Vijverberg S.J.H., Arets H.G.M., de Boer A., Turner S., Devereux G., Arabkhazaeli A., Soares P., Mukhopadhyay S., Garssen J., et al. . 2017. Early life antibiotic use and the risk of asthma and asthma exacerbations in children. Pediatr. Allergy Immunol. 28:430–437. 10.1111/pai.12725 [DOI] [PubMed] [Google Scholar]
- Alard J., Lehrter V., Rhimi M., Mangin I., Peucelle V., Abraham A.L., Mariadassou M., Maguin E., Waligora-Dupriet A.J., Pot B., et al. . 2016. Beneficial metabolic effects of selected probiotics on diet-induced obesity and insulin resistance in mice are associated with improvement of dysbiotic gut microbiota. Environ. Microbiol. 18:1484–1497. 10.1111/1462-2920.13181 [DOI] [PubMed] [Google Scholar]
- Alisi A., Bedogni G., Baviera G., Giorgio V., Porro E., Paris C., Giammaria P., Reali L., Anania F., and Nobili V.. 2014. Randomised clinical trial: The beneficial effects of VSL#3 in obese children with non-alcoholic steatohepatitis. Aliment. Pharmacol. Ther. 39:1276–1285. 10.1111/apt.12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allais L., Kerckhof F.M., Verschuere S., Bracke K.R., De Smet R., Laukens D., Van den Abbeele P., De Vos M., Boon N., Brusselle G.G., et al. . 2016. Chronic cigarette smoke exposure induces microbial and inflammatory shifts and mucin changes in the murine gut. Environ. Microbiol. 18:1352–1363. 10.1111/1462-2920.12934 [DOI] [PubMed] [Google Scholar]
- Amar J., Chabo C., Waget A., Klopp P., Vachoux C., Bermúdez-Humarán L.G., Smirnova N., Bergé M., Sulpice T., Lahtinen S., et al. . 2011. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol. Med. 3:559–572. 10.1002/emmm.201100159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D., Oh S.F., Olszak T., Neves J.F., Avci F.Y., Erturk-Hasdemir D., Lu X., Zeissig S., Blumberg R.S., and Kasper D.L.. 2014. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 156:123–133. 10.1016/j.cell.2013.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andoh A., Nishida A., Takahashi K., Inatomi O., Imaeda H., Bamba S., Kito K., Sugimoto M., and Kobayashi T.. 2016. Comparison of the gut microbial community between obese and lean peoples using 16S gene sequencing in a Japanese population. J. Clin. Biochem. Nutr. 59:65–70. 10.3164/jcbn.15-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anhê F.F., Roy D., Pilon G., Dudonné S., Matamoros S., Varin T.V., Garofalo C., Moine Q., Desjardins Y., Levy E., and Marette A.. 2015. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut. 64:872–883. 10.1136/gutjnl-2014-307142 [DOI] [PubMed] [Google Scholar]
- Arrieta M.C., Stiemsma L.T., Dimitriu P.A., Thorson L., Russell S., Yurist-Doutsch S., Kuzeljevic B., Gold M.J., Britton H.M., Lefebvre D.L., et al. CHILD Study Investigators . 2015. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 7:307ra152 10.1126/scitranslmed.aab2271 [DOI] [PubMed] [Google Scholar]
- Atarashi K., Tanoue T., Shima T., Imaoka A., Kuwahara T., Momose Y., Cheng G., Yamasaki S., Saito T., Ohba Y., et al. . 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 331:337–341. 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K., Tanoue T., Oshima K., Suda W., Nagano Y., Nishikawa H., Fukuda S., Saito T., Narushima S., Hase K., et al. . 2013. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 500:232–236. 10.1038/nature12331 [DOI] [PubMed] [Google Scholar]
- Bäckhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P., Li Y., Xia Y., Xie H., Zhong H., et al. . 2015. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 17:690–703. 10.1016/j.chom.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Bansal T., Alaniz R.C., Wood T.K., and Jayaraman A.. 2010. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. USA. 107:228–233. 10.1073/pnas.0906112107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercik P., Park A.J., Sinclair D., Khoshdel A., Lu J., Huang X., Deng Y., Blennerhassett P.A., Fahnestock M., Moine D., et al. . 2011. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut-brain communication. Neurogastroenterol. Motil. 23:1132–1139. 10.1111/j.1365-2982.2011.01796.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagi E., Franceschi C., Rampelli S., Severgnini M., Ostan R., Turroni S., Consolandi C., Quercia S., Scurti M., Monti D., et al. . 2016. Gut Microbiota and Extreme Longevity. Curr. Biol. 26:1480–1485. 10.1016/j.cub.2016.04.016 [DOI] [PubMed] [Google Scholar]
- Blekhman R., Goodrich J.K., Huang K., Sun Q., Bukowski R., Bell J.T., Spector T.D., Keinan A., Ley R.E., Gevers D., and Clark A.G.. 2015. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 16:191 10.1186/s13059-015-0759-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N.A., Chung J., Battaglia T., Henderson N., Jay M., Li H., Lieber A.D., Wu F., Perez-Perez G.I., Chen Y., et al. . 2016. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 8:343ra82 10.1126/scitranslmed.aad7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonder M.J., Kurilshikov A., Tigchelaar E.F., Mujagic Z., Imhann F., Vila A.V., Deelen P., Vatanen T., Schirmer M., Smeekens S.P., et al. . 2016. The effect of host genetics on the gut microbiome. Nat. Genet. 48:1407–1412. 10.1038/ng.3663 [DOI] [PubMed] [Google Scholar]
- Brahe L.K., Le Chatelier E., Prifti E., Pons N., Kennedy S., Blædel T., Håkansson J., Dalsgaard T.K., Hansen T., Pedersen O., et al. . 2015. Dietary modulation of the gut microbiota--a randomised controlled trial in obese postmenopausal women. Br. J. Nutr. 114:406–417. 10.1017/S0007114515001786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J.A., Forsythe P., Chew M.V., Escaravage E., Savignac H.M., Dinan T.G., Bienenstock J., and Cryan J.F.. 2011. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. USA. 108:16050–16055. 10.1073/pnas.1102999108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyner N.M., Michon C., de Sousa C.S., Vilas Boas P.B., Chain F., Azevedo V.A., Langella P., and Chatel J.M.. 2017. Microbial anti-inflammatory molecule (MAM) from Faecalibacterium prausnitzii shows a protective effect on DNBS and DSS-induced colitis model in mice through inhibition of NF-κB pathway. Front. Microbiol. 8:114 10.3389/fmicb.2017.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhurst M.J., Leung J.M., Kashyap V., McCune J.M., Mahadevan U., McKerrow J.H., and Loke P.. 2010. IL-22+ CD4+ T cells are associated with therapeutic trichuris trichiura infection in an ulcerative colitis patient. Sci. Transl. Med. 2:60ra88 10.1126/scitranslmed.3001500 [DOI] [PubMed] [Google Scholar]
- Brodin P., Jojic V., Gao T., Bhattacharya S., Angel C.J., Furman D., Shen-Orr S., Dekker C.L., Swan G.E., Butte A.J., et al. . 2015. Variation in the human immune system is largely driven by non-heritable influences. Cell. 160:37–47. 10.1016/j.cell.2014.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.S., Sourander A., Hinkka-Yli-Salomäki S., McKeague I.W., Sundvall J., and Surcel H.M.. 2014. Elevated maternal C-reactive protein and autism in a national birth cohort. Mol. Psychiatry. 19:259–264. 10.1038/mp.2012.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie C.G., Bucci V., Stein R.R., McKenney P.T., Ling L., Gobourne A., No D., Liu H., Kinnebrew M., Viale A., et al. . 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 517:205–208. 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffington S.A., Di Prisco G.V., Auchtung T.A., Ajami N.J., Petrosino J.F., and Costa-Mattioli M.. 2016. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell. 165:1762–1775. 10.1016/j.cell.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero S., Kim S., Carter R.A., Leiner I.M., Sušac B., Miller L., Kim G.J., Ling L., and Pamer E.G.. 2017. Cooperating commensals restore colonization resistance to vancomycin-resistant Enterococcus faecium. Cell Host Microbe. 21:592–602.e4. 10.1016/j.chom.2017.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabana M.D., McKean M., Caughey A.B., Fong L., Lynch S., Wong A., Leong R., Boushey H.A., and Hilton J.F.. 2017. Early probiotic supplementation for eczema and asthma prevention: a randomized controlled trial. Pediatrics. 140:e20163000 10.1542/peds.2016-3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cait A., Hughes M.R., Antignano F., Cait J., Dimitriu P.A., Maas K.R., Reynolds L.A., Hacker L., Mohr J., Finlay B.B., et al. . 2018. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. 11:785–795. 10.1038/mi.2017.75 [DOI] [PubMed] [Google Scholar]
