Abstract
Plant science is an important, rapidly developing area of study. Within plant science, one area of study that has grown tremendously with recent technological advances, such as mass spectrometry, is the field of plant-omics; however, plant peptidomics is relatively underdeveloped in comparison with proteomics and metabolomics. Endogenous plant peptides can act as signaling molecules and have been shown to affect cell division, development, nodulation, reproduction, symbiotic associations, and defense reactions. There is a growing need to uncover the role of endogenous peptides on a molecular level. Mass spectrometric imaging (MSI) is a valuable tool for biological analyses as it allows for the detection of thousands of analytes in a single experiment and also displays spatial information for the detected analytes. Despite the prediction of a large number of plant peptides, their detection and imaging with spatial localization and chemical specificity is currently lacking. Here we analyzed the endogenous peptides and proteins in Medicago truncatula using matrix-assisted laser desorption/ionization (MALDI)–MSI. Hundreds of endogenous peptides and protein fragments were imaged, with interesting peptide spatial distribution changes observed between plants in different developmental stages.
Keywords: plant science, Medicago truncatula, MALDI, orbitrap, mass spectrometry, imaging, peptides, proteins, peptidomics
Graphic Abstract
INTRODUCTION
Plant sciences play a significant role in mitigating three main challenges facing humanity in the 21st century, viz. food, energy, and environment (including pollution and climate change).1 These challenges are intricately linked to each other. For instance, climate change affects crop yield, which consequently affects food and energy supply. Besides food, plants are also used for the production of therapeutic and antimicrobial products that are used to treat various human diseases such as cancer, Alzheimer’s disease, and high cholesterol.2–6 Plants are also used for biofuel production and thereby act as “CO2 mitigators” due to the lower carbon footprint of plant-derived energy compared with energy produced from petroleum or natural gas.2 While significant progress has been made in the field of plant sciences, there remains a huge potential for improvement, which needs to be achieved to feed the burgeoning human population in the face of dwindling availability of arable land and water. For instance, despite the availability of close to 400 000 species of flowering plants, only a small fraction, about 200 species, have been domesticated for food and feed purposes. Among these, only 12 species contribute >75% of the food consumed across the world.2,7 Furthermore, with the vagaries of climate, plants are subjected to new or increased incidence of biotic and abiotic stresses. While it is true that the traditional method of plant genetics has led to most of the crop varieties we use today, relying on these traditional techniques alone will not satisfy our future needs for food, energy, and a stable environment. To produce better crops for the future, it is imperative that we understand the molecular processes that govern plant growth and development and their responses to the environment. The advent of “omics” technologies has facilitated our understanding of the molecular underpinnings of these complex traits like never before.
Among the omics technologies, the field of plant proteomics is rapidly growing and focuses on the study of proteins and enzymes expressed within various plant tissues. Limitations of more traditional gel electrophoresis-based proteomic methods include diffculty in analyzing highly basic or acidic proteins, bias toward more abundant proteins, and limited dynamic range. These limitations make low abundance proteins diffcult to detect.8,9 Mass spectrometry is an advantageous technique for plant proteomic analyses due the higher sensitivity, selectivity, and structural determination capabilities of this technique.10
Branching off of proteomics, plant peptidomics, or the study of the endogenous peptides produced by a plant, is a relatively new and underdeveloped field.11–14As signaling molecules, plant peptides have been shown to affect cell division, development, nodulation, reproduction, symbiotic associations, and defense reactions.14–18 Secreted peptides can act at low nano-molar concentrations, and the mature plant peptides are usually processed from larger polypeptides that undergo extensive proteolysis and posttranslational modifications (PTMs);17 therefore, the discovery and identification of bioactive signaling peptides represent a significant analytical challenge.
Mass spectrometric imaging (MSI) is a valuable tool for biological analyses because it allows for molecular analysis of tissue while retaining information about the spatial distribution of different analytes found within the tissue.19 In an MSI experiment, a laser is fired at the tissue sample in a predefined raster pattern, resulting in an array of mass spectra that can be compiled into a 2D distribution map for each mass measured. An advantage of MSI is that it lends itself to discovery experiments as it allows for the mass analysis of thousands of analytes in a single experiment and provides spatial information for the detected ions.
In the past decade, MSI has been increasingly utilized for the analysis of plant metabolomics.19–25 However, as previously mentioned, plant peptidomics is a relatively under-explored area in mass spectrometry and especially mass spectrometry imaging. A literature search resulted in one report of matrix-assisted laser desorption/ionization (MALDI)-MSI being used for the analysis of cyclotides in petunias.26 Additionally, while MS analysis is the gold standard for proteomic analysis, very few reports use MALDI–MSI for plant proteomics. Studies using MALDI–MSI for plant proteomics include a known allergenic protein in peaches shown to be exclusively found in the outer skin of the peach,27 a lipid-transfer protein imaged in tomato seeds,28 and proof-of-principle images of proteins in soybean cotyledons or barley grains.22,29 So far, no further applications of MALDI–MSI to plant proteomics have been reported.
Here we present a study using MALDI–MSI to investigate endogenous peptides and proteins in the model legume, Medicago truncatula (Medicago). This legume forms a symbiotic association with rhizobial bacteria that are housed in plant-derived organs, the root nodules. Inside these nodules, rhizobia are enclosed in unique structures, symbiosomes. These symbiosomes provide a conducive environment for the oxygen-sensitive rhizobial nitrogenase enzyme, thereby enabling the fixation of atmospheric nitrogen into a plant-available form.30,31 Nodule formation requires substantial energy expenditure by the plants and is therefore under tight regulation by local and systemic endogenous signals.32,33 Nodule development is also affected by environmental factors and, in particular, when nitrogen is available in the soil. Several signaling peptides that play a critical role in nodulation have been identified. For instance, a number of CLAVATA/ESR-related (CLE) peptides such as MtCLE12 and 13 of Medicago, LjCLE-RS1 and 2 of Lotus japonicas, and GmRIC1 and 2 of soybean act systemically as negative regulators of nodulation.33–37 These CLE peptides are induced upon rhizobial inoculation and are predicted to be the root-derived signals that, upon perception in the shoots, lead to the production of the shoot-derived inhibitor (SDI) that is transported to the roots, ultimately affecting nodulation. Recent evidence suggests that cytokinins are one of the shoot-derived inhibitors.38 Besides CLE, peptides such as nodule-specific cysteine-rich (NCR) peptides, C-terminally encoded peptide (CEP), rapid alkalinization factor (RALF), and devil/rotundifolia (ROT)-four-like (DVL/RTFL) play roles in a wide range of functions including bacteroid differentiation, nodule development, and infection thread formation and progression.39–42 Because of the importance of these peptides in legume symbiosis, we used MALDI–MSI to analyze 1 week old Medicago seedling roots and mature Medicago roots and root nodules. MSI was performed using both of the most common MALDI matrices, 2,5-dihydroxybenzoic acid (DHB) and α-cyano-4-hydroxycinnamic acid (CHCA), in the mass range from m/z 900–4000.43 In addition to studying the Medicago peptidome and proteome at different ages, wild-type plants were also compared with well-characterized Medicago mutants that are known to lack certain classes of peptides (dnf1–1) or overexpress certain classes of peptides (35S:MtCLE13). In addition to the MALDI–MSI experiments, parallel ESI–MS experiments were conducted to obtain accurate mass and high-quality tandem mass information for the m/z values detected via MSI. De novo sequencing was performed on the peptides detected with ESI using PEAKS software.44 Hundreds of endogenous peptides and protein fragments were imaged and showed interesting spatial distribution differences between plants at different growth stages and between wild-type and the various mutants.
