Purkinje cells rapidly degenerate in pcd mice due to loss of function of tubulin deglutamylase cytosolic carboxypeptidase 1 (CCP1), which promotes mitochondrial fusion in a Drosophila model, CCP1 null cells, and pcd neurons. The CCP1-mediated increase in mitochondrial motility accounts for CCP1-dependent mitochondrial fusion, underscoring the linkage between regulation of mitochondria dynamics, mitochondria motility, and Purkinje neuron survival.
Abstract
A perplexing question in neurodegeneration is why different neurons degenerate. The Purkinje cell degeneration (pcd) mouse displays a dramatic phenotype of degeneration of cerebellar Purkinje cells. Loss of CCP1/Nna1 deglutamylation of tubulin accounts for pcd neurodegeneration, but the mechanism is unknown. In this study, we modulated the dosage of fission and fusion genes in a Drosophila melanogaster loss-of-function model and found that mitochondrial fragmentation and disease phenotypes were rescued by reduced Drp1. We observed mitochondrial fragmentation in CCP1 null cells and in neurons from pcd mice, and we documented reduced mitochondrial fusion in cells lacking CCP1. We examined the effect of tubulin hyperglutamylation on microtubule-mediated mitochondrial motility in pcd neurons and noted markedly reduced retrograde axonal transport. Mitochondrial stress promoted Parkin-dependent turnover of CCP1, and CCP1 and Parkin physically interacted. Our results indicate that CCP1 regulates mitochondrial motility through deglutamylation of tubulin and that loss of CCP1-mediated mitochondrial fusion accounts for the exquisite vulnerability of Purkinje neurons in pcd mice.
Introduction
One of the most perplexing questions in the neurodegenerative disease field is why different types of neurons degenerate in different disorders. The Purkinje cell degeneration (pcd) mouse model offers an opportunity to investigate this conundrum as pcd mice display a dramatic phenotype of postdevelopmental neurodegeneration of cerebellar Purkinje cells. The cerebellar architecture of pcd mice is perfectly normal at 2 wk of age (P14), with healthy-appearing Purkinje cells and no evidence of neurological abnormality. However, soon thereafter, pcd mice develop a wobbly gait, which progresses to visibly severe ataxia by P35. During this time frame, cerebellar Purkinje cells rapidly degenerate as >99% of all Purkinje cell neurons are absent from the cerebellum by P35 (Mullen et al., 1976). While Purkinje cell degeneration and death indicate that this neuron cell type is exquisitely vulnerable in pcd, other populations of neurons, including cerebellar granule neurons (CGNs), thalamic neurons, retinal photoreceptors, and mitral neurons of the olfactory bulb, also degenerate in pcd mice but at a much slower rate over the course of 1 year. No other neuron populations are affected, and pcd mice do not exhibit any nonneurological phenotypes except for sterility in males (Mullen et al., 1976).
We became interested in the molecular genetic basis of pcd, which exhibits recessive inheritance, and after linkage mapping and positional cloning, we identified loss of function of the Nna1 gene as the cause of this classic spontaneous mouse mutant (Fernandez-Gonzalez et al., 2002). The Nna1 protein is 1,218 amino acids in length and contains a highly conserved zinc carboxypetidase domain (Fernandez-Gonzalez et al., 2002; Chakrabarti et al., 2006). We and others found that pcd neurodegenerative phenotypes could be rescued by transgenic complementation with normal Nna1 protein but not with an enzymatically dead version of Nna1 protein (Wang et al., 2006; Chakrabarti et al., 2008), highlighting the crucial role of this enzymatic function in preventing the neurodegeneration. In addition to being highly conserved in species ranging from Caenorhabditis elegans to humans, comparative sequence analysis in the protein database revealed that Nna1 is one member of a family of six cytosolic carboxypeptidases (CCPs) and is now commonly (and hereafter) referred to as CCP1 (Kalinina et al., 2007). Although it was initially thought that CCP1 detyrosinates tubulin (Kalinina et al., 2007), a subsequent study demonstrated that CCP1 is instead a deglutamylase, regulating a specific posttranslational modification (PTM) known as polyglutamylation (Rogowski et al., 2010). Polyglutamylation of tubulin in microtubules (MTs) has emerged as an important regulatory PTM, especially in the central nervous system, where MTs in neurons carry long glutamate tails on α-tubulin (Audebert et al., 1994). Indeed, regulation of polyglutamylation is linked to MT metabolism by the protein spastin, which when absent causes a neurodegenerative disorder known as hereditary spastic paraplegia (Fonknechten et al., 2000). Studies of pcd mice revealed that loss of function of CCP1 deglutamylation of tubulin accounts for the degeneration of Purkinje cell neurons, as depletion of the tubulin-specific neuronal glutamylase TTLL1 was sufficient to prevent Purkinje cell neuron demise in pcd mice (Rogowski et al., 2010). The mechanistic basis by which tubulin hyperglutamylation results in the specific dysfunction and degeneration of cerebellar Purkinje cell neurons, however, is yet to be defined.
In a previous study, we linked pcd neurodegeneration to mitochondrial dysfunction by delineating mitochondrial disease phenotypes in a loss-of-function Drosophila melanogaster model that exhibited greatly reduced expression of the single Drosophila Nna1/CCP1 orthologue NnaD and by documenting altered bioenergetics and mitochondrial morphology defects in the cerebellum of pcd mice (Chakrabarti et al., 2010). We also observed increased mitochondrial autophagy (mitophagy) in the cerebellum of pcd mice (Chakrabarti et al., 2009). As mitochondrial dynamics controls mitochondrial function and has been implicated in the regulation of mitophagy (Twig et al., 2008), and deletion of the profusion factor mitofusin 2 (Mfn2) yields striking Purkinje cell degeneration in mice (Chen et al., 2007), we considered a role for altered mitochondrial dynamics in pcd neurodegeneration by modulating the dosage of mitochondrial fission and fusion genes in the NnaD loss-of-function Drosophila model. These studies indicated that reduced NnaD expression promoted mitochondrial fragmentation, which could be rescued by reduced expression of the profission factor Drp1 and that NnaD–Drp1 double mutant flies exhibited marked improvements in NnaD loss-of-function disease phenotypes. We developed primary cell line models for CCP1 loss of function in retinal epithelial cells, observed similar abnormalities in mitochondrial morphology and function in these cell lines and in primary cerebellar neurons from pcd mice, and documented reduced mitochondrial fusion in cells lacking CCP1. As mitochondrial motility is likely a prerequisite for proper mitochondrial dynamics (Liu et al., 2009; Misko et al., 2010), we examined the effect of hyperglutamylation of tubulin on MT-mediated mitochondrial motility in pcd cerebellar neurons and noted reduced anterograde axonal transport and markedly reduced retrograde axonal transport. Further studies of CCP1 revealed that mitochondrial stress promotes the Parkin-dependent turnover of CCP1 along with other mitochondrial fusion factors and that CCP1 and Parkin physically interact. Our results indicate that CCP1 is a key node in the regulation of mitochondrial motility through the deglutamylation of tubulin and that loss of CCP1-mediated mitochondrial fusion accounts for the exquisite vulnerability of Purkinje cell neurons in pcd mice.
Results
Increased mitochondrial fragmentation underlies Drosophila NnaD loss-of-function phenotypes
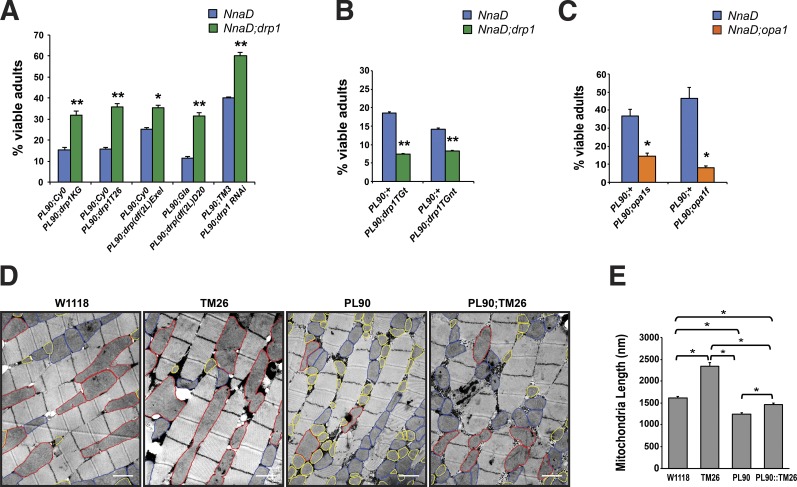
To delineate the molecular basis of pcd, we previously studied a Drosophila model of CCP1 loss of function as flies possess only a single CCP1/Nna1 orthologue known as NnaD (Rodriguez de la Vega et al., 2007). As the NnaD gene resides on the Drosophila X chromosome, we obtained a Drosophila line carrying a P element insertion in the NnaD gene (Bourbon et al., 2002). When we characterized this Drosophila line, which we designated as NnaDPL90, we determined that NnaD expression is reduced to ∼8% of normal levels in male NnaDPL90 flies and observed frequent larval lethality (Chakrabarti et al., 2010). Escaper male NnaDPL90 flies exhibited droopy wings and retinal degeneration, accompanied by mitochondrial morphology defects including mitochondrial fragmentation. In light of these results and increased mitophagy in pcd cerebellum (Chakrabarti et al., 2009), we hypothesized that altered mitochondrial dynamics might play a role in CCP/Nna loss-of-function phenotypes. We thus crossed the NnaDPL90 line with various Drosophila lines carrying different Drp1 loss-of-function alleles including two P element insertion lines, a large deletion mutant encompassing the entire Drp1 gene, and a Drp1 RNA interference expressing line as Drp1 promotes mitochondrial fission. In all crosses, we documented a significant rescue of NnaDPL90 larval lethality (Fig. 1 A). We also obtained Drosophila lines that overexpress Drp1 or exhibit partial loss of function of Opa1, which encodes a protein that promotes mitochondrial fusion (Song et al., 2009). When we crossed the NnaDPL90 flies with Drp1 overexpressers or Opa1 hypomorphs, we observed an enhancement of NnaDPL90 larval lethality (Fig. 1 B). To determine whether rescue of larval lethality in NnaDPL90–loss-of-function Drp1 double mutant flies is accompanied by changes in mitochondrial morphology, we performed ultrastructural analysis of flight muscle sections obtained from WT, NnaDPL90, Drp1TM26, and NnaDPL90–Drp1TM26 double mutant flies. As expected, we found that the mitochondrial network in NnaDPL90 flies was substantially fragmented, and individual mitochondria appeared much smaller than in WT flies, while Drp1TM26 flies displayed obviously elongated mitochondria (Fig. 1 C). We then measured mitochondrial length for each of these four genotypes, and when we calculated mean mitochondrial length and compared them, we found that the length of mitochondria in NnaDPL90–Drp1TM26 flies was significantly increased in comparison with NnaDPL90 flies but markedly reduced in comparison with Drp1TM26 flies (Fig. 1 D). Indeed, mean mitochondrial length in NnaDPL90–Drp1TM26 flight muscle was comparable with mitochondrial length in WT flight muscle (Fig. 1 D), indicating that altered mitochondrial dynamics, and in particular reduced mitochondrial fusion, is a feature of the CCP/Nna loss-of-function phenotype. To determine whether reduced dosage of mammalian Drp1 might rescue pcd phenotypes, we crossed pcd5J mice with Drp1 heterozygous knockout mice that display a 30% reduction in Drp1 expression levels (Fig. S1 A) and derived pcd5J/5J;Drp1+/− mice. Derivation of double homozygous null mice was precluded by the fact that Drp1 knockout mice are embryonic lethal (Ishihara et al., 2009; Wakabayashi et al., 2009). We found that pcd5J/5J;Drp1+/− mice exhibited neurological abnormalities, ataxia, and cerebellar degeneration that were identical in timing and severity to disease phenotypes in pcd5J homozygous mice (Fig. S1, B and C).