- Caporaso J.G., Lauber C.L., Costello E.K., Berg-Lyons D., Gonzalez A., Stombaugh J., Knights D., Gajer P., Ravel J., Fierer N., et al. . 2011. Moving pictures of the human microbiome. Genome Biol. 12:R50 10.1186/gb-2011-12-5-r50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Careaga M., Rogers S., Hansen R.L., Amaral D.G., Van de Water J., and Ashwood P.. 2017. Immune endophenotypes in children with autism spectrum disorder. Biol. Psychiatry. 81:434–441. 10.1016/j.biopsych.2015.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cekanaviciute E., Yoo B.B., Runia T.F., Debelius J.W., Singh S., Nelson C.A., Kanner R., Bencosme Y., Lee Y.K., Hauser S.L., et al. . 2017. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. USA. 114:10713–10718. 10.1073/pnas.1711235114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan L., Chai J.N., Tianero M.D., Di Luccia B., Ahern P.P., Merriman J., Cortez V.S., Caparon M.G., Donia M.S., Gilfillan S., et al. . 2017. Lactobacillus reuteri induces gut intraepithelial CD4+CD8αα+ T cells. Science. 357:806–810. 10.1126/science.aah5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P.V., Hao L., Offermanns S., and Medzhitov R.. 2014. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA. 111:2247–2252. 10.1073/pnas.1322269111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Ringel-Kulka T., Heikamp-de Jong I., Ringel Y., Carroll I., de Vos W.M., Salojärvi J., and Satokari R.. 2016. Discordant temporal development of bacterial phyla and the emergence of core in the fecal microbiota of young children. ISME J. 10:1002–1014. 10.1038/ismej.2015.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimerel C., Emery E., Summers D.K., Keyser U., Gribble F.M., and Reimann F.. 2014. Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine L cells. Cell Reports. 9:1202–1208. 10.1016/j.celrep.2014.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.M., Ma J., Prince A.L., Antony K.M., Seferovic M.D., and Aagaard K.M.. 2017. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 23:314–326. 10.1038/nm.4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H., Pamp S.J., Hill J.A., Surana N.K., Edelman S.M., Troy E.B., Reading N.C., Villablanca E.J., Wang S., Mora J.R., et al. . 2012. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 149:1578–1593. 10.1016/j.cell.2012.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarlo E., Heinonen T., Herderschee J., Fenwick C., Mombelli M., Le Roy D., and Roger T.. 2016. Impact of the microbial derived short chain fatty acid propionate on host susceptibility to bacterial and fungal infections in vivo. Sci. Rep. 6:37944 10.1038/srep37944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson M.J., Jeffery I.B., Conde S., Power S.E., O’Connor E.M., Cusack S., Harris H.M.B., Coakley M., Lakshminarayanan B., O’Sullivan O., et al. . 2012. Gut microbiota composition correlates with diet and health in the elderly. Nature. 488:178–184. 10.1038/nature11319 [DOI] [PubMed] [Google Scholar]
- Cohen L.J., Esterhazy D., Kim S.H., Lemetre C., Aguilar R.R., Gordon E.A., Pickard A.J., Cross J.R., Emiliano A.B., Han S.M., et al. . 2017. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature. 549:48–53. 10.1038/nature23874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M.C., Rautava S., Aakko J., Isolauri E., and Salminen S.. 2016. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 6:23129 10.1038/srep23129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman R.J., and Rubin D.T.. 2014. Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. J. Crohn’s Colitis. 8:1569–1581. 10.1016/j.crohns.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello E.K., Lauber C.L., Hamady M., Fierer N., Gordon J.I., and Knight R.. 2009. Bacterial community variation in human body habitats across space and time. Science. 326:1694–1697. 10.1126/science.1177486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotillard A., Kennedy S.P., Kong L.C., Prifti E., Pons N., Le Chatelier E., Almeida M., Quinquis B., Levenez F., Galleron N., et al. ANR MicroObes consortium . 2013. Dietary intervention impact on gut microbial gene richness. Nature. 500:585–588. 10.1038/nature12480 [DOI] [PubMed] [Google Scholar]
- Cusick M.F., Libbey J.E., and Fujinami R.S.. 2012. Molecular mimicry as a mechanism of autoimmune disease. Clin. Rev. Allergy Immunol. 42:102–111. 10.1007/s12016-011-8294-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao M.C., Everard A., Aron-Wisnewsky J., Sokolovska N., Prifti E., Verger E.O., Kayser B.D., Levenez F., Chilloux J., Hoyles L., et al. MICRO-Obes Consortium . 2016. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 65:426–436. 10.1136/gutjnl-2014-308778 [DOI] [PubMed] [Google Scholar]
- Dasgupta S., Erturk-Hasdemir D., Ochoa-Reparaz J., Reinecker H.C., and Kasper D.L.. 2014. Plasmacytoid dendritic cells mediate anti-inflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host Microbe. 15:413–423. 10.1016/j.chom.2014.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E., Ling A.V., Devlin A.S., Varma Y., Fischbach M.A., et al. . 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 505:559–563. 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Besten G., Bleeker A., Gerding A., van Eunen K., Havinga R., van Dijk T.H., Oosterveer M.H., Jonker J.W., Groen A.K., Reijngoud D.J., and Bakker B.M.. 2015. Short-chain fatty acids protect against high-fat diet–induced obesity via a PPARγ-dependent switch from lipogenesis to fat oxidation. Diabetes. 64:2398–2408. 10.2337/db14-1213 [DOI] [PubMed] [Google Scholar]
- Desai M.S., Seekatz A.M., Koropatkin N.M., Kamada N., Hickey C.A., Wolter M., Pudlo N.A., Kitamoto S., Terrapon N., Muller A., et al. . 2016. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 167:1339–1353.e21. 10.1016/j.cell.2016.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello M.G., Costello E.K., Contreras M., Magris M., Hidalgo G., Fierer N., and Knight R.. 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA. 107:11971–11975. 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello M.G., De Jesus-Laboy K.M., Shen N., Cox L.M., Amir A., Gonzalez A., Bokulich N.A., Song S.J., Hoashi M., Rivera-Vinas J.I., et al. . 2016. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 22:250–253. 10.1038/nm.4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson G.P., Lee S.M., and Mazmanian S.K.. 2016. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14:20–32. 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia M.S., and Fischbach M.A.. 2015. HUMAN MICROBIOTA. Small molecules from the human microbiota. Science. 349:1254766 10.1126/science.1254766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durack J., Boushey H.A., and Lynch S.V.. 2016. Airway Microbiota and the Implications of Dysbiosis in Asthma. Curr. Allergy Asthma Rep. 16:52 10.1007/s11882-016-0631-8 [DOI] [PubMed] [Google Scholar]
- Durack J., Kimes N.E., Lin D.L., Rauch M., McKean M., McCauley K., Panzer A.R., Mar J.S., Cabana M.D., and Lynch S.V.. 2018. Delayed gut microbiota development in high-risk for asthma infants is temporarily modifiable by Lactobacillus supplementation. Nat. Commun. 9:707 10.1038/s41467-018-03157-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elazab N., Mendy A., Gasana J., Vieira E.R., Quizon A., and Forno E.. 2013. Probiotic administration in early life, atopy, and asthma: a meta-analysis of clinical trials. Pediatrics. 132:e666–e676. 10.1542/peds.2013-0246 [DOI] [PubMed] [Google Scholar]
- Estruch R., Ros E., Salas-Salvadó J., Covas M.I., Corella D., Arós F., Gómez-Gracia E., Ruiz-Gutiérrez V., Fiol M., Lapetra J.; PREDIMED Study Investigators, et al. . 2018. Primary Prevention of Cardiovascular Disease with a Mediterranean Diet Supplemented with Extra-Virgin Olive Oil or Nuts. N. Engl. J. Med. 378:e34 10.1056/NEJMoa1800389 [DOI] [PubMed] [Google Scholar]
- Everard A., Belzer C., Geurts L., Ouwerkerk J.P., Druart C., Bindels L.B., Guiot Y., Derrien M., Muccioli G.G., Delzenne N.M., et al. . 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA. 110:9066–9071. 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith J.J., Guruge J.L., Charbonneau M., Subramanian S., Seedorf H., Goodman A.L., Clemente J.C., Knight R., Heath A.C., Leibel R.L., et al. . 2013. The long-term stability of the human gut microbiota. Science. 341:1237439 10.1126/science.1237439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D., Coughlin L.A., Neubauer M.M., Kim J., Kim M.S., Zhan X., Simms-Waldrip T.R., Xie Y., Hooper L.V., and Koh A.Y.. 2015. Activation of HIF-1α and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat. Med. 21:808–814. 10.1038/nm.3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorak R.N., Feagan B.G., Hotte N., Leddin D., Dieleman L.A., Petrunia D.M., Enns R., Bitton A., Chiba N., Paré P., et al. . 2015. The probiotic VSL#3 has anti-inflammatory effects and could reduce endoscopic recurrence after surgery for Crohn’s disease. Clin. Gastroenterol. Hepatol. 13:928–35.e2. 10.1016/j.cgh.2014.10.031 [DOI] [PubMed] [Google Scholar]
- Ferretti P., Pasolli E., Tett A., Asnicar F., Gorfer V., Fedi S., Armanini F., Truong D.T., Manara S., Zolfo M., et al. . 2018. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 24:133–145.e5. 10.1016/j.chom.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegold S.M., Dowd S.E., Gontcharova V., Liu C., Henley K.E., Wolcott R.D., Youn E., Summanen P.H., Granpeesheh D., Dixon D., et al. . 2010. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 16:444–453. 10.1016/j.anaerobe.2010.06.008 [DOI] [PubMed] [Google Scholar]
