Di Carlo et al. discuss how the regulation/dysregulation of Polycomb group proteins contributes to hematopoiesis and hematological disorders.
Abstract
Epigenetic mechanisms are crucial for sustaining cell type–specific transcription programs. Among the distinct factors, Polycomb group (PcG) proteins are major negative regulators of gene expression in mammals. These proteins play key roles in regulating the proliferation, self-renewal, and differentiation of stem cells. During hematopoietic differentiation, many PcG proteins are fundamental for proper lineage commitment, as highlighted by the fact that a lack of distinct PcG proteins results in embryonic lethality accompanied by differentiation biases. Correspondingly, proteins of these complexes are frequently dysregulated in hematological diseases. In this review, we present an overview of the role of PcG proteins in normal and malignant hematopoiesis, focusing on the compositional complexity of PcG complexes, and we briefly discuss the ongoing clinical trials for drugs targeting these factors.
Introduction
Adult blood cell production is a hierarchical process that takes place in the bone marrow, where low proliferating hematopoietic stem cells (HSCs) both self-renew and differentiate into every mature blood cell type. Based on reconstitution ability, HSCs can be subgrouped into a small fraction of quiescent, long-term (LT)-HSCs and a more active group of short-term (ST)-HSCs (Smith et al., 1991; Osawa et al., 1996; Yang et al., 2005). According to a classical hematopoietic lineage differentiation model, these populations give rise to multipotent progenitors (MPPs) that lack self-renewal ability, enter the cell cycle more frequently, and are primed for differentiation (Morrison et al., 1997). Either common lymphoid or common myeloid progenitors (CLPs and CMPs, respectively) arise from the commitment of MPPs. CLPs further differentiate to produce T and B cells as well as natural killer and dendritic cells. CMPs produce megakaryocytes and erythrocytes (with a common progenitor, megakaryocyte–erythroid progenitor [MEP]) along with granulocytes and macrophages (from the granulocyte–monocyte progenitor cell [GMP]; Kondo et al., 1997; Akashi et al., 2000; Na Nakorn et al., 2002). However, a large body of evidence is now challenging this classical view of differentiation (Woolthuis and Park, 2016).
The entire differentiation process is highly regulated by both extrinsic and intrinsic factors, the latter being mainly represented by epigenetic regulators of gene expression. Indeed, genome-wide sequencing approaches show that epigenetic regulators are frequently mutated in hematological malignancies (Plass et al., 2013), making it important that we obtain a better understanding of their roles in both physiological and malignant hematopoiesis. Numerous proteins of the Polycomb (Pc) and Trithorax (Trx) complexes have been identified among these epigenetic factors. These two complexes play crucial roles in gene expression regulation in mammals. The Pc repressive complexes 1 and 2 (PRC1 and PRC2) enforce gene silencing through chromatin compaction and repressive histone posttranslational modifications (Schuettengruber et al., 2017). Their activity is counteracted by the Trx complexes, which deposit activating histone marks and thus allow high levels of transcription (see text box).
Trx group (TrxG) proteins
In Drosophila melanogaster, Pc-mediated repression of the Hox gene cluster is counteracted by the activity of the Trx gene and TrxG proteins. The mixed-lineage leukemia (MLL) gene is a mammalian homologue of Trx and was first identified as frequently involved in chromatin rearrangements in infant leukemia patients (Ziemin-van der Poel et al., 1991; Rowley, 1993; Mbangkollo et al., 1995). MLL has seven paralogues in mammals (MLL1–5 and SETd1A/B). Analogous to PRC1/2, MLL assembles distinct complexes around the set of evolutionarily conserved core subunits WDR5, RbBP5, ASH2L, and DPY30 (WRAD; Nakamura et al., 2002). These proteins are necessary to enhance MLL histone methyltransferase activity and to regulate MLL complex recruitment to chromatin (Bochyńska et al., 2018). Association of additional subunits such as the histone demethylase UTX (specific for H3K27) extends the catalytic repertoire of the complex by simultaneously providing erasing of H3K27me3 repressive marks deposited by EZH2 and deposition of the H3K4me3 activating mark by MLL (Agger et al., 2007; Lan et al., 2007).
In hematopoiesis, MLL is necessary for self-renewal in adult (but not fetal) hematopoietic stem/progenitor cells (HSPCs; Jude et al., 2007; McMahon et al., 2007; Gan et al., 2010) as well as for proliferation and lymphopoiesis by maintaining proper expression of HOX genes (Yu et al., 1995; Yagi et al., 1998; Ayton et al., 2001; Ernst et al., 2004). However, the catalytic activity of MLL seems to be dispensable (Terranova et al., 2006; Mishra et al., 2014). Heterozygous translocations involving the MLL gene are found in a very high percentage of infant leukemia patients affected by either acute myeloid leukemia (AML; >35%) or acute lymphoblastic leukemia (ALL; >70%). In >90% of the cases, the breakpoint region is localized between exon 9 and intron 11 (Meyer et al., 2018), resulting in the production of chimeric gain-of-function (GOF) proteins containing an N-terminal truncated form of MLL. To date, 135 distinct translocation partner genes (TPGs) have been described. The five most common TPGs (AF4, AF9, ENL, AF10, and ELL) account for ∼80% of the translocations (Meyer et al., 2018). All of these gene products belong to multiprotein complexes involved in transcription elongation either in the super elongation complex (SEC), the DOT1L complex (DotCom), or both (Okada et al., 2005; Lin et al., 2010; Mohan et al., 2010). Molecular mechanisms behind MLL chimera–mediated leukemogenesis are not yet fully understood; however, this process seems to involve the aberrant expression of the HOXA9 and MEIS1 genes, two master regulators of myeloid lineage. Both of these genes are targeted by SEC and DotCom (Okada et al., 2005; Lin et al., 2010) as well as by WT MLL and MLL chimeras in leukemic cells (Milne et al., 2005; Faber et al., 2009). HOXA9 and MEIS1 expression is necessary for survival of leukemic cells, and their overexpression in normal HSPCs is sufficient to induce leukemic transformation (Kroon et al., 1998; Zeisig et al., 2004; Faber et al., 2009). In line with a GOF scenario, most of the proteins involved in physiological regulation of these loci are necessary for MLL chimera–mediated leukemogenesis, including WT MLL, its interactor menin (Yokoyama et al., 2005), and the SEC subunits pTEFb and DOT1L (Okada et al., 2005; Krivtsov et al., 2008). Indeed, drugs targeting the WRAD–MLL interaction (Karatas et al., 2013; Senisterra et al., 2013; Cao et al., 2014) or the menin–MLL interaction (Grembecka et al., 2012; Shi et al., 2012; Borkin et al., 2015) or that inhibit the DOT1L H3K79 methyltransferase activity (Cai et al., 2015) have been shown to be effective in arresting proliferation of leukemic cells.
MLL2 and -3 appear to play an oncogenic role in AML (Chen et al., 2014, 2017; Santos et al., 2014). Conversely, these proteins seem to act as tumor suppressors in B cells and derived lymphomas (Ortega-Molina et al., 2015; Zhang et al., 2015). In line with this, loss-of-function (LOF) mutations of MLL2 and -3 are found at relatively high frequencies in diffuse large B cell lymphoma (DLBCL), follicular lymphoma (FL), and ALL (Morin et al., 2011; Lohr et al., 2012; Zhang et al., 2013; Green et al., 2015; Lindqvist et al., 2015; Neumann et al., 2015). UTX, another important accessory factor, is also found mutated in various types of leukemia (van Haaften et al., 2009; Jankowska et al., 2011; Mar et al., 2012).
In this review, we discuss the importance of the Pc complexes in normal hematopoiesis, with a particular focus on the specific subunits and complexes involved in the distinct differentiation steps. We also review the roles played by gain-of-function (GOF) and loss-of-function (LOF) mutations of Pc group (PcG) proteins responsible for altered epigenetic landscapes in hematological disorders. Finally, we focus on drugs designed to target PcG proteins, with the aim of counteracting aberrant epigenetic regulation in hematological disorders.
Composition and function of Pc complexes
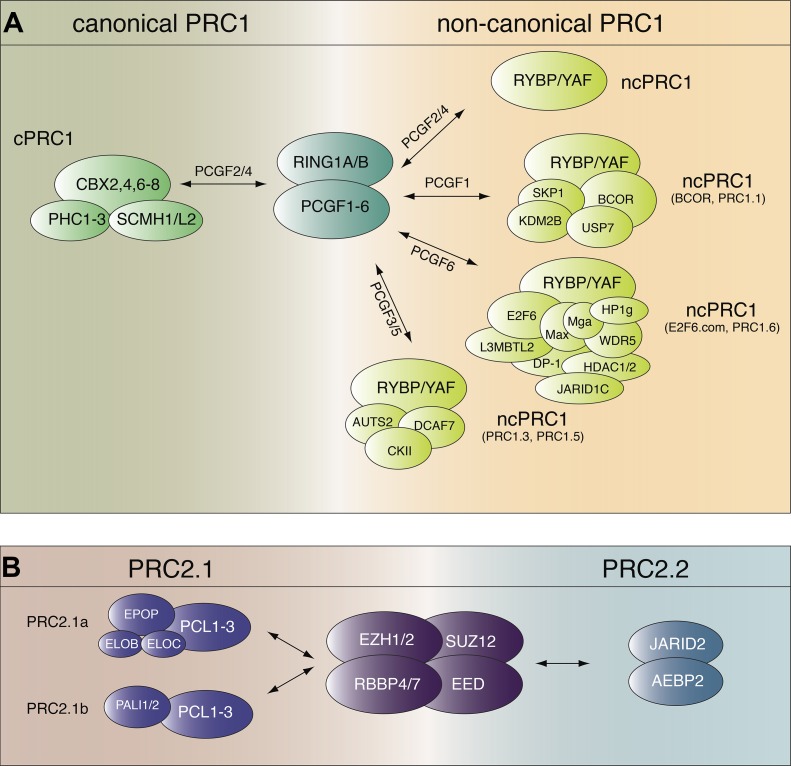
Mutations in Drosophila that are associated with sex comb development were first isolated in the 1940s and termed extra sex combs (esc) and Pc (Slifer, 1942; Lewis and Mislove, 1947). More than 30 yr later, esc and Pc gene products were identified as negative regulators of the homeotic gene Ultrabithorax (Ubx; Lewis, 1978; Struhl, 1981). Genes whose mutations give rise to developmental defects resembling those of esc and Pc were thereafter termed PcG genes (Jürgens, 1985). The proteins encoded by PcG genes were described as part of two distinct multiprotein complexes, PRC1 and PRC2 (Shao et al., 1999; Kuzmichev et al., 2002), which are highly conserved in mammals (Kuzmichev et al., 2002; Levine et al., 2002). Gene silencing by these complexes is associated with their ability to catalyze posttranslational modifications of histone tails, namely histone H2A monoubiquitylation for PRC1 and histone H3 lysine 27 methylation for PRC2 (Cao et al., 2002; Wang et al., 2004a). In both cases, the enzymatic activity is endowed in the core subunits, around which different sets of accessory factors assemble to modulate catalysis and to regulate PRC1 and -2 recruitment to chromatin. The six subtypes of PRC1 (PRC1.1−6) are specified by the incorporation of one of the six PcG ring finger (PCGF) proteins: NSPC1/PCGF1, MEL-18/PCGF2, PCGF3, BMI-1/PCGF4, PCGF5, or MBLR/PCGF6 (Fig. 1 A; Gao et al., 2012). PRC2 has two main configurations, PRC2.1 and PRC2.2 (Fig. 1 B; Beringer et al., 2016). A list of PcG proteins along with their reported function is shown in Table 1 (see also Aranda et al., 2015; Holoch and Margueron, 2017; Schuettengruber et al., 2017).
Figure 1.
Mammalian PRC1/2 compositional complexity. (A) PcG proteins RING1A/B and PCGF1−6 compose a core around which accessory subunits associate. cPRC1 incorporates one PHC and one CBX protein. Noncanonical PRC1 (ncPRC1) complexes incorporate RYBP/YAF2 along with specific sets of additional proteins. (B) PRC2 shares a similar organization, with a tetrameric core composed of EZH1/2, SUZ12, EED, and RBBP4/7. Association with PCL proteins defines a PRC2.1 subtype that can associate with either EPOP or PALI1/2 (PRC2.1a/b). Conversely, association with AEBP2 and JARID2 defines a PRC2.2 subtype.
Table 1. PcG proteins and their molecular functions.
| Complex | Protein | Function |
|---|---|---|
| Core PRC1 | RING1A/B | H2A monoubiquitylation and nucleosome binding |
| PCGF1−6 | Stimulation of enzymatic activity | |
| PRC1.2, 4 | CBX2,4,6−8 | H3K9/K27me3 binding |
| PHC1−3 | Oligomerization and chromatin compaction | |
| SCMH1/L2 | Histone methyl–lysine binding and RNA binding | |
| PRC1.1, 3, 5, 6 | RYBP/YAF2 | DNA binding (unspecific) and interaction with YY1 |
| PRC1.1 | BCOR/BCORL1 | Scaffold |
| KDM2B | H3K36 demethylation and DNA binding (unmethylated CpG islands) | |
| SKP1 | Ubiquitin ligase and interaction with CUL1 | |
| USP7 | Stimulation of enzymatic activity | |
| PRC1.3, 5 | DCAF7 | Scaffold |
| CK2 | Inhibition of enzymatic activity | |
| AUTS2/FBRS/FBSL | Transcription activation | |
| PRC1.6 | WDR5 | Scaffold |
| L3MBTL2 | Histone methyl–lysine binding and chromatin compaction | |
| HP1γ/CBX3 | H3K9me3 binding | |
| JARID1C | H3K4me2/3 demethylase | |
| G9a | H3K9 methyltransferase | |
| HDAC1/2 | Histone deacetylase | |
| DP-1 | DNA binding (E2F recognition site) | |
| E2F6 | DNA binding (E2F recognition site) | |
| MAX | DNA binding (E-boxes) | |
| MGA | DNA binding (E-boxes) | |
| Core PRC2 | EZH1/2 | H3K27 methyltransferase |
| SUZ12 | DNA/RNA binding | |
| EED | H3K27me3 binding | |
| RBBP4/7 | Histones binding | |
| PRC2.1 | PCL1/PHF1 | H3K36me2/3 binding, DNA binding (unspecific), and stimulation of enzymatic activity |
| PCL2/MTF2 | H3K36me2/3 binding and DNA binding (unmethylated CpG islands) | |
| PCL3/PHF19 | H3K36me2/3 binding | |
| EPOP | Inhibition of enzymatic activity and interaction with Elongin B and C | |
| PALI1/2 | Stimulation of enzymatic activity | |
| PRC2.2 | JARID2 | DNA/RNA binding, H2Aub binding, and stimulation of enzymatic activity |
| AEBP2 | DNA binding, H2Aub binding, and stimulation of enzymatic activity |
In the classical model of recruitment for these two complexes, the H3K27me3 mark is deposited by PRC2, which is in turn recognized by chromobox homolog (CBX) proteins contained in PRC1.2/4 (also termed canonical PRC1 [cPRC1]; Wang et al., 2004b). However, noncanonical PRC1s (PRC1.1, 3, 5, and 6), which contain RING1- and YY1-binding protein (RYBP), rely on an alternative, H3K27me3-independent mode of recruitment (Tavares et al., 2012). Moreover, PRC2.2 is able to recognize ubiquitylated H2A (H2Aub; Cooper et al., 2016), suggesting that there is more than a single way of crosstalk between PcG proteins.