EXPERIMENTAL PROCEDURES
Plant Growth and Inoculation with Rhizobia
Medicago truncatula seeds of wild-type (cv. Jemalong A17), dnf1–1, and 35S:CLE13 were scarified with pure sulfuric acid for 8 min and sterilized with 8% (w/v) calcium hypochlorite solution for 2 min and germinated on agar supplemented with gibberellic acid. Freshly germinated, 1 day old seedlings were transferred to square plates containing nitrogen-free modified Fåhraeus medium,45 after which the seedlings were grown in a growth chamber on modified Fåhraeus medium that was overlaid with germination paper. For nodule sampling, 10 seedlings were placed per plate, and the root part of the plate was covered with aluminum foil. The plates were placed vertically on a shelf at room temperature under fluorescent light. After 5 days of growth, the roots were inoculated with Sinorhizobium meliloti Rm1021 (wild-type)46 at an OD600 of 0.1 and then returned to the light shelf. At 3 weeks post-inoculation, nodules were separated from the roots and ground to a fine powder in liquid nitrogen. For seedling sampling, 100 1 day old seedlings were transferred to a plate containing nitrogen-free modified Fåhraeus medium. After 7 days, entire seedlings were collected, immediately frozen, and ground with liquid nitrogen.
Sample Preparation for MALDI
For the mature plants, root nodules with approximately 2 to 3 mm of surrounding root were excised from the plant. The excised tissue was placed in a plastic cup, covered with gelatin (100 mg/mL in double-distilled water), and frozen gently with dry ice. For the seedlings, 2 to 3 cm long portions of the root were cut from the seedling, embedded in gelatin, and frozen as detailed above. The frozen tissue was then sectioned into 16 μm slices using a cryostat at –20 °C and thaw-mounted onto standard glass microscope slides. A TM Sprayer (HTX Technologies, Carrboro, NC) was used to apply MALDI-matrix to the samples. The TM Sprayer method for DHB (40 mg/mL DHB in 50:50 water: methanol) was as follows: 80 °C, 0.1 mL/min flow rate, 24 passes-rotate and offset, 3 mm spacing, and a velocity of 1250 mm/min. The TM Sprayer method for CHCA (10 mg/mL CHCA in 50:50 water/acetonitrile) was as follows: 90 °C, 0.2 mL/min flow rate, eight passes-rotate and offset, 3 mm spacing, and a velocity of 1100 mm/min. DHB and CHCA were purchased from Sigma-Aldrich (St. Louis, MO).
MALDI-Orbitrap MSI
A high-resolution, accurate mass (≤5 ppm error) MALDI-LTQ Orbitrap hybrid mass spectrometer (Thermo Scientific, Waltham, MA) was used for MSI of both mature nodules and 1 week old seedlings. Multiple technical replicates of three or more biological replicates were imaged in the positive ion mode using a mass range of m/z 900–4000 and a mass resolution of 60 000. Mass spectra were collected across the surface of the sample with a raster step size of 75 μm. Peptide images were extracted automatically using MSiReader.47 In brief, the plant tissue was selected as the “region of interest” and the matrix was selected as the reference region. Masses from the matrix region were removed, and a list of m/z values detected in the plant samples was automatically generated. Ion images for each of the masses in the list were automatically extracted using the “generate an image for each peak in a list” function in MSiReader. Each extracted image was then manually confirmed as a true ion image, excluding isotope and matrix ion images.
Tissue Extraction
Approximately 50–100 root nodules with 2 to 3 mm of surrounding root were detached from mature Medicago plants or ∼50 1 week old Medicago seedlings were removed from the growth media and placed into a prechilled mortar. The tissue was flash-frozen with liquid nitrogen and ground to powder with the mortar and pestle. The powder was transferred to a prechilled 13 mL PTFE-coated centrifuge tube. The endogenous peptides were extracted with 3:1:4 methanol/chloroform:water (v/v), followed by brief vortexing and centrifugation for 10 min at 4 °C and 4700 rpm. The resulting aqueous supernatant was collected and dried in a SpeedVac. An additional four parts methanol were added to the remaining solution, followed by brief vortexing and centrifuging for 5 min at 4 °C and 4700 rpm. The organic layer was removed from the protein pellet, and both fractions were dried in a SpeedVac. The samples were stored at −80 °C until analysis.
Q-Exactive for ESI–MS
To acquire LC–ESI–MS/MS data, Medicago root nodule or seedling extracts were initially subjected to SCX fractionation on a Waters Alliance HPLC using a PolySulfethyl A column (2.1 mm internal diameter × 200 mm length, 5 μm particle size with 300 Å pore size; PolyLC, Columbia, MD). The mobile phases were (A) 10 mM ammonium formate in 75% water/25% acetonitrile at pH ∼6.8 and (B) 500 mM ammonium formate in 75% water/25% acetonitrile at pH 3. The samples were separated over 80 min under the following conditions: 0–15 min, 0% B; 15–45 min, 0–50% B; 45–55 min, 50–100% B; 55–65 min, 100% B; 65–65.5 min, 100–0% B, and finally re-equilibration at 0% B for 14.5 min. The column temperature was 30 °C, the flow rate was 0.2 mL/min, and the injection volume was 100 μL. Fractions were collected every 6 min between 10–70 min of the gradient. The fractions were combined and dried down three times with pure water to remove excess salts. Following SCX fractionation, the samples were resuspended in either water (aqueous fractions) or acetonitrile (organic and protein fractions) to a final concentration of 0.34 μg/μL (a total of 1.2 μg loaded onto the column). The samples were then separated on a NanoAcquity UPLC apparatus (Waters, Milford, MA) using a self-packed column (75 μm internal diameter ×160 mm length, 1.7 μm particle size with 130 Å pore size). The mobile phases were (A) water with 0.1% formic acid and (B) acetonitrile with 0.1% formic acid. The samples were separated over 108 min under the following conditions: 0–2 min, 0–4% B; 2–70 min, 4–30% B; 70–71 min, 30–75% B; 71–81 min, 75% B; 81–82 min, 75–95% B; 82–92 min, 75–95% B; 92–93 min, 95–0% B. The system was re-equilibrated at 0% B for 15 min. The flow rate was 0.3 mL/min, and the injection volume was 3.5 μL. The samples were kept at 4 °C during the analysis. MS/MS data were acquired in positive ion mode on an ESI-Q-Exactive Orbitrap mass spectrometer (Thermo Scientific). A top-15 data-dependent analysis (DDA) method was used with the full MS scan range from m/z 300– 2000; an isolation window of 2.0; an intensity threshold of 5.0 × 102 for triggering MS2; exclusion of 1, 8, >8 charged species; a normalized collision energy of 30; a dynamic exclusion of 30 s; an MS1 resolution of 35 000; and an MS2 resolution of 17 500.
MS/MS spectra were de novo sequenced and matched to proteins using PEAKS software (Bioinformatics Solutions) with a parent mass tolerance of 20.0 ppm, fragment tolerance of 0.01 Da, no enzyme, and five variable post-translational modifications (amidation, oxidation (M), hydroxylation, arabinosylation, and acetylation (protein N-term)). Three variable PTMs were allowed per peptide, and spectra were matched against the NCBI Medicago database. The ALC cutoff score was set at 50% for de novo sequenced peptides, and an FDR of 0.1% was used for protein matches.