- Fonseca W., Lucey K., Jang S., Fujimura K.E., Rasky A., Ting H.A., Petersen J., Johnson C.C., Boushey H.A., Zoratti E., et al. . 2017. Lactobacillus johnsonii supplementation attenuates respiratory viral infection via metabolic reprogramming and immune cell modulation. Mucosal Immunol. 10:1569–1580. 10.1038/mi.2017.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K., Hildebrand F., Nielsen T., Falony G., Le Chatelier E., Sunagawa S., Prifti E., Vieira-Silva S., Gudmundsdottir V., Pedersen H.K., et al. MetaHIT consortium . 2015. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 528:262–266. 10.1038/nature15766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen F., van Beek A.A., Borghuis T., Meijer B., Hugenholtz F., van der Gaast-de Jongh C., Savelkoul H.F., de Jonge M.I., Faas M.M., Boekschoten M.V., et al. . 2017. The impact of gut microbiota on gender-specific differences in immunity. Front. Immunol. 8:754 10.3389/fimmu.2017.00754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa E.A., Huang K., Meadow J.F., Gevers D., Lemon K.P., Bohannan B.J.M., and Huttenhower C.. 2015. Identifying personal microbiomes using metagenomic codes. Proc. Natl. Acad. Sci. USA. 112:E2930–E2938. 10.1073/pnas.1423854112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K.E., and Lynch S.V.. 2015. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe. 17:592–602. 10.1016/j.chom.2015.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K.E., Demoor T., Rauch M., Faruqi A.A., Jang S., Johnson C.C., Boushey H.A., Zoratti E., Ownby D., Lukacs N.W., and Lynch S.V.. 2014. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc. Natl. Acad. Sci. USA. 111:805–810. 10.1073/pnas.1310750111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K.E., Sitarik A.R., Havstad S., Lin D.L., Levan S., Fadrosh D., Panzer A.R., LaMere B., Rackaityte E., Lukacs N.W., et al. . 2016. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 22:1187–1191. 10.1038/nm.4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.C., Olson C.A., and Hsiao E.Y.. 2017. Interactions between the microbiota, immune and nervous systems in health and disease. Nat. Neurosci. 20:145–155. 10.1038/nn.4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y., Obata Y., Fukuda S., Endo T.A., Nakato G., Takahashi D., Nakanishi Y., Uetake C., Kato K., Kato T., et al. . 2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 504:446–450. 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- Garibay-Nieto N., Queipo-García G., Alvarez F., Bustos M., Villanueva E., Ramírez F., León M., Laresgoiti-Servitje E., Duggirala R., Macías T., et al. . 2017. Effects of conjugated linoleic acid and metformin on insulin sensitivity in obese children: Randomized clinical trial. J. Clin. Endocrinol. Metab. 102:132–140. [DOI] [PubMed] [Google Scholar]
- Gensollen T., Iyer S.S., Kasper D.L., and Blumberg R.S.. 2016. How colonization by microbiota in early life shapes the immune system. Science. 352:539–544. 10.1126/science.aad9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geva-Zatorsky N., Sefik E., Kua L., Pasman L., Tan T.G., Ortiz-Lopez A., Yanortsang T.B., Yang L., Jupp R., Mathis D., et al. . 2017. Mining the human gut microbiota for immunomodulatory organisms. Cell. 168:928–943.e11. 10.1016/j.cell.2017.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D., Kugathasan S., Denson L.A., Vázquez-Baeza Y., Van Treuren W., Ren B., Schwager E., Knights D., Song S.J., Yassour M., et al. . 2014. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 15:382–392. 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J.K., Waters J.L., Poole A.C., Sutter J.L., Koren O., Blekhman R., Beaumont M., Van Treuren W., Knight R., Bell J.T., et al. . 2014. Human genetics shape the gut microbiome. Cell. 159:789–799. 10.1016/j.cell.2014.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V., Prieto P.A., Vicente D., Hoffman K., Wei S.C., et al. . 2018. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 359:97–103. 10.1126/science.aan4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosalbes M.J., Llop S., Vallès Y., Moya A., Ballester F., and Francino M.P.. 2013. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin. Exp. Allergy. 43:198–211. 10.1111/cea.12063 [DOI] [PubMed] [Google Scholar]
- Guo C.J., Chang F.Y., Wyche T.P., Backus K.M., Acker T.M., Funabashi M., Taketani M., Donia M.S., Nayfach S., Pollard K.S., et al. . 2017. Discovery of reactive microbiota-derived metabolites that inhibit host proteases. Cell. 168:517–526.e18. 10.1016/j.cell.2016.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier S., Lawson M.A., Slack E., Kirundi J.K., Stoel M., Heikenwalder M., Cahenzli J., Velykoredko Y., Balmer M.L., Endt K., et al. . 2010. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 328:1705–1709. 10.1126/science.1188454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth M.R., Monticelli L.A., Fung T.C., Ziegler C.G.K., Grunberg S., Sinha R., Mantegazza A.R., Ma H.-L., Crawford A., Angelosanto J.M., et al. . 2013. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 498:113–117. 10.1038/nature12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister E.B., Riehle K., Luna R.A., Weidler E.M., Rubio-Gonzales M., Mistretta T.A., Raza S., Doddapaneni H.V., Metcalf G.A., Muzny D.M., et al. . 2015. Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome. 3:36 10.1186/s40168-015-0101-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L.V., Wong M.H., Thelin A., Hansson L., Falk P.G., and Gordon J.I.. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 291:881–884. 10.1126/science.291.5505.881 [DOI] [PubMed] [Google Scholar]
- Hsiao E.Y., McBride S.W., Hsien S., Sharon G., Hyde E.R., McCue T., Codelli J.A., Chow J., Reisman S.E., Petrosino J.F., et al. . 2013. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 155:1451–1463. 10.1016/j.cell.2013.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao K.-C., Ponsonby A.-L., Axelrad C., Pitkin S., Tang M.L.K., Burks W., Donath S., Orsini F., Tey D., Robinson M., and Su E.L.. PPOIT Study Team . 2017. Long-term clinical and immunological effects of probiotic and peanut oral immunotherapy after treatment cessation: 4-year follow-up of a randomised, double-blind, placebo-controlled trial. Lancet Child Adolesc. Health. 1:97–105. 10.1016/S2352-4642(17)30041-X [DOI] [PubMed] [Google Scholar]
- Hunter T.M., Boytsov N.N., Zhang X., Schroeder K., Michaud K., and Araujo A.B.. 2017. Prevalence of rheumatoid arthritis in the United States adult population in healthcare claims databases, 2004-2014. Rheumatol. Int. 37:1551–1557. 10.1007/s00296-017-3726-1 [DOI] [PubMed] [Google Scholar]
- Hwang I.K., Yoo K.Y., Li H., Park O.K., Lee C.H., Choi J.H., Jeong Y.G., Lee Y.L., Kim Y.M., Kwon Y.G., and Won M.H.. 2009. Indole-3-propionic acid attenuates neuronal damage and oxidative stress in the ischemic hippocampus. J. Neurosci. Res. 87:2126–2137. 10.1002/jnr.22030 [DOI] [PubMed] [Google Scholar]
- Imhann F., Bonder M.J., Vich Vila A., Fu J., Mujagic Z., Vork L., Tigchelaar E.F., Jankipersadsing S.A., Cenit M.C., Harmsen H.J., et al. . 2016. Proton pump inhibitors affect the gut microbiome. Gut. 65:740–748. 10.1136/gutjnl-2015-310376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal R., Anand S., Ounpuu S., Islam S., Zhang X., Rangarajan S., Chifamba J., Al-Hinai A., Keltai M., and Yusuf S.. INTERHEART Study Investigators . 2008. Dietary patterns and the risk of acute myocardial infarction in 52 countries: results of the INTERHEART study. Circulation. 118:1929–1937. 10.1161/CIRCULATIONAHA.107.738716 [DOI] [PubMed] [Google Scholar]
- Ivanov I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V., et al. . 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139:485–498. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M.A., Jeffery I.B., Beaumont M., Bell J.T., Clark A.G., Ley R.E., O’Toole P.W., Spector T.D., and Steves C.J.. 2016. Signatures of early frailty in the gut microbiota. Genome Med. 8:8 10.1186/s13073-016-0262-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson H.E., Abrahamsson T.R., Jenmalm M.C., Harris K., Quince C., Jernberg C., Björkstén B., Engstrand L., and Andersson A.F.. 2014. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 63:559–566. 10.1136/gutjnl-2012-303249 [DOI] [PubMed] [Google Scholar]
- Johnson C.C., Ownby D.R., Alford S.H., Havstad S.L., Williams L.K., Zoratti E.M., Peterson E.L., and Joseph C.L.. 2005. Antibiotic exposure in early infancy and risk for childhood atopy. J. Allergy Clin. Immunol. 115:1218–1224. 10.1016/j.jaci.2005.04.020 [DOI] [PubMed] [Google Scholar]
- Kaikiri H., Miyamoto J., Kawakami T., Park S.B., Kitamura N., Kishino S., Yonejima Y., Hisa K., Watanabe J., Ogita T., et al. . 2017. Supplemental feeding of a gut microbial metabolite of linoleic acid, 10-hydroxy-cis-12-octadecenoic acid, alleviates spontaneous atopic dermatitis and modulates intestinal microbiota in NC/nga mice. Int. J. Food Sci. Nutr. 68:941–951. 10.1080/09637486.2017.1318116 [DOI] [PubMed] [Google Scholar]