PRC1 and -2 are responsible for repressing pluripotency genes during embryonic stem cell (ESC) differentiation in both mouse and human (Boyer et al., 2006). For both complexes, changes in the expression and arrangement of the different subunits occur along the differentiation pathways, suggesting that their dynamic expression is relevant for committing cells to a specific fate (Morey et al., 2012, 2015; Kloet et al., 2016). Notably, however, the influence of PcG is not limited to early developmental stages but extends to various subtypes of adult stem cells (Aloia et al., 2013; Schuettengruber et al., 2017).
PcG proteins in hematopoiesis
Canonical PRC1
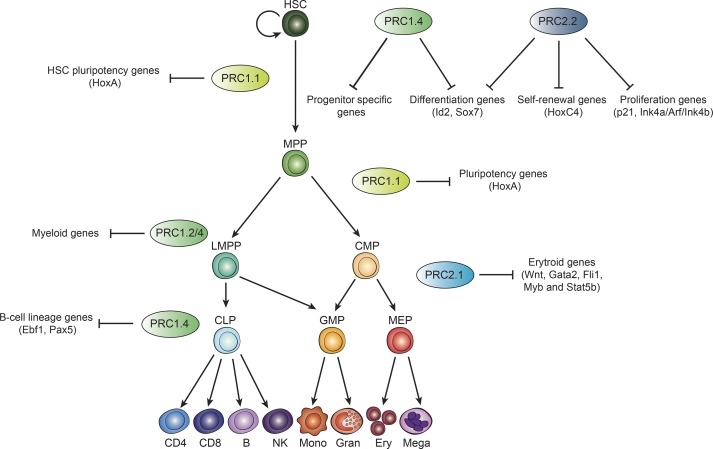
The B cell–specific Moloney murine leukemia virus integration site 1 (BMI-1/PCGF4) was first identified as an oncogene in MYC-mediated lymphomagenesis (Haupt et al., 1991; van Lohuizen et al., 1991) and has since been thoroughly studied in both normal and malignant hematopoiesis. PRC1 containing BMI-1 (PRC1.4) appears to be responsible for both the commitment of mesoderm layer to primitive HSC formation and the maintenance of LT-HSC self-renewal and proliferation capacities. Specifically, BMI-1 overexpression in ESCs leads to enhanced proliferation of embryoid body–derived primitive HSCs (Ding et al., 2012). Moreover, HSCs that overexpress BMI-1 display increased proliferation and self-renewal rates both in mouse models and human cell models (Iwama et al., 2004; Rizo et al., 2008). In accordance with this, BMI-1–depleted mice show defects in self-renewal and increased apoptosis of HSCs (Park et al., 2003; Iwama et al., 2004; Oguro et al., 2006; Liu et al., 2009; Rizo et al., 2009). In particular, PRC1.4 enables HSCs to overcome senescence and apoptosis by repressing the Ink4a/Arf (Cdkn2a) locus as well as by preventing DNA damage (Fig. 2).
Figure 2.
PRC1/2 in normal adult hematopoiesis. Schematic representation of the hematopoietic differentiation according to the classical model. PRC complexes that were described to have a role in regulating gene expression at specific stages are shown along with their reported genomic targets. Adapted from Corces et al. (2016).
In addition to its role in HSCs, PRC1.4 seems to have a fundamental function in regulating lymphoid specification by preventing B cell lineage commitment. Specifically, RING1B and BMI-1 are localized at the bivalent promoters of B-lineage master regulators Ebf1 and Pax5, and their depletion in T cells leads to accelerated activation of these transcription factors, resulting in T-to-B cell conversion (Oguro et al., 2010; Ikawa et al., 2016). Notably, MEL-18/PCGF2 seems to play an opposite role, which is however limited to adult hematopoiesis: mice lacking MEL-18 show no defects in fetal liver cell proliferation (Iwama et al., 2004) but display spleen and thymus hypoplasia at birth as well as perinatal lethality, which are associated with defects in B cell production (Akasaka et al., 1997; Tetsu et al., 1998). Moreover, HSCs from Mel-18−/− mice are more quiescent and less proliferative than those from WT mice. These studies point to complementary functions for PRC1.2 and PRC1.4 in regulating HSC self-renewal and proliferation as well as in maintaining the balance between B and T cells in lymphoid lineage (Fig. 2).
Along with the BMI-1/MEL-18 duality, cPRC1 activity in hematopoiesis is modulated by incorporation of alternative CBX proteins. In particular, LT-HSCs seem to preferentially express and incorporate CBX7; in mice models, CBX7 overexpression results in an enhanced self-renewal ability and overproliferation of HSCs, eventually leading to T cell leukemia/lymphoma in transplanted mice, while its depletion has the opposite effect (Scott et al., 2007; Klauke et al., 2013). In HSCs, PRC1 that contains CBX7 is located on genes that are progressively up-regulated during the HSC-to-progenitor transition, consistent with the rapid down-regulation of CBX7 during this phase (Klauke et al., 2013). Indeed, concomitant posttranscriptional up-regulation of CBX8 at the progenitor stage results in retargeting PRC1 to myeloid-specific genes (Klauke et al., 2013), suggesting that CBX8 plays a specific role at the level of MPPs and during lymphoid differentiation. This dynamics resembles that observed in ESCs: CBX7, which is responsible for maintaining the pluripotent state, is progressively down-regulated and then replaced with CBX2/4 during differentiation (Morey et al., 2012). In contrast, CBX8 appears dispensable for HSC activity. Recent evidence points to a fundamental function for the CBX8-containing PRC1, which works together with PRC2 to determine B cell germ cell formation (Béguelin et al., 2013, 2016, 2017; Caganova et al., 2013), suggesting that it is functional more for lineage commitment than for HSC maintenance (Tan et al., 2011). CBX2 impairs HSC and progenitor proliferation by regulating p21 expression in human cells (van den Boom et al., 2013). CBX2 also has a specific role in committing cells toward B-lymphoid lineage as irradiated mice transplanted with HSCs overexpressing CBX2 are only able to reconstitute B cells (Coré et al., 1997; Klauke et al., 2013). Analogously, CBX4 seems to play a role in differentiation rather than in maintaining pluripotency: depletion of this protein results in arrest of T cell development shortly after birth as a result of impaired thymic epithelial cell proliferation (Liu et al., 2013). Altogether, these studies reveal nonredundant roles for CBX proteins, with CBX7 sustaining LT- and ST-HSC proliferation and self-renewal, and CBX2, -4, and -8 mainly playing specific roles during hematopoietic lineage commitment but unable to functionally compensate for each other. These differences could be explained by differential recruitment mechanisms; however, mechanistic insights that could support this hypothesis are still missing (Fig. 2).
PHC1 is essential for PRC1 functioning in hematopoiesis, and in particular in B cell development. Knockout (KO) of Phc1 in mice results in impaired B cell development and perinatal lethality (Takihara et al., 1997; Tokimasa et al., 2001). Defects in B cell maturation are also visible in Phc1 heterozygous mice. Moreover, Phc1-deficient HSCs are not able to reconstitute blood in transplanted irradiated mice (Ohta et al., 2002; Kim et al., 2004). Although mechanistic insight is still lacking about the role of this protein (as well as its paralogues PHC2/3) in hematopoiesis, evidence suggests a role for PRC1.2/4 in regulating lymphopoiesis.
Noncanonical PRC1
PCGF1-containing noncanonical PRC1 (PRC1.1) seems to be involved in allowing hematopoietic progenitor cells to escape from pluripotency. Thus, PCGF1 is necessary for shutting down the HSC pluripotency program by repressing HoxA genes, thereby priming them for further commitment by the master regulator RUNX1 (Ross et al., 2012). Upon PCGF1 depletion, HSCs are biased toward the myeloid lineage (van den Boom et al., 2013). Ectopic overexpression of the H3K36me2 demethylase KDM2B (also in PRC1.1) increases T-lymphoid commitment in a way that is dependent on its demethylase activity, while KDM2B depletion results in myeloid skewing; this is comparable with that observed for PCGF1 (Andricovich et al., 2016). Similarly, overexpression of BCL6 corepressor (BCOR; another PRC1.1 member) in myeloid-committed cells impairs proliferation by repressing HoxA genes, while mutations in BCOR give a proliferative advantage for this lineage (Cao et al., 2016). In patients with X-linked oculo-facio-cardio-dental (OFCD) syndrome, 90–100% of white blood cells undergo inactivation of the X chromosome containing the BCOR-mutated allele, indicating that BCOR-expressing cells have a proliferative disadvantage and cannot fully contribute to hematopoiesis; this was confirmed in a chimeric mouse model (Ng et al., 2004; Wamstad et al., 2008). These observations reinforce the notion that PRC1.1 activity is specifically needed to commit progenitors toward lymphopoiesis (Fig. 2).
The roles played by remaining PRC1 complexes (PRC1.3/5/6) in hematopoiesis still have not been fully addressed. However, results for PCGF5 and PCGF6 suggest that PRC1.5 and -1.6, respectively, do not play a major role in hematopoiesis (van den Boom et al., 2013; Si et al., 2016).
PRC2
The PRC2 components EZH2 and SUZ12 are highly expressed in both fetal and adult bone marrow, while EZH1 is preferentially expressed in adult HSCs (Lessard et al., 1998, 1999; Mochizuki-Kashio et al., 2011; Xie et al., 2014). Consistent with these patterns of expression, EZH1 KO mice do not display defects in primitive HSCs, while adults show impaired B cell development (Hidalgo et al., 2012). Conversely, EZH2 KO mice display embryonic lethality, while EZH2 inactivation at the adult stage produces defects in B cell maturation. This suggests that EZH1 can compensate for EZH2 loss, thereby maintaining self-renewal capacity, only at the HSC stage (Su et al., 2003; Mochizuki-Kashio et al., 2011); similar results have been observed for other types of stem cells (Fig. 2; Shen et al., 2008; Ezhkova et al., 2009, 2011).
As depletion of EZH2 (or of any PRC2 core component) leads to early embryonic lethality (Faust et al., 1995; O’Carroll et al., 2001; Su et al., 2003; Pasini et al., 2004; Mochizuki-Kashio et al., 2011), the roles of these proteins in hematopoiesis were addressed using lineage-specific KOs and heterozygous models. Heterozygous depletion of EZH2, SUZ12, or embryonic ectoderm development (EED) increases hematopoietic stem/progenitor cell (HSPC) activity, suggesting that PRC2 has an antiproliferative effect on HSPCs and thus an opposite role with respect to BMI-1–PRC1 (Lessard et al., 1999; Majewski et al., 2008, 2010). Nonetheless, more recent studies have shown that hematopoiesis-specific KO of either SUZ12 or EED results in HSC exhaustion at the fetal or adult stage (likely depending on the developmental stage at which the KO is induced) rather than hyperproliferation, arguing for a dosage-dependent effect of PRC2 on HSC activity (Xie et al., 2014; Lee et al., 2015; Yu et al., 2017). PRC2 also plays a key role in the lymphoid branch: lineage-specific dissection revealed that SUZ12 is essential for T and B cell maturation but dispensable for proper myelopoiesis (Lee et al., 2015). Moreover, EZH2 is needed to prevent aberrant activation of naive T cells toward Th1/2 by repressing crucial regulators (e.g., Il10, Ifng, and Gata3; Zhang et al., 2014). In B cells, EZH2 is necessary for Igh rearrangement (Su et al., 2003) and germinal center (GC) formation by silencing p21/p27 and Blimp1 loci (Béguelin et al., 2013, 2016, 2017; Caganova et al., 2013). Indeed, the GC reaction is accompanied by a marked up-regulation of all PRC2 core components as well as of PHF19, suggesting a possible role for this accessory subunit in modulating PRC2 activity in this process (Béguelin et al., 2016; Ning et al., 2018). For PRC2 accessory factors, JARID2 knockdown in HSPCs phenocopies that of SUZ12, resulting in higher repopulating capacity in competitive transplants. Accordingly, JARID2 chromatin localization in HSPCs largely overlaps that of SUZ12 and H3K27me3 on genes associated with self-renewal in fetal HSCs. Conversely, depletion of PHF1, MTF2, or PHF19 does not affect HSPC proliferation (Kinkel et al., 2015), suggesting that HSPCs mainly rely on PRC2.2 activity. In the myeloid lineage, MTF2 is necessary for proper PRC2.1 targeting at master regulators of erythrocyte maturation such as the Wnt signaling pathway and its downstream targets Gata2, Fli1, Myb, and Stat5b (Fig. 2; Rothberg et al., 2018). Altogether, these studies prove that PRC2 is necessary for long-term maintenance of hematopoiesis and maturation of lymphoid lineage as well as for erythropoiesis, and they point toward functional roles of specific accessory subunits in recruiting PRC2 to specific genomic targets at each differentiation step.