RESULTS AND DISCUSSION
MALDI-Orbitrap MS Imaging
This study utilized wild-type Medicago and well-characterized Medicago mutants dnf1–1 and 35S:MtCLE13.48,49 A complete summary of the putative peptide m/z values detected can be found in the Supporting Information (Tables S1, S2, S3, and S4); m/z values detected in multiple samples were averaged in the reported lists. These tables detail a list of m/z values detected in each replicate, a comparison of the peptides detected using CHCA compared with DHB, and a comparison of the peptides detected in the seedling roots compared with the mature roots and nodules.
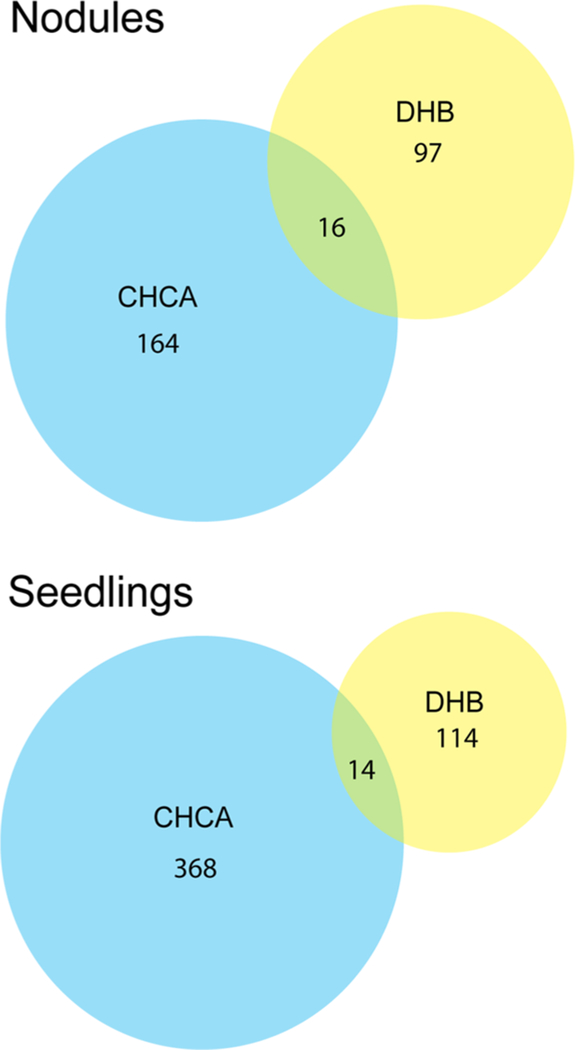
CHCA and DHB were chosen as complementary matrices for MALDI–MSI. Photographs showing the difference between application of DHB and CHCA on root nodules can be found in Figure S1. There was a surprisingly small percentage of peptide masses detected using both DHB and CHCA, as shown in the Venn diagrams in Figure 1. A greater number of peptides was detected using CHCA as the matrix in comparison with DHB. Sinapic acid (SA), a matrix more often used to analyze higher molecular weight species such as peptides and proteins, was also used but did not show improved signal or detectability compared with CHCA. Representative images of putative peptides detected with CHCA or DHB matrices in the mature Medicago roots and root nodules are shown in Figure 2. These representative images display ions with different spatial distributions in the plant root and nodules, which could provide further insight into the function of these putative peptides/proteins within the plant.
Figure 1.
Comparison of the numbers of endogenous peptides detected using either CHCA or DHB as MALDI matrices in both mature Medicago roots/root nodules and seedling roots.
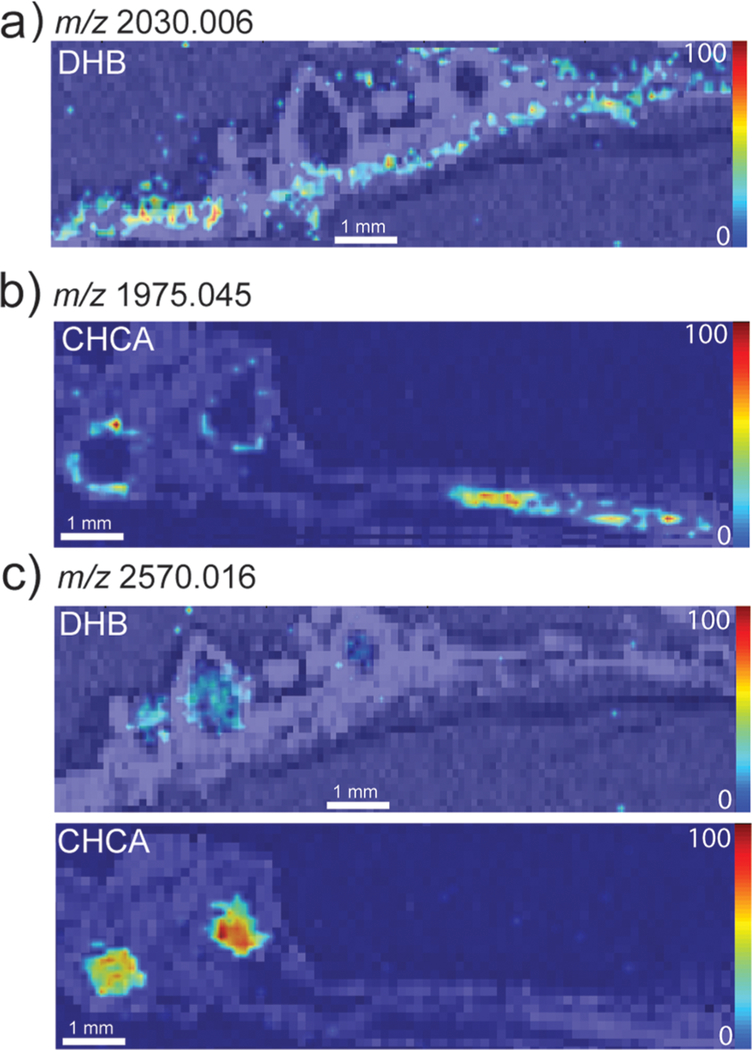
Figure 2.
Representative images of putative peptides detected with CHCA or DHB matrices in mature Medicago roots and root nodules show peptides that are differentially located within the root and nodules of the plant. (a) m/z 2030.006 was only detected using DHB as the matrix and is localized to the plant root. (b) m/z 1975.045 was only detected using CHCA as the matrix and is localized to the plant root and outer portion of the nodules. (c) m/z 2570.016 was detected using both DHB and CHCA as the matrix and is localized to the nodules. Intensity scale = high (red) to low (blue).