Figure 1.
Drosophila NnaD loss-of-function phenotypes and mitochondrial fragmentation can be modified by modulating the dosage of fission and fusion genes. (A) Percentage of viable adults in NnaDPL90 fly lines and NnaDPL90 fly lines carrying various drp1 loss-of-function alleles, where drp1 expression is reduced by at least 70%. n ≥ 300 flies per group. (B) Percentage of viable adults in NnaDPL90 fly lines and NnaDPL90 fly lines crossed with drp1 transgenic lines. n ≥ 100 flies per group. (C) Percentage of viable adults in NnaDPL90 fly lines and NnaDPL90 fly lines crossed with opa1 loss-of-function mutations. n ≥ 200 flies per group. (D) Transmission EM of Drosophila flight muscle from WT (W1118), drp1 loss-of-function (TM26), NnaDPL90(PL90), and NnaDPL90;drp1TM26 double mutant flies (PL90;TM26). Normal-sized mitochondria are outlined in blue, enlarged mitochondria are outlined in red, and fragmented mitochondria are outlined in yellow. Bars, 1 µm. (E) Quantification of mitochondrial length from (D). χ2 analysis (A–C) or ANOVA with post hoc Tukey test (E): *, P < 0.05; **, P < 0.01. Number of mitochondria analyzed per genotype were as follows: W1118 = 355, TM26 = 192, PL90 = 492, and PL90;TM26 = 416. Error bars are SEM.
CCP1 knockout cells and pcd neurons exhibit mitochondrial fragmentation and dysfunction
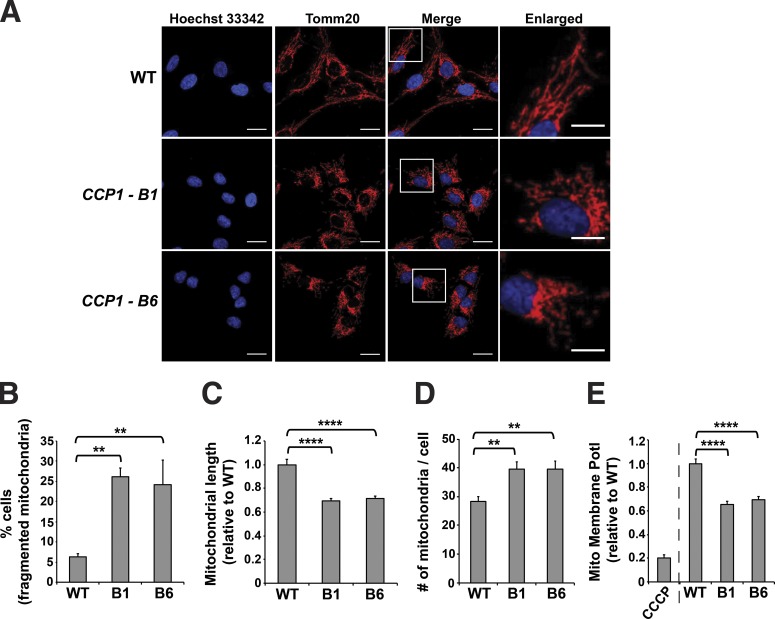
To understand the role of CCP1 in mitochondrial regulation, we performed CRISPR-Cas9 genome editing in retinal pigmented epithelial (hTert-RPE-1) cells as this cell type is affected in the retinal degeneration in pcd mice. We derived two distinct null RPE cell lines, CCP1-B1 and CCP1-B6 (Fig. S2), and we found that both CCP1 null RPE cell lines exhibited normal growth rates. To assess mitochondrial morphology in these lines, we immunostained with anti-Tomm20 antibody and noted that both the CCP1-B1 line and the CCP1-B6 line displayed a marked propensity to mitochondrial fragmentation (Fig. 2, A and B). When we measured mitochondrial length, we observed that mitochondria in the CCP1-B1 and CCP1-B6 cell lines are markedly shorter than mitochondria in WT RPE1 cells and that these cell lines contained many more individual mitochondria per cell (Fig. 2, C and D). Furthermore, mitochondrial membrane potential was markedly reduced in both CCP1-B1 and CCP1-B6 RPE cells in comparison with WT RPE1 cells (Fig. 2 E).
Figure 2.
Retinal pigmented epithelial cells lacking CCP1 display mitochondrial fragmentation and mitochondrial dysfunction. (A) Immunofluorescence staining of mitochondria in WT RPE1 cells and in two different CCP1 null RPE1 cell lines. Red indicates Tomm20; blue indicates Hoechst 33342. Bars: 20 µm (main images); 10 µm (insets). (B) Cells with a fragmented mitochondrial network from A. (C) Length of individual mitochondria from A. (D) Number of mitochondria per cell from A. (E) Mitochondrial membrane potential (Mito Membrane Potl) determined by JC-1 assay. The CCCP treatment of WT cells served as a positive control. For B–E, ANOVA with post hoc Tukey test: **, P < 0.01; ****, P < 0.0001. n = 119–150 cells per genotype. Error bars are SEM.
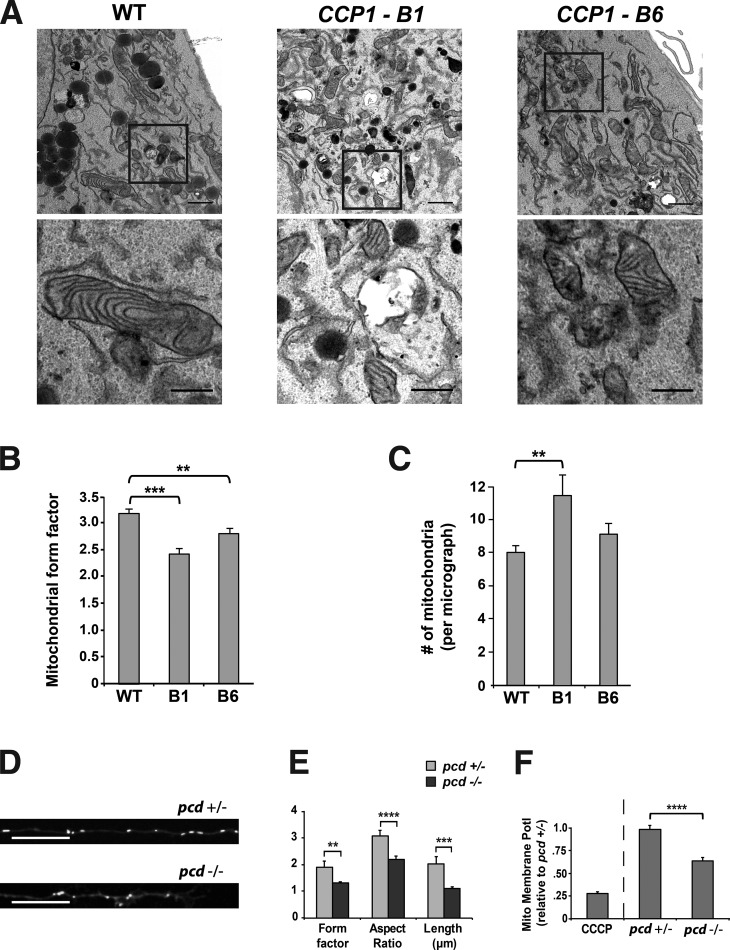
To more precisely characterize mitochondrial morphology changes in CCP1 loss-of-function cell lines, we pursued ultrastructural analysis of the CCP1-B1 and CCP1-B6 cell lines together with WT control RPE cells. Inspection of the resulting electron micrographs revealed that mitochondria in both CCP1 null RPE cell lines are smaller, more circular, and more abundant than mitochondria in WT RPE cells (Fig. 3 A). To objectively quantify these mitochondrial morphology changes, we determined the mitochondrial form factor to gauge circularity and found that mitochondria in both CCP1 null RPE cell lines are significantly more circular than mitochondria in WT RPE cells (Fig. 3 B). We also counted the number of mitochondria per micrograph, and while we noted that there were significantly more mitochondria in the CCP1-B1 cell line in comparison with control RPE cells, the difference between the CCP1-B6 cell line and control RPE cells only constituted a trend (Fig. 3 C). In addition to characterizing cell line models for CCP1 loss of function, we studied mitochondrial morphology in cultured CGNs from WT and pcd5J mice by transfecting an EGFP fluorophore fused to a mitochondrial-targeting sequence derived from Cox8. We then analyzed mitochondrial morphology by determining mitochondrial length, form factor, and aspect ratio, and we found that mitochondria from pcd5J/5J mice are significantly smaller, more circular, and less elongated than mitochondria from pcd+/5J mice (Fig. 3, D and E). We also documented a marked reduction in mitochondrial membrane potential in cerebellar neurons from pcd5J/5J mice in comparison with cerebellar neurons from pcd+/5J mice (Fig. 3 F). All of these findings indicate that loss of CCP1 function promotes mitochondrial fragmentation and dysfunction.
Figure 3.