Oncogenic functions of PcG proteins
PRC1
Numerous PcG proteins have been linked to hematological diseases (Table S1). Early on, BMI-1 was identified as a protooncogene that cooperates with MYC in repressing the Ink4a/Arf gene locus (Haupt et al., 1991; van Lohuizen et al., 1991; Jacobs et al., 1999). Ectopic expression of BMI-1 in the lymphoid compartment is also sufficient to perturb normal lymphogenesis, giving rise to B and T cell lymphomas in mice (Alkema et al., 1997). A role for BMI-1 has also been proposed in leukemia in which LOF of the gene in mice delays the onset of primary leukemia and blocks the development of secondary leukemia, probably due to cancer stem cell exhaustion (Jacobs et al., 1999; Park et al., 2003; Rizo et al., 2009). In pediatric acute lymphoblastic leukemia (ALL), Bmi-1 mRNA is expressed at high levels and correlates with poor prognosis, while it is significantly decreased in patients in complete remission (Peng et al., 2017). Indeed, BMI-1 expression has been proposed as a molecular marker to follow disease progression in B cell lymphomas (Raaphorst et al., 2000; Beà et al., 2001; van Kemenade et al., 2001; van Galen et al., 2007), myelodysplastic syndromes (MDSs), and leukemia; in all cases, its expression correlates with reduced survival and poor prognosis (Sawa et al., 2005; Mihara et al., 2006; Chowdhury et al., 2007; Mohty et al., 2007; Saudy et al., 2014; Peng et al., 2017).
Another PRC1 component associated with cell transformation and lymphomas is CBX7 (Klauke et al., 2013). Under normal conditions, CBX7 is highly expressed in HSCs and GCs, where B cells proliferate and maturate. However, in vivo experiments have demonstrated a role for CBX7 in initiating T cell lymphomas and, in cooperation with MYC, in accelerating aggressive B cell lymphomagenesis through the regulation of the Ink4a/Arf locus (similar to BMI-1; Scott et al., 2007; Klauke et al., 2013).
PRC2
Together with BMI-1, EZH2 is the most studied PcG protein that has been determined to have a strong link with cancer. EZH2 is overexpressed or amplified in several distinct hematological disorders as well as in solid tumors (Piunti and Pasini, 2011). EZH2 plays a pivotal role in controlling the correct formation of GCs. While its deletion suppresses GC formation, expression of mutant EZH2 with hypermethylation activity causes GC hyperplasia, due at least in part to a greater repression of PRC2 target genes such as p21 (Cdkn1a) and Ink4a/Arf/Ink4b (Béguelin et al., 2013; Caganova et al., 2013). Additionally, EZH2 cooperates with BCL6, a transcriptional repressor involved in the GC reaction, to recruit a PRC1–BCOR–CBX8 complex to repress gene expression, thus regulating GC formation and lymphomagenesis (Hatzi and Melnick, 2014; Béguelin et al., 2016). These roles are in line with evidence linking high levels of EZH2 expression with poor prognosis and survival outcome, both of which are dependent on its enzymatic activity in B cell lymphomas (Raaphorst et al., 2000; van Kemenade et al., 2001; Visser et al., 2001; Sneeringer et al., 2010; Okosun et al., 2014).
EZH2 GOF mutations have also been identified in non–Hodgkin lymphomas (NHLs) and solid tumors. Mutations of the tyrosine 641 (Y641F/N/S/H/C) are found in 22% of GC B cells and DLBCLs as well as in 7% of FLs, where they are considered an early clonal event leading to the disease (Morin et al., 2010; Caganova et al., 2013; Okosun et al., 2014). These mutations occur in the EZH2 SET domain and alter the substrate-binding pocket. They were initially believed to be LOF mutations as mutated EZH2 prefers substrates with a higher state of methylation (H3K27me0:me1:me2 kcat/Km ratio = 1:2:13) as compared with the WT one (H3K27me0:me1:me2 kcat/Km ratio = 9:6:1), suggesting a decreased capacity to deposit the correct mark (Sneeringer et al., 2010; McCabe et al., 2012). However, these mutations are always heterozygous; thus, while the WT form is responsible for mono- and dimethylation, the mutated isoform enhances the di- to trimethylation conversion. The result of this cooperation is an aberrant, strong overall increase in H3K27me3 (Morin et al., 2010; Sneeringer et al., 2010; Yap et al., 2011; Béguelin et al., 2013; Bödör et al., 2013). Two additional EZH2 point mutations, A677G and A687V, occur less frequently (in 1–2% of lymphoma patients), and only A687V shows a slight preference for methylating H3K27me2; both mutations result in decreased H3K27me2 levels and a hypertrimethylation phenotype (Majer et al., 2012; McCabe et al., 2012; Ott et al., 2014).
Posttranscriptional mechanisms can also alter EZH2 protein levels. For instance, a molecular circuit with a potential role in Burkitt’s lymphoma has been proposed in which EZH2 is negatively regulated by miR-26a; when MYC is present at high levels, it represses miR-26a, leading to increased EZH2 expression (Sander et al., 2008).
The scenario is even more complex in leukemias in which fusion proteins with oncogenic activities act together with PRC1 and PRC2 complexes. PML-RARα and PLZF-RARα fusion proteins interact with SUZ12 and BMI-1, respectively, to tether Pc complexes to retinoic acid response elements. In both cases, depletion of PcG proteins decreases the oncogenic potential by promoting cellular differentiation (Villa et al., 2007; Boukarabila et al., 2009). Likewise, in MLL-AF9 acute myeloid leukemia (AML), EED is necessary for leukemia initiation and progression, likely due to derepression of the Ink4a/Arf tumor suppressor locus. However, other studies examining the role of EZH2 suggest that EZH1 can partially compensate for its function (Neff et al., 2012; Tanaka et al., 2012; Shi et al., 2013). CBX8 has an important role in MLL-AF9 leukemia as well through its direct interactions with AF9 and TIP60 proteins, which regulate proliferation and survival of leukemic cells in a PRC1-independent way (Tan et al., 2011).
Tumor-suppressive functions of PcG proteins
PRC1
Various PcG proteins also have been shown to act as tumor suppressors (Table S2). For instance, BMI-1 is not only crucially involved in HSC maintenance and differentiation (Jacobs et al., 1999; Park et al., 2003; Rizo et al., 2009) but also has a role as a tumor suppressor. Its genetic ablation promotes myeloid malignancies (primary myelofibrosis [PMF]) through direct derepression of a cohort of genes including that of Hmga2, a well-known oncogene usually expressed at high levels in PMF (Oguro et al., 2012). Likewise, PHC1 has a role as a tumor suppressor in the proper B cell maturation and differentiation, and its expression is lost in leukemic cells from pediatric patients with ALL (Tokimasa et al., 2001).
BCOR and BCORL1 (proteins that cooperate in recruiting the complex to CpG islands) are frequently mutated in myeloid malignancies. Several deletions and mutations affecting the mRNA levels of these factors have been identified in patients with MDS; these account for 4.2% and 0.8% of the cases for BCOR and BCORL1, respectively (Damm et al., 2013). Notably, both proteins are also often down-regulated in cytogenetically normal AML patients (in 4–6% of cases); this down-regulation is associated with poor prognosis. In AML, BCOR disruptive alterations frequently occur together with DNMT3A mutations, suggesting a crosstalk between these two epigenetic factors (Grossmann et al., 2011; Li et al., 2011).
PRC2
EZH2 acts as a tumor suppressor in myeloid malignancies such as MDS and myeloproliferative neoplasms (MPNs). It is a frequent target of chromosomal deletions and missense and frameshift mutations, which have an adverse effect on survival (Ernst et al., 2010; Nikoloski et al., 2010; Bejar et al., 2011; Mochizuki-Kashio et al., 2015; Shirahata-Adachi et al., 2017; Gangat et al., 2018). Missense mutations usually affect EZH2 regions involved in protein–protein interactions or the catalytic pocket, suggesting that the functional integrity of the complex is crucial for PRC2 tumor suppressor functions in these malignancies. EZH2 levels can also be altered by indirect effects. For instance, mutations in the splicing factors SF3B1 and SRSF2 occur in 24% and 14% of MSD cases, respectively, and are considered an early event in disease progression (Yoshida et al., 2011; Papaemmanuil et al., 2013). In mice, a premature termination codon (in a cassette exon) is introduced into EZH2 by a mutant SRSF2 (with P95H), which leads to nonsense mediated decay of EZH2 (Kim et al., 2015).
Other PRC2 components are also mutated in myeloid disorders, although to a lesser extent. Mutations in SUZ12 or EED lead to reduced EZH2 methyltransferase activity in vitro (Score et al., 2012), and JARID2 mutations can potentially alter PRC2 targeting, suggesting that distinct genetic alterations can affect the same pathway (Score et al., 2012). Accordingly, SUZ12 mutations found in patients with MDS or MPN usually affect its VEFS domain, which is necessary for SUZ12’s interaction with EZH2 (Brecqueville et al., 2012; Score et al., 2012). Likewise, point mutations in EED can alter its protein stability or its interaction with EZH2 (Lessard et al., 1999; Score et al., 2012; Ueda et al., 2012).
Alterations in these epigenetic modifiers can have wide-ranging effects through modulation and aberrant interactions with other transcription factors and epigenetic regulators. For instance, RUNX1, a master regulator of hematopoietic cell differentiation, is mutated in ∼25% of MDS cases that present EZH2 deletions (Bejar et al., 2011). RUNX1 collaborates with EZH2, and loss of these two factors causes ineffective hematopoiesis and initiation and propagation of an MDS phenotype (Sashida et al., 2014). However, RUNX1 mutants recruit PRC1 to PRC2 target genes such as HOXA9, a gene that is usually activated in high-risk MDS and MDS/AML, thus preventing progression to AML (Sashida et al., 2014).
LOF mutations of PRC2 members also have roles in leukemia outcome and progression: inactivating mutations affecting EZH2, EED, and SUZ12 correlate with poor prognosis in both T cell ALL (T-ALL) and early T cell precursor (ETP) ALL (Ntziachristos et al., 2012; Simon et al., 2012; Zhang et al., 2012). Likewise, deletions affecting JARID2 have been associated to various types of leukemia (Su et al., 2015). EZH2 and SUZ12 are misregulated in 25% of all T-ALL cases, and 65% of these mutations associate with an oncogenic increase of NOTCH1. In T-ALL, NOTCH1 binding sites and the PRC2-deposited H3K27me3 mark overlap, suggesting that the absence of PRC2 can reinforce altered NOTCH1 signaling (Ntziachristos et al., 2012). For both EZH2 and EED, a role in ETP-ALL development has also been proposed: they cooperate with the mutated form of GTPase NRAS (Q61K) to enhance cell growth and survival signaling (Danis et al., 2016).
PRC2 components act as tumor suppressors also in AML, in which deletions of PRC2 genes (EZH2, JARID2, SUZ12, and AEBP2) have been identified in 35% of AML patients with a previous history of MPN/MDS (Puda et al., 2012). These mutations alter the correct enzymatic activity of the complex, thus facilitating leukemia progression (Puda et al., 2012). Accordingly, in vivo experiments confirmed that an EED missense mutation (I363M) found in AML affects the region close to the aromatic cage, altering the correct deposition of the H3K27me3 mark. This mutation also increases the susceptibility to leukemia in cooperation with other genetic alterations such as EVI1 (myeloid leukemia) and RUNX1 (T cell leukemia; Ueda et al., 2016).
Targeting PcG proteins in hematologic cancers
Considering the strong link between PcG protein alterations and hematological diseases, major efforts have been directed to developing compounds that aim to restore the correct levels of these chromatin modifiers for disease treatment (Bhaumik et al., 2007; Copeland et al., 2009). Importantly, directly targeting epigenetic regulation has the advantage of being more plastic than therapies that aim to correct the patient’s genomic DNA.
For PRC1, inhibitors targeting BMI-1 have been developed that provide good responses in distinct tumor types. The first BMI-1 inhibitor identified was PTC-209, which is able to lower BMI-1 transcript levels without affecting those of RING1B or CBX7. In models of human colorectal cancer, it reduces the number of functional cancer-initiating cells, resulting in a strong reduction of tumorigenic potential in xenograft models (Kreso et al., 2014). PTC-209 has shown promising results both in primary AML and chronic myeloid lymphoma (CML) cell lines: it induces the expression of CDKN2A and CCNG2, two direct targets, leading to significant arrest in G1 and apoptosis in both (Mourgues et al., 2015; Nishida et al., 2015).
The first BMI-1 inhibitor to enter clinical trials was PTC596 (Nishida et al., 2017). PTC596 increases BMI-1 protein degradation by enhancing CDK1 association with BMI-1, followed by phosphorylation at two N-terminal sites. In the AML cell line, PTC596 induces p53-independent apoptosis through MCL-1 down-regulation. Even more promisingly, it has shown antileukemia activity in vivo (in xenograft models; Nishida et al., 2017), and it has been recently tested for patients with advanced solid tumors (NCT02404480).
Several EZH2 inhibitors as well as compounds that disrupt the PRC2 complex have been developed over the years that have distinct selectivity/specificity. Some of these are in preclinical or clinical trials (Lund et al., 2014; Kim and Roberts, 2016). The first compound, DZNep (developed in 1986), is an S-adenosyl-l-homocysteine (SAH) hydrolase inhibitor that causes an increase of the cellular levels of SAH, which in turn blocks methyltransferase activity. This compound is not specific for EZH2, has a short half-life, and is toxic in animal models (Glazer et al., 1986; Miranda et al., 2009; Sandow et al., 2017). Several inhibitors have been further developed that have greatly increasing selectivity toward EZH2. For instance, GSK126 and EPZ005687 have been tested in lymphomas carrying EZH2-activating mutations and have been found to reduce tumor growth and increase survival in xenograft mouse models in a dose-dependent way (Knutson et al., 2012; McCabe et al., 2012; Verma et al., 2012). A further compound, EI1, does not alter EZH2 protein levels but rather reduces H3K27me2 and H3K27me3 levels by competing with the cofactor SAM. In DCBCL-carrying EZH2 mutations, EI1 reduces cell growth, apoptosis, and induction of genes involved in memory B cell differentiation (Qi et al., 2012).