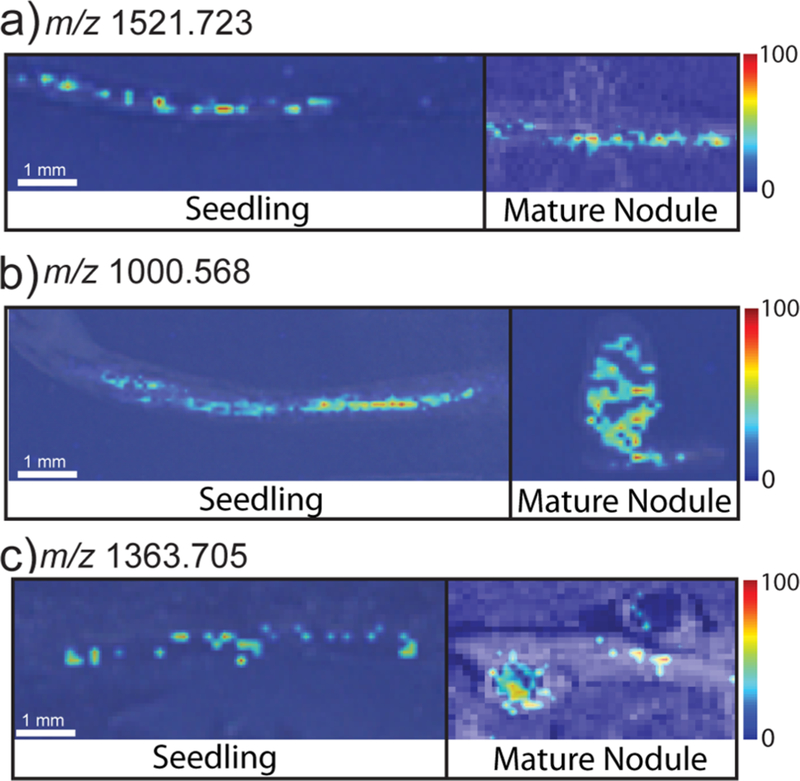
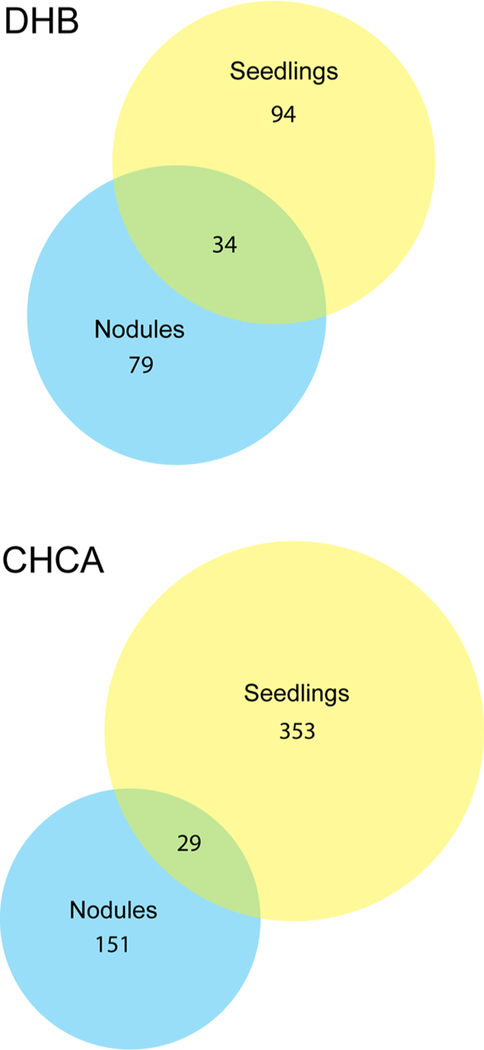
Putative peptides in Medicago roots were also compared in different stages of plant development. Figure 3 presents representative putative peptide images showing distinct distribution patterns in the Medicago seedlings and the mature roots and root nodules. Some of the detected species show similar distribution patterns when we compare the seedlings to the mature plants (Figure 3a); however, other putative peptides seem to shift their localization from the root to the root nodules as they develop into older plants (Figure 3b,c). This implies that these putative peptides/proteins may play a role in nodule development or other nodule-related processes. Figure 4 shows Venn diagrams comparing the numbers of putative peptides detected in the seedlings compared to the mature plants. Interestingly, a greater number of peptides was detected in the young seedling plants compared with the mature plants, regardless of the MALDI matrix used for ionization, and there was little overlap between the peptides detected in the plants in either stage of development.
Figure 3.
Representative putative peptide images showing distinct distribution patterns in the Medicago seedlings and the mature roots and root nodules. (a) m/z 1521.723 shows a similar distribution pattern in both the seedlings and the mature plants. (b) m/z 1000.568 and (c) m/z 1363.705 represent two of the detected peptides that show a shift in their localization from the root to the root nodules as they develop into older plants.
Figure 4.
Comparison of the endogenous peptides detected in Medicago roots in different stages of development. The diagrams show the numbers of peptides present uniquely in the young seedlings compared with the mature plants as well as the number of detected peptides that are present in both stages of development. The results are shown for the peptides detected using both the DHB and CHCA matrices.
In addition to comparing wild-type Medicago roots/root nodules in two different stages of development, wild-type plants were also compared to two different mutants. In the first mutant line, 35S:MtCLE13, the CLAVATA3/endosperm-surrounding region (CLE) family of peptides is overexpressed; therefore, this mutant was thought to be a good model for MSI method development to diminish the challenge of trying to detect low-concentration peptides. Figure 5 displays putative peptides that were detected in the 35S:MtCLE13 plants but not in the wild-type plants. It is thought that these putative peptides could belong to the CLE peptide family but are too low in concentration to be detected via MALDI–MSI in the wild-type plants. We also compared the wild-type plants to dnf1–1 mutants, which develop stunted, nonfunctional nodules. Figure 6 shows putative peptides that were detected in the wild-type plants but were absent from the dnf1–1 plants. This suggests that these putative peptides may play a role in nodule development or function.
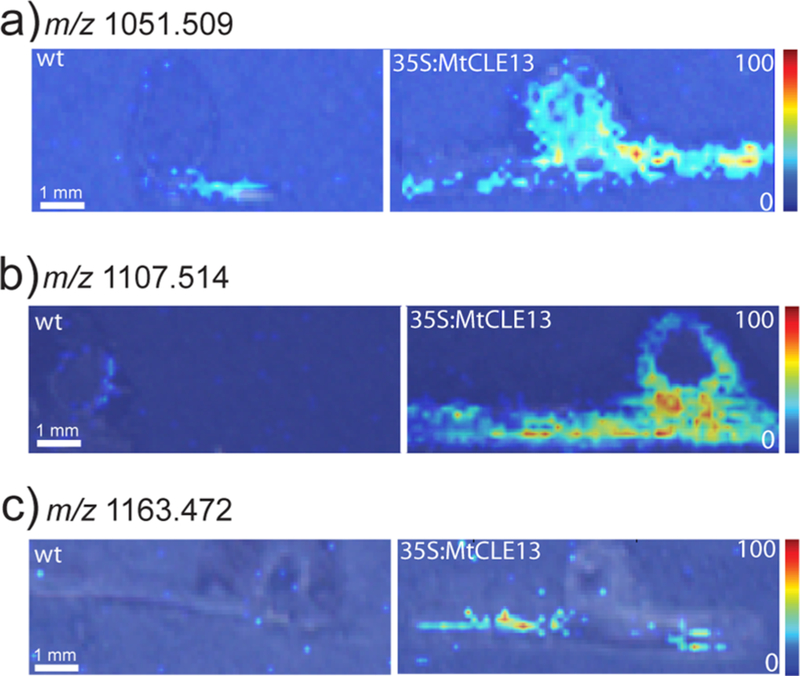
Figure 5.