- Kaisar M.M.M., Pelgrom L.R., van der Ham A.J., Yazdanbakhsh M., and Everts B.. 2017. Butyrate conditions human dendritic cells to prime Type 1 regulatory T cells via both histone deacetylase inhibition and G protein-coupled receptor 109A signaling. Front. Immunol. 8:1429 10.3389/fimmu.2017.01429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallaur A.P., Reiche E.M.V., Oliveira S.R., Simão A.N.C., Pereira W.L.C.J., Alfieri D.F., Flauzino T., Proença C.M., Lozovoy M.A.B., Kaimen-Maciel D.R., and Maes M.. 2017. Genetic, immune-inflammatory, and oxidative stress biomarkers as predictors for disability and disease progression in multiple sclerosis. Mol. Neurobiol. 54:31–44. 10.1007/s12035-015-9648-6 [DOI] [PubMed] [Google Scholar]
- Kamiya T., Tang C., Kadoki M., Oshima K., Hattori M., Saijo S., Adachi Y., Ohno N., and Iwakura Y.. 2018. β-Glucans in food modify colonic microflora by inducing antimicrobial protein, calprotectin, in a Dectin-1-induced-IL-17F-dependent manner. Mucosal Immunol. 11:763–773. 10.1038/mi.2017.86 [DOI] [PubMed] [Google Scholar]
- Kang D.-W., Adams J.B., Gregory A.C., Borody T., Chittick L., Fasano A., Khoruts A., Geis E., Maldonado J., McDonough-Means S., et al. . 2017. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome. 5:10 10.1186/s40168-016-0225-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl J.P., Meydani M., Barnett J.B., Vanegas S.M., Barger K., Fu X., Goldin B., Kane A., Rasmussen H., Vangay P., et al. . 2017. Fecal concentrations of bacterially derived vitamin K forms are associated with gut microbiota composition but not plasma or fecal cytokine concentrations in healthy adults. Am. J. Clin. Nutr. 106:1052–1061. 10.3945/ajcn.117.155424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson F.H., Tremaroli V., Nookaew I., Bergström G., Behre C.J., Fagerberg B., Nielsen J., and Bäckhed F.. 2013. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 498:99–103. 10.1038/nature12198 [DOI] [PubMed] [Google Scholar]
- Kelly C.J., Zheng L., Campbell E.L., Saeedi B., Scholz C.C., Bayless A.J., Wilson K.E., Glover L.E., Kominsky D.J., Magnuson A., et al. . 2015. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 17:662–671. 10.1016/j.chom.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshteli A.H., Millan B., and Madsen K.L.. 2017. Pretreatment with antibiotics may enhance the efficacy of fecal microbiota transplantation in ulcerative colitis: a meta-analysis. Mucosal Immunol. 10:565–566. 10.1038/mi.2016.123 [DOI] [PubMed] [Google Scholar]
- Khan S., and Jena G.. 2016. Sodium butyrate reduces insulin-resistance, fat accumulation and dyslipidemia in type-2 diabetic rat: A comparative study with metformin. Chem. Biol. Interact. 254:124–134. 10.1016/j.cbi.2016.06.007 [DOI] [PubMed] [Google Scholar]
- Khosravi A., Yáñez A., Price J.G., Chow A., Merad M., Goodridge H.S., and Mazmanian S.K.. 2014. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 15:374–381. 10.1016/j.chom.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kim H., Yim Y.S., Ha S., Atarashi K., Tan T.G., Longman R.S., Honda K., Littman D.R., Choi G.B., and Huh J.R.. 2017. Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature. 549:528–532. 10.1038/nature23910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeth R.A., Wang Z., Levison B.S., Buffa J.A., Org E., Sheehy B.T., Britt E.B., Fu X., Wu Y., Li L., et al. . 2013. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19:576–585. 10.1038/nm.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kootte R.S., Levin E., Salojärvi J., Smits L.P., Hartstra A.V., Udayappan S.D., Hermes G., Bouter K.E., Koopen A.M., Holst J.J., et al. . 2017. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab. 26:611–619.e6. 10.1016/j.cmet.2017.09.008 [DOI] [PubMed] [Google Scholar]
- Korpela K., Salonen A., Virta L.J., Kekkonen R.A., Forslund K., Bork P., and de Vos W.M.. 2016. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat. Commun. 7:10410 10.1038/ncomms10410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpela K., Costea P., Coelho L.P., Kandels-Lewis S., Willemsen G., Boomsma D.I., Segata N., and Bork P.. 2018. Selective maternal seeding and environment shape the human gut microbiome. Genome Res. 28:561–568. 10.1101/gr.233940.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic A.D., Gevers D., Siljander H., Vatanen T., Hyötyläinen T., Hämäläinen A.M., Peet A., Tillmann V., Pöhö P., Mattila I., et al. DIABIMMUNE Study Group . 2015. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 17:260–273. 10.1016/j.chom.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudelka M.R., Hinrichs B.H., Darby T., Moreno C.S., Nishio H., Cutler C.E., Wang J., Wu H., Zeng J., Wang Y., et al. . 2016. Cosmc is an X-linked inflammatory bowel disease risk gene that spatially regulates gut microbiota and contributes to sex-specific risk. Proc. Natl. Acad. Sci. USA. 113:14787–14792. 10.1073/pnas.1612158114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas B., Richard M.L., Leducq V., Pham H.P., Michel M.L., Da Costa G., Bridonneau C., Jegou S., Hoffmann T.W., Natividad J.M., et al. . 2016. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 22:598–605. 10.1038/nm.4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., Almeida M., Arumugam M., Batto J.M., Kennedy S., et al. MetaHIT consortium . 2013. Richness of human gut microbiome correlates with metabolic markers. Nature. 500:541–546. 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- Lee B.K., Magnusson C., Gardner R.M., Blomström Å., Newschaffer C.J., Burstyn I., Karlsson H., and Dalman C.. 2015. Maternal hospitalization with infection during pregnancy and risk of autism spectrum disorders. Brain Behav. Immun. 44:100–105. 10.1016/j.bbi.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Cella M., McDonald K.G., Garlanda C., Kennedy G.D., Nukaya M., Mantovani A., Kopan R., Bradfield C.A., Newberry R.D., and Colonna M.. 2011. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 13:144–151. 10.1038/ni.2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin A.M., Sitarik A.R., Havstad S.L., Fujimura K.E., Wegienka G., Cassidy-Bushrow A.E., Kim H., Zoratti E.M., Lukacs N.W., Boushey H.A., et al. . 2016. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci. Rep. 6:31775 10.1038/srep31775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M., Thaiss C.A., Zeevi D., Dohnalová L., Zilberman-Schapira G., Mahdi J.A., David E., Savidor A., Korem T., Herzig Y., et al. . 2015. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. 163:1428–1443. 10.1016/j.cell.2015.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E., Bäckhed F., Turnbaugh P., Lozupone C.A., Knight R.D., and Gordon J.I.. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA. 102:11070–11075. 10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Jia H., Cai X., Zhong H., Feng Q., Sunagawa S., Arumugam M., Kultima J.R., Prifti E., Nielsen T., et al. MetaHIT Consortium . 2014. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32:834–841. 10.1038/nbt.2942 [DOI] [PubMed] [Google Scholar]
- Li J., Lin S., Vanhoutte P.M., Woo C.W., and Xu A.. 2016a Akkermansia muciniphila protects against atherosclerosis by preventing metabolic endotoxemia-induced inflammation in Apoe−/− mice. Circulation. 133:2434–2446. 10.1161/CIRCULATIONAHA.115.019645 [DOI] [PubMed] [Google Scholar]
- Li Q., Han Y., Dy A.B.C., and Hagerman R.J.. 2017. The Gut Microbiota and Autism Spectrum Disorders. Front. Cell. Neurosci. 11:120 10.3389/fncel.2017.00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.S., Zhu A., Benes V., Costea P.I., Hercog R., Hildebrand F., Huerta-Cepas J., Nieuwdorp M., Salojärvi J., Voigt A.Y., et al. . 2016b Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science. 352:586–589. 10.1126/science.aad8852 [DOI] [PubMed] [Google Scholar]
- Lim S.M., and Kim D.H.. 2017. Bifidobacterium adolescentis IM38 ameliorates high-fat diet-induced colitis in mice by inhibiting NF-κB activation and lipopolysaccharide production by gut microbiota. Nutr. Res. 41:86–96. 10.1016/j.nutres.2017.04.003 [DOI] [PubMed] [Google Scholar]
- Lim E.S., Zhou Y., Zhao G., Bauer I.K., Droit L., Ndao I.M., Warner B.B., Tarr P.I., Wang D., and Holtz L.R.. 2015. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med. 21:1228–1234. 10.1038/nm.3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.M., Jeong J.J., Woo K.H., Han M.J., and Kim D.H.. 2016. Lactobacillus sakei OK67 ameliorates high-fat diet-induced blood glucose intolerance and obesity in mice by inhibiting gut microbiota lipopolysaccharide production and inducing colon tight junction protein expression. Nutr. Res. 36:337–348. 10.1016/j.nutres.2015.12.001 [DOI] [PubMed] [Google Scholar]