CCP1 is required for maintenance of mitochondrial network morphology and mitochondrial function in retinal pigmented epithelial cells and pcd neurons. (A) Representative electron micrographs of mitochondria in WT RPE1 cells and in two different CCP1 null RPE1 cell lines. Top (lower power): bars, 1 µm. Bottom panels present higher magnification of respective insets; bars, 0.5 µm. (B) We calculated the mitochondrial form factor (perimeter2/4 × π × area) from electron micrographs shown in A. (C) We counted the number of mitochondria from electron micrographs shown in A. n ≥ 30 micrographs per cell line, and n ≥ 218 mitochondria per cell line. (D) Representative images of cerebellar granule cell neurons from pcd heterozygous null or pcd homozygous null mice transfected with mito-EGFP. Bars, 20 µm. (E) Determination of mitochondrial shape (form factor and aspect ratio) and mitochondrial length in mito-EGFP expressing cerebellar granule cell neurons shown in D. (F) Mitochondrial membrane potential (Mito Membrane Potl) determined by JC-1 assay for cerebellar granule cell neurons in D. ANOVA with post hoc Tukey test: **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. n = 25–29 neurons per genotype. Error bars are SEM.
CCP1 loss of function results in decreased mitochondrial fusion in living cells
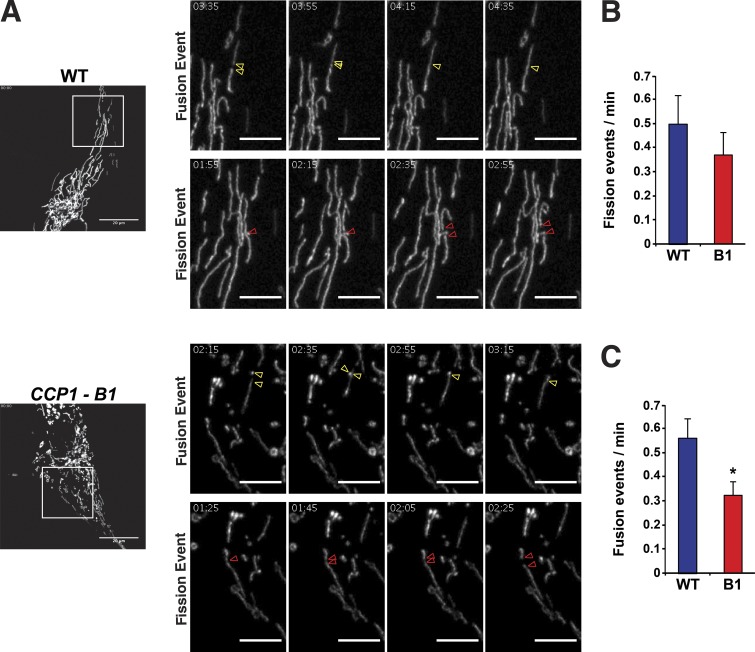
As alterations in mitochondrial morphology in CCP1 null RPE cells and pcd neurons suggest a defect in mitochondrial fusion upon loss of CCP1 function, we chose to directly examine mitochondrial fission and fusion rates in the CCP1-B1 cell line with the photoactivatable fluorescent protein mEOS.2 linked to a mitochondrial targeting sequence (mito-mEOS.2). After transient transfection of mito-mEOS.2 into the CCP1-B1 cell line and WT RPE cells, we performed live-cell imaging and scored individual mitochondrial fission or fusion events (Fig. 4 A). We observed similar rates of mitochondrial fission between WT RPE cells and CCP1 null RPE cells (Fig. 4 B); however, when we counted individual mitochondrial fusion events, we detected a significant reduction in the rate of mitochondrial fusion in CCP1 null RPE cells (Fig. 4 C). These results are consistent with the mitochondrial morphology changes observed in CCP1 null RPE cells and provide direct evidence for CCP1 positive regulation of mitochondrial fusion.
Figure 4.
Retinal pigmented epithelial cells lacking CCP1 undergo reduced mitochondrial fusion. (A) Representative examples of mitochondrial fission and fusion events in WT RPE1 cells and in a CCP1 null (CCP1-B1) RPE1 cell line. Red arrowheads indicate a fission event, while yellow arrowheads highlight a fusion event. Left (lower power): bars, 20 µm. All other panels present higher magnification of respective insets followed at 20-s intervals; bars, 5 µm. (B) We counted mitochondrial fission events from live cell imaging in A and calculated the fission rate as fission events per minute for WT RPE1 cells and in a CCP1 null (B1) RPE1 cell line. (C) We counted mitochondrial fusion events from live-cell imaging in A and calculated the fusion rate as fusion events per minute for WT RPE1 cells and in a CCP1 null (B1) RPE1 cell line. t test: *, P < 0.05. n ≥ 10 cells per genotype, and n = 3 independent experiments. Error bars are SEM.
CCP1 requires its enzymatic activity to promote mitochondrial fusion
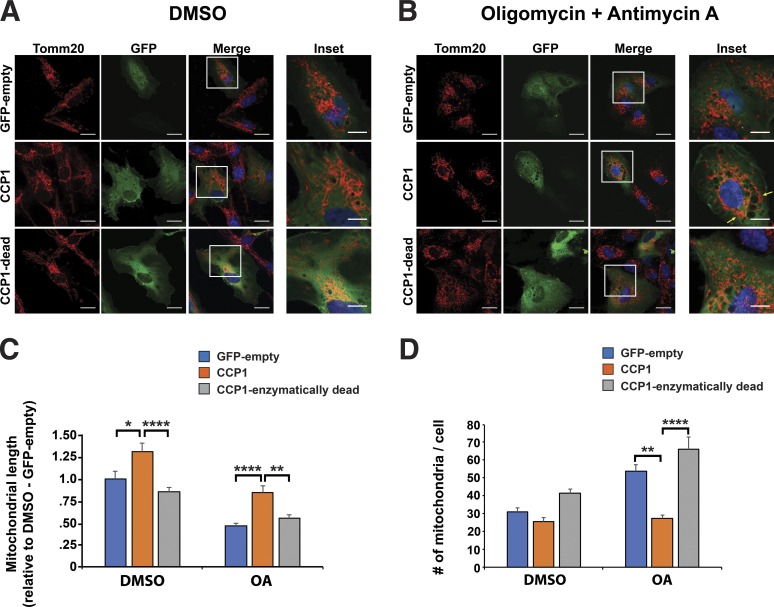
We next chose to determine whether CCP1 can promote expansion of the mitochondrial tubular network and if CCP1 requires its deglutamylase activity to enhance this mitochondrial fusion–dependent activity. To do this, we transfected WT RPE cells with empty vector, CCP1, or enzymatically dead CCP1 and monitored mitochondrial morphology and the status of the mitochondrial network under mitochondrial stress conditions by treating with oligomycin + antimycin A (OA) and then immunostaining with anti-Tomm20 antibody (Fig. 5, A and B). We confirmed that the OA stress treatment induced mitochondrial fragmentation but without killing RPE cells (Fig. 5 B). We found that overexpression of CCP1 dramatically increased mean mitochondrial length in RPE cells upon treatment with OA, but overexpression of enzymatically dead CCP1 did not significantly improve mean mitochondrial length (Fig. 5 C). The ability of enzymatically intact CCP1 to promote an increase in mitochondrial length was also observed in WT RPE cells treated with vehicle (Fig. 5 C). When we quantified the number of mitochondria per cell, we noted that CCP1 overexpression rescued the fragmented mitochondrial network in OA-treated RPE cells as CCP1-expressing RPE cells subjected to mitochondrial stress displayed a marked reduction in number of mitochondria per cell (Fig. 5 D). Indeed, the number of mitochondria per cell in the CCP1-expressing RPE cells treated with OA was comparable with the number of mitochondrial per cell in RPE cells exposed to vehicle and transfected with an empty vector (Fig. 5 D).
Figure 5.
Enzymatically intact CCP1 prevents mitochondrial fragmentation. (A) Immunofluorescence staining of WT RPE1 cells expressing either empty GFP vector, CCP1, or enzymatically dead CCP1 and treated with vehicle (DMSO). Red indicates Tomm-20, green indicates GFP, and blue indicates Hoechst 33342. Bars: 20 µm (main images); 10 µm (insets). (B) Immunofluorescence staining of WT RPE1 cells expressing either empty GFP vector, CCP1, or enzymatically dead CCP1 and treated with oligomycin (2.5 µM) and antimycin A (10 µM; OA) for 60 min before fixation. Yellow arrows indicate regions rescued from mitochondrial fragmentation. Bars, 20 µm (main images); 10 µm (insets). (C) Cells with a fragmented mitochondria network from A and B. (D) Number of mitochondria per cell from A and B. ANOVA with post hoc Tukey test: *, P < 0.05; **, P < 0.01; ****, P < 0.0001. n ≥ 50 cells per condition. Error bars are SEM.
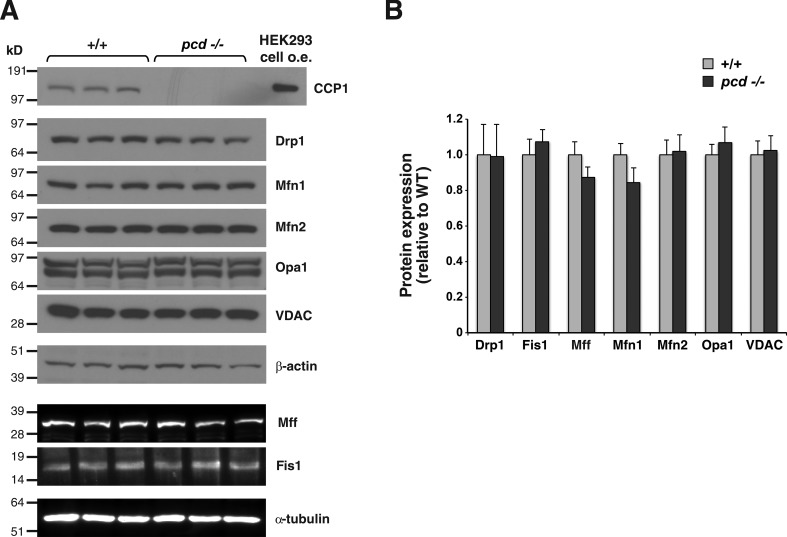
To determine whether CCP1 promotion of mitochondrial fusion involves modulation of the protein expression levels of mitochondrial fission and fusion factors, we dissected out the cerebellum from 5.5-mo-old pcd5J/5J mice and control WT mice, and after isolating protein lysates from these cerebellar samples, we surveyed the expression of mitochondrial fission and fusion factors by immunoblot analysis. While we confirmed absence of CCP1 expression in pcd5J/5J mice, we did not observe any noticeable differences in protein expression levels of mitochondrial fission and fusion factors upon inspection of immunoblots (Fig. 6 A). When we quantified the expression levels of mitochondrial fission and fusion factors by densitometry analysis of the resulting immunoblots, we noted comparable levels of protein expression, with no significant differences detected between pcd5J/5J mice and control, WT mice (Fig. 6 B).