Putative peptides that were detected in the 35S:MtCLE13 plants (demonstrating overexpression of CLAVATA3/endosperm-surrounding region (CLE) peptides) but are present in much lower concentrations in the wild-type (wt) plants. (a) m/z 1051.509 is located in both the plant root and nodule. (b) m/z 1107.514 is distributed to the plant root and outer nodule. (c) m/z 1163.472 is localized to the plant root.
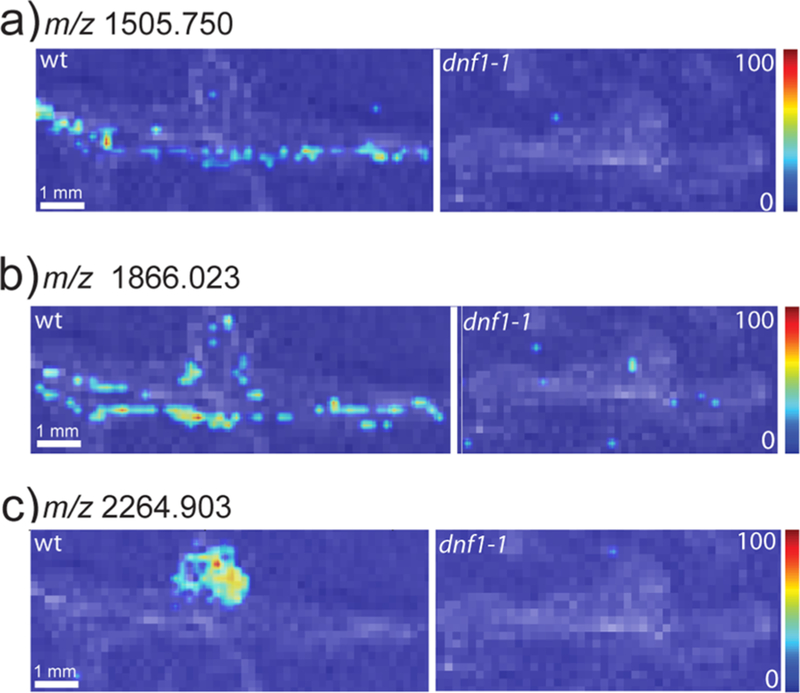
Figure 6.
Putative peptides that were detected in the wild-type (wt) plants but were absent from the dnf1–1 plants. dnf1–1 mutants develop stunted, nonfunctional nodules, suggesting that these putative peptides may play a role in nodule development or function. (a) m/z 1505.750 is localized to the root. (b) m/z 1866.023 is distributed to the plant root and outer nodule. (c) m/z 2264.903 is localized to the plant nodule.
Peptide/Protein Identification
Peptides and proteins can sometimes be identified by accurate-mass-matching; however, the more widely accepted approach for identification is to match MS/MS data to sequenced genomes or by de novo sequencing with software packages like PEAKS.44 Furthermore, MALDI generally produces poor MS/MS fragmentation due to limited sample amounts and the fact that typically only singly-charged ions are generated. Therefore, MALDI–MSI results were matched to LC–MS/MS results for identifications as described below. Table 1 details a list of m/z values detected via MALDI–MSI and LC–MS/MS that were able to be de novo sequenced using PEAKS. Since MALDI–MSI typically generates +1 charged ions and LC–MS typically generates +2 or +3 charged peptide ions, the molecular weight of each peptide detected was calculated from the acquired m/z to compare data across ionization methods. Using the molecular weights, the PEAKS de novo sequencing data generated from LC–MS/MS experiments was searched for peptide masses detected via MALDI–MSI. Since these calculations were made and multiple ionization sources were used, a mass error of <10 ppm was allowed for confident peptide identification via de novo sequencing. The annotated MS/MS spectra used for de novo sequencing can be found in the Supporting Information (Figure S2).
Table 1.
List of m/z Values That Were De Novo Sequenced Using PEAKS
| de novo sequenced peptides (molecular weight) | |||
|---|---|---|---|
| MALDI measured |
LC–MS measured |
Δppm | Peptide sequence |
| 908.4234 | 908.4240 | 0.66 | FGGSTVEVN |
| 962.4813 | 962.4743 | 7.31 | TVNEEKLM |
| 990.5163 | 990.5134 | 2.89 | PSPPLRGEP |
| EPPPHVTSK | |||
| 1006.5114 | 1006.5195 | 8.09 | TVGKGAHAGPP |
| 1008.4694 | 1008.4624 | 6.98 | DDARPPQGGP |
| 1021.5575 | 1021.5556 | 1.85 | AVGKDYRLT |
| 1068.5733 | 1068.5791 | 5.42 | CKVWPLPGK |
| CKVWPPLGK | |||
| 1094.5280 | 1094.5244 | 3.28 | PQTEAPAVGAP |
| 1106.5081 | 1106.4993 | 7.92 | YNDQDTPVR |
| 1106.5255 | 1106.5356 | 9.10 | TDSSAPGGFLR |
| 1112.5974 | 1112.5938 | 3.21 | STGGVAAPRAVQ |
| TSGVQAPRGPK | |||
| 1159.6191 | 1159.6084 | 9.20 | TEAATATPAVTK |
| 1170.6070 | 1170.6133 | 5.38 | VLDPGDSDLLK |
| LVDPGDSDLLK | |||
| GPDDSDLVLLK | |||
| DPGDSDLVLLK | |||
| DPGSDDLVLLK | |||
| 1175.5925 | 1175.5935 | 0.88 | TGAEGKVHSYK |
| 1179.6039 | 1179.6135 | 8.10 | KAPPPVADDTK |
| 1186.5763 | 1186.5830 | 5.68 | TVGNGPVEASGLS |
| 1192.5108 | 1192.5107 | 0.09 | HGGTEDPVTSGH |
| 1300.6790 | 1300.6775 | 1.12 | QSSHSPVLVKGF |
| 1300.6790 | 1300.6697 | 7.12 | TVGAVDAVTLMPQ |
| 1308.6282 | 1308.6211 | 5.45 | SYFANAQPQQR |
| 1342.6713 | 1342.6663 | 3.72 | QSVKMTNAHSLQ |
| 1376.7577 | 1376.7598 | 1.55 | VSLALVCSPVPHR |
| 1728.9683 | 1728.9597 | 4.95 | KPLNVELGFKAVAAGLC |
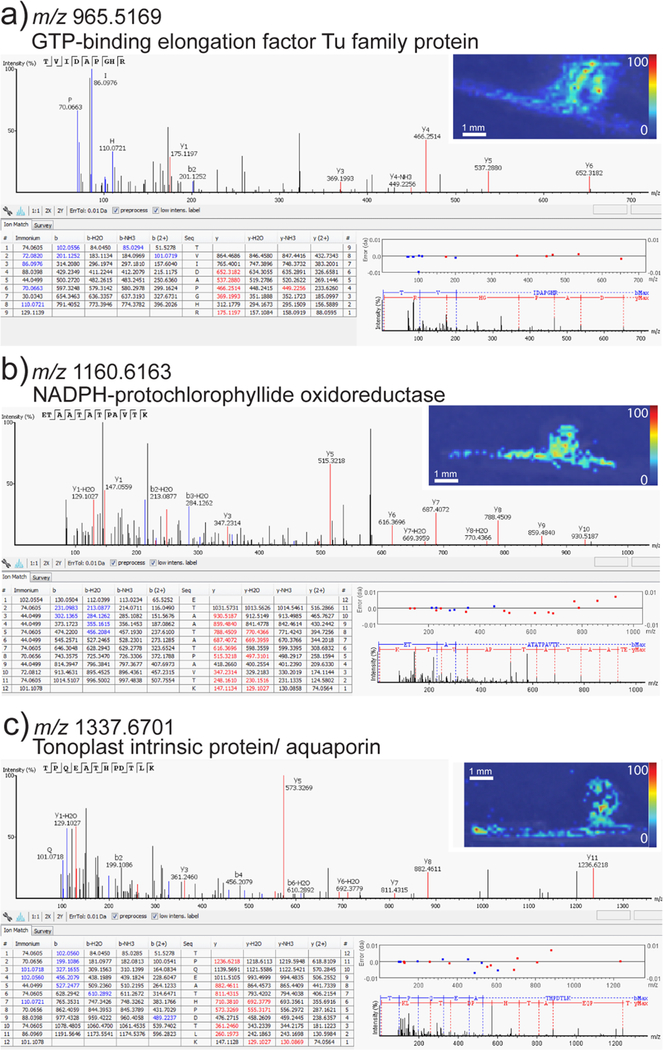
In addition to de novo sequencing, PEAKS also allows for MS/MS matching to sequenced genomes. PEAKS was used to generate a mass list from the LC–MS/MS data of unique peptide masses that matched proteins from the genome data within a 0.1% false discovery rate. A .fasta file of the complete Medicago truncatula proteome was generated using the NCBI NR Database and used for peptide identification. Using this method, 10 imaged peptides were identified. Example images and corresponding LC–MS/MS spectra for these unique peptides are shown in Figure 7 (all additional unique peptides with corresponding LC–MS/MS spectra are shown in the Supporting Information, Figure S3). Peptides corresponding to the ferritin, an iron storage protein, were identified. Iron is an essential component of nitrogenase, the bacterial enzyme that converts atmospheric dinitrogen to ammonia.50 Depending upon the stage of nodule development the concentration and distribution of iron vary. In addition to accumulating in infected cells, ferritin also accumulates in uninfected cells within nodules. This increased concentration of ferritin may facilitate iron incorporation into nitrogenase.51,52 Besides ferritin, we also detected aquaporins, which are the predominant members of major intrinsic proteins (MIPs) commonly implicated in the transport of water, glycerol, and ammonia.53 These plant aquaporins are divided into five subfamilies, including the tonoplast intrinsic proteins (TIPs). At least seven different TIPs have been identified in Medicago and, except for TIP1g, TIPs are expressed at low levels in 14 day old nodules.54 The exception, TIP1g, is localized