- Livanos A.E., Greiner T.U., Vangay P., Pathmasiri W., Stewart D., McRitchie S., Li H., Chung J., Sohn J., Kim S., et al. . 2016. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat. Microbiol. 1:16140 10.1038/nmicrobiol.2016.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston G., Matias M., Calcagno V., Barbera C., Combe M., Leibold M.A., and Mouquet N.. 2012. Competition-colonization dynamics in experimental bacterial metacommunities. Nat. Commun. 3:1234 10.1038/ncomms2239 [DOI] [PubMed] [Google Scholar]
- Lloyd-Price J., Abu-Ali G., and Huttenhower C.. 2016. The healthy human microbiome. Genome Med. 8:51 10.1186/s13073-016-0307-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K., Abo R.P., Schlieper K.A., Graffam M.E., Levine S., Wishnok J.S., Swenberg J.A., Tannenbaum S.R., and Fox J.G.. 2014. Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environ. Health Perspect. 122:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn M.A., Tumes D.J., Choo J.M., Sribnaia A., Blake S.J., Leong L.E.X., Young G.P., Marshall H.S., Wesselingh S.L., Rogers G.B., and Lynn D.J.. 2018. Early-Life Antibiotic-Driven Dysbiosis Leads to Dysregulated Vaccine Immune Responses in Mice. Cell Host Microbe. 23:653–660.e5. 10.1016/j.chom.2018.04.009 [DOI] [PubMed] [Google Scholar]
- Magnúsdóttir S., Ravcheev D., de Crécy-Lagard V., and Thiele I.. 2015. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front. Genet. 6:148 10.3389/fgene.2015.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyari M., and Koch-Henriksen N.. 2017. The incidence of multiple sclerosis in Danish women has duplicated over the last sixty years. J. Neurol. Sci. 381:782 10.1016/j.jns.2017.08.2207 [DOI] [Google Scholar]
- Mar J.S., LaMere B.J., Lin D.L., Levan S., Nazareth M., Mahadevan U., and Lynch S.V.. 2016. Disease severity and immune activity relate to distinct interkingdom gut microbiome states in ethnically distinct Ulcerative Colitis patients. MBio. 7:e01072-16 10.1128/mBio.01072-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin I.A., Goertz J.E., Ren T., Rich S.S., Onengut-Gumuscu S., Farber E., Wu M., Overall C.C., Kipnis J., and Gaultier A.. 2017. Microbiota alteration is associated with the development of stress-induced despair behavior. Sci. Rep. 7:43859 10.1038/srep43859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson V., Fessler J., Bao R., Chongsuwat T., Zha Y., Alegre M.-L., Luke J.J., and Gajewski T.F.. 2018. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 359:104–108. 10.1126/science.aao3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice C.F., Haiser H.J., and Turnbaugh P.J.. 2013. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 152:39–50. 10.1016/j.cell.2012.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Han Y., Srisai D., Belakhov V., Farias M., Xu Y., Palmiter R.D., Baasov T., and Wu Q.. 2016. New inducible genetic method reveals critical roles of GABA in the control of feeding and metabolism. Proc. Natl. Acad. Sci. USA. 113:3645–3650. 10.1073/pnas.1602049113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura T., Rizzello F., Helwig U., Poggioli G., Schreiber S., Talbot I.C., Nicholls R.J., Gionchetti P., Campieri M., and Kamm M.A.. 2004. Once daily high dose probiotic therapy (VSL#3) for maintaining remission in recurrent or refractory pouchitis. Gut. 53:108–114. 10.1136/gut.53.1.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto J., Mizukure T., Park S.B., Kishino S., Kimura I., Hirano K., Bergamo P., Rossi M., Suzuki T., Arita M., et al. . 2015. A gut microbial metabolite of linoleic acid, 10-hydroxy-cis-12-octadecenoic acid, ameliorates intestinal epithelial barrier impairment partially via GPR40-MEK-ERK pathway. J. Biol. Chem. 290:2902–2918. 10.1074/jbc.M114.610733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moor K., Diard M., Sellin M.E., Felmy B., Wotzka S.Y., Toska A., Bakkeren E., Arnoldini M., Bansept F., Co A.D., et al. . 2017. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature. 544:498–502. 10.1038/nature22058 [DOI] [PubMed] [Google Scholar]
- Morland C., Frøland A.S., Pettersen M.N., Storm-Mathisen J., Gundersen V., Rise F., and Hassel B.. 2018. Propionate enters GABAergic neurons, inhibits GABA transaminase, causes GABA accumulation and lethargy in a model of propionic acidemia. Biochem. J. 475:749–758. 10.1042/BCJ20170814 [DOI] [PubMed] [Google Scholar]
- Nagpal R., Tsuji H., Takahashi T., Kawashima K., Nagata S., Nomoto K., and Yamashiro Y.. 2016. Sensitive Quantitative Analysis of the Meconium Bacterial Microbiota in Healthy Term Infants Born Vaginally or by Cesarean Section. Front. Microbiol. 7:1997 10.3389/fmicb.2016.01997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S., Bouladoux N., Linehan J.L., Han S.J., Harrison O.J., Wilhelm C., Conlan S., Himmelfarb S., Byrd A.L., Deming C., et al. . 2015. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 520:104–108. 10.1038/nature14052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T., Chen T.H., Narala S., Chun K.A., Two A.M., Yun T., Shafiq F., Kotol P.F., Bouslimani A., Melnik A.V., et al. . 2017. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 9:eaah4680 10.1126/scitranslmed.aah4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natividad J.M.M., Hayes C.L., Motta J.-P., Jury J., Galipeau H.J., Philip V., Garcia-Rodenas C.L., Kiyama H., Bercik P., and Verdu E.F.. 2013. Differential induction of antimicrobial REGIII by the intestinal microbiota and Bifidobacterium breve NCC2950. Appl. Environ. Microbiol. 79:7745–7754. 10.1128/AEM.02470-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor G.T., Lynch S.V., Bloomberg G.R., Kattan M., Wood R.A., Gergen P.J., Jaffee K.F., Calatroni A., Bacharier L.B., Beigelman A., et al. . 2018. Early-life home environment and risk of asthma among inner-city children. J. Allergy Clin. Immunol. 141:1468–1475. 10.1016/j.jaci.2017.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odamaki T., Kato K., Sugahara H., Hashikura N., Takahashi S., Xiao J.Z., Abe F., and Osawa R.. 2016. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 16:90 10.1186/s12866-016-0708-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J., Byrd A.L., Deming C., Conlan S., Kong H.H., and Segre J.A.. NISC Comparative Sequencing Program . 2014. Biogeography and individuality shape function in the human skin metagenome. Nature. 514:59–64. 10.1038/nature13786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnmacht C., Park J.H., Cording S., Wing J.B., Atarashi K., Obata Y., Gaboriau-Routhiau V., Marques R., Dulauroy S., Fedoseeva M., et al. . 2015. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORγt+ T cells. Science. 349:989–993. 10.1126/science.aac4263 [DOI] [PubMed] [Google Scholar]
- Ottman N., Reunanen J., Meijerink M., Pietilä T.E., Kainulainen V., Klievink J., Huuskonen L., Aalvink S., Skurnik M., Boeren S., et al. . 2017. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS One. 12:e0173004 10.1371/journal.pone.0173004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ownby D.R., Johnson C.C., and Peterson E.L.. 2002. Exposure to dogs and cats in the first year of life and risk of allergic sensitization at 6 to 7 years of age. JAMA. 288:963–972. 10.1001/jama.288.8.963 [DOI] [PubMed] [Google Scholar]
- Pan W.H., Sommer F., Falk-Paulsen M., Ulas T., Best P., Fazio A., Kachroo P., Luzius A., Jentzsch M., Rehman A., et al. . 2018. Exposure to the gut microbiota drives distinct methylome and transcriptome changes in intestinal epithelial cells during postnatal development. Genome Med. 10:27 10.1186/s13073-018-0534-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandiyan P., Conti H.R., Zheng L., Peterson A.C., Mathern D.R., Hernández-Santos N., Edgerton M., Gaffen S.L., and Lenardo M.J.. 2011. CD4(+)CD25(+)Foxp3(+) regulatory T cells promote Th17 cells in vitro and enhance host resistance in mouse Candida albicans Th17 cell infection model. Immunity. 34:422–434. 10.1016/j.immuni.2011.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K., and Bassler B.L.. 2016. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14:576–588. 10.1038/nrmicro.2016.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson C.C., Dahlquist G.G., Gyürüs E., Green A., and Soltész G.. EURODIAB Study Group . 2009. Incidence trends for childhood type 1 diabetes in Europe during 1989-2003 and predicted new cases 2005-20: a multicentre prospective registration study. Lancet. 373:2027–2033. 10.1016/S0140-6736(09)60568-7 [DOI] [PubMed] [Google Scholar]
- Pedersen H.K., Gudmundsdottir V., Nielsen H.B., Hyotylainen T., Nielsen T., Jensen B.A.H., Forslund K., Hildebrand F., Prifti E., Falony G., et al. MetaHIT Consortium . 2016. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 535:376–381. 10.1038/nature18646 [DOI] [PubMed] [Google Scholar]