Figure 6.
Loss of CCP1 does not affect the protein expression levels of mitochondrial fission and fusion factors in pcd mice. (A) Immunoblot analysis of cerebellar protein lysates from 5.5-mo-old pcd homozygous null mice (−/−) and WT littermate controls for CCP1 and various mitochondrial fission and fusion factors. HEK293 cell overexpression (o.e.) of CCP1 served as a positive control. β-Actin and α-tubulin served as respective loading controls for immunoblots shown above. (B) Expression levels of the different mitochondrial fission and fusion factors were quantified by densitometry. Two-tailed t test: P = NS; n = 3 mice per genotype. Error bars are SEM.
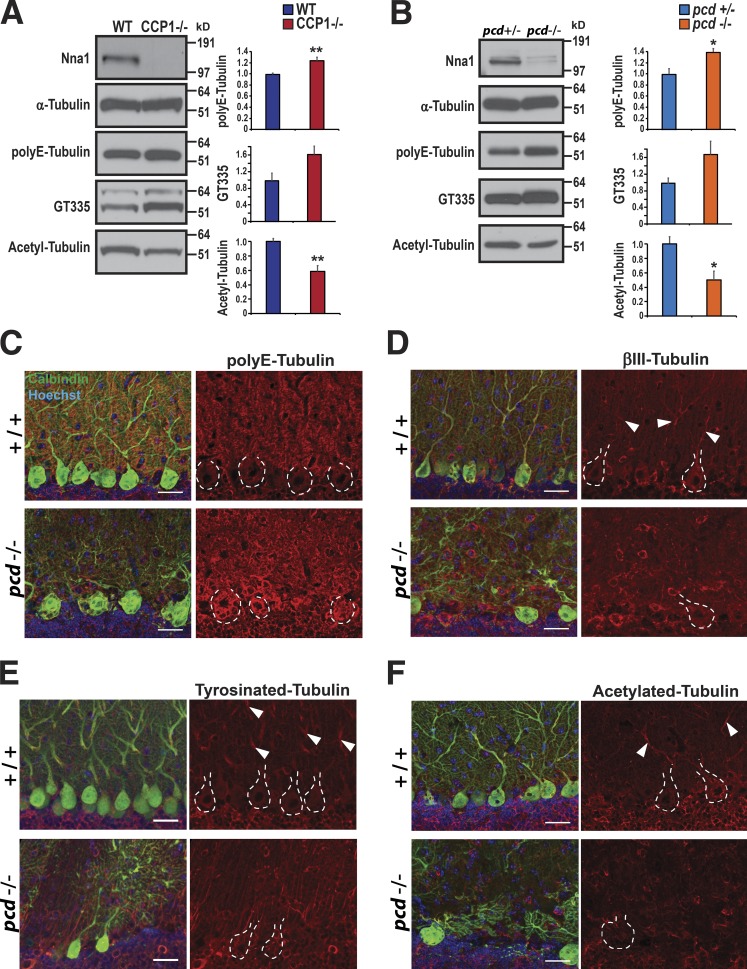
Tubulin PTMs are altered in pcd
An important turning point in the study of pcd was the realization that CCP1 protein possesses deglutamylase activity and that failure of proper tubulin deglutamylation contributes to the pathogenesis of pcd neurodegeneration (Rogowski et al., 2010). MTs are composed of arrays of tubulin polymers, which are subject to various PTMs, of which polyglutamylation plays a key regulatory role in MT stability and metabolism (Wloga et al., 2010). We examined the status of tubulin PTMs both in the CCP1-B1 and CCP1-B6 null RPE cell lines and in cultured primary cerebellar neurons from pcd mice and confirmed increased polyglutamylation of α-tubulin in the absence of CCP1 by immunoblot analysis but also detected a decrease in acetylated α-tubulin (Fig. 7, A and B). To further examine the relevance of altered regulation of tubulin glutamylation in pcd cerebellum and its subcellular distribution in degenerating Purkinje neurons, we immunostained cerebellar sections from P25 pcd mice, when Purkinje cell degeneration is prominent. As expected, we confirmed increased accumulation of polyglutamylated tubulin in pcd Purkinje neurons, especially in Purkinje cell soma (Fig. 7 C). However, when we compared Purkinje cell dendrites from P25 pcd and WT controls, we observed obvious differences in the patterns, distributions, and staining intensities of βIII-tubulin, tyrosinated tubulin, and acetylated tubulin. While we observed numerous examples of continuous segments of βIII-tubulin immunoreactivity in Purkinje cell dendrites in normal mice, such βIII-tubulin staining was noticeably absent from Purkinje cell dendrites of pcd mice (Fig. 7 D). Similarly, we detected segments of tyrosinated tubulin and acetylated tubulin staining in Purkinje cell dendrites of control mice, but this staining pattern was lacking in Purkinje cell dendrites from pcd mice (Fig. 7, E and F). When we quantified the immunostaining results for these tubulin PTMs, we observed significant alterations between the pcd mice and WT littermate controls (Fig. S3). Alterations in the levels of tubulin PTMs and an absence of linear staining arrays of modified tubulins upon visual inspection of pcd Purkinje cell dendrites suggested the existence of a possible defect in MT function in neuron processes in the cerebellum of pcd mice.
Figure 7.
Loss of CCP1 results in altered PTM of tubulin. (A) Immunoblot analysis of α-tubulin, polyE-tubulin, glutamylated tubulin (GT335), and acetylated tubulin in WT RPE1 cells and in CCP1 null (CCP1−/−) RPE1 cell lines. Expression levels of the different tubulins were quantified by densitometry. n = 4 mice per genotype. (B) Immunoblot analysis of α-tubulin, polyE-tubulin, glutamylated tubulin (GT335), and acetylated tubulin in cerebellar granule cell neurons from pcd heterozygote null and pcd homozygous null mice. Two-tailed t test: *, P < 0.05; **, P < 0.01. n = 3 replicates. (C–F) Cerebellar sections from 23-d-old pcd homozygous (−/−) mice and WT (+/+) littermate controls were immunostained for polyE-tubulin (C), βIII-tubulin (D), tyrosinated tubulin (E), or acetylated tubulin (F). Red indicates tubulin of interest, green indicates calbindin, and blue indicates Hoechst 33342. Dashed lines outline Purkinje cell soma, and white arrowheads indicate linear arrays of tubulin. Bars, 25 µm. Error bars are SEM.
pcd neurons display a specific defect in the axonal transport of mitochondria
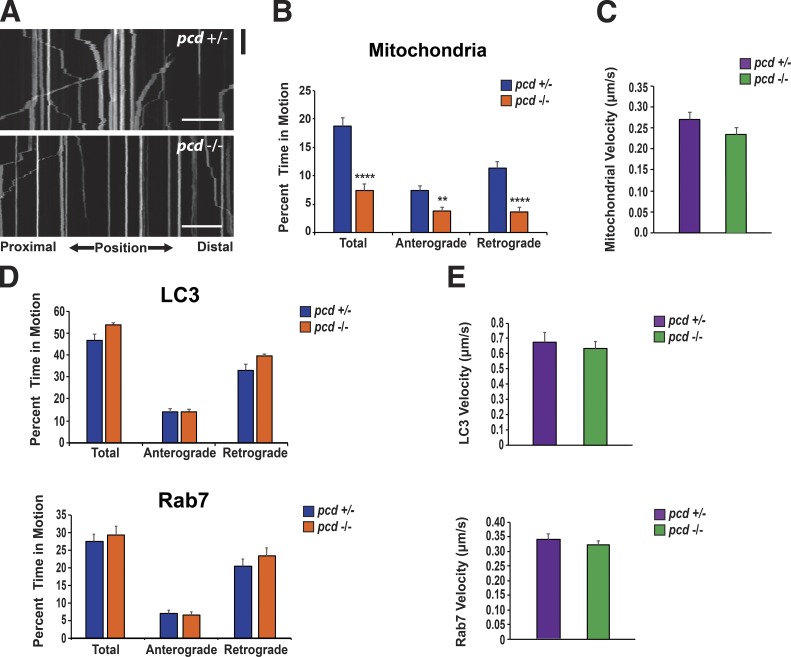
MTs mediate a variety of functions in the cell. In neurons, MTs are essential for proper transport of materials back and forth along axons and long neurite processes, and in the case of mitochondria, there exists a transport system in which MTs interact with kinesin and dynein to promote anterograde and retrograde transport, respectively (Goldstein and Yang, 2000). As mitochondrial motility has been linked to mitochondrial dynamics, we hypothesized that CCP1 loss of function may interrupt the proper transport of mitochondria along neuron axons in pcd mice. To assess mitochondrial transport, we transfected CGNs from pcd5J homozygous and heterozygous mice with mito-EGFP and performed kymograph analysis of mitochondrial movements (Fig. 8 A). We found that mitochondria from pcd5J/5J neurons spent significantly more time paused between anterograde movements than mitochondria from pcd+/5J neurons (Fig. 8 B). When we evaluated retrograde transport, we documented an even greater difference between mitochondria from pcd5J/5J neurons and pcd+/5J neurons as mitochondria from pcd5J/5J neurons spent dramatically more time paused between retrograde movements than mitochondria from pcd+/5J neurons (Fig. 8 B). Mitochondria from pcd5J/5J neurons, however, moved at a similar velocity to mitochondria from pcd+/5J neurons (Fig. 8 C). To determine whether the transport defect in pcd5J/5J neurons is specific to mitochondrial motility or simply reflects a general defect of MT transport, we measured transport rates and kinetics for two additional cargoes, LC3 and Rab7. In both cases, we measured comparable pause times between anterograde movements and between retrograde movements, regardless of pcd genotype (Fig. 8 D). We also recorded similar velocities for LC3 and Rab7 transport in pcd5J/5J and pcd+/5J neurons (Fig. 8 E). As polyglutamylation is known to be enriched in tubulin associated with centromeres (Wolff et al., 1992), we investigated whether CCP1 null RPE cells showed any signs of altered initiation or propagation of nascent MT tracks. To mark the growing + tip ends of MTs, we transiently transfected WT and CCP1 null RPE1 cells with EB1-GFP. We then compared the overall comet number, speed, distance, and duration but detected no differences in any of these parameters between WT and CCP1 null RPE cells (Fig. S4). Taken together, these results indicate that loss of CCP1 function does not alter MT generation but instead leads to a selective defect in MT-mediated transport of mitochondria.