to the tonoplast in the infection zone (the zone where rhizobia are released into nodule cells) of nodules, whereas in the nitrogen-fixation zone TIP1g is redirected toward the symbiosome membrane. Knocking down the expression of TIP1g affected symbiosome maturation to the nitrogen-fixing stage, and it is hypothesized that this may be the result of altered water availability.54 Further analysis will be necessary to identify the TIP we detected here, but based on the expression pattern of Medicago TIPs, it is highly likely that it is TIP1g. In addition to these peptides, a wound-induced basic family protein was identified. Although the function of this protein is still unknown, it should be noted that this family of proteins is known to be upregulated in soybean nodules.55 A complete list of the imaged unique peptides is shown in Table 2.
Figure 7.
Example images and corresponding LC–MS/MS spectra for unique peptides of known Medicago proteins. The MS/MS with annotated de novo sequencing, the predicted protein, and the corresponding MS image are shown.
Table 2.
List of m/z Values of Unique Peptides for Which MS Images Were Acquired
| [M + H] | protein group |
protein accession | peptide | protein description |
|---|---|---|---|---|
| 965.51694 | 47 | gi|657381377, gi|657381378 | C.TVIDAPGHR.D | GTP-binding elongation factor Tu family protein |
| 47 | gi|357496973 | C.TVIDAPGHR.D | elongation factor 1-alpha | |
| 1132.53084 | 215 | gi|657377737 | K.ANENKPVMTE | wound-inducible basic family protein |
| 1160.61634 | 43 | gi|657399288 | A.ETAATATPAVTK.S | NADPH-protochlorophyllide oxidoreductase |
| 1162.59554 | 31 | gi|357474991 | M.A(+42.01)ASGEEKKIST.S | low-temperature inducible |
| 31 | gi|355508836 | M.A(+42.01)ASGEEKKIST.S | nodulin-like protein | |
| 31 | gi|388522163 | M.A(+42.01)ASGEEKKIST.S | unknown | |
| 1171.62114 | 71 | gi|388504426, gi|217073834, gi|388496542 | S.IVDPGDSDIIK.T | unknown |
| 71 | gi|657371310, gi|355496648 | S.IVDPGDSDIIK.T | ribosomal protein L7Ae/L30e/ S12e/Gadd45 family protein |
|
| 71 | gi|357502689 | S.IVDPGDSDIIK.T | 60S ribosomal protein L30 | |
| 1180.62144 | 148 | gi|388522541, gi|388492578 | K.AVAPPPVADDTK.A | unknown |
| 148 | gi|657373135 | K.AVAPPPVADDTK.A | carboxy-terminal region remorin | |
| 1293.67314 | 149 | gi|388507838, gi|217073043 | T.GVIFEPFEEVK.K | unknown |
| 149 | gi|357468557 | T.GVIFEPFEEVK.K | ferritin-3 | |
| 149 | gi|355505618 | T.GVIFEPFEEVK.K | ferritin | |
| 149 | gi|657385619, gi|355518020, gi|355498374 | T.GVLFEPFEEVK.K | ferritin | |
| 149 | gi|388491178, gi|217073544, gi|388499902, gi|217073522 | T.GVLFEPFEEVK.K | unknown | |
| 149 | gi|357492793 | T.GVLFEPFEEVK.K | ferritin-2 | |
| 149 | gi|357506141 | T.GVIFEPFEEVK.K | ferritin-1 | |
| 1337.67014 | 61 | gi|657379753 | G.TPQEATHPDTLK.A | tonoplast intrinsic protein |
| 61 | gi|32363409, gi|9716259 | G.TPQEATHPDTLK.A | probable aquaporin TIP-type | |
| 1483.75454 | 13 | gi|357471525, gi|355507102, gi|404332436, gi|404332513, gi| 355501595, gi|357512583, gi|404332359, gi|357502811, gi| 124360830, gi|355496709 |
A.AFRVSPQPGVPAEE.A | ribulose biphosphate carboxylase large chain domain protein |
| 13 | gi|543174105, gi|153012229 | A.AFRVSPQPGVPAEE.A | ribulose-1,5-biphosphate carboxylase/oxygenase large subunit (chloroplast) |
|
| 1976.02434 | 11 | gi|355501329, gi|357512051 | N.NKNNPNLFNNLVYTPLT.I | basic 7S globulin-like protein |
| 11 | gi|87240526 | N.NKNNPNLFNNLVYTPLT.I | peptidase A1, pepsin |
CONCLUSIONS AND FUTURE DIRECTIONS
We have demonstrated the benefits of using MALDI and ESI for the complementary detection and identification of endogenous peptides and protein fragments in plants. Using both CHCA and DHB as MALDI matrices for MSI greatly increased the coverage of peptides/protein fragments that were detected. We noticed interesting differences in the overall numbers of peptides detected between seedlings and mature plants. In addition to the difference in overall peptides, we also observed changes in the spatial distributions of peptides detected in both the seedlings and mature plants. Because of the low concentrations of endogenous peptides in wild-type plant tissues, peptide enrichment strategies might be a valuable next step for targeting and detecting specific classes of peptides in a biologically relevant manner. Other sample preparation approaches are in development for reducing the number of protein fragments that are detected and improving the detection of endogenous peptides. Additional biological studies examining the peptides/protein fragments detected in this study could reveal more insights into the functions of these peptides within the plant in different stages of development.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by funding from the National Science Foundation (NSF) Division of Integrative Organismal Systems (IOS) RESEARCH PGR award #1546742, University of Wisconsin-Madison Graduate School and the Wisconsin Alumni Research Foundation (WARF), a Romnes Faculty Research Fellowship, and a Vilas Distinguished Achievement Professorship to L.L., and an NSF grant to JMA (NSF#0701846). E.G. acknowledges an NSF Graduate Research Fellowship (DGE-1256259). The MALDI-Orbitrap and Q-Exactive instruments were purchased through an NIH shared instrument grant (NCRR S10RR029531).