An important breakthrough in EZH2 inhibitors came with the development of orally bioavailable inhibitors. The first one developed was UNC1999, which can block both EZH2 and EZH1, making it advantageous for treating cancers that rely on both enzymes. Accordingly, UNC1999 reduces global levels of H3K27 trimethylation/dimethylation (H3K27me3/2), thus inducing apoptosis and differentiation of MLL-rearranged acute leukemia cells. Moreover, in a MLL-AF9 mouse model, UNC1999 gives rise to a phenotype similar to that of EED KO, altering the correct deposition of the H3K27me3 mark and affecting CDKN2A levels, with strong effects on the mouse survival (Konze et al., 2013; Xu et al., 2015). Another potent EZH2 inhibitor, EPZ6438 (Tazemetostat), is being tested in several clinical trials for treating B cell lymphomas and solid tumors (NCT02220842, NCT03456726, NCT01897571, NCT03028103, NCT03009344, and NCT03010982; Knutson et al., 2013, 2014a; Italiano et al., 2018); one is already in phase 2 for patients with DLBCL and FL, in order to test the efficacy and safety of this compound either alone or in combination with prednisolone (NCT01897571). OR-S1 and OR-S2 are methyltransferase inhibitors that are highly specific for EZH1 and EZH2, and their efficacies have been tested in preclinical studies in AML murine models (MOZ-TIF2 and MLL-AF10) in which they lead to a complete remission of AML (Honma et al., 2017; Fujita et al., 2018).
Another possible therapeutic approach is to target the stability of the PRC2 complex, which disassembles in the absence of a core subunit, to reduce or eliminate its methyltransferase activity. The drug SAH-EZH2A was modeled on the α-helical domain of EZH2 that interacts with EED, a stabilized α-helix of EZH2 peptide capable of disassembling the PRC2 complex and impairing its function by impeding their association. This reduces H3K27me3 levels and increase cell differentiation in MLL-AF9 leukemia cell lines (Kim et al., 2013). Two more compounds (EED226 and A-395) have been developed that impair PRC2 function by targeting EED; specifically, EED226 disrupts the integrity of the complex, while A-395 prevents H3K27me3 recognition, and both cause tumor regression in xenograft mouse models of DLBCL (He et al., 2017; Huang et al., 2017). Finally, GNA002, a gambogenic acid derivative, covalently binds to the Cys668 residue of EZH2, causing its proteasome-mediated degradation and consequent PRC2 disassembly. In xenograft models, GNA002 reduces tumor growth (Wang et al., 2017).
Drug resistance is a major issue in addressing cancer treatments, considering that cell populations vary greatly and are continuously evolving. Two EZH2 amino acid substitutions (Y111L and Y661D) were identified after EI1 inhibitor treatment; these mutations cooperate in conferring acquired resistance in EZH2-mutated lymphoma models (Gore et al., 2006; Gibaja et al., 2016). In particular, Y111L was able to restore PRC2 activity and methylation levels in the presence of distinct PRC2 inhibitors (Gibaja et al., 2016). Acquired resistance has also been observed to be associated with EZH2 protein levels in AML patients: many patients have low EZH2 protein levels after chemotherapy, which correlates with poor prognosis. AML cell lines treated with PKC412, a kinase inhibitor, can develop drug resistance due at least in part to EZH2 protein degradation. This EZH2 reduction in turn alters gene expression of various factors associated to the HOX genes. Interestingly, knocking down HOXB7 and HOXA9 proteins in AML cell lines partially rescues sensitivity to drugs. Furthermore, a combination of Ara-C and bortezomib, used both ex vivo on primary AML samples and in vivo in AML patients, rescues EZH2 protein levels and reduces levels of immature blasts from peripheral blood (Göllner et al., 2017).
Many other combination of treatments have been proposed for lymphomas (Zhao et al., 2013; Knutson et al., 2014b; Béguelin et al., 2016), leukemias (Kowolik et al., 2016; Wen et al., 2018), and myelomas (Bolomsky et al., 2016; Alzrigat et al., 2017). In general, these show synergistic effects, leading to increased apoptosis and reduced tumor burdens. Overall, these reports provide an encouraging avenue that warrants continued work on identifying additional compounds and on studying more thoroughly different combinations of therapies as a way to achieve better and more durable antitumor effects.
Conclusions
Hematological diseases are characterized by lower levels of genetic mutations but higher levels of alterations of epigenetic factors as compared with other diseases (Haladyna et al., 2015). These alterations (GOF, LOF, and aberrant recruitment of complexes) greatly affect gene expression and play a major role in hematopoietic malignancies. Of note, both overexpression and LOF of the PcG proteins are strongly correlated with cancers. These apparently contradictory observations could be due to the distinct roles played by the complexes during the differentiation process. Additionally, they could also be due to misregulation of these proteins causing a general alteration of gene expression that, together with the distinct tumor niches, can lead to very distinct outcomes. This topic is of particular interest for therapeutics: development of new molecules with increased selectivity and decreased toxicity should be encouraged, but we should keep in mind that we still lack knowledge about many biological processes. In any case, an accurate patient selection will be mandatory to avoid secondary health problems. Another possible caveat is the risk of development of drug resistances (Gibaja et al., 2016); nevertheless, with the use of combinational therapies, this phenomenon can be greatly reduced and controlled.
Results from GOF studies of PRC1 and PRC2 have highlighted that mutations in distinct proteins (especially for PRC2 subunits) down-regulate common genes that have tumor suppressor functions such as the CDKN2A locus. Distinct complexes can, of course, affect the same pathway; however, it is crucial to point out that most of our knowledge focuses on BMI-1 and EZH2 and that PcG proteins can potentially form numerous distinct complexes with many different targets. We now need to focus on studying the other subunits in order to clarify their contribution to normal and malignant hematopoiesis.
Supplementary Material
Acknowledgments
We thank P. Vizan, S. Aranda, and M. Garcia for critical reading of the manuscript; all the members of Di Croce laboratory for helpful discussions; and V.A. Raker for scientific editing. We apologize to colleagues whose work has not been cited due to space limitations.
Support to the Di Croce laboratory comes from grants from the Spanish Ministry of Economy, Industry, and Competitiveness (BFU2016-75008-P), the European Regional Development Fund (FEDER), Fundacion Vencer El Cancer, and Agència de Gestió d’Ajuts Universitaris i de Recerca; to V. Di Carlo from an ImPuLSe Marie Curie Postdoctoral Fellowship of the European Union Seventh Framework Program (FP7/2007-2013; grant 608959); and I. Mocavini from an FPI fellowship. We acknowledge support of the Spanish Ministry of Economy, Industry, and Competitiveness through the Instituto de Salud Carlos III and for the European Molecular Biology Laboratory partnership; Centro de Excelencia Severo Ochoa; and the Centres de Recerca de Catalunya Program/Generalitat de Catalunya.
The authors declare no competing financial interests.
References
- Agger K., Cloos P.A.C., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., and Helin K.. 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 449:731–734. 10.1038/nature06145 [DOI] [PubMed] [Google Scholar]
- Akasaka T., Tsuji K., Kawahira H., Kanno M., Harigaya K., Hu L., Ebihara Y., Nakahata T., Tetsu O., Taniguchi M., and Koseki H.. 1997. The role of mel-18, a mammalian Polycomb group gene, during IL-7-dependent proliferation of lymphocyte precursors. Immunity. 7:135–146. 10.1016/S1074-7613(00)80516-6 [DOI] [PubMed] [Google Scholar]
- Akashi K., Traver D., Miyamoto T., and Weissman I.L.. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197. 10.1038/35004599 [DOI] [PubMed] [Google Scholar]
- Alkema M.J., Jacobs H., van Lohuizen M., and Berns A.. 1997. Pertubation of B and T cell development and predisposition to lymphomagenesis in Emu Bmi1 transgenic mice require the Bmi1 RING finger. Oncogene. 15:899–910. 10.1038/sj.onc.1201262 [DOI] [PubMed] [Google Scholar]
- Aloia L., Di Stefano B., and Di Croce L.. 2013. Polycomb complexes in stem cells and embryonic development. Development. 140:2525–2534. 10.1242/dev.091553 [DOI] [PubMed] [Google Scholar]
- Alzrigat M., Párraga A.A., Majumder M.M., Ma A., Jin J., Österborg A., Nahi H., Nilsson K., Heckman C.A., Öberg F., et al. . 2017. The polycomb group protein BMI-1 inhibitor PTC-209 is a potent anti-myeloma agent alone or in combination with epigenetic inhibitors targeting EZH2 and the BET bromodomains. Oncotarget. 8:103731–103743. 10.18632/oncotarget.21909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andricovich J., Kai Y., Peng W., Foudi A., and Tzatsos A.. 2016. Histone demethylase KDM2B regulates lineage commitment in normal and malignant hematopoiesis. J. Clin. Invest. 126:905–920. 10.1172/JCI84014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda S., Mas G., and Di Croce L.. 2015. Regulation of gene transcription by Polycomb proteins. Sci. Adv. 1:e1500737 10.1126/sciadv.1500737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayton P., Sneddon S.F., Palmer D.B., Rosewell I.R., Owen M.J., Young B., Presley R., and Subramanian V.. 2001. Truncation of the MLL gene in exon 5 by gene targeting leads to early preimplantation lethality of homozygous embryos. Genesis. 30:201–212. 10.1002/gene.1066 [DOI] [PubMed] [Google Scholar]
- Beà S., Tort F., Pinyol M., Puig X., Hernández L., Hernández S., Fernández P.L., van Lohuizen M., Colomer D., and Campo E.. 2001. BMI-1 gene amplification and overexpression in hematological malignancies occur mainly in mantle cell lymphomas. Cancer Res. 61:2409–2412. [PubMed] [Google Scholar]
- Béguelin W., Popovic R., Teater M., Jiang Y., Bunting K.L., Rosen M., Shen H., Yang S.N., Wang L., Ezponda T., et al. . 2013. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 23:677–692. 10.1016/j.ccr.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béguelin W., Teater M., Gearhart M.D., Calvo Fernández M.T., Goldstein R.L., Cárdenas M.G., Hatzi K., Rosen M., Shen H., Corcoran C.M., et al. . 2016. EZH2 and BCL6 Cooperate to Assemble CBX8-BCOR Complex to Repress Bivalent Promoters, Mediate Germinal Center Formation and Lymphomagenesis. Cancer Cell. 30:197–213. 10.1016/j.ccell.2016.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béguelin W., Rivas M.A., Calvo Fernández M.T., Teater M., Purwada A., Redmond D., Shen H., Challman M.F., Elemento O., Singh A., and Melnick A.M.. 2017. EZH2 enables germinal centre formation through epigenetic silencing of CDKN1A and an Rb-E2F1 feedback loop. Nat. Commun. 8:877 10.1038/s41467-017-01029-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R., Stevenson K., Abdel-Wahab O., Galili N., Nilsson B., Garcia-Manero G., Kantarjian H., Raza A., Levine R.L., Neuberg D., and Ebert B.L.. 2011. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 364:2496–2506. 10.1056/NEJMoa1013343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer M., Pisano P., Di Carlo V., Blanco E., Chammas P., Vizán P., Gutiérrez A., Aranda S., Payer B., Wierer M., and Di Croce L.. 2016. EPOP Functionally Links Elongin and Polycomb in Pluripotent Stem Cells. Mol. Cell. 64:645–658. 10.1016/j.molcel.2016.10.018 [DOI] [PubMed] [Google Scholar]
- Bhaumik S.R., Smith E., and Shilatifard A.. 2007. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 14:1008–1016. 10.1038/nsmb1337 [DOI] [PubMed] [Google Scholar]
- Bochyńska A., Lüscher-Firzlaff J., and Lüscher B.. 2018. Modes of Interaction of KMT2 Histone H3 Lysine 4 Methyltransferase/COMPASS Complexes with Chromatin. Cells. 7:17 10.3390/cells7030017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bödör C., Grossmann V., Popov N., Okosun J., O’Riain C., Tan K., Marzec J., Araf S., Wang J., Lee A.M., et al. . 2013. EZH2 mutations are frequent and represent an early event in follicular lymphoma. Blood. 122:3165–3168. 10.1182/blood-2013-04-496893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolomsky A., Schlangen K., Schreiner W., Zojer N., and Ludwig H.. 2016. Targeting of BMI-1 with PTC-209 shows potent anti-myeloma activity and impairs the tumour microenvironment. J. Hematol. Oncol. 9:17 10.1186/s13045-016-0247-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkin D., He S., Miao H., Kempinska K., Pollock J., Chase J., Purohit T., Malik B., Zhao T., Wang J., et al. . 2015. Pharmacologic inhibition of the Menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell. 27:589–602. 10.1016/j.ccell.2015.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukarabila H., Saurin A.J., Batsché E., Mossadegh N., van Lohuizen M., Otte A.P., Pradel J., Muchardt C., Sieweke M., and Duprez E.. 2009. The PRC1 Polycomb group complex interacts with PLZF/RARA to mediate leukemic transformation. Genes Dev. 23:1195–1206. 10.1101/gad.512009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., et al. . 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 441:349–353. 10.1038/nature04733 [DOI] [PubMed] [Google Scholar]