Figure 8.
Mitochondrial transport is reduced in pcd CGNs. (A) Kymograph analysis of mitochondrial movements in mito-EGFP expressing cerebellar granule cell neurons from pcd heterozygous (+/−) mice and pcd homozygous (−/−) mice. Black bar is 60 s. Bars, 20 µm. (B) Markedly increased pausing of mitochondria in cerebellar neurons from pcd homozygous (−/−) mice. Two-tailed t test: **, P < 0.01; ****, P < 0.0001. n ≥ 25 axons per genotype. (C) Mean velocity of mitochondria in cerebellar neurons from pcd heterozygous (+/−) mice and pcd homozygous (−/−) mice. (D) Results of kymograph analysis of LC3 movement and Rab7 movement in cerebellar granule cell neurons from pcd heterozygous (+/−) mice and pcd homozygous (−/−) mice, expressing either LC3-GFP or Rab7-GFP. Movements of LC3 and Rab7 were similar in pcd+/− and pcd−/− neurons. For LC3-GFP, n = 28 axons per genotype. For Rab7-GFP, n ≥ 15 axons per genotype. (E) Mean velocities of LC3 and Rab7 in cerebellar neurons from pcd+/− and pcd−/− mice. Two-tailed t test: P = NS. Error bars are SEM.
Parkin degrades CCP1 in a proteasome-dependent manner
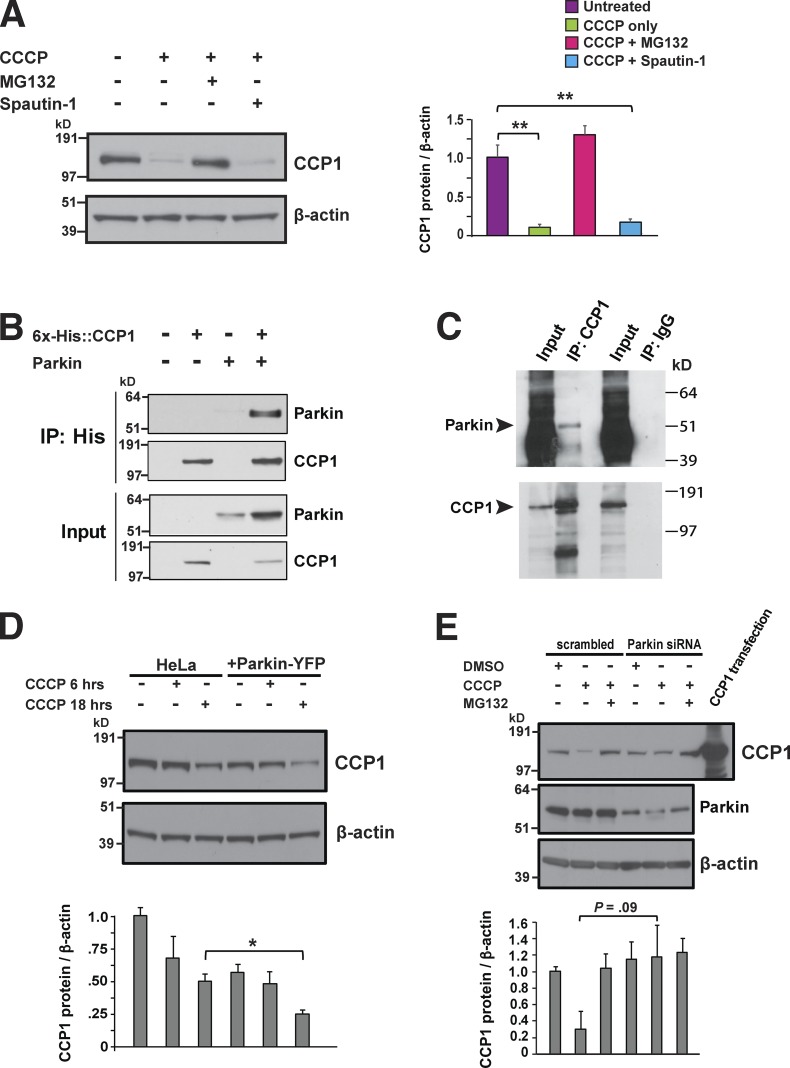
If CCP1 deglutamylation of tubulin promotes mitochondrial motility to enhance mitochondrial fusion and bolster mitochondrial quality control, then we hypothesized that CCP1 might be degraded under conditions of mitochondrial stress. To test this hypothesis, we subjected primary granule cell neurons to mitochondrial stress and found that CCP1 is indeed degraded under these conditions and that the degradation of CCP1 requires the ubiquitin–proteasome system (Fig. 9 A). As Parkin has been shown to mediate the proteasome-dependent degradation of target proteins that promote mitochondrial fusion and mitochondrial motility upon PINK1 stabilization at the mitochondrial outer membrane (Narendra et al., 2008; Matsuda et al., 2010), we performed a cotransfection coimmunoprecipitation analysis of CCP1 and Parkin and found evidence for a physical interaction (Fig. 9 B). We then pursued in vitro transcription translation of CCP1 and Parkin and again detected evidence of a physical interaction (Fig. 9 C), suggesting that CCP1 and Parkin may directly interact. When we overexpressed Parkin in HeLa cells, which normally do not express Parkin, we observed that CCP1 undergoes significant degradation (Fig. 9 D). To assess the physiological significance of Parkin-dependent degradation of CCP1, we again cultured primary granule cell neurons under conditions of mitochondrial stress but did so in the presence or absence of Parkin knockdown, and we noted that CCP1 degradation was obviously reduced upon Parkin knockdown, though the level of reduction only constituted a strong trend (Fig. 9 E). Taken together, our results indicate that CCP1 protein stability is linked to mitochondrial health and likely regulated by the PINK1/Parkin mitochondrial surveillance pathway.
Figure 9.
CCP1 is degraded by the ubiquitin–proteasome system in response to mitochondrial insult. (A) Immunoblot analysis of CCP1 expression in cerebellar granule cell neurons treated as indicated (CCCP at 5 µm, MG132 at 10 µm, and Spautin-1 at 10 µm), with densitometry analysis of CCP1 expression relative to β-actin expression for n = 3 replicates, shown on right. ANOVA with post hoc Tukey test: **, P < 0.01. (B) Cotransfection coimmunoprecipitation of 6×His-tagged CCP1 and Parkin-YFP. (C) The CCP1 antibody pulldown of in vitro transcription–translation generated CCP1 and Parkin. The IgG-only pulldown serves as a negative control. IP, immunoprecipitation. (D) Immunoblot analysis of CCP1 expression in HeLa cells or HeLa cells stably expressing Parkin-YFP after being treated with CCCP (5 µm) as indicated, with densitometry analysis of CCP1 expression relative to β-actin expression for n = 3 replicates shown at the bottom. Parkin expression significantly enhanced CCP1 degradation upon CCCP treatment for 18 h. Two-tailed t test: *, P < 0.05. (E) Cerebellar granule cell neurons were treated as indicated (CCCP at 5 µm and MG132 at 10 µm) in the presence of either a control scrambled siRNA or Parkin siRNA, and the resulting protein lysates were immunoblotted for CCP1, Parkin, and β-actin. Densitometry analysis of CCP1 expression relative to β-actin expression is shown at the bottom and reveals a strong trend toward reduced CCP1 degradation upon Parkin knockdown. Error bars are SEM.
Discussion
In this study, we provide evidence that CCP1 functions in an evolutionarily conserved manner to maintain mitochondrial quality control. Based upon our previous work in a Drosophila model of CCP/Nna protein loss of function and in pcd mice (Chakrabarti et al., 2010), we proposed that CCP1/Nna1 is required for normal mitochondrial function and bioenergetics. Mitochondrial dysfunction is a common theme in the field of neurodegeneration, and there are numerous examples of recessive inherited cerebellar ataxias, where the defective gene product performs a vital function required for mitochondrial health (Fogel and Perlman, 2007). Our results indicate that CCP1 should now join the list of critically important nuclear-encoded genes whose protein products mediate an irreplaceable function in neuronal mitochondria.
Another important theme in neurodegeneration is the essential role of the MT machinery in the transport of vital cargoes along the extensive lengths of axons and processes that comprise the unique architecture of neurons. Indeed, there are many examples of neurodegenerative disorders where an inherited defect in a molecular motor, MT processing enzyme, or cytoskeletal component is sufficient to produce a neurological disease phenotype in humans (Chen and Chan, 2009). The transport of various cargoes back and forth along axons has been carefully studied, and this work has revealed a transport machinery consisting of MTs connected to a molecular motor protein, kinesin or dynein, to permit delivery of cargoes in either the anterograde (to nerve terminals) or retrograde (to the cell body) direction (Saxton and Hollenbeck, 2012). Although the mechanism by which neurons regulate the transport of most cargoes is still unclear, the basis for axonal transport of mitochondria has been well worked out and involves the interaction of Miro1/Miro2 anchored to the mitochondrial membrane with an adaptor protein, Milton, that couples kinesin and dynein/dynactin to mitochondria. Over the last decade, it has become increasingly clear that this mitochondrial transport machinery is inextricably linked with the mitochondrial fusion machinery, suggesting regulatory cross-talk between mitochondrial dynamics and mitochondrial motility as overexpression of Miro or Milton can increase mitochondrial fusion, while the profusion factor Mfn2 is required for axonal transport of mitochondria and physically interacts with the Miro–Milton protein complex (Saxton and Hollenbeck, 2012).