- Pianta A., Arvikar S.L., Strle K., Drouin E.E., Wang Q., Costello C.E., and Steere A.C.. 2017. Two rheumatoid arthritis-specific autoantigens correlate microbial immunity with autoimmune responses in joints. J. Clin. Invest. 127:2946–2956. 10.1172/JCI93450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planer J.D., Peng Y., Kau A.L., Blanton L.V., Ndao I.M., Tarr P.I., Warner B.B., and Gordon J.I.. 2016. Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature. 534:263–266. 10.1038/nature17940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plovier H., Everard A., Druart C., Depommier C., Van Hul M., Geurts L., Chilloux J., Ottman N., Duparc T., Lichtenstein L., et al. . 2017. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 23:107–113. 10.1038/nm.4236 [DOI] [PubMed] [Google Scholar]
- Poutahidis T., Kearney S.M., Levkovich T., Qi P., Varian B.J., Lakritz J.R., Ibrahim Y.M., Chatzigiagkos A., Alm E.J., and Erdman S.E.. 2013. Microbial symbionts accelerate wound healing via the neuropeptide hormone oxytocin. PLoS One. 8:e78898 10.1371/journal.pone.0078898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor D.M., and Relman D.A.. 2017. The Landscape ecology and microbiota of the human nose, mouth, and throat. Cell Host Microbe. 21:421–432. 10.1016/j.chom.2017.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., et al. MetaHIT Consortium . 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 464:59–65. 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., Liang S., Zhang W., Guan Y., Shen D., et al. . 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 490:55–60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- Quévrain E., Maubert M.A., Michon C., Chain F., Marquant R., Tailhades J., Miquel S., Carlier L., Bermúdez-Humarán L.G., Pigneur B., et al. . 2016. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut. 65:415–425. 10.1136/gutjnl-2014-307649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan D., Bowcutt R., Lee S.C., Tang M.S., Kurtz Z.D., Ding Y., Honda K., Gause W.C., Blaser M.J., Bonneau R.A., et al. . 2016. Helminth infection promotes colonization resistance via type 2 immunity. Science. 352:608–612. 10.1126/science.aaf3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan K.J., Pedicord V.A., Wang Y.C., Kim B., Lu Y., Shaham S., Mucida D., and Hang H.C.. 2016. A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science. 353:1434–1437. 10.1126/science.aaf3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijnders D., Goossens G.H., Hermes G.D., Neis E.P., van der Beek C.M., Most J., Holst J.J., Lenaerts K., Kootte R.S., Nieuwdorp M., et al. . 2016. Effects of gut microbiota manipulation by antibiotics on host metabolism in obese humans: A randomized double-blind placebo-controlled trial. Cell Metab. 24:63–74. 10.1016/j.cmet.2016.06.016 [DOI] [PubMed] [Google Scholar]
- Richard C., Wadowski M., Goruk S., Cameron L., Sharma A.M., and Field C.J.. 2017. Individuals with obesity and type 2 diabetes have additional immune dysfunction compared with obese individuals who are metabolically healthy. BMJ Open Diabetes Res. Care. 5:e000379 10.1136/bmjdrc-2016-000379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura V.K., Faith J.J., Rey F.E., Cheng J., Duncan A.E., Kau A.L., Griffin N.W., Lombard V., Henrissat B., Bain J.R., et al. . 2013. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 341:1241214 10.1126/science.1241214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano K.A., Martinez-Del Campo A., Kasahara K., Chittim C.L., Vivas E.I., Amador-Noguez D., Balskus E.P., and Rey F.E.. 2017. Metabolic, epigenetic, and transgenerational effects of gut bacterial choline consumption. Cell Host Microbe. 22:279–290.e7. 10.1016/j.chom.2017.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooks M.G., and Garrett W.S.. 2016. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16:341–352. 10.1038/nri.2016.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi O., van Berkel L.A., Chain F., Tanweer Khan M., Taverne N., Sokol H., Duncan S.H., Flint H.J., Harmsen H.J.M., Langella P., et al. . 2016. Faecalibacterium prausnitzii A2-165 has a high capacity to induce IL-10 in human and murine dendritic cells and modulates T cell responses. Sci. Rep. 6:18507 10.1038/srep18507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild D., Weissbrod O., Barkan E., Kurilshikov A., Korem T., Zeevi D., Costea P.I., Godneva A., Kalka I.N., Bar N., et al. . 2018. Environment dominates over host genetics in shaping human gut microbiota. Nature. 555:210–215. 10.1038/nature25973 [DOI] [PubMed] [Google Scholar]
- Routy B., Le Chatelier E., Derosa L., Duong C.P.M., Alou M.T., Daillère R., Fluckiger A., Messaoudene M., Rauber C., Roberti M.P., et al. . 2018. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 359:91–97. 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- Roy S., and Trinchieri G.. 2017. Microbiota: a key orchestrator of cancer therapy. Nat. Rev. Cancer. 17:271–285. 10.1038/nrc.2017.13 [DOI] [PubMed] [Google Scholar]
- Ruiz V.E., Battaglia T., Kurtz Z.D., Bijnens L., Ou A., Engstrand I., Zheng X., Iizumi T., Mullins B.J., Müller C.L., et al. . 2017. A single early-in-life macrolide course has lasting effects on murine microbial network topology and immunity. Nat. Commun. 8:518 10.1038/s41467-017-00531-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S.L., Gold M.J., Hartmann M., Willing B.P., Thorson L., Wlodarska M., Gill N., Blanchet M.R., Mohn W.W., McNagny K.M., and Finlay B.B.. 2012. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 13:440–447. 10.1038/embor.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samah S., Ramasamy K., Lim S.M., and Neoh C.F.. 2016. Probiotics for the management of type 2 diabetes mellitus: A systematic review and meta-analysis. Diabetes Res. Clin. Pract. 118:172–182. 10.1016/j.diabres.2016.06.014 [DOI] [PubMed] [Google Scholar]
- Schaefer A.L., Greenberg E.P., Oliver C.M., Oda Y., Huang J.J., Bittan-Banin G., Peres C.M., Schmidt S., Juhaszova K., Sufrin J.R., and Harwood C.S.. 2008. A new class of homoserine lactone quorum-sensing signals. Nature. 454:595–599. 10.1038/nature07088 [DOI] [PubMed] [Google Scholar]
- Scharschmidt T.C., Vasquez K.S., Truong H.A., Gearty S.V., Pauli M.L., Nosbaum A., Gratz I.K., Otto M., Moon J.J., Liese J., et al. . 2015. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity. 43:1011–1021. 10.1016/j.immuni.2015.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharschmidt T.C., Vasquez K.S., Pauli M.L., Leitner E.G., Chu K., Truong H.-A., Lowe M.M., Sanchez Rodriguez R., Ali N., Laszik Z.G., et al. . 2017. Commensal microbes and hair follicle morphogenesis coordinately drive Treg migration into neonatal skin. Cell Host Microbe. 21:467–477.e5. 10.1016/j.chom.2017.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher J.U., Sczesnak A., Longman R.S., Segata N., Ubeda C., Bielski C., Rostron T., Cerundolo V., Pamer E.G., Abramson S.B., et al. . 2013. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2:e01202 10.7554/eLife.01202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M., Smeekens S.P., Vlamakis H., Jaeger M., Oosting M., Franzosa E.A., Ter Horst R., Jansen T., Jacobs L., Bonder M.J., et al. . 2016. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell. 167:1125–1136.e8. 10.1016/j.cell.2016.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M., Franzosa E.A., Lloyd-Price J., McIver L.J., Schwager R., Poon T.W., Ananthakrishnan A.N., Andrews E., Barron G., Lake K., et al. . 2018. Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat. Microbiol. 3:337–346. 10.1038/s41564-017-0089-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D., Iverson K.D., Petrosino J.F., and Schloss S.J.. 2014. The dynamics of a family’s gut microbiota reveal variations on a theme. Microbiome. 2:25 10.1186/2049-2618-2-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger M., Everard A., Gómez-Valadés A.G., Matamoros S., Ramírez S., Delzenne N.M., Gomis R., Claret M., and Cani P.D.. 2015. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci. Rep. 5:16643 10.1038/srep16643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnupf P., Gaboriau-Routhiau V., Gros M., Friedman R., Moya-Nilges M., Nigro G., Cerf-Bensussan N., and Sansonetti P.J.. 2015. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature. 520:99–103. 10.1038/nature14027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder B.O., Birchenough G.M.H., Ståhlman M., Arike L., Johansson M.E.V., Hansson G.C., and Bäckhed F.. 2018. Bifidobacteria or fiber protects against diet-induced microbiota-mediated colonic mucus deterioration. Cell Host Microbe. 23:27–40.e7. 10.1016/j.chom.2017.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer M., Makki K., Storelli G., Machuca-Gayet I., Srutkova D., Hermanova P., Martino M.E., Balmand S., Hudcovic T., Heddi A., et al. . 2016. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science. 351:854–857. 10.1126/science.aad8588 [DOI] [PubMed] [Google Scholar]