- Brecqueville M., Rey J., Bertucci F., Coppin E., Finetti P., Carbuccia N., Cervera N., Gelsi-Boyer V., Arnoulet C., Gisserot O., et al. . 2012. Mutation analysis of ASXL1, CBL, DNMT3A, IDH1, IDH2, JAK2, MPL, NF1, SF3B1, SUZ12, and TET2 in myeloproliferative neoplasms. Genes Chromosomes Cancer. 51:743–755. 10.1002/gcc.21960 [DOI] [PubMed] [Google Scholar]
- Caganova M., Carrisi C., Varano G., Mainoldi F., Zanardi F., Germain P.-L., George L., Alberghini F., Ferrarini L., Talukder A.K., et al. . 2013. Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. J. Clin. Invest. 123:5009–5022. 10.1172/JCI70626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S.F., Chen C.-W., and Armstrong S.A.. 2015. Drugging Chromatin in Cancer: Recent Advances and Novel Approaches. Mol. Cell. 60:561–570. 10.1016/j.molcel.2015.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F., Townsend E.C., Karatas H., Xu J., Li L., Lee S., Liu L., Chen Y., Ouillette P., Zhu J., et al. . 2014. Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol. Cell. 53:247–261. 10.1016/j.molcel.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q., Gearhart M.D., Gery S., Shojaee S., Yang H., Sun H., Lin D.C., Bai J.W., Mead M., Zhao Z., et al. . 2016. BCOR regulates myeloid cell proliferation and differentiation. Leukemia. 30:1155–1165. 10.1038/leu.2016.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., and Zhang Y.. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 298:1039–1043. 10.1126/science.1076997 [DOI] [PubMed] [Google Scholar]
- Chen C., Liu Y., Rappaport A.R., Kitzing T., Schultz N., Zhao Z., Shroff A.S., Dickins R.A., Vakoc C.R., Bradner J.E., et al. . 2014. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell. 25:652–665. 10.1016/j.ccr.2014.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Anastassiadis K., Kranz A., Stewart A.F., Arndt K., Waskow C., Yokoyama A., Jones K., Neff T., Lee Y., and Ernst P.. 2017. MLL2, Not MLL1, Plays a Major Role in Sustaining MLL-Rearranged Acute Myeloid Leukemia. Cancer Cell. 31:755–770. 10.1016/j.ccell.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury M., Mihara K., Yasunaga S., Ohtaki M., Takihara Y., and Kimura A.. 2007. Expression of Polycomb-group (PcG) protein BMI-1 predicts prognosis in patients with acute myeloid leukemia. Leukemia. 21:1116–1122. 10.1038/sj.leu.2404623 [DOI] [PubMed] [Google Scholar]
- Cooper S., Grijzenhout A., Underwood E., Ancelin K., Zhang T., Nesterova T.B., Anil-Kirmizitas B., Bassett A., Kooistra S.M., Agger K., et al. . 2016. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun. 7:13661 10.1038/ncomms13661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland R.A., Solomon M.E., and Richon V.M.. 2009. Protein methyltransferases as a target class for drug discovery. Nat. Rev. Drug Discov. 8:724–732. 10.1038/nrd2974 [DOI] [PubMed] [Google Scholar]
- Corces M.R., Buenrostro J.D., Wu B., Greenside P.G., Chan S.M., Koenig J.L., Snyder M.P., Pritchard J.K., Kundaje A., Greenleaf W.J., et al. . 2016. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48:1193–1203. 10.1038/ng.3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coré N., Bel S., Gaunt S.J., Aurrand-Lions M., Pearce J., Fisher A., and Djabali M.. 1997. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development. 124:721–729. [DOI] [PubMed] [Google Scholar]
- Damm F., Chesnais V., Nagata Y., Yoshida K., Scourzic L., Okuno Y., Itzykson R., Sanada M., Shiraishi Y., Gelsi-Boyer V., et al. . 2013. BCOR and BCORL1 mutations in myelodysplastic syndromes and related disorders. Blood. 122:3169–3177. 10.1182/blood-2012-11-469619 [DOI] [PubMed] [Google Scholar]
- Danis E., Yamauchi T., Echanique K., Zhang X., Haladyna J.N., Riedel S.S., Zhu N., Xie H., Orkin S.H., Armstrong S.A., et al. . 2016. Ezh2 Controls an Early Hematopoietic Program and Growth and Survival Signaling in Early T Cell Precursor Acute Lymphoblastic Leukemia. Cell Reports. 14:1953–1965. 10.1016/j.celrep.2016.01.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Lin Q., Ensenat-Waser R., Rose-John S., and Zenke M.. 2012. Polycomb group protein Bmi1 promotes hematopoietic cell development from embryonic stem cells. Stem Cells Dev. 21:121–132. 10.1089/scd.2010.0539 [DOI] [PubMed] [Google Scholar]
- Ernst P., Fisher J.K., Avery W., Wade S., Foy D., and Korsmeyer S.J.. 2004. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev. Cell. 6:437–443. 10.1016/S1534-5807(04)00061-9 [DOI] [PubMed] [Google Scholar]
- Ernst T., Chase A.J., Score J., Hidalgo-Curtis C.E., Bryant C., Jones A.V., Waghorn K., Zoi K., Ross F.M., Reiter A., et al. . 2010. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 42:722–726. 10.1038/ng.621 [DOI] [PubMed] [Google Scholar]
- Ezhkova E., Pasolli H.A., Parker J.S., Stokes N., Su I.-H., Hannon G., Tarakhovsky A., and Fuchs E.. 2009. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 136:1122–1135. 10.1016/j.cell.2008.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E., Lien W.-H., Stokes N., Pasolli H.A., Silva J.M., and Fuchs E.. 2011. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 25:485–498. 10.1101/gad.2019811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber J., Krivtsov A.V., Stubbs M.C., Wright R., Davis T.N., van den Heuvel-Eibrink M., Zwaan C.M., Kung A.L., and Armstrong S.A.. 2009. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 113:2375–2385. 10.1182/blood-2007-09-113597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust C., Schumacher A., Holdener B., and Magnuson T.. 1995. The eed mutation disrupts anterior mesoderm production in mice. Development. 121:273–285. [DOI] [PubMed] [Google Scholar]
- Fujita S., Honma D., Adachi N., Araki K., Takamatsu E., Katsumoto T., Yamagata K., Akashi K., Aoyama K., Iwama A., and Kitabayashi I.. 2018. Dual inhibition of EZH1/2 breaks the quiescence of leukemia stem cells in acute myeloid leukemia. Leukemia. 32:855–864. 10.1038/leu.2017.300 [DOI] [PubMed] [Google Scholar]
- Gan T., Jude C.D., Zaffuto K., and Ernst P.. 2010. Developmentally induced Mll1 loss reveals defects in postnatal haematopoiesis. Leukemia. 24:1732–1741. 10.1038/leu.2010.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangat N., Mudireddy M., Lasho T.L., Finke C.M., Nicolosi M., Szuber N., Patnaik M.M., Pardanani A., Hanson C.A., Ketterling R.P., and Tefferi A.. 2018. Mutations and prognosis in myelodysplastic syndromes: karyotype-adjusted analysis of targeted sequencing in 300 consecutive cases and development of a genetic risk model. Am. J. Hematol. 93:691–697. 10.1002/ajh.25064 [DOI] [PubMed] [Google Scholar]
- Gao Z., Zhang J., Bonasio R., Strino F., Sawai A., Parisi F., Kluger Y., and Reinberg D.. 2012. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell. 45:344–356. 10.1016/j.molcel.2012.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibaja V., Shen F., Harari J., Korn J., Ruddy D., Saenz-Vash V., Zhai H., Rejtar T., Paris C.G., Yu Z., et al. . 2016. Development of secondary mutations in wild-type and mutant EZH2 alleles cooperates to confer resistance to EZH2 inhibitors. Oncogene. 35:558–566. 10.1038/onc.2015.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer R.I., Hartman K.D., Knode M.C., Richard M.M., Chiang P.K., Tseng C.K.H., and Marquez V.E.. 1986. 3-Deazaneplanocin: a new and potent inhibitor of S-adenosylhomocysteine hydrolase and its effects on human promyelocytic leukemia cell line HL-60. Biochem. Biophys. Res. Commun. 135:688–694. 10.1016/0006-291X(86)90048-3 [DOI] [PubMed] [Google Scholar]
- Göllner S., Oellerich T., Agrawal-Singh S., Schenk T., Klein H.-U., Rohde C., Pabst C., Sauer T., Lerdrup M., Tavor S., et al. . 2017. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat. Med. 23:69–78. 10.1038/nm.4247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore S.D., Baylin S., Sugar E., Carraway H., Miller C.B., Carducci M., Grever M., Galm O., Dauses T., Karp J.E., et al. . 2006. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 66:6361–6369. 10.1158/0008-5472.CAN-06-0080 [DOI] [PubMed] [Google Scholar]
- Green M.R., Kihira S., Liu C.L., Nair R.V., Salari R., Gentles A.J., Irish J., Stehr H., Vicente-Dueñas C., Romero-Camarero I., et al. . 2015. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc. Natl. Acad. Sci. USA. 112:E1116–E1125. 10.1073/pnas.1501199112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grembecka J., He S., Shi A., Purohit T., Muntean A.G., Sorenson R.J., Showalter H.D., Murai M.J., Belcher A.M., Hartley T., et al. . 2012. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat. Chem. Biol. 8:277–284. 10.1038/nchembio.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann V., Tiacci E., Holmes A.B., Kohlmann A., Martelli M.P., Kern W., Spanhol-Rosseto A., Klein H.-U., Dugas M., Schindela S., et al. . 2011. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood. 118:6153–6163. 10.1182/blood-2011-07-365320 [DOI] [PubMed] [Google Scholar]
- Haladyna J.N., Yamauchi T., Neff T., and Bernt K.M.. 2015. Epigenetic modifiers in normal and malignant hematopoiesis. Epigenomics. 7:301–320. 10.2217/epi.14.88 [DOI] [PubMed] [Google Scholar]
- Hatzi K., and Melnick A.. 2014. Breaking bad in the germinal center: how deregulation of BCL6 contributes to lymphomagenesis. Trends Mol. Med. 20:343–352. 10.1016/j.molmed.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y., Alexander W.S., Barri G., Klinken S.P., and Adams J.M.. 1991. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E μ-myc transgenic mice. Cell. 65:753–763. 10.1016/0092-8674(91)90383-A [DOI] [PubMed] [Google Scholar]
- He Y., Selvaraju S., Curtin M.L., Jakob C.G., Zhu H., Comess K.M., Shaw B., The J., Lima-Fernandes E., Szewczyk M.M., et al. . 2017. The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex. Nat. Chem. Biol. 13:389–395. 10.1038/nchembio.2306 [DOI] [PubMed] [Google Scholar]
- Hidalgo I., Herrera-Merchan A., Ligos J.M., Carramolino L., Nuñez J., Martinez F., Dominguez O., Torres M., and Gonzalez S.. 2012. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell. 11:649–662. 10.1016/j.stem.2012.08.001 [DOI] [PubMed] [Google Scholar]
- Holoch D., and Margueron R.. 2017. Mechanisms Regulating PRC2 Recruitment and Enzymatic Activity. Trends Biochem. Sci. 42:531–542. 10.1016/j.tibs.2017.04.003 [DOI] [PubMed] [Google Scholar]
- Honma D., Kanno O., Watanabe J., Kinoshita J., Hirasawa M., Nosaka E., Shiroishi M., Takizawa T., Yasumatsu I., Horiuchi T., et al. . 2017. Novel orally bioavailable EZH1/2 dual inhibitors with greater antitumor efficacy than an EZH2 selective inhibitor. Cancer Sci. 108:2069–2078. 10.1111/cas.13326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Zhang J., Yu Z., Zhang H., Wang Y., Lingel A., Qi W., Gu J., Zhao K., Shultz M.D., et al. . 2017. Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy. J. Med. Chem. 60:2215–2226. 10.1021/acs.jmedchem.6b01576 [DOI] [PubMed] [Google Scholar]
- Ikawa T., Masuda K., Endo T.A., Endo M., Isono K., Koseki Y., Nakagawa R., Kometani K., Takano J., Agata Y., et al. . 2016. Conversion of T cells to B cells by inactivation of polycomb-mediated epigenetic suppression of the B-lineage program. Genes Dev. 30:2475–2485. 10.1101/gad.290593.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiano A., Soria J.-C., Toulmonde M., Michot J.-M., Lucchesi C., Varga A., Coindre J.-M., Blakemore S.J., Clawson A., Suttle B., et al. . 2018. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol. 19:649–659. 10.1016/S1470-2045(18)30145-1 [DOI] [PubMed] [Google Scholar]
- Iwama A., Oguro H., Negishi M., Kato Y., Morita Y., Tsukui H., Ema H., Kamijo T., Katoh-Fukui Y., Koseki H., et al. . 2004. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity. 21:843–851. 10.1016/j.immuni.2004.11.004 [DOI] [PubMed] [Google Scholar]
- Jacobs J.J.L., Scheijen B., Voncken J.-W., Kieboom K., Berns A., and van Lohuizen M.. 1999. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 13:2678–2690. 10.1101/gad.13.20.2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowska A.M., Makishima H., Tiu R.V., Szpurka H., Huang Y., Traina F., Visconte V., Sugimoto Y., Prince C., O’Keefe C., et al. . 2011. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 118:3932–3941. 10.1182/blood-2010-10-311019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jude C.D., Climer L., Xu D., Artinger E., Fisher J.K., and Ernst P.. 2007. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 1:324–337. 10.1016/j.stem.2007.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G. 1985. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature. 316:153–155. 10.1038/316153a0 [DOI] [Google Scholar]
- Karatas H., Townsend E.C., Cao F., Chen Y., Bernard D., Liu L., Lei M., Dou Y., and Wang S.. 2013. High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction. J. Am. Chem. Soc. 135:669–682. 10.1021/ja306028q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.H., and Roberts C.W.M.. 2016. Targeting EZH2 in cancer. Nat. Med. 22:128–134. 10.1038/nm.4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Ilagan J.O., Liang Y., Daubner G.M., Lee S.C.-W., Ramakrishnan A., Li Y., Chung Y.R., Micol J.-B., Murphy M.E., et al. . 2015. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell. 27:617–630. 10.1016/j.ccell.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Sawada A., Tokimasa S., Endo H., Ozono K., Hara J., and Takihara Y.. 2004. Defective long-term repopulating ability in hematopoietic stem cells lacking the Polycomb-group gene rae28. Eur. J. Haematol. 73:75–84. 10.1111/j.1600-0609.2004.00268.x [DOI] [PubMed] [Google Scholar]