Footnotes
Notes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.6b00471.
REFERENCES
- (1).Ehrhardt DW; Frommer WB New technologies for 21st century plant science. Plant Cell 2012, 24 (2), 374–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Moshelion M; Altman A Current challenges and future perspectives of plant and agricultural biotechnology. Trends Biotechnol 2015, 33 (6), 337–42. [DOI] [PubMed] [Google Scholar]
- (3).Dey A; Mukherjee A Therapeutic potential of bryophytes and derived compounds against cancer. Journal of Acute Disease 2015, 4 (3), 236–248. [Google Scholar]
- (4).Perumal Samy R.; Gopalakrishnakone P Therapeutic Potential of Plants as Anti-microbials for Drug Discovery. Evidence-based complementary and alternative medicine: eCAM 2010, 7 (3), 283–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Glickman-Simon R; Schneider C Homeopathy for Depression, Music for Postoperative Recovery, Red Yeast Rice for High Cholesterol, Acupuncture for Seasonal Allergic Rhinitis, and Ginger for Osteoarthritis. Explore (NY) 2016, 12 (4), 287–91. [DOI] [PubMed] [Google Scholar]
- (6).Mikulass KR; Nagy K; Bogos B; Szegletes Z; Kovacs E; Farkas A; Varo G; Kondorosi E; Kereszt A Antimicrobial nodule-specific cysteine-rich peptides disturb the integrity of bacterial outer and inner membranes and cause loss of membrane potential. Ann. Clin. Microbiol. Antimicrob 2016, 15 (1), 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Food and Agriculture Organization of the United Nations Statistics Division.
- (8).Roe MR; Griffin TJ Gel-free mass spectrometry-based high throughput proteomics: tools for studying biological response of proteins and proteomes. Proteomics 2006, 6 (17), 4678–87. [DOI] [PubMed] [Google Scholar]
- (9).Zhang YY; Fonslow BR; Shan B; Baek MC; Yates JR Protein Analysis by Shotgun/Bottom-up Proteomics. Chem. Rev 2013, 113 (4), 2343–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Jorrin-Novo JV; Pascual J; Sanchez-Lucas R; Romero-Rodriguez MC; Rodriguez-Ortega MJ; Lenz C; Valledor L Fourteen years of plant proteomics reflected in Proteomics: Moving from model species and 2DE-based approaches to orphan species and gel-free platforms. Proteomics 2015, 15 (5–6), 1089–1112. [DOI] [PubMed] [Google Scholar]
- (11).Verhaert P; Uttenweiler-Joseph S; de Vries M; Loboda A; Ens W; Standing KG Matrix-assisted laser desorption/ionization quadrupole time-of-flight mass spectrometry: an elegant tool for peptidomics. Proteomics 2001, 1 (1), 118–31. [DOI] [PubMed] [Google Scholar]
- (12).Buchberger A; Yu Q; Li L Advances in Mass Spectrometric Tools for Probing Neuropeptides. Annu. Rev. Anal. Chem 2015, 8, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Dallas DC; Guerrero A; Parker EA; Robinson RC; Gan JN; German JB; Barile D; Lebrilla CB Current peptidomics: Applications, purification, identification, quantification, and functional analysis. Proteomics 2015, 15 (5–6), 1026–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Matsubayashi Y; Sakagami Y Peptide Hormones in Plants. In Annual Review of Plant Biology; Annual Reviews: Palo Alto, CA, 2006; Vol. 57, pp 649–674. [DOI] [PubMed] [Google Scholar]
- (15).Lindsey K Plant peptide hormones: The long and the short of it. Curr. Biol 2001, 11 (18), R741–R743. [DOI] [PubMed] [Google Scholar]
- (16).Kondo T; Sawa S; Kinoshita A; Mizuno S; Kakimoto T; Fukuda H; Sakagami Y A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 2006, 313 (5788), 845–848. [DOI] [PubMed] [Google Scholar]
- (17).Farrokhi N; Whitelegge JP; Brusslan JA Plant peptides and peptidomics. Plant Biotechnol. J 2008, 6 (2), 105–134. [DOI] [PubMed] [Google Scholar]
- (18).Yamaguchi Y; Huffaker A Endogenous peptide elicitors in higher plants. Curr. Opin. Plant Biol 2011, 14 (4), 351–357. [DOI] [PubMed] [Google Scholar]
- (19).Balluff B; Schone C; Hofler H; Walch A MALDI imaging mass spectrometry for direct tissue analysis: technological advancements and recent applications. Histochem. Cell Biol 2011, 136 (3), 227–44. [DOI] [PubMed] [Google Scholar]
- (20).Gemperline E; Jayaraman D; Maeda J; Ane JM; Li L Multifaceted investigation of metabolites during nitrogen fixation in Medicago via high resolution MALDI-MS imaging and ESI-MS. J. Am. Soc. Mass Spectrom 2015, 26 (1), 149–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Lee YJ; Perdian DC; Song ZH; Yeung ES; Nikolau BJ Use of mass spectrometry for imaging metabolites in plants. Plant J 2012, 70 (1), 81–95. [DOI] [PubMed] [Google Scholar]
- (22).Kaspar S; Peukert M; Svatos A; Matros A; Mock HP MALDI-imaging mass spectrometry - An emerging technique in plant biology. Proteomics 2011, 11 (9), 1840–1850. [DOI] [PubMed] [Google Scholar]
- (23).Gemperline E; Li L MALDI-mass spectrometric imaging for the investigation of metabolites in Medicago truncatula root nodules. J. Visualized Exp 2014, No. 85, 51434. [DOI] [PMC free article] [PubMed]
- (24).Ye H; Gemperline E; Venkateshwaran M; Chen R; Delaux PM; Howes-Podoll M; Ane JM; Li L MALDI mass spectrometry-assisted molecular imaging of metabolites during nitrogen fixation in the Medicago truncatula-Sinorhizobium meliloti symbiosis. Plant J 2013, 75 (1), 130–45. [DOI] [PubMed] [Google Scholar]
- (25).Bjarnholt N; Li B; D’Alvise J; Janfelt C Mass spectrometry imaging of plant metabolites - principles and possibilities. Nat. Prod. Rep 2014, 31 (6), 818–837. [DOI] [PubMed] [Google Scholar]
- (26).Poth AG; Mylne JS; Grassl J; Lyons RE; Millar AH; Colgrave ML; Craik DJ Cyclotides Associate with Leaf Vasculature and Are the Products of a Novel Precursor in Petunia (Solanaceae). J. Biol. Chem 2012, 287 (32), 27033–27046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Cavatorta V; Sforza S; Mastrobuoni G; Pieraccini G; Francese S; Moneti G; Dossena A; Pastorello EA; Marchelli R Unambiguous characterization and tissue localization of Pru P 3 peach allergen by electrospray mass spectrometry and MALDI imaging. J. Mass Spectrom 2009, 44 (6), 891–7. [DOI] [PubMed] [Google Scholar]