- Sefik E., Geva-Zatorsky N., Oh S., Konnikova L., Zemmour D., McGuire A.M., Burzyn D., Ortiz-Lopez A., Lobera M., Yang J., et al. . 2015. Mucosal immunology. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science. 349:993–997. 10.1126/science.aaa9420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segovia S.A., Vickers M.H., Gray C., Zhang X.D., and Reynolds C.M.. 2017. Conjugated linoleic acid supplementation improves maternal high fat diet-induced programming of metabolic dysfunction in adult male rat offspring. Sci. Rep. 7:6663 10.1038/s41598-017-07108-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R., Fuchs S., and Milo R.. 2016. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 164:337–340. 10.1016/j.cell.2016.01.013 [DOI] [PubMed] [Google Scholar]
- Shin N.-R., Lee J.-C., Lee H.-Y., Kim M.-S., Whon T.W., Lee M.-S., and Bae J.-W.. 2014. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 63:727–735. 10.1136/gutjnl-2012-303839 [DOI] [PubMed] [Google Scholar]
- Simeoli R., Mattace Raso G., Pirozzi C., Lama A., Santoro A., Russo R., Montero-Melendez T., Berni Canani R., Calignano A., Perretti M., and Meli R.. 2017. An orally administered butyrate-releasing derivative reduces neutrophil recruitment and inflammation in dextran sulphate sodium-induced murine colitis. Br. J. Pharmacol. 174:1484–1496. 10.1111/bph.13637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A.K., Hollander G.A., and McMichael A.. 2015. Evolution of the immune system in humans from infancy to old age. Proc. Biol. Sci. 282:20143085 10.1098/rspb.2014.3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Gurav A., Sivaprakasam S., Brady E., Padia R., Shi H., Thangaraju M., Prasad P.D., Manicassamy S., Munn D.H., et al. . 2014. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 40:128–139. 10.1016/j.immuni.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie C.S., Sauk J., Gevers D., Friedman J., Sung J., Youngster I., Hohmann E.L., Staley C., Khoruts A., Sadowsky M.J., et al. . 2018. Strain Tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe. 23:229–240.e5. 10.1016/j.chom.2018.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P.M., Howitt M.R., Panikov N., Michaud M., Gallini C.A., Bohlooly-Y M., Glickman J.N., and Garrett W.S.. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 341:569–573. 10.1126/science.1241165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders A.M., Langley S.A., Kim Y.M., Brislawn C.J., Noecker C., Zink E.M., Fansler S.J., Casey C.P., Miller D.R., Huang Y., et al. . 2016. Influence of early life exposure, host genetics and diet on the mouse gut microbiome and metabolome. Nat. Microbiol. 2:16221 10.1038/nmicrobiol.2016.221 [DOI] [PubMed] [Google Scholar]
- Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermúdez-Humarán L.G., Gratadoux J.-J., Blugeon S., Bridonneau C., Furet J.-P., Corthier G., et al. . 2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA. 105:16731–16736. 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H., Leducq V., Aschard H., Pham H.P., Jegou S., Landman C., Cohen D., Liguori G., Bourrier A., Nion-Larmurier I., et al. . 2017. Fungal microbiota dysbiosis in IBD. Gut. 66:1039–1048. 10.1136/gutjnl-2015-310746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg E.D., Smits S.A., Tikhonov M., Higginbottom S.K., Wingreen N.S., and Sonnenburg J.L.. 2016. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 529:212–215. 10.1038/nature16504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood A., Midha V., Makharia G.K., Ahuja V., Singal D., Goswami P., and Tandon R.K.. 2009. The probiotic preparation, VSL#3 induces remission in patients with mild-to-moderately active ulcerative colitis. Clin. Gastroenterol. Hepatol. 7:1202–1209.e1. 10.1016/j.cgh.2009.07.016 [DOI] [PubMed] [Google Scholar]
- Stokholm J., Thorsen J., Chawes B.L., Schjørring S., Krogfelt K.A., Bønnelykke K., and Bisgaard H.. 2016. Cesarean section changes neonatal gut colonization. J. Allergy Clin. Immunol. 138:881–889.e2. 10.1016/j.jaci.2016.01.028 [DOI] [PubMed] [Google Scholar]
- Stokholm J., Blaser M.J., Thorsen J., Rasmussen M.A., Waage J., Vinding R.K., Schoos A.M.M., Kunøe A., Fink N.R., Chawes B.L., et al. . 2018. Maturation of the gut microbiome and risk of asthma in childhood. Nat. Commun. 9:141 10.1038/s41467-017-02573-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strati F., Cavalieri D., Albanese D., De Felice C., Donati C., Hayek J., Jousson O., Leoncini S., Renzi D., Calabrò A., and De Filippo C.. 2017. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome. 5:24 10.1186/s40168-017-0242-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S., Huq S., Yatsunenko T., Haque R., Mahfuz M., Alam M.A., Benezra A., DeStefano J., Meier M.F., Muegge B.D., et al. . 2014. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 510:417–421. 10.1038/nature13421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T.G., Sefik E., Geva-Zatorsky N., Kua L., Naskar D., Teng F., Pasman L., Ortiz-Lopez A., Jupp R., Wu H.-J.J., et al. . 2016. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl. Acad. Sci. USA. 113:E8141–E8150. 10.1073/pnas.1617460113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C., Kamiya T., Liu Y., Kadoki M., Kakuta S., Oshima K., Hattori M., Takeshita K., Kanai T., Saijo S., et al. . 2015. Inhibition of Dectin-1 signaling ameliorates colitis by inducing Lactobacillus-mediated regulatory T cell expansion in the intestine. Cell Host Microbe. 18:183–197. 10.1016/j.chom.2015.07.003 [DOI] [PubMed] [Google Scholar]
- Theriot C.M., Koenigsknecht M.J., Carlson P.E. Jr., Hatton G.E., Nelson A.M., Li B., Huffnagle G.B., Z Li J., and Young V.B.. 2014. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 5:3114 10.1038/ncomms4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevaranjan N., Puchta A., Schulz C., Naidoo A., Szamosi J.C., Verschoor C.P., Loukov D., Schenck L.P., Jury J., Foley K.P., et al. . 2017. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe. 21:455–466.e4. 10.1016/j.chom.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiemann S., Smit N., Roy U., Lesker T.R., Gálvez E.J.C., Helmecke J., Basic M., Bleich A., Goodman A.L., Kalinke U., et al. . 2017. Enhancement of IFNγ production by distinct commensals ameliorates Salmonella-induced disease. Cell Host Microbe. 21:682–694.e5. 10.1016/j.chom.2017.05.005 [DOI] [PubMed] [Google Scholar]
- Thompson S.V., Hannon B.A., An R., and Holscher H.D.. 2017. Effects of isolated soluble fiber supplementation on body weight, glycemia, and insulinemia in adults with overweight and obesity: a systematic review and meta-analysis of randomized controlled trials. Am. J. Clin. Nutr. 106:1514–1528. 10.3945/ajcn.117.163246 [DOI] [PubMed] [Google Scholar]
- Ticinesi A., Milani C., Lauretani F., Nouvenne A., Mancabelli L., Lugli G.A., Turroni F., Duranti S., Mangifesta M., Viappiani A., et al. . 2017. Gut microbiota composition is associated with polypharmacy in elderly hospitalized patients. Sci. Rep. 7:11102 10.1038/s41598-017-10734-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L.C., Wang Y., Wang Z.B., Liu W.Y., Sun S., Li L., Su D.F., and Zhang L.C.. 2016. Propionate ameliorates dextran sodium sulfate-induced colitis by improving intestinal barrier function and reducing inflammation and oxidative stress. Front. Pharmacol. 7:253 10.3389/fphar.2016.00253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshimitsu T., Ozaki S., Mochizuki J., Furuichi K., and Asami Y.. 2017. Effects of Lactobacillus plantarum strain OLL2712 culture conditions on the anti-inflammatory activities for murine immune cells and obese and Type 2 diabetic mice. Appl. Environ. Microbiol. 83:e03001-16 10.1128/AEM.03001-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremlett H., Fadrosh D.W., Faruqi A.A., Hart J., Roalstad S., Graves J., Spencer C.M., Lynch S.V., Zamvil S.S., and Waubant E.. US Network of Pediatric MS Centers . 2016a Associations between the gut microbiota and host immune markers in pediatric multiple sclerosis and controls. BMC Neurol. 16:182 10.1186/s12883-016-0703-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremlett H., Fadrosh D.W., Faruqi A.A., Zhu F., Hart J., Roalstad S., Graves J., Lynch S., and Waubant E.. US Network of Pediatric MS Centers . 2016b Gut microbiota in early pediatric multiple sclerosis: a case-control study. Eur. J. Neurol. 23:1308–1321. 10.1111/ene.13026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A., Gollwitzer E.S., Yadava K., Sichelstiel A.K., Sprenger N., Ngom-Bru C., Blanchard C., Junt T., Nicod L.P., Harris N.L., and Marsland B.J.. 2014. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20:159–166. 10.1038/nm.3444 [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E., Sogin M.L., Jones W.J., Roe B.A., Affourtit J.P., et al. . 2009. A core gut microbiome in obese and lean twins. Nature. 457:480–484. 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpin W., Espin-Garcia O., Xu W., Silverberg M.S., Kevans D., Smith M.I., Guttman D.S., Griffiths A., Panaccione R., Otley A., et al. GEM Project Research Consortium . 2016. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat. Genet. 48:1413–1417. 10.1038/ng.3693 [DOI] [PubMed] [Google Scholar]