- Kim W., Bird G.H., Neff T., Guo G., Kerenyi M.A., Walensky L.D., and Orkin S.H.. 2013. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat. Chem. Biol. 9:643–650. 10.1038/nchembio.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkel S.A., Galeev R., Flensburg C., Keniry A., Breslin K., Gilan O., Lee S., Liu J., Chen K., Gearing L.J., et al. . 2015. Jarid2 regulates hematopoietic stem cell function by acting with polycomb repressive complex 2. Blood. 125:1890–1900. 10.1182/blood-2014-10-603969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauke K., Radulović V., Broekhuis M., Weersing E., Zwart E., Olthof S., Ritsema M., Bruggeman S., Wu X., Helin K., et al. . 2013. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat. Cell Biol. 15:353–362. 10.1038/ncb2701 [DOI] [PubMed] [Google Scholar]
- Kloet S.L., Makowski M.M., Baymaz H.I., van Voorthuijsen L., Karemaker I.D., Santanach A., Jansen P.W.T.C., Di Croce L., and Vermeulen M.. 2016. The dynamic interactome and genomic targets of Polycomb complexes during stem-cell differentiation. Nat. Struct. Mol. Biol. 23:682–690. 10.1038/nsmb.3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson S.K., Wigle T.J., Warholic N.M., Sneeringer C.J., Allain C.J., Klaus C.R., Sacks J.D., Raimondi A., Majer C.R., Song J., et al. . 2012. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat. Chem. Biol. 8:890–896. 10.1038/nchembio.1084 [DOI] [PubMed] [Google Scholar]
- Knutson S.K., Warholic N.M., Wigle T.J., Klaus C.R., Allain C.J., Raimondi A., Porter Scott M., Chesworth R., Moyer M.P., Copeland R.A., et al. . 2013. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc. Natl. Acad. Sci. USA. 110:7922–7927. 10.1073/pnas.1303800110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson S.K., Kawano S., Minoshima Y., Warholic N.M., Huang K.-C., Xiao Y., Kadowaki T., Uesugi M., Kuznetsov G., Kumar N., et al. . 2014a Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol. Cancer Ther. 13:842–854. 10.1158/1535-7163.MCT-13-0773 [DOI] [PubMed] [Google Scholar]
- Knutson S.K., Warholic N.M., Johnston L.D., Klaus C.R., Wigle T.J., Iwanowicz D., Littlefield B.A., Porter-Scott M., Smith J.J., Moyer M.P., et al. . 2014b Synergistic Anti-Tumor Activity of EZH2 Inhibitors and Glucocorticoid Receptor Agonists in Models of Germinal Center Non-Hodgkin Lymphomas. PLoS One. 9:e111840 10.1371/journal.pone.0111840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Weissman I.L., and Akashi K.. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91:661–672. 10.1016/S0092-8674(00)80453-5 [DOI] [PubMed] [Google Scholar]
- Konze K.D., Ma A., Li F., Barsyte-Lovejoy D., Parton T., Macnevin C.J., Liu F., Gao C., Huang X.-P., Kuznetsova E., et al. . 2013. An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem. Biol. 8:1324–1334. 10.1021/cb400133j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowolik C.M., Lin M., Xie J., Overman L.E., and Horne D.A.. 2016. NT1721, a novel epidithiodiketopiperazine, exhibits potent in vitro and in vivo efficacy against acute myeloid leukemia. Oncotarget. 7:86186–86197. 10.18632/oncotarget.13364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreso A., van Galen P., Pedley N.M., Lima-Fernandes E., Frelin C., Davis T., Cao L., Baiazitov R., Du W., Sydorenko N., et al. . 2014. Self-renewal as a therapeutic target in human colorectal cancer. Nat. Med. 20:29–36. 10.1038/nm.3418 [DOI] [PubMed] [Google Scholar]
- Krivtsov A.V., Feng Z., Lemieux M.E., Faber J., Vempati S., Sinha A.U., Xia X., Jesneck J., Bracken A.P., Silverman L.B., et al. . 2008. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 14:355–368. 10.1016/j.ccr.2008.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E., Krosl J., Thorsteinsdottir U., Baban S., Buchberg A.M., and Sauvageau G.. 1998. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 17:3714–3725. 10.1093/emboj/17.13.3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., and Reinberg D.. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16:2893–2905. 10.1101/gad.1035902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F., Bayliss P.E., Rinn J.L., Whetstine J.R., Wang J.K., Chen S., Iwase S., Alpatov R., Issaeva I., Canaani E., et al. . 2007. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 449:689–694. 10.1038/nature06192 [DOI] [PubMed] [Google Scholar]
- Lee S.C.W., Miller S., Hyland C., Kauppi M., Lebois M., Di Rago L., Metcalf D., Kinkel S.A., Josefsson E.C., Blewitt M.E., et al. . 2015. Polycomb repressive complex 2 component Suz12 is required for hematopoietic stem cell function and lymphopoiesis. Blood. 126:167–175. 10.1182/blood-2014-12-615898 [DOI] [PubMed] [Google Scholar]
- Lessard J., Baban S., and Sauvageau G.. 1998. Stage-specific expression of polycomb group genes in human bone marrow cells. Blood. 91:1216–1224. [PubMed] [Google Scholar]
- Lessard J., Schumacher A., Thorsteinsdottir U., van Lohuizen M., Magnuson T., and Sauvageau G.. 1999. Functional antagonism of the Polycomb-Group genes eed and Bmi1 in hemopoietic cell proliferation. Genes Dev. 13:2691–2703. 10.1101/gad.13.20.2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine S.S., Weiss A., Erdjument-Bromage H., Shao Z., Tempst P., and Kingston R.E.. 2002. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 22:6070–6078. 10.1128/MCB.22.17.6070-6078.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E.B. 1978. A gene complex controlling segmentation in Drosophila. Nature. 276:565–570. 10.1038/276565a0 [DOI] [PubMed] [Google Scholar]
- Lewis E.B., and Mislove R.F.. 1947. New mutants report. Drosophila Information Service. 21:69. [Google Scholar]
- Li M., Collins R., Jiao Y., Ouillette P., Bixby D., Erba H., Vogelstein B., Kinzler K.W., Papadopoulos N., and Malek S.N.. 2011. Somatic mutations in the transcriptional corepressor gene BCORL1 in adult acute myelogenous leukemia. Blood. 118:5914–5917. 10.1182/blood-2011-05-356204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Smith E.R., Takahashi H., Lai K.-C., Martin-Brown S., Florens L., Washburn M.P., Conaway J.W., Conaway R.C., and Shilatifard A.. 2010. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell. 37:429–437. 10.1016/j.molcel.2010.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist C.M., Nordlund J., Ekman D., Johansson A., Moghadam B.T., Raine A., Övernäs E., Dahlberg J., Wahlberg P., Henriksson N., et al. . 2015. The mutational landscape in pediatric acute lymphoblastic leukemia deciphered by whole genome sequencing. Hum. Mutat. 36:118–128. 10.1002/humu.22719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Liu Y.-F., Du Y.-R., Mardaryev A.N., Yang W., Chen H., Xu Z.-M., Xu C.-Q., Zhang X.-R., Botchkarev V.A., et al. . 2013. Cbx4 regulates the proliferation of thymic epithelial cells and thymus function. Development. 140:780–788. 10.1242/dev.085035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cao L., Chen J., Song S., Lee I.H., Quijano C., Liu H., Keyvanfar K., Chen H., Cao L.-Y., et al. . 2009. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 459:387–392. 10.1038/nature08040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr J.G., Stojanov P., Lawrence M.S., Auclair D., Chapuy B., Sougnez C., Cruz-Gordillo P., Knoechel B., Asmann Y.W., Slager S.L., et al. . 2012. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. USA. 109:3879–3884. 10.1073/pnas.1121343109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund K., Adams P.D., and Copland M.. 2014. EZH2 in normal and malignant hematopoiesis. Leukemia. 28:44–49. 10.1038/leu.2013.288 [DOI] [PubMed] [Google Scholar]
- Majer C.R., Jin L., Scott M.P., Knutson S.K., Kuntz K.W., Keilhack H., Smith J.J., Moyer M.P., Richon V.M., Copeland R.A., and Wigle T.J.. 2012. A687V EZH2 is a gain-of-function mutation found in lymphoma patients. FEBS Lett. 586:3448–3451. 10.1016/j.febslet.2012.07.066 [DOI] [PubMed] [Google Scholar]
- Majewski I.J., Blewitt M.E., de Graaf C.A., McManus E.J., Bahlo M., Hilton A.A., Hyland C.D., Smyth G.K., Corbin J.E., Metcalf D., et al. . 2008. Polycomb repressive complex 2 (PRC2) restricts hematopoietic stem cell activity. PLoS Biol. 6:e93 10.1371/journal.pbio.0060093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski I.J., Ritchie M.E., Phipson B., Corbin J., Pakusch M., Ebert A., Busslinger M., Koseki H., Hu Y., Smyth G.K., et al. . 2010. Opposing roles of polycomb repressive complexes in hematopoietic stem and progenitor cells. Blood. 116:731–739. 10.1182/blood-2009-12-260760 [DOI] [PubMed] [Google Scholar]
- Mar B.G., Bullinger L., Basu E., Schlis K., Silverman L.B., Döhner K., and Armstrong S.A.. 2012. Sequencing histone-modifying enzymes identifies UTX mutations in acute lymphoblastic leukemia. Leukemia. 26:1881–1883. 10.1038/leu.2012.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbangkollo D., Burnett R., McCabe N., Thirman M., Gill H., Yu H., Rowley J.D., and Diaz M.O.. 1995. The human MLL gene: nucleotide sequence, homology to the Drosophila trx zinc-finger domain, and alternative splicing. DNA Cell Biol. 14:475–483. 10.1089/dna.1995.14.475 [DOI] [PubMed] [Google Scholar]
- McCabe M.T., Ott H.M., Ganji G., Korenchuk S., Thompson C., Van Aller G.S., Liu Y., Graves A.P., Della Pietra A. III, Diaz E., et al. . 2012. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 492:108–112. 10.1038/nature11606 [DOI] [PubMed] [Google Scholar]
- McMahon K.A., Hiew S.Y.-L., Hadjur S., Veiga-Fernandes H., Menzel U., Price A.J., Kioussis D., Williams O., and Brady H.J.M.. 2007. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 1:338–345. 10.1016/j.stem.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Meyer C., Burmeister T., Gröger D., Tsaur G., Fechina L., Renneville A., Sutton R., Venn N.C., Emerenciano M., Pombo-de-Oliveira M.S., et al. . 2018. The MLL recombinome of acute leukemias in 2017. Leukemia. 32:273–284. 10.1038/leu.2017.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara K., Chowdhury M., Nakaju N., Hidani S., Ihara A., Hyodo H., Yasunaga S., Takihara Y., and Kimura A.. 2006. Bmi-1 is useful as a novel molecular marker for predicting progression of myelodysplastic syndrome and patient prognosis. Blood. 107:305–308. 10.1182/blood-2005-06-2393 [DOI] [PubMed] [Google Scholar]
- Milne T.A., Martin M.E., Brock H.W., Slany R.K., and Hess J.L.. 2005. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 65:11367–11374. 10.1158/0008-5472.CAN-05-1041 [DOI] [PubMed] [Google Scholar]
- Miranda T.B., Cortez C.C., Yoo C.B., Liang G., Abe M., Kelly T.K., Marquez V.E., and Jones P.A.. 2009. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol. Cancer Ther. 8:1579–1588. 10.1158/1535-7163.MCT-09-0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra B.P., Zaffuto K.M., Artinger E.L., Org T., Mikkola H.K.A., Cheng C., Djabali M., and Ernst P.. 2014. The histone methyltransferase activity of MLL1 is dispensable for hematopoiesis and leukemogenesis. Cell Reports. 7:1239–1247. 10.1016/j.celrep.2014.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki-Kashio M., Mishima Y., Miyagi S., Negishi M., Saraya A., Konuma T., Shinga J., Koseki H., and Iwama A.. 2011. Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood. 118:6553–6561. 10.1182/blood-2011-03-340554 [DOI] [PubMed] [Google Scholar]
- Mochizuki-Kashio M., Aoyama K., Sashida G., Oshima M., Tomioka T., Muto T., Wang C., and Iwama A.. 2015. Ezh2 loss in hematopoietic stem cells predisposes mice to develop heterogeneous malignancies in an Ezh1-dependent manner. Blood. 126:1172–1183. 10.1182/blood-2015-03-634428 [DOI] [PubMed] [Google Scholar]
- Mohan M., Herz H.-M., Takahashi Y.-H., Lin C., Lai K.C., Zhang Y., Washburn M.P., Florens L., and Shilatifard A.. 2010. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes Dev. 24:574–589. 10.1101/gad.1898410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohty M., Yong A.S.M., Szydlo R.M., Apperley J.F., and Melo J.V.. 2007. The polycomb group BMI1 gene is a molecular marker for predicting prognosis of chronic myeloid leukemia. Blood. 110:380–383. 10.1182/blood-2006-12-065599 [DOI] [PubMed] [Google Scholar]
- Morey L., Pascual G., Cozzuto L., Roma G., Wutz A., Benitah S.A., and Di Croce L.. 2012. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 10:47–62. 10.1016/j.stem.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Morey L., Santanach A., Blanco E., Aloia L., Nora E.P., Bruneau B.G., and Di Croce L.. 2015. Polycomb Regulates Mesoderm Cell Fate-Specification in Embryonic Stem Cells through Activation and Repression Mechanisms. Cell Stem Cell. 17:300–315. 10.1016/j.stem.2015.08.009 [DOI] [PubMed] [Google Scholar]
- Morin R.D., Johnson N.A., Severson T.M., Mungall A.J., An J., Goya R., Paul J.E., Boyle M., Woolcock B.W., Kuchenbauer F., et al. . 2010. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 42:181–185. 10.1038/ng.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin R.D., Mendez-Lago M., Mungall A.J., Goya R., Mungall K.L., Corbett R.D., Johnson N.A., Severson T.M., Chiu R., Field M., et al. . 2011. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 476:298–303. 10.1038/nature10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S.J., Wandycz A.M., Hemmati H.D., Wright D.E., and Weissman I.L.. 1997. Identification of a lineage of multipotent hematopoietic progenitors. Development. 124:1929–1939. [DOI] [PubMed] [Google Scholar]