In this study, we characterized the mitochondrial dysfunction in pcd mice and in the NnaDPL90 loss-of-function fly model to ascertain the nature of the mitochondrial defects that occur when CCP/Nna protein function is lost. Based upon studies of RPE cell lines lacking CCP1, NnaDPL90 fly flight muscle, and primary CGNs from pcd mice, we documented a shared process of mitochondrial fragmentation. We found that expression of enzymatically intact CCP1 could rescue mitochondrial fragmentation elicited by mitochondrial metabolic stress in mammalian cells and that reduced dosage of the fission factor Drp1 could rescue mitochondrial fragmentation in NnaDPL90 flies. We then directly monitored mitochondrial fission and fusion events in WT RPE cells and in CCP1 null RPE cells and observed a marked reduction in mitochondrial fusion in RPE cells lacking CCP1. These findings indicate that CCP1 promotes mitochondrial fusion, but how? We considered the possibility that CCP1 modulates the expression or stability of mitochondrial fission and fusion factors, but immunoblot analysis of cerebellar protein lysates revealed similar levels of these proteins in pcd5J/5J mice and WT control mice. Numerous studies have suggested a functional link between mitochondrial axonal transport and mitochondrial dynamics as overexpression of Miro or Milton results in increased mitochondrial fusion (Fransson et al., 2006; Saotome et al., 2008; Koutsopoulos et al., 2010). As mitochondrial motility is mediated by the MT system in concert with specific motor proteins and tubulin glutamylation is a regulatory PTM affecting MT function (Larcher et al., 1996; Bonnet et al., 2001), we reasoned that CCP1 deglutamylation of tubulin promotes effective MT-mediated mitochondrial motility, thereby enhancing mitochondrial fusion. We thus examined mitochondrial axonal transport in pcd cerebellar neurons and documented impaired anterograde and markedly reduced retrograde mitochondrial transport, consistent with CCP1 regulation of mitochondrial motility. The importance of mitochondrial motility regulation for robust mitochondrial fusion is supported by previous work showing that PINK1 stabilization on the outer membrane of damaged mitochondria targets Miro for Parkin-dependent degradation to render such mitochondria incapable of readily undergoing mitochondrial fusion (Wang et al., 2011). As Parkin recruitment to damaged mitochondria unleashes a wave of degradation of profusion and promotility factors, we examined CCP1 turnover under conditions of mitochondrial stress and observed prominent Parkin-dependent degradation of CCP1, and we also uncovered evidence for a physical interaction between CCP1 and Parkin. Taken together, our findings favor a model in which CCP1 deglutamylation of tubulin primes MTs for effective interaction with the mitochondrial transport machinery to permit motile mitochondria to more easily engage in the mitochondrial fusion process in neurons.
Another aspect of this work is its relevance to our understanding of cell type–specific neurodegeneration. Purkinje cells are among the most electrically active of all neuron subtypes and have extensively subdivided dendrites that form a massive network of synaptic contacts as well as long axonal projections with numerous neuron targets. This cytoarchitecture demands that Purkinje cells are capable of continuously producing enormous quantities of ATP, and therefore, such neurons cannot tolerate a drop in energy output. Previous work has shown that knockout of Mfn2 in mice results in a postdevelopmental degeneration of Purkinje cell neurons, though knockout of Mfn1 does not yield Purkinje cell loss (Chen et al., 2007). While both Mfn1 and Mfn2 promote mitochondrial fusion, Mfn2 is also required for axonal transport of mitochondria (Misko et al., 2010), suggesting that mitochondrial motility is especially important for Purkinje cells. Our discovery of a role for CCP1 in regulating mitochondrial motility highlights the physiological relevance of heightened mitochondrial quality control in Purkinje cell neurons and may explain why pcd mice exhibit a cataclysmic process of Purkinje cell neuron degeneration and death that play out over the course of only a few weeks.
Materials and methods
Drosophila experimentation
Drosophila stocks were obtained from the Bloomington Drosophila stock center (strain/allele as indicated) and the Vienna Drosophila RNAi Center (UAS-Drp1-RNAi; 44155) and were maintained on regular cornmeal molasses food. All crosses were performed at 25°C. Drosophila insect flight muscle was prepared for EM analysis by first fixing the flies using 2.5% glutaraldehyde + 2% PFA in 0.1 M sodium cacodylate, pH 7.4, for 4 h on ice. The muscle tissue was then extracted and washed with ice-cold 0.1 M sodium cacodylate 3 × 10 min on ice followed by postfixation in 1% osmium tetroxide and 0.8% potassium ferrocyanide in 0.1 M sodium cacodylate for 3 h on ice. After three washes in ice-cold double-distilled H2O for 10 min each, the tissue was stained in 2% uranyl acetate for 2 h. Samples were dehydrated in an ethanol series of ice-cold 20, 50, 70, and 90% and then three washes in 100% ethanol at RT for 10 min each. Samples were infiltrated in 50% ethanol/50% Durcupan ACM (Fluka/Sigma-Aldrich) for 3 h at RT with agitation, followed by three changes of 100% Durcupan for 8 h each at RT with agitation. The Durcupan resin was polymerized at 60°C for 2 d under vacuum. Sectioning was performed using a Leica ultramicrotome. Ultrathin (80-nm) sections were poststained with uranyl acetate (5 min) and lead salts (2 min) before imaging using an FEI spirit transmission electron microscope operated at 120 kV. The magnification was 4,400× corresponding with a pixel resolution of 2.9 nm. Representative panels have dimensions of 13.7 µm × 17.0 µm. Mitochondrial number per area, length, and volume density (volume fraction) were measured on the tissue images as described previously (Guo et al., 2013). Briefly, mitochondrial volume density was measured using stereology performed with the aid of Photoshop (Adobe). Point counting was used to determine the mitochondrial volume densities by overlaying a grid on each digitized image. Mitochondria and cytoplasm lying under intercepts were counted. The relative volume of mitochondria was expressed as the ratio of intercepts coinciding with this organelle to the intercepts coinciding with cytoplasm and reported as a percentage. Mitochondrial lengths were measured on these same images using the ImageJ line segment tool (National Institutes of Health). Mitochondrial number per area was measured by counting all the mitochondria in an image and dividing by the cytoplasmic area shown in the image as measured by the ImageJ area tool. For a quick visualization aid of the distribution of mitochondrial lengths, each mitochondrion in an image was traced with either a yellow, blue, or red outline indicating small, larger, and largest mitochondria, respectively. The binning for the three colors was as follows. Because one SD is 34% from the mean, the middle bin (blue) contained the mitochondria falling 0.5× the SD from the total mean of the mitochondrial length measurements on either side. Thus, 34% of the mitochondria fall in the blue bin centered on the mean, and the yellow and red bins each had nearly 33% of the mitochondria at either end of the distribution.
Mouse breeding and behavioral analysis
All mice experiments were conducted in accordance with University of California, San Diego, and Duke University Institutional Animal Care and Use Committee and Animal Care Panel guidelines. The Drp1+/− mice were obtained (Wakabayashi et al., 2009) and bred with pcd5J heterozygotes. The resulting F1 progeny were genotyped as previously described upon weaning at P21 (Chakrabarti et al., 2006). Behavioral data were collected at 4, 6, and 9 wk of age. Weight measurements were taken at all three time points. To assess motor and neurological phenotypes, mice were subjected to a neurological function screening battery at 4 and 6 wk of age (Guyenet et al., 2010). This neurological assessment characterizes four neurological phenotypes—(1) hind limb clasping, (2) ledge test, (3) gait analysis, and (4) presence or absence of kyphosis—and generates a composite score for each animal. Mice motor performance was also evaluated using the accelerating rotarod analysis at 9 wk of age, accelerating from 4 to 60 rpm over 4 min. Mice were given one practice day and then were assayed over four consecutive days. On each day, mice were subjected to five independent trials with at least 5 min rest in between, and the best four times were recorded and averaged.
Cell line model derivation and analysis
The hTert-RPE-1 cells (ATCC CRL-4000) were cultured in DMEM/F12 media lacking Hepes (Gibco) supplemented with 10% FBS in a 37°C humidified incubator with 5% CO2. All transfections were performed using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. The Nna1-gRNA, 5′-GGTGGAGGTCGAAGAGTGA-3′, was selected using ECRISP (http://e-crisp-test.dkfz.de/E-CRISP/designcrispr.html) using the input ENSEMBL code for NNA1 gene and confirmed using BLAST against the human mRNA database. A restriction enzyme digestion with EcoRI/PstI was done on lyophilized gRNA resuspended at 20 ng/µl in NEB3 buffer for 1 h at 37°C and a pUC57 vector, followed by purification with QIAquick PCR purification kit (Qiagen). After purification, the digested PCR products were ligated with T4 DNA ligase (New England Biolabs) and transformed into competent DH5α cells. Genomic DNA was isolated (Qiagen), and proper cloning was confirmed through sequencing (Eton Bioscience). The RPE1 cells were transformed with the gRNA plasmid and a Cas9-GFP plasmid. After 48 h, cells were sorted for high GFP expression using FACS (BD Influx Flow Cytometer) in order to generate clonal populations. Clones were then screened for Nna1 protein expression via Western blot. Candidates lacking any noticeable Nna1 protein expression had their DNA extracted and sequenced to confirm the mutagenesis at the genetic level. For mitochondrial stress experiments, WT and nna1Δ RPE1 cells were exposed to either DMSO or 2.5 µM oligomycin (50-230-6538; Thermo Fisher Scientific) and 10 µM antimycin A (A8674; Sigma-Aldrich) for 1 h. Mitochondrial membrane potential (Δψm) was determined using the JC-1 aggregate assay (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Immunoblot analysis
For isolation of protein from brain tissue, 5-mo-old pcd+/− and pcd−/− cerebella were isolated and snap frozen in liquid nitrogen. Approximately 5× volume of radioimmunoprecipitation assay buffer (Pierce) containing protease inhibitors and phosphatase inhibitors (Roche) was added to the cerebellar tissue. The tissue was passed through a 26-g needle 10× and then centrifuged at 12,000 rpm for 15 min at 4°C. Cell culture samples were washed once with ice-cold PBS and lysed by passing the cell suspension through a 26-g needle 10× before centrifugation. Protein concentration was measured using a bicinchoninic acid assay (Pierce). 20 µg protein from each sample was run on SDS-PAGE and then transferred to polyvinylidene difluoride membranes (EMD Millipore). Membranes were blocked in PBST with 5% milk or 5% BSA for 1 h and then probed with various antibodies (α-tubulin[Ms], DM1A, Cell Signaling Technology; tyrosinated tubulin[Ms], T9028, Sigma-Aldrich; polyglutamylated tubulin[Ms], T9822, Sigma-Aldrich; pan-polyglutamylation[Ms], AG-20B-0020-C100, AdipoGen; acetylated tubulin[Ms], D20G3, Sigma-Aldrich; CCP1[Rb], 14067-1-AP, Proteintech; Drp1[Ms], 611738, BD; Fis1[Rb], ALX-210-1037, Enzo Scientific; Mff[Rb], Ab81127, Abcam; Mfn1[Ms], Ab57602, Abcam; Mfn2[Ms], Ab56889, Abcam; Opa1[Ms], 612606, BD; β-actin[Ms], Ab8227, Abcam; VDAC[Rb], PA1-954A, Invitrogen; and Parkin[Ms], sc-32282, Santa Cruz) overnight at 4°C at a dilution of 1:1,000. Secondary HRP-conjugated anti-mouse or anti-rabbit antibodies (Santa Cruz) were applied for 1 h at RT at a dilution of 1:5,000 in PBST. Blots were developed using ECL (HyGlo Quickspray). Densitometry analysis was performed using ImageJ. All data shown are the average of three independent experiments.