- (28).Bencivenni M; Faccini A; Zecchi R; Boscaro F; Moneti G; Dossena A; Sforza S Electrospray MS and MALDI imaging show that non-specific lipid-transfer proteins (LTPs) in tomato are present as several isoforms and are concentrated in seeds. J. Mass Spectrom 2014, 49 (12), 1264–1271. [DOI] [PubMed] [Google Scholar]
- (29).Grassl J; Taylor NL; Millar AH Matrix-assisted laser,desorption/ionisation mass spectrometry imaging and its development for plant protein imaging. Plant Methods 2011, 7 (1), 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Mus F; Crook MB; Garcia K; Garcia Costas A.; Geddes BA; Kouri ED; Paramasivan P; Ryu MH; Oldroyd GE; Poole PS; Udvardi MK; Voigt CA; Ane JM; Peters JW Symbiotic Nitrogen Fixation and the Challenges to Its Extension to Nonlegumes. Appl. Environ. Microbiol 2016, 82 (13), 3698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Venkateshwaran M; Volkening JD; Sussman MR; Ane JM Symbiosis and the social network of higher plants. Curr. Opin. Plant Biol 2013, 16 (1), 118–27. [DOI] [PubMed] [Google Scholar]
- (32).McKay IA; Djordjevic MA Production and Excretion of Nod Metabolites by Rhizobium leguminosarum bv. trifolii Are Disrupted by the Same Environmental Factors That Reduce Nodulation in the Field. Appl. Environ. Microbiol 1993, 59 (10), 3385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Reid DE; Ferguson BJ; Gresshoff PM Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Mol. Plant-Microbe Interact 2011, 24 (5), 606–18. [DOI] [PubMed] [Google Scholar]
- (34).Mortier V; Den Herder G; Whitford R; Van de Velde W; Rombauts S; D’Haeseleer K; Holsters M; Goormachtig S CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol 2010, 153 (1), 222–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Saur IM; Oakes M; Djordjevic MA; Imin N Crosstalk between the nodulation signaling pathway and the autoregulation of nodulation in Medicago truncatula. New Phytol 2011, 190 (4), 865–74. [DOI] [PubMed] [Google Scholar]
- (36).Okamoto S; Ohnishi E; Sato S; Takahashi H; Nakazono M; Tabata S; Kawaguchi M Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol 2009, 50 (1), 67–77. [DOI] [PubMed] [Google Scholar]
- (37).Lim CW; Lee YW; Hwang CH Soybean nodule-enhanced CLE peptides in roots act as signals in GmNARK-mediated nodulation suppression. Plant Cell Physiol 2011, 52 (9), 1613–27. [DOI] [PubMed] [Google Scholar]
- (38).Sasaki T; Suzaki T; Soyano T; Kojima M; Sakakibara H; Kawaguchi M Shoot-derived cytokinins systemically regulate root nodulation. Nat. Commun 2014, 5, 4983. [DOI] [PubMed] [Google Scholar]
- (39).Kondorosi E; Mergaert P; Kereszt A A paradigm for endosymbiotic life: cell differentiation of Rhizobium bacteria provoked by host plant factors. Annu. Rev. Microbiol 2013, 67, 611–28. [DOI] [PubMed] [Google Scholar]
- (40).Bedinger PA; Pearce G; Covey PA RALFs: peptide regulators of plant growth. Plant Signaling Behav 2010, 5 (11), 1342–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Imin N; Mohd-Radzman NA; Ogilvie HA; Djordjevic MA The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. J. Exp. Bot 2013, 64 (17), 5395–409. [DOI] [PubMed] [Google Scholar]
- (42).Valdivia ER; Hertweck KL; Cho SK; Walker JC DVL/RTFL. In Handbook of Biologically Active Peptides, 2nd ed.; Elsevier, Inc: 2013; pp 15–19. [Google Scholar]
- (43).Chen R; Cape SS; Sturm RM; Li L Mass spectrometric imaging of neuropeptides in decapod crustacean neuronal tissues. Methods Mol. Biol 2010, 656, 451–63. [DOI] [PubMed] [Google Scholar]
- (44).Ma B; Zhang K; Hendrie C; Liang C; Li M; Doherty-Kirby A; Lajoie G PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom 2003, 17 (20), 2337–42. [DOI] [PubMed] [Google Scholar]
- (45).Catoira R; Galera C; de Billy F; Penmetsa R; Journet E; Maillet F; Rosenberg C; Cook D; Gough C; Denarie J Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 2000, 12 (9), 1647–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Oke V; Long SR Bacteroid formation in the rhizobium-legume symbiosis. Curr. Opin. Microbiol 1999, 2 (6), 641–6. [DOI] [PubMed] [Google Scholar]
- (47).Robichaud G; Garrard KP; Barry JA; Muddiman DC MSiReader: an open-source interface to view and analyze high resolving power MS imaging files on Matlab platform. J. Am. Soc. Mass Spectrom 2013, 24 (5), 718–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Mitra RM; Long SR Plant and bacterial symbiotic mutants define three transcriptionally distinct stages in the development of the Medicago truncatula/Sinorhizobium meliloti symbiosis. Plant Physiol 2004, 134 (2), 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Wang D; Griffitts J; Starker C; Fedorova E; Limpens E; Ivanov S; Bisseling T; Long SR A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis. Science 2010, 327 (5969), 1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Brear EM; Day DA; Smith PM Iron: an essential micronutrient for the legume-rhizobium symbiosis. Front. Plant Sci 2013, 4, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Lucas MM; Van de Sype G; Herouart D; Hernandez MJ; Puppo A; de Felipe MR Immunolocalization of ferritin in determinate and indeterminate legume root nodules. Protoplasma 1998, 204 (1), 61–70. [Google Scholar]
- (52).Ragland M; Theil EC Ferritin (mRNA, protein) and iron concentrations during soybean nodule development. Plant Mol. Biol 1993, 21 (3), 555–60. [DOI] [PubMed] [Google Scholar]
- (53).Chaumont F; Tyerman SD Aquaporins: highly regulated channels controlling plant water relations. Plant Physiol 2014, 164 (4), 1600–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Gavrin A; Kaiser BN; Geiger D; Tyerman SD; Wen Z; Bisseling T; Fedorova EE Adjustment of host cells for accommodation of symbiotic bacteria: vacuole defunctionalization, HOPS suppression, and TIP1g retargeting in Medicago. Plant Cell 2014, 26 (9), 3809–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Li X; Xu J; Yu G; Luo L A wound-induced small polypeptide gene family is upregulated in soybean nodules. Chin. Sci. Bull 2013, 58 (9), 1003–1009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.