- Mourgues L., Imbert V., Nebout M., Colosetti P., Neffati Z., Lagadec P., Verhoeyen E., Peng C., Duprez E., Legros L., et al. . 2015. The BMI1 polycomb protein represses cyclin G2-induced autophagy to support proliferation in chronic myeloid leukemia cells. Leukemia. 29:1993–2002. 10.1038/leu.2015.112 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Mori T., Tada S., Krajewski W., Rozovskaia T., Wassell R., Dubois G., Mazo A., Croce C.M., and Canaani E.. 2002. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell. 10:1119–1128. 10.1016/S1097-2765(02)00740-2 [DOI] [PubMed] [Google Scholar]
- Na Nakorn T., Traver D., Weissman I.L., and Akashi K.. 2002. Myeloerythroid-restricted progenitors are sufficient to confer radioprotection and provide the majority of day 8 CFU-S. J. Clin. Invest. 109:1579–1585. 10.1172/JCI0215272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff T., Sinha A.U., Kluk M.J., Zhu N., Khattab M.H., Stein L., Xie H., Orkin S.H., and Armstrong S.A.. 2012. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc. Natl. Acad. Sci. USA. 109:5028–5033. 10.1073/pnas.1202258109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M., Vosberg S., Schlee C., Heesch S., Schwartz S., Gökbuget N., Hoelzer D., Graf A., Krebs S., Bartram I., et al. . 2015. Mutational spectrum of adult T-ALL. Oncotarget. 6:2754–2766. 10.18632/oncotarget.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D., Thakker N., Corcoran C.M., Donnai D., Perveen R., Schneider A., Hadley D.W., Tifft C., Zhang L., Wilkie A.O.M., et al. . 2004. Oculofaciocardiodental and Lenz microphthalmia syndromes result from distinct classes of mutations in BCOR. Nat. Genet. 36:411–416. 10.1038/ng1321 [DOI] [PubMed] [Google Scholar]
- Nikoloski G., Langemeijer S.M.C., Kuiper R.P., Knops R., Massop M., Tönnissen E.R.L.T.M., van der Heijden A., Scheele T.N., Vandenberghe P., de Witte T., et al. . 2010. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat. Genet. 42:665–667. 10.1038/ng.620 [DOI] [PubMed] [Google Scholar]
- Ning F., Wang C., Niu S., Xu H., Xia K., and Wang N.. 2018. Transcription factor Phf19 positively regulates germinal center reactions that underlies its role in rheumatoid arthritis. Am. J. Transl. Res. 10:200–211. [PMC free article] [PubMed] [Google Scholar]
- Nishida Y., Maeda A., Chachad D., Ishizawa J., Qiu Y.H., Kornblau S.M., Kimura S., Andreeff M., and Kojima K.. 2015. Preclinical activity of the novel B-cell-specific Moloney murine leukemia virus integration site 1 inhibitor PTC-209 in acute myeloid leukemia: Implications for leukemia therapy. Cancer Sci. 106:1705–1713. 10.1111/cas.12833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y., Maeda A., Kim M.J., Cao L., Kubota Y., Ishizawa J., AlRawi A., Kato Y., Iwama A., Fujisawa M., et al. . 2017. The novel BMI-1 inhibitor PTC596 downregulates MCL-1 and induces p53-independent mitochondrial apoptosis in acute myeloid leukemia progenitor cells. Blood Cancer J. 7:e527 10.1038/bcj.2017.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P., Tsirigos A., Van Vlierberghe P., Nedjic J., Trimarchi T., Flaherty M.S., Ferres-Marco D., da Ros V., Tang Z., Siegle J., et al. . 2012. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat. Med. 18:298–303. 10.1038/nm.2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Carroll D., Erhardt S., Pagani M., Barton S.C., Surani M.A., and Jenuwein T.. 2001. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 21:4330–4336. 10.1128/MCB.21.13.4330-4336.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro H., Iwama A., Morita Y., Kamijo T., van Lohuizen M., and Nakauchi H.. 2006. Differential impact of Ink4a and Arf on hematopoietic stem cells and their bone marrow microenvironment in Bmi1-deficient mice. J. Exp. Med. 203:2247–2253. 10.1084/jem.20052477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro H., Yuan J., Ichikawa H., Ikawa T., Yamazaki S., Kawamoto H., Nakauchi H., and Iwama A.. 2010. Poised lineage specification in multipotential hematopoietic stem and progenitor cells by the polycomb protein Bmi1. Cell Stem Cell. 6:279–286. 10.1016/j.stem.2010.01.005 [DOI] [PubMed] [Google Scholar]
- Oguro H., Yuan J., Tanaka S., Miyagi S., Mochizuki-Kashio M., Ichikawa H., Yamazaki S., Koseki H., Nakauchi H., and Iwama A.. 2012. Lethal myelofibrosis induced by Bmi1-deficient hematopoietic cells unveils a tumor suppressor function of the polycomb group genes. J. Exp. Med. 209:445–454. 10.1084/jem.20111709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H., Sawada A., Kim J.Y., Tokimasa S., Nishiguchi S., Humphries R.K., Hara J., and Takihara Y.. 2002. Polycomb group gene rae28 is required for sustaining activity of hematopoietic stem cells. J. Exp. Med. 195:759–770. 10.1084/jem.20011911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Feng Q., Lin Y., Jiang Q., Li Y., Coffield V.M., Su L., Xu G., and Zhang Y.. 2005. hDOT1L links histone methylation to leukemogenesis. Cell. 121:167–178. 10.1016/j.cell.2005.02.020 [DOI] [PubMed] [Google Scholar]
- Okosun J., Bödör C., Wang J., Araf S., Yang C.-Y., Pan C., Boller S., Cittaro D., Bozek M., Iqbal S., et al. . 2014. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 46:176–181. 10.1038/ng.2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Molina A., Boss I.W., Canela A., Pan H., Jiang Y., Zhao C., Jiang M., Hu D., Agirre X., Niesvizky I., et al. . 2015. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat. Med. 21:1199–1208. 10.1038/nm.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M., Hanada K., Hamada H., and Nakauchi H.. 1996. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 273:242–245. 10.1126/science.273.5272.242 [DOI] [PubMed] [Google Scholar]
- Ott H.M., Graves A.P., Pappalardi M.B., Huddleston M., Halsey W.S., Hughes A.M., Groy A., Dul E., Jiang Y., Bai Y., et al. . 2014. A687V EZH2 is a driver of histone H3 lysine 27 (H3K27) hypertrimethylation. Mol. Cancer Ther. 13:3062–3073. 10.1158/1535-7163.MCT-13-0876 [DOI] [PubMed] [Google Scholar]
- Papaemmanuil E., Gerstung M., Malcovati L., Tauro S., Gundem G., Van Loo P., Yoon C.J., Ellis P., Wedge D.C., Pellagatti A., et al. Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium . 2013. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 122:3616–3627, quiz :3699. 10.1182/blood-2013-08-518886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.K., Qian D., Kiel M., Becker M.W., Pihalja M., Weissman I.L., Morrison S.J., and Clarke M.F.. 2003. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 423:302–305. [DOI] [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Jensen M.R., Lazzerini Denchi E., and Helin K.. 2004. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 23:4061–4071. 10.1038/sj.emboj.7600402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H.-X., Liu X.-D., Luo Z.-Y., Zhang X.-H., Luo X.-Q., Chen X., Jiang H., and Xu L.. 2017. Upregulation of the proto-oncogene Bmi-1 predicts a poor prognosis in pediatric acute lymphoblastic leukemia. BMC Cancer. 17:76 10.1186/s12885-017-3049-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piunti A., and Pasini D.. 2011. Epigenetic factors in cancer development: polycomb group proteins. Future Oncol. 7:57–75. 10.2217/fon.10.157 [DOI] [PubMed] [Google Scholar]
- Plass C., Pfister S.M., Lindroth A.M., Bogatyrova O., Claus R., and Lichter P.. 2013. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat. Rev. Genet. 14:765–780. 10.1038/nrg3554 [DOI] [PubMed] [Google Scholar]
- Puda A., Milosevic J.D., Berg T., Klampfl T., Harutyunyan A.S., Gisslinger B., Rumi E., Pietra D., Malcovati L., Elena C., et al. . 2012. Frequent deletions of JARID2 in leukemic transformation of chronic myeloid malignancies. Am. J. Hematol. 87:245–250. 10.1002/ajh.22257 [DOI] [PubMed] [Google Scholar]
- Qi W., Chan H., Teng L., Li L., Chuai S., Zhang R., Zeng J., Li M., Fan H., Lin Y., et al. . 2012. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc. Natl. Acad. Sci. USA. 109:21360–21365. 10.1073/pnas.1210371110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaphorst F.M., van Kemenade F.J., Blokzijl T., Fieret E., Hamer K.M., Satijn D.P.E., Otte A.P., and Meijer C.J.L.M.. 2000. Coexpression of BMI-1 and EZH2 polycomb group genes in Reed-Sternberg cells of Hodgkin’s disease. Am. J. Pathol. 157:709–715. 10.1016/S0002-9440(10)64583-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo A., Dontje B., Vellenga E., de Haan G., and Schuringa J.J.. 2008. Long-term maintenance of human hematopoietic stem/progenitor cells by expression of BMI1. Blood. 111:2621–2630. 10.1182/blood-2007-08-106666 [DOI] [PubMed] [Google Scholar]
- Rizo A., Olthof S., Han L., Vellenga E., de Haan G., and Schuringa J.J.. 2009. Repression of BMI1 in normal and leukemic human CD34(+) cells impairs self-renewal and induces apoptosis. Blood. 114:1498–1505. 10.1182/blood-2009-03-209734 [DOI] [PubMed] [Google Scholar]
- Ross K., Sedello A.K., Todd G.P., Paszkowski-Rogacz M., Bird A.W., Ding L., Grinenko T., Behrens K., Hubner N., Mann M., et al. . 2012. Polycomb group ring finger 1 cooperates with Runx1 in regulating differentiation and self-renewal of hematopoietic cells. Blood. 119:4152–4161. 10.1182/blood-2011-09-382390 [DOI] [PubMed] [Google Scholar]
- Rothberg J.L.M., Maganti H.B., Jrade H., Porter C.J., Palidwor G.A., Cafariello C., Battaion H.L., Khan S.T., Perkins T.J., Paulson R.F., et al. . 2018. Mtf2-PRC2 control of canonical Wnt signaling is required for definitive erythropoiesis. Cell Discov. 4:21 10.1038/s41421-018-0022-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley J.D. 1993. Rearrangements involving chromosome band 11Q23 in acute leukaemia. Semin. Cancer Biol. 4:377–385. [PubMed] [Google Scholar]
- Sander S., Bullinger L., Klapproth K., Fiedler K., Kestler H.A., Barth T.F.E., Möller P., Stilgenbauer S., Pollack J.R., and Wirth T.. 2008. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 112:4202–4212. 10.1182/blood-2008-03-147645 [DOI] [PubMed] [Google Scholar]
- Sandow J.J., Infusini G., Holik A.Z., Brumatti G., Averink T.V., Ekert P.G., and Webb A.I.. 2017. Quantitative proteomic analysis of EZH2 inhibition in acute myeloid leukemia reveals the targets and pathways that precede the induction of cell death. Proteomics Clin. Appl. 11:1700013 10.1002/prca.201700013 [DOI] [PubMed] [Google Scholar]
- Santos M.A., Faryabi R.B., Ergen A.V., Day A.M., Malhowski A., Canela A., Onozawa M., Lee J.-E., Callen E., Gutierrez-Martinez P., et al. . 2014. DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature. 514:107–111. 10.1038/nature13483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashida G., Harada H., Matsui H., Oshima M., Yui M., Harada Y., Tanaka S., Mochizuki-Kashio M., Wang C., Saraya A., et al. . 2014. Ezh2 loss promotes development of myelodysplastic syndrome but attenuates its predisposition to leukaemic transformation. Nat. Commun. 5:4177 10.1038/ncomms5177 [DOI] [PubMed] [Google Scholar]
- Saudy N.S., Fawzy I.M., Azmy E., Goda E.F., Eneen A., and Abdul Salam E.M.. 2014. BMI1 gene expression in myeloid leukemias and its impact on prognosis. Blood Cells Mol. Dis. 53:194–198. 10.1016/j.bcmd.2014.07.002 [DOI] [PubMed] [Google Scholar]
- Sawa M., Yamamoto K., Yokozawa T., Kiyoi H., Hishida A., Kajiguchi T., Seto M., Kohno A., Kitamura K., Itoh Y., et al. . 2005. BMI-1 is highly expressed in M0-subtype acute myeloid leukemia. Int. J. Hematol. 82:42–47. 10.1532/IJH97.05013 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Bourbon H.-M., Di Croce L., and Cavalli G.. 2017. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell. 171:34–57. 10.1016/j.cell.2017.08.002 [DOI] [PubMed] [Google Scholar]
- Score J., Hidalgo-Curtis C., Jones A.V., Winkelmann N., Skinner A., Ward D., Zoi K., Ernst T., Stegelmann F., Döhner K., et al. . 2012. Inactivation of polycomb repressive complex 2 components in myeloproliferative and myelodysplastic/myeloproliferative neoplasms. Blood. 119:1208–1213. 10.1182/blood-2011-07-367243 [DOI] [PubMed] [Google Scholar]
- Scott C.L., Gil J., Hernando E., Teruya-Feldstein J., Narita M., Martínez D., Visakorpi T., Mu D., Cordon-Cardo C., Peters G., et al. . 2007. Role of the chromobox protein CBX7 in lymphomagenesis. Proc. Natl. Acad. Sci. USA. 104:5389–5394. 10.1073/pnas.0608721104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senisterra G., Wu H., Allali-Hassani A., Wasney G.A., Barsyte-Lovejoy D., Dombrovski L., Dong A., Nguyen K.T., Smil D., Bolshan Y., et al. . 2013. Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5. Biochem. J. 449:151–159. 10.1042/BJ20121280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z., Raible F., Mollaaghababa R., Guyon J.R., Wu C.T., Bender W., and Kingston R.E.. 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 98:37–46. 10.1016/S0092-8674(00)80604-2 [DOI] [PubMed] [Google Scholar]