Immunohistochemistry
Once anesthetized with a cocktail of ketamine and xylene, P25 or 4-mo-old mice underwent cardiac perfusion with PBS followed by fixation using 4% PFA. Once fully fixed, the brain was removed and placed in 4% PFA overnight. The PFA was then decanted and cryoprotected with 15% sucrose in PBS for 24 h and then 30% sucrose in PBS for an additional 24 h. The brains were embedded in optimal cutting temperature compound and sectioned using a Leica cryostat (Scottish Centre for Regenerative Medicine Histology Core Facility) into 30–50-µm sections. The sections were blocked for 1 h using PBS containing 10% normal goat serum, 0.3% Triton X-100, and 1% BSA. Purkinje cells were immunolabeled using anti-calbindin (D28K[Ms]; Sigma-Aldrich), and gliosis was monitored using anti–Glial Fibrillary Acidic Protein (GFAP[Rb]; G9269; Sigma-Aldrich). Antibodies for immunostaining of tubulin including α-tubulin, polyglutamylated tubulin, tyrosinated tubulin, and acetylated tubulin are the same antibodies detailed previously for immunoblot analysis. Alexa fluorophore secondary antibodies were used (Alexa fluor 488 and Alexa fluor 594; Thermo Fisher Scientific) to label, and Hoechst 33342 (Life Technologies) was used to counterstain nuclei. Sections were mounted using Fluoromount G (Electron Microscopy Services) and imaged using a Zeiss 780 laser scanning confocal microscope and Zen Black software (Salk Biophotonics Core Facility). Images were collected using a 63×, 1.4 NA objective and either GaAsP or photomultiplier tube detectors, depending on fluorophore wavelength.
The RPE1 cells were fixed in 4% PFA in PBS, then permeabilized using 0.3% Triton X-100 and blocked with 2% BSA for 15 min. Primary antibodies against Tomm20 (F-10[Ms], FL-145[Rb], and GFP[Rb], Santa Cruz; A6455, Thermo Fisher Scientific) or tubulin (same antibodies as previously described) were added at a dilution of 1:250 to 1:500 and incubated overnight at 4°C. Alexa fluorophore secondary antibodies (Life Technologies) were added for 1 h at RT, and Hoechst 33324 (Thermo Fisher Scientific) was added for 10 min. After washing with PBS, cells were mounted using Fluoromount G (cat. OB100-01; Thermo Fisher Scientific), covered, and sealed with nail polish. Images were collected using a Zeiss LSM 780 microscope at 63× magnification. The Z-stacks were obtained and compressed into single images. Analysis of mitochondria was performed as previously described using ImageJ (Dickey and Strack, 2011).
Mitochondrial morphology analysis
Two independent ImageJ-based computational methods were employed to evaluate mitochondrial morphology in confocal images. The first method uses a plugin to mask and measure the signal information, generating data concerning the perimeter, area, and length of the signal in a given region of interest, which is typically a single cell (Dickey and Strack, 2011). The second approach consisted of isolating a single cell, generating a thresholded image of that cell, and then using the analyze particles function in ImageJ to count each discrete mitochondria. Schematic flowthrough and examples can be found in the supplemental information (Fig. S5). For mitochondrial length determination, all cells were included in the analysis.
WT RPE cells and CP1 null RPE1 cell lines were grown on Aclar coverslips, fixed in 2% PFA + 2.5% glutaraldehyde in 0.15 M sodium cacodylate buffer + 2 mM CaCl2, postfixed in buffered 1% osmium tetroxide with 1.5% potassium cyanide, stained en bloc in 1% uranyl acetate, and dehydrated in graded ethanol immersion with 100% acetone. Infiltration was microwave processed in Eponate-12 resin. After cutting 70-nm sections with a Leica UC7 ultramicrotome, we examined the resulting sections in a Zeiss Libra 120 energy filter transmission electron microscope with a 2K charge-coupled device camera. Mitochondria were identified by visual inspection and traced for size and shape measurements by an individual who was blinded to the genotype of the cell line. We analyzed ≥30 micrographs per cell line for a total of ≥218 mitochondria per cell line in two independent experiments.
Primary cerebellar granule neuron studies
Primary mouse CGNs were isolated from P6 animals as previously described (Krämer and Minichiello, 2010). Cells were plated on either live imaging chamber slides (Lab-TekII Chambered #1.5 coverglass) or plastic dishes previously coated with poly-D-lysine (Sigma-Aldrich). Primary neuron cultures were incubated at 37°C with 5% CO2 either for 5 d for transport experiments or for 14 d before drug exposure experiments. For drug-exposure assays, cells were treated with either DMSO, 5 µM m-chlorophenylhydrazone (CCCP), 10 µM MG-132, 10 µM Spautin-1, 2.5 µM oligomycin, 2.5 µM antimycin A, 5 µg/ml brefeldin A, or 0.5 mM hydroxyurea for 2 h before harvesting protein. For Parkin knockdown experiments, two independent Silencer Select siRNA (Thermo Fisher Scientific) were obtained (s10043 and s10044; 4427037; Ambion). Cells were treated according to the manufacturer’s instructions and maintained for 3 d before analysis.
Live-cell imaging
24 h after initial plating of either RPE1 cells or CGNs, cells were transfected with the desired fluorescent protein. RPE1 cells were transfected with EB1-GFP, while CGNs were transfected with either mito-EGFP, LC3-GFP, or Rab7-EGFP using Lipofectamine 3000 per the manufacturer’s instructions. The RPE1 cells were incubated for 24 h before imaging, while CGNs were allowed to express the fluorescent fusion proteins for 4 d and then imaged in standard RPE1 cell media. For axonal transport experiments, axons were first identified using a brightfield image of the GFP-positive neuron. Axons were selected based on a three-point set of criteria: (1) neurite must emerge from cell body at roughly a 90° angle, (2) neurite must be exceptionally thin, and (3) there must be a lack, or very low number, of branch points or spines. For live imaging of organelle trafficking and EB1 comets we used a Zeiss CSU Spinning Disk confocal microscope, with a Yokogawa spinning disk scan head and electron-multiplying charge-coupled device camera. Images were collected using a 63× 1.4 NA objective using 1-s intervals for 4 min total with temperature set to 37° and 5% humidity. For neuronal transport experiments, before collecting time-lapse data, brightfield and GFP fluorescence were imaged in order to effectively identify axons and their overall directionality. Analysis of EB1-GFP comets was conducted using the TrackMate ImageJ plugin (Tinevez et al., 2017). Analysis of axonal transport was performed using a previously developed ImageJ macro (Pekkurnaz et al., 2014).
For analysis of the rates of mitochondrial fission and fusion, 24 h after initial plating of RPE1 cells, cells were transfected with the mito-mEOS.2 expression construct (Addgene). After 30 h of mito-mEOS.2 expression, we imaged living RPE cells in standard growth media using an LSM 880 confocal microscope with an Airyscan detector (Zeiss) at 37°C and 5% CO2 for at least 5 min at 2.5-s intervals. Images were acquired using a 63×, 1.4 NA oil immersion objective. The default Airyscan algorithm was used to process the original images. An individual blinded to the genotype of the cell line counted individual mitochondrial fission and fusion events. We analyzed ≥10 cells in three independent experiments.
Statistical analysis
All data were prepared for analysis with standard spreadsheet software (Microsoft Excel). Unless otherwise specified, all data shown represents the mean ± SEM. Data distribution was assumed to be normal, but this was not formally tested. We performed statistical analysis using Prism 7.0 (GraphPad Software), Excel, or the VassarStats Website for Statistical Computation (http://vassarstats.net/), and the level of significance (α) was always set to 0.05. For ANOVA, if statistical significance was achieved, we performed post hoc analysis to account for multiple comparisons.
Online supplemental material
Fig. S1 shows that dosage reduction of Drp1 does not rescue pcd phenotypes. Fig. S2 shows the validation of the CCP1 knockout RPE1 cell lines. Fig. S3 shows that loss of CCP1 results in significant changes in the levels of tubulin PTMs. Fig. S4 shows that an Eb1-GFP comet assay indicates normal nascent MT formation in CCP1 null cells. Fig. S5 illustrates the algorithms employed to quantify mitochondrial morphology characteristics and detect mitochondrial fragmentation.
Supplementary Material
Acknowledgments
This work was supported by funding from the National Institutes of Health (R01 EY014997 and R01 AG033082 to A.R. La Spada and T32 DA007315 to S. Gilmore-Hall).
The authors declare no competing financial interests.
Author contributions: S. Gilmore-Hall and A.R. La Spada provided the conceptual framework for the study. R. Zahra and A.R. La Spada designed the Drosophila experimentation, which was performed by R. Zahra and G. Perkins. J. Kuo and J.M. Ward generated the CCP1 null retinal pigmented epithelial cells, and A.R. La Spada and S. Gilmore-Hall designed the experiments to analyze mitochondrial morphology and function in CCP1 null cells and pcd neurons and to evaluate fission and fusion factors, tubulin PTMs, and mitochondrial transport, with assistance from R.S. Morrison; S. Gilmore-Hall performed all of these experiments. J. Kuo and S. Gilmore-Hall evaluated CCP1 protein turnover in experiments designed by A.R. La Spada, J. Kuo, and S. Gilmore-Hall. S. Gilmore-Hall and A.R. La Spada performed the analysis of experiments, assembled figures, and wrote the manuscript with input from all the authors. A.R. La Spada supervised the overall research and obtained funding to support this research project, together with S. Gilmore-Hall and J. Kuo.