- Uusitalo U., Liu X., Yang J., Aronsson C.A., Hummel S., Butterworth M., Lernmark Å., Rewers M., Hagopian W., She J.X., et al. TEDDY Study Group . 2016. Association of early exposure of probiotics and islet autoimmunity in the TEDDY study. JAMA Pediatr. 170:20–28. 10.1001/jamapediatrics.2015.2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nood E., Vrieze A., Nieuwdorp M., Fuentes S., Zoetendal E.G., de Vos W.M., Visser C.E., Kuijper E.J., Bartelsman J.F.W.M., Tijssen J.G.P., et al. . 2013. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368:407–415. 10.1056/NEJMoa1205037 [DOI] [PubMed] [Google Scholar]
- Vatanen T., Kostic A.D., d’Hennezel E., Siljander H., Franzosa E.A., Yassour M., Kolde R., Vlamakis H., Arthur T.D., Hämäläinen A.M., et al. DIABIMMUNE Study Group . 2016. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 165:842–853. 10.1016/j.cell.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh M., Mukherjee S., Wang H., Li H., Sun K., Benechet A.P., Qiu Z., Maher L., Redinbo M.R., Phillips R.S., et al. . 2014. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 41:296–310. 10.1016/j.immuni.2014.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verster A.J., Ross B.D., Radey M.C., Bao Y., Goodman A.L., Mougous J.D., and Borenstein E.. 2017. The landscape of type VI secretion across human gut microbiomes reveals its role in community composition. Cell Host Microbe. 22:411–419.e4. 10.1016/j.chom.2017.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viladomiu M., Hontecillas R., and Bassaganya-Riera J.. 2016. Modulation of inflammation and immunity by dietary conjugated linoleic acid. Eur. J. Pharmacol. 785:87–95. 10.1016/j.ejphar.2015.03.095 [DOI] [PubMed] [Google Scholar]
- Vrieze A., Van Nood E., Holleman F., Salojärvi J., Kootte R.S., Bartelsman J.F., Dallinga-Thie G.M., Ackermans M.T., Serlie M.J., Oozeer R., et al. . 2012. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 143:913–6.e7. 10.1053/j.gastro.2012.06.031 [DOI] [PubMed] [Google Scholar]
- Wang L., Christophersen C.T., Sorich M.J., Gerber J.P., Angley M.T., and Conlon M.A.. 2012. Elevated fecal short chain fatty acid and ammonia concentrations in children with autism spectrum disorder. Dig. Dis. Sci. 57:2096–2102. 10.1007/s10620-012-2167-7 [DOI] [PubMed] [Google Scholar]
- Wang Y., Begum-Haque S., Telesford K.M., Ochoa-Repáraz J., Christy M., Kasper E.J., Kasper D.L., Robson S.C., and Kasper L.H.. 2014a A commensal bacterial product elicits and modulates migratory capacity of CD39(+) CD4 T regulatory subsets in the suppression of neuroinflammation. Gut Microbes. 5:552–561. 10.4161/gmic.29797 [DOI] [PubMed] [Google Scholar]
- Wang Z., Klipfell E., Bennett B.J., Koeth R., Levison B.S., Dugar B., Feldstein A.E., Britt E.B., Fu X., Chung Y.-M., et al. . 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 472:57–63. 10.1038/nature09922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Friedrich C., Hagemann S.C., Korte W.H., Goharani N., Cording S., Eberl G., Sparwasser T., and Lochner M.. 2014b Regulatory T cells promote a protective Th17-associated immune response to intestinal bacterial infection with C. rodentium. Mucosal Immunol. 7:1290–1301. 10.1038/mi.2014.17 [DOI] [PubMed] [Google Scholar]
- Wang Z., Roberts A.B., Buffa J.A., Levison B.S., Zhu W., Org E., Gu X., Huang Y., Zamanian-Daryoush M., Culley M.K., et al. . 2015. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell. 163:1585–1595. 10.1016/j.cell.2015.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler M.L., Limon J.J., Bar A.S., Leal C.A., Gargus M., Tang J., Brown J., Funari V.A., Wang H.L., Crother T.R., et al. . 2016. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe. 19:865–873. 10.1016/j.chom.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer D.A., Luck H., Tsai S., and Winer S.. 2016. The intestinal immune system in obesity and insulin resistance. Cell Metab. 23:413–426. 10.1016/j.cmet.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Wu G.D., Compher C., Chen E.Z., Smith S.A., Shah R.D., Bittinger K., Chehoud C., Albenberg L.G., Nessel L., Gilroy E., et al. . 2016. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut. 65:63–72. 10.1136/gutjnl-2014-308209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.J., Ivanov I.I., Darce J., Hattori K., Shima T., Umesaki Y., Littman D.R., Benoist C., and Mathis D.. 2010. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 32:815–827. 10.1016/j.immuni.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Guo R., Zhong H., Feng Q., Lan Z., Qin B., Ward K.J., Jackson M.A., Xia Y., Chen X., et al. . 2016. Shotgun metagenomics of 250 adult twins reveals genetic and environmental impacts on the gut microbiome. Cell Syst. 3:572–584.e3. 10.1016/j.cels.2016.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto-Hanada K., Yang L., Narita M., Saito H., and Ohya Y.. 2017. Influence of antibiotic use in early childhood on asthma and allergic diseases at age 5. Ann. Allergy Asthma Immunol. 119:54–58. 10.1016/j.anai.2017.05.013 [DOI] [PubMed] [Google Scholar]
- Yan J., Herzog J.W., Tsang K., Brennan C.A., Bower M.A., Garrett W.S., Sartor B.R., Aliprantis A.O., and Charles J.F.. 2016. Gut microbiota induce IGF-1 and promote bone formation and growth. Proc. Natl. Acad. Sci. USA. 113:E7554–E7563. 10.1073/pnas.1607235113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano J.M., Yu K., Donaldson G.P., Shastri G.G., Ann P., Ma L., Nagler C.R., Ismagilov R.F., Mazmanian S.K., and Hsiao E.Y.. 2015. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 161:264–276. 10.1016/j.cell.2015.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M., Magris M., Hidalgo G., Baldassano R.N., Anokhin A.P., et al. . 2012. Human gut microbiome viewed across age and geography. Nature. 486:222–227. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi D., Korem T., Zmora N., Israeli D., Rothschild D., Weinberger A., Ben-Yacov O., Lador D., Avnit-Sagi T., Lotan-Pompan M., et al. . 2015. Personalized nutrition by prediction of glycemic responses. Cell. 163:1079–1094. 10.1016/j.cell.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Zelante T., Iannitti R.G., Cunha C., De Luca A., Giovannini G., Pieraccini G., Zecchi R., D’Angelo C., Massi-Benedetti C., Fallarino F., et al. . 2013. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 39:372–385. 10.1016/j.immuni.2013.08.003 [DOI] [PubMed] [Google Scholar]
- Zenewicz L.A., Yin X., Wang G., Elinav E., Hao L., Zhao L., and Flavell R.A.. 2013. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J. Immunol. 190:5306–5312. 10.4049/jimmunol.1300016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Derrien M., Levenez F., Brazeilles R., Ballal S.A., Kim J., Degivry M.-C., Quéré G., Garault P., van Hylckama Vlieg J.E.T., et al. . 2016. Ecological robustness of the gut microbiota in response to ingestion of transient food-borne microbes. ISME J. 10:2235–2245. 10.1038/ismej.2016.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhang D., Jia H., Feng Q., Wang D., Liang D., Wu X., Li J., Tang L., Li Y., et al. . 2015. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 21:895–905. 10.1038/nm.3914 [DOI] [PubMed] [Google Scholar]
- Zhao L., Zhang F., Ding X., Wu G., Lam Y.Y., Wang X., Fu H., Xue X., Lu C., Ma J., et al. . 2018. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 359:1151–1156. 10.1126/science.aao5774 [DOI] [PubMed] [Google Scholar]
- Zheng L., Kelly C.J., Battista K.D., Schaefer R., Lanis J.M., Alexeev E.E., Wang R.X., Onyiah J.C., Kominsky D.J., and Colgan S.P.. 2017. Microbial-derived butyrate promotes epithelial barrier function through IL-10 receptor-dependent repression of Claudin-2. J. Immunol. 199:2976–2984. 10.4049/jimmunol.1700105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Gregory J.C., Org E., Buffa J.A., Gupta N., Wang Z., Li L., Fu X., Wu Y., Mehrabian M., et al. . 2016. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. 165:111–124. 10.1016/j.cell.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Winter M.G., Byndloss M.X., Spiga L., Duerkop B.A., Hughes E.R., Büttner L., de Lima Romão E., Behrendt C.L., Lopez C.A., et al. . 2018. Precision editing of the gut microbiota ameliorates colitis. Nature. 553:208–211. 10.1038/nature25172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L., Ma Y., Raoult D., Kroemer G., and Gajewski T.F.. 2018. The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Science. 359:1366–1370. 10.1126/science.aar6918 [DOI] [PubMed] [Google Scholar]
- Zmora N., Zilberman-Schapira G., Suez J., Mor U., Dori-Bachash M., Bashiardes S., Kotler E., Zur M., Regev-Lehavi D., Brik R.B., et al. . 2018. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell. 174:1388–1405.e21. 10.1016/j.cell.2018.08.041 [DOI] [PubMed] [Google Scholar]
- Zou J., Chassaing B., Singh V., Pellizzon M., Ricci M., Fythe M.D., Kumar M.V., and Gewirtz A.T.. 2018. Fiber-mediated nourishment of gut microbiota protects against diet-induced obesity by restoring IL-22-mediated colonic health. Cell Host Microbe. 23:41–53.e4. 10.1016/j.chom.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]