- Shen X., Liu Y., Hsu Y.-J., Fujiwara Y., Kim J., Mao X., Yuan G.-C., and Orkin S.H.. 2008. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell. 32:491–502. 10.1016/j.molcel.2008.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A., Murai M.J., He S., Lund G., Hartley T., Purohit T., Reddy G., Chruszcz M., Grembecka J., and Cierpicki T.. 2012. Structural insights into inhibition of the bivalent menin-MLL interaction by small molecules in leukemia. Blood. 120:4461–4469. 10.1182/blood-2012-05-429274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wang E., Zuber J., Rappaport A., Taylor M., Johns C., Lowe S.W., and Vakoc C.R.. 2013. The Polycomb complex PRC2 supports aberrant self-renewal in a mouse model of MLL-AF9;Nras(G12D) acute myeloid leukemia. Oncogene. 32:930–938. 10.1038/onc.2012.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahata-Adachi M., Iriyama C., Tomita A., Suzuki Y., Shimada K., and Kiyoi H.. 2017. Altered EZH2 splicing and expression is associated with impaired histone H3 lysine 27 tri-Methylation in myelodysplastic syndrome. Leuk. Res. 63:90–97. 10.1016/j.leukres.2017.10.015 [DOI] [PubMed] [Google Scholar]
- Si S., Nakajima-Takagi Y., Aoyama K., Oshima M., Saraya A., Sugishita H., Nakayama M., Ishikura T., Koseki H., and Iwama A.. 2016. Loss of Pcgf5 Affects Global H2A Monoubiquitination but Not the Function of Hematopoietic Stem and Progenitor Cells. PLoS One. 11:e0154561 10.1371/journal.pone.0154561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C., Chagraoui J., Krosl J., Gendron P., Wilhelm B., Lemieux S., Boucher G., Chagnon P., Drouin S., Lambert R., et al. . 2012. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 26:651–656. 10.1101/gad.186411.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slifer E.H. 1942. A mutant stock of Drosophila with extra sex-combs. J. Exp. Zool. 90:31–40. 10.1002/jez.1400900103 [DOI] [Google Scholar]
- Smith L.G., Weissman I.L., and Heimfeld S.. 1991. Clonal analysis of hematopoietic stem-cell differentiation in vivo. Proc. Natl. Acad. Sci. USA. 88:2788–2792. 10.1073/pnas.88.7.2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneeringer C.J., Scott M.P., Kuntz K.W., Knutson S.K., Pollock R.M., Richon V.M., and Copeland R.A.. 2010. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl. Acad. Sci. USA. 107:20980–20985. 10.1073/pnas.1012525107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G. 1981. A gene product required for correct initiation of segmental determination in Drosophila. Nature. 293:36–41. 10.1038/293036a0 [DOI] [PubMed] [Google Scholar]
- Su C.-L., Deng T.-R., Shang Z., and Xiao Y.. 2015. JARID2 inhibits leukemia cell proliferation by regulating CCND1 expression. Int. J. Hematol. 102:76–85. 10.1007/s12185-015-1797-x [DOI] [PubMed] [Google Scholar]
- Su I.-H., Basavaraj A., Krutchinsky A.N., Hobert O., Ullrich A., Chait B.T., and Tarakhovsky A.. 2003. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 4:124–131. 10.1038/ni876 [DOI] [PubMed] [Google Scholar]
- Takihara Y., Tomotsune D., Shirai M., Katoh-Fukui Y., Nishii K., Motaleb M.A., Nomura M., Tsuchiya R., Fujita Y., Shibata Y., et al. . 1997. Targeted disruption of the mouse homologue of the Drosophila polyhomeotic gene leads to altered anteroposterior patterning and neural crest defects. Development. 124:3673–3682. [DOI] [PubMed] [Google Scholar]
- Tan J., Jones M., Koseki H., Nakayama M., Muntean A.G., Maillard I., and Hess J.L.. 2011. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell. 20:563–575. 10.1016/j.ccr.2011.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Miyagi S., Sashida G., Chiba T., Yuan J., Mochizuki-Kashio M., Suzuki Y., Sugano S., Nakaseko C., Yokote K., et al. . 2012. Ezh2 augments leukemogenicity by reinforcing differentiation blockage in acute myeloid leukemia. Blood. 120:1107–1117. 10.1182/blood-2011-11-394932 [DOI] [PubMed] [Google Scholar]
- Tavares L., Dimitrova E., Oxley D., Webster J., Poot R., Demmers J., Bezstarosti K., Taylor S., Ura H., Koide H., et al. . 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 148:664–678. 10.1016/j.cell.2011.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova R., Agherbi H., Boned A., Meresse S., and Djabali M.. 2006. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc. Natl. Acad. Sci. USA. 103:6629–6634. 10.1073/pnas.0507425103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetsu O., Ishihara H., Kanno R., Kamiyasu M., Inoue H., Tokuhisa T., Taniguchi M., and Kanno M.. 1998. mel-18 negatively regulates cell cycle progression upon B cell antigen receptor stimulation through a cascade leading to c-myc/cdc25. Immunity. 9:439–448. 10.1016/S1074-7613(00)80627-5 [DOI] [PubMed] [Google Scholar]
- Tokimasa S., Ohta H., Sawada A., Matsuda Y., Kim J.Y., Nishiguchi S., Hara J., and Takihara Y.. 2001. Lack of the Polycomb-group gene rae28 causes maturation arrest at the early B-cell developmental stage. Exp. Hematol. 29:93–103. 10.1016/S0301-472X(00)00620-2 [DOI] [PubMed] [Google Scholar]
- Ueda T., Sanada M., Matsui H., Yamasaki N., Honda Z.I., Shih L.-Y., Mori H., Inaba T., Ogawa S., and Honda H.. 2012. EED mutants impair polycomb repressive complex 2 in myelodysplastic syndrome and related neoplasms. Leukemia. 26:2557–2560. 10.1038/leu.2012.146 [DOI] [PubMed] [Google Scholar]
- Ueda T., Nakata Y., Nagamachi A., Yamasaki N., Kanai A., Sera Y., Sasaki M., Matsui H., Honda Z., Oda H., et al. . 2016. Propagation of trimethylated H3K27 regulated by polycomb protein EED is required for embryogenesis, hematopoietic maintenance, and tumor suppression. Proc. Natl. Acad. Sci. USA. 113:10370–10375. 10.1073/pnas.1600070113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boom V., Rozenveld-Geugien M., Bonardi F., Malanga D., van Gosliga D., Heijink A.M., Viglietto G., Morrone G., Fusetti F., Vellenga E., and Schuringa J.J.. 2013. Nonredundant and locus-specific gene repression functions of PRC1 paralog family members in human hematopoietic stem/progenitor cells. Blood. 121:2452–2461. 10.1182/blood-2012-08-451666 [DOI] [PubMed] [Google Scholar]
- van Galen J.C., Muris J.J.F., Oudejans J.J., Vos W., Giroth C.P.E., Ossenkoppele G.J., Otte A.P., Raaphorst F.M., and Meijer C.J.L.M.. 2007. Expression of the polycomb-group gene BMI1 is related to an unfavourable prognosis in primary nodal DLBCL. J. Clin. Pathol. 60:167–172. 10.1136/jcp.2006.038752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haaften G., Dalgliesh G.L., Davies H., Chen L., Bignell G., Greenman C., Edkins S., Hardy C., O’Meara S., Teague J., et al. . 2009. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat. Genet. 41:521–523. 10.1038/ng.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kemenade F.J., Raaphorst F.M., Blokzijl T., Fieret E., Hamer K.M., Satijn D.P., Otte A.P., and Meijer C.J.. 2001. Coexpression of BMI-1 and EZH2 polycomb-group proteins is associated with cycling cells and degree of malignancy in B-cell non-Hodgkin lymphoma. Blood. 97:3896–3901. 10.1182/blood.V97.12.3896 [DOI] [PubMed] [Google Scholar]
- van Lohuizen M., Verbeek S., Scheijen B., Wientjens E., van der Gulden H., and Berns A.. 1991. Identification of cooperating oncogenes in E μ-myc transgenic mice by provirus tagging. Cell. 65:737–752. 10.1016/0092-8674(91)90382-9 [DOI] [PubMed] [Google Scholar]
- Verma S.K., Tian X., LaFrance L.V., Duquenne C., Suarez D.P., Newlander K.A., Romeril S.P., Burgess J.L., Grant S.W., Brackley J.A., et al. . 2012. Identification of Potent, Selective, Cell-Active Inhibitors of the Histone Lysine Methyltransferase EZH2. ACS Med. Chem. Lett. 3:1091–1096. 10.1021/ml3003346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa R., Pasini D., Gutierrez A., Morey L., Occhionorelli M., Viré E., Nomdedeu J.F., Jenuwein T., Pelicci P.G., Minucci S., et al. . 2007. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 11:513–525. 10.1016/j.ccr.2007.04.009 [DOI] [PubMed] [Google Scholar]
- Visser H.P.J., Gunster M.J., Kluin-Nelemans H.C., Manders E.M.M., Raaphorst F.M., Meijer C.J.L.M., Willemze R., and Otte A.P.. 2001. The Polycomb group protein EZH2 is upregulated in proliferating, cultured human mantle cell lymphoma. Br. J. Haematol. 112:950–958. 10.1046/j.1365-2141.2001.02641.x [DOI] [PubMed] [Google Scholar]
- Wamstad J.A., Corcoran C.M., Keating A.M., and Bardwell V.J.. 2008. Role of the transcriptional corepressor Bcor in embryonic stem cell differentiation and early embryonic development. PLoS One. 3:e2814 10.1371/journal.pone.0002814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., and Zhang Y.. 2004a Role of histone H2A ubiquitination in Polycomb silencing. Nature. 431:873–878. 10.1038/nature02985 [DOI] [PubMed] [Google Scholar]
- Wang L., Brown J.L., Cao R., Zhang Y., Kassis J.A., and Jones R.S.. 2004b Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell. 14:637–646. 10.1016/j.molcel.2004.05.009 [DOI] [PubMed] [Google Scholar]
- Wang X., Cao W., Zhang J., Yan M., Xu Q., Wu X., Wan L., Zhang Z., Zhang C., Qin X., et al. . 2017. A covalently bound inhibitor triggers EZH2 degradation through CHIP-mediated ubiquitination. EMBO J. 36:1243–1260. 10.15252/embj.201694058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen S., Wang J., Liu P., Li Y., Lu W., Hu Y., Liu J., He Z., and Huang P.. 2018. Novel combination of histone methylation modulators with therapeutic synergy against acute myeloid leukemia in vitro and in vivo. Cancer Lett. 413:35–45. 10.1016/j.canlet.2017.10.015 [DOI] [PubMed] [Google Scholar]
- Woolthuis C.M., and Park C.Y.. 2016. Hematopoietic stem/progenitor cell commitment to the megakaryocyte lineage. Blood. 127:1242–1248. 10.1182/blood-2015-07-607945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Xu J., Hsu J.H., Nguyen M., Fujiwara Y., Peng C., and Orkin S.H.. 2014. Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell. 14:68–80. 10.1016/j.stem.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., On D.M., Ma A., Parton T., Konze K.D., Pattenden S.G., Allison D.F., Cai L., Rockowitz S., Liu S., et al. . 2015. Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood. 125:346–357. 10.1182/blood-2014-06-581082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi H., Deguchi K., Aono A., Tani Y., Kishimoto T., and Komori T.. 1998. Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood. 92:108–117. [PubMed] [Google Scholar]
- Yang L., Bryder D., Adolfsson J., Nygren J., Månsson R., Sigvardsson M., and Jacobsen S.E.. 2005. Identification of Lin(-)Sca1(+)kit(+)CD34(+)Flt3- short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. 105:2717–2723. 10.1182/blood-2004-06-2159 [DOI] [PubMed] [Google Scholar]
- Yap D.B., Chu J., Berg T., Schapira M., Cheng S.-W.G., Moradian A., Morin R.D., Mungall A.J., Meissner B., Boyle M., et al. . 2011. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 117:2451–2459. 10.1182/blood-2010-11-321208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A., Somervaille T.C.P., Smith K.S., Rozenblatt-Rosen O., Meyerson M., and Cleary M.L.. 2005. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 123:207–218. 10.1016/j.cell.2005.09.025 [DOI] [PubMed] [Google Scholar]
- Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M., et al. . 2011. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 478:64–69. 10.1038/nature10496 [DOI] [PubMed] [Google Scholar]
- Yu B.D., Hess J.L., Horning S.E., Brown G.A., and Korsmeyer S.J.. 1995. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 378:505–508. 10.1038/378505a0 [DOI] [PubMed] [Google Scholar]
- Yu W., Zhang F., Wang S., Fu Y., Chen J., Liang X., Le H., Pu W.T., and Zhang B.. 2017. Depletion of polycomb repressive complex 2 core component EED impairs fetal hematopoiesis. Cell Death Dis. 8:e2744 10.1038/cddis.2017.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisig B.B., Milne T., García-Cuéllar M.-P., Schreiner S., Martin M.-E., Fuchs U., Borkhardt A., Chanda S.K., Walker J., Soden R., et al. . 2004. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol. Cell. Biol. 24:617–628. 10.1128/MCB.24.2.617-628.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ding L., Holmfeldt L., Wu G., Heatley S.L., Payne-Turner D., Easton J., Chen X., Wang J., Rusch M., et al. . 2012. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 481:157–163. 10.1038/nature10725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Grubor V., Love C.L., Banerjee A., Richards K.L., Mieczkowski P.A., Dunphy C., Choi W., Au W.Y., Srivastava G., et al. . 2013. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA. 110:1398–1403. 10.1073/pnas.1205299110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Dominguez-Sola D., Hussein S., Lee J.-E., Holmes A.B., Bansal M., Vlasevska S., Mo T., Tang H., Basso K., et al. . 2015. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat. Med. 21:1190–1198. 10.1038/nm.3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Kinkel S., Maksimovic J., Bandala-Sanchez E., Tanzer M.C., Naselli G., Zhang J.-G., Zhan Y., Lew A.M., Silke J., et al. . 2014. The polycomb repressive complex 2 governs life and death of peripheral T cells. Blood. 124:737–749. 10.1182/blood-2013-12-544106 [DOI] [PubMed] [Google Scholar]
- Zhao X., Lwin T., Zhang X., Huang A., Wang J., Marquez V.E., Chen-Kiang S., Dalton W.S., Sotomayor E., and Tao J.. 2013. Disruption of the MYC-miRNA-EZH2 loop to suppress aggressive B-cell lymphoma survival and clonogenicity. Leukemia. 27:2341–2350. 10.1038/leu.2013.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemin-van der Poel S., McCabe N.R., Gill H.J., Espinosa R. III, Patel Y., Harden A., Rubinelli P., Smith S.D., LeBeau M.M., Rowley J.D., et al. . 1991. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc. Natl. Acad. Sci. USA. 88:10735–10739. 10.1073/pnas.88.23.10735 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.