References
- Audebert S., Koulakoff A., Berwald-Netter Y., Gros F., Denoulet P., and Eddé B.. 1994. Developmental regulation of polyglutamylated alpha- and beta-tubulin in mouse brain neurons. J. Cell Sci. 107:2313–2322. [DOI] [PubMed] [Google Scholar]
- Bonnet C., Boucher D., Lazereg S., Pedrotti B., Islam K., Denoulet P., and Larcher J.C.. 2001. Differential binding regulation of microtubule-associated proteins MAP1A, MAP1B, and MAP2 by tubulin polyglutamylation. J. Biol. Chem. 276:12839–12848. 10.1074/jbc.M011380200 [DOI] [PubMed] [Google Scholar]
- Bourbon H.M., Gonzy-Treboul G., Peronnet F., Alin M.F., Ardourel C., Benassayag C., Cribbs D., Deutsch J., Ferrer P., Haenlin M., et al. . 2002. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech. Dev. 110:71–83. 10.1016/S0925-4773(01)00566-4 [DOI] [PubMed] [Google Scholar]
- Chakrabarti L., Neal J.T., Miles M., Martinez R.A., Smith A.C., Sopher B.L., and La Spada A.R.. 2006. The Purkinje cell degeneration 5J mutation is a single amino acid insertion that destabilizes Nna1 protein. Mamm. Genome. 17:103–110. 10.1007/s00335-005-0096-x [DOI] [PubMed] [Google Scholar]
- Chakrabarti L., Eng J., Martinez R.A., Jackson S., Huang J., Possin D.E., Sopher B.L., and La Spada A.R.. 2008. The zinc-binding domain of Nna1 is required to prevent retinal photoreceptor loss and cerebellar ataxia in Purkinje cell degeneration (pcd) mice. Vision Res. 48:1999–2005. 10.1016/j.visres.2008.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti L., Eng J., Ivanov N., Garden G.A., and La Spada A.R.. 2009. Autophagy activation and enhanced mitophagy characterize the Purkinje cells of pcd mice prior to neuronal death. Mol. Brain. 2:24 10.1186/1756-6606-2-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti L., Zahra R., Jackson S.M., Kazemi-Esfarjani P., Sopher B.L., Mason A.G., Toneff T., Ryu S., Shaffer S., Kansy J.W., et al. . 2010. Mitochondrial dysfunction in NnaD mutant flies and Purkinje cell degeneration mice reveals a role for Nna proteins in neuronal bioenergetics. Neuron. 66:835–847. 10.1016/j.neuron.2010.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., and Chan D.C.. 2009. Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Hum. Mol. Genet. 18(R2):R169–R176. 10.1093/hmg/ddp326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., McCaffery J.M., and Chan D.C.. 2007. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 130:548–562. 10.1016/j.cell.2007.06.026 [DOI] [PubMed] [Google Scholar]
- Dickey A.S., and Strack S.. 2011. PKA/AKAP1 and PP2A/Bβ2 regulate neuronal morphogenesis via Drp1 phosphorylation and mitochondrial bioenergetics. J. Neurosci. 31:15716–15726. 10.1523/JNEUROSCI.3159-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez A., La Spada A.R., Treadaway J., Higdon J.C., Harris B.S., Sidman R.L., Morgan J.I., and Zuo J.. 2002. Purkinje cell degeneration (pcd) phenotypes caused by mutations in the axotomy-induced gene, Nna1. Science. 295:1904–1906. 10.1126/science.1068912 [DOI] [PubMed] [Google Scholar]
- Fogel B.L., and Perlman S.. 2007. Clinical features and molecular genetics of autosomal recessive cerebellar ataxias. Lancet Neurol. 6:245–257. 10.1016/S1474-4422(07)70054-6 [DOI] [PubMed] [Google Scholar]
- Fonknechten N., Mavel D., Byrne P., Davoine C.S., Cruaud C., Bönsch D., Samson D., Coutinho P., Hutchinson M., McMonagle P., et al. . 2000. Spectrum of SPG4 mutations in autosomal dominant spastic paraplegia. Hum. Mol. Genet. 9:637–644. 10.1093/hmg/9.4.637 [DOI] [PubMed] [Google Scholar]
- Fransson S., Ruusala A., and Aspenström P.. 2006. The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem. Biophys. Res. Commun. 344:500–510. 10.1016/j.bbrc.2006.03.163 [DOI] [PubMed] [Google Scholar]
- Goldstein L.S., and Yang Z.. 2000. Microtubule-based transport systems in neurons: The roles of kinesins and dyneins. Annu. Rev. Neurosci. 23:39–71. 10.1146/annurev.neuro.23.1.39 [DOI] [PubMed] [Google Scholar]
- Guo Y., Darshi M., Ma Y., Perkins G.A., Shen Z., Haushalter K.J., Saito R., Chen A., Lee Y.S., Patel H.H., et al. . 2013. Quantitative proteomic and functional analysis of liver mitochondria from high fat diet (HFD) diabetic mice. Mol. Cell. Proteomics. 12:3744–3758. 10.1074/mcp.M113.027441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyenet S.J., Furrer S.A., Damian V.M., Baughan T.D., La Spada A.R., and Garden G.A.. 2010. A simple composite phenotype scoring system for evaluating mouse models of cerebellar ataxia. J. Vis. Exp. (39):1787 10.3791/1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Nomura M., Jofuku A., Kato H., Suzuki S.O., Masuda K., Otera H., Nakanishi Y., Nonaka I., Goto Y., et al. . 2009. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 11:958–966. 10.1038/ncb1907 [DOI] [PubMed] [Google Scholar]
- Kalinina E., Biswas R., Berezniuk I., Hermoso A., Aviles F.X., and Fricker L.D.. 2007. A novel subfamily of mouse cytosolic carboxypeptidases. FASEB J. 21:836–850. 10.1096/fj.06-7329com [DOI] [PubMed] [Google Scholar]
- Koutsopoulos O.S., Laine D., Osellame L., Chudakov D.M., Parton R.G., Frazier A.E., and Ryan M.T.. 2010. Human Miltons associate with mitochondria and induce microtubule-dependent remodeling of mitochondrial networks. Biochim. Biophys. Acta. 1803:564–574. 10.1016/j.bbamcr.2010.03.006 [DOI] [PubMed] [Google Scholar]
- Krämer D., and Minichiello L.. 2010. Cell Culture of Primary Cerebellar Granule Cells. In: Ward A., Tosh D. (eds). Mouse Cell Culture. Methods in Molecular Biology (Methods and Protocols), vol 633 Humana Press; 10.1007/978-1-59745-019-5_17 [DOI] [PubMed] [Google Scholar]
- Larcher J.C., Boucher D., Lazereg S., Gros F., and Denoulet P.. 1996. Interaction of kinesin motor domains with alpha- and beta-tubulin subunits at a tau-independent binding site. Regulation by polyglutamylation. J. Biol. Chem. 271:22117–22124. 10.1074/jbc.271.36.22117 [DOI] [PubMed] [Google Scholar]
- Liu X., Weaver D., Shirihai O., and Hajnóczky G.. 2009. Mitochondrial ‘kiss-and-run’: Interplay between mitochondrial motility and fusion-fission dynamics. EMBO J. 28:3074–3089. 10.1038/emboj.2009.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., et al. . 2010. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189:211–221. 10.1083/jcb.200910140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misko A., Jiang S., Wegorzewska I., Milbrandt J., and Baloh R.H.. 2010. Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. J. Neurosci. 30:4232–4240. 10.1523/JNEUROSCI.6248-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen R.J., Eicher E.M., and Sidman R.L.. 1976. Purkinje cell degeneration, a new neurological mutation in the mouse. Proc. Natl. Acad. Sci. USA. 73:208–212. 10.1073/pnas.73.1.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D.F., and Youle R.J.. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183:795–803. 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekkurnaz G., Trinidad J.C., Wang X., Kong D., and Schwarz T.L.. 2014. Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase. Cell. 158:54–68. 10.1016/j.cell.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez de la Vega M., Sevilla R.G., Hermoso A., Lorenzo J., Tanco S., Diez A., Fricker L.D., Bautista J.M., and Avilés F.X.. 2007. Nna1-like proteins are active metallocarboxypeptidases of a new and diverse M14 subfamily. FASEB J. 21:851–865. 10.1096/fj.06-7330com [DOI] [PubMed] [Google Scholar]
- Rogowski K., van Dijk J., Magiera M.M., Bosc C., Deloulme J.C., Bosson A., Peris L., Gold N.D., Lacroix B., Bosch Grau M., et al. . 2010. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell. 143:564–578. 10.1016/j.cell.2010.10.014 [DOI] [PubMed] [Google Scholar]
- Saotome M., Safiulina D., Szabadkai G., Das S., Fransson A., Aspenstrom P., Rizzuto R., and Hajnóczky G.. 2008. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc. Natl. Acad. Sci. USA. 105:20728–20733. 10.1073/pnas.0808953105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton W.M., and Hollenbeck P.J.. 2012. The axonal transport of mitochondria. J. Cell Sci. 125:2095–2104. 10.1242/jcs.053850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Ghochani M., McCaffery J.M., Frey T.G., and Chan D.C.. 2009. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol. Biol. Cell. 20:3525–3532. 10.1091/mbc.e09-03-0252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez J.Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., and Eliceiri K.W.. 2017. TrackMate: An open and extensible platform for single-particle tracking. Methods. 115:80–90. 10.1016/j.ymeth.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. . 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27:433–446. 10.1038/sj.emboj.7601963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi J., Zhang Z., Wakabayashi N., Tamura Y., Fukaya M., Kensler T.W., Iijima M., and Sesaki H.. 2009. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J. Cell Biol. 186:805–816. 10.1083/jcb.200903065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Parris J., Li L., and Morgan J.I.. 2006. The carboxypeptidase-like substrate-binding site in Nna1 is essential for the rescue of the Purkinje cell degeneration (pcd) phenotype. Mol. Cell. Neurosci. 33:200–213. 10.1016/j.mcn.2006.07.009 [DOI] [PubMed] [Google Scholar]
- Wang X., Winter D., Ashrafi G., Schlehe J., Wong Y.L., Selkoe D., Rice S., Steen J., LaVoie M.J., and Schwarz T.L.. 2011. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 147:893–906. 10.1016/j.cell.2011.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D., Dave D., Meagley J., Rogowski K., Jerka-Dziadosz M., and Gaertig J.. 2010. Hyperglutamylation of tubulin can either stabilize or destabilize microtubules in the same cell. Eukaryot. Cell. 9:184–193. 10.1128/EC.00176-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff A., de Néchaud B., Chillet D., Mazarguil H., Desbruyères E., Audebert S., Eddé B., Gros F., and Denoulet P.. 1992. Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 59:425–432. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.